- 1National Institutes for Food and Drug Control, State Food and Drug Administration, Beijing, China
- 2Department of Chinese Medicine Chemistry, Beijing University of Chinese Medicine, Beijing, China
Since the cost of Ophiocordyceps sinensis has increased dramatically and the counterfeits may have adverse effect to health, a rapid and precise species-level DNA barcoding identification system could be a potent approach and significantly enhance the regulatory capacity. The discrimination power of three subunits sequences from nuclear ribosomal RNA gene cluster were determined by Simpson’s index of discrimination using 43 wild O. sinensis fruiting bodies, pure cultures, commercial mycelium fermented powder and counterfeits. The internal transcribed spacer (ITS) sequences showed the highest variance and discrimination power among 43 samples, as determined by Simpson’s index of discrimination (D = 0.972), followed by large subunit (LSU; D = 0.963) and small subunit (SSU; D = 0.921). ITS-2 sequences showed the highest discrimination power for 43 samples among ITS-1, ITS-2, and 5.8S region of ITS sequences. All O. sinensis samples were grouped into a unique ITS sequence cluster under 95% similarity and two O. sinensis samples and six non-O. sinensis samples showed false claims. Our data showed that the ITS region could provide accurate species identification for O. sinensis samples, especially when macroscopic and microscopic method could not be applied in the highly processed commercial products. Since the authentication of O. sinensis related products is essential to ensure its safety and efficacy, identification of O. sinensis through ITS sequence comparison or unique PCR amplification of the species specific target, such as the ITS region, should be considered in the next revision of Chinese pharmacopeia.
Introduction
Ophiocordyceps sinensis, as a well-known Chinese caterpillar fungus, has been used in tonic and healthy food among Asia countries from the 15th century since it contains multiple valuable medicinal components determined by modern pharmacological science (Sun et al., 2017; Tsuk et al., 2017; Wang et al., 2017). O. sinensis is endemic to the Tibetan Plateau, including Tibet, Gansu, Qinghai, Sichuan, and Yunnan province (Yang et al., 2009). The cost of O. sinensis has increased dramatically because of the contradiction between limited natural resource and increasing demand. Its manufacture and sales were strictly regulated by the China Food and Drug Administration (CFDA) since 2016 because the natural fruiting bodies usually contain high amount of arsenic and other heavy metals1. Other Ophiocordyceps related fungi and the conidial form of the artificially cultured O. sinensis fermentation mycelia have also been used as substitutes in Chinese medicine and healthy food (Zhou et al., 2014; Cao et al., 2015).
Because non-O. sinensis species may have adverse effect to health, authentication of O. sinensis related products is essential to ensure its safety and efficacy. Traditionally, the identification of O. sinensis is through its morphological characteristics, but this method could not control the mixed final product authentication. In the last decade, fungi DNA barcoding has become a powerful tool to classify fungal species and provide an optimal option for the contradiction of traditional fungal classification criteria (Jensen et al., 1998; Zhong et al., 2010). Three subunits from nuclear ribosomal RNA gene cluster, including nuclear ribosomal internal transcribed spacer (ITS), large subunit (LSU), and small subunit (SSU) regions in O. sinensis, have been widely used in fungi identification (Xu, 2016) and the ITS region was formally recommended by the International Fungal Barcoding Consortium as the primary fungi barcode (Schoch et al., 2012). But for a specific genus, the discriminatory power of other barcodes might have higher resolving power for species discrimination, for example in lineages outside of Dikarya, ITS showed lower discriminatory power than nSSU and nLSU (Schoch et al., 2012). Identification of O. sinensis through DNA barcoding has been reported (Xiang et al., 2013), but limited data were available to characterize the discrimination power of different nuclear ribosomal DNA barcoding regions for O. sinensis.
In this study, the discrimination power of three subunits sequences from nuclear ribosomal RNA gene cluster were determined by Simpson’s index of discrimination using wild O. sinensis fruiting bodies, pure cultures, commercial mycelium fermented powder and counterfeits.
