Corrigendum: Diverse Mobile Genetic Elements and Conjugal Transferability of Sulfonamide Resistance Genes (sul1, sul2, and sul3) in Escherichia coli Isolates From Penaeus vannamei and Pork From Large Markets in Zhejiang, China
- Key Laboratory of Marine Food Quality and Hazard Controlling Technology of Zhejiang Province, College of Life Sciences, China Jiliang University, Hangzhou, China
High prevalence rates of sulfonamide resistance genes sul1, sul2, and sul3 have been observed in Gram-negative bacteria isolated from humans, domestic animals, and aquaculture species worldwide. We investigated the distribution characteristics, location, conjugative transferability, and genetic environments of sul genes from Escherichia coli isolates collected from Penaeus vannamei and pork samples from three large markets in Zhejiang, China. The prevalence rates of sul genes in sulfonamide-resistant E. coli isolates from P. vannamei and pork samples were 90.0 and 88.6%, respectively, and the prevalence of sul1 and sul2 was significantly higher than that of sul3 (p < 0.05). Twenty-four representative sul-positive E. coli isolates were analyzed in detail. Southern blot hybridization confirmed that sul genes of E. coli isolates were located on plasmids and/or chromosomes. Transfer of resistance through conjugation was observed in all 18 E. coli isolates harboring sul genes on plasmids. Replicon typing identified seven different incompatibility groups and IncF was the dominant replicon type among sul gene-containing plasmids from both sources. PCR walking analysis indicated that 87.5% (35/40) of sul gene-related fragments carried insertion sequences (ISs) belonging to a variety of families in diverse sites, with IS26 occurring most frequently. In addition, the sul1 gene was detected mainly in fragments carrying class 1 integrons. Co-location on the same fragment with resistance genes that may contribute to the persistence and dissemination of sul1 and/or sul2 genes. The diversity of mobile genetic elements and resistance genes adjacent to sul3 was much lower than those adjacent to sul1 and sul2, especially those located in chromosomes, which reduced the transmission potential of the sul3 gene. In conclusion, combined with the results of clonal relatedness analysis by PFGE and MLST of 24 representative E. coli isolates from P. vannamei and pork samples, it showed that a small number of sul genes were vertically transmitted among E. coli from P. vannamei and that horizontal gene transfer was likely the main transmission mechanism of sul genes from both sources. Our results provide important information to better understand the risk of transmission of sul genes from seafood and meat to humans.
Introduction
Sulfonamides, which are fully synthetic antibiotics, have been widely used to treat bacterial and protozoan infections in humans, domestic animals, and aquaculture species (Blahna et al., 2006; Peixoto et al., 2016). Although the use of sulfonamides in human medicine has decreased in developed countries, they are still frequently used in developing Asian countries owing to their low price and availability (Changkaew et al., 2014). In veterinary medicine, sulfonamides are considered “high priority” due to high usage and their high potential to enter the environment (Heuer and Smalla, 2007; Wang et al., 2014).
In the past decade, high prevalence rates of sulfonamide resistance have been observed in mainly Gram-negative bacteria isolated from animals and humans all over the world (Wu et al., 2010; Card et al., 2016; Ben et al., 2017). Resistance to sulfonamides occurs principally through the acquisition of the alternative dihydropteroate synthase (DHPS) gene sul, the product of which has a low affinity for sulfonamides (Changkaew et al., 2014). Unlike resistance genes to other classes of antimicrobials such as tetracycline, which are encoded by many different genes, only three known sulfonamide resistance (sul) genes (sul1, sul2, and sul3) have been identified (Wu et al., 2010; Ben et al., 2017). Based on alignments using MegAlign software, the nucleotide sequences of sul1, sul2, and sul3 are approximately 50% similar (Hsu et al., 2014).
Horizontal or vertical transfer of mobile genetic elements (MGEs) that harbor resistance genes is considered the main mechanism of dissemination of antibiotic resistance genes (Rehman et al., 2017). sul genes have been identified on both chromosomes and plasmids and are often associated with MGEs such as transposons, integrons and insertion sequences (ISs) (Wu et al., 2010). MGEs enable translocation of sul genes between chromosomes and plasmids. In addition, plasmids carrying sul genes can spread among bacteria of the same or different species or genera by conjugation or transformation, thereby disseminating sul genes.
Penaeus vannamei, also known as Pacific whiteleg shrimp, are among the most popular culturable penaeid shrimp in China, and China has been the world’s largest producer of P. vannamei since 2001 (Global Aquaculture Production 1950−2016, FAO)1. Similarly, in China, pork accounts for approximately 64% of all meat production and is consistently the most consumed meat (Ministry of Agriculture and Rural Affairs of the People’s Republic of China, March 2019)2. Moreover, sulfonamides are among the most commonly used antibiotics at swine farms, second only to tetracyclines (Hsu et al., 2014), and are widely used in P. vannamei culture for the prevention and treatment of common bacterial diseases (Gao et al., 2012). Excessive usage of sulfonamides in the shrimp farming and pig breeding industries in China imposes widespread selective pressures on bacteria, leading to the enrichment of sulfonamide resistant strains in shrimp and pork products that are capable of spreading between different environments (Liu et al., 2019; Yuan et al., 2019). In our study, P. vannamei and pork were chosen to study the prevalence of sulfonamide resistance in three large markets in Zhejiang, China to assess (i) whether differences occur in the distribution of sul genes in Escherichia coli isolates from samples of two different host species collected from markets, (ii) whether vertical or horizontal transmission of sulfonamide resistant E. coli occurs between P. vannamei and pork in market environments, despite the different growth conditions of the two animals and (iii) whether regional spreading of sulfonamide resistant E. coli in the same or different host species occurs between different markets.
Escherichia coli are ubiquitous commensal bacteria, and certain strains are zoonotic pathogens (Zhang et al., 2013). E. coli are also considered effective model organisms for studying the essential processes of life due to their fast growth rate, minimal nutritional requirements, comprehensively understood biological processes, and extensively characterized genetics (Jiang et al., 2016; Paraoan et al., 2017). More importantly, E. coli are often used to monitor antimicrobial resistance, as they have a wide range of hosts and can easily acquire resistance genes via horizontal gene transfer (Fang et al., 2019). Many nutritious aquatic and meat products are considered reservoirs of E. coli as well as some antimicrobial resistance genes (Alvarez-Fernandez et al., 2013; Skočková et al., 2015; Boss et al., 2016).
Here we investigated sul genes in E. coli isolates collected from P. vannamei and pork samples from three large markets in Zhejiang Province, China. The clonal relatedness among the representative sul-positive E. coli isolates was analyzed by pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST). In addition, the location, genetic environment, and conjugative transferability of sul genes were further researched to assess the potential transmission ability of sul genes in E. coli from two different animal host species.
