- 1Gaolan Station of Agricultural and Ecological Experiment, Northwest Institute of Eco-Environment and Resources, Chinese Academy of Sciences (CAS), Lanzhou, China
- 2CAS Key Laboratory of Tropical Forest Ecology, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences (CAS), Mengla, China
- 3University of Chinese Academy of Sciences, Beijing, China
- 4Key Laboratory of Desert and Desertification, Northwest Institute of Eco-Environment and Resources, Chinese Academy of Sciences (CAS), Lanzhou, China
Root rot disease is a serious infection leading to production loss of Chinese wolfberry (Lycium barbarum). This study tested the potential for two bacterial biological control agents, Bacillus amyloliquefaciens HSB1 and FZB42, against five fungal pathogens that frequently cause root rot in Chinese wolfberry. Both HSB1 and FZB42 were found to inhibit fungal mycelial growth, in vitro and in planta, as well as to promote the growth of wolfberry seedlings. In fact, a biocontrol experiment showed efficiency of 100% with at least one treatment involving each biocontrol strain against Fusarium oxysporum. Metagenomic sequencing was used to assess bacterial community shifts in the wolfberry rhizosphere upon introduction of each biocontrol strain. Results showed that HSB1 and FZB42 differentially altered the abundances of different taxa present and positively influenced various functions of inherent wolfberry rhizosphere bacteria. This study highlights the application of biocontrol method in the suppression of fungal pathogens that cause root rot disease in wolfberry, which is useful for agricultural extension agents and commercial growers.
Introduction
Chinese wolfberry (Lycium barbarum) is a deciduous perennial plant of economic importance that grows well in the northwest, arid regions of China due to its salt tolerance, drought resistance, and fast-growing qualities (Byambasuren et al., 2019; Wang et al., 2019b). In China, wolfberry is used in traditional medicine because of its high content in bioactive secondary metabolites and its multitude of benefits to human health (Bucheli et al., 2011; Byambasuren et al., 2019; Wang et al., 2019a). Currently, the total planting area of wolfberry plants in China represents more than 1.33 × 105 ha (Meng et al., 2019; Wang et al., 2019a). Unfortunately, wolfberry yield has been severely impacted by root rot, one of the most widespread and destructive soil borne diseases. In our previous study, we investigated root rot disease in Chinese wolfberry. Fusarium species were the most abundant among all isolated fungal pathogens, and Fusarium infected plants were characterized by yellow leaves, necrosis, death and rotten roots (Uwaremwe et al., 2021). It can be difficult to identify, measure and manage root rot disease in a nursery setting because pathogens may easily and quickly spread from plant to plant causing widespread death of seedlings (Omukhua and Godwin-Egein, 2011). Various fungicides are known to be effective against soil borne disease (Dhahira-Beevi and Qadri, 2010). However, an increasing use of chemical treatments causes several negative effects such as environmental pollution, imbalance in the soil ecosystem, potential threat to silkworms, and development of pathogen resistance (Compant et al., 2005; O’Brien, 2017). Thus, biocontrol using antagonistic microorganisms is a safer alternative to reduce the use of chemicals in agriculture, and it is considered as a promising approach for the management of soil borne diseases (Dhahira-Beevi and Qadri, 2010; Shahid and Khan, 2016; Singh et al., 2017; Alamri et al., 2019). Biocontrol Agents (BCAs) are potentially beneficial microorganisms including fungi, viruses and a group of bacteria called plant growth promoting rhizobacteria (PGPR) (Pal and McSpadden, 2006; Mota et al., 2017; O’Brien, 2017; Köhl et al., 2019). Members of Bacillus spp. and Pseudomonas spp. (bacteria), and Trichoderma spp. (fungi), have demonstrated abilities to suppress several soil borne plant pathogens, including species of Streptomyces and Fusarium, while also promoting plant growth (Singhai et al., 2011; Meng et al., 2013; Saravanakumar et al., 2017). Thus, they can serve simultaneously as both biopesticide and biofertilizer. PGPR colonize the root surface and the closely adhering soil interface (i.e., the rhizosphere) and some of them can also enter the root interior as endophytes (Compant et al., 2005; Beneduzi et al., 2012; Lyu et al., 2019). PGPR enhance nutrient availability, stimulate growth hormones, and suppress disease prevalence (Passari et al., 2018; Lyu et al., 2019; Rajaofera et al., 2020). Moreover, PGPR can suppress a broad range of pathogenic microbes including viruses, bacteria and fungi (Mazhar et al., 2016). Additionally, PGPR can improve plant health by acting as antagonists of pathogens using mechanisms such as solubilizing Fe and P, N fixation, or production of antibiotic compounds or hormones (Labuschagne et al., 2010; Ambreen et al., 2012; Beneduzi et al., 2012; Salomon et al., 2017). Disease suppression mechanisms include antibiosis, Induced Systemic Resistance (ISR), high affinity siderophore production, competition for nutrient and niches, and production of lytic enzymes (Rajiv et al., 2017; Salomon et al., 2017). The predominant genera of PGPR are Pseudomonas and Bacillus (Mazhar et al., 2016; Passari et al., 2018; Hashem et al., 2019). Members of genus Bacillus have been reported to be effective PGPR in a wide range of plants, and this genus is one of the principal PGPR groups known for their application as BCAs against several pathogenic fungi (Zhao et al., 2013; Leila et al., 2015; Ge et al., 2016; Mazhar et al., 2016; Hashem et al., 2019; Tiwari et al., 2019). Many Bacillus species are commonly isolated endophytes and are known to impart biological control against various diseases (Melnick et al., 2013). Bacillus cereus S42, isolated from Nicotiana glauca organs, suppressed Fusarium wilt in tomato (Aydi-Ben et al., 2016). Endophytic B. subtilis strain E1R displayed a biocontrol efficacy against wheat powdery mildew (Gao et al., 2015). Bacillus methylotrophicus strain NKG-1, isolated from the rhizosphere of a Pinus koraiensis in a dormant volcano in southern China, exhibited significant antifungal and pro-fertilization activities on tomato plants (Ge et al., 2016). Bacillus subtilis SQR9 showed potential to control Fusarium wilt in cucumber plants by root colonization (Cao et al., 2011). Es-soufi et al. (2020) showed that B. amyloliquefaciens Bc2 is a potent biocontrol agent against strawberry anthracnose. B. amyloliquefaciens Q-426 displayed a potential biocontrol ability against Fusarium oxysporum f. sp. spinaciae (Zhao et al., 2013). The importance of the composition of the rhizosphere microbiome on plant health and productivity has been increasingly recognized. Some studies revealed that the application of BCAs belonging to Bacillus spp. or Trichoderma spp. suppressed soil borne plant diseases and altered the composition of the rhizosphere microbial community in banana, cucumber, and potato (Shen et al., 2015; Han et al., 2019; Wang et al., 2019c).
Here, we showed F. oxysporum root rot disease incidence and severity in Chinese wolfberry was preventable by implementing a biocontrol strategy. With application of each of two bacterial BCAs (B. amyloliquefaciens strains HSB1 and FBZ42), we observed (1) inhibited growth of fungal mycelia along with enhanced plant growth, and (2) disease suppression through each BCA altering the bacterial composition of the wolfberry rhizosphere. Therefore, these strains offer potential as safe BCAs to protect Chinese wolfberry while also ensuring a good yield of plant material.
Materials and Methods
Fungal Pathogens and Bacterial Antagonists Used in This Study
We used five fungal pathogens, including Fusarium oxysporum, F. solani, F. chlamydosporum, F. tricinctum and Alternaria alternata, that were previously identified as root-rot-causing pathogens in wolfberry plants and they were preserved in glycerol at −80°C (Uwaremwe et al., 2021). The five fungi were cultivated at 25°C for 7 days on potato dextrose agar (PDA) that included 20% potato infusion, 2% dextrose, and 1.5% agar obtained from Qingdao Hope Bio-Technology Co., Ltd., in China. Afterward, a small block of mycelium agar was cut and placed into the center of a fresh PDA plate. One of the BCAs used in the current study was Bacillus amyloliquefaciens strain FBZ42, a commercial strain from the company ABiTEP GmbH that was donated by the Bacillus Genetic Stock Center (BGSC). This strain has shown both plant growth promotion and disease suppression potential for different plants (Kröber et al., 2014). The second bacterial BCA, HSB1, was an uncharacterized endophytic bacterium we isolated from wolfberry root tissues while conducting this study. Briefly, wolfberry roots were cleaned using 75% ethanol for 30 s and immediately transferred to 3% sodium hypochlorite for 5 min, and finally washed three times with sterile water. They were finally cut into small, thin blocks 0.5 cm × 0.5 cm, and placed on NA (Nutrient Agar) culture medium obtained from Qingdao Hope Bio-Technology Co., Ltd., in China, followed by incubation at 37°C. The FZB42 and HSB1 cells were grown and maintained in Luria Bertani liquid medium at 4°C for further experiment.
In vitro Antifungal Assay
In the plate confrontation assay, a small block of agar covered with fungal mycelia was excised and placed onto the center of a fresh PDA plate and incubated for three additional days to ensure fungal colonization of the new plate. Afterward, FZB42 and HSB1 were added to Luria Bertani liquid medium (30 mL) and incubated for overnight at 37°C with shaking at 150 rpm. Then, 5 μl of either FZB42 or HSB1 cells were point inoculated on the PDA plate 2.5 cm away from the fungus. Their antifungal activity was evaluated by comparing fungal mycelium growth in the presence of each bacterial BCA, and plates that were inoculated with fungi alone (control plates), after 7 days of incubation at 25°C as reported in Cao et al. (2018).
Strain HSB1 was identified using 16S rRNA gene sequences as previously reported (Mignard and Flandrois, 2006; Janda and Abbott, 2007), after it had already displayed antifungal activity. The resulting 16S rRNA gene sequence was compared in a BLAST search to those in the National Library of Medicine (Bethesda, United States) database (Altschul et al., 1997). Phylogenetic analysis of HSB1 was performed using MEGA 5.5 (Kumar et al., 2008) and the relationships between HSB1 and other Bacillus sequences were analyzed using the neighbor-joining method (Saitou and Nei, 1987; Uwaremwe et al., 2021). Bootstrap values for the neighbor-joining tree were calculated for 1,000 replicates (Felsenstein, 1985; Uwaremwe et al., 2021).
Effects of FZB42 and HSB1 on the Growth of Wolfberry Seedlings
In addition to assessing their antifungal abilities, FZB42 and HSB1 were tested twice for their potential to promote plant growth of Chinese wolfberry seedlings under laboratory conditions. The first assay, conducted from August to October 2019, compared the plant growth promotion potentials of HSB1 and FZB42. To prepare bacterial suspensions, each BCA was first added to Luria Bertani liquid medium (30 mL) and incubated for 12 h at 35°C with shaking at 150 rpm until the logarithmic growth phase was reached. Afterward, all cells were harvested by centrifugation at 5,000 rpm for 5 min, the supernatant was discarded, and the pellets were washed and resuspended in sterile distilled water to obtain initial bacterial population densities of 6 × 109 (FZB42) and 4 × 109 (HSB1) colony forming units (CFU) mL–1. Finally, sterilized distilled water was used to make the final suspension at 5 × 108 CFU mL–1 for both HSB1 and FZB42 following the protocol of Cheng et al. (2019). Lycium barbarum “Ningxia N1” seeds obtained from the Institute of Plant Protection, Ningxia Academy of Agricultural and Forestry Sciences, China, were planted in three large plastic pots (40 cm × 60 cm) containing autoclaved pindstrup substrate (pH 5.5–6) obtained from market. Each pot was amended with 2 L of bacterial suspension, with FZB42 as treatment 1, HSB1 as treatment 2, and water control as treatment 3. When each plant had 3–4 leaves, seedlings were individually transferred to small plastic pots (3 seedlings/pot). In each pot was a mixture of autoclaved sand obtained from the Tengger desert in China (latitude 37°30′ to 40° north × longitude 102°20′ to 106° east), soil obtained from Gaolan county in Gansu province (latitude 36°05′–36°51′ north × longitude 103°32′–104°14′ east) and pindstrup substrate in a 1:1:1 ratio (vol/vol/vol), that was irrigated with 50 ml of each treatment suspension (FZB42, HSB1 or water). Root length (cm) and stem height (cm) were recorded for inoculated and non-inoculated seedlings at 60, 68, and 70 days after planting. The second assay was conducted from May through the end of June 2020, and involved HSB1 alone. L. barbarum “Ningxia N1” seeds were planted in a large plastic pot containing autoclaved pindstrup substrate as in the first assay, and the pot was amended with 2 L of water. When each had 3–4 leaves, seedlings were individually transferred to small plastic pots (1 seedling/pot) filled with a mixture of autoclaved sand, soil and pindstrup substrate (1:1:1 ratio, vol/vol/vol), and immediately irrigated with water. When seedlings reached 35 days of growth, HSB1 was inoculated at a concentration of 2 × 107 CFU mL–1. Shoot weight (g) and root weight (g) were recorded for inoculated and non-inoculated seedlings every 7 days after inoculation. The two experiments were conducted according to a randomized complete design composed of three replicates for each treatment and water control.
