- Department of General Surgery, State Key Laboratory of Complex Severe and Rare Diseases, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
Pancreatic ductal adenocarcinoma (PDAC) is a highly malignant tumor with especially poor prognosis. However, the molecular mechanisms of pancreatic oncogenesis and malignant progression are not fully elucidated. Epithelial-mesenchymal transition (EMT) process is important to drive pancreatic carcinogenesis. Recently, long noncoding RNAs (lncRNAs) and circular RNAs(circRNAs) have been characterized to participate in EMT in PDAC, which can affect the migration and invasion of tumor cells by playing important roles in epigenetic processes, transcription, and post-transcriptional regulation. LncRNAs can act as competing endogenous RNAs (ceRNA) to sequester target microRNAs(miRNAs), bind to the genes which localize physically nearby, and directly interact with EMT-related proteins. Currently known circRNAs mostly regulate the EMT process in PDAC also by acting as a miRNA sponge, directly affecting the protein degradation process. Therefore, exploring the functions of lncRNAs and circRNAs in EMT during pancreatic cancer might help pancreatic cancer treatments.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is one of the malignant tumors with poor prognosis, and currently, the only possible cure is through surgical removal. The relatively low overall survival (11%) is caused by the deep-seated position of the pancreas, the lack of appropriate diagnostic approaches, difficulties in performing a tissue biopsy, and its low response rate to radiotherapy or chemotherapy (1). Most pancreatic cancer patients are diagnosed at an advanced stage with metastasis, and therefore surgical management is unavailable for over 80% of patients (2). Metastatic pancreatic cancer has poor prognosis with a survival rate of 5% at 5 years (3, 4). For pancreatic cancer patients undergoing surgery, 5-year survival of them is also lower than 30%, which is critically ascribed to the distance metastases after surgery (5). Therefore, understanding the mechanism of PDAC metastasis is pivotal for the development of prognostic biomarkers and therapeutic targets.
Epithelial-mesenchymal transition (EMT) is a critical pathophysiological step that associates PDAC cells with invasion, migration, and acquisition of stem cell-like phenotype (6). During the process of EMT, PDAC cells change the protein expression with the loss of epithelial markers such as E-cadherin, cytokeratins, claudin, and occludin, and the elevation of mesenchymal markers such as N-cadherin, vimentin, and fibronectin (7). These changes lead to the loss of cellular polarity, disruption of normal cell adhesion, remodeling of the cytoskeleton, and changes in cell apoptosis, which collectively translate into enhanced invasion, migration, and metastasis (8). The mechanism behind this process is complex and involves different EMT-inducible transcription factors, such as SNAIL, SLUG, ZEB1, ZEB2, and Twist (9). Although the entire process of EMT is not yet completely understood, recent studies have shown that a variety of non-coding RNAs (ncRNAs), which include circRNA and lncRNAs are involved in regulating the EMT process of pancreatic cancer cells (10–14).
Long noncoding RNAs (lncRNAs) are operationally defined as RNA transcripts longer than 200 nucleotides, and current evidence showed that they could not encode peptides, since their locations most in the nucleus with low expression levels and little primary sequence conservation. These molecules serve vital roles in gene regulation, including modulating gene activating and silencing, alternative splicing, post-translational regulating, and chromosome inactivation, and perform these functions through different pathways to contribute not only to the development and metabolism but also to the cancer progression (15). Circular RNAs (circRNAs) have emerged as a large category of non-coding RNA molecules, many of which are highly abundant, conserved, and play important physiological and pathophysiological roles. Besides, these molecules also have exceptional promise as diagnostic, prognostic, and predictive biomarkers (16). CircRNA and lncRNAs not only regulate the intracellular EMT pathway but also can be encapsulated by extracellular vesicles (EVs), such as exosomes, thus regulating the whole EMT pathway of multiple cells in the microenvironment of pancreatic cancer (17, 18).
In this review, we focused on recent literature findings and review the mechanism of circRNA and lncRNA involved in the regulation of EMT in pancreatic cancer.
EMT in Pancreatic Cancer
EMT is a dynamic and reversible process through which epithelial cells present a mesenchymal-like phenotype, which is defined by changes in cell surface markers, cell morphology, and migration functions (19). It is now widely accepted that EMT is associated with invasion, dissemination, metastasis, as well as a poor diagnosis in epithelial cancers, including pancreatic cancer (20, 21). In the past few decades, besides the multi-faceted consideration of EMT-related invasiveness, the switch of cell surface markers from E-cadherin to N-cadherin and vimentin has been increasingly used to monitor the EMT process during cancer development as the hallmarks (22).The process of EMT contains the decomposition of epithelial cell-cell contacts, the loss of cell polarity, and the acquisition of mesenchymal features (Figure 1).
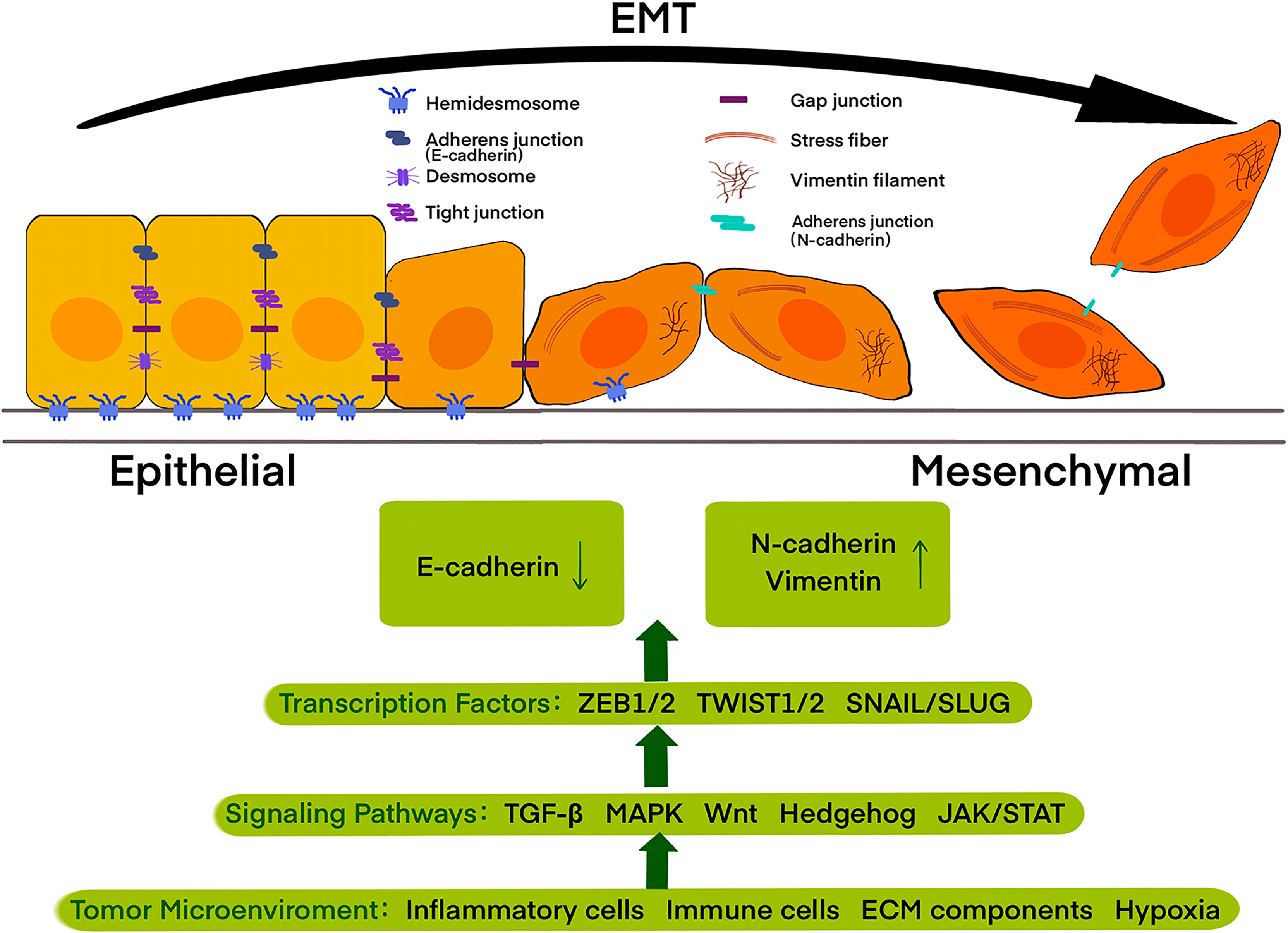
Figure 1 Epithelial cells with apical-basal polarity are tethered to the basement membrane by hemidesmosomes and held together by adherens junctions, tight junctions, and desmosomes, while mesenchymal cells display an extensively reorganized cytoskeleton with front-to-back polarity. Cells during epithelial-mesenchymal transition (EMT) undergo a left to right transition, with the loss of epithelial features accompanied by the acquisition of mesenchymal features. Tumor microenvironment including inflammatory cells, immune cells, ECM components, and also physical constraints, such as hypoxia, trigger signaling pathways, including the TGF-β, MAPK, Wnt, Hedgehog, and JAK/STAT, thus activating EMT-promoting transcription factors of the TWIST, SNAIL and ZEB families and further regulate the expression of E-cadherin, N-cadherin, and vimentin.
E- and N-cadherin both belong to type-I classical cadherins with similar sizes of 120 kDa and130 kDa. but different structures. Cadherin cytoplasmic tails attach with β-catenin which connects to α-catenin, thus forming the cadherin-catenin adhesion complex (23). E-cadherin loss is described as a key event of EMT, leading to decreased cell-cell adhesion, permitting the separation of individual cells from the primary tumor mass, and therefore represents invasion and metastasis in early tumor progression. In contrast with E-cadherin, N-cadherin is prevalent in non-epithelial tissues and is expressed in different types of cells such as neural cells, endothelial cells, stromal cells, and osteoblasts (24). N-cadherin is upregulated while E-cadherin is downregulated during EMT in cancers and this “cadherin switch”, which is regulated by several factors and pathways, is associated with enhanced migratory and invasive traits, which caused inferior patient survival rate (25). Vimentin is a type III intermediate filament (IF) protein, which supports other cellular organelles in EMT due to its viscoelastic attributes and directs cell migration through forming cellular protrusions and increasing the migratory ability of cells (26).
The EMT process is orchestrated by a suite of transcription factors (such as TWIST, SNAIL, SLUG, and ZEB1) (27). SNAIL1 (also known as SNAIL) and SNAIL2 (also known as SLUG) can activate the EMT programming during development and cancer. They repress epithelial genes expression by binding to E-box sequences in the proximal promoter region of the E-cadherin gene (28, 29). TWIST expression not only down-regulates E-cadherin expression as SNAIL does, but also induces N-cadherin expression. TWIST1 and TWIST2 form homodimers and also heterodimers, regulate E-box DNA binding and transcription regulation through mediating histone mono-methylation on different positions of E-cadherin and N-cadherin promoters (30, 31). Zinc finger E-box binding homeobox 1 (ZEB1) (also known as δEF1) and ZEB2 (also known as SMAD interacting protein 1 (SIP1)), members of the ZEB family, are characterized by two separated highly conserved clusters of zinc fingers. ZEB1 and ZEB2 have been found to bind CACCT sequences, including E2 boxes of the E-cadherin promoter, by their zinc finger clusters. Thus they can repress the transcription activity of the E-cadherin promoter (32).
