- Department of Respiratory Medicine, Xiangya Hospital of Central South University, Changsha, China
Introduction: Epidermal growth factor receptor (EGFR) 19del and L858R mutation are known as “common mutations” in non-small cell lung cancer (NSCLC) and predict sensitivities to EGFR tyrosine kinase inhibitors (TKIs), whereas 20ins and T790M mutations confer drug-resistance to EGFR-TKIs. The role of the remaining uncommon EGFR mutations remains elusive.
Methods: We retrospectively screened a group of NSCLC patients with uncommon EGFR mutations other than 20ins and T790M. The mutation patterns, use of different generations of EGFR-TKIs, and concurrent genetic alterations were analyzed. Meanwhile, a cohort of patients with single 19del or L858R were included for comparison.
Results: A total of 180/1,300 (13.8%) patients were identified. There were 102 patients with advanced or recurrent NSCLC that received first-line therapy of gefitinib/erlotinib/icotinib and afatinib and were eligible for analysis. The therapeutic outcomes among patients with common mutations (EGFRcm, n = 97), uncommon mutation plus common mutations (EGFRum+EGFRcm, n = 52), complex uncommon mutations (complex EGFRum, n = 22), and single uncommon mutations (single EGFRum, n = 28) were significantly different (ORRs: 76.3%, 61.5%, 54.5%, and 50.0%, respectively, p = 0.023; and mPFS: 13.3, 14.7, 8.1, and 6.0 months, respectively, p = 0.004). Afatinib showed superior efficacy over gefitinib/erlotinib/icotinib in EGFRcm (ORR: 81.0% vs. 75.0%, p = 0.773; mPFS: 19.1 vs. 12.0m, p = 0.036), EGFRum+EGFRcm (ORR: 100% vs. 54.5%, p = 0.017; mPFS: NE vs. 13.6m, p = 0.032), and single EGFRum (ORR: 78.6% vs. 21.4%, p = 0.007; mPFS: 10.1 vs. 3.0m, p = 0.025) groups. Comprehensive genomic profiling by Next Generation Sequencing encompassing multiple cancer-related genes was performed on 51/102 patients; the mPFS of patients without co-mutation (n = 16) and with co-mutations of tumor-suppressor genes (n = 31) and driver oncogenes (n = 4) were 31.1, 9.2, and 12.4 months, respectively (p = 0.046). TP53 mutation was the most common co-alteration and showed significantly shorter mPFS than TP53 wild-type patients (7.0 vs. 31.1m, p < 0.001). Multivariate analysis revealed that concurrent 19del/L858R and tumor-suppressor gene alterations independently predicted better and worse prognosis in patients with uncommon mutations, respectively.
Conclusions: Uncommon EGFR mutations constitute a highly heterogeneous subgroup of NSCLC that confer different sensitivities to EGFR-TKIs with regard to the mutation patterns. Afatinib may be a better choice for most uncommon EGFR mutations. Concurrent 19del/L858R and tumor-suppressor gene alterations, especially TP53, can be established as prognostic biomarkers.
Introduction
Epidermal growth factor receptor (EGFR) mutation is the most common oncogenic alteration in NSCLC, occurring in about 50% of Asian (1) and 10–15% of Caucasian patients (2). Exon 19 deletions and exon 21 L858R substitutions are known as “common mutations” as they account for approximately 85–90% of all EGFR mutations and predict responses to EGFR-TKIs (3). EGFR mutations other than 19del and L858R are known as “uncommon mutations”, which can occur alone or coexisted with other EGFR mutations (termed “complex mutations”) (4, 5). These uncommon mutations constitute a highly heterogeneous group with varied responses to EGFR-TKIs which have not been fully elucidated. The presence of drug-resistant mutations including exon 20 insertions (20ins) and T790M mutation usually showed poor responses to both first- and second-generation EGFR-TKIs (6, 7). As for uncommon EGFR mutations other than 20ins and T790M, G719X, L861Q, and S768I are frequently observed and showed sensitivities to EGFR-TKIs, with ORR and mPFS of 41.6% and 7.7 months, though not as favorable as common mutations (8). Nevertheless, evidence on the clinical responses of other uncommon EGFR mutations remains elusive.
Prospective data regarding the activity of EGFR-TKIs against uncommon EGFR mutations are limited because only a few randomized clinical trials of EGFR-TKIs involved patients with uncommon mutations (9–12). Researchers analyzed the clinical data from the NEJ002 study involving 10 participants harboring uncommon mutations, and the results showed that gefitinib was ineffective against G719X or L861Q mutation (13). In contrast, several retrospective studies observed moderate efficacy of first-generation EGFR-TKIs against uncommon mutations, with ORR ranging from 13.27% to 48.40%, and mPFS ranging from 5.0 to 7.7 months (8, 14, 15). Meanwhile, increasing evidence has suggested improved efficacy of second-generation EGFR-TKIs on patients with uncommon mutations. A combined post-hoc analysis based on data from serial LUX-Lung trials reported high efficacy of afatinib against certain types of uncommon EGFR mutations, especially G719X, L861Q, and S768I (6). Based on these findings, afatinib was approved by the US Food and Drug Administration (FDA) in 2018 for patients with advanced NSCLC harboring these mutations. The widespread access of the highly sensitive Next Generation Sequencing (NGS) technology in clinical practice and the implementation of liquid-based mutation detection assays can identify an expanded spectrum of uncommon EGFR mutations of which the optimal treatment regimen is warranted further investigation.
The improvement of molecular detection technologies could identify both targetable driver mutations and some other concurrent alterations that may potentially be served as predictive markers for the efficacy of EGFR-TKIs treatment. An increasing number of studies reported that co-occurring abnormalities such as mutations in TP53 and RB1, and amplification in MET and ERBB2 were associated with significantly shorter PFS in patients with common EGFR mutations when they received targeted therapy (16, 17). However, it remains unclear whether these co-mutations would affect the clinical outcomes of patients harboring uncommon EGFR mutations.
In the present study, we investigated the therapeutic outcomes of NSCLC patients harboring uncommon EGFR mutations other than 20ins and T790M who received first-line therapy of gefitinib/erlotinib/icotinib and afatinib. The study also evaluated a number of clinical variables as predictive or prognostic factors including mutation patterns, use of different generations of EGFR-TKIs, and additional concurrent genetic alterations.
Materials and Methods
Patients
From February 2016 to June 2020, 1,300 patients with histologically or cytologically confirmed NSCLC who had EGFR mutations were retrospectively screened. One hundred eighty patients with uncommon EGFR mutations other than exon 20 insertions and T790M mutation were identified. Of which, 102 patients with advanced or recurrent disease who received gefitinib/erlotinib/icotinib or afatinib as first-line therapy were eligible for survival analysis (Figure 1). Besides, a cohort consisting of 97 NSCLC patients with single common EGFR mutations who received first-line EGFR-TKIs therapy during the same period were enrolled for comparison. The study was approved by the Medical Ethics Committee of Xiangya Hospital, Central South University (IRB (S) No.201907700).
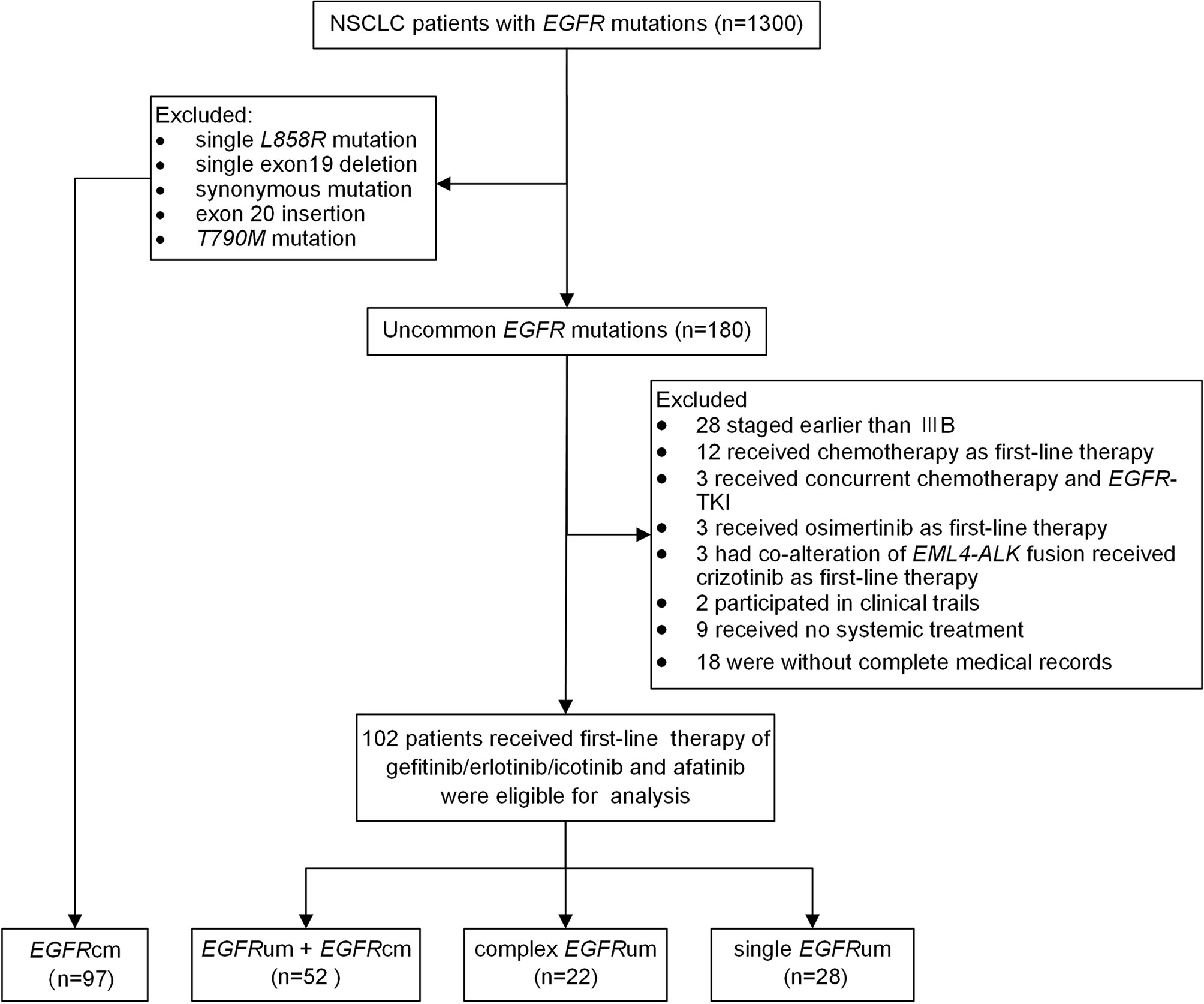
Figure 1 The flow chart for patient inclusion. NSCLC, non-small cell lung cancer; EGFR, epidermal growth factor receptor; TKI, tyrosine kinase inhibitor; EGFRcm, common EGFR mutations; EGFRum, uncommon EGFR mutations.
