- 1Department of Pharmacy, Shandong Provincial Hospital Affiliated to Shandong First Medical University, Jinan, China
- 2Department of Pharmacy, The First Affiliated Hospital of Medical College of Shantou University, Shantou, China
- 3Department of Pharmacy, The First Affiliated Hospital of Shandong First Medical University, Jinan, China
Breast cancer is the most common malignancy among women worldwide, excluding non-melanoma skin cancer. It is now well understood that breast cancer is a heterogeneous entity that exhibits distinctive histological and biological features, treatment responses and prognostic patterns. Therefore, the identification of novel ideal diagnostic and prognostic biomarkers is of utmost importance. Long non-coding RNAs (lncRNAs) are commonly defined as transcripts longer than 200 nucleotides that lack coding potential. Extensive research has shown that lncRNAs are involved in multiple human cancers, including breast cancer. LncRNAs with dysregulated expression can act as oncogenes or tumor-suppressor genes to regulate malignant transformation processes, such as proliferation, invasion, migration and drug resistance. Intriguingly, the expression profiles of lncRNAs tend to be highly cell-type-specific, tissue-specific, disease-specific or developmental stage-specific, which makes them suitable biomarkers for breast cancer diagnosis and prognosis.
Introduction
Breast cancer is a major public health dilemma on a global scale. Inherited and acquired genetic as well as epigenetic alterations have been extensively demonstrated as the driving events of breast cancer (1). Breast cancer is a heterogeneous entity, and different subtypes exhibit distinctive histological and biological features, treatment responses and prognostic patterns (2). Despite substantial advancement in early detection and management, breast cancer remains the second-leading cause of cancer-related death among women worldwide (3). Moreover, advanced or metastatic breast cancer is almost incurable by current systemic treatment options (4). As such, one key challenge in breast cancer therapy is to identify novel reliable diagnostic and prognostic biomarkers.
Only approximately 2% of the human genome is composed of protein-coding transcripts, indicating that the majority of transcripts are non-coding RNAs (ncRNAs) (5). NcRNAs are broadly divided into small ncRNAs (20~200 nucleotides) and long ncRNAs (lncRNAs, >200 nucleotides) (6). LncRNAs were previously considered “transcriptional noise” due to the lack of a significant open reading frame (7). However, increasing evidence indicates that lncRNAs are involved in different biological and pathological processes, including cell apoptosis, differentiation and autophagy (8). Over the last decade, high-throughput and next-generation sequencing technologies have allowed the study of RNAs in an unbiased manner. These technological advances contribute to an explosion of genomic information and increase the ability to identify novel lncRNAs.
More importantly, accumulating evidence suggests that lncRNAs are involved in various human cancers, including breast cancer. Approximately 1,900 lncRNAs are dysregulated in breast cancer (9), and their levels may be associated with distinct clinical outcomes. Dysregulated lncRNAs can act as oncogenes or tumor suppressors to control breast cancer pathophysiology and should be investigated to obtain a better understanding of their roles in breast cancer biology and determine their suitability as diagnostic and prognostic biomarkers. Therefore, this study aims to review the current knowledge about lncRNAs and evaluate their potential roles as molecular markers in breast cancer.
Breast Cancer
Breast cancer is the most common cancer among women worldwide, excluding non-melanoma skin cancer (10). In 2018, approximately 2.1 million cases of breast cancer were newly diagnosed, and approximately 626,679 patients died that same year (10). The well-established risk factors for breast cancer are race, family history of cancer, genetic susceptibility, modifiable exposures, environmental factors and unhealthy lifestyles (11). The incidence of breast cancer varies worldwide and is higher in high-income countries than in low-income countries. However, the death rate in lower-income regions is higher due to the lack of early diagnosis and limited access to treatment (12).
Breast cancer is a complex, heterogeneous disease, and different subtypes have distinct clinical presentations and therapeutic responses. Breast cancer is defined into 3 clinically relevant subtypes according to estrogen receptor (ER) expression, progesterone receptor (PR) expression and human epidermal growth factor 2 (ERBB2; formerly HER2) gene amplification: ER-positive/PR-positive, HER2/ERBB2-positive, and triple-negative (lacking expression of all three molecular markers) (2). Triple-negative breast cancer is an aggressive subtype and accounts for 15% of breast tumors (13). It has been well established that triple-negative tumors have a relatively high mitotic activation index, prominent lymphocytic infiltrate, high incidence of distant relapse, and poor clinical outcomes (14). Breast cancer is staged I-IV, and stage IV has distant metastases at diagnosis. Metastatic breast cancer remains essentially incurable, and the therapeutic goals are symptom palliation and prolonging life. The median overall survival (OS) of stage IV breast cancer patients for the triple-negative and HER2/ERBB2-positive subtypes was approximately 1 year and 5 years, respectively (15). As such, detecting breast cancer at an early stage is of paramount importance.
Discovery, Classification, and Function of LNCRNAS
LncRNAs are commonly defined as RNA transcripts larger than 200 nucleotides. Once considered junk DNA, lncRNAs have recently attracted wide attention as crucial regulators in a diverse array of biological processes. Over the past decade, technical advancements in high-throughput sequencing have greatly streamlined the process of identifying all forms of RNAs. To date, more than 58,000 lncRNAs have been identified, and approximately 30,000 lncRNA transcripts have been curated in GENCODE v29 (16).
Like other RNAs, lncRNAs mainly consist of four core nucleotides (17). LncRNAs exhibit the same characteristics as mRNA transcripts, i.e., they are RNA polymerase II–transcribed, 5nscribedrase II–aracteristics as mR (18). Almost all lncRNAs are localized in the cell nucleus, but some lncRNAs can be exported to the cytoplasm (19). With respect to their genomic location, lncRNAs can be classified into intronic, intergenic, sense, antisense and bidirectional loci (6). Unlike the well-studied miRNAs, lncRNA homologs exhibit weak or untraceable primary sequence conservation (20). Importantly, lncRNAs have evolutionarily conserved promoters, suggesting the importance of lncRNA regulation. Furthermore, compared to small ncRNAs, lncRNAs have highly conserved secondary and tertiary structures, which are considered their major functional units (21).
LncRNAs have been identified as indispensable regulatory elements in multiple cell processes, such as chromatin modification and transcriptional and post-transcriptional regulation (22). There is accumulating evidence that the biological functions of lncRNAs depend strictly on their subcellular location (23). Nuclear lncRNAs can function as cis- and trans-acting elements to modulate chromatin remodeling (24). Some lncRNAs may recruit DNA or chromatin regulatory complexes to regulate the epigenetic silencing or activation of target loci by altering histone modifications or DNA methylation patterns (25) (Figure 1A). Intriguingly, lncRNAs have dynamic and flexible biophysical structures, which confer lncRNAs the ability to serve as scaffolds and allow the assembly of various proteins (26) (Figure 1B). Besides, nuclear lncRNAs can also modulate messenger RNA (mRNA) alternative spicing (27) (Figure 1C). In the cytoplasm, lncRNAs may act as miRNA sponges or competitive endogenous RNAs (ceRNAs) to prevent miRNA binding with target mRNAs, hence regulating the expression of downstream target genes at the post-transcriptional level (28) (Figure 1D). Recent studies have shown that lncRNAs interact with some proteins to regulate their stability and posttranslational modifications (29) (Figure 1E). Furthermore, cytoplasmic lncRNAs transcribed from enhancer regions can stabilize mRNAs by recruiting specific proteins (30) (Figure 1F). More interestingly, some lncRNAs even hold the potential to encode functional small peptides (31) (Figure 1G). Collectively, lncRNAs exert regulatory roles by directly or indirectly interacting with DNA, RNA, or proteins, and the functions of lncRNAs are still under investigation.
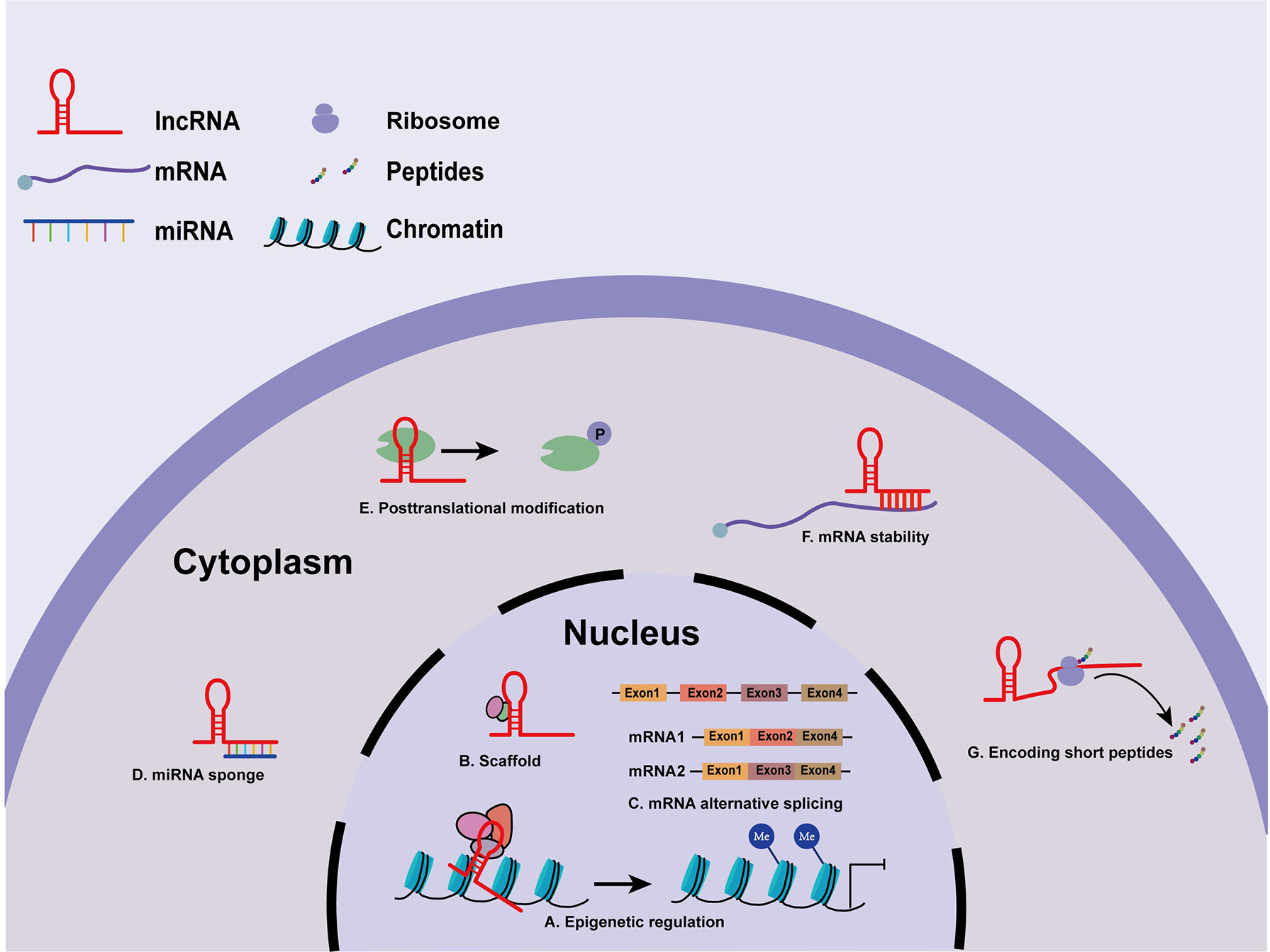
Figure 1 Molecular mechanisms of lncRNAs. (A) LncRNAs modulate histone modification and chromatin remodeling. (B) LncRNAs serve as scaffolds for various proteins. (C) Nuclear lncRNAs participate in mRNA processing. (D) LncRNAs act as miRNA sponges or ceRNAs in regulating miRNAs. (E) LncRNAs regulate the stability and posttranslational modifications of some proteins. (F) lncRNAs stabilize mRNAs. (G) lncRNAs encode functional small peptides.
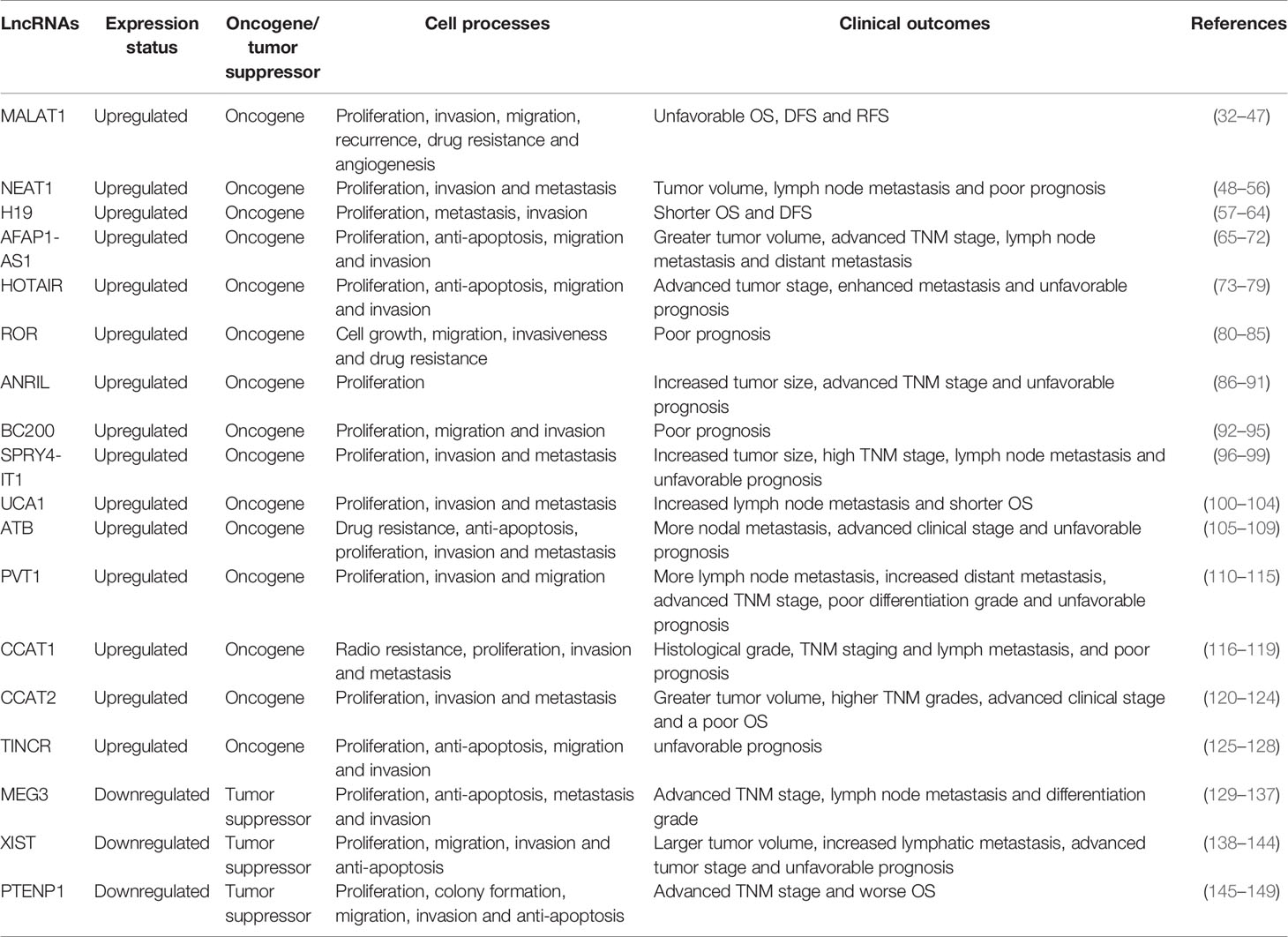
Recent studies have shed light on the regulatory effects of lncRNAs in human cancers. Importantly, some aberrant lncRNAs hold the promise to serve as ideal biomarkers in certain cancers. Here, main lncRNAs with diagnostic and prognostic values in breast cancer are summarized in Table 1.
