- 1College of Public Health Sciences, Chulalongkorn University, Bangkok, Thailand
- 2Diagnosis and Monitoring of Animal Pathogen Research Unit, Chulalongkorn University, Bangkok, Thailand
- 3Department of Veterinary Microbiology, Faculty of Veterinary Science, Chulalongkorn University, Bangkok, Thailand
- 4International Health Policy Program, Ministry of Public Health, Nonthaburi, Thailand
- 5Jockey Club College of Veterinary Medicine and Life Sciences, City University of Hong Kong, Kowloon Tong, Hong Kong
- 6School of Veterinary Medicine, Murdoch University, Perth, WA, Australia
Antimicrobial resistance (AMR) is a critical challenge worldwide as it impacts public health, especially via contamination in the food chain and in healthcare-associated infections. In relation to farming, the systems used, waste management on farms, and the production line process are all determinants reflecting the risk of AMR emergence and rate of contamination of foodstuffs. This review focuses on South East Asia (SEA), which contains diverse regions covering 11 countries, each having different levels of development, customs, laws, and regulations. Routinely, here as elsewhere antimicrobials are still used for three indications: therapy, prevention, and growth promotion, and these are the fundamental drivers of AMR development and persistence. The accuracy of detection of antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARG) depends on the laboratory standards applicable in the various institutes and countries, and this affects the consistency of regional data. Enterobacteriaceae such as Escherichia coli and Klebsiella pneumoniae are the standard proxy species used for indicating AMR-associated nosocomial infections and healthcare-associated infections. Pig feces and wastewater have been suspected as one of the hotspots for spread and circulation of ARB and ARG. As part of AMR surveillance in a One Health approach, clonal typing is used to identify bacterial clonal transmission from the production process to consumers and patients – although to date there have been few published definitive studies about this in SEA. Various alternatives to antibiotics are available to reduce antibiotic use on farms. Certain of these alternatives together with improved disease prevention methods are essential tools to reduce antimicrobial usage in swine farms and to support global policy. This review highlights evidence for potential transfer of resistant bacteria from food animals to humans, and awareness and understanding of AMR through a description of the occurrence of AMR in pig farm food chains under SEA management systems. The latter includes a description of standard pig farming practices, detection of AMR and clonal analysis of bacteria, and AMR in the food chain and associated environments. Finally, the possibility of using alternatives to antibiotics and improving policies for future strategies in combating AMR in a SEA context are outlined.
Introduction
Antimicrobial resistance (AMR) in bacterial species and strains is a critical global threat, and has been monitored in animals, humans and the environment through international collaborations aimed at AMR reduction (Gerbin, 2014; Lechner et al., 2020; Singh et al., 2021). The main factors contributing to the emergence of AMR are overuse and misuse of antimicrobials, and the lack of quality control of antimicrobials and active pharmaceutical ingredients in the market (Simba et al., 2016; Tangcharoensathien et al., 2018; Barroga et al., 2020). Also, the multi-faceted nature of the problem includes misconceptions about antibiotic types and AMR; lack of diagnostic facilities; limited availability of alternatives to antibiotics as a means to establish antibiotic-free farms; and insufficient training of veterinarians on AMR and antibiotic prescribing (Lekagul et al., 2021). Low- and middle-income countries (LMICs), including those in South East Asia (SEA) are crucial in the global response to AMR due to their diverse medicine regulation and access to antimicrobials. Furthermore, folk conceptions or unexpected practices around antimicrobials in these regions can complicate AMR communication efforts and entail unforeseen consequences (Haenssgen et al., 2019). Nevertheless, it is encouraging that countries such as Denmark and the Netherlands that both have massive pig production in recent years have achieved tremendous reductions in antimicrobial usage while sustaining peak production (Dall, 2019). Comparable results have been accomplished in Belgium, France, Sweden, and the United Kingdom (More, 2020). In Brazil, one of the biggest pork producers in the world, the increased demand for meat production has led to ongoing environmental problems, such as soil and water contamination by pathogenic and/or resistant microorganisms (Silva et al., 2015; Brisola et al., 2019). In Russia, relatively high levels of MDR Escherichia coli that are resistant to critically important antimicrobials such as colistin, cefotaxime, and ciprofloxacin have been recorded (Makarov et al., 2020). A study in New Zealand also found that the widespread use of oral antimicrobials within pig production was a significant risk factor for development of AMR and MDR E. coli (Riley et al., 2020). The rate of antibiotic resistance differs considerably from country to country, depending upon the amount of usage. In the EU, the lowest levels of AMR E. coli isolates were found in countries where lower antimicrobial usage was practiced, such as in Norway, Sweden, and Finland, whereas countries with high levels of use such as Spain, Portugal, and Belgium had relatively higher levels of AMR E. coli (Holmer et al., 2019).
Nosocomial infections or healthcare-associated infections fall under the political concerns of the World Health Organization (WHO). Nosocomial infections account for 7 and 10% of all infections in developed and developing countries, respectively (Khan et al., 2017; World Health Organization [WHO], 2019). Enterobacteriaceae such as Escherichia coli and Klebsiella pneumoniae have been identified as the most common resistant bacteria found in healthcare-associated infections (Malchione et al., 2019; Shrestha et al., 2019). The possibility that AMR colonization in humans can be related to livestock and environmental sources was shown in a recently published One Health AMR risk assessment (Opatowski et al., 2020). WHO emphasizes the importance of antimicrobial classes (agents) that are used in humans being targeted for antimicrobial susceptibility surveillance in livestock, including aminoglycosides (gentamicin), carbapenems (meropenem), cephalosporins (3rd, 4th, and 5th generation: ceftriaxone, cefepime, ceftaroline, ceftobiprole), glycopeptides (vancomycin), amoxicillin-clavulanic-acid, polymyxins (colistin), and quinolones (ciprofloxacin). Due to high prevalence and incidence of AMR rates in the pig industries in South-East Asian (ASEAN) countries, a regional database of AMR surveillance has been developed (Nguyen N. T. et al., 2016; Nhung et al., 2016; Lugsomya et al., 2018a, b; Mobasseri et al., 2019; Vounba et al., 2019; Khine et al., 2020; World Health Organization [WHO], 2020; Dawangpa et al., 2021). It is important to emphasize that SEA is a diverse region, where different levels of development, customs, laws, and regulations among different countries in the region. Pork is one of the most common protein sources and is widely produced for local consumption, with some export also occurring. Hence it is an important target for AMR surveillance.
Antimicrobial resistance surveillance through the pig production cycle, covering from the neonatal to the slaughtering periods has demonstrated the possibility of transmission of AMR bacteria in the food chain, and has provided a modicum of comparative evidence for nosocomial infection through genetic fingerprinting of isolates using multi-locus sequence typing (MLST) and whole-genome sequencing (Mather et al., 2018). A plasmid carrying mcr-1 conferring colistin resistance was first identified in China in 2014 (Liu et al., 2016). To date, up to 10 types of the mcr gene family harbored in Enterobacteriaceae have been reported in clinical samples and in animals and human carriers (Xu et al., 2021). Colistin has been extensively used in pig production, often as a growth promoter until the 2017 prohibition of its use under regional policy. However, a high prevalence of colistin-resistant Klebsiella pneumoniae was found in healthy people living in rural communities in Thailand, Vietnam, and Laos with no history of colistin therapy (Olaitan et al., 2014; Yamaguchi et al., 2020). A 10–22% prevalence of the mcr-1 and mcr-3 genes has been reported in E. coli isolated from pig and chicken farms in Vietnam and Thailand (Malhotra-Kumar et al., 2016; Khine et al., 2020) and in Klebsiella pneumoniae from farmed pigs in Malaysia (Mobasseri et al., 2019). The clonal similarity of a colistin-resistant E. coli isolate from a local pig with a human isolate was demonstrated in Laos (Olaitan et al., 2015). Overall, these occurrences confirm the interchangeability of colistin-resistant bacteria between humans and farm animals that use colistin as a feed additive. Cross-species transmission is a suspected course of distribution.
Similar clonal types of multidrug-resistant Salmonella Typhimurium and E. coli have been found in humans and pigs in SEA and in the United States (Oloya et al., 2009; Nhung et al., 2016; Mather et al., 2018; Sudatip et al., 2021). Pigs and the environment can be potential sources for the dissemination of AMR to humans; however, there have been a few longitudinal studies monitoring linkages of AMR strains among pigs, the environment, and consumers/patients. Genetic similarity between bacterial clones from farmed pigs and pork was not found in any comprehensive monitoring study in Thailand (Lugsomya et al., 2018b). To our knowledge, to date there has been no unequivocal direct evidence of transmission of AMR bacteria to human patients from farms or the slaughtering process in SEA.
This article aims to improve awareness and understanding of AMR through a description of the occurrence of AMR in pig farm and food chains under AMR management systems in SEA: this includes a description of standard farming practices; detection of AMR and clonal analysis of bacteria; AMR in the food chain, wastewater, and associated environments, the possibilities of AMR transfer from farming and slaughtering practices to humans. Finally, the possibility of reducing AMR by using alternatives to antibiotics and improving policies are discussed.
Standard Farming Systems in Sea
Asia is the largest producer of pigs in the world (58.4%), and within SEA the top three pig producing countries are Myanmar, Vietnam, and the Philippines. In contrast, Thailand exports the most pork raised under a high standard of farming and food safety conditions (FAOSTAT, 2019). Pig production systems in SEA vary greatly, from backyard pigs held in small-scale often peri-urban farms to semi-commercial units and large intensive units. For example, 80% of pig farms in Cambodia involve smallholders, while 80% in Thailand are intensive farms. However, even pig farming in peri-urban areas operates under a high standard of biosecurity similar to that in large commercial approved farms (World Organization of Animal Health, 2020).
According to the ASEAN Good Animal Husbandry Practice Guidelines (ASEAN GAHP), pig farms should be located in areas with an appropriate clean water source, with physical barriers to reduce the risk of contamination arising from visitors and animal access. Quarantine pens should be located in a separate area and must be prepared for sick pigs and pig carcass to limit pathogen spread. Housing and equipment providing good hygiene and ventilation with easy-to-clean pig manure systems are recommended, along with good personal hygiene management for workers, including a pre-entry farm protocol which includes facilities for showering and hair washing, and changing into protective farm clothes and footwear before accessing the animal housing. Wastewater and manure should be kept in a closed area well away from the animal house, and the farm should possess a treatment system that limits odor and hazards such as biogas. Moreover, a veterinarian must authorize a program of vaccination and antimicrobial and chemical use for disease prevention and treatment. Vaccination is a potential method to prevent certain infectious diseases and is established as an important alternative to antibiotics (Founou et al., 2016). In many countries the governments produce and promote vaccines against the Foot and Mouth Diseases virus and Classical Swine Fever virus, as these are very important pathogens. While commercial and autogenous vaccines against pathogenic bacteria are alternatives to control infections, their efficacy varies considerably. To date, vaccines against bacteria such as E. coli, Lawsonia intracellularis, and Actinobacillus pleuropneumonia have been commercialized and used worldwide (FAO, 2014; Founou et al., 2016).
In 2013 the total amount of antimicrobials used for food-producing animal in SEA was 2,950 tons: antimicrobial classes included penicillin (666 tons), tetracyclines (484 tons), quinolones (321 tons), sulfonamides (317 tons) and macrolides (281 tons). Thailand consumed the largest amount of antimicrobials (531 tons in 2013), and the amount has subsequently increased (3,816.3 tons in 2018), probably due to the growing number of food-producing animals (Van Boeckel et al., 2017; Ministry of Public Health, 2020). In practice, antimicrobials are used for three indications: therapy, prevention, and growth promotion. Therapeutic use involves a short duration of treatment with a high dose of antimicrobial given only to sick pigs. Prophylactic administration is used to prevent disease occurring where it is routinely expected to occur (e.g., to control diarrhea after weaning), and the same amount and duration of administration is applied to all susceptible pigs during this period. Lastly, growth promotor use involves administering a lower than recommended therapeutic dose, long-term for all pigs on-farm (FAO, 2014). The intention is to improve growth rate by controlling subclinical infections and improving digestive efficacy. Antimicrobial growth promotion use has been banned in Thailand and Vietnam (Coyne et al., 2019), while Indonesia, Myanmar, and Timor-Leste policies promote a decrease in antimicrobial use in response to WHO announcements (Cardinal et al., 2020). Furthermore, the use of colistin as a feed additive has been restricted in Malaysia and Thailand (Olaitan et al., 2021), and nitrofuran has been banned in the Philippines and Thailand (Islam et al., 2014).
Implementation of standard biosecurity and management systems in pig farms is an essential part of veterinary preventive medicine, and promotes improved pig production, and reduced antimicrobial consumption in pigs, leading to fewer AMR bacteria on pig farms which otherwise might enter the food chain (Founou et al., 2016).
Antimicrobial Usage and Relationship to Antimicrobial Resistance
The key driver of resistance in bacterial population is considered to be the extensive use of antimicrobial agents which created a selection pressure on susceptible bacteria (Aarestrup et al., 2008; Koningstein et al., 2010; Lunha et al., 2020). Frequency, amount, duration, and combined use of antimicrobial agents are important factors related to the emergence of both ARB and MDR bacteria (McEwen, 2006; Martinez and Baquero, 2009). Additionally, there have been many reports on a positive association between the amount of antimicrobial use and the prevalence of ARB in both animals and human (Lazarus et al., 2015). A recent study from Thailand reported the detection of high frequencies of ARB isolates from pigs, particularly resistance against tetracycline, which is also reported as an antimicrobial agent that has had long term and extensive use in livestock both in Thailand and throughout SEA (Van Boeckel et al., 2017; Lunha et al., 2020). Nevertheless, it is very important that monitoring of AMU using standardized systems for monitoring programs for AMR data is continued, and that the quantification and temporal trends in AMU are continued to be recorded in order to obtain evidence between the linkage of AMU and AMR so as to further support policy and decision makers to understand and fight against AMR (Aarestrup, 2005; Magouras et al., 2017).
Detection of AMR and Clonal Analysis
At present, the main approaches to detect antibiotic resistance bacteria (ARB) and antibiotic resistance genes (ARG) from bacteria in hosts and environments are primarily categorized into the conventional culture-based methods, and molecular biology-based methods. Clone typing by core-gene multilocus sequence typing (MLST) is used to determine the molecular epidemiology of bacteria in the family Enterobacteriaceae.
Conventional Culture-Based Methods
This approach requires isolating and growing the microorganisms of interest on a nutrient medium. For samples from wastewaters and associated environments the membrane filtration method is frequently applied along with selective media supplemented with the antibiotics of interest for isolation, and enumeration of individual ARB (Rizzo et al., 2013). The antibiotic resistance pattern of individual pure isolates can be further investigated by performing antibiotic susceptibility testing, which depends on the phenotypic traits of individual isolates. The frequently applied resistance susceptibility testing methods are of four types, including diffusion methods (Stokes method and Kirby-Bauer method), dilution methods (Minimum inhibitory concentration such as broth and agar dilution), diffusion and dilution methods (E-test method and gradient diffusion method), and automated instrument methods (such as Vitek 2 system by bioMérieux and Sensititre ARIS 2X by Trek Diagnostic Systems) (Jorgensen and Ferraro, 2009; McLain et al., 2016; Benkova et al., 2020). The results from antibiotic susceptibility testing should be interpreted following standardized values, which allow qualitative assessment distinction between susceptible, intermediate, and resistant isolates (Clinical & Laboratory Standards Institute: CLSI Guidelines; EUCAST: AST of bacteria; Jorgensen and Ferraro, 2009).
Nevertheless, the conventional culture-based methods are time-consuming and somewhat labor-intensive. Investigators are required to constantly attain a reliable and up-to-date interpretation of the resistance susceptibility testing. Besides, only a minority of bacteria are culturable and suitable for the conventional culture-based methods, which are presumed to be only a tiny fraction of the total bacteria present (Kummerer, 2004; Bouki et al., 2013; Rizzo et al., 2013).
