- 1The College of Basic Medicine, Shaanxi University of Chinese Medicine, Xianyang, China
- 2The Second Affiliated Hospital, Shaanxi University of Chinese Medicine, Xianyang, China
- 3The Affiliated Hospital, Shaanxi University of Chinese Medicine, Xianyang, China
- 4The College of Pharmacy, Shaanxi University of Chinese Medicine, Xianyang, China
- 5Department of Neurology, The Second Affiliated Hospital of Xi'an Medical University, Xi'an, China
Background: Adenylyl cyclase type 9 (ADCY9) modulates signal transduction by producing the second messenger cyclic AMP. It has been reported that ADCY9 gene polymorphisms were associated with cancer development. The aim of this study was to investigate whether ADCY9 gene polymorphisms could contribute to the susceptibility of hepatocellular carcinoma (HCC) in the Chinese Han population.
Methods: In the present study, five single-nucleotide polymorphisms (SNPs) in ADCY9 were genotyped using Agena MassARRAY platform in 876 subjects from China. Logistic regression was used to assess the effects of SNPs on HCC risk. Associations were also evaluated for HCC risk stratified by age and gender. False discovery rate (FDR) was used to correct multiple testing.
Results: After adjusting for age and gender, we found a significant relationship between heterozygous genotypes of rs2531995 and HCC risk (OR = 1.34, 95% CI = 1.01–1.77, p = 0.045). ADCY9 rs2230742 had a strong relationship with lower risk of HCC in allele (p = 0.006), co-dominant (p = 0.023), dominant (p = 0.010), and additive (p = 0.006) models. Stratified analysis showed that rs879620 increased HCC risk and rs2230742 was associated with lower risk of HCC in the individuals aged 55 or younger, rs2531992 significantly decreased HCC risk in the elder group (age > 55). For women, rs2230742 and rs2230741 were significantly associated with HCC risk in multiple models (p < 0.05). FDR analysis showed that rs2230742 could protect individuals from HCC risk in the allele model (FDR-p = 0.030). In addition, haplotype analysis indicated that Crs879620Ars2230742Ars2230741 haplotype was a protective factor for HCC (OR = 0.67, 95% CI = 0.50–0.89, p = 0.007, FDR-p = 0.028).
Conclusion: Our findings suggest that ADCY9 gene polymorphisms are associated with HCC risk in the Chinese Han population.
Introduction
Hepatocellular carcinoma (HCC) is the most common malignant tumor in the liver (1). Although mechanism of hepatocellular carcinogenesis is still not fully clear, it is widely recognized that HCC development is the consequence of complex interactions between the genome, lifestyle, and environment (2). Genome-wide association studies have highlighted that genetic variants might play a vital role in HCC susceptibility (3). It is reported that HCC is the fifth most common cancer and the third leading cause of cancer mortality worldwide (4). The prevalence of HCC is the highest in East, Southeast Asia, and Sub-Saharan Africa, especially in China, which accounts for ~50% of all cases (5, 6). Moreover, the prevalence of HCC is increasing with age and in men (7, 8). Hence, understanding the effects of genetic variants in different populations is highly important for studying the mechanisms of HCC and applying these results to risk prediction.
The adenylyl cyclase type 9 (ADCY9) gene belongs to the adenylyl cyclase (AC) gene family, which produces the second messenger cyclic AMP (adenosine-3′,5′-monophosphate) in response to G protein-coupled receptors (GPCRs) activation and codes for the protein AC type 9, an integral membrane protein composed of 12 transmembrane segments (9, 10). Previous studies have shown that ADCY9 gene polymorphisms were related to the development of many diseases, including cardiovascular diseases, mood disorders, malaria, asthma, and allergy (2, 10–12). The potential function of ADCY9 in cancer development was also reported. Yongzhen et al. found that ADCY9 mutation altered bladder cancer development (13). The involvement of ADCY9 in phospholipase C signaling has effects on colorectal cancer progression and metastasis (14). Elevated expression of ADCY9 is a potential prognostic biomarker for patients with colon cancer (9). Nonetheless, there were no studies that focused on the relationship between ADCY9 gene polymorphisms and HCC risk.
Hence, we hypothesized that ADCY9 gene polymorphisms could play an important role in the progression of HCC. This case–control study was conducted to assess the effects of five single-nucleotide polymorphisms (SNPs) in ADCY9 gene on the risk of HCC among the Chinese Han population. To the best of our knowledge, this is the first study to investigate the impact of ADCY9 SNPs for HCC susceptibility in the Chinese Han population.
