- 1Urology Department, The Affiliated Hospital of Qingdao University, Qingdao, China
- 2Laboratory of Medical Biology, Medical Research Center, The Affiliated Hospital of Qingdao University & The Biomedical Sciences Institute of Qingdao University (Qingdao Branch of SJTU Bio-X Institutes), Qingdao University, Qingdao, China
- 3Nursing Department, The Shengli College, China University of Petroleum, Dongying, China
Objectives: To evaluate copy number alterations (CNAs) in genes associated with penile cancer (PeC) and determine their correlation and prognostic ability with PeC.
Methods: Whole-exome sequencing was performed for tumor tissue and matched normal DNA of 35 patients diagnosed with penile squamous cell carcinoma from 2011 to 2016. Somatic CNAs were detected using the Genome Analysis Toolkit (GATK). Retrospective clinical data were collected and analyzed. All the data were statistically analyzed using SPSS 16.0 software. The cancer-specific survival rates were estimated by Kaplan-Meier curves and compared with the log-rank test.
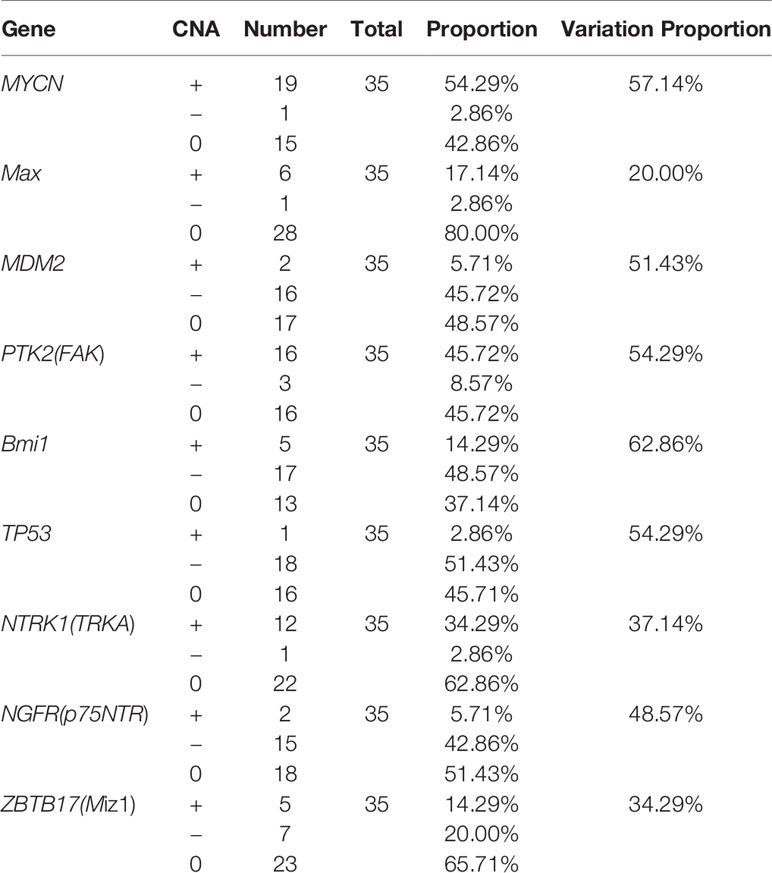
Results: CNAs in the MYCN gene was detected in 19 (amplification: 54.29%) patients. Other CNAs gene targets were FAK (amplification: 45.72%, deletion: 8.57%), TP53 (amplification: 2.86%, deletion: 51.43%), TRKA (amplification: 34.29%, deletion: 2.86%), p75NTR (amplification: 5.71%, deletion: 42.86%), Miz-1 (amplification: 14.29%, deletion: 20.00%), Max (amplification: 17.14%, deletion: 2.86%), Bmi1 (amplification:14.29%, deletion: 48.57%), and MDM2 (amplification: 5.71%, deletion: 45.72%). The CNAs in MYCN and FAK correlated significantly with patient prognosis (P<0.05). The 3-year Recurrence-free survival rate was 87.10% among patients followed up. The 5-year survival rate of patients with MYCN amplification was 69.2%, compared to 94.4% in the non-amplification group. The 5-year survival rate of patients with FAK amplification was 65.6%, compared to 94.7% in the non-amplification group. The PPI network showed that TP53 and MYCN might play meaningful functional roles in PeC.
Conclusion: MYCN and FAK amplification and TP53 deletion were apparent in PeC. MYCN and TP53 were hub genes in PeC. MYCN and FAK amplification was also detected and analyzed, and the findings indicated that these two genes are predictors of poor prognosis in PeC.
Introduction
Penile cancer (PeC) is a rare and aggressive malignant tumor that accounts for less than 1% of carcinomas in males in the United States (1). The regional differences in incidence are significant, with the high incidence in the developing countries (2.8–6.8 per 100,000), where the low rate of neonatal circumcision and socioeconomic conditions make the patients vulnerable to a variety of risk factors. Various have been identified, including lack of circumcision, phimosis, smoking, balanitis, obesity, lichen sclerosus, and psoralen UV-A phototherapy, contributing to the courses of PeC. Moreover, human papilloma virus (HPV) has been linked to nearly 40%–50% of cases (2), and the molecular mechanism. Several studies have detected somatic changes that arise in (3–5), but few of them were based on whole-exome sequencing (5). Our previous study has performed a whole-exome sequencing analysis of PeC, and identified recurrent mutations in 11 genes (6).
Copy number alterations (CNAs) are somatic changes that cause the amplification or deletion of DNA fragment (7, 8), and represent the most common alterations of cancer cells (7, 9, 10). They contribute to both onset and progression of cancer by inappropriate activation of proto-oncogenes and/or inactivation of tumor suppressor genes (11, 12). Similar to genome-wide association studies (GWAS) that help find single nucleotide polymorphisms associated with disease phenotypes, GWAS can be extended to CNAs to help find structural variations associated with human traits and diseases (13). To date, the CNAs of PIK3CA, IL-22 and MYC have been reported in PeC. PIK3CA copy number amplification was found to have no prognostic value for cancer-specific survival (14). The function of IL-22 copy number amplifications in PeC is not clear (15). However, MYC amplification increases during PeC progression (16–18).
MYCN, an MYC family member, is a proto-oncogene that is mainly expressed in primarily neuronal cell lineages during embryogenesis and could be involved in tumorigenesis when uninhibited (19). The MYCN oncogene is amplified in approximately 20% of neuroblastomas (NBs) (20). MYCN belongs to the Myc/Max/Mad/Mnt network of proteins that regulate proliferation, apoptosis, and differentiation (21), which were amplified in neuroblastoma, small-cell lung cancer (22) and hepatic cancer (23), respectively.