Materials and Methods
Shrimp and Pork Sampling and E. coli Isolation
A total of 180 P. vannamei and 180 pork samples were collected from three different open markets (n = 60 per market) in Zhejiang Province, China between June 2017 and June 2018 (Table 1). Samples were collected from each market every 4 months (June 2017, October 2017, February 2018, and June 2018). A total of 90 samples were collected from the three markets at each visit, including 15 P. vannamei and 15 pork samples from each market. The methods for sample treatment and E. coli isolation and identification are described in detail in our previously published studies (Cheng et al., 2019; Fang et al., 2019). Following the previous protocol, one E. coli isolate was obtained from each sample (Changkaew et al., 2014; Boss et al., 2016; Cheng et al., 2019; Fang et al., 2019). Colonies were stored at −80°C until further examination.
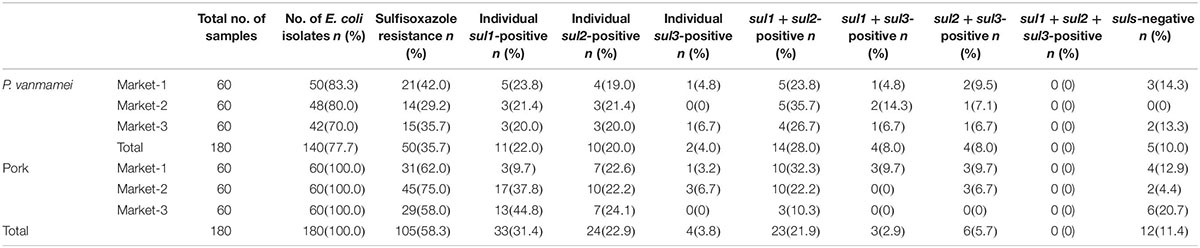
Table 1. Distribution characteristics of sulfonamide resistance (sul) genes present in Escherichia coli isolated from Penaeus vanmamei and pork samples from three different open markets.
Phenotypic Screening for Sulfonamide Resistance
All confirmed E. coli isolates collected from P. vannamei and pork samples were assessed for susceptibility to sulfisoxazole using the disk diffusion method on Mueller-Hinton (MH) agar (Oxoid Ltd., Basingstoke, United Kingdom) in accordance with the guidelines of the (Clinical and Laboratory Standards Institute [CLSI], 2017). According to CLSI guidelines, sulfisoxazole can be used to test all known types of sulfonamide resistance. Sulfisoxazole (300 μg) antibiotic disks were obtained from Hangzhou Microbial Reagent Co., Ltd. (Hangzhou, China). E. coli isolates were incubated overnight at 37°C and diluted to a turbidity equivalent to a 0.5 McFarland standard. The inoculated MH agar plates were then incubated at 37°C for 24 h, and the size of the zone of inhibition was used to classify isolates as susceptible, intermediate or resistant according to CLSI guidelines. E. coli ATCC 25922 was used as a sensitivity control.
Detection of sul Genes
Polymerase chain reaction (PCR) was used to detect the presence of sul genes in all sulfisoxazole-resistant E. coli isolates. The boiling method described in our previous studies was used to obtain genomic DNA templates (Cheng et al., 2019). The primers used to detect sul genes are shown in Table 2. Each PCR amplification reaction was performed in a 25 μL mixture containing 400 nM each primer, 10 × PCR buffer, 200 mM each deoxynucleotide triphosphate (dNTP), 5 U Ex-Taq DNA polymerase (Takara-Bio, Beijing, China) and 250 ng DNA as the template. PCR amplification was initiated by incubating the reaction mixture at 94°C for 1 min; followed by 30 cycles at 98°C for 30 s, annealing at 56°C (sul2) or 55°C (sul1 and sul3) for 30 s, and extension at 72°C for 30 s; and a final extension at 72°C for 10 min. PCR products (5 μL) were mixed with 1 μL 6 × loading buffer dye and analyzed by electrophoresis on a 1.2% agarose gel. Positive and negative controls were included in all PCR reactions.
Clonal Analysis by Molecular Typing
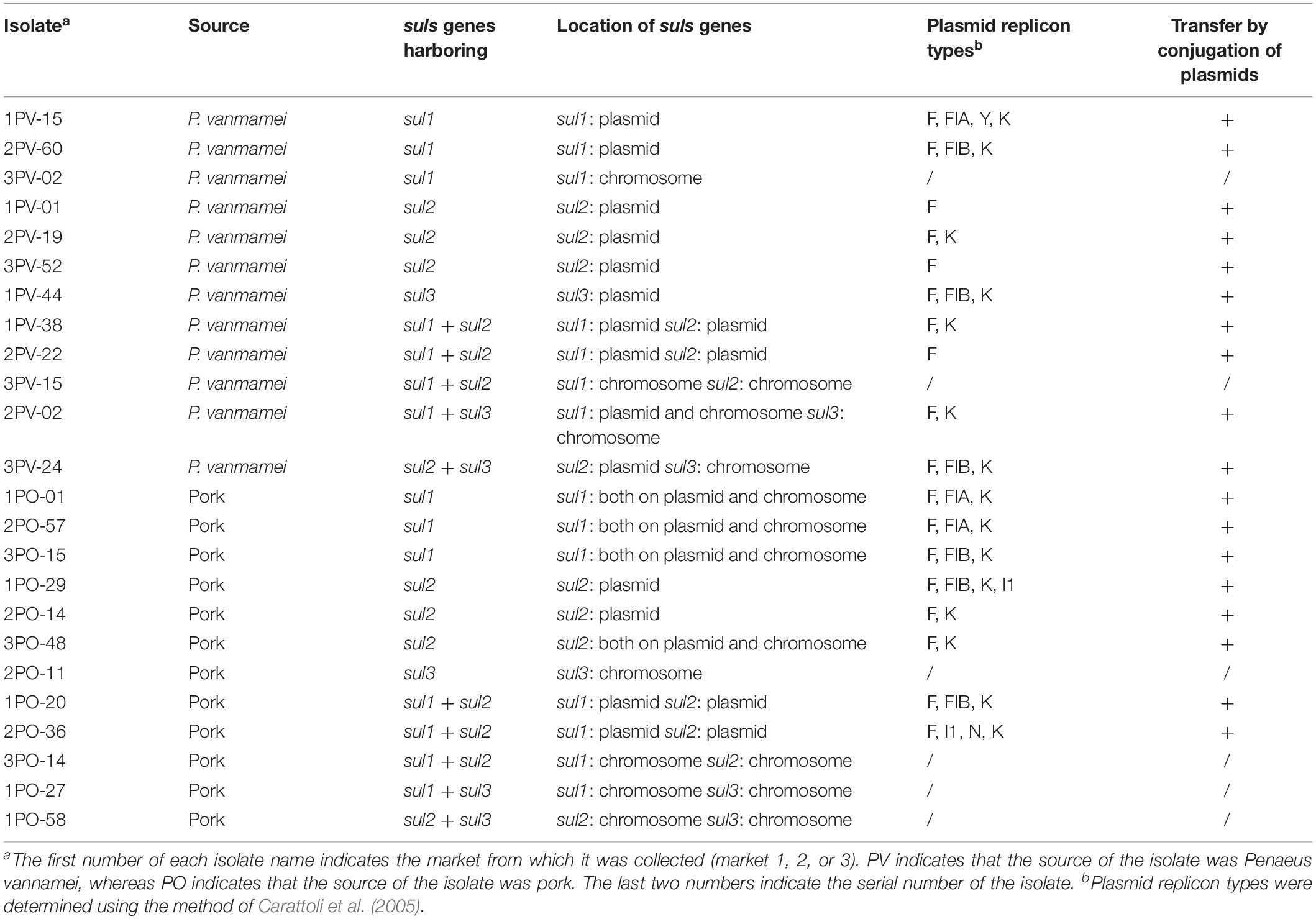
Twelve sul-positive E. coli isolates from P. vannamei and 12 sul-positive E. coli isolates from pork samples were selected from all sul-positive E. coli isolates and further analyzed by XbaI-PFGE typing. The selection criteria for these 24 strains were based on the distribution characteristics and isolation rates of each sulfonamide resistance gene and included all sulfonamide resistance gene arrays (for a detailed description see Table 3). PFGE patterns were compared using BioNumerics Version 7.6 (Applied Maths, Inc., Austin, TX, United States) with a similarity cutoff of 90% to indicate identical groups. The same isolates were also analyzed by MLST using seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA) and the primers and protocol specified in the online MLST database for E. coli3.