EMT is also controlled by a complicated network of signaling pathways, including the transforming growth factor-β (TGF-β) signaling pathway, the mitogen-activated protein kinase (MAPK) signaling pathway, the Wnt signaling pathway, the hedgehog (HH) signaling pathway, the Hippo-Yes-associated protein (YAP) signaling pathway, the Janus kinase (JAK)/signal transducer and activator of transcription (STAT) signaling pathway, etc (33). Type 1 TGF-β receptor (TβR1) needs be phosphorylating by type 2 TGF-β receptor and activated by TGF-β signals thus initiating classical TGF-β/Smad signaling. This signaling continues to phosphorylate Smad2 and Smad3, which activates the transcriptions of SNAI1 and SLUG by binding directly to their promoters (34, 35). TGF-β can also activate ERK, Jun N-terminal kinase (JNK), MAPK through the β-catenin/T-cell factor (TCF) pathway (36). MAPK signaling in turn activates the TGF-β signaling pathway, probably because of the co-stimulation, leading to repression decreasing in E-cadherin and increasing in N-cadherin and vimentin (37). In addition, MAPK can stabilize SNAIL1 and increase its activity by inhibiting glycogen synthase kinase-3β (GSK-3β), which can phosphorylate SNAIL and induce SNAIL inactivation and degradation (38, 39). In canonical Wnt signaling, Wnt ligands bind and inhibit GSK3β, hence preventing β-catenin and SNAIL phosphorylation, ubiquitylation and degradation thus promoting EMT (40). In HH signaling, the ligand binds to patched (PTC) receptors to activate glioma-associated oncogene (GLI) family transcription factors (41). SHH/GLI1 expression induced upregulation of expression of S100A4, a member of the S100 gene family, which was found to promote the expression of TWIST and SNAIL during EMT but with an unclear mechanism (42). The Hippo-YAP was found to increase the expression of ZEB2 by binding to the promoter of ZEB2 (43). In JAK/STAT pathway, STAT1 is known to involve in anti-tumor immunity, while STAT3 is found to act as a transcriptional activator through binding to the promoter of LIV-1 genes in a tyrosine phosphorylation-dependent manner, resulting in the overexpression of LIV-1, which can stabilize Snail by inactivating of GSK3β, leading to EMT progress (44).
EMT is also regulated by various pro-invasion signals of the tumor microenvironment (TME), which is defined as a cellular and physical environment surrounding the tumor cells, including inflammatory cells, immune cells, ECM components, and so on (45). During tumor programming, tumor cells recruit fibroblasts and immune cells which in turn secrete cytokines to impact tumor development and have been discovered to directly regulate the EMT process. TGF-β, secreted by tumor-associated platelets, fibroblasts, and tumor cells, can induce SNAIL1 and SNAIL2 via SMAD signaling in PDAC cells (46). Tumor necrosis factor-α (TNFα) is shown to activate NFκB signaling pathway thus inducing the expression of EMT transcription factors, including TWIST1, SNAIL2, and ZEB1/2 (47–49). Interleukins released by endothelial cells, immune cells, and fibroblasts can also contribute to EMT. Interleukin 6 (IL-6) increased TWIST1 and SNAIL1 expressions and promotes EMT (50). Several extracellular matrix (ECM) proteins, including Fibronectin, Collagen-I, and Hyaluronan are also implicated in controlling EMT. Fibronectin can induce SNAIL1 expression partially through binding to integrin receptors, and can also cooperate with TGFβ to activate the downstream ERK/MAPK kinases thus inducing EMT (51). Collagen-I can stabilize SNAIL1 by activating downstream SRC/ERK2 to promote tumor cell EMT (52). Hyaluronan can bind to tumor cells and activate LOX expression, which is a copper-dependent amine oxidase that catalyzes the cross-linking of collagens and elastins in the ECM, in turn, upregulates TWIST1 expression to promote EMT (52).
Hypoxia condition is another element affecting EMT, which is found to be related to the ability of tumor invasion and metastasis and increases tumor glycolysis and angiogenesis by inducing relevant gene expressions through hypoxia-inducible factors (HIFs) (53). Hypoxia-inducible factor-1α (HIF-1α) can increase SNAIL1 protein stability, resulting in the suppression of E-cadherin (54). Besides, HIF-1α directly binds to the TWIST1 promoter to induce TWIST1 expression (55). In addition, together with TGF-β, HIF1α can promote SNAIL1 nuclear translocation and induce EMT (56).
EMT process has been involved in not only invasion and migration but also resistance to chemotherapy and radiotherapy. It was proved that cells undergoing EMT increased chemoresistance and radioresistance by acquiring stem-like properties, enhancing survival pathways, preventing apoptosis, and activating signaling pathways involved in DNA damage repair and cell cycle progression (57–60).
Mesenchymal-like tumor cells produced by EMT have shown the characteristics of self-renewal, radiation resistance, and drug resistance of tumor stem cells. Therefore, the study of EMT will not only help to deepen the understanding of tumor progression, but also help tumor treatment.
lncRNA
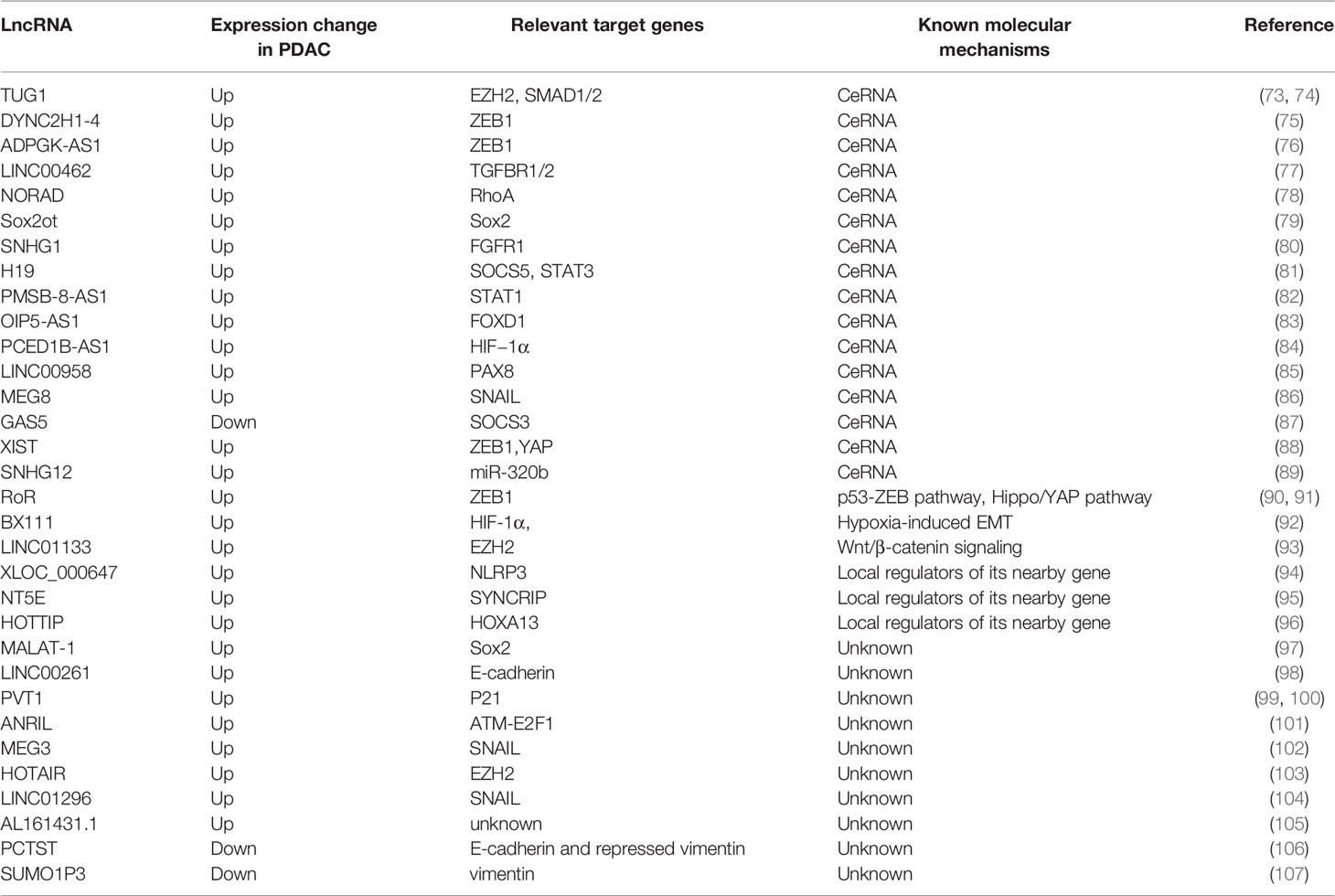
lncRNAs are RNA transcripts longer than 200nt without protein-coding potential. They are recognized to govern substantial biological processes including cell growth, differentiation, and proliferation (61–63). Accumulated evidence demonstrated that lncRNAs are aberrantly expressed in different cancers (64, 65). Some of them are also involved in oncogenic or tumor-suppressive pathways and have diagnostic and prognostic values (66, 67). Recent studies have confirmed the importance of lncRNAs in tumor growth and metastasis, including lncRNA-XIST in gastric cancer, lncRNA-MALAT1 in colorectal cancer, and lncRNA-PVT1 in gastric cancer, etc (68–70). LncRNAs can regulate gene expressions in different ways, including chromosome remodeling, transcription, as well as post-transcriptional modification (71, 72). Nowadays, increasing data suggests that lncRNAs play important roles in the EMT progression of PDAC (Figure 2). The expression change in PDAC, molecular mechanisms, and related genes of these lncRNAs are summarized in Table 1.
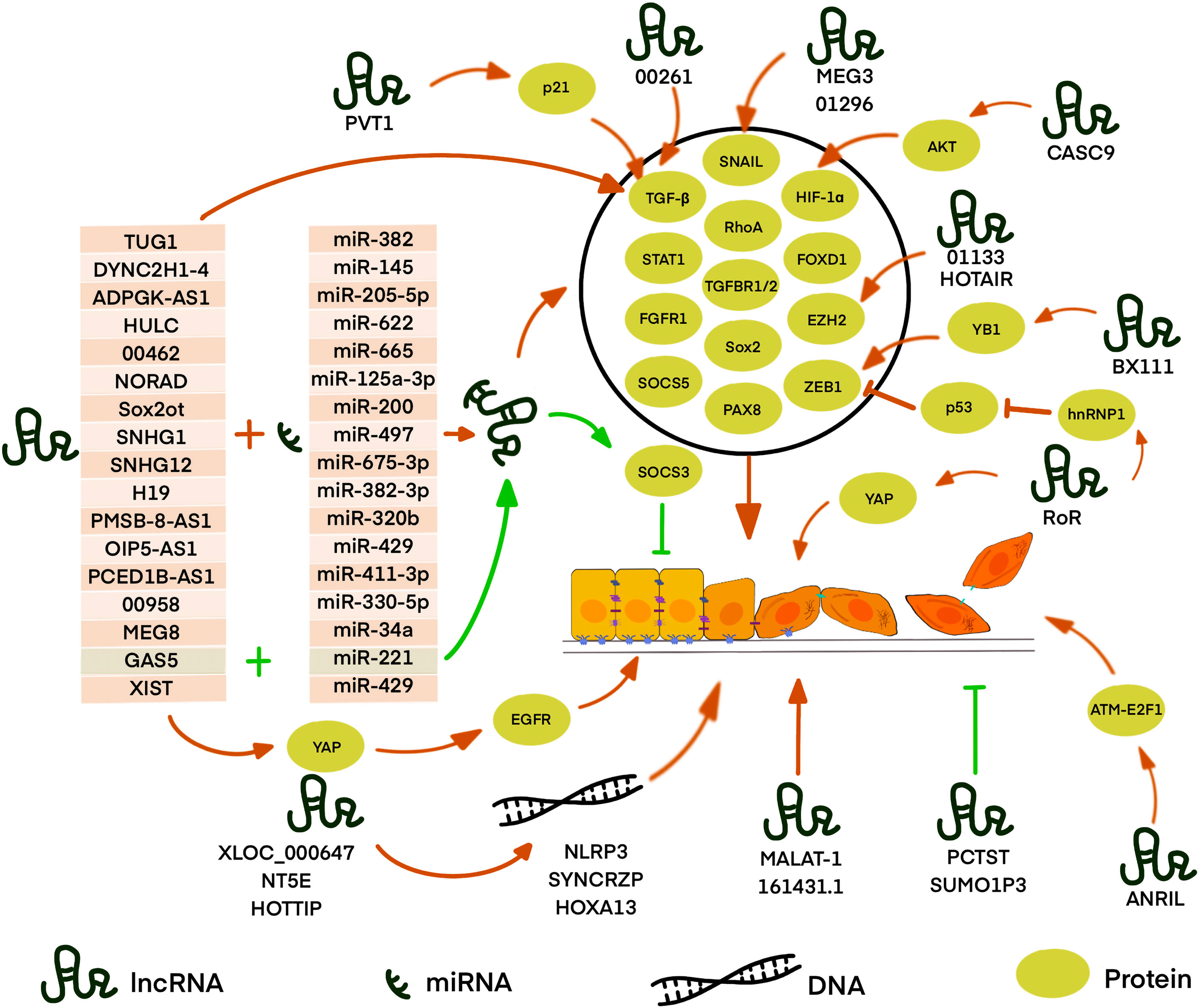
Figure 2 LncRNA is involved in EMT in PDAC through different pathways. Most known LncRNA can act as competing endogenous RNAs (ceRNA) to sequester target miRNA away from mRNA or DNA thus regulating the expression of downstream proteins. LncRNAs, such as XLOC_000647, NE5T, and HOTTIP, can regulate gene expression by binding with local chromatin architectures which localize physically nearby. LncRNAs,such as RoR, BX111, LINC01133, and CASC9, can directly regulate or interact with EMT-related proteins or further affect the expression of its downstream targets. However, some lncRNAs, such as MALAT-1, TUG1, LINC00261, PVT I, ANRIL, MEG3, HOTAIR, SUMO1P3, and LINC01296, play an important role in the EMT process with unclear mechanisms. Red arrows: the pathway promotes EMT, green arrows: the pathway inhibits EMT, the black circle: the proteins regulated by ceRNA and positively regulates EMT.
