- 1Department of Clinical Laboratory, State Key Laboratory of Complex Severe and Rare Diseases, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
- 2Beijing Key Laboratory for Mechanisms Research and Precision Diagnosis of Invasive Fungal Diseases, Beijing, China
- 3Nanjing Hospital of Chinese Medicine Affiliated to Nanjing University of Chinese Medicine, Nanjing, China
- 4School of Biomedical Sciences, Charles Sturt University, Orange, NSW, Australia
- 5NSW Health Pathology, Regional and Rural, Orange Hospital, Orange, NSW, Australia
- 6Department of Infectious Disease, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences, Beijing, China
Streptococcus pneumoniae is a common human pathogen that can cause severe invasive pneumococcal diseases (IPDs). Penicillin-binding proteins (PBPs) are the targets for β-lactam antibiotics (BLAs), which are the common empirical drugs for treatment of pneumococcal infection. This study investigated the serotype distribution and antibiotic resistance patterns of S. pneumoniae strains causing IPD in China, including exploring the association between penicillin (PEN) susceptibility and PBPs variations. A total of 300 invasive S. pneumoniae isolates were collected from 27 teaching hospitals in China (2010-2015). Serotypes were determined by Quellung reaction. Serotypes 23F and 19F were the commonest serotypes in isolates from cerebrospinal fluid (CSF), whilst serotypes 19A and 23F were most commonly seen in non-CSF specimens. Among the 300 invasive S. pneumoniae strains, only one strain (serotype 6A, MIC = 0.25 μg/ml) with PEN MIC value ≤ 0.25 μg/ml did not have any substitutions in the PBPs active sites. All the strains with PEN MIC value ≥ 0.5 μg/ml had different substitutions within PBPs active sites. Substitutions in PBP2b and PBP2x active sites were common in low-level penicillin-resistant S. pneumoniae (PRSP) strains (MIC = 0.5 μg/ml), with or without PBP1a substitution, while all strains with PEN MIC ≥ 1 μg/ml had substitutions in PBP1a active sites, accompanied by PBP2b and PBP2x active site substitutions. Based on the three PBPs substitution combinations, a high degree of diversity was observed amongst the isolates. This study provides some new insights for understanding the serology and antibiotic resistance dynamics of S. pneumoniae causing IPD in China. However, further genomic studies are needed to facilitate a comprehensive understanding of antibiotic resistance mechanisms of S. pneumoniae.
Introduction
Streptococcus pneumoniae is one of the most common Gram-positive cocci that is mainly transmitted through the respiratory tract. The organism usually colonizes the human nasopharynx and can migrate to the middle ear and lungs causing local non-invasive pneumococcal disease (NIPD) such as otitis media and pneumonia in immune-deficient people (Lynch and Zhanel, 2009; Henriques-Normark and Tuomanen, 2013). Data from the World Health Organization (WHO) shows that pneumonia killed 808, 694 children under five years old in 2017, accounting for 15% of all deaths in children. S. pneumoniae is the most common pneumonia pathogen in children worldwide, with a mortality rate in children much higher than other diseases such as AIDS, malaria, and measles (Lynch and Zhanel, 2009). In Europe and America, S. pneumoniae is also the most common cause of community-acquired pneumonia in adults (World Health Organization [WHO], 2007). In addition to respiratory tract infections, S. pneumoniae can also migrate to the blood and brain and cause severe invasive pneumococcal disease (IPD), such as bacteremia, meningitis etc., causing a huge economic and medical burden on both developed and developing countries (Mehr and Wood, 2012).
The major empirical antimicrobial drugs used in the treatment of S. pneumoniae infections are β-lactam antibiotics (BLAs), which act on the bacterial cell wall. Penicillin-binding proteins (PBPs) are crucial enzymes in the biosynthesis of peptidoglycan (PG), a major cell wall component that surrounds the cytoplasmic membrane and is required to maintain the shape and osmotic stability of bacteria (Hakenbeck et al., 2012). The target of BLAs are PBPs, and function by covalently binding to the active site serine of PBPs through the β-lactam ring, thereby interfering with the synthesis of bacterial cell walls and eventually leading to bacterial cell death. With the widespread use of antibiotics, penicillin-intermediate S. pneumoniae (PISP) and penicillin-resistant S. pneumoniae (PRSP), commonly referred to as penicillin-non-susceptible S. pneumoniae (PNSP), have emerged and are detected continually worldwide. Data from the Asian Network for Surveillance of Resistant Pathogens (ANSORP) shows that the isolation rate of PNSP from 2012 to 2017 (9.0%) was significantly higher than that from 2008 to 2009 (4.9%), and the detection rates of PNSP from patients in China was 1.9% (2012-2017, oral breakpoint) (Kim et al., 2012, 2020).
The main mechanism of BLAs resistance by S. pneumoniae is through PBPs substitutions. Alterations in PBPs via substitutions reduce their reactivity for β-lactam attachment to the binding site and thereby reduce their effectiveness (Zapun et al., 2008). S. pneumoniae has six PBPs, but only three PBPs, PBP2x, PBP2b and PBP1a play a main role in BLAs resistance. Alterations in all other PBPs have been described occasionally (Zapun et al., 2008). Mosaicity is the product of homologous recombination that causes the sequence diversity in S. pneumoniae. In most resistant clinical S. pneumoniae isolates, the sequencing revealed that the mosaic genes encode PBP2x, PBP2b, and PBP1a (Laible et al., 1991; Martin et al., 1992; Sibold et al., 1994). The active sites of PBPs comprises three conserved sequences SXXK, SXN and KT(S)G. The serine of the SXXK motif is the active site residue that reacts with BLAs. They are located in PBP2x: 337STMK340, 395SSN397, 547KTG549; in PBP2b: 386STMK389, 443SSN445, and 615KTG617; and in PBP1a: 370STMK373, 428SRN430, 557KTG559 (Hakenbeck et al., 2012). Changes in these active site motifs and their adjacent sequences lead to a decrease in the affinity of PBPs to penicillin resulting in antibiotic resistance (Zhanel et al., 2006; Hakenbeck et al., 2012). Few studies have been carried out to understand the association between penicillin susceptibility and PBPs variations in S. pneumoniae isolates from patients with IPD in mainland China.
Due to the widespread use of antimicrobial drugs, and the fastidious nature of S. pneumoniae, the number of isolates from IPD specimens is very low, and hence a scarcity of relevant research studies (Xue et al., 2010; Yao et al., 2011; Quan-Cheng et al., 2016). This study aimed to analyze the serotype distribution and antibiotic resistance pattern of S. pneumoniae strains causing IPD in China, and to explore the association between penicillin susceptibility and PBPs variations.
Materials and Methods
Bacterial Isolates
A total of 300 non-duplicate invasive S. pneumoniae isolates from 27 teaching hospitals in 13 provinces of China (2010-2015) were studied (Supplementary Table 1). The isolates were transported to the Department of Clinical Laboratory in Peking Union Medical College Hospital for re-identification and further analysis. The most common specimen type was blood, accounting for 72.7% (218/300) of the isolates, followed by cerebrospinal fluid (CSF) (19.0%, 57/300) and pleural effusion (5.7%, 17/300). Other specimen types included ascites, joint drainages, pleural drainage and lung tissue, each accounting for ≤ 2.0% of the isolates. The majority of the patients were males, accounting for 65.7% (197/300) of the isolates. The average age of patients was 46 ± 26.69 years old.
Serotyping
All the S. pneumoniae isolates were serotyped by Quellung reaction (Maha et al., 2014). The serotype was first determined by latex agglutination test using the checkerboard typing system. Then specific type antiserum was mixed with the bacterial suspension to determine the final serotype. Capsular swelling was observed under oil immersion microscope. If the test strain was negative with all antisera, it was classified as non-typeable (NT).
Antimicrobial Susceptibility Testing
The minimum inhibitory concentrations (MICs) of S. pneumoniae against penicillin (PEN), amoxicillin/clavulanic (AMC), cefuroxime (CXM), ceftriaxone (CRO), cefepime (FEP), ertapenem (ETP), imipenem (IPM), meropenem (MEM), levofloxacin (LEV), trimethoprim/sulfamethoxazole (SXT), clindamycin (DA), clarithromycin (CLA), erythromycin (E), linezolid (LZD) and vancomycin (VA), were determined by broth microdilution method as recommended by the Clinical and Laboratory Standards Institute (CLSI) M07-A10 (Clinical and Laboratory Standards Institute (CLSI), 2012). S. pneumoniae ATCC49619 and Escherichia coli ATCC25922 were used as the quality control strains, and were tested along each batch. Results were considered valid when the MIC values of the quality control strains were within the expected range. Antimicrobial susceptibility testing results were interpreted according to CLSI 2019 guidelines (Clinical and Laboratory Standards Institute (CLSI), 2019).
Penicillin-Binding Protein Gene Amplification and Sequencing
The isolates were cultured on blood agar plates and incubated overnight at 35°C in a 5% CO2 atmosphere. DNA was extracted using the AxyGen amp DNA Mini Extraction Kit (Axygen, United States) according to the manufacturer’s instructions. The final pure DNA was stored at −20°C until use. The nucleotide sequence of an around1-kb region encoding the penicillin-binding domain of pbp2x, pbp2b and pbp1a genes were amplified and sequenced based on published primers (Table 1; Zhanel et al., 2006). PCR products were sent to Beijing RuiBiotech Co., Ltd., for sequencing.

Table 1. Primers for amplification and sequencing for the region encoding the penicillin-binding domain of pbp2x, pbp2b and pbp1a genes.
CLC Sequence Viewer software (CLC, Denmark) was used to manually correct the sequencing peaks and sequences to ensure the sequencing quality, and a two-way splicing was carried out. The spliced sequences were translated to simulated protein sequences and compared with PBP2x, PBP2b, and PBP1a corresponding to S. pneumoniae reference strain R6 (PSSP, GenBank accession No. NC003098) for variation analysis.
Data Analysis and Statistical Analysis
Differences in antimicrobial susceptibility were analyzed by MIC range, MIC50 and MIC90, and statistical analysis was performed by chi-square test or Fisher’s exact probability test using SPSS software (version 22.0, SPSS Inc., Chicago, IL, United States). P value < 0.05 was considered statistically significant.
