Comparative Genomic Analysis of Two Serotype 1/2b Listeria monocytogenes Isolates from Analogous Environmental Niches Demonstrates the Influence of Hypervariable Hotspots in Defining Pathogenesis
- 1Teagasc Food Research Centre, Fermoy, Ireland
- 2Department of Biological Sciences, Cork Institute of Technology, Bishopstown, Ireland
- 3CSIRO Agriculture and Food, Werribee, VIC, Australia
The vast majority of clinical human listeriosis cases are caused by serotype 1/2a, 1/2b, 1/2c, and 4b isolates of Listeria monocytogenes. The ability of L. monocytogenes to establish a systemic listeriosis infection within a host organism relies on a combination of genes that are involved in cell recognition, internalization, evasion of host defenses, and in vitro survival and growth. Recently, whole genome sequencing and comparative genomic analysis have proven to be powerful tools for the identification of these virulence-associated genes in L. monocytogenes. In this study, two serotype 1/2b strains of L. monocytogenes with analogous isolation sources, but differing infection abilities, were subjected to comparative genomic analysis. The results from this comparison highlight the importance of accessory genes (genes that are not part of the conserved core genome) in L. monocytogenes pathogenesis. In addition, a number of factors, which may account for the perceived inability of one of the strains to establish a systemic infection within its host, have been identified. These factors include the notable absence of the Listeria pathogenicity island 3 and the stress survival islet, of which the latter has been demonstrated to enhance the survival ability of L. monocytogenes during its passage through the host intestinal tract, leading to a higher infection rate. The findings from this research demonstrate the influence of hypervariable hotspots in defining the physiological characteristics of a L. monocytogenes strain and indicate that the emergence of a non-pathogenic isolate of L. monocytogenes may result from a cumulative loss of functionality rather than by a single isolated genetic event.
Introduction
Listeria monocytogenes is a Gram-positive, facultatively anaerobic food-borne pathogen, and is the causative agent of the bacterial disease listeriosis in humans and animals. Recent figures demonstrate that approximately 99% of all human listeriosis cases arise due to the consumption of contaminated food produce (1), with serotypes 1/2a, 1/2b, 1/2c, and 4b implicated as the source of infection in 95% of these cases (2). Its psychrotrophic nature coupled with its tolerance of low pH and high salt concentrations (3) allows the bacterium to survive in refrigerated foods and reach levels required for human infection, if the food can support growth. L. monocytogenes is also commonly found in farm environments and silage in particular (4), and as such contaminated feeds represent a similar vector for food-borne transmission of the bacterium to animals used in food production. Its ability to cause a systemic infection in humans and animals alike is reliant on a combination of physical attributes, including resistance to environmental stresses and a capacity for virulence and survival within the host.
Traditionally, genetic relationships between L. monocytogenes strains are elucidated either by pulsed-field gel electrophoresis (PFGE) involving macrorestriction of genomic DNA to generate an associated DNA fingerprint (5) or by multilocus sequence typing (MLST) where specific sequences from a number of housekeeping genes are analyzed (6). These approaches are limited, however, in that they provide little insight into the pan genome of L. monocytogenes isolates. Comparative genome analysis has emerged as a robust tool for evaluating underlying genetic properties of bacterial strains, such as their evolutionary relationships, pathogenic potential, antibiotic resistances, and niche adaptation capabilities. With regard to Listeria, comparative genomics has proven to be particularly effective in determining the basis behind a number of observed phenotypic characteristics of L. monocytogenes, including the putative identification of many virulence genes responsible for L. monocytogenes pathogenesis on the basis of their relative absence in strains of the non-pathogenic Listeria innocua (7–9). Also, recent comparative analysis of two persistent L. monocytogenes strains that were isolated from separate fish processing plants almost 6 years apart (10) identified an extremely close relationship between their genomes. As such, it was proposed that strains with specific genetic traits may be selected for within a given environmental niche, providing a potential insight into the mechanisms of persistence of L. monocytogenes. Persistence is defined as the regular re-isolation of a given strain from the same environment over the course of several months or years. Comparative genomics has also been used to analyze L. monocytogenes isolates associated with listeriosis outbreaks (11, 12), to understand the unique genomic properties harbored by these strains contributing to systemic infection, and to determine the most efficient manner in which to analyze the epidemiological traits of future outbreak strains (13).
From an evolutionary perspective, one particular study involving a range of L. monocytogenes genomes of differing lineage and serotype demonstrated that this bacterial species, like others, has a highly conserved set of genes shared by all sequenced strains known as the “core genome” (14). While this core genome is common to all strains, subtyping methods (such as PFGE, MLST, and ribotyping) have demonstrated that examined L. monocytogenes isolates form a structured population consisting of a number of different evolutionary lineages (15). The majority of tested strains of serotypes 1/2a, 1/2b, 1/2c, 3a, 3b, 3c, 4b, and 4e cluster to evolutionary lineages I and II. Flagellar type “a” isolates such as serotypes 1/2a and 3a cluster to lineage II along with serotype 1/2c and 3c isolates, while flagellar type “b” isolates such as the 1/2b, 3b, and 4b serotypes all cluster within lineage I along with serotype 4d and 4e isolates (15). Two other evolutionary lineages have also been discovered and characterized. Lineage III contains serotype 4a, 4c, and a small number of 4b isolates (16) and represents a sister group to lineage I (15). Lineage IV, which was originally thought to represent a subgroup of lineage III (IIIB), is the most recently discovered (17), though only a limited number of isolates belonging to this lineage have been characterized to date. In general, the genomes of lineage I strains of L. monocytogenes (serotypes 1/2b, 3b, 4b, and 7) share a much higher degree of sequence similarity and exhibit a much lower degree of recombination than their lineage II and III counterparts (18, 19). Indeed, lineage I strains of L. monocytogenes predominantly differ from one another only in terms of their serotype, sequence type, prophage compositions, and a small fraction of chromosomal genes (12–23% of the total genome) that are collectively known as the accessory genome (14). Accessory genes are not as highly conserved as the core genes and in many cases can be strain-specific. Furthermore, while these accessory genes are located throughout the L. monocytogenes genome, their distribution is not entirely random. In certain chromosomal regions, accessory gene accumulations occur as a result of prophage acquisition (14), while other regions exhibit a non-random accumulation of these genes and are therefore denoted “hypervariable hotspots,” with nine such genomic regions recently defined in L. monocytogenes (19).
In this study, the genomes of two serotype 1/2b isolates of L. monocytogenes were subjected to comparative analysis in order to determine if there is a link between their core and accessory genome contents and their phenotypic characteristics. The two strains differed in their infection abilities. One of the isolates, strain DPC6895, was incapable of establishing a systemic infection within its animal host, despite it representing one of the four L. monocytogenes serotypes responsible for the vast majority of listeriosis cases (2, 20). Instead, this isolate caused a subclinical infection (21), and such subclinical infections generally go undetected, resulting in a potential public health hazard. On the other hand, strain FSL J2-064 did cause a systemic infection within its animal host. The aim of this research was to focus on a broad comparison of genes responsible for infection, intracellular survival, and proliferation within the host, in an attempt to discover a genomic basis for the perceived attenuation of pathogenesis in strain DPC6895 compared to strain FSL J2-064, and to evaluate the importance of the accessory genome in L. monocytogenes virulence and disease manifestation.
