Corrigendum: Dicer-Like Genes Are Required for H2O2 and KCl Stress Responses, Pathogenicity and Small RNA Generation in Valsa mali
- College of Plant Protection and State Key Laboratory of Crop Stress Biology for Arid Areas, Northwest A&F University, Yangling, China
Valsa mali (V. mali) is the causative agent of apple tree Valsa canker, which heavily damages the production of apples in China. However, the biological roles of the RNA interfering (RNAi) pathway in the pathogenicity of V. mali remain unknown. Dicer-like proteins (DCLs) are important components that control the initiation of the RNAi pathway. In this study, VmDCL1 and VmDCL2 were isolated and functionally characterized in V. mali. VmDCL1 and VmDCL2 are orthologous in evolution to the DCLs in Cryphonectria parasitica. The deletion of VmDCL1 and VmDCL2 did not affect vegetative growth when the mutants (ΔVmDCL1, ΔVmDCL2 and ΔVmDCL1DCL2) and wild type strain 03–8 were grown on a PDA medium at 25°C in the dark. However, the colony of ΔVmDCL1 increased by 37.1% compared to the 03–8 colony in a medium containing 0.05% H2O2 3 days after inoculation, and the growth of ΔVmDCL1 was significantly inhibited in a medium containing 0.5 M KCl at a ratio of 25.7%. Meanwhile, in the presence of 0.05% H2O2, the growth of ΔVmDCL2 decreased by 34.5% compared with the growth of 03–8, but ΔVmDCL2 grew normally in the presence of 0.5 M KCl. More importantly, the expression of VmDCL2 was up-regulated 125-fold during the pathogen infection. In the infection assays using apple twigs, the pathogenicity of ΔVmDCL2 and ΔVmDCL1DCL2 was significantly reduced compared with that of 03–8 at a ratio of 24.7 and 41.3%, respectively. All defective phenotypes could be nearly rescued by re-introducing the wild type VmDCL1 and VmDCL2 alleles. Furthermore, the number and length distribution of unique small RNAs (unisRNAs) in the mutants and 03–8 were analyzed using deep sequencing. The number of unisRNAs was obviously lower in ΔVmDCL1, ΔVmDCL2 and ΔVmDCL1DCL2 than that in 03–8, and the length distribution of the sRNAs also markedly changed after the VmDCLs were deleted. These results indicated that VmDCLs function in the H2O2 and KCl stress response, pathogenicity and generation of sRNAs.
Introduction
RNA interference (RNAi) is a gene silencing mechanism at the post-transcriptional level. It is triggered by the presence of double-stranded RNA molecules that are homologous to the corresponding target gene, thereby resulting in the degradation of the messenger RNA and attenuation of the target gene expression (Pratt and MacRae, 2009). This mechanism was first observed in petunia plants; in an attempt to over-express the enzyme chalcone synthase, the plants showed a purple pigment (Napoli et al., 1990). This process was called post-transcriptional gene silencing. The first study to report RNA silencing in fungi was performed in Neurospora crassa, and alterations of albino gene resulted in an albino phenotype (Romano and Macino, 1992). This phenomenon was termed “quelling.” Subsequently, Fire et al. (1998) found RNA mediated genetic interference in Caenorhabditis elegans. This phenomenon was named RNAi. However, RNAi in animals is equivalent to post-transcriptional gene silencing in plants or gene quelling in fungi since they all work using a series of basic components, such as Dicer, Argonaute and RNA-dependent RNA polymerase (Fagard et al., 2000; Ipsaro and Joshua-Tor, 2015; Armas-Tizapantzi and Montiel-González, 2016). In the RNAi pathway, Dicer cleaves long dsRNA into short RNA duplexes of approximately 21–25 nucleotide (nt) small non-coding RNAs (sRNAs) (Zamore et al., 2000; Bernstein et al., 2001; Chapman and Carrington, 2007). Subsequently, these sRNAs are incorporated into the RISC to target the corresponding cognate mRNA for its destruction or translational suppression as sequence-specific guides (Hammond et al., 2000; Kim et al., 2010). Therefore, Dicer protein-mediated RNA cleavage is the initiation step of RNAi, and Dicer is essential for the generation and action of sRNAs.
Dicer or Dicer-like (DCL) protein is an endonuclease belonging to the RNase III family and contains a typical structure with an RNA helicase domain and a PAZ domain at the N′ terminal and two RNase III domains and a double-stranded RNA binding domain (dsRBD) at the C terminal (MacRae et al., 2006). Genome-wide sequencing projects have revealed that DCLs are evolutionarily conserved in a wide range of eukaryotic genomes (Kadotani et al., 2004). The fungal kingdom comprises an enormous diverse group of organisms, and the structure and number of DCLs also showed a conserved evolution among different species of fungi (Nakayashiki et al., 2006; Nunes et al., 2011b). However, the biological functions of fungal DCLs are diverse among different species. There have been some reports regarding the function of Dicers in sRNA generation. In 2004, dcl-2 was shown to be indispensable in the siRNA generation process in Magnaporthe grisea (Magnaporthe oryzae) (Kadotani et al., 2004). In N. crassa, the generation of siRNA was not markedly affected in the single-knockout mutant of Dicer-1 and Dicer-2 but was abolished in the double-knockout mutant (Catalanotto et al., 2004). Knocking out Dicer genes could also affect the generation and accumulation of sRNAs in Mucor circinelloides (Nicolás et al., 2007, 2010; de Haro et al., 2009). Additionally, fungal Dicers have been shown to be associated with growth, development, environmental stress and pathogenicity. In Mucor circinelloides, the silencing of Dicers caused altered phenotypes, such as changes in morphology, growth rate, asexual sporulation and autolysis (Nicolás et al., 2007, 2015). Knocking out the DCLs in Trichoderma atroviride also resulted in significant changes in morphology, growth rate and sporulation (Carreras-Villaseñor et al., 2013). Segers et al. (2007) found that the interruption of the dc1-1 and dcl-2 genes in Cryphonectria parasitica produces strains that are highly susceptible to hypovirus infection, which was the first report regarding the role of fungal immunity against a virus. In Magnaporthe oryzae, only a growth inhibition was found in the Modcl2 mutant, and the pathogenicity of the Modcl1, Modcl2 and Modcl1dcl2 mutants was not affected (Kadotani et al., 2004). Meanwhile, the virulence of Sporisorium reilianum DCL mutants was also not significantly changed (Schirawski et al., 2010). More importantly, when both Dicer genes were double-knocked out in Botrytis cinerea, the mutants showed a weakened pathogenicity (Weiberg et al., 2013). These results suggest that the fungal Dicers play important and various roles in specific physiological, developmental, and pathogenic processes. In-depth studies investigating the functions of the Dicer genes will lay the foundation for the exploration of the RNAi mechanism of fungi.
Valsa mali (V. mali) is one of the most important plant pathogens, causing canker disease in apple trees, which severely restricts the development of the apple industry and has been the key target of prevention and treatment in apple production in China (Cao et al., 2009; Wang et al., 2014). The lack of information regarding the molecular pathogenic mechanism in this fungus is the primary limitation in disease management. Therefore, the exploration of the pathogenic mechanism of V. mali has become extremely urgent. V. mali has a relatively small genome that was recently sequenced, and the major components of the RNAi pathway, such as Dicer, AGO, and RdRP, have been isolated (Yin et al., 2015). More importantly, most of these components exhibit regulated expression profiles during the V. mali infection process (Ke et al., 2014). In this study, we demonstrated that the Dicer genes of V. mali were responsible for the growth, development, and pathogenic processes and sRNAs generation based on gene knock-out mutants and high-throughput sequencing. This study laid a foundation for the comprehensive exploration of the pathogenesis mechanism of V. mali.
Materials and Methods
Fungus Strains and Growth Conditions
The V. mali wide type strain 03–8 used in this study was deposited in the Laboratory of Pathogen Biology and Integrated Control of Fruit Tree Diseases, College of Plant Protection, Northwest A&F University, Yangling, Shaanxi, China. The wild type and mutant strains generated in this study were maintained on a potato dextrose agar (PDA, 20% potato, 2% glucose, and 1.5% agar) medium at 25°C in the dark. Liquid potato dextrose broth (PDB) was used as the growth medium in the mycelium collection.
Sequence Characterization and Phylogenetic Analysis
The VmDCL sequences were isolated from the available V. mali genome database using the DCL sequences of M. oryzae and N. crassa as the probe sequences for the in silico cloning. The domain architecture was analyzed using InterProScan1. A phylogenetic comparison of the deduced protein sequences and the corresponding proteins from other species was performed with DNAMAN version 6.0.