Oncogenic LNCRNAS
Metastasis-Associated Lung Adenocarcinoma Transcript 1
MALAT1, located in the nucleus, is approximately 8,000 nucleotides in length. MALAT1 is among the most conserved and extremely abundant lncRNAs in different tissues, suggesting that it may have vital biological functions (32, 33). MALAT1 was initially identified as a tumor promoter in non-small cell lung cancer (34). To date, MALAT1 overexpression has been shown in multiple human cancers, such as ovarian, bladder and colorectal cancers (35–37). It has been demonstrated that serum MALAT1 levels in breast cancer patients are markedly higher than those in patients with benign breast disease (38). Huang et al. indicated that the expression of MALAT1 was elevated in breast cancer patients compared to healthy cases. Moreover, silencing of MALAT1 significantly hindered angiogenesis via upregulation of miR-145 expression (39). It has also been found that MALAT1 promotes epithelial-mesenchymal transition (EMT) mainly by regulating the miR-204/ZEB2 axis (40). Another study demonstrated that MALAT1 could enhance the proliferation and invasion of breast cancer cells by altering the histone 3 lysine 4 (H3K4) epigenotype to activate the EEF1A1 promoter. Downregulation of MALAT1 expression may significantly reduce the promoter activity of EEF1A1, suggesting a novel MALAT1-mediated epigenetic mechanism of EEF1A1 regulation (41). Zhao et al. indicated that a high concentration of 17β-estradiol inhibited the proliferation, invasion and metastasis of breast cells via downregulation of MALAT1 expression (42). In breast cancer, early postoperative fever indicates unfavorable clinical outcomes. Li et al. demonstrated that overexpression of MALAT1 in breast cancer patients with early postoperative fever was significantly related to inflammatory responses and lung metastasis (43). In addition, Wang et al. indicated that elevated MALAT1 levels were inversely correlated with OS in invasive ductal carcinoma (44). Furthermore, a meta-analysis showed that high expression of MALAT1 in breast cancer is correlated with unfavorable disease-free survival (DFS) and recurrence-free survival (RFS) (45). In recent years, accumulating evidence has shown that the aberrant expression of MALAT1, especially in serum/plasma, may serve as a suitable biomarker in various human cancer entities (46, 47).
Nuclear Enriched Abundant Transcript 1
NEAT1 has two variants (NEAT1-1 and NEAT1-2), and it is indispensable for paraspeckle integrity (150). In the last few years, several studies indicating the involvement of NEAT1 in breast cancer have been published, and NEAT1 is closely related to different miRNAs. Jiang et al. indicated that high levels of NEAT1 in diverse breast cancer cell lines were linked to aggressive progression and unfavorable prognosis. They also found that miR-448, an inhibitor of cancer cell growth, was inhibited by NEAT1 and consequently led to increased expression of zinc-finger E-box binding protein 1 (ZEB1), an oncogene (48). Zhang et al. demonstrated that ectopic expression of NEAT1 was related to tumor volume and lymph node metastasis, while silencing of NEAT1 expression resulted in decreased proliferation and migration in breast cancer cell lines (49). In addition, overexpression of NEAT1 leads to the downregulated expression of miR-133b, which is a known inhibitor of tumorigenesis, consequently resulting in enhanced migration and invasion (50). Ke et al. indicated that downregulation of NEAT1 expression by miR-548 could abrogate proliferation and induce apoptosis in breast cancer. They also found that fused in sarcoma (FUS), a nuclear RNA binding protein, directly interacted with NEAT1, and the role of NEAT1 in cancer cell survival was mediated by FUS (51). Enhancer of zest homolog 2 (EZH2), known as a molecular marker of aggressive malignancies, is a target of miR-101. Qian et al. showed that NEAT1 promoted the growth of breast cancer cells via miR-101-dependent EZH2 regulation (52). In addition, NEAT1 could increase the expression of high mobility group AT-hook 2 (HMGA2) by sponging miR-211, thereby enhancing the invasiveness of breast cancer cells (53). High levels of NEAT1, miR-21, and RRM2 have been observed in different breast cancer cell lines, and their elevated levels correlate with poor clinical outcomes, suggesting that the NEAT1/miR-21-RRM2 signaling axis contributes to breast cancer development (54). Moreover, NEAT1 acts as a sponge for 146b-5p to promote the proliferation, migration, and metastasis of breast cancer cells (55). Zhou et al. showed that NEAT1 could coordinate various miRNAs in different breast cancer subtypes and thus exert diverse regulatory functions (56). In conclusion, these data revealed various mechanisms of NEAT1 in the regulation of breast cancer and suggested that NEAT1 might function as a potential biomarker in breast cancer.
H19
H19, an imprinted gene, is located on chromosome 11p15.5 (151). H19 is abundantly expressed during embryogenesis, and the expression of H19 is repressed upon birth, except for basal expression in adult tissues, such as lung, skeletal muscle and mammary gland (152). The first evidence that H19 has a pivotal role in breast cancer was provided by Adriaenssens et al., who found that H19 overexpression was significantly associated with ER/PR status and tumor progression (57). Matouk et al. found that H19 suppressed the expression of E-cadherin, a representative inhibitor of EMT, and promoted metastasis via regulation of Slug in breast cancer (58). H19 functions as a molecular sponge of miR‐152 to upregulate the expression of the DNA methyltransferase DNMT1, thus facilitating the proliferation and invasiveness of breast cancer cells (59). It has also been demonstrated that H19 can sponge miR-200b/c and let-7b differently to enhance EMT and mesenchymal-epithelial transition (MET) (60). Additionally, downregulation of H19 expression could result in S-phase arrest of breast cancer cells, suggesting its role in regulating cell cycle progression (61). H19 could serve as a precursor for miR-675, which is encoded by the exon of H19. Vennin et al. found that high expression of H19 upregulated miR-675 expression, negatively regulating E3 ubiquitin ligases (c-Cbl and Cbl-b) to enhance the aggressiveness of breast cancer cells (62). Zhang et al. showed that high plasma H19 levels were correlated with ER/PR status and lymph node metastasis (63). Moreover, elevated levels of H19 have been significantly associated with unfavorable OS and DFS, particularly in the triple-negative subtype (64). A meta-analysis showed that dysregulated H19 expression correlated with poor differentiation, high tumor stage, early distant metastasis, and lymph node involvement in multiple cancers (153). In sum, current evidence establishes H19 as a potential breast cancer biomarker.
Actin Filament Associated Protein 1 Antisense RNA1
AFAP1-AS1 is 6,810 bp long and is located on human chromosome 4p16.1 (154). Further studies have shown that AFAP1-AS1 expression is markedly upregulated in breast cancer tissues and cell lines and predicts poor clinical outcomes (65). In addition, AFAP1-AS1 overexpression in cancers correlates with greater tumor volume, advanced tumor-node-metastasis (TNM) staging, lymph node metastasis and distant metastasis (65). Intriguingly, elevated levels of AFAP1-AS1 are more common in triple-negative breast cancer (66). Previous experiments suggested that AFAP1-AS1 could promote tumorigenesis by interfering with AFAP1 expression (67). However, Dianatpour et al. demonstrated that high levels of AFAP1-AS1 had no regulatory effect on AFAP1 expression in breast cancer patients (68). Consistent with this study, Ma et al. indicated that silencing of AFAP1-AS1 exerted no effects on AFAP1 expression or actin filament integrity (69). Such discrepancies among different cancers need to be further elucidated. Ki-67, a nuclear antigen, is not expressed in quiescent cells (70). Downregulation of AFAP1-AS1 expression was detected in all of the Ki-67-negative samples, suggesting that AFAP1-AS1 might be implicated in cell proliferation (68). Moreover, AFAP1-AS1 regulates the wnt/β-catenin pathway, facilitating the expression of c-Myc and EMT-associated transcription factors to promote tumorigenesis and induce EMT (66). Furthermore, AFAP1-AS1 directly binds to miR-497-5p to upregulate the expression of Septin 2, a well-known oncogene. Depletion of AFAP1-AS1 inhibits proliferation and migration and induces apoptosis in breast cancer (71). In triple-negative breast cancer, AFAP1-AS1 sponges miR-154 to coordinate the expression of MutT homolog-1, which in turn induces cellular proliferation and invasion (72). Hence, the dysregulated expression of AFAP1-AS1 and its molecular mechanisms identify it as a putative biomarker and actionable target in breast cancer.
HOX Transcript Antisense RNA
HOTAIR is a 2,158-bp lncRNA located on human chromosome 12q13.13 between the HOXC11 and HOXC12 genes (155). Ectopic HOTAIR expression has been implicated in a variety of cancers, such as pancreatic, colorectal and non-small-cell lung cancers (156–158). HOTAIR expression seems to be elevated in cancer tissues compared to paired non-cancerous tissues, and high expression of HOTAIR has been associated with an enhanced proliferation rate, advanced tumor stage, elevated risk of metastasis, and unfavorable prognosis (73, 74). Gupta et al. indicated that HOTAIR expression was upregulated in primary breast tumors and metastases and that dysregulation of HOTAIR in primary tumors correlated with metastasis and poor prognosis (75). On the other hand, treatment with transforming growth factor-β1 (TGF-β1) upregulates the expression of HOTAIR and contributes to EMT. TGF-β1-induced EMT is reversed by HOTAIR knockdown, suggesting that the effect of TGF-β1 on EMT is, at least partly, mediated through HOTAIR (76). Mechanistically, depletion of HOTAIR inhibits the growth, invasion and migration of breast cancer cells through downregulation of p53 expression (77). In addition, HOTAIR sequesters miR-206 to enhance the expression of Bcl-w, an anti-apoptotic protein, thereby promoting the proliferation of breast cancer cells (78). Moreover, HOTAIR could recruit polycomb repressive complex 2 (PRC2), known as a transcriptional corepressor, to facilitate epigenetic gene silencing (75). However, another study demonstrated that the oncogenic role of HOTAIR in breast cancer cells may be independent of PRC2. Instead, the recruitment of PRC2 seemed to be a consequence of gene silencing (79). These contradictory findings on HOTAIR have caused confusion about its role in breast cancer. Hence, further studies are needed to elucidate the interaction between HOTAIR and PRC2. Overall, the involvement of HOTAIR in these signaling pathways contributes to the progression of breast cancer, and HOTAIR might be utilized as a new predictive and prognostic biomarker in breast cancer.
Regulator of Reprogramming
ROR is a 2.6-kb intergenic transcript located on chromosome 18q21.31 (159). ROR was initially discovered as a promoter of the reprogramming process, and it was shown to contribute to the maintenance of pluripotent and embryonic stem cells via inhibition of cellular stress signaling pathways (159). Elevated ROR expression has been detected across cancer cell lines (160). Recent studies have shown that ROR promotes EMT in various cancers (161, 162). Accordingly, ROR overexpression induces EMT and promotes cell growth, migration and invasiveness in breast cancer (80–82). Functionally, ROR regulates the TGF-β pathway to promote breast cancer progression, whereas suppression of ROR inhibits tumor growth (80). In addition, ROR acts as a ceRNA for miR-205 to upregulate the expression of a miR-205 target gene, the EMT inducer ZEB2 (81). Zhou et al. showed that ROR sponged miR-194-3p and upregulated the expression of a miR-194-3p target, the methyl-CpG-binding protein 2 (MECP2) gene, to decrease the sensitivity of breast cancer cells to rapamycin (82). Moreover, ROR activated the MAPK/ERK pathway and upregulated the expression of dual specificity phosphatase 7 (DUSP7), an ERK-specific phosphatase, thereby facilitating estrogen-independent proliferation of breast cancer cells (83). Furthermore, silencing ROR reversed gemcitabine-induced apoptosis and autophagy in MDA-MB-231 cell lines. Mechanistically, ROR decreased acetylated histone H3 at the miR-34a promoter and resulted in increased expression of autophagy-related genes and decreased expression of p62 (84). ROR polymorphisms could influence cancer susceptibility. For instance, Luo et al. indicated that the TT genotype of ROR rs4801078 correlated with elevated ROR mRNA levels and an increased risk of breast cancer (85). In summary, these studies identified ROR as an oncogene in human cancers and established it as a potential cancer biomarker.
Antisense Non-Coding RNA in the INK4 Locus
ANRIL is a 3.8-kb-long transcript consisting of 19 exons (163). It is located on the human chromosome 9p21 locus, which contains three genes: CDKN2A (encoding p14ARF and p16INK4a) and CDKN2B (encoding p15INK4b) (164). ANRIL was initially discovered in the hereditary cutaneous melanoma-neural system tumor syndrome family with a large germline deletion of the entire CDKN2A and CDKN2B gene cluster (163). ANRIL expression has been reported to be upregulated in many malignancies, such as colorectal (165), gastric (166), and brain cancers (167). Some studies have shown that the ectopic expression of ANRIL is associated with increased tumor size, advanced TNM stage, and poor clinical outcomes (168). Elevated ANRIL expression has been found in breast cancer, particularly in triple-negative breast cancer (86, 87). ANRIL was also included in a three-ncRNA signature, which was proposed to distinguish triple-negative breast cancer from other subtypes (87). ANRIL promotes tumorigenesis in triple-negative breast cancer by directly binding to miR-199a (88). In breast cancer, ANRIL was found to be predominantly located in the nucleus, and nuclear ANRIL positively correlated with periostin expression, suggesting that the subcellular localization of ANRIL impacts cancer progression (89). Moreover, ANRIL coordinates the expression of adjacent tumor-associated genes to promote carcinogenesis. ANRIL could bind to and recruit PRC2 to attenuate the expression of p15INK4b (169). Furthermore, 9p21 polymorphisms have been implicated in cancer susceptibility. In breast cancer patients, the rs11515 CG genotype was more common and correlated with increased ANRIL expression and decreased p16INK4a expression (90). Another study showed that ANRIL was linked to breast cancer susceptibility at the haplotype level and that haplotype analysis was more efficient than single nucleotide polymorphism (SNP) analyses (91). Hence, targeting ANRIL could provide novel insight into breast cancer treatment.
Brain Cytoplasmic 200
BC200 is a 200-nucleotide-long transcript that is also known as brain cytoplasmic RNA 1 (BCYRN1) (170). BC200 is expressed exclusively to the nervous system, where it acts as a translational modulator (171). In 1997, abnormal expression of BC200 was found in diverse human cancers, such as breast, cervix, lung and ovary cancers (172). BC200 is overexpressed in proliferating cultured cells regardless of their origin. Knockdown of BC200 leads to decreased cell viability through regulation of growth arrest and induction of apoptosis (173). In non-small-cell lung cancer, BC200 increased the expression of matrix metalloproteases (MMPs), MMP-9 and MMP-13, resulting in enhanced invasion and migration (174). In addition, BC200 increased the expression of MMP-9 in colon cancer (175). In cervical cancer, BC200 competitively binds with miRNA-138, which leads to the enhancement of cell proliferation and metastasis (176). Moreover, elevated expression of BC200 has also been detected in luminal and triple-negative breast cancer cell lines. High BC200 levels could lead to increased cell viability, growth, migration, and invasion in vitro as well as to increased tumor size in vivo (177). Intriguingly, the expression of BC200 in ER-positive tumors was higher than that in ER-negative tumors. Mechanistically, BC200 binds to B-cell leukemia/lymphoma-x (Bcl-x) pre-mRNA to coordinate its alternative splicing, which results in suppressed expression of Bcl-xS and overexpression of Bcl-xL (92). Furthermore, BC200 RNA was reported to be significantly expressed in invasive breast cancer tissues but was not detectable in benign tumor tissues (93). Lacoangeli et al. also showed that plasma BC200 RNA levels were markedly elevated in invasive breast cancer patients compared to healthy subjects (94, 174), indicating that BC200 is a noninvasive molecular marker for invasive breast cancer detection.
