- 1Department of Medical Biochemistry and Biophysics, Division of Vascular Biology, Karolinska Institute, Stockholm, Sweden
- 2MediCity Research Laboratory, University of Turku, Turku, Finland
Breast cancer progression toward metastatic disease is linked to re-activation of epithelial–mesenchymal transition (EMT), a latent developmental process. Breast cancer cells undergoing EMT lose epithelial characteristics and gain the capacity to invade the surrounding tissue and migrate away from the primary tumor. However, less is known about the possible role of EMT in providing cancer cells with properties that allow them to traffic to distant sites. Given the fact that pro-metastatic cancer cells share a unique capacity with immune cells to traffic in-and-out of blood and lymphatic vessels we hypothesized that tumor cells undergoing EMT may acquire properties of immune cells. To study this, we performed gene-profiling analysis of mouse mammary EpRas tumor cells that had been allowed to adopt an EMT program after long-term treatment with TGF-β1 for 2 weeks. As expected, EMT cells acquired traits of mesenchymal cell differentiation and migration. However, in addition, we found another cluster of induced genes, which was specifically enriched in monocyte-derived macrophages, mast cells, and myeloid dendritic cells, but less in other types of immune cells. Further studies revealed that this monocyte/macrophage gene cluster was enriched in human breast cancer cell lines displaying an EMT or a Basal B profile, and in human breast tumors with EMT and undifferentiated (ER−/PR−) characteristics. The results identify an EMT-induced monocyte/macrophage gene cluster, which may play a role in breast cancer cell dissemination and metastasis.
Introduction
Metastasis is a complex multistep process whereby pro-metastatic cancer cells manage to break through cellular and molecular barriers and adapt to foreign microenvironments as they traffic to, and colonize distant organs (1, 2). The journey starts at primary tumor sites, where cancer cells invading through the basement membrane and into the surrounding tissue may migrate away from the primary tumor. For further trafficking and completion of the metastatic process, they need to intravasate into blood and lymphatic vessels, and stay alive in the circulation. Finally, they need to extravasate from vessels and propagate at distant sites. Based on both clinical observations and experimental data, it appears that only a minor fraction of cancer cells actually display all these pro-metastatic capabilities (1–4). Yet, it is not clear what these properties are, or how they are induced and maintained throughout the metastatic journey.
Epithelial–mesenchymal transition is considered to play an important role in the initial parts of the metastatic cascade – providing cancer cells with invasive, migratory, and cancer stem cell properties (5–7). Epithelial–mesenchymal transition (EMT) is a plastic process, and migratory cancer cells may cycle between epithelial and mesenchymal states (8, 9). Yet, less is known about the role of EMT in regulating trafficking of tumor cells in-and-out of blood and lymphatic vessels. The term “mesenchymal” is a rather unspecific definition. Although it may adequately describe the elongated, fibroblast-like morphology, and the invasive and migratory properties of EMT cells, it may not be sufficient to fully describe the properties of pro-metastatic EMT cells capable of disseminating through vessels and tissues. Clearly, fibroblasts and other type of normal mesenchymal cells do not migrate similar to metastatic cancer cells. Instead, the only cells that actually do traffic in our bodies similar to metastatic cancer cells are immune cells. In fact, immune cells are highly specialized in migrating through vessels and various tissue compartments as they migrate to different sites during inflammation.
Recently, it has been reported that circulating tumor cells (CTCs) in breast cancer patients may display epithelial, or mesenchymal/EMT properties, or a combination of both (10). Interestingly, the presence of CTCs with EMT properties was associated with worse prognosis. This suggests that migratory tumor cells that are capable of maintaining their EMT phenotype, not only within the primary tumor and its surroundings, but also as they migrate further and eventually reach the circulation, may represent a pool of pro-metastatic tumor cells.
We hypothesized that breast cancer cells not only being able to undergo EMT, but to sustain an EMT program, might develop properties that may be used at later steps of the metastatic process. In particular, we were interested to determine whether such EMT cells would acquire immune cell properties, which might be used for trafficking through the vasculature and dissemination to distant sites. To begin to address this, we performed gene expression profiling of mammary tumor EpRas cells that had been allowed to adopt an EMT program after long-term treatment with transforming growth factor beta 1 (TGF-β1) for 2 weeks. TGF-β1 is an inflammatory cytokine and a potent inducer of EMT (11). We found that long-term EMT cells indeed display properties of mesenchymal cells (fibroblasts). However, in addition, we identified a cluster of EMT-induced genes enriched in monocyte-derived immune cells including macrophages, mast cells, and myeloid dendritic cells (DCs). This cluster was enriched in human breast cancer cell lines classified as Basal B cells, and in human breast tumors negative for estrogen and progesterone receptors. It will be of interest for further studies to determine the role of genes within this cluster in breast cancer metastasis.
Materials and Methods
Cell Culture
Mouse mammary EpRas tumor cells were a kind gift from the laboratory of H. Beug (Wien, Austria) and were generated through stable overexpression of the Ha-Ras oncogene in mouse mammary EpH4 epithelial cells (12). EpRas cells were cultured as adherent epithelial cells in DMEM/F12 medium supplemented with 10% fetal calf serum (FCS), 1% penicillin/streptomycin, and 1% l-glutamine. Cells were treated with 5 ng/ml recombinant human TGF-β1 (Peprotech Nordic, Stockholm, Sweden) for 48 h, or 2 ng/ml for 7 or 14 days (long-term). During treatment for 7 and 14 days cells were split every 3 days and new TGF-β1 was added in new DMEM/F12 medium supplemented with 10% FCS, 1% penicillin/streptomycin, and 1% l-glutamine. Cells were culture in a humidified incubator (37°C, 5% CO2). For gene expression analysis by microarray and q-PCR, 5 × 105 cells of treated and non-treated cells were seeded in six-well plates 48 h prior to cells lysis.
Immunofluorescence Analysis
Cells grown on coverslips were fixed in absolute ethanol for 20 min. Samples were blocked in incubation buffer (5% normal goat serum and 0.1% BSA in PBS) for 1 h at room temperature. Primary antibody was diluted in 0.1% BSA/PBS and incubated 1 h at room temperature. The following primary antibodies and dilutions were used: mouse anti-E-cadherin (1:1000; clone 36, BD Biosciences, Stockholm, Sweden) and vimentin (1:1000; clone VI-10, Abcam, Cambridge, UK). Cells were washed five times in PBS/BSA and further incubated with secondary antibodies (1:1000 dilution) for 30 min and mounted in vectashield mounting media supplemented with DAPI (Vector Laboratories Ltd, Peterborough, UK). Immunoflouresent staining was assessed using a Nikon Eclipse microscope equipped for immunofluorescence.
Gene Expression Analysis (Q-PCR)
Total RNA was extracted using an RNeasy mini kit (Qiagen, Valencia, CA, USA) according to manufacturer’s instructions. An amount of 2 μg of total RNA per sample was used for first-strand cDNA synthesis using the iScript cDNA synthesis kit (BioRad, Solna, Sweden). For q-PCR analysis, 5 ng of cDNA was used for PCR amplification by KAPA SYBR fast q-PCR kit (Kapa Biosystems, Wilmington, MA, USA) with validated QuantiTect primers (Qiagen). The following genes were analyzed: Cdh1 (primer: QT00121163), Ocln (primer: QT00111055), Cdh5 (primer: QT00110467), Snai1 (primer: QT00240940), Snai2 (primer: QT00098273), Zeb1 (primer: QT00105385), Zeb2 (primer: QT00148995), and Twist1 (primer: QT00097223). The PCR was carried out as follows: 3 min at 95°C followed by 35 cycles of 3 s at 95°C, 20 s at 55°C, and 2 s extension step at 72°C in RotorGene RG-3000A PCR system.