Biocontrol Experiment
In addition to the in vitro biocontrol experiments, and in planta biocontrol experiment was conducted whereby HSB1 and FZB42 were used as BCAs against a strain of F. oxysporum that was previously isolated from Chinese wolfberry and identified based on ITS and TEF (Genbank reference: MN959986 and MT811807) (Uwaremwe et al., 2021). L. Barbarum seedlings were obtained from seed germination. Briefly, seeds were surface sterilized with 75% ethanol for 30 s, sodium hypochlorite for 5 min, and finally washed with sterile distilled water five times. They were planted in two different large plastic pots containing autoclaved pindstrup substrate amended with tap water. All pots were kept in laboratory conditions at a temperature of 25°C, with humidity between 75 and 90%, and an alternating cycle of 16 h light / 8 h dark having a total light intensity of 800 μMol/m2/s. After 20 days, germinated seedlings were individually transplanted into different plug trays containing a mixture of soil, sand and pindstrup substrate in equal ratio (1:1:1 ratio, vol/vol/vol) autoclaved two times to ensure complete disinfection. At the 4–5 leaf developmental stage, individual seedlings were transplanted into their own large plastic pots containing autoclaved soil, sand and pindstrup substrate.
For inoculum preparation, HSB1 and FZB42 were prepared as previously described in the plant growth promotion experiment. To obtain the fungal inoculum, F. oxysporum was cultured on petri dishes containing PDA and incubated at 25°C for 10–15 days. A conidial suspension was prepared by pouring 30 mL of sterile distilled water into each of the petri dishes and dislodging spores with a sterile toothbrush. The initial concentration of conidia in the suspension was determined using a hemocytometer, and the final inoculum concentration was adjusted to 5 × 107 conidia ml–1. Two methods of inoculation were used for this experiment. The first method involved inoculating seedlings with F. oxysporum 5 days before either FZB42 or HSB1 was introduced. The second inoculation procedure method involved inoculating FZB42 or HSB1 first, followed by a supplementary inoculation after 7 days to ensure their colonization as reported in Gadhave et al. (2018). After another 10 days, the BCA-treated seedlings were inoculated with F. oxysporum. In total, eight treatments were used: (1) CK, untreated seedlings (Control), (2) F. oxysporum alone, (3) FZB42 alone, (4) HSB1 alone, (5) F. oxysporum + FZB42, (6) F. oxysporum + HSB1, (7) FZB42 + F. oxysporum and (8) HSB1 + F. oxysporum. Seedlings were inoculated by pouring 40 ml of the prepared inoculum onto the soil surface. The control treatments consisted of an equivalent volume of sterile distilled water. This experiment was conducted according to a randomized complete design composed of three replicates for each treatment and water control.
Disease incidence (DI) was calculated according to the formula developed by Sharma and Kolte (1994) as the percentage of infected seedlings out of the total of all treated seedlings for each treatment, according to the following formula:
Disease severity (DS) of the foliage was evaluated using a rating scale from 0 to 4 as reported in Han et al. (2019). Based on different stages of root rot, 0 = seedlings with no symptoms, 1 = leaf yellowing, 2 = necrosis, 3 = wilting and 4 = leaf loss. The DS values were obtained from the averages of these scores. Biocontrol efficiency (BE) was calculated according to a formula by Wang et al. (2016) and Cheng et al. (2019) as follows:
Metagenomic Sequencing of Bacterial Community
Twenty-five days after F. oxysporum inoculation, rhizosphere soils for the three replicate pots were collected from all treatments (24 samples in total) and sieved (2 mm). Briefly, the roots were lightly shaken to remove loosely attached soil. The soil that was still tightly adhering to the roots was harvested as rhizosphere soil and frozen at −80°C for DNA extraction following the protocol of Wu et al. (2016). Total soil DNA was extracted using a Qiagen DNeasy PowerSoil Kit following all steps provided in the kit handbook1. Genomic DNA concentration and purity were measured using a Qubit fluorometer (Thermo Fisher Scientific, United States).
Bacterial community composition was assessed by sequencing the V1-V9 region of the 16S rRNA gene using PCR primers 27F (5′- AGRGTTTGATYNTGGCTCAG-3′) and 1492R (5′- TASGGHTACCTTGTTASGACTT-3′) as reported in Větrovsky and Baldrian (2013). PCR conditions were initiated at 95°C for 5 min, followed by 25 cycles of denaturation at 95°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 1 min, followed by a final elongation at 72°C for 7 min, and then hold at 4°C. The PCR products were pooled and visualized on 1% agarose gels, purified using a MinElute PCR Purification Kit according to the manufacturer’s instructions, and quantified using QuantiFluorTM-ST (Promega, United States). High-throughput sequencing was carried out on the PacBio Sequel II platform (BioMarker Technologies Co., Ltd., China). The original subreads were first corrected to generate circular consensus sequences (CCS) (SMRT Link, version 8.0), and then Lima software (v1.7.0) was used to identify the CCS of different samples through their barcoded sequences, and UCHIME1 (version 8.1) was used to remove the chimera bodies for high quality CCS sequences. Using USEARCH 4 (version 10.22) with a cut-off of 97% similarity, the Operational Taxonomic Units (OTUs) were clustered and the taxonomic classifications were performed using RDP Classifier (Version 2.2, based on Bergey’s taxonomy) with the classification threshold set at 0.5.
Putative bacterial metagenomic functions were inferred using a phylogenetic investigation of communities by reconstruction with unobserved states (PICRUSt) on the 16S rRNA gene abundance data as reported by Langille et al. (2013). Using functions within the PICRUSt pipeline, the OTU-table was normalized and used for metagenome inferences involving the KEGG (Kyoto Encyclopedia of Genes and Genomes) orthologs (KOs). The predicted functions were then collapsed into hierarchical KEGG pathways using the “categorize by function” step in the PICRUSt pipeline as performed by Wilkinson et al. (2017).
Statistical Analysis
Plant growth promotion data were analyzed using the analysis of variance (ANOVA) procedure of SAS 8.1 software (SAS Institute Inc., Cary, NC, United States). Differences between treatments were assessed at each time point by Fisher’s protected least significance difference (LSD) test at 0.05 levels. Kruskal–Wallis one-way analysis of variance by ranks was used for comparing DI and DS. The rarefaction curve, corresponding to observed OTUs at different sequencing depths, was examined using QIIME software to determine whether the depth was reasonable. Chao1 and abundance-based coverage estimator (ACE) indices were used to calculate the evenness of each sample (Bokulich et al., 2013), while the Shannon and Simpson indices were used to measure diversity (Hong et al., 2015). Beta diversity among samples was determined by principal component analysis (PCA) using R software2. Significant differences in bacterial community composition between paired samples were determined using the Metastats analysis and Mothur program which counted taxa in five classified levels (Lu et al., 2016). The BE values were not subjected to statistical analysis.
Results
Molecular Identification of Bacterial Strain HSB1 and Phylogenetic Analysis
16S rRNA was used to identify the genus and species of HSB1. This sequence has been accessioned to GenBank (MT626060). The BLAST result showed 99.93% homology with a strain of Bacillus amyloliquefaciens. The phylogenetic analysis showed HSB1 clustering with other B. amyloliquefaciens, B. methylotrophicus, and B. velezensis strains, which are known to share identity and a most recent common ancestor (Figure 1a). B. mojavensis strains were used as out-group (Figure 1b).
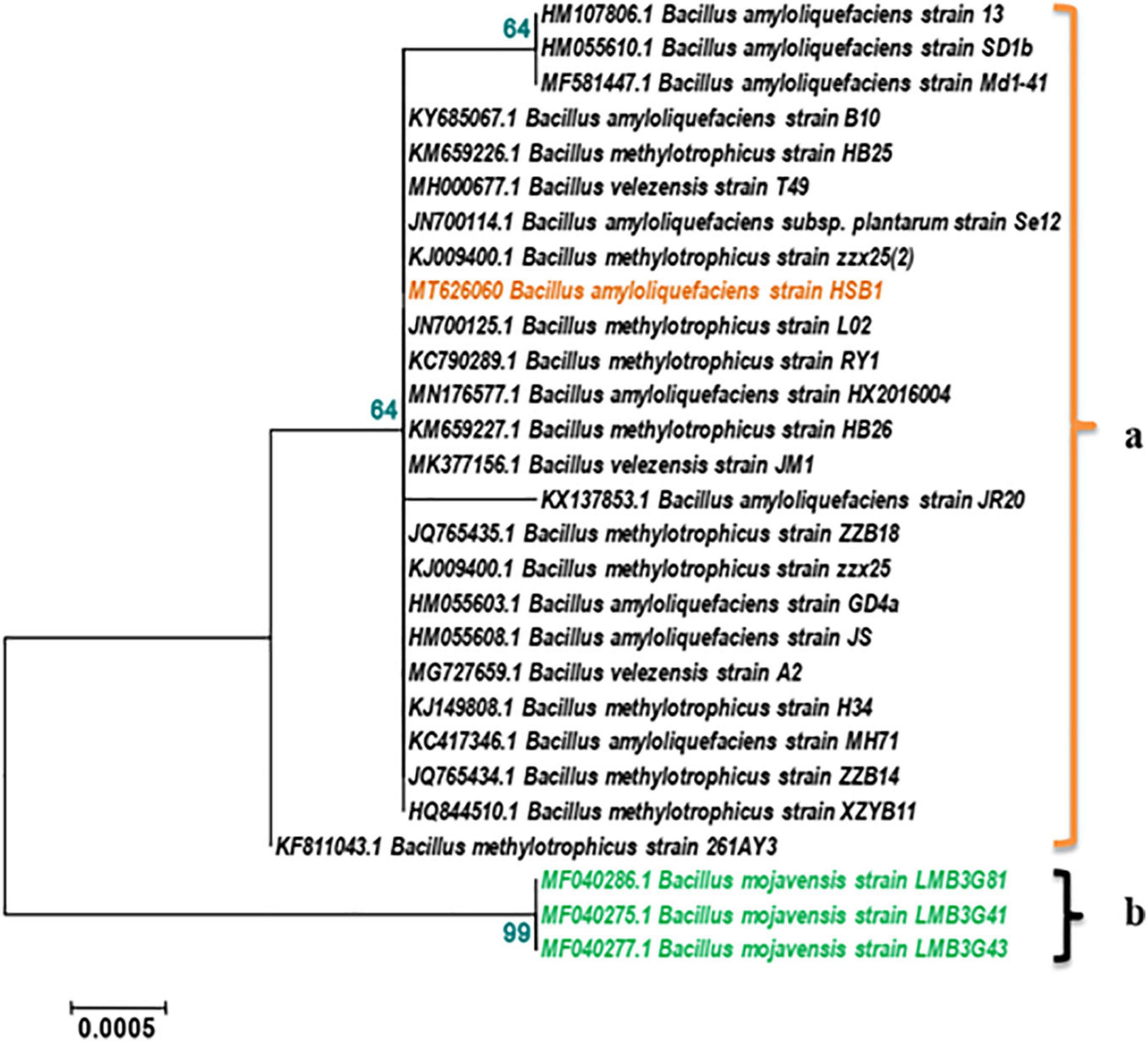
Figure 1. Phylogenetic tree based on 16S rRNA gene showing relationships between Bacillus strain HSB1 and other Bacillus strains/species derived from NCBI accessions. The tree was inferred using the neighbor-joining method and MEGA 5.5 software with 1000 bootstrap replicates (bootstrap values are shown next to nodes). The strain HSB1 clustered with B. amyloliquefaciens, B. methylotrophicus, and B. velezensis strains, (a). B. mojavensis strains were used as out-group (b).
In vitro Antifungal Assay
The potential of the two B. amyloliquefaciens strains, HSB1 and FBZ42, to inhibit the five root rot fungal pathogens (F. oxysporum, F. solani, F. chlamydosporum, F. tricinctum and A. alternata) was assessed using dual culture technique. The results showed that both HSB1 and FZB42 inhibited mycelial growth of all five fungal pathogens compared to the control (petri plates without bacterial served as control (Figure 2). All five fungal pathogens were inhibited (up to 100%). Due to its faster growth rate compared to the Fusarium strains tested, A. alternata was the first to show inhibition from exposure to our BCAs (Figures 2A–E).
