Integrating the Ecosystem Services Framework to Define Dysbiosis of the Breastfed Infant Gut: The Role of B. infantis and Human Milk Oligosaccharides
- 1Evolve BioSystems, Inc., Davis, CA, United States
- 2Department of Food Science and Technology, University of Nebraska, Lincoln, NE, United States
Mounting evidence supports a connection between the composition of the infant gut microbiome and long-term health. In fact, aberrant microbiome compositions during key developmental windows in early life are associated with increased disease risk; therefore, making pertinent modifications to the microbiome during infancy offers significant promise to improve human health. There is growing support for integrating the concept of ecosystem services (the provision of benefits from ecosystems to humans) in linking specific microbiome functions to human well-being. This framework is widely applied in conservation efforts of macro-ecosystems and offers a systematic approach to guide restoration actions aimed to recover critical ecological functions. The aim of this work is to apply the ecosystem services framework to integrate recent studies demonstrating stable alteration of the gut microbiome of breastfed infants when Bifidobacterium longum subsp. infantis EVC001, a gut symbiont capable of efficiently utilizing human milk oligosaccharides into organic acids that are beneficial for the infant and lower intestinal pH, is reintroduced. Additionally, using examples from the literature we illustrate how the absence of B. infantis results in diminished ecosystem services, which may be associated with health consequences related to immune and metabolic disorders. Finally, we propose a model by which infant gut dysbiosis can be defined as a reduction in ecosystem services supplied to the host by the gut microbiome rather than merely changes in diversity or taxonomic composition. Given the increased interest in targeted microbiome modification therapies to decrease acute and chronic disease risk, the model presented here provides a framework to assess the effectiveness of such strategies from a host-centered perspective.
Introduction
Disruption to the composition and function of the early life gut microbiome is now recognized for its role in irregular immune development (1, 2), metabolic disorders (3) and inflammation (4, 5). Several of these phenotypes have been reconstructed using animal models or epidemiological approaches providing a compelling link between aberrant microbiome development in early life and these negative health outcomes (6–9). Thus, if pandemic non-communicable diseases such as type 1 diabetes, obesity, allergy, and asthma are associated with impaired microbiomes during infancy, as suggested by emerging evidence (3, 10–15), then relevant modulation of the microbiome in early life provides a compelling solution for addressing the increasing public health burden associated with these diseases. However, evaluative parameters to identify desirable microbiome compositions and their potential interrelationship with health, are currently lacking.
The application of methods derived from ecological theory and evolutionary biology have been fundamental to elucidating the factors that shape the microbiome throughout the lifespan. In this work, we apply concepts from the “ecosystem services” framework (16) to guide the ecological assessment of the breastfed infant gut microbiome from a host-centered perspective. We first describe the ecological processes that shape and define the composition of the microbiome in early life. This description is centered on the hypothesis that human hosts select, via diet (human milk), for the enrichment of specialized symbionts that fulfill beneficial functions underlying the provision of ecosystem services that contribute to their fitness and well-being. We then propose a model in which the absence of these beneficial functions and the consequential reduction in one or more ecosystem services can be defined as dysbiosis. To demonstrate the applicability of the model, the discussion is centered on the coevolution of specialized bifidobacteria, namely B. infantis, for which clinical evidence is available (17). Finally, we summarize, as evidence for this model, large cohort studies indicating the absence of bifidobacteria in early life is associated with negative health outcomes.
Ecological Processes Shaping the Composition of the Breastfed Infant Gut Microbiome
Immediately following birth, the neonatal intestine becomes rapidly colonized by microbes from the mother and the surrounding environment. Infants delivered by cesarean section are more likely to become colonized by environmental microorganisms from the maternal skin, healthcare staff and hospital surfaces. Vaginally delivered infants come in contact with bacteria from mother's vaginal canal and the fecal microbiota (18, 19). From this initial load of microbes, the allochthonous, vaginally-derived and environmental species are then rapidly replaced by organisms adapted to the gut (20–23); however, the microbiome differences based on delivery mode persist over time (19). Nutritional resources that reach the gut are another major factor influencing the neonatal gut microbiome, in terms of both composition and function. In exclusively breastfed infants, human milk oligosaccharides (HMOs) represent the main nutritional resources for bacteria in the gut. As a result, the gut microbiome of exclusively breastfed infants exhibits lower alpha diversity and higher abundance of specialized taxa able to metabolize HMOs, namely bifidobacteria (24–27). In the absence of specialized infant-associated species of bifidobacteria, HMOs are under-utilized, resulting in excess resources with profound impacts on ecosystem function (Figure 1). Cessation of breastfeeding and the introduction of solid foods represent a major shift in the nutritional resource landscape and a more functionally complex community of microbes is then required to deplete the greater variety of dietary substrates reaching the large intestine (Figure 1).
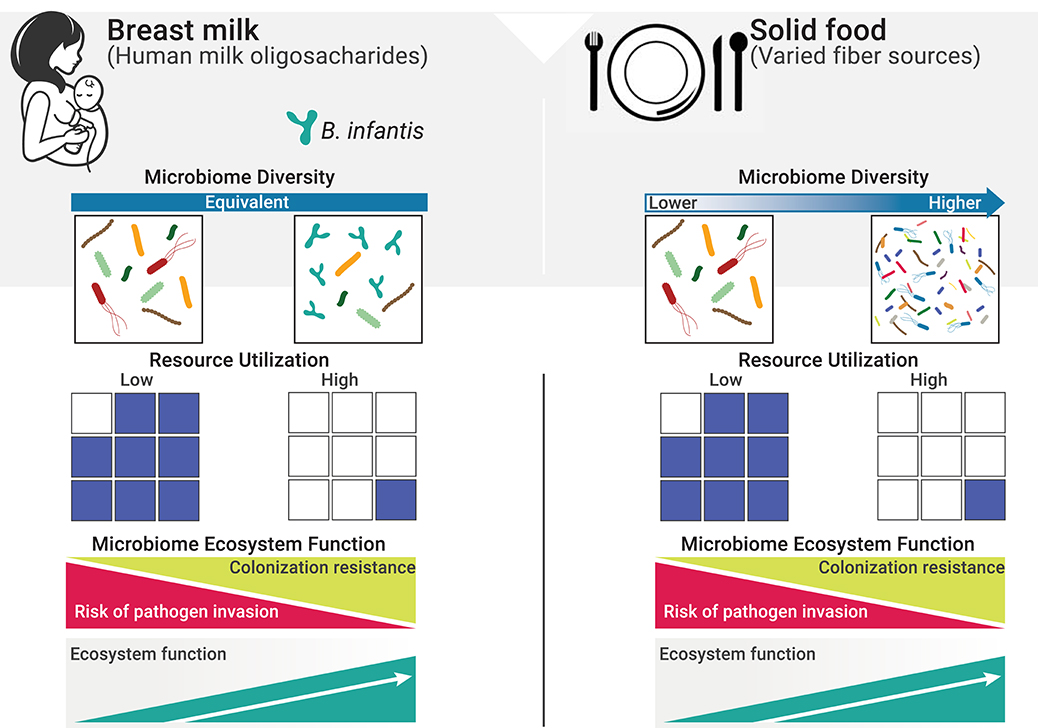
Figure 1. Resource utilization and diversity in the gut in determining invasion resistance. The resource landscape in the human gut is vastly influenced by diet. One fundamental function of the microbiome is to keep potential pathogens at bay by direct competition for space and resources (i.e., co-lonization resistance, a regulating service). Excess resources (blue squares) represent open niche opportunities and increase the risk of colonization by invasive species, including pathogens. When resources are efficiently utilized, the risk of successful invasion is greatly reduced due to the lack of available resources to sustain growth (28). On a solid food diet, a more diverse composite of species is required to deplete the greater variety of resources reaching the gut (29). However, in early life, and while diet is restricted to mother's milk, resource utilization is independent from diversity and will only reach maximum levels when specialized species able to efficiently consume HMOs are present.
Additional ecological events, including random processes, ultimately influence the overall composition of the infant gut microbiome; however, initial microbiome inoculation based on birth mode, and the subsequent environmental selection through the provision of selective substrates from human milk, are the two major ecological processes shaping the gut microbiome of breastfed infants (30–33).
