Campylobacter jejuni acquire new host-derived CRISPR spacers when in association with bacteriophages harboring a CRISPR-like Cas4 protein
- Division of Food Sciences, School of Biosciences, University of Nottingham, Loughborough, UK
Incorporation of self-derived CRISPR DNA protospacers in Campylobacter jejuni PT14 occurs in the presence of bacteriophages encoding a CRISPR-like Cas4 protein. This phenomenon was evident in carrier state infections where both bacteriophages and host are maintained for seemingly indefinite periods as stable populations following serial passage. Carrier state cultures of C. jejuni PT14 have greater aerotolerance in nutrient limited conditions, and may have arisen as an evolutionary response to selective pressures imposed during periods in the extra-intestinal environment. A consequence of this is that bacteriophage and host remain associated and able to survive transition periods where the chances of replicative success are greatly diminished. The majority of the bacteriophage population do not commit to lytic infection, and conversely the bacterial population tolerates low-level bacteriophage replication. We recently examined the effects of Campylobacter bacteriophage/C. jejuni PT14 CRISPR spacer acquisition using deep sequencing strategies of DNA and RNA-Seq to analyze carrier state cultures. This approach identified de novo spacer acquisition in C. jejuni PT14 associated with Class III Campylobacter phages CP8/CP30A but spacer acquisition was oriented toward the capture of host DNA. In the absence of bacteriophage predation the CRISPR spacers in uninfected C. jejuni PT14 cultures remain unchanged. A distinct preference was observed for incorporation of self-derived protospacers into the third spacer position of the C. jejuni PT14 CRISPR array, with the first and second spacers remaining fixed. RNA-Seq also revealed the variation in the synthesis of non-coding RNAs with the potential to bind bacteriophage genes and/or transcript sequences.
Introduction
The functional role of CRISPR-Cas systems in Bacteria and Archaea is recognized as providing the benefit of acquired immunity to the prokaryotic kingdom. In order to counter the constant threats posed by mobile genetic elements the CRISPR-Cas system has evolved to survey, acquire, adapt, and destroy invading DNA in a sequence-specific manner. The invading elements may be in the form of plasmids or bacteriophages, which if not dealt with in a timely fashion could lead to the demise of the infected cell/bacterial population. The major components found in all CRISPR-Cas systems are the CRISPR-associated (Cas) proteins, Cas1 DNA endonuclease and Cas2 RNA endonuclease (Koonin and Krupovic, 2015; Selle and Barrangou, 2015). These proteins operate during the acquisition stage to generate protospacers (captured DNA) which are subsequently incorporated into the CRISPR array. Recently, analysis of mobile genetic elements known as transposons revealed a possible evolutionary origin for Cas1 protein. Named casposons, these transposable genetic elements were found to possess Cas1 DNA endonuclease domains as well as hallmarks of eukaryotic DNA transposons (B family DNA polymerase genes and terminal inverted repeats). Integration and excision of the casposon is proposed to be mediated by the encoded Cas1 endonuclease domain in a similar manner to how CRISPR spacer acquisition occurs in prokaryotes (Krupovic et al., 2014). Transcription of integrated spacers leads to the generation of CRISPR RNAs (crRNAs) that serve as RNA guides for the interference step (Hochstrasser and Doudna, 2015). Various other proteins combine to produce the interference effect, and the presence or absence of these proteins has allowed the classification of CRISPR-Cas systems into Type IA-U, Type IIA-C, Type IIIA-D, Type IV, and Type V (Makarova et al., 2015).
Type II CRISPR-Cas systems have been shown to consist of several different permutations of genes encoding Cas1, Cas2, Csn2, Cas4, and Cas9 proteins (Figure 1). These proteins work in conjunction with trans-activated CRISPR RNAs (tracrRNAs), which are encoded within the sequence environment adjacent to the CRISPR array (Jiang and Doudna, 2015). The global gastrointestinal pathogen Campylobacter jejuni possesses a Type II-C CRISPR system. A recent survey of the available C. jejuni genome sequences reported that the Type II-C CRISPR system was almost universally present, with the exception that it was absent in representatives of the MLST clonal complex 42 (Pearson et al., 2015). Analysis of the distribution of CRISPR arrays in C. jejuni (n = 3746) and Campylobacter coli (n = 486) genome sequences indicated that CRISPR systems of C. coli strains isolated from agricultural environments were more closely related to those of C. jejuni rather than the CRISPR system encoded by C. coli isolates of non-agricultural origin. It is suggested that the shared environmental niche has driven the acquisition of a specific CRISPR system within and between circulating C. jejuni and C. coli (Pearson et al., 2015). Due to the almost ubiquitous distribution of CRISPRs and their associated genes within C. jejuni it may prove that these systems can be adopted as a means of typing different strains. However, for C. coli, the limited distribution of the CRISPR systems would prove to be a barrier to any successful CRISPR-based typing scheme (de Cárdenas et al., 2015).
