- 1School of Life Sciences, Northwestern Polytechnical University, Xi'an, China
- 2Key Laboratory of Algal Biology, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China
- 3Donghu Experimental Station of Lake Ecosystems, State Key Laboratory of Freshwater Ecology and Biotechnology of China, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China
Pili are found on the surface of many bacteria and play important roles in cell motility, pathogenesis, biofilm formation, and sensing and reacting to environmental changes. Cell motility in the model cyanobacterium Synechocystis sp. PCC 6803 relies on expression of the putative pilA9-pilA10-pilA11-slr2018 operon. In this study, we identified the antisense RNA PilR encoded in the noncoding strand of the prepilin-encoding gene pilA11. Analysis of overexpressor [PilR(+)] and suppressor [PilR(−)] mutant strains revealed that PilR is a direct negative regulator of PilA11 protein. Although overexpression of PilR did not affect cell growth, it greatly reduced levels of pilA11 mRNA and protein and decreased both the thickness and number of pili, resulting in limited cell motility and small, distinct colonies. Suppression of PilR had the opposite effect. A hypothetical model on the regulation of pilA9-pilA10-pilA11-slr2018 operon expression by PilR was proposed. These results add a layer of complexity to the mechanisms controlling pilA11 gene expression and cell motility, and provide novel insights into how sRNA and the intergenic region secondary structures can work together to discoordinatly regulate target gene in an operon in cyanobacterium.
Introduction
Cyanobacteria are ancient organisms that perform oxygenic photosynthesis (Waterbury et al., 1985). According to endosymbiotic theory, plant chloroplasts originated from cyanobacteria (or a cyanobacteria-like organism) through primary endosymbiosis. Many cyanobacteria move by gliding, swimming, or twitching (Waterbury et al., 1985; Häder, 1987). Gliding motility is a slow, uniform, forward motion, which parallel to the cell's longitudinal axis on a solid surface (Häder, 1987). This type of motion is occasionally interrupted by reversals in filamentous cyanobacteria such as Phormidium uncinatum and Anabaena variabilis (Häder, 1987). Several marine species of unicellular Synechococcus show swimming motility through liquids at a rate of 25 μm s−1(Waterbury et al., 1985). Twitching motility is small and intermittent translocation on a solid surface with frequent changes in direction (Henrichsen, 1972). Synechocystis sp. PCC 6803, a model unicellular cyanobacterium, exhibits twitching motility on an agar plate or glass slide (Stanier et al., 1971; Ng et al., 2003).
The pilus is a hair-like appendage found on the surface of many single-celled prokaryotes and has emerged as an efficient device for cell motility, pathogenesis (Herrington et al., 1988; Strom and Lory, 1993; Sauer et al., 2000), biofilm formation (Pratt and Kolter, 1998; Barken et al., 2008), and environmental sensing (Kawagishi et al., 1996). The genomic sequencing of Synechocystis sp. PCC 6803 was finished in 1996 (Kaneko et al., 1996). Since then, a number of genes (known as the pil genes) involved in pilus biogenesis, cell motility, and transformation competency have been revealed by mutational analysis (Bhaya et al., 1999, 2000, 2001; Yoshihara et al., 2001), which show homology to type IV pili biogenesis genes in many Gram-negative bacteria. Nonflagellar appendages of Gram-negative bacteria can be categorized into five major classes based on their biosynthetic pathway (Fronzes et al., 2008; Lo et al., 2013): chaperone–usher pili (Sauer et al., 1999; Waksman and Hultgren, 2009; Busch and Waksman, 2012; Geibel and Waksman, 2014; Pham et al., 2016), curli (Olsén et al., 1989; Barnhart and Chapman, 2006; Green et al., 2016), type IV pili (Bhaya et al., 2000; Merz et al., 2000; Maier et al., 2002; Busch and Waksman, 2012; Busch et al., 2015), type III secretion needle (Roine et al., 1997; Kubori et al., 1998), and type IV secretion pili (Seubert et al., 2003; Schröder and Lanka, 2005). Eleven pilA-like genes are contained in Synechocystis genome, which encode a prepilin peptide with a characteristic sequence (Yoshihara et al., 2002; Yoshimura et al., 2002). Of these genes, pilA10, pilA11, and slr2018 function in cell motility (Bhaya et al., 2001), and pilA1 is essential for the formation of thick and thin pili (Bhaya et al., 2000; Yoshihara et al., 2001).
Transcriptome analyses have identified numerous noncoding transcripts in bacteria, mainly trans-encoded RNAs and cis-antisense RNAs (asRNAs) (Waters and Storz, 2009). Cis-encoded asRNA transcripts appear to be dominant in several cyanobacteria. For example, asRNAs respectively comprise 26 and 39% of all genes in Synechocystis sp. PCC 6803 (Georg et al., 2009; Mitschke et al., 2011a) and Anabaena sp. PCC 7120 (Mitschke et al., 2011b). Chromosomally encoded asRNAs may play important roles in the regulatory networks of cyanobacteria. During the past decade, numerous newly discovered asRNAs have been shown to be involved in a wide range of processes (Kopf and Hess, 2015), including stress responses, photoprotection, low carbon responses, and carbon assimilation (Dühring et al., 2006; Eisenhut et al., 2012; Sakurai et al., 2012; Hu et al., 2017).
The transcriptome analysis using differential RNA sequencing in Synechocystis sp. PCC 6803 (Xu et al., 2014; Hu et al., 2017) revealed a low-abundance asRNA encoded in the noncoding strand of pilA11, which was named PilR. In this study, we determined the molecular functions of PilR by identifying its target gene, pilA11. An analysis of mutants with either elevated or reduced levels of PilR expression showed that PilR plays a key role in pilus formation and cell motility by negatively regulating pilA11 expression.
Results
Characterization of PilR
Differential RNA sequencing of the Synechocystis sp. PCC 6803 (hereafter Synechocystis) transcriptome detected an asRNA, designated PilR, with a transcription start site at the 3′ end of the pilA11 gene, but on its complementary strand. PilR was determined to be 210 long using Northern blot (Figure 1A) and RACE analyses, and its transcription start site was mapped to nucleotide nt758305 in the sequenced genome. PilR extends from positions 677–886 in the coding sequence of pilA11 (Figure 1B), and can be folded into two extended stem regions, with a terminal loop at each ending as predicted by the mfold software (http://www.bioinfo.rpi.edu/applications/mfold/; Figure 1C), such loops structures are believed to be involved in RNA–RNA interactions and therefore functionally related to the hypothetical trans-acting function (Dühring et al., 2006).
