Corrigendum: Dynamic nature of SecA and Its associated proteins in Escherichia coli
Corrigendum: Dynamic nature of SecA and Its associated proteins in Escherichia coli
- Department of Radiation Genetics, Graduate School of Medicine, Kyoto University, Kyoto, Japan
Mechanical properties such as physical constraint and pushing of chromosomes are thought to be important for chromosome segregation in Escherichia coli and it could be mediated by a hypothetical molecular “tether.” However, the actual tether that mediates these features is not known. We previously described that SecA (Secretory A) and Secretory Y (SecY), components of the membrane protein translocation machinery, and AcpP (Acyl carrier protein P) were involved in chromosome segregation and homeostasis of DNA topology. In the present work, we performed three-dimensional deconvolution of microscopic images and time-lapse experiments of these proteins together with MukB and DNA topoisomerases, and found that these proteins embraced the structures of tortuous nucleoids with condensed regions. Notably, SecA, SecY, and AcpP dynamically localized in cells, which was interdependent on each other requiring the ATPase activity of SecA. Our findings imply that the membrane protein translocation machinery plays a role in the maintenance of proper chromosome partitioning, possibly through “tethering” of MukB [a functional homolog of structural maintenance of chromosomes (SMC) proteins], DNA gyrase, DNA topoisomerase IV, and SeqA (Sequestration A).
Introduction
In eukaryotes, mitotic spindles attach to kinetochores of chromosomes to assure bi-oriental segregation of sister chromosomes into the two dividing daughter cells. In prokaryotes, however, no such structure has been identified, and the mechanisms mediating faithful chromosome segregation in bacteria remain elusive. On the one hand, Jun and Mulder (2006) proposed entropic forces as driving forces for compartmentalization of polymeric sister chromosomes in the cylindrical cell periphery, a concept that has been confounded by a number of discrepancies (Odijk, 1998; Cunha et al., 2001; Wiggins et al., 2010; Hadizadeh Yazdi et al., 2012; Fisher et al., 2013). On the other hand, Bates and Kleckner (2005) proposed that the principal mechanism of chromosome segregation involves a mechanical pushing force from coherent chromosomes followed by the release from a tether. These models raise the question of whether the nature of the energy required for segregation depends on passive entropic forces or active mechanisms involving ATPases. Recently, four-dimensional imaging of the Escherichia coli nucleoid revealed surprising dynamics such as individualization, radial confinement, and longitudinal density spots moving back and forth that can affect segregation of tortuously structured sister chromosomes (Fisher et al., 2013). More specifically, (I) sister chromosomes occupy their respective spaces due to initial bundling of chromosomes; (II) molecular tethers minimize radial confinement of chromosomes; (III) stressing and releasing cycles of this “tether” allow chromosomes with longitudinal density spots to move back and forth, acting as “grease.” Bundling, confinement, and stressing of chromosomes are all mediated by this “tether,” followed by release from the same “tether,” illustrating the mechanical forces required for chromosome segregation. Therefore, progression through the steps of chromosome segregation may involve cyclic accumulation and release of intranucleoid mechanical stress mediated by the hypothetical molecular “tether.” However, the actual molecular “tether” has not been identified.
One of the clues for identifying the actual molecular tether is that the molecular tether is likely to be one or more proteins involved in chromosome positioning and partitioning. In addition, the molecular tether may also show dynamic localization as is seen with the nucleoid. In E. coli, MukB (“mukaku” means anucleate in Japanese) (Niki et al., 1992) is a functional homolog of SMC (structural maintenance of chromosomes) and is involved in activities of chromosome organization such as positioning and partitioning, i.e., proper localization of the oriC region, nucleoids, and replication forks. MukB localizes equidistantly as clusters along the long cell axis in living cells (Hiraga, 2000; Ohsumi et al., 2001; Adachi et al., 2008; Nolivos and Sherratt, 2014). Similarly, SeqA (Sequestration A) also appears to be spatially regulated. SeqA foci localize equidistantly along the long cell axis, and this ordered localization is hypothesized to be important for chromosome organization (Hiraga et al., 1998, 2000; Onogi et al., 2002; Yamazoe et al., 2005). These proteins might interact with molecular tether. Furthermore, MukB molecules form one to three foci in a living cell together with other mobile MukB molecules that show dynamic localization, suggesting roles for MukB in chromosome positioning/partitioning and/or condensation (Badrinarayanan et al., 2012). The dynamics of MukB thus indicates its interaction with molecular tether.