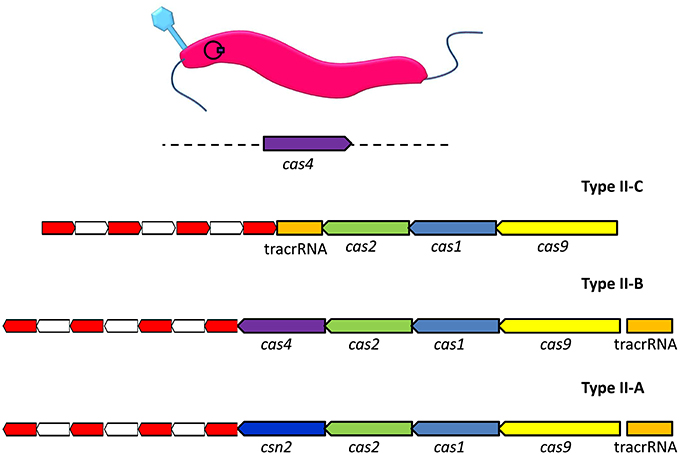
Figure 1. Organization of the genomic loci of representative Type II CRISPR-Cas systems. The type II-C arrangement represents the Campylobacter jejuni carrier state with the addition of bacteriophage encoded cas4 in trans. The red and white sections represent, respectively, direct repeats and spacer sequences. The analogous genes in the Type II systems are labeled and color coded, where Type II-B is Legionella pneumophila and Type II-A Streptococcus thermophilus.
By definition Type II-C CRISPR systems encode Cas1 and Cas2 nucleases for protospacer acquisition, tracrRNA which acts as a guide for targeting recognized foreign DNA, and Cas 9 protein which provides the immunity function (Chylinski et al., 2013; Plagens et al., 2015). The production of antisense CRISPR RNA requires the presence of host-encoded RNase III in Type II-C CRISPR systems. Subsequent reinfection events trigger the RNA-mediated targeting of Cas9 endonuclease to foreign DNA. This is facilitated by recognition of the dsRNA molecule that arises following complementary base pairing of tracrRNA to the 3′-end of the processed antisense crRNA (Chylinski et al., 2014).
The discovery of CRISPR-Cas systems has benefited molecular biology and the life sciences tremendously. Targeted genome editing utilizing CRISPR Cas9-based technologies has led to a revolution in functional genomics and how genes may be analyzed (Hsu et al., 2014). Ultimately the development of next generation technologies may well lead to major breakthroughs in our understanding of gene expression in health and disease (Sontheimer and Barrangou, 2015). The ability of Cas9 protein to introduce double-stranded breaks in target DNA sequences is a key component of CRISPR-based technologies. The precision afforded by Cas9-mediated DNA editing has provoked intense research in the fields of gene therapy (Barrangou, 2015). By combining Cas9 with a guide RNA, a chimeric RNA molecule consisting of tracrRNA:CRISPR RNA, it is possible to introduce breaks in dsDNA molecules in a highly-reproducible precise manner. DNA cleavage is achieved via the action of two Cas9 nuclease domains (HNH and RuvC-like), and with the aid of bespoke guide RNA transcripts it was found possible to target any given DNA sequence (Jinek et al., 2012).
However, functional studies of CRISPR-Cas systems in prokaryotes suggest there is a degree of complexity as to how they operate in nature. The evolutionary consequences of possessing CRISPR-Cas systems have required that both prokaryotes and phages adopt mechanisms capable of countering the potentially deleterious effects of CRISPR immunity. Recently it was shown that within a population of S. epidermidis RP62a, and in the presence of an invading plasmid, a sub-fraction of the culture was actively deleting its Type III-A CRISPR system. The obvious benefit to the mutant population is that it may acquire beneficial traits associated with the uptake of the plasmid. Conversely, the population which retains the system would survive plasmid-invasion by CRISPR-mediated immunity if uptake was proven to be deleterious (Jiang et al., 2013).
It is becoming evident that the evasion of CRISPR-mediated immunity is another key mechanism involved in the phage/bacteria co-evolutionary arms race that should be added to the established mechanisms including restriction–modification systems, alteration of outer membrane proteins countered by changes to adsorption apparatus, and abortive infection mechanisms (Plagens et al., 2015). For bacteriophages, the constant selective pressures imposed on their replicative success drive this evolutionary process, which has resulted in the emergence of a diverse array of anti-CRISPR mechanisms. Phage-encoded genes have the ability to inactivate the Type I-E and I-F CRISPR systems of Pseudomonas aeruginosa (Bondy-Denomy et al., 2013; Pawluk et al., 2014). The development of single nucleotide polymorphisms in protospacer adjacent motif (PAM) sequences prevents targeting of phage 2972 by the Type II-A CRISPR system of Streptococcus thermophilus DGCC7710 (Sun et al., 2013). In the presence of constant CRISPR-mediated selective pressure, phage 2972 was found to accumulate mutations in the genomic regions that were selectively targeted by the S. thermophilus DGCC7710 CRISPR system (Paez-Espino et al., 2015). Myoviridae phages which infect Vibrio cholerae have been found to encode a complete Type I-F CRISPR system. Acquisition of the system resulted in the ability to overcome V. cholerae anti-phage responses encoded within a pathogenicity island (Seed et al., 2013). As more CRISPR-Cas systems are studied within the Bacteria and Archaea it will be interesting to see what other resistance mechanisms emerge.
