- 1Division of Bacterial Diseases, Center for Laboratory Control of Infectious Diseases, Korea Centers for Disease Control and Prevention, Chungju, South Korea
- 2Infectious Diseases Team, Seoul Metropolitan Government Research Institute of Public Health and Environment, Seoul, South Korea
- 3Microbiology Team, Gyeonggi-do Institute of Health and Environment, Suwon, South Korea
- 4Division of Antimicrobial Resistance, National Institute of Health, Center for Infectious Diseases Research, Centers for Disease Control and Prevention, Chungcheongbuk-do, South Korea
The emergence of third-generation cephalosporin resistance in Escherichia coli is increasing at an alarming rate in many countries. Thus, the aim of this study was to analyze co-infecting blaCTX-M-producing pathogenic E. coli isolates linked to three school outbreaks. Among 66 E. coli isolates, 44 were identified as ETEC O25, an ETEC isolate serotype was O2, and the other 21 were confirmed as EAEC O44. Interestingly, six patients were co-infected with EAEC O44 and ETEC O25. For these isolates, molecular analysis [antibiotic susceptibility testing, identification of the β-lactamase gene, multilocus sequence typing (MLST), and pulsed-field gel electrophoresis (PFGE)] was performed for further characterization. In addition, the transmission capacity of blaCTX-M genes was examined by conjugation experiments. Whole-genome sequencing (WGS) was performed on representative EAEC O44 and ETEC O25 isolates associated with co-infection and single-infection. All isolates were resistant to cefotaxime and ceftriaxone. All EAEC isolates carried the blaCTX-M-14 gene and all ETEC isolates the blaCTX-M-15 gene, as detected by multiplex PCR and sequencing analysis. Sequence type and PFGE results indicated three different patterns depending on the O serotype. WGS results of representative isolates revealed that the ETEC O25 strains harbored blaCTX-M-15 located on IncK plasmids associated with the ΔblaTEM-blaCTX-M-15-orf477 transposon. The representative EAEC O44 isolates carried blaCTX-M-14 on the chromosome, which was surrounded by the ISEcp1-blaCTX-M-14-IS903 transposon. To the best of our knowledge, this is the first report of co-infection with chromosomally located blaCTX-M-14 and plasmid-encoding blaCTX-M-15 in pathogenic E. coli. Our findings indicate that resistance genes in clinical isolates can spread through concurrent combinations of chromosomes and plasmids.
Introduction
Pathogenic Escherichia coli is a cause of gastroenteritis, including foodborne outbreaks, worldwide. Most E. coli infections are self-limiting, and sometimes require antimicrobial treatment. For treatment of E. coli infections, antibiotics such as third-generation cephalosporins and fluoroquinolones are prescribed (Kim et al., 2014b).
However, the emergence of antimicrobial resistance is increasing at an alarming rate in many countries. Furthermore, third-generation cephalosporin resistance, including resistance against cefotaxime and ceftriaxone, has been steadily reported in recent years. Additionally, many countries have rapidly experienced the dissemination of extended-spectrum-β-lactamase (ESBL)-producing Enterobacteriaceae isolates, particularly E. coli (Oteo et al., 2012).
Extended-spectrum-β-lactamase-producing Enterobacteriaceae appeared in the 1990s, and these bacteria have steadily become prevalent, being primarily associated with the blaCTX-M-14 and blaCTX-M-15 genes in the Republic of Korea (Naseer and Sundsfjord, 2011; Shin et al., 2011, 2012).
The first finding of CTX-M-14 and CTX-M-15 in clinical E. coli isolates was reported in 2001 and 2005 (Pai et al., 2001; Kim et al., 2005), respectively. Since then, a growing trend of CTX-M-producing clinical E. coli isolates has been observed (Kim et al., 2016, 2019).
The spread of CTX-M enzyme-coding capacity occurs due to the mobilizing ability of their insertion sequences and integrons (Oteo et al., 2010). Moreover, a majority of CTX-M-producing isolates can transfer ESBL plasmids between bacteria of the same and/or different species via horizontal transmission (Baker et al., 2018). Moreover, a number of chromosomally located CTX-M genes in E. coli have been reported in several studies, indicating that the CTX-M genes can be transferred via transposons or insertion sequences into the chromosome (Hamamoto and Hirai, 2019).
In this study, we describe blaCTX-M-14‐ and blaCTX-M-15-producing pathogenic E. coli associated with three school outbreaks in distinct regions in the Republic of Korea. In particular, several patients were co-infected with CTX-M-14-producing EAEC and CTX-M-15-producing ETEC during this outbreak period. Thus, the aim of this study was to analyze co-infecting blaCTX-M-producing pathogenic E. coli, including their resistance genes, genetic environments, and plasmid profiles.
Materials and Methods
Bacterial Isolates
In August 2017, local public laboratories reported an outbreak of acute diarrheal illness in three local high schools (school A in Siheung, school B in Gwangju, and school C in Seoul). These outbreaks affected 634 persons from the three different schools during the period 14 August–25 August. A total of 250 stool samples (216 from case patients and 34 from food handlers) and 140 environmental samples, including 92 preserved food products consumed during the outbreak period, 22 cooking utensils, and 16 drinking water samples, were collected and tested for bacteriological and virological assessments (Shin et al., 2015). A total of 66 stool samples were positive for E. coli, whereas the environmental samples and stool samples from staff of the three schools were negative for any pathogenic bacteria or virus.
The isolated E. coli strains were pathotyped by an 8-plex real-time PCR kit (Kogene Biotech, Seoul, South Korea) to detect specific virulence genes, such as VT1 and VT2 for enterohemorrhagic E. coli (EHEC), LT, and sequence type (ST) for enterotoxigenic E. coli (ETEC), eaeA and bfpA for enteropathogenic E. coli (EPEC), ipaH for enteroinvasive E. coli (EIEC), and aggR for enteroaggregative E. coli (EAEC), according to the manufacturer’s instructions (Shin et al., 2016). All isolates were serotyped by a slide agglutination test using E. coli antiserum for the O antigens (Denka Seiken, Tokyo, Japan).
Antimicrobial Susceptibility Testing and Identification of the β-Lactamase Gene
Antimicrobial susceptibility testing of the 66 E. coli isolates was performed using the broth microdilution method with customized Sensititre KRCDC1F panels (TREK Diagnostic Systems, East Grinstead, United Kingdom) in accordance with the guidelines established by the Clinical and Laboratory Standards Institute (CLSI). The antimicrobial agents tested were ampicillin, azithromycin, amoxicillin/clavulanic acid, cefoxitin, ceftazidime, ceftriaxone, cefotaxime, imipenem, gentamicin, amikacin, streptomycin, tetracycline, nalidixic acid, ciprofloxacin, trimethoprim/sulfamethoxazole, and chloramphenicol.
