- 1School of Life Sciences, Jilin University, Changchun, China
- 2Key Laboratory for Molecular Enzymology and Engineering, The Ministry of Education, School of Life Sciences, Jilin University, Changchun, China
- 3National Engineering Research Center for Strategic Drugs, Beijing Institute of Pharmacology and Toxicology, Beijing, China
- 4State Key Laboratory of Toxicology and Medical Countermeasures, Beijing Institute of Pharmacology and Toxicology, Beijing, China
- 5No. 971 Hospital of People’s Liberation Army Navy, Qingdao, China
Bacterial drug resistance caused by overuse and misuse of antibiotics is common, especially in clinical multispecies infections. It is of great significance to discover novel agents to treat clinical bacterial infections. Studies have demonstrated that autoinducer-2 (AI-2), a signal molecule in quorum sensing (QS), plays an important role in communication among multiple bacterial species and bacterial drug-resistance. Previously, 14 AI-2 inhibited compounds were selected through virtual screening by using the AI-2 receptor protein LuxP as a target. Here, we used Vibrio harveyi BB170 as a reporter strain for the preliminary screening of 14 inhibitors and compound Str7410 had higher AI-2 QS inhibition activity (IC50 = 0.3724 ± 0.1091 μM). Then, co-culture of Pseudomonas aeruginosa PAO1 with Staphylococcus aureus ATCC 25923 was used to evaluate the inhibitory effects of Str7410 on multispecies infection in vitro and in vivo. In vitro, Str7410 significantly inhibited the formation of mixed bacterial biofilms. Meanwhile, the combination of Str7410 with meropenem trihydrate (MEPM) significantly improved the susceptibility of mixed-species-biofilm cells to the antibiotic. In vivo, Str7410 significantly increased the survival rate of wild-type Caenorhabditis elegans N2 co-infected by P. aeruginosa PAO1 and S. aureus ATCC 25923. Real-time quantitative PCR analysis showed that Str7410 reduced virulence factor (pyocyanin and elastase) production and swarming motility of P. aeruginosa PAO1 by downregulating the expression of QS-related genes in strain PAO1 in co-culture with S. aureus ATCC 25923. Compound Str7410 is a candidate agent for treating drug-resistant multispecies infections. The work described here provides a strategy for discovering novel antibacterial drugs.
Introduction
Most bacterial diseases are caused by infection with multiple species of bacteria, for example, cystic fibrosis (CF) and chronic wounds (Harrison, 2007; DeLeon et al., 2014; Blanchard and Waters, 2019). On co-infection by multiple bacteria, different strains interact with each other and contribute to the pathogenesis of the disease. The pathogenic mechanisms resulting from the interaction of multiple microbes are often different from those of individual species (Baldan et al., 2014).
Pseudomonas aeruginosa and Staphylococcus aureus are the two most important pathogens in CF and chronic wounds (DeLeon et al., 2014; Blanchard and Waters, 2019). Interactions between P. aeruginosa and S. aureus have been the focus of several studies on multiple-strain infections. Notably, P. aeruginosa significantly increased the production of S. aureus biofilms and resistance of S. aureus to vancomycin on co-culture in vitro (Yang et al., 2011; Orazi and O’Toole, 2017). In a wound model co-infected by P. aeruginosa and S. aureus, the resistance of the bacteria to antibiotics was significantly increased, and the toxicity of P. aeruginosa was also increased (Dalton et al., 2011; DeLeon et al., 2014). Moreover, after co-infection by P. aeruginosa and S. aureus, the healing of pig epithelial cell wounds (Pastar et al., 2013) and mouse wounds (Korgaonkar et al., 2013) was significantly delayed, and S. aureus promoted the pathogenicity of P. aeruginosa. Korgaonkar et al., 2013 showed that peptidoglycan produced by S. aureus can significantly increase the lethality of P. aeruginosa toward Drosophila and promote the production of P. aeruginosa virulence factors such as pyocyanin and elastase. In the sputum of CF patients, P. aeruginosa and other bacteria, such as Streptococcus and Staphylococcus, can communicate with each other through the autoinducer-2 (AI-2) quorum sensing (QS) system (Duan et al., 2003). This indicates that different bacteria in multispecies infections can increase in pathogenicity through QS.
Quorum sensing is a signaling mechanism that regulates the life activities of bacteria by transmitting signals through the synthesis, release, and acceptance of autoinducers (AIs). Bacteria sense the density of the surrounding population via AI molecules, and this regulates gene expression (Gokalsin et al., 2017). AI-2 is an intra- and interspecies signal molecule “a common language” for bacterial interaction. AI-2 QS was first discovered and characterized in the Gram-negative marine bacterium Vibrio harveyi (Bassler et al., 1994). Bioluminescence of V. harveyi is regulated by QS. V. harveyi produces AI-2 signal molecule through LuxS protease and after AI-2 binds to LuxP receptor, the LuxP–AI-2 complex then interacts with LuxQ phosphokinase in the membrane, and finally LuxR receptor protein regulates the production of bioluminescence (Roy et al., 2011). Therefore, V. harveyi is often used as a test species for laboratory AI-2 QS inhibitors (QSIs) research (Lowery et al., 2008; Collins et al., 2015). Research found >40 Gram-positive and Gram-negative bacterial species can communicate using AI-2 as a signaling molecule (Mok et al., 2003). AI-2-mediated QS plays a critical role in the interaction among multiple strains, and it has also been shown that AI-2 is closely related to the formation of mixed biofilms and gene regulation among multiple strains (Roy et al., 2011).
As a Gram-positive pathogen, S. aureus has two types of QS system. One is the autoinducing peptide signaling molecule-mediated agr system, and the other is the LuxS/AI-2 system (Sifri, 2008). Staphylococcus aureus is able to produce AI-2 signaling molecule, which is regulated by the luxS gene. However, no potential AI-2 receptor (such as the LuxPQ receptor of Vibrio harveyi or the LsrABC transporter of Salmonella enterica serovar Typhimurium) has been found by searching for established AI-2 receptors in S. aureus genomes (Zhao et al., 2010). The LuxS/AI-2 system can negatively regulate the formation of S. aureus biofilms (Ma et al., 2017), and loss of the luxS gene leads to a decrease in susceptibility to cell wall synthesis inhibitor antibiotics (Wang et al., 2019). Because of the dual function of LuxS and the absence of genomic evidence of established AI-2 receptors, the AI-2 QS function in S. aureus needs further study (Zhao et al., 2010).
Pseudomonas aeruginosa lacks the luxS gene and cannot produce AI-2 signaling molecule. However, P. aeruginosa regulates the production of a variety of virulence factors and biofilms through the las, rhl, pqs, and iqs QS systems, which destroys tissues and induces inflammation, leading to impaired immune mechanisms, in an infected patient (Van Delden and Iglewski, 1998). Therefore, studies in P. aeruginosa have focused on QSIs that target QS mediated by acyl-homoserine lactone (AHL) signaling molecules (Chbib, 2020; Jiang et al., 2020). Nevertheless, P. aeruginosa can sense the AI-2 signaling molecule produced by other bacteria, such as Escherichia coli (Roy et al., 2011), Salmonella typhimurium (Roy et al., 2011), Streptococcus mitis (Wang et al., 2016b), and S. aureus (Hotterbeekx et al., 2017), which regulates the production of virulence factors and biofilms and increases its pathogenicity (Li et al., 2015; Wang et al., 2016b). Recent studies have shown that C1-alkyl AI-2 analogs reduced V. harveyi QS-associated bioluminescence (Lowery et al., 2009), and analogs of 4,5-dihydroxy-2,3-pentanedione (DPD), a precursor substance of AI-2 signaling molecule, like butyl and pentyl-DPD, were shown to inhibit pyocyanin production by P. aeruginosa by 50% (Ganin et al., 2009). Inhibiting the AI-2 QS system using a QSI is a new strategy to treat bacterial infections caused by P. aeruginosa and S. aureus.
In our previous study, 14 AI-2 QSI compounds were selected through virtual screening. In this study, V. harveyi BB170 was used as a reporter strain for the preliminary screening. Compound Str7410, which had the highest AI-2 QS inhibition activity, was chosen for further research. We then co-cultured P. aeruginosa PAO1 with S. aureus ATCC 25923 to analyze the inhibitory effects of compound Str7410 on multispecies infections in vitro and in vivo. The effects of the compound on biofilm formation and of the combination of Str7410 with antibiotics on drug resistance were determined in vitro. We tested the effects of Str7410 on P. aeruginosa PAO1 virulence factor (pyocyanin and elastase) production, swarming motility, and the expression of QS-related genes in co-culture of P. aeruginosa and S. aureus. We used Caenorhabditis elegans N2 as an in vivo model to test the effects of Str7410 on survival rates on co-infection of the nematode with P. aeruginosa and S. aureus. Finally, we preliminarily evaluated the inhibitory mechanism of interspecies QS by the compound. This study provides a new strategy for the treatment of clinical multispecies infections.
Materials and Methods
Synthesis of Compound Str7410
All the chemical reagents used in the synthesis reactions were analytical-grade and available from commercial resources without further purification. All the reactions were monitored by analytical thin-layer chromatography (TLC) using silica gel TLC plates (GF254). Silica gel (200–300 mesh) was used for chromatography. The 1H and 13C nuclear magnetic resonance (NMR) spectroscopy were recorded on a JEOLECA400 spectrometer, with TMS as an internal standard at ambient temperature. All coupling constants were reported in Hertz. Proton coupling patterns were described as singlet (s), doublet (d), triplet (t), quartet (q), multiplet (m), and broad (br). High-resolution mass spectra (HRMS) were obtained by electrospray ionization (ESI) using an Agilent TOF G6230A mass spectrometer. The synthesis of compound Str7410 was shown in Scheme 1. Intermediates 1, 2, and 3 were prepared by the following routes.
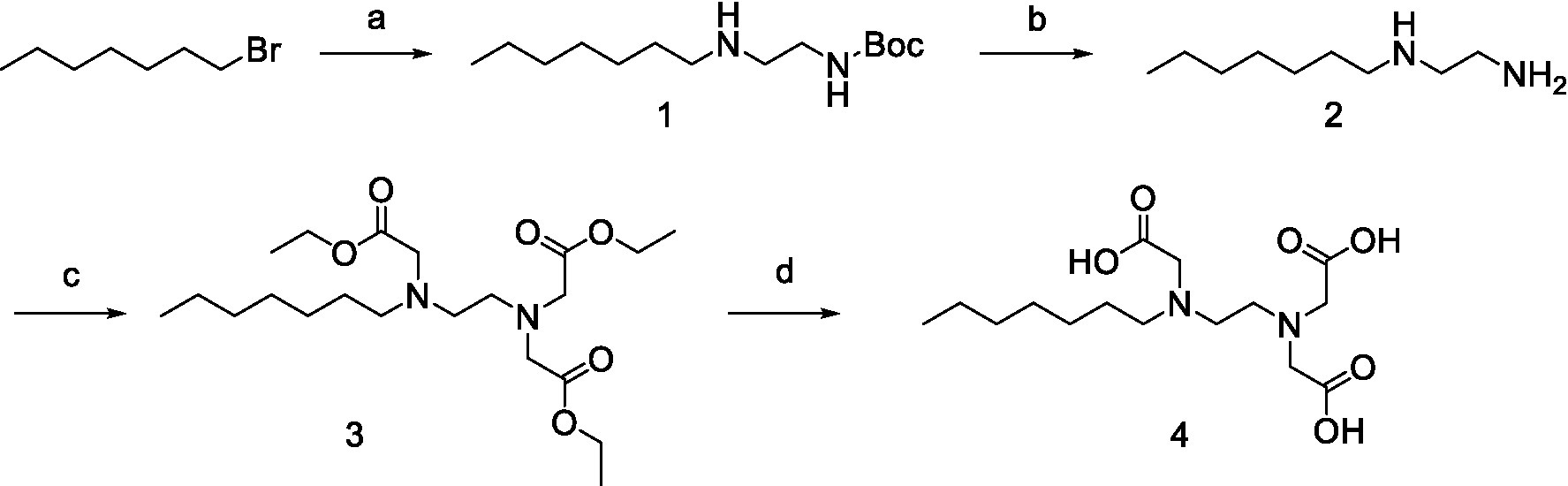
Scheme 1. Synthetic route toward the target compound Str7410. ɑReagents and conditions. (A) N-tert-butoxycarbonyl-1,2-ethylenediamine, K2CO3, MeCN, 65°C, and 63% yield; (B) trifluoroacetic acid, CH2Cl2, room temperature, and 100% yield; and (C) K2CO3, ethyl 2-bromoacetate, MeCN, 45°C, and 56% yield; (D) concentrated hydrochloric acid, 100°C, and 54% yield.