Materials and Methods
Study Subjects
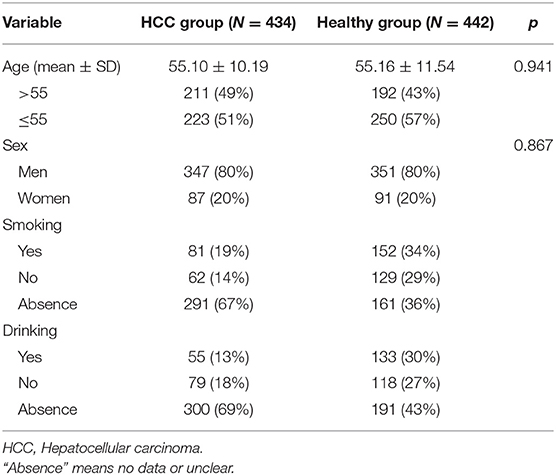
A total of 434 HCC patients and 442 age- and gender-matched healthy individuals were enrolled in this study. The patients were diagnosed as HCC by histology or pathology in the hospital, and the controls were healthy individuals without family history diseases, cardiovascular diseases, autoimmune diseases, respiratory diseases, cancers, or other severe diseases derived randomly from the same hospital. None of the patients had received radiation, chemotherapy, or surgical therapy before joining our study. Characteristic information was obtained from their medical records, including age, gender, smoking, and drinking status.
DNA Extraction and Genotyping
Peripheral blood samples were collected from the participants and were stored at −80°C until analysis. Genomic DNA was extracted from EDTA-containing blood via a blood DNA kit (GoldMag Co. Ltd., Xi'an, China). DNA concentration was measured by Nanodrop 2000 (Thermo Scientific, Waltham, Massachusetts, USA) (15). Combined with previous studies and the criteria of minor allele frequency (MAF) ≥ 0.05, five SNPs (rs2531995, rs879620, rs2230742, rs2230741, and rs2531992) were selected in this study based on the dbSNP database (https://www.ncbi.nlm.nih.gov/snp/) and HapMap database (http://www.hapmap.org). Primers were designed by MassARRAY Assay Design 3.0 software and were listed in Supplemental Table 1. SNP genotyping was performed by the MassARRAY iPLEX platform (Agena Bioscience, San Diego, CA, USA) (16). Finally, the data analysis was accomplished by Agena Bioscience TYPER version 4.0 software (17).
Genetic Models
For most SNPs, they often contain two types of alleles: a minor allele “A” with low frequency and a wild allele “B” with high frequency. Four genetic models were identified based on the alleles: co-dominant model BB vs. AB vs. AA, dominant BB vs. AB+AA, recessive BB+AB vs. AA, and log-additive: for each A increase. These models were applied to further detect the risk effect of minor allele on disease in a population. In the manuscript, we defined the allele with low frequency as the minor allele “A,” and the other was the wild allele “B.” Furthermore, four genetic models (codominant: BB vs. AB vs. AA, dominant: BB vs. AB+AA, recessive: BB+AB vs. AA, and log-additive: for each A increase) were employed using SNPstats software (https://www.snpstats.net/start.htm) to estimate the relationship between each SNP and EC risk.
Statistical Analysis
The statistical analyses were performed using the SPSS 17.0 (IBM®, Armonk, New York, USA). The Hardy–Weinberg equilibrium (HWE) in the healthy control was assessed by Fisher exact test. A chi-square test was performed to compare the categorical variable—sex. We used t-test to assess the difference in age. Association analysis based on logistic regression was performed by estimating odds ratios (OR) and 95% confidence intervals (CI) with multiple models for each SNP. We used Power and Sample Size Calculation software (http://sampsize.sourceforge.net/iface/s3.html#ccp) to calculate the power of the significant difference. False discovery rate (FDR) was used to correct multiple testing. The haplotype analysis and linkage disequilibrium (LD) were conducted by PLINK software and Haploview software (version 4.2) (18). p < 0.05 was considered statistically significant in all tests. In addition, HaploReg v4.1 (https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php) was used to predict the possible functional effects on the selected SNPs.
Results
Characteristics of the Participants
The characteristics of the 876 subjects (434 cases and 442 controls) are shown in Table 1. There was no significant difference in age (cases: 55.10 ± 10.19, controls: 55.16 ± 11.54; p = 0.941) and gender (cases: 80% men; controls: 80% men) between two groups. In addition, smoking and drinking status of all participants are presented in Table 1.
