- Department of Food Science and Biotechnology, Institute of Life Sciences and Resources, Kyung Hee University, Yongin, South Korea
Bifidobacterium species are used as probiotics to provide beneficial effects to humans. These effects are specific to some species or subspecies of Bifidobacterium. However, some Bifidobacterium species or subspecies are not distinguished because similarity of 16S rRNA and housekeeping gene sequences within Bifidobacterium species is very high. In this study, we developed a real-time polymerase chain reaction (PCR) assay to rapidly and accurately detect 22 Bifidobacterium species by selecting genetic markers using comparative genomic analysis. A total of 210 Bifidobacterium genome sequences were compared to select species- or subspecies-specific genetic markers. A phylogenetic tree based on pan-genomes generated clusters according to Bifidobacterium species or subspecies except that two strains were not grouped with their subspecies. Based on pan-genomes constructed, species- or subspecies-specific genetic markers were selected. The specificity of these markers was confirmed by aligning these genes against 210 genome sequences. Real-time PCR could detect 22 Bifidobacterium specifically. We constructed the criterion for quantification by standard curves. To further test the developed assay for commercial food products, we monitored 26 probiotic products and 7 dairy products. Real-time PCR results and labeling data were then compared. Most of these products (21/33, 63.6%) were consistent with their label claims. Some products labeled at species level only can be detected up to subspecies level through our developed assay.
Introduction
Probiotics are living microorganisms that provide health benefits such as improving digestive health and preventing infectious diarrhea, irritable bowel syndrome, and inflammatory bowel disease of hosts (O’Callaghan and van Sinderen, 2016; Floch, 2018; Shehata et al., 2019). Health benefits of probiotics are species- or strain- specific. Not all lactic acid bacteria are considered as probiotics (Pinto-Sanchez et al., 2017). Bifidobacterium is one important member of probiotics that has benefits such as anti-cancer effects (Inoue et al., 2009) and reducing cholesterol level (Zhang et al., 2016) for the host. Bifidobacterium is Gram-positive, non-motile, and catalase-negative lactic acid bacterium that survives in the intestine of human. Bifidobacterium species in human gut microbiota include Bifidobacterium longum, Bifidobacterium bifidum, Bifidobacterium breve, Bifidobacterium adolescentis, Bifidobacterium dentium, and Bifidobacterium pseudocatenulatum (Junick and Blaut, 2012). Bifidobacterium pseudolongum and Bifidobacterium thermophilum previously considered to be of animal origin have been isolated from baby feces and human adults, respectively (von Ah et al., 2007; Turroni et al., 2009; Junick and Blaut, 2012).
Bifidobacterium species have universally different functions according to subspecies. For instance, Bifidobacterium animalis subsp. lactis has a strong anti-inflammatory effect to improve the immune system (Weizman et al., 2005), whereas B. animalis subsp. animalis cannot grow in milk (Masco et al., 2004). B. longum also has different types of glycolytic enzymes according to its subspecies (LoCascio et al., 2010; Lewis et al., 2016). Hence, differentiating Bifidobacterium subspecies is necessary. Furthermore, presenting correct species in probiotic products is critical for providing correct information to consumers and claiming health benefits of the product (Shehata et al., 2019). Recently, some studies have shown mislabeling issues such as absence of some species, inaccurate taxonomy information, and undeclared species (Lewis et al., 2016; Morovic et al., 2016) of commercial probiotic products. However, there is no reliable detection method to distinguish different species and subspecies of Bifidobacterium.
Polymerase chain reaction (PCR)-based methods have been widely used to detect bacterial strains in probiotics, dairy products, meat products, and seafood (Binetti et al., 2008; Cammà et al., 2012; Kim et al., 2019). In particular, the 16S rRNA gene has been used as a useful target gene for bacterial identification. However, the resolution of this gene among closely related species is low (Junick and Blaut, 2012). To differentiate Bifidobacterium species, more distinguishable identification markers need to be found because 16S rRNA genes of Bifidobacterium species share high similarities (mean, 95%) (Ventura et al., 2006; Junick and Blaut, 2012). Housekeeping genes such as recA (Ventura and Zink, 2003), tuf (Ventura and Zink, 2003), atpD (Ventura et al., 2004a), groEL (Zhu et al., 2003), and groES (Ventura et al., 2004b) have been used as alternative genetic markers for the discrimination of Bifidobacterium. Although these genes have been demonstrated to have a relatively higher resolution than 16S rRNA gene, similar species and subspecies are still indistinguishable. Thus, those genes can only be applied to limited species (Lawley et al., 2017).
Whole-genome sequencing (WGS) is a powerful method for identifying unique genes through bioinformatics (Chen et al., 2010; Mellmann et al., 2017). Comparative genomics has been performed for pathogenic bacteria and lactic acid bacteria using various algorithms (Lugli et al., 2017; Zhang et al., 2019). But, studies on development of specific primers of probiotic species based on comparative genomics have not been widely conducted. The objective of the present study was to develop a real-time PCR assay using comparative genomics known to be able to detect highly specific genetic markers and bacterial strains very quickly. A brief description of the method is as follows: specific genetic markers were selected using comparative genomics from 210 Bifidobacterium genomes, and species- or subspecies-specific primers were designed based on identified markers. Real-time PCR assay was then applied for quantitative identification of 22 Bifidobacterium species, which is mainly found in intestine of human and food samples such as probiotic or dairy products and difficult to differentiate by conventional methods. Furthermore, label claims of commercial probiotics and dairy products were verified using the developed real-time PCR assay.
Materials and Equipment
Bacterial Strains
Forty-one Bifidobacterium species or subspecies strains, 11 Lactobacillus species, 1 Lactococcus species, and 2 Enterococcus species obtained from Korean Agricultural Culture Collection (KACC, Jeonju, South Korea), Korean Collection for Type Cultures (KCTC, Daejeon, South Korea), and Korean Culture Center of Microorganisms (KCCM, Seoul, South Korea) were used to confirm the specificity of the developed real-time PCR (Table 1).
Equipment and Software
Anvi’o, Bacterial Pan Genome Analysis pipeline (BPGA), USEARCH, and Basic Local Alignment Search Tool (BLAST) software were used for comparative genomics to select specific genetic genes for Bifidobacterium species or subspecies. 7500 Real-time PCR System (Applied Biosystems, Foster City, CA, United States) and 7500 software were used for the specificity and accuracy of species- or subspecies-specific primers.
Methods
Cultivation and Genomic DNA Extraction of Bifidobacterium Strains
Bifidobacterium strains were cultured in Bifidobacterium broth (MB cell, Seoul, South Korea) and BL broth (MB cell, Seoul, South Korea) at 37°C for 48 h under anaerobic condition. Other lactic acid bacterial strains were cultured in MRS broth (Difco, Becton Dickinson, Sparks, MD, United States) at 30°C for 48 h under anaerobic condition (Kim et al., 2020). All strains were stored in 30% (v/v) glycerol (Bioshop, Burlington, ON, Canada) at −80°C until use. To extract genomic DNA, all cultured bacterial cells were collected by centrifugation at 16,200 × g for 3 min. DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany) was used to extract total genomic DNAs from all strains following the manufacturer’s protocol for Gram-positive bacteria. Purity and concentration of extracted bacterial DNA were measured using a MaestroNano® spectrophotometer (Maestrogen, Las Vegas, NV, United States).
