- 1Department of Pharmacology and Pharmaceutical Sciences, School of Pharmacy, University of Southern California, Los Angeles, CA, United States
- 2Biotechnology and Planetary Protection Group, Jet Propulsion Laboratory, California Institute of Technology, Pasadena, CA, United States
- 3Department of Immunology and Theranostics, Beckman Research Institute of City of Hope, Duarte, CA, United States
- 4Department of Microbiology and Plant Pathology, Institute for Integrative Genome Biology, University of California, Riverside, Riverside, CA, United States
- 5Ecology Department, Lawrence Berkeley National Laboratory, Berkeley, CA, United States
- 6Department of Chemistry, Dornsife College of Letters, Arts, and Sciences, University of Southern California, Los Angeles, CA, United States
Due to immense phenotypic plasticity and adaptability, Aspergillus niger is a cosmopolitan fungus that thrives in versatile environments, including the International Space Station (ISS). This is the first report of genomic, proteomic, and metabolomic alterations observed in A. niger strain JSC-093350089 grown in a controlled experiment aboard the ISS. Whole-genome sequencing (WGS) revealed that ISS conditions, including microgravity and enhanced irradiation, triggered non-synonymous point mutations in specific regions, chromosomes VIII and XII of the JSC-093350089 genome when compared to the ground-grown control. Proteome analysis showed altered abundance of proteins involved in carbohydrate metabolism, stress response, and cellular amino acid and protein catabolic processes following growth aboard the ISS. Metabolome analysis further confirmed that space conditions altered molecular suite of ISS-grown A. niger JSC-093350089. After regrowing both strains on Earth, production of antioxidant—Pyranonigrin A was significantly induced in the ISS-flown, but not the ground control strain. In summary, the microgravity and enhanced irradiation triggered unique molecular responses in the A. niger JSC-093350089 suggesting adaptive responses.
Introduction
The International Space Station (ISS) is a research facility orbiting at an approximate altitude of 250 miles that is utilized to study physiological, psychological, and immunological responses of humans living in isolation (Mehta et al., 2004; Crucian et al., 2013; Cucinotta, 2014; Benjamin et al., 2016; Ombergen et al., 2017). However, the distinct ISS environment, which includes microgravity and enhanced irradiation, affects the metabolism of all living organisms aboard the ISS including humans. There is a growing body of research that focuses on molecular characterization of animal (Ijiri, 2003; Tavella et al., 2012), plant (Link et al., 2003; Driss-Ecole et al., 2008; Kittang et al., 2014), and microbial (Rabbow et al., 2003; Benoit et al., 2006) responses to the conditions encountered in the ISS. Among the most studied microorganisms are various species of bacteria (Klaus et al., 1997; Weng et al., 1998; Nickerson et al., 2000; Vaishampayan et al., 2012), yeast (Takahashi et al., 2001; Purevdorj-Gage et al., 2006; Altenburg et al., 2008; Liu et al., 2008; Crabbé et al., 2013), and black fungi (Onofri et al., 2008, 2012, 2015). However, there are few reported studies that characterize the molecular responses of filamentous fungi (Romsdahl et al., 2018, 2019, 2020; Blachowicz et al., 2019b).
Filamentous fungi are producers of a myriad of bioactive compounds or secondary metabolites (SMs). These SMs often confer environmental advantage, which facilitate survival in hostile niches despite not being directly essential for survival (Keller et al., 2005; Fox and Howlett, 2008; Brakhage and Schroeckh, 2011; Rohlfs and Churchill, 2011; Brakhage, 2013). SMs span from potent bioactive molecules used in the drug discovery processes (Borel et al., 1995; Elander, 2003; Mulder et al., 2015) or other branches of the industry (Vandenberghe et al., 2000; Rodríguez Couto and Toca Herrera, 2006; Piscitelli et al., 2010; Dhillon et al., 2011) to health hazardous toxins (Barnes, 1970; Shephard, 2008; Hof and Kupfahl, 2009; Eaton and Groopman, 2013). Altered production of various SMs is one potential mechanism of fungal adaptation to extreme environments. For example, increased production of melanin, a pigment with UV protective properties, was observed in fungi isolated from Chernobyl nuclear power plant (Dadachova et al., 2008) and “Evolution Canyon” (Singaravelan et al., 2008). One such highly melanized fungal species is Aspergillus niger.
Industrially important A. niger (Schuster et al., 2002) has been isolated from various ecological niches, including decaying leaves (Nikolcheva et al., 2003), common households (Adams et al., 2013; Barberán et al., 2015), and the ISS (Checinska et al., 2015). The A. niger strain JSC-093350089 isolated from the surface of the US compartment of the ISS was previously characterized using multi-omics techniques. Performed analyses revealed genetic variance typical for the A. niger clade, increased abundance of proteins involved in starvation response, oxidative stress, and cell wall modulation (Romsdahl et al., 2018), and alteration in SM production levels when compared to well-studied A. niger ATCC 1015 strain (Romsdahl et al., 2019). However, definite ascribing of observed molecular alterations to the ISS environment was not possible, since the strains were not grown in microgravity using a controlled experiment with ground counterparts. Nevertheless, in-depth characterization of ISS-isolated JSC-093350089 A. niger provided insight into potential space-induced molecular phenotypes.
This study is the first report of the multi-omics characterization of A. niger JSC-093350089 grown aboard the ISS and compared to ground controls. To study the impact of the enhanced irradiation and microgravity on JSC-093350089, the strain was transported to and grown aboard the ISS. Upon return to Earth, ISS-grown samples, along with ground controls, were immediately processed for metabolomic, proteomic, and genomic analyses with the aim of obtaining important insights into the adaptive responses of A. niger to space conditions. In addition, ISS-grown samples were regrown on Earth to identify any conserved molecular alterations.
Materials and Methods
Isolation and Identification of Aspergillus niger
Procedures to isolate and identify A. niger collected from the ISS were described previously (Romsdahl et al., 2018). In brief, sterile swabs soaked in saline solution were used to sample the ISS surface and transported to Earth. Particles retrieved from the swab were spread into potato dextrose agar (PDA) plates and any growing colonies were purified, collected, and further analyzed. One of the collected isolates was identified as A. niger via ITS region sequencing, which was subsequently confirmed via whole-genome sequencing (WGS).
Growth Conditions
JSC-093350089 was cultivated on glucose minimal medium (GMM) agar plates (6 g/l NaNO3, 0.52 g/l KCl, 0.52 g/l MgSO4·7H2O, 1.52 g/l KH2PO4, 10 g/l D-glucose, and 15 g/l agar supplemented with 1 ml/l of Hutner’s trace elements) covered with a cellophane membrane. Each of 10 prepared Petri plates (D = 10 cm) was inoculated with 1 × 107 conidia/plate. Subsequently, plates were sealed with 3 M™ Micropore™ Surgical Tape (VWR International, Radnor, PA, United States) and placed in four Biological Research in Canister (BRIC) systems (three and two plates/BRIC). BRICs were divided into two groups, which were exact mimics, and transferred to 4°C. The exact timeline of the experiment is presented in Supplementary Figure 1. The whole experiment lasted 42 days from preparing the payload by a science team prior to launch till the handout of the ISS-grown samples back to the science team after the flight. The first group of BRICs was sent to the ISS and continuously kept at 4°C (1–20 days) prior to being transferred to ambient temperature for the active growth phase ~22°C for 12 days (21–32 days). After that time BRICs were stored at 4°C before returning to Earth (33–42 days). Upon arrival to Earth, BRICs were turned over to the science team for the downstream analyses, which commenced immediately. The second group of BRICs, treated as controls, was kept on Earth at Kennedy Space Center (KSC) and mimicked the ISS experiment timeline with roughly 2 h of delay. BRICs containing control samples were shipped from KSC to research team along with the ISS-grown samples. Lastly, for an additional secondary metabolite analysis, 1 × 107 conidia/plate of the ISS- and ground-grown JSC-093350089 were grown on GMM medium at 28°C for 5 days.
Genomic DNA Extraction and Whole-Genome Sequencing
Mycelia and conidia were collected from ground-and ISS-grown JSC-09335008 GMM agar plates. DNA was extracted using the Power Soil DNA Isolation Kit (Mo Bio Laboratories, Carlsbad, California, United States) following the manufacturers protocol. Extracted DNA was checked for quality using Qubit 2.0 Fluorometer and used for paired-end library preparation with TruSeq Nano DNA Library Preparation Kit (Illumina, San Diego, California, United States) followed by WGS at the Duke Center for Genomic and Computational Biology. Samples were sequenced using a HiSeq 4,000 Illumina Sequencer generating 101 base long reads.
Genetic Variation Identification
Illumina sequence reads were trimmed using Trimmomatic v 0.36 (Bolger et al., 2014) and checked for quality using FastQC v 0.11.5 (Andrews, 2010). The genome and annotation files for A. niger CBS 513.88, (Pel et al., 2007) were downloaded from the FungiDB web portal (Stajich et al., 2012). Reads were mapped to CBS 513.88 the reference genome using the Burrows-Wheeler Aligner (BWA) software package v 0.7.12 (Li and Durbin, 2009) and further processed with SAMtools v 1.6 to generate sorted BAM files (Li et al., 2009). SNPs and INDELs were identified using GATK v 3.7 (DePristo et al., 2011). Duplicates were marked using Picard-tools MarkDuplicates1 to remove PCR artifacts. Sequence reads containing putative INDELs were realigned using GATK’s IndelRealigner to generate an updated BAM file. Variants within each sample were called using GATK’s Haplotype Caller. GATK’s VariantFiltration was used to filter each VCF file based on stringent cutoffs for quality and coverage {SNPs: QD < 2.0, MQ < 40.0, QUAL < 100, FS > 60.0, MQRankSum < −12.5, SOR > 4.0, ReadPosRankSum < −8.0; Indels: QD < 2.0, FS > 200.0, MQRankSum < −12.5, SOR > 4, InbreedingCoeff < −0.8, ReadPosRankSum < −20.0}, so that only high-quality variants remained.
Protein Extraction
Mycelia and conidia from GMM agar plates were collected and stored at –80°C prior to protein extraction. Proteins were extracted with the lysis buffer consisting of 100 mm triethylammonium bicarbonate (TEAB) with 1:100 Halt Protease Inhibitor Cocktail (Thermo Scientific, Rockford, IL) and 200 μg/ml phenylmethylsulfonyl fluoride (Sigma-Aldrich, St. Louis, MO, United States). Mycelia and conidia were homogenized by bead beating using Precellys 24 homogenizer (Bertin, Rockville, MD). The lysed fungal material was centrifuged at 17,000 g for 15 min and the protein concentration in the supernatants was measured by the Bradford assay (Bio-Rad Laboratories, Inc. Hercules, CA, United States).
