- 1Liziping Giant Panda’s Ecology and Conservation Observation and Research Station of Sichuan Province (Science and Technology Department of Sichuan Province), China West Normal University, Nanchong, China
- 2Key Laboratory of Southwest China Wildlife Resources Conservation (Ministry of Education), China West Normal University, Nanchong, China
- 3Liziping National Nature Reserve Administration, Ya’an, China
The giant panda has developed a series of evolutionary strategies to adapt to a bamboo diet. The abundance and diversity of the phyllosphere microbiome change dramatically depending on the season, host species, location, etc., which may, in turn, affect the growth and health of host plants. However, few studies have investigated the factors that influence phyllosphere bacteria in bamboo, a staple food source of the giant panda. Amplicon sequencing of the 16S rRNA gene was used to explore the abundance and diversity of phyllosphere bacteria in three bamboo species (Arundinaria spanostachya, Yushania lineolate, and Fargesia ferax) over different seasons (spring vs. autumn), elevation, distance from water, etc., in Liziping National Nature Reserve (Liziping NR), China. And whole-genome shotgun sequencing uncovered the differences in biological functions (KEGG and Carbohydrate-Active enzymes functions) of A. spanostachya phyllosphere bacteria between spring and autumn. The results showed that the abundance and diversity of F. ferax phyllosphere bacteria were greater than that of the other two bamboo species in both seasons. And three kinds of bamboo phyllosphere bacteria in autumn were significantly higher than in spring. The season was a more important factor than host bamboo species in determining the community structure of phyllosphere bacteria based on the (un)weighted UniFrac distance matrix. The composition, diversity, and community structure of phyllosphere bacteria in bamboo were primarily affected by the season, species, altitude, tree layer, and shrub layer. Different bacterial communities perform different functions in different bamboo species, and long-term low temperatures may shape more varied and complex KEGG and Carbohydrate-Active enzymes functions in spring. Our study presented a deeper understanding of factors influencing the bacterial community in the bamboo phyllosphere. These integrated results offer an original insight into bamboo, which can provide a reference for the restoration and management of giant panda bamboo food resources in the Xiaoxiangling mountains.
Introduction
Large numbers of different kinds of microorganisms within the phyllosphere constitute a stable and complex micro-ecosystem (Mei, 1991; Li, 1998; Shi and Zhang, 2007; Vorholt, 2012). Different hosts have different microbial communities (Yadav et al., 2008; Leveau, 2015), including bacteria, archaea, fungi, and protists (Newton et al., 2010). Bacteria are the most abundant microorganism in the phyllosphere (Lindow and Brandl, 2003) and Proteobacteria are the dominant phylum (Delmotte et al., 2009).
Numerous environmental factors determine phyllosphere community composition, such as plant species, temperature, humidity, nutrient availability (on the plant surface), sun/UV exposure levels, and even the underlying soil geochemistry (Lindow and Brandl, 2003; Leveau, 2015). Ecological factors and plant attributes, e.g., wood density, leaf mass per unit area, and leaf nitrogen and phosphorus concentration, may also affect the microbial community structure in the phyllosphere (Steven et al., 2014).
The phyllosphere microbial community represents a wide range of primitive symbiotic relationships (Fedorov et al., 2011; Partida and Martin, 2011). Phyllosphere microbiota can affect plant yield (e.g., via nutrients or hormones) (Gourion et al., 2006; Reed et al., 2010) and plant health in a variety of ways (Balint-Kurti et al., 2010; Gerd et al., 2011). For example, Beauveria fungus on rice leaf can protect rice enzyme activity and is an environmentally friendly microbe (Du et al., 2014). Some phyllosphere microbes are beneficial to plants via nitrogen fixation (Leveau, 2015), growth promotion (Bringel and Couée, 2015), acting as barriers against pathogen invasion, and decomposition of residual pesticides (Venkatachalam et al., 2016).
Bamboo is a perennial evergreen plant belonging to the Gramineae family and Bambusoideae subfamily and is an important forest resource all over the world. It has the characteristics of being widely distributed, fast-growing and high-yield, and strong regeneration ability. Therefore, it has considerable economic, ecological, and social benefits (Shanmughavel and Francis, 1997; Gupta and Kumar, 2008; Chang and Chiu, 2015; Zhang and Xue, 2018). Bamboo leaves are an important habitat for many microorganisms, with microbial abundance and diversity acting as an indicator of forest health. However, few studies have investigated the environmental factors that influence phyllosphere bacterial composition in bamboo. Using the traditional cultivation method, Zhang et al. (2014) found that the composition of the phyllosphere microbial community differed between bamboo species and seasons. The species composition and frequency of leaf endophytic bacteria and fungi have also been found to differ between bamboo species (Helander et al., 2013), while endophytic fungi from fish-scale bamboo (Phyllostachys heteroclada) differs between the branch and leaf tissue (Zhou et al., 2017). Significant differences in bacterial richness and diversity were also observed between different bamboo species using high throughput amplicon sequencing (Jin et al., 2020). However, there is a lack of research on what factors influence the composition and diversity in the bamboo phyllosphere microbiome.
The giant panda (Ailuropoda melanoleuca) belongs to a carnivorous clade (Wei et al., 1999), and yet has an exclusively herbivorous diet and specializes in the consumption of bamboo leaves throughout the year (Hu et al., 1985; Zhao et al., 2013). In the long evolutionary process, the giant panda developed a series of foraging strategies to adapt to a bamboo diet, such as seasonal vertical migration, selection of habitat, and feeding point (Hong et al., 2015, 2016). However, in the different seasons of different mountain systems, the diet of giant pandas does vary. For example, in the Qinling mountains, giant pandas mainly feed on bamboo leaves during the non-bamboo shoot seasons (Pan, 2001; Wu et al., 2017), while bamboo leaves of the Xiaoxiangling mountains account for more than half of the panda feces in summer and autumn (Wei et al., 1999). However, leaves usually contribute to intestinal diseases of giant pandas in captivity, and bacterial infections are an important cause of intestinal diseases. Previous research has shown that Escherichia coli and Klebsiella pneumoniae could cause giant pandas’ diarrhea and septicemia (Zhang and Chen, 1997; Xiong et al., 1999). However, there are few studies concentrating on the phyllosphere microbiome in the bamboo species foraged by wild giant pandas.
In this study, we investigate the phyllosphere bacterial community of bamboo species frequently used as a food source by giant pandas, using Next Generation Sequencing Technology (NGS) in Liziping National Nature Reserve (Liziping NR), Sichuan, China. The specific goals include (i) comparing interspecies differences and seasonal diversity of phyllosphere bacterial communities in bamboo; (ii) exploring the ecological factors that influence the phyllosphere bacteria community changes; and (iii) revealing the functional differences of phyllosphere bacterial communities among different bamboo species/different seasons. Our integrated results can provide a reference for the restoration and management of giant panda bamboo food resources in the Xiaoxiangling mountains.
Materials and Methods
Study Area
Liziping NR is located in Shimian County, Sichuan Province, China, and is in the middle and upper reaches of the Dadu River, on the southwestern edge of the Sichuan Basin and southeast of Gongga Mountain (E102°10′33′′-E102°29′07′′, N28°51′02′′ -N29°08′42′′) (Hong et al., 2015). The reserve covers an area of 47,940 km2 and ranges from 1,330 to 4,550 m above sea level, with uneven ridges and narrow valleys. The annual average temperature and rainfall are 11.7–14.4°C and 800–1250 mm, respectively (Xei and Hong, 2020). As the altitude increases, the vegetation in the reserve transitions from evergreen broad-leaved forest to deciduous broad-leaved forest, then coniferous and broad-leaved mixed forest, coniferous forest to alpine thickets, and alpine rocky beech. According to the Fourth National Survey of giant pandas, there were 22 giant pandas in the reserve in 2015 (Sichuan Forestry Bureau, 2015). To strengthen the small isolated populations of giant pandas, the Chinese government has initiated the Captive Giant Panda Release Program (CGPRP). To date, one giant panda rescued from the wild and nine giant pandas from captivity have been released into this nature reserve (Hong et al., 2012). There are seven bamboo species (three genera) consumed by the giant panda within the Xiaoxiangling Mountains. The species with the largest distribution is Arundinaria spanostachya, which accounts for 38.08% of the total area of giant panda feeding bamboos in these mountains. Next is Yushania lineolate and Fargesia ferax, which account for 28.02 and 12.48% of the giant panda’s feeding bamboo, respectively (Sichuan Forestry Bureau. 2015). A. spanostachya mainly grows above 2,500 m a.s.l, while Y. lineolate and F. ferax occur below 2,800 m a.s.l. Giant pandas in this reserve prefer to eat A. spanostachya throughout the whole year, some Y. lineolate in winter, and occasionally F. ferax (Hong et al., 2015, 2016; Xei and Hong, 2020).
Experimental Design
We conducted wild surveys and sampling during May (spring) and October (autumn) in 2020. First, we set up four transects in A. spanostachya, Y. lineolate, and F. ferax bamboo forests, respectively. The transects were set from low altitude to high altitude, and the distance between them was no less than 200 m. Secondly, we set up three to five survey plots (20 m × 20 m) in each transect, with the altitude distance of adjacent survey plots on the same transect no less than 50 m. We then recorded and measured bamboo species, latitude and longitude, altitude, and other related variables in the tree and shrub layer. Moreover, one bamboo plot (1 m × 1 m) was set up in the center of each survey plot, and another two bamboo plots (1 m × 1 m) were set up east and south, 5 m from the center point of each survey plot. Finally, the related variables within the bamboo layer were also measured and recorded (Supplementary Table 1).
Sample Collection and DNA Extraction
In each survey plot, one mixed bamboo leaf sample (not less than 200 g) was collected with sterile gloves, immediately transported to the laboratory (less than 2 h), and stored at −20°C for further processing within 48 h. Each 200 g sample was aseptically transferred into a Ziplock bag (24 cm × 35 cm) containing 200 ml sterile precooled TE-buffer (10 mM Tris, 1 mM EDTA, and pH 7.5) supplemented with 0.05% Tween-80 (Hong et al., 2017). Leaf surfaces were washed to collect the microbial population by 5 min of shaking, vortexing, and sonication of each sample in the TE-buffer, with the Ziplock bag kept in ice water (∼4°C) for each processing step (Hong et al., 2017). The cell suspension was separated from the leaf material by filtration through three-layer sterile nylon mesh. Sonication was performed at a frequency of 40 kHz in an ultrasonic cleaning bath (Shanghai Kudos Instrument Co., Shanghai, China) to dislodge the microbes from the leaf surface. Following filtration, cell suspensions were placed in four 50 ml tubes/sample, and cells were pelleted using centrifugation at 2,000 × g for 15 min at 4°C. Cell pellets from multiple tubes were pooled into 2.0-ml reaction tubes and washed twice with TE-buffer with Tween-80. Cell pellets were immediately frozen at −80°C until DNA extraction.
