- 1Beijing Laboratory for Food Quality and Safety, Beijing, China
- 2College of Food Science and Nutritional Engineering, China Agricultural University, Beijing, China
G-protein-coupled receptors (GPCRs) are transmembrane receptors involved in transducing signals from the external environment inside the cell, which enables fungi to coordinate cell transport, metabolism, and growth to promote their survival, reproduction, and virulence. There are 14 classes of GPCRs in fungi involved in sensing various ligands. In this paper, the synthesis of mycotoxins that are GPCR-mediated is discussed with respect to ligands, environmental stimuli, and intra-/interspecific communication. Despite their apparent importance in fungal biology, very little is known about the role of ochratoxin A (OTA) biosynthesis by Aspergillus ochraceus and the ligands that are involved. Fortunately, increasing evidence shows that the GPCR that involves the AF/ST (sterigmatocystin) pathway in fungi belongs to the same genus. Therefore, we speculate that GPCRs play an important role in a variety of environmental signals and downstream pathways in OTA biosynthesis. The verification of this inference will result in a more controllable GPCR target for control of fungal contamination in the future.
Introduction
Ochratoxin A (OTA), which is a type of mycotoxin produced mainly by various Aspergillus and Penicillium species (Frisvad et al., 2019), was classified as a group IIB carcinogen by the International Agency for Research on Cancer (IARC) (International Agency For Research On Cancer, 1993). The latest in vitro toxicology experiment showed that alteration of DNA methylation occurred? in OTA-induced G0/G1 phase arrest (Zhang et al., 2019). Numerous cell and animal experiments have reported that OTA exposure can cause many toxicological effects, which include teratogenicity, carcinogenicity, mutagenicity, hepatotoxicity, and was a main causative agent of human Balkan endemic nephropathy (Pfohl-Leszkowicz and Manderville, 2012; Zheng et al., 2013; Qi et al., 2014; Zhang et al., 2014; Zhao et al., 2017; Hou et al., 2020). OTA widely contaminates feed and food commodities, and it is accumulated through the food chain in animals (Wenying et al., 2018). Global warming promotes the poleward movement of toxigenic fungi, causes contamination in previously unsuitable geographic regions, and exacerbates this threat (Bebber et al., 2013). OTA pollution occurs at various stages of agricultural production (i.e., plant growth, harvest, processing, transportation, and storage), and the occurrence of toxigenic fungi and the biosynthesis of OTA was influenced significantly at different stages of synthesis by external environmental factors, signaling molecules, and trans-kingdom communication.
Sensing and responding to environmental fluctuations are crucial to the survival of microorganisms. GPCRs are ubiquitous and the largest family of transmembrane receptors in both prokaryotes and eukaryotes. They contain seven transmembrane domains (TMDs) and are connected by alternating intracellular coils (IC1-IC3) and extracellular loops (EC1-EC3) (Baldwin, 1993). The extracellular amino acid? fragments sense the environment by interacting with a diverse array of ligands, then transmit this interaction through protein folding modifications to the intracellular carboxy-terminal end that recognizes the cognate heterotrimeric G proteins (Gαβγ) (Ballesteros et al., 2001; Shapiro et al., 2002). Sensitization of a GPCR by ligands results in the exchange of GTP for GDP on the Gα subunit, which leads to the dissociation of Gβγ; both GTP-Gα and Gβγ dimers can interact with respective effectors to activate or inhibit specific downstream pathways (Khan et al., 2013; Brown et al., 2018). In fungi, GPCR-regulated signaling pathways that include the cAMP-activated Protein Kinase A (PKA) pathway (Xue et al., 2006), mitogen-activated protein kinases (MAPK) cascades pathway (Chen and Thorner, 2007; Atoui et al., 2008; Hamel et al., 2012; Ma and Li, 2013), and the phospholipase C (PLC) pathway (Ansari et al., 1999) influence gene expression to regulate cell growth, morphogenesis, mating, stress responses, and metabolism in a complex and intersecting way (Rispail et al., 2009).
The regulator of G protein signaling (RGS) is a GTPase activating protein (GAP), which accelerates the decomposition of GTP-binging Gα by enhancing Gα subunit GTPase activity that negatively regulates G protein signal transduction (Kasahara et al., 2000; McPherson et al., 2018). Another G protein signal-regulating protein, Phosducin-like protein (PhLP), is involved in the G protein signal transduction pathway by positively regulating Gβγ subunits (Lukov et al., 2014). Significant differences exist in the abundance and diversity of these receptors in fungi and the potential ligands that they detect. A variety of GPCRs relates to OTA biosynthesis, which involves a wide range of environmental signals and downstream pathways (Figure 1). Because of their cell surface location and central mediating role, GPCRs are specific targets to control the biosynthesis of fungal toxins and to intervene in fungal disease and mycotoxin contamination.
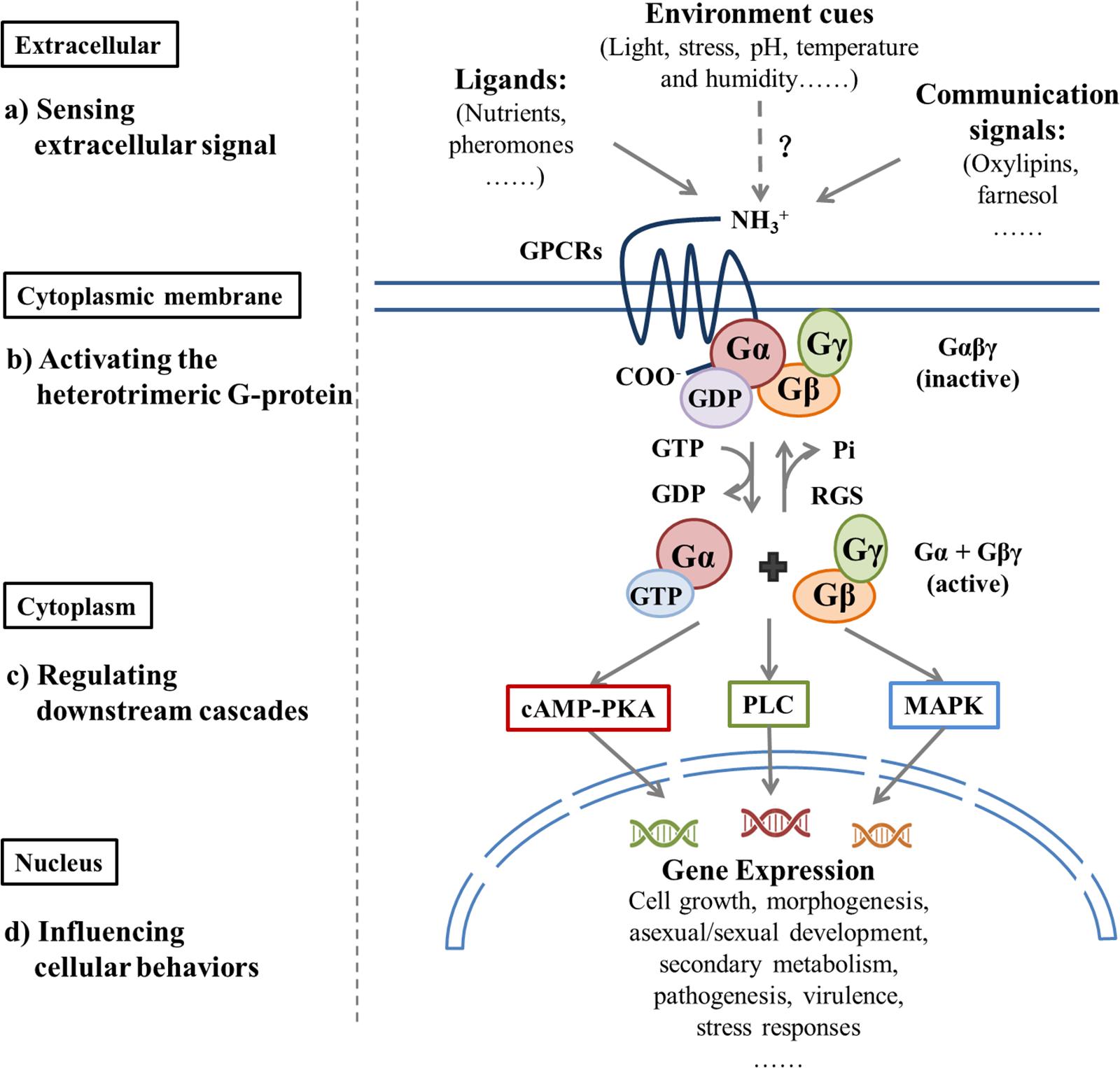
Figure 1. Extracellular signals regulate downstream pathways by activating G protein signaling and, thus, influence cellular behavior.| Different classes of GPCRs sense various extracellular factors, which include ligands, environmental cues, and communication signals that bind to the GPCR; this causes GDP-GTP exchange on the Gα protein and dissociation of Gα and Gβγ. Both the Gα-GTP and Gβγ dimers may trigger respective downstream signal cascades, which include the cAMP-activated Protein Kinase A (PKA) pathway, mitogen-activated protein kinase (MAPK) cascades pathway, and the phospholipase C (PLC) pathway. This process influences cellular growth, reproduction, metabolism, virulence, and stress responses. GTP hydrolysis by the Gα subunit results in the reassociation of Gα-GDP with the Gβγ dimer and GPCR, which completes the G protein cycle. The Regulator of G-protein Signaling (RGS) proteins can inactivate G protein signaling rapidly by increasing intrinsic GTPase activity of GTP-bound Gα subunits. In this figure, dashed arrows with the question mark (?) indicates hypothetical interactions.
The well-researched GPCR-mediated, G-protein signaling pathway, which regulates fungal behavior in response to a variety of signals, is highly conserved in animals, plants, and microorganisms. This phenomenon and the mechanism of model strains have been studied thoroughly enough to provide the guiding ideology and practical basis for research on GPCRs in fungi. The bovine rhodopsin and human beta2-adrenergic GPCRs were crystallized and based on homology and structural similarity, which served as models for the structure of other members of the GPCR family (Stenkamp et al., 2002; Rasmussen et al., 2007). Although GPCR signaling is essential for fungal biology, the identified interactions of GPCR–G-protein, the studies of receptor binding ligands, and the resolved GPCR crystal structures are only the beginning of what we need to know. Perfecting the above information will prove vital in understanding the upstream ligand–receptor relationship of the OTA synthesis regulatory pathways and the development of novel fungal GPCR-targeting of mycotoxin control. This paper reviews the current understanding of fungal GPCR-mediated signaling pathways that regulate fungal behavior and OTA synthesis; this has important theoretical significance for improving our knowledge about the function of GPCRs and the synthesis of secondary metabolites. It is expected that we will eventually use GPCR as targets to prevent and control the harm caused by fungal toxins.
GPCRs Sense Nutrients and Pheromones
Fungal adaptation to distinct microenvironments, which include hosts, natural habitats, and culture media, is essential for their success (Brown and Goldman, 2016). G-protein signaling pathways play a vital role in sensing external ligands, which include nutrients, hormones, proteins, and peptides (including pheromones), ions, hydrophobic surfaces, and light (Kochman, 2014). This enable fungi to coordinate function, metabolism, and development that, in turn, promotes their survival, propagation, and virulence (Van Dijck et al., 2017). At present, based on structural similarity and homology, fungal GPCRs are classified into 14 categories, which include six original classes and eight novel classes (Supplementary Table 1; Lafon et al., 2006; Li et al., 2007; Zheng et al., 2010; Grice et al., 2013; Cabrera et al., 2015; Brown et al., 2018; Nadarajah et al., 2018). The model filamentous ascomycete fungus A. nidulans possesses 86 putative GPCRs, which are classified into 16 receptors, named GprA∼GprP and NopA, constitute nine categories of GPCRs; only a few have been characterized functionally (Lafon et al., 2006; Krishnan et al., 2012; Brown et al., 2015).
Many studies have found that GPCR-mediated perception of signaling molecules, especially pheromones, nutritions and oxylipins, are closely related to fungal reproduction and the production of secondary metabolites, which seems to imply that there are cross-effects of different GPCRs. This complex regulatory network provides numerous targets for controlling OTA biosynthesis.
Perception of Nutrients
G-protein-coupled receptors sense nutrients, and then they influence fungal development and metabolism. An enormous amount of basic insights into nutrient utilization by fungi has been established. The interaction of GPCR protein Gpr1 with Gα protein Gpa2 is necessary for the stimulation of cAMP synthesis by sugars. In the model organism Saccharomyces cerevisiae, Gpr1 is a high-affinity sucrose and a low-affinity glucose sensor. The addition of glucose to a glucose starvation culture triggers a rapid and transient increase in the cAMP level, which sets off a PKA-mediated protein phosphorylation cascade. Deletion of Gpr1 and/or Gpa2 affected a variety of physiological events controlled by cAMP-dependent protein kinases (cAPKs), such as mobilization of trehalose and glycogen and a rapid loss of stress resistance (Rolland et al., 2000; Lemaire et al., 2004). The absence of Gpr1 can be rescued by the constitutively activated Gpa2Val–132 allele, which supports the interaction of GPCR and its cognate G protein in regulation (Kraakman et al., 2010). GprC and GprD (Class III) in Aspergilli are analogous to Gpr1 in S. cerevisiae, which is involved in the sensing of sugar and oxylipin (De et al., 2013). Deletion of the gprC or gprD gene in A. fumigatus affected the ability of strains to produce several toxic secondary metabolites and then affected the growth and pathogenicity in model murine infections (Gehrke et al., 2010). In A. flavus, deletion of genes gprC and/or gprD resulted in alteration of quorum sensing (QS), sporulation, sclerotia formation, and biosynthesis of aflatoxin (AF) (Affeldt et al., 2012). GprK (ClassVI) in A. fumigatus was involved in sensing external carbon sources, which is a more complex sensor protein that contains a RGS domain in addition to the 7-TM domains. Overexpression of gprK significantly activated toxin in the biosynthesis-related cAMP-PKA pathway, then activated the MAPK signaling pathway, which partially regulates the expression of genes that code for catalase and superoxide dismutase and, thus, indirectly affects secondary metabolism.
