- 1Key Laboratory of Food Nutrition and Safety, Ministry of Education, Tianjin University of Science and Technology, Tianjin, China
- 2Beijing Advanced Innovation Center for Food Nutrition and Human Health, Beijing Technology and Business University, Beijing, China
Cronobacter sakazakii is an important foodborne pathogen that causes neonatal meningitis and sepsis, with high mortality in neonates. However, very little information is available regarding the pathogenesis of C. sakazakii at the genetic level. In our previous study, a cellulose biosynthesis-related gene (bcsR) was shown to be involved in C. sakazakii adhesion/invasion into epithelial cells. In this study, the detailed functions of this gene were investigated using a gene knockout technique. A bcsR knockout mutant (ΔbcsR) of C. sakazakii ATCC BAA-894 showed decreased adhesion/invasion (3.9-fold) in human epithelial cell line HCT-8. Biofilm formation by the mutant was reduced to 50% of that exhibited by the wild-type (WT) strain. Raman spectrometry was used to detect variations in biofilm components caused by bcsR knockout, and certain components, including carotenoids, fatty acids, and amides, were significantly reduced. However, another biofilm component, cellulose, was increased in ΔbcsR, suggesting that bcsR negatively affects cellulose biosynthesis. This result was also verified via RT-PCR, which demonstrated up-regulation of five crucial cellulose synthesis genes (bcsA, B, C, E, Q) in ΔbcsR. Furthermore, the expression of other virulence or biofilm-related genes, including flagellar assembly genes (fliA, C, D) and toxicity-related genes (ompA, ompX, hfq), was studied. The expression of fliC and ompA in the ΔbcsR mutant was found to be remarkably reduced compared with that in the wild-type and the others were also affected excepted ompX. In summary, bcsR is a negative regulator of cellulose biosynthesis but positively regulates biofilm formation and the adhesion/invasion ability of C. sakazakii.
Introduction
Cronobacter sakazakii is a Gram-negative, non-spore-forming, rod-shaped bacterium within the genus of Cronobacter spp., which is a group of emerging opportunistic pathogens associated with meningitis, septicemia, and necrotizing enterocolitis in neonates and infants. The mortality among infected infants is as high as 40–80% (Iversen and Forsythe, 2003). In addition, some patients who survive the disease suffer from mental or physical developmental delay and quadriplegia (Lai, 2001; Holy and Forsythe, 2014). Cronobacter spp. also infect elderly and immunocompromised adults. Unlike neonatal infection, Cronobacter is associated with bacteremia, osteomyelitis, splenic abscesses, pneumonia, conjunctivitis, wound infections, and urinary tract infections in adults (Blackwood and Hunter, 2016).
C. sakazakii has the capacity to adhere to human intestinal epithelial and brain microvascular endothelial cells (Quintero et al., 2011). The first step before colonization and infection for most pathogens is adherence to host cell surfaces. Bacterial variants that do not express adherence factors are unable to adhere and initiate infections; thus, adherence is important for pathogenicity (Cleary et al., 2004). Adherence to intestinal epithelial cells is a key step that determines whether an infection becomes refractory to treatment because C. sakazakii entries into intestinal epithelial cells is followed by entrance into the bloodstream, invasion of brain microvascular endothelial cells, and survival in the cerebrospinal fluid (Mange et al., 2006; Nair et al., 2009).
The ability of bacteria to attach to a surface and form biofilms facilitates the development of a continuous source of contamination and maintains disease incidence (Kives et al., 2006). A biofilm is generally defined as a structured community of bacterial cells that are adherent to a zoetic or abiotic surface and acts as an adhesive foundation and defense barrier (Aparna and Yadav, 2008). Cronobacter spp. are capable of attaching to different surfaces to form biofilms to resist multiple stress conditions, improve adherence, and increase pathogenesis (Lehner et al., 2005; Kim et al., 2006). Therefore, the investigation of biofilm characteristics, such as formation capacity and biochemical components, is helpful to better understand the mechanisms underlying C. sakazakii adherence, invasion of epithelial cells, and pathogenesis.
Some virulence factors involved in the interaction between C. sakazakii and host cells have been reported, including outer membrane protein A (OmpA) (Nair and Venkitanarayanan, 2007) and extracellular polysaccharides (EPS) (Iversen et al., 2004). However, very little information is available regarding C. sakazakii adhesion pathogenesis at the genetic level. In our previous work, we demonstrated the involvement of a cellulose production-related gene, bcsR, in interactions between C. sakazakii and intestinal epithelial cells using a random transposon insertion mutant library (Du et al., 2016). Cellulose (poly-β-(1 → 4)-D-glucose) is the most abundant biopolymer in nature. Most bacterial cellulose-producing genes encoding proteins involved in the UDP-glucose polymerization process are organized in a bacterial cellulose synthesis operon (bcs) (Wong et al., 1990; Recouvreux et al., 2008). The cellulose gene cluster, comprising 9 genes (bcsCZBAQEFG and bcsR), is present in nearly all Cronobacter strains (Ogrodzki and Forsythe, 2015). Among these 9 genes, bcsR is a short gene encoding a 62-amino-acid protein. Some studies have reported a role for bcsR in cellulose production in certain bacteria (Grimm et al., 2008; Serra et al., 2013). However, the role of this gene in the adhesion process is unclear.
In this study, we investigated the function of bcsR in C. sakazakii ATCC BAA-894 by constructing a gene mutant. The ability of the mutant to invade/adhere to HCT-8 cells was investigated and compared with that of the wild-type (WT) strain. Additionally, we assessed biofilm formation ability and performed Raman spectroscopy to analyze differences between the biochemical components in the WT and ΔbcsR mutant biofilms. This work helps to elucidate the role of bcsR in cellulose biosynthesis and C. sakazakii pathogenesis.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions
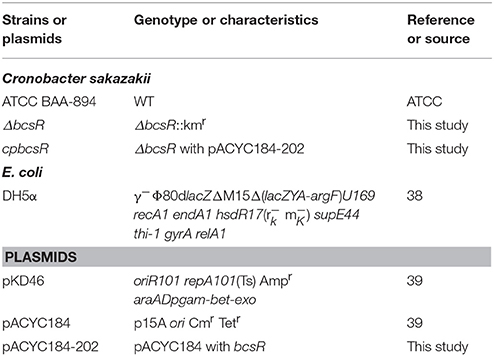
All bacterial strains and plasmids used in this study are listed in Table 1. C. sakazakii ATCC BAA-894 (WT and mutant strains) was routinely grown at 37°C in Tryptic Soy Broth (TSB; Difco, MD, USA) and on Tryptic Soy Agar (TSA; Difco, MD, USA) under aerobic conditions with constant shaking, unless indicated otherwise. When necessary, ampicillin, kanamycin, and chloramphenicol were used at 100, 100, and 10 μg/ml, respectively.
