Application Potential of CTHRC1 as a Diagnostic and Prognostic Indicator for Colon Adenocarcinoma
- 1Department of Oncology and Southwest Cancer Centre, Southwest Hospital, Third Military Medical University (Army Medical University), Chongqing, China
- 2Department of Dermatology, The First Affiliated Hospital of Chongqing Medical University, Chongqing, China
Colon adenocarcinoma (COAD), ranking third in incidence and second in mortality, is one of the most common cancer types in the world. The initial stages of COAD usually show no obvious clinical symptoms; moreover, effective screening or diagnostic indicators with high sensitivity and specificity are lacking, which often leads to missed treatment opportunities. Collagen triple helix repeat containing 1 (CTHRC1) is a glycosylated protein secreted during tissue repair, which reduces collagen matrix deposition and promotes cell migration. Under physiological conditions, the expression of CTHRC1 is conducive to wound healing; however, the pathological overexpression of CTHRC1 promotes tumour growth and proliferation. In this study, we evaluated the application potential of CTHRC1 as an early diagnosis and prognostic survival monitoring biomarker for COAD in addition to unravelling its molecular mechanism in the development of COAD and exploring new therapeutic targets. Therefore, various tumour databases were used to investigate the expression of CTHRC1 in COAD at the mRNA and protein levels. CTHRC1 expression was found to be significantly increased in COAD, regardless of clinical cancer stage, age, sex or race. Moreover, CTHRC1 expression was significantly correlated with poor prognosis and positively correlated with CD8+ T cell, CD4+ T cell, neutrophil, macrophage and dendritic cell infiltration. The relevant function pathways and neighbouring proteins to CTHRC1 in COAD were identified as ROR2, VAPA, LY6E and several collagen family proteins. Therefore, this study suggests that CTHRC1 is a potential diagnostic and prognostic biomarker for patients with COAD.
Introduction
A recent study by the American Cancer Society reported that colorectal cancer has the third highest (10.0%) incidence rate after female breast cancer (11.7%) and lung cancer (11.4%) (Sung et al., 2021). The incidence rate of colorectal cancer in developed countries is approximately four times higher than that in developing countries (Fidler et al., 2016). Based on the current prediction models, the global incidence of colorectal cancer has been estimated to reach approximately 2.2 million new cases/year by 2030, accounting for 20% of all patients with cancer (Araghi et al., 2018). Colorectal carcinoma has the second-highest death rate of 9.4% after lung cancer. The official statistics on the prognosis of patients with colorectal cancer published by the American Cancer Society report that the 5 year survival rate is approximately 64% (Sung et al., 2021). Specifically, early diagnosis or detection of colorectal cancer at stage I, stage IIA or IIB increases the 5 years survival rate to approximately 90%; however, delayed diagnosis decreases the survival rate of patients with pathological stage IV tumour to approximately 14% (Pilonis et al., 2020). Although the current screening of colorectal cancer has been strengthened, resulting in a slight decrease in late diagnosis case numbers, up to 90% of cases are diagnosed after symptoms appear (Vanessa and Karen, 2016).
The occurrence of colorectal cancer is attributed to complex genetic and environmental factor interactions, involving multiple genes at different stages. The main pathways include the chromosomal instability pathway, CpG island methylator phenotype pathway, microsatellite instability pathway and the serrated pathway (Harrison and Benziger, 2011). Colon adenocarcinoma (COAD) is the main manifestation of colorectal cancer, with more than 80% of colorectal cancers being diagnosed as COAD (Li and Gu, 2005). The initial stage of COAD usually shows no obvious clinical symptoms, and the lack of effective screening or highly sensitive and specific diagnostic indicators often lead to missed treatment opportunities (Garborg et al., 2013). Therefore, it is crucial to establish an effective screening mechanism to improve the early diagnosis rate.
Collagen triple helix repeat containing 1 (CTHRC1) protein was first identified in the injured arteries of rats as an extracellular secretory protein, which is expressed in the injured part and smooth muscle cells of neointima, mainly promoting the growth and proliferation of newly generated cells (Leclair and Lindner, 2007). CTHRC1 regulates the occurrence and development of cervical, pancreatic and liver carcinoma by participating in cell proliferation, cell migration, type I collagen synthesis and damaged vascular repair (Tameda et al., 2014). Studies have shown that promoting CTHRC1 expression increased the migration and invasion of primary gastrointestinal stromal tumour cells, whereas silencing CTHRC1 expression inhibited the epithelial–mesenchymal transformation of glioblastoma cells (Ma et al., 2014). CTHRC1 promotes the proliferation of colorectal cancer by activating the Wnt/PCP signalling pathway (Yang et al., 2015). Additionally, CTHRC1 plays an important role in the pathogenesis of systemic lupus erythematosus and other diseases (Wu et al., 2018).
This study, therefore, aimed to evaluate the application potential of CTHRC1 as an early diagnosis and prognostic survival monitoring biomarker for COAD. Additionally, the molecular mechanism of COAD occurrence and development along with various novel therapeutic targets were explored. Therefore, the expression of CTHRC1 in COAD at the mRNA and protein levels was investigated using various tumour databases. By evaluating the expression of CTHRC1 in patients with COAD under different physiological and pathological conditions, the application potential of CTHRC1 as a diagnostic indicator was determined. Moreover, the effects of different expression levels and genetic mutations of CTHRC1 on COAD survival rate was analysed, including the immune infiltration of CTHRC1 in COAD. Further, the associated proteins and pathways of CTHRC1 in tumorigenesis are also discussed.
