- 1Department of Environmental Sciences, Connecticut Agricultural Experiment Station, New Haven, CT, United States
- 2Center for Vector Biology and Zoonotic Diseases, Connecticut Agricultural Experiment Station, New Haven, CT, United States
The increasing availability of modern research tools has enabled a revolution in studies of non-model organisms. Yet, one aspect that remains difficult or impossible to control in many model and most non-model organisms is the presence and composition of the host-associated microbiota or the microbiome. In this review, we explore the development of axenic (microbe-free) mosquito models and what these systems reveal about the role of the microbiome in mosquito biology. Additionally, the axenic host is a blank template on which a microbiome of known composition can be introduced, also known as a gnotobiotic organism. Finally, we identify a “most wanted” list of common mosquito microbiome members that show the greatest potential to influence host phenotypes. We propose that these are high-value targets to be employed in future gnotobiotic studies. The use of axenic and gnotobiotic organisms will transition the microbiome into another experimental variable that can be manipulated and controlled. Through these efforts, the mosquito will be a true model for examining host microbiome interactions.
The Origins of an Axenic Mosquito Model
The term axenic refers to the growth of a single strain or species entirely free from contamination of any other organisms. The axenic state has also been referred to as “germ free.” We discourage this practice as it tends to implicate the microbes associated with a host with disease, ignoring the myriad of commensal and beneficial associations between microorganisms and their host. In its original usage, the term axenic was generally applied to cultures of bacteria or single-celled eukaryotes. As early as 1885, Louis Pasteur hypothesized the potential of an axenic animal host, although he was of the belief that the resulting axenic animal would not be viable (Pasteur, 1885). His views seemed to be borne out in 1896 when the production of axenic guinea pigs failed to survive past 13 days (Nuttall and Thierfelder, 1896). An axenic fly (Calliphora vomitoria) was first reported by Wollman in 1911, raising the potential for producing axenic insects (Wollman, 1911). With advancements in nutrition, handling facilities, and aseptic techniques, species, such as mice and fruit flies, are now routinely reared for multiple generations under axenic conditions, demonstrating that the axenic state is not a death sentence for the host organism (Sang and King, 1961; Smith et al., 2007; Douglas, 2018).
In 1930s, researchers turned their efforts to rear axenic mosquitoes, reporting the production of aseptic Aedes aegypti larvae in sterile media (Trager, 1935a,b, 1937). A series of studies would go on to refine the axenic techniques and were generally focused on determining the nutritional requirements of mosquito larvae (Lea et al., 1956; Singh and Brown, 1957; Akov, 1962; Lang et al., 1972; Rosales-Ronquillo et al., 1973). From the earliest studies, it was recognized that the microbiome was a potential source of essential nutrients. In 1935, Trager reported that sterile larval rearing media needed to be supplemented with particular nutrients, reporting that an essential ingredient in the media formulation was “heat-and alkali-stable…it seems to belong to the B group of vitamins” (Trager, 1935b). In fact, it was these studies that established a diet of liver and yeast extract for larval rearing, which is employed in many mosquito laboratories to this day. However, these studies occurred before the molecular revolution in microbiology in which we learned that the majority of microorganisms in the environment are often recalcitrant to laboratory cultivation (Hug, 2018; Lewis et al., 2021). Thus, there was some concern as to whether reports of axenic mosquitoes were reliable, as they may have been colonized by some of this biological “dark matter” (Rinke et al., 2013). Then, in 2014, a study called into question whether mosquitoes could be reared axenically at all, and proposed a potentially novel mechanism for the role of microbes in mosquito biology (Coon et al., 2014, 2017).
Microbial Induced Hypoxia and Larval Development
It was recently reported that removing the mosquito larval microbiome by surface sterilizing eggs resulted in the production of axenic larvae that would expire in the first instar stage of development. However, larval development could be rescued if the larvae were provided with a live culture of a laboratory strain of Escherichia coli (Coon et al., 2014). In a follow-up experiment, the researchers performed a transposon mutant screen to identify the genetic determinants allowing for E. coli to rescue larval development. Through this screen, they identified that axenic larvae colonized by mutants in cydB and cydD, which encode cytochrome bd oxidase, failed to develop (Coon et al., 2017). The respiratory oxidase cytochrome bd allows E. coli to grow in oxygen-replete conditions, with maximal activity at oxygen concentrations of 25–50 nM (D’mello et al., 1996). This suggested an important role for this terminal electron transporter in bacteria-host interactions. Coon et al. observed that the guts of larvae colonized by wild-type E. coli showed a cyclical behavior in oxic conditions over development, with declining oxygen levels prior to molting. When colonized by the cydB/cydD mutants, gut anoxia did not manifest, and the larvae failed to develop. Furthermore, expression of mosquito-encoded hypoxia-inducible transcription factor was associated with larval development, supporting a connection between anoxia and larval growth (Valzania et al., 2018a). Therefore, the hypoxic model proposes that E. coli cytochrome bd mutants do not scavenge O2 from the gut lumen, thereby not producing anoxic conditions, and thus, the larvae failed to develop (Coon et al., 2017). This would require that microbes are not just a passive source of nutrients to the larvae, but a living respiring microbial population was necessary for larval development. These observations would make mosquitoes unique from other insects, like Drosophila, which are routinely reared in an axenic state and would seem to support Pasteur’s original thesis that some hosts may require a living microbiome for survival.
Reemergence of Axenic Mosquito Models
In 2018, we reported the generation of axenic Ae. aegypti larvae that were capable of developing into adults. The sterility of the mosquito host was verified by both culture-dependent and culture-independent methods (Correa et al., 2018). More recently, Romoli et al. described a method to produce axenic Ae. aegypti larvae through a process of “transient colonization.” Briefly, axenic larvae are colonized by a genetically modified strain of E. coli that can be removed during larval development, referred to as decolonization. The decolonized larvae go on to produce axenic adult mosquitoes (Romoli et al., 2021).
The description of axenic mosquitos seems to contradict the microbial-driven hypoxia model described above, which would require a living functional microbiome. Yet, several lines of evidence suggest that the hypoxic model may be incorrect. First, there was no difference in oxygen concentration between colonized larvae and decolonized axenic larvae (Romoli et al., 2021). Here, we show that axenic Ae. aegypti mosquitoes raised in the complete absence of a microbiome and stained with a fluorescent dye as a hypoxia marker maintain anoxic conditions in the gut (Figure 1). This indicates that mosquitoes themselves are capable of scavenging oxygen from the gut lumen. We thusly propose an alternative explanation to the observation that E. coli cytochrome bd mutants are unable to rescue larval development. Cytochrome bd mutants of E. coli demonstrate slower growth and produce lower biomass than their wild-type counterparts (Goojani et al., 2020). This suggests that the reproduction rate of cytochrome bd mutants may not be sufficient to support larval growth, not their ability to drawdown oxygen. In this view, the defects in the electron transport chain of E. coli lead to slower growth, which is then insufficient to support the high levels of metabolism required for larval development. A similar phenomenon has been observed in Drosophila, where it was reported that the quantity of bacteria present determined fly development and longevity rather than a particular microbial species or traits. In fact, the best predictor of how well a microbe would affect Drosophila development was how well that microbe grew on fruit fly culture medium (Keebaugh et al., 2018). This may also explain why E. coli K12, a laboratory-adapted bacterium, is still able to rescue larval development (Correa et al., 2018). E. coli K12 maintains high growth levels in larval growth media and thus can act as a source of larval nutrition, despite presumably losing many characteristics that would normally facilitate host colonization (Liu and Reeves, 1994; Browning et al., 2013).
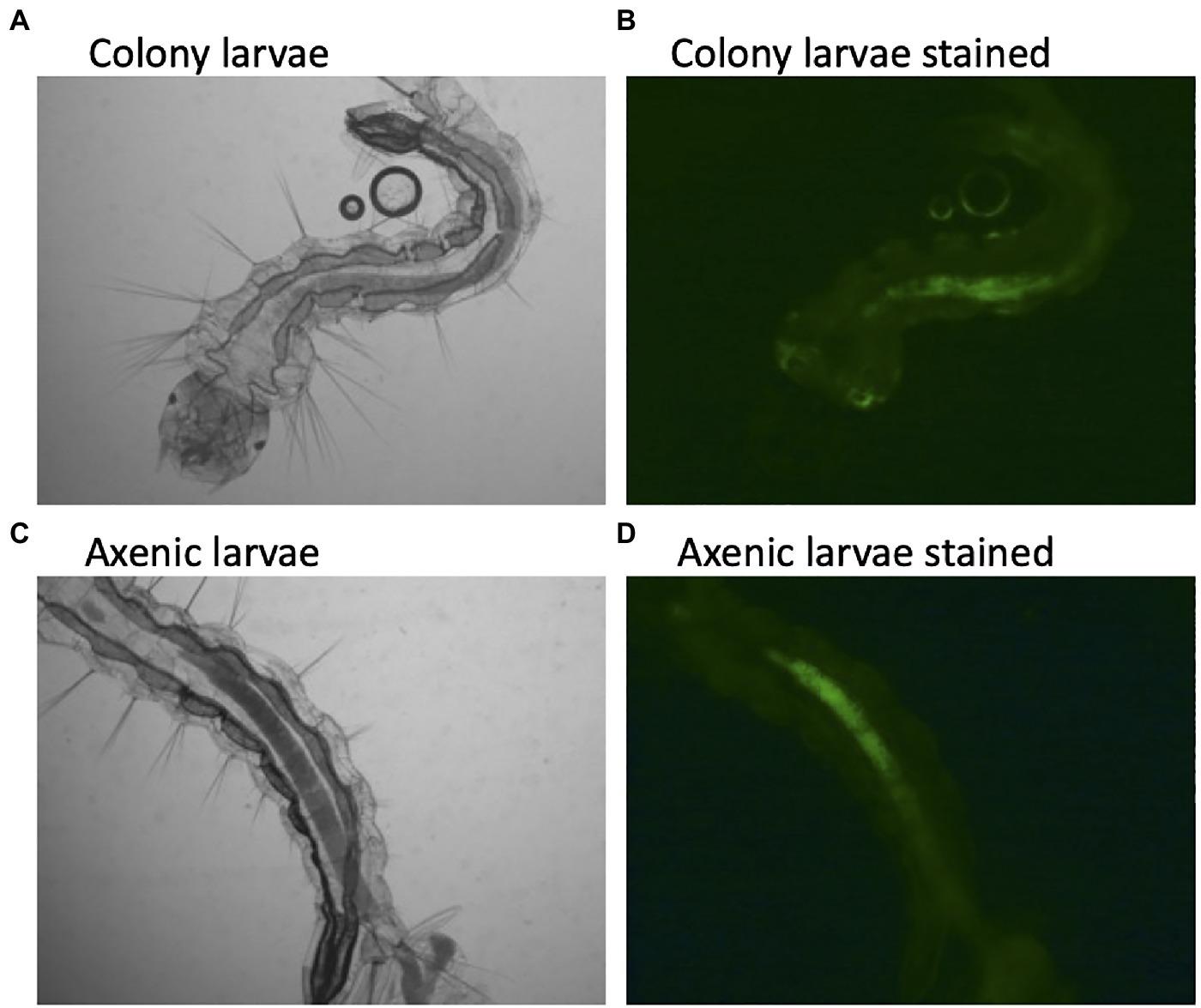
Figure 1. Hypoxic conditions in the guts of conventionally reared and axenic Aedes aegypti larvae. Larvae were reared to the third instar of development and stained with Image-iT Hypoxia Reagent, which fluoresces in anoxic conditions (<5% oxygen). Axenic larvae were reared in the dark to protect light sensitive B-vitamins in the culture medium. A subset of axenic larvae were subsequently verified to be free of microbial contamination by both culture-dependent and culture-independent (16S rRNA and fungal rRNA gene PCR) methods.
Data from the newly developed axenic models also support a nutritional role of the microbiome. For instance, larvae reared on a high-density diet of liver and yeast extract have to be reared in the dark to protect a light-sensitive component in the food, presumably a B vitamin (Hyde et al., 2019a). Transcriptional data from decolonized axenic larvae showed that folate biosynthesis, folate transport, and thiamine metabolism were all upregulated in comparison with their microbially colonized cohorts, additionally pointing to a B vitamin deficiency in axenic larvae (Romoli et al., 2021). Furthermore, supplementing the diet of axenic larvae with folate increased larval survival 4-fold (Romoli et al., 2021). Finally, using data from the axenic and decolonized mosquito models, Wang et al. verified and replicated the observation that supplementing the diet of axenic larvae with B vitamins, in this case riboflavin, can rescue development. They further showed that axenic larvae undergo gut hypoxia when reared under appropriate conditions, verifying that microorganisms are not required to produce anoxic conditions (Wang et al., 2021a). Thus, axenic studies of mosquito larvae have come full circle, highlighting the role of the microbiome in supplying essential nutrients, B-vitamins, such as folate and riboflavin, as the key function of the microbiome in mosquito larval development.
Are Microbes Essential for Larval Development?