Biological Considerations in Defining a Healthy Infant Gut Microbiome
Identifying a healthy gut microbiome in both infants and adults has proven to be a major challenge to the scientific and medical fields (34). Historically, diversity has been speculated to maximize functionality, in a generalization of the “insurance hypothesis” (35–38). However, diversity indices are of limited value alone and have proven insufficient to determine ecosystem functionality, or to categorize microbial ecosystems as healthy or unhealthy (37, 39) (see Box 1 for an in-depth discussion on the limitations of diversity). Moreover, taxonomic composition can be highly variable among individuals, while functions encoded by the gut microbiome are remarkably coherent (45) and breastfed infants across different geographies develop a common microbial functional core (15, 32, 33). This implies hosts are under a strong pressure to select high-fidelity microbial partners to maintain key ecosystem functions (38), and that breast milk establishes key niches that can only be occupied by specialized taxa (46) (Figure 1). Furthermore, given the host and its microbiome operate as a highly interconnected and co-evolved ecosystem in which interactions among members and community characteristics are governed by the principles of community ecology, we argue the evaluation of gut microbiomes can only be successful if based on ecological and evolutionary criteria. To this end, the ecosystem services framework has been implemented to link ecological processes of macro-ecosystems with elements of human well-being (47) and has recently been adapted to value the services of gut microbial ecosystems from a host-centered perspective (16, 48, Box 2). Therefore, we propose to use this framework to guide the assessment of the infant gut microbiome and to determine the ecological conditions within the gut that may increase host health and, ultimately, fitness.
Box 1. Diversity: How is it measured and what does it mean?
There are two main types of diversity computed in microbial ecology studies, particularly as it pertains to microbiome profiling: alpha diversity and beta diversity.
Alpha diversity refers to the measure of diversity within a specific ecological community or locality in a given sample. Depending on the metric used, this index describes either species richness (i.e., the number of different species in a community); or both species richness and the evenness (i.e., the distribution of the species' abundances in the community) (40). There are several metrics to determine alpha diversity, each different in their sensitivity to richness and evenness (41). Depending on the index used, it is possible that no change in alpha diversity may be detected despite the presence of highly divergent community compositions (Figure 2).
Beta diversity is a measure of diversity between samples. It answers the questions: How different is the microbial composition in one sample or group of samples compared to others? How many species are shared between samples? Similar to alpha diversity, there are different metrics to establish beta diversity. Some methods are purely qualitative based on presence/absence of species, while others include a quantitative component and take into account a phylogenetic distance between species. Each method presents its own inherent biases and sensitivity capturing changes in community composition.
Uses and limitations of diversity in microbial ecology
Diversity is speculated to maximize the functionality in a generalization of the “insurance hypothesis” (35, 36), which suggests that stabilization of communities against decline in function is improved by increasing diversity (42, 43). Thus, higher diversity is often assumed to be desirable. However, unless substantial functional redundancy exists in a microbial community, any loss in key functional species will likely alter the capacity of the microbiome to support ecosystem services (44). Further, a reduction in diversity is not necessarily unfavorable to the host, especially when it is a consequence of the selective enrichment of health-promoting symbionts.
Another inherent challenge exists in the lack of an accepted, absolute value of diversity for a given community. Moreover, as previously discussed [see (39) for an excellent discussion on the matter], diversity is relative and always constrained by method of measurement. In fact, different indices vary in their sensitivity to species richness and evenness, and inferences made can differ widely depending on the measure chosen. Thus, caution must be exercised when drawing conclusions from any one diversity index and when comparing findings across studies.
Overall, simplifying the microbiome to a measure of biodiversity has obvious limitations as it does not reflect composition or function, or relevant ecosystem properties such as stability, productivity or invisibility. We and others (37, 39) argue that the continued use of this index, without context of function, distracts the field from the development of relevant hypotheses to gain insight into the underlying ecological mechanisms driving patterns and processes in microbial communities and their potential relationship to host health.
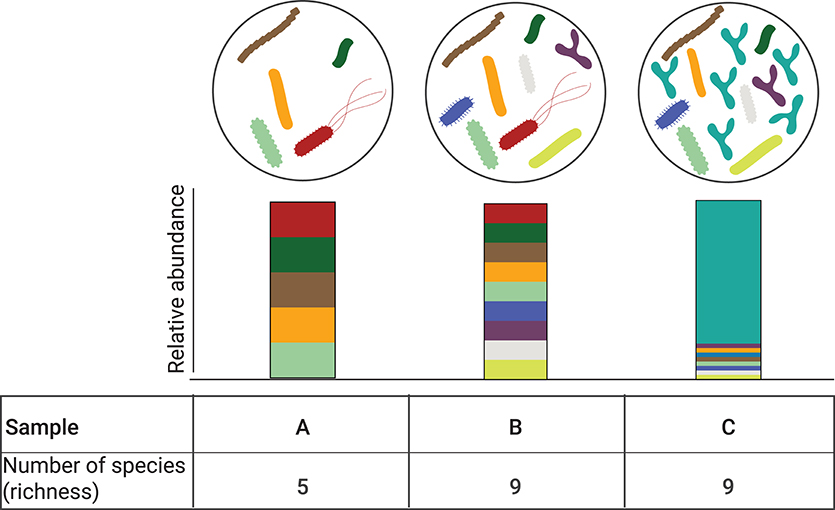
Figure 2. Alpha diversity is independent of relative abundance. Three different bacterial communities are depicted (A–C). Corresponding relative abundance of the individual species in the bacterial comminutes is represented by the stacked bar graphs. Bacterial communities B and C have the same number of observed species (n = 9) but their relative abundance is different, with community C being dominated by one species. While the alpha diversity can be computed with different metrics, when accounting for community richness, communities B and C species have the same alpha diversity.
Box 2. Advantages of an ecosystem services paradigm to evaluate the infant gut microbiome.
The application of concepts drawn from applied macroecology research has provided important insights into the mechanisms shaping the gut microbiome, especially as it relates to how microbial communities assemble, function and evolve (38, 49–53), and how these processes influence human health (54, 55).
Unlike abiotic geographies for macroecology, hosts have faced millennia of coevolution to shape the populations of microbes that colonize them. Exquisitely specific mechanisms to select for specific microbial symbionts have been described for plants (56) invertebrates (squid, insects) (57), and vertebrates (58–60). Selective pressures have shaped these interactions between host and microbe over time, and in the gut microbiome, toward selection for the key ecosystem services that improve host health (i.e., fitness). Evaluation of the infant gut microbiome through the lens of ecosystem services will facilitate the identification of key ecosystem “service providers” as those species whose functions are critical for the delivery of a given service. Colonization resistance and access to specialized foods or diets (provisioning services) are examples of ecosystem services where research may offer clues as to how services in the gut microbiome have been maximized by host-microbe interactions under strong selective pressures.
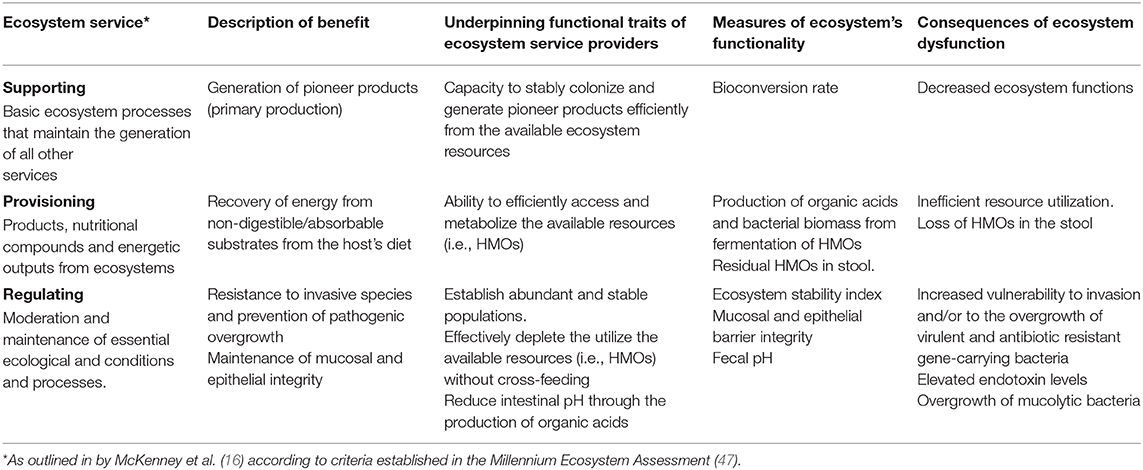
The ecosystem services framework is widely applied to evaluate terrestrial and marine ecosystems (47) and was recently adapted to evaluate the mammalian gut (16). Viewed through the lens of ecosystems services, the goods and services humans obtain from their microbiomes can be categorized as supporting, provisioning, or regulating (Table 1). Provisioning services are those obtained directly from the production of goods, e.g., microbial production of vitamins, antimicrobials, organic and short-chain fatty acids. Regulating services are those involved in maintaining stable ecosystem conditions, e.g., resistance to pathogen invasion. Supporting services are those necessary for the production and maintenance of all other ecosystem services, e.g., generation of pioneer products.
