- 1Department of Oncology, Taizhou People’s Hospital, Taizhou, China
- 2Graduate school, Dalian Medical University, Dalian, China
- 3Department of Neurosurgery, Taizhou People’s Hospital, Taizhou, China
- 4Department of Medical Oncology, Cancer Center, State Key Laboratory of Biotherapy, West China Hospital, Sichuan University, Chengdu, China
- 5Department of Radiation Oncology, The First Affiliated Hospital of Nanjing Medical University, Nanjing, China
N6-methyladenosine (m6A) is the most abundant internal modification in eukaryotic messenger RNAs (mRNAs). m6A RNA methylation is involved in all stages of RNA life cycle, from RNA processing, nuclear output, translation regulation to RNA degradation, indicating that m6A has various functions affecting RNA metabolism positively or negatively. Reading proteins are vital in regulating the translation and stability of m6A mRNAs positively or negatively. Recent studies have enhanced the understanding of the molecular mechanism of the YT521-B homology (YTH) domain family and the modification of m6A. This study aimed to review the specific mechanisms, functions, and interactions of the YTH domain protein family. It also discussed future research directions, thus providing new ideas for the clinical diagnosis and targeted therapy of cancer.
Introduction
As early as in the 1970s, researches were conducted on N6-methyladenosine (m6A) RNA (1). The m6A RNA modification is the most common internal modification of mammalian mRNAs, which occurs on the sixth N atom of base A (2). However, substantial evidence was lacking due to the immaturity of the technology and the instability of methylation modification at that time. Exploration of the genome-wide distribution of m6A remained unclear until 2011 when the first RNA demethylase named the fat mass- and obesity-associated protein (FTO) was identified (3), and the m6A RNA modification was found to be a dynamic and reversible process. Current knowledge of m6A has been established. The dynamic modification of m6A RNA methylation is completed by methyltransferase (“writers”) and demethyltransferase (“erasers”). Reading proteins (“readers”) recognize the m6A-modified mRNA and regulate the expression of downstream genes.
Surprisingly, an increasing number of studies have documented that m6A modification is crucial in regulating tumor function, and the dynamic imbalance of m6A modification can lead to the occurrence and development of tumor. Studies at the cellular level have shown that the dynamic modification of mRNAs is important in regulating mRNA splicing, output, stability, translation, and hence gene expression (4, 5). By extension, studies at the tissue and organ levels have demonstrated that m6A modification controls many biological processes, including spermatogenesis (6), neurogenesis (7), sex determination (8, 9), and stem cell self-renewal and life span (10, 11). Thus, the imbalance of the m6A pathway is often found in lung cancer (12), breast cancer (13), glioblastoma (14–16), acute myeloid leukemia (17), liver cancer (18), and other tumors. The regulatory mechanism of m6A and its biological functions have become a research hotspot in the field of RNA. But the influence of m6A on these biological processes and performing different functions in different settings still need further in-depth study.
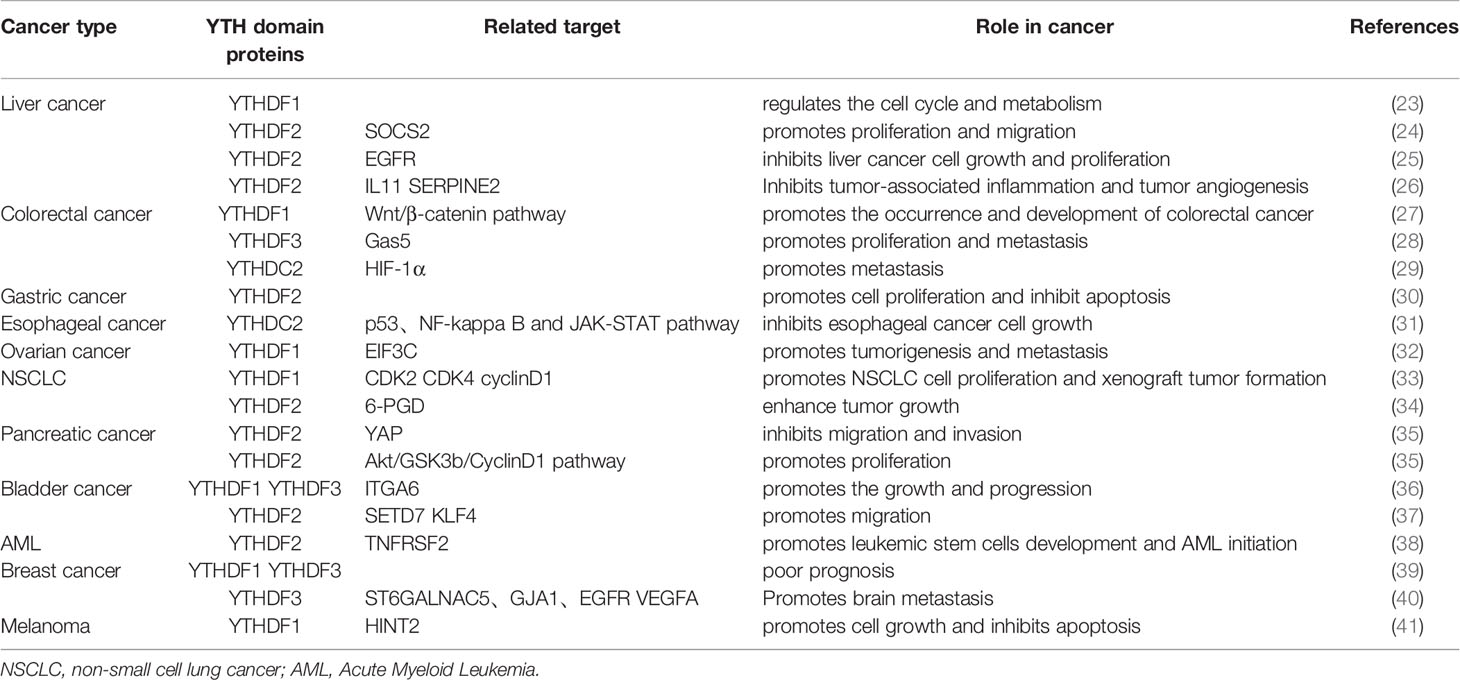
Methyltransferase and demethyltransferase make the mRNA dynamically and reversibly modified by m6A, while “readers” can recognize the methylated RNA and realize its function. Moreover, several different “readers” of m6A, including members of the YTH N6-Methyladenosine RNA Binding Proteins (YTHDFs) and YTH domain containing (YTHDCs) families, can recognize the modification of m6A and realize many biological functions specifically (2, 19–22) (Table 1 and Figure 1).
