- 1Department of Pathology, Shanghai Children's Medical Center, Shanghai Jiao Tong University School of Medicine, Shanghai, China
- 2Department of General Surgery/Surgical Oncology Center, Shanghai Children's Medical Center, Shanghai Jiao Tong University School of Medicine, Shanghai, China
- 3State Key Laboratory for Oncogenes and Related Genes, Key Laboratory of Gastroenterology and Hepatology, Ministry of Health, Division of Gastroenterology and Hepatology, Renji Hospital, Shanghai Cancer Institute, Shanghai Institute of Digestive Disease, Shanghai Jiao Tong University School of Medicine, Shanghai, China
- 4Department of Hematology and Oncology, Shanghai Children's Medical Center, Shanghai Jiao Tong University School of Medicine, Shanghai, China
- 5Shanghai Children's Medical Center, Pediatric Translational Medicine Institute, Shanghai Jiao Tong University School of Medicine, Shanghai, China
- 6Department of Infectious Diseases, Shanghai Children's Medical Center, Shanghai Jiaotong University School of Medicine, Shanghai, China
- 7Department of Rehabilitation, Xinhua Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China
Neuroblastoma (NB) is the most common pediatric extra-cranial solid tumor with heterogeneous characteristics, and the prognosis of patients with high-risk NB is usually poor. Discovery of novel biomarkers for early detection and investigation of the underlying mechanisms governing invasion and metastasis of NB are urgently needed. Recently, exosomal microRNAs (miRNAs) have been shown to play vital regulatory or communication roles in the process of various types of cancers. However, the roles and mechanisms of exosomal miRNAs in NB remain unknown. Thus, the present study aims to investigate the detailed functions of tumor-derived exosomal miRNAs in progression and migration of NB in vivo and in vitro. By examining different exosomal miRNA expression profiles in the plasma of NB patients, we identified that the expression of hsa-miR199a-3p from exosomes was significantly upregulated, which was correlated with the severity of NB patients. Furthermore, we confirmed that exosomal hsa-miR199a-3p could facilitate proliferation and migration of NB via regulating NEDD4 expression. In summary, our data, for the first time, revealed that exosomal hsa-miR199a-3p could promote tumor proliferation and migration via decreasing NEDD4 expression in NB, suggesting that exosomal hsa-miR199a-3p may be applicated as a fast, easy, and non-invasive detection biomarker and contribute to the development of novel therapeutic strategies for NB in the future.
Introduction
Neuroblastic tumors (NTs), including neuroblastoma (NB), ganglioneuroblastoma, intermixed (GNBi), ganglioneuroblastoma, nodular (GNBn), and ganglioneuroma (GN), are the most common pediatric extra-cranial solid tumors (1). They arise from the embryonic sympathoadrenal neural crest and range from immature to differentiated mature tumors. Based on the International Neuroblastoma Pathology Classification (INPC), GNBi, and GN are relatively differentiated mature variants of NT, while NB represents the immature cancer type (2). As the third most frequent childhood cancer, NB accounts for 7% of the total cases of cancer diagnosed among children aged birth to 14 years old (3). More remarkably, NB is the primary cause of death from pediatric cancers in children between 1 and 5 years old and leads to approximately 13% of pediatric cancer mortality (4). However, the survival rates of NB at different risk categories are markedly variable. The 5-year survival rates of NB patients with low- to intermediate-risk are over 90%, compared to only 40–50% in patients with high-risk NB (5). Because of rapid invasion and metastasis, the prognosis of NB patients with high-risk NB is poor even using some aggressive therapeutic strategies. Easy and fast detection in NB patients can contribute to their diagnosis and monitoring responses to therapy. Therefore, discovery of novel diagnostic and prognostic biomarkers in NB and comprehensive investigation of the mechanisms governing its invasion and metastasis are urgently needed.
Recently, there have been renewed interests in the extracellular vehicles (EVs) because of their numerous functions in physiology and pathology. Exosomes, small EVs (40–100 nm in diameter) formed inside the endosomal compartments, contain various RNAs such as mRNAs and miRNAs. Although the biological functions of exosomal miRNAs are not yet completely clear, they have been reported to play vital roles in the regulation of tumorigenesis and tumor development (6–8). A recent study has demonstrated that MYCN-amplified NB cells can secrete some exosomal miRNAs which may play potential roles in cancer progression via the change of tumor environment (9). Moreover, circulating exosomal miRNA-21 and miRNA-155 have been reported to be involved in NB resistance to chemotherapy through a novel exosomal miR-21/TLR8-NF-κB/exosomal miR-155/TERF1 signaling pathway (10). However, few studies have detected the specific circulating exosomal miRNA profiles in the peripheral blood samples from patients with high-risk NB and investigated the mechanisms of specific exosomal miRNAs in the NB invasion and metastasis.
In the present study, an extensive investigation on the circulating exosomal miRNA profiles of NB patients paired with GNBi and healthy individuals was performed using miRNA-sequencing. The purpose of this study is to identify suitable circulating exosomal miRNAs as novel biomarkers that can improve early diagnosis in NB, and to investigate the underlying molecular mechanisms involved in these exosomal miRNAs.
Materials and Methods
Patients and Clinical Samples
A total of 17 NB patients, 6 GNBi patients as well as 7 age- and sex-matched healthy individuals were recruited in Shanghai Children's Medical Center (SCMC) from January 2017 to September 2017. According to the INPC, all enrolled patients were newly diagnosed as NB and GNBi with histological provement, and had not received chemotherapy or radiotherapy. The clinic-pathological characteristics of all the patients were listed in the Tables S1, S2.
Two milliliter of peripheral blood was collected in EDTA anticoagulant tubes. Plasma was obtained after a 20-min centrifugation at 1,500 × g and a second 10-min centrifugation at 2,000 × g to remove the blood cells. The plasma samples were apportioned in aliquots and stored at −80°C until further use.
The study was approved by the Institutional Review Board and the Ethics Committee of SCMC (SCMCIRB-K2017042), and written informed consent was obtained from the parents of each participant.
Cell Culture
Three human NB cell lines [i.e., SK-N-SH, SH-SY5Y, SK-N-BE(2)] and human umbilical vein endothelial cell line (HUVEC), human embryonic kidney cell line (HEK293), and human fibroblast cell line (MRC5) were used in this study. All cell lines were obtained from Cell Bank of Chinese Academy of Sciences (Shanghai, China) except for MRC5, which was kindly provided by Dr. Jing Ye (Ruijin Hospital, Shanghai Jiao Tong University School of Medicine). The cells were cultured in exosome-free conditioned medium containing Dulbecco's modified Eagle's medium (DMEM) or Dulbecco's modified Eagle's Medium: Nutrient Mixture F-12 (DMEM/F12) supplemented with 10% fetal bovine serum (FBS) and incubated at 37°C in a humidified incubator with 5% CO2. The FBS was depleted of bovine exosomes by overnight ultracentrifugation at 120,000 × g followed by filtration with 0.22-μm filter.
Isolation of Exosomes From Plasma Samples
Plasma exosomes were isolated and purified using the exoRNeasy serum/plasma midi kit (Qiagen, USA) according to the manufacturer's protocol. Briefly, 1 volume of buffer XBP was mixed with the pre-filtered plasma sample and added onto the exoEasy spin column. After centrifugation, the pelleted fraction was re-suspended in nuclease-free water for later analysis or subjected to RNA extraction immediately. All centrifugation steps were performed at 4°C.
Isolation of Exosomes From Cell Culture Supernatants
Exosomes from cell culture supernatants were isolated as described previously (11). Briefly, conditioned medium was harvested 4 days after cell seeding. After centrifugation at 2,000 × g and 10,000 × g to remove the cells and cellular debris, respectively, the supernatant was centrifuged at 100,000 × g for 70 min to collect the exosome fraction. The crude exosome pellets were washed in phosphate buffer saline (PBS), centrifuged at 100,000 × g for 1 h, and was then re-suspended in PBS. All centrifugation steps were performed at 4°C.
