- 1Shanxi Key Laboratory of Minor Crops Germplasm Innovation and Molecular Breeding, College of Life Sciences, Shanxi Agricultural University, Jinzhong, China
- 2Experimental Teaching Center, Shanxi Agricultural University, Jinzhong, China
- 3State Key Laboratory of Rice Biology, Institute of Insect Sciences, College of Agriculture and Biotechnology, Zhejiang University, Hangzhou, China
Many fusion tags have been developed to improve the expression of recombinant proteins. Besides the translocation of cargo proteins, the signal peptides (SPs) of some secretory proteins, such as the ssTorA and Iasp, have been used as an inclusion body tag (IB-tag) or the recombinant expression enhancer in the cytosol of E. coli. In this study, the approach to utilize the SP of Vip3A (Vasp) from Bacillus thuringiensis (Bt) as a fusion tag was investigated. The results showed that either the Vasp or its predicted N- (VN), H- (VH), and C-regions (VC), as well as their combinations (VNH, VNC, and VHC), were able to significantly enhance the production yield of eGFP. However, the hydrophobic region of the Vasp (VH and/or VC) made more than half of the eGFP molecules aggregated (VeGFP, VHeGFP, VCeGFP, VNHeGFP, VNCeGFP, and VHCeGFP). Interestingly, the addition of the Bt trigger factor (BtTF) led to the neutralization of the negative impact and solubilization of the fusion proteins. Therefore, the coexpression of Vasp or its derivates with the chaperone BtTF could be a novel dual-enhancement system for the production yield and solubility of recombinant proteins. Notably, EcTF was unable to impact the solubility of Vasp or its derivates guided proteins, suggesting its different specificities on the recognition or interaction. Additionally, this study also suggested that the translocation of Vip3 in the host cell would be regulated by the BtTF-involved model.
Introduction
Fusion tags have been widely used for producing recombinant proteins. A fusion tag can be derived from an artificial polypeptide, a partial fragment, or a whole natural protein. Many fusion tags have been applied to facilitate protein purification or improve protein production yield, solubility, and folding (reviewed in Costa et al., 2014; Rosano and Ceccarelli, 2014; Paraskevopoulou and Falcone, 2018; Vandemoortele et al., 2019; Ki and Pack, 2020). Some of them are versatile. For instance, the well-known solubility enhancer tags such as maltose-binding protein (MBP) (Maina et al., 1988), glutathione S-transferase (GST) (Smith and Johnson, 1988), small ubiquitin-related modifier (SUMO) (Marblestone et al., 2006), and Fasciola hepatica 8-kDa antigen (Fh8) (Costa et al., 2013) are also used in affinity purification. Signal peptides (SPs) of many secretory proteins have also been used as fusion tags to direct the recombinant proteins to different cellular locations (reviewed in Overton, 2014; Rosano and Ceccarelli, 2014; Gupta and Shukla, 2016; Malik, 2016; Kleiner-Grote et al., 2018). Recently, novel applications of the SP tags were developed. For instance, the SP of trimethylamine N-oxide reductase (torA) in E. coli was reported to be an inclusion body tag (IB-tag) that produced some recombinant proteins in insoluble form in E. coli cytosol (Jong et al., 2017). The SP of Cry1Ia toxin (Iasp) of Bacillus thuringiensis (Bt) can not translocate recombinant proteins through the cell membrane in high efficiency but enhance their expression level significantly (Gao et al., 2020). The produced fusion fluorescent indicators such as IeGFP and ImCherry showed better performance than the individually expressed eGFP and mCherry in E. coli and Bt strains. Notably, SPs are ubiquitous in nature and are worthy of deeper exploration.
Vip3 toxins were produced by Bt cells during the vegetative stage of growth. The first vip3 gene was reported in 1996, and since then, many homologs were collected (Crickmore et al., 2020). At present, the action mode of the Vip3 toxins still remains elusive. Two ways were proposed to solve the problem: apoptosis induced by the Vip3A protoxin and cell perforation mediated by the activated Vip3A toxin (reviewed in Chakroun et al., 2016; Chakrabarty et al., 2020; Syed et al., 2020). Several pieces of research concluded that there was no significant cross-resistance between the Vip3 and Cry families, suggesting the different receptors for both toxins when acted inside the target insects (reviewed in Chakroun et al., 2016; Chakrabarty et al., 2020; Syed et al., 2020). Recently, the determination of the three-dimensional structure (3D-structure) of the protease-activated Vip3A and Vip3B at higher resolution revealed a homotetramer architecture containing a long coiled-coil that explained the structural basis of the perforation mode (Núñez-Ramírez et al., 2020; Zheng et al., 2020). Vip3 proteins share a similar overall tetrameric organization, especially for the domains I, II, and III at the N terminus, but the last two domains (IV and V) show slight variation in position and orientation that would result from the greater diversity of the primary structure at the C-terminus of the Vip3 family (Chakroun et al., 2016).
The N terminus of Vip3, including the predicted SP region (Vsp), was conserved (Chakroun et al., 2016). Due to the deficiency of the peptidase recognition site, the exact length of Vsp is still unknown (Estruch et al., 1996; Doss et al., 2002; Chen et al., 2003; Rang et al., 2005). The N-terminal 33 amino acids (AAs) are the longest version of Vsp (Rang et al., 2005) in which three classic regions can be roughly distinguished, i.e., the positively charged N-region (from M1 to R11), followed by the hydrophobic residues (H-region, from A12 to F20), and the C-region (from N21 to I33). It is noteworthy that the C-region is rich in hydrophilic AAs, but the counterpart of Vsp has a highly amphiphilic nature resulting from the alternative arrangement of the hydrophilic and hydrophobic AAs. The alternative arrangement in this region facilitates the formation of a four-helix bundle, transporting protons and divalent metals through the membrane, by contacting with the lipid bilayer of targeted cells in the final architecture of Vip3 tetramer (Núñez-Ramírez et al., 2020).
Interactions between the nascent peptide and the signal peptide recognition proteins or the molecular chaperones prevent the recombinant protein from aggregation, thus facilitating the translocation. At present, the translocation pathway of Vip3 is still unclarified. Given that the strongly conserved motif (S/T-RRXFLK) in SP of the twin-arginine translocation system (Tat system) is not present in Vsp, Vip3 proteins might translocate via the other single membrane-spanning secretion system, the Sec pathway (reviewed in Berks, 2015; Costa et al., 2015; Green and Mecsas, 2016; Anné et al., 2017). In contrast to most secretory proteins, Vsp is not removed after Vip3 protein translocation. The hydrophobic regions in SPs would interact with the corresponding regions of the client proteins and therefore negatively affect its folding. For instance, the existence of SP of MBP or pelB (pectate lyase B) at the N-terminus interfered with the thermodynamic stability of mature MBP or thioredoxin (Trx) (Beena et al., 2004; Krishnan et al., 2009; Kulothungan et al., 2009; Singh et al., 2013). This observation would explain the capability of inducing inclusion body formation of the SPs described above (Jong et al., 2017). This study will explore the effects of the predicted SP of Vip3A protein (Vasp) and its derivates on the expression of enhanced green fluorescent protein (eGFP) and provide a novel solution for enhancing the production yield and solubility of recombinant proteins, simultaneously.
Materials and Methods
Bacterial Strains and Growth Conditions
All plasmids and strains used in this study are listed in Supplementary Table S1. Unless otherwise noted, the E. coli cells were incubated in Luria-Bertani medium (LB medium) with 50 μg/ml ampicillin for pMD19, pHT304 and its derived vectors, or 50 μg/ml kanamycin for pET28a and its derived vectors at 37°C shaking with 200 rpm.
Sequence Analysis of the SPs of Vip3 Proteins
For phylogenetic analysis, the Vip3 protein sequences were retrieved from Bacterial Pesticidal Protein Resource Center (https://www.bpprc.org/) for extracting the corresponding SP sequences (Crickmore et al., 2020). Then, the peptide alignment was conducted using MAFFT software (v7.453) (Katoh and Standley, 2013) with default parameters, and the result was graphically enhanced by ESPript (v3.0) (Robert and Gouet, 2014).