Tandem Mass Tag (TMT) Labeling
A 100 μg proteins from each sample were precipitated in 20% trichloroacetic acid (TCA) at 4°C. Protein pellets were washed with ice-cold acetone and re-suspended in 25 μl TEAB (100 mM) and 25 μl 2,2,2-trifluoroethanol (TFE). Proteins were reduced with 1 μl of tris(2-carboxyethyl)phosphine (TCEP, 500 mM), alkylated with iodoacetamide (IAA, 30 mM), and digested with 2.5 μg/sample of trypsin/lysC (Promega, Madison, WI, United States) overnight at 37°C. The digested peptides were quantified using the Pierce Quantitative Colorimetric Peptide Assay (Thermo Scientific, Waltham, MA, United States). 40 μg of peptides from each specific sample was labeled with the Thermo Scientific TMTsixplex Isobaric Mass Tagging Kit (JSC-E1 (ground 1) with TMT6-128, JSC-E2 (ground 2) with TMT6-130, JSC-S1 (ISS 1) with TMT6-129, JSC-S2 (ISS 2) with TMT6-131) according to the manufacturer’s protocol. All labeled-peptide mixtures were combined into a single tube, mixed, and fractionated using the Thermo Scientific Pierce High pH Reversed-Phase Peptide Fractionation Kit. While this kit usually uses eight fractions with step elution of up to 50% acetonitrile, ninth fraction was added eluting at 100% acetonitrile. Nine fractionated samples were dried using a SpeedVac concentrator and re-suspended in 1% (v/v) formic acid prior to liquid chromatography with tandem mass spectrometry (LC-MS/MS) analysis.
LC–MS/MS Analysis
The samples were analyzed on an Orbitrap Fusion Tribrid mass spectrometer with an EASY-nLC 1,000 Liquid Chromatograph, a 75 μm × 2 cm Acclaim PepMap100 C18 trapping column, and a 75 μm × 25 cm PepMap RSLC C18 analytical column, and an Easy-Spray ion source (Thermo Scientific, Waltham, MA, United States). The peptides were eluted at 45°C with a flow rate of 300 nl/min over a 110 min gradient, from 3 to 30% solvent B (100 min), 30–50% solvent B (3 min), 50–90% solvent B (2 min), and 90% solvent B (2 min). The solvent A was 0.1% formic acid in water and the solvent B was 0.1% formic acid in acetonitrile.
The full MS survey scan (m/z 400–1,500) was acquired at a resolution of 120,000 and an automatic gain control (AGC) target of 2 × 105 in the Orbitrap with the 50 ms maximum injection time for MS scans. Monoisotopic precursor ions were selected with charge states 2–7, a ± 10 ppm mass window, and 70 s dynamic exclusion. The MS2 scan (m/z 400–2000) was performed using the linear ion trap with the 35% collision-induced dissociation (CID) energy. The ion trap scan rate was set to “rapid,” with an AGC target of 4 × 103, and a 150 ms maximum injection time. Ten fragment ions from each MS2 experiment were subsequently selected for an MS3 experiment. The MS3 scan (m/z 100–500) was performed to generate the TMT reporter ions in the linear ion trap using higher-energy collisional dissociation (HCD) at a 55% collision energy, a rapid scan rate and an AGC target of 5 × 103, and a maximum injection time of 250 ms.
Proteome Data Analysis
All MS data (MS1, MS2, and MS3) were searched using the Proteome Discoverer (version 2.2.0.388, Thermo Scientific) with the Sequest-HT searching engines against an Aspergillus niger CBS 513.88 database containing 10,549 sequences (NCBI). The searches were performed with the following parameters: 2 maximum missed cleavage sites, 6 minimum peptide length, 5 ppm tolerance for precursor ion masses, and 0.6 Dalton tolerance for fragment ion masses. The static modification settings included carbamidomethyl of cysteine residues, and dynamic modifications included oxidation of methionine, TMT6plex modification of lysine ε-amino groups and peptide N-termini, and acetyl modification of protein N-terminus. A false discovery rate (FDR) of 1% was set using a target-decoy database search. The reporter ions integration tolerance was 0.5 Da and the co-isolation threshold was 75%. The average signal-to-noise threshold of all reporter peaks was bigger than 10. The total intensity of a reporter ion for a protein was calculated based on the sum of all detected reporter ions of associated peptides from that protein. The ratios between an abundance of a reporter and the average abundance of all reporters were used to estimate the abundance ratio of each protein.
For statistical analysis, the sum of reporter ion intensities for each protein was Log2 transformed and the biological duplicate and technical triplicate measurement for each protein was averaged. Only the proteins that were identified and quantified in both biological and technical replicates were used for the analysis. Student t-test was performed to identify proteins with changed abundance. Proteins with value of p <0.05 were further evaluated for increased and decreased abundance using a cut-off value of ≥ ± 1 fold (Log2) change.
Secondary Metabolite Extraction and Analysis
Upon return to Earth single plates from both ground- and ISS-kept BRICs were used to collect agar plugs in triplicate for secondary metabolite (SM) extraction. Plugs were extracted with 3 ml of methanol and 1:1 methanol/dichloromethane each followed by 1 h sonication. Crude extract was evaporated in vacuo to yield a residue that was then suspended in 1 ml of 20% dimethyl sulfoxide/methanol and 10 μl was examined by high-performance liquid chromatography-photodiode array detection-mass spectrometry (HPLC-DAD-MS) analysis. HPLC–MS was carried out using ThermoFinnigan LCQ Advantage ion trap mass spectrometer with an RP C18 column (Alltech Prevail C18 3 mm 2.1 × 100 mm) at a flow rate 125 μl/min. The solvent gradient for LC–MS was 95% acetonitrile/H2O (solvent B) in 5% acetonitrile/H2O (solvent A) both containing 0.05% formic acid, as follows: 0% solvent B from 0 min to 5 min, 0 to 100% solvent B from 5 min to 35 min, 100% solvent B from 35 min to 40 min, 100 to 0% solvent B from 40 min to 45 min, and re-equilibration with 0% solvent B from 45 min to 50 min. The SM profiles of ISS- and ground-grown JSC-093350089, which were regrown at 28°C were obtained following the procedure described above.
SM Statistical Analysis
To compare the yields of produced SMs in ISS-grown, ground-grown, and regrown samples, the area under the electrospray ionization curve (ESI) was integrated for each compound. SM data collected from three independent biological replicates of ISS- and ground-grown, and regrown JSC-093350089 were used for testing statistical significance of production yields of identified SMs by Welch’s corrected t-test. The data are presented as column charts with corresponding error bars. Data analysis was conducted using GraphPad Prism version 7.
Results
Genome Variation in the ISS-Grown JSC-093350089 Aspergillus niger
The genomes of ISS-flown and ground-grown JSC-093350089 were compared to identify occurring genetic variations. Obtained reads were aligned to the CBS 513.88 reference genome and single-nucleotide polymorphisms (SNPs) present in the ground control were filtered. This revealed presence of 375 SNPs and 620 INDELs that occurred because of the exposure to conditions aboard the ISS (Table 1). All identified genetic variations are summarized in Supplementary Tables 1 and 2, presenting SNPs and INDELs, respectively. Distribution of non-synonymous point mutations among genes is presented in Table 2. Interestingly, about 80% of these mutations occurred in chromosome VIII and 13% occurred in chromosome XII, while the remaining 7% were distributed among other chromosomes (Supplementary Figure 2A). The majority of missense point mutations (75%) were observed within genes of unknown function. However, several characterized genes containing missense SNPs have DNA-binding activity (An06g01180, An08g11890, and An12g00840), DNA polymerase activity (An08g11520), protein kinase and transferase activity (An08g12110), phospholipase activity (An08g12250), and chromosome anchoring RacA protein binding activity (An12g06420; Table 2). Additional, mutations like one stop lost, one start gained and one 5 prime untranslated region (UTR) mutation were observed. Most of the observed SNPs (~55%) and INDELs (71%) were located in intergenic regions (Table 1). Interestingly, unlike SNPs, INDELs were distributed throughout all chromosomes. However, similarly to SNPs, the highest number of INDELS was found in chromosomes VIII and XII. Among observed INDELS 109 caused frameshift, 14 lead to disruptive inframe deletion, and a few caused start lost and stop gained (Supplementary Table 2; Supplementary Figure 2B).
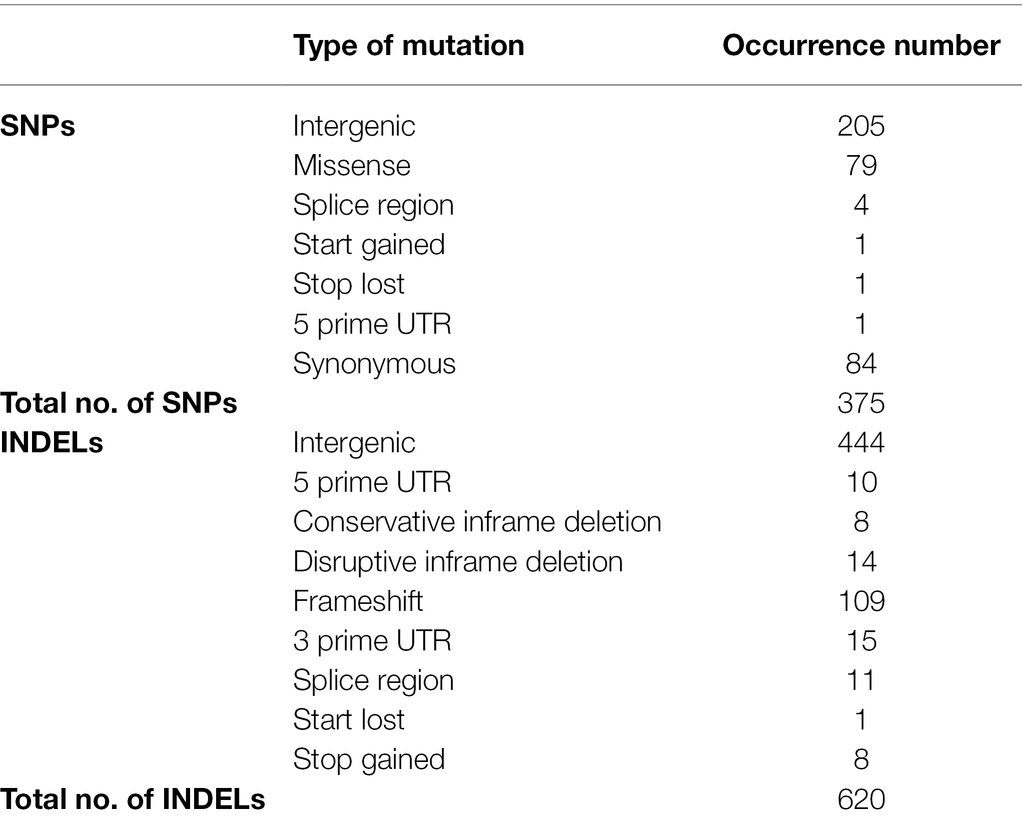
Table 1. Summary of genetic variations observed in ISS-grown JSC-093350089 when compared to ground control.