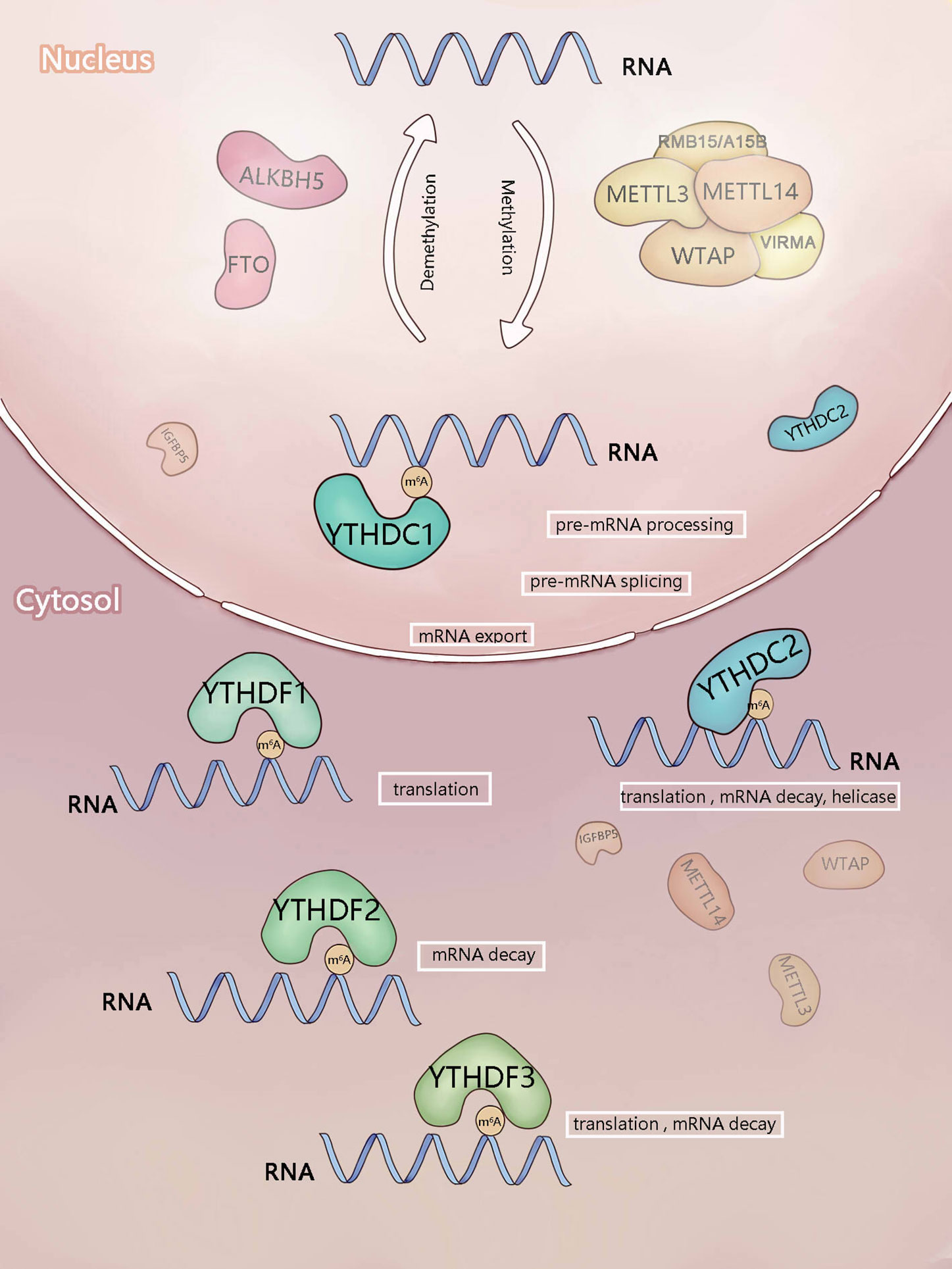
Figure 1 Components involved in m6A modification. m6A is mainly added by methyltransferase complex composed of methyltransferase-like3 (METTL3), METTL14),WTAP, RBM15/RBM15B, and VIRMA (writers). ALKBH5 and FTO (erasers) can remove m6A as demethyltransferase. In the nucleus, m6A can be recognized and regulated by YTHDC1 (reader) to regulate RNA splicing and mRNA export. In the cytosol, YTHDF1-3 and YTHDC2 (readers) recognize m6A, then they regulate the translation and stability of mRNA.
The YTH domain family proteins comprise reading proteins widely distributed in the human body, and these “readers” are the major genes that mediate the function of m6A methylated RNAs. In this review, we summarize the up-to-date knowledge of ways the YTH domain family proteins affect biological functions in cancer and discuss their prospects as m6A regulators in cancer therapy.
Mechanism of the YTH Domain Family Proteins
The YTH domain recognizes the m6A modification through a conserved aromatic ring. This is an RNA-binding domain dependent on m6A modification (42). Reading proteins recognize and read the information of m6A RNA in a methylation-dependent manner. YTHDC1-2 and YTHDF1-3 are the major Intracellular proteins in the human body. YTHDC2 is localized in the cytoplasm of meiotic spermatocytes, and YTHDF1-3 mainly recognizes the information of m6A methylation in the cytoplasm (21, 24, 44). These five proteins not only contain the same domain but also have their special domains that determine their different roles.
YTHDF1
YTHDF1 promotes the translation of m6A RNA. Studies have shown that the main role of YTHDF1 is to accelerate the translation of m6A mRNA by promoting the ribosome assembly of m6A mRNA and interacting with the initiation factor, thereby regulating the translation kinetics (22) (Figure 1). For instance, YTHDF1 can increase the translation of Eukaryotic translation initiation factor 3 subunit C (ELF3C) and promote the overall translation output by combining with the m6A-modified ELF3C mRNA, thus promoting the occurrence and metastasis of ovarian cancer (32). In bladder cancer cells, YTHDF1 preferentially binds to the m6A mRNA of oncogene CUB domain containing protein 1 (CDCP1) to promote its translation, leading to the malignant transformation of urinary tract epithelium and occurrence of bladder cancer (45). YTHDF1 promotes the translation of Histidine triad nucleotide binding protein 2 (HINT2)-methylated mRNA but reduces the m6A modification in ocular melanoma (41). The extremely high or low level of m6A modification can lead to the occurrence of tumors.
YTHDF1 can selectively recognize the m6A RNA and bind to the GRAC (R is G or A) site, but not to other motifs near the m6A site (22). Snail family transcriptional repressor, the key transcription factor of EMT, has m6A methylation in its coding sequence (CDS) region. YTHDF1 is more likely to combine with the m6A RNA in CDS region to promote Snail translation (46). However, the binding sites of ELF3C mRNA and YTHDF1 are mainly located in coding region and untranslated regions in ovarian cancer cells (32). Furthermore, The YTHDF1-binding sites in dendritic cells are also enriched near the stop codon and 3’ untranslated regions (3’ UTRs) (47). A study on mouse hippocampus found that the binding sites of YTHDF1 were mainly located near the stop codons and 3’-UTRs (48). The methylation level of oncogene CDCP1 in the 3’ UTR significantly increased in bladder cancer cells (45).
Intriguingly, YTHDF1 is vital in promoting the translation initiation and translation elongation of m6A RNA. The binding of YTHDF1 to targeted RNA is selective. How this property is acquired and how YTHDF1 modifies downstream signaling pathways need further investigation.
YTHDF2
Main function of YTHDF2 is to regulate the stability of m6A RNA. YTHDF2 mainly controls the life span and stability of target mRNA (Figure 1). The carboxyl-terminal domain of YTHDF2 is responsible for binding to the m6A RNA, and the amino-terminal domain is responsible for locating the YTHDF2 mRNA complex to the RNA decay site and transforming the m6A RNA from the translation state to the degradation state (21). Moreover, YTHDF2 destroys and further degrades the target mRNA by recruiting C-C motif chemokine receptor 4 (CCR4), not the deacetylase complex (49). It recognizes the m6A modification site in the 3’ UTR of Epidermal growth factor receptor (EGFR) mRNA, promotes its degradation (25), and plays an anti-tumor role. It inhibits the proliferation of hematopoietic stem cells by degrading the mRNA of self-renewal-related genes of hematopoietic stem cells (50). Furthermore, it targets Autophagy related 5 (ATG5) and Autophagy related 7 (ATG7) transcripts with a high level of m6A, resulting in mRNA degradation and thus reducing autophagy and adipogenesis (33). These studies suggest the involvement of YTHDF2 in biological processes by destroying the stability of key gene transcripts. In the heat shock stress response, YTHDF2 could promote the independent translation from the cytoplasmic localization to the protection of m6A at the 5’ UTRs end of the nucleus.
YTHDF3
YTHDF3 can promote the translation and degradation of mRNA. YTHDF3 can promote the translation and degradation of m6A RNA (Figure 1). The function of YTHDF3 is both similar to and different from that of YTHDF2. YTHDF3 can locate the mRNA from the translation site to the explanation site by recognizing the m6A mRNA, and regulate its own mRNA translation to increase protein expression by itself (21, 51). Moreover, YTHDF3 can also interact with ribosomes and take part in the initial stage of translation (52). Further studies have shown that YTHDF3 induces the translation of m6A mRNA by binding with YTHDF1 and Eukaryotic translation initiation factor 4A3 (EIF4A3). The YTH domains at the C-terminus of YTHDF2 and YTHDF3 bind to the conserved G(m6A)C core motif, while the N-terminal domain is responsible for the migration of protein mRNA complexes to RNA decay sites (53).