Molecular Biology-Based Methods
This approach allows the identification of deoxyribonucleic acid (DNA) targets with and without the requirement to culture individual isolates. Methods such as Polymerase Chain Reaction (PCR) amplification and use of DNA probes are well known for detecting resistance genes and genetic elements. With the application of multiplex PCR, various ARGs can be screened and detected simultaneously (Luby et al., 2016; Anjum et al., 2017; Preena et al., 2020). In comparison, quantitative PCR (qPCR) has been applied as a quantifying method for tracking and tracing ARGs from various pathogenic bacteria in different environments from municipal wastewater to surface water. However, the nucleic acid extraction method and its product quality are critical for PCR gene detection, since wastewater and sludge samples contain much indeterminate substance (Obst et al., 2006; Lupo et al., 2012; Rizzo et al., 2013). Additionally, several typing methods are used for determining and characterizing the diversity of resistant strains and clones of individual isolates, such as pulsed-field gel electrophoresis (PFGE), restriction fragment length polymorphism (RFLP), random amplified polymorphic DNA (RAPD), and amplified ribosomal DNA restriction analysis (ARDRA) (Hamelin et al., 2006; Faria et al., 2009; Li et al., 2009; Brooks et al., 2014; Papadopoulos et al., 2016). Unfortunately, there is limited availability of specific primer sets, especially for PCR and qPCR methods. These molecular techniques often encounter an absence of standardization and validation of interlaboratory tests (Rizzo et al., 2013).
Most recently, molecular high throughput (HPT) and next-generation sequencing (metagenomic analysis, transcriptome, and resistome) are among the techniques that facilitate studies of the microbial community and their diverse genomes and gene expression without a requirement for culture. Moreover, these techniques are very valuable since they can compute numerous target genes without knowledge of the bacteria or genes in the samples, are not limited by specific primer sets, and are even faster to perform than the conventional molecular methods such as PCR and qPCR (Rizzo et al., 2013; Limayem et al., 2019; Lira et al., 2020). They also provide a deeper insight into antibiotic resistance conditions related to both vertical and horizontal gene transfer occurrences (Von Wintersdorff et al., 2016).
Therefore, molecular biology-based methods make possible the detection of ARB and ARG that cannot be cultured and express in individual isolates and provide faster detection of ARB and ARG with high specificity and sensitivity compared with conventional culture-based methods. Nevertheless, these methods cannot separate between living and non-living microorganisms. They are also less useful for identifying ARB and resistance patterns compared to the conventional culture-based methods.
Clonal Analysis of AMR Strains
Bacterial strains identified using DNA fingerprinting techniques can be assigned as clones representing the ancestor’s phylogenetic root tree. In general, bacteria are highly diverse genetically, even at the species-level, because of inter-and intraspecies gene interchange leading to diversification of genotype in a particular niche of a bacterial clone (a clonal complex). For AMR, clonal relations are used as a tool to indicate the phenomenon of bacterial transmission or “spillover” between sources: humans to animals, animals to the environment, or environment to humans or vice versa (Spratt, 2004). The diversification of bacterial clones depends on the extent of gene recombination and varies by bacterial type. Some species, e.g., Salmonella enterica members, are stable clones, whereas in some species, e.g., Helicobacter pylori, these may be transient or genetical diverse without phenotypical change (Selander et al., 1990; Kennemann et al., 2011). Index genetic parameters are used to assign the clones and clonal complexes. In the 1980s to 2000s, the best results were achieved using pulsed-field gel electrophoresis and multilocus sequence typing (Schwartz and Cantor, 1984; Maiden et al., 1998). Since the Whole Genome Sequencing (WGS) era, WGS-based strain typing has been increasingly used to analyze bacterial pathogens and AMR bacteria in the public health field and for implementing disease control strategies (Snitkin et al., 2012; Harris et al., 2013; Walker et al., 2013). Core genome MLST (cgMLST) has become a standard tool for WGS-based strain typing as it has high accuracy and epidemiological concordance. Outcomes from cgMLST are analyzed by comparison of shared common genes sets from genomes within the same species, which can be used to determine the source and routes of transmission, trace cross-contamination of healthcare-associated pathogens, and identify antibiotic-resistant lineages or subpopulations (Dekker and Frank, 2016; Mellmann et al., 2016; Nadon et al., 2017). From each species-specific cgMLST scheme, there is the joint-calculated parameter called the “relatedness threshold.” If cgMLST allelic mismatches between two bacterial stains are less than (or equal to) the relatedness threshold, there are clonal relations between them (Miro et al., 2020). The relatedness threshold is calculated using an algorithm in the Ridom SeqSphere + online package (Calibrating the cgMLST Complex Type Threshold1). The relatedness threshold of clonality from cgMLST diversity depends on insight of species-specific population genetics (i.e., the relative impact of mutation and recombination on genetic variation) (Schurch et al., 2018), as shown in Table 1.
For clonal relationships, the horizontal genetic transmission of mobile genetic elements (plasmids, integrative conjugative elements, transposons) and their relatedness are important parameters indicating the transmission of AMR characteristics (Conlan et al., 2014, 2016; Sheppard et al., 2016). There are not many reports of clonal typing during AMR surveillance in a One Health approach. Epidemiological data and sources of bacterial strains defining clonal relationships in Thailand and Vietnam are shown in Table 2; however, insights into bacterial clonal transmission from the production process to consumers and patients remain incomplete.
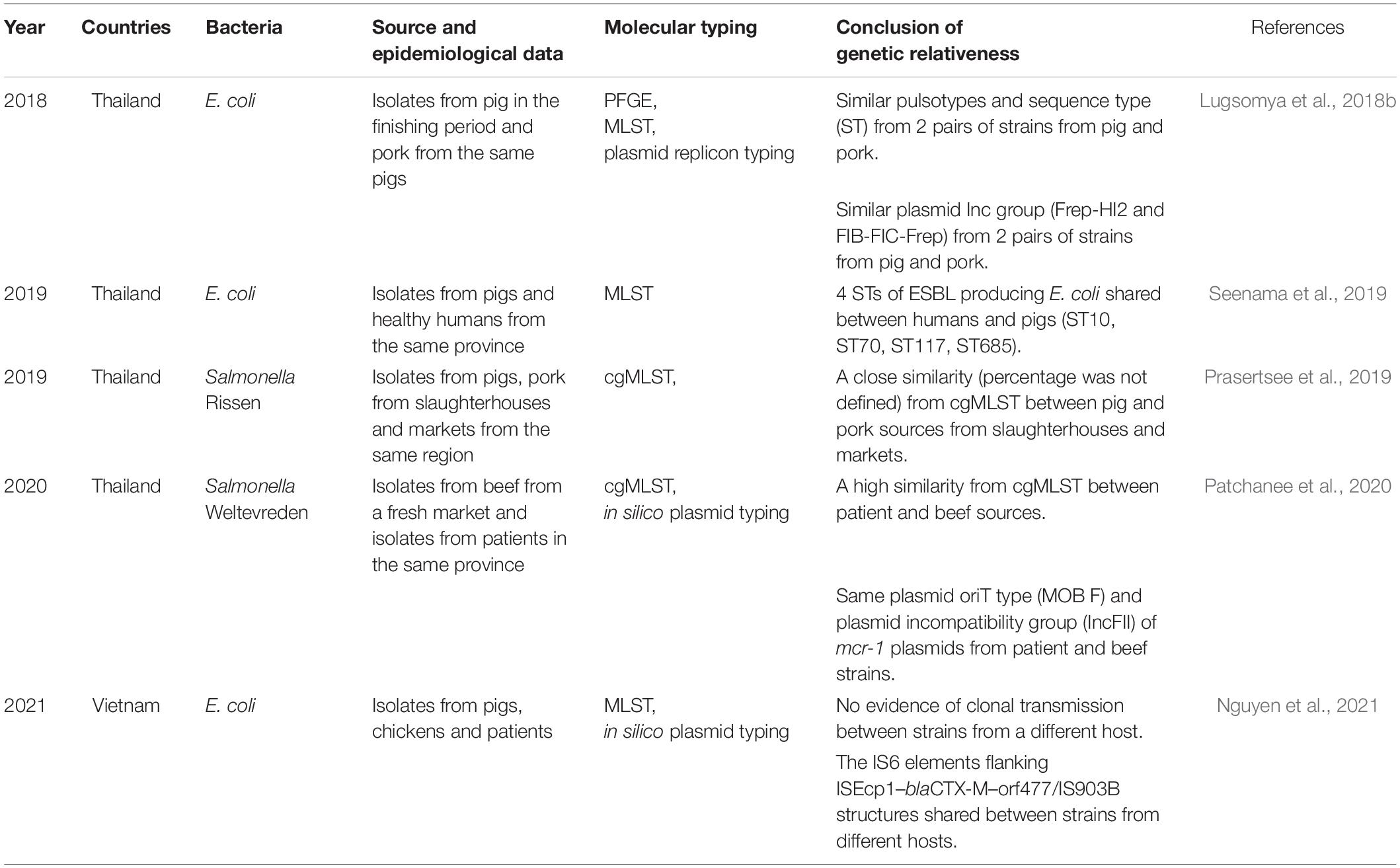
Table 2. Lists of clonal relationships of AMR strains from South East Asia Countries in the One Health paradigm.
Possibility of AMR Transfer From Pigs to Humans
Humans can be exposed to antibiotic-resistant bacteria, both pathogenic and commensal bacteria, by direct transmission or through food or the environment. AMR problems are global issues since resistant bacteria can occur from any sector and spread intra-species, inter-species, and across borders (Kempf et al., 2013; Daniel et al., 2015). Certain antibiotics, particularly the Critically Important Antimicrobials to human medicine are applied in livestock farming. Inappropriate use of antibiotics in animals accelerates selective pressure and is a cause of antimicrobial cross-resistance in human medicine (Marshall and Levy, 2011). Moreover, the genes encoding antibiotic resistance in bacteria from animals can be transferred to bacteria that are pathogenic to humans (Archawakulathep et al., 2014). Many studies have reported the emergence of ARB from pigs and spread to humans; for example, quinolone resistant Salmonella spreading from pigs to human in Taiwan, and nourseothricin resistant E. coli from pigs to pig farmers, families, municipal communities, and patients with urinary tract infections (Witte, 1998; Aarestrup, 2005). Since livestock farms are a hotspot for resistant bacteria, workers on farms and in the food chain are potentially exposed to AMR bacteria from animals.
AMR in Livestock
The AMR situation is critical in LMICs, including in the SEA. Although there is social and economic progress in those regions, the average population is still in poverty (Singh, 2017). Moreover, inadequate sanitation, poor quality control in slaughterhouses, and conditions where people live adjacent to animals (backyard farming) accelerate the spreading of resistant pathogens and ARG (Cook et al., 2017). Self-medication is cheaper in those regions, and antibiotics are easily accessible over the counter, leading to an increase in antimicrobial use not only by farmers but also by people without proper diagnosis (Nguyen et al., 2013). The major potential pathways for spreading of resistant bacteria and/or genes between food animals and humans could be horizontal transmission or clonal transfer of resistant bacteria (Lipsitch et al., 2002; Chang et al., 2015). An association between clonal types of human cephalosporin-resistant E. coli with those from food animals linked through food products has been reported (Lazarus et al., 2015).
Additionally, a study in Denmark found the possible transmission of sulfonamide resistant E. coli via the food chain to a healthy human who did not receive antibiotic therapy (Hammerum et al., 2006; Wu et al., 2010). The occurrence of resistant bacteria carrying mobile genetic elements in livestock or food of animal origins is concerning. The spread of antibiotic-resistance genes via horizontal gene transmission could lead to acquisition of multidrug-resistant (MDR) bacteria. MDR infections in humans can cause treatment failure and high mortality, especially if resistance is to a drug of last resort. Worrisomely, there are growing cases of colistin-resistant Enterobacteriaceae and sporadic cases of carbapenem-resistant cases in pigs from some Asian countries.
A study in Vietnam found that almost half of the pig farms in Bac Ninh province had colistin-resistant E. coli, and farmworkers also harbored such bacteria (Dang et al., 2020). Moreover, colistin-resistant E. coli have been detected in humans from rural areas of Vietnam (5–71.4%) (Trung et al., 2017; Kawahara et al., 2019). The mcr genes are currently globally distributed and are mostly detected from livestock origins followed by from humans and meat products. In SEA, mcr-1 positive E. coli has been detected in Cambodia, Vietnam, Malaysia, Singapore, and Thailand (Olaitan et al., 2016; Paveenkittiporn et al., 2017; Runcharoen et al., 2017; Srijan et al., 2018). On the other hand, K. pneumoniae isolates harboring mcr-1 genes have been reported in Laos and Singapore (Rolain et al., 2016; Teo et al., 2016). Moreover, ESBL-producing K. pneumoniae clones ST307, ST2958 and ST2959 that were resistant to β-lactams, aminoglycosides, fluoroquinolones, macrolide, lincosamide and streptogramins, rifampicin, sulfonamides, trimethoprim, phenicols and tetracycline, have been observed to be shared between both pig and human-sources within and across abattoirs. K. pneumoniae has been suggested to be a potential unheeded reservoir of resistance in the food chain that impacts on public health (Founou et al., 2018).
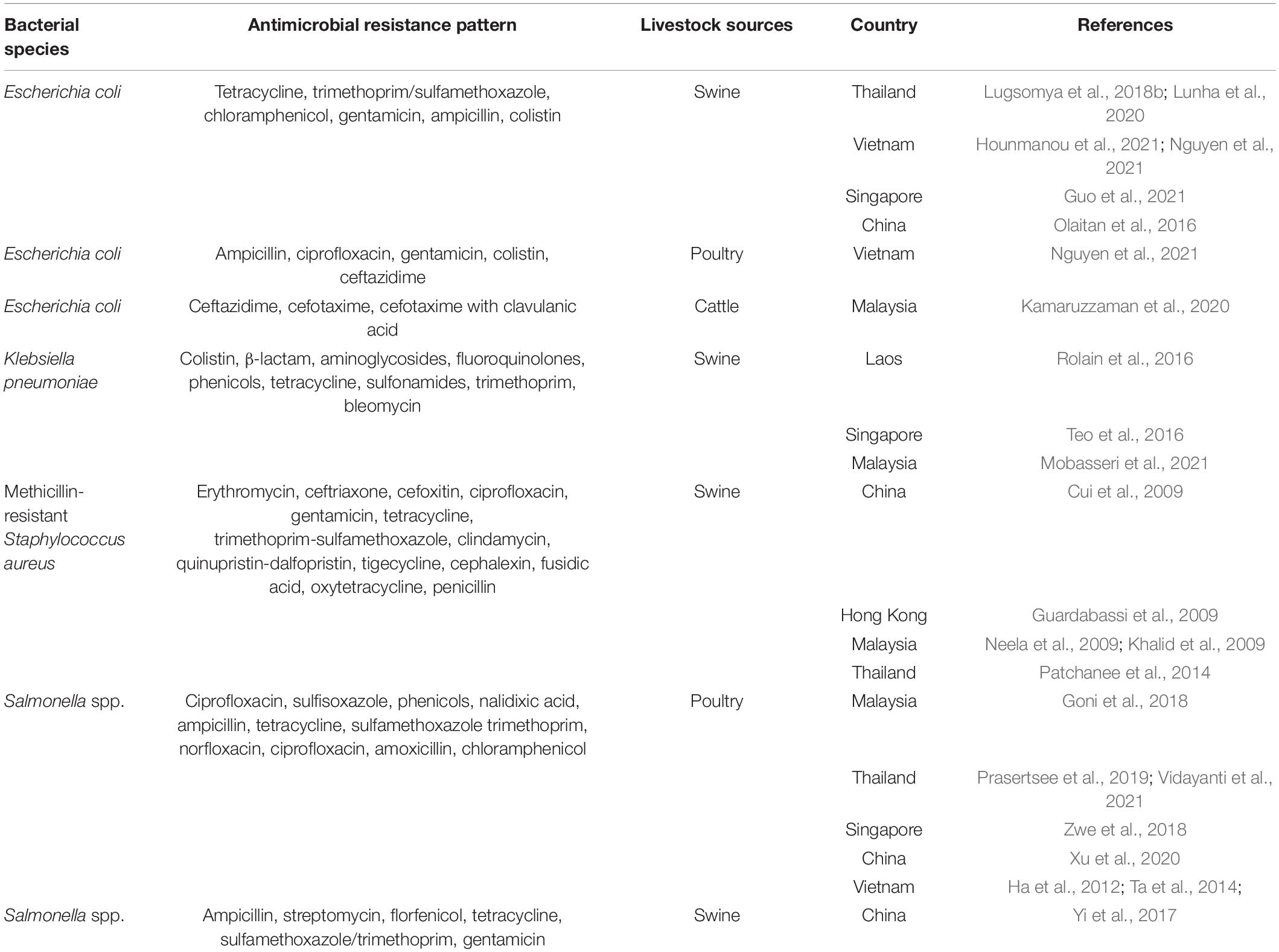
In addition to enteric bacteria, livestock-associated methicillin resistant Staphylococcus aureus (LA-MRSA) is another resistant bacterium of substantial health concern. LA-MRSA has been found worldwide, predominantly among people involved in livestock farming (Smith et al., 2009; Frana et al., 2013). These bacteria can be transmitted to humans in proximity to MRSA-harboring animal skin, especially from pigs (Smith and Pearson, 2011). There are several reports regarding the occurrence of MRSA cases in Asian countries, including in Malaysia (5.5%), China (15%), and Northern Thailand (2.53%) (Patchanee et al., 2014). The spa type t4358-SCCmec V is the most abundant clone in Malaysia, whilst it is t337-SCCmec IX in Thailand. In northeastern Thailand patients were identified whose MRSA pulso type pattern (PFGE) was related to that of MRSA discovered in a diseased pig in the same area (Lulitanond et al., 2013). A similar case providing evidence for the potential for transmitting resistant clones from pigs to humans occurred where an isolate from a human and two isolates from pigs shared SCCmec IX-S. haemolyticus (Sinlapasorn et al., 2015). The occurrence of livestock associated ARB in SEA is shown in Table 3.