SPRY4 Intronic Transcript 1
SPRY4-IT1 is a 708-bp transcript located on chromosome 5 (95). It has specific secondary structures, which are possibly related to its functional properties (178). SPRY4-IT1 was initially identified as an oncogene in melanoma (178). To date, dysregulation of SPRY4-IT1 has been detected in multiple cancers, such as colorectal (179), non-small-cell lung (180), and breast cancers (96). Some studies have shown that upregulated SPRY4-IT1 expression decreased apoptosis and increased proliferation and migration (97). Expression profile analysis of breast cancer samples revealed that the expression of SPRY4-IT1 was upregulated, and SPRY4-IT1 had a good specificity value (96). Interestingly, the expression level of SPRY4-IT1 in ER‐negative tumors is higher than that in ER‐positive tumors, suggesting that estradiol expression may inversely correlate with SPRY4-IT1 expression (98). Functionally, deletion of SPRY4-IT1 induced G0/G1 cell cycle arrest and apoptosis of breast cancer cells by downregulating the expression of the oncogene zinc finger 703 (ZNF703) (98). Moreover, an N-terminal polypeptide derived from vMIP-II (NT21MP) downregulated SPRY4-IT1 expression, and the oncogenic role of SPRY4-IT1 was compromised by depletion of SKA2, suggesting that the antitumor activity of NT21MP was, at least partly, mediated through the SPRY4-IT1/SKA2 signaling pathway (99). Xiang et al. indicated that high SPRY4-IT1 levels correlated with increased tumor size, high TNM stage, lymph node metastasis and unfavorable clinical outcomes (181). The aforementioned findings indicate that SPRY4-IT1 may serve as a potential biomarker for the diagnosis and prognosis of breast cancer.
Urothelial Carcinoma Associated 1
UCA1 is a 1,442-bp transcript located on chromosome 19p13.12 (182, 183). UCA1 was first identified in bladder cancer and is considered a novel oncogenic lncRNA (184). UCA1 is ubiquitously expressed in embryonic tissues but not in normal adult tissues except for the heart and spleen (185). UCA1 expression is significantly upregulated in many types of cancers, and high levels of UCA1 are associated with enhanced cell proliferation, invasion and metastasis (186). For instance, UCA1 was shown to promote both the proliferation and migration of lung cancer cells by targeting the miR-193a/HMGB1 axis (187). In addition, Luo et al. confirmed that UCA1 enhanced invasion and EMT by suppressing the expression of miR-143 in bladder cancer (188). Li et al. examined the strong association between UCA1 and protein tyrosine phosphatase 1B (PTP1B). Their results showed that the regulation of PTP1B by UCA1 was involved in the proliferation of breast cancer cells (100). Moreover, high expression of UCA1 activated the wnt/β‐catenin signaling pathway, enhanced the nuclear translocation of β‐catenin and promoted invasion in breast cancer. In addition, knockdown of UCA1 inhibited the EMT process by downregulating the expression of β‐catenin and its downstream targets MMP‐7 and cyclin D1 (101). It has also been shown that the lncRNA AC026904 and UCA1 cooperatively increase Slug expression at both the transcriptional and post-transcriptional levels, thereby inducing EMT and metastasis in breast cancer (102). Furthermore, it has been demonstrated that higher levels of UCA1 are associated with shorter OS and increased lymph node metastasis in multiple human cancers (103, 104).
Activated by Transforming Growth Factor β
ATB is a 2,446-bp non-polyadenylated lncRNA located on human chromosome 14 (189). Numerous studies have evaluated the function of ATB in tumorigenesis. ATB was initially discovered as an oncogene in hepatocellular carcinoma, and high levels of ATB were associated with poor clinical outcomes (189). As a mediator of TGF‐β, ATB can regulate different transcription factors to induce the invasion-metastasis cascade. ATB was reported to be highly expressed in breast cancer tissues compared with non-cancerous tissues and the investigated cell lines, and this increase in ATB levels was associated with more nodal metastasis, advanced clinical stage and unfavorable prognosis (105, 106). In addition, the serum level of ATB was significantly elevated in breast cancer patients and could serve as a novel diagnostic biomarker for stage I-II breast cancer patients (107). Functionally, ATB increased the expression of Twist by sponging the miR‐200 family, consequently inducing EMT (105). Furthermore, downregulation of ATB expression could promote E-cadherin expression and suppress EMT by targeting miR-141-3p (106). Moreover, highly expressed ATB could act as a ceRNA for miR-200c and upregulate the expression of the miR-200c target genes ZEB1 and ZNF-217 to promote invasiveness and trastuzumab resistance in HER2-positive breast cancer (108). Intriguingly, the oncogenic role of ATB has been disputed by conflicting studies. For instance, ATB acts as a tumor suppressor in pancreatic cancer (190). Similarly, Nikpayam et al. showed that ATB expression was significantly downregulated in most breast cancer tissues compared with adjacent non-cancerous tissues (109). Both upregulation and downregulation of ATB expression have been indicated to contribute to tumorigenesis, suggesting that ATB might play distinct roles in different cancers or even different cancer subtypes. Further mechanistic studies should be focused on elucidating the role of ATB in cancer pathology.
Plasmacytoma Variant Translocation 1
PVT1, an intergenic lncRNA, is located on chromosome 8q24.21 adjacent to c-Myc (191). PVT1 is highly expressed in cancer tissues compared with non-cancerous tissues and in cancer cell lines (192, 193). Co-amplification of adjacent PVT1 and Myc has been found in many human cancers. PVT1 increases Myc protein levels in 8q24-gain cancers, while either Myc or PVT1 fails to measurably promote cancer (194). Moreover, depletion of PVT1 resulted in decreased c-Myc expression and increased apoptosis of cancer cells (195). PVT1 could also enhance the stability of Kruppel-like factor 5 (KLF5) and increase the expression of β-catenin, an important downstream effector of KLF5, to promote tumorigenesis in triple-negative breast cancer (110). Several studies have shown the connection between PVT1 and different miRNAs in breast cancer. PVT1 functions as a sponge to regulate miR-543 (111), which is a known tumor suppressor miRNA in breast cancer (112). The tumorigenic potency of PVT1 could be abrogated by miR-543 overexpression, and loss of PVT1 is associated with inhibition of growth, increased apoptosis, and decreased tumor size (111). A cluster of oncogenes (miR-1204, 1205, 1206, 1207-3p, 1207-5p, 1208) at the 8q24.21 locus is regulated by PVT1 (193). For instance, PVT1 upregulates the expression of miR-1207-5p to repress the expression of signal transducer and activator of transcription 6 (STAT6) and cyclin inhibitors, thus enhancing cell proliferation and colony formation in breast cancer (113). Additionally, miR-1204 overexpression contributes to the proliferation, invasion and EMT of breast cancer cells both in vitro and in vivo (114). Furthermore, a meta-analysis carried out by Lu et al. indicated that high PVT1 expression correlated with more lymph node metastasis, increased distant metastasis, advanced TNM stage, poor differentiation grade and unfavorable prognosis but not with tumor volume (115). Thus, PVT1 could act as a useful molecular marker for breast cancer.
Colon Cancer Associated Transcript 1
CCAT1, initially identified in colon cancer, is mapped to the 8q.24.2 locus and is ~2,628 nucleotides long (196). The 8q.24.2 locus contains only a few protein-coding genes and is often referred to as a ‘gene desert’ (197). CCAT1 expression is consistently upregulated in multiple types of cancers and correlates with poor prognosis (198). Han et al. found that CCAT1 was overexpressed in triple-negative breast cancer tissues compared to adjacent normal tissues and in a panel of triple-negative breast cancer cell lines in comparison to normal breast epithelial cell lines (116). CCAT1 has been shown to act as a decoy to inhibit the expression of several miRNAs. Loss of CCAT1 resulted in the upregulation of miR-218 expression and the simultaneous inhibition of a miR-218 target gene, zinc finger protein ZFX, resulting in inhibited cell proliferation, migration, and invasion. Moreover, silencing of miR-218, in turn, can block the tumor suppressive effect of CCAT1 knockdown, suggesting that CCAT1 may promote breast carcinogenesis through regulation of the miR-218/ZFX axis (116). Another study showed that the expression of CCAT1 was higher in radioresistant breast cancer tissues than in radiosensitive breast cancer tissues. Depletion of CCAT1 dramatically decreased the colony formation rate and promoted apoptosis by directly interacting with miR-148b. The authors concluded that loss of CCAT1 might enhance the radiosensitivity of breast cancer cells by downregulating miR-148b expression (117). CCAT1 could function as a regulator of wnt/β-catenin signaling pathway in cervical cancer (199) and non-small-cell lung cancer (200). Consistent with these studies, Tang et al. indicated that CCAT1 coordinated miR-204/211, miR-148a/152 and annexin A2 to hyperactivate the wnt/β-catenin signaling pathway, consequently promoting the proliferation and metastasis of breast cancer stem cells (118). Overexpression of CCAT1 in breast cancer has been related to histological grade, TNM staging and lymph metastasis, and it is also an independent predictor of OS and progression-free survival (PFS) (119). The ubiquitous nature of CCAT1 upregulation in cancers shows promise for future discovery of diagnostic biomarkers and pharmaceutical targets for cancer control.
Colon Cancer Associated Transcript 2
CCAT2 is 1,752 bp in length and is located within the chromosome 8q24.21 gene desert adjacent to Myc (201). Amplification of the oncogenes in the 8q24.21 region has been found in numerous human cancers. High CCAT2 levels positively correlated with Myc levels in colon and colorectal cancer (202, 203). Accordingly, CCAT2 could upregulate the expression of Myc in breast cancer, suggesting that the amplification of CCAT2 and Myc might occur simultaneously (120). Huang et al. explored the expression of CCAT2 in 33 cancer types and 13,285 tumor patients. The study revealed that CCAT2 was substantially overexpressed in cancer tissues compared to paired normal tissues, and this increase in CCAT2 levels correlated with a greater tumor volume, higher TNM grades, advanced clinical stage and a poor OS in patients. In addition, CCAT2 expression was mainly upregulated in stage II tumor pathology, followed by stage III, indicating that CCAT2 could be used for the early detection of cancers (121). Moreover, CCAT2 expression levels in metastatic breast cancer were higher than those in non-metastatic breast cancer. Downregulation of CCAT2 expression significantly inhibited the expression of TGF-β, Smad2 and α-SMA, thereby inducing apoptosis and G0/G1 cell cycle arrest (122). Deng et al. indicated that CCAT2 knockdown suppressed the expression of cell cycle-related proteins and G0/G1 phase arrest in breast cancer cells (204). They also found that CCAT2 interacted with EZH2, a marker of aggressive breast cancer (123) and abrogated the expression of P15 (204). It has been shown that Notch signaling could be activated and upregulated in breast cancer (205). Xu et al. demonstrated the strong association between CCAT2 and Notch 2 in triple-negative breast cancer (124). Functionally, CCAT2 promoted the growth, invasion and migration of breast cancer stem cells by sponging miR-205, which targets Notch 2 (124). Overall, accumulating evidence suggests that CCAT2 is an oncogene and could serve as a useful biomarker and therapeutic target for breast cancer treatment.
Tissue Differentiation-Induced Non-Coding RNA
TINCR is highly expressed in keratinocytes and is essential for normal epidermal differentiation (206). It is a 3,733-nucleotide long transcript located on chromosome 19p13 (207). Aberrant TINCR expression has been implicated in multiple human cancers. TINCR expression is upregulated in gastric, gladder and breast cancer but downregulated in glioma and prostate cancer (208). In recent years, several studies have been performed on the contribution of TINCR to breast cancer. Liu et al. indicated that TINCR was activated by transcription factor specificity protein 1 (SP1) in breast cancer (125). Consistent with this observation, Xu et al. showed that SP1 could bind to the putative GC-rich motifs of TINCR to upregulate the expression of TINCR in gastric cancer (209). In addition, TINCR overexpression competed with miR-7 and facilitated KLF4 expression, which in turn regulated cell proliferation, migration, and invasion in breast cancer (125). Insulin-like growth factor receptor 1 (IGFR-1), a tyrosine kinase cell surface receptor, is involved in the development and progression of breast cancer (210). Guo et al. showed that TINCR played an oncogenic role in breast cancer through regulation of the miR-589-3p/IGF1R axis (126). Moreover, the expression of TINCR was higher in trastuzumab-resistant tissues than in sensitive tissues owing to enhanced histone acetylation of the TINCR promoter. Functionally, TINCR promoted the expression of HER-2 by sponging miR-125b, consequently conferring trastuzumab resistance (127). Moreover, TINCR promoted EMT via downregulation of Snail-1 expression, while enhanced Snail-1 expression reversed EMT suppression induced by TINCR silencing in trastuzumab-resistant cell lines (127). Furthermore, Kaplan-Meier survival curves showed that high levels of tissue TINCR correlated with unfavorable prognosis in breast cancer (126). Wang et al. found that circulating TINCR was dramatically elevated in breast cancer, particularly in the aggressive triple-negative subtype. The authors further noted that serum TINCR levels were associated with unfavorable prognosis, suggesting that TINCR could serve as a novel biomarker for breast cancer therapy (128).
Tumor Suppressor LNCRNAS
Maternally Expressed Gene 3
MEG3 is an imprinted gene from the maternal allele mapped to the human chromosome 14q32.3 region (211). The transcript contains 10 exons and approximately 12 alternative splicing isoforms, some of which are expressed in a tissue- or cell-type-specific manner (211). MEG3 was the first lncRNA to be identified as a tumor suppressor in the inhibition of cancer cell growth (212). A loss of MEG3 expression has been found across human cancer cell lines, and decreased MEG3 levels significantly correlate with TNM stage, lymph node metastasis and differentiation grade (129, 130). Loss of MEG3 expression also predicts shorter OS, PFS, distant metastasis-free survival (DMFS), and disease-specific survival (DSS) (130–132). Zhang et al. showed that ectopic MEG3 overexpression promoted breast cancer progression by upregulating the expression of the endoplasmic reticulum stress-related proteins NF−κB and p53 (133). Mechanistically, MEG3 can bind directly to the p53 promoter and increase the transcriptional activity of p53, thus regulating the expression of p53 target genes (134). In addition, MEG3 deactivated the AKT/mTOR signaling pathway by sponging miR-21, while miR-21 overexpression partially abolished the tumor suppressive function of MEG3 in breast cancer cells (135). Moreover, elevated expression of MEG3 can inhibit cell invasion, proliferation, and apoptosis induction (213, 214), indicating that MEG3 might be a novel therapeutic target for cancers. SNPs mainly refer to a set of DNA sequence polymorphisms based on single nucleotide variations at the genomic level (215). It has been reported that SNPs are linked to genetic susceptibility to cancer (216). Ali et al. indicated that MEG3 rs7158663 G > A with the mutant A allele correlated with decreased serum MEG3 expression and unfavorable clinical outcomes in an Egyptian population (136). Additionally, the GG genotype of rs3087918 could influence the secondary structure of MEG3 and decrease the susceptibility to breast cancer risk in Chinese women (137). Hence, MEG3 could be a suitable biomarker candidate for clinical cancer management.