Nucleic Acid Isolation and Manipulation
The fungal genomic DNA was extracted using the CTAB method (Möller et al., 1992). For the RNA isolation, apple twigs infected with V. mali strain 03–8 were sampled at 48 hpi (hours post inoculation). RNA samples from 03–8 mycelium and apple twigs infected by 03–8 were isolated using the TRIzolTM Reagent (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s instructions. Contaminated genomic DNA was removed using DNase I (Invitrogen, Carlsbad, CA, United States). DNA/RNA degradation was monitored on 1% agarose gels. A NanoDropTM 1000 spectrophotometer (Thermo Fisher Scientific, United States) was used to assess the DNA/RNA purity, concentration and integrity.
cDNA Synthesis and qRT-PCR Analysis
In total, three micrograms of total RNA were used to synthesize the first-strand cDNA using an RT-PCR system (Thermo Fisher Scientific, United States) according to the manufacturer’s instructions. The quantitative PCR amplifications were performed on a CFX96 Real-Time System (Bio-Rad) with SYBR Green (Invitrogen). The Glyceraldehyde-6-phosphate dehydrogenase gene (G6PDH) of V. mali was used as the internal control gene (Yin et al., 2013). The relative expression levels of each gene were calculated by the 2-ΔΔCT method (Livak and Schmittgen, 2001). Three biological replicates were performed for each experiment. All primers are listed in Supplementary Table S1.
Gene Deletion and Complementation
The vector pBIG2RHPH2-GFP-GUS, which contains a hygromycin resistance gene, was used to construct the gene deletion vector. The double-joint PCR approach was used for the vector construction and generating the gene knock-out mutants (Yu et al., 2004). For the single gene knock-out mutants, the strategy consisted of replacing the VmDCLs with the hygromycin-phosphotransferase (hph) cassette, which was amplified from PHIG2RHPH2-GFP-GUS. The upstream and downstream flanking sequences were amplified with the 1F/2R and 3F/4R primers, respectively. After ligation with the hph cassette, the ligated PCR product was transformed into 03–8 according to the method described by Gao et al. (2011). Positive transformants were selected using hygromycin B at a final concentration of 100 μg/mL. The putative knockout mutants identified by screening with the primers 5F and 6R were further analyzed by PCR with the primers 7F and H855R and H856F and 8R to confirm the gene replacement events (Supplementary Figure S1). For the double gene knock-out mutants, the single gene knock-out mutant (ΔVmDCL2) was used to generate the protoplast, and another vector containing a geneticin resistance gene NEO was used to replace the ORF of the second gene. The detailed method used for the mutant generation is similar to that described above. The putative mutants were further confirmed by a Southern blot analysis with probes for the HYG/NEO fragments according to the manufacturer’s instructions of the DIG DNA Labeling and Detection Kit II (Roche, Mannheim, Germany). For the complementation assays, the VmDCLs were amplified using the primer pairs Com-F and Com-R. The resulting PCR products were cloned into the vector pFL2 using the yeast gap repair approach (Bruno et al., 2004; Zhou et al., 2011). The resulting fusion constructs rescued from the Trp+ yeast transformants were confirmed by sequencing and transformed into their respective V. mali deletion mutant. The geneticin-resistant transformants expressing the complementing constructs were identified by PCR using the primer pairs Com-F and Com-R. The primers used for the gene deletion and complementation are listed in Supplementary Table S1.
Phenotypic Analysis of the VmDCL Mutants
To detect the growth rate of the colonies, VmDCL mutant strains (ΔVmDCLs) and 03–8 mycelium were placed onto the center of a PDA medium plate and were cultured in the dark at 25°C. The size of the colonies was examined after 24, 48, and 72 h. To observe the morphology of the colonies and hyphae, the colonies were photographed, and the hyphae morphology was observed using samples from the colony edge of each strain cultured in the dark at 25°C for 2 days under a microscope. To assay the stress response defects in the ΔVmDCLs, mycelium of the ΔVmDCLs and 03–8 were placed onto a PDA medium plate containing 0.05% H2O2 and 0.5 M KCl, and then, the plates were cultured in the dark at 25°C. The size and morphology of the colonies were examined after 3 days. The significance was analyzed using SPSS18 software. All experiments were performed in triplicate with three Petri dishes in each replication.
Pathogenicity Test
‘Fuji’ apple (Malus domestica Borkh. cv ‘Fuji’) twigs were used to detect the pathogenicity of the ΔVmDCLs and 03–8. The test was conducted using the detailed methods described by Wei et al. (2010). The statistical significance of the differences was analyzed using SPSS18 software. The infection assays were repeated three times with six twigs per replicate.
Library Preparation and RNA Deep Sequencing
Small RNA libraries were prepared following the detailed protocol provided by the genome sequencing company (Novogene, China). Three individual biological replicates of each sample (ΔVmDCL1, ΔVmDCL2, ΔVmDCL1DCL2 and 03–8) were prepared, and the three sample replicates were mixed for the RNA isolation. For the small RNA library construction, three micrograms of total RNA from each sample were used as the input material. The sequencing libraries were generated using the NEBNext® Multiplex Small RNA Library Prep Set for Illumina® (NEB, United States) following the manufacturer’s recommendations, and index codes were added to attribute sequences to each sample. The library quality was assessed on an Agilent Bioanalyzer 2100 system using DNA High Sensitivity Chips. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using the TruSeq SR Cluster Kit v3-cBot-HS (Illumia) according to the manufacturer’s instructions. After the cluster generation, the library preparations were sequenced on an Illumina HiSeq 2500/2000 platform, and 50 bp single-end reads were generated.
Sequence Data Analysis
Raw data (raw reads) in the fastq format were first processed through custom Perl and python scripts. During this step, clean data (clean reads) were obtained by removing reads containing ploy-N, reads with 5′ adapter contaminants, reads without 3′ adapters or insert tags, reads containing ploy A, T, G or C, and low-quality reads from the raw data. The clean reads were mapped onto the reference sequence using Bowtie (Langmead et al., 2009) without a mismatch to confirm the sequence accuracy. The number of total unique sRNAs from the different samples was calculated to compare the difference among the ΔVmDCLs and 03–8. The common and specific sequences among the ΔVmDCLs and 03–8 were also compared. To preliminarily estimate the sRNAs varieties, the length distribution of each sample was also analyzed.
Results
Protein Sequence and Phylogenetic Tree Analysis
In total, there are two DCL homologous sequences in V. mali. The genes were designated VmDCL1 and VmDCL2, and the sequences were deposited in GenBank with accession numbers KUI68257.1 and KUI73784.1, respectively. The InterProScan analyses showed that the deduced VmDCL1 protein contains five domains, which are known as RNA helicase domains (DEAD and hel C), dsRBD and Ribonuclease domains (RNase III a and RNase III b) and are characteristic of DCL proteins. Compared with VmDCL1, VmDCL2 also contains an additional special domain, i.e., DSRM (Figure 1A). In addition, the phylogenetic analysis suggests that the VmDCLs are clustered with the DCLs from fungi as expected, and they are close to the DCLs from Cryphonectria parasitica in evolution (Figure 1B).
