- 1Engineering Research Center of Ministry of Education on Food Synthetic Biotechnology, and School of Biotechnology, Jiangnan University, Wuxi, China
- 2Science Center for Future Foods, School of Biotechnology, Jiangnan University, Wuxi, China
- 3Changsha Hospital for Maternal & Child Health Care, Changsha, China
- 4Hunan Kerey Pharmaceutical Co., Ltd., Shaoyang, China
- 5Jiangsu Provisional Research Center for Bioactive Product Processing Technology, Jiangnan University, Wuxi, China
7-Dehydrocholesterol (7-DHC) is a widely used sterol and a precursor of several costly steroidal drugs. In this study, 7-DHC biosynthesis pathway was constructed and modified in Saccharomyces cerevisiae. Firstly, the biosynthesis pathway was constructed by knocking out the competitive pathway genes ERG5 and ERG6 and integrating two DHCR24 copies from Gallus gallus at both sites. Then, 7-DHC titer was improved by knocking out MOT3, which encoded a transcriptional repressor for the 7-DHC biosynthesis pathway. Next, by knocking out NEM1 and PAH1, 7-DHC accumulation was improved, and genes upregulation was verified by quantitative PCR (qPCR). Additionally, tHMG1, IDI1, ERG2, ERG3, DHCR24, POS5, and CTT1 integration into multi-copy sites was used to convert precursors to 7-DHC, and increase metabolic flux. Finally, qPCR confirmed the significant up-regulation of key genes transcriptional levels. In a 96 h shaker flask fermentation, the 7-DHC titer was 649.5 mg/L by de novo synthesis. In a 5 L bioreactor, the 7-DHC titer was 2.0 g/L, which was the highest 7-DHC titer reported to date. Our study is of great significance for the industrial production of 7-DHC and steroid development for medical settings.
Introduction
7-Dehydrocholesterol (7-DHC) is a sterol found in animals, which can be directly converted to vitamin D3 after UV irradiation. Vitamin D3 is not only used to prevent and treat rickets and other bone diseases, but also improves immunity and reduces the cardiovascular disease risk (Guo et al., 2018). Vitamin D3 is widely used in biomedicine, feedstuffs, and other fields. 7-DHC is a precursor for 25-hydroxyvitamin D3 production, which is the active form of vitamin D3 and widely used to treat patients with severe liver and kidney disease (Warnke et al., 2016; Tang et al., 2020). Because current price of 25-hydroxyvitamin D3 is high and productivity is low, improving 7-DHC supplies is vital for steroids production.
Due to the wide applications and increasing demand for 7-DHC, its production in Saccharomyces cerevisiae is attractive. Su et al. (2015) ameliorated redox imbalance using a cofactor regeneration strategy and generated a titer of 44.49 mg/L (± 9.63 mg/L) in a 5 L bioreactor. Guo et al. (2018). increased precursor supply by overexpressing all mevalonate (MVA) pathway genes, and found the DHCR24 from Gallus gallus was the most suitable gene to product 7-DHC in S. cerevisiae. Finally, the 7-DHC titer peaked at 1.07 g/L in a 5 L bioreactor. Guo et al. (2021) used the modular organelle localization of pathway genes to generate a 7-DHC titer of 360.6 mg/L in shaker flasks after 100 h. Qu et al. (2022) inhibited the expression of ERG6 using CRISPRi, and the S. cerevisiae Ty1 transposon was used to increase the copies of key genes. The final 7-DHC titer was 365.5 mg/L in shaker flasks and 1,328 mg/L in a 3 L bioreactor.
7-DHC is similar to ergosterol in S. cerevisiae, which is produced by ergosterol pathway modification, therefore S. cerevisiae is an excellent host for 7-DHC production (Shen et al., 2020; Xu and Li, 2020). Hmg1p (HMG-CoA reductase) and Idi1p (isoprenoid diphosphate isomerase) are the rate-limiting enzymes in the MVA pathway (Xia et al., 2022). Up-regulated tHMG1 (truncated HMG1) and IDI1 can increase squalene accumulation (Veen et al., 2003; Kwak et al., 2017). The endoplasmic reticulum (ER) is a critical organelle for 7-DHC biosynthesis (Hu et al., 2017; Jorda and Puig, 2020). Dhcr24p is the only exogenous enzyme of 7-DHC biosynthesis (Figure 1A). Mot3p is a transcriptional repressor of some genes under aerobic and hypoxia conditions (Hong et al., 2019) and directly inhibits ERG2 and ERG11 expression under hypoxia (Sertil et al., 2003; Montanes et al., 2011; Martinez-Montanes et al., 2013). Hongay et al. (2002) and Montanes et al. (2011) showed that Mot3p inhibited ERG2 and ERG9 transcription in S. cerevisiae. Under hypoxic conditions, ERG2 mRNA levels were inhibited more than 10-fold. PAH1 encodes a phosphatidic acid phosphatase, and its knockout improved ER-localized protein expression (Park et al., 2015; Arendt et al., 2017). Pah1p is dephosphorylated by the catalytic subunit (NEM1) of NEM1-SPO7 phosphatase (Su et al., 2014; Mirheydari et al., 2020), therefore, NEM1 knockout may physically expand the ER. It can also expand the ER by overexpressing INO2 (Carman and Han, 2009; Cirigliano et al., 2019; Kim et al., 2019). Upc2p and Ecm22p are involved in ergosterol regulation by binding to the ERG gene promoters (Davies et al., 2005; Yang et al., 2015; Joshua and Hofken, 2017). Iron is an important trace element for 7-DHC biosynthesis, e.g., the squalene monooxygenase Erg1p, the lanosterol C-14 demethylase Erg11p, the sterol C-4 methylase Erg25p, and the sterol C-5 desaturase Erg3p all require iron as a cofactor (Jorda et al., 2021).