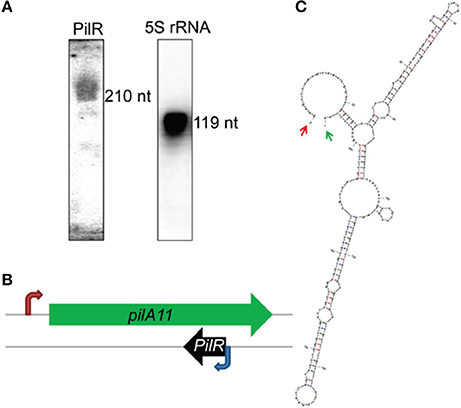
Figure 1. Characterization of the antisense RNA PilR in Synechocystis sp. PCC 6803. (A) Typical detection pattern of PilR and 5S rRNA by Northern blot analysis. Five micrograms of total RNA were loaded for detection of 5S rRNA. Fifty micrograms of total RNA were used to detect PilR. (B) Location of the pilA11 gene within the genome. The red arrow marks the transcription initiation site. The blue arrow marks the PilR transcription initiation site detected by the RACE experiments. (C) The RNA secondary structure prediction for PilR. Arrows point to the experimentally detected 5′ (red) and 3′ (green) ends.
We further constructed background control, PilR overexpression [PilR(+)], and PilR suppression [PilR(−)] strains (Supplementary Figure S1) for investigating the relationship between PilR and its target gene, pilA11. PilR appears to be a negative regulator of pilA11 expression during the exponential growth phase, as revealed through qRT-PCR and immunoblot analyses of these strains (Figures 2A,E). As shown in Figure 2A, the levels of asRNA PilR were 4.28-fold higher in the PilR(+) strain than in the control, and 0.25-fold lower in the PilR(−) strain than in the control (Figure 2A, white speckled bars; **P < 0.01). By contrast, the levels of pilA11 mRNA were 0.45-fold lower in the PilR(+) strain than in the control, and 2.06-fold higher in the PilR(−) strain than in the control (Figure 2A, gray bars, **P < 0.01). Similarly, the levels of PilA11 protein were 0.55-fold lower in the PilR(+) strain than in the control, and 1.25-fold higher in the PilR(−) strains than in the control (Figure 2E).
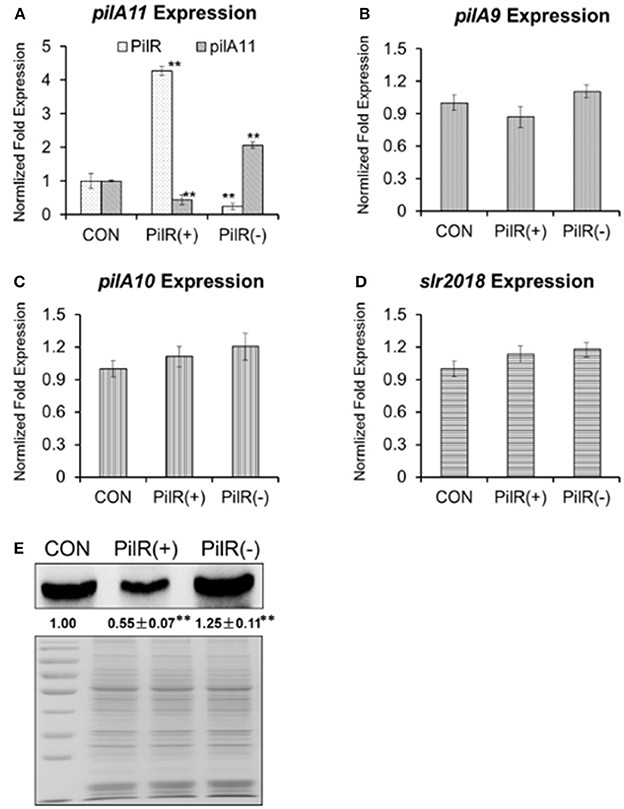
Figure 2. The influence of PilR on pilA9–pilA10–pilA11–slr2018 operon transcription and PilA11 protein. The expression levels of (A) PilR (white) and pilA11 mRNA (gray), (B) pilA9 mRNA (gray), (C) pilA10 mRNA (gray), and (D) slr2018 mRNA (gray) in cells with normal (control), overexpressed [PilR(+)], and suppressed [PilR(-)] levels of PilR in BG11 medium as measured by qRT–PCR. All data are shown as means ± SD (n = 5). (E) Immunoblot of PilA11 protein isolated from total protein of control, PilR(+), and PiIR(−) mutant cells in BG11 medium. Significant differences between the control (CON) and test values were determined using a one–way ANOVA. **P < 0.01 vs. CON.
As previous investigations on the expression levels of the six genes (slr1667, slr1668, pilA9, pilA10, pilA11, and slr2018) strongly suggested that pilA9, pilA10, pilA11, and slr2018 constitute one operon (Kamei et al., 2001a; Yoshimura et al., 2002; Panichkin et al., 2006) to verify that PilR affects pilA11 but no other genes in the pilA9-pilA10-pilA11-slr2018 operon, pilA9, pilA10, and slr2018 transcript levels were also analyzed by qRT-PCR in the mutant strains (Figures 2B–D). We found that the mRNA levels of pilA9 were 0.87-fold lower in the PilA(+) strain than in the control, and 1.10-fold higher in the PilA(−) strain (Figure 2B, P > 0.05); those of pilA10 were 1.11-fold higher in the PilA(+) strain than in the control, and 1.20-fold higher in the PilA(−) strain (Figure 2C, P > 0.05); and those of slr2018 were 1.13-fold higher in the PilA(+) strain than in the control, and 1.18-fold higher in the PilA(−) strain (Figure 2D, P > 0.05). These results suggest that PilR has no significant effect on the stability of the mRNA portions that encode pilA9, pilA10, and slr2018, providing strong evidence that PilR, despite its relatively low steady-state expression level, negatively regulates the amount of pilA11 mRNA and PilA11 protein in Synechocystis.
PilA11 Localization
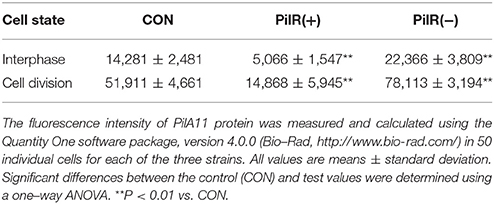
To visualize the influence of PilR on PilA11 protein, we observed the in vivo localization of PilA11 protein in control, PilR(+), and PiIR(−) strains using immunofluorescence and confocal microscopy. PilR(+) cells exhibited weaker fluorescence compared to the control strain, but PilR(−) cells had significantly enhanced fluorescence compared to the control (Figures 3, 4; Table 1, **P < 0.01), in agreement with their protein levels (Figure 2E). When enlarged and visualized at the single cell level, the fluorescence signal could be observed clearly at the cell surface of all strains (Figure 3), as expected given its role in cell motility. We also observed high levels of fluorescence in all three strains in dividing cells (Figure 4, Supplementary Figure S2), suggesting that PilA11 protein is enriched at the cell surface during division regardless of PilR levels.
