- 1College of Enology, Northwest A&F University, Yangling, China
- 2Shandong Engineering and Technology Research Center for Ecological Fragile Belt of Yellow River Delta, Binzhou, China
- 3Heyang Experimental and Demonstrational Stations for Grape, Weinan, China
- 4Shaanxi Engineering Research Center for Viti-Viniculture, Yangling, China
Oenococcus oeni can survive well in wine (an acid-stress environment) and dominate malolactic fermentation (MLF). To demonstrate a possible role of argininosuccinate synthase gene (argG) in the acid tolerance response of O. oeni, a related argG gene was inserted into a plasmid pMG36e and heterologously expressed in Lactobacillus plantarum SL09, a wine isolate belonging to a species of relevant importance in MLF. The expression levels of the argG gene in L. plantarum were analyzed by RT-qPCR, argininosuccinate synthase (ASS) activity and cell properties (amino acids, pH, H+-ATPase activity, and ATP levels) were determined at pH 3.7 in comparison with that at pH 6.3. Results showed that the recombinant strain L. plantarum SL09 (pMG36eargG) exhibited stronger growth performance compared with the control strain (without argG gene), and the expression levels of hsp1, cfa, atp, the citrate and malate metabolic genes were apparently increased under acid stress. In addition, the recombinant strain exhibited 11.0-, 2.0-, 1.9-fold higher ASS activity, H+-ATPase activity and intracellular ATP level, compared with the corresponding values for control strain during acid-stresses condition, which may take responsible for the acid tolerance enhancement of the recombinant strain. This is the first work report on heterologous expression of argG gene, and the results presented in this study will be beneficial for the research on acid stress response of O. oeni.
Introduction
Oenococcus oeni, an important lactic acid bacteria (LAB), is critical for winemaking owing to the ability of deacidification and stabilization of wine through malolactic fermentation (Betteridge et al., 2015; Peng et al., 2018; Romero et al., 2018). The optimal pH for O. oeni growth is 4.8–5.5, however, wine is a harsh environment, with high acidity (pH 3.0–3.5), which is considered as a major stress for O. oeni growth (Fortier et al., 2003; Rosi et al., 2003; Guzzo, 2011). O. oeni has a certain acid resistance mechanism because it survives well in a wine environment and play a crucial role in winemaking (Dimopoulou et al., 2018; Lorentzen and Lucas, 2019).
Significant efforts have been made in order to reveal the mechanism by which O. oeni tolerates acid stress. A potential method used by bacteria is the use of a H+-ATPase to pump H+ out of the cell and thus increase the acid tolerance (Fortier et al., 2003). The citrate metabolism affects the acid tolerance of O. oeni, owing to its end products (Augagneur et al., 2007), with Margalef-Català et al. (2016) finding that the genes involved in glutamine and glutamate metabolism were upregualted of O. oeni under stress. Darsonval et al. (2018) verified that the CtsR is the master regulator of stress-response in O. oeni. In addition, the arginine catabolism through the arginine deiminase (ADI) pathway is known to enhance acid tolerance by converting arginine into an alkaline product and by increasing external pH (Tonon et al., 2001; Arena and Manca de Nadra, 2005; Araque et al., 2016). Moreover, the argG and argH gene involved in acid tolerance response of Lactobacillus casei (Quivey et al., 2000), which were responsible for encoding argininosuccinate synthase (ASS) and argininosuccinate lyase (ASL), respectively. Of these, ASS is considered as the rate-limiting enzyme for arginine biosynthesis (Lemke and Howell, 2002), we observed that the argG gene of O. oeni SD-2a was apparently over-expressed after acid shock in our previous research (Liu et al., 2017), and functioned as a core regulatory gene during acid stress response according to its place in a gene co-expression network. However, the potential role of the argG gene on O. oeni acid stress has not been elucidated.
Oenococcus oeni seems to be good model for the study of mechanisms involved in stress resistance, but there are difficulties to genetically modify the O. oeni cells even using the current molecular biology technologies (Beltramo et al., 2004; Assad-Garcia et al., 2008; Darsonval et al., 2016). Currently, more attention was turned to other LABs as a model to explore O. oeni, Schümann et al. (2012) expressed O. oeni mle in L. plantarum WCFS1, and the recombinant L. plantarum cells expressing MLE accelerate the malolactic fermentation. Weidmann et al. (2017) reported that the production of Lo18 from O. oeni expressed in L. lactis improved tolerance to heat and acid stress, suggesting Lo18 may play a role in cytoplasmic protein and membrane stabilization during stress. These studies suggested that achieving heterologous expression in other LABs can be used to explore the stress response mechanism of O. oeni. In addition, Lactobacillus plantarum is able to conduct MLF, and some L. plantarum strains are commercially used MLF starter (Du Toit et al., 2011; Capozzi et al., 2012; Valdés La Hens et al., 2015; Berbegal et al., 2016).
Therefore, in the present work, the argG was heterologously expressed in L. plantarum and the expression of argG gene was detected by RT-qPCR and ASS activity. Moreover, the growth of recombinant (pMG36eargG) and control strain (pMG36e) were measured under acid stress (pH 3.0–4.0), and cell properties (amino acids, pH, H+-ATPase activity, and ATP levels) were also determined at pH 3.7 and 6.3 to study the heterologous expression of the argG gene in the recombinant L. plantarum strain.
Materials and Methods
Strains and Growth Conditions
The industrial O. oeni strain, SD-2a was isolated from a local wine region in Shandong Province, China (Wang et al., 2015), and preserved in China General Microbiological Culture Collection Center (CGMCC 0715). L. plantarum SL09 was isolated from red wine and identified by 16S rRNA gene analysis (Supplementary Table S1; Lee et al., 2016; Wang et al., 2016).
Oenococcus oeni SD-2a was cultured at 28°C in FMATB (5 g/L glucose, 5 g/L D, L-malate, 5 g/L yeast extract, 10 g/L peptone, 0.2 g/L MgSO4⋅7H2O, 0.05 g/L MnSO4⋅4H2O, 0.5 g/L cysteine/HCl, and 250 mL fresh tomato juice, pH 4.8). L. plantarum SL09 was cultured at 37°C in MRS broth, and Escherichia coli DH5α grew at 37°C in Luria-Bertani (LB) medium. Agar plates were prepared with 15 g/L agar. The culture of L. plantarum SL09 strain with plasmid pMG36e or pMG36eargG required the addition of erythromycin (Solarbio, Beijing, China), with a final concentration of 100 μg/ml, meanwhile, the E. coli DH5α strain with plasmid pMG36e or pMG36eargG required the addition of erythromycin (Solarbio, Beijing, China) with a final concentration to 200 μg/ml.
Data Acquisition and Gene Screening
The changes in the transcriptome of O. oeni SD-2a during acid shock were studied previously (Liu et al., 2017), the RNA-seq data were downloaded from Sequence Read Archive (SRA) database with an accession number of SRP105332. The gene co-expression network was constructed in this study using the differential expression of genes and the Pearson model to calculate the co-expression coefficient and P-value between genes. Genes with a high co-expression coefficient and different expression levels (more than 2 folds) were selected.