In the case of cefotaxime‐ or ceftriaxone-resistant isolates, the presence of ESBL genes was confirmed by multiplex PCR for TEM, SHV, CMY, OXA, DHA, and CTX-M types, and CTX-M-type genes were characterized by sequencing analysis of the amplicons (Kim et al., 2009).
Multilocus Sequence Typing
All isolates were assigned by multilocus sequence typing (MLST) as previously described (Wirth et al., 2006). Seven housekeeping genes (adkA, fumC, gyrB, icd, mdh, purA, and recA) were sequenced following the protocols specified on the E. coli MLST website.1 The primer sequences are available at http://enterobase.warwick.ac.uk/species/ecoli/download_7_gene. Seven different gene fragments of each isolate were assigned an allele number, and the sequence type (ST) was determined by each unique combination of seven allelic profiles.
Pulsed-Field Gel Electrophoresis
All E. coli isolates were analyzed by pulsed-field gel electrophoresis (PFGE) after XbaI digestion, according to the PulseNet International protocol.2 Fragments of XbaI-digested DNA were separated using a CHEF-Mapper system (Bio-Rad, Hercules, CA, United States) at 6 V/cm, with a linear increase in switching times from 2.16 to 54.17 s over 18 h at 14°C. Genetic similarities between the PFGE patterns were calculated with BioNumerics v7.6 (Applied-Maths, Sint-Martens-Latem, Belgium) using the Dice coefficient with a 1.5% band tolerance and the unweighted-pair group method using arithmetic averages (UPGMA).
Whole-Genome Sequencing
To determine genetic characteristics, representative isolates were selected according to their serotypes, antimicrobial resistance patterns, sequence types, PFGE results, and blaCTX-M genes. Genomic DNA was isolated using a Blood and Tissue kit (Qiagen, Stockach, Germany) according to the manufacturer’s protocol. DNA purity was quantified using a NanoDrop 2000 spectrophotometer (Thermo Fisher, DE, United States) and a Qubit 4 fluorometer using a high-sensitivity kit (Invitrogen, CA, United States). Short-read sequencing libraries were prepared with an Illumina Nextera Flex library preparation kit (Fonteneau et al., 2017). Sequencing was performed using a MiSeq sequencer (Illumina, San Diego, CA, United States) to generate 250-bp paired-end reads according to the manufacturer’s instructions. A long-read MinION sequencing library was prepared by using the ligation sequencing kit (SQK-LSK109) according to the manufacturer’s protocol for genomic DNA. Sequencing was carried out using a version R9.4.1 flow cell (FLO-MIN 106D).
Data Analysis and Molecular Characterization
The raw sequences generated by Illumina MiSeq were quality filtered using FastQC, with the average quality set at Q30. The contigs of genomic sequences were de novo assembled with a minimum contig size threshold of 200 bp using SPAdes assembler v3.9.0 (Abdalhamid et al., 2019). The raw data generated by the MinION instrument were processed, and base calling was performed using guppy software version 2.3.7; long-read assembly was performed using CLC genomics workbench 20.0.3. Subsequently, a hybrid de novo assembly of Illumina and Nanopore reads was performed using the long read support tool within the CLC genomic workbench 20.0.3. Genome annotations were performed using Rapid Annotation using Subsystem Technology (RAST). Assembled sequences were analyzed using bioinformatics web tools available from the Center for Genomic Epidemiology (CGE) website3 to detect resistance genes (ResFinder 3.2) and identify plasmid replicon types (PlasmidFinder 2.1). These whole-genome sequence data have been deposited in the National Center for Biotechnology Information (NCBI) under the Bio-Project PRJNA595397. The GenBank accession numbers are listed in Supplementary Table 1.
Mating Experiments and Plasmid Analysis
Strains exhibiting cefotaxime resistance were examined by conjugation experiments using azide-resistant E. coli J53 as the recipient strain to confirm the transmission capacity of blaCTX-M. Transconjugants were selected on MacConkey agar plates (Difco, United States) supplemented with cefotaxime (1 μg/L) and sodium azide (200 μg/L), and putative transconjugants were confirmed by antimicrobial susceptibility tests. In addition, transconjugants were selected according to their incompatibility group determined by PCR-based replicon typing (PBRT) and their acquisition of the blaCTX-M gene. Whole-genome DNA from transconjugants was digested with S1 endonuclease (Thermo Fisher Scientific, MA, United States) and incubated at 37°C for 30 min to estimate their plasmid sizes. DNA fragments were separated by PFGE through a CHEF-Mapper system for 14 h at 6 V/cm, with initial and final pulse times of 1 and 25 s.
Results
Bacterial Isolates
The RT-PCR results for 66 E. coli strains indicated that 45 carried the ST gene and were classified as ETEC (45, 68.2%); 21 strains carried the aggR gene and were confirmed as EAEC (21, 31.8%). A total of 44 ETEC strains were classified as O-type O25, and the serotype of one ETEC strain was O2; the other 21 strains were confirmed as O44. Additionally, we confirmed that six patients were co-infected with ETEC O25 and EAEC O44.
Antimicrobial Susceptibility Testing and ESBL Gene Analysis
All of the E. coli isolates were resistant to cefotaxime and ceftriaxone. In addition, the EAEC O44 isolates were resistant to tetracycline, chloramphenicol, azithromycin, and trimethoprim/sulfamethoxazole. The ETEC O25 isolates were resistant to azithromycin, and ETEC O2 isolates were resistant to tetracycline, chloramphenicol, and trimethoprim/sulfamethoxazole. The co-infection isolates from the six patients displayed the same resistance patterns as the ETEC O25 and EAEC O44 isolates (Table 1). PCR analysis showed that all EAEC O44 isolates harbored blaCTX-M-14 and that both ETEC O25 and O2 isolates carried blaCTX-M-15.