The DNA extraction was performed using the E.Z.N.A.TM Soil DNA Kit (Omega, Norcross, GA, United States) as described with slight modifications. Frozen cell pellets were resuspended in 1 ml of kit-supplied SLX Mlus buffer with 500 mg of glass beads, and cell lysis was performed at 65 Hz for 90 s. The cell debris suspension was immediately processed following the instructions in the kit manual. Finally, the total DNA was obtained from the column by two sequential elutions with 50 μl elution buffer.
16S rRNA Gene V3–V4 Amplification, Quantification, and Sequencing
The sequencing of the V3–V4 hypervariable region of the 16S rRNA gene was performed for bacterial identification (Caporaso et al., 2011; Lundberg et al., 2013). Thirty-five cycles of polymerase chain reaction (PCR) amplification of the target marker genes were performed. Error-correcting 12-bp barcoded primers specific to each sample were used to permit multiplexing of samples. Polymerase chain reaction products from all samples were quantified using the PicoGreen dsDNA assay and pooled together in equimolar concentrations. Each library was submitted to Mega Biotech on the Illumina MiSeq PE300 platform.
Sequence Data Analysis
The raw 16S rRNA gene sequencing reads were demultiplexed, quality-filtered by fastp version 0.20.0 (Chen et al., 2018), and merged by FLASH version 1.2.7 (Magoč and Salzberg, 2011) with the following criteria: (i) the 300 bp reads were truncated at any site receiving an average quality score of <20 over a 50 bp sliding window, and the truncated reads shorter than 50 bp were discarded. Reads containing ambiguous characters were also discarded; (ii) only overlapping sequences longer than 10 bp were assembled according to their overlapped sequence. The maximum mismatch ratio of the overlap region was 0.2. Reads that could not be assembled were discarded; (iii) samples were distinguished according to the barcode and primers. The sequence direction was adjusted using exact barcode matching with two nucleotide mismatches in primer matching.
Operational taxonomic units (OTUs) with 97% similarity cut-off (Stackebrandt and Goebel, 1994; Edgar, 2013) were clustered using UPARSE version 7.1, and chimeric sequences were identified and removed. The taxonomy of each OTU representative sequence was analyzed by RDP Classifier version 2.2 (Wang et al., 2007) against the 16S rRNA database (e.g., Silva v138) using a confidence threshold of 0.7. Non-target sequences, including mitochondrial and chloroplast sequences, were also removed by QIIME from the final OTU data set. To better convey the biological information in these samples, the average relative abundance of the bacterial community was visualized by bar chart at the level of phylum and genus.
Statistical Analysis
First, Two-way ANOVA (species × season) were used to exam the differences in bacterial abundance between dominant bacteria abundance at both phylum and genus levels. We used mothur software (version v.1.30.1) to calculate the Sobs and Shannon indices for each sample, to estimate the species abundance and diversity of phyllosphere bacteria between the different bamboo species and seasons, and used one-way ANOVA to identify any significant differences. We then used PCoA based on the weighted UniFrac and unweighted UniFrac method distance matrix to evaluate the differences in the microbial community structure between the bamboo species and seasons. We also used the permutational MANOVA (PERMANOVA) to analyze the effect of different bamboo species and seasons on the phyllosphere bacterial community, based on Bray–Curtis distance matrices with a substitution test to analyze the statistical significance of the division.
Second, Spearman’s correlation heatmap analysis was performed to examine the relationship between the relative abundance of bacterial taxa and the environmental factors described in Supplementary Table 1. To explore the spatial association between each environmental factor and phyllosphere bacterial community, Mantel tests were performed to examine the spatial correlation between each environmental factor distance matrix and phyllosphere bacterial UniFrac distance matrix based on OUT level. Redundancy analysis (RDA) was used to detect the relationship between environmental factors, samples and microflora, or directly detect the relationship between two pairs. Linear regression was used to evaluate the relationship between environmental factors and the results of either Alpha (Sobs and Shannon indices) or Beta diversity analysis (Bray–Curtis distance).
Finally, the PICRUSt prediction was used to predict the functional composition of all phyllosphere bacteria in the different bamboo species. The greengene id corresponding to each OTU, the COG, and KEGG functions of the OTU were annotated to obtain the function level of COG and KEGG, and the abundance information for each function in different samples.
Metagenome Sequencing and Analysis
The raw reads from metagenome sequencing were used to generate clean reads by removing adaptor sequences, trimming and removing low-quality reads (reads with N bases, a minimum length threshold of 50 bp, and a minimum quality threshold of 20) using the fastp1 (Chen et al., 2018) (version 0.20.0) on the free online platform of Majorbio Cloud Platform (cloud.majorbio.com). The clean reads were mapped to the host reference genome using BWA2 (Li and Durbin, 2009) (version 0.7.9a) to identify and remove the host-originated reads. These high-quality reads were then assembled to contigs using MEGAHIT (Li et al., 2015) (parameters: kmer_min = 47, kmer_max = 97, step = 10)3 (version 1.1.2), which makes use of succinct de Bruijn graphs. Contigs with the length being or over 300 bp were selected as the final assembling result. Open reading frames (ORFs) in contigs were identified using MetaGene (Noguchi et al., 2006). The predicted ORFs with a length equal to or greater than 100 bp were retrieved and translated into amino acid sequences using the NCBI translation table4.
A non-redundant gene catalog was constructed using CD-HIT5 (Fu et al., 2012) (version 4.6.1) with 90% sequence identity and 90% coverage. Reads after quality control were mapped to the non-redundant gene catalog with 95% identity using SOAPaligner (Li et al., 2008) (version 2.21), and gene abundance in each sample was evaluated. Representative sequences of non-redundant gene catalog were annotated based on the NCBI NR database using blastp as implemented in DIAMOND v0.9.19 with an e-value cutoff of 1e-5 using Diamond6 (Buchfink et al., 2015) (version 0.8.35) for taxonomic annotations. For functional analyses, KEGG annotation was conducted using Diamond (Buchfink et al., 2015) (see Text Footnote 8, version 0.8.35) against the Kyoto Encyclopedia of Genes and Genomes database7 (version 94.2) with an e-value cutoff of 1e-5. Carbohydrate-active enzymes annotation was conducted using hmmscan against CAZy database8 with an e-value cutoff of 1e-5. Abundant different features of KEGG Level 2 and CAZyme were determined using linear discriminant analysis (LDA) effect size (LEfSe) and setting 2 as the threshold on the logarithmic LDA score (Segata et al., 2011).
Results
Community Composition Among Different Bamboo Species in Spring and Autumn
Up to 19 samples of each bamboo species were collected in each study season (Table 1). After removing the mitochondrial and chloroplast sequences, a total of 1,233,917 effective target 16S rRNA reads were obtained. The average sequence of each sample was 12,217 ± 3,037. Clustered by 97% similarity, all samples had a total of 2,564 OTUs. A total of 1,378 OTUs were shared between the three bamboo species, and the bacterial OTUs of each in autumn were significantly higher than in spring (Figure 1 and Supplementary Figure 1). Fargesia ferax had a higher number of OTUs than the other two species in both seasons (Figure 1A).
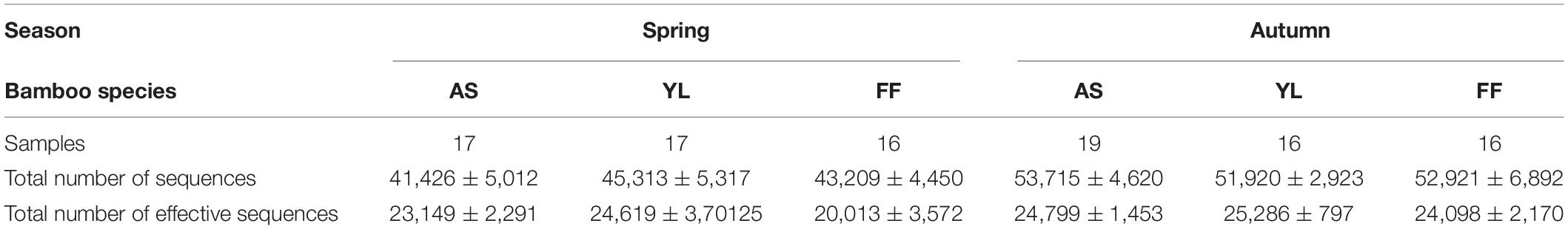
Table 1. Statistics table of each sample and sequence (AS, A. spanostachya; YL, Y. lineolate; and FF, F. ferax) (Mean ± SD).
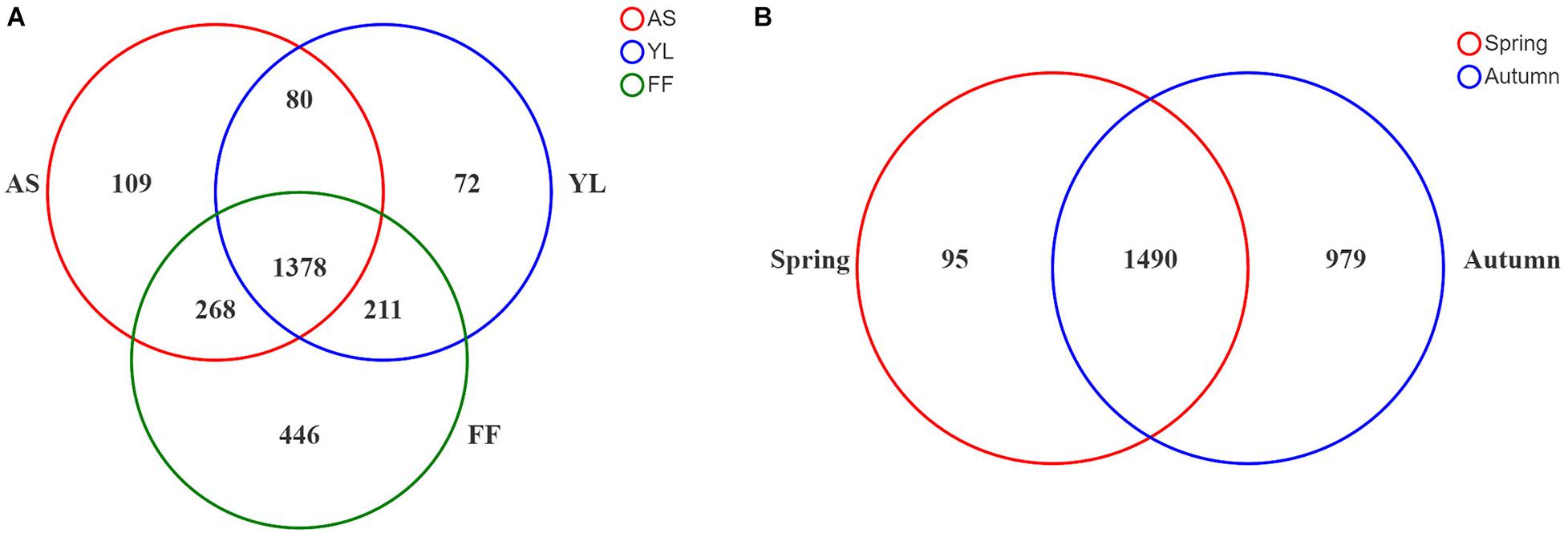
Figure 1. Venn diagram showing overlap/non-overlap of phyllosphere bacteria OTUs among three bamboo species (A) and two seasons (B). AS, A. spanostachya; YL, Y. lineolate; FF, F. ferax.