Materials and Methods
Sample Collection
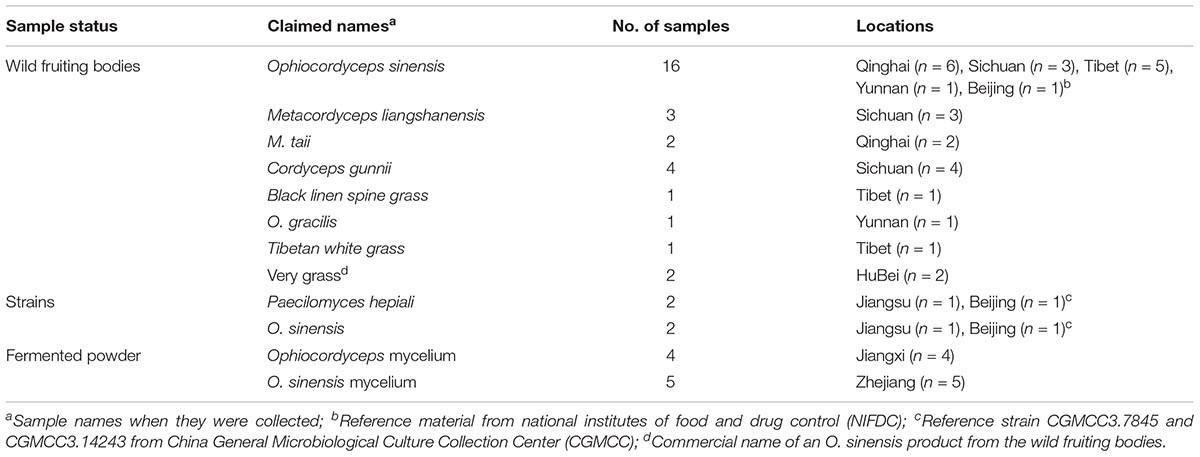
From January 2015 to December 2016, a total of 40 Ophiocordyceps related samples were collected from Sichuan (n = 10), Qinghai (n = 8), Tibet (n = 7), Zhejiang (n = 5), Jiangxi (n = 4), Hubei (n = 2), Jiangsu (n = 2), and Yunnan (n = 2) provinces. An Ophiocordyceps reference material was obtained from the National Institutes of Food and Drug Control (NIFDC), and two reference strains were obtained from the China General Microbiological Culture Collection Center (CGMCC) (Table 1). The reference strains were stored in Brucella broth (BD, Beijing, China) with 50% glycerol at −80°C. Prior to testing, the reference strains were cultured on potato dextrose agar at 18 or 25°C until sufficient growth was obtained.
PCR Amplification and Sequence Analysis
The total genomic DNA from 43 Ophiocordyceps-related samples, including 40 Ophiocordyceps related samples, two reference strains and one reference material, was extracted using a DNeasy plant mini kit (Qiagen, Germany), and the DNA concentrations were quantified using a Qubit fluorometer (Invitrogen, Shanghai, China). Three nuclear ribosomal gene regions, including the SSU, LSU, and ITS regions, were amplified by PCR, as described previously (Schoch et al., 2012). The following primers were used: LSU-F: ACCCGCTGAACTTAAGC, LSU-R: TCCTGAGGGAAACTTCG; SSU-F: GTAGTCATATGCTTGTCTC, SSU-R: CTTCCGTCAATTCCTTTAAG; ITS-F: TCCTCCGCTTATTGATATGC, ITS-R: GGAAGTAAAAGTCGTAACAAGG (Schoch et al., 2012). All PCR products were cloned into the pMD18-T plasmid (Takara Biotechnology Corp., Dalian, China) for sequence analysis at TianyiHuiyuan Biotechnology Corp. The obtained sequences were analyzed using the Sequencher 4.6 software (Gene Codes Corp., Ann Arbor, MI, United States). The search for homologous sequences was performed using the BLAST program at the US National Center for Biotechnology Information (NCBI) website2. The nucleotide sequences identified in this study were deposited into the GenBank.
Comparison of the Discriminatory Power of Three Nuclear Ribosomal Genes
Multiple DNA sequence comparisons were performed with the BioNumerics 7.6 software (Applied Maths, Belgium), and the phylogenetic tree indicating relative genetic similarity was constructed on the basis of the neighbor-joining method. The discriminatory power of the SSU, LSU, and ITS sequence variations were compared, and Simpson’s index of diversity (D) was calculated using the following equation as described previously (Hunter and Gaston, 1988; Dillon et al., 1993):
where N is the total number of Ophiocordyceps-related samples in the study, s is the total number of types described by each method, and nj is the number of samples belonging to the jth type. DNA sequence types were defined as DNA sequences sharing 100% similarity and clusters were defined as ≥95% similarity (C1, C2, C3, …).
