Autopromotion of K-Ras4B Feedback Activation Through an SOS-Mediated Long-Range Allosteric Effect
- 1Department of Pharmacy, Ruijin Hospital, Shanghai Jiao Tong University, School of Medicine, Shanghai, China
- 2School of Chemistry and Chemical Engineering, Shaoxing University, Shaoxing, China
- 3Department of Pathophysiology, Key Laboratory of Cell Differentiation and Apoptosis of Chinese Ministry of Education, School of Medicine, Shanghai Jiao Tong University, Shanghai, China
- 4The Charles Perkins Centre, University of Sydney, Sydney, NSW, Australia
- 5Medicinal Chemistry and Bioinformatics Center, Shanghai Jiao Tong University, School of Medicine, Shanghai, China
The Ras-specific guanine nucleotide exchange factors Son of Sevenless (SOS) regulates Ras activation by converting inactive GDP-bound to active GTP-bound states. The catalytic activity of Ras is further allosterically regulated by GTP−Ras bound to a distal site through a positive feedback loop. To address the mechanism underlying the long-range allosteric activation of the catalytic K-Ras4B by an additional allosteric GTP–Ras through SOS, we employed molecular dynamics simulation of the K-Ras4BG13D•SOScat complex with and without an allosteric GTP-bound K-Ras4BG13D. We found that the binding of an allosteric GTP−K-Ras4BG13D enhanced the affinity between the catalytic K-Ras4BG13D and SOScat, forming a more stable conformational state. The peeling away of the switch I from the nucleotide binding site facilitated the dissociation of GDP, thereby contributing to the increased nucleotide exchange rate. The community networks further showed stronger edge connection upon allosteric GTP−K-Ras4BG13D binding, which represented an increased interaction between catalytic K-Ras4BG13D and SOScat. Moreover, GTP−K-Ras4BG13D binding transmitted allosteric signaling pathways though the Cdc25 domain of SOS that enhanced the allosteric regulatory from the K-Ras4BG13D allosteric site to the catalytic site. This study may provide an in-depth mechanism for abnormal activation and allosteric regulation of K-Ras4BG13D.
1 Introduction
As the central control element of the signal transduction pathway, GTPases and their related regulatory factors and effectors are involved in almost all aspects of cell biology [(Colicelli, 2004; Mitin et al., 2005; Bos et al., 2007)]. Ras superfamily proteins, the classical members of small GTPases, possess a shared biochemical activity: GTP (guanosine triphosphate) binding and hydrolysis. Three major isoforms, including H-Ras, K-Ras, and N-Ras, are closely related due to high overall sequence identity [(Prior et al., 2012), (Cox et al., 2014)]. Among them, K-Ras covers 85% mutated isoforms in Ras-driven cancers [(Fernandez-Medarde and Santos, 2011; Stephen et al., 2014)]. Ras transforms between inactive GDP-bound and active GTP-bound states, where only the Ras−GTP complex can activate downstream signaling pathways via high-affinity binding to its effectors (Bourne et al., 1990; Qiu et al., 2021a). The oncogenic forms of Ras are manifested as the impaired GTPase cycle because of the exchange dysregulation of Ras–GTP hydrolysis/guanine nucleotide (Lu et al., 2015; Parker and Mattos, 2018). The continuous activation of Ras contributes to several malignant phenotypes such as cell differentiation and proliferation (Drosten et al., 2010). The nucleotide binding pocket of Ras consists of three functional regions including the P-loop [residues (Milburn et al., 1990; Vetter and Wittinghofer, 2001; Drosten et al., 2010; Goitre et al., 2014; Lu et al., 2015; Simanshu et al., 2017; Parker and Mattos, 2018; Lu et al., 2019a)], switch I [residues (Sondermann et al., 2004; Freedman et al., 2006; Ni et al., 2017; Ni et al., 2018; Lu et al., 2019b; Ni et al., 2020; Fan et al., 2021; Li et al., 2021; Moghadamchargari et al., 2021)], and switch II [residues (Boykevisch et al., 2006; Newman, 2006; Krissinel and Henrick, 2007; Eargle and Luthey-Schulten, 2012; Vo et al., 2016; Li et al., 2018; Bandaru et al., 2019; Wang et al., 2019; Li et al., 2020; Aledavood et al., 2021; Feng et al., 2021; Foutch et al., 2021; He et al., 2021; Liang et al., 2021; Liu et al., 2021; Marasco et al., 2021; Ni et al., 2021; Okeke et al., 2021)] (Milburn et al., 1990; Vetter and Wittinghofer, 2001; Goitre et al., 2014). The structures of GDP-bound and GTP-bound forms mainly differ by the conformation of switches I and II, both of which are highly dynamic regions. The activation of Ras requires guanine nucleotide exchange factors (GEFs), facilitating the exchange of GDP to GTP, while its inactivation is terminated by GTPase-activating proteins (GAPs) stimulating the hydrolysis of intrinsic GTP (Simanshu et al., 2017; Lu et al., 2019a; Jang et al., 2020).
The multiprotein, Son of Sevenless (SOS), is a GEF consisting of ∼1,330 residues (Gureasko et al., 2008; Rojas et al., 2011). The core catalytic region of SOS, which is required for Ras-specific nucleotide exchange activity, consists of a Ras exchanger motif (Rem) domain and a Cdc25 domain, termed SOScat (residues 551–1050) (Chardin et al., 1993). Grb2 recruits SOS to the plasma membrane to initiate SOS activation [(Kazemein Jasemi et al., 2021)]. In general, the interaction of SOS with Ras–GDP promotes the disassociation of Ras and GDP molecules, and the nucleotide-free Ras is accessible to bind more cellular GTP (Lu et al., 2016a). It has been proposed that the catalyzed conversion of Ras by SOS covers a multi-step mechanism, which can be roughly divided into the following processes: 1) the exchange reaction is initiated by the formation of a low-affinity complex containing an inactive Ras−GDP and activated SOS; 2) the Ras active site opens widely and expels GDP, thereby forming a high-affinity binary Ras•SOS complex. Ras has a picomolar high affinity to both GDP and GTP, and the tightly bound nucleotides must turn loose for their fast dissociation (Goody et al., 1991). According to experimental evidence, the reverse isomerization reaction from high- to low-affinity bound conformation of the Ras–GDP complex may be one rate-limiting step during the SOS-catalyzed exchange (Lenzen et al., 1998). The structure of the Ras•SOScat complex showed that the helical hairpin region protruding from the main body of the Cdc25 domain of the SOScat is inserted between the switch I and II regions of Ras protein, opening the Ras nucleotide binding site for GDP release (Hall et al., 2001). In addition, Ras is unstable in the absence of nucleotide, and SOS stabilizes its nucleotide-free form by forming a large interface with the switch II, which provides the main anchor for the interactions of SOS with Ras. The formation of this extensive interface is probably crucial for stabilizing the unstable nucleotide-free Ras and protect it from unfolding (Cherfils and Zeghouf, 2013); 3) since the cellular concentration of GTP is high, GTP binds to the nucleotide-free Ras (Buday and Downward, 2008); and 4) after completing the exchange from GDP to GTP, the binary GTP-bound Ras complex releases from the SOS, yielding the active GTP-bound Ras form. Removal of the bound nucleotide resulted in a stable complex, which was key for obtaining many crystal structures of nucleotide-free Ras•SOS complexes and had been deeply investigated during the last decades (Cherfils and Zeghouf, 2013).
Multiple crystal structures of Ras under different nucleotide-bound states with SOS have been determined, providing comprehensive details on the activation mechanism of Ras (Chardin et al., 1993; Bos et al., 2007). Interestingly, Margarit et al. (2003) reported an H-RasA59G•SOScat structural complex and found an additional Ras−GTP located at the Rem and Cdc25 domains of SOScat. The resulting mutant structure is an Ras•SOScat•Ras−GTP ternary complex, consisting of two H-RasA59G molecules (one is nucleotide-free catalytic H-RasA59G and the other is GTP-bound allosteric H-RasA59G) and one SOScat molecule (Margarit et al., 2003). Given that SOS is allosterically activated by Ras−GTP, multiple studies have revealed a conformational switch of SOS induced by Ras−GTP and the change of catalytic activity in the ternary system (Freedman et al., 2006). A positive feedback mechanism has been reported in the activation of Ras by SOS (Sondermann et al., 2004). However, the interaction between the oncogenic Ras mutants and SOS remains insufficiently characterized, and the precise structural details of the positive feedback mechanism are still unknown.
Recently, a structure of the K-Ras4BG13D•SOScat•K-Ras4BG13D−GTP complex was resolved. Biochemical data implied that K-Ras4BG13D−GTP binding allosterically increased the nucleotide exchange rate of K-Ras4B at the active site and led to the reposition of the switch I and II regions of K-Ras4BG13D (Moghadamchargari et al., 2021). This brought a partial understanding of the interaction between oncogenic Ras mutants and SOS. However, the mechanism underlying the long-range allosteric activation of the catalytic K-Ras4B by an additional allosteric GTP–Ras through SOS remains poorly understood. Thus, further exploration of the allosteric regulation of Ras–GTP can uncover more information pertaining to the activation mechanism of Ras and may provide an avenue for drug discovery (Lu et al., 2019b; Fan et al., 2021).