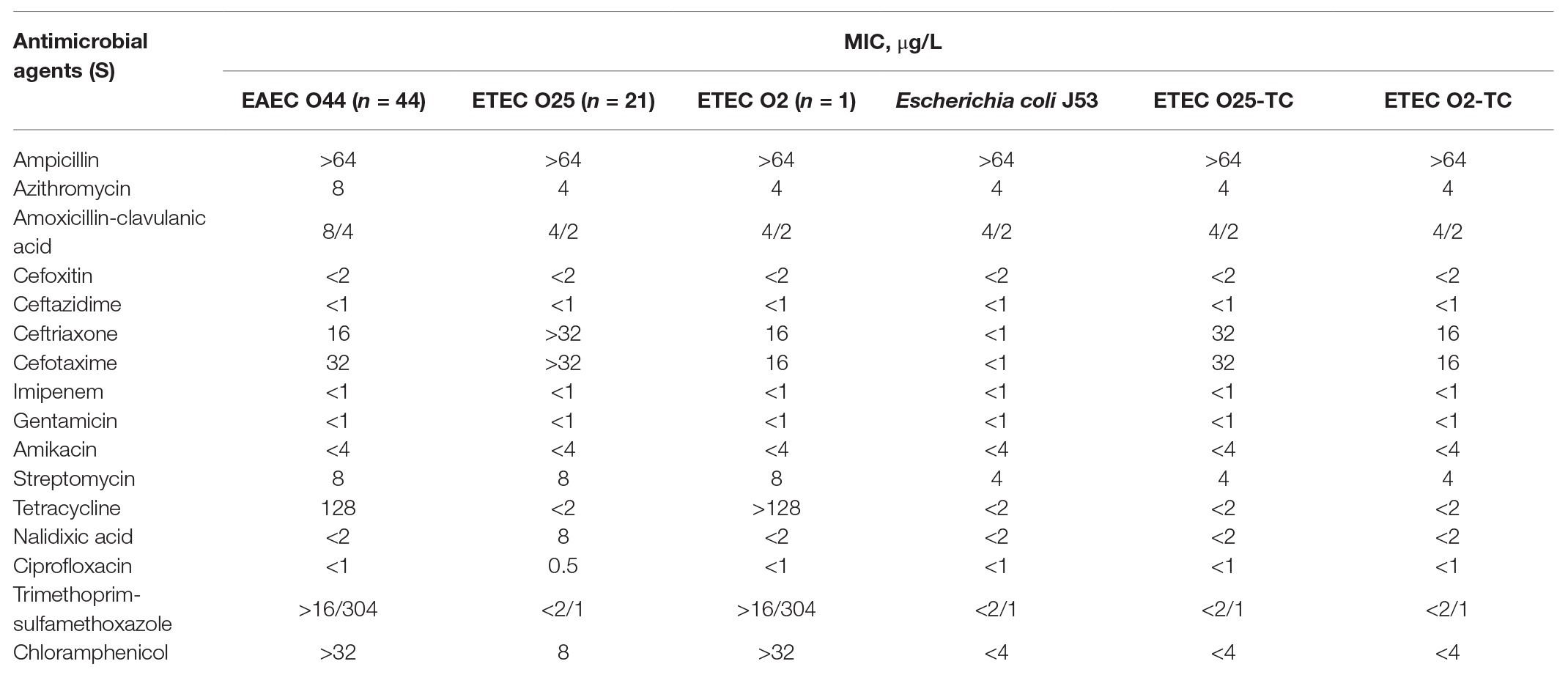
Table 1. Antimicrobial susceptibility profiles of EAEC O44, ETEC O25, and ETEC O2, representative strains of three school outbreaks, and the transconjugant strains ETEC O25-TC and ETEC O2-TC.
MLST and PFGE Analysis
The identified MLSTs of the EAEC O44, ETEC O25, and O2 strains were ST414, ST1492, and ST95, respectively. All ETEC O25 isolates displayed an identical XbaI digestion pattern (ETCX01.232), except for a strain with a different serotype, which was an ETEC O2 isolate exhibiting an ETCX01.231 XbaI digestion pattern (Figure 1). All of the EAEC O44 isolates showed the same XbaI digestion pattern: EACX01.213 (Figure 1). These three patterns were identified as new patterns that do not match any known PFGE cluster in the PulseNet Korea database of E. coli strains.
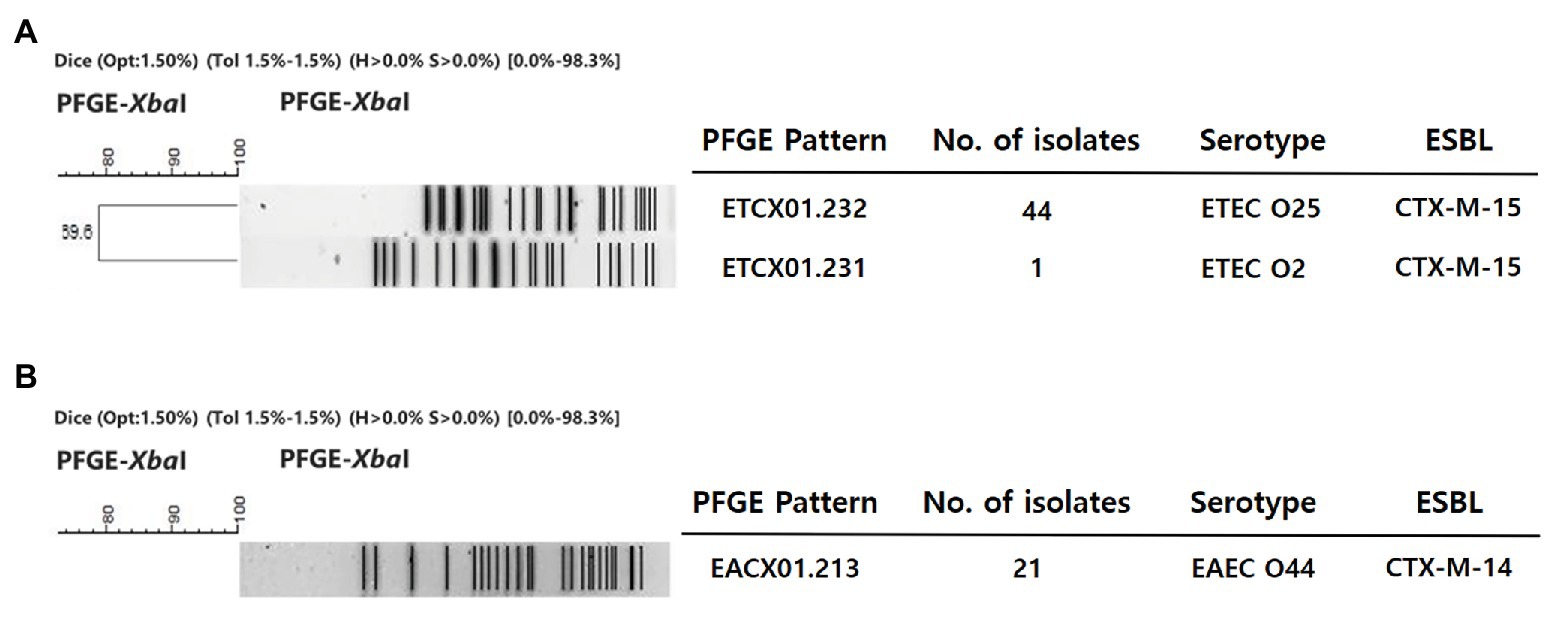
Figure 1. Dendrogram of the XbaI-pulsed-field gel electrophoresis (PFGE) patterns of enterotoxigenic E. coli (ETEC) and enteroaggregative E. coli (EAEC) strains isolated from the clinic. This dendrogram was constructed with BioNumerics v5.1 (Applied-Maths, Belgium) using the unweighted-pair group method with arithmetic means and a Dice coefficient (1.5% optimization and 1.5% position tolerance). (A) ETEC. (B) EAEC.
