- 1State Key Laboratory of Crop Stress Biology for Arid Areas, College of Plant Protection, Northwest A&F University, Yangling, China
- 2NIAB East Malling Research, East Malling, United Kingdom
The Valsa canker, caused by Valsa mali (V. mali), is a destructive disease of apple in Eastern Asia. Effector proteins are important for fungal pathogenicity. We studied a candidate effector VmPxE1 isolated based on the genome information of V. mali. By using the yeast invertase secretion assay system, VmPxE1 was shown to contain a signal peptide with secretory functions. VmPxE1 can suppress BCL-2-associated X protein (BAX)-induced cell death with a high efficacy of 92% in Nicotiana benthamiana. The expression of VmPxE1 was upregulated during the early infection stage and deletion of VmPxE1 led to significant reductions in virulence on both apple twigs and leaves. VmPxE1 was also shown to target an apple ascorbate peroxidase (MdAPX1) by the yeast two-hybrid screening, bimolecular fluorescence complementation and in vivo co-immunoprecipitation. Sequence phylogenetic analysis suggested that MdAPX1 was an ascorbate peroxidase belonging to a subgroup of heme-dependent peroxidases of the plant superfamily. The ectopic expression of MdAPX1 in the mutant of VmPxE1 significantly enhanced resistance to H2O2, while the presence of VmPxE1 seems to disturb MdAPX1 function. The present results provide insights into the functions of VmPxE1 as a candidate effector of V. mali in causing apple canker.
Introduction
Secreted by bacteria, oomycetes, and fungi, effectors are defined as small rich cysteine secreted proteins, contributing to the pathogen virulence (Vleeshouwers and Oliver, 2014). Pathogen and host have been co-evolving, resulting in the establishment of multi-layered pathogen offense and host defense systems. Pathogen-associated molecular patterns (PAMPs) are recognized by pattern recognition receptors (PRRs), and then PAMP-triggered immunity (PTI) of the host is induced (Boller and He, 2009; Albert, 2013). Under the pressure from host defense, pathogens secrete effectors to suppress host defense, leading to effector-triggered susceptibility (ETS). ETS results in production of plant resistance proteins (R proteins) and leads to the second layer of immune response, the effector-triggered immunity (ETI), leading to heavy oxidative burst and hypersensitive reaction (HR) of the host (Jones and Dangl, 2006; Dodds and Rathjen, 2010).
Filamentous pathogenic effectors have been reported to interfere with several aspects of host immunity (Rovenich et al., 2014). Some act as inhibitors of proteases, such as Pit2 from Ustilago maydis and EPI10 from Phytophthora infestans (Tian et al., 2005; Mueller et al., 2013). Some influence enzymes related to the ROS pathway. For example, two cytoplasmic effectors of Phytophthora sojae interact with catalases to regulate H2O2 concentration (Zhang et al., 2015). An U. maydis effector Pep1 targets a maize peroxidase POX12 in vivo and suppresses the early immune responses of maize (Hemetsberger et al., 2012). Some effectors may competitively bind defense-related proteins to disturb the host-recognition system, such as PsXLP1, from P. sojae, acting as a decoy to shield the true virulence factor (Ma et al., 2017), and Ecp6 from Cladosporium fulvum, targeting chitin to suppress the chitin-induced immunity (de Jonge et al., 2010). Moreover, the resistance signal pathway could also be disturbed by pathogenic effectors. For instance, the effector HaRxL44 from Hyaloperonospora arabidopsidis breaks down the SA-triggered immunity (Caillaud et al., 2013) and the U. maydis effector Cmu1 interdicts SA biosynthesis (Djamei et al., 2011). Thus, pathogenic fungi have effectors that could function via several mechanisms to defeat/avoid host defense systems.
Valsa mali is a necrotrophic fungus belonging to Ascomycete and causes Valsa canker on apple, a destructive disease of apple in the Eastern Asia. In China, this disease resulted in significant economic losses (Lee et al., 2006; Wang et al., 2011). Previous research identified 193 candidate effector proteins (CEPs) with unknown functions and predicted 779 secreted proteins of V. mali with rich cysteine residues (average length of 233 amino acids) (Yin et al., 2015). The ability to suppress BAX triggered PCD is an important initial criterion for screening pathogenic effectors (Wang et al., 2011a). BAX is a member of the Bcl-2 family proteins, triggering cell death when expressed in plants. The cell death-promoting function of BAX in plants correlated with the upregulated expression of the defense-related protein PR1, which strongly suggests that BAX activates an endogenous cell-death program, one specific host defense reaction (Lacomme and Santa Cruz, 1999). Using this initial screening system for BAX responses, eight effectors have been identified and one of them (VmEP1) shown to be an important virulence factor of V. mali (Li et al., 2015). There are probably other V. mali effectors yet to be identified (Ke et al., 2014; Yin et al., 2015). Further research is needed to identify other effectors and study their functions.
In this study, we identified VmPxE1 as a cell death suppressor, contributing to the full virulence of V. mali and directly targeting a peroxidase of apple tree.
Materials and Methods
Strains and Culture Conditions
Valsa mali strain 03-8 was cultured at 25°C in the dark on potato dextrose agar (PDA) medium. Nicotiana benthamiana plants were maintained at 25°C with a daily 13 h:11 h light:dark regime. Agrobacterium tumefaciens strain GV3101 used for agro-infiltration experiments was cultured on Luria-Bertani medium at 28°C. Escherichia coli strain JM109 used for storing and propagating plasmids was cultured on Luria-Bertani medium at 37°C. All the strains are stored in the Laboratory of Integrated Management of Plant Diseases at the College of Plant Protection, Northwest A&F University, Yangling, China.
Construction of V. mali cDNA Libraries
Total RNA was extracted using the RNeasy Micro kit (Qiangen, Shenzhen, China) according to the manufacturer’s protocol from (a) V. mali mycelia grown on PDA medium for 3 days, and (b) apple twig tissues of Malus domestica borkh. cv. ‘Fuji’ inoculated with V. mali mycelium plugs [0 h (i.e. immediately before inoculation), 6, 12, 18, 24, 36, 48, 72 h post inoculation (hpi)]. First-strand cDNA was synthesized by the RevertAidTM First Strand cDNA Synthesis Kit (Fermentas, Shenzhen, China) according to the manufacturer’s protocol.
Plasmid Constructs
Targeted genes were amplified from the cDNA library using the Ex-taq mix DNA polymerase (Takara, Dalian, China). For A. tumefaciens infiltration assays in N. benthamiana, PCR products were cloned into the corresponding vectors (Giraldo and Valent, 2013) with the restriction enzyme digestion and T4 DNA ligase (Takara, Dalian, China). Primers used for plasmid constructs are listed in Supplementary Table S1. Sequences of all plasmids were confirmed by Sangon Biotech, Shanghai.
Sequence Analyses
In our previous study, the whole genome shotgun sequences of V. mali were deposited at DDBJ/EMBL/GenBank under the accession JUIY01000000. The secretome and CEPs of V. mali were predicted from materials obtained in previous studies (Li et al., 2015; Yin et al., 2015). Pfam1 was used to predict protein domain structure, and SignalP 4.12 to signal peptides. An unrooted phylogenetic tree was constructed by MEGA5 with the neighbor-joining method.
Agrobacterium tumefaciens Infiltration Assays
Agro-infiltration assays were carried out following the previously described procedure (Dou et al., 2008). Strain GV3101 carrying an expression plasmids was grown in LB medium containing kanamycin (50 μg/ml) for transient expression; cells were re-suspended in 10 mM MgCl2 (pH 5.6) with the suspension adjusted to an OD600 of 0.5. Bacteria were infiltrated with a syringe to the upper leaves of 5-week-old N. benthamiana plants. As a control, plants were infiltrated with bacteria carrying an empty pGR106 vector. Symptoms were assessed 4–5 days after infiltration. Each assay was performed three times.
Secretory Function Validation of Putative N-Terminal Signal Peptide of VmPxE1
The yeast invertase secretion assay was performed to validate the function of the putative N-terminal signal peptide of VmPxE1. The predicted signal peptide of VmPxE1 was cloned to pSUC2 and transformed into yeast strain YTK12 with the lithium acetate method (Geitz et al., 1995). The CMD-W medium (0.08% tryptophan dropout supplement, 2.5% sucrose, 0.65% yeast N base without amino acids, 0.1% glucose, and 2% agar) was used to select YTK12 colonies with pSUC2 empty vector or pSUC2-VmPxE1sigp. For validating invertase secretion, positive colonies on the CMD-W medium were transferred to YPRAA plates (1% yeast extract, 2% raffinose, 2% peptone, and 2 mg/ml antimycin A) containing raffinose as the only carbohydrate source. The untransformed YTK12 colonies, YTK12 colonies transformed with an empty pSUC2 vector, and the non-secreted Mg87 protein from Magnaporthe oryzae were used as negative controls (Gu et al., 2011); the YTK12 colonies carrying the signal peptide of Avr1b from oomycete was used as a positive control (Shan et al., 2004; Gu et al., 2011).