SecA (Secretory A) plays a role in the translocation of membrane proteins through attaching to and dismantling from the Sec pore complex of the inner cell membrane, shuttling between inner cell membrane and cytosol, thus maintaining homeostatic conditions in cells via membrane transport machinery (Cabelli et al., 1991; Robson and Collinson, 2006). Secretory Y (SecY) is a component of the membrane protein translocation channel (Robson and Collinson, 2006). SecY and SecB interact with SecA and stimulate the ATPase activity of SecA (Miller et al., 2002; Natale et al., 2004). Regarding the function of the ATPase of SecA, “Push and Slide” mechanism, in which translocating peptide is pushed toward outer direction during ATP-bound phase in immobile form, followed by ATP hydrolysis and investing sliding mobility to the peptide. Finally, ADP is converted to ATP and the cycle is repeated again and again (Bauer et al., 2014). Previously we have described that SecA, SecY, and Acyl carrier protein P (AcpP) are involved in chromosome partition and that inactivation of SecA causes disorder in decatenation and maintenance of superhelicity on plasmid DNA (Adachi et al., 2014). AcpP, which is an acyl carrier protein of ACP complex, acts as a source of acyl groups inside the cell of E. coli. Acylated proteins are transferred to membrane by lipid-acyl group interaction. For example, bacterial toxin Haemolysin is distributed to cell membrane by this process (Issartel et al., 1991; also refer Towler and Gordon, 1988; Cronan and Rock, 1996). Therefore, it can be speculated that AcpP relates protein dynamics closely related to the inner cell membrane. Furthermore, AcpP is copurified with MukB (Niki et al., 1992), suggesting the role of AcpP on chromosome segregation. It is notable that AcpP binds to both SecA and MukB, In addition, we have described that a network involving the interaction of SecA/SecY-AcpP-MukB-Topo IV (DNA topoisomerase IV, ParC/ParE complex)/SeqA-DNA gyrase (GyrA/GyrB complex) may be important for chromosome positioning and partitioning (separation of chromosomes) (Adachi et al., 2014).
In the present work, we examined subcellular localization of these proteins and found that these proteins trace tortuous nucleoids, and were dynamic. The dynamic localization of SecA was ceased by inactivation of SecY and AcpP. Conversely, dynamic localization of SecY was ceased by inactivation of SecA. We revealed that the ATPase activity of SecA was essential for dynamic localization of these proteins. Integrating the present results with our previous work that SecA/SecY/AcpP genetically interact with MukB and DNA topoisomerases (Adachi et al., 2014), we hypothesize that SecA/SecY/AcpP is the core component of the dynamic property of the chromosome segregation proteins in E. coli.
Materials and Methods
Bacterial Strains and Plasmids
The strains and plasmids used in this study are shown in Tables S1 and S2, respectively. Derivatives with the azide-resistant secA204 mutation were obtained by P1 transduction (using P1vir) of W208 and selection and purification with 3–4 mM sodium azide. Derivatives with the mukB-gfpUV4 mutation were obtained by co-transduction of KAT1 with cat and selection and purification with 5 μg/ml chloramphenicol. Fab+ derivatives of the NAD strains were obtained by co-transduction of TL212 with fab+ and the zce-726::Tn10 marker from TL212 and selection and purification with 7.5 μg/ml tetracycline. We also created the MQ574 strain by P1vir transduction of JC12334 with the tna-300::Tn10 mutation [linked to gyrB(Ts)] and selection and purification with 7.5 μg/ml tetracycline. Derivatives with the gyrB(Ts) mutation were obtained by P1vir transduction of MQ574.
Media
Medium L (Hiraga et al., 1989) consists of 1% Bactotryptone (Difco, Sparks, MD), 0.5% yeast extract (Difco), and 0.5% NaCl (pH 7.4). Medium C consists of synthetic medium M9 (Miller, 1992) supplemented with 0.5% glycerol, 50 μg/ml L-threonine, L-leucine, L-proline, L-arginine, L-histidine, L-tryptophan, and 5 μg/ml vitamin B1. L-Glutamate (50 μg/ml) and chloramphenicol (5 μg/ml) were added to the cultures of strains when necessary.