Synthesis of Tert-Butyl (2-(Heptylamino)Ethyl)Carbamate (1)
To a solution of N-tert-butoxycarbonyl-1,2-ethylenediamine (4.49 g, 28.0 mmol) in CH3CN (60 ml) was added K2CO3 (1.55 g, 11.2 mmol), then the mixture was heated to 65°C. At this temperature, 1-bromoheptane (1 g, 5.6 mmol) in CH3CN (10 ml) was added dropwise to the mixture. The mixture was stirred at 80°C for 6 h and cooled to room temperature. The solvent was concentrated in vacuo. The residue was dissolved in CHCl3, washed with brine, dried over Na2SO4, and concentrated in vacuo. The residue was purified by flash silica gel chromatography (1% triethylamine, petroleum ether: ethyl acetate = 1: 3) to afford 1 as colorless oil (0.91 g, 63%). 1H NMR [400 MHz, dimethylsulfoxide (DMSO)-d6] δ 6.69 (t, J = 6.8 Hz, 1H), 2.98 (q, J = 6.2 Hz, 2H), 2.53–2.48 (m, 2H), 2.46 (t, J = 7.0 Hz, 2H), 1.37 (s, 11H), 1.25 (s, 8H), and 0.86 (t, J = 6.8 Hz, 3H). 13C NMR (101 MHz, DMSO) δ 156.14, 77.94, 49.52, 31.83, 30.10, 29.21, 28.75, 27.29, 22.59, and 14.46. HRMS (ESI) of C14H30N2O2 [M + H]+ calcd., 259.2381; found 259.2380.
Synthesis of N1-Heptylethane-1,2-Diamine (2)
To a cold (0°C) solution of trifluoroacetic acid (2.5 ml) and CH2Cl2 (7.5 ml) was added tert-butyl (2-(heptylamino)ethyl)carbamate (0.5 g, 1.9 mmol), then the mixture was warmed to room temperature and stirred for 2 h. The solution was concentrated in vacuo and used directly in the next step without further purification.
Synthesis of Diethyl 2,2'-((2-((2-Ethoxy-2-Oxoethyl)(Heptyl)Amino)Ethyl)Azanediyl)Diacetate (3)
To a solution of N1-heptylethane-1,2-diamine (0.3 g, 1.9 mmol) in CH3CN (40 ml) was added K2CO3 (1.05 g, 7.6 mmol) and ethyl 2-bromoacetate (0.98 g, 5.9 mmol). Then, the mixture was stirred at 45°C for 3 h and cooled to room temperature. The solvent was concentrated in vacuo. The residue was dissolved in CHCl3, washed with brine, dried over Na2SO4, and concentrated in vacuo. The residue was purified by flash silica gel chromatography (1% triethylamine, CH2Cl2: CH3OH = 20: 1) to afford 3 as colorless oil (0.44 g, 56%). 1H NMR (400 MHz, DMSO-d6) δ 4.09 (t, J = 7.1 Hz, 6H), 3.57 (s, 4H), 3.37 (s, 2H), 2.93 (d, J = 5.0 Hz, 2H), 2.89 (d, J = 4.9 Hz, 2H), 2.86–2.81 (m, 2H), 1.50 (m, 2H), 1.27 (s, 8H), 1.21 (t, J = 7.1 Hz, 9H), and 0.87 (d, J = 7.1 Hz, 3H). 13C NMR (101 MHz, DMSO) δ 171.47, 169.40, 60.46, 56.23, 55.06, 54.58, 52.49, 50.32, 31.64, 28.90, 26.73, 25.54, 22.53, 14.60, and 14.43. HRMS (ESI) of C21H40N2O6 [M + H]+ calcd., 417.2959; found 417.2960.
Synthesis of 2,2'-((2-((Carboxymethyl)(Heptyl)Amino)Ethyl)Azanediyl)Diacetic Acid (4)
Diethyl 2,2'-((2-((2-ethoxy-2-oxoethyl)(heptyl)amino)ethyl)azanediyl) diacetate (0.2 g, 0.5 mmol) was added to concentrated hydrochloric acid (15 ml). The mixture was heated to 100°C and stirred for 12 h. Then, the residue was cooled to room temperature and concentrated in vacuo. The residue was added to concentrated hydrochloric acid (15 ml), and the mixture was stirred at 100°C for 12 h and concentrated in vacuo. The residue was recrystallized with ethanol (5 ml) and methyl tert-butyl ether (MTBE, 45 ml), filtered, and washed with MTBE to afford 4 as white solid (86.2 mg, 54%). 1H NMR (400 MHz, DMSO-d6) δ 4.23 (s, 2H), 3.63 (s, 4H), 3.37 (t, J = 5.6 Hz, 2H), 3.26–3.19 (m, 2H), 3.16 (t, J = 5.9 Hz, 2H), 1.71–1.59 (m, 2H), 1.26 (s, 8H), and 0.89–0.83 (m, 3H). 13C NMR (101 MHz, DMSO) δ 172.64, 168.12, 55.11, 54.97, 53.39, 51.70, 49.28, 31.51, 28.67, 26.34, 23.68, 22.49, and 14.44. HRMS (ESI) of C15H28N2O6 [M + H]+ calcd., 333.2020; found 333.2019.
Bacterial Strains and Growth Media
Vibrio harveyi BB170 (ATCC BAA-1117), S. aureus ATCC 25923, P. aeruginosa PAO1, and E. coli OP50 were used in this study. AB medium contained 0.3 M NaCl, 0.05 M MgSO4, and 2% acid-hydrolyzed casein (without vitamins), adjusted to pH 7.5 with 1 M KOH. After autoclaving at 121°C, 10 ml of 1 M potassium phosphate buffer (K2HPO4/KH2PO4, pH 7.0), 10 ml of 0.1 M sterile arginine solution (L-Arg), and 20 ml of 50% sterile glycerol were added to the medium (per 1 L). LB medium contained 1% tryptone, 1% NaCl, and 0.5% yeast extract. TSB medium contained 1.5% tryptone, 0.5% soy peptone, and 0.5% NaCl (pH = 7.2 ± 0.2). PB medium contained 2% peptone, 1% K2SO4, and 0.3% MgCl2. PTSB medium contained 5% peptone, 1.7% tryptone, 0.5% NaCl, 0.25% K2HPO4, 0.25% glucose, and 0.3% soy peptone. PGS-agar plates contained 1% peptone, 0.15 M sorbitol, 1% NaCl, 1% glucose, and 1.7% agar. Swimming solid medium contained 0.8% nutrient broth, 0.5% glucose, and 0.3% agar. Swarming solid medium contained 0.8% nutrient broth, 0.5% glucose, and 0.5% agar.
Test compounds were dissolved in DMSO to a stock concentration of 65 mM. Antibiotics [tobramycin base, meropenem trihydrate (MEPM), ceftazidime, amikacin, colistin sulfate, and ciprofloxacin] were purchased from Sigma and stored at 6.4 mg/ml at −20°C.
Half-Maximal Inhibitory Concentration of Compounds in Assay of Vibrio harveyi BB170 QS
The concentration of compounds that resulted in 50% maximum V. harveyi BB170 bioluminescence (the IC50 value) was tested as described previously (Collins et al., 2015). Vibrio harveyi BB170 was grown in AB medium (14 h, 30°C) to OD600 nm = 1.5. The cells were diluted into fresh AB medium (1:2,500) and added (100 μl/well) to serially diluted test compounds in AB medium (100 μl/well); the final DMSO concentration was 0.5%. The plate was incubated at 30°C for 8 h, and then the bioluminescence (OD460 nm) and cell density (OD600 nm) were measured. The values of OD460 nm and OD600 nm were normalized and IC50 values were calculated using GraphPad Prism 5 software.
Growth Assays
Pseudomonas aeruginosa PAO1 and S. aureus ATCC 25923 were inoculated into LB medium, respectively, and cultured at 37°C at 150 rpm until OD600 nm = 0.5. Then, the cultures were diluted with fresh LB medium to OD600 nm = 0.05. Next, the bacteria were cultured in the presence of compound Str7410 (80, 40, 20, 10, and 1 μM), and OD600 nm was measured at intervals of 2 h for up to 24 h using a spectrophotometer (UV-1800, Bio-Rad Smart Spec Plus, United States). All experiments were performed three times, independently (Wang et al., 2016b).
Biofilm Quantification Assays
Quantitative analysis of biofilms used crystal violet assay (Armbruster et al., 2016). First, we used fresh LB medium to dilute P. aeruginosa PAO1 and S. aureus ATCC 25923 cultures in the logarithmic growth phase to OD600 nm = 0.05, and then mixed them in equal proportions in the same volume and added them to a 96-well plate (100 μl/well, three parallel wells for each group). The final test compound concentrations were 80, 40, 20, 10, and 1 μM. The 96-well plate was incubated at 37°C for 24 h, then the bacterial culture was removed, and the plate was washed three times with phosphate-buffered saline (PBS) and dried. Next, 0.1% (w/v) crystal violet solution was added for 10 min. After washing three times with PBS, the crystal violet-stained biofilms were air-dried. To quantify the biofilm biomass, the crystal violet was removed by adding 150 μl of 33% glacial acetic acid, and OD595 nm of the solubilized dye was determined using a Biotek multifunction microplate reader. The biofilm inhibition rate was defined as (OD595 blank − OD595 sample)/OD595 blank × 100%. Each crystal violet assay was run in triplicate, with a minimum of three replicates per assay.
Determination of Susceptibility to Antibiotics
The determination of minimal inhibitory concentrations (MICs) was performed as described previously (Peeters et al., 2009; Guo et al., 2016). Pseudomonas aeruginosa PAO1 and S. aureus ATCC 25923 were, respectively, cultured in LB medium to OD600 nm = 0.8. Pseudomonas aeruginosa PAO1 alone or in co-culture with S. aureus ATCC 25923 (mixed in equal proportions) were diluted 1,000-fold with fresh LB medium and added to a 96-well plate. According to the broth dilution method, Antibiotics concentrations ranged from 64 to 0.125 μg/ml. Finally, the plates were incubated at 37°C for 18 h, and OD590 nm was determined using a Biotek multifunction microplate reader.
Determination of minimum biofilm inhibitory concentrations (MBICs) of antibiotics (Wang et al., 2016a). Based on the above method, 100 μl of diluted bacteria was added to each well of a 96-well plate and cultured at 37°C for 48 h. After removing the bacterial culture, the biofilms were washed twice using PBS. Antibiotics and fresh LB medium were added to the 96-well plate, which was incubated at 37°C for 24 h. The lowest drug concentration without turbidity well is MBIC value of antibiotics.
