Legume Plant Peptides as Sources of Novel Antimicrobial Molecules Against Human Pathogens
- 1Institute of Plant Biology, Biological Research Centre, ELKH, Szeged, Hungary
- 2Department of Medical Chemistry, Albert Szent-Györgyi Medical School, University of Szeged, Szeged, Hungary
Antimicrobial peptides are prominent components of the plant immune system acting against a wide variety of pathogens. Legume plants from the inverted repeat lacking clade (IRLC) have evolved a unique gene family encoding nodule-specific cysteine-rich NCR peptides acting in the symbiotic cells of root nodules, where they convert their bacterial endosymbionts into non-cultivable, polyploid nitrogen-fixing cells. NCRs are usually 30–50 amino acids long peptides having a characteristic pattern of 4 or 6 cysteines and highly divergent amino acid composition. While the function of NCRs is largely unknown, antimicrobial activity has been demonstrated for a few cationic Medicago truncatula NCR peptides against bacterial and fungal pathogens. The advantages of these plant peptides are their broad antimicrobial spectrum, fast killing modes of actions, multiple bacterial targets, and low propensity to develop resistance to them and no or low cytotoxicity to human cells. In the IRLC legumes, the number of NCR genes varies from a few to several hundred and it is possible that altogether hundreds of thousands of different NCR peptides exist. Due to the need for new antimicrobial agents, we investigated the antimicrobial potential of 104 synthetic NCR peptides from M. truncatula, M. sativa, Pisum sativum, Galega orientalis and Cicer arietinum against eight human pathogens, including ESKAPE bacteria. 50 NCRs showed antimicrobial activity with differences in the antimicrobial spectrum and effectivity. The most active peptides eliminated bacteria at concentrations from 0.8 to 3.1 μM. High isoelectric point and positive net charge were important but not the only determinants of their antimicrobial activity. Testing the activity of shorter peptide derivatives against Acinetobacter baumannii and Candida albicans led to identification of regions responsible for the antimicrobial activity and provided insight into their potential modes of action. This work provides highly potent lead molecules without hemolytic activity on human blood cells for novel antimicrobial drugs to fight against pathogens.
Introduction
The WHO priority pathogens, known also as ESKAPE bacteria, are A. baumannii, Pseudomonas aeruginosa, Klebsiella pneumoniae, Escherichia coli and Enterobacter spp. at the highest priority, followed by Enterococcus faecium, Staphylococcus aureus, Helicobacter pylori, Salmonella spp., Campylobacter and Neisseria gonorrhoeae, which are prone to escape from conventional antibiotics and treatment (Bhatia et al., 2021). A. baumannii infections are rising at a faster rate than other ESKAPE bacteria, making carbapenem-resistant A. baumannii the number one target for antibiotic development (Harding et al., 2018; Kyriakidis et al., 2021). The WHO also lists various Candida species as prominent fungal pathogens (Oliveira et al., 2021), since invasive Candida infections cause hundreds of thousands of deaths each year. Multidrug-resistant strains are rapidly appearing and spreading around the world, threatening to render our antifungal arsenal unusable. Hospital-acquired infections have made everyday surgeries such as cesarean sections, hip replacements, cancer treatments and organ transplants more dangerous (Fernando et al., 2017). In the current COVID-19 pandemic, hospitalized patients are at increased risk of respiratory bacterial coinfection caused by Acinetobacter (Rangel et al., 2021) and of severe Candida infections because of their impaired immune systems (Moser et al., 2021).
To be effective against these drug-resistant bacteria and fungi, new antimicrobials are required with novel modes of action that have little or no toxicity for human cells. Antimicrobial peptides (AMP), which are host defense agents (Zasloff, 2002; Egorov et al., 2005; Campos et al., 2018; Li et al., 2021), are promising candidates for novel types of effective antimicrobials based on their broad antimicrobial spectrum, different mechanisms of action, and low tendency for resistance development (Magana et al., 2020). The weaknesses of certain natural AMPs can be their lower stability and incidental toxicity, which can be overcome with designed modifications of synthetic peptides (Büyükkiraz and Kesmen, 2021).
AMPs have a wide variety of molecular configurations, but the majority are linear peptides found in plants, insects and mammals. The amphiphilic nature of peptides and the presence of positively charged residues enable their attachment and insertion into the bacterial membrane, resulting in membrane permeabilization and their antibacterial action (Hancock, 1997; Yamaguchi and Huffaker, 2011). The antifungal effect of AMPs is mostly attributed to permeabilization and/or the formation of holes in the fungal membranes (Cesare et al., 2020).
Most known AMPs are of animal origin (Wang et al., 2016), however, plants have evolved an extremely rich source of AMPs that act as the most prominent defense barriers. The largest group of plant AMPs are cysteine-rich defensins characterized by four conserved disulfide bridges, positive net charge and amphipathic character and capability to interact with microbial membranes, and by entering the cells to inhibit translation and enzymatic activity (Maróti et al., 2015; Tam et al., 2015).
In the Inverted Repeat Lacking Clade (IRLC) of legumes, in addition to defensins, a related family of peptides has evolved that encodes nodule specific, cysteine-rich NCR peptides that play essential roles in symbiosis (Mergaert et al., 2003; Nallu et al., 2013; Montiel et al., 2017). Legume plants in symbiosis with their rhizobium bacterium partner form nitrogen-fixing root nodules. The primary role of the NCR peptides is to convert bacteria into large polyploid, non-cultivable nitrogen-fixing bacteroids in the nodule cells (Van de Velde et al., 2010). This is a terminal differentiation process as bacteroids permanently lose their cell division potential, therefore some of the peptides must be involved in inhibiting the growth of bacteria and may exert antibacterial activity in vitro. In M. truncatula ∼700 genes code for secreted NCR peptides that are targeted to the bacteroids via the secretory pathway. Mature MtNCR peptides range in size from 24 to 65 amino acids, but most of them composed of 30–50 amino acids and contain 4 or 6 cysteine residues at conserved positions in a highly variable amino acid sequence (Mergaert et al., 2003; Mergaert, 2018; Lima et al., 2020; Roy et al., 2020). Until now, only a few NCRs have been tested for antimicrobial activity against human, animal and plant pathogenic bacterial and fungal strains (Tiricz et al., 2013; Balogh et al., 2014; Ördögh et al., 2014; Mikuláss et al., 2016; Farkas et al., 2017; Montiel et al., 2017; Velivelli et al., 2020; Isozumi et al., 2021; Szerencsés et al., 2021). In this respect, the most studied NCR is the highly cationic NCR247 (pI: 10.15) and its derivatives, which are highly effective against ESKAPE pathogens, in most cases outperforming levofloxacin, a third-generation fluoroquinolone antibiotic under laboratory conditions (Jenei et al., 2020). The regions responsible for the antimicrobial activity of NCRs were delineated by synthesizing their shorter and substituted derivatives, which in some cases even had increased activity (Jenei et al., 2020; Isozumi et al., 2021; Szerencsés et al., 2021). The reduced forms of NCR247 and NCR335 were found to be more effective in vitro against the natural symbiont Sinorhizobium meliloti Rm2011 than the oxidized variants forming the disulfide bridges (Ribeiro et al., 2017). The reduced NCR247 was also more effective against E. coli than any of the three disulfide regioisomers and activity was retained even when all four cysteines in NCR247 were replaced with serine (Shabab et al., 2016).
IRLC includes 56 genera and more than 4,000 species (Duan et al., 2021), and presumably all species have NCRs as it was confirmed in a few selected species. The genus Astragalus, with 2,300–2,500 members, has the highest number of species among angiosperms (Duan et al., 2021). In A. canadensis 108 NCRs have been identified (Montiel et al., 2017), while in the members of the Vicioid subclade 200–700 NCRs (Ištvánek et al., 2014; Montiel et al., 2017), thus, it is reasonable to assume that IRLC species in total express hundreds of thousand different NCRs, providing almost unlimited amount of bioactive peptides and likely many AMPs. However, not all NCRs have antimicrobial activity as it would not be compatible with the symbiotic functions and maintaining the endosymbionts alive. Antimicrobial activity of MtNCRs was reported for cationic peptides with positive net charge (NC) except for the anionic NCR211 with −2.1 net charge (Kim et al., 2015).