Clinical Data Collection and Efficacy Evaluation
Clinical data and therapeutic information were collected and analyzed, including age, sex, smoking status, Eastern Cooperative Oncology Group (ECOG) performance status, histologic type, EGFR mutation pattern, concurrent genetic alterations, EGFR-TKI use, and treatment outcomes. The disease stages were defined according to the eighth edition of the Lung Cancer Stage Classification System. EGFR-TKI treatment was initiated as per the physicians’ decision. Imaging examinations including chest Computed Tomography scanning (showing the liver and adrenal glands), brain Magnetic Resonance Imaging, and whole-body bone scan were performed every 8–12 weeks as a routine clinical procedure or as needed otherwise, to evaluate the treatment response and disease progression. The tumor response was assessed according to the Response Evaluation Criteria in Solid Tumours (RECIST; version 1.1) (18), including complete response (CR), partial response (PR), stable disease (SD), and progressive disease (PD). The objective response rate (ORR) was defined as the percentage of patients who achieved CR or PR. Progression-free survival (PFS) was measured as the period from the initiation of treatment to disease progression or death from any cause. Patients who had not experienced progression at the data cutoff date (January 15, 2021) or missing at the follow-up were censored.
EGFR Mutation Testing
EGFR mutation status was assessed by amplification refractory mutation system (ARMS) or next generation sequencing (NGS). All samples were obtained prior to EGFR-TKIs treatment. ARMS was performed using tissue specimens, according to the protocol of the ADx-ARMS kit (Amoy Diagnostics, Xiamen, China), which is designed to identify a total of 29 EGFR mutations occurring within exons 18–21. For NGS assay, tumor tissues or plasma cell-free DNA (cfDNA) were available for targeted sequencing of genomic alterations using commercial gene panels (Supplementary Tables 1, 2). The detailed procedures and conditions followed previously established protocols (19, 20).
For 102 uncommon EGFR-mutant patients included, 83 (81.4%) were detected by NGS. Of which, 67 were profiled with tumor tissue specimens and 16 with plasma samples. The remaining 19 (18.6%) patients were confirmed by ARMs assay. The corresponding detection methods and sample types for each patient with uncommon EGFR mutations were shown in Supplementary Table 3. For 97 common EGFR mutated cases, 89 (91.8%) were tested using NGS, and 8 (8.2%) were determined by ARMS.
Statistical Analysis
The chi-square test was used to compare qualitative data, and data with an expected frequency of <5 were analyzed using Fisher’s exact test. Mann–Whitney U test was applied to analyze continuous variables. Survival curves were plotted by the Kaplan–Meier method, and differences of median PFS among the subgroups were analyzed using the log-rank test. Univariate and multivariate Cox proportional hazards regression was performed to evaluate independent prognostic factors associated with PFS. Two-sided P values < 0.05 were considered statistically significant. All analyses were performed using SPSS 26 (IBM SPSS Statistics. Inc., Chicago, IL, USA).
Results
Patients’ Characteristics
A total of 1,300 patients with NSCLC and EGFR mutations were screened. There were 180 (13.8%) patients identified with uncommon EGFR mutations other than 20ins and T790M, including 79 (44%) patients with a coexisting common EGFR mutation (19del or L858R), 39 (22%) patients with complex uncommon EGFR mutations, and 62 (34%) patients with single uncommon EGFR mutations such as G719X (n = 9, 5%), L861Q (n = 16, 9%), S768I (n = 4, 2%), and other single uncommon mutations (n = 33, 18%) (Supplementary Figure 1). Noteworthily, 60/180 (33%) patients detected using the NGS method were found to have uncommon EGFR mutations occurring outside the most common 18–21 exons of the tyrosine kinase domain. Among them, 20 cases carried single mutation and 40 cases had complex mutations. Of the 40 complex mutations, 35 patients were combined with a common EGFR mutation.
Among the 180 patients with uncommon EGFR mutations other than 20ins and T790M, 102 patients with advanced or recurrent disease received first-line therapy of gefitinib/erlotinib/icotinib and afatinib and were eligible for analysis. Figure 1 shows the flow chart of patient inclusion. The median age at the initiation of EGFR-TKIs was 60 years old. The majority of the patients were female (55.9%), never smokers (66.7%), diagnosed with lung adenocarcinoma (92.2%), with ECOG performance status scored 0–1 (74.5%), and without brain metastases (70.4%). A total of 97 patients with single 19del (50/97, 51.5%) or L858R (47/97, 48.5%) were included as common EGFR mutations (EGFRcm) for comparison. A significant difference in the use of first-line EGFR-TKIs was observed between the common mutations group and uncommon mutations group. No significant differences were observed for other baseline characteristics (Supplementary Table 4).
According to mutation patterns, patients with uncommon EGFR mutations were further grouped as follows: uncommon mutation plus common mutations (EGFRum+EGFRcm, n = 52), complex uncommon mutations (complex EGFRum, n = 22), and single uncommon mutations (single EGFRum, n = 28). There were 67 (65.7%) patients who received the first-generation TKIs (gefitinib/erlotinib/icotinib) and 35 (34.3%) patients who received the second-generation TKI (afatinib). Significant different EGFR mutation patterns were observed between the two generations of TKI cohorts, as there were more EGFRum+EGFRcm patients receiving gefitinib/erlotinib/icotinib but more complex EGFRum patients receiving afatinib (Supplementary Table 5). In the EGFRcm group, all baseline characteristics were comparable between the two treatment cohorts (Supplementary Table 5).
Therapeutic Outcomes Among Patients With Different EGFR Mutation Patterns
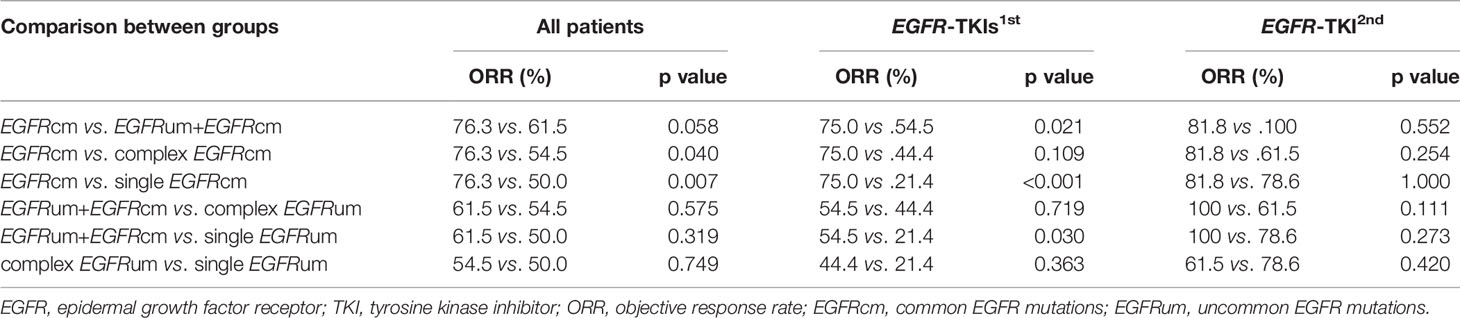
At the time of data cutoff (January 15, 2021), the median follow-up was 26.0 months. The ORRs in patient subgroups of EGFRcm, EGFRum+EGFRcm, complex EGFRum, and single EGFRum were 76.3%, 61.5%, 54.5%, and 50.0%, respectively (p = 0.023). There was no significant difference in ORRs between EGFRcm and EGFRum+EGFRcm groups. The EGFRcm group had a significantly higher ORR than complex EGFRum (76.3% vs. 54.5%, p = 0.040) and single EGFRum groups (76.3% vs. 50.0%, p = 0.007).
In the gefitinib/erlotinib/icotinib cohort, the therapeutic responses remained significantly distinct among the four mutation groups, with ORRs of 75.0% in EGFRcm, 54.5% in EGFRum+EGFRcm, 44.4% in complex EGFRum, and 21.4% in single EGFRum, respectively (p < 0.001). Further analysis showed that the ORR of EGFRcm group was significantly higher than that of EGFRum+EGFRcm (75.0% vs. 54.5%, p = 0.021) and single EGFRum (75.0% vs. 21.4%, p<0.001) groups. Meanwhile, the ORR of EGFRum+EGFRcm group was also significantly higher than that of single EGFRum group (54.5% vs. 21.4%, p = 0.030). In the afatinib cohort, no significant differences in ORRs were observed among these groups. The clinical responses to EGFR-TKIs in patients with different mutation patterns were summarized in Table 1.