Results
Serotype Distribution
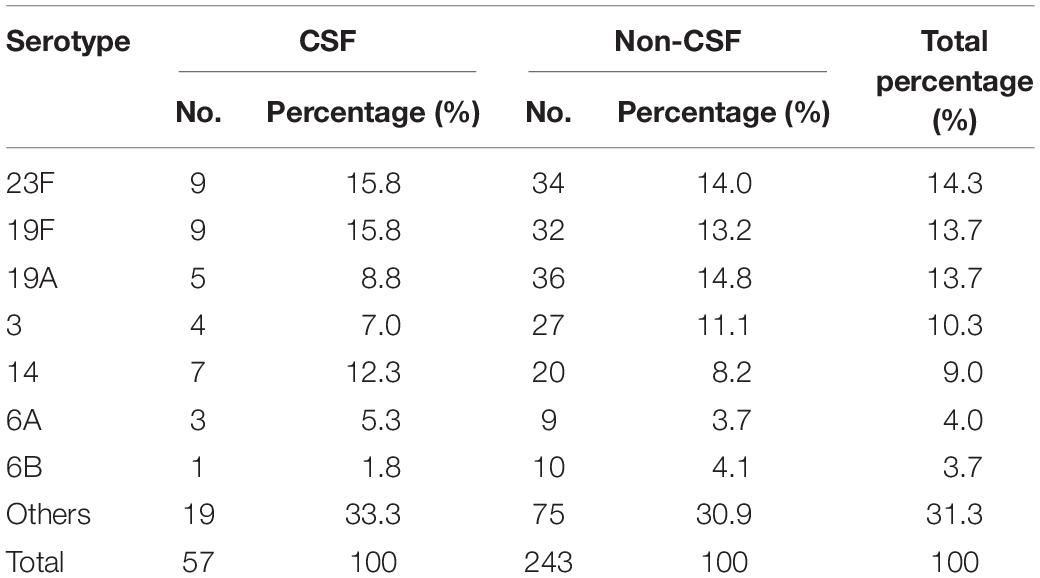
Based on the Quellung reaction, a total of 299 S. pneumoniae isolates were identified to the serotype level accurately and one strain was considered as non-typeable (NT). Among the 299 serotypeable isolates, 41 serotypes were detected. The top five serotypes were: 23F (14.3%, 43/300), 19F (13.7%, 41/300), 19A (13.7%, 41/300), 3 (10.3%, 31/300) and 14 (9.0%, 27/300). The main serotypes in CSF isolates were 23F and 19F, both accounting for 15.8% (9/57) each, whilst in non-CSF specimens, serotypes 19A and 23F were the most common, accounting for 14.8% (36/243) and 14.0% (34/243) of the isolates, respectively (Table 2).
Antimicrobial Susceptibility
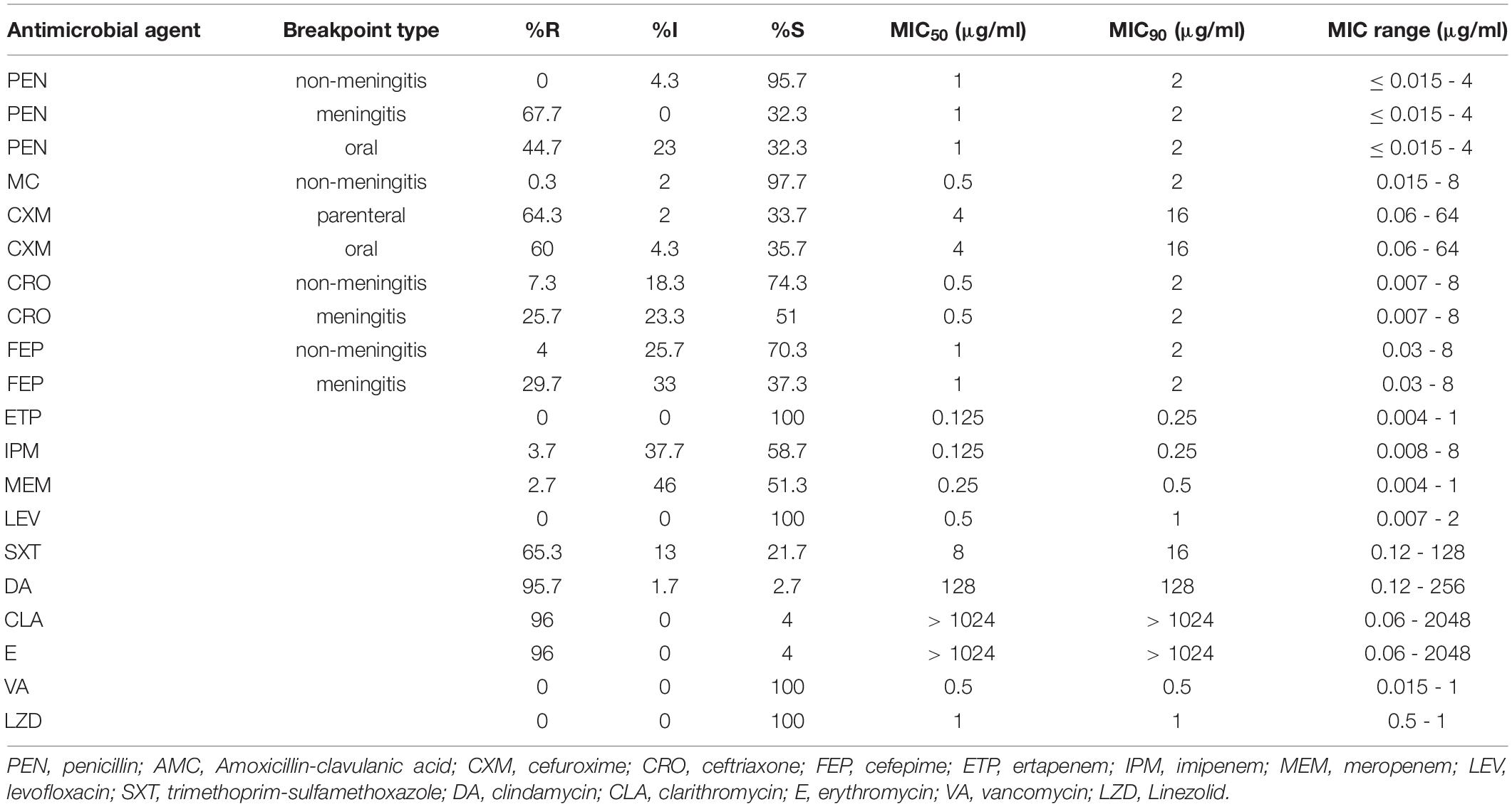
Concerning non-BLAs drugs, all the S. pneumoniae isolates were susceptible to LEV, VA and LZD, with MIC90 values of 1 μg/ml, 0.5 μg/ml, and 1 μg/ml. The prevalence of resistance of the isolates to SXT was 65.3%. Resistance rates to CLA, E and DA were extremely high, all above 90%. PISP accounted for 4.3% of the isolates based on the non-meningitis (R ≥ 8 μg/ml) breakpoint while none, 67.7% and 44.7% of the isolates were classified as PRSP based on non-meningitis, meningitis (R ≥ 0.12 μg/ml) and oral administration (R ≥ 2 μg/ml) breakpoints, respectively. Susceptibility to AMC was about 97.7%. The prevalence of resistance of the isolates to the second-generation cephalosporins CXM was about ≥60%. The third- and fourth-generation cephalosporins CRO and FEP had the same MIC90 value of 2 μg/ml. The resistance rates of the isolates to IPM and MEM was only 3.7 and 2.7%, respectively, but the intermediate rates were much higher at 37.7 and 46.0%, respectively (Table 3).
Association Between Serotypes, Penicillin Susceptibility and Variations in Penicillin-Binding Proteins Active Sites
Among the 300 isolates studied, 106 isolates, including all strains of serotypes1-5, 6C, 7F, 9A, 9N, 9V, 8, 17, 34, 10A, 11A, 12F, 15F, 17A, 18C, 24F, 25A, 25F, 28A, and 28F, one strain each of serotypes 6A, 7C, 19A and 23F, and two strains each of serotypes 6B, 13, 29, and 15A, exhibited PEN MIC values of ≤0.25 μg/ml. None of these isolates had PBPs substitution in the three conserved motifs and nearby sites, except one isolate of serotype 6A (PEN MIC = 0.25 μg/ml), in which TAA6A substitution was detected in the active sites of PBP2b. The remaining 194 isolates all had different amino acid substitutions in the PBPs active sites. In the PBP2x conserved motifs or nearby sites, 181 isolates had T338A substitution (threonine → alanine), 175 isolates had L546V substitution (leucine → valine), and 39 isolates had M339F substitution (methionine → phenylalanine), 4 isolates had H394L substitution (histidine → leucine), and the substitution rates were 60.3, 58.3, 13, and 1.3%, respectively. In the PBP2b conserved motifs or nearby sites, 190 isolates had T446A substitution (threonine → alanine), 3 isolates had T446S substitution (threonine → serine), and 72 isolates had A619G substitution (alanine → glycine), with substitution rates of 63.3, 1, and 24%, respectively. In the PBP1a conserved motifs or nearby sites, 53 isolates had T371A substitution (threonine → alanine), 122 isolates had T371S substitution (threonine → serine), and 175 isolates had P432T substitution (proline → threonine), with substitution rates of 17.7, 40.7, and 58.3%, respectively (Table 4).
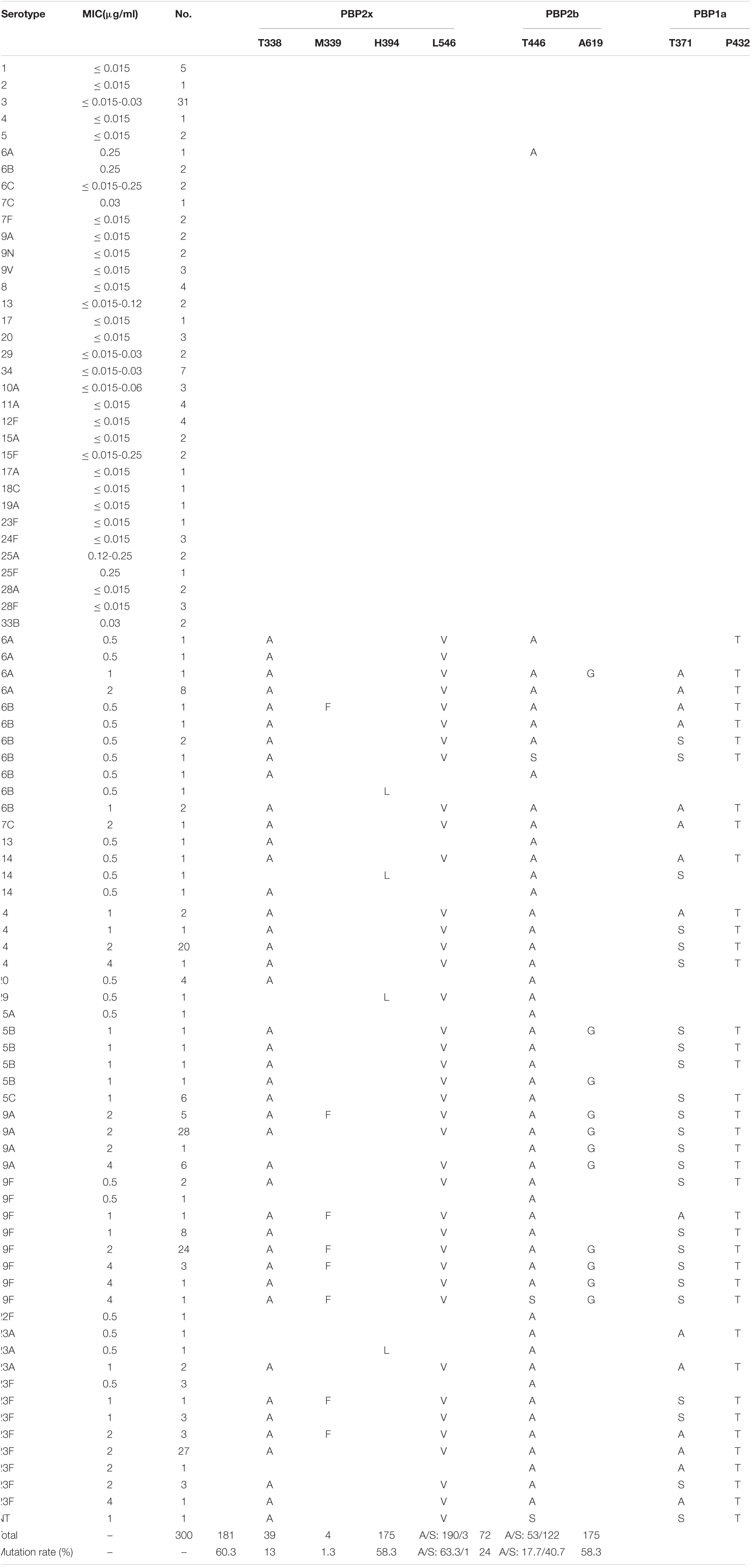
Table 4. Association of serotypes, penicillin susceptibility and variations of conserved motifs forming or surrounding active penicillin-binding proteins (PBPs) binding sites in PBP2x, PBP2b, PBP1a among 300 S. pneumoniae isolates.