Unfortunately, the role of farm animals in the emergence and dissemination of resistant bacteria or genes to humans is still controversial (Marshall and Levy, 2011). Further studies focusing on genomic data of resistant bacteria and nation-wide epidemiological approaches are required to explore the transmission of AMR from animals to humans.
AMR in the Food Chain
As discussed, possible transmission routes of ARB and ARGs to humans may be from direct contact with colonized animals or from the consumption of foodstuffs that are contaminated with ARB/ARGs along the food chain (Founou et al., 2016; Thapa et al., 2020). There are different potential pathways for contamination of ARB and ARGs along the food chain. ARB may disseminate to crops and plants from the utilization of contaminated irrigation water, mostly from manure discharges of both human and animals and from irrigated soil (Gatica and Cytryn, 2013). Whilst livestock products such as pork, beef and poultry can be contaminated with ARB during the slaughtering processes, aquaculture products may become contaminate with ARB and ARGs that have already been circulating in aquatic environments (Bridier et al., 2019; Amarasiri et al., 2020). The phenotype of AMR bacteria may be influenced by food preservation stresses such as decreasing pH to under 5.0, or increasing salt to more than 4.5%, which has been shown to increase phenotypic antibiotic resistance in E. coli, S. enterica serovar Typhimurium, and S. aureus (McMahon et al., 2007). In addition, contamination by ARB and ARGs may occur during transportation and distribution of food products, or occur as a post-contamination event after food processing, or from a cross-contamination as a result of inappropriate food handlings which are among the most common contamination pathways along the food chain (Verraes et al., 2013; Holzel et al., 2018).
AMR in Wastewater and Associated Environments
Alarmingly, there have been increasing reports about the existence of AMR in the environment, especially resulting from the activities of both humans and animal industries (Prestinaci et al., 2015). The concern focuses mainly on the increase in ARB and ARG resulting from the dissemination of antibiotics in the environment (Kraemer et al., 2019; Serweciñska, 2020). Wastewater has been suspected as being one of the hotspots for the spread and circulation of ARB and ARG (Rizzo et al., 2013; Pazda et al., 2019). Human and animal excreta have been demonstrated to contain large amount of antibiotics which are distributed to municipal wastewater where the antibiotics are absorbed into sewage sludge (Nagulapally et al., 2009; Singer et al., 2016). Sludge (biosolids) is produced from the wastewater treatment processes in the wastewater treatment plant, and contains a large amount of non-degraded fat, oil, and protein. This composition contributes to poor retrieval of water-soluble antimicrobial agents such as ciprofloxacin from sludge (Gerba and Pepper, 2009; Singer et al., 2016; Serweciñska, 2020). Therefore, it is no surprise that ARB and ARG have been detected mainly in samples from wastewater and associated environments such as groundwater and surface water (Barancheshme and Munir, 2018; Amarasiri et al., 2020; Sabri et al., 2020).
AMR in the Slaughtering Process
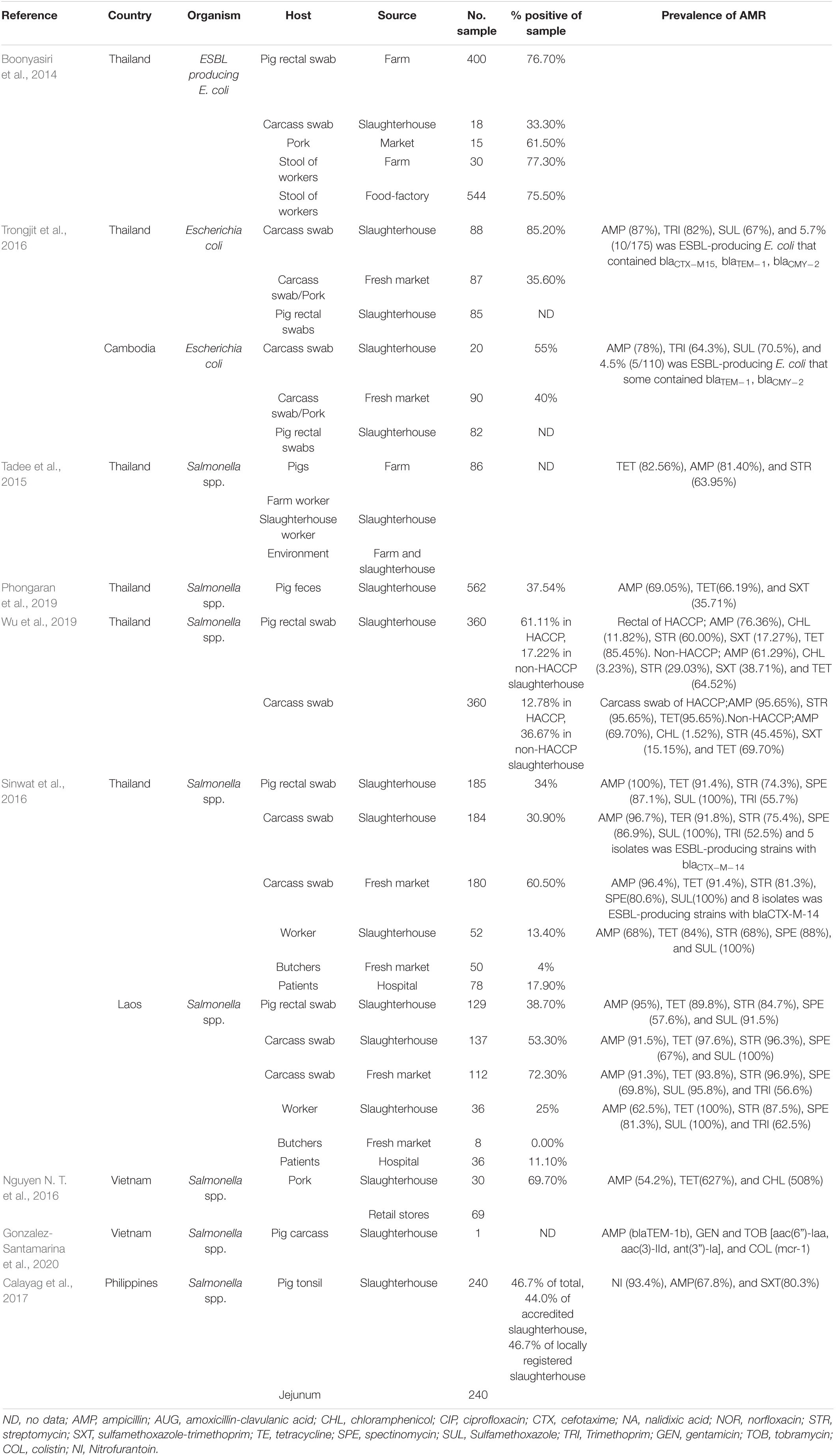
The risk of AMR bacterial contamination in the food chain is associated with AMR commensal bacteria circulating on carcasses at slaughterhouses (EFSA, 2016). In addition, the direct exposure of workers to resistant bacteria can happen throughout the production cycle to the slaughtering process and during food preparation (You et al., 2016). The exposure of slaughterhouse workers to extended spectrum beta lactamase (ESBL)-producing Enterobacteriaceae has received global attention (Wadepohl et al., 2020). Substantial data relating to the association between ESBL producing Enterobacteriaceae in pigs, farmworkers and slaughterhouse workers has been reviewed (Alali et al., 2008; Hammerum et al., 2014; Huijbers et al., 2014; Dohmen et al., 2015). In the Netherlands, workers were most likely to be exposed to ESBL producing E. coli with bla CTX–M–1 in the early slaughtering process (before chilling of carcasses), at a prevalence of 4.8% (Dohmen et al., 2017). In comparison, a 4.5% prevalence of ESBL/AmpC producers was found in residents living in the vicinity of livestock farms in the Netherlands (Wielders et al., 2017). Exposure to ESBL carriage is especially high in the slaughtering process, particularly during removal of visceral organs and handling of the throat area where there is massive colonization with enteric bacteria (Lowe et al., 2011; Lăpuşan et al., 2012). Moreover, a high percentage (50%, n = 59) of MDR E. coli have been recovered from slaughterhouse workers, where the rates in Nigeria were more elevated in butchers than in cleaners (Aworh et al., 2021). In slaughterhouses in Thailand, ESBL-producing E. coli were detected in 76.7% of pig feces, 33.3% of fresh pork samples, and 75% of workers (Boonyasiri et al., 2014). In a comparative study between Thailand and Cambodia, the prevalence of ESBL-producing E. coli in pig rectal and carcass swabs were similar, at only 5.3–5.6%. Among these, ESBL-producing E. coli containing blaTEM–1 and blaCMY–2 were found in both countries, but the blaCTX–M–15 gene was detected only in the Thai isolates (Trongjit et al., 2016).
The occurrence of multidrug-resistance bacteria circulating in slaughterhouses also has been reporting in many studies in SEA. Salmonella spp. are frequently used as an indicator of pathogen and AMR carriage, reflecting management standards. In Thailand, salmonella isolates with high levels of resistance to tetracycline, ampicillin, sulfonamide-trimethoprim, and streptomycin, at approximately 63–82%, can be found in pigs, workers, and the environment from farms to slaughterhouses (Tadee et al., 2015; Phongaran et al., 2019). Salmonella Rissen was found to be a common serovar in a comprehensive survey of pig farms, slaughterhouses, and retail outlets in Northern Thailand (Phongaran et al., 2019). Moreover, Wu et al. (2019) compared AMR salmonella contamination in pig rectal swabs and carcasses between slaughterhouses with and without a hazard analysis critical control point (HACCP) system. Both systems had a high level of indirect contamination, but the HACCP system decreased salmonella contamination in carcasses and reduced the number of AMR patterns (Wu et al., 2019). In a comparison between pig slaughterhouses in Thailand and Laos, the prevalence of salmonella contamination of pig carcasses in the two countries were 30.9 and 53.3%, respectively. Salmonella spp. from both countries were highly resistant to sulfonamide (98.3%), ampicillin (91%), and tetracycline (92.5%). These isolates contained class1 integrons carrying resistance cassette dfrA12-aadA2. Interestingly, five Salmonella isolates from Thai pig carcasses were producers of extended-spectrum □-lactamase (ESBL) and harbored blaCTX–M–14, whereas isolations from Laos were negative. The study reflected differences in the hygienic procedures of slaughterhouses and antibiotics uses in the two countries (Sinwat et al., 2016). In Vietnam, salmonella isolated from pork slaughterhouses and retail shops were highly resistant to ampicillin, tetracycline, and chloramphenicol (Nguyen D. T. et al., 2016), and Salmonella Rissen isolated from a pig carcass in 2013 showed phenotypical colistin resistance, with a plasmid-mediated mcr-1 gene (Gonzalez-Santamarina et al., 2020). In the Philippines, 46.7% of pig tonsil and jejunum samples from three below-standard slaughterhouses were positive for Salmonella enterica, rates higher than those from five national accredited slaughterhouses, although there was no significant difference in prevalence. Most isolates (n = 183) were resistant to nitrofurantoin (93.4%), ampicillin (67.8%), and sulfa-trimethoprim (80.3%) (Calayag et al., 2017), even though nitrofurantoin use in livestock had been banned for a decade. However, it needs to be determined whether the source of contamination was from farming practices or occurred in the slaughtering process. The occurrence of ARB in the slaughtering process is summarized in Table 4.
Overall, the risk of bacterial contamination in the slaughtering process and of pig products is critical, and there is still a lack of comprehensive monitoring throughout the system. Operation under HACCP programs and good manufacturing and sanitation practice (GMP) are intended to control meat safety (Samelis, 2006; Wu et al., 2019), but a reduction in salmonella and AMR bacteria prevalence is not relevant to its implementation in SEA. Decontamination applications post-slaughter aim to reduce bacterial contamination in pork, and include physical applications such as hot water washing, chilling, freezing, and high-pressure processing. Secondly, chemical applications such as treatment with organic acid solutions, or electrolyzed or ozonated water are recommended. Lastly, biological applications include bacteriophages and wrapping film with postbiotics containing lactic acid bacteria. However, all applications must take into account meat quality and the physiology of the end product (Bolumar et al., 2020; Li et al., 2020; Shafipour Yordshahi et al., 2020; Aydin Demirarslan et al., 2021).
Alternatives to Antibiotics
The use of antibiotics on farms increase the possibility of the AMR transmission from livestock production systems to consumers. There have been many attempts to replace antibiotic use on farms. This review outlines two applications for reduction of antibiotic use, including for growth promotion and disease prevention. An overview of possible antibiotic alternatives for growth promotion derived from previous studies is presented in Supplementary Table 1 (Cha et al., 2012; Kim et al., 2014; Yoon et al., 2014; Zeng et al., 2014; Devi et al., 2015; Gois et al., 2016; Hossain et al., 2016; Lee et al., 2016; Peng et al., 2016; Sbardella et al., 2016; Tang et al., 2016; Upadhaya et al., 2016; Wan et al., 2016; Lan et al., 2017; Lei et al., 2017; Ma et al., 2017; Pan et al., 2017; Wu et al., 2017; Huang et al., 2018; Lei et al., 2018; Samolińska et al., 2018; Sayan et al., 2018; Seo et al., 2018; Xu et al., 2018; Pei et al., 2019; San Andres et al., 2019; Wang et al., 2019; Duarte et al., 2020; Petry et al., 2020; Ren et al., 2020; Satessa et al., 2020; van der Peet-Schwering et al., 2020; Wei et al., 2020; Zhang et al., 2020; Sun et al., 2021; Wang et al., 2021). These vary considerably from chemical compounds, biologically active substances, and microbially derived products. These could perform several functions, including eliminating pathogenic microbes, modulating gut microbial communities, strengthening intestinal integrity, enhancing growth performances, or diminishing morbidity from other causes (Cheng et al., 2014; Liao and Nyachoti, 2017; Zeineldin et al., 2019).
Although these alternatives could be used as antibiotic replacements in terms of in-feed growth promotion, they are not suitable for therapy. In addition, proper disease prevention measures, including vaccination and enhanced biosecurity, are needed to help reduce antibiotic usage. Vaccines trigger a protective immunity that simulates the effects of a natural infection without the negative consequences. Vaccination has been successfully used in animals (Rose and Andraud, 2017). In livestock production, vaccination has been extensively used for disease prevention caused by pathogenic bacteria or viruses, and as such it is an important substitute for antibiotic usage (Meeusen et al., 2007). Remarkably, even protection against viral infections may reduce antibiotic use by decreasing the risk of misdiagnosis and treatment of secondary bacterial infections (Potter et al., 2008). For example, vaccination against Porcine Circovirus Type 2 (PCV-2), which is an immune-suppressive viral infection leading to secondary bacterial infections, resulted in a significant diminution of antibiotic use in swine farms (Raith et al., 2016). Similarly, vaccination against Porcine Reproductive and Respiratory Syndrome (PRRS) virus on pig farms reduced antibiotic consumption (Van Looveren et al., 2015). Amongst important bacterial pathogens of swine, vaccination against Lawsonia intracellularis, which causes severe ileitis, has been reported to reduce oxytetracycline medication in pigs (Bak and Rathkjen, 2009), whilst immunization against the respiratory pathogen Actinobacillus pleuropneumoniae also reduced administration of antibiotics (Kruse et al., 2015).