Construction of a bcsR Gene Deletion Mutant
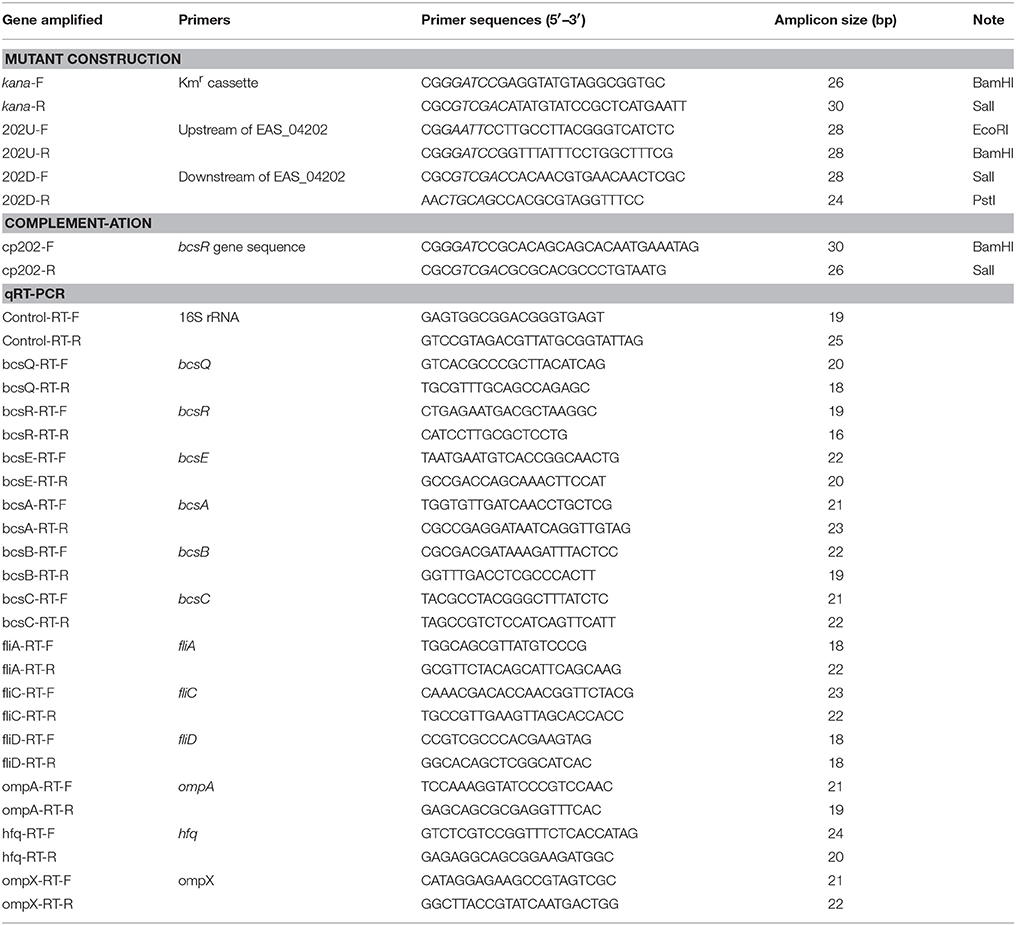
Site-specific mutation of C. sakazakii ATCC BAA-894 was performed using the Lambda-Red recombination method described by Geng et al. (2009). Briefly, the kanamycin resistance cassette from plasmid pET-26b was amplified using primers kana-F/kana-R. Two primer pairs, 202U-F/202U-R and 202D-F/202D-R, were used to amplify the upstream and downstream DNA sequences of the bcsR gene from the whole genome sequence of C. sakazakii ATCC BAA-894, obtained from GenBank (Grim et al., 2012). The amplified upstream and downstream DNA fragments of the bcsR gene and kanamycin resistance cassette were treated with appropriate restriction enzymes and subsequently cloned into the pMD18-T vector to produce pMD-202 (kana), respectively, which was then transferred into E. coli DH5α. The segment consisting of bcsR upstream-kana-bscR downstream was cut from the vector and transformed into the wild-type (WT) strain C. sakazakii ATCC BAA-894 (harboring the pKD46 plasmid) via electroporation, and the kanamycin-resistant transformant, i.e., the 202::kana mutant (ΔbcsR), was selected. All primer sequences are listed in Table 2.
Complementation Study
A complementation plasmid, containing the bcsR-coding sequence and its own promoter, was constructed. The bcsR gene was amplified by PCR using the primers 202 cp-F/202 cp-R (restriction sites were introduced into the primers) from C. sakazakii ATCC BAA-894 genomic DNA. The product was cloned into pACYC184 at appropriate restriction sites and then transferred into the mutant to generate ΔbcsR harboring pACYC184-202, cpbcsR. Nucleotide sequencing was performed to verify the sequence of the bcsR-coding region in the recombinant plasmid and complemented strain.
Growth Curves
C. sakazakii strain ATCC BAA-894 (WT) and the ΔbcsR and cpbcsR strains were cultured overnight in TSB medium at 37°C and subcultured as a 1% overnight culture in 100 ml of TSB medium. The cultures were incubated with shaking at 200 rpm per minute at 37°C for 14 h in a 100-ml conical flask. The optical density was measured at 600 nm (OD600) per hour.
Scanning Electron Microscopy (SEM)
SEM was performed to examine the morphological differences between the C. sakazakii ATCC BAA-894 (WT), ΔbcsR, and cpbcsR strains (Wang et al., 2011). Bacterial strains were grown to the logarithmic phase and collected via centrifugation, and the pellets were fixed with 2.5% glutaraldehyde overnight at 4°C. The cells were washed three times with distilled water (5 min), followed by dehydration with an ethanol series (25, 50, 70, 80, 90, and 100%). The cells were further dried via vacuum freeze-drying for 1.5–2.5 h. The dehydrated bacterial powder was observed with a Hitachi SU1510 scanning electron microscope using an accelerating voltage of 5 kV (Hitachi, Tokyo, Japan).
Adhesion/Invasion Assay
HCT-8 cells, derived from adenocarcinomas in a human colon and rectum (Tompkins et al., 1974; White et al., 1996), have been successfully used as an infection model to investigate pathogenesis of bacteria in previous studies (Luck et al., 2005; Pradel et al., 2015; Zargar et al., 2015). To evaluate bacterial invasion into HCT-8 cells (ATCC CCL-244; American Type Culture Collection, Manassas, Virginia), an invasion assay was conducted as described by Rogers et al. (2012), with modifications. Briefly, bacteria were incubated overnight aerobically at 37°C, transferred into fresh TSB medium as 1% inoculum and grown to 1 × 108 CFU at 37°C with constant shaking. C. sakazakii cells were harvested by centrifugation at 3,000 g for 5 min, washed and resuspended in RPMI-1640 (Gibco, Invitrogen, USA). HCT-8 cells were grown in RPMI-1640 supplemented with 10% fetal bovine serum (FBS) (Gibco, Invitrogen, USA) in six-well tissue culture plates. The C. sakazakii cells were applied to the HCT-8 cell monolayer at a multiplicity of infection (MOI) of 100. After incubation for 1, 2, 3, or 4 h in the presence of 5% CO2, the cells were washed three times with PBS and lysed in 1 ml of 0.1% Triton X-100 for 10 min. The bacteria were serially diluted in PBS and plated on TSA for quantification.
Analysis of Biofilm Formation Capacity
The experiment was performed as previously described (Du et al., 2012), with modifications. C. sakazakii was inoculated into 5 ml of TSB and incubated at 37°C with aeration until the cell density reached 107 CFU/ml. One hundred-microliter portions were loaded into a 96-well polystyrene plate and incubated at 37°C for 48 h without shaking. To fix the biofilms, 200 μl of 99% methanol was added for 15 min, the supernatants were removed, and the plates were air-dried. Subsequently, 200 μl of a 0.1% crystal violet (CV) solution was added. After 30 min, the excess CV was removed, and the plates were washed with normal saline (0.9% NaCl). Finally, the bound CV was released by adding 200 μl of 95% ethanol. The absorbance was measured at 570 nm using a Sunrise basic microplate reader (Tecan, Austria).