X-Inactive Specific Transcript
XIST, 17 kb in length, is located at the X-inactivation center (217). During primary embryogenesis, XIST recruits multiple factors to orchestrate X chromosome inactivation (218). Recent studies have identified associations between aberrant XIST expression and breast cancer. Zheng et al. indicated that XIST expression was drastically downregulated in breast cancer tissues and cell lines. The authors also found that XIST sponged miR-155, which in turn upregulated the expression of caudal-type homeobox 1 (CDX1) and inhibited the progression of breast cancer (138). In addition, Liu et al. also found that XIST functioned as a ceRNA for miR-362-5p and thus inhibited its repressive effect on ubiquitin-associated protein 1 (UBAP1), consequently inhibiting breast cancer progression (139). BRCA1 is a high-penetrance gene in which loss-of-function mutations predispose patients to breast and ovarian cancers (140, 141). Sirchia et al. indicated that BRCA1 participates in XIST regulation on the active X chromosome as well as XIST dysregulation and drives tumorigenesis in breast cancer. Mechanistically, BRCA1 knockdown resulted in enhanced XIST expression, promoter demethylation and X chromosome inactivation (142). However, another study suggested the potential oncogenic role of XIST in breast cancer. Zong et al. showed that XIST knockdown dramatically reduced characteristics associated with breast cancer, such as cell proliferation, anti-apoptosis, invasion, and migration activities. Functionally, XIST induced sponging of miR-125b-5p and removed the inhibitory effect of this miRNA on NLRC5, a breast cancer promotor, thus promoting the malignancy of breast cancer cells (143). In addition, a meta-analysis carried out by Zhu et al. demonstrated that XIST was overexpressed in multiple cancers and that elevated XIST levels correlated with larger tumor volume, increased lymphatic metastasis, advanced tumor stage and unfavorable clinical outcomes (144). XIST could serve as an oncogene or tumor suppressor, and further studies are still needed to elucidate the roles of XIST in cancer biology.
Growth Arrest−Specific Transcript 5
GAS5, a well-known tumor suppressor, is located on chromosome 1q25 (219). Abnormal expression levels of GAS5 have been reported in different cancer types (220–222). For example, GAS5 has been shown to promote proliferation by regulating miR-22 and its downstream target transcripts in gastric cancer (223). GAS5 can also promote cell invasion and migration by targeting miR-196a and the PI3K/Akt/mTOR signaling pathway in oesophageal squamous cell carcinoma (224, 225). In ovarian cancer, loss of GAS5 is related to increased tumor volume and advanced tumor stage (226, 227). In addition, the expression of GAS5 is significantly downregulated in breast cancer tissues compared with adjacent non-cancerous tissues (228). Larger tumor volume, advanced lymph node metastasis, and estrogen receptor negativity in breast cancer cells are the outcomes of GAS5 downregulation (229). In HER2-positive breast cancer, silencing of GAS5 contributes to trastuzumab resistance. Mechanistically, GAS5 serves as a molecular sponge of miR-21 to increase the expression of phosphatase and tensin homologs (PTEN) and alleviate trastuzumab resistance (230). Zhang et al. demonstrated the reciprocal inhibition between miR-21 and GAS5 in breast cancer. MiR-21 downregulated GAS5 expression, while silencing of GAS5 increased miR-21 expression (231). Jing et al. found that GAS5 expression was significantly downregulated by Notch‐1 and that decreased GAS5 levels were involved in the proliferation of breast cancer (232). Thus, these studies demonstrate that GAS5 could be an attractive biomarker candidate in cancer therapy.
Phosphatase and Tensin Homolog Pseudogene 1
As a pseudogene of PTEN, PTENP1 has a highly homologous region upstream of the 3′UTR of PTEN (233). To date, aberrant expression of PTENP1 has been found in various malignancies, including breast cancer (73). Low levels of PTENP1 have been shown to be associated with increased proliferation, migration, invasion and colony formation, as well as decreased apoptosis, in breast cancer (73, 145, 146). PTENP1 has been implicated in the regulation of the PI3K/Akt signaling pathway, which plays a pivotal role in tumorigenesis and tumor development, particularly in breast cancer (147). Chen et al. indicated that PTENP1 suppressed breast cancer cell proliferation and migration via regulation of Akt and cell cycle-related proteins (145). In addition, PTENP1-induced sponging of miR-19b resulted in increased expression of PTEN (73, 146) and decreased expression of p-PI3K, PI3K and p-Akt, thereby inhibiting cell proliferation and migration (73). Moreover, the regulatory effect of PTENP1 on the PI3K/Akt signaling pathway can be reversed by the overexpression of miR-19b (73). Furthermore, Gao et al. showed that PTENP1 inversely correlated with miR-20, a known oncogenic mRNA. PTENP1 acts as a decoy for miR-20 to derepress its inhibitory effect on PTEN, ultimately attenuating the activation of the PI3K/Akt pathway (148). They also found that low expression of PTENP1 and PTEN was associated with advanced TNM stage and worse OS (148). Interestingly, the involvement of PTENP1 in breast cancer biology may depend on the hormone receptor status. PTENP1 overexpression was linked to decreased PTEN expression and increased proliferation in ER-positive cells, while increased PTEN expression and inhibited tumorigenesis were observed in ER-negative cells (149). Hence, PTEN may represent a promising biomarker for breast cancer.
Circulating LNCRNAS as Biomarkers in Cancer
Biomarker is defined as “a characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes or pharmacologic responses to a therapeutic intervention” by the US NIH’s Biomarkers Definition Working Group and the Biomarkers Consortium (1). A tumor marker is any specific molecule indicating the presence or progression of human cancers. Tumor biomarkers can be either found in body fluids or tumor tissues. Biomarkers in body fluids (especially those in blood serum) are readily measured, and their diagnostic performances have been confirmed in multiple cancers. Carcinoembryonic antigen (CEA) and cancer antigen 15-3 (CA15-3) in serum have been approved by the US Food and Drug Administration (FDA) as biomarkers for breast cancer. Notwithstanding CEA and CA15-3 are widely used in diagnosis of breast cancer, they bear some limitations, mainly regarding to low sensitivity and specificity. Therefore, it is critical to discover novel molecular markers with improved diagnostic value.
Dysregulated lncRNAs in primary tumor tissues could be mirrored in different body fluids, such as blood plasma, urine and saliva (234–236). Many studies have revealed that lncRNAs remain stable while circulating in body fluids even under extreme conditions, further enhancing their competitive advantage of being good diagnostic tools. In recent years, several circulating lncRNAs have been proved as suitable diagnostic and prognostic markers in various cancer types, such as prostate cancer antigen 3 (PCA3) and MALAT1. lncRNA PCA3 in urine samples has received the approval of the FDA as a diagnostic molecule for prostate cancer. Intriguingly, lncRNA PCA3 is much more specific and sensitive than prostate-specific antigen, the conventional gold standard for prostate cancer. A meta-analysis carried out by Xue et al. has determined the diagnostic value of PCA3 for the detection of prostate cancer, with sensitivity and specificity of 62% and 75%, respectively (235). Also, plasma H19 holds great potential as an independent biomarker for gastric cancer due to its high diagnostic performance (sensitivity 82.9%; specificity 72.9%) (234). In addition, serum MALAT1 has proven its diagnostic value for breast cancer (sensitivity 83.7%; specificity 81.2%) (237). More importantly, lncRNA-based detection method is noninvasive, convenient and inexpensive when compared to the traditional biopsies.
Prospects and Challenges
Since lncRNAs are dysregulated in cancers, the functional lncRNAs may be targeted to halt the process of carcinogenesis. LncRNAs targeting strategies can be achieved by antisense oligonucleotides (ASOs), RNA interference (RNAi) and clustered regularly-interspaced, CRISPR-Cas9, etc. It was suggested that ASOs mainly targeted the lncRNAs retaining in the nucleus, whereas RNAi predominantly targeted the lncRNAs in the cytoplasm (238). CRISPR-Cas9, a precise versatile toolkit, could target lncRNAs at high throughput, representing a major technology breakthrough in gene editing (239).
Although lncRNAs hold potential to serve as ideal diagnostic biomarkers and therapeutic targets, some challenges need to be addressed and resolved in the future. First, the poor consequence conversation of lncRNAs has complicated the pre-clinical studies across different species. In addition, lncRNAs have secondary and tertiary structures, which could lead to ineffectiveness of the lncRNAs targeting therapeutics. Secondly, the lncRNA-based treatment may perturb other genes. Thus, there is a risk of off-target effects and new strategies should be developed to maximize the on-target efficacy. Thirdly, the concentration of circulating lncRNAs may be below the detection limit of the existing equipment, such as NanoDrop spectrophotometer. It is expected that in the near future, more sensitive detection instruments will open a new window for lncRNA quantification. Fourthly, a single lncRNA may not be feasible for cancer diagnosis. Xie et al. suggested that a diagnostic panel for NSCLC possessed higher specificity (79.2%) and sensitivity (77.1%) when compared to any single molecular marker, such as CEA and lncRNA ANRIL (240). Indeed, biomarkers in a panel can complement each other, contributing to enhanced diagnostic performance.
Concluding Remarks
As indicated in this review, lncRNAs have gained considerable attention as pivotal regulators in various physiological and pathophysiological events. Altered expression levels of lncRNAs have been reported in multiple human cancers, including breast cancer. It has become clear that lncRNAs with dysregulated expression drive the initiation and progression of cancers via interactions with other types of RNA molecules, DNA and proteins. Intriguingly, lncRNAs are differentially regulated in diverse cancers or even cancer subtypes and show a significant association with pathological features and clinical prognosis. Regarding the aberrant expression of lncRNAs and the underlying mechanisms, lncRNAs may act as suitable diagnostic and prognostic biomarkers in breast cancer. Furthermore, lncRNAs could be targeted to reverse the process of carcinogenesis and represent valuable therapeutic targets for cancer treatment. LncRNA-based tests and therapy are promising strategies that deserve extensive research and thorough exploration in the future.
Author Contributions
WZ and CL were responsible for the study conception and design. CL drafted the manuscript. DW and PW contributed to the significant portions of the manuscript. WZ and YZ revised and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (Grant No. 81800643).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Dumitrescu RG. Interplay Between Genetic and Epigenetic Changes in Breast Cancer Subtypes. Methods Mol Biol (2018) 1856:19–34. doi: 10.1007/978-1-4939-8751-1_2
2. Harbeck N, Gnant M. Breast Cancer. Lancet (2017) 389(10074):1134–50. doi: 10.1016/s0140-6736(16)31891-8
3. Ferlay J, Colombet M, Soerjomataram I, Dyba T, Randi G, Bettio M, et al. Cancer Incidence and Mortality Patterns in Europe: Estimates for 40 Countries and 25 Major Cancers in 2018. Eur J Cancer (2018) 103:356–87. doi: 10.1016/j.ejca.2018.07.005
4. Senkus E, Lacko A. Over-Treatment in Metastatic Breast Cancer. Breast (2017) 31:309–17. doi: 10.1016/j.breast.2016.06.024
5. Huang Z, Zhou JK, Peng Y, He W, Huang C. The Role of Long Noncoding RNAs in Hepatocellular Carcinoma. Mol Cancer (2020) 19(1):77. doi: 10.1186/s12943-020-01188-4
6. Takeiwa T, Ikeda K, Mitobe Y, Horie-Inoue K, Inoue S. Long Noncoding RNAs Involved in the Endocrine Therapy Resistance of Breast Cancer. Cancers (Basel) (2020) 12(6):1424. doi: 10.3390/cancers12061424
7. Gutschner T, Diederichs S. The Hallmarks of Cancer: A Long Non-Coding RNA Point of View. RNA Biol (2012) 9(6):703–19. doi: 10.4161/rna.20481
8. Wu H, Yang L, Chen LL. The Diversity of Long Noncoding RNAs and Their Generation. Trends Genet (2017) 33(8):540–52. doi: 10.1016/j.tig.2017.05.004
9. Sun M, Gadad SS, Kim DS, Kraus WL. Discovery, Annotation, and Functional Analysis of Long Noncoding RNAs Controlling Cell-Cycle Gene Expression and Proliferation in Breast Cancer Cells. Mol Cell (2015) 59(4):698–711. doi: 10.1016/j.molcel.2015.06.023
10. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492
11. Coughlin SS. Epidemiology of Breast Cancer in Women. Adv Exp Med Biol (2019) 1152:9–29. doi: 10.1007/978-3-030-20301-6_2
12. Torre LA, Siegel RL, Ward EM, Jemal A. Global Cancer Incidence and Mortality Rates and Trends–An Update. Cancer Epidemiol Biomarkers Prev (2016) 25(1):16–27. doi: 10.1158/1055-9965.EPI-15-0578
13. Denkert C, Liedtke C, Tutt A. Von Minckwitz G. Molecular Alterations in Triple-Negative Breast Cancer—the Road to New Treatment Strategies. Lancet (2017) 389(10087):2430–42. doi: 10.1016/s0140-6736(16)32454-0
14. Mueller C, Haymond A, Davis JB, Williams A, Espina V. Protein Biomarkers for Subtyping Breast Cancer and Implications for Future Research. Expert Rev Proteomics (2018) 15(2):131–52. doi: 10.1080/14789450.2018.1421071
15. Waks AG, Winer EP. Breast Cancer Treatment: A Review. JAMA (2019) 321(3):288–300. doi: 10.1001/jama.2018.19323
16. Lim LJ, Wong SYS, Huang F, Lim S, Chong SS, Ooi LL, et al. Roles and Regulation of Long Noncoding RNAs in Hepatocellular Carcinoma. Cancer Res (2019) 79(20):5131–9. doi: 10.1158/0008-5472.CAN-19-0255
17. Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, et al. The RNA Modification Database, RNAMDB: 2011 Update. Nucleic Acids Res (2011) 39(Database issue):D195–201. doi: 10.1093/nar/gkq1028
18. Freedman JE, Miano JM, National Heart L, Blood Institute Workshop P. Challenges and Opportunities in Linking Long Noncoding RNAs to Cardiovascular, Lung, and Blood Diseases. Arterioscler Thromb Vasc Biol (2017) 37(1):21–5. doi: 10.1161/ATVBAHA.116.308513
19. Wang KC, Chang HY. Molecular Mechanisms of Long Noncoding RNAs. Mol Cell (2011) 43(6):904–14. doi: 10.1016/j.molcel.2011.08.018
20. Ulitsky I. Evolution to the Rescue: Using Comparative Genomics to Understand Long Non-Coding RNAs. Nat Rev Genet (2016) 17(10):601–14. doi: 10.1038/nrg.2016.85
21. Peng L, Jiang J, Tang B, Nice EC, Zhang YY, Xie N. Managing Therapeutic Resistance in Breast Cancer: From the lncRNAs Perspective. Theranostics (2020) 10(23):10360–77. doi: 10.7150/thno.49922
22. Hauptman N, Glavac D. Long Non-Coding RNA in Cancer. Int J Mol Sci (2013) 14(3):4655–69. doi: 10.3390/ijms14034655
23. Chen LL. Linking Long Noncoding RNA Localization and Function. Trends Biochem Sci (2016) 41(9):761–72. doi: 10.1016/j.tibs.2016.07.003
24. Zhang XZ, Liu H, Chen SR. Mechanisms of Long Non-Coding RNAs in Cancers and Their Dynamic Regulations. Cancers (Basel) (2020) 12(5):1245. doi: 10.3390/cancers12051245
25. Dutta A, Roy A, Chatterjee S. Long Noncoding RNAs in Cancer Immunity: A New Avenue in Drug Discovery. Drug Discovery Today (2021) 26(1):264–72. doi: 10.1016/j.drudis.2020.07.026
26. Yoon JH, Abdelmohsen K, Kim J, Yang X, Martindale JL, Tominaga-Yamanaka K, et al. Scaffold Function of Long Non-Coding RNA HOTAIR in Protein Ubiquitination. Nat Commun (2013) 4:2939. doi: 10.1038/ncomms3939
27. Kopp F, Mendell JT. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell (2018) 172:393–407. doi: 10.1016/j.cell.2018.01.011
28. Tay Y, Rinn J, Pandolfi PP. The Multilayered Complexity of ceRNA Crosstalk and Competition. Nature (2014) 505(7483):344–52. doi: 10.1038/nature12986
29. Huarte M. The Emerging Role of lncRNAs in Cancer. Nat Med (2015) 21(11):1253–61. doi: 10.1038/nm.3981
30. Quinodoz S, Guttman M. Long Noncoding RNAs: An Emerging Link Between Gene Regulation and Nuclear Organization. Trends Cell Biol (2014) 24(11):651–63. doi: 10.1016/j.tcb.2014.08.009
31. Huang JZ, Chen M, Chen D, Gao XC, Zhu S, Huang H, et al. A Peptide Encoded by a Putative lncRNA HOXB-AS3 Suppresses Colon Cancer Growth. Mol Cell (2017) 68:171–84. doi: 10.1016/j.molcel.2017.09.015
32. Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G, et al. The Noncoding RNA MALAT1 Is a Critical Regulator of the Metastasis Phenotype of Lung Cancer Cells. Cancer Res (2013) 73(3):1180–9. doi: 10.1158/0008-5472.CAN-12-2850
33. Zhang X, Hamblin MH, Yin KJ. The Long Noncoding RNA Malat1: Its Physiological and Pathophysiological Functions. RNA Biol (2017) 14(12):1705–14. doi: 10.1080/15476286.2017.1358347
34. Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, et al. MALAT-1, a Novel Noncoding RNA, and Thymosin Beta4 Predict Metastasis and Survival in Early-Stage non-Small Cell Lung Cancer. Oncogene (2003) 22(39):8031–41. doi: 10.1038/sj.onc.1206928
35. Fan C, Yuan Q, Liu G, Zhang Y, Yan M, Sun Q, et al. Long Non-Coding RNA MALAT1 Regulates Oxaliplatin-Resistance via miR-324-3p/ADAM17 Axis in Colorectal Cancer Cells. Cancer Cell Int (2020) 20:473. doi: 10.1186/s12935-020-01549-5
36. Li C, Cui Y, Liu LF, Ren WB, Li QQ, Zhou X, et al. High Expression of Long Noncoding RNA MALAT1 Indicates a Poor Prognosis and Promotes Clinical Progression and Metastasis in Bladder Cancer. Clin Genitourin Cancer (2017) 15(5):570–6. doi: 10.1016/j.clgc.2017.05.001
37. Liu S, Jiang X, Li W, Cao D, Shen K, Yang J. Inhibition of the Long Non-Coding RNA MALAT1 Suppresses Tumorigenicity and Induces Apoptosis in the Human Ovarian Cancer SKOV3 Cell Line. Oncol Lett (2016) 11(6):3686–92. doi: 10.3892/ol.2016.4435
38. Miao YF, Fan RG, Chen LG, Qian HX. Clinical Significance of Long Non-Coding RNA MALAT1 Expression in Tissue and Serum of Breast Cancer. Ann Clin Lab Sci (2016) 46(4):418–24.
