- 1State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, National Clinical Research Center for Infectious Diseases, National Medical Center for Infectious Diseases, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, The First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, China
- 2Department of Infectious Diseases, ShuLan (Hangzhou) Hospital Affiliated to Zhejiang Shuren University Shulan International Medical College, Hangzhou, China
- 3The Second Clinical Medical College of Zhejiang Chinese Medical University, Hangzhou, China
- 4Research Center for Air Pollution and Health, Zhejiang University, Hangzhou, China
Introduction: Submassive hepatic necrosis (SMHN, defined as necrosis of 15–90% of the entire liver on explant) is a likely characteristic pathological feature of ACLF in patients with hepatitis B cirrhosis. We aimed to comprehensively explore microbiome and bile acids patterns across enterhepatic circulation and build well-performing machine learning models to predict SMHN status.
Methods: Based on the presence or absence of SMHN, 17 patients with HBV-related end-stage liver disease who received liver transplantation were eligible for inclusion. Serum, portal venous blood, and stool samples were collected for comparing differences of BA spectra and gut microbiome and their interactions. We adopted the random forest algorithm with recursive feature elimination (RF-RFE) to predict SMHN status.
Results: By comparing total BA spectrum between SMHN (−) and SMHN (+) patients, significant changes were detected only in fecal (P = 0.015). Compared with the SMHN (+) group, the SMHN (−) group showed that UDCA, 7-KLCA, 3-DHCA, 7-KDCA, ISOLCA and α-MCA in feces, r-MCA, 7-KLCA and 7-KDCA in serum, γ-MCA and 7-KLCA in portal vein were enriched, and TUDCA in feces was depleted. PCoA analysis showed significantly distinct overall microbial composition in two groups (P = 0.026). Co-abundance analysis showed that bacterial species formed strong and broad relationships with BAs. Among them, Parabacteroides distasonis had the highest node degree. We further identified a combinatorial marker panel with a high AUC of 0.92.
Discussion: Our study demonstrated the changes and interactions of intestinal microbiome and BAs during enterohepatic circulation in ACLF patients with SMHN. In addition, we identified a combinatorial marker panel as non-invasive biomarkers to distinguish the SMHN status with high AUC.
Introduction
Although the definition of acute-on-chronic liver failure (ACLF) remains controversial, it is recognized as the most serious complication of acute decompensated (AD) cirrhosis with a 28-day mortality of 40–50% when it occurs (Li et al., 2016). Deranged systemic inflammatory responses are one of the core clinical elements of ACLF (Laleman et al., 2011; Olson and Kamath, 2011). Although AD itself has high mortality rates, the pro-gression to ACLF increases this mortality dramatically4; ACLF is the main cause of death in decompensated cirrhosis5. Even after active treatment, some patients will develop ACLF on the basis of AD, while some patients will not. The underlying pathogenesis is still unclear. From AD to ACLF, the differences in intestinal flora between the two groups of patients are worth further exploration.
Several studies have reported specific microbiome patterns in the stool of patients with ACLF (Bajaj et al., 2014; Zhang et al., 2019). The potential pathogens of Pasteurellaceae, Streptococcaceae, and Enterococcaceae were more abundant in the ACLF group than in the health control (HC) group, and the relative abundance of Bacteroidaceae, Ruminococcaceae, and Lachnospiraceae was decreased (Chen et al., 2015). In addition, Lachnospiraceae might be a biomarker of ACLF, whereas the relative abundance of Pasteurellaceae could predict the mortality rate (Chen et al., 2015). Bile acids (BAs) are the end products of hepatic cholesterol metabolism, synthesized in the liver, and transported across the canalicular membrane of hepatocytes as primary conjugated BAs into the biliary system, which ultimately drains them into the small intestine. Within the intestine, gut microbiota modulates the biological activity of BAs through enzymatic modifications, producing secondary BAs. Recent data highlighted the role of BAs as essential mediators of the gut–liver crosstalk (Wahlstrom et al., 2016; Schneider et al., 2018). For instance, BAs regulate specific host metabolic pathways and modulate inflammatory responses via G protein-coupled and nuclear receptors, such as the G protein-coupled BA receptor 1 (TGR5) and farnesoid X-activated receptor (FXR), respectively (Aguiar Vallim et al., 2013; Chiang, 2013). However, the differences in the microenvironment in gut and BA profiles between ACLF and AD patients are largely unknown. Is it possible that the difference in intestinal microflora between patients with the two diseases leads to a different prognosis for the disease?
A recent prospective cohort study reported that submassive hepatic necrosis (SMHN) is a likely characteristic pathological feature of ACLF in patients with hepatitis B cirrhosis (Li et al., 2015). Compared with AD patients who did not have SMHN, patients with SMHN had a higher level of anti-inflammatory cytokines, as shown by serum cytokine, gene expression, and immunohistochemical studies (Li et al., 2015). However, these reports did not investigate intestinal microecology and bile acid metabolism as well as their relationship on the basis of pathology.
In this study, we collect serum, portal venous blood, and stool and liver tissue samples from patients with end-stage liver disease on the day of transplantation. BA spectrum analysis, metagenomics sequencing, and histopathological typing were performed to fully elucidate the changes in bile acid composition in decompensated cirrhosis with different pathological conditions (with or without SMHN) and the interaction between BA and gut microbiota across enterohepatic circulation.
Methods
Participants
This was a cross-sectional study conducted at a Liver Transplantation Center at the Shulan (Hangzhou) Hospital, Zhejiang Shuren University School of Medicine from 2018 to 2019. All patients or their representatives provided written informed consent to participate in this study. A total of 33 patients with chronic liver disease for an AD on the hospitalized waiting list were included in this study. AD was defined as described previously (Li et al., 2015). Since this study was focused on patients with chronic HBV infection, 16 patients were excluded either because they had another single etiology (e.g., alcohol, drug, and autoimmune) or because they were co-infected with HCV or human immunodeficiency virus. Clinical/biochemical characteristics were collected systematically during hospitalization. In addition, two common scoring systems [Child–Pugh (Pugh et al., 1973) and MELD (Kamath et al., 2007)] associated with the clinical prognosis were calculated. This study was approved by the Ethics Committee of Shulan (Hangzhou) Hospital, Zhejiang Shuren University School of Medicine.