The development of axenic larvae seems to suggest that living microorganisms are dispensable to larval growth. Yet, the microbes that make up the microbiome are clearly providing high amounts of biomass, food, and essential vitamins. It is unlikely any of these nutrients would be present in the environment in sufficient quantities to support larval growth in the absence of living microbes. In this regard, a living microbiome is likely required to support mosquito development in the wild. The fact that axenic larvae took longer to develop and produced smaller adults indicates that the axenic diet has not yet been optimized (Correa et al., 2018). In addition, several nutrients, such as amino acids and vitamin mixes, were lethal to the larvae when provided in high concentrations in the rearing water, which was only overcome when nutrients were provided in a semi-solid agar plug (Correa et al., 2018). This provides information on how mosquito larvae feed and acquire nutrients in the environment. Larval nutrition has long been recognized as a potential target for mosquito control and influences the development and fitness of adult mosquitoes (Souza et al., 2019; Dittmer and Gabrieli, 2020). Thus, the axenic model will continue to be important in defining and characterizing the nutritional requirements of mosquito larvae and the potential for identifying diet or microbial-based larval control programs.
The Role of the Microbiome in Adult Mosquitoes
The microbiome of mosquito larvae is thought to be largely acquired from their aquatic environment (Coon et al., 2016b; Saab et al., 2020). As mosquitoes are holometabolous, going through a complete metamorphosis from larvae to adults through a “resting” pupal phase (Figure 2), larval nutrition and health have direct consequences on the fitness of the adult (Moller-Jacobs et al., 2014; Linenberg et al., 2016). Presumably, this means the status of the larval microbiome will also affect traits of the adult mosquito. Evidence suggests that many microbes are lost during metamorphosis when mosquitoes develop from larvae to pupae and emerge as adults (Lindh et al., 2008; Chavshin et al., 2015). The adult mosquito is exposed to new food sources, such as flower nectar, as well as being more mobile and able to visit other locations, and the anautogenous female mosquito requires a blood meal for reproduction (Figure 2). Thus, the microbiome of adult mosquitoes differs from larvae, in both composition and diversity (Muturi et al., 2017; Wang et al., 2018; Hyde et al., 2019b). Yet, no serious detrimental effects of the axenic state on adult Ae. aegypti mosquitoes have been documented. Axenic mosquitoes had similar, if not slightly longer lifespans than their bacterially colonized cohorts, and female mosquitoes took a blood meal, laid a similar number of eggs, and the eggs of the axenic Ae. aegypti females gave rise to viable offspring (Correa et al., 2018). Similarly, there were no detrimental effects of the axenic state on longevity of fecundity of decolonized mosquitoes (Romoli et al., 2021). Additionally, transcriptomic analysis of axenic adult mosquitoes showed a muted response in terms of gene expression change between axenic and conventionally reared mosquitoes, indicating similar physiological states for colonized and axenic mosquitoes (Hyde et al., 2020). Thus, it appears that the axenic state is not an obvious burden for the adult mosquito, at least in the case of Ae. aegypti.
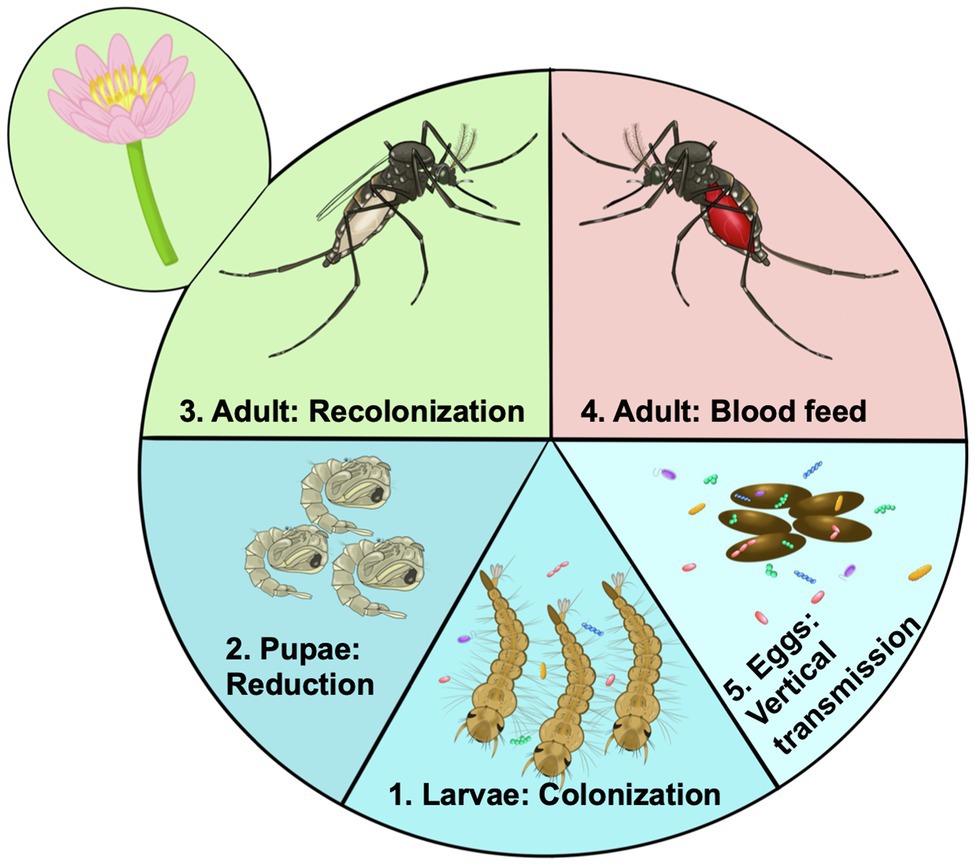
Figure 2. Acquisition and transmission of the microbiome over the life cycle of the mosquito. 1. Microorganisms from the aquatic environment colonize the larvae. These organisms provide essential nutrients such as B vitamins. 2. During pupation there is a large reduction for or even elimination of the microbiome. 3. After emergence mosquitoes are exposed to rearing water potentially being recolonized by aquatic bacteria, but are also exposed to new sources of colonizing bacteria such as flower nectar. 4. Certain members of the microbiome may facilitate blood digestion by female mosquitoes becoming more abundant in the gut. Other bacteria may be adapted to survive the high nutrients and reactive oxygen species. 5. Through colonizing the female ovaries microbes may be deposited on the egg surface, ensuring they are available to colonize newly hatched larvae, there by continuing the colonization cycle.
Mosquitoes pose a significant and continued public health threat worldwide as they are responsible for transmitting numerous pathogenic viruses and parasites, such as dengue virus, West Nile virus, malaria parasites, and filarial nematodes, resulting in millions of infections each year. As such, there has been considerable research interest into the interactions between the mosquito microbiota and the pathogens they carry. Different microbial taxa have shown both positive and negative associations with the vectorial capacity of mosquitoes (Cirimotich et al., 2011; Guégan et al., 2018; Romoli and Gendrin, 2018; Caragata et al., 2019). Additionally, the composition and membership of the microbiome have been associated with a long list of other mosquito phenotypes. These include insecticide resistance, lifespan, and fecundity (Minard et al., 2013a; Dada et al., 2018; Wang et al., 2021b). Yet, all these studies have one thing in common, and they assume an interaction between microbiome members and host phenotypes. In this regard, these associations are not addressable with an axenic model, which can only investigate phenotypes in the presence or absence of a microbiome. What is required to link the status of the microbiome to host phenotypes is a model in which the diversity and composition of the microbiome can be mechanistically controlled and altered.
A Defined Microbiome: Gnotobiotic Mosquitoes
The ability to perform the microbiome presence/absence studies is important, but they are binary in nature and, therefore, somewhat limited. The true power of axenic models is their ability to be manipulated so that defined microbiomes, whether it be an individual or multiple community members, can be introduced, thereby allowing the systematic examination of the effects that specific community members have on host biology. These systems, referred to as gnotobiotics, allow microbiome research to move beyond correlational studies to hypothesis-driven examinations of causation.
A gnotobiotic host is one in which every living organism in association with the host is defined. Consequently, gnotobiotic systems are limited by the availability of both an axenic host and microbiome community members that can be cultured. Despite these limitations, our ability to decipher the complexity of host-microbiome interactions has accelerated in recent years through advancements in genomics, proteomics, metabolomics, and culturing techniques. Initial studies in axenic flies revealed that developmental delays could be avoided through the introduction of a single community member and that certain community members were more effective at counteracting these delays (Bakula, 1969; Storelli et al., 2011). Functional characterization of these observations through a transposon mutagenesis screen and metagenome-wide association analysis determined that Drosophila larval development and adult fly metabolic homeostasis were significantly affected by the bacterial community members’ ability to synthesize pyrroloquinoline quinone, which is an important modulator of the flies insulin/insulin-like growth factor signaling (Shin et al., 2011; Chaston et al., 2014).
Monoculture gnotobiotics are a very useful tool for elucidating the contributions that the microbiota has on host development and physiology but are reductionist and cannot measure the influence of intra-community interactions on the system at large. Early on it was demonstrated that the microbiomes influence on host biology can be greater than the sum of its parts (Schaedler et al., 1965). Axenic flies colonized with communities of varying complexity reveal that some phenotypic traits are highly influenced by community interactions (Newell and Douglas, 2014; Gould et al., 2018). For instance, axenic Drosophila showed prolonged development times and elevated triglyceride contents in comparison with conventionally reared bacterial colonized flies. When the flies were recolonized with individual members cultivated from the microbiome, there were strain specific responses of the flies in triglyceride levels and longevity, but it was only when strains of both Acetobacter and Lactobacillus (the dominant members of the Drosophila microbiome) were presented to the axenic flies that the flies phenotypes returned to wildtype levels (Newell and Douglas, 2014).
Another approach for examining community scale effects and interactions is through transfer studies. While not truly gnotobiotic, transfer studies in which the microbiome of an individual with a specific disease state is introduced into an axenic host have become a useful tool for examining microbiome correlates of disease (Vrieze et al., 2012; Fei and Zhao, 2013; Schulz et al., 2014). For instance, the now famous study documents that transferring the microbiome from lean and obese mice results in increased capacity for energy harvest and weight gain for the mice receiving the “obese microbiome” (Turnbaugh et al., 2006). These systems have the benefit of more realistically capturing the complexity of interactions associated with the microbiome but suffer from an incomplete understanding of the members within the community and their individual contributions to host phenotypes.
Until recently, mosquito-microbiome studies have been limited due to the lack of an axenic model. Consequently, most studies have relied upon the use of antibiotics to clear the resident bacteria in order to interrogate microbiome-mosquito interactions. Using this approach, several groups have reported that the microbiome plays a role in modulating gut immunity thereby effecting susceptibility to viral and parasitic pathogens (Xi et al., 2008; Dong et al., 2009; Kalappa et al., 2018; Wu et al., 2019). Similarly, antibiotic clearance of the microbiota has demonstrated a role for the microbiome in mosquito metabolism and sensitivity to insecticides (Xiao et al., 2017; Barnard et al., 2019; Chabanol et al., 2020). These studies, like the presence/absence studies used in axenic models, offer generalizable insights into the effects of the microbiota on host phenotypes. While these studies support that the presence of a microbiome is linked to mosquito phenotypes, it is difficult to parse out microbiome impacts from those potentially associated with sustained antibiotic exposure. It has been demonstrated in mammalian systems that antibiotics can induce immunologic and metabolic changes in the host, inhibit eukaryotic translation, and alter mitochondrial function (Kalghatgi et al., 2013; Badal et al., 2015; Moullan et al., 2015; Yang et al., 2017; Gopinath et al., 2018). In addition to potential side effects, it has been shown that antibiotics do not eliminate resident microbiota, but rather cause a dysbiosis, as some members of the mosquito microbiome likely harbor antibiotic resistance (Hughes et al., 2014; Schubert et al., 2015; Hyde et al., 2019b). This may also explain contrasting results between studies that employ antibiotic clearance, as the net result may be an altered microbial composition rather than a comparison between the presence and absence of a microbiome. For instance, Xi et al. (2008) reported a significant reduction in dengue virus infection after antibiotic treatment of Ae. aegypti mosquitoes, whereas Audsley et al. reported no effect of antibiotic treatment on the permissiveness of Ae. aegypti to infection by dengue virus (Audsley et al., 2017). In addition, several studies have employed antibiotic clearance in attempts to recapitulate monoculture gnotobiotics in mosquitoes, by “clearing” the resident microbiota with antibiotics and then exposing the mosquitoes to a bacterium of interest. These studies demonstrate that specific bacterial community members may affect mosquito susceptibility to pathogens and host physiology (Dong et al., 2009; Apte-Deshpande et al., 2012; Ramirez et al., 2012; Hughes et al., 2014; Xiao et al., 2017; Wu et al., 2019). However, there is no way to separate the possible influence of antibiotics on these traits, or little to no verification that the mosquitoes are true gnotobiotics.
Recent efforts have been made to examine mosquito-microbiome interactions using axenic mosquitoes. Introduction of individual bacterial isolates at the larval stage revealed strain-specific effects on larval survivorship and development time as well as adult mosquito biometrics, such as body size and reproductive fitness as well as susceptibility to dengue virus and Zika virus (Coon et al., 2016a; Dickson et al., 2017; Correa et al., 2018; Carlson et al., 2020; Giraud et al., 2021). It has also been demonstrated that simplified communities can successfully colonize both axenic larvae and adult mosquitoes (Correa et al., 2018). Correa et al. found that the composition and function of the microbiome may be important determinants of phenotypic plasticity observed between individual mosquitoes. The generation of axenic/gnotobiotic mosquito models now make it possible to systematically interrogate the effects of the microbiome on mosquito biology without the use of antibiotics.