Genetic Profiles of E. coli Isolates
EAEC O44 and ETEC O25 co-infecting and single-infecting isolates were selected for further experiments and divided into two types according to serotypes, antimicrobial resistance patterns, sequence types, PFGE results, and blaCTX-M genes. Twelve isolates were chosen for the comparison of the genetic characteristics of co-infecting and single-infecting isolates: six EAEC O44 isolates and six ETEC O25 isolates, including eight co-infecting isolates from four patients.
To obtain a highly accurate closed complete genome sequence of the six EAEC O44 and six ETEC O25 strains, hybrid de novo assembly was performed using Nanopore long reads and Illumina short reads. The selected representative EAEC O44 and ETEC O25 isolates showed an identical genetic characteristic. The representative EAEC O44 strains were found to contain a 5.3-Mb circularized chromosome and three plasmids ranging from 90,954 to 99,334 bp. The blaCTX-M-14 genes mediating resistance in representative EAEC O44 isolates are located on the chromosome, and additional resistance genes, including for tetracyclines [tet(A)], phenicols (catA1), macrolides [mdf(A)], trimethoprim (dfrA5), and sulfonamides (sul1), are also present on this chromosome. Three plasmids were confirmed to be IncFIC(FII) plasmids with no resistance genes (Table 2).
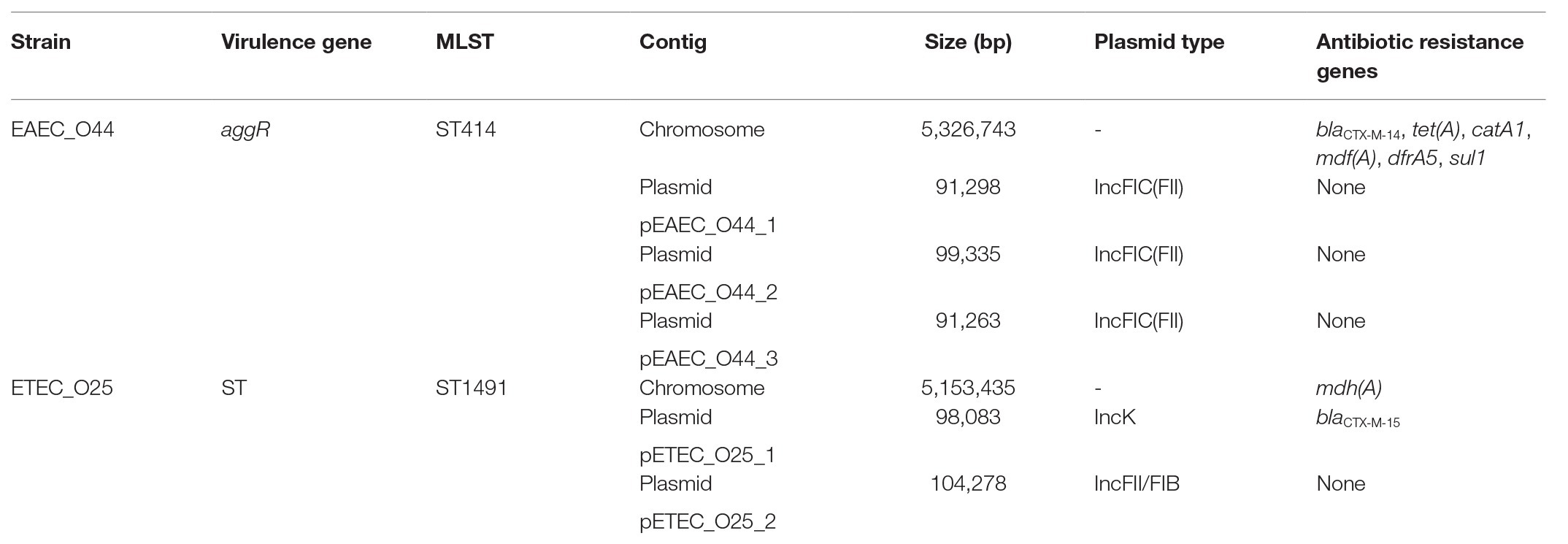
Table 2. Overview of antibiotic resistance genes, virulence genes and multilocus sequence typing (MLST) detected for representative EAEC O44 and ETEC O25 isolates associated with co-infection and single-infection.
The selected ETEC O25 strains exhibit a circular chromosome and two plasmids of 98,076 and 104,278 bp. Each plasmid type was confirmed to be IncK (repZ) and IncF (repFIB/repFII) based on the plasmid replicons by PlasmidFinder. The IncK replicon plasmid of a selected ETEC O25 isolate carries a resistance gene for β-lactams, blaCTX-M-15, at positions 9,141–10,016 bp; additional resistance genes, such as mdh(A) for macrolides, were detected on the chromosome. The IncF replicon plasmid carries two replicons genes (repFIB and repFII) in a same plasmid and no antibiotic resistance genes (Table 2).
Analysis of Regions Surrounding blaCTX-M
Combining long-read and short-read sequencing results indicated that the representative EAEC O44 genome is 5,326,743 bp and that the ISEcp1-blaCTX-M-14-IS903 transposon is located between 702,200 and 705,073 bp on the chromosome (Figure 2). The EAEC O44 isolates carry the ISEcp1-blaCTX-M-14-IS903 transposon (2,874 bp), in which ISEcp1 is located upstream of the start codon of the blaCTX-M-14 gene and the IS903 sequence in the downstream region (Figure 3).
