- 1School of Life Science, Inner Mongolia University, Hohhot, China
- 2Inner Mongolia Academy of Agricultural and Animal Husbandry Sciences, Hohhot, China
- 3Key Laboratory of Black Soil Protection and Utilization (Hohhot), Ministry of Agriculture and Rural Affairs, Hohhot, China
- 4Inner Mongolia Key Laboratory of Degradation Farmland Ecological Restoration and Pollution Control, Hohhot, China
- 5College of Agronomy, Inner Mongolia Agricultural University, Hohhot, China
Drought is the most prevalent environmental stress in crop production, posing a significant danger to food security. Microorganisms in the crop root zone affect crop growth and development, enhance effective nutrient use, and resist adversity hazards. To analyze the changes and functional differences of root space microbial (endosphere-rhizosphere-bulk soil) communities in spring wheat under drought stress. In this study, the root, rhizosphere, and bulk soil of the drought-tolerant group (DTG, three varieties) and drought-sensitive group (DSG, three varieties) were collected. The control (CK, 25–28%), moderate drought (MD, 15–18%), and severe drought (SD, 9–12%) were analyzed by high-throughput sequencing and bioinformatics. The results showed significant differences in the diversity of Bacteria and Fungi in the root space of spring wheat under drought stress (P < 0.05), with the drought-tolerant group exhibiting higher microbial diversity. The microbial community change in spring wheat root space was mainly determined by the niche differentiation of endosphere, rhizosphere, and bulk soil and declined from endosphere to bulk soil due to drought. The antagonism between microbial and root-space species increased, and the community’s complexity and stability deteriorated. Enriching drought-resistant preference groups like Actinobaciota, Variovorax, Streptomyces, and Conocybe altered the structure and function of the microbial community in the root space of spring wheat. Spring wheat’s root space Bacteria and Fungi have different strategies to respond to drought.
1. Introduction
Microorganisms are abundant in soil and are influenced by elements including vegetation type, nutrients, and climate (Leff et al., 2015; Prober et al., 2015; Zhou et al., 2016). Soil microflora microbial communities are micro-ecosystems that regulate and drive several biological processes in the global macro-ecosystem (Bhattacharjee et al., 2022). Earth Microbiome Project (EMP) (Gilbert et al., 2014; Locey and Lennon, 2016; Thompson et al., 2017) showed that the diversity, structure, and function of microbial community were closely correlated with soil environment (Human Microbiome Project Consortium, 2012; Amir et al., 2017). The interaction between crops, soil microenvironment, and soil microorganism maintains the agroecosystem’s equilibrium (Xu et al., 2018). Crop root-space microorganisms include root endosphere (inside the root), rhizosphere (soil close enough to the root system to be affected by the release of root exudates), and bulk soil (soil areas other than the rhizosphere) communities (Naylor and Coleman-Derr, 2017; Francioli et al., 2021), These microorganisms can affect the crops’ growth and development, enhance the effective utilization of nutrients, and resist adversity (Berendsen et al., 2012; Bulgarelli et al., 2013) to improve soil productivity and crop yield. The crops can selectively “acclimate” certain beneficial Bacteria through root exudates, thus enhancing plant growth and development (Reinhold-Hurek et al., 2015; Zhalnina et al., 2018). Improving crop resilience and mitigating adversity hazards through microorganisms is an important means of sustainable agriculture (Zhou et al., 2017).
Drought is the most prevalent environmental stress in crop production, and climate change and global warming are accelerating the occurrence of severe droughts, causing serious ecological and food security problems (Lesk et al., 2016; Webber et al., 2018; Yang N. et al., 2021). Drought will not only affect crop growth and development of crops and the microbial community structure, but it will also increase soil heterogeneity, limit nutrient transport, and raise soil oxygen concentration, resulting in a substantial fall in soil microbial biomass (Xu et al., 2018). Studies have shown that drought can significantly change the microflora structure of Bacteria and Fungi in soil and promote the microorganisms’ enrichment with higher drought tolerance (Ochoa-Hueso et al., 2018). Specific recombination of rice root microbiota distribution was found after drought stress. The proportion of monoderm bacteria such as Actinobacteria, Chloroflexi, and aerobic Firmicutes is substantially higher in root endosphere communities than in rhizosphere populations (Santos-Medellín et al., 2017, 2021). Li et al. (2022) suggested that, under water stress, the relative abundance of Gemmatimonas in rhizosphere soil is positively related to the soil water content, while the relative abundance of drought-tolerant microbiota such as Azospirillum and Bradyrhizobium remained unchanged with the change of soil water content, which potentially used inert strategy to adapt to different levels of water content to resist drought hazard. Several studies have demonstrated that, under drought, plant root microbiota favors migration to gram-positive populations, displacing most gram-negative lineages. The enrichment occurred mostly in the plants’ root endosphere and could also be observed in the rhizosphere. Moreover, this enrichment has also been shown to be positively correlated with the intensity and duration of drought and to disappear rapidly after rehydration (Edwards et al., 2015; Naylor et al., 2017; Fitzpatrick et al., 2018; Ault, 2020). Therefore, to investigate crop resilience mechanisms, it is crucial to comprehend the precise drought-induced alterations in the microbiota of the root space.
Plants benefit from various soil microorganisms, among which root-space microorganisms are vital in plant health, and certain microbiota may mitigate plant drought stress (Santos-Medellín et al., 2021). Plant growth-promoting rhizobacteria (PGPR) can directly or indirectly boost plant growth under stress, inhibit plant pathogenic Bacteria, and enhance plant tolerance to drought by stimulating nitrogen fixation, increasing nutrient absorption, and improving soil properties (Glick, 1995a,1995b; Babalola, 2010). A four-species bacterial population (Stenotrophomonas rhizophila, Xanthomonas retroflexus, Microbacterium oxydans, and Paenibacillus amylolyticus, termed SPMX) significantly increased the survival rate of Arabidopsis plants under drought stress and induced drought resistance in plants, which resulted from the potential overall emergence property of SPMX (Yang Y. et al., 2021). Some Fungi colonized in the plants’ endosphere can improve the hosts’ defense against abiotic and biotic stresses and promote plant growth (Feng et al., 2017; Latz et al., 2018). In the rhizosphere, beneficial Fungi, Arbuscular mycorrhizal fungi (AMFs), can form a connection between the host and soil by an external hyphal network, hence enhancing the adaptability of host plants, increasing their nutrient absorption and improving their productivity under drought stress (Juroszek and Tiedemann, 2013; Kwak and Weller, 2013). Wheat (Triticum aestivum L.) is one of China’s three main foods, and its planting area and production are second only to corn and rice. Spring wheat is mainly grown in northern China, with a wide distribution range and a long planting history. However, drought has caused a serious yield reduction. Soil water stress affects changes in fungal and bacterial communities in the spring wheat microbiome, but has a smaller effect on plant genotypes, and communities sensitive to water stress differ significantly among different wheat genotypes (Azarbad et al., 2020). Breitkreuz et al. (2021) conducted drought stress treatment on two winter wheat varieties with different soil types. It was found that the composition of the bacterial community in rhizosphere soil was strongly affected by drought and soil types, and drought reduced the diversity of rhizosphere Bacteria, increasing the relative abundance of drought-tolerant Bacteria. Jochum et al. (2019) to improve the seedling growth of wheat under extreme drought conditions, a microbial community that delays the dry time of wheat seedlings was obtained through 6 rounds of artificial selection using HMME, and these microbiomes mediate changes in the rhizosphere environment, thereby improving the adaptive ability of plants to drought stress. Mavrodi et al. (2018), research results showed that indigenous Phz+ rhizobacteria that produce the antibiotic phenazine-1-carboxylic acid (PCA) contribute to the natural inhibition of soil-borne pathogens in wheat. The previous studies on wheat drought resistance mainly focused on the rhizosphere or only concentrated on one aspect of soil microorganisms (Bacteria, Fungi). However, the changes in microbiota and their interactions in wheat root-space (endosphere-rhizosphere-bulk soil) are rarely reported. Therefore, understanding the response of root-space microorganisms to drought stress in drought-tolerant spring wheat would aid in elucidating the potential causes of the spring wheat’s drought resistance.
Three drought-tolerant spring wheat varieties (Dingxi 40, Longmai 36, and Longmai 33) and three drought-sensitive spring wheat varieties (Non-gmai 2, Bamai 12, and Bafeng 5) were used in this study. Rainproof sheds and regulated hydration were used to achieve water content control. Using high-throughput sequencing technology and bioinformatics, we primarily explored the changes in spring wheat root-space microorganisms. We analyzed the crucial function of several edaphic conditions in creating root-space microorganisms. The following hypotheses were proposed: (1) From the endosphere to the bulk soil, the effect of drought stress on root-space microbial diversity declines; (2) Drought impairs the diversity and stability of root-space microorganisms; (3) Critical bacterial and fungal communities from different locations of the root-space (endosphere-rhizosphere-bulk soil) exhibit different response toward drought.
2. Materials and methods
2.1. Field experiment design
The field experiment was initiated in 2022 at Inner Mongolia Autonomous Region Academy of Agriculture and Animal Husbandry Tenihe Soil Management and Ecological Restoration Scientific Observation and Experimental Station (Tenihe Farm), Inner Mongolia Autonomous Region, China (49°55′N, 120°48′E, 650 m above sea level). The region has a moderately temperate semi-arid continental steppe climate with an average annual precipitation of 373–474 mm. The daily average temperature and precipitation changes during the whole growth period of spring wheat in 2022 are shown in Supplementary Figure 1. The soil type of the cultivated land at the experimental station was black calcium soil, and the water holding capacity in the field was 30–35%, which belonged to the representative of the black soil in the western foothills of the Daxing’anling Mountains in the typical Inner Mongolia agro-pastoral staggered belt. Pre-sowing 0–20 cm soil nutrients: available nitrogen 37.26 mg ⋅ kg–1, available potassium 195.83 mg ⋅ kg–1, available phosphorus 21.25 mg ⋅ kg–1, organic matter 57.82 g ⋅ kg–1, and pH 6.8.
It was a two-factor experiment combining three water treatments and two groups of spring wheat varieties. Using the plant water gradient division method proposed by Hsiao (1973), the main factor was three kinds of water treatments: In the 0–20 cm soil layer, control (CK) the soil mass water content was 25–28% (accounting for 70–80% of the field water-holding capacity); moderate drought (MD) the soil mass water content was 15–18% (accounting for 40–50% of the field water-holding capacity); severe drought (SD) the soil mass water content was 9–12% (accounting for 25–35% of the field water-holding capacity) (Ji et al., 2021). The secondary factor was three cultivars (DX40, LM36, and LM33) in the drought-tolerant group (DTG) and three cultivars (NM2, BM12, and BF5) in the drought-sensitive Group (DSG), for various details in Supplementary Table 1. There were 18 treatments; the drought treatment group and the control group were conducted in the dry shed, and the soil mass water content was controlled through rain prevention and water replenishment in the dry shed, which was mechanically sown on May 9, 2022, with a seeding volume of 300 kg ⋅ hm–2. The experiment adopted a random block design (Supplementary Figure 2). Each treatment had three replicate plots with a size of 3 × 3 m, a total of 54 plots. The spacing between the plots was 0.5 m, and a protection row with a width of 1 m was set. The drought treatment lasted 30 days from the beginning of the spring wheat jointing period (July 4) to the end of the flowering period (August 2). On July 28, the soil mass water content reached the preset range, and after 5 days (August 2), the soil mass water content of SD, MD, and CK treatment was 9.6, 15.4, and 25.9% (Supplementary Table 8), respectively, and sample collection began. Details of soil mass water content monitoring are shown in Supplementary Figure 3.
