- State Key Laboratory of Agricultural Microbiology, College of Life Science and Technology, Huazhong Agricultural University, Wuhan, China
Acquisition of spacers confers the CRISPR–Cas system with the memory to defend against invading mobile genetic elements. We previously reported that the CRISPR-associated factor Csa3a triggers CRISPR adaptation in Sulfolobus islandicus. However, a feedback regulation of CRISPR adaptation remains unclear. Here we show that another CRISPR-associated factor, Csa3b, binds a cyclic oligoadenylate (cOA) analog (5′-CAAAA-3′) and mutation at its CARF domain, which reduces the binding affinity. Csa3b also binds the promoter of adaptation cas genes, and the cOA analog enhances their binding probably by allosteric regulation. Deletion of the csa3b gene triggers spacer acquisition from both plasmid and viral DNAs, indicating that Csa3b acted as a repressor for CRISPR adaptation. Moreover, we also find that Csa3b activates the expression of subtype cmr-α and cmr-β genes according to transcriptome data and demonstrate that Csa3b binds the promoters of cmr genes. The deletion of the csa3b gene reduces Cmr-mediated RNA interference activity, indicating that Csa3b acts as a transcriptional activator for Cmr-mediated RNA interference. In summary, our findings reveal a novel pathway for the regulation of CRISPR adaptation and CRISPR–Cmr RNA interference in S. islandicus. Our results also suggest a feedback repression of CRIPSR adaptation by the Csa3b factor and the cOA signal produced by the Cmr complex at the CRISPR interference stage.
Introduction
CRISPR–Cas is a prokaryotic immune system that defends bacteria and archaea against invasive plasmids and viruses (Barrangou et al., 2007; Makarova et al., 2011). CRISPR–Cas adaptive immunity occurs in three stages: acquisition of de novo spacers (also known as adaptation), crRNA biogenesis, and targeting and cleavage of invasive nucleic acids (Brouns et al., 2008; Westra et al., 2012). According to the latest classification scheme, CRISPR–Cas systems are categorized into two main classes, each subdivided into three types. Class 1 systems (types I, III, and IV) contain multiple protein subunits in their effector complexes, while class 2 systems (types II, V, and VI) carry a single large protein in their effector complexes (Koonin and Makarova, 2013; Makarova et al., 2015).
Spacer acquisition into CRISPR arrays constitutes the first stage of the immune reaction (Brouns et al., 2008; Westra et al., 2012). Short DNA fragments, called protospacers, are taken from the invasive genetic elements and integrated into the CRISPR arrays at the leader proximal end facilitated by the conserved core proteins Cas1 and Cas2 (Makarova et al., 2011) and, in some cases, additional subunits such as Cas4 family proteins (Kieper et al., 2018; Lee et al., 2018; Shiimori et al., 2018; Zhang et al., 2019). The first successful demonstration of spacer acquisition under laboratory conditions was based on Streptococcus thermophilus subtype II-A system (Barrangou et al., 2007), after which more studies have focused mainly on type I systems (Sternberg et al., 2016), including Sulfolobus subtype I-A systems (Erdmann and Garrett, 2012; Erdmann et al., 2014b; Liu et al., 2015), Haloarcula hispanica subtype I-B system (Li et al., 2014a, b), Escherichia coli subtype I-E system (Swarts et al., 2012; Yosef et al., 2012, 2013), and Pseudomonas aeruginosa (Cady et al., 2012) and Pectobacterium atrosepticum (Richter et al., 2014) subtype I-F systems.
Both the adaptation and the interference stages of CRISPR–Cas systems are known to be regulated at the transcriptional level. In particular, the regulation of cas genes in subtype I-E CRISPR–Cas systems has been studied in great detail. In the E. coli subtype I-E system, the heat-stable nucleoid-structuring protein (H-NS) (Pul et al., 2010) and the cAMP receptor protein (CRP) (Westra et al., 2010; Yang et al., 2014) repress CRISPR-based immunity, and H-NS-mediated repression may be relieved by LeuO, a LysR-type transcription factor (Westra et al., 2010), and by BaeR, an envelope stress-activated regulator (Perez-Rodriguez et al., 2011). Besides transcriptional regulations, the CRISPR ribonucleases are allosterically regulated by small molecules. It has been reported that the type III effector complex synthesizes cyclic oligoadenylates (cOAs) upon binding to target RNA and that the cOAs activate the ribonuclease activity of the CARF domain containing proteins, including the subtype III-A associated Csm6 protein (Kazlauskiene et al., 2017; Niewoehner et al., 2017) and the subtype III-B Csx1 protein (Han et al., 2018). For example, 5′-CAAAA-3′ can act as the signaling molecule to activate the ribonuclease activity of Csx1 from Sulfolobus (Han et al., 2017), suggesting that the oligonucleotides generated during RNA metabolism play an important role in the regulation of CRISPR functions. cOAs can be degraded by a ring nuclease, switching off the antiviral state mediated by the CARF domain-containing ribonucleases (Athukoralage et al., 2018; Jia et al., 2019). However, there are other CARF domain families in addition to the Csm6 and the Csx1 families (Makarova et al., 2014). Csa3, the most frequent CARF family linked to type I system and subtype I-A in particular, lacks a C-terminal catalytic effector domain but is fused with a HTH domain (Makarova et al., 2014). Csa3 family proteins are involved in the regulation of CRISPR adaptation and interference in Sulfolobus islandicus (Liu et al., 2015, 2017; He et al., 2017). The Csa3a factor triggers CRISPR adaptation (Liu et al., 2015), and the Csa3b factor represses the expression of Cascade genes of subtype I-A system (He et al., 2017). However, whether Csa3 proteins allosterically regulate CRISPR functions remain unclear.
In the model archaeon S. islandicus, the regulatory mechanisms of cas genes differ markedly from those of bacteria which employ global transcriptional factors (Patterson et al., 2017). Two CRISPR-linked factors, Csa3a and Csa3b, are employed for CRISPR–Cas regulation. The Csa3a regulator triggers CRISPR de novo spacer acquisition by activating adaptation genes (Liu et al., 2015). Furthermore, the Csa3a regulator couples the transcriptional activation of adaptation genes, CRISPR arrays, and DNA repair genes in order to efficiently target invading nucleic acids (Liu et al., 2017). Csa3b binds to the promoter region of the CRISPR–interference gene cassette and facilitates binding of the CRISPR interference complex to the promoter region, leading to the repression of subtype I-A cas expression (He et al., 2017). Both Csa3a and Csa3b regulators carry a CARF domain (Lintner et al., 2011). However, whether Csa3b regulates other CRISPR–Cas modules except for I-A interference module and whether signaling molecules are involved in the regulation of the function of the Csa3b factor have not yet been reported. Thus, in this work, we have studied the in vivo function of the Csa3b factor in CRISPR adaptation and Cmr-mediated RNA interference in S. islandicus.
Materials and Methods
Strains, Growth, and Transformation of Sulfolobus
S. islandicus strains, including the genetic host E233S (ΔpyrEFΔlacS, termed wildtype in this work) (Deng et al., 2009), a csa3b deletion strain (Δcsa3b) (He et al., 2017), and these strains carrying the csa3b overexpression vector were cultured in SCV medium or ACV inducible medium at 78°C (Peng et al., 2009). The S. islandicus cells were transformed by electroporation, and the resulting transformants were selected on two-layer phytal gel plates, as described previously (Deng et al., 2009). The S. islandicus E233S wild-type (wt) strain carrying the empty pSeSD plasmid and the wt strain carrying the Csa3b overexpression plasmid were grown in ACVy medium (Peng et al., 2009, 2012), and growth curves were obtained by measuring their optical densities at 600 nm (OD600). The E. coli DH5α cells used for DNA cloning were cultured at 37°C in Luria–Bertani medium, and ampicillin was added to the culture at a final concentration of 100 μg/ml, where required.
Plasmid Construction, Gene Deletion, and Protein Purification
The csa3b deletion plasmid was constructed by cloning the mini-CRISPR sequence comprising “repeat–spacer–repeat,” where the spacer matches the csa3b gene and the upstream and downstream homologous arms of the csa3b gene into the E. coli–Sulfolobus shuttle vector pSeSD. The csa3b genes were cloned into pET30a and the Sulfolobus expression vector pSeSD, respectively. Mutations were introduced into the csa3b genes by inverse PCR, with mutations on the primer sequences using the E. coli and Sulfolobus expression plasmids as the templates, respectively, by PCR amplification. Then, the PCR products were separated from gel and purified. The purified PCR products were 5′ phosphorylated by T4 kinase and self-ligated by T4 DNA ligase, respectively, giving the mutant overexpression plasmids. All the primers used are listed in Supplementary Table 1. The purification of Csa3b protein heterogeneously expressed in E. coli was carried out as described previously (Liu et al., 2015).