In this work we tested the antimicrobial activity of 104 chemically synthetized NCR peptides; 78 NCRs from M. truncatula and 26 peptides from other IRLC legumes; from P. sativum, G. orientalis, C. arietinum and M. sativa against ESKAPE pathogens. From the latter group, shorter peptide derivatives were also synthesized and tested against A. baumannii and C. albicans.
Materials and Methods
Chemical Synthesis of Peptides
The NCR peptides and their derivatives (Table 1 and Supplementary Table S1) were chemically synthesized with–COOH or–CONH2 C-terminus. Peptides with–COOH -terminus were purchased from ProteoGenix (France), GenicBio Limited (People’s Republic of China), BBI Life Sciences Corporation (People’s Republic of China). Peptides with C-terminal–CONH2 group were synthesized using an automated peptide synthesizer (CEM Liberty Blue) and TentaGel S RAM resin (loading of amino groups 0.23 mmol/g) according to the standard protocol of solid-phase peptide synthesis (SPPS) and as described before (Jenei et al., 2020). The isoelectric point (pI) and net charge (NC) of the peptides were calculated with https://pepcalc.com/. The calculation of other physicochemical properties were done with DBAASP v3 (Pirtskhalava et al., 2021) and AMP predictions were carried out with CS-AMPPred (Porto et al., 2012), CAMPR3 (Waghu et al., 2016; Waghu and Idicula-Thomas, 2020), Antimicrobial Peptide Scanner vr.2 (Veltri et al., 2018), DBAASP v3 (Pirtskhalava et al., 2021) and Sense the moment (Porto et al., 2022).
Antimicrobial Activity Assay
The following pathogenic bacterial strains were used: The Gram-positive Enterococcus faecalis (ATCC 29212), Staphylococcus aureus (HNCMO112011), Listeria monocytogenes (ATCC 19111) and the Gram-negative Pseudomonas aeruginosa (ATCC 27853), Escherichia coli (ATCC 8739), Salmonella enterica (ATCC 13076), Klebsiella pneumoniae (NCTC 13440), Acinetobacter baumannii (ATCC 17978) obtained from the ATCC (United States) and NCTC (National Collection of Type Cultures–England). For the antifungal experiment the Candida albicans W01 strain (Ördögh et al., 2014) was used. Antimicrobial activities of peptides against ∼107 log phase bacteria were tested in Potassium-Phosphate Buffer (PPB, pH 7.4) as described by Jenei et al. (2020) while anti-Candida assays were done according to Szerencsés et al. (2021). The two-fold dilution series of peptides ranged from 25 μM to 0.125 μM, while that of the antibiotics ampicillin (Merck) and miconazole (Duchefa Biochemie) from 10.24 mM to 0.1 μM. The lowest concentration of the antimicrobial agents, which completely eliminated viable bacteria or C. albicans were considered as the minimal bactericidal concentration (MBC) and minimal fungicidal concentration (MFC).
Biofilm Degradation Assay
To test the possible effect of NCRs on the already formed biofilms, A. baumannii and C. albicans (at starting OD600 = 0.1) were first incubated in 96 well plates containing 200 μl LB broth or YPD broth, respectively at 37°C for 48 h to allow biofilm formation (Kanugala et al., 2019). After formation of the biofilm 160 µl were gently discarded from the wells and then NCR solutions were added at 25, 50 and 100 µM final concentrations in the volume of 200 µl for overnight treatment (Montiel et al., 2017). Then, the unattached suspending cells were discarded and the remaining biofilms were washed twice with PBS. The bound biofilms were stained with 35 μl of 0.1% methanolic crystal violet for an hour at room temperature. Excess crystal violet was removed by washing the wells twice with distilled water. The stained biofilms were dried overnight at room temperature and then solubilized with 200 µl of 95% ethanol and mixing. The quantity of biofilms was measured by the absorbance of the samples at 570 nm. Untreated biofilms of A. baumannii and C. albicans were taken as controls corresponding to 100% of biofilms. All experiments were performed in duplicate and mean values and standard deviations (SD) were calculated.
Membrane Permeability Assay
Microbial cell membrane damage provoked by the peptides was assayed with Live/Dead staining. A. baumannii and C. albicans were grown as for the antimicrobial activity assay and washed gently with PPB. Cells were resuspended in PPB, and diluted to OD600 = 0.1 and treated with peptides at various concentrations (as indicated in the Figures 3, 4 legends) at room temperature for 30 min. Untreated cells served as negative control. Cells were subsequently stained with 5 μM SYTO and 5 μM propidium iodide (PI). After 10 min incubation in the dark, 5 μl of the cell suspensions were spotted on microscope slide and covered with 2% (w/v) thin agar slices, and then observed with Olympus Fluoview FV 1000 confocal laser microscope at ×60 magnification using for SYTO9 488 nm laser for excitation, and 500–530 nm for emission. Excitation and emission wavelengths were 543 and 555–655 nm for PI, respectively. Sequential scanning was used to avoid crosstalk of the fluorescent dyes.
Scanning Electron Microscopy
A. baumannii and C. albicans were grown and treated the same way as for the membrane permeability assay. Cells were then fixed with 2.5% (v/v) glutaraldehyde in phosphate buffered saline (PBS, pH 7.4). 5 μl of the above bacterial and yeast suspensions were spotted on a silicon disk coated with 0.01% Poly-L-Lysine. The filters were washed twice with PBS and gradually dehydrated with ethanol series (30, 50, 70, 80%, and three times 100% ethanol, each for minimum 30 min). The samples were dried with a critical point dryer (K850: Quorum Technologies Ltd.), followed by 12 nm gold coating and observed under a JEOL JSM-7100F/LV scanning electron microscope.
In Vitro Transcription/Translation Assay
RTS 100 E. coli HY Kit (biotechrabbit Gmbh, Germany, catalog number: BR1400102) was used for the in vitro transcription/translation assay, according to the protocol. pIVEX2.3 control vector was used to produce GFP protein and to measure the translational efficiency. The amount of GFP produced was measured using a Hidex Plate Reader (Hidex Sense Microplate Reader with Plate Reader Software version 5064). The translational inhibitors, streptomycin and MtNCR247 as well as MtNCR001 without effect on translation (Farkas et al., 2014) were used as controls. All peptides and streptomycin were used at 100 µM concentration.
Gel Retardation Assay
Interactions of peptides with DNA was visualized with gel retardation assay (Hsu et al., 2005). 100 ng Lambda DNA/HindIII marker (ThermoFisher Scientific) was incubated with 0, 5 or 10 µM synthetic NCRs or NCR derivatives in water for 30 min at room temperature and then loaded into 1% agarose gel. Electrophoresis of DNA fragments separated the mobile HindIII-digested DNA fragments while the aggregated DNA remained in the loading wells.
Hemolysis Assay
Human blood was purchased from the Regional Blood Centre in Szeged. The use of human blood for the hemolysis assay has been authorized by the Regional Hungarian Ethics Committee and approved by the Ethics Review Sector of DG RTD (European Commission) in connection with EK’s ERC AdG SymBiotics. The cells from 10 ml of EDTA-blood were centrifuged at 1,500 × g for 1 min and washed several times in TBS buffer (10 mM Tris, pH = 7.2, 150 mM NaCl) until the supernatant became colorless. The cells were then resuspended in 12 ml TBS buffer and 100 μl of this cell suspension was incubated for 1 h at 37°C in the presence of NCR peptides (0.4–100 μM). The cells were centrifuged at 1,500 g for 1 min and the supernatants were transferred into sterile 96-well plates and the hemoglobin release was measured at OD560 (Hidex Sense Microplate Reader with Plate Reader Software version 5064). Cells in TBS buffer were used to determine baseline OD values, while 0.5% Triton X-100 (Serva) added at the same time as NCRs represented 100% of hemolysis. The hemolytic activity was calculated as % of red blood cell disruption relative to the positive control sample lysed with detergent Triton X-100.