For the minimum biofilm eradication concentrations (MBECs) of compound for biofilm cells (Tsukatani et al., 2016, 2020), the 96-well plate containing 100 μl diluted bacterial solution of each well was incubated at 37°C for 24 h. After removing the bacterial culture, the biofilms were washed three times using PBS, then 180 μl reaction solution of 2-fold serial dilutions of compound (range 1,024–1 μg/ml) in fresh LB medium added to 96-well plate. After 20 h challenge at 37°C, the reaction solution were removed and washed three times with sterile PBS, then 150 μl fresh LB medium added to each well of 96-well plate and incubated for a further 24 h at 37°C. After removing the LB medium, the biofilms were washed three times using PBS and dried, and then 150 μl 0.1% (w/v) crystal violet solution was added for 10 min. After washing three times with PBS, adding 150 μl of 33% glacial acetic acid to 96-well plate, and the OD460 nm was measured using a Biotek multifunction microplate reader. The MBEC represents the lowest concentration of constituents for which the biofilm eradication activity was >99%. Biofilm eradication activity (%) was determined using the following formula:
Treatment of biofilms and quantification of cells was performed as follows. Dual-species biofilms were grown in 96-well plate. The liquid medium was decanted every 2 days leaving the attached cells at the bottom of the plate, and fresh LB medium was added (Armijo et al., 2020). After 6 days, the liquid culture was removed and the biofilms were slowly washed twice with PBS. In the test compound + antibiotic group, the test compound (1 μl, 10 μM final concentration), antibiotic (1 μl of MEPM, 0.5 μg/ml final concentration), and LB medium (98 μl) were added to each well. In the test compound or antibiotic alone groups, 99 μl of LB medium and 1 μl of test compound or MEPM were added, respectively. In the antibiotic + sonication group, bacteria in the biofilm were detached by sonication (40 kHz, 300 W, and 5 min) before adding 99 μl of LB medium and 1 μl of MEPM. In the control group, 100 μl of LB medium was added. After incubation for 3.5 h, biofilm cells were detached by sonication (40 kHz, 300 W, and 5 min) and the number of colony-forming units was determined by plating the resulting suspension (Brackman et al., 2011; Guo et al., 2016).
Caenorhabditis elegans Survival Assay
Pseudomonas aeruginosa PAO1 and S. aureus ATCC 25923 were cultured to OD600 nm = 2.0 at 37°C in TSB medium, respectively. Then, after mixing in equal proportions, 20 μl of culture were spread on PGS-agar plates containing antibiotic or test compound. The plates were placed in an incubator at 37°C and cultured for 18 h to allow the formation of a bacterial lawn. Then, 50 synchronized nematodes (L4 stage of wild-type C. elegans N2) were selected and placed on a plate. The plates were incubated at 20°C and the number of living and dead nematodes was counted every 24 h using a stereomicroscope. A nematode was defined as dead when it no longer responded to touch. Any nematodes that died as a result of getting stuck to the wall of the plate were excluded from the analysis. Survival curves were drawn for analysis (Rajkumari et al., 2018).
Virulence Factor Quantification
Pyocyanin was quantified as follows: P. aeruginosa PAO1 and S. aureus ATCC 25923 were, respectively, cultured in LB medium at 37°C, 150 rpm, to the logarithmic growth phase. After mixing in equal proportions, the culture was diluted tenfold with fresh PB medium. Bacterial cultures (5 ml) with and without the test compound (40, 20, and 10 μM) were incubated at 37°C and 150 rpm for 16 h. To extract pyocyanin, the culture was centrifuged at 1,665 × g for 10 min, and the supernatant was collected and extracted with 3 ml of chloroform. Then, 1 ml of 0.2 M HCl was mixed with the chloroform layer, and the upper (aqueous) phase was collected by centrifugation at 1,665 × g for 10 min. The absorbance was measured at 520 nm (Essar et al., 1990). The inhibition rate of pyocyanin production was calculated as (OD520 blank − OD520 sample)/OD520 blank × 100%.
Elastase was determined using the Elastin-Congo red assay with modifications, and the final absorbance was read at 495 nm (Pearson et al., 1997). Pseudomonas aeruginosa PAO1 and S. aureus ATCC 25923 were, respectively, cultured in PTSB medium at 37°C and 150 rpm to the logarithmic growth phase. After mixing in equal proportions, co-cultures were divided into the experimental group and the control group, and then incubated at 37°C and 150 rpm for 18 h. Then, the cultures were centrifuged at 9,590 × g for 5 min and the supernatant was collected. The bacteria were filtered through a 0.22-μm pore-size filter. Then, 200 μl of the filtrate was added to 1 ml of Elastin-Congo red reaction solution [20 mg/ml Congo red, 0.1 M Tris–HCl (pH 7.2), and 1 mM CaCl2], and the mixture was shaken for 18 h at 37°C. Stop-reaction solution (ethylenediaminetetraacetic acid, 0.1 ml, and 0.12 M) was added and the mixture was incubated on ice for 5 min, and then centrifuged at 9,590 × g for 10 min. The supernatant was collected and OD495nm was determined.
Swarming Motility Assay
Pseudomonas aeruginosa PAO1 was shaken in LB medium at 37°C until OD600 nm = 1.0. Bacterial culture (1 μl) was inoculated into the center of cooled swimming solid medium and further cultured at 30°C for 16 h. Staphylococcus aureus ATCC 25923 was shaken at 37°C in LB medium to OD600 nm = 0.5. The culture was centrifuged at 9,590 × g for 5 min, and the supernatant was filtered (0.22-μm pore-size). The filtered supernatant was added to thawed swarming solid medium at a ratio of 1:10, mixed, poured into a plate, and left to stand for 8 h to dry. Finally, colonies were picked from the P. aeruginosa PAO1 swimming solid medium, added to the center of the swarming solid plate, and cultured at 30°C for 16 h (Tremblay and Deziel, 2008; Pallett et al., 2019).
Quantitative Reverse Transcriptase PCR
Pseudomonas aeruginosa PAO1 and S. aureus ATCC 25923 cultured overnight for 16 h were diluted with fresh LB medium to OD600 nm = 0.05. After mixing in equal proportions, they were divided into a control group and an experimental group (with test compound added) and grown at 37°C and 150 rpm for 10 h. Pseudomonas aeruginosa PAO1 was also grown in monoculture. After centrifugation (9,590 × g, 5 min) to retain the bacterial cells, a one-step cDNA kit was used [5× All-In-One RT MasterMix (With Excellent Lysis Kit, abm, Canada) for cDNA Extraction]; the DNA was stored at −20°C.
Quantitative reverse transcriptase PCR (qRT-PCR) was performed with a Roche LightCycler® 480 instrument and EvaGreen 2× qPCR MasterMix. The 20-μl reaction system contained 0.6 μl each of forward and reverse primers, 1 μl template cDNA, 10 μl EvaGreen 2× qPCR MasterMix, and 7.8 μl nuclease-free H2O. The primer sets used for these analyses are listed in Table 1. A no-template control was included during each qPCR experiment. The thermal cycling program started with 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. After qPCR amplification, the comparative threshold method (ΔΔCt analysis) was applied to evaluate relative changes in gene expression. The experiment used GraphPad Prism 8 software for mapping (Conway et al., 2012). ΔΔCt = ΔΔCt, sample – ΔΔCt, reference.
Statistical Analysis
All assays were performed with three replicates and the values obtained are expressed as the mean ± SD. Differences in data were compared with the untreated control at each time-point and considered significant when p < 0.05 (*p < 0.05, **p < 0.01, and ***p < 0.001) by one-way ANOVA with t-tests using GraphPad Prism 8 software.
Results
Synthesis of Str7410
Synthesis of compound Str7410 was accomplished in four steps from the commercially-available 1-bromoheptane (Scheme 1). Single substitution of N-tert-butoxycarbonyl-1,2-ethylenediamine with 1-bromoheptane generated intermediate 1. Removal of the Boc amino-protecting group followed by reaction with ethyl 2-bromoacetate afforded intermediate 3. Acidic hydrolysis twice afforded the desired compound Str7410.
Effect of Compound Str7410 on Vibrio harveyi AI-2 QS
We used V. harveyi BB170 (a luxN mutant) as a reporter strain to evaluate the AI-2 QS inhibitory activity of test compounds. The IC50 values of compounds were tested in terms of V. harveyi BB170 bioluminescence. Bromo-furanone C56 (Zang et al., 2009) was used as the positive control. As shown in Table 2, Str7410 had the best inhibitory effect on AI-2 QS of V. harveyi BB170 (IC50 = 0.3724 ± 0.1091 μM). Based on this, compound Str7410 was selected for further research.
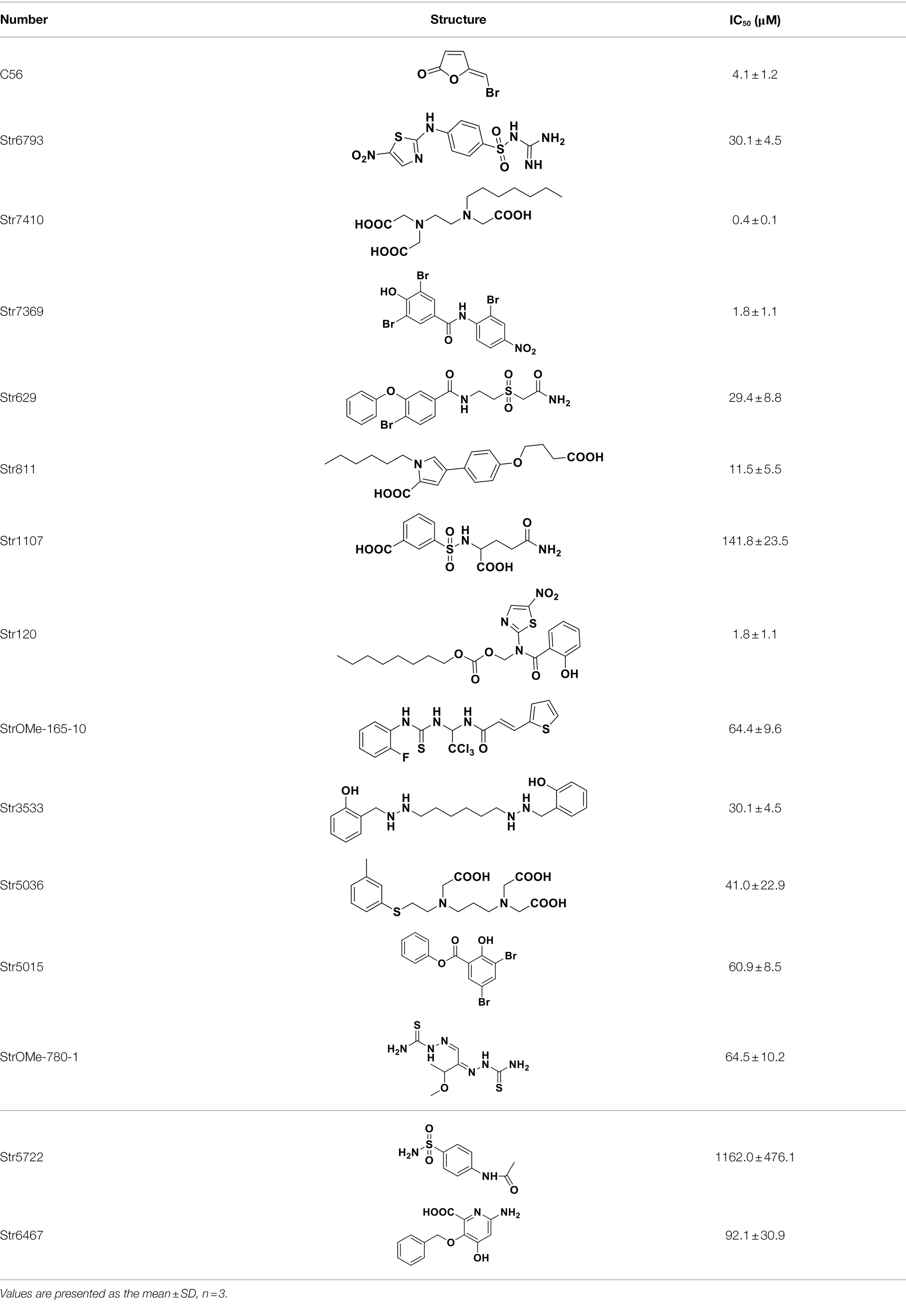
Table 2. Anti-Vibrio harveyi BB170 autoinducer-2 (AI-2) quorum sensing (QS) activities of test compounds and its structure.