Transcriptome Analysis
Strains (two biological repeats for each strain) for transcriptome analysis were cultured to log phase (OD600 = 0.2), after which 1-ml culture of each strain was transferred to 100 ml fresh ACVy medium in 250-ml flasks. Then, total RNA was isolated using Trizol reagent (Invitrogen, Carlsbad, CA, United States) from exponentially growing Sulfolobus cultures (OD600 = 0.2) in ACVy medium for the induction of the csa3b gene under control of the araS promoter, as described previously (Liu et al., 2015). The genomic DNA in the total RNA sample was removed using DNase I (Roche, Basel, Switzerland). The quality and the quantity of the purified total RNA were determined by measuring the absorbance at 260 and 280 nm by using a NanoDrop ND-1000 spectrophotometer (Labtech, Wilmington, MA, United States). Total RNA integrity was verified by electrophoresis on a 1.5% agarose gel. A total of 3 μg RNA per sample was used as input material for cDNA library preparations. Sequencing libraries were generated using the NEBNext Ultra™ RNA Library Prep Kit for Illumina (NEB, United States), following the manufacturer’s recommendations, and index codes were added to assign sequences to each sample. First-strand cDNA synthesis was performed using random hexamer primers and M-MuLV Reverse Transcriptase (RNase H–). Second-strand cDNA synthesis was subsequently performed using DNA polymerase I and RNase H, which was followed by 15 cycles of PCR enrichment. Sequencing was performed with an Illumina HiSeq2000 instrument. Raw data were initially processed to obtain clean reads by removing adapter sequences and low-quality bases. The clean reads were aligned to the reference genome sequence of S. islandicus REY15A (GenBank accession no. NC_017276). An index of the reference genome was built using Bowtie software v2.0.6, and paired-end clean reads were aligned to the reference genome using TopHat software v2.0.9. HTSeq software v0.6.1 was used to count the number of reads mapped to each gene, following which the reads per kilobase per million mapped reads (RPKM) for each gene were calculated based on the length of the gene and the number of reads mapped to the gene. Each strain was sequenced in duplicate. To investigate the expression level of each gene in different groups, transcript expression levels were expressed as the RPKM. Next, p-values were used to identify differentially expressed genes (DEGs) between the two groups using the chi-square test (2 × 2), and the significance threshold of the p-value in multiple tests was set based on the false discovery rate (FDR). Fold-changes [log2(RPKM1/RPKM2)] were also estimated according to normalized gene expression levels. The threshold for DEGs was set at p < 0.01 and log2 fold-change > 1 (FDR < 0.05). The transcriptome data were deposited in the GEO database under accession number PRJNA511557, and the DEGs are listed in Supplementary Table 2.
Synthesis of the Oligonucleotide
The oligonucleotide 5′-CAAAA-3′ was synthesized in Tsingke (Beijing, China).
Localized Surface Plasmon Resonance Technology for Analysis of the Interactions of the Oligonucleotide, Csa3b Protein, and Promoter DNAs
The equilibrium-binding constants (KD) of the interaction between Csa3b protein and oligonucleotide 5′-CAAAA-3′ and the promoter DNAs and the oligonucleotide and the promoter DNAs were determined by Open SPR (Nicoya, Canada), respectively. All the steps were performed according to the manufacturer’s protocol. Briefly, Csa3b family proteins (40 μg/ml) were covalently immobilized on COOH-sensor chips (Nicoya, Canada) by EDC/NHS chemistry. The 5′-end of the promoter DNA was marked with sulfydryl and then the DNA was labeled on the chip. Then, the oligonucleotide 5′-CAAAA-3′, promoter DNA, or proteins were continuously diluted into several different concentrations using the running buffer and injected into the chip from low to high concentrations. Meanwhile, bovine serum albumin was used as a negative control. In each cycle, a 250-μl sample was flowed through the chip for 5 min at a constant flow rate of 20 μl/min. After detection, 0.02% sodium dodecyl sulfate (SDS) was added to dissociate the binding. Finally, the kinetic parameters of the binding reactions were calculated and analyzed by Trace Drawer software (Ridgeview Instruments AB, Sweden) and one-to-one fitting model.
PCR Amplification and Sequencing of the Leader-Proximal CRISPR Regions
The S. islandicus E233S (wt) and the csa3b deletion strains carrying empty pSeSD vector or infected with STSV2 virus were cultured in SCV medium at 78°C. Samples of each culture (0.1 ml) were taken every day (for cells carrying the pSeSD plasmid) or every 3 days (for cells carrying the STSV2 virus), and total DNA from these cells was used as the PCR template. The leader-proximal region of CRISPR locus 1 was amplified via PCR using Taq polymerase, with forward primer CRISPR-F and reverse primer CRISPR1S5-R for locus 1 (Supplementary Table 1). The PCR products were separated on 1.5% agarose gel and visualized via ethidium bromide staining. The purified PCR products of the expanded bands were cloned into the T-vector (Clone Smarter TA Cloning Kit, Wuhan, China), following the manufacturer’s instructions. Subsequently, the ligation products were transformed into E. coli DH5α cells. The plasmids from single colonies were purified and sequenced at Tsingke (Wuhan, China).
DNase I Footprinting Analysis
DNase I footprinting was carried out as reported previously (Liu et al., 2015). For the preparation of fluorescent FAM-labeled probes, the promoter region was PCR amplified with 2 × TOLO HIFI DNA polymerase premix (TOLO Biotech, Shanghai) from the plasmids carrying the promoter DNAs of the SiRe_0760 and SiRe_0890 genes using primers of 5′-FAM labeled-M13F and M13R primers, respectively. The FAM-labeled probes were purified with the Wizard® SV Gel and PCR Clean-Up System (Promega, United States) and were quantified with NanoDrop 2000C (Thermo, United States). For each DNase I footprinting assay, 400-ng probes were incubated with different amounts of Csa3b protein in a total volume of 40 μl. After incubation for 30 min at 30°C, 10-μl solution containing 0.015 unit DNase I (Promega) and 100 nmol freshly prepared CaCl2 was added, and further incubation was performed at 37°C for 1 min. The reaction was stopped by adding 140 μl DNase I stop solution (200 mM unbuffered sodium acetate, 30 mM EDTA, and 0.15% SDS). The samples were firstly extracted with phenol/chloroform and then precipitated with ethanol. The pellets were dissolved in 30 μl MiniQ water. The samples were analyzed in a 3730 DNA Analyzer (Applied Biosystems, Foster City, CA, United States), and the electropherograms were aligned with GeneMapper v3.5 (Applied Biosystems).
STSV2 Infection Into S. islandicus
The CRISPR deletion strain of S. islandicus REY15A infected with Sulfolobus tengchongensis spindle-shaped virus STSV2 was used to prepare the STSV2 virus particle for infection into S. islandicus wild-type or csa3b deletion strains, as described previously (Erdmann et al., 2014a). When S. islandicus wild-type and Δcsa3b strains grew to mid-log phase (OD600 = 0.6), cells were harvested from a 15-ml culture and suspended in 1 ml SCV medium for infection. One microliter of concentrated STSV2 was mixed with the prepared cell culture, and the mixed culture was incubated for 2 h at 78°C. After that, the infected cells were transferred to 50 ml of pre-heated (78°C) SCV medium and incubated for 3 days at 78°C. During cultivation, 1 ml was taken from each culture every 12 h, and the OD600 values were measured. Then, 1 ml was taken as well from each culture and transferred into 50 ml fresh SCV medium every 3 days.
Determination of in vivo RNA Interference Efficiency by β-Galactosidase Activity Assay
The plasmids, pLacS-ck and pAC-SS1-LacS, were constructed as described previously (Peng et al., 2015). The plasmid pLacS-ck carries the lacS gene of S. islandicus Rey15A with the native promoter and terminator. Plasmid pAC-SS1-LacS carries the lacS gene of S. islandicus Rey15A and an artificial CRISPR array with the spacer from lacS (promoter–repeat–spacerlacS–repeat). Both plasmids were transformed into S. islandicus E233S (wild-type) and the csa3b gene deletion strain (Δcsa3b). Selected transformants were cultured in SCVy medium, and cells were harvested at OD600 = 0.2–0.3. These cells were sonicated and used for β-galactosidase activity assay, as reported previously (Peng et al., 2009).
Determination of in vivo DNA Interference Efficiency Using Plasmid Challenging Assay
The challenging plasmid, pS10i, carries a protospacer targeted only by the subtype III-B Cmr complexes through base-pairing with spacer 10 in CRISPR locus 2 in S. islandicus REY15A (Deng et al., 2013). The control plasmids pSeSD and pS10i (500 ng of each plasmid) were transformed into S. islandicus E233S (wild-type) and the Δcsa3b strain, and the transformation efficiencies were compared.
Results
The CRISPR-Associated Factor Csa3b Bound a Cyclic Oligoadenylate Analog
The Crenarchaeon S. islandicus strain REY15A encodes a subtype I-A adaptation module, a subtype I-A interference module, and two subtype III-B interference modules (Deng et al., 2013; Figure 1A). The csa3 genes, neighboring the adaptation cas genes and subtype I-A interference genes, encode the transcription regulators (Csa3a and Csa3b), each carrying a CARF domain (Lintner et al., 2011). The Csa3 CARF domain carries two conserved motifs: motif 1 seems to be specific to Csa3 family protein and motif 2 is conserved in Csa3, a ring nuclease (SiRe_0244), Csx1, and Csm6 family proteins (Lintner et al., 2011; Athukoralage et al., 2018; Jia et al., 2019; Molina et al., 2019; Figure 1B).