Based on the serotype and PEN MIC distribution among the isolates, ten isolates had only one PBP gene active site or nearby sites substitution, including two isolates each of serotype 6A and 6B, one isolate each of serotypes 19F, 22 F and 23A, and three isolates of serotype 23F. Seven of these isolates had PBP2b active sites or nearby sites substitutions; two had only PBP2x active sites or nearby sites substitutions, while none had any substitution in single PBP1a active sites or nearby sites. Save for one isolate of serotype 6A with PEN MIC of 0.25 μg/ml, all these strains had an MIC of 0.5 μg/ml. Thirteen isolates had substitutions in two of the PBPs gene active sites or nearby sites, including four isolates of serotype 20, two isolates of serotype 23A, and one isolate each for serotypes 6B, 13, 14, 29, 15B, 19A, and 23F. Among them, three isolates had substitutions in both PBP2b and PBP1a genes (serotypes 19A, 23F, and 23A), and the rest had substitution in PBP2x and PBP2b. The PEN MIC of most isolates in the group with two substitutions in the PBPs gene active sites or nearby sites was 0.5 μg/ml. The remaining 171 isolates had substitutions in the active sites or nearby sites of all the three PBPs genes, mainly distributed in serotypes 14 (n = 26), 19A (n = 39), 19F (n = 40) and 23F (n = 38).
Taken together, all isolates of serotypes 1, 2, 3, 4, 5, 6C, 7F, 9A, 9N, 9V, 8, 17, 34, 10A, 11A, 12F, 15F, 17A, 18C, 24F, 25A, 25F, 28A, 28F, and 33B, had no amino acid changes in the three active sites or nearby sites of PBPs (MIC ≤ 0.25 μg/ml), and all isolates of serotypes 14, 15B, 15C, 19F, 22F, and 23A, had at least one substitution in the active sites or nearby sites of PBPs (0.5 μg/ml ≤ MIC ≤ 4 μg/ml).
Association Between Specimen Types and Variations in Penicillin-Binding Proteins Active Sites
Among the 300 invasive S. pneumoniae isolates, 57 (19.0%) were isolated from CSF, and the rest (81.0%) from other sterile body fluids, mostly in blood. Antimicrobial susceptibilities were interpreted according to meningitis and non-meningitis break points. PRSP and PSSP from CSF accounted for 70.2 and 29.8% of the isolates, respectively. PISP and PSSP from non-CSF accounted for 4.5 and 95.5% of the isolates, respectively. Although the proportion of PRSP derived from CSF was significantly higher than that from non-CSF sources (P < 0.0001), the proportion of CSF derived isolates with PEN MIC ≥ 1μg/ml (59.6%) was similar to that from non-CSF sources (54.3%) (P = 0.5644) (Figure 1).
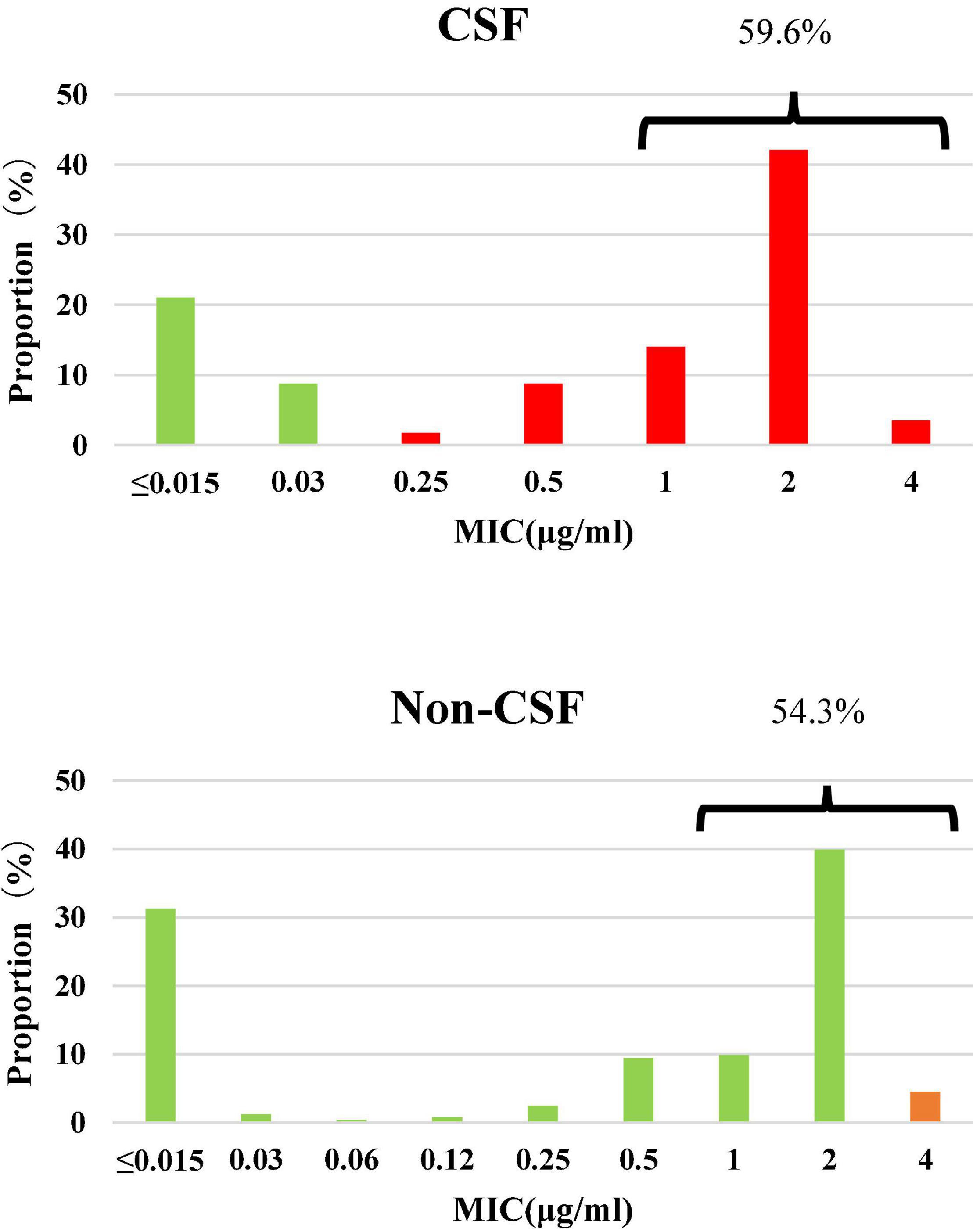
Figure 1. Susceptibility and MIC distribution against penicillin in 300 S. pneumoniae isolates. Green: susceptible, orange: intermediate, red: resistant. CSF: PSSP 29.8%, PISP 0%, PRSP 70.2%; Non-CSF: PSSP 95.5%, PISP 4.5%, PRSP 0%.
Analysis of PBPs active sites of strains from CSF showed that all PSSP, and one strain of PRSP, with PEN MIC of 0.25 μg/ml, had no substitution in the PBPs active sites. PBP2x and PBP2b substitutions were common in low-level PRSP strains (MIC = 0.5μg/ml), with or without PBP1a substitution. The strains with PEN MIC ≥ 1 μg/ml all had substitutions in the PBP1a active site or nearby sites, accompanied by substitutions in the PBP2x and PBP2b regions (Table 5).
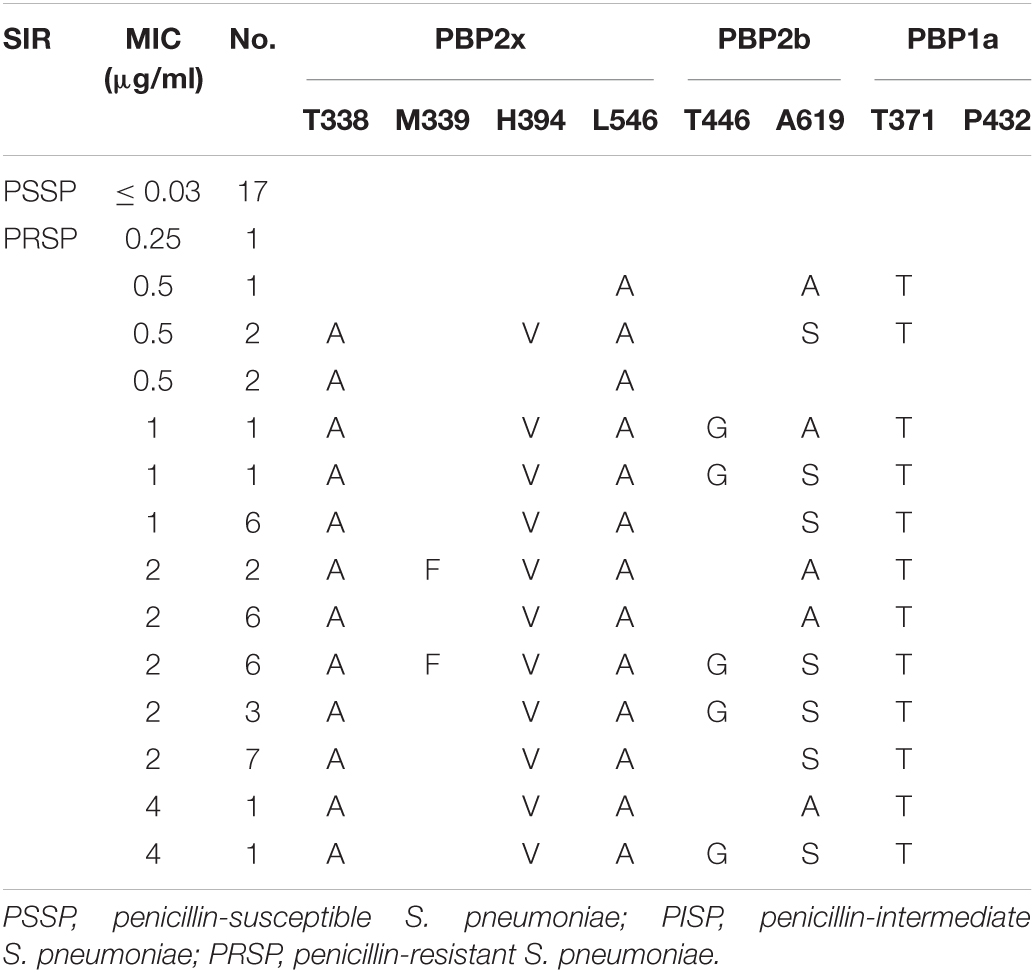
Table 5. Association of penicillin susceptibility and variations of conserved motifs forming or surrounding active penicillin-binding proteins (PBPs) binding sites in PBP2x, PBP2b and PBP1a in S. pneumoniae isolates from CSF.