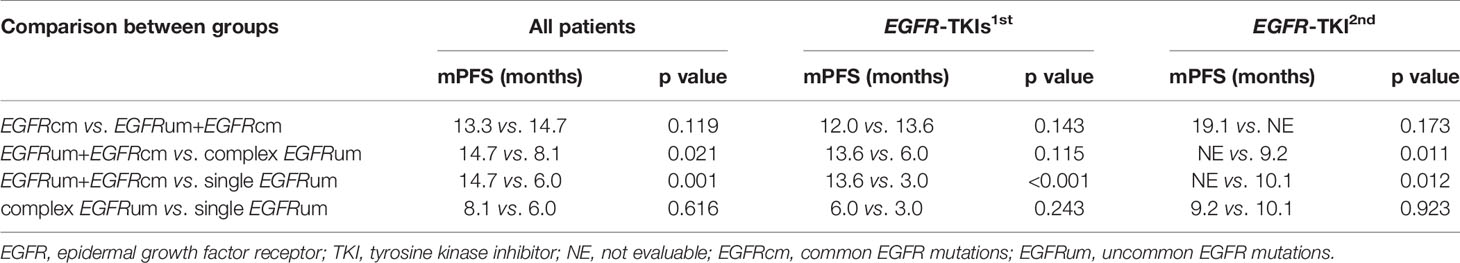
For survival analysis, the mPFS in the EGFRcm, EGFRum+EGFRcm, complex EGFRum, and single EGFRum groups were 13.3 (95% CI 11.1–15.4), 14.7 (95% CI 12.5–16.8), 8.1 (95% CI 4.1–12.0), and 6.0 (95% CI 4.2–7.8) months, respectively (P = 0.004; Figure 2A). Furthermore, no significant difference in mPFS was found between the EGFRcm and EGFRum+EGFRcm groups. The mPFS of patients in the EGFRum+EGFRcm group was significantly longer than those in the complex EGFRum (14.7 vs. 8.1m, HR 1.924, 95% CI 1.105–3.351, p = 0.021) and single EGFRum (14.7 vs. 6.0m, HR 2.335, 95% CI 1.402–3.888, p = 0.001) groups.
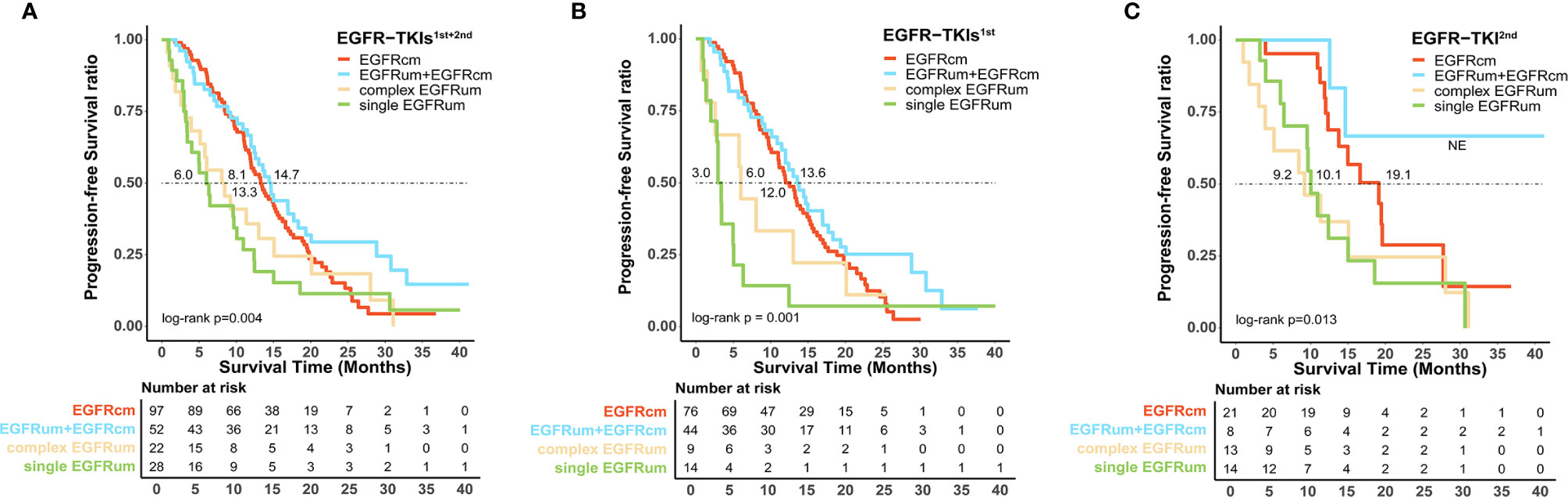
Figure 2 Patients harboring uncommon EGFR mutations with different mutation patterns exhibited diverse survival outcomes to the first-line therapy of EGFR-TKIs. Kaplan–Meier curves for progression-free survival in patients with different mutation patterns (A) in the first and second generations of EGFR-TKIs cohort; (B) in the first generation EGFR-TKIs cohort; and (C) in the second generation EGFR-TKI cohort. EGFRcm, common EGFR mutations; EGFRum, uncommon EGFR mutations.
We further performed a separate analysis in patients treated with different generations of EGFR-TKIs; the PFS curves of the four mutation groups in the two generations of TKI cohorts were shown in Figures 2B, C, respectively. In the gefitinib/erlotinib/icotinib cohort, the mPFS in EGFRcm and EGFRum+EGFRcm groups were comparable, while the mPFS of the EGFRum+EGFRcm group was significantly longer than that of the single EGFRum group (13.6 vs. 3.0m, HR 3.400, 95% CI 1.771–6.527, p < 0.001). In the afatinib cohort, although the mPFS in EGFRum+EGFRcm group was not yet reached, it remained statistically insignificant compared with the mPFS of EGFRcm group and prominently longer than those of complex EGFRum (NE vs. 9.2m, HR 6.397, 95% CI 1.411–28.995, p = 0.011) and single EGFRum (NE vs. 10.1m, HR 6.036, 95% CI 1.332–27.364, p = 0.012) groups. Table 2 demonstrates the comparison of mPFS in patients with different mutation patterns. In addition, the PFS time for each specific mutation pattern was displayed in Figure 3.
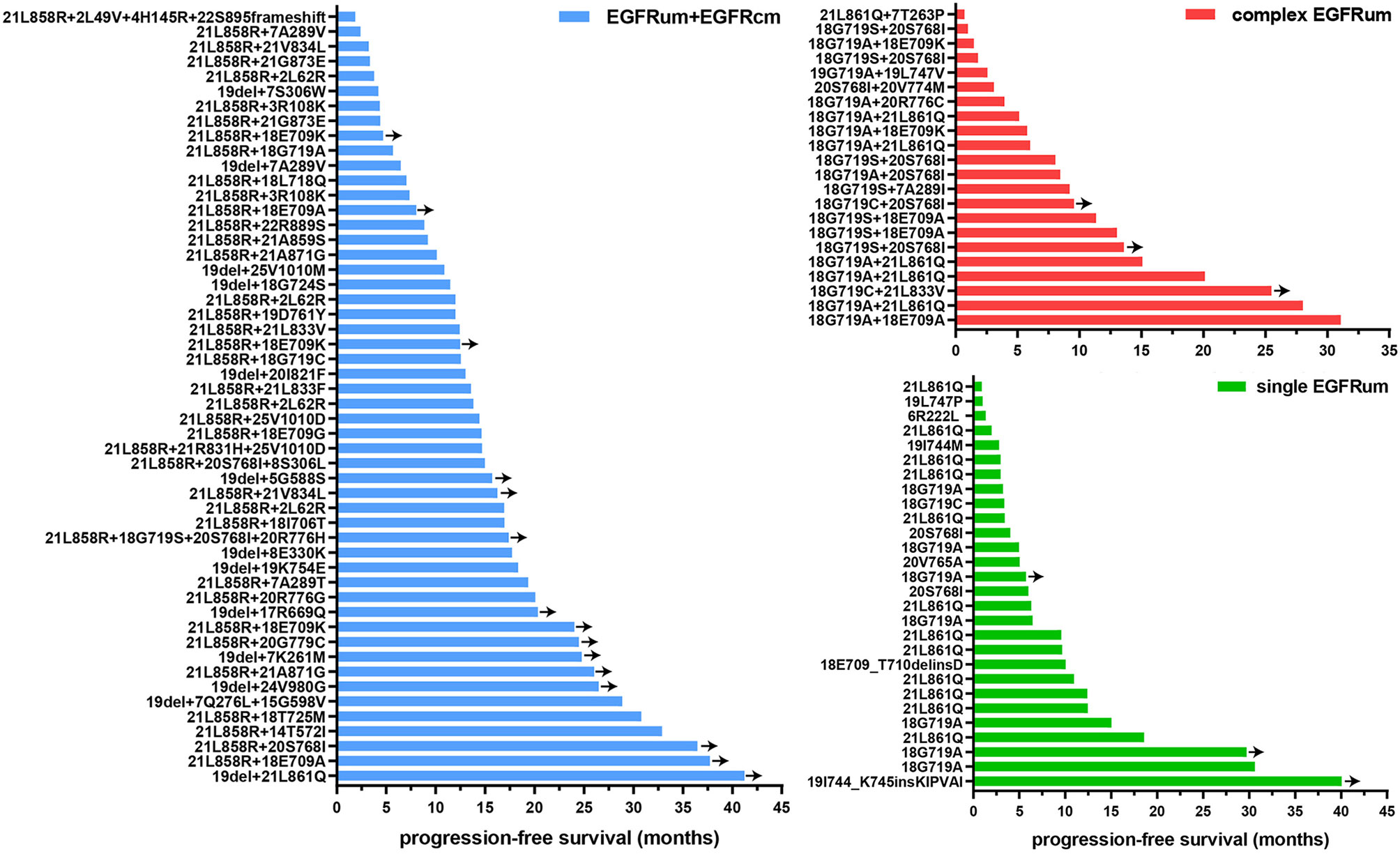
Figure 3 The progression-free survival (PFS) time for patients with specific mutation patterns received EGFR-TKIs as first-line therapy. Y-axis denotes mutation pattern, and x-axis indicates PFS. EGFRcm, common EGFR mutations; EGFRum, uncommon EGFR mutations.