2.2. Sample collection
According to the sample collection methods of Bulgarelli et al. (2012) and Bai et al. (2022), the samples from this experiment were collected. The specific method was as follows: in each plot, we adopted the 5-point sampling method, the topsoil was removed on the soil surface at the sampling point, and the soil drill was used to take soil samples of 0–20 cm soil layer, and repeat thrice for each treatment. After the soil sample was fully and evenly mixed, it was screened with 1 mm sterile stainless-steel mesh and divided into two parts. A portion of it was placed in a sterile centrifuge tube, frozen in liquid nitrogen, and stored at −80°C for bulk soil microbial diversity sequencing. The remaining portion was shade-dried and placed in a plastic bag containing desiccant for soil microbiological and chemical analysis. Five representative spring wheat plants were randomly selected in the plot; the bulk soil was removed from the spring wheat’s root, the root was shaken to remove the loose soil at the root, and then a sterile brush was used to remove rhizosphere soil from the root surface. After the spring wheat rhizosphere soil samples repeated thrice in the same treatment were fully mixed and evenly, they were screened with 0.5 mm sterile stainless-steel mesh to remove obvious plant roots, and animal remains, and other residues. Afterward, the screened rhizosphere soil was placed in a sterile centrifuge tube, frozen in liquid nitrogen, and stored at −80°C for rhizosphere microbial diversity sequencing. The remaining wheat roots were rinsed with sterile water 3–5 times, dried with filter paper, and cut with sterile scissors. The roots were placed in a sterile centrifuge tube, frozen with liquid nitrogen, and stored at −80°C for subsequent detection of endosphere microbial analysis. Endosphere, rhizosphere, bulk soil Bacteria, and Fungi were sequenced using 16S rRNA and ITS amplicon sequencing techniques. A total of 162 samples [18 treatments × 3 sample types (root, rhizosphere, and bulk soil) × 3 replicates = 162] were collected.
2.3. Microbial DNA extraction, PCR amplification, and amplicon sequencing
According to E.Z.N.A.® soil DNA Kit (Omega Bio-tek, Norcross, GA, U.S.), total microbial genome DNA was extracted from spring wheat root and soil samples under different drought treatments. The total DNA quality and concentration were determined by 1% agarose gel electrophoresis and NanoDrop2000 (Thermo Scientific), an ultramicro nucleic acid quantifier.
The hypervariable region V4 of the bacterial 16S rRNA gene was amplified with primer pairs 515F (5′-GTGCCAGCMGCCGC GGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) primers (Caporaso et al., 2012). PCR amplification reaction system: 4 μL 5 × FastPfu buffer, 2 μL 2.5 mM dNTPs, 0.8 μL Each Primer (5 μM), 0.4 μL FastPfu polymerase, 0.2 μL BSA, 10 ng of template DNA, and ddH2O to a final volume of 20 μL. ITS1F (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS2R (5′-GCTGCGTTCTTCATCGATGC-3′) primers were used to amplify the fungus ITS sequence (Adams et al., 2013). PCR amplification reaction system: 2 μL 10× Buffer, 2 μL 2.5 mM dNTPs, 0.8 μL Each Primer (5 μM), 0.2 μL rTaq Polymerase, 0.2 μL BSA, 10 ng of template DNA, and ddH2O to a final volume of 20 μL. PCR amplification condition: 1 × (initial denaturation 95°C 3 min); 27 × (denaturing 95°C 30 s; annealing 55°C 30 s; extension 72°C 45 s); 1 × (single extension 72°C for 10 min). The sample was then kept at 4°C (PCR instrument: ABI GeneAmp® 9700). Purification and quantification were performed using the AxyPrep DNA gel extraction kit (AxyPrep Biosciences, Union City, CA, USA) and quantum™Fluorometer (Promega, USA), respectively. Sequencing was performed by Illumina’s Miseq PE300 platform (Majorbio Bio-Pharm Technology Co., Ltd., Shanghai, China).
2.4. Microbial data analysis
The raw sequencing data were processed with fastp (version 0.19.6; Chen et al., 2018) and software splicing FLASH (version 1.2.11; Magoč and Salzberg, 2011), screening out low-quality Reads sequences and splicing the remaining two-terminal sequence data into Tags. Using UPARSE software (version 7.1; Stackebrandt and Goebel, 1994; Edgar, 2013) According to 97% similarity, the sequences after quality control splicing were clustered by operational taxonomic unit (OTU), and chimeras were removed. The OTU matrix was normalized using the trimmed mean of M values (TMM) method in the R-package edgeR (Robinson et al., 2010). Using an RDP classifier (version 2.11; Wang et al., 2007) compared the Silva 16 S rRNA (Quast et al., 2013) and UNITE ITS (Rustgi et al., 2017) gene databases to classify OTUs, and the confidence threshold was 70%. Each sample’s community composition was counted at different species classification levels.
2.5. Statistical analysis
The Bacteria and Fungi sequencing data were analyzed on the Majorbio Cloud platform.1 The Shannon index of Alpha diversity was calculated using Mothur (v1.30.1; Schloss et al., 2009) software.2 Kruskal-Wallis rank sum test was used to analyze the difference between Alpha diversity groups, and GraphPad Prism 9 was used for drawing. One-way ANOVA and linear mixed model (LMM) were used to analyze the effects of drought stress on bacterial and fungal α diversity. R (version 3.3.1) PCoA statistical analysis (bray-Curtis) was used to test the similarity of microbial community structure in spring wheat root space under drought stress, and the PERMANOVA non-parametric test was used to determine if the difference between drought treatment and spring wheat root space microbial community groups was significant (Oksanen et al., 2013). Moreover, linear discriminant analysis effect Size (LEfSe) (Segata et al., 2011)3 (LDA > 3.2, P < 0.05) was used to determine the microbial groups with significant differences in genus abundance between different groups. The VPA analysis in the vegan package of R (version 3.3.1) was used to quantitatively evaluate the individual and joint interpretation of soil microbial traits and soil chemical traits on the microbial community differences in spring wheat root space under drought stress. Based on spearman correlation |r| > 0.5 p < 0.01, the top 30 genera of Bacteria and Fungi were screened for correlation network analysis in spring wheat root space under drought stress (Segata et al., 2011). The network analysis tool (Assenov et al., 2008) in Cytoscape was used to calculate the node connectivity (Degree), transitivity, clustering coefficient, the average shortest path of the network, and other attributes obtain the pertinent information between microorganism groups and samples and draw a correlation network diagram.
The structural equation model (SEM) was used to explore the possible direct and indirect effects of drought treatment, spatial position, and spring wheat variety category on soil factors and bacterial and fungal diversity (Shannon). CK, MD, and SD treatments used 25, 15, and 9 as soil mass water content variables, and endosphere, rhizosphere, and bulk soil used 1, 2 and 3 as spatial position variables. Moreover, DTG and DSG used 1, 2 as spring wheat variety category variables. Alpha diversity of Fungi and Bacteria was measured by the Shannon index. The SEM model was fitted using IBM SPSS Amos 22.0 (USA). Chi-square (P > 0.05), the goodness of fit index (GFI > 0.90), and root mean square error of approximation (RMSEA < 0.001) (Grace and Keeley, 2006; Bai et al., 2022). FAPROTAX (Louca et al., 2016) and FUNGuild (Nguyen et al., 2016) tools were employed to predict the bacterial and fungal sequencing results function.
3. Results
3.1. Changes in microbial diversity
This study employed 16S rRNA and ITS amplicon sequencing techniques were used to systematically analyze the bacterial and fungal community structure in the endosphere (RE), rhizosphere (RS), and bulk soil (BS) of spring wheat under drought stress to clarify the drought stress effect on the spatial microbial diversity of spring wheat roots. Moreover, it was further determined whether the spatial microbial diversity of spring wheat roots in drought-tolerant group (DTG) and drought-sensitive group (DSG) was different in response to drought stress. All samples contained 23,559,191 bacterial and 22,157,557 fungal high-quality sequences (Supplementary Table 2). The bacterial and fungal sequences were clustered into 24,336 and 8,338 OTUs, with an average length of 253.03 and 245.70 bp, respectively. The dilution curve of each sample tends to be flat as sequencing data increases, and the amount of sequencing data is reasonable (Supplementary Figure 4).
There were significant differences in the Shannon index of Bacteria and Fungi in root space (RE, RS, and BS) of spring wheat under drought stress (P < 0.05). The bacterial diversity of the rhizosphere and bulk soil was significantly higher than the endosphere under different drought treatments. The fungal diversity was BS > RS > RE, and the Shannon index of endosphere Bacteria and Fungi was the lowest (Figure 1). Moreover, through single factor variance and LMM analysis, it was found that the interaction of Drought treatment × Variety category × Spatial position had a significant effect on fungal community diversity (P < 0.01). There were significant differences in bacterial diversity between drought treatment, rhizosphere, and bulk soil (P < 0.01). The effect of spatial location on bacterial and fungal Alpha diversity was greater than drought treatment (Supplementary Table 3).
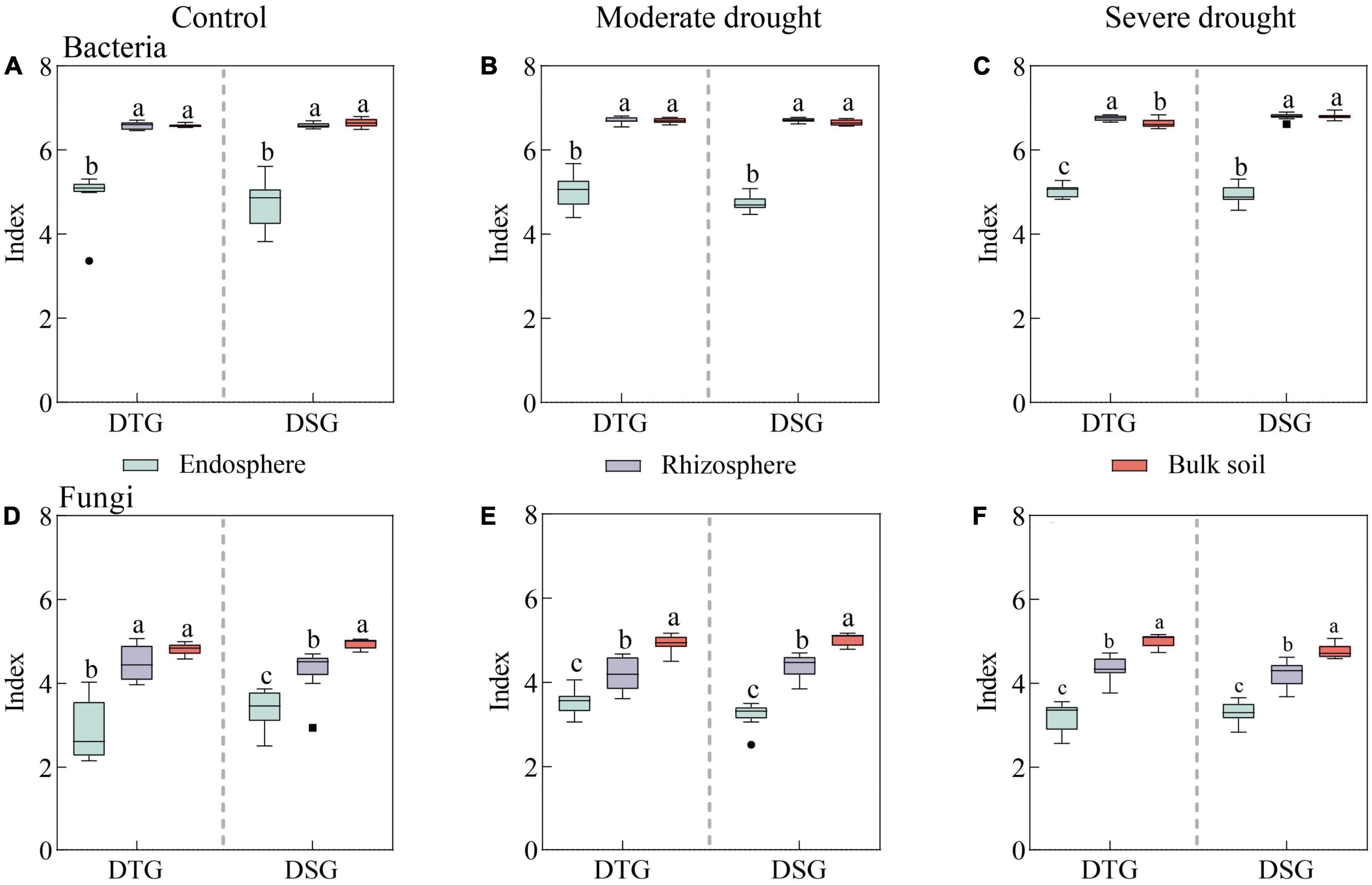
Figure 1. Shannon diversity index (RE, RS, and BS) of Bacteria and Fungi in the rhizosphere of spring wheat under the same drought stress. Shannon index of RE Bacteria (A), Shannon index of RS Bacteria (B), Shannon index of BS Bacteria (C), Shannon index of RE Fungi (D), Shannon index of RS Fungi (E), Shannon index of BS Fungi (F). Endosphere (RE), Rhizosphere (RS), Bulk soil (BS). Drought-tolerant group (DTG) and Drought-sensitive group (DSG).