Genomic DNA Extraction of Commercial Products
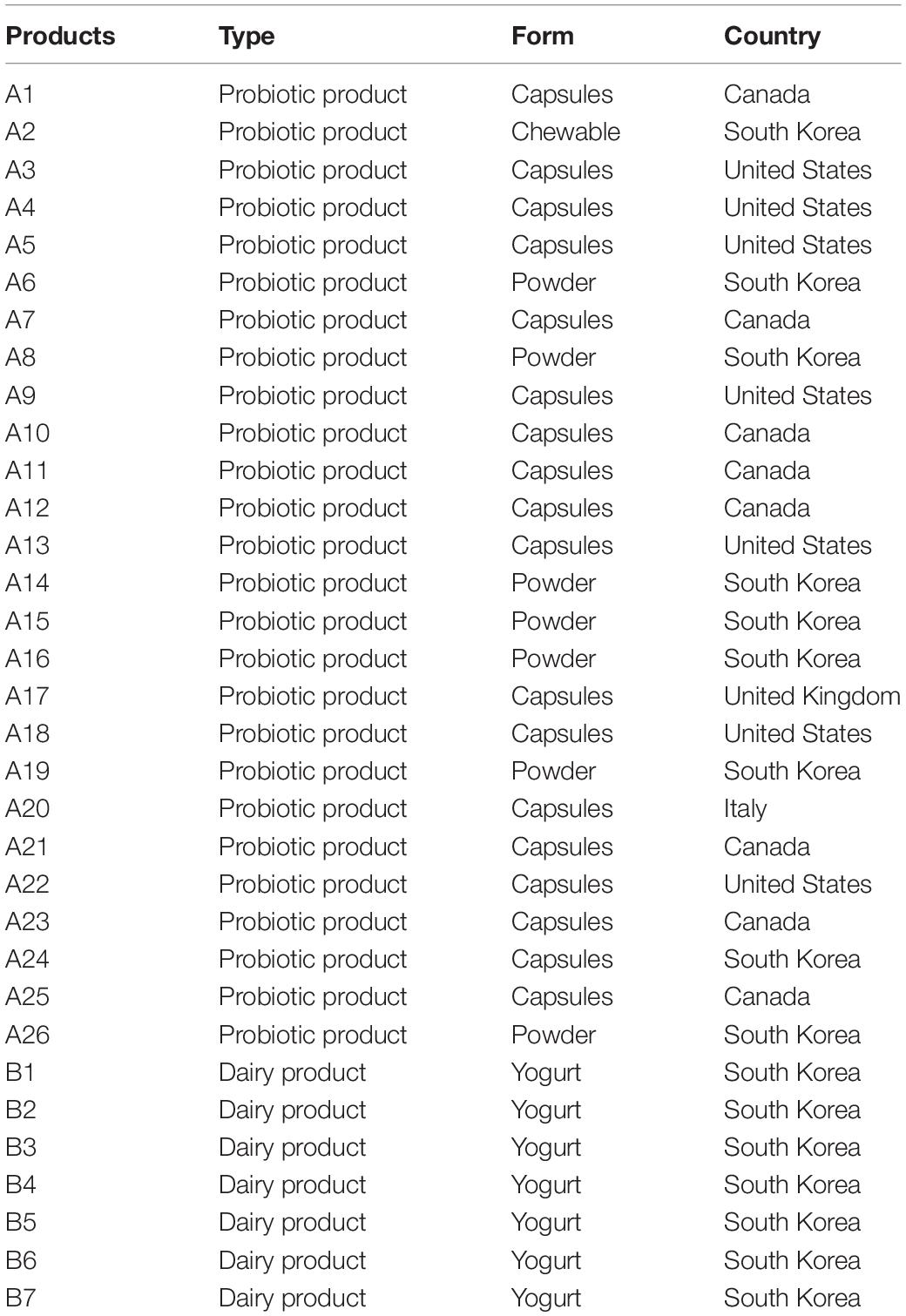
Commercial products used in this study are listed in Table 2. These products were classified from A1 to A26 for probiotic products and from B1 to B7 for dairy products. Twenty-six probiotic products and 7 dairy products were purchased from markets worldwide (South Korea: 16, United States: 7, Canada: 8, United Kingdom: 1, Italy: 1). These probiotic products included 18 capsules, 7 powders, and 1 chewable. Total genomic DNAs of probiotic products were extracted according to a previous study (Kim et al., 2017). One-hundred milligrams of probiotic product were aliquoted and dissolved in 300 μL of lysis buffer following the manufacture’s instruction (DNeasy Blood and Tissue kit, Qiagen). Purity and concentration of extracted probiotic DNAs were measured as previously mentioned.
Comparative Genomic Analysis of Bifidobacterium Species or Subspecies
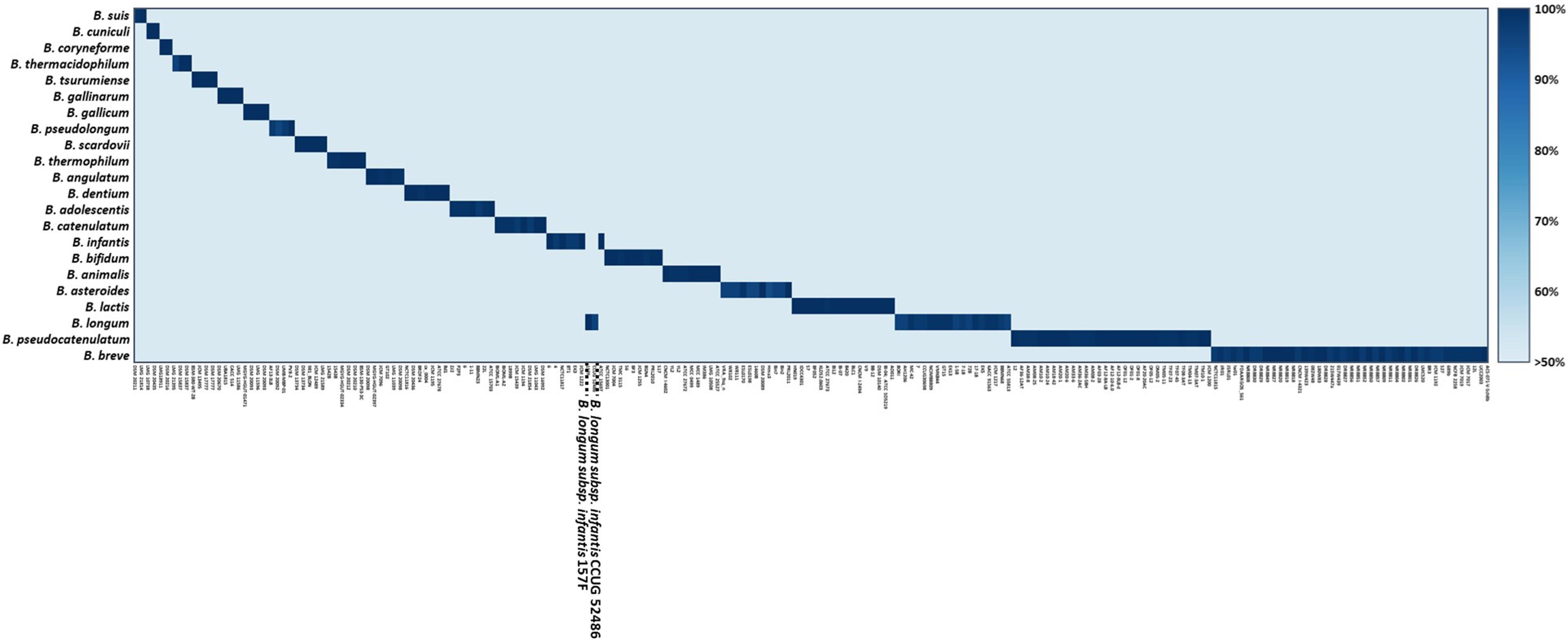
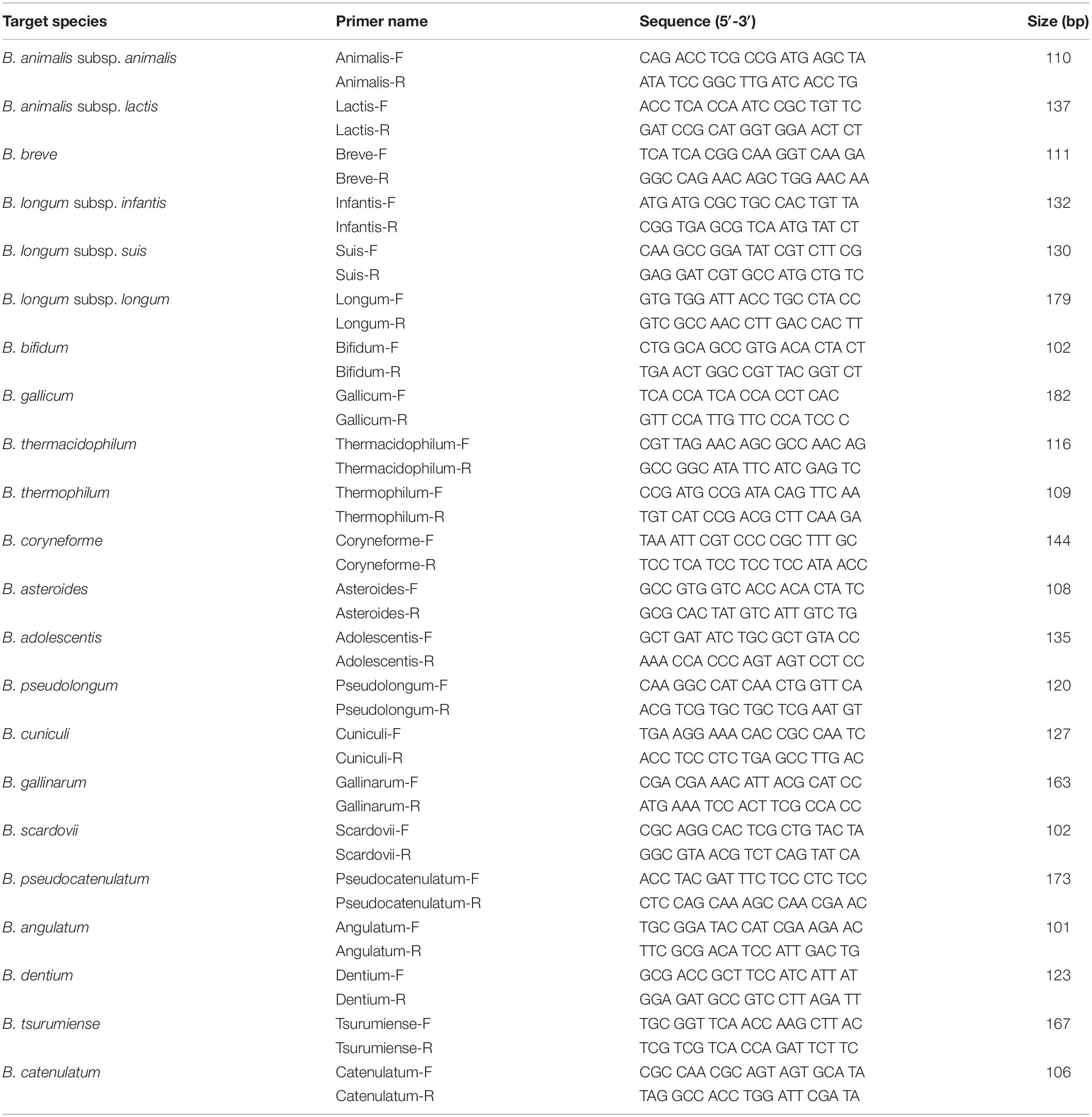
All Bifidobacterium genome sequences were downloaded from the National Center for Biotechnology Information (NCBI)1, including 110 complete genomes, 52 scaffolds, and 31 contigs (Supplementary Table S1). To avoid drawing incorrect conclusions from the genomic analysis due to mislabeled genomes, a total of 210 Bifidobacterium genomes were evaluated using phylogenetic trees based on pan and core genes. A phylogenetic tree based on the pan-genome was constructed using Anvi’o version 6.0 publically available software according to the workflow for pan-genomics (Eren et al., 2015). Genome sequences obtained from NCBI were stored in Anvi’o storage for genomes to build a genome database. Pan-genome analysis was performed with the genome database. A phylogenetic tree was constructed according to pan gene cluster frequencies. Also, a phylogenetic tree based on core genes was constructed using BPGA version 1.3. The core genes were aligned using MUSCLE in BPGA, and a neighbor-joining phylogenetic tree was constructed. To select Bifidobacterium species- or subspecies-specific genetic markers, the core genome common to each species or subspecies was constructed. Core genomes were then compared to explore candidate genetic markers using BPGA version 1.3 with default identity value (Chaudhari et al., 2016). Final candidates for species- or subspecies-specific genetic markers