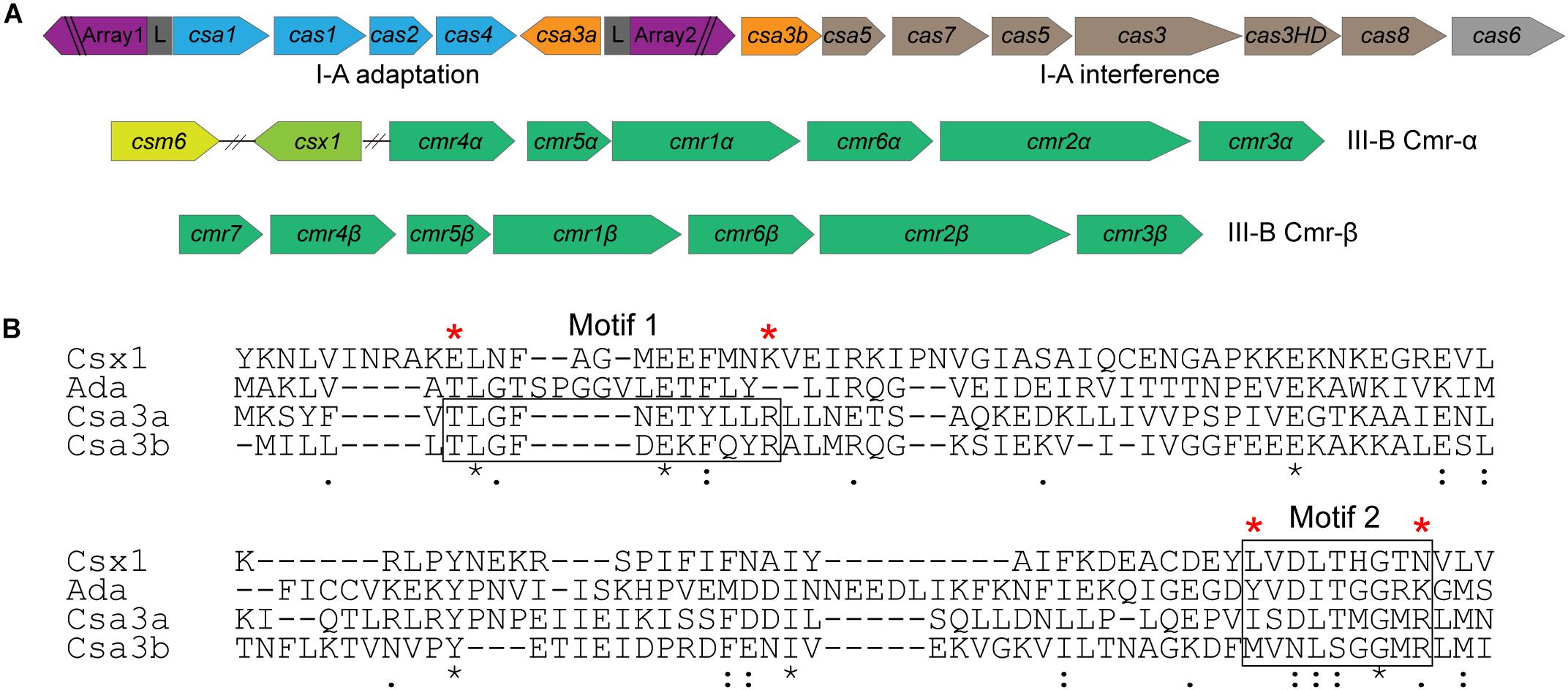
Figure 1. Csa3b is a CRISPR-associated factor carrying a CARF domain. (A) Organization of CRISPR–Cas subtype I-A adaptation and interference modules and subtype III-B (Cmr-α and Cmr-β) interference modules in Sulfolobus islandicus REY15A. (B) Alignment of the CARF domains of Csx1 (SiRe_0884), a ring nuclease (SiRe_0244), Csa3a (SiRe_0764), and Csa3b (SiRe_0765) in S. islandicus REY15A. Two conserved motifs found for Csa3 family proteins (Lintner et al., 2011) were boxed. Csa3b mutation (Csa3bM) was generated by changing the amino acids marked with a red star to Ala. The “*” means that amino acids are conserved in the CARF domain.
Csa3b, the CRISPR-associated transcriptional regulator carrying a CARF domain (Figure 1B), was previously reported to repress the expression of Sulfolobus subtype I-A interference genes (He et al., 2017). In this study, we further analyzed the Csa3b regulatory effects in S. islandicus. The growth curves indicated that Csa3b overexpression strongly inhibited cell growth, while csa3b deletion had no effect on cell growth relative to the wild-type cells (Figure 2A). However, overexpressing Csa3b CARF mutant (Csa3bM, in which the Ala residues were used to replace the star-marked amino acid residues at both motifs 1 and 2) showed no inhibition to cell growth (Figure 2A), indicating that the conserved motifs shown in Figure 1B were important for the regulatory effects of Csa3b. We further tested whether Csa3b bound the cOA analog, an oligonucleotide 5′-CAAAA-3′ which showed the allosteric regulation of Csx1 activity in vitro (Han et al., 2017). A localized surface plasmon resonance (LSPR) analysis demonstrated that Csa3b bound the oligonucleotide in vitro (Figure 2B), while a mutation at the Csa3b CARF domain slightly reduced its affinity to the oligonucleotide (Figure 2C), as revealed by the KD values (46.10 ± 8.14 vs. 123.00 ± 29.90 μM).
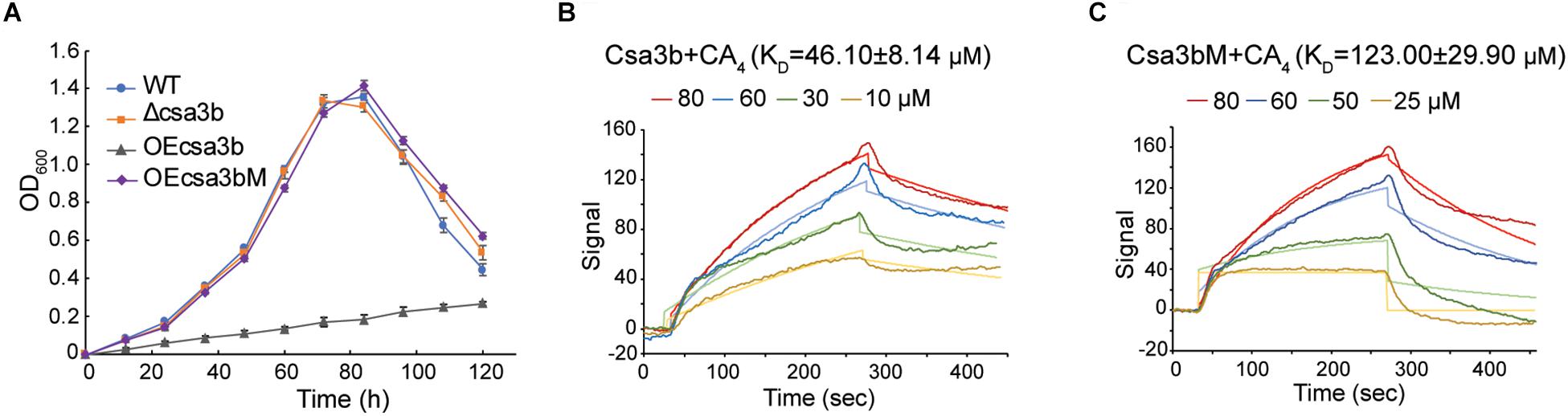
Figure 2. Csa3b binds a cyclic oligoadenylate analog. (A) Growth curves of wild type (wt), csa3b or csa3b CARF mutant overexpression and csa3b deletion strains. (B,C) Localized surface plasmon resonance analysis of interaction between fixed Csa3b or Csa3b mutant proteins and CA4 signals. The KD value and the CA4 concentrations for each assay were indicated.
Csa3b Regulated the Expression of cmr Genes
We further analyzed DEGs between the Sulfolobus csa3b overexpression strain and the wild-type strain by comparative transcriptome analysis. All DEGs are listed in Supplementary Table 1. Compared with the wild-type strain, the cas6 gene (SiRe_0772) probably co-transcribed with the subtype I-A interference genes, which were down-regulated, in line with a previous report that Csa3b acted as a repressor of subtype I-A interference genes (He et al., 2017). Interestingly, both the CRISPR–Cas subtype III-B interference gene cassettes, cmr-α (SiRe_0890 ∼ SiRe_0892) and cmr-β (SiRe_0599 ∼ SiRe_0603) genes, were upregulated in the csa3b overexpression strain (Table 1), suggesting that Csa3b acted as an activator for the regulation of subtype III-B Cmr-α and Cmr-β systems. Furthermore, adaptation cas genes were significantly upregulated in the csa3b deletion strain as revealed by a transcriptome data reported previously (He et al., 2017), suggesting that Csa3b could act as a transcriptional repressor for the regulation of CRISPR adaptation genes. Furthermore, the DNA double-strand break repair genes, including nurA, rad50, mre11, and herA (SiRe_0061 ∼ SiRe_0064), as well as the genes encoding two subunits of DNA polymerase II (PolB2, SiRe_0614, and SiRe_0615) that function in DNA repair, were significantly upregulated in the csa3b overexpression strain (Table 1). The nurA-rad50-mre11-herA operon is essential for S. islandicus REY15A (Zhang et al., 2013), while the polB2 genes neighboring an IS transposase gene is non-essential (unpublished data). This result infers that the csa3b gene might play an important role in the regulation of DNA damage repair systems in S. islandicus.
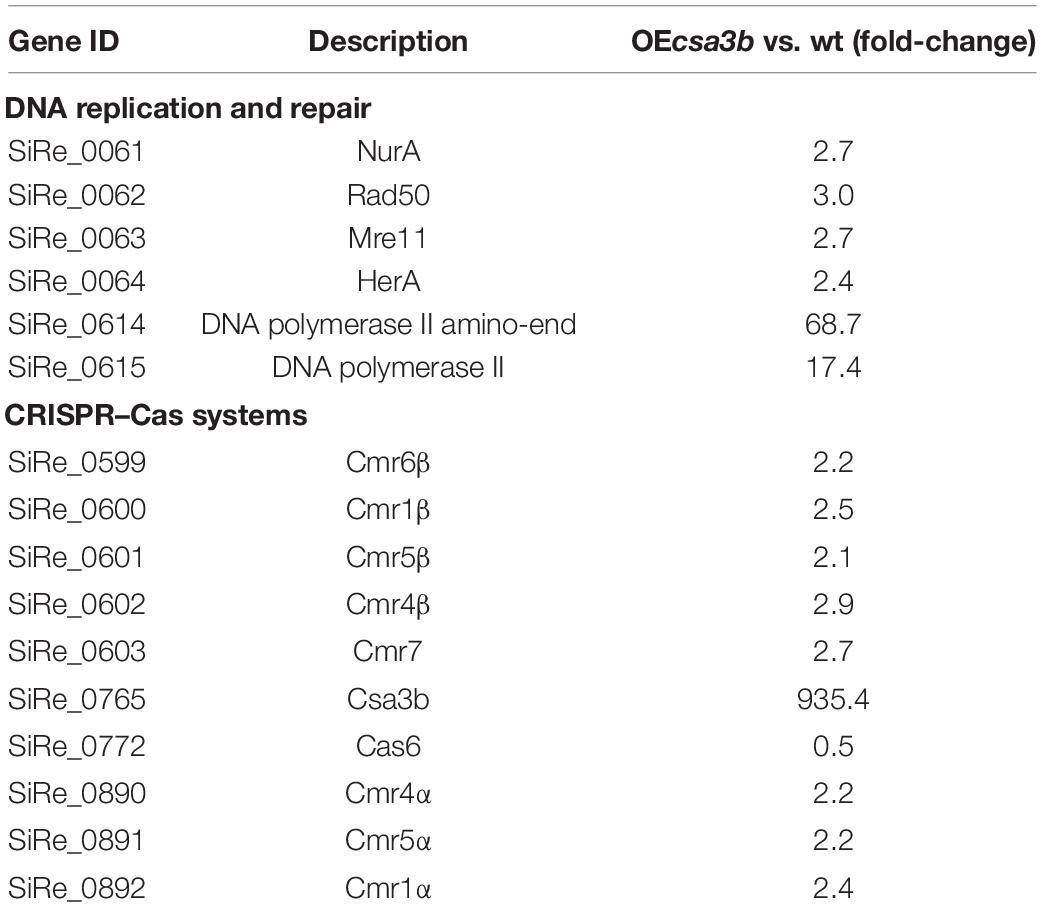
Table 1. Differentially expressed genes in Sulfolobus islandicus csa3b overexpression strain vs. wild-type strain as identified by transcriptome analysis.