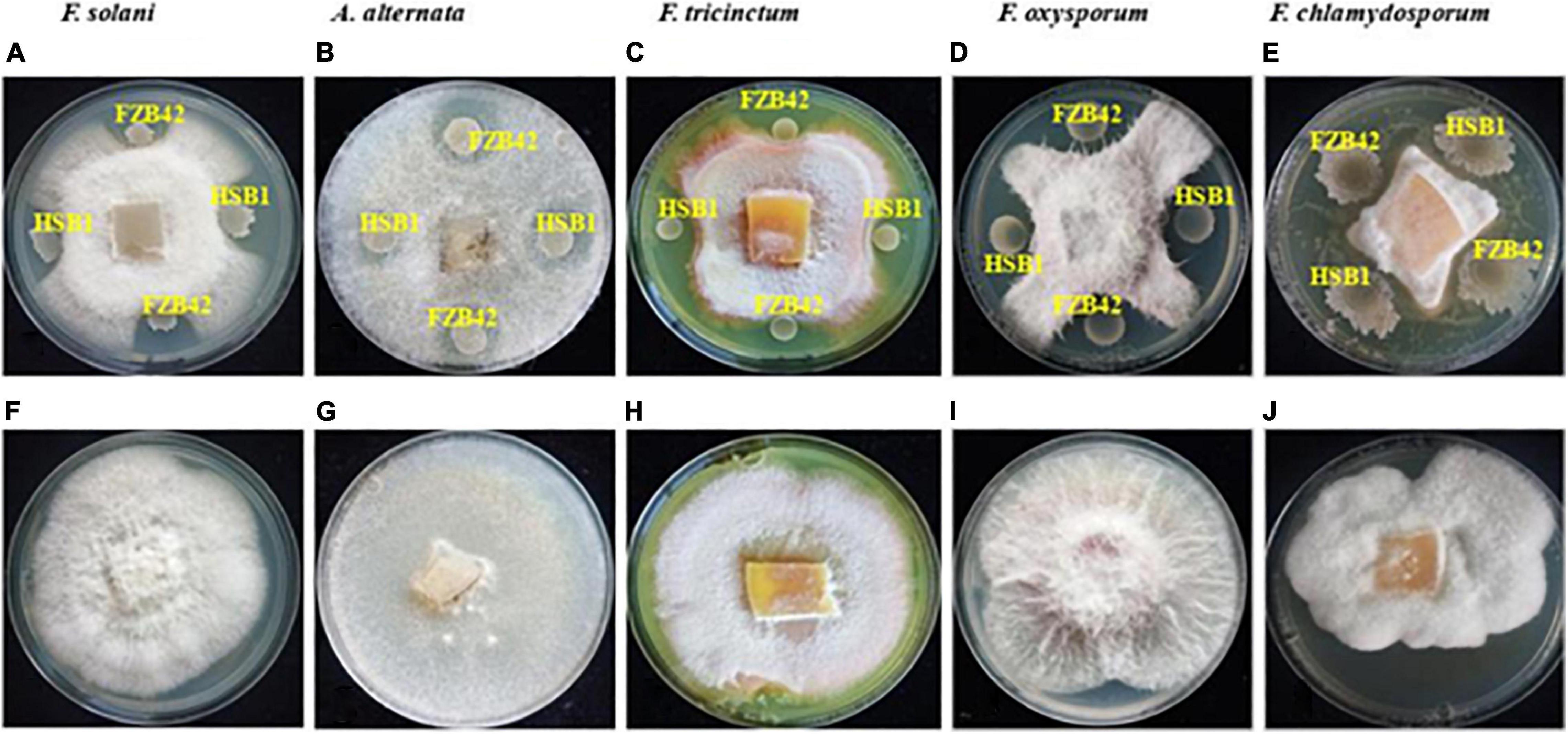
Figure 2. Plate assay showing antagonistic activity of both B. amyloliquefaciens FZB42 and HSB1 against each of five fungal pathogens of Chinese wolfberry: F. solani, A. alternata, F. tricinctum, F. oxysporum and F. chlamydosporum (A–E). Panels (F–J) are the respective controls.
Effects of FZB42 and HSB1 on the Growth of Wolfberry Seedlings
This set of experiments showed that the two bacterial strains, FZB42 and HSB1, had different effects on the development of wolfberry seedlings. Both strains promoted plant growth in some way compared to the CK. In the first assay comparing HSB1 to FZB42, all seedlings were growing at same rate during first 60 days (Supplementary Figures 1A–C). In the days after, both FZB42 and HSB1 increased stem length compared to the water control. At 68 and 78 day time points, the greatest stem length was observed in HSB1-treated seedlings (Figure 3A). Measurements showed that both FZB42 and HSB1 increased root length in 60-day-old seedlings compared to the control. However, HSB1 had significantly increased the root length compared to both the FZB42 and control treatments, with FZB42-treated roots having shorter root length than even the water control treatment (Figure 3B). In the second assay, HSB1-treated seedlings were healthier and taller compared to control treatment (Supplementary Figures 1D,E). Results showed that HSB1 increased wolfberry seedlings shoot weight at the 7, 14, and 21 days post-inoculation time points compared to the control. There was a significant difference between HSB1 and control shoot weights (Figures 4A,B). HSB1also increased seedling root weight compared to control treatment. There was a significant difference between HSB1 and control at 14 and 21 day time after inoculation (Figures 4C,D).
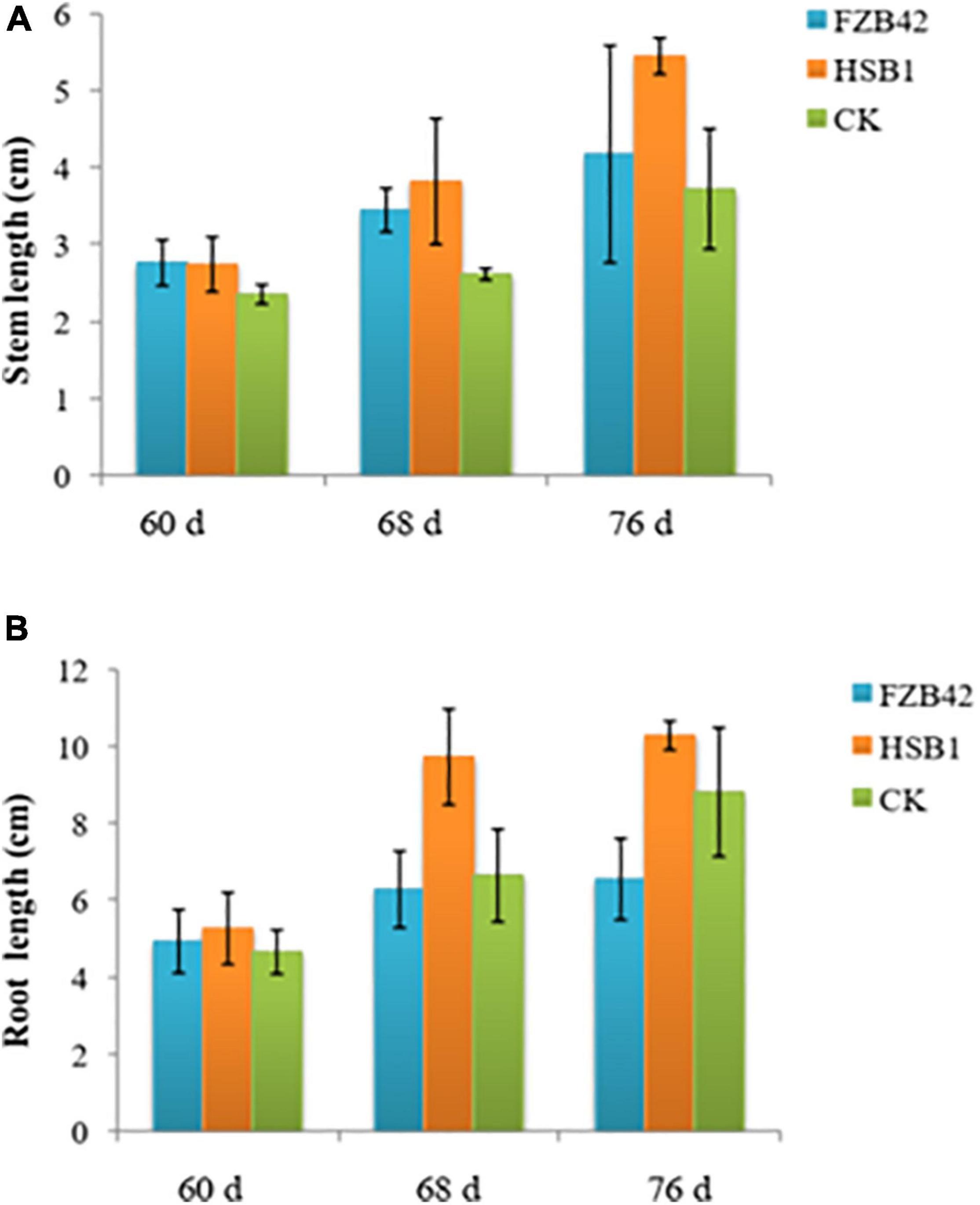
Figure 3. Bar graphs showing changes in stem length (A), and root length (B) of wolfberry seedlings whose soil was treated with Bacillus amyloliquefaciens strains (FZB42 or HSB1) compared to a water control (CK). Measurement time points are based on growth at 60, 68, and 76 day. Error bars represent standard deviation of three replicates.
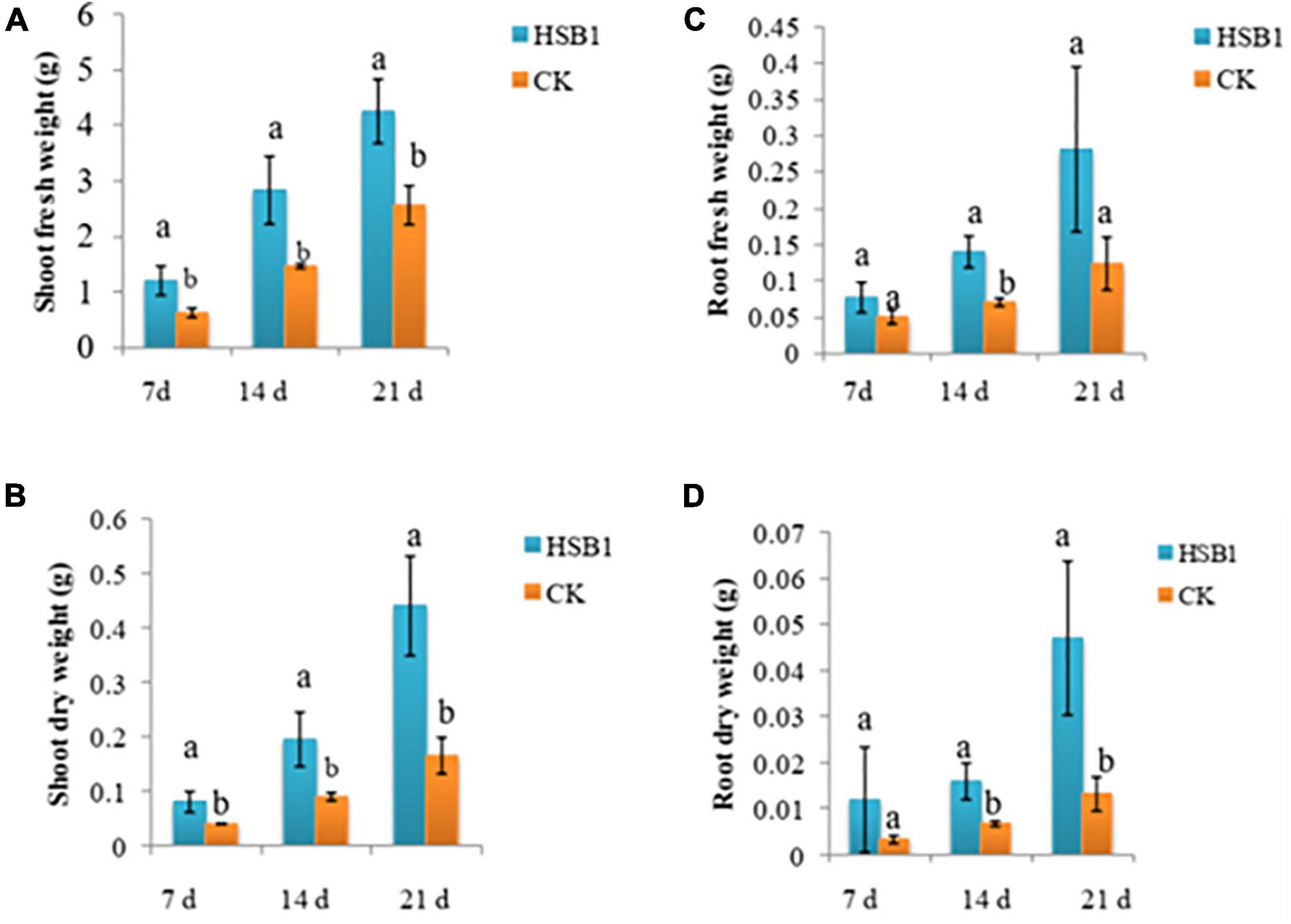
Figure 4. Bar graphs showing changes in shoot weight (A,B), and root weight (C,D) of wolfberry seedlings whose soil was treated with Bacillus amyloliquefaciens strains (FZB42 or HSB1) compared to a water control (CK). Measurement time points are based on days after inoculation growth at 7, 14, and 21 days after inoculation. Error bars represent standard deviation of three replicates. Letters above each bar indicate significant differences from the control (CK) (p < 0.05).