Effect of Compound Str7410 on Bacterial Growth
The main difference between QSIs and antibiotics is that QSIs inhibit the production of virulence factors without killing pathogenic bacteria, so that the bacteria do not develop drug-resistance mutations (Allen et al., 2014). Therefore, we analyzed the effect of the putative QSI compound Str7410 on the growth of P. aeruginosa PAO1 and S. aureus ATCC 25923. As Figure 1 shows, compound Str7410 affected the growth of neither bacterium.
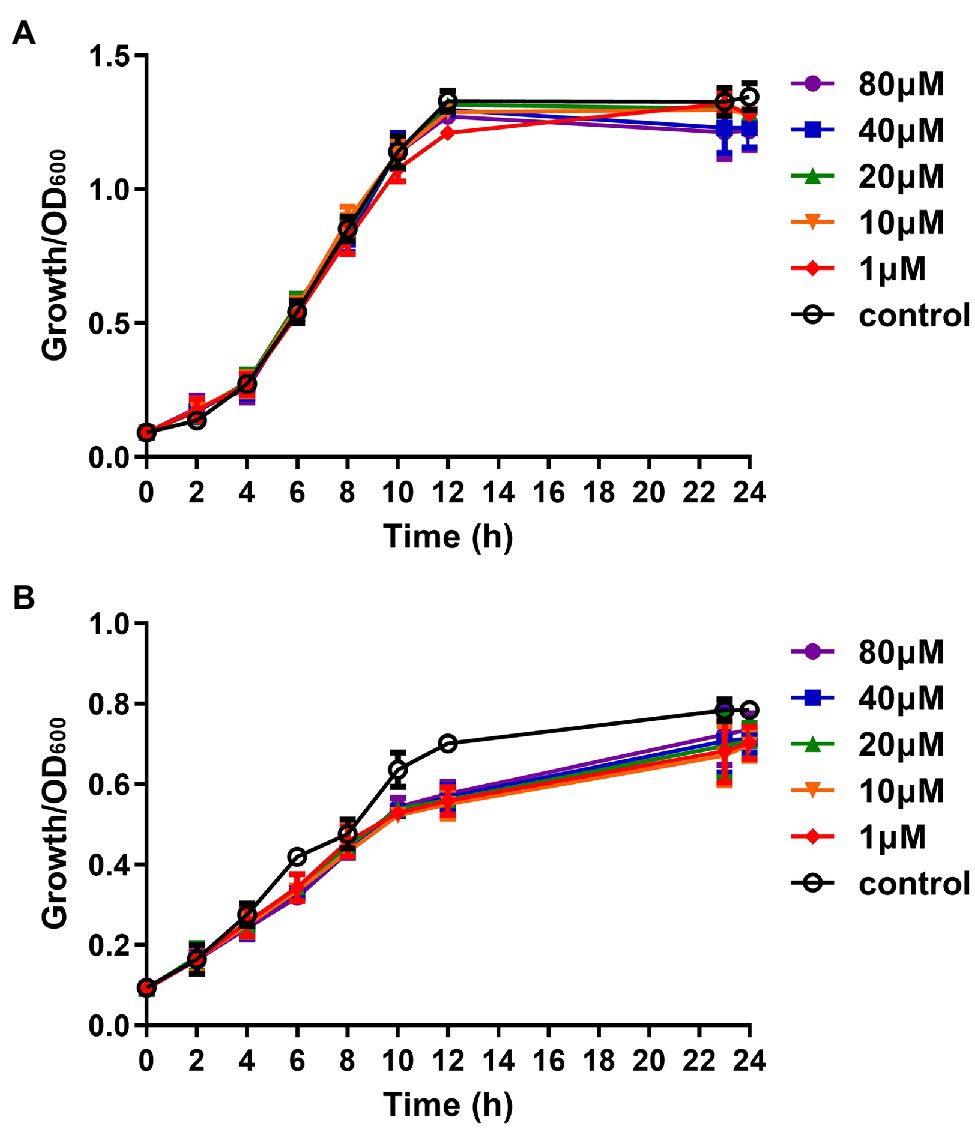
Figure 1. Effect of compound Str7410 (1–80 μM) on the growth of Pseudomonas aeruginosa PAO1 and Staphylococcus aureus ATCC 25923. (A) Pseudomonas aeruginosa PAO1 growth curves. (B) Staphylococcus aureus ATCC 25923 growth curves. Values are presented as the mean ± SD (n = 3).
Effect of Compound Str7410 on Biofilm Formation
Bacterial resistance to antibiotics is related to the formation of biofilms (Kalia, 2013). We used the crystal violet assay to evaluate the effect of compound Str7410 on formation of biofilms by P. aeruginosa PAO1 and S. aureus ATCC 25923 mixed in equal proportion. As shown in Figure 2, significantly more biofilm was formed by the mixture of species than by either species alone. When compound Str7410 was added to the co-culture, the formation of biofilm was significantly reduced [range 9.75% (p < 0.05)–48.40% (p < 0.001)]. The inhibitory effect of compound Str7410 was concentration-dependent. When the concentration of Str7410 was 40 μM, the inhibition rate of biofilm formation was 40.58% (p < 0.01).
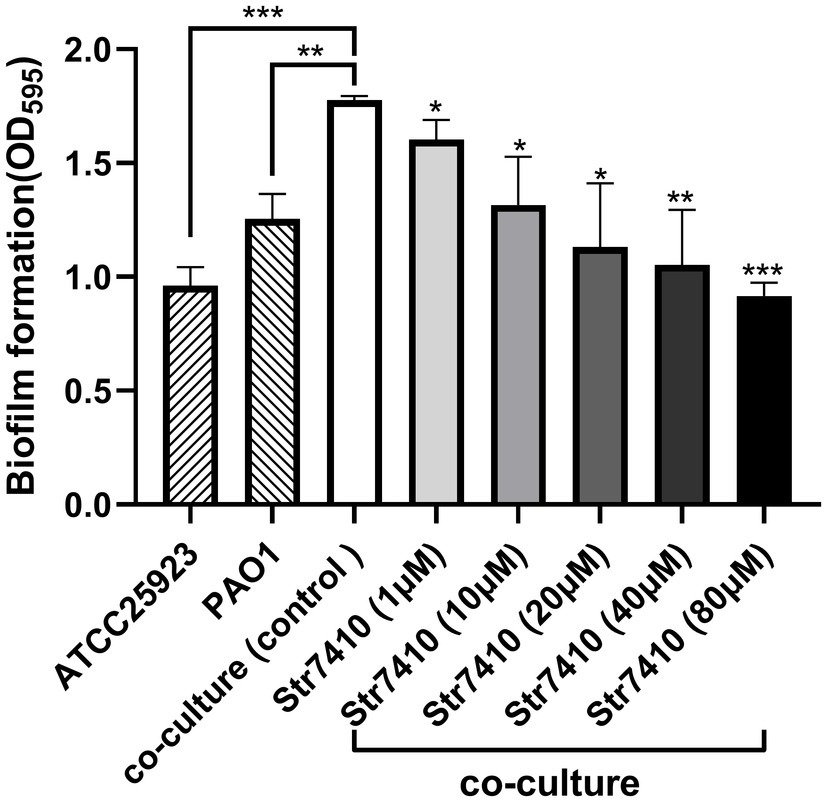
Figure 2. Crystal violet assay of biofilm formation by Pseudomonas aeruginosa PAO1 and Staphylococcus aureus ATCC 25923 alone and in co-culture after 24 h, and the inhibitory effect of compound Str7410 on biofilm formation by the mixed culture. Values are the mean ± SD (n = 3). Differences in data were compared with the untreated control at each time-point and considered significant when p < 0.05 (*p < 0.05, **p < 0.01, and ***p < 0.001) by one-way ANOVA with t-tests.
Effect of Compound Str7410 on the Antibiotic Susceptibility of Biofilm Cells
First, the MIC and MBIC values of antibiotics for single and mixed bacteria were determined when P. aeruginosa PAO1 was cultured alone or in co-culture with S. aureus ATCC 25923. As shown in Table 3, compared with planktonic cells, biofilm cells showed significantly increased resistance to antibiotics. The MBIC values of MEPM against biofilm cells increased 32 and 16-fold, respectively, compared with the MIC values for planktonic cells, when P. aeruginosa PAO1 was cultured alone and co-cultured with S. aureus ATCC 25923. Moreover, planktonic cells or biofilm cells from co-cultures of P. aeruginosa PAO1 and S. aureus ATCC 25923 showed significantly increased resistances to some antibiotics (ceftazidime, colistin sulfate, ciprofloxacin, and MEPM) compared with that of P. aeruginosa PAO1 monoculture. These data indicate that cultures of multiple bacterial species and the formation of biofilms can increase resistance to antibiotics. We also evaluated the MBEC of the compound Str7410 against the formed biofilm. As shown in Supplementary Table S1, the compound Str7410 had no eradication effect on the formed biofilm.
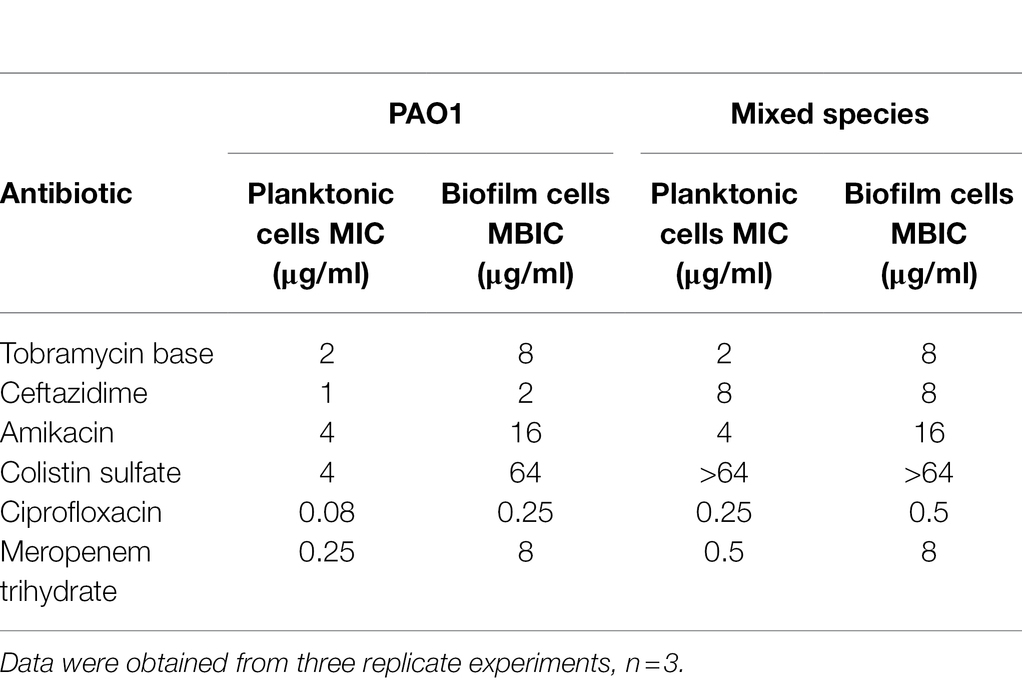
Table 3. Minimal inhibitory concentration (MIC) and minimum biofilm inhibitory concentration (MBIC) of antibiotics toward planktonic and biofilm cells of Pseudomonas aeruginosa PAO1 alone or in co-culture with Staphylococcus aureus ATCC 25923.
Next, we evaluated the effects of compound Str7410 on the susceptibility of mixed-species biofilms to antibiotics. We tested the inhibitory effects of MEPM and compound Str7410 alone, and MEPM combined with compound Str7410. MEPM in combination with sonication was used as a reference treatment. As Figure 3 shows, compound Str7410 when applied alone had no inhibitory effect on biofilms of mixed P. aeruginosa PAO1 and S. aureus ATCC 25923 cells. Treatment with MEPM alone and MPEM combined with sonication significantly decreased the number of biofilm cells (p < 0.05 and p < 0.01, respectively). The combination of MEPM with Str7410 also had a significant inhibitory effect on biofilm cells (p < 0.01). Notably, compound Str7410 increased the inhibitory effect of MEPM toward biofilm cells (p < 0.05), and the level of inhibition by Str7410 + MEPM was greater than that by MEPM + sonication.