RNA Extraction and Transcript Level Analysis
VmPxE1 transcript level was measured by qRT-PCR in apple twig tissues inoculated with V. mali strain 03-8 sampled at 0 (immediately before inoculation), 6, 12, 18, 24, 36, 48, and 72 hpi. Total RNA was extracted using the RNAeasy R Plant Mini Kit (Qiagen, Shenzhen, China) following the recommended protocol. First-strand cDNA was synthesized using a RT-PCR system (Promega, Madison, WI, United States) following the manufacturer’s instructions. SYBR green qRT-PCR assays were performed to quantify transcript levels; G6PDH of V. mali was chosen as a housekeeping gene (Yin et al., 2013). There were three biological replicates for each treatment. Primers used for qRT-PCR are given in Supplementary Table S2.
Generation of VmPxE1 Mutants
A reaction with three components using the Neo gene as a selective marker was performed for single gene deletion. The Neo gene was amplified with primers Neo-F and Neo-R from PFL2. The upstream and downstream flanking sequences of VmPxE1 were amplified using primer pairs 1F/2R, 3F/4R, respectively. Then, deletion cassette for homologous recombination was generated by double-joint PCR as described previously (Yu et al., 2004). The primer pair CF/CR was used for nest-PCR and produced the gene-replacement construct. Protoplasts of V. mali were prepared and then the gene-replacement construct was transformed into the protoplasts as previously described (Gao et al., 2011). Each putative single gene deletion mutant was verified by PCR using four primer pairs (5F/6R, 7F/NeoR, NeoF/8R, and NeoF/NeoR) to detect the target gene, upstream-neo fusion segment, neo-downstream fusion segment, and the neo gene, respectively. Southern blot hybridization using the DIG DNA Labeling and Detection Kit II (Roche, Mannheim, Germany) was performed to confirm deletion mutants. All primers used for gene deletion are given in Supplementary Table S1.
Complementation of the Deletion Mutants
The entire VmPxE1 gene with upstream 2000 bp was amplified with primers pDL2-VmPxE1-F and pDL2-VmPxE1-R cloned into plasmid pDL2 with the yeast gap repair approach (Bruno et al., 2004; Zhou et al., 2012). The resulting construct, pDL2-VmPxE1, was transformed into protoplasts of the VmPxE1 gene deletion mutants. Complementation was selected by geneticin (G418) and hygromycin, and confirmed by PCR using the primer pair 5F/6R.
Pathogenicity, Conidiation, and Vegetative Growth of Mutants
For pathogenicity assay, detached apple (M. domestica borkh. cv. ‘Fuji’) twigs and leaves were inoculated with the deletion mutant ΔVmPxE1, complementation mutant ΔVmPxE1/VmPxE1 and wild type following the method previously described (Oliva et al., 2010). The experiments were repeated twice with fifteen replicates in each repeat experiment. Vegetative growth and conidiation was examined at 3 and 40 days, respectively. Data were analyzed by Student’s t-test using the SAS software package (SAS Institute, Cary, NC, United States).
Yeast Two-Hybrid Screening
The yeast two-hybrid system was performed for screening VmPxE1 interacting proteins (Ito et al., 2001). The VmPxE1 coding region was cloned into the bait vector pGBKT7 without the signal peptide sequence and pGBKT7-VmPxE1 was transformed into yeast strain AH109. Yeast cells carrying pGBKT7-VmPxE1 were transformed with cDNA library that was constructed into the prey vector pGADT7 using mRNA isolated from the junction of diseased (infected by V. mali wild type strain 03-8) and healthy twigs. Candidate clones growing on the SD/-Leu-Trp-His medium were picked to SD/-Leu-Trp-His+X-α-galactosidase medium for confirmation of the interaction.
Bimolecular Fluorescence Complementation
A published BiFC procedure (Waadt et al., 2008) was used. VmPxE1 and MdAPX1 were cloned into the vector pSPYNE(R)173 and the vector pSPYCE(M), respectively. A. tumefaciens strain GV3101 carrying the expression vector pSPYNE(R)173-VmPxE1 was co-injected with A. tumefaciens strain GV3101 carrying pSPYCE(M)-MdAPX1 at the 1:1 ratio into 4-week-old N. benthamiana plants. About 48–60 h after co-agroinfection, live-cell imaging was taken with a two-photon confocal laser scanning microscopy (Olympus FV1000MPE) with the FV10-ASW 3.1 software suite. The assay was repeated twice.
Protein Extraction and Western Blots
About 60–72 h after agro-infection, the treated N. benthamiana leaves were rapidly frozen in liquid nitrogen and triturated in mortar, then total proteins of the leaf tissues were extracted with Plant Total Protein Extraction Kit, following the manufacturer’s instruction (P0028, Beyotime technologies, Shanghai, China).
For the western blot analysis, total proteins from leaves were separated using SDS–PAGE (40V, 8 mA) and transferred onto nitrocellulose membranes (60V, 150 mA). The blot was blocked with Western Quick Block mixture (P0023B, Beyotime Technologies, Shanghai, China) for 1 h at ambient temperature and followed by (1) incubation with the monoclonal antibody (1:500) in WB primary antibody diluent (P0103, Beyotime technologies, Shanghai, China) for 2 h at ambient temperature; (2) being washed with TBS containing 0.1% Tween-20 (TBST), and (3) reacting with goat anti-mouse IgG horseradish peroxidase (HRP) (1:500) (A0216, Beyotime Technologies, Shanghai, China) for 1 h at ambient temperature. Finally, the nitrocellulose membrane was immunostained with 3,30-diaminobenzidine (DAB) for 10 min in the dark before visualization.
In Vivo Co-immunoprecipitation
For the in vivo Co-IP assay, A. tumefaciens strain GV3101 carrying the expression vectors PICH86988-HA-MdAPX1 and pBin-GFP-VmPxE1 was co-agroinfiltrated into N. benthamiana leaves. Harvest proteins 500 μL was added to equilibrated GFP-trap beads (Chromotek) and fully mixed at 4°C overnight on a slow shake incubator, and then centrifuged with the supernatant discarded. Re-suspended GFP-trap beads were washed by pre-cold wash buffer twice and then transferred to 100 μL 2 × SDS loading buffer. The sample was boiled for 10 min to dissociate for immunoblot analysis using the anti-GFP or anti-HA antibody.
Ectopic Expression of MdAPX1 in V. mali
The entire MdAPX1 gene sequence was cloned into plasmid pDL2 with the yeast gap repair approach (Bruno et al., 2004; Zhou et al., 2012). Then PEG-mediate protoplast transformation was performed to transferred pDL2-MdAPX1 into V. mali wild type 03-8 and the VmPxE1 gene deletion mutant ΔVmPxE1 with the hygromycin B phosphotransferase gene (hph) as a selective marker. Transformed colonies were selected by hygromycin B. Then the stress resistance against H2O2 was measured by culturing wild type 03-8, transformed colonies of 03-8/MdAPX1 and ΔVmPxE1/ MdAPX1 on PDA with 0.06% H2O2 at 25°C. Fungal colony size was assessed 3 days after treatment. There were 30 petri dishes per treatment. The expression of MdAPX1 was identified with RT-PCR. The experiment was repeated twice.
Results
VmPxE1 From V. mali Suppressed BAX-Induced Cell Death in N. benthamiana
A protein of unknown functions, VmPxE1 (KUI70334.1) residing on chromosome 6 of V. mali, was found to contain 213 amino acid residues with a predicted signal peptide. Transient expression assay in N. benthamiana leaves with BAX suggested that VmPxE1 is an effective cell death suppressor effector with a cell death suppression ratio of 92% (46/50) (Figure 1). Subsequent RT-PCR and western blot with an anti-eGFP and anti-HA antibody confirmed that VmPxE1, BAX, and eGFP were all expressed in N. benthamiana (Supplementary Figures S1A,B).

FIGURE 1. Transient expression of VmPxE1 in N. benthamiana leaves by agro-infiltration. Symptoms on leaves of N. benthamiana were assessed 5 days after inoculation.
Secretory Function Validation of the Putative N-Terminal Signal Peptide of VmPxE1
The VmPxE1 amino acid sequence contained a predicted signal peptide at the position of 1-19 aa (Figure 2A). The invertase mutant yeast strain YTK12 containing VmPxE1 signal peptide recombinant plasmids could grow on YPRAA medium (with raffinose instead of sucrose) indicating that the invertase was secreted (Figure 2B). This result indicates that the putative N-terminal signal peptide of VmPxE1 was a functional secretion signal peptide.