LncRNA: Binding miRNA as Competing Endogenous RNAs (ceRNA)
One of the most important functions of lncRNA is to compete for the miRNA binding site through partial complementarity as a ceRNA.
When lncRNA act as ceRNA, they can interfere with the process of EMT by affecting transcription factors. Gao et al. investigated that lnc-DYNC2H1-4, as a sponge of miR-145, upregulates ZEB1, which leads to upregulation and downregulation of vimentin and E-cadherin, respectively (75). Due to the discovery of the direct interaction between miR-145 and ZEB1 in many solid cancers, it was suggested that lnc-DYNC2H1-4 participates in EMT by the miR-145/ZEB1 pathway (108–110). Moreover, in vitro and in vivo investigations by Song et al. indicated that lncRNA ADPGK-AS1 inhibits miR-205-5p, which could directly target ZEB1, and therefore induces EMT (76). Also, Shen et al. investigated that lncRNA X-inactive specific transcript (XIST) promotes PDAC cell EMT by acting as a sponge for miR-429, which can directly bind to ZEB1 mRNA, to regulate ZEB1 expression (88). Besides, Zou et al. reported that XIST promotes the expression of YAP, leading to TGF-β1-induced EMT via up-regulation of EGFR (111). Li et al. figured out a similar mechanism that lnc-Sox2ot regulates the expression of Sox2 by binding to the miR-200 family and therefore promote EMT in PDAC, thus produce an effect on TNM stage and overall survival rate of PDAC patients (79). Sox, which contributes to the metastasis in various tumor tissue, has been reported to promote EMT via regulation of the WNT/β-catenin signal (112). L. Wu et al. revealed that lncRNA OIP5-AS1 can sponge miR-429, which targets FOXD1, thereby activating the ERK pathway and EMT in PDAC (83). S. Chen et al. used LINC00958 as a ceRNA to competitively sponge miR-330-5p and regulate paired box 8 (PAX8), and then promote the EMT process in PDAC. However, further studies are required to fully understand the mechanisms of the miR-330-5p/PAX8 axis on the regulation of EMT in PDAC cells (85). Terashima et al. elucidated that lncRNA maternally expressed gene 8 (MEG8) reduces the expression of miRNA-34a gene, thus up-regulated the expression of SNAIL transcription factors, which repressed the expression of E-cadherin and induced EMT (86). However, there are few reports on the mechanism of the interaction between miRNA 34a and SNAIL. A previous study suggested that STAT1 and STAT3 have opposing roles in the tumor process. STAT1 signaling acts as a tumor suppressor by inhibiting angiogenesis, tumor growth, metastasis, and promoting apoptosis. Alternatively, the STAT3 pathway is implicated in oncogenic progression (113). However, H. Zhang et al. demonstrated that lncRNA PMSB8-AS1 promotes EMT progression in PDAC via upregulation of STAT1 by sponging miR-382-3p. They found that STAT1 can transcribe PD-L1, which mediates the inactivation of T cells and promotes invasion and migration, thus participate in EMT process (82). In addition to the transcription factors, some transcription regulatory proteins can also be affected by ceRNA, which in turn affects EMT process. In L. Zhao’s study, lncRNA taurine upregulated gene 1 (TUG1) acts as a molecular sponge for miR-382 and regulates its target, enhancer of zeste homolog 2 (EZH2), which could contribute to the cell proliferation, migration and EMT formation in PDAC (73). A previous study has reported that EZH2 inhibits tumor suppressor genes such as … via trimethylation of H3K27 (114). However, the detailed mechanism of EZH2-regulated EMT needs further study. In addition, some cellular receptors and molecular pathways are also affected by the lncRNA acted as ceRNA. B. Zhou et al. demonstrated that the overexpression of linc00462, which is a target of miR-665, enhancing the expression of TGFBR1 and TGFBR2, thus activating the TGF-β/SMAD2/3 pathway and accelerating EMT processing in PDAC cells (77). Moreover, Li et al. reported that lncRNA NORAD act as a ceRNA to regulate the expression of the small GTP binding protein RhoA by sequestering its inhibitor, miR-125a-3p, thereby promoting hypoxia and EMT (78). RhoA is a member of the Rho GTPase family, which was reported to facilitate the EMT process probably by promoting actin cytoskeleton reorganization, leading to the promotion of cell attachment and motility (115). Besides, S. Chen et al. found that lncRNA small nucleolar RNA host gene 1 (SNHG1) competes for the miR-497 binding site as a ceRNA, thus regulating FGFR1 expression to promote the EMT process, shown by changes in the expression levels of E-cadherin, N-cadherin, and vimentin. The detailed mechanisms should be investigated to fully clarify the contributions of the SNHG1-miR-497-FGFR1 interaction in the activation of EMT in future studies (80). Wang et al. discovered that the lncRNA H19/miR-675-3p signaling axis plays an important role in maintaining the EMT process and promoting cancer cell proliferation of PDAC cells, which possibly by directly targeting suppressor of cytokine signaling 5 (SOCS5) and thus activating the STAT3 pathway (81). SOCS3 is another member of the SOCS family that plays a significant role in EMT progression (116). Liu et al. showed that overexpression of lncRNA growth arrest-specific 5 (GAS5) suppresses the proliferation, migration, gemcitabine resistance, stem cell-like properties, and EMT of PDAC cells by directly binding to and suppressing miR-221 and enhancing SOCS3 expression, which can inhibit the EMT and tumor stem cell accumulation. They suggested that lncGAS5 acts as a tumor suppressor (87). Previous studies reported that overexpression of SOCS3 suppresses JAK2/STAT3-signaling activation thus inhibiting the JAK2/STAT3-mediated EMT in breast cancer (117, 118). Thus there is supposed to be the same pathway in PDAC. In Y. Zhang et al.’s study, lncRNA PC−esterase domain containing 1B−antisense RNA 1 (PCED1B−AS1) was shown to target miR−411−3p, resulting in upregulation of HIF−1α, thus participating in promoting proliferation, invasion, and EMT in PDAC (84). Some lncRNAs as ceRNAs can mediate EMT by playing other roles.Cao et al. revealed that lncRNA SNHG12 promotes PDAC cells invasion and EMT by absorbing miR-320b, which was affirmed as a pivotal element in other cancers that suppressed proliferation, migration, invasion, and EMT. Thus in-depth studies are needed to further indicate the related other mechanisms of the miR-320b/EMT axis (89). In general, lncRNA can act as ceRNA to regulate EMT process through various signaling pathways and downstream molecules.
LncRNAs Function as Stabilizating, Regulating, Remodeling DNA, RNA, and Protein
LncRNAs also function in chromatin and genomic structural remodeling, RNA stabilization, transcriptional regulation, and protein stability (119, 120).
Zhan et al. found that lncRNA regulator of reprogramming (RoR) increases levels of mesenchymal markers N-cadherin, and decreases levels of epithelial markers E-cadherin and induces EMT, and EMT-associated cell proliferation, invasion, and tumourigenicity in PC probably by ZEB1 pathway with the mechanism still unclear (90). A previous study showed that lnc-RoR can suppress p53 translation through direct interaction with heterogeneous nuclear ribonucleoprotein I (hnRNP I), which can bind to p53 mRNA to stimulate p53 translation (121). P53 was found to transactivate the miR-200c and miR-192, which can bind to ZEB1/2 mRNA, thus repressing ZEB1/2 translation (122). Therefore, they suggested that lnc-ROR repressed EMT in PC probably by inhibiting p53-ZEB pathway. Besides, in endometrial cancer, p53 was found to promote the expression of ZEB1 by directly binding to the promoter of miR-130b, the negative regulator of ZEB1, and inhibiting its transcription (123). Besides, W. Chen et al.’s study demonstrated that lnc-RoR overexpression activates the Hippo/YAP pathway and then promotes EMT (91). However, the mechanism by which lnc‐RoR affects YAP needs further exploration. Additionally, Deng et al. found that lncRNA BX111 recruited transcriptional factor Y-box protein (YB1) to its promoter region thus activating the transcription of ZEB1. They also revealed that BX111 transcription is induced by HIF-1α, which contributed to the hypoxia-induced EMT (92). Moreover, Y. Liu et al. evaluated that LINC01133 can promote EMT and metastasis in PDAC by interacting with EZH2 and epigenetically regulating axis inhibition protein 2 (AXIN2), directly activating β-catenin, thus influencing Wnt/β-catenin signaling (93). Hu’s results indicated that overexpression of lncRNA XLOC_000647 results in down-regulating the expression of NOD-like receptor family pyrin domain-containing 3(NLRP3) and inhibition of EMT-induced cell invasion (94). lncRNA XLOC_000647 might act as local regulators of its nearby gene, NLRP3, which was located at 25 kb of the XLOC_000647 downstream. Recent studies have validated that NLRP3 can activate inflammatory cytokines and promotes the metastasis of melanoma cells and hepatocellular carcinoma cells (124, 125). Thus, lncRNA XLOC_000647 might involve in EMT via NLRP3, though the molecular mechanism of the regulation in PDAC needs further definition. Similarly, P. Zhang et al.’s results showed that lncNT5E also acts as a significant modulator of its neighboring genes synaptotagmin‐binding cytoplasmic RNA‐interacting protein (SYNCRIP), which effectively regulated EMT by decreasing E‐cadherin and increasing vimentin and N‐cadherin. Nevertheless, the molecular mechanism of SYNCRIP in EMT warrants further investigation (95). In addition, Z. Li et al. found that lnc-HOTTIP contributes to EMT through upregulating HOXA13, which is located in physical continuity with HOTTIP in pancreatic cancer cells. Accompanied by EMT process, it is also revealed that HOTTIP promoted PDAC cell proliferation, invasion, and chemoresistance through regulating HOXA13. HOXA13, as a marker of gut primordial posteriorization during development, promotes lymph node metastasis and EMT, but still needs more studies of its mechanisms (96).
LncRNAs With Unclear Mechanisms
There are still many lncRNAs play a vital part in the EMT process with unclear mechanisms that need further studies.