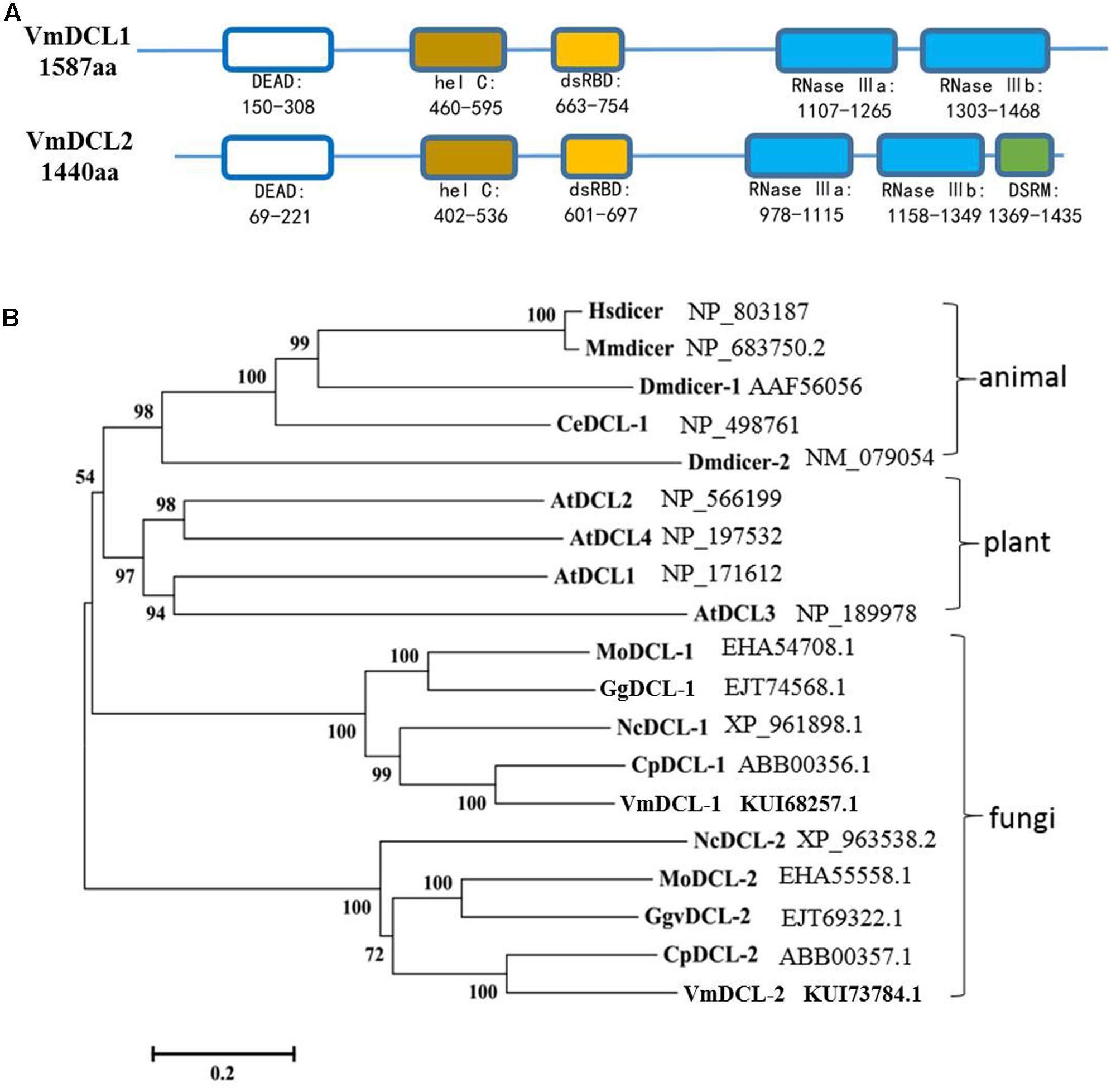
FIGURE 1. Domain structure and phylogeny of the VmDCLs. (A) Typical domains of the VmDCLs. VmDCL1 and VmDCL2 both contain five conserved domains, including DEAD, hel C, dsRBD, RNase IIIa and RNase IIIb. VmDCL2 also has a special domain, i.e., DSRM that is responsive to the generation of miRNAs. The number under the structures indicates the amino acid location of each domain. (B) Phylogenetic tree constructed via the multiple alignment program DNAMAN using DCLs sequences from plants, animals and fungi. VmDCLs clustered with other DCLs from fungi and shared a common ancestor with DCLs from plants and animals. Entries are labeled with the gene name in different species and the corresponding GenBank accession number. Hsdicer, Dicer of Homo sapiens, Mmdicer, Dicer of Mus musculus, Dmdicer, Dicer of Drosophila melanogaster, CeDCL, Dicer of Caenorhabditis elegans, AtDCL, Dicer of Arabidopsis thaliana, MoDCL, Dicer of Magnaporthe oryzae, GgDCL, Dicer of Gaeumannomyces graminis var. tritici, NcDCL, Dicer of Neurospora crassa, CpDCL, Dicer of Cryphonectria parasitica, VmDCL, Dicers of Valsa mali.
Acquisition of the VmDCL Mutants
As shown in Supplementary Figure S2, the VmDCL1 single-gene deletion mutants (ΔVmDCL1), VmDCL2 single-gene deletion mutants (ΔVmDCL2) and the VmDCL1DCL2 double-gene deletion mutants (ΔVmDCL1DCL2) were identified via screening with different PCR primers. The results indicated that we obtained putative knockout mutants of VmDCL1, VmDCL2 and VmDCL1DCL2. Subsequently, all putative knockout mutants were further verified by Southern blot hybridization. Furthermore, no hybridization signal was detected in the wild type strain 03–8 using the HYG probe amplified with the primers HYG/F and HYG/R, but there was a 4.3 and 6.0 Kb band in the VmDCL1 and VmDCL2 mutants, respectively; meanwhile, a NEO probe amplified with the primers NEO/F and NEO/R was used to hybridize with the genomic DNA of ΔVmDCL1DCL2, and a single corresponding 3.5 Kb band was detected. Thus, the single copy mutants of VmDCL1, VmDCL2 and VmDCL1DCL2 were generated successfully (Figure 2).
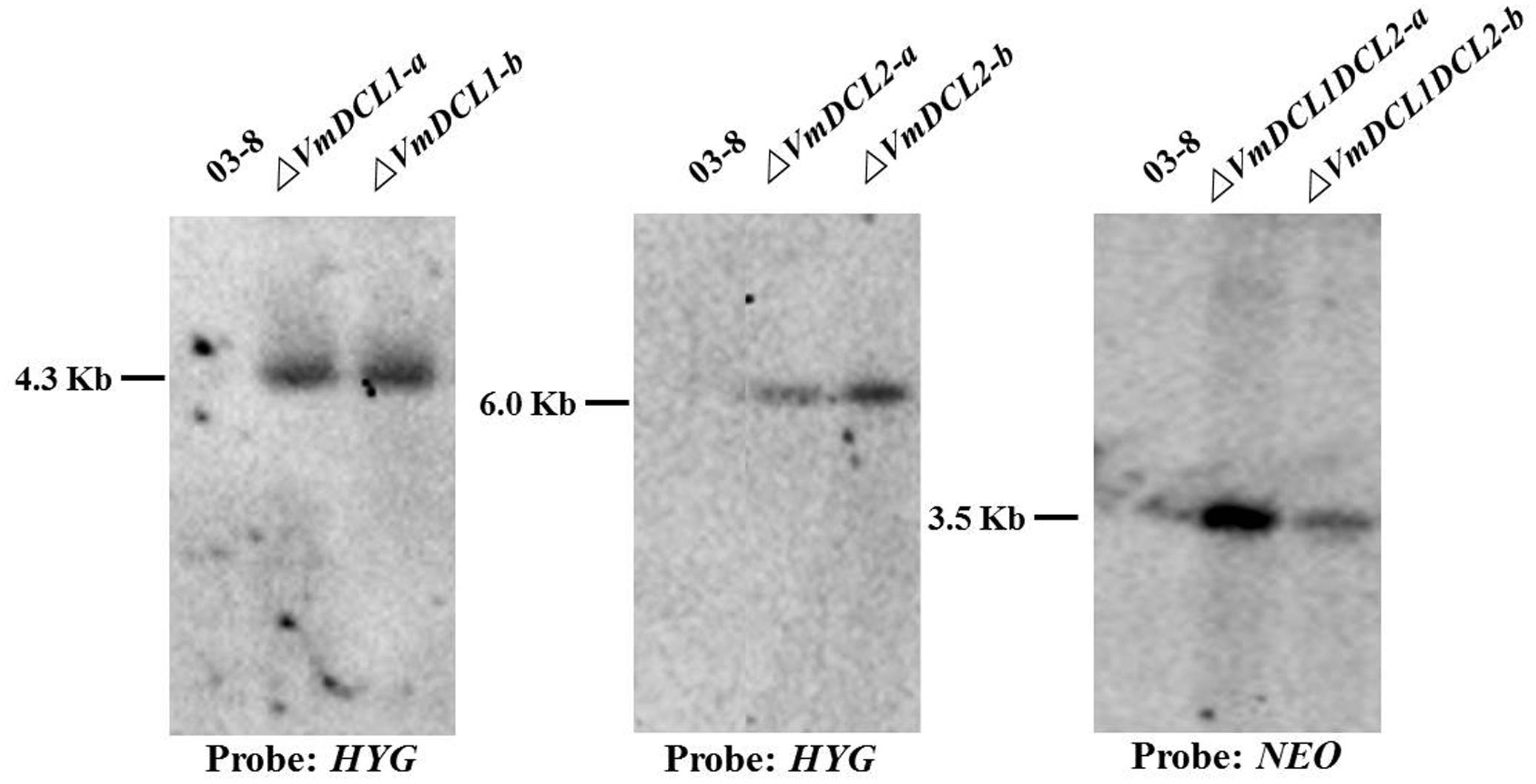
FIGURE 2. Construction of the VmDCL deletion mutants. Two candidate positive transformants of each mutant (ΔVmDCL1-a and ΔVmDCL1-b; ΔVmDCL2-a and ΔVmDCL2-b; and ΔVmDCL1DCL2-a and ΔVmDCL1DCL2-b) were used for Southern blotting. For ΔVmDCL1, genomic DNA from 03 to 8 and mutants was isolated and digested with restriction enzyme Cla I; for ΔVmDCL2, genomic DNA from 03 to 8 and ΔVmDCL2 was isolated and digested with restriction enzyme Sal I; for ΔVmDCL1DCL2, genomic DNA from 03 to 8 and ΔVmDCL2 was isolated and digested with restriction enzyme Hind III. DNA was hybridized with the probes HYG (ΔVmDCL1 and ΔVmDCL2) and NEO (ΔVmDCL1DCL2).