DNA Extraction and Plasmid Construction
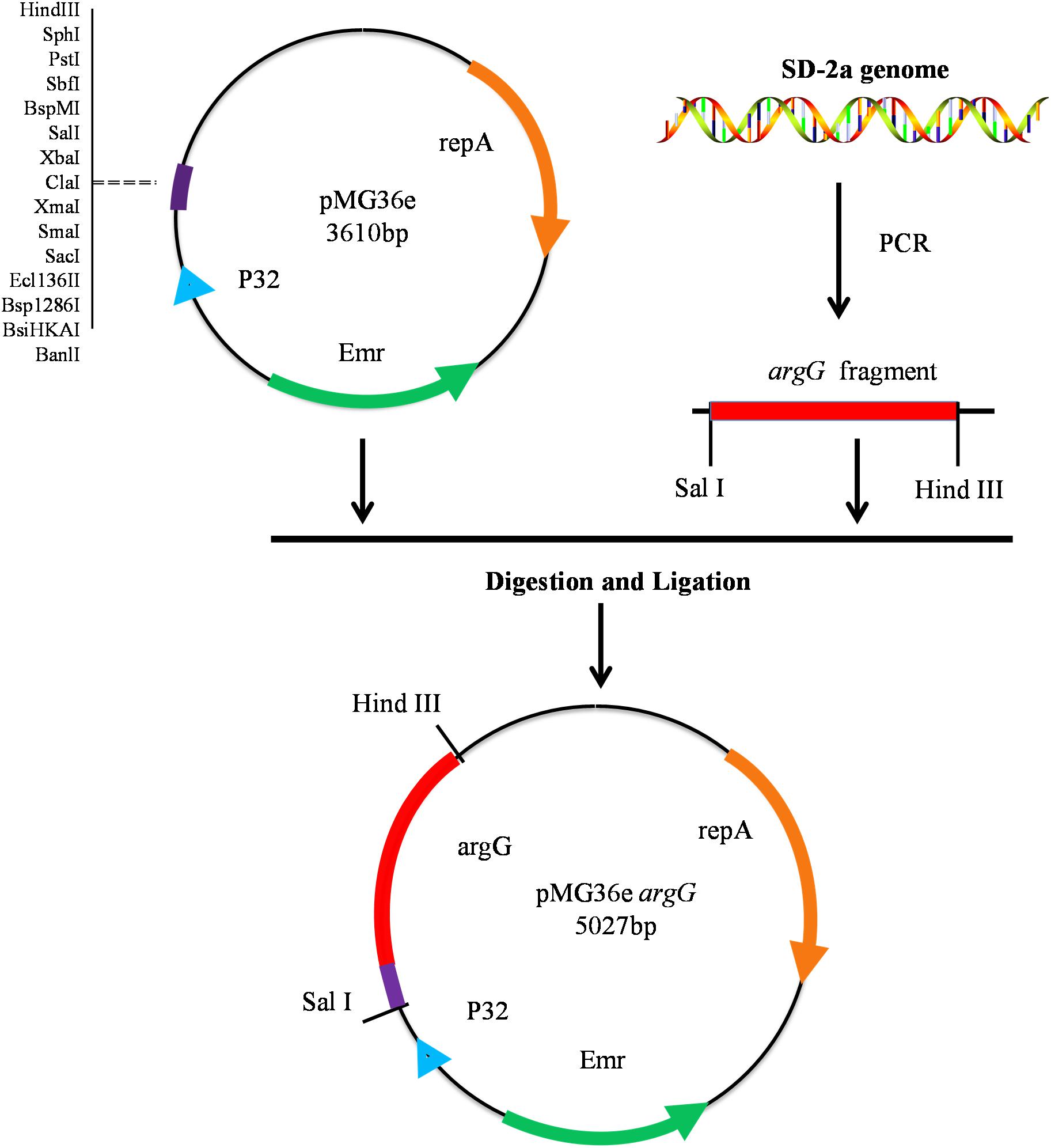
Oenococcus oeni SD-2a was grown in FMATB medium to an OD600 nm of 1.0 (5 × 108 CFU/mL), then harvested by centrifugation at 12,000 × g for 2 min. DNA extraction was performed using the TIANamp Bacteria DNA Kit (Tiangen, Beijing, China) according to the manufacturer’s instructions. The argG gene from O. oeni SD-2a was amplified from genomic DNA using primers argG-F and argG-R (Table 1) to introduce the restriction site with PrimeSTAR® HS DNA Polymerase (Takara). Subsequently, the PCR product and the vector pMG36e were digested by Sal I and Hind III. Both fragments were ligated and the resulting plasmid transformed into chemically competent E. coli DH5α cells according to the method recommended by the manufacturer (Takara) (Figure 1). Positive colony PCR amplified constructs were verified by sequencing, performed by a commercial provider, and the plasmid pMG36eargG was extracted using the Plasmid Mini Kit I (omega).
Cell Preparation and Electroporation
The overnight culture of L. plantarum SL09 was inoculated into 10 ml MRS broth supplemented with 4% glycine and incubated at 37°C to OD600 nm≈0.4. The cells were harvested by centrifugation, washed two times with 10 ml of sterile electroporation buffer (5 mM potassium phosphate, 0.5 mM MgCl2 and 0.5 M sucrose). Then, the cells were gently resuspended in 0.2 ml of electroporation buffer, and 100 μl of the solution was mixed together with 1 μg of plasmid DNA (pMG36e or pMG36eargG), transferred to a sterile 2-mm Gene Pulser cuvette (Bio-Rad) and left on ice for 5 min. Electroporation was performed with a Bio-Rad pulse gene controller (4 ms at 2.0 kV). The cells were immediately rescued into 1.8 ml of MRS supplemented with 0.3 M sucrose and incubated for 3 h at 37°C and plated onto MRS containing erythromycin (100 μg/ml). All strains and plasmids used in this study are listed in Table 1.
Stress Challenges in L. plantarum
Stress Experiment
An overnight culture of SL09 (pMG36eargG) and SL09 (pMG36e) grown in MRS (pH 6.3) medium at 37°C was used to inoculate (1%, v/v) into fresh MRS media (pH 6.3), and cultured to an OD600 nm of 1.0. The culture was then inoculated (1%, v/v, without washing treatment) into fresh normal MRS (pH 6.3) or acid-stressed MRS pH 3.0–pH 4.0 (gradient 0.1) then cultured at 37°C to investigated the growth performance of strains by measuring absorbance at 600 nm and counting plates colonies. The pH of the medium was adjusted by 1 M HCL using pH meter (INESA Scientific Instrument Co., Ltd., Shanghai, China). All growth experiments were carried out in triplicate, and all the culture added with 100 μg/mL erythromycin.
ASS Activity Assay and Determination of Amino Acids in Cells
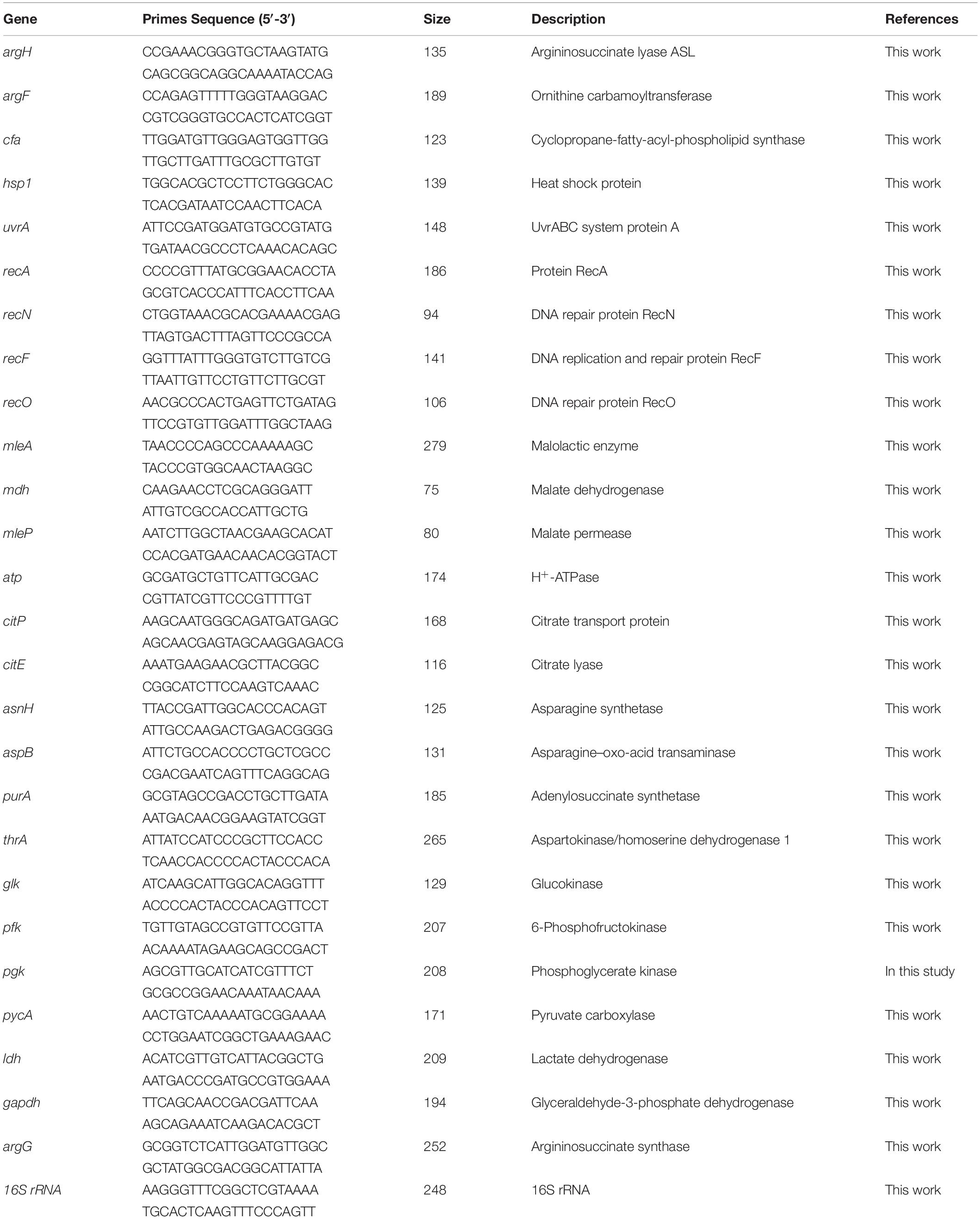
The samples used in these assays were taken from cultures under pH 3.7 and pH 6.3, respectively. L. plantarum SL09 (pMG36e) and recombinant SL09 (pMG36eargG) were cultivated to logarithmic phase (8 h for pH 6.3, 36 h for pH 3.7). The cells were harvested by centrifugation at 12,000 × g for 10 min, washed twice with Tris–Hcl buffer, ground with liquid nitrogen, resuspended in Tris–Hcl buffer (50 mmol/L, pH 7.4), and then centrifuged at 12,000 × g, 4°C for 10 min to obtain the cell supernatant. Reaction mixtures included 100 μl of cell supernatant, 100 μl of 50 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (pH 7.5), 16 mM ATP, 30 mM citrulline, 90 mM aspartic acid, and 5 mM MgCl2. Reactions were incubated at 27°C for 60 min and terminated with 70% (w/v) trichloroacetic acid. Final reaction supernatants were obtained by centrifugation at 5,000 × g, 4°C for 2 min. The supernatants added 300 μL O-phthalaldehyde and 600 μL borate buffer, then mixed at room temperature, filtered the mixture using 0.45 μm filtration, incubated in the dark strictly for 15 min, then applied to HPLC according to Hu et al. (2014) to obtain the concentration of other amino acids. Units of ASS activity (U) were expressed as micromoles of citrulline consumed per minute at 27°C (Cruz et al., 2007).