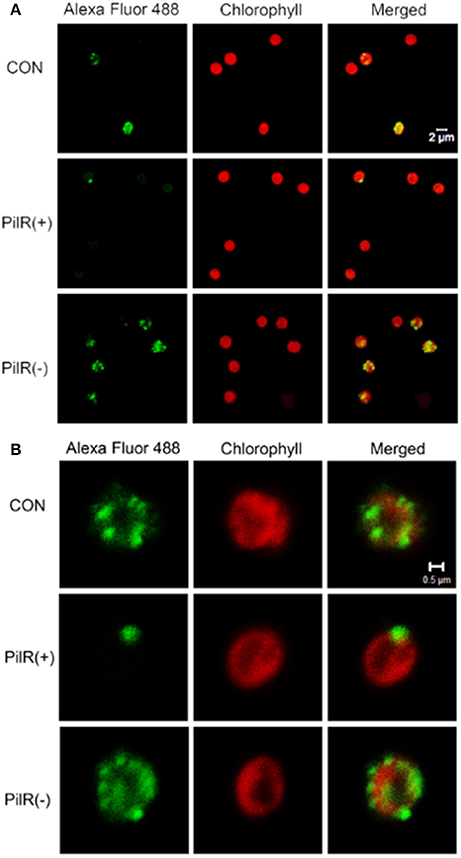
Figure 3. Fluorescence images showing the control, PilR(+), and PiIR(−) cells labeled with PilA11–Alexa Fluor 488. (A) A group of individual cells. (B) A single cell. The merged panel shows images of cells labeled with PilA11 (green) and chlorophyII (red) merged together. White bars = 2 and 0.5 μm.
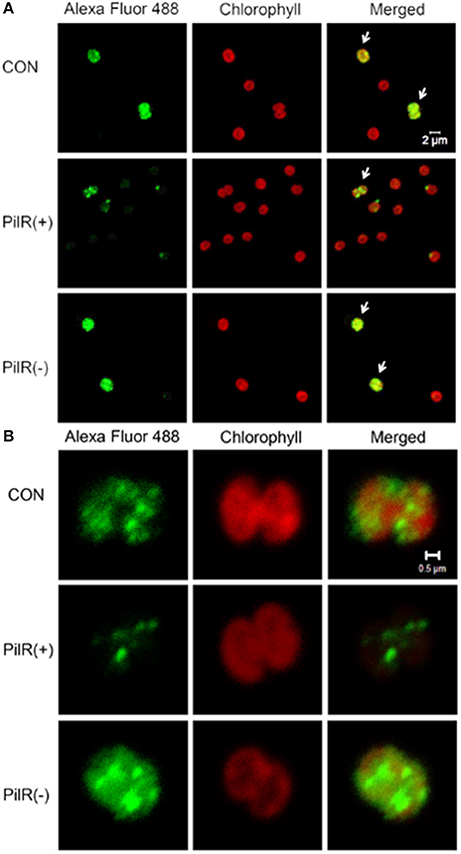
Figure 4. Up–regulated expression of PilA11 protein during cell division. (A) A group of individual cells. (B) A single cell. Fluorescence images of control and PilR(+)/(−) mutant cells labeled with PilA11–Alexa Fluor 488. Arrows indicate cells undergoing division. White bars = 2 and 0.5 μm.
Pilus Formation and Cell Motility of the PilR Mutant Strains
Previous studies showed that PilA11 was an essential protein for cell motility and thick pili formation (Bhaya et al., 2001; Panichkin et al., 2006). To test the effect of PilR on PilA11 protein function, we examined cell motility and thick pili formation in the PilR strains. Cyanobacterial cells were grown under normal conditions, and cells were maintained in the presence of 20 μg·mL−1 kanamycin. However, for eliminating possible phenotypic alterations due to the antibiotic, the final cultures without kanamycin were used in the experiments.
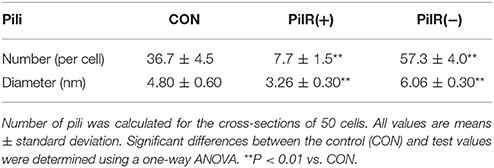
We examined control, PilR(+), and PiIR(−) cells with an electron microscope to investigate whether changes in PilR expression affected the formation of pili. As shown in Figure 5, the pili of PilR(+) cells were significantly thinner and fewer than the well-developed, normal pili of the control cells. Pili of PilR(−) cells were thicker and denser than those of the control. The number and diameter of pili of fifty individual cells each of the control, PilR(+), and PiIR(−) strains were examined and the results confirmed that PilR overexpression or suppression significantly affected pilus formation (Table 2).
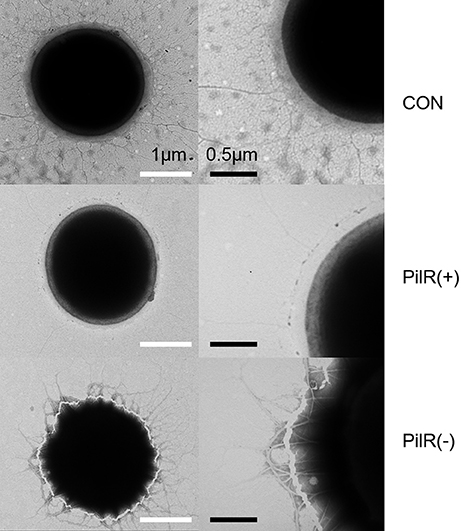
Figure 5. Electron micrograph of control, PilR(+), and PiIR(−) cells. Cells were processed by negative staining techniques and examined with an electron microscope. The pili of PilR(+) cells were thinner than those of the control cells, and the pili of PilR(−) cells were thicker than those of the control cells. White bars = 1 μm; black bars = 0.5 μm.
To investigate the effects of PilR on cell growth and motility in Synechocystis, we analyzed the three strains under normal conditions in liquid culture or on agar plates. As shown in Figure 6A and Table 3, when cultured in liquid BG11, the growth rate and pigmentation were not affected in either the PilR(+) or PilR(−) strains. We examined the effects of PilR overexpression or suppression on motility by monitoring the colonies shape formed on agar plates. The colonies of the PilR(−) strain were larger and more diffuse than the control colonies, whereas those of the PilR(+) strain were smaller and more centralized (Figure 6B).