Raman Spectroscopy Analysis
Raman spectroscopy analyses were performed to examine the biochemical profiles of C. sakazakii biofilms according to methods described in a previous report (Du et al., 2016), with modifications. Briefly, 100 μl of each logarithmic phase bacterial culture was dropped onto a Memberance Filter (Millipore, Ireland), and the filter was placed on a TSA plate containing appropriate antibiotics. The plates were incubated at 37°C for 72 h, and the media were changed every 24 h. Spectroscopic analyses were performed using a Renishaw inVia Raman system (Renishaw, Gloucestershire, UK) equipped with a 785-nm near-infrared diode laser and a Leica microscope (Leica Biosystems, Wetzlar, Germany). Raman spectra were collected using a WITec alpha300 Raman microscope (WITec, Ulm, Germany) equipped with a UHTS-300 spectrometer (Lu et al., 2011a; Wang et al., 2015). Wavenumbers from 400 to 1,800 cm−1 were selected for Raman-based chemometric analyses. Peaks were assigned based on methods described in a previous study (Du et al., 2016). The area and height of each Raman band were calculated using MATLAB, and significant (P < 0.05) band variations were characterized and analyzed. Twenty spectra were collected for each strain in triplicate at minimum. Principal component analysis (PCA) was performed to segregate the biofilms obtained from the WT, mutant, and complementation C. sakazakii strains by creating a two- or three-dimensional image (Lu et al., 2012).
qRT-PCR
Quantitative RT-PCR assays were performed using SYBR® Premix Ex Taq II (Takara Bio Inc.), following the manufacturer's instructions, to investigate the transcriptional levels of various cellulose synthesis, biofilm-related, or virulence genes (Jing et al., 2016). Corresponding primers were designed based on the genome sequence of C. sakazakii ATCC BAA-894 (GenBank accession number CP000783.1; Table 2). Total bacterial RNA was isolated using an E.Z.N.A.™ Bacterial RNA Kit (Omega, USA). The 2−ΔΔCT value method was used to compare the expression of genes from different samples (Schmittgen and Livak, 2008). The 16S rRNA gene was used as an internal control for within-sample normalization of mRNA abundance (Choi et al., 2015). All real-time PCR reactions were performed using the Mastercycler ep gradient realplex system (Eppendorf, Germany). A reaction mixture lacking cDNA was used as the negative control.
Bacterial RNA was isolated from the WT strain (ATCC BAA-894), the 202 deletion mutant (ΔbcsR), or the complementation strain harboring the pACYC184-202 plasmid (cpbcsR). To obtain relative mRNA expression values on the y-axis, the mRNA level for each gene was divided by the mRNA level of the 16S rRNA-coding gene. The mRNA expression values were further normalized to the transcription levels exhibited by the WT strain. The means and standard deviations from three independent experiments are shown. Asterisks indicate significant differences (* P < 0.05).
Statistical Analysis
Statistical analyses were carried out using Origin 8.0 (OriginLab Co., Northampton, MA) and MATLAB (MathWorks, Natick, MA, USA). Each experiment was independently repeated a minimum of three times to ensure reproducibility. All results were analyzed through the Duncan test and analysis of variance (ANOVA), a collection of statistical models used to analyze the differences among group means and their associated procedures (Tjur, 2005). The data are represented as the mean and standard deviation, and statistical analyses were performed using SPSS 19.0 (SPSS Inc., Chicago, IL, USA). Differences were considered statistically significant at P < 0.05.
Results
Construction and Morphological Characteristics of the ΔbcsR Mutant in C. sakazakii ATCC BAA-894
The C. sakazakii gene (ESA_RS19400) homologous to the bcsR gene is located in a clockwise orientation in the genome of C. sakazakii ATCC BAA-894 and is also found in other Cronobacter spp. The gene (ESA_RS19400) exhibited high protein sequence similarity to the bcsR gene of Kosakonia arachidis (67%). However, the function of this gene is not yet known in C. sakazakii. To understand the roles of bcsR in pathogenesis of C. sakazakii ATCC BAA-894, we generated a mutant in which the entire bcsR gene was replaced by kanamycin resistance gene insertion using the λRed recombination technique. The ΔbcsR (kana) mutant was verified by PCR (Figure 1) and nucleotide sequencing. To further verify that the bcsR gene was knocked out, quantitative RT-PCR assays were performed, employing the bcsRY F/R primers and using WT, ΔbcsR or cpbcsR genomic DNA as the template. The results are shown in Figure 6. The expression of the bcsR gene was significantly reduced (26.57-fold change) in the ΔbcsR mutant when compared with that in the WT strain, which demonstrated that the bcsR gene was deleted.
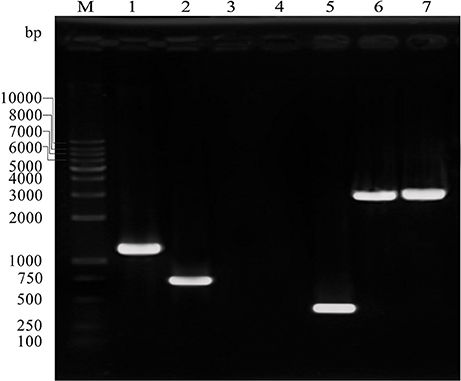
Figure 1. PCR verification of the ΔbcsR mutant strain using specific primers targeting sequences outside of homologous fragments and within the resistance gene. Sample 1, amplified with Kan F and KanR, using ΔbcsR as the template, 1,174 bp; sample 2, amplified with cp202-VF and cp202-VR, using cp202 as the template, 738 bp; sample 3, amplified with 202F and 202R, using ΔbcsR as the template, 0 bp; sample 4, amplified with KanF and KanR, using WT as the template, 0 bp; sample 5, amplified with 202F and 202R, using WT as the template, 275 bp; sample 6, amplified with 202-VF and Kan-VR, using ΔbcsR as the template, 2,989 bp; sample 7, amplified with Kan-VF and 202-VR, using ΔbcsR as the template, 3047 bp; M, DL10000 marker. Electrophoresis was performed using 1% agarose.
The deletion mutant showed a similar growth rate to that of the WT strain (Figure 2A). However, the complementation strain (cpbcsR) demonstrated a slightly lower growth rate than that of the WT in the early stages of growth. In addition, similar morphological characteristics were observed in SEM images of the WT, ΔbcsR, and cpbcsR strains (Figure 2B). Thus, bcsR gene knockout did not affect bacterial growth and morphology in TSB medium.
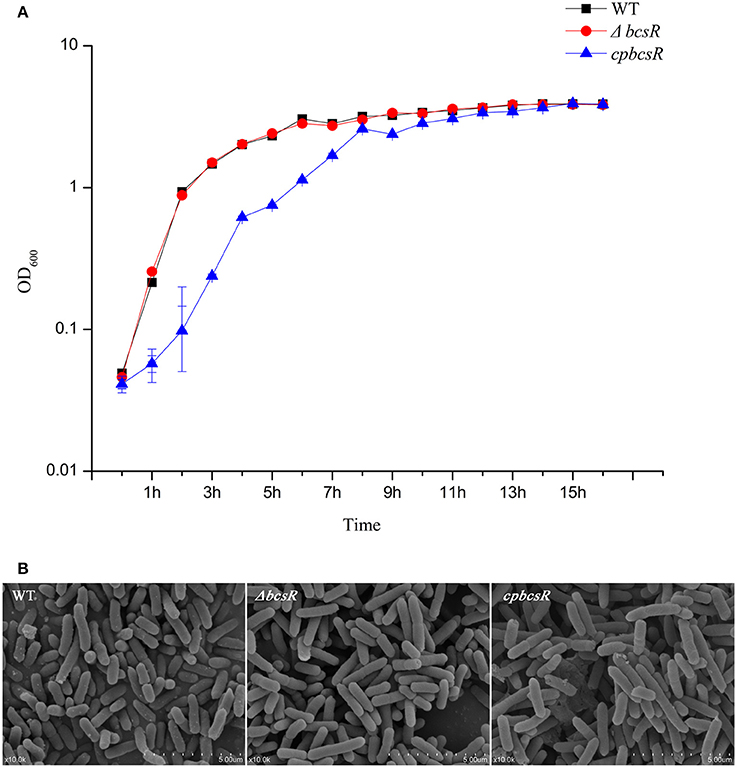
Figure 2. (A) Growth curves of C. sakazakii WT, ΔbcsR, and cpbcsR strains grown in the TSB broth at 37°C. The OD600 of each strain was measured hourly. Error bars represent the standard error of the mean (SEM) from three independent biological replicates. (B) SEM of biofilms formed on glass by WT and mutant strains. WT: WT ATCC BAA-894; ΔbcsR: mutant strain; cpbcsR: complementation strain.