Measurement of pHi
Cells used for pHi measurement were cultivated to logarithmic phase (8 h for pH 6.3, 36 h for pH 3.7). The pHi was assayed by the fluorescence method using 2′, 7′-Bis-(2-Carboxyethyl)-5-(and-6)-Carboxy- fluorescein, acetoxymethyl ester (BCECF AM) as the fluorescent probe (Breeuwer et al., 1996; Zhang et al., 2007).
Determination of H+-ATPase and Intracellular ATP Concentration
Intracellular H+-ATPase activity was measured using the H+-ATPase assay kit (Beyotime Biotechnology, Shanghai, China). Enzyme activity units (U) were defined as the amount of enzyme required to oxidate 1 μmol NADH per minute at 37°C, pH 7.5. ATP concentrations were determined using a Firefly Luciferase ATP Assay Kit (Beyotime Biotechnology, Shanghai, China).
RT-qPCR
Total RNA was extracted using the RNAprep pure Cell/Bacteria Kit (Tiangen, Beijing, China) following the manufacturer’s instructions. The quality of the RNA samples was verified on a 1% (v/v) agarose gel, and the concentration of RNA was determined by measuring the A260 nm using a BioDrop μLITE Spectrophotometer (Tamar Laboratory Supplies LTD., Cambridge, United Kingdom). Next, cDNA was synthesized using the Thermo Fisher Scientific RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, MA, United States). RT-qPCR was conducted according to the instructions of ChamQTM SYBR qPCR Master Mix (Vazyme, Nanjing, China). The L. plantarum SL09 16S rRNA gene was used as the housekeeping gene, and the SL09 (pMG36e) served as the control strain and was cultivated at the same pH value (3.7 and 6.3) (Tang et al., 2017). The primers used for RT-qPCR are described in Table 2. The results were analyzed using the comparative critical threshold (2–ΔΔCT) method in which the amount of target RNA was adjusted to a reference signal (internal target RNA) as described previously (Livak and Schmittgen, 2001).
Statistical Analysis
The activity of ASS and H+-ATPase, the concentration of amino acids, and the pHi and ATP levels were all determined in triplicate for each pH growth condition tested. A one-way analysis of variance (ANOVA) with Duncan test was performed using SPSS 19.0 software (SPSS Inc., Chicago, IL, United States) to investigate the significance of differences.
Results and Discussion
Gene Screening
The argG gene (orf00834) is a core regulatory gene during acid stress response according to the co-expression network (Figure 2), and the argG gene was over-expressed (2.94 folds) after acid shock (pH 3.0) 1 h. This gene was selected for this assay.
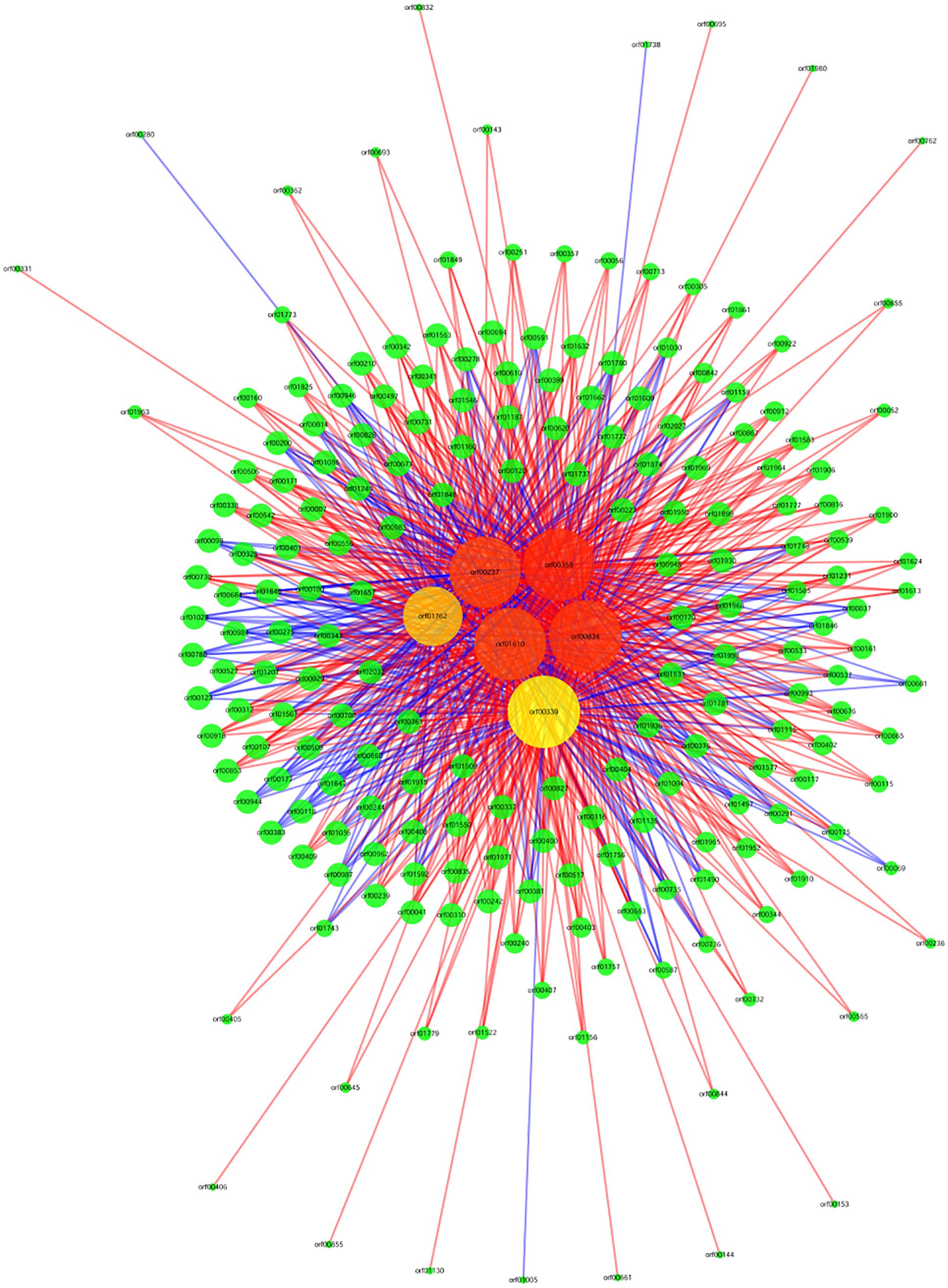
Figure 2. Gene co-expression network. Genes from O. oeni SD-2a after acid shock at 1 h and 0 h. Cycle nodes represent genes, the size of nodes represents the power of the interrelation among the nodes, and the edges between two nodes represent the interactions between genes. The more edges of a gene indicate that more genes connect to it, indicating it has a more central role within the network. A straight line represents a mutual relationship, red indicates that the correlation coefficient is positive, and blue indicates that the correlation coefficient is negative.
Growth Resistance of L. plantarum Under Different pH Conditions
To investigate the influence of ASS on the resistance of transformed cells toward acidity, the growth of each strain was evaluated at OD600 nm and counted plates colonies under different pH conditions (Figure 3 and Supplementary Figure S2). Despite their similar growth performance at pH 6.3, the growth of recombinant strain L. plantarum SL09 (pMG36eargG) was more robust than that of the control L. plantarum SL09 (pMG36e) under acid stress conditions (pH 3.7, 3.3, and 3.2). As the pH decreased from 6.3 to 3.2, the maximum OD600 nm of both strains gradually decreased. The maximum OD600 nm of SL09 (pMG36eargG) was significantly higher than that of the control strain at pH 3.7. This difference was more obvious for the cells grown at pH 3.3, where the maximum OD600 nm of the recombinant strain was 5-fold higher than that of the control strain. A similar result was also observed at pH 3.2, where only the SL09 strain (pMG36eargG) grew well. The results of plate counting shown similar performance as results of the OD600 nm measurement.