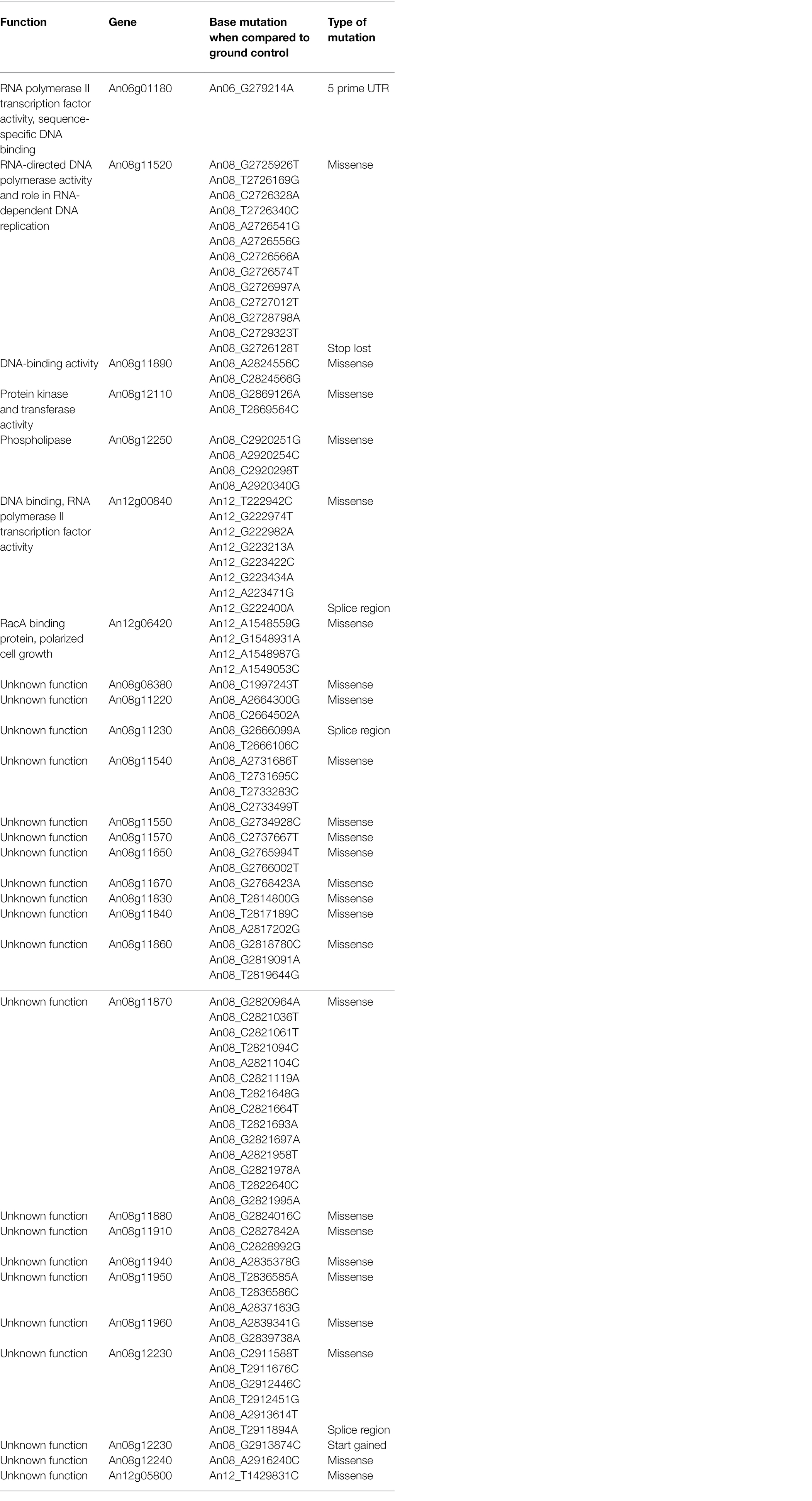
Table 2. Single-nucleotide polymorphisms (SNPs) in ISS-grown JSC-093350089 when compared to ground control.
Proteomic Characterization of ISS-Grown JSC-093350089 Aspergillus niger
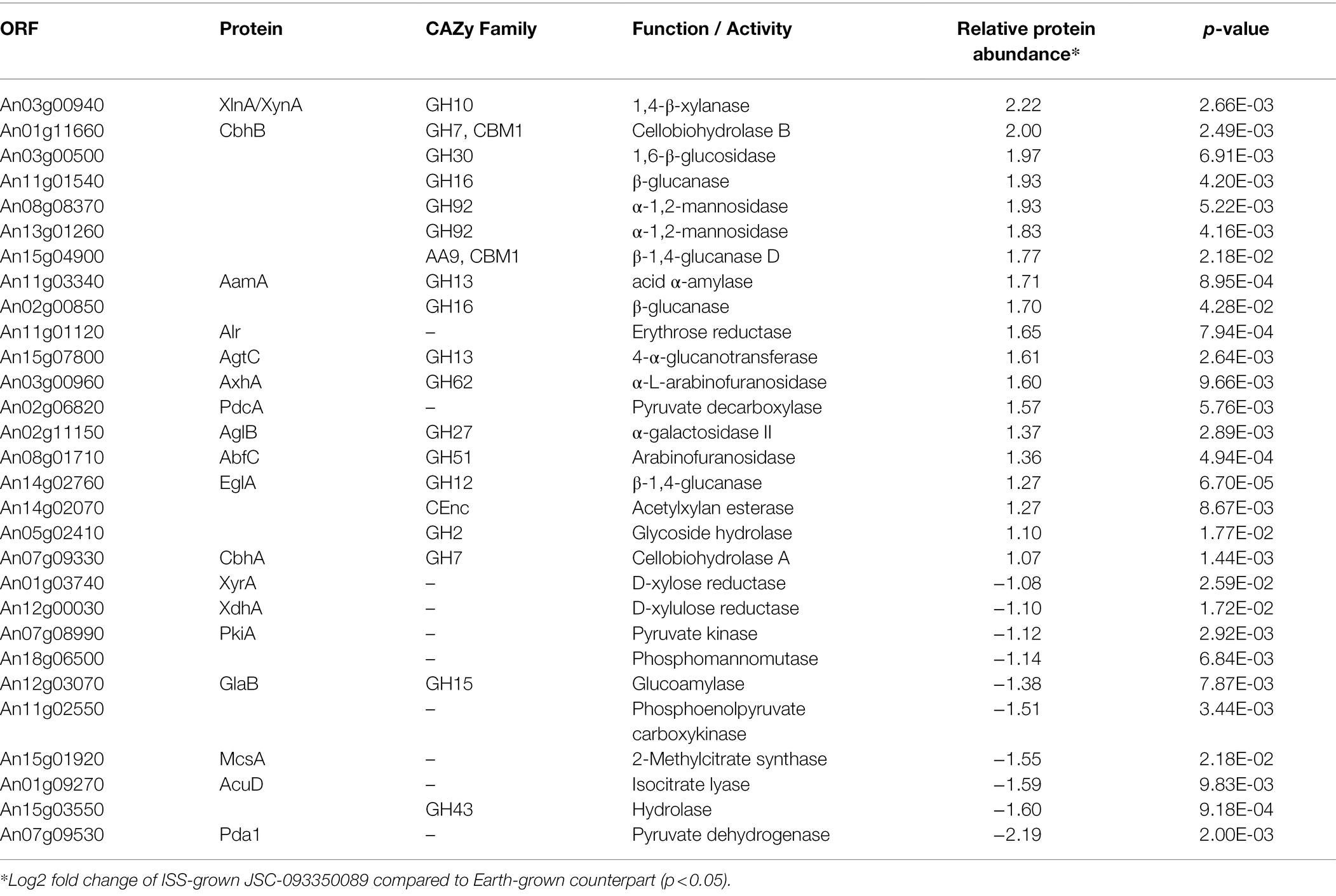
Differentially expressed proteins in ISS-grown JSC-093350089 strain were investigated following the extraction of total protein from two biological replicates of ISS-grown and ground control counterpart strains. Due to the low yields of extracted proteins, biological replicates were combined and divided into two parts that were then TMT labeled and subtracted to analysis via LC–MS/MS followed by spectrum/sequence matching using A. niger CBS 513.88 protein database (NCBI). Protein abundance ratios in ISS-grown JSC-093350089 were normalized to Earth-grown counterparts, which enabled identification of 70 up- and 142 downregulated proteins (fold change (FC) > ∣2∣, p < 0.05) in response to space conditions (Supplementary Tables 3 and 4, respectively). AspGD Gene Ontology (GO) Slim terms (Cerqueira et al., 2014) were used to study the distribution of differentially expressed proteins in ISS-grown JSC-093350089 when compared to ground controls (Figure 1). Among differentially expressed proteins, 29 were involved in carbohydrate metabolism, 18 in stress responses, 21 in amino acid metabolism, and 15 in protein catabolic processes (Figure 1). Protein GO term enrichment analysis was conducted using FungiDB (Stajich et al., 2012), which revealed that significantly over-represented upregulated biological processes included carbohydrate metabolic processes (28% of all upregulated proteins) and stress response (10%), whereas significantly over-represented downregulated processes included cellular amino acid metabolic processes (13%), proteasomal ubiquitin-independent (10%) and dependent processes (10%), and proteasomal protein catabolic processes (10%; Supplementary Table 5).
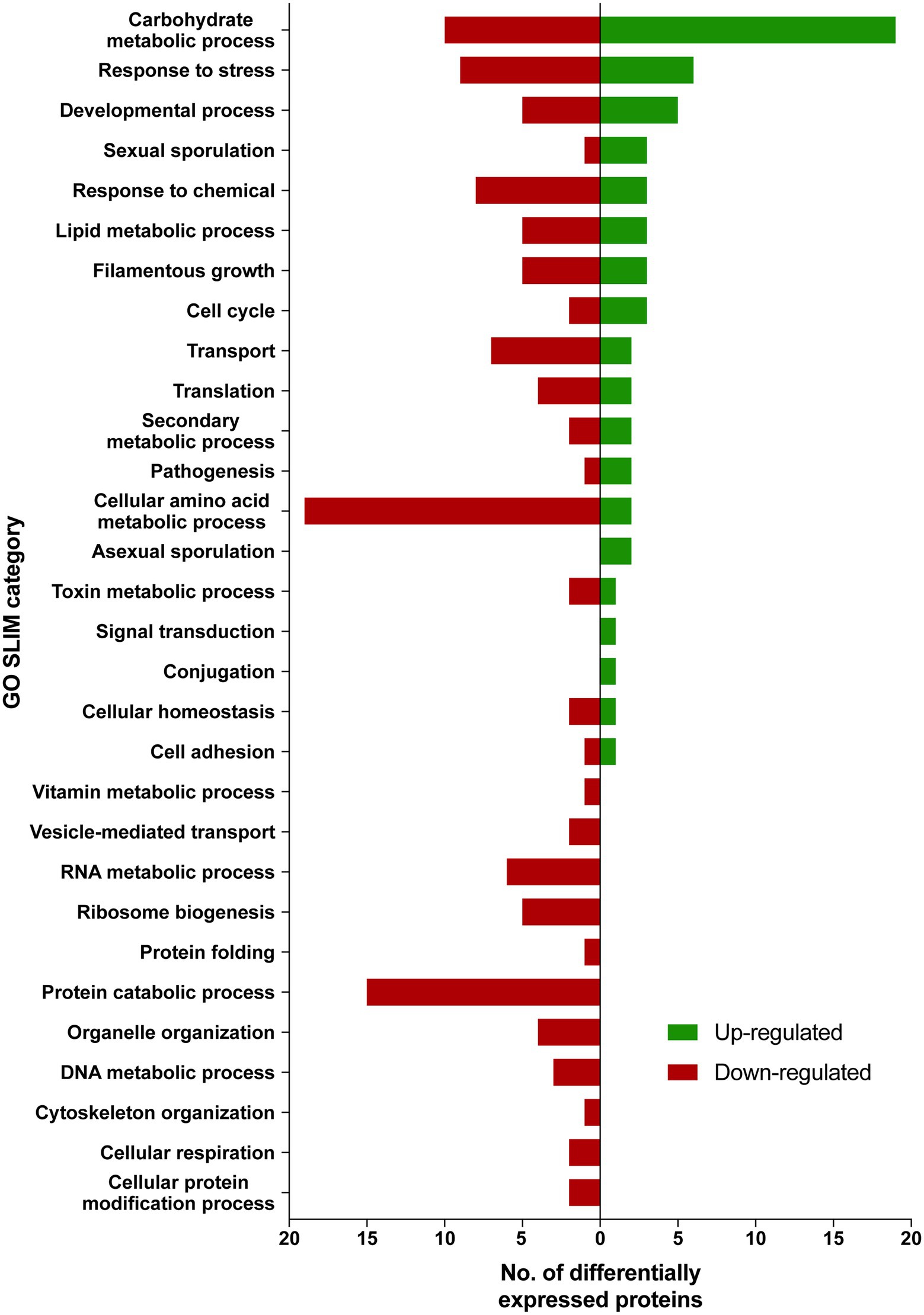
Figure 1. AspGD GO Slim terms of differentially expressed proteins in ISS-grown JSC-093350089. Differentially expressed proteins in FC > |2|, p < 0.05 were mapped to terms representing various biological processes using AspGD Gene Ontology (GO) Slim Mapper.