Immunofluorescence Microscopy
Indirect immunofluorescence microscopy of E. coli cells was performed as described previously (Hiraga et al., 1998; Hirano et al., 1998). Rabbit anti-SecA/SecY antisera were gifts from Dr. Koreaki Ito (Kyoto Sangyo University). Rabbit anti-AcpP antiserum was a gift from Dr. Charles Rock (St. Jude Children's Research Hospital). Mouse anti-GFP monoclonal antibody is commercially available (Roche Diagnostics, K.K., Tokyo, Japan). We used goat anti-rabbit/mouse IgG antiserum conjugated to Cy3 (GE Healthcare, U.K. Ltd., Little Chalfont, UK) as the secondary antibody. For optical sectioning, an OLYMPUS BX61 microscope with an OLYMPUS UPlanApo ×100/1.35 oil objective lens (OLYMPUS, Corp., Tokyo, Japan) connected to CoolSNAPHQ (NIPPON ROPER, K.K., Chiba, Japan) was equipped with MetaMorph (Universal Imaging, Corp., Downingtown, PA) to obtain series of Z-sections with a fixed spacing of 0.1 μm. Each stack of 26 optically sectioned fluorescent images was deconvoluted through 50 iterations using the three-dimensional deconvolution function in the AutoQuant X version 1.4.1 program (Media Cybernetics, Inc., Silver Spring, MD). Three-dimensional images were then obtained by Volume Viewer function of 3D plug-in in ImageJ software.
Time-Lapse Experiments for GFPUV4-Fused Proteins in Living Cells
We used cultures grown in medium C at 30°C unless otherwise indicated. IPTG (isopropyl ß-D-1-thiogalactopyranoside, 0.1 mM) was added and incubated for 1 h when we observed a GFPuv4 (uv4 is a GFP variant with strong fluorescence and an altered spectrum; Ito et al., 1999) signal in living cells. Ten microliters of the cell suspension was fixed on a 1% agarose/medium-coated glass slide, covered with a cover glass, and sealed with vacuum grease. Images of the fluorescence signal were observed using an OLYMPUS BX51 fluorescence microscope with an OLYMPUS UApo ×2×150/1.45 oil objective lens (OLYMPUS, Corp.) connected to Cascade II 512 (Photometrics, Tucson, AZ) equipped with MetaMorph (Universal Imaging, Corp.). The obtained fluorescence signals were averaged over cell width and plotted as histograms along long cell axes by ImageJ. All time-lapse images were exposed for 100 ms to capture the indicated images of Figure 2 with 45-s intervals for approximately 3 min or of Supplementary Movies with 3 s/frame for approximately 5 min.
Conventional Fluorescence Microscopy
Microscopy was performed as described in Adachi et al. (2008).
Run-Off Replication Method to Analyze the DNA Content Per Cell Using Flow Cytometry
The procedure was performed as previously described (Adachi et al., 2008).
FRAP (Fluorescence Recovery after Photobleaching) Microscopy
Images of fluorescence signals were observed using an OLYMPUS Fluoview™ FV1000 confocal fluorescence microscope connected to OLYMPUS IX81 with an OLYMPUS PlanSApo ×60/1.35 oil objective lens (OLYMPUS, Corp.) in a pre-warmed chamber. The images were taken with FLUOVIEW Ver. 1.6 software and were simultaneously subjected twice to Kalman line function. For setting up the conditions, see Supplementary Text.
Results
Subcellular Localization of SecA, SecY, and AcpP Proteins Embracing Nucleoids
We used PA340 cells expressing proteins fused to GFPuv4. The GFPuv4-fused proteins are inducible by 0.1 mM IPTG. All strains that harbored plasmids carrying genes encoding GFPuv4-fused proteins grew normally and similarly and the localizations of GFPuv4-fused proteins were indistinguishable in the presence or absence (under leaky expression status) of 0.1 mM IPTG, suggesting that IPTG induction of GFPuv4-fused proteins did not affect localization of protein molecules or cell growth. Importantly, we confirmed that all the GFPuv4-fused proteins described in this manuscript corrected the defects of the corresponding temperature sensitive or deletion mutations in E. coli, showing that these fusion proteins were functional.