Conformational dynamics of signaling proteins are vital for the realization of their biological functions (Ni et al., 2017; Ni et al., 2018; Ni et al., 2020; Lu et al., 2021a; Qiu et al., 2021b; Li et al., 2021). In this study, using explicit molecular dynamics (MD) simulations, we investigated the mechanism of allosteric activation of the catalytic Ras located at the SOScat active site induced by the allosteric Ras–GTP binding at the SOScat distal site. We established the K-Ras4BG13D•SOScat binary system and the K-Ras4BG13D•SOScat•K-Ras4BG13D–GTP ternary system to explore conformational changes upon allosteric K-Ras4BG13D–GTP binding. The results mainly focused on dynamic conformational changes of the catalytic K-Ras4BG13D and allosteric pathways. In both structural and energetic aspects, binding of K-Ras4BG13D–GTP enhanced the affinity between the catalytic K-Ras4BG13D and SOScat, yielding a more stable structural state. Furthermore, the variation of the K-Ras4BG13D switch I region caused the expansion of the nucleotide binding pocket, which contributed to the increased nucleotide exchange rate upon K-Ras4BG13D–GTP binding. This study may provide insights into the allosteric regulation of the K-Ras4BG13D•SOScat complex by K-Ras4BG13D–GTP.
2 Materials and Methods
2.1 Construction of Simulated Systems
Two simulated systems were established, including the K-Ras4BG13D•SOScat binary system and the K-Ras4BG13D•SOScat•K-Ras4BG13D–GTP ternary system, based on the K-Ras4BG13D•SOScat•K-Ras4BG13D–GppNHp crystal structure (PDB ID: 7KFZ) (Moghadamchargari et al., 2021). The missing residues were remodeled using Discovery Studio and GppNHp was replaced by a GTP molecule. By deleting K-Ras4BG13D–GTP in the ternary system, the K-Ras4BG13D•SOScat binary system was extracted from the K-Ras4BG13D•SOScat•K-Ras4BG13D–GTP complex.
2.2 MD Simulations
The initial parameter files for minimization and simulation were prepared using Amber18 package with the ff14SB force field (Maier et al., 2015) and the general Amber force field (GAFF) (Wang et al., 2004; Lu et al., 2016b; Wang et al., 2021a). Both complexes were solvated in a truncated octahedron transferable intermolecular potential three-point (TIP3P) water box, and then Na+ and Cl−counterions were added to neutralize the system and to mimic a simulated body fluid (Jorgensen et al., 1983), (Wang et al., 2022). Next, both systems were processed by two rounds of energy minimizations using steepest descent and conjugate gradient minimization steps. After that, both systems’ temperatures increased from 0 to 300 K in 300 ps in a canonical ensemble (NVT), followed by the equilibration runs of 700 ps in the NVT ensemble. Finally, three independent rounds of 1 μs MD simulations were performed for both systems in the isothermal and isobaric ensembles (NPT) with the periodic boundaries condition. In the course of MD simulations, long-range electrostatic interactions were calculated using the particle mesh Ewald (PME) method [(York et al., 1994), (Pal et al., 2021)], while short-range electrostatic interactions and van der Waals interactions were defined using a cutoff distance of 10 Å. Covalent bonds involving hydrogen were constrained using the SHAKE method [(Ryckaert et al., 1977; Byun et al., 2020; Liang et al., 2020)].
2.3 Principal Component Analysis
PCA was applied to capture the essential motions and characterize the overall dominant conformational transitions in two systems [(Hernández-Alvarez et al., 2021)]. In order to describe the motions of the system, the covariance matrix of Cα atoms was diagonalized to create a new set of eigenvectors (also called PC) in PCA. The eigenvalue of each PC was related to the mean square fluctuation of the PC projected by the trajectory of the entire system. Hence, the first ranked PC (PC1) corresponded to the most dominant amplitude movement within the system, and the system dynamics projected along PC1 was defined as “essential dynamics” [(Amadei et al., 1993)]. In this work, PCA results were conducted on the 2D plane according to the PC1 and PC2 to determine the major conformational dynamics of K-Ras4BG13D•SOScat. After that, cluster analyses were used to extract the most representative conformations from PC1 to PC2, which were superimposed using all Cα atoms prior to eliminate the overall rotation and transition [(Shao et al., 2007)].
2.4 Molecular Mechanics Poisson–Boltzmann Surface Area Calculations
The binding free energies between K-Ras4BG13D and SOScat in both systems were calculated using the MM/PBSA plugin from MMPBSA.py in Amber18 package (Chong et al., 2009; Shibata et al., 2020; Wang et al., 2020; An et al., 2021). The binding free energy
According to the second thermodynamic law,
For the
The non-polar component
The conformation entropy component (
2.5 Dynamic Network Analysis
In order to characterize the inter-residue correlations, all protein Cα atoms were calculated using the correlation coefficient Cij, which reflected the correlated degree of motions of Cα atoms (Wang et al., 2019; Aledavood et al., 2021; He et al., 2021; Marasco et al., 2021; Okeke et al., 2021). Cij was calculated as follows:
where ri and rj represent the positions of the ith and jth Cα atoms.
To calculate the community network via the NetworkView plugin in VMD (Eargle and Luthey-Schulten, 2012), the Cα atom of each residue was recognized as a node, and edges were formed when two nodes stayed within a cutoff distance of 4.5 Å for at least 75% of the simulation time. To weight edges and describe the edge distance, the correlation data can be further calculated using the following equation:
The community network was defined and optimized using the Girvan–Newman algorithm based on the information of betweenness (Newman, 2006), (Liu et al., 2021). In addition, the shortest paths were calculated using the Floyd–Warshall algorithm as cross-community communication (He et al., 2021). Suboptimal pathways were defined as within 20 Å to the optimal pathway (69).
3 Results
3.1. RMSD and RMSF Analysis
To capture the dynamic conformational changes, we conducted three independent rounds of 1 μs MD simulations on both binary and ternary complexes. Root-mean-square deviations (RMSDs) of Cα atoms in each residue of K-Ras4BG13D•SOScat were analyzed in the two systems relative to the initial structures, which reflected the conformational dynamics over simulations. K-Ras4BG13D•SOScat in both systems reached equilibrium after ∼200ns MD simulations. The RMSD values of K-Ras4BG13D•SOScat in the binary and ternary systems were 3.65 ± 0.26 Å and 2.55 ± 0.27 Å, respectively, which indicated some conformational discrepancies between two systems (Figure 1A). Meanwhile, we separately calculated the RMSDs of K-Ras4BG13D and SOScat. As shown in Figures 1B,C, the RMSD of SOScat in the ternary system was notably lower than that in the binary system, while the RMSD of K-Ras4BG13D showed slight conformational differences between two systems. Then, we further explored the specific functional regions of K-Ras4BG13D and SOScat. Focusing on the switch I and switch II regions, we found that the discrepancies of K-Ras4BG13D mainly contributed by the switch II region (Figures 1D,E). However, the RMSD of the Cdc25 domain of SOScat showed significant discrepancies between two systems, which led to the overall conformational differences in the K-Ras4BG13D•SOScat complex (Figure 1F). Furthermore, the differences of fluctuations among individual residues in local regions were revealed by the root-mean-square fluctuations (RMSFs) of Cα atoms of K-Ras4BG13D (Figure 1G) and SOScat (Figure 1H). Notably, RMSF analysis showed that the switch II region of K-Ras4BG13D exhibited lower flexibilities in response to allosteric K-Ras4BG13D−GTP binding, while little distinctions were observed in the switch I and P-loop of K-Ras4BG13D and the SOScat. Taken together, these data indicated higher stability and more constrained conformation of the switch II region in the ternary complex than in the binary complex. This may partially be due to the increased interaction between the catalytic K-Ras4BG13D and SOScat upon the allosteric K-Ras4BG13D–GTP binding.
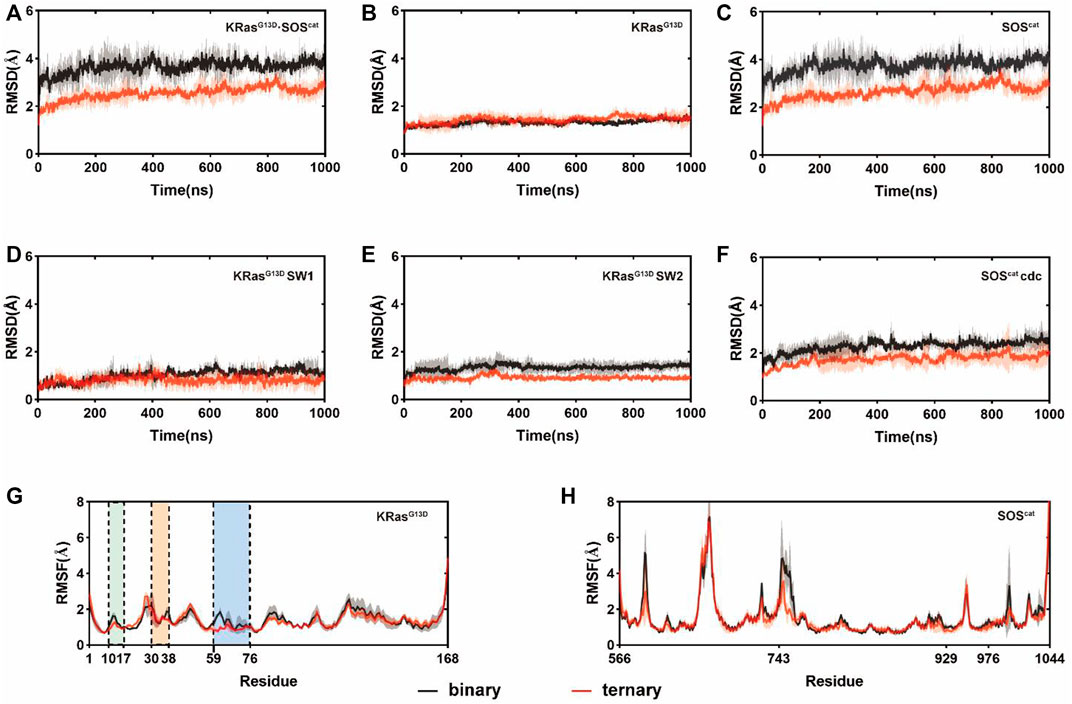
FIGURE 1. Conformational dynamics of the K-Ras4BG13D•SOScat complex in the binary and ternary systems. RMSDs of Cα atoms in the binary (black) and ternary (red) systems within (A) all K-Ras4BG13D•SOScat complex residues; (B) K-Ras4BG13D residues; (C) SOScat residues; (D) K-Ras4BG13D switch I residues; (E) K-Ras4BG13D switch II residues, and (F) SOScat Cdc25 domain residues. RMSFs of Cα atoms in the binary (black) and ternary (red) systems within (G) K-Ras4BG13D residues and (H) SOScat residues. The P-loop, the switch I and switch II regions are marked by green, orange, and blue backgrounds, respectively. Gray and red transparencies represent the standard deviations.