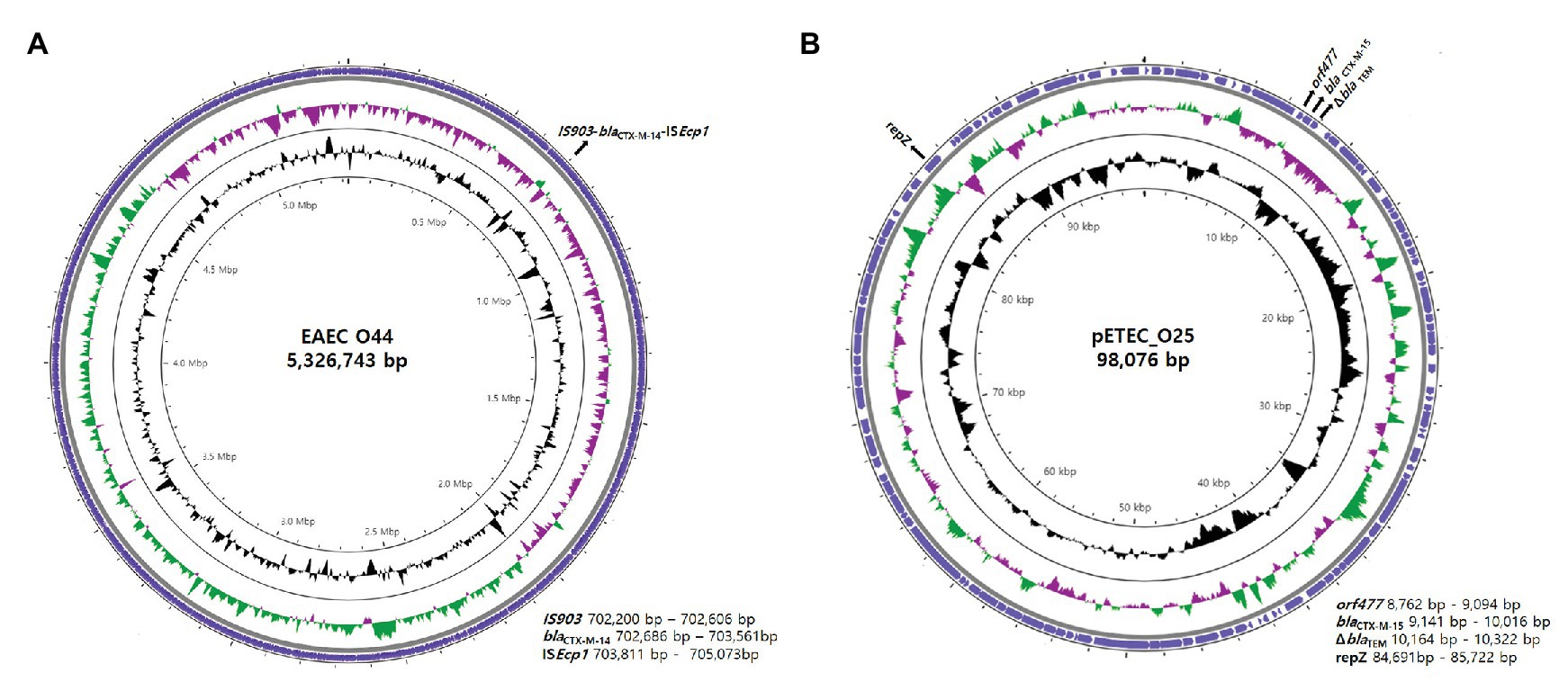
Figure 2. Genetic overview of EAEC O44 isolates carrying chromosomally located blaCTX-M-14 and ETEC O25 isolates harboring plasmid-encoded blaCTX-M-15. Circular genome map representation. The outermost ring represents the blast results for EAEC O44 and ETEC O25. G + C content, black peak; G + C positive skew, green peak; G + C negative skew, purple peak. (A) EAEC_O44: the EAEC O44 genome is 5,326,743 bp, and the ISEcp1-blaCTX-M-14-IS903 transposon (2,874 bp) is located between 702,200 and 705,073 bp on the chromosome. (B) pETEC_O25: pETEC_O25 is 98,076 bp in size, and the ΔblaTEM-blaCTX-M-15-orf477 transposon (1,540 bp) was found on the IncK plasmid between positions 8,762 and 10,322 bp.
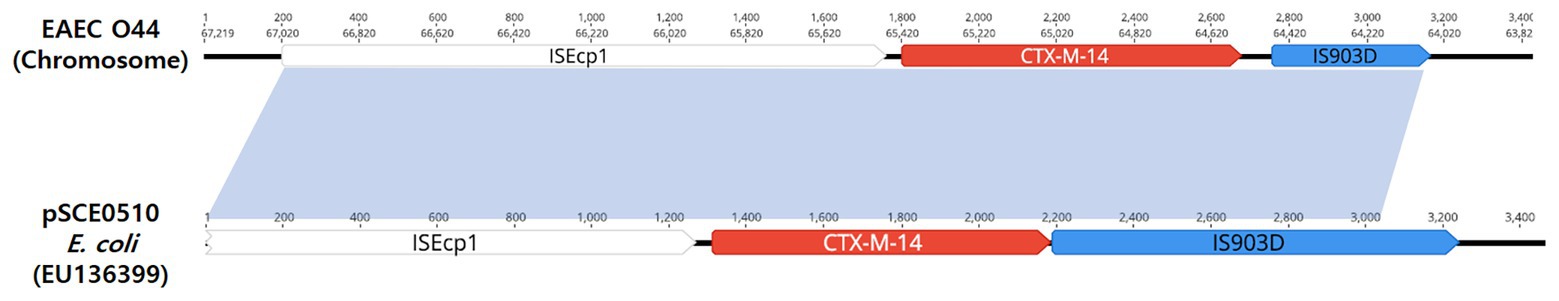
Figure 3. Schematic representation of the genetic environment surrounding the blaCTX-M-14 gene. The hatched box and the arrow indicate the genetic structure ISEcp1-blaCTX-M-14-IS903.
The results for selected ETEC O25 isolates indicated the transposon region (ΔblaTEM-blaCTX-M-15-orf477) to be located on the IncK plasmid between positions 8,762 and 10,322 bp (Figure 2).
The representative isolates of blaCTX-M-15-producing ETEC O25 include the ΔblaTEM-blaCTX-M-15-orf477 transposon (1,540 bp) as the genetic structure. Analysis of the region flanking blaCTX-M-15 revealed an orf477 sequence upstream and downstream of the blaCTX-M-15 gene, and the spacer region between the inverted repeat (IR) sequences of ISEcp1 and a truncated blaTEM gene was observed (Figure 4).