Characterization of Isolated Exosomes
To confirm the purification of the isolated exosomes, transmission electron microscopy (TEM), Nanosight analysis, flow cytometry analysis, and Western blotting were performed. In Brief, 10 μL of isolated exosomes was mounted on copper grids and fixed with 1% glutaraldehyde. The grids were stained with 3% (v/v) phosphotungstic acid in ddH2O and the samples were examined immediately with JEM1230 TEM (JEOL, Japan) at a voltage of 80 kV. Moreover, the size distribution of the exosomes was evaluated with Zeta sizer Nano series-Nano-ZS (Malvern Instruments, UK) using Nanoparticle Tracking Analysis (NTA) software v2.3 (NanoSight Ltd, UK). Furthermore, flow cytometry analysis with an Accuri C6 flow cytometer (BD Instruments, USA) was performed to detect exosomal specific surface antigens (i.e., CD63 and CD81). At last, exosome-specific markers CD63 and calnexin were identified with Western blotting. Briefly, 10 μg of the exosomal proteins were separated on 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gels and transferred to polyvinylidene difluoride (PVDF) membrane (GE Healthcare, USA). Blocked with 5% non-fat milk in PBST (PBS containing 0.1% TWEEN-20), the membrane was incubated with the primary antibodies against CD63 (Abcam, ab68418) and calnexin (NOVUS, NB100-1974), respectively, at 4°C overnight. The membrane was then washed 3 times with PBST and exposed to horseradish peroxidase (HRP)-conjugated donkey anti-mouse IgG (1:1000 dilution, Santa Cruz Biotechnology) for 1 h at room temperature. After 3-time washes with PBST, antibody binding was detected by enhanced chemiluminescence substrates (Millipore, USA) and visualized with ImageQuant LAS 4000 mini densitometer (GE Healthcare Life Sciences, USA).
Extraction of RNA From Exosomes
The exosomal RNAs were extracted using Qiazol and RNeasy MinElute Spin Column (Qiagen, USA) following the manufacturer's user guide. RNA concentration was determined using Nanodrop (Thermo Fisher Scientific, USA). The content and quality of exosomal RNAs were determined using the 2100 Bioanalyzer instrument (Agilent technologies, USA).
miRNA Library Construction and Sequencing Data Analysis
Deep sequencing was performed on all exosomal RNAs from the plasma samples. 18–30 nt miRNAs were first size-selected with PAGE gel and reverse transcribed to cDNA through adaptor-specific primers. PCR amplification was then carried out and eluted in nuclease-free H2O. The quality and yield of miRNA libraries were assessed using the Agilent 2100 Bioanalyzer instrument. The PCR products of 100–200 bp were then separated from primer-dimers and other products with PAGE gel.
The expression levels of miRNAs in the constructed libraries were quantified using the BGIseq-500 platform (BGI, China). The raw data cleaning analysis included removing the low-quality tags and 5' adaptor contaminants from the 50-nt tags. Based on miRbase and Rfam databases, the clean tags were annotated into different categories to predict the novel miRNAs and seed edit of potential known miRNAs. Prediction for target miRNAs and the target genes accordingly were finally performed using Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis.
Verification of miRNA Expression With Quantitative Real Time PCR (qRT-PCR)
Qiagen miScript SYBR Green PCR assay was used for miRNA quantification in accordance with the manufacturer's instructions. In brief, the reverse transcription of miRNAs from the plasma samples, cell culture supernatants and tissues were performed with miScript II RT kit. The specific primers of miRNAs were designed by Qiagen, and exogenous cel-miR-39-3p was used as control. qRT-PCR was performed in triplicate using the CFX Connect™ Real-Time System (Bio-Rad, USA) under the following conditions: 95°C for 15 min, 40 amplification cycles of 94°C for 15 s, 55°C for 30 s, and 70°C for 30 s. The data are presented as 2−ΔΔCT. In the analysis of exosomal miRNAs, the first sample collected in the healthy controls was used as 1 for normalization. In the analysis of miRNAs of the tumor tissues, the first sample collected in the low-risk patients was used as 1 for normalization.
Cell Transfection
The specific mimics for hsa-miR199a-3p and negative control were designed and synthetized by Ribobio (Ribo, China). The sequence of hsa-miR199a-3p mimic was sense 5′-ACAGUAGUCUGCACAUUGGUUA-3′ and antisense 5′-UAACCAAUGUGCAGACUACUGU-3′. Sequence of miRNA mimic used as control miRNA was: sense 5′-UUUGUACUACACAAAAGUACUG-3′, antisense 5′-CAGUACUUUUGUGUAGUACAAA-3′. The hsa-miR199a-3p mimic and negative control were transfected into SK-N-SH cells using riboFECT™ CP transfection reagent (Ribo, China) following the manufacturer's instructions. In addition, NEDD4 cDNA with HA tag at the C-terminus was synthesized and constructed into pcDNA3.1 vector (Generay Biotech Co., China). Both pcDNA3.1-NEDD4 and empty pcDNA3.1 plasmids were transfected into cells with Lipo3000 transfection reagent (Thermo Fisher Scientific, USA) following the manufacturer's instructions.
BrdU Assay
The effects of the exosomal miRNAs on cell proliferation were also detected using the bromodeoxyuridine (BrdU) cell proliferation assay kit (Roche, USA) following the manufacturer's instructions. Briefly, the cells were firstly incubated with 10 μM BrdU labeling solution for 2 h, and were then fixed and incubated with anti-BrdU-POD for 2 h after removal of the BrdU labeling solution. The absorbance of each well was measured with a Multilabel Counter and normalized to that of the cells at 0 h.
MTS Assay
The effects of the exosomal miRNAs on cell proliferation were also evaluated with 3-(4,5-dimethylthiazol-2yl)-2,5-dipheiltertrazolium bromide (MTS) Cell Proliferation Assay (Promega, USA) following the manufacturer's instructions. In brief, 0, 24, 48, and 72 h after miRNA transfection, 20 μL of MTS was added to each well, and the plate was detected by a Multilabel Counter (Bio-Rad Laboratories, USA) after 3 h of incubation. Absorbance at 490 nm of each well was measured and normalized to that of the cells at 0 h.
Cell Migration Assay
The effects of the exosomal miRNAs on cell migration were assessed using Transwell migration chamber with 8.0 μm pore size polycarbonate membrane (Corning, USA). In brief, 50,000 cells were seeded in the Transwell upper chamber with 200 μL Opti-MEM medium (Gibco, USA). The lower chamber was filled with 200 μL culture medium supplemented with 20% FBS. After 24, 48, and 72 h, the filters were washed, fixed and stained with Crystal Violet (Sigma-Aldrich, USA). The number of migrated cells onto the lower surface of the filter was counted in five different fields at a 400 × magnification.
Luciferase Assay
Following transfections of the reporter plasmid, dual luciferase reporter assays (Promega, USA) were carried out according to the manufacturer's instructions. Luciferase activity was measured using a Multilabel Counter (Bio-Rad Laboratories, USA).
Statistical Analysis
All statistical analysis was performed with IBM SPSS 24.0 statistical software (SPSS Inc., USA) and graphs were generated using GraphPad PRISM 6.0 (GraphPad Software, Inc., USA). The differential expression levels of exosomal miRNAs between different groups were compared using Wilcoxon rank-sum test, and continuous variables were analyzed using the Student's t-test. p < 0.05 was considered as statistically significant.
Results
Exosomal hsa-miR199a-3p Is Highly Expressed in the Plasma of NB Patients
Isolated from the plasma of NB patients, the EVs were identified with transmission electron microscope and Nanoparticle Tracking Analysis (Figures 1A,B). In addition, results from flow cytometry analysis demonstrated the existence of exosomal markers CD63 and CD81 (Figure 1C). The presence of the exosomal marker CD63 but the absence of endoplasmic reticulum compartment-specific marker calnexin was also confirmed by Western blotting (Figure 1D). All these results indicated that the isolated EVs were purified exosomes.