3.2 Principal Component Analysis
Subsequently, we used principal component analysis (PCA) for characterizing and comparing the dominant conformations among different systems. It provides more objective information to describe the overall conformational changes in the system. The conformational landscapes of K-Ras4BG13D•SOScat differed obviously in the presence or absence of an allosteric K-Ras4BG13D–GTP (Figure 2). Consistent with the trends shown by the RMSD and RMSF results, the conformational landscapes of the ternary system appeared to be more convergent. This indicated that the K-Ras4BG13D•SOScat complex exhibited reduced conformational dynamics upon K-Ras4BG13D–GTP binding, further suggesting the regulatory role of K-Ras4BG13D–GTP to stabilize the conformational state of K-Ras4BG13D•SOScat.
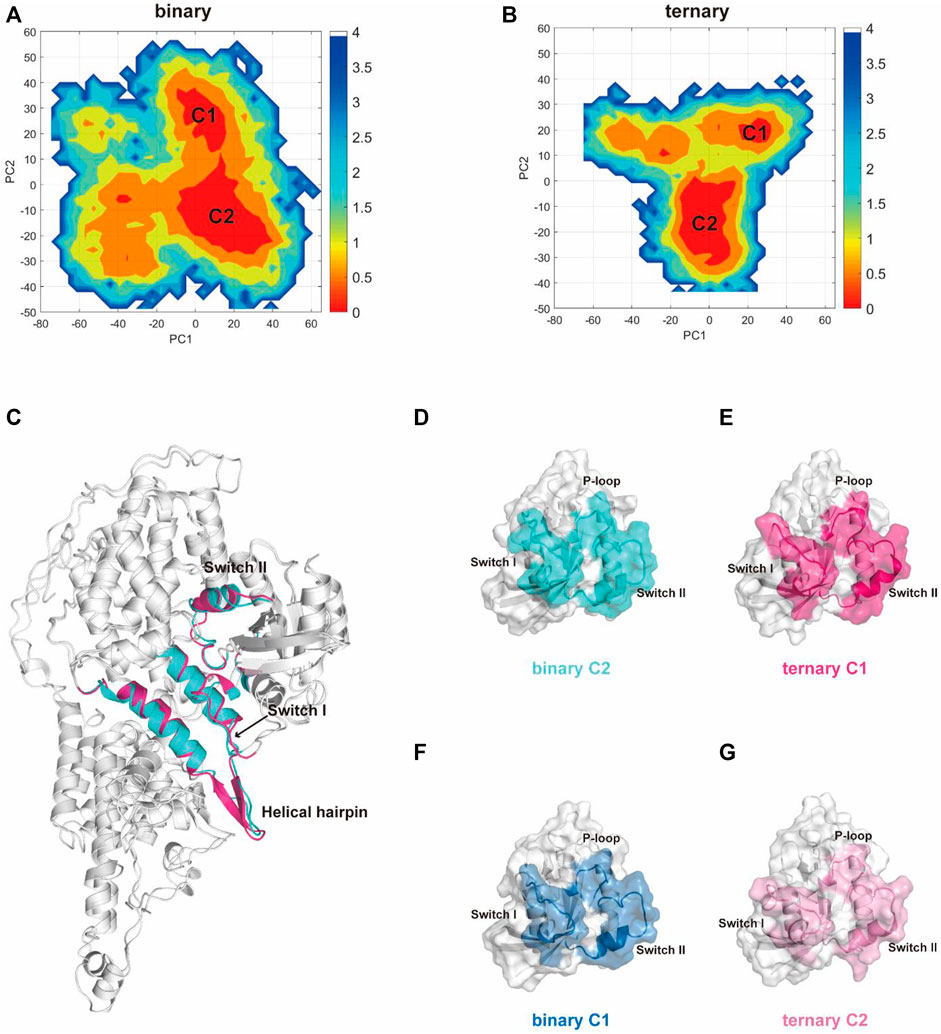
FIGURE 2. PCA analysis of the K-Ras4BG13D•SOScat complex and the representative structures of clusters C1 and C2 in the two systems. Projection of trajectories along with the first two collective principal components (PC1 and PC2) of K-Ras4BG13D•SOScat in the binary (A) and ternary (B) systems. (C) Cartoon representations of K-Ras4BG13D•SOScat in the binary C2 (cyan) and ternary C1 (hot pink). (D–G) Representative structures of K-Ras4BG13D in the binary C2 (cyan), ternary C1 (hot pink), binary C1 (blue), and ternary C2 (light pink).
There were two clusters in both binary and ternary systems. Representative structures of each cluster in the two systems were extracted using the cluster analysis, and RMSD of Cα atoms was calculated to compare the overall structural dynamics of every cluster. As shown in Table 1, the difference between the RMSD values of K-Ras4BG13D•SOScat in the binary C2 and ternary C1 was the smallest, and their structures shared similar overall conformations through superimposing and comparing the dominant conformers among all clusters (Figure 2C). In addition, the RMSD in the C2 of the ternary system was notably higher than that of the other three clusters, which reflected that a new conformation with considerable changes was formed in the ternary system upon K-Ras4BG13D–GTP binding. Significantly, we compared the overall structural similarity of K-Ras4BG13D and found a conspicuous transformation of C2 in the ternary system on the switch I and II regions (Figures 2D–G). These results indicated that K-Ras4BG13D–GTP binding initiated the conformational transitions of K-Ras4BG13D•SOScat from the binary system toward the ternary state with significant switch I and II conformational changes. Since both switch I and II regions participate in the interaction of K-Ras4BG13D with SOScat, we further monitored the difference of the K-Ras4BG13D•SOScat interfacial interaction in both binary and ternary systems. Proteins, Interfaces, Structures, and Assemblies (PISA) analyses (Krissinel and Henrick, 2007) of representative structures among every cluster showed that the binding of K-Ras4BG13D–GTP significantly strengthened the interaction of SOScat with K-Ras4BG13D through the formation of more intermolecular hydrogen bonds and salt bridges at the interface in the ternary system (Table 1).

TABLE 1. Summary of RMSDs of Cα atoms, salt bridge, and hydrogen bond numbers along the K-Ras4BG13D•SOScat interface in the binary and ternary systemsa.
3.3 Binding Free Energy Analysis
To evaluate the influence of allosteric K-Ras4BG13D–GTP on the binding free energies between the catalytic K-Ras4BG13D and SOScat, the molecular mechanics Poisson–Boltzmann surface area (MM/PBSA) was employed and the binding free energy (
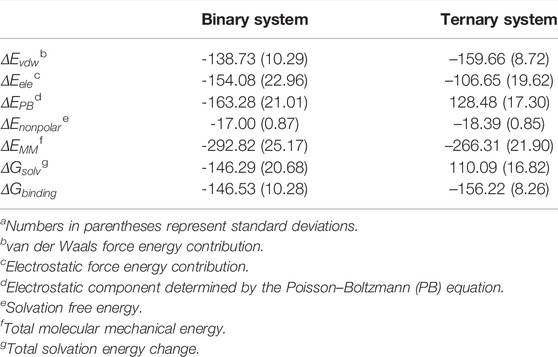
TABLE 2. Binding free energy (kcal/mol) analysis between K-Ras4BG13D and SOScat.a
Furthermore, the
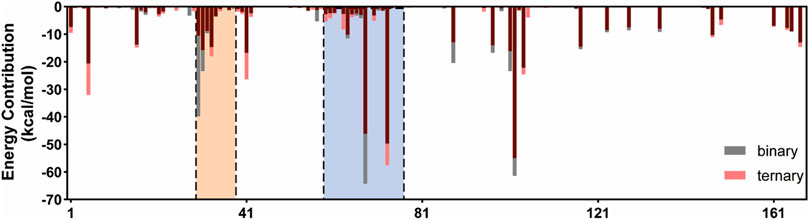
FIGURE 3. Binding free energy decomposition of the residues of Ras in the K-Ras4BG13D•SOScat complex. The switch I and switch II regions of K-Ras4BG13D are marked with orange and blue backgrounds, respectively.
3.4 Dynamics of the Interface Domain Between K-Ras4BG13D and SOScat
We further extracted the representative structures from the equilibrium stage and superimposed them between the two systems. In the switch I region of K-Ras4BG13D, there found the predominant conformational variations between the binary and ternary systems (Figure 4A). The switch I region in the ternary system distinctly stretched away from the rest of Ras and formed significant displacement for ∼6.0 Å. This indicated that the nucleotide binding pocket was expanded in the ternary system, providing space for GDP dissociation. This notion was supported by the distance distributions of Cα atoms among three pairs of inter-residue distances (G12–P34, G12–G60, and G13–E31 residues). To further evaluate the detailed contributions of P-loop, switch I and II regions to the opening of the nucleotide binding pocket, the inter-residue distances were calculated and the probability distributions were shown (Figure 4B). The distances of G12–P34 and G12–G60 pairs described the size of the phosphate binding site and D13–E31 pairs described the size of the ribose binding site (Wang et al., 2021a). The average distances of G12–P34 and D13–E31 pairs significantly increased in the ternary system, while the G12–G60 distance pair showed no obvious distinctions between the two systems. The three pairs of residue distances indicated that the space of the phosphate and ribose binding sites enlarged in the ternary system. The expanded nucleotide binding pocket could result in an increased rate of the nucleotide exchange.