After annotating the representative sequence of OTU with species clustering, all bacterial OTUs were sorted into 24 phyla and 614 genera. The most dominant phylum abundance was Proteobacteria, accounting for over 60% of the bacterial relative abundance observed across all samples (Figure 2A). The remaining bacteria were distributed among the phyla Acidobacteria, Bacteroidota, Actinobacteria, Planctomycetota, Myxococcota, and others. In spring, the relative abundance of phyla Bacteroidota, Actinobacteriota, and Myxococcota in the F. ferax phyllosphere were significantly higher than that of A. spanostachya and Y. lineolate. However, the relative abundance of phylum Acidobacteriota in the F. ferax phyllosphere was significantly lower than that of A. spanostachya and Y. lineolate. In autumn, the relative abundance of phyla Actinobacteriota and Myxococcota in the F. ferax phyllosphere were significantly higher than that of A. spanostachya and Y. lineolate. The relative abundance of phyla Bacteroidota in Y. lineolate and Planctomycetota in the A. spanostachya phyllosphere was significantly lower than that of the other two bamboo species. No difference was found in the relative abundance of dominant phyla in the F. ferax phyllosphere between seasons, except Planctomycetota. The relative abundance of phylum Acidobacteriota in the A. spanostachya and Y. lineolate phyllospheres were lower in autumn than spring, and the relative abundance of the phyla Bacteroidota, Actinobacteriota, and Myxococcota all increased from spring to autumn (Table 2).
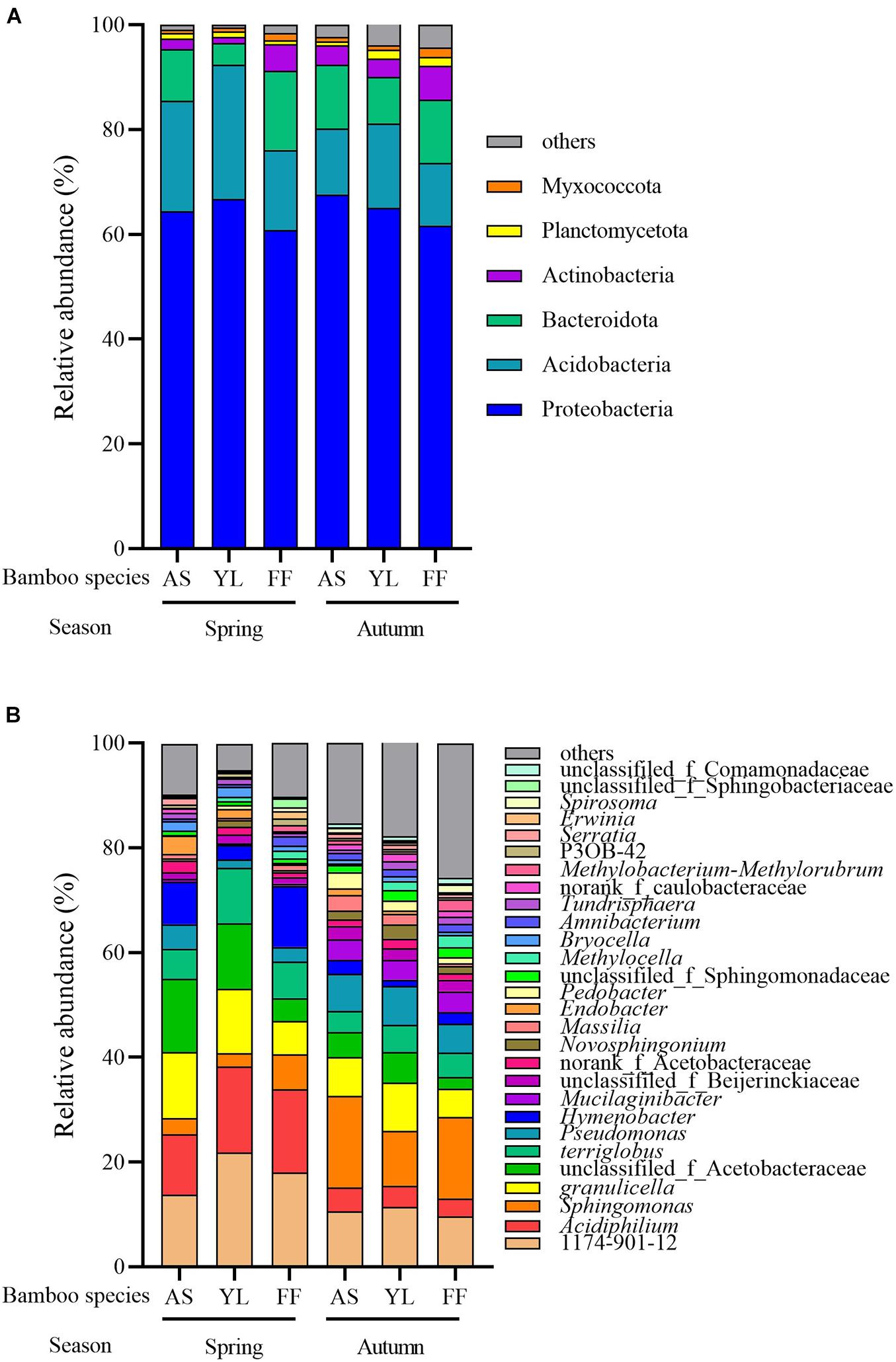
Figure 2. Relative abundance of different phyllosphere bacterial phyla (A) and genus (B) among A. spanostachya (AS), Y. lineolate (YL), and F. ferax (FF) in spring and autumn.
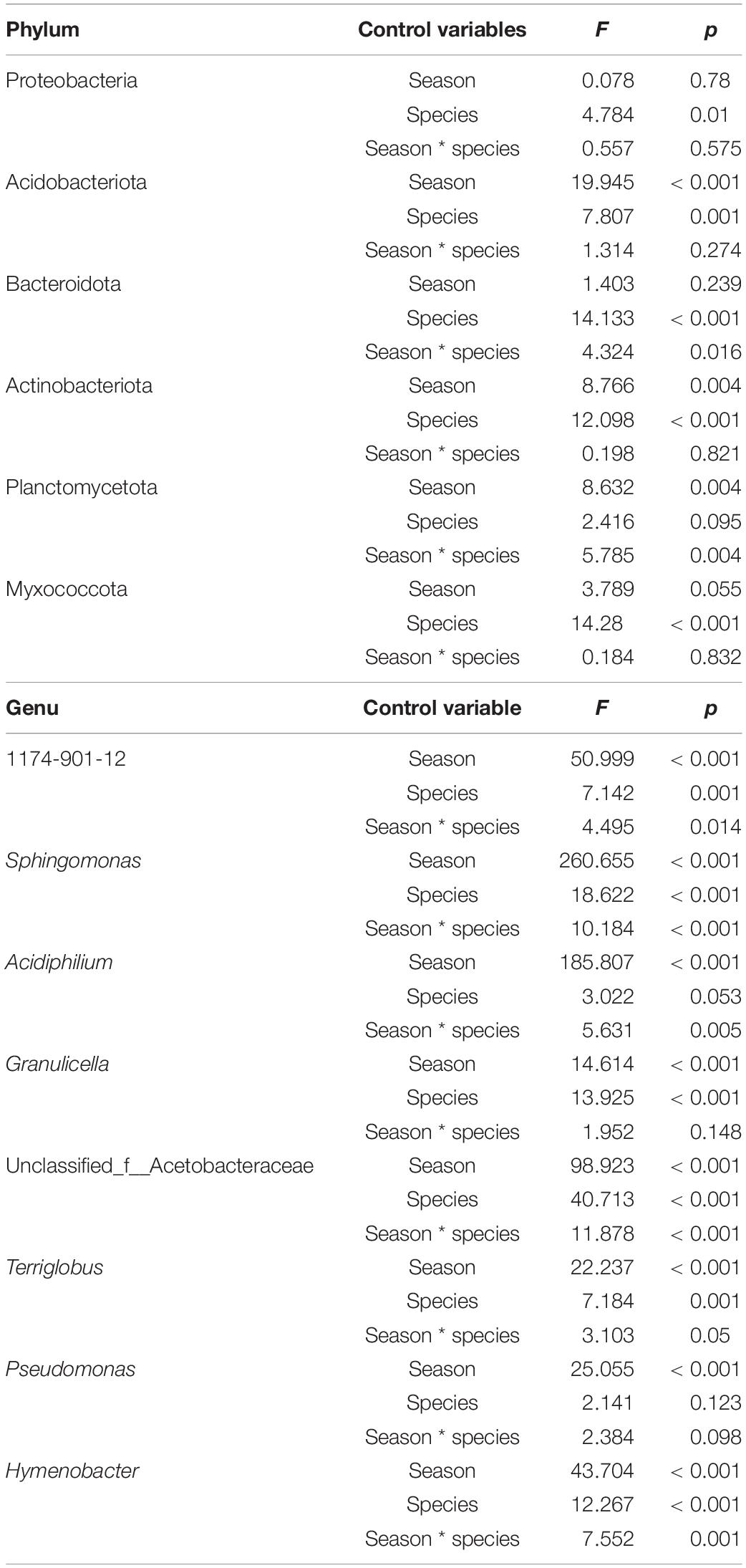
Table 2. Two-way ANOVA (species × season) examining the differences in bacterial abundance between dominant bacteria abundance at both phylum and genus levels.
The relative abundances of genera 1174-901-12, Acidiphilium, Sphingomonas, unclassified_f__Acetobacteraceae, Terriglobus, Granulicella, Pseudomonas, and Hymenobacter revealed them to be the dominant bacteria in the phyllosphere of all three bamboo species, and the differences among the top eight total abundances of genera were calculated. In spring, the relative abundance of 1174-901-12, Acidiphilium, and Terriglobus were highest in the Y. lineolate phyllosphere. The relative abundance of Hymenobacter and Sphingomonas was highest in the F. ferax phyllosphere, but the opposite pattern was observed for Granulicella. In autumn, the lowest relative abundance of Acidiphilium and Granulicella existed in the F. ferax phyllosphere, and that of Hymenobacter and Sphingomonas existed in the Y. lineolate phyllosphere. For all three bamboo species, similar relative abundance patterns were found for the seven most dominant bacteria genus in each phyllosphere. The relative abundance of 1174-901-12, Acidiphilium, Hymenobacter, Granulicella, and Terriglobus decreased from spring to autumn but increased for Sphingomonas and Pseudomonas (Figure 2B and Table 2).