Materials and Methods
Oncomine Analysis
The Oncomine platform (https://www.oncomine.org/) is a publicly accessible, online tumour-related gene microarray database that collects disease-related gene expression profiles and relevant clinical information. The expression level of CTHRC1 in different cancers was investigated via Oncomine. When compared to corresponding normal tissues, the transcriptional levels were considered statistically significant at fold change >1.5 and p-value < 0.001. The threshold value of gene rank was set to “top 10%”, and the data type was set to “mRNA” (Rhodes et al., 2004).
TIMER 2.0 Analysis
TIMER 2.0 (http://timer.comp-genomics.org/) was employed to investigate the expression levels of CTHRC1 in various tumour tissues. Additionally, data of 32 tumour types from more than 10,000 samples were collected from the TCGA database and used for immune infiltration analysis via TIMER 2.0, which ascertains the abundance of tumour infiltrates based on gene expression levels. CTHRC1 was chosen as the input and tumour cells were detected under the Immune Association module. B cells, CD8+ T cells, CD4+ T cells, neutrophils, macrophages and dendritic cells were selected as the test types based on the study by Li et al. and Danaher et al. (Li et al., 2016; Danaher et al., 2017). Gene expression values were converted to Log2 RNA-Seq by Expectation-Maximization values.
Human Protein Atlas Analysis
The Human Protein Atlas (https://www.proteinatlas.org) is an online dataset that collects the expression characteristics of various functional proteins via immunohistochemistry from tumours and corresponding normal tissues (Asplund et al., 2012). The Human Protein Atlas was used to compare the expression of CTHRC1 proteins in normal and COAD tissues using the images of immunohistochemical staining.
UALCAN Analysis
UALCAN (http://ualcan.path.uab.edu) is an open-access web platform that contains cancer-related clinical data, which can be obtained from the TCGA database. This was used to compare the CTHRC1 expression levels between the COAD and normal tissues, along with the relationship between gene expression and pathologic features in these tissues (Chandrashekar et al., 2017). In the website, TCGA was chosen, and the corresponding tumour type was selected. The student’s t-test was used to compare the transcription levels of CTHRC1 between the tissue types, and p < 0.05 was considered statistically significant.
GEPIA Analysis
GEPIA (http://gepia.cancer-pku.cn/index.html) was used to analyse the relationship between CTHRC1 expression and overall survival (OS) or disease-free survival (DFS) prognosis in patients with COAD based on the parameters of hazard ratios and log-rank p-values. After logging onto the database, Single Gene Analysis was firstly chosen. After entering CTHRC1, “survival plots” was selected. Survival analysis was performed using the following parameters: Group Cut-off: Median; Hazards Ratio: Yes; 95% Confidence Interval: Yes (Tang et al., 2017).
cBioPortal Analysis
cBioPortal (http://www.cbioportal.org/) was used to analyse the alteration frequency of CTHRC1 gene mutations. Putative copy-number calls on 478 cases were determined using GISTIC 2.0. In the module Comparison/Survival, the influence of the alterations on prognostic survival in patients with COAD was analysed using default parameters (Gao et al., 2013).
Functional Analysis
GeneMANIA was used to identify the physical interaction and co-expression of CTHRC1 with 20 related proteins using the Homo sapiens datasets with default parameters (Mostafavi et al., 2008). GO enrichment and KEGG pathway analyses (FDR cutoff <0.05) of related gene were conducted using ShinyGO v0.741 (http://bioinformatics.sdstate.edu/go/).
Results
Expression of CTHRC1 in COAD
RNA-seq data extracted from TCGA database showed a consistent trend of abnormally high CTHRC1 expression in more than 16 types of tumour tissues compared with the corresponding normal tissues, such as COAD, breast invasive carcinoma and stomach adenocarcinoma (Figure 1A). Similarly, Oncomine analysis of the pathological samples showed that the transcriptional levels of CTHRC1 mRNA were significantly up-regulated in various cancer types including colorectal cancer (Figure 1B). Further comparison of the expression levels of CTHRC1 protein between the normal and COAD tissue using immunohistochemical data from the Human Protein Atlas dataset showed that CTHRC1 protein expression in COAD was consistent with the mRNA detected (Figure 2). These findings strongly suggest the positive role of CTHRC1 in COAD tumorigenesis.
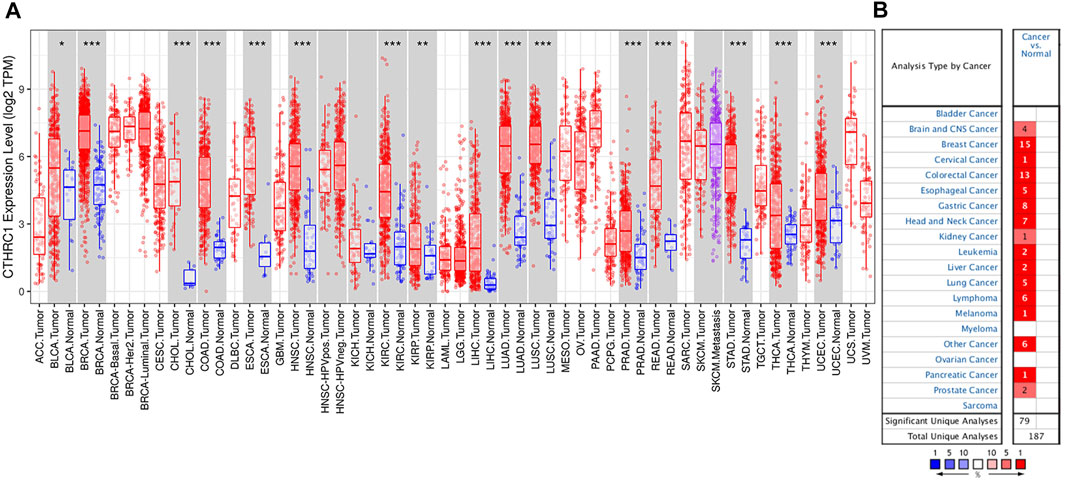
FIGURE 1. Expression levels of CTHRC1 in different types of tumour tissues. (A) CTHRC1 expression in various tumour tissues via TIMER analysis. (B) CTHRC1 expression in different cancer types via Oncomine analysis (*p < 0.05; **p < 0.01; ***p < 0.001).