So far applications of CRISPR technologies and studies aimed at modifying lytic phages seem to be lacking behind the new-found roles for Cas9-technologies in genome engineering. Recently however an elegant study involving the above-mentioned S. thermophilus DGCC7710 and phage 2972 showed how Cas9-technologies can be used for the study of lytic phages (Martel and Moineau, 2014). The Cas9 protein encoded within the Type II-A CRISPR system of S. thermophilus DGCC7710 is already widely used in genome engineering (Jinek et al., 2012). Initially point-mutations were introduced into specific regions of the phage 2972 genome with an extremely high degree of efficiency. Introduction of a small 2 bp deletion in orf39 led to a frameshift and successful inactivation of the target gene. Two further large deletions in orf39 were also possible and as adjudged by smaller plaque sizes in phage 2972 orf39 mutants it was concluded that this gene may be important for fitness of the phage. Of significant interest one of the main concepts to emerge from the study was the successful deletion of an open reading frame (orf33) previously shown to be dispensable, and its replacement with a bacterial gene. The gene introduced was the lactococcal methyltransferase LladCHIA which was found to be fully functional following insertion in phage 2972. It was observed that immunity to restriction via in vivo methylation of phage DNA was accomplished by phages bearing the introduced gene (Martel and Moineau, 2014). The prospect of efficient engineering of lytic phages capable of overcoming restriction-modification systems of any given host may be of exceptional use to industries such as phage therapeutics. Innate resistance to phages is often mediated via restriction–modification and alleviating this obstacle may allow advances to be made in trying to find novel phage-derived treatments for bacterial pathogens of humans, animals, and plants.
Aside from their classical function of providing a protective mechanistic response to invading genetic elements, non-canonical roles for CRISPR-Cas systems have been proposed. The ability to respond to invasive DNA elements such as plasmids and phage genomes has led to the discovery that stress responses triggered by disruption of membrane integrity can lead to activation of CRISPR-Cas systems (Ratner et al., 2015). Several important observations have delineated environmental signals that can be utilized by bacteria to regulate the expression of CRISPR-Cas systems. Transcriptional activation of CRISPR systems in Salmonella enterica and Escherichia coli can be mediated by the global DNA binding histone-like nucleoid structural protein (H-NS). Under normal circumstances H-NS represses CRISPR-Cas transcription; however in response to breaches in the integrity of the cell membrane H-NS becomes inactivated leading to transcription of the CRISPR-Cas system, and enabling the bacteria to respond to lethal infection or the acquisition of selfish genetic elements that can either compromise the microorganisms competitive ability or threaten the integrity of the genome (Westra et al., 2010; Medina-Aparicio et al., 2011).
Interestingly, other functional concepts have emerged from analyzing CRISPR-Cas biology. For the intracellular human pathogen Francisella novicida, the ability to survive exposure to potentially lethal insults such as antibiotics, membrane degradation, or perturbation, and host immunological responses is essential. Not only does F. novicida have to contend with cationic antimicrobial peptides, it is also required to have the ability to survive within the macrophage following phagocytosis wherein the bacterium needs to escape the phagosome in order to replicate in the macrophage cytosolic compartment. Intriguingly the Cas9 protein encoded within the Type II-B CRISPR system of F. novicida appears to be essential to overcome several barriers to successful completion of the intracellular lifecycle. These actions include resistance to the antibiotic polymyxin B (functionally akin to host cationic antimicrobial peptides), evasion of toll-like receptor 2 signaling and dampening of the inflammasome were all found to be reliant on the presence of a functioning Cas9 protein, tracrRNA and a scaRNA (Sampson et al., 2014).
The Campylobacter Bacteriophage Carrier State
Analysis of bacteriophages that target Campylobacter spp. has led to many interesting insights into how they interact with their bacterial hosts. In some cases, what could be deemed as typical predator/prey behavior associated with lytic phages is foregone and is replaced by an equilibrium known as the carrier state. The nature of the carrier state is such that rather than committing to lysis the phage population is stably maintained in association with its host. Unlike temperate phages which commit to lysogeny and integrate their genomes into that of their host, Campylobacter phages remain associated as freely-replicating infective particles which propagate on a small fraction of the host population (Siringan et al., 2014; Brathwaite et al., 2015). This phenomenon has precedents in the literature with reports of bacteriophage carrier states being identified in Shigella dysenteriae (Li et al., 1961), Brucella abortus (Jones et al., 1962), Proteus mirabilis (Coetzee and Hawtree, 1962), mycobacteria (Baess, 1971), and most recently using phage P22 and Salmonella typhimurium LT2 (Cenens et al., 2013).
KEY CONCEPT 1. Bacteriophage Carrier State
The carrier state describes mixtures of bacteria and of bacteriophages that remain associated in a stable equilibrium. Carrier state cultures contain subpopulations of bacteria that either support bacteriophage replication or are resistant but can act as continuous founders of the sensitive subpopulation.
Analysis of carrier state cultures of C. jejuni PT14 confirmed that the phages and bacterial host remain associated in stable ratios even following serial passage representing multiple generations. Initial identification of the bacteriophage carrier state in C. jejuni PT14 arose following studies that were initiated to monitor the ability of Class III Campylobacter phages to disperse Campylobacter biofilms (Siringan et al., 2011). Bacteria recovered from the biofilms were found to spontaneously produce phage following disruption of the structure prompting further investigation of the origins of the phage burst. A combination of PFGE and Southern blot analysis was used in order to discount the unlikely event that otherwise virulent bacteriophage had formed lysogens by integration of Campylobacter phage genomic DNA. Non-integrated phage genomic DNAs (band size ~140 kb) were visible on PFGE gels containing SmaI-digested C. jejuni genomic DNA migrating alongside undigested C. jejuni DNA. Hybridization of Southern blots with Campylobacter phage specific probes for CP8 and CP30A confirmed that the 140 kb bands represented phage DNA and that the SmaI-fragments arising from bacterial chromosomal DNA contained no phage-hybridizing DNA sequences. This approach revealed that the phage genomes were present in carrier-state cultures as non-integrated genetic elements and absent from control cultures. In vivo studies showed that the carrier state bacteria were unable to colonize the poultry gut following analysis of the caecal contents of experimentally inoculated chickens (Siringan et al., 2014). Observations of planktonic growth phase-dependent parameters indicated that C. jejuni PT14 growing in the presence of bacteriophages CP8 and CP30A showed reduced fitness in vitro. Initially, bacteria growing during the early-logarithmic growth phase were found to be non-motile in carrier state cultures compared to non-carrier state cultures (uninfected controls). Coincident with the impairment in motility, bacteria isolated from early-logarithmic growth cultures displayed a vastly reduced ability to adhere to and infect HCA-7 colonic epithelial cells (Brathwaite et al., 2015). Transcriptional studies of the carrier state cultures revealed significant reduction in the expression of the flaA gene encoding the major flagellin protein of C. jejuni. Instead the carrier state cultures show increased σ54-dependent expression of the flaB gene, which encodes for a second normally minor flagellin protein (Brathwaite et al., 2015). Translation of the alternative flagellin genes results in proteins with similar amino acid sequences; it is therefore likely that the change in regulation is accompanied with other functional changes in the control of motility.