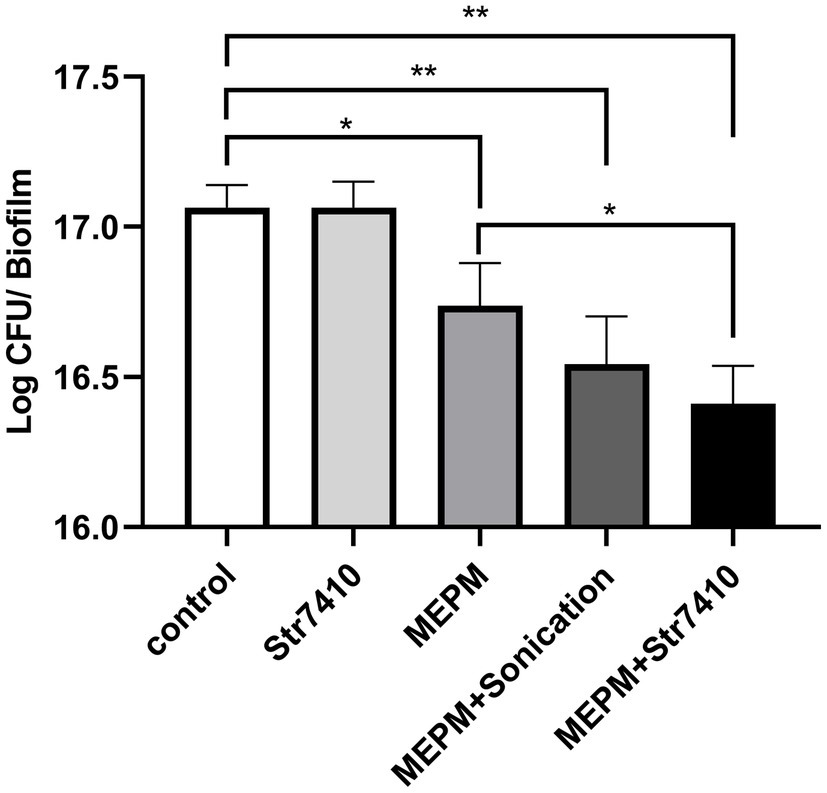
Figure 3. Effect of compound Str7410 on the number of cells [determined as log colony-forming units (CFU)] in biofilms formed from mixed Pseudomonas aeruginosa PAO1 and Staphylococcus aureus ATCC 25923 cells. Treatments: Str7410 (10 μM) alone, meropenem trihydrate (MEPM, 0.5 μg/ml) alone, and MEPM combined with sonication or Str7410. Values are presented as the mean ± SD (n = 3). Differences in data were compared with the untreated control at each time-point and considered significant when p < 0.05 (*p < 0.05, and **p < 0.01) by one-way ANOVA with t-tests.
Effect of Compound Str7410 on Survival of Infected Caenorhabditis elegans
We used wild-type C. elegans N2 as a model to evaluate the effect of compound Str7410 on survival rates following co-infection with P. aeruginosa PAO1 and S. aureus ATCC 25923 in vivo.
After 240 h of incubation, the survival rate of nematodes decreased by 2.55% for C. elegans co-infected with P. aeruginosa PAO1 and S. aureus ATCC 25923 compared with that of nematodes infected with P. aeruginosa PAO1 alone (Figure 4). This indicates that the pathogenicity caused by multispecies infection was greater than that by a single bacterial species.
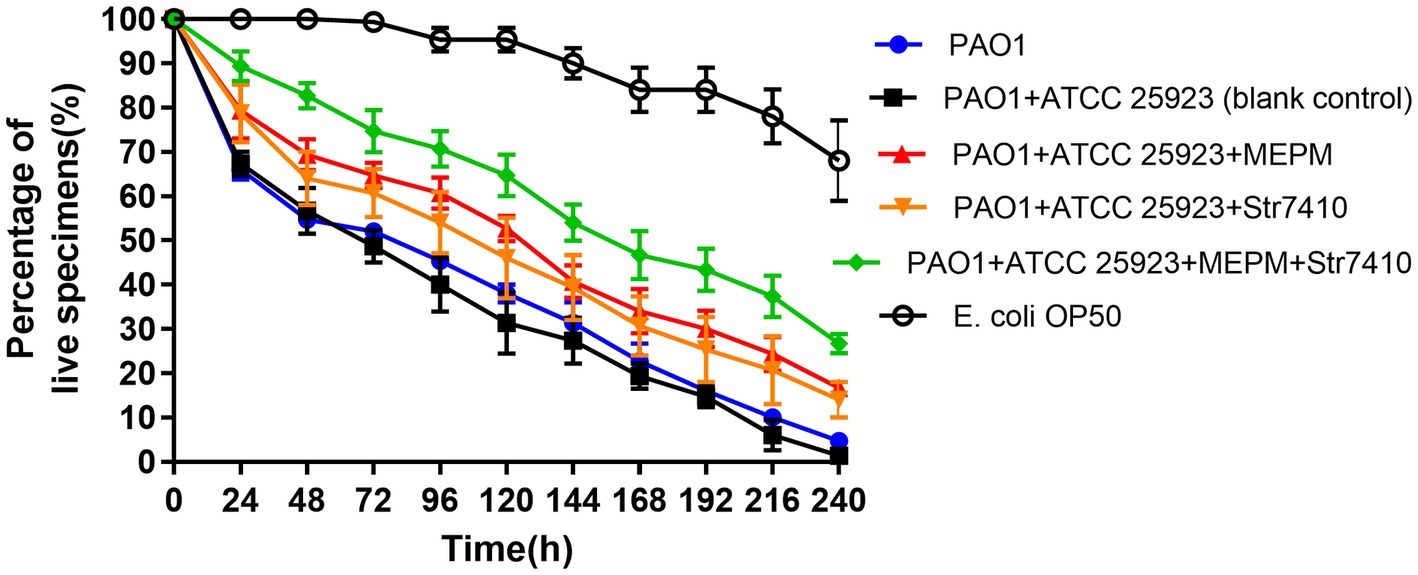
Figure 4. Effect of treatment with compound Str7410 (40 μM) and the antibiotic MEPM (0.5 μg/ml) alone or combination on survival of Caenorhabditis elegans N2 co-infected with Pseudomonas aeruginosa PAO1 and Staphylococcus aureus ATCC 25923. Escherichia coli OP50 was used as a negative control. Values are presented as the mean ± SD (n = 3).
Compared with the positive control group co-infected by P. aeruginosa PAO1 and S. aureus ATCC 25923, the survival rate of the infected group treated with compound Str7410 (40 μM) increased by 12.67% (p < 0.05), and the survival rate of the infected group treated with MEPM (0.5 μg/ml) increased by 14.67% (p < 0.001). Furthermore, the survival rate of the group treated with the combination of Str7410 and MEPM increased by 12.67% (p < 0.05) and 10.67% (p < 0.05) compared with the Str7410-alone and MEPM-alone treatments, respectively (Figure 4). Although compound Str7410 inhibited the growth of neither P. aeruginosa PAO1 nor S. aureus ATCC 25923 (e.g., Figure 1), it decreased the pathogenicity of these bacteria by inhibiting interspecific QS. In addition, the combination of compound Str7410 with MEPM increased the therapeutic effect of MEPM.
Effect of Compound Str7410 on Virulence Factors of Pseudomonas aeruginosa PAO1
The effect of Str7410 on virulence factors of P. aeruginosa PAO1 in multispecies infection was investigated. We quantitatively analyzed production of the virulence factors pyocyanin and elastase by P. aeruginosa PAO1 when this strain was cultured alone or co-cultured with S. aureus ATCC 25923. As Figure 5 shows, the production of pyocyanin and elastase was significantly increased (p < 0.001 and p < 0.05, respectively) when P. aeruginosa PAO1 was co-cultured with S. aureus ATCC 25923 compared with P. aeruginosa PAO1 monoculture. However, treatment with Str7410 significantly reduced the production of pyocyanin and elastase in co-cultures compared with the untreated controls; this effect of Str7410 was concentration dependent. When the concentration of Str7410 was 40 μM, the inhibition of production of pyocyanin and elastase was 22.07% (p < 0.01) and 24.85% (p < 0.05), respectively, compared with no treatment with Str7410.
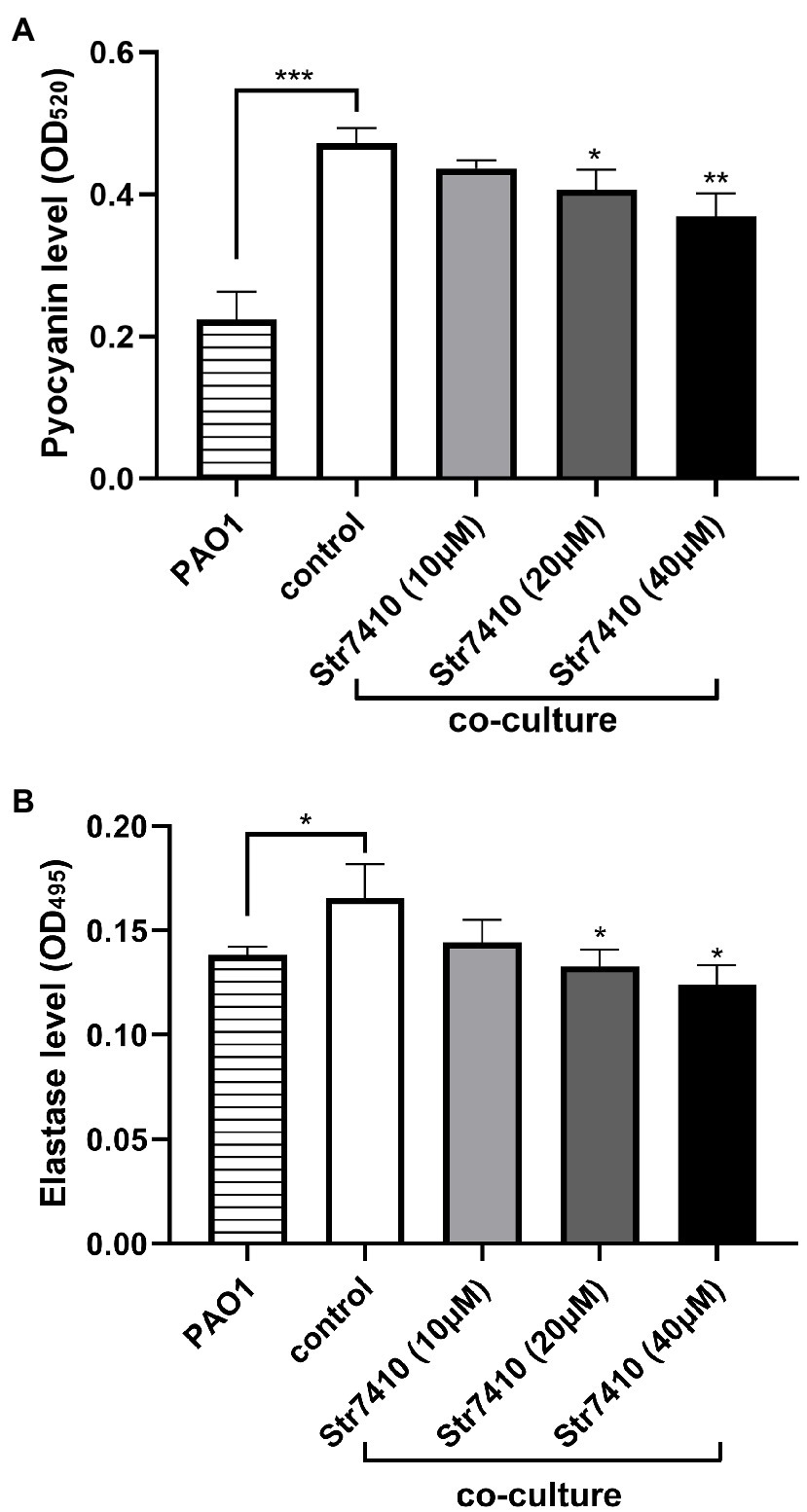
Figure 5. Production of virulence factors by Pseudomonas aeruginosa PAO1 in monoculture or co-culture with Staphylococcus aureus ATCC 25923, and the inhibitory effect of compound Str7410 on virulence factor production in co-culture. (A) Pyocyanin; (B) Elastase. Values are presented as mean ± SD (n = 3). Differences in data were compared with the untreated control at each time-point and considered significant when p < 0.05 (*p < 0.05, **p < 0.01, and ***p < 0.001) by one-way ANOVA with t-tests.