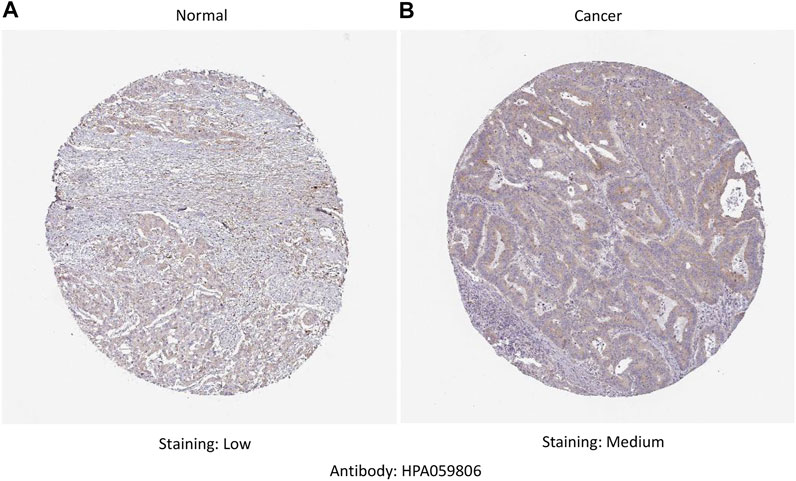
FIGURE 2. Analysis of CTHRC1 protein expression in colon adenocarcinoma tissue collected from the Human Protein Atlas dataset. (A) The control (normal) tissue with low staining; (B) The colon adenocarcinoma tissue with medium staining. Primary antibody: HPA059806.
Expression of CTHRC1 in Patients With COAD Under Different Physiological or Pathological States
UALCAN analysis showed that the expression level of CTHRC1 was significantly higher in the patients with primary COAD than that in normal tissues (Figure 3A). Notably, this abnormally high expression pattern is generally applicable to patients with different clinicopathological characteristics, such as clinical cancer stage, age, sex or race. However, no significant difference in expression level was observed among patients diagnosed with different states, apart from the high expression in patients aged 21–40 years (Figures 3B–F). Importantly, CTHRC1 showed a strong abnormal expression in patients with early stage COAD, i.e., CTHRC1 has shown significant high expression in stage I COAD, which substantiates the potential role of CTHRC1 as an early diagnostic biomarker for COAD.
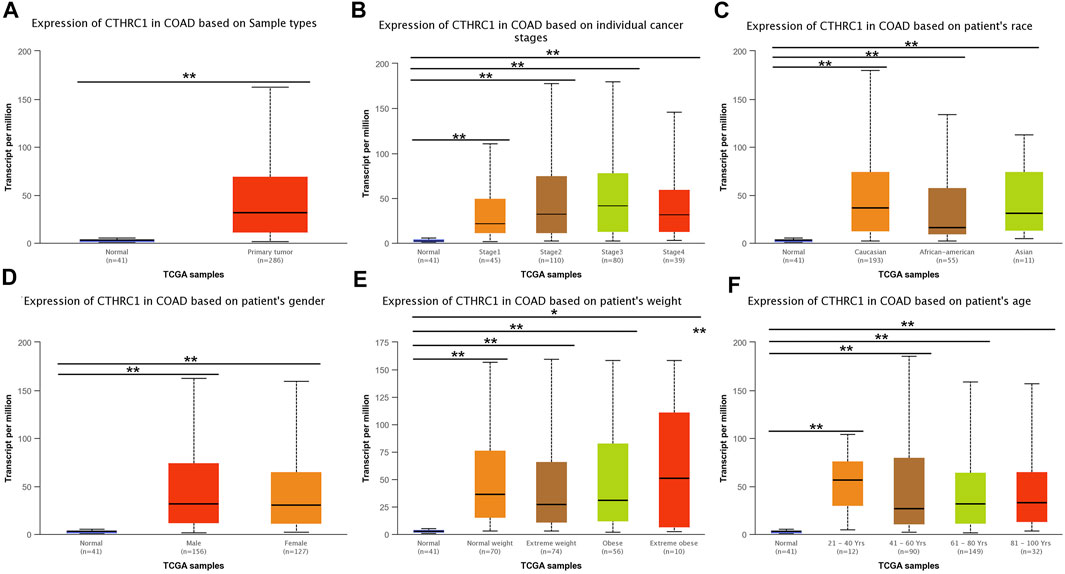
FIGURE 3. Analysis of CTHRC1 expression in patients with colon adenocarcinoma (COAD) under different physiological or pathological states using UALCAN. (A–F) CTHRC1 expression in COAD patients of various physiological and pathological states (* denotes 0.01 < p < 0.05, ** denotes p < 0.01).