Because knowledge of the pathogenesis and carcinogenesis of PeC remains limited. In the present study, we performed a whole-exome sequencing analysis of PeC in Han Chinese patients to search for the relationship between CNAs and clinical characters in penile cancer.
Materials and Methods
Sample Source and Clinical Characteristics
Fresh PeC and blood samples from 35 PeC patients diagnosed from 2011 to 2016 were collected in the Affiliated Hospital of Qingdao University and frozen at constant temperature of −80°C. The patients were followed up by telephone and outpatient service to assess their health. All the tumor samples used for DNA extraction and exome sequencing were confirmed to have 80% tumor content by an experienced pathologist. PSCC diagnoses were made according to the clinical history, physical examination, and biopsy results. Primary treatment for PSCC included partial or radical penectomy with concomitant inguinal lymph node dissection (ILD), which included ipsilateral or ilioinguinal lymphadenectomy via contralateral superficial inguinal or ilioinguinal dissection according to the clinical condition. Recurrence-free survival was defined as the period from the time of present surgery in our hospital to tumor recurrence (or last follow-up visit).
Clinical information for 35 patients, including gender, age, patient number, sample acquisition method, tumor size, pathological subtype, differentiated degree, local infiltration status, WHO grade, and follow-up results (recurrence and survival), was collected and summarized in Table S1. Informed consent was obtained from all human subjects, and our study was approved by the Affiliated Hospital of Qingdao University Ethics Committee.
Whole-Exome Sequencing
In order to capture the tumor and blood exonic region, library preparation was performed with Agilent SureSelect Human All Exon V5+UTR Kits (Santa Clara, CA) according to the manufacturer’s guidelines. Samples were deeply sequenced on the Illumina HiSeq2500 platform. The mean per-target depth of coverage across all targets was 92, with 89% of targets sequenced to an average of 10× or greater.
CNAs Detection
Raw reads from each library were quality controlled with FastQC, trimmed using Trimmomatic and then mapped to the reference human genome (NCBI build 38, hg 38) by a Burrows Wheeler Alignment (BWA) tool v 0.7.17 (24) with the BWA-maximal exact match algorithm and default parameters. PCR duplicates were flagged with Picard, and the outputs were locally realigned using the Indel Realignment tool of the Genome Analysis Toolkit (GATK) version 4.1.2.0 (25). After local realignment, the BaseRecalibrator tool from GATK was used for recalibration. CNAs in normal samples were compared to matching tumor samples using a relative coverage method performed in GATK. GISTIC version 2.0 (http://archive.broadinstitute.org/cancer/cga/gistic) analysis was performed to identify significantly recurrent copy number amplification and deletion at focal level. A log2 ratio above 0.1 was considered as “amplification,” and a log2 ratio below -0.1 was considered as “deletion.” (https://gatkforums.broadinstitute.org/firecloud/discussion/8254/gistic2-0)
KEGG Pathway Analysis
The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database can be seen as a set of orthologue group tables including category pathways, subcategory pathways and secondary pathways, which are often encoded by positionally coupled genes on the chromosome and much meaningful in predicting gene functions (3). To analyze the possible relationship between related genes and PeC, we characterized somatic mutations of this pathway for PeC, which was absent in KEGG. One canonical signaling pathway, MYCN/Max, was found to be altered at varying frequencies in the different cancer types analyzed by KEGG (https://www.kegg.jp/kegg-bin/highlight_pathway?scale=1.0&map=map05202&keyword=MYCN).
Analysis of the PPI Network
The STRING database (http://string-db.org/) is a precomputed global resource for predicting functional associations between proteins. In this paper, the STRING online tool was applied to analyze the protein-protein interactions (PPIs) of the CNA genes in PeC with the threshold of combined score >0.4.
Statistical Analysis
All the data collected were statistically analyzed by SPSS 16.0 software. We divided the patients into two groups based on the CNA results: the normal target gene group and the abnormal target gene amplification group. Patient status information was obtained through outpatient or telephone follow-up. Chi-square analysis and Kaplan-Meier survival curves were used to compare the five-year survival rates among different groups, and significant differences were determined using the log-rank test. All P values were two-sided, and a P value < 0.05 was considered to indicate statistical significance. Figures were edited with Origin 8 software.
Results
Clinical Characteristics and Gene Variations
The median age of the 35 patients participating in exome sequencing analysis was 63 years (range 27–86 years). Twenty-three patients (65.71%) had redundant prepuce, 9 patients (25.71%) had phimosis, and nobody had been circumcised. All patients were diagnosed with penile squamous cell carcinoma. The majority of primary lesions presented on the corpus cavernosum (n = 16; 45.71%) and occasionally involved the adjacent structures (submucosa [n = 5; 14.29%], dartos [n = 1; 2.86%], subcutaneous soft tissue [n = 1; 2.86%]), and 5 specimens could not be defined. Sixteen (45.71%) patients were diagnosed with pT stage expressing T2 disease. The proportion of histologically well to high and moderately differentiated cases were 57.14% and 31.43%, separately. The proportion of samples with low differentiation was 8.82%, and 1 case was unknown. Partial penectomy was performed in the majority of cases (n = 29; 82.86%). Five patients also underwent inguinal lymph node dissection in addition to surgical removal of the lesion, and the result showed that 2 cases were positive. The median follow-up time was 60 months, and the number of patients lost to follow-up was 4. Recurrence occurred in four patients within 3 years after surgery and The 3-year Recurrence-free survival rate was 87.10%. The overall 3- and 5-year cancer-specific survival (CSS) rates were 87.10% and 83.87%, respectively.
For HPV detection, only 30 samples were tested for HPV and five were not. Among 30 PSCC samples, six were found to be HPV-positive. Moreover, five were HPV type 16 and one appeared HPV types 18 and 81. Clinicopathologic data were obtained from electronic clinical medical records and are shown in Supporting Information Table S1.
The average sequencing depth was 120× for the tumor samples and 70× for matched normal blood samples, with ≥10× coverage for 89.0% of the target regions in tumor samples and 88.7% of the target regions in blood samples (Figure 1 and Table S1).