The Basic Information and Potential Function of the Selected SNPs
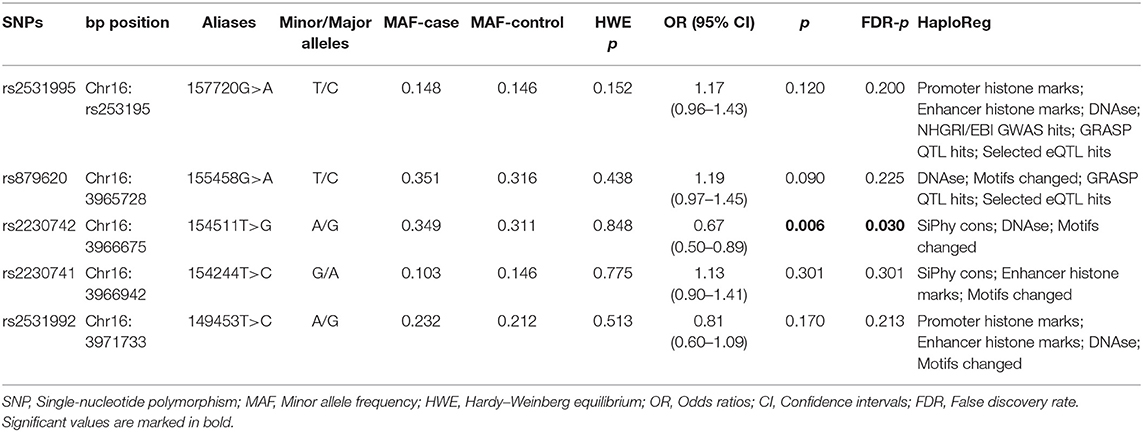
As shown in Table 2, the genotypic frequencies of the ADCY9 gene polymorphisms among the controls were in accord with HWE (p > 0.05) and the MAF of ADCY9 gene polymorphisms were larger than 0.05. In allelic tests, we found ADCY9 rs2230742 significantly decreased the risk of HCC (OR = 0.67, 95% CI = 0.50–0.89, p = 0.006, FDR-p = 0.030). However, there was no significant association between other SNPs and risk of HCC. Besides, these selected SNPs had association with “Promoter histone marks,” “Enhancer histone mark,” “SiPhy cons,” “DNAse,” “Motifs changed,” “NHGRI/EBI GWAS hits,” “GRASP QTL hits,” and “Selected eQTL hits” by online tool.
Association Between ADCY9 Gene Polymorphisms and Risk of HCC
Among the five SNPs in ADCY9 gene, rs2531995 and rs2230742 were significantly associated with HCC risk (Table 3). In contrast to homozygous wild-type alleles, heterozygote T/C of rs2531995 remarkably increased the risk of HCC (adjusted OR = 1.34, 95% CI = 1.01–1.77, p = 0.045). rs2230742 was identified to decrease the HCC risk in codominant (adjusted OR = 0.68, 95% CI = 0.49–0.95, p = 0.023), dominant (adjusted OR = 0.66, 95% CI = 0.48–0.90, p = 0.010), and additive (adjusted OR = 0.67, 95% CI = 0.50–0.89, p = 0.006) models. The other three SNPs showed no significant evidence of an association with HCC.
Stratified Analysis Based on Age and Gender
To further assess the five potential susceptible polymorphisms to the risk of HCC, a stratified analysis was performed by subgroups of participants' age and gender (Tables 4, 5). In the subgroup of age > 55, rs2531992 was associated with decreased HCC risk in additive (adjusted OR = 0.64, 95% CI = 0.42–0.99, p = 0.044) and allele (adjusted OR = 0.64, 95% CI = 0.42–0.99, p = 0.044) models. Furthermore, in the group of age ≤ 55, rs879620 was associated with higher risk of HCC in multiple models (dominant: adjusted OR = 1.50, 95% CI = 1.04–2.18, p =0.031; additive: adjusted OR = 1.35, 95% CI = 1.04–1.76, p = 0.026; allele: adjusted OR = 1.35, 95% CI =1.03–1.76, p = 0.028), whereas rs2230742 was related to lower risk of HCC. For the subgroup of gender, rs2230742 also significantly decreased HCC risk in the women group (dominant: adjusted OR = 0.43, 95% CI = 0.21–0.88, p =0.022; additive: adjusted OR = 0.42, 95% CI = 0.21–0.83, p = 0.012; allele: adjusted OR = 0.43, 95% CI = 0.22–0.83, p = 0.011). The rs2230741 polymorphism showed an increased relationship with HCC risk in dominant (adjusted OR = 1.91, 95% CI = 1.04–3.50, p = 0.037) and additive (adjusted OR = 1.78, 95% CI = 1.04–3.06, p = 0.037) models, and the minor allele G of rs2230741 was also related to increasing HCC risk (adjusted OR = 1.69, 95% CI = 1.01–2.80, p = 0.043) among women. It revealed that the effects of ADCY9 gene polymorphism on HCC risk were dependent on age or gender. Nevertheless, there was no significant association of these five polymorphisms with the risk of HCC in the men group. After FDR correction, no remarkable linkages were shown in the subgroups, suggesting that the differences in age or gender may not affect the relationship of ADCY9 gene polymorphism and the susceptibility of HCC.