Prognostic Value of CTHRC1 Expression in Patients With COAD
The GEPIA dataset was employed to assess the prognostic value of CTHRC1 expression in patients with COAD. CTHRC1 proves to be a promising prognostic indicator due to the high expression level of CTHRC1, which indicates poor OS and DFS prognosis with a same high hazard ratio of 1.8 (Figure 4). Investigation of the alteration frequency of CTHRC1 using the TCGA database revealed approximately 7.1% gene alteration in 634 colorectal cancer cases (Figure 5A,B). However, these mutations did not significantly affect the OS and DFS of patients (Figure 5C,D), indicating that CTHRC1 has considerable stability as a prognostic indicator.
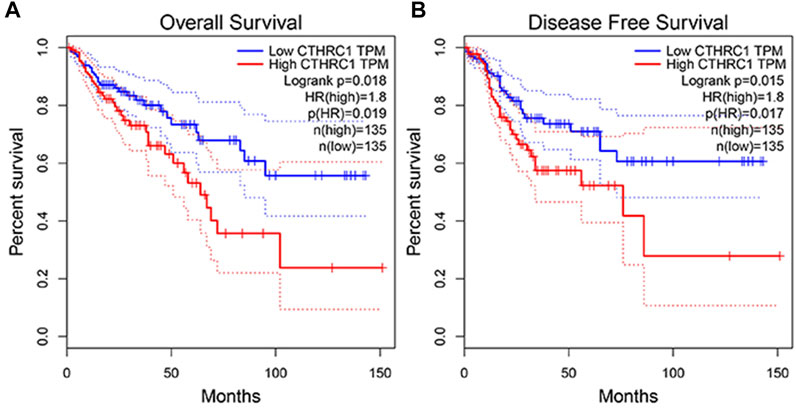
FIGURE 4. Correlation analysis between CTHRC1 expression and prognostic survival in patients with colon adenocarcinoma (COAD) using GEPIA. (A) Correlation analysis between CTHRC1 expression and overall survival in patients with COAD; (B) Correlation analysis between CTHRC1 expression and disease free survival in patients with COAD.
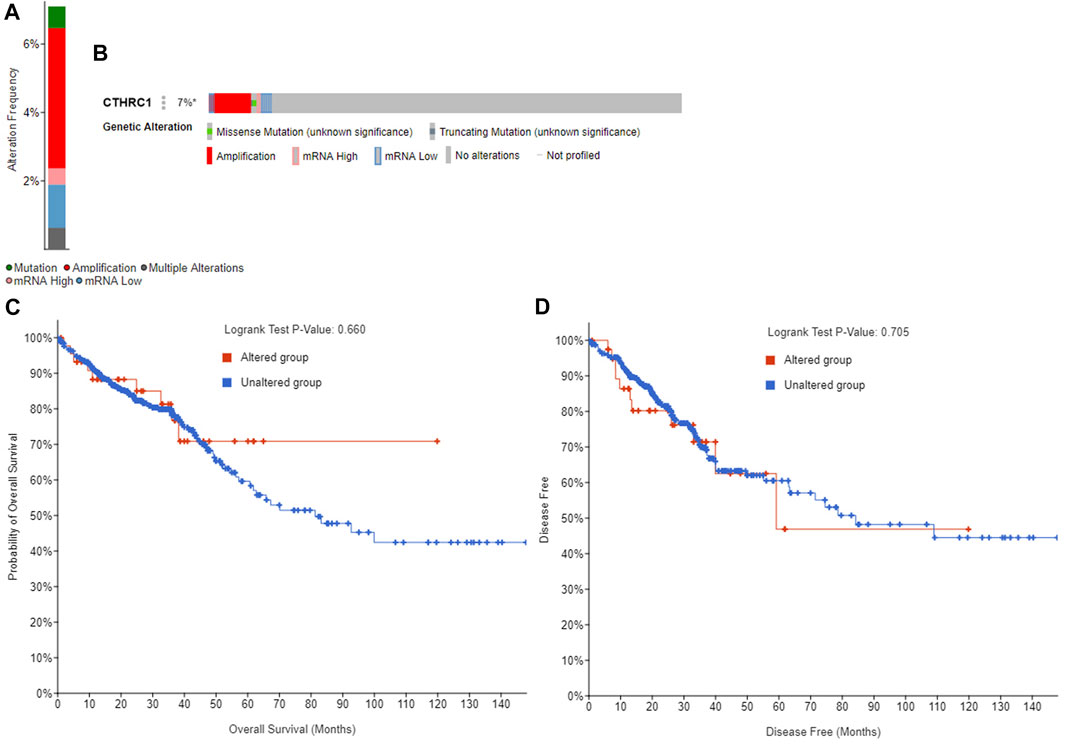
FIGURE 5. Alteration frequency analysis of CTHRC1 and its influence on prognosis in patients with colorectal cancer using cBioPortal. (A) Alteration frequency of CTHRC1 in patients with colorectal cancer. (B) Effect of CTHRC1 alteration on the overall survival of COAD patients. (C) Effect of CTHRC1 alteration on the disease free survival of COAD patients. (D) Effect of CTHRC1 alteration on the disease free survival of COAD patients.