Materials and Methods
Input Strains for Comparative Analysis
The two L. monocytogenes strains examined in this study were of the 1/2b serotype. Strain DPC6895 was originally isolated from raw milk expressed by a cow with subclinical bovine mastitis (20, 21), while strain FSL J2-064 is a bovine clinical isolate (22, 23), but of a ribotype (or restriction digest fingerprint) that is also commonly found among food isolates (DUP-1052), and is associated with human disease (24). The genomes of both strains are available from public databases. The Whole Genome Shotgun project for L. monocytogenes strain DPC6895 was deposited at DDBJ/EMBL/GenBank and is available for download under the accession number LABG00000000. The version described in this paper is version LABG01000000. The genome sequence of L. monocytogenes strain FSL J2-064 is available from GenBank under the accession number NC_021824.
Identification of Strain-Specific Genes in Each of the Input Genomes
Whole genome comparisons were undertaken using BLAST Ring Image Generator (25) and Mauve (26), in order to visually identify unique genomic regions belonging to each of the strains. Genes within these regions were then confirmed to be strain-specific to each particular isolate through BLAST comparisons of their translated protein sequences against the genome of the other isolate, using RAST (27, 28).
Detection of Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)/CRISPR-Associated (Cas) Systems and Prophage Identification
Clustered regularly interspaced short palindromic repeat clusters in each genome were identified using CRISPRFinder (29), with flanking sequences of these clusters subsequently scanned for the presence Cas gene sequences. Viable and cryptic prophages within each of these genomes were detected using the PHAST tool (30).
Linear Comparisons and Identification of Hypervariable Hotspots
Linear comparisons of genes and gene clusters were prepared with Artemis (31) and subsequently visualized using the EasyFig software (32). Hypervariable hotspot locations in each genome were determined via BLAST analysis using conserved core gene identifiers and previously defined hypervariable hotspot locations for L. monocytogenes strain SLCC2755, which was used as a reference (19).
Results and Discussion
General Features of the L. monocytogenes Serotype 1/2b Strains
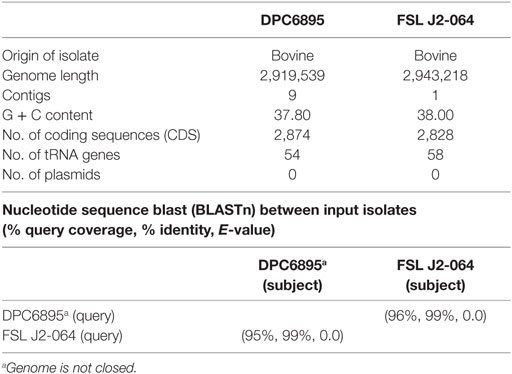
The main features of both strains used in this study (Table 1), as well as the locations of their respective hypervariable hotspots (Table S1 in Supplementary Material) are shown. Both genomes were similar in length (2.9–3.0 Mb) and had a G + C content of 37.8–38%, which is within the range typically observed for strains of L. monocytogenes. Neither of these strains contained any plasmids.
Comparative Genomic Analysis of Strains DPC6895 and FSL J2-064
Strains DPC6895 and FSL J2-064 were both obtained from bovine sources. While strain FSL J2-064 was originally isolated from a diseased animal (22, 23), strain DPC6895 was notably unable to establish a systemic clinical listeriosis infection in its respective host (20), though this strain did survive numerous antibiotic treatments and continued to be detected in milk expressed from the host for a prolonged period of time. While the host’s immune system may have been a contributing factor, comparative analysis of these two strains was undertaken in order to determine the extent of genetic diversity between them and to identify genomic characteristics, which may account for the observed physiological differences. L. monocytogenes strain DPC6895 was determined to contain a total of 123 genes that were not present in the genome of FSL J2-064 (Figure 1; Table S2 in Supplementary Material), while strain FSL J2-064 contained a total of 121 genes that were absent from the DPC6895 genome (Figure 2; Table S3 in Supplementary Material).
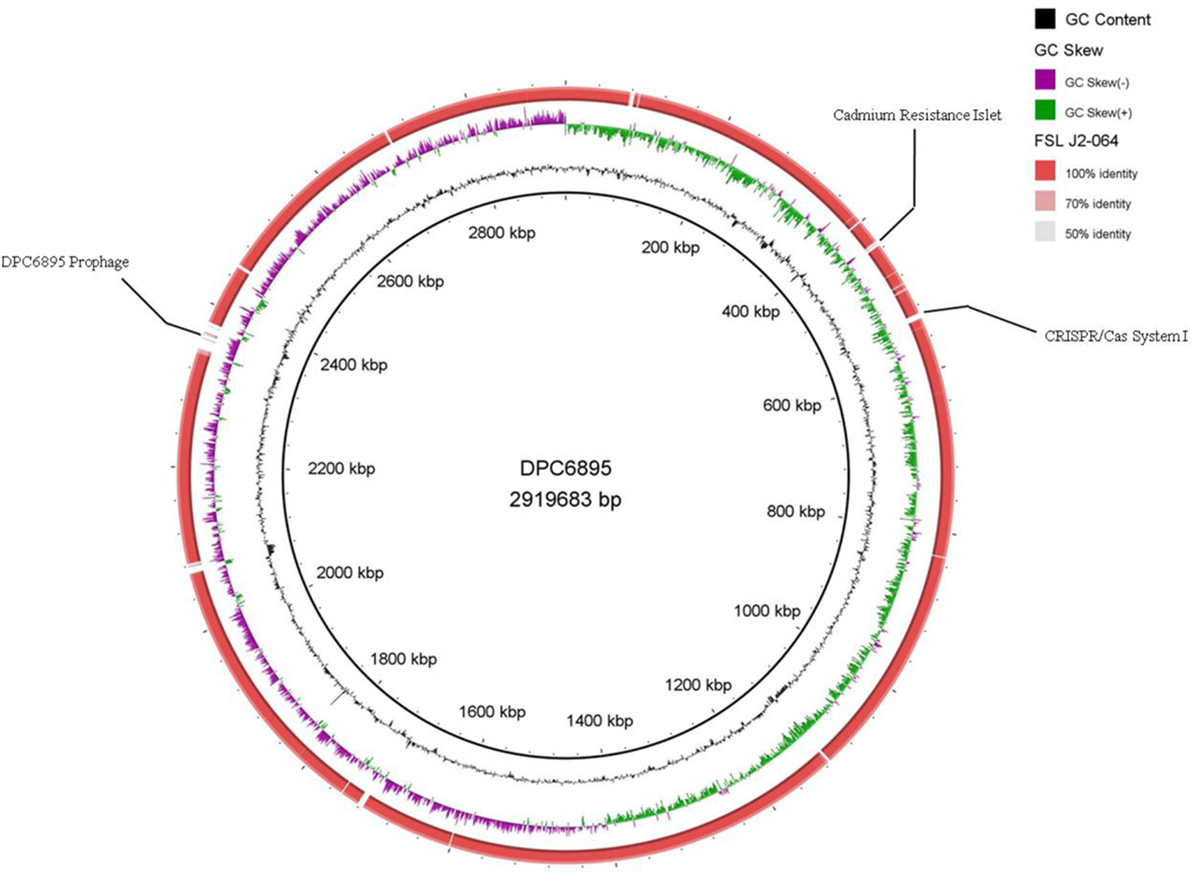
Figure 1. Circular map of Listeria monocytogenes strain FSL J2-064, using strain DPC6895 as a reference genome. The inner ring denotes the DPC6895 genome with corresponding genetic coordinates. The outer ring denotes the genome of strain FSL J2-064 (red).