In bacterial diversity, compared to CK, there was no significant difference in the Shannon diversity index of spring wheat RE between the drought-tolerant and sensitive group under moderate drought (MD) and severe drought (SD) treatments (P > 0.05) (Supplementary Figure 5A). The Shannon index of RS and BS of spring wheat in the drought-tolerant and the drought-sensitive groups increased slightly with the drought increase (Supplementary Figures 5B, C). Compared to CK, the RE Shannon diversity index of spring wheat in the drought-tolerant group under MD and SD treatments increased initially and then declined (P < 0.05) in terms of fungal diversity (Supplementary Figure 5D). There was no significant difference in RS fungal diversity between the drought-tolerant and the sensitive groups (P > 0.05) (Supplementary Figure 5E). Under SD stress, the drought-sensitive group’s lowest spring wheat BS Shannon index was 4.76 (Supplementary Figure 5F). Drought stress significantly affected the microbial diversity in the root space of spring wheat. Compared to the drought-sensitive group, the drought-tolerant group had higher microbial diversity.
PCoA and PERMANOVA tests revealed significant differences in bacterial and fungal community structure between three water treatments and spatial root positions (P < 0.001). CK samples were significantly distinct from MD and SD samples. Endosphere samples are significantly separated from bulk soil and rhizosphere samples (Figure 2). Furthermore, the PERMANOVA test confirmed that the effect of different drought treatments on fungal (R2 = 0.17) community composition was greater than Bacteria (R2 = 0.05). The effects of drought treatment on bulk soil (R2 = 0.54), rhizosphere (R2 = 0.45), and endosphere (R2 = 0.28) fungal communities gradually diminished, although the effect on the rhizosphere (R2 = 0.39) bacterial community was the greatest (Supplementary Table 4).
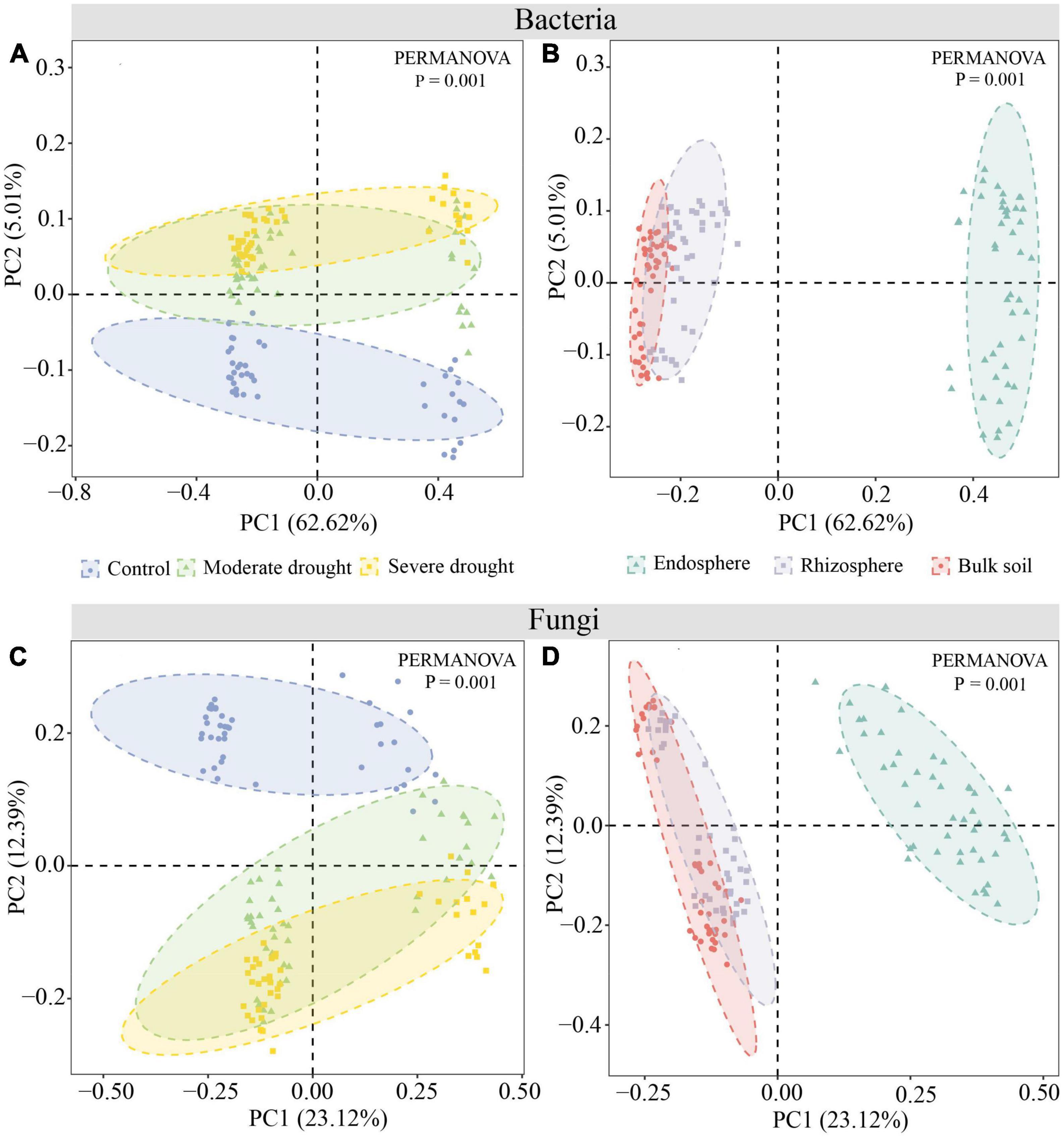
Figure 2. PCoA and Bray-Curtis similarity indexes were used to compare the microbial diversity of spring wheat root space under different drought treatments at the OTU classification level. Bacteria (A,B), Fungi (C,D).
3.2. Changes in microbial community composition
Analysis of the detected OTUs revealed that drought treatment had significant effects on the endosphere, rhizosphere, and bulk soil community compositions of spring wheat in the drought-tolerant and the drought-sensitive groups (P < 0.05), with the endosphere community composition being more sensitive to drought stress (Supplementary Figures 6, 7). Under drought stress, the relative abundance of Bacteroidota and Proteobacteria in the bacterial community composition of spring wheat root space gradually increased from bulk soil to endosphere. In contrast, the relative abundance of Firmicutes showed the opposite trend. Among the top 10 bacterial phyla, Patescibacteria was only enriched in the endosphere. Compared with CK, the relative abundance of Actinobacteriota in the endosphere Bacteria of spring wheat in the drought-tolerant group increased by 39.17 and 61.48% under MD and SD treatments, respectively. While the relative abundance in the drought-sensitive group gradually decreased by 20.68 and 12.71%, respectively. Under drought stress, the relative abundance of Proteobacteria and Planctomycetota in the endosphere and rhizosphere of spring wheat in the drought-sensitive group grew progressively. However, the drought-tolerant group exhibited the reverse pattern (Supplementary Figure 6). At the genus level, the relative abundance of Variovorax in the endosphere of spring wheat in the drought-tolerant and drought-sensitive groups increased significantly under drought stress. In contrast, the relative abundance of Lechevalieria, Streptomyces, Rhizobacter, and TM7a increased significantly in the drought-tolerant group, and the relative abundance of Pseudomonas increased significantly in the drought-sensitive group. The relative abundance of Bacillus in bulk soil and rhizosphere declined steadily with increasing drought, and spring wheat from the drought-sensitive group decreased the most under the SD treatment (RS: 37.42%; BS: 32.01%) (Supplementary Figure 7).
Consistent with the results of bacterial community composition, drought stress also caused significant differences (P < 0.05) in the fungal community composition of spring wheat’s root space (RE, RS, and BS). Under drought stress, Ascomycota, Basidiomycota, and Mortierellomycota were the dominant phyla (Supplementary Figure 6). Compared to CK, the relative abundance of Basidiomycota, Schizothecium, and Conocybe in spring wheat rhizosphere and bulk soil increased gradually under MD and SD treatments. The relative abundance of the pathogenic fungus Parastagonospora in the endosphere increased with the drought stress increase, particularly under SD treatment; the sensitive group of spring wheat increased by 986.03% (Supplementary Figure 7). In conclusion, drought stress will alter the microbial community composition in the spring wheat’s root area. Actinobacteriota and Variovorax are more resistant to drought. The drought-tolerant group strongly prefers Lechevalieria, Streptomyces, Rhizobacter, and TM7a, while the drought-sensitive group prefers Proteobacteria, Planctomycetota, and Pseudomonas.
3.3. Analysis of differences between microbiomes
Through Lefse analysis, the microbial community differences in root space between the drought-resistant and drought-sensitive groups under drought treatment were explored. At the level of bacterial genus, the biomarkers of endosphere, rhizosphere, and bulk soil of spring wheat in the drought-tolerant and drought-sensitive groups decreased gradually, namely RE-RS-BS: 35-25-14 (Figure 3A; Supplementary Figures 8A, C) and RE-RS-BS: 39-25-9 (Supplementary Figures 9A, C, E), respectively. These biomarkers differed significantly in response to drought treatment (P < 0.05). Bacillus was a prevalent biomarker for CK. Variovorax, Streptomyces, and Nocardioides are common biomarkers in the endosphere and rhizosphere of MD and SD spring wheat. Flavobacterium and Pedobacter were common biomarkers in the rhizosphere of MD spring wheat. In bulk soil, Blastococcus under MD treatment and Massilia and Thermomonas under SD treatment were unique biomarkers for the drought-tolerance group. Norank _ f _ Vicinamibacteraceae and Norank _ f _ Nitrososphaeraceae are unique biomarkers for the drought-sensitive group under MD and SD treatment, respectively. At the fungal genus level, the two groups (DTG and DSG) of spring wheat bulk soil had the most biomarkers, 51 (Figure 3B; Supplementary Figures 8B, D) and 55 (Supplementary Figures 9B, D, F), respectively. Schizothecium is a frequent root space fungus biomarker in spring wheat. Consequently, the biomarkers of Bacteria and Fungi in the root space of spring wheat responded differentially to various drought conditions. Under drought stress, Variovorax, Streptomyces, and other genera in the endosphere and rhizosphere are key biomarkers.