Jiao F. et al. reported that lncRNA metastasis-associated lung adenocarcinoma transcript-1 (MALAT-1) is associated with EMT in PDAC. They found that MALAT-1 expression increases N-cadherin, vimentin, Slug, and Snail, while it down-regulates the expression of E-cadherin. The function analysis also revealed that EMT phenotype could make further efforts to cell proliferation, migration, invasion and cancer stem-like properties in vitro (97). Though there was evidence indicating that MALAT1 promotes the proliferation in many solid tumors, the detailed mechanisms of MALAT-1 regulating EMT in PDAC require further exploration (126–128). Qin et al. found that lncRNA TUG1 can increase the phosphorylation of Smad2 and Smad3 as well as levels of TGF-β and TGF-β receptor protein. Thus they believed that lncRNA TUG1 promotes cell proliferation, migration, and the EMT process of pancreatic cancer cell via the TGF-β/Smad signaling pathway regardless of the mechanism was still unclear (74). Moreover, Dorn et al. validated the importance of LINC00261 in EMT progression, partly due to its effect on E-cadherin, and it also regulated CDH1 and TGF-β (98). Wu et al. & Zhang et al. illustrated that the overexpression of lncRNA plasmacytoma variant translocation I (PVT I) in PDAC heightens cell proliferation and EMT via the TGF-β/Smad pathway by downregulating p21 with an unclear molecular mechanism. Besides, p21 can also directly regulate ZEB1 and Snail expression in PDAC cells (99, 100, 103). Chen et al. substantiated that lncRNA ANRIL leads to the activation of the ATM-E2F1 signaling pathway, and E-cadherin expression decreased but N-cadherin and vimentin expressions increased, thus resulting in the promotion of EMT of PDAC cells (101). Ma et al. exposited that differential expression levels of lnc MEG3, which locates near MEG8 in human chromosome 14q32.3 region, regulates the mRNA and protein levels of Snail and EMT induction and stem cell properties in PDAC. It is speculated that Snail might be a downstream target of MEG3 that alters EMT progression (102, 129, 130). However, more studies are needed to define how Snail is regulated by MEG3 in PDAC. Y. Tang et al. expounded that the silencing of lncRNA HOX antisense intergenic RNA (HOTAIR) could prohibit the Wnt/β-catenin signaling pathway-related gene expression thereby upregulating E-cadherin, downregulating N-cadherin and inhibiting EMT. However, a more specific mechanism of how HOTAIR acts in the Wnt/β-catenin signaling pathway in PDAC is waiting to be discovered (103). C. Li et al. found that HOTAIR guided EZH2 to the promoter of miR-34a resulting in the repression of miR-34a, and the EZH2-interacting region of HOTAIR was essential in promoting cell proliferation (131). Moreover, it was approved that HOTAIR exhibits pro-oncogenic activity and acts as a negative prognostic factor in pancreatic cancer (132). Wang et al. confirmed that lncRNA-PCTST is a potential tumor suppressor in PDAC. Its overexpression increases E-cadherin and repressed vimentin expression, thus inhibiting EMT in vitro (106). Similarly, Tian et al. observed that downregulation of lncRNA small ubiquitin-like modifier 1 pseudogene 3 (SUMO1P3) increases the expression of epithelial cadherin, and decreased the expression of neuronal cadherin, vimentin, and β-catenin, thus suppressing the EMT progression PDAC. But the pathway involved remains unknown (107). Moreover, Yuan et al. observed that upregulation of LINC01296 promotes the Snail-mediated process of EMT in PDAC (104). Besides, Ma et al. found increased E-cadherin accompanied by decreased N-cadherin and vimentin in lncRNA AL161431.1 knockdown PDAC cells, indicating that lncRNA AL161431.1 plays an important role in the EMT process. But the detailed mechanism of the pathway remained unknown (105).
In conclusion, lncRNAs play an important role in the EMT of PDAC. Ongoing endeavors to understand the functions of lncRNAs will help the understanding of EMT and provide opportunities to find better lncRNA-based therapeutic strategies for PDAC.
CircRNA
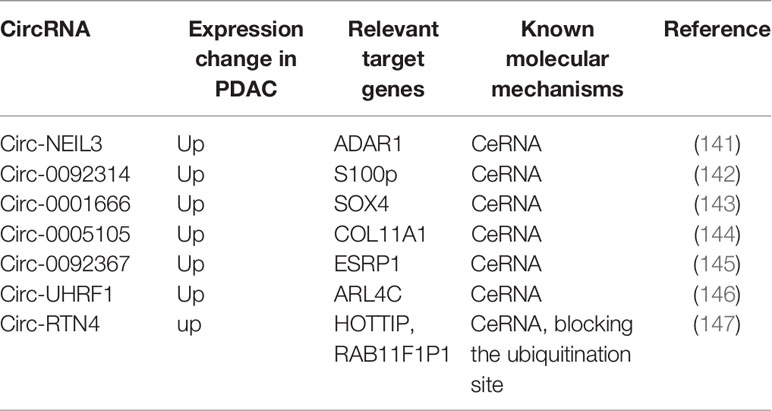
Circular RNAs (circRNAs) are a class of ncRNA family and belong to the long-size RNAs family, which are characterized by their closed continuous loop without 5′-3′ polarity and poly(A) tail (133). Studies have demonstrated that circRNAs can involve in DNA, RNA, and also protein synthesis and gene expression (134, 135). For example, circRNAs can function as miRNA sponges or bind to other molecules to regulate mRNA expression (136, 137). CircRNA could also interact with proteins via dissociating bindings between proteins, cementing interactions between proteins, sequestering proteins from DNA, RNA, or other proteins. Besides, circRNAs might recruit proteins to chromatin, and then translocate or redistribute proteins (138). CircRNAs are also involved in multiple disease progression, especially in cancer (139, 140). Recently, some evidence suggests that circRNAs play an essential role in PDAC progression and EMT (Figure 3). The expression change in PDAC, molecular mechanisms, and related genes of these circRNAs are summarized in Table 2.
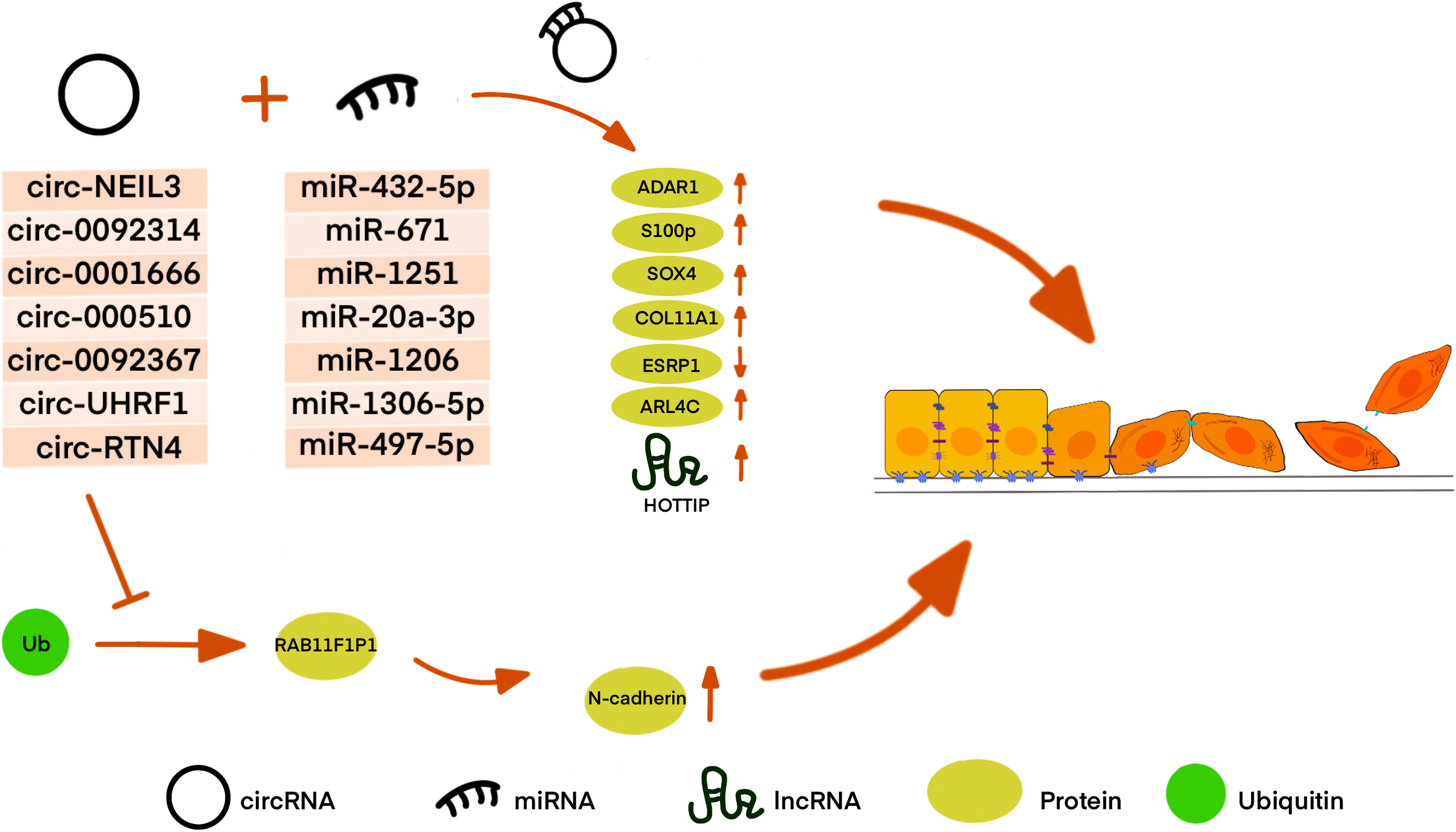
Figure 3 CircRNA affects EMT in many different ways in PDAC. Most CircRNAs can bind to miRNA as a sponge to regulate downstream protein or lncRNA. Circ-RTN4 is also found to regulate protein by hindering protein ubiquitination and degradation as well as acting as ceRNA.
CircRNA Binding to miRNA as a Sponge to Inhibit miRNA Levels
For instance, P. Shen et al. demonstrated that circNEIL3 is upregulated in PDAC and is shown to regulate the expression level of ADAR1 by sponging miR-432-5p, thus inducing RNA editing of GLI1, which was previously reported to have an increased ability to activate some transcriptional targets, including Snail. ADAR1 edited the GLI1 RNA by targeting the domain in the C-terminus of GLI1, thus altering the secondary structure of the GLI1 protein. Therefore, circNEIL3 induces the EMT phenotype in PDAC cells by activating the ADAR1/GLI1/Snail signal pathway (141, 148). Moreover, Q. Shen et al. found that overexpression of circ_0092314 in PDAC tissues and cells might repress miR-671, relieving its suppression of the downstream target S100P, which acts as an EMT activator in PDAC cells by activating the AKT pathway (142). Zhang et al. investigated that circ_0001666 acts as a sponge of miR-1251, thus increasing the expression of sex-determining region Y-related (SRY) high-mobility group box 4 (SOX4) (143). SOX4 can regulate EMT by acting as a component of TGF-β signaling, enhancing the transcription of ZEB, Snail, and TWIST, and targeting growth factor receptors such as the epidermal growth factor receptor (EGFR) (149). G. Ma et al. found that circ-000510 serves as a ceRNA of miR-20a-3p and indirectly increased collagen type XI alpha 1 (COL11A1) expression, which can enhance EMT probably by the remodeling of extracellular matrix, and further impact cellular proliferation and invasion, leading to poor prognosis of patients with PDAC (144). S. Yu et al. determine that circ_0092367 regulated the expression of epithelial splicing regulator protein 1 (ESRP1) through sponging miR-1206. ESRP1, which was verified as a tumor suppressor and a prognostic factor, can also induce E-cadherin, and reduce vimentin and N-cadherin levels in PDAC. Although the mechanism remains unclear, it was identified that circ_0092367 drives EMT properties in PDAC cells by regulating the miR-1206/ESRP1 axis. In this study, they confirmed that circ_0092367 also mediates gemcitabine resistance, and its expression was positively correlated with lymph node metastasis and tumor stage and negatively associated with outcome of PDAC patients, demonstrating that circ_0092367 may be a potential prognostic biomarker for PDAC (145). Similarly, W. Liu et al. suggested that circUHRF1 directly represses the level of miR-1306-5p, thus upregulating ADP ribosylation factor like GTPase 4 C (ARL4C), which is the target of miR-1306-5p. Vimentin was downregulated in ARL4C knockdown PDAC cells, while E-cadherin was upregulated, but the mechanism was still unknown. Therefore, an in-depth investigation was required into the role of the circUHRF1/miR-1306-5p/ARL4C axis in the EMT process (146).
More Roles of CircRNA in EMT
Although the circRNAs-miRNAs interaction is the most reported function of circRNAs in PDAC, some circRNAs are found to have additional roles.
Wong et al. identified circRTN4 interacts with RAB11FIP1 by blocking its ubiquitination site Lys578 to suppress the degradation of RAB11FIP1 and stabilize it, which can participate in the vesicle trafficking to regulate cadherin recycling. It has been provided that RAB11FIP1, which also known as Rab coupling protein (RCP), induced Slug expression through the β1 integrin/integrin linked kinase (ILK)/EGFR/Ras/NF-κB signaling pathway, thus promoting the expression of Slug, Snai1, Zeb1, Twist, and N-cadherin for EMT in PDAC. Their results also suggested that circRTN4 may function as a sponge of miR-497-5p, which can also bind with lnc-HOTTIP. Thus circRTN4 can also contribute to the EMT via the miR-497-5p/HOTTIP/HOXA13 pathway in PDAC (147, 150).