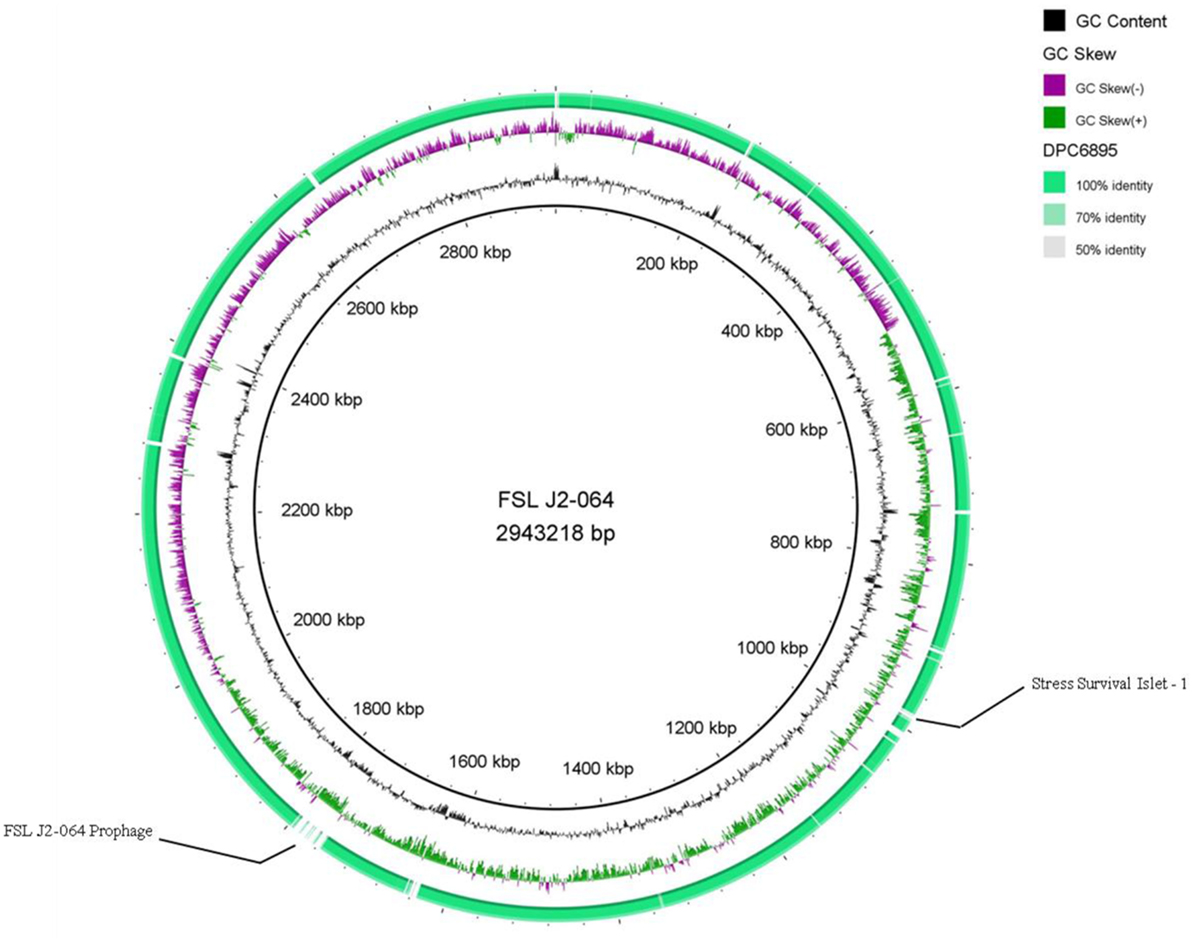
Figure 2. Circular map of Listeria monocytogenes strain DPC6895, using strain FSL J2-064 as a reference genome. The inner ring denotes the FSL J2-064 genome with corresponding genetic coordinates. The outer ring denotes the genome of strain DPC6895 (green).
Strain-Specific Genes in L. monocytogenes Strain DPC6895
The strain-specific genes in DPC6895 predominantly had functions, which contributed to an enhanced survival ability of this strain in a number of unfavorable environmental conditions. First, a number of these strain-specific genes had annotated functions in bacteriophage resistance. L. monocytogenes utilizes a number of biological systems in order to achieve resistance to bacteriophage infection. Foremost among these are the CRISPR sequences together with adjacent Cas genes and the restriction modification (RM) systems, which are widely distributed among prokaryotes (33). CRISPR/Cas genes comprise the adaptive immune system in many bacterial species including L. monocytogenes and have a role in defense of the bacterial cell against invading bacteriophages or plasmid-derived elements (34). Immunity against foreign invasion in bacteria is achieved first by integration of a small piece of viral or plasmid DNA (known as a spacer sequence) into the CRISPR locus. During infection, CRISPR-RNAs are transcribed, which guide the Cas proteins to target DNA that matches these spacer sequences that are then cleaved (35). RM systems are also used by bacteria in order to protect themselves from foreign invading DNA, of which there are three distinct classical types in addition to several atypical systems, which differ from one another in terms of their composition and cofactor requirements (36). Each of the different classical RM system types have been previously observed in L. monocytogenes (12, 37–39). Strain DPC6895 contained one CRISPR cluster, which consisted of 22 highly conserved direct repeat (DR) regions interspersed with 21 spacer sequences (Table 2). Flanking this CRISPR cluster were a total of seven open reading frames (ORFs) with annotated functions such as Cas genes. BLASTn analyses of the spacer sequences identified homologies to a number of different temperate serovar 1/2-specific L. monocytogenes phages including A006, A118, and LP-101, suggesting that this system has a functional role in resistance to infection from these particular siphoviruses. These homologies indicate a role for this particular system in expanding the immunity of these two particular strains to cover a range of both lytic and temperate L. monocytogenes phages. The presence of this CRISPR/Cas system may enhance the capacity of strain DPC6895 to withstand a wider array of extracellular threats posed by bacteriophages. No definitive CRISPR/Cas systems were detected in strain FSL J2-064; however, this strain contained all three subunits of a type I RM system, suggesting a difference between these strains in terms of their mechanisms of phage resistance. Strain DPC6895 contains two genes with 100% nucleotide identity to the R and M subunits of this system in FSL J2-064 but does not harbor the third S subunit, and as such this system is presumed to be non-functional in strain DPC6895.

Table 2. Clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated (Cas) systems in each of the Listeria monocytogenes isolates.