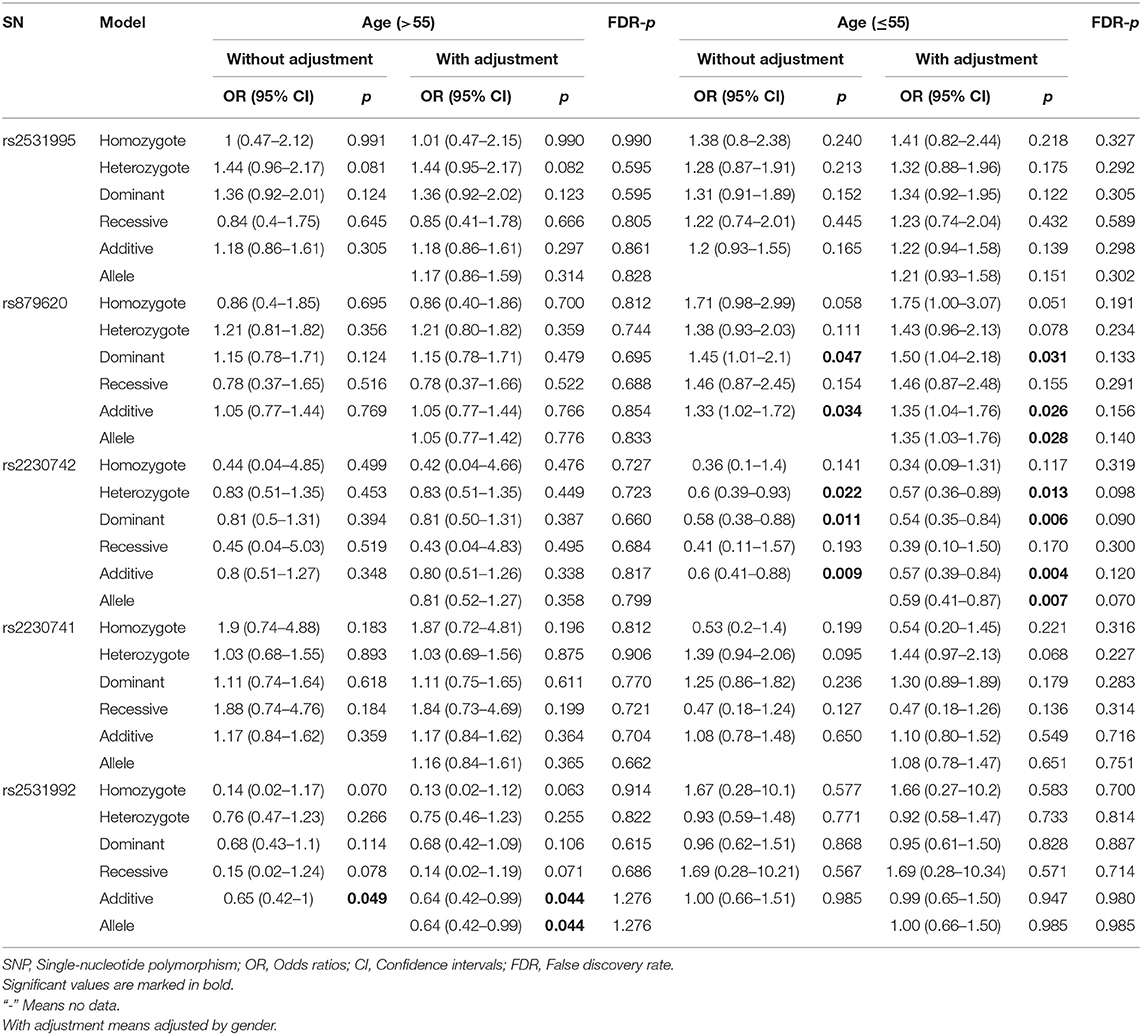
Table 4. The association between ADCY9 gene polymorphisms and hepatocellular carcinoma risk in subgroup of age.