Csa3b Regulator Bound the Promoter of CRISPR Adaptation cas Genes
Previous transcriptome data suggested that Csa3b represses the expression of adaptation cas genes (He et al., 2017). Here a footprinting assay was carried out to determine the binding of Csa3b regulator on the csa1 promoter. The region from −168 to −128, relative to the start codon, of the promoter sequence was shown to be protected by Csa3b (Figure 3A). This protected sequence is similar to the Csa3b binding sequence on the csa5 promoter identified previously (He et al., 2017), located upstream of the Csa3a binding site on csa1 promoter (Figure 3A). These results, in combination with the previously reported transcriptome data that the deletion of csa3b up-regulated the expression of adaptation cas genes (He et al., 2017), indicated that Csa3b specifically bound the adaptation cas promoter to inhibit their transcription. Furthermore, DNA fragments of csa1 promoter, controlling the transcription of adaptation cas operon (Liu et al., 2015), and the Csa3b protein were used for LSPR analysis. Csa3b bound the full-length csa1 promoter in the LSPR analysis with the KD value of 4.24 ± 0.15 μM (Figure 3B). Importantly, the addition of 20 μM of the cOA analog 5′-CAAAA-3′ strongly enhanced the binding between csa1 promoter DNA and Csa3b (4.24 ± 0.15 vs. 0.30 ± 1.30 × 10–3 μM, Figures 3B,C), inferring that cOA might enhance the repression effect of Csa3b factor on adaptation cas genes.
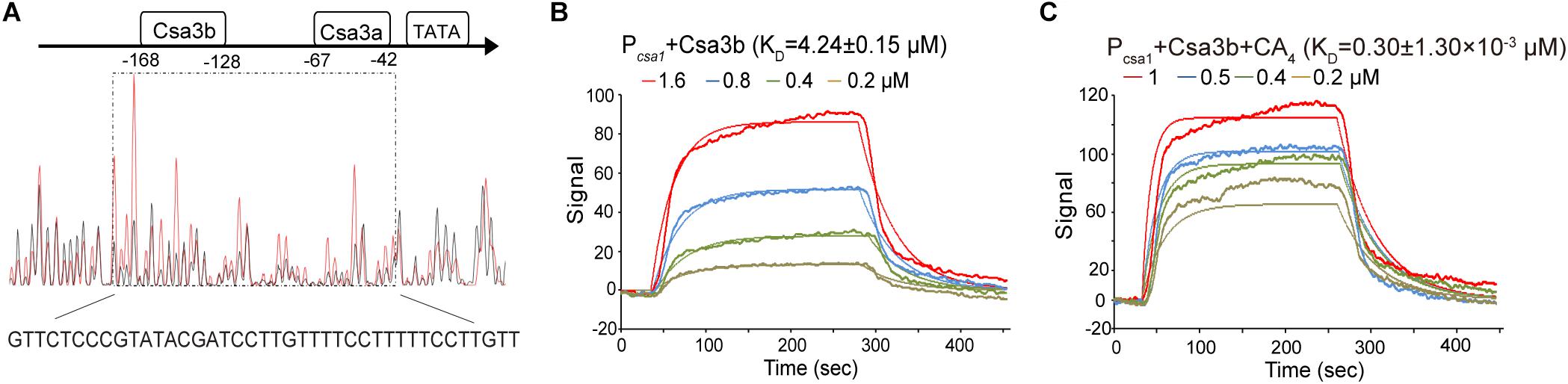
Figure 3. The oligonucleotide 5′-CAAAA-3′ enhances the binding between Csa3b and csa1 promoter. (A) DNase I footprinting assay with FAM-labeled coding strand of csa1 promoter in the presence (red peaks) or the absence (black peaks) of Csa3b. The protected region was boxed and the sequence was shown below the peaks. The Csa3a and Csa3b binding sites on the csa1 promoter, related to start codon, were indicated. (B,C) Localized surface plasmon resonance analysis of interaction between the fixed csa1 promoter DNA and the Csa3b regulator in the absence or the presence of 20 μM CA4. The KD value and the Csa3b concentrations for each assay were indicated.
The csa3b Deletion Strain Sampled Spacers From Mobile Genetic Elements
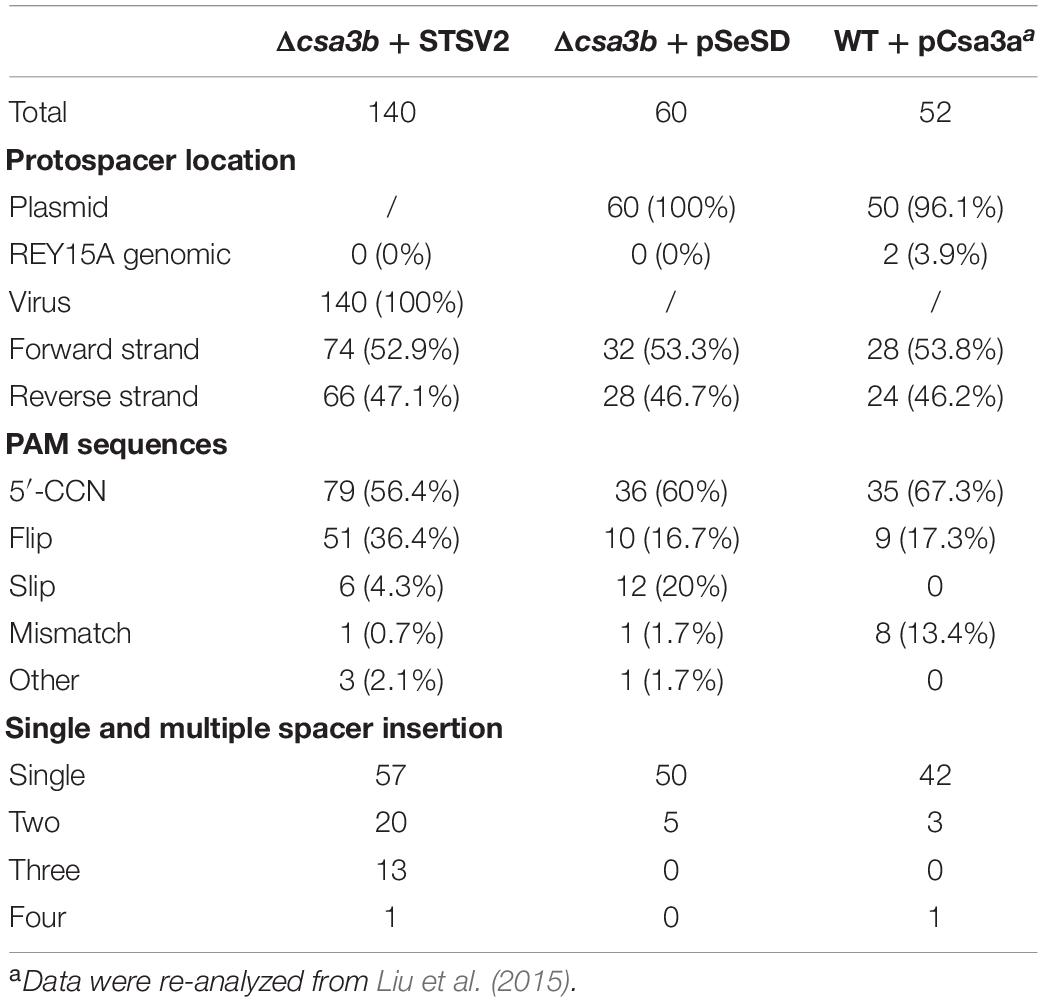
The wild-type and csa3b deletion cells carrying empty vector pSeSD were cultured for 18 days, and the cells were sampled every day for amplification of the leader-proximal region. No expanded band was detected in wild-type cells carrying pSeSD through the cultivation, while a weak expanded band was detected in the csa3b deletion strain carrying pSeSD at the beginning of cultivation (Figure 4A). Sequencing of the expanded bands from the csa3b deletion strain cells sampled at the first day revealed that most of the new spacers were adapted from the empty vector pSeSD (Table 2). This result indicated that Csa3b acted as a repressor which inhibited CRISPR acquisition in S. islandicus.