Second, a number of the strain-specific genes in DPC6895 had annotated functions associated with antibiotic and heavy metal resistance. L. monocytogenes has previously been demonstrated to have quite a broad spectrum of resistance to numerous antibiotics and antimicrobial agents (2, 40) in addition to exhibiting an elevated tolerance to heavy metals (41). The two strains in this study were analyzed for the presence of antibiotic and antimicrobial resistance genes and for heavy metal transporters. The results of this analysis (Table S4 in Supplementary Material) demonstrated that each of the genomes harbor specific resistance genes to a number of antibiotics, including the β-lactams, quinolone, fosfomycin, lincomycin, vancomycin, and tetracycline. Additionally, these strains also contained a number of antimicrobial and quaternary ammonium compound resistance genes including mdrL and lde, which are believed to be associated with increased tolerance of L. monocytogenes to benzalkonium chloride (42). Furthermore, lde is also thought to function in resistance of L. monocytogenes to fluoroquinolone (43). While both strains encoded many non-specific multidrug resistance transporters, strain DPC6895 harbored one additional multidrug transporter (locus tag TZ05_2661c), which was absent from strain FSL J2-064. The exact function of this particular transporter has not yet been fully elucidated, but subsequent BLASTp analysis identified a conserved domain within the translated protein product of this gene, which has a putative function in resistance to the lantibiotic gallidermin (44), suggesting a potentially similar role for this gene in each of these strains. As previously stated, L. monocytogenes strain DPC6895 was originally isolated from raw milk expressed by a cow with subclinical bovine mastitis. Following the confirmation of a subclinical infection, the infected cow was medically treated with subsequent intramammary injections of Synulox LC, Tylosin, and oxytetracycline. However, the infected animal’s milk continued to test positive for L. monocytogenes despite the intervention with antibiotic treatments (20). Synulox LC contains the antibiotics clavulanic acid and amoxicillin, which are both β-lactams, while oxytetracycline is an antibiotic that is related to tetracycline, and tylosin is a macrolide antibiotic. Analysis of the DPC6895 genome identified a total of eight genes encoding proteins with associated functions in resistance to β-lactams, while a gene associated with tetracycline resistance was also identified. The presence of the aforementioned mdrL gene in strain DPC6895 may account for its perceived resistance to Tylosin, given that previous research has demonstrated that disruption of this particular gene resulted in a higher susceptibility to macrolides (45).
In terms of heavy metal resistance, both of the strains contained a number of non-specific heavy metal transporters and specific lead/cadmium/zinc resistance genes. Interestingly, strain DPC6895 was also found to contain a novel 6.5 kb “islet” consisting of six genes, which included a heavy metal transporting ATPase, and a cadmium efflux system accessory protein (Table S5 in Supplementary Material). The G + C content of this islet was 35.9%, indicating that it is possibly of plasmid origin. This six-gene islet has not previously been observed in L. monocytogenes, with the only known Listeria homologs to these genes found in the recently sequenced L. innocua strain MOD1_LS888 (46). Linear alignments between these genomic regions identified a shared 99% nucleotide sequence identity, while also demonstrating that this islet is absent in strain FSL J2-064 and two other L. monocytogenes serotype 1/2b genomes that were available on Genbank (Figure 3). The product of one of the genes in this cluster, namely TZ05_0424, shares 100% amino acid sequence identity with the Staphylococcus aureus transposase Tn552 (47), suggesting strain DPC6895 may have acquired this cadmium resistance islet through a horizontal gene transfer event.
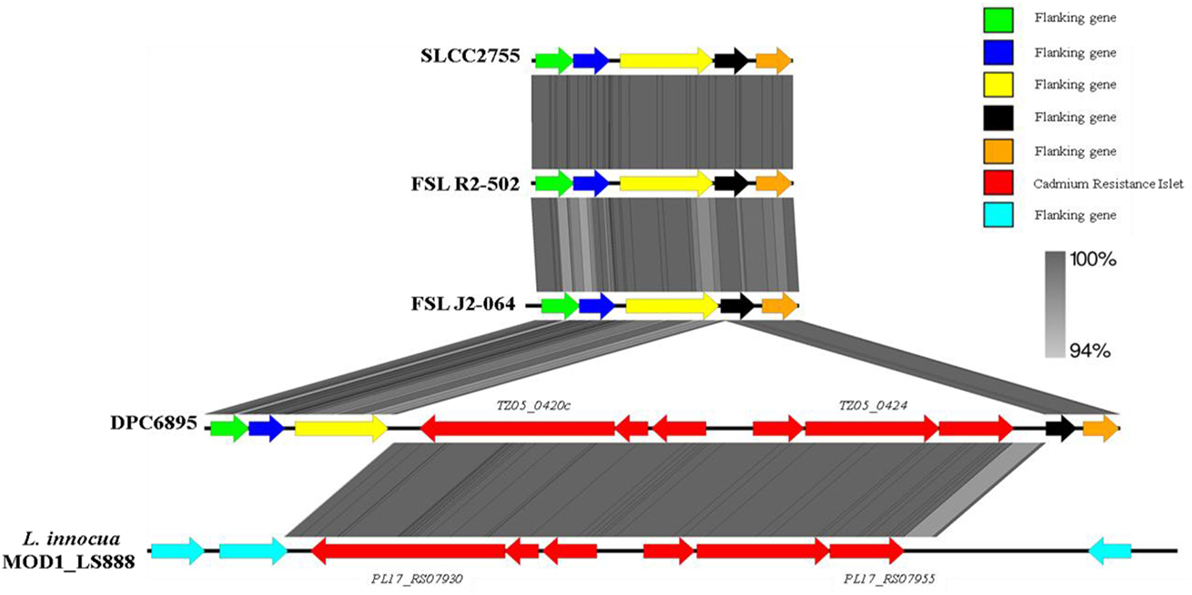
Figure 3. A linear comparison of the cadmium resistance islet identified in strain DPC6895 with the corresponding genomic region from strain FSL J2-064, two other serotype 1/2b Listeria monocytogenes isolates, and an additional Listeria innocua isolate, indicating the % amino acid identities between proteins in these clusters.
Finally, a number of the strain-specific genes in DPC6895 had annotated functions associated with peptide transport. The oligopeptide permease (opp) operon in L. monocytogenes consists of five genes (oppA, oppB, oppC, oppD, and oppF) that are essential for growth at low temperatures and contribute to intracellular growth of this bacterium (48). Comparative analysis of each of the isolates in this study identified the presence of this operon in both of the genomes (data not shown). However, in addition to the oligopeptide transporter operon, strain DPC6895 also contained a unique 5 kb cluster of genes (TZ05_2018–2022) within hypervariable hotspot 9, which BLASTp analysis indicated as a dipeptide transport system (dppABCDF) (Table S2 in Supplementary Material). The role of this system in strain DPC6895 is unclear. However, previous research has indicated that the presence of dipeptide transporters may confer a selective advantage on L. monocytogenes, given the fact that unlike numerous competing bacteria within an environment, it would not need to expend vast amounts of energy on protease synthesis (49). In addition, the presence of a dipeptide transporter would allow the organism to thrive in food samples, which may be deficient in free amino acids but rich in peptides. The presence of this system in strain DPC6895, therefore, could allow it to proliferate in what would be otherwise considered unfavorable environmental conditions.