Campylobacter Phage Cas4 Protein
Recently we reported the presence of a conserved Cas4-like ortholog in the genomes of both Class II and Class III Campylobacter phages (Hooton and Connerton, 2014). Cas4 proteins are reported as playing a role during the acquisition of protospacers in Type II-B CRISPR systems. The structural basis of Cas4 monomer is reliant on the presence of four conserved cysteines which together provide the basis for formation of a 4Fe-4S cluster. This structural arrangement often referred to as an “iron-staple” is exemplified in AddB nuclease of Bacillus subtilis and RecB nuclease of E. coli. A decameric toroidal configuration forms the quaternary structure of the protein which is observed to consist of five Cas4 dimers (Lemak et al., 2013). The in vitro activity of Cas4 protein isolated from the archaeon Sulfolobus solfataricus SSO0001 is reported to be a 5′–3′ single stranded DNA exonuclease. Specifically, the Cas4 protein was proposed to function in the generation of protospacer 3′ single stranded DNA ends that have the potential to undergo recombination with the CRISPR array (Zhang et al., 2012). Furthermore, the Cas4 protein was also shown to have the ability to perform ATP-independent DNA unwinding against single stranded DNA (Lemak et al., 2013). Recently, it was reported that a Cas4 protein isolated from the hyperthermophilic archaeon Pyrobaculum calidifontis contained a 2Fe-2S cluster (Lemak et al., 2014). In the case of 4Fe-4S Cas4 proteins the four conserved cysteines are absolutely required for maintaining structural integrity of the iron sulfur cluster and activity of the protein (Zhang et al., 2012). For 2Fe-2S Cas4, the nuclease function of the protein was found to remain intact following mutation of these residues, even though the iron sulfur cluster was lost (Lemak et al., 2014). Various nuclease domains/motifs are present in Cas4 proteins including RecB-like nuclease domains, PD(D/E)XK nuclease domain, and a QXXXY motif. In silico analysis of Cas4-like proteins in Campylobacter phages showed that all the domains and motifs described above are present. The conserved cysteine residues were found to be intact and even though very little amino acid homology is observed between intervening sequences in other Campylobacter phages, the core nuclease domains are also present. Analysis of phylogenetic trees drawn using Cas4 primary sequence data for several prokaryotes and Campylobacter phages highlighted the relatedness between their respective proteins. The addition of a phage-encoded Cas4 protein into the apparatus of the C. jejuni PT14 Type II-C CRISPR system (Cas1, Cas2, Cas9, and tracrRNA) creates a putative system that phenotypically resembles Type II-B CRISPR systems. Prior to analysis it could be hypothesized that if the carrier state was to function in a manner that allowed the survival of phages and their bacterial hosts then the host CRISPR system must be in some way inactive, or functioning in an atypical fashion. By combining the deep sequencing strategies of whole genome sequencing and RNA-Seq it was possible to identify rare molecular events which took place during carrier state associations between C. jejuni PT14 and Campylobacter phages CP8 and CP30A.
A phylogenetic tree drawn from the alignment of amino acid sequences of Class II and Class III Campylobacter phage Cas4-like proteins with a diverse selection of bacterial and archael Cas4 proteins is shown in Figure 2. The Campylobacter phages are found to cluster together based on their genome size such that Class II (170–190 kb) and Class III (110–150 kb) phages have their own branches. C. coli phages Cpt10 and vBCcoM-IBB_35 are also observed to cluster on their own sub-branch of the Class II lineage. A selection of Campylobacter spp. Cas4 proteins were included in the analysis in order to evaluate if the Campylobacter phage-encoded Cas4 protein originated within the bacterial genus. Analysis of aligned amino acid sequences shows that conservation between phage and bacterial Cas4 protein appears restricted to the conserved cysteine residues and nuclease domains found within the proteins.
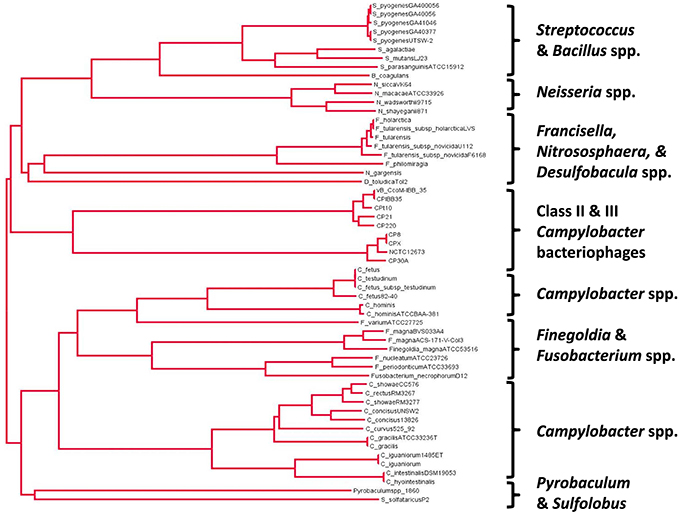
Figure 2. Phylogenetic analysis of the Cas4 protein family related to the proteins encoded in class II and III Campylobacter bacteriophages using a neighbor-joining tree methodology (ClustalW2 alignment and FigTree v1.4.2.).