Correlation Analysis Between CTHRC1 Expression and Immune Cell Infiltration in COAD Tissue
TIMER 2.0 was used to analyse the correlation between CTHRC1 expression and immune cell infiltration levels, including B cells, CD8+ T cells, CD4+ T cells, neutrophils, macrophages and dendritic cells. CTHRC1 expression was significantly positively correlated with all the test immune cells except B cells (p = 0.0644). Among them, macrophages had the strongest correlation with CTHRC1, with a high partial correlation coefficient of 0.608 and a p-value of 3.36e−42 (Figure 6). To clarify the immune response mechanism induced by CTHRC1 and develop new immunotherapeutic targets, the expression correlation between CTHRC1 and immune cell subsets was investigated. CTHRC1 was significantly correlated with most of the corresponding biomarker genes of the test immune cell subsets (Supplementary Table S1).
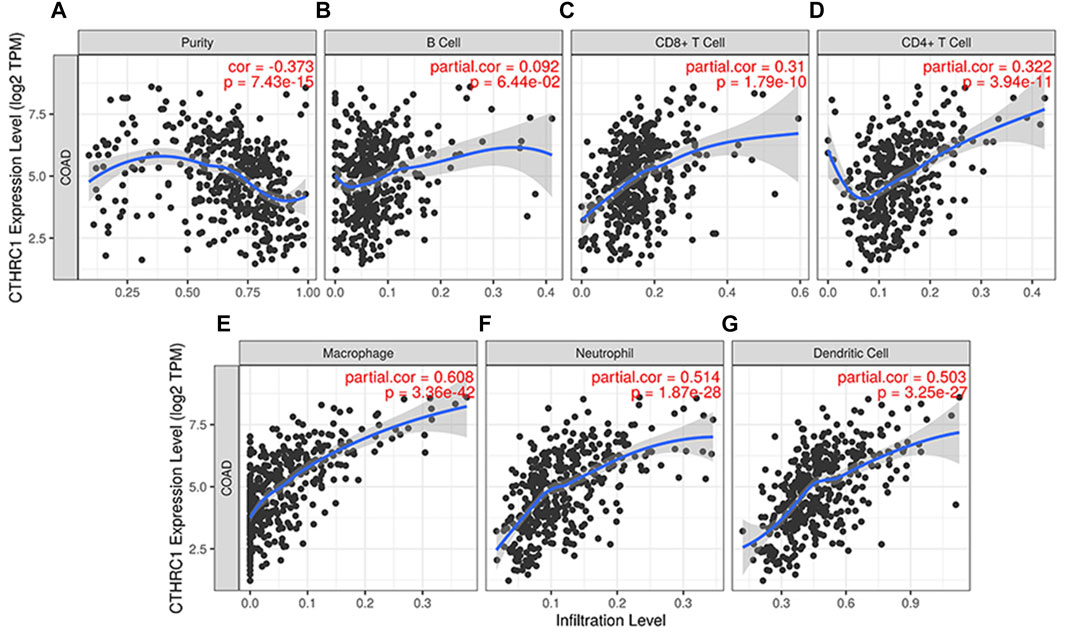
FIGURE 6. Correlation between CTHRC1 expression and immune cell infiltration levels in colon adenocarcinoma tissue analysed via TIMER 2.0. (A–G) The correlation between CTHRC1 expression and immune cell infiltration in COAD patients (n = 458).
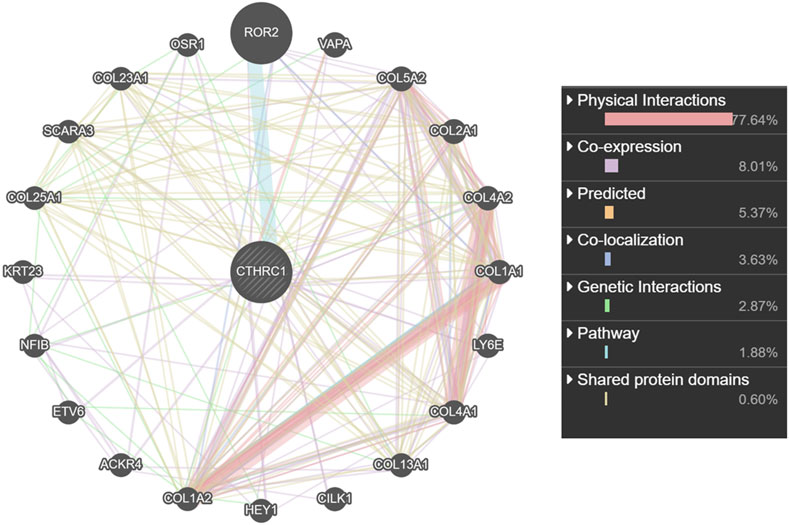
Co-Expression and Interaction Analysis of CTHRC1 in COAD Tissue
CTHRC1 was co-expressed with ROR2, VAPA, LY6E and several collagen family proteins via GeneMANIA analysis (Figure 7). These associated molecules are mainly involved in collagen fibril, extracellular matrix/structure and external encapsulating structure organisations of biological processes; the collagen type I trimer, fibrillar collagen trimer and banded collagen fibril of cellular component construction; the platelet-derived growth factor binding, FFAT motif binding, extracellular matrix structural constituent conferring tensile strength and Wnt-protein binding of molecular function. Moreover, KEGG analysis using ShinyGO showed that CTHRC1 and its related proteins were predominantly enriched in the signalling pathways of AGE-RAGE, Relaxin and Pl3K-Akt; the pathological processes of small cell lung cancer, amoebiasis and human papillomavirus infection; and the physiological functions of protein digestion and absorption, ECM-receptor interaction, focal adhesion and platelet activation (Figure 8).