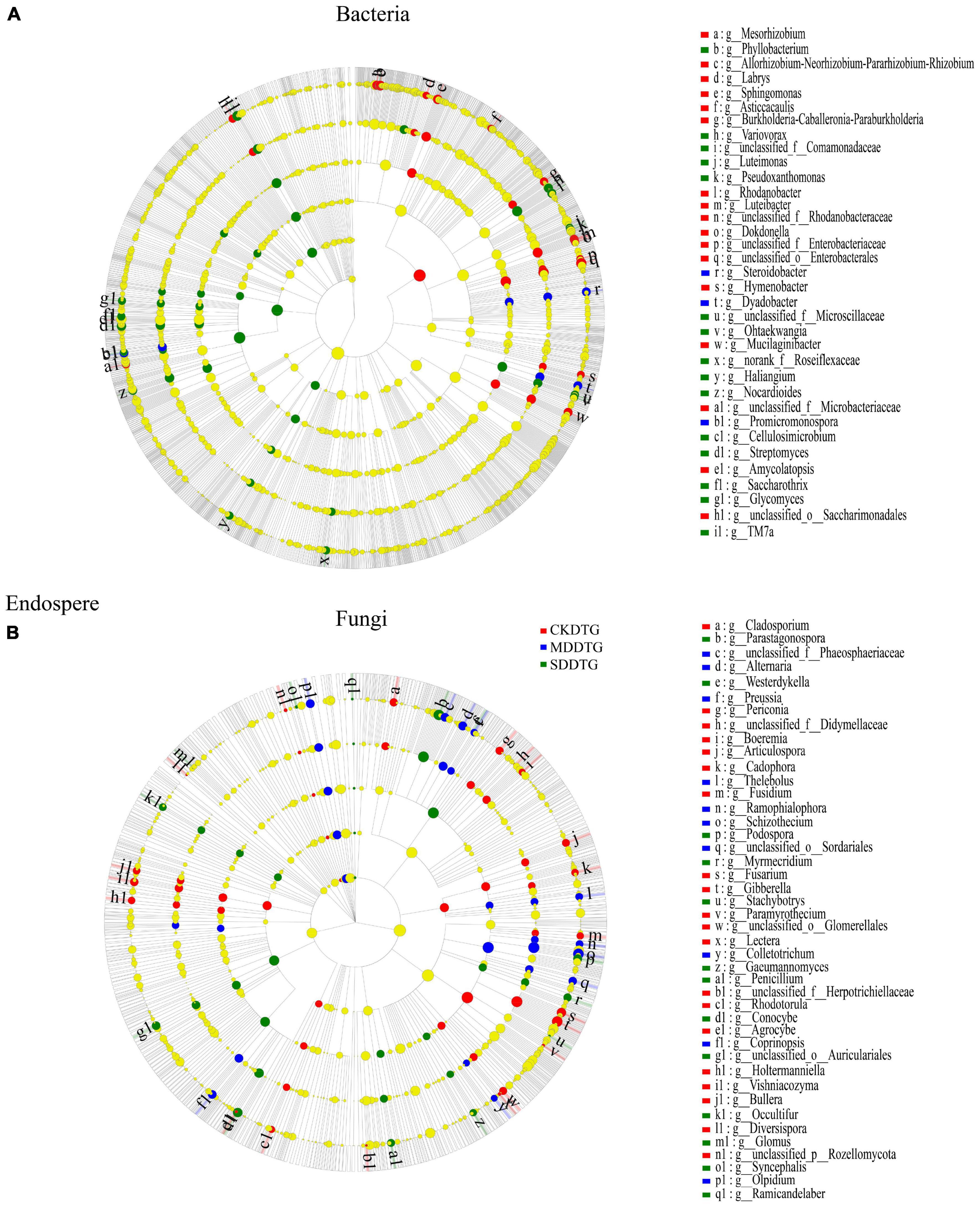
Figure 3. LEfSe analysis of different species between Bacteria (A) and Fungi (B) in the endosphere of the drought-tolerant group (DRG) spring wheat under drought treatment. Following the species at the phylum, class, order, family, and genus levels is a cladogram from the inner circle to the outer circle. The figure’s letters indicate the species names in the right legend. Different color nodes represent the microbial populations significantly enriched by different treatments, and the differences between groups are significant. The yellow nodes represented the microbial populations with no significant difference between groups (P < 0.05, LDA score 3.2).
3.4. Relationship between microbial community and environmental factors
Spearman correlation analysis was used to analyze the correlation between the endosphere, rhizosphere, bulk soil microbial community, and soil chemical and microbial properties of spring wheat under drought stress in the drought-tolerant group (DTG) and drought-sensitive group (DSG). The results showed that there were six phyla in the spring wheat’s root space in the drought-tolerant group (DTG) that were significantly positively correlated with soil indicators (P < 0.05). They were Actinobacteriota, Proteobacteria, Acidobacteriota, Chloroflexi, Bacteroidota and Cyanobacteria. The relative abundance of Actinobacteriota in the endosphere was significantly positively correlated with pH, TK, CAT, and URE. The relative abundance of Proteobacteria was positively correlated with MBC, SC, SOC, MBN, and MBP. Significantly good correlations were found between the relative abundance of Cyanobacteria in the endosphere of spring wheat in the two groups (DTG and DSG) and ALP (Supplementary Figure 10A). Basidiomycota, Olpidiomycota, Mortierellomycota, Chytridiomycota, and Ascomycota were significantly positively correlated with soil indicators (P < 0.05) at the phylum level. The relative abundance of Basidiomycota in the rhizosphere and bulk soil was significantly positively correlated with TK except in the endosphere of DSG spring wheat (Supplementary Figures 10B, C).
VPA analysis showed that soil microbiological properties (SMP) were important driving factors for the changes of Bacteria and Fungi communities in the rhizosphere and bulk soil of spring wheat under drought stress (Figure 4; Supplementary Table 5). Soil microbiological properties (SMP) explained 13.24% (Figure 4A) and 14.60% (Supplementary Table 5) of the differences in bacterial community composition between rhizosphere and bulk soil, respectively, while they explained 16.35% (Figure 4B) and 9.97% (Supplementary Table 5) of the differences in fungal community composition, respectively. However, the soil chemical characteristics can only partially explain the discrepancies between the rhizosphere and bulk soil microbial communities (Supplementary Table 5). Afterward, we used the structural equation model (SEM) to analyze the possible pathways of direct and indirect effects of drought treatment, spatial position, and spring wheat variety category on soil factors and bacterial and fungal diversity (Shannon) (Figure 5; Supplementary Table 6). Drought stress had the greatest negative effect on soil chemical properties (−0.945) and soil microbiological properties (−0.941). The spatial position (RE-RS-BS) had the greatest positive effect on the Alpha diversity of Bacteria (0.827) and Fungi (0.878). The spring wheat variety category had the greatest positive effect on soil chemical properties (0.170) (Figure 5). Drought stress harmed bacterial (−0.154) and fungal (−0.282) Alpha diversity. It was found that drought stress had a considerable impact on the Alpha diversity of microorganisms in the root space of spring wheat and that this effect diminished from the endosphere to the bulk soil (Figure 5).
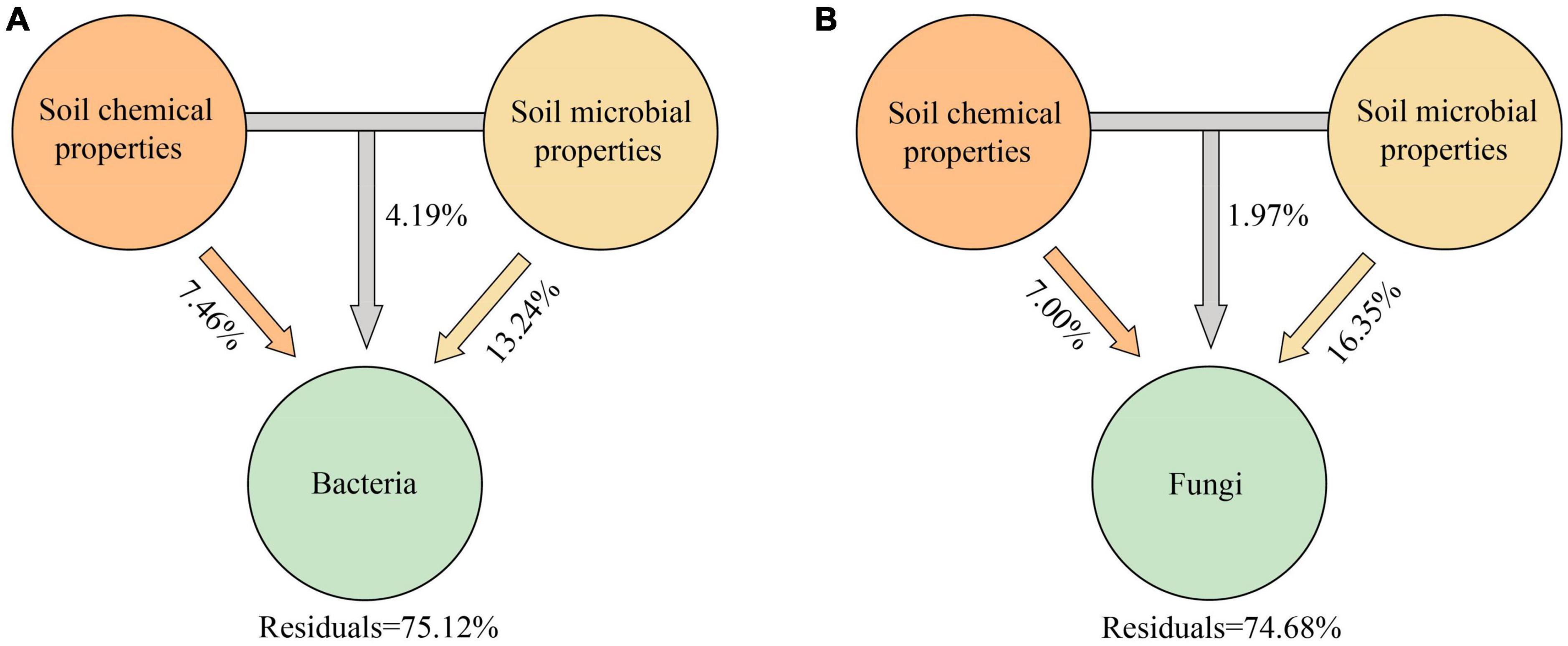
Figure 4. Variance partitioning analysis (VPA) of the effects of soil chemical and microbial properties on rhizosphere Bacteria (A) and Fungi (B) communities. Soil chemical properties (SCP) include pH; SOC, soil organic carbon content; TP, total phosphorus content; TK, total potassium content. Soil microbiological properties (SMP) include CAT, soil catalase activity; SC, soil invertase activity; URE, soil urease activity; ALP, soil alkaline phosphatase activity; MBC, soil microbial biomass carbon; MBN, soil microbial biomass nitrogen; MBP, soil microbial biomass phosphorus.
