- 1Department of Lung Cancer Center, West China Hospital, Sichuan University, Chengdu, China
- 2Department of Otorhinolaryngology, Head & Neck Surgery, West China Hospital, Sichuan University, Chengdu, China
- 3Intensive Care Unit, West China Hospital, Sichuan University, Chengdu, China
- 4Department of Laboratory Medicine, West China Hospital, Sichuan University, Chengdu, China
Background: Lung cancer is the most common malignant tumor worldwide. Accumulating results have shown that long non-coding RNAs (lncRNAs) play a key role in tumorigenesis.
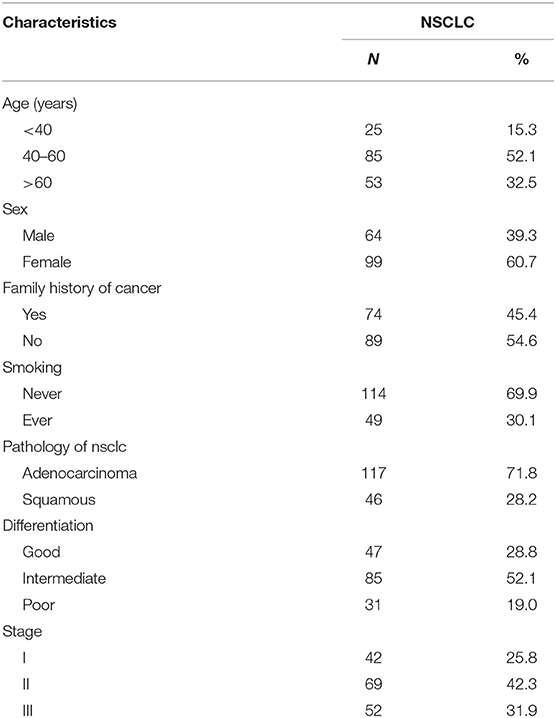
Patients and Methods: A total of 163 tumor tissues were collected from non-small cell lung cancer (NSCLC) patients from West China Hospital of Sichuan University. LincRNA00494 is a novel lncRNA, and its expression and biological effect in NSCLC were reported in this study. NSCLC cell lines were used in this study.
Results: LincRNA00494 is mainly distributed in the cytoplasm. LincRNA00494 was downregulated in the tumor tissues compared with the adjacent non-tumor tissues. LincRNA00494 expression was positively correlated with SRCIN1 expression (R = 0.57, P < 0.05). Silencing of LincRNA00494 in the cell lines substantially decreased SRCIN1 expression at the mRNA and protein levels, whereas overexpression of LincRNA00494 enhanced the SRCIN1 levels. miR-150-3p significantly decreased the luciferase signals of LincRNA00494 and SRCIN1 reporters. After transfection with miR-150-3p mimics and miR-150-3p inhibitor, overexpression of LincRNA00494 decreased the proliferation of the H358 (36%) and H1299 (29%) cell lines compared with that of the control cells, as shown by CCK-8 assays, whereas silencing LincRNA00494 promoted the proliferation of the H358 (47%) and H1299 (35%) cells. Tumor growth from LincRNA00494-overexpressing xenografts was significantly decreased; additionally, LincRNA00494 silencing substantially increased tumor growth compared with that of the control cells.
Conclusions: Functional experiments revealed that LincRNA00494 inhibited NSCLC cell proliferation, which might be related to the suppression of SRCIN1, a tumor suppressor gene, by acting as a decoy for miR-150-3p. The data showed that LincRNA00494 might have antineoplastic effects during NSCLC tumorigenesis through its role as a ceRNA.
Introduction
Lung cancer is the leading cause of cancer-related deaths worldwide (1). Due to the difficulty in early diagnosis and the poor chemotherapy response, the 5-years survival rate of lung cancer has remained at ~15% (2). Identification of for new biomarkers for the occurrence and development of non-small cell lung cancer (NSCLC) is urgently needed. Although the functions of protein-coding genes in the development of lung cancer have been extensively studied in recent decades, protein-coding genes comprise <2% of the human genome. Approximately 85% of human genomic sequences are transcribed into non-coding RNAs that are categorized into new and poorly understood RNA families (3, 4). Recent studies have shown the relationship between long non-coding RNAs (lncRNAs) and cancer subtypes, such as esophageal squamous cell carcinoma (ESCC), colorectal cancer (CRC), gastric cancer (GC), and NSCLC (5). In addition, lncRNAs may play an important role in cancer development by modulating various biological processes, including chromatin remodeling, transcription and post-transcriptional regulation. However, little is known about the specific mechanisms of lncRNAs in NSCLC.