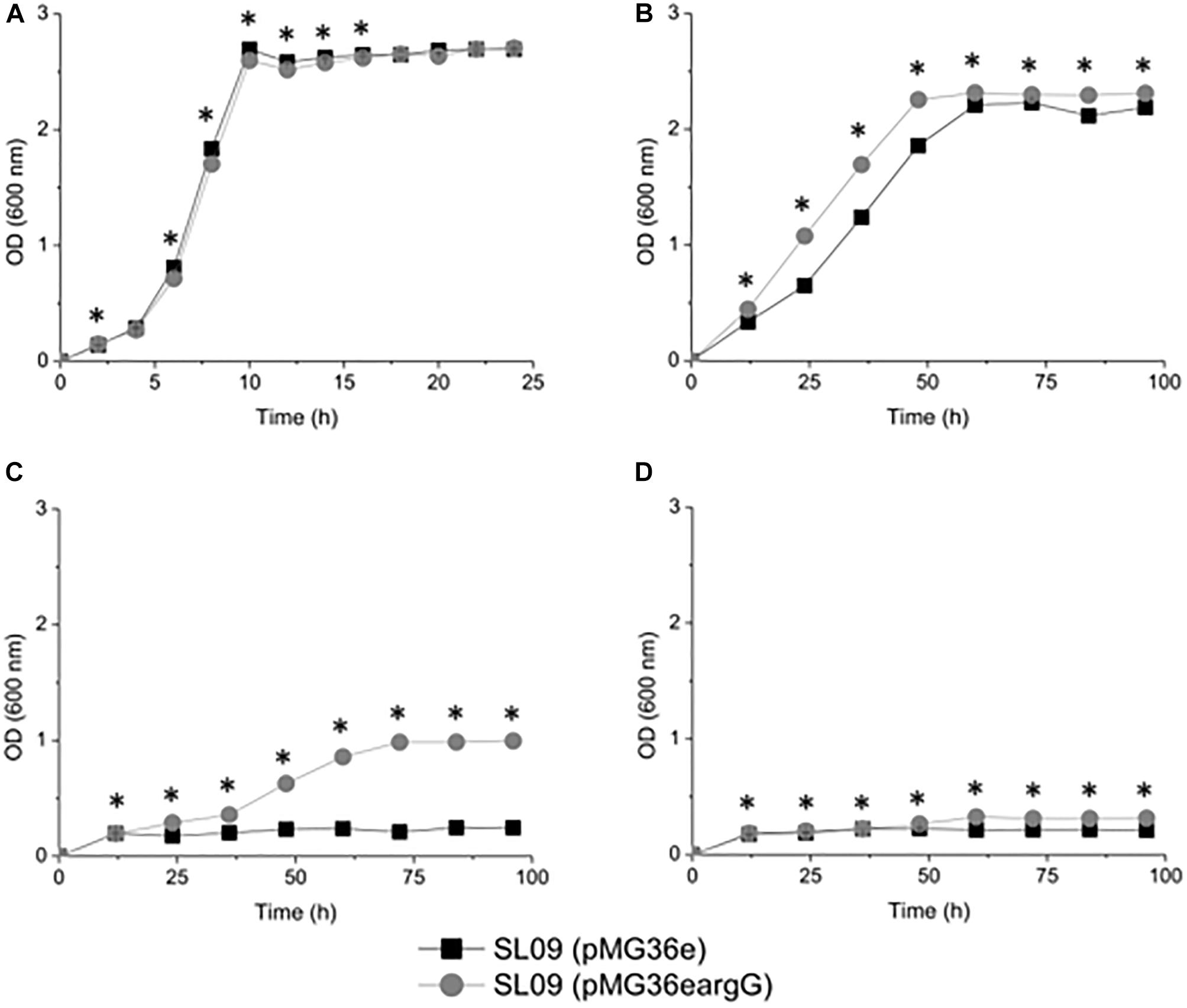
Figure 3. Growth curve of the recombination strain and the control strain under different pH conditions. (A) pH 6.3, (B) pH 3.7, (C) pH 3.3, and (D) pH 3.2. Values presented are the mean of three independent experiments. *Difference significant at 95% confidence level.
The results indicated that the SL09 (pMG36eargG) strain displayed stronger resistance to acid stress than SL09 (pMG36e). Although the acidic environment still inhibited cell growth, the introduction of the argG gene dramatically enhanced the acid tolerance of L. plantarum, allowing these bacteria to survive at a lower pH, one which would normally reduce growth.
ASS Activity Assay and Effect on Intracellular Amino Acids
To verify the heterologous expression of the argG gene, the transcriptional level of argG gene in recombinant and control L. plantarum was analyzed. (The RNA quality was shown in Supplementary Figure S1). As shown in Figure 4A, the expression level of argG was detected in the recombinant strain, (pMG36eargG) with strain SL09 (pMG36e) as control, and the relative expression level was significantly higher under acid stress conditions (pH 3.7). Figure 5A shows the ASS activity of both strains under the favorable and acid stress conditions (pH 6.3 and pH 3.7, respectively). Indeed, the recombinant strain exhibited higher ASS activity than did the control strain, especially under acid stress (pH 3.7, 11-fold difference). From pH 6.3 to pH 3.7, the ASS activity of the control strain was decreased by 61%, but the ASS activity of SL09 (pMG36eargG) increased by 260%. The improvement of ASS activity at pH 3.7 demonstrated that acid stress induced the high-efficiency expression of the argG gene in the recombinant strain. In arginine biosynthesis, ASS acts as the rate-limiting enzyme encoded by argG gene (Lemke and Howell, 2002). Indeed, as shown in Figure 5B, the amount of arginine synthesized was elevated, which may be attributed to the increased ASS activity level. Based on these findings, the acid tolerance enhancement of recombinant strain benefited from the heterologous expression of the argG gene that regulates ASS in the arginine deiminase pathway (ADI pathway).
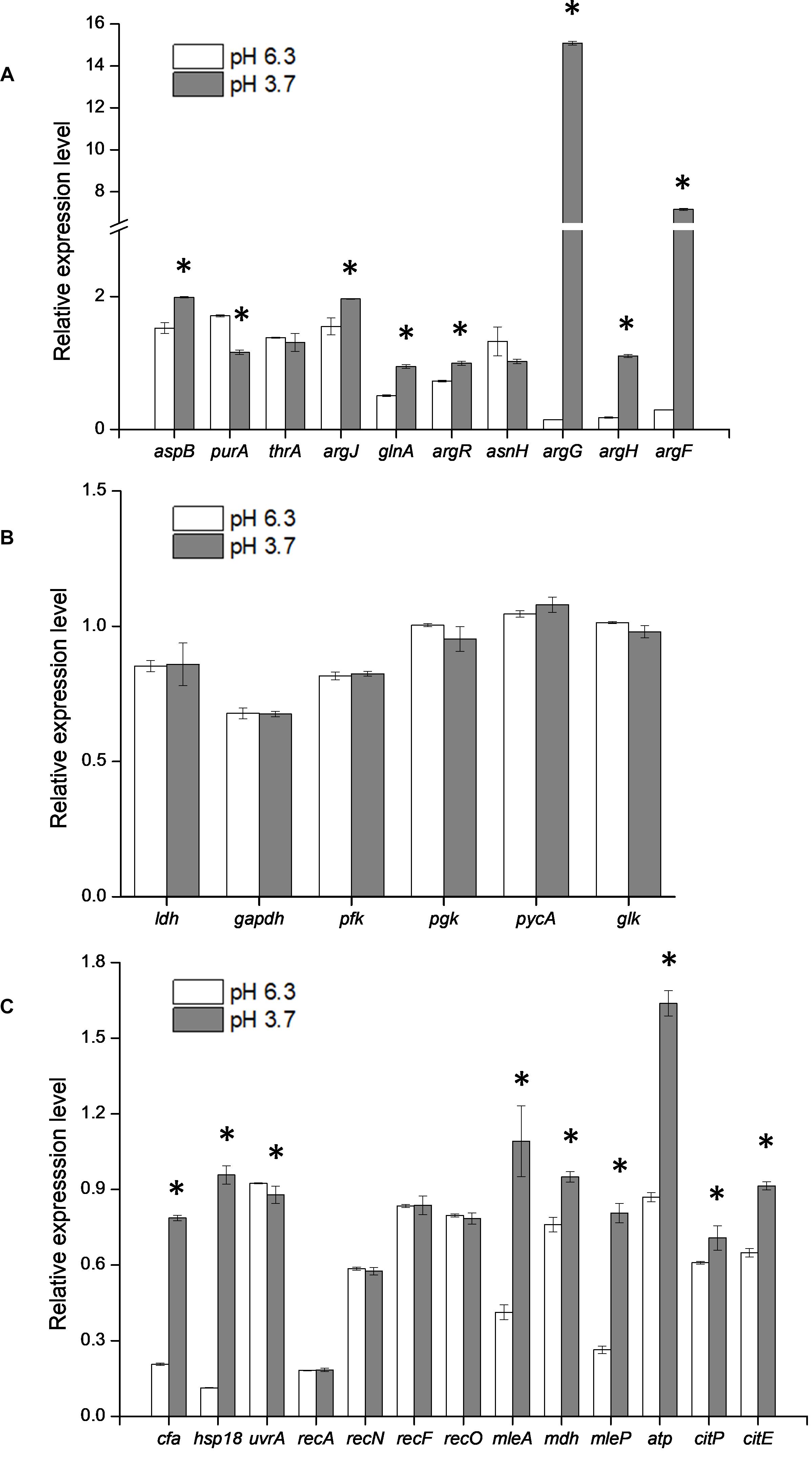
Figure 4. Effect of argG gene heterologous expression on the transcription of amino acid metabolic genes (A), glycolytic genes (B), and other stress response genes (C). *Difference significant at 95% confidence level.