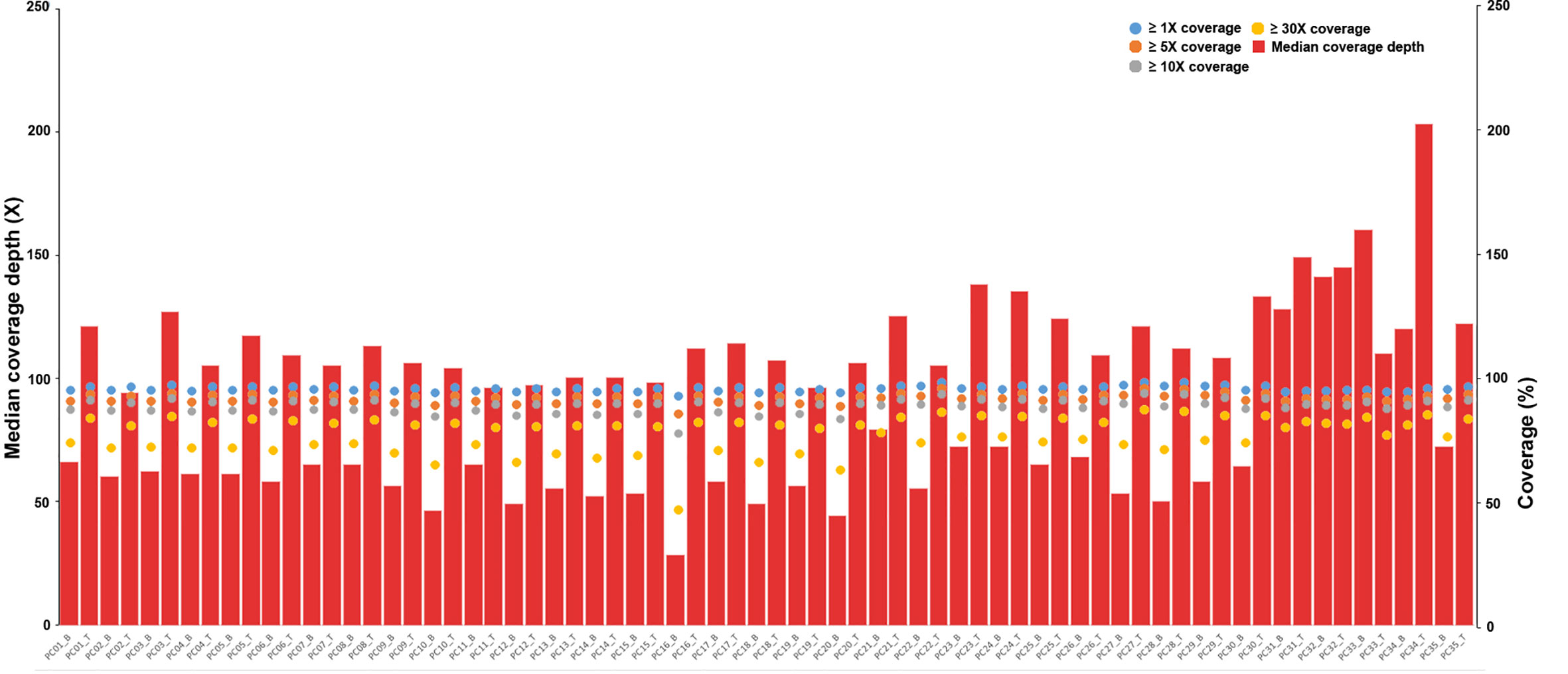
Figure 1 Depth and coverage of 35 paired samples. The histograms represent the average sequencing depth of each sample, and the specific values are shown on the left axis. The scatter plot shows the distribution of all samples under coverage values of 1×, 5×, 10× and 30×, and the specific values refer to the right axis.
Mutational Features and Pathway Alterations Relative to PeC
The mutational features of the PeC sample were also assessed. These features can be indicative of specific mutagenic mechanisms promoting tumorigenesis. A gene set analysis was performed with KEGG to determine pathways associated with PeC. In our PeC samples, this pathway had at least one alteration, and most alterations were found in 19 tumors (54.29% of samples). The relative pathway targets involve 10 kinds of proteins (Figure 2A). Nine target proteins were detected in PeC samples, except for sp-1 in the known pathway. Most mutations caused amplification of the affected genes, including MYCN, Max, FAK, SP-1 and TRKA, while MDM2, Bmi1,TP53, p75NTR, and Miz-1were more likely to exhibits downregulation levels.
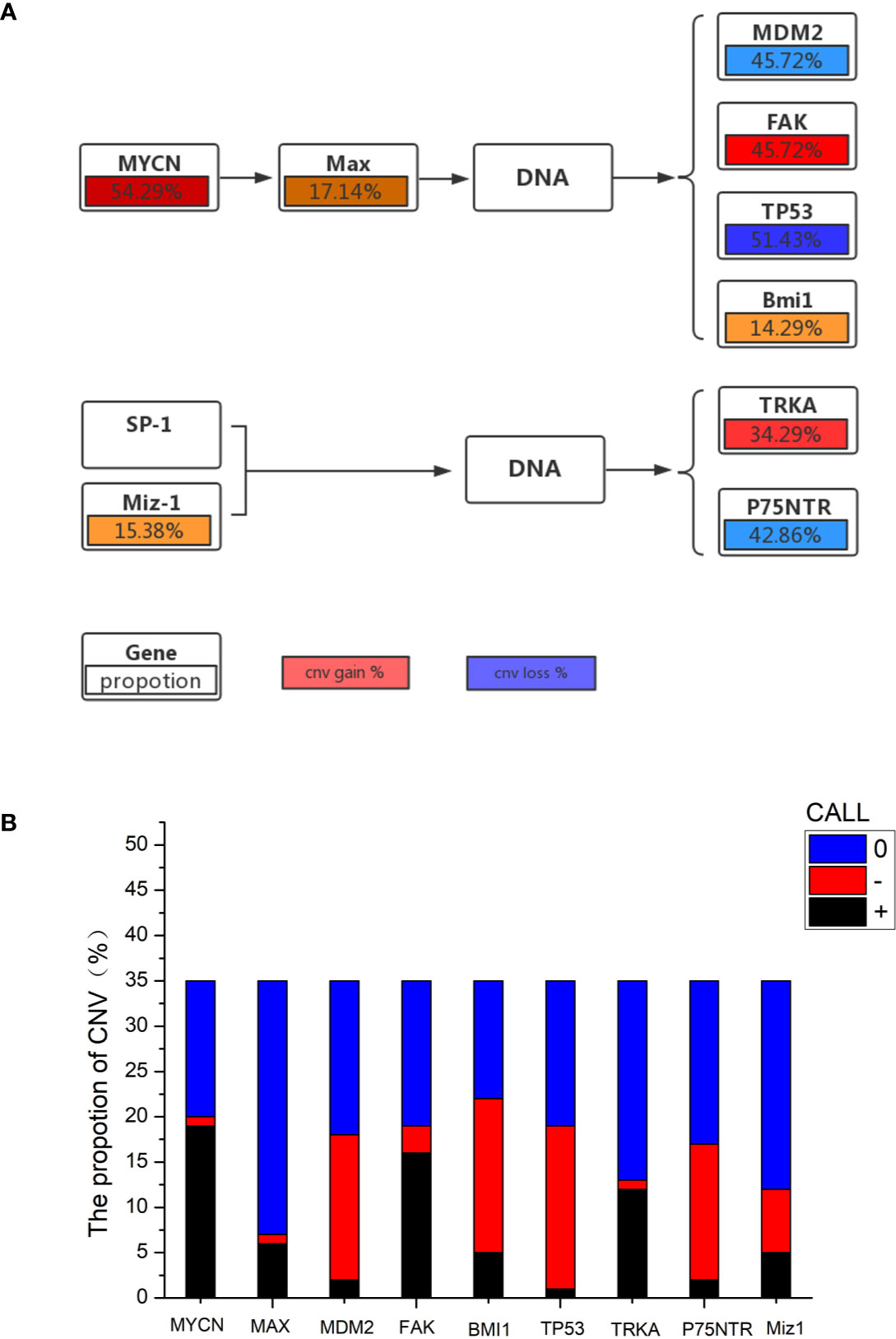
Figure 2 (A) Proportion of gene copy number variation in the KEGG pathway. (B) Proportion of gene copy number variation: 0: normal gene number; –: gene copy number deletion. +: gene copy number amplification.