Results
Discriminatory Power Comparison of Three Nuclear Ribosomal Genes
All 43 samples were successfully amplified by ITS, LSU, and SSU universal primers. Overall, the ITS sequences showed the highest variance and discrimination power among 43 samples, as determined by Simpson’s index of discrimination (D = 0.972), followed by LSU (D = 0.963) and SSU (D = 0.921) (Table 2). The size of the largest ITS sequence type included five samples compared to eight samples of LSU and SSU sequences. Seven, four and three clusters were identified for ITS, LSU, and SSU sequences using 95% similarity as the cut-off value, respectively. The largest cluster of ITS, LSU, and SSU sequences included 24 samples of one species, 29 samples of two species and 40 samples of four species, respectively (Figure 1–3).
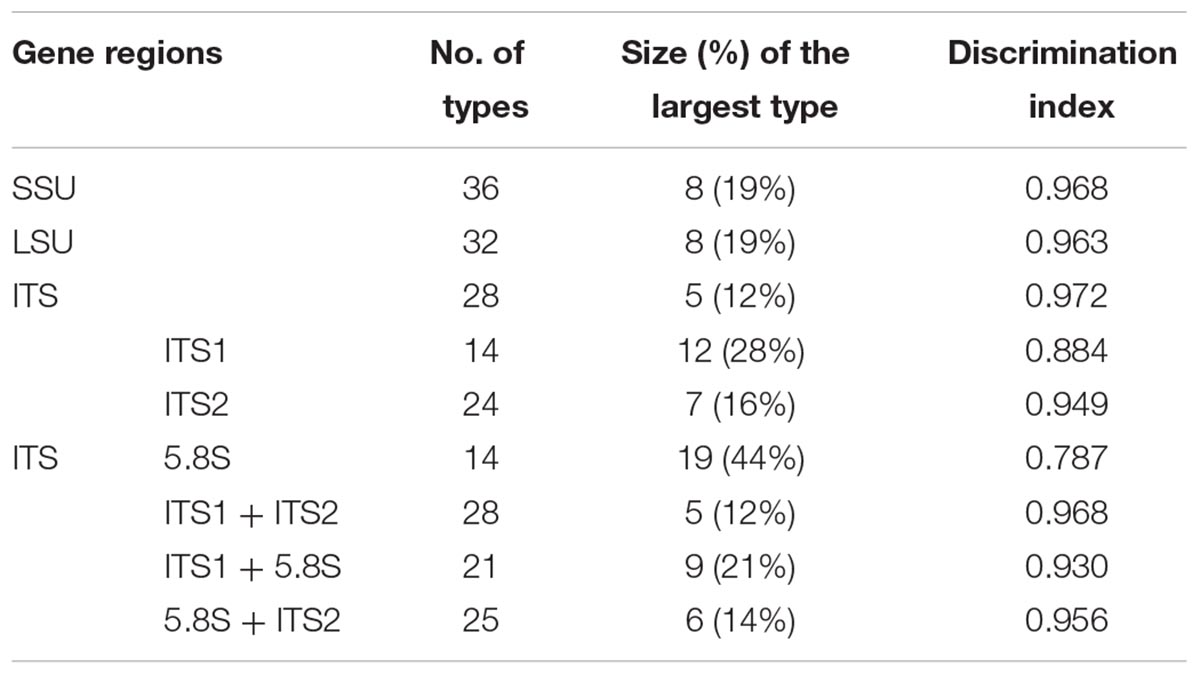
TABLE 2. Discriminatory power evaluation of three nuclear ribosomal RNA genes for 43 Ophiocordyceps related samples.

FIGURE 1. Discrimination of 43 Ophiocordyceps related samples through the internal transcribed spacer (ITS) sequences.