Location and Transferability of sul-Genes
The locations of sul genes were determined by Southern blotting with digoxigenin (DIG)-labelled sul1, sul2, and sul3 probes. Plasmid and genomic DNA were extracted using an EndoFree Plasmid Maxi Kit (Qiagen, Germantown, MD, United States) and a Bacteria Genomic DNA Extraction Kit (Biotek, Beijing, China), respectively. Plasmid and genomic DNA were digested with BamHI (Fermentas, Waltham, MA, United States) at 37°C for 5 h and then used for Southern blotting. Southern blotting, including DIG-DNA labeling, determination of labeling efficiency, DNA transfer, DNA fixation, hybridization and immunological detection, was performed using a DIG High Prime DNA Labeling and Detection Starter Kit II (Roche Applied Sciences, Penzberg, Germany).
Conjugation assays were performed to investigate the transferability of sul genes. The 18 E. coli isolates that were found by location analysis to harbor sul genes in plasmids were used as donor strains. The rifampin-resistant E. coli strain NK5449 was used as the recipient strain. The donor strain and the recipient strain were mixed at a ratio of 1:4 (v/v) and incubated overnight at 37°C in LB broth. Transconjugants were selected on LB agar plates containing 1 mg/mL sulfisoxazole and 1 mg/mL rifampicin. Plasmid DNA of transconjugants was extracted and the presence of sul genes was confirmed by PCR analysis.
Replicon Typing of Plasmids
The plasmid replicon types of all 18 transconjugants mentioned above were determined using the previously described PCR-based replicon typing (PBRT) method targeting 18 replicon types (Carattoli et al., 2005). Plasmids of all 18 transconjugants harboring sul genes were confirmed by Southern blotting using sul gene probes.
Genetic Context Analysis of sul-Genes
The genetic context of the sul genes of the 24 sul-positive E. coli isolates mentioned above was assessed using the PCR walking method with a Universal GenomeWalkerTM 2.0 Kit (Clontech, Mountain View, CA, United States) and the primers listed in Table 4. The upstream adaptor primer AP-F and the downstream sul gene primers sul1R-1, sul2R-1, and sul3R-1 targeted the upstream genetic context of the sul1, sul2, and sul3 genes, respectively. The downstream adaptor primer AP-R and the upstream sul gene primers sul1F-1, sul2F-1, and sul3F-1 targeted the downstream genetic context of the sul1, sul2, and sul3 genes, respectively. PCR products were purified using an Axyprep DNA Gel Extraction Kit (Axygen, Hangzhou, China) and sequenced by Sangon Biotech Co., Ltd. (Shanghai, China). Annotation analysis of the nucleotide sequences was performed using the BLAST program4 and ISs were analyzed using the ISfinder program5. Schematic diagrams of the genetic contexts of sul genes were drawn using Inkscape software Version 0.92.3.
Results and Discussion
sul Gene Distribution in E. coli
As shown in Table 1, 35.7% (50/180) of the E. coli isolates collected from P. vannamei samples were sulfisoxazole resistant, and 58.3% (105/180) of the E. coli isolates collected from pork samples were sulfisoxazole resistant. E. coli isolates from pork and aquatic food products often display resistance against sulfonamide, as this antibiotic has been widely used to treat bacterial and protozoan infections in swine and aquaculture. Even in the absence of selective pressure, sulfonamide-resistant bacteria can remain stable in the environment for 5–10 years and persist longer than sulfonamide itself (Gao et al., 2012).
As shown in Supplementary Figure S1, sul1, sul2, and sul3 were detected by PCR. The distribution characteristics of E. coli sul genes were similar between the two sources. Among all 50 sulfisoxazole-resistant E. coli isolates from P. vannamei samples, 90.0% (45/50) harbored sul genes, and among all 105 sulfisoxazole-resistant E. coli isolates from pork samples, 88.6% (93/105) harbored sul genes. The prevalence of sul genes was nearly as high as the rate of sulfisoxazole resistance in E. coli isolates. The remainder of sulfisoxazole resistance in sul-negative isolates was likely due to mechanisms such as mutations in the chromosomal DHPS gene flop (Changkaew et al., 2014).
The prevalence rates of individual sul1 and sul2 genes in E. coli, although variable among sampling sites, were consistently higher than that of individual sul3 gene (p < 0.05). Moreover, the prevalence of combined sul1 and sul2 genes was significantly higher than that of other gene combinations at each sampling point. This is consistent with trends previously reported in the literature (Antunes et al., 2005; Hammerum et al., 2006; Byrne-Bailey et al., 2008; Hoa et al., 2008; Gao et al., 2012; Lopes et al., 2015; Ma̧ka et al., 2015), although other studies have reported higher prevalence of sul3 in Salmonella and E. coli isolates from pigs and poultry sources in Canada (Kozak et al., 2009) and Thailand (Chuanchuen and Padungtod, 2009). These two studies also reported isolates carrying sul1, sul2, and sul3 genes, a combination that was not detected in our study. This discrepancy may be due to differences in the transmission ability of the three genes in the E. coli isolates we selected, a possibility that is discussed in the following sections.
Clonal Relatedness of sul-Positive E. coli Strains
Among the 24 sul-positive E. coli isolates, PFGE analysis identified 19 pulsotypes that could be grouped into 18 groups. In addition, MLST analysis identified 19 distinct sequence types (STs). The results of PFGE and MLST analyses are summarized in Figure 1.