Opportunities and Challenges for Axenic and Gnotobiotic Mosquitoes
The advent of the axenic/gnotobiotic mosquitoes opens up a wide range of questions that can be addressed by the research community. The method for generating and rearing axenic mosquitoes has been published and is achievable by any laboratory with the ability to maintain aseptic conditions, sterilize the required equipment, and perform basic microbiology (see Hyde et al., 2019a for a detailed protocol). To date, axenic rearing from larvae to adults has only been reported for Ae. aegypti, but gnotobiotic Aedes atropalpus, Aedes albopictus, Anopheles gambiae, Culex quinquefasciatus, and Toxorhynchites amboinensis have all been reported (Coon et al., 2016a,b, 2020; Valzania et al., 2018b). In this respect, gnotobiotic models may be achievable for a wider range of host species. Implementing these gnotobiotic studies will require an appropriate mosquito host and bacterial strains capable of supporting larval development.
Perhaps, one of the greatest opportunities for gnotobiotics is the potential for standardizing mosquito studies. It stands to reason that different research laboratories employing varied diets, rearing conditions, and being geographically separated are likely to harbor differing microbiome compositions in the mosquitoes they rear. Thus, at least a portion of variability reported between studies is likely due to heterogeneity in microbiome composition and structure between laboratories. Gnotobiotic models allow for standardization of the microbiome and transitioning the microbiome to a controlled variable. The creation of a defined tractable model microbiome, similar to the altered Schaedler flora employed in gnotobiotic mouse studies (Biggs et al., 2017), would be a resource that could be shared among researchers, and act as a baseline to investigate the consequences of microbiome manipulation on mosquito phenotypes. This will require identifying those bacteria that would serve the greatest utility for a defined microbiome.
A Path Forward: A Most Wanted List for Microbes in Gnotobiotic Studies
In general, the diversity of the microbiome in an individual mosquito is rather low, being comprised of ~10–50 bacterial species (Minard et al., 2013a). Yet, there are certain microbial members that appear to be commonly associated with mosquitoes or that have been correlated to particular phenotypic outcomes, which makes them particularly attractive targets to investigate in a gnotobiotic model. Below, we list five microbiome members that are high-value targets for future gnotobiotic studies (Figure 3). This is by no means an extensive list of microbiome members that may play a role in mosquito physiology but is a review of some of the mosquito microbiome members that show the greatest promise for untangling host microbe interactions or potential microbes that could be employed in microbial-based mosquito borne disease control. Notably, not included on the list is the bacterial endosymbiont Wolbachia. This maternally inherited organism may be the most well-studied mosquito-associated bacterium, with over 100 years of active study (Kaur et al., 2021). However, the difficulty in growth and maintenance of Wolbachia in pure culture, its biology, host range, and inheritance makes it a difficult organism to employ in gnotobiotic studies (Voronin et al., 2010; Hughes et al., 2012). Despite the myriad of effects Wolbachia plays in mosquito biology, reproduction, and control of mosquito carried pathogens (Turley et al., 2009; Hancock et al., 2011; Iturbe-Ormaetxe et al., 2011; Jiggins, 2017), it does not make the list.
Genus Asaia
Bacteria in the genera Asaia (phylum Proteobacteria) are acetic acid bacteria within the family Acetobacteraceae. Acetic acid bacteria are differentiated from other bacteria as they are obligate aerobes that oxidize sugars, sugar alcohols, and ethanol with the production of acetic acid as the major end product (Raspor and Goranovič, 2008). Bacteria of the genus Asaia colonize multiple insects, across multiple orders, such as the Diptera, Hymenoptera, Hemiptera, and Homoptera (Crotti et al., 2010). This includes mosquitoes within Aedes sp., Anopheles sp., and Culex sp. (Crotti et al., 2009; Freece et al., 2014; Ramos-Nino et al., 2020). Asaia is among the numerically dominant bacterial populations that colonize mosquitoes as assessed by sequencing surveys (Chouaia et al., 2010; Damiani et al., 2010) and can be found in larval and adult mosquitoes and multiple tissues (e.g., the adult gut, testes, ovaries, and salivary glands; Favia 2007, 2008; Mancini, 2018). Geographically dispersed, Asaia has been identified in mosquitoes from Brazil (Oliveira et al., 2020), Canada (Novakova et al., 2017), Iran (Rami et al., 2018), Italy (Alfano et al., 2019), Kenya (Osei-Poku et al., 2012), Madagascar (Minard et al., 2013b), and the United States (Muturi et al., 2017), among others. Species of Asaia are often found in flower nectar, the food source of newly emerged mosquitoes (Bassene et al., 2020). They can also spread through mosquito populations by paternal transmission during mating (Damiani et al., 2008). Asaia may use their colonization of the female reproductive tract to ensure vertical transmission through a process of egg smearing, whereby they colonize the egg surface in order to be ingested by newly hatched larvae (Damiani et al., 2010). The importance of the bacteria to the larvae is demonstrated by the fact that reduction of Asaia bacterial load by treatment with antibiotics slows larval growth. Subsequently, supplementing larval diets with Asaia bacteria accelerated larval development time (Mitraka et al., 2013). This suggests that these bacteria are able to meet the nutritional needs of developing larvae (Chouaia et al., 2012). Evolutionary analysis of Asaia genomes indicates that these organisms have underwent a process of genome reduction as they became associated with insects, yet preserved an insecticide degrading gene, pyrethroid hydrolase (Comandatore et al., 2021). Thus, beyond providing a source of nutrition to developing larvae, Asaia may play a role in protecting mosquitoes from insecticides.
Asaia bacteria appear to interact with the endosymbiotic bacteria and parasites carried by mosquitoes. When the mosquito microbiome was supplemented with Asaia bacteria, there was an observed impediment to the vertical transmission of Wolbachia, a mosquito endosymbiont, suggesting inter-species competition (Hughes et al., 2014). Additionally, several studies have documented a negative relationship between Asaia bacteria and Plasmodium, the causative agent of malaria (Capone et al., 2013; Cappelli et al., 2019a). It is thought that Asaia bacteria prime an immune response that prevents the malarial parasite from developing in the mosquito (Cappelli et al., 2019a). In this regard, the Asaia-mosquito symbiosis may be a relationship with significant public health implications.
These data and observations show that Asaia sp. colonize multiple mosquito hosts as well as other sugar-feeding insects. Asaia sp. are present across multiple life stages, in multiple tissues, and are reliably inherited between mosquito generations. Asaia bacteria are also amenable to genetic modification and mosquito recolonization, which makes them an attractive tool for studying the genetic determinants of mosquito colonization (Favia et al., 2007). Additionally, because bacteria in the genus Asaia are generally non-pathogenic they are attractive targets for paratransgenic strategies for mosquito vector control. Thus, Asaia is on the list of most wanted as they represent an ideal model bacterium to study an apparent beneficial relationship between a bacterium and the mosquito host.
Serratia marcescens
Serratia (phylum Proteobacteria) is a genus of facultatively anaerobic bacteria within the family Yersiniaceae. Bacteria in the genus Serratia are common among the bacteria that make up the mosquito microbiome (Sharma et al., 2020). Several different species of Serratia have been identified in mosquitoes, including Serratia odorifera (Apte-Deshpande et al., 2012), Serratia nematodiphila (Patil et al., 2012), and Serratia fonticola (Chen and Walker, 2020, 14). Yet, one bacterium in particular has received a considerable research focus as a mosquito associate, Serratia marcescens.
Serratia marcescens is a cosmopolitan bacterium with multiple environmental reservoirs, including soil and water (Abreo and Altier, 2019). It is often found in hospital settings and is a significant cause of nosocomial infections (Mahlen, 2011; Khanna et al., 2013). Mosquitoes are also an environmental reservoir. Strains of S. marcescens have been found to colonize mosquito larvae, the adult midgut, female ovaries, and male accessory glands, and on the surface of newly laid eggs (Tchioffo et al., 2016; Wang et al., 2017). Various isolates of Serratia demonstrate larvicidal activity. A strain of S. nematodiphila demonstrated high mortality to several mosquitoes species: C. quinquefasciatus (100%), Anopheles stephensi (95%), and Ae. aegypti (91%) after 48 h of exposure (Patil et al., 2012). Similarly, gnotobiotic Ae. aegypti mosquito larvae colonized with a strain of S. marcescens experienced >85% mortality, and the surviving larvae took approximately twice as long to develop (Correa et al., 2018). The characteristic red coloration of S. marcescens may play a role in its antagonism to larval development. Prodigiosin, the red pigment produced by S. marcescens shows larvicidal activity when introduced to larvae (Patil et al., 2011; Suryawanshi et al., 2015).
In contrast, S. marcescens seems to play a beneficial or at least neutral role in the adult mosquito. Gnotobiotic adult Ae. aegypti mosquitoes colonized by S. marcescens showed no increase in mortality (Correa et al., 2018). Similarly, no fitness defects were noted for adult An. gambiae or Culex pipiens mosquitoes colonized by S. marcescens (Koosha et al., 2019; Ezemuoka et al., 2020). In fact, S. marcescens may participate in mosquito blood digestion. The genome of S. marcescens encodes several genes for heme uptake and storage, as well as demonstrating alpha-hemolytic activity, i.e., the complete lysis of blood cells (Chen et al., 2017). Yet, fewer females infected with S. marcescens took blood meals in comparison with their uninfected cohorts (Kozlova et al., 2021).
Serratia marcescens has also been shown to interact with the capacity of mosquitoes to transmit disease. For instance by inhibiting Plasmodium development in the mosquito, thereby reducing the spread of malaria (Seitz et al., 1987; Bahia et al., 2014; Bai et al., 2019). Yet, there is high intra-specific diversity between S. marcescens strains capable of inhibiting Plasmodium. This indicates the anti-parasitic ability is likely due to a small number of genetic determinants, such as flagellum biosynthesis, that may not be conserved among all members of the species (Bando et al., 2013). Genome sequencing of mosquito-associated strains of S. marcescens showed various virulence factors and antibiotic production which may be involved in controlling pathogen infection of the mosquito (Chen et al., 2017). A recently described class of natural products, the stephensiolides, was isolated from a mosquito-associated Serratia. It is posited that these compounds have antimicrobial properties, which may also antagonize Plasmodium, as well as facilitate motility and transfer of bacterial cells within and between mosquitoes (Ganley et al., 2018). Additionally, colonization of the mosquito with S. marcescens induces gene expression changes in pathways, such as peptidoglycan recognition receptors that may enhance anti-parasitic immune responses that act to inhibit Plasmodium (Stathopoulos et al., 2014). In contrast, mosquito colonization by S. marcescens has been reported to increase the infection rate of mosquitoes by particular arboviruses. Specifically, S. marcescens colonization has been linked to increases in the infection of mosquitoes with dengue-2 and chikungunya virus (Apte-Deshpande et al., 2012). This increased permissiveness to viral infection is thought to occur through bacterial excretion of a protein SmEnhancin, which interferes with mucins of the mosquito gut epithelia, allowing dissemination of viral particles (Wu et al., 2019). In this regard, S. marcescens appears to inhibit or facilitate the vector competence of mosquitoes depending on the pathogen under consideration.
Serratia marcescens is on the target of most wanted mosquito which associates to study in gnotobiotic systems due to its complex interactions with the mosquito host. Colonization outcomes vary widely between larvae and adults. The potential relationships and mechanisms driving interactions between S. marcescens and the pathogens vectored by mosquitoes vary from the general, such as a priming of the immune system to the specific production of metabolites that aid or hinder pathogen infection or spread. In this respect, there is a rich list of potential host microbe interactions and phenotypic outcomes to characterize in regard to the association between S. marcescens and mosquitoes.
Elizabethkingia anophelis
Elizabethkingia (phylum Bacteroidetes) is a genus within the family Weeksellaceae. This bacterium is unique on this list as its origins are from mosquitoes, being first isolated from the midguts of An. gambiae (Kämpfer et al., 2011). However, the genus Elizabethkingia is considered to be ubiquitous in the environment. Although genomic-based analyses suggest that the mosquito-associated strains form a distinct evolutionary sublineage within the Elizabethkingia anophelis species complex (Breurec et al., 2016).
Elizabethkingia anophelis is an opportunistic human pathogen, causing pneumonia, bacteremia, neonatal meningitis, nosocomial bacteremia, and neutropenic fever (Lau et al., 2016). Cases of bacteremia are often associated with poor clinical outcomes, with mortality rates as high as 23.5% (Lau et al., 2016). Cases of E. anophelis infections have been reported from Singapore, Hong Kong, Taiwan, and the United States (Janda and Lopez, 2017). Because of the association between E. anophelis and mosquitoes, it was initially posited that mosquitoes may be a vector (Frank et al., 2013). Subsequent studies suggest that E. anophelis cases are far more likely to be hospital acquired infections, although sporadic, community-acquired cases have been reported (Hayek et al., 2013; Perrin et al., 2017; Lee et al., 2021). One characteristic that makes E. anophelis infections particularly challenging in the clinical environment is that the bacterium is resistant to multiple antibiotics including cephalosporins, aminoglycosides, and carbapenems (González and Vila, 2012; Breurec et al., 2016). A notable 112 predicted proteins identified in the genome of mosquito-associated strains of E. anophelis were annotated to features involved in resistance to antibiotics or other toxic compounds (Kukutla et al., 2013). Indeed, in recent surveys of antibiotic-resistant bacteria in the mosquito microbiome, isolates of Elizabethkingia were identified to possess multi-drug resistance against ampicillin, carbenicillin, gentamycin, tetracycline, and kanamycin (Hyde et al., 2019b; Ganley, 2020). It is not clear what, if any, role this multi-drug resistance may play in colonizing the mosquito host, other than a potential fitness advantage against other bacteria making up the mosquito microbiome.