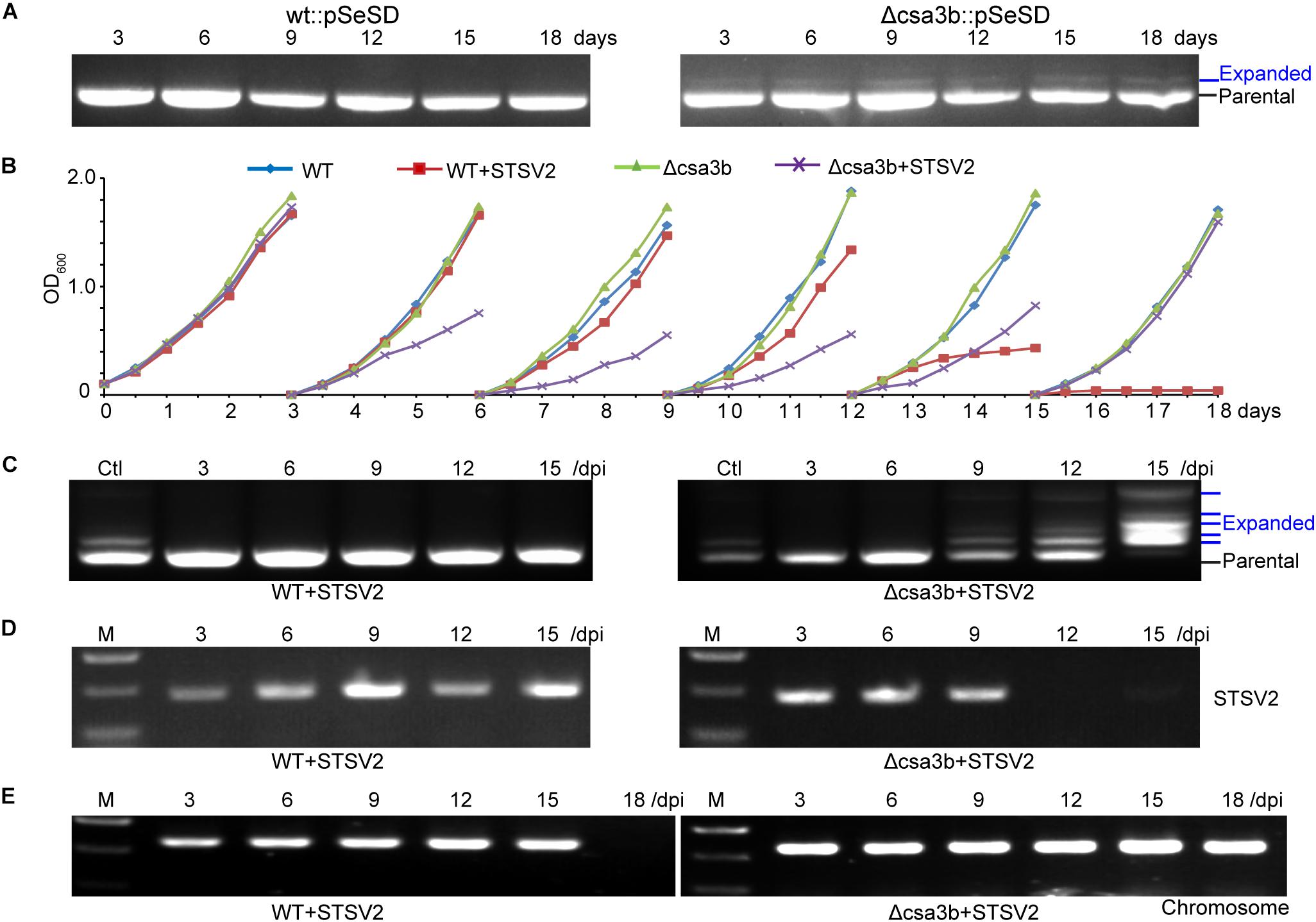
Figure 4. The deletion of the csa3b gene triggered spacer acquisition from plasmid and viral DNA. (A) PCR analysis of leader-proximal regions of CRISPR locus 1 of wt and csa3b deletion strains carrying the empty vector pSeSD. (B) Growth curves wt and csa3b deletion strains with or without infection of the Sulfolobus virus STSV2 in sucrose basic medium SCV at 78°C. Every third day following growth, 1 ml of cells was transferred into 50 ml of preheated fresh SCV medium. (C) PCR analysis of leader-proximal regions of CRISPR locus 1 of wt and csa3b deletion strains infected with STSV2. Multiple bands in addition to the parental band indicated the integration of new spacers. Ctl, positive control (Csa3a overexpression sample). (D) Semi-quantitative PCR analysis indicated the presence of STSV2 DNA in infected cells using gp17 as a marker. Sample collection times were indicated. (E) Semi-quantitative PCR analysis indicates the presence of host genomic DNA using tfe (transcriptional factor E) gene as the marker. Cells were collected immediately before each transfer and used as the PCR templates. All results represent three independent experiments of virus infection assays.
STSV2 is a single-tailed fusiform virus which infects Sulfolobus species, including S. islandicus REY15A (Erdmann et al., 2014a). STSV2 could not induce Sulfolobus CRISPR spacer acquisition in basic sugar medium SCV (Leon-Sobrino et al., 2016). In this study, STSV2 was used to infect the S. islandicus wild-type and csa3b gene deletion strains in SCV medium to see whether Csa3b was involved in the regulation of CRISPR immunity adaptation in the presence of the virus. At the first round of cultivation (first 3 days), the growth curves for all strains were similar and did not display any growth retardation due to STSV2 infection (Figure 4B). However, in the second and third rounds of cultivation (3rd–9th days), the STSV2-infected csa3b deletion strain grew much slower than the non-virus-infected wild-type and csa3b deletion strains as well as the STSV2-infected wild-type cells (Figure 4B). In the fourth round of cultivation (9th–12th days), the growth retardation of the STSV2-infected csa3b deletion strain stopped and, surprisingly, the growth of the STSV2-infected wild type cells became slower than that of the non-virus-infected wild-type and csa3b deletion strains (Figure 4B). Importantly, in the fifth round of cell cultivation (12th–15th days), the growth retardation of the STSV2-infected csa3b deletion strain was reduced, while the growth of the wild-type cells was inhibited by STSV2 infection (Figure 4B). Finally, in the last round of cultivation (15th–18th days), the growth curve of the STSV2-infected csa3b deletion strain recovered as did that of the non-virus-infected strains (Figure 4B). However, the growth of the wild-type cells was completely inhibited by STSV2 (Figure 4B). The virus infection experiments revealed that the STSV2 may completely inhibit the growth of wild-type cells during several rounds of cultivation in basic sugar medium and that the host CRISPR–Cas did not appear to function well against STSV2. However, the csa3b deletion strain underwent rounds of growth retardation and recovery (Figure 4B), indicating that some hidden mechanisms were involved in countering the virus (see below). The repeat experiments also showed similar growth curves.
PCR amplification of the leader proximal regions of the wild-type cells infected with STSV2 sampled at different time points showed no expanded bands as revealed by agarose gel electrophoresis, suggesting a weakness of spacer acquisition (Figure 4C), while the csa3a gene overexpression strain, as the positive control, showed expanded bands (Figure 4C). However, spacer acquisition occurred in the STSV2-infected csa3b deletion strain on day 9 (Figure 4C). Moreover, at 15 days, parental bands were not detected for either CRISPR array, and multiple expanded bands appeared according to the agarose gel analysis, indicating that a hyperactive spacer acquisition occurred (Figure 4C). As we know, host self-DNA is sampled into CRISPR arrays as new spacers during CRISPR adaptation in Sulfolobus (Liu et al., 2017), leading to self-immunity. This could probably explain the growth retardation at the early stage of STSV2 infection (Figure 4B). The semi-quantitative PCR of STSV2 gp17 gene did not detect viral DNA following spacer acquisition from STSV2 after day 12 (Figure 4D), while the PCR amplification signal of genomic csa3a in STSV2-infected wild-type cells disappeared in the last rounds of cultivation (Figure 4E). This result correlated with the growth curves and the PCR analyses of spacer acquisition. In summary, the deletion of csa3b relieved the repression of the adaptation cas genes and thereby induced hyperactive spacer acquisition from viral DNA.
The expanded bands representing the integrated spacers in the virus infection experiment were cloned and sequenced. In total, 56.4% of protospacers had a 5′-end CCN PAM, while 36.4% of protospacers had a 3′-end NGG motif, suggesting that these spacers were selected from protospacers with 5′-end CCN PAM but were inverted during integration (called flip mode) (Table 2). Similar characters of new spacers were found from the csa3b deletion strain carrying the empty vector (Table 2). In contrast, for spacer acquisition triggered by Csa3a overexpression, only 67.3% of protospacers carried a 5′-end CCN PAM, while 15.4% exhibited a 5′-end CDN or DCN PAM (D = A, T, or G) (Table 2; Liu et al., 2015). A re-analysis of the protospacers lacking 5′-end CCN, CNN, or NCN PAMs from the latter experiment identified a 3′-end NGG motif in 17.3% of the total protospacers (Table 2), suggesting that these spacers were inverted during integration. This result also revealed the differences between spacer integration models (precise, flipped, or slipped integration) with csa3b deletion as opposed to csa3a overexpression cells during CRISPR adaptation, suggesting that Csa3a might be involved in facilitating precise spacer integration.
Csa3b Regulator Bound the Promoters of CRISPR Subtype III-B cmr Genes
Upregulation of cmr-α and cmr-β by csa3b overexpression enabled putative Csa3b-binding sites to be identified in the promoter sequences of the first genes, cmr4α and cmr7, of the operons (Table 1). To test this, a DNase I footprinting assay was carried out to determine the binding between cmr4 promoter and to identify the precise binding region of Csa3b regulator on this promoter. The region from −230 to 190, relative to the start codon, of the promoter sequence was shown to be protected by Csa3b (Figure 5A). This sequence is similar to the Csa3b binding sequence on the csa5 promoter identified previously (He et al., 2017). This result indicated that Csa3b specifically bound cmr gene promoter. Furthermore, LSPR assay was used to study the interaction between Csa3b regulator and the promoter of cmr genes (cmr4 promoter). A strong affinity between Csa3b and cmr4 promoter DNA was detected, as revealed by the KD value of 17.20 ± 0.38 nM (Figure 5B). The cmr7 promoter, controlling the transcription of cmr-β genes, also carries a putative Csa3b binding motif at the region of −83 to −45 related to the start codon (Supplementary Figure 1). However, DNase I footprinting failed to identify the protected region on the cmr7 promoter (data not shown). In the electrophoretic mobility shift assay experiment, Csa3b formed a sharp retarded band with the full-length cmr7 promoter (P1, −221 to −1) (Supplementary Figure 1). The signal intensity of the retarded band was slightly reduced in the presence of a two- and fourfold excess of the cold DNA probe (Supplementary Figure 1). Mutations at the putative binding site completely abolished the signal of the retarded band (Supplementary Figure 1). These results, in combination with the transcriptome data (Table 1), indicated that Csa3b bound the promoter sequences of cmr genes to activate their transcription.