Biocontrol Experiment
As a baseline, DI and DS were assessed at 15, 21, and 25 day post-inoculation with F. oxysporum alone. The highest DI (42%) and DS (1.7) were recorded for day 15 (Table 1 and Figure 5). In the foliage, the disease usually was recognized by the yellowing, necrosis and wilting of leaves, followed by complete leaf loss (Table 1 and Supplementary Figure 2). At the beginning, infected seedlings were characterized by severe yellowing of leaves and the disease symptoms continually increased throughout the experiment (Supplementary Figures 2A,B and Table 1). Assessment conducted at 21 and 25 day after F. oxysporum inoculation revealed that the greatest respective DI (65 and 84%) and DS (1.28 and 1.45) values were also recorded in F. oxysporum alone (Figures 5B,C,E,F). Unexpectedly, no DI and DS were recorded in treatments where F. oxysporum was inoculated before HSB1 (FO + HSB1) or where FZB42 was inoculated before F. oxysporum (FZB42 + FO) treatments throughout the experiment (BE = 100%); all seedlings had zero disease symptoms in the foliage (Supplementary Figures 2G,H). Additionally, control plants treated with water (CK), FZB42 alone, as well as HSB1 alone did not also show any disease symptoms (Supplementary Figures 2E,F,I). Brown lesions were observed on root surfaces of seedlings treated with FO alone or HSB1 + FO (Supplementary Figures 3A,C), whereas no lesions were observed on root surfaces of seedlings having the other treatments (Supplementary Figures 3B,D–H).
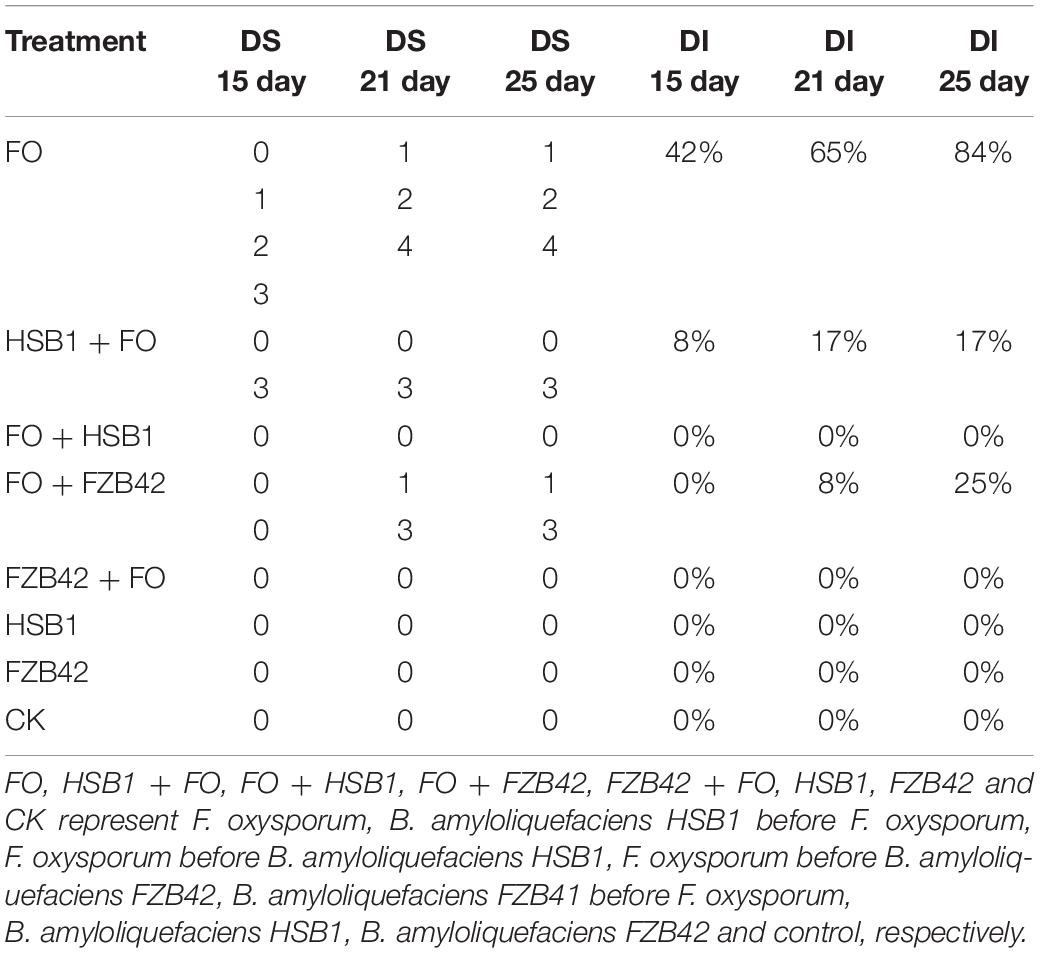
Table 1. Disease severity (DS) scores and averaged percentages of disease incidence (DI) observed with each treatment.
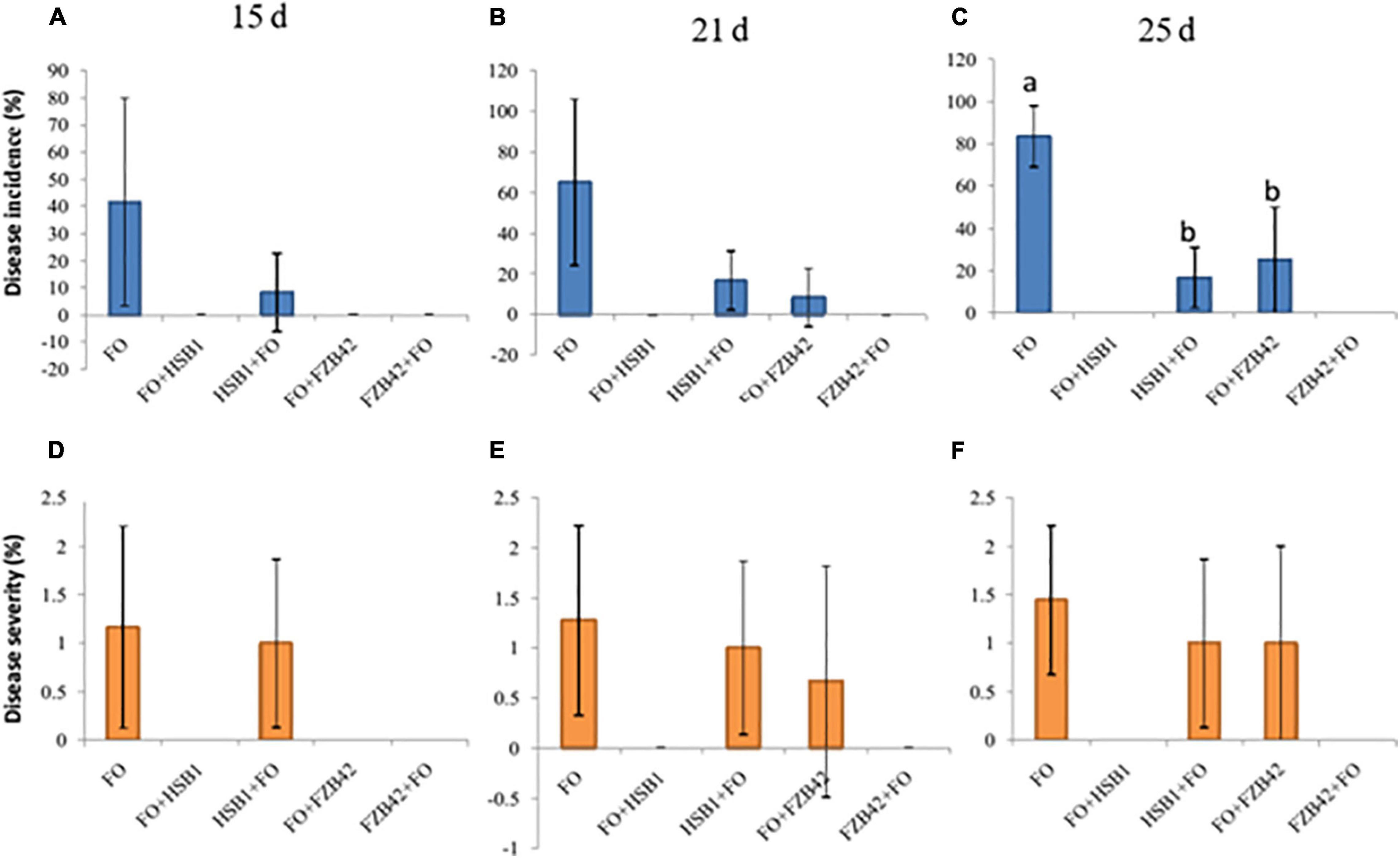
Figure 5. Bar charts showing disease incidence (A–C), blue bars) and disease severity (D–F), orange bars) for all treatments recorded at 15, 21, and 25 days time points. Error bars represent standard error of three replicateds. Letters above each bar indicate significant differences in disease incidence at 25 days between F. oxysporum, HSB1 + F. oxysporum, and F. oxysporum + FZB42, and treatments (p < 0.05).
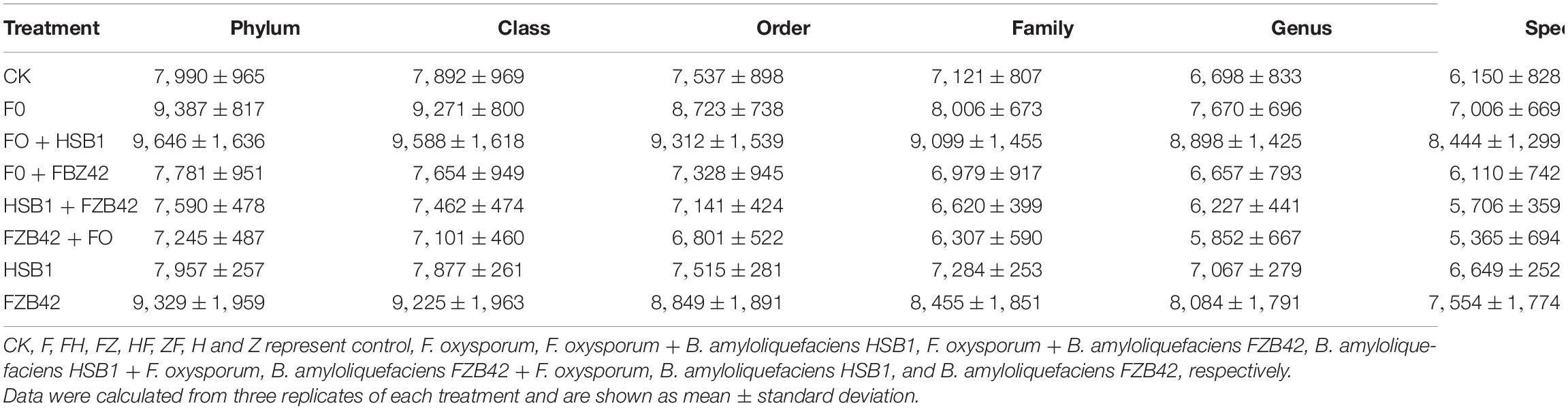
Metagenomic Sequencing of Bacterial Community
A total of 211,372 16S rRNA V1-V9 gene sequences were analyzed across 24 rhizosphere soil samples, with an average of 8,807 ± 959 sequences per soil sample (Supplementary Table 1). Based on a threshold of 97% shared nucleotide identity, these sequences were grouped by OTU, yielding 8,366, 8,259, 7,901, 7,484, 7,145, and 6,623 OTUs at the phylum, class, order, family, genus, and species levels, respectively (Table 2). The sequencing depth was analyzed to identify new taxa. A rarefaction curve analysis at 3% dissimilarity for the bacterial community revealed that the sharp of the curve was increasing and depth did not reach saturation, indicating that a greater sequencing depth was needed (Figure 6A). However, the data were sufficient for showing differences among the treatments and suggested that BCA application increased bacterial diversity. The richness indices (ACE and Chao1), and diversity indices (Shannon and Simpson) were further estimated and presented in Table 3. Chao1 and ACE indices were higher in HSB1 + FO and FZB42 + FO treatments compared to FO alone. The lowest values were recorded in HSB1 alone and FO + HSB1 treatments. In addition, Simpson index was higher in HSB1 alone and FO + HSB1 treatments compared to the FO alone treatment and CK, whereas the Shannon index was higher for FO + FZB42, FZB42 + FO, and HSB1 + FO treatments. PCA comparison of changes in soil bacterial community across different treatment groups showed that the first two principle components could explain 42.82 and 17.92% of the total variation. The bacterial communities differed according to treatment, indicating the effects of F. oxysporum and the two Bacillus strains on bacterial composition in the wolfberry rhizosphere (Figure 6B).