Even in the absence of carbon sources, ΔgprK mutants showed a severe morphological alteration, a change in asexual reproduction, and a reduction in tolerance to different stressing factors, which included oxidative stress and destruction of the ability to produce gliotoxin (Jung et al., 2016). Both GprM (ClassVII, homologous to rat growth hormone-releasing factor receptors) and GprJ (Class IV, nitrogen source sensors) were involved in regulating the production of DHN-melanin in A. fumigatus, and this regulation partially occurred through the activation of MpkA (Manfiolli et al., 2019; Filho et al., 2020). GprH (ClassV), a common glucose and tryptophan sensor, has been found in many filamentous fungi, which regulates hyphal growth and sexual reproduction during carbon starvation in A. nidulans. In cultures supplemented with glucose and tryptophan, the deletion of gprH caused a decrease in cAMP-PKA activity under stress conditions (Brown et al., 2015), which suggested that a single GPCR interacted with multiple ligands. In Cryptococcus neoforme, Gpr4 was similar to Gpr1 in S. cerevisiae and to GprH in A. nidulans, which induced the cAMP–PKA pathway. Glucose, but not methionine, acted on Δgpr4 mutants to cause a change in the cAMP level, and the deficiencies in capsule formation and mating defects were reverted by cAMP supplementation. Therefore, Gpr4 protein has been proposed to be a sensor of amino acids, particularly for methionine (Xue et al., 2006).
There are also examples of other types of nutrient sources that are sensed and regulated by fungal GPCR. Four cAMP-type GPCR genes (gpr1 to gpr4) in Trichoderma atroviride belong to class V, and expression of all four gpr genes increased after carbon starvation. Gpr1 protein is essential to conidium germination and mycelial growth. Expression of gpr3 and gpr4 responded to exogenous cAMP, and the addition of hyphal fragments and cellulosic material also increased the expression of gpr4 (Brunner et al., 2008). The Pth11 type GPCR of Neurospora crassa was involved in the detection of cellulose materials, plant cell walls, or their degradation products, which triggered a response that favored fungal infection of plant hosts (Cabrera et al., 2015; Thieme et al., 2017).
The above results showed that fungi can sense a variety of nutrients through GPCRs, and they can regulate cell growth, development, immune evasion, invasiveness, mycotoxin production, chemotropism, and virulence. Disrupted nutrient-sensing GPCRs could be targeted to reduce fungal virulence and mycotoxin contamination.
Perception of Pheromones
Pheromones are another type of typical GPCR ligand that influences fungal reproduction and virulence. The benefits of rapid evolution that is driven by sexual reproduction of virulence and resistance are essential for promoting genetic diversity and evolutionary competition with hosts. Pheromones secreted by one type of mating cell and recognized by the opposite type of mating cell trigger downstream signal transduction cascades that lead to cell mating, and they also play a potential role in the production of secondary metabolites (Karlson and Lüscher, 1959; Herskowitz, 1995; Xue et al., 2008; Kim and Borkovich, 2010). In S. cerevisiae, the GPCR sex pheromone sensor proteins Ste2 and Ste3 detect the a and α sex peptide pheromones, respectively, which can activate the MAPK cascade to result in cell cycle arrest and cell fusion with the opposite mating type (Burkholder and Hartwell, 1985; Kuchler et al., 1989; Herskowitz, 1995; Chen and Thorner, 2007). PcPRE1 and PcPRE2, which are homologs of Ste2 and Ste3 pheromone receptors involved in mating, have been found in P. notatum, P. chrysogenum, and Acremonium chrysogenum; previously, it was thought that these fungi were incapable of sexual reproduction (Paoletti et al., 2005; Poggeler et al., 2008; Bohm et al., 2013; Terfehr et al., 2014).
The mating-type loci MAT1-1-1 and MAT1-1-2 were found in A. chrysogenum, and the mating-type genes of the recombinant strains control conidia formation, hyphal differentiation, and penicillin production. In the Δmat1 mutant, the expression of biosynthetic genes in penicillin decreased because the pellet size and structure were affected by hyphal differentiation (Poggeler et al., 2008). In A. nidulans, GprA and GprB are required for sexual development (without vegetative growth), which include formation of self-fertilized fruiting bodies, asexual development, Hul̈le cell production, and heterothallic sexual development (Han et al., 2004). Single ΔgprA and ΔgprB mutants and double ΔgprAΔgprB mutants were defective in sexual reproduction. Interestingly, GprD-mediated carbon sensing also acted as a repressor of sexual development through the PKA pathway; when grown in the presence of glucose, GprD promoted hyphal growth and conidial germination. The sexual developmental activator NsdD functioned downstream of GprA or GprB, whereas GprD-mediated attenuation of sexual development functioned upstream of GprA and GprB (Han et al., 2004; Seo et al., 2004). On the other hand, GprB and GprD affected QS, spore and sclerotia formation, mycotoxin production, and other metabolic pathways (De et al., 2013). The putative carbon and amino acid receptor GprH worked upstream of the cAMP-PKA pathway, which promoted glucose uptake and hyphal growth and repressed sexual development during carbon starvation.
Cross-Talk and Interaction Among GPCR-Mediated Pathways
Ligands, GPCRs, and downstream regulatory pathways are not strictly corresponding; this cross-over internal relationship increases the possibility of GPCR as targets for regulating cell behavior. Nutritional state and the perception of sexual partners regulate sexual developmentbecause nutrient limitation reduces pheromone signaling and, in turn, mating efficiency. Nutrient and pheromone GPCR pathways significantly influence the next step of cell growth or reproduction as cell cycle arrest by integrating environmental and internal signals.
In C. albicans, the carbon source sensing protein CaGpa2 not only regulated the cAMP level, but it also inhibited pheromone-mediated cell cycle arrest; the absence of CaGpa2 caused pheromone hypersensitivity and mating (Dignard et al., 2008). CaGpa2 was also important for activating the mating MAPK pathway, which showed a link between the nutrient-sensing pathway and the pheromone-responsive pathway (Bennett and Johnson, 2010). The A. nidulans AnGprD protein regulates hyphal growth and conidial germination and represses sexual development during growth on glucose. In A. fumigatus, although GrpC and GprD are similar to the S. cerevisiae glucose receptor Gpr1, experiments showed that they were not involved in glucose and cAMP sensing. AfuGprC and AfuGprD proteins regulated growth, morphogenesis, reactive oxygen species (ROS), and temperature tolerance, and virulence in a murine model of pulmonary aspergillosis, although it had an opposing influence on the transcriptional regulation of primary and secondary metabolism (Gehrke et al., 2010). In A. flavus, inactivating each of 15 GPCR proteins separately showed mixed effects on the response to carbon sources, nitrogen sources, lipid molecules, environment stress signals, cell growth, conidiation, production of secondary metabolites, and virulence; two or more these responses were altered in several null mutants (Cabrera et al., 2015). It seems that GPCR-mediated signal transduction pathways cross each other in downstream cascades, which resulted in different outcomes? that were expected for a single GPCR target (Martín et al., 2019). Therefore, signals generated by distinct GPCR-mediated nutrient and pheromone-sensing pathways are potentially integrated into one biological outcome through downstream, dual-function signaling elements. In addition, GPCRs are adapted to detect multiple environmental cues and to bind multiple ligands to induce different signaling pathways. Thus, the interlinked signaling pathways are modulated differentially and fine-tuned to regulate multiple aspects of fungal development, metabolism, and virulence.
Potential Role of GPCRs in a Fungal-Sensing Environment
The secondary metabolism biosynthetic genes in the fungal genome are clustered on the chromosome, which contain several key structural genes that encode multimodular enzymes that belong to the polyketide synthases (PKSs) or non-ribosomal peptide synthetases (NRPSs) families. These enzymes facilitate the construction of the main scaffold structure of many secondary metabolites. Additional enzymes introduced various modifications to the original structure (Alkhayyat and Yu, 2014). This complex process requires one or more cluster-specific transcription factors (TFs) to regulate, which operates downstream of G-protein signaling. The regulation of global TFs related to environmental signals is also at an upper level of cluster-specific regulation.
For example, the most studied pathway-specific TF, AflR, which regulates the AF/sterigmatocystin (ST) biosynthetic gene cluster, was affected by CreA (carbon source), AreA (nitrogen source), Velvet (light), PacC (pH), and other global TFs (Macheleidt et al., 2016; Caceres et al., 2020; Gil-Serna et al., 2020). Other environmental factors, such as temperature, humidity, and osmotic pressure, also have a comprehensive influence on microbial behavior. High-similarity, specific gene clusters were found in several common OTA-produced Aspergillus and Penicillium species, which contained four highly conserved biosynthetic genes that encoded polyketide synthase (PKS), non-ribosomal peptide synthetase (NRPS), halogenate or chloroperoxidase (HAL/CHL), cytochrome P450 (P450), and a bZIP transcription factor (Antonia et al., 2016; Ferrara et al., 2016; Gil-Serna et al., 2018; Yan et al., 2018). bZIP transcription factor is supposed to be a specific regulator for secondary metabolite biosynthesis and the hub that accommodates various inputs that lead to a single output, and it altered the expression of all four biosynthetic genes (pks, nrps, p450, hal) (Figure 2; Reverberi et al., 2008; Nadia, 2015; Yan et al., 2018). Based on a similar mechanism of secondary metabolite production, we speculate that environmental signals may regulate OTA biosynthetic gene clusters in a cluster-specific, transcription factor-dependent manner; complex and integrated GPCRs likely play an important transfer role in this process.
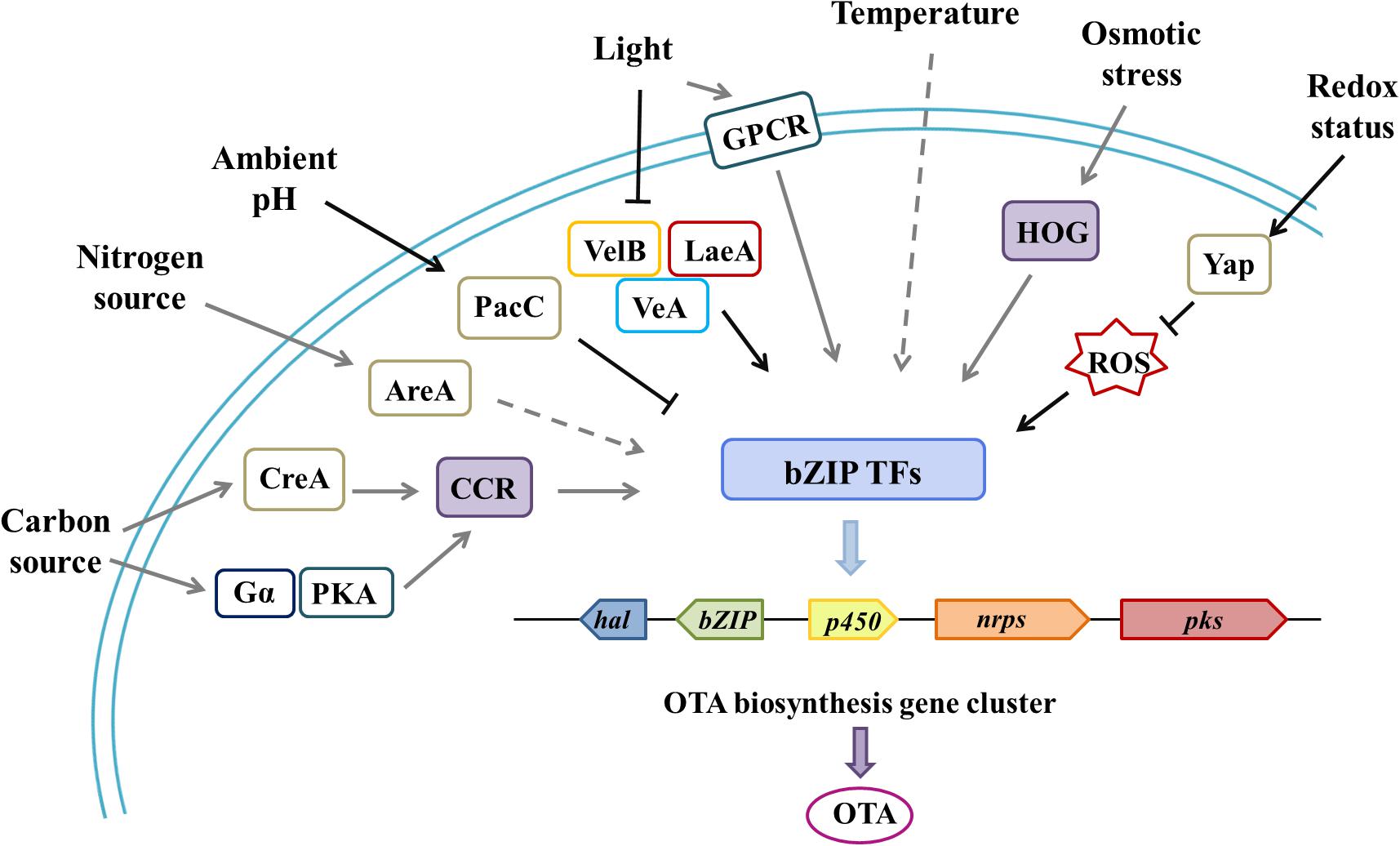
Figure 2. Environmental factors and associated regulatory elements that affect OTA biosynthesis through cluster-specific transcription factors.| Through various global transcription factors or specific pathways, related environmental cues affect the regulation of OTA gene clusters, which contain four highly conserved biosynthetic genes (pks, nrps, hal/chl, p450) and a cluster-specific, transcription factor gene (bZIP). bZIP plays a pivotal role in processing various environmental signals for a carbon source (CreA transcription factor and the carbon catabolite repression pathway), a nitrogen source (AreA transcription factor and the nitrogen catabolite repression pathway), light (Velvet complex proteins, VelB-VeA-LaeA, and the global regulator VosA), temperature and humidity, redox status (Yap transcription factor and ROS), and osmotic stress (the high osmolarity glycerol system). At the same time, there are also some examples where G protein pathways are partly involved in the perception of environmental cues, such as Gα(GanB) and PKA (PkaA) in A. nidulans and GPCR (NopA) in A. fumigatus. In this figure, the black solid arrows indicate connections verified in OTA-producing fungi; the gray solid arrows indicate connections that have been verified in other toxin-producting fungi, but not verified in OTA-producting fungi; and the gray dashed arrows indicate possible, yet unproven, connections.