Effect of Compound Str7410 on Swarming Motility of Pseudomonas aeruginosa PAO1
Bacterial motility is important for pathogenicity and antibiotic resistance. Swarming motility, an important virulence factor of P. aeruginosa, is mainly regulated by QS-related genes, such as lasB and pvdQ (Tremblay and Deziel, 2008). Furthermore, S. aureus culture supernatant can promote the swarming motility of P. aeruginosa PAO1 under normoxia (Pallett et al., 2019). Similarly, in this study, culture supernatant of S. aureus ATCC 25923 promoted the swarming motility of P. aeruginosa PAO1 (Figures 6A,B). However, when compound Str7410 was added, the swarming motility was significantly inhibited (Figures 6B,C).
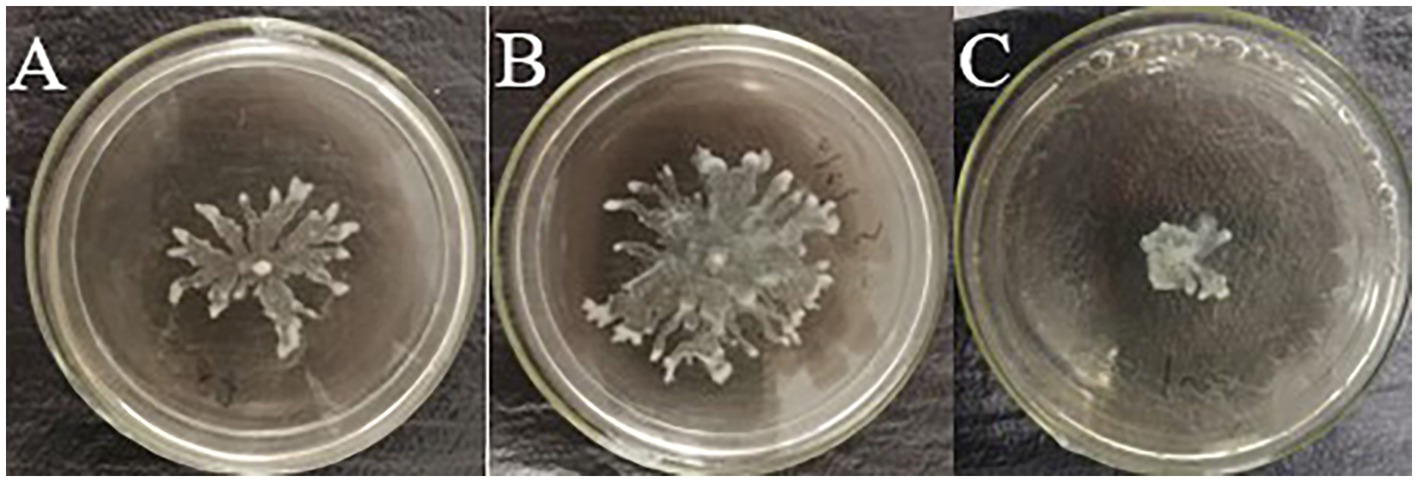
Figure 6. Effect of compound Str7410 on swarming motility of Pseudomonas aeruginosa PAO1. (A) Negative control (P. aeruginosa PAO1 alone). (B) Blank control (P. aeruginosa PAO1 with supernatant of Staphylococcus aureus ATCC 25923 culture). (C) Treatment group (P. aeruginosa PAO1 treated with supernatant of S. aureus ATCC 25923 culture and 10 μM Str7410).
Expression of QS-Related Genes in Pseudomonas aeruginosa PAO1 in Mixed Culture
From the data above, we found that S. aureus ATCC 25923 can increase the production of virulence factors and the swarming motility of P. aeruginosa PAO1 on co-culture of strains PAO1 and ATCC 25923. Compound Str7410 significantly inhibited this. These findings indicate that in multispecies infections, bacteria can enhance pathogenicity through their interactions, potentially via AI-2 QS systems. Therefore, we quantified the relative expression of QS-related genes of P. aeruginosa PAO1 in co-culture of P. aeruginosa PAO1 with S. aureus ATCC 25923. As shown in Figure 7, the expression of PAO1 QS-related genes (lasI, lasR, rhlI, rhlR, pqsH, and pqsR) and virulence factor-related genes (elastase, lasB; pyocyanin, and phzH) was significantly upregulated in the co-culture group compared with that in P. aeruginosa PAO1 monoculture. When Str7410 (40 μM) was added, the expression of all the strain PAO1 QS genes was significantly downregulated in co-culture compared with that in the absence of Str7410.
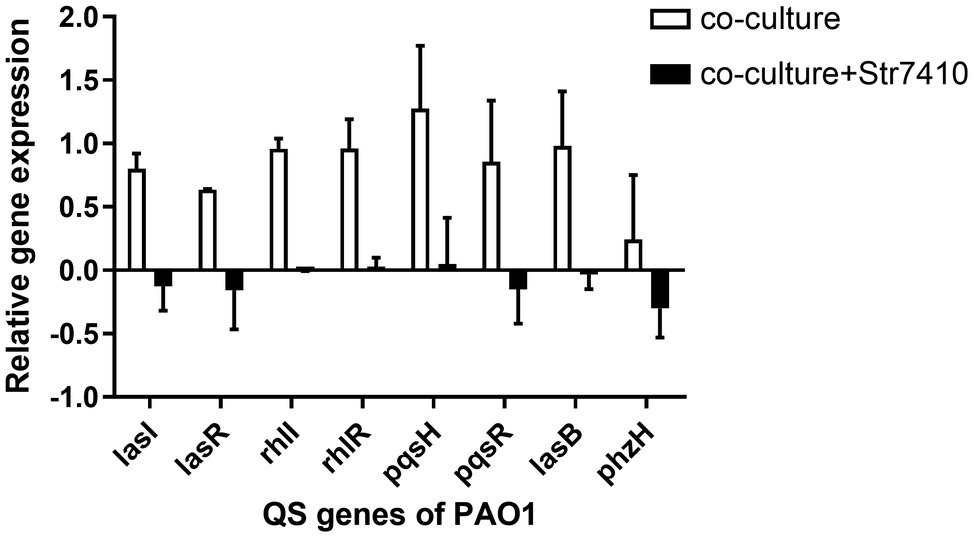
Figure 7. Expression on quorum sensing (QS) and virulence factor-related genes of Pseudomonas aeruginosa PAO1 when strain PAO1 was co-cultured with Staphylococcus aureus ATCC 25923 compared with PAO1 monoculture, and the effect of compound Str7410 (40 μM) on the expression of PAO1 QS and virulence factor-related genes in co-culture. Values are presented as the mean ± SD (n = 3).
Discussion
Bacterial infections, such as chronic wound infections and lung infections in patients with CF, are often caused by infection with multiple bacterial species (DeLeon et al., 2014; Nguyen and Oglesby-Sherrouse, 2016). Pseudomonas aeruginosa is a major opportunistic pathogen in hospitals, and nearly 80% of 18-year-old CF patients suffer chronic colonization and infection by P. aeruginosa (Duan et al., 2003). Staphylococcus aureus is another main bacterium in CF and chronic wounds. Staphylococcus aureus can interact with P. aeruginosa to promote the development of disease (Baldan et al., 2014). Studies demonstrated that P. aeruginosa and S. aureus compete with each other when co-cultured in vitro, and P. aeruginosa can produce 4-hydroxy-2-heptylquinoline-N-oxide (HQNO) through the pqs system to inhibit the aerobic respiration of S. aureus (Williams and Camara, 2009). In addition, virulence factors produced by P. aeruginosa, such as pyocyanin, have a significant inhibitory effect on the aerobic respiration and growth of S. aureus (Essar et al., 1990). However, although P. aeruginosa inhibits the growth of S. aureus in the planktonic state, a mixture of the two strains coordinates in the formation of biofilms and resistance to antibiotics. Staphylococcus aureus can avoid inhibition by P. aeruginosa by forming small colony variants, while the extracellular polymeric substances produced by P. aeruginosa can protect S. aureus from the killing effect of antibiotics (Atalla et al., 2011). Furthermore, P. aeruginosa and S. aureus co-culture promotes the production of virulence factors, which increase bacterial pathogenicity (Dalton et al., 2011; Orazi and O’Toole, 2017). Studies have indicated that QS mediated by AI-2 signaling molecules, a general communication system between bacteria, plays an important role in bacterial pathogenicity and drug resistance (Duan et al., 2003). In infectious disease, multiple species can coordinate with each other via QS, promoting bacterial pathogenicity and drug resistance. Pseudomonas aeruginosa can sense AI-2 signaling molecules produced by Streptococcus mitis, Staphylococcus aureus, and so on, leading to increased production of its own virulence factors and the formation of biofilms (Duan et al., 2003; Li et al., 2015; Wang et al., 2016b). Therefore, research into inhibitors of AI-2-mediated QS is a new strategy to target multispecies infections.
In previous study, we used the AI-2 signal molecule receptor LuxP as a target to screen 8,600 small molecule compounds from an in-house compound library using virtual screening techniques, and identified 14 AI-2 QSI compounds (Yuan, 2019). Wild-type V. harveyi senses AHL signal molecules as well as AI-2 signal molecules. To eliminate interference from AHLs, V. harveyi strain BB170 was used as the reporter here; this strain lacks the LuxN receptor for AHLs, but contains the LuxP receptor to sense AI-2 (Ren et al., 2001). As shown in Table 2, compound Str7410 had the best inhibitory effect on AI-2 QS by V. harveyi BB170 (IC50 = 0.3724 ± 0.1091 μM). Thus, we selected compound Str7410 as the test compound to evaluate its inhibitory effect on interspecific QS when P. aeruginosa PAO1 and S. aureus ATCC 25923 were co-cultured.
Because of abuse of antibiotics, bacterial drug resistance is increasing. In multibacterial infections, pathogenic bacteria can communicate with each other through AI-2 QS to promote pathogenicity and drug resistance (Roy et al., 2011). Biofilms, produced via QS, are an important reason for bacterial drug resistance. Pseudomonas aeruginosa can sense AI-2 produced by other bacteria, which regulates its production of virulence factors and formation of biofilms (Li et al., 2015; Wang et al., 2016b). Exogenous AI-2 can significantly increase P. aeruginosa biofilm formation (Li et al., 2017). However, an AI-2 QSI (D-ribose) affected the sensation of PAO1 to AI-2 and decreased the biomass of a mixed bacterial biofilm (Wang et al., 2016b). In this study, we found that P. aeruginosa PAO1 and S. aureus ATCC 25923 formed a mixed-species biofilm when co-cultured (Figure 2). Compound Str7410 inhibited QS between P. aeruginosa and S. aureus, thereby reducing the biofilm formation.
Nearly 80% of human infections are related to biofilms (Romero et al., 2008; Yan and Bassler, 2019). Meanwhile, the results of the present study showed that, compared with monoculture, co-culturing multiple species significantly reduced the effect of antibiotics (Table 3). In addition, compared with planktonic cells, biofilm formation significantly increased bacterial resistance to antibiotics (Table 3). Thus, the prevention and treatment of biofilms is a key concern of clinical staff and researchers (Stewart and Costerton, 2001). Studies have found that QSI can increase the susceptibility of pathogenic biofilm cells to antibiotics (Brackman et al., 2011; Furiga et al., 2015; Topa et al., 2020). Our study confirmed that compound Str7410 improved the susceptibility of a mixed-strain biofilm to the antibiotic MEPM, and that compound Str7410 can promote the inhibitory activities of antibiotics toward biofilm cells while having no effect on bacterial growth (Figure 1) and no eradication effect on the formed biofilm (Supplementary Table S1). Therefore, combining QSI with antibiotics is a new, and potentially highly effective, strategy for effective use of antibiotics against biofilm cells.
