Expression and Gene Regulation Network of Adenosine Receptor A2B in Lung Adenocarcinoma: A Potential Diagnostic and Prognostic Biomarker
- 1Shenzhen Hospital, Southern Medical University, Shenzhen, China
- 2Department of Radiation Oncology, Harbin Medical University Cancer Hospital, Harbin, China
- 3The First Affiliated Hospital, Biomedical Translational Research Institute and Guangdong Province Key Laboratory of Molecular Immunology and Antibody Engineering, Jinan University, Guangzhou, China
- 4NHC Key Laboratory of Hormones and Development, Tianjin Key Laboratory of Metabolic Diseases, Chu Hsien-I Memorial Hospital and Tianjin Institute of Endocrinology, Tianjin Medical University, Tianjin, China
Adenosinereceptor A2B (ADORA2B) encodes a protein belonging to the G protein–coupled receptor superfamily. Abnormal expression of ADORA2B may play a pathophysiological role in some human cancers. We investigated whether ADORA2B is a potential diagnostic and prognostic biomarker for lung adenocarcinoma (LUAD). The expression, various mutations, copy number variations, mRNA expression levels, and related network signaling pathways of ADORA2B were analyzed using bioinformatics-related websites, including Oncomine, UALCAN, cBioPortal, GeneMANIA, LinkedOmics, KM Plotter, and TIMER. We found that ADORA2B was overexpressed and amplified in LUAD, and a high ADORA2B expression predicted a poor prognosis for LUAD patients. Pathway analyses of ADORA2B in LUAD revealed ADORA2B-correlated signaling pathways, and the expression level of ADORA2B was associated with immune cell infiltration. Furthermore, ADORA2B mRNA and protein levels were significantly higher in human LUAD cell lines (A549 cells and NCl-H1299 cells) than in normal human bronchial epithelial (HBE) cells, and the transcript levels of genes positively or negatively correlated with ADORA2B were consistent and statistically significant. siRNA transfection experiments and functional experiments further confirmed these results. In vitro results were also consistent with those of bioinformatics analysis. Our findings provide a foundation for studying the role of ADORA2B in tumorigenesis and support the development of new drug targets for LUAD.
Introduction
Lung adenocarcinoma (LUAD) is a common pathological form of lung cancer and is associated with high mortality, leading to more than one million deaths worldwide every year (TCGA, 2018). LUAD is characterized by the presence of heterogeneous tumors that originate in the small airway and spread to the surrounding lung tissue (Yu et al., 2010; Russell et al., 2011). Multiple organ metastasis is the leading cause of death in LUAD patients. Within a few months after diagnosis, metastasis to different organs can occur rapidly. Traditional modalities, such as radiotherapy and chemotherapy, are ineffective for LUAD (Zagryazhskaya et al., 2015), and targeted gene therapy holds potential for the clinical management of LUAD. Epidermal growth factor receptor (EGFR) mutation and anaplastic lymphoma kinase (ALK) fusion are the common gene abnormalities reported in LUAD. Personalized therapy targeting these genes has become a well-designed standard treatment, with erlotinib and gefitinib being the main drugs for LUAD treatment (Yu et al., 2013). Furthermore, mutations in Kirsten rat sarcoma viral oncogene (KRAS), human epidermal growth factor receptor 2 (HER2), and V-raf murine sarcoma viral oncogene homolog B1 (BRAF) as well as other fusions of driver genes, such as rearranged during transfection (RET) and ROS proto-oncogene 1 (ROS1), have been reported in LUAD (Roman et al., 2018; Pillai et al., 2017; Mazieres et al., 2020; Drilon, et al., 2020; Shaw, et al., 2017). These driver genes vary depending on race, sex, and smoking status. The pathogenesis of LUAD is complex and mainly involves cell cycle regulation and signal transduction, reflected in altered gene function at different developmental stages of the disease. New drug targets for LUAD should be identified by screening gene networks associated with tumor formation and progression.
Most of the extracellular adenosine is released from adenine nucleotides in response to various stimuli, including mechanical stress, osmotic stimulation, inflammation, and tissue damage (Borea et al., 2016; Covarrubias et al., 2016; Hamidzadeh and Mosser, 2016). Extracellular adenosine binds to four adenosine receptor subtypes, namely, A1, A2A, A2B, and A3, each of which exhibits unique pharmacological and coupling effects and tissue distribution characteristics. Adenosine receptor A2B (ADORA2B) encodes an adenosine receptor belonging to the G protein–coupled receptor superfamily and is highly expressed in various carcinomas, which leads to the promotion of carcinoma cell proliferation and metastasis (Kasama et al., 2015; Vecchio et al., 2016). ADORA2B also regulates the tumor microenvironment and is involved in peripheral vascular growth, inflammatory cell infiltration, fibroblast proliferation, and extracellular matrix accumulation (Csoka et al., 2012). Therefore, ADORA2B regulates tumor progression and metastasis and could serve as a useful target for cancer therapy or combination therapy.
Recent studies have suggested that ADORA2B is an indispensable target in the regulation of both acute and chronic pulmonary diseases, such as pulmonary hypertension, chronic obstructive pulmonary disease, and pulmonary fibrosis (Karmouty-Quintana et al., 2013; Philip et al., 2017; Mertens et al., 2018; Tian et al., 2021). LUAD is one of the most lethal pulmonary diseases, with the highest mortality rate; however, the role of ADORA2B in LUAD is still unknown. In this study, we analyzed the expression and mutations of ADORA2B in patients with LUAD using data obtained from various public databases. We also analyzed the genomic variations, survival rates, and functional networks associated with ADORA2B in LUAD. Additionally, human LUAD and normal bronchial epithelial cell lines were used to confirm the role of ADORA2B in LUAD. siRNA transfection experiments and functional experiments further confirmed the obtained results. Therefore, we used bioinformatics analysis in combination with in vitro experiments to comprehensively investigate the role of ADORA2B in LUAD.
Materials and Methods
Oncomine Analysis
The mRNA expression of ADORA2B in LUAD was analyzed using the Oncomine database (www.oncomine.org). LUAD-based databases established from the Okayama Lung, Landi Lung, Selamat Lung, Stearman Lung, Beer Lung, Su Lung, Bhattacharjee Lung, and Garber Lung datasets were used for this analysis (Bhattacharjee et al., 2001; Garber et al., 2001; Beer et al., 2002; Stearman et al., 2005; Su et al., 2007; Landi et al., 2008; Okayama et al., 2012; Selamat et al., 2012). The difference in the expression of ADORA2B between LUAD tissues and para-carcinoma tissues was considered statistically significant at p < 0.01.
UALCAN Analysis
Multiple clinical and pathological features related to the mRNA expression of ADORA2B in LUAD were analyzed by UALCAN (http://ualcan.path.uab.edu). ADORA2B expression was analyzed by TCGA Level 3 RNA sequencing in UALCAN, and the clinical data were analyzed to determine ADORA2B expression levels in different tumor subgroups (Chandrashekar et al., 2017).
cBioPortal Analysis
cBioPortal (http://cbioportal.org) was used to analyze ADORA2B alterations observed in The Cancer Genome Atlas (TCGA) LUAD samples. By analyzing various types of mutations, copy number variations, and mRNA expression levels, the tab OncoPrint displayed an overview of the genetic changes in each sample as gene mutations and heat maps of ADORA2B expression. The tab plots displayed an overview of the data type of clinical attributes and mutations, including clinical attributes and group mutations by mutation type.
GeneMANIA Analysis
GeneMANIA (http://genemania.org/) is a web interface that uses large sets of functional association data to identify single genes related to a set of input genes, generating hypotheses on gene function, analyzing gene lists, and prioritizing genes for functional assays. GeneMANIA was used to construct the ADORA2B software for the analysis (Warde-Farley et al., 2010).
LinkedOmics Analysis
The LinkFinder module of LinkedOmics (http://www.linkedomics.org/login.php) was used to identify differentially expressed genes related to ADORA2B (n = 515) in the TCGA LUAD section. The search and target datasets were obtained by RNA-seq, and the results were analyzed with the Pearson correlation coefficient. The data from the LinkFinder results were labeled and sorted before enrichment analysis was performed for Gene Ontology, KEGG, PANTHER, and Reactome pathway analyses.
Survival Analysis
We used the Kaplan–Meier (KM) Plotter (http://kmplot.com), an online database that contains gene expression data and survival information of 865 LUAD patients, to analyze the prognostic value of ADORA2B in LUAD. The patient samples were separated into two groups by median expression (high expression and low expression) to analyze the overall survival (OS) with hazard ratios (HRs) with 95% confidence intervals and log-rank p-values.
Immune Infiltration Analysis
Tumor Immune Estimation Resource (TIMER, https://cistrome.shinyapps.io/timer/) is a web resource for systematic analysis of the infiltration levels of different subsets of immune cells in different types of cancers. We analyzed the expression of ADORA2B in LUAD in relation to tumor purity and the abundance of immune infiltrating cells including B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells. We also analyzed the relationship between the gene copy number variation and the abundance of immune infiltrating cells.
Cell Culture
A549 cells (human LUAD, ATCC®CCL-185™), NCl-H1299 cells (human LUAD, ATCC®CRL-5803™), and HBE cells (human bronchial epithelial cells, ATCC®CRL-2741™) were obtained from ATCC (Manassas, VA, United States). A549 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Grand Island, NY, United States) containing 10% fetal bovine serum (FBS, Gibco), whereas the culture medium for HBE cells contained 20% FBS. NCl-H1299 cells were cultured in RPMI-1640 medium (Gibco, Grand Island, NY, United States) containing 10% FBS. All the cells were incubated in a humidified incubator at 5% CO2 and 37°C. The cells that passaged less than five with >70% confluency were used for experiments.