Biosecurity and farm management are essential parts of disease prevention that can enhance health status and significantly diminish the risk of exposure of pigs to pathogens (Postma et al., 2016b). These practices have been applied in varied species, production systems, and for different pathogens (Dahiya et al., 2006). Practices such as all-in all-out housing systems, reduced stocking rates, increased ventilation and improved waste management all help to improve the health status of farms. Improvements in biosecurity have been widely accepted as an effective tool for protecting from introduction of diseases into farms (Lewerin et al., 2015). Postma et al. (2016a) found that better biosecurity resulted in a lower antibiotic usage from birth to slaughter in pig herds. Similarly, Raasch et al. (2018) and Caekebeke et al. (2020) reported the same relationship between increased biosecurity in pig farms and reduced antibiotic use.
In summary, various means are available to reduce antibiotic use on farms. The addition of some alternatives and improved disease prevention in practice are important tools to reduce antimicrobial usage in swine farm and support global policy. Their increased use in the SEA region is to be encouraged.
Policy and Strategies on AMR in Food Animals in the Association of South East Asian Nation (Asean)2
The importance of AMR as a human security threat has been recognized worldwide since the 1990s. In response the World Health Organization arranged a series of consultative meetings and developed recommendations for action. The culmination of this work was the 2001 WHO global strategy for containment of AMR (Sack et al., 2001). Later, in 2015, the Global Action Plan on AMR (GAP-AMR) was adopted by WHO member states at the World Health Assembly, the Food and Agricultural Organization Governing Conference and the World Assembly of World Organisations for Animal Health (World Health Organization [WHO], 2015). Member States committed to develop multi-sectoral national action plans on AMR and endorsed a ‘One Health’ approach to facilitate collaboration among various sectors and actors in the defense of human, animal, and environmental health. Through global political declarations, a common direction to tackle AMR was seen across countries and sectors (United Nations [UN], 2016).
Most countries in the ASEAN region have taken action to tackle AMR through the development of their National AMR action plan. According to the Global Database for the 2019, Tripartite AMR Country Self-assessment Survey (TrACSS), all eleven countries in the ASEAN region have developed a National action plan (NAP) which the government has approved (World Health Organization [WHO], 2018). The One Health approach has been adopted as a key principle for multi-sectoral coordination, and it was enshrined in all NAPs (Chua et al., 2021). Other policies for controlling and optimizing the use of antimicrobials have been developed through legal provisions and program implementation, including surveillance of AMR and antimicrobial consumption in both humans and the animal sectors.
Reviews of policy interventions that address AMR in food animal production indicate that there are many existing regulations for controlling veterinary medicines, including antimicrobials, in ASEAN countries. However, they are relatively varied across countries (Goutard et al., 2017). Policies of banning antibiotics for growth promotion in food animals have been enforced in some countries, such as Singapore and Thailand (Chua et al., 2021) while others have yet to enforce the ban. Thailand has restricted the use of antimicrobials for growth promotion since 2015 (Sommanustweechai et al., 2018). In 2019, some antimicrobial classes were assigned the highest priority, with critically important antimicrobials for human medicine only being available for food animals through veterinary prescription (notably polymyxins, third and fourth generation cephalosporins, macrolides, and fluoroquinolones) (Scott et al., 2019). However, a few SEA countries, including Vietnam and Myanmar, have neither laws nor regulations on the prescription and sale of antimicrobials for animal use (World Health Organization [WHO], 2018). In terms of antimicrobial stewardship programs, NAPs in most countries in the Region cover both human and animal health sectors, except for Cambodia and Malaysia. Indonesia and Thailand set codes of practices for control of veterinary drug use. Likewise, Brunei has set up national guidelines for prudent use of antimicrobial in livestock (Goutard et al., 2017).
Surveillance on AMR and antimicrobial consumption is a vital component of GAP-AMR. The WHO Global Antimicrobial Resistance and Use Surveillance System (GLASS) and Organisation for Animal Health global database on antimicrobials in animals were established to help provide evidence on the emergence of resistance and trends in antimicrobial use globally (World Health Organization [WHO], 2017). The 2019 TrACSS reported that most countries in the ASEAN region have a national surveillance system for AMR in food animals, with only a few countries such as Laos and Myanmar having inadequate laboratory capacity to perform AMR surveillance. Only Thailand and Malaysia have established systems for monitoring antimicrobial consumption in animals, in which are recorded data on the total quantity of antimicrobials sold for/used in animals nationally, by antimicrobial class, by species, method of administration, and by type of use. Malaysia and Thailand share their reports with the OIE annually (Chua et al., 2021). Thailand has successfully published One Health Reports on AMR, antimicrobial consumption, and public knowledge and awareness on AMR in 2017 and 2018, contributing to an evidence-based policy decision (NSC, 2020a, b). The 2019 version will be launched in April 2021.
Despite existing national commitments, implementation of AMR policy remains a significant challenge, particularly with the monitoring of AMR and AMU in the human and animal sectors. The monitoring systems are hampered by lack of institutional capacity, inadequate investment in human resources, and the need to strength data platforms for routine monitoring. Significant financial boosts are required to support these areas.
Conclusion
This paper addressed the cross-species transmission of AMR and ARGs in the SEA. The importance of pig production in SEA has encouraged particular attention to this industry, but similar problems occur with other animal industries (chicken, fish etc.). Due to the limited number AMR studies, it is difficult to quantify the extent of the problem in the region, although some countries have better data available than others. Certainly, there is reason to believe that transmission of resistant bacteria and ARGs from pigs at the farm and through the slaughtering process provide risks to the human population, either through direct contact or via contaminated environments or pork products. Efforts should be made to reduce antimicrobial use at the farm level by improving farm hygiene and the use of alternatives to antibiotics. The AMR policy in the region has been implemented, although progress in this area has varied considerably between different countries due to different implementation’s capacities. Additional funding to support AMR surveillance and improved control in SEA is required.
Author Contributions
NP: original idea, study design, and supervision of writing. WS: AMR distribution in food chain, wastewater, and detection. PA and PP: alternatives to antibiotics and data analysis. AL: policy and strategy controlling AMR in SEA. IS and NK: risk of AMR in pig production cycle and slaughtering. KL: molecular epidemiology analysis. DH: study design, supervision, and proofreading of the writing. All authors contributed to the article and approved the submitted version.
Funding
The open access fee for this article was paid by Office of Research Affairs, Chulalongkorn University. Ratchadaphisek Sompote Endowment Fund, Chulalongkorn University (CUGR63295301).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are gratefully to the CHE-TRF Senior Research Fund (RTA6280013) and Pathogen Bank, Faculty of Veterinary Science, Chulalongkorn University, Bangkok, Thailand for supporting work undertaken in this area.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.689015/full#supplementary-material
Supplementary Table 1 | Summary of available alternatives to antibiotics in swine production.
Footnotes
- ^ https://www.ridom.de/u/Server_Database_Initialization.html
- ^ The scope of this article covers 11 member countries of the Association of Southeast Asian Nations (ASEAN). They are: Brunei Darussalam, Cambodia, Indonesia, Lao PDR, Malaysia, Myanmar, the Philippines, Singapore, Thailand, and Vietnam.
References
Aarestrup, F. M. (2005). Veterinary drug usage and antimicrobial resistance in bacteria of animal origin. Basic Clin. Pharmacol. Toxicol. 96, 271–281.
Aarestrup, F. M., Wegener, H. C., and Collignon, P. (2008). Resistance of bacteria of the food chain: epidemiology and control strategies. Expert Rev. Anti Infect. Ther. 6, 733–750. doi: 10.1586/14787210.6.5.733
Alali, W. Q., Scott, H. M., Harvey, R. B., Norby, B., Lawhorn, D. B., and Pillai, S. D. (2008). Longitudinal study of antimicrobial resistance among Escherichia coli isolates from integrated multisite cohorts of humans and swine. Appl. Environ. Microbiol. 74, 3672–3681. doi: 10.1128/aem.02624-07
Amarasiri, M., Sano, D., and Suzuki, S. (2020). Understanding human health risks caused by antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARG) in water environments: current knowledge and questions to be answered. Crit. Rev. Environ. Sci. Technol. 50, 2016–2059. doi: 10.1080/10643389.2019.1692611
Anjum, M. F., Zankari, E., and Hasman, H. (2017). Molecular methods for detection of antimicrobial resistance. Microbiol. Spectr. 5, 5–6. doi: 10.1128/microbiolspec.arba-0011-2017
Archawakulathep, A., Kim, C. T. T., Meunsene, D., Handijatno, D., Hassim, H. A., Rovira, H. R., et al. (2014). Perspectives on antimicrobial resistance in livestock and livestock products in ASEAN countries. Thai J. Vet. Med. 44, 5–13.
Aworh, M. K., Abiodun-Adwusi, D., Mba, N., Helwigh, B., and Hendriksen, R. S. (2021). Prevalence and risk factors for faecal carriage of multidrug resistant Escherichia coli among slaughterhouse workers. Sci. Rep. 11:13362. doi: 10.1038/s41598-021-92819-3
Aydin Demirarslan, Ö., Alasalvar, H., and Yildirim, Z. (2021). Biocontrol of Salmonella Enteritidis on chicken meat and skin using lytic SE-P3, P16, P37, and P47 bacteriophages. LWT Food Sci. Technol. 137:110469. doi: 10.1016/j.lwt.2020.110469
Bak, H., and Rathkjen, P. H. (2009). Reduced use of antimicrobials after vaccination of pigs against porcine proliferative enteropathy in a Danish SPF herd. Acta Vet. Scand. 51:1.
Barancheshme, F., and Munir, M. (2018). Strategies to combat antibiotic resistance in the wastewater treatment plants. Front. Microbiol. 8:2603. doi: 10.3389/fmicb.2017.02603
Barroga, T. R. M., Morales, R. G., Benigno, C. C., Castro, S. J. M., Caniban, M. M., Cabullo, M. F. B., et al. (2020). Antimicrobials used in backyard and commercial poultry and swine farms in the philippines: a qualitative pilot study. Front. Vet. Sci. 7:329. doi: 10.3389/fvets.2020.00329
Bartels, M. D., Larner-Svensson, H., Meiniche, H., Kristoffersen, K., Schonning, K., Nielsen, J. B., et al. (2015). Monitoring meticillin resistant Staphylococcus aureus and its spread in Copenhagen, Denmark, 2013, through routine whole genome sequencing. Euro Surveill. 20:21112.
Benkova, M., Soukup, O., and Marek, J. (2020). Antimicrobial susceptibility testing: currently used methods and devices and the near future in clinical practice. J. Appl. Microbiol. 129, 806–822. doi: 10.1111/jam.14704
Bolumar, T., Orlien, V., Bak, K. H., Aganovic, K., Sikes, A., Guyon, C., et al. (2020). “Chapter 10 - High-pressure processing (HPP) of meat products: Impact on quality and applications,” in Present and Future of High Pressure Processing, eds F. J. Barba, C. Tonello-Samson, E. Puértolas, and M. Lavilla (Amsterdam: Elsevier), 221–244. doi: 10.1016/b978-0-12-816405-1.00010-8
Boonyasiri, A., Tangkoskul, T., Seenama, C., Saiyarin, J., Tiengrim, S., and Thamlikitkul, V. (2014). Prevalence of antibiotic resistant bacteria in healthy adults, foods, food animals, and the environment in selected areas in Thailand. Pathog. Glob. Health 108, 235–245. doi: 10.1179/2047773214y.0000000148
Bouki, C., Venieri, D., and Diamadopoulos, E. (2013). Detection and fate of antibiotic resistant bacteria in wastewater treatment plants: a review. Ecotoxicol. Environ. Saf. 91, 1–9. doi: 10.1016/j.ecoenv.2013.01.016
Bridier, A., Le Grandois, P., Moreau, M. H., Prenom, C., Le Roux, A., Feurer, C., et al. (2019). Impact of cleaning and disinfection procedures on microbial ecology and Salmonella antimicrobial resistance in a pig slaughterhouse. Sci. Rep. 9:12947.
Brisola, M. C., Crecencio, R. B., Bitner, D. S., Frigo, A., Rampazzo, L., Stefani, L. M., et al. (2019). Escherichia coli used as a biomarker of antimicrobial resistance in pig farms of Southern Brazil. Sci. Total Environ. 647, 362–368. doi: 10.1016/j.scitotenv.2018.07.438
Brooks, J. P., Adeli, A., and McLaughlin, M. R. (2014). Microbial ecology, bacterial pathogens, and antibiotic resistant genes in swine manure wastewater as influenced by three swine management systems. Water Res. 57, 96–103. doi: 10.1016/j.watres.2014.03.017
Caekebeke, N., Jonquiere, F. J., Ringenier, M., Tobias, T. J., Postma, M., Van Den Hoogen, A., et al. (2020). Comparing farm biosecurity and antimicrobial use in high-antimicrobial-consuming broiler and pig farms in the Belgian–Dutch border region. Front. Vet. Sci. 7:558455. doi: 10.3389/fvets.2020.558455
Calayag, A. M. B., Paclibare, P. A. P., Santos, P. D. M., Bautista, C. A. C., and Rivera, W. L. (2017). Molecular characterization and antimicrobial resistance of Salmonella enterica from swine slaughtered in two different types of Philippine abattoir. Food Microbiol. 65, 51–56. doi: 10.1016/j.fm.2017.01.016
Cardinal, K. M., Pires, P. G. D. S., and Ribeiro, A. M. L. R. (2020). Promotor de crescimento na produção de frangos e suínos. Pubvet 14, 1–6. doi: 10.31533/pubvet.v14n3a532.1-11
Cha, S. B., Yoo, A. N., Lee, W. J., Shin, M. K., Jung, M. H., Shin, S. W., et al. (2012). Effect of bacteriophage in enterotoxigenic Escherichia coli (ETEC) infected pigs. J. Vet. Med. Sci. 74, 1037–1039. doi: 10.1292/jvms.11-0556
Chang, Q., Wang, W., Regev-Yochay, G., Lipsitch, M., and Hanage, W. P. (2015). Antibiotics in agriculture and the risk to human health: How worried should we be? Evol. Appl. 8, 240–247. doi: 10.1111/eva.12185
Cheng, G., Hao, H., Xie, S., Wang, X., Dai, M., Huang, L., et al. (2014). Antibiotic alternatives: The substitution of antibiotics in animal husbandry? Front. Microbiol. 5:217. doi: 10.3389/fmicb.2014.00217
Chua, A. Q., Verma, M., Hsu, L. Y., and Legido-Quigley, H. (2021). An analysis of national action plans on antimicrobial resistance in Southeast Asia using a governance framework approach. Lancet Reg. Health West. Pac. 7:100084. doi: 10.1016/j.lanwpc.2020.100084
Cody, A. J., Mccarthy, N. D., Jansen Van Rensburg, M., Isinkaye, T., Bentley, S. D., Parkhill, J., et al. (2013). Real-time genomic epidemiological evaluation of human Campylobacter isolates by use of whole-genome multilocus sequence typing. J. Clin. Microbiol. 51, 2526–2534. doi: 10.1128/jcm.00066-13
Conlan, S., Park, M., Deming, C., Thomas, P. J., Young, A. C., Coleman, H., et al. (2016). Plasmid dynamics in KPC-Positive Klebsiella pneumoniae during long-term patient colonization. mBio 7:e00742-16.
Conlan, S., Thomas, P. J., Deming, C., Park, M., Lau, A. F., Dekker, J. P., et al. (2014). Single-molecule sequencing to track plasmid diversity of hospital-associated carbapenemase-producing Enterobacteriaceae. Sci. Transl. Med. 6:254ra126. doi: 10.1126/scitranslmed.3009845
Cook, E. A., De Glanville, W. A., Thomas, L. F., Kariuki, S., Bronsvoort, B. M., and Fevre, E. M. (2017). Working conditions and public health risks in slaughterhouses in western Kenya. BMC Public Health 17:14. doi: 10.1186/s12889-016-3923-y
Coyne, L., Arief, R., Benigno, C., Giang, V. N., Huong, L. Q., Jeamsripong, S., et al. (2019). Characterizing antimicrobial use in the livestock sector in three South East Asian Countries (Indonesia, Thailand, and Vietnam). Antibiotics 8:33. doi: 10.3390/antibiotics8010033
Cui, S., Li, J., Hu, C., Jin, S., Li, F., Guo, Y., et al. (2009). Isolation and characterization of methicillin-resistant Staphylococcus aureus from swine and workers in China. J. Antimicrob. Chemother. 64, 680–683. doi: 10.1093/jac/dkp275
Dahiya, J. P., Wilkie, D. C., Van Kessel, A. G., and Drew, M. D. (2006). Potential strategies for controlling necrotic enteritis in broiler chickens in post-antibiotic era. Anim. Feed Sci. Technol. 129, 60–88. doi: 10.1016/j.anifeedsci.2005.12.003
Dall, C. (2019). Antibiotic Resistance in Farm Animals Tied to Global Hot Spots. Available online at: https://www.cidrap.umn.edu/news-perspective/2019/09/antibiotic-resistance-farm-animals-tied-global-hot-spots (accessed March 26, 2021).