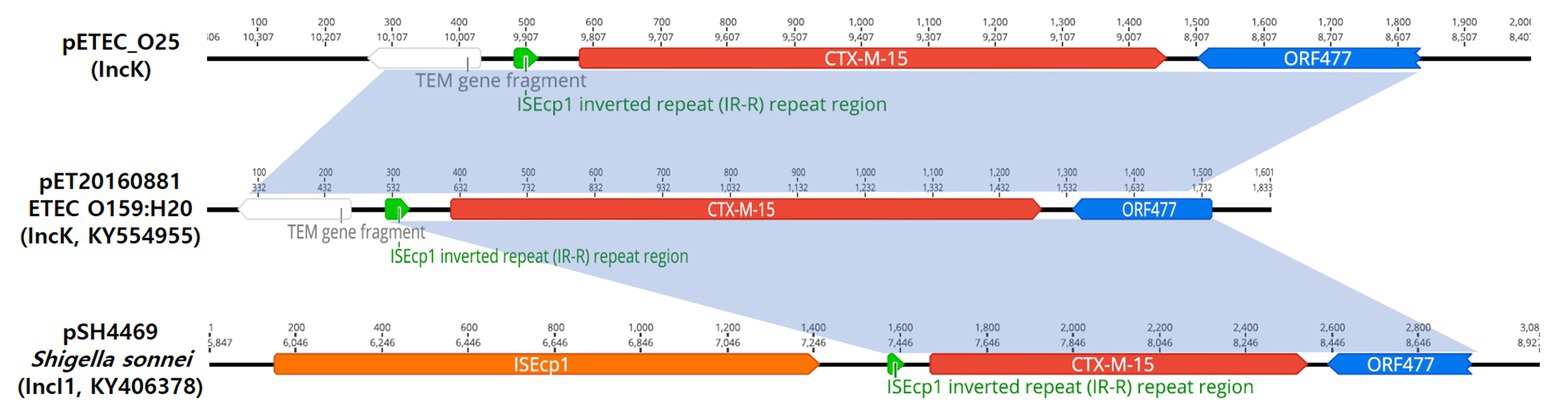
Figure 4. Schematic representation of the genetic environment surrounding the blaCTX-M-15 gene on three plasmids. The hatched box and the arrow indicate the inverted repeat (IR) of the ISEcp1 element and the transcriptional start site for the blaCTX-M-15 gene, respectively.
Transmission of the blaCTX-M Gene
Conjugation experiments were performed with all cefotaxime‐ and ceftriaxone-resistant isolates, which served as donors and E. coli J53 as the recipient. The results indicated that both ETEC O2 and ETEC O25 isolates were able to transfer resistance to recipient E. coli cells. According to the minimal inhibitory concentrations (MICs) of the antimicrobials, the ETEC transconjugants exhibited resistance to cefotaxime and ceftriaxone (Table 1). Additionally, PCR amplification and sequencing results confirmed that the blaCTX-M-15 gene was present in all of the ETEC transconjugants. Analysis of PBRT of all transconjugants demonstrated that the CTX-M-15-producing ETEC isolates carried two replicons of IncK and IncF (FII and FIB). These results supported by S1-PFGE analysis showed two plasmids ranging from 78 to 104 kb. Based on WGS analysis, two plasmids were present in the ETEC O25 strains, IncFII, FIB (104 kb), and IncK (98 kb), with blaCTX-M-15 located on the IncK plasmid. Conversely, the mating experiments with EAEC isolates failed to yield transconjugants.
Discussion
According to surveillance for foodborne and waterborne infection outbreaks during the period of 2013–2017 in the Republic of Korea, an estimated 7,600 cases occurred annually, among which 8.7% were associated with multiple pathogen outbreaks involving co-infection (Korea Centers for Disease Control & Prevention, 2013, 2014, 2015, 2016, 2017). In addition, several cases of co-infection with dual pathogens have been steadily reported in other countries (Wensley and Coole, 2013; Ahmed et al., 2014).
In this study, we describe co-infection with chromosomally located blaCTX-M-14‐ and plasmid-encoding blaCTX-M-15-producing pathogenic E. coli associated with three outbreaks. These outbreaks occurred at three schools located in distinct regions in the Republic of Korea. Based on an epidemiological investigation, the main cause of this outbreak was linked to the consumption of imported food (kimchi) supplied by the same food company. This assumption was supported by an outcome questionnaire that indicated that food (kimchi) was significantly related to illness. The results of laboratory tests suggested that these strains might have been introduced into the common source of infection. As follow-up actions in response to the described outbreak, the local public health laboratories launched an additional investigation of the food company. Unfortunately, however, food items contaminated by pathogenic E. coli were not identified in this investigation.
Interestingly, these outbreaks have been associated with EAEC O44 and ETEC O25 as co-infecting and single-infecting isolates. These isolates were divided into two types based on O serotype, antimicrobial resistance patterns, MLST, XbaI digestion patterns, and blaCTX-M genes. Additionally, by combining long-read and short-read sequencing results, we confirmed identical results for the selected representative co-infecting and single-infecting EAEC O44 and ETEC O25 isolates.
Each representative EAEC O44 and ETEC O25 strain was confirmed to produce the blaCTX-M-14 gene located on the chromosome and carry the blaCTX-M-15 gene on the IncK plasmid. Additionally, several patients were identified with a co-infection of both blaCTX-M-14-producing EAEC O44 and blaCTX-M-15-producing ETEC O25. The difference between the EAEC O44 and ETEC O25 strains was that the latter transmitted resistance via horizontal transmission between the chromosome and resident plasmids with a K replicon.
The prevalence of chromosomally located blaCTX-M-14 genes in clinical E. coli isolates from Europe and Asian countries has been observed in previous studies (Hawkey and Jones, 2009; Rodríguez et al., 2014; Hamamoto et al., 2016). Notably, chromosomal integration of blaCTX-M-14 genes in E. coli strains has been described in the Republic of Korea (Figure 3; Kim et al., 2011). These results support the hypothesis that the blaCTX-M-14 gene was chromosomally integrated into E. coli strains via transposable elements, such as ISEcp1-like elements (Hawkey and Jones, 2009). However, the phenomenon of dissemination of the chromosomal location of the blaCTX-M-14 gene in Enterobacteriaceae, including E. coli isolates, remains unclear, as has been the case in previous studies (Rodríguez et al., 2014; Hamamoto et al., 2016).
In contrast, the representative isolates of blaCTX-M-15-possessing ETEC O25 were found to harbor IncK plasmids. The blaCTX-M-15 gene is located between 9,141 and 10,016 bp on the plasmid, with an orf477 sequence downstream, but none of the transposable elements frequently associated with blaCTX-M were observed in the upstream region. The overall genetic structure of the selected ETEC O25 isolates (ΔblaTEM-blaCTX-M-15-orf477) has been described to be similar to that of pET20160881 (IncK, GenBank accession number KY554955; Figure 4) and pSH4469 (IncI1, GenBank accession number KY406378; Figure 4; Kim et al., 2014a, 2017). The plasmid backbone shares high identity (99.8 and 98.7%, respectively) with these plasmids. Interestingly, these plasmids were isolated from outbreaks in 2008 and 2016 in different regions of the Republic of Korea and share the same plasmid backbone and genetic environment as blaCTX-M-15.
We observed this transposable unit (blaCTX-M-15-orf477) to be present among the same or different bacterial species. These results suggest that the plasmid genetic structure may be used as a successful genetic vehicle for dissemination among the same bacterial species or even other species.