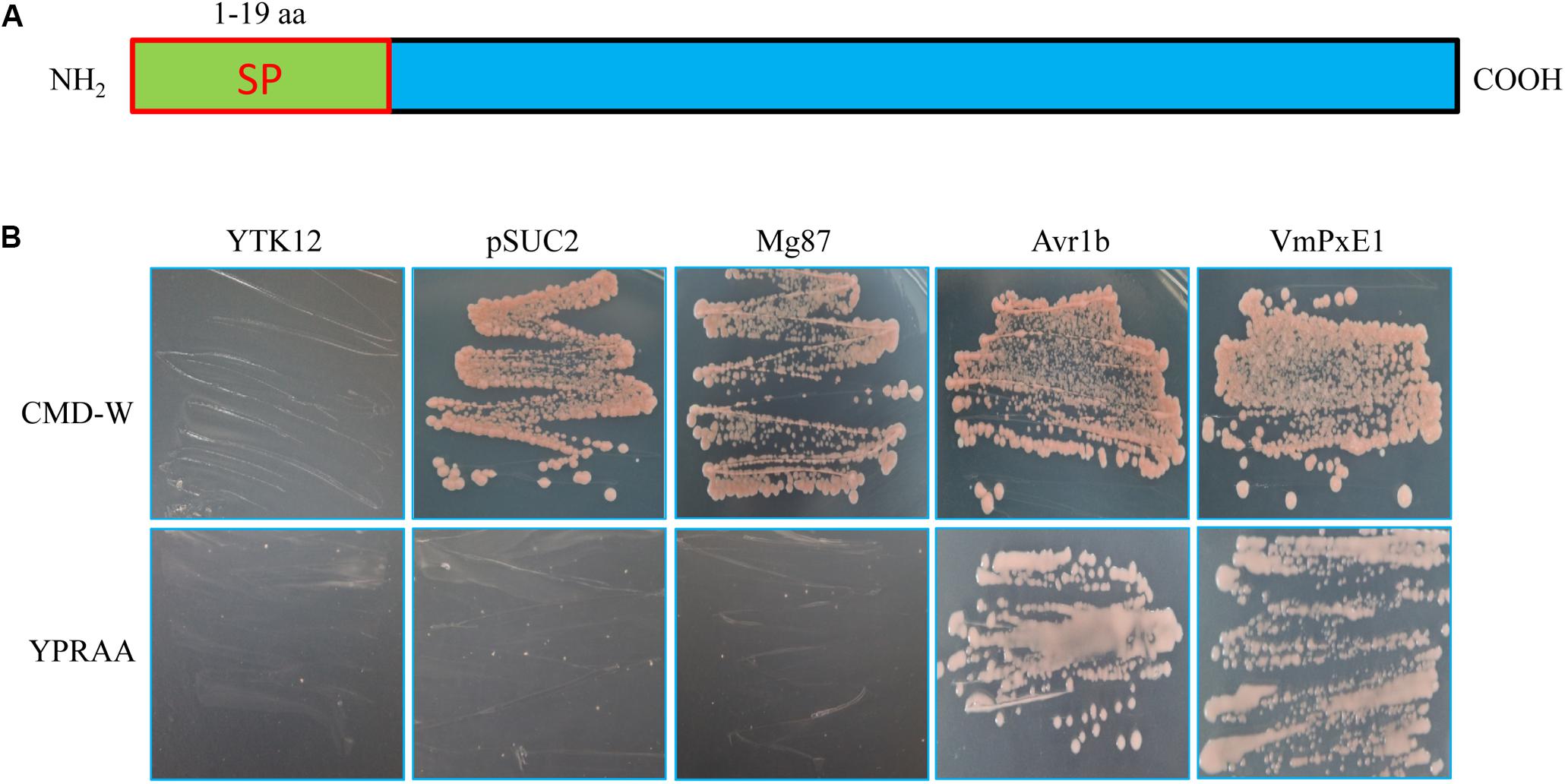
FIGURE 2. Secretory function validation of the putative N-terminal signal peptide of VmPxE1 using the yeast invertase secretion assay: (A) a schematic diagram of putative VmPxE1 signal peptide; (B) the sequence of the putative VmPxE1 signal peptide fused in-frame to the invertase sequence in the pSUC2 vector and then transformed into yeast strain YTK12. The empty pSUC2 and the non-secreted Mg87 protein from Magnaporthe oryzae were used as negative controls and the secreted effector Avr1b from oomycete as a positive control. Only the yeast strains that are able to secrete invertase can grow on both CMD-W and YPRAA media.
Transcription Level of Effector Gene VmPxE1
VmPxE1 expression was quantified at 0 (immediately before inoculation), 6, 12, 18, 24, 36, 48, and 72 hpi on apple twigs with G6PDH as a housekeeping gene. VmPxE1 was upregulated for all sampling points. The expression of VmPxE1 was increased significantly at 6, 12, 18, 24, and 36 hpi (Figure 3).
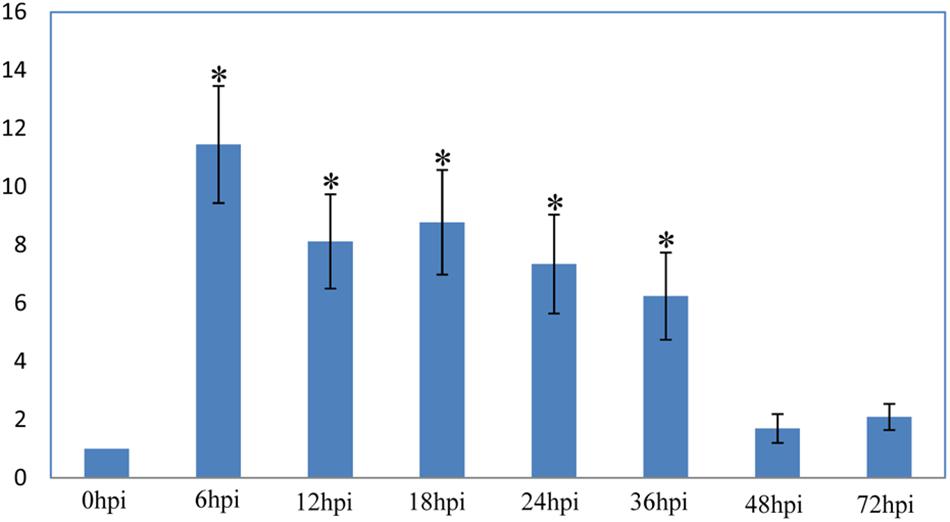
FIGURE 3. Relative expression level of VmPxE1 at 0, 6, 12, 18, 24, 36, 48, and 72 h post inoculation of apple twigs and leaves with G6PDH as a housekeeping gene. Results were presented as a mean fold change in the expression relative to the expression at 0 h. Significant difference in pathogenicity was indicated with asterisks (P < 0.05). Each experiment was repeated twice and error bars indicate SEM.
The VmPxE1 Is Not a Necessary Growth Factor but a Virulence Factor of V. mali
To estimate the contribution of VmPxE1 to virulence, this gene was deleted (Supplementary Figure S2A). The deletion mutant ΔVmPxE1 was obtained and further confirmed by PCR analysis (Supplementary Figure S2C) and Southern hybridization (Supplementary Figure S2B). The complementation mutant ΔVmPxE1/VmPxE1 was generated and confirmed by PCR analysis (Supplementary Figure S2D).
In vitro inoculation showed that deletion of VmPxE1 did not affect vegetative growth (Figure 4A) and sporulation of V. mali (Figure 4B). However, ΔVmPxE1 showed significant reduction in virulence on both apple twigs and leaves (Figure 5A), about 42.1 and 38.9% on leaves and twigs, respectively (Figure 5B). The complementation mutant ΔVmPxE1/VmPxE1 had the similar level of infection as the wild type 03-8.