were verified using BLAST against 57,122,612 sequences, including sequences of other lactic acid bacteria. Then 22 genetic markers and 210 genome sequences were aligned with UBLAST algorithm with USEARCH version 9.0 (Edgar, 2010). The alignment of genetic markers to genomes is shown in a heatmap (Figure 2). Also, the presence/absence of genes is easily skewed when the selected genetic marker is variable, so for all genetic markers their locations were verified, such as whether they are located in prophage genomes and plasmids or are really part of the core genome of that species using PlasmidFinder version 2.1 and BLAST analysis. Species- and subspecies-specific primers were designed based on selected genetic markers using Primer Designer (Scientific and Education Software, Chapel Hill, NC, United States). All 22 primer pairs were developed to be less than 200 bp in size to increase the amplification efficiency (Kim et al., 2020) suitable for the application of processed food products. All primers were synthesized by Bionics (Seoul, South Korea).
Specificity and Standard Quantification Using Real-Time PCR Assay
To confirm the specificity of designed primers, real-time PCR was performed for 41 Bifidobacterium strains and 14 non-Bifidobacterium strains using a 7500 Real-time PCR System (Applied Biosystems, Foster City, CA, United States). The reaction mixture consisted of 10 μL of 2 × Thunderbird SYBR® qPCR Mix (Toyobo, Osaka, Japan), 20 ng of template DNA, 0.5 μM of each primer pair, and deionized-distilled water to have a total volume of 20 μL. Each target was amplified with the following conditions: initiation at 95°C for 2 min for one single step, followed by 35 cycles at 95°C for 5 s and 60°C for 30 s. A melting curve was generated in the range of 95°C for 15 s, 60°C for 1 min, 95°C for 30 s, and 60°C for 15 s. Standard curve was obtained according to previously reported methods (Gómez-Rojo et al., 2015; Kim et al., 2020). Briefly, Bifidobacterium strains at 8 × 105 to 8 × 109 CFU/mL as determined by plate counting on Bifidobacterium agar (MB cell, Seoul, South Korea) were subjected to DNA extraction. Amplification was repeated three times. Standard curves for quantification were obtained by plotting Ct values against the number of bacteria per reaction (log CFU/mL). Results of real-time PCR assay were evaluated using 7500 software v2.0.6 (Applied Biosystems).