Carbon/Nitrogen Sources
Fungi can utilize a diverse array of nutrient sources, which include hexose, pentose, and complex saccharides. This nutritional plasticity may be reflected, therefore, in the dramatic expansion of putative GPCRs (Wang et al., 2020). A wider evaluation of the function of all 15 classical GPCRs in A. flavus revealed multiple GPCRs that were potentially involved in carbon sensing because the individual absence of the receptors GprA, GprC, GprJ, GprK, or GprR impaired growth on several carbon sources (Dierking and Pita, 2020). In addition to the GPCR mentioned above (Section “Perception of Nutrients”), nutrients also regulate life activities through global transcription factors. In the presence of favored carbon sources that can be metabolized rapidly to provide quick energy, fungi will repress the utilization of alternative, less preferred carbon sources, which is known as carbon catabolite repression (CCR) (Ruijter and Visser, 1997). The CCR mechanism is regulated in part by the C2H2 zinc-finger global transcription factor CreA, which represses the expression of genes required for catabolism of less preferable carbon sources, gluconeogenic genes, and nutrient acquisition genes in response to carbon starvation (Adnan et al., 2018). CreA up-regulated AF/ST biosynthesis by affecting the regulatory factor AflR (Hicks et al., 2001). Notably, loss of CreA in A. nidulans, Fusarium oxysporum, P. chrysogenum, and Colletotrichum gloeosporoides was fatal, but it did not affect the activity of N. crassa, T. reesei, and A. fumigatus (Ries et al., 2018). Recent studies revealed that Gα (GanB) and PKA (PkaA) participated in CreA-independent CCR, then regulated mycotoxin synthesis at the transcriptional level in A. nidulans (Kunitake et al., 2019; Figure 2). This study indicated that the G-protein signaling pathway and the global regulator pathway partially overlapped in their perception of a carbon source.
Nitrogen is another important nutrient that affects mycotoxin synthesis. In A. carbonarius, inorganic nitrogen and ammonium chloride strongly reduced the OTA level, but organic nitrogen promoted OTA yield (Tudzynski, 2014). Similarly, there was a nitrogen catabolite repression (NCR) for nitrogen sources, which was affected by the C2H2 zinc-finger, global transcription factor AreA (Wong et al., 2008). However, AreA contributed to, but was not essential for, virulence of A. fumigatus (Kmetzsch et al., 2011).
Light
Photon is a type of ligand of rhodopsin-like GPCRs, which is a special animal neuronal photoreceptor that converts light signals into electrical signals. After photons are absorbed by rhodopsin, the voltage of the photoreceptor cell membrane is changed, thereby regulating life activities, and G protein plays a role in the conduction and amplification of this photoelectric signal (Nagy, 1991). In microorganisms, the bacterial rhodopsins bind retinal and act as light-driven proton pumps that pump protons from intracellular to extracellular regions? areas? to form a proton gradient, which is used by ATPase to synthesize ATP (Ignatov and Mosin, 2014). One class of fungal GPCR that is similar to bacterial rhodopsin has been identified. In Neurospora crassa, the NOP-1 receptor that is homologous to the bacterial opsin was verified and classified as a class IX GPCR. Results from Northern analysis supported light-based regulation of nop-1 gene expression, and the NOP-1 protein functions as rhodopsin in N. crassa photobiology; (Bieszke et al., 1999) a unique homolog of NOP-1 was also identified in Aspergillus species (Supplementary Table 1).
The heterotrimeric Velvet complexes (VelB-VeA-LaeA) is a light-dependent regulator in fungi, which contain transcription factors (i.e., VelB, VelC, and VosA), and the global regulator (VeA) interacts with methyltransferase (LaeA) to regulate fungal development and secondary metabolism (Dasgupta et al., 2016; Zhang et al., 2018; Gil-Serna et al., 2020). VeA regulates the synthesis of ST in A. nidulans and AF in A. flavus (Eom et al., 2018); the ΔveA and ΔvelB mutations impair ST production in A. nidulans (Kim et al., 2020) and OTA production in A. carbonarius (Crespo-Sempere et al., 2013) and A. ochraceus (Wang et al., 2019). The LaeA protein acts as a positive regulator on conidia production, OTA biosynthesis, and oxidative stress tolerance in A. niger (Ruitao et al., 2019). VeA is not a light sensor, but shows light-dependent mobility. Under light exposure, it occurs abundantly in the cytoplasm and is associated with filamentous bodies, but in the dark it is transferred into the nuclei by integrins. There, it is functionally active and interacts in concert with other proteins to form the velvet complex; it supports sexual reproduction and secondary metabolism in the dark and sporulation under light conditions (Calvo et al., 2016). Studies in A. nidulans linked the light-regulated Velvet pathway and GPCR, in which VeA regulators act as the bond. GprH, GprI (Class V), and GprM (Class VII) receptors collectively represent a carbon starvation-induced, nutrient sensing mechanism, which sensed glucose and propagated signals through the light-responsive VeA pathways and cAMP-PKA pathways to promote vegetative growth. GprH coordinated sexual development by regulating VeA nuclear localization and activity and then it repressed sexual development and ST production (Dos Reis et al., 2019).
Interestingly, different wavelengths are sensed by different light-receptors and have almost the opposite effect on mycotoxin production. This similar phenomenon was found in Cherax quadricarinatus because the activation of the Gq protein was related to the wavelength for light stimulation (Yan et al., 2003). Major OTA-produced Aspergillus and Penicillium spp. exposed to white light consistently reduced OTA production, whereas Fusarium spp. produced more fumonisin under light conditions (Fanelli et al., 2016). In A. ochraceus, A. niger, and Penicillium spp., red and blue wavelengths reduced OTA biosynthesis by modulating the level of expression of ochratoxin polyketide synthase. However, the red light for A. carbonarius and the yellow and green light for A. steynii caused increased OTA production (Fanelli et al., 2016). Different wavelengths of light are known to be accepted by different protein receptors, but the association with GPCRs needs to be explored further.
Stress
Fungi have sophisticated signaling cascades to sense and to respond to different stressors, which include UV irradiation, temperature, osmotic shock, high salt, oxidative or nitrosative damage, and exposure to antifungal drugs (Arroyo et al., 2009). Mainly involved is the high osmolarity glycerol (HOG) system and its activated MAPK pathway. In addition to stress control, the fungal HOG pathway regulates cell-cycle progression, sexual development, and morphological differentiation. Cryptoccus neoformans has developed a specialized HOG pathway. Hog1 of C. neoformans is constitutively phosphorylated under normal conditions and represses sexual reproduction and the synthesis of capsule and melanin. In P. verrucosum, P. nordicum, and A. carbonarius, osmotic stress is associated with severe changes in OTA biosynthesis through the HOG pathway (De Assis et al., 2020). Metabolic reactions are triggered in part by cells in response to oxidative stress (redox state). In A. parasiticus, oxidative stress represented by intracellular ROS accumulation, enhanced AF production, but the application of antioxidants, such as butanol, reduced AF production (Jayashree and Subramanyam, 2000; Reverberi et al., 2005). The transcription factor Yap1 in S. cerevisiae controlled toxins produced in response to oxidative stress. Deletion of the Yap-1 homologous gene Aoyap1 in A. ochraceus damaged the activity of superoxide dismutases and catalases and, therefore, did not effectively scavenge high-level ROS to maintain the redox balance, which triggered the biosynthesis of OTA (Reverberi et al., 2012). Yap1 orthologs are coregulators of oxidative stress response with secondary metabolism in other aspergilli, such as napA in A. nidulans, Apyap1 in A. parasiticus, and Afap1 in A. flavus (Reverberi et al., 2007, 2008, 2012; Yin et al., 2013; Guan et al., 2019; Mendoza-Martinez et al., 2020). On the other hand, ROS catalyzed the production of oxylipin non-enzymatically, which is the ligand of receptors GprC and GprD, and they affected cellular activity that included toxin synthesis (described below). However, evidence for a strong link between the redox state and GPCR is still lacking.
pH
G-protein-coupled receptors are mainly located on the surface of cell membranes and are regularly exposed to a wide range of dynamic pH microenvironments. Human OGR1 and GPR4 are typical proton-sensing, G-protein coupled receptors involved in pH homeostasis. GPR4, GPR65, and GPR68 receptors can be activated directly by pH and elicit cAMP formation (Rowe et al., 2020). However, no fungal proton-sensing GPCRs have been reported. In fungi, the perception of pH is closely related to the PacC pH adaptation signaling pathway. The dedicated proteins PalH, PalI, PalF, PalC, PalA, and PalB transmit environmental pH changes to transcription factor PacC (Cornet and Gaillardin, 2014). PacC effectively inhibits the expression of the known acid-expressed genes and undergoes pH-dependent proteolytic cleavage under alkaline ambient pH conditions, and then it regulates neutral-alkaline sensing in filamentous fungi (Tilburn et al., 1995; Penalva et al., 2008; Bignell, 2012). PacC is involved in the synthesis of secondary metabolites in fungi that activates penicillin biosynthesis at an alkaline pH (Brakhage, 1998), but it inhibits the biosynthesis of AF in A. parasiticus, ST in A. nidulans, OTA in A. ochraceus, fumonisin in F. verticillioides (Keller et al., 1997; O’callaghan et al., 2006), and gliotoxin in A. fumigatus (Kapetanakou et al., 2009). The regulation of pH in OTA synthesis is related to pks genes in the OTA biosynthesis gene cluster. Expression of the otapksPN gene in P. nordicum has been reported to be lower under acidic conditions below pH 5 (Geisen, 2004). Currently, there is no evidence that the pH sensor interacts with GPCRs in fungi. However, the structure of sensing external signals contained in pH sensors is similar to GPCR, which contains 7TMDs and a cytoplasmic C-terminus (Herranz et al., 2005; Hervas-Aguilar et al., 2010), and both regulate OTA synthesis at the transcriptional level. Therefore, we speculate that there is a certain connection between potential GPCRs and pH sensing in fungi, just like human pH-sensing GPCRs.
Temperature and Humidity
Temperature and humidity both are key factors in the environmental regulation of fungal growth and secondary metabolites (MaríN et al., 1998; Tai et al., 2020; Wang et al., 2020). The minimum temperature for fungal growth increases with the enhanced inhibition of other environmental factors on growth, and mycotoxin production increases with the humidity increasing at the same temperature. However, there was not always a positive correlation between water activity and mycotoxin production in different cultures. The temperature range for mycotoxin production is stricter than for fungal growth. For example, suboptimal growth conditions that enhanced mycotoxin production is the optimum temperature for the biosynthesis of AF (i.e., 30°C), but the optimum temperature for Aspergillus growth is 37°C (Marc et al., 2002). The RNA-seq results of Yu et al. (year?) showed that most of the genes involved in AF synthesis, which included the cluster-specific regulatory genes aflR and aflS, were highly upregulated at low temperatures (Yu et al., 2011). Affeldt et al. (year?) annotated 15 GPCR genes in the A. flavus genome. The expression of gprC, gprD, gprF, gprG, and gprO were significantly different at three temperatures (20°C, 28°C, and 37°C). The deletion of either gprC or gprD resulted in a severe, temperature-dependent growth defect that was independent of the carbon source in A. fumigatus (Gehrke et al., 2010; Han et al., 2019). Knockout of gprD at a low temperature (20°C) increased the yield of AF, but there was no significant difference between wild-type and mutant under 28°C and 37°C (Han et al., 2019); this may have been due to the existence of alternative pathways to maintain normal biological functions.
The type of GPCR receptor and their ligands are being explored continuously, and the crossover phenomenon suggests that we cannot completely separate the G-protein signaling pathway and environmental factors perception for the regulation of OTA synthesis. Macroscopically, the dominant influence of temperature and humidity on OTA yield has been observed, and oxidative stress also plays a significant role; common stimuli, such as light and pH, also play a role. Indeed, studies have linked this regulation with GPCR. Although a large number of environmental factors influence the phenotypic phenomena associated with OTA biosynthesis, further confirmation of these is lacking. GPCRs play a critical role in perceiving these environmental stimuli and regulating the appropriate signaling pathways. However, the importance of GPCRs and the functional connections in regulating secondary metabolism are unknown.