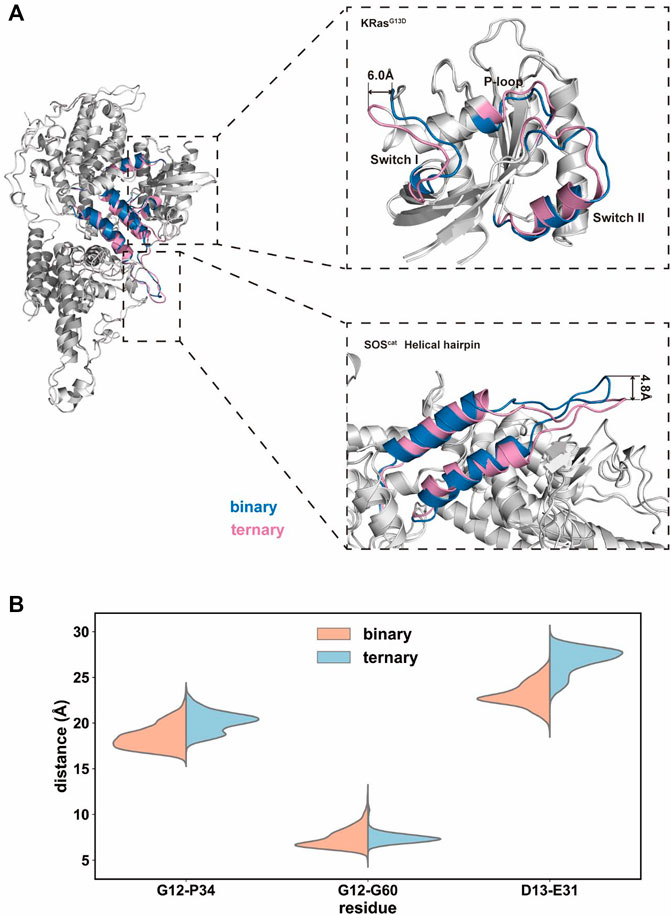
FIGURE 4. Conformational dynamics of the K-Ras4BG13D•SOScat complex in the binary and ternary systems. (A) Representative structure of the K-Ras4BG13D•SOScat complex conformation in the binary (blue) and ternary (pink) systems. (B) Distance distributions of Cα atoms between the G12–P34, G12–G60, and G13–E31 residue pairs.
Meanwhile, a prominent difference was found in the conformation of the helical hairpin between the two structures of SOScat (Figure 4A). The helical hairpin was shifted away from the active site of SOS in the ternary system, and the interdomain distance increased about ∼4.8 Å between the two systems. Consistent with previous evidence, the helical hairpin region of the nucleotide-free Ras–SOS structure constricted the site where SOS attracts the switch II region of nucleotide-free Ras (Freedman et al., 2006; Bandaru et al., 2019). In fact, the combination of Ras–GTP and SOS promoted the rotation and opening of the helical hairpin, thereby freeing the catalytic site where Ras binds to.
3.5 Insights Into the K-Ras4BG13D•SOScat Interfacial Residues
Structural comparison and free energy analysis revealed that allosteric K-Ras4BG13D–GTP binding altered the principal conformations of switch I and switch II and enhanced the K-Ras4BG13D•SOScat interaction. The detailed differences in the binding process (
There were constantly five hydrogen bonds in the binary systems for the interaction between the switch I region and SOScat domains (Figure 5). Residues Y32 and D30 at the active site on the switch I region of the nucleotide-free K-Ras4bG13D engage in polar interactions with residues N547 and K566 of SOScat and residues K566 of SOScat, respectively. Nevertheless, upon allosteric K-Ras4BG13D–GTP binding, both residues D30 and K566 were shifted away from each other, especially the significant clockwise rotation of K566, which disrupted the hydrogen bond between D30 and K566. This suggested that binding of K-Ras4BG13D–GTP impaired the restriction of SOScat on the switch I region, which may promote the opening of nucleotide binding pocket.
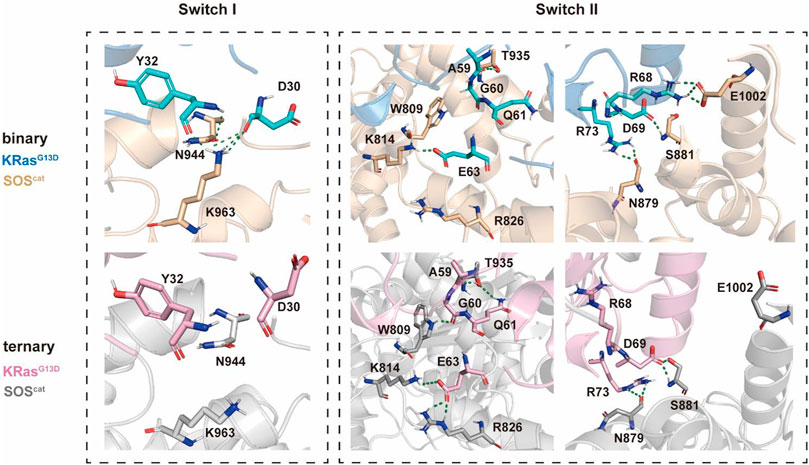
FIGURE 5. Detailed interaction patterns between K-Ras4BG13D and SOScat interfacial residues. Hydrogen bonds are marked with green dotted lines.
Consistent with the previous findings that switch II provided the main anchor for the interaction of SOS with Ras, we found more hydrogen bonds formed between the switch II region and SOScat in the ternary system. In the presence of K-Ras4BG13D–GTP at the allosteric site of SOScat, residues A59, G60, Q61, and E63 formed significantly more hydrogen bonds with residues T935, W809, K814, and R826 compared to the binary system. Meanwhile, numbers of hydrogen bonds formed at residues R68, D69, and R73 were similar between two systems. The residues of the switch II region underwent significant conformational changes, especially in which residues E63 and R73 rotated counterclockwise and displaced toward the direction of SOScat. These observations indicated that binding of K-Ras4BG13D–GTP at the distal site allosterically altered the conformations of key interfacial residues, thereby enhancing the interaction between the switch II region of K-Ras4BG13D and SOScat. The increased affinity of Ras at the catalytic site may promote the nucleotide exchange rate.
3.6 Allosteric Signaling Pathways Within K-Ras4BG13D•SOScat
3.6.1 Dynamic Cross-Correlation Matrices
We further explored how the signal triggered by K-Ras4BG13D–GTP binding at the allosteric site could allosterically regulate the activation of the catalytic K-Ras4BG13D and the interaction patterns between K-Ras4BG13D and SOScat. To determine the dynamic variation of Ras in the two systems, we analyzed the inter-residue correlations using the dynamic cross-correlation matrix calculations. Correlation coefficients were calculated among the related motions between each Cα atom in the whole trajectory composing the dynamic cross-correlation matrices, which reflected the relationship among different domains [(Ni et al., 2021), (Feng et al., 2021)]. As shown in Figures 6A,B, compared to the binary system, the intramolecular anticorrelations of K-Ras4BG13D were weakened in the ternary system, while the correlation of intramolecular motions were slightly strengthened. In each system, C1 represented the correlated movement of the helical hairpin domain of the SOScat to the switch I and switch II regions of the K-Ras4BG13D, while K-Ras4BG13D–GTP binding at the allosteric site effectively impaired the C1 correlation. Meanwhile, we investigated the correlation of the switch II region with the interface residues between SOScat and K-Ras4BG13D–GTP. C2 showed the enhanced anticorrelated movement of the switch II with the interface residues. It suggested that the binding of K-Ras4BG13D–GTP to the allosteric site of SOScat resulted in the varied interface residues in the binding site of SOScat that remotely transmitted to regulate the catalytic activity of K-Ras4BG13D.