There were 24/102 patients that had uncommon mutations occurring beyond the exon 18–21 tyrosine kinase domain. Of them, 21 individuals had a coexisting 19del/L858R and were classified into the EGFRum+EGFRcm group for analysis. Comparing the survival outcomes of the 21 patients to those who had single common mutations, the mPFS were 13.8 (95% CI 8.5–19.1) and 13.3 (95% CI 11.1–15.4) months, showing no statistically significant difference.
Clinical Efficacy Between First-Line Therapy of Gefitinib/Erlotinib/Icotinib and Afatinib
Comparing the first- and second-generation EGFR-TKIs, patients with common EGFR mutations receiving afatinib exhibited a comparable ORR (81.0% vs. 75.0%, p = 0.773) but significantly longer mPFS (19.1 vs. 12.0m, HR 0.533, 95% CI 0.293–0.971, p = 0.036, Figure 4A) than those receiving gefitinib/erlotinib/icotinib. For patients with uncommon EGFR mutations, the ORR of afatinib therapy was significantly higher than that of gefitinib/erlotinib/icotinib (77.1% vs. 46.3%, p = 0.003), but there was no significant difference in mPFS between the two treatment cohorts (12.4 vs. 10.9m, p = 0.333). In subgroup analysis of uncommon EGFR mutations, afatinib was associated with significantly favorable ORRs and mPFS than gefitinib/erlotinib/icotinib in patient subgroups of EGFRum+EGFRcm (100% vs. 54.5%, p = 0.017; NE vs. 13.6m, HR 0.235, 95% CI 0.056–0.989, p = 0.032, Figure 4B) and single EGFRum (78.6% vs. 21.4%, p = 0.007; 10.1 vs. 3.0m, HR 0.410, 95% CI 0.183–0.918, p = 0.025, Figure 4D). In the complex EGFRum group, no significant differences in ORRs (61.5% vs. 44.4%, p = 0.666) and mPFS were found between the two generations of TKIs (9.2 vs. 6.0m, p = 0.451, Figure 4C). The clinical efficacy between first-line therapy of gefitinib/erlotinib/icotinib and afatinib was shown in Table 3.
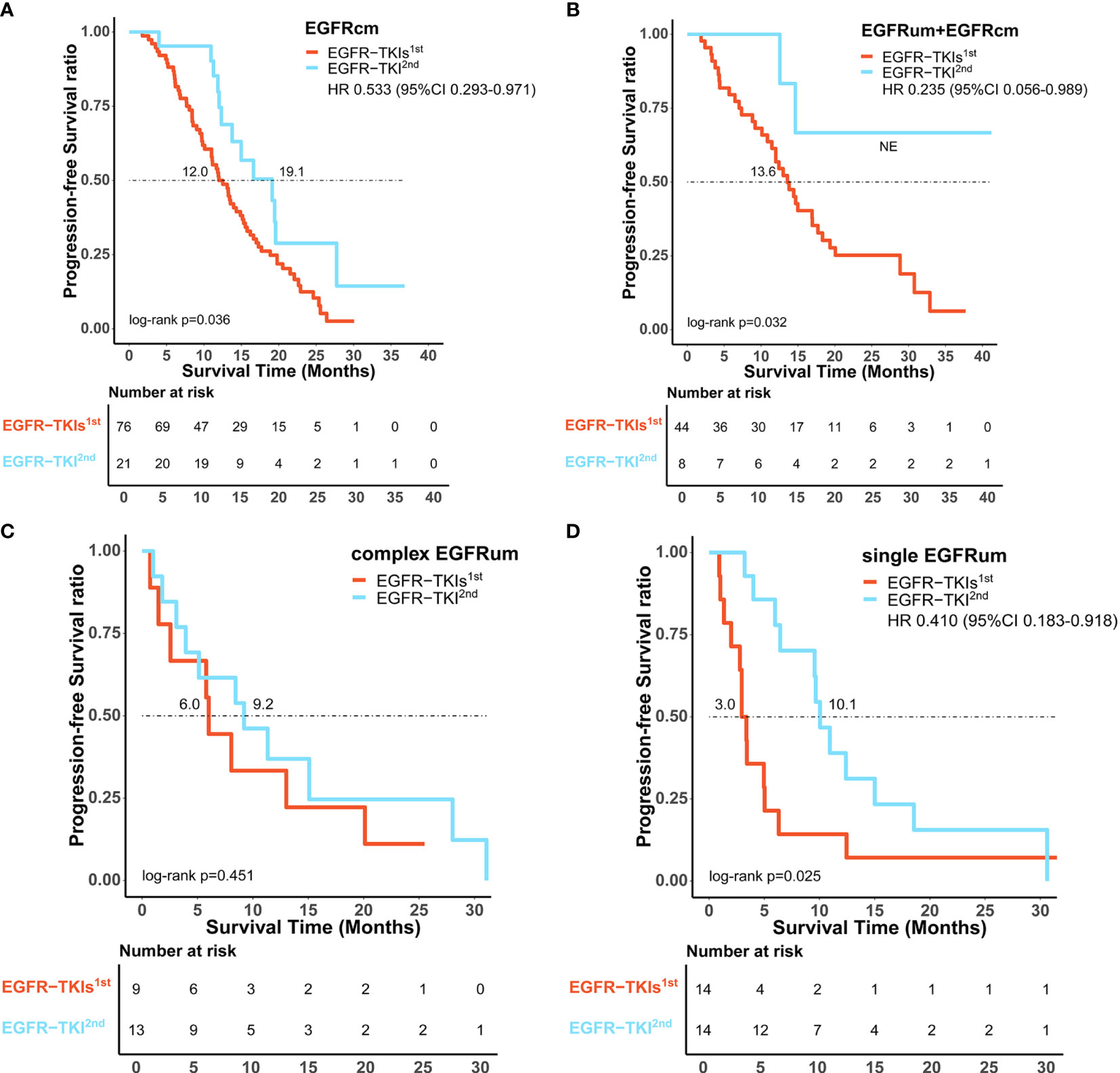
Figure 4 Comparison of survival outcomes between first-line therapy of gefitinib/erlotinib/icotinib (EGFR-TKIs1st) and afatinib (EGFR-TKI2nd). Afatinib showed significantly improved progression-free survival benefit over gefitinib/erlotinib/icotinib in patients subtypes of EGFRcm (A), EGFRum+EGFRcm (B), and single EGFRum (D), but comparable median progression-free survival in the complex EGFRum group (C). EGFRcm, common EGFR mutations; EGFRum, uncommon EGFR mutations; HR, hazard ratio; 95% CI, 95% confidence interval.
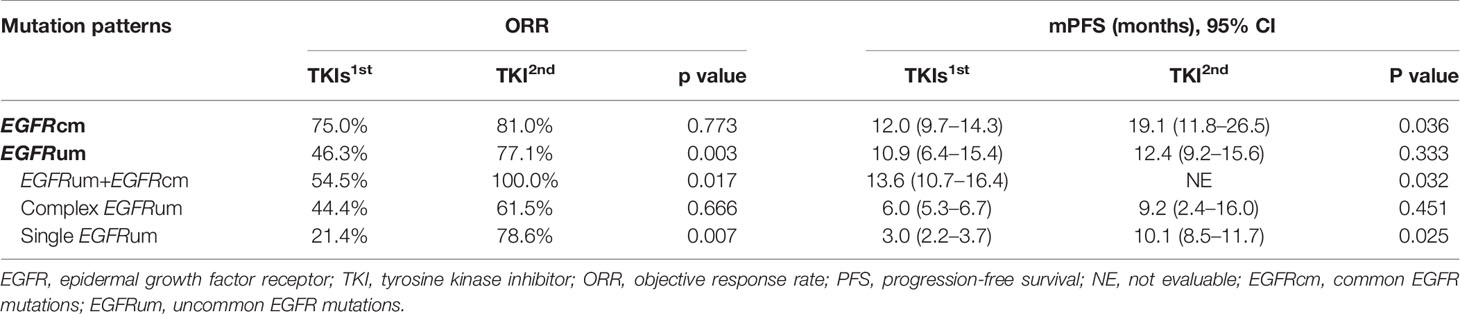
Table 3 Clinical efficacy between first-line therapy of gefitinib/erlotinib/icotinib and afatinib in patients with different EGFR mutation patterns.