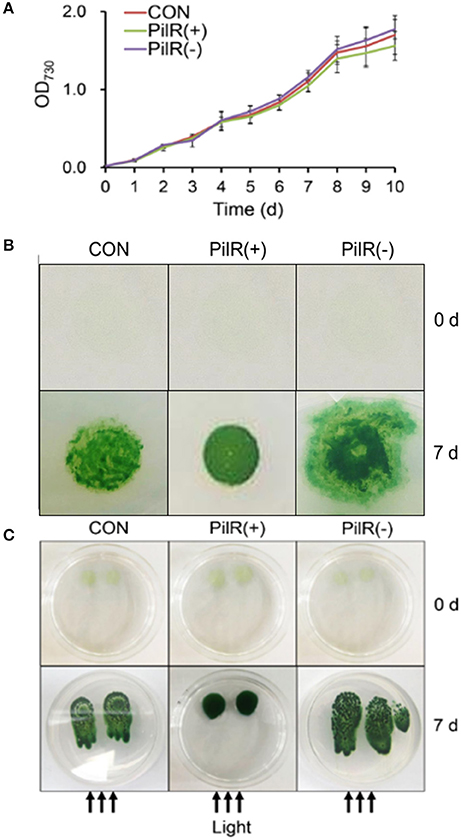
Figure 6. The effect of PilR on growth and motility in Synechocystis sp. PCC 6803. (A) Growth of cells in liquid culture. OD730, optical density at 730 nm. (B) Exponentially growing cells (200 μL) were spotted onto a 1% agar BG11 plate. The plates were placed under an incandescent light source of 30 μmol photons·m−2·s−1 and allowed to grow for 7 days. (C) Comparison of the agar surface-based phototaxis assays. Cells were applied to the surface of 0.5% agar with a micropipette and placed in front of a unidirectional light source of 30 μmol photons·m−2·s−1 for 7 days. The arrows indicate the direction of light.
We then performed agar surface-based phototaxis assays to clarify the differences in motility between the control, PilR(+), and PiIR(−) strains (Figure 6C). When the cells were dot plated and exposed to a unidirectional light source for 7 days, the control and PilR(−) mutant strains showed positive phototactic movement, with the latter having a stronger phenotype. The PilR(+) mutant strain showed almost no sign of phototactic movement (Figure 6C), suggesting that PilR expression influences Synechocystis cell motility in a concentration-dependent manner.
Discussion
Many species of cyanobacteria move by gliding, twitching, or swimming. Unlike Escherichia coli and Chlamydomonas reinhardtii, which use flagella, cyanobacteria use a pilus apparatus for motility (Waterbury et al., 1985; Häder, 1987). The oxygenic phototrophic cyanobacterium Synechocystis exhibits twitching motility (Stanier et al., 1971; Ng et al., 2003). Two morphologically distinct pilus types, thick and thin, exist in wild-type Synechocystis cells. Thick pili, possibly encoded by pilA1, are similar to type IV pili in many functional and morphological characteristics. Thin pili with smaller diameter are shorter than typical type IV pili (Bhaya et al., 2000). Type IV pilus biogenesis requires a complex polypeptides assemblage, located in the cytoplasmic membrane, the periplasm, or the outer membrane, for post-translational modification (e.g., PilD), assembly and export (e.g., PilC, PilQ) (Bhaya et al., 2000).. Type IV pili subunits (i.e., PilA) have a conserved, hydrophobic α-helix domain at the N-terminus, which consists of 20–25 amino acids that forms the hydrophobic pilus core (Proft and Baker, 2009). pilA1 is responsible for the structure, motility, and transformation efficiency of thick pilus (Bhaya et al., 1999; Yoshihara et al., 2001). Mutants with disrupted pilA2, which encodes a second pilin-like protein, are still motile with normal cell-surface pili morphology and density. By contrast, inactivation of pilD, which encodes the leader peptidase, or pilC, which encodes a protein required for pilus assembly, abolishes cell motility and causes the absence of both pilus morphotypes (Bhaya et al., 2000). In our study, suppression of pilA11 by overexpression of its antisense RNA PilR [PilR(+)] leads to thin, sparse pili, whereas PilR suppression [PilR(−)] has the opposite effect (Figure 5). This indicates that pilA11 may also contribute to thick pilus biogenesis and, indeed, the ratio of thick pili to all pili was altered in the PilR mutant strains.
A locus containing five genes (pilA9-pilA10-pilA11-slr2018-slr2019) was discovered in Synechocystis in an analysis of transposon-generated mutants (Bhaya et al., 2001). The protein encoded by pilA10 shows weak similarity to members of the PilA-like protein family (Bhaya et al., 1999), but the proteins encoded by pilA11 and slr2018 lack obvious functional motifs and have no clear homologs in protein databases. We observed that expression of pilA11, but not pilA9, pilA10, or slr2018, is negatively affected by its antisense sRNA PilR (Figure 2), and that overexpression of PilR disturbs the biogenesis of the thick pilus morphotype (Figure 5), which affects cell motility (Figure 6). These results suggest that PilR negatively regulates pilA11 and has an important function in cell motility.
SpkA and the ATPase PilT are required for motility in Synechocystis (Kamei et al., 2001b; Okamoto and Ohmori, 2002). Bhaya et al. showed that the pilA10, pilA11, and slr2018 genes were essential for cell motility (Bhaya et al., 2001). This study found that on 1% agar-solidified plates the pilA11 asRNA overexpressor PilR(+) colonies are small and distinct, whereas the suppressor PilR(−) colonies are large and diffuse (Figure 6B). spkA::Cmr mutant cells, in which the pilA9-pilA10-pilA11-slr2018 operon is down-regulated, also form distinct colonies similar to PilR(+) (Panichkin et al., 2006). The observed phenotype of PilR(+) mutant strains is consistent with a previous analysis showing that the putative pilA9-pilA10-pilA11-slr2018 operon in Synechocystis might be involved in the formation of thick pili (Panichkin et al., 2006). However, the effect that expression of the operon has on Synechocystis cells has not been deciphered until now.
sRNAs regulate gene expression from polycistronic messages through a variety of mechanisms (Balasubramanian and Vanderpool, 2013), which can alter expression of select genes in an operon by inhibiting the translation of genes or by altering the stability of mRNA, resulting in discoordinate regulation of the target mRNA (Møller et al., 2002; Kalamorz et al., 2007; Desnoyers et al., 2009). Alternatively, they can affect expression of all genes in an operon by sRNA-mRNA interactions, causing coordinate regulation (Rice and Vanderpool, 2011; Lu et al., 2012). The 210 nt sRNA PilR was identified as an asRNA by repressing expression of pilA11 gene, but not pilA9, pilA10, or slr2018, in Synechocystis (Figure 2). It is speculated that PilR prevents pilA11 gene expression by selective mRNA degradation. This type of regulation represses pilA11 gene expression, but allows continued synthesis of other Type IV pilin-like proteins.