The copy number amplification of MYCN was 19 (54.29%), which is the largest number of mutations in the analysis. The remaining targets were FAK (amplification: 45.72%, deletion: 8.57%), TP53 (amplification: 2.86%, deletion: 51.43%), TRKA (amplification: 34.29%, deletion: 2.86%), p75NTR (amplification: 5.71%, deletion: 42.86%), Miz-1 (amplification: 14.29%, deletion: 20.00%), Max (amplification: 17.14%, deletion: 2.86%), Bmi1(amplification:14.29%, deletion: 48.57%), and MDM2 (amplification: 5.71%, deletion: 45.72%). Therein, the TP53 and p75NTR mutations were inactivating, and the remaining targets were amplified. Detailed information about the samples for the exome sequencing analysis can be found in Supporting Information Table 1 and Figure 2B.
Correlation and Prognosis Between CNAs and PeC
The 35 patients’ follow-up information was obtained through outpatient and telephone visits. The rate of lost follow-up was 11.4%, and four patients could not be contacted. Finally, only 31 patients were included to analyze the correlation between prognosis and gene mutations. According to the results of the Kaplan-Meier survival curve, the five-year survival rates of the MYCN amplification and MYCN non-amplification groups were 69.2% and 94.4%, respectively. Simultaneously, the P value from the log-rank test was 0.047<0.05, which means that the difference between the groups with significantly correlation and MYCN was an independent prognostic factor of PeC (Figure 3A). Analogously, the five-year survival rates of the FAK non-amplification group and the FAK amplification group were 94.7% and 65.6%, respectively. The P value from the log-rank test was 0.032<0.05, which means that the difference between the groups was statistically significant, and FAK amplification could act as an independent prognostic factor of PeC (Figure 3B). However, the five-year survival rates of the normal TP53 group and TP53 inactivation group were 88.9% and 76.9%, respectively. The P value from the log-rank test was 0.329<0.05, which means that the difference between the groups without correlation and TP53 inactivation was not the prognostic value of PeC (Figure 3C).
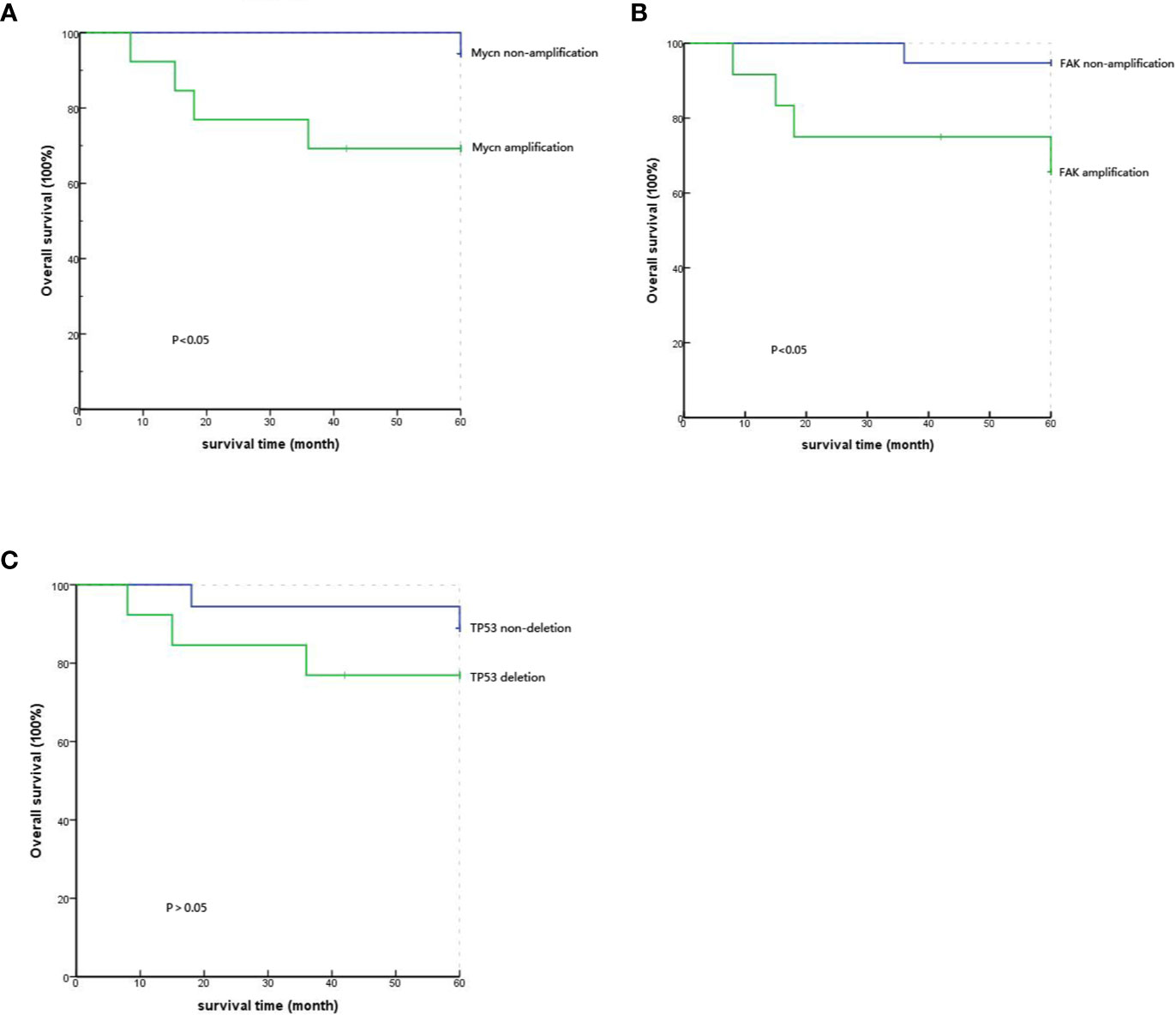
Figure 3 Comparison of the 5-year survival rate with different gene variations by Kaplan-Meier analysis. (A) Comparison of the 5-year survival rate between MYCN amplification and MYCN non-amplification showed significant difference (P=0.047). (B) Comparison of the 5-year survival rate between FAK amplification and FAK non-amplification showed a significant difference (P=0.032). (C) Comparison of the 5-year survival rate between TP53 deletion and TP53 nondeletion showed no significant difference (P=0.329).