Application of the Developed Real-Time PCR Assay for Probiotic Products
Probiotic products were monitored to detect 22 Bifidobacterium species or subspecies using the real-time PCR developed in this study (Supplementary Figure S1). For the application, 20 ng of DNA from each probiotic product was added to each well of a 96-well plate containing 2 × Thunderbird SYBR® qPCR Mix (Toyobo, Osaka, Japan) and the species- or subspecies-specific primers. Real-time PCR was performed using a 7500 Real-Time PCR system (Applied Biosystems). PCR conditions were the same as indicated above in the “Specificity and Standard Quantification using Real-Time PCR Assay” section.
Results
Comparative Genomic Analysis of Bifidobacterium
Species- or subspecies-specific genetic markers were selected using comparative genomic analysis for 210 Bifidobacterium genomes. Candidate genetic markers for targets were selected by comparing core genomes with non-target pan-genome. To select specific genetic markers for the target, candidate genetic markers were blasted against 57,122,612 sequences, including sequences of other lactic acid bacteria. A phylogenetic tree was constructed based on the pan-genome for Bifidobacterium. Phylogeny showed that most genomes (n = 208) shared the same lineage according to their species or subspecies type (Figure 1). In contrast, B. longum subsp. infantis CCUG 52486 and 157F were more closely related to B. longum subsp. longum group than to B. longum subsp. infantis. The phylogenetic tree constructed by core genomes also showed the same clusters, where these two B. longum subsp. infantis genomes were clustered into B. longum subsp. longum (Supplementary Figure S2).
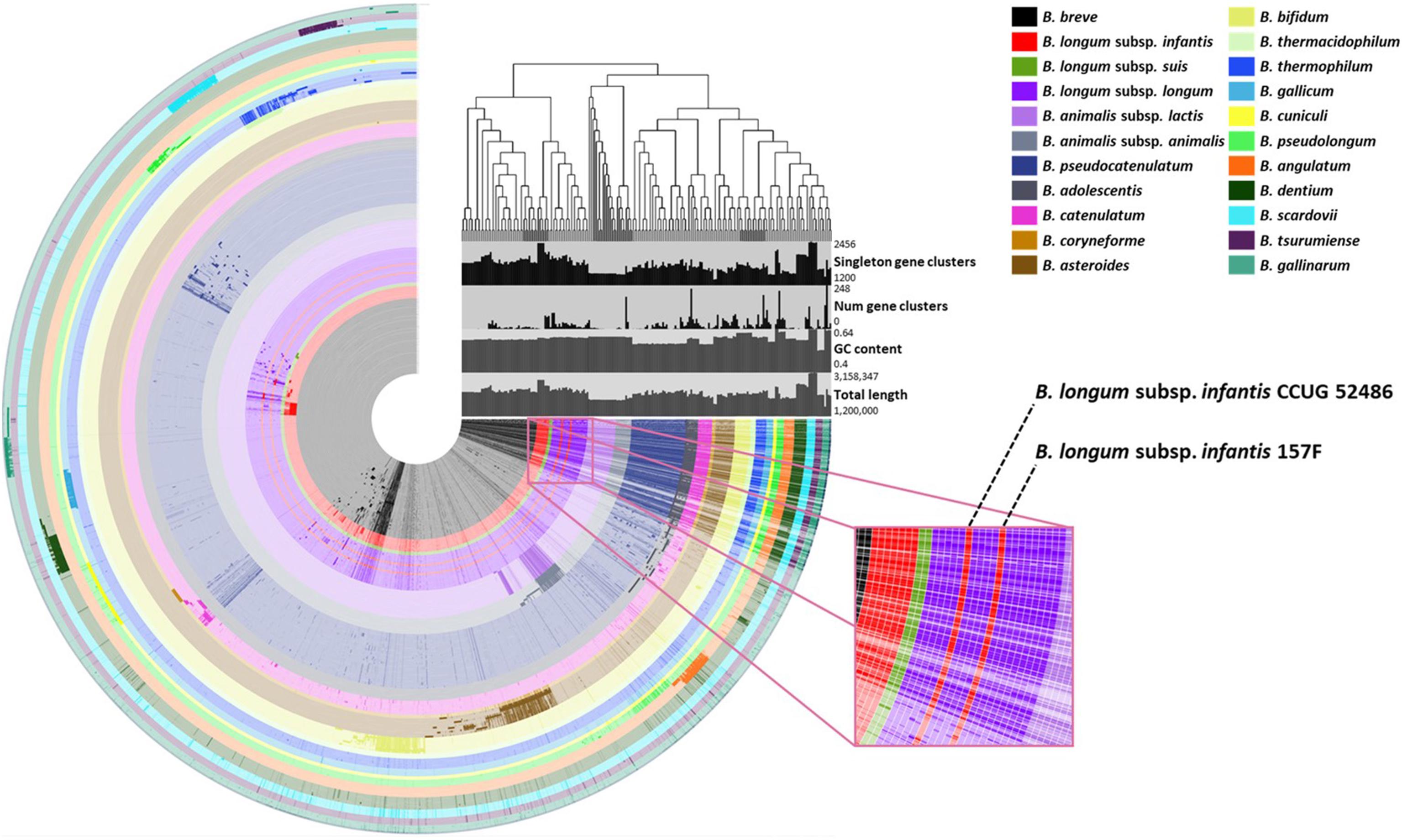
Figure 1. Pan-genomic phylogenetic tree of the Bifidobacterium. The figure shows that each ring represents Bifidobacterium genome and each layer displays the pan-genome distribution. The dark and bright colors of each ring indicate the presence and absence of core genes, respectively.
A total of 372,743 genes yielded a pan-genome of 21,669 genes. The core genome had 250 genes. The accessory genome had 15,429 genes. The unique genome had 7,170 genes. The unique genome was divided into genetic markers common to the same species or subspecies. The specificity of identified genetic markers was confirmed by BLAST. Most of these genomes (208/210, 99.05%) shared 90–100% sequence identities within genetic markers of the same species or subspecies and 0–50% sequence identities against other species. Information of these genes is shown in Table 3. These identified genetic markers shared more than 90% sequence identities against each target genome except two B. longum subsp. infantis strains. These two strains were classified into B. longum subsp. longum according to our pan-genome analysis (Figure 2). The genetic marker for B. longum subsp. infantis such as ABC transporter permease (accession no. WP012576966.1) was present in 7 out of 9 strains (except CCUG 52486 and 157F). Instead, B. longum subsp. infantis CCUG 52486 and 157F had a bacterial Ig-like domain-containing protein (WP013141462.1), a genetic marker for B. longum subsp. longum. We confirmed that in these two B. longum subsp. infantis genomes, the genetic marker of B. longum subsp. longum was not present in their plasmids but on the chromosome, by blasting the contigs against the reported plasmid sequences. As well as, all genetic markers identified in this study were not located in plasmids or phage proteins and present in chromosome, meaning that these genetic markers are not variable and are part of the core genome. Based on these results, species- or subspecies-specific primers were designed and used for further studies (Table 4).