FIGURE 8. Enrichment analysis of the Kyoto Encyclopedia of Genes and Genomes functional pathways of CTHRC1 and their related proteins using ShinyGO. (A) GO enrichment analysis of CTHRC1 in biological process of COAD. (B) GO enrichment analysis of CTHRC1 in cellular components of COAD. (C) GO enrichment analysis of CTHRC1 in molecular function of COAD. (D) KEGG pathway enrichment analysis of CTHRC1 in COAD.
Discussion
Biomarkers are key tools for early diagnosis, prediction of survival and prognosis, and evaluation of treatment responses. Effective biomarkers benefit clinical decision-making and improve the patient’s survival rate and life quality (Ogunwobi et al., 2020). With the development of omics technology, many potential indicator genes have been screened for various diseases, such as adenomatous polyposis, transforming growth factor β and several tumour suppressors, which are widely used in the auxiliary diagnosis of tumorigenesis (Fodde et al., 2001; Gyorffy et al., 2013; Seoane and Gomis, 2017). However, the clinical application of these indicator genes is limited due to the lack of sufficient systematic research.
CTHRC1 is a glycosylated protein secreted during tissue repair, which functions by reducing the collagen matrix deposition, thereby promoting cell migration. The expression of CTHRC1 under physiological conditions promotes wound healing; however, the pathological overexpression of CTHRC1 promotes tumour cell growth and invasion (Cheng et al., 2019). Studies have shown that CTHRC1 promotes colorectal cancer metastasis by inducing the Wnt/PCP signal transduction (Yang et al., 2015). In hepatocellular carcinoma, supressing CTHRC1 expression can inhibit integrin β, and thereby inhibiting cell migration and invasion and inducing apoptosis (Zhou et al., 2019). Additionally, CTHRC1 promotes the invasion of human epithelial ovarian cancer cells by activating the epidermal growth factor receptor signalling pathway (Ye et al., 2016). In short, the tumour promoting mechanism of CTHRC1 involves multiple targets.
The occurrence and development of COAD have the common characteristics of tumorigenesis. It promotes the massive proliferation of cells by avoiding growth inhibitory factors and apoptosis-related genes and the production of tumour blood vessels to induce tumour development. Tumorigenesis mechanisms are also attributed to unstable gene expression, tumour microenvironment change, genetic diversity and inflammatory factor activation (Goubran et al., 2014). To understand the mechanism of CTHRC1 in COAD and evaluate its potential as a diagnostic and prognostic indicator in patients with COAD, various online public databases were used to systematically investigate CTHRC1 expression profiles in COAD tissue, its impact on patient survival and immunity, and its related functional pathways and associated proteins. The consistently high expression of CTHRC1 in patients with COAD under different physiological and pathological states reflects its stability as a diagnostic indicator. Moreover, CTHRC1 was highly expressed in at least 16 tumour types (Tang et al., 2006), which broadened the application of CTHRC1 in early disease detection. Tumours usually have the characteristics of high metastasis tendency. Hence, patients aged 21–40 years and/or with Stage I COAD are more likely to obtain satisfying treatment responses; however, they are not easily diagnosed at the early stages. Notably, CTHRC1 was obviously highly-expressed in these patients, which further highlights its value as an indicator gene. Further analysis of the prognostic value revealed that the characteristics of the high hazard ratio of CTHRC1 expression and significant differences in prognostic survival indicate the superiority of CTHRC1 as a prognostic biomarker, even the high mutation frequency would not offset its indicating effect on prognosis. Therefore, these data emphasise the high value of CTHRC1 as a diagnostic and prognostic indicator for patients with COAD.
During tumorigenesis, due to the intervention of non-coding RNA, such as microRNA, the expression level of mRNA may be inconsistent with that of the associated protein (Macfarlane and Murphy, 2010). Taking this into account, the gene and protein levels were detected separately, revealing that in transcriptional and translational level, CTHRC1 was both highly expressed in COAD tissues. Since the time-consuming, laborious and expensive method of protein level detection, the follow-up investigation is mainly based on the analysis of mRNA level, which ensures the effectiveness and improves the convenience of CTHRC1 for future clinical research and application.
The response mechanism of the immune system in tumorigenesis has always been a popular research topic. The fluctuation of the tumour microenvironment is strongly related to the progress and treatment of the tumour. Understanding the tumour microenvironment provides an insight into tumour diagnosis, therapeutic targets and prognostic biomarkers (Goubran et al., 2014). The correlation analysis between CTHRC1 level and B cell infiltration showed no significant correlation, which suggests that COAD avoided the immune effect of B cells. However, this observation needs further verification by deeper and profound studies. CTHRC1 showed a close correlation (>0.5) with the infiltration levels of macrophages, neutrophils, dendritic cells and immune cell biomarker genes, which could be used as early screening targets for immunotherapy.
Through the enrichment analyses of GO and KEGG, the mechanism of CTHRC1 in biological processes, not only its cellular components and molecular function were defined, but also the specific regulatory pathways (such as AGE-RAGE, Relaxin and Pl3K signalling pathways) and action targets (including protein digestion and absorption, ECM-receptor interaction, focal adhesion and platelet activation) were identified, most of which are classical regulatory models that promote tumorigenesis or are involved in tumour development (Feng et al., 2009; Bao et al., 2019; Waghela et al., 2021). Therefore, unravelling the tumour promoting mechanism of CTHRC1could narrow the scope of further research and drug screening.