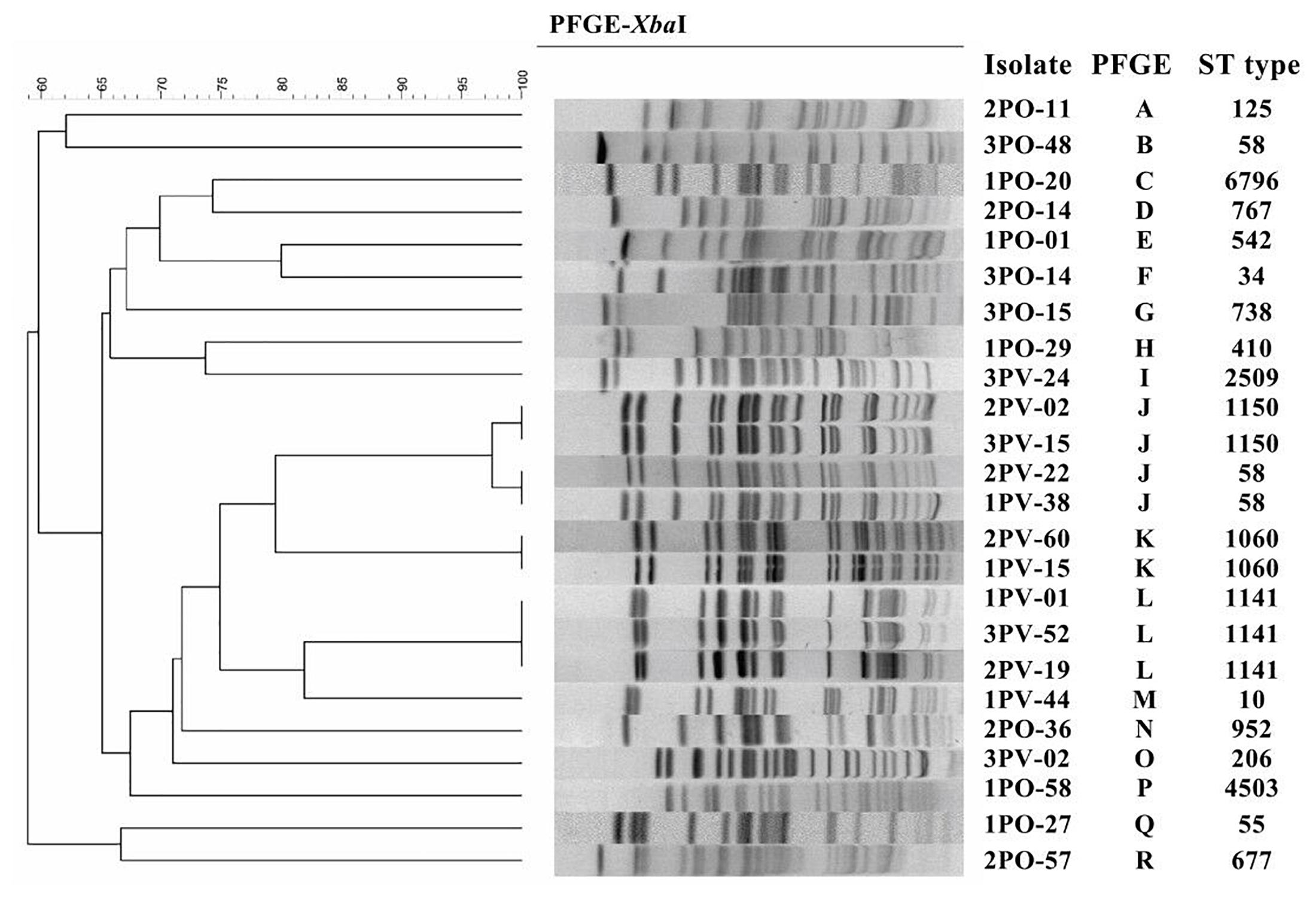
Figure 1. Pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST) of 12 sul-positive Escherichia coli isolates from Penaeus vannamei and 12 sul-positive E. coli isolates from pork products.
PFGE results showed that, of the 12 strains detected in P. vannamei samples, 2PV-02 and 3PV-15 had 100% similar fingerprint patterns, 2PV-22 and 1PV-38 had 100% similar fingerprint patterns, 2PV-60 and 1PV-15 had 100% similar fingerprint patterns, and 1PV-01, 3PV-52, and 2PV-19 had 100% similar fingerprint patterns. The remaining three strains (3PV-24, 1PV-44, and 3PV-02) each had unique pulsotypes. These results suggest potential clonal dissemination of sul-positive E. coli isolates from P. vannamei samples. Although the E. coli isolates in this study were recovered from three different large open markets on different sampling days, all P. vannamei at these markets were reared in the same large-scale freshwater shrimp farm in Zhejiang Province. Therefore, high clonality of E. coli strains circulating on the shrimp farm was likely. Additionally, similarities between E. coli strains isolated from different large open markets indicates regional spreading of clones, which may have occurred by several possible pathways, including human contact, water sources, animals and fomites (Alves et al., 2018). On contrary, all 12 strains collected from pork samples had different pulsotypes, demonstrating wide genomic diversity. The possible non-clonal dissemination of sul genes in E. coli isolates from pork samples might be effected by MGEs via horizontal gene transfer (HGT) (Zhao et al., 2018). However, further genomic analysis is necessary to confirm this hypothesis.
Based on the results of MLST combined with the results of PFGE, the four group J strains belonged to two different STs, while the remaining MLST and PFGE patterns were highly correlated. Moreover, different pulsotypes and STs were identified in isolates from P. vannamei samples and pork samples. These differences might be related to the different regions where the aquatic and pork products from each large market originated, different conditions at each farm and various transportation routes of the two types of products (Alves et al., 2018).
Localization of sul Genes and Characterization of sul Gene-Harboring Plasmids
Southern blot assays showing the locations of sul1, sul2, and sul3 are shown in Supplementary Figure S2. Location of sul genes in the 24 E. coli strains is summarized in Table 3. Among the 12 E. coli strains detected in P. vannamei samples, eight strains carried sul genes only on plasmids, two strains carried sul genes only on chromosomes and two strains carried sul genes on both plasmids and chromosomes. Among the 12 E. coli strains isolated from pork samples, four strains carried sul genes only on plasmids, four strains carried sul genes only on chromosomes and four strains carried sul genes on both plasmids and chromosomes.
Several researchers have highlighted the vital role of plasmids in the carriage and dissemination of sul genes (Wu et al., 2010; Ma̧ka et al., 2015). In our study, 18 E. coli isolates harboring sul genes on plasmids were selected for conjugation assays, with a conjugation rate of 100% (Table 3). Replicon typing identified seven different incompatibility groups (F, FIA, FIB, Y, K, I1, and N) for the plasmids (Table 3). Overall, IncF (carrying F alone or in combination with FIA or/and FIB as a multireplicon) seemed to be the most common replicon associated with sul genes among E. coli isolates from both P. vannamei and pork samples. IncF plasmids are thought to have a narrow host range and to be conjugative. IncF plasmids seem to be well adapted to E. coli, as they are found in more than 50% of E. coli strains from different sources (Wu et al., 2010; Hayashi et al., 2018). Of the 24 E. coli isolates in this study, 18 were found to harbor IncF plasmids. Therefore, sul genes located on IncF plasmids may be disseminated in E. coli by conjugation.