RNA Extraction and Quantitative Real-time PCR (qRT-PCR)
RNA from A549, NCl-H1299, and HBE cells was isolated using TRIzol Reagent (Invitrogen, Carlsbad, CA, United States) according to the manufacturer's instructions. cDNA synthesis was performed using a cDNA First Strand Synthesis Kit (ABclonal, Wuhan, China), and mRNA expression was quantified by qRT-PCR using the SYBR Green Fast qPCR Mix (ABclonal). Relative mRNA expression levels were normalized to those of β-actin and measured by the comparative Ct (2−ΔΔCt) method (n = 6 samples/group). Primer sequences used in this study are listed in Supplementary Table S1.
siRNA Transfection
ADORA2B siRNAs and negative control siRNA (50 nM) were obtained from GenePharma (Shanghai, China). The siRNAs were transfected with Lipofectamine 2000 (Invitrogen) for 4 h, following the manufacturer’s protocol. ADORA2B knockdown efficiencies were verified by RT-PCR. The siRNA sequences are listed in Supplementary Table S1.
Western Blotting
A549, NCl-H1299, and HBE cells were cultured as previously described (Cell Culture). Total protein was extracted using RIPA buffer (Solarbio, Beijing, China). Protein concentrations were measured using a BCA protein assay kit (Solarbio). Protein extracts (50 μg/well) were separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred onto polyvinylidene fluoride membranes. The membranes were blocked with 5% skim milk for 2 h and incubated with anti-β-actin (Bioss, bs-0061R, Beijing, China), anti-ADORA2B (Bioss, bs-5900R), anti-cyclin D1 (ABclonal, A2708), anti-PCNA (Proteintech, 10205-2-AP, Wuhan, China), anti-N-cadherin (ABclonal, A0433), and anti-vimentin (ABclonal, A2584) antibodies overnight. The next day, the membranes were incubated with a HRP-conjugated secondary antibody and goat anti-rabbit IgG antibody (ABclonal) for 1 h. A western blot detection ECL kit (Advansta, Menlo Park, United States) was used to detect protein bands. Fold-changes in ADORA2B protein expression were normalized to those of β-actin.
Cell Proliferation Assay
Cell proliferation was measured using cell counting kit-8 (CCK-8, Beyotime, Beijing, China). A549 cells (3000 cells/well) were plated in a 96-well plate and transfected with ADORA2B siRNA-2 and negative control siRNA as previously described (siRNA Transfection). The cell proliferation rate was detected with CCK-8 at 24, 48, and 72 h. The absorbance at 450 nm was measured using a microplate reader.
Cell Migration Assay
The cells were transfected with ADORA2B siRNA-2 and negative control siRNA using Lipofectamine 2000 as previously described. Transfected cells were grown to 100% confluence, and then wounds were created using pipette tips. Serum-free medium was used instead of complete medium to eliminate the effects of cell proliferation. Images were captured at 0 and 48 h using a microscope. The cell migration rate was calculated by ImageJ software.
Statistical Analysis
Data are presented as the mean ± standard error of the replicates. Comparisons between two groups were performed using Student's t-test. GraphPad Prism 8.0 software (GraphPad, Inc., La Jolla, CA, United States) was used for statistical analysis. Results with p < 0.05 were considered statistically significant (*p < 0.05; **p < 0.01; ***p < 0.001).
Results
Expression of ADORA2B in LUAD
We determined ADORA2B transcript levels in LUAD patients using data obtained from TCGA and Gene Expression Omnibus (GEO). Data from the Oncomine database showed that the expression of ADORA2B mRNA was significantly upregulated in LUAD tissues when compared with that in para-carcinoma tissues (p < 0.01). ADORA2B ranked in the top 33% of genes with high mRNA expression, with fold-difference values <2 (Figures 1A–H).
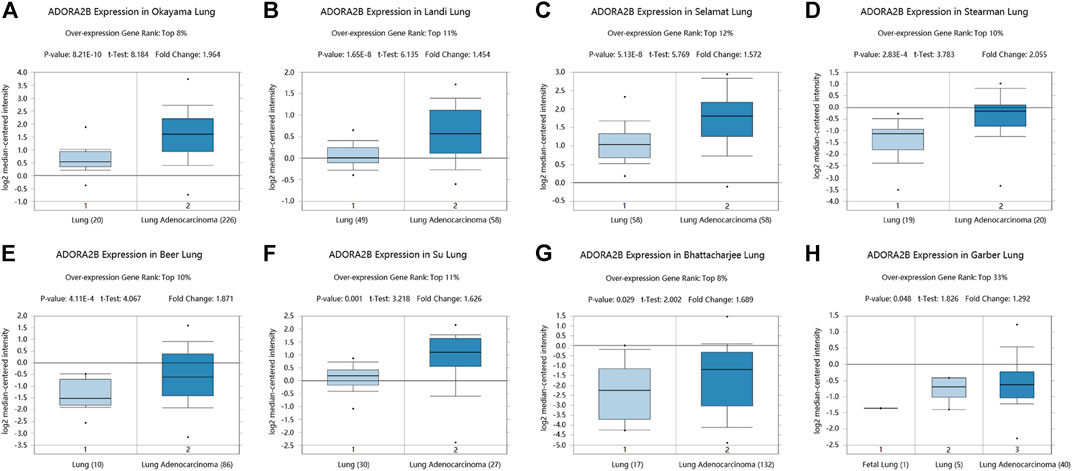
FIGURE 1. ADORA2B transcription in LUAD. ADORA2B mRNA copy numbers were significantly higher in LUAD in (A) Okayama Lung, (B) Landi Lung, (C) Selamat Lung, (D) Stearman Lung, (E) Beer Lung, (F) Su Lung, (G) Bhattacharjee Lung, and (H) Garber Lung datasets than in para-carcinoma tissues.
Similarly, analysis of multiple clinical and pathological features of LUAD patients from the TCGA database revealed elevated ADORA2B mRNA expression. Moreover, ADORA2B mRNA expression was significantly higher in LUAD tissues than in para-carcinoma tissues in subgroup analyses based on sample type, individual cancer stage, ethnicity, gender, age, smoking habits, nodal metastasis status, and TP53 mutation status (Figures 2A–H).
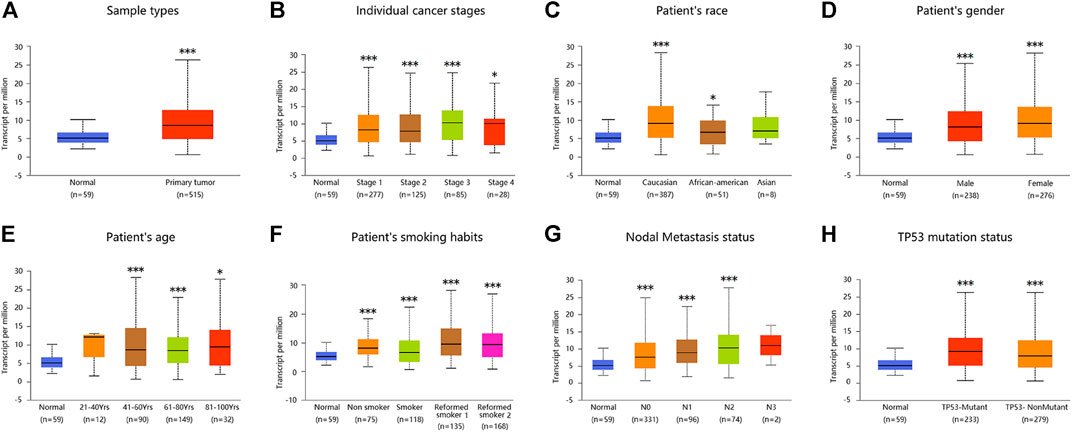
FIGURE 2. ADORA2B expression level in subgroups of patients with LUAD. Relative expression of ADORA2B in normal and different classifications of LUAD tissues. (A) Sample types, (B) individual cancer stages, (C) patient’s race, (D) patient’s gender, (E) patient’s age, (F) patient’s smoking habits, (G) nodal metastasis status, and (H) TP53 mutation status. *p < 0.05 and ***p < 0.001 are statistically significant.
Frequency and Type of ADORA2B Alterations in LUAD
Next, we used the cBioPortal database to evaluate the types and frequencies of ADORA2B alterations in LUAD tissues based on sequencing data from patients with LUAD obtained from TCGA’s Pan-Cancer Atlas database. ADORA2B was altered in 30 out of 503 (6%) patients with LUAD (Figure 3A). These genetic alterations included missense mutations (1, 0.2%), amplification (3, 0.6%), deep deletion (4, 0.8%), mRNA high (2, 0.4%), and mRNA low (20, 4%). The patient order in OncoPrint is shown in Supplementary Table S2. Analysis included the type of clinical attribute and mutation, fraction of genome altered, and mutation type, including missense (VUS), not mutated, not profiled for mutations, amplification, gain, diploid, shallow deletion, deep deletion, and not profiled for CNA (Figure 3B). mRNA low, shallow deletion, and diploid were the most common types of ADORA2B genetic alterations found in LUAD.
