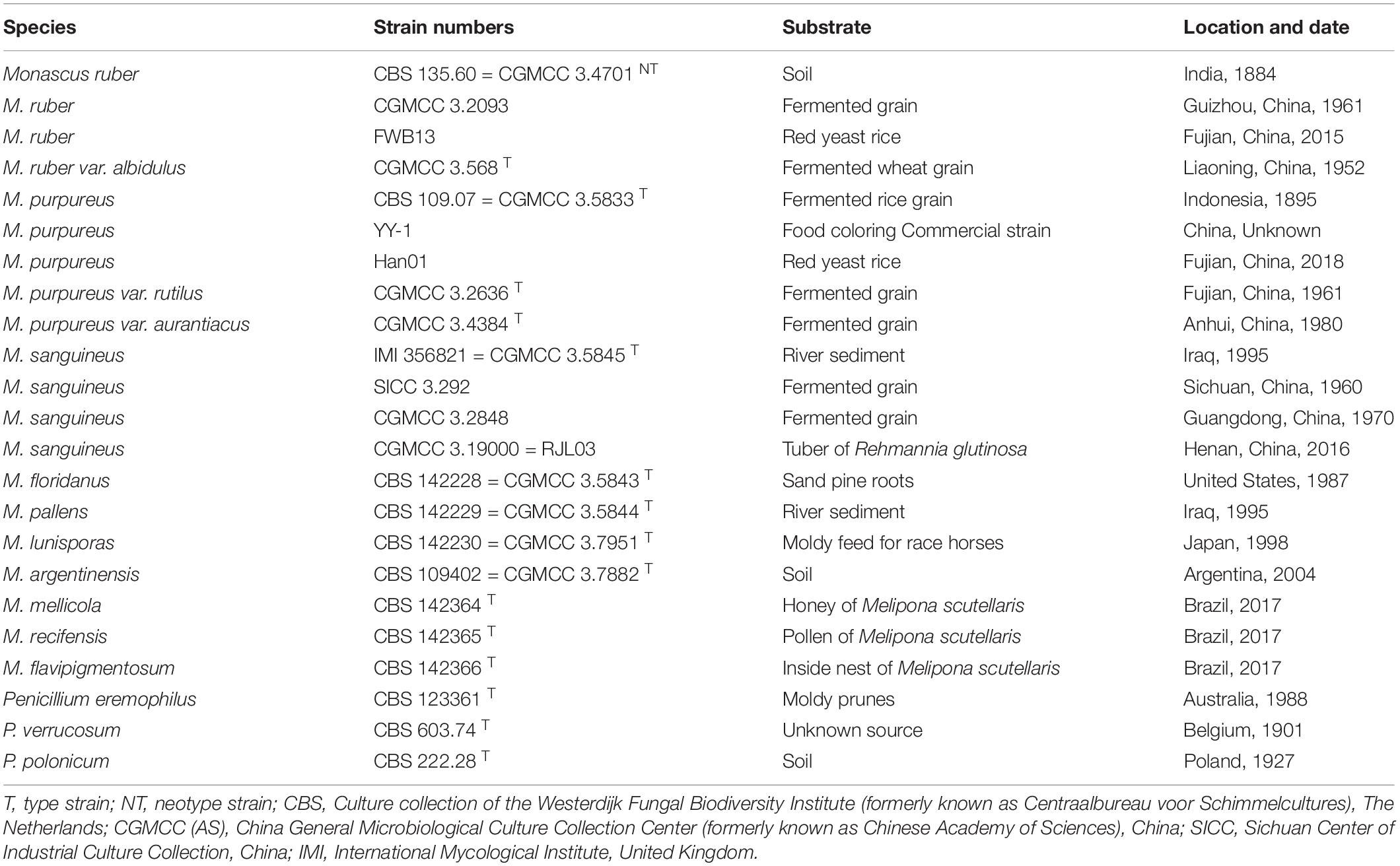
- 1College of Life Science and Engineering, Northwest Minzu University, Lanzhou, China
- 2Center for Life Sciences, School of Life Sciences, Yunnan University, Kunming, China
The genus Monascus has important economic and ecological values. In 2016, we isolated a strain M. sanguineus. After studying the phylogenetic relationship of Monascus, we believe that M. sanguineus is an independent species and speculate that it is a natural nothospecies. Recently, the morphological characteristics and sequences of seven genes (ITS, LSU, β-tubulin, calmodulin, RNA polymerase II subunit, β-ketoacyl synthase, and mating-type locus 1-1) of 15 Monascus strains were analyzed, including sequencing of multiple clones of five protein genes in four M. sanguineus strains. Two types of haplotypes (A and B) were observed in the five protein genes of M. sanguineus. Haplotype A was closely related to M. ruber, and haplotype B may be derived from an unknown Monascus species. The results demonstrated that M. sanguineus including type strains may be a natural nothospecies. This study laid the foundation for further exploration of the M. sanguineus genome, and the study may be of significant importance for the Monascus fermentation industry.