Of the two plasmids from E. coli isolates from P. vannamei that carried both sul1 and sul2 genes, one harbored the F replicon alone and the other harbored an F-K multireplicon. Of the two plasmids from E. coli isolates from pork samples, one harbored an F-FIB-K multireplicon and the other harbored an F-I1-N-K multireplicon. The I1 and N replicons were found only in E. coli isolates from pork samples. However, due to the limited number of strains obtained from each source, testing of additional strains is required to determine whether specific replicon profiles are associated with P. vannamei or pork.
Overall, no clear association was identified between replicon types and specific sul genes and/or sample sources. However, the localization of sul genes on wide-spread conjugative replicons such as IncF and other multireplicons very likely contributes to the dissemination of sulfonamide resistance (Wu et al., 2010).
Genetic Environments of sul Genes
In this study, we analyzed the structural characteristics of the regions approximately 5 kb upstream and 5 kb downstream of sul genes in 24 E. coli isolates using the PCR walking method. Representative primary PCR products are shown in Supplementary Figure S3. The genetic environments of sul1, sul2, and sul3 genes in 24 E. coli isolates were shown in Figure 2.
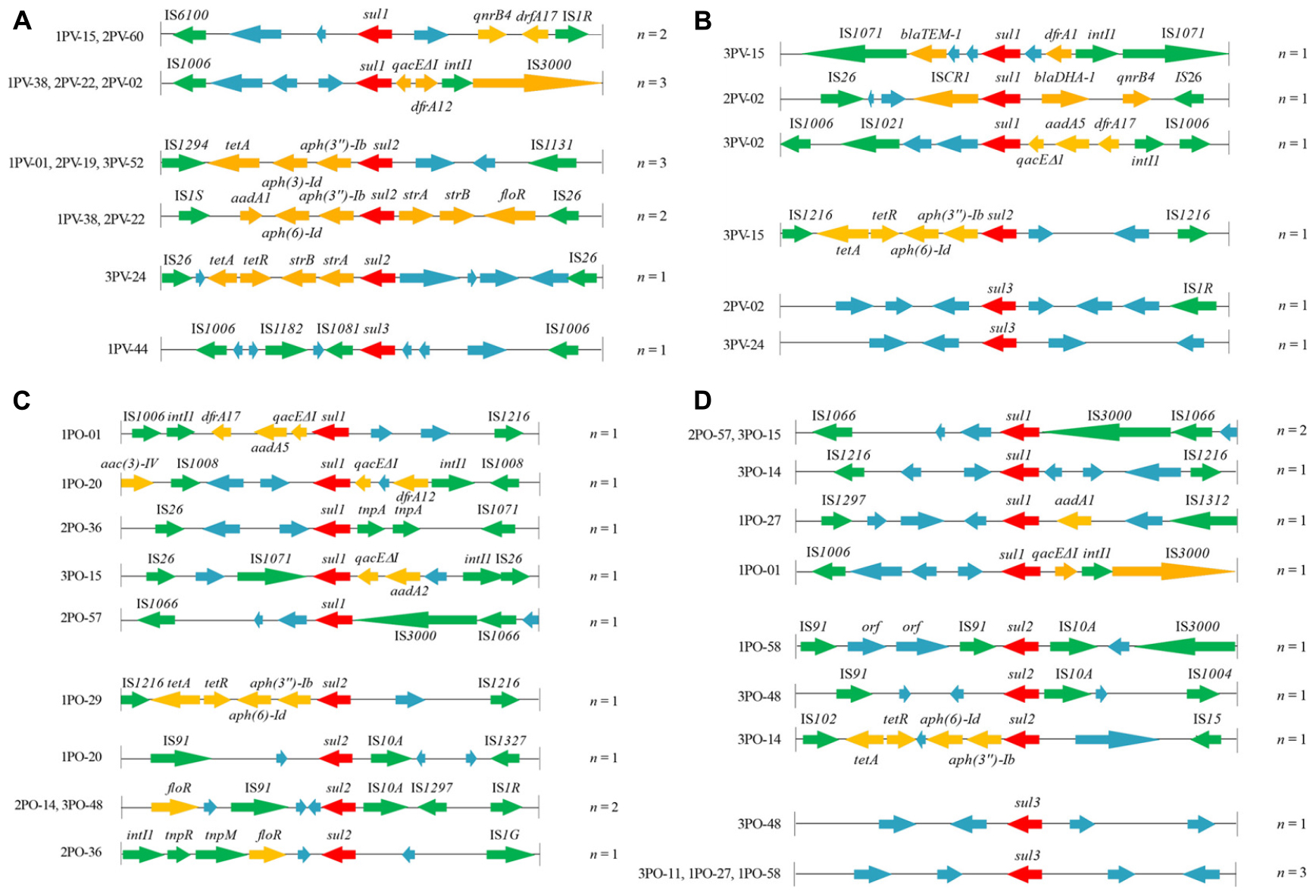
Figure 2. Genetic organization of sul gene-associated regions in (A) plasmids of 12 sul-positive Escherichia coli isolates from Penaeus vannamei; (B) chromosomes of 12 sul-positive E. coli isolates from P. vannamei; (C) plasmids of 12 sul-positive E. coli isolates from pork products; and (D) chromosomes of 12 sul-positive E. coli isolates from pork products presented with their isolate numbers. The orientation of each gene and insertion element is indicated by arrows. The same units are shown in the same color. The same functional units or unknown functional units are shown in the same color (red, sul genes; yellow, antibiotic resistance genes other than sul genes; green, mobile genetic elements; blue, unknown functional unit). Names of sequence units are indicated above or below the arrows, and sequence units with unknown functions have been left blank.
ISs are the simplest autonomous MGEs capable of altering the expression of neighboring genes (Zhao et al., 2018). In the present study, 87.5% (35/40) of sul gene-related fragments were found to carry ISs belonging to a variety of families in diverse sites. All were intact with their inverted repeats (IRR and IRL) still identifiable. Among these, the IS6 family element IS26, which has been reported to play a vital role in the dissemination of resistance determinants in Gram-negative bacteria (Partridge et al., 2018), was the most frequently occurring IS among the 24 E. coli strains. Moreover, sul2 genes on both plasmids (1PO-20, 2PO-14, and 3PO-48) and chromosomes (1PO-58 and 3PO-48) of E. coli isolates from pork samples carried IS91 and IS10A, which suggested that IS91-sul2-IS10A may mediate the dissemination of sul2 genes in E. coli in pork. Other IS families found in our study, such as IS6100, ISIR, and IS1006, may also have the ability to transfer sulfonamide resistance genes by “cut-and-paste” or “copy-out-paste-in” mechanisms (Partridge et al., 2018). In addition, IS common regions (ISCRs) are complex genetic elements that can integrate non-genetic cassette antibiotic resistance genes (Cheng et al., 2019). ISCR1 elements located on the chromosome of the 2PV-02 isolate from P. vannamei were closely linked to the sul1 gene. ISCR1 appears to be responsible for capturing and transferring many different antibiotic resistance genes and is often found adjacent to the ori end of ISCR1 in complex class 1 integrons (Toleman et al., 2006; Partridge et al., 2018). However, in the present study, ISCR1 was located downstream of the sul1 gene; therefore, its potential role in the sul1 gene transfer requires further study. Moreover, researchers have reported that ISCR2 is associated with several different resistance genes, in particular sul2 in the Glsul2 genomic island (Partridge et al., 2018). Thus, more genetic environments of sul genes must be analyzed to confirm the relationship between sul genes and ISCR elements.