39. Huang XJ, Xia Y, He GF, Zheng LL, Cai YP, Yin Y, et al. MALAT1 Promotes Angiogenesis of Breast Cancer. Oncol Rep (2018) 40(5):2683–9. doi: 10.3892/or.2018.6705
40. Wang Y, Zhou Y, Yang Z, Chen B, Huang W, Liu Y, et al. MiR-204/ZEB2 Axis Functions as Key Mediator for MALAT1-Induced Epithelial-Mesenchymal Transition in Breast Cancer. Tumor Biol (2017) 39(7):1010428317690998. doi: 10.1177/1010428317690998
41. Li XL, Chen NF, Zhou L, Wang C, Wen X, Jia L, et al. Genome-Wide Target Interactome Profiling Reveals a Novel EEF1A1 Epigenetic Pathway for Oncogenic lncRNA MALAT1 in Breast Cancer. Am J Cancer Res (2019) 9(4):714–29.
42. Zhao Z, Chen C, Liu Y, Wu C. 17beta-Estradiol Treatment Inhibits Breast Cell Proliferation, Migration and Invasion by Decreasing MALAT-1 RNA Level. Biochem Biophys Res Commun (2014) 445(2):388–93. doi: 10.1016/j.bbrc.2014.02.006
43. Li ZH, Liang X, Liu Y, Fu SK, Tu JH, Hu YY, et al. LncRNA MALAT1 Promotes Relapse of Breast Cancer Patients With Postoperative Fever. Am J Transl Res (2018) 10(10):3186–97.
44. Wang Z, Katsaros D, Biglia N, Shen Y, Fu Y, Loo LWM, et al. High Expression of Long Non-Coding RNA MALAT1 in Breast Cancer is Associated With Poor Relapse-Free Survival. Breast Cancer Res Treat (2018) 171(2):261–71. doi: 10.1007/s10549-018-4839-2
45. Wang Y, Zhang Y, Hu K, Qiu J, Hu Y, Zhou M, et al. Elevated Long Noncoding RNA MALAT-1 Expression Is Predictive of Poor Prognosis in Patients With Breast Cancer: A Meta-Analysis. Biosci Rep (2020) 40(8):BSR20200215. doi: 10.1042/BSR20200215
46. Li ZX, Zhu QN, Zhang HB, Hu Y, Wang G, Zhu YS. MALAT1: A Potential Biomarker in Cancer. Cancer Manag Res (2018) 10:6757–68. doi: 10.2147/CMAR.S169406
47. Zhao Y, Yu YQ, You S, Zhang CM, Wu L, Zhao W, et al. Long Non-Coding RNA MALAT1 as a Detection and Diagnostic Molecular Marker in Various Human Cancers: A Pooled Analysis Based on 3255 Subjects. Onco Targets Ther (2020) 13:5807–17. doi: 10.2147/OTT.S250796
48. Jiang X, Zhou Y, Sun AJ, Xue JL. NEAT1 Contributes to Breast Cancer Progression Through Modulating miR-448 and ZEB1. J Cell Physiol (2018) 233(11):8558–66. doi: 10.1002/jcp.26470
49. Zhang M, Wu WB, Wang ZW, Wang XH. lncRNA NEAT1 is Closely Related With Progression of Breast Cancer via Promoting Proliferation and EMT. Eur Rev Med Pharmacol Sci (2017) 21(5):1020–6.
50. Li X, Deng S, Pang X, Song Y, Luo S, Jin L, et al. LncRNA NEAT1 Silenced miR-133b Promotes Migration and Invasion of Breast Cancer Cells. Int J Mol Sci (2019) 20(15):3616. doi: 10.3390/ijms20153616
51. Ke H, Zhao L, Feng X, Xu H, Zou L, Yang Q, et al. NEAT1 Is Required for Survival of Breast Cancer Cells Through FUS and miR-548. Gene Regul Syst Bio (2016) 10(Suppl 1):11–7. doi: 10.4137/GRSB.S29414
52. Qian K, Liu G, Tang Z, Hu Y, Fang Y, Chen Z, et al. The Long Non-Coding RNA NEAT1 Interacted With miR-101 Modulates Breast Cancer Growth by Targeting EZH2. Arch Biochem Biophys (2017) 615:1–9. doi: 10.1016/j.abb.2016.12.011
53. Li X, Wang S, Li Z, Long X, Guo Z, Zhang G, et al. The lncRNA NEAT1 Facilitates Cell Growth and Invasion via the miR-211/HMGA2 Axis in Breast Cancer. Int J Biol Macromol (2017) 105(Pt 1):346–53. doi: 10.1016/j.ijbiomac.2017.07.053
54. Quan D, Chen K, Zhang J, Guan Y, Yang D, Wu H, et al. Identification of lncRNA NEAT1/miR-21/RRM2 Axis as a Novel Biomarker in Breast Cancer. J Cell Physiol (2020) 235(4):3372–81. doi: 10.1002/jcp.29225
55. Li S, Hao J, Hong Y, Mai J, Huang W. Long Non-Coding RNA NEAT1 Promotes the Proliferation, Migration, and Metastasis of Human Breast-Cancer Cells by Inhibiting miR-146b-5p Expression. Cancer Manag Res (2020) 12:6091–101. doi: 10.2147/CMAR.S252295
56. Zhou S, Wang L, Yang Q, Liu H, Meng Q, Jiang L, et al. Systematical Analysis of lncRNA-mRNA Competing Endogenous RNA Network in Breast Cancer Subtypes. Breast Cancer Res Treat (2018) 169(2):267–75. doi: 10.1007/s10549-018-4678-1
57. Adriaenssens E, Dumont L, Lottin S, Bolle D, Leprêtre A, Delobelle A, et al. H19 Overexpression in Breast Adenocarcinoma Stromal Cells Is Associated With Tumor Values and Steroid Receptor Status But Independent of P53 and Ki-67 Expression. Am J Pathol (1998) 153(5):1597–607. doi: 10.1016/s0002-9440(10)65748-3
58. Matouk IJ, Raveh E, Abu-lail R, Mezan S, Gilon M, Gershtain E, et al. Oncofetal H19 RNA Promotes Tumor Metastasis. Biochim Biophys Acta (2014) 1843(7):1414–26. doi: 10.1016/j.bbamcr.2014.03.023
59. Li Z, Li Y, Li Y, Ren K, Li X, Han X, et al. Long Non-Coding RNA H19 Promotes the Proliferation and Invasion of Breast Cancer Through Upregulating DNMT1 Expression by Sponging miR-152. J Biochem Mol Toxicol (2017) 31(9):e21933. doi: 10.1002/jbt.21933
60. Zhou W, Ye XL, Xu J, Cao MG, Fang ZY, Li LY, et al. The lncRNA H19 Mediates Breast Cancer Cell Plasticity During EMT and MET Plasticity by Differentially Sponging miR-200b/C and Let-7b. Sci Signal (2017) 10(483):eaak9557. doi: 10.1126/scisignal.aak9557
61. Berteaux N, Lottin S, Monte D, Pinte S, Quatannens B, Coll J, et al. H19 mRNA-Like Noncoding RNA Promotes Breast Cancer Cell Proliferation Through Positive Control by E2F1. J Biol Chem (2005) 280(33):29625–36. doi: 10.1074/jbc.M504033200
62. Vennin C, Spruyt N, Dahmani F, Julien S, Bertucci F, Finetti P, et al. H19non Coding RNA-Derived miR-675 Enhances Tumorigenesis and Metastasis of Breast Cancer Cells by Downregulating C-Cbl and Cbl-B. Oncotarget (2015) 6(30):29209–23. doi: 10.18632/oncotarget.4976
63. Zhang K, Luo Z, Zhang Y, Zhang L, Wu L, Liu L, et al. Circulating lncRNA H19 in Plasma as a Novel Biomarker for Breast Cancer. Cancer Biomark (2016) 17(2):187–94. doi: 10.3233/CBM-160630
64. Shima H, Kida K, Adachi S, Yamada A, Sugae S, Narui K, et al. Lnc RNA H19 is Associated With Poor Prognosis in Breast Cancer Patients and Promotes Cancer Stemness. Breast Cancer Res Treat (2018) 170(3):507–16. doi: 10.1007/s10549-018-4793-z
65. Liu RH, Wang MY, Chen LY, Yin ZJ, Ji QK, Wang YY, et al. Meta-Analysis of the Prognostic Value of Long Non-Coding RNA AFAP1-AS1 for Cancer Patients in China. Oncotarget (2018) 9(8):8100–10. doi: 10.18632/oncotarget.23568
66. Zhang K, Liu P, Tang H, Xie X, Kong Y, Song C, et al. AFAP1-AS1 Promotes Epithelial-Mesenchymal Transition and Tumorigenesis Through Wnt/beta-Catenin Signaling Pathway in Triple-Negative Breast Cancer. Front Pharmacol (2018) 9:1248. doi: 10.3389/fphar.2018.01248
67. Liu FT, Xue QZ, Zhu PQ, Luo HL, Zhang Y, Hao T. Long Noncoding RNA AFAP1-AS1, a Potential Novel Biomarker to Predict the Clinical Outcome of Cancer Patients: A Meta-Analysis. Onco Targets Ther (2016) 9:4247–54. doi: 10.2147/OTT.S107188
68. Dianatpour A, Faramarzi S, Geranpayeh L, Mirfakhraie R, Motevaseli E, Ghafouri-Fard S. Expression Analysis of AFAP1-AS1 and AFAP1 in Breast Cancer. Cancer Biomark (2018) 22(1):49–54. doi: 10.3233/CBM-170831
69. Ma D, Chen C, Wu J, Wang H. Wu D. Up-Regulated lncRNA AFAP1-AS1 Indicates a Poor Prognosis and Promotes Carcinogenesis of Breast Cancer. Breast Cancer (2019) 26(1):74–83. doi: 10.1007/s12282-018-0891-3
70. Kwok TC, Rakha EA, Lee AH, Grainge M, Green AR, Ellis IO, et al. Histological Grading of Breast Cancer on Needle Core Biopsy: The Role of Immunohistochemical Assessment of Proliferation. Histopathology (2010) 57(2):212–9. doi: 10.1111/j.1365-2559.2010.03620.x
71. Cai B, Wang X, Bu Q, Li P, Xue Q, Zhang J, et al. LncRNA AFAP1-AS1 Knockdown Represses Cell Proliferation, Migration, and Induced Apoptosis in Breast Cancer by Downregulating SEPT2 Via Sponging miR-497-5p. Cancer Biother Radiopharm (2020). doi: 10.1089/cbr.2020.3688
72. Zhang X, Zhou Y, Mao F, Lin Y, Shen S, Sun Q. lncRNA AFAP1-AS1 Promotes Triple Negative Breast Cancer Cell Proliferation and Invasion via Targeting miR-145 to Regulate MTH1 Expression. Sci Rep (2020) 10(1):7662. doi: 10.1038/s41598-020-64713-x
73. Choudhari R, Sedano MJ, Harrison AL, Subramani R, Lin KY, Ramos EI, et al. Long Noncoding RNAs in Cancer: From Discovery to Therapeutic Targets. Adv Clin Chem (2020) 95:105–47. doi: 10.1016/bs.acc.2019.08.003
74. Tang Q, Hann SS. HOTAIR: An Oncogenic Long Non-Coding RNA in Human Cancer. Cell Physiol Biochem (2018) 47(3):893–913. doi: 10.1159/000490131
75. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long Non-Coding RNA HOTAIR Reprograms Chromatin State to Promote Cancer Metastasis. Nature (2010) 464(7291):1071–6. doi: 10.1038/nature08975
76. Padua Alves C, Fonseca AS, Muys BR, de Barros ELBR, Burger MC, de Souza JE, et al. Brief Report: The lincRNA Hotair Is Required for Epithelial-to-Mesenchymal Transition and Stemness Maintenance of Cancer Cell Lines. Stem Cells (2013) 31(12):2827–32. doi: 10.1002/stem.1547
77. Yu Y, Lv F, Liang D, Yang Q, Zhang B, Lin H, et al. HOTAIR may Regulate Proliferation, Apoptosis, Migration and Invasion of MCF-7 Cells Through Regulating the P53/Akt/JNK Signaling Pathway. BioMed Pharmacother (2017) 90:555–61. doi: 10.1016/j.biopha.2017.03.054
78. Ding W, Ren J, Ren H, Wang D. Long Noncoding RNA HOTAIR Modulates MiR-206-Mediated Bcl-W Signaling to Facilitate Cell Proliferation in Breast Cancer. Sci Rep (2017) 7(1):17261. doi: 10.1038/s41598-017-17492-x
79. Portoso M, Ragazzini R, Brencic Z, Moiani A, Michaud A, Vassilev I, et al. PRC2 is Dispensable for HOTAIR-Mediated Transcriptional Repression. EMBO J (2017) 36(8):981–94. doi: 10.15252/embj.201695335
80. Hou L, Tu J, Cheng F, Yang H, Yu F, Wang M, et al. Long Noncoding RNA ROR Promotes Breast Cancer by Regulating the TGF-Beta Pathway. Cancer Cell Int (2018) 18:142. doi: 10.1186/s12935-018-0638-4
81. Hou P, Zhao Y, Li Z, Yao R, Ma M, Gao Y, et al. LincRNA-ROR Induces Epithelial-to-Mesenchymal Transition and Contributes to Breast Cancer Tumorigenesis and Metastasis. Cell Death Dis (2014) 5:e1287. doi: 10.1038/cddis.2014.249
82. Zhou Q, Guo J, Huang W, Yu X, Xu C, Long X. Linc-ROR Promotes the Progression of Breast Cancer and Decreases the Sensitivity to Rapamycin Through miR-194-3p Targeting MECP2. Mol Oncol (2020) 14(9):2231–50. doi: 10.1002/1878-0261.12700
83. Peng WX, Huang JG, Yang L, Gong AH, Mo YY. Linc-RoR Promotes MAPK/ERK Signaling and Confers Estrogen-Independent Growth of Breast Cancer. Mol Cancer (2017) 16(1):161. doi: 10.1186/s12943-017-0727-3
84. Chen YM, Liu Y, Wei HY, Lv KZ, Fu PF. Large Intergenic Non-Coding RNA-ROR Reverses Gemcitabine-Induced Autophagy and Apoptosis in Breast Cancer Cells. Oncotarget (2016) 7(37):59604–17. doi: 10.18632/oncotarget.10730