bcsR Affects Adhesion/Invasion
In our previous work, we showed that the bcsR gene in C. sakazakii ATCC BAA-894 strain was involved in interactions with intestinal epithelial cells (Du et al., 2016). To explore the virulence-related functions of the bcsR gene in C. sakazakii ATCC BAA-894, an adhesion/invasion assay was performed at different time points (1, 2, 3, or 4 h) using HCT-8 cells (Figure 3). The invasion rate of the ΔbcsR mutant was significantly lower than that of the WT at 1, 2, 3, and 4 h in the invasion assay. Although the adhesion/invasion ability of the complementation strain did not reach that of the wild-type strain, it showed a significantly higher adhesion/invasion ability than the ΔbcsR mutant strain. The results suggested that the phenotypic defect of the ΔbcsR mutant was indeed attributable to the lack of bcsR, which led to a decreased adhesion/invasion ability in HTC-8 cells (Figure 3). Thus, we speculated that bcsR might positively regulate C. sakazakii adhesion/invasion in HCT-8 cells.
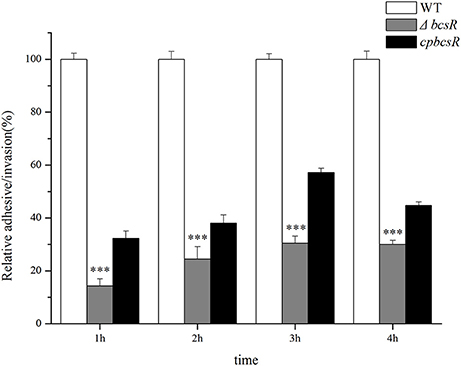
Figure 3. Absence of the bcsR gene impairs epithelial cell invasion at different times. Percent invasion for the mutant and complementation strains relative to the WT strain; ***p < 0.001.
Estimation of Biofilm Formation Ability
To investigate whether the bcsR gene contributes to biofilm formation of C. sakazakii ATCC BAA-894, a CV staining assay was carried out. The ΔbcsR mutant showed a significant decrease (1.5-fold change) in biofilm formation compared with that in the WT, and the complementation strain demonstrated similar biofilm formation to that in the WT strain (Figure 4). The results suggest the bcsR gene is a positive effector in biofilm formation.
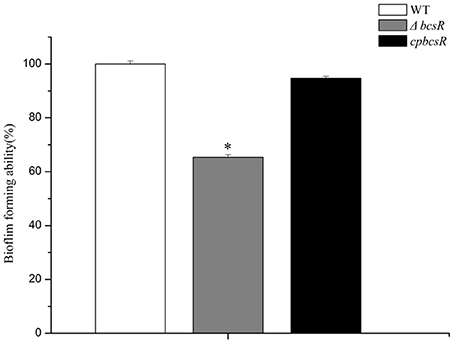
Figure 4. Biofilm formation by the WT, ΔbcsR, and cpbcsR strains. Percentages of biofilm formation for the mutant and complementation strains relative to the WT strain are shown. The error bars represent the mean standard deviation; *P < 0.05.
Differences in Biofilm Composition between the bcsR Mutant and WT Strains
To further investigate variations in the biochemical components of the biofilms formed by the tested strains, Raman spectroscopic analyses were performed. A PCA model was established to differentiate between the WT, mutant, and complementation strain. There were clear segregations between the WT, mutant, and complementation strains (Figure 5A), whose biochemical components exhibited variations.
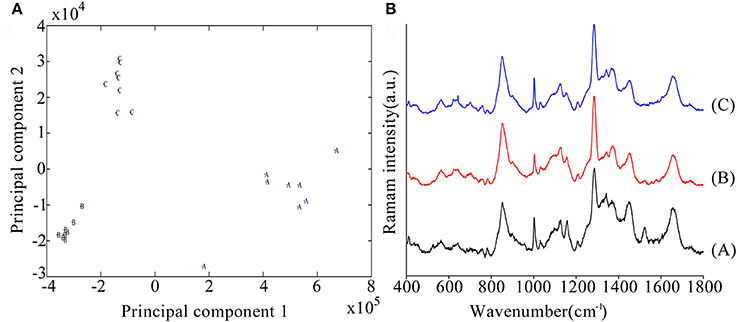
Figure 5. (A) Principal component analysis (PCA) model based on Raman spectral features. A: Wild-type strain, B: mutant strains, C: complementation strains. (B) Comparison of C. sakazakii biofilm spectral features between the WT (A), mutant (B), and complementation strains (C) using Raman spectroscopy.
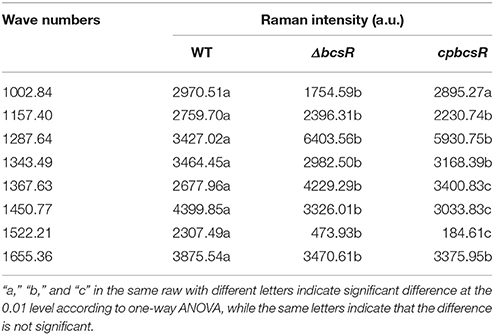
A comparison of Raman spectra indicated that two peaks were higher (1,287 and 1,367 cm−1) and six peaks were lower (1,002, 1,157, 1,343, 1,450, 1,522, and 1,655 cm−1) in the mutant than in the WT strain, and these differences were significant (Table 3). The 1,287 and 1,367 cm−1 peaks were assigned to cytosine and cellulose respectively (De Gelder et al., 2007). The 1,002 cm−1 peak reflected the characteristics of phenylalanine (Movasaghi et al., 2007). The 1,157 and 1,522 cm−1 peaks were assigned to carotenoids (Feng et al., 2014). The 1,450 cm−1 peak was assigned to fatty acids (De Gelder et al., 2007). And the 1,655 cm−1 peak was assigned to amide I and amide III (Lu et al., 2011b).
To clearly express the differences between the wild-type, mutant, and complementation strains at various points, we present the Raman intensities at each Raman wavenumber in Table 3 and indicate the differences between each of the strains. The Figure S1 shows the distribution of Raman peaks.
qRT-PCR Analysis
The expression of several genes (bcsR, bcsQ, bcsE, bcsA, bcsB, bcsC, fliA, fliC, fliD, ompA, hfq, and ompX) involved in cellulose synthesis, flagella, and toxicity, was detected in the bcsR mutant and WT strain by qRT-PCR.