YTHDC1
YTHDC1 regulates the splicing of RNA. The YTH domain in YTHDC1 specifically recognizes the m6A modification and preferentially recognizes the G(m6A)C sequence (54). Then, YTHDC1 combines with the m6A-modified pre-RNAs to recruit the splicing factor Serine and arginine rich splicing factor 3 (SRSF3) but blocks the binding of Serine and arginine rich splicing factor 10 (SRSF10) to nuclear spots, thus promoting the splicing of exon inclusion bodies and mRNA output from the nucleus to the cytoplasm (20, 55) (Figure 1). YTHDC1 can recognize the m6A-modified chromosome-related regulatory RNAs (CarRNAs) [including promoter-related RNA (PaRNA), enhancer RNA (ERNA), and transposable element transcript RNA (repetitive RNA)], promote the decay and interpretation of CarRNAs, and regulate the state and transcription of chromatin.
YTHDC2
YTHDC2 has many functions. YTHDC2 regulates the stability of m6A mRNA by recognizing the modification of m6A and recruiting the RNA degradation mechanism. It can also use its unique RNA-binding domain to build a bridge between m6A mRNA and ribosome and thus promote their effective translation. Among the YTH domain proteins, YTHDC2 is the only one containing the helicase domain; it is involved in meiosis (43, 56, 57). In addition, YTHDC2 is a protein with multiple domains. It may have complex functions in different cell environments, such as promoting translation, regulating the stability of transcripts, and serving as helicase (Figure 1). The interaction and regulation of these three functions remains unknown. Therefore, the contribution of these different functions in each field needs to be examined in more detail.
Interactions in YTH Domain Family
In addition to their respective targeted mRNA sequences, YTHDF1 and YTHDF2 share a large group of targeted mRNA sequences, which can be regulated according to different cell signals. YTHDF1 can bind to the common sequence earlier (22). These results indicate that YTHDF1 promotes the translation of targeted mRNA, and YTHDF2 controls its life span. YTHDF1 and YTHDF2 regulate protein production and promote gene expression. The synergistic effect of YTHDF3 and YTHDF1 promotes protein synthesis and also affects the degradation of methylated mRNA mediated by YTHDF2. YTHDF3 may interact with the target mRNA before YTHDF1 and YTHDF2 (53). The three YTHDF proteins coordinate with each other to promote protein expression and have a certain degree of time sequence. The interaction between YTH domain family proteins can be regulated by m6A modification. Recent research confirms that m6A-modified mRNAs promote liquid-liquid phase separation (LLPS) of YTHDF proteins. On the contrary, mRNA-YTHDF complexes can regulate the translation efficiency and mRNA stability of m6A-modified mRNA (58).
The most widely studied and consensual mechanisms for YTHDF proteins were their ability to recognize m6A-mRNA, promote its degradation (YTHDF2/3) and translation (YTHDF1/3) specifically. A recent study has shown that no evidence demonstrates YTHDF proteins can promotes translation directly (59). In contrast, a finding suggests the opposite. m6A-Quantitative trait loci (QTL) mapping data analysis reveals that YTHDF proteins participate in different translational efficiency in a context-dependent manner. The investigators concluded that the translational efficiency depends on not only the sequence context surrounding the m6A site but also the binding site of other YTHDF proteins, rather than promoting translation simply (60). This is the first use of m6A-QTLs as a molecular genetic analysis tool for studies on m6A mechanisms. In our view, there are several potential explanations for this discrepancy. By using only YTHDF paralogs and different combinations of YTHDFs knockdown in HeLa cells, YTHDF proteins are believed to lead to mRNA degradation in the former study (59). This is inconsistent with our notion. Perhaps, additional experiments of this view are first required.
YTHDC1 is active in the nucleus, while YTHDC2 mainly functions in the cytoplasm. They are distributed in different functional areas and do not interfere with each other. Some functions of YTHDC2 are similar to that of the YTHDF proteins. However, a synergistic effect between them is still unclear.
Techniques and Research Tools
YTH protein family regulates gene expression via selective combination of gene-specific m6A mRNA sequences and YTH proteins. Currently, there are several experimental approaches that can be used to elucidate the underlying mechanisms. To date, crosslinking and immunoprecipitation sequencing (CLIP-Seq) has been used to high-throughput approach of RNA binding protein interactions (61). However, the RNA-protein cross-linking efficiency of CLIP remains poor due to the low crosslink identification. Based on this, photoactivatable ribonucleoside-enhanced-crosslinking immunoprecipitation (PAR-CLIP) (62) and individual-nucleotide resolution CLIP (iCLIP) (63, 64) are optimized to give precise binding sites of YTH domains and transcripts (21, 22).
Moreover, Some databases help us to understand interactions within m6A reader proteins. RMBase, a new database, provides map of m6A modifications from epitranscriptome sequencing data (65, 66). Reliable m6A sites and transcriptional profiles were pooled by m6A-Atlas. m6A-Atlas database enables prediction of the biological function of every m6A site and inferences of pathogenesis of m6A-evoked disease (67). The M6A2Target database is a specialist database for estimating the prediction target of m6A modification (68). RNAWRE is the first database for finding effectors of RNA modifications (69). Knowing these database can help us gain a deeper insight of m6A modifications.
Function of YTH Domain Family Proteins in Malignant Tumors
Liver Cancer
The analysis of The Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO) databases and the immunohistochemical analysis have shown that YTHDF1 is upregulated, associated with poor overall survival (OS) and pathologic staging of liver cancer. YTHDF1 is an independent adverse prognostic factor involved in regulating the cell cycle and metabolism of liver cancer (23, 70, 71). Therefore, it is considered to be a cancer-promoting factor. The expression of YTHDF2 in Hepatocellular Carcinoma (HCC) increases, which is closely related to the malignant degree of HCC. Research shows that miR-145 can regulate the expression of YTHDF2 and thus inhibit the proliferation of HCC cells (72). Another study showed that m6A modifications of Cytokine Signaling 2 (SOCS2) mRNA can be facilitated by methyltransferase-like3 (METTL3), and the degradation process of SOCS2 can be accelerated by YTHDF2 during HCC progression (24). Other studies have shown that YTHDF2 can be regarded as a tumor suppressor, inhibiting the growth of hepatoma cells. YTHDF2 promotes mRNA degradation of EGFR by binding to m6A site within the EGFR 3’UTR, simultaneously inhibiting proliferation of HCC cells. YTHDF2 inhibits inflammasome-mediated tumor vascularization and malignancy by degradation of m6A-containing serpin family E member 2 (SERPINE2) and interleukin-11 (IL-11) mRNA (25, 26). Also, YTHDF2 has a contradictory role in different research backgrounds. Future studies should demonstrate the specific effect of YTHDF2 on liver cancer and the involved signaling pathway, and also determine whether YTHDF2 is a tumor suppressor or a cancer-promoting factor. The participation of other members of the YTH family, such as YTHDC1 and YTHDC2, in the development of liver cancer has not yet been explored.
Colorectal Cancer
Through the analysis of cancer tissues and in vitro experiments, some researchers found that YTHDF1 was related to the behavior of a variety of malignant tumors and affected the sensitivity of colorectal cancer to chemotherapeutic medicines; also, the carcinogenic factor c-myc could regulate the expression of YTHDF1 (73). The results of Bai et al. also suggested that the high expression of YTHDF1 in colorectal cancer promoted the occurrence and development of colorectal cancer through Wnt/β-catenin pathway (27). Therefore, YTHDF1 can be used as a cancer-promoting factor. Low expression of YTHDF1 was found to be correlated with the infiltration of CD8+ T cells in colorectal carcinoma (CRC) tissue (47). The data suggest that YTHDF1 may have contributed to the regulation of tumor immune microenvironment. Another study demonstrates that lncRNA Gas5 inhibits cellular proliferation and metastasis of CRC by promoting Yes-associated protein (YAP) phosphorylation and YAP-mediated YTHDF3 transcription. Moreover, YTHDF3 binds to the m6A-modified lncRNA Gas5 and inhibits lncRNA Gas5 decay, thus generating a negative feedback loop (28). The high expression of YTHDC2 is related to the malignancy of colon cancer and contributes to tumor metastasis (29). However, YTHDC2 does not play the role of a reading protein and has nothing to do with the modification of m6A. YTHDC2 promotes the translation of hypoxia-inducible factor-1 α (HIF-1α) and the metastasis of colorectal cancer cells by playing the role of helicase.