LincRNA00494 (located at 20q13.13: 48359911…48370638, 10.1 kb) is a novel long intergenic non-protein coding gene, and its function has not been fully elucidated. LincRNA00494 showed low expression in esophageal cancer in a previous screen (6). Furthermore, we independently verified LincRNA00494 in NSCLC. LincRNA00494 was also found to be poorly expressed in NSCLC tissues. In the present study, we demonstrated that LincRNA00494 was downregulated in NSCLC tissues compared with the corresponding adjacent non-tumor tissues.
SRCIN1, a tumor suppressor gene, was reported to be inhibited by multiple microRNAs (miRNAs). MiRNA150 had a significant effect on SRCIN1 (7). LincRNAs can act by binding miRNAs. The aim of this study was to determine whether there is a targeted binding relationship between LincRNA00494 and miRNA150. Furthermore, a mechanistic investigation revealed that LincRNA00494 might suppress NSCLC cell proliferation by decoying miR-150-3p, which targets SRCIN1, a tumor suppressor in the progression of cancers (8, 9). Our findings might reveal the underlying mechanism by which aberrant LincRNA00494 expression promotes NSCLC tumorigenesis.
Patients and Methods
Study Subjects
A total of 163 tumor and adjacent tissue samples were collected from patients with NSCLC at the West China Hospital of Sichuan University. After recruitment, every participant underwent an interview involving questionnaires, and each patient provided informed consent. The study protocols were approved by the Medical Ethics Committee of Sichuan University. The clinical characteristics of all the patients are listed in Table 1.
Cell Culture
The NSCLC cell lines H358, HCC827, and H1299 were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Cell culture procedures were performed as previously described (10). Briefly, the cell lines were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum, penicillin and streptomycin in a humidified 5% CO2 atmosphere at 37°C.
Northern Blot Analysis and RNA in situ Hybridization Assay of Tumor Cells
In this study, we performed northern blotting to confirm the size of LincRNA00494. LincRNA00494 and vimentin gene expression in tumor cells was detected by RNA in situ hybridization using CanPatrolTM (SurExam Biotech, Guangzhou, China).
PCR and siRNA Knockdown
RNA from the cells and tissues was isolated using TRIzol reagent. All protocols were based on the manufacturer's instructions. An ABI Prism 7500 sequence detection system (Applied Biosystems, USA) was used to test the level of LincRNA00494. GAPDH was used as an internal standard control. In this study, all PCR assays were performed in triplicate (11). The LincRNA00494 primers for qPCR were as follows: GGTCTGGTGTGGAGACAGTG and AGCTTGCAGCCAAGAAAAGC (reverse). Mature miR-150 and SRCIN1 expression were detected by a quantitative real-time PCR assay with miR-150-specific and SRCIN1 primers and a TaqMan probe, as previously reported (7). We applied specialized kits (Sengbio, Inc., Beijing, China) to perform siRNA knockdown of LincRNA00494.
Reporter Plasmid Construction
The method for reporter plasmid construction was described in a previous study (12). psiCHECK2 (Clontech) was used to construct the plasmids psiCHECK2-LincRNA00494 (the plasmid containing LincRNA00494) and psiCHECK2-SRCIN1-3′UTR. DNA sequencing was used to verify the constructs.
Transfection and Luciferase Assay
Lipofectamine 2000 (Invitrogen, CA, USA) was used to transfect the H358 and H1299 cell lines with reporter plasmids. All procedures were based on the manufacturer's instructions. As described previously, with minor modifications, the Dual-Luciferase Reporter assay system (Promega, Madison, WI, USA) was used to measure luciferase activity (13). We carried out two independent experiments, and each group included three replicates.
Actinomycin D Assay
We also used Lipofectamine 2000 (Invitrogen) to transfect the H358 and H1299 cell lines. Moreover, the cell lines were co-transfected with miR-150 for 24 h and were exposed to actinomycin D (Sigma, St Louis, MO). As previously described, the stable expression of LincRNA00494 was analyzed using qRT-PCR (10).
Western Blot
Consistent with previous experimental procedures, Western blot analysis was conducted to assess SRCIN1 expression (10). Protein was extracted from the cell lines, and the immunoprecipitation samples were prepared using detergent-containing lysis buffer. Total protein (60 μg) was subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore). The membranes were incubated with primary antibodies against SRCIN1 (Cell Signaling Technology, dilution: 1:1,000) and β-actin (Proteintech, dilution: 1:1,000) overnight at 4°C, and the proteins were detected with a Phototope horseradish peroxidase Western blot detection kit (Thermo Fisher).