The mRNA levels of the cellulose synthase operon genes bcsQ, bcsE, bcsA, bcsB, and bcsC increased by 2.68-, 1.41-, 3.91-, 3.27-, and 3.28-fold, respectively, in the ΔbcsR strain compared with those in the WT strain (Figure 6). The expression of fliA, fliC, and fliD, which are involved in regulating flagellar assembly, was reduced by 2.17-, 2.70-, and 1.79-fold, respectively, in the bcsR mutant compared with that in the WT strain (Figure 6). We also examined the expression levels of several toxicity-related genes, including ompA, ompX, and hfq, which have previously been associated with toxicity in C. sakazakii, in the bcsR mutant and WT strain. The mRNA levels of ompA and hfq genes in the ΔbcsR mutant decreased by 1.80- and 1.92-fold, respectively, compared with those in the WT strain (Figure 6). However, ompX transcription was not affected by the lack of bcsR. Thus, the bcsR gene down-regulated cellulosic synthesis in C. sakazakii ATCC BAA-894, in agreement with the Raman spectra data.
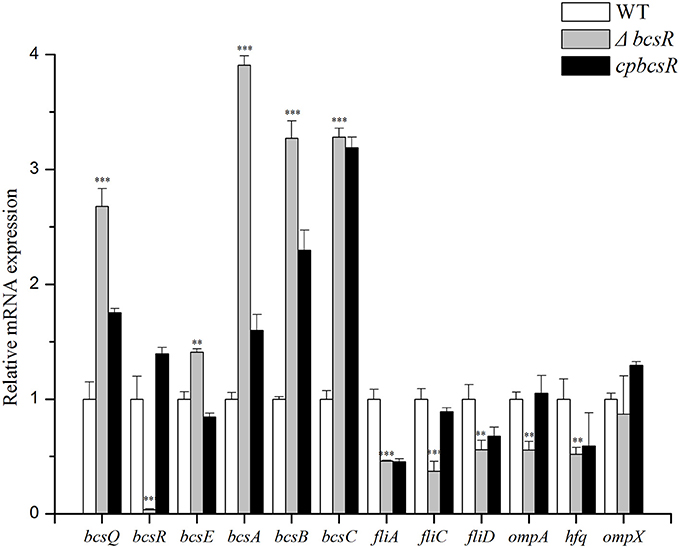
Figure 6. Transcription levels of 9 genes in C. sakazakii were determined by qRT-PCR. Experiments were repeated three times. The error bars represent the mean standard deviation; *P < 0.05, **P < 0.01, and ***P < 0.001.
Discussion
Bacterial pathogens must bind to epithelial cell surfaces prior to successful invasion. Therefore, adhesion and invasion into host tissue cells play essential roles in the virulence of most pathogenic bacteria. In our previous study, a cellulose biosynthesis-related gene was shown to be involved in C. sakazakii adhesion to epithelial cells. The bcsR gene was observed downstream of the bcs operon, which encodes enzymes or other proteins responsible for cellulose biosynthesis. However, the function of this gene in cellulose biosynthesis is unknown (Grimm et al., 2008). In this study, our data indicated that bcsR is a positive regulator of C. sakazakii adhesion/invasion into HCT-8 cells and biofilm formation.
Bacterial cells typically adhere to and interact with surfaces before eventually forming biofilms. Biofilms are defined as sessile communities embedded in polymeric substances produced by bacteria and are primarily composed of polysaccharides, proteins, and nucleic acids (Kim et al., 2006; Kolter and Greenberg, 2006; Serra et al., 2013). The ability to form a biofilm is a crucial trait that enhances bacterial resistance to environmental stresses and provides protection against drugs and disinfectants (Lehner et al., 2005; Furukawa et al., 2006). Acting as adhesive foundations and defense barriers, biofilms also play an important role in protecting embedded cells against detachment caused by flow shear. Potential bacterial toxicity may be enhanced when cells are present in biofilms, which are heterogeneously sessile bacterial communities that adhere to each other and to solid surfaces (Wang et al., 2012). Kunyanee et al. (2016) reported the important role of Burkholderia pseudomallei biofilms in bacterial pathogenesis in human epithelial cells with respect to initial attachment and invasion. In the ΔbcsR mutant of C. sakazakii, the decrease of biofilm formation suggests that this gene may affect adhesion/invasion by regulating biofilm synthesis.
According to the Raman spectra results, bands representing carotenoids, fatty acids and amides in the mutant exhibited significant decreases compared with those in the WT strain, indicating that bcsR may be positively associated with carotenoids, fatty acids and amides. The biofilms of certain Cronobacter strains contain high levels of carotenoids (Du et al., 2012). Therefore, bcsR may be positively associated with biofilm formation through the up-regulation of carotenoid, fatty acid, and amide biosynthesis in C. sakazakii.
The Raman spectra results also indicated that the band representing cellulose was greatly increased in the mutant compared with that in the WT strain, suggesting that bcsR may be a negative regulator of cellulose biosynthesis. Cellulose is an extracellular matrix component present in C. sakazakii biofilms (Grimm et al., 2008). Bacterial cellulose synthase is a multicomponent protein complex encoded in an operon containing at least three genes: bcsA, bcsB, and bcsC (Keiski et al., 2010). bcsA, bcsB, and bcsC encode for the cellulose synthase catalytic subunit, a cyclic-di-GMP binding protein and the cellulose oxidoreductase enzyme, respectively (Hu et al., 2015). The bcsABC genes in Cronobacter are necessary for cellulose production and are involved in biofilm formation and cell-cell aggregation (Hu et al., 2015). However, as a cellulose synthase operon gene, the function of bcsR remains even more elusive (Grimm et al., 2008). In this study, the decrease of biochemical components (including carotenoids, fatty acids, and amides) potentially explains the decline in biofilm formation after bcsR gene deletion in C. sakazakii. Interestingly, cellulose synthesis (Figure 5B) and cellulose synthase operon gene (bcsABC) expression levels significantly increased in the bcsR mutant (Figure 6). We hypothesize that bcsR affects the synthesis of other biofilm components during increased cellulose synthesis. In Salmonella enteritidis, the synthesis of colanic acid, lipopolysaccharide, enterobacterial common antigen, and cellulose are affected by the cellulose operon; these elements are highly important for biofilm formation (Solano et al., 2002). Cellulose is composed of β-D-1,4-glucan chains, and the precursor molecule is uridine diphosphate glucose (UDP-glucose) (Ross et al., 1991), which is also the substrate for the synthesis of colanic acid, lipopolysaccharide, and the enterobacterial common antigen. Excessive cellulose synthesis affects the synthesis of the other three components and thus affects biofilm synthesis (Solano et al., 2002). Channeling copious amounts of UDP-glucose to cellulose biosynthesis leads to the reprogramming of cellular metabolism to favor gluconeogenesis, which is a metabolic pathway that results in the generation of glucose from certain non-carbohydrate carbon substrates (Romling and Galperin, 2015). Furthermore, cellulose synthesis is energy-consuming and is affected by cellulose synthase activity. Cellulose synthase is specifically activated by the unique nucleotide cyclic diguanylic acid, which is synthesized from GTP by diguanylate cyclase. The synthesis of other biofilm components is also energy-consuming (Mika and Hengge, 2013). Therefore, excessive energy consumption for cellulose synthesis may not be conducive to the formation of other biofilm components. Consistent with our results, we speculate that excessive cellulose synthesis affects the synthesis of other components, thereby affecting biofilm formation.