Community Diversity Among Different Bamboo Species in Spring and Autumn
The Sobs and Shannon indices for the bacterial community in the F. ferax phyllosphere were higher than that of A. spanostachya and Y. lineolate in both seasons. However, in spring the Sobs and Shannon indices in Y. lineolate were lower than that of F. ferax and A. spanostachya. The phyllosphere bacterial community of A. spanostachya showed the lowest Sobs and Shannon indices in autumn (Figure 3 and Supplementary Table 2). These indices were also found to increase significantly from spring to autumn in all bamboo species.
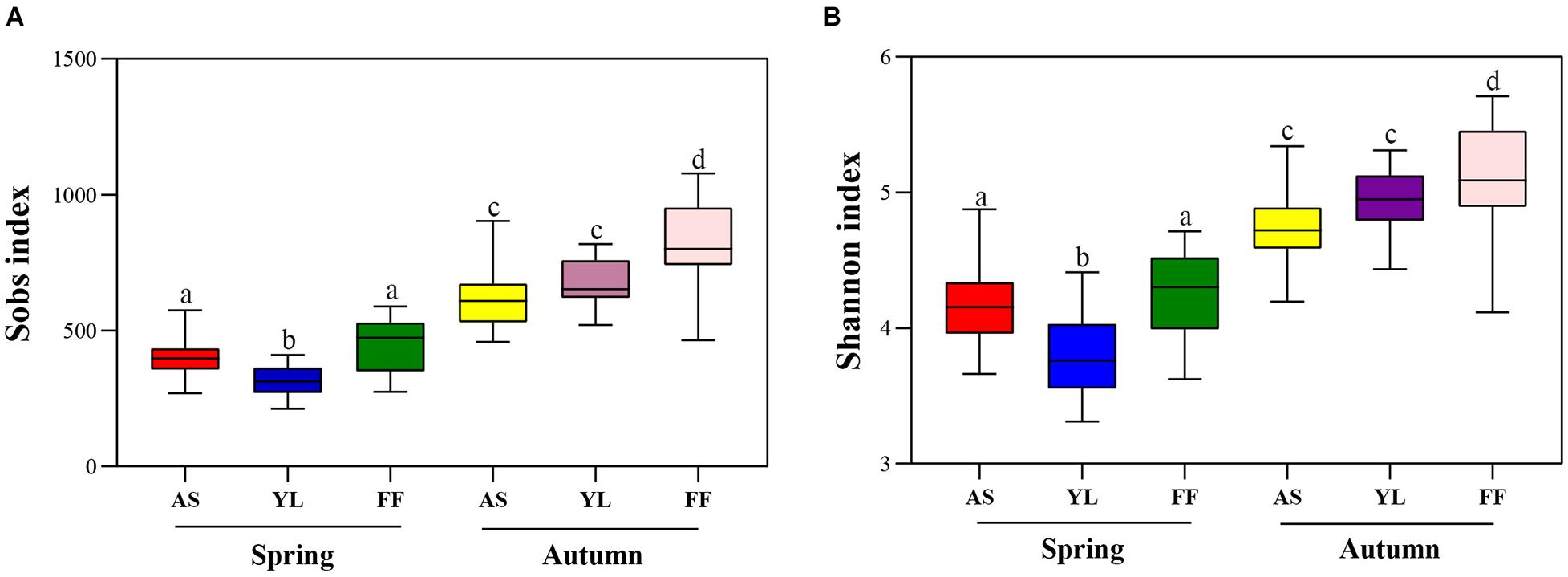
Figure 3. The observed species index (Sobs index) (A) and Shannon index (B) of phyllosphere bacterial community among A. spanostachya (AS), Y. lineolate (YL), and F. ferax (FF) in spring and autumn. One-way ANOVA was used to compare the significance. Different letters indicate significant differences.
The results of the PCoA revealed that samples were clustered by species and season. Samples of A. spanostachya and Y. lineolate were also clustered together away from that of F. ferax in the same season. The PCoA based on unweighted UniFrac distinguished samples better than that based on weighted UniFrac (Figure 4). The PERMANOVA revealed significant differences in phyllosphere bacterial community based on Bray–Curtis distance matrixes between the three bamboo species in spring and autumn (Table 3).
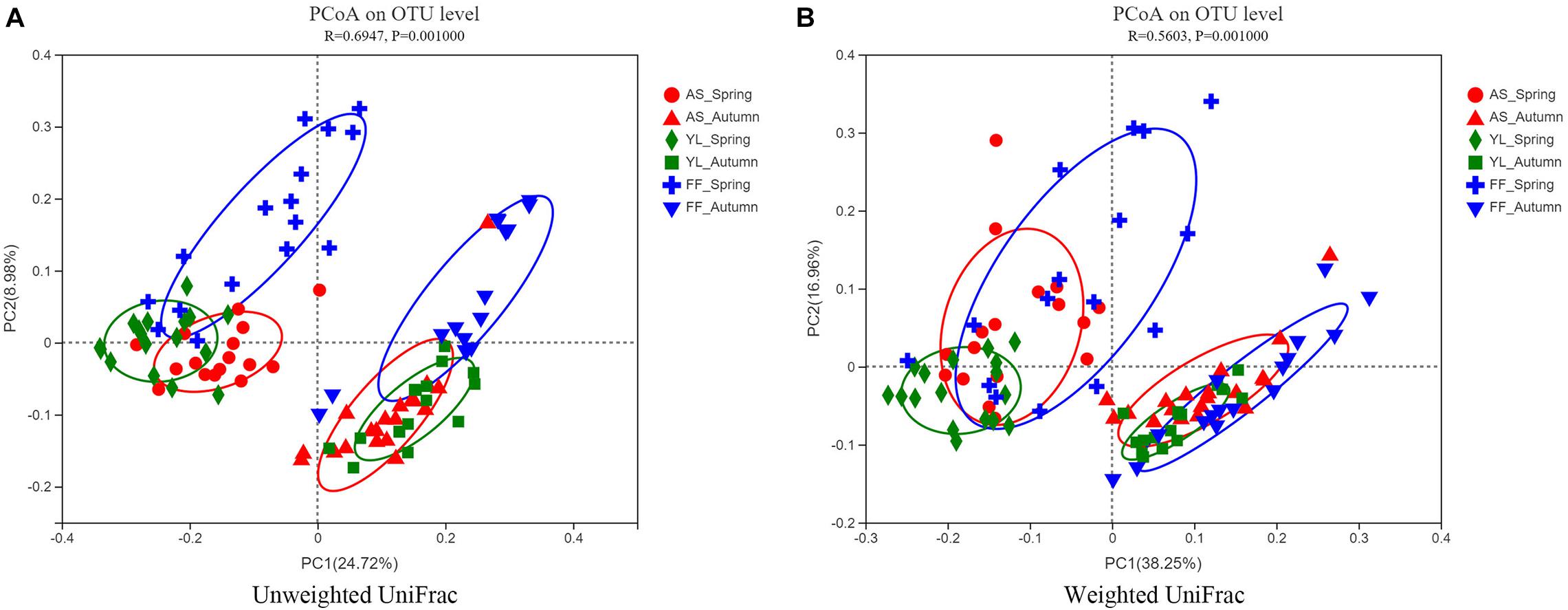
Figure 4. Principal coordinate analysis (PCoA) plots of phyllosphere bacterial communities using the unweighted UniFrac method (A) and weighted UniFrac method (B) at OTU level among A. spanostachya (AS), Y. lineolate (YL), and F. ferax (FF) in spring and autumn.
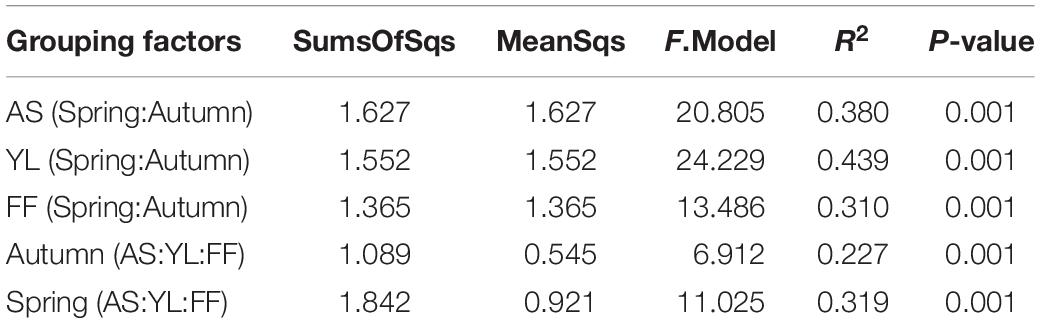
Table 3. PERMANOVAs exploring the effects of different bamboo species (AS, A. spanostachya; YL, Y. lineolate; and FF, F. ferax) and seasons (Spring and Autumn) on the phyllosphere bacteria community based on Bray–Curtis distance matrixes.
Ecological Factors Influencing Bamboo Phyllosphere Bacterial Community
The results of Mantel analysis revealed that elevation had the greatest impact on the phyllosphere bacterial community (R = 0.313, P = 0.001). And elevation, distance from water, trees height, trees diameter at breast height, shrubs coverage, shrubs numbers, mean height of bamboo, and mean base diameter of bamboo also had significant impacts on the phyllosphere bacterial community (Supplementary Table 3). Furthermore, RDA analysis showed that the number of trees, canopy density, bamboo coverage, and bamboo deaths also influenced the bacterial community in samples (Figure 5 and Supplementary Table 4). However, distance from water and shrubs coverage had no significant impact on bacterial community in samples (Figure 5 and Supplementary Table 4).