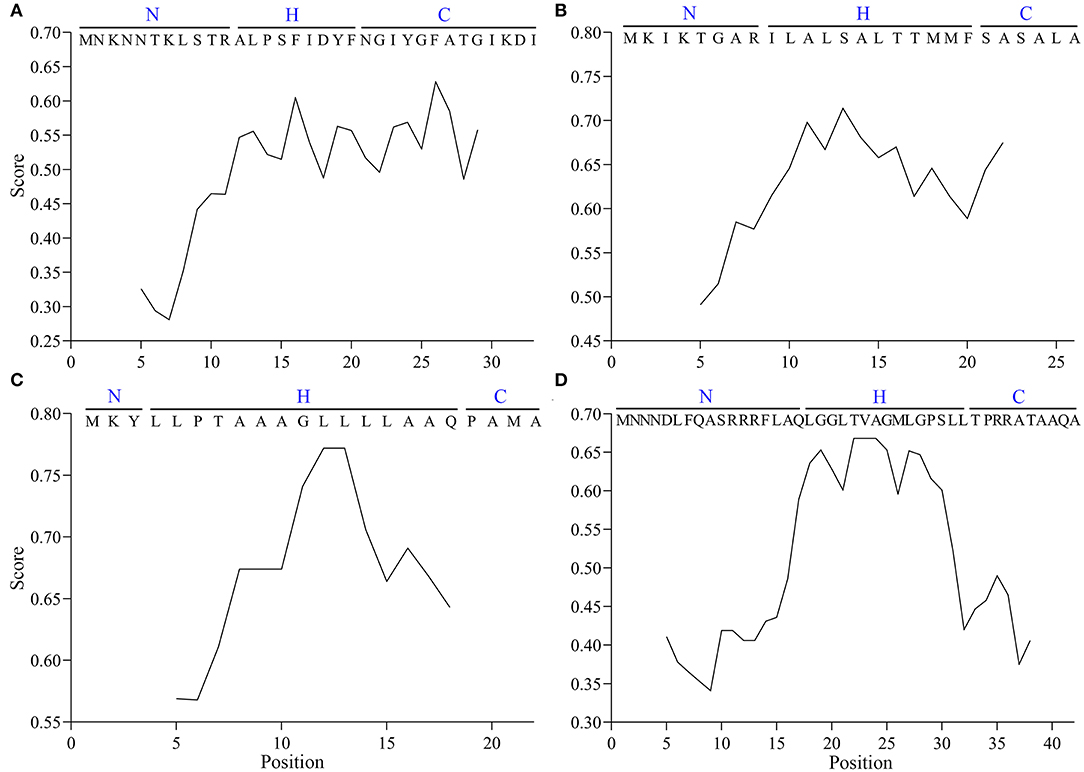
The hydropathicity scale of the SP sequences of Vip3Aa1, MBP, pelB, and TorA was computed by the online program ProtScale (https://web.expasy.org/protscale/) with the default parameters except that the scale values were normalized (Kyte and Doolittle, 1982). The output was plotted by GraphPad Prism (v9.0.0).
Construction of Expression Vectors
pET Expression Constructions
The Vasp-encoding sequence (primers VEGFP-F and VEGFP-fuR) was fused to the 5′ end of the egfp gene (primers VEGFP-fuF and I/EGFP-R) by overlapping PCR using primers VEGFP-F and I/EGFP-R to produce the Vegfp gene. The Vegfp gene was TA-cloned into the pMD19 vector (Takara, Beijing, China) for sequencing. The correct Vegfp sequence was cloned into the BamH I/Xho I restriction site of the pET28aDel plasmid to form p28aD-VeGFP. Then, the Vasp-encoding sequence in p28aD-VeGFP was replaced by the synthesized fragments to produce the p28aD-VNeGFP, p28aD-VNHeGFP, p28aD-VNCeGFP, p28aD-VHeGFP, p28aD-VHCeGFP, and p28aD-VCeGFP. The BtCsaA promoter (PBtCsaA) guided chaperone genes encoding SecA (BtSecA), TF (BtTF), and CsaA (BtCsaA) of Bt and SecB of E. coli (EcSecB) as well as its mutants (SecB7577, SecB142, and SecB142-7577) were artificially synthesized and cloned into the Xho I/Blp I site of the p28aD-VeGFP vector. The PBtCsaA-Bttig sequence was also inserted into the same site of the p28aD-VNeGFP, p28aD-VNHeGFP, p28aD-VNCeGFP, p28aD-VHeGFP, p28aD-VHCeGFP, and p28aD-VCeGFP, respectively.
The genes that include human growth differentiating factor-8 (GDF8), Setaria italica cytochrome P450 71A1-like (71A1), ent-Cassadiene C2-hydroxylase (ECH), and cinnamate beta-D-glucosyltransferase (CGT) were artificially synthesized and cloned into the pET28aDel derived plasmid containing PBtCsaA-Bttig sequence.
These plasmids were individually transformed into E. coli BL21-star (DE3) strain by calcium chloride (CaCl2) transformation.
pHT304 Expression Constructions
The Vegfp gene was amplified by PCR using the p28aD-VeGFP plasmids as the template (primers VEGFP-F and 304I/EGFP-R) and was TA-cloned into the pMD19 vector. After sequencing, the BamH I/Sac I fragment of the Vegfp gene was inserted into the corresponding site of the pAc-eGFP vector. The plasmid was designated as pAc-VeGFP and was transformed into E. coli MC4100 strain.
Proteins Expression and Samples Preparation
E. coli MC4100 Strain
The recombinant protein expression protocol in E. coli MC4100 was the same as in E. coli TG1 that was described previously (Gao et al., 2020). Briefly, the single colony of each E. coli strain harboring p304ΔSacI or its derived vectors was inoculated into 3 ml LB medium and incubated overnight. Each strain was inoculated into 8 ml fresh LB medium with an equally initial amount. Three replicates were tested for each strain. At 8 or 12 h after inoculation, 3 ml of cell cultures for each replicate was taken for protein analysis.
E. coli BL21-Star (DE3) Strain
The overnight incubated cells of each BL21-star (DE3) strain in 3 ml LB medium were transferred into fresh LB medium (1:1,000, v/v) and continuously cultivated in the same condition. IPTG (isopropyl β-D-1-thiogalactopyranoside, 1 mM or other concentrations indicated) was used to induce the recombinant protein expression when the optical density at 600 nm (OD600) of the cell culture reached 0.8. After induction at 16°C, the cells of each sample were harvested after centrifuging at 8,000 rpm. The collected cells were washed with deionized water three times and resuspended in 2/5 of the initial volume of ice-cold PBS buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4, pH 7.4). The resuspended cells were disrupted by ultrasonic treatment in ice-cold conditions. A fraction of the cell lysate was pipetted into a new centrifuge tube (total protein sample). After centrifuging at 12,000 rpm at 4°C for 15 min, the remaining lysates were separated. The precipitate was resuspended by the same volume of PBS buffer as the corresponding supernatant. These samples were prepared for sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) analysis.
SDS-PAGE Analysis
The sample preparation and SDS-PAGE analysis referred to the method described previously (Gao et al., 2020). Briefly, each protein sample was mixed with one-fourth volume of 5× SDS gel-loading buffer and boiled for 5 min. The samples were centrifuged at 12,000 rpm for 5 min and then loaded onto SDS-PAGE gels for separation. The separated proteins were stained by Coomassie bright blue and the gels were photographed and analyzed by Bio-Rad Image-Lab-Software (v6.0.1). For western blot analysis, the separated proteins in the gel were transformed to the nitrocellulose membrane and then incubated with the rabbit antiserum against IeGFP protein and the horseradish peroxidase-conjugated goat antirabbit IgG (H + L) antibody (MultiSciences, Hangzhou, China) successively. The target bands were visualized using eECL western blot kit (Cowin Biotech, Jiangsu, China).
Fluorescence Localization
The E. coli cells were prepared by the method described previously (Gao et al., 2020). Briefly, the harvested cells corresponding to 1 A600 unit were washed two times by PBS buffer and immersed in 600 μl of the fixing solution (2% paraformaldehyde, 2.5% glutaraldehyde in PBS buffer). After 45-min incubation at room temperature, the cells were washed three times with PBS and finally resuspended in 100 μl PBS buffer. The suspension was detected by an inverted confocal microscope (Leica SP8, Leica Microsystems, Wetzlar, Germany) with a 63-time oil immersion objective. The outputs were recorded with Leica Application Suite X (v3.1.5).
Results
Sequence Conservation and Hydrophobicity Scale of Vsp
A total of 108 Vip3 proteins were collected in the Bacterial Pesticidal Protein Resource Center (Crickmore et al., 2020), but only 26 patterns were found in the predicted Vsp region (Figure 1; Supplementary Table S3). For instance, there were 49 Vip3 proteins that shared the same AA sequences with Vip3Aa1 at N-terminus. The alignment results indicated the high conservation of Vsps, especially in the predicted C-region, which is also the part of helix α1 (from I23 to K39) of Vip3 protein (Núñez-Ramírez et al., 2020). It was observed that three Vip3 proteins, including Vip3Aa31, Vip3Aa32, and Vip3Aa29, were more divergent in the predicted H-region compared to other members. The nine AAs extensions were observed in the predicted N-region of Vip3Bc1, whereas only two extra AAs were found in Vip3Ai1.