Introduction
Monascus spp. are filamentous fungi first described by a French scientist Van Tieghem (1884). Having medicinal as well as edible uses, Monascus has been used in China for nearly 2000 years. Its use can be dated back to the Han dynasty (BC 202–AD 220). As a characteristic species, Monascus-fermented rice (also known as red yeast rice, a rice-based fermentation product) is widely consumed throughout East Asia and has a profound impact on local life and culture. Monascus has received worldwide attention because of its diverse products and rich beneficial metabolites. Its distribution ranges from the Korean Peninsula to the Malay Archipelago, and it is even spread globally (Lee and Pan, 2011, 2012; Shi and Pan, 2012; Chen et al., 2015). In industrial production, Monascus has three principal applications or products: starter culture (as a starter in various food fermentations), Monascus-fermented rice (as a food supplement), and functional food, which are widely used in brewing, food coloring, and healthcare industries. Monascus is an important source of numerous types of hydrolytic enzymes required in fermentation of foods (including red rice wine, red rice vinegar, Chinese spirits, fish paste, and fermented tofu). The beneficial secondary metabolites produced by Monascus mainly include Monascus pigments (food colorants and condiments), biofunctional components (including monacolins, γ-amino butyric acid, and dimerumic acid) (Feng et al., 2012; Hsu and Pan, 2012; Vendruscolo et al., 2014; Mérillon and Ramawat, 2017), and citrinin (safety disputed because of renal toxicity) (Kim et al., 2010; Li et al., 2012, 2015). More than one billion people have been estimated to eat Monascus-fermented products daily, with the most popular product being Monascus-fermented rice (Yang et al., 2015). Additionally, researchers have found that Monascus has a mutually beneficial symbiotic relationship with some bees, indicating its important ecological value (Menezes et al., 2015; Barbosa et al., 2017).
The genus Monascus belongs to the family Aspergillaceae and order Eurotiales. It contains 36 species; however, many of them are considered synonymous (Shao et al., 2011, 2014). Based on the study by Barbosa et al. (2017), we preliminarily unified the phylogenetic relationships of the species within the genus Monascus and confirmed that Monascus includes the Rubri and Floridani sections. The Rubri section consists of three species and three varieties; the Floridani section consists of seven species, and additionally, one species speculated to be a natural nothospecies was found (He et al., 2018). In this study, to improve the phylogenetic relationship within Monascus and identify the parents of suspected nothospecies, we analyzed the sequences of the internal transcribed spacer (ITS), large subunit (LSU), beta-tubulin (BenA), calmodulin (CaM), RNA polymerase II subunit (RPB2), beta-ketoacyl synthase (pksKS), and mating-type locus 1-1 (MAT1-1) in 15 strains of Monascus. Based on a polyphasic approach combining sequence data with macroscopic and microscopic characters, it is speculated that M. sanguineus may be a natural nothospecies; M. ruber is one of its parents, and the other parent may be an unknown species.
Materials and Methods
Strains
The strain M. sanguineus (CGMCC 3.19000 = RJL03) was isolated from the medicinal plant Rehmannia glutinosa. M. purpureus (Han01) was isolated from the commercially available Fujian (China) red yeast rice. The strain M. sanguineus (SICC 3.292) was purchased from the Sichuan Center of Industrial Culture Collection (SICC,1, China), and the remaining 12 Monascus strains were purchased from the China General Microbiological Culture Collection Center (CGMCC,2, China).