Cell Viability Analysis
We used the Cell Counting Kit-8 system (Dojindo Laboratory, Kumamoto, Japan) to determine the cell viability, and all procedures were performed according to the manufacturer's instructions (13). There were six replicates for each group, and all experiments were repeated at least three times.
Xenograft Growth of the NSCLC Cells in Nude Mice
Five-weeks-old female BALB/c nude mice were injected subcutaneously with 0.1 ml of cell suspension (with 1 × 106 cells) containing H358 and H1299 control cells, LincRNA00494-silenced cells or LincRNA00494-overexpressing cells into the back flank. The tumors were measured every 2 days, and their volumes were calculated according to the following formula: volume = length × width2 × 0.5. This study was carried out in accordance with the principles of the Basel Declaration and the guidelines of the Institutional Animal Care and Use Committee of Sichuan University. The protocol was approved by the Institutional Animal Care and Use Committee of Sichuan University.
Statistical Analysis
Analysis of variance and linear regression were used to detect the correlation between the expression of LincRNA00494 and SRCIN1 in the NSCLC tissue. The differences between the two groups were assessed using paired Student's t-tests. A p-value < 0.05 was considered significant.
Results
Cellular Characterization of LincRNA00494
To determine the subcellular localization of LincRNA00494, we detected the mRNA levels of U6 and GAPDH via RT-qPCR. In the H358 and H1299 cell lines, RT-qPCR analysis revealed that 17.2 and 14.8% of the LincRNA00494 transcripts were detected in the nuclear fraction, respectively, and 85.7 and 87.4% of these transcripts were found in the cytoplasmic fraction (Figure 1A). FISH shows that LincRNA00494 (red) were detected by RNA in situ hybridization using CanPatrolTM (Surexam Biotech, Guangzhou, China) in cytoplasm (Figure 3A). Meanwhile, PhyloCSF was utilized to examine the coding potential of LincRNA00494, and the PhyloSCF score was −149.3492, which indicated the low coding potential of LincRNA00494. Northern blot analysis showed that LincRNA00494 was 10 kb (Figure 1B).
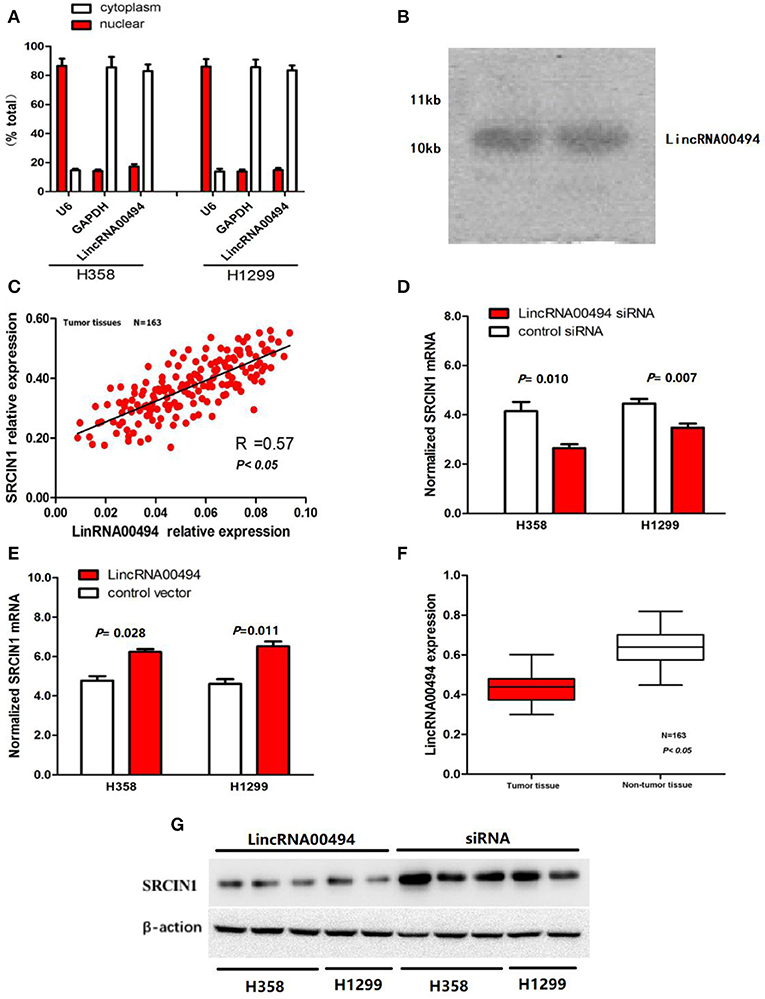
Figure 1. Cellular and molecular characterization of LincRNA00494. (A) The levels of the nuclear control transcript (U6), the cytoplasmic control transcript (GAPDH mRNA) and LincRNA00494 were assessed by RT-qPCR. Data are shown as the mean ± SEM. (B) A northern blot shows that LincRNA00494 is 10 kb. (C) The linear correlations between the LincRNA00494 expression and the SRCIN1 mRNA levels were tested. The relative expression value was normalized to the GAPDH expression level. (D,E) LincRNA00494 expression significantly affected SRCIN1 mRNA expression. Knockdown of LincRNA00494 decreased the SRCIN1 expression, while ectopic expression of LincRNA00494 increased the SRCIN1 mRNA levels. (F) LincRNA00494 was expressed at a lower level in the NSCLC tissues than in the matched tumor-adjacent tissues. The expression level of LincRNA00494 was analyzed by RT-qPCR and normalized to the GAPDH level. Data are represented as the mean ± SEM from three independent experiments. (G) The protein levels of SRCIN1 were assessed by Western blots of H358 cells and H1299 cells.