KEY CONCEPT 2. Phage-encoded Cas4 nuclease
Cas4 proteins have a functional role in the protospacer acquisition of Type II-B CRISPR systems. An analogous role for a phage-encoded Cas4 could confer a phage activated CRISPR immunity and/or autoimmunity in host bacteria with alternative Type II CRISPR systems.
C. jejuni Spacer Acquisition
The unexpected finding of a Cas4-like protein in Campylobacter phages prompted us to analyze the CRISPR array of C. jejuni PT14 specifically during carrier state infections. Bioinformatic analysis of Cas4-like proteins of Class II and III Campylobacter phages showed they were conserved throughout, and the proteins all bore the hallmarks of Cas4 proteins. For the analysis two well characterized phages that were identified as encoding a Cas4-like protein (Class III Campylobacter phages CP8 and CP30A) were selected on the basis of their ability to establish carrier state cultures with C. jejuni PT14, and that the host CRISPR array did not already feature spacers directed at these phages (Siringan et al., 2014; Brathwaite et al., 2015). However, evidence suggests that it is possible to acquire spacer sequences directed at the genomes of the phages CP8 and CP30A since corresponding spacer DNA sequences have been noted in CRISPR arrays of C. jejuni genomes deposited in public databases (Pearson et al., 2015). Using these two phages it was possible to observe de novo spacer acquisition but the spacers consisted solely of host-derived sequences. In the absence of phages CP8 or CP30A no alterations were observed in the C. jejuni PT14 CRISPR array with the native self-derived spacers remaining fixed.
KEY CONCEPT 3. Activated self-spacer acquisition
If the CRISPR array can fix host spacer sequences then the interference mechanism can target the host genome itself resulting in cell death, a situation termed CRISPR-mediated autoimmunity.
To try to gain an understanding of the impact of carrier state infection upon C. jejuni PT14 an in-depth molecular analysis of the Type II-C CRISPR array was performed. Encoded upstream of the gene moe2 (A911_07320) in C. jejuni PT14—Acc. No. CP003831 (Brathwaite et al., 2013) is a CRISPR array containing four 36 bp direct repeats with three intervening 30 bp spacer sequences. Analysis of the whole genome sequence of C. jejuni PT14 revealed that the 30 bp spacers found within the CRISPR array were representative of self-genes, rather than the expected plasmid or phage sequences. Intriguingly two of the spacers have homology with genes that are associated with externally presented proteins—peptidoglycan-associated lipoprotein Omp18 (18/30 nucleotides—A911_00540) and apolipoprotein N-acyltransferase (16/30 nucleotides—A911_05300). Analysis of RNA-Seq reads for these sequences indicated that apolipoprotein N-acyltransferase is transcribed at very low levels in C. jejuni PT14. This may allow the bacterium to evade immunological detection by toll like receptor 2 (TLR2) specifically by down-regulating the production of stimulatory acylated lipoproteins. Further spacer sequences were identified which matched the sequences of an isoleucyl tRNA synthetase (15/30 nucleotides—A911_05135), and a di/tripeptide transporter (12/30 nucleotides—A911_03195). The acquisition of spacer sequences of host-origin in carrier state cultures was found to be in stark contrast to the fixed nature of the C. jejuni PT14 CRISPR array. For CP8 carrier state cultures two of the spacers acquired showed complete matches to genes which are associated with motility and capsular polysaccharide biosynthesis. The motility-associated spacer was found to target the gene encoding the motility-associated protein PseE (30/30 nucleotides—A911_06495) which may have some bearing on the lack of motility observed in C. jejuni PT14 carrier state cultures. Loss of invasion and adherence capabilities of carrier state cultures may also be influenced by acquisition of the spacer targeting the capsular polysaccharide biosynthesis gene (30/30 nucleotides—A911_06909). A similar scenario was observed for CP30A carrier state cultures in which several different spacers were observed to target genes exclusively of host origin with perfect or near-perfect matches to the C. jejuni PT14 genome. The most notable acquisition was multiple spacers targeting the genes ADP-heptose-lipooligosaccharide-heptosyltransferase I and II.
The terminal direct repeat of the CRISPR array is positioned 144 bp upstream of the genes cas2 (A911_07325), cas1 (A911_07330), and cas9 (A911_07335) which are accompanied by tracrRNA. The generation of pre-crRNAs occurs via transcription from a −10 transcriptional start site (5′-TAAAAAT-3′) located 116 bp upstream of the CRISPR array. Independent promoters with similarity to extended −10 transcriptional start sites (5′-GGTAAAAAT-3′) are also found preceding each direct repeat. A hallmark of Type II CRISPR systems is the co-ordination of endogenous RNase III/tracrRNA during processing to produce mature crRNAs during the biogenesis step (Deltcheva et al., 2011; Dugar et al., 2013) Transcriptomic analysis of early exponential phase C. jejuni PT14 carrier state cultures showed that several genes associated with RNA biogenesis and RNA processing are differentially upregulated including RNase III (Brathwaite et al., 2015).