Cultivation and Morphological Analyses
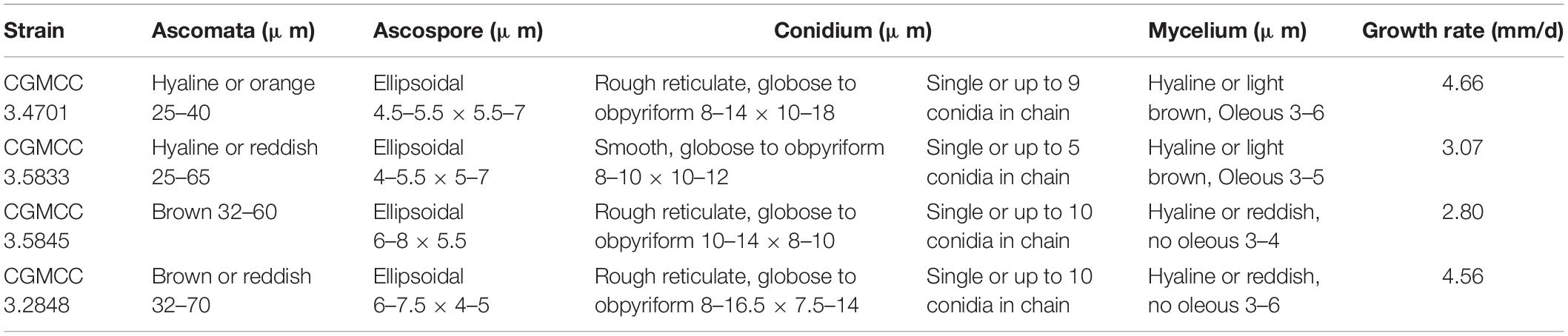
For observing the colonial morphology, the 15 strains (Table 1) were cultured in three points on potato dextrose agar (PDA), malt extract agar (MEA), and Czapek yeast extract agar (CYA) plates at 30°C for 7 days. Macroscopic characteristics such as soluble pigments, color of the mycelium, and obverse and reverse colony colors were studied. Single colony diameters were measured after incubation for 7 days, and the average growth rate was calculated. The hyphae were observed by the transparent tape method using an optical microscope. A drop of lactophenol cotton blue stain was placed on a glass slide. The hyphae were adhered to a transparent tape. Ethanol was added dropwise to the surface of the hyphae, and they were placed on the slide with the drop of stain. The stain solution was added dropwise on the surface of the tape, which was later covered with a coverslip. Scanning electron microscopy revealed that the strains were cultured by insert coverslip. After the hyphae were climbed, the coverslips were gently removed for treatment. The coverslips with hyphae were fixed in 2.5% glutaraldehyde for more than 4 h and further rinsed 3 times with phosphate buffer. The coverslips with samples were dehydrated with graded concentrations of ethanol (50, 70, 80, 90, 95, and 100%) for 20 min for each concentration, transferred to pure isoamyl acetate for 1 h, and coated with gold–palladium. After pretreatment of the samples, morphological characteristics such as size, shape, and pigmentation of conidia, conidiophores, ascomata, asci, and ascospores were observed under the BME Biooptical microscope (Shanghai Leica Microsystems Co., Ltd., China) and SM-5600LV low vacuum scanning electron microscope (Japan Electronic Co., Ltd., Japan).
DNA Extraction, Amplification, Cloning, and Sequencing
Strains were grown on MEA for 7–14 days prior to DNA extraction. Genomic DNA was extracted using Fungi Genomic DNA Extraction Kit (Beijing Solarbio Science and Technology Co., Ltd., China) as per the manufacturer’s instructions. DNA was amplified through polymerase chain reaction (PCR) using seven pairs of primers for seven genes (Supplementary Table 1). According to our study, cloning of PCR products was not required except for five protein genes in four M. sanguineus strains. At least 10 clones were randomly selected for each sample using the blue–white selection system, and both regular and clones were sequenced (Sangon Biotech Co., Ltd., Shanghai, China).
Sequence Alignment and Phylogenetic Analyses
Contings were assembled using the forward and reverse sequences with the SeqMan v.7.1.0. Analysis of homology of amplified products was studied using Blastn. Further, the sequences generated in this study were submitted to GenBank via the web tool BankIt or Sequin program. Sequence datasets were generated by combining the 197 newly generated sequences and 44 sequences that we deposited from GenBank (Supplementary Table 2). Sequence alignments were performed in MAFFT3 and were manually optimized using MEGA 7. The best substitution model for each partition was inferred with the program MrModeltest 2.3. Phylogenetic trees were constructed through maximum likelihood (ML) analysis in raxmlGUI 1.5 using the GTRGAMMA substitution model and 1000 bootstrap replicates. Bayesian inference (BI) in MrBayes v.3.2.1 was performed using the Markov Chain Monte Carlo (MCMC) algorithm. Sequence format conversion was performed using Mesquite 3.10. Individual alignments were concatenated using Sequence Matrix v.1.7.8 for multilocus phylogenetic analyses. Each gene was analyzed separately, and further, two sequences with the highest rate of the two haplotypes were selected. Data partitioning was performed to construct a multigene phylogenetic tree.