Serum Stability Assay
Peptides were incubated in 10% Fetal Bovine Serum at 37°C for 18 h. Full length NCRs were used at 25 μM, while short derivatives at 50 μM. The serum proteins and peptides were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE, 12% bis-tris gel, MES buffer pH 7.3) and visualized by Coomassie blue staining. NCRs with StrepII tag at their C-terminus were also detected by Western blot analysis using Precision Protein StrepTactin-HRP conjugate antibody (BioRad) and Clarity Western ECL (BioRad) for chemiluminescence detection according to the supplier.
Results
Antimicrobial Activity of NCR Peptides Requires Positive Net Charge and Depends on the Amino Acid Sequence
We investigated the antimicrobial activity of 78 NCR peptides from M. truncatula and 12 NCRs from other IRLC legumes as well as their shorter derivatives against E. coli, S. enteritidis, P. aeruginosa, L. monocytogenes, S. aureus, E. faecalis, K. pneumoniae and A. baumannii (Supplementary Table S1). In the selection of the M. truncatula peptides, we considered their symbiotic expression levels but also their physicochemical properties. The antimicrobial activity of NCRs has so far been attributed to cationic peptides, with the exception of one anionic peptide. To investigate whether other anionic NCRs are antimicrobial, in addition to the cationic peptides, neutral and anionic NCRs were also included covering a broad range of peptides with different net charges (NC: from 7.8 to −10.2) and isoelectric points (pI: from 10.69 to 2.95). 50 peptides had positive net charge and pI > 7.0, while 28 peptides had negative net charge and pI < 7.0 (Supplementary Table S1A). Of the other legumes, only cationic NCR peptides were studied (Supplementary Table S1B).
The minimal bactericidal concentrations (MBCs) were determined after 3 h of treatment of ∼107 log phase bacteria with 2-fold serial dilution of peptides from 25 µM to 0.8 µM. Only 25 of the 50 cationic M. truncatula peptides had activity at ≤25 µM (Supplementary Table S1). NCR073, NCR336 and NCR358 were the most active peptides capable of killing all tested bacteria. Of the 3 peptides, the less cationic peptides, NCR073 and NCR336 were somewhat more effective than the highly cationic NCR358. Interestingly, peptides with almost identical net charge and pI to the fully active NCR073 and NCR336 were inactive or only partially active, confirming high importance of the amino acid sequence. NCR377 and NCR645 could also kill all bacteria except E. faecalis. NCR280 and NCR700 were active against all strains except S. aureus and E. faecalis. NCR055 was also effective against six bacterial strains but not against S. aureus and K. pneumonia. Likewise, NCR384 was able to kill all tested bacteria except E. faecalis and K. pneumoniae. NCR183, NCR299, NCR471 and NCR520 were effective against five or four strains, while NCR135 against three strains. Three peptides were only active against two bacterial strains; NCR649 against L. monocytogenes and A. baumannii, NCR281 against L. monocytogenes and P. aeruginosa and NCR350 against A. baumannii and P. aeruginosa. NCR035 was only effective against E. coli, while NCR030 and NCR032 only against A. baumannii. None of the anionic NCR peptides affected the survival of the tested bacterial strains. However, the neutral peptide, NCR147 showed activity against four strains: E. coli, P. aeruginosa, L. monocytogenes and A. baumannii.
Of the other legumes, CaNCR13, CaNCR63, MsNCR443, MsNCR463, PsNCR349 and PsNCR352 had broad-spectrum activity, with the exception of MsNCR463, which was only effective against P. aeruginosa at 25 µM (Supplementary Table S1B). In MsNCR463 the positively charged amino acid residues accumulated at the C-terminal half of the molecule and we assumed that the relative inefficiency of the full length MsNCR463 might be caused by the N-terminal region. Therefore, we synthetized and tested two C-terminal derivatives of MsNCR463; MsNCR46316-35 and MsNCR46317-30. Likewise, nine lysine residues (K) were present in the C-terminal part of PsNCR349, that could be responsible for the antimicrobial activity, therefore, PsNCR34926-50 and PsNCR349331-50 were also synthetized. On the contrary, the positively charged residues spread over the whole sequence in CaNCR63, indicating that likely the full-length sequence is necessary for the activity, though it could not be excluded that shorter derivatives of CaNCR63 were also active. From CaNCR63 we synthetized both the N-terminal sequence, CaNCR631-20 and a C-terminal fragment, CaNCR6315-34. The MBC and the minimal fungicide concentration (MFC) of these peptides were determined against A. baumannii and C. albicans as examples of top bacterial and fungal pathogens on the WHO priority list (Table 1). All peptides were effective against both A. baumannii (MBCs: 1.6–6.25 µM) and C. albicans (MFCs: 1.6–25 µM), but the most active ones were the C-terminal derivatives of MsNCR463; MsNCR46316-35 and MsNCR46317-30 (MBC: 1.6 and 3.1 µM, MFC: 3.1 µM), which have the highest pI (11.79 and 12.44) of the tested NCRs. PsNCR349 and its C terminal derivative PsNCR34926-50 were similarly effective against A. baumannii (MBC: 3.125 µM) and C. albicans (MFC: 3.125 µM). The even shorter PsNCR34931-50, had slightly reduced activity against A. baumannii (MBC: 6.25 µM), but showed the best activity against C. albicans (MFC: 1.6 µM). The MBC value of the other peptides (CaNCR13, CaNCR63, CaNCR631-20, CaNCR6315-34, MsNCR443, PsNCR34931-50, PsNCR352) was 6.25 µM.
In terms of pathogens, E. faecalis was the most resistant to NCRs, as only nine peptides (NCR055, NCR073, NCR183, NCR299, NCR336, NCR358, NCR520, GoNCR308, PsNCR351) were able to kill this Gram-positive bacterium. K. pneumoniae was sensitive to 13 NCRs, S. aureus to 18 NCRs, S. enterica to 21 NCRs, L. monocytogenes to 24 NCRs, A. baumannii to 27 NCRs, E. coli to 33 NCRs and P. aeruginosa to 36 NCRs (Supplementary Table S1).
Physicochemical Properties Playing Role in the Antimicrobial Activity of NCRs
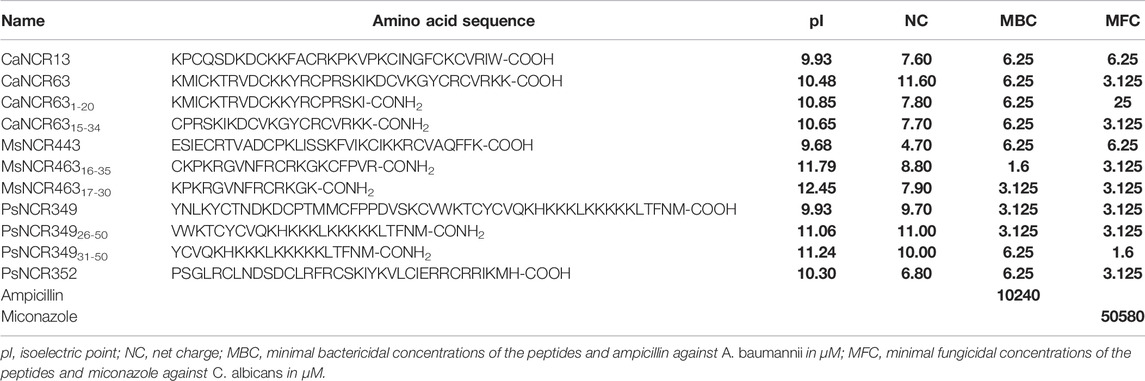
Based on our experiments, the net charge and pI were not the only determinants of the antimicrobial activity. AMP predictions for all peptides by eight different tools (Supplementary Table S1) gave different and only partially overlapping results, and none were fully consistent with our experimental data, as several NCRs, with high pI and net charge predicted as AMPs, were inactive. Therefore, other physicochemical properties of the peptides may play a role in the antimicrobial property. We used DBAASP v3 database that provides tools for multifactor analysis of physicochemical properties of peptides (Pirtskhalava et al., 2021). The normalized hydrophobic moment, normalized hydrophobicity, net charge, pI, penetration depth, tilt angle, disordered conformation propensity, linear moment, propensity to in vitro aggregation, angle subtended by the hydrophobic residues, amphiphilicity index and propensity to PPII coil of all peptides is shown in Supplementary Table S1. Comparing these properties of the active and inactive peptides revealed significant differences in normalized hydrophobicity, net charge, pI, disordered conformation propensity, amphiphilicity index and to a lesser extent in linear moment and angle subtended by the hydrophobic residues (Supplementary Figure S1). The active NCRs are positively charged, cationic and disordered peptides, but less hydrophobic and more amphiphilic than the inactive peptides. The plot of pI and net charge and the amphiphilicity index and disordered conformation propensity of each peptide together with the activity shown in Figure 1. As it was evident from the antimicrobial tests, high pI and net charge were required for antimicrobial activity but in some cases they alone were insufficient (Figure 1A). NCRs with net charge higher than 2 can be antimicrobial, but NCRs with net charge higher than 5 were all active, with the exception of NCR192. Moreover, the disordered nature and higher amphiphilicity index of active NCRs (Figure 1B) indicate higher possibility for antimicrobial activity likely due to adoption of secondary structures at membrane-water interphases (Vishnepolsky and Pirtskhalava, 2014; Pirtskhalava et al., 2021).