Esophageal and Gastric Cancer
In vitro studies have shown that the knockdown of YTHDF2 can inhibit the proliferation of gastric cancer cells and promote their apoptosis (30). YTHDF2 can be used as a new target to study the mechanism of gastric cancer. As a suppressor of esophageal cancer, YTHDC2 is lowly expressed in esophageal cancer. Furthermore, high expression of YTHDC2 can inhibit the growth of tumor cells in vitro. A single nucleotide polymorphism named Rs2416282 is localized at YTHDC2 promoter and regulates YTHDC2 expression. In addition, Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis about the regulation of esophageal squamous cell carcinoma (ESCC) by YTHDC2 revealed that YTHDC2 is associated with p53, NF-kappa B and JAK-STAT signaling pathways (31).
Ovarian Cancer
The public database of ovarian cancer and in vitro and in vivo experiments showed that the expression of YTHDF1 was related to the stage and prognosis of ovarian cancer. Eukaryotic Translation Initiation Factor 3 Subunit C (EIF3C) exerts its oncogenic function by promoting its translation in an m6A-dependent manner that lead to tumor growth and metastasis in ovarian cancer (32). The occurrence of ovarian cancer is related to the modification of m6A. YTHDF1 can be used as a new target of ovarian cancer-targeted diagnosis and treatment. The effect of other m6A-modified related proteins on ovarian cancer can be explored in future studies.
Lung Cancer
Studies on nonsmall cell lung cancer (NSCLC) have shown that the high expression of YTHDF1 can promote the development of tumors by promoting protein translation of cyclin E-associated kinase 2 (CDK2), cyclin-dependent kinase 4 (CDK4) and cyclin D1 mRNAs. Furthermore, low expression of YTHDF1 can reduce the sensitivity of cancer tissue to cisplatin was attributed to the Keap1-Nrf2-AKR1C1 axis (Kelch-like ECH-associated protein 1, NF-E2 p45-related factor 2 and antioxidant genes including aldo-keto reductases 1C1) (33). This study suggested that YTHDF1 was related to lung cancer.
The role of YTHDF1 in cancer occurrence and treatment remains controversial. YTHDF1 is involved in biological processes via different molecular mechanisms. It is related to the sensitivity to cisplatin. However, whether ITF1 has the same effect as other chemotherapy medicines is still unclear. All these problems need exploration. YTHDF2 binds to the m6A modification site of 6-phosphogluconate dehydrogenase (6PGD) 3’-UTR and facilitate 6PGD mRNA translation in lung cancer cells. Based on this, the authors suggest that YTHDF2 can promote the growth of lung cancer by affecting the pentose phosphate pathway (PPP) (34). Recent studies have shown that YTHDF1 and YTHDF2 are highly expressed in lung adenocarcinoma and related to the prognosis of patients, but the specific mechanism remains unclear (74). Data from The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) database show that both YTHDF1 and YTHDF2 were highly expressed in lung adenocarcinoma (75). Alkylation Repair Homolog 5 (ALKBH5) inhibits tumor growth and metastasis by reducing m6A modification of YAP and suppressing activity of miR-107/Large Tumor Suppressor Kinase 2 (miR-107/LATS2)-mediated YAP. YTHDF proteins may play a critical role in this process (76).
Pancreatic Cancer
A study on pancreatic cancer demonstrated the high expression of YTHDF2. YTHDF2 inhibited adhesion, invasion, migration, and Epithelial-Mesenchymal Transition (EMT) through the YAP signal and promoted the proliferation of pancreatic cancer cells through the Serine/threonine-protein kinases/Glycogen synthase kinase 3 beta/Cyclin D1 (AKT/GSK3B/CCND1) pathway (35). YTHDF2 has dual effects on pancreatic cancer. YTHDF2 can be used as a new marker of pancreatic cancer, but the specific mechanism needs to be clarified. YTHDC2 is also considered to be a frequently mutated gene in patients with pancreatic cancer. We conclude that YTHDC2 may be associated with pancreatic cancer susceptibility (77).
Bladder Cancer
The combination of YTHDF1 and YTHDF3 promotes the translation of Integrin subunit alpha 6 (ITGA6) mRNA and the development of bladder cancer (36). YTHDF2 promotes the mRNA degradation of tumor suppressor genes SET domain containing lysine methyltransferase 7 (SETD7) and Kruppel Like Factor 4 (KLF4) by identifying METTL3-mediated m6A modification, thus inducing bladder cancer progression (37). METTL3/YTHDF2-SETD7/KLF4 m6A axis plays an important role in bladder carcinogenesis. Many reading proteins of the YTH family are related to bladder cancer; they regulate the tumor tissue by influencing different target genes downstream positively or negatively.
Leukemia
YTHDF2 is involved in regulating the self-renewal of hematopoietic stem cells (HSCs) (50, 78). The abnormal expression of YTHDF2 may relate to some hematologic neoplasms. One study has demonstrated that inhibited YTHDF2 selectively compromises acute myeloid leukemia (AML) (38). YTHDF2 can be used as a potential target in the treatment of leukemia.
Breast Cancer
It has been shown that higher YTHDF1 and YTHDF3 expressions were correlated with a shorter survival in breast cancer (BC) (39), and the importance of YTHDF3 as an independent risk factor for BC has been a topic of significant research. Another study has proposed that overexpressed YTHDF1 and YTHDF3 promotes translation of oncogenes in a m6A-dependent manner in BC, resulting in tumor progression and inferior outcomes (51). YTHDF3 expression in BC brain metastases is higher than that in BC and nontumor tissues, and facilitates expressions of ST6 N-Acetylgalactosaminide Alpha-2,6-Sialyltransferase 5 (ST6GALNAC5), gap junction proteinα1 (GJA1), EGFR, and vascular endothelial growth factor A (VEGFA) in a m6A-dependent manner during the regulation of multiple steps in brain metastases (40).
Melanoma
Initial study on melanoma demonstrated that YTHDF1 can promote translation by binding Histidine Triad Nucleotide Binding Protein 2 (HINT2) transcripts, that in turn promotes tumor cell proliferation, inhibits apoptosis and confers a poor prognosis (41). Of interest, the increased m6A methylation of important oncogenes in FTO knockdown cells also promoted YTHDF2-induced degradation in melanoma, whereas melanoma cells appear to be more sensitive to IFN-γ and enhance the efficacy of melanoma immunotherapy (79).
The abnormal expression of YTH family proteins in various tumor tissues is related to the occurrence of tumors and promotes the growth and metastasis of tumors (Figure 2). The role of the YTH domain protein family in tumor target tissue is preliminarily understood; it can be used as a potential for later research. However, different mechanisms exist in different research backgrounds. Hence, extensive investigation is needed to clarify the specific mechanism underlying the involvement of YTH family proteins in tumors as the target of treatment. The available findings suggested that the mRNA and protein levels of YTH family proteins could be simultaneously examined. The questions to be addressed were as follows: Whether the involvement of reading proteins varies in different tumors at the RNA level due to different splicing and maturation processes controlled by upstream signals; whether the binding to the m6A mRNA is related to the surrounding environment at the protein level, leading to the different roles of m6A in different stages of tumor development; The relationship between YTH family proteins and cancer is still unknown to a large extent. These findings provide novel ideas for the study of epigenetics of cancer and hence may help find new targets for antitumor therapy.