Results and Discussion
Evidence for M. sanguineus as a Natural Nothospecies
Our analysis revealed two well-supported sections (Rubri and Floridani) in Monascus. Seven lineages are present in the section Floridani and these lineages are treated as separate species. M. purpureus, M. sanguineus, and M. ruber are located in the section Rubri. Also, the results of this study demonstrated that four M. sanguineus strains including the type strain may be natural nothospecies (not found in previous research data). Additionally, two types of haplotypes (A and B) were found after cloning and sequencing of five protein genes. Haplotype A was closely related to M. ruber. Haplotype B may be derived from an unknown Monascus species (Figure 1 and Supplementary Figure 1). Haplotype B had a much higher red-pigment-producing ability than its suspected parent M. ruber, but its growth rate was lower than that of M. ruber. Thus, it was speculated that the yet unknown parent (Monascus sp.) of haplotype B confers the ability to produce red pigment. Additionally, the heterozygosity of the four M. sanguineus strains was notably different. For example, we had not cloned haplotype A of the strain CGMCC 3.5845; all SNPs were only observed at the corresponding sites in the direct sequencing of the five protein genes, and the two types of haplotypes between each hybrid strain were not completely consistent. For example, the results of cloning and sequencing demonstrated that the β-tubulin (BenA) gene of strain CGMCC 3.2848 had five haplotypes A and five haplotypes B (Supplementary Figure 2). For better understanding, an example of yeast (Saccharomyces) can be considered, which is similar to the M. sanguineus hybrid and has been studied in detail. The allopolyploid hybrid S. pastorianus was once considered the synonym of its parent strain S. cerevisiae; however, the parent strain S. eubayanus, with its most important low-temperature fermentation characteristics, was discovered after a long period (Bing et al., 2014; Wendland, 2014). Simple tests, such as DAPI staining to examine karyotypes and qPCR to assess fold changes in gene copy number, can be used to analyze the difference between chromosome ploidy in M. sanguineus and M. ruber (Waalwijk et al., 2018). It is speculated that in the previous study, the ribosomal ITS and LSU gene sequences could not distinguish between the hybrids M. ruber and Monascus sp., probably because in the chromosome of the hybrid, the chromosome containing the ribosomal RNA gene (rDNA) cluster from the M. ruber parent was substantially lost or reduced in length after hybridization. Referring to this example, to reflect the characteristics of M. sanguineus as a hybrid, we describe it as an independent species (M. sanguineus). The official DNA barcode for the fungal ITS region can recognize all Monascus species (including M. purpureus and M. sanguineus), but larger sequence variations can be observed in the BenA gene. Therefore, we suggest that the BenA gene can be used as a secondary barcode for the identification of Monascus species, besides the methods based on morphological features (Barbosa et al., 2017).