GPCRs Mediate Trans-Kingdom Communication
The natural interactions of fungi with bacteria, and cross-talks between infecting fungi and their infected hosts, induce many response signals that are sensed by fungi. Fungal G-protein signaling pathways that regulate cell behavior are also influenced by signaling molecules derived within and between fungal species (Fischer and Keller, 2016; Van Dijck et al., 2017), which implies that communication between species is partly mediated through GPCRs. This suggests that further study of these communication events is necessary to find targets to control mycotoxin biosynthesis.
Fungal Quorum Sensing
Fungal QS is one of the main mechanisms for intra- and interspecific communication. Hormone-like small molecular compounds known as quorum-sensing molecules (QSMs) are secreted by fungi and enter into other cells through specific transporters and activate the expression of corresponding genes in a density-dependent manner within cells?. The QS mechanism plays a role in monitoring population density and regulating the physiology of fungal cells, that affect growth, sexual/asexual reproduction, apoptosis, secondary metabolism, and pathogenesis. This mechanism causes fungi to coordinate their actions and to enhance their survival, host immune evasion, and infection ability to adapt to environmental changes. Currently, verified fungal QSMs include alcohols, oxylipins, small molecule peptide pheromones, and certain volatile substances (Wongsuk et al., 2016; Barriuso et al., 2018; Padder et al., 2018). The perception of pheromones and their effects on fungal biology are mediated by GPCRs, and oxylipin is also a class of ligand for GPCRs.
Oxylipins, which is a large family of enzymatic or non-enzymatic oxidation products of fatty acids, are common signaling molecules in animals, plants, and microorganisms (Wasternack and Feussner, 2018). Treatment of A. nidulans wild-type with exogenous oxylipins resulted in cAMP accumulation, but this could be prevented in the absence of the gprD gene (De et al., 2013). Indeed, studies on both the GprD protein of A. nidulans and GprC and/or GprD proteins of A. fumigatus revealed that oxylipins were likely to be a type of ligand of these fungal GPCRs and to activate the cAMP pathway (Affeldt et al., 2012).
9S-Hydroxyoctadecadienoic acid (9-HODE), 13S-Hydroxyoctadecadienoic acid (13-HODE), and their derivatives that are derived from linoleic acid, act as crucial signals and elicitors of secondary metabolites in fungi and plants. 9(S)-HODE inhibited A. ochraceus sporulation and promoted the production of OTA, but 13(S)-HODE promoted the sporulation and inhibited the production of OTA (Reverberi et al., 2010). 13(S)-HPODE inhibited the expression of mycotoxin synthesis genes of A. parasiticus (ver-1) and A. nidulans (stcU) and significantly reduced the production of AFB1 and ST, although the inhibition effect at the same concentration of 9(S)-HPODE was not obvious (Burow et al., 1997).
Precocious sexual inducer (PSI) factors are mixed oxylipin signals produced by PpoA-C oxygenases, which are the main oxidases that regulate oxylipin synthesis in Aspergillus species, and the ratio of psiA-C determines if fungi enter sexual or asexual development (Fischer and Keller, 2016). A. fumigatus and A. flavus can produce the secreted PpoA oxylipin 5,8-dihydroxyoctadecadienoic acid (5,8-diHODE), which use a model of an autocrine-like mechanism to regulate hyphal branching. Also, the rice blast pathogen Magnaporthe grisea produces the branching-inducing oxylipin 7,8-diHODE. When exposed to exogenous 5,8-diHODE, M. grisea germlings differentiated predominantly into appressoria, the infectious structure required for plant penetration (Niu et al., 2020). On the other hand, M. grisea can produce the autocrine signal 7,8-diHODE to induce branching, and it is proposed that there is a cross-genera recognition of fungal dihydroxyl oxylipins between Aspergillus and M. grisea, where the 5,8-diHODE acts as a paracrine signaling molecule between cells of some fungal species.
Knocking out the ppo genes in A. nidulans reduced the production of oxylipins and then it disrupted the balance of sexual/asexual sporulation, although the double ΔppoAΔppoC mutants and triple ΔppoA–C mutants lost their ability to produce the mycotoxin ST, but showed an overproduction of the antibiotic penicillin. It also weakened the fungal ability to colonize in peanuts and maize hosts (Tsitsigiannis and Keller, 2010). These phenotypes were similar to the constitutively activated Gα, AnFadAG42R, which suppressed the ST inducer gene AnAflR, but enhanced the penicillin biosynthetic gene AnIpnA that was mediated through the cAMP-PKA pathway (Tag et al., 2010). Disruption of Ppo orthologs also reduced T2 production in Fusarium sporotrichioides (Mcdonald et al., 2003). After knocking out all five dioxygenase genes (ppoA-D and lox), A. flavus lost its density-dependent regulation of sporulation and AF production; both the ΔppoC and Δlox mutant strains produced high levels of AF at any population density (Brown et al., 2009). Oxylipins added at different concentrations exogenously affected the normal morphological development of AF. In addition, when fungi were exposed to oxidative stress, such as an increased ROS level, this led to the synthesis of oxylipins and induced toxin production. Therefore, the QS mechanism represented by fungal oxylipins can impact fungal sporulation, mycotoxin production, and virulence in a density-dependent regulation that is partly mediated by GPCRs.
Interspecies Fungal Communication
Fungi communicate with other fungi and bacteria to produce signaling molecules that also regulate their growth form and virulence through G-protein signaling pathways. For example, in co-cultivation of C. albicans and A. nidulans, farnesol produced by the former affected the latter by inhibiting its growth and/or inducing apoptosis (Semighini et al., 2006). Exposure of A. nidulans to farnesol did not influence the emergence of the germ-tube, but relied on G-protein signaling to affect mitochondrial function and ROS production; what was dependent? dependent on autophagy and PKC signaling, which caused cell apoptosis. Experiments have shown that the ΔflbA mutation affected activity of the Gα protein FadA that caused a significant increase in the sensitivity of farnesol, which revealed the signal transduction role of the FadA-G protein complex in this fungal interspecies communication process (Semighini, 2006; Savoldi et al., 2010). Conversely, another QSM pantothenic acid has positive impacts on fungal growth. Both C. neoformans-conditioned media, which contained C. neoformans autocrine pantothenic acid, and exogenous pantothenic acid increased the growth of C. albicans and S. cerevisiae (Albuquerque et al., 2014).
Fungi and bacteria can interact through signaling molecules. Pseudomonas aeruginosa is often found with C. albicans in mixed mammalian infections; the former can grow on and damage filamentous hyphae, but not budding yeast cells, and the bacterial QSM homoseryl lactone inhibits filamentation (Hogan et al., 2004; Cugini et al., 2010). Recent studies have shown that in mixed C. albicans and S. aureus biofilm, farnesol produced by the former induced ROS in the latter bacterium, which resulted in the up-regulation of drug efflux pumps that protected bacterial cells from antibiotic damage (Kong et al., 2017).
Host–Pathogen Communication
There is a complex GPCR-mediated communication between fungi and their hosts, in which oxylipins may play a completely different, but crucial role (Figure 3). For example, Gpr1 of S. cerevisiae showed structural and functional homology with mammalian GPCRs, and 9(S)-HODE and other oxidized free fatty acids bound to the mammalian G2A (oxylipins receptor), which inhibited cellular cAMP accumulation and MAP kinase activation (Obinata et al., 2005). Further, different eicosanoids mediated inflammatory responses through GPCRs in mammals (Dennis and Norris, 2015). CaGpr1 in C. albicans detected l-lactate released by the gut microorganism Lactobacillus reuteri, which promoted fungal β-glucan masking and evaded the mammalian immune system (Ballou et al., 2016). C. albicans can secrete QSMs tyrosol and farnesol; the former hindered the killing of mammalian host neutrophils by inhibiting ROS production (Josef et al., 2010), and the latter induced macrophage apoptosis (Abe et al., 2010). In the entomopathogenic Metarhizium species, deletion of the GPCR MrGpr8 (Class XIV Pth11-like GPCR) substantially impaired the nucleus translocation in MAPK, which resulted in the failure of appressorium to form on different substrates and the loss of virulence during topical infection of insects. The ΔMrGpr8 mutants could not be rescued with the addition of cAMP for appressorium formation (Shang et al., 2020). This model recognizes that the G protein signaling pathway can integrate intra- and interspecific signal transmission and cellular activity. In humans and mice, it is well-established that GPCRs detect microbial-derived signals and that the microbiota can affect host physiology through GPCRs (Dierking and Pita, 2020).
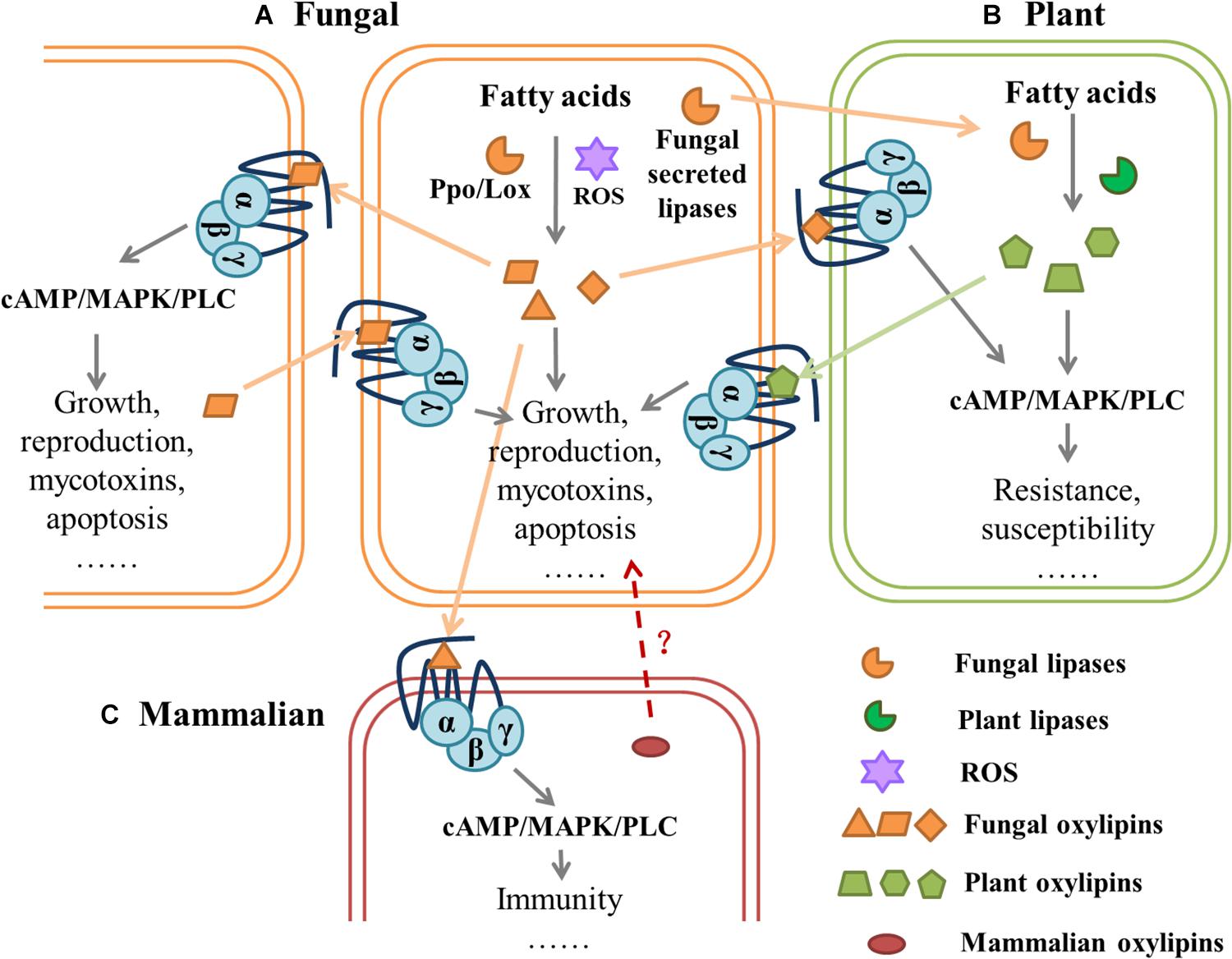
Figure 3. Hypothetical model of mediated communication of G-protein-coupled receptors between fungi and their hosts.| (A) Fungal lipoxygenase (Lox) and psi-producing oxygenases (Ppo) or reactive oxygen species (ROS) can catalyze the oxidation of fatty acids (FAs) to oxylipins, which affect fungal behavior through downstream pathways. Different species of fungi can sense oxylipins secreted in vitro by GPCRs, which regulate cell growth, reproduction, mycotoxins, and apoptosis. (B) Fungal lipases are secreted into plant cells where fatty acid substrates are cleaved and processed by fungi- secreted lipoxygenases and/or plant lipoxygenases for oxylipin production. Plant-produced oxylipins are perceived and exploited by fungi to regulate GPCR-mediated behavior. (C) In mammalian cells, different signals from microbial-derived oxylipids, mediate inflammatory responses, and fungi can affect host physiology through GPCRs. It is also possible that mammalian oxylipins affect fungal activity. All fungal lipases and oxylipins are orange, all plant lipases and oxylipins are green, mammalian oxylipins are red. Dashed arrows with the question mark (?) indicates hypothetical interactions, and solid arrows represent proven interactions.