Growth Rate and Morphology of the Colony and Hypha Showed No Obvious Change in the Mutants Compared with Those in 03–8
To investigate whether VmDCLs play a critical function in the growth and development of V. mali, the growth rate and morphology of the colony and hypha of the mutants (ΔVmDCL1, ΔVmDCL2 and ΔVmDCL1DCL2) were compared with those of 03–8. No obvious difference in the colonial morphology was observed between the mutants and 03–8 when they were cultured on a PDA medium at 25°C in the dark for 2 days (Supplementary Figure S3a). Furthermore, the hypha morphology did not change obviously (Supplementary Figure S3b). In addition, the growth rate of 03–8, ΔVmDCL1, ΔVmDCL2 and ΔVmDCL1DCL2 was 2.50, 2.48, 2.52, and 2.47 cm/d, respectively, also showing no statistically significant difference (Supplementary Figure S3c).
VmDCLs Were Involved in the Stress Responses
To determine whether VmDCLs were involved in the stress responses, H2O2 and KCl were added to the PDA medium. In the presence of 0.05% H2O2, ΔVmDCL2 grew more slowly than 03–8 3 days after the inoculation, but the colony of ΔVmDCL1 was much larger than the colony of 03–8. Interestingly, the colony of ΔVmDCL1DCL2 showed no obvious change compared with that of 03–8 (Figures 3A,B). In the presence of 0.5 M KCl, the colony growth of only ΔVmDCL1 was significantly inhibited, while there was no significant difference among 03–8, ΔVmDCL2 and ΔVmDCL1DCL2 (Figures 3A,C). The complemented strain showed a nearly normal growth compared with 03–8 under the H2O2 and KCl treatment conditions. These results suggest that the VmDCLs may be important but play distinct roles in the response to oxidative and osmotic stresses.
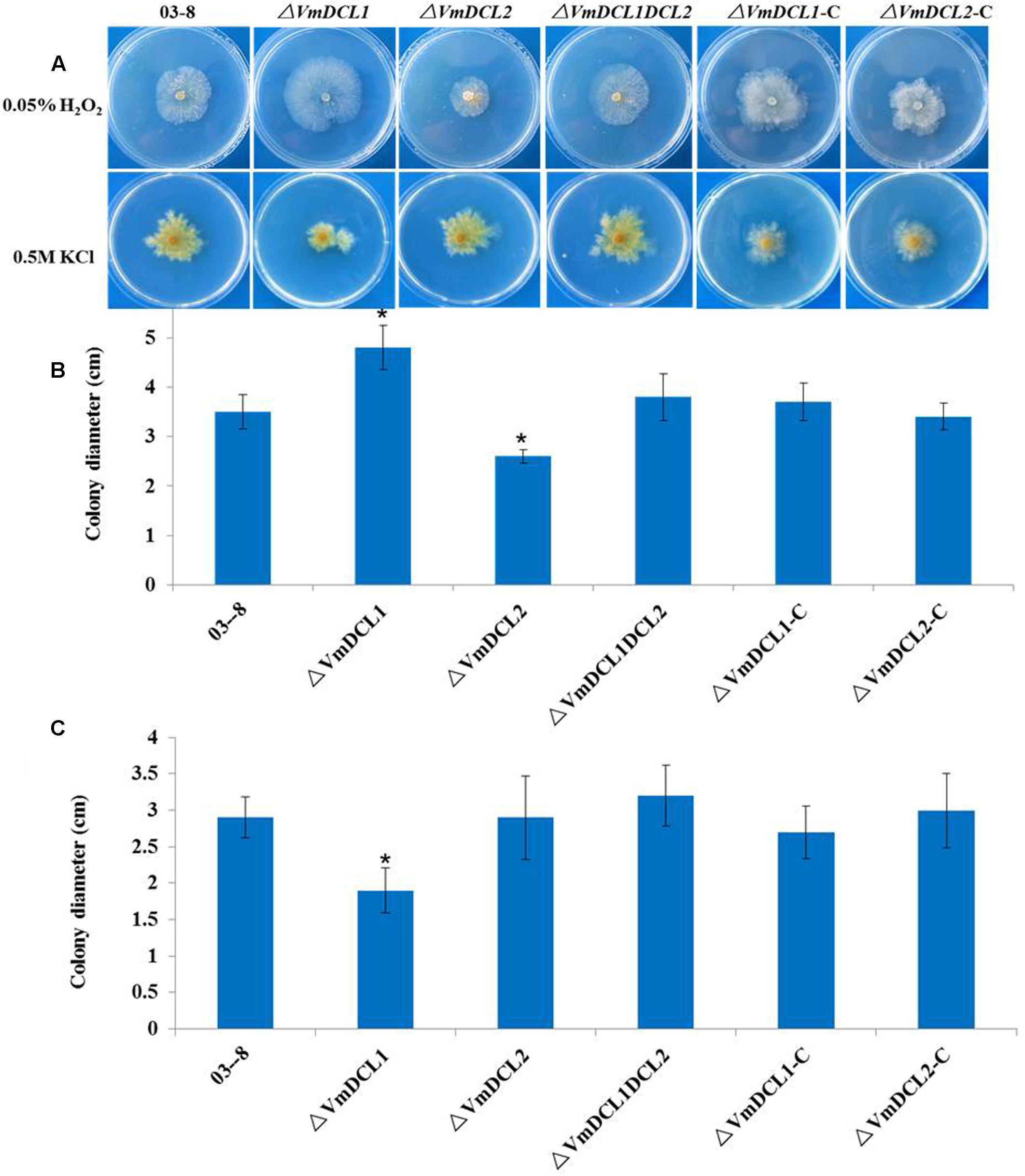
FIGURE 3. Abiotic stress responses of VmDCL mutants and 03–8. 03–8, ΔVmDCL1, ΔVmDCL2, ΔVmDCL1DCL2 and the complementary mutants (ΔVmDCL1-C andΔVmDCL2-C) were cultured on PDA supplemented with 0.05% H2O2 (A,B) and 0.5 M KCl (A,C). Photographs were taken after 3 days of incubation at 25°C. All experiments were performed in triplicate in three Petri dishes in each replication. The bars indicate the standard deviation of the mean of three replicates. The stars above the bars indicate significantly different means (LSD test, P < 0.05).
VmDCLs Play Distinct Roles in the Pathogenicity of V. mali
The expression analysis of the VmDCLs during the V. mali infection in apple twig showed that compared with the control (mycelium), VmDCL2 was up-regulated and peaked (125-fold) at 48 hpi. However, VmDCL1 expressed stably with no significant change and only exhibited a 1.7-fold increase at 48 hpi (Figure 4A). The pathogenicity of ΔVmDCLs and 03–8 was further analyzed. Compared with 03–8, the pathogenicity of ΔVmDCL1 showed no significant change, while the ΔVmDCL2 was defective in plant infections with a smaller lesion. Furthermore, the pathogenicity markedly decreased when both VmDCL1 and VmDCL2 were knocked out together. Under the same conditions, the pathogenicity of the complemented transformants ΔVmDCL1-C and ΔVmDCL2-C was similar to that of 03–8 in the twig infection assays (Figures 4B,C). These results suggest that VmDCL2 plays a critical role during the infection process in twigs, and the detailed mechanism will be a key scientific problem to be explored in the future.