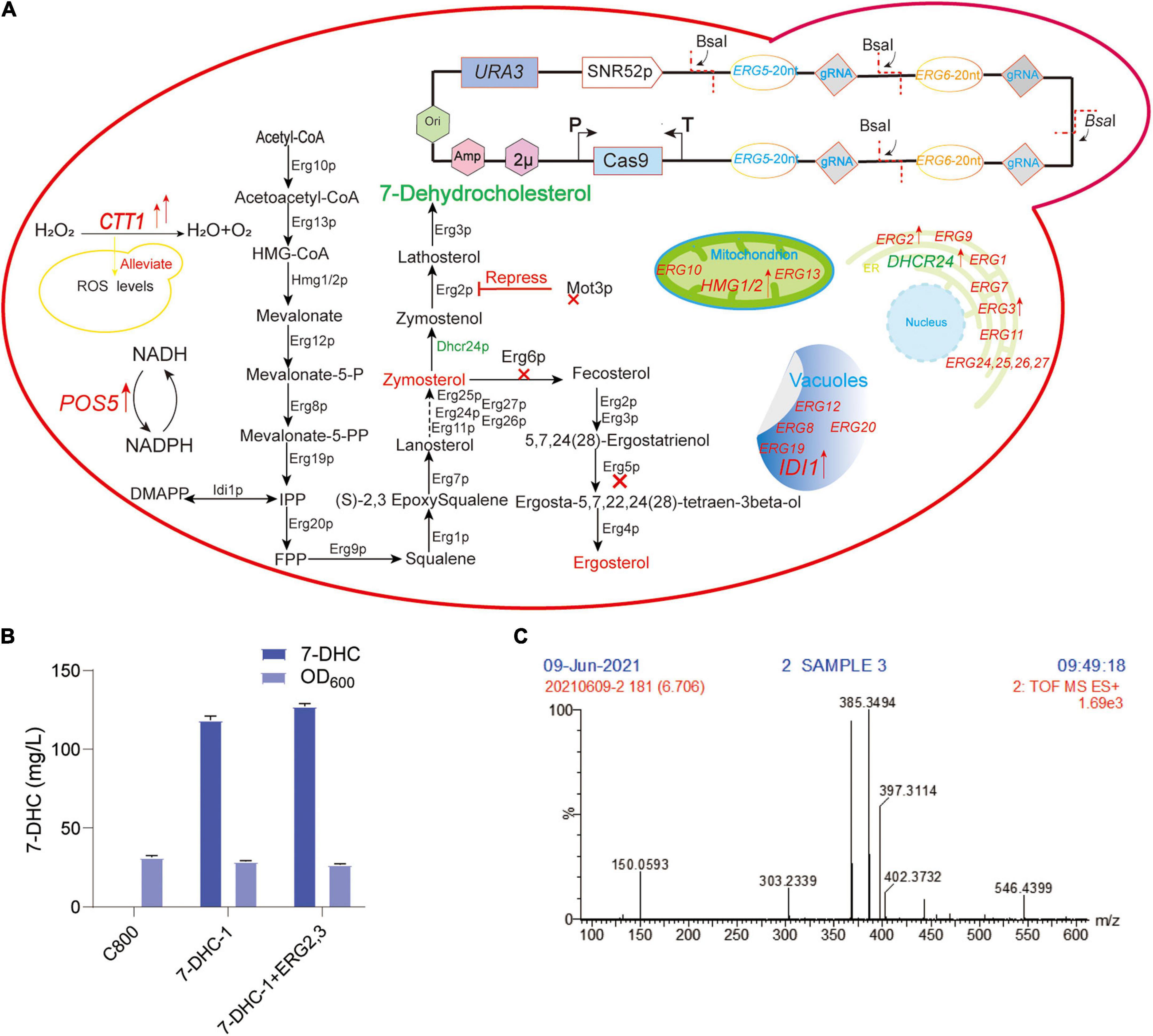
Figure 1. Preliminary 7-DHC synthesis in S. cerevisiae. (A) 7-DHC metabolic pathway in S. cerevisiae; (B) 7-DHC titer in preliminary synthesis in S. cerevisiae; (C) 7-DHC identification by LC-MS.
Considering the current research status, the production capacity of S. cerevisiae must be improved to generate industrial 7-DHC levels for clinicians and patients. Due to its complex metabolic pathway, poor precursor conversion, and accumulation of intermediate sterols which impact on 7-DHC titer, driving pathway genes to convert most precursors to 7-DHC is required. DHCR24 from Gallus gallus (Guo et al., 2018) was selected to generate 7-DHC. As MOT3 inhibits ERG2 expression, it was knocked out to enhance pathway gene expression. NEM1 was also knocked out to enhance ER-localized gene expression. To reduce reactive oxygen species (ROS) in the cytoplasm, CTT1 (cytoplasmic catalase) was overexpressed (Liu et al., 2021). Also, by enhancing POS5 (mitochondrial NADH kinase) expression, NADPH supplies were increased. Finally, “push and pull” 7-DHC biosynthesis was achieved using the multi-copy site integration of tHMG1, IDI, ERG2, ERG3, POS5, DHCR24, and CTT1. The coordinated expression of all metabolic pathway genes was identified by qPCR. Finally, by de novo 7-DHC biosynthesis, the final 7-DHC titer was 649.5 mg/L (96 h) in shaker flasks, and 2.0 g/L in a 5 L bioreactor.
Materials and methods
Chemicals and standards
Standard 7-DHC was purchased from Shanghai Yuanye Co., Ltd., China, and ether and other chemicals were purchased from Shanghai Sinopharm Group, China. Enzymes for Golden Gate Assembly were purchased from NEB (Beijing).
Strains, plasmids, and genes
S. cerevisiae strain C800 (CEN.PK2-1D; MATα; ura3-52; his3Δ1; trp1- 289; leu2-3,112; MAL2-8C; SUC2; gal80: KanMX) was used as the starting strain. Escherichia coli JM109 was used for plasmid construction and preservation. Heterologous, codon-optimized genes were synthesized by Sangon Biotech (Shanghai, China).
Plasmid and strain construction
All fragments were cloned into a vector using the Gibson assembly kit (Gibson et al., 2009). Primers are listed in Supplementary Tables 1, 2. Engineered yeast was generated by episomal plasmid expression and genome integration. The yeast used in this study are listed in Supplementary Table 3.
This study used multiple gene editing technologies to knock out genes. Editor Benchling1 was used to design sgRNA, and the Golden Gate method (Apel et al., 2017) was used to construct multi-gene editing plasmids. The efficient S. cerevisiae transformation method was used for yeast transformations, and plasmids and fragments were transformed into yeast cells for expression and genome modification (Gietz and Woods, 2002).
Multi-copy site integration
Histidine, tryptophan, and uracil tags were integrated with gene fragments, and relevant deficient yeast nitrogen base (YNB) culture medium was used to screen high-copy strains expressing integrated genes (Maury et al., 2016). Strains with the highest 7-DHC accumulation were screened in shaking flasks.
Strain cultivation
Plasmid cloning and DNA extraction were performed using E. coli JM109 cells cultured in Luria-Bertani medium (10 g/L tryptone, 5 g/L yeast extract, and 10 g/L NaCl). Yeast expressing auxotrophic markers was selected on synthetic medium (20 g/L glucose, 1.74 g/L amino acid-free YNB, 5 g/L ammonium sulfate, 0.05 g/L leucine, 0.05 g/L histidine, 0.05 g/L tryptophan, and 0.05 g/L uracil). Engineered yeast strains were cultured in YPD (20 g/L glucose, 10 g/L yeast extract, and 20 g/L peptone). Selected colonies were cultured in YPD medium at 30°C and 220 rpm for 16–20 h, then an inoculum was added to 250 mL flasks containing 25 mL YPD medium. The initial OD600 was 0.2.
For 5 L fermentations, an engineered colony strain was inoculated into 15 mL YPD medium in a 250 mL shaker flask and cultured for 24 h at 220 rpm, 30°C. An inoculum (1%) was then transferred to a 250 mL flask containing 25 mL YPD medium (seed solution) and cultured for 17 h at 30°C and 220 rpm. Then, the culture was inoculated into a 5 L fermenter containing 2.5 L YPD medium at a 5% inoculum and a feed-batch fermentation initiated at 30°C, using 3 M NaOH to maintain the pH at 5.5. The feed medium (500 mL) was composed of: 400 g/L glucose, 18 g/L KH2PO4, 7g/L K2SO4, 0.56 g/L Na2SO3, 20 mL/L trace element A (comprising 5.75 g/L ZnSO4⋅7H2O, 0.32 g/L MnCl2⋅4H2O, 0.47 g/L CoCl2⋅6H2O, 0.48 g/L NaMoO4⋅2H2O, 2.9 g/L CaCl2⋅2H2O, 2.8 g/L FeSO4⋅7H2O, and 80 mL 0.5M EDTA, adjusted to pH 8.0), 24 mL/L trace element B (comprising 0.05 g/L biotin, 1 g/L calcium pantothenate, 1 g/L nicotinic acid, 25 g/L myo-inositol, 1 g/L thiamine HCl, 1 g/L pyridoxal HCl, and 0.02 g/L p-aminobenzoic acid), 1.0 g FeSO4⋅7H2O and 250 mL ethanol. The dissolved oxygen concentration was maintained at 25% by adjusting stirring rates (200–800 rpm). The feeding medium rate was 6–8 mL/h.