Plant hosts secrete a variety of elicitors that induce defense responses against the infecting fungi, but fungal signal molecules also modulate host immunity and damage host cells (Ariyo et al., 1997; Liu et al., 2001; Benz et al., 2014). The plant oxylipin jasmonate inhibited fungal reproduction and secondary metabolism as a defense against necrotrophic fungal pathogens (Calvo et al., 1999), and it can also promote F. oxysporum infection (Thatcher et al., 2009). Plant oxylipins seem to have a completely different effect on fungi. Linoleic acid and 9S-HPODE promoted mycotoxin synthesis in Aspergillus, whereas 13S-HPODE inhibited it (Burow et al., 1997). A. ochraceus prefers to infect crops that contain more fatty acid and produce more OTA (Caiyan et al., 2017), which is speculated to be related to the oxidation of plant fatty acids to produce oxylipins. Who proposed? proposed a hypothetical model that the host-derived oxylipins can bind with fungal GPCR to regulate growth, sporulation, and synthesis of mycotoxin; on the other hand, these oxylipins may stimulate biosynthesis of fungal oxylipins. The psiB factor in Aspergillus is also derived from linoleic acid.
The complementation of ΔppoAΔppoC mutants with the maize ZmLOX3 gene restored conidiation and, thus, the ability of plant oxylipins to mimic or to interfere with fungal signaling may be due to structural similarities (Brodhagen et al., 2010). Destroying the ZmLOX3 gene of maize caused a deficiency in 9-LOX derivatives, which compromised conidiation, pathogenicity, and mycotoxin production of maize pathogenic Fusarium verticillioides, and this promoted resistance to other fungal pathogens (Wilson et al., 2001). On the contrary, maize that lacked the lox3 gene were more susceptible to Aspergillus infection, and AF contamination was more serious, which indicated that host oxylipins promoted pathogenesis in addition to resisting pathogenic (Reverberi et al., 2010; Yan et al., 2015).
Another mechanism proposed in F. graminearum is that Aspergillus can secrete lipases and LOX in host cells to regulate host lipid metabolism by cleaving off free fatty acids and oxidizing them to produce oxylipins. Fungi can then use host-derived oxylipins to facilitate invasive growth, spores, and mycotoxin production (Voigt et al., 2010). Similarly, host-derived 3-hydroxyoxylipin promoted the ability of C. albicans to grow and become more virulent within mammalian cells, whereas salicylic acid treatment inhibited fungal development and biofilm formation (Tsitsigiannis and Keller, 2007).
In summary, QS plays an important role in intra- and interspecies communication, which impact fungal development, mycotoxin regulation, and disease. The key role of oxylipins and their status as GPCR ligands suggest that these mechanisms are related to the G-protein signaling pathway, but the receptors capable of sensing these QSMs and the specific mechanisms remain to be discovered.
Conclusion
Biological detoxification methods have greater safety, availability, and cost-effectiveness than physical and chemical detoxification methods. Currently, research advances in OTA biodetoxification has been made in degradation, adsorption, or enzymatic degradation[9]. With the development of bioinformatics and molecular biology, researchers have turned to studies of molecular biological mechanisms to crack the code of fungal physiology. Fungal GPCRs have been proposed as targets for controlling mycotoxins. The importance of the GPCR signaling pathway to fungal biology and virulence is underexplored, and only a limited number of receptors have been shown to regulate OTA synthesis directly. However, there are a large number of unclassified orphan GPCRs. The ligands of these orphan GPCRs are unidentified, and their physiological role is yet to be determined, which implies a huge number of possible targets link the perception of extracellular signals with mycotoxin synthesis, such as pH-sensing GPCRs. Another area worthy of further study is that of quorum sensing inhibition through the blockage of signal production (i.e., Quorum Quenching), which limits fungal growth and mycotoxin production by affecting intercellular communication. This can be achieved partly by inhibiting the GPCR’s reception of QSMs such as oxylipins (Turan and Engin, 2018). Therefore, fungal-specific GPCRs represent promising and unexplored targets to potentially intervene or to reduce the impact of mycotoxin contamination and fungal diseases. Deeper understanding of fungal GPCRs will enhance our ability to develop novel strategies in agricultural and clinical settings to promote human, animal, plant, and even ecosystem health.
Author Contributions
JG contributed to conception and design of the study, and wrote the first draft of the manuscript. XX wrote sections of the manuscript. KH final approval of the version to be published. ZL wrote sections of the manuscript and final approval of the version to be published. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (No. 31671947).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Thomas A. Gavin, Professor Emeritus, Cornell University, for help with editing this paper.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.631392/full#supplementary-material
Supplementary Table 1 | Classification of G-protein-coupled receptors in fungi (Martín et al., 2019).
References
Abe, S., Tsunashima, R., Iijima, R., Yamada, T., and Maruyama, N. (2010). Suppression of anti-Candida activity of macrophages by a quorum-sensing molecule, farnesol, through induction of oxidative stress [J]. Microbiol. Immunol. 53, 323–330. doi: 10.1111/j.1348-0421.2009.00128.x
Adnan, M., Zheng, W. H., Islam, W., Arif, M., Abubakar, Y. S., Wang, Z. H., et al. (2018). Carbon catabolite repression in filamentous fungi [J]. Int. J. Mol. Sci. 19:48. doi: 10.3390/ijms19010048
Affeldt, K. J., Brodhagen, M., and Keller, N. P. (2012). Aspergillus oxylipin signaling and quorum sensing pathways depend on g protein-coupled receptors [J]. Toxins 4, 695–717. doi: 10.3390/toxins4090695
Albuquerque, P., Nicola, A. M., Nieves, E., Paes, H. C., Williamson, P. R., Silva-Pereira, I., et al. (2014). Quorum sensing-mediated, cell density-dependent regulation of growth and virulence in Cryptococcus neoformans [J]. mBio 5:e0986-13.
Alkhayyat, F., and Yu, J. H. (2014). Upstream regulation of mycotoxin biosynthesis [J]. Adv. Appl. Microbiol. 86, 251–278. doi: 10.1016/b978-0-12-800262-9.00005-6
Ansari, K., Martin, S., Farkasovsky, M., Ehbrecht, I. M., and Kuntzel, H. (1999). Phospholipase C binds to the receptor-like GPR1PROTEIN and controls pseudohyphal differentiation in Saccharomyces cerevisiae [J]. J. Biol. Chem. 274:30052. doi: 10.1074/jbc.274.42.30052
Antonia, S., Proctor, R. H., Massimiliano, M., Miriam, H., Antonia, G., Logrieco, A. F., et al. (2016). Variation in fumonisin and ochratoxin production associated with differences in biosynthetic gene content in Aspergillus niger and A. welwitschiae isolates from multiple crop and geographic origins [J]. Front. Microbiol. 7:1412. doi: 10.3389/fmicb.2016.01412
Ariyo, B. T., And, C. B., and Keshaverz, T. (1997). Alginate oligosaccharides as enhancers of penicillin production in cultures of penicillium chrysogenum [J]. Biotechnol. Bioeng. 53, 17–20. doi: 10.1002/(sici)1097-0290(19970105)53:1<17::aid-bit3>3.0.co;2-1
Arroyo, J., Bermejo, C., Garcia, R., and Rodriguez-Pena, J. M. (2009). Genomics in the detection of damage in microbial systems: cell wall stress in yeast [J]. Clin. Microbiol. Infect. 15, 44–46. doi: 10.1111/j.1469-0691.2008.02676.x
Atoui, A., Bao, D. P., Kaur, N., Grayburn, W. S., and Calvo, A. M. (2008). Aspergillus nidulans natural product biosynthesis is regulated by mpkB, a putative pheromone response mitogen-activated protein kinase [J]. Appl. Environ. Microbiol. 74, 3596–3600. doi: 10.1128/aem.02842-07
Baldwin, J. M. (1993). The probable arrangement of the helices in G protein-coupled receptors [J]. EMBO J. 12, 1693–1703. doi: 10.1002/j.1460-2075.1993.tb05814.x
Ballesteros, J. A., Jensen, A. D., Liapakis, G., Rasmussen, S. G. F., and Javitch, J. A. (2001). Activation of the β2-adrenergic receptor involves disruption of an ionic lock between the cytoplasmic ends of transmembrane segments 3 and 6 [J]. J. Biol. Chem. 276:29171. doi: 10.1074/jbc.m103747200
Ballou, E. R., Avelar, G. M., Childers, D. S., Mackie, J., Bain, J. M., Wagener, J., et al. (2016). Lactate signalling regulates fungal β-glucan masking and immune evasion [J]. Nat. Microbiol. 2:16238.
Barriuso, J., Hogan, D. A., Keshavarz, T., and Martinez, M. J. (2018). Role of quorum sensing and chemical communication in fungal biotechnology and pathogenesis [J]. FEMS Microbiol. Rev. 42, 627–638. doi: 10.1093/femsre/fuy022
Bebber, D. P., Ramotowski, M. A. T., and Gurr, S. J. (2013). Crop pests and pathogens move polewards in a warming world [J]. Nat. Clim. Change 3, 985–988. doi: 10.1038/nclimate1990
Bennett, R. J., and Johnson, A. D. (2010). The role of nutrient regulation and the Gpa2 protein in the mating pheromone response of C. albicans [J]. Mol. Microbiol. 62, 100–119. doi: 10.1111/j.1365-2958.2006.05367.x
Benz, J. P., Chau, B. H., Zheng, D., Bauer, S., Glass, N. L., and Somerville, C. R. A. (2014). comparative systems analysis of polysaccharide-elicited responses in Neurospora crassa reveals carbon source-specific cellular adaptations [J]. Mol. Microbiol. 91, 275–299. doi: 10.1111/mmi.12459
Bieszke, J. A., Braun, E. L., Bean, L. E., Kang, S. C., Natvig, D. O., and Borkovich, K. A. (1999). The nop-1 gene of Neurospora crassa encodes a seven transmembrane helix retinal-binding protein homologous to archaeal rhodopsins [J]. Proc. Natl. Acad. Sci. U.S.A. 96, 8034–8039. doi: 10.1073/pnas.96.14.8034
Bignell, E. (2012). The molecular basis of pH sensing, signaling, and homeostasis in fungi [J]. Adv. Appl. Microbiol. 79, 1–18. doi: 10.1016/b978-0-12-394318-7.00001-2
Bohm, J., Hoff, B., O’gorman, C. M., Wolfers, S., Klix, V., Binger, D., et al. (2013). Sexual reproduction and mating-type-mediated strain development in the penicillin-producing fungus Penicillium chrysogenum [J]. Proc. Natl. Acad. Sci. U.S.A. 110, 1476–1481. doi: 10.1073/pnas.1217943110
Brakhage, A. A. (1998). Molecular regulation of beta-lactam biosynthesis in filamentous fungi [J]. Microbiol. Mol. Biol. Rev. 62, 547–585. doi: 10.1128/mmbr.62.3.547-585.1998
Brodhagen, M., Tsitsigiannis, D. I., Hornung, E., Goebel, C., and Keller, N. P. (2010). Reciprocal oxylipin-mediated cross-talk in the Aspergillus-seed pathosystem [J]. Mol. Microbiol. 67, 378–391. doi: 10.1111/j.1365-2958.2007.06045.x
Brown, N. A., Dos, Reis TF, Ries, L. N., Caldana, C., Mah, J. H., Yu, J. H., et al. (2015). G-protein coupled receptor-mediated nutrient sensing and developmental control in Aspergillus nidulans [J]. Mol. Microbiol. 98, 420–439. doi: 10.1111/mmi.13135
Brown, N. A., and Goldman, G. H. (2016). The contribution of Aspergillus fumigatus stress responses to virulence and antifungal resistance [J]. J. Microbiol. 54, 243–253. doi: 10.1007/s12275-016-5510-4
Brown, N. A., Schrevens, S., Dijck, P. V., and Goldman, G. H. (2018). Fungal G-protein-coupled receptors: mediators of pathogenesis and targets for disease control [J]. Nat. Microbiol. 3, 402–414. doi: 10.1038/s41564-018-0127-5
Brown, S. H., Scott, J. B., Bhaheetharan, J., Sharpee, W. C., and Keller, N. P. (2009). Oxygenase coordination is required for morphological transition and the host-fungus interaction of Aspergillus flavus [J]. Mol. Plant Microbe Interact. 22:882. doi: 10.1094/mpmi-22-7-0882
Brunner, K., Omann, M., Pucher, M. E., Delic, M., Lehner, S. M., Domnanich, P., et al. (2008). Trichoderma G protein-coupled receptors: functional characterisation of a camp receptor-like protein from Trichoderma atroviride [J]. Curr. Genet. 54, 283–299. doi: 10.1007/s00294-008-0217-7
Burkholder, A. C., and Hartwell, L. H. (1985). The yeast alpha-factor receptor: structural properties deduced from the sequence of the STE2 gene [J]. Nuclie Acids Res. 13, 8463–8475. doi: 10.1093/nar/13.23.8463
Burow, G. B., Nesbitt, T. C., Dunlap, J., and Keller, N. P. (1997). Seed lipoxygenase products modulate Aspergillus mycotoxin biosynthesis [J]. Mol. Plant Microbe Interact. 10, 380–387. doi: 10.1094/mpmi.1997.10.3.380
Cabrera, I. E., Pacentine, I. V., Lim, A., Guerrero, N., Krystofova, S., Li, L., et al. (2015). Global analysis of predicted G protein coupled receptor genes in the filamentous fungus, Neurospora crassa [J]. G3 Genes Genomes Genet. 5, 2729–2743. doi: 10.1534/g3.115.020974
Caceres, I., Al Khoury, A., El Khoury, R., Lorber, S., Oswald, I. P., El Khoury, A., et al. (2020). Aflatoxin biosynthesis and genetic regulation: a review [J]. Toxins 12:150. doi: 10.3390/toxins12030150
Caiyan, L., Yanmin, S., Lu, X., Kunlun, H., and Zhihong, L. (2017). Initial spore density has an influence on ochratoxin a content in Aspergillus ochraceus CGMCC 3.4412 in PDB and its interaction with seeds [J]. Toxins 9:146. doi: 10.3390/toxins9040146
Calvo, A. M., Hinze, L. L., Gardner, H. W., and Keller, N. P. (1999). Sporogenic effect of polyunsaturated fatty acids on development of Aspergillus spp [J]. Applenvironmicrobiol 65, 3668–3673. doi: 10.1128/aem.65.8.3668-3673.1999
Calvo, A. M., Lohmar, J. M., Ibarra, B., and Satterlee, T. (2016). 18 Velvet Regulation of Fungal Development [M]. Berlin: Springer International Publishing.