The Predictive Value of Concurrent Genetic Alterations in Patients With Advanced NSCLC Harboring Uncommon EGFR Mutations
In addition to EGFR, information on concurrent genetic alterations was available in 51 patients with uncommon EGFR mutations who were identified by NGS using large sequencing gene panels. Genomic aberrations identified in these patients were indicated in Figure 5. There were 62.7% (32/51) of patients that had co-mutations of tumor-suppressor genes, including TP53 (30/51, 59%), RB1 (5/51, 10%), and PTEN (4/51, 8%). Co-mutations of driver oncogenes were found in 4/51 (8%) patients, including 1 patient with MET amplification, 2 with ERBB2 amplification, and 1 with KRAS amplification. ALK, ROS1, BRAF, and RET alterations were not found because of the limited sample size. According to identified co-mutations, the patients were divided into three subgroups including subgroup A (n = 16) without co-mutation, subgroup B (n = 31) with co-mutations of tumor-suppressor genes (TP53, RB1, PTEN), and subgroup C (n = 4) with co-mutations of driver oncogenes, irrespective of tumor-suppressor gene alterations (MET, ERBB2, KRAS). Corresponding mPFS in these subgroups were 31.1 months (95% CI 6.8–55.3), 9.2 months (95% CI 5.6–12.8), and 12.4 months (95% CI 0.0–28.1), respectively (p = 0.046; Figure 6A). The mPFS of subgroup B was significantly shorter than that of subgroup A (HR 2.657, 95% CI 1.187-5.949, p = 0.014), while no significant difference of mPFS was observed in subgroup C compared with subgroups A and B.
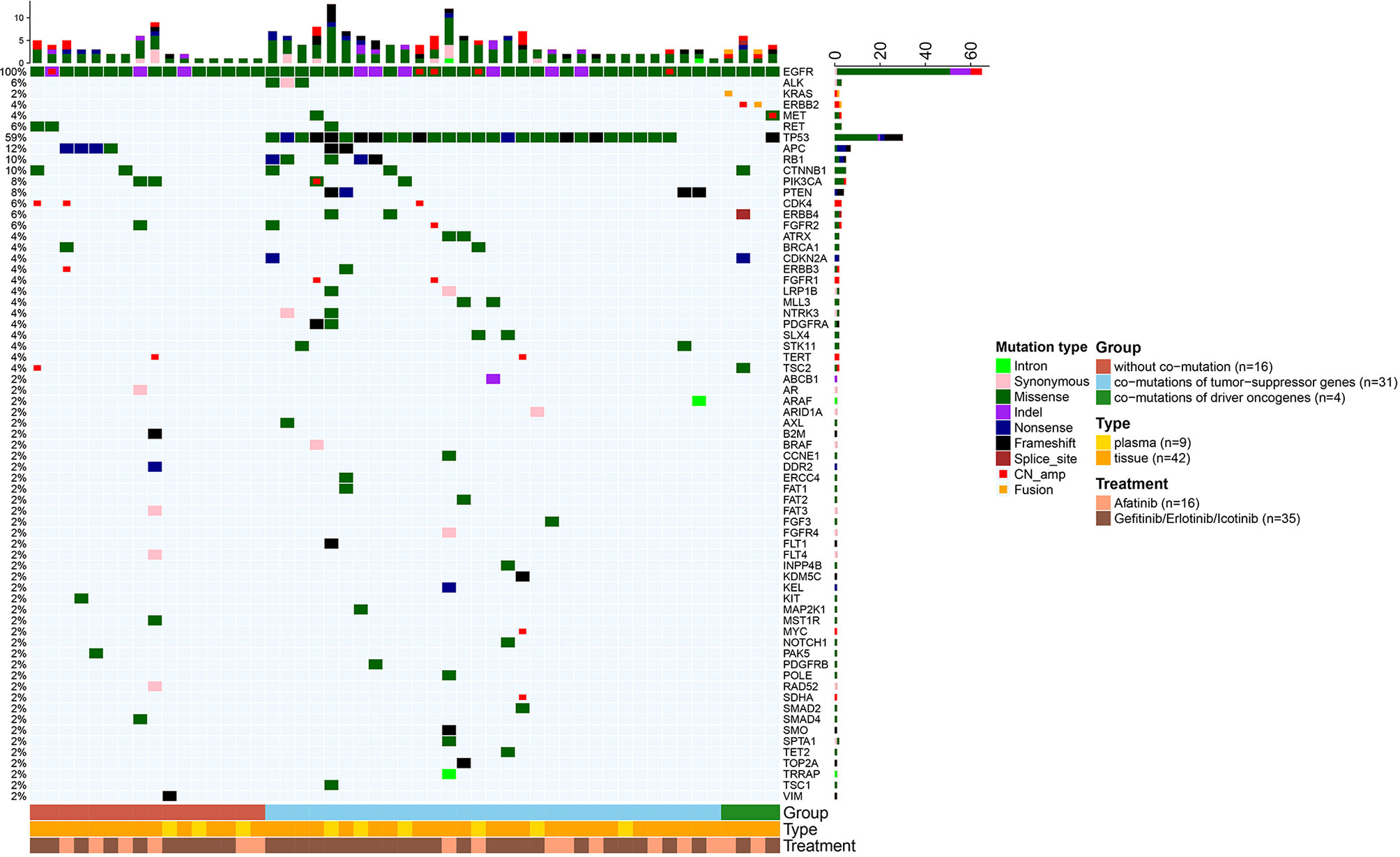
Figure 5 Heatmaps of genetic alterations of the 51 uncommon EGFR mutated patients who received NGS for testing multiple cancer-related genes. Patients were stratified into three subgroups according to the identified concurrent genetic alterations.
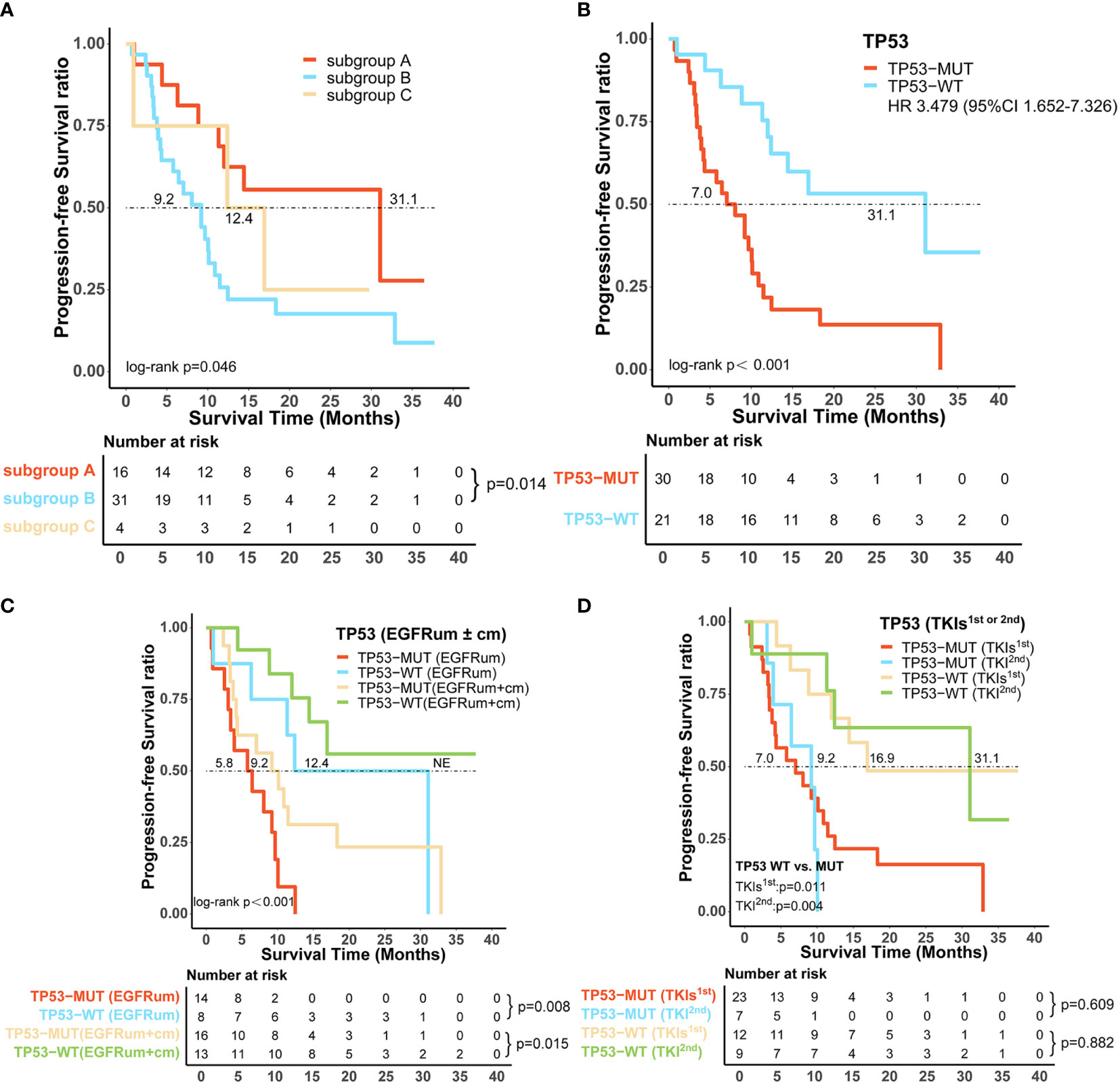
Figure 6 Certain co-alterations were associated with inferior survival outcomes in patients with uncommon EGFR mutations treated with EGFR-TKIs. (A) Progression-free survival (PFS) among patients without co-mutation (subgroup A), with co-mutations of tumor-suppressor genes (subgroup B) and driver oncogenes (subgroup C). (B) TP53-mutated (TP53-MUT) patients showed significantly shorter median PFS than TP53 wild-type (TP53-WT) patients. The predictive value of TP53 co-mutations in the survival outcomes of patients harboring uncommon EGFR mutations (C) with or without a combined common EGFR mutation, (D) receiving different generations of EGFR-TKIs. EGFRcm, common EGFR mutations; EGFRum, uncommon EGFR mutations; HR, hazard ratio; 95% CI, 95% confidence interval.