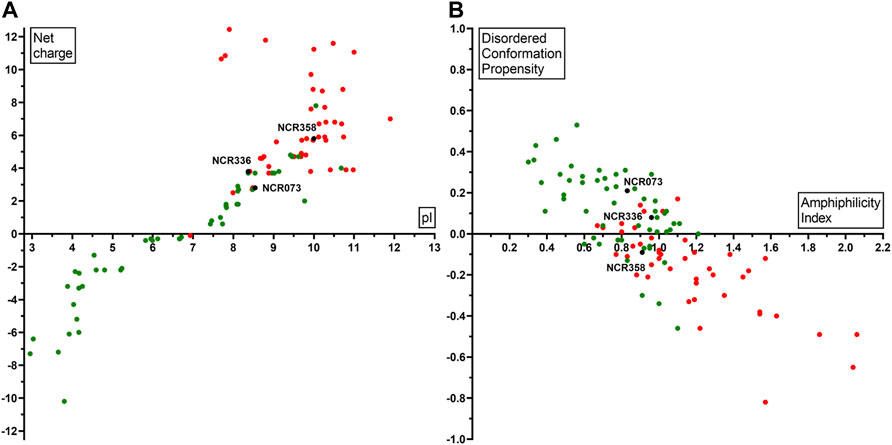
FIGURE 1. Physicochemical properties of NCRs in correlation with their antimicrobial activity (A) pI (X axis) and net charge (Y axis) (B) Amphiphilicity Index (X axis) and Disordered conformation propensity (Y axis) in correlation with the antimicrobial activity. Black dots show NCR073, NCR336, NCR358 which are active against all the eight tested pathogens. Red dots represent NCRs which are active against at least one pathogen. Green dots indicate the inactive NCRs.
NCRs Degrade Biofilms of C. albicans
Due to the protective effect of the matrix, pathogens in the biofilm are less accessible to antimicrobials and are therefore more resistant. As the biofilms are the main sources of persistent or recurrent infections, we tested whether NCRs could degrade the already formed biofilms. Biofilms of A. baumannii and C. albicans were treated overnight with 25, 50 and 100 µM concentrations of the peptides listed in Table 1. The quantity of biofilms remaining after the peptide treatments were compared to the control untreated biofilms (Figure 2). The peptides had only a mild effect on biofilms of A. baumannii while they were more effective in destroying the C. albicans biofilms reducing the biofilm quantity by 50–60%.
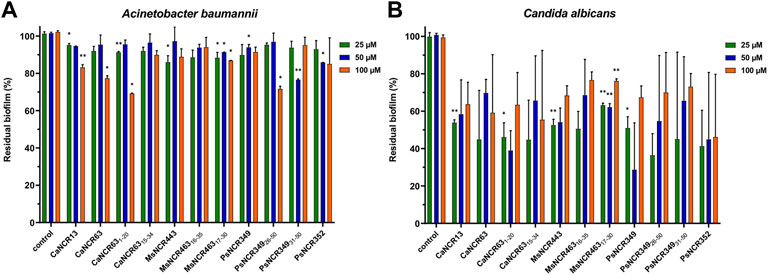
FIGURE 2. Effect of the peptides on biofilms of (A) A. baumannii and (B) C. albicans. The quantity of residual biofilms after overnight treatment with NCRs at 25, 50 or 100 μM compared to the untreated controls. The values are the means of two experiments, the bars indicate SD. *: p < 0.05, **: p < 0.01 as determined by one-way ANOVA.
NCR Peptides Cause Membrane Permeability, Morphological Changes and Cell Death
Live/Dead staining of microbes with the membrane-permeable SYTO9 dye, which emits green fluorescence and propidium iodide (PI), which only enters the damaged dead cells and emits red fluorescence, allows investigation of membrane damage caused by the NCR peptides. In addition, morphological changes were also observed by scanning electron microscopy (SEM). A. baumannii (Figure 3) and C. albicans (Figure 4) cells were treated for 30 min with different concentrations of NCRs as indicated in the figure legends and then were either co-stained with SYTO9/PI dyes and observed under confocal microscopy, or the cells were fixed and observed with SEM. The untreated A. baumannii showed green fluorescence after the SYTO9/PI staining, while the treatment of A. baumannii with any of the peptides resulted in varying degrees of red fluorescence (Figure 3). Only red dead cells were observed after the treatment with CaNCR13, MsNCR443, PsNCR34926-50 and PsNCR34931-50 (Figures 3B,F,J,K). The few green, live cells observed among the red, dead cells might indicate a slower killing action of peptides CaNCR63, MsNCR46316-35, MsNCR46317-30 and PsNCR349 (Figures 3C,G–I), which can be even more pronounced in the case of PsNCR352 (Figure 3L) based on the generally yellowish staining of cells.
FIGURE 3. Viability, membrane permeability and morphology of A. baumannii cells after NCR treatment (A–L) SYTO9/PI staining observed with confocal microscope, (M–X) Cell morphology observed with SEM. (A,M) untreated control, (B,N) CaNCR13, 3.125 µM, (C,O) CaNCR63, 3.125 µM, (D,P) CaNCR631-20, 3.125 µM, (E,Q) CaNCR6315-34, 3.125 µM, (F,R) MsNCR443, 3.125 µM, (G,S) MsNCR46316-35, 1.6 µM, (H,T) MsNCR46317-30, 3.125 µM, (I,U) PsNCR349, 1.6 µM, (J,V) PsNCR34926-50, 3.125 µM, (K,W) PsNCR34931-50, 3.125 µM, (L,X) PsNCR352, 1.6 µM.
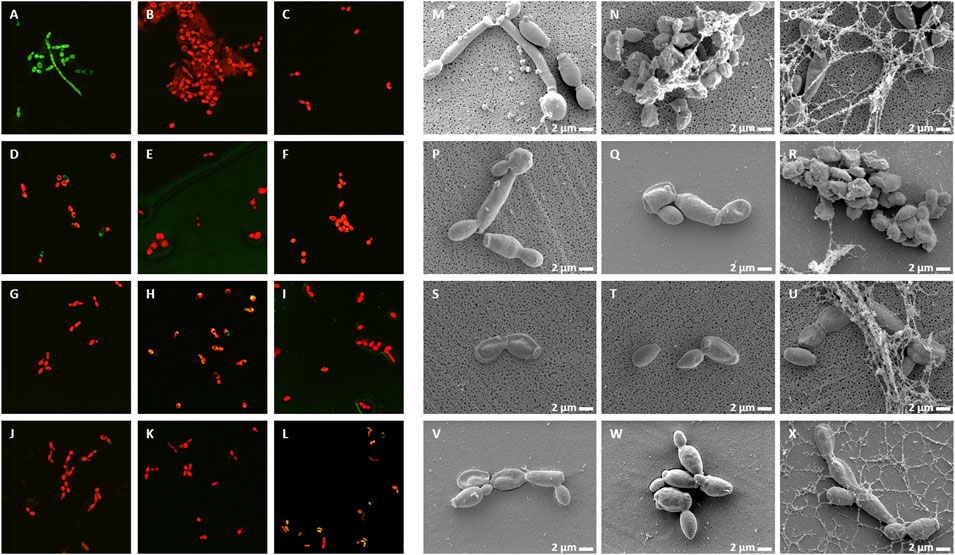
FIGURE 4. Viability, membrane permeability and morphology of C. albicans cells after NCR treatment (A–L) SYTO9/PI staining observed with confocal microscope, (M–X) Cell morphology observed with SEM (A,M) untreated control, (B,N) CaNCR13, 3.125 µM, (C,O) CaNCR63, 3.125 µM, (D,P) CaNCR631-20, 25 μM, (E,Q) CaNCR6315-34, 3.125 µM, (F,R) MsNCR443, 3.125 µM, (G,S) MsNCR46316-35, 1.6 µM, (H,T) MsNCR46317-30, 1.6 µM, (I,U) PsNCR349, 1.6 µM, (J,V) PsNCR34926-50, 3.125 µM, (K,W) PsNCR34931-50, 3.125 µM, (L,X) PsNCR352, 1.6 µM.