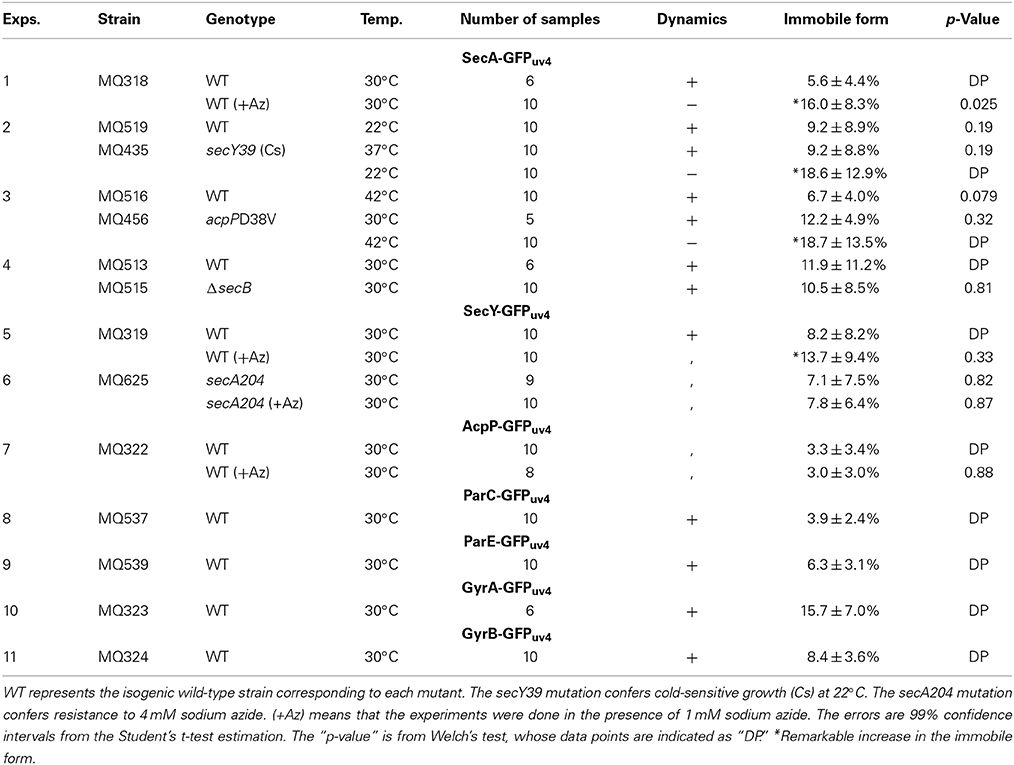
Cells grown in rich medium L were fixed and stained for immunofluorescence using anti-GFP monoclonal antibody. To obtain images with high-resolution without out-of-focus fluorescence, methanol-fixed and immunostained cells were analyzed by three-dimensional deconvolution. Imaging of living cells is not feasible for 3D deconvolution, because of rapid movements of protein molecules within the time range of obtaining Z-stacked images as described in the following section. First, 3D deconvolution of DAPI (4,6′-diamidino-2-phenylindole)-stained cells showed that nucleoids exhibited a tortuous structure (Figures 1A,A',B,B'; Movies S1, S3) as reported previously by Hadizadeh Yazdi et al. (2012) and Fisher et al. (2013). We next examined the subcellular localization of SecA-GFPuv4. Three-dimensional Deconvolution of SecA-GFPuv4 images revealed that the protein was localized with condensed regions tracing over, complementing or in parallel with nucleoid structures (Figures 1B,B'; Movies S2–S4). Although the condensed regions of SecA-GFPuv4 overlapped with the nucleoid, patterns of the condensed regions were not entirely the same as those of nucleoids (Figures 1B,B'; Movies S2, S4). Furthermore, SecY-GFPuv4 (Figures 1C,C'; Movies S5–S7) and AcpP-GFPuv4 (Figures 1D,D'; Movies S8–S10) were also localized as tortuous structures embracing nucleoids with asymmetric condensed regions. However, the patterns of the condensed regions were not entirely the same as those of the nucleoids. In contrast, GFPuv4 (not fused to a protein) was intensively localized in cytosolic areas with absolute symmetry and was faint in regions occupied by nucleoids apparently different from more uniform distribution without deconvolution (Figures 1E,E'; Movies S11–S13). This is, to our knowledge, the first deconvolved image of non-fused GFP in E. coli, which are supposed to reflect more precise information, such as exclusion of GFPuv4 from tightly packed nucleoid. As a control, cells without GFPuv4 did not exhibit a fluorescence signal (Figures 1F,F', Movies S14, S15).