Western Blot
Cells were lysed in RIPA buffer [50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS supplemented with protease inhibitors (Complete, Roche, Basel, Switzerland)]. Further, samples were boiled in Laemmli sample buffer (BioRad), separated by SDS–PAGE under reducing conditions, and transferred to a nitrocellulose membrane using the iBlot (Invitrogen, Carlsbad, CA, USA) dry transfer. The membrane was blocked using Blocking reagent (Roche) for 1 h and then incubated with primary antibodies over night at 4°C. Following antibodies were used: rabbit-α-LSP1 (1:500, HPA019693, Atlas, Stockholm), chicken-α Cotl1 (1:500) (13), rabbit-α-Dock8 (1:500, HPA003218, Atlas, Stockholm), and rabbit-α-Calnexin (1:1000) (14). Further secondary antibodies, α-mouse and α-rabbit Horseradish peroxidase IgG (1:8000, #7074, Cell Signaling Technology, Danvers, MA, USA), were incubated for 1 h at room temperature. All antibodies were diluted in blocking reagent (Roche). Immunoreactive bands were visualized by chemiluminescence (Roche) and developed using a LAS1000 system (Fuji Photo Film Co.).
Microarray Analysis
Microarray analysis was performed at the core facility for Bioinformatics and Expression Analysis (BEA) at the Karolinska Institutet. Total RNA was prepared from triplicate samples of EpRas cells that were either left untreated, or treated for long-term (14 days) with 2 ng/ml of TGFβ1 (Peprotech), and purified using RNeasy Mini Kit (Qiagen), supplemented with RNase-Free DNase (Qiagen). One hundred fifty nanogram of total RNA per sample was used to synthesize cDNA using a cDNA synthesis kit (Ambion/Life Technologies, Stockholm, Sweden). A GeneChip Sense Target Labeling kit (Affymetrix) was used to fragment and biotin label single stranded cDNA, which was probed on a MoGene-1_1-st-v1 array (Affymetrix) using an Affymterix Gene Titan instrument. Data were analyzed using the Robust Multi-array Average (RMA) algorithm. Gene expression data presented in this manuscript have been deposited to the Gene Expression Omnibus (GEO) database (Accession number: GSE59922).
Cluster Analysis of Genes Regulated during Long-Term Induced EMT
Cluster analysis of genes upregulated or downregulated more than 1.5-fold was performed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID)1, which is based on functional annotation of genes. Clusters with a P ≤ 0.05 were considered as significantly enriched.
Meta-Analysis of the Expression of Long-Term Induced EMT Genes in Immune Cells
All 515 genes significantly upregulated by >1.5-fold in long-term EMT cells were analyzed for their degree of expression in fibroblasts (NIH3T3 and mouse embryonic fibroblasts) and different types of immune cells using the BioGPS database2. Gene expression was defined as “enriched” if the level in a certain cell type was higher than the average level for all 96 human tissues and cell lines included in the database. The following type of immune cells were included: Dendritic plasmacytoid (B220+), DCs lymphoid (CD8α+), DCs myeloid (CD8α−), Macrophage bone marrow, Macrophage (peri LPS thio 0 h), Mast cells, Granulocytes (Mac1+/Gr1+), NK cells, T cells FoxP3+, B cells GL7 positive Alum, B cells GL7 positive KLH, B cells (GL7 negative KLH), B cells (GL7 negative Alum), B cells marginal zone, T cells (CD4+), and T cells (CD8+).
One hundred twenty-three genes were found to be enriched in myeloid DCs, Macrophages, Mast cells, and Granulocytes, but not in fibroblasts, and were clustered into a monocyte/macrophage group, which was further analyzed.
Meta-Analysis of the Expression of Long-Term Induced EMT Genes in Human Breast Cancer Cell Lines
The 123 genes induced in long-term EMT cells and enriched in the monocyte/macrophage gene cluster were analyzed for their expression in datasets from 51 human breast cancer cell lines available from the European Bioinformatics Institute at the European Molecular Biology Laboratory (EMBL-EBI)3. The E-GEOD-41313 51 database, which contains microarray profiles (Affymetrix HT HG-U133+ PM Array Plate) of 51 human breast cancer cell lines, was used.
First, all tumor cell lines in the database were analyzed for the expression of EMT markers. The ratio between the average expression values of the mesenchymal markers TWIST, SNAIL1, and VIM, and the epithelial markers OCLN, CLDN3, and CDH1, was calculated for the 51 cell lines. Cell lines with a mesenchymal/epithelial ratio higher than 1.1 were defined as tumors displaying EMT characteristics. The average expression of the monocyte/macrophage gene cluster was compared between the EMT and non-EMT groups of cell lines. The expression value for each gene was normalized to the average expression of that gene in all samples. Statistical differences with P ≤ 0.05 were defined as significant.
Second, 38 of the human breast cancer cell lines were grouped into Luminal, Basal A, and Basal B cells, according to a previous classification (15). The average expression of the monocyte/macrophage gene cluster was compared between the different classes of cell lines and P ≤ 0.05 was considered as a significant difference.
Meta-Analysis of the Expression of Long-Term Induced EMT Genes in Human Breast Tumors
Genes within the monocyte/macrophage gene cluster were analyzed for their expression in a database (E-GEOD-36771) of microarray datasets (Affymetrix GeneChip Human Genome U133 Plus 2.0) from 107 human breast tumors (EMBL-EBI).
First, the average expression of genes in the monocyte/macrophage cluster was calculated for each tumor sample. Second, the average values were compared to ER/PR status, and the expression of EMT markers (using the same strategy as described for the tumor cell lines). P ≤ 0.05 was considered as a significant difference.
Statistical Analysis
Data represent means ± SEM with three independent experiments in triplicate. Statistical analyses were determined by using Student’s t test. P < 0.05 was considered as significant difference between the groups. In experiments with multiple groups, Bonferroni’s test for multiple comparisons was used, and P ≤ 0.05 was considered as significant.
Results
Long-Term Induction of EMT
Mouse mammary EpRas tumor cells were treated with 5 ng/ml of TGF-β1 for 48 h, or with 2 ng/ml for 7 days or 14 days (long-term) to study the effect on the commonly used EMT markers E-cadherin and vimentin. Immunoblotting analysis showed slight reduction in E-cadherin levels and increased expression of vimentin in EpRas cells that had been treated with TGF-β1 for 48 h, compared to non-treated cells (Figure 1A). After 7 and 14 days of TGF-β1 treatment, changes in E-cadherin and vimentin levels were more pronounced (Figure 1B; Figure S1 in Supplementary Material). After 14 days, E-cadherin was no longer detected in TGF-β1 treated cells. Microscopic analysis demonstrated that these cells had adopted an elongated, spindle-like shaped morphology, a characteristic feature of EMT (Figure 1C). Furthermore, immunofluorescence staining showed that E-cadherin was absent at cell–cell junctions while vimentin was localized in the cytoplasm in these long-term treated cells. Together, these results indicated that EpRas cells that had been treated with TGF-β1 for 48 h were in the induction phase of EMT and thus, displayed both epithelial and mesenchymal properties. In contrast, EpRas cells that had been treated with TGF-β1 for 14 days had lost epithelial features indicating that they had been reprogramed into mesenchymal-like cells. Based on these results, we decided to focus on profiling long-term EMT cells to identify their properties, based on gene expression.