Elizabethkingia anophelis shows differential preferences for mosquito hosts. The bacterium showed high colonization rates for An. gambiae and A. stephensi but was rarely detected in Aedes triseriatus (Chen et al., 2015). Like other bacteria on this list, Elizabethkingia sp. can be found in high numbers in the mosquito ovaries and may be vertically transmitted from mother to offspring (Akhouayri et al., 2013). Yet, colonization by Elizabethkingia sp. may induce melanotic lesions in the fat bodies of mosquito larvae and adults, suggesting a potential antagonistic relationship (Akhouayri et al., 2013). E. anophelis may be particularly important in mosquito blood digestion. Cell counts of E. anophelis increased approximately 3-fold in the guts of post-blood fed mosquitoes, and animal erythrocytes promoted E. anophelis growth in cell culture (Chen et al., 2015, 2020). The bacterium also displays hemolytic activity and encodes several hemolysins that may participate in the digestion of erythrocytes in the mosquito gut, along with antioxidant genes, which could provide defense against the oxidative stress that is associated with blood digestion (Kukutla et al., 2014). Mosquitoes colonized with E. anophelis produced more eggs than did those treated with erythromycin or with a standard microbiome, suggesting E. anophelis may increase mosquito fecundity, potentially by increasing available nutrients from the blood meal (Chen et al., 2020).
Elizabethkingia anophelis makes this list as it is a true mosquito associate that may shed light on specific genomic adaptations for colonizing the mosquito host. Furthermore, with the high levels of antibiotic resistance, E. anophelis is a model to study competition among microbiome members. Finally, E. anophelis appears to play a significant role in blood digestion a key point in the mosquito lifecycle.
Genus Chromobacterium
The genus Chromobacterium (phylum Proteobacteria) is facultatively anaerobic bacteria withing the family Neisseriaceae. Like the other bacteria on this list, species of Chromobacterium are abundant in the environment and can be readily isolated from soils and water sources (Batista and da Silva Neto, 2017). Much of the research interest in Chromobacterium has been driven by the biotechnological and pharmaceutical importance of secondary metabolites produced by the strains, which include antibiotics, quorum sensing molecules, lipopolysaccharides, and the pigment violacein that gives Chromobacterium violaceum its characteristic purple coloration (McClean et al., 1997; Durán and Menck, 2008; Kothari et al., 2017).
Chromobacterium strains are not particularly abundant in the mosquito microbiome, although they are among bacterial members commonly identified in several species of mosquito (Minard et al., 2013a). Instead, the research focuses on the Chromobacterium predominately derives for their role in control of mosquito populations and their interactions with mosquito vector competence. Various strains have shown detrimental effects of mosquito survival, lifespan, blood feeding, and fecundity (Gnambani et al., 2020). Colonization of Ae. aegypti or An. gambiae with a strain of Chromobacterium isolated from mosquito midguts resulted in rapid mortality of both larvae and adults (Ramirez et al., 2014). Larvae exposed to sublethal doses of the bacterium had lengthened developed time, suggesting chronic effects of even small populations of the bacteria (Short et al., 2018a). In the adult mosquito, C. violaceum exposure decreased the proportion of females seeking a blood meal, significantly reduced the numbers of eggs laid, and reduced hatches from the resulting eggs (Gnambani et al., 2020). A preparation of a strain of Chromobacterium with no living cells maintained strong lethal effects in mosquitoes, indicating this bacterium was producing one or several bioactive compounds (Caragata et al., 2020). Transcriptional analysis of mosquitoes with chromobacterium exposure revealed gene expression changes in pathways related to detoxification, xenobiotic response, and stress response, similar to that of an insecticide exposure (Short et al., 2018b). The same strain was also observed to produce hydrogen cyanide in larval water at sufficient concentrations to induce larval mortality, offering another possible mechanism for larvicidal activity (Short et al., 2018a). The effects of chromobacteria exposure can be transgenerational with the offspring of exposed females showing developmental delays and increased mortality (Short et al., 2018b). The genome of Chromobacterium vaccinii, another bacterium with potential roles in mosquito biocontrol, encodes several genes for virulence factors that may explain their toxicity, and these include siderophores, production of hydrogen cyanide, as well as multiple chitinase genes (Vöing et al., 2020).
Chromobacterium sp. are among the microbes that show inhibitory activity against the pathogens carried by mosquitoes. Chromobacteria cell extracts and cultures show anti-pathogen activity outside of the mosquito host, indicating the potential production of secreted metabolites inhibiting pathogen growth (Ramirez et al., 2014). For example, violacein, the violet pigment compound produced by many species of Chromobacteria, is potent antimicrobial with antiparasitic activities against Plasmodium (Lopes et al., 2009). A specific compound produced by Chromobacterium sp. Panama, romidepsin a histone deacetylase inhibitor, also showed high activity against Plasmodium (Saraiva et al., 2018b). Additionally, the Chromobacterium sp. Panama produces a protease that attacks the envelope protein of dengue virus, thereby blocking its ability to bind to and infect cells (Saraiva et al., 2018a). Thus, these observations point to the wealth of chemical compounds produced by species of Chromobacterium and their potential to influence the biology, behavior, and vector competence of their mosquito hosts.
The genus Chromobacterium makes the list based on the assortment of potentially bioactive compounds that are produced by these bacteria. In this regard, these bacteria offer a unique insight into the chemical ecology of the mosquito microbiome.
Microbial Eukaryotes
The microbiome of mosquitoes consists of more than just bacteria. Yet, there is a significant knowledge gap concerning the single-celled eukaryotes that inhabit the mosquito microbiome.
Fungal diseases are common in insects, including mosquitoes. These entomopathogenic fungi have been extensively described elsewhere (e.g., Scholte et al., 2004; Kanzok and Jacobs-Lorena, 2006; Shen et al., 2020). However, it is not so clear that these organisms can be considered part of the normal microflora of the mosquito microbiome. Far less is known of the commensal fungi that are common residents of the microbiome and are the next set of organisms on the most wanted list. A survey of culturable fungal isolates among laboratory reared and field caught mosquitoes found fungal isolates in the class Microbotryomycetes to be common among field caught mosquitoes, but absent in the microbiome of laboratory-reared mosquitoes (Hyde et al., 2019b). This suggests fungi may play an important role in the microbiome, but normal colony conditions may not be favorable to commensal fungi (Hyde et al., 2019b). As to the role fungi may play in the environment, certain fungi may exert their effect on the microbiome by attracting gravid females to a breeding site. Particularly, yeasts produce CO2 and other volatile compounds through fermentation, which can signal to the mosquito, a suitable habitat with sufficient sugar and microbial resources to support larval development (Malassigné et al., 2020). In this manner, these fungi can ensure colonization of the newly hatched larvae (Reeves, 2004). Gnotobiotic larvae colonized by Saccharomyces cerevisiae develop normally, indicating yeast can supply all the required nutrients for larval development (Correa et al., 2018). Certain fungi, such as Cladosporium, Aspergillus, Ampullimonas, and Cyberlindnera, actively participate in digesting the fructose that mosquitoes ingest from flower nectar (Guégan et al., 2020). Thus, fungi appear to play roles in both larval and adult nutrition. Other yeasts, such as Wickerhamomyces anomalus, colonize the reproductive organs of both male and female mosquitoes, indicating the potential for vertical transmission between generations, suggesting a stable multi-generational association (Ricci et al., 2011). Several fungi have also shown potential in inhibiting the pathogens carried by mosquitoes. For instance, mosquitoes colonized by microsporidian fungi demonstrated a significant decline in Plasmodium development in the mosquito, potentially through priming the mosquito immune response to the malarial parasite (Bargielowski and Koella, 2009; Herren et al., 2020). Similarly, protein toxins produced by the yeast Wickerhamomyces anomalus inhibit Plasmodium and other entomopathogenic fungi (Cappelli et al., 2019b). In contrast, fungi of the species Talaromyces may promote mosquito infection by dengue virus through suppression of the digestive enzyme trypsin (Angleró-Rodríguez et al., 2017). In this regard, fungi clearly have the potential to play a multitude of roles in the mosquito microbiome, yet remain an enigma.
Beyond the protist parasite Plasmodium, there is very little knowledge concerning whether mosquitoes harbor a stable population of protists in their microbiome (Guégan et al., 2018). Belda et al. employed a method based on peptide-nucleic acid clamps to suppress amplification of host DNA and specifically interrogate the eukaryotic members of the mosquito microbiome (Belda et al., 2017; Taerum et al., 2020). The eukaryotic microbiome of larval samples was dominated by the Ichthyosporea group, a lineage of unicellular organisms that includes parasites and commensals of a wide range of animals (Glockling et al., 2013). Similarly, metabarcoding and sequencing of the 18S rRNA genes from mosquitoes in Thailand identified Ascogregarina as the dominant microbial eukaryote in the mosquito microbiome (Thongsripong et al., 2018). This protist has been shown to have a range of fitness consequences on host mosquitoes ranging from detrimental to neutral (Erthal et al., 2012). Yet, the organism displays the hallmarks of a parasitic infection as oocysts are ingested from the larval water, enter epithelial cells, and use host cell mitochondria to supply the energy required to mature (Chen and Wu, 1997). Trypanosoma brucei, a protist parasite and causative agent of trypanosomiases, normally carried by tsetse flies, can survive in mosquito midguts for up to 48 h. Co-infection of mosquitoes with Trypanosoma and Plasmodium increased the malarial parasite load in the mosquito, potentially increasing the risk of malarial spread (Dieme et al., 2020). Thus, there is evidence that the microbiome of the mosquito may host a population of protists, but their roles and interactions with the host are essentially undescribed.
The microbial eukaryotes carried by mosquitoes represent a virtually uncharted territory for discovery in the mosquito microbiome. As such, they are the final members to make the list of high-value targets for gnotobiotic studies.
Community Ecology and Microbiome Interactions
Bacteria are unlikely to find themselves in the mosquito as a monoculture. Instead, mosquitoes are colonized by a community of interacting individuals and populations.
Microbial interactions may be parasitic, where one organism benefits at the cost of another: mutualistic, such that both organisms benefit, or commensal, when one organism benefits at no cost or benefit to the other. These interactions are facilitated by mechanisms, such as metabolite exchange, cross-feeding, and antibiotic production (Phelan et al., 2012; Pacheco and Segrè, 2019). Thus, the properties of the community are determined by the separate functional contributions from each species and their interactions. In this respect, the attributes of a community are difficult to predict from the traits of its members when they are reared in a mono-culture (Sanchez-Gorostiaga et al., 2019). It is increasingly apparent that the microbiome of mosquitoes acts as a community, rather than a collection of individuals. Co-occurrence networks based on bacterial census data identified multiple pairwise and higher order interactions, indicating an interwoven and linked microbial community (Hegde et al., 2018). More direct evidence of microbial interactions within the mosquito has also been documented. For example, when a strain of S. marcescens was introduced to Ae. aegypti larvae as a monoculture, 89% of the larvae died. The same strain inoculated in a simple three-member community reduced mortality to 50%. Yet, Serratia was identified among all of the assayed mosquitoes, suggesting that the larvicidal activity of Serratia is attenuated by the presence of other microbes (Correa et al., 2018). Microbial interactions also influence the digestion of mosquito food sources. The fructose that mosquitoes obtain through feeding on flower nectar can be digested in a trophic interaction involving both fungi and bacteria (Guégan et al., 2020). In another example, it was shown that when E. anophelis was co-cultured with a strain of Pseudomonas in the midguts of mosquitoes, E. anophelis upregulated gene products for heme degradation. This activity presumably facilitates blood digestion in the mosquito but also produced a metabolite of the class biliverdin, which may inhibit Pseudomonas growth. In this manner, E. anophelis gains a competitive advantage and may indirectly benefit the mosquito host (Ganley, 2020). These observations all point to the importance of viewing the mosquito microbiome as a community and taking a population ecology viewpoint when linking the status of the mosquito microbiome to host phenotypes. Thus, an important step going forward will be to employ gnotobiotic mosquitoes as a resource to characterize microbiome interaction networks.
Conclusion and Perspectives
The development of axenic and gnotobiotic mosquito models offers the potential to transform the study of the mosquito microbiome. These models transition microbiome studies from correlational associations between microbes and their host to controlled experiments that can systematically manipulate the composition, genetics, and biochemistry of the of the microbiome. In this manner, the mechanistic underpinnings of the relationship between the mosquito and its microflora can begin to be uncovered. The wealth of studies that have already linked the microbiome to mosquito biology and the diseases they carry have already provided an abundance of hypotheses to test and will be an excellent foundation for future studies. We have provided a list of potential microbiome members that represent particularly high-value targets for future gnotobiotic studies, but they are only the forefront of a broad field of investigation. Recently, researchers proposed the formation of a “Mosquito Microbiome Research Consortium” and laid out recommendations for best practices for collecting, analyzing, and sharing mosquito microbiome data (Dada et al., 2021). We propose that properly designed and controlled axenic and gnotobiotic studies should be central pillars to a unified effort to disentangle the role of the microbiome in mosquito biology and microbe-mosquito control programs.