PPI Network Constructed for CNA Genes in PeC
We constructed a PPI network of proteins encoded by CNA genes in PeC based on the PPI network, and the present study identified the top 2 hub genes: TP53 and MYCN (Figure 4). These two genes might play meaningful functional roles in PeC.
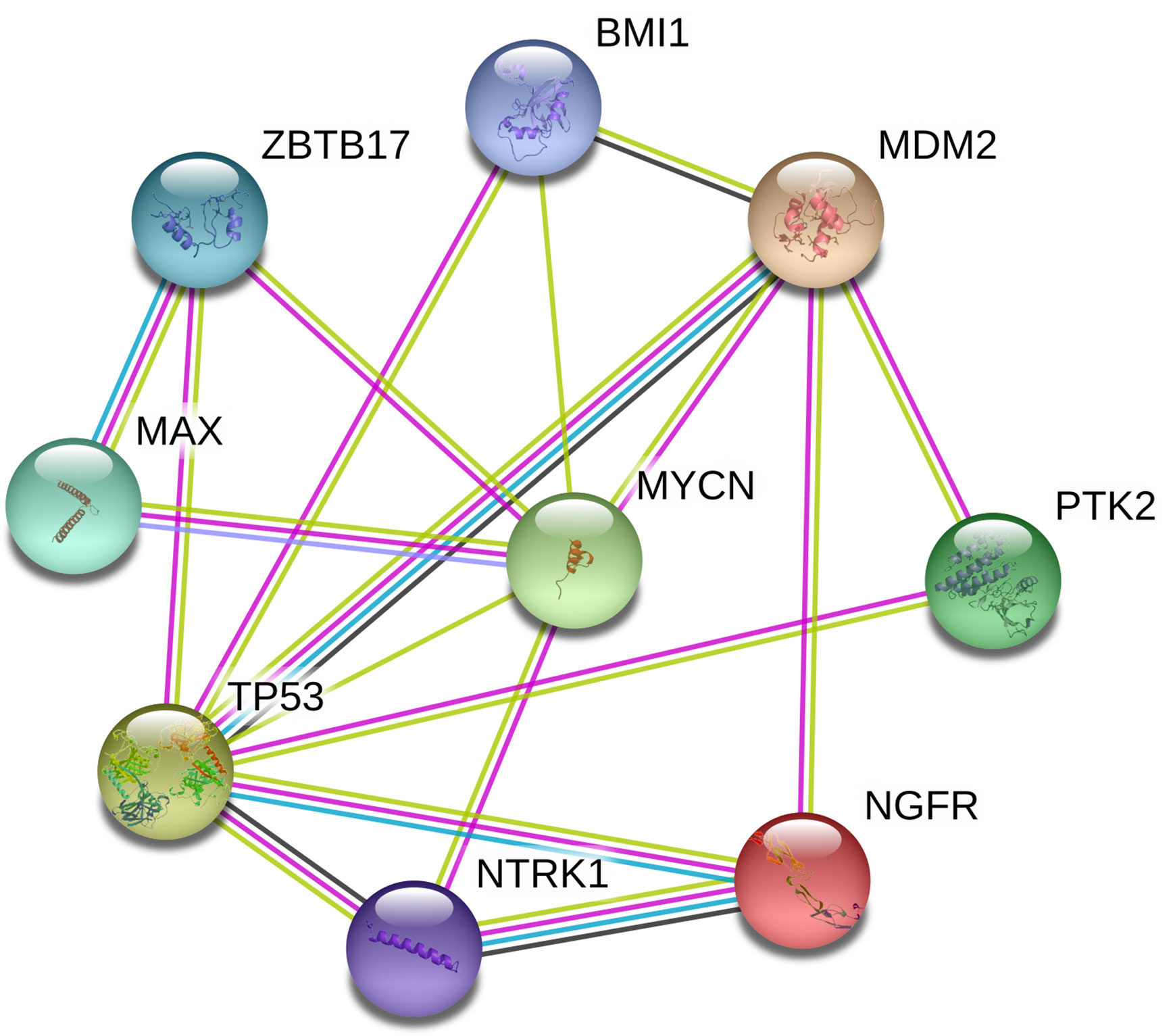
Figure 4 Protein-protein interaction network of copy number alteration (CNA) genes in penile cancer (PeC). Circles represent genes, lines represent the interaction of proteins encoded by the genes, and the results within the circle represent the structures of the proteins. The line color represents evidence of the interaction between the proteins.
Discussion
PeC is a rare malignancy in the developed world but is much more common in developing countries. The genetic and molecular basis of PeC is still poorly understood (5), and further understanding of these aspects is important to improve our ability to diagnose, treat and prevent PeC. In our previous study (6), we characterized the PSCC genomic landscape using whole-exome sequencing. Of the 30 paired blood and tumor samples, recurrent mutations were identified in 11 genes; we also observed the frequently altered pathways for potential. The present study was based on the previous study in the samples and another five samples was added to the present study but not done HPV detection. SNPs have been reported in the previous study (6), so we’re not going to repeat it here.
Overall survival and recurrence-free survival in our study were higher than in Jong Won’ s study (26), the possible reason for which was that the pathological stage of the patients in our study were all≤T2, while the pathological stage ≥T3 accounts for 30.2% in Jong Won’ s study resulting to the lower OS and RFS. The patients in our study had earlier stages of pathology, perhaps because modern people in our region are more concerned about their health.
Currently, significant poor prognosticator in patients with penile cancer include lymph node positivity (27), metastatic nodes≥ 4 (28), AJCC stage ≥ III disease (26), pathologic stage of the primary tumor (29), histologic grade<G1 (30, 31), a tumor thickness≥ 5 mm (32), vertical growth pattern (33), age<53, Lactate Dehydrogenase (LDH)<188.5 U/L and Platelet-lymphocyte Ratio (PLR)<133.5 (34), p53 positivity (31), Human papillomavirus infection (35). However, there are few studies on the prognosis of penile cancer at the gene level, so our study also studied the gene types of prognostic factors later.