Dang, S. T. T., Truong, D. T. Q., Olsen, J. E., Tran, N. T., Truong, G. T. H., Vu, H. T. K., et al. (2020). Research note: occurrence of mcr-encoded colistin resistance in Escherichia coli from pigs and pig farm workers in Vietnam. FEMS Microbes 1:xtaa003.
Daniel, D. S., Lee, S. M., Dykes, G. A., and Rahman, S. (2015). Public health risks of multiple-drug-resistant Enterococcus spp. in Southeast Asia. Appl. Environ. Microbiol. 81:6090. doi: 10.1128/aem.01741-15
Dawangpa, A., Lertwatcharasarakul, P., Ramasoota, P., Boonsoongnern, A., Ratanavanichrojn, N., Sanguankiat, A., et al. (2021). Genotypic and phenotypic situation of antimicrobial drug resistance of Escherichia coli in water and manure between biogas and non-biogas swine farms in central Thailand. J. Environ. Manage. 279:111659. doi: 10.1016/j.jenvman.2020.111659
de Been, M., Pinholt, M., Top, J., Bletz, S., Mellmann, A., Van Schaik, W., et al. (2015). Core genome multilocus sequence typing scheme for high- resolution typing of Enterococcus faecium. J. Clin. Microbiol. 53, 3788–3797. doi: 10.1128/jcm.01946-15
Dekker, J. P., and Frank, K. M. (2016). Next-generation epidemiology: using real-time core genome multilocus sequence typing to support infection control Policy. J. Clin. Microbiol. 54, 2850–2853. doi: 10.1128/jcm.01714-16
Devi, S. M., Park, J. W., and Kim, I. H. (2015). Effect of plant extracts on growth performance and insulin-like growth factor 1 secretion in growing pigs. Rev. Bras. Zootec. 44, 355–360. doi: 10.1590/s1806-92902015001000003
Dohmen, W., Bonten, M. J., Bos, M. E., Van Marm, S., Scharringa, J., Wagenaar, J. A., et al. (2015). Carriage of extended-spectrum beta-lactamases in pig farmers is associated with occurrence in pigs. Clin. Microbiol. Infect. 21, 917–923. doi: 10.1016/j.cmi.2015.05.032
Dohmen, W., VAN Gompel, L., Schmitt, H., Liakopoulos, A., Heres, L., Urlings, B. A., et al. (2017). ESBL carriage in pig slaughterhouse workers is associated with occupational exposure. Epidemiol. Infect. 145, 2003–2010. doi: 10.1017/s0950268817000784
Duarte, M. E., Tyus, J., and Kim, S. W. (2020). Synbiotic effects of enzyme and probiotics on intestinal health and growth of newly weaned pigs challenged with enterotoxigenic F18(+) Escherichia coli. Front. Vet. Sci. 7:573. doi: 10.3389/fvets.2020.00573
EFSA (2016). The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2014. EFSA J. 14:3590.
FAO (2014). REPORT on Antimicrobial Usage and Alternatives for Prophylaxis and Performance Enhancement in Pig Populations in East and Southeast Asia. Rome: FAO.
FAOSTAT (2019). FAO Stat. Available online at: http://www.fao.org/faostat/en/#compare (accessed March 26, 2021).
Faria, C., Vaz-Moreira, I., Serapicos, E., Nunes, O. C., and Manaia, C. M. (2009). Antibiotic resistance in coagulase negative staphylococci isolated from wastewater and drinking water. Sci. Total Environ. 407, 3876–3882. doi: 10.1016/j.scitotenv.2009.02.034
Founou, L. L., Founou, R. C., Allam, M., Ismail, A., Djoko, C. F., and Essack, S. Y. (2018). Genome sequencing of extended-spectrum beta-Lactamase (ESBL)-Producing Klebsiella pneumoniae isolated from pigs and abattoir workers in Cameroon. Front. Microbiol. 9:188. doi: 10.3389/fmicb.2018.00188
Founou, L. L., Founou, R. C., and Essack, S. Y. (2016). Antibiotic resistance in the food chain: a developing country-perspective. Front. Microbiol. 7:1881. doi: 10.3389/fmicb.2016.01881
Frana, T. S., Beahm, A. R., Hanson, B. M., Kinyon, J. M., Layman, L. L., Karriker, L. A., et al. (2013). Isolation and characterization of methicillin-resistant Staphylococcus aureus from pork farms and visiting veterinary students. PLoS One 8:e53738. doi: 10.1371/journal.pone.0053738
Gatica, J., and Cytryn, E. (2013). Impact of treated wastewater irrigation on antibiotic resistance in the soil microbiome. Environ. Sci. Pollut. Res. Int. 20, 3529–3538. doi: 10.1007/s11356-013-1505-4
Gerba, C. P., and Pepper, I. L. (2009). Wastewater treatment and biosolids reuse. Environ. Microbiol. 503–530. doi: 10.1016/B978-0-12-370519-8.00024-9
Gerbin, C. S. (2014). Enhancing US-Japan cooperation to combat antimicrobial resistance. Biosecur. Bioterror. 12, 337–345. doi: 10.1089/bsp.2014.0034
Gois, F. D., Cairo, P. L. G., De Souza Cantarelli, V., Do Bomfim Costa, L. C., Fontana, R., Allaman, I. B., et al. (2016). Effect of Brazilian red pepper (Schinus terebinthifolius Raddi) essential oil on performance, diarrhea and gut health of weanling pigs. Livest. Sci. 183, 24–27. doi: 10.1016/j.livsci.2015.11.009
Goni, A. M., Effarizah, M. E., and Rusul, G. (2018). Prevalence, antimicrobial resistance, resistance genes and class 1 integrons of Salmonella serovars in leafy vegetables, chicken carcasses and related processing environments in Malaysian fresh food markets. Food Control 91, 170–180. doi: 10.1016/j.foodcont.2018.02.039
Gonzalez-Santamarina, B., Busch, A., Garcia-Soto, S., Abdel-Glil, M. Y., Linde, J., Fries, R., et al. (2020). Draft genome sequence of multi-resistant Salmonella enterica subsp. enterica serovar Rissen strain 19CS0416 isolated from Vietnam reveals mcr-1 plasmid mediated resistance to colistin already in 2013. J. Genomics 8, 76–79. doi: 10.7150/jgen.42790
Goutard, F. L., Bordier, M., Calba, C., Erlacher-Vindel, E., Gochez, D., De Balogh, K., et al. (2017). Antimicrobial policy interventions in food animal production in South East Asia. BMJ 358:j3544. doi: 10.1136/bmj.j3544
Guardabassi, L., O’Donoghue, M., Moodley, A., Ho, J., and Boost, M. (2009). Novel lineage of Methicillin-resistant Staphylococcus aureus, Hong Kong. Emerg. Infect. Dis. 15, 1998–2000.
Guo, S., Aung, K. T., Leekitcharoenphon, P., Tay, M. Y., Seow, K. L., Zhong, Y., et al. (2021). Prevalence and genomic analysis of ESBL-producing Escherichia coli in retail raw meats in Singapore. J. Antimicrob. Chemother. 76, 601–605. doi: 10.1093/jac/dkaa461
Ha, T., Hirai, T., Thi, N., and Yamaguchi, R. (2012). Antibiotic resistance profiles of Salmonella serovars isolated from retail pork and chicken meat in North Vietnam. Int. J. Food Microbiol. 156, 147–151. doi: 10.1016/j.ijfoodmicro.2012.03.016
Haenssgen, M. J., Charoenboon, N., Zanello, G., Mayxay, M., Reed-Tsochas, F., Lubell, Y., et al. (2019). Antibiotic knowledge, attitudes and practices: new insights from cross-sectional rural health behaviour surveys in low-income and middle-income South-East Asia. BMJ Open 9:e028224. doi: 10.1136/bmjopen-2018-028224
Hamelin, K., Bruant, G., El-Shaarawi, A., Hill, S., Edge, T. A., Bekal, S., et al. (2006). A virulence and antimicrobial resistance DNA microarray detects a high frequency of virulence genes in Escherichia coli Isolates from Great Lakes recreational waters. Appl. Environ. Microbiol. 72, 4200–4206. doi: 10.1128/AEM.00137-06
Hammerum, A. M., Larsen, J., Andersen, V. D., Lester, C. H., Skovgaard Skytte, T. S., Hansen, F., et al. (2014). Characterization of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli obtained from Danish pigs, pig farmers and their families from farms with high or no consumption of third-or fourth-generation cephalosporins. J. Antimicrob. Chemother. 69, 2650–2657. doi: 10.1093/jac/dku180
Hammerum, A. M., Sandvang, D., Andersen, S. R., Seyfarth, A. M., Porsbo, L. J., Frimodt-Moller, N., et al. (2006). Detection of sul1, sul2 and sul3 in sulphonamide resistant Escherichia coli isolates obtained from healthy humans, pork and pigs in Denmark. Int. J. Food Microbiol. 106, 235–237. doi: 10.1016/j.ijfoodmicro.2005.06.023
Harris, S. R., Cartwright, E. J., Torok, M. E., Holden, M. T., Brown, N. M., Ogilvy-Stuart, A. L., et al. (2013). Whole-genome sequencing for analysis of an outbreak of meticillin-resistant Staphylococcus aureus: a descriptive study. Lancet Infect. Dis. 13, 130–136. doi: 10.1016/s1473-3099(12)70268-2
Higgins, P. G., Prior, K., Harmsen, D., and Seifert, H. (2017). Development and evaluation of a core genome multilocus typing scheme for whole-genome sequence-based typing of Acinetobacter baumannii. PLoS One 12:e0179228. doi: 10.1371/journal.pone.0179228
Holmer, I., Salomonsen, C. M., Jorsal, S. E., Astrup, L. B., Jensen, V. F., Hog, B. B., et al. (2019). Antibiotic resistance in porcine pathogenic bacteria and relation to antibiotic usage. BMC Vet. Res. 15:449. doi: 10.1186/s12917-019-2162-8
Holzel, C. S., Tetens, J. L., and Schwaiger, K. (2018). Unraveling the role of vegetables in spreading antimicrobial-resistant bacteria: a need for quantitative risk assessment. Foodborne Pathog. Dis. 15, 671–688. doi: 10.1089/fpd.2018.2501
Hossain, M. M., Park, J. W., and Kim, I. H. (2016). δ-Aminolevulinic acid, and lactulose supplements in weaned piglets diet: effects on performance, fecal microbiota, and in-vitro noxious gas emissions. Livest. Sci. 183, 84–91. doi: 10.1016/j.livsci.2015.11.021
Hounmanou, Y. M. G., Bortolaia, V., Dang, S. T. T., Truong, D., Olsen, J. E., and Dalsgaard, A. (2021). ESBL and AmpC β-lactamase encoding genes in E. coli from pig and pig farm workers in Vietnam and their association with mobile genetic elements. Front. Microbiol. 12:506. doi: 10.3389/fmicb.2021.629139
Huang, G., Li, X., Lu, D., Liu, S., Suo, X., Li, Q., et al. (2018). Lysozyme improves gut performance and protects against enterotoxigenic Escherichia coli infection in neonatal piglets. Vet. Res. 49:20.
Huijbers, P. M., Graat, E. A., Haenen, A. P., Van Santen, M. G., Van Essen-Zandbergen, A., Mevius, D. J., et al. (2014). Extended-spectrum and AmpC beta-lactamase-producing Escherichia coli in broilers and people living and/or working on broiler farms: prevalence, risk factors and molecular characteristics. J. Antimicrob. Chemother. 69, 2669–2675. doi: 10.1093/jac/dku178
Islam, M. J., Lisa, A. A., Reza, A. H. M. M., Reza, M. S., Absar Khan, M. N., and Kamal, M. (2014). Source identification and entry pathways of banned antibiotics nitrofuran and chloramphenicol in shrimp value chain of Bangladesh. Eurasian J. Biosci. 8, 71–83. doi: 10.5053/ejobios.2014.8.0.7
Jorgensen, J. H., and Ferraro, M. J. (2009). Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin. Infect. Dis. 49, 1749–1755. doi: 10.1086/647952
Kamaruzzaman, E. A., Abdul Aziz, S., Bitrus, A. A., Zakaria, Z., and Hassan, L. (2020). Occurrence and characteristics of extended-spectrum β-Lactamase-Producing Escherichia coli from dairy cattle, milk, and farm environments in Peninsular Malaysia. Pathogens 9:1007. doi: 10.3390/pathogens9121007
Kawahara, R., Khong, D. T., Le, H. V., Phan, Q. N., Nguyen, T. N., Yamaguchi, T., et al. (2019). Prevalence Of mcr-1 among cefotaxime-resistant commensal Escherichia coli in residents of Vietnam. Infect. Drug Resist. 12, 3317–3325. doi: 10.2147/idr.s224545
Kempf, I., Fleury, M. A., Drider, D., Bruneau, M., Sanders, P., Chauvin, C., et al. (2013). What do we know about resistance to colistin in Enterobacteriaceae in avian and pig production in Europe? Int. J. Antimicrob. Agents 42, 379–383. doi: 10.1016/j.ijantimicag.2013.06.012
Kennemann, L., Didelot, X., Aebischer, T., Kuhn, S., Drescher, B., Droege, M., et al. (2011). Helicobacter pylori genome evolution during human infection. Proc. Natl. Acad. Sci. U.S.A. 108, 5033–5038.
Khalid, K. A., Zakaria, Z., Toung, O. P., and McOrist, S. (2009). Low levels of methicillin-resistant Staphylococcus aureus in pigs in Malaysia. Vet. Rec. 164, 626–627.
Khan, H. A., Baig, F. K., and Mehboob, R. (2017). Nosocomial infections: epidemiology, prevention, control and surveillance. Asian Pac. J. Trop. Biomed. 7, 478–482. doi: 10.2307/30141390
Khine, N. O., Lugsomya, K., Kaewgun, B., Honhanrob, L., Pairojrit, P., Jermprasert, S., et al. (2020). Multidrug resistance and virulence factors of Escherichia coli harboring plasmid-mediated colistin resistance: mcr-1 and mcr-3 Genes in Contracted Pig Farms in Thailand. Front. Vet. Sci. 7:582899. doi: 10.3389/fvets.2020.582899
Kim, K., Ingale, S., Kim, J., Lee, S., Lee, J., Kwon, I., et al. (2014). Bacteriophage and probiotics both enhance the performance of growing pigs but bacteriophage are more effective. Anim. Feed Sci. Technol. 196, 88–95. doi: 10.1016/j.anifeedsci.2014.06.012
Koningstein, M., Simonsen, J., Helms, M., and Mølbak, K. (2010). The interaction between prior antimicrobial drug exposure and resistance in human Salmonella infections. J. Antimicrob. Chemother. 65, 1819–1825. doi: 10.1093/jac/dkq176
Kraemer, S. A., Ramachandran, A., and Perron, G. G. (2019). Antibiotic pollution in the environment: from microbial ecology to public policy. Microorganisms 7:180. doi: 10.3390/microorganisms7060180
Kruse, A. B., Nielsen, L. R., and Alban, L. (2015). “Vaccination against Actinobacillus pleuropneumoniae as an alternative strategy to antimicrobial use in Danish pig herds,” in Proceedings of the Safe Pork Conference, Porto, 357–359.
