- 1Nuclear-Agricultural Sciences, Zhejiang University, Hangzhou, China
- 2Institute of Disease Control and Prevention, Academy of Military Medical Sciences, Beijing, China
Shigella flexneri serotype 2 variant (II:3,4,7,8) was isolated in 2008 and first reported in China in 2013. In the present study, epidemiological surveillance from 2003 to 2013 in China suggested that this serotype first appeared in Guangxi in 2003; it then emerged in Shanghai and Xinjiang in 2004 and in Henan in 2008. Of the 1813 S. flexneri isolates, 58 S. flexneri serotype 2 variant strains were identified. Serotype 2 variant has emerged as a prominent serotype in recent years, with 2a (32.6%), X variant (25.2%), 1a (9.4%), X (6.3%), 2b (5.4%), and 1b (3.6%). According to phenotypic and genotypic analysis, the serotype 2 variant originated from 2a to 2b. A higher antibiotic resistance rate was observed between 2009 and 2013 than that between 2003 and 2008. Among 22 cephalosporin-resistant isolates, blaTEM-1, blaOXA-1, blaCTX-3, blaCTX-14, and blaCTX-79 were detected. Among 22 fluoroquinolone-resistant isolates, a Ser80Ile mutation in parC was present in all of the isolates. Moreover, 21 isolates had three gyrA point mutations (Ser83Leu, His211Tyr, Asp87Asn, or Gly) and one isolate had two gyrA point mutations (Ser83Leu and His211Tyr). The prevalence of His211Tyr in the fluoroquinolone-resistant isolates is concerning, and the mutation was first reported in China. Besides, 22 isolates harbored the aac(6′)-Ib-cr gene, and two isolates harbored qnrS1. In view of the increased epidemic frequency and multidrug-resistant strain emergence, continuous surveillance will be needed to understand the actual disease burden and provide guidance for shigellosis.
Introduction
Shigellosis, caused by Shigella species, is recognized as a major public health burden and continues to be an important cause of diarrheal diseases. Worldwide, Shigella episodes were estimated to be 164.7 million annually, of which, 163.2 million were in developing countries, resulting in 1.1 million deaths (Kotloff et al., 1999). Epidemics generally occur in underdeveloped and developing countries with poor sanitary conditions where transmission from person to person is common, or when food or water is contaminated by the organism (Qu et al., 2012). Despite economic and public health improvements, outbreaks of shigellosis are still reported regularly (Huang et al., 2005; Gaynor et al., 2009; Kuo et al., 2009; Qiu et al., 2011). In China, shigellosis has been ranked third in morbidity, and it has become the number one cause of disease-related death in children (Mathers et al., 2009).
Based on biochemical and serological properties, the genus Shigella is divided into four species or subgroups: S. flexneri, S. dysenteriae, S. boydii, and S. sonnei. Among these subgroups, the epidemic subgroup S. sonnei caused diarrhea in industrialized countries and S. flexneri in developing countries (Kotloff et al., 1999). A previous study showed that the annual shigellosis morbidity rate was 20.28 cases per 100,000 people in mainland China from the national surveillance data of 2009, and S. flexneri (67.3%) and S. sonnei (32.7%) were two major causative species (Sui et al., 2010). S. flexneri is divided into at least 20 serotypes (serotypes 1a, 1b, 1c, 1d, 2a, 2b, 2 variant, 3a, 3b, 4a, 4av, 4b, 5a, 5b, X, Xv, Y, Yv, F6, and 7b) based on the combinations of antigenic determinants present on the O antigen of the cell envelope lipopolysaccharide (Qiu et al., 2013; Sun et al., 2013). Serotypes 1c, 4av, 7b, 1d, Yv, 2 variant and Xv are newly reported serotypes in recent years, and some have caused epidemic-level disease (Talukder et al., 2001; Stagg et al., 2008; Ye et al., 2010). Serotype 1c, first appeared in Bangladesh in 1989, was subsequently found to be prevalent in Bangladesh, Egypt, and Vietnam (Carlin et al., 1989; El-Gendy et al., 1999; Stagg et al., 2008). Serotype Xv was first identified in 2000 and later become the predominant serotype in Henan Province between 2002 and 2006. Serotype Xv was also the most prevalent serotype in Gansu and Anhui in 2007 (Ye et al., 2010). Therefore, the rapid expansion and spread of novel serotypes pose a severe threat to public health in areas where shigellosis is endemic.
The S. flexneri serotype 2 variant (II:3,4,7,8) was first reported by Qiu et al. in 2013 (Qiu et al., 2013). During our routine surveillance of bacillary dysentery from 2003 to 2013, a total of 58 serotype 2 variant strains were identified. This novel serotype first appeared in Guangxi in 2003, and then emerged in other provinces. However, no study has yet extensively characterized the newly emerging strains of serotype 2 variant; therefore, an extensive study is needed to determine the emergence, antimicrobial resistance pattern, and epidemic trends of the serotype 2 variant.
Materials and Methods
Bacterial Isolates, Serotyping, and Biochemical Characterization
Fresh stool samples from diarrhea patients with clinically suspected dysentery were collected in sentinel hospitals. Samples were cultured for Shigella by streaking directly onto Salmonella–Shigella agar and incubated at 37°C for 18 h. Resultant Shigella colonies (colorless, semitransparent, smooth, and moist circular colonies) were routinely grown in a 37°C incubator in Luria–Bertani agar plates (Qu et al., 2012). Then, the strains were submitted to our laboratory for further confirmation. This study was approved by the ethics committee of the Academy of Military Medical Sciences (China), and written approval was also obtained from the patients involved in this study. The isolates were confirmed using API 20E test strips (bioMerieux Vitek, Marcy-l'Etoile, France) following the manufacturer's recommendations. Serotypes of Shigella isolates were further determined with two serotyping kits: a commercially available kit (Denka Seiken, Tokyo, Japan) and monoclonal antibody reagents (Reagensia AB, Stockholm, Sweden).
Multilocus Sequence Typing (MLST)
MLST of these isolates was carried out using the protocols described at http://www.shigatox.net/ecmlst/cgi-bin/index. The PCR amplification conditions were as follows: 94°C for 5 min; 33 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min; and 72°C for 10 min with Ex Taq DNA polymerase (Takara, Dalian, China). The PCR amplicons of 15 housekeeping genes were sequenced, and sequences were edited using SeqMan 7.0. Then, the sequences were uploaded to the EcMLST website for comparison, which allowed us to determine the sequence type.