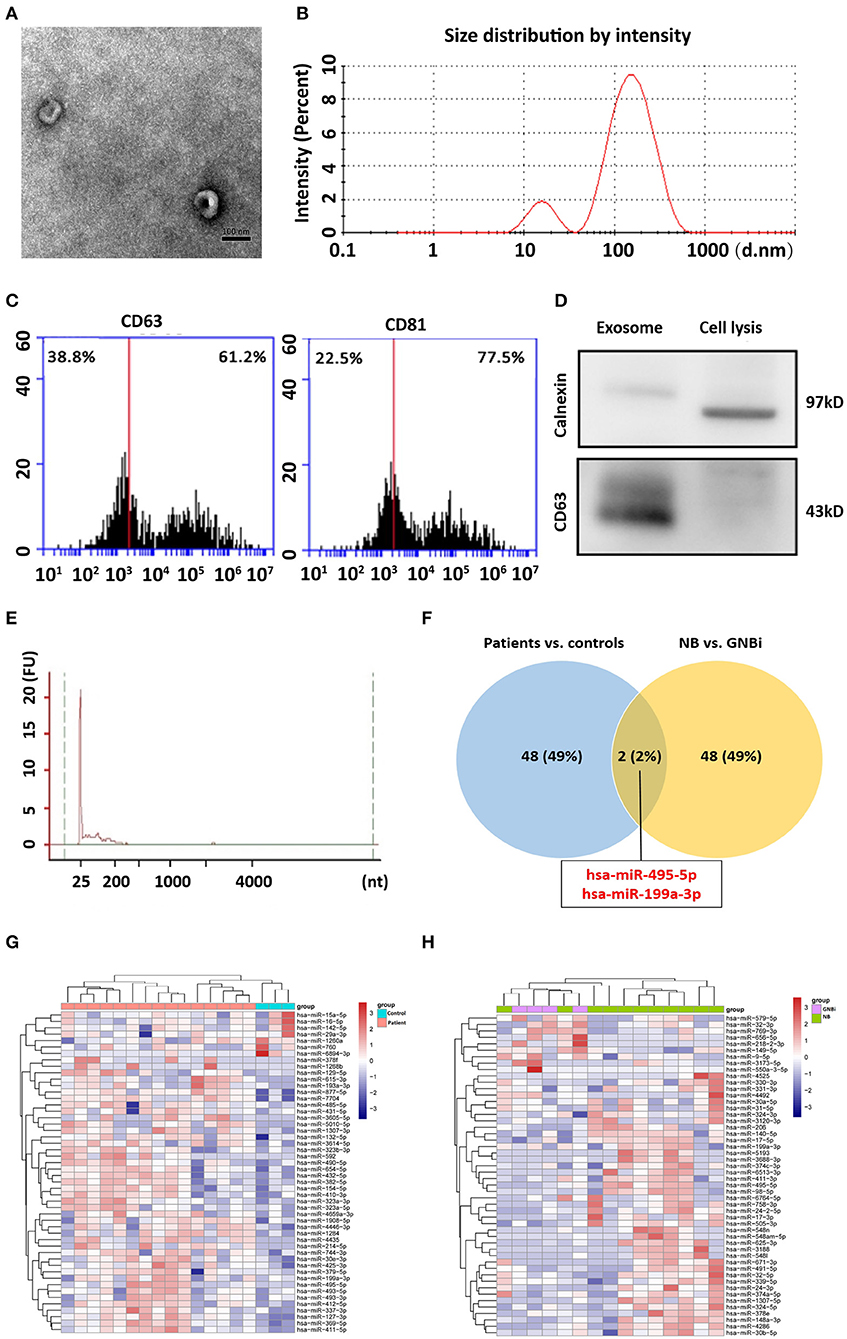
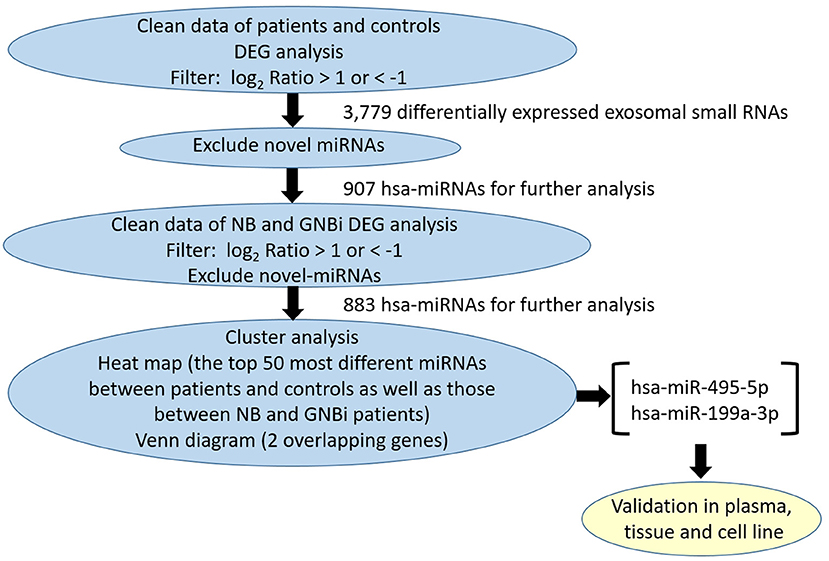
Figure 1. Identification of plasma exosomes and differentially expressed exosomal miRNAs. (A) Transmission electron microscopy demonstrated the presence of exosome vesicles with typical cup-shape morphology (size bar 100 nm). (B) Analysis of exosomes isolated from plasma by Nanoparticle Tracking Analysis. The average size was 79.42 nm and the peak was 146.4 nm. (C) 61.2% particles isolated from the plasma samples showed CD63 positive and 77.5% showed CD81 positive as detected by flow cytometry. (D) Confirmation of the exosomes markers with Western blotting indicated the presence of CD63 but the absence of calnexin in exosomes. (E) Bioanalzyer trace analysis revealed abundant miRNAs present in the exosomes isolated from the plasma samples. The exosomes presented in (A–E) were obtained from NB patient 1. (F) A Venn diagram showed that hsa-miR199a-3p and hsa-miR-495-5p overlapped in the top 50 most different miRNAs between NB/GNBi patients vs. healthy individuals and NB vs. GNBi patients (red color indicates up-regulation). (G,H) Cluster analysis of exosomal miRNAs among NB, GNBi, and healthy individuals. The top 50 most different miRNAs between NB/GNBi and healthy controls (G) as well as those between NB and GNBi patients (H) were presented.
Bioanalyzer trace analysis revealed abundant miRNAs present in the exosomes isolated from the plasma samples (Figure 1E). Afterwards, next-generation sequencing was performed on these exosomal miRNA samples from NB, GNBi and healthy individuals, respectively. The criteria used to screen differences in miRNA encapsulation in the plasma exosomes between NB/GNBi patients and healthy individuals, as well as those between NB patients and GNBi patients were based on differentially expressed gene (DEG) analysis (log2 Ratio > 1 or < −1). A total of 3,779 differentially expressed exosomal small RNAs met these criteria and were selected for further analysis, among which 3,248 small RNAs were up-regulated and 531 small RNAs were down-regulated in the NB/GNBi patients. A Venn diagram showed that only hsa-miR199a-3p and hsa-miR-495-5p overlapped in the top 50 most different miRNAs between NB/GNBi patients vs. healthy individuals and NB vs. GNBi patients (Figure 1F). Besides, two heat maps of these 50 selected miRNAs demonstrated a gene cluster and sample cluster according to the levels of exosomal miRNAs in the plasma among NB, GNBi and healthy individuals (Figures 1G,H). The screening procedures used to identify the target exosomal miRNAs are shown in Figure 2. qRT-PCR was performed to validate the 2 selected exosomal miRNA candidates in other 8 newly diagnosed NB/GNBi patients and 4 healthy volunteers. We confirmed that only the expression levels of hsa-miR199a-3p, but not the hsa-miR-495-5p were significantly higher in the plasma exosomes from NB patients (p = 0.042; Figure 3A), suggesting that plasma exosomal hsa-miR199a-3p may play a vital role in NB progression.