Kummerer, K. (2004). Resistance in the environment. J. Antimicrob. Chemother. 54, 311–320. doi: 10.1093/jac/dkh325
Lan, R., Li, T., and Kim, I. (2017). Effects of xylanase supplementation on growth performance, nutrient digestibility, blood parameters, fecal microbiota, fecal score and fecal noxious gas emission of weaning pigs fed corn-soybean meal-based diet. Anim. Sci. J. 88, 1398–1405. doi: 10.1111/asj.12771
Lăpuşan, A., Mihaiu, L., Mihaiu, M., Sorin, D., Mihaiu, R., Cordiş, I., et al. (2012). The importance of pig tonsils removal for the final assessment of the carcasses’ hygiene quality. Sci. Works 58, 347–352.
Lazarus, B., Paterson, D. L., Mollinger, J. L., and Rogers, B. A. (2015). Do human extraintestinal Escherichia coli infections resistant to expanded-spectrum cephalosporins originate from food-producing animals? A systematic review. Clin. Infect. Dis. 60, 439–452. doi: 10.1093/cid/ciu785
Lechner, I., Freivogel, C., Stark, K. D. C., and Visschers, V. H. M. (2020). Exposure pathways to antimicrobial resistance at the human-animal interface-A qualitative comparison of Swiss expert and consumer opinions. Front. Public Health 8:345. doi: 10.3389/fpubh.2020.00345
Lee, S., Hosseindoust, A., Goel, A., Choi, Y., Kwon, I. K., and Chae, B. (2016). Effects of dietary supplementation of bacteriophage with or without zinc oxide on the performance and gut development of weanling pigs. Ital. J. Anim. Sci. 15, 412–418. doi: 10.1080/1828051x.2016.1188676
Lei, X. J., Park, J. W., Baek, D. H., Kim, J. K., and Kim, I. H. (2017). Feeding the blend of organic acids and medium chain fatty acids reduces the diarrhea in piglets orally challenged with enterotoxigenic Escherichia coli K88. Anim. Feed Sci. Technol. 224, 46–51. doi: 10.1016/j.anifeedsci.2016.11.016
Lei, X. J., Zhang, W. L., Cheong, J. Y., Lee, S. I., and Kim, I. H. (2018). Effect of antibiotics and synbiotic on growth performance, nutrient digestibility, and faecal microbial shedding in growing-finishing pigs. J. Appl. Anim. Res. 46, 1202–1206. doi: 10.1080/09712119.2018.1484359
Lekagul, A., Tangcharoensathien, V., Liverani, M., Mills, A., Rushton, J., and Yeung, S. (2021). Understanding antibiotic use for pig farming in Thailand: a qualitative study. Antimicrob. Resist. Infect. Control 10:3.
Lewerin, S. S., Österberg, J., Alenius, S., Elvander, M., Fellström, C., Tråvén, M., et al. (2015). Risk assessment as a tool for improving external biosecurity at farm level. BMC Vet. Res. 11:171. doi: 10.1186/s12917-015-0477-7
Li, D., Yang, M., Hu, J., Zhang, J., Liu, R., Gu, X., et al. (2009). Antibiotic-resistance profile in environmental bacteria isolated from penicillin production wastewater treatment plant and the receiving river. Environ. Microbiol. 11, 1506–1517. doi: 10.1111/j.1462-2920.2009.01878.x
Li, H., Sun, X., Liao, X., and Gänzle, M. (2020). Control of pathogenic and spoilage bacteria in meat and meat products by high pressure: challenges and future perspectives. Compr. Rev. Food Sci. Food Saf. 19, 3476–3500. doi: 10.1111/1541-4337.12617
Liao, S. F., and Nyachoti, M. (2017). Using probiotics to improve swine gut health and nutrient utilization. Anim. Nutr. 3, 331–343. doi: 10.1016/j.aninu.2017.06.007
Limayem, A., Wasson, S., Mehta, M., Pokhrel, A. R., Patil, S., Nguyen, M., et al. (2019). High-throughput detection of bacterial community and its drug-resistance profiling from local reclaimed wastewater plants. Front. Cell. Infect. Microbiol. 9:303. doi: 10.3389/fcimb.2019.00303
Lipsitch, M., Singer, R. S., and Levin, B. R. (2002). Antibiotics in agriculture: When is it time to close the barn door? Proc. Natl. Acad. Sci. U.S.A. 99, 5752–5754. doi: 10.1073/pnas.092142499
Lira, F., Vaz-Moreira, I., Tamames, J., Manaia, C. M., and Martínez, J. L. (2020). Metagenomic analysis of an urban resistome before and after wastewater treatment. Sci. Rep. 10, 1–9. doi: 10.1038/s41598-020-65031-y
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism mcr-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/s1473-3099(15)00424-7
Llarena, A. K., Taboada, E., and Rossi, M. (2017). Whole-genome sequencing in epidemiology of Campylobacter jejuni infections. J. Clin. Microbiol. 55, 1269–1275. doi: 10.1128/jcm.00017-17
Lowe, B. A., Marsh, T. L., Isaacs-Cosgrove, N., Kirkwood, R. N., Kiupel, M., and Mulks, M. H. (2011). Microbial communities in the tonsils of healthy pigs. Vet. Microbiol. 147, 346–357. doi: 10.1016/j.vetmic.2010.06.025
Luby, E., Ibekwe, A. M., Zilles, J., and Pruden, A. (2016). Molecular methods for assessment of antibiotic resistance in agricultural ecosystems: prospects and challenges. J. Environ. Qual. 45, 441–453. doi: 10.2134/jeq2015.07.0367
Lugsomya, K., Chatsuwan, T., Niyomtham, W., Tummaruk, P., Hampson, D. J., and Prapasarakul, N. (2018a). Routine prophylactic antimicrobial use is associated with increased phenotypic and genotypic resistance in commensal Escherichia coli isolates recovered from Healthy Fattening Pigs on Farms in Thailand. Microb. Drug Resist. 24, 213–223. doi: 10.1089/mdr.2017.0042
Lugsomya, K., Yindee, J., Niyomtham, W., Tribuddharat, C., Tummaruk, P., Hampson, D. J., et al. (2018b). Antimicrobial resistance in Commensal Escherichia coli isolated from pigs and pork derived from farms either routinely using or not using in-feed antimicrobials. Microb. Drug Resist. 24, 1054–1066. doi: 10.1089/mdr.2018.0154
Lulitanond, A., Ito, T., Li, S., Han, X., Ma, X. X., Engchanil, C., et al. (2013). ST9 MRSA strains carrying a variant of type IX SCCmec identified in the Thai community. BMC Infect. Dis. 13:214. doi: 10.1186/1471-2334-13-214
Lunha, K., Leangapichart, T., Jiwakanon, J., Angkititrakul, S., Sunde, M., Järhult, J. D., et al. (2020). Antimicrobial resistance in fecal escherichia coli from humans and pigs at farms at different levels of intensification. Antibiotics 9:662. doi: 10.3390/antibiotics9100662
Lupo, A., Coyne, S., and Berendonk, T. U. (2012). Origin and evolution of antibiotic resistance: the common mechanisms of emergence and spread in water bodies. Front. Microbiol. 3. doi: 10.3389/fmicb.2012.00018
Ma, X., Zhang, S., Pan, L., and Piao, X. (2017). Effects of lysozyme on the growth performance, nutrient digestibility, intestinal barrier, and microbiota of weaned pigs fed diets containing spray-dried whole egg or albumen powder. Can. J. Anim. Sci. 97, 466–475.
Magouras, I., Carmo, L. P., Stärk, K. D., and Schüpbach-Regula, G. (2017). Antimicrobial usage and-resistance in livestock: where should we focus? Front. Vet. Sci. 4:148. doi: 10.3389/fvets.2017.00148
Maiden, M. C., Bygraves, J. A., Feil, E., Morelli, G., Russell, J. E., Urwin, R., et al. (1998). Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U.S.A. 95, 3140–3145. doi: 10.1073/pnas.95.6.3140
Makarov, D. A., Ivanova, O. E., Karabanov, S. Y., Gergel, M. A., and Pomazkova, A. V. (2020). Antimicrobial resistance of commensal Escherichia coli from food-producing animals in Russia. Vet. World 13, 2053–2061. doi: 10.14202/vetworld.2020.2053-2061
Malchione, M. D., Torres, L. M., Hartley, D. M., Koch, M., and Goodman, J. L. (2019). Carbapenem and colistin resistance in Enterobacteriaceae in Southeast Asia: review and mapping of emerging and overlapping challenges. Int. J. Antimicrob. Agents 54, 381–399. doi: 10.1016/j.ijantimicag.2019.07.019
Malhotra-Kumar, S., Xavier, B. B., Das, A. J., Lammens, C., Hoang, H. T., Pham, N. T., et al. (2016). Colistin-resistant Escherichia coli harbouring mcr-1 isolated from food animals in Hanoi, Vietnam. Lancet Infect. Dis. 16, 286–287. doi: 10.1016/s1473-3099(16)00014-1
Marshall, B. M., and Levy, S. B. (2011). Food animals and antimicrobials: impacts on human health. Clin. Microbiol. Rev. 24, 718–733. doi: 10.1128/cmr.00002-11
Martinez, J. L., and Baquero, F. (2009). “Antibiotics and the evolution of antibiotic resistance,” in Encyclopedia of Life Sciences (Chichester: John Wiley & Sons, Ltd). doi: 10.1002/9780470015902.a0021782
Mather, A. E., Phuong, T. L. T., Gao, Y., Clare, S., Mukhopadhyay, S., Goulding, D. A., et al. (2018). New variant of multidrug-resistant Salmonella enterica Serovar Typhimurium associated with invasive disease in Immunocompromised Patients in Vietnam. mBio 9:e01056-18.
McEwen, S. A. (2006). Antibiotic use in animal agriculture: what have we learned and where are we going? Anim. Biotechnol. 17, 239–250. doi: 10.1080/10495390600957233
McLain, J. E., Cytryn, E., Durso, L. M., and Young, S. (2016). Culture-based methods for detection of antibiotic resistance in agroecosystems: advantages, challenges, and gaps in knowledge. J. Environ. Qual. 45, 432–440. doi: 10.2134/jeq2015.06.0317
McMahon, M. A., Xu, J., Moore, J. E., Blair, I. S., and Mcdowell, D. A. (2007). Environmental stress and antibiotic resistance in food-related pathogens. Appl. Environ. Microbiol. 73, 211–217. doi: 10.1128/aem.00578-06
Meeusen, E. N. T., Walker, J., Peters, A., Pastoret, P.-P., and Jungersen, G. (2007). Current status of veterinary vaccines. Clin. Microbiol. Rev. 20, 489–510.
Mellmann, A., Bletz, S., Boking, T., Kipp, F., Becker, K., Schultes, A., et al. (2016). Real-time genome sequencing of resistant bacteria provides precision infection control in an institutional setting. J. Clin. Microbiol. 54, 2874–2881. doi: 10.1128/jcm.00790-16
Ministry of Public Health (2020). Thailand’s One Health Report on Antimicrobial Consumption and Antimicrobial Resistance in 2018”. (Nonthaburi: Ministry of Public Health).
Miro, E., Rossen, J. W. A., Chlebowicz, M. A., Harmsen, D., Brisse, S., Passet, V., et al. (2020). Core/whole genome multilocus sequence typing and core genome SNP-Based Typing of OXA-48-Producing Klebsiella pneumoniae clinical isolates from spain. Front. Microbiol. 10:2961. doi: 10.3389/fmicb.2019.02961
Mobasseri, G., Teh, C. S. J., Ooi, P. T., and Thong, K. L. (2019). The emergence of colistin-resistant Klebsiella pneumoniae strains from swine in Malaysia. J. Glob. Antimicrob. Resist. 17, 227–232. doi: 10.1016/j.jgar.2018.12.015
Mobasseri, G., Thong, K. L., and Teh, C. S. J. (2021). Genomic analysis revealing the resistance mechanisms of extended-spectrum β-lactamase-producing Klebsiella pneumoniae isolated from pig and humans in Malaysia. Int. Microbiol. 24, 243–250. doi: 10.1007/s10123-021-00161-5
More, S. J. (2020). European perspectives on efforts to reduce antimicrobial usage in food animal production. Ir. Vet. J. 73:2.
Nadon, C., Van Walle, I., Gerner-Smidt, P., Campos, J., Chinen, I., Concepcion-Acevedo, J., et al. (2017). PulseNet International: vision for the implementation of whole genome sequencing (WGS) for global food-borne disease surveillance. Euro Surveill. 22:30544.
Nagulapally, S R., Ahmad, A., Henry, A., Marchin, G. L., Zurek, L., and Bhandari, A. (2009). Occurrence of Ciprofloxacin-, Trimethoprim-Sulfamethoxazole-, and Vancomycin-Resistant bacteria in a municipal wastewater treatment plant. Water Environ. Res. 81, 82–90. doi: 10.2175/106143008X304596
Neela, V., Zafrul, A. M., Mariana, N. S., van Belkum, A., Liew, Y. K., and Rad, E. G. (2009). Prevalence of ST9 methicillin-resistant Staphylococcus aureus among pigs and pig handlers in Malaysia. J. Clin. Microbiol. 47, 4138–4140. doi: 10.1128/jcm.01363-09
Nguyen, D. T., Kanki, M., Nguyen, P. D., Le, H. T., Ngo, P. T., Tran, D. N., et al. (2016). Prevalence, antibiotic resistance, and extended-spectrum and AmpC β-lactamase productivity of Salmonella isolates from raw meat and seafood samples in Ho Chi Minh City, Vietnam. Int. J. Food Microbiol. 236, 115–122. doi: 10.1016/j.ijfoodmicro.2016.07.017
Nguyen, K. V., Thi Do, N. T., Chandna, A., Nguyen, T. V., Pham, C. V., Doan, P. M., et al. (2013). Antibiotic use and resistance in emerging economies: a situation analysis for Viet Nam. BMC Public Health 13:1158. doi: 10.1186/1471-2458-13-1158
Nguyen, M. N., Hoang, H. T. T., Xavier, B. B., Lammens, C., Le, H. T., Hoang, N. T. B., et al. (2021). Prospective One Health genetic surveillance in Vietnam identifies distinct blaCTX-M-harbouring Escherichia coli in food-chain and human-derived samples. Clin. Microbiol. Infect. doi: 10.1016/j.cmi.2021.01.006
Nguyen, N. T., Nguyen, H. M., Nguyen, C. V., Nguyen, T. V., Nguyen, M. T., Thai, H. Q., et al. (2016). Use of Colistin and Other critical antimicrobials on pig and chicken farms in Southern Vietnam and its association with resistance in commensal Escherichia coli Bacteria. Appl. Environ. Microbiol. 82, 3727–3735. doi: 10.1128/aem.00337-16
Nhung, N. T., Cuong, N. V., Thwaites, G., and Carrique-Mas, J. (2016). Antimicrobial usage and antimicrobial resistance in animal production in southeast asia: a review. Antibiotics 5:37. doi: 10.3390/antibiotics5040037
NSC (2020a). Thailand’s First One Health Report on antimicrobial Consumption and Antimicrobial Resistance in 2017. Nonthaburi: NSC.
NSC (2020b). Thailand’s One Health Report on Antimicrobial Consumption and Antimicrobial Resistance in 2018. Nonthaburi: NSC.
Obst, U., Schwartz, T., and Volkmann, H. (2006). Antibiotic resistant pathogenic bacteria and their resistance genes in bacterial biofilms. Int. J. Artif. Organs. 29, 387–394. doi: 10.1177/039139880602900408
Olaitan, A. O., Chabou, S., Okdah, L., Morand, S., and Rolain, J. M. (2016). Dissemination of the mcr-1 colistin resistance gene. Lancet Infect. Dis. 16, 289–290.
Olaitan, A. O., Dandachi, I., Baron, S. A., Daoud, Z., Morand, S., and Rolain, J.-M. (2021). Banning colistin in feed additives: a small step in the right direction. Lancet Infect. Dis. 21, 29–30. doi: 10.1016/s1473-3099(20)30915-4
Olaitan, A. O., Diene, S. M., Kempf, M., Berrazeg, M., Bakour, S., Gupta, S. K., et al. (2014). Worldwide emergence of colistin resistance in Klebsiella pneumoniae from healthy humans and patients in Lao PDR, Thailand, Israel, Nigeria and France owing to inactivation of the PhoP/PhoQ regulator mgrB: an epidemiological and molecular study. Int. J. Antimicrob. Agents 44, 500–507. doi: 10.1016/j.ijantimicag.2014.07.020
Olaitan, A. O., Thongmalayvong, B., Akkhavong, K., Somphavong, S., Paboriboune, P., Khounsy, S., et al. (2015). Clonal transmission of a colistin-resistant Escherichia coli from a domesticated pig to a human in Laos. J. Antimicrob. Chemother. 70, 3402–3404.