Author Contributions
BS and DB contributed to manuscript conception and writing. JH contributed to data for Figure 1 and manuscript editing. JL contributed to figure production. All authors contributed to the article and approved the submitted version.
Funding
JH was supported by a Louis A Magnarelli grant to BS and DB.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abreo, E., and Altier, N. (2019). Pangenome of Serratia marcescens strains from nosocomial and environmental origins reveals different populations and the links between them. Sci. Rep. 9:46. doi: 10.1038/s41598-018-37118-0
Akhouayri, I. G., Habtewold, T., and Christophides, G. K. (2013). Melanotic pathology and vertical transmission of the gut commensal Elizabethkingia meningoseptica in the major malaria vector Anopheles gambiae. PLoS One 8:e77619. doi: 10.1371/journal.pone.0077619
Akov, S. (1962). A qualitative and quantitative study of the nutritional requirements of Aedes aegypti L. larvae. J. Insect Physiol. 8, 319–335. doi: 10.1016/0022-1910(62)90035-5
Alfano, N., Tagliapietra, V., Rosso, F., Manica, M., Arnoldi, D., Pindo, M., et al. (2019). Changes in microbiota across developmental stages of Aedes koreicus, an invasive mosquito vector in Europe: indications for microbiota-based control strategies. Front. Microbiol. 10:2832. doi: 10.3389/fmicb.2019.02832
Angleró-Rodríguez, Y. I., Talyuli, O. A., Blumberg, B. J., Kang, S., Demby, C., Shields, A., et al. (2017). An Aedes aegypti-associated fungus increases susceptibility to dengue virus by modulating gut trypsin activity. eLife 6:e28844. doi: 10.7554/eLife.28844
Apte-Deshpande, A., Paingankar, M., Gokhale, M. D., and Deobagkar, D. N. (2012). Serratia odorifera a midgut inhabitant of Aedes aegypti mosquito enhances its susceptibility to dengue-2 virus. PLoS One 7:e40401. doi: 10.1371/journal.pone.0040401
Audsley, M. D., Ye, Y. H., and McGraw, E. A. (2017). The microbiome composition of Aedes aegypti is not critical for Wolbachia-mediated inhibition of dengue virus. PLoS Negl. Trop. Dis. 11:e0005426. doi: 10.1371/journal.pntd.0005426
Badal, S., Her, Y. F., and Maher, L. J. (2015). Nonantibiotic effects of fluoroquinolones in mammalian cells. J. Biol. Chem. 290, 22287–22297. doi: 10.1074/jbc.M115.671222
Bahia, A. C., Dong, Y., Blumberg, B. J., Mlambo, G., Tripathi, A., BenMarzouk-Hidalgo, O. J., et al. (2014). Exploring Anopheles gut bacteria for Plasmodium blocking activity. Environ. Microbiol. 16, 2980–2994. doi: 10.1111/1462-2920.12381
Bai, L., Wang, L., Vega-Rodríguez, J., Wang, G., and Wang, S. (2019). A gut symbiotic bacterium Serratia marcescens renders mosquito resistance to Plasmodium infection through activation of mosquito immune responses. Front. Microbiol. 10:1580. doi: 10.3389/fmicb.2019.01580
Bakula, M. (1969). The persistence of a microbial flora during postembryogenesis of Drosophila melanogaster. J. Invertebr. Pathol. 14, 365–374. doi: 10.1016/0022-2011(69)90163-3
Bando, H., Okado, K., Guelbeogo, W. M., Badolo, A., Aonuma, H., Nelson, B., et al. (2013). Intra-specific diversity of Serratia marcescens in Anopheles mosquito midgut defines Plasmodium transmission capacity. Sci. Rep. 3:1641. doi: 10.1038/srep01641
Bargielowski, I., and Koella, J. C. (2009). A possible mechanism for the suppression of Plasmodium berghei development in the mosquito Anopheles gambiae by the microsporidian Vavraia culicis. PLoS One 4:e4676. doi: 10.1371/journal.pone.0004676
Barnard, K., Jeanrenaud, A. C. S. N., Brooke, B. D., and Oliver, S. V. (2019). The contribution of gut bacteria to insecticide resistance and the life histories of the major malaria vector Anopheles arabiensis (Diptera: Culicidae). Sci. Rep. 9:9117. doi: 10.1038/s41598-019-45499-z
Bassene, H., Niang, E. H. A., Fenollar, F., Doucoure, S., Faye, O., Raoult, D., et al. (2020). Role of plants in the transmission of Asaia sp., which potentially inhibit the Plasmodium sporogenic cycle in Anopheles mosquitoes. Sci. Rep. 10:7144. doi: 10.1038/s41598-020-64163-5
Batista, J. H., and da Silva Neto, J. F. (2017). Chromobacterium violaceum pathogenicity: updates and insights from genome sequencing of novel Chromobacterium species. Front. Microbiol. 8:2213. doi: 10.3389/fmicb.2017.02213
Belda, E., Coulibaly, B., Fofana, A., Beavogui, A. H., Traore, S. F., Gohl, D. M., et al. (2017). Preferential suppression of Anopheles gambiae host sequences allows detection of the mosquito eukaryotic microbiome. Sci. Rep. 7:3241. doi: 10.1038/s41598-017-03487-1
Biggs, M. B., Medlock, G. L., Moutinho, T. J., Lees, H. J., Swann, J. R., Kolling, G. L., et al. (2017). Systems-level metabolism of the altered Schaedler flora, a complete gut microbiota. ISME J. 11, 426–438. doi: 10.1038/ismej.2016.130
Breurec, S., Criscuolo, A., Diancourt, L., Rendueles, O., Vandenbogaert, M., Passet, V., et al. (2016). Genomic epidemiology and global diversity of the emerging bacterial pathogen Elizabethkingia anophelis. Sci. Rep. 6:30379. doi: 10.1038/srep30379
Browning, D. F., Wells, T. J., França, F. L. S., Morris, F. C., Sevastsyanovich, Y. R., Bryant, J. A., et al. (2013). Laboratory adapted Escherichia coli K-12 becomes a pathogen of Caenorhabditis elegans upon restoration of O antigen biosynthesis. Mol. Microbiol. 87, 939–950. doi: 10.1111/mmi.12144
Capone, A., Ricci, I., Damiani, C., Mosca, M., Rossi, P., Scuppa, P., et al. (2013). Interactions between Asaia, Plasmodium and Anopheles: new insights into mosquito symbiosis and implications in malaria symbiotic control. Parasit. Vectors 6:182. doi: 10.1186/1756-3305-6-182
Cappelli, A., Damiani, C., Mancini, M. V., Valzano, M., Rossi, P., Serrao, A., et al. (2019a). Asaia activates immune genes in mosquito eliciting an anti-Plasmodium response: implications in malaria control. Front. Genet. 10:836. doi: 10.3389/fgene.2019.00836
Cappelli, A., Valzano, M., Cecarini, V., Bozic, J., Rossi, P., Mensah, P., et al. (2019b). Killer yeasts exert anti-plasmodial activities against the malaria parasite Plasmodium berghei in the vector mosquito Anopheles stephensi and in mice. Parasit. Vectors 12:329. doi: 10.1186/s13071-019-3587-4
Caragata, E. P., Otero, L. M., Carlson, J. S., Dizaji, N. B., and Dimopoulos, G. (2020). A nonlive preparation of Chromobacterium sp. Panama (Csp_P) is a highly effective larval mosquito biopesticid. Appl. Environ. Microbiol. 86:e00240-2. doi: 10.1128/AEM.00240-20
Caragata, E. P., Tikhe, C. V., and Dimopoulos, G. (2019). Curious entanglements: interactions between mosquitoes, their microbiota, and arboviruses. Curr. Opin. Virol. 37, 26–36. doi: 10.1016/j.coviro.2019.05.005
Carlson, J. S., Short, S. M., Angleró-Rodríguez, Y. I., and Dimopoulos, G. (2020). Larval exposure to bacteria modulates arbovirus infection and immune gene expression in adult Aedes aegypti. Dev. Comp. Immunol. 104:103540. doi: 10.1016/j.dci.2019.103540
Chabanol, E., Behrends, V., Prévot, G., Christophides, G. K., and Gendrin, M. (2020). Antibiotic treatment in Anopheles coluzzii affects carbon and nitrogen metabolism. Pathogens 9:679. doi: 10.3390/pathogens9090679
Chaston, J. M., Newell, P. D., and Douglas, A. E. (2014). Metagenome-wide association of microbial determinants of host phenotype in Drosophila melanogaster. MBio 5:e01631-14. doi: 10.1128/mBio.01631-14
Chavshin, A., Oshaghi, M., Vatandoost, H., Yakhchali, B., Zarenejad, F., and Terenius, O. (2015). Malpighian tubules are important determinants of Pseudomonas transstadial transmission and longtime persistence in Anopheles stephensi. Parasit. Vectors 8:36. doi: 10.1186/s13071-015-0635-6
Chen, S., Bagdasarian, M., and Walker, E. D. (2015). Elizabethkingia anophelis: molecular manipulation and interactions with mosquito hosts. Appl. Environ. Microbiol. 81, 2233–2243. doi: 10.1128/AEM.03733-14
Chen, S., Blom, J., and Walker, E. D. (2017). Genomic, physiologic, and symbiotic characterization of Serratia marcescens strains isolated from the mosquito Anopheles stephensi. Front. Microbiol. 8:1483. doi: 10.3389/fmicb.2017.01483
Chen, S., Johnson, B. K., Yu, T., Nelson, B. N., and Walker, E. D. (2020). Elizabethkingia anophelis: physiologic and transcriptomic responses to iron stress. Front. Microbiol. 11:804. doi: 10.3389/fmicb.2020.00804
Chen, S., and Walker, E. D. (2020). Genome sequence of Serratia fonticola strain S14, isolated from the mosquito Aedes triseriatus. Microbiol. Resour. Announc. 9, e00099–e00020. doi: 10.1128/MRA.00099-20
Chen, W.-J., and Wu, S.-T. (1997). Ultrastructure of infection, development and gametocyst formation of Ascogregarina taiwanensis (Apicomplexa: Lecudinidae) in its mosquito host, Aedes albopictus (Diptera: Culicidae). J. Eukaryot. Microbiol. 44, 101–108. doi: 10.1111/j.1550-7408.1997.tb05945.x
Chouaia, B., Rossi, P., Epis, S., Mosca, M., Ricci, I., Damiani, C., et al. (2012). Delayed larval development in Anopheles mosquitoes deprived of Asaia bacterial symbionts. BMC Microbiol. 12:S2. doi: 10.1186/1471-2180-12-S1-S2
Chouaia, B., Rossi, P., Montagna, M., Ricci, I., Crotti, E., Damiani, C., et al. (2010). Molecular evidence for multiple infections as revealed by typing of Asaia bacterial symbionts of four mosquito species. Appl. Environ. Microbiol. 76, 7444–7450. doi: 10.1128/AEM.01747-10
Cirimotich, C. M., Dong, Y., Clayton, A. M., Sandiford, S. L., Souza-Neto, J. A., Mulenga, M., et al. (2011). Natural microbe-mediated refractoriness to Plasmodium infection in Anopheles gambiae. Science 332, 855–858. doi: 10.1126/science.1201618
Comandatore, F., Damiani, C., Cappelli, A., Ribolla, P. E. M., Gasperi, G., Gradoni, F., et al. (2021). Phylogenomics reveals that Asaia symbionts from insects underwent convergent genome reduction, preserving an insecticide-degrading gene. mBio 12:e00106-21. doi: 10.1128/mBio.00106-21
Coon, K. L., Brown, M. R., and Strand, M. R. (2016a). Gut bacteria differentially affect egg production in the anautogenous mosquito Aedes aegypti and facultatively autogenous mosquito Aedes atropalpus (Diptera: Culicidae). Parasit. Vectors 9:375. doi: 10.1186/s13071-016-1660-9
Coon, K. L., Brown, M. R., and Strand, M. R. (2016b). Mosquitoes host communities of bacteria that are essential for development but vary greatly between local habitats. Mol. Ecol. 25, 5806–5826. doi: 10.1111/mec.13877
Coon, K. L., Valzania, L., Brown, M. R., and Strand, M. R. (2020). Predaceous Toxorhynchites mosquitoes require a living gut microbiota to develop. Proc. R. Soc. B Biol. Sci. 287:20192705. doi: 10.1098/rspb.2019.2705
Coon, K. L., Valzania, L., McKinney, D. A., Vogel, K. J., Brown, M. R., and Strand, M. R. (2017). Bacteria-mediated hypoxia functions as a signal for mosquito development. PNAS 114, E5362–E5369. doi: 10.1073/pnas.1702983114
