Germline recombination by conditional gene targeting with Parvalbumin-Cre lines
- 1Department of Molecular and Cellular Biology, Center for Brain Science, Harvard University, Cambridge, MA, USA
- 2Department of Neurology, F. M. Kirby Neurobiology Center, Harvard Medical School, Boston Children's Hospital, MA, USA
Conditional gene targeting allows us to study gene function in specific tissues or cell types. This is commonly achieved by Cre DNA recombinase and its 34–base pair target sequences called loxP sites. Through the efforts of individual labs and large-scale projects, a sizable collection of Cre mouse lines has been generated to express or delete specific genes in a wide range of cell types throughout the nervous system (Madisen et al., 2010; Taniguchi et al., 2011). Typically, the specificity of Cre transgene expression is controlled by tissue or cell-type promoters. However, increasing evidence has revealed that the desired Cre expression pattern is not always guaranteed (Schmidt-Supprian and Rajewsky, 2007).
Here, we show an example of the undesired recombination in a Cre line whose expression is controlled by the parvalbumin (Pvalb) promoter. Currently, two Pvalb-Cre lines are available: the Pvalb-IRES-Cre (Hippenmeyer et al., 2005) and Pvalb-2A-Cre (Madisen et al., 2010) in which either IRES-Cre or 2A-Cre sequence is inserted by knock-in into the Pvalb 3′ untraslated region. Both Cre lines have been widely used to induce or delete genes of interest in parvalbumin (PV)-positive GABAergic neurons in the brain. However, PV is expressed not only in brain but also in other tissues, including testis in particular (Kagi et al., 1987), suggesting that undesired germline recombination could occur in the Pvalb-Cre testis.
To examine recombination, we crossed Ai32 (ChR2 (H134R)-EYFP) line whose EYFP expression can be detected after recombination (Madisen et al., 2012) with either the Pvalb-IRES-Cre or Pvalb-2A-Cre line. In the Pvalb-2A-Cre+/− background, strong EYFP expression was observed in Leydig cells (Figures 1A,C, arrows) and spermatids in the seminiferous tubules (Figures 1A,C, asterisks) in which PV was expressed (Figure 1B). In contrast, in the Pvalb-IRES-Cre+/− background, although EYFP was slightly expressed in Leydig cells, EYFP was absent from spermatids (Figures 1D,E). This probably reflects weaker expression of Cre under control of the IRES as compared to the 2A sequence (Madisen et al., 2010).
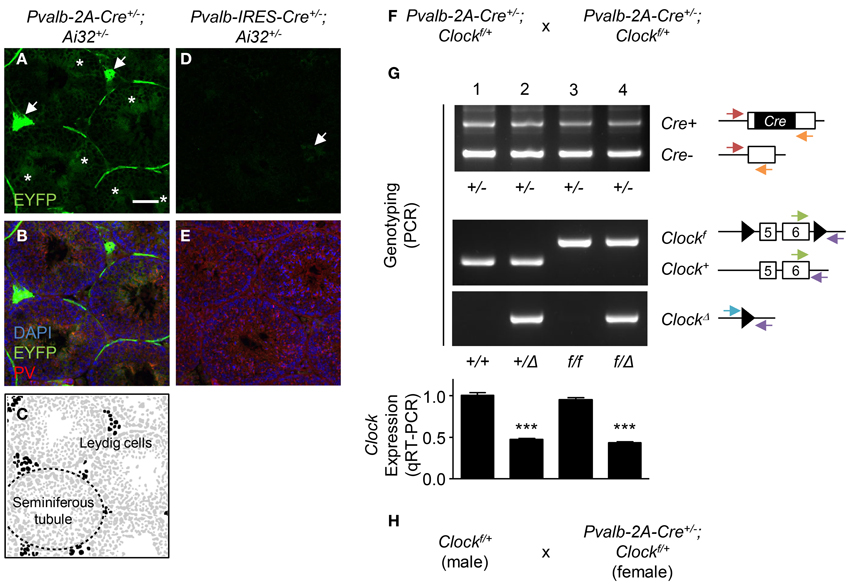
Figure 1. Germline recombination in Pvalb-2A-Cre testis. (A–E) Expression of EYFP in Pvalb-2A-Cre+/−; Ai32+/− (A–C) or Pvalb-IRES-Cre+/−; Ai32+/− testes (D,E). Testes were co-stained with DAPI and anti-parvalbumin (PV) antibody (B,E). One of the seminiferous tubules (dashed line) and Leydig cells (black) are depicted (C). Arrows, EYFP-expressing Leydig cells. Asterisks, EYFP-expressing spermatids. Scale bar, 100 μm. (F) Breeding strategy to generate PV-cell specific Clock conditional knock-out mice using Pvalb-2A-Cre line. (G) Genotyping and qRT-PCR analysis of the progeny obtained by (F). Genotyping was performed to detect Cre positive (Cre+) or negative (Cre−), floxed Clock (Clockf) or wild-type (Clock+), or deletion of Clock (ClockΔ). Arrows indicate location and direction of primers for PCR genotyping. Expression of Clock in neocortex was analyzed by qRT-PCR (graph, 8–12 mice each). ***P < 0.001 (One-Way ANOVA, Dunnett's post-hoc analysis). Values are mean ± s.e.m. (H) Modified breeding strategy to avoid germline recombination when generating PV-cell specific Clock conditional knock-out mice.