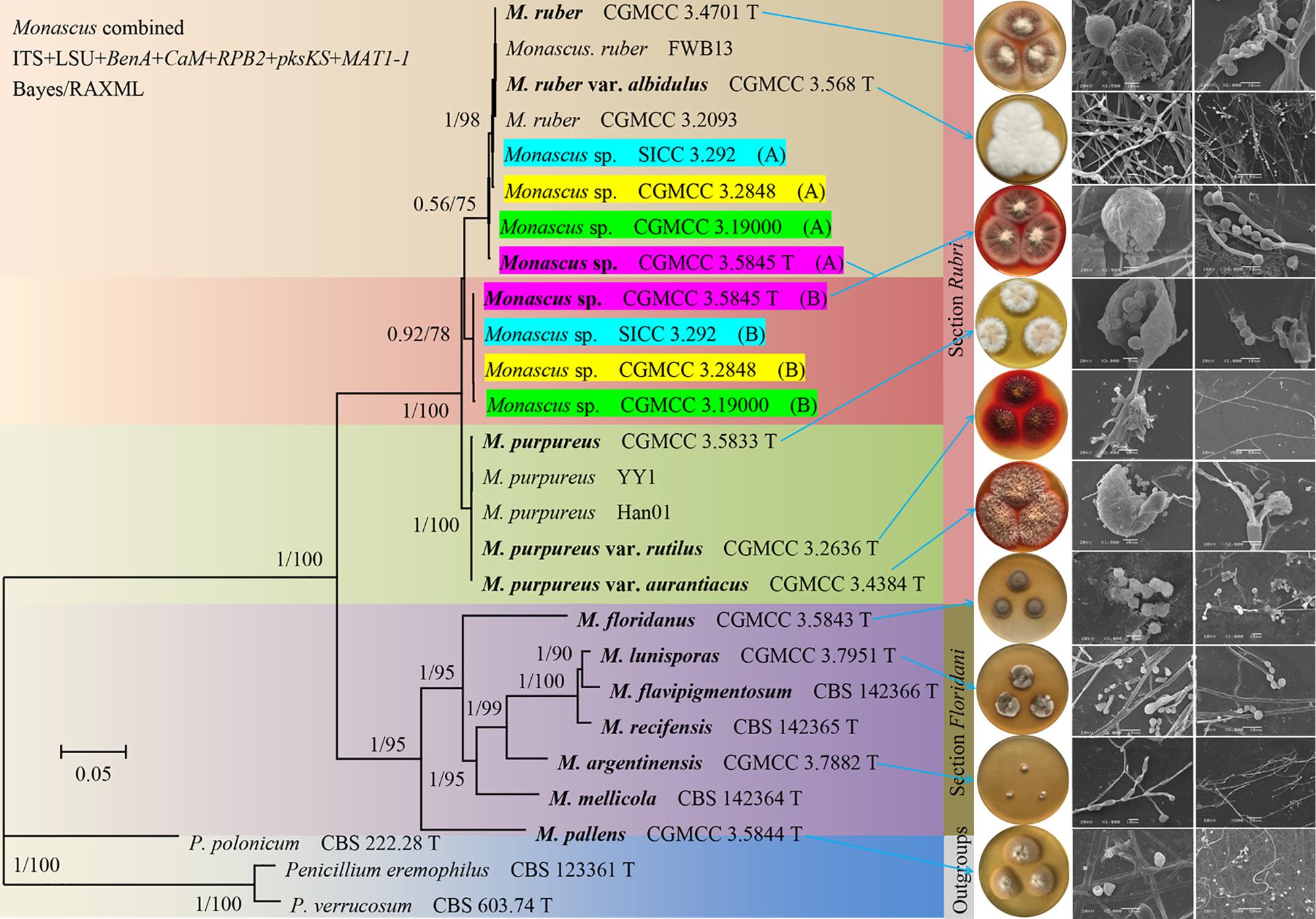
Figure 1. Phylogeny of Monascus. Phylogenetic tree constructed from Bayesian analysis of the concatenated sequences of five protein genes (BenA, CaM, RPB2, pksKS, and MAT1-1) and two ribosomal RNA genes (ITS and LSU) with a total length of 6,983 bp. Posterior probabilities from Bayesian analysis/bootstrap percentages from 1,000 replicates of maximum likelihood analysis are shown on major branches. The right side of the figure is the cultural characters and scanning electron microscope structural of the strains indicated by the arrow. For the sake of illustration, the M. sp. CGMCC 3. 5845 (A) in the figure are manually supplemented.
Natural hybridization is very common in flowering plants. Approximately 25% of plant species are reported to hybridize with other species (Arnold, 1992; Mallet, 2005). Natural hybridization plays an important role in speciation, genetic exchange, and adaptive evolution. However, it can also lead to the extinction of rare species or generate super invasive species (Hegarty and Hiscock, 2007; Zhuang et al., 2019). M. sanguineus (CGMCC 3.5845) was first isolated from the sediments of the Arab River in Iraq in 1995 (Cannon et al., 1995) and was later reported in an Indian plant Punica granatum (Rashmi and Padmavathi, 2014). The SICC 3.292 strain was isolated from fermented grain in Sichuan Province in 1960, and the RJL03 strain was isolated from the plant Rehmannia glutinosa in our laboratory (He et al., 2019). Four M. sanguineus strains from different sources used in this study showed hybridization, indicating that hybrids may not have occurred by artificial breeding but by natural hybridization. However, the origin of its formation still needs to be studied. M. sanguineus was considered to be the synonym species of M. purpureus. This study demonstrated that M. ruber may be one of its parents, and there are some significant differences among the four M. sanguineus strains. We compared the morphological characteristics of two different M. sanguineus strains (CGMCC 3.5845 and CGMCC 3.2848) with M. ruber (CGMCC 3.4701) and M. purpureus (CGMCC 3.5833); the results showed that the morphological structure of the ascomata, ascospores, and conidium of the two hybrid strains was different from that of M. ruber and M. purpureus, but it was close to that of M. ruber (Table 2). Additionally, a significant difference was observed between the two hybrids; the three M. sanguineus strains grew faster than the type strain CGMCC 3.5845 (Supplementary Figures 3, 4).