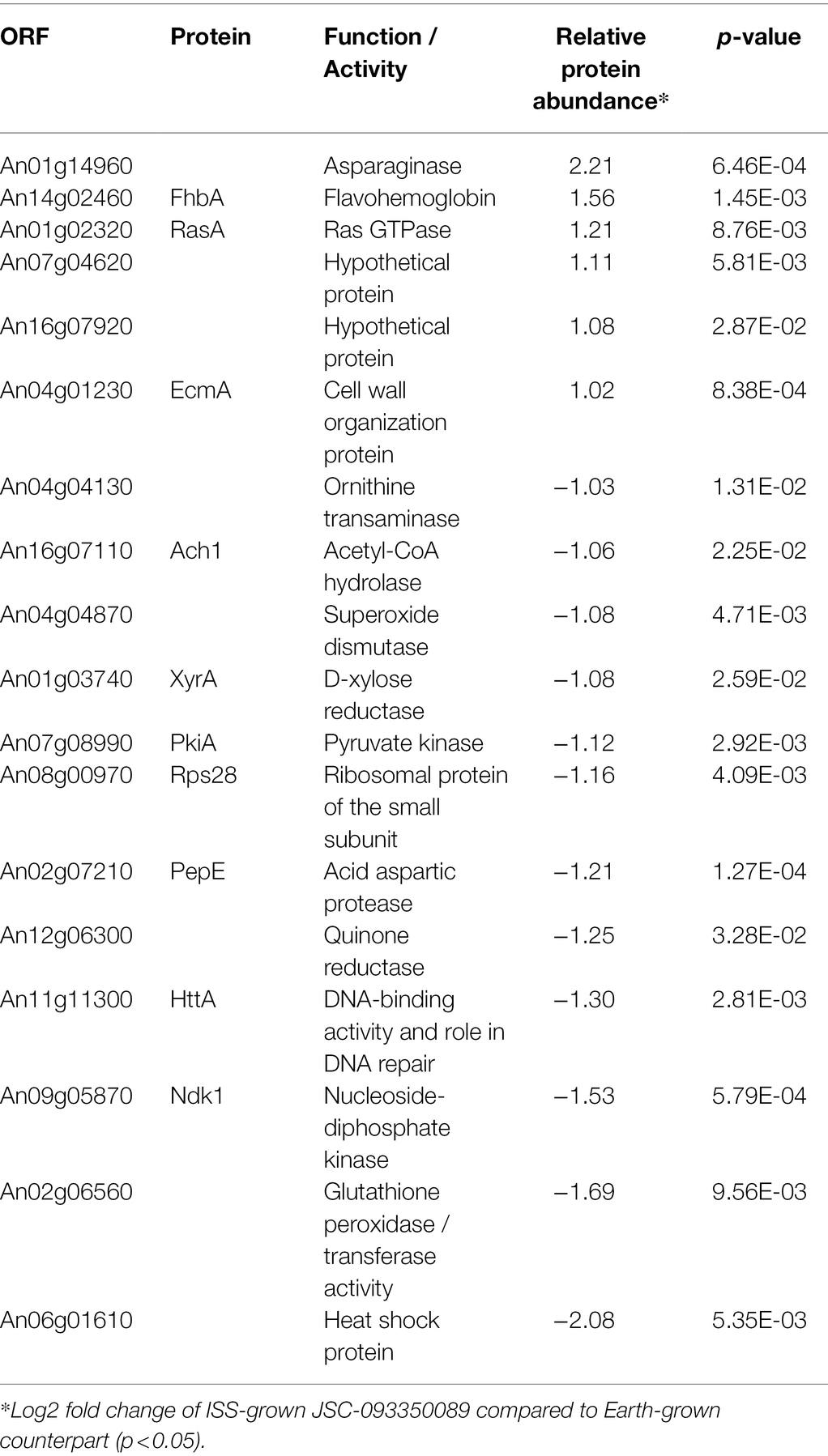
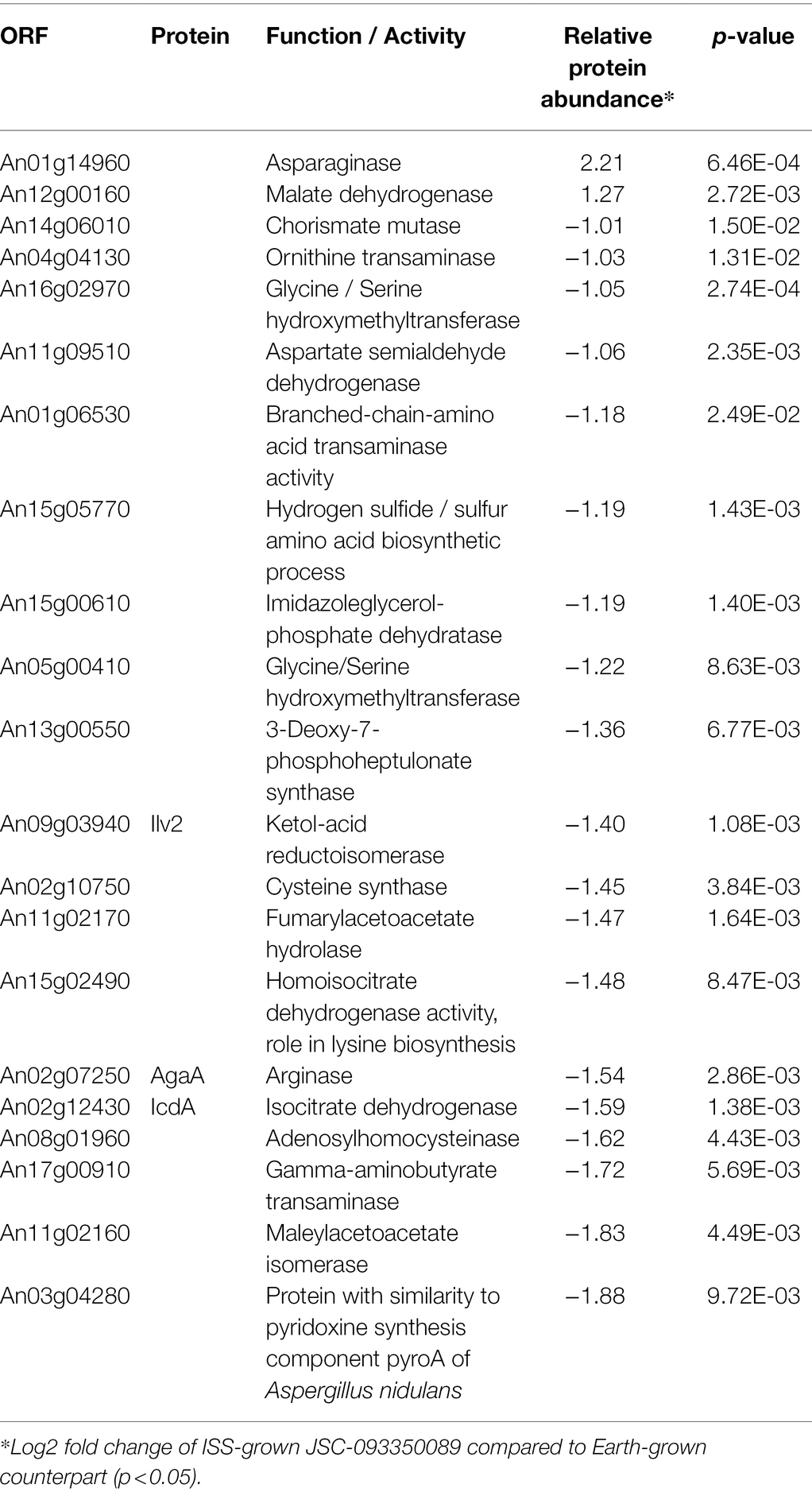
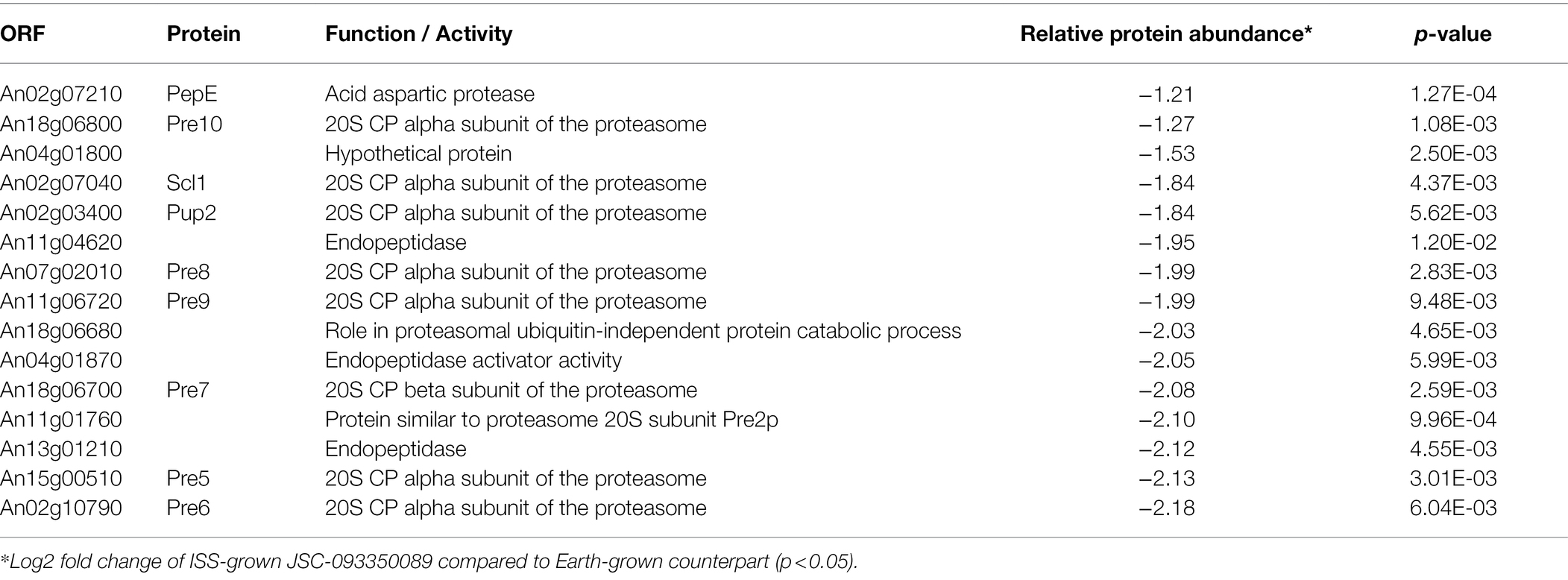
The majority of differentially expressed proteins in ISS-grown JSC-093350089 A. niger were involved in carbohydrate metabolism (Table 3). Interestingly, eight of these genes, including cellobiohydrolases A and B (An07g09330 and An01g11660), XlnA 1,4-β-xylanase (An03g00940), and D-xylose reductases XyrA and XdhA (An01g03740 and An12g00030) were regulated by XlnR. XlnR is a transcriptional regulator involved in degradation of polysaccharides, xylan, cellulose, and D-xylose (Hasper et al., 2000). β-glucanases An11g01540 and An02g00850, which are involved in carbon starvation response in A. niger (Nitsche et al., 2012), were at minimum 3-fold upregulated in ISS-grown strain. α-1,2-mannosidases An08g08370, An13g01260 were at least 3.5-fold upregulated, whereas pyruvate decarboxylase PdcA (An02g06820) was nearly 3-fold upregulated. Pyruvate kinase PkiA (An07g08990), pyruvate dehydrogenase Pda1 (An07g09530), and isocitrate lyase AcuD (An01g09270) were at least 2-fold less abundant in ISS-grown samples. Several proteins involved in the stress response were differentially expressed in ISS-grown JSC-093350089 (Table 4). Proteins exhibiting at least 2-fold upregulation included cell wall organization protein EcmA (An04g01230) and An16g07920, whose orthologs play a role in salt stress response. Downregulated stress response proteins included heat shock protein An06g01610, DNA-binding protein HttA (An11g11300), and quinone reductase An12g06300. Lastly, a variety of proteins involved in cellular amino acid processes (Table 5), and protein catabolic processes (Table 6) were downregulated.