Figure 1. The alignment of the representative Vsp sequences. The predicted N-, H-, and C-regions of SP are indicated. The numbers of genes sharing the corresponding sequences are indicated in parentheses.
Given the high frequency of the Vip3Aa1 SP sequence in the Vip3 family, it was considered a representative pattern and designated as Vasp in this study. The hydrophobicity scale of Vasp was determined, and the differences with the common SPs of MBP, pelB, and TorA were revealed (Figure 2). The classic distribution of AA residues in the latter three SPs, including the positively charged N-region, the hydrophobic H-region, and the hydrophilic C-region, was observed. However, in Vasp, the H- and C-regions were ambiguous in which the hydrophilic and hydrophobic AAs were arranged alternatively, resulting in a relatively amphiphilic nature (Núñez-Ramírez et al., 2020). Notably, the hydrophobicity level of the H- and C-regions (0.45–0.65 in Figure 2A) matched with the H-region of the MBP (0.55–0.75 in Figure 2B), pelB (0.55–0.80 in Figure 2C), and TorA's (0.55–0.70 in Figure 2D) SPs, respectively.
Vasp Enhanced the Expression Level of eGFP in E. coli MC4100 Strain
The effect of Vasp on the eGFP protein (VeGFP) was investigated using the constitutive promoter of the cry1Ac gene (Pac) in E. coli MC4100 cells. The expression cassette of the Vegfp gene was similar to the Iegfp described previously (Gao et al., 2020). The results showed that the expression level of the eGFP was improved slightly by Vasp at 8 (~1.14-folds) and 12 h (~2.05-folds) after inoculation (Figure 3). The observation prompts that Vasp would be also used as another production-enhancement fusion tag such as Iasp.
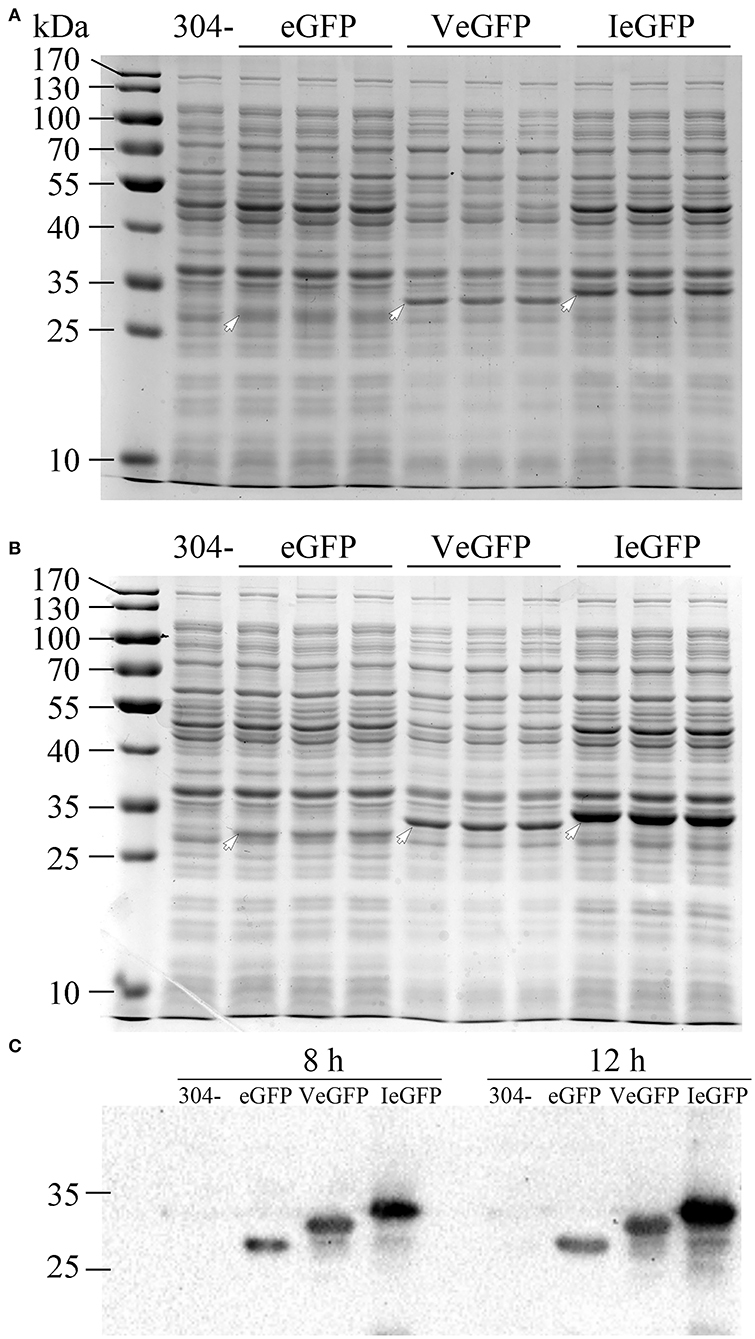
Figure 3. The expression of VeGFP in the E. coli MC4100 strain controlled by the constitutive promoter Pac. The samples of eGFP (27.9 kDa), VeGFP (lane 2, 31.8 kDa), and IeGFP (lane 3, 33.1 kDa) were prepared at 8 (A) and 12 h (B) after inoculation, respectively. Lane “–” is the negative control prepared from M304 cells. Lane “M” is the molecular weight standard. The arrows indicate the target bands of the recombinant proteins. The target proteins were also verified by western blot (C).
Bt Trigger Factor Improved the Solubility of the VeGFP
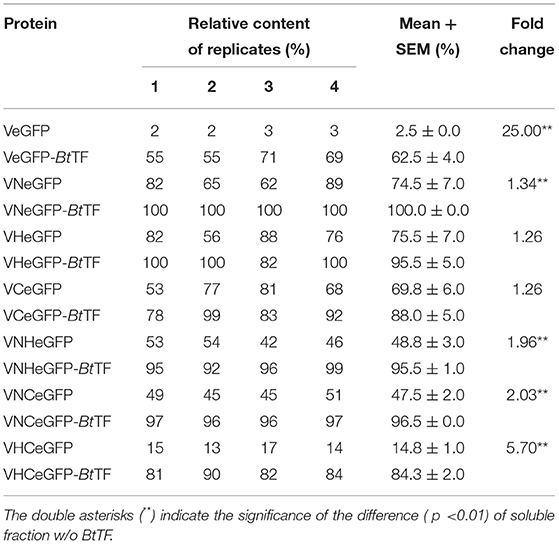
Previously, the reservation of SP was reported to affect the thermodynamic stability of the client protein, such as MBP and Trx (Beena et al., 2004; Krishnan et al., 2009; Kulothungan et al., 2009; Singh et al., 2013). In this study, the negative impact of Vasp on the solubility of eGFP was also observed in the BL21-star (DE3) strain (Figure 4), i.e., only 2.5% of products were soluble (Table 1). Since the Tat-pathway was excluded according to the conserved motif analysis, the performances of several chaperones in the Sec pathway in the Bt strain including BtSecA, Bt trigger factor (BtTF), and also BtCsaA and its counterpart in E. coli (EcSecB) on the aggregation-prone protein VeGFP were investigated. Each chaperone gene is located at the 3′ end of the BtCsaA promoter (PBtCsaA) that follows the Vegfp gene directly (Supplementary Figure S1A). Controlled by the regulation structure, the expression of the VeGFP and BtTF was detected simultaneously after the IPTG induction (Supplementary Figure S1B). The results showed that both BtCsaA and its counterpart EcSecB were unable to prevent the VeGFP from aggregation (Figure 4). BtSecA, a translocation ATPase in Sec pathway, also failed to keep the VeGFP soluble. To exclude the possible interference resulting from the interaction between EcSecB and EcSecA, the variants of EcSecB were tested. EcSecB mutant (SecB7577) with substitutions at two positions, L75Q and E77V, still bound the client proteins but showed a marked reduction in its binding affinities for EcSecA, leading to the disruption of translocation (Fekkes et al., 1998; Sala et al., 2017). The 13 AAs at the C-terminus decreased the dimerization efficiency of SecA (Randall et al., 2005). In this study, the SecB7577, the 13 AAs deficiency mutant at C-terminus (SecB142), and even their combination SecB142-7577 did not impact the solubility of VeGFP positively. Interestingly, only the chaperone BtTF made VeGFP solubilized.