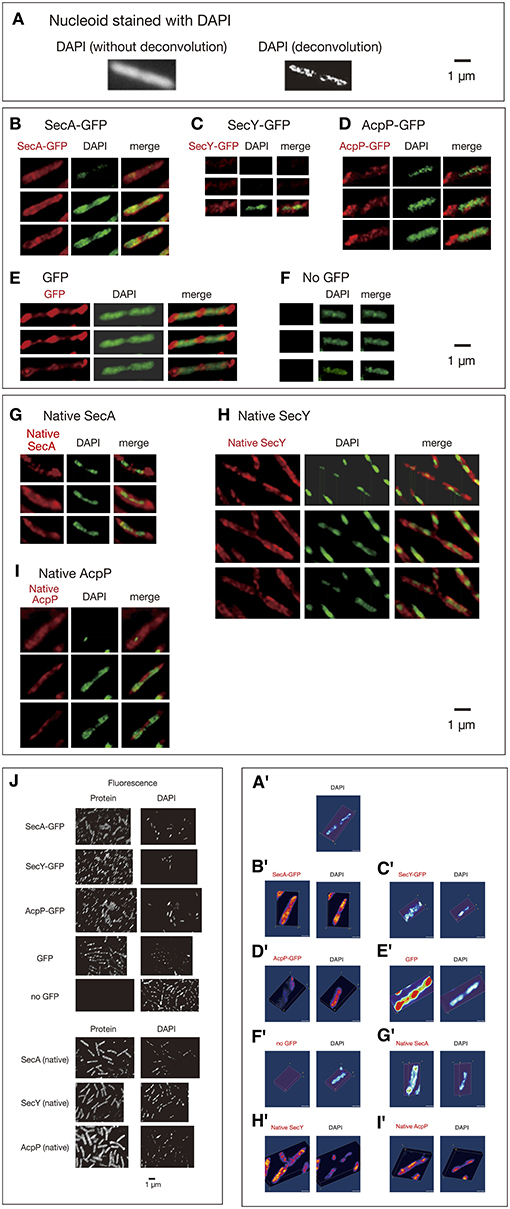
Figure 1. Three-dimensional deconvoluted images of various immunostained proteins. Cells were grown in medium L at 30°C and fixed with 70% methanol. Red, immunofluorescence of GFP or native proteins. Green, DAPI staining. Merge, merged images of red and green images. Note that synthesized yellow signal is only recognizable with eyes when the ratio of each signal is between ~4:6 and 6:4. More difference is recognizable as mere green or red. (A) Images of a DAPI-stained wild-type cell before and after 3D deconvolution in the Z-axis. (B–F) Fixed cells expressing GFPuv4-fused proteins or GFPuv4 protein (not fused with any protein) were immunostained using an anti-GFP monoclonal antibody. Fluorescence images of the proteins were analyzed with 3D deconvolution at 0.5-μm intervals in the Z-axis. Images of DAPI-stained nucleoids were also analyzed with 3D deconvolution. The yellow arrowheads indicate the clustered localization of protein molecules. (B) SecA-GFPuv4, (C) SecY-GFPuv4, (D) AcpP-GFPuv4, (E) GFPuv4 (not fused with any protein), (F) No GFPuv4. (G–I) Deconvoluted images of native proteins. Wild-type cells (PA340) were grown in medium L at 37°C, fixed, and immunostained using rabbit antisera for native proteins. Images of DAPI-stained nucleoids were also deconvoluted. (G) Native SecA, (H) Native SecY, (I) Native AcpP, (J) Three-dimensional deconvolution images of immunostained cells in wider sights corresponding to (B)–(I). (A'–I') Three-dimensional images corresponding to (A)–(I).
Additionally, we confirmed that the native SecA, SecY, and AcpP proteins without GFPuv4, which were immunostained with specific rabbit antisera for each protein, were also localized as asymmetric tortuous structures with condensed regions complementing or in parallel with nucleoids similar to the tagged proteins (Figures 1G,G',H,H',I,I'; Movies S16–S24). Although in Swulius and Jensen (2012) artificial helices of YFP-tagged MreB was reported, our results for native antisera suggest that the localization of GFPuv4-fused proteins was not an artifact due to GFPuv4 tagging. In all of the images, the localizations of the proteins extended out of nucleoids regions, including polar localization. Cells of wider sights are shown in Figure 1J.