Small RNAs
Differences in the expression of non-coding RNAs were evident in the carrier state cultures compared to wild type C. jejuni PT14, most notably the accumulation of sRNAs. RNA-seq analysis identified sRNAs with heterogeneity at their termini implying they were generated by RNase processing or perhaps mis-processing. For example, PT14CP30 carrier state cultures contained an over-represented sRNA mapping to an intergenic genomic sequence between tRNAs (A911_t08346–A911_t08348), and exhibiting a 40/51 bp match to the CP30 phage genome. Association between complementary regions of bacterial and phage RNAs could lead to the preservation of the paired regions and exoribonuclease processing of the unpaired ends leading to the observed terminal heterogeneity. Other antisense sRNAs of chromosomal origin have the potential to bind phage transcripts (>25 nucleotide sequence matches) and act as trans-encoded inhibitory RNAs to suppress phage expression. Duplex RNA structures would also require cleavage by RNase III. Carrier state cultures exhibit a coordinate increase in the transcription of genes encoding ribosomal proteins that would also require an increase in RNase III to process rRNAs for the formation of ribosomes (Brathwaite et al., 2015). However, increased levels of RNase III in the carrier state cultures could lead to dysregulation of any regulatory RNAs that have the potential to fine tune regulation in a species that is noted for its minimal quota of regulatory genes. Non-coding RNAs CJnc10, CJnc60, CJnc160, and CJnc170 reported for C. jejuni NCTC11168 (Dugar et al., 2013) were conserved in C. jejuni PT14 but of these CJnc10 and CJnc170 RNAs were all but absent in the carrier state cultures. CJnc10 and CJnc170 were recently reported to be expressed from σ28 promoters and to effect translation of σ54-dependent transcripts through conserved complementary sequences in their 5′ untranslated regions (Le et al., 2015).
KEY CONCEPT 4. Small RNA effectors of carrier state gene expression
Antisense sRNAs have the potential to bind phage or host transcripts and act as trans-encoded inhibitory RNAs to suppress gene expression.
Future Perspectives
The consequences of expressing a phage-encoded Cas4-like protein in C. jejuni are intriguing and merit further study. The proposed role of Cas4-like protein as a mediator of the incorporation of host-derived CRISPR spacers would require controlled gene expression to suppress CRISPR-mediated autoimmunity. However, we note that Figure 2 contains alternative Campylobacter species in the second major clade that carry cas4-like coding sequences. These are also subject to modifications in genomic content, for example Campylobacter fetus subsp. fetus features a CRISPR array but this is partially deleted in C. fetus subsp. venerealis (Ali et al., 2012). The relationships between CRISPR-cas related functions of the genus will require further functional analysis.
The intricate relationship between Campylobacters and their associated bacteriophage has resulted in insurance mechanisms that allow for long-term replicative success of predator and prey. Short-term effects such as transient bacterial resistance to virulent phage infection following Mu prophage-driven genomic rearrangements may skew the replication bias toward the bacterium (Scott et al., 2007), albeit with a reduction in poultry colonization potential. However, restoration to phage sensitivity allows the wildtype population to recolonize poultry at the expected levels. In the carrier state virulent bacteriophage replication is accommodated whilst a subpopulation of bacteria remain resistant. The carrier state bacteria themselves are poor colonizers of poultry but are able to deliver bacteriophage to sensitive pre-colonized inhabitants (Siringan et al., 2014). Population level studies of bacteriophage/host interactions may shed yet further light on the nature of the success of Campylobacter as a major zoonotic foodborne pathogen.
Author Contributions
SH, KB, and IC reviewed experimental and computational data. SH and IC wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Support from the BBSRC of the UK is gratefully acknowledged. KB was the recipient of a studentship from the Ministry of Education Barbados.
Author Biography

References
Ali, A., Soares, S. C., Santos, A. R., Guimarães, L. C., Barbosa, E., Almeida, S. S., et al. (2012). Campylobacter fetus subspecies: comparative genomics and prediction of potential virulence targets. Gene 508, 145–156. doi: 10.1016/j.gene.2012.07.070
Baess, I. (1971). Report on a pseudolysogenic mycobacterium and a review of the literature concerning pseudolysogeny. Acta Pathol. Microbiol. Scand. B Microbiol. Immunol. 79, 428–434. doi: 10.1111/j.1699-0463.1971.tb00084.x
Barrangou, R. (2015). The roles of CRISPR-Cas systems in adaptive immunity and beyond. Curr. Opin. Immunol. 32, 36–41. doi: 10.1016/j.coi.2014.12.008