Antimicrobial Susceptibility Testing
The antimicrobial MICs of 21 antimicrobials including ceftazidime (CAZ), ceftriaxone (CRO), cefepime (FEP), cefoperazone (CFP), cefazolin (CFZ), cefoxitin (FOX), imipenem (IPM), nitrofurantoin (NIT), piperacillin (PIP), ampicillin (AMP), ticarcillin (TIC), tetracycline (TE), tobramycin (TO), gentamicin (GEN), amikacin (AK), aztreonam (ATM), chloramphenicol (C), ticarcillin/clavulanic acid (TIM), levofloxacin (LEV), norfloxacin (NOR), and trimethoprim/sulfamethoxazole (SXT) were determined by broth microdilution using a 96-well microtiter plate (Sensititre, Thermo Fisher Scientific Inc., West Sussex, United Kingdom) according to the manufacturer's instructions, and MIC values follow Clinical and Laboratory Standards Institute recommendations. An Escherichia coli (ATCC 25922) strain was used as the quality control strain.
Pulsed-Field Gel Electrophoresis (PFGE)
All isolates of S. flexneri were analyzed by PFGE according to the standard protocol for S. flexneri developed by the Centers for Disease Control and Prevention of the USA. Salmonella enterica serotype Braenderup H9812 was digested with XbaI and used as molecular weight standard. Slices of agarose plugs were digested with NotI (Takara, Dalian, China) at 37°C for 3 h. Electrophoresis was carried out in 1% agarose SeaKem Gold gel (Lonza, Rockland, ME, USE) with the CHEF Mapper system (Bio-Rad) with the following run parameters: 6 V/cm and a linear increase in switching times from 2.16 to 54.17 s over a period of 20 h. The interpretation of the PFGE patterns was performed with BioNumerics software version 6.0 (Applied Maths, Sint-Martens-Latem, Belgium). A tree indicating relative genetic similarity was constructed based on the unweighted pair group method of averages and a position tolerance of 1.2%.
PCR Amplification of the Antibiotic-Resistant Determinants and Integrons
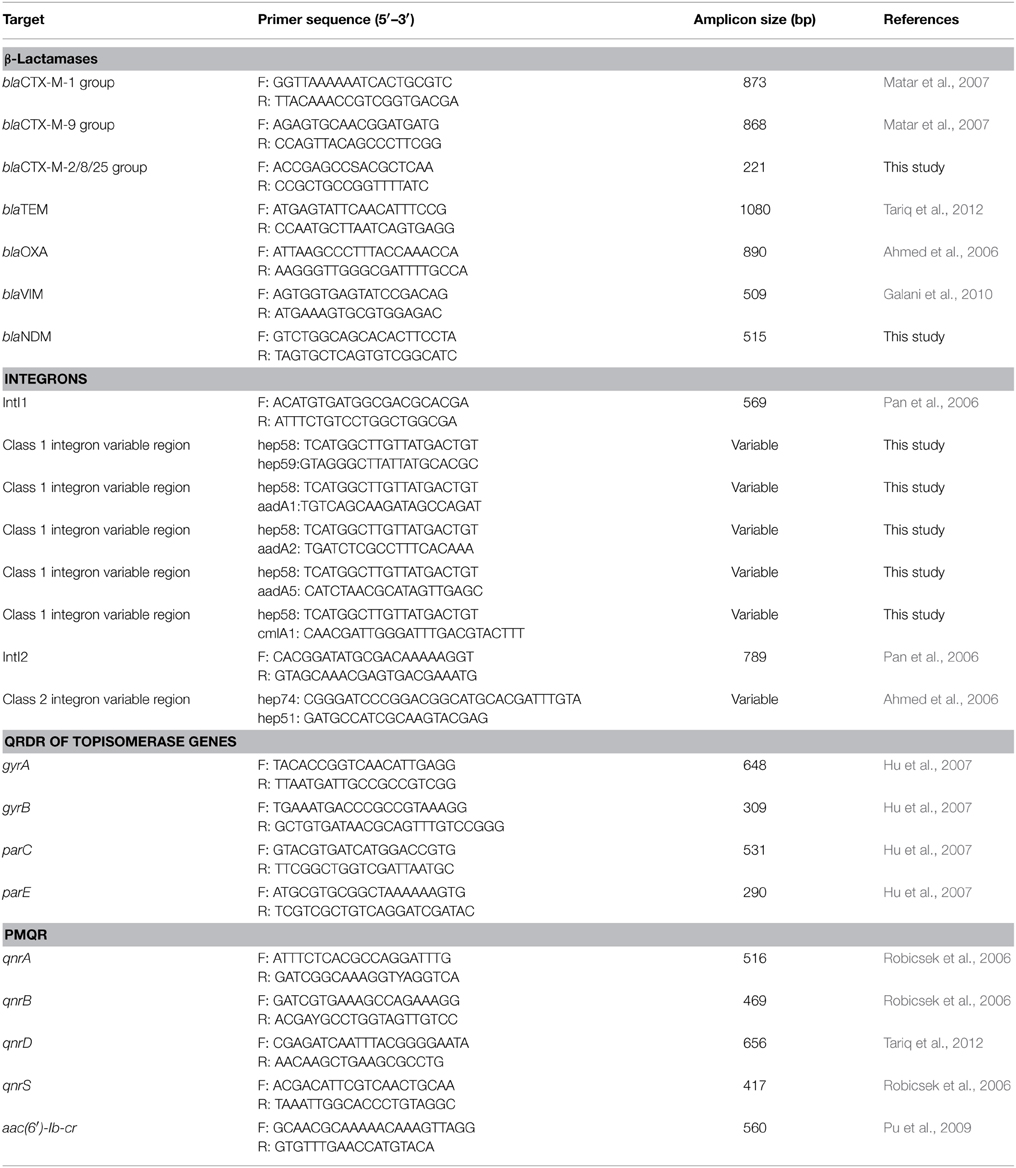
To understand the underlying mechanism conferring resistance to third-generation cephalosporins, PCR assays (Ahmed et al., 2006; Pan et al., 2006; Matar et al., 2007; Galani et al., 2010; Tariq et al., 2012) were used to detect the presence of β-lactamase genes such as blaSHV, blaTEM, blaOXA, and blaCTX−M. The variable regions of class 1 integrons and class 2 integrons were also amplified with primers listed in Table 1. The primers hep58 and hep59, hep74 and hep51 were used to detect the variable regions of class 1 integrons and class 2 integrons, respectively. The reverse primers aadA1, aadA2, aadA5, and cmlA1, together with the forward primer hep58, were used to amplify the gene cassettes of class 1 integron-positive strains. The forward primer hep74 and reverse primer hep51 were used to amplify the gene cassettes of class 2 integron-positive strains. To understand the underlying mechanism conferring resistance to quinolones, plasmid-mediated quinolone resistance determinants (PMQRs) such as qnrA, qnrB, qnrD, qnrS, and aac(6′)-Ib-cr and quinolone resistance–determining regions (QRDRs) of the DNA gyrase (gyrA) and topoisomerase IV (parC) genes were amplified (Robicsek et al., 2006; Hu et al., 2007; Pu et al., 2009; Tariq et al., 2012). Purified PCR fragments were sequenced and analyzed by comparison with sequences in GenBank.