Histology
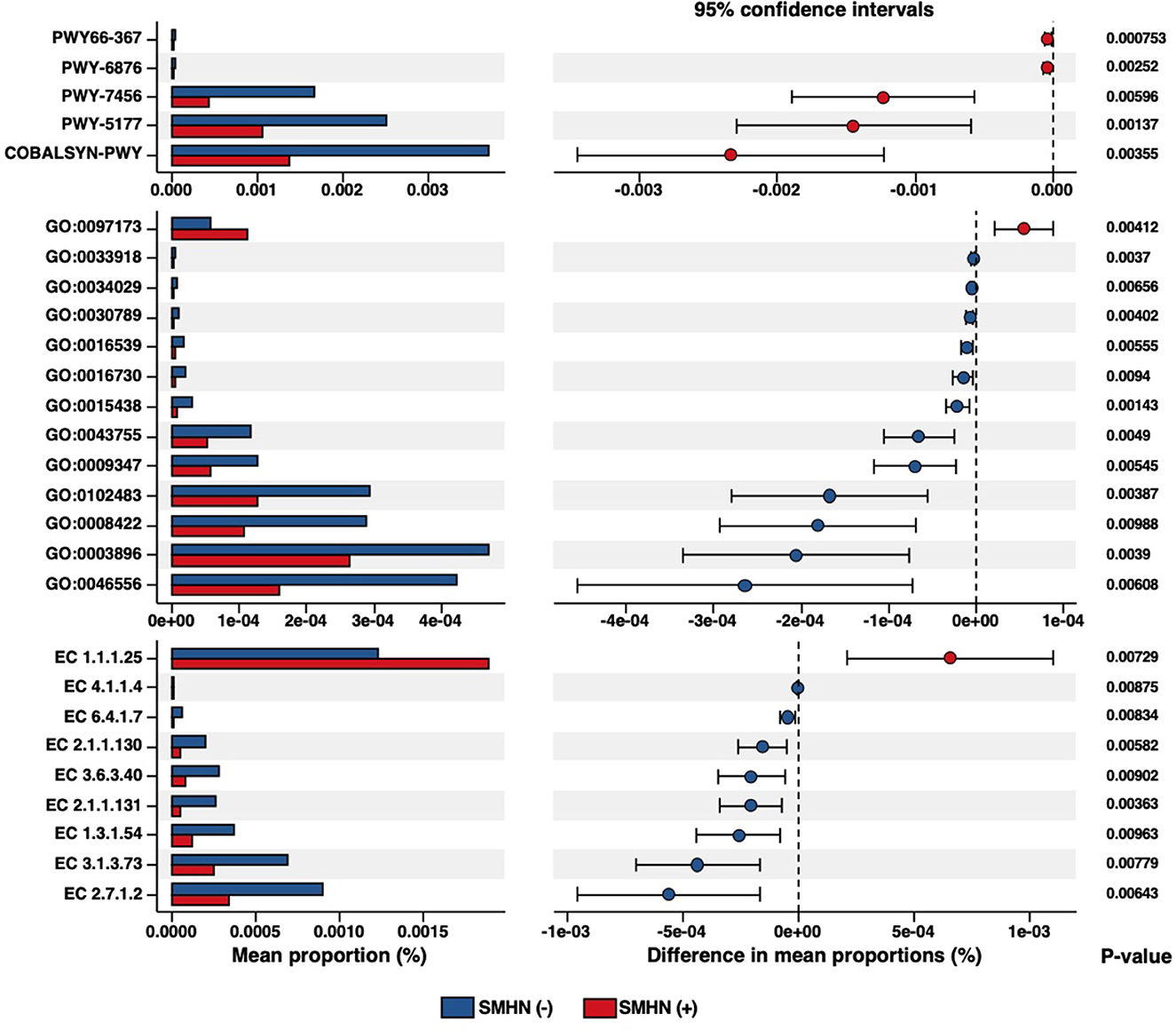
After total hepatectomy for subsequent LT, two wax specimens of the left liver and right liver and one specimen of the caudate lobe were collected from each patient for histological examination. SMHN was defined as necrosis of 15–90% of the entire liver on explant. The presence of SMHN was determined according to the Iskak score (Ishak et al., 1995). After histological evaluation by two experienced pathologists, patients were divided into two groups, with or without SMHN.
Statistical analysis
Statistical analysis was performed using R software (R Foundation for Statistical Computing, v. 4.0.3; https://www.r-project.org/). The group differences of baseline information in the training and validation cohorts were assessed using the chi-square test or the Fisher exact test for categorical variables and the t-test or the Mann–Whitney U-test for continuous variables, as appropriate. The Kruskal–Wallis test was used for comparing more than two groups. Multivariate hypothesis testing for BAs profile and microbiome composition data was carried out via permutational analysis of variance (PERMANOVA) for comparisons between the SMHN (+) and SMHN(-) groups. P-values of < 0.05 were considered significant. *P < 0.05, **P < 0.01, and ***P < 0.001.
Results
Clinical characteristics of the patients
A total of 17 patients comprising ACLF (n = 6) and AD (n = 11) were eligible for inclusion (Supplementary Figure S1). All ACLF patients were diagnosed as SMHN (+) and further confirmed by histology. In total, two out of 11 clinical decompensation cirrhosis patients were diagnosed as SMHN (+), whereas nine patients were not diagnosed as SMHN (+). Thus, eight patients were included in the SMHN (+) group. Nine out of 11 patients were enrolled in the SMHN(-) group. Two patients in the SMHN(-) group had concurrent small hepatocellular carcinoma, which met the Milan criteria.
As shown in Table 1, the median ages of SMHN (+) and SMHN(-) patients were 41.8 ± 12.4 and 52.8 ± 8.9 years, and 94% were men. Compared with the SMHN(-) group, patients with SMHN (+) had more advanced liver disease, higher frequencies of ascites and HE, more marked impairment of liver function tests, and higher Child-Pugh scores and model for end-stage liver disease (MELD) scores.