To further elucidate the discoordinate regulation mechanism of the cis-type PilR transcript to pilA11 gene, a closer sequence inspection is needed in both target gene pilA11 and asRNA PilR, their cleavage by RNases. By sequence analysis, a strong secondary structure (Figure 7A) in the intergenic region between the pilA11 and slr2018 cistrons was predicted. Add up to the present understanding of the function of PilR in the regulation of pilA11 gene expression, a hypothetical model on the regulation of pilA9-pilA10-pilA11-slr2018 operon expression by PilR was proposed (Figure 7). Normally, single-stranded pilA9-pilA10-pilA11-slr2018 mRNA is translated into three type IV pilin-like protein, PilA9, PilA10, PilA11 and the unknown protein Slr2018 (Figure 7B, upper panel). While the degradation of pilA11 mRNA is typically triggered by sRNA PilR base pairing to the target mRNA, and results in translational silencing and promoted degradosome-dependent endonucleolytic cleavage (Kaberdin et al., 2011; Balasubramanian and Vanderpool, 2013) of the pilA11 portion of the mRNA, presumably by RNase E (Figure 7B, middle panel). The degradosome is stopped and discharged by the hairpin structure (Figure 7A) between the pilA11 and slr2018 cistrons and thus selectively prevents the slr2018 mRNA from degradation (Figure 7B, lower panel). The suggested model provides novel insights into how sRNA and the intergenic region secondary structures can work together to discoordinatly regulate target gene in an operon in Synechocystis (Figures 2A,E).
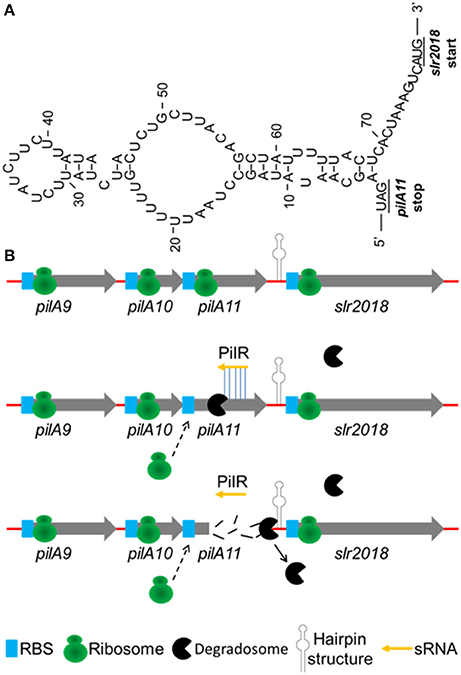
Figure 7. Hypothetical model on the regulation of pilA9-pilA10-pilA11-slr2018 operon expression by PilR. (A) A secondary structure of the intergenic region between pilA11 and slr2018 cistrons (represented by hairpin structure in panel B) as determined by the mfold software (http://www.bioinfo.rpi.edu/applications/mfold/). (B) Normally, single-stranded pilA9-pilA10-pilA11-slr2018 mRNA is translated into three type 4 pilin-like protein, PilA9, PilA10, PilA11 and the unknown protein Slr2018 (upper panel). While the degradation of pilA11 mRNA is typically triggered by sRNA PilR base pairing to the target mRNA, and results in translational silencing and promoted degradosome-dependent endonucleolytic cleavage of the pilA11 portion of the mRNA (middle panel). The degradosome is stopped and discharged by the hairpin structure between the pilA11 and slr2018 cistrons and thus selectively prevents the slr2018 mRNA from degradation (lower panel).
Cis-encoded asRNA transcripts typically regulate target gene expression either negatively, as is the case for IsrR (Dühring et al., 2006) and As1_flv4 (Eisenhut et al., 2012), or positively, as is the case for PsbA2R, PsbA3R (Sakurai et al., 2012), and RblR (Hu et al., 2017). These asRNAs are involved in various processes (Kopf and Hess, 2015), but prior to this study, no asRNAs related to cell motility were known. Our results show that the asRNA PilR is encoded by the antisense strand of the pilA11 gene and negatively regulates its expression through complementary base pairing. Downregulation of this member of the putative pilA9-pilA10-pilA11-slr2018 operon reduces the number and thickness of the pili, affecting cell motility. These results add a new layer of complexity to the regulatory mechanisms controlling pilA11 gene expression and function and hence cell motility.
Experimental Procedures
Strains and Growth Conditions
Wild-type Synechocystis was cultured as described previously (Hu et al., 2017). Solid medium supplemented with 1% (w/v) or 0.5% (w/v) agar is used to observe colony morphology for motility evaluation. After generating mutant strains (see below), kanamycin (20 μg/mL) was added to the growth medium to identify the transformed cells. Antibiotics were excluded during phenotyping to avoid interactions. The colonies were trained for 7 days under lateral illumination with a white fluorescent lamp (~30 μmol photons·m−2·S−1).
RNA Extraction and Northern Blot Analysis
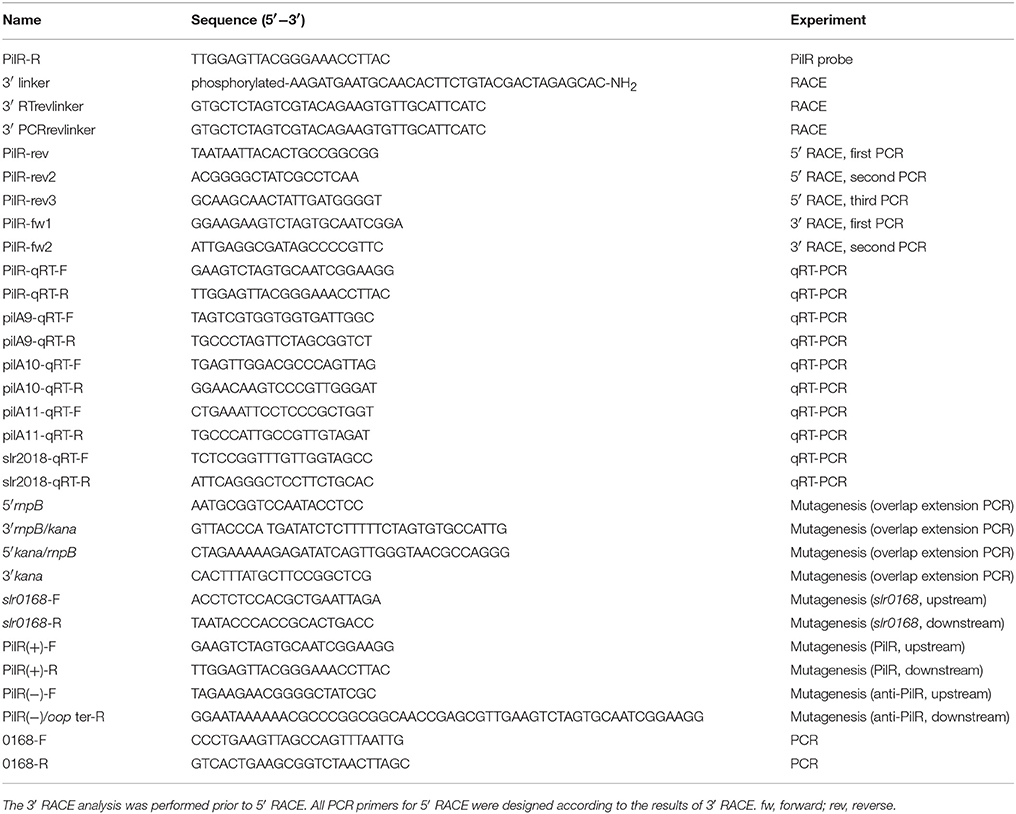
Total RNA extraction was performed as described previously (Hu et al., 2017). Northern blot analysis was performed as previously described (Hu et al., 2014). [γ-32P] ATP (PerkinElmer, USA) was used for the labeling of probes. DNA oligonucleotides used for the Northern blot analysis are listed in Table 4.