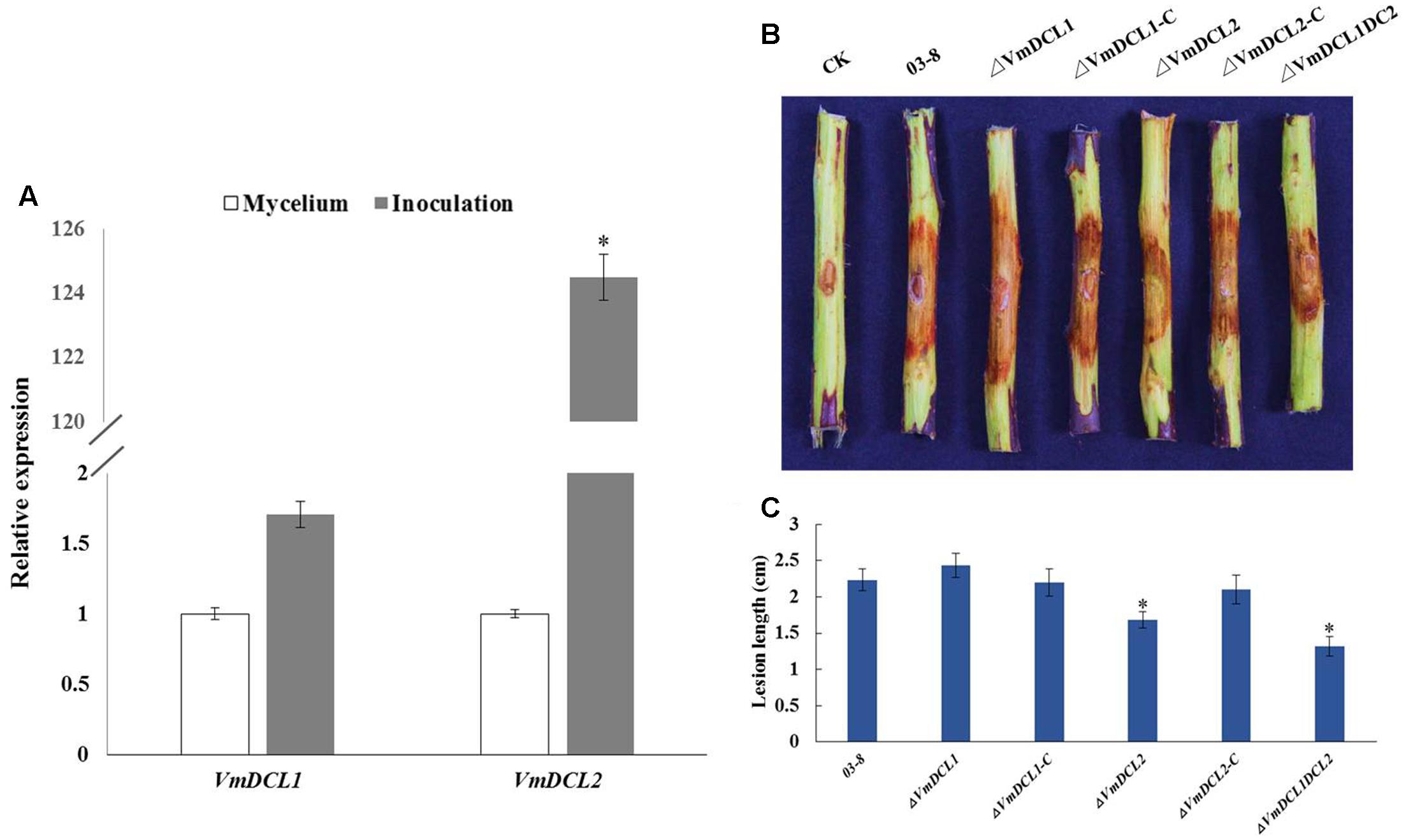
FIGURE 4. Phenotypical analysis of VmDCL mutants and 03–8. (A) Transcript levels of VmDCL1 and VmDCL2 at 48 hpi compared to those of the control (mycelium) using G6PDH for normalization. The results were calculated using the comparative threshold (2-ΔΔCT) method. Vertical bars represent the standard deviation. The mean values and standard deviations were calculated using data from three independent replicates. (B) ‘Fuji’ apple (Malus domestica Borkh. cv ‘Fuji’) twigs were used to determine the pathogenicity of the mutants relative to that of 03–8. Photographs were taken at 3 days after inoculation. (C) Lesion diameters of twigs were measured 3 days after the inoculation. The bars indicate the standard deviation of the mean of three biological replicates. The stars above the bars indicate significantly different means (LSD test, P < 0.05).
VmDCLs Were Associated with sRNA Generation
To comprehensively examine the sRNA abundance in strains of 03–8 (MVM), ΔVmDCL1 (MD1), ΔVmDCL2 (MD2) and ΔVmDCL1DCL2 (MD12), four sRNA libraries were constructed and sequenced using an Illumina Analyzer. The distinct libraries generated 12818591, 6927296, 11565746 and 8990152 raw reads. After removing the low-quality and non-sense sequences (<15 nt), in total, 1364324, 788373, 834923 and 708262 unisRNAs were isolated (data not shown). As shown in Figure 5, the number of unisRNAs was markedly smaller in the libraries of MD1, MD2 and MD12 than that in the library of MVM, indicating that the deletion of the VmDCLs could affect the generation of sRNAs in V. mali. In addition, the length distribution of the unisRNAs was further analyzed (18–30 nt long), and the length distribution also markedly changed when the VmDCLs were deleted. When VmDCL1 was knocked out, the abundance of sRNAs consisting of 29 nt showed the most obvious increase (Figures 6A,B). Meanwhile, the abundance of sRNAs consisting of 18–20 nt noticeably increased, and the abundance of sRNAs consisting of 21–22 nt noticeably decreased when VmDCL2 was knocked out (Figures 6A,C). Additionally, in the ΔVmDCL1DCL2, the abundance of sRNAs consisting of 21 nt noticeably decreased, and the abundance of sRNAs consisting of 29 nt noticeably increased (Figures 6A,D). The other sRNA abundances were also affected in varying degrees between the mutants and 03–8. The results indicated that the VmDCLs could affect the generation of sRNAs in V. mali, and their function is likely redundant. Furthermore, there are at least two pathways (i.e., DCL-dependent and DCL-independent) for sRNA generation in V. mali. The detailed mechanism will be explored in further studies.
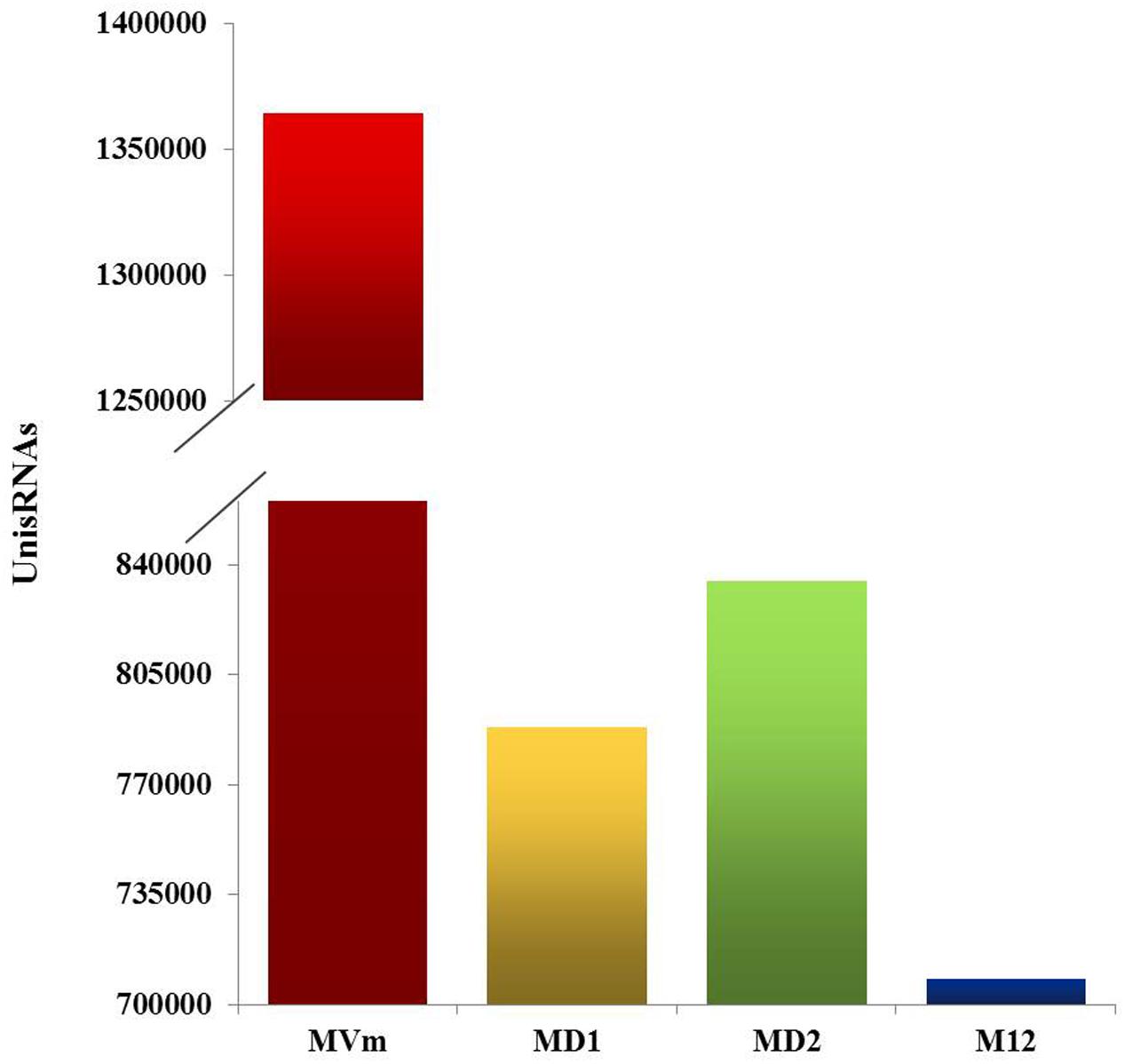
FIGURE 5. Quantification of the unisRNAs quantity in the VmDCL mutants and 03–8. Total number of uniRNAs in 03–8 and each mutants was analyzed, respectively. MVm, 03–8; MD1, ΔVmDCL1; MD2, ΔVmDCL2; MD12, ΔVmDCL1DCL2.