Analysis of PBPs active sites or nearby sites of strains derived from non-CSF sources revealed that all PSSP with PEN MIC ≤ 0.25 μg/ml had no substitutions in PBPs active sites or nearby sites, except one serotype 6A strain, in which a T446A substitution nearby the active site of PBP2b was detected. Various substitutions in the PBPs active sites or nearby sites of PSSP strains with PEN MIC ≥ 0.25 μg/ml were detected. Similar to CSF derived strains, the substitutions in active sites or nearby sites of PBP2x and PBP2b were common in strains with low PEN MIC level, with or without PBP1a active site substitution. In contrast, strains with PEN MIC ≥ 1 μg/ml all had substitutions in PBP1a active sites or nearby sites, accompanied by PBP2x or PBP2b substitutions (Table 6).

Table 6. Association of penicillin susceptibility and variations of conserved motifs forming or surrounding active penicillin-binding proteins (PBPs) binding sites in PBP2x, PBP2b, PBP1a in S. pneumoniae isolates from non-CSF specimens.
Variations of the pbp2x Gene
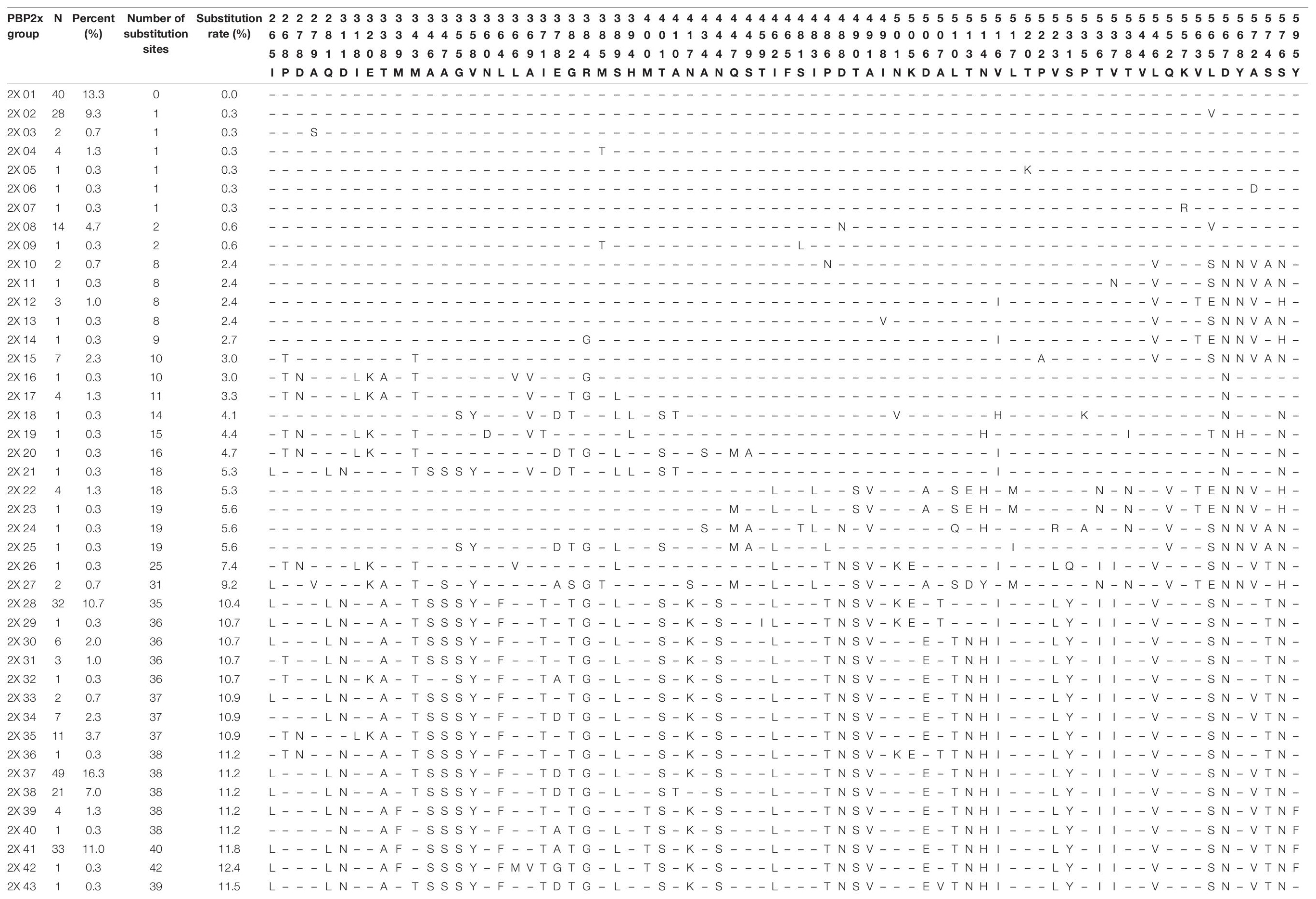
Compared with the reference strain R6 (GenBank accession No. NC003098), a total of 338 amino acids from positions 259 to 596 of the PBP2x protein were analyzed. A total of 98 different substitutions at 73 amino acid positions were detected in the PBP2x sequence, among which D567N (Asp → Asn) was the most common substitution, accounting for 69.3% (208/300), followed by D488N (Asp → Asn) and S576N (Ser → Asn), each accounting for 64.0% (192/300). Strains with L565S (Leu → Ser) and R384G (Arg → Gly) substitutions each accounted for 62.7% (188/300) and 61.3% (184/300), respectively. According to the distribution of various substitutions in PBP2x, all strains could be divided into 43 groups. The number of substitution sites in each group ranged from 1 to 42, accounting for 0-12.4% of the total number of amino acids analyzed. Group 2X37 was the commonest, accounting for 16.3% of all strains (49/300). There were 38 substitutions detected in all strains within the group, and the substitution rate was 11.2%. The second common group was 2X01, accounting for 13.3% of all strains (40/300), and the PBP2x sequence of strains in this group was exactly the same as that of R6 strain. There were 33 strains in the 2X41 group, accounting for 11.0% of the strains. Forty substitutions were detected in all strains of this group and the substitution rate was 11.8% (Table 7). Compared with PBP2b and PBP1a sequences, PBP2x had the least number of total substitutions (98 vs. 112 vs. 105). According to the distribution of PEN MIC, the number of detectable substitution sites in strains with PEN MIC ≤ 0.25 μg/ml ranged from 10-32, while for strains with PEN MIC = 0.5, 1, 2, and 4 μg/ml the number of detectable substitutions were 86, 46, 57, and 48, respectively (Supplementary Table 2).
Variations of the pbp2b Gene
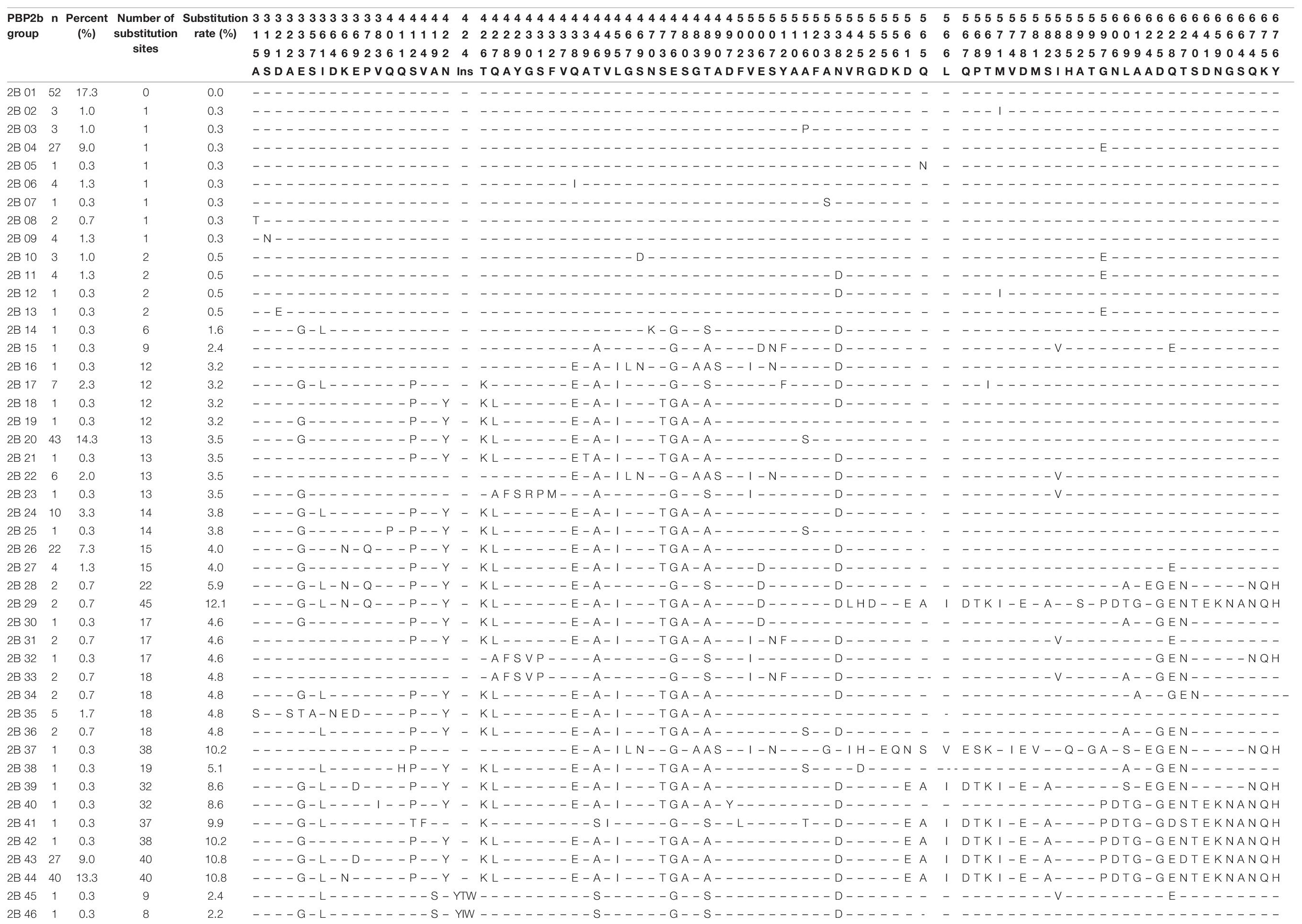
Compared with the reference strain R6 (GenBank accession No. NC003098), a total of 372 amino acids from positions 305 to 676 of the PBP2b protein were analyzed for the isolates. A total of 111 different substitutions and two insertion variations at 88 amino acid positions were detected in the PBP2b sequence. The two insertion variations were only found in two strains, both located between amino acids 424-425, and the insertion sequences were YIW (Tyr–Ile-Try) and YTW (Tyr-Thr-Try). The most common substitution was E476G (Glu → Gly), accounting for 64.7% (194/300) of the isolates, followed by T446A (Thr → Ala), Q438E (Gln → Glu), L455I (Leu → Ile), which accounted for 63.3% (190/300), 61.7% (185/300) and 61.0% (183/300), respectively. According to the distribution of various substitutions in PBP2b, all strains could be divided into 46 groups. Except for group 2B45 and 2B46, each containing one insertion variation, the other groups all had amino acid substitutions in PBP2b protein. The number of substituted amino acid sites ranged from 0 to 45, accounting for 0-12.1% of the total number of amino acids (n = 372) analyzed. Group 2B01 was the most common, accounting for 17.3% of the strains (52/300), and had exactly the same sequence of PBP2b as reference strain R6. The second common group was 2B20, accounting for 14.3% of the strains (43/300). A total of 13 amino acid substitutions were detected in the PBP2b sequence of the strains in this group, and the substitution rate was 3.5% (Table 8). According to the distribution of PEN MICs, the number of substitution sites in strains with MIC ≤ 0.12μg/ml were less than ten, while the number of detectable substitution sites in strains with MIC = 0.25, 0.5, 1, 2, and 4μg/ml were 12, 75, 61, 52, and 54, respectively (Supplementary Table 3).