Comparison of multi-omic profile differences between the SMHN(-) and SMHN(+) groups
The Shannon diversity index, Simpson diversity index, and inverse Simpson diversity index revealed a significant difference between the two groups, with greater bacterial alpha diversity detected in the SMHN(-) patients (Figure 1A). PCoA analysis reviewing that bacterial signatures between the two groups were significantly different (PERMANOVA, P = 0.026; Figure 1B).
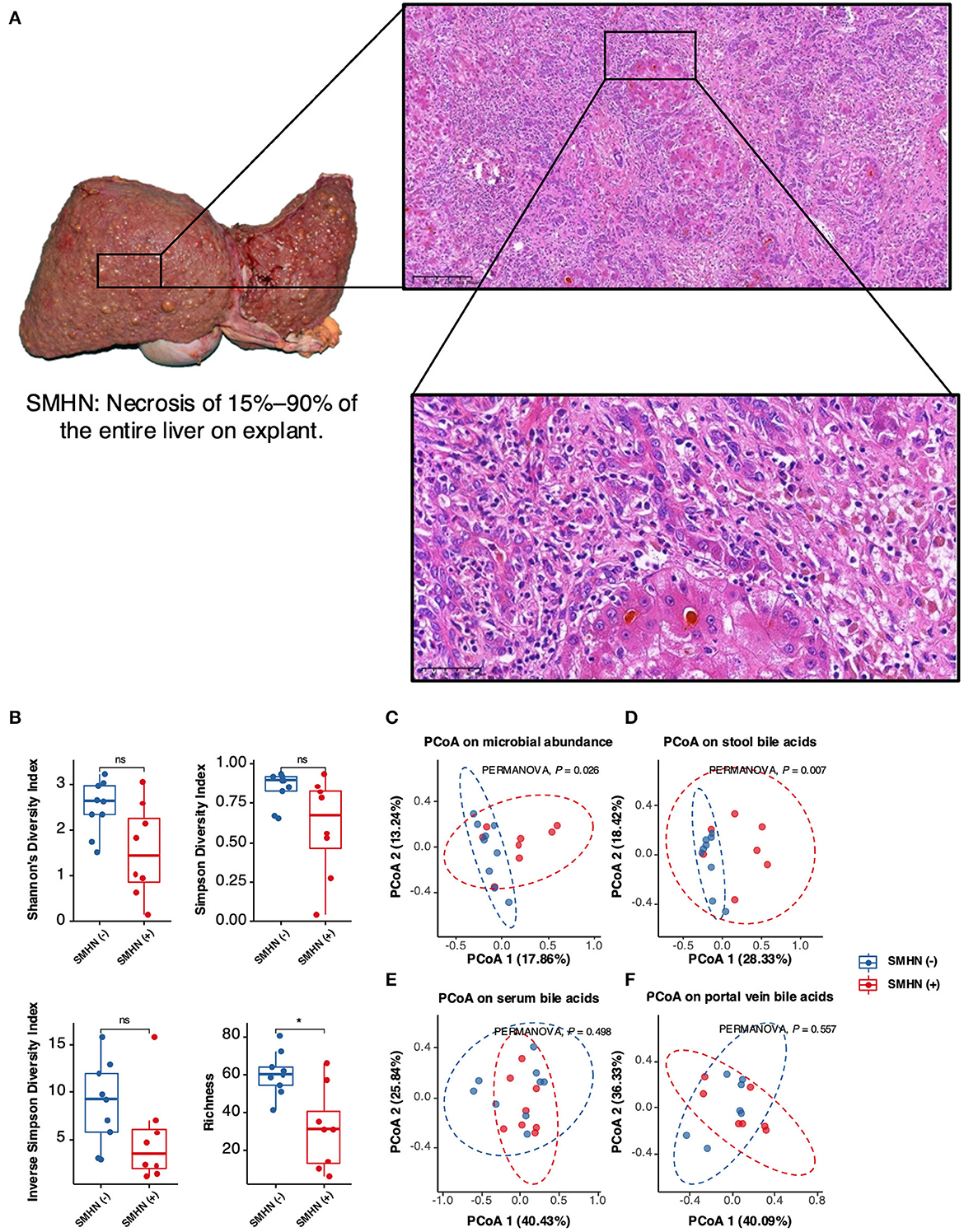
Figure 1. Multi-omic characteristics in SMHN(-) versus SMHN (+). (A) Alpha diversity in SMHN(-) and SMHN (+) patients. *P < 0.05. (B–E) Principal coordinates analysis (PCoA) based on bacterial signatures, stool BAs, serum BAs, and portal vein BAs (Bray–Curtis distance).
Together, we identified 18 discriminative bacterial species between the SMHN(-) and SMHN(+) groups, with the LEfSe algorithm (P < 0.05; Figure 2A; Supplementary Table S1). Compared with the subjects in the SMHN(+) group, SMHN(-) patients were characterized by 16 enriched species mainly belonging to the orders of Bacteroidales, Clostridiales, and Lactobacillales and two depleted species (Abiotrophia_sp_HMSC24B09, Actinomyces_sp_oral_taxon_181). To further explore these findings, we additionally included healthy group samples for multi-class comparisons. Abiotrophia_sp_HMSC24B09 and Actinomyces_sp_oral_taxon_181 remained significantly enriched in the SMHN(+) group, while Ruminococcus_torques, Eubacterium_ramulus, and Coprococcus_comes were significantly decreased compared with subjects in the SMHN(-) group and the healthy control group (Supplementary Table S2). LDA analysis at all taxonomic levels further showed that the SMHN(-) subjects were enriched by two classes, two orders, and four families, whereas the SMHN (+) subjects were only enriched by two families (Aerococcaceae and Enterococcaceae) (Figure 2B; Supplementary Table S3).
Figure 2. The bacterial species, portal vein BAs, serum BAs, and fecal BAs that discriminate SMHN(-) from SMHN (+). (A) LDA analysis showed significant bacteria species differences between SMHN(-) and SMHN (+). (B) A cladogram of different taxonomic compositions in SMHN(-) and SMHN (+). (C–E) Significant different BAs in stool, serum, and portal vein. *P < 0.05; **P < 0.01.