SEM revealed uniform size, morphology and smooth cell envelop of A. baumannii control cells (Figure 3M). This cell form was preserved in the case of PsNCR352 (Figure 3X) in line with the slower death indicated by the SYTO9/PI staining. The treatment by MsNCR46316-35 and MsNCR46317-30 caused budding of vesicles (Figures 3S,T). The treatment with CaNCR63, CaNCR631-20 and CaNCR6315-34 caused rough cell surface, lysis and flattening of the cells (Figures 3O–Q). Only dead cells were visible after treatment with CaNCR13, PsNCR349 and PsNCR34931-50 (Figures 3N,U,W). Filament-like structures, observed also by confocal microscopy by attachment of cells at their poles, were formed in the presence of CaNCR6315-34, MsNCR443, MsNCR46316-35, MsNCR46317-30 and PsNCR34926-50 (Figures 3Q–V).
Treatment of C. albicans with any of these peptides resulted in PI staining of the cells while in the untreated control culture, all cells were alive and showed green fluorescence (Figure 4A–L). A few viable green cells were, however, observed in the CaNCR631-20 sample (Figure 4D) in line with its highest MFC value. SEM observation revealed both yeast and hyphal forms in the control sample (Figure 4M). The CaNCR631-20 treated sample was the most similar to the control (Figure 4P) where a few living cells were observed with confocal microscopy (Figure 4D). Flattening of cells was visible after CaNCR63, MsNCR46316-35, MsNCR46317-30, PsNCR349 and PsNCR352 treatments (Figures 4O,S,T,U,X) and aggregation of shrunken cells in a spider net like matrix in the samples treated with CaNCR13, CaNCR63, MsNCR443 and PsNCR352 (Figures 4N,O,R,X).
NCR Peptides Have No or Only Weak Effects on Bacterial Translation
There is growing evidence that AMPs in addition to their membrane interactions can also have intracellular bacterial targets and affect various cellular functions. Previously it was shown that MtNCR247 inhibits the bacterial translation, thus here we investigated if any of the peptides in Table 1 have similar functions. The effect of peptides was studied using an E. coli in vitro coupled transcription/translation assay, which measures the translational efficiency by GFP production (Figure 5). As controls, we included streptomycin and MtNCR247 as inhibitors of translation, as well as MtNCR001 without effect (Farkas et al., 2014). In agreement with previous data, streptomycin inhibited strongly and MtNCR247 to lesser extent the GFP production. The tested peptides resulted in GFP production comparable to the water and MtNCR001 controls indicating that these peptides have no significant effects on translation.
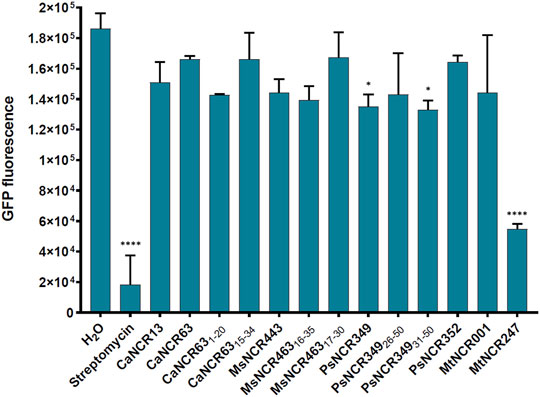
FIGURE 5. GFP production by E. coli in vitro coupled transcription-translation assay in the presence of 100 µM NCR or streptomycin. GFP fluorescence represents the average values of 2 technical parallels with their respective standard deviations (SD). *: p < 0.05, ****: p < 0.0001, as determined by one-way ANOVA.
NCR Peptides Can Form Complexes With DNA
In the natural symbiotic system NCRs provoke amplification of bacterial DNA via endoreduplication cycles (Mergaert et al., 2006). While the functions of NCRs in this process is still unknown we investigated whether the peptides could interact with DNA using HindIII-digested λ DNA as test molecule (Figure 6). The peptides were added to the DNA at 5 μM and 10 μM concentrations for 30 min and then loaded into agarose gel. Separation of the λ HindIII-fragments by gel electrophoresis occurred in the control untreated samples, while the DNA remained in the wells in the presence of 10 μM CaNCR63, CaNCR6315-34, MsNCR46316-35, PsNCR349, PsNCR34926-50 and PsNCR34931-50. While these peptides aggregated the DNA fragments, CaNCR13, MsNCR443 and PsNCR352 did not. The N-terminal fragment of CaNCR63, CaNCR631-20 was less efficient than either the full-length peptide or the C-terminal CaNCR6315-34, which retained completely the DNA-binding ability. CaNCR631-20 and CaNCR6315-34 have similar pI and net charge values, indicating the contribution of the sequence downstream of the 15th amino acid to the DNA-binding. In PsNCR349 the positively charged amino acid residues are at the C-terminal part, accordingly the sequence from 31 to 50 was sufficient for aggregation of DNA. PsNCR352 was less active in DNA-binding than PsNCR349, which may be due to its lower net charge (6.80). MsNCR46317-30 was less efficient than MsNCR46316-35 indicating that the last five amino acids are necessary to the DNA-interaction.
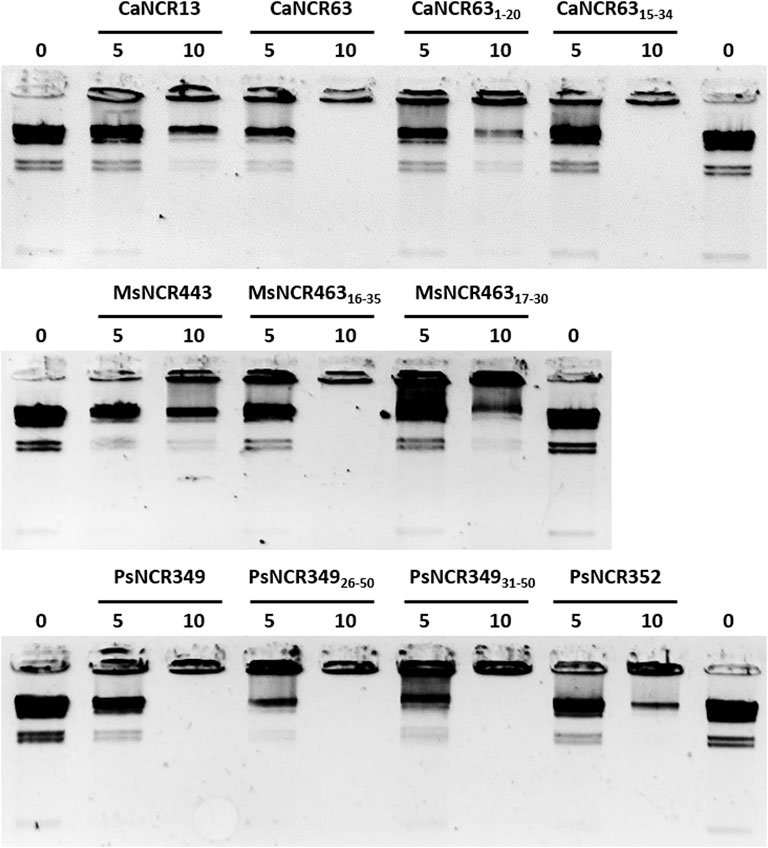
FIGURE 6. Aggregation of DNA by NCR peptides. Lambda HindIII digested DNA was incubated without (0) or with NCR peptides at 5 or 10 μM concentration.