Urgent Need to Study the M. sanguineus Genome
With the sequencing of the M. pilosus genome in 2004, the study on Monascus has entered the era of genomics (data not shown). Genomic sequencing of M. ruber (M7, NRRL 1597) and M. purpureus (NRRL 1596, YY-1) was completed. The average genome size was found to be 24.04 Mb, containing 7 or 8 chromosomes. The sketch genome coverage rate was approximately 95.6%. This gave us a deeper understanding of the physiology of Monascus and revealed new methods for strain improvement (Chen et al., 2015; Yang et al., 2015; Ding et al., 2016). However, only a few genes have been sequenced in M. sanguineus, and characteristics of the whole genome (genome size, chromosome number, and precise structure) remain undiscovered. We suggest that first, the genome of M. sanguineus should be sequenced, its ploidy should be determined, and comparative genomics studies should be conducted to compare its genomic sequence with that of other Monascus species (especially M. ruber and M. purpureus). Further, the structural features of two subgroups of hybrids (A and B subgenomes), the homology of the M. sanguineus subgroup and M. sanguineus should be analyzed, and it should be determined whether M. ruber is the ancestor of the M. sanguineus subgroup A while investigating the donor parent of subgroup B. In addition to its economic value, Monascus genomics research plays an important role in recognizing structural variability, integrating phenotype–genotype association, understanding the origin and evolution of the Monascus genome, and elucidating the genetic structure of some important traits. A comparative genomic study of Monascus species will enrich the knowledge about Monascus genetics and biology. Additionally, as an industrial strain, Saccharomyces has been studied in more detail; hence, the related research cases can be assessed in the study of Monascus genomics.
Monascus fermentation and its applications are the driving forces for research on Monascus. This study demonstrated that M. sanguineus may be a natural hybrid; we suggest to describe it as an independent species, and sequence analysis of its whole genome should be performed. In recent years, to analyze the regulation of secondary metabolites in Monascus, molecular biological studies such as those related to genes (clusters) of the main secondary metabolites of Monascus, biosynthetic pathways, and regulatory mechanism have made significant progress (Chen et al., 2017). At present, we can improve the production of beneficial metabolites of Monascus through strain selection, optimization of fermentation conditions, and genetic modification and effectively eliminate or reduce the content of citrinin in Monascus products. The completion of Monascus genome sequencing will greatly promote the studies on various aspects of Monascus, by which we can better understand the fermentation characteristics of different strains and molecular mechanisms underlying metabolite production and clarify the breeding and genetic transformation of Monascus strains. This will be beneficial for the Monascus fermentation industry. During the era of molecular biology, the aim was to establish the connection between Monascus genetics and biological performance. The studies on Monascus genomics will eventually boost Monascus research to a global level with clear goals. In the foreseeable future, the combination of genomics and molecular biology techniques will play a major role in Monascus research (Chen et al., 2015).
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
YH and JL conceived and designed the experiment. YH, QC, SG, and TS carried out the experiment and performed the analysis. YH wrote the manuscript. JL and SH revised the manuscript. All authors discussed the results and commented on the manuscript.
Funding
This work was supported by the Laboratory Open Project of Northwest Minzu University (SYSKF-2018157); the National Trainning Program of Innovation and Entrepreneurship for Undergraduates (201810742120); and the Fundamental Research Funds for the Central Universities (31920190204).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully thank Peijie Han (Institute of Microbiology, Chinese Academy of Sciences, China) for his help during the research and TopEdit (www.topeditsci.com) for English language editing of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.614910/full#supplementary-material
Supplementary Figure 1 | Single gene phylogenetic trees of the ITS, LSU, BenA, CaM, RPB2, pksKS, and MAT1-1 gene regions of species from Monascus.
Supplementary Figure 2 | Two haplotypes of the BenA gene of four M. sanguineus strains (only inconsistent sequences are retained after alignment).