Specificity and Quantification of the Developed Real-Time PCR Assay
The specificity of the developed real-time PCR assay was confirmed with 41 Bifidobacterium strains and 14 non-Bifidobacterium strains. As a result, all primer sets specific for each Bifidobacterium species/subspecies in silico showed detectable amplicons, with Ct values between 11 and 16 against target strains, whereas those from all non-targets did not generate any positive signal (Figure 3 and Supplementary Table S2). To quantify the number of bacteria and to confirm the accuracy of real-time PCR, a standard curve was obtained using template DNA of Bifidobacterium at a range of 8 × 105 to 8 × 109 CFU/mL in triplicates. This range included the number of bacteria labeled on probiotic products used. Slope for standard curves of B. animalis subsp. lactis, B. bifidum, B. breve, and B. longum subsp. infantis mainly used in probiotic products were −3.499, −3.134, −3.275, and −3.552, respectively. All R2 values (correlation coefficients) were ≥ 0.997 (Figure 4). Results of the slope, R2 value, and efficiency of remaining primers are shown in Supplementary Table S3. According to the efficiency of quantitative real-time PCR, R2 values ≥ 0.98 are considered as reliable (Broeders et al., 2014). Thus, the real-time PCR developed in this study was confirmed to be highly accurate and efficient.

Figure 3. Specificity of species- and subspecies-specific primer pairs which were mainly used in probiotic products against 41 strains. (A) Specificity of B. animalis subsp. lactis specific primer pair, amplification curve: B. animalis subsp. lactis KACC 16638, LI 001941, LI 001942, LI 000026, LI 000004, LI 000019, and LI 000062; (B) Specificity of Bifidobacterium bifidum specific primer pair, amplification curve: B. bifidum KCTC 3418, KCTC 3440, LI 000058, LI 000061, and LI 000063; (C) Specificity of Bifidobacterium breve specific primer pair, amplification curve: B. breve KACC 16639, KCTC 3419, and LI 000070; (D) Specificity of B. longum subsp. infantis specific primer pair, amplification curve: B. longum subsp. infantis KCTC 3249, LI 000033, LI 000261, and LI 000262.
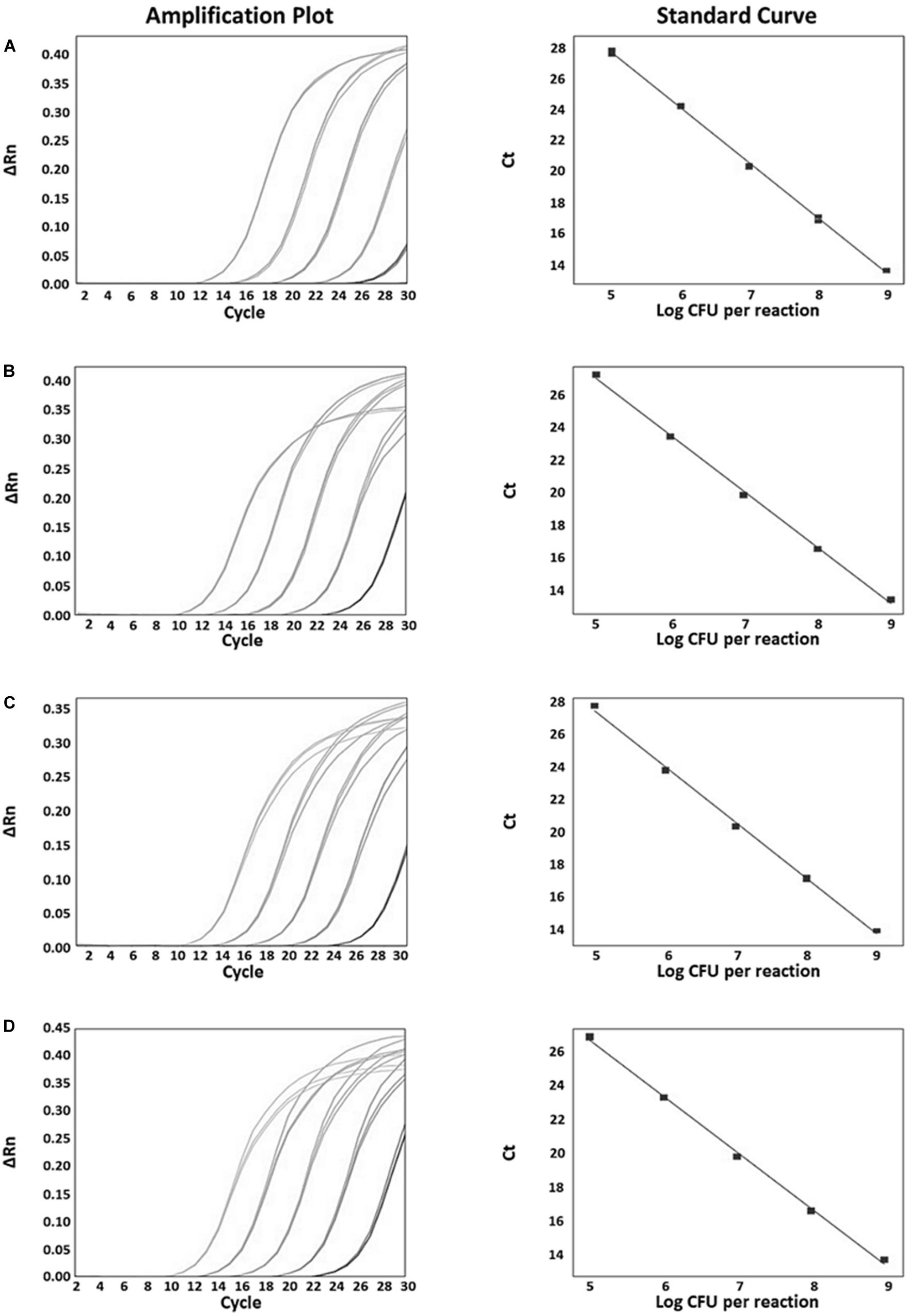
Figure 4. Real-time PCR amplification plots and standard curves which were mainly used in probiotic products for quantitative evaluation. (A) B. animalis subsp. lactis amplification plot (left), standard curve between 20 and 0.002 ng (y = –3.564x + 18.03, R2 = 0.998, Eff% = 90.788, right); (B) B. bifidum amplification plot (left), standard curve (y = –3.438x + 17.713, R2 = 0.998, Eff% = 95.359, right); (C) B. breve amplification plot (left), standard curve (y = –3.448x + 18.169, R2 = 0.998, Eff% = 94.987, right); (D) B. longum subsp. infantis amplification plot (left), standard curve (y = –3.312x + 17.727, R2 = 0.998, Eff% = 100.424, right).
Monitoring of Probiotic and Dairy Products Using the Real-Time PCR Developed
Commercially available probiotic and dairy products were used to verify whether the real-time PCR developed in this study could be applicable to quantify and identify probiotics in food products (Supplementary Figure S1). A total of 33 commercial probiotic and dairy products containing Bifidobacterium were monitored. Obtained results were compared with product label claims. Results of 21 products were identical to their label claims. In particular, probiotic strains of eight products that were only labeled at the species level such as B. longum and B. animalis were able to be analyzed up to subspecies level using our real-time PCR assay (Table 5). For the remaining four products (B4 to B7) labeled as “Lactic acid bacteria or Bifidus,” this real-time PCR assay was able to detect Bifidobacterium at the subspecies level. Based on the standard quantitative curve for each Bifidobacterium species or subspecies obtained by plotting Ct values against the number of bacteria per reaction, the number of Bifidobacterium species or subspecies present in the food products was estimated to be within the range of 8 × 105 to 8 × 109 CFU/mL. Thus, the real-time PCR method developed in this study could accurately detect and quantify Bifidobacterium strains contained in probiotic and dairy products at species level and subspecies level.