Our results provide the first detection of a MYCN CNA in the somatic mutant spectrum of PeC in Chinese men through whole-exome sequencing. In our sample, the MYCN amplification detection rate was 54.29%; MYCN is the type of gene in which we detected the highest mutation rate. In addition, the CNAs associated with FAK amplification and TP53 deletion were found in more than one-third of the total samples tested. Furthermore, variations in other genes, such as Max, Miz-1, Bmi1, and p75NTR, were found in more than 10% of the samples.
The MYCN oncogene plays an important role in human tumorigenesis and has been proven to bind to gene promoters for various oncogenes involved in multiple life activities (36) and to increase the expression of many downstream targets. Previous data have indicated that the primary function of MYCN is as a transcription factor known to specifically bind the DNA E-box sequence CACGTG (37). A later study supports a dual role for MYCN. Murphy et al. showed that MYCN binds more often to the CATGTG E-box sequence, and MYCN binding is associated with DNA hypermethylation and can therefore also serve as an intermediary for chromatin structure-mediated regulation of various cellular processes, including cell growth, cell proliferation and cell differentiation (38). As a hub gene identified by our PPI study, MYCN plays an important regulatory role in its related pathways.
A previous study (39) found that MYCN amplification is one of the most significant prognostic indicators of neuroblastoma and is associated with rapid tumor progression and poor prognosis. Similarly, our data analysis found that MYCN was correlated with the prognosis of PeC, MYCN amplification indicated a poor prognosis. Lloveras B’s study (40) found that the MYC copy number amplification was significantly associated with poor outcome (mortality, node metastasis and/or recurrence) in PeC. As a member of the MYC family of proto-oncogenes, MYCN is more likely to appear as an independent prognostic indicator of PeC if the sample size is increased.
Studies (36) found that in neuroblastoma, the relationship between MYCN amplification and cell activity and aggressiveness suggests a potential relationship between focal adhesion kinase (FAK) and MYCN, since FAK is a key protein involved in cell activity.
FAK is a nonreceptor protein tyrosine kinase that targets focal adhesion and controls multiple cellular signaling pathways, including proliferation and survival (41). The inhibition of FAK activation has been found to affect a number of cellular pathways (36). FAK overexpression has also been shown in human sarcoma and melanoma tumors (42). A previous study (43) found that MYCN binds to the FAK promoter in vitro and in vivo, resulting in upregulation of FAK with electrophoretic mobility shift, chromatin immunoprecipitation (ChIP), and dual luciferase assays. Therefore, it is well proven that MYCN-regulated FAK intervention affects the prognosis of patients, which is consistent with our finding that FAK amplification could be an independent prognostic factor of PeC.
Cloning and evaluation of the FAK promoter has shown that it has many binding sites for various oncogenes, such as TP53 (44). The tumor protein TP53 (TP53 or TTP53) was the first tumor suppressor gene, discovered in 1979, and is the guardian of the genome (45). TP53 is the most widely studied tumor suppressor gene, playing an important role in inhibiting tumor development. The function of the TP53 gene is to inhibit cell proliferation in response to DNA damage. By regulating target genes, TP53 induces a variety of cellular responses, including growth arrest, senescence, and apoptosis (46, 47). However, the mutated TP53 protein loses its protection against genomic functions, including the ability to inhibit cell proliferation and induce apoptosis, when mutated (48). Mutations in the TP53 gene occur in most malignant tumors, such as lung cancer (49) and breast cancer (50). At the genetic level, carcinogenesis is a multistep process in which both oncogene activation and tumor suppressor gene inactivation are involved. Examination of the samples revealed a large number of SNP mutations in TP53 in PeC, and the most common mutation classifications were missense mutation and nonsense mutation. These may account for the role of TP53 in the molecular etiology of PeC.
In this study, a large number of gene variants of the samples were found in the MYCN/Max pathway in PeC, especially in MYCN. In addition, the MYCN and FAK CNA is associated with the prognosis of PeC, and its high expression level indicates a poor prognosis. However, mutations in TP53 were not found to be related to prognosis, perhaps because our sample size was insufficient; it is necessary to carry out relevant tests with a larger sample size in the future.
Data Availability Statement
According to national legislation/guidelines, specifically the Administrative Regulations of the People’s Republic of China on Human Genetic Resources (http://www.gov.cn/zhengce/content/2019-06/10/content_5398829.htm, http://english.www.gov.cn/policies/latest_releases/2019/06/10/content_281476708945462.htm), no additional raw data is available at this time. Data of this project can be accessed after an approval application to the China National Genebank (CNGB, https://db.cngb.org/cnsa/). Please refer to https://db.cngb.org/, or email: CNGBdb@cngb.org for detailed application guidance. The accession code CNP0001368 should be included in the application.
Ethics Statement
The studies involving human participants were reviewed and approved by the Affiliated Hospital of Qingdao University Ethics Committee. The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author Contributions