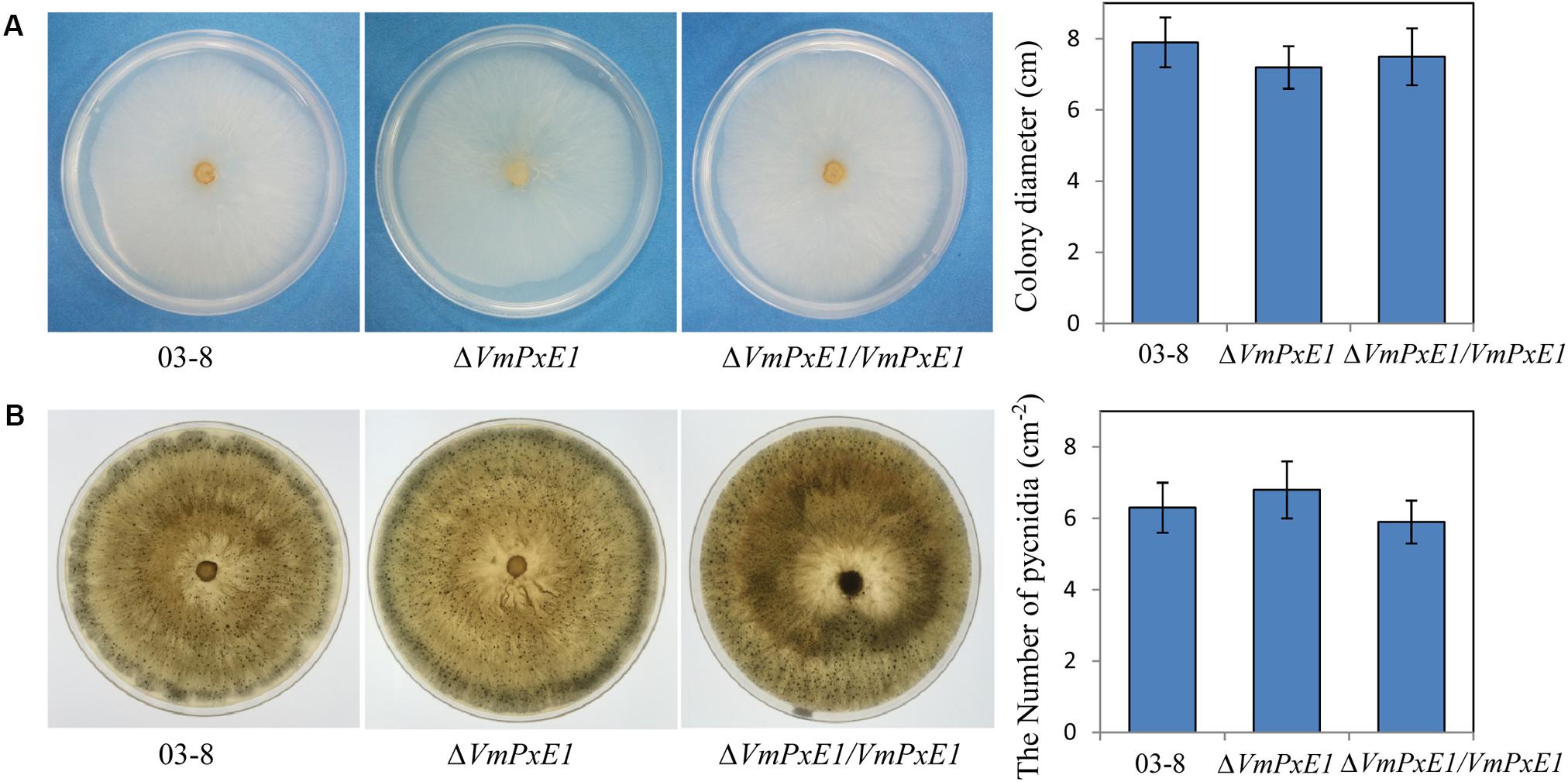
FIGURE 4. Vegetative growth and sporulation of V. mali wild type strain 03-8, ΔVmPxE1 and ΔVmPxE1/VmPxE1 on PDA for 3 days (A) or 40 days (B) at 25°C. Colony diameter was measured by crossing method and pycnidia were counted per square centimeter (cm-2). Bars indicate SD of the mean of 30 individual dishes.
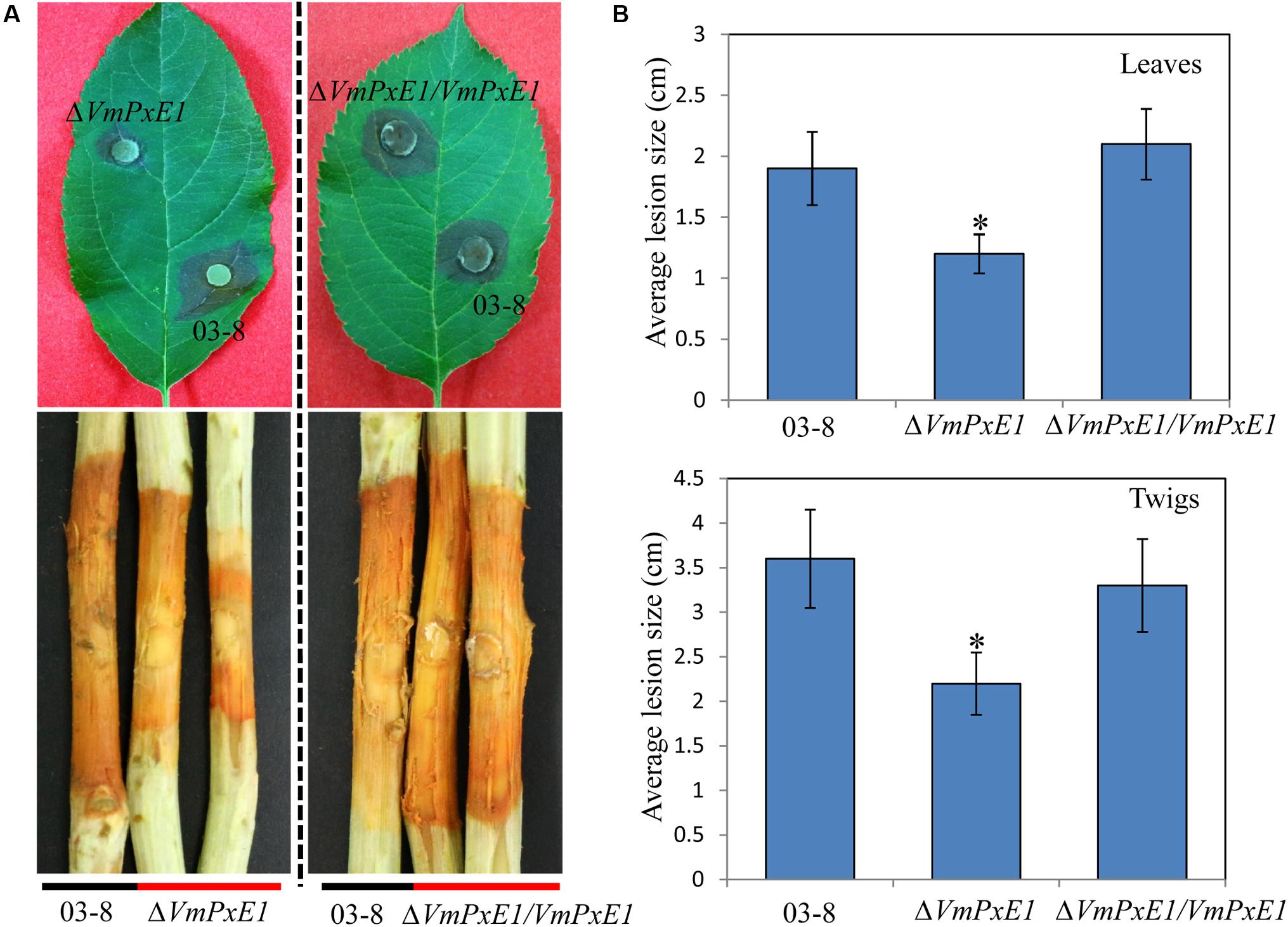
FIGURE 5. Pathogenicity of V. Mali wild type strain 03-8, deletion mutant ΔVmPxE1 and ΔVmPxE1 complementation mutant (ΔVmPxE1/VmPxE1) when inoculated onto leaves and twigs of Malus domestica borkh. cv. ‘Fuji.’ (A) Symptom on twigs and leaves after inoculation. (B) Average lesion size of scabs on twigs and leaves. Observation was made 60 and 120 h after inoculation for leaves and twigs, respectively. Asterisk indicates significant (P < 0.05) differences. Bars indicate SD of the mean of 30 individual host plants.
VmPxE1 Interacted With MdAPX1, an Apple Ascorbate Peroxidase
To identify the target of VmPxE1, the GAL4 yeast two-hybrid system was performed using a V. mali-apple cDNA library. A potential gene segment was captured after screening tests on SD-Trp-Leu-His medium. The sequence was predicted as an apple ascorbate peroxidase protein, named as MdAPX1 (Malus domestica ascorbate peroxidase 1). Its intact coding sequence was constructed into the prey vector pGADT7. Auxotroph yeast strain AH109 was able to recover growth on SD-Trp-Leu-His when synchronously transformed with pGBKT7-VmPxE1 and pGADT7-MdAPX1 (Figure 6A). The dichotomous YFP segments were recovered and yellow fluorescence signal was detected in leaf cells of N. benthamiana, indicating that MdAPX1 could interact with VmPxE1 (Figure 6B). HA-MdAPX1 and GFP-VmPxE1 were transiently co-expressed in N. benthamiana, and total proteins went through GFP-trap gel beads for the specific adsorption. The immunoblotting showed that HA-MdAPX1 was present in the final GFP-VmPxE1-precipitated immunocomplex (Figure 6C), suggesting that MdAPX1 is a potential target of VmPxE1. Phylogenetic analysis showed that MdAPX1 (XP008344846.1) is an L-ascorbic peroxidase, belonging to a subgroup of heme-dependent peroxidases of the plant superfamily (Figure 7).