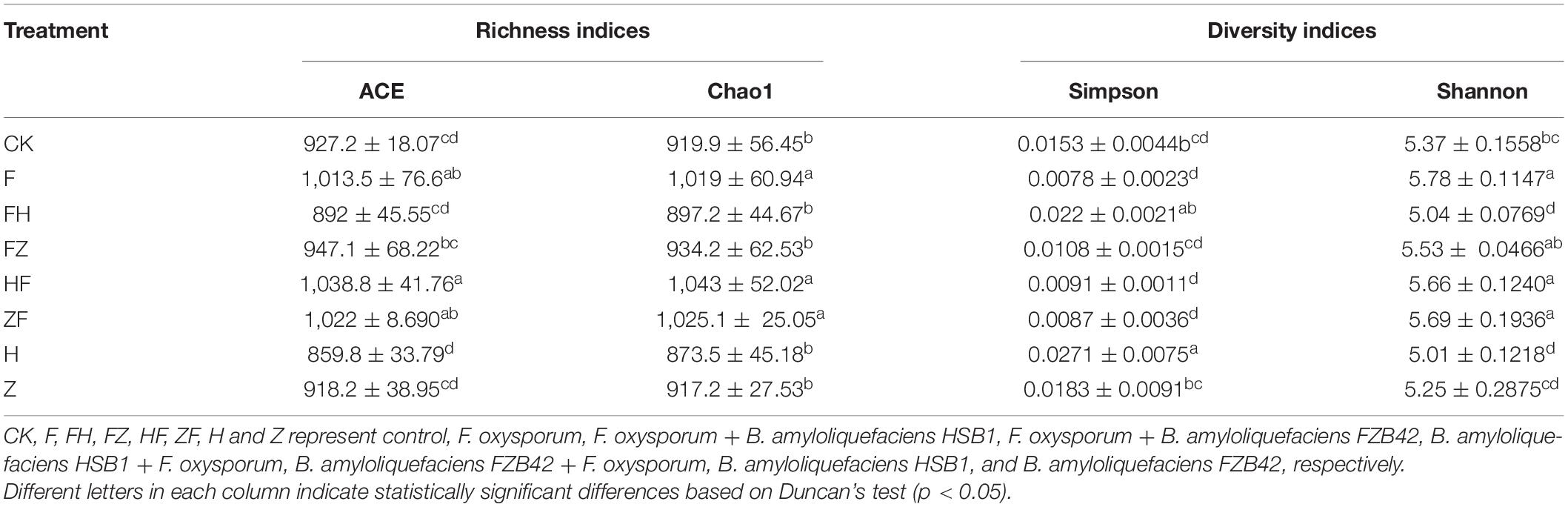
Table 3. The mean of the ACE, Chao1, and Simpson and Shannon indices of rhizosphere soil treatments with BCA and F. oxysporum at 97% similarity.
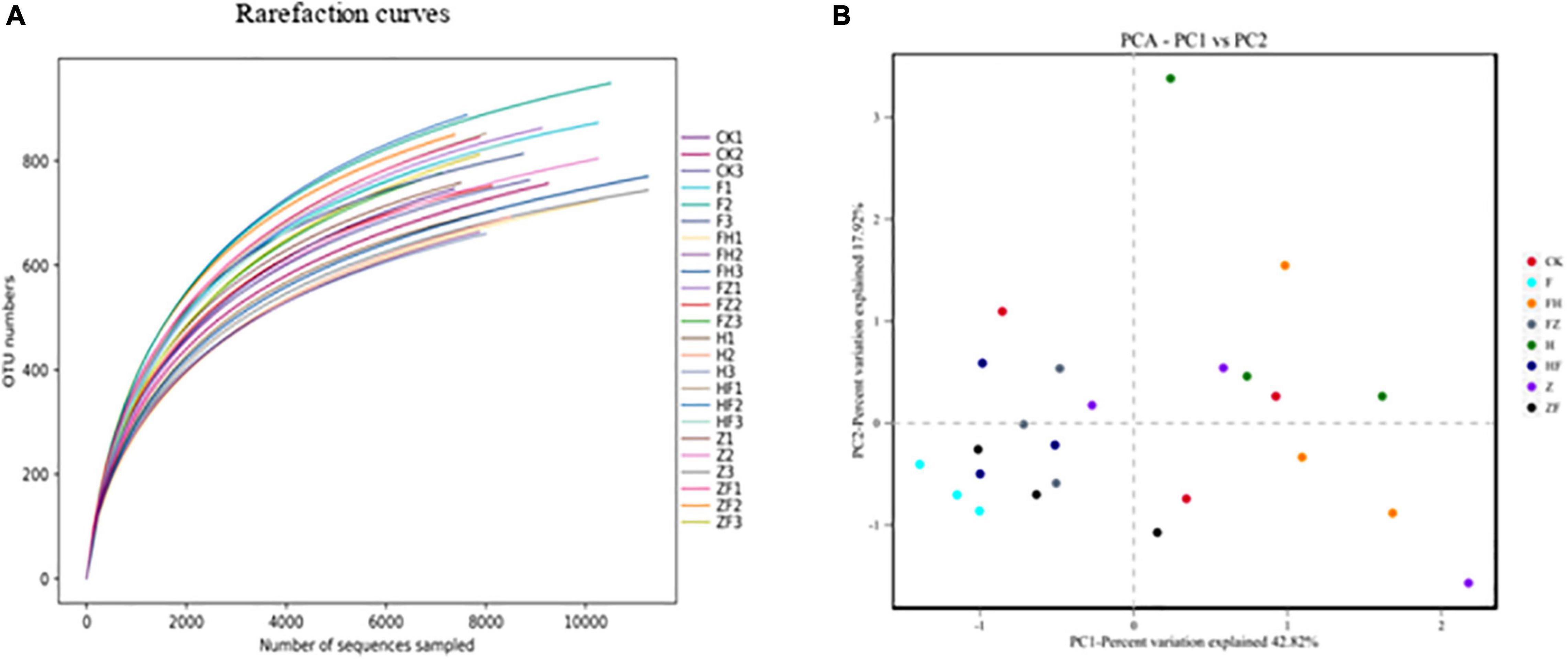
Figure 6. Rarefaction curves (A) showing the relationship between sequence number per sample and observed OTUs for different treatment replicates. The length of the curve reflects sequencing depth (i.e., a longer curve indicates a greater depth), while the smoothness reflects the effect of sequencing depth on sample diversity. Principal component analysis (B) based on the distance matrix calculated using the Bray-Curtis algorithm for soil samples collected from different treatments: CK, control; F, F. oxysporum alone; FH, F. oxysporum + HSB1; FZ, F. oxysporum + FZB42; H, HSB1 alone; HF, HSB1 + F. oxysporum; Z, FZB42 alone; ZF, FZB42 + F. oxysporum.
The average number of microbial groups at phylum, class, order, family, genus and species levels were 24, 34, 73, 105, 192, and 239, respectively (Supplementary Table 2). All samples showed similar phylum and genus composition, but differed in terms of the relative abundances of other taxonomic groups (Figure 7). Of the 10 most abundant phyla across all samples, Proteobacteria and Bdellovibrionota were the least abundant. Although phylum Proteobacteria predominated, each treatment affected the relative abundance of this phylum in the overall rhizosphere bacterial composition (Figure 7A). The 10 most abundant genera included Massilia followed by Arenimonas, Pelomonas, Gemmatimonas, Vicinamibacter, Comamonas, Pseudoxanthomonas, Pseudomonas, Pedosphaera and Piscinibacter (Figure 7B). Consistent with our phylum level observations, relative abundances of Massilia differed by treatment (Figure 6B and Supplementary Table 3). Finally, LEfSe analysis was used to detect taxa with significantly different abundances between the FO alone, FO + HSB1, FO + FZB42, HSB1 alone, and FZB42 + FO treatments. The most differentially abundant bacterial taxa in the rhizosphere samples belonged to the Proteobacteria phylum (Supplementary Figure 4).
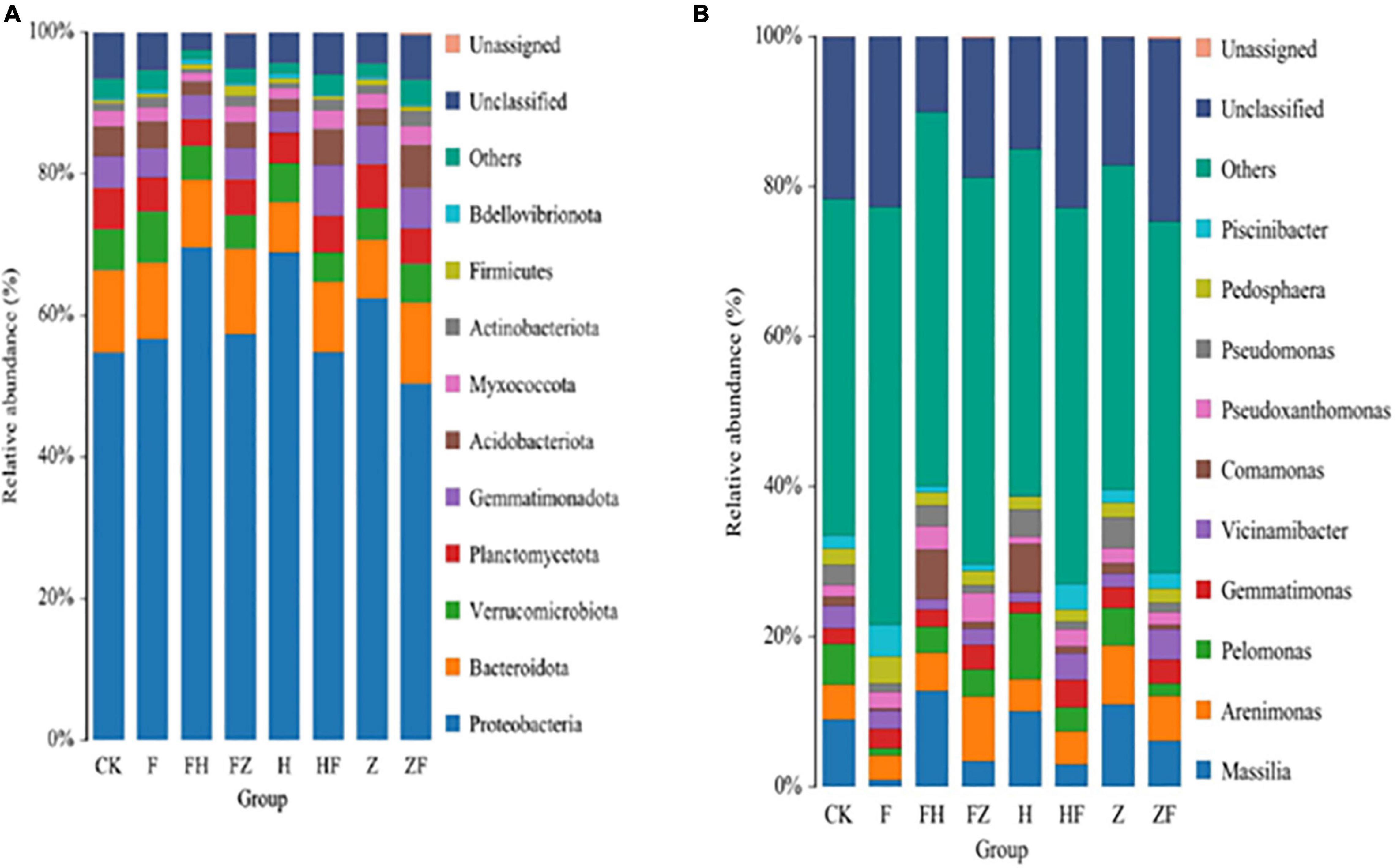
Figure 7. Stacked bar charts showing the relative abundances (in different colors) of the top 10 classified bacterial phyla (A) and genera (B) detected in soil samples subjected to different treatments: CK, control; F, F. oxysporum alone; FH, F. oxysporum + HSB1; FZ, F. oxysporum + FZB42; H, HSB1 alone; HF, HSB1 + F. oxysporum; Z, FZB42 alone; ZF, FZB42 + F. oxysporum. Relative abundance was based on the proportional frequencies of DNA sequences classified at the phylum and genus levels. Length of a color bar correlates with amount of abundance. Data were averaged from three replicates of each treatment.
KEGG ortholog predictions were performed on the 16S rRNA data using PICRUSt. We conducted comparisons between different treatments, including CK vs. HSB1, F. oxysporum vs. HSB1, and HSB1 vs. F. oxysporum + FZB42 (Figure 7). Seven pathways related to lipid transport and metabolism, transcription, energy production and conversion, amino acid transport and metabolism, inorganic ion transport and metabolism, secondary metabolite biosynthesis, transport and catabolism, and “function unknown” were overrepresented in the HSB1 alone sample compared to the CK (Figure 8A). Comparison between F. oxysporum and HSB1 showed that pathways related to membrane transport, cellular community-prokaryotes, and lipid metabolism were overrepresented in the HSB1 alone sample (Figure 8B). In comparing HSB1 to FO + FZB42, pathways related to metabolism of cofactors and vitamins, translation, glycan biosynthesis and metabolism, replication and repair, global and overview maps were overrepresented in the FO + FZB42 sample (Figure 8C).