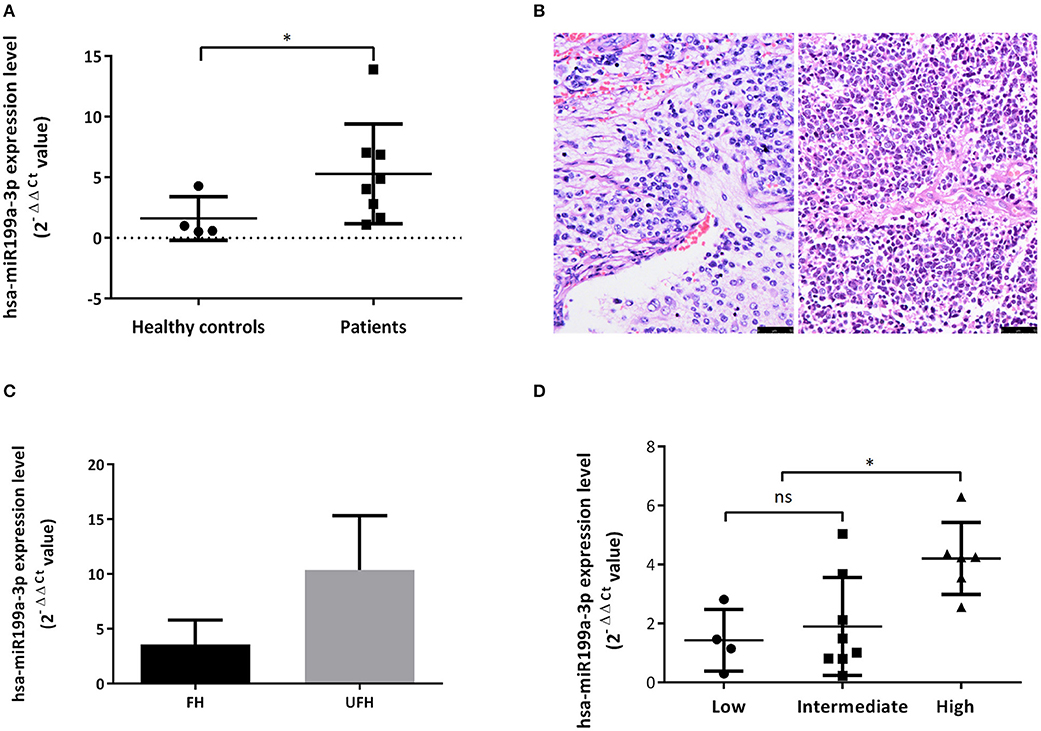
Figure 3. Exosomal hsa-miR199a-3p was highly expressed in the plasma of NB patients and predicted high risk of NB patients. (A) The exosomal hsa-miR199a-3p expression levels were significantly higher in the patients with NB/GNBi compared with healthy controls as determined by qRT-PCR. (B) Representative images of FH (left) and UFH (right) neuroblastoma tissues were presented after H&E staining (size bar 50 μm). (C) Exosomal hsa-miR199a-3p levels in the plasma of the UFH patients (N = 2) were 2-fold more than those of the FH patients (N = 6). (D) hsa-miR199a-3p in the fresh tumor tissues were higher in the COG high-risk group compared with the intermediate- and low-risk group as determined by qRT-PCR. *p < 0.05; ns, not significant.
Upregulated hsa-miR199a-3p Predicts High Risk of NB Patients
To explore whether exosomal hsa-miR199a-3p expression correlates with the severity of NB patients, we divided the patients into groups with favorable histology (FH) and unfavorable histology (UFH) according to the INPC (Figure 3B). The clinico-pathological characteristics of the patients in both groups were comparable (Table S2). Under such classification, the exosomal hsa-miR199a-3p levels in the plasma of patients with UFH were 2-fold higher than those patients with FH (Figure 3C), suggesting that upregulated exosomal hsa-miR199a-3p may serve as an independent risk factor. In addition, the expression of hsa-miR199a-3p in the fresh tumor tissues from 18 patients with NB and GNB were detected. The characteristics of these 18 patients were listed in the Table S3. Even in the fresh tissues, the expression levels of hsa-miR199a-3p were significantly higher in the Children's Oncology Group (COG) high-risk patients than those in the intermediate- and low-risk patients (Figure 3D).
Exosomal hsa-miR199a-3p Facilitates Proliferation and Migration of NB Cells
To verify whether the significantly higher exosomal hsa-miR199a-3p identified in the plasma of NB patients were due to the high expression of hsa-miR199a-3p in NB tumors, we analyzed the levels of hsa-miR199a-3p in three NB cell lines, including SK-N-SH cells, SH-SY5Y and SK-N-BE(2) and the exosomes secreted from these cells. HUVEC, human fetal lung fibroblast cells (MRC-5), and human embryonic kidney 293 cells (HEK 293) were used as normal controls. As determined by qRT-PCR, the expression levels of hsa-miR199a-3p in both the SK-N-SH cells (309.50 ± 39.17 vs. 1.05 ± 0.02, p = 0.0014) and the exosomes secreted from SK-N-SH (1,087 ± 178.4 vs. 0.90 ± 0.08, p = 0.0037) were significantly higher than those in HUVEC cells and their exosomes. Compared to HUVEC, similar phenomena were also observed in SH-SY5Y cells (45.5 ± 4.38, p = 0.0005) and SK-N-BE(2) cells (111.5 ± 13.53, p = 0.0005), as well as the exosomes [(38.02 ± 3.613, p = 0.0005 in SH-SY5Y and 666.1 ± 193.5, p = 0.012 in SK-N-BE(2)] secreted from the cells (Figures 4A,B). In contrast, no significant difference of hsa-miR199a-3p has been observed among the three non-NB cell lines, either in the cells or in the exosomes. These results show that compared to the non-NB cell lines, hsa-miR199a-3p were significantly higher expressed in the NB cells and their exosomes, suggesting that the remarkably abundant exosomal hsa-miR199a-3p levels in the plasma of NB patients may be due to its high expression in NB tumors.
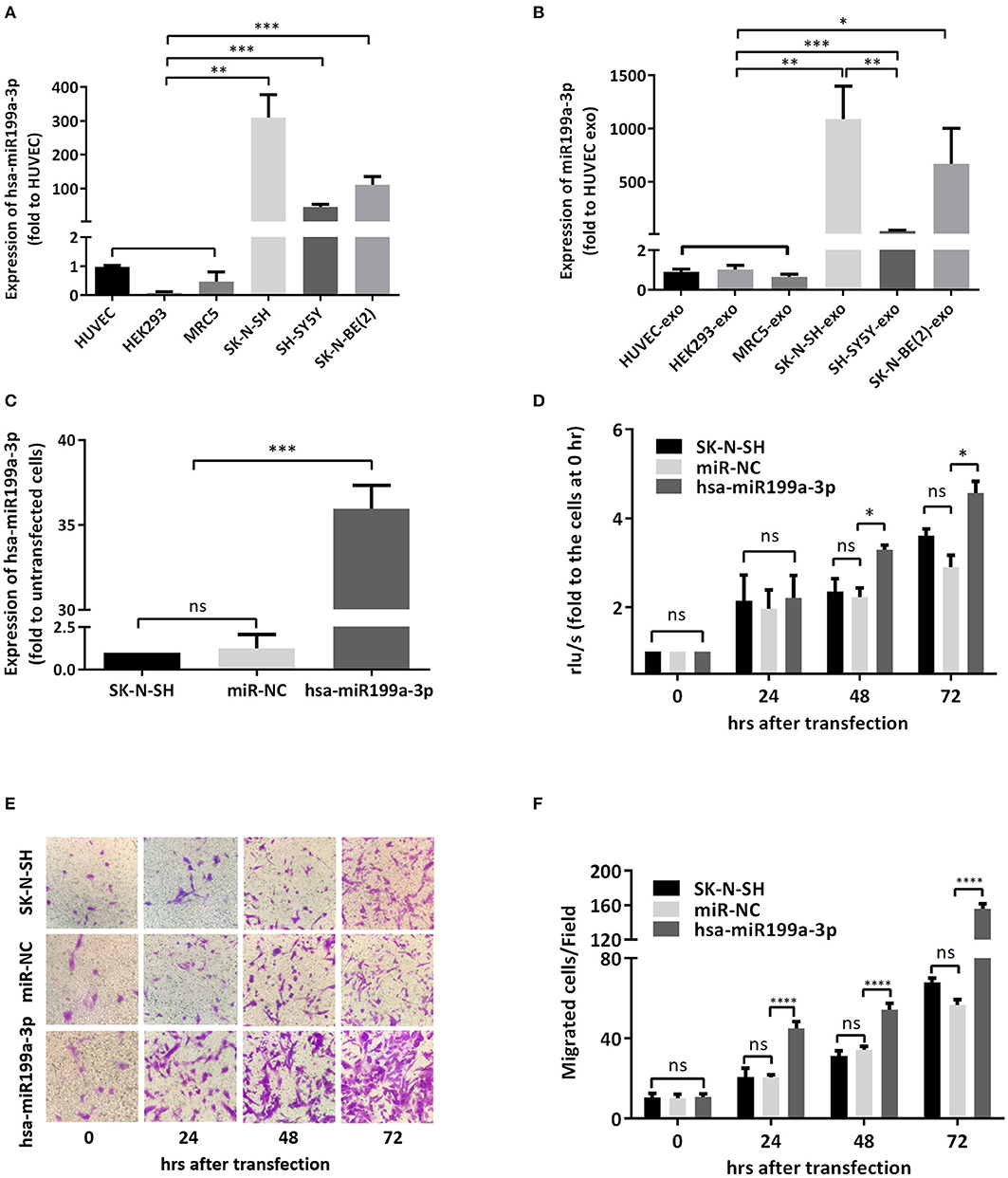
Figure 4. hsa-miR199a-3p facilitated proliferation and migration of NB cells. (A,B) The expression levels of hsa-miR199a-3p in the NB cells [SK-N-SH, SH-SY5Y, SK-N-BE(2)] (A) and in the exosomes secreted from these cells (B) were significantly higher than those in the normal control cells (HUVEC, HEK293, MRC5), as detected by qRT-PCR. (C) hsa-miR199a-3p was up-regulated in SK-N-SH cells after transfection with the designed hsa-miR199a-3p mimic. (D) BrdU cell proliferation assay demonstrated that over-expression of hsa-miR199a-3p can promote the proliferation of SK-N-SH cell lines 48 and 72 h after transfection. (E,F) Transwell assay revealed the up-regulation of exosomal hsa-miR199a-3p can promote the migration of SK-N-SH cells 24, 48, and 72 h after transfection. Representative photographs were shown in (E) and quantification of the migrated cells per field was shown in (F). All the data were presented as the mean ± SD from three independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns, not significant. exo, exosome; NC, negative controls.