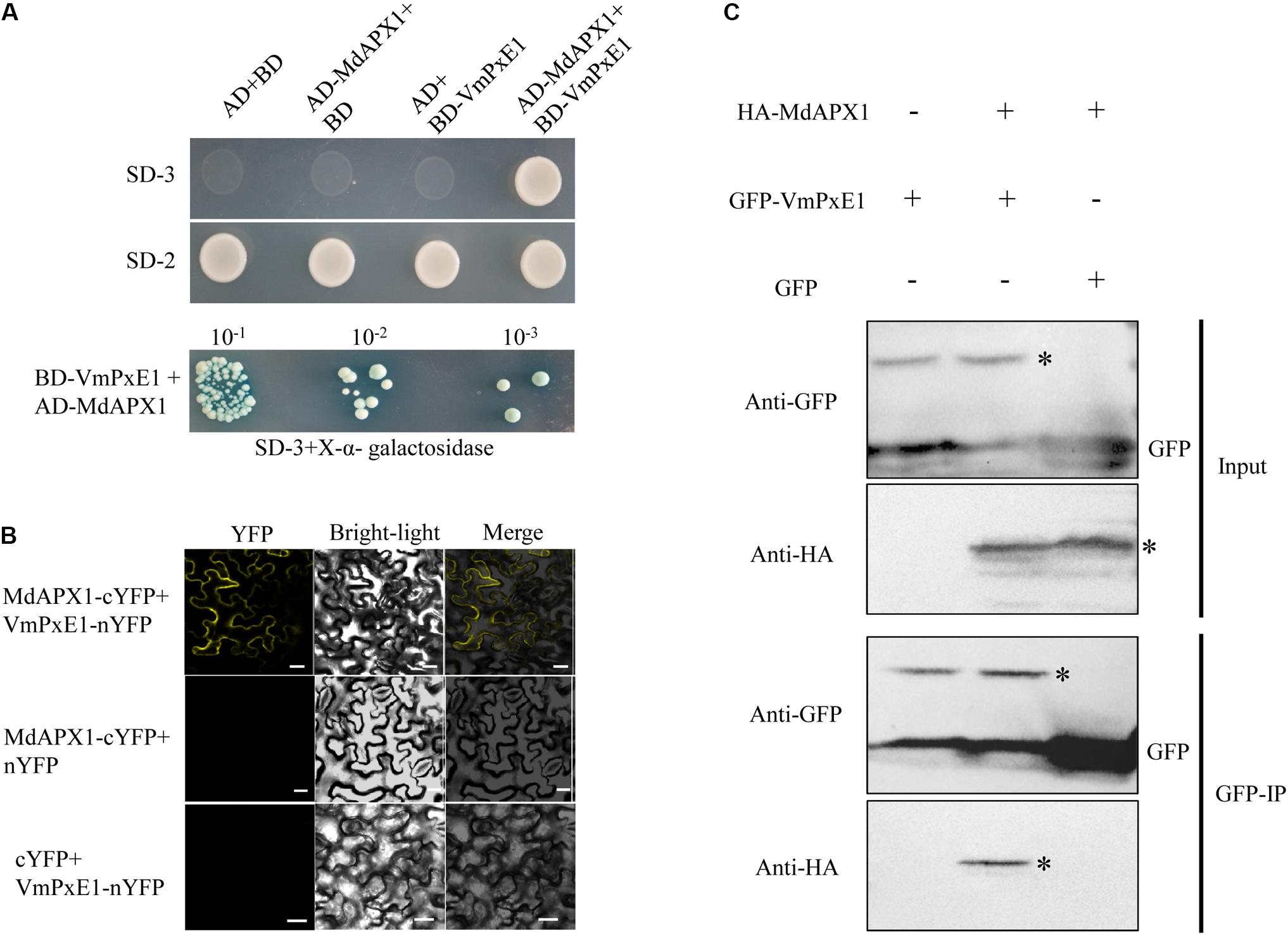
FIGURE 6. Interaction of VmPxE1 with MdAPX1. (A) Yeast cells were cultured on SD-Trp-Leu-His and SD-Trp-Leu as control. Positive yeast clones were cultured on SD-3+X-α-galactosidase with ladder concentration of yeast suspension (10-1, 10-2, and 10-3) for further confirmation. (B) Bimolecular fluorescence complementation showed that VmPxE1 interacted with MdAPX1 in leaf cells of N. benthamiana. VmPxE1–nYFP and MdAPX1–cYFP were co-expressed in N. benthamiana by agro-infiltration. The yellow fluorescence was observed 48–72 h post inoculation (Bars = 20 μm). (C) In vivo Co-IP assay of HA: MdAPX1 and GFP: VmPxE1 (without the signal peptide sequence). Both genes were expressed in N. benthamiana leaves using agro-infection. The input experiment was performed by western blot with the HA antibody and GFP antibody to confirm the expression of the two proteins. The mixed proteins were blended with GFP-trap agarose beads. The final eluent was analyzed by immunoblot using above-mentioned antibodies to detect VmPxE1 and MdAPX1. Asterisk represents target band.
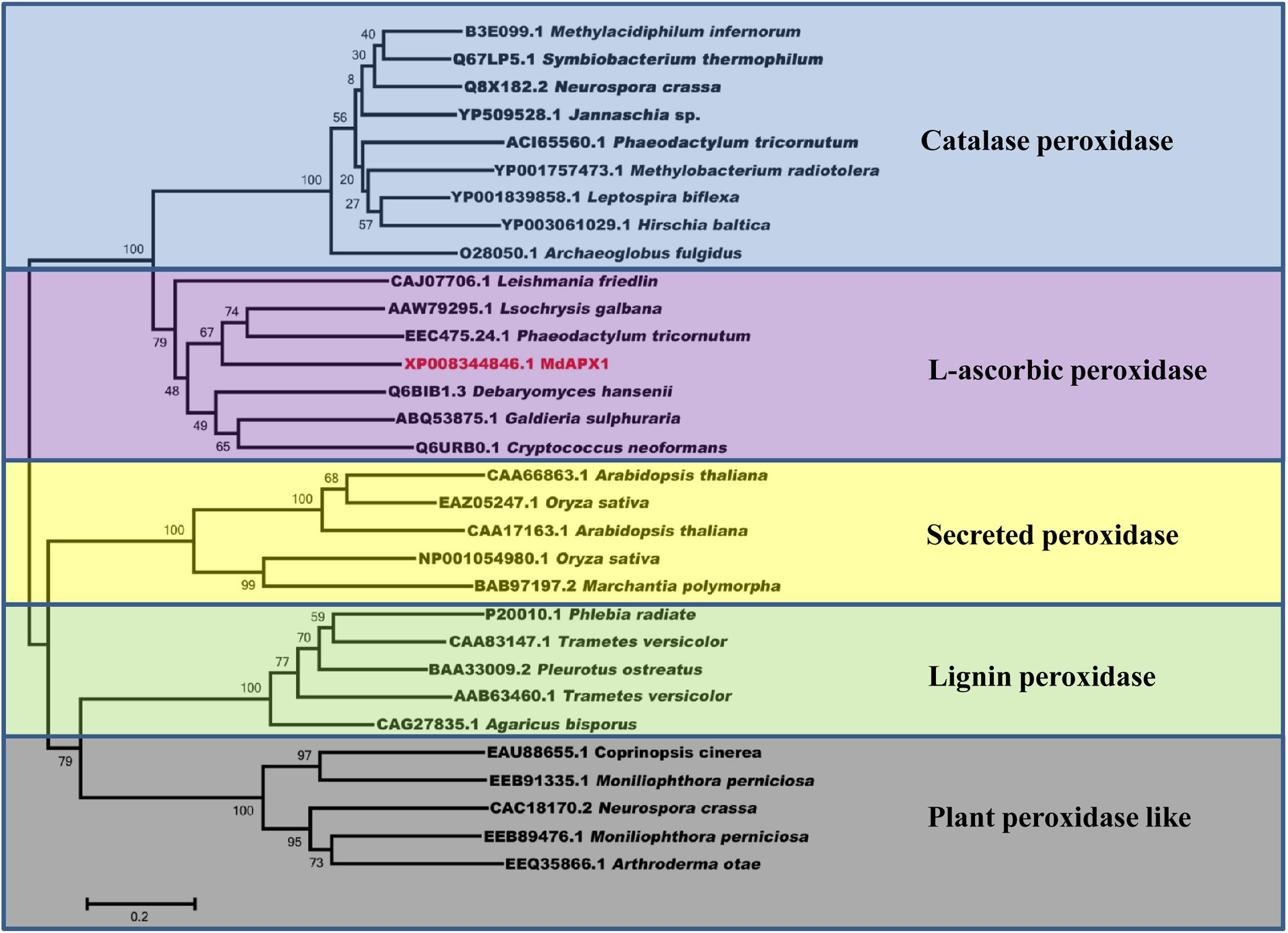
FIGURE 7. Sequence phylogenetic analysis of MdAPX1 showed that MdAPX1 was an L-ascorbic peroxidase, belonging to a subgroup of heme-dependent peroxidases of the plant superfamily. The unrooted phylogram was constructed based on the NJ method with 1000 bootstrap replicates.