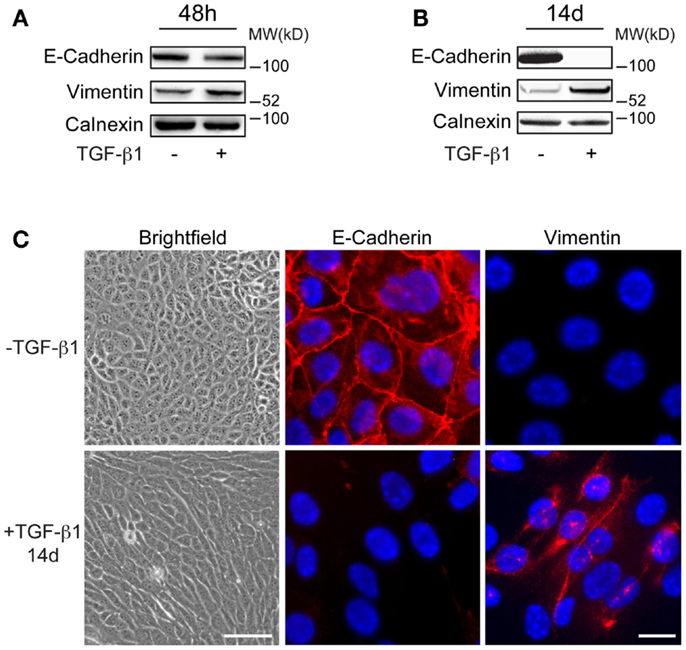
Figure 1. Characteristics of long-term EMT cells. (A,B) Western blot analysis showing more prominent changes in E-cadherin and vimentin expression in EpRas tumor cells after 14 days (B) compared to 48 h (A) of TGF-β1 exposure. Calnexin was used as a internal loading control. (C) Brightfield and immunofluorescence images showing that EpRas cells exposed to TGF-β1 for 14 days acquire characteristic features of EMT including an elongated, fibroblast-like morphology (left panels), loss of E-cadherin at cell–cell junctions (middle panels), and cytoplasmic staining of vimentin (right panels). Scale bars: 20 μm (left panels); 5 μm (middle and right panels).
Long-Term EMT Cells Display Mesenchymal Properties
Microarray-based gene expression profiling of triplicate samples revealed that 515 genes were significantly upregulated, and 463 genes downregulated by more than 1.5-fold in long-term EMT cells compared to control cells (Figure 2A). The whole array set can be accessed at GEO database (Accession number: GSE59922). The 100 most upregulated and downregulated genes are displayed in Tables S1 and S2 in Supplementary Material). As expected, epithelial genes known to be inactivated during EMT in breast cancer cells, such as Cdh1 (E-cadherin), Ocln (Occludin), Epcam, Cldn7 (Claudin 7), Id1, Krt7, Esr1, Cav2, Dsp, Gata3, and Cadm1 were among the downregulated genes. Reciprocally, known EMT-related genes, such as Tnc, Mmp9, Jag1, Itgb3, Cdh5, Sema7a, Ctsw, and Itg5 were among the genes significantly upregulated in long-term EMT cells. Changes in the expression of several of these genes were validated by q-PCR (Figure S2A in Supplementary Material).
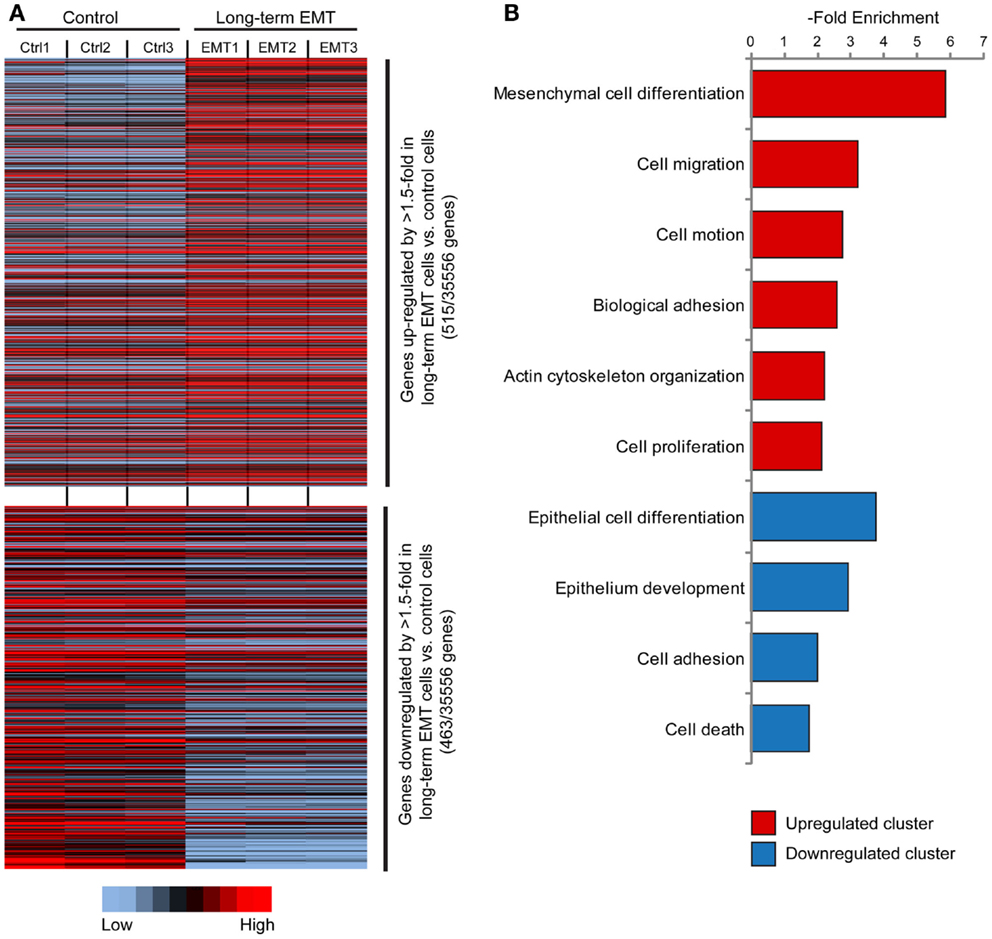
Figure 2. Gene expression profiling of long-term EMT cells. (A) Heat map showing genes significantly (P ≤ 0.05) upregulated (515 genes, upper part) or downregulated (463 genes, lower part) by >1.5-fold in long-term EMT vs. control EpRas cells. (B) Results from cluster analysis of genes significantly upregulated or downregulated in long-term EMT cells.