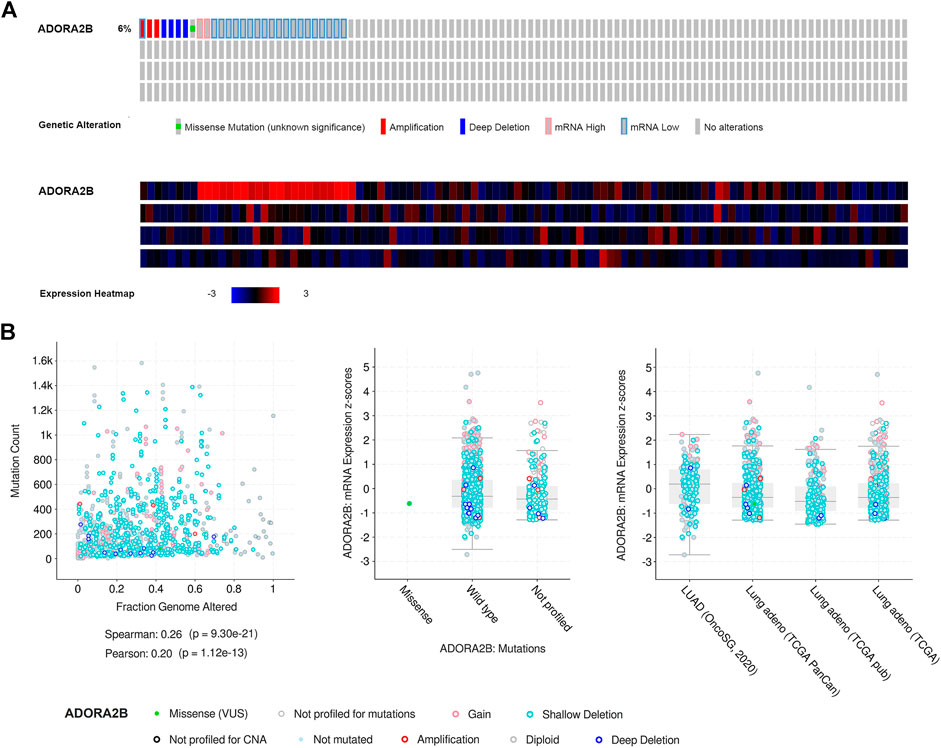
FIGURE 3. Frequency and type of ADORA2B alterations in LUAD. (A) OncoPrint of ADORA2B alterations in LUAD (cBioPortal) and (B) plots of ADORA2B alterations in LUAD with clinical attributes and mutations (cBioPortal).
Expression of ADORA2B Is Associated With the Survival of LUAD Patients
Previous studies have demonstrated that ADORA2B participates in the proliferation and metastasis of carcinomas (Kasama et al., 2015; Vecchio et al., 2016). Thus, we further examined the prognostic value of ADORA2B in LUAD patients. LUAD patients with a higher expression of ADORA2B exhibited poor OS according to the KM Plotter database. Survival analysis showed that the mean OS time in the low ADORA2B expression group was longer than that in the high ADORA2B expression group (p = 0.034) (Figure 4A). The mean OS times for 12 (1 year), 36 (3 years), and 60 months (5 years) in the low ADORA2B expression group were significantly longer than those in the high expression group (Figures 4B–D). These results indicated that ADORA2B was significantly associated with the prognosis of LUAD patients and that a high ADORA2B expression predicted a poor prognosis in LUAD patients.
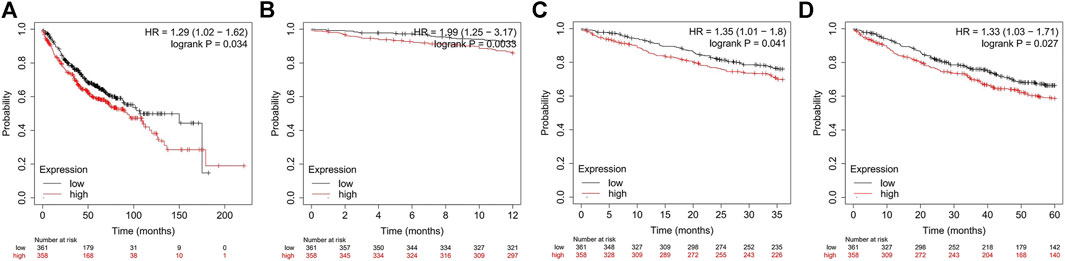
FIGURE 4. The expression of ADORA2B is associated with LUAD patient survival. (A) Mean OS analysis of LUAD patients with high and low ADORA2B expression OS analysis of LUAD patients with high and low ADORA2B expressions for (B) 12 months (1 year), (C) 36 months (3 years), and (D) 60 months (5 years).
Interaction Network of ADORA2B
An ADORA2B correlation network was constructed to determine the potential mutual effects between ADORA2B and cancer-related targets. The results indicated that ADORA2B could interact with netrin 1 receptor (DCC), solute carrier family 9 member A3 regulator 2 (SLC9A3R2), zinc finger protein 267 (ZNF267), and tumor protein D52-like 2 (TPD52L2); ADORA2B was co-expressed with potassium inwardly rectifying channel subfamily J member 3 (KCNJ3), angio-associated migratory cell protein (AAMP), TPD52L2, and solute carrier family 29 member 2 (SLC29A2); and ADORA2B was co-located with AAMP, peroxisomal biogenesis factor 6 (PEX6), SLC29A2, ADORA2A, and diphthamide biosynthesis 1 (DPH1). ADORA2B was identified to interact with DCC and shared protein domains with its isoform ADORA2A (Figure 5A).
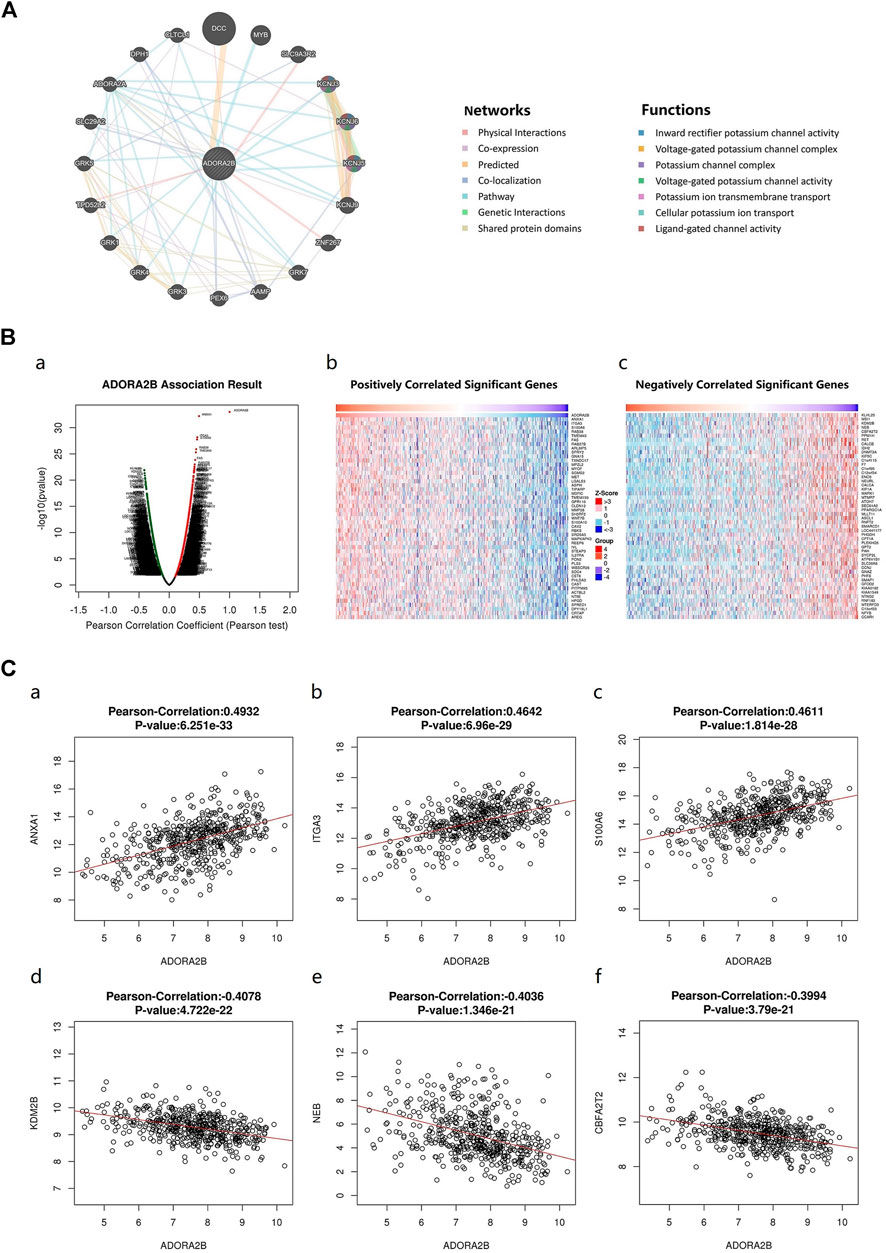
FIGURE 5. Genes associated with ADORA2B expression. (A) Biological interaction network of ADORA2B. (B) Differentially expressed genes correlated with ADORA2B (LinkedOmics). (a) Correlations between ADORA2B and differentially expressed genes in LUAD were analyzed with Pearson’s correlation coefficient. (b) Heat map of genes positively correlated with ADORA2B in LUAD. (c) Heat map of genes negatively correlated with ADORA2B in LUAD. (C) Gene expression correlation analysis of ADORA2B and ADORA2B-correlated genes (LinkedOmics). Pearson correlation of ADORA2B expression with that of (a) ANXA1, (b) ITGA3, (c) S100A6, (d) KDM2B, (e) NEB, and (f) CBFA2T2 (n = 515).