Results
Bacteria Isolation and Biochemical Characterization
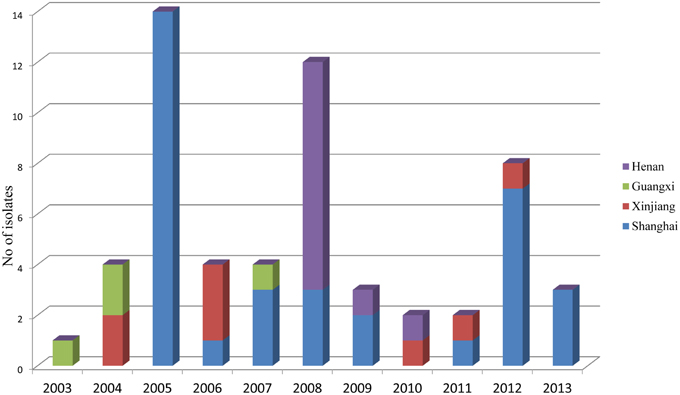
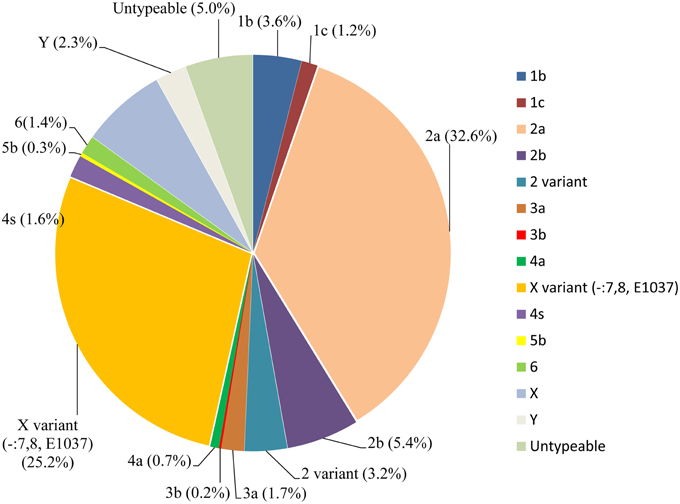
During the routine surveillance of shigellosis in our laboratory, we screened 1813 S. flexneri isolates collected from the eastern, western, southern, northern and central regions of China during 2003–2013. In the 11-year surveillance of Shigella infections, a total of 58 S. flexneri serotype 2 variant isolates were identified. The serotype 2 variant first appeared in Guangxi Province in 2003, then emerged in Shanghai City and Xinjiang Province in 2004, and it was found in Henan Province in 2008. Among the 58 serotype 2 variant isolates, 34 isolates were collected from Shanghai City during the period 2005–2013, 12 isolates were collected from Henan Province during the period 2008–2010, eight isolates were collected from Xinjiang Province during the period 2004–2012, and four isolates were collected from Guangxi Province during the period 2003–2007 (Figure 1; see also Table S1 in the Supplemental Material). Notably, serotype 2 variant has emerged as a common serotype (3.2%), with 2a (32.6%), X variant (25.2%), 1a (9.4%), X (6.3%), 2b (5.4%) and 1b (3.6%) during 2003–2013 (Figure 2). In addition, all examined strains of S. flexneri serotype 2 variant displayed the same typical biochemical features of Shigella species with the ability to ferment glucose, arabinose, mannitol, and melibiose.
Antimicrobial Susceptibility Testing
Fifty-eight S. flexneri serotype 2 variant isolates were tested for MICs with 21 antimicrobials. Resistance to ampicillin was the most common (57, 98.3%), followed by ticarcillin (56, 96.6%), tetracycline (55, 94.8%), chloramphenicol (54, 93.1%), trimethoprim/sulfamethoxazole (25, 43.1%), norfloxacin (22, 37.9%), cefazolin (22, 37.9%), ceftriaxone (20, 34.5%), cefoperazone (17, 29.3%), piperacillin (14, 24.1%), tobramycin (7, 12.1%), gentamicin (5, 8.6%), ticarcillin/clavulanic acid (3, 5.2%), cefoxitin (3, 5.2%), nitrofurantoin (2, 3.4%), aztreonam (2, 3.4%), and levofloxacin (1, 1.7%). None of the isolates were resistant to cefepime, imipenem, ceftazidime, and amikacin. In addition, strains intermediately resistant to ceftazidime, cefoperazone, levofloxacin, norfloxacin, piperacillin, tobramycin, gentamicin, aztreonam, and ticarcillin/clavulanic acid were also observed (Table 2). Furthermore, 57 of 58 S. flexneri serotype 2 variant isolates exhibited multidrug-resistant (MDR; resistant to three or more classes of antimicrobials), and only one strain from Shanghai was not. All of the isolates had 26 antibiotic-resistance profiles, dominated by resistance to TE/TIC/AMP/C (14/58, 24.1%), followed by TE/TIC/AMP/C/SXT (8/58, 13.8%), TE/TIC/AMP/C/SXT/NOR (5/58, 8.6%), and TE/TIC/AMP/C/NOR (3/58, 5.2%) (Table 3).