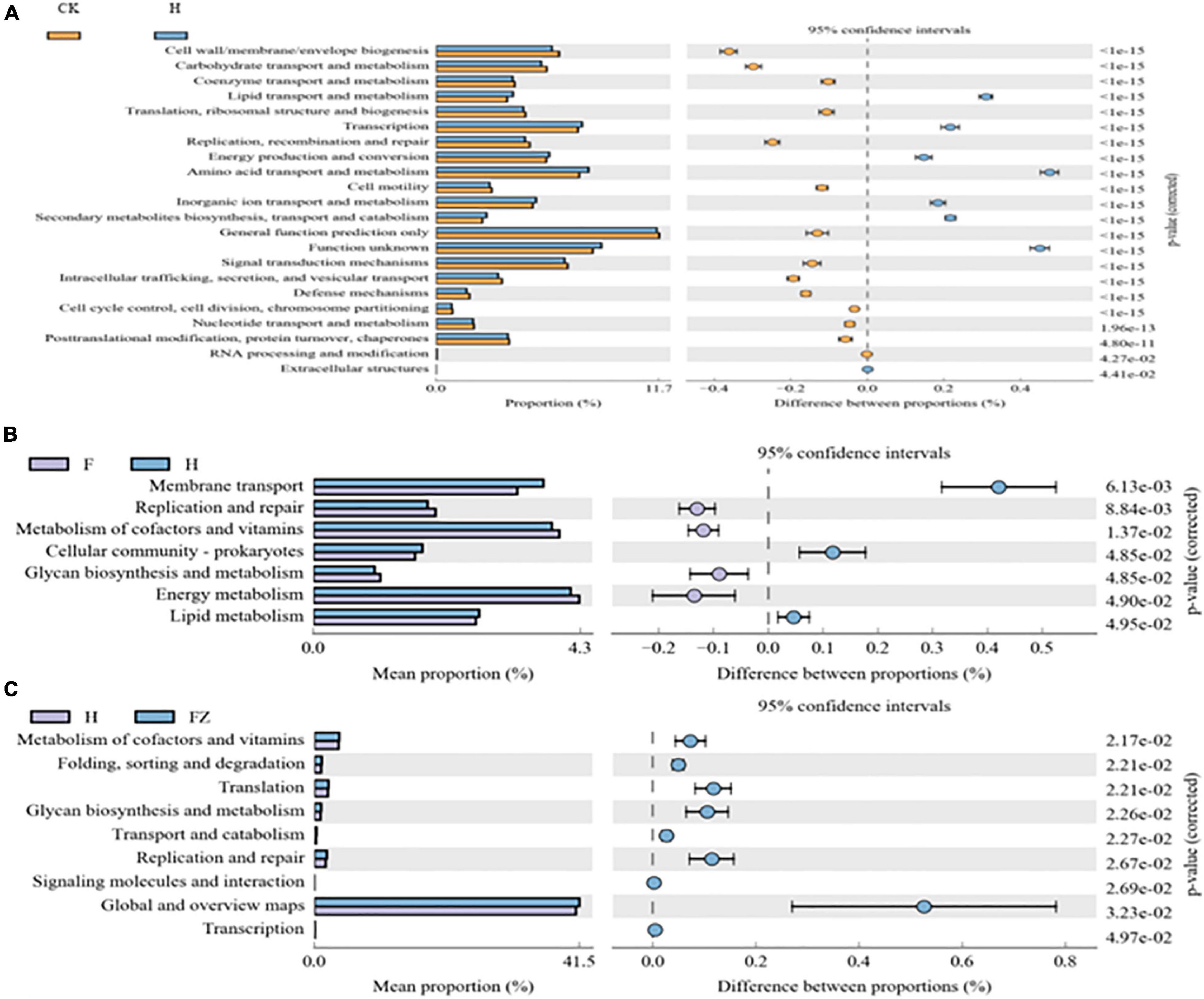
Figure 8. Metagenome comparisons, as predicted by PICRUSt, showing significant differences in the functionality of microbial genes detected in soil samples collected from selected treatments: CK, control; F, F. oxysporum alone, H, HSB1 alone, and FZ, F. oxysporum + FZB42. (A): significant comparisons between CK and H treatments, (B): significant comparisons between F and H treatments, and (C): significant comparisons between H FZ treatments.
Discussion
Currently, use of chemical pesticides represents one of the biggest ecological concerns, therefore biological control using beneficial microorganisms is considered a promising approach to manage soil borne diseases (Dhahira-Beevi and Qadri, 2010; Shahid and Khan, 2016; Singh et al., 2017; Alamri et al., 2019). Bacteria of the genus Bacillus are good candidates for use as BCAs (Falcäo et al., 2014; Chowdhury et al., 2015; Jiang et al., 2015). Therefore, a deep understanding of biocontrol mechanisms will help us to assess and to enhance the biological control of various diseases caused by soil borne pathogens in agriculture. This study determined the potential of two bacterial strains, belonging to the BCA genus Bacillus, to prevent root rot disease in wolfberry plants. Our results revealed that both HSB1 and FZB42 inhibited the mycelial growth of five fungal pathogens, which may be due to the production of antifungal secondary metabolites. This result is in line with previous studies showing the inhibition of fungal mycelial growth by different antagonistic bacterial strains. Falcäo et al. (2014) reported the antifungal activity of an endophytic bacterium, B. subtilis ALB629, that inhibited the mycelial growth of F. solani and Colletotrichum gossypii. Two other Bacillus strains, B. subtilis GM2 and B. subtilis GM5, isolated from the rhizosphere of potato roots have shown an ability to inhibit growth of different phytopathogenic fungi including A. alternata TP 712, F. solani, F. oxysporum, F. redolens and Colletotrichum coccodes 14raKK6 (Mardanova et al., 2017). B. amyloliquefaciens JDF35 was reported to inhibit the growth of F. oxysporum f. sp. niveum which causes wilt disease of watermelon (Zhao et al., 2017). The growth of Colletotrichum gloeosporioides and F. oxysporum was also reported to be inhibited by Bacillus species (BT42) isolated from the Coffea arabica rhizosphere (Kejela et al., 2016).
Beyond disease prevention, the application of HSB1 and FZB42 to wolfberry seeds resulted in seedlings with significantly stimulated the growth of shoots and/or roots. This finding is in agreement with previous studies that assessed plant-growth-promoting activity of various Bacillus strains in different plants (Falcäo et al., 2014; Yuan et al., 2015; Kejela et al., 2016). The addition of HSB1 and FZB42 before or after F. oxysporum inoculation significantly reduced DI and DS in wolfberry seedlings. Several studies reported the efficient application of Bacillus species as BCAs in the suppression of different pathogens causing diseases in plants (Zhang and Xue, 2010; Zhao et al., 2017; Kulimushi et al., 2018; Gautam et al., 2019).
Rhizosphere microbial communities play important roles in plant health and disease prevention (Dudenhöffer et al., 2016; Gu et al., 2016). An analysis based on richness indices (Chao1 and ACE, and Shannon), revealed that HSB1 and FZB42 altered the bacterial diversity of the rhizosphere in different ways. The inoculation of HSB1 5 days before F. oxysporum decreased Chao1 and ACE indices, while increasing the Simpson index, suggesting it negatively affected soil bacterial richness. However, inoculation of HSB1 and FZB42 17 days before F. oxysporum greatly increased the richness and Shannon indices, indicating that these BCAs positively affected soil bacterial richness. The abundances of rhizosphere microorganisms can be perturbed by biotic and abiotic factors (Ding et al., 2014; Huang et al., 2017). For instance, the introduction of additional bacteria and fungi can change native community structure (Karpouzas et al., 2011). The analysis of bacterial community composition and structure revealed that all treatments harbored structurally distinct taxa. Previous studies reported Proteobacteria, Acidobacteria, Actinobacteria, Bacteroidetes and Gemmatimonadetes as the predominant phyla in most rhizosphere soils (Zhu et al., 2013; Huang et al., 2017). Proteobacteria is the most abundant phylum in various soils (Shang et al., 2016; Wan et al., 2017; Han et al., 2019). This was also the predominant phylum represented in our study. Bacteria belonging to Proteobacteria generally expand faster by absorbing root-associated carbon substrates. Consequently, their abundance is positively proportional with carbon availability (Cleveland et al., 2006; Fierer et al., 2007). We also found that the relative abundance of Proteobacteria was elevated in rhizosphere soils in which F. oxysporum was inoculated first, more significantly in the FO + HSB1 treatment compared to FO alone. A similar result was obtained by Wan et al. (2017) after first inoculating FO, followed by BCA inoculation 7 days later. Assessment of DI showed asymptomatic seedlings in the FO + HSB1 treatment, indicating that the increased abundance of Proteobacteria may be associated with seedling protection and growth enhancement. Bacteroidota was the second-most abundant phylum in this study, a result confirmed by Han et al. (2019). The relative abundance of Actinobacteriota was found reduced in the FO alone treatment compared to HSB1 + FO, FZB42 + FO and FO + FZB42 treatments. This finding is consistent with previous reports, in which the Actinobacteriota phylum was found to be associated with disease suppression, due to its higher abundance in many disease-suppressive soils (Hunter et al., 2006; Fu et al., 2016). Proteobacteria was also reported to be highly abundant in disease-suppressive soil, because this group is known to produce high levels of secondary metabolites that inhibit pathogens (Mendes et al., 2011; Rosenzweig et al., 2012). Hence, a great abundance of Proteobacteria and Actinobacteria in soils should correlate with higher disease suppression ability.
Massilia is a rhizosphere-inhabiting and root-colonizing bacterium that associates with various plant species (Mendes et al., 2011; Rosenzweig et al., 2012). In our study, it showed to be most abundant in all treatments although its relative abundance varied markedly. It was found to be highly represented in FO + HSB1 treatment (DI and DS = 0) compared to F. oxysporum alone. A reduction in the relative abundance of Massilia was also reported in F. oxysporum treatment (Wan et al., 2017). Cretoiu et al. (2013) reported that genus Massilia suppresses soil borne diseases, thus the inoculation of HSB1 and FZB42 may synergistically protect wolfberry seedlings from root rot. Arenimonas, the second-most abundant genus, was highly represented in the FO + FZB42 treatment, and less abundant in the FO alone treatment. Jeong et al. (2016) reported a novel species of the genus Arenimonas, isolated from estuary sediment, to be oxidase- and catalase-positive. Genus Pseudomonas was highly represented in FZB42 alone and HSB1 alone treatments. This genus is known to display an ability to suppress different pathogens through various mechanisms such as production of antimicrobial compounds, induction of systemic resistance, promotion of plant growth, production of siderophores, and sequestration of nutrients (Li et al., 2012; Kong et al., 2016; Ma et al., 2017). Functional predictions associated with a microbiome are key to understanding the way the microbial community interacts with its environment. The recently-developed PICRUSt program was previously shown to be effective at obtaining functional predictions from 16S rRNA taxonomic data (Langille et al., 2013). Therefore, with the help of PICRUSt, we were able to gain functional insights into the bacterial community within the wolfberry rhizosphere. We noticed that HSB1 and FZB42 application significantly affected the function of the rhizosphere bacteria supporting a result reported by Han et al. (2019). Mendes et al. (2014) reported that the function of membrane transport may be associated with plant growth promotion and nutrition in the soybean rhizosphere. Plant growth promoting rhizobacteria are known to produce secondary metabolites antagonistic to various soil borne pathogens. In this study, introduction of HSB1 and FZB42 increased biosynthesis of secondary metabolites. A study by Berg et al. (2007) demonstrated that energy metabolism and signal transduction may improve resistance to Fusarium wilt in banana. Our findings will help us to develop an environmentally friendly and potent method to combat different pathogens responsible for root rot disease in Chinese wolfberry.
Conclusion
The endophytic bacterium, B. amyloliquefaciens HSB1, isolated from wolfberry root tissues and characterized in this study, as well as B. amyloliquefaciens FZB42, efficiently inhibited several root rot pathogens of Chinese wolfberry plants. Their abilities to concomitantly enhance plant growth and the presence of other beneficial microbes showed their potential as suitable BCAs. However, a field study involving these potential BCAs is needed to support our laboratory findings.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
CU: experiments, data processing, interpretation, writing, and submitting. LY, YW, YT, XZ, YL, QZ, and YZ: experiments. RW: project coordination and supervising. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key Research and Development Project (2018YFE0127200), International Science and Technology Cooperation Base of Gansu Province (2020-0413- GHC-0036) and the Gansu Province Science and Technology Major Special Plan (21ZD4NA019).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.782523/full#supplementary-material
Footnotes
References
Alamri, S. A. M., Hashem, M., Moustafa, Y. S., Nafady, N. A., and Abo-Elyousr, K. A. M. (2019). Biological control of root rot in lettuce caused by Exserohilum rostratum and Fusarium oxysporum via induction of the defense mechanism. Biol. Control. 128, 76–84. doi: 10.1016/j.biocontrol.2018.09.014
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSIBLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3404. doi: 10.1093/nar/25.17.3389
Ambreen, A., Hisamuddin, Merajul, I. R., Saad, J. A., and Rushda, S. (2012). Plant growth promoting Rhizobacteria: An overview. J. Nat. Prod. Plant Resour. 2, 19–31.