Metabolomic Characterization of ISS-Grown JSC-093350089 Aspergillus niger
Alterations in SM production in response to ISS conditions were assessed by extracting organic compounds from three biological replicates of ISS- and ground-grown JSC-093350089. The extracts were analyzed using HPLC-DAD-MS. Compounds were identified based on mass, UV absorption, and retention time, which were in agreement with literature (Chiang et al., 2011; Figure 2). Assessment of SM production yields revealed slight decrease in production of bicoumanigrin A, fonsecinones B and C, aurasperones C and B, and kotanin in ISS-grown isolate. Production levels of pestalamide B, nigerazine B, and nigragillin exhibited a reduction of approximately 50% when compared to control strains.
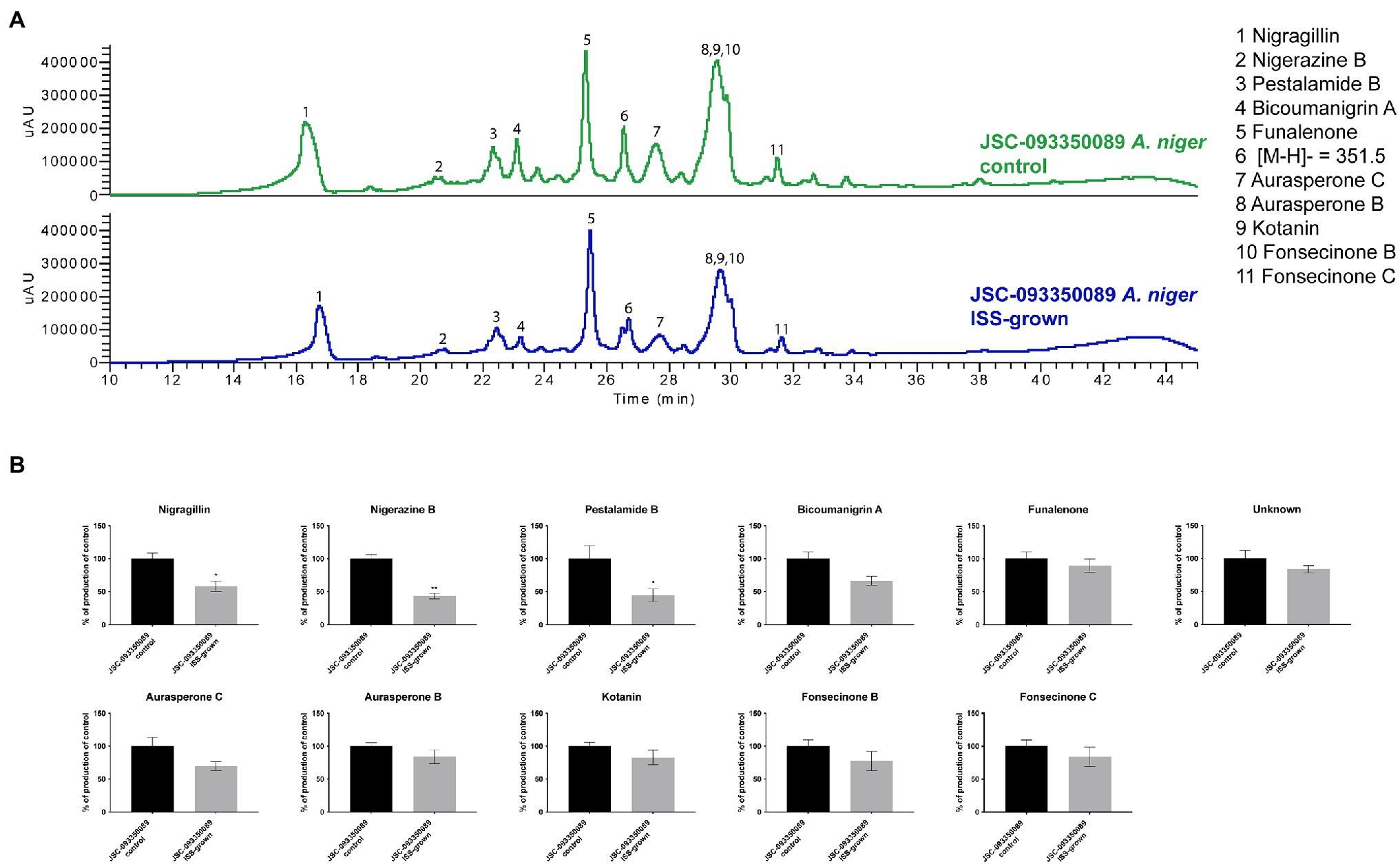
Figure 2. Secondary metabolite production of ISS-grown JSC-093350089 when compared to ground controls. (A) Secondary metabolite profiles of ISS- and ground-grown JSC-093350089 when grown on GMM. (B) Metabolite quantification showing the percent change for each metabolite in relation to ground-grown JSC-093350089; significance was determined using Welch’s t-test. “*” means and indicates statistical significance.
Both ISS- and ground-grown JSC-093350089 were regrown at 28°C and SMs production was evaluated. Due to the difference in growth temperature, obtained SM profiles of regrown isolates differed from those grown on the ISS (~22°C; Figure 3), as the trend in the production of SMs was the opposite of the one observed immediately after growth on the ISS. The majority of identified compounds showed slight increase in the production levels. Further, statistically significant increase in the production of the antioxidant pyranonigrin A (Miyake et al., 2007) and kotanin was observed when compared to ground controls.
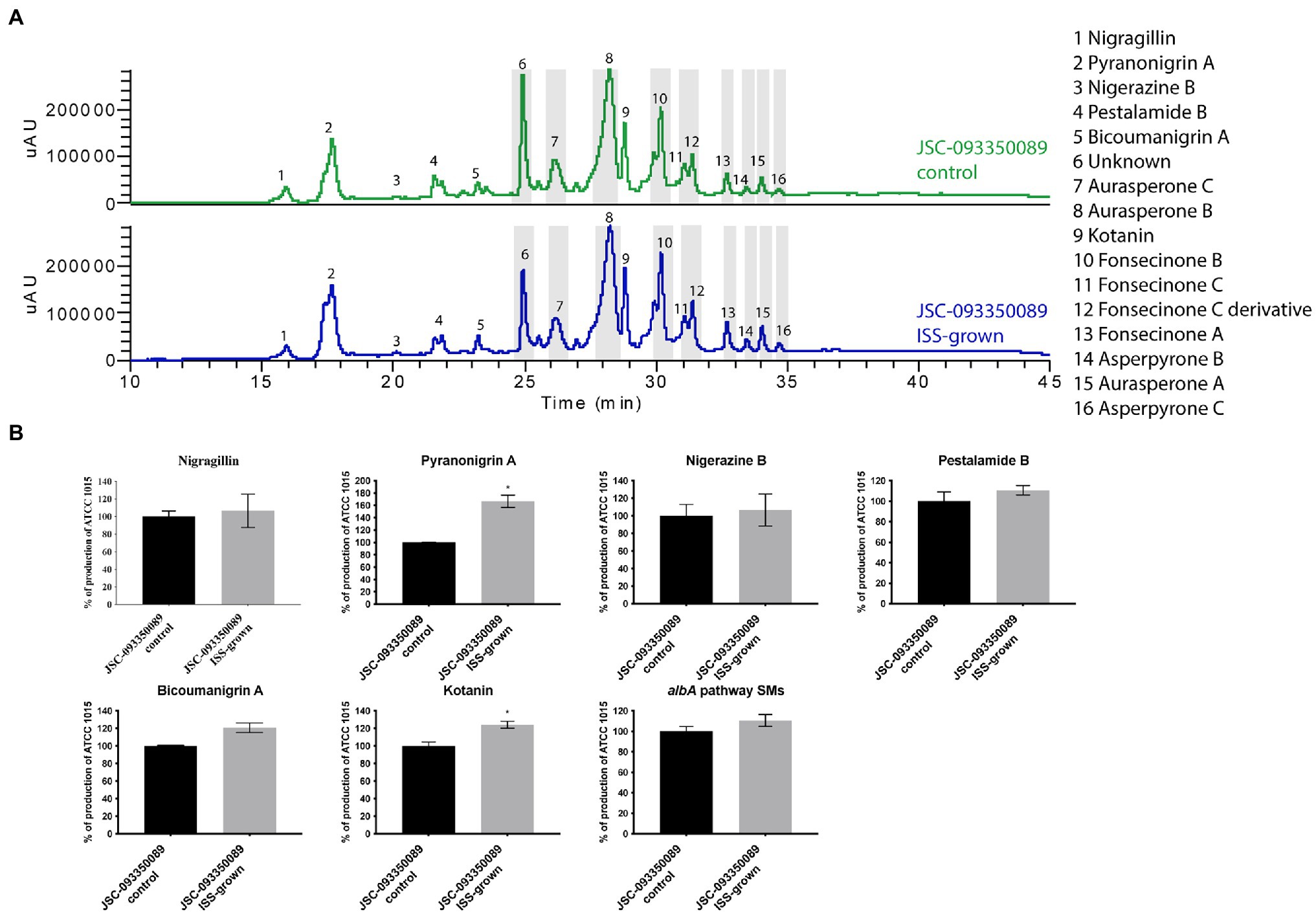
Figure 3. Secondary metabolite production of regrown ISS- and ground-grown JSC-093350089. (A) Secondary metabolite profiles of regrown ISS- and ground-grown JSC-093350089 when grown on GMM. (B) Metabolite quantification showing the percent change for each metabolite in relation to regrown ground-grown JSC-093350089; significance was determined using Welch’s t-test. “*” means and indicates statistical significance.