Variations of the pbp1a Gene
Compared with the reference strain R6 (GenBank accession No. NC003098), a total of 344 amino acids from positions 312 to 655 of the PBP1a protein were analyzed for the studied isolates. A total of 105 different substitutions at 85 amino acid positions were detected in PBP1a. The E388D substitutions (Glu → Asp) was detected in all the 300 isolates. Apart from this, the most common substitution was S540T (Ser → Thr), accounting for 66.7% (200/300), followed by N546G (Asn → Gly), A550P (Gly → Pro), T574N (Thr → Asn), S575T (Ser → Thr), Q576G (Gln → Gly), F577Y (Phe → Tyr), N609D (Asn → Asp), each being detected in 60.0% (180/300) of the strains. According to the distribution of various substitutions in PBP1a, all strains could be divided into 25 groups. The number of substituted amino acids in each group ranged from 1 to 55, accounting for 0.3 −16.0% of the total number of amino acids analyzed. Based on the above substitution classification, group 1A15 was the most common, accounting for 28.7% (86/300) of the strains. All strains in this group had 43 amino acid substitution sites, with the substitution rate of 12.5% of the total number of amino acids analyzed, followed by group 1A04, accounting for 14.7% (44/300) of all the strains (Table 9). Based on the distribution of PEN MIC, the number of substitution sites in strains with MIC ≤ 0.12 μg/ml were less than ten. The number of substitution sites in strains with MIC = 0.25, 0.5, 1, 2, and 4 μg/ml were 17, 97, 88, 71, and 68, respectively (Supplementary Table 4).
Association Between Serotypes, Penicillin Susceptibility and Penicillin-Binding Proteins Substitution Patterns
Based on a comprehensive analysis of the substitutions of PBP2x, PBP2b and PBP1a, the 300 S. pneumoniae isolates could be divided into 101 PBPs substitution combinations: PBP001- PBP101 (Table 10).
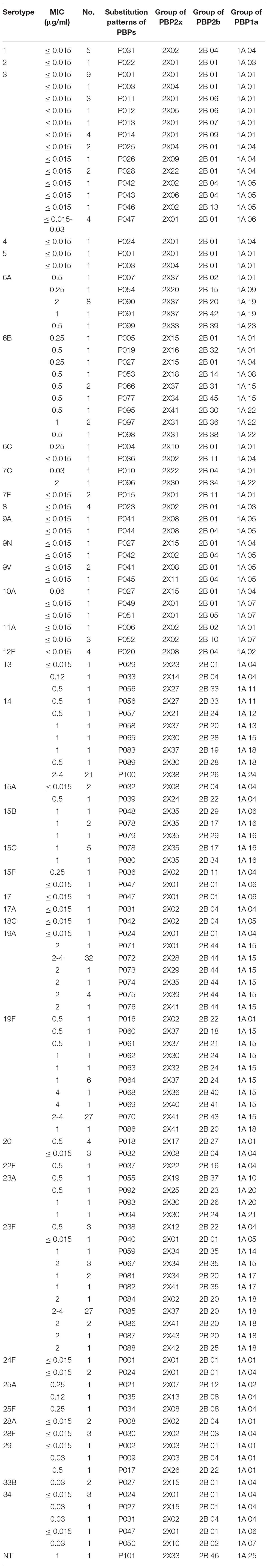
Table 10. Association of serotypes, penicillin susceptibility and substitution patterns of variations in PBP2x, PBP2b and PBP1a in 300 S. pneumoniae isolates.
Serotype 3 had the most PBPs substitution combinations, with a total of 13 groups. Except for the P047 combination, in which strains had PEN MICs ranging between 0.015-0.03 μg/ml, strains in the other 12 groups all had a PEN MIC of ≤0.015 μg/ml. The next serotype with the most PBPs substitution combinations was 23F with 11combinations. Save for the PEN MIC of 3 strains in group P038 and 1 strain in group P040 which were 0.5 μg/ml and ≤ 0.015 μg/ml, respectively, the PEN MIC of 42 strains in the other nine PBPs substitution combinations were between 1- 4 μg/ml. A total of 10, 8, 7, 7, 5, 5 PBPs substitution combinations were detected in serotypes 19F, 6B, 14, 19A, 6A, and 34, respectively. The number of the corresponding substitution combinations in other serotypes were ≤4. The strains of serotypes 1 (n = 5, P031), 2 (n = 1, P022), 4 (n = 1, P024), 7F (n = 2, P015), 8 (n = 4, P023), 12F (n = 4, P020), 17 (n = 1, P047), 17A (n = 1, P047), 18C (n = 1, P042), 28A (n = 2, P008), 28F (n = 3, P030) all had a PEN MIC ≤ 0.015 μg/ml and all strains within one serotype belonged to the same PBPs substitution combination. Serotypes 22F, 25F, and NT only had one strain in each group and PEN MICs were 0.5, 0.25, and 1 μg/ml respectively, corresponding to the unique substitution combination P037, P034, and P101, respectively.
Discussion
In this study, 300 S. pneumoniae strains isolated from IPD were analyzed for serotype distribution, antimicrobial susceptibility and PBPs substitutions. To our knowledge, this is the largest multicenter study involving invasive S. pneumoniae strains in China, in terms of number and diversity of origin of the isolates studied. Based on the Quellung reaction, 40 different serotypes were detected amongst 299 typeable S. pneumoniae strains, with the five most common serotypes being 23F, 19A, 19F, 3, and 14. The serotype distribution in the current study is consistent with those from other studies in China, although the isolation rates varied (Zhao et al., 2013, 2020).
All strains were susceptible to ETP, LEV, LZD, and VA, which is consistent with most studies in our region and abroad (Zhanel et al., 2006; Zhao et al., 2013), (Zhou et al., 2016; Cai et al., 2018; Fu et al., 2019; Golden et al., 2019; Shi et al., 2019). AMC was the second most active antimicrobial drug with a non-susceptibility rate of 2.3%, which is significantly lower than that reported in the national data for 2006 – 2008 in which the non- susceptibility rate was 5.3%, and the one from Beijing for the period 2012 and 2017 (10.2%) (Zhanel et al., 2006; Shi et al., 2019). A meta-analysis of invasive S. pneumoniae strains from Chinese children (2000 to 2016) also showed a very high proportion of AMC resistance among the isolates at 16.1%, which may be due to the higher proportion of PCV7-related serotype strains in these studies of 60.8%, compared to 42.3% in the present study (Fu et al., 2019). It has been previously reported that invasive S. pneumoniae strains of PCV7-related serotypes are more resistant to AMC than non-PCV7 serotype strains (15.7% vs. 1.7, P = 0.013) (Liu et al., 2010).
PRSP accounted for 70.2% of CSF derived strains in this study, which is lower than in previous studies based on invasive S. pneumoniae strains from children only, ranging from 76.6 to 95.7%, but higher than that reported in isolates from IPD cases in both adults and children, at 51.5% (Xue et al., 2010; Zhou et al., 2016; Shi et al., 2019). No PRSP was detected in strains from non-CSF specimens, and PISP accounted for 4.5% of the isolates, which was higher than that reported in IPD isolates from both adults and children at 3.8% (Zhao et al., 2013), but significantly lower than reported in children isolates from Shenyang (2010 to 2014), in which the resistance rate was very high at 32.3% (Fu et al., 2019). According to the oral breakpoint interpretation for all strains studied, PISP and PRSP accounted for 23.0% and 44.7% of the isolates, respectively, which were higher than reported in the comprehensive meta-analysis study of invasive S. pneumoniae strains from children (oral break points PISP: 42.6%, PRSP: 32%) (Fu et al., 2019). Data from the SENTRY program of the global multi-center surveillance study (1997-2016) showed that the Asia-Pacific region had the lowest S. pneumoniae penicillin susceptibility rate (52.4%, oral breakpoint) but the highest multi-drug (49.8%) and pan-drug resistance (17.3%) rates, compared with North America, Europe, and Latin America. However, the SENTRY program did not include data from mainland China (Sader et al., 2019). Previous data based on 881 S. pneumoniae isolates from 23 teaching hospitals across China from 2011 to 2016, showed that PRSP accounted for 51.6% of the isolates (oral breakpoint), a rate which is slightly higher than in the present study, but only 11.6% of the isolates were considered invasive in that previous study (Zhao et al., 2017).
The resistance mechanism of S. pneumoniae to penicillin and other BLAs is mainly affected through the modification of PBPs (Hakenbeck et al., 2012). There are 6 different PBP types described in S. pneumoniae, including PBP1a, PBP1b, PBP2a, PBP2b, PBP2x and PBP3. They can be divided into three categories according to their molecular weights and functions; type A: high molecular weight PBP containing PBP1a, PBP1b and PBP2a with transglycosidase activity and transpeptidase activity; type B: high molecular weight PBPs containing PBP2b and PBP2x with transpeptidase activity; type C: low molecular weight PBPs containing PBP3 with carboxypeptidase activity (Zapun et al., 2008; Hakenbeck et al., 2012).