Chen, R. E., and Thorner, J. (2007). Function and regulation in MAPK signaling pathways: lessons learned from the yeast Saccharomyces cerevisiae [J]. Biochim. Biophys. Acta 1773:1311. doi: 10.1016/j.bbamcr.2007.05.003
Cornet, M., and Gaillardin, C. (2014). pH signaling in human fungal pathogens: a new target for antifungal strategies (vol 13, pg 342, 2014) [J]. Eukaryotic Cell 13:691. doi: 10.1128/ec.00073-14
Crespo-Sempere, A., Marin, S., Sanchis, V., and Ramos, A. J. (2013). VeA and LaeA transcriptional factors regulate ochratoxin A biosynthesis in Aspergillus carbonarius [J]. Int. J. Food Microbiol. 166, 479–486. doi: 10.1016/j.ijfoodmicro.2013.07.027
Cugini, C., Calfee, M. W., Farrow, J. M., Morales, D. K., and Hogan, D. A. (2010). Farnesol, a common sesquiterpene, inhibits PQS production in Pseudomonas aeruginosa [J]. Mol. Microbiol. 65, 896–906. doi: 10.1111/j.1365-2958.2007.05840.x
Dasgupta, A., Fuller, K. K., Dunlap, J. C., and Loros, J. J. (2016). Seeing the world differently: variability in the photosensory mechanisms of two model fungi [J]. Environ. Microbiol. 18, 5–20. doi: 10.1111/1462-2920.13055
De, S. W. R., Rezende, M. E., Graciele, K. N., Marcela, S., Goldman, M. H. S., Fernando, R., et al. (2013). Identification of metabolic pathways influenced by the G-protein coupled receptors GprB and GprD in Aspergillus nidulans [J]. PLoS One 8:e62088. doi: 10.1371/journal.pone.0062088
De Assis, L. J., Silva, L. P., Liu, L., Schmitt, K., Valerius, O., Braus, G. H., et al. (2020). The high osmolarity glycerol mitogen-activated protein kinase regulates glucose catabolite repression in filamentous fungi [J]. PLos Genet. 16:e1008996. doi: 10.1371/journal.pgen.1008996
Dennis, E. A., and Norris, P. C. (2015). Eicosanoid storm in infection and inflammation (vol 15, pg 511, 2015) [J]. Nat. Rev. Immunol. 15:511. doi: 10.1038/nri3859
Dierking, K., and Pita, L. (2020). Receptors mediating host-microbiota communication in the metaorganism: the invertebrate perspective [J]. Front. Immunol. 11:1251. doi: 10.3389/fimmu.2020.01251
Dignard, D., Andre, D., and Whiteway, M. (2008). Heterotrimeric G-protein subunit function in Candida albicans: both the alpha and beta subunits of the pheromone response G protein are required for mating [J]. Eukaryotic Cell 7, 1591–1599. doi: 10.1128/ec.00077-08
Dos Reis, T. F., Mellado, L., Lohmar, J. M., Silva, L. P., Zhou, J. J., Calvo, A. M., et al. (2019). GPCR-mediated glucose sensing system regulates light-dependent fungal development and mycotoxin production [J]. PLoS Genet. 15:e1008419. doi: 10.1371/journal.pgen.1008419
Eom, T. J., Moon, H., Yu, J. H., and Park, H. S. (2018). Characterization of the velvet regulators in Aspergillus flavus [J]. J. Microbiol. 56, 893–901. doi: 10.1007/s12275-018-8417-4
Fanelli, F., Geisen, R., Schmidt-Heydt, M., Logrieco, A. F., and Mule, G. (2016). Light regulation of mycotoxin biosynthesis: new perspectives for food safety [J]. World Mycotoxin J. 9, 129–145. doi: 10.3920/wmj2014.1860
Ferrara, M., Perrone, G., Gambacorta, L., Epifani, F., Solfrizzo, M., and Gallo, A. (2016). Identification of a halogenase involved in the biosynthesis of ochratoxin a in Aspergillus carbonarius [J]. Appl. Environ. Microbiol. 82, 5631–5641. doi: 10.1128/aem.01209-16
Filho, A. P. D. C., Brancini, G. T. P., De Castro, P. A., Valero, C., Ferreira Filho, J. A., Silva, L. P., et al. (2020). Aspergillus fumigatus G-protein coupled receptors GprM and GprJ are important for the regulation of the cell wall integrity pathway, secondary metabolite production, and virulence [J]. mBio 11:e02458-20.
Fischer, G. J., and Keller, N. P. (2016). Production of cross-kingdom oxylipins by pathogenic fungi: An update on their role in development and pathogenicity [J]. J. Microbiol. 54, 254–264. doi: 10.1007/s12275-016-5620-z
Frisvad, J. C., Hubka, V., Ezekiel, C. N., Hong, S. B., Novakova, A., Chen, A. J., et al. (2019). Taxonomy of Aspergillus section Flavi and their production of aflatoxins, ochratoxins and other mycotoxins [J]. Stud. Mycol. 93, 1–63. doi: 10.1016/j.simyco.2018.06.001
Gehrke, A., Heinekamp, T., Jacobsen, I. D., and Brakhage, A. A. (2010). Heptahelical receptors GprC and GprD of Aspergillus fumigatus are essential regulators of colony growth, hyphal morphogenesis, and virulence [J]. Appl. Environ. Microbiol. 76, 3989–3998. doi: 10.1128/aem.00052-10
Geisen, R. (2004). Molecular monitoring of environmental conditions influencing the induction of ochratoxin A biosynthesis genes in Penicillium nordicum [J]. Mol. Nutr. Food Res. 48, 532–540. doi: 10.1002/mnfr.200400036
Gil-Serna, J., GarcíA-Díaz, M., González-JaéN, M. T., Vázquez, C., and Pati, O. B. (2018). Description of an orthologous cluster of ochratoxin A biosynthetic genes in Aspergillus and Penicillium species. A comparative analysis [J]. Int. J. Food Microbiol. 268, 35–43. doi: 10.1016/j.ijfoodmicro.2017.12.028
Gil-Serna, J., Vazquez, C., and Patino, B. (2020). Genetic regulation of aflatoxin, ochratoxin A, trichothecene, and fumonisin biosynthesis: A review [J]. Int. Microbiol. 23, 89–96. doi: 10.1007/s10123-019-00084-2
Gomar-Alba, M., Jiménez-Martí, E., and Del Olmo, M. (2012). The Saccharomyces cerevisiae Hot1p regulated gene YHR087W (HGI1) has a role in translation upon high glucose concentration stress [J]. BMC Mol. Biol. 13:19. doi: 10.1186/1471-2199-13-19
Grice, C. M., Bertuzzi, M., and Bignell, E. M. (2013). Receptor-mediated signaling in Aspergillus fumigatus [J]. Front. Microbiol. 4:26. doi: 10.3389/fmicb.2013.00026
Guan, X., Zhao, Y., Liu, X., Shang, B., Xing, F., Zhou, L., et al. (2019). The bZIP transcription factor Afap1 mediates the oxidative stress response and aflatoxin biosynthesis in Aspergillus flavus [J]. Rev. Argentina Microbiol. 51, 292–301. doi: 10.1016/j.ram.2018.07.003
Hamel, L. P., Nicole, M. C., Duplessis, S., and Ellis, B. E. (2012). Mitogen-activated protein kinase signaling in plant-interacting fungi: distinct messages from conserved messengers [J]. Plant Cell 24, 1327–1351. doi: 10.1105/tpc.112.096156
Han, G., Zhao, K., Yan, X., Xiang, F., and Tao, F. (2019). Differential regulation of mycelial growth and aflatoxin biosynthesis by Aspergillus flavus under different temperatures as revealed by strand-specific Rna-Seq [J]. MicrobiologyOpen 8:e897.
Han, K. H., Seo, J. A., and Yu, J. H. A. (2004). putative G protein-coupled receptor negatively controls sexual development in Aspergillus nidulans [J]. Mol. Microbiol. 51, 1333–1345. doi: 10.1111/j.1365-2958.2003.03940.x
Herranz, S., Rodriguez, J. M., Bussink, H. J., Sanchez-Ferrero, J. C., Arst, H. N. Jr., Penalva, M. A., et al. (2005). Arrestin-related proteins mediate pH signaling in fungi [J]. Proc. Natl. Acad. Sci. U.S.A. 102, 12141–12146. doi: 10.1073/pnas.0504776102
Herskowitz, I. (1995). MAP kinase pathways in yeast: for mating and more [J]. Cell 80, 187–197. doi: 10.1016/0092-8674(95)90402-6
Hervas-Aguilar, A., Galindo, A., and Penalva, M. A. (2010). Receptor-independent Ambient pH signaling by ubiquitin attachment to fungal arrestin-like PalF [J]. J. Biol. Chem. 285, 18095–18102. doi: 10.1074/jbc.m110.114371
Hicks, J., Lockington, R. A., Strauss, J., Dieringer, D., Kubicek, C. P., Kelly, J., et al. (2001). RcoA has pleiotropic effects on Aspergillus nidulans cellular development [J]. Mol. Microbiol. 39, 1482–1493.
Hogan, D. A., Vik, A., and Kolter, R. A. (2004). Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology [J]. Mol. Microbiol. 54, 1212–1223.
Hou, L. L., Le, G. N., Lin, Z. M., Qian, G., Gan, F., Gu, C., et al. (2020). Nontoxic concentration of ochratoxin A decreases the dosage of cyclosporine A to induce chronic nephropathy model via autophagy mediated by toll-like receptor 4 [J]. Cell Death Dis. 11:153.
Ignatov, I., and Mosin, O. (2014). Visual perception and electromagnetic conception for the eyesight. rhodopsin and bacteriorhodopsin in nano- and biotechnologies [J]. J. Health Med. Nurs. 4:14947.
Inadome, H., Noda, Y., Adachi, H., and Yoda, K. (2005). Immunoisolaton of the yeast golgi subcompartments and characterization of a novel membrane protein, Svp26, discovered in the Sed5-containing compartments [J]. Mol. Cell. Biol. 25, 7696–7710.
International Agency For Research On Cancer (1993). IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, Vol. 56. Some Naturally Occurring Substances: Food Items and Constituents, Heterocyclic Aromatic Amines and Mycotoxins [M]. Lyon: IARC.
Jayashree, T., and Subramanyam, C. (2000). Oxidative stress as a prerequisite for aflatoxin production by Aspergillus parasiticus [J]. Free Radic. Biol. Med. 29, 981–985.
Josef, C., Vassilia, V., and Ilja, B. (2010). 2,4-(Hydroxyphenyl)-ethanol, an antioxidative agent produced by Candida spp., impairs neutrophilic yeast killing in vitro [J]. FEMS Microbiol. Lett. 2, 319–325.
Jung, M. G., Kim, S. S., Yu, J. H., and Shin, K. S. (2016). Characterization of gprK encoding a putative hybrid G-protein-coupled receptor in Aspergillus fumigatus [J]. PLoS One 11:e0161312. doi: 10.1371/journal.pone.0161312
Kapetanakou, E., Panagou, E. Z., Gialitaki, M., Drosinos, E. H., and Skandamis, P. N. (2009). Evaluating the combined effect of water activity, pH and temperature on ochratoxin A production by Aspergillus ochraceus and Aspergillus carbonarius on culture medium and Corinth raisins [J]. Food Control 20, 725–732.
Karlson, P., and Lüscher, M. (1959). Pheromones’: a new term for a class of biologically active substances [J]. Nature 183, 55–56.
Kasahara, S., Wang, P., and Nuss, D. L. (2000). Identification of bdm-1, a gene involved in G protein β-subunit function and α-subunit accumulation [J]. Proc. Natl. Acad. Sci. U.S.A. 97, 412–417.
Keller, N. P., Nesbitt, C., Sarr, B., Phillips, T. D., and Burow, G. B. (1997). pH regulation of sterigmatocystin and aflatoxin biosynthesis in Aspergillus spp [J]. Phytopathology 87, 643–648.
Khan, S. M., Sleno, R., Gora, S., Zylbergold, P., Laverdure, J.-P., Labbe, J.-C., et al. (2013). The expanding roles of Gβγ subunits in G protein–coupled receptor signaling and drug action [J]. Pharmacol. Rev. 65, 545–577.
Kim, H., and Borkovich, K. A. A. (2010). pheromone receptor gene, pre-1, is essential for mating type-specific directional growth and fusion of trichogynes and female fertility in Neurospora crassa [J]. Mol. Microbiol. 52, 1781–1798.