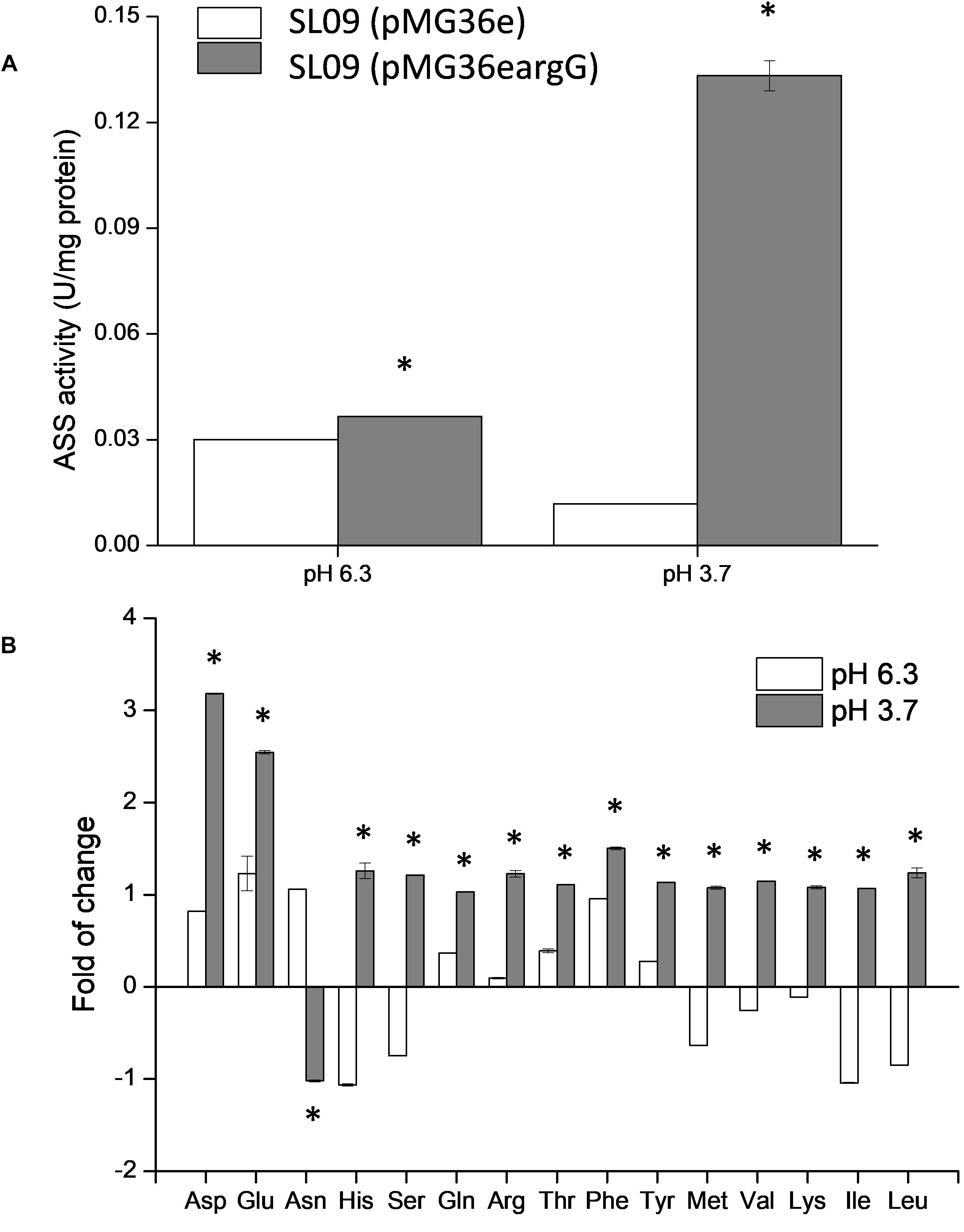
Figure 5. Effect of argG gene heterologous expression on ASS activity (A) and intracellular amino acids (B). The fold change of intracellular amino acids in the recombinant strain were determined relative to the levels in SL09 (pMG36e) at pH 6.3 and pH 3.7. *Difference significant at 95% confidence level.
Since the metabolism of amino acids is complex and consists of multiple interactions (Fernández and Zúñiga, 2006), the impact of heterologous expression of the argG gene on amino acid metabolic genes can be seen at the transcriptional level (Figure 4A). The expressions of aspB, thrA, glnA, argR, argG, argH, and argF were significantly higher when SL09 (pMG36eargG) was exposed to acid stress than under the control condition, while the expression of purA and asnH was decreased. We observed that the genes involved in the ADI pathway were upregulated while the genes converting aspartate into adenylosuccinate and asparagine were downregulated, which is beneficial to the accumulation of aspartate, an arginine precursor. Investigating further, the levels of intracellular amino acids in the recombinant strain were compared to those in the control strain at pH 6.3 and pH 3.7. As is shown in Figure 5B, the heterologous expression of argG gene increases the concentrations of aspartate, glutamate, glutamine, arginine, and threonine under acid stress, most of them are related to the ADI pathway, which was in accordance with the RT-qPCR results.
In this study, the heterologous expression of argG gene tilted amino acid metabolism toward ADI pathway, which can produce alkaline products to neutralize H+ (Tonon and Lonvaud-Funel, 2000), meanwhile, putrescine could be formed (Henríquez-Aedo et al., 2016). Putrescine is one of the biogenic amine present in wine, one which will affect the quality and safety of wine. The relationship of heterologous expression of argG gene and content of putrescine needs to be explored it next step. In addition, Bourdineaud et al. (2002) found that arginine stimulated pre-adaption of O. oeni to wine stress at the start of wine-making, which may be related to the expression level of stress response genes, including ftsH, omrA, and arcR, which were higher when the medium contained arginine. Subsequent study showed that arginine combined with fructose triggered the expression of ftsH, omrA, and arcR genes (Bourdineaud, 2006).
ASS Effect on Glycolysis Pathway and Other Response Genes
Glycolysis is the major pathway that produces energy for LAB growth, except for the amino acid metabolism pathway. There are many genes involved glycolysis, such as the gapdh, ldh, pfk, pgk, pycA, and glk genes (Jan et al., 2013). Additionally, other stress response genes together with the genes in glycolysis pathway were chosen to investigate the influence induced by the expression of argG gene (Jobin et al., 1997; Grandvalet et al., 2008; Figures 4B,C). The expression level of all genes in this study was calculated based the expression of the control strain SL09 (pMG36e). Compared with pH 6.3, the expression level of some genes (gapdh, ldh, pfk, pgk, pycA, and glk) in glycolysis pathway were not significantly changed at pH 3.7. These results suggested that the heterologous expression of argG in L. plantarum did not have significant effects on the glycolysis pathway under acid stress. This also recommended that those genes, such as ldh, can be used as internal control genes for RT-qPCR experiments in L. plantarum (Fiocco et al., 2008; Duary et al., 2010). Furthermore, at pH 3.7, the expression level of cfa, hsp1, mleA, mdh, mleP, atp, citP, and citE were higher than pH 6.3, displaying an increase between 1.0- and 8.5-fold. The cell membrane is the first barrier against an external unfavorable environment for LAB. Maintenance of the quality of the cell membrane is improved by the increased expression of the cfa and hsp1 genes. In O. oeni, the CFA encoded by the cfa gene could reduce the effects of stress on the membrane, since cyclopropane rings restrict the overall mobility and disorder of acyl chains more than the cis double bonds. Additionally, in O. oeni, the hsp18 gene encodes a small heat shock protein (sHSP), which contributes to the maintenance of membrane integrity under stress conditions by preventing the thermal aggregation of cellular proteins (Maitre et al., 2014). There are three genes which encode for small heat shock protein in L. plantarum. The hsp1 gene was involved in controlling and improving membrane fluidity, and the hsp3 gene may be responsible for the induction of thermotolerance. However, the deletion of hsp2 did not significantly impair resistance to heat and other stresses (Arena et al., 2019). In this study, we investigated the expression level of hsp1 genes which may be related to acid-stress response of L. plantarum. The consumption of H+ is another response to acid stress, and the recombinant strain at pH 3.7 showed a higher expression level of atp gene than pH 6.3. H+-ATPase is encoded by the atp gene, an enzyme can synthesize ATP using the H+ from the extracellular space into the cell.
The mleP, mleA, and mdh gene were important genes, responsible for malate metabolism (Augagneur et al., 2007, 2016), and citP and citE gene played a major role in citrate metabolism (Augagneur et al., 2007), the expression of these genes was enhanced and was beneficial in improving strain acid tolerance. The metabolism of L-malate and citrate does not directly provide an energy source, but decarboxylation and the efflux of metabolites generate a proton motive force that can be used to drive ATP synthesis by H+-ATPase (Olguín et al., 2010).
The effects of heterologous expression of the argG gene on SL09 at pH 3.7 are presented in Figure 6. The heterologous expression of argG gene did not affect the expression of genes related to DNA damage repair, because the uvrA, recA, recN, recF, and recO, were not significantly improved at pH 3.7,but may have stimulated the expression of hsp1, cfa, atp, and the malate and citrate metabolic genes under the acid condition. The functions of these genes include maintenance of the quality of the cell membrane, to exclude H+ and produce more ATP.