Three types of PBPs play a major role in BLAs resistance, namely PBP2x, PBP2b, and PBP1a (Zapun et al., 2008). Normally, BLAs can combine with serine at the PBPs active site to form serine esters, thereby inhibiting the synthesis of bacterial cell walls, and leading to bacterial death. When there is PBPs substitution, the reactivity for BLAs drugs is reduced resulting in limited effectiveness of the drugs and eventual development of drug resistance (Zapun et al., 2008). Studies have shown that substitutions in the active sites of PBP2x and PBP2b, and their adjacent sites only cause low levels of BLAs resistance (Laible et al., 1991; Grebe and Hakenbeck, 1996; Sifaoui et al., 1996). However, combination substitutions involving PBP1a and those of PBP2b and PBP2x, can cause high levels of BLAs resistance, but PBP1a substitution alone cannot cause an increase in BLAs resistance levels (Zerfass et al., 2009). Substitutions in the PBP2b active sites mainly cause bacteria to be resistant to penicillin, but have nothing to do with cephalosporin resistance. On the contrary, substitutions in the PBP2x active sites are mainly related to cephalosporin resistance, and can result in high-level resistance to third-generation cephalosporin when combined with PBP1a substitutions (Zapun et al., 2008; Hakenbeck et al., 2012).
Although many studies have been performed on PBPs, both domestically in Asia and abroad, there are limited studies on S. pneumoniae isolates from IPD in mainland China. Our study mainly focused on the gene substitutions of pbp2x, pbp2b, and pbp1a among 300 invasive S. pneumoniae strains and their relationship with PEN MIC and serotype. In this study, the variations in the PBPs gene active sites were highly diverse, and the number of substitutions were higher than those in previous studies (Zhou et al., 2016; Chu et al., 2018).
Among the invasive S. pneumoniae isolates studied, strains with PEN MIC ≤ 0.25 μg/ml had no PBPs active site substitutions, except for one strain of serotype 6A (MIC = 0.25 μg/ml). On the other hand, strains with PEN MIC ≥ 0.5 μg/ml all had PBPs active site substitutions, while strains with high PEN MIC levels (MIC ≥ 1 μg/ml) all had PBP1a substitutions, which were accompanied by PBP2x and PBP2b substitutions. Even if the antimicrobial susceptibility phenotype of invasive S. pneumoniae derived from non-CSF was interpreted as susceptible by the meningitis breakpoint, the strains still had substitutions in different PBPs active sites. Thus the PBPs substitution of the strains was mainly related to PEN MICs, and less influenced by specimen type and antibiotic breakpoints.
We analyzed a total of 338 amino acids from positions 259-596 of PBP2x, and 98 different substitutions at 73 amino acid positions were detected. According to the distribution of PEN MIC, the number of substitution sites of strains with PEN MIC ≤ 0.12 μg/ml were more than that of PBP1a and PBP2b, ranging from 10-32. With the increase in PEN MIC, the number of substitution sites increased also, and the strains with MIC = 0.5 μg/ml had the maximum number of substitutions. In the study of Zhou et al., the average number of substitutions in PSSP strains was much higher for PBP2x than for PBP2b (29.26 vs. 8.22). Interestingly, they also found a significantly higher number of substitution sites in PRSP (65.55 ± 2.93) and PISP (63.37 ± 2.51) compared to PSSP (29.26 ± 27.88) (Zhou et al., 2016). Thus, it could be presumed that the MIC value of PEN was associated with the number of substitution sites in PBP2x. Specifically, common substitutions in PBP2x active sites included T338A, M339F, H394L, and L546V, which have also been reported in previous studies (Smith and Klugman, 1995; Zhou et al., 2016; Cai et al., 2018). In the present study, 181 strains had the T338A substitution and 39 strains also contained the M339F substitution. The PEN MIC of most strains was 2-4 μg/ml. The M339F substitution was first discovered in a highly resistant clinical strain in France, and then successively detected in the USA and Japan (Coffey et al., 1995; Asahi et al., 1999; Nagai et al., 2002). Previous crystal structure studies showed that the active center 337STMK of PBP2x which had the T338A and M339F double-site substitution was deformed. Furthermore, the serine active site Ser337 also presented another conformational change, resulting in reduction in the acylation efficiency of penicillin and cefotaxime by more than 20 times (Lu et al., 2001; Chesnel et al., 2003).
T338A and M339F substitutions have also been described in research reports from Taiwan, Japan, South Africa and other places, and were related to resistance in penicillin, amoxicillin and third-generation cephalosporins (Smith and Klugman, 1995; Du Plessis et al., 2002; Sanbongi et al., 2004; Davies et al., 2012; Liu et al., 2016; Zhou et al., 2016). Previous studies have shown that the L546V substitution is associated with high level resistance in BLAs, especially cephalosporins, but this substitution has also been reported in PSSP and cephalosporin-susceptible strains, suggesting that a single L546V substitution is not sufficient enough to cause high level resistance of BLAs (Nichol et al., 2002; Davies et al., 2012; Liu et al., 2016). Both crystal structure studies in vitro and transformation experiments in vivo have confirmed that R384G could affect the susceptibility of bacterial strains to penicillin and cefotaxime (Smith and Klugman, 2005; Maurer et al., 2008; Liu et al., 2016). A crystal structure study of PBP2x has found that the amino acid substitutions at position 371 and 384 affected the mobility of loop between amino acids 365-394 and were important for BLAs resistance (Carapito et al., 2006). In our study, strains with PBP2b sequences belonging to Group 2X27−2X43 all harbor the T338A substitution along with some other relevant substitutions, such as I371T, R384G, and M400T, but it is unknown which substitutions contribute to the resistance of the strains. Further experiments are needed to figure out the inner association between resistance and various substitutions. The Q552E substitution was located near the third catalytic motif 547KTG549 at the end of strand β3 loop, thus substitutions can led to an increase of the acylation efficiency for BLAs (Pernot et al., 2004; Zapun et al., 2008). In previous studies, Q552E was widely reported while in this study Q552V substitution was frequently detected in strains with PEN MIC ≤ 0.5 μg/ml. D567N substitution was previously detected in S. pneumoniae strains with high penicillin resistance in Taiwan, but has also been described in PSSP, which is consistent with the findings of this study (Liu et al., 2016).
A total of 372 amino acids from positions 305-676 of PBP2b were analyzed, and a total of 111 substitutions and 2 insertion variations at 88 amino acid sites were detected. Similar to PBP1a, the number of substitution sites in strains with PEN MIC ≤ 0.12 μg/ml was no more than 10. Furthermore, the number of substitution sites increased with MIC increase, but the most substitution sites were detected in strains with PEN MIC = 0.5 μg/ml. The two insertion variations YIW and YTW were found in two strains, located between 386SVVK and 443SSN in the active site of PBP2b, which have never been reported elsewhere. Previously, Japanese researchers reported that there was a duplication of the amino acid sequence WYT at the positions 429-431 between the amino acid positions 431-432 of PBP2b (Yamane et al., 1996). The insertion sequence was detected in 13 strains, of which 10 were of serotype 19, and three were serotype 6. In that study, the MIC of PEN and cefotaxime of S. pneumoniae strains carrying the above insertion variation (WYT amino acid sequence) were 0.125-2 μg/ml and 0.063-1 μg/ml, respectively (Yamane et al., 1996). In the present study, the two strains were of serotype 6B and NT and the PEN MIC were 0.5 μg/ml and 1 μg/ml respectively.
Previous studies have shown that the insertion of amino acid sequence between the conserved sequences of PBP2b in Neisseria gonorrhoeae reduced its affinity to PEN, further studies are needed to confirm its function in S. pneumoniae (Brannigan et al., 1990). Substitutions in the PBP2b active sites included T446A, T446S, and A619G, which have been reported in previous studies (Yamane et al., 1996; Sanbongi et al., 2004; Granger et al., 2005; Izdebski et al., 2008; Tian et al., 2008). In vitro experiments have shown that the affinity of T446A substitution strain to penicillin was 60% lower than that of the wild strain (Pagliero et al., 2004). However, researchers in the United States (USA), South Korea and Canada also found this substitution site in PSSP strains (Nagai et al., 2002; Baek et al., 2004; Granger et al., 2006), a finding which was also confirmed in one serotype 6A PSSP strain derived from blood (MIC = 0.25 μg/ml) in the present study. The A619G substitution has also been reported in strains in United States, Spain, Mexico, and other regions, and has been shown to be associated with high level amoxicillin resistance (Kosowska et al., 2004; Cafini et al., 2006). Other common substitutions identified in this study included E476G, Q438E, and L455I. Among them, Q438E and E476G have been reported in studies in Taiwan, France and Japan, and these substitutions were related increased levels of bacterial resistance to BLAs (Sanbongi et al., 2004; Chesnel et al., 2005; Liu et al., 2016). To the best of our knowledge, the L455I substitution is reported for the first time in this study.
A total of 344 amino acids from positions 312-655 of PBP1a were analyzed, and 105 different substitutions at 85 amino acid positions were detected. Based on PEN MIC distribution, isolates with MIC ≤ 0.12 μg/ml had no more than 10 substitution sites, and the substitution sites increased with increasing MIC. However, the maximum number of substitution sites was found in strains with MIC = 0.5 μg/ml. Substitutions in the PBP1a active sites, including T371A, T371S, and P432T, have been reported in multinational studies, and are related to high level resistance in penicillin and cephalosporin (Smith and Klugman, 1998; Liu et al., 2016; Zhou et al., 2016; Chu et al., 2018). In vitro site-directed mutagenesis experiments showed that T371A substitution reduces the acylation efficiency of PBP1a to cefotaxime and penicillin by 2.4 and 26 times, respectively, and TSQF574-577NTGY substitution reduces the acylation efficiency of PBP1a to cefotaxime and penicillin by 5.5 and 49 times, respectively (Job et al., 2008). In vivo transformation studies showed that only when the two substitutions exist at the same time, the resistance level of substitution strains to BLAs would increase significantly (Smith and Klugman, 1998; Job et al., 2008). In addition, the sites with high substitution rates in this study included S540T, N546G, A550P, T574N, S575T, Q576G, F577Y, and N609D, and part of these sites have been reported in some strains in Shenyang (Zhou et al., 2016).
Classification of isolates based on combination of substitution patterns in the three PBPs revealed a high degree of diversity among the isolates. Strains with the same serotype and PEN MIC exhibited different PBPs substitution combinations due to differences in the three PBPs substitution sites. For example, most strains of serotype 3 had a PEN MIC ≤ 0.015 μg/ml, but there were as many as 13 corresponding PBPs substitution combinations, although the difference between the different PBPs sites was small. Likewise, isolates with similar PBPs substitution combinations did not necessarily have the same PEN MIC levels, and this was reflected in serotypes 14, 19A, 19F, and 23F. The substitution combinations P100, P072, P070, and P085 were detected in isolates with PEN MICs between 2-4 μg/ml. A similar finding has also been reported previously, where five strains of serotype 23F with exactly the same PBPs substitutions and murM had PEN MICs ranging between 0.25-2 μg/ml (Chesnel et al., 2005).