Interestingly, Zeb1 and Zeb2 were the only known EMT-inducing transcription factors that were upregulated in long-term EMT cells. In contrast, none of the master regulators of EMT – Snai1, Snai2, and Twist1, nor the EMT promoting factors Tcf3/4, Lef1, Foxc2, and Ets1 were upregulated. In addition, the expression of known epigenetic regulators of EMT including Hdac factors (Hdac1-6) and Mcm factors (Mcm 2-7) was unchanged in long-term EMT cells. Expression of Snai1, Snai2, and Twist1 instead appeared to peak at earlier time points as assessed by q-PCR analysis of EpRas cells treated for 48 h or 7 days with TGF-β1 (Figure S2B in Supplementary Material).
Cluster analysis showed that the induced genes were associated with mesenchymal cell differentiation, cell migration, and motility (Figure 2B; Table S3 in Supplementary Material). In addition, genes encoding proteins involved in non-epithelial cell–cell adhesion including various integrins (Itga2, Itga5, Itga7, and Itgb3) and cadherins (Cdh5, Cdh10, and Cdh17), as well as other cell adhesion molecules, such as Ncam1, Alcam, and Mcam, were induced. As expected, downregulated genes were linked to epithelial differentiation or development, and cell adhesion. Some downregulated genes were also associated with positive regulation of cell death.
Identification of a Monocyte/Macrophage Gene Cluster Induced in Long-Term EMT Cells
We used the BioGPS database to compare the expression of the 515 induced EMT genes in both mesenchymal cells (NIH3T3 and Mouse Embryonic Fibroblasts), and different types of immune cells. As expected, a large cluster of genes was enriched in fibroblasts (Figure 3). In addition, we found another cluster of 123 genes that was significantly enriched in monocyte-derived macrophages, mast cells, and DCs of a myeloid type, but not in B cells, T cells, other type of DCs, or granulocytes.
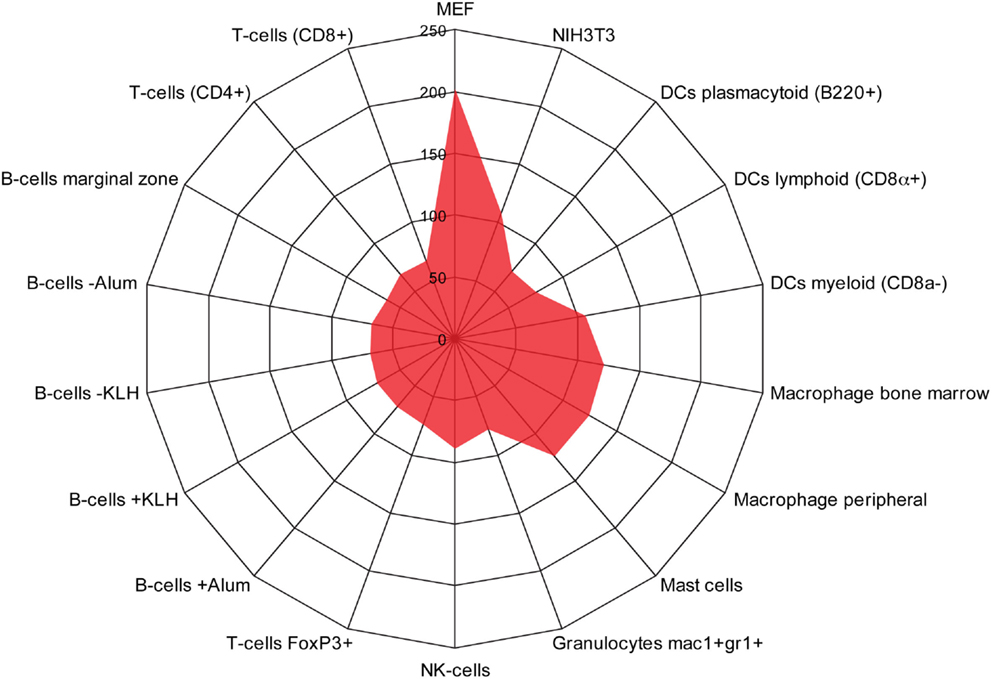
Figure 3. Identification of a monocyte/macrophage cluster in long-term EMT cells. Radar chart showing the number of genes upregulated in long-term EMT cells and enriched in mesenchymal cells [NIH3T3 and mouse embryonic fibroblasts (MEF)] or various types of immune cells. The highest number of genes was found in MEFs (202) followed by macrophages (bone marrow: 124, peripheral: 121), mast cells (124), NIH3T3 cells (108), and dendritic cells myeloid (106). Other immune cells had lower numbers that pended between 63 and 89. Thus, in addition to mesenchymal properties, properties of monocyte-derived cells were enriched in long-term EMT cells.
The largest pool of genes in this monocyte/macrophage cluster encodes plasma membrane proteins. Some of these, such as Cd53, Gab2, Glipr1, Laptm5, Lsp1, and Rap2a were enriched in different types of monocyte-derived cells (macrophages, mast cells, and myeloid DCs) (Table 1). Others, such as Cnnm4, Kctd13, Fyn, Dag1a, and Sema6d were more restrictively enriched in some of the cell types. A similar type of variation in enrichment between different genes and different type of immune cells was seen for the second and third largest sets of genes, which encode nuclear and cytosolic proteins, respectively. Western blot analysis was performed to validate changes in gene expression for one gene from each of the groups encoding plasma membrane (Lsp1), cytosol (Dock8), and cytoskeletal (Cotl1) proteins (Figure S3 in Supplementary Material).
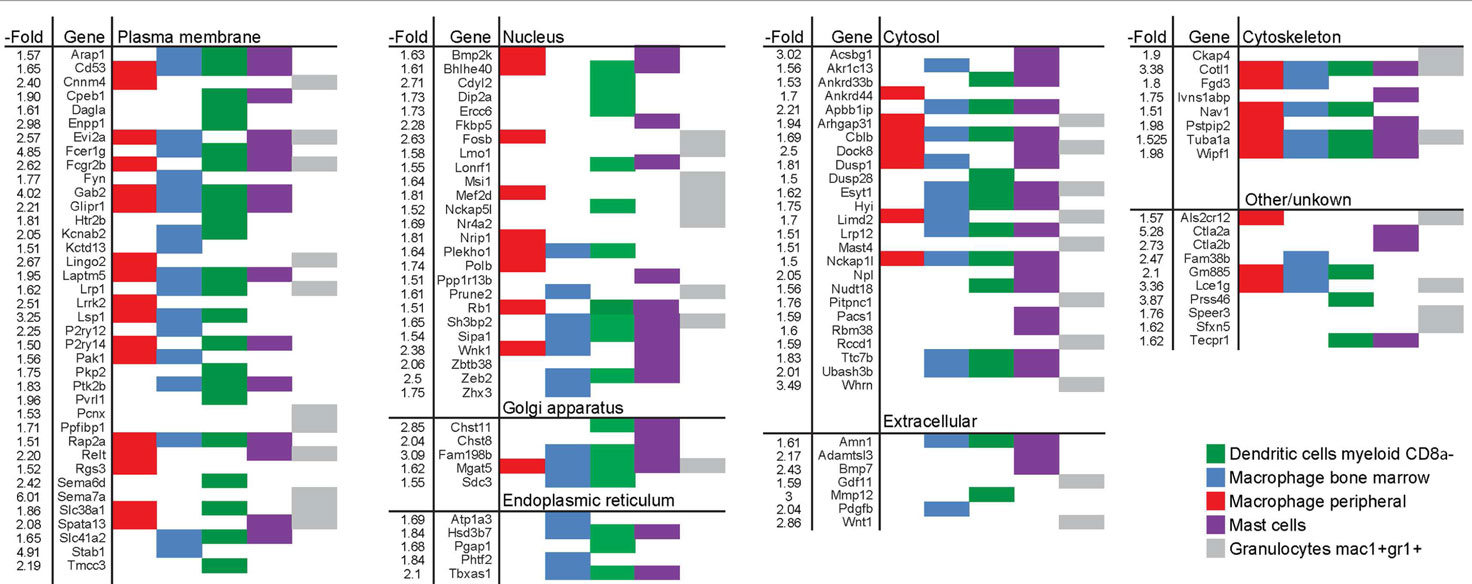
Table 1. List of genes induced by >1.5-fold in long-term EpRas EMT cells and enriched in human myeloid cells.