To further evaluate the functions of exosomal hsa-miR199a-3p in NB progression, hsa-miR199a-3p mimic was synthesized and transfected into SK-N-SH cells. After transfection, the expression level of exosomal hsa-miR199a-3p from SK-N-SH cell culture supernatants was significantly increased (Figure 4C). Moreover, BrdU cell proliferation assay showed that hsa-miR199a-3p overexpression promoted significant proliferation of NB cells at 48 (3.29 ± 0.010 vs. 2.23 ± 0.21, p = 0.0431) and 72 h (4.57 ± 0.26 vs. 2.90 ± 0.27, p = 0.0462) after transfection (Figure 4D). Similar results have also been observed when detected by MTS assay (data not shown). In addition, as detected in the Transwell assay, up-regulation of hsa-miR199a-3p significantly increased the migration of NB cells at different time points after transfection (45.00 ± 1.52 migrated cells/field vs. 20.40 ± 0.68, 54.20 ± 1.43 vs. 34.20 ± 0.86, and 56.60 ± 1.21 vs. 147.40 ± 3.06 at 24, 48, and 72 h respectively, p < 0.0001; Figures 4E,F). To further validate the role of exosomal hsa-miR199a-3p in promoting proliferation and migration in NB, exosomes obtained from the culture supernatant of SK-N-SH cells, which contain the highest level of exosomal hsa-miR199a-3p among the three NB cell lines, were used to treat the SH-SY5Y cells, which contain the lowest level of exosomal hsa-miR199a-3p. As expected, hsa-miR199a-3p-enriched exosomes could facilitate the proliferation and migration of SH-SY5Y cells (Figure 5). Taken together, the data indicate that upregulation of exosomal hsa-miR199a-3p significantly increases the proliferation and migration of NB cells.
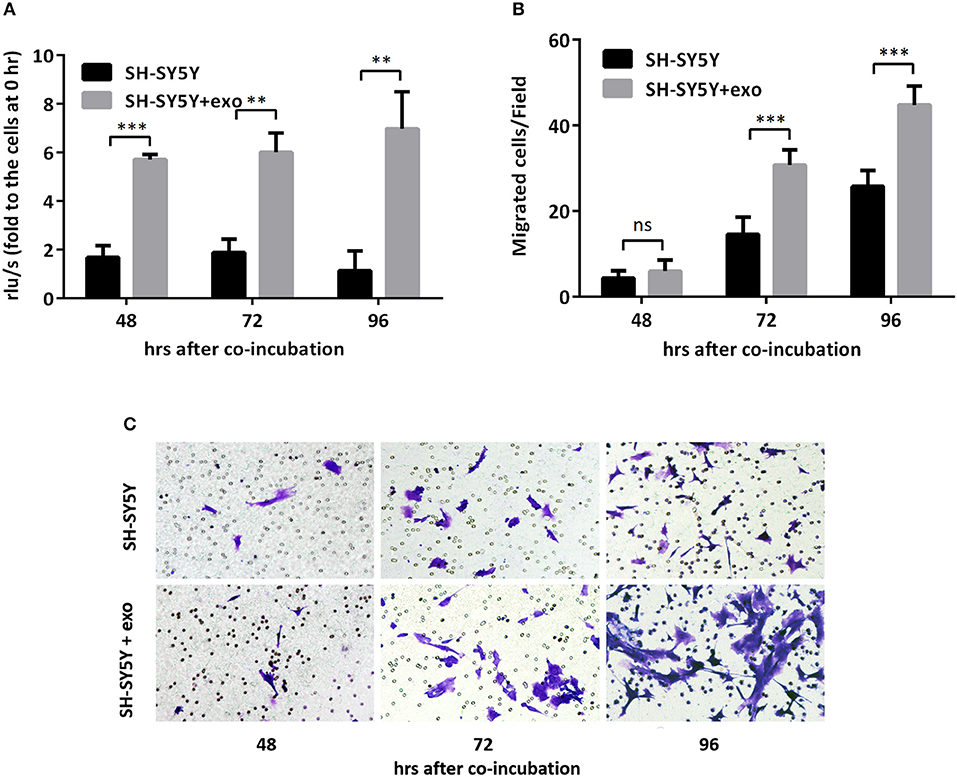
Figure 5. Exosomal hsa-miR199a-3p from SK-N-SH cells facilitated proliferation and migration of NB cells. (A) BrdU cell proliferation assay demonstrated that exosomal hsa-miR199a-3p obtained from SK-N-SH cells could promote SH-SY5Y cell proliferation 48, 72, and 96 h after co-incubation. (B,C) Transwell assay revealed exosomal hsa-miR199a-3p obtained from SK-N-SH cells could promote SH-SY5Y cell migration 72 and 96 h after co-incubation. Quantification of the migrated cells per field was shown in (B) and representative photographs were shown in (C). All the data were presented as the mean ± SD from three independent experiments. **p < 0.01; ***p < 0.001; ns, not significant. exo, exosome.