Supplementary Figure 3 | Cultural characters of 15 Monascus strains cultured on MEA, PDA, and CYA media at 30°C for 7 days.
Supplementary Figure 4 | Mean growth rates (mm/d) of 15 Monascus strains cultured on PDA, MEA and CYA media at 30°C for 7 days.
Supplementary Table 1 | Genes sequenced and primers for PCR.
Supplementary Table 2 | GenBank accession numbers of sequences used in the molecular study.
Footnotes
References
Arnold, M. L. (1992). Natural hybridization as an evolutionary process. Annu. Rev. Ecol. Evol. S. 85, 1893–1896. doi: 10.1146/annurev.es.23.110192.001321
Barbosa, R. N., Leong, S. L., Vinnere-Pettersson, O., Chen, A. J., Souza-Motta, C. M., Frisvad, J. C., et al. (2017). Phylogenetic analysis of Monascus, and new species from honey, pollen and nests of stingless bees. Stud. Mycol. 86, 29–51. doi: 10.1016/j.simyco.2017.04.001
Bing, J., Han, P., Liu, W., Wang, Q., and Bai, F. (2014). Evidence for a Far East Asian origin of lager beer yeast. Curr. Biol. 24, R380–R381. doi: 10.1016/j.cub.2014.04.031
Cannon, P. F., Abdullah, S. K., and Abbas, B. A. (1995). Two new species of Monascus from Iraq, with a key to known species of the genus. Mycol. Res. 99, 659–662. doi: 10.1016/s0953-7562(09)80523-6
Chen, W., Chen, R., Liu, Q., He, Y., He, K., Ding, X., et al. (2017). Orange, Red, Yellow: Biosynthesis of Azaphilone Pigments in Monascus Fungi. Chem. Sci. 8, 4917–4925. doi: 10.1039/c7sc00475c
Chen, W., He, Y., Zhou, Y., Shao, Y., Feng, Y., Li, M., et al. (2015). Edible filamentous fungi from the species Monascus: early traditional fermentations, modern molecular biology, and future genomics. Compr. Rev. Food Sci. F. 14, 555–567. doi: 10.1111/1541-4337.12145
Ding, X., Chen, W., and Chen, F. (2016). Karyotype analysis and chromosome localization of pigment and citrinin biosynthesis gene clusters of Monascus ruber M7. Mycosystema 35, 309–16. doi: 10.13346/j.mycosystema.140287
Feng, Y., Shao, Y., and Chen, F. (2012). Monascus pigments. Appl. Microbiol. Biot. 96, 1421–1440. doi: 10.1007/s00253-012-4504-3
He, Y., Gao, D., Gan, S., Sun, T., Cai, K., and Liu, J. (2019). Isolation and identification of a red pigment producer endophytic fungus Monascus sanguineus from Rehmannia glutinosa Libosch. J. Zhejiang Univ. 45, 1–7. doi: 10.3785/j.issn.1008-9209.2018.02.022
He, Y., Ran, Q., Liu, Y., Lan, Q., and Liu, J. (2018). Phylogenetic relationship of the genus Monascus in China based on multi-gene sequence analysis. Mycosystema 37, 1154–1169. doi: 10.13346/j.mycosystema.180091
Hegarty, M., and Hiscock, S. (2007). Polyploidy: Doubling up for Evolutionary Success. Curr. Biol. 17, R927–R929. doi: 10.1016/j.cub.2007.08.060
Hsu, W. H., and Pan, T. M. (2012). Monascus purpureus-fermented products and oral cancer: a review. Appl. Microbiol. Biot. 93, 1831–1842. doi: 10.1007/s00253-012-3891-9
Kim, J. Y., Kim, H. J., Oh, J. H., and Lee, I. (2010). Characteristics of monascus sp. isolated from monascus fermentation products. Food Sci. Biotechnol. 19, 1151–1157. doi: 10.1007/s10068-010-0164-1
Lee, C. L., and Pan, T. M. (2011). Red mold fermented products and Alzheimer’s disease: a review. Appl. Microbiol. Biot. 91, 461–469. doi: 10.1007/s00253-011-3413-1
Lee, C. L., and Pan, T. M. (2012). Benefit of Monascus-fermented products for hypertension prevention: a review. Appl. Microbiol. Biot. 94, 1151–1161. doi: 10.1007/s00253-012-4076-2
Li, Y. P., Tang, X., Wu, W., Xu, Y., Huang, Z. B., and He, Q. H. (2015). The ctnG gene encodes carbonic anhydrase involved in mycotoxin citrinin biosynthesis from Monascus aurantiacus. Food Addit. Contam. B 32, 577–583. doi: 10.1080/19440049.2014.990993
Li, Y., Zhou, Y. C., Yang, M. H., and Ou-Yang, Z. (2012). Natural occurrence of citrinin in widely consumed traditional Chinese food red yeast rice, medicinal plants and their related products. Food Chem. 132, 1040–1045. doi: 10.1016/j.foodchem.2011.11.051
Mallet, J. (2005). Hybridization as an invasion of the genome. Trends Ecol. Evol. 20, 229–237. doi: 10.1016/j.tree.2005.02.010
Menezes, C., Vollet-Neto, A., Marsaioli, A. J., Zampieri, D., Fontoura, I. C., Luchessi, A. D., et al. (2015). A Brazilian social bee must cultivate fungus to survive. Curr. Biol. 25, 2851–2855. doi: 10.1016/j.cub.2015.09.028
Mérillon, J., and Ramawat, K. G. (2017). Fungal Metabolites. Switzerland: Springer International Publishing, 821–851.