Bondy-Denomy, J., Pawluk, A., Maxwell, K. L., and Davidson, A. R. (2013). Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 493, 429–432. doi: 10.1038/nature11723
Brathwaite, K. J., Siringan, P., Connerton, P. L., and Connerton, I. F. (2015). Host adaption to the bacteriophage carrier state of Campylobacter jejuni. Res. Microbiol. 166, 504–515. doi: 10.1016/j.resmic.2015.05.003
Brathwaite, K. J., Siringan, P., Moreton, J., Wilson, R., and Connerton, I. F. (2013). Complete genome sequence of universal bacteriophage host strain Campylobacter jejuni subsp. jejuni PT14. Genome Announc. 1:e00969–13. doi: 10.1128/genomea.00969-13
Cenens, W., Mebrhatu, M. T., Makumi, A., Ceyssens, P. J., Lavigne, R., Van Houdt, R., et al. (2013). Expression of a novel P22 ORFan gene reveals the phage carrier state in Salmonella typhimurium. PLoS Genet. 9:e1003269. doi: 10.1371/journal.pgen.1003269
Chylinski, K., Le Rhun, A., and Charpentier, E. (2013). The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems. RNA Biol. 10, 726–737. doi: 10.4161/rna.24321
Chylinski, K., Makarova, K. S., Charpentier, E., and Koonin, E. V. (2014). Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res. 42, 6091–6105. doi: 10.1093/nar/gku241
Coetzee, J. N., and Hawtree, A. O. (1962). A change in phenotype associated with the bacteriophage carrier statein a strain of Proteus mirabilis. Nature 194, 1196–1197 doi: 10.1038/1941196b0
de Cárdenas, I., Fernández-Garayzábal, J. F., de la Cruz, M. L., Domínguez, L., Ugarte-Ruiz, M., and Gómez-Barrero, S. (2015). Efficacy of a typing scheme for Campylobacter based on the combination of true and questionable CRISPR. J. Mic. Methods 119, 147–153. doi: 10.1016/j.mimet.2015.10.020
Deltcheva, E., Chylinski, K., Sharma, C. M., Gonzales, K., Chao, Y., Pirzada, Z. A., et al. (2011). CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471, 602–607. doi: 10.1038/nature09886
Dugar, G., Herbig, A., Förstner, K. U., Heidrich, N., Reinhardt, R., Nieselt, K., et al. (2013). High-resolution transcriptome maps reveal strain-specific regulatory features of multiple Campylobacter jejuni isolates. PLoS Genet. 9:e1003495. doi: 10.1371/journal.pgen.1003495
Hochstrasser, M. L., and Doudna, J. A. (2015). Cutting it close: CRISPR-associated endoribonuclease structure and function. Trends Biochem. Sci. 40, 58–66. doi: 10.1016/j.tibs.2014.10.007
Hooton, S. P., and Connerton, I. F. (2014). Campylobacter jejuni acquire new host-derived CRISPR spacers when in association with bacteriophages harboring a CRISPR-like Cas4 protein. Front. Microbiol. 5:744. doi: 10.3389/fmicb.2014.00744
Hsu, P. D., Lander, E. S., and Zhang, F. (2014). Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278. doi: 10.1016/j.cell.2014.05.010
Jiang, F., and Doudna, J. A. (2015). The structural biology of CRISPR-Cas systems. Curr. Opin. Struct. Biol. 30, 100–111. doi: 10.1016/j.sbi.2015.02.002
Jiang, W., Maniv, I., Arain, F., Wang, Y., Levin, B. R., and Marraffini, L. A. (2013). Dealing with the evolutionary downside of CRISPR immunity: bacteria and beneficial plasmids. PLoS Genet. 9:e1003844. doi: 10.1371/journal.pgen.1003844
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
Jones, L. M., McDuff, C. R., and Wilson, J. B. (1962). Phenotypic alterations in the colonial morphology of Brucella abortus due to a bacteriophage carrier state. J. Bacteriol. 83, 860–866.
Koonin, E. V., and Krupovic, M. (2015). Evolution of adaptive immunity from transposable elements combined with innate immune systems. Nat. Rev. Genet. 16, 184–192. doi: 10.1038/nrg3859
Krupovic, M., Makarova, K. S., Forterre, P., Prangishvili, D., and Koonin, E. V. (2014). Casposons: a new superfamily of self-synthesizing DNA transposons at the origin of prokaryotic CRISPR-Cas immunity. BMC Biol. 12:36. doi: 10.1186/1741-7007-12-36
Le, M. T., Van Veldhuizen, M., Porcelli, I., Bongaerts, R. J., Gaskin, D. J., Pearson, B. M., et al. (2015). Conservation of sigma28-dependent non-coding RNA paralogs and predicted sigma54-dependent targets in thermophilic Campylobacter species. PLoS ONE 10:e0141627. doi: 10.1371/journal.pone.0141627
Lemak, S., Beloglazova, N., Nocek, B., Skarina, T., Flick, R., Brown, G., et al. (2013). Toroidal structure and DNA cleavage by the CRISPR-associated [4Fe-4S] cluster containing Cas4 nuclease SSO0001 from Sulfolobus solfataricus. J. Am. Chem. Soc. 135, 17476–17487. doi: 10.1021/ja408729b
Lemak, S., Nocek, B., Beloglazova, N., Skarina, T., Flick, R., Brown, G., et al. (2014). The CRISPR-associated Cas4 protein Pcal_0546 from Pyrobaculum calidifontis contains a [2Fe-2S] cluster: crystal structure and nuclease activity. Nucleic Acids Res. 42, 11144–11155. doi: 10.1093/nar/gku797
Li, K., Barksdale, L., and Garmise, L. (1961). Phenotypic alterations associated with the bacteriophage carrier state of Shigella dysenteriae. J. Gen. Microbiol. 24, 355–367. doi: 10.1099/00221287-24-3-355