The LinkedOmics database was used to identify RNA-seq genes co-expressed with ADORA2B in LUAD. A volcano plot indicated that the expression of 5,246 genes was negatively correlated with ADORA2B expression, whereas the expression of 3,447 genes was positively correlated with ADORA2B expression. We confirmed the top 50 genes that negatively or positively interacted with ADORA2B (Figure 5B and Supplementary Table S3). The results revealed that multiple differentially expressed genes were correlated with ADORA2B expression.
Pearson's correlation coefficient analysis revealed a positive correlation between ADORA2B expression levels and those of annexin A1 (ANXA1), integrin subunit alpha 3 (ITGA3), and S100 calcium binding protein A6 (S100A6) and a negative correlation between the expression of ADORA2B and that of lysine demethylase 2B (KDM2B), nebulin (NEB), and CBFA2/RUNX1 partner transcriptional co-repressor 2 (CBFA2T2) (Figure 5C).
Enrichment Analysis of ADORA2B Functional Networks in LUAD
Three independent ontologies (biological process, cellular component, and molecular function) were analyzed by gene set enrichment analysis (GSEA). The results indicated that ADORA2B-associated differentially expressed genes were involved in several biological processes (protein targeting, neutrophil mediated immunity, response to IFN-γ, etc.), cellular components (ribosome, cell-substrate junction, secretory granule membrane, etc.), and molecular functions (structural constituent of ribosome, cell adhesion molecule binding, enzyme inhibitor activity, etc.) (Figure 6A).
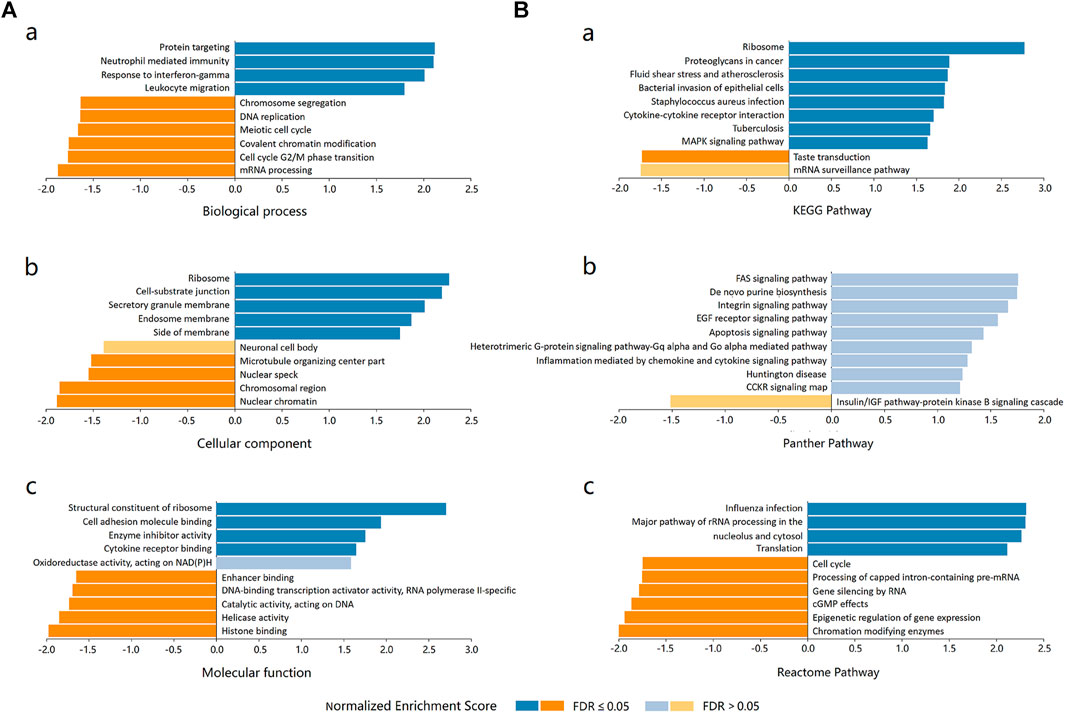
FIGURE 6. Enrichment analysis of ADORA2B functional networks in LUAD. (A) Enriched Gene Ontology annotations of ADORA2B-correlated genes in LUAD (LinkedOmics). (a) Biological process. (b) Cellular component. (c) Molecular function. (B) Enrichment pathway analysis of ADORA2B-correlated genes in LUAD (LinkedOmics). (a) KEGG pathway. (b) PANTHER pathway. (c) Reactome pathway.
The differentially expressed genes associated with ADORA2B were then evaluated for potential functional pathways using KEGG (ribosome, proteoglycans in cancer, fluid shear stress, and atherosclerosis, etc.), PANTHER (FAS, EGF receptor, apoptosis, etc.), and Reactome (influenza infection, major pathway of rRNA processing in the nucleolus and cytosol, cell cycle, etc.) pathway analyses (Figure 6B).
Relationship Between ADORA2B Expression and Infiltrating Immune Cells in LUAD
The tumor microenvironment is a very complex system composed of a variety of cells, enzymes, cytokines, and metabolites and is characterized by low oxygen, low pH, and high pressure. An important immunosuppressive mechanism is mediated by the CD73–adenosine receptor metabolic signaling pathway (Chen et al., 2020). To comprehensively investigate the role of ADORA2B in LUAD, we selected the TIMER database to analyze the association of ADORA2B expression levels with subsets of infiltrating immune cells. The expression of ADORA2B was significantly correlated with the infiltration of B cells, macrophages, neutrophils, and dendritic cells (Figure 7A). In addition, the copy number variation (CNV) of ADORA2B was significantly correlated with the infiltration levels of macrophages (Figure 7B).
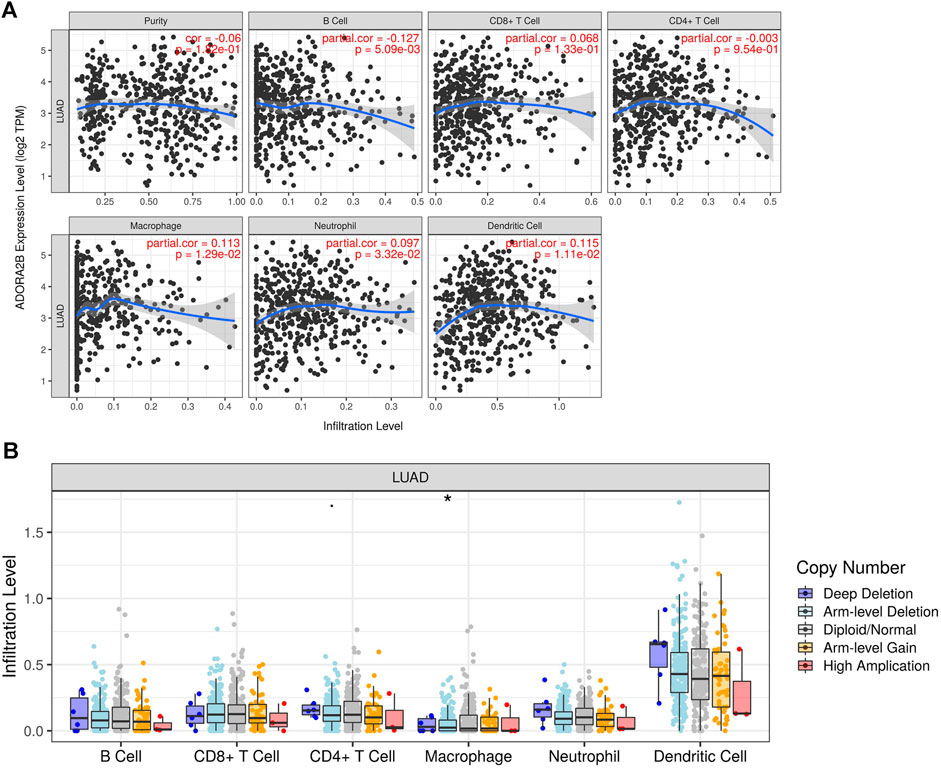
FIGURE 7. Correlation of ADORA2B expression with immune cell infiltration in LUAD. (A) Correlation of ADORA2B with tumor purity and infiltration of different subsets of immune cells. (B) Effect of copy number variation of ADORA2B on the distribution of various immune cells. *p < 0.05 is statistically significant.
Expression Levels of ADORA2B and Correlated Genes in Human LUAD Cells
We found that compared with that in normal lung tissues, the expression level of ADORA2B was significantly elevated in tumor tissues of LUAD patients with higher morbidity. To confirm that ADORA2B expression is related to LUAD, we investigated the ADORA2B gene and protein expression in human LUAD (A549 and NCl-H1299) and normal human bronchial epithelial (HBE) cell lines. Western blotting analysis indicated that the relative ADORA2B protein expression levels in A549 cells and NCl-H1299 cells were significantly higher than those in HBE cells (Figures 8A,B). Next, we examined the mRNA expression level of ADORA2B by qRT-PCR and found that the relative ADORA2B mRNA expression level was consistent with the protein expression level in the three cell lines (Figure 8C).