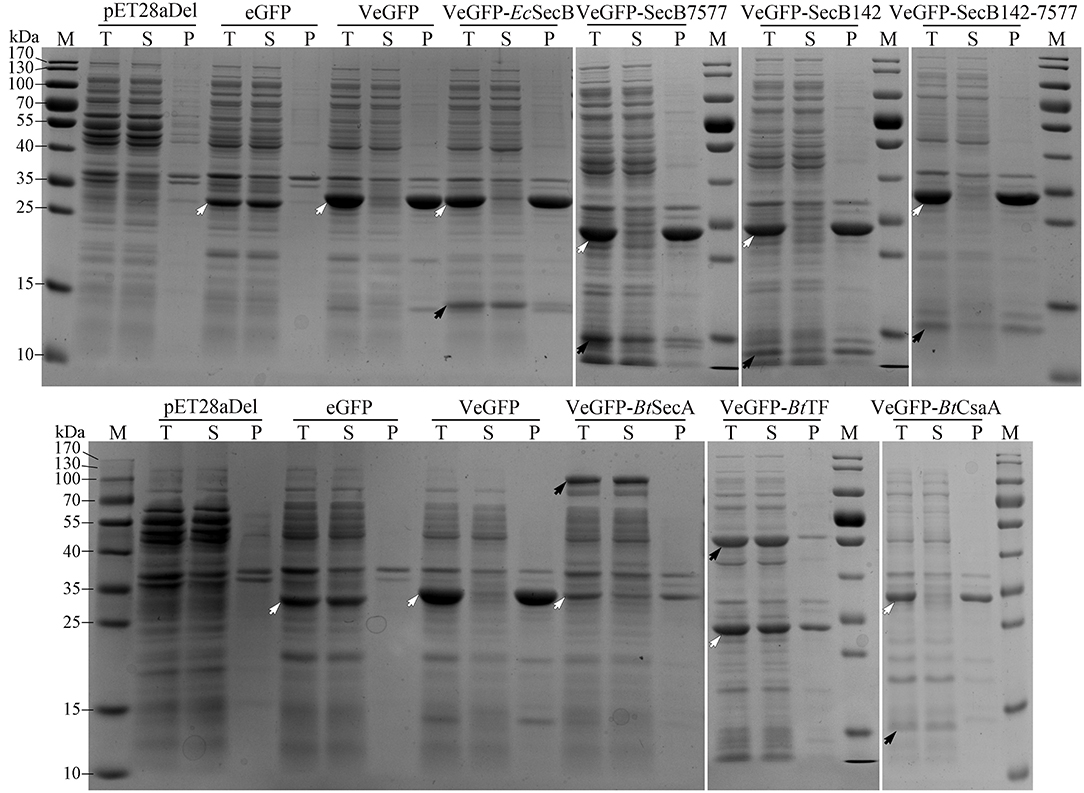
Figure 4. Coexpression analysis of VeGFP with several chaperones. For each sample, the total proteins (T), the soluble component after cell lysis (S), and the precipitates (P) were loaded, respectively. The black arrows indicate the corresponding chaperones (EcSecB, 17.3 kDa; SecB7577, 17.3 kDa; SecB142, 15.9 kDa; SecB142-7577, 15.9 kDa; BtCsaA, 11.9 kDa; BtSecA, 95.0 kDa; BtTF, 47.3 kDa). The hollowed arrows indicate the target recombinant proteins.
The polar localization of VeGFP products in the BL28-VeGFP strain was observed by confocal microscope (Figures 5a–c). The light spots in each cell indicated the corresponding inclusion body cores (Rinas et al., 2017). However, when expressed together with BtTF (BL28-VeGFP-BtTF strain), the clear fluorescent signal inside the cell emerged evenly without the disappearence of the polar bright spots (Figures 5d–f). The observations were in line with the SDS-PAGE analysis and suggested that the accumulation of BtTF made part of the VeGFP products solubilized and folded correctly in the cytosol.
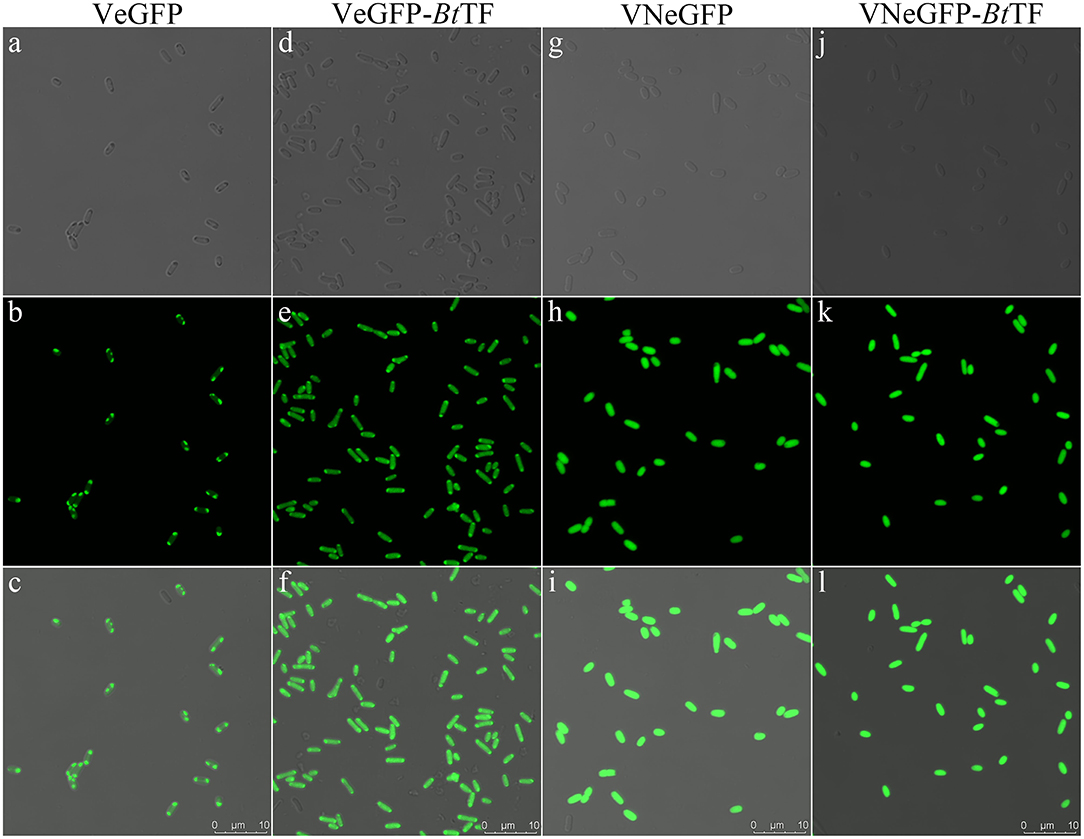
Figure 5. The fluorescent signal distribution of VeGFP and VNeGFP in E. coli BL21-star (DE3) cells. The corresponding cells expressing VeGFP (a–c) or VNeGFP (g–h) individually, or with BtTF d–f for VeGFP and j–l for VNeGFP) were observed using an inverted confocal microscope (Leica SP8). For each sample, the image in the third row was merged with the corresponding bright-field image in the first row and the fluorescent one in the second row. The scale bar represents 10 μm.
Three Regions of Vasp Boosted the Production Yield of eGFP With Distinct Solubility
To confirm the crucial segment enhancing the expression level of eGFP in Vasp, the predicted N- (VaspN, VN), H- (VaspH, VH), and C-regions (VaspC, VC), as well as their combinations, were used to guide the fluorescent protein, respectively (Figure 6A). As a result, all of these extra peptides boosted the production yield of the eGFP (~2.49- to 3.20-fold enhancement compared to eGFP, Figure 6B; Supplementary Table S4). However, their solubility varied widely (Figure 7A; Table 1). For instance, deletion of H- (VNCeGFP, 47.5%) or C-region (VNHeGFP, 48.8%) both recovered the solubility partially of VeGFP (2.5%). The individual H- or C-region produced 75.5 or 69.8% soluble recombinant proteins (VHeGFP and VCeGFP), respectively. When both the hydrophobic regions were removed (VNeGFP), most of the product molecules (74.5%) were also kept soluble, and even distribution of the fluorescent signal was observed inside the cells (Figures 5g–i). The loss of the N-region (VHCeGFP) led to 14.8% of the products being soluble. All of these results indicated the negative effects of the extra fragments originated from Vasp on the solubility of the client protein. These data revealed the negative correlation between the sequence integrity of Vasp and the solubility of the corresponding fusion fluorescent proteins, and interestingly, the decrements of the solubility impacted by the combination of VN with any one of the other two regions (VH or VC) were cumulative. Approximately a 51.2% decrease in VNHeGFP solubility would result from the addition of the impact of VN (25.5%) and VH (24.5%). Surprisingly, the synergetic effects were observed when the VH and VC coexisted. Approximately 85.2% insoluble fraction is far more than the cumulative effect of the VH (24.5%) and VC (30.2%). A similar result was observed on the maximal reduction in solubility for VeGFP (97.5%) compared to the cumulative effect of the three individual regions.