85. Luo C, Cao J, Peng R, Guo Q, Ye H, Wang P, et al. Functional Variants in Linc-ROR are Associated With mRNA Expression of Linc-ROR and Breast Cancer Susceptibility. Sci Rep (2018) 8(1):4680. doi: 10.1038/s41598-018-22881-x
86. Iranpour M, Soudyab M, Geranpayeh L, Mirfakhraie R, Azargashb E, Movafagh A, et al. Expression Analysis of Four Long Noncoding RNAs in Breast Cancer. Tumor Biol (2016) 37(3):2933–40. doi: 10.1007/s13277-015-4135-2
87. Liu M, Xing LQ, Liu YJ. A Three-Long Noncoding RNA Signature as a Diagnostic Biomarker for Differentiating Between Triple-Negative and Non-Triple-Negative Breast Cancers. Med (Baltimore) (2017) 96(9):e6222. doi: 10.1097/MD.0000000000006222
88. Xu ST, Xu JH, Zheng ZR, Zhao QQ, Zeng XS, Cheng SX, et al. Long Non-Coding RNA ANRIL Promotes Carcinogenesis via Sponging miR-199a in Triple-Negative Breast Cancer. BioMed Pharmacother (2017) 96:14–21. doi: 10.1016/j.biopha.2017.09.107
89. Mehta-Mujoo PM, Cunliffe HE, Hung NA, Slatter TL. Long Non-Coding RNA ANRIL in the Nucleus Associates With Periostin Expression in Breast Cancer. Front Oncol (2019) 9:885. doi: 10.3389/fonc.2019.00885
90. Royds JA, Pilbrow AP, Ahn A, Morrin HR, Frampton C, Russell IA, et al. The Rs11515 Polymorphism Is More Frequent and Associated With Aggressive Breast Tumors With Increased ANRIL and Decreased P16 (INK4a) Expression. Front Oncol (2015) 5:306. doi: 10.3389/fonc.2015.00306
91. Khorshidi HR, Taheri M, Noroozi R, Sarrafzadeh S, Sayad A, Ghafouri-Fard S. ANRIL Genetic Variants in Iranian Breast Cancer Patients. Cell J (2017) 19(Suppl 1):72–8. doi: 10.22074/cellj.2017.4496
92. Singh R, Gupta SC, Peng WX, Zhou N, Pochampally R, Atfi A, et al. Regulation of Alternative Splicing of Bcl-X by BC200 Contributes to Breast Cancer Pathogenesis. Cell Death Dis (2016) 7(6):e2262. doi: 10.1038/cddis.2016.168
93. Iacoangeli A, Lin Y, Morley EJ, Muslimov IA, Bianchi R, Reilly J, et al. BC200 RNA in Invasive and Preinvasive Breast Cancer. Carcinogenesis (2004) 25(11):2125–33. doi: 10.1093/carcin/bgh228
94. Iacoangeli A, Adzovic L, Chen EQ, Latif Cattie R, Soff GA. Tiedge H. Regulatory BC200 RNA in Peripheral Blood of Patients With Invasive Breast Cancer. J Investig Med (2018) 66(7):1055–63. doi: 10.1136/jim-2018-000717
95. Nagini S. Breast Cancer: Current Molecular Therapeutic Targets and New Players. Anti-Cancer Agents Medicinal Chem (2017) 17(2):152–63. doi: 10.2174/1871520616666160502122724
96. Mohebi M, Sattari A, Ghafouri-Fard S, Modarressi MH, Kholghi-Oskooei V, Taheri M. Expression Profiling Revealed Up-Regulation of Three lncRNAs in Breast Cancer Samples. Exp Mol Pathol (2020) 117:104544. doi: 10.1016/j.yexmp.2020.104544
97. Li J, Chen Y, Chen Z, He A, Xie H, Zhang Q, et al. SPRY4-IT1: A Novel Oncogenic Long Non-Coding RNA in Human Cancers. Tumor Biol (2017) 39(6):1010428317711406. doi: 10.1177/1010428317711406
98. Shi Y, Li J, Liu Y, Ding J, Fan Y, Tian Y, et al. The Long Noncoding RNA SPRY4-IT1 Increases the Proliferation of Human Breast Cancer Cells by Upregulating ZNF703 Expression. Mol Cancer (2015) 14:51. doi: 10.1186/s12943-015-0318-0
99. Wu H, Wang Y, Chen T, Li Y, Wang H, Zhang L, et al. The N-Terminal Polypeptide Derived From vMIP-II Exerts Its Anti-Tumor Activity in Human Breast Cancer by Regulating lncRNA SPRY4-It1. Biosci Rep (2018) 38(5):BSR20180411. doi: 10.1042/BSR20180411
100. Li Y, Zeng Q, Qiu J, Pang T, Xian J, Zhang X. Long Non-Coding RNA UCA1 Promotes Breast Cancer by Upregulating PTP1B Expression via Inhibiting miR-206. Cancer Cell Int (2019) 19:275. doi: 10.1186/s12935-019-0958-z
101. Yang YT, Wang YF, Lai JY, Shen SY, Wang F, Kong J, et al. Long Non-Coding RNA UCA1 Contributes to the Progression of Oral Squamous Cell Carcinoma by Regulating the WNT/beta-Catenin Signaling Pathway. Cancer Sci (2016) 107(11):1581–9. doi: 10.1111/cas.13058
102. Li GY, Wang W, Sun JY, Xin B, Zhang X, Wang T, et al. Long Non-Coding RNAs AC026904.1 and UCA1: A “One-Two Punch” for TGF-Beta-Induced SNAI2 Activation and Epithelial-Mesenchymal Transition in Breast Cancer. Theranostics (2018) 8(10):2846–61. doi: 10.7150/thno.23463
103. Hong HH, Hou LK, Pan X, Wu CY, Huang H, Li B, et al. Long Non-Coding RNA UCA1 is a Predictive Biomarker of Cancer. Oncotarget (2016) 7(28):44442–7. doi: 10.18632/oncotarget.10142
104. Wang YH, Wang F, Zhang L, Lu JC. Long Non-Coding RNA UCA1 can Predict Tumor Lymph Node Metastasis. Tumor Biol (2017) 39(5):1010428317706208. doi: 10.1177/1010428317706208
105. Li RH, Chen M, Liu J, Shao CC, Guo CP, Wei XL, et al. Long Noncoding RNA ATB Promotes the Epithelial-Mesenchymal Transition by Upregulating the miR-200c/Twist1 Axe and Predicts Poor Prognosis in Breast Cancer. Cell Death Dis (2018) 9(12):1171. doi: 10.1038/s41419-018-1210-9
106. Zhang Y, Li J, Jia S, Wang Y, Kang Y, Zhang W. Down-Regulation of lncRNA-ATB Inhibits Epithelial-Mesenchymal Transition of Breast Cancer Cells by Increasing miR-141-3p Expression. Biochem Cell Biol (2019) 97(2):193–200. doi: 10.1139/bcb-2018-0168
107. El-Ashmawy NE, Hussien FZ, El-Feky OA, Hamouda SM, Al-Ashmawy GM. Serum LncRNA-ATB and FAM83H-AS1 as Diagnostic/Prognostic Non-Invasive Biomarkers for Breast Cancer. Life Sci (2020) 259:118193. doi: 10.1016/j.lfs.2020.118193
108. Shi SJ, Wang LJ, Yu B, Li YH, Jin Y, Bai XZ. LncRNA-ATB Promotes Trastuzumab Resistance and Invasion-Metastasis Cascade in Breast Cancer. Oncotarget (2015) 6(13):11652–63. doi: 10.18632/oncotarget.3457
109. Nikpayam E, Soudyab M, Tasharrofi B, Sarrafzadeh S, Iranpour M, Geranpayeh L, et al. Expression Analysis of Long Non-Coding ATB and Its Putative Target in Breast Cancer. Breast Dis (2017) 37(1):11–20. doi: 10.3233/BD-160264
110. Tang J, Li Y, Sang Y, Yu B, Lv D, Zhang W, et al. LncRNA PVT1 Regulates Triple-Negative Breast Cancer Through KLF5/beta-Catenin Signaling. Oncogene (2018) 37(34):4723–34. doi: 10.1038/s41388-018-0310-4
111. Wang H, Huang Y, Yang Y. LncRNA PVT1 Regulates TRPS1 Expression in Breast Cancer by Sponging miR-543. Cancer Manag Res (2020) 12:7993–8004. doi: 10.2147/CMAR.S263383
112. Chen P, Xu W, Luo Y, Zhang Y, He Y, Yang S, et al. MicroRNA 543 Suppresses Breast Cancer Cell Proliferation, Blocks Cell Cycle and Induces Cell Apoptosis via Direct Targeting of ERK/MAPK. Onco Targets Ther (2017) 10:1423–31. doi: 10.2147/OTT.S118366
113. Yan C, Chen Y, Kong W, Fu L, Liu Y, Yao Q, et al. PVT1-Derived miR-1207-5p Promotes Breast Cancer Cell Growth by Targeting STAT6. Cancer Sci (2017) 108(5):868–76. doi: 10.1111/cas.13212
114. Liu X, Bi L, Wang Q, Wen M, Li C, Ren Y, et al. miR-1204 Targets VDR to Promotes Epithelial-Mesenchymal Transition and Metastasis in Breast Cancer. Oncogene (2018) 37(25):3426–39. doi: 10.1038/s41388-018-0215-2
115. Lu D, Luo P, Wang Q, Ye Y, Wang B. lncRNA PVT1 in Cancer: A Review and Meta-Analysis. Clin Chim Acta (2017) 474:1–7. doi: 10.1016/j.cca.2017.08.038
116. Han C, Li X, Fan Q, Liu G, Yin J. CCAT1 Promotes Triple-Negative Breast Cancer Progression by Suppressing miR-218/ZFX Signaling. Aging (Albany NY) (2019) 11(14):4858–75. doi: 10.18632/aging.102080
117. Lai Y, Chen Y, Lin Y, Ye L. Down-Regulation of LncRNA CCAT1 Enhances Radiosensitivity via Regulating miR-148b in Breast Cancer. Cell Biol Int (2018) 42(2):227–36. doi: 10.1002/cbin.10890
118. Tang T, Guo C, Xia T, Zhang R, Zen K, Pan Y, et al. LncCCAT1 Promotes Breast Cancer Stem Cell Function Through Activating WNT/beta-Catenin Signaling. Theranostics (2019) 9(24):7384–402. doi: 10.7150/thno.37892
119. Zhang XF, Liu T, Li Y, Li S. Overexpression of Long Non-Coding RNA CCAT1 Is a Novel Biomarker of Poor Prognosis in Patients With Breast Cancer. Int J Clin Exp Pathol (2015) 8(8):9440–5.
120. Redis RS, Sieuwerts AM, Look MP, Tudoran O, Ivan C, Spizzo R, et al. CCAT2, A Novel Long Non-Coding RNA in Breast Cancer: Expression Study and Clinical Correlations. Oncotarget (2013) 4(10):1748–62. doi: 10.18632/oncotarget.1292
121. Huang B, Yu M, Guan R, Liu D. Hou B. A Comprehensive Exploration of the lncRNA CCAT2: A Pan-Cancer Analysis Based on 33 Cancer Types and 13285 Cases. Dis Markers (2020) 2020:5354702. doi: 10.1155/2020/5354702
122. Wu ZJ, Li Y, Wu YZ, Wang Y, Nian WQ, Wang LL, et al. Long Non-Coding RNA CCAT2 Promotes the Breast Cancer Growth and Metastasis by Regulating TGF-β Signaling Pathway. Eur Rev Med Pharmacol Sci (2017) 21(4):706–14.
123. Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, et al. EZH2 is a Marker of Aggressive Breast Cancer and Promotes Neoplastic Transformation of Breast Epithelial Cells. Proc Natl Acad Sci USA (2003) 100(20):11606–11. doi: 10.1073/pnas.1933744100
124. Xu Z, Liu C, Zhao Q, Lu J, Ding X, Luo A, et al. Long Non-Coding RNA CCAT2 Promotes Oncogenesis in Triple-Negative Breast Cancer by Regulating Stemness of Cancer Cells. Pharmacol Res (2020) 152:104628. doi: 10.1016/j.phrs.2020.104628
125. Liu Y, Du Y, Hu X, Zhao L, Xia W. Up-Regulation of ceRNA TINCR by SP1 Contributes to Tumorigenesis in Breast Cancer. BMC Cancer (2018) 18(1):367. doi: 10.1186/s12885-018-4255-3
126. Guo F, Zhu X, Zhao Q, Huang Q. Mir5893p Sponged by the lncRNA TINCR Inhibits the Proliferation, Migration and Invasion and Promotes the Apoptosis of Breast Cancer Cells by Suppressing the Akt Pathway via IGF1R. Int J Mol Med (2020) 46(3):989–1002. doi: 10.3892/ijmm.2020.4666
127. Dong H, Hu J, Zou K, Ye M, Chen Y, Wu C, et al. Activation of LncRNA TINCR by H3K27 Acetylation Promotes Trastuzumab Resistance and Epithelial-Mesenchymal Transition by Targeting MicroRNA-125b in Breast Cancer. Mol Cancer (2019) 18(1):3. doi: 10.1186/s12943-018-0931-9
128. Wang X, Li S, Xiao H, Deng X. Serum lncRNA TINCR Serve as a Novel Biomarker for Predicting the Prognosis in Triple-Negative Breast Cancer. Technol Cancer Res Treat (2020) 19:1533033820965574. doi: 10.1177/1533033820965574
129. Zhang X, Zhou Y, Mehta KR, Danila DC, Scolavino S, Johnson SR, et al. A Pituitary-Derived MEG3 Isoform Functions as a Growth Suppressor in Tumor Cells. J Clin Endocrinol Metab (2003) 88(11):5119–26. doi: 10.1210/jc.2003-030222
130. Zhang JJ, Guo SH, Jia BQ. Down-Regulation of Long Non-Coding RNA MEG3 Serves as an Unfavorable Risk Factor for Survival of Patients With Breast Cancer. Eur Rev Med Pharmacol Sci (2016) 20(24):5143–7.