In brief, circRNAs play a pivotal role in the EMT of PDAC. Evidence has revealed that lots of circRNAs are changed in PDAC during the EMT process, but the functions and roles of these circRNAs are still unclear. Therefore, comprehensive studies are required to determine the functions and expression of circRNAs in EMT.
Conclusion and Perspective
Pancreatic cancer is known to have poor prognosis, mostly due to lack of effective diagnosis at the early stage of tumor development and effective therapy. Although surgical resection of pancreatic cancers diagnosed at an early stage remains the potentially curative option for PDAC, it is only possible in a small subset of patients.
Although the functions of miRNA, a major class of ncRNA, have been extensively reported in the past decades, we are beginning to explore the implications of lncRNA and circRNA in PDAC. More and more lncRNAs and circRNAs with key functions in PDAC are discovered these years, and most of them majorly regulate a multi-step process of cancer development including EMT. LncRNAs and circRNAs are found to have oncogene as well as tumor suppressor roles and contribute to tumor progression and EMT. Mechanistically, lncRNAs can act as ceRNAs that ‘sponge’ microRNAs, bind with some EMT-related proteins, or directly regulate genes located physically nearby. CircRNAs are also known to act as ceRNAs, or regulate protein degradation. It is increasingly evident that multiple functions of lncRNAs and circRNAs are undefined. There are still challenges in the molecular mechanisms of EMT in PDAC regulated by lncRNAs and circRNAs which need an in-depth investigation. Some of those RNAs may be potential biomarkers to predict staging and metastasis of PDAC in the future. More research on that could direct new therapies and tools for liquid biopsy to patient stratification and individualized precision treatment. Understanding the regulation of lncRNA and circRNA expressions during the progression of EMT in PDAC would provide a new paradigm of clinical therapy.
Author Contributions
WW directed and guided this study. XY and CQ collected related literature and drafted this manuscript. BZ, YW, ZL, THL, and TYL made critical revisions to this manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China Grants (81773215, 82173074).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
ARL4C, ADP ribosylation factor like GTPase 4 C; AXIN2, axis inhibition protein 2; circRNA, circular RNA; COL11A1, collagen type XI alpha 1; ceRNA, competing endogenous RNA; EGFR, epidermal growth factor receptor; EMT, epithelial-mesenchymal transition;ESRP1, epithelial splicing regulator protein 1; EVs, extracellular vesicles; EZH2, enhancer of zeste homolog 2; GAS5, growth arrest-specific 5; GLI, glioma-associated oncogene; GSK-3β, glycogen synthase kinase-3β; HH, hedgehog; hnRNP I, heterogeneous nuclear ribonucleoprotein I; HIF, hypoxia-inducible factor; HIF-1α, hypoxia-inducible factor-1α; HOTAIR, HOX antisense intergenic RNA; IL-6, interleutin 6; IF, intermediate filament; JAK, Janus kinase; JNK, jun N-terminal kinase; lncRNA, long noncoding RNA; MALAT-1, metastasis-associated lung adenocarcinoma transcript-1; MAPK, mitogen-activated protein kinase; MEG8, maternally expressed gene 8; NLRP3, NOD-like receptor family pyrin domain-containing 3; ncRNA, non-coding RNA; PAX8, paired box 8; PTC, patched; PCED1B−AS1, PC−esterase domain containing 1B−antisense RNA 1; PVT I, plasmacytoma variant translocation I; RoR, regulator of reprogramming; SNHG12, small nuclear RNA host gene 12; SOCS5, suppressor of cytokine signaling 5; SOX4, sex-determining region Y-related high-mobility group box 4; SRY, sex-determining region Y-related; STAT, signal transducer and activator of transcription; SUMO1P3, small ubiquitin-like modifier 1 pseudogene 3; SYNCRIP, synaptotagmin‐binding cytoplasmic RNA‐interacting protein; TCF, T-cell factor; TGF-β, transforming growth factor-β; TME, tumor microenvironment; TNFα, tumor necrosis factor-α; TUG1, taurine upregulated gene 1; TβR1, type 1 TGF-β receptor; XIST, X-inactive specific transcript; YAP, Yes-associated protein; YB1, Y-box protein; ZEB, zinc finger E-box binding homeobox.
References
1. Huang J, Lok V, Ngai CH, Zhang L, Yuan J, Lao XQ, et al. Worldwide Burden of, Risk Factors for, and Trends in Pancreatic Cancer. Gastroenterology (2021) 160(3):744–54. doi: 10.1053/j.gastro.2020.10.007
2. Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic Cancer. Lancet (2016) 388(10039):73–85. doi: 10.1016/S0140-6736(16)00141-0
3. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2022. CA Canc J Clin (2022) 72(1):7–33. doi: 10.3322/caac.21708
4. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer Statistics in China, 2015. CA Canc J Clin (2016) 66(2):115–32. doi: 10.3322/caac.21338
5. Strobel O, Neoptolemos J, Jäger D, Büchler MW. Optimizing the Outcomes of Pancreatic Cancer Surgery. Nat Rev Clin Oncol (2019) 16(1):11–26. doi: 10.1038/s41571-018-0112-1
6. Yang J, Antin P, Berx G, Blanpain C, Brabletz T, Bronner M, et al. Guidelines and Definitions for Research on Epithelial-Mesenchymal Transition. Nat Rev Mol Cell Biol (2020) 21(6):341–52. doi: 10.1038/s41580-020-0237-9
7. Chaffer CL, San Juan BP, Lim E, Weinberg RA. EMT, Cell Plasticity and Metastasis. Canc Metastasis Rev (2016) 35(4):645–54. doi: 10.1007/s10555-016-9648-7
8. Derynck R, Weinberg RA. EMT and Cancer: More Than Meets the Eye. Dev Cell (2019) 49(3):313–6. doi: 10.1016/j.devcel.2019.04.026
9. Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell (2016) 166(1):21–45. doi: 10.1016/j.cell.2016.06.028
10. Lee HY, Son SW, Moeng S, Choi SY, Park JK. The Role of Noncoding RNAs in the Regulation of Anoikis and Anchorage-Independent Growth in Cancer. Int J Mol Sci (2021) 22(2). doi: 10.3390/ijms22020627
11. Limb C, Liu DSK, Veno MT, Rees E, Krell J, Bagwan IN, et al. The Role of Circular RNAs in Pancreatic Ductal Adenocarcinoma and Biliary-Tract Cancers. Cancers (Basel) (2020) 12(11). doi: 10.3390/cancers12113250
12. Tang J, Li Y, Wang J, Wen Z, Lai M, Zhang H. Molecular Mechanisms of microRNAs in Regulating Epithelial-Mesenchymal Transitions in Human Cancers. Cancer Lett (2016) 371(2):301–13. doi: 10.1016/j.canlet.2015.11.043
13. Taniue K, Akimitsu N. The Functions and Unique Features of LncRNAs in Cancer Development and Tumorigenesis. Int J Mol Sci (2021) 22(2). doi: 10.3390/ijms22020632
14. Fesler A, Ju J. Development of microRNA-Based Therapy for Pancreatic Cancer. J Pancreatol (2019) 2(4):147–51. doi: 10.1097/JP9.0000000000000029
15. Liu SJ, Dang HX, Lim DA, Feng FY, Maher CA. Long Noncoding RNAs in Cancer Metastasis. Nat Rev Canc (2021) 21(7):446–60. doi: 10.1038/s41568-021-00353-1
16. Kristensen LS, Jakobsen T, Hager H, Kjems J. The Emerging Roles of circRNAs in Cancer and Oncology. Nat Rev Clin Oncol (2022) 19(3):188–206. doi: 10.1038/s41571-021-00585-y
17. Li Y, Zhao J, Yu S, Wang Z, He X, Su Y, et al. Extracellular Vesicles Long RNA Sequencing Reveals Abundant mRNA, circRNA, and lncRNA in Human Blood as Potential Biomarkers for Cancer Diagnosis. Clin Chem (2019) 65(6):798–808. doi: 10.1373/clinchem.2018.301291
18. Tong Y, Liu X, Xia D, Peng E, Yang X, Liu H, et al. Biological Roles and Clinical Significance of Exosome-Derived Noncoding RNAs in Bladder Cancer. Front Oncol (2021) 11:704703. doi: 10.3389/fonc.2021.704703
19. Yang J, Antin P, Berx G, Blanpain C, Brabletz T, Bronner M, et al. Author Correction: Guidelines and Definitions for Research on Epithelial-Mesenchymal Transition. Nat Rev Mol Cell Biol (2021) 22(12):834. doi: 10.1038/s41580-021-00428-9
20. Shibue T, Weinberg RA. EMT, CSCs, and Drug Resistance: The Mechanistic Link and Clinical Implications. Nat Rev Clin Oncol (2017) 14(10):611–29. doi: 10.1038/nrclinonc.2017.44
21. Soundararajan R, Fradette JJ, Konen JM, Moulder S, Zhang X, Gibbons DL, et al. Targeting the Interplay Between Epithelial-To-Mesenchymal-Transition and the Immune System for Effective Immunotherapy. Cancers (Basel) (2019) 11(5). doi: 10.3390/cancers11050714
22. Beuran M, Negoi I, Paun S, Ion AD, Bleotu C, Negoi RI, et al. The Epithelial to Mesenchymal Transition in Pancreatic Cancer: A Systematic Review. Pancreatology (2015) 15(3):217–25. doi: 10.1016/j.pan.2015.02.011
23. Seddiki R, Narayana G, Strale PO, Balcioglu HE, Peyret G, Yao M, et al. Force-Dependent Binding of Vinculin to α-Catenin Regulates Cell-Cell Contact Stability and Collective Cell Behavior. Mol Biol Cell (2018) 29(4):380–8. doi: 10.1091/mbc.E17-04-0231
24. van Roy F, Berx G. The Cell-Cell Adhesion Molecule E-Cadherin. Cell Mol Life Sci (2008) 65(23):3756–88. doi: 10.1007/s00018-008-8281-1
25. Mrozik KM, Blaschuk OW, Cheong CM, Zannettino ACW, Vandyke K. N-Cadherin in Cancer Metastasis, its Emerging Role in Haematological Malignancies and Potential as a Therapeutic Target in Cancer. BMC Canc (2018) 18(1):939. doi: 10.1186/s12885-018-4845-0
26. Usman S, Waseem NH, Nguyen TKN, Mohsin S, Jamal A, Teh MT, et al. Vimentin is at the Heart of Epithelial Mesenchymal Transition (EMT) Mediated Metastasis. Cancers (Basel) (2021) 13(19). doi: 10.3390/cancers13194985
27. Lamouille S, Xu J, Derynck R. Molecular Mechanisms of Epithelial-Mesenchymal Transition. Nat Rev Mol Cell Biol (2014) 15(3):178–96. doi: 10.1038/nrm3758
28. Batlle E, Sancho E, Francí C, Domínguez D, Monfar M, Baulida J, et al. The Transcription Factor Snail is a Repressor of E-Cadherin Gene Expression in Epithelial Tumour Cells. Nat Cell Biol (2000) 2(2):84–9. doi: 10.1038/35000034
29. Cano A, Pérez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, et al. The Transcription Factor Snail Controls Epithelial-Mesenchymal Transitions by Repressing E-Cadherin Expression. Nat Cell Biol (2000) 2(2):76–83. doi: 10.1038/35000025