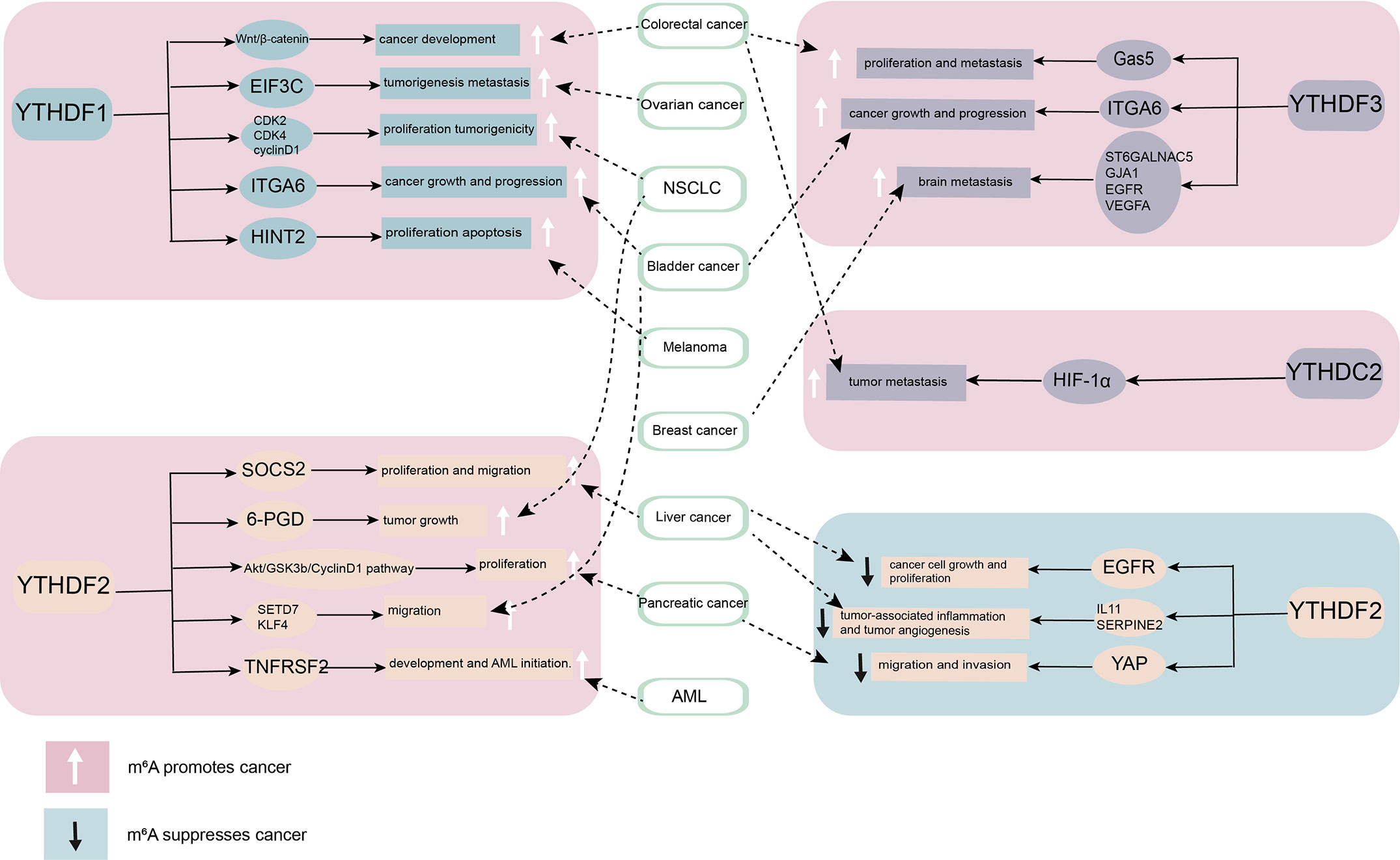
Figure 2 The role of YT521-B homology (YTH) domain family proteins in cancer. By combining different downstream targets, YTH domain family proteins promote or inhibit the occurrence and development of cancer. YTHDF1, YTHDF3, and YTHDC2 mainly play a role as cancer promoting factor. YTHDF2 plays diverse roles in different cancers.
These studies show not only that YTH protein family has significant effects on regulating the development and function of tumor, but also that YTH protein family can influence the antineoplastic treatment. How YTH protein family might impact cancer immunotherapy will be important to examine in the future. In the current study, YTHDF1 have the potential to identify and translate m6A-modified lysosomal protease, which suppress cross-presentation activity in dendritic cells (DC) and weakens the sensitization to CD8+ T cells. This could suggest a potential underlying mechanism of immune escape (47). Taken together, the current study suggests that YTHDF1 may be harnessed as a new immunotherapeutic target to improve patient outcomes with cancer. In another study conducted in head and neck squamous cell carcinoma implicates that YTH domain protein family are related to tumor immune microenvironment (TIME), whereas PD-L1 upregulation is associated with m6A methylation and patients with high-risk scores may receive more benefit from radiotherapy (80). Meanwhile YTHDF2 also appear to exert negative regulation in macrophages-induced inflammatory responses (81) (Figure 3).
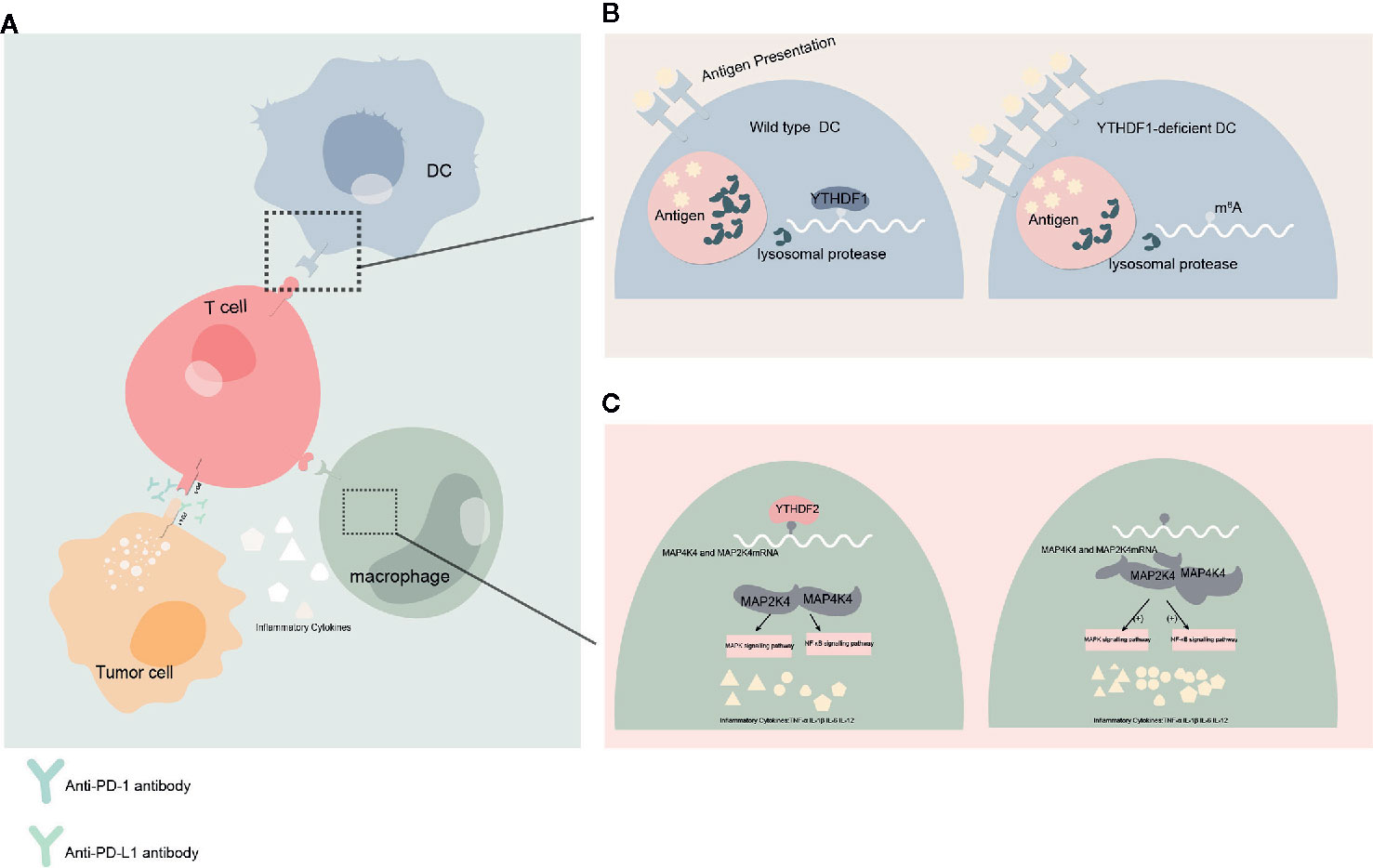
Figure 3 Effect of YTHDFs on tumor immune microenvironment (TIME) (A) Dendritic cells (DC) presents the antigen to the surface of T cells and initiates the adaptive immune response. Programmed cell death protein 1 (PD-1)/programmed death-ligand 1 (PD-L1) is a negative immune checkpoint pathway that inhibit immune responses and promotes immune escape of tumor cells. This process can be blocked by anti-PD-1/PD-L1 antibodies. Macrophages can affect anti-tumor immunity by secreting inflammatory factors. (B) When YTHDF1 is deficient, the translation efficiency of lysosomal protease transcripts decreases, the degradation of antigens is reduced, more antigens are involved in antigen presentation, and the immune response of T cells is enhanced. This process may affect the efficacy of PD-L1 checkpoint inhibitors. (C) In macrophages, YTHDF2 knockdown promotes the translation and stability of MAP2K4 and MAP4K4 mRNA, which activates MAPK and NF-κB signaling pathways and promotes the expression of pro-inflammatory cytokines. These inflammatory factors can participate in the regulation of immune response and affect the efficacy of anti-tumor immunity.