YY, CG, and ZL contributed conception and design of the study. YY and YC organized the database. MW performed the statistical analysis. YY wrote the first draft of the manuscript. CG, YC, MW, and JZ wrote sections of the manuscript. XM, SL, and HY read and revised the manuscript. HN provided approval for publication of the content. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (82071750, 81772713, 81472411, 81372752, 81401899, 81871055, 81701321, 81421061, 31325014, 81130022, 21375139, 31571012 and 81501154), Taishan Scholar Program of Shandong Province (tsqn20161077), National Basic Research Program of China (973 Program; 2015CB559100), National Key R&D Program of China (2016YFC0903402, 2016YFC1306903, 2016YFC0902403, and 2017YFC0908105), Program of Shanghai Academic Research Leader (15XD1502200), Natural Science Foundation of Shandong Province (ZR2014HM088 and ZR2016HQ18), Major Science and technology innovation project of Shandong Province (2019JZZY021002), Key projects of Qingdao Science and Technology Program (18-6-1-64-nsh), Key Research and Development Program of Shandong Province (2018GSF118197), China Postdoctoral Science Foundation (2017M622144), Qingdao Postdoctoral Application Research Project, and Qingdao Young Scientist Applied Basic Research Fund (15-9-1-51-jch and 15-9-1-105-jch).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.596261/full#supplementary-material
References
1. Douglawi A, Masterson TA. PeC epidemiology and risk factors: a contemporary review. Curr Opin Urol (2018) 29(2):1. doi: 10.1097/MOU.0000000000000581
2. Yu YB, Wang YH, Yang XC, Zhao Y, Niu HT. The relationship between human papillomavirus and PeC over the past decade: a systematic review and meta-analysis. Asian J Androl (2019) 21(4):375–80. doi: 10.4103/aja.aja_39_19
3. Andersson P, Kolaric A, Windahl T, Kirrander P, Söderkvist P, Karlsson MG. PIK3CA, HRAS and KRAS gene mutations in human penile cancer. J Urol (2008) 179(5):2030–4. doi: 10.1016/j.juro.2007.12.040
4. Annunziata C, Buonaguro L, Losito S. Somatic mutations of STK11 gene in human papillomavirus positive and negative penile cancer. Infect Agents Cancer (2013) 8:6. doi: 10.1186/1750-9378-8-2
5. Feber A, Worth DC, Chakravarthy A, De WP, Shah K, Arya M, et al. CSN1 Somatic Mutations in Penile Squamous Cell Carcinoma. Cancer Res (2016) 76(16):4720–7. doi: 10.1158/0008-5472.CAN-15-3134
6. Wang Y, Wang K, Chen Y, Zhou J, Liang Y, Yang X, et al. Mutational landscape of penile squamous cell carcinoma in a Chinese population. Int J Cancer (2019) 145(5):1280–9. doi: 10.1002/ijc.32373
7. Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The landscape of somatic copy-number alteration across human cancers. Nature (2010) 463(7283):899–905. doi: 10.1038/nature08822
8. Redon R. Global variation in copy number in the human genome. Nature (2006) 444(7118):444–54. doi: 10.1038/nature05329
9. Kim TM, Xi CH, Luquette LJ, Park RW, Johnson MD, Park PJ. Functional genomic analysis of chromosomal aberrations in a compendium of 8000 cancer genomes. Genome Res (2013) 23(2):217–27. doi: 10.1101/gr.140301.112
10. Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature (2015) 518(7540):495–501. doi: 10.1038/nature14169
11. The Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature (2012) 489(7417):519–25. doi: 10.1038/nature11404
12. Nik-Zainal S, Van Loo P, Wedge DC, Alexandrov LB, Greenman CD, Lau KW, et al. The Life History of 21 Breast Cancers. Cell (2012) 149(5):14. doi: 10.1016/j.cell.2012.04.023
13. Larsen SJ, do Canto LM, Rogatto SR, Jan B. CoNVaQ: a web tool for copy number variation-based association studies. BMC Genomics (2018) 19(1):369. doi: 10.1186/s12864-018-4732-8
14. Adimonye A, Stankiewicz E, Touche S, Kudahetti S, Tinwell B, Corbishley C, et al. The Prognostic Value of PIK3CA Copy Number Gain in PeC. Urology (2018) S0090-4295(18)30560-0. doi: 10.1016/j.urology.2018.03.056
15. Marchi FA, Martins DC, Barros-Filho MC, Kuasne H, Busso LA, Brentani H, et al. Multidimensional integrative analysis uncovers driver candidates and biomarkers in penile carcinoma. Sci Rep (2017) 7:11. doi: 10.1038/s41598-017-06659-1
16. Kayes O, Minhas S. Biomarker discovery in PeC. J Urol (2012) 188(5):1660–1. doi: 10.1016/j.juro.2012.08.056
17. Masferrer E, Ferrándiz-Pulido C, Lloveras B, Masferrer-Niubò M, Espinet B, Salido M, et al. MYC Copy Number Gains are Associated with Poor Outcome in Penile Squamous Cell Carcinoma. J Urol (2012) 188(5):1965–71. doi: 10.1016/j.juro.2012.07.003
18. Busso-Lopes AF, Marchi FA, Kuasne H, Scapulatempo-Neto C, Trindade-Filho JCS, De Jesus CMN, et al. Genomic Profiling of Human Penile Carcinoma Predicts Worse Prognosis and Survival. Cancer Prev Res (2015) 8(2):149–56. doi: 10.1158/1940-6207.CAPR-14-0284
19. Lovén J, Zinin N, Wahlström T, Muller I, Brodin P, Fredlund E, et al. MYCN-regulated microRNAs repress estrogen receptor-alpha (ESR1) expression andneuronal differentiation in human neuroblastoma. Proc Natl Acad Sci USA (2010) 107(4):1553–8. doi: 10.1073/pnas.0913517107
20. Matthay KK, Maris JM, Schleiermacher G, Nakagawara A, Mackall CL, Diller L, et al. Neuroblastoma. Nat Rev Dis Primers (2016) 2:16078. doi: 10.1038/nrdp.2016.78
21. Smith AG, Popov N, Imreh M, Axelson HK, Henriksson M. Expression and DNA-binding activity of MYCN/Max and Mnt/Max during induced differentiation of human neuroblastoma cells. J Cell Biochem (2004) 92(6):1282–95. doi: 10.1002/jcb.20121
22. Ruizpérez MV, Albihn A, Arsenianhenriksson M. MYC Oncogene. In: Schwab M, editor. Encyclopedia of Cancer. Berlin, Heidelberg:Springer Berlin Heidelberg (2017). p.2976–82.
23. Yasukawa K, Liew LC, Hagiwara K, Hironaka-Mitsuhashi A, Qin XY, Furutani Y, et al. MicroRNA-493-5p-mediated repression of the MYCNoncogene inhibits hepatic cancer cell growth and invasion. Cancer Sci (2020) 111(3):869–80. doi: 10.1111/cas.14292
24. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheelertransform. Bioinformatics (2009) 25(14):1754–60. doi: 10.1093/bioinformatics/btp324
25. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res (2010) 20(9):1297–303. doi: 10.1101/gr.107524.110
26. Kim JW, Kim YS, Ko WJ, Choi YD, Hong SJ, Chung BH, et al. Prognostic Factors of Penile Cancer and the Efficacy of Adjuvant Treatment after Penectomy: Results from a Multi-institution Study. J Korean Med Sci (2018) 33(37):e233. doi: 10.3346/jkms.2018.33.e233