Discussion
It is critical to study molecular changes occurring in living organisms to understand the adaptation mechanisms allowing for surviving in extreme environments. One of such scientifically intriguing environments is the ISS, which is characterized by the presence of enhanced irradiation and microgravity. Due to its uniqueness, the ISS is under constant microbial monitoring, which allows for the isolation of wide array of microorganisms that often-become subjects of scientific investigations (Checinska et al., 2015; Knox et al., 2016; Checinska Sielaff et al., 2017; Venkateswaran et al., 2017; Bijlani et al., 2021). However, like in the case of the A. niger strain JSC-093350089 (Romsdahl et al., 2018, 2020), these investigations are more of a descriptive nature as definitive ascribing of observed molecular changes requires precisely controlled experiments. Therefore, to further investigate the differences in JSC-093350089 that were observed when compared to a “terrestrial” strain, the isolate was sent to the ISS in a planned experiment. Genomic, proteomic, and metabolomic alterations occurring in ISS-grown samples were analyzed following sample return and compared to ground-grown counterparts.
Genome analysis of ISS-grown JSC-093350089 revealed the introduction of SNPs and INDELs in response to space conditions. Interestingly, the majority of observed non-synonymous SNPs and INDELS were located within chromosomes VIII and XII, which suggests that only selected regions of the genome undergo positive selection to confer selective advantage while adapting to the space environment. This is in agreement with previous reports of space-induced genetic variations, as ISS-grown Aspergillus nidulans (Romsdahl et al., 2019) and spaceflight-grown Staphylococcus aureus (Guo et al., 2015) both exhibited genetic mutations that occurred in specific clustered regions of the genome. Although the functions of many genes containing non-synonymous SNPs were unknown, several of these genes possessed transposable element and DNA-binding activity. One such gene, An08g11520, was an analogue of transposon I factor and has RNA-directed DNA polymerase activity, which is consistent with genetic changes observed in transposable element genes in both A. nidulans (Romsdahl et al., 2019) and S. aureus (Guo et al., 2015). Alterations in transposable element genes likely influence their activity and lead to the introduction of variations within the genome in response to environmental stress (Capy et al., 2000; Muszewska et al., 2017). The results from this study further underscore the significant role of transposable elements in adaptation to the spacecraft environment. Future studies should investigate the functions of uncharacterized genes containing non-synonymous SNPs, as such knowledge may provide key information on how fungi adapt to space conditions. Noteworthy, when radioadapted strain of Exophiala dermatitidis and the non-radioadapted control strain were exposed to Polonium-210, a mostly transcriptomic rather than genomic response to radiation was observed in radioadapted strain. This suggests that strains previously exposed to irradiation respond to subsequent exposures in a unique way (Malo et al., 2021). Based on the observations reported for E. dermatitidis, it is plausible that A. niger JSC-093350089 strain response to the ISS environment was also unique, as it was previously isolated from the ISS and likely radioadapted itself. However, such assumption may not be confirmed in the current study, as there is no available not-radioadapted control for the A. niger JSC-093350089 strain. Future studies should be warranted to investigate whether A. niger JSC-093350089 strain’s response to the ISS environment changes with consecutive exposures to the ISS environment when compared to the original ISS isolate. Finally, it has been previously reported that in Aspergillus genome intragenic regions contain high-level developmental and metabolic transcriptional regulators (Noble and Andrianopoulos, 2013; Lim et al., 2021). Therefore, the high occurrence of SNPs and INDELS in the intergenic regions may contribute to developing the space environment-induced phenotype in A. niger JSC-093350089.
Investigation into the proteome of ISS- and ground-grown JSC-093350089 revealed that space conditions induce changes in protein expression when compared to ground controls. Observed, alterations in proteins involved in carbohydrate metabolism upon exposure to space conditions stay in agreement with previous studies of A. niger JSC-093350089 (Romsdahl et al., 2018), and other filamentous fungi, including Aspergillus fumigatus (Blachowicz et al., 2019a,b) and A. nidulans (Romsdahl et al., 2019). Similarly, a black fungus Knufia chersonesos exposed to low-shear simulated microgravity (LSSMG) also exhibited differential expression of proteins involved in carbohydrate metabolism (Tesei et al., 2021). On the other hand, a yeast, Candida albicans, exposed to spaceflight environment on the “SJ-10” satellite for 12 days showed decreased abundance of proteins involved in carbon metabolism, suggesting that yeast and filamentous fungi differ in adaptive responses to space environment (Wang et al., 2020). Additionally, ISS-grown JSC-093350089 exhibited upregulation of starvation-induced glycoside hydrolases, including AamA, and CbhB. This further confirms that changes in carbohydrate metabolism, may be a result of adaptation to scarce nutrient availability on ISS surfaces, due to the maintained cleaning regimen. Further, differential expression of XlnR-regulated proteins appears to be a key adaptive response to microgravity and enhanced irradiation, as both this and the previous study characterizing the space-induced molecular phenotype of JSC-093350089 (Romsdahl et al., 2018) revealed differential expression of various XlnR-regulated proteins. Activation of XlnR-regulated metabolic pathways leads to use of versatile carbon sources (Hasper et al., 2000), which may facilitate survival in hostile space environments. A few proteins upregulated in ISS-grown JSC-093350089 were involved in stress response, including the cell wall organization protein EcmA and flavohemoglobin, whose analogue in Aspergillus flavus has been reported to have a function correlated with hyphal growth phenotype (te Biesebeke et al., 2010). These observations further suggest that modulation of fungal growth and cell organization is an essential response to space conditions.
Metabolomic analysis of ISS-grown JSC-09335008 showed alterations in secondary metabolite production in response to the space environment. This observation was not surprising, as fungi often respond to versatile environmental conditions by altering the type and yield of produced SMs (Brakhage, 2013). Interestingly, ISS-grown JSC-09335008 showed decreased production of all SMs, which is the opposite production pattern observed during the initial characterization of the metabolome of JSC-09335008 when compared to a “terrestrial” strain (Romsdahl et al., 2020). This discrepancy may be related to the fact that metabolomic profile of ISS-isolated JSC-09335008 was compared to the well-studied ATCC 1015 strain, rather than a “proper” JSC-09335008 ground control. Further investigation of the space environment-induced SM profile of the JSC-09335008 strain should be conducted to confirm whether decreased production of pestalamide B, nigerazine B (alkaloid), and nigragillin (alkaloid) are important biological adaptations. Given that sending experiments to the ISS is not readily available, it will be critical to use more easily accessible microgravity simulators, like High Aspect Ratio Vessels—HARV, or random positioning machines—RPMs to gain more insights in the space-induced phenotype of A. niger JSC-09335008 strain.
Finally, to gain insight into observed differences in acquired SMs profiles between current and previous study (Romsdahl et al., 2020) further experiment was conducted. Both ISS- and ground-grown JSC-09335008 were regrown in the same conditions (28°C for 5 days) as used in the previous study, which resulted in observing similar trends in SM production. After regrowing at 28°C, the ISS-grown JSC-09335008 produced higher yields of all SMs when compared to the regrown ground control, including approximately 60% increased production of the antioxidant pyranonigrin A. Pyranonigrin A was previously proposed to have a radioprotective nature, as pyranonigrin A-deficient JSC-09335008 strain was more sensitive to UVC exposure than the wild type JSC-09335008 strain (Romsdahl et al., 2020). Interestingly, pyranonigrin A production was not detected in the ISS- and ground-grown samples following the experiment on the ISS where growth temperature was about at 22°C. Due to this temperature-dependent discrepancy in observed SM profiles, the most important question to address is whether pyranonigrin A truly provides A. niger protection while in space. Such protection could potentially have various biotechnological applications for use of pyranonigrin A, including within human space programs and cancer therapies. Therefore, studies confirming pyranonigrin A potential as a radioprotective agent should be warranted. Finally, future studies should examine production of pyranonigrin A under various temperatures aboard the ISS, as well as its protective nature within the space environment to definitively answer this question.
This study is the first report of the multi-omics response of A. niger to space conditions during a controlled experiment, which enhances our understanding of its space-induced phenotype. Such understanding may be translated to development of protective measures for both astronauts and the spacecraft during future manned space explorations, as A. niger is ubiquitous fungus present in many human-occupied closed habitats (Checinska et al., 2015; Blachowicz et al., 2017). Lastly, a thorough understanding of the space-induced secondary metabolomic alterations of industrially important A. niger may result in creating a potent producer of compounds of interest during space voyages.
Data Availability Statement
Raw WGS reads for JSC-093350089 ISS- and ground-grown are available in the NCBI SRA, under accession numbers SAMN25997338 and SAMN25997339 and BioProject accession number PRJNA807647. Proteomics data is accessible through Massive with the dataset identifier MSV000088986.
Author Contributions
AB drafted the manuscript, contributed to sample processing, and conducted data analysis and interpretation. JR contributed to sample processing and data interpretation. AC and MK conducted protein sample processing, LC–MS analyses, and data processing. SM and JS contributed to genome analysis. TT designed the study and drafted the manuscript. KV and CW conceptualized the project, coordinated the flight experiment, designed the study, interpreted the data, and drafted the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the Center for the Advancement of Science in Space (CASIS) and sponsored by the International Space Station U.S. National Laboratory under grant/agreement number UA-2015-207 awarded to KV that funded a portion of the fellowship for AB. The funders had no role in study design, data collection and interpretation, the writing of the manuscript, or the decision to submit the work for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor DT declared a past co-authorship with the authors AC, MK, JS, and KV.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Part of the research described in this publication was carried out at the Jet Propulsion Laboratory, California Institute of Technology, under a contract with NASA. We would like to thank astronaut Jeff Williams for handling the samples aboard the ISS and CASIS Team for coordinating this effort.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.893071/full#supplementary-material
Footnotes
References
Adams, R. I., Miletto, M., Taylor, J. W., and Bruns, T. D. (2013). Dispersal in microbes: fungi in indoor air are dominated by outdoor air and show dispersal limitation at short distances. ISME J. 7:1460. doi: 10.1038/ismej.2013.84
Altenburg, S. D., Nielsen-Preiss, S. M., and Hyman, L. E. (2008). Increased filamentous growth of Candida albicans in simulated microgravity. Genomics Proteomics Bioinformatics 6, 42–50. doi: 10.1016/S1672-0229(08)60019-4
Andrews, S. (2010). FastQC: a quality control tool for high throughput sequence data. Available at: https://www.scienceopen.com/document?vid=de674375-ab83-4595-afa9-4c8aa9e4e736 (Accessed October 31, 2018).