Oloya, J., Doetkott, D., and Khaitsa, M. L. (2009). Antimicrobial drug resistance and molecular characterization of Salmonella isolated from domestic animals, humans, and meat products. Foodborne Pathog. Dis. 6, 273–284. doi: 10.1089/fpd.2008.0134
Opatowski, L., Opatowski, M., Vong, S., and Temime, L. (2020). A one-health quantitative model to assess the risk of antibiotic resistance acquisition in asian populations: impact of exposure through food, water, livestock and humans. Risk Anal. doi: 10.1111/risa.13618
Pan, L., Zhao, P. F., Ma, X. K., Shang, Q. H., Xu, Y. T., Long, S. F., et al. (2017). Probiotic supplementation protects weaned pigs against enterotoxigenic Escherichia coli K88 challenge and improves performance similar to antibiotics. J. Anim. Sci. 95, 2627–2639. doi: 10.2527/jas.2016.1243
Papadopoulos, T., Zdragas, A., Mandilara, G., Vafeas, G., Giantzi, V., Petridou, E., et al. (2016). Characterization of Salmonella isolates from municipal sewage, patients, foods, and animals in Greece using antimicrobial susceptibility testing and pulsed field gel electrophoresis. Int. J. One Heal. 2, 12–18. doi: 10.14202/IJOH.2016.12-18
Patchanee, P., Chokesajjawatee, N., Santiyanont, P., Chuammitri, P., Deeudom, M., Monteith, W., et al. (2020). Multiple clones of colistin-resistant Salmonella enterica carrying mcr-1 plasmids in meat products and patients in Northern Thailand. bioRxiv [Preprint]. doi: 10.1101/2020.12.08.415869
Patchanee, P., Tadee, P., Arjkumpa, O., Love, D., Chanachai, K., Alter, T., et al. (2014). Occurrence and characterization of livestock-associated methicillin-resistant Staphylococcus aureus in pig industries of northern Thailand. J. Vet. Sci. 15, 529–536. doi: 10.4142/jvs.2014.15.4.529
Paveenkittiporn, W., Kerdsin, A., Chokngam, S., Bunthi, C., Sangkitporn, S., and Gregory, C. J. (2017). Emergence of plasmid-mediated colistin resistance and New Delhi metallo-beta-lactamase genes in extensively drug-resistant Escherichia coli isolated from a patient in Thailand. Diagn. Microbiol. Infect. Dis. 87, 157–159. doi: 10.1016/j.diagmicrobio.2016.11.005
Pazda, M., Kumirska, J., Stepnowski, P., and Mulkiewicz, E. (2019). Antibiotic resistance genes identified in wastewater treatment plant systems – a review. Sci. Total Environ. 697. doi: 10.1016/j.scitotenv.2019.134023
Pei, X., Xiao, Z., Liu, L., Wang, G., Tao, W., Wang, M., et al. (2019). Effects of dietary zinc oxide nanoparticles supplementation on growth performance, zinc status, intestinal morphology, microflora population, and immune response in weaned pigs. J. Sci. Food Agric. 99, 1366–1374. doi: 10.1002/jsfa.9312
Peng, Z., Wang, A., Xie, L., Song, W., Wang, J., Yin, Z., et al. (2016). Use of recombinant porcine β-defensin 2 as a medicated feed additive for weaned piglets. Sci. Rep. 6:26790.
Petry, A. L., Huntley, N. F., Bedford, M. R., and Patience, J. F. (2020). Xylanase increased the energetic contribution of fiber and improved the oxidative status, gut barrier integrity, and growth performance of growing pigs fed insoluble corn-based fiber. J. Anim. Sci. 98:skaa233.
Phongaran, D., Khang-Air, S., and Angkititrakul, S. (2019). Molecular epidemiology and antimicrobial resistance of Salmonella isolates from broilers and pigs in Thailand. Vet. World 12, 1311–1318. doi: 10.14202/vetworld.2019.1311-1318
Postma, M., Backhans, A., Collineau, L., Loesken, S., Sjölund, M., Belloc, C., et al. (2016a). Evaluation of the relationship between the biosecurity status, production parameters, herd characteristics and antimicrobial usage in farrow-to-finish pig production in four EU countries. Porcine Health Manage. 2:9.
Postma, M., Backhans, A., Collineau, L., Loesken, S., Sjölund, M., Belloc, C., et al. (2016b). The biosecurity status and its associations with production and management characteristics in farrow-to-finish pig herds. Animal 10, 478–489. doi: 10.1017/s1751731115002487
Potter, A., Gerdts, V., and Littel-Van Den Hurk, S. (2008). Veterinary vaccines: Alternatives to antibiotics? Anim. Health Res. Rev. 9, 187–199. doi: 10.1017/s1466252308001606
Prasertsee, T., Chuammitri, P., Deeudom, M., Chokesajjawatee, N., Santiyanont, P., Tadee, P., et al. (2019). Core genome sequence analysis to characterize Salmonella enterica serovar Rissen ST469 from a swine production chain. Int. J. Food Microbiol. 304, 68–74. doi: 10.1016/j.ijfoodmicro.2019.05.022
Preena, P. G., Swaminathan, T. R., Rejish Kumar, V. J., and Bright Singh, I. S. (2020). Unravelling the menace: detection of antimicrobial resistance in aquaculture. Lett. Appl. Microbiol. 71, 26–38. doi: 10.1111/lam.13292
Prestinaci, F., Pezzotti, P., and Pantosti, A. (2015). Antimicrobial resistance: a global multifaceted phenomenon. Pathog. Glob. Health 109, 309–318. doi: 10.1179/2047773215Y.0000000030
Raasch, S., Postma, M., Dewulf, J., Stärk, K. D. C., and Grosse Beilage, E. (2018). Association between antimicrobial usage, biosecurity measures as well as farm performance in German farrow-to-finish farms. Porcine Health Manage. 4:30.
Raith, J., Trauffler, M., Firth, C. L., Lebl, K., Schleicher, C., and Köfer, J. (2016). Influence of porcine circovirus type 2 vaccination on the level of antimicrobial consumption on 65 Austrian pig farms. Vet. Rec. 178:504. doi: 10.1136/vr.103406
Ren, P., Chen, J., Wedekind, K., Hancock, D., and Vázquez-Añón, M. (2020). Interactive effects of zinc and copper sources and phytase on growth performance, mineral digestibility, bone mineral concentrations, oxidative status, and gut morphology in nursery pigs. Transl. Anim. Sci. 4, 783–798. doi: 10.1093/tas/txaa083
Riley, C. B., Chidgey, K. L., Bridges, J. P., Gordon, E., and Lawrence, K. E. (2020). Isolates, antimicrobial susceptibility profiles and multidrug resistance of bacteria cultured from pig submissions in New Zealand. Animals 10:1427. doi: 10.3390/ani10081427
Rizzo, L., Manaia, C., Merlin, C., Schwartz, T., Dagot, C., Ploy, M. C., et al. (2013). Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Sci. Total Environ. 447, 345–360. doi: 10.1016/j.scitotenv.2013.01.032
Rolain, J. M., Kempf, M., Leangapichart, T., Chabou, S., Olaitan, A. O., Le Page, S., et al. (2016). Plasmid-Mediated mcr-1 Gene in Colistin-resistant clinical isolates of Klebsiella pneumoniae in France and Laos. Antimicrob. Agents Chemother. 60, 6994–6995. doi: 10.1128/aac.00960-16
Rose, N., and Andraud, M. (2017). The use of vaccines to control pathogen spread in pig populations. Porcine Health Manage. 3:8.
Runcharoen, C., Raven, K. E., Reuter, S., Kallonen, T., Paksanont, S., Thammachote, J., et al. (2017). Whole genome sequencing of ESBL-producing Escherichia coli isolated from patients, farm waste and canals in Thailand. Genome Med. 9:81.
Sabri, N. A., Schmitt, H., Van Der Zaan, B., Gerritsen, H. W., Zuidema, T., Rijnaarts, H. H. M., et al. (2020). Prevalence of antibiotics and antibiotic resistance genes in a wastewater effluent-receiving river in the Netherlands. J. Environ. Chem. Eng. 8:102245. doi: 10.1016/j.jece.2018.03.004
Sack, D. A., Lyke, C., McLaughlin, C., Suwanvanichkij, V., and World Health Organization (2001). Antimicrobial Resistance in Shigellosis, Cholera and Campylobacteriosis (No. WHO/CDS/CSR/DRS/2001.8). Geneva: World Health Organization.
Samelis, J. (2006). “9 - Managing microbial spoilage in the meat industry,” in Food Spoilage Microorganisms, ed. C. D. W. Blackburn (Sawston: Woodhead Publishing), 213–286. doi: 10.1533/9781845691417.2.213
Samolińska, W., Kowalczuk-Vasilev, E., and Grela, E. R. (2018). Comparative effect of different dietary inulin sources and probiotics on growth performance and blood characteristics in growing-finishing pigs. Arch. Anim. Nutr. 72, 379–395. doi: 10.1080/1745039x.2018.1505147
San Andres, J. V., Mastromano, G. A., Li, Y., Tran, H., Bundy, J. W., Miller, P. S., et al. (2019). The effects of prebiotics on growth performance and in vitro immune biomarkers in weaned pigs. Transl. Anim. Sci. 3, 1315–1325. doi: 10.1093/tas/txz129
Satessa, G., Kjeldsen, N., Mansouryar, M., Hansen, H., Bache, J., and Nielsen, M. (2020). Effects of alternative feed additives to medicinal zinc oxide on productivity, diarrhoea incidence and gut development in weaned piglets. Animal 14, 1638–1646. doi: 10.1017/s1751731120000154
Sayan, H., Assavacheep, P., Angkanaporn, K., and Assavacheep, A. (2018). Effect of Lactobacillus salivarius on growth performance, diarrhea incidence, fecal bacterial population and intestinal morphology of suckling pigs challenged with F4+ enterotoxigenic Escherichia coli. Asian Australas. J. Anim. Sci. 31, 1308–1314. doi: 10.5713/ajas.17.0746
Sbardella, M., Perina, D., Andrade, C., Santos, C., Cairo, P., Marques, E., et al. (2016). Effects of dietary hop β-acids or colistin on the performance, nutrient digestibility, and intestinal health of weanling pigs. Anim. Feed Sci. Technol. 217, 67–75. doi: 10.1016/j.anifeedsci.2016.04.007
Schurch, A. C., Arredondo-Alonso, S., Willems, R. J. L., and Goering, R. V. (2018). Whole genome sequencing options for bacterial strain typing and epidemiologic analysis based on single nucleotide polymorphism versus gene-by-gene-based approaches. Clin. Microbiol. Infect. 24, 350–354. doi: 10.1016/j.cmi.2017.12.016
Schwartz, D. C., and Cantor, C. R. (1984). Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell 37, 67–75. doi: 10.1016/0092-8674(84)90301-5
Scott, H. M., Acuff, G., Bergeron, G., Bourassa, M. W., Gill, J., Graham, D. W., et al. (2019). Critically important antibiotics: criteria and approaches for measuring and reducing their use in food animal agriculture. Ann. N. Y. Acad. Sci. 1441, 8–16. doi: 10.1111/nyas.14058
Seenama, C., Thamlikitkul, V., and Ratthawongjirakul, P. (2019). Multilocus sequence typing and bla ESBL characterization of extended-spectrum beta-lactamase-producing Escherichia coli isolated from healthy humans and swine in Northern Thailand. Infect. Drug Resist. 12, 2201–2214. doi: 10.2147/idr.s209545
Selander, R. K., Beltran, P., Smith, N. H., Helmuth, R., Rubin, F. A., Kopecko, D. J., et al. (1990). Evolutionary genetic relationships of clones of Salmonella serovars that cause human typhoid and other enteric fevers. Infect. Immun. 58, 2262–2275. doi: 10.1128/iai.58.7.2262-2275.1990
Seo, B.-J., Song, E.-T., Lee, K., Kim, J.-W., Jeong, C.-G., Moon, S.-H., et al. (2018). Evaluation of the broad-spectrum lytic capability of bacteriophage cocktails against various Salmonella serovars and their effects on weaned pigs infected with Salmonella Typhimurium. J. Vet. Med. Sci. 80, 851–860. doi: 10.1292/jvms.17-0501
Serweciñska, L. (2020). Antimicrobials and antibiotic-resistant bacteria: a risk to the environment and to public health. Water 12:3313. doi: 10.3390/w12123313
Shafipour Yordshahi, A., Moradi, M., Tajik, H., and Molaei, R. (2020). Design and preparation of antimicrobial meat wrapping nanopaper with bacterial cellulose and postbiotics of lactic acid bacteria. Int. J. Food Microbiol. 321:108561. doi: 10.1016/j.ijfoodmicro.2020.108561
Sheppard, A. E., Stoesser, N., Wilson, D. J., Sebra, R., Kasarskis, A., Anson, L. W., et al. (2016). Nested Russian doll-like genetic mobility drives rapid dissemination of the Carbapenem resistance Gene blaKPC. Antimicrob. Agents Chemother. 60, 3767–3778. doi: 10.1128/aac.00464-16
Shrestha, R., Koju, P., Xinliang, L., Maharjan, B., Madhup, S., Shah, P., et al. (2019). Health care associated infection and trend of antimicrobial resistance in tertiary care hospital -A study in low income setting. Kathmandu Univ. Med. J. 17, 329–335.
Silva, C., De Oliveira, A., Bezerra, P., and Evangelista, J. (2015). Escherichia coli in pig farming. Clinical aspects. A review. Rev. Bras. Hig. Sanid. Anim. 9, 288–293.
Simba, D., Kakoko, D., Semali, I., Kessy, A., and Embrey, M. (2016). Household knowledge of antimicrobials and antimicrobial resistance in the wake of an Accredited Drug Dispensing Outlet (ADDO) Program Rollout in Tanzania. PLoS One 11:e0163246. doi: 10.1371/journal.pone.0163246
Singer, A. C., Shaw, H., Rhodes, V., and Hart, A. (2016). Review of antimicrobial resistance in the environment and its relevance to environmental regulators. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01728
Singh, P. K. (2017). One Health approach to tackle antimicrobial resistance in South East Asia. BMJ 358:j3625. doi: 10.1136/bmj.j3625
Singh, S., Charani, E., Devi, S., Sharma, A., Edathadathil, F., Kumar, A., et al. (2021). A road-map for addressing antimicrobial resistance in low- and middle-income countries: lessons learnt from the public private participation and co-designed antimicrobial stewardship programme in the State of Kerala, India. Antimicrob. Resist. Infect. Control 10:32.
Sinlapasorn, S., Lulitanond, A., Angkititrakul, S., Chanawong, A., Wilailuckana, C., Tavichakorntrakool, R., et al. (2015). SCCmec IX in meticillin-resistant Staphylococcus aureus and meticillin-resistant coagulase-negative staphylococci from pigs and workers at pig farms in Khon Kaen, Thailand. J. Med. Microbiol. 64, 1087–1093. doi: 10.1099/jmm.0.000119
Sinwat, N., Angkittitrakul, S., Coulson, K. F., Pilapil, F., Meunsene, D., and Chuanchuen, R. (2016). High prevalence and molecular characteristics of multidrug-resistant Salmonella in pigs, pork and humans in Thailand and Laos provinces. J. Med. Microbiol. 65, 1182–1193. doi: 10.1099/jmm.0.000339
Smith, T. C., and Pearson, N. (2011). The emergence of Staphylococcus aureus ST398. Vector Borne Zoonotic Dis. 11, 327–339.