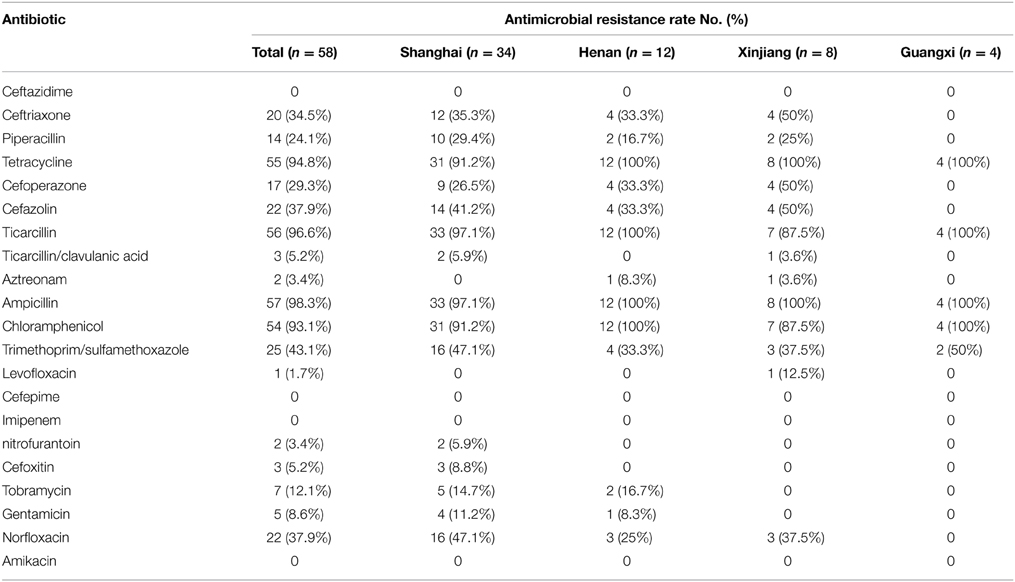
Table 2. Comparison and antimicrobial susceptibility to 21 antibiotics among S. flexneri serotype 2 variant isolates from different regions.
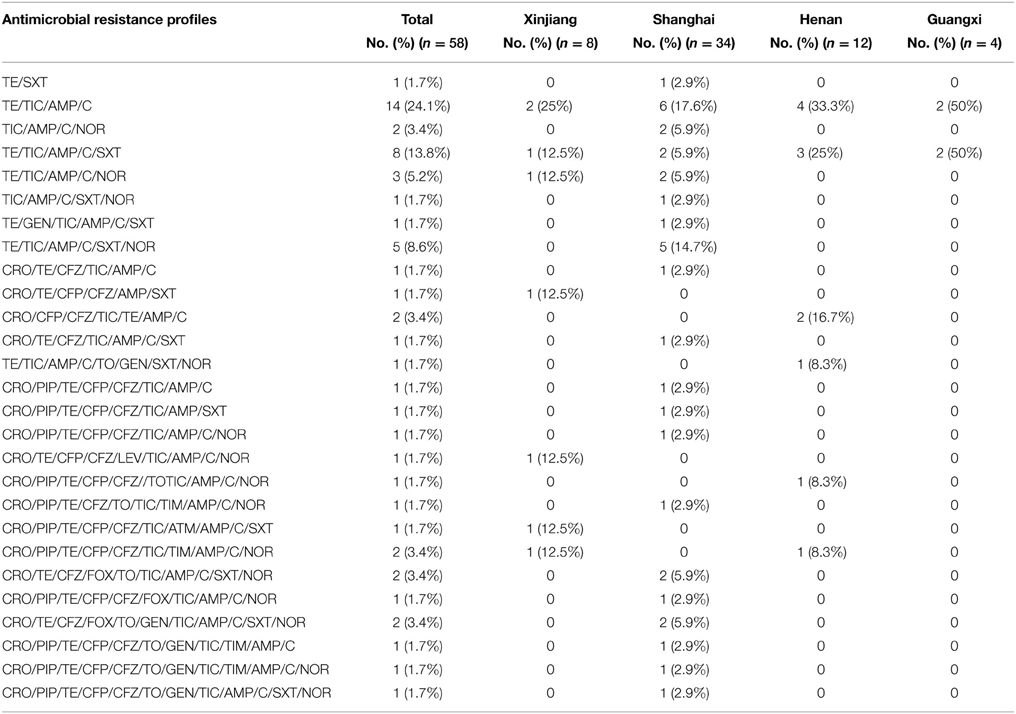
Table 3. Dominant antimicrobial resistance profiles of 58 Shigella flexneri serotype 2 variant isolates from different regions of China.
Moreover, notable differences of antibiotic resistance profiles were also observed during the periods 2003–2008 and 2009–2013. Of the 58 serotype 2 variant strains, the resistance rate to cephalosporin and quinolones was 33.3% and 17.9% during 2003–2008, respectively, which increased to 47.4% and 78.9% during 2009–2013. The resistance rate to both cephalosporin and quinolones was 5.1% during 2003–2008, which increased to 42.1% during 2009–2013 (Table 4).
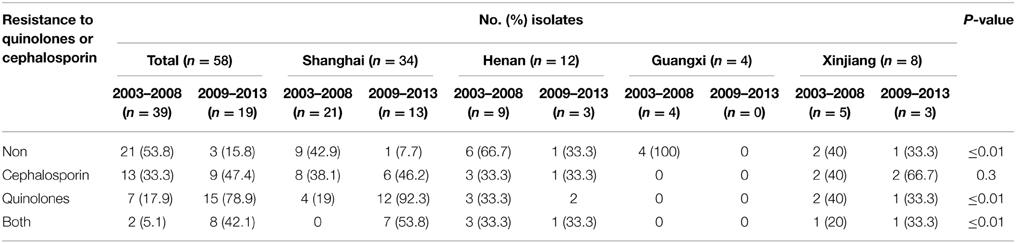
Table 4. Resistance to quinolones or cephalosporin of 58 S. flexneri serotype 2 variant isolates from different regions of China during 2003–2008 and 2009–2013.
MLST and PFGE Analysis
To determine the phylogenetic relationship of these serotype 2 variant strains, an extended MLST scheme of 15 genes was performed. We found that all the isolates in this study belonged to the same sequence type, which was designated ST100. ST100 also contained multiple other serotypes such as 1a, 1c, 2a, 2b, and Y, suggesting that ST100 has been circulating among different S. flexneri serotypes for a long time in China. PFGE was further performed to determine genetic relatedness among the isolates of serotype 2 variant and other serotypes. With a similarity of 80%, all the serotype 2 variant strains except two isolates (SH05Sh584 and SH12Sh282) formed a single cluster (containing 2a and 2b isolates). This result suggested that the serotype 2 variant strains were closely related to serotype 2a and 2b. Besides, the 58 S. flexneri serotype 2 variant isolates generated 45 PFGE patterns (Figure 3), suggesting a high genetic diversity among the serotype 2 variant strains.