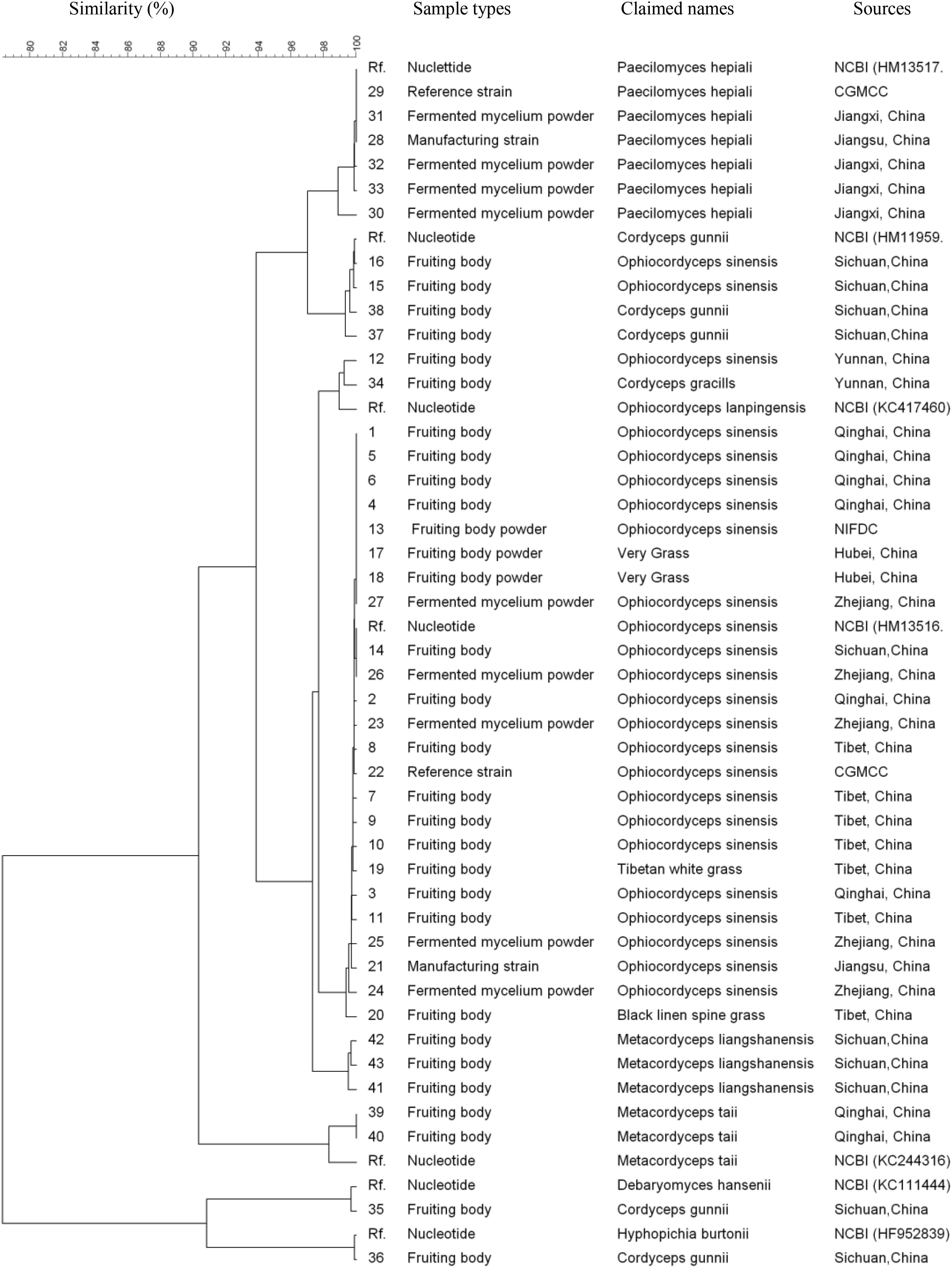
FIGURE 2. Discrimination of 43 Ophiocordyceps related samples through the large subunit (LSU) sequences.
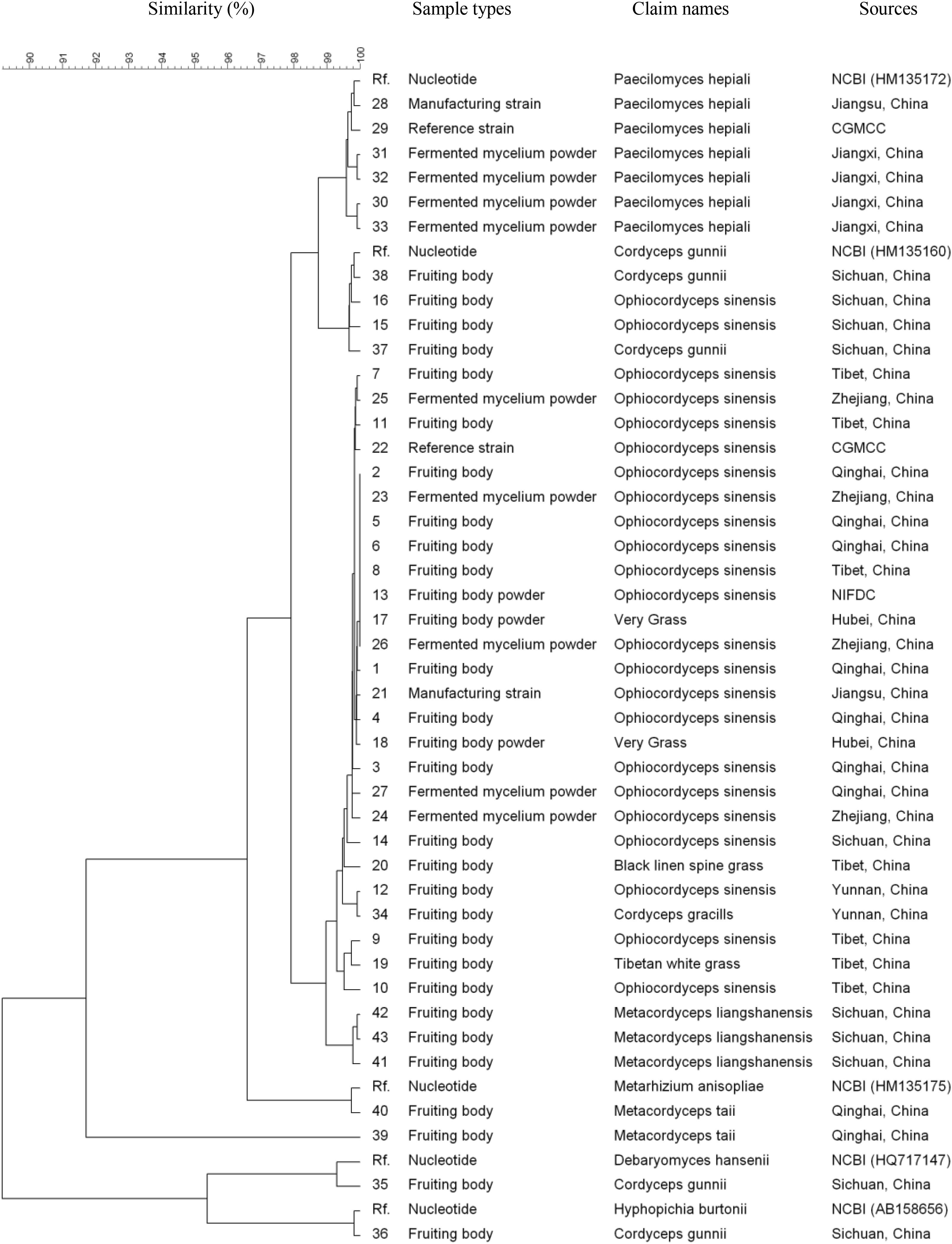
FIGURE 3. Discrimination of 43 Ophiocordyceps related samples through the small subunit (SSU) sequences.
Among ITS-1, ITS-2 and 5.8S region of ITS sequences, ITS-2 sequences showed the highest discrimination power for 43 samples, as determined by Simpson’s index of discrimination (D = 0.949), followed by ITS-1 (D = 0.884) and 5.8 S (D = 0.787).
Species Identification Through ITS Sequences
Among 18 wild fruiting body samples claimed as O. sinensis, the ITS sequences of 15 samples showed 99–100% homology with the O. sinensis sequences deposited in the GenBank and two samples from Sichuan province showed 100% homology with the Cordyceps gunnii sequences deposited in the GenBank, one sample from Yunnan showed 100% homology with the O. lanpingensis sequences deposited in the GenBank. The ITS sequences of one Tibetan white grass sample and one Black linen spine grass sample showed 98–100% homology with the O. sinensis sequences deposited in the GenBank.
The ITS sequences of three Metacordyceps liangshanensis wild fruiting body samples, two O. hawkesii wild fruiting body samples and one O. gracilis wild fruiting body sample showed 95–100% homology with Aspergillus pseudoglaucus, Hyphopichia burtonii and O. lanpingensis sequences deposited in the GenBank, respectively. The ITS sequences of two M. taii wild fruiting body samples and two O. gunnii wild fruiting body samples showed 99% homology with the M. taii or O. gunnii sequences deposited in the GenBank, respectively. The ITS sequences of five O. sinensis mycelium fermented samples and two O. sinensis strains showed 99–100% homology with the O. sinensis sequence deposited in the GenBank. The ITS sequences of four Ophiocordyceps mycelium fermented samples and two Paecilomyces hepiali strains showed 99–100% homology with the P. hepiali sequence deposited in the GenBank.