Based on the qRT-PCR results, the expression of flagella (FliA, FliC, and FliD) and outer membrane proteins was reduced in the bcsR mutant. Therefore, we speculate that bcsR may be a global regulator of biofilm, flagella, and outer membrane protein biosynthesis, similar to CodY, which is a global transcriptional regulator that represses toxin gene expression by binding with high affinity to the tcdR promoter region (Girinathan et al., 2017). The fliA, fliC, and fliD genes regulate flagellar assembly (Chevance and Hughes, 2008). The fliA gene encodes an alternative sigma factor that regulates the transcription of class III flagellar genes, including filament structure genes and genes in the chemosensory pathway (Ohnishi et al., 1990; Chevance and Hughes, 2008; Choi et al., 2015). Because the fliC gene is a class III flagellar gene, and fliD encodes a late-secretion substrate of the filament-capping protein, free σ28 (a flagellar-specific sigma factor), which transcribes class III promoters, the enhanced expression of fliC and fliD may be attributable to large amounts of FliA, suggesting the bcsR gene affects the expression of class III flagellar genes via fliA (Chevance and Hughes, 2008). The three flagellar genes are reportedly involved in biofilm formation by C. sakazakii (Barken et al., 2008; Hartmann et al., 2010; Ye et al., 2016), consistent with our findings. Kim et al. (2010) reported important roles for OmpA and OmpX of Cronobacter in the invasion of Caco-2 and INT-407 cells. Hfq, oligomerized into a hexameric ring structure, is a posttranscriptional global regulator involved in the biosynthesis of OMPs, quorum sensing, stress responses, or metabolism and adhesin-mediating interactions with host tissue (Kakoschke et al., 2016). In our qTR-PCR analysis, the genes encoding OmpA, OmpX, and Hfq were all down-regulated in the mutant, suggesting their positive correlation with bacterial adhesion/invasion into epithelial cells. Based on the findings in this study, bcsR plays significant roles in biofilm formation and adhesion/invasion by regulating flagellar and toxicity-related genes.
Interestingly, in bcsR complementation strains, certain functions were restored, but bcsR expression levels only barely reached that of the wild strain. To explore whether this result was due to the gene polarity effects, we investigated the expression of genes flanking bscR in the WT, mutant, and complementation strains through qRT-PCR analysis (Figure 6). The expression of bcsQ and bcsE was investigated. bcsQ and bcsE, are the cellulose synthase operon genes that are closest to bcsR in the downstream region and upstream region, respectively. In the complemented strain, although the expression of the flanking genes was not recovered to wild-type levels, no significant difference was observed in the wild-type, mutant, and complemented strains (fold change <2). The lack of complete recovery of flanking gene expression may have been caused by the differences in the efficiency of bcsR gene expression in the complemented strain and the wild-type strain, considering that the bcsR gene was located in the pACYC184 plasmid in the complemented strain. This phenomenon was previously reported in studies performed by Kim et al. (2015) and Schilling and Gerischer (2009). Additionally, Kim et al. (2015) reported that failure of the pHFQ plasmid (a pACYC184 derivative with its own hfq promoter) to complement the hypermotility of the hfq strain might be caused by an imbalance of Hfq production from the low-copy-number pACYC184 plasmid. Therefore, we cannot completely exclude that the deletion of the bcsR gene did not affect the expression of other genes (fliA, fliC, fliD, ompA, and hfq) via polar effect. However, we are inclined to conclude that bcsR was a negative regulator for cellulose biosynthesis and we confirmed that cellulose biosynthesis was negatively related to biofilm formation and the epithelial cells adhesion/invasion capability of C. sakazakii.
Conclusion
In this study, a gene knockout technique was employed to demonstrate the positive role of the bcsR gene in C. sakazakii adhesion/invasion in epithelial cells and biofilm formation. However, bcsR was found to act as a negative regulator of cellulose biosynthesis. Raman spectrometry and qRT-PCR results verified positive regulation of pathogenesis and negative regulation of cellulose biosynthesis by bcsR in C. sakazakii. This study significantly contributes to our understanding of the detailed functions of the bcsR gene in bacteria.
Author Contributions
JG contributed significantly to the conceived, designed, and performed the experiments, and the writing and editing of paper. PL, XD, and SW contributed significantly to conceived and designed of the work. ZH, RX, and BL mainly made a great contribution in performing the experiments. They all involved in the process of experimental design and the writing of paper. All authors agree to be accountable for the content of the work contributed to the conception of the study.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by the Tianjin Research Program of the Application Foundation and Advanced Technology (Project No. 15JCQNJC44600) and the Ministry of Science and Technology of the People's Republic of China (Project No. 2014BAD04B03).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01839/full#supplementary-material
Figure S1. Raman peak distribution of C. sakazakii biofilm.
References
Aparna, M. S., and Yadav, S. (2008). Biofilms: microbes and disease. Braz. J. Infect. Dis. 12, 526–530. doi: 10.1590/S1413-86702008000600016
Barken, K. B., Pamp, S. J., Yang, L., Gjermansen, M., Bertrand, J. J., Klausen, M., et al. (2008). Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 10, 2331–2343. doi: 10.1111/j.1462-2920.2008.01658.x
Blackwood, B. P., and Hunter, C. J. (2016). Cronobacter spp. Microbiol. Spectr. 4:EI10-0002-2015. doi: 10.1128/microbiolspec.EI10-0002-2015
Chevance, F. F. V., and Hughes, K. T. (2008). Coordinating assembly of a bacterial macromolecular machine. Nat. Rev. Microbiol. 6, 455–465. doi: 10.1038/nrmicro1887
Choi, Y., Kim, S., Hwang, H., Kim, K. P., Kang, D. H., and Ryu, S. (2015). Plasmid-encoded MCP is involved in virulence, motility, and biofilm formation of Cronobacter sakazakii ATCC 29544. Infect. Immun. 83, 197–204. doi: 10.1128/IAI.02633-14
Cleary, J., Lai, L. C., Shaw, R. K., Straatman-Iwanowska, A., Donnenberg, M. S., Frankel, G., et al. (2004). Enteropathogenic Escherichia coli (EPEC) adhesion to intestinal epithelial cells: role of bundle-forming pili (BFP), EspA filaments and intimin. Microbiology 150, 527–538. doi: 10.1099/mic.0.26740-0