Strain-Specific Genes in L. monocytogenes Strain FSL J2-064
The strain-specific genes in FSL J2-064 predominantly had functions, which contribute to enhancing the pathogenicity of this isolate. First, a number of the strain-specific genes in FSL J2-064 had annotated functions associated with virulence. L. monocytogenes requires a wide array of genes in order to successfully establish a systemic infection within a host organism. These genes, termed “virulence factors,” have functions in a number of different biological processes throughout the infection cycle, including host interaction, internalization, host defense evasion, and in vitro proliferation. A large family of leucine-rich proteins, known as the internalins, are important virulence factors involved in host interaction and internalization of pathogenic strains (50). The internalins are classified into four general types on the basis of their specific surface binding domains (51). Type I are known as the LPXTG internalins due to the presence of this sorting signal motif and are covalently anchored to the bacterial cell surface by another virulence factor known as Sortase A. Type II are the GW and WxL internalins, of which just two members (including inlB) have been classified to date. Both members of this subfamily display a C-terminal domain that directs a non-covalent association with the L. monocytogenes cell surface (51). Type III internalins lack a cell wall-anchoring domain and are secreted by the bacterium. They are thought to promote the cell-to-cell spread of L. monocytogenes by relieving the cortical tension of the host cell and enhancing the ability of the bacterium to protrude into the plasma membrane (52). A fourth type of internalin, which contains an atypical leucine-rich repeat region, has also been recently described (53) with lmo0460 as the sole representative member. The genomes of both strains in this study were examined for the presence of internalin and internalin-like genes. A similar complement of internalins was observed in each input strain (Table S6 in Supplementary Material). Strain FSL J2-064 contained 20 type I internalins, while strain DPC6895 contains 21 (TZ05_2026c is novel to this strain). In addition, both isolates contained a virtually identical set of type II and type III internalins. Five homologs of the L. monocytogenes strain EGDe type IV internalin lmo0460 were identified in strain FSL J2-064, localized within hypervariable hotspots 7 and 9. Though the precise function of these type IV internalin proteins has yet to be fully established, they are known to be present in many strains of L. monocytogenes, but absent from non-pathogenic Listeria species such as L. innocua, and thus may have a role in L. monocytogenes virulence. Interestingly, strain DPC6895 lacked any identifiable homolog to the recently described type IV internalin of L. monocytogenes, and as such, the absence of a type IV internalin in strain DPC6895 may be a contributing factor to its perceived attenuated virulence. Further research, however, is necessary to fully establish their functional role in infection. Additionally, two homologs of the internalin-like gene lmo0463 of L. monocytogenes strain EGDe (hypervariable hotspots 7 and 9, respectively) were identified in strain FSL J2-064 but were absent once again from strain DPC6895. Likewise, their precise role in L. monocytogenes virulence remains unclear.
Second, the stress survival islet (SSI-1) of L. monocytogenes was identified to be present in the genome of strain FSL J2-064 but is absent from that of strain DPC6895 (Figure 4). This islet is an 8.7 kb region of DNA located between lmo0443 and lmo0449 of strain EGDe and contains five genes that have been previously implicated to assist in the survival of the bacterium under suboptimal environmental conditions (54). Included within this cluster are the pva gene, which has a role in resistance of L. monocytogenes to acute toxicity from bile and bile salts (55), and the gadD1 and gadT1 genes, which contribute to the efficient growth of the bacterium at low pH (56). The corresponding region in strain DPC6895 was identified to contain a single gene encoding a protein that is 100% identical to LMOf2365_0481 of L. monocytogenes serotype 4b strain F2365. The presence of this 182 amino acid protein in strain DPC6895 represents a common feature of islet-negative strains of L. monocytogenes (54). Although this particular gene has been observed to be highly conserved within islet-negative strains of L. monocytogenes, the function of its hypothetical protein product has not yet been established. Prior research has shown that strains, which do not harbor SSI-1 were observed to grow less efficiently under acidic stresses (12), suggesting that the presence of this particular islet within the genome is beneficial in promoting survival in unfavorable acidic environments posed by the stomach and intestinal tract of a host organism. Additionally, the observed differences in growth efficiency between islet-positive and islet-negative strains of L. monocytogenes indicate that the presence of this islet could confer an enhanced ability to proliferate within an organism and lead to an overall higher rate of infection. Therefore, the absence of SSI-1 in DPC6895 may have been a contributing factor in the observed inability of this isolate to establish a clinical infection in its host (20). A number of the strain-specific genes in FSL J2-064 also had annotated functions associated with iron uptake that were absent in strain DPC6895, including the twin-arginine translocase system (57). Given the known association between iron uptake and L. monocytogenes virulence (58), the absence of these genes in strain DPC6895 provides another potential insight into its inability to cause a systemic infection.
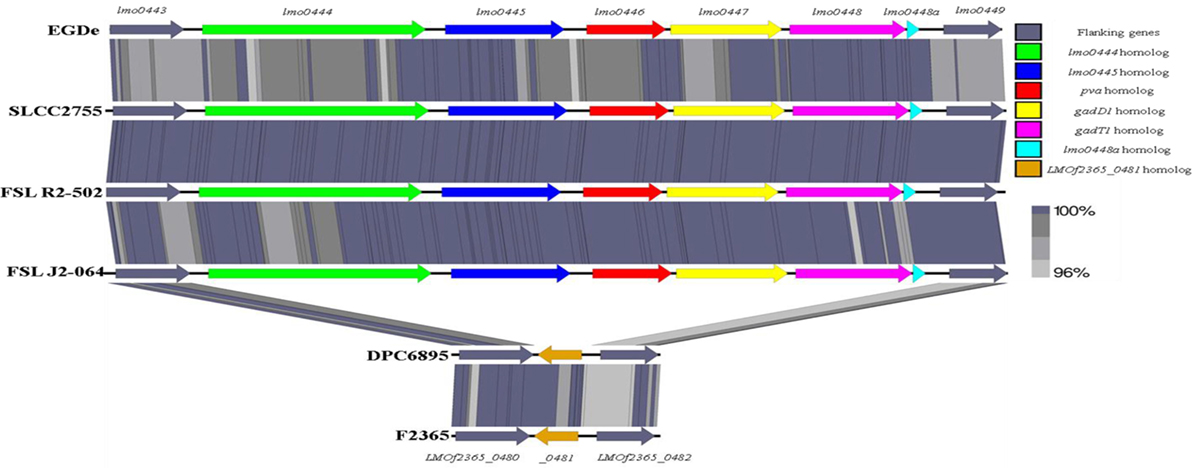
Figure 4. Linear comparison of the stress survival islet-1 in six different Listeria monocytogenes strains, with the % amino acid identities between proteins in these clusters indicated. The islet is present in strain FSL J2-064, along with the other serotype 1/2b strains FSL R2-502 and SLCC2755, but is absent from strain DPC6895. Instead, the corresponding genomic region in strain DPC6895 displays homology to gene LMOf2365_0481 of L. monocytogenes strain f2365.
Finally, the genome of strain FSL J2-064 contained an intact copy of the comK gene, while a prophage insertion (contig 7, position 257959–311605) interrupted the comK gene in strain DPC6895. The entire comK gene sequence in DPC6895 is instead represented by two separate ORFs; TZ05_2272 and TZ05_2336, which together share 100% nucleotide sequence identity with the N- and C-terminal regions of the comK gene in FSL J2-064, respectively. A prophage insertion into the comK gene of L. monocytogenes is a common observation, as this gene represents a “hotspot” for integration of the serotype 1/2-specific bacteriophage A118 and other related phages (11, 13, 59). The phage insertion into this gene may hold downstream consequences for the pathogenic potential of strain DPC6895, as the comK gene has recently been shown to have an important role in phagosomal escape of L. monocytogenes during infection (60). As such, this interruption to the comK gene may be a contributing factor to the attenuated virulence of strain DPC6895. Interestingly however, the same research demonstrated that the comK prophage in L. monocytogenes strain 10403S excises during bacterial phagocytosis resulting in a reactivation of this gene and the production of a functional ComK protein product, and such an occurrence, therefore, must also be considered a possibility in strain DPC6895. Further investigation is required in order to fully understand the consequences of this prophage insertion.