One main advantage of applying the ecosystem service model to evaluate the infant gut microbiome is that it facilitates the systematic identification of key “service providers” whose functional traits underpin the delivery of a given service (61, 62). By explicitly linking functional traits to ecosystem service delivery, it is possible to assign “functional importance” and “irreplaceability” indices, and correspondingly, predict the extent to which the loss of key “service provider” species can impact the ecological processes that sustain ecosystem functioning (35, 62).
In the following sections we outline three key ecosystem services: (1) supporting services; (2) provisioning services; and (3) regulating services underlying the relationship between the breastfed infant and the gut microbiome based on previously defined criteria (16). Specifically, we discuss the ability of B. infantis to efficiently utilize resources (i.e., HMOs) and produce organic acids as key functional traits that sustain the provision of these services.
Supporting and Provisioning Services of the Gut Microbiome in Early Life
Organic acids, including short chain fatty acids (SCFA), are the major metabolic products of anaerobic microbial fermentation in the gut and have demonstrated roles in human health (63, 64). In the breastfed infant gut, fermentation of HMOs into lactate and acetate depends critically on specialized primary degrader organisms that have the metabolic machinery to capture and metabolize these complex compounds (46). This process generates pioneer products (supporting service; Table 1) and releases energy that is otherwise inaccessible to the infant (provisioning service; Table 1). Selected strains of bifidobacteria and Bacteroides metabolize HMOs (24), but only B. infantis contains complete pathways enabling intracellular HMO-transport and degradation. Consequently, it is the only organism with the demonstrated capacity to significantly increase the production of lactate and acetate in the breastfed infant gut while simultaneously decreasing residual HMOs in the stool of breastfed infants (17, 65). In fact, in the absence of B. infantis, high concentrations of these HMOs are expelled into the stools of infants (1, 17, 66–68) which is a clear indication of low utilization of these resources (i.e., HMOs) in the gut, even when compared to infants colonized by other bifidobacteria. This observation highlights the importance of B. infantis in providing and provisioning services that underlie the overall function of the infant gut microbiome ecosystem (Table 1).
In addition to their role as pioneer substrates in the gut, organic acids and SCFA can enter circulation and directly affect the adipose tissue, brain, and liver (69–71). Acetate has been proposed to have an important role in inducing anti-inflammatory effects via the modulation of regulatory T-cells and anti-inflammatory cytokines (70), as well as improve mucosal epithelial integrity in the gut leading to protection from infectious disease in animal models (64). Lactate crosses the blood-brain barrier and functions as a modulator of neural activity, and is actively transported by gut epithelial cells (72–74). Acetate and lactate are also precursors of butyrate, which has anti-tumorigenic and anti-inflammatory properties and provides energy to gut epithelial cells (64). Overall, these microbially-produced organic acids have a major influence on host physiology. Thus, the presence of taxa able to efficiently metabolize HMOs into these key metabolites is critical to the delivery of fundamental ecosystem services that can affect the short- and long-term health of the growing infant.
Colonization Resistance and Stability are Critical Regulating Services of the Infant Gut Microbiome
One of the critical functions of the gut microbiome is to protect the immunologically naïve infant from acquiring exogenous pathogens and to prevent the overgrowth of opportunistic commensals (10, 75), a process known as colonization resistance (76, 77). Direct competition for resources, metabolic exclusion by production of organic acids, and indirect stimulation of the mucosal barrier system are well-characterized mechanisms by which the microbiome provides the host with this regulatory service (78). More competition for resources increases ecological stability at the expense of diversity by favoring the growth of specialized taxa, and limits the ability of invading microbes to establish and replicate (79). Thus, increased stability is central to the delivery of this regulatory service (Table 1) as stable ecosystems are inherently more resistant to external disturbances (42). In a clinical study, it was shown that colonization with B. infantis EVC001 significantly increases the stability of the infant microbiome (17). Moreover, consumption of HMOs by B. infantis produces acidic end-products mainly lactate and acetate, thereby altering the intestinal environment to prohibit the growth of pH-sensitive populations (e.g., Enterobacteriaceae and Clostridia) (69, 80, 81) including known enteric pathogens (17, 82), many of which carry antibiotic resistant genes (83–85). Further, the resulting high abundance of bifidobacteria contributes to maintaining intestinal barrier function through the production of acetate and tryptophan metabolites, and the reduction of mucus-eroding bacteria (86–89). Thus, the regulating services infants obtain from a microbiome abundantly colonized by B. infantis represents an archetypal model of protection, in which the host selects (via HMOs) microbial taxa most adept at strengthening epithelial defenses as well as creating biotic (i.e., competition for resources) and abiotic (i.e., pH) resistance barriers against invasion (28). A conceptual depiction of these concepts is shown in Figures 1, 3.
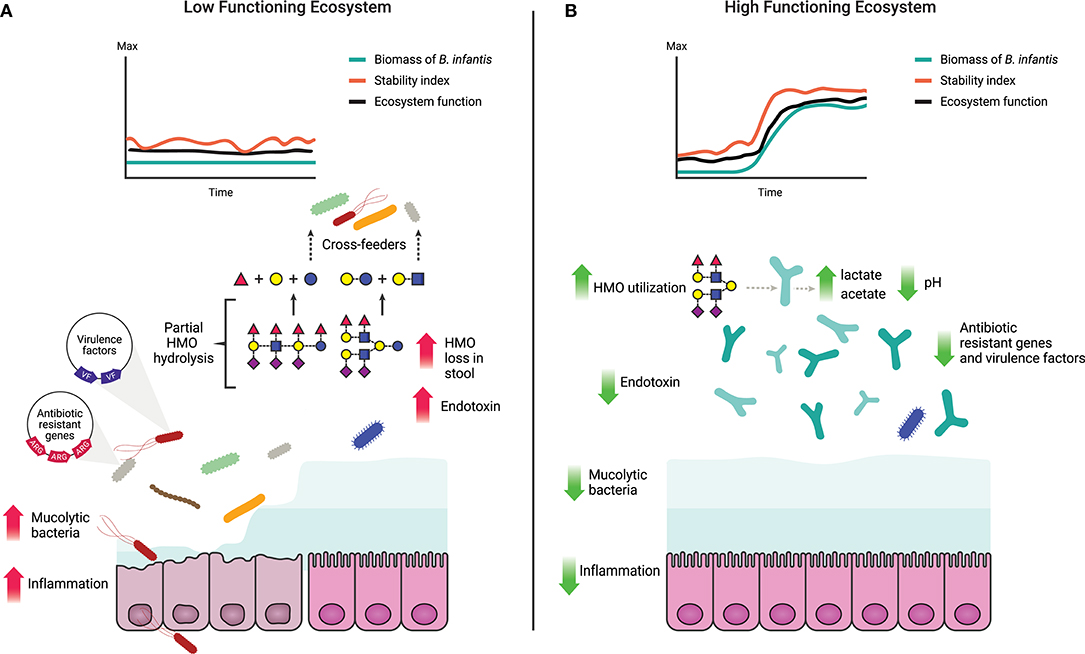
Figure 3. Characteristics of a high and a low functioning infant gut microbiome based on delivery of ecosystem services. (A) A low functioning infant gut microbiome ecosystem where the resources (HMOs) are inefficiently utilized leading to their loss in the stool (17) and potential cross-feeding to non-adapted opportunistic taxa. Prevalence of non-adapted opportunistic taxa leads to loss of ecosystem stability and decreased ecosystem function along with increased abundance of virulence factors and antibiotic resistance genes (82, 84, 85). Overgrowth of mucus-degrading bacteria and elevated levels of endotoxin compromise the intestinal barrier leading to chronic enteric inflammation and/or increased susceptibility to bacterial translocation (5, 89, 90). (B) A high functioning gut microbiome where ecosystem functions and stability are maintained over time, corollary to an increase in biomass of B. infantis. HMOs are efficiently utilized by B. infantis and converted into cell biomass, organic and SCFA. Production of organic and SCFA reduces the luminal pH (17) creating an unfavorable environment for opportunistic taxa, including virulent, antibiotic resistant, and mucolytic bacteria.