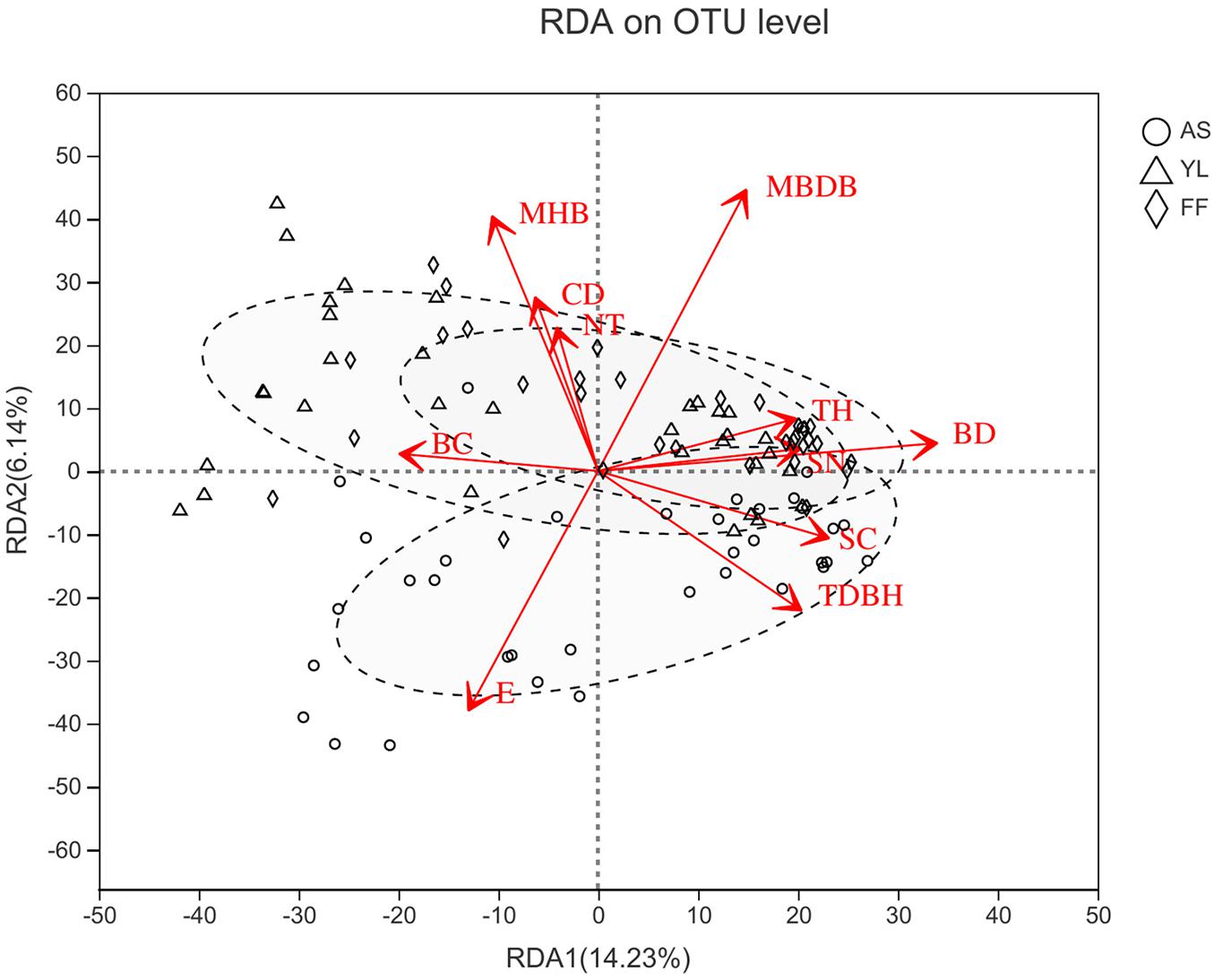
Figure 5. Redundancy analysis (RDA) between environmental factors and phyllosphere bacterial communities at OTU level among A. spanostachya (AS), Y. lineolate (YL), and F. ferax (FF). E, elevation; TH, trees height; TDBH, trees diameter at breast height; SN, shrubs numbers; MHB, mean height of bamboo; MBDB, mean base diameter of bamboo. CD, canopy density; BC, bamboo coverage; BD, bamboo deaths; NT, number of trees.
Spearman’s correlation heatmap analysis revealed that Proteobacteria (P < 0.05) and Acidobacteria (P < 0.05) abundance were positively correlated with elevation (E), while Actinobacteria (P < 0.001) and Myxococcota (P < 0.001) abundance were negatively correlated with elevation (E). Actinobacteria (P < 0.001) and Myxococcota (P < 0.001) abundance were positively correlated with the mean base diameter of bamboo (MBDB). Acidobacteria (P < 0.01) abundance was positively correlated with distance from water (DW), but Actinobacteria (P < 0.01) and Bacteroidota (P < 0.05) were negatively correlated with distance from water (DW). Myxococcota (P < 0.001) abundance was positively correlated with the mean height of bamboo (MHB) (Figure 6A). At the genus level, 1174-901-12 and Acidiphilium were both negatively correlated with tree height (TH), trees diameter at breast height (TDBH), shrub coverage (SC), and shrubs number (SN). Positive correlations were observed between Sphingomonas and shrubs number (SN), tree height (TH), and mean base diameter of bamboo (MBDB). Granulicella showed significant correlations with elevation (E) and distance from water (DW), but negative correlations with the mean base diameter of bamboo (MBDB) (Figure 6B).
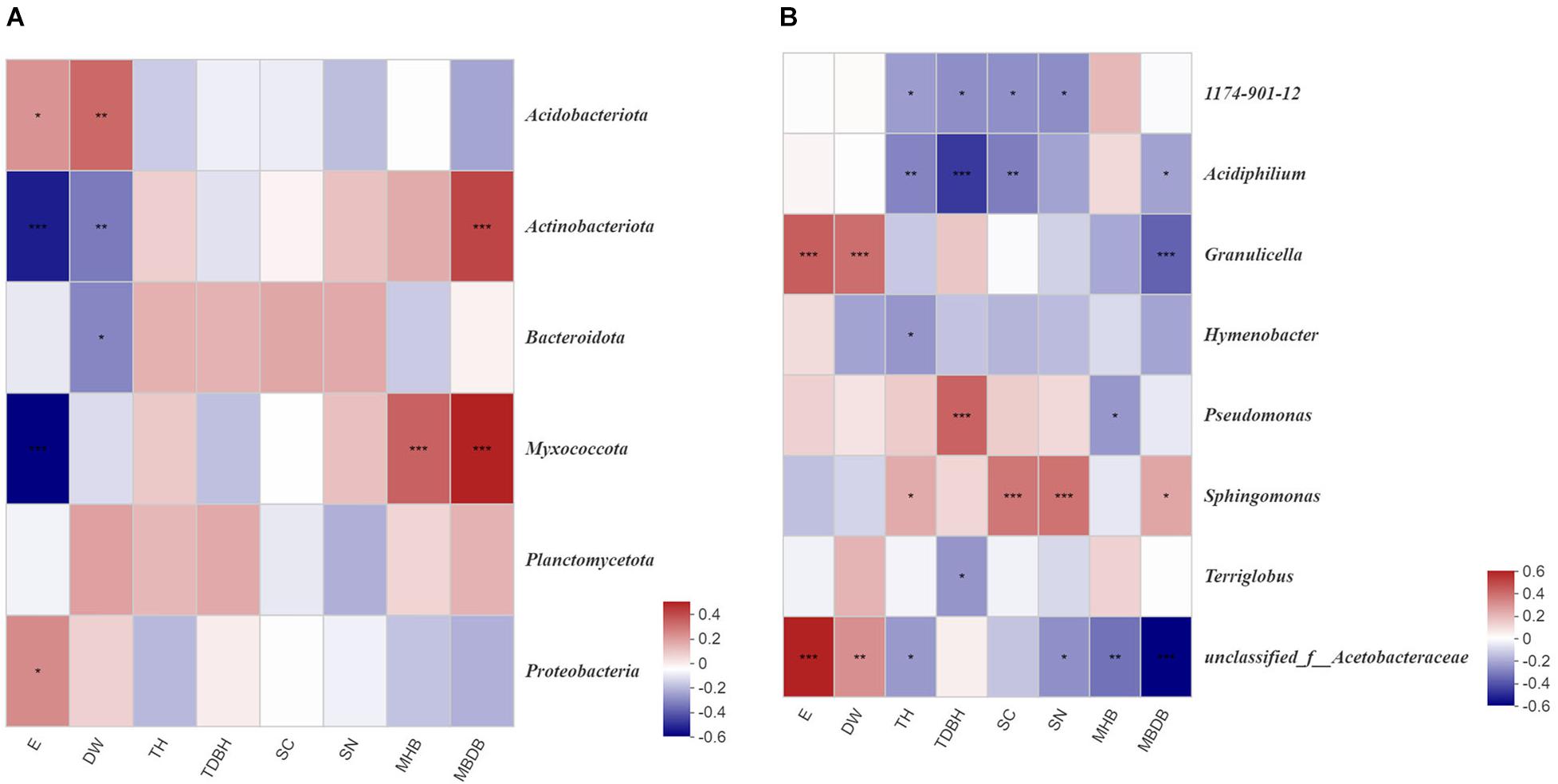
Figure 6. Correlation heatmap of bamboo environmental factors and read numbers at the phylum (A) and genus (B) level for bacteria. E, elevation; DW, distance from water; TH, trees height; TDBH, trees diameter at breast height; SC, shrubs coverage; SN, shrubs numbers; MHB, mean height of bamboo; MBDB, mean base diameter of bamboo. The color intensity in each panel indicates the relative correlation between bamboo environmental factors and read numbers of each group. *0.01 < P < 0.05, **0.001 < P < 0.01, ***P < 0.001.
Further linear regression found that elevation (E), bamboo deaths (BD), mean base diameter of bamboo (MBDB), bamboo coverage (BC), and tree height (TH) had a significant impact on the Sobs and Shannon indices (Supplementary Table 5). Elevation (E) had the highest influence on the Sobs index, followed by bamboo deaths (BD), mean base diameter of bamboo (MBDB), bamboo coverage (BC), and tree height (TH). The highest ecological influence factor for the Shannon index and Bray–Curtis distance was bamboo deaths (BD). Furthermore, shrub coverage (SC) and shrubs number (SN) all had a significant impact on bacterial community structure in the bamboo phyllosphere based on Bray–Curtis distance matrixes (Supplementary Table 5).
Function Differences Among Different Bamboo Species/Different Seasons
All three bamboo phyllosphere bacterial communities had similar COG function classification patterns as generated by PICRUSt in both spring and autumn (Supplementary Figure 2). There were higher relative abundance sequences related to cell wall/membrane/envelope biogenesis, amino acid transport, and metabolism. The relative abundance of transporter function (4.17%) was the highest in all three bamboo species, followed by ABC transporters (2.55%), DNA repair and recombination proteins (2.37%), and two-component systems (2.23%) (Supplementary Figure 2). Prediction software PICRUSt enriched 22 categorizable dominant pathways (relative abundance > 1%) in the level 3 KEGG pathway. Among them, nine pathways had significant differences between bamboo species (P < 0.05) (Figure 7 and Supplementary Table 6). It is worth noting that there were significant differences in the relative abundance of the bacterial secretion system, the secretion system, and the two-component system. Oxidative phosphorylation in energy metabolism, porphyrin, and chlorophyll metabolism were also different, and the gene relative abundance for these functions for F. ferax was significantly lower than that of the other two species (Figure 7 and Supplementary Table 6).