Kim, H., Wright, S. J., Park, G., Ouyang, S., Krystofova, S., and Borkovich, K. A. (2012). Roles for receptors, pheromones, G proteins, and mating type genes during sexual reproduction in Neurospora crassa [J]. Genetics 190, 1389–1404.
Kim, J., Bortz, E., Zhong, H., Leeuw, T., Leberer, E., Vershon, A. K., et al. (2000). Localization and signaling of G(beta) subunit Ste4p are controlled by a-factor receptor and the a-specific protein Asg7p [J]. Mol. Cell. Biol. 20:8826.
Kim, M. J., Lee, M. K., Pham, H. Q., Gu, M. J., Zhu, B. H., Son, S. H., et al. (2020). The velvet regulator VosA governs survival and secondary metabolism of sexual spores in Aspergillus nidulans [J]. Genes 11:103.
Kmetzsch, L., Staats, C. C., Simon, E., Fonseca, F. L., Oliveira, D. L., Joffe, L. S., et al. (2011). The Gata-type transcriptional activator Gat1 regulates nitrogen uptake and metabolism in the human pathogen Cryptococcus neoformans [J]. Fungal Genet. Biol. 48, 192–199.
Kochman, K. (2014). Superfamily of G-protein coupled receptors (GPCRS) - extraordinary and outstanding success of evolution [J]. Postepy Higieny I Medycyny Doswiadczalnej 68, 1225–1237.
Kong, E. F., Tsui, C., Kucharíková, S., Dijck, P. V., and Jabra-Rizk, M. A. (2017). Modulation of Staphylococcus aureus response to antimicrobials by the Candida albicans quorum sensing molecule farnesol [J]. Antimicrob. Agents Chemother. 61:e01573-17.
Kraakman, L., Lemaire, K., Ma, P., Teunissen, A. W., and Thevelein, J. M. (2010). A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the camp pathway during the transition to growth on glucose [J]. Mol. Microbiol. 32, 1002–1012.
Krishnan, A., Almen, M. S., Fredriksson, R., and Schioth, H. B. (2012). The origin of Gpcrs: identification of mammalian like rhodopsin, adhesion, glutamate and frizzled GPCRs in fungi [J]. PLoS One 7:e29817. doi: 10.1371/journal.pone.0029817
Kuchler, K., Sterne, R. E., and Thorner, J. (1989). Saccharomyces cerevisiae STE6 gene product: a novel pathway for protein export in eukaryotic cells [J]. EMBO J. 8, 3973–3984.
Kunitake, E., Li, Y., Uchida, R., Nohara, T., Asano, K., Hattori, A., et al. (2019). CreA-independent carbon catabolite repression of cellulase genes by trimeric G-protein and protein kinase A in Aspergillus nidulans [J]. Curr. Genet. 65, 941–952.
Lafon, A., Han, K.-H., Seo, J.-A., Yu, J.-H. Y., and D’enfert, C. G. - (2006). protein and CAMP-mediated signaling in aspergilli: A genomic perspective [J]. Fungal Genet. Biol. 43, 490–502.
Lemaire, K., De Velde, S. V., Van Dijck, P., and Thevelein, J. M. (2004). Glucose and sucrose act as agonist and mannose as antagonist ligands of the G protein-coupled receptor Gpr1 in the yeast Saccharomyces cerevisiae [J]. Mol. Cell 16, 293–299.
Li, L., and Borkovich, K. A. G. P. R. - (2006). 4 is a predicted G-protein-coupled receptor required for carbon source-dependent asexual growth and development in Neurospora crassa [J]. Eukaryotic Cell 5, 1287–1300.
Li, L., Wright, S. J., Krystofova, S., Park, G., and Borkovich, K. A. (2007). Heterotrimeric G protein signaling in filamentous fungi [J]. Annu. Rev. Microbiol. 61:423.
Liu, G., Casqueiro, J., Gutiérrez, S., Kosalková, K., and MartíN, J. F. (2001). Elicitation of penicillin biosynthesis by alginate in Penicillium chrysogenum, Exerted on pcbAB, pcbC, and penDE genes at the transcriptional level [J]. J. Microbiol. Biotechnol. 11, 812–818.
Lorenz, M. C., and Pan, X. (2000). The G protein-coupled receptor Gpr1 is a nutrient sensor that regulates pseudohypal [J]. Genetics 154, 609–622.
Lukov, G. L., Hu, T., Mclaughlin, J. N., Hamm, H. E., and Willardson, B. M. (2014). Phosducin-like protein acts as a molecular chaperone for G protein βγ dimer assembly [J]. EMBO J. 24, 1965–1975.
Ma, D., and Li, R. (2013). Current Understanding of HOG-MAPK Pathway in Aspergillus fumigatus [J]. Mycopathologia 175, 13–23.
Macheleidt, J., Mattern, D. J., Fischer, J., Netzker, T., Weber, J., Schroeckh, V., et al. (2016). Regulation and role of fungal secondary metabolites [J]. Annu. Rev. Genet. 50, 371–392.
Manfiolli, A. O., Siqueira, A. F. S., Reis, A. T. F. D., Dijck, A. P. V., and Schrevens, C. S. (2019). Mitogen-activated protein kinase cross-talk interaction modulates the production of melanins in Aspergillus fumigatus [J]. mBio 10:e00215-19.
Marc, Y. L., Huchet, V., Bourgeois, C. M., Guyonnet, J. P., Mafart, P., and Thuault, D. (2002). Modelling the growth kinetics of Listeria as a function of temperature, pH and organic acid concentration [J]. Int. J. Food Microbiol. 73, 219–237.
MaríN, S., Companys, E., Sanchis, V., Ramos, A. J., and Magan, N. (1998). Effect of water activity and temperature on competing abilities of common maize fungi [J]. Mycol. Res. 102, 959–964.
Martín, J. F., van den Berg, M. A., Ver Loren van Themaat, E., and Liras, P. (2019). Sensing and transduction of nutritional and chemical signals in filamentous fungi: Impact on cell development and secondary metabolites biosynthesis [J]. Biotechnol. Adv. 37:107392.
Mcdonald, T., Devi, T., Shimizu, K., Sim, S., and Keller, N. (2003). Signaling events connecting mycotoxin biosynthesis and sporulation in Aspergillus and Fusarium spp [J]. JSM Mycotoxins 2003(Suppl.3), 139–147.
McPherson, K. B., Leff, E. R., Li, M. H., Meurice, C., Tai, S., Traynor, J. R., et al. (2018). Regulators of G-protein signaling (RGS) proteins promote receptor coupling to G-protein-coupled inwardly rectifying potassium (GIRK) channels [J]. J. Neurosci. Off. J. Soc. Neurosci. 38, 8737–8744.
Mendoza-Martinez, A. E., Cano-Dominguez, N., and Aguirre, J. (2020). Yap1 homologs mediate more than the redox regulation of the antioxidant response in filamentous fungi [J]. Fungal Biol. 124, 253–262.
Nadarajah, K., Kumar, I. S., Sangapillai, V., and Omar, N. S. (2018). Recent advances in understanding the fungal G-protein-coupled receptors [J]. Malaysian J. Microbiol. 14, 611–623. doi: 10.21161/mjm.1461823
Nadia, P. (2015). Mycotoxins are a component of Fusarium graminearum stress-response system [J]. Front. Microbiol. 6:1234. doi: 10.3389/fmicb.2015.01234
Nagy, K. (1991). Biophysical processes in invertebrate photoreceptors - recent progress and a critical overview based on limulus photoreceptors [J]. Q. Rev. Biophys. 24, 165–226. doi: 10.1017/S0033583500003401
Niu, M., Steffan, B. N., Fischer, G. J., Venkatesh, N., Raffa, N. L., Wettstein, M. A., et al. (2020). Fungal oxylipins direct programmed developmental switches in filamentous fungi [J]. Nat. Commun. 11:5158. doi: 10.1038/s41467-020-18999-0
Obinata, H., Hattori, T., Nakane, S., Tatei, K., and Izumi, T. (2005). Identification of 9-hydroxyoctadecadienoic acid and other oxidized free fatty acids as ligands of the G protein-coupled receptor G2A [J]. J. Biol. Chem. 280, 40676–40683. doi: 10.1074/jbc.M507787200
O’callaghan, J., Stapleton, P. C., and Dobson, A. D. (2006). Ochratoxin A biosynthetic genes in Aspergillus ochraceus are differentially regulated by pH and nutritional stimuli [J]. Fungal Genet. Biol. 43, 213–221. doi: 10.1016/j.fgb.2005.11.005
Padder, S. A., Prasad, R., and Shah, A. H. (2018). Quorum sensing: a less known mode of communication among fungi [J]. Microbiol. Res. 210:51. doi: 10.1016/j.micres.2018.03.007
Paoletti, M., Rydholm, C., Schwier, E. U., Anderson, M. J., Szakacs, G., Lutzoni, F., et al. (2005). Evidence for sexuality in the opportunistic fungal pathogen Aspergillus fumigatus [J]. Curr. Biol. 15, 1242–1248. doi: 10.1016/j.cub.2005.05.045
Penalva, M. A., Tilburn, J., Bignell, E., and Arst, H. N. (2008). Ambient pH gene regulation in fungi: making connections [J]. Trends Microbiol. 16, 291–300. doi: 10.1016/j.tim.2008.03.006
Pfohl-Leszkowicz, A., and Manderville, R. A. (2012). An update on direct genotoxicity as a molecular mechanism of ochratoxin a carcinogenicity [J]. Chem. Res. Toxicol. 25, 252–262. doi: 10.1021/tx200430f
Poggeler, S., Hoff, B., and Kuck, U. (2008). Asexual cephalosporin C producer Acremonium chrysogenum carries a functional mating type locus [J]. Appl. Environ. Microbiol. 74, 6006–6016. doi: 10.1128/AEM.01188-08
Qi, X., Yang, X., Chen, S., He, X., Dweep, H., Guo, M., et al. (2014). Ochratoxin A induced early hepatotoxicity: new mechanistic insights from microRNA, mRNA and proteomic profiling studies [J]. Sci. Rep. 4:5163. doi: 10.1038/srep05163
Raisley, B., Zhang, M., Hereld, D., and Hadwiger, J. A. A. (2004). camp receptor-like G protein-coupled receptor with roles in growth regulation and development [J]. Dev. Biol. 265, 433–445. doi: 10.1016/j.ydbio.2003.09.035
Rasmussen, S. G., Choi, H. J., Rosenbaum, D. M., Kobilka, T. S., Thian, F. S., Edwards, P. C., et al. (2007). Crystal structure of the human beta2 adrenergic G-protein-coupled receptor [J]. Nature 450, 383–387. doi: 10.1038/nature06325
Reverberi, M., Fabbri, A. A., Zjalic, S., Ricelli, A., Punelli, F., and Fanelli, C. (2005). Antioxidant enzymes stimulation in Aspergillus parasiticus by Lentinula edodes inhibits aflatoxin production [J]. Appl. Microbiol. Biotechnol. 69, 207–215. doi: 10.1007/s00253-005-1979-1
Reverberi, M., Gazzetti, K., Punelli, F., Scarpari, M., Zjalic, S., Ricelli, A., et al. (2012). Aoyap1 regulates OTA synthesis by controlling cell redox balance in Aspergillus ochraceus [J]. Appl. Microbiol. Biotechnol. 95, 1293–1304. doi: 10.1007/s00253-012-3985-4
Reverberi, M., Punelli, F., Scarpari, M., Camera, E., Zjalic, S., Ricelli, A., et al. (2010). Lipoperoxidation affects ochratoxin A biosynthesis in Aspergillus ochraceus and its interaction with wheat seeds [J]. Appl. Microbiol. Biotechnol. 85, 1935–1946. doi: 10.1007/s00253-009-2220-4
Reverberi, M., Zjalic, S., Punelli, F., Ricelli, A., Fabbri, A. A., and Fanelli, C. (2007). Apyap1 affects aflatoxin biosynthesis during Aspergillus parasiticus growth in maize seeds [J]. Food Addit. Contaminants A Chem. Anal. Control Exposure Risk Assess. 24, 1070–1075. doi: 10.1080/02652030701553244
Reverberi, M., Zjalic, S., Ricelli, A., Punelli, F., and Camera, E. (2008). Modulation of antioxidant defense in Aspergillus parasiticus is involved in aflatoxin biosynthesis: a role for the ApyapA gene [J]. Eukaryotic Cell 7, 988–1000. doi: 10.1128/EC.00228-07
Ries, L. N. A., Beattie, S., Cramer, R. A., and Goldman, G. H. (2018). Overview of carbon and nitrogen catabolite metabolism in the virulence of human pathogenic fungi [J]. Mol. Microbiol. 107, 277–297. doi: 10.1111/mmi.13887
Rispail, N., Soanes, D. M., Ant, C., Czajkowski, R., Grünler, A., Huguet, R., et al. (2009). Comparative genomics of MAP kinase and calcium-calcineurin signalling components in plant and human pathogenic fungi [J]. Fungal Genet. Biol. Fg B 46, 287–298. doi: 10.1016/j.fgb.2009.01.002
Rolland, F., De Winde, J. H., Lemaire, K., Boles, E., Thevelein, J. M., and Winderickx, J. (2000). Glucose-induced cAMP signalling in yeast requires both a G-protein coupled receptor system for extracellular glucose detection and a separable hexose kinase-dependent sensing process [J]. Mol. Microbiol. 38, 348–358. doi: 10.1046/j.1365-2958.2000.02125.x
Rowe, J., Kapolka, N., Taghon, G., Morgan, W., and Isom, D. (2020). Mechanistic Insights into pH-sensing GPCRs [J]. FASEB J. 34:1. doi: 10.1096/fasebj.2020.34.s1.06154
Ruijter, G. J. G., and Visser, J. (1997). Carbon repression in aspergilli [J]. FEMS Microbiol. Lett. 151, 103–114. doi: 10.1111/j.1574-6968.1997.tb12557.x
Ruitao, X., Haoyu, C., Meirong, C., Mengwei, Z., and Jian, Z. (2019). LaeA regulates the morphological development, ochratoxin synthesis and oxidation tolerance of Aspergillus niger [J]. Food Res. Dev. 40, 189–195.