Integrating Ecosystem Services in the Evaluation of the Infant Gut Microbiome
The application of traditional concepts from macroecology has proven successful in providing relevant insight into the ecological dynamics that govern the human microbiome (38, 49). According to ecological theory, ecosystem productivity can be measured by total biomass and by changes in the concentration of a limiting substrate (35, 62, 91). In the gut, dietary and host-derived carbohydrates are the primary resources for microbial metabolism (31, 32). Productivity of the ecosystem can thus be determined based on the efficiency of their utilization, and in concert, determining bacterial biomass (Table 1; Figure 3). Together, these two functions offer complementary and independent approaches to monitor productivity and to identify states in which the delivery of the ecosystem services is maximized. Thus, by combining evaluations of ecosystem productivity and the generation of ecosystem services we propose a model for the definition of dysbiosis of the breastfed infant gut as a low-functioning ecosystem, in which the gut microbiome community is characterized by (1) low stability even without perturbations (e.g., diet change or antibiotics); (2) high susceptibility to invasion by external taxa; and (3) low utilization of the available resources (i.e., HMOs). The alternative to dysbiosis or a “healthy” state is characterized as being a high-functioning ecosystem when the gut microbiome community is: (1) stable over time, (2) resistant to invasion by allochthonous bacteria; and (3) shown to exhibit a high conversion of HMOs to pioneer products and biomass of benefit to the host (Figure 3). Overall, by focusing on function, this model is agnostic to method and index of choice and provides a quantifiable and objective approach to evaluate the microbiome.
Application of the Model to Evaluate the Symbiosis Between B. Infantis and the Breastfed Infant
Humans live in symbiosis with the composite of microbial inhabitants residing in their intestinal tracts but the contribution of specific species to the overall ecosystem function and terms of the individual symbiotic relationships, which can range from commensal to parasitic, are less understood. Considering the ecosystem services the infant host obtains from selectively favoring the growth of B. infantis, it is evident that the symbiotic relationship is mutualistic. Free selectively consumed resources like HMOs are extremely rare in nature and the composition of HMOs is unique among mammals (92, 93). Of the thousands of species able to colonize the human gut, only a very limited number of species have the molecular machinery to utilize them (24). Within the genus bifidobacteria, only B. infantis encodes the complete set of genes required to transport, and intracellularly deconstruct and metabolize all the chemical structures found among HMOs (65), thus indicating maintenance of these genes is under strong selection. Indeed, since its discovery, B. infantis has so far been exclusively found in association with human beings (94) and phylogenetic analysis indicates humans and bifidobacteria have co-speciated (95). Taken together, the association of B. infantis and the breastfed infant host presents strong characteristics of an exclusive symbiotic alliance that has persisted over evolutionary timescales, whereby the human host requires the symbiont to access a significant portion of its diet (i.e., HMOs), while concurrently the symbiont benefits from the nutritional niche provided by the host. This concept is congruent with well-established models of coevolved symbioses (57, 96, 97).
Interdependent biological alliances are best understood in binary symbiotic models (57). One invariable lesson from decades of research in these model systems has been that aposymbiosis (i.e., the absence of the symbiont) can represent a major stressor to the host and often results in physiological and developmental deficiencies. For example in the well-characterized Squid-Vibrio model, external perturbations are markedly different between apo- and symbiotic squids (98, 99). Indeed, the presence of V. fischeri may help modulate the host stress responses (100). Similarly, the removal of nutritional symbionts (i.e., symbiotic bacteria that help their animal partners digest, absorb, and metabolize complex nutrients) is known to pose appreciable fitness costs to the host (96, 101, 102). Notably, the removal of vertically transmitted (from parent to offspring) nutritional symbionts has been shown to have the greatest negative impact on host fitness (102), which bears surprising parallels to the conspicuous depletion of B. infantis among infants with severe acute malnutrition (103) and with the inverse correlation between fecal pH and stunting (104). Further examples include aposymbiotic pea aphids which have reduced growth rates, attain a lower adult size, and are reproductively sterile (101) and fruit flies, for which the presence of the facultative symbiont Lactobacillus plantarum is critical to the growth and maturation of larvae ingesting nutritionally suboptimal diets (9). Together, these examples demonstrate broadly that the disruption of ancient symbiotic associations can have negative implications on the host.
All data indicate human infants have evolved to partner with key symbiotic gut bacteria specialized in metabolizing host-provided resources in the form of HMOs; however, it appears over time the role of B. infantis and the impact of its absence from the infant gut have become obscured, likely because the generational loss of B. infantis predates the advent of high resolution tools to investigate the gut microbiome. For instance, substantial fecal excretion of HMOs and high fecal pH are not considered abnormal, and considerable instability of the gut microbial ecosystem is considered normal in early life (33). However, historical records suggest bifidobacteria was once more prevalent among infant populations in developed nations than what contemporary reports indicate (105), and correlative evidence from large cohort studies suggest absence of this key symbiont comes with important negative acute and chronic health consequences during a critical developmental stage (2, 4, 103, 104, 106).
What are the Acute and Chronic Health Consequences of the Absence of B. Infantis in the Infant Gut?
The importance of individual species to ecosystem function, and ultimately to the services, can become apparent through their loss. There is growing appreciation that interventions known to disrupt microbiome development may lead to the extinction of certain taxa across entire populations (107). Widespread antibiotic use, cesarean section delivery, and formula feeding are associated with altered gut microbiome compositions and subsequent negative health outcomes, including obesity and autoimmune diseases (3, 108, 109). In particular, the increased prevalence of these dietary and medical interventions has been associated with the decline of Bifidobacterium over the past century (20, 21, 60, 105, 110, 111). We pose the loss of critical functions in the gut resulting from the decline in the prevalence of B. infantis may have selected for microbiota that lack the resilience and stability during critical stages of immune and metabolic development. In fact, lower abundance of bifidobacteria has been associated with greater risk for developing colic, atopic dermatitis, asthma, food allergies, type I diabetes and chronic inflammation (2, 10, 11, 15, 112). Additionally, infants lacking B. infantis show signs of chronic enteric inflammation during the first 60 days of life (5), which has been directly linked to an increased risk of certain chronic disorders such as atopy and asthma later in life (113).
Interestingly, in geographic locations where breastfeeding rates are high and vaginal birth is widespread, Bifidobacterium is normally abundant in infant microbiomes (66, 114, 115). In contrast, the gut communities of infants in developed countries are largely unstable and highly variable (25, 111) and the distribution of Bifidobacterium is notably bimodal (26). This variation is clearly evident in a recent comparison of the gut microbiome of infants in geographically similar but developmentally diverse locations in which the level of Bifidobacterium was found to be higher in infants in more resource-limited locations, which correlated with decreased incidence of autoimmune and allergic diseases (2). Together these findings raise the question of whether the modern infant gut microbiome has been fundamentally altered from that of our ancestors and how the loss of key symbiotic species and the resulting disruption in immune development could be connected to the increased incidence of metabolic, autoimmune, and allergic diseases observed in developed countries today.
Fecal pH is another factor that has changed significantly over the past century and is consistent with the loss of Bifidobacterium (105). Fecal pH values directly correlates with the bacterial species colonizing the infant gut, particularly pertaining is the direct association between lower fecal pH and significantly decreased abundance of potentially harmful bacterial populations (i.e., Clostridiaceae, Enterobacteriaceae, Peptosteptoccocaceae, and Veillonellaceae) (105). These findings are intriguing, as an abundance of specific Enterobacteriaceae species induce gut inflammation (21), which has been positively associated with colic and crying in infants (116, 117). These adverse conditions may be due to the fact that Enterobacteriaceae-derived lipopolysaccharides induce stronger inflammatory activity compared with other lipopolysaccharide-producing bacteria (2, 118). In addition, lower fecal pH has been shown to be associated with better anthropometric growth scores (104) and improved thymic growth, a sign of immune system development (1). This may partially explain why B. infantis-colonized infants exhibit more robust vaccine responses (66, 119) and why there is a reduced incidence of autoimmune diseases in populations colonized with high levels of Bifidobacterium (2). Taken together, these data indicate functions provided by key symbiotic partners (i.e., B. infantis) during infancy have a strong impact on development and conversely, the absence of these taxa can have negative health consequences, underscoring the need to restore specific beneficial taxa to the infant gut.