The overall fecal bile acid signatures of the SMHN(-) group were significantly different from that in the SMHN(+) group (PERMANOVA, P = 0.007; Figure 1C). However, no significant changes were detected in serum BAs and portal vein BAs (Figures 1D, E). Based on the Spearman correlation analysis of BAs from serum, porta vein, and stool, we found that there existed significant positive correlations among them (i.e., serum vs. portal vein: R = 0.95, P = 2.2e-16; portal vein vs. stool: R = 0.22, P = 4.5e-05; and serum vs. stool: R = 0.22, P = 6.7e-05).
Compared with the SMHN(+) group, the SMHN(-) group displayed significant enrichments in six stool BAs (i.e., UDCA, 7-KLCA, 3-DHCA, 7- KDCA, IsoLCA, and α-MCA), three serum BAs (r-MCA, 7-KLCA, and 7-KDCA), two portal vein BAs (i.e., γ-MCA and 7-KLCA), and depletion in TUDCA in stool samples (Figures 2C–E).
Compared with the healthy controls, we found that most of the detected significant BAs (i.e., UDCA, γ-MCA, 7-KLCA, 7-KDCA, and α-MCA) were higher in the SMHN(-) patients but lower in the SMHN (+) patients, although they all showed a similar pattern. There were also some bile acids that showed a trend of increasing (i.e., TUDCA) or decreasing (i.e., AlloCA) gradients from healthy controls to SMHN(-) and SMHN (+) patients (Supplementary Figure S2). Significant differences in the total fecal BA concentrations were detected between healthy controls and SMHN(-) or SMHN(+) patients as well as between SMHN(-) and SMHN(+) patients (Supplementary Figure S3). Furthermore, we found a significantly higher concentration of the secondary bile acids and bile acid metabolites in the SMHN(-) stool samples compared to the SMHN(+) group and a similar pattern for the primary BAs in the stool (Wilcoxon, P = 0.07; Supplementary Figure S4), indicating that intestinal microbiome mediates the disorder of BAs in SMHN(+) patients.
We, next, explored the direct roles of the gut microbiome in modulating BA metabolism. According to the previous report, Clostridium and Eubacterium contain an enzyme for 7α-dehydroxylation (Jia et al., 2018), which impacts the concentrations of 7-KLCA and 7-KDCA in the intestine. These BAs can return to the liver through the portal vein and then finally enter into the systemic circulation (Supplementary Figure S5).
Co-abundance analysis among the bacteria and BAs
We, next, explored the correlations of abundances for these differential gut bacterial species and BAs in enterohepatic circulation (Figure 3). In the SMHN(-) group, compared to the SMHN (+) group, we observed four enriched bacterial species from the order Bacteroidales, five enriched bacterial species from the order Clostridiales, four enriched bacterial species from the order Lactobacillales, and one depleted species from the order Lactobacillales.
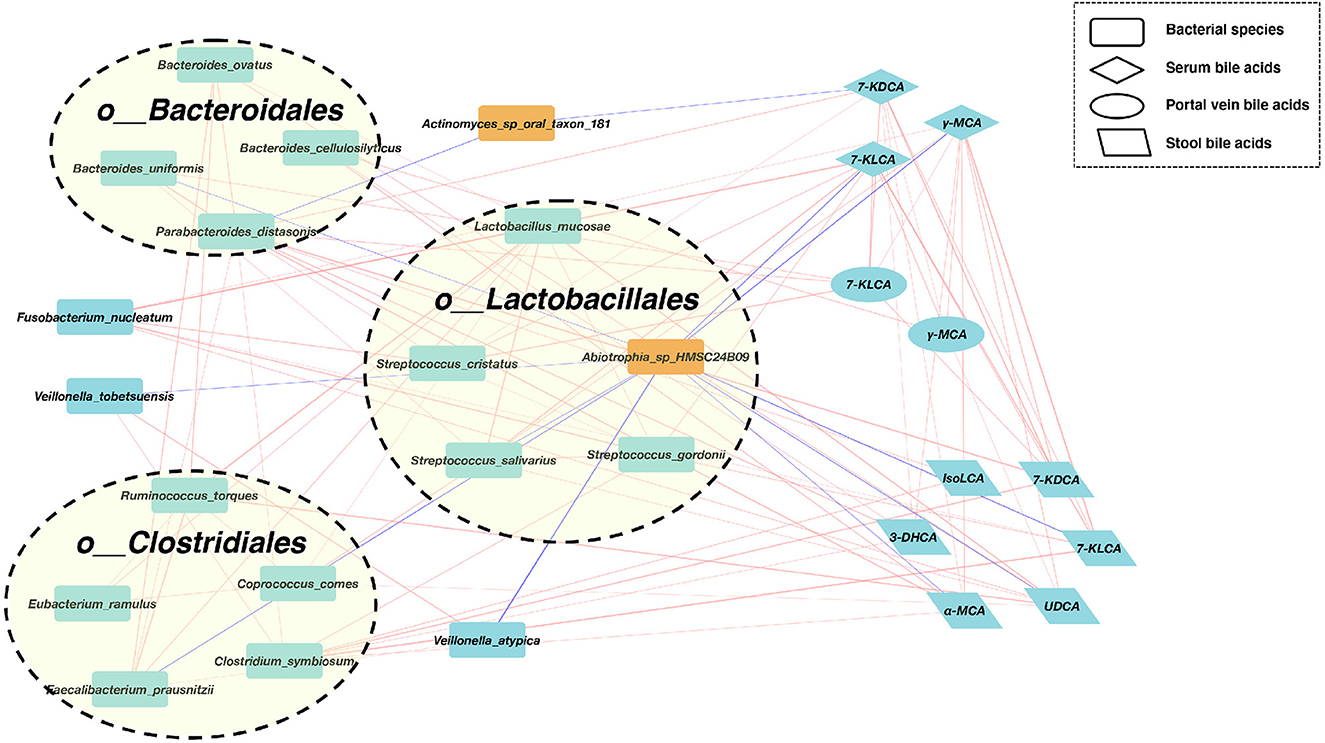
Figure 3. A co-abundance network constructed from the relative abundances of differential bacterial species, portal vein BAs, serum Bas, and fecal BAs in SMHN(-) vs. SMHN (+). Correlation networks in patients reaching outcomes filtered at P < 0.05 between microbiota (orange nodes) and metabolites (cyan-blue nodes). Red edges indicate negative while blue indicates positive correlation.