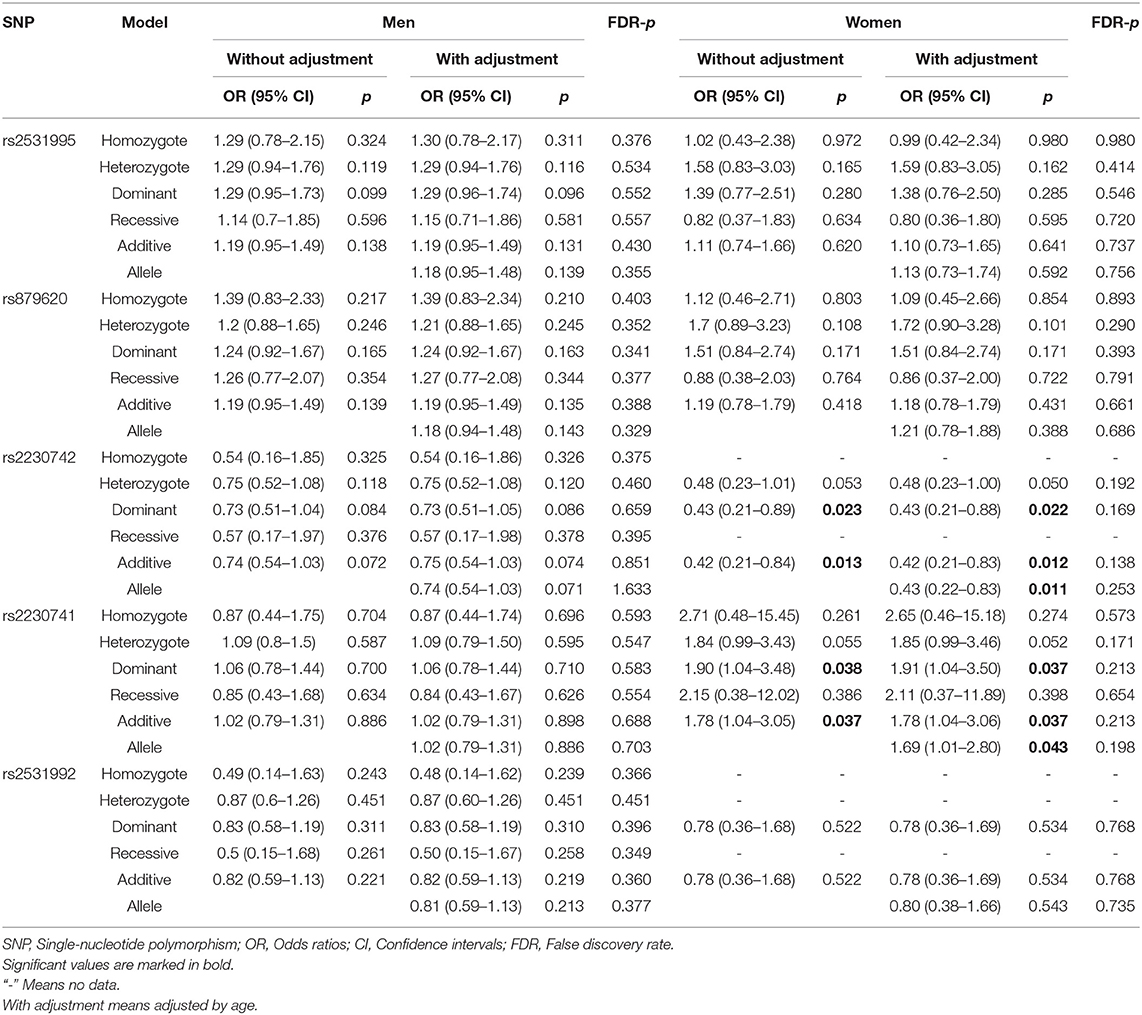
Table 5. The association between ADCY9 gene polymorphisms and hepatocellular carcinoma risk in subgroup of gender.
Haplotype Analysis of Polymorphisms in ADCY9
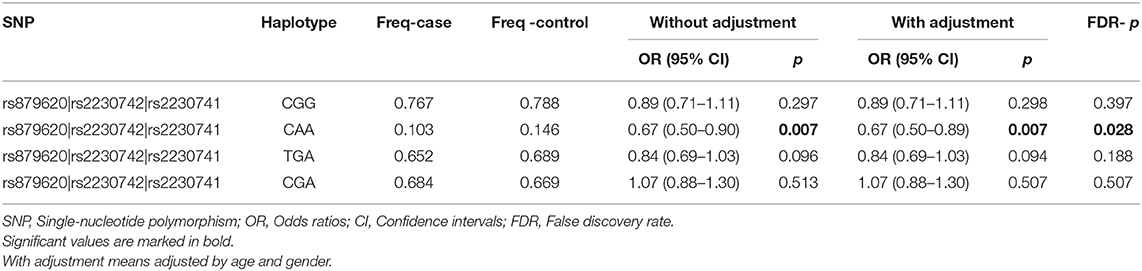
Then, we performed the LD and haplotype analysis of these five polymorphisms to HCC risk. As shown in Figure 1, one block including rs879620, rs2230742, and rs2230741 was detected. In Table 6, the results indicated that the haplotype C-A-A (rs879620, rs2230742, and rs2230741) was associated with the decreased risk of HCC (adjusted OR = 0.67, 95% CI = 0.50–0.89, p = 0.007, FDR-p = 0.028).