TP53 was the most frequently identified co-occurring genomic alteration; we further investigated the role of TP53 mutation in patients with uncommon EGFR mutations and found that TP53 mutated (TP53-MUT) patients had a significantly shorter mPFS than those of TP53 wild-type (TP53-WT) (mPFS:7.0 vs. 31.1m, HR 3.479, 95% CI 1.652–7.326, p < 0.001, Figure 6B). Subsequently, we stratified the 51 patients with uncommon EGFR mutations based on whether they had a combined common EGFR mutation; the results showed that the TP53-MUT group was associated with significantly worse mPFS regardless of the presence of common EGFR mutation (EGFRum+EGFRcm: 9.2m vs. NE, HR 3.378, 95% CI 1.194–9.556, p = 0.015; EGFRum only: 5.8 vs. 12.4m, HR 4.594, 95% CI 1.39–15.22, p = 0.008, Figure 6C). Likewise, patients with concomitant TP53 mutation demonstrated significantly shorter mPFS than those with TP53 wild-type both on gefitinib/erlotinib/icotinib (7.0 vs. 16.9m, HR 3.113, 95% CI 1.238–7.825, p = 0.011, Figure 6D) and afatinib therapy (9.2 vs. 31.1m, HR:13.685, 95% CI 1.532–122.215, p = 0.004, Figure 6D). Furthermore, for TP53 co-altered patients, no significant difference in mPFS was found between the two generations of EGFR-TKIs regimens (7.0 vs. 9.2m, p = 0.609, Figure 6D).
Prognostic Factors for the Clinical Outcomes of Patients Harboring Uncommon EGFR Mutations
The Cox regression model included variables such as age, sex, smoking status, histological types, Eastern Cooperative Oncology Group (ECOG) performance status, brain metastasis at the initiation of therapy, EGFR-TKI treatment, mutation patterns, and co-mutations. Univariate analysis was performed to screen variables for multivariate analysis. In 102 patients with uncommon EGFR mutations, multivariate analysis indicated that patients with a concomitant common EGFR mutation (19del/L858R) were associated with longer PFS (p = 0.003, HR 0.500, 95% CI: 0.315–0.792), while patients with non-adenocarcinoma and poor ECOG PS score were associated with shorter PFS (p = 0.004, HR 3.221, 95% CI 1.447–7.168; and p = 0.005, HR 2.111, 95% CI 1.258–3.541, respectively) (Table 4). In 51 patients who had uncommon EGFR mutations with comprehensive tumor genomic information, the multivariate analysis additionally identified that co-mutations of tumor-suppressor genes were also independently associated with poorer PFS (p = 0.001, HR 3.545, 95% CI 1.632–7.703) (Supplementary Table 6).

Table 4 Cox regression analysis for prognostic factors in the 102 advanced NSCLC patients harboring uncommon EGFR mutations who received EGFR-TKIs as first-line therapy.
Discussion
In this study, we enrolled a group of NSCLC patients with uncommon EGFR mutations and performed a comprehensive analysis focusing on the mutation patterns, use of different generations of EGFR-TKIs, and concurrent genetic alterations. Our results suggested that uncommon EGFR-mutant NSCLC can be further stratified into various mutation subgroups which exhibit distinct therapeutic responses and survival outcomes to EGFR-TKIs. The second-generation TKI afatinib showed improved therapeutic effects than the first-generation TKI on patient’s subtypes of common mutations, uncommon mutation plus common mutations, and single uncommon mutations. Co-occurring tumor-suppressor gene alterations, especially TP53, are associated with poor survival outcomes in patients with uncommon EGFR mutations treated with EGFR-TKIs. To the best of our knowledge, this is the first study to explore the predictive value of concurrent genetic alterations focusing on uncommon EGFR-mutant NSCLC populations.
Patients with uncommon EGFR mutations showed variable ORRs and PFS among different mutation patterns. In the study by Keam et al., patients with coexisting uncommon mutations and 19del/L858R showed comparable sensitivities to EGFR-TKIs with single common mutations (ORR of 68.8% and mPFS of 8.1 months) which were higher than patients harboring uncommon mutations without concomitant 19del/L858R (ORR of 25% and mPFS of 1.4 months) (21). Herein, we observed 44% of patients had uncommon mutations coexisting with 19del/L858R. In consistency with previous studies (5, 22), these patients showed similar therapeutic outcomes with patients harboring common mutations only and better outcomes than patients harboring single or complex uncommon mutations. In the complex uncommon mutations group, ORR was 54.5% and mPFS was 8.1 months, which was close to the results of Zhang et al. with an ORR of 71.0% and mPFS of 9.6 months (4). These findings suggested that complex uncommon mutations may also be effective targets for EGFR-TKI therapy. As for single uncommon mutations, earlier published data showed that patients of this subtype treated with first-generation TKIs had a significantly shorter mPFS than those of complex uncommon mutations (6.5 vs. 11.9 months, p = 0.010) (8). In the current study, however, no significant difference in mPFS was found between the two mutation subtypes, and larger datasets are required to validate this observation.
Owing to the advantage of the intact exon coverage of EGFR in NGS assays and the increase of sequencing depth, an increasing number of uncommon EGFR variants of unknown significance have been detected. In this study, 60 patients with uncommon EGFR mutations occurring beyond the exon 18–21 tyrosine kinase domain were identified. The biological function of these mutations and their sensitivities to EGFR-TKIs remain unclear while considering the large proportion; we did not exclude these patients. It should be noted, however, that most of these patients (21/24, 87.5%) met the inclusion criteria harboring a combined 19del/L858R and were classified into the EGFRum+EGFRcm group for analysis. The clinical outcomes of the 21 patients showed no statistically significant difference compared with those who had 19del/L858R alone. This indicates that the co-existence of uncommon EGFR mutations happening outside the exon 18–21 tyrosine kinase domain would not affect the sensitivities of common EGFR mutations to EGFR-TKIs.
Unlike the first-generation TKIs which reversibly bind to the ATP site of EGFR, afatinib is an oral pan-HER blocker that irreversibly binds to ErbB1 (EGFR), ErbB2 (HER2), and ErbB4, and inhibits signaling from all of these receptors (23). Therefore, the antitumor activity of afatinib should be more potent pharmacologically. The LUX-Lung 7 trial is the first prospective clinical trial to evaluate the clinical efficacy of afatinib and gefitinib as first-line therapy in patients with advanced NSCLC harboring common EGFR mutations. The results showed that afatinib demonstrated a statistically improved ORR and mPFS than gefitinib (ORR: 70.0% vs. 56.0%, p = 0.0083; mPFS:11.0 vs.10.9months, p = 0.017) (24). Consistently, we also observed superior efficacy in patients with common EGFR mutations using afatinib, with a significantly improved mPFS of 19.1 months. However, there have been no prospective studies compare these two generations of EGFR-TKIs in patients with uncommon mutations. Preclinical evidence suggests that afatinib has broad activity against uncommon EGFR mutations, with IC50 values much lower than those of first-generation EGFR-TKIs (25, 26). These in vitro preclinical data seem to be supported and reflected in the clinic. As shown by the results from a pooled analysis, afatinib demonstrated encouraging efficacy toward uncommon EGFR mutations, especially in patients whose tumors harbored major uncommon mutations (G719X, L861Q, and S768I, with or without any other mutation except T790M or an exon 20 insertion) and complex mutations, with ORR of 60.0% and 77.1%, respectively, and median time to treatment failure (TTF) of 10.8 months and 14.7 months, respectively (27). Shen et al. compared the effects of the two-generations of EGFR-TKIs and found that afatinib was more effective than gefitinib/erlotinib in the treatment of patients harboring uncommon EGFR mutations (mPFS:11.0 vs.3.6 months, p = 0.030), particularly for those lacking a combination of 19del or L858R (mPFS:18.3 vs. 2.8 months, p = 0.070) (28). In a recently published retrospective study, afatinib as first-line therapy was reported to have a significantly greater therapeutic response than gefitinib or erlotinib (ORR:60.6% vs. 35.8%, p = 0.036) and a trend towards longer mPFS but not archived threshold of statistical significance (mPFS:8.8 vs. 12.0 months, p = 0.163) (29). Likewise, in the present study, for the whole group of uncommon EGFR mutations, afatinib achieved a significantly higher ORR but a comparable mPFS compared to gefitinib/erlotinib/icotinib. In subgroup analysis, afatinib was associated with significantly higher ORRs and longer mPFS in patient subgroups of uncommon mutation plus common mutations and single uncommon mutations. For complex uncommon mutations, these two treatment cohorts were similar. These observations indicated that afatinib could be a better choice in patients harboring common mutations and most uncommon mutations.
Previous reports suggested that concurrent genetic alterations within EGFR genomic aberrations play a role in the molecular resistance mechanisms to EGFR-TKI therapy, which might explain the shorter PFS in some patients (16, 17). The BENEFIT trial, for example, observed that patients with only common EGFR mutations had significantly longer mPFS than those possessing concurrent tumor-suppressor genes or other driver oncogene alterations (30). In the current study, we investigated the co-occurring genomic alterations in uncommon EGFR-mutant populations and explored their potential impact on the therapeutic outcomes of EGFR-TKIs. Similarly, our data suggested that patients carrying uncommon EGFR mutations with concurrent tumor-suppressor genes aberrations are associated with inferior outcomes on EGFR-TKIs therapy. As for patients harboring additional driver oncogene alterations, the mPFS was 12.39 months, which was comparable to the other two groups. However, this observation is not conclusive, due to the small number of cases. Further investigations with larger sample size are warranted to confirm our results.