Monocyte/Macrophage Gene Cluster is Linked to EMT in Human Breast Cancer
Next, we performed studies to determine whether the monocyte/macrophage gene cluster, which we found was induced in long-term EMT cells, could be linked to EMT in human breast cancer cells and tumors. For this, we took advantage of published microarray-based datasets available through the EMBL-EBI database. First, we compared the expression levels of genes within the monocyte/macrophage cluster in 51 human breast cancer cell lines. We found that the average expression of the genes was significantly (P ≤ 0.001) higher in cell lines that displayed features of EMT, compared to non-EMT cell lines (Figure 4A). In addition, we compared the expression of the gene cluster in breast cancer cell lines previously classified as Basal B, Basal A, or Luminal cells (15). In these earlier studies, it was found while luminal breast cancer cells appeared most differentiated, Basal cells were less differentiated. In particular, Basal B cells were found to display EMT features. We found that the monocyte/macrophage gene cluster was significantly more expressed in cells classified in Basal B compared to Luminal or Basal A cell lines (P ≤ 0.01) (Figure 4B).
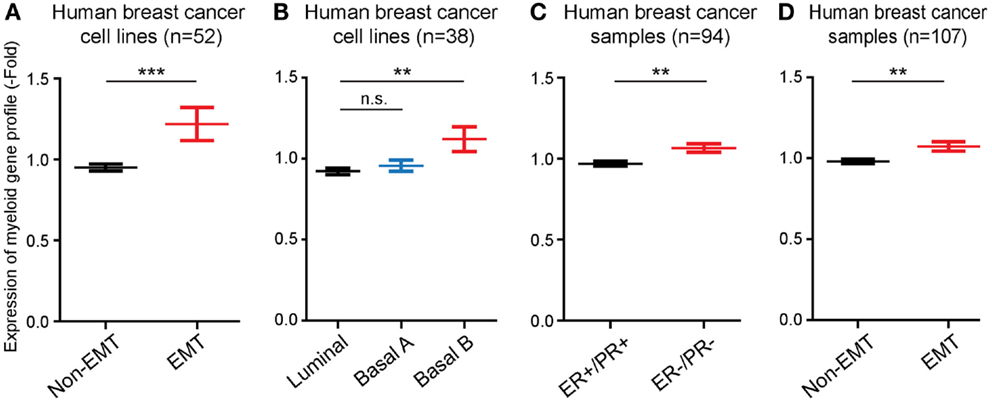
Figure 4. The EMT-induced monocyte/macrophage cluster is linked to EMT in human breast cancer. (A,B) Bar graphs showing that the average expression level of the EMT-induced monocyte/macrophage cluster was increased in human breast cancer cell lines displaying EMT vs. non-EMT properties, and in basal B cells compared to luminal and basal A cells. (C,D) Bar graphs showing that the average expression level of the EMT-induced monocyte/macrophage cluster was increased in human breast cancer samples negative for estrogen and progesterone receptors (ER−/PR−) compared to ER+/PR+ tumors, and in tumors displaying EMT vs. non-EMT properties.
Finally, we also compared the expression of the EMT-induced monocyte/macrophage gene cluster in samples of human breast cancer. The gene cluster was significantly more expressed (P ≤ 0.01) in estrogen receptor and progesterone receptor negative tumors (ER−/PR−), which are classified as the most aggressive and least differentiated breast tumors (16), compared to ER+/PR+ tumors (Figure 4C), as well as in EMT tumors compared to non-EMT tumors (P ≤ 0.01) (Figure 4D).
Discussion
We tested the hypothesis that breast cancer cells that are reprogramed through TGF-β1-induced EMT acquire properties of immune cells. To study this, we performed microarray-based gene profiling of tumor cells that had been allowed to adopt an EMT program for 14 days. In addition to the expected mesenchymal traits that were induced in EMT cells, we identified a gene cluster, which is enriched in monocyte-derived immune cells including macrophages, mast cells, and myeloid DCs.
Compared to most previous studies of gene expression changes during TGF-β1-induced EMT, which have focused on the first 24–72 h (17, 18), we decided to profile tumor cells that had been allowed to adopt an EMT phenotype for 14 days. There were several reasons behind this approach. First, since the early phase of TGF-β1-induced EMT is paralleled by growth inhibitory and apoptotic responses (19), we figured that many gene expression changes observed during this phase might not be primarily related to the EMT response. However, with time, growth inhibitory and apoptotic responses are released while EMT proceeds (20, 21). Thus, we figured that profiling the late phase of EMT might give valuable information about the transcriptional changes involved in reprograming tumor epithelial cells into mesenchymal cells. In comparison, reprograming of differentiated mouse and human cells into pluripotent stem cells takes weeks, rather than days (22, 23). Second, since recent results show that tumor cells maintaining an EMT profile in the circulation are the most capable ones in forming distant metastasis (10), we speculated that cells capable of maintaining EMT for a sustained period might display properties that could be important for later steps of the metastatic process including intra- and extravasation.
As expected, cluster analysis showed that known epithelial genes and cell adhesion molecules were the most downregulated in long-term EMT cells. In addition, genes encoding positive regulators of apoptosis were downregulated. We find this interesting since EMT has been linked to survival and resistance to apoptotic stimuli in breast cancer (24). In contrast, master regulators of EMT including Snai1, Snai2, and Twist1 were not upregulated in long-term EMT cells, but peaked at earlier time points. Among known EMT inducers only Zeb2, which encodes Smad-interacting protein-1 (Sip1) (25, 26), was upregulated in long-term EMT cells. This was somewhat surprising given the fact that these EMT inducers are known to be rapidly induced and vital for the induction of EMT (7, 27–33). However, other results show that after the induction of EMT, the expression of EMT inducers, such as Snai1, declines to almost baseline levels (34).
Together with our data, this suggests that while Snail1 and other EMT-inducing factors are critical for the early phase of EMT, other mechanisms come into play at later stages of EMT, as cells are reprogramed into mesenchymal cells. Such mechanisms are likely to play important roles in maintaining cells in an EMT state but might also provide EMT cells with properties used at later stages of the metastatic process. In line with this, we found that several of the most upregulated genes in long-term EMT cells including Tnc, Mmp9, Jag1, and Ret, are known to promote breast cancer metastasis (35–38).