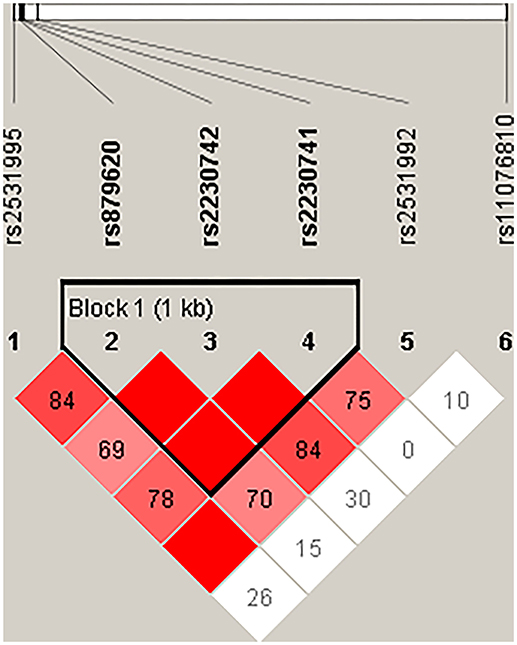
Figure 1. Haplotype block map for the SNPs of ADCY9. Block includes rs879620, rs2230742, and rs2230741. The LD between two SNPs is standardized D′.
Discussion
In the present study, we focused on the Chinese Han population and found that the AA, AA-AG genotype, and the A allele of rs2230742 could decrease the risk of HCC. Moreover, rs2230742 was related to HCC risk in the subgroup of younger participants (age ≤ 55 years old) and women, indicating that subjects with A allele of rs2230742 are less likely to have HCC. After FDR analysis, rs2230742 was still significantly associated with lower risk of HCC in the allele model. It means that rs2230742 may be a potential protective factor for HCC and help to guide treatment, and rs2230742 possibly affects the susceptibility of HCC by associating with “SiPhy cons,” “DNAse,” and “Motifs changed.” However, there were no significant linkages between other polymorphisms and HCC susceptibility after FDR correction. Besides that, Crs879620Ars2230742Ars2230741 haplotype could protect individuals from HCC (OR = 0.67, 95% CI = 0.50–0.89, p = 0.007, FDR-p = 0.028). These results suggested that ADCY9 gene polymorphisms might be involved in the susceptibility of HCC in the Chinese Han population.
ADCY9 is a widely distributed adenylyl cyclase, which catalyzes the formation of cyclic AMP from ATP. Human ADCY9 is stimulated by beta-adrenergic receptor activation but is insensitive to forskolin, calcium, and somatostatin (10). Defects in ADCY9 gene can lead to immune-mediated diseases (14). Nevertheless, the role of ADCY9 in tumorigenesis is still not clear. ADCY9 expression was found to be significantly different in endometrial cancer when compared to the controls, which might be involved in the pathogenesis of this cancer (19). In addition, ADCY9 is a known target of microribonucleic acids-−142-3p, which is associated with the invasiveness of breast cancer cells (20). Previous studies also reported that the SNPs in ADCY9 were associated with stroke, malaria, medicine responses, and cancer (9, 21–23). Rs2230739 of ADCY9 was involved in various pathways and processes, which might contribute to the susceptibility of pancreatic cancer (24). However, no significant associations were reported between ADCY9 and diseases. Our study firstly demonstrated that ADCY9 gene polymorphisms were associated with HCC risk, especially rs2230742 significantly altered the susceptibility of HCC, and it confirmed that ADCY9 was involved in cancer development in previous studies (9). However, further studies on the molecular function of ADCY9 SNPs should be performed to decipher its role in HCC.
Age and gender are considered as risk factors in the development of cancer, including HCC. The incidence of HCC increases with age, and it is the highest in individuals around the age of 70 (25). It also provided that males have higher liver cancer rates than females (26). Then, we did stratification analysis by age and gender. We found that ADCY9 gene polymorphisms altered HCC susceptibility in the subgroups except for men. Among them, rs2230742 significantly decreased HCC risk in the subgroup of women and individuals younger than 55 years old. Rs2531992 was also associated with a decreased risk of HCC for the elderly people (age > 55). Nevertheless, rs879620 and rs2230741 were associated with higher risk of HCC for the individuals aged 55 or younger and women, respectively. It suggests that the influence of ADCY9 gene polymorphisms on HCC risk is age- and gender-dependent, which may be helpful for the individual treatment of HCC in the Chinese Han population. After FDR correction, no significant associations were observed in the subgroups. It suggests that some positive findings might be caused by false positives. In the future, more studies are required to verify the influences of age and gender on the association of ADCY9 gene polymorphisms with HCC risk.
We further conducted haplotype analysis in order to demonstrate whether the interaction of these five SNPs has effect on HCC risk. Analysis of the results indicates that the ADCY9 haplotype CAA (rs879620, rs2230742, and rs2230741) is associated with a decreased risk of HCC, which may suggest that these SNPs work together. There is also a probability that this haplotype is a genetic marker for a rare mutation among the Chinese Han population.