Antimicrobial NCRs Do Not Provoke Notable Hemolysis and Are Stable in the Presence of Serum
While AMPs can be very effective against microbes, they should not be toxic for human cells. The potential hemolytic activity of the legume peptides listed in Table 1 was tested in hemolysis assay (Jenei et al., 2020). Human red blood cells were co-incubated with the peptides used in two-fold dilutions from 100 to 0.2 µM or with 0.5% Triton X-100 provoking complete hemolysis. Disruption of red blood cells was measured by spectrophotometry and compared to samples lysed with the detergent Triton X-100 (Supplementary Figure S2). None of the peptides caused hemolysis in the studied concentration range, except MsNCR465, provoking mild hemolysis at 50 and 100 μM concentrations, far above the MBCs of this peptide.
We also tested if NCRs remain stable in the presence of serum. 18 antimicrobial NCRs and NCR derivatives were incubated overnight at 37°C with Fetal Bovine Serum (FBS). The amount of peptides with or without FBS was comparable based on Coomassie Blue staining of SDS-PAGE gels (Supplementary Figure S3). To confirm the results, NCR183 and NCR055 carrying a StrepII tag at their C-terminus were also visualized by Western blot analysis using anti-StrepII-tag antibody and in line with the results of Coomassie Blue staining both of them were stable in the presence of FBS.
Discussion
Species belonging to the IRLC clade of legumes likely produce hundreds of thousands of NCR peptides in symbiosis mostly having roles in terminal differentiation of their rhizobium bacterium partner. In recent years, many studies have referred to NCR peptides as AMPs being effective against several pathogens (Mergaert, 2018; Jenei et al., 2020; Lima et al., 2020; Velivelli et al., 2020; Isozumi et al., 2021). Until now, only M. truncatula NCRs were tested and except for the anionic NCR211 (Kim et al., 2015) all NCRs with antimicrobial properties were cationic. MtNCRs above pI 9.5 were also effective against Candida species (Ördögh et al., 2014; Szerencsés et al., 2021). According to the sequence analysis, NCRs represent a novel class of AMPs (Lima et al., 2020).
In this work we addressed three main questions: i) are all cationic NCRs antimicrobial agents? ii) do the non-cationic NCRs have antimicrobial activities? and iii) do cationic NCRs from other IRLC legumes have antimicrobial activities?
The first and second questions were answered by studying 78 MtNCR peptides varying in their charge from −10.2 to 7.8 and isoelectric points from 2.95 to 10.69 against eight pathogenic bacteria including ESKAPE strains. 50 peptides had pI above 7.0, and 25 of them had antibacterial activities. Among the eight AMP prediction tools used in this study, CS-AMPPred developed to predict the antimicrobial activity of cysteine-stabilized peptides (Porto et al., 2012) and CAMPR3 using a large dataset of AMPs (Waghu et al., 2016; Waghu and Idicula-Thomas, 2020), were relatively the most accurate in finding active NCRs (Supplementary Table S1). However, the measured efficiency of these AMP predictions appears to be lower for NCRs than for other AMPs. The most important determinant of NCR activity was the high net charge and all tested NCRs with net charge higher than 5 (except NCR192) were active against at least one pathogenic bacteria. Previous work demonstrated the importance of pI values above 9.5 in antifungal activities (Ördögh et al., 2014) and here we show that pI > 9.5 is not an absolute requirement for the antibacterial effect as 12 peptides were effective with lower pI values (7.99–9.49), as was the neutral NCR147 peptide (pI 6.93). However, high positive charge and pI alone did not guarantee activity, as several peptides with such properties were inactive. The amino acid sequence and thereby other physicochemical properties of peptides could also contribute and be crucial for the antimicrobial activity. For example, the disordered conformation and higher amphiphilicity index show correlation with the NCRs’ antimicrobial activity. It will need further research to find out the structure-function relationship, why certain peptides with similar physicochemical properties were active or inactive in our screen. Three peptides were effective against all bacterial strains, eleven NCRs had broad range of activities, while seven NCRs could only kill one or two strains and three of them only A. baumannii. Having peptides with either broad or highly specific activities could be important for therapeutic applications.
One drawback of using cationic AMPs is their sensitivity to divalent cations; Ca2+, Mg2+, which can decrease their activity by inhibiting their interactions with microbial membranes. Therefore, non-cationic NCRs, likely with different modes of action, could be even more suited for therapeutic applications. We have tested 28 non-cationic NCR peptides and except for the neutral NCR147 (pI: 6.93, NC: −0.1) none of the peptides had antimicrobial activity. In this test, NCR211, with reported antibacterial activity against the symbiotic bacterium partner (Kim et al., 2015), was also inactive and even by repeating the described conditions the antibacterial activity of NCR211 could not be confirmed. It will be interesting to find more neutral NCRs with antimicrobial activities and to discover the mode of their killing actions.
To answer the third question, by analyzing 12 cationic NCRs from four legume species, we confirmed that NCRs from different IRLC legumes have antimicrobial activities. The activity of five broad spectrum IRLC NCRs and their six derivatives was tested against A. baumannii and C. albicans. All peptides were effective against A. baumannii and all except one against C. albicans with 1.6–6.25 µM MBC/MFC (Table 1). Moreover, the peptides were able to degrade C. albicans biofilm that can have a particular importance in therapeutic applications (Figure 1). Although understanding their exact mode of action has not been the subject of the present work and requires further research, this work indicates that their antimicrobial effects are primarily based on their interaction with microbial membranes. Within 30 min both A. baumannii and C. albicans cells were killed as it was demonstrated by confocal microscopy and SEM (Figures 2, 3). However, it is likely that the killing action of certain NCRs can even be faster as it was observed for other MtNCRs (Jenei et al., 2020). The NCRs may also enter the cells and have intracellular targets. In our experiments the tested NCRs had no significant effect on the in vitro coupled transcription/translation (Figure 4), but they provoked aggregation of DNA with different efficacy (Figure 5). These NCRs even at high 100 μM did not provoke hemolysis of human blood cells (Figure 6).
Our work confirmed that cationic NCRs from other IRLC legumes also have antimicrobial activities, of which MsNCR46317-30 and PsNCR349331-50 are outstanding with their efficiency and short length (14 and 20 amino acids), which makes their synthesis cost effective. We proved with MtNCR147 that neutral NCRs can be antimicrobial agents as well. Thus, the 4000 IRLC species can provide an uncountable source of antimicrobial peptides. The peptides selected for this study provide already a list of potential candidates for further drug development and clinical trials.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
Antimicrobial tests were done by BR and SJ. Biofilm degradation assay was done by BR. Confocal microscopy was done by RL, BR, and MA. Scanning electron microscopy was done by BR and MA. In vitro transcription and translation assay was done by HT. Gel retardation assay was done by RL. Hemolytic assay was done by BR and SJ. Serum stability assay was done by ET. Peptides were designed by GT, ÉK and synthesized by DH, and GT. Experiments were designed and evaluated by ÉK and GE. RL, BR, HT, GE, and ÉK wrote the manuscript.
Funding
Research has been supported by the Hungarian National Office for Research, Development and Innovation (NKFIH) Frontline Research project KKP129924 and the Balzan research grant to ÉK.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2022.870460/full#supplementary-material
Supplementary Figure S1 | Physicochemical properties of active and inactive NCRs calculated by DBAASP v3 (normalized hydrophobic moment, normalized hydrophobicity, penetration depth, tilt angle, disordered conformation propensity, linear moment, propensity to in vitro aggregation, angle subtended by the hydrophobic residues, amphiphilicity index, propensity to PPII coil) and PepCalc.com (net charge, pI). Active NCRs have antimicrobial activity against at least one pathogen. Graphs represent mean ± SD. *: p < 0.05, ****: p < 0.0001, as determined by t-test.
Supplementary Figure S2 | Hemolytic activity of NCRs. Human red blood cells were treated with different NCRs (0.4–100 µM). Hemolysis of red blood cells with 0.5% Triton-X represents 100% hemolytic activity.