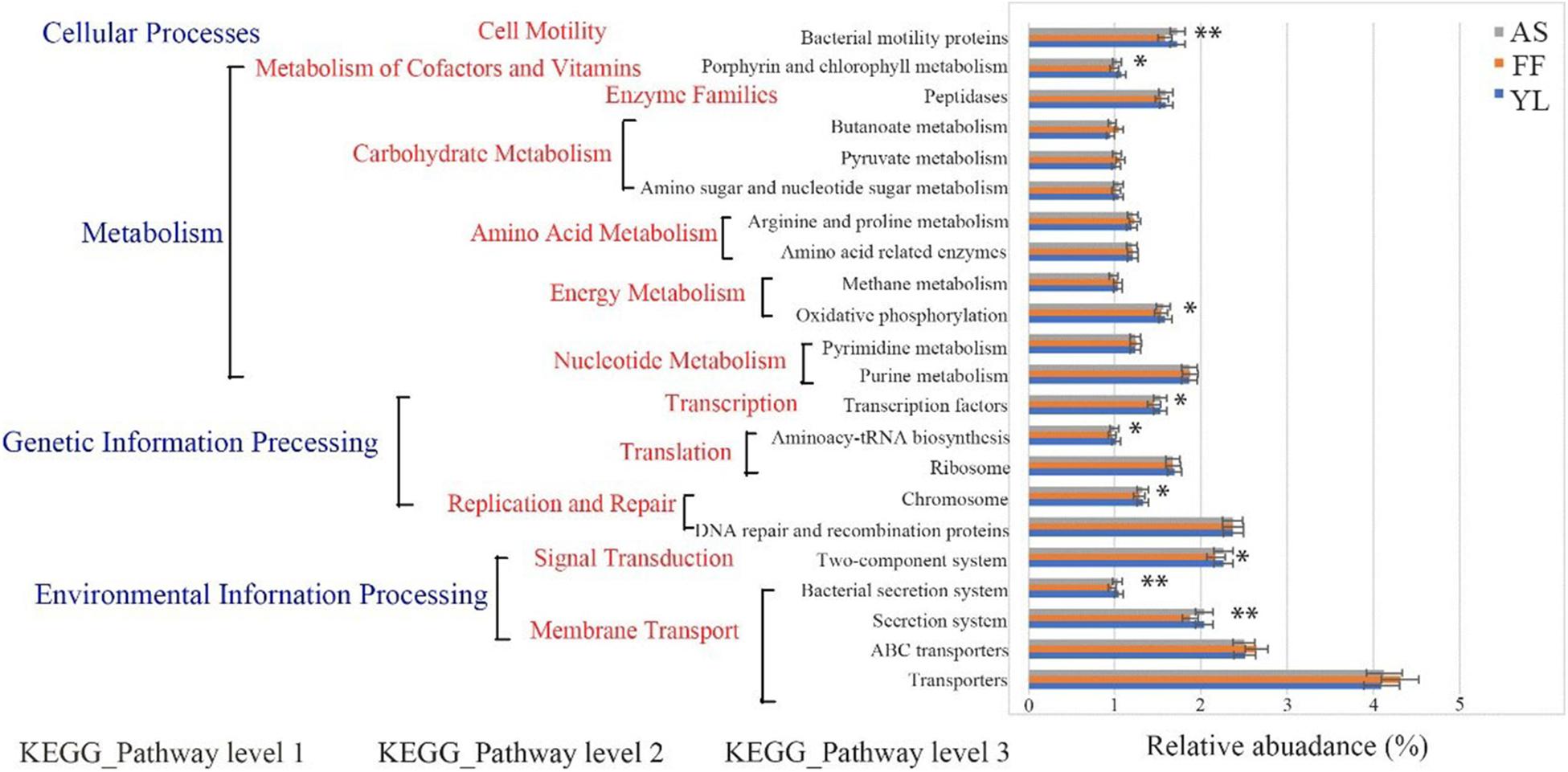
Figure 7. Functional predictions for phyllosphere bacterial communities with significantly different KEGG pathways (P < 0.05) for the three bamboo species (AS, FF, and YL). KEGG pathways at Level 1, Level 2, and Level 3 are represented. *0.01 < P < 0.05, **0.001 < P < 0.01.
To explore the functional potential of phyllosphere bacteria in bamboo between different seasons, a total of eight bamboo samples from A. spanostachya (spring, four samples; autumn, four samples) were used to perform metagenomic sequencing. After filtering out low-quality and host genomic reads, 129.50 Gb of high-quality reads were obtained, with an average of 16.18 Gb for each sample. In total, 3.91 million contigs and 2,473 million bp of assembly sequence were obtained with an average contig N50 of 5,238 bp (Supplementary Table 7). A non-redundant gene catalog containing 2,206,289 entries was constructed, with an average of 275,786 entries per sample (Supplementary Table 7). The representative sequences of the non-redundant gene catalog were annotated to obtain five domains, and the bacterial domain was further studied in this paper. The metagenomic analysis confirmed 456 KOs, including 44 KEGG Level 2 categories in bacteria. A. spanostachya in spring displayed high abundances in KEGG Level 2 categories of signal transduction, amino acid metabolism, lipid metabolism, infectious disease: bacterial, xenobiotics biodegradation and metabolism, aging, cell growth and death, cancer: overview, cancer: specific types, endocrine system, infectious disease: viral, cardiovascular disease, and drug resistance: antineoplastic (Figure 8A), whereas cellular community-eukaryotes, immune system, nucleotide metabolism, biosynthesis of other secondary metabolites, glycan biosynthesis and metabolism, and cellular community-prokaryotes exhibited higher abundance in autumn (Figure 8A). According to the LEfse results of the CAZyme, four families had significantly higher relative abundance in spring, including glycoside hydrolases (GH2, GH3, and GH130), carbohydrate esterases (CE9 and CE10), and glycosyl transferases (GT51 and GT2_Glyco_trans_2_3) and auxiliary activities (AA3, AA12, and AA3_2); interestingly, in autumn, glycosyl transferases (GTs) showed the main difference (Figure 8B).
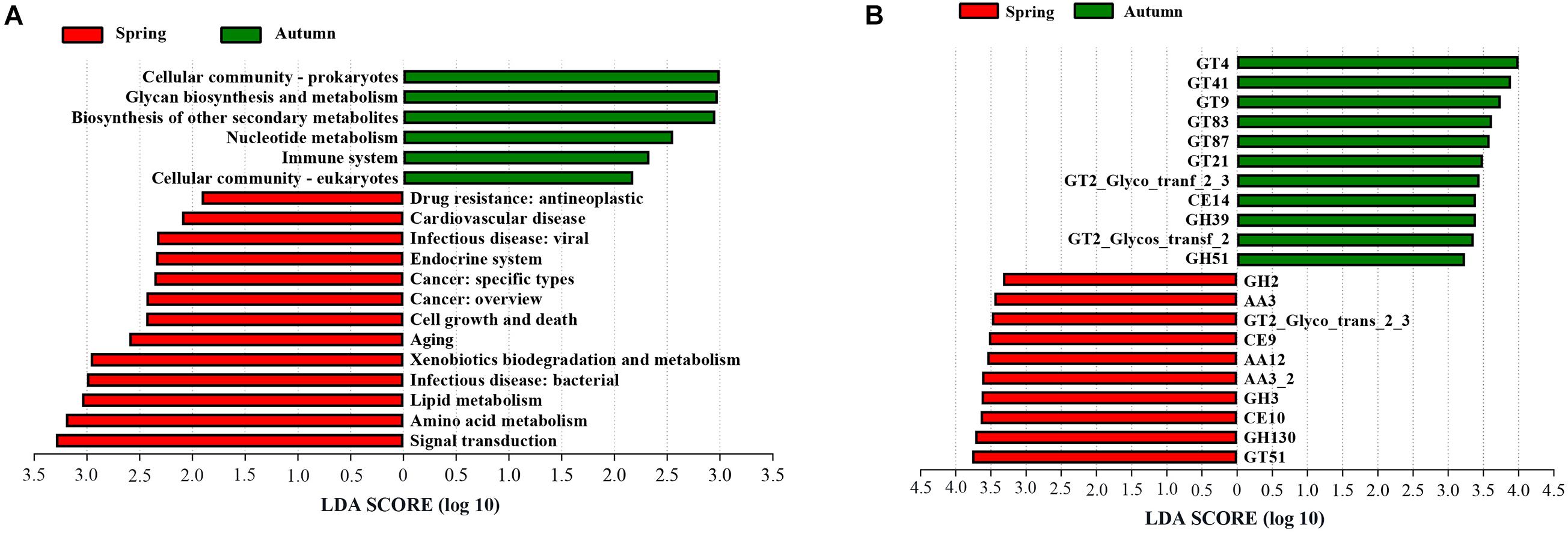
Figure 8. Linear discriminant analysis (LDA) effect size (LEfSe) analysis of KEGG level 2 (A) and CAZy (B) between A. spanostachya in spring and autumn. GT, glycosyl transferases; CE, carbohydrate esterases; GH, glycoside hydrolases; AA, auxiliary activities. LDA > 2.
Discussion
This study revealed that the bacterial OTU richness of the three bamboo species in autumn was significantly higher than that in spring. The Phyllosphere of F. ferax had a greater diversity of bacterial OTUs than that of A. spanostachya and Y. lineolate. The dominant phyllosphere bacteria of the three bamboo species are Proteobacteria, Acidobacteria, Bacteroides, and Actinomycetes. In a warm and humid climate, the diversity and richness of phyllosphere bacteria in the three bamboo species in spring were significantly higher than that in autumn. The overall importance of seasonality to the structure and composition of the phyllosphere microbial community has been confirmed by many studies (Thompson et al., 1993; Copeland et al., 2015). Proteobacteria, Acidobacteria, Bacteroides, and Actinomycetes are often detected in a variety of forests, indicating that these organisms have a wide ecological range and an ability to adapt to many environments (Laforest et al., 2016; Feng et al., 2019). In this study, the relative abundance of the Proteobacteria in the three bamboo species was all above 60%, and the differences between the bamboo species were not significant, indicating that Proteobacteria played a dominant role in the phyllosphere microbial community. Its variation in turn may impact the health of the staple food bamboo foraged by giant panda around Xiaoxiangling mountains.
The bacterial diversity and abundance of all three bamboo species in autumn were significantly higher than that in spring, which was similar to the result of Zheng (2011), who found that the number of phyllosphere microbial communities of Pinus tabulaeformis varied significantly between different seasons, with the largest diversity and abundance in autumn, followed by summer and the least in spring. Higher temperatures and humidity in summer and autumn resulted in higher species diversity and richness than in spring, while higher elevations and longer periods of low temperatures in winter could result in lower species diversity and richness than in spring (Laforest et al., 2016).
The Mantel test found that elevation, distance from water, tree diameter at breast height, mean height of bamboo, mean base diameter of bamboo, tree height, shrub coverage, and the number of shrubs all significantly affected the phyllosphere microbial community (Supplementary Table 3). Jackson et al. (2010) found that the changes in the phyllosphere bacterial community in resurgent ferns were related to rainfall and humidity. Similarly, Laforest et al. (2016) found that the host species, habitat, and climate (average summer temperature and precipitation) drove the phyllosphere bacterial community structure in temperate trees. In this study, elevation had the strongest relationship with phyllosphere microbial community. Elevation was significantly and positively correlated to many bacteria phyla abundance, such as Proteobacteria (Figure 6). However, with an increase in elevation, both the Shannon and Sobs indices declined (Supplementary Figure 6). Higher elevation generally resulted in lower temperature, which can limit the fluidity of microbial cell membranes and proteins, which is not conducive to microbial reproduction and growth (Zhang et al., 2014).