Our study also had some limitations. First, this is a single central study, so selection bias is inevitable. Then, we did not analyze the influence of lifestyle factors and other risk factors because of lacking related information. Hence, further studies are necessary to confirm the association between ADCY9 gene polymorphisms and HCC risk.
Conclusion
To sum up, the present study suggests that the ADCY9 gene polymorphisms (rs2531995 and rs2230742) are associated with HCC susceptibility in the Chinese Han population and may be involved in tumor development. Additionally, the relationships of ADCY9 gene polymorphisms and HCC susceptibility are age- and gender-dependent; it may guide us to individual treatment. Further functional studies and large population with more races are needed to confirm the influence of ADCY9 variants on HCC risk.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Ethics Statement
This study was reviewed and approved by the Ethics Committee of Shaanxi University of Chinese Medicine (application number: SQ2020024), and written informed consents were collected from all participates. The research protocol was completed according to the Declaration of Helsinki.
Author Contributions
XC and CJ designed and supervised this study. YJ, XF, GW, and XW mainly performed this study. HS analyzed the data. FZ wrote this manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the grant from: National Natural Science Foundation of China (81774132), Science and Technology Program of Shaanxi Province (2020SF-324), and Subject Innovation Team of Shaanxi University of Chinese Medicine (No. 2019-YS05).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We sincerely thank all participants involved in this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.01450/full#supplementary-material
References
1. Ye S, Zhao S-Y, Hu S-G, Li T, Xu Q-R, Yang H-M, et al. TP53 and RET may serve as biomarkers of prognostic evaluation and targeted therapy in hepatocellular carcinoma. Oncol Rep. (2017) 37:2215–26. doi: 10.3892/or.2017.5494
2. Toyota T, Hattori E, Meerabux J, Yamada K, Saito K, Shibuya H, et al. Molecular analysis, mutation screening, and association study of adenylate cyclase type 9 gene (ADCY9) in mood disorders. Am J Med Genet. (2002) 114:84–92. doi: 10.1002/ajmg.10117
3. Nahon P, Zucman-Rossi J. Single nucleotide polymorphisms and risk of hepatocellular carcinoma in cirrhosis. J Hepatol. (2012) 57:663–74. doi: 10.1016/j.jhep.2012.02.035
4. Mary Maluccio MD, Anne Covey MD. Recent progress in understanding, diagnosing, and treating hepatocellular carcinoma †. Ca A Cancer J Clin. (2012) 62:394–9. doi: 10.3322/caac.21161
5. Msph LAT, Bray F, Siegel RL, Jacques Ferlay ME, Lortet-Tieulent J, Dvm AJ. Global cancer statistics 2012. Ca A Cancer J Clin. (2015) 65:87–108. doi: 10.3322/caac.21262
6. Barbaric J, Sekerija M, Agius D, Coza D, Dimitrova N, Demetriou A, et al. Disparities in melanoma incidence and mortality in South-Eastern Europ e: increasing incidence and divergent mortality patterns. Is progress around the corner? Europ J Cancer. (2016) 55:47–55. doi: 10.1016/j.ejca.2015.11.019
7. Mcglynn KA, Petrick JL, London WT. Global epidemiology of hepatocellular carcinoma: an emphasis on demographic and regional variability. Clin Liver Dis. (2015) 19:223–38. doi: 10.1016/j.cld.2015.01.001
8. Galun D, Basaric D, Zuvela M, Bulajic P, Bogdanovic A, Bidzic N, et al. Hepatocellular carcinoma: from clinical practice to evidence-based treatment protocols. World J Hepatol. (2015) 7:2274–91. doi: 10.4254/wjh.v7.i20.2274
9. Yi H, Wang K, Jin J-F, Jin H, Yang L, Zou Y, et al. Elevated adenylyl cyclase 9 expression is a potential prognostic biomarker for patients with colon cancer. Med Sci Monitor Int Med J Exp Clin Res. (2018) 24:19–25. doi: 10.12659/MSM.906002
10. Teixeira HM, Alcantara-Neves NM, Barreto M, Figueiredo CA, Costa RS. Adenylyl cyclase type 9 gene polymorphisms are associated with asthma and allergy in Brazilian children. Mol Immunol. (2017) 82:137. doi: 10.1016/j.molimm.2017.01.001