We further found that Abiotrophia_sp_HMSC24B09 enriched in the SMHN (+) group exhibited negative correlations with four bacterial species (Bacteroides uniformis, Streptococcus salivarius, Faecalibacterium prausnitzii, and Veillonella atypica) and five BAs (7-KLCA and γ-MCA in serum, 7-KLCA, UDCA, and α-MCA in stool). Another depleted species in the SMHN(-) group was Actinomyces_sp_oral_taxon_181, which was negatively correlated with Parabacteroides distasonis and 7-KDCA in serum.
To identify key regulators involved in modulating the bacterial and BA processes, we calculated all node degrees of the differential bacteria–BA network. Defining a hub as having a node degree of >5, we identified 21 hub nodes, including 12 species, five stool BAs, three serum BAs, and one portal vein BA (Supplementary Table S4). Notably, P. distasonis, a member of the Bacteroidales order, exhibited the highest node degree of 12, with positive correlations with five bacteria species (i.e., Bacteroides ovatus, Bacteroides uniformis, Faecalibacterium prausnitzii, Clostridium symbiosum, and Streptococcus cristatus), 7-KDCA in serum and portal vein, and four stool BAs (IsoLCA, 3-DHCA, α-MCA, and 7-KDCA). Among BAs, 7-KLCA and γ-MCA in serum had the highest node degree. Serum 7-KDCA showed positive correlations with four species (Fusobacterium nucleatum, Streptococcus cristatus, Streptococcus salivarius, and Streptococcus gordonii), two portal vein BAs (7-KLCA and γ-MCA), and three stool BAs (7-KLCA, 7-KDCA, and UDCA). The γ-MCA in serum showed positive correlations with two species (Fusobacterium nucleatum and Streptococcus salivarius), two portal vein BAs (7-KLCA, γ-MCA), and five stool BAs (7-KLCA, 7-KDCA, UDCA, α-MCA, and 3-DHCA).
Association of microbial function alterations with SMHN status
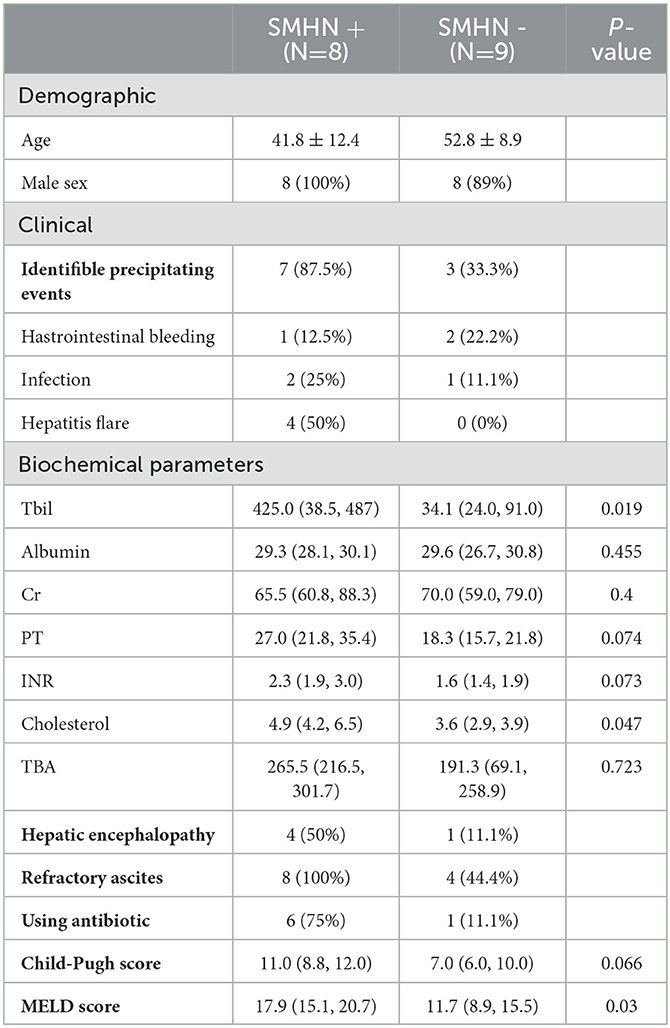
To further explore the SMHN status associated with functional dysbiosis, we performed functional profiling, which revealed a total of five differential Metacyc pathways, 13 differential GO terms, nine differential enzymes, seven differential KO identifiers, 120 differential eggnog accessions, and 37 differential Pfam accessions (Figure 4, Supplementary Tables S5–S7).
The differential MetaCyc pathways were mainly involved in Ketogenesis (PWY66-367), isopropanol biosynthesis (PWY-6876), β-(1, 4)-mannan degradation (PWY-7456), glutaryl-CoA degradation (PWY-5177), and adenosylcobalamin salvage from cobinamide I (COBALSYN-PWY). Compared with SMHN (+) patients, SMHN(-) patients were characterized by 10 enriched GO molecular function terms. The N-acetylmuramic acid catabolic process (GO: 0097173) displayed enrichment in SMHN (+) patients, and intein-mediated protein splicing (GO: 0016539) displayed enrichment in SMHN(-) patients.
Combinatorial biomarkers for discriminating SMHN(-) from SMHN (+)
The potential value of gut metagenomic and BA markers was further investigated in SMHN (+) diagnosis using machine learning models based on multi-omics data. A recursive feature elimination procedure with a random forest algorithm was used to identify the representative features (Supplementary Figures S9, S10). The random forest algorithm was also used to develop prediction models. We found that individual marker panels could discriminate patients with SMHN(-) from SMHN (+) subjects with an area under the curve (AUC) ranging from 0.60 to 0.88 (bacterial species: AUC = 0.88; Stool BAs: AUC = 0.88; Serum BAs: AUC = 0.85; and Portal vein BAs: AUC = 0.60; Figure 5A).