Oxygen content in tumor cells effects radiosensitivity directly. It has been demonstrated that YTHDF1 is associated with hypoxia adaptation (33). Moreover, m6A-modified METTL3 is involved in the influence of DNA damage response upon UV radiation (82). These results suggest that m6A modifications might also influence the radiosensitivity through DNA damage and/or DNA repair. Therefore, the mechanisms of YTH family proteins effect on radiosensitivity are ongoing in our laboratory
Future Prospects
RNA m6A modification is a dynamic and reversible modification catalyzed by methyltransferase, demethylase, and reading proteins. Readers can link RNA modification at specific sites with specific regulatory functions in cells. Although the recent progress of YTH family proteins in cancer has been summarized previously in review form (83), we conclude by summarizing the most important findings about mechanisms, functions, and interactions of YTH protein family from a different perspective.
The research on the role of YTH domain family proteins in cancer has greatly advanced, but is still associated with various challenges. First, the mechanism of YTH domain family proteins in cancer research is still unclear to a large extent. Second, whether YTH domain family proteins can be used as potential targets for cancer diagnosis and treatment, as well as their specificity and sensitivity, needs to be further explored. Also, some studies have shown that the YTH family proteins and their related new signaling pathways can be used. However, the lack of large-sample clinical studies and the side effects of targeted therapy are unknown.
With recent striking combination treatment effects of cancer-immunotherapy (40, 47), identifying m6A-related immunotherapy-sensitive tumor types and understanding how m6A modifications influence immunotherapy sensitivity and radiosensitivity are urgently needed.
The continuous progress of high-throughput sequencing technology, RNA-Seq technology, and immunoprecipitation technology has provided a deeper understanding of the m6A modification pathway and indicated an important biological function of m6A by regulating RNA stabilization, localization, transportation, splicing, and translation. The research on the involvement of YTH domain family proteins in cancer and other diseases has provided new ideas and laid a theoretical foundation for developing a therapeutic strategy for the use of YTH domain family proteins in cancer.
Author Contributions
YX and WZ contributed to the drafting and editing of the manuscript, and shared the first authorship. QG and JH designed, revised, and finalized the manuscript. HL, FS, XY, SD, XS, LO, and DC participated in the drafting and editing of the manuscript. XS assisted in the revision of the manuscript. YX contributed to literature search. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Innovative Team of Jiangsu Province (CXTDA2017042), China Postdoctoral Science Foundation (2019M663505), and Taizhou science and technology support plan (Social Development) project (TS201903).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci U S A (1974) 71:3971–75. doi: 10.1073/pnas.71.10.3971
2. Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA Modifications in Gene Expression Regulation. Cell (2017) 169:1187–200. doi: 10.1016/j.cell.2017.05.045
3. Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol (2011) 7:885–7. doi: 10.1038/nchembio.687
4. Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet (2014) 15:293–306. doi: 10.1038/nrg3724
5. Roundtree IA, He C. RNA epigenetics–chemical messages for posttranscriptional gene regulation. Curr Opin Chem Biol (2016) 30:46–51. doi: 10.1016/j.cbpa.2015.10.024
6. Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell (2013) 49:18–29. doi: 10.1016/j.molcel.2012.10.015
7. Yoon KJ, Ringeling FR, Vissers C, Jacob F, Pokrass M, Jimenez-Cyrus D, et al. Temporal Control of Mammalian Cortical Neurogenesis by m6A Methylation. Cell (2017) 171:877–89.e17. doi: 10.1016/j.cell.2017.09.003
8. Haussmann IU, Bodi Z, Sanchez-Moran E, Mongan NP, Archer N, Fray RG, et al. m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature (2016) 540:301–4. doi: 10.1038/nature20577
9. Lence T, Akhtar J, Bayer M, Schmid K, Spindler L, Ho CH, et al. m6A modulates neuronal functions and sex determination in Drosophila. Nature (2016) 540:242–7. doi: 10.1038/nature20568
10. Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell (2014) 15:707–19. doi: 10.1016/j.stem.2014.09.019
11. Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol (2014) 16:191–8. doi: 10.1038/ncb2902
12. Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol Cell (2016) 62:335–45. doi: 10.1016/j.molcel.2016.03.021
13. Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I, et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA. Proc Natl Acad Sci U S A (2016) 113:E2047–56. doi: 10.1073/pnas.1602883113
14. Visvanathan A, Patil V, Arora A, Hegde AS, Arivazhagan A, Santosh V, et al. Essential role of METTL3-mediated m6A modification in glioma stem-like cells maintenance and radioresistance. Oncogene (2018) 37:522–33. doi: 10.1038/onc.2017.351
15. Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, et al. m6A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell (2017) 31:591–606.e6. doi: 10.1016/j.ccell.2017.02.013
16. Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, et al. m6A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep (2017) 18:2622–34. doi: 10.1016/j.celrep.2017.02.059
17. Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, et al. FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N6-Methyladenosine RNA Demethylase. Cancer Cell (2017) 31:127–41. doi: 10.1016/j.ccell.2016.11.017
18. Ma JZ, Yang F, Zhou CC, Liu F, Yuan JH, Wang F, et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N6 -methyladenosine-dependent primary MicroRNA processing. Hepatology (2017) 65:529–43. doi: 10.1002/hep.28885
19. Meyer KD, Jaffrey SR. Rethinking m6A Readers, Writers, and Erasers. Annu Rev Cell Dev Biol (2017) 33:319–42. doi: 10.1146/annurev-cellbio-100616-060758
20. Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell (2016) 61:507–19. doi: 10.1016/j.molcel.2016.01.012
21. Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature (2014) 505:117–20. doi: 10.1038/nature12730
22. Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell (2015) 161:1388–99. doi: 10.1016/j.cell.2015.05.014
23. Zhao X, Chen Y, Mao Q, Jiang X, Jiang W, Chen J, et al. Overexpression of YTHDF1 is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Biomark (2018) 21:859–68. doi: 10.3233/CBM-170791
24. Chen M, Wei L, Law CT, Tsang FH, Shen J, Cheng CL, et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology (2018) 67:2254–70. doi: 10.1002/hep.29683
25. Zhong L, Liao D, Zhang M, Zeng C, Li X, Zhang R, et al. YTHDF2 suppresses cell proliferation and growth via destabilizing the EGFR mRNA in hepatocellular carcinoma. Cancer Lett (2019) 442:252–61. doi: 10.1016/j.canlet.2018.11.006
26. Hou J, Zhang H, Liu J, Zhao Z, Wang J, Lu Z, et al. YTHDF2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma. Mol Cancer (2019) 18:163. doi: 10.1186/s12943-019-1082-3
27. Bai Y, Yang C, Wu R, Huang L, Song S, Li W, et al. YTHDF1 Regulates Tumorigenicity and Cancer Stem Cell-Like Activity in Human Colorectal Carcinoma. Front Oncol (2019) 9:332. doi: 10.3389/fonc.2019.00332
28. Ni W, Yao S, Zhou Y, Liu Y, Huang P, Zhou A, et al. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m6A reader YTHDF3. Mol Cancer (2019) 18:143. doi: 10.1186/s12943-019-1079-y
29. Tanabe A, Tanikawa K, Tsunetomi M, Takai K, Ikeda H, Konno J, et al. RNA helicase YTHDC2 promotes cancer metastasis via the enhancement of the efficiency by which HIF-1α mRNA is translated. Cancer Lett (2016) 376:34–42. doi: 10.1016/j.canlet.2016.02.022
30. Zhang J, Pi J, Liu Y, Yu J, Feng T. Knockdown of YTH N6-methyladenosine RNA binding protein 2 (YTHDF2) inhibits proliferation and promotes apoptosis in MGC-803 gastric cancer cells. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. (2017) 33:1628–34.