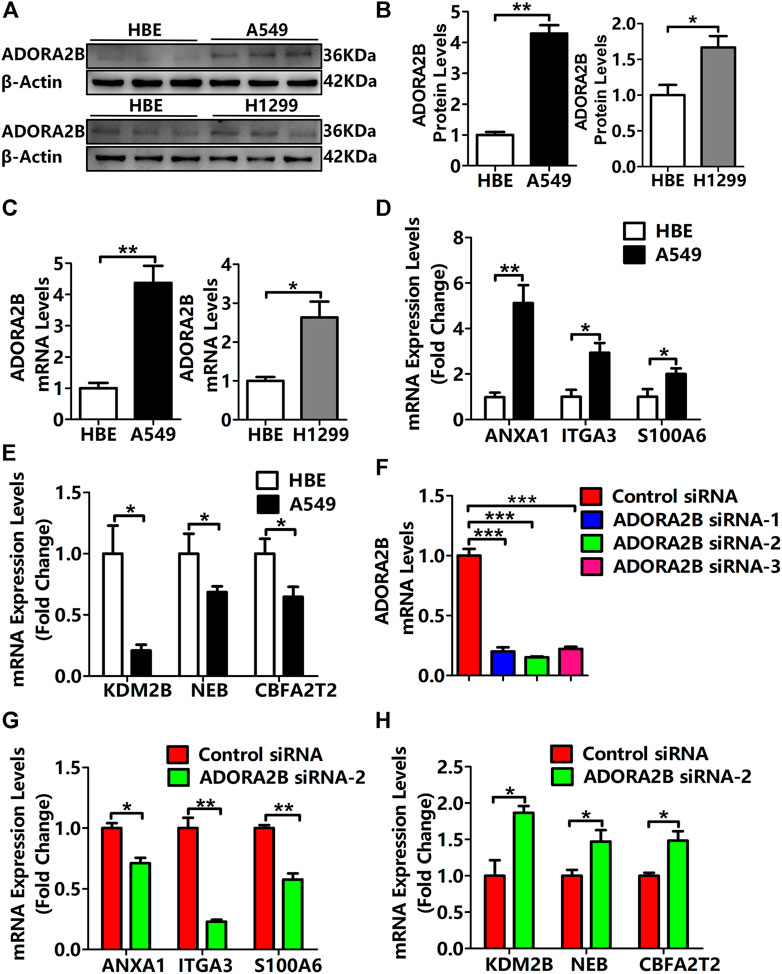
FIGURE 8. Expression levels of ADORA2B and correlated genes in human LUAD cells. (A) Relative protein expression of ADORA2B was determined by western blotting. (B) Protein quantitative statistics. (C) Relative mRNA expression of ADORA2B was determined by qRT-PCR. mRNA expression levels of genes (D) positively and (E) negatively correlated with ADORA2B. (F) mRNA expression levels of ADORA2B after siRNA transfection. mRNA expression levels of genes (G) positively and (H) negatively correlated with ADORA2B after siRNA transfection. *p < 0.05, **p < 0.01, and ***p < 0.001 are statistically significant.
As shown in Figure 5C, LinkedOmics was used to identify the differentially expressed genes related to ADORA2B. The expression levels of ADORA2B-associated genes were positively (ANXA1, ITGA3, and S100A6) or negatively (KDM2B, NEB, and CBFA2T2) correlated with ADORA2B expression in LUAD. To verify these results, we measured the mRNA expression levels of these genes. The expression levels of ANXA1, ITGA3, and S100A6 were significantly higher in A549 cells than in HBE cells and were positively correlated with ADORA2B expression levels (Figure 8D). In contrast, the expression levels of KDM2B, NEB, and CBFA2T2 were significantly lower in A549 cells than in HBE cells and negatively correlated with ADORA2B expression (Figure 8E). These results were consistent with those from our bioinformatics analyses.
To further confirm these findings, specific ADORA2B siRNAs were transfected into A549 cells to inhibit ADORA2B expression. First, we verified the knockdown efficiencies of ADORA2B by qRT-PCR and then selected siRNA-2, which had the highest knockdown efficiency, for subsequent experiments (Figure 8F). Then, we measured the mRNA expression levels of genes that were positively or negatively correlated with ADORA2B expression. The expression levels of ANXA1, ITGA3, and S100A6 were significantly lower in the ADORA2B siRNA group than in the control siRNA group and positively correlated with ADORA2B expression (Figure 8G). The expression levels of KDM2B, NEB, and CBFA2T2 were significantly higher in the ADORA2B siRNA group than in the control siRNA group and negatively correlated with ADORA2B expression (Figure 8H). Taken together, ADORA2B was highly expressed in human LUAD A549 cells and the expression levels of ANXA1, ITGA3, and S100A6 were positively correlated with ADORA2B expression, while those of KDM2B, NEB, and CBFA2T2 were negatively correlated with ADORA2B expression. In vitro experimental results were consistent with those of the bioinformatics analyses.
Knockdown of ADORA2B Inhibited the Proliferation and Metastasis of LUAD Cells
Previous studies have demonstrated that ADORA2B participates in the proliferation and metastasis of carcinomas, such as prostate and oral cancers (Kasama et al., 2015; Vecchio et al., 2016). To further clarify the role of ADORA2B in the progression of LUAD, we performed related functional experiments to verify the function of ADORA2B. First, cell proliferation was measured using CCK-8. Compared with that of the control siRNA group, the cell viability of the ADORA2B siRNA group was significantly decreased at 24, 48, and 72 h after transfection (Figure 9A). Second, the cell migration rate was evaluated by the wound healing assay, the results of which showed that knockdown of ADORA2B reduced the migration capacity of A549 cells after 48 h of induction, partially supporting the role of ADORA2B in promoting metastasis in LUAD (Figure 9B). We further examined the protein expression levels of markers of carcinoma cell proliferation and metastasis after ADORA2B knockdown. Western blotting analysis indicated that the relative protein expression levels of proliferative (cyclin D1 and PCNA) and metastatic (N-cadherin and vimentin) markers in the ADORA2B siRNA group were significantly lower than those in the control siRNA group (Figures 9C,D). These results demonstrated that knockdown of ADORA2B inhibited the proliferation and metastasis of LUAD cells and that ADORA2B might participate in the proliferation and metastasis of LUAD cells.
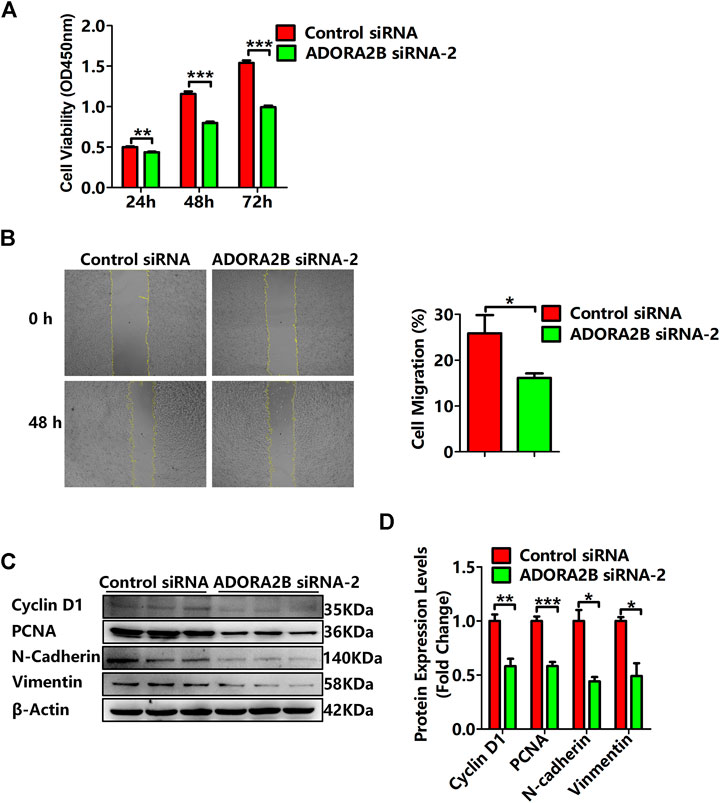
FIGURE 9. Knockdown of ADORA2B inhibited the proliferation and metastasis of LUAD cells. (A) The cell viability of LUAD cells transfected with control or ADORA2B siRNA was detected by the CCK-8 assay. (B) The wound healing assay was used to examine the effect of ADORA2B siRNA on A549 cell migration for 48 h (magnification: ×40). (C) Relative protein expressions of proliferative (cyclin D1 and PCNA) and metastatic (N-cadherin and vimentin) markers were determined by western blotting. (D) Protein quantitative statistics. *p < 0.05, **p < 0.01, and ***p < 0.001 are statistically significant.
Discussion
Adenosine is a purinergic molecule that regulates tissue damage and repair. Adenosine receptors belong to the G-protein–coupled receptor superfamily and include A1, A2A, A2B, and A3, which are widely distributed in different human tissues. The expression levels and affinities of adenosine receptors can vary across different tissues and cell types. In addition, these adenosine receptors have different phosphorylation abilities and sensitivities to the G-proteins with which they are conjugated. The A1 receptor is conjugated with the Gi/G0 protein, while the A3 receptor is conjugated with the Gi protein to inhibit the activity of adenylate cyclase and reduce the level of cAMP. In contrast, A2A and A2B receptors are conjugated with Gs proteins to activate the activity of adenylate cyclase and increase the level of cAMP. Low concentrations usually activate the A2A receptor, while higher concentrations of adenosine activate the A2B receptor, which suggests that the basal level of adenosine mainly activates the A2A receptor in a physiological state and that the A2B receptor is activated only when the adenosine level rises above physiological levels or reaches a pathological level (Vecchio et al., 2018; Fenouillet et al., 2019).