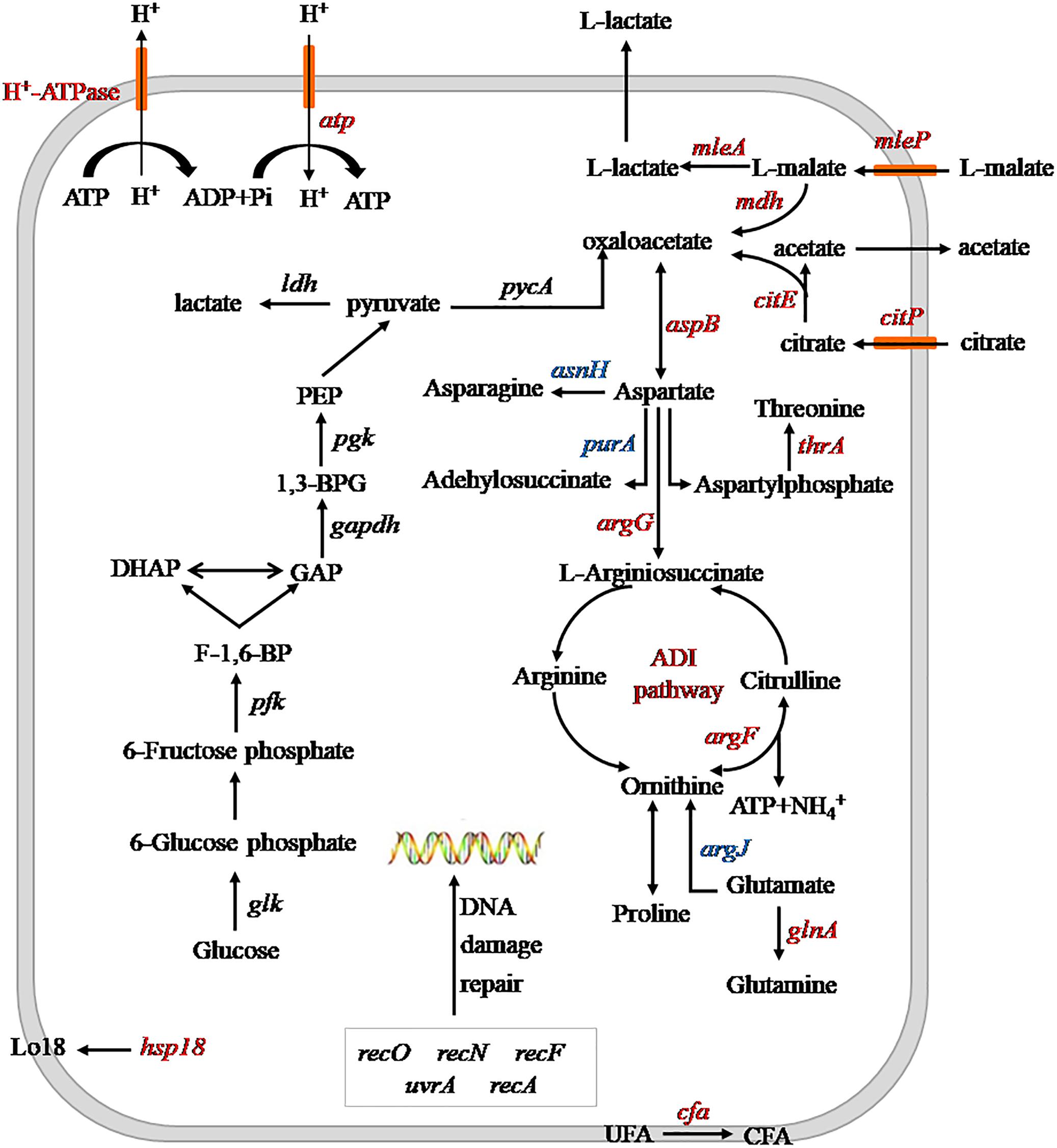
Figure 6. Schematic diagram of heterologous expression of the argG gene in SL09 at pH 3.7. Genes are in italics, the red color represents increased expression of genes or enzyme activity, the blue color represents decreased expression of genes. PEP, phosphoenolpyruvate; 1,3-BPG, 1,3- diphosphoglycerate; GAP, glyceraldehyde phosphate; DHAP, dihydroxyacetone phosphate; F-1,6-BP, fructose diphosphate; UFA unsaturated fatty acids; and CFA, cyclopropane fatty acids; ADI, arginine deiminase.
ASS Effect on pHi, H+-ATPase Activity and Intracellular ATP Level
The pHi, H+-ATPase activity and intracellular ATP level of both strain were measured at pH 3.7 and pH 6.3, respectively (Figures 7A–C), to further investigate the effect of heterologous expression of the argG gene on acid stress resistance and the ASS effect on other stress response genes.
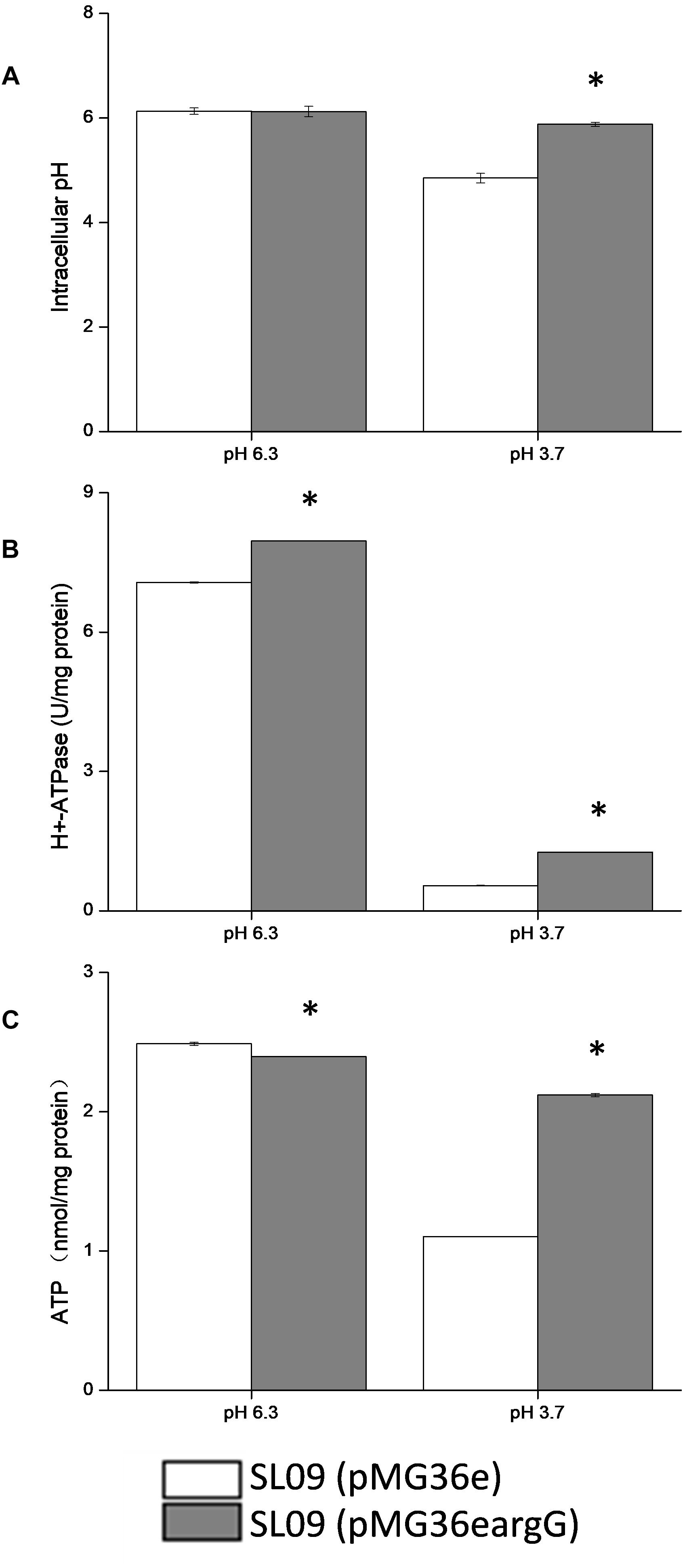
Figure 7. Effect of argG gene heterologous expression on intracellular pH (A), H+-ATPase activity (B), and intracellular ATP level (C). *Difference significant at 95% confidence level.
The intracellular pH of both strains did not show a significant difference at pH 6.3. Compared with pH 6.3, the pHi of the recombinant strain and the control strain were decreased at pH 3.7, the intracellular pH of the recombinant strain stabilized at 5.83, and the control strain decreased to 4.75. Obviously, the pHi of the recombinant strain declined less than the control strains. Although the H+-ATPase activity of both strains decreased at pH 3.7, but the H+-ATPase activity of the recombinant strain was two-fold higher than the control strain (pMG36e). Similarly, the ATP level of SL09 (pMG36eargG) and SL09 (pMG36e) were both decreased at pH 3.7, with the recombinant strain maintaining a higher ATP level (88.8%) than the wild-type strain (44.6%). These results were in accordance with the RT-qPCR results.