Influence of Hypervariable Hotspots on the Virulence of Strains DPC6895 and FSL J2-064
As previously mentioned, the absence of type IV internalins (all of which are located within hypervariable hotspots in the L. monocytogenes genome) may be a contributory factor to the inability of strain DPC6895 to establish a systemic infection in the host. In addition, Listeria pathogenicity island 3 (LIPI-3) is a relatively recently discovered pathogenicity island, which has been identified in a subset of atypical L. innocua isolates (61) and a number of lineage I strains of L. monocytogenes. LIPI-3 contributes to virulence and intracellular survival of the pathogen (62) and is located within hypervariable hotspot 8. The main function of this island is the production of a second L. monocytogenes hemolysin, namely listeriolysin S, which is induced under oxidative stress conditions (62). LIPI-3 consists of eight lls genes flanked on either side of the island by two related glyoxalase-encoding genes. Comparative analyses with other serotype 1/2b strains of L. monocytogenes identified that LIPI-3 was not found in either of the bovine isolates DPC6895 or FSL J2-064 (Figure 5), though homologs of the flanking glyoxalase-encoding genes were identified. The high variability generally observed within hypervariable hotspots of the L. monocytogenes genome may account for the absence of this island in these strains. While the presence of LIPI-3 does not appear to be essential in order to establish a systemic infection, the absence of this island may hinder the ability of a particular strain in the establishment of a systemic infection within the host.
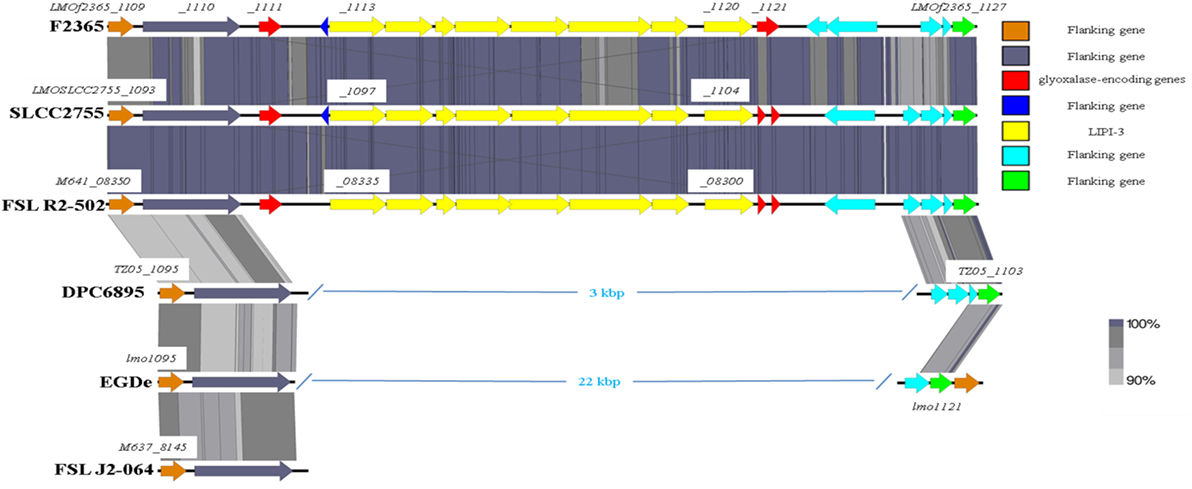
Figure 5. Comparison of the Listeria pathogenicity island 3 between the two bovine isolates and a number of other L. monocytogenes strains.
Conclusion
The results of this study demonstrate the high degree of variability that exists between the accessory genomes of closely related L. monocytogenes isolates. The hypervariable hotspots found in various areas of the genome may be crucial in defining the physiological characteristics of a particular strain, as evidenced by the presence of important gene clusters such as the type IV internalins and the LIPI-3 within these regions. L. monocytogenes strain DPC6895 was shown to be missing some of the key factors that are associated with in vivo survival and virulence, including SSI-1 and LIPI-3, providing insights into the inability of this strain to establish a systemic infection in its host. The results highlight a number of potentially crucial factors for L. monocytogenes virulence within the accessory genome and suggest that bacterial pathogenesis in L. monocytogenes relies on the cumulative effect of a number of genetic factors rather than any single attribute alone. From a regulatory perspective, differentiation of virulent from non-virulent strains is crucially important. As used in this study, whole genome sequencing can be employed as a tool to explore this differentiation.
Author Contributions
ACasey carried out the laboratory work; KJ, OM, ACoffey, ACasey, and EF were involved in obtaining funding, designing the experiments, interpreting the results, and writing the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This work was supported by The Department of Agriculture, Food and the Marine under the Food Institutional Research Measure, grant number 11/F/008.
Supplementary Material
The Supplementary Material for this article can be found online at https://www.frontiersin.org/article/10.3389/fnut.2016.00054/full#supplementary-material.
References
1. Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson M-A, Roy SL, et al. Foodborne illness acquired in the United States – major pathogens. Emerg Infect Dis (2011) 17:7–15. doi: 10.3201/eid1707.110572
2. Khen B, Lynch O, Carroll J, Mcdowell D, Duffy G. Occurrence, antibiotic resistance and molecular characterization of Listeria monocytogenes in the beef chain in the Republic of Ireland. Zoonoses Public Health (2015) 62:11–7. doi:10.1111/zph.12106
3. Cole M, Jones M, Holyoak C. The effect of pH, salt concentration and temperature on the survival and growth of Listeria monocytogenes. J Appl Bacteriol (1990) 69:63–72. doi:10.1111/j.1365-2672.1990.tb02912.x
4. Ho A, Ivanek R, Gröhn Y, Nightingale K, Wiedmann M. Listeria monocytogenes fecal shedding in dairy cattle shows high levels of day-to-day variation and includes outbreaks and sporadic cases of shedding of specific L. monocytogenes subtypes. Preventive Vet Med (2007) 80:287–305. doi:10.1016/j.prevetmed.2007.03.005
5. Herschleb J, Ananiev G, Schwartz DC. Pulsed-field gel electrophoresis. Nat Protoc (2007) 2:677–84. doi:10.1038/nprot.2007.94
6. Ragon M, Wirth T, Hollandt F, Lavenir R, Lecuit M, Le Monnier A, et al. A new perspective on Listeria monocytogenes evolution. PLoS Pathog (2008) 4:e1000146. doi:10.1371/journal.ppat.1000146
7. Buchrieser C, Rusniok C, Kunst F, Cossart P, Glaser P; Listeria Consortium. Comparison of the genome sequences of Listeria monocytogenes and Listeria innocua: clues for evolution and pathogenicity. FEMS Immunol Med Microbiol (2003) 35:207–13. doi:10.1016/S0928-8244(02)00448-0
8. Camejo A, Carvalho F, Reis O, Leitão E, Sousa S, Cabanes D. The arsenal of virulence factors deployed by Listeria monocytogenes to promote its cell infection cycle. Virulence (2011) 2:379–94. doi:10.4161/viru.2.5.17703
9. Stachowiak R, Jagielski T, Roeske K, Osińska O, Gunerka P, Wiśniewski J, et al. Lmo0171, a novel internalin-like protein, determines cell morphology of Listeria monocytogenes and its ability to invade human cell lines. Curr Microbiol (2015) 70:267–74. doi:10.1007/s00284-014-0715-4
10. Holch A, Webb K, Lukjancenko O, Ussery D, Rosenthal BM, Gram L. Genome sequencing identifies two nearly unchanged strains of persistent Listeria monocytogenes isolated at two different fish processing plants sampled 6 years apart. Appl Environ Microbiol (2013) 79:2944–51. doi:10.1128/AEM.03715-12
11. Nelson KE, Fouts DE, Mongodin EF, Ravel J, Deboy RT, Kolonay JF, et al. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res (2004) 32:2386–95. doi:10.1093/nar/gkh562
12. Rychli K, Müller A, Zaiser A, Schoder D, Allerberger F, Wagner M, et al. Genome sequencing of Listeria monocytogenes “Quargel” listeriosis outbreak strains reveals two different strains with distinct in vitro virulence potential. PLoS One (2014) 9:e89964. doi:10.1371/journal.pone.0089964
13. Gilmour MW, Graham M, Van Domselaar G, Tyler S, Kent H, Trout-Yakel KM, et al. High-throughput genome sequencing of two Listeria monocytogenes clinical isolates during a large foodborne outbreak. BMC Genomics (2010) 11:120. doi:10.1186/1471-2164-11-120
14. Den Bakker HC, Desjardins CA, Griggs AD, Peters JE, Zeng Q, Young SK, et al. Evolutionary dynamics of the accessory genome of Listeria monocytogenes. PLoS One (2013) 8:e67511. doi:10.1371/journal.pone.0067511
15. Orsi RH, Den Bakker HC, Wiedmann M. Listeria monocytogenes lineages: genomics, evolution, ecology, and phenotypic characteristics. Int J Med Microbiol (2011) 301:79–96. doi:10.1016/j.ijmm.2010.05.002
16. Wiedmann M, Bruce JL, Keating C, Johnson AE, McDonough PL, Batt CA. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect Immun (1997) 65:2707–16.