Quantitative PCR
A fresh single yeast colony was inoculated into 5 mL YPD medium, cultured to the logarithmic growth phase for secondary transfer for 15–19 h, 1 mL cells aspirated and centrifuged, and a total RNA extraction kit (TaKaRa, Beijing, China) used to extract RNA. RNA was reverse transcribed to cDNA using a PrimeScript™ RT reagent kit with a gDNA eraser kit (TaKaRa). Quantitative PCR (qPCR) was conducted using the SYBR Premix Ex Taq II kit (TaKaRa). A LightCycler 480 II Real-Time PCR instrument (Roche Applied Science, Mannheim, Germany) was used for amplifications. ACT1 functioned as an internal reference gene, and related gene transcription was determined using the 2–ΔΔCt method (Chen et al., 2022).
Processing and analysis methods
First, a potassium hydroxide ethanol saponification solution, with a mass fraction 30% (90% ethanol), was prepared. Next, 1 mL fermentation medium was centrifuged at 12,000 rpm for 8 min, the supernatant removed, resuspended in 1 mL saponification solution, and heated to 86–88°C for 3.5–4.0 h to undergo a reflux saponification reaction. Ether was the extractant (Wang et al., 2018). To a high performance liquid chromatography system (Waters, Milford MA, United States), a C18 column (30 m × 0.25 mm, 0.25 μm film thickness) was attached and maintained at 30°C. 7-DHC was separated in 100% methanol (mobile phase) at 280 nm at 1 mL/min in a separation time of 20 min (Xia et al., 2022). LC-MS method was used to identify 7-DHC (Sun et al., 2021).
Results
Constructing the 7-dehydrocholesterol pathway in Saccharomyces cerevisiae
In our previous study, the S. cerevisiae C800 host strain (CEN.PK2-1D; MATα; ura3-52; his3Δ1; trp1-289; leu2-3,112; MAL2-8C; SUC2; gal80:KanMX), which tolerated high glucose titer, was suitable for the de novo synthesis of exogenous sterols (Gao et al., 2020; Shi et al., 2021). To achieve de novo 7-DHC biosynthesis, DHCR24 (heterologous Δ24-dehydrocholesterol reductase) was introduced to S. cerevisiae. In this study, two DHCR24 copies were separately integrated into ERG5 and ERG6 sites, generating strain 7-DHC-1 (Figure 1A), which produced 118.5 mg/L 7-DHC in shaker flasks within 72 h (Figure 1B). ERG2 and ERG3 were overexpressed in 7-DHC-1 using the episomal expression vector pY26-TEF-GPD. However, this overexpression did not significantly increase the 7-DHC titer (Figure 1B). 7-DHC product was identified by liquid chromatography-mass spectroscopy (LC-MS) (Figure 1C).
The effects of MOT3 knockout on 7-dehydrocholesterol biosynthesis
ERG2 was previously identified as a key gene for 7-DHC and other sterol biosynthesis (Hongay et al., 2002). Similarly, some genes were found to inhibit ERG2 expression (Hongay et al., 2002; Hong et al., 2019). For example, MOT3, which was identified as a transcriptional repressor for sterol biosynthesis pathways, inhibited ERG2 expression under hypoxia. Thus, in this study, the knockout of MOT3 in the 7-DHC-1 strain, could upregulate ERG2 and ERG11 transcription levels (Figure 2A), and could also increase the 7-DHC titer to 159.3 mg/L. This titer was increased by 24% when compared to the 7-DHC-1 strain. To further enhance precursor supplies for 7-DHC, one copy each of ERG2 and ERG3 were integrated at the MOT3 site in the 7-DHC-1 strain, to generate the 7-DHC-2 strain. The qPCR results showed that ERG2 transcriptional level was up-regulated, as were ERG1, ERG3, and ERG11 levels (Figure 2B). The 7-DHC titer in the 7-DHC-2 strain was further increased from 128.7 mg/L to 220.2 mg/L within 72 h in shaker flasks (Figure 2C).
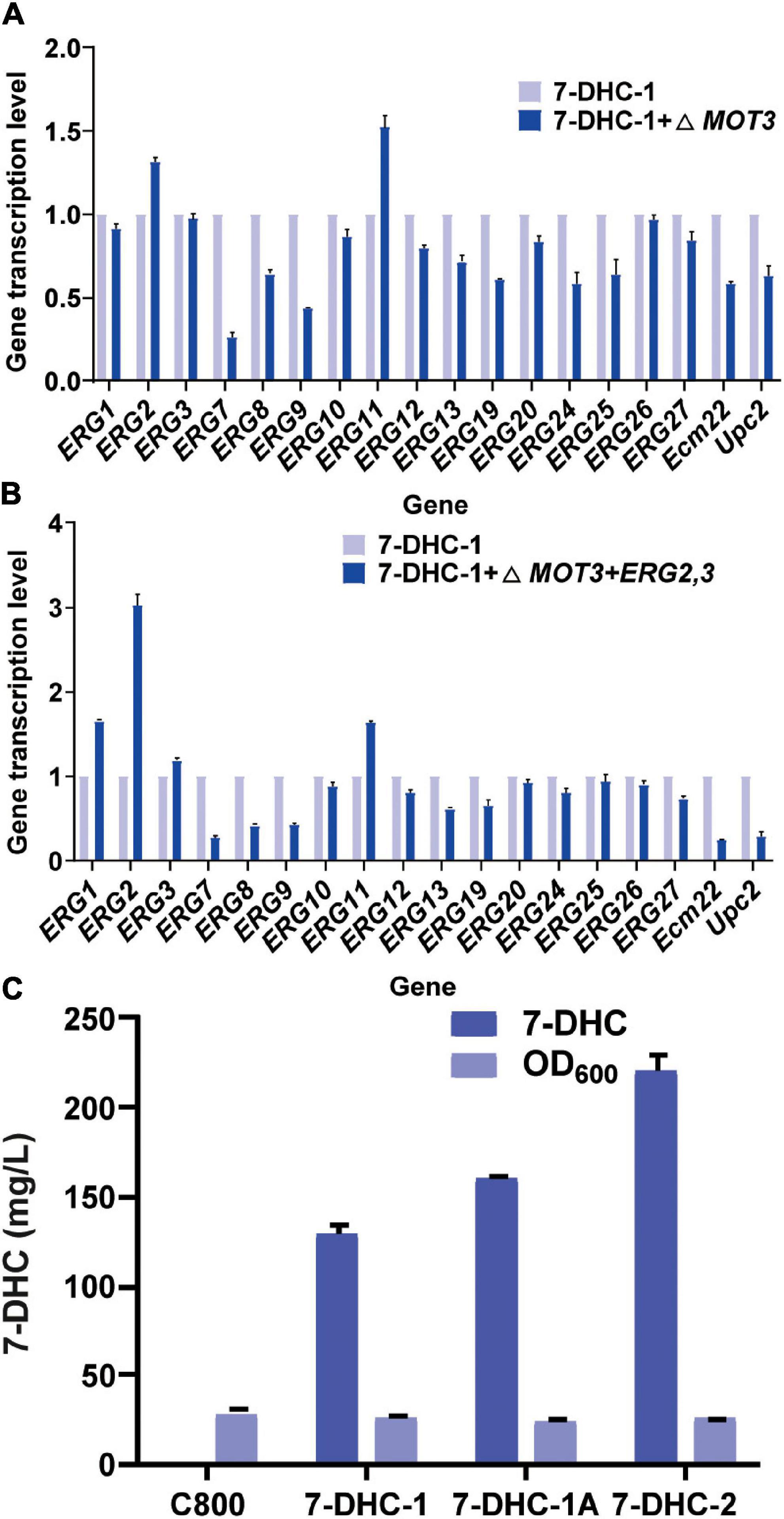
Figure 2. The knockout of related repressor genes in the 7-DHC synthesis pathway in S. cerevisiae. (A) qPCR validation of pathway genes after knockout of the hypoxia repressor MOT3 in S. cerevisiae; (B) qPCR validation of pathway genes after one-copy integration of the ERG2 and ERG3 expression cassette at the MOT3 site; (C) 7-DHC titers in 7-DHC-1, 7-DHC-1A (7-DHC-1 + ΔMOT3), and 7-DHC-2 (7-DHC-1 + ΔMOT3 + ERG2,3) strains.