Rashmi, D., and Padmavathi, T. (2014). Statistical optimization of pigment production by Monascus sanguineus under stress condition. Prep. Biochem. Biotech. 44, 68–79. doi: 10.1080/10826068.2013.792097
Shao, Y., Lei, M., Mao, Z., Zhou, Y., and Chen, F. (2014). Insights into Monascus biology at the genetic level. Appl. Microbiol. Biot. 98, 3911–3922. doi: 10.1007/s00253-014-5608-8
Shao, Y., Xu, L., and Chen, F. (2011). Genetic diversity analysis of Monascus strains using SRAP and ISSR markers. Mycoscience 52, 224–233. doi: 10.1007/s10267-010-0087-y
Shi, Y. C., and Pan, T. M. (2012). Red mold, diabetes, and oxidative stress: a review. Appl. Microbiol. Biot. 94, 47–55. doi: 10.1007/s00253-012-3957-8
Van Tieghem, M. (1884). Monascus, genre nouveau de l’ordre des Ascomycetes. Bull. Soc. Bot. Fr. 31, 226–231. doi: 10.1080/00378941.1884.10828230
Vendruscolo, F., Tosin, I., Admir José, G., Schmidell, W., and Ninow, J. L. (2014). Antimicrobial activity of Monascus pigments produced in submerged fermentation. J. Food Process. Pres. 38, 1860–1865. doi: 10.1111/jfpp.12157
Waalwijk, C., Taga, M., Zheng, S. L., Proctor, R. H., and Vaughan, M. M. (2018). Karyotype evolution in fusarium. IMA Fungus 9, 13–26. doi: 10.5598/imafungus.2018.09.01.02
Wendland, J. (2014). Lager yeast comes of age. Eukaryotic Cell 13, 1256–1265. doi: 10.1128/EC.00134-14
Yang, Y., Liu, B., Du, X., Li, P., Liang, B., Cheng, X., et al. (2015). Complete genome sequence and transcriptomics analyses reveal pigment biosynthesis and regulatory mechanisms in an industrial strain, Monascus purpureus YY-1. Sci. Rep. 5:8331. doi: 10.1038/srep08331
Keywords: Monascus sanguineus, phylogeny, morphology, nothospecies, fungi, natural pigment, industrial strain
Citation: He Y, Liu J, Chen Q, Gan S, Sun T and Huo S (2020) Monascus sanguineus May Be a Natural Nothospecies. Front. Microbiol. 11:614910. doi: 10.3389/fmicb.2020.614910
Received: 07 October 2020; Accepted: 24 November 2020;
Published: 22 December 2020.
Edited by:
Wanping Chen, Georg-August-University Goettingen, GermanyReviewed by:
Yanchun Shao, Huazhong Agricultural University, ChinaJiao Liu, Hubei Academy of Agricultural Sciences, China
Copyright © 2020 He, Liu, Chen, Gan, Sun and Huo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yatao He, hyt@mail.ynu.edu.cn; Shengdong Huo, huoshd@xbmu.edu.cn
†These authors have contributed equally to this work and share first authorship
 Yatao He
Yatao He Junlin Liu1†
Junlin Liu1†