Changes in the phyllosphere microbial community could impact the degradation and absorption of plant nutrients and the metabolism of enzymes (Fazal et al., 2021). In this study, we used PICRUSt to predict the function of phyllosphere bacteria of three bamboo species foraged by giant pandas. Our results have shown that the relative abundance of gene function in the membrane transport secretion system, signal transduction, and oxidative phosphorylation metabolism in the third level of the KEGG pathway for F. ferax was lower than that of the other two bamboo species (Figure 7). This implied that some phyllosphere bacteria flora in F. ferax at lower altitude need less metabolic activity to maintain the physiological activities relative to that in high-altitude A. spanostachya and Y. lineolate phyllosphere. It is worth noting that the relative abundances of gene functions about transporters and ABC transporters in the bamboo phyllosphere bacterial community were the most expressed pathways in membrane transport. Hamana et al. (2012) revealed that ABC transporters can protect animals from the barrier of toxic substances. In addition, genes related to replication and repair may help reduce damage to biomolecules and may help bamboo adapt to high-altitude environments.
Different microbial compositions, diversity, and community structure imply different functional characteristics. Further metagenomic analysis uncovered the significant differences in biological functions (KEGG and Carbohydrate-Active enzymes functions) of A. spanostachya phyllosphere bacteria between spring and autumn (Figure 8). In spring, despite the pathways of human diseases (infectious disease: bacterial and viral, cancer: overview and specific types, cardiovascular disease, and drug resistance: antineoplastic) being more abundant, the raised biological functions including the immune system and biosynthesis of other secondary metabolites in autumn may neutralize these negative effects. Abundant xenobiotics biodegradation and metabolism function be conducive to A. spanostachya phyllosphere health. More abundant nucleotide metabolism, glycan biosynthesis and metabolism, and cellular community indicated stronger bacterial cell viability and cell proliferation in autumn. The LEfse results of the CAZyme support an important role for the metabolism of carbohydrates in bacterial survival in the plant phyllosphere (Ryffel et al., 2015; Müller et al., 2016). GHs families can break down complex carbohydrates (Lee et al., 2013) and played a crucial role in the processing of various exogenous and endogenous glycoconjugate in human gut microbiota (Pellock et al., 2018). Our results father indicated a diversification in carbohydrate metabolism pathways existing in A. spanostachya phyllosphere bacterial microorganisms in spring, which may be due to harsh environment, such as low temperatures in lengthy winter (Laforest et al., 2016).
Food microbes can affect the gut microbes of animals (Kohl and Dearing, 2014; Kohl et al., 2017). Jin et al. (2020) found significant associations of certain bacteria and fungi between bamboo and the gut of giant panda. The diversity of bamboo bacteria was also positively correlated with that of gut bacteria in giant panda. Giant pandas prefer to consume bamboo that grows naturally at high altitudes, probably because the total number of endophytic bacteria in high altitudes tends to be lower (Helander et al., 2013). There is a lot of work needed to fully understand the relationship between the food microbes and gut microbes of pandas. To explore the relevance of these genes and their functions in the environmental adaptability of giant pandas and bamboo, further research is needed to determine whether there are specific enzymes coded in phyllosphere microorganisms related to digestion in giant pandas’ gut.
Conclusion
In this study, high-throughput sequencing was used to explore phyllosphere bacterial communities in three bamboo species (A. spanostachya, Y. lineolate, and F. ferax) foraged by giant panda in different seasons (spring vs. autumn), in Liziping National Nature Reserve (Liziping NR), China. Our findings suggested that the diversity of F. ferax phyllosphere bacterial species was greater than that of the other two bamboo species in both seasons, indicating that low altitude may be a promoter of bamboo phyllosphere microbial richness and diversity, of course, strong human interference at low altitudes was also a factor. Besides, phyllosphere bacterial diversity was also significantly higher in autumn than in spring, implying that season-related changes in environmental factors (e.g., temperature and moisture) may influence bacterial communities. In autumn, there was a lower abundance and diversity of phyllosphere bacteria at higher altitudes, and giant pandas chose more high-altitude bamboo species like A. spanostachya as their staple food. In spring, a lower abundance and diversity of bamboo phyllosphere bacteria was found, and giant pandas chose some low-altitude bamboo species like Y. lineolate as their staple food. Long-term low temperatures may shape more varied and complex KEGG and Carbohydrate-Active enzymes functions in spring. Interestingly, the feeding of giant pandas on bamboo species was consistent with low bacterial concentrations in bamboo leaves. Was this related to the giant panda gut microbes’ adaptability? This needs rigorous controlled experimental approaches to investigate. All these findings may provide a reference for the restoration and management of giant panda bamboo dietary and food resources in this area, especially for those small isolated populations of giant pandas in Xiaoxiangling mountains.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA718425.
Author Contributions
MH, ZZ, and JL conceived and designed the study. JL, WL, JX, YY, JW, LK, and YL collected data and samples in the field. MH, JL, JX, YY, and JW processed samples in the lab. MH, JL, and YY analyzed the data. MH, ZZ, JL, and LW wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was financially supported by the National Natural Science Foundation of China (No. 31900337), the Second Qinghai-Tibet Plateau Comprehensive Scientific Survey (2019QZKK05010502), and the Fundamental Research Funds of China West Normal University (18B025, 18Q040, and 20B006).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank all staff of the Liziping National Nature Reserve Administration who assisted with the data collection and experiments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.748141/full#supplementary-material
Footnotes
- ^ https://github.com/OpenGene/fastp
- ^ http://bio-bwa.sourceforge.net
- ^ https://github.com/voutcn/megahit
- ^ http://www.ncbi.nlm.nih.gov/Taxonomy/taxonomyhome.html/index.cgi?chapter=tgencodes#SG1
- ^ http://www.bioinformatics.org/cd-hit/
- ^ https://github.com/bbuchfink/diamond
- ^ http://www.genome.jp/kegg/
- ^ http://www.cazy.org/
References
Balint-Kurti, P., Simmons, S. J., Blum, J. E., Ballaré, C. L., and Stapleton, A. E. (2010). Maize leaf epiphytic bacteria diversity patterns are genetically correlated with resistance to fungal pathogen infection. Mol. Plant Microbe Interact. 23, 473–484. doi: 10.1094/MPMI-23-4-0473
Bringel, F., and Couée, I. (2015). Pivotal roles of phyllosphere microorganisms at the interface between plant functioning and atmospheric trace gas dynamics. Front. Microbiol. 6:486. doi: 10.3389/fmicb.2015.00486
Buchfink, B., Xie, C., and Huson, D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat Methods 12, 59–60. doi: 10.1038/nmeth.3176
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversityat a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108, 4516–4522. doi: 10.1073/pnas.1000080107
Chang, E. H., and Chiu, C. Y. (2015). Changes in soil microbial community structure and activity in a cedar plantation invaded by moso bamboo. Appl. Soil Ecol. 91, 1–7. doi: 10.1016/j.apsoil.2015.02.001
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Copeland, J. K., Yuan, L., Layeghifard, M., Wang, P. W., and Guttman, D. S. (2015). Seasonal community succession of the phyllosphere microbiome. Molecular plant-microbe interactions. Mol. Plant Microbe Interact. 28, 274–285. doi: 10.1094/MPMI-10-14-0331-FI
Delmotte, N., Knief, C., Chaffron, S., Innerebner, G., Roschitzki, B., Schlapbach, R., et al. (2009). Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc. Natl. Acad. Sci. U.S.A. 106, 16428–16433. doi: 10.1073/pnas.0905240106
Du, W., Jiang, P., Wang, Y., Lv, L., Wang, H., Bu, Y., et al. (2014). Effects of Beauveria bassiana on paddy antioxidant enzymes activities and phyllosphere microbial diversity. Acta Ecol. Sin. 34, 6975–6984. doi: 10.5846/stxb201303050350
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Fazal, U. R., Aftab, S., Maria, K., Muhammad, A., and Yousaf, G. (2021). Chemistry of microbial life in phyllosphere: a review. Green Rep. 2, 1–8. doi: 10.36686/Ariviyal.GR.2021.02.04.009
Fedorov, D. N., Doronina, N. V., and Trotsenko, Y. A. (2011). Phytosymbiosis of aerobic methylobacteria: new facts and views. Microbiology 80, 435–446. doi: 10.1134/S0026261711040047
Feng, X., Cao, X., Ju, W., and Liu, K. (2019). A study on community characteristics of forest soil bacteria in Lushan National Nature Reserve. For. Resour. Manag. 6, 101–107. doi: 10.13466/j.cnki.lyzygl.2019.06.018
Fu, L., Niu, B., Zhu, Z., Wu, S., and Li, W. (2012). CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152. doi: 10.1093/bioinformatics/bts565
Gerd, I., Claudia, K., and Vorholt, J. A. (2011). Protection of Arabidopsis thaliana against leaf-pathogenic Pseudomonas syringae by Sphingomonas strains in a controlled model system. Appl. And Environ. Microbiol. 77, 3202–3210. doi: 10.1128/AEM.00133-11
Gourion, B., Rossignol, M., and Vorholt, J. A. (2006). A proteomic study of Methylobacterium extorquens reveals a response regulator essential for epiphytic growth. Proc. Natl. Acad. Sci. U.S.A. 103, 13186–13191. doi: 10.1073/pnas.0603530103
Gupta, A., and Kumar, A. (2008). Potential of bamboo in sustainable development. Asia Pacif. Bus. Rev. 4, 100–107. doi: 10.1177/097324700800400312
Hamana, K., Itoh, T., Sakamoto, M., and Hayashi, H. (2012). Covalently linked polyamines in the cell wall peptidoglycan of the anaerobes belonging to the order Selenomonadales. J. Gen. Appl. Microbiol. 58, 339–347. doi: 10.2323/jgam.58.339
Helander, M., Jia, R., Huitu, O., Sieber, T. N., Jia, J., Niemelä, P., et al. (2013). Endophytic fungi and silica content of different bamboo species in giant panda diet. Symbiosis 61, 13–22. doi: 10.1007/s13199-013-0253-z
Hong, M. S., Peng, G. X., Keyhani, N. O., and Xia, Y. X. (2017). Application of the entomogenous fungus, Metarhizium anisopliae for leafroller (Cnaphalocrocis medinalis) control and its effect on rice phyllosphere microbial diversity. Appl. Microbiol. Biotechnol. 101, 6793–6807. doi: 10.1007/s00253-017-8390-6
Hong, M., Wang, J., Yang, Z., Gu, X., and Zhang, Z. (2012). Comparison of microhabitat structure of giant panda in primary forest and secondary forest. J. China West Normal Univ. (Natural Science Edition) 33, 356–361. doi: 10.16246/j.issn.1673-5072.2012.04.022
Hong, M., Wei, W., Yang, Z., Yuan, S., Yang, X., Gu, X., et al. (2016). Effects of timber harvesting on Arundinaria spanostachya bamboo and feeding-site selection by giant pandas in Liziping Nature Reserve, China. For. Ecol. Manag. 373, 74–80. doi: 10.1016/j.foreco.2016.04.039
Hong, M., Yuan, S., Yang, Z., Yang, X., Gu, X., Huang, F., et al. (2015). Comparison of microhabitat selection and trace abundance of giant pandas between primary and secondary forests in Liziping Nature Reserve, China: effects of selective logging. Mamm. Biol. 80, 373–379. doi: 10.1016/j.mambio.2015.05.003
Hu, J., Schaller, G. B., Wenshi, P., and Jing, Z. (1985). The Giant Pandas of Wolong. Sichuan Science and Technology Press, Chendu. 244.