hsa-miR199a-3p Inhibits NEDD4 Expression by Binding to Its mRNA in NB Cells
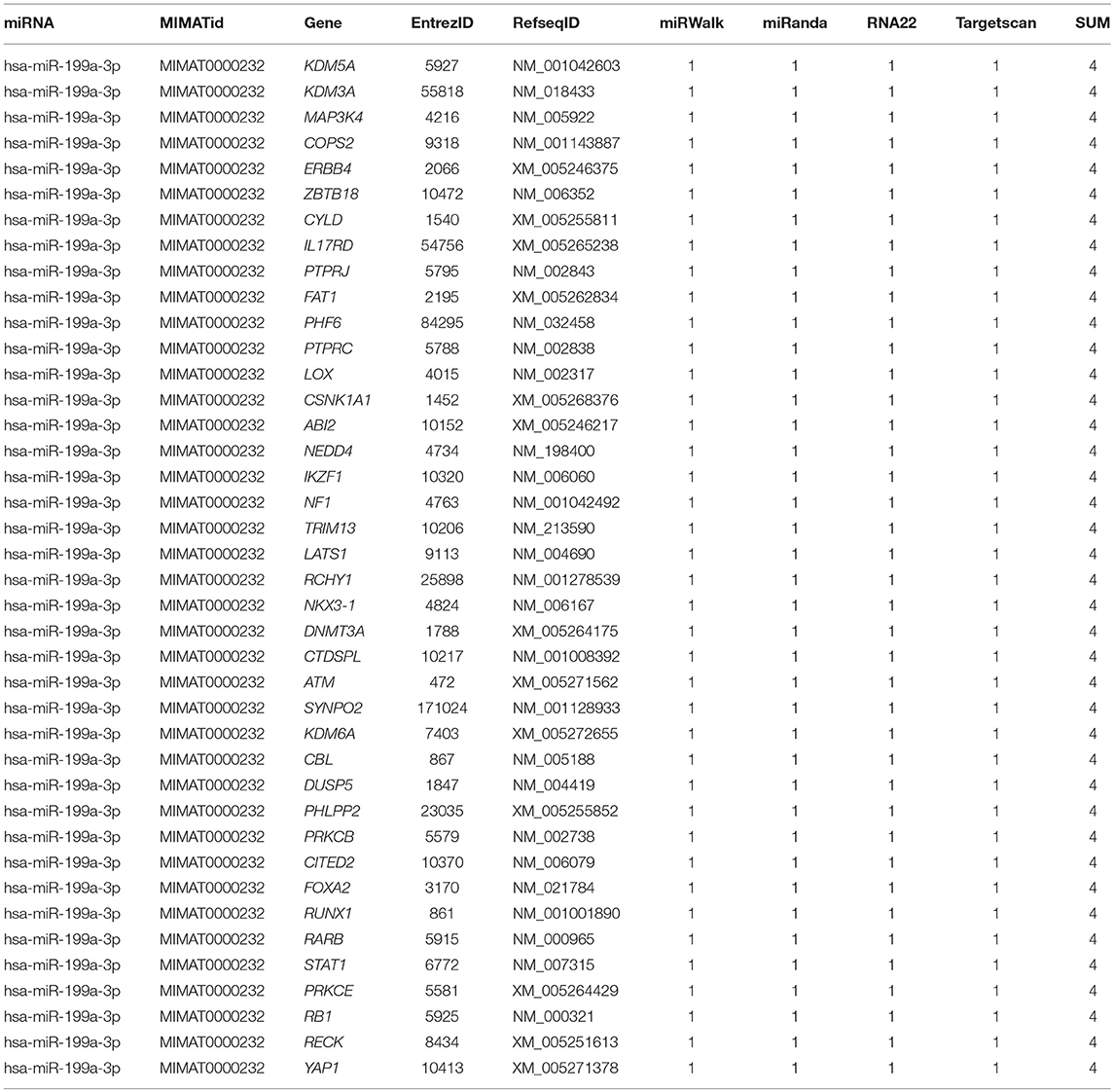
To identify the direct target of exosomal hsa-miR199a-3p, five miRNA target- predicting databases (miRWalk, miRanda, RNA22, Targetscan, and Tumor Suppressor Gene Database) were used. Combining the overlapped predicted targets across the five databases, 40 potential target genes of hsa-miR199a-3p were identified (listed in Table 1). Among these 40 predicted targets, NEDD4, a tumor suppressor gene, was selected because of its known biological functions related to NB progression. To confirm the interaction between hsa-miR199a-3p and NEDD4, the hsa-miR199a-3p mimic and wild-type NEDD4 overexpression plasmid, which contains the 3′-untranslated regions (UTR) of NEDD4, were transfected separately or in combination into SK-N-SH cells. As determined by qRT-PCR, the expression of NEDD4 were negatively associated with hsa-miR199a-3p (Figures 6A,B), suggesting that NEDD4 may be taken as the acting target of the antagonist for hsa-miR199a-3p in NB. In addition, Western blotting showed the expression of NEDD4 was down-regulated in SK-N-SH cells after transfection with hsa-miR199a-3p mimic (Figure 6C). To further demonstrate the direct regulation of NEDD4 by hsa-miR199a-3p, the pmiR-GLO vectors with or without wild-type or mutant 3′-UTR of NEDD4 mRNA were constructed (Figure 6D). In NEDD4 associated assays, the luciferase activity of the NEDD4 wild-type group was significantly inhibited by hsa-miR199a-3p (Figure 6E). These results indicate that hsa-miR199a-3p may inhibit NEDD4 expression by binding to the 366-373 site of the 3′-UTR of NEDD4 mRNA in NB cells.
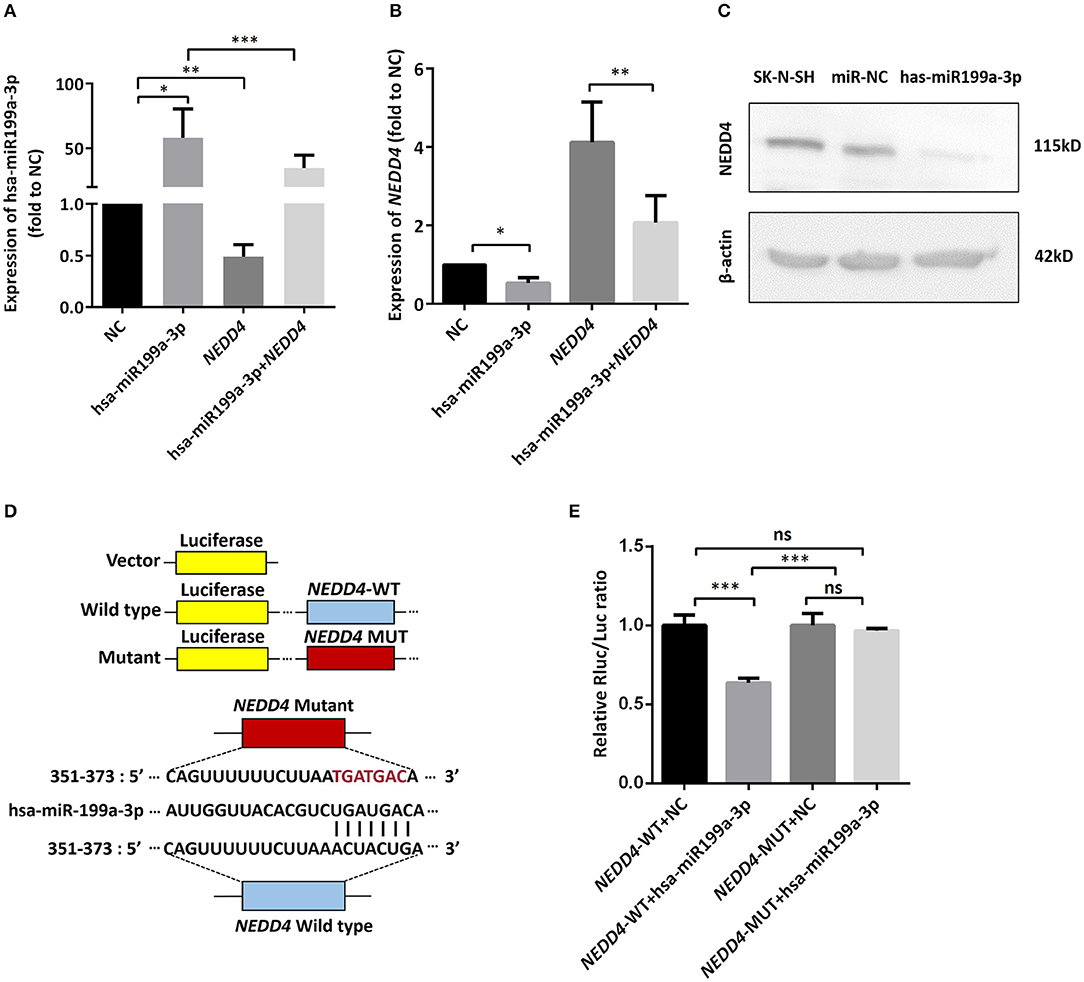
Figure 6. hsa-miR199a-3p inhibited NEDD4 expression by binding to its mRNA in NB cells. (A,B) Expression of hsa-miR199a-3p (A) and the mRNA levels of NEDD4 (B) after transfection with hsa-miR199a-3p mimic and NEDD4 overexpression plasmid separately or in combination, as determined by qRT-PCR. The data showed that the expression of hsa-miR199a-3p was negatively correlated with the NEDD4 expression levels. (C) Western blotting showed the expression of NEDD4 was down-regulated in SK-N-SH cells after transfection with hsa-miR199a-3p mimic. The figure was a representative from three independent experiments. (D) Vectors with or without wild type or mutant 3′-untranslated regions (UTR) of NEDD4 mRNA and diagram of the binding site of 3′-UTR of NEDD4 mRNA and hsa-miR199a-3p. (E) Data from luciferase assay showed that in NEDD4 associated assays, the luciferase activity of the NEDD4 wild-type group was significantly inhibited by hsa-miR199a-3p. The data were presented as the mean ± SD from three independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001; ns, not significant. NC, negative controls; WT, wild-type; MUT, mutant.