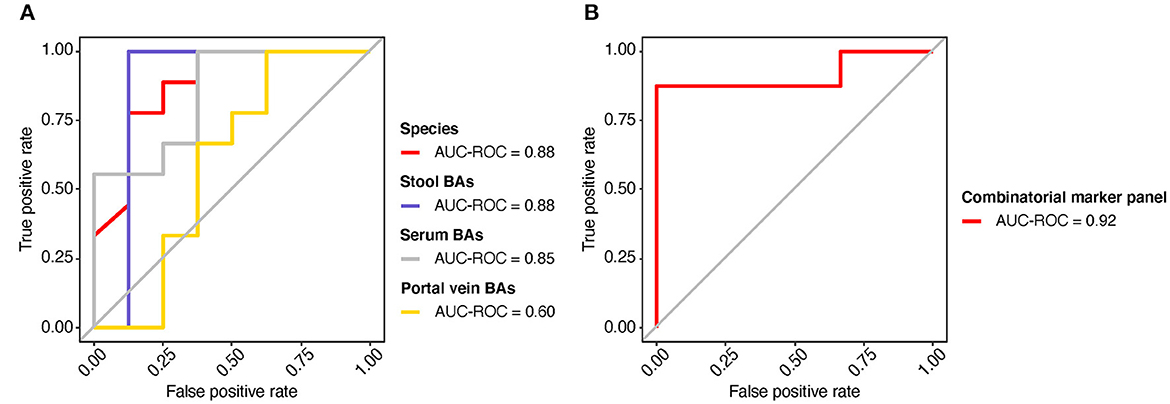
Figure 5. Predicting the SMHN status from multi-omic features. (A) ROC curves for the models based on bacterial species, stool BAs, serum BAs, and portal vein BAs. (B) ROC curve for the combinatorial marker panel.
Among the bacterial characteristics selected in the model, 11 bacterial species overlapped with the results of the LDA analysis (Supplementary Figure S9). Furthermore, we found that a combinatorial marker panel of multi-omic biomarkers enabled discriminating SMHN(-) from SMHN (+) subjects with a high classification power (AUC = 0.92, Figure 5B). The markers used in the combinatorial model include nine bacterial species, 15 stool BAs, five serum BAs, and one portal vein BAs (Supplementary Figure S11).
Discussion
In this study, we presented the landscapes and interaction networks of differential gut bacterial species, stool bile acids, serum bile acids, and portal vein bile acids in decompensated cirrhosis patients with different pathological conditions (with or without SMHN). We identified a combinatorial marker panel that can distinguish the SMHN status with a high AUC. These findings lay the foundation for understanding the roles of the overall gut–liver ecosystem in submassive hepatic necrosis, which further supports the notion that ACLF is a separate entity from liver cirrhosis with a distinct pathophysiology.
Previous reports have commonly focused on using plasma metabolite signatures, potentially deriving from gut microbiota, to differentiate ACLF from AD (Claria et al., 2016, 2019; Horvatits et al., 2017; Moreau et al., 2020). These studies clearly demonstrate that there is a bioenergetics failure coupled with a systemic inflammatory response in ACLF patients compared to those with decompensated cirrhosis. In the current study, since we performed bile acid quantification on serum, portal vein blood, and stool samples, it allowed us to study the flow of bile acids in the hepatoenteric circulation much more straightforwardly and explore the BA correlations in different sampling sites. We found there were significant differences in stool bile acid composition between the two groups. Gut microbes are involved in the metabolic process of secondary bile acids and play a key role in submassive hepatic necrosis. For example, UDCA can be produced by bacterial epimerization of the primary bile acid chenodeoxycholic acid and signifies the presence of specific beneficial Clostridium and Ruminococcus species (Winston and Theriot, 2020). Such species can also form 7-KLCA or 7-KDCA and secondary bile acids, and their relative reduction likely represents a bacterial functional alteration that predisposes patients to mortality and ACLF (Doden et al., 2018). These bile acids that differ in the intestine will further flow back to the liver through the portal vein, where BA-mediated effects may impact disease progression. Previous studies have found that BAs seem to directly activate inflammatory pathways in hepatocytes during cholestasis by stimulating the production of proinflammatory mediators (Gujral et al., 2004; Allen et al., 2011). Furthermore, BA signaling via FXR and TGR5 activation may attenuate inflammatory damage in the liver at a lower concentration (Xu et al., 2012; Shaik et al., 2015).
It is now widely accepted that gut bacteria can significantly shape several metabolic pathways in the host. Previous studies mainly characterized the gut microbiome in patients with cirrhosis using 16S rRNA gene amplicon sequencing technology with a phylogenetic resolution at the genus level or the family level (Bajaj et al., 2014, 2017, 2018, 2019; Chen et al., 2015). Consistent with our findings, some bacterial species from Bacteroides, Clostridiales, and Lactobacillales are the main bacterial orders of the gut microbiota involved in BA metabolism (Kullak-Ublick et al., 2004; Sayin et al., 2013). Moreover, P. distasonis (belonging to order Bacteroides), the species with the highest node degree in our differential species–BA network, can generate the succinate and secondary bile acids and plays key roles in the modulation of host metabolism. It has been reported that treatment with live P. distasonis can alter the bile acid profiles of mice, increasing the levels of LCA and UDCA, and in turn, reducing hyperlipidemia by activating the FXR pathway, and as a result, repairing gut barrier integrity (Wang et al., 2019). Previous studies have found that after ceftriaxone treatment, UB. ceftriaxensis and P. distasonis were driving two observed monodominance community states that peaked at 92 and 95% relative abundance, respectively. P. distasonis, the main driver of the second postantibiotic response stage, has been associated with faster microbiome recovery after antibiotic interventions50. The predominance of P. distasonis means the production of more secondary bile acids and the re-establishment of intestinal homeostasis. The difference in P. distasonis between ACLF and AD patients might be the main reason for the different progress of the two diseases. These findings could form the basis of future clinical trials focusing on changing the bile acids or the intestinal flora, such as the use of prebiotics, probiotics, and even fecal transplantation in hospitalized patients with liver cirrhosis. In addition, Bacteroides ovatus and Bacteroides uniformis (both belonging to order Bacteroides) were found to present at higher levels in healthy controls than in ulcerative colitis (UC) or irritable bowel syndrome (IBS) patients, suggesting a possible protective role played by this group of bacteria (Noor et al., 2010; Lopez-Siles et al., 2017). Overall, the disruption of bile acid metabolism caused by intestinal microbes may affect not only gut microbiota but also the whole circulatory system of patients.