Barberán, A., Dunn, R. R., Reich, B. J., Pacifici, K., Laber, E. B., Menninger, H. L., et al. (2015). The ecology of microscopic life in household dust. Proc. R. Soc. B 282:20151139. doi: 10.1098/rspb.2015.1139
Barnes, J. M. (1970). Aflatoxin as a health Hazard*. J. Appl. Bacteriol. 33, 285–298. doi: 10.1111/j.1365-2672.1970.tb02200.x
Benjamin, C. L., Stowe, R. P., St. John, L., Sams, C. F., Mehta, S. K., Crucian, B. E., et al. (2016). Decreases in thymopoiesis of astronauts returning from space flight. JCI Insight. 1:e88787. doi: 10.1172/jci.insight.88787
Benoit, M. R., Li, W., Stodieck, L. S., Lam, K. S., Winther, C. L., Roane, T. M., et al. (2006). Microbial antibiotic production aboard the international Space Station. Appl. Microbiol. Biotechnol. 70, 403–411. doi: 10.1007/s00253-005-0098-3
Bijlani, S., Singh, N. K., Eedara, V. V. R., Podile, A. R., Mason, C. E., Wang, C. C. C., et al. (2021). Methylobacterium ajmalii sp. nov., isolated From the international Space Station. Front. Microbiol. 12:639396. doi: 10.3389/fmicb.2021.639396
Blachowicz, A., Chiang, A. J., Elsaesser, A., Kalkum, M., Ehrenfreund, P., Stajich, J. E., et al. (2019a). Proteomic and Metabolomic characteristics of Extremophilic Fungi Under simulated Mars conditions. Front. Microbiol. 10:1013. doi: 10.3389/fmicb.2019.01013
Blachowicz, A., Chiang, A. J., Romsdahl, J., Kalkum, M., Wang, C. C. C., and Venkateswaran, K. (2019b). Proteomic characterization of Aspergillus fumigatus isolated from air and surfaces of the international Space Station. Fungal Genet. Biol. 124, 39–46. doi: 10.1016/j.fgb.2019.01.001
Blachowicz, A., Mayer, T., Bashir, M., Pieber, T. R., De León, P., and Venkateswaran, K. (2017). Human presence impacts fungal diversity of inflated lunar/Mars analog habitat. Microbiome 5:62. doi: 10.1186/s40168-017-0280-8
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Borel, J. F., Kis, Z. L., and Beveridge, T. (1995). “The History of the Discovery and Development of Cyclosporine (Sandimmune®),” in The Search for Anti-Inflammatory Drugs: Case Histories from Concept to Clinic. eds. V. J. Merluzzi and J. Adams (Boston, MA: Birkhäuser Boston), 27–63.
Brakhage, A. A. (2013). Regulation of fungal secondary metabolism. Nat. Rev. Microbiol. 11, 21–32. doi: 10.1038/nrmicro2916
Brakhage, A. A., and Schroeckh, V. (2011). Fungal secondary metabolites – strategies to activate silent gene clusters. Fungal Genet. Biol. 48, 15–22. doi: 10.1016/j.fgb.2010.04.004
Capy, P., Gasperi, G., Biémont, C., and Bazin, C. (2000). Stress and transposable elements: co-evolution or useful parasites? Heredity 85, 101–106. doi: 10.1046/j.1365-2540.2000.00751.x
Cerqueira, G. C., Arnaud, M. B., Inglis, D. O., Skrzypek, M. S., Binkley, G., Simison, M., et al. (2014). The Aspergillus genome database: multispecies curation and incorporation of RNA-Seq data to improve structural gene annotations. Nucleic Acids Res. 42, D705–D710. doi: 10.1093/nar/gkt1029
Checinska, A., Probst, A. J., Vaishampayan, P., White, J. R., Kumar, D., Stepanov, V. G., et al. (2015). Microbiomes of the dust particles collected from the international Space Station and spacecraft assembly facilities. Microbiome 3:50. doi: 10.1186/s40168-015-0116-3
Checinska Sielaff, A., Kumar, R. M., Pal, D., Mayilraj, S., and Venkateswaran, K. (2017). Solibacillus kalamii sp. nov., isolated from a high-efficiency particulate arrestance filter system used in the international Space Station. Int. J. Syst. Evol. Microbiol. 67, 896–901. doi: 10.1099/ijsem.0.001706
Chiang, Y.-M., Meyer, K. M., Praseuth, M., Baker, S. E., Bruno, K. S., and Wang, C. C. C. (2011). Characterization of a polyketide synthase in Aspergillus Niger whose product is a precursor for both dihydroxynaphthalene (DHN) melanin and naphtho-γ-pyrone. Fungal Genet. Biol. 48, 430–437. doi: 10.1016/j.fgb.2010.12.001
Crabbé, A., Nielsen-Preiss, S. M., Woolley, C. M., Barrila, J., Buchanan, K., McCracken, J., et al. (2013). Spaceflight enhances cell aggregation and random budding in Candida albicans. PLoS One 8:e80677. doi: 10.1371/journal.pone.0080677
Crucian, B., Stowe, R., Mehta, S., Uchakin, P., Quiriarte, H., Pierson, D., et al. (2013). Immune system dysregulation occurs during short duration spaceflight on board the space shuttle. J. Clin. Immunol. 33, 456–465. doi: 10.1007/s10875-012-9824-7
Cucinotta, F. A. (2014). Space radiation risks for astronauts on multiple international Space Station missions. PLoS One 9:e96099. doi: 10.1371/journal.pone.0096099
Dadachova, E., Bryan, R. A., Howell, R. C., Schweitzer, A. D., Aisen, P., Nosanchuk, J. D., et al. (2008). The radioprotective properties of fungal melanin are a function of its chemical composition, stable radical presence and spatial arrangement. Pigment Cell Melanoma Res. 21, 192–199. doi: 10.1111/j.1755-148X.2007.00430.x
DePristo, M. A., Banks, E., Poplin, R. E., Garimella, K. V., Maguire, J. R., Hartl, C., et al. (2011). A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498. doi: 10.1038/ng.806
Dhillon, G. S., Brar, S. K., Verma, M., and Tyagi, R. D. (2011). Utilization of different agro-industrial wastes for sustainable bioproduction of citric acid by Aspergillus niger. Biochem. Eng. J. 54, 83–92. doi: 10.1016/j.bej.2011.02.002
Driss-Ecole, D., Legué, V., Carnero-Diaz, E., and Perbal, G. (2008). Gravisensitivity and automorphogenesis of lentil seedling roots grown on board the international Space Station. Physiol. Plant. 134, 191–201. doi: 10.1111/j.1399-3054.2008.01121.x
Eaton, D. L., and Groopman, J. D. (2013). The Toxicology of Aflatoxins: Human Health, Veterinary, and Agricultural Significance. United States: Elsevier.
Elander, R. P. (2003). Industrial production of β-lactam antibiotics. Appl. Microbiol. Biotechnol. 61, 385–392. doi: 10.1007/s00253-003-1274-y
Fox, E. M., and Howlett, B. J. (2008). Secondary metabolism: regulation and role in fungal biology. Curr. Opin. Microbiol. 11, 481–487. doi: 10.1016/j.mib.2008.10.007
Guo, J., Han, N., Zhang, Y., Wang, H., Zhang, X., Su, L., et al. (2015). Use of genome sequencing to assess nucleotide structure variation of Staphylococcus aureus strains cultured in spaceflight on Shenzhou-X, under simulated microgravity and on the ground. Microbiol. Res. 170, 61–68. doi: 10.1016/j.micres.2014.09.001
Hasper, A. A., Visser, J., and Graaff, L. H. D. (2000). The Aspergillus niger transcriptional activator XlnR, which is involved in the degradation of the polysaccharides xylan and cellulose, also regulates d-xylose reductase gene expression. Mol. Microbiol. 36, 193–200. doi: 10.1046/j.1365-2958.2000.01843.x
Hof, H., and Kupfahl, C. (2009). Gliotoxin in Aspergillus fumigatus: an example that mycotoxins are potential virulence factors. Mycotoxin Res. 25, 123–131. doi: 10.1007/s12550-009-0020-4
Ijiri, K. (2003). Life-cycle experiments of medaka fish aboard the international space station. Adv. Space Biol. Med. 9, 201–216. doi: 10.1016/S1569-2574(03)09008-7
Keller, N. P., Turner, G., and Bennett, J. W. (2005). Fungal secondary metabolism — from biochemistry to genomics. Nat. Rev. Microbiol. 3, 937–947. doi: 10.1038/nrmicro1286
Kittang, A.-I., Iversen, T.-H., Fossum, K. R., Mazars, C., Carnero-Diaz, E., Boucheron-Dubuisson, E., et al. (2014). Exploration of plant growth and development using the European modular cultivation system facility on the international Space Station. Plant Biol. 16, 528–538. doi: 10.1111/plb.12132
Klaus, D., Simske, S., Todd, P., and Stodieck, L. (1997). Investigation of space flight effects on Escherichia coli and a proposed model of underlying physical mechanisms. Microbiology 143, 449–455. doi: 10.1099/00221287-143-2-449
Knox, B. P., Blachowicz, A., Palmer, J. M., Romsdahl, J., Huttenlocher, A., Wang, C. C. C., et al. (2016). Characterization of Aspergillus fumigatus isolates from air and surfaces of the international Space Station. Clin. Vacc. Immun. 1, e00227–e00216. doi: 10.1128/mSphere.00227-16
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinforma. Oxf. Engl. 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Lim, C. S., Weinstein, B. N., Roy, S. W., and Brown, C. M. (2021). Analysis of fungal genomes reveals commonalities of intron gain or loss and functions in intron-poor species. Mol. Biol. Evol. 38, 4166–4186. doi: 10.1093/molbev/msab094
Link, B. M., Durst, S. J., Zhou, W., and Stankovic, B. (2003). Seed-to-seed growth of Arabidopsis Thaliana on the international space station. Adv. Space Res. 31, 2237–2243. doi: 10.1016/S0273-1177(03)00250-3
Liu, H.-Z., Wang, Q., Liu, X.-Y., and Tan, S.-S. (2008). Effects of spaceflight on polysaccharides of Saccharomyces cerevisiae cell wall. Appl. Microbiol. Biotechnol. 81, 543–550. doi: 10.1007/s00253-008-1692-y
Malo, M. E., Schultzhaus, Z., Frank, C., Romsdahl, J., Wang, Z., and Dadachova, E. (2021). Transcriptomic and genomic changes associated with radioadaptation in Exophiala dermatitidis. Comput. Struct. Biotechnol. J. 19, 196–205. doi: 10.1016/j.csbj.2020.12.013