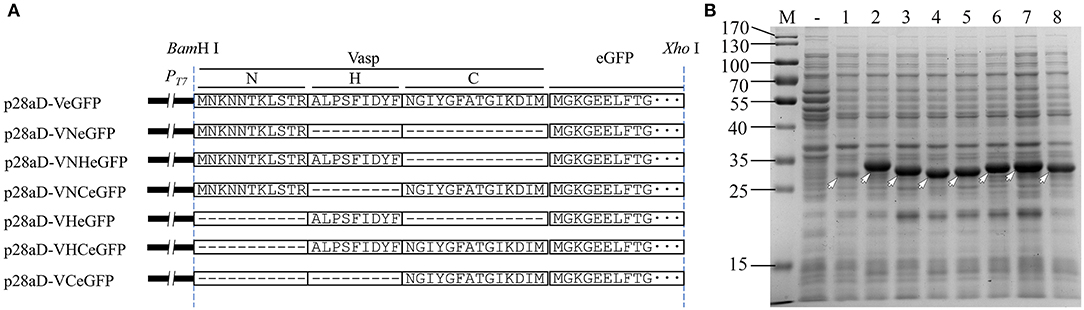
Figure 6. Diagram of Vasp variants (A) and their effect (B) on the expression of eGFP. In panel B, the target bands of eGFP (lane 1), VeGFP (lane 2), VNeGFP (lane 3, 29.2 kDa), VHeGFP (lane 4, 29.1 kDa), VCeGFP (lane 5, 29.5 kDa), VNHeGFP (lane 6, 30.3 kDa), VNCeGFP (lane 7, 30.7 kDa), and VHCeGFP (lane 8, 30.6 kDa) are indicated by hollowed arrows, respectively. Lane “–” is the negative control prepared from BL28aD cells. Lane “M” is the molecular weight standard.
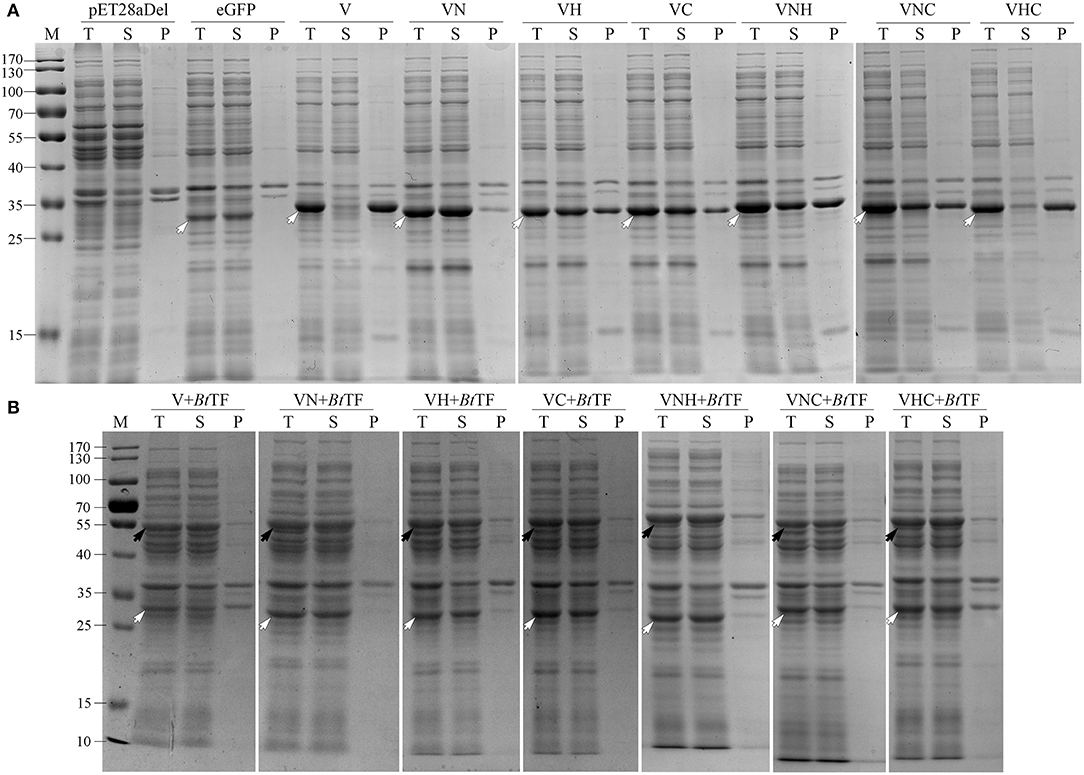
Figure 7. The solubility analysis of the fusion fluorescent proteins with (A) or without (B) the coexpression of BtTF. For each sample, the total proteins (T), the soluble component after cell lysis (S), and the precipitates (P) were loaded. The black arrows indicate the target bands of BtTF and the hollowed arrows indicate the target recombinant proteins including eGFP, VeGFP (V), VNeGFP (VN), VHeGFP (VH), VCeGFP (VC), VNHeGFP (VNH), VNCeGFP (VNC), and VHCeGFP (VHC). Lane “M” is the molecular weight standard.
BtTF Improved the Solubility of the Fusion Fluorescent Proteins
The coexpressed chaperone BtTF also made the variants of VeGFP soluble (Figure 7B; Table 1). The soluble fraction of VeGFP coexpressed with BtTF (VeGFP+BtTF) increased by 25 fold (2.5 vs. 62.5%). Similarly, the solubility of VHCeGFP, VNHeGFP, VNCeGFP, VHeGFP, and VCeGFP proteins expressed in the E. coli BL21-star (DE3) strain was improved by 5.70-, 1.96-, 2.03-, 1.26-, and 1.26-fold by BtTF, respectively. Interestingly, the BtTF was able to make all of the tested VNeGFP molecules soluble (1.34-folds, Figures 5j–l, 7B; Table 1). These data also indicated the positive correlation between the sequence integrity of Vasp and the interaction strength with BtTF.
The Novel Expression System Was Also Applicable to Other Recombinant Proteins
According to the results described above, the combination of BtTF and Vasp or its variants would be used as a novel expression system. The expression vectors were redesigned for adding the encoding sequences of His-tag, TEV protease, and enterokinase recognition sites at the 3′ end of VN or VNH-encoding sequences (Figure 8A, p28aD-VNhteGDF8-BtTF and p28aD-VNHhteGDF8-BtTF). The expression cassette structures were designated as the VNhte-BtTF or VNHhte-BtTF, respectively. Additionally, three VN-encoding sequences were arranged in a tandem array in the p28aD-V3NhteGDF8-BtTF plasmid (the V3Nhte-BtTF cassette). The optimum concentration of IPTG (0.05 mM) for the novel vectors was identified by expressing the VNhteGDF8 protein in the E. coli BL21-star (DE3) strain harboring the p28aD-VNhteGDF8-BtTF plasmid at low temperature (16°C) with 150 rpm (Supplementary Figure S2).
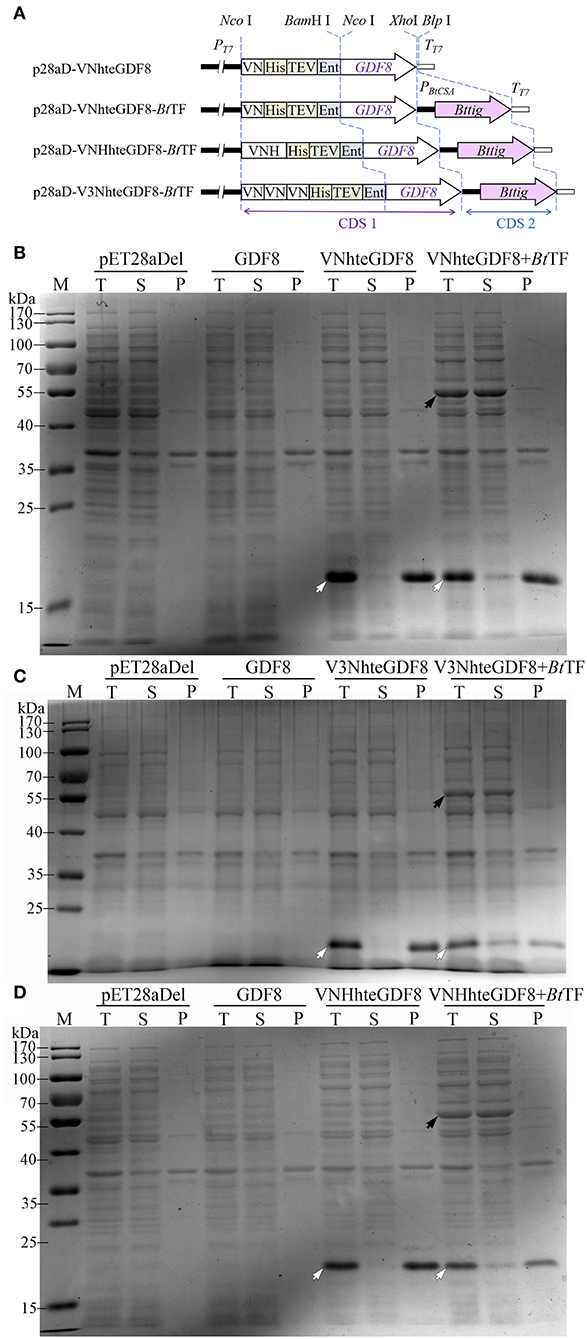
Figure 8. The effect of the Vasp derivates and BtTF on the expression of GDF8 protein. (A) The diagram of expression cassettes of VNhteGDF8, VNHhteGDF8, and V3NhteGDF8. The expression of VNhteGDF8 (B, 16.9 kDa), V3NhteGDF8 (C, 19.2 kDa), and VNHhteGDF8 (D, 18.0 kDa) was analyzed by SDS-PAGE. For each sample, the total proteins (T), the soluble component after cell lysis (S), and the precipitates (P) were loaded. Lane “M” is the molecular weight standard. The black arrow indicates the BtTF and the hollowed arrow indicates the recombinant proteins.