Increasing evidence suggests that adenosine receptors are potential diagnostic biomarkers for some diseases, inflammation, and certain types of cancers (Feoktistov and Biaggioni, 2011; Koeppen et al., 2011; Fishman et al., 2012). Additionally, adenosine receptors are upregulated in various tumors (Fishman et al., 2009). ADORA2B plays an important role in the occurrence, development, and metastasis of human tumors and is expressed in a variety of tumor cells. In the present study, to observe the functional network of ADORA2B in LUAD, we performed bioinformatics analyses to analyze transcriptional sequencing data from clinical sample data obtained from the Gene Expression Omnibus (GEO) and TCGA databases. To determine the role of ADORA2B in LUAD, we evaluated the expression pattern of several genes expressed in the disease. The analysis of the Oncomine database revealed fold-changes in gene expression levels that were similar to those reported in previous LUAD studies. According to the mRNA expression level, ADORA2B ranked among the top 8–33% of all upregulated genes expressed in LUAD, indicating that ADORA2B is significantly overexpressed in LUAD (Figure 1). Survival analysis showed that the OS time (including the overall, one-year, three-year, and five-year survival rates) in the low ADORA2B expression group was significantly longer than that in the high ADORA2B expression group. These results indicated that a high ADORA2B expression predicted a poor prognosis for LUAD patients (Figure 4).
Further analysis using the UALCAN database showed that the mRNA expression levels of ADORA2B were significantly higher in LUAD samples than in para-carcinoma tissues. Furthermore, the expression varied depending on the cancer stage, age, smoking habit, nodal metastasis status, and TP53 mutation status. In addition, the copy number of ADORA2B was significantly increased in LUAD (Figure 2). These associations indicated the impact of ADORA2B expression on human health or the types of biological and social factors that contribute to the pathological process of LUAD.
Cancer progression involves a series of histopathological processes (Xie et al., 2017; Chen et al., 2016) and changes in the functions of specific genes that have potential inhibitory or carcinogenic effects on cancer progression (Cui et al., 2018; Klonowska et al., 2016). Therefore, in the present study, we used the web-based tool, cBioPortal, to detect amplification, mutations, and mRNA expression of ADORA2B in LUAD. The major types of ADORA2B alterations were mRNA low, deep deletion, and amplification. The fraction of genome altered and mutation type, including not mutated, not profiled for mutations, amplification, gain, diploid, shallow deletion, and deep deletion, were analyzed. These mutations may play key roles in cancer progression and prognosis (Figure 3). Tumorigenesis is an extremely complex process, and new intricate protein–protein interactions and altered signaling pathways accelerate the pathological process of malignant tumors. ADORA2B was reported to be involved in tumor-cell proliferation, tumor-cell metastasis, and tumor microenvironment changes (Desmet et al., 2013; Kasama et al., 2015; Vecchio et al., 2016). To further clarify the role of ADORA2B in the progression of LUAD, we performed related functional experiments to verify the function of ADORA2B. The results indicated that knockdown of ADORA2B inhibited the proliferation and metastasis of LUAD cells and that ADORA2B might participate in the proliferation and metastasis of LUAD cells (Figure 9).
To identify the functional proteins in LUAD, we used the GeneMANIA web server. Interacting proteins regulate tumor progression. To determine the hierarchy of interacting proteins, we used the LinkedOmics database and drew a heat map of differentially expressed genes correlated with ADORA2B expression. In LUAD, ADORA2B was predicted to be associated with several ADORA2B-associated genes. Among them, ANXA1 was highly expressed in various malignant tumors (Huang et al., 2015; Okano et al., 2015), and upregulation of ANXA1 levels in the serum was related to the pathological grade and clinical stage of patients with particular forms of LUAD. Knockout of ANXA1 expression inhibited the proliferation, migration, and invasion of lung carcinoma cells (Fang et al., 2016), while the expression level of ITGA3 can be used as a diagnostic and prognostic marker for several malignant tumors (Koshizuka et al., 2017; Jiao et al., 2019; Ramovs et al., 2019). S100A6, which regulates cytoskeleton protein dynamics, cell proliferation, cell differentiation, calcium metabolism, ubiquitination, and acetylation, was found to promote the proliferation, invasion, migration, and angiogenesis of lung carcinoma cells by inhibiting acetylation of p53 when overexpressed (Li et al., 2019). KDM2B can reduce the proliferation of carcinoma cells by inhibiting the expression of oncogenes that play important roles in the self-renewal, differentiation, and apoptosis of stem cells (Hong et al., 2016; Goyama and Kitamura, 2017). Therefore, the findings of the present study validated the significance of ADORA2B in LUAD (Figure 5). ADORA2B interacts with these proteins that regulate important cellular processes associated with cancers and diseases.
Enrichment analysis of target gene sets using GSEA can identify important networks involved in protein targeting, mRNA processing, and transcription factors. ADORA2B was revealed to regulate the FAS signaling pathway, EGF receptor signaling pathway, and apoptosis signaling pathway (Figure 6). The CD73–adenosine receptor signaling pathway can promote the immunosuppressive effect of the tumor microenvironment, which is closely associated with a poor prognosis of multiple tumors (Chen et al., 2020). Immune cell infiltration analysis results suggested that the expression level of ADORA2B was associated with infiltrating immune cells in LUAD, including B cells, macrophages, neutrophils, and dendritic cells (Figure 7). These results revealed the indispensable role of ADORA2B in the development of LUAD.
The results of our preliminarily bioinformatics analysis suggested that ADORA2B can be used as a biomarker for LUAD, and we further confirmed the role of ADORA2B in LUAD through in vitro experiments. Human LUAD– and bronchial epithelial cell–derived cells lines were cultured in vitro, and mRNA and protein levels of ADORA2B were analyzed. Our results showed that both the transcription and protein levels of ADORA2B were significantly higher in A549 and NCl-H1299 cells than in HBE cells. The expression of genes positively (ANXA1, ITGA3, and S100A6) or negatively (KDM2B, NEB, and CBFA2T2) correlated with ADORA2B was consistent with the transcription level of ADORA2B. siRNA ADORA2B transfection experiments further confirmed these results (Figure 8). Our in vitro experiments confirmed the predictive and diagnostic role of ADORA2B in LUAD, but there were some limitations of this study. We tried to obtain human LUAD tissues to verify our in vitro results; however, it is difficult to acquire human LUAD tissues owing to ethical and technical reasons. If we get an opportunity to acquire LUAD tissues in the future, we will further investigate the role of ADORA2B in LUAD.
Conclusion
In conclusion, we used several public databases to extract data related to mRNA expression and mutations of ADORA2B and to the expression of genes associated with ADORA2B in LUAD. Furthermore, we predicted the differentially expressed genes correlated with ADORA2B expression in LUAD. Survival analysis showed that the OS time in the low ADORA2B expression group was longer than that in the high ADORA2B expression group. These results indicated that a high expression of ADORA2B predicted a poor prognosis for LUAD patients. Enrichment analysis of target gene sets using GSEA revealed important networks of the ADORA2B signaling pathway. Immune infiltration analysis results suggested that the expression level of ADORA2B was associated with immune cell infiltration in LUAD. In vitro experiments revealed that the relative ADORA2B expression levels in LUAD cells were significantly higher than those in normal bronchial epithelial cells. Knockdown of ADORA2B inhibited the proliferation and metastasis of LUAD cells, indicating that ADORA2B might participate in the proliferation and metastasis of LUAD cells. Taken together, our results revealed the significance of ADORA2B expression and its interaction networks in LUAD; the results of our in vitro experiments were consistent with those of the bioinformatics analysis. Although A549, NCl-H1299, and HBE cell lines were mainly used in the current experiments, we will verify these results with human and murine LUAD tissues in future studies. Our findings provide valuable insights into ADORA2B as a potential diagnostic and prognostic marker for LUAD and thus provide a foundation for further research focusing on LUAD biomarker development.
Data Availability Statement
Okayama Lung data were downloaded from the Oncomine database under Series accession number GSE31210 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE31210). Landi Lung data were downloaded from the Oncomine database under Series accession number GSE10072 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE10072). Selamat Lung data were downloaded from the Oncomine database under Series accession number GSE32863 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE32863). Stearman Lung data were downloaded from the Oncomine database under Series accession number GSE2514 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE2514). Beer Lung data were downloaded from the Oncomine database under the Array Type: HumanGeneFL Array, Measured 5,338 genes, 7,133 reporters (Beer, D. G., Kardia, S. L., Huang, C. C., Giordano, T. J., Levin, A. M., Misek, D. E., et al., 2002). Gene expression profiles predict survival of patients with lung adenocarcinoma (Nat. Med. 8, 816–824. doi: 10.1038/nm733). Su Lung data were downloaded from the Oncomine database under Series accession number GSE7670 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE7670). Bhattacharjee Lung data were downloaded from the Oncomine database under the Array Type: Human Genome U95A-Av2 Array, Measured 8,603 genes, 12,651 reporters (Bhattacharjee, A., Richards, W. G., Staunton, J., Li, C., Monti, S., Vasa, P., et al., 2001). Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses (Proc. Natl. Acad. Sci. U S A98, 13790–13795. doi: 10.1073/pnas.191502998). Garber Lung data were downloaded from the Oncomine database under Series accession number GSE3398 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE3398). OncoPrint data were downloaded from the cBioPortal database Shortened URL https://bit.ly/3oTsOd5. Mutation data were downloaded from the cBioPortal database Shortened URL https://bit.ly/2ObTF7t. Clinical attribute data were downloaded from the cBioPortal database Shortened URL https://bit.ly/3rnelHU. LinkedOmics data were downloaded from the LinkedOmics database under ID number ID-62189.