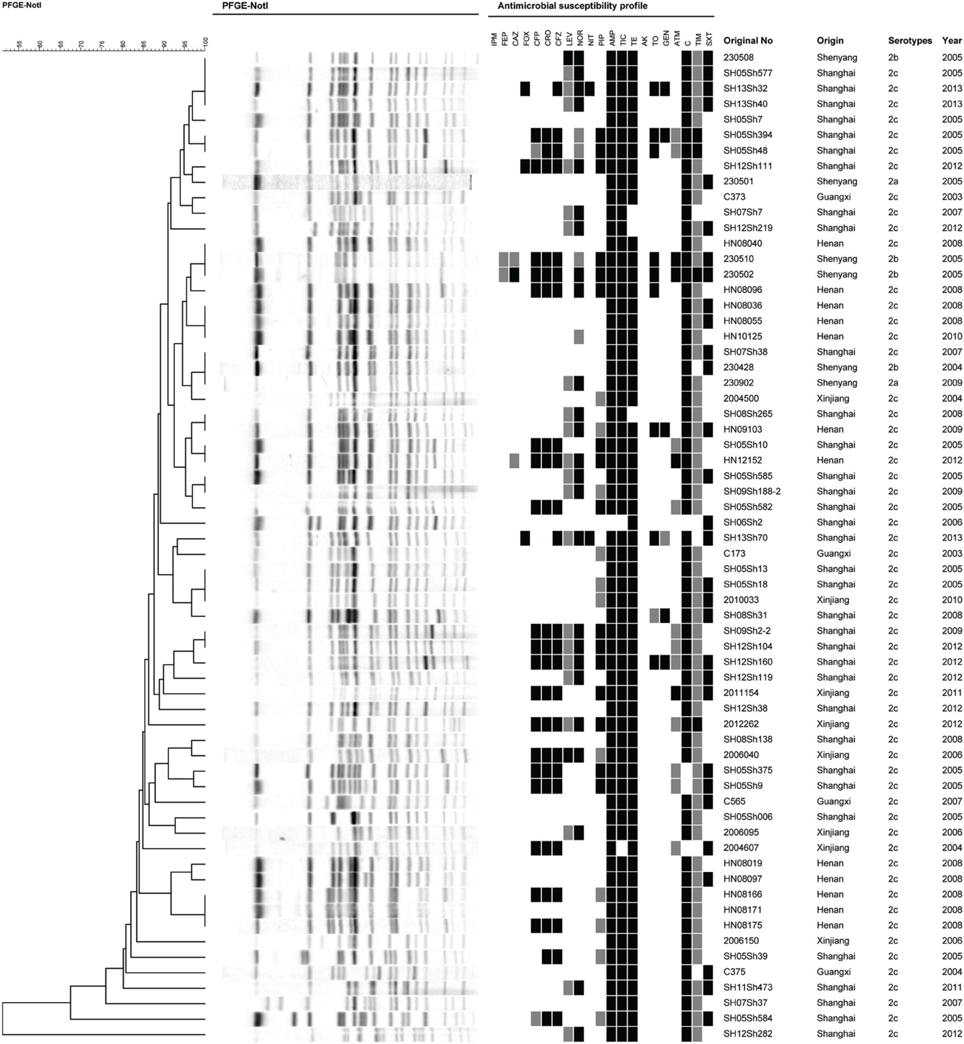
Figure 3. Pulsed-field gel electrophoresis dendrogram and the antibiotic resistance profile of S. flexneri serotype 2 variant, 2a, and 2b isolates. The original number, origin, serotype, and isolation year are indicated for each strain.
Molecular Analysis of Antibiotic-Resistance Determinants and Integrons
Twenty-two of 58 isolates showing resistance to cephalosporin were selected to test for antibiotic-resistance determinants and integrons, including the blaVIM, blaNDM, blaSHV, blaTEM, blaOXA, blaCTX−M, intI1, and intI2 gene regions (Table 5). PCR screenings showed that all strains were negative for blaSHV, blaVIM, and blaNDM, but positive for blaTEM, blaOXA, and blaCTX−M. Sequencing results using primers amplifying the whole blaTEM gene showed 100% identity with the blaTEM−1 gene. All except two isolates from Shanghai and one isolate from Xinjiang harbored blaOXA−1. Besides, 18 isolates contained the blaCTX−M gene; nine isolates from Shanghai, two from Xinjiang, and one from Henan harbored blaCTX−M−14; three isolates from Shanghai and one isolate from Henan harbored blaCTX−M−3; and one isolate from Xinjiang and one isolate from Henan harbored blaCTX−M−79. All isolates harbored class 1 integrons with blaOXA−30 and aadA1 gene cassettes, and one isolate from Shanghai harbored aacA4 and cmlA1 gene cassettes. All of the isolates harbored class 2 integrons following dfrA1, sat1, and aadA1 gene cassettes. In 22 quinolone-resistant isolates, no point mutations in QRDRs of gyrB and parE were found, but point mutations in QRDRs of gyrA and parC were identified in each resistant isolate. All of the norfloxacin-resistant isolates had the gyrA mutation of Ser83Leu and the parC mutations of Ser80Ile and His211Tyr in our study. Interestingly, the strain 2006040 (the only isolate resistant to levofloxacin) from Xinjiang and strain HN08096 from Henan had the gyrA mutation of Asp87Asn, while the rest of the strains except strain HN09103 had the gyrA mutation of Asp87Gly. Only two species of PMQR determinants [qnrS and aac(6′)-Ib-cr] were identified. All of the isolates harbored the aac(6′)-Ib-cr gene, and only two isolates from Shanghai harbored qnrS. Sequencing analysis determined that the two qnrS genes were qnrS1 (Table 6).

Table 5. Antibiogram and molecular analysis of the antibiotic-resistance determinants and integrons of S. flexneri serotype 2 variant isolates with resistance to cephalosporin.
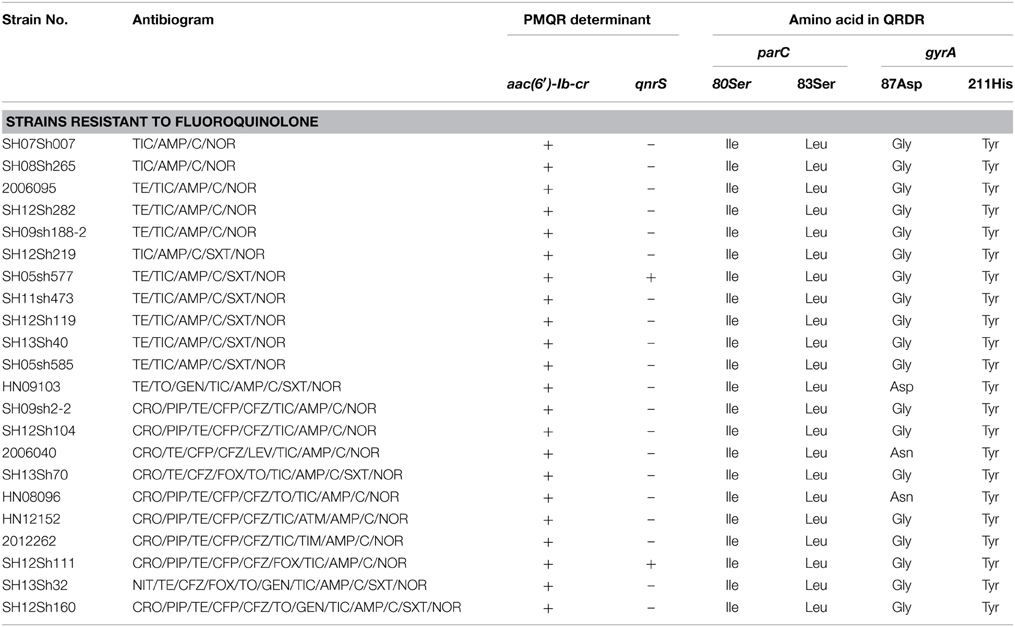
Table 6. Antibiogram and molecular analysis of the antibiotic-resistance determinants and integrons of S. flexneri serotype 2 variant isolates with resistance to fluoroquinolone.