Engineering organelle-related genes to enhance 7-dehydrocholesterol accumulation
Previous studies reported that engineering cell organelles improved sterol accumulation (Arendt et al., 2017). Therefore, several genes related to ER and vacuole were engineered to evaluate their effects on 7-DHC accumulation. NEM1 is reportedly responsible for PAH1 dephosphorylation, which affects membrane areas of the ER and lipid metabolism. It was speculated that ER membrane expansion would enhance the expression of ER-localized genes. NEM1 was knocked out in 7-DHC-2 to generate 7-DHC-3, which produced up to 279.8 mg/L 7-DHC within 72 h in shaker flasks, and was 27.1% higher than 7-DHC-2 (Figure 3A). qPCR results showed that NEM1 knockout enhanced the transcriptional levels of ER localization genes (Figure 3B). Furthermore, PAH1 and DGK1 knockout in 7-DHC-3 expanded the ER, and generated 7-DHC-3-1 (ΔPAH1), 7-DHC-3-2 (ΔDGK1), and 7-DHC-3-3 (ΔPAH1 and ΔDGK1) strains. PAH1 and DGK1 knockout did not significantly increase 7-DHC accumulation, while the DGK1 knockout reduced 7-DHC accumulation. The PEP4 knockout in 7-DHC-3, which was responsible for vacuole expansion and heterologous gene expression, generated strain 7-DHC-3-4, which produced 284.4 mg/L 7-DHC and was slightly higher (9.3 mg/L) than 7-DHC-3. However, qPCR results (Figure 3C) showed PEP4 knockout caused transcriptional level downregulation of all pathway genes except ERG9. Thus, strain 7-DHC-3 was selected as the platform strain for further study.
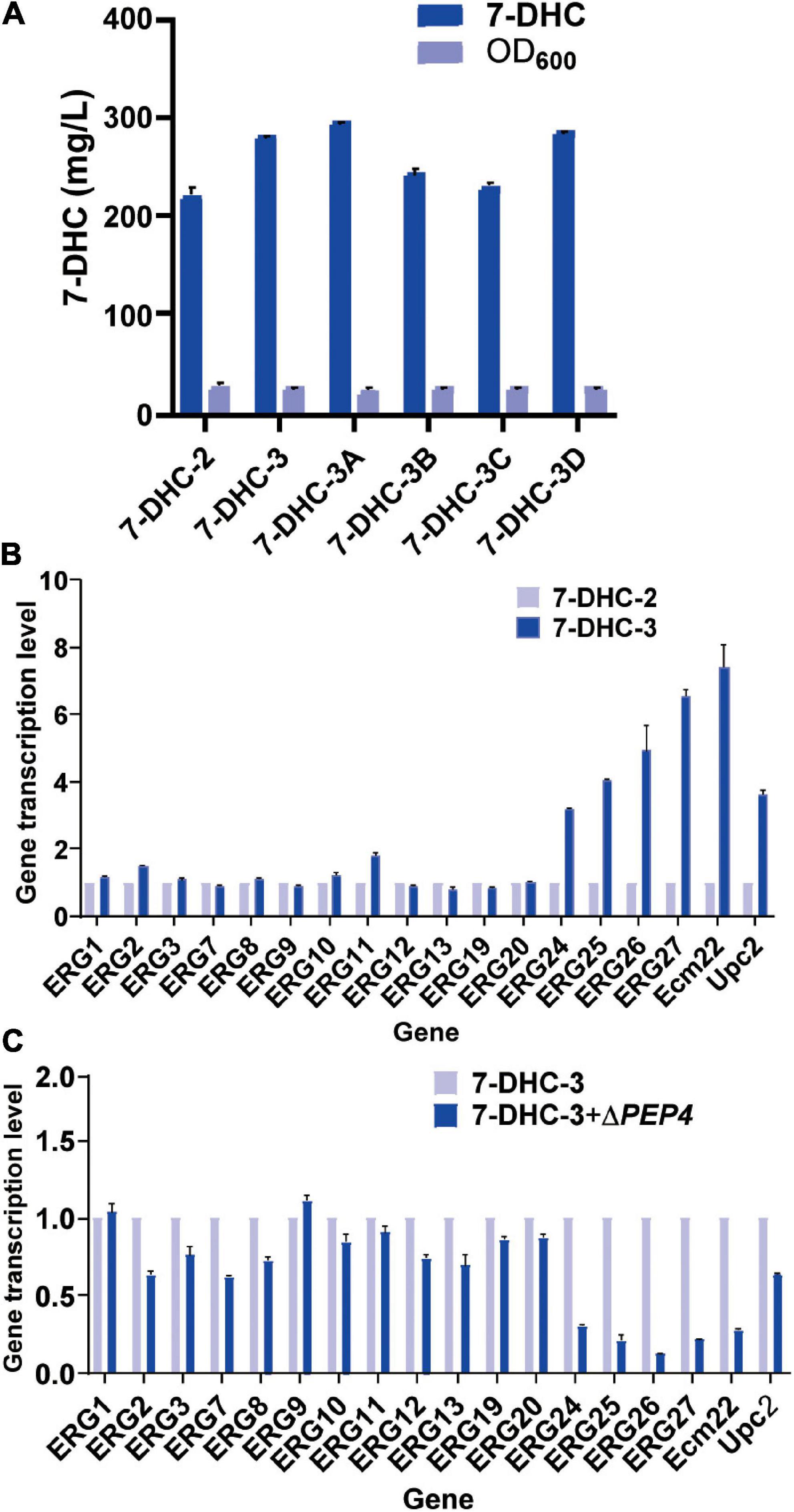
Figure 3. Engineering organelle related genes in S. cerevisiae. (A) 7-DHC titers in 7-DHC-2, 7-DHC-3 (7-DHC-2 + ΔNEM1), 7-DHC-3A (7-DHC-3 + ΔPAH1), 7-DHC-3B (7-DHC-3 + ΔDGK1), 7-DHC-3C (7-DHC-3 + ΔPAH1 + ΔDGK1), 7-DHC-3D (7-DHC-3 + ΔPEP4) strains. (B) qPCR validation of pathway genes after NEM1 knockout in S. cerevisiae. (C) qPCR validation of pathway genes after PEP4 knockout in S. cerevisiae.