In the present study, the sul1 gene was detected mostly in fragments carrying the intI1 gene of class 1 integrons (9/18), which are recognized as playing an essential role in the dissemination of multiple antimicrobial resistance genes (Lopes et al., 2015). Class 1 integrons are MGEs and usually carry the sul1 gene in their 3’ conserved region along with qacΔE1, which encodes a semifunctional derivate of the quaternary ammonium compounds resistance gene qacE (Sandvang et al., 2002). aadA and drf genes are also prevalent in class 1 integrons adjacent to qacΔE1 and sul1 genes, forming a highly stable, low cost structure with reported prevalence rates in E. coli of 63% (Lacotte et al., 2017; Fang et al., 2019). In addition, in the present study, the sul2 gene was commonly found to be part of a cassette structure linked with the streptomycin resistance genes strA and strB in the plasmid of E. coli isolates from P. vannamei, a feature that has been reported in many other studies in both E. coli and Salmonella (Enne et al., 2004; Bean et al., 2009; Lopes et al., 2015; Ma̧ka et al., 2015). This cassette is not recognized as an MGE in its own right but has been associated with other MGEs, such as IS1294, IS1131, and IS1S. More importantly, co-location of resistance genes in the same plasmid or fragment, such as genes adjacent to sul1 found to encode for quinolone resistance (qnrB4) or β-lactamase resistance (blaTEM-1 and blaDHA-1) and genes adjacent to sul2 found to encode for tetracycline resistance (tetA and tetR), aminoglycoside resistance (aph and aad) and amphenicol resistance (floR), likely contribute to the persistence and dissemination of sul1 and/or sul2 genes under the selective pressure of other commonly used antibiotics. This co-selection effect has also been reported in other MGEs of class 2 integrons of E. coli isolates (Alonso et al., 2018). However, the prevalence of sul3 is reportedly much lower than that of sul1 and sul2 (Byrne-Bailey et al., 2008; Lopes et al., 2015; Ma̧ka et al., 2015), and sul3 was rarely detected in the E. coli isolates from P. vannamei and pork samples in our study. In this study, the diversity of MGEs and resistance genes adjacent to sul3 was much lower than those adjacent to sul1 and sul2, especially among those located in chromosomes, which reduced the transmission potential of the sul3 gene. However, other studies have found that sul3-carrying plasmids are conjugative and associated with class 1 integrons, and can replace the sul1 gene to form atypical class 1 integrons. This underscores the potential of sul3 to become more widespread in the future (Wu et al., 2010).
Finally, since sul gene-related fragments in E. coli isolates from P. vannamei and pork samples were linked to various MGEs, which facilitate intracellular and intercellular mobility, in both plasmids and chromosomes, clonal similarities between E. coli isolates from P. vannamei samples might be coincidental.
In conclusion, sul genes were widely distributed in E. coli isolates from both P. vannamei and pork samples, with sul1 and sul2 being more prevalent than sul3. Because sul genes are located on both plasmids and chromosomes, diverse gene transfer mechanisms can contribute to the efficient spread of these genes to novel species. This was demonstrated by the presence of conjugative and/or mobilizable plasmids of diverse replicon types harboring sul genes and the abundant MGEs located in the flanking regions of these genes. In addition, co-location of resistance genes likely contributed to the persistence and dissemination of sul genes under the selective pressure of commonly used antibiotics. Our results provide important information to gain a better understanding of the risk of transmission of sul genes from seafood and meat to humans.
Data Availability
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
HJ completed the clonal analysis by molecular typing, location and transferability of sul-genes, and replicon typing of plasmids, and prepared the manuscript. HC completed the genetic context analysis of sul-genes and prepared the figures and tables. YL, SY, and TY completed the shrimp and pork sampling, E. coli isolation, and the phenotypic screening of sulfonamide resistance and detection of sul-genes. JF and CZ designed the project, completed the data analysis, and revised the manuscript.
Funding
This research was supported by the National Natural Science Foundation of China under Grant No. 31801655, Zhejiang Provincial Natural Science Foundation of China under Grant No. LQ18C200004, and National Students’ platform for innovation and entrepreneurship training program under Grant No. 201810356032.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01787/full#supplementary-material
FIGURE S1 | Detection of sul genes in sulfisoxazole-resistant E. coli isolates (partial) by PCR. (A) detection of sul1; (B) detection of sul2; (C) detection of sul3. M, DL2000 marker (100, 250, 500, 750, 1000, and 2000 bp). P, positive control; N, negative control; 1−21, sulfisoxazole-resistant E. coli isolates.
FIGURE S2 | Southern blot analysis of the locations of sul genes in E. coli isolates. (A) Analysis of the location of sul1. 1, chromosomal DNA of 1PO-01 isolate; 2, plasmid DNA of 1PO-01 isolate; 3, chromosomal DNA of 2PO -57 isolate; 4, plasmid DNA of 2PO-57 isolate; 5, chromosomal DNA of 3PO-15 isolate; 6, plasmid DNA of 3PO-15 isolate; 7, chromosomal DNA of 1PO-20 isolate; 8, plasmid DNA of 1PO-20 isolate. (B) Analysis of the location of sul2. 1, plasmid DNA of 1PV-01 isolate; 2, plasmid DNA of 2PV-19 isolate; 3, plasmid DNA of 3PV-52 isolate; 4, plasmid DNA of 1PV-38 isolate; 5, plasmid DNA of 2PV-22 isolate; 6, plasmid DNA of 3PV-15 isolate; 7, plasmid DNA of 3PV-24 isolate. (C) Analysis of the location of sul3. 1, chromosomal DNA of 1PO-58 isolate; 2, chromosomal DNA of 1PO-27 isolate; 3, chromosomal DNA of 2PO- 11 isolate; 4, chromosomal DNA of 3PV-24 isolate; 5, chromosomal DNA of 2PV-02 isolate; 6, chromosomal DNA of 1PV-44 isolate. M, DNA molecular weight marker II, digoxigenin-labeled (Roche, Penzberg, Germany) with the following base pair lengths: 125, 564, 2027, 2322, 4361, 6557, 9416, and 23,130 bp.
FIGURE S3 | Primary PCR products of sul gene flanking sequences obtained using the PCR walking method. (A) Products of sul1 gene flanking sequences. (B) Products of sul2 gene flanking sequences. (C) Products of sul3 gene flanking sequences. M: DL5000 marker (100, 250, 500, 750, 1000, 2000, 3000, and 5000 bp). 1−7, products of sul gene upstream sequences (DNA template is DraI library, EcoRV library, HpaI library, PvuII library, SmaI library, StuI library, and SspI library). 8−14, products of sul gene downstream sequences (DNA template is DraI library, EcoRV library, HpaI library, PvuII library, SmaI library, StuI library, and SspI library).