31. Yang N, Ying P, Tian J, Wang X, Mei S, Zou D, et al. Genetic variants in m6A modification genes are associated with esophageal squamous-cell carcinoma in the Chinese population. Carcinogenesis (2020) 41:761–68. doi: 10.1093/carcin/bgaa012
32. Liu T, Wei Q, Jin J, Luo Q, Liu Y, Yang Y, et al. The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res (2020) 48:3816–31. doi: 10.1093/nar/gkaa048
33. Shi Y, Fan S, Wu M, Zuo Z, Li X, Jiang L, et al. YTHDF1 links hypoxia adaptation and non-small cell lung cancer progression. Nat Commun (2019) 10:4892. doi: 10.1038/s41467-019-12801-6
34. Sheng H, Li Z, Su S, Sun W, Zhang X, Li L, et al. YTH domain family 2 promotes lung cancer cell growth by facilitating 6-phosphogluconate dehydrogenase mRNA translation. Carcinogenesis (2020) 41:541–50. doi: 10.1093/carcin/bgz152
35. Chen J, Sun Y, Xu X, Wang D, He J, Zhou H, et al. YTH domain family 2 orchestrates epithelial-mesenchymal transition/proliferation dichotomy in pancreatic cancer cells. Cell Cycle (2017) 16:2259–71. doi: 10.1080/15384101.2017.1380125
36. Jin H, Ying X, Que B, Wang X, Chao Y, Zhang H, et al. N6-methyladenosine modification of ITGA6 mRNA promotes the development and progression of bladder cancer. EBioMedicine (2019) 47:195–207. doi: 10.1016/j.ebiom.2019.07.068
37. Xie H, Li J, Ying Y, Yan H, Jin K, Ma X, et al. METTL3/YTHDF2 m6 A axis promotes tumorigenesis by degrading SETD7 and KLF4 mRNAs in bladder cancer. J Cell Mol Med (2020) 24:4092–104. doi: 10.1111/jcmm.15063
38. Paris J, Morgan M, Campos J, Spencer GJ, Shmakova A, Ivanova I, et al. Targeting the RNA m6A Reader YTHDF2 Selectively Compromises Cancer Stem Cells in Acute Myeloid Leukemia. Cell Stem Cell (2019) 25:137–48.e6. doi: 10.1016/j.stem.2019.03.021
39. Liu L, Liu X, Dong Z, Li J, Yu Y, Chen X, et al. N6-methyladenosine-related Genomic Targets are Altered in Breast Cancer Tissue and Associated with Poor Survival. J Cancer (2019) 10:5447–59. doi: 10.7150/jca.35053
40. Chang G, Shi L, Ye Y, Shi H, Zeng L, Tiwary S, et al. YTHDF3 Induces the Translation of m6A-Enriched Gene Transcripts to Promote Breast Cancer Brain Metastasis. Cancer Cell (2020) 38:857–71. doi: 10.1016/j.ccell.2020.10.004
41. Jia R, Chai P, Wang S, Sun B, Xu Y, Yang Y, et al. m6A modification suppresses ocular melanoma through modulating HINT2 mRNA translation. Mol Cancer (2019) 18:161. doi: 10.1186/s12943-019-1088-x
42. Xu C, Liu K, Ahmed H, Loppnau P, Schapira M, Min J. Structural Basis for the Discriminative Recognition of N6-Methyladenosine RNA by the Human YT521-B Homology Domain Family of Proteins. J Biol Chem (2015) 290:24902–13. doi: 10.1074/jbc.M115.680389
43. Wojtas MN, Pandey RR, Mendel M, Homolka D, Sachidanandam R, Pillai RS. Regulation of m6A Transcripts by the 3’→5’ RNA Helicase YTHDC2 Is Essential for a Successful Meiotic Program in the Mammalian Germline. Mol Cell (2017) 68:374–87.e12. doi: 10.1016/j.molcel.2017.09.021
44. Bailey AS, Batista PJ, Gold RS, Chen YG, de Rooij DG, Chang HY, et al. The conserved RNA helicase YTHDC2 regulates the transition from proliferation to differentiation in the germline. Elife (2017) 6:e26116. doi: 10.7554/eLife.26116
45. Yang F, Jin H, Que B, Chao Y, Zhang H, Ying X, et al. Dynamic m6A mRNA methylation reveals the role of METTL3-m6A-CDCP1 signaling axis in chemical carcinogenesis. Oncogene (2019) 38:4755–72. doi: 10.1038/s41388-019-0755-0
46. Lin X, Chai G, Wu Y, Li J, Chen F, Liu J, et al. RNA m6A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of Snail. Nat Commun (2019) 10:2065. doi: 10.1038/s41467-019-09865-9
47. Han D, Liu J, Chen C, Dong L, Liu Y, Chang R, et al. Anti-tumour immunity controlled through mRNA m6A methylation and YTHDF1 in dendritic cells. Nature (2019) 566:270–4. doi: 10.1038/s41586-019-0916-x
48. Shi H, Zhang X, Weng YL, Lu Z, Liu Y, Lu Z, et al. m6A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature (2018) 563:249–53. doi: 10.1038/s41586-018-0666-1
49. Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, et al. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun (2016) 7:12626. doi: 10.1038/ncomms12626
50. Li Z, Qian P, Shao W, Shi H, He XC, Gogol M, et al. Suppression of m6A reader Ythdf2 promotes hematopoietic stem cell expansion. Cell Res (2018) 28:904–17. doi: 10.1038/s41422-018-0072-0
51. Anita R, Paramasivam A, Priyadharsini JV, Chitra S. The m6A readers YTHDF1 and YTHDF3 aberrations associated with metastasis and predict poor prognosis in breast cancer patients. Am J Cancer Res (2020) 10:2546–54.