However, there were some limitations regarding this study. First, the sample size of our cohort was relatively small, and thus, our developed model requires further validation in independent samples. Second, although we first detected disturbances of the overall gut–liver ecology implicated in submassive hepatic necrosis and provided a comprehensive and multilevel understanding of the role of the disturbed gut microbiome, we did not investigate the specific mechanisms underlying such a disturbance using animal models. Third, how intestinal flora and metabolites induce SMHN in HBV-related cirrhotic livers was not answered by our data.
To the best of our knowledge, this is the first study to comprehensively explore microbiome and BA patterns across the enterohepatic circulation. Especially, the characteristics of bile acids from the portal vein in patients with or without SMHN. For the first time, our study revealed that P. distasonis may play a role in the prognosis of ACLF and AD. Based on multi-omics data, using the random forest algorithm, we developed a combinatorial marker panel with a high AUC value. These findings provide new directions to uncover pathogenesis and develop novel diagnostic strategies for submassive hepatic necrosis.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
XZ and TZ conducted the pathological experiments. RW participated in tissue and clinical data collection. ZB and ZY participated in data analysis. HG, ML, RW, ZB, and ZY participated in manuscript writing and editing. YZ and HG conceived the study and were involved in every step of this study. All authors approved the manuscript as submitted.
Funding
This study was supported by the Chinese Nature Science Foundation (grant no. 81790634), the Zhejiang Non-public Medical Specialty Program, the National Key Research and Development Program of China (2021YFA1301104), and the Research Project of Jinan Microecological Biomedicine Shandong Laboratory.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer NJ declared a shared parent affiliation with the authors ZB, RW, SS, ML, and ZY to the handling editor at the time of review.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1185993/full#supplementary-material
Abbreviations
ACLF, acute-on-chronic liver failure; ALF, acute liver failure; AD, acute decompensation; SMHN, submassive hepatic necrosis; HBV, hepatitis B virus; AUC, area under curve; UPLC-MS, ultraperformance liquid chromatography-tandem mass spectrometry; MELD, model for end-stage liver disease; BAs, bile acids; RF-RFE, random forest with recursive feature elimination.
References
Aguiar Vallim, D., Tarling, T. Q. E. J., and Edwards, P. A. (2013). Pleiotropic roles of bile acids in metabolism. Cell Metab. 17, 657–669. doi: 10.1016/j.cmet.2013.03.013
Allen, K., Jaeschke, H., and Copple, B. L. (2011). Bile acids induce inflammatory genes in hepatocytes: a novel mechanism of inflammation during obstructive cholestasis. Am. J. Pathol. 178, 175–186. doi: 10.1016/j.ajpath.2010.11.026
Bajaj, J. S., Fagan, A., Sikaroodi, M., White, M. B., Sterling, R. K., Gilles, H., et al. (2017). Liver transplant modulates gut microbial dysbiosis and cognitive function in cirrhosis. Liver Transpl. 23, 907–914. doi: 10.1002/lt.24754
Bajaj, J. S., Heuman, D. M., Hylemon, P. B., Sanyal, A. J., White, M. B., Monteith, P., et al. (2014). Altered profile of human gut microbiome is associated with cirrhosis and its complications. J Hepatol 60, 940–947. doi: 10.1016/j.jhep.2013.12.019
Bajaj, J. S., Thacker, L. R., Fagan, A., White, M. B., Gavis, E. A., Hylemon, P. B., et al. (2018). Gut microbial RNA and DNA analysis predicts hospitalizations in cirrhosis. JCI Insight 3, 19. doi: 10.1172/jci.insight.98019
Bajaj, J. S., Vargas, H. E., Reddy, K. R., Lai, J. C., O'leary, J. G., and Tandon, P. (2019). Association between intestinal microbiota collected at hospital admission and outcomes of patients with cirrhosis. Clin. Gastroenterol. Hepatol 17, 756–765. e753. doi: 10.1016/j.cgh.2018.07.022
Chen, Y., Guo, J., Qian, G., Fang, D., Shi, D., Guo, L., et al. (2015). Gut dysbiosis in acute-on-chronic liver failure and its predictive value for mortality. J. Gastroenterol. Hepatol. 30, 1429–1437. doi: 10.1111/jgh.12932
Chiang, J. Y. (2013). Bile acid metabolism and signaling. Compr. Physiol. 3, 1191–1212. doi: 10.1002/cphy.c120023
Claria, J., Moreau, R., Fenaille, F., Amoros, A., Junot, C., Gronbaek, H., et al. (2019). Orchestration of tryptophan-kynurenine pathway, acute decompensation, and acute-on-chronic liver failure in cirrhosis. Hepatology 69, 1686–1701. doi: 10.1002/hep.30363
Claria, J., Stauber, R. E., Coenraad, M. J., Moreau, R., Jalan, R., Pavesi, M., et al. (2016). Systemic inflammation in decompensated cirrhosis: characterization and role in acute-on-chronic liver failure. Hepatology 64, 1249–1264. doi: 10.1002/hep.28740
Doden, H., Sallam, L. A., Devendran, S., Ly, L., Doden, G., Daniel, S. L., et al. (2018). Metabolism of oxo-bile acids and characterization of recombinant 12alpha-hydroxysteroid dehydrogenases from bile acid 7alpha-dehydroxylating human gut bacteria. Appl Environ Microbiol 84, 18. doi: 10.1128/AEM.00235-18
Gujral, J. S., Liu, J., Farhood, A., Hinson, J. A., and Jaeschke, H. (2004). Functional importance of ICAM-1 in the mechanism of neutrophil-induced liver injury in bile duct-ligated mice. Am. J. Physiol. Gastrointest. Liver Physiol. 286, G499–507. doi: 10.1152/ajpgi.00318.2003