Footnotes
- ^ http://www.fao.org/fishery/statistics/global-aquaculture-production/query/en?tdsourcetag=s_pctim_aiomsg/en
- ^ http://www.moa.gov.cn/govpublic/SYJ/201509/t20150907_4819267.htm
- ^ http://enterobase.warwick.ac.uk/species/ecoli/allele_st_search
- ^ https://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^ http://www-is.biotoul.fr/
References
Alonso, C. A., Cortés-Cortés, G., Maamar, E., Massó, M., Rocha-Gracia, R. D. C., Torres, C., et al. (2018). Molecular diversity and conjugal transferability of class 2 integrons among Escherichia coli isolates from food, animal and human sources. Int. J. Antimicrob. Agents 51, 905–911. doi: 10.1016/j.ijantimicag.2018.02.001
Alvarez-Fernandez, E., Cancelo, A., Diaz-Vega, C., Capita, R., and Alonso-Calleja, C. (2013). Antimicrobial resistance in E. coli isolates from conventionally and organically reared poultry: a comparison of agar disc diffusion and Sensi Test Gram-negative methods. Food Control 30, 227–234. doi: 10.1016/j.foodcont.2012.06.005
Alves, T. S., Lara, G. H. B., Maluta, R. P., Ribeiro, M. G., and Leite, D. S. (2018). Carrier flies of multidrug-resistant Escherichia coli as potential dissemination agent in dairy farm environment. Sci. Total Environ. 633, 1345–1351. doi: 10.1016/j.scitotenv.2018.03.304
Antunes, P., Machado, J., Sousa, J. C., and Peixe, L. (2005). Dissemination of sulfonamide resistance genes (sul1, sul2, and sul3) in Portuguese Salmonella enterica strains and relation with integrons. Antimicrob. Agents Chemother. 49, 836–839. doi: 10.1128/AAC.49.2.836-839.2005
Bean, D. C., Livermore, D. M., and Hall, L. M. C. (2009). Plasmids imparting sulfonamide resistance in Escherichia coli: implications for persistence. Antimicrob. Agents Chemother. 53, 1088–1093. doi: 10.1128/AAC.00800-08
Ben, W. W., Wang, J., Pan, X., and Qiang, Z. M. (2017). Dissemination of antibiotic resistance genes and their potential removal by on-farm treatment processes in nine swine feedlots in Shandong Province, China. Chemosphere 167, 262–268. doi: 10.1016/j.chemosphere.2016.10.013
Blahna, M. T., Zalewski, C. A., Reuer, J., Kahlmeter, G., Foxman, B., and Marrs, C. F. (2006). The role of horizontal gene transfer in the spread of trimethoprim-sulfamethoxazole resistance among uropathogenic Escherichia coli in Europe and Canada. J. Antimicrob. Chemother. 57, 666–672. doi: 10.1093/jac/dkl020
Boss, R., Overesch, G., and Baumgartner, A. (2016). Antimicrobial resistance of Escherichia coli, enterococci, Pseudomonas aeruginosa, and Staphylococcus aureus from raw fish and seafood imported into Switzerland. J. Food Prot. 79, 1240–1246. doi: 10.4315/0362-028X.JFP-15-463
Byrne-Bailey, K. G., Gaze, W. H., Kay, P., Boxall, A. B., Hawkey, P. M., and Wellington, E. M. (2008). Prevalence of sulfonamide resistance genes in bacterial isolates from manured agricultural soils and pig slurry in the United Kingdom. Antimicrob. Agents Chemother. 53, 696–702. doi: 10.1128/AAC.00652-07
Carattoli, A., Bertini, A., Villa, L., Falbo, V., Hopkins, K. L., and Threlfall, E. J. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63, 219–228. doi: 10.1016/j.mimet.2005.03.018
Card, R., Vaughan, K., Bagnall, M., Spiropoulos, J., Cooley, W., Strickland, T., et al. (2016). Virulence characterisation of Salmonella enterica isolates of differing antimicrobial resistance recovered from UK livestock and imported meat samples. Front. Microbiol. 7:640. doi: 10.3389/fmicb.2016.00640
Changkaew, K., Utrarachkij, F., Siripanichgon, K., Nakajima, C., Suthienkul, O., and Suzuki, Y. (2014). Characterization of antibiotic resistance in Escherichia coli isolated from shrimps and their environment. J. Food Prot. 77, 1394–1401. doi: 10.4315/0362-028X.JFP-13-510
Cheng, H., Jiang, H., Fang, J., and Zhu, C. (2019). Antibiotic resistance and characteristics of integrons in Escherichia coli isolated from Penaeus vannamei at a freshwater shrimp farm in Zhejiang Province, China. J. Food Prot. 82, 470–478. doi: 10.4315/0362-028X.JFP-18-444
Chuanchuen, R., and Padungtod, P. (2009). Antimicrobial resistance genes among Salmonella enterica isolates from poultry and swine in Thailand. J. Vet. Med. Sci. 71, 1349–1355. doi: 10.1292/jvms.001349
Clinical and Laboratory Standards Institute [CLSI] (2017). Performance Standards for Antimicrobial Susceptibility Testing, 27th Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
Enne, V. I., Bennett, P. M., Livermore, D. M., and Hall, L. M. C. (2004). Enhancement of host fitness by the sul2-coding plasmid p9123 in the absence of selective pressure. J. Antimicrob. Chemother. 53, 958–963. doi: 10.1093/jac/dkh217
Fang, J., Shen, Y., Qu, D., and Han, J. (2019). Antimicrobial resistance profiles and characteristics of integrons in Escherichia coli strains isolated from a large-scale centralized swine slaughterhouse and its downstream markets in Zhejiang, China. Food Control 95, 215–222. doi: 10.1016/j.foodcont.2018.08.003
Gao, P., Mao, D., Luo, Y., Wang, L., Xu, B., and Xu, L. (2012). Occurrence of sulfonamide and tetracycline-resistant bacteria and resistance genes in aquaculture environment. Water Res. 46, 2355–2364. doi: 10.1016/j.watres.2012.02.004
Hammerum, A. M., Sandvang, D., Andersen, S. R., Seyfarth, A. M., Porsbo, L. J., Frimodt-Møller, N., et al. (2006). Detection of sul1, sul2 and sul3 in sulphonamide resistant Escherichia coli isolates obtained from healthy humans, pork and pigs in Denmark. Int. J. Food Microbiol. 106, 235–237. doi: 10.1016/j.ijfoodmicro.2005.06.023
Hayashi, W., Ohsaki, Y., Taniguchi, Y., Koide, S., Kawamura, K., Suzuki, M., et al. (2018). High prevalence of bla CTX-M-14 among genetically diverse Escherichia coli recovered from retail raw chicken meat portions in Japan. Int. J. Food Microbiol. 284, 98–104. doi: 10.1016/j.ijfoodmicro.2018.08.003
Heuer, H., and Smalla, K. (2007). Manure and sulfadiazine synergistically increased bacterial antibiotic resistance in soil over at least two months. Environ. Microbiol. 9, 657–666. doi: 10.1111/j.1462-2920.2006.01185.x