131. Cui X, Jing X, Long C, Tian J, Zhu J. Long Noncoding RNA MEG3, a Potential Novel Biomarker to Predict the Clinical Outcome of Cancer Patients: A Meta-Analysis. Oncotarget (2017) 8(12):19049–56. doi: 10.18632/oncotarget.14987
132. Cui X, Yi Q, Jing X, Huang Y, Tian J, Long C, et al. Mining Prognostic Significance of MEG3 in Human Breast Cancer Using Bioinformatics Analysis. Cell Physiol Biochem (2018) 50(1):41–51. doi: 10.1159/000493956
133. Zhang Y, Wu J, Jing H, Huang G, Sun Z, Xu S. Long Noncoding RNA MEG3 Inhibits Breast Cancer Growth via Upregulating Endoplasmic Reticulum Stress and Activating NF-kappaB and P53. J Cell Biochem (2019) 120(4):6789–97. doi: 10.1002/jcb.27982
134. Sun L, Li Y, Yang B. Downregulated Long Non-Coding RNA MEG3 in Breast Cancer Regulates Proliferation, Migration and Invasion by Depending on P53’s Transcriptional Activity. Biochem Biophys Res Commun (2016) 478(1):323–9. doi: 10.1016/j.bbrc.2016.05.031
135. Zhu M, Wang X, Gu Y, Wang F, Li L, Qiu X. MEG3 Overexpression Inhibits the Tumorigenesis of Breast Cancer by Downregulating miR-21 Through the PI3K/Akt Pathway. Arch Biochem Biophys (2019) 661:22–30. doi: 10.1016/j.abb.2018.10.021
136. Ali MA, Shaker OG, Alazrak M, AbdelHafez MN, Khalefa AA, Hemeda NF, et al. Association Analyses of a Genetic Variant in Long Non-Coding RNA MEG3 With Breast Cancer Susceptibility and Serum MEG3 Expression Level in the Egyptian Population. Cancer Biomark (2020) 28(1):49–63. doi: 10.3233/CBM-191072
137. Zheng Y, Wang M, Wang S, Xu P, Deng Y, Lin S, et al. LncRNA MEG3 Rs3087918 was Associated With a Decreased Breast Cancer Risk in a Chinese Population: A Case-Control Study. BMC Cancer (2020) 20(1):659. doi: 10.1186/s12885-020-07145-0
138. Zheng R, Lin S, Guan L, Yuan H, Liu K, Liu C, et al. Long Non-Coding RNA XIST Inhibited Breast Cancer Cell Growth, Migration, and Invasion via miR-155/CDX1 Axis. Biochem Biophys Res Commun (2018) 498(4):1002–8. doi: 10.1016/j.bbrc.2018.03.104
139. Liu B, Luo C, Lin H, Ji X, Zhang E, Li X. Long Noncoding RNA XIST Acts as a ceRNA of miR-362-5p to Suppress Breast Cancer Progression. Cancer Biother Radiopharm (2020) 36(6):456–66. doi: 10.1089/cbr.2019.3481
140. Jara L, Morales S, de Mayo T, Gonzalez-Hormazabal P, Carrasco V, Godoy R. Mutations in BRCA1, BRCA2 and Other Breast and Ovarian Cancer Susceptibility Genes in Central and South American Populations. Biol Res (2017) 50(1):35. doi: 10.1186/s40659-017-0139-2
141. Sharma B, Preet Kaur R, Raut S, Munshi A. BRCA1 Mutation Spectrum, Functions, and Therapeutic Strategies: The Story So Far. Curr Probl Cancer (2018) 42(2):189–207. doi: 10.1016/j.currproblcancer.2018.01.001
142. Sirchia SM, Tabano S, Monti L, Recalcati MP, Gariboldi M, Grati FR, et al. Misbehaviour of XIST RNA in Breast Cancer Cells. PloS One (2009) 4(5):e5559. doi: 10.1371/journal.pone.0005559
143. Zong YY, Zhang YZ, Hou DS, Xu JW, Cui FY, Qin YJ, et al. The lncRNA XIST Promotes the Progression of Breast Cancer by Sponging miR-125b-5p to Modulate NLRC5. Am J Transl Res (2020) 12(7):3501–11.
144. Zhu J, Kong F, Xing L, Jin Z, Li Z. Prognostic and Clinicopathological Value of Long Noncoding RNA XIST in Cancer. Clin Chim Acta (2018) 479:43–7. doi: 10.1016/j.cca.2018.01.005
145. Chen S, Wang Y, Zhang JH, Xia QJ, Sun Q, Li ZK, et al. Long Non-Coding RNA PTENP1 Inhibits Proliferation and Migration of Breast Cancer Cells via AKT and MAPK Signaling Pathways. Oncol Lett (2017) 14(4):4659–62. doi: 10.3892/ol.2017.6823
146. Li RK, Gao J, Guo LH, Huang GQ, Luo WH. PTENP1 Acts as a ceRNA to Regulate PTEN by Sponging miR-19b and Explores the Biological Role of PTENP1 in Breast Cancer. Cancer Gene Ther (2017) 24(7):309–15. doi: 10.1038/cgt.2017.29
147. Suter R, Marcum JA. The Molecular Genetics of Breast Cancer and Targeted Therapy. Biologics (2007) 1(3):241–58.
148. Gao X, Qin T, Mao J, Zhang J, Fan S, Lu Y, et al. PTENP1/miR-20a/PTEN Axis Contributes to Breast Cancer Progression by Regulating PTEN via PI3K/AKT Pathway. J Exp Clin Cancer Res (2019) 38(1):256. doi: 10.1186/s13046-019-1260-6
149. Yndestad S, Austreid E, Skaftnesmo KO, Lonning PE, Eikesdal HP. Divergent Activity of the Pseudogene PTENP1 in ER-Positive and Negative Breast Cancer. Mol Cancer Res (2018) 16(1):78–89. doi: 10.1158/1541-7786.MCR-17-0207
150. Klec C, Prinz F, Pichler M. Involvement of the Long Noncoding RNA NEAT1 in Carcinogenesis. Mol Oncol (2019) 13(1):46–60. doi: 10.1002/1878-0261.12404
151. Gabory A, Ripoche MA, Yoshimizu T, Dandolo L. The H19 Gene: Regulation and Function of a Non-Coding RNA. Cytogenet Genome Res (2006) 113(1-4):188–93. doi: 10.1159/000090831
152. Lottin S, Adriaenssens E, Dupressoir T, Berteaux N, Montpellier C, Coll J, et al. Overexpression of an Ectopic H19 Gene Enhances the Tumorigenic Properties of Breast Cancer Cells. Carcinogenesis (2002) 23(11):1885–95. doi: 10.1093/carcin/23.11.1885
153. Yu H, Li S, Wu SX, Huang S, Li S, Ye L. The Prognostic Value of Long Non-Coding RNA H19 in Various Cancers: A Meta-Analysis Based on 15 Studies With 1584 Patients and the Cancer Genome Atlas Data. Med (Baltimore) (2020) 99(2):e18533. doi: 10.1097/MD.0000000000018533
154. Zhang F, Li J, Xiao H, Zou Y, Liu Y, Huang W. AFAP1-AS1: A Novel Oncogenic Long Non-Coding RNA in Human Cancers. Cell Prolif (2018) 51(1):e12397. doi: 10.1111/cpr.12397
155. Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs. Cell (2007) 129(7):1311–23. doi: 10.1016/j.cell.2007.05.022
156. Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, et al. HOTAIR Is a Negative Prognostic Factor and Exhibits Pro-Oncogenic Activity in Pancreatic Cancer. Oncogene (2013) 32(13):1616–25. doi: 10.1038/onc.2012.193
157. Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, et al. Long Noncoding RNA HOTAIR Regulates Polycomb-Dependent Chromatin Modification and is Associated With Poor Prognosis in Colorectal Cancers. Cancer Res (2011) 71(20):6320–6. doi: 10.1158/0008-5472.CAN-11-1021
158. Nakagawa T, Endo H, Yokoyama M, Abe J, Tamai K, Tanaka N, et al. Large Noncoding RNA HOTAIR Enhances Aggressive Biological Behavior and is Associated With Short Disease-Free Survival in Human Non-Small Cell Lung Cancer. Biochem Biophys Res Commun (2013) 436(2):319–24. doi: 10.1016/j.bbrc.2013.05.101
159. Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, et al. Large Intergenic Non-Coding RNA-RoR Modulates Reprogramming of Human Induced Pluripotent Stem Cells. Nat Genet (2010) 42(12):1113–7. doi: 10.1038/ng.710
160. Chen W, Yang J, Fang H, Li L, Sun J. Relevance Function of Linc-ROR in the Pathogenesis of Cancer. Front Cell Dev Biol (2020) 8:696. doi: 10.3389/fcell.2020.00696
161. Huang J, Zhang A, Ho TT, Zhang Z, Zhou N, Ding X, et al. Linc-RoR Promotes C-Myc Expression Through hnRNP I and AUF1. Nucleic Acids Res (2016) 44(7):3059–69. doi: 10.1093/nar/gkv1353
162. Zhan HX, Wang Y, Li C, Xu JW, Zhou B, Zhu JK, et al. LincRNA-ROR Promotes Invasion, Metastasis and Tumor Growth in Pancreatic Cancer Through Activating ZEB1 Pathway. Cancer Lett (2016) 374(2):261–71. doi: 10.1016/j.canlet.2016.02.018
163. Pasmant E, Laurendeau I, Heron D, Vidaud M, Vidaud D, Bieche I. Characterization of a Germ-Line Deletion, Including the Entire INK4/ARF Locus, in a Melanoma-Neural System Tumor Family: Identification of ANRIL, an Antisense Noncoding RNA Whose Expression Coclusters With ARF. Cancer Res (2007) 67(8):3963–9. doi: 10.1158/0008-5472.CAN-06-2004
164. Rivandi M, Khorrami MS, Fiuji H, Shahidsales S, Hasanzadeh M, Jazayeri MH, et al. The 9p21 Locus: A Potential Therapeutic Target and Prognostic Marker in Breast Cancer. J Cell Physiol (2018) 233(7):5170–9. doi: 10.1002/jcp.26332
165. Zhang Z, Feng L, Liu P, Duan W. ANRIL Promotes Chemoresistance via Disturbing Expression of ABCC1 by Regulating the Expression of Let-7a in Colorectal Cancer. Biosci Rep (2018) 38(6):BSR20180620. doi: 10.1042/BSR20180620
166. Zhang EB, Kong R, Yin DD, You LH, Sun M, Han L, et al. Long Noncoding RNA ANRIL Indicates a Poor Prognosis of Gastric Cancer and Promotes Tumor Growth by Epigenetically Silencing of miR-99a/miR-449a. Oncotarget (2014) 5(8):2276–92. doi: 10.18632/oncotarget.1902
167. Paul Y, Thomas S, Patil V, Kumar N, Mondal B, Hegde AS, et al. Genetic Landscape of Long Noncoding RNA (lncRNAs) in Glioblastoma: Identification of Complex lncRNA Regulatory Networks and Clinically Relevant lncRNAs in Glioblastoma. Oncotarget (2018) 9(51):29548–64. doi: 10.18632/oncotarget.25434
168. Lou N, Liu G, Pan Y. Long Noncoding RNA ANRIL as a Novel Biomarker in Human Cancer. Future Oncol (2020) 16(35):2981–95. doi: 10.2217/fon-2020-0470
169. Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, et al. Long Non-Coding RNA ANRIL Is Required for the PRC2 Recruitment to and Silencing of P15(INK4B) Tumor Suppressor Gene. Oncogene (2011) 30(16):1956–62. doi: 10.1038/onc.2010.568
170. Samson J, Cronin S, Dean K. BC200 (BCYRN1) - The Shortest, Long, Non-Coding RNA Associated With Cancer. Noncoding RNA Res (2018) 3(3):131–43. doi: 10.1016/j.ncrna.2018.05.003
171. Shin H, Kim Y, Kim M, Lee Y. BC200 RNA: An Emerging Therapeutic Target and Diagnostic Marker for Human Cancer. Mol Cells (2018) 41(12):993–9. doi: 10.14348/molcells.2018.0425
172. Chen W, Böcker W, Brosius J, Tiedge H. Expression of Neural BC200 RNA in Human Tumors. J Pathol (1997) 183(3):345–51. doi: 10.1002/(sici)1096-9896(199711)183:3<345::aid-path930>3.0.co;2-8
173. Booy EP, McRae EK, Koul A, Lin F, McKenna SA. The Long Non-Coding RNA BC200 (BCYRN1) Is Critical for Cancer Cell Survival and Proliferation. Mol Cancer (2017) 16(1):109. doi: 10.1186/s12943-017-0679-7
174. Hu T, Lu YR. BCYRN1, a C-MYC-Activated Long Non-Coding RNA, Regulates Cell Metastasis of Non-Small-Cell Lung Cancer. Cancer Cell Int (2015) 15:36. doi: 10.1186/s12935-015-0183-3
175. Wu K, Xu K, Liu K, Huang J, Chen J, Zhang J, et al. Long Noncoding RNA BC200 Regulates Cell Growth and Invasion in Colon Cancer. Int J Biochem Cell Biol (2018) 99:219–25. doi: 10.1016/j.biocel.2018.04.001
176. Peng J, Hou F, Feng J, Xu SX, Meng XY. Long Non-Coding RNA BCYRN1 Promotes the Proliferation and Metastasis of Cervical Cancer via Targeting microRNA-138 In Vitro and In Vivo. Oncol Lett (2018) 15(4):5809–18. doi: 10.3892/ol.2018.8015
177. Barton M, Santucci-Pereira J, Vaccaro OG, Nguyen T, Su Y, Russo J. BC200 Overexpression Contributes to Luminal and Triple Negative Breast Cancer Pathogenesis. BMC Cancer (2019) 19(1):994. doi: 10.1186/s12885-019-6179-y
178. Khaitan D, Dinger ME, Mazar J, Crawford J, Smith MA, Mattick JS, et al. The Melanoma-Upregulated Long Noncoding RNA SPRY4-IT1 Modulates Apoptosis and Invasion. Cancer Res (2011) 71(11):3852–62. doi: 10.1158/0008-5472.CAN-10-4460
179. Cao D, Ding Q, Yu W, Gao M, Wang Y. Long Noncoding RNA SPRY4-IT1 Promotes Malignant Development of Colorectal Cancer by Targeting Epithelial-Mesenchymal Transition. Onco Targets Ther (2016) 9:5417–25. doi: 10.2147/OTT.S111794
180. Sun M, Liu XH, Lu KH, Nie FQ, Xia R, Kong R, et al. EZH2-Mediated Epigenetic Suppression of Long Noncoding RNA SPRY4-IT1 Promotes NSCLC Cell Proliferation and Metastasis by Affecting the Epithelial-Mesenchymal Transition. Cell Death Dis (2014) 5:e1298. doi: 10.1038/cddis.2014.256
181. Xiang YQ, Chen YZ, Shi YX, Wu XB, Hao RT, Li Q, et al. Upregulation of the Long Non-Coding RNA SPRY4-IT1 Predicts Poor Prognosis in Breast Cancer. Int J Clin Exp Pathol (2019) 12(3):1003–8.
182. Huang J, Zhou N, Watabe K, Lu Z, Wu F, Xu M, et al. Long Non-Coding RNA UCA1 Promotes Breast Tumor Growth by Suppression of P27 (Kip1). Cell Death Dis (2014) 5:e1008. doi: 10.1038/cddis.2013.541
183. Wang X, Fa XE. Knockdown of UCA1 Inhibits Viability and Glycolysis by Suppressing PKM2 Expression Through the mTOR Pathway in Non-Small Cell Lung Cancer Cells. RSC Adv (2018) 8(19):10610–9. doi: 10.1039/c8ra00860d
184. Wang Y, Chen W, Yang C, Wu W, Wu S, Qin X, et al. Long Non-Coding RNA UCA1a(CUDR) Promotes Proliferation and Tumorigenesis of Bladder Cancer. Int J Oncol (2012) 41(1):276–84. doi: 10.3892/ijo.2012.1443
185. Xie XJ, Li X, Wang F, Chen W. Cellular Localization and Tissue Expression Pattern of UCA1, A Non-Coding RNA. Nan Fang Yi Ke Da Xue Xue Bao (2010) 30(1):57–60.