Similar cases have been reported to be associated with co-infection of ESBL-producing pathogenic E. coli in other countries, such as Bangladesh and China (Ahmed et al., 2014; Xu and He, 2019). These findings indicate that co-infecting pathogenic E. coli strains and Enterobacter cloacae or uropathogenic E. coli isolated from urinary tract infection (UTI) patients also produce ESBL genes.
To the best of our knowledge, this is the first known report of co-infection by two types of blaCTX-M-producing pathogenic E. coli from clinical isolates in the Republic of Korea. Our findings suggest that the primary transmission route of resistance genes in human isolates can occur through concurrent combinations of chromosomes and plasmids. The prevalence of blaCTX-M-carrying isolates might represent a serious problem for public health and may lead to compromised efficacy of widely used broad-spectrum cephalosporins for the treatment. To prevent the spread of antimicrobial resistance genes via chromosome and plasmid transmission, monitoring antimicrobial susceptibilities for pathogenic E. coli strains and maintaining measures such as standard precautions for contaminant sources are needed.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
This study uses strains obtained from local public health departments in the Republic of Korea. The Ethics committee of the first affiliated Korea Centers for Disease Control and Prevention decided that Institutional Review Board approval was not required, because patient information was collected anonymously and confidential patient information was not included.
Author Contributions
JP and JuK conceived of the study, and participated in its design and draft the manuscript. JiK, Y-HJ and NP collected samples and identified isolates. JP, ES, AP, SK, and HJ carried out the experiments and analyzed the data. J-hC, KH, and KL contributed to experiment conception. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by a grant from the Korea Centers for Disease Control and Prevention (4847-311-210/4845-300-210).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.545591/full#supplementary-material
Supplementary Table 1 | Information for the pathogenic E. coli isolates used in this study.
Footnotes
References
Abdalhamid, B., Mccutchen, E. L., Bouska, A. C., Weiwei, Z., Loeck, B., Hinrichs, S. H., et al. (2019). Whole genome sequencing to characterize Shiga toxin-producing Escherichia coli O26 in a public health setting. J. Infect. Public Health 12, 884–889. doi: 10.1016/j.jiph.2019.06.008
Ahmed, D., Wahid, S. U. H., Sadique, T., Sultana, N., Islam, N., Halim, F., et al. (2014). Recurrent urinary tract infection due to co-infection with extended spectrum β-lactamase-producer uropathogenic Escherichia coli and enteroaggregative E. coli. JMM Case Rep. 1, 1–5. doi: 10.1099/jmmcr.0.001404
Baker, S., Thomson, N., and Holt, K. E. (2018). Genomic insights into the emergence and spread of antimicrobial-resistant bacterial pathogens. Science 360, 733–738. doi: 10.1126/science.aar3777
Fonteneau, L., Da Silva, N. J., Fabre, L., Ashton, P., Torpdahl, M., Müller, L., et al. (2017). Multinational outbreak of travel-related Salmonella Chester infections in Europe, summers 2014 and 2015. Euro Surveill. 22, 1–11. doi: 10.2807/1560-7917.ES.2017.22.7.3046
Hamamoto, K., and Hirai, I. (2019). Characterisation of chromosomally-located blaCTX-M and its surrounding sequence in CTX-M-type extended-spectrum β-lactamase-producing Escherichia coli isolates. J. Glob. Antimicrob. Resist. 17, 53–57. doi: 10.1016/j.jgar.2018.11.006
Hamamoto, K., Ueda, S., Toyosato, T., Yamamoto, Y., and Hirai, I. (2016). High prevalence of chromosomal blaCTX-M-14 in Escherichia coli isolates possessing blaCTX-M-14. Antimicrob. Agents Chemother. 60, 2582–2584. doi: 10.1128/AAC.00108-16
Hawkey, P. M., and Jones, A. M. (2009). The changing epidemiology of resistance. J. Antimicrob. Chemother. 64, i3–i10. doi: 10.1093/jac/dkp256
Kim, J., Bae, I. K., Jeong, S. H., Chang, C. L., Lee, C. H., and Lee, K. (2011). Characterization of IncF plasmids carrying the blaCTX-M-14 gene in clinical isolates of Escherichia coli from Korea. J. Antimicrob. Chemother. 66, 1263–1268. doi: 10.1093/jac/dkr106
Kim, J., Jeon, S., Rhie, H., Lee, B., Park, M., Lee, H., et al. (2009). Rapid detection of extended spectrum β-lactamase (ESBL) for Enterobacteriaceae by use of a multiplex PCR-based method. Infect. Chemother. 41:181. doi: 10.3947/ic.2009.41.3.181
Kim, K. G., Jeong, J., Kim, M. J., Park, D. W., Shin, J. H., Park, H. J., et al. (2019). Prevalence and molecular epidemiology of ESBLs, plasmid-determined AmpC-type β-lactamases and carbapenemases among diarrhoeagenic Escherichia coli isolates from children in Gwangju, Korea: 2007-16. J. Antimicrob. Chemother. 74, 2181–2187. doi: 10.1093/jac/dkz175
Kim, J. S., Kim, J., Jeon, S. E., Kim, S. J., Kim, N. O., Hong, S., et al. (2014a). Complete nucleotide sequence of the IncI1 plasmid pSH4469 encoding CTX-M-15 extended-spectrum β-lactamase in a clinical isolate of Shigella sonnei from an outbreak in the Republic of Korea. Int. J. Antimicrob. Agents 44, 533–537. doi: 10.1016/j.ijantimicag.2014.08.007
Kim, J. S., Kim, J., Kim, S. J., Jeon, S. E., Oh, K. H., Cho, S. H., et al. (2014b). Characterization of CTX-M-type extended-spectrum beta-lactamase-producing diarrheagenic Escherichia coli isolates in the republic of Korea during 2008-2011. J. Microbiol. Biotechnol. 24, 421–426. doi: 10.4014/jmb.1401.01023
Kim, J., Lim, Y., Jeong, Y., and Seol, S. (2005). Enterobacteriaceae clinical isolates in Korea occurrence of CTX-M-3, CTX-M-15, CTX-M-14, in Enterobacteriaceae clinical isolates in Korea. Antimicrob. Agents Chemother. 49, 1572–1575. doi: 10.1128/AAC.49.4.1572
Kim, J. S., Park, J., Shin, E., Kim, S., and Oh, S. (2017). Outbreak of CTX-M-15-producing enterotoxigenic Escherichia coli O159:H20 in the Republic of Korea in 2016. Antimicrob. Agents Chemother. 61:e00339–17. doi: 10.1128/AAC.00339-17
Kim, S., Sung, J. Y., Cho, H. H., Kwon, K. C., and Koo, S. H. (2016). Characteristics of the molecular epidemiology of CTX-M-producing Escherichia coli isolated from a tertiary hospital in Daejeon, Korea. J. Microbiol. Biotechnol. 26, 1643–1649. doi: 10.4014/jmb.1603.03063
Korea Centers for Disease Control & Prevention (2013). Epidemiological Investigation of Infectious Diseases in Korea Annual Report 2013. Available at: http://www.cdc.go.kr/npt/biz/npp/portal/nppPblctDtaView.do?pblctDtaSeAt=5&pblctDtaSn=17 (Accessed October 22, 2020).