The monocyte/macrophage cluster, which we identified, contains genes encoding proteins with variable intracellular localization and molecular function. Some of these proteins have been linked to cancer cell invasion, migration, and metastasis. For example, Fyn encodes a member of the Src family of protein kinases that are critical for macrophage migration (39), which promotes invasion and migration in breast cancer (40). Enpp1 encodes an ecto-nucleotide pyrophosphatase/phosphodiesterase (ENPP), which is expressed in osteoclasts, and which promotes calcification of bone (41), and facilitates breast cancer metastasis to the bone (42). Both Fyn and Enpp1 have been linked to resistance to tamoxifen treatment (43, 44). Gab2, which encodes a scaffolding adapter protein known to regulate EGF receptor signaling through ERK, is critical for the phagocytic function of macrophages (45), and was recently shown to promote breast cancer invasion and metastasis (46). Glipr1, which encodes glioma pathogenesis-related protein-1, has been linked to macrophage differentiation (47), and the invasive potential of melanoma cells (48).
Other genes within the monocyte/macrophage cluster have known roles in immune cells, but have been less studied in cancer. Cd53 encodes a tetraspanin protein, which is among the most induced genes in macrophages exposed to lipopolysaccharide (49); Fcer1g encodes a high affinity receptor for IgE, which is critical for activation of mast cells and plays a key role in allergic inflammation (50). Lrrk2, which encodes leucine-rich repeat kinase 2, may play a role in monocyte maturation and variants of this gene is associated with an increased risk of both Parkinson’s and Crohn’s disease (51–53). P2ry12 encodes a purinergic G protein-coupled receptor, which promotes macrophage chemotaxis and microglial activation (54, 55). Relt and Nrip1, which encode a member of the tumor necrosis factor receptor superfamily, and nuclear receptor-interacting protein-1, respectively, are both involved in activating NF-KappaB (56, 57). Tbxas1, Dock8, and Wipf1, which encode thromboxane A synthase 1, dedicator of cytokinesis 8, and WAS/WASL-interacting protein family member 1, respectively, are expressed in myeloid DCs and important for their polarization and migration (58–60). Some of the genes, such as Fcgr2b, which encodes the cell surface protein CD32; Lsp1, which encodes the lymphocyte-specific protein-1; and Stab1, which encodes the scavenger receptor Stabilin-1/CLEVER-1, are known to regulate monocyte/macrophage migration and endothelial cell interactions (61–63).
Interestingly, some of the genes in the monocyte/macrophage cluster have been linked to cellular stemness. In our microarray data, genes encoding cancer stem cell markers, such as CD44 and CD133, were not significantly upregulated. However, other genes known to regulate cellular stemness were induced. Msi1, which encodes Musashi1, an RNA binding protein, and Wnt1, are both master regulators of self-renewal in multiple stem cell populations (64, 65). Sipa1 encodes a Rap1 GTPase-activating protein essential for proliferative control of hematopoietic progenitors (66). Upregulation of Cdyl2 has been found in CD133-positive stem cells (67). Further studies are warranted to elucidate whether any of these genes contribute with stem cell properties to EMT cells.
In summary, our results indicate that tumor cells undergoing EMT in response to TGF-β1 do not simply turn into typical mesenchymal cells, such as fibroblasts. Rather, they acquire features of immune cells, in particular hematopoietic cells derived from myeloid precursors. The increased expression of genes in the monocyte/macrophage cluster in more aggressive breast cancer cell lines and tumors (EMT+, Basal B+, and ER−PR−) indicate that these genes might play roles in breast cancer metastasis. Clearly, further studies are needed to analyze the role of genes within the monocyte/macrophage cluster in providing cancer EMT cells with invasive and migratory properties. Such studies will include loss- and gain-of-function experiments to determine to the role of genes in providing EMT cells with the capacity to perform different steps of the metastatic process.
Author Contributions
Study concept and design: Joel Johansson, Vedrana Tabor, Jonas Fuxe; acquisition and analysis of data: Joel Johansson, Vedrana Tabor, Anna Wikell, Jonas Fuxe; drafting of the manuscript: Joel Johansson, Vedrana Tabor, Jonas Fuxe; and funding: Jonas Fuxe; study supervision: Jonas Fuxe.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Olof Rådmark at the Department of Medical Biochemistry and Biophysics at Karolinska Institutet for kindly providing us with the Cotl1/CPL antibody. Vedrana Tabor was supported by a grant from the Swedish Cancer Society. Jonas Fuxe was supported by project grants from the Swedish Research Council, the Swedish Cancer Society, and the Strategic Cancer (StratCan) Programme at Karolinska Institutet.
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/Journal/10.3389/fonc.2015.00003/abstract
Footnotes
References
1. Chiang AC, Massague J. Molecular basis of metastasis. N Engl J Med (2008) 359(26):2814–23. doi:10.1056/NEJMra0805239
2. Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science (2011) 331(6024):1559–64. doi:10.1126/science.1203543
3. Jahroudi N, Greenberger JS. The role of endothelial cells in tumor invasion and metastasis. J Neurooncol (1995) 23(2):99–108. doi:10.1007/BF01053415
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
4. Kang Y, Pantel K. Tumor cell dissemination: emerging biological insights from animal models and cancer patients. Cancer Cell (2013) 23(5):573–81. doi:10.1016/j.ccr.2013.04.017
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
5. Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest (2009) 119(6):1420–8. doi:10.1172/JCI39104
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
6. Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell (2008) 133(4):704–15. doi:10.1016/j.cell.2008.03.027
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
7. Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell (2009) 139(5):871–90. doi:10.1016/j.cell.2009.11.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
8. Nieto MA. Epithelial plasticity: a common theme in embryonic and cancer cells. Science (2013) 342(6159):1234850. doi:10.1126/science.1234850
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
9. Bednarz-Knoll N, Alix-Panabieres C, Pantel K. Plasticity of disseminating cancer cells in patients with epithelial malignancies. Cancer Metastasis Rev (2012) 31(3–4):673–87. doi:10.1007/s10555-012-9370-z
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
10. Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science (2013) 339(6119):580–4. doi:10.1126/science.1228522
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
11. Fuxe J, Karlsson MC. TGF-beta-induced epithelial-mesenchymal transition: a link between cancer and inflammation. Semin Cancer Biol (2012) 22(5–6):455–61. doi:10.1016/j.semcancer.2012.05.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
12. Oft M, Peli J, Rudaz C, Schwarz H, Beug H, Reichmann E. TGF-beta1 and Ha-Ras collaborate in modulating the phenotypic plasticity and invasiveness of epithelial tumor cells. Genes Dev (1996) 10(19):2462–77. doi:10.1101/gad.10.19.2462
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
13. Rakonjac M, Fischer L, Provost P, Werz O, Steinhilber D, Samuelsson B, et al. Coactosin-like protein supports 5-lipoxygenase enzyme activity and up-regulates leukotriene A4 production. Proc Natl Acad Sci U S A (2006) 103(35):13150–5. doi:10.1073/pnas.0605150103
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
14. Andersson AM, Pettersson RF. Targeting of a short peptide derived from the cytoplasmic tail of the G1 membrane glycoprotein of Uukuniemi virus (Bunyaviridae) to the Golgi complex. J Virol (1998) 72(12):9585–96.
15. Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell (2006) 10(6):515–27. doi:10.1016/j.ccr.2006.10.008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
16. Ismail-Khan R, Bui MM. A review of triple-negative breast cancer. Cancer Control (2010) 17(3):173–6.
17. Zavadil J, Bitzer M, Liang D, Yang YC, Massimi A, Kneitz S, et al. Genetic programs of epithelial cell plasticity directed by transforming growth factor-beta. Proc Natl Acad Sci U S A (2001) 98(12):6686–91. doi:10.1073/pnas.111614398
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
18. Xie L, Law BK, Aakre ME, Edgerton M, Shyr Y, Bhowmick NA, et al. Transforming growth factor beta-regulated gene expression in a mouse mammary gland epithelial cell line. Breast Cancer Res (2003) 5(6):R187–98. doi:10.1186/bcr640
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
19. Brown KA, Aakre ME, Gorska AE, Price JO, Eltom SE, Pietenpol JA, et al. Induction by transforming growth factor-beta1 of epithelial to mesenchymal transition is a rare event in vitro. Breast Cancer Res (2004) 6(3):R215–31. doi:10.1186/bcr762
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
20. Gal A, Sjoblom T, Fedorova L, Imreh S, Beug H, Moustakas A. Sustained TGF beta exposure suppresses Smad and non-Smad signalling in mammary epithelial cells, leading to EMT and inhibition of growth arrest and apoptosis. Oncogene (2008) 27(9):1218–30. doi:10.1038/sj.onc.1210741
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
21. Johansson J, Berg T, Kurzejamska E, Pang MF, Tabor V, Jansson M, et al. MiR-155-mediated loss of C/EBPbeta shifts the TGF-beta response from growth inhibition to epithelial-mesenchymal transition, invasion and metastasis in breast cancer. Oncogene (2013) 32(50):5614–24. doi:10.1038/onc.2013.322
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
22. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell (2006) 126(4):663–76. doi:10.1016/j.cell.2006.07.024
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
23. Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell (2007) 131(5):861–72. doi:10.1016/j.cell.2007.11.019
24. Toft DJ, Cryns VL. Minireview: basal-like breast cancer: from molecular profiles to targeted therapies. Mol Endocrinol (2011) 25(2):199–211. doi:10.1210/me.2010-0164
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
25. Vandewalle C, Comijn J, De Craene B, Vermassen P, Bruyneel E, Andersen H, et al. SIP1/ZEB2 induces EMT by repressing genes of different epithelial cell-cell junctions. Nucleic Acids Res (2005) 33(20):6566–78. doi:10.1093/nar/gki965
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
26. Bindels S, Mestdagt M, Vandewalle C, Jacobs N, Volders L, Noel A, et al. Regulation of vimentin by SIP1 in human epithelial breast tumor cells. Oncogene (2006) 25(36):4975–85. doi:10.1038/sj.onc.1209511
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
27. Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, et al. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol (2000) 2(2):84–9. doi:10.1038/35000034
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
28. Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, Del Barrio MG, et al. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol (2000) 2(2):76–83. doi:10.1038/35010506
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
29. Casas E, Kim J, Bendesky A, Ohno-Machado L, Wolfe CJ, Yang J. Snail2 is an essential mediator of Twist1-induced epithelial mesenchymal transition and metastasis. Cancer Res (2011) 71(1):245–54. doi:10.1158/0008-5472.CAN-10-2330
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
30. Hajra KM, Chen DY, Fearon ER. The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res (2002) 62(6):1613–8.
31. Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer (2007) 7(6):415–28. doi:10.1038/nrc2131
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
32. Vincent T, Neve EP, Johnson JR, Kukalev A, Rojo F, Albanell J, et al. A SNAIL1-SMAD3/4 transcriptional repressor complex promotes TGF-beta mediated epithelial-mesenchymal transition. Nat Cell Biol (2009) 11(8):943–50. doi:10.1038/ncb1905
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
33. Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell (2004) 117(7):927–39. doi:10.1016/j.cell.2004.06.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
34. Shirakihara T, Saitoh M, Miyazono K. Differential regulation of epithelial and mesenchymal markers by deltaEF1 proteins in epithelial mesenchymal transition induced by TGF-beta. Mol Biol Cell (2007) 18(9):3533–44. doi:10.1091/mbc.E07-03-0249
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
35. Van Kempen LC, Coussens LM. MMP9 potentiates pulmonary metastasis formation. Cancer Cell (2002) 2(4):251–2. doi:10.1016/S1535-6108(02)00157-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
36. Sethi N, Dai X, Winter CG, Kang Y. Tumor-derived JAGGED1 promotes osteolytic bone metastasis of breast cancer by engaging notch signaling in bone cells. Cancer Cell (2011) 19(2):192–205. doi:10.1016/j.ccr.2010.12.022
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
37. Gattelli A, Nalvarte I, Boulay A, Roloff TC, Schreiber M, Carragher N, et al. Ret inhibition decreases growth and metastatic potential of estrogen receptor positive breast cancer cells. EMBO Mol Med (2013) 5(9):1335–50. doi:10.1002/emmm.201302625
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
38. Oskarsson T, Acharyya S, Zhang XH, Vanharanta S, Tavazoie SF, Morris PG, et al. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat Med (2011) 17(7):867–74. doi:10.1038/nm.2379
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
39. Baruzzi A, Iacobucci I, Soverini S, Lowell CA, Martinelli G, Berton G. c-Abl and Src-family kinases cross-talk in regulation of myeloid cell migration. FEBS Lett (2010) 584(1):15–21. doi:10.1016/j.febslet.2009.11.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
40. Yadav V, Denning MF. Fyn is induced by Ras/PI3K/Akt signaling and is required for enhanced invasion/migration. Mol Carcinog (2011) 50(5):346–52. doi:10.1002/mc.20716
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
41. Hajjawi MO, Macrae VE, Huesa C, Boyde A, Millan JL, Arnett TR, et al. Mineralisation of collagen rich soft tissues and osteocyte lacunae in Enpp1 mice. Bone (2014) 69C:139–47. doi:10.1016/j.bone.2014.09.016
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
42. Lau WM, Doucet M, Stadel R, Huang D, Weber KL, Kominsky SL. Enpp1: a potential facilitator of breast cancer bone metastasis. PLoS One (2013) 8(7):e66752. doi:10.1371/journal.pone.0066752
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
43. Elias D, Vever H, Laenkholm AV, Gjerstorff MF, Yde CW, Lykkesfeldt AE, et al. Gene expression profiling identifies FYN as an important molecule in tamoxifen resistance and a predictor of early recurrence in patients treated with endocrine therapy. Oncogene (2014). doi:10.1038/onc.2014.138
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
44. Umar A, Kang H, Timmermans AM, Look MP, Meijer-Van Gelder ME, Den Bakker MA, et al. Identification of a putative protein profile associated with tamoxifen therapy resistance in breast cancer. Mol Cell Proteomics (2009) 8(6):1278–94. doi:10.1074/mcp.M800493-MCP200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
45. Gu H, Botelho RJ, Yu M, Grinstein S, Neel BG. Critical role for scaffolding adapter Gab2 in Fc gamma R-mediated phagocytosis. J Cell Biol (2003) 161(6):1151–61. doi:10.1083/jcb.200212158
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
46. Ke Y, Wu D, Princen F, Nguyen T, Pang Y, Lesperance J, et al. Role of Gab2 in mammary tumorigenesis and metastasis. Oncogene (2007) 26(34):4951–60. doi:10.1038/sj.onc.1210315
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
47. Gingras MC, Margolin JF. Differential expression of multiple unexpected genes during U937 cell and macrophage differentiation detected by suppressive subtractive hybridization. Exp Hematol (2000) 28(1):65–76. doi:10.1016/S0301-472X(99)00126-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
48. Awasthi A, Woolley AG, Lecomte FJ, Hung N, Baguley BC, Wilbanks SM, et al. Variable expression of GLIPR1 correlates with invasive potential in melanoma cells. Front Oncol (2013) 3:225. doi:10.3389/fonc.2013.00225
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
49. Kim TR, Yoon JH, Kim YC, Yook YH, Kim IG, Kim YS, et al. LPS-induced CD53 expression: a protection mechanism against oxidative and radiation stress. Mol Cells (2004) 17(1):125–31.