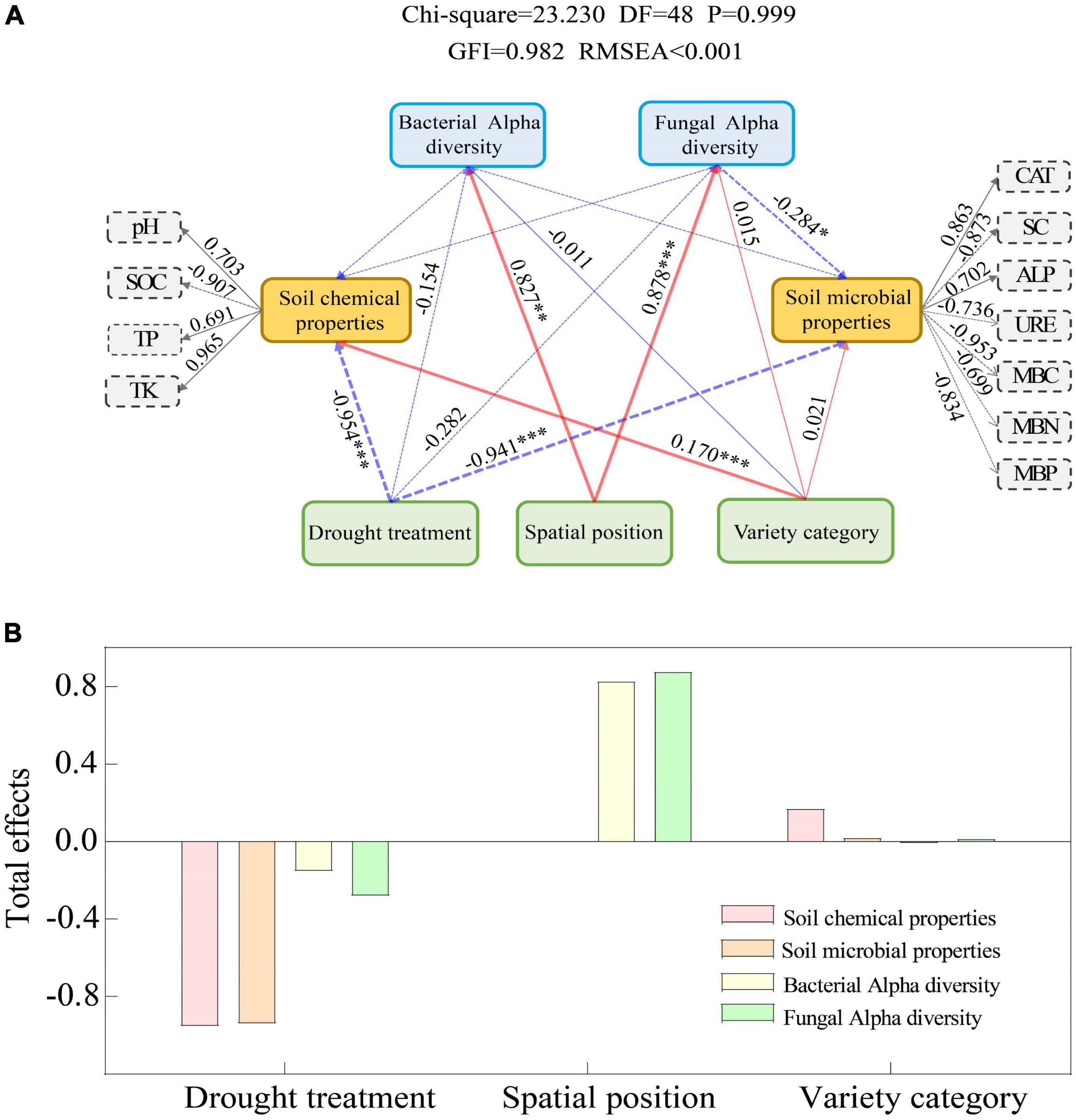
Figure 5. Structural equation model (SME) for direct and indirect effects of drought treatment, spatial position, and spring wheat variety category on soil factors and bacterial and fungal diversity (Shannon) (A). Standardized total effect (B). GIF > 0.9, RMSEA < 0.001 indicated that the model fit well. The red arrow indicates a significant positive impact, the blue arrow indicates a significant negative impact, the thickness of the arrow line indicates the size of the path coefficient in the model, and the dotted line indicates no significant impact; * means P < 0.05, ** means P < 0.01, *** means P < 0.001.
3.5. Species correlation network analysis
To further clarify the effect of drought stress on the microbial community in the spring wheat’s root space, we selected the top 30 species at the genus classification level for microbial network construction (Spearman correlation, P < 0.01). The results showed that the network density of bacterial and fungal communities in the root space of the drought-tolerant group of spring wheat under drought stress was BS > RS > RE (Figure 6; Supplementary Figure 11). The maximum network density of the Bacteria rhizosphere in the drought-sensitive group was 0.483, and Fungi results were consistent with the drought-tolerant group (Supplementary Table 7). The positive correlation ratio between rhizosphere and bulk soil bacterial communities in spring wheat 2 groups (DTG and DSG) reduced compared to the endosphere (Figure 6).
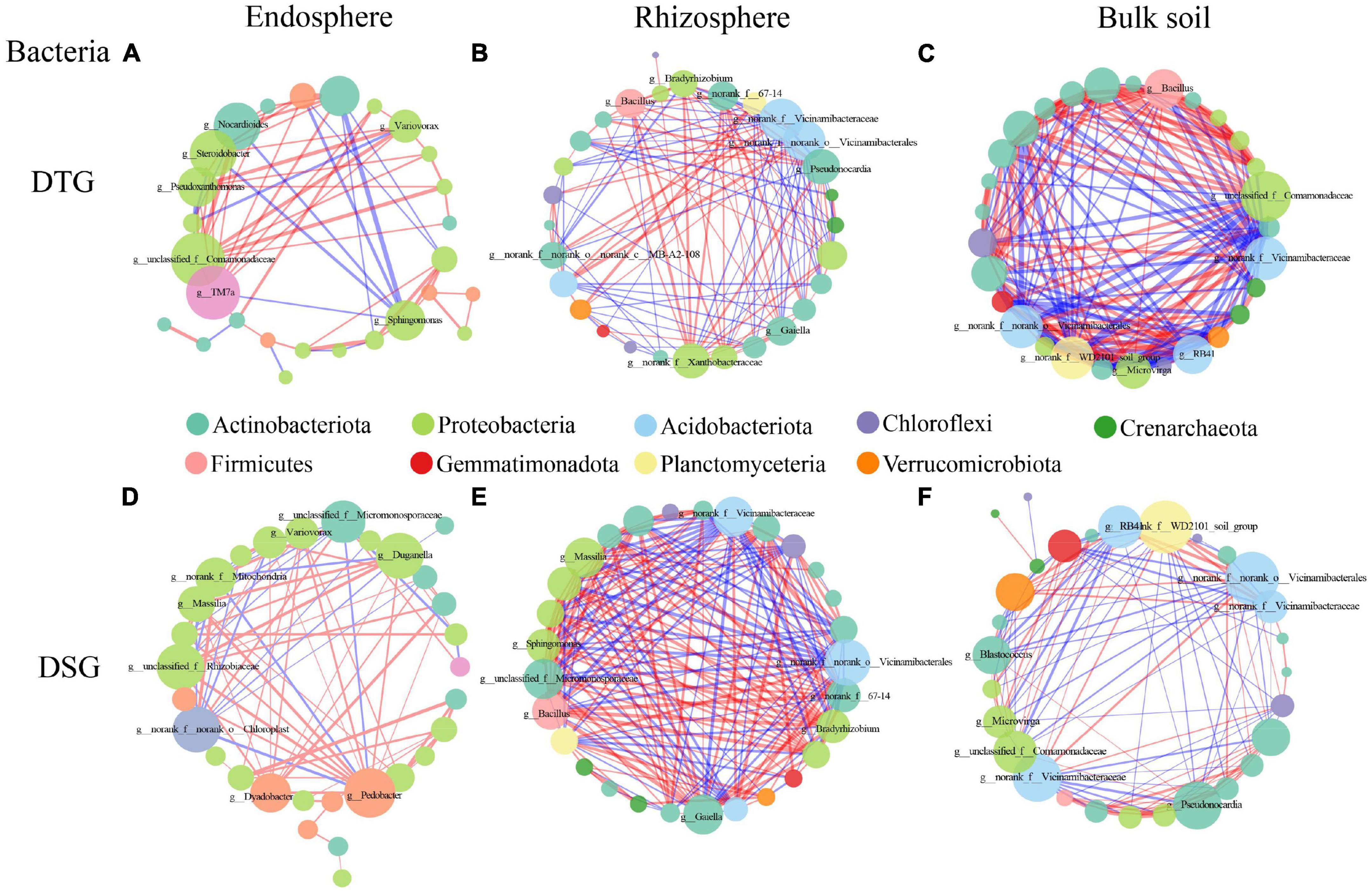
Figure 6. The bacterial genus level correlation network diagram of two groups (DTG and DSG) of spring wheat under drought stress. The red connection indicates a positive correlation, the blue connection indicates a negative correlation, the connection’s thickness represents the correlation coefficient’s size, and the number of lines indicates the degree of connection between nodes.
In Bacteria, TM7a (10) and Variovorax (7) in the DTG endosphere; Pseudonocardia (15) and Bacillus (12) in the rhizosphere; Bacillus (18) and Microvirga (17) in the bulk soil (Figures 6A–C); in the DSG, Pedobacter (13) in the endosphere; Massilia (20) and Bacillus (19) in the rhizosphere; Blastococcus (12) and Microvirga (11) in the bulk soil (Figures 6D–F), had the highest degree and positive correlation are both high in the whole network. In Fungi, in the DTG, Schizothecium (endosphere, 10), Occultifur (rhizosphere, 11), Coniochaeta (bulk soil, 18) (Supplementary Figures 11A–C); In DSG, Parastagonospora (endosphere, 9), Subulicystidium (rhizosphere, 15), Coniophora (bulk soil, 19) had the highest degree and the highest sensitivity in the whole network. Most of them were pathogenic Fungi (Supplementary Figures 11D–F). Therefore, Proteobacteria, Actinobacteriota, Firmicutes, Patescibacteria, and Bacteroidota were the essential core Bacteria of drought stress in the root-space microorganisms of spring wheat 2 groups (DTG and DSG).
3.6. Microbial community function prediction
FAPROTAX and FUNGuild tools were used to predict the bacterial and fungal communities’ function in spring wheat root space under drought stress and to explore the functional differences of microbial communities in spring wheat root space under drought stress. The results exhibited that nitrogen fixation was significantly reduced in DTG and DSG (RE-RS-BS) under drought stress in Bacteria (Figure 7). The abundance of nitrate reduction in DTG and DSG spring wheat increased significantly in the endosphere (Figure 7A). Compared to CK, SD dramatically boosted the nitrification abundance of DTG spring wheat by 52% in the rhizosphere (Figure 7B). In bulk soil, the ureolysis abundance of DTG and DSG increased significantly, and DTG increased the most (Figure 7C).
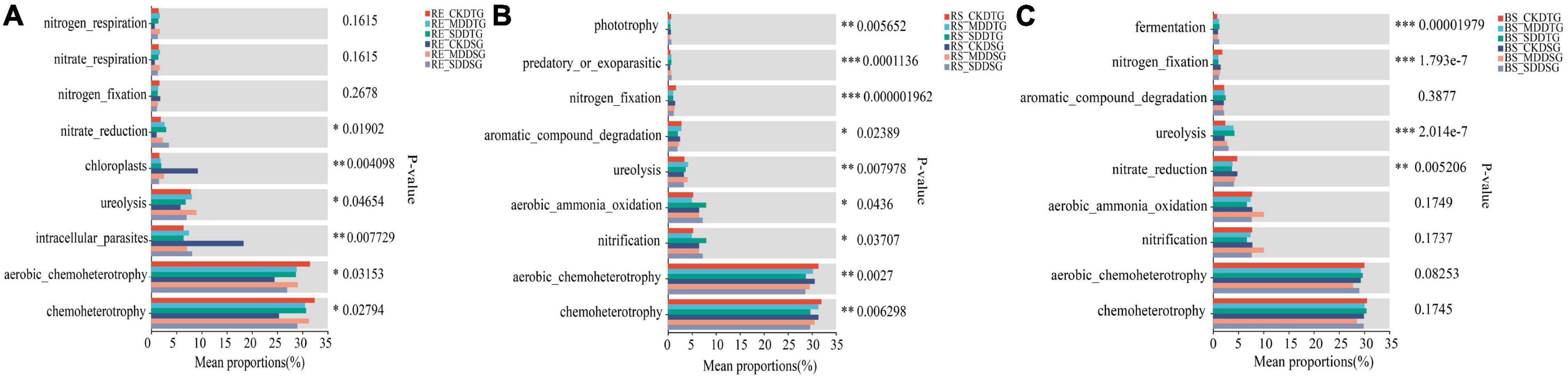
Figure 7. FAPROTAX was used to predict the function of the bacterial community in the root space of spring wheat under drought stress. Endosphere bacterial function prediction (A); Rhizosphere bacterial function prediction (B); Bulk soil bacterial function prediction (C). *P < 0.05, **P < 0.01, and ***P < 0.001.