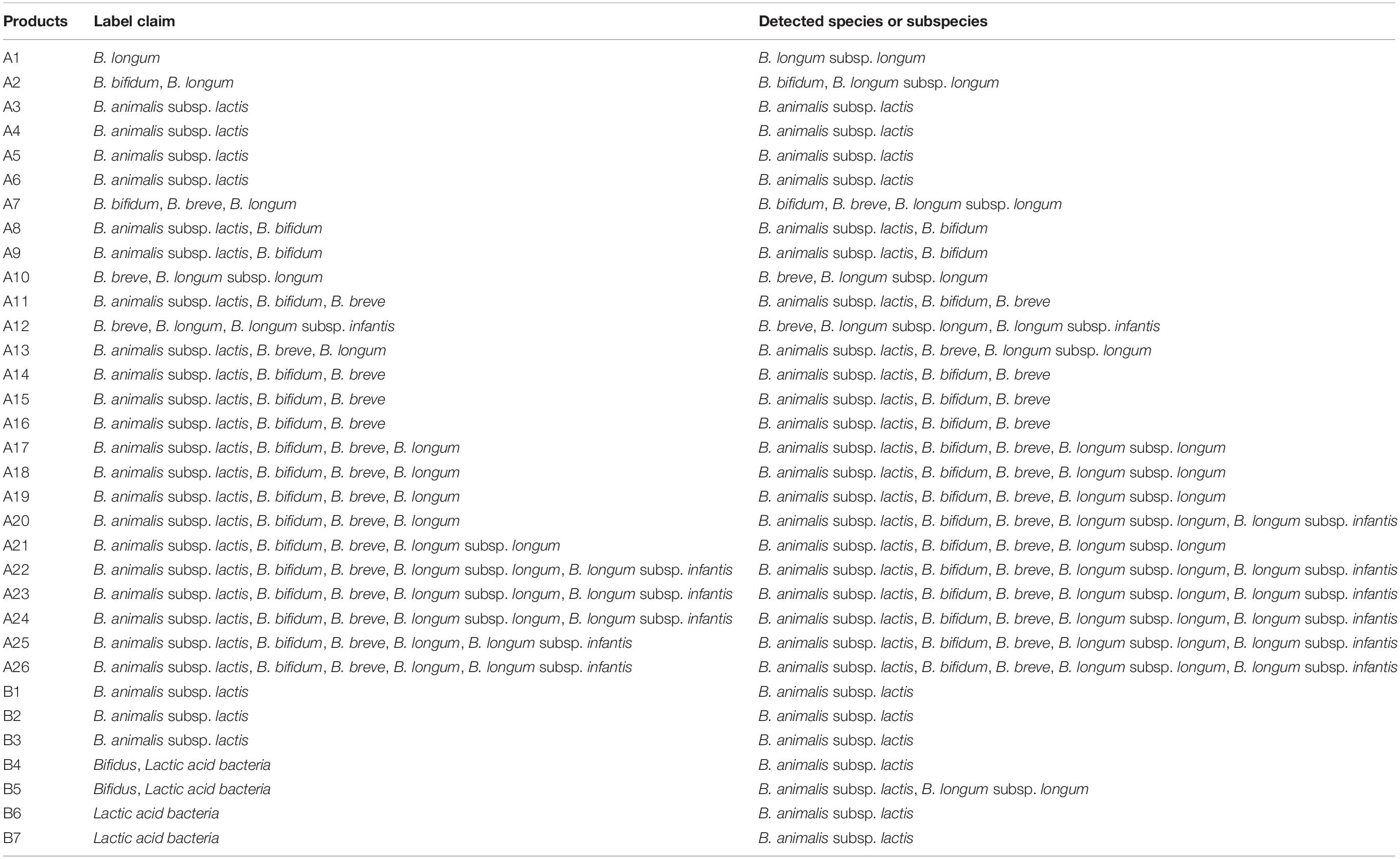
Table 5. Monitoring of commercial probiotic and dairy products for the verification of the developed 96-well plate.
Discussion
Bifidobacterium subspecies (B. animalis subsp. animalis or B. animalis subsp. lactis and B. longum subsp. longum or B. longum subsp. infantis) are known to be similar to each other. However, these subspecies have different functions such as having ability to grow in milk or expressing enzymes (Masco et al., 2004). To distinguish these species or subspecies, previous studies have targeted marker genes such as 16S rRNA and tuf genes. However, it is difficult to distinguish subspecies by using these genes because of their highly similar sequences (Tannock et al., 2013; Kurakawa et al., 2015). Some researchers have screened specific genes through genomic analysis to distinguish Bifidobacterium subspecies. Lawley et al. (2017) have reported the identification of functional gene targets for the differentiation of B. longum subsp. longum and B. longum subsp. infantis based on comparative genomic analysis. However, these functional genes they identified showed some limitations. For example, B. longum subsp. infantis specific sialidase gene (accession no. ACJ53406.1) was limited to some B. longum subsp. infantis strains, but not all subspecies. It was also found to be present in B. bifidum. In addition, B. longum subsp. longum specific kinase gene (accession no. AAN24115.1) was present in many Bifidobacterium species such as B. adolescentis and B. dentium. Because of the limited number of genomes (n = 2) used in their analysis, these identified genes could not be applied to distinguish all Bifidobacterium species.
To overcome limitations of previous studies, we identified genetic markers with large-scale Bifidobacterium genome sequences (n = 210). All genetic markers obtained through comparative genomic analysis were confirmed to be specific by in silico analysis. We also confirmed that some genomes deposited in NCBI were misclassified. Previous studies have also reported that taxonomy information for similar species in the NCBI is incorrect (Kim et al., 2020). For Bifidobacterium, this is the first report to confirm the incorrect classification of genomes in NCBI. Inaccuracies of genomic information may contribute to difficulty in developing methods to distinguish Bifidobacterium. Our results suggest that B. longum subsp. infantis CCUG 52486 and 157F are B. longum subsp. longum.