The nematode C. elegans is often used in in vivo models to study the pathogenicity of bacteria, for example, Serratia marcescens, P. aeruginosa, and Staphylococcus aureus (Powell and Ausubel, 2008; Brooks et al., 2009). The pathogenicity of co-infection with P. aeruginosa PAO1 and S. aureus ATCC 25923 toward C. elegans N2 was greater than that of either bacterial species alone. Compound Str7410 decreased the pathogenicity of the bacteria by inhibiting interspecific QS, thereby increasing the survival rate of infected nematodes (Figure 4). Thus, compound Str7410 can achieve a therapeutic effect in multispecies infections. Moreover, the combination of compound Str7410 with MEPM significantly increased the therapeutic effect of MEPM in infected nematodes.
We also evaluated the effect of compound Str7410 on the inhibition of interspecies QS via analysis of virulence factors of P. aeruginosa. Pyocyanin is a blue–green phenazine pigment, which has a number of physiological roles, such as facilitating biofilm development and influencing colony formation, and it is also one of the causes of pulmonary infections caused by P. aeruginosa (Wilson et al., 1988; Dietrich et al., 2006; Zhu et al., 2019). Pseudomonas aeruginosa regulates the production of pyocyanin through two QS pathways, the rhl and pqs systems (Diggle et al., 2003). Elastase destroys cellular tissues in infected hosts (Ben Haj Khalifa et al., 2011). Elastase is regulated by the QS-related lasB gene. Bacterial motility plays a critical role in pathogenicity and antibiotic resistance. Pseudomonas aeruginosa swarming motility is mainly regulated by QS-related genes such as lasB and pvdQ (Tremblay and Deziel, 2008). Pseudomonas aeruginosa can sense exogenous AI-2, which enhances the production of the virulence factors pyocyanin and elastase (Li et al., 2015). The peptidoglycan produced by S. aureus can also enhance the production of pyocyanin and elastase by P. aeruginosa (Korgaonkar et al., 2013). Under normoxia, the supernatant of S. aureus can promote the swarming motility of P. aeruginosa PAO1 (Pallett et al., 2019). In this study, we confirmed that the amount of pyocyanin and elastase produced by P. aeruginosa PAO1 significantly increased when P. aeruginosa and S. aureus were co-cultured (Figure 5), and S. aureus ATCC 25923 culture supernatant promoted the swarming motility of strain PAO1 (Figure 6B). Compound Str7410 significantly inhibited the virulence factor production and swarming motility of P. aeruginosa PAO1 during co-culture of P. aeruginosa PAO1 and S. aureus ATCC 25923 (Figures 5, 6C). We suggest that compound Str7410 reduced the production of virulence factors and swarming motility by inhibiting the sensation of P. aeruginosa PAO1 to AI-2 signaling molecules.
As a general QS system possessed by both Gram-positive and Gram-negative bacteria, AI-2 QS boosts the coordination and communication between bacteria in bacterial infection (Duan et al., 2003). Woods et al. (2018) found that there is an interaction between P. aeruginosa and S. aureus in co-cultured bacterial biofilms, and S. aureus can increase the expression of QS-related genes of P. aeruginosa. Moreover, the presence of exogenous AI-2 signal molecules can upregulate the expression of P. aeruginosa QS-related genes (Li et al., 2015). In this study (Figure 7), S. aureus ATCC 25923 significantly upregulated the expression of P. aeruginosa PAO1 QS-related genes (lasI, lasR, rhlI, rhlR, pqsH, and pqsR) and some QS target genes such as the virulence factor-related genes lasB and phzH. However, when compound Str7410 was added to the co-cultures, it reduced the pathogenicity of P. aeruginosa PAO1 by inhibiting the sensation of strain PAO1 to interspecific QS, thereby downregulating the expression of QS-related genes and the production of virulence factors.
In conclusion, this study focused on the inhibitory effects of a compound identified by virtual screening on the QS system of co-cultured Gram-positive bacterium S. aureus and Gram-negative bacterium P. aeruginosa in vitro and in vivo. Compound Str7410 was a potent AI-2 QSI. The combination of compound Str7410 with the antibiotic MEPM significantly increased the susceptibility of mixed-species-biofilm cells to the antibiotic. Preliminary mechanistic studies showed that Str7410 inhibited interspecific QS by inhibiting the AI-2 sensation of P. aeruginosa PAO1 and downregulating the expression of QS-related genes. This study provides a new strategy for the discovery of antibiotics in theory and a strategy for the clinical treatment of multidrug-resistant bacterial infections in practice.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding authors.
Author Contributions
FL and JX conceived and designed the experiments. KJ, YX, BY, YY, MZ, RL, HW, LW, and YZ performed the experiments. KJ and YX created the figures, analyzed the data, and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Science and Technology Major Project of China (2018ZX09711003-003).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.791802/full#supplementary-material
References
Allen, R. C., Popat, R., Diggle, S. P., and Brown, S. P. (2014). Targeting virulence: can we make evolution-proof drugs? Nat. Rev. Microbiol. 12, 300–308. doi: 10.1038/nrmicro3232
Armbruster, C. R., Wolter, D. J., Mishra, M., Hayden, H. S., Radey, M. C., Merrihew, G., et al. (2016). Staphylococcus aureus protein A mediates interspecies interactions at the cell surface of Pseudomonas aeruginosa. MBio 7, e00538–e00516. doi: 10.1128/mBio.00538-16
Armijo, L. M., Wawrzyniec, S. J., Kopciuch, M., Brandt, Y. I., Rivera, A. C., Withers, N. J., et al. (2020). Antibacterial activity of iron oxide, iron nitride, and tobramycin conjugated nanoparticles against Pseudomonas aeruginosa biofilms. J. Nanobiotechnol. 18:35. doi: 10.1186/s12951-020-0588-6
Atalla, H., Gyles, C., and Mallard, B. (2011). Staphylococcus aureus small colony variants (SCVs) and their role in disease. Anim. Health Res. Rev. 12, 33–45. doi: 10.1017/S1466252311000065
Baldan, R., Cigana, C., Testa, F., Bianconi, I., De Simone, M., Pellin, D., et al. (2014). Adaptation of Pseudomonas aeruginosa in cystic fibrosis airways influences virulence of Staphylococcus aureus in vitro and murine models of co-infection. PLoS One 9:e89614. doi: 10.1371/journal.pone.0089614
Bassler, B. L., Wright, M., and Stiverman, M. R. (1994). Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 13, 273–286. doi: 10.1111/j.1365-2958.1994.tb00422.x
Ben Haj Khalifa, A., Moissenet, D., Vu Thien, H., and Khedher, M. (2011). Virulence factors in Pseudomonas aeruginosa: mechanisms and modes of regulation. Ann. Biol. Clin. 69, 393–403. doi: 10.1684/abc.2011.0589
Blanchard, A. C., and Waters, V. J. (2019). Microbiology of cystic fibrosis airway disease. Semin. Respir. Crit. Care Med. 40, 727–736. doi: 10.1055/s-0039-1698464
Brackman, G., Cos, P., Maes, L., Nelis, H. J., and Coenye, T. (2011). Quorum sensing inhibitors increase the susceptibility of bacterial biofilms to antibiotics in vitro and in vivo. Antimicrob. Agents Chemother. 55, 2655–2661. doi: 10.1128/AAC.00045-11
Brooks, K. K., Liang, B., and Watts, J. L. (2009). The influence of bacterial diet on fat storage in C. elegans. PLoS One 4:e7545. doi: 10.1371/journal.pone.0007545
Chbib, C. (2020). Impact of the structure-activity relationship of AHL analogues on quorum sensing in gram-negative bacteria. Bioorg. Med. Chem. 28:115282. doi: 10.1016/j.bmc.2019.115282
Collins, K. C., Tsuchikama, K., Lowery, C. A., Zhu, J., and Janda, K. D. (2015). Dissecting AI-2-mediated quorum sensing through C5-analogue synthesis and biochemical analysis. Tetrahedron 72, 3593–3598. doi: 10.1016/j.tet.2015.08.063
Conway, C. A., Esiobu, N., and Lopez, J. V. (2012). Co-cultures of Pseudomonas aeruginosa and Roseobacter denitrificans reveal shifts in gene expression levels compared to solo cultures. Sci. World J. 2012:120108. doi: 10.1100/2012/120108
Dalton, T., Dowd, S. E., Wolcott, R. D., Sun, Y., Watters, C., Griswold, J. A., et al. (2011). An in vivo polymicrobial biofilm wound infection model to study interspecies interactions. PLoS One 6:e27317. doi: 10.1371/journal.pone.0027317
DeLeon, S., Clinton, A., Fowler, H., Everett, J., Horswill, A. R., and Rumbaugh, K. P. (2014). Synergistic interactions of Pseudomonas aeruginosa and Staphylococcus aureus in vivo an in vitro wound model. Infect. Immun. 82, 4718–4728. doi: 10.1128/IAI.02198-14
Dietrich, L. E., Price-Whelan, A., Petersen, A., Whiteley, M., and Newman, D. K. (2006). The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol. Microbiol. 61, 1308–1321. doi: 10.1111/j.1365-2958.2006.05306.x
Diggle, S. P., Winzer, K., Chhabra, S. R., Worrall, K. E., Camara, M., and Williams, P. (2003). The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol. Microbiol. 50, 29–43. doi: 10.1046/j.1365-2958.2003.03672.x
Duan, K., Dammel, C., Stein, J., Rabin, H., and Surette, M. G. (2003). Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol. Microbiol. 50, 1477–1491. doi: 10.1046/j.1365-2958.2003.03803.x
Essar, D. W., Eberly, L., Hadero, A., and Crowford, I. P. (1990). Identification and characterization of genes for a second Anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two Anthranilate synthases and evolutionary implications. J. Bacteriol. 172, 884–900. doi: 10.1128/jb.172.2.884-900.1990
Furiga, A., Lajoie, B., Hage, E. I. S., Baziard, G., and Roques, C. (2015). Impairment of Pseudomonas aeruginosa biofilm resistance to antibiotics by combining the drugs with a new quorum-sensing inhibitor. Antimicrob. Agents Chemother. 60, 1676–1686. doi: 10.1128/AAC.02533-15
Ganin, H., Tang, X., and Meijler, M. M. (2009). Inhibition of Pseudomonas aeruginosa quorum sensing by AI-2 analogs. Bioorg. Med. Chem. Lett. 19, 3941–3944. doi: 10.1016/j.bmcl.2009.03.163
Gokalsin, B., Aksoydan, B., Erman, B., and Sesal, N. C. (2017). Reducing virulence and biofilm of Pseudomonas aeruginosa by potential quorum sensing inhibitor carotenoid: Zeaxanthin. Microb. Ecol. 74, 466–473. doi: 10.1007/s00248-017-0949-3