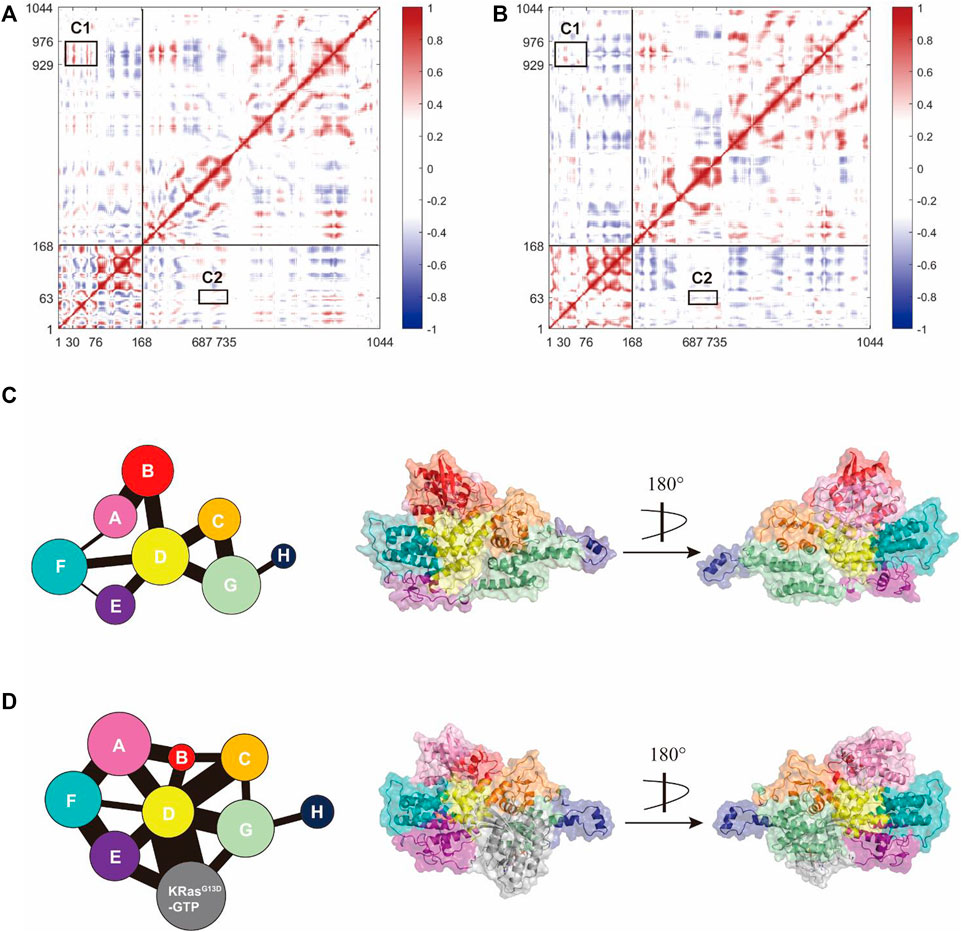
FIGURE 6. K-Ras4BG13D and SOScat allosteric interactions. The dynamic cross-correlation matrix of the binary (A) and ternary (B) systems. Positive regions (red) represent correlated motions, whereas negative regions (blue) represent anticorrelated motions. C1 shows correlations of the helical hairpin domain with the switch I and switch II region, and C2 shows anticorrelated movement of the switch II with the SOScat•K-Ras4BG13D–GTP interface residues. Correlated motions with absolute values < 0.3 were neglected and shown in white. Map of the community network in the binary (C) and ternary (D) systems. Areas of the circles represent the numbers of residues in corresponding communities and the widths of sticks connecting communities represent the intercommunity connections.
3.6.2 Community Network Analysis
Subsequently, we focused on the allosteric network from the allosteric K-Ras4BG13D–GTP to the catalytic K-Ras4BG13D in both binary and ternary systems. Given that a node was defined within a cutoff distance of 4.5 Å between the two Cα atoms populating at least 75% of the MD trajectory, these residues were categorized into the same community and were regarded as a synergistic functional unit within the protein structure (Foutch et al., 2021). There were eight communities for the K-Ras4BG13D•SOScat complex in both systems and each community was represented by colored circles, whose area was proportional to the amounts of residues contained, connecting by sticks of different width proportional to the value of edge connectivity (Figures 6C,D). In general, their distributions in the two systems were similar, but the eight communities on the K-Ras4BG13D•SOScat interface and within Ras presented prominent differences. In the binary system, the K-Ras4BG13D•SOScat interfacial residues mainly constituted the Communities B and D, while it was formed by the Communities A, B, C, D, and F in the ternary system. This suggested that considerable conformational changes occurred in a portion of residues along the K-Ras4BG13D•SOScat interface upon the allosteric K-Ras4BG13D–GTP bidning throughout simulations. Particularly, in the ternary system, the marked change in the K-Ras4BG13D was peeling away of the whole switch I region from the rest of Ras and formed the Community B with a proportion of residues from the SOScat helical hairpin domain, implying the increased distance between the switch I region and the rest of Ras upon the K-Ras4BG13D–GTP binding. It should be noted that the complete switch II region was incorporated into the Community D with partial SOScat residues, suggesting that they were in close proximity and have more interactions.
On the other hand, the edge connectivity among communities which implied the interactions between communities changed considerably upon K-Ras4BG13D–GTP binding. We mainly studied the community cross talk along the K-Ras4BG13D and SOScat interface. In the ternary system, the binding of K-Ras4BG13D–GTP introduced a new strong connection between K-Ras4BG13D and SOScat. The Communities A and B derived from the majority of the residues from K-Ras4BG13D were in direct and strong edge connections with the Communities C and D (main residues from the SOScat helical hairpin domain). Moreover, the direct information flow between the Communities A and G, representing the partial interfacial residues from SOScat, completely disappeared in the binary system. This indicated that the emerging strong edge connection may strengthen the interaction between K-Ras4BG13D and SOScat, thereby reinforced the allosteric regulation from SOScat toward the catalytic K-Ras4BG13D.
3.6.3 Allosteric Pathway Analysis
Moreover, PISA analyses involving the representative structures of the ternary system showed the critical roles of R694 and W729, I752 and I922 from SOScat upon the K-Ras4BG13D–GTP binding, since they participated in the formation of hydrogen bonds or salt bridges at the interface. We next calculated the optimal and suboptimal pathways, followed by analysis of the potential allosteric relationship from the three interfacial residues in SOScat down to the switch II region via NetworkView plugin in the VMD tool. As listed in Table 3, the pathways from K-Ras4BG13D–GTP•SOScat interface toward the switch I and switch II regions in the ternary system presented shorter lengths of the optimal pathway, that is, to say, less residues were involved in the optimal pathway and more suboptimal pathways were formed relative to the binary system. The characterization of these promoted connections highlighted the Community D (αB, αD helix, and αH helix of the Cdc25 domain) and the Community E (especially αC helix of Cdc25 domain) as a core transmission hub. Taken together, these indicated that allosteric K-Ras4BG13D–GTP binding exerted an extensive and reinforced allosteric regulation on the catalytic K-Ras4BG13D through the Cdc25 domain of the SOScat.
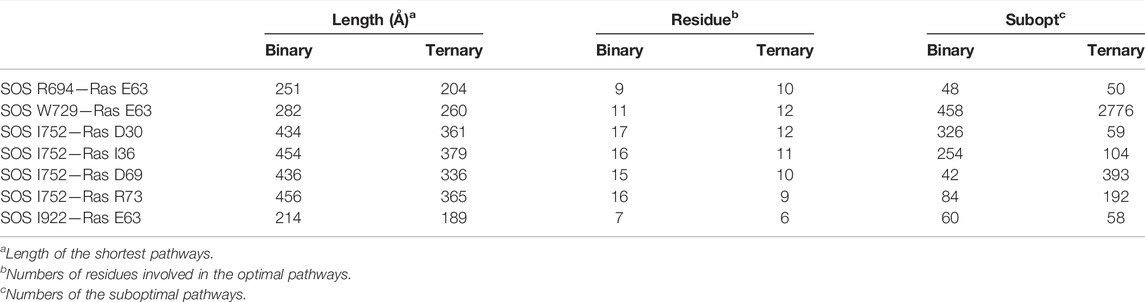
TABLE 3. Allosteric pathway analysis between the SOScat•K-Ras4BG13D–GTP interface and the switch I and switch II regions of K-Ras4BG13D.
4 Discussion
MD simulations were performed in the binary (K-Ras4BG13D•SOScat) and ternary (K-Ras4BG13D•SOScat•K-Ras4BG13D–GTP) systems to explore the underlying mechanisms driving allosteric activation of the catalytic K-Ras4BG13D through distal binding of K-Ras4BG13D–GTP at the allosteric site of SOScat. On the whole, less fluctuations of overall residues and more concentrated conformational landscapes distribution were found in the ternary system. From the perspective of structure, we found that the switch I region of K-Ras4BG13D distinctly stretched away from the rest of K-Ras4BG13D upon K-Ras4BG13D–GTP binding, which caused the expanded nucleotide binding pocket. From the perspective of energy, allosteric K-Ras4BG13D–GTP binding increased the binding free energy between the catalytic K-Ras4BG13D and SOScat by enhancing the interactions between the switch II region of K-Ras4BG13D and SOScat. We revealed the detailed mechanism of the activation process of K-Ras4BG13D with structural transformations. Meanwhile, we proposed potential pathways induced by the allosteric K-Ras4BG13D–GTP binding to convey the information of K-Ras4B activation over a long-range distance.
SOS-mediated positive feedback had been proposed to dynamically regulate Ras signaling since 2003. The SOS-catalyzed nucleotide exchange by fluorescence spectroscopy showed that H-Ras–GTP markedly increased the rate of nucleotide release from H-Ras stimulated by SOScat (Vo et al., 2016). Recently, Moghadamchargari et al. found that K-Ras4BG13D–GTP can allosterically increase the nucleotide exchange rate of K-Ras4B at the active site >2-fold compared to the K-RasWT–GTP (Moghadamchargari et al., 2021). Moreover, the positive feedback loop exists between H-Ras–GTP and SOS, increases the amplitude and duration of Ras activation after the stimulation of EGF, and leads to the higher activity of downstream proteins (Boykevisch et al., 2006). It was characterized by the sustained EGF-induced ERK phosphorylation and enhanced serum response element (SRE)-dependent transcription (Lu et al., 2016a). These data supported the positive feedback activation of SOS, but previous studies mainly focused on the activation process of SOS by Ras–GTP. Analysis of the resulting structure revealed that the binding of H-RasY64A–GppNHp at the distal binding site of SOScat had a significant impact on the conformational change of the Rem domain. This domain rotated by less than 10° relative to the Cdc25 domain, and the rotation changed the affinity of the helical hairpin of SOScat with the switch I region of nucleotide-free H-RasWT in the active site (Hall et al., 2001; Boykevisch et al., 2006). This structural feature was verified by mutant residues in the helical hairpin, which was able to stabilize the catalytically competent open conformation (Bandaru et al., 2019). We also observed a similar transformation of SOScat that the helical hairpin was moved away from the active site of SOScat upon K-Ras4BG13D–GTP binding. This suggested that the nucleotide-free Ras binding to the active site of SOScat required accommodated space providing by the pulling away of the helical hairpin.