Compared to CK, DTG, and DSG, spring wheat symbiotroph (Arbuscular Mycorrhiza) under MD and SD treatment considerably increased in (RS-BS) (Supplementary Figures 12C–F). In the endosphere, DSG Arbuscular Mycorrhiza of spring wheat decreased significantly, and in the rhizosphere, DSG increased by 1,036–1,463% (Supplementary Figures 12B, D). Fungal Parasite-Plant Pathogen-Plant Saprotroph increased significantly in (RE-RS-BS) (Supplementary Figures 12A, C, E). It suggested that Bacteria and Fungi in the root space of spring wheat responded to dryness using various tactics under drought stress.
4. Discussion
4.1. Drought-tolerant spring wheat can maintain relatively high microbial diversity
This study explored the microbial diversity of spring wheat roots under drought stress in two groups (DTG and DSG). The bacterial diversity of the rhizosphere and bulk soil was found to be significantly higher than the endosphere under different drought treatments. The fungal diversity was BS > RS > RE, and the Shannon index of endosphere Bacteria and Fungi was the lowest. The results of this study were also confirmed in the study of Xu et al. (2018). Drought stress would significantly reduce the bacterial diversity of the crop endosphere, while the effect on the bulk soil was insignificant. The effect of drought on fungal diversity was consistent with the bacterial diversity results, showing a decreasing trend from the endosphere, rhizosphere, and bulk soil. These results also indicate that the endosphere microbial community of spring wheat is more sensitive to drought stress and more susceptible to drought. Moreover, the Beta diversity PERMANOVA test showed that different drought treatments affected fungal (R2 = 0.17) community composition more than Bacteria (R2 = 0.05). Dassen et al. (2017) found that fungal communities are more sensitive to vegetation changes than bacterial communities, and Fungi are also the main consumers of underground plant-derived carbon (Ballhausen and de Boer, 2016). Our results also showed that Fungi are more sensitive to drought than bacteria. It is hypothesized that drought may alter the physiological metabolites of spring wheat and the carbon level of the soil, making fungus more vulnerable to drought. However, the precise reaction mechanism requires additional study.
Our study also found that the drought-tolerant group had higher microbial diversity than the drought-sensitive group under drought stress. Plant species or genotypes can choose different soil communities (Berendsen et al., 2012). Root exudates are vital in the selection of rhizosphere microbial communities (Williams and de Vries, 2020), and rhizosphere microorganisms prefer root exudates (Iannucci et al., 2013). When plants are harmed by adversity, a beneficial rhizosphere microbial community can be selected by modifying the type and amount of root exudates to help the plants survive adversity. There were significant differences in root exudates among different varieties of the same crop (Pang et al., 2021), which may be one of the reasons for the high microbial diversity of drought-tolerant spring wheat in this study.
4.2. Drought-induced enrichment of drought-resistant microbial community in spring wheat
Changes in microbial communities between compartments associated with plant roots are mostly governed by the host selectivity, which is influenced by the environment. Therefore, it has a strong driving effect on shaping the root-related microbial community (Chaparro et al., 2014; Reinhold-Hurek et al., 2015; Wagner et al., 2016). This experiment demonstrated that drought stress alters the microbial community composition in the root space of spring wheat. Among them, Actinobacteriota and Variovorax were significantly enriched in the endosphere, and the relative abundance increased with the deepening of drought stress. The drought-tolerant group strongly preferred Streptomyces and other Bacteria, while the drought-sensitive group preferred Pseudomonas and other Bacteria. Variovorax and Streptomyces were significant biomarkers in the roots and rhizosphere of spring wheat under drought stress. Numerous studies have shown that Actinobacteriota is widely enriched in numerous plants under drought stress (Naylor and Coleman-Derr, 2017; Santos-Medellín et al., 2017; Fitzpatrick et al., 2018), and Actinobacteriota was rapidly enriched in the roots, with the highest relative abundance.
Furthermore, it has been reported that the relative abundance of Streptomyces in Actinobacteriota continues to rise under drought stress, which may provide long-term memory stress protection for plants after drought alleviation (Santos-Medellín et al., 2021). Our study found that the relative abundance of Streptomyces in the endosphere of spring wheat in the drought-tolerant group gradually increased with the deepening of drought, and was significantly higher than that in the drought-sensitive group. This also suggests that Streptomyces has a predilection for drought-tolerant types and is more likely to favor them. This conjecture has also been confirmed in the studies of Fitzpatrick et al. (2018) and Santos-Medellín et al. (2021). Variovorax can protect plants from drought stress (Qi et al., 2022). Finkel et al. (2020) found that the widely distributed genus Variovorax can produce and degrade auxin, which can maintain normal root development through the rhizosphere environment.
Moreover, they further determined the core position of Variovorax in the plant-microorganism-microorganism interaction network. Variovorax biomarker levels in drought-tolerant and drought-sensitive spring wheat rhizosphere were considerably different. Simultaneously the relative abundance of Variovorax was significantly increased under drought stress in the spring wheat’s roots in both groups. Based on our findings and earlier research on the genus Variovorax, we believe the genus Variovorax may play a significant role in enhancing spring wheat drought tolerance.
We also found that the relative abundance of Basidiomycota, Schizothecium, and Conocybe in spring wheat rhizosphere and bulk soil increased significantly under drought stress. Parastagonospora was only enriched in the endosphere, and its abundance increased with the drought stress aggravation. The increased relative abundance of Conocybe is beneficial to maize growth under drought stress (Yang et al., 2022). In the experimental studies of long-term rotation (Neupane et al., 2021) and organic fertilizer (Ding et al., 2017), it was found that the relative abundance of Schizothecium in soil increased significantly, and the increase of the abundance of the Bacteria could alleviate the harm of other pathogens to crops. Parastagonospora is the pathogen of wheat glume blight. It produces cell necrosis in wheat plants carrying the dominant susceptible gene by secreting some effectors, thus providing nutrition for its growth and reproduction (Dagvadorj et al., 2022). Therefore, the effect of drought stress on the fungal community in the spring wheat’s root space will result in the coexistence of beneficial and harmful Bacteria. However, harmful Bacteria appear more inclined to enrich the root.
Additionally, we found that the effect of drought stress on the crop endosphere was greater than on the rhizosphere and bulk soil. We do not have concrete proof that this outcome is related to the enrichment of related diseases such as Parastagonospora, which is a problem that must be addressed in future studies on the crop-microorganism collaborative drought resistance mechanism. In summary, the spring wheat’s root space under drought stress will enrich drought-related microbial communities such as Variovorax, Streptomyces, and Conocybe.
4.3. Root niche of spring wheat determines the difference in microbial response to drought stress
In this study, the effect of spring wheat root spatial position on bacterial and fungal Alpha diversity was greater than drought treatment. Soil microbial properties were related to the changes of bacterial and fungal communities in the rhizosphere and bulk soil of spring wheat under drought stress. In contrast, endosphere bacterial community composition responded more to soil chemical properties (pH, SOC, TK, and TP). Ladau et al. (2018) showed that soil physical and chemical properties (pH, organic carbon, C: N ratios) were closely related to soil microbial community structure and richness, and the distribution of soil characteristics lagged behind climate change, which in turn led to the lag of Bacteria and Archaea. Different drought treatment methods will affect soil matrix potential and change soil physical properties. In PEG or pot experiments, when the water treatment reached −1.5Mpa, the soil adsorption capacity was large, the plant was at a permanent wilting point, the growth was stagnant, and the microorganisms were difficult to survive. However, for this study, there is a difference in water gradient between soil layers in the field. Whether 0–20 cm of the soil mass water content of 9–12% reaches the permanent wilting point needs to be further judged by plant height, plant physiology, differential genes, and other indicators. Furthermore, (Yang Y. et al., 2021) found that soil microbial biomass was vital in solution under drought stress, which was conducive to the microorganisms’ survival under high osmotic pressure. The species correlation network revealed that the network density of bacterial and fungal communities in the root space of the drought-tolerant spring wheat under drought stress was BS > RS > RE; in the drought-sensitive group, the network density of Bacteria rhizosphere was the highest, and the results of Fungi were consistent with the drought-tolerant group. Xiong et al. (2021) also found that the complexity of the soil network gradually decreased from bulk soil-rhizosphere-endosphere, which was consistent with the results of this study. A rise in drought may result in more antagonistic or competitive biological interactions (Wang et al., 2018; Jiao et al., 2022). The same results were obtained in this study. Drought stress increased the negative correlation between spring wheat’s root space species, which was more intuitive in fungal communities. This may be because drought exacerbates the continual decline of water, which decreases species stability and increases species competition, while species filling the small global network limits the efficiency of resource transfer (Ghoul and Mitri, 2016; Morriën et al., 2017; Jiao et al., 2022). Through the species attributes analysis, it was found that the connectivity number and positive correlation were higher. The root space Bacteria were clustered in Proteobacteria, Actinobacteriota, Firmicutes, Patescibacteria, and Bacteroidota. The above bacteria may be the key core Bacteria of drought stress.
Functional prediction analysis showed that in Bacteria, the abundance of nitrification and ureolysis in drought-tolerant spring wheat increased significantly in endosphere and bulk soil, respectively. The abundance of nitrogen fixation in group 2 (DTG and DSG) was significantly reduced. In fungi, the symbiotroph forms of DTG and DSG spring wheat (Arbuscular Mycorrhiza) under drought stress increased significantly in the endosphere and bulk soil. Fungal Parasite-Plant Pathogen-Plant Saprotroph both increased significantly in (RE-RS-BS). Nitrification is a crucial part of the soil nitrogen cycle, facilitated by nitrifying Bacteria. In addition to autotrophic nitrifying Bacteria, heterotrophic Bacteria, and Actinomycetes can oxidize ammonium salt into nitrite and nitric acid. The heterotrophic bacteria’ ammonium oxidation efficiency is low, but their acid resistance and tolerance to hostile environments are strong. It also contributes to nitrification (Nelson et al., 2016; Kuypers et al., 2018). Microbially induced calcium carbonate precipitation (MICP) can be promoted by ureolysis, which is an innovative method for enzymatic hydrolysis of urea to improve soil (Lauchnor et al., 2015; Hamdan et al., 2017). Xun et al. (2021) showed that functional genes related to soil nitrogen and phosphorus metabolism were the most critical factors in community stability. Bacterial groups represented by nitrogen and phosphorus metabolism may be the core members to maintain the stability of soil microbial communities. Arbuscular mycorrhizal fungi (AMF) are multifunctional symbionts with important ecological significance for most terrestrial plants (Gorzelak et al., 2017; Gu et al., 2020; Jiang et al., 2020; Zhang et al., 2020; Kim et al., 2022). AMF can use fungal hyphae to obtain soil water and nutrients such as phosphorus (P), nitrogen (N), and trace elements for plant absorption (Bulgarelli et al., 2013; Pellegrino et al., 2019; Jiang et al., 2020). The prediction results of microbial community function also showed that Bacteria and Fungi in the spring wheat’s root space under drought stress would respond to drought through different strategies. However, because this study only explored the function of microorganisms at the level of functional prediction, there were still certain limitations, and additional research on microbial functions was still necessary. In conclusion, the changes in the microbial community in the spring wheat’s root space were mainly determined by the niche differentiation of the endosphere, rhizosphere, and bulk soil. Enriching drought-resistant preference groups like Actinobacteriota, Variovorax, and Streptomyces could significantly change the structure and function of the microbial community in spring wheat’s root space under stress.
5. Conclusion
This study detected significant differences in bacterial and fungal diversity in the root space of spring wheat under drought stress (P < 0.05). Moreover, the effects on microbial diversity in the endosphere, rhizosphere, and bulk soil gradually weakened. Compared with the drought-sensitive group, the drought-tolerant group had higher microbial diversity. The endosphere microbial community composition was more responsive to drought stress. Actinobacteriota and Streptomyces preferred the drought-tolerant group, while Proteobacteria and Pseudomonas preferred the drought-sensitive group. The genus Variovorax is vital in assisting spring wheat drought resistance in the rhizosphere and bulk soil. The antagonism between the endosphere and spring wheat species grew, but the community’s complexity and stability diminished. The functional prediction showed that drought stress induced Bacteria to tend to nitrogen cycle (nitrification and ureolysis) related functional taxa enrichment, and Fungi tended to symbiotroph (Arbuscular Mycorrhiza) related functional taxa enrichment. Therefore, under drought stress, Bacteria and Fungi in the spring wheat’s root space may respond with distinct strategies. Of course, these strategies may include the response of microbial network structure to drought, the regulation of microorganisms by host plants, and the resistance of aboveground and underground communities to drought. Future research should further explore the mechanism of crop-soil-microorganism collaborative drought resistance in farmland.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA943479.