Relevance, Applications, and Limitations of the Model
With the growing recognition of the role of the microbiome in human health, the incorporation of microbiome-based diagnostics will inevitably become routine. In fact, a number of commercial tests are currently available to the general public and physicians are increasingly requested to interpret test reports. However, we currently lack a “gold standard” for what constitutes a healthy microbiome. Here, we proposed an anthropocentric model whereby gut microbiome function is determined in terms of ecosystem services that ultimately benefit the infant. Thus, microbiome composition can be evaluated objectively with regard to its contribution to host health, facilitating interpretation by health professionals. Furthermore, linking functional traits to specific ecosystem services may assist both the development of prognostic tools of infant microbiome function and probiotic interventions aimed at restoring the ecosystem services of the infant gut microbiome.
However, it is important to recognize that this model is limited to conditions in which the nutrient landscape in the gut is shaped by a single nutritional resource (i.e., HMOs) and will have to be re-validated for conditions known to shift the type and amount of resources as well as the distribution of biomass. Such conditions include, the introduction of complementary foods, formula feeding, antibiotic use and other microbiome-modifying practices. Moreover, the principles on which this model is based may be affected by stochastic events including niche pre-emption (i.e., “first come, first served”) driven by priority effects (120). We also recognize the overall dynamics of the infant microbiome involve complex intra and inter-species interactions which are not considered in our model. For example, the ecological relevance of Bifidobacterium species other than B. infantis, which are known to have limited capacity to metabolize HMOs but are found in the stools of infants, is currently unknown (121). Additionally, future models should aim to integrate the non-bacterial microbial inhabitants of the microbiome (e.g., virus, archaea, fungi, and other eukaryotes) which are increasingly recognized as important functional components. Nevertheless, the ecological principles presented here, can be broadly applicable to other host species, and evaluation of additional body sites, and can be adapted to inform the selection of taxa that may be relevant for health in other stages of life.
Lastly, we hope this work encourages the field to propose analogous models that incorporate ecological theory and testable frameworks to identify microbiome characteristics that are conducive to health or disease.
Author Contributions
RD, BH, GC, and SF drafted and wrote this manuscript. All authors are responsible for idea conception, critical evaluation, and manuscript review.
Conflict of Interest
RD, BH, GC, and SF are employed by Evolve BioSystems, Inc.
References
1. Indrio F, Ladisa G, Mautone A, Montagna O. Effect of a fermented formula on thymus size and stool pH in healthy term infants. Pediatr Res. (2007) 62:98–100. doi: 10.1203/pdr.0b013e31806772d3
2. Vatanen T, Kostic AD, d'Hennezel E, Siljander H, Franzosa EA, Yassour M, et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell. (2016) 165:842–53. doi: 10.1016/j.cell.2016.04.007
3. Mahana D, Trent CM, Kurtz ZD, Bokulich NA, Battaglia T, Chung J, et al. Antibiotic perturbation of the murine gut microbiome enhances the adiposity, insulin resistance, and liver disease associated with high-fat diet. Genome Med. (2016) 8:48. doi: 10.1186/s13073-016-0297-9
4. Rhoads JM, Collins J, Fatheree NY, Hashmi SS, Taylor CM, Luo M, et al. Infant colic represents gut inflammation and dysbiosis. J Pediatr. (2018) 203:55–61.e53. doi: 10.1016/j.jpeds.2018.07.042
5. Henrick BM, Chew S, Casaburi G, Brown HK, Frese SA, Zhou Y, et al. Colonization by B. infantis EVC001 modulates enteric inflammation in exclusively breastfed infants. Pediatr Res. (2019) 86:749–57. doi: 10.1038/s41390-019-0533-2
6. Hosokawa T, Kikuchi Y, Shimada M, Fukatsu T. Symbiont acquisition alters behaviour of stinkbug nymphs. Biol Lett. (2008) 4:45–8. doi: 10.1098/rsbl.2007.0510
7. Weiss BL, Maltz M, Aksoy S. Obligate symbionts activate immune system development in the tsetse fly. J Immunol. (2012) 188:3395–403. doi: 10.4049/jimmunol.1103691
8. Trasande L, Blustein J, Liu M, Corwin E, Cox LM, Blaser MJ. Infant antibiotic exposures and early-life body mass. Int J Obes. (2013) 37:16–23. doi: 10.1038/ijo.2012.132
9. Storelli G, Strigini M, Grenier T, Bozonnet L, Schwarzer M, Daniel C, et al. Drosophila perpetuates nutritional mutualism by promoting the fitness of its intestinal symbiont Lactobacillus plantarum. Cell Metab. (2018) 27:362–77 e368. doi: 10.1016/j.cmet.2017.11.011
10. Kalliomaki M, Kirjavainen P, Eerola E, Kero P, Salminen S, Isolauri E. Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J Allergy Clin Immunol. (2001) 107:129–34. doi: 10.1067/mai.2001.111237
11. Fujimura KE, Lynch SV. Microbiota in allergy and asthma and the emerging relationship with the gut microbiome. Cell Host Microbe. (2015) 17:592–602. doi: 10.1016/j.chom.2015.04.007
12. Livanos AE, Greiner TU, Vangay P, Pathmasiri W, Stewart D, McRitchie S, et al. Antibiotic-mediated gut microbiome perturbation accelerates development of type 1 diabetes in mice. Nat Microbiol. (2016) 1:16140. doi: 10.1038/nmicrobiol.2016.140
13. Arrieta MC, Arevalo A, Stiemsma L, Dimitriu P, Chico ME, Loor S, et al. Associations between infant fungal and bacterial dysbiosis and childhood atopic wheeze in a nonindustrialized setting. J Allergy Clin Immunol. (2018) 142:424–34 e410. doi: 10.1016/j.jaci.2017.08.041
14. Insel R, Knip M. Prospects for primary prevention of type 1 diabetes by restoring a disappearing microbe. Pediatr Diabetes. (2018) 19:1400–6. doi: 10.1111/pedi.12756
15. Vatanen T, Franzosa EA, Schwager R, Tripathi S, Arthur TD, Vehik K, et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature. (2018) 562:589–94. doi: 10.1038/s41586-018-0620-2
16. McKenney EA, Koelle K, Dunn RR, Yoder AD. The ecosystem services of animal microbiomes. Mol Ecol. (2018) 27:2164–72. doi: 10.1111/mec.14532
17. Frese SA, Hutton AA, Contreras LN, Shaw CA, Palumbo MC, Casaburi G, et al. Persistence of supplemented Bifidobacterium longum subsp. infantis EVC001 in breastfed infants. mSphere. (2017) 2:e00501-17. doi: 10.1128/mSphere.00501-17
18. Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. (2010) 107:11971–5. doi: 10.1073/pnas.1002601107
19. Reyman M, van Houten MA, van Baarle D, Bosch A, Man WH, Chu M, et al. Impact of delivery mode-associated gut microbiota dynamics on health in the first year of life. Nat Commun. (2019) 10:4997. doi: 10.1038/s41467-019-13014-7
20. Makino H, Kushiro A, Ishikawa E, Kubota H, Gawad A, Sakai T, et al. Mother-to-infant transmission of intestinal bifidobacterial strains has an impact on the early development of vaginally delivered infant's microbiota. PLoS ONE. (2013) 8:e78331. doi: 10.1371/journal.pone.0078331
21. Rutayisire E, Huang K, Liu Y, Tao F. The mode of delivery affects the diversity and colonization pattern of the gut microbiota during the first year of infants' life: a systematic review. BMC Gastroenterol. (2016) 16:86. doi: 10.1186/s12876-016-0498-0
22. Ferretti P, Pasolli E, Tett A, Asnicar F, Gorfer V, Fedi S, et al. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe. (2018) 24:133–45 e135. doi: 10.1016/j.chom.2018.06.005