References
Balogh, E., Mosolygó, T., Tiricz, H., Szabó, Á., Karai, A., Kerekes, F., et al. (2014). Anti-Chlamydial Effect of Plant Peptides. Acta Microbiol. Immunol. Hung. 61, 229–239. doi:10.1556/AMicr.61.2014.2.12
Bhatia, P., Sharma, A., George, A. J., Anvitha, D., Kumar, P., Dwivedi, V. P., et al. (2021). Antibacterial Activity of Medicinal Plants Against ESKAPE: An Update. Heliyon 7, e06310. doi:10.1016/j.heliyon.2021.e06310
Büyükkiraz, M. E., and Kesmen, Z. (2021). Antimicrobial Peptides (AMPs): A Promising Class of Antimicrobial Compounds. J. Appl. Microbiol. 132, 1573–1596. doi:10.1111/jam.15314
Campos, M. L., de Souza, C. M., de Oliveira, K. B. S., Dias, S. C., and Franco, O. L. (2018). The Role of Antimicrobial Peptides in Plant Immunity. J. Exp. Bot. 69, 4997–5011. doi:10.1093/jxb/ery294
Cesare, G. B. D., Cristy, S. A., Garsin, D. A., and Lorenz, M. C. (2020). Antimicrobial Peptides: A New Frontier in Antifungal Therapy. mBio 11 (6), e02123–20. doi:10.1128/mBio.02123-20
Duan, L., Li, S.-J., Su, C., Sirichamorn, Y., Han, L.-N., Ye, W., et al. (2021). Phylogenomic Framework of the IRLC Legumes (Leguminosae Subfamily Papilionoideae) and Intercontinental Biogeography of Tribe Wisterieae. Mol. Phylogenet. Evol. 163, 107235. doi:10.1016/j.ympev.2021.107235
Egorov, T. A., Odintsova, T. I., Pukhalsky, V. A., and Grishin, E. V. (2005). Diversity of Wheat Anti-Microbial Peptides. Peptides 26, 2064–2073. doi:10.1016/j.peptides.2005.03.007
Farkas, A., Maróti, G., Dürgő, H., Györgypál, Z., Lima, R. M., Medzihradszky, K. F., et al. (2014). Medicago Truncatula Symbiotic Peptide NCR247 Contributes to Bacteroid Differentiation through Multiple Mechanisms. Proc. Natl. Acad. Sci. 111, 5183–5188. doi:10.1073/pnas.1404169111
Farkas, A., Maróti, G., Kereszt, A., and Kondorosi, É. (2017). Comparative Analysis of the Bacterial Membrane Disruption Effect of Two Natural Plant Antimicrobial Peptides. Front. Microbiol. 8, 51. doi:10.3389/fmicb.2017.00051
Fernando, S. A., Gray, T. J., and Gottlieb, T. (2017). Healthcare-Acquired Infections: Prevention Strategies. Int. Med. J. 47, 1341–1351. doi:10.1111/imj.13642
Harding, C. M., Hennon, S. W., and Feldman, M. F. (2018). Uncovering the Mechanisms of Acinetobacter Baumannii Virulence. Nat. Rev. Microbiol. 16, 91–102. doi:10.1038/nrmicro.2017.148
Hsu, C.-H., Chen, C., Jou, M.-L., Lee, A. Y.-L., Lin, Y.-C., Yu, Y.-P., et al. (2005). Structural and DNA-Binding Studies on the Bovine Antimicrobial Peptide, Indolicidin: Evidence for Multiple Conformations Involved in Binding to Membranes and DNA. Nucleic. Acids. Res. 33, 4053–4064. doi:10.1093/nar/gki725
Isozumi, N., Masubuchi, Y., Imamura, T., Mori, M., Koga, H., and Ohki, S. (2021). Structure and Antimicrobial Activity of NCR169, a Nodule-Specific Cysteine-Rich Peptide of Medicago Truncatula. Sci. Rep. 11, 9923. doi:10.1038/s41598-021-89485-w
Ištvánek, J., Jaroš, M., Křenek, A., and Řepková, J. (2014). Genome Assembly and Annotation for Red Clover (Trifolium Pratense; Fabaceae). Am. J. Bot. 101, 327–337. doi:10.3732/ajb.1300340
Jenei, S., Tiricz, H., Szolomájer, J., Tímár, E., Klement, É., Al Bouni, M. A., et al. (2020). Potent Chimeric Antimicrobial Derivatives of the Medicago Truncatula NCR247 Symbiotic Peptide. Front. Microbiol. 11, 270. doi:10.3389/fmicb.2020.00270
Kanugala, S., Jinka, S., Puvvada, N., Banerjee, R., and Kumar, C. G. (2019). Phenazine-1-Carboxamide Functionalized Mesoporous Silica Nanoparticles as Antimicrobial Coatings on Silicone Urethral Catheters. Sci. Rep. 9, 6198. doi:10.1038/s41598-019-42722-9
Kim, M., Chen, Y., Xi, J., Waters, C., Chen, R., and Wang, D. (2015). An Antimicrobial Peptide Essential for Bacterial Survival in the Nitrogen-Fixing Symbiosis. Proc. Natl. Acad. Sci. 112, 15238–15243. doi:10.1073/pnas.1500123112
Kyriakidis, I., Vasileiou, E., Pana, Z. D., and Tragiannidis, A. (2021). Acinetobacter Baumannii Antibiotic Resistance Mechanisms. Pathogens 10, 373. doi:10.3390/pathogens10030373
Li, J., Hu, S., Jian, W., Xie, C., and Yang, X. (2021). Plant Antimicrobial Peptides: Structures, Functions, and Applications. Bot. Stud. 62, 5. doi:10.1186/s40529-021-00312-x
Lima, R. M., Kylarová, S., Mergaert, P., and Kondorosi, É. (2020). Unexplored Arsenals of Legume Peptides with Potential for Their Applications in Medicine and Agriculture. Front. Microbiol. 11, 1307. doi:10.3389/fmicb.2020.01307
Magana, M., Pushpanathan, M., Santos, A. L., Leanse, L., Fernandez, M., Ioannidis, A., et al. (2020). The Value of Antimicrobial Peptides in the Age of Resistance. Lancet Infect. Dis. 20, e216–e230. doi:10.1016/S1473-3099(20)30327-3
Maróti, G., Downie, J. A., and Kondorosi, É. (2015). Plant Cysteine-Rich Peptides that Inhibit Pathogen Growth and Control Rhizobial Differentiation in Legume Nodules. Curr. Opin. Plant Biol. 26, 57–63. doi:10.1016/j.pbi.2015.05.031
Mergaert, P., Nikovics, K., Kelemen, Z., Maunoury, N., Vaubert, D., Kondorosi, A., et al. (2003). A Novel Family in Medicago Truncatula Consisting of More Than 300 Nodule-Specific Genes Coding for Small, Secreted Polypeptides with Conserved Cysteine Motifs. Plant Physiol. 132, 161–173. doi:10.1104/pp.102.018192
Mergaert, P. (2018). Role of Antimicrobial Peptides in Controlling Symbiotic Bacterial Populations. Nat. Prod. Rep. 35, 336–356. doi:10.1039/C7NP00056A
Mergaert, P., Uchiumi, T., Alunni, B., Evanno, G., Cheron, A., Catrice, O., et al. (2006). Eukaryotic Control on Bacterial Cell Cycle and Differentiation in the Rhizobium–Legume Symbiosis. Proc. Natl. Acad. Sci. 103, 5230–5235. doi:10.1073/pnas.0600912103
Mikuláss, K. R., Nagy, K., Bogos, B., Szegletes, Z., Kovács, E., Farkas, A., et al. (2016). Antimicrobial Nodule-Specific Cysteine-Rich Peptides Disturb the Integrity of Bacterial Outer and Inner Membranes and Cause Loss of Membrane Potential. Ann. Clin. Microbiol. Antimicrob. 15, 43. doi:10.1186/s12941-016-0159-8
Montiel, J., Downie, J. A., Farkas, A., Bihari, P., Herczeg, R., Bálint, B., et al. (2017). Morphotype of Bacteroids in Different Legumes Correlates with the Number and Type of Symbiotic NCR Peptides. Proc. Natl. Acad. Sci. 114, 5041–5046. doi:10.1073/pnas.1704217114