Discussion
In our study, the discrimination power of three subunits sequences from nuclear ribosomal RNA gene cluster were determined by Simpson’s index of discrimination using O. sinensis fruiting bodies, pure cultures, commercial mycelium fermented powder and counterfeits, the ITS region showed the highest discrimination power for 43 tested samples which was similar as shown in previous DNA barcode study for fungi (Schoch et al., 2012). All O. sinensis samples were grouped into a unique ITS sequence cluster under 95% similarity and two O. sinensis samples and six non-O. sinensis samples showed false claims. Our data showed that the ITS region could provide accurate species identification for O. sinensis samples, especially when macroscopic and microscopic method could not be applied in mixed commercial products.
Previous studies have compared the discriminatory power of different barcoding candidates in O. sinensis and its counterfeit identification (Zhang et al., 2009; Xiang et al., 2013). The probability of correct identification to species level of each barcoding amplicon wasn’t the same for different species (Xu, 2016). Most studies did the research through tree based method, such as maximum parsimony, neighbor-joining, and maximum likelihood analysis (Zhang et al., 2009; Quan et al., 2014). No study has compared the discriminatory power of different barcoding candidates in O. sinensis through a generally acknowledged index. Simpson’s index of diversity has been widely used for the typing method evaluation in molecular epidemiological research (Demczuk et al., 2017; Ramonaite et al., 2017). In this study, the ITS sequence showed the highest discrimination power among ITS, LSU and SSU region determined by Simpson’s index for 43 tested samples. The ITS sequence difference among samples from different species was significantly higher than those among samples from the same species. The length of ITS region is approximately 600 bp and consists of two variable spacers, ITS-1 and ITS-2, which are separated by the highly conserved 5.8S rRNA gene (Schoch et al., 2012). Although both ITS1 and ITS2 are the most common metagenomics sequencing amplicons in fungal community studies (Tonge et al., 2014; Motooka et al., 2017), our data showed ITS-2 had a higher discriminatory power than ITS-1 and should be the optimal metagenomics sequencing target for O. sinensis mixed samples in the future studies.
Since P. hepiali was recovered from the natural O. sinensis fruiting bodies as an endoparasitic fungus (Yang et al., 2008), it is also widely used in Asian countries for various potential pharmacological activities similar to O. sinensis (Thakur et al., 2011; Wang et al., 2016). In this study, 17 ITS clones from individual fruiting bodies were sequenced and no P. hepiali sequence was found which further confirmed that P. hepiali was not the dominant fungi in the natural O. sinensis fruiting bodies (Yang et al., 2008).
Ophiocordyceps sinensis parasitizes more than 50 different species of caterpillar larvae including Hapialus and Thitarodes distributed in the Tibetan Plateau (Wang and Yao, 2011). The morphology identification of O. sinensis fruiting bodies highly relies on experience and is hard to establish new species morphology recognition criteria (Xiang et al., 2013). Furthermore, it takes more than 10 years to train a traditional fungal taxonomist, the number of these experts decreased dramatically in the past decades. In recent years, ITS region has become the consensus primary fungal barcode (Schoch et al., 2012). Our data showed ITS could differentiate O. sinensis from other non-O. sinensis samples at cluster level of 95% similarity, much better than the other two barcoding candidates in this study because of the high similarity among different species. A unique O. sinensis cluster was identified from samples of different sources in this study. Even the closest ITS gene sequence cluster to O. sinensis which was O. lanpingensis showed higher than 7% variation.
In this study, we found two O. sinensis samples and six non-O. sinensis samples had false claims through ITS sequencing. C. gunnii, a synonym of C. hawkesii that parasitizes the larvae of Napialus hunanensis, is a natural counterfeit. Although the morphology is similar, the ITS sequence showed high variation that further confirmed the necessity to establish ITS identification criteria for O. sinensis. Besides, ITS sequence has multiple copies in a single cell which makes it possible to amplify this gene from tiny amounts of samples. Although different O. sinensis PCR identification methods have been developed (Peng et al., 2013; Hou et al., 2017), none of them have been accepted by the Chinese pharmacopeia.
Conclusion
The ITS region showed the highest discrimination power as determined by Simpson’s index of diversity among 43 tested samples which could identify O. sinensis to species level. Since the authentication of O. sinensis related products is essential to ensure its safety and efficacy, O. sinensis identification either through ITS sequence variation comparison or unique PCR amplification of the species specific target, such as the ITS region, should be considered in the next revision of Chinese pharmacopeia, especially for those highly processed products.
Author Contributions
SC designed this study. PZ and XR carried out the experimental work and data analysis. SK and FW collected and analyzed the samples. BL and SM guided the experiments.