27. Lopes A, Hidalgo GS, Kowalski LP, Torloni H, Rossi BM, Fonseca FP. Prognostic Factors in Carcinoma of the Penis: Multivariate Analysis of 145 Patients Treated with Amputation and Lymphadenectomy. J Urol (2015) 156(5):1637. doi: 10.1016/S0022-5347(01)65471-5
28. Pandey D, Mahajan V, Kannan RR. Prognostic factors in node-positive carcinoma of thepenis. J Surg Oncol (2010) 93(2):133–8. doi: 10.1002/jso.20414
29. Lont AP, Kroon BK, Horenblas S, Gallee MPW, Berkhof J, Meijer CJLM, et al. Presence of high-risk human papillomavirus DNA in penile carcinoma predicts favorable outcome in survival. Int J Cancer (2010) 119(5):1078–81. doi: 10.1002/ijc.21961
30. Ficarra V, Zattoni F, Cunico SC, Galetti TP, Luciani L, Fandella A, et al. Lymphatic and vascular embolizations are independent predictive variables of inguinal lymph node involvement in patients with squamous cell carcinoma of the penis. Cancer (2010) 103(12):2507–16. doi: 10.1002/cncr.21076
31. Lopes A, Bezerra ALR, Pinto CAL, Serrano SV, De Mello CA, Villa LL, et al. p53 as a New Prognostic Factor for Lymph Node Metastasis in Penile Carcinoma: Analysis of 82 Patients Treated with Amputation and Bilateral Lymphadenectomy. J Urol (2002) 168(1):81–6. doi: 10.1016/S0022-5347(05)64836-7
32. Slaton JW, Morgenstern N, Levy DA, Santos MJ, Tamboli P, Ro JY, et al. Tumor Stage, Vascular Invasion and The Percentage of Poorly Differentiated Cancer: Independent Prognosticators for Inguinal Lymph Node Metastasis In Penile Squamous Cancer. J Urol (2001) 165(4):1138–42. doi: 10.1016/S0022-5347(05)66450-6
33. Villavicencio H, Rubio-Briones J, Regalado R, Chéchile G, Palou J. Grade, local stage and growth pattern as prognostic factors in carcinoma of the penis. Eur Urol (1997) 32(4):442–7. doi: 10.1159/000480804
34. Hu C, Bai Y, Li J, Zhang G, Yang L, Bi C, et al. Prognostic value of systemic inflammatory factors NLR, LMR, PLR and LDH in penile cancer. BMC Urol (2020) 20(1):57. doi: 10.1186/s12894-020-00628-z
35. Bezerra AL, Lopes A, Santiago GH, Ribeiro KC, Latorre MR, Villa LL. Human papillomavirus as a prognostic factor in carcinoma of thepenis. Cancer (2001)91(12):2315–21. doi: 10.1002/1097-0142(20010615)91:12<2315::AID-CNCR1263>3.0.CO;2-C
36. Megison ML, Stewart JE, Nabers HC, Gillory LA, Beierle EA. FAK inhibition decreases cell invasion, migration and metastasis in MYCN amplified neuroblastoma. Clin Exp Metastasis (2012) 30(5):555–68. doi: 10.1007/s10585-012-9560-7
37. Yang J, Sung E, Donlin-Asp PG, Corces VG. A subset of Drosophila Myc sites remain associated with mitotic chromosomes colocalized with insulator proteins. Nat Commun (2013) 4:1464. doi: 10.1038/ncomms2469
38. Murphy DM, Buckley PG, Bryan K, Sudipto D, Leah A, Foley NH, et al. Global MYCN transcription factor binding analysis in neuroblastoma reveals association with distinct E-box motifs and regions of DNA hypermethylation. PLoS One (2009) 4(12):e8154. doi: 10.1371/journal.pone.0008154
39. Fulda S, Poremba C, Berwanger B, Hacker S, Eilers M, Christiansen H, et al. Loss of Caspase-8 Expression Does Not Correlate with MYCN Amplification, Aggressive Disease, or Prognosis in Neuroblastoma. Cancer Res (2006) 66(20):10016–23. doi: 10.1158/0008-5472.CAN-05-4079
40. Masferrer E, Ferrándiz-Pulido C, Lloveras B, Masferrer-Niubò M, Espinet B, Salido M, et al. MYC Copy Number Gains are Associated with Poor Outcome in Penile Squamous Cell Carcinoma. J Urol (2012) 188(5):1965–71. doi: 10.1016/j.juro.2012.07.003
41. Dunty JM, Schaller MD, Gabarra-Niecko V. FAK regulates biological processes important for the pathogenesis of cancer. Cancer Metastasis Rev (2003) 22(4):359–74. doi: 10.1023/A:1023725029589
42. Hanks SK, Ryzhova L, Shin NY, Brabek J. Focal adhesion kinase signaling activities and their implications in the control of cell survival and motility. Front Biosci (2003) 8(4):d982–96. doi: 10.2741/1114
43. Beierle EA, Trujillo A, Nagaram A, Kurenova EV, Finch R, Ma X, et al. N-MYC regulates focal adhesion kinase expression in human neuroblastoma. J Biol Chem (2007) 282(17):12503–16. doi: 10.1074/jbc.M701450200
44. Beierle EA. MYCN, neuroblastoma and focal adhesion kinase (FAK). Front Biosci (Elite Ed) (2011) 3(2):421–33. doi: 10.2741/e257
45. Saha MN, Qiu L, Chang H. Targeting TP53by small molecules in hematological malignancies. J Hematol Oncol (2013) 6(1):23. doi: 10.1186/1756-8722-6-23
46. Ma JT, Han CB, Zhao JZ, Jing W, Zhou Y, Huang L-T, et al. Synergistic cytotoxic effects of recombinant human adenovirus TP53and radiation at various time points in A549 lung adenocarcinoma cells. Oncol Lett (2012) 4(3):529–33. doi: 10.3892/ol.2012.747
47. Guan Y-s, Liu Y, Zou Q, He Q, La Z, Yang L, et al. Adenovirus-mediated wild-type TP53gene transfer in combination with bronchial arterial infusion for treatment of advanced non-small-cell lung cancer, one year follow-up. J Zhejiang Universityence B (2009) 10(5):331–40. doi: 10.1631/jzus.B0820248
48. Rousseau B, Jacquot C, Palabe JL, Malleter M, Roussakis C. TP53transcription factor for the NEDD9/HEF1/Cas-L gene: potential targets in Non-Small Cell Lung Cancer treatment. Sci Rep (2015) 5:10356. doi: 10.1038/srep10356
49. Liu K, Gao W, Lin J. Effect of the TP53α gene on thechemosensitivity of the H1299 human lung adenocarcinoma cell line. Oncol Lett (2017) 14(2):1411–8. doi: 10.3892/ol.2017.6356
Keywords: penile cancer, copy number alterations, MYCN, FAK, TP53
Citation: Yu Y, Gao C, Chen Y, Wang M, Zhang J, Ma X, Liu S, Yuan H, Li Z and Niu H (2020) Copy Number Analysis Reveal Genetic Risks of Penile Cancer. Front. Oncol. 10:596261. doi: 10.3389/fonc.2020.596261
Received: 18 August 2020; Accepted: 10 November 2020;
Published: 14 December 2020.
Edited by:
Adam R. Metwalli, Howard University Hospital, United StatesReviewed by:
Mohummad Minhaj Siddiqui, University of Maryland School of Medicine, United StatesPatrick T. Gomella, National Institutes of Health (NIH), United States
Copyright © 2020 Yu, Gao, Chen, Wang, Zhang, Ma, Liu, Yuan, Li and Niu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiqiang Li, lizqsjtu@163.com; Haitao Niu, niuht0532@126.com
†These authors have contributed equally to this work
 Yongbo Yu
Yongbo Yu Chengwen Gao1,2†
Chengwen Gao1,2†