On the other hand, more attention was focused on the process of Ras activation. Liao TJ et al. found the interaction of k-ras4b4B-GTP with SOS1 at the allosteric site induces a local conformation change at the catalytic site, facilitating the accommodation of the inactive Ras (Liao et al., 2018). According to the results, binding of allosteric K-Ras4BG13D–GTP may affect the rate-limiting step of the SOS-catalyzed nucleotide change. The switch I region of the catalytic K-Ras4BG13D in the ternary system was distinctly away from the rest of K-Ras4BG13D, and the three increased pairwise distances describing the nucleotide binding pocket both suggested the enlarged space of GDP phosphate and the ribose binding site in the ternary system. The opening of nucleotide binding site would weaken the binding affinity of GDP to the K-Ras4BG13D, promoting the release of GDP and the subsequent rebinding of GTP to the K-Ras4B active site.
In order to effectively inhibit the activation of Ras catalyzed by SOS, small molecules or peptides that bind to the Ras–SOS interface can be designed based on the Ras–SOS protein–protein interaction (Lu et al., 2016c; Lu et al., 2016d; Ni et al., 2019a; Yang et al., 2021). This may be a potential therapeutic strategy for the treatment of Ras-driven cancer (Ostrem and Shokat, 2016; Ni et al., 2019b; Lu et al., 2021b; Wang et al., 2021b). For instance, it has been reported that nSH3/cSH3 binding peptides, which effectively interrupt the Grb2–SOS interaction, can serve as tumor suppressors (Liao et al., 2020a), (Liao et al., 2020b). However, another interesting phenomenon is that small-molecule Ras•SOS disruptors fail to dissociate K-Ras4BG13D•SOScat complexes. We may explain this evidence from the perspective of binding free energy. Due to the formation of the K-Ras4BG13D•SOScat•K-Ras4BG13D–GTP ternary complex, allosteric K-Ras4BG13D–GTP binding exerted increased binding free energy between K-Ras4BG13D and SOScat. We also provided structural details to explain the observed higher binding affinity of K-Ras4BG13D for SOScat in response to allosteric K-Ras4BG13D–GTP binding. Upon K-Ras4BG13D–GTP binding, there were more salt bridges formed between the switch II region and SOScat. Furthermore, the enhanced interaction between K-Ras4BG13D and SOScat provided a basis for allosteric regulation within the ternary system. The allosteric propagation pathway was found from the K-Ras4BG13D–GTP binding site to the K-Ras4BG13D functional region. This indicated that K-RasG13D–GTP at the distal site of SOScat may regulate K-Ras4BG13D catalytic activity using allosteric modulation.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
Conceptualization, SL and YL; methodology, XH, KD and YW; validation, XL, YW and DN; formal analysis, XH and JF; investigation, XH and YW; resources, SL; data curation, XH and JF; writing—original draft preparation, XH; writing—review and editing, SL and XB; visualization, SL and YL; supervision, SL and YL; project administration, SL and funding acquisition, SL and XB. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Natural Science Foundation of China (No. 22077082).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aledavood, E., Forte, A., Estarellas, C., and Javier Luque, F. (2021). Structural Basis of the Selective Activation of Enzyme Isoforms: Allosteric Response to Activators of β1- and β2-containing AMPK Complexes. Comput. Struct. Biotechnol. J. 19, 3394–3406. doi:10.1016/j.csbj.2021.05.056
Amadei, A., Linssen, A. B. M., and Berendsen, H. J. C. (1993). Essential Dynamics of Proteins. Proteins 17 (4), 412–425. doi:10.1002/prot.340170408
An, X., Bai, Q., Bing, Z., Liu, H., and Yao, X. (2021). Insights into the Molecular Mechanism of Positive Cooperativity between Partial Agonist MK-8666 and Full Allosteric Agonist AP8 of hGPR40 by Gaussian Accelerated Molecular Dynamics (GaMD) Simulations. Comput. Struct. Biotechnol. J. 19, 3978–3989. doi:10.1016/j.csbj.2021.07.008
Bandaru, P., Kondo, Y., and Kuriyan, J. (2019). The Interdependent Activation of Son-Of-Sevenless and Ras. Cold Spring Harb Perspect. Med. 9 (2), a031534. doi:10.1101/cshperspect.a031534
Bos, J. L., Rehmann, H., and Wittinghofer, A. (2007). GEFs and GAPs: Critical Elements in the Control of Small G Proteins. Cell 129 (5), 865–877. doi:10.1016/j.cell.2007.05.018
Bourne, H. R., Sanders, D. A., and McCormick, F. (1990). The GTPase Superfamily: a Conserved Switch for Diverse Cell Functions. Nature 348 (6297), 125–132. doi:10.1038/348125a0
Boykevisch, S., Zhao, C., Sondermann, H., Philippidou, P., Halegoua, S., Kuriyan, J., et al. (2006). Regulation of Ras Signaling Dynamics by Sos-Mediated Positive Feedback. Curr. Biol. 16 (21), 2173–2179. doi:10.1016/j.cub.2006.09.033
Buday, L., and Downward, J. (2008). Many Faces of Ras Activation. Biochim. Biophys. Acta (Bba) - Rev. Cancer 1786 (2), 178–187. doi:10.1016/j.bbcan.2008.05.001
Byun, J. A., VanSchouwen, B., Akimoto, M., and Melacini, G. (2020). Allosteric Inhibition Explained through Conformational Ensembles Sampling Distinct "mixed" States. Comput. Struct. Biotechnol. J. 18, 3803–3818. doi:10.1016/j.csbj.2020.10.026
Chardin, P., Camonis, J. H., Gale, N. W., van Aelst, L., Schlessinger, J., Wigler, M. H., et al. (1993). Human Sos1: a Guanine Nucleotide Exchange Factor for Ras that Binds to GRB2. Science 260 (5112), 1338–1343. doi:10.1126/science.8493579
Cherfils, J., and Zeghouf, M. (2013). Regulation of Small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev. 93 (1), 269–309. doi:10.1152/physrev.00003.2012
Chong, L. T., Pitera, J. W., Swope, W. C., and Pande, V. S. (2009). Comparison of Computational Approaches for Predicting the Effects of Missense Mutations on P53 Function. J. Mol. Graphics Model. 27 (8), 978–982. doi:10.1016/j.jmgm.2008.12.006
Colicelli, J. (2004). Human RAS Superfamily Proteins and Related GTPases. Sci. STKE 2004 (250), RE13. doi:10.1126/stke.2502004re13
Cox, A. D., Fesik, S. W., Kimmelman, A. C., Luo, J., and Der, C. J. (2014). Drugging the Undruggable RAS: Mission Possible? Nat. Rev. Drug Discov. 13 (11), 828–851. doi:10.1038/nrd4389
Drosten, M., Dhawahir, A., Sum, E. Y. M., Urosevic, J., Lechuga, C. G., Esteban, L. M., et al. (2010). Genetic Analysis of Ras Signalling Pathways in Cell Proliferation, Migration and Survival. EMBO J. 29 (6), 1091–1104. doi:10.1038/emboj.2010.7
Eargle, J., and Luthey-Schulten, Z. (2012). NetworkView: 3D Display and Analysis of Protein{middle Dot}RNA Interaction Networks. Bioinformatics 28 (22), 3000–3001. doi:10.1093/bioinformatics/bts546
Fan, J., Liu, Y., Kong, R., Ni, D., Yu, Z., Lu, S., et al. (2021). Harnessing Reversed Allosteric Communication: A Novel Strategy for Allosteric Drug Discovery. J. Med. Chem. 64 (24), 17728–17743. doi:10.1021/acs.jmedchem.1c01695
Feng, L., Lu, S., Zheng, Z., Chen, Y., Zhao, Y., Song, K., et al. (2021). Identification of an Allosteric Hotspot for Additive Activation of PPARγ in Antidiabetic Effects. Sci. Bull. 66 (15), 1559–1570. doi:10.1016/j.scib.2021.01.023
Fernandez-Medarde, A., and Santos, E. (2011). Ras in Cancer and Developmental Diseases. Genes & Cancer 2 (3), 344–358. doi:10.1177/1947601911411084
Foutch, D., Pham, B., and Shen, T. (2021). Protein Conformational Switch Discerned via Network Centrality Properties. Comput. Struct. Biotechnol. J. 19, 3599–3608. doi:10.1016/j.csbj.2021.06.004
Freedman, T. S., Sondermann, H., Friedland, G. D., Kortemme, T., Bar-Sagi, D., Marqusee, S., et al. (2006). A Ras-Induced Conformational Switch in the Ras Activator Son of Sevenless. Proc. Natl. Acad. Sci. 103 (45), 16692–16697. doi:10.1073/pnas.0608127103
Goitre, L., Trapani, E., Trabalzini, L., and Retta, S. F. (2014). The Ras Superfamily of Small GTPases: the Unlocked Secrets. Methods Mol. Biol. 1120, 1–18. doi:10.1007/978-1-62703-791-4_1