50. Yu M, Tsai M, Tam SY, Jones C, Zehnder J, Galli SJ. Mast cells can promote the development of multiple features of chronic asthma in mice. J Clin Invest (2006) 116(6):1633–41. doi:10.1172/JCI25702
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
51. Manolio TA. Genomewide association studies and assessment of the risk of disease. N Engl J Med (2010) 363(2):166–76. doi:10.1056/NEJMra0905980
52. Nalls MA, Plagnol V, Hernandez DG, Sharma M, Sheerin UM, Saad M, et al. Imputation of sequence variants for identification of genetic risks for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet (2011) 377(9766):641–9. doi:10.1016/S0140-6736(10)62345-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
53. Thevenet J, Pescini Gobert R, Hooft Van Huijsduijnen R, Wiessner C, Sagot YJ. Regulation of LRRK2 expression points to a functional role in human monocyte maturation. PLoS One (2011) 6(6):e21519. doi:10.1371/journal.pone.0021519
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
54. Kronlage M, Song J, Sorokin L, Isfort K, Schwerdtle T, Leipziger J, et al. Autocrine purinergic receptor signaling is essential for macrophage chemotaxis. Sci Signal (2010) 3(132):ra55. doi:10.1126/scisignal.2000588
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
55. Haynes SE, Hollopeter G, Yang G, Kurpius D, Dailey ME, Gan WB, et al. The P2Y12 receptor regulates microglial activation by extracellular nucleotides. Nat Neurosci (2006) 9(12):1512–9. doi:10.1038/nn1805
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
56. Sica GL, Zhu G, Tamada K, Liu D, Ni J, Chen L. RELT, a new member of the tumor necrosis factor receptor superfamily, is selectively expressed in hematopoietic tissues and activates transcription factor NF-kappaB. Blood (2001) 97(9):2702–7. doi:10.1182/blood.V97.9.2702
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
57. Zschiedrich I, Hardeland U, Krones-Herzig A, Berriel Diaz M, Vegiopoulos A, Muggenburg J, et al. Coactivator function of RIP140 for NFkappaB/RelA-dependent cytokine gene expression. Blood (2008) 112(2):264–76. doi:10.1182/blood-2007-11-121699
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
58. Harada Y, Tanaka Y, Terasawa M, Pieczyk M, Habiro K, Katakai T, et al. DOCK8 is a Cdc42 activator critical for interstitial dendritic cell migration during immune responses. Blood (2012) 119(19):4451–61. doi:10.1182/blood-2012-01-407098
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
59. Hata M, Takahara S, Tsuzaki H, Ishii Y, Nakata K, Akagawa KS, et al. Expression of Th2-skewed pathology mediators in monocyte-derived type 2 of dendritic cells (DC2). Immunol Lett (2009) 126(1–2):29–36. doi:10.1016/j.imlet.2009.07.008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
60. Chou HC, Anton IM, Holt MR, Curcio C, Lanzardo S, Worth A, et al. WIP regulates the stability and localization of WASP to podosomes in migrating dendritic cells. Curr Biol (2006) 16(23):2337–44. doi:10.1016/j.cub.2006.10.037
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
61. Devaraj S, Davis B, Simon SI, Jialal I. CRP promotes monocyte-endothelial cell adhesion via Fcgamma receptors in human aortic endothelial cells under static and shear flow conditions. Am J Physiol Heart Circ Physiol (2006) 291(3):H1170–6. doi:10.1152/ajpheart.00150.2006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
62. Palani S, Maksimow M, Miiluniemi M, Auvinen K, Jalkanen S, Salmi M. Stabilin-1/CLEVER-1, a type 2 macrophage marker, is an adhesion and scavenging molecule on human placental macrophages. Eur J Immunol (2011) 41(7):2052–63. doi:10.1002/eji.201041376
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
63. Liu L, Cara DC, Kaur J, Raharjo E, Mullaly SC, Jongstra-Bilen J, et al. LSP1 is an endothelial gatekeeper of leukocyte transendothelial migration. J Exp Med (2005) 201(3):409–18. doi:10.1084/jem.20040830
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
64. Sutherland JM, Mclaughlin EA, Hime GR, Siddall NA. The Musashi family of RNA binding proteins: master regulators of multiple stem cell populations. Adv Exp Med Biol (2013) 786:233–45. doi:10.1007/978-94-007-6621-1_13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
65. Katoh M. WNT signaling pathway and stem cell signaling network. Clin Cancer Res (2007) 13(14):4042–5. doi:10.1158/1078-0432.CCR-06-2316
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
66. Ishida D, Kometani K, Yang H, Kakugawa K, Masuda K, Iwai K, et al. Myeloproliferative stem cell disorders by deregulated Rap1 activation in SPA-1-deficient mice. Cancer Cell (2003) 4(1):55–65. doi:10.1016/S1535-6108(03)00163-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: epithelial–mesenchymal transition, breast cancer, immune cells, monocytes, macrophages, gene expression profiling, properties
Citation: Johansson J, Tabor V, Wikell A, Jalkanen S and Fuxe J (2015) TGF-β1-induced epithelial–mesenchymal transition promotes monocyte/macrophage properties in breast cancer cells. Front. Oncol. 5:3. doi: 10.3389/fonc.2015.00003
Received: 01 August 2014; Accepted: 08 January 2015;
Published online: 26 January 2015.
Edited by:
Adetunji T. Toriola, Washington University School of Medicine, USAReviewed by:
Hugo Vankelecom, Catholic University of Leuven, BelgiumWagner Ricardo Montor, Faculdade de Ciências Médicas da Santa Casa de São Paulo, Brazil
Copyright: © 2015 Johansson, Tabor, Wikell, Jalkanen and Fuxe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jonas Fuxe, Department of Medical Biochemistry and Biophysics, Division of Vascular Biology, Karolinska Institute, Stockholm, SE-17177, Sweden e-mail: jonas.fuxe@ki.se
†Joel Johansson and Vedrana Tabor have contributed equally to this work.
 Joel Johansson
Joel Johansson Vedrana Tabor
Vedrana Tabor Anna Wikell1
Anna Wikell1 Sirpa Jalkanen
Sirpa Jalkanen Jonas Fuxe
Jonas Fuxe