Hoa, P. T. P., Nonaka, L., Viet, P. H., and Suzuki, S. (2008). Detection of the sul1, sul2, and sul3 genes in sulfonamide-resistant bacteria from wastewater and shrimp ponds of north Vietnam. Sci. Total Environ. 405, 377–384. doi: 10.1016/j.scitotenv.2008.06.023
Hsu, J. T., Chen, C. Y., Young, C. W., Chao, W. L., Li, M. H., Liu, Y. H., et al. (2014). Prevalence of sulfonamide-resistant bacteria, resistance genes and integron-associated horizontal gene transfer in natural water bodies and soils adjacent to a swine feedlot in northern Taiwan. J. Hazard. Mater. 277, 34–43. doi: 10.1016/j.jhazmat.2014.02.016
Jiang, H., Li, P., and Gu, Q. (2016). Heterologous expression and purification of plantaricin NC8, a two-peptide bacteriocin against Salmonella spp. from Lactobacillus plantarum ZJ316. Protein Expres. Purif. 127, 28–34. doi: 10.1016/j.pep.2016.06.013
Kozak, G. K., Pearl, D. L., Julia, P., Reid-Smith, R. J., Deckert, A., and Boerlin, P. (2009). Distribution of sulfonamide resistance genes in Escherichia coli and Salmonella isolates from swine and chickens at abattoirs in Ontario and Québec. Can. Appl. Environ. Microbiol. 75, 5999–6001. doi: 10.1128/AEM.02844-08
Lacotte, Y., Ploy, M. C., and Raherison, S. (2017). Class 1 integrons are low-cost structures in Escherichia coli. ISME J. 11, 1535–1544. doi: 10.1038/ismej.2017.38
Liu, Z., Klümper, U., Shi, L., Ye, L., and Li, M. (2019). From pig breeding environment to subsequently produced pork: comparative analysis of antibiotic resistance genes and bacterial community composition. Front. Microbiol. 10:43. doi: 10.3389/fmicb.2019.00043
Lopes, G. V., Pissetti, C., da Cruz Payão Pellegrini, D., da Silva, S. L., and Cardoso, M. (2015). Resistance phenotypes and genotypes of Salmonella enterica subsp. enterica isolates from feed, pigs, and carcasses in Brazil. J. Food Prot. 78, 407–413. doi: 10.4315/0362-028X.JFP-14-274
Ma̧ka, Ł, Maćkiw, E., Ścieżyńska, H., Modzelewska, M., and Popowska, M. (2015). Resistance to sulfonamides and dissemination of sul genes among Salmonella spp. isolated from food in Poland. Foodborne Pathog. Dis. 12, 383–389. doi: 10.1089/fpd.2014.1825
Paraoan, C. E. M., Rivera, W. L., and Vital, P. G. (2017). Detection of Class I and II integrons for the assessment of antibiotic and multidrug resistance among Escherichia coli isolates from agricultural irrigation waters in Bulacan, Philippines. J. Environ. Sci. Heal. B 52, 306–313. doi: 10.1080/03601234.2017.1281647
Partridge, S. R., Kwong, S. M., Firth, N., and Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31:e00088-17. doi: 10.1128/CMR.00088-17
Peixoto, P. S., Tóth, I. V., Segundo, M. A., and Lima, J. L. (2016). Fluoroquinolones and sulfonamides: features of their determination in water. A review. Int. J. Environ. Anal. Chem. 96, 185–202. doi: 10.1080/03067319.2015.1128539
Rehman, M. U., Zhang, H., Huang, S., Iqbal, M. K., Mehmood, K., Luo, H., et al. (2017). Characteristics of integrons and associated gene cassettes in antibiotic-resistant Escherichia coli isolated from free-ranging food animals in China. J. Food Sci. 82, 1902–1907. doi: 10.1111/1750-3841.13795
Sandvang, D., Diggle, M., and Platt, D. J. (2002). Translocation of integron-associated resistance in a natural system: acquisition of resistance determinants by Inc P and Inc W plasmids from Salmonella enterica Typhimurium DT104. Microb. Drug Resist. 8, 151–160. doi: 10.1089/107662902760326850
Skočková, A., Koláèková, I., Bogdanovièová, K., and Karpíšková, R. (2015). Characteristic and antimicrobial resistance in Escherichia coli from retail meats purchased in the Czech Republic. Food Control 47, 401–406. doi: 10.1016/j.foodcont.2014.07.034
Toleman, M. A., Bennett, P. M., and Walsh, T. R. (2006). ISCR elements: novel gene-capturing systems of the 21st century? Microbiol. Mol. Biol. Rev. 70, 296–316. doi: 10.1128/MMBR.00048-05
Wang, N., Yang, X., Jiao, S., Zhang, J., Ye, B., and Gao, S. (2014). Sulfonamide-resistant bacteria and their resistance genes in soils fertilized with manures from Jiangsu Province, Southeastern China. PLoS One 9:e112626. doi: 10.1371/journal.pone.0112626
Wu, S., Dalsgaard, A., Hammerum, A. M., Porsbo, L. J., and Jensen, L. B. (2010). Prevalence and characterization of plasmids carrying sulfonamide resistance genes among Escherichia coli from pigs, pig carcasses and human. Acta Vet. Scand. 52:47. doi: 10.1186/1751-0147-52-47
Yuan, J., Ni, M., Liu, M., Zheng, Y., and Gu, Z. (2019). Occurrence of antibiotics and antibiotic resistance genes in a typical estuary aquaculture region of Hangzhou Bay, China. Mar. Pollut. Bull. 138, 376–384. doi: 10.1016/j.marpolbul.2018.11.037
Zhang, R. Q., Ying, G. G., Su, H. C., Zhou, L. J., and Liu, Y. S. (2013). Antibiotic resistance and genetic diversity of Escherichia coli isolates from traditional and integrated aquaculture in South China. J. Environ. Sci. Health B 48, 999–1013. doi: 10.1080/03601234.2013.816611
Keywords: sulfonamide resistance genes, Escherichia coli, mobile genetic elements, insertion sequences, conjugation
Citation: Jiang H, Cheng H, Liang Y, Yu S, Yu T, Fang J and Zhu C (2019) Diverse Mobile Genetic Elements and Conjugal Transferability of Sulfonamide Resistance Genes (sul1, sul2, and sul3) in Escherichia coli Isolates From Penaeus vannamei and Pork From Large Markets in Zhejiang, China. Front. Microbiol. 10:1787. doi: 10.3389/fmicb.2019.01787
Received: 06 April 2019; Accepted: 19 July 2019;
Published: 02 August 2019.
Edited by:
Kwangcheol Casey Jeong, University of Florida, United StatesReviewed by:
Dongjin Park, University of Nebraska-Lincoln, United StatesSi Hong Park, Oregon State University, United States
Copyright © 2019 Jiang, Cheng, Liang, Yu, Yu, Fang and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiehong Fang, figo0726@163.com; Cheng Zhu, pzhch@cjlu.edu.cn
 Han Jiang
Han Jiang Hui Cheng
Hui Cheng