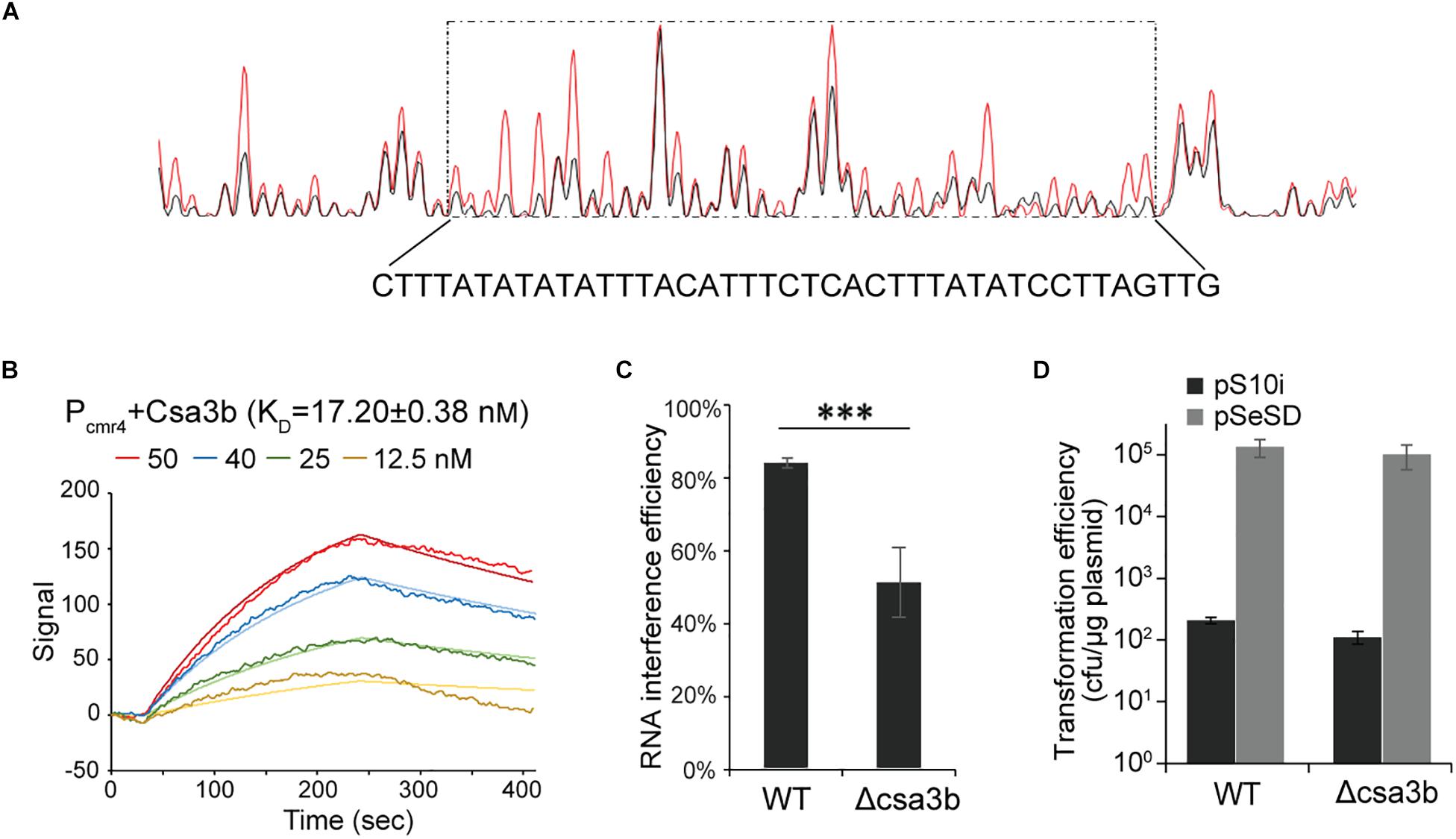
Figure 5. Csa3b bound the cmr promoter and enhance RNA cleavage activity. (A) DNase I footprinting assay with FAM-labeled coding strand of cmr4 promoter in presence (red peaks) or absence (black peaks) of Csa3b. The protected region (−230 to 190, related to start codon) was boxed and the sequence was shown below the peaks. (B) Localized surface plasmon resonance analysis of interaction between fixed cmr4 promoter DNA and Csa3b. KD value and Csa3b concentrations were indicated. (C) RNA interference efficiency against plasmid expressed lacS gene of S. islandicus wt and csa3b deletion strains determined by β-galactosidase assay. (D) DNA interference activities of CRISPR–Cmr system in S. islandicus wt and csa3b deletion strains analyzed by calculation of transformation efficiency of the challenging plasmid carrying protospacers paired with spacer 10 from the CRISPR array 2. For RNA and DNA interference assays, all experiments were performed in triplicate and the results are presented as mean ± SD. The error bars indicate SD. The “∗∗∗” means significant difference.
Csa3b Enhances the RNA Interference Activity of Subtype III-B Cmr Systems
Csa3b can bind directly to the promoters of two cmr cassettes and activates their transcription in S. islandicus (Figure 5, Supplementary Figure 1, and Table 1). Here the RNA and DNA interference activities of CRISPR–Cmr systems in S. islandicus wild-type (wt) and csa3b gene deletion (Δcsa3b) strains were studied. The RNA interference plasmid, carrying the “repeat–spacer–repeat” element used to target the lacS transcript from a plasmid-encoded lacS gene, was transformed into S. islandicus wt and Δcsa3b strains. In the wt cells, the RNA interference efficiency was over 80%, according to the LacS activity assay (Figure 5C). However, ca. 50% of lacS transcript was cleaved, which was significantly lower than that of the wt strain (Figure 5C). This result revealed that Csa3b was important for the regulation of subtype III-B-mediated RNA interference in S. islandicus.
In order to study the effect of csa3b deletion on DNA interference, the Cmr-mediated transcription-dependent DNA interference was analyzed using an RNA/DNA targeting plasmid. In detail, the spacer 10 sequence of CRISPR locus 2 was cloned into the pSeSD shuttle vector. This produced the DNA interference plasmid pS10i, which will only be cleaved by the Cmr systems with the transcription-dependent DNA cleavage activity due to the lacking CCN PAM sequence required for subtype I-A system (Deng et al., 2013). The plasmid transformation efficiencies of both wild-type and Δcsa3b cells were very low (Figure 5D), compared with the transformation efficiency of the empty vector pSeSD, suggesting that Csa3b has less effect on the regulation of Cmr-mediated DNA interference. These results suggested that the cmr genes were expressed in the absence of the csa3b gene, and their expression level was probably sufficient for efficient Cmr-mediated DNA interference in this study using a plasmid as the targeted mobile genetic element (MGE).
Discussion
Previously, we demonstrated that Csa3a transcriptionally activated the expression of adaptation cas genes and CRISPR RNAs for de novo spacer acquisition and target interference (Liu et al., 2015, 2017; Zhang et al., 2019). Csa3a carries a CARF domain and was previously suggested to bind a ligand for its regulatory effect (Lintner et al., 2011). However, this ligand was not identified until recently, where cOAs were found to be bound by the CARF domain of Csm6 (Kazlauskiene et al., 2017; Niewoehner et al., 2017) or Csx1 (Han et al., 2018) to regulate their ribonuclease activity. However, whether cOAs regulate the regulatory effect of Csa3a on CRISPR adaptation remains unclear. In this study, we found that another CRISPR-associated factor, Csa3b, specifically bound the promoter of adaptation cas genes (Figure 3), and the deletion of csa3b gene triggers CRISPR spacer acquisition from both plasmid and viral DNA (Figure 4). Moreover, the interaction between csa1 promoter DNA and Csa3b was strongly enhanced in the presence of the cOA analog (5′-CAAAA-3′) according to the LSPR analysis (Figures 3B,C), suggesting the allosteric regulation on DNA binding.
Supported by the findings of the current study and our previous findings (Liu et al., 2015, 2017), as well as by a recent report of He et al. (2017), we summarize an integrative regulation model mediated by Csa3a and Csa3b regulators for the transcriptional regulation of CRISPR spacer acquisition. In the absence of MGEs, Csa3b acts as a repressor to repress subtype I-A interference and adaptation modules (Figure 6; He et al., 2017). The repression of CRISPR adaptation avoids the uptake of self-DNA into CRISPR arrays at this stage in the absence of invasive genetic elements. As previously revealed, the CRISPR–Cas system adapted spacers from both genome and invasive DNA in Sulfolobus subtype I-A system (Liu et al., 2017). Csa3b also acts as an activator of subtype III-B cmr genes (Figure 6). Upon invasion of MGEs, the Cascade complex is released from interference cas gene promoter and binds the target DNA via crRNA base pairing. Thus, the transcription of interference cas genes is derepressed (He et al., 2017). Csa3b still binds the promoters of cmr genes and activates their expression, leading to yielding more signaling cOA molecules if Cmr complex cleaves target RNA. At this stage, the expression of Csa3a regulator is activated by invasive genetic elements via an unknown mechanism (Leon-Sobrino et al., 2016) to trigger CRIPSR adaptation (Liu et al., 2015). Csa3a also binds the promoters of CRISPR arrays and activates the transcription of CRISPR RNAs for interference at this stage (Liu et al., 2017; Figure 6). The activation of Cmr expression by Csa3b could further provide more cOAs. At the final stage of CRIPSR immunity, cOAs enhance the binding ability of Csa3b with the promoter of adaptation cas genes to inhibit their transcription, providing feedback regulation of CRIPSR adaptation (Figure 6).