Coon, K. L., Vogel, K. J., Brown, M. R., and Strand, M. R. (2014). Mosquitoes rely on their gut microbiota for development. Mol. Ecol. 23, 2727–2739. doi: 10.1111/mec.12771
Correa, M. A., Matusovsky, B., Brackney, D. E., and Steven, B. (2018). Generation of axenic Aedes aegypti demonstrate live bacteria are not required for mosquito development. Nat. Commun. 9:4464. doi: 10.1038/s41467-018-07014-2
Crotti, E., Damiani, C., Pajoro, M., Gonella, E., Rizzi, A., Ricci, I., et al. (2009). Asaia, a versatile acetic acid bacterial symbiont, capable of cross-colonizing insects of phylogenetically distant genera and orders. Environ. Microbiol. 11, 3252–3264. doi: 10.1111/j.1462-2920.2009.02048.x
Crotti, E., Rizzi, A., Chouaia, B., Ricci, I., Favia, G., Alma, A., et al. (2010). Acetic acid bacteria, newly emerging symbionts of insects. Appl. Environ. Microbiol. 76, 6963–6970. doi: 10.1128/AEM.01336-10
D’mello, R., Hill, S., and Poole, R. K. (1996). The cytochrome bd quinol oxidase in Escherichia coli has an extremely high oxygen affinity and two oxygen-binding haems: implications for regulation of activity in vivo by oxygen inhibition. Microbiology 142, 755–763. doi: 10.1099/00221287-142-4-755
Dada, N., Jupatanakul, N., Minard, G., Short, S. M., Akorli, J., and Villegas, L. M. (2021). Considerations for mosquito microbiome research from the mosquito microbiome consortium. Microbiome 9:36. doi: 10.1186/s40168-020-00987-7
Dada, N., Sheth, M., Liebman, K., Pinto, J., and Lenhart, A. (2018). Whole metagenome sequencing reveals links between mosquito microbiota and insecticide resistance in malaria vectors. Sci. Rep. 8:2084. doi: 10.1038/s41598-018-20367-4
Damiani, C., Ricci, I., Crotti, E., Rossi, P., Rizzi, A., Scuppa, P., et al. (2008). Paternal transmission of symbiotic bacteria in malaria vectors. Curr. Biol. 18, R1087–R1088. doi: 10.1016/j.cub.2008.10.040
Damiani, C., Ricci, I., Crotti, E., Rossi, P., Rizzi, A., Scuppa, P., et al. (2010). Mosquito-bacteria symbiosis: the case of Anopheles gambiae and Asaia. Microb. Ecol. 60, 644–654. doi: 10.1007/s00248-010-9704-8
Dickson, L. B., Jiolle, D., Minard, G., Moltini-Conclois, I., Volant, S., Ghozlane, A., et al. (2017). Carryover effects of larval exposure to different environmental bacteria drive adult trait variation in a mosquito vector. Sci. Adv. 3:e1700585. doi: 10.1126/sciadv.1700585
Dieme, C., Zmarlak, N. M., Brito-Fravallo, E., Travaillé, C., Pain, A., Cherrier, F., et al. (2020). Exposure of Anopheles mosquitoes to trypanosomes reduces reproductive fitness and enhances susceptibility to Plasmodium. PLoS Negl. Trop. Dis. 14:e0008059. doi: 10.1371/journal.pntd.0008059
Dittmer, J., and Gabrieli, P. (2020). Transstadial metabolic priming mediated by larval nutrition in female Aedes albopictus mosquitoes. J. Insect Physiol. 123:104053. doi: 10.1016/j.jinsphys.2020.104053
Dong, Y., Manfredini, F., and Dimopoulos, G. (2009). Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog. 5:e1000423. doi: 10.1371/journal.ppat.1000423
Douglas, A. E. (2018). The Drosophila model for microbiome research. Lab Anim. 47, 157–164. doi: 10.1038/s41684-018-0065-0
Durán, N., and Menck, C. F. M. (2008). Chromobacterium violaceum: a review of pharmacological and industiral perspectives. Crit. Rev. Microbiol. 27, 201–222. doi: 10.1080/20014091096747
Erthal, J. A., Soghigian, J. S., and Livdahl, T. (2012). Life cycle completion of parasite Ascogregarina taiwanensis (Apicomplexa: Lecudinidae) in non-native host Ochlerotatus japonicus (Diptera: Culicidae). J. Med. Entomol. 49, 1109–1117. doi: 10.1603/ME12018
Ezemuoka, L. C., Akorli, E. A., Aboagye-Antwi, F., and Akorli, J. (2020). Mosquito midgut Enterobacter cloacae and Serratia marcescens affect the fitness of adult female Anopheles gambiae s.l. PLoS One 15:e0238931. doi: 10.1371/journal.pone.0238931
Favia, G., Ricci, I., Damiani, C., Raddadi, N., Crotti, E., Marzorati, M., et al. (2007). Bacteria of the genus Asaia stably associate with Anopheles stephensi, an Asian malarial mosquito vector. Proc. Natl. Acad. Sci. 104, 9047–9051. doi: 10.1073/pnas.0610451104
Favia, G., Ricci, I., Marzorati, M., Negri, I., Alma, A., Sacchi, L., et al. (2008). “Bacteria of the genus Asaia: a potential paratransgenic weapon against malaria,” Transgenesis and the Management of Vector-Borne Disease. Vol. 627. ed. S. Aksoy (New York, NY: Springer), 49–59.
Fei, N., and Zhao, L. (2013). An opportunistic pathogen isolated from the gut of an obese human causes obesity in germfree mice. ISME J. 7, 880–884. doi: 10.1038/ismej.2012.153
Frank, T., Gody, J. C., Nguyen, L. B. L., Berthet, N., Fleche-Mateos, A. L., Bata, P., et al. (2013). First case of Elizabethkingia anophelis meningitis in the Central African Republic. Lancet 381:1876. doi: 10.1016/S0140-6736(13)60318-9
Freece, C. D., Damiani, C., Valzano, M., D’amelio, S., Cappelli, A., Ricci, I., et al. (2014). Detection and isolation of the α-proteobacterium Asaia in Culex mosquitoes. Med. Vet. Entomol. 28, 438–442. doi: 10.1111/mve.12045
Ganley, J. G. (2020). Coculturing of mosquito-microbiome bacteria promotes heme degradation in Elizabethkingia anophelis. Chembiochem 21, 1279–1284. doi: 10.1002/cbic.201900675
Ganley, J. G., Carr, G., Ioerger, T. R., Sacchettini, J. C., Clardy, J., and Derbyshire, E. R. (2018). Discovery of antimicrobial lipodepsipeptides produced by a Serratia sp. within mosquito microbiomes. Chembiochem 19, 1590–1594. doi: 10.1002/cbic.201800124
Giraud, E., Varet, H., Legendre, R., Sismeiro, O., Aubry, F., Dabo, S., et al. (2021). Mosquito-bacteria interactions during larval development trigger metabolic changes with carry-over effects on adult fitness. bioRxiv [Preprint]. doi: 10.1101/2021.05.20.444942
Glockling, S. L., Marshall, W. L., and Gleason, F. H. (2013). Phylogenetic interpretations and ecological potentials of the Mesomycetozoea (Ichthyosporea). Fungal Ecol. 6, 237–247. doi: 10.1016/j.funeco.2013.03.005
Gnambani, E. J., Bilgo, E., Sanou, A., Dabiré, R. K., and Diabaté, A. (2020). Infection of highly insecticide-resistant malaria vector Anopheles coluzzii with entomopathogenic bacteria Chromobacterium violaceum reduces its survival, blood feeding propensity and fecundity. Malar. J. 19:352. doi: 10.1186/s12936-020-03420-4
González, L. J., and Vila, A. J. (2012). Carbapenem resistance in Elizabethkingia meningoseptica is mediated by metallo-β-lactamase BlaB. Antimicrob. Agents Chemother. 56, 1686–1692. doi: 10.1128/AAC.05835-11
Goojani, H. G., Konings, J., Hakvoort, H., Hong, S., Gennis, R. B., Sakamoto, J., et al. (2020). The carboxy-terminal insert in the Q-loop is needed for functionality of Escherichia coli cytochrome bd-I. Biochim. Biophys. Acta Bioenerg. 1861:148175. doi: 10.1016/j.bbabio.2020.148175
Gopinath, S., Kim, M. V., Rakib, T., Wong, P. W., van Zandt, M., Barry, N. A., et al. (2018). Topical application of aminoglycoside antibiotics enhances host resistance to viral infections in a microbiota-independent manner. Nat. Microbiol. 3, 611–621. doi: 10.1038/s41564-018-0138-2
Gould, A. L., Zhang, V., Lamberti, L., Jones, E. W., Obadia, B., Korasidis, N., et al. (2018). Microbiome interactions shape host fitness. PNAS 115, E11951–E11960. doi: 10.1073/pnas.1809349115
Guégan, M., Minard, G., Tran, F.-H., Fel, B., Hay, A.-E., Simon, L., et al. (2020). Who is eating fructose within the Aedes albopictus gut microbiota? Environ. Microbiol. 22, 1193–1206. doi: 10.1111/1462-2920.14915
Guégan, M., Zouache, K., Démichel, C., Minard, G., Tran Van, V., Potier, P., et al. (2018). The mosquito holobiont: fresh insight into mosquito-microbiota interactions. Microbiome 6:49. doi: 10.1186/s40168-018-0435-2
Hancock, P. A., Sinkins, S. P., and Godfray, H. C. J. (2011). Strategies for introducing Wolbachia to reduce transmission of mosquito-borne diseases. PLoS Negl. Trop. Dis. 5:e1024. doi: 10.1371/journal.pntd.0001024
Hayek, S. S., Abd, T. T., Cribbs, S. K., Anderson, A. M., Melendez, A., Kobayashi, M., et al. (2013). Rare Elizabethkingia meningosepticum meningitis case in an immunocompetent adult. Emerging Microbes Infect. 2, 1–4. doi: 10.1038/emi.2013.16
Hegde, S., Khanipov, K., Albayrak, L., Golovko, G., Pimenova, M., Saldaña, M. A., et al. (2018). Microbiome interaction networks and community structure from laboratory-reared and field-collected Aedes aegypti, Aedes albopictus, and Culex quinquefasciatus mosquito vectors. Front. Microbiol. 9:2160. doi: 10.3389/fmicb.2018.02160
Herren, J. K., Mbaisi, L., Mararo, E., Makhulu, E. E., Mobegi, V. A., Butungi, H., et al. (2020). A microsporidian impairs Plasmodium falciparum transmission in Anopheles arabiensis mosquitoes. Nat. Commun. 11:2187. doi: 10.1038/s41467-020-16121-y
Hug, L. A. (2018). Sizing up the uncultured microbial majority. mSystems 3:e00185-18. doi: 10.1128/mSystems.00185-18
Hughes, G. L., Dodson, B. L., Johnson, R. M., Murdock, C. C., Tsujimoto, H., Suzuki, Y., et al. (2014). Native microbiome impedes vertical transmission of Wolbachia in Anopheles mosquitoes. Proc. Natl. Acad. Sci. 111, 12498–12503. doi: 10.1073/pnas.1408888111
Hughes, G. L., Pike, A. D., Xue, P., and Rasgon, J. L. (2012). Invasion of Wolbachia into Anopheles and other insect germlines in an ex vivo organ culture system. PLoS One 7:e36277. doi: 10.1371/journal.pone.0036277
Hyde, J., Correa, M. A., Brackney, D. E., and Steven, B. (2019a). Generation and rearing of axenic Aedes aegypti mosquitoes. [Preprint]. doi: 10.21203/rs.2.17705/v1
Hyde, J., Correa, M. A., Hughes, G. L., Steven, B., and Brackney, D. E. (2020). Limited influence of the microbiome on the transcriptional profile of female Aedes aegypti mosquitoes. Sci. Rep. 10:10880. doi: 10.1038/s41598-020-67811-y
Hyde, J., Gorham, C., Brackney, D. E., and Steven, B. (2019b). Antibiotic resistant bacteria and commensal fungi are common and conserved in the mosquito microbiome. PLoS One 14:e0218907. doi: 10.1371/journal.pone.0218907
Iturbe-Ormaetxe, I., Walker, T., and O’ Neill, S. L. (2011). Wolbachia and the biological control of mosquito-borne disease. EMBO Rep. 12, 508–518. doi: 10.1038/embor.2011.84
Janda, J. M., and Lopez, D. L. (2017). Mini review: new pathogen profiles: Elizabethkingia anophelis. Diagn. Microbiol. Infect. Dis. 88, 201–205. doi: 10.1016/j.diagmicrobio.2017.03.007
Jiggins, F. M. (2017). The spread of Wolbachia through mosquito populations. PLoS Biol. 15:e2002780. doi: 10.1371/journal.pbio.2002780
Kalappa, D. M., Subramani, P. A., Basavanna, S. K., Ghosh, S. K., Sundaramurthy, V., Uragayala, S., et al. (2018). Influence of midgut microbiota in Anopheles stephensi on Plasmodium berghei infections. Malar. J. 17:385. doi: 10.1186/s12936-018-2535-7