We found that TP53 mutation was the most frequently identified co-occurring genomic alteration in uncommon EGFR-mutant NSCLC, with an incidence rate of 59% (30/51), which is consistent with the reported 40–60% incidence of TP53 co-mutations in EGFR mutation-positive patients in previous studies (17, 31, 32). The promoting effect of TP53 mutations on early tumor progression of various malignancies, including NSCLC, has been confirmed by multiple reports (33, 34). In the present study, we noted that patients with advanced NSCLC carrying uncommon EGFR mutations with co-mutations of TP53 were associated with a markedly shorter time to disease progression on initial EGFR-TKIs therapy. Further analysis showed that afatinib did not provide a survival benefit in patients with co-altered TP53 compared with gefitinib/erlotinib/icotinib. However, the potential reasons that underlie the negative prognostic value of TP53 mutations have not been well elucidated. It has been reported that the somatic mutations of TP53 are related to the inactivation of P53 protein, which causes impaired tumor suppressor functions in anti-proliferation and apoptosis regulation, and is also associated with genomic instability and defects in DNA damage repair (35, 36). In addition, emerging evidence suggests that TP53-mutant lung cancers exhibited remarkably increased somatic mutation burden and higher expression of immune checkpoints such as PD-L1 (37). These findings indicate that TP53 mutations may be somehow involved in the tumor adaptive immune escape, which might contribute to a favorable response toward immune checkpoint inhibitors but potential resistance to non-immunotherapy including molecular targeted therapy. Identification of additional co-mutations with predictive values using comprehensive tumor genomic profiling may help to tailor personalized therapeutic strategies to overcome primary resistance. However, there are currently no approved agents that specifically target TP53 in NSCLC. Clinical trials assessing the efficacy of combination therapy are under investigation (38, 39), which may be a promising option for the treatment of EGFR-mutant NSCLC with co-alterations.
After adjustment for potential confounding factors, our results of multivariate analysis showed that concomitant common EGFR mutation is a predictor for better therapeutic outcomes in patients with uncommon EGFR mutations, which is consistent with the result from a prior study (29). Meanwhile, the presence of concurrent tumor-suppressor gene alterations is an independent risk factor for poor PFS. These observations further illustrated the predictive value of mutation patterns and certain concurrent alterations for patients with uncommon mutations.
Limitations
There are some limitations in our study. Firstly, the single-center and retrospective design would involve potential biases. Secondly, although the majority of patients included in this study were tested using NGS assay, some uncommon EGFR-mutant variants beyond the 29 identifiable EGFR mutations may be missed for patients analyzed by ARMS. Thirdly, due to the limited number of cases in which multigene sequencing was performed, we failed to analyze more genetic co-alterations other than TP53 mutations individually. Finally, we were unable to assess the overall survival time due to incomplete follow-up data after referral and the fact that a certain number of patients had not yet reached the endpoint events at the time of the data cutoff.
Conclusion
The clinical outcomes of uncommon EGFR mutations are closely related to the mutation patterns, use of different generations of EGFR-TKIs, and concurrent genetic alterations. Patients carrying uncommon EGFR mutations coupled with 19del/L858R are correlated with better efficacy than other mutation patterns. Afatinib provides improved therapeutic outcomes for most uncommon EGFR mutations and therefore could be a recommended option for these patients. The co-mutations of tumor-suppressor genes especially TP53, can serve as a predictive factor for poor prognosis.
Data Availability Statement
The data presented in the study are deposited in the CNGB Sequence Archive (CNSA) of China National GeneBank DataBase (CNGBdb) repository (https://db.cngb.org/cnsa/), accession number CNP0001988.
Ethics Statement
The studies involving human participants were reviewed and approved by The Medical Ethics Committee of Xiangya Hospital, Central South University (IRB (S) No.201907700). Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author Contributions
JT participated in the study design, data collection and analysis, and drafted the manuscript. CH contributed to the study design, data acquisition, and manuscript review. PD contributed to study design, data interpretation, overall review, and funding acquisition. RW participated in data curation and visualization. LC, ML, HY, QG, JA, and JJ contributed to data acquisition. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (81502699,81600025); the National Key R&D Program of China (2016YFC1303300); the National Multidisciplinary Cooperative Diagnosis and Treatment Capacity Building Project for Major Diseases (Lung Cancer); and Xiangya clinical big data project of Central South University (Clinical big data project of lung cancer).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.646577/full#supplementary-material
References
1. Shi Y, Au JS, Thongprasert S, Srinivasan S, Tsai CM, Khoa MT, et al. A Prospective, Molecular Epidemiology Study of EGFR Mutations in Asian Patients With Advanced Non-Small-Cell Lung Cancer of Adenocarcinoma Histology (PIONEER). J Thorac Oncol (2014) 9(2):154–62. doi: 10.1097/jto.0000000000000033
2. Douillard J-Y, Ostoros G, Cobo M, Ciuleanu T, Cole R, McWalter G, et al. Gefitinib Treatment in EGFR Mutated Caucasian NSCLC Circulating-Free Tumor DNA as a Surrogate for Determination of EGFR Status. J Thorac Oncol (2014) 9(9):1345–53. doi: 10.1097/jto.0000000000000263
3. Shigematsu H, Gazdar AF. Somatic Mutations of Epidermal Growth Factor Receptor Signaling Pathway in Lung Cancers. Int J Cancer (2006) 118(2):257–62. doi: 10.1002/ijc.21496
4. Zhang B, Wang S, Qian J, Yang W, Qian F, Lu J, et al. Complex Epidermal Growth Factor Receptor Mutations and Their Responses to Tyrosine Kinase Inhibitors in Previously Untreated Advanced Lung Adenocarcinomas. Cancer (2018) 124(11):2399–406. doi: 10.1002/cncr.31329
5. Kobayashi S, Canepa HM, Bailey AS, Nakayama S, Yamaguchi N, Goldstein MA, et al. Compound EGFR Mutations and Response to EGFR Tyrosine Kinase Inhibitors. J Thorac Oncol (2013) 8(1):45–51. doi: 10.1097/JTO.0b013e3182781e35
6. Yang JCH, Sequist LV, Geater SL, Tsai C-M, Mok TSK, Schuler M, et al. Clinical Activity of Afatinib in Patients With Advanced Nn-Small-Cell Lung Cancer Harbouring Uncommon EGFR Mutations: A Combined Post-Hoc Analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung 6. Lancet Oncol (2015) 16(7):830–38. doi: 10.1016/s1470-2045(15)00026-1
7. Yasuda H, Park E, Yun CH, Sng NJ, Lucena-Araujo AR, Yeo WL, et al. Structural, Biochemical, and Clinical Characterization of Epidermal Growth Factor Receptor (EGFR) Exon 20 Insertion Mutations in Lung Cancer. Sci Transl Med (2013) 5(216):216ra177. doi: 10.1126/scitranslmed.3007205
8. Chiu CH, Yang CT, Shih JY, Huang MS, Su WC, Lai RS, et al. Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitor Treatment Response in Advanced Lung Adenocarcinomas With G719X/L861Q/S768I Mutations. J Thorac Oncol (2015) 10(5):793–9. doi: 10.1097/jto.0000000000000504
9. Wu YL, Zhou C, Hu CP, Feng J, Lu S, Huang Y, et al. Afatinib Versus Cisplatin Plus Gemcitabine for First-Line Treatment of Asian Patients With Advanced Non-Small-Cell Lung Cancer Harbouring EGFR Mutations (LUX-Lung 6): An Open-Label, Randomised Phase 3 Trial. Lancet Oncol (2014) 15(2):213–22. doi: 10.1016/s1470-2045(13)70604-1
10. Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, et al. Gefitinib or Chemotherapy for Non-Small-Cell Lung Cancer With Mutated EGFR. N Engl J Med (2010) 362(25):2380–88. doi: 10.1056/NEJMoa0909530
11. Sequist LV, Yang JC, Yamamoto N, O’Byrne K, Hirsh V, Mok T, et al. Phase III Study of Afatinib or Cisplatin Plus Pemetrexed in Patients With Metastatic Lung Adenocarcinoma With EGFR Mutations. J Clin Oncol (2013) 31(27):3327–34. doi: 10.1200/jco.2012.44.2806
12. Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, et al. Gefitinib or Carboplatin-Paclitaxel in Pulmonary Adenocarcinoma. N Engl J Med (2009) 361(10):947–57. doi: 10.1056/NEJMoa0810699