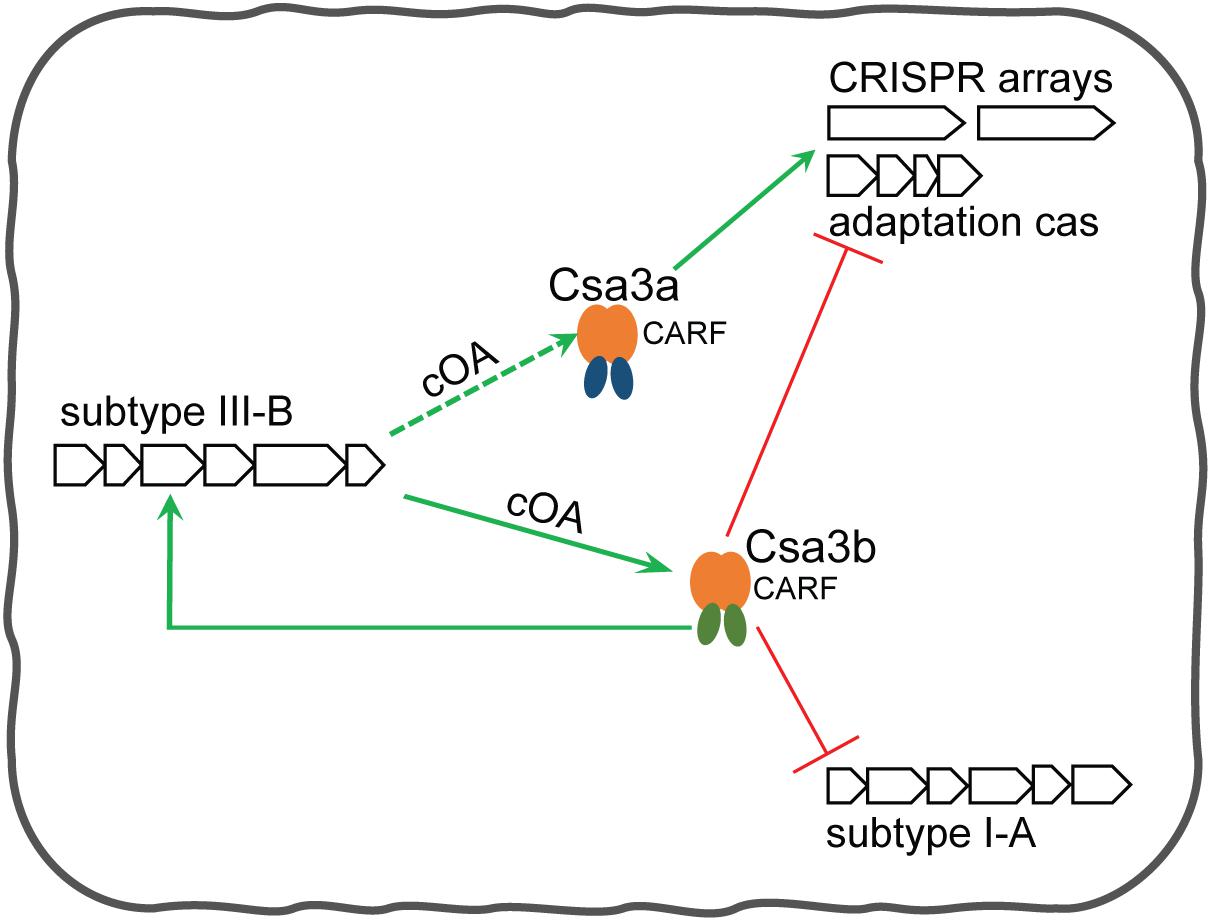
Figure 6. Csa3b-mediated regulation model for CRISPR spacer acquisition and target interferences in Sulfolobus. In the absence of mobile genetic elements (MGEs), (1) Csa3b together with Cascade binds the promoter of subtype I-A interference cas genes to repress their expression, and (2) Csa3b binds the promoter of adaptation cas genes to repress their transcription and (3) binds the promoters of subtype III-B cmr genes to activate their transcription. In the presence of invasive genetic elements, (1) Csa3b binds the promoters of cmr genes and activates their expression, (2) Cascade is released from the promoter of subtype I-A interference genes and their expression is de-repressed (He et al., 2017), and (3) another CRISPR-associated factor, Csa3a, is activated via an unknown mechanism upon MGE invasion. Csa3a activates the expression of the adaptation module (Liu et al., 2015) and enhances CRISPR transcription (Liu et al., 2017) for CRISPR immunity. Csa3b-activated Cmr expression would enhance the synthesis of signaling molecules cOAs. These signaling molecules could further enhance the binding between Csa3b and adaptation cas promoter through allosteric regulation, providing feedback regulation for repression of the adaptation module at the late stage of the host-MGE interaction. Since Csa3a factor also carries a CARF domain, its regulatory effects could also be regulated by the signaling molecules.
This hypothesis is reinforced by the transcriptome data from virus-infected S. islandicus cells (Leon-Sobrino et al., 2016). In that study, new spacers were detected approximately 7 days post-infection, when the transcript levels of csa3a increased fourfold while those of csa3b did not change significantly in rich medium (Leon-Sobrino et al., 2016). However, beyond this time point, csa3a transcription decreased and csa3b transcription increased, probably due to the cells needing to restrict the functions of the CRISPR–Cas system in order to avoid excessive adaptation (Leon-Sobrino et al., 2016). However, spacer acquisition was quite low in non-rich (SCV) medium where no change in the expression of csa3a and csa3b was evident in STSV2-infected cells (Leon-Sobrino et al., 2016). These results strongly support our hypothesis of Csa3a- and Csa3b-mediated transcriptional regulation of CRISPR adaptation.
Data Availability Statement
The datasets generated for this study can be found in the online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
QY, XZ, JL, and ZZe conducted the experiments. ZZh, TL, and YL analyzed the data. QY, WH, and NP designed the experiments. QY and NP wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (Nos. 91751104 and 31671291 to NP and No. 31900400 to TL), the National Postdoctoral Program for Innovative Talents (No. BX20180112 to TL), and the Fundamental Research Funds for the Central Universities (No. 2662019PY028 to NP). Funding for open access charge was provided by the National Natural Science Foundation of China (No. 31671291).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.02038/full#supplementary-material
References
Athukoralage, J. S., Rouillon, C., Graham, S., Gruschow, S., and White, M. F. (2018). Ring nucleases deactivate type III CRISPR ribonucleases by degrading cyclic oligoadenylate. Nature 562, 277–280. doi: 10.1038/s41586-018-0557-5
Barrangou, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. doi: 10.1126/science.1138140
Brouns, S. J., Jore, M. M., Lundgren, M., Westra, E. R., Slijkhuis, R. J., Snijders, A. P., et al. (2008). Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321, 960–964. doi: 10.1126/science.1159689
Cady, K. C., Bondy-Denomy, J., Heussler, G. E., Davidson, A. R., and O’toole, G. A. (2012). The CRISPR/Cas adaptive immune system of Pseudomonas aeruginosa mediates resistance to naturally occurring and engineered phages. J. Bacteriol. 194, 5728–5738. doi: 10.1128/jb.01184-12
Deng, L., Garrett, R. A., Shah, S. A., Peng, X., and She, Q. (2013). A novel interference mechanism by a type IIIB CRISPR-Cmr module in Sulfolobus. Mol. Microbiol. 87, 1088–1099. doi: 10.1111/mmi.12152
Deng, L., Zhu, H., Chen, Z., Liang, Y. X., and She, Q. (2009). Unmarked gene deletion and host-vector system for the hyperthermophilic crenarchaeon Sulfolobus islandicus. Extremophiles 13, 735–746. doi: 10.1007/s00792-009-0254-2
Erdmann, S., Chen, B., Huang, X., Deng, L., Liu, C., Shah, S. A., et al. (2014a). A novel single-tailed fusiform Sulfolobus virus STSV2 infecting model Sulfolobus species. Extremophiles 18, 51–60. doi: 10.1007/s00792-013-0591-z
Erdmann, S., Le Moine Bauer, S., and Garrett, R. A. (2014b). Inter-viral conflicts that exploit host CRISPR immune systems of Sulfolobus. Mol. Microbiol. 91, 900–917. doi: 10.1111/mmi.12503
Erdmann, S., and Garrett, R. A. (2012). Selective and hyperactive uptake of foreign DNA by adaptive immune systems of an archaeon via two distinct mechanisms. Mol. Microbiol. 85, 1044–1056. doi: 10.1111/j.1365-2958.2012.08171.x
Han, W., Pan, S., Lopez-Mendez, B., Montoya, G., and She, Q. (2017). Allosteric regulation of Csx1, a type IIIB-associated CARF domain ribonuclease by RNAs carrying a tetraadenylate tail. Nucleic Acids Res. 45, 10740–10750. doi: 10.1093/nar/gkx726
Han, W., Stella, S., Zhang, Y., Guo, T., Sulek, K., Peng-Lundgren, L., et al. (2018). A Type III-B Cmr effector complex catalyzes the synthesis of cyclic oligoadenylate second messengers by cooperative substrate binding. Nucleic Acids Res. 46, 10319–10330. doi: 10.1093/nar/gky844
He, F., Vestergaard, G., Peng, W., She, Q., and Peng, X. (2017). CRISPR-Cas type I-A Cascade complex couples viral infection surveillance to host transcriptional regulation in the dependence of Csa3b. Nucleic Acids Res. 45, 1902–1913. doi: 10.1093/nar/gkw1265
Jia, N., Jones, R., Yang, G., Ouerfelli, O., and Patel, D. J. (2019). CRISPR-Cas III-A Csm6 CARF domain is a ring nuclease triggering stepwise cA4 cleavage with ApA>p formation terminating RNase activity. Mol. Cell 75, 944–956. doi: 10.1016/j.molcel.2019.06.014
Kazlauskiene, M., Kostiuk, G., Venclovas, C., Tamulaitis, G., and Siksnys, V. (2017). A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science 357, 605–609. doi: 10.1126/science.aao0100
Kieper, S. N., Almendros, C., Behler, J., Mckenzie, R. E., Nobrega, F. L., Haagsma, A. C., et al. (2018). Cas4 facilitates PAM-compatible spacer selection during CRISPR adaptation. Cell Rep. 22, 3377–3384. doi: 10.1016/j.celrep.2018.02.103
Koonin, E. V., and Makarova, K. S. (2013). CRISPR-Cas: evolution of an RNA-based adaptive immunity system in prokaryotes. RNA Biol. 10, 679–686. doi: 10.4161/rna.24022
Lee, H., Zhou, Y., Taylor, D. W., and Sashital, D. G. (2018). Cas4-dependent prespacer processing ensures high-fidelity programming of CRISPR arrays. Mol. Cell 70, 48–59. doi: 10.1016/j.molcel.2018.03.003
Leon-Sobrino, C., Kot, W. P., and Garrett, R. A. (2016). Transcriptome changes in STSV2-infected Sulfolobus islandicus REY15A undergoing continuous CRISPR spacer acquisition. Mol. Microbiol. 99, 719–728. doi: 10.1111/mmi.13263