Discussion
The emergence of MDR strains has become a serious problem and has complicated the selection of empirical treatment for shigellosis. In the present study, the serotype 2 variant isolates were highly resistant to ampicillin, chloramphenicol, tetracycline, and ticarcillin, and 98.3% of these isolates showed MDR profiles. One of the reasons for the rapid accumulation of resistance is excessive or inappropriate use of antibiotics in outpatients in China (Pickering, 2004; Zhang et al., 2011).
Our study further indicates that current resistance patterns have changed and that empirical therapy should keep pace with these changes. According to previous results, fluoroquinolones and third-generation cephalosporins are probably the best choice for empiric treatment of severe gastrointestinal infections caused by pathogenic bacteria (Gendrel et al., 2008). However, the resistance rates of these isolates to fluoroquinolones and third-generation cephalosporins in the serotype 2 variant strains have increased at an alarming rate, and continuous surveillance of the resistance pattern will be essential to choose the appropriate antimicrobial therapy.
The increasing antibiotic resistance rate led us to study the genetics and mechanisms of antibiotic resistance. The horizontal transfer of integrons may account for the dissemination of resistance genes, and resistance to some antibiotics is associated with the presence of class 1 and class 2 integrons containing resistance gene cassettes (Ke et al., 2011). In the current study, blaOXA−30 and aadA1 were detected in the gene cassettes of class 1 integrons, and dfrA1+sat1+aadA1 was detected in the gene cassettes of class 2 integrons. In addition, of the 22 cephalosporin-resistant isolates, 100% harbored the blaTEM−1 resistance gene, 81.8% harbored the blaCTX resistance gene, and 90.9% harbored the blaOXA resistance gene. Additionally, the strong ability to acquire resistance genes facilitated the survival of bacteria under the pressure of various antibiotics. The blaTEM−1 gene was detected in all of the ESBL-producing isolates; this gene exists at high frequencies in antibiotic-resistant bacteria across the globe and confers resistance to penicillin and other β-lactamase antibiotics (Barlow and Hall, 2003). OXA-type β-lactamases confer resistance to ampicillin and cephalothin, and they are characterized by high hydrolytic activity against oxacillin and cloxacillin (Bradford, 2001). Sequencing analysis showed that all of the blaOXA genes were blaOXA−1, indicating the host specificity for this subtype in S. flexneri, which is consistent with the result of a previous study (Siu et al., 2000).
Fluoroquinolone resistance in Enterobacteriaceae has been mostly attributed to modifications in the QRDRs of the gyrA and parC genes and/or PMQR determinants such as qnr and [aac(6′)-Ib-cr] (Folster et al., 2011). In the current study, 95% of the quinolone-resistant Shigella isolates had at least three point mutations in gyrA (Ser83Leu, Asp87Gly/Asn, and His211Tyr) and parC (Ser80Ile). Furthermore, different amino acid substitutions at the same position may result in different quinolone susceptibility levels (Ruiz, 2003). The His211Tyr mutation in gyrA was detected in resistant S. flexneri serotype 2a strains from Bangladesh (Azmi et al., 2014). Notably, all of the quinolone-resistant isolates in our study had the gyrA mutation of His211Tyr. PMQR determinants are located on mobile genetic elements (i.e., plasmids), which may allow for dissemination among Shigella (Folster et al., 2011). The presence of the qnr gene can facilitate the selection of chromosomal mutations that cause quinolone resistance (Jacoby et al., 2003; Martinez-Martinez et al., 2003). Worldwide, S. flexneri serotypes 1a, 2a, 2b, and 4c carrying the qnrS gene were reported with low incidence (Hata et al., 2005; Pu et al., 2009). In our study, only two strains from Shanghai carried the qnrS1 gene with a percentage of 3.4%, and one of the qnrS-positive strains was the only isolate that was resistant to levofloxacin. The aac(6′)-Ib-cr gene is identified in many Enterobacteriaceae and is responsible for low-level resistance to fluoroquinolones (Frasson et al., 2011). The aac(6′)-Ib-cr–positive S. flexneri 2a strain was first isolated in 1998 (Pu et al., 2009), and all of the 17 quinolone-resistant isolates were aac(6′)-Ib-cr–positive, suggesting that the aac(6′)-Ib-cr gene had been present in China for many years.
Serotype conversion is a major mechanism for S. flexneri to escape protective host immune responses, which may drive the emergence of novel and atypical serotypes under pressure (Allison and Verma, 2000). Based on phenotypic and genotypic analysis, the serotype 2 variant is likely to originate from serotypes 2a and 2b, which have experienced changes in their genome to adapt to altered environmental conditions. The serotype 2 variant was circulated as a major serotype during our 11-year study period, and the widespread dissemination of this serotype poses a serious threat to public health not only in China but also in the world.
In summary, studies on the serotype 2 variant have become very important owing to the increased epidemic frequency and the prevalence of MDR strains. In view of the fact that this serotype was prevalent in several provinces in China, it is likely to provoke a major crisis because MDR clones can and may have already spread to other countries. Therefore, continuous surveillance will be needed to determine the distribution and resistance development of this serotype so as to understand the actual disease burden and provide guidance for the clinical treatment of shigellosis.