In the conditional knock-out strategy, such recombination in testis should be avoided because it could induce a “total” knock-out of the gene. One example of this undesired recombination by the Pvalb-2A-Cre line is shown in Figures 1F,G. To knock-out the Clock gene, Pvalb-2A-Cre+/−; Clockf/+ mice were crossed with each other (Figure 1F), and genotypes of its progeny were determined for the Cre and Clock alleles (Figure 1G). In some offspring, undesired deletion of the Clock gene (ClockΔ) was detected (Figure 1G, lane 2 and 4).
In our experience, such undesired recombination happened in 50% of the progeny using this strategy (24 out of 48 mice). Quantitative RT-PCR (qRT-PCR) analysis of brain samples further confirmed deletion of the Clock gene with expression decreased by half in the recombined offspring (Figure 1G, graph, lane 2 and 4), while only ~5% decrease of Clock expression was seen in the expected conditional progeny (Figure 1G, graph, lane 3). To minimize this undesired recombination, breeding Cre-negative males (Clockf/+) with Cre-positive females (Pvalb-2A-Cre+/−; Clockf/+) was performed (Figure 1H). In this strategy, the undesired recombination frequency became less than 5%, although it was not completely abolished for unknown reasons.
Such germline recombination is difficult to track in the literature, but it has been noted across several Cre lines widely used in the field of neuroscience, including Camk2a-Cre (Friedel et al., 2011), Nestin-Cre (Friedel et al., 2011; Harno et al., 2013), Emx1-Cre (Zeller et al., 2008), and Pcp2-Cre (Tsai et al., 2012). Given this situation, these could simply be the tip of an iceberg. In order to avoid undesired outcomes, here we propose several precautionary measures:
First, expression of Cre outside the region of interest, especially in the germline, should be investigated. For several Cre lines, expression patterns are available in common databases such as The Jackson Laboratory Cre Repository (http://cre.jax.org/data.html) or Cre Portal (http://www.creportal.org/). It should be noted that the recombination pattern has to be evaluated for each floxed line, because recombination efficiency depends upon the region on the chromosome. Second, a breeding strategy should be set up in a way that minimizes the occurrence of germline recombination. Third, deletion of the floxed genes must be analyzed by genotyping as shown in Figure 1G, which is often omitted in the standard conditional knock-out strategy. This is particularly important to avoid misinterpretation of phenotypes due to unexpected recombination in other cells than the desired target.
Materials and Methods
Mice
C57BL/6J (JAX no. 000664), Pvalb-2A-Cre (JAX no. 012358) (Madisen et al., 2010), Pvalb-IRES-Cre (JAX no. 008069) (Hippenmeyer et al., 2005), Ai32 (JAX no. 012569) (Madisen et al., 2012), and Clockflox (JAX no. 010490) (Debruyne et al., 2006) mice were purchased from Jackson Laboratory. All mice were on a C57BL/6J background. For examining recombination in testis, Pvalb-2A-Cre+/+ or Pvalb-IRES-Cre+/+ mice were crossed with Ai32+/− mice to generate Pvalb-2A-Cre+/−; Ai32+/− or Pvalb-IRES-Cre+/−; Ai32+/− mice. For examining deletion and expression level of Clock gene, Pvalb-2A-Cre+/+ mice were crossed with Clockf/f mice to generate Pvalb-2A-Cre+/−; Clockf/+ mice, and these progeny were then crossed with each other. Animal housing and experimental procedures were approved (AEP28-19) and followed guidelines of the Harvard University Institutional Animal Care and Use Committee.
Immunohistochemistry
Testes were decapsulated and embedded in OCT compound (Sakura Finetek USA Inc), quickly frozen and sectioned on a Cryostat (Leica). Cryo- sections (7 μm) were fixed in 4% (wt/vol) paraformaldehyde in 0.1 M phosfate buffer for 15 min at room temperature, washed with PBS thrice, and blocked with buffer (10% normal goat serum and 0.1% Triton X-100 (vol/vol) in PBS) for 1 h at room temperature. Incubation with rabbit anti-parvalbumin (Swant, 1:1000) was performed in blocking buffer overnight at 4°C, followed by incubation with Alexa Fluor 488 Goat Anti-Rabbit IgG (Molecular Probes, 1:400) in blocking buffer overnight at 4°C. Stained sections were mounted with 4′,6-diamidino-2-phenylindole (DAPI) and visualized by confocal microscopy (FV1000, Olympus).