Funding
This study was supported by grant (2014ZX09304-307-02) from the Ministry of Science and Technology of the People’s Republic of China.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We sincerely thank Dr. Kai Cao from EP Technology (Shanghai) Co., Ltd. and Prof. Yijian Yao from the Institute of Microbiology and Chinese Academy of Sciences for their expert technical assistance.
Footnotes
References
Cao, L., Ye, Y., and Han, R. (2015). Fruiting body production of the medicinal Chinese caterpillar mushroom, Ophiocordyceps sinensis (ascomycetes), in artificial medium. Int. J. Med. Mushrooms 17, 1107–1112. doi: 10.1615/IntJMedMushrooms.v17.i11.110
Demczuk, W., Sidhu, S., Unemo, M., Whiley, D. M., Allen, V. G., Dillon, J. R., et al. (2017). Neisseria gonorrhoeae sequence typing for antimicrobial resistance, a novel antimicrobial resistance multilocus typing scheme for tracking global dissemination of N. gonorrhoeae strains. J. Clin. Microbiol. 55, 1454–1468. doi: 10.1128/JCM.00100-17
Dillon, J. A., Rahman, M., and Yeung, K. H. (1993). Discriminatory power of typing schemes based on Simpson’s index of diversity for Neisseria gonorrhoeae. J. Clin. Microbiol. 31, 2831–2833.
Hou, F. X., Cao, J., Wang, S. S., Wang, X., Yuan, Y., Peng, C., et al. (2017). Development and evaluation of a rapid PCR detection kit for Ophiocordyceps sinensis. Zhongguo Zhong Yao Za Zhi 42, 1125–1129. doi: 10.19540/j.cnki.cjcmm.20170217.011
Hunter, P. R., and Gaston, M. A. (1988). Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J. Clin. Microbiol. 26, 2465–2466.
Jensen, A. B., Gargas, A., Eilenberg, J., and Rosendahl, S. (1998). Relationships of the insect-pathogenic order Entomophthorales (Zygomycota, fungi) based on phylogenetic analyses of nuclear small subunit ribosomal DNA sequences (SSU rDNA). Fungal Genet. Biol. 24, 325–334.
Motooka, D., Fujimoto, K., Tanaka, R., Yaguchi, T., Gotoh, K., Maeda, Y., et al. (2017). Fungal ITS1 deep-sequencing strategies to reconstruct the composition of a 26-species community and evaluation of the gut Mycobiota of healthy Japanese individuals. Front. Microbiol. 8:238. doi: 10.3389/fmicb.2017.00238
Peng, Q., Zhong, X., Lei, W., Zhang, G., and Liu, X. (2013). Detection of Ophiocordyceps sinensis in soil by quantitative real-time PCR. Can. J. Microbiol. 59, 204–209. doi: 10.1139/cjm-2012-0490
Quan, Q. M., Chen, L. L., Wang, X., Li, S., Yang, X. L., Zhu, Y. G., et al. (2014). Genetic diversity and distribution patterns of host insects of caterpillar fungus Ophiocordyceps sinensis in the Qinghai-Tibet Plateau. PLoS One 9:e92293. doi: 10.1371/journal.pone.0092293
Ramonaite, S., Tamuleviciene, E., Alter, T., Kasnauskyte, N., and Malakauskas, M. (2017). MLST genotypes of Campylobacter jejuni isolated from broiler products, dairy cattle and human campylobacteriosis cases in Lithuania. BMC Infect. Dis. 17:430. doi: 10.1186/s12879-017-2535-1
Schoch, C. L., Seifert, K. A., Huhndorf, S., Robert, V., Spouge, J. L., Levesque, C. A., et al. (2012). Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc. Natl. Acad. Sci. U.S.A. 109, 6241–6246. doi: 10.1073/pnas.1117018109
Sun, S., Xie, J., Peng, T., Shao, B., Zhu, K., Sun, Y., et al. (2017). Broad-spectrum immunoaffinity cleanup for the determination of aflatoxins B1, B2, G1, G2, M1, M2 in Ophiocordyceps sinensis and its pharmaceutical preparations by ultra performance liquid chromatography tandem mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 106, 112–118. doi: 10.1016/j.jchromb.2017.10.013