30. Yang F, Sun L, Li Q, Han X, Lei L, Zhang H, et al. SET8 Promotes Epithelial-Mesenchymal Transition and Confers TWIST Dual Transcriptional Activities. EMBO J (2012) 31(1):110–23. doi: 10.1038/emboj.2011.364
31. Yang MH, Hsu DS, Wang HW, Wang HJ, Lan HY, Yang WH, et al. Author Correction: Bmi1 is Essential in Twist1-Induced Epithelial-Mesenchymal Transition. Nat Cell Biol (2019) 21(4):533. doi: 10.1038/s41556-019-0290-9
32. Comijn J, Berx G, Vermassen P, Verschueren K, van Grunsven L, Bruyneel E, et al. The Two-Handed E Box Binding Zinc Finger Protein SIP1 Downregulates E-Cadherin and Induces Invasion. Mol Cell (2001) 7(6):1267–78. doi: 10.1016/S1097-2765(01)00260-X
33. Loh CY, Chai JY, Tang TF, Wong WF, Sethi G, Shanmugam MK, et al. The E-Cadherin and N-Cadherin Switch in Epithelial-to-Mesenchymal Transition: Signaling, Therapeutic Implications, and Challenges. Cells (2019) 8(10). doi: 10.3390/cells8101118
34. Feng XH, Derynck R. Specificity and Versatility in Tgf-Beta Signaling Through Smads. Annu Rev Cell Dev Biol (2005) 21:659–93. doi: 10.1146/annurev.cellbio.21.022404.142018
35. Xu J, Lamouille S, Derynck R. TGF-Beta-Induced Epithelial to Mesenchymal Transition. Cell Res (2009) 19(2):156–72. doi: 10.1038/cr.2009.5
36. Derynck R, Zhang YE. Smad-Dependent and Smad-Independent Pathways in TGF-Beta Family Signalling. Natur (2003) 425(6958):577–84. doi: 10.1038/nature02006
37. Grände M, Franzen A, Karlsson JO, Ericson LE, Heldin NE, Nilsson M. Transforming Growth Factor-Beta and Epidermal Growth Factor Synergistically Stimulate Epithelial to Mesenchymal Transition (EMT) Through a MEK-Dependent Mechanism in Primary Cultured Pig Thyrocytes. J Cell Sci (2002) 115(Pt 22):4227–36. doi: 10.1242/jcs.00091
38. Marchetti A, Colletti M, Cozzolino AM, Steindler C, Lunadei M, Mancone C, et al. ERK5/MAPK is Activated by TGFbeta in Hepatocytes and Required for the GSK-3beta-Mediated Snail Protein Stabilization. Cell Signal (2008) 20(11):2113–8. doi: 10.1016/j.cellsig.2008.08.002
39. Zhou BP, Deng J, Xia W, Xu J, Li YM, Gunduz M, et al. Dual Regulation of Snail by GSK-3beta-Mediated Phosphorylation in Control of Epithelial-Mesenchymal Transition. Nat Cell Biol (2004) 6(10):931–40. doi: 10.1038/ncb1173
40. Niehrs C. The Complex World of WNT Receptor Signalling. Nat Rev Mol Cell Biol (2012) 13(12):767–79. doi: 10.1038/nrm3470
41. Briscoe J, Thérond PP. The Mechanisms of Hedgehog Signalling and its Roles in Development and Disease. Nat Rev Mol Cell Biol (2013) 14(7):416–29. doi: 10.1038/nrm3598
42. Xu X, Su B, Xie C, Wei S, Zhou Y, Liu H, et al. Sonic Hedgehog-Gli1 Signaling Pathway Regulates the Epithelial Mesenchymal Transition (EMT) by Mediating a New Target Gene, S100A4, in Pancreatic Cancer Cells. PloS One (2014) 9(7):e96441. doi: 10.1371/journal.pone.0096441
43. Gao Y, Zhang W, Han X, Li F, Wang X, Wang R, et al. YAP Inhibits Squamous Transdifferentiation of Lkb1-Deficient Lung Adenocarcinoma Through ZEB2-Dependent DNp63 Repression. Nat Commun (2014) 5:4629. doi: 10.1038/ncomms5629
44. Jin W. Role of JAK/STAT3 Signaling in the Regulation of Metastasis, the Transition of Cancer Stem Cells, and Chemoresistance of Cancer by Epithelial-Mesenchymal Transition. Cells (2020) 9(1). doi: 10.3390/cells9010217
45. Jung HY, Fattet L, Yang J. Molecular Pathways: Linking Tumor Microenvironment to Epithelial-Mesenchymal Transition in Metastasis. Clin Cancer Res (2015) 21(5):962–8. doi: 10.1158/1078-0432.CCR-13-3173
46. Brandl M, Seidler B, Haller F, Adamski J, Schmid RM, Saur D, et al. IKK(α) Controls Canonical TGF(ß)-SMAD Signaling to Regulate Genes Expressing SNAIL and SLUG During EMT in Panc1 Cells. J Cell Sci (2010) 123(Pt 24):4231–9. doi: 10.1242/jcs.071100
47. Chua HL, Bhat-Nakshatri P, Clare SE, Morimiya A, Badve S, Nakshatri H. NF-kappaB Represses E-Cadherin Expression and Enhances Epithelial to Mesenchymal Transition of Mammary Epithelial Cells: Potential Involvement of ZEB-1 and ZEB-2. Oncogene (2007) 26(5):711–24. doi: 10.1038/sj.onc.1209808
48. Li CW, Xia W, Huo L, Lim SO, Wu Y, Hsu JL, et al. Epithelial-Mesenchymal Transition Induced by TNF-α Requires NF-κb-Mediated Transcriptional Upregulation of Twist1. Cancer Res (2012) 72(5):1290–300. doi: 10.1158/0008-5472.CAN-11-3123
49. Storci G, Sansone P, Mari S, D'Uva G, Tavolari S, Guarnieri T, et al. TNFalpha Up-Regulates SLUG via the NF-Kappab/HIF1alpha Axis, Which Imparts Breast Cancer Cells With a Stem Cell-Like Phenotype. J Cell Physiol (2010) 225(3):682–91. doi: 10.1002/jcp.22264
50. Sullivan NJ, Sasser AK, Axel AE, Vesuna F, Raman V, Ramirez N, et al. Interleukin-6 Induces an Epithelial-Mesenchymal Transition Phenotype in Human Breast Cancer Cells. Oncogene (2009) 28(33):2940–7. doi: 10.1038/onc.2009.180
51. Park J, Schwarzbauer JE. Mammary Epithelial Cell Interactions With Fibronectin Stimulate Epithelial-Mesenchymal Transition. Oncogene (2014) 33(13):1649–57. doi: 10.1038/onc.2013.118
52. Zhang K, Corsa CA, Ponik SM, Prior JL, Piwnica-Worms D, Eliceiri KW, et al. The Collagen Receptor Discoidin Domain Receptor 2 Stabilizes SNAIL1 to Facilitate Breast Cancer Metastasis. Nat Cell Biol (2013) 15(6):677–87. doi: 10.1038/ncb2743
53. Lu X, Kang Y. Hypoxia and Hypoxia-Inducible Factors: Master Regulators of Metastasis. Clin Cancer Res (2010) 16(24):5928–35. doi: 10.1158/1078-0432.CCR-10-1360
54. Imai T, Horiuchi A, Wang C, Oka K, Ohira S, Nikaido T, et al. Hypoxia Attenuates the Expression of E-Cadherin via Up-Regulation of SNAIL in Ovarian Carcinoma Cells. Am J Pathol (2003) 163(4):1437–47. doi: 10.1016/S0002-9440(10)63501-8
55. Yang MH, Wu MZ, Chiou SH, Chen PM, Chang SY, Liu CJ, et al. Direct Regulation of TWIST by HIF-1alpha Promotes Metastasis. Nat Cell Biol (2008) 10(3):295–305. doi: 10.1038/ncb1691
56. Mak P, Leav I, Pursell B, Bae D, Yang X, Taglienti CA, et al. ERbeta Impedes Prostate Cancer EMT by Destabilizing HIF-1alpha and Inhibiting VEGF-Mediated Snail Nuclear Localization: Implications for Gleason Grading. Cancer Cell (2010) 17(4):319–32. doi: 10.1016/j.ccr.2010.02.030
57. Dong B, Li S, Zhu S, Yi M, Luo S, Wu K. MiRNA-Mediated EMT and CSCs in Cancer Chemoresistance. Exp Hematol Oncol (2021) 10(1):12. doi: 10.1186/s40164-021-00206-5
58. Lee SY, Jeong EK, Ju MK, Jeon HM, Kim MY, Kim CH, et al. Induction of Metastasis, Cancer Stem Cell Phenotype, and Oncogenic Metabolism in Cancer Cells by Ionizing Radiation. Mol Cancer (2017) 16(1):10. doi: 10.1186/s12943-016-0577-4
59. Mitra A, Mishra L, Li S. EMT, CTCs and CSCs in Tumor Relapse and Drug-Resistance. Oncotarget (2015) 6(13):10697–711. doi: 10.18632/oncotarget.4037
60. Olivares-Urbano MA, Griñán-Lisón C, Marchal JA, Núñez MI. CSC Radioresistance: A Therapeutic Challenge to Improve Radiotherapy Effectiveness in Cancer. Cells (2020) 9(7). doi: 10.3390/cells9071651
61. Hirano T, Yoshikawa R, Harada H, Harada Y, Ishida A, Yamazaki T. Long Noncoding RNA, CCDC26, Controls Myeloid Leukemia Cell Growth Through Regulation of KIT Expression. Mol Cancer (2015) 14:90. doi: 10.1186/s12943-015-0364-7
62. Kong R, Zhang EB, Yin DD, You LH, Xu TP, Chen WM, et al. Long Noncoding RNA PVT1 Indicates a Poor Prognosis of Gastric Cancer and Promotes Cell Proliferation Through Epigenetically Regulating P15 and P16. Mol Cancer (2015) 14:82. doi: 10.1186/s12943-015-0355-8
63. Rinn JL, Chang HY. Genome Regulation by Long Noncoding RNAs. Annu Rev Biochem (2012) 81:145–66. doi: 10.1146/annurev-biochem-051410-092902
64. Gibb EA, Brown CJ, Lam WL. The Functional Role of Long non-Coding RNA in Human Carcinomas. Mol Cancer (2011) 10:38. doi: 10.1186/1476-4598-10-38
65. Prensner JR, Chinnaiyan AM. The Emergence of lncRNAs in Cancer Biology. Cancer Discovery (2011) 1(5):391–407. doi: 10.1158/2159-8290.CD-11-0209
66. Li J, Chen Z, Tian L, Zhou C, He MY, Gao Y, et al. LncRNA Profile Study Reveals a three-lncRNA Signature Associated With the Survival of Patients With Oesophageal Squamous Cell Carcinoma. Gut (2014) 63(11):1700–10. doi: 10.1136/gutjnl-2013-305806
67. Zeng S, Xiao YF, Tang B, Hu CJ, Xie R, Yang SM, et al. Long Noncoding RNA in Digestive Tract Cancers: Function, Mechanism, and Potential Biomarker. Oncologist (2015) 20(8):898–906. doi: 10.1634/theoncologist.2014-0475
68. Chen DL, Ju HQ, Lu YX, Chen LZ, Zeng ZL, Zhang DS, et al. Long non-Coding RNA XIST Regulates Gastric Cancer Progression by Acting as a Molecular Sponge of miR-101 to Modulate EZH2 Expression. J Exp Clin Cancer Res (2016) 35(1):142. doi: 10.1186/s13046-016-0420-1
69. Huang T, Liu HW, Chen JQ, Wang SH, Hao LQ, Liu M, et al. The Long Noncoding RNA PVT1 Functions as a Competing Endogenous RNA by Sponging miR-186 in Gastric Cancer. BioMed Pharmacother (2017) 88:302–8. doi: 10.1016/j.biopha.2017.01.049
70. Ji Q, Zhang L, Liu X, Zhou L, Wang W, Han Z, et al. Long non-Coding RNA MALAT1 Promotes Tumour Growth and Metastasis in Colorectal Cancer Through Binding to SFPQ and Releasing Oncogene PTBP2 From SFPQ/PTBP2 Complex. Br J Cancer (2014) 111(4):736–48. doi: 10.1038/bjc.2014.383
71. Batista PJ, Chang HY. Long Noncoding RNAs: Cellular Address Codes in Development and Disease. Cell (2013) 152(6):1298–307. doi: 10.1016/j.cell.2013.02.012
72. Ponting CP, Oliver PL, Reik W. Evolution and Functions of Long Noncoding RNAs. Cell (2009) 136(4):629–41. doi: 10.1016/j.cell.2009.02.006
73. Zhao L, Sun H, Kong H, Chen Z, Chen B, Zhou M. The Lncrna-TUG1/EZH2 Axis Promotes Pancreatic Cancer Cell Proliferation, Migration and EMT Phenotype Formation Through Sponging Mir-382. Cell Physiol Biochem (2017) 42(6):2145–58. doi: 10.1159/000479990
74. Qin CF, Zhao FL. Long non-Coding RNA TUG1 can Promote Proliferation and Migration of Pancreatic Cancer via EMT Pathway. Eur Rev Med Pharmacol Sci (2017) 21(10):2377–84.