LincRNA00494 Was Downregulated in the NSCLC Tissues
To investigate the potential key role of LincRNA00494 in NSCLC, we detected the expression levels of LincRNA00494 in 163 pairs of NSCLC and adjacent non-tumor tissues via RT-qPCR. The detailed clinical features are presented in Table 1. The results showed that LincRNA00494 was strongly downregulated in the tumor tissues compared with the adjacent non-tumor tissues (Figure 1F), which suggested that LincRNA00494 might have an antineoplastic effect during NSCLC tumorigenesis.
Association of LincRNA00494 and SRCIN1 in the NSCLC Tissues
According to the results mentioned above, we tested the correlation between LincRNA00494 and SRCIN1 in the 163 NSCLC tumor tissues. The expression of LincRNA00494 was positively correlated with the expression of SRCIN1 (R = 0.57, P < 0.05, Figure 1C). Next, we disturbed endogenous LincRNA00494 expression by using gene overexpression and knockdown to investigate its effects on SRCIN1 expression. The results showed that when LincRNA00494 was silenced in the two cell lines, SRCIN1 was substantially decreased at the mRNA and protein levels. In contrast, the overexpression of LincRNA00494 increased the SRCIN1 expression level (Figures 1D,E,G).
LincRNA00494 Regulated the SRCIN1 Expression Levels by Competing With miR-150
miR-150-3p was predicted to be the target of both LincRNA00494 and SRCIN1. We cloned the 3′ untranslated region (UTR) of SRCIN1 and LincRNA00494 into the psiCHECK2 vector and cotransfected these reporters with miR-150-3p mimics in the NSCLC cells to verify the role of miR-150-3p in the two NSCLC cell lines. miR-150-3p notably decreased the luciferase activity of SRCIN1 and LincRNA00494, as shown in Figure 2A. Moreover, we measured the mRNA levels of SRCIN1 and LincRNA00494 in the NSCLC cells after treatment with miR-150-3p mimics. As shown in Figure 2B, the SRCIN1 and LincRNA00494 levels were notably decreased. Furthermore, the expression levels of SRCIN1 and miR-150-3p in the tumor tissues of the 163 NSCLC patients were detected, and we identified a negative relationship between the miR-150-3p and SRCIN1 levels (R = 0.49, P = 0.002; Figure 2C). Subsequently, we cotransfected psiCHECK2-SRCIN1 3′UTR with LincRNA00494 siRNA. The results demonstrated that the knockdown of LincRNA00494 had a negative effect on the luciferase intensity in the H358, H1299, and HCC827 cells (Figure 2D). All these results showed that miR-150-3p could target both LincRNA00494 and SRCIN1. Thus, miR-150-3p, as a molecular decoy, regulates SRCIN1 expression via LincRNA00494.