Under the optimum condition, the significant enhancement of the expression level of the GDF8 fusion proteins was identified for all three novel vectors (Figures 8B–D). However, without the Vasp derivate sequences, GDF8 cannot be detected by SDS-PAGE, which was consistent with the previous observation (Gao et al., 2020). Notably, the coexistence of BtTF and the Vasp-related sequences (VN, VNH, or V3N) made the GDF8 soluble, albeit in a small percentage. The triple-arranged VN (V3N) was unable to improve the expression level of GDF8 further compared to the single one. Compared to the common solubility enhancer tags, such as MBP, Trx, and GST, the Vasp and its variant VN brought out a better production yield for eGFP (Supplementary Figure S3).
A total of three proteins of Setaria italica including the cytochrome P450 71A1-like (71A1), ent-cassadiene C2-hydroxylase (ECH), and cinnamate beta-D-glucosyltransferase (CGT) were also expressed successfully in soluble form by the VNhte-BtTF cassette (Supplementary Figure S4). Additionally, when located at the C-terminal of the recombinant proteins, VN failed to enhance the expression level of GDF8 and ECH proteins (Supplementary Figure S5).
EcTF Cannot Improve the Solubility of the VN Guided Recombinant Proteins
Trigger factor of E. coli (EcTF) has a low identity with BtTF (29.4%, Supplementary Figure S6). Therefore, the effect of EcTF encoded by the extra chaperone plasmid pTf16 on the solubility of the fusion proteins VNhteGDF8, V3NhteGDF8, and VeGFP was investigated. Notably, in the VNhte-BtTF-like cassettes, the rapid expression of the recombinant proteins and BtTF was almost synchronous (Supplementary Figure S1) after adding IPTG. The pTf16 involved dual-plasmid system made it easy to produce the recombinant proteins and EcTF asynchronously. The results showed that, when the host cells only harbored the pTf16 plasmid, the production of EcTF was high after induction of L-arabinose. The coexistence with the second plasmid significantly reduced the accumulation of EcTF, but the products can also be detected by SDS-PAGE analysis. Unfortunately, the extra produced EcTF molecules were unable to enhance the solubility of VeGFP, VNhteGDF8, and V3NhteGDF8 (Supplementary Figure S7), which revealed the different specificities between EcTF and BtTF.
Discussion
Due to the lack of the recognition site of signal peptidase, the length of Vsp has not been determined precisely. This study used the longest version (33 AAs) of Vasp to develop a novel protein expression system. The result showed that Vasp exerted a positive effect on the expression level of eGFP. Previously, we reported a novel fusion tag Iasp and proposed new insights into the abundant SPs of natural secretory proteins that would be an ideal resource for fusion tags (Gao et al., 2020). This study provided another example. However, the hydrophobic region of SPs negatively affected the thermodynamic stability of the cargo proteins, leading to the formation of inclusion bodies (Beena et al., 2004; Krishnan et al., 2009; Kulothungan et al., 2009; Singh et al., 2013). This drawback cannot be ignored unless the kind of the tags used was similar to the IB-tag, such as the SP of torA (Jong et al., 2017). This study identified two ways to at least partially avoid or solve the problem.
The N-region of Vasp (VN) could be considered as the polycationic tag harboring two lysine residues and one arginine residue. The net positive charged VN enhanced the expression level of eGFP with a slight compromise in its solubility. The polycationic tags containing the polylysine or polyarginine residues have been used as the protein solubility enhancers for almost three decades (reviewed in Paraskevopoulou and Falcone, 2018). The net charges are positively associated with the solubilizing effect and the polyarginine tags perform better than the polylysine tags, especially those located at the C-terminus of the partner protein. Unfortunately, the C-terminal location of VN failed to enhance the expression level of GDF8 and ECH proteins. Interestingly, the improvement in expression levels on recombinant proteins of most polycationic tags was negligible, such as diarginine (R2), hexaarginine (R6), decaarginine (R10), and decalysine (K10) (Jung et al., 2011). Since almost all of the classic SPs are comprised of a net positively charged region at the N-terminus (Owji et al., 2018), there must be a large number of VN-like candidates that would be used as the production yield and solubility dual-enhancers directly.
The negative impact of the regions of Vasp on the solubility of eGFP cannot be neutralized by EcSecB, a common chaperone in the Sec pathway (Chatzi et al., 2013). EcSecB also assists in the folding of some cytosolic clients such as barnase and MBP (Randall and Hardy, 2002; Ullers et al., 2004). EcSecA plays a crucial role in transferring the preprotein into the inner membrane channel formed by the EcSecYEG complex (Eser and Ehrmann, 2003). This protein binds to either the SPs or the mature domains of the client proteins and even interacts with the nascent polypeptides without the participation of the EcSecB or EcTF (Huber et al., 2017). The roles and their action orders of the crucial chaperones involved in the Sec pathway are intricate but employ different routes to the same destination (summarized in De Geyter et al., 2020). Briefly, the nascent peptides are recognized and bound by EcSecB or EcSecA. The binding prevents the aggregation of the client proteins and facilitates the next interactions. For instance, the EcSecB:client complex targets the free or bound EcSecA on EcSecYEG translocon directly. The binding of EcSecA to the nascent peptide was also reported, and the EcSecA:client will be anchored by the EcSecYEG translocon directly or after combining with EcSecB. CsaA is the counterpart of SecB in many gram-positive bacteria, such as Bacillus subtilis and Bt, as well as in most of archaea (Müller et al., 2000a,b; Sala et al., 2013). In B. subtilis, BsCsaA is a replacement of SecB to transport some preproteins such as ProOmpA and PrePhoB. In the present study, the effect of BtCsaA on the VeGFP was investigated, but no visible change in the client solubility was obtained. Neither EcSecB nor BtSecA was able to prevent the products of VeGFP from aggregation. To exclude the possible interference resulting from the interaction between EcSecB and EcSecA in vivo, the performances of the variants of EcSecB including SecB7577 and SecB142, as well as their combination SecB142-7577, were tested. Unfortunately, all of them also failed to keep the VeGFP soluble.
There should be another interaction model accounting for the translocation of Vip3. TF is a versatile chaperone, which either participates in the folding processes or anti-aggregation of several proteins (Patzelt et al., 2001; Bhandari and Houry, 2015; Saio et al., 2018; De Geyter et al., 2020), or surprisingly regulates the degradation of some newly synthesized proteins (Rizzolo et al., 2021). More importantly, it is a crucial chaperone in the Sec pathway. Recently, the following TF-associated models were elaborated in E. coli (Müller et al., 2000a; De Geyter et al., 2020): (1) TF-only model, named by the present study: The interaction of the client protein with TF (TF:client) is enough for settling down on the SecYEG-SecA translocase; (2) TF:bound-SecB model: The TF:client targets the translocase bound with the SecB already; (3) TF:free-SecB model: the TF:client is recognized and bound by the free SecB in the cytosol, and afterward, the triple complex is transported to the translocase; and (4) free-SecB:TF model: The SecB:client combination described above can interact with TF prior to reaching the location at the translocase. The contribution of BtTF to the solubilization of VeGFP and its variants supported the models and implicated that the combination between Vasp and BtTF would be preferential. It is important to note that TF is also associated with the ribosome located near the exit tunnel and plays a crucial role in the de novo folding of many cytosolic proteins by interaction with the nascent peptides (reviewed in Bhandari and Houry, 2015). Interestingly, EcTF, which shares a low identity with BtTF, did not affect the solubility of Vasp or its variants guided recombinant proteins. The interactions between Vasp or its variants and BtTF are worthy of further investigation.