Exosomal hsa-miR199a-3p Promotes Proliferation and Migration via NEDD4 in NB Cells
To further verify whether the effects of exosomal hsa-miR199a-3p on the proliferation and migration in NB are mediated by targeting NEDD4, rescue assays were performed. Overexpression of NEDD4 alone in SK-N-SH cells resulted in a significant decrease in cell proliferation and migration 96 and 72 h after transfection, respectively. In contrast, simultaneous overexpression of NEDD4 and the hsa-miR199a-3p mimic in SK-N-SH cells can restore their proliferation and migration capacity (Figure 7). Taken together, our data indicate that hsa-miR199a-3p promotes proliferation and facilitates migration by regulating NEDD4 expression.
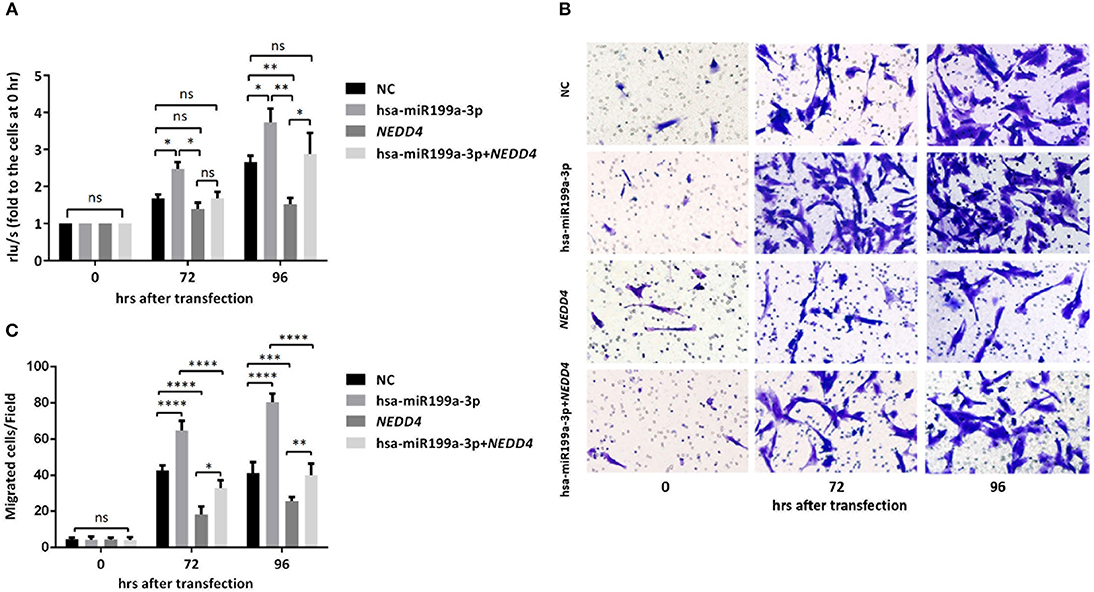
Figure 7. hsa-miR199a-3p promoted proliferation and migration via NEDD4 in NB cells. (A) Rescue assays showed that overexpression of hsa-miR199a-3p mimic can restore the proliferation of SK-N-SH cells 96 h after transfection by antagonizing the inhibitory effects of NEDD4. (B,C) Overexpression of hsa-miR199a-3p mimic can inhibit migration capacity of SK-N-SH cells 72 and 96 h after transfection by antagonizing the inhibitory effects of NEDD4. Representative photographs were shown in B and quantification of the migrated cells per field was shown in (C). All the data were presented as the mean ± SD from three independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns, not significant.
Discussion
Fast, easy and non-invasive detection in NB patients is important for guiding its therapeutic strategy and assessing prognosis. So far, the biopsy of tumor tissues or bone marrows from patients suspected with NB is the golden standard for NB diagnosis (12, 13), but it is not specific enough to identify histology for the tumor heterogeneous in some cases. Although several advanced investigations that include next-generation sequencing of tumor cells and immunological assessments to define targets in the tumor microenvironment can contribute to the diagnosis and staging of NB, these detective methods are invasive, time-consuming and also difficult to repeat the procedure (14, 15). More recently, there has been a growing number of publications focusing on the function of exosomes in body fluids as an early and non-invasive diagnostic biomarker in malignant tumors. For example, serum exosomal hsa-miR-18a, hsa-miR-221, hsa-miR-222, and hsa-miR-224 have been reported as novel biomarkers for hepatocellular carcinoma (HCC) (16), and these serum exosomal miRNAs are better in distinguishing HCC from chronic hepatitis B or liver cirrhosis compared to the corresponding serum circulating miRNAs. In another study, the test of glypican-1 (GPC1) protein on cancer-cell-derived exosomes may serve as a potential diagnostic and screening tool to detect early stages of pancreatic cancer (17). Because tumor-derived exosomes have been reported to carry a cargo of molecules and factors (DNAs, miRNAs, lncRNAs and proteins) that are able to transfer information from the parent tumor cells to other cells present within and outside the tumor microenvironment (18), they are potential biomarkers for the early diagnosis of cancers. In this study, the plasma exosomal hsa-miR199a-3p was firstly reported to be upregulated in NB patients and correlated with high-risk NB, suggesting that the plasma exosomal hsa-miR199a-3p may be a suitable diagnostic and prognostic biomarker for NB in clinic without invasive operation.
Although there is no report about circulating exosomal hsa-miR199a-3p, circulating hsa-miR199a-3p has been demonstrated to play vital roles in the progression of malignant tumors. Interestingly, hsa-miR199a-3p shows opposite effects in distinct cancers (19). For instance, hsa-miR199a-3p was observed significantly lower in the tumor tissues compared with their adjacent non-tumor tissues in patients with HCC, and further research found that hsa-miR199a-3p can suppress tumor growth, migration, invasion, and angiogenesis via targeting VEGFA, VEGFR1, VEGFR2, HGF, and MMP2 proteins in HCC (20). Moreover, hsa-miR199a-3p can also restrain migration and invasion of breast cancer by downregulating PAK4/MEK/ERK signaling pathway (21). However, hsa-miR199a-3p was found to be up-regulated in gastric cancer, biliary tract cancer, and esophageal adenocarcinoma as an oncogene (22–24). In the current study, we reported that exosomal hsa-miR199a-3p can contribute to promote tumor proliferation and migration, indicating that hsa-miR199a-3p was identified as an oncogene in NB.
NEDD4 is an HECT domain ubiquitin E3 ligase that play a critical role in the regulation of membrane receptors and endocytic machinery components (25). It is believed to be a potential dual regulator of PTEN, which exerts a tumor suppressive function by catalyzing PTEN mono-ubiquitination and regulating PTEN nuclear translocation (26). Furthermore, NEDD4 can interact with Myc via ubiquitination and degradation of Myc protein directly. Myc protein was considered as a predictor for poor prognosis in patients with NB. Repression of NEDD4 protein leads to the upregulation of Aurora A gene, which is important to stabilize Myc protein in NB (27). In this study, we observed that hsa-miR199a-3p caused the downregulation of NEDD4 in NB cells and consequently promoted tumor proliferation and migration. To our best knowledge, this is the first study demonstrating that upregulation of exosomal hsa-miR199a-3p in NB can promote proliferation and migration via the inhibition of NEDD4 expression in vivo and in vitro and lead to a poor prognosis.
There are also some limitations to the current study that need to be considered. Firstly, because of the particularity of NB, this study did not include suitable neuron-derived normal cells as controls. Second, this study is a cross-sectional study. Longitudinal studies focusing on the different periods of NB will have an important role in terms of disease progression. In addition, the sample size was limited, so larger research studies from different populations need to be conducted to confirm the findings.
In conclusion, this study reveals that exosomal hsa-miR199a-3p can promote tumor proliferation and migration via decreasing NEDD4 expression in NB patients and cells. Our study elucidates a new molecular mechanism concerning the initiation and progression of NB, which indicates that exosomal hsa-miR199a-3p can be applicated as a fast, easy and non-invasive detection biomarker and also can contribute to investigate novel therapeutic strategies for NB.