Author Contributions
YS and JL designed the study, performed the data analysis, and aided in writing the manuscript. JZ, ZZ, and ZW completed the data download and analysis and in vitro experiments. ZJ and ZM evaluated and reviewed the manuscript structure, ideas, and science. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant Number 31800722 to ZM), Tianjin Natural Science Foundation (Grant Number 19JCQNJC11400 to ZM), China Postdoctoral Science Foundation (Grant Number 2019M660030 to YS), Tianjin Health Commission Science and Technology Talent Cultivation Project (Grant Number KJ20040 to ZM), Postdoctoral Fund of the First Affiliated Hospital, Jinan University (Grant Number 809036 to ZJ), and HAI YAN Science Foundation of Harbin Medical University Cancer Hospital (Grant Numbers JJQN 2021-14 and JJMS 2021-30 to JL). We are grateful to the data contributors of Oncomine, UALCAN, cBioPortal, GeneMANIA, LinkedOmics, KM Plotter, and TIMER for providing access to all the data required for this study. We are also grateful to Editage for editing the revised manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2021.663011/full#supplementary-material
Abbreviations
AAMP, angio-associated migratory cell protein; ADORA2B, adenosine receptor A2B; ALK, anaplastic lymphoma kinase; ANXA1, annexin A1; CBFA2T2, CBFA2/RUNX1 partner transcriptional co-repressor 2; BRAF, V-raf murine sarcoma viral oncogene homolog B1; DCC, netrin 1 receptor; DPH, diphthamide biosynthesis 1; EGFR, epidermal growth factor receptor; GEO, Gene Expression Omnibus; HER2, human epidermal growth factor receptor 2; ITGA3, integrin subunit alpha 3; KCNJ3, potassium inwardly rectifying channel subfamily J member 3; KDM2B, lysine demethylase 2B; KRAS, Kirsten rat sarcoma viral oncogene; LUAD, lung adenocarcinoma; NEB, nebulin; OS, overall survival; PEX6, peroxisomal biogenesis factor 6; RET, rearranged during transfection; ROS1, ROS proto-oncogene 1; SLC29A2, solute carrier family 29 member 2; SLC9A3R2, solute carrier family 9 member A3 regulator 2; S100A6, S100 calcium binding protein A6; TCGA, The Cancer Genome Atlas; TPD52L2, tumor protein D52-like 2; ZNF267, zinc finger protein 267.
References
Beer, D. G., Kardia, S. L. R., Huang, C.-C., Giordano, T. J., Levin, A. M., Misek, D. E., et al. (2002). Gene-expression Profiles Predict Survival of Patients with Lung Adenocarcinoma. Nat. Med. 8, 816–824. doi:10.1038/nm733
Bhattacharjee, A., Richards, W. G., Staunton, J., Li, C., Monti, S., Vasa, P., et al. (2001). Classification of Human Lung Carcinomas by mRNA Expression Profiling Reveals Distinct Adenocarcinoma Subclasses. Proc. Natl. Acad. Sci. 98, 13790–13795. doi:10.1073/pnas.191502998
Borea, P. A., Gessi, S., Merighi, S., and Varani, K. (2016). Adenosine as a Multi-Signalling Guardian Angel in Human Diseases: when, where and How Does it Exert its Protective Effects?. Trends Pharmacol. Sci. 37, 419–434. doi:10.1016/j.tips.2016.02.006
Cancer Genome Atlas Research Network (2018). Author Correction: Comprehensive Molecular Profiling of Lung Adenocarcinoma. Nature 559, E12. doi:10.1038/s41586-018-0228-6
Chandrashekar, D. S., Bashel, B., Balasubramanya, S. A. H., Creighton, C. J., Ponce-Rodriguez, I., Chakravarthi, B. V. S. K., et al. (2017). UALCAN: a portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 19, 649–658. doi:10.1016/j.neo.2017.05.002
Chen, S., Akdemir, I., Fan, J., Linden, J., Zhang, B., and Cekic, C. (2020). The Expression of Adenosine A2B Receptor on Antigen-Presenting Cells Suppresses CD8+ T-Cell Responses and Promotes Tumor Growth. Cancer Immunol. Res. 8, 1064–1074. doi:10.1158/2326-6066.CIR-19-0833
Chen, W.-C., Wang, C.-Y., Hung, Y.-H., Weng, T.-Y., Yen, M.-C., and Lai, M.-D. (2016). Systematic Analysis of Gene Expression Alterations and Clinical Outcomes for Long-Chain Acyl-Coenzyme A Synthetase Family in Cancer. PLoS One 11, e0155660. doi:10.1371/journal.pone.0155660
Covarrubias, R., Chepurko, E., Reynolds, A., Huttinger, Z. M., Huttinger, R., Stanfill, K., et al. (2016). Role of the CD39/CD73 Purinergic Pathway in Modulating Arterial Thrombosis in Mice. Arterioscler Thromb. Vasc. Biol. 36, 1809–1820. doi:10.1161/ATVBAHA.116.307374
Csóka, B., Selmeczy, Z., Koscsó, B., Németh, Z. H., Pacher, P., Murray, P. J., et al. (2012). Adenosine Promotes Alternative Macrophage Activation via A2A and A2B Receptors. FASEB j. 26, 376–386. doi:10.1096/fj.11-190934
Cui, X., Jing, X., Yi, Q., Long, C., Tan, B., Li, X., et al. (2018). Systematic Analysis of Gene Expression Alterations and Clinical Outcomes of STAT3 in Cancer. Oncotarget 9, 3198–3213. doi:10.18632/oncotarget.23226
Desmet, C. J., Gallenne, T., Prieur, A., Reyal, F., Visser, N. L., Wittner, B. S., et al. (2013). Identification of a Pharmacologically Tractable Fra-1/ADORA2B axis Promoting Breast Cancer Metastasis. Proc. Natl. Acad. Sci. 110, 5139–5144. doi:10.1073/pnas.1222085110
Drilon, A., Oxnard, G. R., Tan, D. S. W., Loong, H. H. F., Johnson, M., Gainor, J., et al. (2020). Efficacy of Selpercatinib in RET Fusion-Positive Non-small-cell Lung Cancer. N. Engl. J. Med. 383, 813–824. doi:10.1056/NEJMoa2005653
Fang, Y., Guan, X., Cai, T., Long, J., Wang, H., Xie, X., et al. (2016). Knockdown of ANXA1 Suppresses the Biological Behavior of Human NSCLC Cells In Vitro. Mol. Med. Rep. 13, 3858–3866. doi:10.3892/mmr.2016.5022
Fenouillet, E., Mottola, G., Kipson, N., Paganelli, F., Guieu, R., and Ruf, J. (2019). Adenosine Receptor Profiling Reveals an Association between the Presence of Spare Receptors and Cardiovascular Disorders. Int. J. Mol. Sci. 20, 5964. doi:10.3390/ijms20235964
Feoktistov, I., and Biaggioni, I. (2011). Role of Adenosine A2B Receptors in Inflammation. Adv. Pharmacol. 61, 115–144. doi:10.1016/B978-0-12-385526-8.00005-9
Fishman, P., Bar-Yehuda, S., Liang, B. T., and Jacobson, K. A. (2012). Pharmacological and Therapeutic Effects of A3 Adenosine Receptor Agonists. Drug Discov. Today 17, 359–366. doi:10.1016/j.drudis.2011.10.007
Fishman, P., Bar-Yehuda, S., Synowitz, M., Powell, J. D., Klotz, K. N., Gessi, S., et al. (2009). Adenosine Receptors and Cancer. Handb. Exp. Pharmacol. 193, 399–441. doi:10.1007/978-3-540-89615-9_14
Garber, M. E., Troyanskaya, O. G., Schluens, K., Petersen, S., Thaesler, Z., Pacyna-Gengelbach, M., et al. (2001). Diversity of Gene Expression in Adenocarcinoma of the Lung. Proc. Natl. Acad. Sci. 98, 13784–13789. doi:10.1073/pnas.241500798
Goyama, S., and Kitamura, T. (2017). Epigenetics in normal and Malignant Hematopoiesis: An Overview and Update 2017. Cancer Sci. 108, 553–562. doi:10.1111/cas.13168
Hamidzadeh, K., and Mosser, D. M. (2016). Purinergic Signaling to Terminate TLR Responses in Macrophages. Front. Immunol. 7, 74. doi:10.3389/fimmu.2016.00074
Hong, X., Xu, Y., Qiu, X., Zhu, Y., Feng, X., Ding, Z., et al. (2016). MiR-448 Promotes Glycolytic Metabolism of Gastric Cancer by Downregulating KDM2B. Oncotarget 7, 22092–22102. doi:10.18632/oncotarget.8020
Huang, Y., Zhang, C., Chen, C., Sun, S., Zheng, H., Wan, S., et al. (2015). Investigation of Circulating Antibodies to ANXA1 in Breast Cancer. Tumor Biol. 36, 1233–1236. doi:10.1007/s13277-014-2751-x
Jiao, Y., Li, Y., Liu, S., Chen, Q., and Liu, Y. (2019). ITGA3 Serves as a Diagnostic and Prognostic Biomarker for Pancreatic Cancer. Ott 12, 4141–4152. doi:10.2147/OTT.S201675
Karmouty-Quintana, H., Weng, T., Garcia-Morales, L. J., Chen, N.-Y., Pedroza, M., Zhong, H., et al. (2013). Adenosine A2B Receptor and Hyaluronan Modulate Pulmonary Hypertension Associated with Chronic Obstructive Pulmonary Disease. Am. J. Respir. Cel Mol. Biol. 49, 1038–1047. doi:10.1165/rcmb.2013-0089OC
Kasama, H., Sakamoto, Y., Kasamatsu, A., Okamoto, A., Koyama, T., Minakawa, Y., et al. (2015). Adenosine A2b Receptor Promotes Progression of Human Oral Cancer. BMC Cancer 15, 563. doi:10.1186/s12885-015-1577-2
Klonowska, K., Czubak, K., Wojciechowska, M., Handschuh, L., Zmienko, A., Figlerowicz, M., et al. (2016). Oncogenomic Portals for the Visualization and Analysis of Genome-wide Cancer Data. Oncotarget 7, 176–192. doi:10.18632/oncotarget.6128
Koeppen, M., Eckle, T., and Eltzschig, H. K. (2011). Interplay of Hypoxia and A2B Adenosine Receptors in Tissue protection. Adv. Pharmacol. 61, 145–186. doi:10.1016/B978-0-12-385526-8.00006-0
Koshizuka, K., Hanazawa, T., Kikkawa, N., Arai, T., Okato, A., Kurozumi, A., et al. (2017). Regulation of ITGA 3 by the Anti‐tumor miR‐199 Family Inhibits Cancer Cell Migration and Invasion in Head and Neck Cancer. Cancer Sci. 108, 1681–1692. doi:10.1111/cas.13298
Landi, M. T., Dracheva, T., Rotunno, M., Figueroa, J. D., Liu, H., Dasgupta, A., et al. (2008). Gene Expression Signature of Cigarette Smoking and its Role in Lung Adenocarcinoma Development and Survival. PLoS One 3, e1651. doi:10.1371/journal.pone.0001651
Li, P., Lv, X., Zhang, Z., and Xie, S. (2019). S100A6/miR193a Regulates the Proliferation, Invasion, Migration and Angiogenesis of Lung Cancer Cells through the P53 Acetylation. Am. J. Transl. Res. 11, 4634–4649.