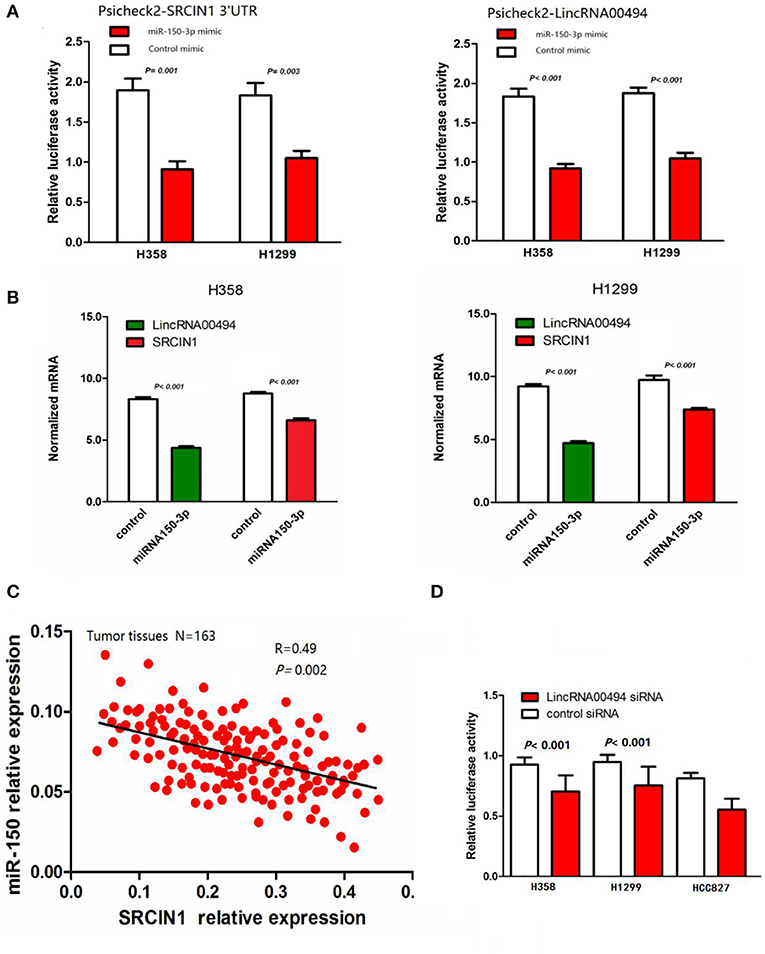
Figure 2. The relationships among miR-150-3p, LincRNA00494 and SRCIN1. (A) Both LincRNA00494 and SRCIN1 are targeted by miR-150-3p. miR-150-3p significantly decreased the luciferase signals of both LincRNA00494 and SRCIN1. (B) The SRCIN1 and LincRNA00494 levels were significantly decreased. Data are shown as the mean ± SEM (normalized to GAPDH). (C) The negative correlations between the LincRNA00494 expression levels and the SRCIN1 mRNA levels were tested. (D) The reporter vector was cotransfected into the NSCLC cells with LincRNA00494 siRNA or control siRNA. The luciferase signal was substantially decreased.
LincRNA00494 Modulated Tumor Cell Growth
Next, we investigated the effect of miR-150-3p on the RNA stability of LincRNA00494. We transfected miR-150-3p mimics and miR-150-3p inhibitor into the cells, and LincRNA00494 was downregulated due to the inhibition of RNA synthesis by actinomycin D in the presence of miR-150-3p (Figure 3B). As shown in Figure 3C, the proliferation of the H358 (36%) and H1299 (29%) cell lines decreased after overexpression of LincRNA00494. Silencing LincRNA00494 promoted the proliferation of the H358 (47%) and H1299 (35%) cells.
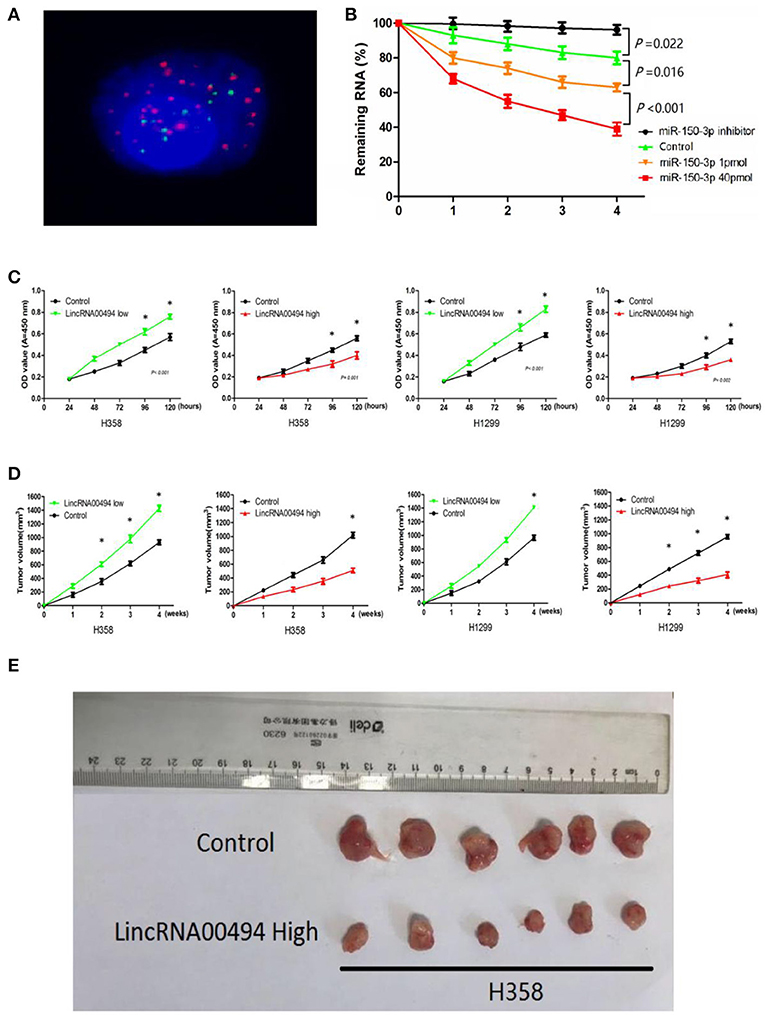
Figure 3. LincRNA00494 mediated proliferation of the NSCLC cells. (A) FISH showed that LincRNA00494 was mostly located in the cytoplasm, with a small portion in the nucleus. Green indicates vimentin; red indicates LincRNA00494. (B) Cells were harvested, and the RNA stability of LincRNA00494 was analyzed by RT-qPCR relative to time 0 after new RNA synthesis was blocked with actinomycin D; the data are shown as the mean ± SEM (normalized to GAPDH). (C) H358 and H1299 cells were seeded in 96-well plates after transfection, and the cell proliferation was assessed daily for 5 days using the CCK-8 assay. Six replicates were performed for each group, and the experiment was repeated three times. Data are shown as the mean ± SEM. (D,E) The data show the tumor volumes of the xenografts in each group 4 weeks after subcutaneously implantation of stable NSCLC cells. The mean tumor volumes from six nude mice from each group are shown at different time points.
LincRNA00494 Accelerated the Tumor Growth of the Xenografts
For confirmation of the importance of LincRNA00494 in tumor growth, we performed subcutaneous injection of NSCLC cells for the generation of xenografts. As shown in Figures 3D,E, the overexpression of LincRNA00494 decreased the growth of xenografts; additionally, LincRNA00494 silencing substantially increased the tumor growth compared with that of the control cells.
Discussion