Ectopic Expression of MdAPX1
The ectopic expression of MdAPX1 in wild type 03-8 (03-8/MdAPX1) and VmPxE1 gene deletion mutant strain (ΔVmPxE1/MdAPX1) was successful (Supplementary Figure S3). The growth of both the strains was significantly suppressed on PDA containing 0.06% H2O2. The transformed strain ΔVmPxE1/MdAPX1 exhibited stronger resistance to H2O2 than the 03-8/MdAPX1 strain. These results suggested that MdAPX1 enhanced the resistance of only ΔVmPxE1 to H2O2 and that the presence of VmPxE1 seems to have disturbed MdAPX1 function (Figure 8).
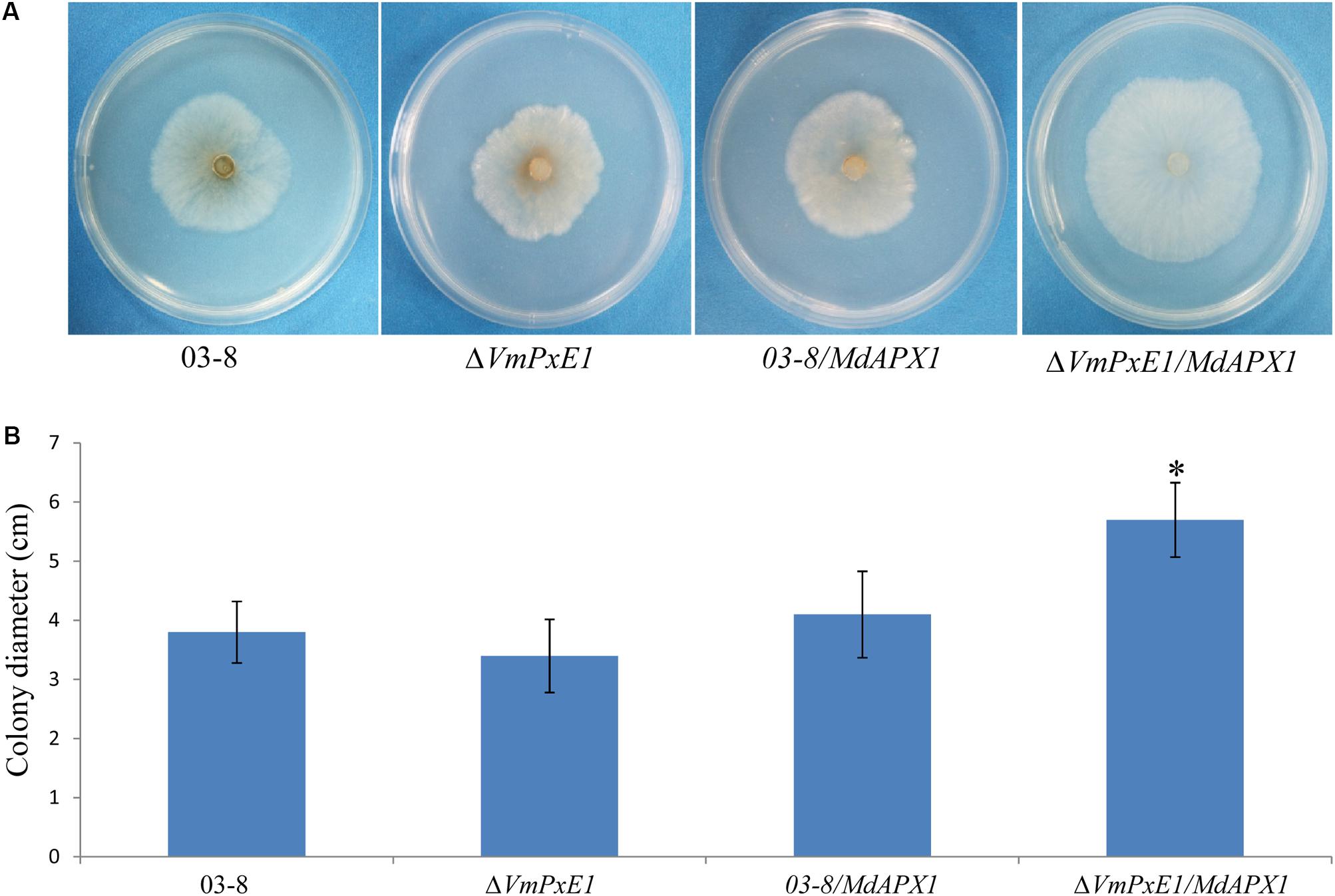
FIGURE 8. Ectopic expression of MdAPX1 in V. mali wild type strain 03-8 and VmPxE1 gene deletion mutant ΔVmPxE1. (A) Colony of strains on PDA supplemented with 0.06% H2O2, 3 dpi at 25°C. (B) Colony diameters. Bars indicate SD of the mean of 30 dishes and asterisk above the bar indicate significantly difference from 03-8 (P < 0.05).
Discussion
We demonstrated that a cell death suppressor effector of V. mali, VmPxE1, was upregulated during the early stage of fungal infection of apple twigs and leaves, and identified one potential target of VmPxE1.
VmPxE1 from V. mali, a nectrophic pathogen, could suppress the BAX-induced PCD, which implies that VmPxE1 is involved in plant immune response, resulting in weakened host defense. This is supported by the fact that deletion of VmPxE1 reduced virulence of V. mali but did not affect fungal vegetative growth and sporulation. Plant cell death induced by hypersensitive responses is believed to be detrimental for biotrophic pathogens, because of the reduction of vivosphere and restriction of hyphae extension (Gilchrist, 1998). On the other hand, plant cell death could facilitate the infection of necrotrophic pathogens and hence presence of effectors inducing cell death may be important for necrotrophic pathogens during the early infection process (Govrin and Levine, 2000; Wang et al., 2014). On the contrary, cell death suppressor effectors are vitally important for biotrophic and hemibiotrophic pathogens (Sharpee et al., 2017). However, the present study suggested that suppressing cell death may also play an important role in the infection by V. mali. Therefore, V. mali may not need to kill host cells rapidly but control cell death via these suppressors to enable successful colonization over time.
The target of VmPxE1 was identified to be an ascorbate peroxidase MdAPX1 from apple. Ascorbate peroxidase utilizes ascorbic acid as a specific electron donor and scavenges H2O2. During the plant–pathogen interaction, oxidative burst is concurrent and accompanied with the production of reactive oxygen species (ROS) (Apel and Hirt, 2004; Daudi et al., 2012). The ROS act as antimicrobial molecules and are also important as a signal related to plant disease resistance (Suzuki et al., 2011; Gilroy et al., 2014). However, excessive ROS would be phytotoxic. Plants possess a complex mechanism to balance the pernicious and beneficial effects of ROS (Daudi et al., 2012). Peroxidases act as an equalizer beam in the defense system of plant by using phenols, amine, or heterocyclic compound as hydrogen donor and preferring H2O2 as reaction substrates. Therefore, peroxidases could regulate H2O2 and concentrations of hydrogen donors (Welinder, 1992; Hiner et al., 2002; Yoshida et al., 2003; Camejo et al., 2016). Changes in H2O2 concentration could induce pathogenesis-related proteins and affect plant signals of disease resistance such as salicylic acid (Klessig and Malamy, 1994; Passardi et al., 2005), and is closely associated with hypersensitive cell death (Yoda et al., 2003). Apple tree possesses a large and complete peroxidase system that is well documented to be involved in its defense against pathogens (Camejo et al., 2016). The increased peroxidase activity in apple enhances resistance to Glomerella leaf spot (Araujo and Stadnik, 2013). Peroxidases increased activity associated with increased apple resistance to apple ring spot, blue mold, and apple scab (Valentines et al., 2005; Rachenko et al., 2014). These evidences suggest that this group of enzymes may play an important role in plant resistance to pathogens. Thus, it is not surprising that pathogens have to deal with peroxidases of its host. For instance Ustilago maydis effector Pep1 targets a maize peroxidase POX12 in vivo and suppresses the early immune responses of maize (Hemetsberger et al., 2012). Moreover, ascorbate peroxidase is reported to be closely related to plant resistance (Sarowar et al., 2005) and to act as a defense enzyme, enhancing systemic acquired resistance (Kvaratskhelia et al., 1997). We may, therefore, speculate that MdAPX1 is involved in the apple defense system against pathogens. For successful infection, MdAPX1 needs to be suppressed. Ectopic expression of MdAPX1 in ΔVmPxE1 (ΔVmPxE1/MdAPX1) showed stronger resistance to H2O2 than ectopic expression of MdAPX1 in wild type 03-8 (03-8/MdAPX1), indicating that VmPxE1 interfered the function of MdAPX1. Further research is needed to confirm whether and, if so, how VmPxE1 disrupts the function of MdAPX1.