The promoting effect of CTHRC1 on tumour metastasis and proliferation could be considered a contributing factor to its abnormal high expression in various cancers including COAD. After systematic analysis, this study recommends CTHRC1 as a biomarker gene for the early diagnosis and prognostic monitoring of COAD. This study aims to provide a base for future research, regarding the molecular mechanism and therapeutics development for COAD. However, verification of the clinical applications is still lacking although this study uses many databases for comprehensive analyses and comparison.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
HL and CP designed the research; CP, CS, and HW analyzed the data; CP wrote the manuscript. All authors read and approved the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2022.849771/full#supplementary-material
References
Araghi, M., Soerjomataram, I., Jenkins, M., Brierley, J., Morris, E., Bray, F., et al. (2018). Global Trends in Colorectal Cancer Mortality: Projections to the Year 2035. Int. J. Cancer 144 (12), 2992–3000. doi:10.1002/ijc.32055
Asplund, A., Edqvist, P.-H. D., Schwenk, J. M., and Pontén, F. (2012). Antibodies for Profiling the Human Proteome-The Human Protein Atlas as a Resource for Cancer Research. Proteomics 12, 2067–2077. doi:10.1002/pmic.201100504
Bao, Y., Wang, L., Shi, L., Yun, F., Liu, X., Chen, Y., et al. (2019). Transcriptome Profiling Revealed Multiple Genes and ECM-Receptor Interaction Pathways that May Be Associated with Breast Cancer. Cell Mol Biol Lett 24, 38. doi:10.1186/s11658-019-0162-0
Chandrashekar, D. S., Bashel, B., Balasubramanya, S. A. H., Creighton, C. J., Ponce-Rodriguez, I., Chakravarthi, B. V. S. K., et al. (2017). UALCAN: a portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 19, 649–658. doi:10.1016/j.neo.2017.05.002
Cheng, X.-N., Shao, M., and Shi, D.-L. (2019). Collagen Triple helix Repeat Containing 1a (Cthrc1a) Regulates Cell Adhesion and Migration during Gastrulation in Zebrafish. Exp. Cell Res. 381 (1), 112–120. doi:10.1016/j.yexcr.2019.04.033
Danaher, P., Warren, S., Dennis, L., D’Amico, L., White, A., Disis, M. L., et al. (2017). Gene Expression Markers of Tumor Infiltrating Leukocytes. J. Immunotherapy Cancer 5, 18–32. doi:10.1186/s40425-017-0215-8
Feng, S., Agoulnik, I. U., Li, Z., Han, H. D., Lopez-Berestein, G., Sood, A., et al. (2009). Relaxin/RXFP1 Signaling in Prostate Cancer Progression. Ann. N Y Acad. Sci. 1160, 379–380. doi:10.1111/j.1749-6632.2008.03793.x
Fidler, M. M., Soerjomataram, I., and Bray, F. (2016). A Global View on Cancer Incidence and National Levels of the Human Development index. Int. J. Cancer 139, 2436–2446. doi:10.1002/ijc.30382
Fodde, R., Smits, R., and Clevers, H. (2001). APC, Signal Transduction and Genetic Instability in Colorectal Cancer. Nat. Rev. Cancer 1 (1), 55–67. doi:10.1038/35094067
Gao, J., Aksoy, B. A., Dogrusoz, U., Dresdner, G., Gross, B., Sumer, S. O., et al. (2013). Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. 6, pl1. doi:10.1126/scisignal.2004088
Garborg, K., Holme, Ø., Løberg, M., Kalager, M., Adami, H. O., and Bretthauer, M. (2013). Current Status of Screening for Colorectal Cancer. Ann. Oncol. 24 (8), 1963–1972. doi:10.1093/annonc/mdt157
Goubran, H. A., Kotb, R. R., Stakiw, J., Emara, M. E., and Burnouf, T. (2014). Regulation of Tumor Growth and Metastasis: the Role of Tumor Microenvironment. Cancer Growth Metastasis 7, 9–18. doi:10.4137/CGM.S11285
Gyorffy, B., Surowiak, P., Budczies, J., and Lánczky, A. (2013). Online Survival Analysis Software to Assess the Prognostic Value of Biomarkers Using Transcriptomic Data in Non-small-cell Lung Cancer. PLoS ONE 8, e82241.