Multi-copy site integration coordinates gene expression
Based on 7-DHC-3, tHMG1 and IDI were integrated at the multi-copy site Ty1(Figure 4A). This 7-DHC-4 strain generated 56.0 mg/L more 7-DHC than 7-DHC-3 (Figure 4B); it produced 317.3 mg/L 7-DHC in shaker flasks within 72 h. Using 7-DHC-3, ERG2, ERG3, CTT1, and DHCR24 were integrated at the multi-copy site Ty2. This 7-DHC-5 strain produced 304.0 mg/L 7-DHC within 72 h in shaker flasks, which was 24.2 mg/L higher than 7-DHC-3 (Figure 4C). Using 7-DHC-5, the rate-limiting genes tHMG1 and IDI were integrated into Ty1. Finally, the 7-DHC-6 strain generated the highest 7-DHC titer in shaker flasks; 440.9 mg/L (72 h) which was 130.9 mg/L higher than 7-DHC-3 (Figure 4E). 7-DHC-6 analysis by qPCR confirmed that ERG1, ERG2, ERG3, ERG7, ERG8, ERG9, ERG10, ERG11, ERG12, ERG13, ERG19, ERG20, ERG24, ERG25, ERG26, ERG27, and transcriptional factors Ecm22p and Upc2p transcriptional levels were significantly higher than 7-DHC-1. Of these genes, ERG2 was up-regulated 14-fold (Figure 4D). Based on 7-DHC-6, ERG2, ERG3, POS5, and DHCR24 were integrated into Ty3. This 7-DHC-7 strain generated a 7-DHC titer of 459.3 mg/L within 72 h in shaker flasks, which was 18.4 mg/L higher than 7-DHC-6 (Figure 4F).
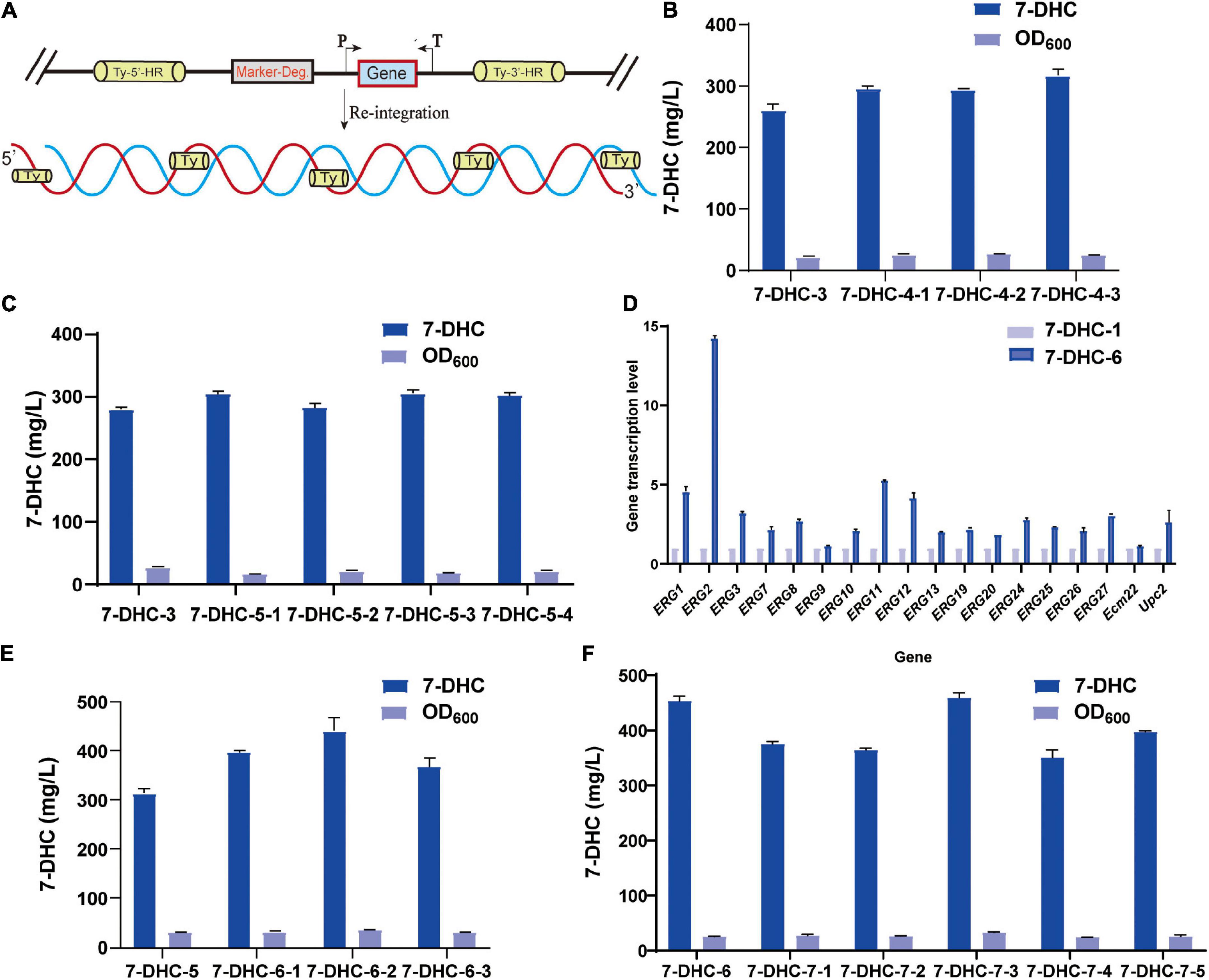
Figure 4. Multi-copy site integration coordinates gene expression. (A) Schematic showing multi-copy site integration. (B) Based on the 7-DHC-3 strain, the multi-copy integration of the rate-limiting genes tHMG1 and IDI at the Ty1 site was completed. (C) Based on the 7-DHC-3 strain, the multi-copy integration of ERG2, ERG3, CTT1, and DHCR24 genes at the Ty2 site was completed. (D) qPCR analyses of the 7-DHC-6-2 strain compared with the 7-DHC-1 strain. (E) Based on the 7-DHC-5 strain, the multi-copy integration of tHMG1 and IDI at the Ty1 site was completed. (F) Based on the 7-DHC-6 strain, the multi-copy integration of ERG2, ERG3, POS5, and DHCR24 at the Ty3 site was completed.
Fermentation optimization of the 7-dehydrocholesterol high-yielding strain
By analyzing the 7-DHC production metabolic pathway in S. cerevisiae, the effects of carbon sources and trace elements were explored on 7-DHC accumulation. Also, by examining the exogenous addition of the carbon sources, glucose and ethanol (Figure 5A), and the trace elements, Mg2+ and Fe2+ (Figure 5B), 7-DHC accumulation peaked when initial glucose was 30 g/L and ferrous sulfate was 0.4 g/L (Figure 5B). 7-DHC accumulation was 649.5 mg/L within 96 h in shaker flasks, and 2.0 g/L (Figures 5C–H) in a 5 L bioreactor.