The intracellular pH affected a variety of biochemical reactions including reactions catalyzed by enzymes, since pHi is crucial for the maintenance of normal physiological activity in cells. The results indicated that the heterologous expression of the argG gene in L. plantarum increased the ability of cells to maintain neutral pH, which may be related to the higher mRNA level of atp, citrate and malate metabolic genes, and H+-ATPase activity under acid conditions. The pHi affected the activity of H+-ATPase, meanwhile the H+-ATPase activity also affected pHi because H+ is pumped out of cells through the H+-ATPase coupled with ATP hydrolysis (O’Sullivan and Condon, 1999). Moreover, citrate and malate metabolism could improve the pHi by using H+ during decarboxylation. The influence of the heterologous expression of the argG gene brought was complex and systematic and may increase the expression genes involved in consumption of H+ indirectly based on results, but further investigation into the relationship between them is needed.
In addition, the heterologous expression of the argG gene in L. plantarum contributed the ATP level in cells suggested by results. ATP levels in cells is a direct energy source, playing a crucial role in maintaining bacteria growth, proliferation, and cellular functions. The content of intracellular ATP was affected by many factors, in this study, the higher H+-ATPase activity of SL09 (pMG36eargG) under pH 3.7 promoted formation of ATP by consuming H+, the increased expression of genes involved malate and citrate of SL09 (pMG36eargG) under pH 3.7 was beneficial to ATP synthesis, and the heterologous expression of the argG gene in L. plantarum improved the concentration of amino acids participated in ADI pathway, which the arginine metabolism through the ADI pathway produces 1 mol of ATP per mol of arginine consumed. The level of intracellular ATP of SL09 (pMG36eargG) was higher than SL09 (pMG36e) according to the results, which will be helpful to the growth of the strain, the maintenance of cell functions, and the instigation of the stress mechanism. Therefore, the SL09 (pMG36eargG) demonstrated better performance than SL09 (pMG36e) under acid stress.
Conclusion
The heterologous expression of argG gene fromO. oeni SD-2a was achieved in L. plantarum SL09. Due to the expression of the argG gene, the acid stress resistance of recombination L. plantarum was improved, mainly affecting the ADI pathway, malate and citrate metabolism, with an increase in pHi, H+-ATPase activity and intracellular ATP levels. Additionally, the heterologous expression of argG gene also stimulated the expression of hsp1, cfa, and other genes related to malate and citrate metabolism, which requires further investigation. This work may be helpful to understand and eventually obtain O. oeni strains with high acid tolerance in winemaking industry.
Data Availability
The datasets generated for this study can be found in Sequence Read Archive (SRA) database, SRP105332.
Author Contributions
HL, LL, and HZ conceived the idea of the study. HZ, LL, SP, and LY designed and carried out the experiments. LL and HZ analyzed the data and wrote the manuscript. HW revised the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 31471708).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Prof. Shuwen Liu for supplying pMG36e plasmid and L. plantarum SL09. We would also like to thank the reviewers for their comments and suggestions, which greatly improved the original version of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01393/full#supplementary-material
References
Araque, I., Gil, J., Carrete, R., Constanti, M., Bordons, A., and Reguant, C. (2016). Arginine deiminase pathway genes and arginine degradation variability in Oenococcus oeni strains. Folia Microbiol. 61, 109–118. doi: 10.1007/s12223-015-0416-419
Arena, M. E., and Manca de Nadra, M. C. (2005). Influence of ethanol and low pH on arginine and citrulline metabolism in lactic acid bacteria from wine. Res. Microbiol. 156, 858–864. doi: 10.1016/j.resmic.2005.03.010
Arena, M. P., Capozzi, V., Longo, A., Russo, P., Weidmann, S., Rieu, A., et al. (2019). The phenotypic analysis of Lactobacillus plantarum shsp mutants reveals a potential role for hsp1 in cryotolerance. Front. Microbiol. 10:838. doi: 10.3389/fmicb.2019.00838
Assad-Garcia, J. S., Bonnin-Jusserand, M., Garmyn, D., Guzzo, J., Alexandre, H., and Grandvalet, C. (2008). An improved protocol for electroporation of Oenococcus oeni ATCC BAA-1163 using ethanol as immediate membrane fluidizing agent. Lett. Appl. Microbiol. 47, 333–338. doi: 10.1111/j.1472-765X.2008.02435.x
Augagneur, Y., Ritt, J. F., Linares, D. M., Remize, F., Tourdot-Maréchal, R., Garmyn, D., et al. (2007). Dual effect of organic acids as a function of external pH in Oenococcus oeni. Arch. Microbiol. 188, 147–157. doi: 10.1007/s00203-007-0230-230
Beltramo, C., Oraby, M., Bourel, G., Garmyn, D., and Guzzo, J. (2004). A new vector, pGID052, for genetic transfer in Oenococcus oeni. FEMS Microbiol. Lett. 236, 53–60. doi: 10.1016/j.femsle.2004.05.029
Berbegal, C., Peña, N., Russo, P., Grieco, F., Pardo, I., Ferrer, S., et al. (2016). Technological properties of Lactobacillus plantarum strains isolated from grape must fermentation. Food Microbiol. 57, 187–194. doi: 10.1016/j.fm.2016.03.002
Betteridge, A., Grbin, P., and Jiranek, V. (2015). Improving Oenococcus oeni to overcome challenges of wine malolactic fermentation. Trends Biotechnol. 33, 547–553. doi: 10.1016/j.tibtech.2015.06.008
Bourdineaud, J. P. (2006). Both arginine and fructose stimulate pH-independent resistance in the wine bacteria Oenococcus oeni. Int. J. Food Microbiol. 107, 274–280. doi: 10.1016/j.ijfoodmicro.2005.09.011
Bourdineaud, J. P., Nehmé, B., and Lonvaud-Funel, A. (2002). Arginine stimulates preadaptation of Oenococcus oeni to wine stress. Sci. Aliment. 22, 113–121. doi: 10.3166/sda.22.113-121
Breeuwer, P., Drocourt, J., Rombouts, F. M., and Abee, T. (1996). A novel method for continuous determination of the intracellular pH in bacteria with the internally conjugated fluorescent probe 5 and 6-carboxyfluorescein succinimidyl ester. Appl. Environ. Microbiol. 62, 178–183. doi: 10.1021/bk-1996-0647.ch017
Capozzi, V., Russo, P., Ladero, V., Fernandez, M., Fiocco, D., Alvarez, M. A., et al. (2012). Biogenic amines degradation by Lactobacillus plantarum: toward a potential application in wine. Front. Microbiol. 3:122. doi: 10.3389/fmicb.2012.00122
Cruz, C., Egsgaard, H., Trujillo, C., Ambus, P., Requena, N., Martins-Loucao, M. A., et al. (2007). Enzymatic evidence for the key role of arginine in nitrogen translocation by arbuscular mycorrhizal fungi. Plant Physiol. 144, 782–792. doi: 10.1104/pp.106.090522
Darsonval, M., Julliat, F., Msadek, T., Alexandre, H., and Grandvalet, C. (2018). CtsR, the master regulator of stress-response in Oenococcus oeni, is a heat sensor interacting with ClpL1. Front. Microbiol. 9:3135. doi: 10.3389/fmicb.2018.03135
Darsonval, M., Msadek, T., Alexandre, H., and Grandvalet, C. (2016). The antisense RNA approach: a new application for in vivo investigation of the stress response of Oenococcus oeni, a wine-associated lactic acid bacterium. Appl. Environ. Microbiol. 82, 18–26. doi: 10.1128/AEM.02495-2415
Dimopoulou, M., Raffenne, J., Claisse, O., Miot-Sertier, C., Iturmendi, N., Moine, V., et al. (2018). Oenococcus oeni exopolysaccharide biosynthesis, a tool to improve malolactic starter performance. Front. Microbiol. 9:1276. doi: 10.3389/fmicb.2018.01276
Du Toit, M., Engelbrecht, L., Lerm, E., and Krieger-Weber, S. (2011). Lactobacillus: the next generation of malolactic fermentation starter cultures—an overview. Food Bioproc. Tech. 4, 876–906. doi: 10.1007/s11947-010-0448-448
Duary, R. K., Batish, V. K., and Grover, S. (2010). Expression of the atpD gene in probiotic Lactobacillus plantarum strains under in vitro acidic conditions using RT-qPCR. Res. Microbiol. 161, 399–405. doi: 10.1016/j.resmic.2010.03.012
Fernández, M., and Zúñiga, M. (2006). Amino acid catabolic pathways of lactic acid bacteria. Crit. Rev. Microbiol. 32:155. doi: 10.1080/10408410600880643
Fiocco, D., Crisetti, E., Capozzi, V., and Spano, G. (2008). Validation of an internal control gene to apply reverse transcription quantitative PCR to study heat, cold and ethanol stresses in Lactobacillus plantarum. World J. Microb. Biot. 24, 899–902. doi: 10.1007/s11274-007-9556-9557
Fortier, L. C., Tourdot-Maréchal, R., Diviès, C., Lee, B. H., and Guzzo, J. (2003). Induction of Oenococcus oeni H+-ATPase activity and mRNA transcription under acidic conditions. FEMS Microbiol. Lett. 222, 165–169. doi: 10.1016/s0378-1097(03)00299-294
Grandvalet, C., Assad-Garcia, J. S., Chu-Ky, S., Tollot, M., Guzzo, J., Gresti, J., et al. (2008). Changes in membrane lipid composition in ethanol- and acid-adapted Oenococcus oeni cells: characterization of the cfa gene by heterologous complementation. Microbiology 154, 2611–2619. doi: 10.1099/mic.0.2007/016238-16230
Guzzo, J. (2011). “Stress responses of Oenococcus oeni,” in Stress Responses of Lactic Acid Bacteria, eds E. Tsakalidou and K. Papadimitriou (Berlin: Springer), 349–365.