The real-time PCR method developed in this study showed high specificity and accuracy. However, the limited information of some species, such as Bifidobacterium coryneforme, Bifidobacterium cuniculi, and B. longum subsp. suis were available in the NCBI (only one or two representatives), thus, we can only include the small number of genomes for those strains. This method was also successfully applied to monitoring of probiotic products. It correctly identified Bifidobacterium species contained in all products. We were also able to analyze these strains up to subspecies level labeled in probiotic products as B. animalis and B. longum, allowing us to better understand the presence of strains contained in probiotic products. A previous study (Patro et al., 2016) using shotgun next-generation sequencing has shown that nine out of ten probiotic products are consistent with their label claims. One product, which was misidentified, contained B. longum subsp. longum instead of B. longum subsp. infantis. They found that these strains were frequently mislabeled in other products (Patro et al., 2016). In another study (Lewis et al., 2016), 16 probiotic products containing Bifidobacterium were monitored by terminal-restriction fragment length polymorphism (T-RFLP) profiling. It was found that only one product was consistent with label claims (Lewis et al., 2016). Our assay can also distinguish between B. longum subsp. longum and B. longum subsp. infantis. The resolution of our method is comparable to shotgun sequencing. It is better than T-RFLP typing based on 16S rRNA gene. To provide more detailed information of commercial probiotic products to consumers, identifying and quantifying bacteria strains in food products is important. However, in this study, since quantification was performed on the DNA isolated from a culture, the concentration of Bifidobacterium may be underestimated when applied to a food matrix, as reported in previous studies (Kralik and Ricchi, 2017; Frentzel et al., 2018). Our real-time PCR assay can be used to differentiate and quantify multiple Bifidobacterium subspecies in food sample. The methods described in this study, such as the identification of genetic marker using pan-genome analysis and the design of specific primers using the selected genetic markers, can be applied to pathogenic bacteria in complex clinical samples and other bacterial strains as well.
Conclusion
In conclusion, genetic markers were identified to distinguish different Bifidobacterium species and subspecies through comparative genomics based on their whole-genome sequences. Although Bifidobacterium species are commonly used in probiotic and dairy products, it is still difficult to distinguish all Bifidobacterium species by conventional detection methods. This study designed specific primers from these identified genetic markers. A real-time PCR assay was developed in this study to accurately and rapidly detect 22 Bifidobacterium in a single 96-well plate. The developed real-time PCR assay can be used to monitor commercial probiotic and dairy products. Our assay can also be used to verify the reliability of claims of probiotic and dairy products. Furthermore, it can be applied to identify Bifidobacterium communities in various food products and environmental samples.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
H-BK, EK, S-MY, and H-YK designed the experiment. H-BK, EK, and SL performed the comparative genomic analysis. H-BK, EK, and S-MY confirmed primer specificity and performed application tests using real-time PCR. H-BK, EK, and M-JK prepared a draft manuscript. H-BK, EK, SL, and H-YK reviewed and edited the manuscript. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.02087/full#supplementary-material
Footnotes
References
Binetti, A. G., Capra, M. L., Alvarez, M. A., and Reinheimer, J. A. (2008). PCR method for detection and identification of Lactobacillus casei/paracasei bacteriophages in dairy products. Int. J. Food Microbiol. 124, 147–153. doi: 10.1016/j.ijfoodmicro.2008.03.006
Broeders, S., Huber, I., Grohmann, L., Berben, G., Taverniers, I., Mazzara, M., et al. (2014). Guidelines for validation of qualitative real-time PCR methods. Trends Food Sci. Technol. 37, 115–126. doi: 10.1016/j.tifs.2014.03.008
Cammà, C., Di Domenico, M., and Monaco, F. (2012). Development and validation of fast real-time PCR assays for species identification in raw and cooked meat mixture. Food Control 23, 400–404. doi: 10.1016/j.foodcont.2011.08.007
Chaudhari, N. M., Gupta, V. K., and Dutta, C. (2016). BPGA- an ultra-fast pan-genome analysis pipeline. Sci. Rep. 6:24373. doi: 10.1038/srep24373
Chen, J., Zhang, L., Paoli, G. C., Shi, C., Tu, S. I., and Shi, X. (2010). A real-time PCR method for the detection of Salmonella enterica from food using a target sequence identified by comparative genomic analysis. Int. J. Food Microbiol. 1373, 168–174. doi: 10.1016/j.ijfoodmicro.2009.12.004
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Eren, A. M., Esen, ÖC., Quince, C., Vineis, J. H., Morrison, H. G., Sogin, M. L., et al. (2015). Anvi’o: an advanced analysis and visualization platform for ’omics data. PeerJ 3:e1319. doi: 10.7717/peerj.1319
Floch, M. H. (2018). The role of prebiotics and probiotics in gastrointestinal disease. Gastroenterol. Clin. North Am. 47, 179–191. doi: 10.1016/j.gtc.2017.09.011
Frentzel, H., Thanh, M. D., Krause, G., Appel, B., and Mader, A. (2018). Quantification and differentiation of Bacillus cereus group species in spices and herbs by real-time PCR. Food Control 83, 99–108. doi: 10.1016/j.foodcont.2016.11.028
Gómez-Rojo, E. M., Romero-Santacreu, L., Jaime, I., and Rovira, J. (2015). A novel real-time PCR assay for the specific identification and quantification of Weissella viridescens in blood sausages. Int. J. Food Microbiol. 215, 16–24. doi: 10.1016/j.ijfoodmicro.2015.08.002
Inoue, Y., Iwabuchi, N., Xiao, J. Z., Yaeshima, T., and Iwatsuki, K. (2009). Suppressive effects of Bifidobacterium breve strain M-16V on T-helper type 2 immune responses in a murine model. Biol. Pharm. Bull. 32, 760–763. doi: 10.1248/bpb.32.760
Junick, J., and Blaut, M. (2012). Quantification of human fecal Bifidobacterium species by use of quantitative real-time PCR analysis targeting the groEL gene. Appl. Environ. Microbiol. 78, 2613–2622. doi: 10.1128/AEM.07749-11
Kim, E., Cho, Y., Lee, Y., Han, S. K., Kim, C. G., Choo, D. W., et al. (2017). A proteomic approach for rapid identification of Weissella species isolated from Korean fermented foods on MALDI-TOF MS supplemented with an in-house database. Int. J. Food Microbiol. 243, 9–15. doi: 10.1016/j.ijfoodmicro.2016.11.027
Kim, E., Kim, H. J., Yang, S. M., Kim, C. G., Choo, D. W., and Kim, H. Y. (2019). Rapid identification of Staphylococcus species isolated from food samples by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J. Microbiol. Biotechnol. 29, 548–557. doi: 10.4014/jmb.1901.01046
Kim, E., Yang, S. M., Cho, E. J., and Kim, H. Y. (2020). Novel real-time PCR assay for Lactobacillus casei group species using comparative genomics. Food Microbiol. 90:103485. doi: 10.1016/j.fm.2020.103485