De Gelder, J., De Gussem, K., Vandenabeele, P., and Moens, L. (2007). Reference database of Raman spectra of biological molecules. J. Raman Spectrosc. 38, 1133–1147. doi: 10.1002/jrs.1734
Du, X. J., Wang, F., Lu, X., Rasco, B. A., and Wang, S. (2012). Biochemical and genetic characteristics of Cronobacter sakazakii biofilm formation. Res. Microbiol. 163, 448–456. doi: 10.1016/j.resmic.2012.06.002
Du, X. J., Zhang, X., Li, P., Xue, R., and Wang, S. (2016). Screening of genes involved in interactions with intestinal epithelial cells in Cronobacter sakazakii. AMB Express 6:74. doi: 10.1186/s13568-016-0246-4
Feng, S., Eucker, T. P., Holly, M. K., Konkel, M. E., Lu, X., and Wang, S. (2014). Investigating the responses of Cronobacter sakazakii to garlic-drived organosulfur compounds: a systematic study of pathogenic-bacterium injury by use of high-throughput whole-transcriptome sequencing and confocal micro-raman spectroscopy. Appl. Environ. Microb. 80, 959–971. doi: 10.1128/AEM.03460-13
Furukawa, S., Kuchma, S. L., and O'Toole, G. A. (2006). Keeping their options open: acute versus persistent infections. J. Bacteriol. 188, 1211–1217. doi: 10.1128/JB.188.4.1211-1217.2006
Geng, S. Z., Jiao, X. A., Pan, Z. M., Chen, X. J., Zhang, X. M., and Chen, X. (2009). An improved method to knock out the asd gene of Salmonella enterica serovar Pullorum. J. Biomed. Biotechnol. 2009:646380. doi: 10.1155/2009/646380
Girinathan, B. P., Monot, M., Boyle, D., McAllister, K. N., Sorg, J. A., Dupuy, B., et al. (2017). Effect of tcdR mutation on sporulation in the epidemic clostridium difficile strain R20291. mSphere 2:e00383–16. doi: 10.1128/mSphere.00383-16
Grim, C. J., Kothary, M. H., Gopinath, G., Jarvis, K. G., Beaubrun, J. J., McClelland, M., et al. (2012). Identification and characterization of Cronobacter iron acquisition systems. Appl. Environ. Microb. 78, 6035–6050. doi: 10.1128/AEM.01457-12
Grimm, M., Stephan, R., Iversen, C., Manzardo, G. G. G., Rattei, T., Riedel, K., et al. (2008). Cellulose as an extracellular matrix component present in Enterobacter sakazakii biofilms. J. Food Prot. 71, 13–18. doi: 10.4315/0362-028X-71.1.13
Hartmann, I., Carranza, P., Lehner, A., Stephan, R., Eberl, L., and Riedel, K. (2010). Genes involved in Cronobacter sakazakii biofilm formation. Appl. Environ. Microb. 76, 2251–2261. doi: 10.1128/AEM.00930-09
Holy, O., and Forsythe, S. (2014). Cronobacter spp. as emerging causes of healthcare-associated infection. J. Hosp. Infect. 86, 169–177. doi: 10.1016/j.jhin.2013.09.011
Hu, L., Grim, C. J., Franco, A. A., Jarvis, K. G., Sathyamoorthy, V., Kothary, M. H., et al. (2015). Analysis of the cellulose synthase operon genes, bcsA, bcsB, and bcsC in Cronobacter species: prevalence among species and their roles in biofilm formation and cell-cell aggregation. Food Microbiol. 52, 97–105. doi: 10.1016/j.fm.2015.07.007
Iversen, C., and Forsythe, S. (2003). Risk profile of Enterobacter sakazakii, an emergent pathogen associated with infant milk formula. Trends Food Sci. Tech. 14, 443–454. doi: 10.1016/S0924-2244(03)00155-9
Iversen, C., Lane, M., and Forsythe, S. J. (2004). The growth profile, thermotolerance and biofilm formation of Enterobacter sakazakii grown in infant formula milk. Lett. Appl. Microbiol. 38, 378–382. doi: 10.1111/j.1472-765X.2004.01507.x
Jing, C. E., Du, X. J., Li, P., and Wang, S. (2016). Transcriptome analysis of Cronobacter sakazakii ATCC BAA-894 after interaction with human intestinal epithelial cell line HCT-8. Appl. Microbiol. Biot. 100, 311–322. doi: 10.1007/s00253-015-7053-8
Kakoschke, T. K., Kakoschke, S. C., Zeuzem, C., Bouabe, H., Adler, K., Heesemann, J., et al. (2016). The RNA chaperone Hfq is essential for virulence and modulates the expression of four adhesins in Yersinia enterocolitica. Sci. Rep. 6:29275. doi: 10.1038/srep29275
Keiski, C. L., Harwich, M., Jain, S., Neculai, A. M., Yip, P., Robinson, H., et al. (2010). AlgK is a TPR-Containing protein and the periplasmic component of a novel exopolysaccharide secretin. Structure 18, 265–273. doi: 10.1016/j.str.2009.11.015
Kim, H., Ryu, J. H., and Beuchat, L. R. (2006). Attachment of and biofilm formation by Enterobacter sakazakii on stainless steel and enteral feeding tubes. Appl. Environ. Microb. 72, 5846–5856. doi: 10.1128/AEM.00654-06
Kim, K., Kim, K. P., Choi, J., Lim, J. A., Lee, J., Hwang, S., et al. (2010). Outer membrane proteins A (OmpA) and X (OmpX) are essential for basolateral invasion of Cronobacter sakazakii. Appl. Environ. Microbiol. 76, 5188–5198. doi: 10.1128/AEM.02498-09
Kim, S., Hwang, H., Kim, K. P., Yoon, H., Kang, D. H., and Ryu, S. (2015). hfq plays important roles in virulence and stress adaptation in Cronobacter sakazakii ATCC 29544. Infect. Immun. 83, 2089–2098. doi: 10.1128/IAI.03161-14
Kives, J., Orgaz, B., and Sanjose, C. (2006). Polysaccharide differences between planktonic and biofilm-associated EPS from Pseudomonas fluorescens B52. Colloids Surf. B Biointerfaces 52, 123–127. doi: 10.1016/j.colsurfb.2006.04.018
Kolter, R., and Greenberg, E. P. (2006). Microbial sciences - The superficial life of microbes. Nature 441, 300–302. doi: 10.1038/441300a
Kunyanee, C., Kamjumphol, W., Taweechaisupapong, S., Kanthawong, S., Wongwajana, S., Wongratanacheewin, S., et al. (2016). Burkholderia pseudomallei biofilm promotes adhesion, internalization and stimulates proinflammatory cytokines in human epithelial A549 cells. PLoS ONE 11:e00025. doi: 10.1371/journal.pone.0160741
Lai, K. K. (2001). Enterobacter sakazakii infections among neonates, infants, children, and adults - case reports and a review of the literature. Medicine 80, 113–122. doi: 10.1097/00005792-200103000-00004
Lehner, A., Riedel, K., Eberl, L., Breeuwer, P., Diep, B., and Stephan, R. (2005). Biofilm formation, extracellular polysaccharide production, and cell-to-cell signaling in various Enterobacter sakazakii strains: aspects promoting environmental persistence. J. Food Protect. 68, 2287–2294. doi: 10.4315/0362-028X-68.11.2287
Lu, X. N., Al-Qadiri, H. M., Lin, M. S., and Rasco, B. A. (2011a). Application of mid-infrared and Raman spectroscopy to the study of bacteria. Food Bioprocess Tech. 4, 919–935. doi: 10.1007/s11947-011-0516-8
Lu, X. N., Rasco, B. A., Jabal, J. M. F., Aston, D. E., Lin, M. S., and Konkel, M. E. (2011b). Investigating antibacterial effects of garlic (Allium sativum) concentrate and garlic-derived organosulfur compounds on Campylobacter jejuni by using Fourier transform infrared spectroscopy, Raman spectroscopy, and electron microscopy. Appl. Environ. Microb. 77, 5257–5269. doi: 10.1128/AEM.02845-10
Lu, X., Samuelson, D. R., Rasco, B. A., and Konkel, M. E. (2012). Antimicrobial effect of diallyl sulphide on Campylobacter jejuni biofilms. J. Antimicrob. Chemother. 67, 1915–1926. doi: 10.1093/jac/dks138