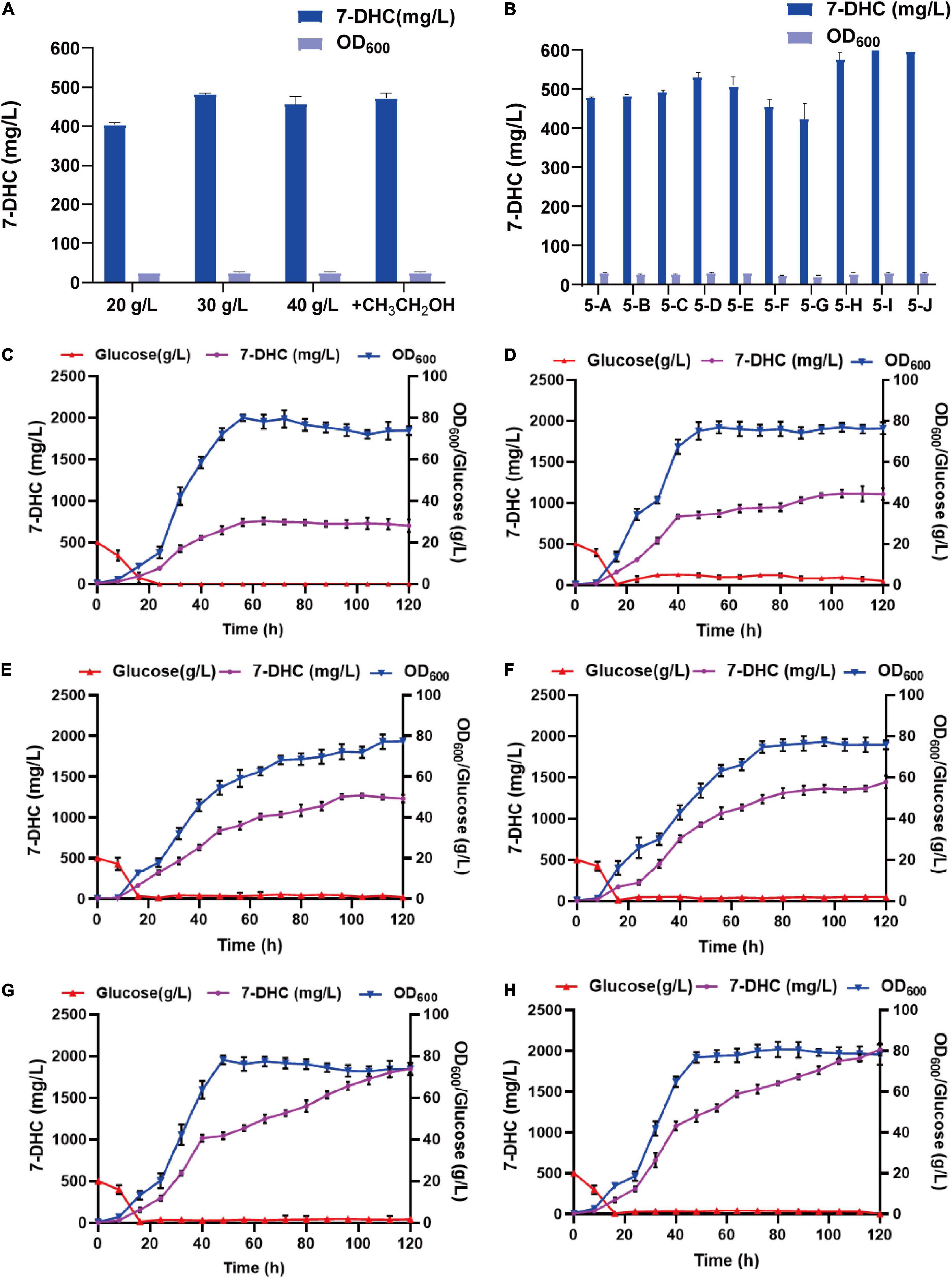
Figure 5. Optimizing 7-DHC production in S. cerevisiae. (A) The effects of glucose and ethanol (20 g/L) on 7-DHC accumulation. The initial concentration of glucose is controlled as 20 g/L, 30 g/L, 40 g/L. Ethanol was added at the 24th hour of fermentation, based on the initial glucose concentration of 20 g/L. (B) The effects of Mg2+ and Fe2+ on 7-DHC accumulation. 7-DHC titers in 5-A (20 g/L glucose), 5-B (20 g/L glucose + 0.05 g/L Fe2+), 5-C (20 g/L glucose + 0.1 g/L Fe2+), 5-D (20 g/L glucose + 0.3 g/L Fe2+), 5-E (20 g/L glucose + 0.5 g/L Fe2+), 5-F (20 g/L glucose + 1 g/L Mg2+), 5-G (20 g/L glucose + 3 g/L Mg2+), 5-H (30 g/L glucose + 0.3 g/L Fe2+), 5-I (30 g/L glucose + 0.4 g/L Fe2+), 5-J (30 g/L glucose + 0.5 g/L Fe2+) strains. (C) 7-DHC production by feed-batch fermentation in a 5 L bioreactor, and the dissolved oxygen concentration was maintained at 40%. (D) 7-DHC production by feed-batch fermentation in a 5 L bioreactor. The dissolved oxygen concentration was maintained at 30% and glucose concentration was maintained at 3–4 g/L. (E) 7-DHC production by feed-batch fermentation in a 5 L bioreactor. The dissolved oxygen concentration was maintained at 30% and glucose concentration was maintained at 0–2 g/L. (F) 7-DHC production by feed-batch fermentation in a 5 L bioreactor. The dissolved oxygen concentration was maintained at 30–10% and glucose concentration was maintained at 0–2 g/L (add ethanol after 72 h). (G) 7-DHC production by feed-batch fermentation in a 5 L bioreactor. The dissolved oxygen concentration was maintained at 25% and glucose concentration was maintained at 0–2 g/L (add ethanol after 72 h). 0.4 g/L FeSO4⋅7H2O was added at the time of inoculation. (H) FeSO4⋅7H2O was added by fed-feed method to the final concentration of 0.4 g/L.
Discussion
In this study, the combinatorial engineering strategies were used to construct several 7-DHC producing strains. Exogenous DHCR24 was introduced, then, the off-pathway genes MOT3 and NEM1 were knocked out, the rate-limiting step genes tHMG1 and IDI were overexpressed to increase precursor supply, and ERG2, ERG3, DHCR24 expression was enhanced to convert precursors to 7-DHC. The multi-copy integration of the cytoplasmic catalase gene CTT1 was used to reduce cytoplasmic ROS levels. The POS5 was overexpressed to increase NADPH supply. Finally, de novo 7-DHC synthesis was achieved in S. cerevisiae; the 7-DHC titer was 649.5 mg/L within 96 h in shaker flasks and 2.0 g/L at 120 h in a 5 L bioreactor.
From previous research, enhancing ER-localized ERG2 and ERG3 expression had no significant effects on 7-DHC accumulation (Guo et al., 2021). However, ERG2 and ERG3 are both key genes for 7-DHC accumulation. MOT3 inhibited ERG2 expression under hypoxic conditions, therefore, in this study, ERG2 and ERG3 were integrated into MOT3 site, which significantly increased the 7-DHC titer. Hongay et al. (2002) showed that S. cerevisiae experienced a > 10-fold inhibition of ERG2 mRNA levels under hypoxic conditions. In MOT3 deletion mutants, ERG2 inhibition was almost completely eliminated under hypoxia. Also, ERG1, ERG3, and ERG11 expression were somewhat up-regulated. Since ERG1, ERG2, ERG3, and ERG11 are key rate-limiting genes in the 7-DHC biosynthesis pathway, the MOT3 knockout benefitted their expression (Sertil et al., 2003; Montanes et al., 2011; Martinez-Montanes et al., 2013).
Increasing ER membrane area benefits the expression of most genes in the 7-DHC biosynthesis pathway. PAH1 knockout reportedly dilated the ER, while NEM1 was responsible for PAH1 dephosphorylation, thereby affecting ER membrane area and lipid metabolism (Carman and Han, 2009; Arendt et al., 2017; Kim et al., 2019). According to Park et al. (2015), DGK1 knockout alleviated the effects of PAH1 knockout on yeast growth. PEP4 encodes a protease, which was shown to mature different vacuolar peptidases; however, the PEP4 knockout positively affected vacuole expansion and heterologous gene expression (Arendt et al., 2017). In this study, after NEM1 was knocked out, the 7-DHC titer increased by 27.1%. Based on this knockout, the 7-DHC titer increased when PAH1 was knocked out, but cell optical density decreased. However, the 7-DHC titer decreased after both DGK1 and PAH1 were knocked out. Therefore, NEM1, PAH1, and DGK1 could not be simultaneously knocked out, as lipid metabolism was seriously affected. After the PEP4 knockout, most 7-DHC pathway genes except ERG9 were down-regulated.