5′- and 3′- Rapid Amplification of cDNA Ends (RACE)
The 5′ end and 3′ end RACE was performed according to Hu et al. (2014). All oligonucleotides and primers used in the RACE analysis are listed in Table 4.
qRT-PCR Validation
The qRT-PCR analysis was done by standard procedure using cDNA as previously described (Hu et al., 2014, 2017). All data are shown as the mean ± SD (n = 5). All primers used for the analysis are listed in Table 4.
Mutagenesis
Mutant strains PilR(+), and PilR(−) were created as previously described (Golden et al., 1987; Hu et al., 2017). All primers used for this analysis are listed in Table 4.
Protein Gel and Immunoblot Analysis
Protein gel and immunoblot analysis were performed as previously described (Hu et al., 2014). The membranes were probed with rabbit primary anti-PilA11 antibodies (1:5,000; QWbio, http://www.qwbio.com Beijing).
Electron Microscopy
Specimens were prepared for electron microscopy using the conventional negative staining procedure (Bhaya et al., 1999) with the following modifications. Briefly, a 200 μL drop of sample solution was adsorbed onto a glow-discharged carbon-coated copper grid for 10 min, stained with two drops of freshly prepared 2% phosphotungstic acid (pH 7.0), and examined using a Hitachi HT7700 microscope.
Immunofluorescence Microscopy
Microscopy analysis of cells grown in liquid BG11 medium was carried out using a confocal scanning microscope (Leica TCS SP8, Wetzlar, Germany). PilA11 localization was performed as previously described (Miyagishima et al., 2005; Zhan et al., 2016). The fluorescence intensity was measured and quantified using the Quantity One software package, version 4.0.0 (Bio-Rad, http://www.bio-rad.com/) in 50 individual cells for each of the three strains.
Measurements of Chlorophyll a and Carotenoid Contents
Chlorophyll a and carotenoid contents were measured as described (Zhang et al., 2013). Chlorophyll a (Ca) and total carotenoid (Cc) contents were calculated according to Equation (1) (Harmut and Lichtenthaler, 1987).
Calculating the Number and Diameter of Pili
Number and diameter of pili were calculated for the cross-sections of 50 cells of electron microscopy using ImageJ software. All values are means ± standard deviation. Significant differences between the control (CON) and test values were determined using a one-way ANOVA. **P < 0.01 vs. CON.
Author Contributions
JH and QW: Designed the study; JH, JZ, HuiC, CH, and QW: Collected, analyzed, and interpreted the data; JH, HuaC, and QW: Wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported jointly by the National Natural Science Foundation of China (31770128, 31700107, and 31700055), the State Key Laboratory of Freshwater Ecology and Biotechnology (Y11901-1-F01), the Fundamental Research Funds for the Central Universities (3102017OQD042), and the China Postdoctoral Science Foundation (2017M610648). We thank Yuan Xiao and Fang Zhou of the Institute of Hydrobiology, CAS for assistance with the electron microscopy and confocal scanning microscopy.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00786/full#supplementary-material
References
Balasubramanian, D., and Vanderpool, C. K. (2013). New developments in post-transcriptional regulation of operons by small RNAs. RNA Biol. 10, 337–341. doi: 10.4161/rna.23696
Barken, K. B., Pamp, S. J., Yang, L., Gjermansen, M., Bertrand, J. J., Klausen, M., et al. (2008). Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 10, 2331–2343. doi: 10.1111/j.1462-2920.2008.01658.x
Barnhart, M. M., and Chapman, M. R. (2006). Curli biogenesis and function. Annu. Rev. Microbiol. 60, 131–147. doi: 10.1146/annurev.micro.60.080805.142106
Bhaya, D., Bianco, N. R., Bryant, D., and Grossman, A. (2000). Type IV pilus biogenesis and motility in the cyanobacterium Synechocystis sp. PCC6803. Mol. Microbiol. 37, 941–951. doi: 10.1046/j.1365-2958.2000.02068.x
Bhaya, D., Takahashi, A., Shahi, P., and Grossman, A. R. (2001). Novel motility mutants of synechocystis strain PCC 6803 generated by in vitro transposon mutagenesis. J. Bacteriol. 183, 6140–6143. doi: 10.1128/JB.183.20.6140-6143.2001
Bhaya, D., Watanabe, N., Ogawa, T., and Grossman, A. R. (1999). The role of an alternative sigma factor in motility and pilus formation in the cyanobacterium Synechocystis sp. strain PCC6803. Proc. Natl. Acad. Sci. U.S.A. 96, 3188–3193. doi: 10.1073/pnas.96.6.3188
Busch, A., Phan, G., and Waksman, G. (2015). Molecular mechanism of bacterial type 1 and P pili assembly. Philos. Trans. A Math. Phys. Eng. Sci. 373:20130153. doi: 10.1098/rsta.2013.0153
Busch, A., and Waksman, G. (2012). Chaperone-usher pathways: diversity and pilus assembly mechanism. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 1112–1122. doi: 10.1098/rstb.2011.0206
Desnoyers, G., Morissette, A., Prévost, K., and Massé, E. (2009). Small RNA-induced differential degradation of the polycistronic mRNA iscRSUA. EMBO J. 28, 1551–1561. doi: 10.1038/emboj.2009.116
Dühring, U., Axmann, I. M., Hess, W. R., and Wilde, A. (2006). An internal antisense RNA regulates expression of the photosynthesis gene isiA. Proc. Natl. Acad. Sci. U.S.A. 103, 7054–7058. doi: 10.1073/pnas.0600927103
Eisenhut, M., Georg, J., Klähn, S., Sakurai, I., Mustila, H., Zhang, P., et al. (2012). The antisense RNA As1_flv4 in the cyanobacterium Synechocystis sp. PCC 6803 prevents premature expression of the flv4-2 operon upon shift in inorganic carbon supply. J. Biol. Chem. 287, 33153–33162. doi: 10.1074/jbc.M112.391755
Fronzes, R., Remaut, H., and Waksman, G. (2008). Architectures and biogenesis of non-flagellar protein appendages in Gram-negative bacteria. EMBO J. 27, 2271–2280. doi: 10.1038/emboj.2008.155
Geibel, S., and Waksman, G. (2014). The molecular dissection of the chaperone-usher pathway. Biochim. Biophys. Acta 1843, 1559–1567. doi: 10.1016/j.bbamcr.2013.09.023
Georg, J., Voss, B., Scholz, I., Mitschke, J., Wilde, A., and Hess, W. R. (2009). Evidence for a major role of antisense RNAs in cyanobacterial gene regulation. Mol. Syst. Biol. 5, 305. doi: 10.1038/msb.2009.63
Golden, S. S., Brusslan, J., and Haselkorn, R. (1987). Genetic engineering of the cyanobacterial chromosome. Method Enzymol. 153, 215–231. doi: 10.1016/0076-6879(87)53055-5
Green, A., Pham, N., Osby, K., Aram, A., Claudius, R., Patray, S., et al. (2016). Are the curli proteins CsgE and CsgF intrinsically disordered? Intrins. Disord. Proteins 4:e1130675. doi: 10.1080/21690707.2015.1130675
Harmut, A., and Lichtenthaler, K. (1987). Chlorophylls and carotenoids: pigments of photosynthetic membranes. Method Enzymol. 148, 350–383. doi: 10.1016/0076-6879(87)48036-1
Henrichsen, J. (1972). Bacterial surface translocation: a survey and a classification. Bacteriol. Rev. 36, 478–503.