Harrison, S., and Benziger, H. (2011). The Molecular Biology of Colorectal Carcinoma and its Implications: a Review. The Surgeon 9 (4), 200–210. doi:10.1016/j.surge.2011.01.011
Leclair, R., and Lindner, V. (2007). The Role of Collagen Triple Helix Repeat Containing 1 in Injured Arteries, Collagen Expression, and Transforming Growth Factor β Signaling. Trends Cardiovasc. Med. 17 (6), 202–205. doi:10.1016/j.tcm.2007.05.004
Li, B., Severson, E., Pignon, J.-C., Zhao, H., Li, T., Novak, J., et al. (2016). Comprehensive Analyses of Tumor Immunity: Implications for Cancer Immunotherapy. Genome Biol. 17 (1), 174–189. doi:10.1186/s13059-016-1028-7
Li, M., and Gu, J. (2005). Changing Patterns of Colorectal Cancer in China over a Period of 20 Years. Wjg 11 (30), 4685–4688. doi:10.3748/wjg.v11.i30.4685
Ma, M.-Z., Zhuang, C., Yang, X.-M., Zhang, Z.-Z., Ma, H., Zhang, W.-M., et al. (2014). CTHRC1 Acts as a Prognostic Factor and Promotes Invasiveness of Gastrointestinal Stromal Tumors by Activating Wnt/PCP-Rho Signaling. Neoplasia 16 (3), 265–278. doi:10.1016/j.neo.2014.03.001
Macfarlane, L.-A., and R. Murphy, P. (2010). MicroRNA: Biogenesis, Function and Role in Cancer. Cg 11 (7), 537–561. doi:10.2174/138920210793175895
Mostafavi, S., Ray, D., Warde-Farley, D., Grouios, C., and Morris, Q. (2008). GeneMANIA: a Real-Time Multiple Association Network Integration Algorithm for Predicting Gene Function. Genome Biol. 9, S4. doi:10.1186/gb-2008-9-s1-s4
Ogunwobi, O. O., Mahmood, F., and Akingboye, A. (2020). Biomarkers in Colorectal Cancer: Current Research and Future Prospects. Ijms 21 (15), 5311. doi:10.3390/ijms21155311
Pilonis, N. D., Bugajski, M., Wieszczy, P., Franczyk, R., Didkowska, J., Wojciechowska, U., et al. (2020). Long-term Colorectal Cancer Incidence and Mortality after a Single Negative Screening Colonoscopy. Ann. Intern. Med. 173 (2), 81–91. doi:10.7326/m19-2477
Rhodes, D. R., Yu, J., Shanker, K., Deshpande, N., Varambally, R., Ghosh, D., et al. (2004). ONCOMINE: a Cancer Microarray Database and Integrated Data-Mining Platform. Neoplasia 6, 1–6. doi:10.1016/s1476-5586(04)80047-2
Seoane, J., and Gomis, R. R. (2017). TGF-β Family Signaling in Tumor Suppression and Cancer Progression. Cold Spring Harb Perspect. Biol. 9 (12), 9a022277. doi:10.1101/cshperspect.a022277
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A. Cancer J. Clin. 71 (3), 209–249. doi:10.3322/caac.21660
Tameda, M., Sugimoto, K., Shiraki, K., Yamamoto, N., Okamoto, R., Usui, M., et al. (2014). Collagen Triple helix Repeat Containing 1 Is Overexpressed in Hepatocellular Carcinoma and Promotes Cell Proliferation and Motility. Int. J. Oncol. 45 (2), 541–548. doi:10.3892/ijo.2014.2445
Tang, L., Dai, D. L., Su, M., Martinka, M., Li, G., and Zhou, Y. (2006). Aberrant Expression of Collagen Triple helix Repeat Containing 1 in Human Solid Cancers. Clin. Cancer Res. 12 (12), 3716–3722. doi:10.1158/1078-0432.ccr-06-0030
Tang, Z., Li, C., Kang, B., Gao, G., Li, C., and Zhang, Z. (2017). GEPIA: a Web Server for Cancer and normal Gene Expression Profiling and Interactive Analyses. Nucleic Acids Res. 45, W98–W102. doi:10.1093/nar/gkx247
Vanessa, B., and Karen, S. (2016). Colorectal Cancer Development and Advances in Screening. Clin. Interventions Aging 11, 967–976.
Waghela, B. N., Vaidya, F. U., Ranjan, K., Chhipa, A. S., Tiwari, B. S., and Pathak, C. (2021). AGE-RAGE Synergy Influences Programmed Cell Death Signaling to Promote Cancer. Mol. Cell Biochem 476, 585–598. doi:10.1007/s11010-020-03928-y
Wu, Q., Yang, Q., and Sun, H. (2018). Collagen Triple Helix Repeat Containing-1: A Novel Biomarker Associated With Disease Activity in Systemic Lupus Erythematosus. Lupus 27(13), 2076–2085. doi:10.1177/0961203318804877
Yang, X. M., You, H. Y., Li, Q., Ma, H., Wang, Y. H., Zhang, Y. L., et al. (2015). CTHRC1 Promotes Human Colorectal Cancer Cell Proliferation and Invasiveness by Activating Wnt/PCP Signaling. Int. J. Clin. Exp. Pathol. 8 (10), 12793–12801.
Ye, J., Chen, W., Wu, Z. Y., Zhang, J. H., Fei, H., Zhang, L. W., et al. (2016). Upregulated CTHRC1 Promotes Human Epithelial Ovarian Cancer Invasion through Activating EGFR Signaling. Oncol. Rep. 36, 3588–3596. doi:10.3892/or.2016.5198
Keywords: collagen triple helix repeat containing 1 (CTHRC1), colon adenocarcinoma (COAD), diagnosis, prognosis, immune infiltration, function pathway
Citation: Pang C, Wang H, Shen C and Liang H (2022) Application Potential of CTHRC1 as a Diagnostic and Prognostic Indicator for Colon Adenocarcinoma. Front. Mol. Biosci. 9:849771. doi: 10.3389/fmolb.2022.849771
Received: 06 January 2022; Accepted: 04 February 2022;
Published: 01 March 2022.
Edited by:
Na Luo, Nankai University, ChinaCopyright © 2022 Pang, Wang, Shen and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chengcheng Shen, shencc2020@163.com; Houjie Liang, lianghoujie@sina.com
 Chen Pang1
Chen Pang1  Chengcheng Shen
Chengcheng Shen Houjie Liang
Houjie Liang