Kalghatgi, S., Spina, C. S., Costello, J. C., Liesa, M., Morones-Ramirez, J. R., Slomovic, S., et al. (2013). Bactericidal antibiotics induce mitochondrial dysfunction and oxidative damage in mammalian cells. Sci. Transl. Med. 5:192ra85. doi: 10.1126/scitranslmed.3006055
Kämpfer, P., Matthews, H., Glaeser, S. P., Martin, K., Lodders, N., and Faye, I. (2011). Elizabethkingia anophelis sp. nov., isolated from the midgut of the mosquito Anopheles gambiae. Int. J. Syst. Evol. Microbiol. 61, 2670–2675. doi: 10.1099/ijs.0.026393-0
Kanzok, S. M., and Jacobs-Lorena, M. (2006). Entomopathogenic fungi as biological insecticides to control malaria. Trends Parasitol. 22, 49–51. doi: 10.1016/j.pt.2005.12.008
Kaur, R., Shropshire, J. D., Cross, K. L., Leigh, B., Mansueto, A. J., Stewart, V., et al. (2021). Living in the endosymbiotic world of Wolbachia: a centennial review. Cell Host Microbe 29, 879–893. doi: 10.1016/j.chom.2021.03.006
Keebaugh, E. S., Yamada, R., Obadia, B., Ludington, W. B., and Ja, W. W. (2018). Microbial quantity impacts Drosophila nutrition, development, and lifespan. iScience 4, 247–259. doi: 10.1016/j.isci.2018.06.004
Khanna, A., Khanna, M., and Aggarwal, A. (2013). Serratia marcescens-a rare opportunistic nosocomial pathogen and measures to limit its spread in hospitalized patients. J. Clin. Diagn. Res. 7, 243–246. doi: 10.7860/JCDR/2013/5010.2737
Koosha, M., Vatandoost, H., Karimian, F., Choubdar, N., Abai, M. R., and Oshaghi, M. A. (2019). Effect of Serratia AS1 (Enterobacteriaceae: Enterobacteriales) on the fitness of Culex pipiens (Diptera: Culicidae) for paratransgenic and RNAi approaches. J. Med. Entomol. 56, 553–559. doi: 10.1093/jme/tjy183
Kothari, V., Sharma, S., and Padia, D. (2017). Recent research advances on Chromobacterium violaceum. Asian Pac J Trop Med 10, 744–752. doi: 10.1016/j.apjtm.2017.07.022
Kozlova, E. V., Hegde, S., Roundy, C. M., Golovko, G., Saldaña, M. A., Hart, C. E., et al. (2021). Microbial interactions in the mosquito gut determine Serratia colonization and blood-feeding propensity. ISME J. 15, 93–108. doi: 10.1038/s41396-020-00763-3
Kukutla, P., Lindberg, B. G., Pei, D., Rayl, M., Yu, W., Steritz, M., et al. (2013). Draft genome sequences of Elizabethkingia anophelis strains R26T and Ag1 from the midgut of the malaria mosquito Anopheles gambiae. Genome Announc. 1:e01030-13. doi: 10.1128/genomeA.01030-13
Kukutla, P., Lindberg, B. G., Pei, D., Rayl, M., Yu, W., Steritz, M., et al. (2014). Insights from the genome annotation of Elizabethkingia anophelis from the malaria vector Anopheles gambiae. PLoS One 9:e97715. doi: 10.1371/journal.pone.0097715
Lang, C. A., Basch, K. J., and Storey, R. S. (1972). Growth, composition and longevity of the axenic mosquito. J. Nutr. 102, 1057–1066. doi: 10.1093/jn/102.8.1057
Lau, S. K. P., Chow, W.-N., Foo, C.-H., Curreem, S. O. T., Lo, G. C.-S., Teng, J. L. L., et al. (2016). Elizabethkingia anophelis bacteremia is associated with clinically significant infections and high mortality. Sci. Rep. 6:26045. doi: 10.1038/srep26045
Lea, A. O., Dimond, J. B., and Delong, D. M. (1956). A chemically defined medium for rearing Aedes aegypti larvae. J. Econ. Entomol. 49, 313–315. doi: 10.1093/jee/49.3.313
Lee, Y.-L., Liu, K.-M., Chang, H.-L., Lin, J.-S., Kung, F.-Y., Ho, C.-M., et al. (2021). A dominant strain of Elizabethkingia anophelis emerged from a hospital water system to cause a three-year outbreak in a respiratory care center. J. Hosp. Infect. 108, 43–51. doi: 10.1016/j.jhin.2020.10.025
Lewis, W. H., Tahon, G., Geesink, P., Sousa, D. Z., and Ettema, T. J. G. (2021). Innovations to culturing the uncultured microbial majority. Nat. Rev. Microbiol. 19, 225–240. doi: 10.1038/s41579-020-00458-8
Lindh, J. M., Borg-Karlson, A.-K., and Faye, I. (2008). Transstadial and horizontal transfer of bacteria within a colony of Anopheles gambiae (Diptera: Culicidae) and oviposition response to bacteria-containing water. Acta Trop. 107, 242–250. doi: 10.1016/j.actatropica.2008.06.008
Linenberg, I., Christophides, G. K., and Gendrin, M. (2016). Larval diet affects mosquito development and permissiveness to Plasmodium infection. Sci. Rep. 6:38230. doi: 10.1038/srep38230
Liu, D., and Reeves, P. R. Y. (1994). Escherichia coli K12 regains its O antigen. Microbiology 140, 49–57. doi: 10.1099/13500872-140-1-49
Lopes, S. C. P., Blanco, Y. C., Justo, G. Z., Nogueira, P. A., Rodrigues, F. L. S., Goelnitz, U., et al. (2009). Violacein extracted from Chromobacterium violaceum inhibits Plasmodium growth in vitro and in vivo. Antimicrob. Agents Chemother. 53, 2149–2152. doi: 10.1128/AAC.00693-08
Mahlen, S. D. (2011). Serratia infections: from military experiments to current practice. Clin. Microbiol. Rev. 24, 755–791. doi: 10.1128/CMR.00017-11
Malassigné, S., Valiente Moro, C., and Luis, P. (2020). Mosquito mycobiota: an overview of non-entomopathogenic fungal interactions. Pathogens 9:564. doi: 10.3390/pathogens9070564
Mancini, M. V., Damiani, C., Accoti, A., Tallarita, M., Nunzi, E., Cappelli, A., et al. (2018). Estimating bacteria diversity in different organs of nine species of mosquito by next generation sequencing. BMC microbiol. 18:126. doi: 10.1186/s12866-018-1266-9
McClean, K. H., Winson, M. K., Fish, L., Taylor, A., Chhabra, S. R., Camara, M., et al. (1997). Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 143, 3703–3711. doi: 10.1099/00221287-143-12-3703
Minard, G., Mavingui, P., and Moro, C. V. (2013a). Diversity and function of bacterial microbiota in the mosquito holobiont. Parasit. Vectors 6:146. doi: 10.1186/1756-3305-6-146
Minard, G., Tran, F. H., Raharimalala, F. N., Hellard, E., Ravelonandro, P., Mavingui, P., et al. (2013b). Prevalence, genomic and metabolic profiles of Acinetobacter and Asaia associated with field-caught Aedes albopictus from Madagascar. FEMS Microbiol. Ecol. 83, 63–73. doi: 10.1111/j.1574-6941.2012.01455.x
Mitraka, E., Stathopoulos, S., Siden-Kiamos, I., Christophides, G. K., and Louis, C. (2013). Asaia accelerates larval development of Anopheles gambiae. Pathog. Global Health 107, 305–311. doi: 10.1179/2047773213Y.0000000106
Moller-Jacobs, L. L., Murdock, C. C., and Thomas, M. B. (2014). Capacity of mosquitoes to transmit malaria depends on larval environment. Parasit. Vectors 7:593. doi: 10.1186/s13071-014-0593-4
Moullan, N., Mouchiroud, L., Wang, X., Ryu, D., Williams, E. G., Mottis, A., et al. (2015). Tetracyclines disturb mitochondrial function across eukaryotic models: a call for caution in biomedical research. Cell Rep. 10, 1681–1691. doi: 10.1016/j.celrep.2015.02.034
Muturi, E. J., Ramirez, J. L., Rooney, A. P., and Kim, C.-H. (2017). Comparative analysis of gut microbiota of mosquito communities in central Illinois. PLoS Negl. Trop. Dis. 11:e0005377. doi: 10.1371/journal.pntd.0005377
Newell, P. D., and Douglas, A. E. (2014). Interspecies interactions determine the impact of the gut microbiota on nutrient allocation in Drosophila melanogaster. Appl. Environ. Microbiol. 80, 788–796. doi: 10.1128/AEM.02742-13
Novakova, E., Woodhams, D. C., Rodríguez-Ruano, S. M., Brucker, R. M., Leff, J. W., Maharaj, A., et al. (2017). Mosquito microbiome dynamics, a background for prevalence and seasonality of West Nile virus. Front. Microbiol. 8:526. doi: 10.3389/fmicb.2017.00526
Nuttall, G. H. F., and Thierfelder, H. (1896). Thierisches Leben ohne Bakterien im Verdauungskanal. Berlin/New York: Walter de Gruyter.
Oliveira, T. M. P., Sanabani, S. S., Sallum, M. A. M., Oliveira, T. M. P., Sanabani, S. S., and Sallum, M. A. M. (2020). Asaia (Rhodospirillales: Acetobacteraceae) and Serratia (Enterobacterales: Yersiniaceae) associated with Nyssorhynchus braziliensis and Nyssorhynchus darlingi (Diptera: Culicidae). Revista Brasileira de Entomologia 64. doi: 10.1590/1806-9665-rbent-2019-0010
Osei-Poku, J., Mbogo, C. M., Palmer, W. J., and Jiggins, F. M. (2012). Deep sequencing reveals extensive variation in the gut microbiota of wild mosquitoes from Kenya. Mol. Ecol. 21, 5138–5150. doi: 10.1111/j.1365-294X.2012.05759.x
Pacheco, A. R., and Segrè, D. (2019). A multidimensional perspective on microbial interactions. FEMS Microbiol. Lett. 366:fnz125. doi: 10.1093/femsle/fnz125
Pasteur, L. (1885). Observations relatives à la note précédente de M. Duclaux. C.R. Acad. Sci 100:68.
Patil, C. D., Patil, S. V., Salunke, B. K., and Salunkhe, R. B. (2011). Prodigiosin produced by Serratia marcescens NMCC46 as a mosquito larvicidal agent against Aedes aegypti and Anopheles stephensi. Parasitol. Res. 109, 1179–1187. doi: 10.1007/s00436-011-2365-9
Patil, C. D., Patil, S. V., Salunke, B. K., and Salunkhe, R. B. (2012). Insecticidal potency of bacterial species Bacillus thuringiensis SV2 and Serratia nematodiphila SV6 against larvae of mosquito species Aedes aegypti, Anopheles stephensi, and Culex quinquefasciatus. Parasitol. Res. 110, 1841–1847. doi: 10.1007/s00436-011-2708-6
Perrin, A., Larsonneur, E., Nicholson, A. C., Edwards, D. J., Gundlach, K. M., Whitney, A. M., et al. (2017). Evolutionary dynamics and genomic features of the Elizabethkingia anophelis 2015 to 2016 Wisconsin outbreak strain. Nat. Commun. 8:15483. doi: 10.1038/ncomms15483
Phelan, V. V., Liu, W.-T., Pogliano, K., and Dorrestein, P. C. (2012). Microbial metabolic exchange—the chemotype-to-phenotype link. Nat. Chem. Biol. 8, 26–35. doi: 10.1038/nchembio.739
Rami, A., Raz, A., Zakeri, S., and Dinparast Djadid, N. (2018). Isolation and identification of Asaia sp. in Anopheles spp. mosquitoes collected from Iranian malaria settings: steps toward applying paratransgenic tools against malaria. Parasit. Vectors 11:367. doi: 10.1186/s13071-018-2955-9
Ramirez, J. L., Short, S. M., Bahia, A. C., Saraiva, R. G., Dong, Y., Kang, S., et al. (2014). Chromobacterium Csp_P reduces malaria and dengue infection in vector mosquitoes and has entomopathogenic and in vitro anti-pathogen activities. PLoS Pathog. 10:e1004398. doi: 10.1371/journal.ppat.1004398
Ramirez, J. L., Souza-Neto, J., Cosme, R. T., Rovira, J., Ortiz, A., Pascale, J. M., et al. (2012). Reciprocal tripartite interactions between the Aedes aegypti midgut microbiota, innate immune system and dengue virus influences vector competence. PLoS Negl. Trop. Dis. 6:e1561. doi: 10.1371/journal.pntd.0001561
Ramos-Nino, M. E., Fitzpatrick, D. M., Eckstrom, K. M., Tighe, S., Hattaway, L. M., Hsueh, A. N., et al. (2020). Metagenomic analysis of Aedes aegypti and Culex quinquefasciatus mosquitoes from Grenada, West Indies. PLoS One 15:e0231047. doi: 10.1371/journal.pone.0231047
Raspor, P., and Goranovič, D. (2008). Biotechnological applications of acetic acid bacteria. Crit. Rev. Biotechnol. 28, 101–124. doi: 10.1080/07388550802046749
Reeves, W. K. (2004). Oviposition by Aedes aegypti (Diptera: Culicidae) in relation to conspecific larvae infected with internal symbiotes. J. Vector Ecol. 29, 159–63.