Goody, R. S., Frech, M., and Wittinghofer, A. (1991). Affinity of Guanine Nucleotide Binding Proteins for Their Ligands: Facts and Artefacts. Trends Biochem. Sci. 16 (9), 327–328. doi:10.1016/0968-0004(91)90134-h
Gureasko, J., Galush, W. J., Boykevisch, S., Sondermann, H., Bar-Sagi, D., Groves, J. T., et al. (2008). Membrane-dependent Signal Integration by the Ras Activator Son of Sevenless. Nat. Struct. Mol. Biol. 15 (5), 452–461. doi:10.1038/nsmb.1418
Hall, B. E., Yang, S. S., Boriack-Sjodin, P. A., Kuriyan, J., and Bar-Sagi, D. (2001). Structure-based Mutagenesis Reveals Distinct Functions for Ras Switch 1 and Switch 2 in Sos-Catalyzed Guanine Nucleotide Exchange. J. Biol. Chem. 276 (29), 27629–27637. doi:10.1074/jbc.m101727200
He, X., Huang, N., Qiu, Y., Zhang, J., Liu, Y., Yin, X. L., et al. (2021). Conformational Selection Mechanism Provides Structural Insights into the Optimization of APC-Asef Inhibitors. Molecules 26 (4), 962. doi:10.3390/molecules26040962
Hernández-Alvarez, L., Oliveira Jr, A. B., Hernández-González, J. E., Chahine, J., Pascutti, P. G., de Araujo, A. S., et al. (2021). Computational Study on the Allosteric Mechanism of Leishmania Major IF4E-1 by 4E-Interacting Protein-1: Unravelling the Determinants of m7GTP Cap Recognition. Comput. Struct. Biotechnol. J. 19, 2027–2044. doi:10.1016/j.csbj.2021.03.036
Jang, H., Zhang, M., and Nussinov, R. (2020). The Quaternary Assembly of KRas4B with Raf-1 at the Membrane. Comput. Struct. Biotechnol. J. 18, 737–748. doi:10.1016/j.csbj.2020.03.018
Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W., and Klein, M. L. (1983). Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 79 (2), 926–935. doi:10.1063/1.445869
Kazemein Jasemi, N. S., Herrmann, C., Magdalena Estirado, E., Gremer, L., Willbold, D., Brunsveld, L., et al. (2021). The Intramolecular Allostery of GRB2 Governing its Interaction with SOS1 Is Modulated by Phosphotyrosine Ligands. Biochem. J. 478 (14), 2793–2809. doi:10.1042/bcj20210105
Krissinel, E., and Henrick, K. (2007). Inference of Macromolecular Assemblies from Crystalline State. J. Mol. Biol. 372 (3), 774–797. doi:10.1016/j.jmb.2007.05.022
Lenzen, C., Cool, R. H., Prinz, H., Kuhlmann, J., and Wittinghofer, A. (1998). Kinetic Analysis by Fluorescence of the Interaction between Ras and the Catalytic Domain of the Guanine Nucleotide Exchange Factor Cdc25Mm. Biochemistry 37 (20), 7420–7430. doi:10.1021/bi972621j
Li, X., Dai, J., Ni, D., He, X., Zhang, H., Zhang, J., et al. (2020). Insight into the Mechanism of Allosteric Activation of PI3Kα by Oncoprotein K-Ras4B. Int. J. Biol. Macromolecules 144, 643–655. doi:10.1016/j.ijbiomac.2019.12.020
Li, X., Wang, C., Peng, T., Chai, Z., Ni, D., Liu, Y., et al. (2021). Atomic-scale Insights into Allosteric Inhibition and Evolutional rescue Mechanism of Streptococcus Thermophilus Cas9 by the Anti-CRISPR Protein AcrIIA6. Comput. Struct. Biotechnol. J. 19, 6108–6124. doi:10.1016/j.csbj.2021.11.010
Li, Y., Cong, Y., Feng, G., Zhong, S., Zhang, J. Z. H., Sun, H., et al. (2018). The Impact of interior Dielectric Constant and Entropic Change on HIV-1 Complex Binding Free Energy Prediction. Struct. Dyn. 5 (6), 064101. doi:10.1063/1.5058172
Liang, S., Wang, Q., Qi, X., Liu, Y., Li, G., Lu, S., et al. (2021). Deciphering the Mechanism of Gilteritinib Overcoming Lorlatinib Resistance to the Double Mutant I1171N/F1174I in Anaplastic Lymphoma Kinase. Front. Cel Dev. Biol. 9, 808864. doi:10.3389/fcell.2021.808864
Liang, Z., Zhu, Y., Long, J., Ye, F., and Hu, G. (2020). Both intra and Inter-domain Interactions Define the Intrinsic Dynamics and Allosteric Mechanism in DNMT1s. Comput. Struct. Biotechnol. J. 18, 749–764. doi:10.1016/j.csbj.2020.03.016
Liao, T.-J., Jang, H., Fushman, D., and Nussinov, R. (2018). Allosteric KRas4B Can Modulate SOS1 Fast and Slow Ras Activation Cycles. Biophysical J. 115 (4), 629–641. doi:10.1016/j.bpj.2018.07.016
Liao, T.-J., Jang, H., Fushman, D., and Nussinov, R. (2020). SOS1 Interacts with Grb2 through Regions that Induce Closed nSH3 Conformations. J. Chem. Phys. 153 (4), 045106. doi:10.1063/5.0013926
Liao, T.-J., Jang, H., Nussinov, R., and Fushman, D. (2020). High-Affinity Interactions of the nSH3/cSH3 Domains of Grb2 with the C-Terminal Proline-Rich Domain of SOS1. J. Am. Chem. Soc. 142 (7), 3401–3411. doi:10.1021/jacs.9b10710
Liu, Q., Wang, Y., Leung, E. L.-H., and Yao, X. (2021). In Silico study of Intrinsic Dynamics of Full-Length Apo-ACE2 and RBD-ACE2 Complex. Comput. Struct. Biotechnol. J. 19, 5455–5465. doi:10.1016/j.csbj.2021.09.032
Lu, S., Banerjee, A., Jang, H., Zhang, J., Gaponenko, V., and Nussinov, R. (2015). GTP Binding and Oncogenic Mutations May Attenuate Hypervariable Region (HVR)-Catalytic Domain Interactions in Small GTPase K-Ras4B, Exposing the Effector Binding Site. J. Biol. Chem. 290 (48), 28887–28900. doi:10.1074/jbc.m115.664755
Lu, S., Chen, Y., Wei, J., Zhao, M., Ni, D., He, X., et al. (2021). Mechanism of Allosteric Activation of SIRT6 Revealed by the Action of Rationally Designed Activators. Acta Pharmaceutica Sinica B 11 (5), 1355–1361. doi:10.1016/j.apsb.2020.09.010
Lu, S., He, X., Ni, D., and Zhang, J. (2019). Allosteric Modulator Discovery: From Serendipity to Structure-Based Design. J. Med. Chem. 62 (14), 6405–6421. doi:10.1021/acs.jmedchem.8b01749
Lu, S., He, X., Yang, Z., Chai, Z., Zhou, S., Wang, J., et al. (2021). Activation Pathway of a G Protein-Coupled Receptor Uncovers Conformational Intermediates as Targets for Allosteric Drug Design. Nat. Commun. 12 (1), 4721. doi:10.1038/s41467-021-25020-9
Lu, S., Jang, H., Gu, S., Zhang, J., and Nussinov, R. (2016). Drugging Ras GTPase: a Comprehensive Mechanistic and Signaling Structural View. Chem. Soc. Rev. 45 (18), 4929–4952. doi:10.1039/c5cs00911a
Lu, S., Jang, H., Muratcioglu, S., Gursoy, A., Keskin, O., Nussinov, R., et al. (2016). Ras Conformational Ensembles, Allostery, and Signaling. Chem. Rev. 116 (11), 6607–6665. doi:10.1021/acs.chemrev.5b00542
Lu, S., Jang, H., Nussinov, R., and Zhang, J. (2016). The Structural Basis of Oncogenic Mutations G12, G13 and Q61 in Small GTPase K-Ras4B. Sci. Rep. 6, 21949. doi:10.1038/srep21949
Lu, S., Jang, H., Zhang, J., and Nussinov, R. (2016). Inhibitors of Ras-SOS Interactions. ChemMedChem 11 (8), 814–821. doi:10.1002/cmdc.201500481
Lu, S., Ni, D., Wang, C., He, X., Lin, H., Wang, Z., et al. (2019). Deactivation Pathway of Ras GTPase Underlies Conformational Substates as Targets for Drug Design. ACS Catal. 9 (8), 7188–7196. doi:10.1021/acscatal.9b02556
Maier, J. A., Martinez, C., Kasavajhala, K., Wickstrom, L., Hauser, K. E., and Simmerling, C. (2015). ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theor. Comput. 11 (8), 3696–3713. doi:10.1021/acs.jctc.5b00255
Marasco, M., Kirkpatrick, J., Nanna, V., Sikorska, J., and Carlomagno, T. (2021). Phosphotyrosine Couples Peptide Binding and SHP2 Activation via a Dynamic Allosteric Network. Comput. Struct. Biotechnol. J. 19, 2398–2415. doi:10.1016/j.csbj.2021.04.040
Margarit, S. M., Sondermann, H., Hall, B. E., Nagar, B., Hoelz, A., Pirruccello, M., et al. (2003). Structural Evidence for Feedback Activation by Ras·GTP of the Ras-specific Nucleotide Exchange Factor SOS. Cell 112 (5), 685–695. doi:10.1016/s0092-8674(03)00149-1
Milburn, M. V., Tong, L., deVos, A. M., Brünger, A., Yamaizumi, Z., Nishimura, S., et al. (1990). Molecular Switch for Signal Transduction: Structural Differences between Active and Inactive Forms of Protooncogenic Ras Proteins. Science 247 (4945), 939–945. doi:10.1126/science.2406906
Mitin, N., Rossman, K. L., and Der, C. J. (2005). Signaling Interplay in Ras Superfamily Function. Curr. Biol. 15 (14), R563–R574. doi:10.1016/j.cub.2005.07.010
Moghadamchargari, Z., Shirzadeh, M., Liu, C., Schrecke, S., Packianathan, C., Russell, D. H., et al. (2021). Molecular Assemblies of the Catalytic Domain of SOS with KRas and Oncogenic Mutants. Proc. Natl. Acad. Sci. U S A. 118 (12), e2022403118. doi:10.1073/pnas.2022403118