In summary, we investigated the variations in pbp2x, pbp2b, and pbp1a genes, and serotype distribution of IPD S. pneumoniae isolates collected between 2010 and 2015 in China. We analyzed the serotype distribution, resistance to PEN and PBPs substitutions amongst these strains. There was a great diversity detected in PBPs substitutions patterns among the strains, suggesting that the PEN MIC level of S. pneumoniae may be affected by several other factors. Therefore, a comprehensive understanding of antibiotic resistance mechanism of S. pneumoniae needs to be further examined at the genomic level.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
YX and ZL conceived and designed the work. MZ, LW, and ZW performed the experiments, data analysis, and wrote the manuscript. YW and TK revised the manuscript. All authors read and approved the final manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by National Key Research and Development Program of China (2017YFC1601502), Special Foundation for National Science and Technology Basic Research Program of China (2019FY101200), Beijing Key Clinical Specialty for Laboratory Medicine - Excellent Project (No. ZK201000), Graduate Innovation Fund of Peking Union Medical College (Grant No. 2017-1002-1-21).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.838790/full#supplementary-material
References
Asahi, Y., Takeuchi, Y., and Ubukata, K. (1999). Diversity of substitutions within or adjacent to conserved amino acid motifs of penicillin-binding protein 2X in cephalosporin-resistant Streptococcus pneumoniae isolates. Antimicrob. Agents Chemother. 43, 1252–1255. doi: 10.1128/aac.43.5.1252
Baek, J. Y., Ko, K. S., Oh, W. S., Jung, S. I., Kim, Y. S., Chang, H. H., et al. (2004). Unique variations of pbp2b sequences in penicillin-nonsusceptible Streptococcus pneumoniae isolates from Korea. J. Clin. Microbiol. 42, 1746–1750. doi: 10.1128/JCM.42.4.1746-1750.2004
Brannigan, J. A., Tirodimos, I. A., Zhang, Q. Y., Dowson, C. G., and Spratt, B. G. (1990). Insertion of an extra amino acid is the main cause of the low affinity of penicillin-binding protein 2 in penicillin-resistant strains of Neisseria gonorrhoeae. Mol. Microbiol. 4, 913–919. doi: 10.1111/j.1365-2958.1990.tb00664.x
Cafini, F., Del Campo, R., Alou, L., Sevillano, D., Morosini, M. I., Baquero, F., et al. (2006). Alterations of the penicillin-binding proteins and murM alleles of clinical Streptococcus pneumoniae isolates with high-level resistance to amoxicillin in Spain. J. Antimicrob. Chemother. 57, 224–229. doi: 10.1093/jac/dki442
Cai, K., Wang, Y., Guo, Z., Xu, X., Li, H., Zhang, Q., et al. (2018). Clinical characteristics and antimicrobial resistance of pneumococcal isolates of pediatric invasive pneumococcal disease in China. Infect. Drug Resist. 11, 2461–2469. doi: 10.2147/IDR.S183916
Carapito, R., Chesnel, L., Vernet, T., and Zapun, A. (2006). Pneumococcal beta-lactam resistance due to a conformational change in penicillin-binding protein 2x. J. Biol. Chem. 281, 1771–1777. doi: 10.1074/jbc.M511506200
Chesnel, L., Carapito, R., Croize, J., Dideberg, O., Vernet, T., Zapun, A., et al. (2005). Identical penicillin-binding domains in penicillin-binding proteins of Streptococcus pneumoniae clinical isolates with different levels of beta-lactam resistance. Antimicrob. Agents Chemother. 49, 2895–2902. doi: 10.1128/AAC.49.7.2895-2902.2005
Chesnel, L., Pernot, L., Lemaire, D., Champelovier, D., Croizé, J., Dideberg, O., et al. (2003). The structural modifications induced by the M339F substitution in PBP2x from Streptococcus pneumoniae further decreases the susceptibility to beta-lactams of resistant strains. J. Biol. Chem. 278, 44448–44456. doi: 10.1074/jbc.M305948200
Chu, M. F., Liu, X. X., Zhang, S. N., Huang, Y. Y., Du, P., Wang, L. F., et al. (2018). Molecular characterization and correlation with beta-lactam resistance of Streptococcus pneumoniae isolates in Hangzhou, China. Biomed. Environ. Sci. 31, 389–393. doi: 10.3967/bes2018.050
Clinical and Laboratory Standards Institute (CLSI) (2012). Methods for Antimicrobial Susceptibility Testing of Aerobic Bacteria; Approved Standard Eighth Edition. Document M07-A10. Wayne, PA: Clinical and Laboratory Standards Institute.
Clinical and Laboratory Standards Institute (CLSI) (2019). Performance Standards for Antimicrobial Susceptibility Testing M100. Wayne, PA: Clinical and Laboratory Standards Institute.
Coffey, T. J., Daniels, M., Mcdougal, L. K., Dowson, C. G., Tenover, F. C., Spratt, B. G., et al. (1995). Genetic analysis of clinical isolates of Streptococcus pneumoniae with high-level resistance to expanded-spectrum cephalosporins. Antimicrob. Agents Chemother. 39, 1306–1313. doi: 10.1128/AAC.39.6.1306
Davies, T. A., Flamm, R. K., and Lynch, A. S. (2012). Activity of ceftobiprole against Streptococcus pneumoniae isolates exhibiting high-level resistance to ceftriaxone. Int. J. Antimicrob. Agents 39, 534–538. doi: 10.1016/j.ijantimicag.2012.02.016
Du Plessis, M., Bingen, E., and Klugman, K. P. (2002). Analysis of penicillin-binding protein genes of clinical isolates of Streptococcus pneumoniae with reduced susceptibility to amoxicillin. Antimicrob. Agents Chemother. 46, 2349–2357. doi: 10.1128/AAC.46.8.2349-2357.2002
Fu, J., Yi, R., Jiang, Y., Xu, S., Qin, P., Liang, Z., et al. (2019). Serotype distribution and antimicrobial resistance of Streptococcus pneumoniae causing invasive diseases in China: a meta-analysis. BMC Pediatr. 19:424. doi: 10.1186/s12887-019-1722-1
Golden, A. R., Baxter, M. R., Davidson, R. J., Martin, I., Demczuk, W., Mulvey, M. R., et al. (2019). Comparison of antimicrobial resistance patterns in Streptococcus pneumoniae from respiratory and blood cultures in Canadian hospitals from 2007-16. J. Antimicrob. Chemother. 74(Suppl._4), iv39–iv47. doi: 10.1093/jac/dkz286
Granger, D., Boily-Larouche, G., Turgeon, P., Weiss, K., and Roger, M. (2005). Genetic analysis of pbp2x in clinical Streptococcus pneumoniae isolates in Quebec, Canada. J. Antimicrob. Chemother. 55, 832–839. doi: 10.1093/jac/dki118
Granger, D., Boily-Larouche, G., Turgeon, P., Weiss, K., and Roger, M. (2006). Molecular characteristics of pbp1a and pbp2b in clinical Streptococcus pneumoniae isolates in Quebec, Canada. J. Antimicrob. Chemother. 57, 61–70. doi: 10.1093/jac/dki401
Grebe, T., and Hakenbeck, R. (1996). Penicillin-binding proteins 2b and 2x of Streptococcus pneumoniae are primary resistance determinants for different classes of beta-lactam antibiotics. Antimicrob. Agents Chemother. 40, 829–834. doi: 10.1128/AAC.40.4.829
Hakenbeck, R., Bruckner, R., Denapaite, D., and Maurer, P. (2012). Molecular mechanisms of beta-lactam resistance in Streptococcus pneumoniae. Future Microbiol. 7, 395–410. doi: 10.2217/fmb.12.2
Henriques-Normark, B., and Tuomanen, E. I. (2013). The pneumococcus: epidemiology, microbiology, and pathogenesis. Cold Spring Harb. Perspect. Med. 3:a010215. doi: 10.1101/cshperspect.a010215
Izdebski, R., Rutschmann, J., Fiett, J., Sadowy, E., Gniadkowski, M., Hryniewicz, W., et al. (2008). Highly variable penicillin resistance determinants PBP 2x, PBP 2b, and PBP 1a in isolates of two Streptococcus pneumoniae clonal groups, Poland 23F-16 and Poland 6B-20. Antimicrob. Agents Chemother. 52, 1021–1027. doi: 10.1128/AAC.01082-07
Job, V., Carapito, R., Vernet, T., Dessen, A., and Zapun, A. (2008). Common alterations in PBP1a from resistant Streptococcus pneumoniae decrease its reactivity toward beta-lactams: structural insights. J. Biol. Chem. 283, 4886–4894. doi: 10.1074/jbc.M706181200
Kim, S. H., Chung, D. R., Song, J. H., Baek, J. Y., Thamlikitkul, V., Wang, H., et al. (2020). Changes in serotype distribution and antimicrobial resistance of Streptococcus pneumoniae isolates from adult patients in Asia: emergence of drug-resistant non-vaccine serotypes. Vaccine 38, 6065–6073. doi: 10.1016/j.vaccine.2019.09.065
Kim, S. H., Song, J. H., Chung, D. R., Thamlikitkul, V., Yang, Y., Wang, H., et al. (2012). Changing trends in antimicrobial resistance and serotypes of Streptococcus pneumoniae isolates in Asian countries: an Asian Network for Surveillance of Resistant Pathogens (ANSORP) study. Antimicrob. Agents Chemother. 56, 1418–1426. doi: 10.1128/AAC.05658-11
Kosowska, K., Jacobs, M. R., Bajaksouzian, S., Koeth, L., and Appelbaum, P. C. (2004). Alterations of penicillin-binding proteins 1A, 2X, and 2B in Streptococcus pneumoniae isolates for which amoxicillin MICs are higher than penicillin MICs. Antimicrob. Agents Chemother. 48, 4020–4022. doi: 10.1128/AAC.48.10.4020-4022.2004
Laible, G., Spratt, B. G., and Hakenbeck, R. (1991). Interspecies recombinational events during the evolution of altered PBP 2x genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Mol. Microbiol. 5, 1993–2002. doi: 10.1111/j.1365-2958.1991.tb00821.x
Liu, C. L., Zhao, C. J., Liu, Y. D., and Wang, H. (2010). Study of serotype distribution, antimicrobial resistance patterns and molecular epidemiology in 148 isolates of invasive Streptococcus pneumoniae. Zhonghua Yi Xue Za Zhi 90, 1565–1570.