Makarova, K. S., Wolf, Y. I., Alkhnbashi, O. S., Costa, F., Shah, S. A., Saunders, S. J., et al. (2015). An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 13, 722–736. doi: 10.1038/nrmicro3569
Martel, B., and Moineau, S. (2014). CRISPR-Cas: an efficient tool for genome engineering of virulent bacteriophages. Nucleic Acids Res. 42, 9504–9513. doi: 10.1093/nar/gku628
Medina-Aparicio, L., Rebollar-Flores, J. E., Gallego-Hernández, A. L., Vázquez, A., Olvera, L., Gutierrez-Rios, R. M., et al. (2011). The CRISPR/Cas immune system is an operon regulated by LeuO, H-NS, and leucine-responsive regulatory protein in Salmonella enterica serovar Typhi. J. Bacteriol. 193, 2396–2407. doi: 10.1128/JB.01480-10
Paez-Espino, D., Sharon, I., Morovic, W., Stahl, B., Thomas, B. C., Barrangou, R., et al. (2015). CRISPR immunity drives rapid phage genome evolution in Streptococcus thermophilus. MBio 6:e00262–15. doi: 10.1128/mBio.00262-15
Pawluk, A., Bondy-Denomy, J., Cheung, V. H., Maxwell, K. L., and Davidson, A. R. (2014). A new group of phage anti-CRISPR genes inhibits the type I-E CRISPR-Cas system of Pseudomonas aeruginosa. MBio 5, e00896. doi: 10.1128/mBio.00896-14
Pearson, B. M., Louwen, R., Van Baarlen, P., and Van Vliet, A. H. (2015). Differential distribution of Type II CRISPR-Cas systems in agricultural and nonagricultural Campylobacter coli and Campylobacter jejuni isolates correlates with lack of shared environments. Genome Biol. Evol. 7, 2663–2679. doi: 10.1093/gbe/evv174
Plagens, A., Richter, H., Charpentier, E., and Randau, L. (2015). DNA and RNA interference mechanisms by CRISPR-Cas surveillance complexes. FEMS Microbiol. Rev. 39, 442–463. doi: 10.1093/femsre/fuv019
Ratner, H. K., Sampson, T. R., and Weiss, D. S. (2015). I can see CRISPR now, even when phage are gone: a view on alternative CRISPR-Cas functions from the prokaryotic envelope. Curr. Opin. Infect. Dis. 28, 267–274. doi: 10.1097/QCO.0000000000000154
Sampson, T. R., Napier, B. A., Schroeder, M. R., Louwen, R., Zhao, J., Chin, C. Y., et al. (2014). A CRISPR-Cas system enhances envelope integrity mediating antibiotic resistance and inflammasome evasion. Proc. Natl. Acad. Sci. U.S.A. 111, 11163–11168. doi: 10.1073/pnas.1323025111
Scott, A. E., Timms, A. R., Connerton, P. L., Loc Carrillo, C., Adzfa Radzum, K., and Connerton, I. F. (2007). Genome dynamics of Campylobacter jejuni in response to bacteriophage predation. PLoS Pathog. 3:e119. doi: 10.1371/journal.ppat.0030119
Seed, K. D., Lazinski, D. W., Calderwood, S. B., and Camilli, A. (2013). A bacteriophage encodes its own CRISPR/Cas adaptive response to evade host innate immunity. Nature 494, 489–491. doi: 10.1038/nature11927
Selle, K., and Barrangou, R. (2015). CRISPR-based technologies and the future of food science. J. Food Sci. 80, R2367–2372. doi: 10.1111/1750-3841.13094
Siringan, P., Connerton, P. L., Cummings, N. J., and Connerton, I. F. (2014). Alternative bacteriophage life cycles: the carrier state of Campylobacter jejuni. Open Biol. 4, 130200. doi: 10.1098/rsob.130200
Siringan, P., Connerton, P. L., Payne, R. J., and Connerton, I. F. (2011). Bacteriophage-mediated dispersal of Campylobacter jejuni biofilms. Appl. Environ. Microbiol. 77, 3320–3326. doi: 10.1128/AEM.02704-10
Sontheimer, E. J., and Barrangou, R. (2015). The bacterial origins of the CRISPR genome-editing revolution. Hum. Gene Ther. 26, 413–424. doi: 10.1089/hum.2015.091
Sun, C. L., Barrangou, R., Thomas, B. C., Horvath, P., Fremaux, C., and Banfield, J. F. (2013). Phage mutations in response to CRISPR diversification in a bacterial population. Environ. Microbiol. 15, 463–470. doi: 10.1111/j.1462-2920.2012.02879.x
Westra, E. R., Pul, U., Heidrich, N., Jore, M. M., Lundgren, M., Stratmann, T., et al. (2010). H-NS-mediated repression of CRISPR-based immunity in Escherichia coli K12 can be relieved by the transcription activator LeuO. Mol. Microbiol. 77, 1380–1393. doi: 10.1111/j.1365-2958.2010.07315.x
Keywords: CRISPR, ncRNA, campylobacter, bacteriophage, carrier state lifecycle, RNA-Seq
Citation: Hooton SPT, Brathwaite KJ and Connerton IF (2016) The Bacteriophage Carrier State of Campylobacter jejuni Features Changes in Host Non-coding RNAs and the Acquisition of New Host-derived CRISPR Spacer Sequences. Front. Microbiol. 7:355. doi: 10.3389/fmicb.2016.00355
Received: 14 December 2015; Accepted: 07 March 2016;
Published: 23 March 2016.
Edited by:
Giovanna Suzzi, Università degli Studi di Teramo, ItalyReviewed by:
Rosalba Lanciotti, University of Bologna, ItalyRosanna Tofalo, University of Teramo, Italy
Copyright © 2016 Hooton, Brathwaite and Connerton. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: ian.connerton@nottingham.ac.uk
†Present Address: Steven P. T. Hooton, Functional and Comparative Genomics, Institute of Integrative Biology, University of Liverpool, Liverpool, UK
 Steven P. T. Hooton
Steven P. T. Hooton Kelly J. Brathwaite
Kelly J. Brathwaite Ian F. Connerton
Ian F. Connerton