In the present study, we examined LincRNA00494 in NSCLC and showed that it was dramatically downregulated, indicating the potential antineoplastic function of LincRNA00494 in NSCLC. Our work illustrated the associations among LincRNA00494, miR-150-3p and SRCIN1. Our findings demonstrated the important anticarcinogenic role of LincRNA00494 via decoying miR-150-3p, which could target SRCIN1.
LncRNAs are becoming important factors in various basic biological processes, and an increasing number of studies have proposed that lncRNAs have critical roles in cancer development (14, 15). Numerous lncRNAs have been implicated in lung cancer; however, only a few of these molecules have been characterized and shown to have specific biological functions and potential mechanisms. HOTAIR is a well-known oncogenic lncRNA that is highly expressed in NSCLC, SCLC, and various other human cancers (16). The expression of MALAT1 is positively correlated with the proliferation and metastasis of tumor cells (17). Moreover, the expression of CCAT2 was upregulated in NSCLC tissues compared to paired adjacent normal lung tissues. CCAT2 upregulation was reported in lung adenocarcinoma but not in squamous cell carcinoma (17). LncRNAs can be categorized into oncogenic LncRNAs and tumor suppressor LncRNAs according to their deregulated expression in cancer cells, similar to protein-coding genes. MEG3 is believed to be a tumor suppressor lncRNA because its expression is decreased in various human tumors, including lung cancer tissues (18). TUG1 was significantly downregulated in lung cancer tissues compared with the corresponding normal lung tissues. Downregulation of TUG1 was also positively related to advanced pathological stage, increased tumor size and decreased survival time in both lung squamous cell carcinoma and adenocarcinoma (19). Similarly, SPRY4-IT1 downregulation promoted the migration and invasion of A549 cells in vitro, whereas SPRY4-IT1 overexpression facilitated apoptosis (20). In our study, LincRNA00494 was significantly downregulated in the NSCLC tissues compared to the corresponding non-tumor lung tissues.
Next, we investigated the antineoplastic mechanism of LincRNA00494. Our data showed that the proliferative capacity of NSCLC cells was accelerated after LincRNA00494 silencing. More importantly, our results also suggested that LincRNA00494 acts as a molecular decoy for miR-150-3p. miRNAs are short RNA sequences that negatively regulate gene expression by targeting the 3′UTRs of mRNAs. miRNAs mediate many biological functions in tumors, such as cell proliferation, differentiation, and migration. Furthermore, miRNAs play an important role in gene regulation by targeting many coding and non-coding genes. Numerous reports have suggested that miRNAs mediated their effects by targeting lncRNAs (21). Our experiments demonstrated that lncRNAs may have an effect on their targets by acting as decoys for miRNAs, which is a very important mechanism.