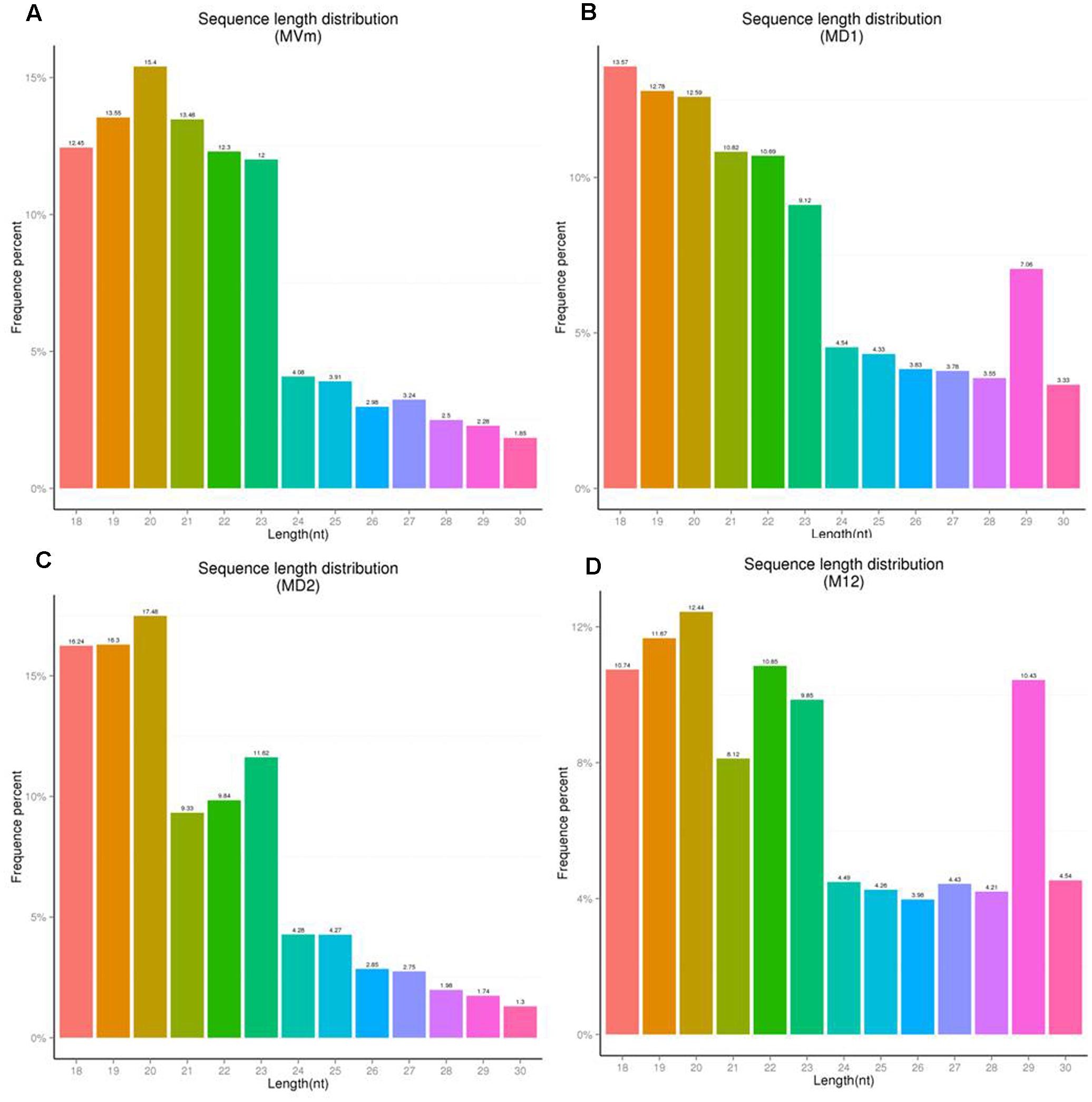
FIGURE 6. Sequence length distribution of unisRNAs in the VmDCL mutants and 03–8. The length distribution of the unisRNAs changed when VmDCL1 (B), ΔVmDCL2 (C) and ΔVmDCL1DCL2 (D) were deleted compared with 03–8 (A). Horizontal axis indicates the sequence length, the vertical axis indicates the proportion of the corresponding reads, and the colors indicate the different lengths. MVm, 03–8; MD1, ΔVmDCL1; MD2, ΔVmDCL2; MD12, ΔVmDCL1DCL2.
Discussion
RNAi, which is mediated by small regulatory RNAs, plays important roles in various biological progresses at the transcriptional and post-transcriptional levels (Pratt and MacRae, 2009). In filamentous fungi, several unique classes of sRNAs have been identified, particularly in N. crassa, although the corresponding targets remain to be fully elucidated (Lee et al., 2009, 2010). In other fungal species, particularly in plant pathogenic fungi, small RNA-mediated gene silencing remains in its initial exploration phase. As increasing numbers of fungal genomes were published and deep sequence technologies and bioinformatics analyses continuously advanced, more fungi were confirmed to possess the RNA silencing mechanism (Nunes et al., 2011a; Zhou J. et al., 2012; Zhou Q. et al., 2012; Kang et al., 2013; Lau et al., 2013; Campo et al., 2016). Information regarding the genome of V. mali, which is an important weak parasitic plant pathogen, has become public, and the major components of the RNAi pathway were also predicted (Yin et al., 2015). However, little is known regarding the function of RNAi pathway in V. mali.
In this study, the ‘switch’ for the generation and action of sRNAs, i.e., Dicer-like genes, in V. mali were isolated and functionally analyzed. Based on the known genome information of V. mali, two DCL genes (VmDCL1 and VmDCL2) were isolated, and the VmDCLs were grouped clearly with DCL members from other fungal species assessed by the phylogenetic analysis. Most of the fungi have two DCL-like proteins, particularly in Ascomycota (Nunes et al., 2011b). In addition, both VmDCL1 and VmDCL2 contain an RNA helicase domain, RNase III domains and double-stranded RNA binding domain, which are the typical domains of DCL proteins (MacRae et al., 2006). However, neither DCL in V. mali contains a PAZ (Piwi/Argonaute/Zwille) domain. Some eukaryotes have been found to lack the PAZ domain, such as Colletotrichum higginsianum, but they still retain the capacity to generate small RNAs (Casas-Mollano et al., 2008; Drinnenberg et al., 2009; Braun et al., 2010; Campo et al., 2016). Furthermore, VmDCL2 contains an additional double-stranded RNA binding motif (DSRM). The DSRM is an important domain of DGCR8, which is required for microRNA (miRNA) processing in the RNAi pathway (Denli et al., 2004; Yeom et al., 2006). Therefore, VmDCL2 may be closely involved in the generation of miRNAs.
The biological function of DCLs in different species is diverse. In previous studies, disruptions of DCL orthologs were sufficient to slow vegetative growth in M. circinelloides, M. oryzae, B. cinerea and T. atroviride (Kadotani et al., 2004; Nicolás et al., 2007; Carreras-Villaseñor et al., 2013; Weiberg et al., 2013). However, the deletion of DCL did not affect the growth of Cryphonectria parasitica and Saccharomyces castellii (Segers et al., 2007; Drinnenberg et al., 2009). To evaluate the function of VmDCLs, mycelial growth and morphology on a PDA medium were measured based on the generation of ΔVmDCL1, ΔVmDCL2 and ΔVmDCL1DCL2. The colony and hypha morphology and mycelial growth were indistinguishable between the mutants and the wild type, suggesting that the VmDCLs are not necessary for vegetative growth. Moreover, ΔVmDCL1 showed an increased adaptability to 0.05% H2O2 and a sensibility to 0.5 M KCl, while ΔVmDCL2 only exhibited an increased sensibility to 0.05% H2O2. The similar phenotype changes were also confirmed in the dcr1, dcr2 and dcr1dcr2 mutants of T. atroviride, which suggested that each DCL controls different biological processes (Carreras-Villaseñor et al., 2013). The VmDCLs play an important but distinct role in the pathogenicity of V. mali. Using qRT-PCR, we found that VmDCL2 was up-regulated significantly during the infection process. Furthermore, we found that the deletion of VmDCL2 and VmDCL1DCL2 resulted in a significant reduction in pathogenicity. Thus, we speculated that VmDCL2 is a critical factor in the pathogenicity of V. mali, and the sRNAs generated by VmDCL2 may be responsive to the pathogenicity. In M. oryzae, no DCLs mutants showed obvious phenotypes associated with pathogenicity (Kadotani et al., 2004). However, the dcl1dcl2 double-mutant of B. cinerea showed a weakened pathogenicity regulated by an sRNA (Weiberg et al., 2013). These results also indicated that the functions of the DCLs were diversified in the evolution of different fungi.