Thakur, A., Hui, R., Hongyan, Z., Tian, Y., Tianjun, C., and Mingwei, C. (2011). Pro-apoptotic effects of Paecilomyces hepiali, a Cordyceps sinensis extract on human lung adenocarcinoma A549 cells in vitro. J. Cancer Res. Ther. 7, 421–426. doi: 10.4103/0973-1482.92007
Tonge, D. P., Pashley, C. H., and Gant, T. W. (2014). Amplicon-based metagenomic analysis of mixed fungal samples using proton release amplicon sequencing. PLoS One 9:e93849. doi: 10.1371/journal.pone.0093849
Tsuk, S., Lev, Y. H., Rotstein, A., Carasso, R., Zeev, A., Netz, Y., et al. (2017). Clinical effects of a commercial supplement of Ophiocordyceps sinensis and Ganoderma lucidum on cognitive function of healthy young volunteers. Int. J. Med. Mushrooms 19, 667–673. doi: 10.1615/IntJMedMushrooms.2017021202
Wang, J., Liu, R., Liu, B., Yang, Y., Xie, J., and Zhu, N. (2017). Systems pharmacology-based strategy to screen new adjuvant for hepatitis B vaccine from traditional Chinese medicine Ophiocordyceps sinensis. Sci. Rep. 7:44788. doi: 10.1038/srep44788
Wang, J., Teng, L., Liu, Y., Hu, W., Chen, W., Hu, X., et al. (2016). Studies on the antidiabetic and antinephritic activities of Paecilomyces hepiali water extract in diet-streptozotocin-induced diabetic sprague dawley rats. J Diabetes Res. 2016:4368380. doi: 10.1155/2016/4368380
Wang, X. L., and Yao, Y. J. (2011). Host insect species of Ophiocordyceps sinensis: a review. Zookeys 127, 43–59. doi: 10.3897/zookeys.127.802
Xiang, L., Song, J., Xin, T., Zhu, Y., Shi, L., Xu, X., et al. (2013). DNA barcoding the commercial Chinese caterpillar fungus. FEMS Microbiol. Lett. 347, 156–162. doi: 10.1111/1574-6968.12233
Yang, J. L., Xiao, W., He, H. X., Zhu, H. X., Wang, S. F., Cheng, K. D., et al. (2008). Molecular phylogenetic analysis of Paecilomyces hepiali and Cordyceps sinensis. Yao Xue Xue Bao 43, 421–426.
Yang, P., Song, P., Sun, S. Q., Zhou, Q., Feng, S., and Tao, J. X. (2009). Differentiation and quality estimation of Cordyceps with infrared spectroscopy. Spectrochim. Acta A Mol. Biomol. Spectrosc. 74, 983–990. doi: 10.1016/j.saa.2009.09.004
Zhang, Y., Xu, L., Zhang, S., Liu, X., An, Z., Wang, M., et al. (2009). Genetic diversity of Ophiocordyceps sinensis, a medicinal fungus endemic to the Tibetan Plateau: implications for its evolution and conservation. BMC Evol. Biol. 9:290. doi: 10.1186/1471-2148-9-290
Zhong, X., Peng, Q., Qi, L., Lei, W., and Liu, X. (2010). rDNA-targeted PCR primers and FISH probe in the detection of Ophiocordyceps sinensis hyphae and conidia. J. Microbiol. Methods 83, 188–193. doi: 10.1016/j.mimet.2010.08.020
Keywords: Ophiocordyceps sinensis, discriminatory analysis, Simpson index of diversity, nuclear ribosomal RNA barcoding sequences, ITS
Citation: Zhang P, Cui S, Ren X, Kang S, Wei F, Ma S and Liu B (2018) Discriminatory Power Evaluation of Nuclear Ribosomal RNA Barcoding Sequences Through Ophiocordyceps sinensis Related Samples. Front. Microbiol. 9:2498. doi: 10.3389/fmicb.2018.02498
Received: 04 March 2018; Accepted: 01 October 2018;
Published: 23 October 2018.
Edited by:
Gustavo Henrique Goldman, Universidade de São Paulo, BrazilReviewed by:
Maryam Dadar, Razi Vaccine and Serum Research Institute, IranBaowei Yang, Northwest A&F University, China
Copyright © 2018 Zhang, Cui, Ren, Kang, Wei, Ma and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shenghui Cui, cuishenghui@aliyun.com Bin Liu, liubinyn67@163.com
 Ping Zhang
Ping Zhang Shenghui Cui
Shenghui Cui Xiu Ren
Xiu Ren Shuai Kang
Shuai Kang Feng Wei
Feng Wei Shuangcheng Ma
Shuangcheng Ma Bin Liu
Bin Liu