Mazieres, J., Cropet, C., Montané, L., Barlesi, F., Souquet, P. J., Quantin, X., et al. (2020). Vemurafenib in Non-small-cell Lung Cancer Patients with BRAFV600 and BRAFnonV600 Mutations. Ann. Oncol. 31, 289–294. doi:10.1016/j.annonc.2019.10.022
Mertens, T. C. J., Hanmandlu, A., Tu, L., Phan, C., Collum, S. D., Chen, N.-Y., et al. (2018). Switching-off Adora2b in Vascular Smooth Muscle Cells Halts the Development of Pulmonary Hypertension. Front. Physiol. 9, 555. doi:10.3389/fphys.2018.00555
Okano, M., Kumamoto, K., Saito, M., Onozawa, H., Saito, K., Abe, N., et al. (2015). Upregulated Annexin A1 Promotes Cellular Invasion in Triple-Negative Breast Cancer. Oncol. Rep. 33, 1064–1070. doi:10.3892/or.2015.3720
Okayama, H., Kohno, T., Ishii, Y., Shimada, Y., Shiraishi, K., Iwakawa, R., et al. (2012). Identification of Genes Upregulated in ALK-Positive and EGFR/KRAS/ALK-negative Lung Adenocarcinomas. Cancer Res. 72, 100–111. doi:10.1158/0008-5472.CAN-11-1403
Philip, K., Mills, W. T., Davies, J., Chen, N. Y., Karmouty‐Quintana, H., Luo, F., et al. (2017). HIF1A Up‐regulates the ADORA2B Receptor on Alternatively Activated Macrophages and Contributes to Pulmonary Fibrosis. FASEB j. 31, 4745–4758. doi:10.1096/fj.201700219R
Pillai, R. N., Behera, M., Berry, L. D., Rossi, M. R., Kris, M. G., Johnson, B. E., et al. (2017). HER2 Mutations in Lung Adenocarcinomas: a Report from the Lung Cancer Mutation Consortium. Cancer 123, 4099–4105. doi:10.1002/cncr.30869
Ramovs, V., Secades, P., Song, J.-Y., Thijssen, B., Kreft, M., and Sonnenberg, A. (2019). Absence of Integrin α3β1 Promotes the Progression of HER2-Driven Breast Cancer In Vivo. Breast Cancer Res. 21, 63. doi:10.1186/s13058-019-1146-8
Román, M., Baraibar, I., López, I., Nadal, E., Rolfo, C., Vicent, S., et al. (2018). KRAS Oncogene in Non-small Cell Lung Cancer: Clinical Perspectives on the Treatment of an Old Target. Mol. Cancer 17, 33. doi:10.1186/s12943-018-0789-x
Russell, P. A., Wainer, Z., Wright, G. M., Daniels, M., Conron, M., and Williams, R. A. (2011). Does Lung Adenocarcinoma Subtype Predict Patient Survival?: a Clinicopathologic Study Based on the New International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society International Multidisciplinary Lung Adenocarcinoma Classification. J. Thorac. Oncol. 6, 1496–1504. doi:10.1097/JTO.0b013e318221f701
Selamat, S. A., Chung, B. S., Girard, L., Zhang, W., Zhang, Y., Campan, M., et al. (2012). Genome-scale Analysis of DNA Methylation in Lung Adenocarcinoma and Integration with mRNA Expression. Genome Res. 22, 1197–1211. doi:10.1101/gr.132662.111
Shaw, A. T., Felip, E., Bauer, T. M., Besse, B., Navarro, A., Postel-Vinay, S., et al. (2017). Lorlatinib in Non-small-cell Lung Cancer with ALK or ROS1 Rearrangement: an International, Multicentre, Open-Label, Single-Arm First-In-Man Phase 1 Trial. Lancet Oncol. 18, 1590–1599. doi:10.1016/S1470-2045(17)30680-0
Stearman, R. S., Dwyer-Nield, L., Zerbe, L., Blaine, S. A., Chan, Z., Bunn, P. A., et al. (2005). Analysis of Orthologous Gene Expression between Human Pulmonary Adenocarcinoma and a Carcinogen-Induced Murine Model. Am. J. Pathol. 167, 1763–1775. doi:10.1016/S0002-9440(10)61257-6
Su, L.-J., Chang, C.-W., Wu, Y.-C., Chen, K.-C., Lin, C.-J., Liang, S.-C., et al. (2007). Selection of DDX5 as a Novel Internal Control for Q-RT-PCR from Microarray Data Using a Block Bootstrap Re-sampling Scheme. BMC Genomics 8, 140. doi:10.1186/1471-2164-8-140
Tian, Z., Dixon, J., Guo, X., Deal, B., Liao, Q., Zhou, Y., et al. (2021). Co-inhibition of CD73 and ADORA2B Improves Long-Term Cigarette Smoke Induced Lung Injury. Front. Physiol. 12, 614330. doi:10.3389/fphys.2021.614330
Vecchio, E. A., Baltos, J. A., Nguyen, A. T. N., Christopoulos, A., White, P. J., and May, L. T. (2018). New Paradigms in Adenosine Receptor Pharmacology: Allostery, Oligomerization and Biased Agonism. Br. J. Pharmacol. 175, 4036–4046. doi:10.1111/bph.14337
Vecchio, E. A., Tan, C. Y. R., Gregory, K. J., Christopoulos, A., White, P. J., and May, L. T. (2016). Ligand-independent Adenosine A2B Receptor Constitutive Activity as a Promoter of Prostate Cancer Cell Proliferation. J. Pharmacol. Exp. Ther. 357, 36–44. doi:10.1124/jpet.115.230003
Warde-Farley, D., Donaldson, S. L., Comes, O., Zuberi, K., Badrawi, R., Chao, P., et al. (2010). The GeneMANIA Prediction Server: Biological Network Integration for Gene Prioritization and Predicting Gene Function. Nucleic Acids Res. 38, W214–W220. doi:10.1093/nar/gkq537
Xie, S., Shen, C., Tan, M., Li, M., Song, X., and Wang, C. (2017). Systematic Analysis of Gene Expression Alterations and Clinical Outcomes of Adenylate Cyclase-Associated Protein in Cancer. Oncotarget 8, 27216–27239. doi:10.18632/oncotarget.16111
Yu, H. A., Arcila, M. E., Rekhtman, N., Sima, C. S., Zakowski, M. F., Pao, W., et al. (2013). Analysis of Tumor Specimens at the Time of Acquired Resistance to EGFR-TKI Therapy in 155 Patients with EGFR-Mutant Lung Cancers. Clin. Cancer Res. 19, 2240–2247. doi:10.1158/1078-0432.CCR-12-2246
Yu, L., Todd, N. W., Xing, L., Xie, Y., Zhang, H., Liu, Z., et al. (2010). Early Detection of Lung Adenocarcinoma in Sputum by a Panel of microRNA Markers. Int. J. Cancer 127, 2870–2878. doi:10.1002/ijc.25289
Keywords: ADORA2B, lung adenocarcinoma, mRNA expression, functional network analysis, verification experiment
Citation: Sui Y, Liu J, Zhang J, Zheng Z, Wang Z, Jia Z and Meng Z (2021) Expression and Gene Regulation Network of Adenosine Receptor A2B in Lung Adenocarcinoma: A Potential Diagnostic and Prognostic Biomarker. Front. Mol. Biosci. 8:663011. doi: 10.3389/fmolb.2021.663011
Received: 02 February 2021; Accepted: 17 June 2021;
Published: 19 July 2021.
Edited by:
Min Zhang, Shanghai First People’s Hospital, ChinaReviewed by:
Jiarong Chen, Jiangmen Central Hospital, ChinaXiaoping Li, Jiangmen Central Hospital, China
Copyright © 2021 Sui, Liu, Zhang, Zheng, Wang, Jia and Meng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhenghu Jia, jiazhenghu86@163.com; Ziyu Meng, mengziyu@tmu.edu.cn
†These authors have contributed equally to this work
 Yutong Sui
Yutong Sui Jiayin Liu2†
Jiayin Liu2†  Zhenghu Jia
Zhenghu Jia Ziyu Meng
Ziyu Meng