To investigate the function of VmDCLs for sRNA generation, we compared the sRNA amounts in mutants and wild type based on high-throughput sequencing. Based on the results, the total sRNA amount was reduced greatly and the number of sRNAs with 21 and 29 nt changed when VmDCL1, VmDCL2 and VmDCL1DCL2 were knocked out. This finding indicated that VmDCLs were related to sRNA biogenesis. However, except for the specific unisRNAs, there were still many common sRNAs in both the wild type and ΔVmDCL1/ΔVmDCL2. DCLs appear to also be functionally redundant in sRNA processing in certain fungi species (Lee et al., 2010; Nicolás et al., 2013; Weiberg et al., 2013). We also found a slew of new sRNAs that could be generated in the VmDCL deletion mutants, and the length distribution of these sRNAs was also different than that in the wild type. This finding confirmed that the function of the VmDCLs in sRNA generation is redundant, and there is more than one pathway responsible for sRNA biogenesis. A new pathway could be activated when the VmDCLs were deleted. In previous studies, at least four different biogenesis pathways of miRNA-like RNAs have been identified in N. crassa, including both DCL-dependent and DCL-independent pathways (Jin and Zhu, 2010; Lee et al., 2010; Li et al., 2010). Thus, whether the phenotypical changes in the VmDCL deletion mutants were associated with the generation of different sRNAs is an interesting scientific question that should be explored.
In summary, we isolated and identified DCLs from V. mali for the first time and investigated their gene function in development, abiotic stress response, and pathogenicity. Another major finding of this study is the functional confirmation of the VmDCLs in the generation of sRNAs. In addition, the possible existence of diverse sRNA biogenesis pathways in V. mali presents a research direction for the identification of sRNA species and the characterization of regulatory sRNAs in pathogenicity.
Author Contributions
HF and LH were responsible for the experimental design, and provided guidance on the whole study. HF wrote the manuscript and LH further revised it. HF, MX, YL, and RD constructed the mutant strains and determined the phenotypes. XG prepared cDNA samples and detected the gene expression. All authors have read and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
The study was funded by the Postdoctoral Science Foundation funded project of Shaanxi Province and China Postdoctoral Science Foundation funded project (NO. 2016T90953 and 2015M580883). We thank Prof. Fengming Song, Zhejing University, for providing the pBIG2RHPH2-GFP-GUS vector.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01166/full#supplementary-material
Footnotes
References
Armas-Tizapantzi, A., and Montiel-González, A. M. (2016). RNAi silencing: a tool for functional genomics research on fungi. Fungal Biol. Rev. 30, 91–100. doi: 10.1016/j.fbr.2016.05.00305.003
Bernstein, E., Caudy, A. A., Hammond, S. M., and Hannon, G. J. (2001). Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366. doi: 10.1038/35053110
Braun, L., Cannella, D., Ortet, P., Barakat, M., Sautel, C. F., Kieffer, S., et al. (2010). A complex small RNA repertoire is generated by a plant/fungal-like machinery and effected by a metazoan-like argonaute in the single-cell human parasite Toxoplasma gondii. PLoS Pathog. 6:e1000920. doi: 10.1371/journal.ppat.1000920
Bruno, K. S., Tenjo, F., Li, L., Hamer, J. E., and Xu, J. R. (2004). Cellular localization and role of kinase activity of PMK1 in Magnaporthe grisea. Eukaryot. Cell 3, 1525–1532. doi: 10.1128/EC.3.6.1525-1532.2004
Campo, S., Gilbert, K. B., and Carrington, J. C. (2016). Small RNA-based antiviral defense in the phytopathoginic fungus Colletotrichum higginsianum. PLoS Pathog. 12:e1005640. doi: 10.1371/journal.ppat.1005640
Cao, K., Guo, L., Li, B., Sun, G., and Chen, H. (2009). Investigations on the occurrence and control of apple canker in China. Plant Prot. 35, 114–116.
Carreras-Villaseñor, N., Esquivel-Naranjo, E. U., Villalobos-Escobedo, J. M., Abreu-Goodger, C., Herrera-Estrella, A., and Herrera-Estrella, A. (2013). The RNAi machinery regulates growth and development in the filamentous fungus Trichoderma atroviride. Mol. Microbiol. 89, 96–112. doi: 10.1111/mmi.12261
Casas-Mollano, J. A., Rohr, J., Kim, E. J., Balassa, E., van Dijk, K., and Cerutti, H. (2008). Diversification of the core RNA interference machinery in Chlamydomonas reinhardtii and the role of DCL1 in transposon silencing. Genetics 179, 69–81. doi: 10.1534/genetics.107.086546
Catalanotto, C., Pallotta, M., ReFalo, P., Sachs, M. S., Vayssie, L., Macino, G., et al. (2004). Redundancy of the two dicer genes in transgene-induced posttranscriptional gene silencing in Neurospora crassa. Mol. Cell Biol. 24, 2536–2545. doi: 10.1128/MCB.24.6.2536-2545.2004
Chapman, E. J., and Carrington, J. C. (2007). Specialization and evolution of endogenous small RNA pathways. Nat. Rev. Genet. 8, 884–896. doi: 10.1038/nrg2179
de Haro, J. P., Calo, S., Cervantes, M., Nicolás, F. E., Torres-Martínez, S., and Ruiz-Vázquez, R. M. (2009). A single dicer gene is required for efficient gene silencing associated with two classes of small antisense RNAs in Mucor circinelloides. Eukaryot. Cell 8, 1486–1497. doi: 10.1128/EC.00191-09
Denli, A. M., Tops, B. B., Plasterk, R. H., Ketting, R. F., and Hannon, G. J. (2004). Processing of primary microRNAs by the microprocessor complex. Nature 432, 231–235. doi: 10.1038/nature03049
Drinnenberg, I. A., Weinberg, D. E., Xie, K. T., Mower, J. P., Wolfe, K. H., Fink, G. R., et al. (2009). RNAi in budding yeast. Science 326, 544–550. doi: 10.1126/science.1176945
Fagard, M., Boutet, S., Morel, J. B., Bellini, C., and Vaucheret, H. (2000). AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc. Natl. Acad. Sci. U.S.A. 97, 11650–11654. doi: 10.1073/pnas.200217597
Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E., and Mello, C. C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. doi: 10.1038/35888
Gao, J., Li, Y., Ke, X., Kang, Z. S., and Huang, L. L. (2011). Development of genetic transformation system of Valsa mali of apple mediated by PEG. Acta Microbiol. Sin. 51, 1194–1199.