Funding
This research was supported by the National Major Scientific and Technological Special Projects for Infectious Diseases during the Twelfth Five-year Plan Period (Nos. 2012ZX10004215 and 2013ZX10004607); the National Nature Science Foundation of China (Nos. 81473024, 81202252, 81373053 and 81371854); and the Beijing Science and Technology Nova program (No. xx2013061).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank all the patients for agreeing to be included in this study.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00435/abstract
References
Ahmed, A. M., Furuta, K., Shimomura, K., Kasama, Y., and Shimamoto, T. (2006). Genetic characterization of multidrug resistance in Shigella spp. from Japan. J. Med. Microbiol. 55, 1685–1691. doi: 10.1099/jmm.0.46725-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Allison, G. E., and Verma, N. K. (2000). Serotype-converting bacteriophages and O-antigen modification in Shigella flexneri. Trends Microbiol. 8, 17–23. doi: 10.1016/S0966-842X(99)01646-7
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Azmi, I. J., Khajanchi, B. K., Akter, F., Hasan, T. N., Shahnaij, M., Akter, M., et al. (2014). Fluoroquinolone resistance mechanisms of Shigella flexneri isolated in Bangladesh. PLoS ONE 9:e102533. doi: 10.1371/journal.pone.0102533
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Barlow, M., and Hall, B. G. (2003). Experimental prediction of the natural evolution of antibiotic resistance. Genetics 163, 1237–1241.
Bradford, P. A. (2001). Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14, 933–951. doi: 10.1128/CMR.14.4.933-951.2001
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Carlin, N. I., Rahman, M., Sack, D. A., Zaman, A., Kay, B., and Lindberg, A. A. (1989). Use of monoclonal antibodies to type Shigella flexneri in Bangladesh. J. Clin. Microbiol. 27, 1163–1166.
El-Gendy, A., El-Ghorab, N., Lane, E. M., Elyazeed, R. A., Carlin, N. I., Mitry, M. M., et al. (1999). Identification of Shigella flexneri subserotype 1c in rural Egypt. J. Clin. Microbiol. 37, 873–874.
Folster, J. P., Pecic, G., Bowen, A., Rickert, R., Carattoli, A., and Whichard, J. M. (2011). Decreased susceptibility to ciprofloxacin among Shigella isolates in the United States, 2006 to 2009. Antimicrob. Agents Chemother. 55, 1758–1760. doi: 10.1128/AAC.01463-10
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Frasson, I., Cavallaro, A., Bergo, C., Richter, S. N., and Palu, G. (2011). Prevalence of aac(6′)-Ib-cr plasmid-mediated and chromosome-encoded fluoroquinolone resistance in Enterobacteriaceae in Italy. Gut Pathog. 3:12. doi: 10.1186/1757-4749-3-12
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Galani, I., Souli, M., Mitchell, N., Chryssouli, Z., and Giamarellou, H. (2010). Presence of plasmid-mediated quinolone resistance in Klebsiella pneumoniae and Escherichia coli isolates possessing blaVIM-1 in Greece. Int. J. Antimicrob. Agents 36, 252–254. doi: 10.1016/j.ijantimicag.2010.05.004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gaynor, K., Park, S. Y., Kanenaka, R., Colindres, R., Mintz, E., Ram, P. K., et al. (2009). International foodborne outbreak of Shigella sonnei infection in airline passengers. Epidemiol. Infect. 137, 335–341. doi: 10.1017/S0950268807000064
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gendrel, D., Cohen, R., European Society for Pediatric Infectious, D., European Society for Gastroenterology, H., and Nutrition (2008). [Bacterial diarrheas and antibiotics: european recommendations]. Arch. Pediatr. 15(Suppl 2), S93–S96. doi: 10.1016/S0929-693X(08)74223-4
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hata, M., Suzuki, M., Matsumoto, M., Takahashi, M., Sato, K., Ibe, S., et al. (2005). Cloning of a novel gene for quinolone resistance from a transferable plasmid in Shigella flexneri 2b. Antimicrob. Agents Chemother. 49, 801–803. doi: 10.1128/AAC.49.2.801-803.2005
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hu, L. F., Li, J. B., Ye, Y., and Li, X. (2007). Mutations in the GyrA subunit of DNA gyrase and the ParC subunit of topoisomerase IV in clinical strains of fluoroquinolone-resistant Shigella in Anhui, China. J. Microbiol. 45, 168–170.
Huang, I. F., Chiu, C. H., Wang, M. H., Wu, C. Y., Hsieh, K. S., and Chiou, C. C. (2005). Outbreak of dysentery associated with ceftriaxone-resistant Shigella sonnei: first report of plasmid-mediated CMY-2-type AmpC beta-lactamase resistance in S. sonnei. J. Clin. Microbiol. 43, 2608–2612. doi: 10.1128/JCM.43.6.2608-2612.2005
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jacoby, G. A., Chow, N., and Waites, K. B. (2003). Prevalence of plasmid-mediated quinolone resistance. Antimicrob. Agents Chemother. 47, 559–562. doi: 10.1128/AAC.47.2.559-562.2003
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ke, X., Gu, B., Pan, S., and Tong, M. (2011). Epidemiology and molecular mechanism of integron-mediated antibiotic resistance in Shigella. Arch. Microbiol. 193, 767–774. doi: 10.1007/s00203-011-0744-3
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kotloff, K. L., Winickoff, J. P., Ivanoff, B., Clemens, J. D., Swerdlow, D. L., Sansonetti, P. J., et al. (1999). Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull. World Health Organ. 77, 651–666.