In S. cerevisiae, Upc2p and Ecm22p are similar zinc-finger transcriptional factors that activate transcription by binding to ERG promoters involved in ergosterol biosynthesis and uptake. The 888th gene mutation of Upc2p and the 790th of Ecm22p both increased transcriptional activation (Davies et al., 2005; Yang et al., 2015). The C-termini of Upc2p and Ecm22p are responsible for ergosterol binding; however, it is unclear if other sterols in S. cerevisiae bind to transcription factor C-termini. Therefore, the effects of two different C-terminal truncated forms of the transcription factor Ecm22p were explored on 7-DHC accumulation. When two Ecm22p forms, Ecm22-1 (1,500 bp) and Ecm22-2 (1,200 bp), were overexpressed, their 7-DHC titers were unchanged in both forms.
As many genes are involved in 7-DHC biosynthesis in S. cerevisiae, the conversion efficiency of precursors directly leads to limited 7-DHC accumulation. Therefore, a strategy was used to enhance the accumulation of 7-DHC by increasing precursor conversion. Firstly, the multi-copy integration of tHMG1 and IDI at Ty1 was engineered to enhance downstream acetyl-CoA metabolism. Also, 7-DHC accumulation was increased after the multi-copy integration of ERG2, ERG3, CTT1, and DHCR24 at Ty2. Finally, the 7-DHC titer at 72 h in shaker flasks was 440.9 mg/L, which was 130.9 mg/L higher than in control strain. This strategy benefitted 7-DHC accumulation in S. cerevisiae. By verifying the 7-DHC-6 strain (qPCR), ERG2 transcriptional levels were increased 14-fold and ERG11 fivefold. MOT3 knockout alleviated metabolic inhibitory effects and significantly promoted the expression of key genes were verified. Finally, POS5 expression was enhanced to further increase 7-DHC titers and cell density. By exploring carbon sources and trace element addition, it was observed that 7-DHC accumulation peaked when the initial glucose concentration was 30 g/L and the initial ferrous sulfate was 0.4 g/L.
Conclusion
In conclusion, this study successfully reengineered the 7-DHC biosynthesis pathway in S. cerevisiae, and used metabolic strategies, including related genes knockout, intracellular ROS reduction, and enhanced precursor supply and conversion, to improve the 7-DHC production. Finally, the 7-DHC titer was 649.5 mg/L in shaker flasks and 2.0 g/L in a 5 L bioreactor. This study lays the foundations for the industrial production of 7-DHC, and provide a platform for the successful production of other steroids in S. cerevisiae.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author/s.
Author contributions
WW: experimental design and implementation, date analysis, and writing. SG: experimental design. QY and AL: data analysis. SY and JZ: reviewing and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key Research and Development Program of China (2019YFA0905300) and the Foundation for Innovative Research Groups of the National Natural Science Foundation of China (32021005).
Conflict of interest
AL was employed by Hunan Kerey Pharmaceutical Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer XC declared a shared affiliation with authors WW, SG, SY, and JZ at the time of the review.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.978074/full#supplementary-material
Footnotes
References
Apel, A. R., d’Espaux, L., Wehrs, M., Sachs, D., Li, R. A., Tong, G. J., et al. (2017). A cas9-based toolkit to program gene expression in Saccharomyces cerevisiae. Nucleic Acids Res. 45, 496–508. doi: 10.1093/nar/gkw1023
Arendt, P., Miettinen, K., Pollier, J., De Rycke, R., Callewaert, N., and Goossens, A. (2017). An endoplasmic reticulum-engineered yeast platform for overproduction of triterpenoids. Metab. Eng. 40, 165–175. doi: 10.1016/j.ymben.2017.02.007
Carman, G. M., and Han, G. S. (2009). Regulation of phospholipid synthesis in yeast. J. Lipid Res. 50, S69–73. doi: 10.1194/jlr.R800043-JLR200
Chen, Y., Zeng, W., Yu, S., Chen, J., and Zhou, J. (2022). Gene co-expression network analysis reveals the positive impact of endocytosis and mitochondria-related genes over nitrogen metabolism in Saccharomyces cerevisiae. Gene 821:146267. doi: 10.1016/j.gene.2022.146267
Cirigliano, A., Macone, A., Bianchi, M. M., Oliaro-Bosso, S., Balliano, G., Negri, R., et al. (2019). Ergosterol reduction impairs mitochondrial DNA maintenance in S. cerevisiae. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1864, 290–303. doi: 10.1016/j.bbalip.2018.12.002
Davies, B. S. J., Wang, H. S., and Rine, J. (2005). Dual activators of the sterol biosynthetic pathway of Saccharomyces cerevisiae: Similar activation/regulatory domains but different response mechanisms. Mol. Cell. Biol. 25, 7375–7385. doi: 10.1128/mcb.25.16.7375-7385.2005
Gao, S., Xu, X. Y., Zeng, W. Z., Xu, S., Lyv, Y. B., Feng, Y., et al. (2020). Efficient biosynthesis of (2S)-eriodictyol from (2S)-naringenin in Saccharomyces cerevisiae through a combination of promoter adjustment and directed evolution. ACS Synth.Biol. 9, 3288–3297. doi: 10.1021/acssynbio.0c00346
Gibson, D. G., Young, L., Chuang, R.-Y., Venter, J. C., Hutchison, C. A., and Smith, H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345. doi: 10.1038/nmeth.1318
Gietz, R. D., and Woods, R. A. (2002). Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Meth. Enzymol. 350, 87–96. doi: 10.1016/s0076-6879(02)50957-5
Guo, X. J., Xiao, W. H., Wang, Y., Yao, M. D., Zeng, B. X., Liu, H., et al. (2018). Metabolic engineering of Saccharomyces cerevisiae for 7-dehydrocholesterol overproduction. Biotechnol. Biofuels 11:14. doi: 10.1186/s13068-018-1194-9
Guo, X. J., Yao, M. D., Xiao, W. H., Wang, Y., Zhao, G. R., and Yuan, Y. J. (2021). Compartmentalized reconstitution of post-squalene pathway for 7-dehydrocholesterol overproduction in Saccharomyces cerevisiae. Front. Microbiol. 12:663973. doi: 10.3389/fmicb.2021.663973
Hong, J., Park, S. H., Kim, S., Kim, S. W., and Hahn, J. S. (2019). Efficient production of lycopene in Saccharomyces cerevisiae by enzyme engineering and increasing membrane flexibility and NAPDH production. Appl. Microbiol. Biotechnol. 103, 211–223. doi: 10.1007/s00253-018-9449-8
Hongay, C., Jia, N., Bard, M., and Winston, F. (2002). Mot3 is a transcriptional repressor of ergosterol biosynthetic genes and is required for normal vacuolar function in Saccharomyces cerevisiae. EMBO J. 21, 4114–4124. doi: 10.1093/emboj/cdf415