23. Duar RM, Kyle D, Tribe RM. Reintroducing B. infantis to the Cesarean-born neonate: an ecologically sound alternative to “vaginal seeding.” FEMS Microbiol Lett. (2020) 367:fnaa032. doi: 10.1093/femsle/fnaa032
24. Marcobal A, Barboza M, Froehlich JW, Block DE, German JB, Lebrilla CB, et al. Consumption of human milk oligosaccharides by gut-related microbes. J Agric Food Chem. (2010) 58:5334–40. doi: 10.1021/jf9044205
25. Zivkovic AM, Lewis ZT, German JB, Mills DA. Establishment of a milk-oriented microbiota (MOM) in early life: how babies meet their MOMs. Funct Food Rev. (2013) 5:3–12. doi: 10.2310/6180.2009.00035
26. Lewis ZT, Mills DA. Differential establishment of bifidobacteria in the breastfed infant gut. Nestle Nutr Inst Workshop Ser. (2017) 88:149–59. doi: 10.1159/000455399
27. Thomson P, Medina DA, Garrido D. Human milk oligosaccharides and infant gut bifidobacteria: molecular strategies for their utilization. Food Microbiol. (2018) 75:37–46. doi: 10.1016/j.fm.2017.09.001
28. Mallon CA, Elsas JDV, Salles JF. Microbial invasions: the process, patterns, and mechanisms. Trends Microbiol. (2015) 23:719–29. doi: 10.1016/j.tim.2015.07.013
29. Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol. (2012) 10:323–35. doi: 10.1038/nrmicro2746
30. Funkhouser LJ, Bordenstein SR. Mom knows best: the universality of maternal microbial transmission. PLoS Biol. (2013) 11:e1001631. doi: 10.1371/journal.pbio.1001631
31. Backhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. (2015) 17:690–703. doi: 10.1016/j.chom.2015.04.004
32. Stewart CJ, Ajami NJ, O'Brien JL, Hutchinson DS, Smith DP, Wong MC, et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature. (2018) 562:583–8. doi: 10.1038/s41586-018-0617-x
33. Derrien M, Alvarez AS, de Vos WM. The gut microbiota in the first decade of life. Trends Microbiol. (2019) 27:997–1010. doi: 10.1016/j.tim.2019.08.001
34. McBurney MI, Davis C, Fraser CM, Schneeman BO, Huttenhower C, Verbeke K, et al. Establishing what constitutes a healthy human gut microbiome: state of the science, regulatory considerations, and future directions. J Nutr. (2019) 149:1882–95. doi: 10.1093/jn/nxz154
35. Balvanera P, Pfisterer AB, Buchmann N, He JS, Nakashizuka T, Raffaelli D, et al. Quantifying the evidence for biodiversity effects on ecosystem functioning and services. Ecol Lett. (2006) 9:1146–56. doi: 10.1111/j.1461-0248.2006.00963.x
36. Lovell R, Wheeler BW, Higgins SL, Irvine KN, Depledge MH. A systematic review of the health and well-being benefits of biodiverse environments. J Toxicol Environ Health B Crit Rev. (2014) 17:1–20. doi: 10.1080/10937404.2013.856361
37. Johnson KV, Burnet PW. Microbiome: Should we diversify from diversity? Gut Microbes. (2016) 7:455–8. doi: 10.1080/19490976.2016.1241933
38. Foster KR, Schluter J, Coyte KZ, Rakoff-Nahoum S. The evolution of the host microbiome as an ecosystem on a leash. Nature. (2017) 548:43–51. doi: 10.1038/nature23292
39. Shade A. Diversity is the question, not the answer. ISME J. (2017) 11:1–6. doi: 10.1038/ismej.2016.118
40. Thukral AK. A review on measurement of Alpha diversity in biology. Agric Res J. (2017) 54:1–10. doi: 10.5958/2395-146X.2017.00001.1
41. Kim B-R, Shin J, Guevarra R, Lee JH, Kim DW, Seol K-H, et al. Deciphering diversity indices for a better understanding of microbial communities. J Microbiol Biotechnol. (2017) 27:2089–93. doi: 10.4014/jmb.1709.09027
42. Allison SD, Martiny JB. Colloquium paper: resistance, resilience, and redundancy in microbial communities. Proc Natl Acad Sci USA. (2008) 105 (Suppl. 1):11512–9. doi: 10.1073/pnas.0801925105
43. McNally L, Brown SP. Building the microbiome in health and disease: niche construction and social conflict in bacteria. Philos Trans R Soc Lond B Biol Sci. (2015) 370:370. doi: 10.1098/rstb.2014.0298
44. Delgado-Baquerizo M, Giaramida L, Reich PB, Khachane AN, Hamonts K, Edwards C, et al. Lack of functional redundancy in the relationship between microbial diversity and ecosystem functioning. J Ecol. (2016) 104:936–46. doi: 10.1111/1365-2745.12585
45. Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. (2012) 486:222–7. doi: 10.1038/nature11053
46. Kirmiz N, Robinson RC, Shah IM, Barile D, Mills DA. Milk glycans and their interaction with the infant-gut microbiota. Annu Rev Food Sci Technol. (2018) 9:429–50. doi: 10.1146/annurev-food-030216-030207
48. Sonnenburg JL, Sonnenburg ED. Vulnerability of the industrialized microbiota. Science. (2019) 366:eaaw9255. doi: 10.1126/science.aaw9255
49. Costello EK, Stagaman K, Dethlefsen L, Bohannan BJ, Relman DA. The application of ecological theory toward an understanding of the human microbiome. Science. (2012) 336:1255–62. doi: 10.1126/science.1224203
50. Martínez I, Maldonado-Gomez MX, Gomes-Neto JC, Kittana H, Ding H, Schmaltz R, et al. Experimental evaluation of the importance of colonization history in early-life gut microbiota assembly. eLife. (2018) 7:e36521. doi: 10.7554/eLife.36521
51. Jeraldo P, Sipos M, Chia N, Brulc JM, Dhillon AS, Konkel ME, et al. Quantification of the relative roles of niche and neutral processes in structuring gastrointestinal microbiomes. Proc Natl Acad Sci USA. (2012) 109:9692–8. doi: 10.1073/pnas.1206721109
52. Robinson CJ, Young VB. Antibiotic administration alters the community structure of the gastrointestinal microbiota. Gut Microbes. (2010) 1:279–84. doi: 10.4161/gmic.1.4.12614
53. Fierer N, Ferrenberg S, Flores GE, González A, Kueneman J, Legg T, et al. From animalcules to an ecosystem: application of ecological concepts to the human microbiome. Annu Rev Ecol Evol Syst. (2012) 43:137–55. doi: 10.1146/annurev-ecolsys-110411-160307
54. Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. (2007) 449:811–8. doi: 10.1038/nature06245
55. Gonzalez A, Clemente JC, Shade A, Metcalf JL, Song S, Prithiviraj B, et al. (2011). Our microbial selves: what ecology can teach us. EMBO Rep. (2011) 12:775–84. doi: 10.1038/embor.2011.137
56. Wang Q, Liu J, Zhu H. Genetic and molecular mechanisms underlying symbiotic specificity in legume-rhizobium interactions. Front Plant Sci. (2018) 9:313. doi: 10.3389/fpls.2018.00313
58. Kohl KD, Weiss RB, Cox J, Dale C, Dearing MD. Gut microbes of mammalian herbivores facilitate intake of plant toxins. Ecol Lett. (2014) 17:1238–46. doi: 10.1111/ele.12329
59. Duar RM, Frese SA, Lin XB, Fernando SC, Burkey TE, Tasseva G, et al. Experimental evaluation of host adaptation of lactobacillus reuteri to different vertebrate species. Appl Environ Microbiol. (2017) 83:e00132-17. doi: 10.1128/AEM.00132-17
60. Turroni F, Milani C, Duranti S, Ferrario C, Lugli GA, Mancabelli L, et al. Bifidobacteria and the infant gut: an example of co-evolution and natural selection. Cell Mol Life Sci. (2018) 75:103–18. doi: 10.1007/s00018-017-2672-0
61. Luck GW, Harrington R, Harrison PA, Kremen C, Berry PM, Bugter R, et al. Quantifying the contribution of organisms to the provision of ecosystem services. Bioscience. (2009) 59:223–35. doi: 10.1525/bio.2009.59.3.7
62. Duncan C, Thompson JR, Pettorelli N. The quest for a mechanistic understanding of biodiversity-ecosystem services relationships. Proc Biol Sci. (2015) 282:20151348. doi: 10.1098/rspb.2015.1348
63. Coppa GV, Gabrielli O, Pierani P, Catassi C, Carlucci A, Giorgi PL. Changes in carbohydrate composition in human milk over 4 months of lactation. Pediatrics. (1993) 91:637–41.