Jackson, E. F., Echlin, H. L., and Jackson, C. R. (2010). Changes in the phyllosphere community of the resurrection fern, Polypodium polypodioides, associated with rainfall and wetting. FEMS Microbiol. Ecol. 58, 236–246. doi: 10.1111/j.1574-6941.2006.00152.x
Jin, L., Wu, D., Li, C., Zhang, A., Xiong, Y., Wei, R., et al. (2020). Bamboo nutrients and microbiome affect gut microbiome of giant panda. Symbiosis 80, 293–304. doi: 10.1007/s13199-020-00673-0
Kohl, K. D., Brun, A., Magallanes, M., Brinkerhoff, J., Laspiur, A., Acosta, J. C., et al. (2017). Gut microbial ecology of lizards: insights into diversity in the wild, effects of captivity, variation across gut regions and transmission. Mol. Ecol. 26, 1175–1189. doi: 10.1111/mec.13921
Kohl, K. D., and Dearing, M. D. (2014). Wild-caught rodents retain a majority of their natural gut microbiota upon entrance into captivity. Environ. Microbiol. Rep. 6, 191–195. doi: 10.1111/1758-2229.12118
Laforest, I., Messier, C., and Kembel, S. W. (2016). Host species identity, site and time drive temperate tree phyllosphere bacterial community structure. Microbiome 4:27. doi: 10.1186/s40168-016-0174-1
Lee, S., Cantarel, B., Henrissat, B., Gevers, D., Birren, B. W., Huttenhower, C., et al. (2013). Gene-targeted metagenomic analysis of glucan-branching enzyme gene profiles among human and animal fecal microbiota. ISME J. 8, 493–503. doi: 10.1038/ismej.2013.167
Leveau, J. (2015). Life of Microbes on Aerial Plant Parts. New York NY: Springer International Publishing, doi: 10.1007/978-3-319-08575-3_4
Li, D., Liu, C. M., Luo, R., Sadakane, K., and Lam, T. W. (2015). MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676. doi: 10.1093/bioinformatics/btv033
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, R., Li, Y., Kristiansen, K., and Wang, J. (2008). SOAP: short oligonucleotide alignment program. Bioinformatics 24, 713–714. doi: 10.1093/bioinformatics/btn025
Li, X. (1998). Relationship between microorganisms and higher plants. J. Hebei North Univ. 2, 64–65.
Lindow, S. E., and Brandl, M. T. (2003). Microbiology of the phyllosphere. Appl. Environ. Microbiol. 69, 1875–1883. doi: 10.1128/AEM.69.4.1875-1883.2003
Lundberg, D. S., Yourstone, S., Mieczkowski, P., Jones, C. D., and Dangl, J. L. (2013). Practical innovations for high-throughput amplicon sequencing. Nat. Methods 10, 999–1002. doi: 10.1038/nmeth.2634
Magoč, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Müller, D. B., Schubert, O. T., Röst, H., Aebersold, R., and Vorholt, J. A. (2016). Systems-level proteomics of two ubiquitous leaf commensals reveals complementary adaptive traits for phyllosphere colonization. Mol. Cell Proteomics 15, 3256–3269. doi: 10.1074/mcp.M116.058164
Newton, A. C., Gravouil, C., and Fountaine, J. M. (2010). Managing the ecology of foliar pathogens: ecological tolerance in crops. Ann. Appl. Biol. 157, 343–359. doi: 10.1111/j.1744-7348.2010.00437.x
Noguchi, H., Park, J., and Takagi, T. (2006). MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res. 34, 5623–5630. doi: 10.1093/nar/gkl723
Partida, M. L., and Martin, H. (2011). The Microbe-Free Plant: Fact or Artifact? Front. Plant Sci. 2:100. doi: 10.3389/FPLS.2011.00100
Pellock, S. J., Walton, W. G., Biernat, K. A., Torres-Rivera, D., Creekmore, B. C., Xu, Y., et al. (2018). Three structurally and functionally distinct β-glucuronidases from the human gut microbe Bacteroides uniformis. J. Biol. Chem. 293, 18559–18573. doi: 10.1074/jbc.RA118.005414
Reed, S. C., Townsend, A. R., Cleveland, C. C., and Nemergut, D. R. (2010). Microbial community shifts influence patterns in tropical forest nitrogen fixation. Oecologia 164, 521–531. doi: 10.1007/s00442-010-1649-6
Ryffel, F., Helfrich, E. J., Kiefer, P., Peyriga, L., Portais, J., Piel, J., et al. (2015). Metabolic footprint of epiphytic bacteria on Arabidopsis thaliana leaves. ISME J. 10, 632–643. doi: 10.1038/ismej.2015.141
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery, and explanation. Genome Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60
Shanmughavel, P., and Francis, K. (1997). Balance and turnover of nutrients in a bamboo plantation (Bambusa bambos) of different ages. Biol. Fert. Soils 25, 69–74. doi: 10.1007/s003740050282
Shi, W., and Zhang, H. (2007). Leaf microenvironment and microbial community. Microbiol. Bull. 4, 761–764. doi: 10.13344/j.microbiol.china.2007.04.015
Sichuan Forestry Bureau (2015). The Giant Panda of Sichuan: Report of the Fourth National Giant Panda Survey. Sichuan Science and Technology Press, Chengdu.
Stackebrandt, E., and Goebel, B. M. (1994). Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44, 846–849. doi: 10.1099/00207713-44-4-846
Steven, K., Timothy, O., Holly, A., Stephen, H., Joseph, W. S., and Jessica, G. (2014). Relationships between phyllosphere bacterial communities and plant functional traits in a neotropical forest. Proc. Natl. Acad. Sci. U.S.A. 111, 13715–13720. doi: 10.1073/pnas.1216057111
Thompson, I. P., Bailey, M. J., Fenlon, J. S., Fermor, T. R., Lilley, A. K., Lynch, J. M., et al. (1993). Quantitative and qualitative seasonal changes in the microbial community from the phyllosphere of sugar beet (Beta vulgaris). Plant Soil 150, 177–191. doi: 10.1007/BF00013015
Venkatachalam, S., Ranjan, K., Prasanna, R., Ramakrishnan, B., Thapa, S., and Kanchan, A. (2016). Diversity and functional traits of culturable microbiome members, including cyanobacteria in the rice phyllosphere. Plant Biol. 18, 627–637. doi: 10.1007/s11274-017-2267-9
Vorholt, J. A. (2012). Microbial life in the phyllosphere. Nat. Rev. Microbiol. 10, 828–840. doi: 10.1038/nrmicro2910
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wei, F., Feng, Z., and Wang, Z. (1999). Habitat selection of giant panda and red panda in Xiangling Mountains. Acta Zool. Sin. 1, 57–63.
Wu, Q., Wang, X., Ding, Y., Hu, Y. B., Nie, Y. G., Wei, W., et al. (2017). Seasonal variation in nutrient utilization shapes gut microbiome structureand function in wild giant pandas. Proc. Biol. Sci. 284:20170955. doi: 10.1098/rspb.2017.0955
Xei, J., and Hong, M. (2020). Effects of ecological factors on growth of Arundinaria spanostachya shoots in Liziping National Nature Reserve, China. Glob. Ecol. Conserv. 23:e01121. doi: 10.1016/J.GECCO.2020.E01121
Xiong, Y., Peng, G., Zhang, G., Li, D., and Wang, P. (1999). Detection of enterotoxin of Escherichia coli EL-1 strain from giant panda by dot enzyme immunobinding assay. Chin. Vet. Sci. Technol. 29:2.
Yadav, R. K. P., Papatheodorou, E. M., Karamanoli, K., Constantinidou, H.-I. A., and Vokou, D. (2008). Abundance and diversity of the phyllosphere bacterial communities of Mediterranean perennial plants that differ in leaf chemistry. Chemoecology 18, 217–226. doi: 10.1007/s00049-008-0408-1
Zhang, C., and Chen, Y. (1997). Pathogenicity of Klebsiella to giant panda. Chin. Vet. Sci. Technol. 27, 40–41.
Zhang, H., and Xue, J. (2018). Spatial pattern and competitive relationships of moso bamboo in a native subtropical rainforest community. Forests 9:774. doi: 10.3390/f9120774
Zhang, X., Xu, S., Li, C., Zhao, L., Feng, H., Yue, G., et al. (2014). The soil carbon/nitrogen ratio and moisture affect microbial community structures in alkaline permafrost-affected soils with different vegetation types on the Tibetan plateau. Res. Microbiol. 165, 128–139. doi: 10.1016/j.resmic.2014.01.002
Zhao, S., Zheng, P., Dong, S., Zhan, X., Wu, Q., Guo, X., et al. (2013). Whole-genome sequencing of giant pandas provides insights into demographic history and local adaptation. Nat. Genetics 45, 67–71. doi: 10.1038/ng.2494
Zheng, Y. (2011). Study on microorganism and bacteriostasis of leaf circumference of Pinus tabulaeformis. Jilin Agric. 10:65.
Keywords: giant panda, phyllosphere bacteria, influencing factors, microbial diversity, metagenome, bamboo
Citation: Long J, Luo W, Xie J, Yuan Y, Wang J, Kang L, Li Y, Zhang Z and Hong M (2021) Environmental Factors Influencing Phyllosphere Bacterial Communities in Giant Pandas’ Staple Food Bamboos. Front. Microbiol. 12:748141. doi: 10.3389/fmicb.2021.748141
Received: 27 July 2021; Accepted: 08 October 2021;
Published: 03 November 2021.
Edited by:
Lifeng Zhu, Nanjing Normal University, ChinaReviewed by:
Wei Zhu, Chengdu Institute of Biology, Chinese Academy of Sciences (CAS), ChinaZhang Shouke, Zhejiang A&F University, China
Copyright © 2021 Long, Luo, Xie, Yuan, Wang, Kang, Li, Zhang and Hong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingsheng Hong, mingshenghong@cqu.edu.cn; Zejun Zhang, zhangzejun66@163.com
†These authors have contributed equally to this work and share first authorship
 Juejie Long1,2†
Juejie Long1,2† Zejun Zhang
Zejun Zhang Mingsheng Hong
Mingsheng Hong