Li, M., Wang, R., and Xiang, H. (2014a). Haloarcula hispanica CRISPR authenticates PAM of a target sequence to prime discriminative adaptation. Nucleic Acids Res. 42, 7226–7235. doi: 10.1093/nar/gku389
Li, M., Wang, R., Zhao, D., and Xiang, H. (2014b). Adaptation of the Haloarcula hispanica CRISPR-Cas system to a purified virus strictly requires a priming process. Nucleic Acids Res. 42, 2483–2492. doi: 10.1093/nar/gkt1154
Lintner, N. G., Frankel, K. A., Tsutakawa, S. E., Alsbury, D. L., Copie, V., Young, M. J., et al. (2011). The structure of the CRISPR-associated protein Csa3 provides insight into the regulation of the CRISPR/Cas system. J. Mol. Biol. 405, 939–955. doi: 10.1016/j.jmb.2010.11.019
Liu, T., Li, Y., Wang, X., Ye, Q., Li, H., Liang, Y., et al. (2015). Transcriptional regulator-mediated activation of adaptation genes triggers CRISPR de novo spacer acquisition. Nucleic Acids Res. 43, 1044–1055. doi: 10.1093/nar/gku1383
Liu, T., Liu, Z., Ye, Q., Pan, S., Wang, X., Li, Y., et al. (2017). Coupling transcriptional activation of CRISPR–Cas system and DNA repair genes by Csa3a in Sulfolobus islandicus. Nucleic Acids Res. 45, 8978–8992. doi: 10.1093/nar/gkx612
Makarova, K. S., Anantharaman, V., Grishin, N. V., Koonin, E. V., and Aravind, L. (2014). CARF and WYL domains: ligand-binding regulators of prokaryotic defense systems. Front. Genet. 5:102. doi: 10.3389/fgene.2014.00102
Makarova, K. S., Haft, D. H., Barrangou, R., Brouns, S. J., Charpentier, E., Horvath, P., et al. (2011). Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 9, 467–477. doi: 10.1038/nrmicro2577
Makarova, K. S., Wolf, Y. I., Alkhnbashi, O. S., Costa, F., Shah, S. A., Saunders, S. J., et al. (2015). An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 13, 722–736. doi: 10.1038/nrmicro3569
Molina, R., Stella, S., Feng, M., Sofos, N., Jauniskis, V., Pozdnyakova, I., et al. (2019). Structure of Csx1-cOA4 complex reveals the basis of RNA decay in Type III-B CRISPR-Cas. Nat. Commun. 10:4302. doi: 10.1038/s41467-019-12244-z
Niewoehner, O., Garcia-Doval, C., Rostol, J. T., Berk, C., Schwede, F., Bigler, L., et al. (2017). Type III CRISPR-Cas systems produce cyclic oligoadenylate second messengers. Nature 548, 543–548. doi: 10.1038/nature23467
Patterson, A. G., Yevstigneyeva, M. S., and Fineran, P. C. (2017). Regulation of CRISPR-Cas adaptive immune systems. Curr. Opin. Microbiol. 37, 1–7. doi: 10.1016/j.mib.2017.02.004
Peng, N., Deng, L., Mei, Y., Jiang, D., Hu, Y., Awayez, M., et al. (2012). A synthetic arabinose-inducible promoter confers high levels of recombinant protein expression in hyperthermophilic archaeon Sulfolobus islandicus. Appl. Environ. Microbiol. 78, 5630–5637. doi: 10.1128/AEM.00855-12
Peng, N., Xia, Q., Chen, Z., Liang, Y. X., and She, Q. (2009). An upstream activation element exerting differential transcriptional activation on an archaeal promoter. Mol. Microbiol. 74, 928–939. doi: 10.1111/j.1365-2958.2009.06908.x
Peng, W., Feng, M., Feng, X., Liang, Y. X., and She, Q. (2015). An archaeal CRISPR type III-B system exhibiting distinctive RNA targeting features and mediating dual RNA and DNA interference. Nucleic Acids Res. 43, 406–417. doi: 10.1093/nar/gku1302
Perez-Rodriguez, R., Haitjema, C., Huang, Q., Nam, K. H., Bernardis, S., Ke, A., et al. (2011). Envelope stress is a trigger of CRISPR RNA-mediated DNA silencing in Escherichia coli. Mol. Microbiol. 79, 584–599. doi: 10.1111/j.1365-2958.2010.07482.x
Pul, Ü, Wurm, R., Arslan, Z., Geißen, R., Hofmann, N., and Wagner, R. (2010). Identification and characterization of E. coli CRISPR-cas promoters and their silencing by H-NS. Mol. Microbiol. 75, 1495–1512. doi: 10.1111/j.1365-2958.2010.07073.x
Richter, C., Dy, R. L., Mckenzie, R. E., Watson, B. N., Taylor, C., Chang, J. T., et al. (2014). Priming in the Type I-F CRISPR-Cas system triggers strand-independent spacer acquisition, bi-directionally from the primed protospacer. Nucleic Acids Res. 42, 8516–8526. doi: 10.1093/nar/gku527
Shiimori, M., Garrett, S. C., Graveley, B. R., and Terns, M. P. (2018). Cas4 nucleases define the PAM, length, and orientation of DNA fragments integrated at CRISPR loci. Mol. Cell 70, 814–824. doi: 10.1016/j.molcel.2018.05.002
Sternberg, S. H., Richter, H., Charpentier, E., and Qimron, U. (2016). Adaptation in CRISPR-cas systems. Mol. Cell 61, 797–808. doi: 10.1016/j.molcel.2016.01.030
Swarts, D. C., Mosterd, C., Van Passel, M. W., and Brouns, S. J. (2012). CRISPR interference directs strand specific spacer acquisition. PLoS One 7:e35888. doi: 10.1371/journal.pone.0035888
Westra, E. R., Pul, U., Heidrich, N., Jore, M. M., Lundgren, M., Stratmann, T., et al. (2010). H-NS-mediated repression of CRISPR-based immunity in Escherichia coli K12 can be relieved by the transcription activator LeuO. Mol. Microbiol. 77, 1380–1393. doi: 10.1111/j.1365-2958.2010.07315.x
Westra, E. R., Swarts, D. C., Staals, R. H., Jore, M. M., Brouns, S. J., and Van Der Oost, J. (2012). The CRISPRs, they are a-changin’: how prokaryotes generate adaptive immunity. Annu. Rev. Genet. 46, 311–339. doi: 10.1146/annurev-genet-110711-155447
Yang, C.-D., Chen, Y.-H., Huang, H.-Y., Huang, H.-D., and Tseng, C.-P. (2014). CRP represses the CRISPR/Cas system in Escherichia coli: evidence that endogenous CRISPR spacers impede phage P1 replication. Mol. Microbiol. 92, 1072–1091. doi: 10.1111/mmi.12614
Yosef, I., Goren, M. G., and Qimron, U. (2012). Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 40, 5569–5576. doi: 10.1093/nar/gks216
Yosef, I., Shitrit, D., Goren, M. G., Burstein, D., Pupko, T., and Qimron, U. (2013). DNA motifs determining the efficiency of adaptation into the Escherichia coli CRISPR array. Proc. Natl. Acad. Sci. U.S.A. 110, 14396–14401. doi: 10.1073/pnas.1300108110
Zhang, C., Tian, B., Li, S., Ao, X., Dalgaard, K., Gokce, S., et al. (2013). Genetic manipulation in Sulfolobus islandicus and functional analysis of DNA repair genes. Biochem. Soc. Trans. 41, 405–410. doi: 10.1042/bst20120285
Keywords: Sulfolobus islandicus, CRISPR adaptation, Cmr, transcriptional regulation, Csa3b
Citation: Ye Q, Zhao X, Liu J, Zeng Z, Zhang Z, Liu T, Li Y, Han W and Peng N (2020) CRISPR-Associated Factor Csa3b Regulates CRISPR Adaptation and Cmr-Mediated RNA Interference in Sulfolobus islandicus. Front. Microbiol. 11:2038. doi: 10.3389/fmicb.2020.02038
Received: 24 May 2020; Accepted: 31 July 2020;
Published: 26 August 2020.
Edited by:
Brian P. Hedlund, University of Nevada, Las Vegas, United StatesReviewed by:
Shiraz Ali Shah, Copenhagen Prospective Studies on Asthma in Childhood (COPSAC), DenmarkZhenfeng Zhang, Institute of Microbiology (CAS), China
Copyright © 2020 Ye, Zhao, Liu, Zeng, Zhang, Liu, Li, Han and Peng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nan Peng, nanp@mail.hzau.edu.cn
 Qing Ye
Qing Ye Xueqiao Zhao
Xueqiao Zhao Jilin Liu
Jilin Liu Zhifeng Zeng
Zhifeng Zeng Zhufeng Zhang
Zhufeng Zhang Tao Liu
Tao Liu Yingjun Li
Yingjun Li Wenyuan Han
Wenyuan Han Nan Peng
Nan Peng