Moser, D., Biere, K., Han, B., Hoerl, M., Schelling, G., Choukér, A., et al. (2021). COVID-19 Impairs Immune Response to Candida Albicans. Front. Immunol. 12, 640644. doi:10.3389/fimmu.2021.640644
Nallu, S., Silverstein, K. A. T., Samac, D. A., Bucciarelli, B., Vance, C. P., and VandenBosch, K. A. (2013). Regulatory Patterns of a Large Family of Defensin-Like Genes Expressed in Nodules of Medicago Truncatula. PLoS One 8, e60355. doi:10.1371/journal.pone.0060355
Oliveira, L. V. N., Wang, R., Specht, C. A., and Levitz, S. M. (2021). Vaccines for Human Fungal Diseases: Close but Still a Long Way to Go. npj Vaccines 6, 33. doi:10.1038/s41541-021-00294-8
Ördögh, L., Vörös, A., Nagy, I., Kondorosi, É., and Kereszt, A. (2014). Symbiotic Plant Peptides Eliminate Candida Albicans Both In Vitro and in an Epithelial Infection Model and Inhibit the Proliferation of Immortalized Human Cells. Biomed. Res. Int. 2014, 1–9. doi:10.1155/2014/320796
Pirtskhalava, M., Amstrong, A. A., Grigolava, M., Chubinidze, M., Alimbarashvili, E., Vishnepolsky, B., et al. (2021). DBAASP v3: Database of Antimicrobial/Cytotoxic Activity and Structure of Peptides as a Resource for Development of New Therapeutics. Nucleic Acids Res. 49, D288–D297. doi:10.1093/nar/gkaa991
Porto, W. F., Ferreira, K. C. V., Ribeiro, S. M., and Franco, O. L. (2022). Sense the Moment: A Highly Sensitive Antimicrobial Activity Predictor Based on Hydrophobic Moment. Biochim. Biophys. Acta Gen. Subj. 1866, 130070. doi:10.1016/j.bbagen.2021.130070
Porto, W. F., Pires, Á. S., and Franco, O. L. (2012). CS-AMPPred: An Updated SVM Model for Antimicrobial Activity Prediction in Cysteine-Stabilized Peptides. PLoS One 7, e51444. doi:10.1371/journal.pone.0051444
Rangel, K., Chagas, T. P. G., and De-Simone, S. G. (2021). Acinetobacter Baumannii Infections in Times of COVID-19 Pandemic. Pathogens 10, 1006. doi:10.3390/pathogens10081006
Ribeiro, C. W., Baldacci-Cresp, F., Pierre, O., Larousse, M., Benyamina, S., Lambert, A., et al. (2017). Regulation of Differentiation of Nitrogen-Fixing Bacteria by Microsymbiont Targeting of Plant Thioredoxin S1. Curr. Biol. 27, 250–256. doi:10.1016/j.cub.2016.11.013
Roy, P., Achom, M., Wilkinson, H., Lagunas, B., and Gifford, M. L. (2020). Symbiotic Outcome Modified by the Diversification from 7 to over 700 Nodule-Specific Cysteine-Rich Peptides. Genes 11, 348. doi:10.3390/genes11040348
Shabab, M., Arnold, M. F. F., Penterman, J., Wommack, A. J., Bocker, H. T., Price, P. A., et al. (2016). Disulfide Cross-Linking Influences Symbiotic Activities of Nodule Peptide NCR247. Proc. Natl. Acad. Sci. 113, 10157–10162. doi:10.1073/pnas.1610724113
Szerencsés, B., Gácser, A., Endre, G., Domonkos, I., Tiricz, H., Vágvölgyi, C., et al. (2021). Symbiotic NCR Peptide Fragments Affect the Viability, Morphology and Biofilm Formation of Candida Species. Int. J. Mol. Sci. 22, 3666. doi:10.3390/ijms22073666
Tam, J., Wang, S., Wong, K., and Tan, W. (2015). Antimicrobial Peptides from Plants. Pharmaceuticals 8, 711–757. doi:10.3390/ph8040711
Tiricz, H., Szűcs, A., Farkas, A., Pap, B., Lima, R. M., Maróti, G., et al. (2013). Antimicrobial Nodule-Specific Cysteine-Rich Peptides Induce Membrane Depolarization-Associated Changes in the Transcriptome of Sinorhizobium Meliloti. Appl. Environ. Microbiol. 79, 6737–6746. doi:10.1128/AEM.01791-13
Van de Velde, W., Zehirov, G., Szatmari, A., Debreczeny, M., Ishihara, H., Kevei, Z., et al. (2010). Plant Peptides Govern Terminal Differentiation of Bacteria in Symbiosis. Science 327, 1122–1126. doi:10.1126/science.1184057
Velivelli, S. L. S., Czymmek, K. J., Li, H., Shaw, J. B., Buchko, G. W., and Shah, D. M. (2020). Antifungal Symbiotic Peptide NCR044 Exhibits Unique Structure and Multifaceted Mechanisms of Action that Confer Plant Protection. Proc. Natl. Acad. Sci. 117, 16043–16054. doi:10.1073/pnas.2003526117
Veltri, D., Kamath, U., and Shehu, A. (2018). Deep Learning Improves Antimicrobial Peptide Recognition. Bioinformatics 34, 2740–2747. doi:10.1093/bioinformatics/bty179
Vishnepolsky, B., and Pirtskhalava, M. (2014). Prediction of Linear Cationic Antimicrobial Peptides Based on Characteristics Responsible for Their Interaction with the Membranes. J. Chem. Inf. Model. 54, 1512–1523. doi:10.1021/ci4007003
Waghu, F. H., Barai, R. S., Gurung, P., and Idicula-Thomas, S. (2016). CAMPR3: A Database on Sequences, Structures and Signatures of Antimicrobial Peptides. Nucleic Acids Res. 44, D1094–D1097. doi:10.1093/nar/gkv1051
Waghu, F. H., and Idicula‐Thomas, S. (2020). Collection of Antimicrobial Peptides Database and its Derivatives: Applications and Beyond. Protein Sci. 29, 36–42. doi:10.1002/pro.3714
Wang, G., Li, X., and Wang, Z. (2016). APD3: The Antimicrobial Peptide Database as a Tool for Research and Education. Nucleic Acids Res. 44, D1087–D1093. doi:10.1093/nar/gkv1278
Yamaguchi, Y., and Huffaker, A. (2011). Endogenous Peptide Elicitors in Higher Plants. Curr. Opin. Plant Biol. Biot. Interact. 14, 351–357. doi:10.1016/j.pbi.2011.05.001
Keywords: legume NCR peptides, antimicrobial activity, ESKAPE pathogens, Acinetobacter baumannii, Candida albicans, mode of action
Citation: Lima RM, Rathod BB, Tiricz H, Howan DHO, Al Bouni MA, Jenei S, Tímár E, Endre G, Tóth GK and Kondorosi É (2022) Legume Plant Peptides as Sources of Novel Antimicrobial Molecules Against Human Pathogens. Front. Mol. Biosci. 9:870460. doi: 10.3389/fmolb.2022.870460
Received: 06 February 2022; Accepted: 18 May 2022;
Published: 09 June 2022.
Edited by:
Elizabete de Souza Cândido, Dom Bosco Catholic University, BrazilReviewed by:
Suzana Meira Ribeiro, Colégio Militar de Curitiba, BrazilApril Hastwell, The University of Queensland, Australia
Karen Garcia Nogueira Oshiro, University of Brasilia, Brazil
Marlon Henrique Cardoso, Universidade Católica Dom Bosco, Brazil
Copyright © 2022 Lima, Rathod, Tiricz, Howan, Al Bouni, Jenei, Tímár, Endre, Tóth and Kondorosi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Éva Kondorosi, eva.kondorosi@gmail.com
†These authors have contributed equally to this work
 Rui M. Lima
Rui M. Lima Balaji Baburao Rathod
Balaji Baburao Rathod Hilda Tiricz
Hilda Tiricz Dian H. O. Howan2
Dian H. O. Howan2  Mohamad Anas Al Bouni
Mohamad Anas Al Bouni Gabriella Endre
Gabriella Endre Gábor K. Tóth
Gábor K. Tóth Éva Kondorosi
Éva Kondorosi