In conclusion, each region of Vasp could be used as a fusion tag to enhance the production yield of eGFP. This may promote a new approach to be developed for the natural secretory peptides. The corresponding region(s) of the Vasp in the fusion proteins also attracted the interaction with BtTF, leading to the higher expression without compromise of the solubility. The solubility enhancement effects of a given fusion tag vary significantly depending on the cargo proteins per se, and unfortunately, the variations are not predictable so far. Therefore, the coexpression of the region(s) of Vasp, especially the VN harboring less hydrophobic residues, and BtTF would be an ideal and predictable method to enhance the production yield and solubility of recombinant proteins simultaneously in the prokaryotic expression system.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
CX, XW, and JG conceived of the presented idea and planned the experiments. JG, CO, JZ, and CX analyzed the data. JG, CO, XW, and CX prepared the manuscript. CO, JZ, YH, QG, TZ, and XL constructed the vectors and analyzed the protein expression. CO, JZ, and XL investigated fluorescent intensity. JG, CO, JZ, and MD took photographs with a confocal microscope. All authors reviewed the manuscript.
Funding
This work was supported by Shanxi Key Laboratory of Minor Crops Germplasm Innovation and Molecular Breeding, Shanxi Agricultural University (Project 202105D121010) and the National Natural Science Foundation of China (Project 31601690).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We wish to thank Prof. Tracy Palmer from the Centre for Bacterial Cell Biology at Newcastle University for her kind gifting of the MC4100 strain.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.892428/full#supplementary-material
References
Anné, J., Economou, A., and Bernaerts, K. (2017). “Protein secretion in gram-positive bacteria: from multiple pathways to biotechnology”, in Protein and Sugar Export and Assembly in Gram-positive Bacteria, eds F. Bagnoli, and R. Rappuoli (Cham: Springer International Publishing), 267–308.
Beena, K., Udgaonkar, J. B., and Varadarajan, R. (2004). Effect of signal peptide on the stability and folding kinetics of maltose binding protein. Biochemistry 43, 3608–3619. doi: 10.1021/bi0360509
Berks, B. C. (2015). The twin-arginine protein translocation pathway. Annu. Rev. Biochem. 84, 843–864. doi: 10.1146/annurev-biochem-060614-034251
Bhandari, V., and Houry, W. A. (2015). “Substrate interaction networks of the Escherichia coli chaperones: trigger factor, DnaK and GroEL”, in Prokaryotic Systems Biology, eds P. N. J. Krogan, and P. M. Babu (Cham: Springer International Publishing), 271–294.
Chakrabarty, S., Jin, M., Wu, C., Chakraborty, P., and Xiao, Y. (2020). Bacillus thuringiensis vegetative insecticidal protein family Vip3A and mode of action against pest Lepidoptera. Pest Manage. Sci. 76, 1612–1617. doi: 10.1002/ps.5804
Chakroun, M., Banyuls, N., Bel, Y., Escriche, B., and Ferre, J. (2016). Bacterial vegetative insecticidal proteins (vip) from entomopathogenic bacteria. Microbiol. Mol. Biol. Rev. 80, 329–350. doi: 10.1128/MMBR.00060-15
Chatzi, K. E., Sardis, M. F., Karamanou, S., and Economou, A. (2013). Breaking on through to the other side: protein export through the bacterial Sec system. Biochem. J. 449, 25–37. doi: 10.1042/BJ20121227
Chen, J., Yu, J., Tang, L., Tang, M., Shi, Y., and Pang, Y. (2003). Comparison of the expression of Bacillus thuringiensis full-length and N-terminally truncated vip3A gene in Escherichia coli. J. Appl. Microbiol. 95, 310–316. doi: 10.1046/j.1365-2672.2003.01977.x
Costa, S., Almeida, A., Castro, A., and Domingues, L. (2014). Fusion tags for protein solubility, purification and immunogenicity in Escherichia coli: the novel Fh8 system. Front. Microbiol. 5, 63. doi: 10.3389/fmicb.2014.00063
Costa, S. J., Almeida, A., Castro, A., Domingues, L., and Besir, H. (2013). The novel Fh8 and H fusion partners for soluble protein expression in Escherichia coli: a comparison with the traditional gene fusion technology. Appl. Microbiol. Biotechnol. 97, 6779–6791. doi: 10.1007/s00253-012-4559-1
Costa, T. R. D., Felisberto-Rodrigues, C., Meir, A., Prevost, M. S., Redzej, A., Trokter, M., et al. (2015). Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat. Rev. Microbiol. 13, 343. doi: 10.1038/nrmicro3456
Crickmore, N., Berry, C., Panneerselvam, S., Mishra, R., Connor, T. R., and Bonning, B. C. (2020). A structure-based nomenclature for Bacillus thuringiensis and other bacteria-derived pesticidal proteins. J. Invertebr. Pathol. 186, 107438. doi: 10.1016/j.jip.2020.107438
De Geyter, J., Portaliou, A. G., Srinivasu, B., Krishnamurthy, S., Economou, A., and Karamanou, S. (2020). Trigger factor is a bona fide secretory pathway chaperone that interacts with SecB and the translocase. EMBO Rep. 21, e49054. doi: 10.15252/embr.201949054
Doss, V., Anup Kumar, K., Jayakumar, R., and Sekara, V. (2002). Cloning and expression of the vegetative insecticidal protein (vip3V) gene of Bacillus thuringiensis in Escherichia coli. Protein Expr. Purif. 26, 82–88. doi: 10.1016/S1046-5928(02)00515-6
Eser, M., and Ehrmann, M. (2003). SecA-dependent quality control of intracellular protein localization. Proc. Natl. Acad. Sci. U. S. A. 100, 13231–13234. doi: 10.1073/pnas.2234410100
Estruch, J., Warren, G., Mullins, M., Nye, G., Craig, J., and Koziel, M. (1996). Vip3A, a novel Bacillus thuringiensis vegetative insecticidal protein with a wide spectrum of activities against lepidopteran insects. Proc. Natl. Acad. Sci. U. S. A. 93, 5389–5394. doi: 10.1073/pnas.93.11.5389
Fekkes, P., de Wit, J. G., van der Wolk, J. P., Kimsey, H. H., Kumamoto, C. A., and Driessen, A. J. (1998). Preprotein transfer to the Escherichia coli translocase requires the co-operative binding of SecB and the signal sequence to SecA. Mol. Microbiol. 29, 1179–1190. doi: 10.1046/j.1365-2958.1998.00997.x
Gao, J., Qian, H., Guo, X., Mi, Y., Guo, J., Zhao, J., et al. (2020). The signal peptide of Cry1Ia can improve the expression of eGFP or mCherry in Escherichia coli and Bacillus thuringiensis and enhance the host's fluorescent intensity. Microb. Cell Fact. 19, 112. doi: 10.1186/s12934-020-01371-8
Green, E. R., and Mecsas, J. (2016). Bacterial secretion systems: an overview. Microbiol. Spectr. 4, VMBF-0012-2015. doi: 10.1128/microbiolspec.VMBF-0012-2015
Gupta, S. K., and Shukla, P. (2016). Advanced technologies for improved expression of recombinant proteins in bacteria: perspectives and applications. Crit. Rev. Biotechnol. 36, 1089–1098. doi: 10.3109/07388551.2015.1084264
Huber, D., Jamshad, M., Hanmer, R., Schibich, D., Döring, K., Marcomini, I., et al. (2017). SecA cotranslationally interacts with nascent substrate proteins in vivo. J. Bacteriol. 199, e00622–e00616. doi: 10.1128/JB.00622-16
Jong, W. S. P., Vikström, D., Houben, D., van den Berg van Saparoea, H. B., de Gier, J.-W., and Luirink, J. (2017). Application of an E. coli signal sequence as a versatile inclusion body tag. Microb. Cell Fact. 16, 50. doi: 10.1186/s12934-017-0662-4
Jung, H. J., Kim, S. K., Min, W. K., Lee, S. S., Park, K., Park, Y. C., et al. (2011). Polycationic amino acid tags enhance soluble expression of Candida antarctica lipase B in recombinant Escherichia coli. Bioprocess Biosystems Eng. 34, 833. doi: 10.1007/s00449-011-0533-z