17. Ward TJ, Ducey TF, Usgaard T, Dunn KA, Bielawski JP. Multilocus genotyping assays for single nucleotide polymorphism-based subtyping of Listeria monocytogenes isolates. Appl Environ Microbiol (2008) 74:7629–42. doi:10.1128/AEM.01127-08
18. Den Bakker HC, Didelot X, Fortes ED, Nightingale KK, Wiedmann M. Lineage specific recombination rates and microevolution in Listeria monocytogenes. BMC Evol Biol (2008) 8:277. doi:10.1186/1471-2148-8-277
19. Kuenne C, Billion A, Mraheil MA, Strittmatter A, Daniel R, Goesmann A, et al. Reassessment of the Listeria monocytogenes pan-genome reveals dynamic integration hotspots and mobile genetic elements as major components of the accessory genome. BMC Genomics (2013) 14:47. doi:10.1186/1471-2164-14-47
20. Hunt K, Drummond N, Murphy M, Butler F, Buckley J, Jordan K. A case of bovine raw milk contamination with Listeria monocytogenes. Ir Vet J (2012) 65:13. doi:10.1186/2046-0481-65-13
21. Casey A, McAuliffe O, Coffey A, Hunt K, Fanning S, Fox E, et al. Complete genome sequence of Listeria monocytogenes strain DPC6895, a serotype 1/2b isolate from bovine raw milk. Genome Announc (2015) 3:e629–615. doi:10.1128/genomeA.00629-15
22. Cai S, Kabuki DY, Kuaye AY, Cargioli TG, Chung MS, Nielsen R, et al. Rational design of DNA sequence-based strategies for subtyping Listeria monocytogenes. J Clin Microbiol (2002) 40:3319–25. doi:10.1128/JCM.40.9.3319-3325.2002
23. Pohl MA, Wiedmann M, Nightingale KK. Associations among Listeria monocytogenes genotypes and distinct clinical manifestations of listeriosis in cattle. Am J Vet Res (2006) 67:616–26. doi:10.2460/ajvr.67.4.616
24. Gray MJ, Zadoks RN, Fortes ED, Dogan B, Cai S, Chen Y, et al. Listeria monocytogenes isolates from foods and humans form distinct but overlapping populations. Appl Environ Microbiol (2004) 70:5833–41. doi:10.1128/AEM.70.10.5833-5841.2004
25. Alikhan N-F, Petty NK, Zakour NLB, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics (2011) 12:402. doi:10.1186/1471-2164-12-402
26. Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res (2004) 14:1394–403. doi:10.1101/gr.2289704
27. Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res (1997) 25:3389–402. doi:10.1093/nar/25.17.3389
28. Aziz RK, Bartels D, Best AA, Dejongh M, Disz T, Edwards RA, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics (2008) 9:75. doi:10.1186/1471-2164-9-75
29. Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res (2007) 35:W52–7. doi:10.1093/nar/gkm360
30. Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. PHAST: a fast phage search tool. Nucleic Acids Res (2011) 39(Web Server issue):W347–52. doi:10.1093/nar/gkr485
31. Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream M-A, et al. Artemis: sequence visualization and annotation. Bioinformatics (2000) 16:944–5. doi:10.1093/bioinformatics/16.10.944
32. Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics (2011) 27:1009–10. doi:10.1093/bioinformatics/btr039
33. Koonin EV, Makarova KS. CRISPR-Cas: an adaptive immunity system in prokaryotes. F1000 Biol Rep (2009) 1:95. doi:10.3410/B1-95
34. Di H, Ye L, Yan H, Meng H, Yamasak S, Shi L. Comparative analysis of CRISPR loci in different Listeria monocytogenes lineages. Biochem Biophys Res Commun (2014) 454:399–403. doi:10.1016/j.bbrc.2014.10.018
35. Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, et al. Evolution and classification of the CRISPR – Cas systems. Nat Rev Microbiol (2011) 9:467–77. doi:10.1038/nrmicro2577
36. Murray NE. Type I restriction systems: sophisticated molecular machines (a legacy of Bertani and Weigle). Microbiol Mol Biol Rev (2000) 64:412–34. doi:10.1128/MMBR.64.2.412-434.2000
37. Den Bakker HC, Cummings CA, Ferreira V, Vatta P, Orsi RH, Degoricija L, et al. Comparative genomics of the bacterial genus Listeria: genome evolution is characterized by limited gene acquisition and limited gene loss. BMC Genomics (2010) 11:688. doi:10.1186/1471-2164-11-688
38. Kim J-W, Dutta V, Elhanafi D, Lee S, Osborne JA, Kathariou S. A novel restriction-modification system is responsible for temperature-dependent phage resistance in Listeria monocytogenes ECII. Appl Environ Microbiol (2012) 78:1995–2004. doi:10.1128/AEM.07086-11
39. Lee S, Ward T, Siletzky R, Kathariou S. Two novel type II restriction-modification systems occupying genomically equivalent locations on the chromosomes of Listeria monocytogenes strains. Appl Environ Microbiol (2012) 78:2623–30. doi:10.1128/AEM.07203-11
40. Lungu B, O’Bryan CA, Muthaiyan A, Milillo SR, Johnson MG, Crandall PG, et al. Listeria monocytogenes: antibiotic resistance in food production. Foodborne Pathog Dis (2011) 8:569–78. doi:10.1089/fpd.2010.0718
41. Xu D, Li Y, Zahid MSH, Yamasaki S, Shi L, Li J-R, et al. Benzalkonium chloride and heavy-metal tolerance in Listeria monocytogenes from retail foods. Int J Food Microbiol (2014) 190:24–30. doi:10.1016/j.ijfoodmicro.2014.08.017
42. Romanova N, Wolffs P, Brovko L, Griffiths M. Role of efflux pumps in adaptation and resistance of Listeria monocytogenes to benzalkonium chloride. Appl Environ Microbiol (2006) 72:3498–503. doi:10.1128/AEM.72.5.3498-3503.2006
43. Godreuil S, Galimand M, Gerbaud G, Jacquet C, Courvalin P. Efflux pump Lde is associated with fluoroquinolone resistance in Listeria monocytogenes. Antimicrob Agents Chemother (2003) 47:704–8. doi:10.1128/AAC.47.2.704-708.2003