Liu, E. Y., Chang, J. C., Lin, J. C., Chang, F. Y., and Fung, C. P. (2016). Important mutations contributing to high-level penicillin resistance in Taiwan(19F)-14, Taiwan(23F)-15, and Spain(23F)-1 of Streptococcus pneumoniae isolated from Taiwan. Microb. Drug Resist. 22, 646–654. doi: 10.1089/mdr.2015.0261
Lu, W. P., Kincaid, E., Sun, Y., and Bauer, M. D. (2001). Kinetics of beta-lactam interactions with penicillin-susceptible and -resistant penicillin-binding protein 2x proteins from Streptococcus pneumoniae. Involvement of acylation and deacylation in beta-lactam resistance. J. Biol. Chem. 276, 31494–31501. doi: 10.1074/jbc.M102499200
Lynch, J. P. III, and Zhanel, G. G. (2009). Streptococcus pneumoniae: epidemiology, risk factors, and strategies for prevention. Semin. Respir. Crit. Care Med. 30, 189–209. doi: 10.1055/s-0029-1202938
Maha, H., Barbara, D. P., and Catherine, S. (2014). Capsular serotyping of Streptococcus pneumoniae using the quellung reaction. J. Vis. Exp. 84:e51208. doi: 10.3791/51208
Martin, C., Sibold, C., and Hakenbeck, R. (1992). Relatedness of penicillin-binding protein 1a genes from different clones of penicillin-resistant Streptococcus pneumoniae isolated in South Africa and Spain. EMBO J. 11, 3831–3836.
Maurer, P., Koch, B., Zerfass, I., Krauss, J., van der Linden, M., Frère, J. M., et al. (2008). Penicillin-binding protein 2x of Streptococcus pneumoniae: three new mutational pathways for remodelling an essential enzyme into a resistance determinant. J. Mol. Biol. 376, 1403–1416. doi: 10.1016/j.jmb.2007.12.058
Mehr, S., and Wood, N. (2012). Streptococcus pneumoniae-a review of carriage, infection, serotype replacement and vaccination. Paediatr. Respir. Rev. 13, 258–264. doi: 10.1016/j.prrv.2011.12.001
Nagai, K., Davies, T. A., Jacobs, M. R., and Appelbaum, P. C. (2002). Effects of amino acid alterations in penicillin-binding proteins (PBPs) 1a, 2b, and 2x on PBP affinities of penicillin, ampicillin, amoxicillin, cefditoren, cefuroxime, cefprozil, and cefaclor in 18 clinical isolates of penicillin-susceptible, -intermediate, and -resistant pneumococci. Antimicrob. Agents Chemother. 46, 1273–1280. doi: 10.1128/AAC.46.5.1273-1280.2002
Nichol, K. A., Zhanel, G. G., and Hoban, D. J. (2002). Penicillin-binding protein 1A, 2B, and 2X alterations in Canadian isolates of penicillin-resistant Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46, 3261–3264. doi: 10.1128/AAC.46.10.3261-3264.2002
Pagliero, E., Chesnel, L., Hopkins, J., Croize, J., Dideberg, O., Vernet, T., et al. (2004). Biochemical characterization of Streptococcus pneumoniae penicillin-binding protein 2b and its implication in beta-lactam resistance. Antimicrob. Agents Chemother. 48, 1848–1855. doi: 10.1128/AAC.48.5.1848-1855.2004
Pernot, J., Chesnel, L., Le Gouellec, A., Croizé, J., Vernet, T., Dideberg, O., et al. (2004). A PBP2x from a clinical isolate of Streptococcus pneumoniae exhibits an alternative mechanism for reduction of susceptibility to β-lactam antibiotics. J. Biol. Chem. 279, 16463–16470. doi: 10.1074/jbc.M313492200
Quan-Cheng, K., Jian-Guo, W., Xiang-Hua, L., and Zhen-Zhen, L. (2016). Inappropriate use of antibiotics in children in China. Lancet 387, 1273–1274. doi: 10.1016/s0140-6736(16)30019-8
Sader, H. S., Mendes, R. E., Le, J., Denys, G., Flamm, R. K., Jones, R. N., et al. (2019). Antimicrobial susceptibility of Streptococcus pneumoniae from North America, Europe, Latin America, and the Asia-Pacific Region: results from 20 years of the SENTRY antimicrobial surveillance program (1997-2016). Open Forum Infect. Dis. 6(Suppl. 1), S14–S23. doi: 10.1093/ofid/ofy263
Sanbongi, Y., Ida, T., Ishikawa, M., Osaki, Y., Kataoka, H., Suzuki, T., et al. (2004). Complete sequences of six penicillin-binding protein genes from 40 Streptococcus pneumoniae clinical isolates collected in Japan. Antimicrob. Agents Chemother. 48, 2244–2250. doi: 10.1128/AAC.48.6.2244-2250.2004
Shi, W., Li, J., Dong, F., Qian, S., Liu, G., Xu, B., et al. (2019). Serotype distribution, antibiotic resistance pattern, and multilocus sequence types of invasive Streptococcus pneumoniae isolates in two tertiary pediatric hospitals in Beijing prior to PCV13 availability. Expert Rev. Vaccines 18, 89–94. doi: 10.1080/14760584.2019.1557523
Sibold, C., Henrichsen, J., König, A., Martin, C., Chalkley, L., and Hakenbeck, R. (1994). Mosaic pbpX genes of major clones of penicillin-resistant Streptococcus pneumoniae have evolved from pbpX genes of a penicillin-sensitive Streptococcus oralis. Mol. Microbiol. 12, 1013–1023. doi: 10.1111/j.1365-2958.1994.tb01089.x
Sifaoui, F., Kitzis, M. D., and Gutmann, L. (1996). In vitro selection of one-step mutants of Streptococcus pneumoniae resistant to different oral beta-lactam antibiotics is associated with alterations of PBP2x. Antimicrob. Agents Chemother. 40, 152–156. doi: 10.1128/AAC.40.1.152
Smith, A. M., and Klugman, K. P. (1995). Alterations in penicillin-binding protein 2B from penicillin-resistant wild-type strains of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 39, 859–867. doi: 10.1128/AAC.39.4.859
Smith, A. M., and Klugman, K. P. (1998). Alterations in PBP 1A essential-for high-level penicillin resistance in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 42, 1329–1333. doi: 10.1128/AAC.42.6.1329
Smith, A. M., and Klugman, K. P. (2005). Amino acid mutations essential to production of an altered PBP 2X conferring high-level beta-lactam resistance in a clinical isolate of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 49, 4622–4627. doi: 10.1128/AAC.49.11.4622-4627.2005
Tian, S. F., Chu, Y. Z., and Chen, B. Y. (2008). Molecular characteristics of penicillin-binding protein 2b, 2x, and 1a sequences in penicillin-nonsusceptible Streptococcus pneumoniae isolates in Shenyang, China. Can. J. Microbiol. 54, 489–494. doi: 10.1139/w08-030
World Health Organization [WHO] (2007). Pneumococcal conjugate vaccine for childhood immunization–WHO position paper. Wkly. Epidemiol. Rec. 82, 93–104.
Xue, L., Yao, K., Xie, G., Zheng, Y., Wang, C., Shang, Y., et al. (2010). Serotype distribution and antimicrobial resistance of Streptococcus pneumoniae isolates that cause invasive disease among Chinese children. Clin. Infect. Dis. 50, 741–744. doi: 10.1086/650534
Yamane, A., Nakano, H., Asahi, Y., Ubukata, K., and Konno, M. (1996). Directly repeated insertion of 9-nucleotide sequence detected in penicillin-binding protein 2B gene of penicillin-resistant Streptococcus pneumoniae. Antimicrob. Agents Chemother. 40, 1257–1259. doi: 10.1128/AAC.40.5.1257
Yao, K. H., Wang, L. B., Zhao, G. M., Zheng, Y. J., Deng, L., Huang, J. F., et al. (2011). Pneumococcal serotype distribution and antimicrobial resistance in Chinese children hospitalized for pneumonia. Vaccine 29, 2296–2301. doi: 10.1016/j.vaccine.2011.01.027
Zapun, A., Contreras-Martel, C., and Vernet, T. (2008). Penicillin-binding proteins and beta-lactam resistance. FEMS Microbiol. Rev. 32, 361–385.
Zerfass, I., Hakenbeck, R., and Denapaite, D. (2009). An important site in PBP2x of penicillin-resistant clinical isolates of Streptococcus pneumoniae: mutational analysis of Thr338. Antimicrob. Agents Chemother. 53, 1107–1115. doi: 10.1128/AAC.01107-08
Zhanel, G. G., Wang, X., Nichol, K., Nikulin, A., Wierzbowski, A. K., Mulvey, M., et al. (2006). Molecular characterisation of Canadian paediatric multidrug-resistant Streptococcus pneumoniae from 1998-2004. Int. J. Antimicrob. Agents 28, 465–471. doi: 10.1016/j.ijantimicag.2006.08.005
Zhao, C., Li, Z., Zhang, F., Zhang, X., Ji, P., Zeng, J., et al. (2017). Serotype distribution and antibiotic resistance of Streptococcus pneumoniae isolates from 17 Chinese cities from 2011 to 2016. BMC Infect. Dis. 17:804. doi: 10.1186/s12879-017-2880-0
Zhao, C., Xie, Y., Zhang, F., Wang, Z., Yang, S., Wang, Q., et al. (2020). Investigation of antibiotic resistance, serotype distribution, and genetic characteristics of 164 invasive Streptococcus pneumoniae from North China between April 2016 and October 2017. Infect. Drug Resist. 13, 2117–2128. doi: 10.2147/IDR.S256663
Zhao, C., Zhang, F., Chu, Y., Liu, Y., Cao, B., Chen, M., et al. (2013). Phenotypic and genotypic characteristic of invasive pneumococcal isolates from both children and adult patients from a multicenter surveillance in China 2005-2011. PLoS One 8:e82361. doi: 10.1371/journal.pone.0082361
Zhou, X., Liu, J., Zhang, Z., Liu, Y., Wang, Y., Liu, Y., et al. (2016). Molecular characteristics of penicillin-binding protein 2b, 2x and 1a sequences in Streptococcus pneumoniae isolates causing invasive diseases among children in Northeast China. Eur. J. Clin. Microbiol. Infect. Dis. 35, 633–645. doi: 10.1007/s10096-016-2582-3
Keywords: Streptococcus pneumoniae, penicillin susceptibility, penicillin-binding protein, serotype, invasive pneumococcal disease
Citation: Zhou M, Wang L, Wang Z, Kudinha T, Wang Y, Xu Y and Liu Z (2022) Molecular Characterization of Penicillin-Binding Protein2x, 2b and 1a of Streptococcus pneumoniae Causing Invasive Pneumococcal Diseases in China: A Multicenter Study. Front. Microbiol. 13:838790. doi: 10.3389/fmicb.2022.838790
Received: 18 December 2021; Accepted: 19 January 2022;
Published: 01 March 2022.
Edited by:
Mattias Collin, Lund University, SwedenReviewed by:
Dalia Denapaite, University of Trento, ItalyAndré Zapun, UMR 5075 Institut de Biologie Structurale (IBS), France
Copyright © 2022 Zhou, Wang, Wang, Kudinha, Wang, Xu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yingchun Xu, xycpumch@139.com; Zhengyin Liu, zhengyinl@hotmail.com
†These authors have contributed equally to this work
 Menglan Zhou
Menglan Zhou Lulu Wang
Lulu Wang Ziran Wang
Ziran Wang Timothy Kudinha
Timothy Kudinha Yao Wang
Yao Wang Yingchun Xu
Yingchun Xu Zhengyin Liu
Zhengyin Liu