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Ki, M., and Pack, S. (2020). Fusion tags to enhance heterologous protein expression. Appl. Microbiol. Biotechnol. 104, 2411–2425. doi: 10.1007/s00253-020-10402-8
Kleiner-Grote, G. R. M., Risse, J. M., and Friehs, K. (2018). Secretion of recombinant proteins from E. coli. Eng. Life Sci. 18, 532–550. doi: 10.1002/elsc.201700200
Krishnan, B., Kulothungan, S. R., Patra, A. K., Udgaonkar, J. B., and Varadarajan, R. (2009). SecB-mediated protein export need not occur via kinetic partitioning. J. Mol. Biol. 385, 1243–1256. doi: 10.1016/j.jmb.2008.10.094
Kulothungan, S. R., Das, M., Johnson, M., Ganesh, C., and Varadarajan, R. (2009). Effect of crowding agents, signal peptide, and chaperone SecB on the folding and aggregation of E. coli maltose binding protein. Langmuir 25, 6637–6648. doi: 10.1021/la900198h
Kyte, J., and Doolittle, R. F. (1982). A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132. doi: 10.1016/0022-2836(82)90515-0
Maina, C. V., Riggs, P. D., Grandea, A. G. III., Slatko, B. E., Moran, L. S., Tagliamonte, J. A., et al. (1988). An Escherichia coli vector to express and purify foreign proteins by fusion to and separation from maltose-binding protein. Gene 74, 365–373. doi: 10.1016/0378-1119(88)90170-9
Malik, A. (2016). Protein fusion tags for efficient expression and purification of recombinant proteins in the periplasmic space of E. coli. 3 Biotech. 6, 44. doi: 10.1007/s13205-016-0397-7
Marblestone, J. G., Edavettal, S. C., Lim, Y., Lim, P., Zuo, X., and Butt, T. R. (2006). Comparison of SUMO fusion technology with traditional gene fusion systems: enhanced expression and solubility with SUMO. Protein Sci. 15, 182–189. doi: 10.1110/ps.051812706
Müller, J. P., Bron, S., Venema, G., and Maarten van Dijl, J. (2000a). Chaperone-like activities of the CsaA protein of Bacillus subtilis. Microbiology 146, 77–88. doi: 10.1099/00221287-146-1-77
Müller, J. P., Ozegowski, J., Vettermann, S., Swaving, J., Van Wely, K. H. M., and Driessen, A. J. M. (2000b). Interaction of Bacillus subtilis CsaA with SecA and precursor proteins. Biochem. J. 348, 367–373. doi: 10.1042/bj3480367
Núñez-Ramírez, R., Huesa, J., Bel, Y., Ferré, J., Casino, P., and Arias-Palomo, E. (2020). Molecular architecture and activation of the insecticidal protein Vip3Aa from Bacillus thuringiensis. Nat. Commun. 11, 3974. doi: 10.1038/s41467-020-17758-5
Overton, T. W. (2014). Recombinant protein production in bacterial hosts. Drug Discov. Today 19, 590–601. doi: 10.1016/j.drudis.2013.11.008
Owji, H., Nezafat, N., Negahdaripour, M., Hajiebrahimi, A., and Ghasemi, Y. (2018). A comprehensive review of signal peptides: structure, roles, and applications. Eur. J. Cell Biol. 97, 422–441. doi: 10.1016/j.ejcb.2018.06.003
Paraskevopoulou, V., and Falcone, F. (2018). Polyionic tags as enhancers of protein solubility in recombinant protein expression. Microorganisms 6, 47. doi: 10.3390/microorganisms6020047
Patzelt, H., Rüdiger, S., Brehmer, D., Kramer, G., Vorderwülbecke, S., Schaffitzel, E., et al. (2001). Binding specificity of Escherichia coli trigger factor. Proc. Natl. Acad. Sci. U. S. A. 98, 14244–14249. doi: 10.1073/pnas.261432298
Randall, L. L., Crane, J. M., Lilly, A. A., Liu, G., Mao, C., Patel, C. N., et al. (2005). Asymmetric binding between SecA and SecB two symmetric proteins: implications for function in export. J. Mol. Biol. 348, 479–489. doi: 10.1016/j.jmb.2005.02.036
Randall, L. L., and Hardy, S. J. S. (2002). SecB, one small chaperone in the complex milieu of the cell. Cell. Mol. Life Sci. 59, 1617–1623. doi: 10.1007/PL00012488
Rang, C., Gil, P., Neisner, N., Van Rie, J., and Frutos, R. (2005). Novel Vip3-related protein from Bacillus thuringiensis. Appl. Environ. Microbiol. 71, 6276–6281. doi: 10.1128/AEM.71.10.6276-6281.2005
Rinas, U., Garcia-Fruitós, E., Corchero, J. L., Vázquez, E., Seras-Franzoso, J., and Villaverde, A. (2017). Bacterial inclusion bodies: discovering their better half. Trends Biochem. Sci. 42, 726–737. doi: 10.1016/j.tibs.2017.01.005
Rizzolo, K., Yu, A. Y. H., Ologbenla, A., Kim, S. R., Zhu, H., Ishimori, K., et al. (2021). Functional cooperativity between the trigger factor chaperone and the ClpXP proteolytic complex. Nat. Commun. 12, 281. doi: 10.1038/s41467-020-20553-x
Robert, X., and Gouet, P. (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324. doi: 10.1093/nar/gku316
Rosano, G. L., and Ceccarelli, E. A. (2014). Recombinant protein expression in Escherichia coli: advances and challenges. Front. Microbiol. 5, 172. doi: 10.3389/fmicb.2014.00172
Saio, T., Kawagoe, S., Ishimori, K., and Kalodimos, C. G. (2018). Oligomerization of a molecular chaperone modulates its activity. Elife 7, e35731. doi: 10.7554/eLife.35731
Sala, A., Calderon, V., Bordes, P., and Genevaux, P. (2013). TAC from Mycobacterium tuberculosis: a paradigm for stress-responsive toxin–antitoxin systems controlled by SecB-like chaperones. Cell Stress Chaperones 18, 129–135. doi: 10.1007/s12192-012-0396-5
Sala, A. J., Bordes, P., Ayala, S., Slama, N., Tranier, S., Coddeville, M., et al. (2017). Directed evolution of SecB chaperones toward toxin-antitoxin systems. Proc. Natl. Acad. Sci. U. S. A. 114, 12584–12589. doi: 10.1073/pnas.1710456114
Singh, P., Sharma, L., Kulothungan, S. R., Adkar, B. V., Prajapati, R. S., Ali, P. S. S., et al. (2013). Effect of signal peptide on stability and folding of Escherichia coli thioredoxin. PLoS ONE 8, e63442. doi: 10.1371/journal.pone.0063442
Smith, D. B., and Johnson, K. S. (1988). Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67, 31–40. doi: 10.1016/0378-1119(88)90005-4
Syed, T., Askari, M., Meng, Z., Li, Y., Abid, M. A., Wei, Y., et al. (2020). Current insights on vegetative insecticidal proteins (vip) as next generation pest killers. Toxins 12, 522. doi: 10.3390/toxins12080522
Ullers, R. S., Luirink, J., Harms, N., Schwager, F., Georgopoulos, C., and Genevaux, P. (2004). SecB is a bona fide generalized chaperone in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 101, 7583–7588. doi: 10.1073/pnas.0402398101
Vandemoortele, G., Eyckerman, S., and Gevaert, K. (2019). Pick a tag and eplore the functions of your pet protein. Trends Biotechnol. 37, 1078–1090. doi: 10.1016/j.tibtech.2019.03.016
Keywords: Vip3, Bacillus thuringiensis, signal peptide, fusion tag, eGFP, Bt trigger factor
Citation: Gao J, Ouyang C, Zhao J, Han Y, Guo Q, Liu X, Zhang T, Duan M, Wang X and Xu C (2022) Coexpressing the Signal Peptide of Vip3A and the Trigger Factor of Bacillus thuringiensis Enhances the Production Yield and Solubility of eGFP in Escherichia coli. Front. Microbiol. 13:892428. doi: 10.3389/fmicb.2022.892428
Received: 09 March 2022; Accepted: 21 June 2022;
Published: 18 July 2022.
Edited by:
Yuxian Xia, Chongqing University, ChinaReviewed by:
Hongbo Li, Jishou University, ChinaMin He, Sichuan Agricultural University, China
Yunjun Sun, Hunan Normal University, China
Copyright © 2022 Gao, Ouyang, Zhao, Han, Guo, Liu, Zhang, Duan, Wang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chao Xu, zjuxuchao@163.com; Xingchun Wang, wxingchun@163.com
†These authors have contributed equally to this work and share first authorship
 Jianhua Gao
Jianhua Gao Chunping Ouyang
Chunping Ouyang Juanli Zhao
Juanli Zhao Yan Han
Yan Han Qinghua Guo
Qinghua Guo Xuan Liu
Xuan Liu Tianjiao Zhang
Tianjiao Zhang Ming Duan
Ming Duan Xingchun Wang
Xingchun Wang Chao Xu
Chao Xu