Conclusion
We identified a cell death suppressor effector VmPxE1 from necrotrophic pathogen V. mali that contributes to fungal virulence and targets a peroxidase MdAPX1 of apple. Although, the putative effector in highly expressed during the apple infective process, VmPxE1 was also expressed in basal conditions and an endogenous function could not be excluded. This data indirectly pointed that VmPxE1 disturbs the function of MdAPX1. Further research is needed to confirm whether and, if so, how VmPxE1 disrupts the function of MdAPX1.
Author Contributions
LH, MZ, and HF contributed to the design of the work. MZ, HF, YZ, LS, and CG performed the experiments. MZ and YZ analyzed the sequencing data. MZ, HF, and XX wrote and revised the manuscript. LH was responsible for all aspects of this study.
Funding
This work was financially supported by the National Natural Science Foundation of China (Grant Nos. 31471732 and 31671982).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Professor Xiaojie Wang (Northwest A&F University, Yangling, China) for providing the plasmids (pSUC2, PGR106, PICH86988, PBin-GFP, pGBKT7, and pGADT7).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00821/full#supplementary-material
FIGURE S1 | (A) RT-PCR detected BAX, VmPxE1 and eGFP expression in N. benthamiana tissues at different locations: VmPxE1 in location 1; eGFP in location 2; VmPxE1 in location 3; BAX in location 3; eGFP in location 4; BAX in location 4; GAPDH. Total RNA was extracted 48 h after the second infiltration. (B) Validation of proteins expression of BAX, HA: VmPxE1 by western blot with respective monoclonal murine antibodies injected in location 3.
FIGURE S2 | Generation and identification of gene VmPxE1 deletion and complementation mutants. (A) Flanks homologous recombination facilitated target gene replacement. The resistance gene cassette denoted by a gray arrow with homologous flank arms replaced the target gene denoted by a blue arrow based on homologous recombination. (B) Southern blot hybridization analysis of VmPxE1 gene deletion mutants using primer NeoF/NeoR with digoxin marked nucleic fragment of Neo gene as hybridization probe. (C) Confirmation of VmPxE1 knockout mutants by PCR analysis with four pairs of primers. The wild type 03-8 was as control. 1: 7F/Neo-CR detected upstream fusion segment. 2: Neo-CF/8R detected downstream fusion segment. 3: 5F/6R detected targeted gene and 4: Neo-CF/Neo-CR detected incoming resistant gene Neo. M: Maker. (D) PCR analysis was performed with primers 5F/6R to identify ΔVmPxE1/VmPxE1 complementation mutant. 1: Wild type 03-8. 2: VmPxE1 gene deletion mutants. 3: ΔVmPxE1/VmPxE1 complementation mutant. M: Maker.
FIGURE S3 | RT-PCR detects MdAPX1 and VmPxE1 in transformed strains 03-8/MdAPX1 and ΔVmPxE1/MdAPX1 after cultured on PDA supplemented with 0.06% H2O2 3 days at 25°C.
TABLE S1 | Primers for cloning gene VmPxE1 to vectors, VmPxE1 gene deletion and PCR analysis in this study. The red color stands for corresponding directing sequences for homologous recombination and yellow color stands for corresponding restriction enzyme cutting site.
TABLE S2 | primers for RT-PCR and qRT-PCR in this study.
Footnotes
References
Albert, M. (2013). Peptides as triggers of plant defence. J. Exp. Bot. 64, 5269–5279. doi: 10.1093/jxb/ert275
Apel, K., and Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. doi: 10.1146/annurev.arplant.55.031903.141701
Araujo, L., and Stadnik, M. J. (2013). Cultivar-specific and ulvan-induced resistance of apple plants to Glomerella leaf spot are associated with enhanced activity of peroxidases. Acta Sci. Agric. 35, 287–293.
Boller, T., and He, S. Y. (2009). Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324, 742–744. doi: 10.1126/science.1171647
Bruno, K. S., Tenjo, F., Li, L., Hamer, J. E., and Xu, J.-R. (2004). Cellular localization and role of kinase activity of PMK1 in Magnaporthe grisea. Eukaryot. Cell 3, 1525–1532. doi: 10.1128/EC.3.6.1525-1532.2004
Caillaud, M. C., Asai, S., Rallapalli, G., Piquerez, S., Fabro, G., and Jones, J. D. (2013). A downy mildew effector attenuates salicylic acid–triggered immunity in Arabidopsis by interacting with the host mediator complex. PLoS Biol. 11:e1001732. doi: 10.1371/journal.pbio.1001909
Camejo, D., Guzmán-Cedeño, A., and Moreno, A. (2016). Reactive oxygen species, essential molecules, during plant-pathogen interactions. Plant Physiol. Biochem. 103, 10–23. doi: 10.1016/j.plaphy.2016.02.035
Daudi, A., Cheng, Z., O’Brien, J. A., Mammarella, N., Khan, S., Ausubel, F. M., et al. (2012). The apoplastic oxidative burst peroxidase in Arabidopsis a major component of pattern-triggered immunity. Plant Cell 24, 275–287. doi: 10.1105/tpc.111.093039
de Jonge, R., van Esse, H. P., Kombrink, A., Shinya, T., Desaki, Y., and Bours, R. (2010). Conserved fungal LysM effector Ecp6 prevents chitin-triggered immunity in plants. Science 329, 953–955. doi: 10.1126/science.1190859
Djamei, A., Schipper, K., Rabe, F., Ghosh, A., Vincon, V., Kahnt, J., et al. (2011). Metabolic priming by a secreted fungal effector. Nature 478, 395–398. doi: 10.1038/nature10454
Dodds, P. N., and Rathjen, J. P. (2010). Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11, 539–548. doi: 10.1038/nrg2812
Dou, D., Kale, S. D., Wang, X., Jiang, R. H., Bruce, N. A., Arredondo, F. D., et al. (2008). RXLR-mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen-encoded machinery. Plant Cell 20, 1930–1947. doi: 10.1105/tpc.107.056093
Gao, J., Li, Y., Ke, X., Kang, Z., and Huang, L. (2011). Development of genetic transformation system of Valsa mali of apple mediated by PEG. Acta Microbiol. Sin. 51, 1194–1199.
Geitz, R. D., Scheistl, R. H., Wellems, A. R., and Woods, R. A. (1995). Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11, 355–360. doi: 10.1002/yea.320110408
Gilchrist, D. (1998). Programmed cell death in plant disease: the purpose and promise of cellular suicide. Annu. Rev. Phytopathol. 36, 393–414. doi: 10.1146/annurev.phyto.36.1.393
Gilroy, S., Suzuki, N., Miller, G., Choi, W.-G., Toyota, M., Devireddy, A. R., et al. (2014). A tidal wave of signals: calcium and ROS at the forefront of rapid systemic signaling. Trends Plant Sci. 19, 623–630. doi: 10.1016/j.tplants.2014.06.013
Giraldo, M. C., and Valent, B. (2013). Filamentous plant pathogen effectors in action. Nat. Rev. Microbiol. 11, 800–814. doi: 10.1038/nrmicro3119
Govrin, E. M., and Levine, A. (2000). The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr. Biol. 10, 751–757. doi: 10.1016/S0960-9822(00)00560-1
Gu, B. A., Kale, S. D., Wang, Q. H., Wang, D. H., Pan, Q. N., Cao, H., et al. (2011). Rust secreted protein Ps87 is conserved in diverse fungal pathogens and contains a RXLR-like motif sufficient for translocation into plant cells. PLoS One 6:e27217. doi: 10.1371/journal.pone.0027217
Hemetsberger, C., Herrberger, C., Zechmann, B., Hillmer, M., and Doehlemann, G. (2012). The Ustilago maydis effector Pep1 suppresses plant immunity by inhibition of host peroxidase activity. PLoS Pathog. 8:e1002684. doi: 10.1371/journal.ppat.1002684
Hiner, A. N., Raven, E. L., Thorneley, R. N., García-Cánovas, F., and Rodríguez-López, J. N. (2002). Mechanisms of compound I formation in heme peroxidases. J. Inorg. Biochem. 91, 27–34. doi: 10.1016/S0162-0134(02)00390-2
Ito, T., Chiba, T., Ozawa, R., Yoshida, M., Hattori, M., and Sakaki, Y. (2001). A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. U.S.A. 98, 4569–4574. doi: 10.1073/pnas.061034498
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Ke, X., Yin, Z., Song, N., Dai, Q., Voegele, R. T., Liu, Y., et al. (2014). Transcriptome profiling to identify genes involved in pathogenicity of Valsa mali on apple tree. Fungal Genet. Biol. 68, 31–38. doi: 10.1016/j.fgb.2014.04.004
Klessig, D. F., and Malamy, J. (1994). The salicylic acid signal in plants. Plant Mol. Biol. 26, 1439–1458. doi: 10.1007/BF00016484
Kvaratskhelia, M., George, S. J., and Thorneley, R. N. (1997). Salicylic acid is a reducing substrate and not an effective inhibitor of ascorbate peroxidase. J. Biol. Chem. 272, 20998–21001. doi: 10.1074/jbc.272.34.20998
Lacomme, C., and Santa Cruz, S. (1999). BAX-induced cell death in tobacco is similar to the hypersensitive response. Proc. Natl. Acad. Sci. U.S.A. 96, 7956–7961. doi: 10.1073/pnas.96.14.7956
Lee, D. H., Lee, S. W., Choi, K. H., Kim, D. A., and Uhm, J. Y. (2006). Survey on the occurrence of apple diseases in Korea from 1992 to 2000. Plant Pathol. J. 22, 375–380. doi: 10.5423/PPJ.2006.22.4.375
Li, Z., Yin, Z., Fan, Y., Xu, M., Kang, Z., and Huang, L. (2015). Candidate effector proteins of the necrotrophic apple canker pathogen Valsa mali can suppress BAX-induced PCD. Front. Plant Sci. 6:579. doi: 10.3389/fpls.2015.00579
Ma, Z., Zhu, L., Song, T., Wang, Y., Zhang, Q., Xia, Y., et al. (2017). A paralogous decoy protects Phytophthora sojae apoplastic effector PsXEG1 from a host inhibitor. Science 355, 710–714. doi: 10.1126/science.aai7919
Mueller, A. N., Ziemann, S., Treitschke, S., Assmann, D., and Doehlemann, G. (2013). Compatibility in the Ustilago maydis-maize interaction requires inhibition of host cysteine proteases by the fungal effector Pit2. PLoS Pathog. 9:e1003177. doi: 10.1371/journal.ppat.1003177
Oliva, R., Win, J., Raffaele, S., Boutemy, L., Bozkurt, T. O., Chaparro-Garcia, A., et al. (2010). Recent developments in effector biology of filamentous plant pathogens. Cell. Microbiol. 12, 705–715. doi: 10.1111/j.1462-5822.2010.01471.x
Passardi, F., Cosio, C., Penel, C., and Dunand, C. (2005). Peroxidases have more functions than a Swiss army knife. Plant Cell Rep. 24, 255–265. doi: 10.1007/s00299-005-0972-6
Rachenko, M. A., Rachenko, E. I., Romanova, I. M., Zhivet’yev, M. A., and Graskova, I. A. (2014). Peroxidase activity and isoforms in the leaves of apple tree varieties differing in scab resistance. J. Stress Physiol. Biochem. 10, 78–83.