Aydi-Ben, A. R., Mokni-Tlili, S., Nefzi, A., Jabnoun-Khiareddine, H., and Daami-Remadi, M. (2016). Biocontrol of Fusarium wilt and growth promotion of tomato plants using endophytic bacteria isolated from Nicotiana glauca organs. Biol. Control. 97, 80–88. doi: 10.1111/jph.12501
Beneduzi, A., Ambrosini, A., and Passaglia, L. M. (2012). Plant growth-promoting rhizobacteria (PGPR): Their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 35, 1044–1051. doi: 10.1590/s1415-47572012000600020
Berg, N. V. D., Berger, D. K., Hein, I., Birch, P. R. J., Wingfield, M. J., and Viljoen, A. (2007). Tolerance in banana to Fusarium wilt is associated with early up-regulation of cell wall-strengthening genes in the roots. Mol. Plant Pathol. 8, 333–341. doi: 10.1111/j.1364-3703.2007.00389.x
Bokulich, N. A., Subramanian, S., Faith, J. J., Gevers, D., Gordon, J. I., Knight, R., et al. (2013). Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10, 57–59. doi: 10.1038/nmeth.2276
Bucheli, P., Vidal, K., Gao, Q., and Zhang, W. (2011). “Biomolecular and clinical aspects of Chinese wolfberry,” in Herbal Medicine: Biomolecular and Clinical Aspects, 2nd Edn, eds I. F. F. Benzie and S. WachtelGalor (Boca Raton, FL: CRC Press), 14.
Byambasuren, S.-E., Wang, J., and Gaudel, G. (2019). Medicinal value of wolfberry (L. barbarum). J. Med. Plants Stud. 7, 90–97.
Cao, Y., Pi, H., Chandrangsu, P., Li, Y., Wang, Y., Han, Z., et al. (2018). Antagonism of two plant-growth promoting Bacillus velezensis isolates against Ralstonia solanacearum and Fusarium oxysporum. Sci. Rep. 8:4360. doi: 10.1038/s41598-018-22782-z
Cao, Y., Zhang, Z., Ling, N., Yuan, Y., Zhen, X., Shen, B., et al. (2011). Bacillussubtilis SQR 9 can control Fusarium wilt in cucumber by colonizing plant roots. Biol. Fertil. Soils. 47, 495–506. doi: 10.1007/s00374-011-0556-2
Cheng, X., Ji, X., Ge, Y., Li, J., Qi, W., and Qiao, K. (2019). Characterization of antagonistic Bacillus methylotrophicus isolated from rhizosphere and its biocontrol effects on maize stalk rot. Phytopathology 109, 571–581.
Chowdhury, S. P., Hartmann, A., Gao, X., and Borriss, R. (2015). Biocontrol mechanism by root associated Bacillus amyloliquefaciens FZB42-a review. Front. Microbiol. 6:780–790. doi: 10.3389/fmicb.2015.00780
Cleveland, C. C., Nemergut, D. R., Schmidt, S. K., and Townsend, A. R. (2006). Increases in soil respiration following labile carbon additions linked to rapid shifts in soil microbial community composition. Biogeochemistry 82, 229–240. doi: 10.1007/s10533-006-9065-z
Compant, S., Duffy, B., Nowak, J., Clement, C., and Barka, E. A. (2005). Use of plant growth promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl. Environ. Microbiol. 71, 4951–4959. doi: 10.1128/AEM.71.9.4951-4959.2005
Cretoiu, M. S., Korthals, G. W., Visser, J. H., and van Elsas, J. D. (2013). Chitin amendment increases soil suppressiveness toward plant pathogens and modulates the actinobacterial and oxalobacteraceal communities in an experimental agricultural field. Appl. Environ. Microbiol. 79, 5291–5301. doi: 10.1128/AEM.01361-13
Dhahira-Beevi, N., and Qadri, S. M. H. (2010). Biological control of mulberry root rot disease (Fusarium spp.) with antagonistic microorganisms. J. Biopestic. 3, 90–92.
Ding, J., Zhang, Y., Zhang, H., Li, X., Sun, Z., Liao, Y., et al. (2014). Effects of Fusarium oxysporum on rhizosphere microbial communities of two cucumber genotypes with contrasting Fusarium wilt resistance under hydroponic condition. Eur. J. Plant Pathol. 140, 643–653. doi: 1007/s10658-014-0494-6
Dudenhöffer, J.-H., Scheu, S., and Jousset, A. (2016). Systemic enrichment of antifungal traits in the rhizosphere microbiome after pathogen attack. J. Ecol. 104, 1566–1575. doi: 10.1111/1365-2745.12626
Es-soufi, R., Tahiri, H., Azaroual, L., Oualkadi, A., Martin, P., Alain, B., et al. (2020). Biocontrol potential of Bacillus amyloliquefaciens Bc2 and Trichoderma harzianum TR against strawberry anthracnose under laboratory and field conditions. Agric. Sci. 11, 260–277. doi: 10.4236/as.2020.113016
Falcäo, L. L., Silva-Werneck, J. O., Vilarinho, B. R., da Silva, J. P., Pomella, A. W. V., and Marcellino, L. H. (2014). Antimicrobial and plant growth-promoting properties of the cacao endophyte Bacillus subtilis ALB629. J. Appl. Microbiol. 116, 1584–1592. doi: 10.1111/jam.12485
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 39, 783–789. doi: 10.1111/j.1558-5646.1985.tb00420.x
Fierer, N., Bradford, M. A., and Jackson, R. B. (2007). Toward an ecological classification of soil bacteria. Ecology 88, 1354–1364. doi: 10.1890/05-1839
Fu, L., Ruan, Y. Z., Tao, C. R., Li, R., and Shen, Q. R. (2016). Continous application of bioorganic fertilizer induced resilient culturable bacteria community associated with banana Fusarium wilt suppression. Sci. Rep. 6:27731. doi: 10.1038/srep27731
Gadhave, K. R., Devlin, P. F., Ebertz, A., Ross, A., Gange, A. C., et al. (2018). Soil Inoculation with Bacillus spp. modifies root endophytic bacterial diversity, evenness, and community composition in a context-specific manner. Microb. Ecol. 76, 741–750. doi: 10.1007/s00248-018-1160-x
Gao, X., Gong, Y., Huo, Y., Han, Q., Kang, Z., and Huang, L. (2015). Endophytic Bacillus subtilis strain E1R-J is a promising biocontrol agent for wheat powdery mildew. Biomed. Res. Int. 2015, 1–8. doi: 10.1155/2015/462645
Gautam, S., Chauhan, A., Sharma, R., Sehgal, R., and Shirko, C. K. (2019). Potential of Bacillus amyloliquefaciens for biocontrol of bacterial canker of tomato incited by Clavibacter michiganensis ssp. michiganensis. Microb. Pathog. 130, 196–203. doi: 10.1016/j.micpath.2019.03.006
Ge, B., Liu, B., Nwet, T. T., Zhao, W., Shi, L., and Zhang, K. (2016). Bacillus methylotrophicus strain NKG-1, isolated from Changbai mountain, China, has potential applications as a biofertilizer or biocontrol agent. PLoS One 11:e0166079. doi: 10.1371/journal.pone.0166079
Gu, Y., Wei, Z., Wang, X., Friman, V.-P., Huang, J., Wang, W., et al. (2016). Pathogen invasion indirectly changes the composition of soil microbiome via shifts in root exudation profile. Biol. Fertil. Soils. 52, 997–1005. doi: 10.1007/s00374-016-1136-2
Han, L., Wang, Z., Li, N., Wang, Y., Feng, J., and Zhang, X. (2019). Bacillus amyloliquefaciens B1408 suppresses Fusarium wilt in cucumber by regulating the rhizosphere microbial community. App. Soil Ecol. 136, 55–66. doi: 10.1016/j.apsoil.2018.12.011
Hashem, A., Tabassum, B., and Fathi Abd_Allah, E. (2019). Bacillus subtilis: A plant-growth promoting rhizobacterium that also impacts biotic stress. Saudi J. Biol. Sci. 26, 1291–1297. doi: 10.1016/j.sjbs.2019.05.004
Hong, C., Si, Y., Xing, Y., and Li, Y. (2015). Illumina MiSeq sequencing investigation on the contrasting soil bacterial community structures in different iron mining areas. Environ. Sc. Pollut. Res. 22, 10788–10799. doi: 10.1007/s11356-015-4186-3
Huang, X., Cui, H., Yang, L., Lan, T., Zhang, J., and Cai, Z. (2017). The microbial changes during the biological control of cucumber damping-off disease using biocontrol agents and reductive soil disinfestation. Bio. Control 62, 97–109. doi: 10.1007/s10526-016-9768-6
Hunter, P. J., Petch, G. M., Calvo-Bado, L. A., Pettitt, T. R., Parsons, N. R., Morgan, J. A. W., et al. (2006). Differences in microbial activity and microbial populations of peat associated with suppression of damping-off disease caused by Pythium sylvaticum. Appl. Environ. Microbiol. 72, 6452–6460. doi: 10.1128/AEM.00313-06
Janda, J. M., and Abbott, S. L. (2007). 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J. Clin. Microbiol. 45, 2761–2764. doi: 10.1128/JCM.01228-07
Jeong, H. I., Jim, H. M., and Jeon, C. O. (2016). Arenimonas aestuarii sp. nov., isolated from estuary sediment. Int. J. Syst. Evol. Microbiol. 66, 1527–1532. doi: 10.1099/ijsem.0.000913
Jiang, C. H., Wu, F., Yu, Z. Y., Xie, P., Ke, H. J., Li, H. W., et al. (2015). Study on screening and antagonistic mechanisms of Bacillus amyloliquefaciens 54 against bacterial fruit blotch (BFB) caused by Acidovorax avenae subsp. citrulli. Microbiol. Res. 170, 95–104. doi: 10.1016/j.micres.2014.08.009
Karpouzas, D. G., Karatasas, A., Spiridaki, E., Rousidou, C., Bekris, F., and Omirou, M. (2011). Impact of a beneficial and of a pathogenic Fusarium strain on the fingerprinting-based structure of microbial communities in tomato (Lycopersicon esculentum Milll.) rhizosphere. Eur. J. Soil Biol. 47, 400–408. doi: 10.1016/j.ejsobi.2011.07.011
Kejela, T., Thakkar, V. R., and Thakor, P. (2016). Bacillus species (BT42) isolated from Coffea arabica L. rhizosphere antagonizes Colletotrichum gloeosporioides and Fusarium oxysporum and also exhibits multiple plant growth promoting activity. BMC Microbiol. 16:277. doi: 10.1186/s12866-016-0897-y
Köhl, J., Kolnaar, R., and Ravensberg, W. J. (2019). Mode of action of microbial biological control agents against plant diseases: relevance beyond efficacy. Front. Plant Sci. 10:845. doi: 10.3389/fpls.2019.00845
Kong, H. G., Kim, N. H., Lee, S. Y., and Lee, S. W. (2016). Impact of a recombinant biocontrol bacterium, Pseudomonas fluorescens pc78, on microbial community in tomato rhizosphere. Plant. Pathol. J. 32, 136–144. doi: 10.5423/ppj.oa.08.2015.0172
Kröber, M., Wibberg, D., Grosch, R., Eikmeyer, F., Verwaaijen, B., and Chowdhury, P. S. (2014). Effect of the strain Bacillus amyloliquefaciens FZB42 on the microbial community in the rhizosphere of lettuce under field conditions analyzed by whole metagenome sequencing. Front. Microbiol. 5:252. doi: 10.3389/fmicb.2014.00252
Kulimushi, P. Z., Basime, G. C., Nachigera, G. M., Thonart, P., and Ongena, M. (2018). Efficacy of Bacillus amyloliquefaciens as biocontrol agent to fight fungal diseases of maize under tropical climates: from lab to field assays in south Kivu. Environ. Sci. Pollut. Res. Int. 25, 29808–22982. doi: 10.1007/s11356-017-9314-9
Kumar, S., Nei, M., Dudley, J., and Tamura, K. (2008). MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinformatics 9, 299–306. doi: 10.1093/bib/bbn017
Labuschagne, N., Pretorius, T., and Idris, A. H. (2010). Plant Growth Promoting Rhizobacteria as Biocontrol Agents Against Soil-Borne Plant Diseases. Plant Growth Health Promot. Bacteria 18, 211–230. doi: 10.1007/978-3-642-13612-2_9
Langille, M. G., Zaneveld, J., Caporaso, J. G., Mcdonald, D., Knights, D., Reyes, J. A., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821. doi: 10.1038/nbt.2676
Leila, B., Abdelwahab, R., Nassira, T., Nabila, K., Meriem, K., and Elhafid, N. (2015). Biological control of Botrytis cinerea by Bacillus sp. strain S7LiBe under abiotic stress. Int. J. Sci. Res. Sci. Technol. 1, 07–14.