52. Li A, Chen YS, Ping XL, Yang X, Xiao W, Yang Y, et al. Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res (2017) 27:444–7. doi: 10.1038/cr.2017.10
53. Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, et al. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res (2017) 27:315–28. doi: 10.1038/cr.2017.15
54. Xu C, Wang X, Liu K, Roundtree IA, Tempel W, Li Y, et al. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat Chem Biol (2014) 10:927–9. doi: 10.1038/nchembio.1654
55. Roundtree IA, Luo GZ, Zhang Z, Wang X, Zhou T, Cui Y, et al. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. Elife (2017) 6:e31311. doi: 10.7554/eLife.31311
56. Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y, et al. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res (2017) 27:1115–27. doi: 10.1038/cr.2017.99
57. Kretschmer J, Rao H, Hackert P, Sloan KE, Höbartner C, Bohnsack MT. The m6A reader protein YTHDC2 interacts with the small ribosomal subunit and the 5’-3’ exoribonuclease XRN1. RNA (2018) 24:1339–50. doi: 10.1261/rna.064238.117
58. Ries RJ, Zaccara S, Klein P, Olarerin-George A, Namkoong S, Pickering BF, et al. m6A enhances the phase separation potential of mRNA. Nature (2019) 571:424–8. doi: 10.1038/s41586-019-1374-1
59. Zaccara S. Jaffrey SR. A Unified Model for the Function of YTHDF Proteins in Regulating m6A-Modified mRNA. Cell (2020) 181:1582–1595.e18. doi: 10.1016/j.cell.2020.05.012
60. Zhang Z, Luo K, Zou Z, Qiu M, Tian J, Sieh L, et al. Genetic analyses support the contribution of mRNA N6-methyladenosine (m6A) modification to human disease heritability. Nat Genet (2020) 52:939–49. doi: 10.1038/s41588-020-0644-z
61. Sanford JR, Wang X, Mort M, Vanduyn N, Cooper DN, Mooney SD, et al. Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts. Genome Res (2009) 19:381–94. doi: 10.1101/gr.082503.108
62. Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell (2010) 141(1):129–41. doi: 10.1016/j.cell.2010.03.009
63. König J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol (2010) 17:909–15. doi: 10.1038/nsmb.1838
64. Huppertz I, Attig J, D’Ambrogio A, Easton LE, Sibley CR, Sugimoto Y, et al. iCLIP: protein-RNA interactions at nucleotide resolution. Methods (2014) 65:274–87. doi: 10.1016/j.ymeth.2013.10.011
65. Sun WJ, Li JH, Liu S, Wu J, Zhou H, Qu LH, et al. RMBase: a resource for decoding the landscape of RNA modifications from high-throughput sequencing data. Nucleic Acids Res (2016) 44(D1):D259–65. doi: 10.1093/nar/gkv1036
66. Xuan JJ, Sun WJ, Lin PH, Zhou KR, Liu S, Zheng LL, et al. RMBase v2.0: deciphering the map of RNA modifications from epitranscriptome sequencing data. Nucleic Acids Res (2018) 46(D1):D327–34. doi: 10.1093/nar/gkx934
67. Tang Y, Chen K, Song B, Ma J, Wu X, Xu Q, et al. m6A-Atlas: a comprehensive knowledgebase for unraveling the N6-methyladenosine (m6A) epitranscriptome. Nucleic Acids Res (2020) 21:gkaa692. doi: 10.1093/nar/gkaa692
68. Deng S, Zhang H, Zhu K, Li X, Ye Y, Li R, et al. M6A2Target: a comprehensive database for targets of m6A writers, erasers and readers. Brief Bioinform (2020) 11:bbaa055. doi: 10.1093/bib/bbaa055
69. Nie F, Feng P, Song X, Wu M, Tang Q, Chen W. RNAWRE: a resource of writers, readers and erasers of RNA modifications. Database (Oxford) (2020) 2020:baaa049. doi: 10.1093/database/baaa049
70. Zhou Y, Yin Z, Hou B, Yu M, Chen R, Jin H, et al. Expression profiles and prognostic significance of RNA N6-methyladenosine-related genes in patients with hepatocellular carcinoma: evidence from independent datasets. Cancer Manag Res (2019) 11:3921–31. doi: 10.2147/CMAR.S191565
71. Qu N, Qin S, Zhang X, Bo X, Liu Z, Tan C, et al. Multiple m6A RNA methylation modulators promote the malignant progression of hepatocellular carcinoma and affect its clinical prognosis. BMC Cancer (2020) 20:165. doi: 10.1186/s12885-020-6638-5
72. Yang Z, Li J, Feng G, Gao S, Wang Y, Zhang S, et al. MicroRNA-145 Modulates N6-Methyladenosine Levels by Targeting the 3’-Untranslated mRNA Region of the N6-Methyladenosine Binding YTH Domain Family 2 Protein. J Biol Chem (2017) 292:3614–23. doi: 10.1074/jbc.M116.749689
73. Nishizawa Y, Konno M, Asai A, Koseki J, Kawamoto K, Miyoshi N, et al. Oncogene c-Myc promotes epitranscriptome m6A reader YTHDF1 expression in colorectal cancer. Oncotarget (2017) 9:7476–86. doi: 10.18632/oncotarget.23554
74. Zhang Y, Liu X, Liu L, Li J, Hu Q, Sun R. Expression and Prognostic Significance of m6A-Related Genes in Lung Adenocarcinoma. Med Sci Monit (2020) 26:e919644. doi: 10.12659/MSM.919644
75. Li F, Wang H, Huang H, Zhang L, Wang D, Wan Y. m6A RNA Methylation Regulators Participate in the Malignant Progression and Have Clinical Prognostic Value in Lung Adenocarcinoma. Front Genet (2020) 11:994:994. doi: 10.3389/fgene.2020.00994
76. Jin D, Guo J, Wu Y, Yang L, Wang X, Du J, et al. m6A demethylase ALKBH5 inhibits tumor growth and metastasis by reducing YTHDFs-mediated YAP expression and inhibiting miR-107/LATS2-mediated YAP activity in NSCLC. Mol Cancer (2020) 19:40. doi: 10.1186/s12943-020-01161-1
77. Fanale D, Iovanna JL, Calvo EL, Berthezene P, Belleau P, Dagorn JC, et al. Germline copy number variation in the YTHDC2 gene: does it have a role in finding a novel potential molecular target involved in pancreatic adenocarcinoma susceptibility? Expert Opin Ther Targets (2014) 18:841–50. doi: 10.1517/14728222.2014.920324
78. Zhang C, Chen Y, Sun B, Wang L, Yang Y, Ma D, et al. m6A modulates haematopoietic stem and progenitor cell specification. Nature (2017) 549:273–6. doi: 10.1038/nature23883
79. Yang S, Wei J, Cui YH, Park G, Shah P, Deng Y, et al. m6A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat Commun (2019) 10:2782. doi: 10.1038/s41467-019-10669-0
80. Yi L, Wu G, Guo L, Zou X, Huang P. Comprehensive Analysis of the PD-L1 and Immune Infiltrates of m6A RNA Methylation Regulators in Head and Neck Squamous Cell Carcinoma. Mol Ther Nucleic Acids (2020) 21:299–314. doi: 10.1016/j.omtn.2020.06.001
81. Yu R, Li Q, Feng Z, Cai L, Xu Q. m6A Reader YTHDF2 Regulates LPS-Induced Inflammatory Response. Int J Mol Sci (2019) 20:1323. doi: 10.3390/ijms20061323
82. Xiang Y, Laurent B, Hsu CH, Nachtergaele S, Lu Z, Sheng W, et al. RNA m6A methylation regulates the ultraviolet-induced DNA damage response. Nature (2017) 543:573–6. doi: 10.1038/nature21671
Keywords: YTH domain, YTHDF1-3, YTHDC1-2, m6A modification, cancer
Citation: Xu Y, Zhang W, Shen F, Yang X, Liu H, Dai S, Sun X, Huang J and Guo Q (2021) YTH Domain Proteins: A Family of m6A Readers in Cancer Progression. Front. Oncol. 11:629560. doi: 10.3389/fonc.2021.629560
Received: 15 November 2020; Accepted: 07 January 2021;
Published: 22 February 2021.
Edited by:
Gelina Kopeina, Lomonosov Moscow State University, RussiaCopyright © 2021 Xu, Zhang, Shen, Yang, Liu, Dai, Sun, Huang and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junxing Huang, hjxtz@sina.cn; Qing Guo, biglemonone@163.com
†These authors have contributed equally to this work
 Yirong Xu1,2†
Yirong Xu1,2† Xi Yang
Xi Yang Xinchen Sun
Xinchen Sun Qing Guo
Qing Guo