Ricci, I., Damiani, C., Scuppa, P., Mosca, M., Crotti, E., Rossi, P., et al. (2011). The yeast Wickerhamomyces anomalus (Pichia anomala) inhabits the midgut and reproductive system of the Asian malaria vector Anopheles stephensi. Environ. Microbiol. 13, 911–921. doi: 10.1111/j.1462-2920.2010.02395.x
Rinke, C., Schwientek, P., Sczyrba, A., Ivanova, N. N., Anderson, I. J., Cheng, J.-F., et al. (2013). Insights into the phylogeny and coding potential of microbial dark matter. Nature 499, 431–437. doi: 10.1038/nature12352
Romoli, O., and Gendrin, M. (2018). The tripartite interactions between the mosquito, its microbiota and Plasmodium. Parasit. Vectors 11:200. doi: 10.1186/s13071-018-2784-x
Romoli, O., Schönbeck, J. C., Hapfelmeier, S., and Gendrin, M. (2021). Production of germ-free mosquitoes via transient colonisation allows stage-specific investigation of host–microbiota interactions. Nat. Commun. 12:942. doi: 10.1038/s41467-021-21195-3
Rosales-Ronquillo, M. C., Simons, R. W., and Silverman, P. H. (1973). Aseptic rearing of Anopheles stephensi (Diptera: Culicidae). Ann. Entomol. Soc. Am. 66, 949–954. doi: 10.1093/aesa/66.5.949
Saab, S. A., Dohna, H. z., Nilsson, L. K. J., Onorati, P., Nakhleh, J., Terenius, O., et al. (2020). The environment and species affect gut bacteria composition in laboratory co-cultured Anopheles gambiae and Aedes albopictus mosquitoes. Sci. Rep. 10:3352. doi: 10.1038/s41598-020-60075-6
Sanchez-Gorostiaga, A., Bajić, D., Osborne, M. L., Poyatos, J. F., and Sanchez, A. (2019). High-order interactions distort the functional landscape of microbial consortia. PLoS Biol. 17:e3000550. doi: 10.1371/journal.pbio.3000550
Sang, J. H., and King, R. C. (1961). Nutritional requirements of axenically cultured Drosophila melanogaster adults. J. Exp. Biol. 38, 793–809. doi: 10.1242/jeb.38.4.793
Saraiva, R. G., Fang, J., Kang, S., Angleró-Rodríguez, Y. I., Dong, Y., and Dimopoulos, G. (2018a). Aminopeptidase secreted by Chromobacterium sp. Panama inhibits dengue virus infection by degrading the E protein. PLoS Negl. Trop. Dis. 12:e0006443. doi: 10.1371/journal.pntd.0006443
Saraiva, R. G., Huitt-Roehl, C. R., Tripathi, A., Cheng, Y.-Q., Bosch, J., Townsend, C. A., et al. (2018b). Chromobacterium spp. mediate their anti- Plasmodium activity through secretion of the histone deacetylase inhibitor romidepsin. Sci. Rep. 8:6176. doi: 10.1038/s41598-018-24296-0
Schaedler, R. W., Dubos, R., and Costello, R. (1965). Association of germfree mice with bacteria isolated from normal mice. J. Exp. Med. 122, 77–82. doi: 10.1084/jem.122.1.77
Scholte, E.-J., Knols, B. G. J., Samson, R. A., and Takken, W. (2004). Entomopathogenic fungi for mosquito control: a review. J. Insect Sci. 4:19. doi: 10.1093/jis/4.1.19
Schulz, M. D., Atay, Ç., Heringer, J., Romrig, F. K., Schwitalla, S., Aydin, B., et al. (2014). High-fat-diet-mediated dysbiosis promotes intestinal carcinogenesis independently of obesity. Nature 514, 508–512. doi: 10.1038/nature13398
Seitz, H. M., Maier, W. A., Rottok, M., and Becker-Feldmann, H. (1987). Concomitant infections of Anopheles stephensi with Plasmodium berghei and Serratia marcescens: additive detrimental effects. Zentralbl. Bakteriol. Mikrobiol. Hyg. Ser. A 266, 155–166. doi: 10.1016/S0176-6724(87)80029-9
Sharma, P., Rani, J., Chauhan, C., Kumari, S., Tevatiya, S., Das De, T., et al. (2020). Altered gut microbiota and immunity defines Plasmodium vivax survival in Anopheles stephensi. Front. Immunol. 11:609. doi: 10.3389/fimmu.2020.00609
Shen, D., Nyawira, K. T., and Xia, A. (2020). New discoveries and applications of mosquito fungal pathogens. Curr. Opin. Insect Sci. 40, 111–116. doi: 10.1016/j.cois.2020.05.003
Shin, S. C., Kim, S.-H., You, H., Kim, B., Kim, A. C., Lee, K.-A., et al. (2011). Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 334, 670–674. doi: 10.1126/science.1212782
Short, S. M., van Tol, S., MacLeod, H. J., and Dimopoulos, G. (2018a). Hydrogen cyanide produced by the soil bacterium Chromobacterium sp. Panama contributes to mortality in Anopheles gambiae mosquito larvae. Sci. Rep. 8:8358. doi: 10.1038/s41598-018-26680-2
Short, S. M., van Tol, S., Smith, B., Dong, Y., and Dimopoulos, G. (2018b). The mosquito adulticidal Chromobacterium sp. Panama causes transgenerational impacts on fitness parameters and elicits xenobiotic gene responses. Parasit. Vectors 11:229. doi: 10.1186/s13071-018-2822-8
Schubert, A. M., Sinani, H., and Schloss, P. D. (2015). Antibiotic-induced alterations of the murine gut microbiota and subsequent effects on colonization resistance against Clostridium difficile. MBio 6:e00974. doi: 10.1128/mBio.00974-15
Singh, K. R. P., and Brown, A. W. A. (1957). Nutritional requirements of Aedes aegypti L. J. Insect Physiol. 1, 199–220. doi: 10.1016/0022-1910(57)90036-7
Smith, K., McCoy, K. D., and Macpherson, A. J. (2007). Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin. Immunol. 19, 59–69. doi: 10.1016/j.smim.2006.10.002
Souza, R. S., Virginio, F., Riback, T. I. S., Suesdek, L., Barufi, J. B., and Genta, F. A. (2019). Microorganism-based larval diets affect mosquito development, size and nutritional reserves in the yellow fever mosquito Aedes aegypti (Diptera: Culicidae). Front. Physiol. 10:152. doi: 10.3389/fphys.2019.00152
Stathopoulos, S., Neafsey, D. E., Lawniczak, M. K. N., Muskavitch, M. A. T., and Christophides, G. K. (2014). Genetic dissection of Anopheles gambiae gut epithelial responses to Serratia marcescens. PLoS Pathog. 10:e1003897. doi: 10.1371/journal.ppat.1003897
Storelli, G., Defaye, A., Erkosar, B., Hols, P., Royet, J., and Leulier, F. (2011). Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab. 14, 403–414. doi: 10.1016/j.cmet.2011.07.012
Suryawanshi, R. K., Patil, C. D., Borase, H. P., Narkhede, C. P., Salunke, B. K., and Patil, S. V. (2015). Mosquito larvicidal and pupaecidal potential of prodigiosin from Serratia marcescens and understanding its mechanism of action. Pestic. Biochem. Physiol. 123, 49–55. doi: 10.1016/j.pestbp.2015.01.018
Taerum, S. J., Steven, B., Gage, D. J., and Triplett, L. R. (2020). Validation of a PNA clamping method for reducing host DNA amplification and increasing eukaryotic diversity in rhizosphere microbiome studies. Phytobiomes J. 4, 291–302. doi: 10.1094/PBIOMES-05-20-0040-TA
Tchioffo, M. T., Boissière, A., Abate, L., Nsango, S. E., Bayibéki, A. N., Awono-Ambéné, P. H., et al. (2016). Dynamics of bacterial community composition in the malaria mosquito’s epithelia. Front. Microbiol. 6:1500. doi: 10.3389/fmicb.2015.01500
Thongsripong, P., Chandler, J. A., Green, A. B., Kittayapong, P., Wilcox, B. A., Kapan, D. D., et al. (2018). Mosquito vector-associated microbiota: metabarcoding bacteria and eukaryotic symbionts across habitat types in Thailand endemic for dengue and other arthropod-borne diseases. Ecol. Evol. 8, 1352–1368. doi: 10.1002/ece3.3676
Trager, W. (1935a). On the nutritional requirements of mosquito larvae (Aëdes aegypti). Am. J. Hyg. 22, 475–493.
Trager, W. (1935b). The culture of mosquito larvae free from living microorganisms. Am. J. Epidemiol. 22, 18–25.
Trager, W. (1937). A growth factor required by mosquito larvae. J. Exp. Biol. 14, 240–251. doi: 10.1242/jeb.14.2.240
Turley, A. P., Moreira, L. A., O’Neill, S. L., and McGraw, E. A. (2009). Wolbachia infection reduces blood-feeding success in the dengue fever mosquito, Aedes aegypti. PLoS Negl. Trop. Dis. 3:e516. doi: 10.1371/journal.pntd.0000516
Turnbaugh, P. J., Ley, R. E., Mahowald, M. A., Magrini, V., Mardis, E. R., and Gordon, J. I. (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031. doi: 10.1038/nature05414
Valzania, L., Coon, K. L., Vogel, K. J., Brown, M. R., and Strand, M. R. (2018a). Hypoxia-induced transcription factor signaling is essential for larval growth of the mosquito Aedes aegypti. Proc. Natl. Acad. Sci. U. S. A. 115, 457–465. doi: 10.1073/pnas.1719063115
Valzania, L., Martinson, V. G., Harrison, R. E., Boyd, B. M., Coon, K. L., Brown, M. R., et al. (2018b). Both living bacteria and eukaryotes in the mosquito gut promote growth of larvae. PLoS Negl. Trop. Dis. 12:e0006638. doi: 10.1371/journal.pntd.0006638
Vöing, K., Harrison, A., and Soby, S. D. (2020). Draft genome sequence of Chromobacterium vaccinii, a potential biocontrol agent against mosquito (Aedes aegypti) larvae. Genome Announc. 2:e00477-15. doi: 10.1128/genomeA.00477-15
Voronin, D., Tran-Van, V., Potier, P., and Mavingui, P. (2010). Transinfection and growth discrepancy of Drosophila Wolbachia strain wMel in cell lines of the mosquito Aedes albopictus. J. Appl. Microbiol. 108, 2133–2141. doi: 10.1111/j.1365-2672.2009.04621.x
Vrieze, A., Van Nood, E., Holleman, F., Salojärvi, J., Kootte, R. S., Bartelsman, J. F. W. M., et al. (2012). Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 143, 913.e7–916.e7. doi: 10.1053/j.gastro.2012.06.031
Wang, S., Dos-Santos, A. L. A., Huang, W., Liu, K. C., Oshaghi, M. A., Wei, G., et al. (2017). Driving mosquito refractoriness to Plasmodium falciparum with engineered symbiotic bacteria. Science 357, 1399–1402. doi: 10.1126/science.aan5478
Wang, Y., Eum, J. H., Harrison, R. E., Valzania, L., Yang, X., Johnson, J. A., et al. (2021a). Riboflavin instability is a key factor underlying the requirement of a gut microbiota for mosquito development. PNAS 118:e2101080118. doi: 10.1073/pnas.2101080118
Wang, X., Liu, T., Wu, Y., Zhong, D., Zhou, G., Su, X., et al. (2018). Bacterial microbiota assemblage in Aedes albopictus mosquitoes and its impacts on larval development. Mol. Ecol. 27, 2972–2985. doi: 10.1111/mec.14732
Wang, Y., Shen, R., Xing, D., Zhao, C., Gao, H., Wu, J., et al. (2021b). Metagenome sequencing reveals the midgut microbiota makeup of Culex pipiens quinquefasciatus and its possible relationship with insecticide resistance. Front. Microbiol. 12:625539. doi: 10.3389/fmicb.2021.625539
Wollman, E. (1911). Sur l’elevage des mouches steriles. Contribution a la connaissance du role des microbes dans les voies digestives. Ann. Inst. Pasteur 25, 79–88.
Wu, P., Sun, P., Nie, K., Zhu, Y., Shi, M., Xiao, C., et al. (2019). A gut commensal bacterium promotes mosquito permissiveness to arboviruses. Cell Host Microbe 25, 101.e5–112.e5. doi: 10.1016/j.chom.2018.11.004
Xi, Z., Ramirez, J. L., and Dimopoulos, G. (2008). The Aedes aegypti toll pathway controls dengue virus infection. PLoS Pathog. 4:e1000098. doi: 10.1371/journal.ppat.1000098
Xiao, X., Yang, L., Pang, X., Zhang, R., Zhu, Y., Wang, P., et al. (2017). A Mesh–Duox pathway regulates homeostasis in the insect gut. Nat. Microbiol. 2, 17020–17012. doi: 10.1038/nmicrobiol.2017.20
Keywords: axenic, gnotobiotic, mosquito, microbiome, model system
Citation: Steven B, Hyde J, LaReau JC and Brackney DE (2021) The Axenic and Gnotobiotic Mosquito: Emerging Models for Microbiome Host Interactions. Front. Microbiol. 12:714222. doi: 10.3389/fmicb.2021.714222
Edited by:
Aram Mikaelyan, North Carolina State University, United StatesReviewed by:
Emily Derbyshire, Duke University, United StatesOttavia Romoli, Institut Pasteur de la Guyane, French Guiana
Copyright © 2021 Steven, Hyde, LaReau and Brackney. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Blaire Steven, blaire.steven@ct.gov
†Present address: Josephine Hyde, Western Australia Department of Biodiversity Conservation and Attractions, Kensington, WA, Australia
 Blaire Steven
Blaire Steven Josephine Hyde
Josephine Hyde Jacquelyn C. LaReau
Jacquelyn C. LaReau Doug E. Brackney
Doug E. Brackney