Kuo, H. W., Kasper, S., Jelovcan, S., Hoger, G., Lederer, I., Konig, C., et al. (2009). A food-borne outbreak of Shigella sonnei gastroenteritis, Austria, 2008. Wien. Klin. Wochenschr. 121, 157–163. doi: 10.1007/s00508-008-1141-7
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Martinez-Martinez, L., Pascual, A., Garcia, I., Tran, J., and Jacoby, G. A. (2003). Interaction of plasmid and host quinolone resistance. J. Antimicrob. Chemother. 51, 1037–1039. doi: 10.1093/jac/dkg157
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Matar, G. M., Jaafar, R., Sabra, A., Hart, C. A., Corkill, J. E., Dbaibo, G. S., et al. (2007). First detection and sequence analysis of the bla-CTX-M-15 gene in Lebanese isolates of extended-spectrum-beta-lactamase-producing Shigella sonnei. Ann. Trop. Med. Parasitol. 101, 511–517. doi: 10.1179/136485907X193860
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mathers, C. D., Boerma, T., and Ma Fat, D. (2009). Global and regional causes of death. Br. Med. Bull. 92, 7–32. doi: 10.1093/bmb/ldp028
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pan, J.-C., Ye, R., Meng, D.-M., Zhang, W., Wang, H.-Q., and Liu, K.-Z. (2006). Molecular characteristics of class 1 and class 2 integrons and their relationships to antibiotic resistance in clinical isolates of Shigella sonnei and Shigella flexneri. J. Antimicrob. Chemother. 58, 288–296. doi: 10.1093/jac/dkl228
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pickering, L. K. (2004). Antimicrobial resistance among enteric pathogens. Semin. Pediatr. Infect. Dis. 15, 71–77. doi: 10.1053/j.spid.2004.01.009
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pu, X. Y., Pan, J. C., Wang, H. Q., Zhang, W., Huang, Z. C., and Gu, Y. M. (2009). Characterization of fluoroquinolone-resistant Shigella flexneri in Hangzhou area of China. J. Antimicrob. Chemother. 63, 917–920. doi: 10.1093/jac/dkp087
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Qiu, S., Wang, Y., Xu, X., Li, P., Hao, R., Yang, C., et al. (2013). Multidrug-resistant atypical variants of Shigella flexneri in China. Emerging Infect. Dis. 19, 1147–1150. doi: 10.3201/eid1907.121221
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Qiu, S., Wang, Z., Chen, C., Liu, N., Jia, L., Liu, W., et al. (2011). Emergence of a novel Shigella flexneri serotype 4s strain that evolved from a serotype X variant in China. J. Clin. Microbiol. 49, 1148–1150. doi: 10.1128/JCM.01946-10
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Qu, F., Bao, C., Chen, S., Cui, E., Guo, T., Wang, H., et al. (2012). Genotypes and antimicrobial profiles of Shigella sonnei isolates from diarrheal patients circulating in Beijing between 2002 and 2007. Diagn. Microbiol. Infect. Dis. 74, 166–170. doi: 10.1016/j.diagmicrobio.2012.06.026
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Robicsek, A., Strahilevitz, J., Sahm, D. F., Jacoby, G. A., and Hooper, D. C. (2006). qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrob. Agents Chemother. 50, 2872–2874. doi: 10.1128/AAC.01647-05
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ruiz, J. (2003). Mechanisms of resistance to quinolones: target alterations, decreased accumulation and DNA gyrase protection. J. Antimicrob. Chemother. 51, 1109–1117. doi: 10.1093/jac/dkg222
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Siu, L. K., Lo, J. Y., Yuen, K. Y., Chau, P. Y., Ng, M. H., and Ho, P. L. (2000). beta-lactamases in Shigella flexneri isolates from Hong Kong and Shanghai and a novel OXA-1-like beta-lactamase, OXA-30. Antimicrob. Agents Chemother. 44, 2034–2038. doi: 10.1128/AAC.44.8.2034-2038.2000
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Stagg, R. M., Cam, P. D., and Verma, N. K. (2008). Identification of newly recognized serotype 1c as the most prevalent Shigella flexneri serotype in northern rural Vietnam. Epidemiol. Infect. 136, 1134–1140. doi: 10.1017/S0950268807009600
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sui, J., Zhang, J., Sun, J., Chang, Z., Zhang, W., and Wang, Z. (2010). Surveillance of bacillary dysentery in China, 2009. Dis. Surveill. 25, 947–950. doi: 10.3784/j.issn.1003-9961.2010.12.006
Sun, Q., Lan, R., Wang, J., Xia, S., Wang, Y., Wang, Y., et al. (2013). Identification and characterization of a novel Shigella flexneri serotype Yv in China. PLoS ONE 8:e70238. doi: 10.1371/journal.pone.0070238
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Talukder, K. A., Dutta, D. K., Safa, A., Ansaruzzaman, M., Hassan, F., Alam, K., et al. (2001). Altering trends in the dominance of Shigella flexneri serotypes and emergence of serologically atypical S. flexneri strains in Dhaka, Bangladesh. J. Clin. Microbiol. 39, 3757–3759. doi: 10.1128/JCM.39.10.3757-3759.2001
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tariq, A., Haque, A., Ali, A., Bashir, S., Habeeb, M. A., Salman, M., et al. (2012). Molecular profiling of antimicrobial resistance and integron association of multidrug-resistant clinical isolates of Shigella species from Faisalabad, Pakistan. Can. J. Microbiol. 58, 1047–1054. doi: 10.1139/w2012-085
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ye, C., Lan, R., Xia, S., Zhang, J., Sun, Q., Zhang, S., et al. (2010). Emergence of a new multidrug-resistant serotype X variant in an epidemic clone of Shigella flexneri. J. Clin. Microbiol. 48, 419–426. doi: 10.1128/JCM.00614-09
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhang, W., Luo, Y., Li, J., Lin, L., Ma, Y., Hu, C., et al. (2011). Wide dissemination of multidrug-resistant Shigella isolates in China. J. Antimicrob. Chemother. 66, 2527–2535. doi: 10.1093/jac/dkr341
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keywords: Shigella flexneri, serotype 2 variant, multidrug-resistant, antibiotic resistance rate, epidemic frequency
Citation: Cui X, Wang J, Yang C, Liang B, Ma Q, Yi S, Li H, Liu H, Li P, Wu Z, Xie J, Jia L, Hao R, Wang L, Hua Y, Qiu S and Song H (2015) Prevalence and antimicrobial resistance of Shigella flexneri serotype 2 variant in China. Front. Microbiol. 6:435. doi: 10.3389/fmicb.2015.00435
Received: 04 March 2015; Accepted: 22 April 2015;
Published: 07 May 2015.
Edited by:
Rafaella Fortini Grenfell E. Queiroz, Fundação Oswaldo Cruz - FIOCRUZ, BrazilReviewed by:
Li Xu, Cornell University, USAMalgorzata Anna Mikaszewska-Sokolewicz, The Medical University of Warsaw, Poland
Copyright © 2015 Cui, Wang, Yang, Liang, Ma, Yi, Li, Liu, Li, Wu, Xie, Jia, Hao, Wang, Hua, Qiu and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuejin Hua, Nuclear-Agricultural Sciences, Zhejiang University, Hangzhou 310029, China, yjhua@zju.edu.cn;
Shaofu Qiu, Institute of Disease Control and Prevention, Academy of Military Medical Sciences, 20 Dongda Street, Fengtai District, Beijing 100071, China, qiushf0613@hotmail.com;
Hongbin Song, Institute of Disease Control and Prevention, Academy of Military Medical Sciences, 20 Dongda Street, Fengtai District, Beijing 100071, China, hongbinsong@263.net
 Xianyan Cui
Xianyan Cui Jian Wang2
Jian Wang2 Yuejin Hua
Yuejin Hua Shaofu Qiu
Shaofu Qiu