186. Yao F, Wang Q, Wu Q. The Prognostic Value and Mechanisms of lncRNA UCA1 in Human Cancer. Cancer Manag Res (2019) 11:7685–96. doi: 10.2147/CMAR.S200436
187. Wu H, Zhou C. Long Non-Coding RNA UCA1 Promotes Lung Cancer Cell Proliferation and Migration via microRNA-193a/HMGB1 Axis. Biochem Biophys Res Commun (2018) 496(2):738–45. doi: 10.1016/j.bbrc.2018.01.097
188. Luo J, Chen J, Li H, Yang Y, Yun H, Yang S, et al. LncRNA UCA1 Promotes the Invasion and EMT of Bladder Cancer Cells by Regulating the miR-143/HMGB1 Pathway. Oncol Lett (2017) 14(5):5556–62. doi: 10.3892/ol.2017.6886
189. Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, et al. A Long Noncoding RNA Activated by TGF-β Promotes the Invasion-Metastasis Cascade in Hepatocellular Carcinoma. Cancer Cell (2014) 25(5):666–81. doi: 10.1016/j.ccr.2014.03.010
190. Qu S, Yang X, Song W, Sun W, Li X, Wang J, et al. Downregulation of lncRNA-ATB Correlates With Clinical Progression and Unfavorable Prognosis in Pancreatic Cancer. Tumor Biol (2016) 37(3):3933–8. doi: 10.1007/s13277-015-4252-y
191. Wan L, Sun M, Liu GJ, Wei CC, Zhang EB, Kong R, et al. Long Noncoding RNA PVT1 Promotes Non-Small Cell Lung Cancer Cell Proliferation Through Epigenetically Regulating LATS2 Expression. Mol Cancer Ther (2016) 15(5):1082–94. doi: 10.1158/1535-7163.MCT-15-0707
192. Derderian C, Orunmuyi AT, Olapade-Olaopa EO, Ogunwobi OO. PVT1 Signaling Is a Mediator of Cancer Progression. Front Oncol (2019) 9:502. doi: 10.3389/fonc.2019.00502
193. Wang W, Zhou R, Wu Y, Liu Y, Su W, Xiong W, et al. PVT1 Promotes Cancer Progression via MicroRNAs. Front Oncol (2019) 9:609. doi: 10.3389/fonc.2019.00609
194. Liu W, Yao D, Huang B. LncRNA PVT1 Promotes Cervical Cancer Progression by Sponging miR-503 to Upregulate ARL2 Expression. Open Life Sci (2021) 16(1):1–13. doi: 10.1515/biol-2021-0002
195. Salehi M, Sharifi M, Bagheri M. Knockdown of Long Noncoding RNA Plasmacytoma Variant Translocation 1 With Antisense Locked Nucleic Acid GapmeRs Exerts Tumor-Suppressive Functions in Human Acute Erythroleukemia Cells Through Downregulation of C-MYC Expression. Cancer Biother Radiopharm (2019) 34(6):371–9. doi: 10.1089/cbr.2018.2510
196. Nissan A, Stojadinovic A, Mitrani-Rosenbaum S, Halle D, Grinbaum R, Roistacher M, et al. Colon Cancer Associated Transcript-1: A Novel RNA Expressed in Malignant and Pre-Malignant Human Tissues. Int J Cancer (2012) 130(7):1598–606. doi: 10.1002/ijc.26170
197. Ozawa T, Matsuyama T, Toiyama Y, Takahashi N, Ishikawa T, Uetake H, et al. CCAT1 and CCAT2 Long Noncoding RNAs, Located Within the 8q.24.21 ‘Gene Desert’, Serve as Important Prognostic Biomarkers in Colorectal Cancer. Ann Oncol (2017) 28(8):1882–8. doi: 10.1093/annonc/mdx248
198. Liu Z, Chen Q, Hann SS. The Functions and Oncogenic Roles of CCAT1 in Human Cancer. BioMed Pharmacother (2019) 115:108943. doi: 10.1016/j.biopha.2019.108943
199. Zhang J, Gao Y. CCAT-1 Promotes Proliferation and Inhibits Apoptosis of Cervical Cancer Cells via the Wnt Signaling Pathway. Oncotarget (2017) 8(40):68059–70. doi: 10.18632/oncotarget.19155
200. Xu C, Xiao G, Zhang B, Wang M, Wang J, Liu D, et al. CCAT1 Stimulation of the Symmetric Division of NSCLC Stem Cells Through Activation of the Wnt Signaling Cascade. Gene Ther (2018) 25(1):4–12. doi: 10.1038/gt.2017.98
201. Fu C, Xu X, Lu W, Nie L, Yin T, Wu D. Increased Expression of Long Non-Coding RNA CCAT2 Predicts Poorer Prognosis in Patients With Hepatocellular Carcinoma. Med (Baltimore) (2019) 98(42):e17412. doi: 10.1097/MD.0000000000017412
202. Ling H, Spizzo R, Atlasi Y, Nicoloso M, Shimizu M, Redis RS, et al. CCAT2, a Novel Noncoding RNA Mapping to 8q24, Underlies Metastatic Progression and Chromosomal Instability in Colon Cancer. Genome Res (2013) 23(9):1446–61. doi: 10.1101/gr.152942.112
203. Shen P, Pichler M, Chen M, Calin GA, Ling H. To Wnt or Lose: The Missing Non-Coding Linc in Colorectal Cancer. Int J Mol Sci (2017) 18(9):2003. doi: 10.3390/ijms18092003
204. Deng X, Zhao Y, Wu X, Song G. Upregulation of CCAT2 Promotes Cell Proliferation by Repressing the P15 in Breast Cancer. BioMed Pharmacother (2017) 91:1160–6. doi: 10.1016/j.biopha.2017.05.030
205. Kontomanolis EN, Kalagasidou S, Pouliliou S, Anthoulaki X, Georgiou N, Papamanolis V, et al. The Notch Pathway in Breast Cancer Progression. Sci World J (2018) 2018:2415489. doi: 10.1155/2018/2415489
206. Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, et al. Control of Somatic Tissue Differentiation by the Long Non-Coding RNA TINCR. Nature (2013) 493(7431):231–5. doi: 10.1038/nature11661
207. Sharma U, Barwal TS, Malhotra A, Pant N, Vivek, Dey D, et al. Long Non-Coding RNA TINCR as Potential Biomarker and Therapeutic Target for Cancer. Life Sci (2020) 257:118035. doi: 10.1016/j.lfs.2020.118035
208. Ghafouri-Fard S, Dashti S, Taheri M, Omrani MD. TINCR: An lncRNA With Dual Functions in the Carcinogenesis Process. Noncoding RNA Res (2020) 5(3):109–15. doi: 10.1016/j.ncrna.2020.06.003
209. Xu TP, Liu XX, Xia R, Yin L, Kong R, Chen WM, et al. SP1-Induced Upregulation of the Long Noncoding RNA TINCR Regulates Cell Proliferation and Apoptosis by Affecting KLF2 mRNA Stability in Gastric Cancer. Oncogene (2015) 34(45):5648–61. doi: 10.1038/onc.2015.18
210. Motallebnezhad M, Aghebati-Maleki L, Jadidi-Niaragh F, Nickho H, Samadi-Kafil H, Shamsasenjan K, et al. The Insulin-Like Growth Factor-I Receptor (IGF-IR) in Breast Cancer: Biology and Treatment Strategies. Tumor Biol (2016) 37(9):11711–21. doi: 10.1007/s13277-016-5176-x
211. Zhang X, Rice K, Wang Y, Chen W, Zhong Y, Nakayama Y, et al. Maternally Expressed Gene 3 (MEG3) Noncoding Ribonucleic Acid: Isoform Structure, Expression, and Functions. Endocrinology (2010) 151(3):939–47. doi: 10.1210/en.2009-0657
212. He Y, Luo Y, Liang B, Ye L, Lu G, He W. Potential Applications of MEG3 in Cancer Diagnosis and Prognosis. Oncotarget (2017) 8(42):73282–95. doi: 10.18632/oncotarget.19931
213. Liu B, Shen ED, Liao MM, Hu YB, Wu K, Yang P, et al. Expression and Mechanisms of Long Non-Coding RNA Genes MEG3 and ANRIL in Gallbladder Cancer. Tumor Biol (2016) 37(7):9875–86. doi: 10.1007/s13277-016-4863-y
214. Zhang J, Yao T, Wang Y, Yu J, Liu Y, Lin Z. Long Noncoding RNA MEG3 Is Downregulated in Cervical Cancer and Affects Cell Proliferation and Apoptosis by Regulating miR-21. Cancer Biol Ther (2016) 17(1):104–13. doi: 10.1080/15384047.2015.1108496
215. Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A Global Reference for Human Genetic Variation. Nature (2015) 526(7571):68–74. doi: 10.1038/nature15393
216. Michailidou K, Beesley J, Lindstrom S, Canisius S, Dennis J, Lush MJ, et al. Genome-Wide Association Analysis of More Than 120,000 Individuals Identifies 15 New Susceptibility Loci for Breast Cancer. Nat Genet (2015) 47(4):373–80. doi: 10.1038/ng.3242
217. Brown CJ, Hendrich BD, Rupert JL, Lafrenière RG, Xing Y, Lawrence J, et al. The Human XIST Gene: Analysis of a 17 Kb Inactive X-Specific RNA That Contains Conserved Repeats and Is Highly Localized Within the Nucleus. Cell (1992) 71(3):527–42. doi: 10.1016/0092-8674(92)90520-m
218. Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell (2016) 29(4):452–63. doi: 10.1016/j.ccell.2016.03.010
219. Yang X, Xie Z, Lei X, Gan R. Long Non-Coding RNA GAS5 in Human Cancer. Oncol Lett (2020) 20(3):2587–94. doi: 10.3892/ol.2020.11809
220. Cheng K, Zhao Z, Wang G, Wang J, Zhu W. lncRNA GAS5 Inhibits Colorectal Cancer Cell Proliferation via the Mir1825p/FOXO3a Axis. Oncol Rep (2018) 40(4):2371–80. doi: 10.3892/or.2018.6584
221. Pickard MR, Mourtada-Maarabouni M, Williams GT. Long Non-Coding RNA GAS5 Regulates Apoptosis in Prostate Cancer Cell Lines. Biochim Biophys Acta (2013) 1832(10):1613–23. doi: 10.1016/j.bbadis.2013.05.005
222. Wang M, Guo C, Wang L, Luo G, Huang C, Li Y, et al. Long Noncoding RNA GAS5 Promotes Bladder Cancer Cells Apoptosis Through Inhibiting EZH2 Transcription. Cell Death Dis (2018) 9(2):238. doi: 10.1038/s41419-018-0264-z
223. Li Y, Gu J, Lu H. The GAS5/miR-222 Axis Regulates Proliferation of Gastric Cancer Cells Through the PTEN/Akt/mTOR Pathway. Dig Dis Sci (2017) 62(12):3426–37. doi: 10.1007/s10620-017-4831-4
224. Wang G, Sun J, Zhao H, Li H. Long Non-Coding RNA (lncRNA) Growth Arrest Specific 5 (GAS5) Suppresses Esophageal Squamous Cell Carcinoma Cell Proliferation and Migration by Inactivating Phosphatidylinositol 3-Kinase (PI3K)/AKT/Mammalian Target of Rapamycin (mTOR) Signaling Pathway. Med Sci Monitor (2018) 24:7689–96. doi: 10.12659/msm.910867
225. Wang K, Li J, Xiong G, He G, Guan X, Yang K, et al. Negative Regulation of lncRNA GAS5 by miR-196a Inhibits Esophageal Squamous Cell Carcinoma Growth. Biochem Biophys Res Commun (2018) 495(1):1151–7. doi: 10.1016/j.bbrc.2017.11.119
226. Gao J, Liu M, Zou Y, Mao M, Shen T, Zhang C, et al. Long Non-Coding RNA Growth Arrest-Specific Transcript 5 is Involved in Ovarian Cancer Cell Apoptosis Through the Mitochondria-Mediated Apoptosis Pathway. Oncol Rep (2015) 34(6):3212–21. doi: 10.3892/or.2015.4318
227. Zhao H, Yu H, Zheng J, Ning N, Tang F, Yang Y, et al. Lowly-Expressed lncRNA GAS5 Facilitates Progression of Ovarian Cancer Through Targeting miR-196-5p and Thereby Regulating HOXA5. Gynecol Oncol (2018) 151(2):345–55. doi: 10.1016/j.ygyno.2018.08.032
228. Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, A Non-Protein-Coding RNA, Controls Apoptosis and Is Downregulated in Breast Cancer. Oncogene (2009) 28(2):195–208. doi: 10.1038/onc.2008.373
229. Gu J, Wang Y, Wang X, Zhou D, Wang X, Zhou M, et al. Effect of the LncRNA GAS5-MiR-23a-ATG3 Axis in Regulating Autophagy in Patients With Breast Cancer. Cell Physiol Biochem (2018) 48(1):194–207. doi: 10.1159/000491718
230. Li W, Zhai L, Wang H, Liu C, Zhang J, Chen W, et al. Downregulation of LncRNA GAS5 Causes Trastuzumab Resistance in Breast Cancer. Oncotarget (2016) 7(19):27778–86. doi: 10.18632/oncotarget.8413
231. Zhang Z, Zhu Z, Watabe K, Zhang X, Bai C, Xu M, et al. Negative Regulation of lncRNA GAS5 by miR-21. Cell Death Differ (2013) 20(11):1558–68. doi: 10.1038/cdd.2013.110
232. Pei J, Wang B. Notch-1 Promotes Breast Cancer Cells Proliferation by Regulating LncRNA Gas5. Int J Clin Exp Med (2015) 8(8):14464–71.
233. Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ. Pandolfi PP. A Coding-Independent Function of Gene and Pseudogene mRNAs Regulates Tumor Biology. Nature (2010) 465(7301):1033–8. doi: 10.1038/nature09144
234. Zhou X, Yin C, Dang Y, Ye F, Zhang G. Identification of the Long Non-Coding RNA H19 in Plasma as a Novel Biomarker for Diagnosis of Gastric Cancer. Sci Rep (2015) 5:11516. doi: 10.1038/srep11516
235. Xue WJ, Ying XL, Jiang JH, Xu YH. Prostate Cancer Antigen 3 as a Biomarker in the Urine for Prostate Cancer Diagnosis: A Meta-Analysis. J Cancer Res Ther (2014) 10 Suppl:C218–21. doi: 10.4103/0973-1482.145881
236. Arantes L, De Carvalho AC, Melendez ME, Lopes Carvalho A. Serum, Plasma and Saliva Biomarkers for Head and Neck Cancer. Expert Rev Mol Diagn (2018) 18(1):85–112. doi: 10.1080/14737159.2017.1404906
237. Zidan HE, Karam RA, El-Seifi OS, Abd Elrahman TM. Circulating Long Non-Coding RNA MALAT1 Expression as Molecular Biomarker in Egyptian Patients With Breast Cancer. Cancer Genet (2018) 220:32–7. doi: 10.1016/j.cancergen.2017.11.005
238. Chery J. RNA Therapeutics_ RNAi and Antisense Mechanisms and Clinical Applications. Postdoc J (2016) 4(7):35–50. doi: 10.14304/surya.jpr.v4n7.5
239. Esposito R, Bosch N, Lanzós A, Polidori T, Pulido-Quetglas C, Johnson R. Hacking the Cancer Genome: Profiling Therapeutically Actionable Long Non-Coding RNAs Using CRISPR-Cas9 Screening. Cancer Cell (2019) 35(4):545–57. doi: 10.1016/j.ccell.2019.01.019
Keywords: long non-coding RNA, breast cancer, biomarker, diagnosis, prognosis
Citation: Lu C, Wei D, Zhang Y, Wang P and Zhang W (2021) Long Non-Coding RNAs as Potential Diagnostic and Prognostic Biomarkers in Breast Cancer: Progress and Prospects. Front. Oncol. 11:710538. doi: 10.3389/fonc.2021.710538
Received: 16 May 2021; Accepted: 09 August 2021;
Published: 30 August 2021.
Edited by:
Desi Shang, Harbin Medical University, ChinaReviewed by:
Zhuojia Chen, Sun Yat-sen University Cancer Center (SYSUCC), ChinaJianwei Sun, Yunnan University, China
Zhizhou Shi, Kunming University of Science and Technology, China
Copyright © 2021 Lu, Wei, Zhang, Wang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wen Zhang, wenzhang2021@163.com
 Cuicui Lu
Cuicui Lu Duncan Wei2
Duncan Wei2 Wen Zhang
Wen Zhang