Henríquez-Aedo, K., Durán, D., Garcia, A., Hengst, M. B., and Aranda, M. (2016). Identification of biogenic amines-producing lactic acid bacteria isolated from spontaneous malolactic fermentation of chilean red wines. LWT Food Sci. Technol. 68, 183–189. doi: 10.1016/j.lwt.2015.12.003
Hu, Z., Li, L., Yuan, Y., and Yue, T. (2014). Ultrasensitive and simultaneous determination of twenty-one amino acids and amines in culture media, red wine and beer. Food Chem. 158, 56–65. doi: 10.1016/j.foodchem.2014.02.098
Jan, M., Christian, S., Anders, K. H., and Peter, R. J. (2013). Engineering strategies aimed at control of acidification rate of lactic acid bacteria. Curr. Opin. Biotech. 24, 124–129. doi: 10.1016/j.copbio.2012.11.009
Jobin, M. P., Delmas, F., Garmyn, D., Divies, C., and Guzzo, J. (1997). Molecular characterization of the gene encoding an 18-kilodalton small heat shock protein associated with the membrane of Leuconostoc oenos. Appl. Environ. Microbiol. 63, 609–614. doi: 10.1089/oli.1.1997.7.55
Lee, D.-E., Lee, J., Kim, Y.-M., Myeong, J.-I., and Kim, K.-H. (2016). Uncultured bacterial diversity in a seawater recirculating aquaculture system revealed by 16S rRNA gene amplicon sequencing. J. Appl. Microbiol. 54, 296–304. doi: 10.1007/s12275-016-5571-5574
Lemke, C. T., and Howell, P. L. (2002). Substrate induced conformational changes in argininosuccinate synthetase. J. Biol. Chem. 277, 13074–13081. doi: 10.1074/jbc.m112436200
Liu, L., Zhao, H., Peng, S., Wang, T., Su, J., Liang, Y., et al. (2017). Transcriptomic analysis of Oenococcus oeni SD-2a response to acid shock by RNA-Seq. Front. Microbiol. 8:1586. doi: 10.3389/fmicb.2017.01586
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lorentzen, M. P. G., and Lucas, P. M. (2019). Distribution of Oenococcus oeni populations in natural habitats. Appl. Microbiol. Biot. 103, 2937–2945. doi: 10.1007/s00253-019-09689-z
Maitre, M., Weidmann, S., Dubois-Brissonnet, F., David, V., Coves, J., and Guzzo, J. (2014). Adaptation of the wine bacterium Oenococcus oeni to ethanol stress: role of the small heat shock protein Lo18 in membrane integrity. Appl. Environ. Microbiol. 80, 2973–2980. doi: 10.1128/AEM.04178-4113
Margalef-Català, M., Araque, I., Bordons, A., Reguant, C., and Bautista-Gallego, J. (2016). Transcriptomic and proteomic analysis of Oenococcus oeni adaptation to wine stress conditions. Front. Microbiol. 7:1554. doi: 10.3389/fmicb.2016.01554
Olguín, N., Bordons, A., and Reguant, C. (2010). Multigenic expression analysis as an approach to understanding the behaviour of Oenococcus oeni in wine-like conditions. Int. J. Food Microbiol. 144, 88–95. doi: 10.1016/j.ijfoodmicro.2010.08.032
O’Sullivan, E., and Condon, S. (1999). Relationship between acid tolerance, cytoplasmic pH, and ATP and H+-ATPase levels in chemostat cultures of Lactococcus lactis. Appl. Environ. Microbiol. 65, 2287–2293.
Papadimitriou, K., Alegría, Á, Bron, P. A., de Angelis, M., Gobbetti, M., Kleerebezem, M., et al. (2016). Stress physiology of lactic acid bacteria. Microbiol. Molecul. Biol. R. 80, 837–890. doi: 10.1128/MMBR.00076-15
Peng, S., Liu, L., Zhao, H., Wang, H., and Li, H. (2018). Selection and validation of reference genes for quantitative real-time PCR normalization under ethanol stress conditions in Oenococcus oeni SD-2a. Front. Microbiol. 9:892. doi: 10.3389/fmicb.2018.00892
Quivey, R. G., Kuhnert, W. L., and Hahn, K. (2000). Adaptation of oral streptococci to low pH. Adv. Microb. Physiol. 42, 239–274. doi: 10.1016/S0065-2911(00)42004-42007
Romero, J., Ilabaca, C., Ruiz, M., and Jara, C. (2018). Oenococcus oeni in chilean red wines: technological and genomic characterization. Front. Microbiol. 9:90. doi: 10.3389/fmicb.2018.00090
Rosi, I., Fia, G., and Canuti, V. (2003). Influence of different pH values and inoculation time on the growth and malolactic activity of a strain of Oenococcus oeni. Aust. J. Grape Wine R. 9, 194–199. doi: 10.1111/j.1755-0238.2003.tb00270.x
Schümann, C., Michlmayr, H., Eder, R., Del Hierro, A. M., Kulbe, K. D., Mathiesen, G., et al. (2012). Heterologous expression of Oenococcus oeni malolactic enzyme in Lactobacillus plantarum for improved malolactic fermentation. AMB Express 2, 1–9. doi: 10.1186/2191-0855-2-19
Tang, W., Xing, Z., Li, C., Wang, J., and Wang, Y. (2017). Molecular mechanisms and in vitro antioxidant effects of Lactobacillus plantarum MA2. Food Chem. 221, 1642–1649. doi: 10.1016/j.foodchem.2016.10.124
Tonon, T., Bourdineaud, J. P., and Lonvaud-Funel, A. (2001). The arcABC gene cluster encoding the arginine deiminase pathway of Oenococcus oeni, and arginine induction of a CRP-like gene. Res. Microbiol. 152, 653–661. doi: 10.1016/S0923-2508(01)01244-X
Tonon, T., and Lonvaud-Funel, A. (2000). Metabolism of arginine and its positive effect on growth and revival of Oenococcus oeni. J. Appl. Microbiol. 89, 526–531. doi: 10.1046/j.1365-2672.2000.01142.x
Valdés La Hens, D., Bravo-Ferrada, B. M., Delfederico, L., Caballero, A. C., and Semorile, L. C. (2015). Prevalence of Lactobacillus plantarum and Oenococcus oeni during spontaneous malolactic fermentation in patagonian red wines revealed by polymerase chain reaction-denaturing gradient gel electrophoresis with two targeted genes. Aust. J. Grape Wine R. 21, 49–56. doi: 10.1111/ajgw.12110
Wang, D., Liu, W., Ren, Y., De, L., Zhang, D., Yang, Y., et al. (2016). Isolation and identification of lactic acid bacteria from traditional dairy products in baotou and bayannur of midwestern inner mongolia and q-PCR analysis of predominant species. Korean J. Food Sci. Anim. Resour. 36, 499–507. doi: 10.5851/kosfa.2016.36.4.499
Wang, T., Li, H., Wang, H., and Su, J. (2015). Multilocus sequence typing and pulsed-field gel electrophoresis analysis of Oenococcus oeni from different wine-producing regions of china. Int. J. Food Microbi. 199, 47–53. doi: 10.1016/j.ijfoodmicro.2015.01.006
Weidmann, S., Maitre, M., Laurent, J., Coucheney, F., Rieu, A., and Guzzo, J. (2017). Production of the small heat shock protein Lo18 from Oenococcus oeni in Lactococcus lactis improves its stress tolerance. Int. J. Food Microbiol. 247, 18–23. doi: 10.1016/j.ijfoodmicro.2016.06.005
Keywords: Oenococcus oeni, heterologous expression, acid stress, argininosuccinate synthase, Lactobacillus plantarum
Citation: Zhao H, Liu L, Peng S, Yuan L, Li H and Wang H (2019) Heterologous Expression of Argininosuccinate Synthase From Oenococcus oeni Enhances the Acid Resistance of Lactobacillus plantarum. Front. Microbiol. 10:1393. doi: 10.3389/fmicb.2019.01393
Received: 24 April 2019; Accepted: 04 June 2019;
Published: 21 June 2019.
Edited by:
Vittorio Capozzi, University of Foggia, ItalyReviewed by:
Liliana Carmen Semorile, Universidad Nacional de Quilmes (UNQ), ArgentinaMaria Dimopoulou, Agricultural University of Athens, Greece
Copyright © 2019 Zhao, Liu, Peng, Yuan, Li and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hua Li, lihuawine@nwafu.edu.cn; lihua@nwsuaf.edu.cn; Hua Wang, wanghua@nwsuaf.edu.cn
†These authors have contributed equally to this work
 Hongyu Zhao
Hongyu Zhao Longxiang Liu
Longxiang Liu Shuai Peng
Shuai Peng Lin Yuan1
Lin Yuan1 Hua Wang
Hua Wang