Li, R., Khafipour, E., Krause, D. O., Entz, M. H., de Kievit, T. R., and Fernando, W. G. (2012). Pyrosequencing reveals the influence of organic and conventional farming systems on bacterial communities. PLoS One 7:e51897. doi: 10.1371/journal.pone.0051897
Lu, Y., Chen, J., Zheng, J., Hu, G., Wang, J., Huang, C., et al. (2016). Mucosal adherent bacterial dysbiosis in patients with colorectal adenomas. Sci. Rep. 6:26337. doi: 0.1038/srep26337
Lyu, D., Backer, R., Robinson, W. G., and Smith, D. L. (2019). Plant growth-promoting rhizobacteria for cannabis production: yield, cannabinoid profile and disease resistance. Front. Microbiol. 10:1761. doi: 10.3389/fmicb.2019.01761
Ma, R., Cao, Y., Cheng, Z., Lei, S., Huang, W., Li, X., et al. (2017). Identification and genomic analysis of antifungal property of a tomato root endophyte Pseudomonas sp. p21. Antonie Van Leeuwenhoek. 110, 387–397. doi: 10.1007/s10482-016-0811-5
Mardanova, A. M., Hadieva, G. F., Lutfullin, M. T., Khilyas, I. V., Minnullina, L. F., Gilyazeva, A. G., et al. (2017). Bacillus subtilis strains with antifungal activity against the phytopathogenic fungi. Agric. Sci. 8, 1–20. doi: 10.4236/as.2017.81001
Mazhar, R., Ilyas, N., Raja, N. I., Saeed, M., Hussain, M., Seerat, W., et al. (2016). Plant growth promoting rhizobacteria: Biocontrol potential for pathogens. Pure Appl. Biol. 5, 1288–1295. doi: 10.19045/bspab.2016.50154
Melnick, R. L., Bailey, B. A., and Backman, P. A. (2013). Bacterial endophytes of perennial crops for management of plant disease. Bact. Agrobiol. 2013, 49–76.
Mendes, L. W., Kuramae, E. E., Navarrete, A. A., van Veen, J. A., and Tsai, S. M. (2014). Taxonomical and functional microbial community selection in soybean rhizosphere. ISME J. 8, 1577–1587. doi: 10.1038/ismej.2014.17
Mendes, R., Kruijt, M., de Bruijn, I., Dekkers, E., van der Voort, M., Schneider, J. H. M., et al. (2011). Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332, 1097–1100. doi: 10.1126/science.1203980
Meng, J., Liu, Z., Gou, C. L., Rogers, K. M., Yu, W. J., Zhang, S. S., et al. (2019). Geographical origin of Chinese wolfberry (goji) determined by carbon isotope analysis of specific volatile compounds. J. Chromatogr. B. 1105, 104–112. doi: 10.1016/j.jchromb.2018.12.011
Meng, Q., Hanson, L. E., Douches, D., and Hao, J. J. (2013). Managing scab diseases of potato and radish caused by Streptomyces spp. using Bacillus amyloliquefaciens BAC03 and other biomaterials. Bio. Control. 67, 373–379. doi: 10.1016/j.biocontrol.2013.09.009
Mignard, S., and Flandrois, J. P. (2006). 16S rRNA sequencing in routine bacterial identification: a 30-month experiment. J. Microbiol. Methods 67, 574–581. doi: 10.1016/j.mimet.2006.05.009
Mota, M. S., Gomes, C. B., Souza Júnior, I. T., and Moura, A. B. (2017). Bacterial selection for biological control of plant disease: criterion determination and validation. Braz. J. Microbiol. 48, 62–70. doi: 10.1016/j.bjm.2016.09.003
O’Brien, P. A. (2017). Biological control of plant diseases. Australas. Plant Pathol. 46, 293–304. doi: 10.1007/s13313-017-0481-4
Omukhua, G. E., and Godwin-Egein, M. I. (2011). Root rot disease of five fruit tree seedlings in the nursery. J. Agric. Soc. Res. 11, 1–5.
Pal, K. K., and McSpadden, B. G. (2006). Biological control of plant pathogens. Plant Health Instr 1:25. doi: 10.1094/PHI-A-2006-1117-02
Passari, A. K., Lalsiamthari, P. C., Zothanpuia, L. V. V., Mishra, V. K., and Yadav, M. K. (2018). Biocontrol of Fusarium wilt of Capsicum annuum by rhizospheric bacteria isolated from turmeric endowed with plant growth promotion and disease suppression potential. Eur. J. Plant Pathol. 150, 831–846. doi: 10.1007/s10658-017-1325-3
Rajaofera, M. J. N., Wang, Y., Jatoi, Z. A., Jin, P., Cui, H., Lin, C., et al. (2020). Bacillus atrophaeus HAB-5 secretion metabolites preventing occurrence of systemic diseases in tobacco plant. Eur. J. Plant Pathol. 156, 159–172. doi: 10.1007/s10658-019-01873-1
Rajiv, P., Anupama, S., Janardan, L., and Dhurva, P. G. (2017). PGPR in biocontrol: mechanisms and roles in disease suppression. Int. J. Agron. Agric. Res. 11, 69–80.
Rosenzweig, N., Tiedje, J. M., Quensen, J. F. III, Meng, Q., and Hao, J. J. (2012). Microbial communities associated with potato common scab-suppressive soil determined by pyrosequencing analyses. Plant Dis. 96, 718–725. doi: 10.1094/PDIS-07-11-0571
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Salomon, M. V., Funes Pinter, I., Piccoli, P., and Bottini, R. (2017). Use of plant growth-promoting rhizobacteria as biocontrol agents: induced systemic resistance against biotic stress in plants. Microb. Appl. 2, 133–152. doi: 10.1007/978-3-319-52669-0_7
Saravanakumar, K., Li, Y., Yu, C., Wang, Q. Q., Wang, M., Sun, J., et al. (2017). Effect of Trichoderma harzianum on maize rhizosphere microbiome and biocontrol of Fusarium stalk rot. Sci. Rep. 7:1771. doi: 1038/s41598-017-01680-w
Shahid, S., and Khan, M. R. (2016). Biological control of root rot on mungbean plants incited by Macrophomina phaseolina through microbial antagonists. J. Plant Pathol. 15, 27–39. doi: 10.3923/ppj.2016.27.39
Shang, Q., Yang, G., Wang, Y., Wu, X., Zhao, X., Hao, H., et al. (2016). Illumina-based analysis of the rhizosphere microbial communities associated with healthy and wilted Lanzhou lily (Lilium davidii var. unicolor) plants grown in the field. World J. Microbiol. Biotechnol. 32:95. doi: 10.1007/s11274-016-2051-2
Sharma, S. R., and Kolte, S. J. (1994). Effects of soil applied NPK fertilizers on severity of blackspot disease (Alternaria brassicae). Plant Soil. 167, 313–320. doi: 10.1007/BF00007958
Shen, Z., Ruan, Y., Chao, X., Zhang, J., Li, R., and Shen, Q. (2015). Rhizosphere microbial community manipulated by 2 years of consecutive biofertilizer application associated with banana Fusarium wilt disease suppression. Biol. Fertil. Soils 51, 553–562. doi: 10.1007/s00374-015-1002-7
Singh, P. C., Shukla, D., Fatima, T., Nautiyal, C. S., and Johri, J. K. (2017). Biological control of Fusarium sp. NBRIPMSF12 pathogenic to cultivated betelvine by Bacillus sp. NBRI-W9, a potential biological control agent. J. Plant Growth Regul. 36, 106–117. doi: 10.1007/s00344-016-9623-0
Singhai, P. K., Sarm, B. K., and Srivastava, J. S. (2011). Biological management of common scab of potato through Pseudomonas species and vermicompost. Bio. Control. 57, 150–157. doi: 10.1016/j.biocontrol.2011.02.008
Tiwari, S., Prasad, V., and Lata, C. (2019). “Bacillus: plant growth promoting bacteria for sustainable agriculture and environment,” in New and Future Developments in Microbial Biotechnology and Bioengineering, eds J. S. Singh and D. P. Singh (Oxford: Elsevier), 43–55. doi: 10.1016/B978-0-444-64191-5.00003-1
Uwaremwe, C., Yue, L., Liu, Y., Tian, Y., Zhao, X., Wang, Y., et al. (2021). Molecular identification and pathogenicity of Fusarium and Alternaria species associated with root rot disease of wolfberry in Gansu and Ningxia provinces, China. Plant Pathol. 70, 397–406. doi: 10.1111/ppa.13285
Větrovsky, T., and Baldrian, P. (2013). The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS One 8:e57923. doi: 10.1371/journal.pone.0057923
Wan, T., Zhao, H., and Wang, W. (2017). Effect of biocontrol agent Bacillus amyloliquefaciens SN16-1 and plant pathogen Fusarium oxysporum on tomato rhizosphere bacterial community composition. Biol. Control. 112, 1–9. doi: 10.1111/jph.12690
Wang, B., Shen, Z., Zhang, F., Raza, W., Yuan, J., Huang, R., et al. (2016). Bacillus amyloliquefaciens strain W19 can promote growth and yield and suppress Fusarium wilt in banana under greenhouse and field conditions. Pedosphere 26, 733–744. doi: 10.1016/S1002-0160(15)60083-2
Wang, Y., Ma, Q., Li, Y., Sun, T., Jin, H., Zhao, C., et al. (2019b). Energy consumption, carbon emissions and global warming potential of wolfberry production in Jingtai Oasis, Gansu Province, China. Environ. Manage. 64, 772–782. doi: 10.1007/s00267-019-01225-z
Wang, Y., Liang, X., Guo, S., Li, Y., Zhang, B., Yin, Y., et al. (2019a). Evaluation of nutrients and related environmental factors for wolfberry (L. barbarum) fruits grown in the different areas of China. Biochem. Syst. Ecol. 86:103916. doi: 10.1016/j.bse.2019.103916
Wang, Z., Li, Y., Zhuang, L., Yu, Y., Liu, J., Zhang, L., et al. (2019c). A rhizosphere derived consortium of Bacillus subtilis and Trichoderma harzianum suppresses common scab of potato and increases yield. Comput. Struct. Biotechnol. J. 17, 645–653. doi: 10.1016/j.csbj.2019.05.003
Wilkinson, T. J., Cowan, A. A., Vallin, H. E., Onime, L. A., Oyama, L. B., Cameron, S. J., et al. (2017). Characterization of the microbiome along the gastrointestinal tract of growing turkeys. Front. Microbiol. 8:1089–1099. doi: 10.3389/fmicb.2017.01089
Wu, K., Fang, Z. Y., Wang, L. L., Yuan, S. F., Guo, R., Shen, B., et al. (2016). Biological potential of bioorganic fertilizer fortified with bacterial antagonist for the control of tomato bacterial wilt and the promotion of crop yields. J. Microbiol. Biotechnol. 26, 1755–1764. doi: 10.4014/jmb.1604.04021
Yuan, J., Yu, L., Ling, N., Raza, W., Shen, Q., and Huang, Q. (2015). Plant-growth-promoting traits and antifungal potential of the Bacillus amyloliquefaciens YL-25. Biocontrol Sci. Technol. 25, 276–290. doi: 10.1080/09583157.2014.971711
Zhang, J. X., and Xue, A. G. (2010). Biocontrol of Sclerotinia stem rot (Sclerotinia sclerotiorum) of soybean using novel Bacillus subtilis strain SB24 under control conditions. Plant Pathol. 59, 382–391. doi: 10.1111/j.1365-3059.2009.02227.x
Zhao, J., Wang, Y., Liang, H., Huang, J., Chen, Z., and Nie, Y. (2017). The rhizosphere microbial community response to a bio-organic fertilizer: finding the mechanisms behind the suppression of watermelon Fusarium wilt disease. Acta Physiol. Plant. 40:17. doi: 10.1007/s11738-017-2581-8
Zhao, P., Quan, C., Wang, Y., Wang, J., and Fan, S. (2013). Bacillus amyloliquefaciens Q-426 as a potential biocontrol agent against Fusarium oxysporum f. sp. spinaciae. J. Basic Microbiol. 54, 448–456. doi: 10.1002/jobm.201200414
Keywords: Bacillus, biocontrol, F. oxysporum, root rot, wolfberry, rhizosphere bacterial community
Citation: Uwaremwe C, Yue L, Wang Y, Tian Y, Zhao X, Liu Y, Zhou Q, Zhang Y and Wang R (2022) An Endophytic Strain of Bacillus amyloliquefaciens Suppresses Fusarium oxysporum Infection of Chinese Wolfberry by Altering Its Rhizosphere Bacterial Community. Front. Microbiol. 12:782523. doi: 10.3389/fmicb.2021.782523
Received: 24 September 2021; Accepted: 25 November 2021;
Published: 05 January 2022.
Edited by:
Tofazzal Islam, Bangabandhu Sheikh Mujibur Rahman Agricultural University, BangladeshReviewed by:
Geromy G. Moore, Southern Regional Research Center (USDA-ARS), United StatesShamim Hasan, University of Bonn, Germany
Copyright © 2022 Uwaremwe, Yue, Wang, Tian, Zhao, Liu, Zhou, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruoyu Wang, wangruoyu@lzb.ac.cn
 Constantine Uwaremwe
Constantine Uwaremwe Liang Yue1,3
Liang Yue1,3 Yuan Tian
Yuan Tian Ruoyu Wang
Ruoyu Wang