64. Fukuda S, Toh H, Taylor TD, Ohno H, Hattori M. Acetate-producing bifidobacteria protect the host from enteropathogenic infection via carbohydrate transporters. Gut Microbes. (2012) 3:449–54. doi: 10.4161/gmic.21214
65. Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, Whitehead TR, et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci USA. (2008) 105:18964–9. doi: 10.1073/pnas.0809584105
66. Huda MN, Lewis Z, Kalanetra KM, Rashid M, Ahmad SM, Raqib R, et al. Stool microbiota and vaccine responses of infants. Pediatrics. (2014) 134:e362–372. doi: 10.1542/peds.2013-3937
67. De Leoz ML, Kalanetra KM, Bokulich NA, Strum JS, Underwood MA, German JB, et al. Human milk glycomics and gut microbial genomics in infant feces show a correlation between human milk oligosaccharides and gut microbiota: a proof-of-concept study. J Proteome Res. (2015) 14:491–502. doi: 10.1021/pr500759e
68. Berding K, Holscher HD, Arthur AE, Donovan SM. Fecal microbiome composition and stability in 4- to 8-year old children is associated with dietary patterns and nutrient intake. J Nutr Biochem. (2018) 56:165–74. doi: 10.1016/j.jnutbio.2018.01.002
69. Cherrington CA, Hinton M, Pearson GR, Chopra I. Short-chain organic acids at ph 5.0 kill Escherichia coli and Salmonella spp. without causing membrane perturbation. J Appl Bacteriol. (1991) 70:161–5. doi: 10.1111/j.1365-2672.1991.tb04442.x
70. Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. (2013) 341:569–73. doi: 10.1126/science.1241165
71. Koh A, De Vadder F, Kovatcheva-Datchary P, Bäckhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. (2016) 165:1332–45. doi: 10.1016/j.cell.2016.05.041
72. Ritzhaupt A, Wood IS, Ellis A, Hosie KB, Shirazi-Beechey SP. Identification and characterization of a monocarboxylate transporter (MCT1) in pig and human colon: its potential to transport L-lactate as well as butyrate. J Physiol. (1998) 513 (Pt 3):719–32. doi: 10.1111/j.1469-7793.1998.719ba.x
73. Bozzo L, Puyal J, Chatton JY. Lactate modulates the activity of primary cortical neurons through a receptor-mediated pathway. PLoS ONE. (2013) 8:e71721. doi: 10.1371/journal.pone.0071721
74. Machler P, Wyss MT, Elsayed M, Stobart J, Gutierrez R, von Faber-Castell A, et al. In vivo evidence for a lactate gradient from astrocytes to neurons. Cell Metab. (2016) 23:94–102. doi: 10.1016/j.cmet.2015.10.010
75. Kearney SM, Gibbons SM, Erdman SE, Alm EJ. Orthogonal dietary niche enables reversible engraftment of a gut bacterial commensal. Cell Rep. (2018) 24:1842–51. doi: 10.1016/j.celrep.2018.07.032
76. van der Waaij D, Berghuis-de Vries JM, Lekkerkerk L-V. Colonization resistance of the digestive tract in conventional and antibiotic-treated mice. J Hyg. (1971) 69:405–11. doi: 10.1017/S0022172400021653
77. Stecher B, Hardt WD. Mechanisms controlling pathogen colonization of the gut. Curr Opin Microbiol. (2011) 14:82–91. doi: 10.1016/j.mib.2010.10.003
78. Lawley TD, Walker AW. Intestinal colonization resistance. Immunology. (2013) 138:1–11. doi: 10.1111/j.1365-2567.2012.03616.x
79. Coyte KZ, Schluter J, Foster KR. The ecology of the microbiome: Networks, competition, and stability. Science. (2015) 350:663–6. doi: 10.1126/science.aad2602
80. Duncan SH, Louis P, Thomson JM, Flint HJ. The role of pH in determining the species composition of the human colonic microbiota. Environ Microbiol. (2009) 11:2112–22. doi: 10.1111/j.1462-2920.2009.01931.x
81. Dohi M, Mougi A. A coexistence theory in microbial communities. R Soc Open Sci. (2018) 5:180476. doi: 10.1098/rsos.180476
82. Casaburi G, Frese SA. Colonization of breastfed infants by Bifidobacterium longum subsp. infantis EVC001 reduces virulence gene abundance. Hum Microb J. (2018) 9:7–10. doi: 10.1016/j.humic.2018.05.001
83. Parnanen K, Karkman A, Hultman J, Lyra C, Bengtsson-Palme J, Larsson DGJ, et al. Maternal gut and breast milk microbiota affect infant gut antibiotic resistome and mobile genetic elements. Nat Commun. (2018) 9:3891. doi: 10.1038/s41467-018-06393-w
84. Taft DH, Liu J, Maldonado-Gomez MX, Akre S, Huda MN, Ahmad SM, et al. Bifidobacterial dominance of the gut in early life and acquisition of antimicrobial resistance. MSphere. (2018) 3:e00441-18. doi: 10.1128/mSphere.00441-18
85. Casaburi G, Duar RM, Vance DP, Mitchell R, Contreras L, Frese SA, et al. Early-life gut microbiome modulation reduces the abundance of antibiotic-resistant bacteria. Antimicrob Resist Infect Control. (2019) 8:1–18. doi: 10.1186/s13756-019-0583-6
86. Aragozzini F, Ferrari A, Pacini N, Gualandris R. Indole-3-lactic acid as a tryptophan metabolite produced by Bifidobacterium spp. Appl Environ Microbiol. (1979) 38:544–6. doi: 10.1128/AEM.38.3.544-546.1979
87. Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. (2011) 469:543–7. doi: 10.1038/nature09646
88. Ehrlich AM, Henrick B, Pacheco A, Taft D, Xu G, Huda N, et al. Bifidobacterium grown on human milk oligosaccharides produce tryptophan metabolite Indole-3-lactic acid that significantly decreases inflammation in intestinal cells in vitro. FASEB J. (2018) 32 (1_Suppl.):lb359.
89. Karav S, Casaburi G, Frese SA. Reduced colonic mucin degradation in breastfed infants colonized by Bifidobacterium longum subsp. infantis EVC001. FEBS Open Bio. (2018) 8:1649–57. doi: 10.1002/2211-5463.12516
90. Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, et al. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell. (2016) 167:1339–53.e1321. doi: 10.1016/j.cell.2016.10.043
91. Kremen C. Managing ecosystem services: what do we need to know about their ecology? Ecol Lett. (2005) 8:468–79. doi: 10.1111/j.1461-0248.2005.00751.x
92. Tao N, Wu S, Kim J, An HJ, Hinde K, Power ML, et al. Evolutionary glycomics: characterization of milk oligosaccharides in primates. J Proteome Res. (2011) 10:1548–57. doi: 10.1021/pr1009367
93. Bode L. Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology. (2012) 22:1147–62. doi: 10.1093/glycob/cws074
94. Mattarelli P, Biavati B. Species in the genus Bifidobacterium. In: Mattarelli P, Biavati B, Holzapfel WH, Wood BJB, editors. The Bifidobacteria and Related Organisms. London, UK: Academic Press (2018). p. 9–48. doi: 10.1016/B978-0-12-805060-6.00002-8
95. Moeller AH, Caro-Quintero A, Mjungu D, Georgiev AV, Lonsdorf EV, Muller MN, et al. Cospeciation of gut microbiota with hominids. Science. (2016) 353:380–2. doi: 10.1126/science.aaf3951
97. Parfrey LW, Moreau CS, Russell JA. Introduction: the host-associated microbiome: pattern, process and function. Mol Ecol. (2018) 27:1749–65. doi: 10.1111/mec.14706
98. Mandel MJ, Schaefer AL, Brennan CA, Heath-Heckman EA, Deloney-Marino CR, McFall-Ngai MJ, et al. Squid-derived chitin oligosaccharides are a chemotactic signal during colonization by Vibrio fischeri. Appl Environ Microbiol. (2012) 78:4620–6. doi: 10.1128/AEM.00377-12
99. Koehler S, Gaedeke R, Thompson C, Bongrand C, Visick KL, Ruby E, et al. The model squid-vibrio symbiosis provides a window into the impact of strain- and species-level differences during the initial stages of symbiont engagement. Environ Microbiol. (2018) 21:3269–83. doi: 10.1111/1462-2920.14392
100. Casaburi G, Goncharenko-Foster I, Duscher AA, Foster JS. Transcriptomic changes in an animal-bacterial symbiosis under modeled microgravity conditions. Sci Rep. (2017) 7:46318. doi: 10.1038/srep46318
101. Douglas AE. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu Rev Entomol. (1998) 43:17–37. doi: 10.1146/annurev.ento.43.1.17
102. Fisher RM, Henry LM, Cornwallis CK, Kiers ET, West SA. The evolution of host-symbiont dependence. Nat Commun. (2017) 8:15973. doi: 10.1038/ncomms15973
103. Subramanian S, Huq S, Yatsunenko T, Haque R, Mahfuz M, Alam MA, et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. (2014) 510:417–21. doi: 10.1038/nature13421