13. Watanabe S, Minegishi Y, Yoshizawa H, Maemondo M, Inoue A, Sugawara S, et al. Effectiveness of Gefitinib Against Non-Small-Cell Lung Cancer With the Uncommon EGFR Mutations G719X and L861Q. J Thorac Oncol (2014) 9(2):189–94. doi: 10.1097/jto.0000000000000048
14. Lei L, Wang W-X, Zhu Y-C, Li J-L, Fang Y, Wang H, et al. Real-World Efficacy and Potential Mechanism of Resistance of Icotinib in Asian Advanced Non-Small Cell Lung Cancer With EGFR Uncommon Mutations: A Multi-Center Study. Cancer Med (2020) 9(1):12–8. doi: 10.1002/cam4.2652
15. Wu J-Y, Yu C-J, Chang Y-C, Yang C-H, Shih J-Y, Yang P-C. Effectiveness of Tyrosine Kinase Inhibitors on "Uncommon" Epidermal Growth Factor Receptor Mutations of Unknown Clinical Significance in Non-Small Cell Lung Cancer. Clin Cancer Res (2011) 17(11):3812–21. doi: 10.1158/1078-0432.Ccr-10-3408
16. Kim Y, Lee B, Shim JH, Lee S-H, Park W-Y, Choi Y-L, et al. Concurrent Genetic Alterations Predict the Progression to Target Therapy in EGFR-Mutated Advanced NSCLC. J Thorac Oncol (2019) 14(2):193–202. doi: 10.1016/j.jtho.2018.10.150
17. Yu HA, Suzawa K, Jordan E, Zehir A, Ni A, Kim R, et al. Concurrent Alterations in EGFR-Mutant Lung Cancers Associated With Resistance to EGFR Kinase Inhibitors and Characterization of MTOR as a Mediator of Resistance. Clin Cancer Res (2018) 24(13):3108–18. doi: 10.1158/1078-0432.Ccr-17-2961
18. Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New Response Evaluation Criteria in Solid Tumours: Revised RECIST Guideline (Version 1.1). Eur J Cancer (2009) 45(2):228–47. doi: 10.1016/j.ejca.2008.10.026
19. Zhai J, Han S, Guo Q, Shan B, Wang J, Guo Y, et al. Identifying Genomic Alterations in Small Cell Lung Cancer Using the Liquid Biopsy of Bronchial Washing Fluid. Front Oncol (2021) 11:647216. doi: 10.3389/fonc.2021.647216
20. Mao X, Zhang Z, Zheng X, Xie F, Duan F, Jiang L, et al. Capture-Based Targeted Ultradeep Sequencing in Paired Tissue and Plasma Samples Demonstrates Differential Subclonal ctDNA-Releasing Capability in Advanced Lung Cancer. J Thorac Oncol (2017) 12(4):663–72. doi: 10.1016/j.jtho.2016.11.2235
21. Keam B, Kim D-W, Park JH, Lee J-O, Kim TM, Lee S-H, et al. Rare and Complex Mutations of Epidermal Growth Factor Receptor, and Efficacy of Tyrosine Kinase Inhibitor in Patients With Non-Small Cell Lung Cancer. Int J Clin Oncol (2014) 19(4):594–600. doi: 10.1007/s10147-013-0602-1
22. Tu HY, Ke EE, Yang JJ, Sun YL, Yan HH, Zheng MY, et al. A Comprehensive Review of Uncommon EGFR Mutations in Patients With Non-Small Cell Lung Cancer. Lung Cancer (2017) 114:96–102. doi: 10.1016/j.lungcan.2017.11.005
23. Solca F, Dahl G, Zoephel A, Bader G, Sanderson M, Klein C, et al. Target Binding Properties and Cellular Activity of Afatinib (BIBW 2992), an Irreversible ErbB Family Blocker. J Pharmacol Exp Ther (2012) 343(2):342–50. doi: 10.1124/jpet.112.197756
24. Park K, Tan EH, O’Byrne K, Zhang L, Boyer M, Mok T, et al. Afatinib Versus Gefitinib as First-Line Treatment of Patients With EGFR Mutation-Positive Non-Small-Cell Lung Cancer (LUX-Lung 7): A Phase 2B, Open-Label, Randomised Controlled Trial. Lancet Oncol (2016) 17(5):577–89. doi: 10.1016/s1470-2045(16)30033-x
25. Banno E, Togashi Y, Nakamura Y, Chiba M, Kobayashi Y, Hayashi H, et al. Sensitivities to Various Epidermal Growth Factor Receptor-Tyrosine Kinase Inhibitors of Uncommon Epidermal Growth Factor Receptor Mutations L861Q and S768I: What Is the Optimal Epidermal Growth Factor Receptor-Tyrosine Kinase Inhibitor? Cancer Sci (2016) 107(8):1134–40. doi: 10.1111/cas.12980
26. Kohsaka S, Nagano M, Ueno T, Suehara Y, Hayashi T, Shimada N, et al. A Method of High-Throughput Functional Evaluation of EGFR Gene Variants of Unknown Significance in Cancer. Sci Transl Med (2017) 9(416):eaan6566. doi: 10.1126/scitranslmed.aan6566
27. Yang JC, Schuler M, Popat S, Miura S, Heeke S, Park K, et al. Afatinib for the Treatment of NSCLC Harboring Uncommon EGFR Mutations: A Database of 693 Cases. J Thorac Oncol (2020) 15(5):803–15. doi: 10.1016/j.jtho.2019.12.126
28. Shen YC, Tseng GC, Tu CY, Chen WC, Liao WC, Chen WC, et al. Comparing the Effects of Afatinib With Gefitinib or Erlotinib in Patients With Advanced-Stage Lung Adenocarcinoma Harboring Non-Classical Epidermal Growth Factor Receptor Mutations. Lung Cancer (2017) 110:56–62. doi: 10.1016/j.lungcan.2017.06.007
29. Chang LC, Lim CK, Chang LY, Chen KY, Shih JY, Yu CJ. Non-Small Cell Lung Cancer Harbouring Non-Resistant Uncommon EGFR Mutations: Mutation Patterns, Effectiveness of Epidermal Growth Factor Receptor-Tyrosine Kinase Inhibitors and Prognostic Factors. Eur J Cancer (2019) 119:77–86. doi: 10.1016/j.ejca.2019.06.025
30. Wang Z, Cheng Y, An T, Gao H, Wang K, Zhou Q, et al. Detection of EGFR Mutations in Plasma Circulating Tumour DNA as a Selection Criterion for First-Line Gefitinib Treatment in Patients With Advanced Lung Adenocarcinoma (BENEFIT): A Phase 2, Single-Arm, Multicentre Clinical Trial. Lancet Respir Med (2018) 6(9):681–90. doi: 10.1016/s2213-2600(18)30264-9
31. Lim SM, Kim HR, Cho EK, Min YJ, Ahn JS, Ahn MJ, et al. Targeted Sequencing Identifies Genetic Alterations That Confer Primary Resistance to EGFR Tyrosine Kinase Inhibitor (Korean Lung Cancer Consortium). Oncotarget (2016) 7(24):36311–20. doi: 10.18632/oncotarget.8904
32. Labbé C, Cabanero M, Korpanty GJ, Tomasini P, Doherty MK, Mascaux C, et al. Prognostic and Predictive Effects of TP53 Co-Mutation in Patients With EGFR-Mutated Non-Small Cell Lung Cancer (NSCLC). Lung Cancer (2017) 111:23–9. doi: 10.1016/j.lungcan.2017.06.014
33. Aisner DL, Sholl LM, Berry LD, Rossi MR, Chen H, Fujimoto J, et al. The Impact of Smoking and TP53 Mutations in Lung Adenocarcinoma Patients With Targetable Mutations-The Lung Cancer Mutation Consortium (LCMC2). Clin Cancer Res (2018) 24(5):1038–47. doi: 10.1158/1078-0432.Ccr-17-2289
34. Petitjean A, Achatz MI, Borresen-Dale AL, Hainaut P, Olivier M. TP53 Mutations in Human Cancers: Functional Selection and Impact on Cancer Prognosis and Outcomes. Oncogene (2007) 26(15):2157–65. doi: 10.1038/sj.onc.1210302
35. Brosh R, Rotter V. When Mutants Gain New Powers: News From the Mutant P53 Field. Nat Rev Cancer (2009) 9(10):701–13. doi: 10.1038/nrc2693
36. Nahar R, Zhai W, Zhang T, Takano A, Khng AJ, Lee YY, et al. Elucidating the Genomic Architecture of Asian EGFR-Mutant Lung Adenocarcinoma Through Multi-Region Exome Sequencing. Nat Commun (2018) 9(1):216. doi: 10.1038/s41467-017-02584-z
37. Dong ZY, Zhong WZ, Zhang XC, Su J, Xie Z, Liu SY, et al. Potential Predictive Value of TP53 and KRAS Mutation Status for Response to PD-1 Blockade Immunotherapy in Lung Adenocarcinoma. Clin Cancer Res (2017) 23(12):3012–24. doi: 10.1158/1078-0432.Ccr-16-2554
38. Zhang Z, Luo F, Zhang Y, Ma Y, Hong S, Yang Y, et al. The ACTIVE Study Protocol: Apatinib or Placebo Plus Gefitinib as First-Line Treatment for Patients With EGFR-Mutant Advanced Non-Small Cell Lung Cancer (CTONG1706). Cancer Commun (2019) 39(1):69. doi: 10.1186/s40880-019-0414-4
39. Nakagawa K, Garon EB, Seto T, Nishio M, Ponce Aix S, Paz-Ares L, et al. Ramucirumab Plus Erlotinib in Patients With Untreated, EGFR-Mutated, Advanced Non-Small-Cell Lung Cancer (RELAY): A Randomised, Double-Blind, Placebo-Controlled, Phase 3 Trial. Lancet Oncol (2019) 20(12):1655–69. doi: 10.1016/s1470-2045(19)30634-5
Keywords: concurrent genetic alterations, tyrosine kinase inhibitors, uncommon EGFR mutations, EGFR, non-small cell lung cancer
Citation: Tan J, Hu C, Deng P, Wan R, Cao L, Li M, Yang H, Gu Q, An J and Jiang J (2021) The Predictive Values of Advanced Non-Small Cell Lung Cancer Patients Harboring Uncommon EGFR Mutations—The Mutation Patterns, Use of Different Generations of EGFR-TKIs, and Concurrent Genetic Alterations. Front. Oncol. 11:646577. doi: 10.3389/fonc.2021.646577
Received: 27 December 2020; Accepted: 29 July 2021;
Published: 26 August 2021.
Edited by:
Francois X. Claret, University of Texas MD Anderson Cancer Center, United StatesReviewed by:
Lin Liu, Nankai University, ChinaHui Guo, First Affiliated Hospital of Xi’an Jiaotong University, China
Tony Y. Hu, Tulane University, United States
Copyright © 2021 Tan, Hu, Deng, Wan, Cao, Li, Yang, Gu, An and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pengbo Deng, yogurt1015@163.com
†These authors have contributed equally to this work
 Jiarong Tan
Jiarong Tan Chengping Hu†
Chengping Hu† Rongjun Wan
Rongjun Wan Jian An
Jian An