Kralik, P., and Ricchi, M. (2017). A basic guide to real time PCR in microbial diagnostics: definitions, parameters, and everything. Front. Microbiol. 8:108. doi: 10.3389/fmicb.2017.00108
Kurakawa, T., Ogata, K., Tsuji, H., Kado, Y., Takahashi, T., Kida, Y., et al. (2015). Establishment of a sensitive system for analysis of human vaginal microbiota on the basis of rRNA-targeted reverse transcription-quantitative PCR. J. Microbiol. Methods 111, 93–104. doi: 10.1016/j.mimet.2015.01.021
Lawley, B., Munro, K., Hughes, A., Hodgkinson, A. J., Prosser, C. G., Lowry, D., et al. (2017). Differentiation of Bifidobacterium longum subspecies longum and infantis by quantitative using functional gene targets. PeerJ 5:e3375. doi: 10.7717/peerj.3375
Lewis, Z. T., Shani, G., Masarweh, C. F., Popovic, M., Frese, S. A., Sela, D. A., et al. (2016). Validating bifidobacterial species and subspecies identity in commercial probiotic products. Pediatr. Res. 79, 445–452. doi: 10.1038/pr.2015.244
LoCascio, R. G., Desai, P., Sela, D. A., Weimer, B., and Mills, D. A. (2010). Broad conservation of milk utilization genes in Bifidobacterium longum subsp. infantis as revealed by comparative genomic hybridization. Appl. Environ. Microbiol. 76, 7373–7381. doi: 10.1128/AEM.00675-10
Lugli, G. A., Milani, C., Turroni, F., Duranti, S., Mancabelli, L., Mangifesta, M., et al. (2017). Comparative genomic and phylogenomic analyses of the Bifidobacteriaceae family. BMC Genomics 18:568. doi: 10.1186/s12864-017-3955-4
Masco, L., Ventura, M., Zink, R., Huys, G., and Swings, J. (2004). Polyphasic taxonomic analysis of Bifidobacterium animalis and Bifidobacterium lactis reveals relatedness at the subspecies level: reclassification of Bifidobacterium animalis as Bifidobacterium animalis subsp. animalis subsp. nov. and Bifidobacterium lactis as Bifidobacterium animalis subsp. lactis subsp. nov. Int. J. Syst. Evol. Microbiol. 54, 1137–1143. doi: 10.1099/ijs.0.03011-0
Mellmann, A., Andersen, P. S., Bletz, S., Friedrich, A. W., Kohl, T. A., Lilje, B., et al. (2017). High interlaboratory reproducibility and accuracy of next-generation-sequencing-based bacterial genotyping in a ring trial. J. Clin. Microbiol. 55, 908–913. doi: 10.1128/JCM.02242-16
Morovic, W., Hibberd, A. A., Zabel, B., Barrangou, R., and Stahl, B. (2016). Genotyping by PCR and high-throughput sequencing of commercial probiotic products reveals composition biases. Front. Microbiol. 7:1747. doi: 10.3389/fmicb.2016.01747
O’Callaghan, A., and van Sinderen, D. (2016). Bifidobacteria and their role as members of the human gut microbiota. Front. Microbiol. 7:925. doi: 10.3389/fmicb.2016.00925
Patro, J. N., Ramachandran, P., Barnaba, T., Mammel, M. K., Lewis, J. L., and Elkins, C. A. (2016). Culture-independent metagenomic surveillance of commercially available probiotics with high-throughput next-generation sequencing. mSphere 1:e00057-16. doi: 10.1128/mSphere.00057-16
Pinto-Sanchez, M. I., Hall, G. B., Ghajar, K., Nardelli, A., Bolino, C., Lau, J. T., et al. (2017). Probiotic Bifidobacterium longum NCC3001 reduces depression scores and alters brain activity: a pilot study in patients with irritable bowel syndrome. Gastroenterology 153, 448–459. doi: 10.1053/j.gastro.2017.05.003
Shehata, H. R., Ragupathy, S., Shanmughanandhan, D., Kesanakurti, P., Ehlinger, T. M., and Newmaster, S. G. (2019). Guidelines for validation of qualitative real-time PCR methods for molecular diagnostic identification of probiotics. J. AOAC Int. 102, 1774–1778. doi: 10.5740/jaoacint.18-0320
Tannock, G. W., Lawley, B., Munro, K., Gowri Pathmanathan, S., Zhou, S. J., Makrides, M., et al. (2013). Comparison of the compositions of the stool microbiotas of infants fed goat milk formula, cow milk-based formula, or breast milk. Appl. Environ. Microbiol. 79, 3040–3048. doi: 10.1128/AEM.03910-12
Turroni, F., Foroni, E., Pizzetti, P., Giubellini, V., Ribbera, A., Merusi, P., et al. (2009). Exploring the diversity of the bifidobacterial population in the human intestinal tract. Appl. Environ. Microbiol. 75, 1534–1545. doi: 10.1128/AEM.02216-08
Ventura, M., Canchaya, C., Del Casale, A., Dellaglio, F., Neviani, E., Fitzgerald, G. F., et al. (2006). Analysis of bifidobacterial evolution using a multilocus approach. Int. J. Syst. Evol. Microbiol. 56, 2783–2792. doi: 10.1099/ijs.0.64233-0
Ventura, M., Canchaya, C., van Sinderen, D., Fitzgerald, G. F., and Zink, R. (2004a). Bifidobacterium lactis DSM 10140: identification of the atp (atpBEFHAGDC) operon and analysis of its genetic structure, characteristics, and phylogeny. Appl. Environ. Microbiol. 70, 3110–3121. doi: 10.1128/AEM.70.5.3110-3121.2004
Ventura, M., Canchaya, C., Zink, R., Fitzgerald, G. F., and van Sinderen, D. (2004b). Characterization of the groEL and groES loci in Bifidobacterium breve UCC 2003: genetic, transcriptional, and phylogenetic analyses. Appl. Environ. Microbiol. 70, 6197–6209. doi: 10.1128/AEM.70.10.6197-6209.2004
Ventura, M., and Zink, R. (2003). Comparative sequence analysis of the tuf and recA genes and restriction fragment length polymorphism of the internal transcribed spacer region sequences supply additional tools for discriminating Bifidobacterium lactis from Bifidobacterium animalis. Appl. Environ. Microbiol. 69, 7517–7522. doi: 10.1128/AEM.69.12.7517-7522.2003
von Ah, U., Mozzetti, V., Lacroix, C., Kheadr, E. E., Fliss, I., and Meile, L. (2007). Classification of a moderately oxygen-tolerant isolate from baby faeces as Bifidobacterium thermophilum. BMC Microbiol. 7:79. doi: 10.1186/1471-2180-7-79
Weizman, Z., Asli, G., and Alsheikh, A. (2005). Effect of a probiotic infant formula on infections in child care centers: comparison of two probiotic agents. Pediatrics 115, 5–9. doi: 10.1542/peds.2004-1815
Zhang, R., He, L., Zhang, L., Li, C., and Zhu, Q. (2016). Screening of cholesterol-lowering Bifidobacterium from Guizhou Xiang Pigs, and evaluation of its tolerance to oxygen, acid, and bile. Korean J. Food Sci. Anim. Resour. 36, 37–43. doi: 10.5851/kosfa.2016.36.1.37
Zhang, X., Payne, M., and Lan, R. (2019). In silico identification of serovar-specific genes for Salmonella serotyping. Front. Microbiol. 10:835. doi: 10.3389/fmicb.2019.00835
Keywords: Bifidobacterium, real-time PCR, pan-genome, whole-genome sequence, probiotic, comparative genomics, identification, detection method
Citation: Kim H-B, Kim E, Yang S-M, Lee S, Kim M-J and Kim H-Y (2020) Development of Real-Time PCR Assay to Specifically Detect 22 Bifidobacterium Species and Subspecies Using Comparative Genomics. Front. Microbiol. 11:2087. doi: 10.3389/fmicb.2020.02087
Received: 05 June 2020; Accepted: 07 August 2020;
Published: 28 August 2020.
Edited by:
Kwangcheol Casey Jeong, University of Florida, United StatesReviewed by:
Raies Mir, United States Department of Agriculture (USDA), United StatesAldert Zomer, Utrecht University, Netherlands
Copyright © 2020 Kim, Kim, Yang, Lee, Kim and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hae-Yeong Kim, hykim@khu.ac.kr
†These authors have contributed equally to this work
 Hyeon-Be Kim
Hyeon-Be Kim Eiseul Kim
Eiseul Kim Seung-Min Yang
Seung-Min Yang Shinyoung Lee
Shinyoung Lee Mi-Ju Kim
Mi-Ju Kim Hae-Yeong Kim
Hae-Yeong Kim