Herrington, D. A., Hall, R. H., Losonsky, G., Mekalanos, J. J., Taylor, R., and Levine, M. M. (1988). Toxin, toxin-coregulated pili, and the toxR regulon are essential for Vibrio cholerae pathogenesis in humans. J. Exp. Med. 168, 1487–1492. doi: 10.1084/jem.168.4.1487
Hu, J., Deng, X., Shao, N., Wang, G., and Huang, K. (2014). Rapid construction and screening of artificial microRNA systems in Chlamydomonas reinhardtii. Plant J. 79, 1052–1064. doi: 10.1111/tpj.12606
Hu, J., Li, T., Xu, W., Zhan, J., Chen, H., He, C., et al. (2017). Small antisense RNA RblR positively regulates RuBisCo in Synechocystis sp. PCC 6803. Front. Microbiol. 8:231. doi: 10.3389/fmicb.2017.00231
Kaberdin, V. R., Singh, D., and Lin-Chao, S. (2011). Composition and conservation of the mRNA-degrading machinery in bacteria. J. Biomed. Sci. 18:23. doi: 10.1186/1423-0127-18-23
Kalamorz, F., Reichenbach, B., März, W., Rak, B., and Görke, B. (2007). Feedback control of glucosamine-6-phosphate synthase GlmS expression depends on the small RNA GlmZ and involves the novel protein YhbJ in Escherichia coli. Mol. Microbiol. 65, 1518–1533. doi: 10.1111/j.1365-2958.2007.05888.x
Kamei, A., Hihara, Y., Yoshihara, S., Geng, X., Kanehisa, M., and Ikeuchi, M. (2001a). Functional analysis of lexA-like gene, sll1626 in Synechocystis sp. PCC 6803 using DNA microarray. Sci. Acc. 3:SA0403733. doi: 10.1071/SA0403733
Kamei, A., Yuasa, T., Orikawa, K., Geng, X. X., and Ikeuchi, M. (2001b). A eukaryotic-type protein kinase, SpkA, is required for normal motility of the unicellular Cyanobacterium synechocystis sp. strain PCC 6803. J. Bacteriol. 183, 1505–1510. doi: 10.1128/JB.183.5.1505-1510.2001
Kaneko, T., Sato, S., Kotani, H., Tanaka, A., Asamizu, E., Nakamura, Y., et al. (1996). Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3, 109–136. doi: 10.1093/dnares/3.3.109
Kawagishi, I., Imagawa, M., Imae, Y., McCarter, L., and Homma, M. (1996). The sodium-driven polar flagellar motor of marine Vibrio as the mechanosensor that regulates lateral flagellar expression. Mol. Microbiol. 20, 693–699. doi: 10.1111/j.1365-2958.1996.tb02509.x
Kopf, M., and Hess, W. R. (2015). Regulatory RNAs in photosynthetic cyanobacteria. FEMS Microbiol. Rev. 39, 301–315. doi: 10.1093/femsre/fuv017
Kubori, T., Matsushima, Y., Nakamura, D., Uralil, J., Lara-Tejero, M., Sukhan, A., et al. (1998). Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280, 602–605. doi: 10.1126/science.280.5363.602
Lo, A. W., Moonens, K., and Remaut, H. (2013). Chemical attenuation of pilus function and assembly in Gram-negative bacteria. Curr. Opin. Microbiol. 16, 85–92. doi: 10.1016/j.mib.2013.02.003
Lu, P., Zhang, Y., Li, L., Hu, Y., Huang, L., Li, Y., et al. (2012). Small non-coding RNA SraG regulates the operon YPK_1206-1205 in Yersinia pseudotuberculosis. FEMS Microbiol. Lett. 331, 37–43. doi: 10.1111/j.1574-6968.2012.02548.x
Maier, B., Potter, L., So, M., Long, C. D., Seifert, H. S., and Sheetz, M. P. (2002). Single pilus motor forces exceed 100 pN. Proc. Natl. Acad. Sci. U.S.A. 99, 16012–16017. doi: 10.1073/pnas.242523299
Merz, A. J., So, M., and Sheetz, M. P. (2000). Pilus retraction powers bacterial twitching motility. Nature 407, 98–102. doi: 10.1038/35024105
Mitschke, J., Georg, J., Scholz, I., Sharma, C. M., Dienst, D., Bantscheff, J., et al. (2011a). An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc. Natl. Acad. Sci. U.S.A. 108, 2124–2129. doi: 10.1073/pnas.1015154108
Mitschke, J., Vioque, A., Haas, F., Hess, W. R., and Muro-Pastor, A. M. (2011b). Dynamics of transcriptional start site selection during nitrogen stress-induced cell differentiation in Anabaena sp. PCC7120. Proc. Natl. Acad. Sci. U.S.A. 108, 20130–20135. doi: 10.1073/pnas.1112724108
Miyagishima, S. Y., Wolk, C. P., and Osteryoung, K. W. (2005). Identification of cyanobacterial cell division genes by comparative and mutational analyses. Mol. Microbiol. 56, 126–143. doi: 10.1111/j.1365-2958.2005.04548.x
Møller, T., Franch, T., Udesen, C., Gerdes, K., and Valentin-Hansen, P. (2002). Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Genes Dev. 16, 1696–1706. doi: 10.1101/gad.231702
Ng, W. O., Grossman, A. R., and Bhaya, D. (2003). Multiple light inputs control phototaxis in Synechocystis sp. strain PCC6803. J. Bacteriol. 185, 1599–1607. doi: 10.1128/JB.185.5.1599-1607.2003
Okamoto, S., and Ohmori, M. (2002). The cyanobacterial PilT protein responsible for cell motility and transformation hydrolyzes ATP. Plant Cell Physiol. 43, 1127–1136. doi: 10.1093/pcp/pcf128
Olsén, A., Jonsson, A., and Normark, S. (1989). Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature 338, 652–655. doi: 10.1038/338652a0
Panichkin, V. B., Arakawa-Kobayashi, S., Kanaseki, T., Suzuki, I., Los, D. A., Shestakov, S. V., et al. (2006). Serine/threonine protein kinase SpkA in Synechocystis sp. strain PCC 6803 is a regulator of expression of three putative pilA operons, formation of thick pili, and cell motility. J. Bacteriol. 188, 7696–7699. doi: 10.1128/JB.00838-06