Luck, S. N., Bennett-Wood, V., Poon, R., Robins-Browne, R. M., and Hartland, E. L. (2005). Invasion of epithelial cells by locus of enterocyte effacement-negative enterohemorrhagic Escherichia coli. Infect. Immun. 73, 3063–3071. doi: 10.1128/IAI.73.5.3063-3071.2005
Mange, J. P., Stephan, R., Borel, N., Wild, P., Kim, K. S., Pospischil, A., et al. (2006). Adhesive properties of Enterobacter sakazakii to human epithelial and brain microvascular endothelial cells. BMC Microbiol. 6:58. doi: 10.1186/1471-2180-6-58
Mika, F., and Hengge, R. (2013). Small regulatory RNAs in the control of motility and biofilm formation in E. coli and Salmonella. Int. J. Mol. Sci. 14, 4560–4579. doi: 10.3390/ijms14034560
Movasaghi, Z., Rehman, S., and Rehman, I. U. (2007). Raman spectroscopy of biological tissues. Appl. Spectrosc. Rev. 42, 493–541. doi: 10.1080/05704920701551530
Nair, M. K. M., and Venkitanarayanan, K. (2007). Role of bacterial OmpA and host cytoskeleton in the invasion of human intestinal epithelial cells by Enterobacter sakazakii. Pediatr. Res. 62, 664–669. doi: 10.1203/PDR.0b013e3181587864
Nair, M. K., Venkitanarayanan, K., Silbart, L. K., and Kim, K. S. (2009). Outer membrane protein A (OmpA) of Cronobacter sakazakii binds fibronectin and contributes to invasion of human brain microvascular endothelial cells. Foodborne Pathog. Dis. 6, 495–501. doi: 10.1089/fpd.2008.0228
Ogrodzki, P., and Forsythe, S. (2015). Capsular profiling of the Cronobacter genus and the association of specific Cronobacter sakazakii and C. malonaticus capsule types with neonatal meningitis and necrotizing enterocolitis. BMC Genomics 16:15. doi: 10.1186/s12864-015-1960-z
Ohnishi, K., Kutsukake, K., Suzuki, H., and Iino, T. (1990). Gene fliA encodes an alternative sigma factor specific for flagellar operons in Salmonella typhimurium. Mol. Gen. Genet. 221, 139–147. doi: 10.1007/BF00261713
Pradel, N., Etienne-Mesmin, L., Thévenot, J., Cordonnier, C., Blanquet-Diot, S., and Livrelli, V. (2015). In vitro adhesion properties of shiga toxinproducing Escherichia coli isolated from cattle, food, and humans. Front. Microbiol. 6:156. doi: 10.3389/fmicb.2015.00156
Quintero, M., Maldonado, M., Perez-Munoz, M., Jimenez, R., Fangman, T., Rupnow, J., et al. (2011). Adherence inhibition of Cronobacter sakazakii to intestinal epithelial cells by prebiotic oligosaccharides. Curr. Microbiol. 62, 1448–1454. doi: 10.1007/s00284-011-9882-8
Recouvreux, D. O. S., Carminatti, C. A., Pitlovanciv, A. K., Rambo, C. R., Porto, L. M., and Antonio, R. V. (2008). Cellulose biosynthesis by the beta-proteobacterium, chromobacterium violaceum. Curr. Microbiol. 57, 469–476. doi: 10.1007/s00284-008-9271-0
Rogers, T. J., Thorpe, C. M., Paton, A. W., and Paton, J. C. (2012). Role of lipid rafts and flagellin in invasion of colonic epithelial cells by shiga-toxigenic Escherichia coli O113:H21. Infect. Immun. 80, 2858–2867. doi: 10.1128/IAI.00336-12
Romling, U., and Galperin, M. Y. (2015). Bacterial cellulose biosynthesis: diversity of operons, subunits, products, and functions. Trends Microbiol. 23, 545–557. doi: 10.1016/j.tim.2015.05.005
Ross, P., Mayer, R., and Benziman, M. (1991). Cellulose biosynthesis and function in bacteria. Microbiol. Rev. 55, 35–58.
Schilling, D., and Gerischer, U. (2009). The Acinetobacter baylyi hfq gene encodes a large protein with an unusual C terminus. J. Bacteriol. 191, 5553–5562. doi: 10.1128/JB.00490-09
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C-T method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Serra, D. O., Richter, A. M., and Hengge, R. (2013). Cellulose as an architectural element in spatially structured Escherichia coli biofilms. J. Bacteriol. 195, 5540–5554. doi: 10.1128/JB.00946-13
Solano, C., Garcia, B., Valle, J., Berasain, C., Ghigo, J. M., Gamazo, C., et al. (2002). Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol. Microbiol. 43, 793–808. doi: 10.1046/j.1365-2958.2002.02802.x
Tjur, T. (2005). Discussion of “Analysis of variance-why it is more important than ever” by A. Gelman. Mathematics 33, 1–53. doi: 10.1214/009053604000001048
Tompkins, W., Watrach, A., Schmale, J., Schultz, R., and Harris, J. (1974). Cultural and antigenic properties of newly established cell strains derived from adenocarcinomas of the human colon and rectum. J. Natl. Cancer Inst. 52:1101. doi: 10.1093/jnci/52.4.1101
Wang, J. P., Xie, X. F., Feng, J. S., Chen, J. C., Du, X. J., Luo, J. Z., et al. (2015). Rapid detection of Listeria monocytogenes in milk using confocal micro-Raman spectroscopy and chemometric analysis. Int. J. Food Microbiol. 204, 66–74. doi: 10.1016/j.ijfoodmicro.2015.03.021
Wang, L., Hu, X., Tao, G., and Wang, X. (2012). Outer membrane defect and stronger biofilm formation caused by inactivation of a gene encoding for heptosyltransferase I in Cronobacter sakazakii ATCC BAA-894. J. Appl. Microbiol. 112, 985–997. doi: 10.1111/j.1365-2672.2012.05263.x
Wang, Y., Zhang, W., Wu, Z., Zhu, X., and Lu, C. (2011). Functional analysis of luxS in Streptococcus suis reveals a key role in biofilm formation and virulence. Vet. Microbiol. 152, 151–160. doi: 10.1016/j.vetmic.2011.04.029
White, L. J., Ball, J. M., Hardy, M. E., Tanaka, T. N., Kitamoto, N., and Estes, M. K. (1996). Attachment and entry of recombinant Norwalk virus capsids to cultured human and animal cell lines. J. Virol. 70, 6589–6597.
Wong, H. C., Fear, A. L., Calhoon, R. D., Eichinger, G. H., Mayer, R., Amikam, D., et al. (1990). Genetic organization of the cellulose synthase operon in Acetobacter xylinum. Proc. Natl. Acad. Sci. U.S.A. 87, 8130–8134. doi: 10.1073/pnas.87.20.8130
Ye, Y., Jiao, R., Gao, J., Li, H., Ling, N., Wu, Q., et al. (2016). Proteins involved in responses to biofilm and planktonic modes in Cronobacter sakazakii. Lwt Food Sci. Technol. 65, 1093–1099. doi: 10.1016/j.lwt.2015.09.039
Keywords: bcsR, Cronobacter sakazakii, cellulose biosynthesis, biofilm formation, adhesion/invasion
Citation: Gao J-x, Li P, Du X-j, Han Z-h, Xue R, Liang B and Wang S (2017) A Negative Regulator of Cellulose Biosynthesis, bcsR, Affects Biofilm Formation, and Adhesion/Invasion Ability of Cronobacter sakazakii. Front. Microbiol. 8:1839. doi: 10.3389/fmicb.2017.01839
Received: 10 April 2017; Accepted: 08 September 2017;
Published: 26 September 2017.
Edited by:
Lanming Chen, Shanghai Ocean University, ChinaReviewed by:
Gergely Maroti, Biological Research Center of the Hungarian Academy of Sciences, HungaryIssmat Kassem, American University of Beirut, Lebanon
Rong Liu, Nanjing Agricultural University, China
Copyright © 2017 Gao, Li, Du, Han, Xue, Liang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuo Wang, s.wang@tust.edu.cn
 Jian-xin Gao
Jian-xin Gao Ping Li1
Ping Li1