Savoldi, M., Malavazi, I., Soriani, F. M., Capellaro, J. L., Kitamoto, K., Ferreira, M. E. D. S., et al. (2010). Farnesol induces the transcriptional accumulation of the Aspergillus nidulans Apoptosis-Inducing Factor (Aif)-like mitochondrial oxidoreductase [J]. Mol. Microbiol. 70, 44–59. doi: 10.1111/j.1365-2958.2008.06385.x
Sekito, T., Nakamura, K., Manabe, K., Tone, J., Sato, Y., Murao, N., et al. (2014). Loss of ATP-dependent lysine uptake in the vacuolar membrane vesicles of Saccharomyces cerevisiae ypq1 mutant [J]. Biosci. Biotechnol. Biochem. 78, 1199–1202. doi: 10.1080/09168451.2014.918489
Semighini, C. P. (2006). Functional characterization of the putative Aspergillus nidulans Poly(ADP-Ribose) polymerase homolog PrpA [J]. Prog. Pharm. Sci. 173, 87–98. doi: 10.1534/genetics.105.053199
Semighini, C. P., Hornby, J. M., Dumitru, R., Nickerson, K. W., and Harris, S. D. (2006). Farnesol-induced apoptosis in Aspergillus nidulans reveals a possible mechanism for antagonistic interactions between fungi [J]. Mol. Microbiol. 59, 753–764. doi: 10.1111/j.1365-2958.2005.04976.x
Seo, J. A., Han, K. H., and Yu, J. H. (2004). The gprA and gprB genes encode putative G protein-coupled receptors required for self-fertilization in Aspergillus nidulans [J]. Mol. Microbiol. 53, 1611–1623. doi: 10.1111/j.1365-2958.2004.04232.x
Shang, J. M., Shang, Y. F., Tang, G. R., and Wang, C. S. (2020). Identification of a key G-protein coupled receptor in mediating appressorium formation and fungal virulence against insects [J]. Sci. China Life Sci. [Epub ahead of print]. doi: 10.1007/s11427-020-1763-1
Shapiro, D. A., Kristiansen, K., Weiner, D. M., Kroeze, W. K., and Roth, B. L. (2002). Evidence for a model of agonist-induced activation of 5-Hydroxytryptamine 2A serotonin receptors that involves the disruption of a strong ionic interaction between helices 3 and 6 [J]. J. Biol. Chem. 277, 11441–11449. doi: 10.1074/jbc.M111675200
Stenkamp, R. E., Teller, D. C., and Palczewski, K. (2002). Crystal structure of rhodopsin: a G-protein-coupled receptor [J]. Chembiochem 3, 963–967. doi: 10.1002/1439-7633(20021004)3:10<963::AID-CBIC963¾3.0.CO;2-9
Tag, A., Hicks, J., Garifullina, G., Ake, C., and Keller, N. G. (2010). -protein signalling mediates differential production of toxic secondary metabolites [J]. Mol. Microbiol. 38, 658–665. doi: 10.1046/j.1365-2958.2000.02166.x
Tai, B. W., Chang, J. H., Liu, Y., and Xing, F. G. (2020). Recent progress of the effect of environmental factors on Aspergillus flavus growth and aflatoxins production on foods [J]. Food Quality Saf. 4, 21–28. doi: 10.1093/fqsafe/fyz040
Terfehr, D., Dahlmann, T. A., Specht, T., Zadra, I., Kurnsteiner, H., and Kuck, U. (2014). Genome sequence and annotation of Acremonium chrysogenum, producer of the β-Lactam antibiotic cephalosporin C [J]. Genome Announc. 2:e00948-14. doi: 10.1128/genomeA.00948-14
Thatcher, L. F., Manners, J. M., and Kazan, K. (2009). Fusarium oxysporum hijacks COI1-mediated jasmonate signaling to promote disease development in Arabidopsis [J]. Plant J. 58, 927–939. doi: 10.1111/j.1365-313X.2009.03831.x
Thieme, N., Wu, V. W., Dietschmann, A., Salamov, A. A., Wang, M., Johnson, J., et al. (2017). The transcription factor PDR-1 is a multi-functional regulator and key component of pectin deconstruction and catabolism in Neurospora crassa [J]. Biotechnol. Biofuels 10:149. doi: 10.1186/s13068-017-0807-z
Tilburn, J., Sarkar, S., Widdick, D. A., Espeso, E. A., Orejas, M., Mungroo, J., et al. (1995). The Aspergillus Pacc zinc-finger transcription factor mediates regulation of both acid-expressed and alkaline-expressed genes by ambient Ph [J]. EMBO J. 14, 779–790. doi: 10.1002/j.1460-2075.1995.tb07056.x
Tsitsigiannis, D. I, and Keller, N. P. (2010). Oxylipins act as determinants of natural product biosynthesis and seed colonization in Aspergillus nidulans [J]. Mol. Microbiol. 59, 882–892. doi: 10.1111/j.1365-2958.2005.05000.x
Tsitsigiannis, D. I., and Keller, N. P. (2007). Oxylipins as developmental and host-fungal communication signals [J]. Trends Microbiol. 15, 109–118. doi: 10.1016/j.tim.2007.01.005
Tudzynski, B. (2014). Nitrogen regulation of fungal secondary metabolism in fungi [J]. Front. Microbiol. 5:656. doi: 10.3389/fmicb.2014.00656
Turan, N. B., and Engin, G. Ö (2018). Quorum quenching [J]. Comprehensive Anal. Chem. 81, 117–149. doi: 10.1016/bs.coac.2018.02.003
Van Dijck, P., Brown, N. A., Goldman, G. H., Rutherford, J., Xue, C. Y., and Van Zeebroeck, G. (2017). Nutrient sensing at the plasma membrane of fungal cells [J]. Microbiol. Spectr. 5:31. doi: 10.1128/9781555819583.ch19
Villa, N. Y., Moussatche, P., Chamberlin, S. G., Kumar, A., and Lyons, T. J. (2011). Phylogenetic and preliminary phenotypic analysis of yeast PAQR receptors: potential antifungal targets [J]. J. Mol. Evol. 73, 134–152. doi: 10.1007/s00239-011-9462-3
Voigt, C. A., Schäfer, W., and Salomon, S. A. (2010). secreted lipase of Fusarium graminearum is a virulence factor required for infection of cereals [J]. Plant J. 42, 364–375. doi: 10.1111/j.1365-313X.2005.02377.x
Wang, G., Wang, Y., Zhang, H., Zhang, C., Yang, B., Huang, S., et al. (2020). Factors that affect the formation of mycotoxins: a literature review [J]. Mygosystema 39, 477–491.
Wang, G., Zhang, H. Y., Wang, Y. L., Liu, F., Li, E. F., Ma, J. N., et al. (2019). Requirement of LaeA, VeA, and VelB on asexual development, ochratoxin a biosynthesis, and fungal virulence in Aspergillus ochraceus [J]. Front. Microbiol. 10:2759. doi: 10.3389/fmicb.2019.02759
Wasternack, C., and Feussner, I. (2018). The oxylipin pathways: biochemistry and function [J]. Annu. Rev. Plant Biol. 69, 363–386. doi: 10.1146/annurev-arplant-042817-040440
Wenying, C., Chen, L., Boyang, Z., Zheng, Z., Yingbin, S., Xin, L., et al. (2018). Advances in biodetoxification of ochratoxin A-a review of the past five decades [J]. Front. Microbiol. 9:1386. doi: 10.3389/fmicb.2018.01386
Wilson, R. A., Gardner, H. W., and Keller, N. P. (2001). Cultivar-dependent expression of a maize lipoxygenase responsive to seed infesting fungi [J]. Mol. Plant Microbe Interact. 14:980. doi: 10.1094/MPMI.2001.14.8.980
Wong, K. H., Hynes, M. J., and Davis, M. A. (2008). Recent advances in nitrogen regulation: a comparison between Saccharomyces cerevisiae and filamentous fungi [J]. Eukaryotic Cell 7, 917–925. doi: 10.1128/EC.00076-08
Wongsuk, T., Pumeesat, P., and Luplertlop, N. (2016). Fungal quorum sensing molecules: role in fungal morphogenesis and pathogenicity [J]. J. Basic Microbiol. 56, 440–447. doi: 10.1002/jobm.201500759
Xue, C., Bahn, Y. S., Cox, G. M., and Heitman, J. G. (2006). protein-coupled receptor Gpr4 senses amino acids and activates the cAMP-PKA pathway in Cryptococcus neoformans [J]. Mol. Biol. Cell 17:667. doi: 10.1091/mbc.e05-07-0699
Xue, C. Y., Hsueh, Y. P., and Heitman, J. (2008). Magnificent seven: roles of G protein-coupled receptors in extracellular sensing in fungi [J]. FEMS Microbiol. Rev. 32, 1010–1032. doi: 10.1111/j.1574-6976.2008.00131.x
Yan, S. J., Liang, Y. T., Zhang, J. D., Chen, Z., and Liu, C. M. (2015). Autoxidated linolenic acid inhibits aflatoxin biosynthesis in Aspergillus flavus via oxylipin species [J]. Fungal Genet. Biol. 81, 229–237. doi: 10.1016/j.fgb.2014.11.005
Yan, W., Liuqing, W., Fan, W., Fei, L., Qi, W., Xiaoling, Z., et al. (2018). A consensus ochratoxin a biosynthetic pathway: insights from the genome sequence of Aspergillus ochraceus and a comparative genomic analysis [J]. Appl. Environ. Microbiol. 84:e01009-18. doi: 10.1128/AEM.01009-18
Yan, X. U., Wei-Jia, Y., Yun-Long, Z., and Jing, X. U. (2003). Identification of the Gq-protein in cherax quadricar-inatus photoreceptor and the influence of wave- length Light on the Content of Gqα [J]. Zool. Res. 24, 602–606.
Yin, W. B., Reinke, A. W., Szilagyi, M., Emri, T., Chiang, Y. M., Keating, A. E., et al. (2013). bZIP transcription factors affecting secondary metabolism, sexual development and stress responses in Aspergillus nidulans [J]. Microbiol. Sgm 159, 77–88. doi: 10.1099/mic.0.063370-0
Yu, J., Fedorova, N. D., Montalbano, B. G., Deepak, B., Cleveland, T. E., Bennett, J. W., et al. (2011). Tight control of mycotoxin biosynthesis gene expression in Aspergillus flavus by temperature as revealed by RNA-Seq [J]. FEMS Microbiol. Lett. 2, 145–149. doi: 10.1111/j.1574-6968.2011.02345.x
Zhang, B., Shen, X. L., Liang, R., Li, Y., Huang, K., Zhao, C., et al. (2014). Protective role of the mitochondrial Lon protease 1 in ochratoxin A-induced cytotoxicity in HEK293 cells [J]. J. Proteom. 101, 154–168. doi: 10.1016/j.jprot.2014.02.017
Zhang, B., Zhu, L., Dai, Y., Li, H., and Xu, W. (2019). An in vitro attempt at precision toxicology reveals the involvement of DNA methylation alteration in ochratoxin A-induced G0/G1 phase arrest [J]. Epigenetics 15, 199–214. doi: 10.1080/15592294.2019.1644878
Zhang, J., Chen, H. Y., Sumarah, M. W., Gao, Q., Wang, D. P., and Zhang, Y. (2018). veA gene acts as a positive regulator of conidia production, ochratoxin A biosynthesis, and oxidative stress tolerance in Aspergillus niger [J]. J. Agric. Food Chem. 66, 13199–13208. doi: 10.1021/acs.jafc.8b04523
Zhao, T., Shen, X. L., Chen, W., Liao, X., Yang, J., Wang, Y., et al. (2017). Advances in research of nephrotoxicity and toxic antagonism of ochratoxin A [J]. Toxin Rev. 36, 39–44. doi: 10.1080/15569543.2016.1243560
Zheng, H., Zhou, L., Dou, T., Han, X., Cai, Y., Zhan, X., et al. (2010). Genome-wide prediction of G protein-coupled receptors in Verticillium spp [J]. Fungal Biol. 114, 359–368. doi: 10.1016/j.funbio.2010.02.008
Keywords: G protein-coupled receptors, ochratoxin A, quorum sensing, oxylipin, trans-kingdom, transcription factor
Citation: Gao J, Xu X, Huang K and Liang Z (2021) Fungal G-Protein-Coupled Receptors: A Promising Mediator of the Impact of Extracellular Signals on Biosynthesis of Ochratoxin A. Front. Microbiol. 12:631392. doi: 10.3389/fmicb.2021.631392
Received: 20 November 2020; Accepted: 21 January 2021;
Published: 12 February 2021.
Edited by:
David L. Moyes, King’s College London, United KingdomReviewed by:
Xuehua Xu, National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH), United StatesMassimo Reverberi, Sapienza University of Rome, Italy
Copyright © 2021 Gao, Xu, Huang and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhihong Liang, lzh105@cau.edu.cn
 Jing Gao
Jing Gao Xinge Xu
Xinge Xu Kunlun Huang2
Kunlun Huang2 Zhihong Liang
Zhihong Liang