Horvatits, T., Drolz, A., Roedl, K., Rutter, K., Ferlitsch, A., Fauler, G., et al. (2017). Serum bile acids as marker for acute decompensation and acute-on-chronic liver failure in patients with non-cholestatic cirrhosis. Liver Int. 37, 224–231. doi: 10.1111/liv.13201
Ishak, K., Baptista, A., Bianchi, L., Callea, F., Groote, D., Gudat, J., et al. (1995). Histological grading and staging of chronic hepatitis. J. Hepatol 22, 696–699. doi: 10.1016/0168-8278(95)80226-6
Jia, W., Xie, G., and Jia, W. (2018). Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat. Rev. Gastroenterol. Hepatol. 15, 111–128. doi: 10.1038/nrgastro.2017.119
Kamath, P. S., Kim, W. R., and Advanced Liver Disease Study, G. (2007). The model for end-stage liver disease (MELD). Hepatology 45, 797–805. doi: 10.1002/hep.21563
Kullak-Ublick, G. A., Stieger, B., and Meier, P. J. (2004). Enterohepatic bile salt transporters in normal physiology and liver disease. Gastroenterology 126, 322–342. doi: 10.1053/j.gastro.2003.06.005
Laleman, W., Verbeke, L., Meersseman, P., Wauters, J., Van Pelt, J., Cassiman, D., et al. (2011). Acute-on-chronic liver failure: current concepts on definition, pathogenesis, clinical manifestations and potential therapeutic interventions. Expert Rev. Gastroenterol. Hepatol. 5, 523–537. quiz 537. doi: 10.1586/egh.11.47
Li, H., Chen, L. Y., Zhang, N. N., Li, S. T., Zeng, B., Pavesi, M., et al. (2016). Characteristics, diagnosis and prognosis of acute-on-chronic liver failure in cirrhosis associated to hepatitis B. Sci. Rep. 6, 25487. doi: 10.1038/srep25487
Li, H., Xia, Q., Zeng, B., Li, S. T., Liu, H., Li, Q., et al. (2015). Submassive hepatic necrosis distinguishes HBV-associated acute on chronic liver failure from cirrhotic patients with acute decompensation. J. Hepatol. 63, 50–59. doi: 10.1016/j.jhep.2015.01.029
Lopez-Siles, M., Duncan, S. H., Garcia-Gil, L. J., and Martinez-Medina, M. (2017). Faecalibacterium prausnitzii: from microbiology to diagnostics and prognostics. ISME J. 11, 841–852. doi: 10.1038/ismej.2016.176
Moreau, R., Claria, J., Aguilar, F., Fenaille, F., Lozano, J. J., Junot, C., et al. (2020). Blood metabolomics uncovers inflammation-associated mitochondrial dysfunction as a potential mechanism underlying ACLF. J. Hepatol .72, 688–701. doi: 10.1016/j.jhep.2019.11.009
Noor, S. O., Ridgway, K., Scovell, L., Kemsley, E. K., Lund, E. K., Jamieson, C., et al. (2010). Ulcerative colitis and irritable bowel patients exhibit distinct abnormalities of the gut microbiota. BMC Gastroenterol. 10, 134. doi: 10.1186/1471-230X-10-134
Olson, J. C., and Kamath, P. S. (2011). Acute-on-chronic liver failure: concept, natural history, and prognosis. Curr. Opin. Crit. Care 17, 165–169. doi: 10.1097/MCC.0b013e328344b42d
Pugh, R. N., Murray-Lyon, I. M., Dawson, J. L., Pietroni, M. C., and Williams, R. (1973). Transection of the oesophagus for bleeding oesophageal varices. Br. J. Surg. 60, 646–649. doi: 10.1002/bjs.1800600817
Sayin, S. I., Wahlstrom, A., Felin, J., Jantti, S., Marschall, H. U., Bamberg, K., et al. (2013). Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 17, 225–235. doi: 10.1016/j.cmet.2013.01.003
Schneider, K. M., Albers, S., and Trautwein, C. (2018). Role of bile acids in the gut-liver axis. J. Hepatol. 68, 1083–1085. doi: 10.1016/j.jhep.2017.11.025
Shaik, F. B., Prasad, D. V., and Narala, V. R. (2015). Role of farnesoid X receptor in inflammation and resolution. Inflamm. Res. 64, 9–20. doi: 10.1007/s00011-014-0780-y
Wahlstrom, A., Sayin, S. I., Marschall, H. U., and Backhed, F. (2016). Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 24, 41–50. doi: 10.1016/j.cmet.2016.05.005
Wang, K., Liao, M., Zhou, N., Bao, L., Ma, K., Zheng, Z., et al. (2019). Parabacteroides distasonis alleviates obesity and metabolic dysfunctions via production of succinate and secondary bile acids. Cell Rep. 26, 222–235. doi: 10.1016/j.celrep.2018.12.028
Winston, J. A., and Theriot, C. M. (2020). Diversification of host bile acids by members of the gut microbiota. Gut Microbes 11, 158–171. doi: 10.1080/19490976.2019.1674124
Xu, Z., Huang, G., Gong, W., Zhou, P., Zhao, Y., Zhang, Y., et al. (2012). FXR ligands protect against hepatocellular inflammation via SOCS3 induction. Cell Signal 24, 1658–1664. doi: 10.1016/j.cellsig.2012.04.015
Keywords: acute-on-chronic liver failure, submassive hepatic necrosis, acute decompensation, bile acids (BAs), gut microbiome
Citation: Bao Z, Wei R, Zheng X, Zhang T, Bi Y, Shen S, Zou P, Zhang J, Yan H, Li MD, Yang Z and Gao H (2023) Landscapes of gut microbiome and bile acid signatures and their interaction in HBV-associated acute-on-chronic liver failure. Front. Microbiol. 14:1185993. doi: 10.3389/fmicb.2023.1185993
Received: 14 March 2023; Accepted: 17 April 2023;
Published: 18 May 2023.
Edited by:
George Grant, University of Aberdeen, United KingdomReviewed by:
Daisuke Tokuhara, Wakayama Medical University, JapanGernot Zollner, University Hospital Graz, Austria
Na Jiao, Zhejiang University, China
Copyright © 2023 Bao, Wei, Zheng, Zhang, Bi, Shen, Zou, Zhang, Yan, Li, Yang and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhongli Yang, zy3p@zju.edu.cn; Hainv Gao, gaohainv@163.com
†These authors have contributed equally to this work
 Zhiwei Bao
Zhiwei Bao Runan Wei1†
Runan Wei1† Yunjiao Bi
Yunjiao Bi Sijia Shen
Sijia Shen Junjie Zhang
Junjie Zhang Huadong Yan
Huadong Yan Ming D. Li
Ming D. Li Zhongli Yang
Zhongli Yang Hainv Gao
Hainv Gao