Hu, Z., He, B., Ma, L., Sun, Y., Niu, Y., and Zeng, B. (2017). Recent advances in ergosterol biosynthesis and regulation mechanisms in Saccharomyces cerevisiae. Indian J. Microbiol. 57, 270–277. doi: 10.1007/s12088-017-0657-1
Jorda, T., and Puig, S. (2020). Regulation of ergosterol biosynthesis in Saccharomyces cerevisiae. Genes 11:18. doi: 10.3390/genes11070795
Jorda, T., Rozes, N., and Puig, S. (2021). Sterol composition modulates the response of Saccharomyces cerevisiae to iron deficiency. J. Fungi 7:901. doi: 10.3390/jof7110901
Joshua, I. M., and Hofken, T. (2017). From lipid homeostasis to differentiation: Old and new functions of the zinc cluster proteins Ecm22, Upc2, Sut1 and Sut2. Int. J. Mol. Sci. 18:17. doi: 10.3390/ijms18040772
Kim, J. E., Jang, I. S., Son, S. H., Ko, Y. J., Cho, B. K., Kim, S. C., et al. (2019). Tailoring the Saccharomyces cerevisiae endoplasmic reticulum for functional assembly of terpene synthesis pathway. Metab. Eng. 56, 50–59. doi: 10.1016/j.ymben.2019.08.013
Kwak, S., Kim, S. R., Xu, H. Q., Zhang, G. C., Lane, S., Kim, H., et al. (2017). Enhanced isoprenoid production from xylose by engineered Saccharomyces cerevisiae. Biotechnol. Bioeng. 114, 2581–2591. doi: 10.1002/bit.26369
Liu, M., Lin, Y. C., Guo, J. J., Du, M. M., Tao, X. Y., Gao, B., et al. (2021). High-level production of sesquiterpene patchoulol in Saccharomyces cerevisiae. ACS Synth. Biol. 10, 158–172. doi: 10.1021/acssynbio.0c00521
Martinez-Montanes, F., Rienzo, A., Poveda-Huertes, D., Pascual-Ahuir, A., and Proft, M. (2013). Activator and repressor functions of the mot3 transcription factor in the osmostress response of Saccharomyces cerevisiae. Eukaryot. Cell 12, 636–647. doi: 10.1128/ec.00037-13
Maury, J., Germann, S. M., Jacobsen, S. A. B., Jensen, N. B., Kildegaard, K. R., Herrgard, M. J., et al. (2016). EasyCloneMulti: A set of vectors for simultaneous and multiple genomic integrations in Saccharomyces cerevisiae. PLoS One 11:e0150394. doi: 10.1371/journal.pone.0150394
Mirheydari, M., Dey, P., Stukey, G. J., Park, Y., Han, G. S., and Carman, G. M. (2020). The Spo7 sequence LLI is required for Nem1-Spo7/Pah1 phosphatase cascade function in yeast lipid metabolism. J. Biol. Chem. 295, 11473–11485. doi: 10.1074/jbc.RA120.014129
Montanes, F. M., Pascual-Ahuir, A., and Proft, M. (2011). Repression of ergosterol biosynthesis is essential for stress resistance and is mediated by the Hog1 MAP kinase and the Mot3 and Rox1 transcription factors. Mol. Microbiol. 79, 1008–1023. doi: 10.1111/j.1365-2958.2010.07502.x
Park, Y., Han, G. S., Mileykovskaya, E., Garrett, T. A., and Carman, G. M. (2015). Altered lipid synthesis by lack of yeast pah1 phosphatidate phosphatase reduces chronological life span. J. Biol. Chem. 290, 25382–25394. doi: 10.1074/jbc.M115.680314
Qu, L., Xiu, X., Sun, G., Zhang, C., Yang, H., Liu, Y., et al. (2022). Engineered yeast for efficient de novo synthesis of 7-dehydrocholesterol. Biotechnol. Bioeng. 119, 1278–1289. doi: 10.1002/bit.28055
Sertil, O., Kapoor, R., Cohen, B. D., Abramova, N., and Lowry, C. V. (2003). Synergistic repression of anaerobic genes by Mot3 and Rox1 in Saccharomyces cerevisiae. Nucleic Acids Res. 31, 5831–5837. doi: 10.1093/nar/gkg792
Shen, B., Zhou, P. P., Jiao, X., Yao, Z., Ye, L. D., and Yu, H. W. (2020). Fermentative production of Vitamin E tocotrienols in Saccharomyces cerevisiae under cold-shock-triggered temperature control. Nat. Commun. 11:14. doi: 10.1038/s41467-020-18958-9
Shi, W. Q., Li, J., Chen, Y. F., Liu, X. H., Chen, Y. F., Guo, X. W., et al. (2021). Metabolic engineering of Saccharomyces cerevisiae for ethyl acetate biosynthesis. ACS Synth. Biol. 10, 495–504. doi: 10.1021/acssynbio.0c00446
Su, W., Xiao, W. H., Wang, Y., Liu, D., Zhou, X., and Yuan, Y. J. (2015). Alleviating redox imbalance enhances 7-dehydrocholesterol production in engineered Saccharomyces cerevisiae. PLoS One 10:e0130840. doi: 10.1371/journal.pone.0130840
Su, W. M., Han, G. S., and Carman, G. M. (2014). Yeast Nem1-Spo7 protein phosphatase activity on Pah1 phosphatidate phosphatase is specific for the Pho85-Pho80 protein kinase phosphorylation sites. J. Biol. Chem. 289, 34699–34708. doi: 10.1074/jbc.M114.614883
Sun, C. C., Li, G. J., Li, H. B., Lyu, Y. B., Yu, S. Q., and Zhou, J. W. (2021). Enhancing flavan-3-ol biosynthesis in Saccharomyces cerevisiae. J. Agric. Food Chem. 69, 12763–12772. doi: 10.1021/acs.jafc.1c04489
Tang, D. D., Liu, W., Huang, L., Cheng, L. M., and Xu, Z. N. (2020). Efficient biotransformation of vitamin D-3 to 25-hydroxyvitamin D-3 by a newly isolated Bacillus cereus strain. Appl. Microbiol. Biotechnol. 104, 765–774. doi: 10.1007/s00253-019-10250-1
Veen, M., Stahl, U., and Lang, C. (2003). Combined overexpression of genes of the ergosterol biosynthetic pathway leads to accumulation of sterols in Saccharomyces cerevisiae. FEMS Yeast Res. 4, 87–95. doi: 10.1016/s1567-1356(03)00126-0
Wang, S. Q., Wang, T., Liu, J. F., Deng, L., and Wang, F. (2018). Overexpression of Ecm22 improves ergosterol biosynthesis in Saccharomyces cerevisiae. Lett. Appl. Microbiol. 67, 484–490. doi: 10.1111/lam.13061
Warnke, M., Jung, T., Dermer, J., Hipp, K., Jehmlich, N., von Bergen, M., et al. (2016). 25-Hydroxyvitamin D-3 synthesis by enzymatic steroid side-chain hydroxylation with water. Angew. Chem. Int. Ed. 55, 1881–1884. doi: 10.1002/anie.201510331
Xia, L., Lv, Y., Liu, S., Yu, S., Zeng, W., and Zhou, J. (2022). Enhancing squalene production in Saccharomyces cerevisiae by metabolic engineering and random mutagenesis. Front. Chem. Eng. 3:790261. doi: 10.3389/fceng.2021.790261
Xu, S. H., and Li, Y. R. (2020). Yeast as a promising heterologous host for steroid bioproduction. J. Ind. Microbiol. Biotechnol. 47, 829–843. doi: 10.1007/s10295-020-02291-7
Keywords: 7-dehydrocholesterol, “push and pull”, multi-copy site integration, metabolic engineering, Saccharomyces cerevisiae
Citation: Wei W, Gao S, Yi Q, Liu A, Yu S and Zhou J (2022) Reengineering of 7-dehydrocholesterol biosynthesis in Saccharomyces cerevisiae using combined pathway and organelle strategies. Front. Microbiol. 13:978074. doi: 10.3389/fmicb.2022.978074
Received: 25 June 2022; Accepted: 21 July 2022;
Published: 09 August 2022.
Edited by:
Jixun Zhan, Utah State University, United StatesCopyright © 2022 Wei, Gao, Yi, Liu, Yu and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingwen Zhou, zhoujw1982@jiangnan.edu.cn
 Wenqian Wei1,2
Wenqian Wei1,2 Song Gao
Song Gao Shiqin Yu
Shiqin Yu Jingwen Zhou
Jingwen Zhou