75. Gao Y, Zhang Z, Li K, Gong L, Yang Q, Huang X, et al. Linc-DYNC2H1-4 Promotes EMT and CSC Phenotypes by Acting as a Sponge of miR-145 in Pancreatic Cancer Cells. Cell Death Dis (2017) 8(7):e2924. doi: 10.1038/cddis.2017.311
76. Song S, Yu W, Lin S, Zhang M, Wang T, Guo S, et al. LncRNA ADPGK-AS1 Promotes Pancreatic Cancer Progression Through Activating ZEB1-Mediated Epithelial-Mesenchymal Transition. Cancer Biol Ther (2018) 19(7):573–83. doi: 10.1080/15384047.2018.1423912
77. Zhou B, Guo W, Sun C, Zhang B, Zheng F. Linc00462 Promotes Pancreatic Cancer Invasiveness Through the miR-665/TGFBR1-TGFBR2/SMAD2/3 Pathway. Cell Death Dis (2018) 9(6):706. doi: 10.1038/s41420-018-0072-3
78. Li H, Wang X, Wen C, Huo Z, Wang W, Zhan Q, et al. Long Noncoding RNA NORAD, a Novel Competing Endogenous RNA, Enhances the Hypoxia-Induced Epithelial-Mesenchymal Transition to Promote Metastasis in Pancreatic Cancer. Mol Cancer (2017) 16(1):169. doi: 10.1186/s12943-017-0738-0
79. Li Z, Jiang P, Li J, Peng M, Zhao X, Zhang X, et al. Tumor-Derived Exosomal lnc-Sox2ot Promotes EMT and Stemness by Acting as a ceRNA in Pancreatic Ductal Adenocarcinoma. Oncogene (2018) 37(28):3822–38. doi: 10.1038/s41388-018-0237-9
80. Chen S, Guo W, Meng M, Wu D, Zhou T, Wang L, et al. LncRNA SNHG1 Promotes the Progression of Pancreatic Cancer by Regulating FGFR1 Expression via Competitively Binding to miR-497. Front Oncol (2022) 12:813850. doi: 10.3389/fonc.2022.813850
81. Wang F, Rong L, Zhang Z, Li M, Ma L, Ma Y, et al. LncRNA H19-Derived miR-675-3p Promotes Epithelial-Mesenchymal Transition and Stemness in Human Pancreatic Cancer Cells by Targeting the STAT3 Pathway. J Cancer (2020) 11(16):4771–82. doi: 10.7150/jca.44833
82. Zhang H, Zhu C, He Z, Chen S, Li L, Sun C. LncRNA PSMB8-AS1 Contributes to Pancreatic Cancer Progression via Modulating miR-382-3p/STAT1/PD-L1 Axis. J Exp Clin Cancer Res (2020) 39(1):179. doi: 10.1186/s13046-020-01687-8
83. Wu L, Liu Y, Guo C, Shao Y. LncRNA OIP5-AS1 Promotes the Malignancy of Pancreatic Ductal Adenocarcinoma via Regulating miR-429/FOXD1/ERK Pathway. Cancer Cell Int (2020) 20:296. doi: 10.1186/s12935-020-01366-w
84. Zhang Y, Ma H, Chen C. Long Noncoding RNA PCED1BAS1 Promotes Pancreatic Ductal Adenocarcinoma Progression by Regulating the Mir4113p/HIF1alpha Axis. Oncol Rep (2021) 46(1). doi: 10.3892/or.2021.8085
85. Chen S, Chen JZ, Zhang JQ, Chen HX, Qiu FN, Yan ML, et al. Silencing of Long Noncoding RNA LINC00958 Prevents Tumor Initiation of Pancreatic Cancer by Acting as a Sponge of microRNA-330-5p to Down-Regulate PAX8. Cancer Lett (2019) 446:49–61. doi: 10.1016/j.canlet.2018.12.017
86. Terashima M, Ishimura A, Wanna-Udom S, Suzuki T. MEG8 Long Noncoding RNA Contributes to Epigenetic Progression of the Epithelial-Mesenchymal Transition of Lung and Pancreatic Cancer Cells. J Biol Chem (2018) 293(47):18016–30. doi: 10.1074/jbc.RA118.004006
87. Liu B, Wu S, Ma J, Yan S, Xiao Z, Wan L, et al. lncRNA GAS5 Reverses EMT and Tumor Stem Cell-Mediated Gemcitabine Resistance and Metastasis by Targeting miR-221/SOCS3 in Pancreatic Cancer. Mol Ther Nucleic Acids (2018) 13:472–82. doi: 10.1016/j.omtn.2018.09.026
88. Shen J, Hong L, Yu D, Cao T, Zhou Z, He S. LncRNA XIST Promotes Pancreatic Cancer Migration, Invasion and EMT by Sponging miR-429 to Modulate ZEB1 Expression. Int J Biochem Cell Biol (2019) 113:17–26. doi: 10.1016/j.biocel.2019.05.021
89. Cao W, Zhou G. LncRNA SNHG12 Contributes Proliferation, Invasion and Epithelial-Mesenchymal Transition of Pancreatic Cancer Cells by Absorbing miRNA-320b. Biosci Rep (2020) 40(6). doi: 10.1042/BSR20200805
90. Zhan HX, Wang Y, Li C, Xu JW, Zhou B, Zhu JK, et al. LincRNA-ROR Promotes Invasion, Metastasis and Tumor Growth in Pancreatic Cancer Through Activating ZEB1 Pathway. Cancer Lett (2016) 374(2):261–71. doi: 10.1016/j.canlet.2016.02.018
91. Chen W, Wang H, Liu Y, Xu W, Ling C, Li Y, et al. Linc-RoR Promotes Proliferation, Migration, and Invasion via the Hippo/YAP Pathway in Pancreatic Cancer Cells. J Cell Biochem (2020) 121(1):632–41. doi: 10.1002/jcb.29308
92. Deng SJ, Chen HY, Ye Z, Deng SC, Zhu S, Zeng Z, et al. Hypoxia-Induced LncRNA-BX111 Promotes Metastasis and Progression of Pancreatic Cancer Through Regulating ZEB1 Transcription. Oncogene (2018) 37(44):5811–28. doi: 10.1038/s41388-018-0382-1
93. Liu Y, Tang T, Yang X, Qin P, Wang P, Zhang H, et al. Tumor-Derived Exosomal Long Noncoding RNA LINC01133, Regulated by Periostin, Contributes to Pancreatic Ductal Adenocarcinoma Epithelial-Mesenchymal Transition Through the Wnt/beta-Catenin Pathway by Silencing AXIN2. Oncogene (2021) 40(17):3164–79. doi: 10.1038/s41388-021-01762-0
94. Hu H, Wang Y, Ding X, He Y, Lu Z, Wu P, et al. Long non-Coding RNA XLOC_000647 Suppresses Progression of Pancreatic Cancer and Decreases Epithelial-Mesenchymal Transition-Induced Cell Invasion by Down-Regulating NLRP3. Mol Cancer (2018) 17(1):18. doi: 10.1186/s12943-018-0761-9
95. Zhang P, Cao M, Zhang Y, Xu L, Meng F, Wu X, et al. A Novel Antisense lncRNA NT5E Promotes Progression by Modulating the Expression of SYNCRIP and Predicts a Poor Prognosis in Pancreatic Cancer. J Cell Mol Med (2020) 24(18):10898–912. doi: 10.1111/jcmm.15718
96. Li Z, Zhao X, Zhou Y, Liu Y, Zhou Q, Ye H, et al. The Long non-Coding RNA HOTTIP Promotes Progression and Gemcitabine Resistance by Regulating HOXA13 in Pancreatic Cancer. J Transl Med (2015) 13:84. doi: 10.1186/s12967-015-0442-z
97. Jiao F, Hu H, Yuan C, Wang L, Jiang W, Jin Z, et al. Elevated Expression Level of Long Noncoding RNA MALAT-1 Facilitates Cell Growth, Migration and Invasion in Pancreatic Cancer. Oncol Rep (2014) 32(6):2485–92. doi: 10.3892/or.2014.3518
98. Dorn A, Glass M, Neu CT, Heydel B, Huttelmaier S, Gutschner T, et al. LINC00261 Is Differentially Expressed in Pancreatic Cancer Subtypes and Regulates a Pro-Epithelial Cell Identity. Cancers (Basel) (2020) 12(5). doi: 10.3390/cancers12051227
99. Wu BQ, Jiang Y, Zhu F, Sun DL, He XZ. Long Noncoding RNA PVT1 Promotes EMT and Cell Proliferation and Migration Through Downregulating P21 in Pancreatic Cancer Cells. Technol Cancer Res Treat (2017) 16(6):819–27. doi: 10.1177/1533034617700559
100. Zhang X, Feng W, Zhang J, Ge L, Zhang Y, Jiang X, et al. Long Noncoding RNA PVT1 Promotes Epithelialmesenchymal Transition via the TGFbeta/Smad Pathway in Pancreatic Cancer Cells. Oncol Rep (2018) 40(2):1093–102. doi: 10.3892/or.2018.6462
101. Chen S, Zhang JQ, Chen JZ, Chen HX, Qiu FN, Yan ML, et al. The Over Expression of Long non-Coding RNA ANRIL Promotes Epithelial-Mesenchymal Transition by Activating the ATM-E2F1 Signaling Pathway in Pancreatic Cancer: An In Vivo and In Vitro Study. Int J Biol Macromol (2017) 102:718–28. doi: 10.1016/j.ijbiomac.2017.03.123
102. He Y, Luo Y, Liang B, Ye L, Lu G, He W. Potential Applications of MEG3 in Cancer Diagnosis and Prognosis. Oncotarget (2017) 8(42):73282–95. doi: 10.18632/oncotarget.19931
103. Tang Y, Song G, Liu H, Yang S, Yu X, Shi L. Silencing of Long Non-Coding RNA HOTAIR Alleviates Epithelial-Mesenchymal Transition in Pancreatic Cancer via the Wnt/beta-Catenin Signaling Pathway. Cancer Manag Res (2021) 13:3247–57. doi: 10.2147/CMAR.S265578
104. Yuan Q, Zhang Y, Feng L, Jiang Y. Upregulated Long Noncoding RNA LINC01296 Indicates a Dismal Prognosis for Pancreatic Ductal Adenocarcinoma and Promotes Cell Metastatic Properties by Affecting EMT. J Cell Biochem (2019) 120(1):552–61. doi: 10.1002/jcb.27411
105. Ma G, Li G, Fan W, Xu Y, Song S, Guo K, et al. The Role of Long Noncoding RNA AL161431.1 in the Development and Progression of Pancreatic Cancer. Front Oncol (2021) 11:666313. doi: 10.3389/fonc.2021.666313
106. Wang X, Li H, Lu X, Wen C, Huo Z, Shi M, et al. Melittin-Induced Long non-Coding RNA NONHSAT105177 Inhibits Proliferation and Migration of Pancreatic Ductal Adenocarcinoma. Cell Death Dis (2018) 9(10):940. doi: 10.1038/s41419-018-0965-3
107. Tian C, Jin Y, Shi S. Long Non-Coding RNA SUMO1P3 may Promote Cell Proliferation, Migration, and Invasion of Pancreatic Cancer via EMT Signaling Pathway. Oncol Lett (2018) 16(5):6109–15. doi: 10.3892/ol.2018.9378
108. Cioce M, Ganci F, Canu V, Sacconi A, Mori F, Canino C, et al. Protumorigenic Effects of Mir-145 Loss in Malignant Pleural Mesothelioma. Oncogene (2014) 33(46):5319–31. doi: 10.1038/onc.2013.476
109. Ren D, Wang M, Guo W, Huang S, Wang Z, Zhao X, et al. Double-Negative Feedback Loop Between ZEB2 and miR-145 Regulates Epithelial-Mesenchymal Transition and Stem Cell Properties in Prostate Cancer Cells. Cell Tissue Res (2014) 358(3):763–78. doi: 10.1007/s00441-014-2001-y
110. Xue M, Pang H, Li X, Li H, Pan J, Chen W. Long non-Coding RNA Urothelial Cancer-Associated 1 Promotes Bladder Cancer Cell Migration and Invasion by Way of the hsa-miR-145-ZEB1/2-FSCN1 Pathway. Cancer Sci (2016) 107(1):18–27. doi: 10.1111/cas.12844
111. Zou L, Chen FR, Xia RP, Wang HW, Xie ZR, Xu Y, et al. Long Noncoding RNA XIST Regulates the EGF Receptor to Promote TGF-β1-Induced Epithelial-Mesenchymal Transition in Pancreatic Cancer. Biochem Cell Biol (2020) 98(2):267–76. doi: 10.1139/bcb-2018-0274