PCR Genotyping
Genomic DNA was extracted from ear punches and PCR genotyping performed using REDExtract-N-Amp Tissue PCR Kit (Sigma). Primers used in this study were described elsewhere (Debruyne et al., 2006).
Real-Time Quantitative RT-PCR
Total RNA was isolated from the neocortex with mirVana miRNA Isolation Kit (Ambion), and any contaminating DNA removed by TURBO DNA-free Kit (Invitrogen) according to manufacturer's instructions. First-strand cDNA was synthesized from total RNA using High Capacity RNA-to-cDNA Kit (Invitrogen) according to manufacturer's instructions. Real-time quantitative PCR was performed using TaqMan Gene Expression Assay (Applied Biosystems) on a StepOnePlus Real-Time PCR System (Applied Biosystems). TaqMan probes used in this study were for Clock exon 5–6 (Mm00455940_g1) and Gapdh (4352932E). Relative expression of target genes was determined by the 2−ΔΔCt method.
Acknowledgments
We thank M. Nakamura for animal maintenance and the National Institute of Mental Health Grant 1P50MH094271 (to Takao K. Hensch) for support.
References
Debruyne, J. P., Noton, E., Lambert, C. M., Maywood, E. S., Weaver, D. R., and Reppert, S. M. (2006). A clock shock: mouse CLOCK is not required for circadian oscillator function. Neuron 50, 465–477. doi: 10.1016/j.neuron.2006.03.041
Friedel, R. H., Wurst, W., Wefers, B., and Kuhn, R. (2011). Generating conditional knockout mice. Methods Mol. Biol. 693, 205–231. doi: 10.1007/978-1-60761-974-1_12
Harno, E., Cottrell, E. C., and White, A. (2013). Metabolic pitfalls of CNS cre-based technology. Cell Metab. 18, 21–28. doi: 10.1016/j.cmet.2013.05.019
Hippenmeyer, S., Vrieseling, E., Sigrist, M., Portmann, T., Laengle, C., Ladle, D. R., et al. (2005). A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol. 3:e159. doi: 10.1371/journal.pbio.0030159
Kagi, U., Berchtold, M. W., and Heizmann, C. W. (1987). Ca2+-binding parvalbumin in rat testis. Characterization, localization, and expression during development. J. Biol. Chem. 262, 7314–7320.
Madisen, L., Mao, T., Koch, H., Zhuo, J. M., Berenyi, A., Fujisawa, S., et al. (2012). A toolbox of Cre-dependent optogenetic transgenic mice for light-induced activation and silencing. Nat. Neurosci. 15, 793–802. doi: 10.1038/nn.3078
Madisen, L., Zwingman, T. A., Sunkin, S. M., Oh, S. W., Zariwala, H. A., Gu, H., et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140. doi: 10.1038/nn.2467
Schmidt-Supprian, M., and Rajewsky, K. (2007). Vagaries of conditional gene targeting. Nat. Immunol. 8, 665–668. doi: 10.1038/ni0707-665
Taniguchi, H., He, M., Wu, P., Kim, S., Paik, R., Sugino, K., et al. (2011). A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron 71, 995–1013. doi: 10.1016/j.neuron.2011.07.026
Tsai, P. T., Hull, C., Chu, Y., Greene-Colozzi, E., Sadowski, A. R., Leech, J. M., et al. (2012). Autistic-like behaviour and cerebellar dysfunction in Purkinje cell Tsc1 mutant mice. Nature 488, 647–651. doi: 10.1038/nature11310
Keywords: Cre, conditional targeting, knock-out mouse, germline, testis, parvalbumin
Citation: Kobayashi Y and Hensch TK (2013) Germline recombination by conditional gene targeting with Parvalbumin-Cre lines. Front. Neural Circuits 7:168. doi: 10.3389/fncir.2013.00168
Received: 16 September 2013; Accepted: 26 September 2013;
Published online: 16 October 2013.
Edited by:
Yoshiyuki Kubota, National Institute for Physiological Sciences, JapanReviewed by:
Uwe Rudolph, McLean Hospital and Harvard Medical School, USACopyright © 2013 Kobayashi and Hensch. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: hensch@mcb.harvard.edu
 Yohei Kobayashi
Yohei Kobayashi Takao K. Hensch
Takao K. Hensch