Author contributions
ZL, XiaoZ, YR, and JF contributed to the conception and design of the study. JF and SW carried out field experiments. YC, XianZ, and JW conducted experimental guidance and help. SW, JM, and YG carried out experimental sample collection and detection. GS, LB, and RX performed the statistical analysis. JF wrote the first draft of the manuscript. All authors contributed to manuscript revision and read and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (31860356 and 32260457); the High-end talent training projects of Grassland Talents in Inner Mongolia Autonomous Region; the Leading Talent Project of “Science and Technology Leading Talent Team Project of Inner Mongolia Autonomous Region” (2022LJRC0010); the Inner Mongolia Autonomous Region Natural Science Foundation Project Key Projects (2022ZD13); the National Key Research and Development Program of China (2022YFD1500901-5); and the Inner Mongolia Agriculture and Animal Husbandry Innovation Fund Project (2022CXJJN08 and 2022CXJJM04).
Acknowledgments
Thanks to Haiming Wu, Lili Zhao, and the de staff of Teni River Soil Management and Ecological Restoration Scientific Observation and Experimental Station (Tenihe Farm) of Inner Mongolia Academy of Agricultural and Animal Husbandry Sciences for their help in the sampling process.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1235708/full#supplementary-material
Footnotes
- ^ https://cloud.majorbio.com
- ^ http://www.mothur.org/wiki/Calculators
- ^ http://huttenhower.sph.harvard.edu/LEfSe
References
Adams, R. I., Miletto, M., Taylor, J. W., and Bruns, T. D. (2013). Dispersal in microbes: Fungi in indoor air are dominated by outdoor air and show dispersal limitation at short distances. ISME J. 7, 1262–1273. doi: 10.1038/ismej.2013.28
Amir, A., McDonald, D., Navas-Molina, J. A., Kopylova, E., Morton, J. T., Zech Xu, Z., et al. (2017). Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems 2, e00191–16. doi: 10.1128/mSystems.00191-16
Assenov, Y., Ramírez, F., Schelhorn, S.-E., Lengauer, T., and Albrecht, M. (2008). Computing topological parameters of biological networks. Bioinformatics 24, 282–284. doi: 10.1093/bioinformatics/btm554
Ault, T. R. (2020). On the essentials of drought in a changing climate. Science 368, 256–260. doi: 10.1126/science.aaz5492
Azarbad, H., Tremblay, J., Giard-Laliberté, C., Bainard, L. D., and Yergeau, E. (2020). Four decades of soil water stress history together with host genotype constrain the response of the wheat microbiome to soil moisture. FEMS Microbiol. Ecol. 96:fiaa098. doi: 10.1093/femsec/fiaa098
Babalola, O. O. (2010). Beneficial bacteria of agricultural importance. Biotechnol. Lett. 32, 1559–1570. doi: 10.1007/s10529-010-0347-0
Bai, L., Zhang, X., Li, B., Sun, F., Zhao, X., Wang, Y., et al. (2022). Fungal communities are more sensitive to nitrogen fertilization than bacteria in different spatial structures of silage maize under short-term nitrogen fertilization. Appl. Soil Ecol. 170, 104275. doi: 10.1016/j.apsoil.2021.104275
Ballhausen, M.-B., and de Boer, W. (2016). The sapro-rhizosphere: Carbon flow from saprotrophic fungi into fungus-feeding bacteria. Soil Biol. Biochem. 102, 14–17. doi: 10.1016/j.soilbio.2016.06.014
Berendsen, R. L., Pieterse, C. M. J., and Bakker, P. A. H. M. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Bhattacharjee, A., Dubey, S., and Sharma, S. (2022). Storage of soil microbiome for application in sustainable agriculture: Prospects and challenges. Environ. Sci. Pollut. Res. Int. 29, 3171–3183. doi: 10.1007/s11356-021-17164-4
Breitkreuz, C., Herzig, L., Buscot, F., Reitz, T., and Tarkka, M. (2021). Interactions between soil properties, agricultural management and cultivar type drive structural and functional adaptations of the wheat rhizosphere microbiome to drought. Environ. Microbiol. 23, 5866–5882. doi: 10.1111/1462-2920.15607
Bulgarelli, D., Rott, M., Schlaeppi, K., Ver Loren van Themaat, E., Ahmadinejad, N., Assenza, F., et al. (2012). Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91–95. doi: 10.1038/nature11336
Bulgarelli, D., Schlaeppi, K., Spaepen, S., Ver Loren van Themaat, E., and Schulze-Lefert, P. (2013). Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 64, 807–838. doi: 10.1146/annurev-arplant-050312-120106
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Huntley, J., Fierer, N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. doi: 10.1038/ismej.2012.8
Chaparro, J. M., Badri, D. V., and Vivanco, J. M. (2014). Rhizosphere microbiome assemblage is affected by plant development. ISME J. 8, 790–803. doi: 10.1038/ismej.2013.196
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Dagvadorj, B., Outram, M. A., Williams, S. J., and Solomon, P. S. (2022). The necrotrophic effector ToxA from Parastagonospora nodorum interacts with wheat NHL proteins to facilitate Tsn1-mediated necrosis. Plant J. 110, 407–418. doi: 10.1111/tpj.15677
Dassen, S., Cortois, R., Martens, H., de Hollander, M., Kowalchuk, G. A., van der Putten, W. H., et al. (2017). Differential responses of soil bacteria, fungi, archaea and protists to plant species richness and plant functional group identity. Mol. Ecol. 26, 4085–4098. doi: 10.1111/mec.14175
Ding, J., Jiang, X., Guan, D., Zhao, B., Ma, M., Zhou, B., et al. (2017). Influence of inorganic fertilizer and organic manure application on fungal communities in a long-term field experiment of Chinese Mollisols. Appl. Soil Ecol. 111, 114–122. doi: 10.1016/j.apsoil.2016.12.003
Edgar, R. C. (2013). UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edwards, J., Johnson, C., Santos-Medellín, C., Lurie, E., Podishetty, N. K., Bhatnagar, S., et al. (2015). Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. U.S.A. 112, E911–E920. doi: 10.1073/pnas.1414592112
Feng, N.-X., Yu, J., Zhao, H.-M., Cheng, Y.-T., Mo, C.-H., Cai, Q.-Y., et al. (2017). Efficient phytoremediation of organic contaminants in soils using plant-endophyte partnerships. Sci. Total Environ. 583, 352–368. doi: 10.1016/j.scitotenv.2017.01.075
Finkel, O. M., Salas-González, I., Castrillo, G., Conway, J. M., Law, T. F., Teixeira, P. J. P. L., et al. (2020). A single bacterial genus maintains root growth in a complex microbiome. Nature 587, 103–108. doi: 10.1038/s41586-020-2778-7
Fitzpatrick, C. R., Copeland, J., Wang, P. W., Guttman, D. S., Kotanen, P. M., and Johnson, M. T. J. (2018). Assembly and ecological function of the root microbiome across angiosperm plant species. Proc. Natl. Acad. Sci. U.S.A. 115, E1157–E1165. doi: 10.1073/pnas.1717617115
Francioli, D., Cid, G., Kanukollu, S., Ulrich, A., Hajirezaei, M.-R., and Kolb, S. (2021). Flooding causes dramatic compositional shifts and depletion of putative beneficial bacteria on the spring wheat microbiota. Front. Microbiol. 12:773116. doi: 10.3389/fmicb.2021.773116
Ghoul, M., and Mitri, S. (2016). The ecology and evolution of microbial competition. Trends Microbiol. 24, 833–845. doi: 10.1016/j.tim.2016.06.011
Gilbert, J. A., Jansson, J. K., and Knight, R. (2014). The earth microbiome project: Successes and aspirations. BMC Biol 12:69. doi: 10.1186/s12915-014-0069-1
Glick, B. R. (1995a). Metabolic load and heterologous gene expression. Biotechnol. Adv. 13, 247–261. doi: 10.1016/0734-9750(95)00004-a
Glick, B. R. (1995b). The enhancement of plant growth by free-living bacteria. Can. J. Microbiol. 41, 109–117. doi: 10.1139/m95-015
Gorzelak, M. A., Pickles, B. J., and Hart, M. M. (2017). Exploring the symbiont diversity of ancient western redcedars: Arbuscular mycorrhizal fungi of long-lived hosts. Mol. Ecol. 26, 1586–1597. doi: 10.1111/mec.14023
Grace, J. B., and Keeley, J. E. (2006). A structural equation model analysis of postfire plant diversity in California shrublands. Ecol. Appl. 16, 503–514. doi: 10.1890/1051-07612006016[0503:asemao]2.0.co;2
Gu, S., Wu, S., Guan, Y., Zhai, C., and Yang, W. (2020). Arbuscular mycorrhizal fungal community was affected by tillage practices rather than residue management in black soil of northeast China. Soil Tillage Res. 198:104552. doi: 10.1016/j.still.2019.104552
Hamdan, N., Kavazanjian, E., Rittmann, B. E., and Karatas, I. (2017). Carbonate mineral precipitation for soil improvement through microbial denitrification. Geomicrobiol. J. 34, 139–146. doi: 10.1080/01490451.2016.1154117
Hsiao, T. C. (1973). Plant responses to water stress. Annu. Rev. Plant. Physiol. 24, 519–570. doi: 10.1146/annurev.pp.24.060173.002511
Human Microbiome Project Consortium (2012). Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. doi: 10.1038/nature11234
Iannucci, A., Fragasso, M., Platani, C., and Papa, R. (2013). Plant growth and phenolic compounds in the rhizosphere soil of wild oat (Avena fatua L.). Front. Plant Sci. 4:509. doi: 10.3389/fpls.2013.00509
Ji, L., Liu, Y., Wang, J., Lu, Z., Zhang, L., and Yang, Y. (2021). Differential variation in non-structural carbohydrates in root branch orders of Fraxinus mandshurica Rupr. Seedlings across different drought intensities and soil substrates. Front. Plant Sci. 12:692715. doi: 10.3389/fpls.2021.692715
Jiang, S., Hu, X., Kang, Y., Xie, C., and Shen, Q. (2020). Arbuscular mycorrhizal fungal communities in the rhizospheric soil of litchi and mango orchards as affected by geographic distance, soil properties and manure input. Appl. Soil Ecol. 152:103593. doi: 10.1016/j.apsoil.2020.103593
Jiao, S., Chu, H., Zhang, B., Wei, X., Chen, W., and Wei, G. (2022). Linking soil fungi to bacterial community assembly in arid ecosystems. iMeta 1:e2. doi: 10.1002/imt2.2
Jochum, M. D., McWilliams, K. L., Pierson, E. A., and Jo, Y.-K. (2019). Host-mediated microbiome engineering (HMME) of drought tolerance in the wheat rhizosphere. PLoS One 14:e0225933. doi: 10.1371/journal.pone.0225933
Juroszek, P., and Tiedemann, A. V. (2013). Climate change and potential future risks through wheat diseases: A review. Eur. J. Plant Pathol. 136, 21–33. doi: 10.1007/s10658-012-0144-9
Kim, K., Neuberger, P., Daly, E. J., Gorzelak, M., and Hernandez-Ramirez, G. (2022). Arbuscular mycorrhizal fungi community linkages to soil nutrient availability across contrasting agroecosystems. Appl. Soil Ecol. 176:104464. doi: 10.1016/j.apsoil.2022.104464
Kuypers, M. M. M., Marchant, H. K., and Kartal, B. (2018). The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 16, 263–276. doi: 10.1038/nrmicro.2018.9