Our work found that SRCIN1 served as a critical component of the LincRNA00494-miRNA network. The protein p140CAP, an important member of the Cap family encoded by SRCIN1, is expressed in various human tissues, and the expression level of p140CAP in tumors is low (8, 22). The silencing of p140CAP, a tumor suppressor gene, promoted tumor cell growth independent of anchoring and enhanced tumor growth and development (8, 23). The Cap protein p140CAP can inhibit the downstream signaling pathway of Src and regulate the activity of focal adhesion kinase and Ras/extracellular signal-related kinases and thus has an anticancer role. Src is a tyrosine kinase that is often overexpressed or abnormally activated in cancer cells. Mounting evidence has demonstrated that Src activity is elevated in lung cancer cells. SRCIN1, a newly identified inhibitor of Src, is the only gene that is negatively regulated by miR-150-3p (7). Through luciferase reporter analysis, we demonstrated that SRCIN1 was repressed by miR-150-3p, and this function was suppressed by the overexpression of LincRNA00494. Furthermore, we found that SRCIN1 expression was gradually improved, along with increased levels of LincRNA00494.
Conclusion
Collectively, our results suggest that LincRNA00494 may enhance SRCIN1 expression by competing with miR-150-3p, thereby mediating NSCLC cell proliferation.
Data Availability Statement
All datasets for this study are included in the article/supplementary material.
Ethics Statement
The studies involving human participants were reviewed and approved by Medical Ethics Committee of Sichuan University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
JD and QZ conceived and designed the experiments. JD, BL, and JL performed the experiments. CL, LT, and DLu analyzed the data. LL, XL, DZ, and XT provided the reagents, materials and analysis tools. JD, DLi, and XQ contributed to the manuscript preparation.
Funding
This work was supported by the National Key Research and Development Program is the key project of International Scientific and Technological Innovation Cooperation between governments (2016YEE0103400), National Natural Science Foundation of China (81572288), Health Commission of Sichuan Province (18PJ432).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank all the patients who participated in this research. This study cited a previous conference abstract by the authors of the study (24) which was published in the website of library.iaslc.org.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. (2017) 67:7–30. doi: 10.3322/caac.21387
2. Polanski J, Jankowska-Polanska B, Rosinczuk J, Chabowski M, Szymanska-Chabowska A. Quality of life of patients with lung cancer. Onco Targets Ther. (2016) 9:1023–8. doi: 10.2147/OTT.S100685
3. Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. (2011) 145:178–81. doi: 10.1016/j.cell.2011.03.014
4. Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. (2014) 15:7–21. doi: 10.1038/nrg3606
5. Xie W, Yuan S, Sun Z, Li Y. Long noncoding and circular RNAs in lung cancer: advances and perspectives. Epigenomics. (2016) 8:1275–87. doi: 10.2217/epi-2016-0036
6. Li J, Chen Z, Tian L, Zhou C, He MY, Gao Y, et al. LncRNA profile study reveals a three-lncRNA signature associated with the survival of patients with oesophageal squamous cell carcinoma. Gut. (2014) 63:1700–10. doi: 10.1136/gutjnl-2013-305806
7. Zhang L, Lin J, Ye Y, Oba T, Gentile E, Lian J, et al. Serum microRNA-150 predicts prognosis for early-stage non-small cell lung cancer and promotes tumor cell proliferation by targeting tumor suppressor gene SRCIN1. Clin Pharmacol Ther. (2018) 103:1061–73. doi: 10.1002/cpt.870
8. Damiano L, Di Stefano P, Camacho Leal MP, Barba M, Mainiero F, Cabodi S, et al. p140Cap dual regulation of E-cadherin/EGFR cross-talk and Ras signalling in tumour cell scatter and proliferation. Oncogene. (2010) 29:3677–90. doi: 10.1038/onc.2010.128