Rovenich, H., Boshoven, J. C., and Thomma, B. P. (2014). Filamentous pathogen effector functions: of pathogens, hosts and microbiomes. Curr. Opin. Plant Biol. 20, 96–103. doi: 10.1016/j.pbi.2014.05.001
Sarowar, S., Kim, E. N., Kim, Y. J., Ok, S. H., Kim, K. D., Hwang, B. K., et al. (2005). Overexpression of a pepper ascorbate peroxidase-like 1 gene in tobacco plants enhances tolerance to oxidative stress and pathogens. Plant Sci. 169, 55–63. doi: 10.1016/j.plantsci.2005.02.025
Shan, W. X., Cao, M., Dan, L. U., and Tyler, B. M. (2004). The Avr1b locus of Phytophthora sojaeencodes an elicitor and a regulator required for Avirulence on soybean plants carrying resistance gene Rps1b. Mol. Plant Microbe Interact. 17, 394–403. doi: 10.1094/MPMI.2004.17.4.394
Sharpee, W., Oh, Y., Yi, M., Franck, W., Eyre, A., Okagaki, L. H., et al. (2017). Identification and characterization of suppressors of plant cell death (SPD) effectors from Magnaporthe oryzae. Mol. Plant Pathol. 18, 850–863. doi: 10.1111/mpp.12449
Suzuki, N., Miller, G., Morales, J., Shulaev, V., Torres, M. A., and Mittler, R. (2011). Respiratory burst oxidases: the engines of ROS signaling. Curr. Opin. Plant Biol. 14, 691–699. doi: 10.1111/mpp.12449
Tian, M., Benedetti, B., and Kamoun, S. (2005). A second Kazal-like protease inhibitor from Phytophthora infestans inhibits and interacts with the apoplastic pathogenesis-related protease P69B of tomato. Plant Physiol. 138, 1785–1793. doi: 10.1104/pp.105.061226
Valentines, M. C., Vilaplana, R., Usall, J., and Larrigaudière, C. (2005). Involvement of enzymatic browning and peroxidase activity as resistance mechanisms in ‘golden delicious’ apples. Acta Hortic. 682, 2041–2048. doi: 10.17660/ActaHortic.2005.682.277
Vleeshouwers, V. G., and Oliver, R. P. (2014). Effectors as tools in disease resistance breeding against biotrophic, hemibiotrophic, and necrotrophic plant pathogens. Mol. Plant Microbe Interact. 27, 196–206. doi: 10.1094/MPMI-10-13-0313-IA
Waadt, R., Schmidt, L. K., Lohse, M., Hashimoto, K., Bock, R., and Kudla, J. (2008). Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 56, 505–516. doi: 10.1111/j.1365-313X.2008.03612.x
Wang, Q., Han, C., Ferreira, A. O., Yu, X., Ye, W., Tripathy, S., et al. (2011a). Transcriptional programming and functional interactions within the Phytophthora sojae RXLR effector repertoire. Plant Cell 23, 2064–2086. doi: 10.1105/tpc.111.086082
Wang, X., Jiang, N., Liu, J., Liu, W., and Wang, G.-L. (2014). The role of effectors and host immunity in plant–necrotrophic fungal interactions. Virulence 5, 722–732. doi: 10.4161/viru.29798
Wang, X., Wei, J., Huang, L., and Kang, Z. (2011). Re-evaluation of pathogens causing Valsa canker on apple in China. Mycologia 103, 317–324. doi: 10.3852/09-165
Welinder, K. G. (1992). Superfamily of plant, fungal and bacterial peroxidases. Curr. Opin. Plant Biol. 2, 388–393. doi: 10.1016/0959-440X(92)90230-5
Yin, Z., Ke, X., Huang, D., Gao, X., Voegele, R. T., Kang, Z., et al. (2013). Validation of reference genes for gene expression analysis in Valsa mali var. mali using real-time quantitative PCR. World J. Microbiol. Biotechnol. 29, 1563–1571. doi: 10.1007/s11274-013-1320-6
Yin, Z., Liu, H., Li, Z., Ke, X., Dou, D., Gao, X., et al. (2015). Genome sequence of Valsa canker pathogens uncovers a potential adaptation of colonization of woody bark. New Phytol. 208, 1202–1216. doi: 10.1111/nph.13544
Yoda, H., Yamaguchi, Y., and Sano, H. (2003). Induction of hypersensitive cell death by hydrogen peroxide produced through polyamine degradation in tobacco plants. Plant Physiol. 132, 1973–1981. doi: 10.1104/pp.103.024737
Yoshida, K., Kaothien, P., Matsui, T., Kawaoka, A., and Shinmyo, A. (2003). Molecular biology and application of plant peroxidase genes. Appl. Microbiol. Biotechnol. 60, 665–670. doi: 10.1007/s00253-002-1157-7
Yu, J.-H., Hamari, Z., Han, K.-H., Seo, J.-A., Reyes-Domínguez, Y., and Scazzocchio, C. (2004). Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41, 973–981. doi: 10.1016/j.fgb.2004.08.001
Zhang, M., Li, Q., Liu, T., Liu, L., Shen, D., Zhu, Y., et al. (2015). Two cytoplasmic effectors of Phytophthora sojae regulate plant cell death via interactions with plant catalases. Plant Physiol. 167, 164–175. doi: 10.1104/pp.114.252437
Keywords: Apple Valsa canker, effector protein, cell death suppressor, virulence factor, peroxidase
Citation: Zhang M, Feng H, Zhao Y, Song L, Gao C, Xu X and Huang L (2018) Valsa mali Pathogenic Effector VmPxE1 Contributes to Full Virulence and Interacts With the Host Peroxidase MdAPX1 as a Potential Target. Front. Microbiol. 9:821. doi: 10.3389/fmicb.2018.00821
Received: 14 December 2017; Accepted: 11 April 2018;
Published: 25 April 2018.
Edited by:
Ivan Baccelli, Consiglio Nazionale delle Ricerche (CNR), ItalyReviewed by:
Birinchi Kumar Sarma, Banaras Hindu University, IndiaSelena Gimenez-Ibanez, Centro Nacional de Biotecnología (CNB), Spain
Copyright © 2018 Zhang, Feng, Zhao, Song, Gao, Xu and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lili Huang, huanglili@nwsuaf.edu.cn
 Mian Zhang
Mian Zhang Hao Feng1
Hao Feng1 Xiangming Xu
Xiangming Xu Lili Huang
Lili Huang