Smith, T. C., Male, M. J., Harper, A. L., Kroeger, J. S., Tinkler, G. P., Moritz, E. D., et al. (2009). Methicillin-resistant Staphylococcus aureus (MRSA) strain ST398 is present in midwestern U.S. swine and swine workers. PLoS One 4:e4258. doi: 10.1371/journal.pone.0004258
Snitkin, E. S., Zelazny, A. M., Thomas, P. J., Stock, F., Group, N. C. S. P., Henderson, D. K., et al. (2012). Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci. Transl. Med. 4:148ra116. doi: 10.1126/scitranslmed.3004129
Sommanustweechai, A., Tangcharoensathien, V., Malathum, K., Sumpradit, N., Kiatying-Angsulee, N., Janejai, N., et al. (2018). Implementing national strategies on antimicrobial resistance in Thailand: potential challenges and solutions. Public Health 157, 142–146. doi: 10.1016/j.puhe.2018.01.005
Spratt, B. G. (2004). Exploring the concept of clonality in bacteria. Methods Mol. Biol. 266, 323–352. doi: 10.1385/1-59259-763-7:323
Srijan, A., Margulieux, K. R., Ruekit, S., Snesrud, E., Maybank, R., Serichantalergs, O., et al. (2018). Genomic Characterization of Nonclonal mcr-1-Positive Multidrug-Resistant Klebsiella pneumoniae from Clinical Samples in Thailand. Microb. Drug Resist. 24, 403–410. doi: 10.1089/mdr.2017.0400
Sudatip, D., Chasiri, K., Kritiyakan, A., Phanprasit, W., Thinphovong, C., Tiengrim, S., et al. (2021). A One Health approach to assessing occupational exposure to antimicrobial resistance in Thailand: the FarmResist project. PLoS One 16:e0245250. doi: 10.1371/journal.pone.0245250
Sun, Y., Duarte, M. E., and Kim, S. W. (2021). Dietary inclusion of multispecies probiotics to reduce the severity of post-weaning diarrhea caused by F18+ E. coli in pigs. Anim. Nutr. 7, 326–333. doi: 10.1016/j.aninu.2020.08.012
Ta, Y. T., Nguyen, T. T., To, P. B., Pham, D. X., Le, T. H., Thi, G. N., et al. (2014). Quantification, serovars, and antibiotic resistance of Salmonella isolated from retail raw chicken meat in Vietnam. J. Food Prot. 77, 57–66. doi: 10.4315/0362-028x.jfp-13-221
Tadee, P., Boonkhot, P., Pornruangwong, S., and Patchanee, P. (2015). Comparative phenotypic and genotypic characterization of Salmonella spp. in pig farms and slaughterhouses in two provinces in northern Thailand. PLoS One 10:e0116581. doi: 10.1371/journal.pone.0116581
Tang, Z., Xu, L., Shi, B., Deng, H., Lai, X., Liu, J., et al. (2016). Oral administration of synthetic porcine beta-defensin-2 improves growth performance and cecal microbial flora and down-regulates the expression of intestinal toll-like receptor-4 and inflammatory cytokines in weaned piglets challenged with enterotoxigenic Escherichia coli. Anim. Sci. J. 87, 1258–1266. doi: 10.1111/asj.12540
Tangcharoensathien, V., Sommanustweechai, A., Chanvatik, S., Kosiyaporn, H., and Tisocki, K. (2018). Addressing the threat of antibiotic resistance in Thailand: monitoring population knowledge and awareness. WHO South East Asia J. Public Health 7, 73–78. doi: 10.4103/2224-3151.239417
Teo, J. Q., Ong, R. T., Xia, E., Koh, T. H., Khor, C. C., Lee, S. J., et al. (2016). mcr-1 in Multidrug-Resistant blaKPC-2-Producing Clinical Enterobacteriaceae Isolates in Singapore. Antimicrob. Agents Chemother. 60, 6435–6437. doi: 10.1128/aac.00804-16
Thapa, S. P., Shrestha, S., and Anal, A. K. (2020). Addressing the antibiotic resistance and improving the food safety in food supply chain (farm-to-fork) in Southeast Asia. Food Control 108:106809. doi: 10.1016/j.foodcont.2019.106809
Trongjit, S., Angkittitrakul, S., and Chuanchuen, R. (2016). Occurrence and molecular characteristics of antimicrobial resistance of Escherichia coli from broilers, pigs and meat products in Thailand and Cambodia provinces. Microbiol. Immunol. 60, 575–585. doi: 10.1111/1348-0421.12407
Trung, N. V., Matamoros, S., Carrique-Mas, J. J., Nghia, N. H., Nhung, N. T., Chieu, T. T., et al. (2017). Zoonotic transmission of mcr-1 Colistin resistance gene from small-scale poultry Farms, Vietnam. Emerg. Infect. Dis. 23, 529–532. doi: 10.3201/eid2303.161553
United Nations [UN] (2016). Draft Political Declaration of the High-Level Meeting of the General Assembly on Antimicrobial Resistance. Available online at: https://www.un.org/pga/71/wp-content/uploads/sites/40/2016/09/DGACM_GAEAD_ESCAB-AMR-Draft-Political-Declaration-1616108E.pdf (accessed May 28, 2021).
Upadhaya, S. D., Lee, K. Y., and Kim, I. H. (2016). Effect of protected organic acid blends on growth performance, nutrient digestibility and faecal micro flora in growing pigs. J. Appl. Anim. Res. 44, 238–242. doi: 10.1080/09712119.2015.1031775
Van Boeckel, T. P., Glennon, E. E., Chen, D., Gilbert, M., Robinson, T. P., Grenfell, B. T., et al. (2017). Reducing antimicrobial use in food animals. Science 357, 1350–1352.
van der Peet-Schwering, C. M. C., Verheijen, R., Jørgensen, L., and Raff, L. (2020). Effects of a mixture of Bacillus amyloliquefaciens and Bacillus subtilis on the performance of growing-finishing pigs. Anim. Feed Sci. Technol. 261:114409. doi: 10.1016/j.anifeedsci.2020.114409
Van Looveren, F., De Jonghe, E., Maass, P., and P, D. B. (2015). “Reduction of antibiotic use after implementation of Ingelvac® PRRS MLV piglet vaccination in a Belgian wean to finish farm,” in Proceedings of the Safe Pork Conference, Porto, 353–355.
Verraes, C., Van Boxstael, S., Van Meervenne, E., Van Coillie, E., Butaye, P., Catry, B., et al. (2013). Antimicrobial resistance in the food chain: a review. Int. J. Environ. Res. Public Health 10, 2643–2669.
Vidayanti, I. N., Sukon, P., Khaengair, S., Pulsrikarn, C., and Angkittitrakul, S. (2021). Prevalence and antimicrobial resistance of Salmonella spp. isolated from chicken meat in upper northeastern Thailand. Vet. Integr. Sci. 19, 121–131.
Vincent, C., Usongo, V., Berry, C., Tremblay, D. M., Moineau, S., Yousfi, K., et al. (2018). Comparison of advanced whole genome sequence-based methods to distinguish strains of Salmonella enterica serovar Heidelberg involved in foodborne outbreaks in Quebec. Food Microbiol. 73, 99–110. doi: 10.1016/j.fm.2018.01.004
Von Wintersdorff, C. J. H., Penders, J., Van Niekerk, J. M., Mills, N. D., Majumder, S., Van Alphen, L. B., et al. (2016). Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 7:173. doi: 10.3389/fmicb.2016.00173
Vounba, P., Arsenault, J., Bada-Alambedji, R., and Fairbrother, J. M. (2019). Pathogenic potential and the role of clones and plasmids in beta-lactamase-producing E. coli from chicken faeces in Vietnam. BMC Vet. Res. 15:106. doi: 10.1186/s12917-019-1849-1
Wadepohl, K., Muller, A., Seinige, D., Rohn, K., Blaha, T., Meemken, D., et al. (2020). Association of intestinal colonization of ESBL-producing Enterobacteriaceae in poultry slaughterhouse workers with occupational exposure-A German pilot study. PLoS One 15:e0232326. doi: 10.1371/journal.pone.0232326
Walker, T. M., Ip, C. L., Harrell, R. H., Evans, J. T., Kapatai, G., Dedicoat, M. J., et al. (2013). Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect. Dis. 13, 137–146. doi: 10.1016/s1473-3099(12)70277-3
Wan, J., Li, Y., Chen, D., Yu, B., Chen, G., Zheng, P., et al. (2016). Recombinant plectasin elicits similar improvements in the performance and intestinal mucosa growth and activity in weaned pigs as an antibiotic. Anim. Feed Sci. Technol. 211, 216–226. doi: 10.1016/j.anifeedsci.2015.12.003
Wang, H., Ha, B. D., and Kim, I. H. (2021). Effects of probiotics complex supplementation in low nutrient density diet on growth performance, nutrient digestibility, faecal microbial, and faecal noxious gas emission in growing pigs. Ital. J. Anim. Sci. 20, 163–170. doi: 10.1080/1828051x.2020.1801358
Wang, W., Van Noten, N., Degroote, J., Romeo, A., Vermeir, P., and Michiels, J. (2019). Effect of zinc oxide sources and dosages on gut microbiota and integrity of weaned piglets. J. Anim. Physiol. Anim. Nutr. 103, 231–241. doi: 10.1111/jpn.12999
Wei, X., Tsai, T., Knapp, J., Bottoms, K., Deng, F., Story, R., et al. (2020). ZnO modulates swine gut microbiota and improves growth performance of nursery pigs when combined with peptide cocktail. Microorganisms 8:146. doi: 10.3390/microorganisms8020146
Wielders, C., Van Hoek, A., Hengeveld, P., Veenman, C., Dierikx, C., Zomer, T., et al. (2017). Extended-spectrum β-lactamase-and pAmpC-producing Enterobacteriaceae among the general population in a livestock-dense area. Clin. Microbiol. Infect. 23, 120.e1–120.e8.
Witte, W. (1998). Medical consequences of antibiotic use in agriculture. Science 279, 996–997. doi: 10.1126/science.279.5353.996
World Health Organization [WHO] (2015). Global Action Plan on Antimicrobial Resistance. Available online at: https://www.who.int/antimicrobial-resistance/global-action-plan/en/ (accessed December 18, 2020).
World Health Organization [WHO] (2017). Global Antimicrobial Resistance Surveillance System (ıGLASS)ı Report: Early Implementation 2016-2017. Available online at: https://apps.who.int/iris/bitstream/handle/10665/259744/9789241513449-eng.pdf (accessed December 17, 2020).
World Health Organization [WHO] (2018). Global Database for the Tripartite Antimicrobial Resistance (AMR) Country Self-assessment Survey (TrACSS). Available online at: https://amrcountryprogress.org/ (accessed March 14, 2021).
World Health Organization [WHO] (2019). Critically Important Antimicrobials for Human Medicine, 6th revision Edn. Available online at: https://www.who.int/foodsafety/publications/antimicrobials-sixth/en/ (accessed March 16, 2021).
World Health Organization [WHO]. (2020). Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report. Available online at: https://www.who.int/glass/resources/publications/early-implementation-report- 2020/en/ (accessed October 6, 2020).
World Organization of Animal Health (2020). Bulletin Panorama. Available online at: https://oiebulletin.com/index.php?panorama=02-2-3-2020-1-value-chain&edition=12827&pdf=panorama&article=13346 (accessed January 26, 2021).
Wu, S., Dalsgaard, A., Hammerum, A. M., Porsbo, L. J., and Jensen, L. B. (2010). Prevalence and characterization of plasmids carrying sulfonamide resistance genes among Escherichia coli from pigs, pig carcasses and human. Acta Vet. Scand. 52:47. doi: 10.1186/1751-0147-52-47
Wu, X., Suksawat, F., Richards, A. L., Phommachanh, S., Phongaran, D., and Angkititrakul, S. (2019). Evaluation of the Containment of Antimicrobial-Resistant Salmonella Species from a Hazard Analysis and Critical Control Point (HACCP) and a Non-HACCP Pig Slaughterhouses in Northeast Thailand. Pathogens 9:20. doi: 10.3390/pathogens9010020
Wu, Y., Pan, L., Shang, Q., Ma, X., Long, S., Xu, Y., et al. (2017). Effects of isomalto-oligosaccharides as potential prebiotics on performance, immune function and gut microbiota in weaned pigs. Anim. Feed Sci. Technol. 230, 126–135. doi: 10.1016/j.anifeedsci.2017.05.013
Xu, T., Zhang, C., Ji, Y., Song, J., Liu, Y., Guo, Y., et al. (2021). Identification of mcr-10 carried by self-transmissible plasmids and chromosome in Enterobacter roggenkampii strains isolated from hospital sewage water. Environ. Pollut. 268:115706. doi: 10.1016/j.envpol.2020.115706
Xu, Y., Liu, L., Long, S., Pan, L., and Piao, X. (2018). Effect of organic acids and essential oils on performance, intestinal health and digestive enzyme activities of weaned pigs. Anim. Feed Sci. Technol. 235, 110–119. doi: 10.1016/j.anifeedsci.2017.10.012
Xu, Y., Zhou, X., Jiang, Z., Qi, Y., Ed-Dra, A., and Yue, M. (2020). Epidemiological investigation and antimicrobial resistance profiles of Salmonella isolated from breeder chicken hatcheries in Henan, China. Front. Cell. Infect. Microbiol. 10:497. doi: 10.3389/fcimb.2020.00497
Yamaguchi, T., Kawahara, R., Hamamoto, K., Hirai, I., Khong, D. T., Nguyen, T. N., et al. (2020). High Prevalence of Colistin-Resistant Escherichia coli with Chromosomally Carried mcr-1 in Healthy Residents in Vietnam. mSphere 5:e00117-20.
Yi, L., Wang, J., Gao, Y., Liu, Y., Doi, Y., Wu, R., et al. (2017). mcr–1–Harboring Salmonella enterica serovar Typhimurium sequence type 34 in pigs, China. Emerg. Infect. Dis. 23, 291–295. doi: 10.3201/eid2302.161543
Yoon, J., Ingale, S., Kim, J., Kim, K., Lee, S., Park, Y., et al. (2014). Effects of dietary supplementation of synthetic antimicrobial peptide-A3 and P5 on growth performance, apparent total tract digestibility of nutrients, fecal and intestinal microflora and intestinal morphology in weanling pigs. Livest. Sci. 159, 53–60. doi: 10.1016/j.livsci.2013.10.025
You, Y., Leahy, K., Resnick, C., Howard, T., Carroll, K. C., and Silbergeld, E. K. (2016). Exposure to pathogens among workers in a poultry slaughter and processing plant. Am. J. Ind. Med. 59, 453–464. doi: 10.1002/ajim.22594
Zeineldin, M., Aldridge, B., and Lowe, J. (2019). Antimicrobial effects on swine gastrointestinal microbiota and their accompanying antibiotic Resistome. Front. Microbiol. 10:1035. doi: 10.3389/fmicb.2019.01035
Zeng, Z., Wang, D., Piao, X., Li, P., Zhang, H., Shi, C., et al. (2014). Effects of adding super dose phytase to the phosphorus-deficient diets of young pigs on growth performance, bone quality, minerals and amino acids digestibilities. Asian Australas. J. Anim. Sci. 27, 237–246. doi: 10.5713/ajas.2013.13370
Zhang, S., Yoo, D. H., Ao, X., and Kim, I. H. (2020). Effects of dietary probiotic, liquid feed and nutritional concentration on the growth performance, nutrient digestibility and fecal score of weaning piglets. Asian Australas. J. Anim. Sci. 33, 1617–1623. doi: 10.5713/ajas.19.0473
Keywords: antibiotic resistance, alternatives to antibiotics, one-health, pig production, policy, slaughtering process, South East Asia
Citation: Sirichokchatchawan W, Apiwatsiri P, Pupa P, Saenkankam I, Khine NO, Lekagul A, Lugsomya K, Hampson DJ and Prapasarakul N (2021) Reducing the Risk of Transmission of Critical Antimicrobial Resistance Determinants From Contaminated Pork Products to Humans in South-East Asia. Front. Microbiol. 12:689015. doi: 10.3389/fmicb.2021.689015
Received: 31 March 2021; Accepted: 21 June 2021;
Published: 27 July 2021.
Edited by:
Zhangqi Shen, China Agricultural University, ChinaReviewed by:
Takehiko Kenzaka, Osaka Ohtani University, JapanShabarinath Srikumar, United Arab Emirates University, United Arab Emirates
Copyright © 2021 Sirichokchatchawan, Apiwatsiri, Pupa, Saenkankam, Khine, Lekagul, Lugsomya, Hampson and Prapasarakul. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nuvee Prapasarakul, Nuvee.p@chula.ac.th
 Wandee Sirichokchatchawan1,2
Wandee Sirichokchatchawan1,2 Prasert Apiwatsiri
Prasert Apiwatsiri Pawiya Pupa
Pawiya Pupa Imporn Saenkankam
Imporn Saenkankam Nwai Oo Khine
Nwai Oo Khine Angkana Lekagul
Angkana Lekagul Kittitat Lugsomya
Kittitat Lugsomya David J. Hampson
David J. Hampson Nuvee Prapasarakul
Nuvee Prapasarakul