Korea Centers for Disease Control & Prevention (2014). Epidemiological Investigation of Infectious Diseases in Korea Annual Report 2014. Available at: http://www.cdc.go.kr/npt/biz/npp/portal/nppPblctDtaView.do?pblctDtaSeAt=5&pblctDtaSn=19 (Accessed October 22, 2020).
Korea Centers for Disease Control & Prevention (2015). Epidemiological Investigation of Infectious Diseases in Korea Annual Report 2015. Available at: http://www.cdc.go.kr/npt/biz/npp/portal/nppPblctDtaView.do?pblctDtaSeAt=5&pblctDtaSn=21 (Accessed October 22, 2020).
Korea Centers for Disease Control & Prevention (2016). Epidemiological Investigation of Infectious Diseases in Korea Annual Report 2016. Available at: http://www.cdc.go.kr/npt/biz/npp/portal/nppPblctDtaView.do?pblctDtaSeAt=5&pblctDtaSn=23 (Accessed October 22, 2020).
Korea Centers for Disease Control & Prevention (2017). Epidemiological Investigation of Infectious Diseases in Korea Annual Report 2017. Available at: http://www.cdc.go.kr/npt/biz/npp/portal/nppPblctDtaView.do?pblctDtaSeAt=5&pblctDtaSn=1010 (Accessed October 22, 2020).
Naseer, U., and Sundsfjord, A. (2011). The CTX-M conundrum: dissemination of plasmids and Escherichia coli clones. Microb. Drug Resist. 17, 83–97. doi: 10.1089/mdr.2010.0132
Oteo, J., Cercenado, E., Fernández-Romero, S., Saéz, D., Padilla, B., Zamora, E., et al. (2012). Extended-spectrum-β-lactamase-producing Escherichia coli as a cause of pediatric infections: report of a neonatal intensive care unit outbreak due to a CTX-M-14-producing strain. Antimicrob. Agents Chemother. 56, 54–58. doi: 10.1128/AAC.05103-11
Oteo, J., Pérez-Vázquez, M., and Campos, J. (2010). Extended-spectrum beta-lactamase producing Escherichia coli: changing epidemiology and clinical impact. Curr. Opin. Infect. Dis. 23, 320–326. doi: 10.1097/QCO.0b013e3283398dc1
Pai, H., Choi, E. -H., Lee, H. -J., Hong, J. Y., and Jacoby, G. A. (2001). Identification of CTX-M-14 extended-spectrum beta-lactamase in clinical isolates of Shigella sonnei, Escherichia coli, and Klebsiella pneumoniae in Korea. J. Clin. Microbiol. 39, 3747–3749. doi: 10.1128/JCM.39.10.3747-3749.2001
Rodríguez, I., Thomas, K., Van Essen, A., Schink, A. K., Day, M., Chattaway, M., et al. (2014). Chromosomal location of blaCTX-M genes in clinical isolates of Escherichia coli from Germany, the Netherlands and the UK. Int. J. Antimicrob. Agents 43, 553–557. doi: 10.1016/j.ijantimicag.2014.02.019
Shin, J., Choi, M. J., and Ko, K. S. (2012). Replicon sequence typing of IncF plasmids and the genetic environments of blaCTX-M-15 indicate multiple acquisitions of blaCTX-M-15 in Escherichia coli and Klebsiella pneumoniae isolates from South Korea. J. Antimicrob. Chemother. 67, 1853–1857. doi: 10.1093/jac/dks143
Shin, J., Kim, D. H., and Ko, K. S. (2011). Comparison of CTX-M-14‐ and CTX-M-15-producing Escherichia coli and Klebsiella pneumoniae isolates from patients with bacteremia. J. Infect. 63, 39–47. doi: 10.1016/j.jinf.2011.05.003
Shin, J., Oh, S. S., Oh, K. H., Park, J. H., Jang, E. J., Chung, G. T., et al. (2015). An outbreak of foodborne illness caused by enteroaggregative Escherichia coli in a high school in South Korea. Jpn. J. Infect. Dis. 68, 514–519. doi: 10.7883/yoken.JJID.2014.460
Shin, J., Yoon, K. -B., Jeon, D. -Y., Oh, S. -S., Oh, K. -H., Chung, G. T., et al. (2016). Consecutive outbreaks of enterotoxigenic Escherichia coli O6 in schools in South Korea caused by contamination of fermented vegetable Kimchi. Foodborne Pathog. Dis. 13, 535–543. doi: 10.1089/fpd.2016.2147
Wensley, A., and Coole, L. (2013). Cohort study of a dual-pathogen point source outbreak associated with the consumption of chicken liver pâté, UK, October 2009. J. Public Health 35, 585–589. doi: 10.1093/pubmed/fdt020
Wirth, T., Falush, D., Lan, R., Colles, F., Mensa, P., Wieler, L. H., et al. (2006). Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60, 1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x
Keywords: co-infection, CTX-M, pathogenic Escherichia coli, chromosomally-located blaCTX-M-14, plasmid-encoding blaCTX-M-15
Citation: Park J, Shin E, Park AK, Kim S, Jeong HJ, Kim JS, Jin Y-H, Park NJ, Chun J-h, Hwang K, Lee KJ and Kim J (2020) Co-infection With Chromosomally-Located blaCTX-M-14 and Plasmid-Encoding blaCTX-M-15 in Pathogenic Escherichia coli in the Republic of Korea. Front. Microbiol. 11:545591. doi: 10.3389/fmicb.2020.545591
Edited by:
Dongsheng Zhou, Beijing Institute of Microbiology and Epidemiology, ChinaReviewed by:
Yahong Liu, South China Agricultural University, ChinaJian Sun, South China Agricultural University, China
Copyright © 2020 Park, Shin, Park, Kim, Jeong, Kim, Jin, Park, Chun, Hwang, Lee and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junyoung Kim, jun49@hanmail.net; kjun49@naver.com
 Jungsun Park1
Jungsun Park1 Ae Kyung Park
Ae Kyung Park Jin Seok Kim
Jin Seok Kim Junyoung Kim
Junyoung Kim