44. Otto M, Peschel A, Götz F. Producer self-protection against the lantibiotic epidermin by the ABC transporter EpiFEG of Staphylococcus epidermidis Tü3298. FEMS Microbiol Lett (1998) 166:203–11. doi:10.1111/j.1574-6968.1998.tb13891.x
45. Mata M, Baquero F, Perez-Diaz J. A multidrug efflux transporter in Listeria monocytogenes. FEMS Microbiol Lett (2000) 187:185–8. doi:10.1111/j.1574-6968.2000.tb09158.x
46. Burall LS, Grim C, Gopinath G, Laksanalamai P, Datta AR. Whole-genome sequencing identifies an atypical Listeria monocytogenes strain isolated from pet foods. Genome Announc (2014) 2:e1243–1214. doi:10.1128/genomeA.01243-14
47. Rowland SJ, Dyke K. Tn552, a novel transposable element from Staphylococcus aureus. Mol Microbiol (1990) 4:961–75. doi:10.1111/j.1365-2958.1990.tb00669.x
48. Borezee E, Pellegrini E, Berche P. OppA of Listeria monocytogenes, an oligopeptide-binding protein required for bacterial growth at low temperature and involved in intracellular survival. Infect Immun (2000) 68:7069–77. doi:10.1128/IAI.68.12.7069-7077.2000
49. Verheul A, Hagting A, Amezaga M-R, Booth IR, Rombouts FM, Abee T. A di- and tripeptide transport system can supply Listeria monocytogenes Scott A with amino acids essential for growth. Appl Environ Microbiol (1995) 61:226–33.
50. Hamon M, Bierne H, Cossart P. Listeria monocytogenes: a multifaceted model. Nat Rev Microbiol (2006) 4:423–34. doi:10.1038/nrmicro1413
51. Bierne H, Cossart P. Listeria monocytogenes surface proteins: from genome predictions to function. Microbiol Mol Biol Rev (2007) 71:377–97. doi:10.1128/MMBR.00039-06
52. Rajabian T, Gavicherla B, Heisig M, Müller-Altrock S, Goebel W, Gray-Owen SD, et al. The bacterial virulence factor InlC perturbs apical cell junctions and promotes cell-to-cell spread of Listeria. Nat Cell Biol (2009) 11:1212–8. doi:10.1038/ncb1964
53. Bécavin C, Bouchier C, Lechat P, Archambaud C, Creno S, Gouin E, et al. Comparison of widely used Listeria monocytogenes strains EGD, 10403S, and EGD-e highlights genomic differences underlying variations in pathogenicity. MBio (2014) 5:e969–914. doi:10.1128/mBio.00969-14
54. Ryan S, Begley M, Hill C, Gahan C. A five-gene stress survival islet (SSI-1) that contributes to the growth of Listeria monocytogenes in suboptimal conditions. J Appl Microbiol (2010) 109:984–95. doi:10.1111/j.1365-2672.2010.04726.x
55. Begley M, Sleator RD, Gahan CG, Hill C. Contribution of three bile-associated loci, bsh, pva, and btlB, to gastrointestinal persistence and bile tolerance of Listeria monocytogenes. Infect Immun (2005) 73:894–904. doi:10.1128/IAI.73.2.894-904.2005
56. Cotter PD, Ryan S, Gahan CG, Hill C. Presence of GadD1 glutamate decarboxylase in selected Listeria monocytogenes strains is associated with an ability to grow at low pH. Appl Environ Microbiol (2005) 71:2832–9. doi:10.1128/AEM.71.6.2832-2839.2005
57. Tiwari KB, Birlingmair J, Wilkinson BJ, Jayaswal RK. The role of the twin-arginine translocase (tat) system in iron uptake in Listeria monocytogenes. Microbiology (2014) 161(Pt 2):264–71. doi:10.1099/mic.0.083642-0
58. Lechowicz J, Krawczyk-Balska A. An update on the transport and metabolism of iron in Listeria monocytogenes: the role of proteins involved in pathogenicity. Biometals (2015) 28(4):587–603. doi:10.1007/s10534-015-9849-5
59. Loessner MJ, Inman RB, Lauer P, Calendar R. Complete nucleotide sequence, molecular analysis and genome structure of bacteriophage A118 of Listeria monocytogenes: implications for phage evolution. Mol Microbiol (2000) 35:324–40. doi:10.1046/j.1365-2958.2000.01720.x
60. Rabinovich L, Sigal N, Borovok I, Nir-Paz R, Herskovits AA. Prophage excision activates Listeria competence genes that promote phagosomal escape and virulence. Cell (2012) 150:792–802. doi:10.1016/j.cell.2012.06.036
61. Clayton EM, Daly KM, Guinane CM, Hill C, Cotter PD, Ross PR. Atypical Listeria innocua strains possess an intact LIPI-3. BMC Microbiol (2014) 14:58. doi:10.1186/1471-2180-14-58
Keywords: comparative genomic analysis, Listeria monocytogenes, pathogenesis, hypervariable hotspots, attenuated virulence, stress survival islet, LIPI-3, DPC6895, serotype 1/2b
Citation: Casey A, Jordan K, Coffey A, Fox EM and McAuliffe O (2016) Comparative Genomic Analysis of Two Serotype 1/2b Listeria monocytogenes Isolates from Analogous Environmental Niches Demonstrates the Influence of Hypervariable Hotspots in Defining Pathogenesis. Front. Nutr. 3:54. doi: 10.3389/fnut.2016.00054
Received: 05 July 2016; Accepted: 07 December 2016;
Published: 21 December 2016
Edited by:
Andrea Gomez-Zavaglia, Center for Research and Development in Food Cryotechnology (CIDCA, CONICET), ArgentinaReviewed by:
David Rodriguez-Lazaro, University of Burgos, SpainLearn-Han Lee, Monash University Malaysia Campus, Malaysia
Gonçalo Nieto Almeida, Catholic University of Portugal, Portugal
Pio Maria Furneri, University of Catania, Italy
Copyright: © 2016 Casey, Jordan, Coffey, Fox and McAuliffe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Olivia McAuliffe, olivia.mcauliffe@teagasc.ie
 Aidan Casey
Aidan Casey Kieran Jordan
Kieran Jordan Aidan Coffey
Aidan Coffey Edward M. Fox
Edward M. Fox Olivia McAuliffe
Olivia McAuliffe