Hammond, S. M., Bernstein, E., Beach, D., and Hannon, G. J. (2000). An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404, 293–296. doi: 10.1038/35005107
Ipsaro, J. J., and Joshua-Tor, L. (2015). From guide to target: molecular insights into eukaryotic RNA-interference machinery. Nat. Struct. Mol. Biol. 22, 20–28. doi: 10.1038/nsmb.2931
Jin, H. L., and Zhu, J. K. (2010). How many ways are there to generate small RNA? Mol. Cell 38, 775–777. doi: 10.1016/j.molcel.2010.06.004
Kadotani, N., Nakayashiki, H., Tosa, Y., and Mayama, S. (2004). One of the two Dicer-like proteins in the filamentous fungi Magnaporthe oryzae genome is responsible for hairpin RNA-triggered RNA silencing and related small interfering RNA accumulation. J. Biol. Chem. 279, 44467–44474. doi: 10.1074/jbc.M408259200
Kang, K., Zhong, J., Jiang, L., Liu, G., Gou, C. Y., Wu, Q., et al. (2013). Identification of microRNA-Like RNAs in the filamentous fungus Trichoderma reesei by solexa sequencing. PLoS ONE 8:e76288. doi: 10.1371/journal.pone.00762880076288
Ke, X., Yin, Z., Song, N., Dai, Q., Voegele, R. T., Liu, Y., et al. (2014). Transcriptome profiling to identify genes involved in pathogenicity of Valsa mali on apple tree. Fungal Genet. Biol. 68, 31–38. doi: 10.1016/j.fgb.2014.04.004
Kim, Y. K., Heo, I., and Kim, V. N. (2010). Modifications of small RNAs and their associated proteins. Cell 143, 703–709. doi: 10.1016/j.cell.2010.11.018
Langmead, B., Trapnell, C., Pop, M., and Salzberg, S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. doi: 10.1186/gb-2009-10-3-r25
Lau, S. K., Chow, W. N., Wong, A. Y., Yeung, J. M., Bao, J., Zhang, N., et al. (2013). Identification of microRNA-like RNAs in mycelial and yeast phases of the thermal dimorphic fungus Penicillium marneffei. PLoS Negl. Trop. Dis. 7:e2398. doi: 10.1371/journal.pntd.0002398
Lee, H. C., Chang, S. S., Choudhary, S., Aalto, A. P., Maiti, M., Bamford, D. H., et al. (2009). qiRNA is a new type of small interfering RNA induced by DNA damage. Nature 459, 274–277. doi: 10.1038/nature08041
Lee, H. C., Li, L., Gu, W., Xue, Z., Crosthwaite, S. K., Pertsemlidis, A., et al. (2010). Diverse pathways generate microRNA-like RNAs and Dicer-independent small interfering RNAs in fungi. Mol. Cell. 38, 803–814. doi: 10.1016/j.molcel.2010.04.005
Li, L., Chang, S. S., and Liu, Y. (2010). RNA interference pathways in filamentous fungi. Cell. Mol. Life Sci. 67, 3849–3863. doi: 10.1007/s00018-010-0471-y
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
MacRae, I. J., Zhou, K., Li, F., Repic, A., Brooks, A. N., Cande, W. Z., et al. (2006). Structural basis for double-stranded RNA processing by Dicer. Science 311, 195–198. doi: 10.1126/science.1121638
Möller, E. M., Bahnweg, G., Sandermann, H., and Geiger, H. H. (1992). A simple and efficient protocol for isolation of high molecular weight DNA from filamentous fungi, fruit bodies, and infected plant tissues. Nucleic Acids Res. 20, 6115–6116. doi: 10.1093/nar/20.22.6115
Nakayashiki, H., Kadotani, N., and Mayama, S. (2006). Evolution and diversification of RNA silencing proteins in fungi. J. Mol. Evol. 63, 127–135. doi: 10.1007/s00239-005-0257-2
Napoli, C., Lemieux, C., and Jorgensen, R. (1990). Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2, 279–289. doi: 10.1105/tpc.2.4.279
Nicolás, F. E., de Haro, J. P., Torres-Martínez, S., and Ruiz-Vázquez, R. M. (2007). Mutants defective in a Mucor circinelloides dicer-like gene are not compromised in siRNA silencing but display developmental defects. Fungal Genet. Biol. 44, 504–516. doi: 10.1016/j.fgb.2006.09.003
Nicolás, F. E., Moxon, S., de Haro, J. P., Calo, S., Grigoriev, I. V., Torres-Martínez, S., et al. (2010). Endogenous short RNAs generated by Dicer 2 and RNA-dependent RNA polymerase 1 regulate mRNAs in the basal fungus Mucor circinelloides. Nucleic Acids Res. 38, 5535–5541. doi: 10.1093/nar/gkq301
Nicolás, F. E., Torres-Martínez, S., and Ruiz-Vázquez, R. M. (2013). Loss and retention of RNA interference in fungi and parasites. PLoS Pathog. 9:e1003089. doi: 10.1371/journal.ppat.1003089
Nicolás, F. E., Vila, A., Moxon, S., Cascales, M. D., Torres-Martınez, S., Ruiz-Vazquez, R. M., et al. (2015). The RNAi machinery controls distinct responses to environmental signals in the basal fungus Mucor circinelloides. BMC Genomics 16:237. doi: 10.1186/s12864-015-1443-2
Nunes, C. C., Gowda, M., Sailsbery, J., Xue, M., Chen, F., Brown, D. E., et al. (2011a). Diverse and tissue-enriched small RNAs in the plant pathogenic fungus, Magnaporthe oryzae. BMC Genomics 12:288. doi: 10.1186/1471-2164-12-288
Nunes, C. C., Sailsbery, J. K., and Dean, R. A. (2011b). Characterization and application of small RNAs and RNA silencing mechanisms in fungi. Fungal Biol. Rev. 25, 172–180. doi: 10.1016/j.fbr.2011.10.001
Pratt, A. J., and MacRae, I. J. (2009). The RNA- induced silencing complex: a versatile gene-silencing machine. J. Biol. Chem. 284, 17897–17901. doi: 10.1074/jbc.R900012200
Romano, N., and Macino, G. (1992). Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol. Microbiol. 6, 3343–3353. doi: 10.1111/j.1365-2958.1992.tb02202.x
Schirawski, J., Mannhaupt, G., Münch, K., Brefort, T., Schipper, K., Doehlemann, G., et al. (2010). Pathogenicity determinants in smut fungi revealed by genome comparison. Science 330, 1546–1548. doi: 10.1126/science.1195330
Segers, G. C., Zhang, X., Deng, F., Sun, Q., and Nuss, D. L. (2007). Evidence that RNA silencing functions as an antiviral defense mechanism in fungi. Proc. Natl. Acad. Sci. U.S.A. 104, 12902–12906. doi: 10.1073/pnas.0702500104
Wang, X. L., Zang, R., Yin, Z., Kang, Z., and Huang, L. (2014). Delimiting cryptic pathogen species causing apple Valsa canker with multilocus data. Ecol. Evol. 4, 1369–1380. doi: 10.1002/ece3.1030
Wei, J. L., Huang, L. L., Gao, Z. P., Ke, X. W., and Kang, Z. S. (2010). Laboratory evaluation methods of apple Valsa canker disease caused by Valsa ceratosperma sensu Kobayashi. Acta Phytopathol. Sin. 40, 14–20.
Weiberg, A., Wang, M., Lin, F. M., Zhao, H., Zhang, Z., Kaloshian, I., et al. (2013). Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342, 118–123. doi: 10.1126/science.12397051239705
Yeom, K. H., Lee, Y., Han, J., Suh, M. R., and Kim, V. N. (2006). Characterization of DGCR8/Pasha, the essential cofactor for Drosha in primary miRNA processing. Nucleic Acids Res. 34, 4622–4629. doi: 10.1093/nar/gkl458gkl458
Yin, Z., Ke, X., Huang, D., Gao, X., Voegele, R. T., Kang, Z., et al. (2013). Validation of reference genes for gene expression analysis in Valsa mali var. mali using real-time quantitative PCR. J. Microbiol. Biotechnol. 29, 1563–1571. doi: 10.1007/s11274-013-1320-6
Yin, Z., Liu, H., Li, Z., Ke, X., Dou, D., Gao, X., et al. (2015). Genome sequence of Valsa canker pathogens uncovers a potential adaptation of colonization of woody bark. New Phytol. 208, 1202–1216. doi: 10.1111/nph.1354413544
Yu, J. H., Hamari, Z., Han, K. H., Seo, J. A., Reyes-Domínguez, Y., and Scazzocchio, C. (2004). Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41, 973–981. doi: 10.1016/j.fgb.2004.08.001
Zamore, P. D., Tuschl, T., Sharp, P. A., and Bartel, D. P. (2000). RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101, 25–33. doi: 10.1016/S0092-8674(00)80620-0
Zhou, J., Fu, Y., Xie, J., Li, B., Jiang, D., Li, G., et al. (2012). Identification of microRNA-like RNAs in a plant pathogenic fungus Sclerotinia sclerotiorum by high-throughput sequencing. Mol. Genet. Genomics 287, 275–282. doi: 10.1007/s00438-012-0678-8
Zhou, Q., Wang, Z., Zhang, J., Meng, H., and Huang, B. (2012). Genome-wide identification and profiling of microRNA-like RNAs from Metarhizium anisopliae during development. Fungal Biol. 116, 1156–1162. doi: 10.1016/j.funbio.2012.09.001
Keywords: Dicer, pathogenesis, RNAi mechanism, small RNA, Valsa mali
Citation: Feng H, Xu M, Liu Y, Dong R, Gao X and Huang L (2017) Dicer-Like Genes Are Required for H2O2 and KCl Stress Responses, Pathogenicity and Small RNA Generation in Valsa mali. Front. Microbiol. 8:1166. doi: 10.3389/fmicb.2017.01166
Received: 22 March 2017; Accepted: 07 June 2017;
Published: 23 June 2017.
Edited by:
Jeremy Astier, Helmholtz Zentrum München, GermanyReviewed by:
Massimo Turina, Consiglio Nazionale delle Ricerche (CNR), ItalyZonghua Wang, Fujian Agriculture and Forestry University, China
Copyright © 2017 Feng, Xu, Liu, Dong, Gao and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lili Huang, huanglili@nwsuaf.edu.cn
 Hao Feng
Hao Feng Ming Xu
Ming Xu Yangyang Liu
Yangyang Liu Ruqing Dong
Ruqing Dong Xiaoning Gao
Xiaoning Gao Lili Huang
Lili Huang