11. Apinjoh TO, Anchang-Kimbi JK, Njua-Yafi C, Ngwai AN, Mugri RN, Clark TG, et al. Association of candidate gene polymorphisms and TGF-beta/IL-10 levels with malaria in three regions of Cameroon: a case-control study. Malaria J. (2014) 13:236. doi: 10.1186/1475-2875-13-236
12. Berinstein E, Levy A. Recent developments and future directions for the use of pharmacogenomics in cardiovascular disease treatments. Expert Opin Drug Metab Toxicol. (2017) 13:973–83. doi: 10.1080/17425255.2017.1363887
13. Zhang Y, Fang L, Zang Y, Xu Z. Identification of core genes and key pathways via integrated analysis of gene expression and dna methylation profiles in bladder cancer. Med Sci Monitor Int Med J Exp Clin Res. (2018) 24:3024. doi: 10.12659/MSM.909514
14. Fang LT, Lee S, Choi H, Kim KK, Jew G, Kang HC, et al. Comprehensive genomic analyses of a metastatic colon cancer to the lung by whole exome sequencing and gene expression analysis. Int J Oncol. (2014). 44:211. doi: 10.3892/ijo.2013.2150
15. Niu D, Ren Y, Xie L, Sun J, Lu W, Hao Y, et al. Association between CCDC132, FDX1 and TNFSF13 gene polymorphisms and the risk of IgA nephropathy. Nephrology. (2015) 20:908–15. doi: 10.1111/nep.12611
16. Kang F, Ma W, Ma X, Shao Y, Yang W, Chen X, et al. Propranolol inhibits glucose metabolism and 18F-FDG uptake of breast cancer through posttranscriptional downregulation of hexokinase-2. J Nucl Med. (2014) 55:439–45. doi: 10.2967/jnumed.113.121327
17. Wang T, Liu JH, Zhang J, Wang L, Chen C, Dai PG. A multiplex allele-specific real-time PCR assay for screening of ESR1 mutations in metastatic breast cancer. Exp Mol Pathol. (2015) 98:152–7. doi: 10.1016/j.yexmp.2015.03.004
18. Luo J, Guo X-R, Tang X-J, Sun X-Y, Yang Z-S, Zhang Y, et al. Intravital biobank and personalized cancer therapy: the correlation with omics. Int J Cancer J Int Du Cancer. (2014) 135:1511–6. doi: 10.1002/ijc.28632
19. Orchel J, Witek L, Kimsa M, Strzalka-Mrozik B, Kimsa M, Olejek A, et al. Expression patterns of kinin-dependent genes in endometrial cancer. Int J Gynecol Cancer. (2012) 22:937–44. doi: 10.1097/IGC.0b013e318259d8da
20. Schwickert A, Weghake E, Brüggemann K, Engbers A, Brinkmann BF, Kemper B, et al. microRNA miR-142-3p inhibits breast cancer cell invasiveness by synchronous targeting of WASL, integrin alpha v, and additional cytoskeletal elements. PLoS ONE. (2015) 10:e0143993. doi: 10.1371/journal.pone.0143993
21. Tardif J-C, Rhéaume E, Lemieux Perreault L-P, Grégoire JC, Zada YF, Asselin G, et al. Pharmacogenomic determinants of the cardiovascular effects of dalcetrapib. Circul Cardiovasc Gen. (2015) 8:372. doi: 10.1161/CIRCGENETICS.114.000663
22. Flanagan JM, Frohlich DM, Howard TA, Schultz WH, Driscoll C, Nagasubramanian R, et al. Genetic predictors for stroke in children with sickle cell anemia. Blood. (2011) 117:6681. doi: 10.1182/blood-2011-01-332205
23. Auburn S, Fry AE, Clark TG, Campino S, Diakite M, Green A, et al. Further evidence supporting a role for Gs signal transduction in severe malaria pathogenesis. PLoS ONE. (2010) 5:e10017. doi: 10.1371/journal.pone.0010017
25. Almahtab M, Uddin H, Akbar SMF. Epidemiology and risk factors of hepatocellular carcinoma in Asia. J GHR. (2014) 3:1019–23.
Keywords: ADCY9, polymorphisms, hepatocellular carcinoma risk, stratification analysis, case–control study
Citation: Chao X, Jia Y, Feng X, Wang G, Wang X, Shi H, Zhao F and Jiang C (2020) A Case–Control Study of ADCY9 Gene Polymorphisms and the Risk of Hepatocellular Carcinoma in the Chinese Han Population. Front. Oncol. 10:1450. doi: 10.3389/fonc.2020.01450
Received: 12 May 2020; Accepted: 08 July 2020;
Published: 25 August 2020.
Edited by:
Aga Syed Sameer, King Saud bin Abdulaziz University for Health Sciences, Saudi ArabiaReviewed by:
Mujeeb Zafar Banday, Government Medical College (GMC), IndiaMuneeb U. Rehman, King Saud University, Saudi Arabia
Copyright © 2020 Chao, Jia, Feng, Wang, Wang, Shi, Zhao and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xu Chao, chaoxu0320@163.com; Chao Jiang, 280165056@qq.com
 Xu Chao
Xu Chao Yong Jia3
Yong Jia3 Fei Zhao
Fei Zhao