104. Hossain MS, Das S, Gazi MA, Alam MA, Haque NMS, Mahfuz M, et al. Association of faecal pH with childhood stunting: results from a cross-sectional study. BMJ Paediatrics Open. (2019) 3:e000549. doi: 10.1136/bmjpo-2019-000549
105. Henrick BM, Hutton AA, Palumbo MC, Casaburi G, Mitchell RD, Underwood MA, et al. Elevated fecal pH indicates a profound change in the breastfed infant gut microbiome due to reduction of Bifidobacterium over the past century. mSphere. (2018) 3:e00041-18. doi: 10.1128/mSphere.00041-18
106. Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. (2015) 7:307ra152. doi: 10.1126/scitranslmed.aab2271
107. Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, Sonnenburg JL. Diet-induced extinctions in the gut microbiota compound over generations. Nature. (2016) 529:212–5. doi: 10.1038/nature16504
108. Blaser MJ. Antibiotic use and its consequences for the normal microbiome. Science. (2016) 352:544–5. doi: 10.1126/science.aad9358
109. Schulfer AF, Battaglia T, Alvarez Y, Bijnens L, Ruiz VE, Ho M, et al. Intergenerational transfer of antibiotic-perturbed microbiota enhances colitis in susceptible mice. Nat Microbiol. (2018) 3:234–42. doi: 10.1038/s41564-017-0075-5
110. Tamburini S, Shen N, Wu HC, Clemente JC. The microbiome in early life: implications for health outcomes. Nat Med. (2016) 22:713–22. doi: 10.1038/nm.4142
111. Lawley B, Munro K, Hughes A, Hodgkinson AJ, Prosser CG, Lowry D, et al. Differentiation of Bifidobacterium longum subspecies longum and infantis by quantitative PCR using functional gene targets. PeerJ. (2017) 5:e3375. doi: 10.7717/peerj.3375
112. Kalliomaki M, Laippala P, Korvenranta H, Kero P, Isolauri E. Extent of fussing and colic type crying preceding atopic disease. Arch Dis Child. (2001) 84:349–50. doi: 10.1136/adc.84.4.349
113. Orivuori L, Mustonen K, de Goffau M, Hakala S, Paasela M, Roduit C, et al. High level of fecal calprotectin at age 2 months as a marker of intestinal inflammation predicts atopic dermatitis and asthma by age 6. Clin Exp Allergy. (2015) 45:928–39. doi: 10.1111/cea.12522
114. Davis JC, Lewis ZT, Krishnan S, Bernstein RM, Moore SE, Prentice AM, et al. Growth and morbidity of gambian infants are influenced by maternal milk oligosaccharides and infant gut microbiota. Sci Rep. (2017) 7:40466. doi: 10.1038/srep40466
115. Lawley B, Otal A, Moloney-Geany K, Diana A, Houghton L, Heath AM, et al. Fecal microbiotas of indonesian and New Zealand children differ in complexity and bifidobacterial taxa during the first year of life. Appl Environ Microbiol. (2019) 85:e01105-19. doi: 10.1128/AEM.01105-19
116. de Weerth C, Fuentes S, de Vos WM. Crying in infants: on the possible role of intestinal microbiota in the development of colic. Gut Microbes. (2013) 4:416–21. doi: 10.4161/gmic.26041
117. Dubois NE, Gregory KE. Characterizing the intestinal microbiome in infantile colic: findings based on an integrative review of the literature. Biol Res Nurs. (2016) 18:307–15. doi: 10.1177/1099800415620840
118. Barron Pastor HJ, Gordon DM. Effects of dispersal limitation in the face of intense selection via dietary intervention on the faecal microbiota of rats. Environ Microbiol Rep. (2016) 8:187–95. doi: 10.1111/1758-2229.12367
119. Huda MN, Ahmad SM, Alam MJ, Khanam A, Kalanetra KM, Taft DH, et al. Bifidobacterium abundance in early infancy and vaccine response at 2 years of age. Pediatrics. (2019) 143:e20181489. doi: 10.1542/peds.2018-1489
120. Sprockett D, Fukami T, Relman DA. Role of priority effects in the early-life assembly of the gut microbiota. Nat Rev Gastroenterol Hepatol. (2018) 15:197–205. doi: 10.1038/nrgastro.2017.173
121. Lawson MA, O'Neill IJ, Kujawska M, Javvadi SG, Wijeyesekera A, Flegg Z, et al. Breast milk-derived human milk oligosaccharides promote Bifidobacterium interactions within a single ecosystem. ISME J. (2020) 14:635–48. doi: 10.1038/s41396-019-0553-2
122. Hooks KB, O'Malley MA. Dysbiosis and its discontents. MBio. (2017) 8. doi: 10.1128/mBio.01492-17
123. Olesen SW, Alm EJ. Dysbiosis is not an answer. Nat Microb. (2016) 1:16228. doi: 10.1038/nmicrobiol.2016.228
124. Brüssow H. Problems with the concept of gut microbiota dysbiosis. Microb Biotechnol. (2020) 13:423–34. doi: 10.1111/1751-7915.13479
Glossary
Allochthonous: An organism whose origin is different than that in which is found.
Biomass: Total quantity of living organisms of a species per unit area of a given habitat.
Dysbiosis: Vague/imprecise term often used to describe a microbiome composition that is different from the control and/or observed in subjects with a particular disease or condition. No scientific consensus has been reached on its definition. The origins and use of the term are reviewed by Hooks and O'Malley (122), and the casual obfuscation of its use to progress in the field, is discussed in Olesen and Alm (123) and Brussow (124).
Ecosystem services: Refers to the benefits to be gained from properly functioning ecosystems.
Ecosystem functioning: Refers to all processing and transport of energy and matter in an ecosystem, integrating multiple individual functions of the ecosystem including the production of biomass, the biochemical cycling of resources and the ability to resist invasion by allochthonous species.
Fitness (ecology): An organism's adaptation to the environment that increases its ability to propagate its genes. Genotypes with higher fitness are therefore selected for in the next generation (see natural selection). Fitness is environment-specific and directly related to the number of offspring produced.
Insurance hypothesis: Suggests that stabilization of communities against decline in function resulting from invasion, species loss, or fluctuations in abiotic features of the environment is improved by increasing diversity, and that diminishing fluctuations over time increases the overall productivity or services provided by the community.
Limiting substrate: Specific resource by which the productivity rate of an ecosystem depends on.
Mutualistic: Exchange of goods and services between species.
Niche (Hutchinsonian): Environmental conditions (biotic and abiotic) of a given habitat under which a species can persist and maintain stable populations without immigration from external sources.
Opportunistic commensal: Otherwise non-harming members of the gut microbiome that bloom upon a disturbance to the ecosystem and exert pathogenicity to the host.
Primary production: First level of nutrient generation in the tropic chain (food web).
Productivity: Rate at which energy is converted to biomass.
Stability (ecology): Measure of the temporal variability of an ecosystem, depends on its resistance to environmental change, and its rate of return to equilibrium following a perturbation (resilience).
Stochastic: With inherent randomness, the opposite of deterministic.
Symbiosis (From Greek: sym “with” and biosis “living”) long-term associations between organisms of distinct genetic makeup.
Keywords: gut microbiome, dysbiosis, human milk oligosaccharides, ecosystem services, microbiome modification, microbial ecology, symbiosis, probiotics
Citation: Duar RM, Henrick BM, Casaburi G and Frese SA (2020) Integrating the Ecosystem Services Framework to Define Dysbiosis of the Breastfed Infant Gut: The Role of B. infantis and Human Milk Oligosaccharides. Front. Nutr. 7:33. doi: 10.3389/fnut.2020.00033
Received: 17 December 2019; Accepted: 05 March 2020;
Published: 14 April 2020.
Edited by:
David A. Sela, University of Massachusetts Amherst, United StatesReviewed by:
Ravinder Nagpal, Wake Forest School of Medicine, United StatesCarlos Gómez-Gallego, University of Eastern Finland, Finland
Copyright © 2020 Duar, Henrick, Casaburi and Frese. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Steven A. Frese, steven.frese@unl.edu
 Rebbeca M. Duar
Rebbeca M. Duar Bethany M. Henrick
Bethany M. Henrick Giorgio Casaburi
Giorgio Casaburi Steven A. Frese
Steven A. Frese