Pham, T., Werneburg, G. T., Henderson, N. S., Thanassi, D. G., and Delcour, A. H. (2016). Effect of chaperone-adhesin complex on plug release by the PapC usher. FEBS Lett. 590, 2172–2179. doi: 10.1002/1873-3468.12257
Pratt, L. A., and Kolter, R. (1998). Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30, 285–293. doi: 10.1046/j.1365-2958.1998.01061.x
Proft, T., and Baker, E. (2009). Pili in Gram-negative and Gram-positive bacteria—structure, assembly and their role in disease. Cell. Mol. Life Sci. 66, 613–635. doi: 10.1007/s00018-008-8477-4
Rice, J. B., and Vanderpool, C. K. (2011). The small RNA SgrS controls sugar–phosphate accumulation by regulating multiple PTS genes. Nucleic Acids Res. 39, 3806–3819. doi: 10.1093/nar/gkq1219
Roine, E., Wei, W., Yuan, J., Nurmiaho-Lassila, E. L., Kalkkinen, N., Romantschuk, M., et al. (1997). Hrp pilus: an hrp-dependent bacterial surface appendage produced by Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. U.S.A. 94, 3459–3464. doi: 10.1073/pnas.94.7.3459
Sakurai, I., Stazic, D., Eisenhut, M., Vuorio, E., Steglich, C., Hess, W. R., et al. (2012). Positive regulation of psbA gene expression by cis-encoded antisense RNAs in Synechocystis sp. PCC 6803. Plant Physiol. 160, 1000–1010. doi: 10.1104/pp.112.202127
Sauer, F. G., Fütterer, K., Pinkner, J. S., Dodson, K. W., Hultgren, S. J., and Waksman, G. (1999). Structural basis of chaperone function and pilus biogenesis. Science 285, 1058–1061. doi: 10.1126/science.285.5430.1058
Sauer, F. G., Mulvey, M. A., Schilling, J. D., Martinez, J. J., and Hultgren, S. J. (2000). Bacterial pili: molecular mechanisms of pathogenesis. Curr. Opin. Microbiol. 3, 65–72. doi: 10.1016/S1369-5274(99)00053-3
Schröder, G., and Lanka, E. (2005). The mating pair formation system of conjugative plasmids-A versatile secretion machinery for transfer of proteins and DNA. Plasmid 54, 1–25. doi: 10.1016/j.plasmid.2005.02.001
Seubert, A., Hiestand, R., de la Cruz, F., and Dehio, C. (2003). A bacterial conjugation machinery recruited for pathogenesis. Mol. Microbiol. 49, 1253–1266. doi: 10.1046/j.1365-2958.2003.03650.x
Stanier, R. Y., Kunisawa, R., Mandel, M., and Cohen-Bazire, G. (1971). Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriol. Rev. 35, 171–205.
Strom, M. S., and Lory, S. (1993). Structure-function and biogenesis of the type IV pili. Ann. Rev. Microbiol. 47, 565–596. doi: 10.1146/annurev.mi.47.100193.003025
Waksman, G., and Hultgren, S. J. (2009). Structural biology of the chaperone-usher pathway of pilus biogenesis. Nat. Rev. Microbiol. 7, 765–774. doi: 10.1038/nrmicro2220
Waterbury, J. B., Willey, J. M., Franks, D. G., Valois, F. W., and Watson, S. W. (1985). A cyanobacterium capable of swimming motility. Science 230, 74–76. doi: 10.1126/science.230.4721.74
Waters, L. S., and Storz, G. (2009). Regulatory RNAs in bacteria. Cell 136, 615–628. doi: 10.1016/j.cell.2009.01.043
Xu, W., Chen, H., He, C.-L., and Wang, Q. (2014). Deep sequencing-based identification of small regulatory RNAs in Synechocystis sp. PCC 6803. PLoS ONE 9:e92711. doi: 10.1371/journal.pone.0092711
Yoshihara, S., Geng, X., and Ikeuchi, M. (2002). pilG Gene cluster and split pilL genes involved in pilus biogenesis, motility and genetic transformation in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 43, 513–521. doi: 10.1093/pcp/pcf061
Yoshihara, S., Geng, X., Okamoto, S., Yura, K., Murata, T., Go, M., et al. (2001). Mutational analysis of genes involved in pilus structure, motility and transformation competency in the unicellular motile cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 42, 63–73. doi: 10.1093/pcp/pce007
Yoshimura, H., Yoshihara, S., Okamoto, S., Ikeuchi, M., and Ohmori, M. (2002). A cAMP receptor protein, SYCRP1, is responsible for the cell motility of Synechocystis sp. PCC 6803. Plant Cell Physiol. 43, 460–463. doi: 10.1093/pcp/pcf050
Zhan, J., Zhu, X., Zhou, W., Chen, H., He, C., and Wang, Q. (2016). Thf1 interacts with PS I and stabilizes the PS I complex in Synechococcus sp. PCC7942. Mol. Microbiol. 102, 738–751. doi: 10.1111/mmi.13488
Keywords: Synechocystis sp. PCC 6803, PilR, pilA11, pili, cell motility
Citation: Hu J, Zhan J, Chen H, He C, Cang H and Wang Q (2018) The Small Regulatory Antisense RNA PilR Affects Pilus Formation and Cell Motility by Negatively Regulating pilA11 in Synechocystis sp. PCC 6803. Front. Microbiol. 9:786. doi: 10.3389/fmicb.2018.00786
Received: 18 February 2018; Accepted: 06 April 2018;
Published: 23 April 2018.
Edited by:
Weiwen Zhang, Tianjin University, ChinaReviewed by:
Jiangxin Wang, Shenzhen University, ChinaTakashi Osanai, Meiji University, Japan
Bao-Sheng Qiu, Central China Normal University, China
Copyright © 2018 Hu, Zhan, Chen, He, Cang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiang Wang, wangqiang@ihb.ac.cn
 Jinlu Hu
Jinlu Hu Jiao Zhan
Jiao Zhan Hui Chen
Hui Chen Chenliu He2
Chenliu He2 Qiang Wang
Qiang Wang