Mehta, S. K., Cohrs, R. J., Forghani, B., Zerbe, G., Gilden, D. H., and Pierson, D. L. (2004). Stress-induced subclinical reactivation of varicella zoster virus in astronauts. J. Med. Virol. 72, 174–179. doi: 10.1002/jmv.10555
Miyake, Y., Ito, C., Itoigawa, M., and Osawa, T. (2007). Isolation of the antioxidant pyranonigrin-A from rice mold starters used in the manufacturing process of fermented foods. Biosci. Biotechnol. Biochem. 71, 2515–2521. doi: 10.1271/bbb.70310
Mulder, K. C. L., Mulinari, F., Franco, O. L., Soares, M. S. F., Magalhães, B. S., and Parachin, N. S. (2015). Lovastatin production: From molecular basis to industrial process optimization. Biotechnol. Adv. 33, 648–665. doi: 10.1016/j.biotechadv.2015.04.001
Muszewska, A., Steczkiewicz, K., Stepniewska-Dziubinska, M., and Ginalski, K. (2017). Cut-and-paste transposons in Fungi with diverse lifestyles. Genome Biol. Evol. 9, 3463–3477. doi: 10.1093/gbe/evx261
Nickerson, C. A., Ott, C. M., Mister, S. J., Morrow, B. J., Burns-Keliher, L., and Pierson, D. L. (2000). Microgravity as a novel environmental signal affecting Salmonella enterica Serovar Typhimurium virulence. Infect. Immun. 68, 3147–3152. doi: 10.1128/IAI.68.6.3147-3152.2000
Nikolcheva, L. G., Cockshutt, A. M., and Bärlocher, F. (2003). Determining diversity of freshwater Fungi on decaying leaves: comparison of traditional and molecular approaches. Appl Env. Microbiol 69, 2548–2554. doi: 10.1128/AEM.69.5.2548-2554.2003
Nitsche, B. M., Jørgensen, T. R., Akeroyd, M., Meyer, V., and Ram, A. F. (2012). The carbon starvation response of Aspergillus niger during submerged cultivation: insights from the transcriptome and secretome. BMC Genomics 13:380. doi: 10.1186/1471-2164-13-380
Noble, L. M., and Andrianopoulos, A. (2013). Fungal genes in context: genome architecture reflects regulatory complexity and function. Genome Biol. Evol. 5, 1336–1352. doi: 10.1093/gbe/evt077
Ombergen, A. V., Demertzi, A., Tomilovskaya, E., Jeurissen, B., Sijbers, J., Kozlovskaya, I. B., et al. (2017). The effect of spaceflight and microgravity on the human brain. J. Neurol. 264, 18–22. doi: 10.1007/s00415-017-8427-x
Onofri, S., Barreca, D., Selbmann, L., Isola, D., Rabbow, E., Horneck, G., et al. (2008). Resistance of Antarctic black fungi and cryptoendolithic communities to simulated space and Martian conditions. Stud. Mycol. 61, 99–109. doi: 10.3114/sim.2008.61.10
Onofri, S., de la Torre, R., de Vera, J.-P., Ott, S., Zucconi, L., Selbmann, L., et al. (2012). Survival of rock-colonizing organisms after 1.5 years in outer space. Astrobiology 12, 508–516. doi: 10.1089/ast.2011.0736
Onofri, S., de Vera, J.-P., Zucconi, L., Selbmann, L., Scalzi, G., Venkateswaran, K. J., et al. (2015). Survival of Antarctic Cryptoendolithic Fungi in simulated Martian conditions On board the international Space Station. Astrobiology 15, 1052–1059. doi: 10.1089/ast.2015.1324
Pel, H. J., de Winde, J. H., Archer, D. B., Dyer, P. S., Hofmann, G., Schaap, P. J., et al. (2007). Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat. Biotechnol. 25, 221–231. doi: 10.1038/nbt1282
Piscitelli, A., Pezzella, C., Giardina, P., Faraco, V., and Sannia, G. (2010). Heterologous laccase production and its role in industrial applications. Bioeng. Bugs 1, 254–264. doi: 10.4161/bbug.1.4.11438
Purevdorj-Gage, B., Sheehan, K. B., and Hyman, L. E. (2006). Effects of low-shear modeled microgravity on cell function, gene expression, and phenotype in Saccharomyces cerevisiae. Appl Env. Microbiol 72, 4569–4575. doi: 10.1128/AEM.03050-05
Rabbow, E., Rettberg, P., Baumstark-Khan, C., and Horneck, G. (2003). The SOS-LUX-LAC-FLUORO-toxicity-test on the international Space Station (ISS). Adv. Space Res. 31, 1513–1524. doi: 10.1016/S0273-1177(03)00086-3
Rodríguez Couto, S., and Toca Herrera, J. L. (2006). Industrial and biotechnological applications of laccases: a review. Biotechnol. Adv. 24, 500–513. doi: 10.1016/j.biotechadv.2006.04.003
Rohlfs, M., and Churchill, A. C. L. (2011). Fungal secondary metabolites as modulators of interactions with insects and other arthropods. Fungal Genet. Biol. 48, 23–34. doi: 10.1016/j.fgb.2010.08.008
Romsdahl, J., Blachowicz, A., Chiang, A. J., Chiang, Y.-M., Masonjones, S., Yaegashi, J., et al. (2019). International Space Station conditions alter genomics, proteomics, and metabolomics in Aspergillus nidulans. Appl. Microbiol. Biotechnol. 103, 1363–1377. doi: 10.1007/s00253-018-9525-0
Romsdahl, J., Blachowicz, A., Chiang, A. J., Singh, N., Stajich, J. E., Kalkum, M., et al. (2018). Characterization of Aspergillus niger isolated from the international Space Station. Microbial systems 3, e00112–e00118. doi: 10.1128/mSystems.00112-18
Romsdahl, J., Blachowicz, A., Chiang, Y.-M., Venkateswaran, K., and Wang, C. C. C. (2020). Metabolomic analysis of Aspergillus niger isolated From the international Space Station reveals enhanced production levels of the antioxidant Pyranonigrin A. Front. Microbiol. 11:931. doi: 10.3389/fmicb.2020.00931
Schuster, E., Dunn-Coleman, N., Frisvad, J., and van Dijck, P. (2002). On the safety of Aspergillus niger – a review. Appl. Microbiol. Biotechnol. 59, 426–435. doi: 10.1007/s00253-002-1032-6
Shephard, G. S. (2008). Impact of mycotoxins on human health in developing countries. Food Addit. Contam. Part A 25, 146–151. doi: 10.1080/02652030701567442
Singaravelan, N., Grishkan, I., Beharav, A., Wakamatsu, K., Ito, S., and Nevo, E. (2008). Adaptive melanin response of the soil fungus Aspergillus niger to UV radiation stress at “evolution canyon”, Mount Carmel Israel. PLoS ONE 3:2993. doi: 10.1371/journal.pone.0002993
Stajich, J. E., Harris, T., Brunk, B. P., Brestelli, J., Fischer, S., Harb, O. S., et al. (2012). FungiDB: an integrated functional genomics database for fungi. Nucleic Acids Res. 40, D675–D681. doi: 10.1093/nar/gkr918
Takahashi, A., Ohnishi, K., Takahashi, S., Masukawa, M., Sekikawa, K., Amano, T., et al. (2001). The effects of microgravity on induced mutation in Escherichia coli and Saccharomyces cerevisiae. Adv. Space Res. 28, 555–561. doi: 10.1016/S0273-1177(01)00391-X
Tavella, S., Ruggiu, A., Giuliani, A., Brun, F., Canciani, B., Manescu, A., et al. (2012). Bone turnover in wild type and Pleiotrophin-transgenic mice housed for three months in the international Space Station (ISS). PLoS One 7:e33179. doi: 10.1371/journal.pone.0033179
te Biesebeke, R., Levasseur, A., Boussier, A., Record, E., van den Hondel, C. A. M. J. J., and Punt, P. J. (2010). Phylogeny of fungal hemoglobins and expression analysis of the Aspergillus oryzae flavohemoglobin gene fhbA during hyphal growth. Fungal Biol. 114, 135–143. doi: 10.1016/j.mycres.2009.08.007
Tesei, D., Chiang, A. J., Kalkum, M., Stajich, J. E., Mohan, G. B. M., Sterflinger, K., et al. (2021). Effects of simulated microgravity on the proteome and Secretome of the Polyextremotolerant black fungus Knufia chersonesos. Front. Genet. 12:638708. doi: 10.3389/fgene.2021.638708
Vaishampayan, P. A., Rabbow, E., Horneck, G., and Venkateswaran, K. J. (2012). Survival of Bacillus pumilus spores for a prolonged period of time in real space conditions. Astrobiology 12, 487–497. doi: 10.1089/ast.2011.0738
Vandenberghe, L. P. S., Soccol, C. R., Pandey, A., and Lebeault, J.-M. (2000). Solid-state fermentation for the synthesis of citric acid by Aspergillus niger. Bioresour. Technol. 74, 175–178. doi: 10.1016/S0960-8524(99)00107-8
Venkateswaran, K., Singh, N. K., Sielaff, A. C., Pope, R. K., Bergman, N. H., Tongeren, S. P., et al. (2017). Non-toxin-producing Bacillus cereus strains belonging to the B. anthracis Clade Isolated from the International Space Station. mSystems 2:17. doi: 10.1128/mSystems.00021-17
Wang, J., Liu, Y., Zhao, G., Gao, J., Liu, J., Wu, X., et al. (2020). Integrated proteomic and metabolomic analysis to study the effects of spaceflight on Candida albicans. BMC Genomics 21:57. doi: 10.1186/s12864-020-6476-5
Keywords: Aspergillus niger, International Space Station, metabolome, proteome, genome
Citation: Blachowicz A, Romsdahl J, Chiang AJ, Masonjones S, Kalkum M, Stajich JE, Torok T, Wang CCC and Venkateswaran K (2022) The International Space Station Environment Triggers Molecular Responses in Aspergillus niger. Front. Microbiol. 13:893071. doi: 10.3389/fmicb.2022.893071
Edited by:
Donatella Tesei, University of Natural Resources and Life Sciences Vienna, AustriaReviewed by:
Ekaterina Dadachova, University of Saskatchewan, CanadaCorrado Nai, FEMS Federation of European Microbiological Societies, Netherlands
Copyright © 2022 Blachowicz, Romsdahl, Chiang, Masonjones, Kalkum, Stajich, Torok, Wang and Venkateswaran. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kasthuri Venkateswaran, kjvenkat@jpl.nasa.gov
 Adriana Blachowicz
Adriana Blachowicz Jillian Romsdahl1
Jillian Romsdahl1 Markus Kalkum
Markus Kalkum Jason E. Stajich
Jason E. Stajich Tamas Torok
Tamas Torok Clay C. C. Wang
Clay C. C. Wang Kasthuri Venkateswaran
Kasthuri Venkateswaran