Newman, M. E. J. (2006). Modularity and Community Structure in Networks. Proc. Natl. Acad. Sci. 103 (23), 8577–8582. doi:10.1073/pnas.0601602103
Ni, D., Wei, J., He, X., Rehman, A. U., Li, X., Qiu, Y., et al. (2020). Discovery of Cryptic Allosteric Sites Using Reversed Allosteric Communication by a Combined Computational and Experimental Strategy. Chem. Sci. 12 (1), 464–476. doi:10.1039/d0sc05131d
Ni, D., Chai, Z., Wang, Y., Li, M., Yu, Z., Liu, Y., et al. (2021). Along the Allostery Stream: Recent Advances in Computational Methods for Allosteric Drug Discovery. Wires Comput. Mol. Sci. doi:10.1002/wcms.1585
Ni, D., Li, X., He, X., Zhang, H., Zhang, J., and Lu, S. (2019). Drugging K-RasG12C through Covalent Inhibitors: Mission Possible? Pharmacol. Ther. 202, 1–17. doi:10.1016/j.pharmthera.2019.06.007
Ni, D., Liu, D., Zhang, J., and Lu, S. (2018). Computational Insights into the Interactions between Calmodulin and the c/nSH2 Domains of P85α Regulatory Subunit of PI3Kα: Implication for PI3Kα Activation by Calmodulin. Ijms 19 (1), 151. doi:10.3390/ijms19010151
Ni, D., Lu, S., and Zhang, J. (2019). Emerging Roles of Allosteric Modulators in the Regulation of Protein‐protein Interactions (PPIs): A New Paradigm for PPI Drug Discovery. Med. Res. Rev. 39 (6), 2314–2342. doi:10.1002/med.21585
Ni, D., Song, K., Zhang, J., and Lu, S. (2017). Molecular Dynamics Simulations and Dynamic Network Analysis Reveal the Allosteric Unbinding of Monobody to H-Ras Triggered by R135K Mutation. Ijms 18 (11), 2249. doi:10.3390/ijms18112249
Okeke, C. J., Musyoka, T. M., Sheik Amamuddy, O., Barozi, V., and Tastan Bishop, Ö. (2021). Allosteric Pockets and Dynamic Residue Network Hubs of Falcipain 2 in Mutations Including Those Linked to Artemisinin Resistance. Comput. Struct. Biotechnol. J. 19, 5647–5666. doi:10.1016/j.csbj.2021.10.011
Ostrem, J. M. L., and Shokat, K. M. (2016). Direct Small-Molecule Inhibitors of KRAS: from Structural Insights to Mechanism-Based Design. Nat. Rev. Drug Discov. 15 (11), 771–785. doi:10.1038/nrd.2016.139
Pal, A., Curtin, J. F., and Kinsella, G. K. (2021). Structure Based Prediction of a Novel GPR120 Antagonist Based on Pharmacophore Screening and Molecular Dynamics Simulations. Comput. Struct. Biotechnol. J. 19, 6050–6063. doi:10.1016/j.csbj.2021.11.005
Parker, J. A., and Mattos, C. (2018). The K-Ras, N-Ras, and H-Ras Isoforms: Unique Conformational Preferences and Implications for Targeting Oncogenic Mutants. Cold Spring Harb Perspect. Med. 8 (8), a031427. doi:10.1101/cshperspect.a031427
Prior, I. A., Lewis, P. D., and Mattos, C. (2012). A Comprehensive Survey of Ras Mutations in Cancer. Cancer Res. 72 (10), 2457–2467. doi:10.1158/0008-5472.can-11-2612
Qiu, Y., Wang, Y., Chai, Z., Ni, D., Li, X., Pu, J., et al. (2021). Targeting RAS Phosphorylation in Cancer Therapy: Mechanisms and Modulators. Acta Pharmaceutica Sinica B 11 (11), 3433–3446. doi:10.1016/j.apsb.2021.02.014
Qiu, Y., Yin, X., Li, X., Wang, Y., Fu, Q., Huang, R., et al. (2021). Untangling Dual-Targeting Therapeutic Mechanism of Epidermal Growth Factor Receptor (EGFR) Based on Reversed Allosteric Communication. Pharmaceutics 13 (5), 747. doi:10.3390/pharmaceutics13050747
Rojas, J. M., Oliva, J. L., and Santos, E. (2011). Mammalian Son of Sevenless Guanine Nucleotide Exchange Factors: Old Concepts and New Perspectives. Genes & Cancer 2 (3), 298–305. doi:10.1177/1947601911408078
Ryckaert, J.-P., Ciccotti, G., and Berendsen, H. J. C. (1977). Numerical Integration of the Cartesian Equations of Motion of a System with Constraints: Molecular Dynamics of N-Alkanes. J. Comput. Phys. 23 (3), 327–341. doi:10.1016/0021-9991(77)90098-5
Shao, J., Tanner, S. W., Thompson, N., and Cheatham, T. E. (2007). Clustering Molecular Dynamics Trajectories: 1. Characterizing the Performance of Different Clustering Algorithms. J. Chem. Theor. Comput. 3 (6), 2312–2334. doi:10.1021/ct700119m
Shibata, T., Iwasaki, W., and Hirota, K. (2020). The Intrinsic Ability of Double-Stranded DNA to Carry Out D-Loop and R-Loop Formation. Comput. Struct. Biotechnol. J. 18, 3350–3360. doi:10.1016/j.csbj.2020.10.025
Simanshu, D. K., Nissley, D. V., and McCormick, F. (2017). RAS Proteins and Their Regulators in Human Disease. Cell 170 (1), 17–33. doi:10.1016/j.cell.2017.06.009
Sondermann, H., Soisson, S. M., Boykevisch, S., Yang, S.-S., Bar-Sagi, D., and Kuriyan, J. (2004). Structural Analysis of Autoinhibition in the Ras Activator Son of Sevenless. Cell 119 (3), 393–405. doi:10.1016/j.cell.2004.10.005
Stephen, A. G., Esposito, D., Bagni, R. K., and McCormick, F. (2014). Dragging Ras Back in the Ring. Cancer Cell 25 (3), 272–281. doi:10.1016/j.ccr.2014.02.017
Vetter, I. R., and Wittinghofer, A. (2001). The Guanine Nucleotide-Binding Switch in Three Dimensions. Science 294 (5545), 1299–1304. doi:10.1126/science.1062023
Vo, U., Vajpai, N., Flavell, L., Bobby, R., Breeze, A. L., Embrey, K. J., et al. (2016). Monitoring Ras Interactions with the Nucleotide Exchange Factor Son of Sevenless (Sos) Using Site-specific NMR Reporter Signals and Intrinsic Fluorescence. J. Biol. Chem. 291 (4), 1703–1718. doi:10.1074/jbc.m115.691238
Wang, J., Wolf, R. M., Caldwell, J. W., Kollman, P. A., and Case, D. A. (2004). Development and Testing of a General Amber Force Field. J. Comput. Chem. 25 (9), 1157–1174. doi:10.1002/jcc.20035
Wang, L., Zheng, G., Liu, X., Ni, D., He, X., Cheng, J., et al. (2019). Molecular Dynamics Simulations Provide Insights into the Origin of Gleevec's Selectivity toward Human Tyrosine Kinases. J. Biomol. Struct. Dyn. 37 (10), 2733–2744. doi:10.1080/07391102.2018.1496139
Wang, Q., Chen, Y., Li, S., Yang, W., Sun, L., Jang, M., et al. (2020). Ca2+-based Allosteric Switches and Shape Shifting in RGLG1 VWA Domain. Comput. Struct. Biotechnol. J. 18, 821–833. doi:10.1016/j.csbj.2020.03.023
Wang, X., Ni, D., Liu, Y., and Lu, S. (2021). Rational Design of Peptide-Based Inhibitors Disrupting Protein-Protein Interactions. Front. Chem. 9, 682675. doi:10.3389/fchem.2021.682675
Wang, Y., Ji, D., Lei, C., Chen, Y., Qiu, Y., Li, X., et al. (2021). Mechanistic Insights into the Effect of Phosphorylation on Ras Conformational Dynamics and its Interactions with Cell Signaling Proteins. Comput. Struct. Biotechnol. J. 19, 1184–1199. doi:10.1016/j.csbj.2021.01.044
Wang, Y., Li, M., Liang, W., Shi, X., Fan, J., Kong, R., et al. (2022). Delineating the Activation Mechanism and Conformational Landscape of a Class B G Protein-Coupled Receptor Glucagon Receptor. Comput. Struct. Biotechnol. J. 20, 628–639. doi:10.1016/j.csbj.2022.01.015
Yang, X., Zhong, J., Zhang, Q., Feng, L., Zheng, Z., Zhang, J., et al. (2021). Advances and Insights of APC-Asef Inhibitors for Metastatic Colorectal Cancer Therapy. Front. Mol. Biosci. 8, 662579. doi:10.3389/fmolb.2021.662579
Keywords: K-Ras4B, SOS, allosteric regulation, molecular dynamics simulation, small-molecule interactions
Citation: He X, Du K, Wang Y, Fan J, Li M, Ni D, Lu S, Bian X and Liu Y (2022) Autopromotion of K-Ras4B Feedback Activation Through an SOS-Mediated Long-Range Allosteric Effect. Front. Mol. Biosci. 9:860962. doi: 10.3389/fmolb.2022.860962
Received: 24 January 2022; Accepted: 01 March 2022;
Published: 08 April 2022.
Edited by:
Alessandro Pandini, Brunel University London, United KingdomReviewed by:
Ruth Nussinov, National Cancer Institute (NIH), United StatesThomas Simonson, École Polytechnique, France
Copyright © 2022 He, Du, Wang, Fan, Li, Ni, Lu, Bian and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shaoyong Lu, lushaoyong@sjtu.edu.cn; Xiaolan Bian, bxl40338@rjh.com.cn; Yaqin Liu, liuyaqing7811@126.com
†These authors have contributed equally to this work
 Xuan He
Xuan He Kui Du2†
Kui Du2†  Shaoyong Lu
Shaoyong Lu