112. Li X, Xu Y, Chen Y, Chen S, Jia X, Sun T, et al. SOX2 Promotes Tumor Metastasis by Stimulating Epithelial-to-Mesenchymal Transition via Regulation of WNT/β-Catenin Signal Network. Cancer Lett (2013) 336(2):379–89. doi: 10.1016/j.canlet.2013.03.027
113. Kachroo P, Lee MH, Zhang L, Baratelli F, Lee G, Srivastava MK, et al. IL-27 Inhibits Epithelial-Mesenchymal Transition and Angiogenic Factor Production in a STAT1-Dominant Pathway in Human Non-Small Cell Lung Cancer. J Exp Clin Cancer Res (2013) 32(1):97. doi: 10.1186/1756-9966-32-97
114. Ma Z, Huang H, Wang J, Zhou Y, Pu F, Zhao Q, et al. Long non-Coding RNA SNHG15 Inhibits P15 and KLF2 Expression to Promote Pancreatic Cancer Proliferation Through EZH2-Mediated H3k27me3. Oncotarget (2017) 8(48):84153–67. doi: 10.18632/oncotarget.20359
115. Xu Z, Gu C, Yao X, Guo W, Wang H, Lin T, et al. CD73 Promotes Tumor Metastasis by Modulating RICS/RhoA Signaling and EMT in Gastric Cancer. Cell Death Dis (2020) 11(3):202. doi: 10.1038/s41419-020-2403-6
116. Chandrashekaran IR, Mohanty B, Linossi EM, Dagley LF, Leung EW, Murphy JM, et al. Structure and Functional Characterization of the Conserved JAK Interaction Region in the Intrinsically Disordered N-Terminus of SOCS5. Biochemistry (2015) 54(30):4672–82. doi: 10.1021/acs.biochem.5b00619
117. Eid RA, Alkhateeb MA, Eleawa S, Al-Hashem FH, Al-Shraim M, El-Kott AF, et al. Cardioprotective Effect of Ghrelin Against Myocardial Infarction-Induced Left Ventricular Injury via Inhibition of SOCS3 and Activation of JAK2/STAT3 Signaling. Basic Res Cardiol (2018) 113(2):13. doi: 10.1007/s00395-018-0671-4
118. Kim MS, Jeong J, Seo J, Kim HS, Kim SJ, Jin W. Dysregulated JAK2 Expression by TrkC Promotes Metastasis Potential, and EMT Program of Metastatic Breast Cancer. Sci Rep (2016) 6:33899. doi: 10.1038/srep33899
119. Mercer TR, Dinger ME, Mattick JS. Long non-Coding RNAs: Insights Into Functions. Nat Rev Genet (2009) 10(3):155–9. doi: 10.1038/nrg2521
120. Ransohoff JD, Wei Y, Khavari PA. The Functions and Unique Features of Long Intergenic Non-Coding RNA. Nat Rev Mol Cell Biol (2018) 19(3):143–57. doi: 10.1038/nrm.2017.104
121. Zhang A, Zhou N, Huang J, Liu Q, Fukuda K, Ma D, et al. The Human Long non-Coding RNA-RoR is a P53 Repressor in Response to DNA Damage. Cell Res (2013) 23(3):340–50. doi: 10.1038/cr.2012.164
122. Kim T, Veronese A, Pichiorri F, Lee TJ, Jeon YJ, Volinia S, et al. P53 Regulates Epithelial-Mesenchymal Transition Through microRNAs Targeting ZEB1 and ZEB2. J Exp Med (2011) 208(5):875–83. doi: 10.1084/jem.20110235
123. Dong P, Karaayvaz M, Jia N, Kaneuchi M, Hamada J, Watari H, et al. Mutant P53 Gain-of-Function Induces Epithelial-Mesenchymal Transition Through Modulation of the miR-130b-ZEB1 Axis. Oncogene (2013) 32(27):3286–95. doi: 10.1038/onc.2012.334
124. Ahmad I, Muneer KM, Tamimi IA, Chang ME, Ata MO, Yusuf N. Thymoquinone Suppresses Metastasis of Melanoma Cells by Inhibition of NLRP3 Inflammasome. Toxicol Appl Pharmacol (2013) 270(1):70–6. doi: 10.1016/j.taap.2013.03.027
125. Fan SH, Wang YY, Lu J, Zheng YL, Wu DM, Li MQ, et al. Luteoloside Suppresses Proliferation and Metastasis of Hepatocellular Carcinoma Cells by Inhibition of NLRP3 Inflammasome. PloS One (2014) 9(2):e89961. doi: 10.1371/journal.pone.0089961
126. Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY, Zhang F, et al. Long non-Coding RNA MALAT-1 Overexpression Predicts Tumor Recurrence of Hepatocellular Carcinoma After Liver Transplantation. Med Oncol (2012) 29(3):1810–6. doi: 10.1007/s12032-011-0004-z
127. Wang J, Su L, Chen X, Li P, Cai Q, Yu B, et al. MALAT1 Promotes Cell Proliferation in Gastric Cancer by Recruiting SF2/ASF. BioMed Pharmacother (2014) 68(5):557–64. doi: 10.1016/j.biopha.2014.04.007
128. Wu XS, Wang XA, Wu WG, Hu YP, Li ML, Ding Q, et al. MALAT1 Promotes the Proliferation and Metastasis of Gallbladder Cancer Cells by Activating the ERK/MAPK Pathway. Cancer Biol Ther (2014) 15(6):806–14. doi: 10.4161/cbt.28584
129. Ma L, Wang F, Du C, Zhang Z, Guo H, Xie X, et al. Long non-Coding RNA MEG3 Functions as a Tumour Suppressor and has Prognostic Predictive Value in Human Pancreatic Cancer. Oncol Rep (2018) 39(3):1132–40. doi: 10.3892/or.2018.6178
130. Zhou Y, Zhang X, Klibanski A. MEG3 Noncoding RNA: A Tumor Suppressor. J Mol Endocrinol (2012) 48(3):R45–53. doi: 10.1530/JME-12-0008
131. Li CH, Xiao Z, Tong JH, To KF, Fang X, Cheng AS, et al. EZH2 Coupled With HOTAIR to Silence MicroRNA-34a by the Induction of Heterochromatin Formation in Human Pancreatic Ductal Adenocarcinoma. Int J Cancer (2017) 140(1):120–9. doi: 10.1002/ijc.30414
132. Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, et al. HOTAIR is a Negative Prognostic Factor and Exhibits Pro-Oncogenic Activity in Pancreatic Cancer. Oncogene (2013) 32(13):1616–25. doi: 10.1038/onc.2012.193
133. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are Abundant, Conserved, and Associated With ALU Repeats. RNA (2013) 19(2):141–57. doi: 10.1261/rna.035667.112
134. Chen YY, Jiang MJ, Tian L. Analysis of Exosomal circRNAs Upon Irradiation in Pancreatic Cancer Cell Repopulation. BMC Med Genomics (2020) 13(1):107. doi: 10.1186/s12920-020-00756-3
135. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The Biogenesis, Biology and Characterization of Circular RNAs. Nat Rev Genet (2019) 20(11):675–91. doi: 10.1038/s41576-019-0158-7
136. Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, et al. Circular RNA: A New Star of Noncoding RNAs. Cancer Lett (2015) 365(2):141–8. doi: 10.1016/j.canlet.2015.06.003
137. Szabo L, Salzman J. Detecting Circular RNAs: Bioinformatic and Experimental Challenges. Nat Rev Genet (2016) 17(11):679–92. doi: 10.1038/nrg.2016.114
138. Zhou WY, Cai ZR, Liu J, Wang DS, Ju HQ, Xu RH. Circular RNA: Metabolism, Functions and Interactions With Proteins. Mol Cancer (2020) 19(1):172. doi: 10.1186/s12943-020-01286-3
139. Rajappa A, Banerjee S, Sharma V, Khandelia P. Circular RNAs: Emerging Role in Cancer Diagnostics and Therapeutics. Front Mol Biosci (2020) 7:577938. doi: 10.3389/fmolb.2020.577938
140. Tabatabaeian H, Peiling Yang S, Tay Y. Non-Coding RNAs: Uncharted Mediators of Thyroid Cancer Pathogenesis. Cancers (Basel) (2020) 12(11). doi: 10.3390/cancers12113264
141. Shen P, Yang T, Chen Q, Yuan H, Wu P, Cai B, et al. CircNEIL3 Regulatory Loop Promotes Pancreatic Ductal Adenocarcinoma Progression via miRNA Sponging and A-To-I RNA-Editing. Mol Cancer (2021) 20(1):51. doi: 10.1186/s12943-021-01333-7
142. Shen Q, Zheng G, Zhou Y, Tong J, Xu S, Gao H, et al. CircRNA Circ_0092314 Induces Epithelial-Mesenchymal Transition of Pancreatic Cancer Cells via Elevating the Expression of S100P by Sponging miR-671. Front Oncol (2021) 11:675442. doi: 10.3389/fonc.2021.675442
143. Zhang R, Zhu W, Ma C, Ai K. Silencing of circRNA Circ_0001666 Represses EMT in Pancreatic Cancer Through Upregulating miR-1251 and Downregulating Sox4. Front Mol Biosci (2021) 8:684866. doi: 10.3389/fmolb.2021.684866
144. Ma G, Li G, Fan W, Xu Y, Song S, Guo K, et al. Circ-0005105 Activates COL11A1 by Targeting miR-20a-3p to Promote Pancreatic Ductal Adenocarcinoma Progression. Cell Death Dis (2021) 12(7):656. doi: 10.1038/s41419-021-03938-8
145. Yu S, Wang M, Zhang H, Guo X, Qin R. Circ_0092367 Inhibits EMT and Gemcitabine Resistance in Pancreatic Cancer via Regulating the miR-1206/ESRP1 Axis. Genes (Basel) (2021) 12(11). doi: 10.3390/genes12111701
146. Liu W, Deng L, Xu A, Xiong X, Tao J, Chang J, et al. Identifying a Novel IRF3/circUHRF1/miR-1306-5p/ARL4C Axis in Pancreatic Ductal Adenocarcinoma Progression. Cell Cycle (2022) 21(4):392–405. doi: 10.1080/15384101.2021.2020450
147. Wong CH, Lou UK, Fung FK, Tong JHM, Zhang CH, To KF, et al. CircRTN4 Promotes Pancreatic Cancer Progression Through a Novel CircRNA-miRNA-lncRNA Pathway and Stabilizing Epithelial-Mesenchymal Transition Protein. Mol Cancer (2022) 21(1):10. doi: 10.1186/s12943-021-01481-w
148. Wang J, Zhang N, Han Q, Lu W, Wang L, Yang D, et al. Pin1 Inhibition Reverses the Acquired Resistance of Human Hepatocellular Carcinoma Cells to Regorafenib via the Gli1/Snail/E-Cadherin Pathway. Cancer Lett (2019) 444:82–93. doi: 10.1016/j.canlet.2018.12.010
149. Hanieh H, Ahmed EA, Vishnubalaji R, Alajez NM. SOX4: Epigenetic Regulation and Role in Tumorigenesis. Semin Cancer Biol (2020) 67(Pt 1):91–104. doi: 10.1016/j.semcancer.2019.06.022
Keywords: long noncoding RNA, circular RNA, epithelial-mesenchymal transition, pancreatic cancer, competing endogenous RNA
Citation: Yang X, Qin C, Zhao B, Li T, Wang Y, Li Z, Li T and Wang W (2022) Long Noncoding RNA and Circular RNA: Two Rising Stars in Regulating Epithelial-Mesenchymal Transition of Pancreatic Cancer. Front. Oncol. 12:910678. doi: 10.3389/fonc.2022.910678
Received: 01 April 2022; Accepted: 09 May 2022;
Published: 03 June 2022.
Edited by:
Jiayin Wang, Xi’an Jiaotong University, ChinaCopyright © 2022 Yang, Qin, Zhao, Li, Wang, Li, Li and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weibin Wang, wwb_xh@163.com
†These authors have contributed equally to this work
 Xiaoying Yang†
Xiaoying Yang† Cheng Qin
Cheng Qin Bangbo Zhao
Bangbo Zhao Tianhao Li
Tianhao Li Zeru Li
Zeru Li Weibin Wang
Weibin Wang