Kwak, Y.-S., and Weller, D. M. (2013). Take-all of wheat and natural disease suppression: A review. Plant Pathol. J. 29, 125–135. doi: 10.5423/PPJ.SI.07.2012.0112
Ladau, J., Shi, Y., Jing, X., He, J.-S., Chen, L., Lin, X., et al. (2018). Existing climate change will lead to pronounced shifts in the diversity of soil prokaryotes. mSystems 3, e167–e118. doi: 10.1128/mSystems.00167-18
Latz, M. A. C., Jensen, B., Collinge, D. B., and Jørgensen, H. J. L. (2018). Endophytic fungi as biocontrol agents: Elucidating mechanisms in disease suppression. Plant Ecol. Divers. 11, 555–567. doi: 10.1080/17550874.2018.1534146
Lauchnor, E. G., Topp, D. M., Parker, A. E., and Gerlach, R. (2015). Whole cell kinetics of ureolysis by Sporosarcina pasteurii. J. Appl. Microbiol. 118, 1321–1332. doi: 10.1111/jam.12804
Leff, J. W., Jones, S. E., Prober, S. M., Barberán, A., Borer, E. T., Firn, J. L., et al. (2015). Consistent responses of soil microbial communities to elevated nutrient inputs in grasslands across the globe. Proc. Natl. Acad. Sci. U.S.A. 112, 10967–10972. doi: 10.1073/pnas.1508382112
Lesk, C., Rowhani, P., and Ramankutty, N. (2016). Influence of extreme weather disasters on global crop production. Nature 529, 84–87. doi: 10.1038/nature16467
Li, W., Li, Y., Lv, J., He, X., Wang, J., Teng, D., et al. (2022). Rhizosphere effect alters the soil microbiome composition and C, N transformation in an arid ecosystem. Appl. Soil Ecol. 170:104296. doi: 10.1016/j.apsoil.2021.104296
Locey, K. J., and Lennon, J. T. (2016). Scaling laws predict global microbial diversity. Proc. Natl. Acad. Sci. U.S.A. 113, 5970–5975. doi: 10.1073/pnas.1521291113
Louca, S., Parfrey, L. W., and Doebeli, M. (2016). Decoupling function and taxonomy in the global ocean microbiome. Science 353, 1272–1277. doi: 10.1126/science.aaf4507
Magoč, T., and Salzberg, S. L. (2011). FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Mavrodi, D. V., Mavrodi, O. V., Elbourne, L. D. H., Tetu, S., Bonsall, R. F., Parejko, J., et al. (2018). Long-term irrigation affects the dynamics and activity of the wheat rhizosphere microbiome. Front. Plant Sci. 9:345. doi: 10.3389/fpls.2018.00345
Morriën, E., Hannula, S. E., Snoek, L. B., Helmsing, N. R., Zweers, H., de Hollander, M., et al. (2017). Soil networks become more connected and take up more carbon as nature restoration progresses. Nat. Commun. 8:14349. doi: 10.1038/ncomms14349
Naylor, D., and Coleman-Derr, D. (2017). Drought stress and root-associated bacterial communities. Front. Plant Sci. 8:2223. doi: 10.3389/fpls.2017.02223
Naylor, D., DeGraaf, S., Purdom, E., and Coleman-Derr, D. (2017). Drought and host selection influence bacterial community dynamics in the grass root microbiome. ISME J. 11, 2691–2704. doi: 10.1038/ismej.2017.118
Nelson, M. B., Martiny, A. C., and Martiny, J. B. H. (2016). Global biogeography of microbial nitrogen-cycling traits in soil. Proc. Natl. Acad. Sci. U.S.A. 113, 8033–8040. doi: 10.1073/pnas.1601070113
Neupane, A., Bulbul, I., Wang, Z., Lehman, R. M., Nafziger, E., and Marzano, S.-Y. L. (2021). Long term crop rotation effect on subsequent soybean yield explained by soil and root-associated microbiomes and soil health indicators. Sci. Rep. 11:9200. doi: 10.1038/s41598-021-88784-6
Nguyen, N. H., Song, Z., Bates, S. T., Branco, S., Tedersoo, L., Menke, J., et al. (2016). FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 20, 241–248. doi: 10.1016/j.funeco.2015.06.006
Ochoa-Hueso, R., Collins, S. L., Delgado-Baquerizo, M., Hamonts, K., Pockman, W. T., Sinsabaugh, R. L., et al. (2018). Drought consistently alters the composition of soil fungal and bacterial communities in grasslands from two continents. Glob. Chang. Biol. 24, 2818–2827. doi: 10.1111/gcb.14113
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., and McGlinn, D. (2013). vegan: Community ecology package. Available online at: https://cran.r-project.org/web/packages/vegan/index.html (accessed March 5, 2023).
Pang, Z., Chen, J., Wang, T., Gao, C., Li, Z., Guo, L., et al. (2021). Linking plant secondary metabolites and plant microbiomes: A review. Front. Plant Sci. 12:621276. doi: 10.3389/fpls.2021.621276
Pellegrino, E., Gamper, H. A., Ciccolini, V., and Ercoli, L. (2019). Forage rotations conserve diversity of arbuscular mycorrhizal fungi and soil fertility. Front. Microbiol. 10:2969. doi: 10.3389/fmicb.2019.02969
Prober, S. M., Leff, J. W., Bates, S. T., Borer, E. T., Firn, J., Harpole, W. S., et al. (2015). Plant diversity predicts beta but not alpha diversity of soil microbes across grasslands worldwide. Ecol. Lett. 18, 85–95. doi: 10.1111/ele.12381
Qi, M., Berry, J. C., Veley, K. W., O’Connor, L., Finkel, O. M., Salas-González, I., et al. (2022). Identification of beneficial and detrimental bacteria impacting sorghum responses to drought using multi-scale and multi-system microbiome comparisons. ISME J. 16, 1957–1969. doi: 10.1038/s41396-022-01245-4
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Reinhold-Hurek, B., Bünger, W., Burbano, C. S., Sabale, M., and Hurek, T. (2015). Roots shaping their microbiome: Global hotspots for microbial activity. Annu. Rev. Phytopathol. 53, 403–424. doi: 10.1146/annurev-phyto-082712-102342
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Rustgi, S., Boex-Fontvieille, E., Reinbothe, C., von Wettstein, D., and Reinbothe, S. (2017). Serpin1 and WSCP differentially regulate the activity of the cysteine protease RD21 during plant development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 114, 2212–2217. doi: 10.1073/pnas.1621496114
Santos-Medellín, C., Edwards, J., Liechty, Z., Nguyen, B., and Sundaresan, V. (2017). Drought stress results in a compartment-specific restructuring of the rice root-associated microbiomes. mBio 8, e00764–17. doi: 10.1128/mBio.00764-17
Santos-Medellín, C., Liechty, Z., Edwards, J., Nguyen, B., Huang, B., Weimer, B. C., et al. (2021). Prolonged drought imparts lasting compositional changes to the rice root microbiome. Nat. Plants 7, 1065–1077. doi: 10.1038/s41477-021-00967-1
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60
Stackebrandt, E., and Goebel, B. M. (1994). Taxonomic note: A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44, 846–849. doi: 10.1099/00207713-44-4-846
Thompson, L. R., Sanders, J. G., McDonald, D., Amir, A., Ladau, J., Locey, K. J., et al. (2017). A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551, 457–463. doi: 10.1038/nature24621
Wagner, M. R., Lundberg, D. S., Del Rio, T. G., Tringe, S. G., Dangl, J. L., and Mitchell-Olds, T. (2016). Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat. Commun. 7:12151. doi: 10.1038/ncomms12151
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wang, S., Wang, S., Han, X., and Deng, Y. (2018). Higher precipitation strengthens the microbial interactions in semi-arid grassland soils. Glob. Ecol. Biogeogr. 27, 570–580.
Webber, H., Ewert, F., Olesen, J. E., Müller, C., Fronzek, S., Ruane, A. C., et al. (2018). Diverging importance of drought stress for maize and winter wheat in Europe. Nat. Commun. 9:4249. doi: 10.1038/s41467-018-06525-2
Williams, A., and de Vries, F. T. (2020). Plant root exudation under drought: Implications for ecosystem functioning. N. Phytol. 225, 1899–1905. doi: 10.1111/nph.16223
Xiong, C., Zhu, Y.-G., Wang, J.-T., Singh, B., Han, L.-L., Shen, J.-P., et al. (2021). Host selection shapes crop microbiome assembly and network complexity. N. Phytol 229, 1091–1104. doi: 10.1111/nph.16890
Xu, L., Naylor, D., Dong, Z., Simmons, T., Pierroz, G., Hixson, K. K., et al. (2018). Drought delays development of the sorghum root microbiome and enriches for monoderm bacteria. Proc. Natl. Acad. Sci. U.S.A. 115, E4284–E4293. doi: 10.1073/pnas.1717308115
Xun, W., Liu, Y., Li, W., Ren, Y., Xiong, W., Xu, Z., et al. (2021). Specialized metabolic functions of keystone taxa sustain soil microbiome stability. Microbiome 9:35. doi: 10.1186/s40168-020-00985-9
Yang, H., Wang, C., Chen, F., Yue, L., Cao, X., Li, J., et al. (2022). Foliar carbon dot amendment modulates carbohydrate metabolism, rhizospheric properties and drought tolerance in maize seedling. Sci. Total Environ. 809:151105. doi: 10.1016/j.scitotenv.2021.151105
Yang, N., Nesme, J., Røder, H. L., Li, X., Zuo, Z., Petersen, M., et al. (2021). Emergent bacterial community properties induce enhanced drought tolerance in Arabidopsis. NPJ Biofilms Microbiomes 7:82. doi: 10.1038/s41522-021-00253-0
Yang, Y., Li, T., Wang, Y., Cheng, H., and An, S. (2021). Negative effects of multiple global change factors on soil microbial diversity. Soil Biol. Biochem. 156:108229. doi: 10.1016/j.soilbio.2021.108229
Zhalnina, K., Louie, K. B., Hao, Z., Mansoori, N., da Rocha, U. N., Shi, S., et al. (2018). Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 3, 470–480. doi: 10.1038/s41564-018-0129-3
Zhang, Y., Ma, Q., Wang, Y., Hu, Z., and Zhang, P. (2020). Variations in soil fungal communities after continuous fertiliser treatment under the ridge and furrow rainfall harvesting system in a semiarid region of China. Soil Res. 58, 161–173. doi: 10.1071/SR18336
Zhou, J., Deng, Y., Shen, L., Wen, C., Yan, Q., Ning, D., et al. (2016). Temperature mediates continental-scale diversity of microbes in forest soils. Nat. Commun. 7:12083. doi: 10.1038/ncomms12083
Keywords: drought stress, spring wheat, root-space microorganism, bacterial diversity, fungal diversity
Citation: Fang J, Wei S, Gao Y, Zhang X, Cheng Y, Wang J, Ma J, Shi G, Bai L, Xie R, Zhao X, Ren Y and Lu Z (2023) Character variation of root space microbial community composition in the response of drought-tolerant spring wheat to drought stress. Front. Microbiol. 14:1235708. doi: 10.3389/fmicb.2023.1235708
Received: 06 June 2023; Accepted: 31 August 2023;
Published: 15 September 2023.
Edited by:
Steffen Kolb, Leibniz Center for Agricultural Landscape Research (ZALF), GermanyReviewed by:
Yong-Xin Liu, Chinese Academy of Agricultural Sciences, ChinaMika Tapio Tarkka, Helmholtz Association of German Research Centres (HZ), Germany
Copyright © 2023 Fang, Wei, Gao, Zhang, Cheng, Wang, Ma, Shi, Bai, Xie, Zhao, Ren and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoqing Zhao, zhaoxq204@163.com; Yongfeng Ren, renyongfeng_1984@163.com; Zhanyuan Lu, lzhy2811@163.com
 Jing Fang1,2,3
Jing Fang1,2,3 Xiaoqing Zhao
Xiaoqing Zhao Zhanyuan Lu
Zhanyuan Lu