9. Damiano L, Le Devedec SE, Di Stefano P, Repetto D, Lalai R, Truong H, et al. p140Cap suppresses the invasive properties of highly metastatic MTLn3-EGFR cells via impaired cortactin phosphorylation. Oncogene. (2012) 31:624–33. doi: 10.1038/onc.2011.257
10. Huang G, Zhu H, Shi Y, Wu W, Cai H, Chen X. cir-ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/beta-catenin pathway. PLoS ONE. (2015) 10:e0131225. doi: 10.1371/journal.pone.0131225
11. Lehmann U, Kreipe H. Real-time PCR analysis of DNA and RNA extracted from formalin-fixed and paraffin-embedded biopsies. Methods. (2001) 25:409–18. doi: 10.1006/meth.2001.1263
12. Hu Y, Wang J, Qian J, Kong X, Tang J, Wang Y, et al. Long noncoding RNA GAPLINC regulates CD44-dependent cell invasiveness and associates with poor prognosis of gastric cancer. Cancer Res. (2014) 74:6890–902. doi: 10.1158/0008-5472.CAN-14-0686
13. Li N, Zhou P, Zheng J, Deng J, Wu H, Li W, et al. A polymorphism rs12325489C>T in the lincRNA-ENST00000515084 exon was found to modulate breast cancer risk via GWAS-based association analyses. PLoS ONE. (2014) 9:e98251. doi: 10.1371/journal.pone.0098251
14. Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. (2007) 129:1311–23. doi: 10.1016/j.cell.2007.05.022
15. Yang X, Song JH, Cheng Y, Wu W, Bhagat T, Yu Y, et al. Long non-coding RNA HNF1A-AS1 regulates proliferation and migration in oesophageal adenocarcinoma cells. Gut. (2014) 63:881–90. doi: 10.1136/gutjnl-2013-305266
16. Sang H, Liu H, Xiong P, Zhu M. Long non-coding RNA functions in lung cancer. Tumour Biol. (2015) 36:4027–37. doi: 10.1007/s13277-015-3449-4
17. Qiu M, Xu Y, Yang X, Wang J, Hu J, Xu L, et al. CCAT2 is a lung adenocarcinoma-specific long non-coding RNA and promotes invasion of non-small cell lung cancer. Tumour Biol. (2014) 35:5375–80. doi: 10.1007/s13277-014-1700-z
18. Zhou Y, Zhang X, Klibanski A. MEG3 noncoding RNA: a tumor suppressor. J Mol Endocrinol. (2012) 48:R45–53. doi: 10.1530/JME-12-0008
19. Zhang EB, Yin DD, Sun M, Kong R, Liu XH, You LH, et al. P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis. (2014) 5:e1243. doi: 10.1038/cddis.2014.201
20. Sun M, Liu XH, Lu KH, Nie FQ, Xia R, Kong R, et al. EZH2-mediated epigenetic suppression of long noncoding RNA SPRY4-IT1 promotes NSCLC cell proliferation and metastasis by affecting the epithelial-mesenchymal transition. Cell Death Dis. (2014) 5:e1298. doi: 10.1038/cddis.2014.256
21. Yu F, Zheng J, Mao Y, Dong P, Lu Z, Li G, et al. Long non-coding RNA growth arrest-specific transcript 5 (GAS5) inhibits liver fibrogenesis through a mechanism of competing endogenous RNA. J Biol Chem. (2015) 290:28286–98. doi: 10.1074/jbc.M115.683813
22. Di Stefano P, Damiano L, Cabodi S, Aramu S, Tordella L, Praduroux A, et al. p140Cap protein suppresses tumour cell properties, regulating Csk and Src kinase activity. EMBO J. (2007) 26:2843–55. doi: 10.1038/sj.emboj.7601724
23. Katoh M, Katoh M. Evolutionary recombination hotspot around GSDML-GSDM locus is closely linked to the oncogenomic recombination hotspot around the PPP1R1B-ERBB2-GRB7 amplicon. Int J Oncol. (2004) 24:757–63. doi: 10.3892/ijo.24.4.757
Keywords: LincRNA00494, ceRNA, non-small cell lung cancer, SRCIN1, LncRNA
Citation: Dong J, Li B, Lin D, Lu D, Liu C, Lu X, Tang X, Li L, Zhu D, Liu J, Qiu X, Tian L and Zhou Q (2020) LincRNA00494 Suppresses Non-small Cell Lung Cancer Cell Proliferation by Regulating SRCIN1 Expression as a ceRNA. Front. Oncol. 10:79. doi: 10.3389/fonc.2020.00079
Received: 31 July 2019; Accepted: 16 January 2020;
Published: 06 February 2020.
Edited by:
Alfons Navarro, University of Barcelona, SpainReviewed by:
Zhe Lei, Soochow University, ChinaMarco Trerotola, Università degli Studi G. d'Annunzio Chieti e Pescara, Italy
Copyright © 2020 Dong, Li, Lin, Lu, Liu, Lu, Tang, Li, Zhu, Liu, Qiu, Tian and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qinghua Zhou, zhouqh@163.com
 Jingsi Dong1
Jingsi Dong1 Bingjie Li
Bingjie Li Daxing Zhu
Daxing Zhu Qinghua Zhou
Qinghua Zhou