Ethics Statement
This study was carried out in accordance with the 1964 Declaration of Helsinki and its later amendments with written informed consent from all subjects. The protocol was approved by the Institutional Review Board and the Ethics Committee of SCMC (SCMCIRB-K2017042), and written informed consent was obtained from the parents of each participant.
Author Contributions
XM, SG, and NS conceived the project, analyzed the data and wrote the manuscript. JM, MX, MY, JH, HC, YG, and CX performed the experiments and analyzed the data.
Funding
This work was financially supported by the National Key R&D Program of China to YG (2018YFC1313000/2018YFC1313005), Shanghai Science and Technology Commission to XM (18ZR1424500), the Science and Technology Commission of Shanghai Municipality to SG (17441903200) and MX (17411950402), Innovation Foundation for Ph.D. Candidate of Shanghai Jiao Tong University School of Medicine to JM (BXJ201740).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the patients and their parents for the support and cooperation in publishing this work.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2019.00459/full#supplementary-material
References
1. Decarolis B, Simon T, Krug B, Leuschner I, Vokuhl C, Kaatsch P, et al. Treatment and outcome of Ganglioneuroma and Ganglioneuroblastoma intermixed. BMC Cancer. (2016) 16:542. doi: 10.1186/s12885-016-2513-9
2. Mora J, Gerald WL. Origin of neuroblastic tumors: clues for future therapeutics. Expert Rev Mol Diagn. (2004) 4:293–302. doi: 10.1586/14737159.4.3.293
3. Ward E, DeSantis C, Robbins A, Kohler B, Jemal A. Childhood and adolescent cancer statistics, 2014. CA Cancer J Clin. (2014) 64:83–103. doi: 10.3322/caac.21219
4. Louis CU, Shohet JM. Neuroblastoma: molecular pathogenesis and therapy. Annu Rev Med. (2015) 66:49–63. doi: 10.1146/annurev-med-011514-023121
5. Ahmed AA, Zhang L, Reddivalla N, Hetherington M. Neuroblastoma in children: update on clinicopathologic and genetic prognostic factors. Pediatr Hematol Oncol. (2017) 34:165–85. doi: 10.1080/08880018.2017.1330375
6. Hannafon BN, Trigoso YD, Calloway CL, Zhao YD, Lum DH, Welm AL, et al. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res. (2016) 18:90. doi: 10.1186/s13058-016-0753-x
7. Felicetti F, De Feo A, Coscia C, Puglisi R, Pedini F, Pasquini L, et al. Exosome-mediated transfer of miR-222 is sufficient to increase tumor malignancy in melanoma. J Transl Med. (2016) 14:56. doi: 10.1186/s12967-016-0811-2
8. Vaksman O, Trope C, Davidson B, Reich R. Exosome-derived miRNAs and ovarian carcinoma progression. Carcinogenesis. (2014) 35:2113–20. doi: 10.1093/carcin/bgu130
9. Haug BH, Hald OH, Utnes P, Roth SA, Lokke C, Flaegstad T, et al. Exosome-like extracellular vesicles from MYCN-amplified neuroblastoma cells contain oncogenic miRNAs. Anticancer Res. (2015) 35:2521–30.
10. Challagundla KB, Wise PM, Neviani P, Chava H, Murtadha M, Xu T, et al. Exosome-mediated transfer of microRNAs within the tumor microenvironment and neuroblastoma resistance to chemotherapy. J Natl Cancer Inst. (2015) 107:djv135. doi: 10.1093/jnci/djv135
11. Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. (2006) Chapter 3:Unit 3 22. doi: 10.1002/0471143030.cb0322s30
12. Burchill SA, Beiske K, Shimada H, Ambros PF, Seeger R, Tytgat GA, et al. Recommendations for the standardization of bone marrow disease assessment and reporting in children with neuroblastoma on behalf of the International Neuroblastoma Response Criteria Bone Marrow Working Group. Cancer. (2017) 123:1095–105. doi: 10.1002/cncr.30380
13. Park JR, Bagatell R, Cohn SL, Pearson AD, Villablanca JG, Berthold F, et al. Revisions to the international neuroblastoma response criteria: a consensus statement from the national cancer institute clinical trials planning meeting. J Clin Oncol. (2017) 35:2580–7. doi: 10.1200/JCO.2016.72.0177
14. Pugh TJ, Morozova O, Attiyeh EF, Asgharzadeh S, Wei JS, Auclair D, et al. The genetic landscape of high-risk neuroblastoma. Nat Genet. (2013) 45:279–84. doi: 10.1038/ng.2529
15. Asgharzadeh S, Salo JA, Ji L, Oberthuer A, Fischer M, Berthold F, et al. Clinical significance of tumor-associated inflammatory cells in metastatic neuroblastoma. J Clin Oncol. (2012) 30:3525–32. doi: 10.1200/JCO.2011.40.9169
16. Sohn W, Kim J, Kang SH, Yang SR, Cho JY, Cho HC, et al. Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Exp Mol Med. (2015) 47:e184. doi: 10.1038/emm.2015.68
17. Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. (2015) 523:177–82. doi: 10.1038/nature14581
18. Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. (2008) 10:619–24. doi: 10.1038/ncb1725
19. Gu S, Chan WY. Flexible and versatile as a chameleon-sophisticated functions of microRNA-199a. Int J Mol Sci. (2012) 13:8449–66. doi: 10.3390/ijms13078449
20. Ghosh A, Dasgupta D, Ghosh A, Roychoudhury S, Kumar D, Gorain M, et al. MiRNA199a-3p suppresses tumor growth, migration, invasion and angiogenesis in hepatocellular carcinoma by targeting VEGFA, VEGFR1, VEGFR2, HGF and MMP2. Cell Death Dis. (2017) 8:e2706. doi: 10.1038/cddis.2017.123
21. Li SQ, Wang ZH, Mi XG, Liu L, Tan Y. MiR-199a/b-3p suppresses migration and invasion of breast cancer cells by downregulating PAK4/MEK/ERK signaling pathway. IUBMB Life. (2015) 67:768–77. doi: 10.1002/iub.1433
22. Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, et al. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. (2010) 11:136–46. doi: 10.1016/S1470-2045(09)70343-2
23. Shigehara K, Yokomuro S, Ishibashi O, Mizuguchi Y, Arima Y, Kawahigashi Y, et al. Real-time PCR-based analysis of the human bile microRNAome identifies miR-9 as a potential diagnostic biomarker for biliary tract cancer. PLoS ONE. (2011) 6:e23584. doi: 10.1371/journal.pone.0023584
24. Feber A, Xi L, Pennathur A, Gooding WE, Bandla S, Wu M, et al. MicroRNA prognostic signature for nodal metastases and survival in esophageal adenocarcinoma. Ann Thorac Surg. (2011) 91:1523–30. doi: 10.1016/j.athoracsur.2011.01.056
25. Boase NA, Kumar S. NEDD4: the founding member of a family of ubiquitin-protein ligases. Gene. (2015) 557:113–22. doi: 10.1016/j.gene.2014.12.020
26. Trotman LC, Wang X, Alimonti A, Chen Z, Teruya-Feldstein J, Yang H, et al. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell. (2007) 128:141–56. doi: 10.1016/j.cell.2006.11.040
Keywords: neuroblastoma, exosome, microRNA, hsa-miR199a-3p, NEDD4, proliferation, migration, biomarker
Citation: Ma J, Xu M, Yin M, Hong J, Chen H, Gao Y, Xie C, Shen N, Gu S and Mo X (2019) Exosomal hsa-miR199a-3p Promotes Proliferation and Migration in Neuroblastoma. Front. Oncol. 9:459. doi: 10.3389/fonc.2019.00459
Received: 09 January 2019; Accepted: 14 May 2019;
Published: 12 June 2019.
Edited by:
Elizabeth R. Lawlor, University of Michigan Health System, United StatesReviewed by:
Joanna Kitlinska, Georgetown University, United StatesYong-mi Kim, Children's Hospital of Los Angeles, United States
Copyright © 2019 Ma, Xu, Yin, Hong, Chen, Gao, Xie, Shen, Gu and Mo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nan Shen, shennan1986@hotmail.com; Song Gu, gusong@shsmu.edu.cn; Xi Mo, Xi.Mo@shsmu.edu.cn
†These authors have contributed equally to this work
 Jing Ma
Jing Ma Min Xu2†
Min Xu2† Minzhi Yin
Minzhi Yin Nan Shen
Nan Shen Xi Mo
Xi Mo