Guo, Q., Wei, Y., Xia, B., Jin, Y., Liu, C., Pan, X., et al. (2016). Identification of a small molecule that simultaneously suppresses virulence and antibiotic resistance of Pseudomonas aeruginosa. Sci. Rep. 6:19141. doi: 10.1038/srep19141
Harrison, F. (2007). Microbial ecology of the cystic fibrosis lung. Microbiology 153, 917–923. doi: 10.1099/mic.0.2006/004077-0
Hotterbeekx, A., Kumar-Singh, S., Goossens, H., and Malhotra-Kumar, S. (2017). In vivo and in vitro interactions between Pseudomonas aeruginosa and staphylococcus spp. Front. Cell. Infect. Microbiol. 7:106. doi: 10.3389/fcimb.2017.00106
Jiang, K., Yan, X., Yu, J., Xiao, Z., Wu, H., Zhao, M., et al. (2020). Design, synthesis, and biological evaluation of 3-amino-2-oxazolidinone derivatives as potent quorum-sensing inhibitors of Pseudomonas aeruginosa PAO1. Eur. J. Med. Chem. 194:112252. doi: 10.1016/j.ejmech.2020.112252
Kalia, V. C. (2013). Quorum sensing inhibitors: an overview. Biotechnol. Adv. 31, 224–245. doi: 10.1016/j.biotechadv.2012.10.004
Korgaonkar, A., Trivedi, U., Rumbaugh, K. P., and Whiteley, M. (2013). Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc. Natl. Acad. Sci. U. S. A. 110, 1059–1064. doi: 10.1073/pnas.1214550110
Li, H., Li, X., Song, C., Zhang, Y., Wang, Z., Liu, Z., et al. (2017). Autoinducer-2 facilitates Pseudomonas aeruginosa PAO1 pathogenicity in vitro and in vivo. Front. Microbiol. 8:1944. doi: 10.3389/fmicb.2017.01944
Li, H., Li, X., Wang, Z., Fu, Y., Ai, Q., Dong, Y., et al. (2015). Autoinducer-2 regulates Pseudomonas aeruginosa PAO1 biofilm formation and virulence production in a dose-dependent manner. BMC Microbiol. 15:192. doi: 10.1186/s12866-015-0529-y
Lowery, C. A., Abe, T., Park, J., Eubanks, L. M., Sawada, D., Kaufmann, G. F., et al. (2009). Revisiting AI-2 quorum sensing inhibitors: direct comparison of alkyl-DPD analogues and a natural product Fimbrolide. J. Am. Chem. Soc. 131, 15584–15585. doi: 10.1021/ja9066783
Lowery, C. A., Park, J., Kaufmann, G. F., and Janda, K. D. (2008). An unexpected switch in the modulation of AI-2-based quorum sensing discovered through synthetic 4,5-Dihydroxy-2,3-pentanedione analogues. J. Am. Chem. Soc. 130, 9200–9201. doi: 10.1021/ja802353j
Ma, R., Qiu, S., Jiang, Q., Sun, H., Xue, T., Cai, G., et al. (2017). AI-2 quorum sensing negatively regulates rbf expression and biofilm formation in Staphylococcus aureus. Int. J. Med. Microbiol. 307, 257–267. doi: 10.1016/j.ijmm.2017.03.003
Mok, K. C., Wingreen, N. S., and Bassler, B. L. (2003). Vibrio harveyi quorum sensing: a coincidence detector for two autoinducers controls gene expression. EMBO J. 22, 870–881. doi: 10.1093/emboj/cdg085
Nguyen, A. T., and Oglesby-Sherrouse, A. G. (2016). Interactions between Pseudomonas aeruginosa and Staphylococcus aureus during co-cultivations and polymicrobial infections. Appl. Microbiol. Biotechnol. 100, 6141–6148. doi: 10.1007/s00253-016-7596-3
Orazi, G., and O’Toole, G. A. (2017). Pseudomonas aeruginosa alters Staphylococcus aureus sensitivity to vancomycin in a biofilm model of cystic fibrosis. Infection 8, e00873–e00817. doi: 10.1128/mBio.00873-17
Pallett, R., Leslie, L. J., Lambert, P. A., Milic, I., Devitt, A., and Marshall, L. J. (2019). Anaerobiosis influences virulence properties of Pseudomonas aeruginosa cystic fibrosis isolates and the interaction with Staphylococcus aureus. Sci. Rep. 9:6748. doi: 10.1038/s41598-019-42952-x
Pastar, I., Nusbaum, A. G., Gil, J., Patel, S. B., Chen, J., Valdes, J., et al. (2013). Interactions of methicillin resistant Staphylococcus aureus USA300 and Pseudomonas aeruginosa in polymicrobial wound infection. PLoS One 8:e56846. doi: 10.1371/journal.pone.0056846
Pearson, J. P., Pesci, E. C., and Iglewski, B. H. (1997). Roles of Pseudomonas aeruginosa las and rhl quorum-sensing Systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 179, 5756–5767. doi: 10.1128/jb.179.18.5756-5767.1997
Peeters, E., Nelis, H. J., and Coenye, T. (2009). In vitro activity of ceftazidime, ciprofloxacin, meropenem, minocycline, tobramycin and trimethoprim/sulfamethoxazole against planktonic and sessile Burkholderia cepacia complex bacteria. J. Antimicrob. Chemother. 64, 801–809. doi: 10.1093/jac/dkp253
Powell, J. R., and Ausubel, F. M. (2008). Models of Caenorhabditis elegans infection by Bacterialand fungal pathogens. Methods Mol. Biol. 415, 403–427. doi: 10.1007/978-1-59745-570-1_24
Rajkumari, J., Borkotoky, S., Murali, A., Suchiang, K., Mohanty, S. K., and Busi, S. (2018). Attenuation of quorum sensing controlled virulence factors and biofilm formation in Pseudomonas aeruginosa by pentacyclic triterpenes, betulin and betulinic acid. Microb. Pathog. 118, 48–60. doi: 10.1016/j.micpath.2018.03.012
Ren, D., Sims, J. J., and Wood, T. K. (2001). Inhibition of biofilm formation and swarming of Escherichia coli by (5Z)-4-bromo-5-(bromomethylene)-3-butyl-2(5H)-furanone. Environ. Microbiol. 3, 731–736. doi: 10.1046/j.1462-2920.2001.00249.x
Romero, R., Schaudinn, C., Kusanovic, J. P., Gorur, A., Gotsch, F., Webster, P., et al. (2008). Detection of a microbial biofilm in intraamniotic infection. Am. J. Obstet. Gynecol. 198, 135.e1–135.e5. doi: 10.1016/j.ajog.2007.11.026
Roy, V., Adams, B. L., and Bentley, W. E. (2011). Developing next generation antimicrobials by intercepting AI-2 mediated quorum sensing. Enzym. Microb. Technol. 49, 113–123. doi: 10.1016/j.enzmictec.2011.06.001
Sifri, C. D. (2008). Healthcare epidemiology: quorum sensing: bacteria talk sense. Clin. Infect. Dis. 47, 1070–1076. doi: 10.1086/592072
Stewart, P. S., and Costerton, J. W. (2001). Antibiotic resistance of bacteria in biofilms. Lancet 358, 135–138. doi: 10.1016/s0140-6736(01)0532-1
Topa, S. H., Palombo, E. A., Kingshott, P., and Blackall, L. L. (2020). Activity of Cinnamaldehyde on quorum sensing and biofilm susceptibility to antibiotics in Pseudomonas aeruginosa. Microorganisms 8:455. doi: 10.3390/microorganisms8030455
Tremblay, J., and Deziel, E. (2008). Improving the reproducibility of Pseudomonas aeruginosa swarming motility assays. J. Basic Microbiol. 48, 509–515. doi: 10.1002/jobm.200800030
Tsukatani, T., Kawaguchi, T., Suenaga, H., Shiga, M., and Ikegami, T. (2016). Rapid and simple determination of minimum biofilm eradication concentration by a colorimetric microbial viability assay based on reduction of a water-tetrazolium salt and combinated effect of antibiotics against microbial biofilm. J. Microb. Biotech. Food 6, 677–680. doi: 10.15414/jmbfs.2016.6.1.677-680
Tsukatani, T., Sakata, F., Kuroda, R., and Akao, R. (2020). Biofilm eradication activity of herb and spice extracts alone and in combination against oral and food-borne pathogenic bacteria. Curr. Microbiol. 77, 2486–2495. doi: 10.1007/s00284-020-02017-z
Van Delden, C., and Iglewski, B. H. (1998). Cell-to-Cell Signaling and Pseudomonas aeruginosa. Infections. Emerg. Infect. Dis. 4, 551–560. doi: 10.3201/eid0404.980405
Wang, S., Chen, X., Wang, H., Xie, S., Wei, H., and Pan, Y. (2016a). In vitro activities of doripenem against Pseudomonas aeruginosa biofilm. Chin. J. Antibiot. 41, 466–476. doi: 10.13461/j.cnki.cja.005756
Wang, Y., Liu, B., Grenier, D., and Yi, L. (2019). Regulatory mechanisms of the LuxS/AI-2 system and bacterial resistance. Antimicrob. Agents Chemother. 63, e01186–e01119. doi: 10.1128/AAC.01186-19
Wang, Z., Xiang, Q., Yang, T., Li, L., Yang, J., Li, H., et al. (2016b). Autoinducer-2 of Streptococcus mitis as a target molecule to inhibit pathogenic multi-species biofilm formation in vitro and in an endotracheal intubation rat model. Front. Microbiol. 7:88. doi: 10.3389/fmicb.2016.00088
Williams, P., and Camara, M. (2009). Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: a tale of regulatory networks and multifunctional signal molecules. Curr. Opin. Microbiol. 12, 182–191. doi: 10.1016/j.mib.2009.01.005
Wilson, R., Sykes, D. A., Watson, D., Rutman, A., Taylor, G. W., and Cole, P. J. (1988). Measurement of Pseudomonas aeruginosa Phenazine pigments in sputum and assessment of their contribution to sputum sol toxicity for respiratory epithelium. Infect. Immun. 56, 2515–2517. doi: 10.1128/iai.56.9.2515-2517.1988
Woods, P. W., Haynes, Z. M., Mina, E. G., and Marques, C. N. H. (2018). Maintenance of S. aureus in co-culture with P. aeruginosa while growing as biofilms. Front. Microbiol. 9:3291. doi: 10.3389/fmicb.2018.03291
Yan, J., and Bassler, B. L. (2019). Surviving as a community: antibiotic tolerance and persistence in bacterial biofilms. Cell Host Microbe 26, 15–21. doi: 10.1016/j.ajog.2007.11.026
Yang, L., Liu, Y., Markussen, T., Hoiby, N., Tolker-Nielsen, T., and Molin, S. (2011). Pattern differentiation in co-culture biofilms formed by Staphylococcus aureus and Pseudomonas aeruginosa. FEMS Immunol. Med. Microbiol. 62, 339–347. doi: 10.1111/j.1574-695X.2011.00820.x
Yuan, B. (2019). Design, Synthesis and Biological Activity of AI-2 Mediated Quorum Sensing Inhibitors in V. harveyi. MA thesis. Academy of Military Sciences, Beijing.
Zang, T., Lee, B. W., Cannon, L. M., Ritter, K. A., Dai, S., Ren, D., et al. (2009). A naturally occurring brominated furanone covalently modifies and inactivates LuxS. Bioorg. Med. Chem. Lett. 19, 6200–6204. doi: 10.1016/j.bmcl.2009.08.095
Zhao, L., Xue, T., Shang, F., Sun, H., and Sun, B. (2010). Staphylococcus aureus AI-2 quorum sensing associates with the KdpDE two-component system to regulate capsular polysaccharide synthesis and virulence. Infect. Immun. 78, 3506–3515. doi: 10.1128/IAI.00131-10
Keywords: Pseudomonas aeruginosa PAO1, Staphylococcus aureus ATCC 25923, AI-2 quorum sensing, ethylene diamine triacetic acid group, interspecies
Citation: Jiang K, Xu Y, Yuan B, Yue Y, Zhao M, Luo R, Wu H, Wang L, Zhang Y, Xiao J and Lin F (2022) Effect of Autoinducer-2 Quorum Sensing Inhibitor on Interspecies Quorum Sensing. Front. Microbiol. 13:791802. doi: 10.3389/fmicb.2022.791802
Edited by:
Rinarani Ray, Maulana Abul Kalam Azad University of Technology, IndiaReviewed by:
Sougata Ghosh, RK University, IndiaTung Truong, University of Pittsburgh, United States
Copyright © 2022 Jiang, Xu, Yuan, Yue, Zhao, Luo, Wu, Wang, Zhang, Xiao and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junhai Xiao, xiaojunhai@139.com; Feng Lin, linfeng@jlu.edu.cn
 Kai Jiang1,2
Kai Jiang1,2 Feng Lin
Feng Lin