Dynamic Localizations of SecA, SecY, and Other Proteins in Living Cells
We next performed time-lapse experiments to analyze living cells expressing GFPuv4-fused proteins growing in poor medium C because the number of condensed regions in cells expressing GFPuv4-fused proteins was lower in poor medium than in rich medium, allowing easier study of the dynamic behavior of the proteins. GFPuv4-fused proteins were induced by incubation with 0.1 mM IPTG for 1 h. Cells were photographed with a short exposure (100 ms) with some of the fluorescence signals. In this exposure, especially highly mobile diffusing molecules were unable to be detected. The fluorescence signals obtained were plotted as histograms at 45-s intervals for approximately 3 min. To monitor rapid movements of protein molecules, obtaining Z-stacked images, which were necessary for three-dimensional deconvolution in living cells, was not feasible. In addition, obtaining phase-contrast images side by side with time-lapse images was not possible because filter exchange was not practical with such high-speed imaging. The localization of fluorescence signals is evaluated by standard deviation (SD) values of relative signal values along long cell axes of 2–12 cells. If the SD value is larger, non-uniform localization of the signals is expected. If smaller, spatially rather uniform signals are expected. The results are summarized in Figure 2.
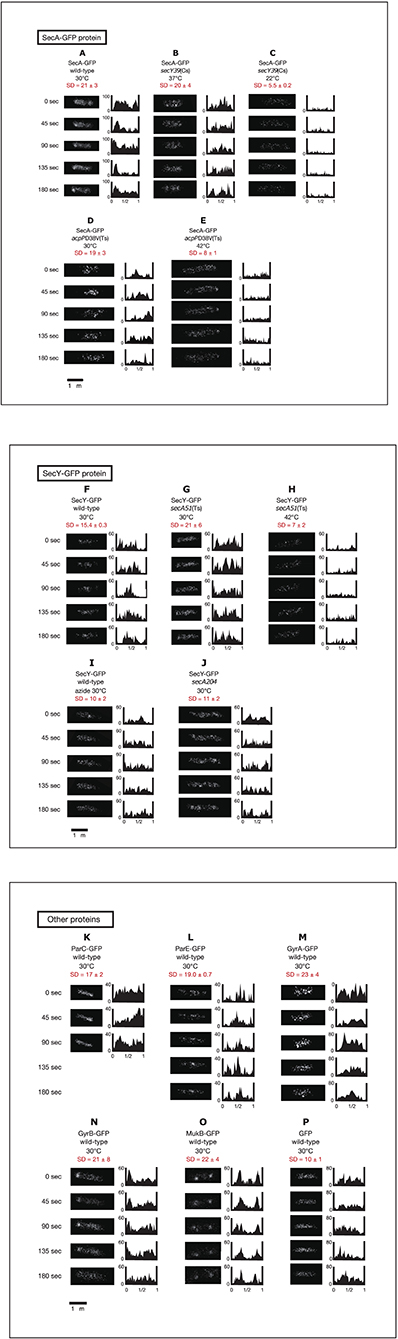
Figure 2. Time-lapse images of GFPuv4-fused proteins reveal the dynamic nature of chromosome partitioning proteins. Cells were grown in medium C and incubated with 0.1 mM IPTG for 1 h. Time-lapse images of fluorescence were taken by exposure for 100 ms. Both images and histograms of signals along long cell axes were obtained at 45-s intervals. The x-axes of histograms are subcellular positions and the y-axes of histograms are relative fluorescence units. Ninety-five Percent confidential SD (standard deviation) values of fluorescence signals along long cell axes in individual cells are also shown. (A) SecA-GFPuv4 in wild-type cells (MQ318) growing at 30°C. (B) SecA-GFPuv4 in secY39(Cs) mutant cells (MQ435) growing at the permissive temperature of 37°C. (C) SecA-GFPuv4 in secY39(Cs) mutant cells (MQ435) growing at the non-permissive temperature of 22°C. (D) SecA-GFPuv4 in acpPD38V(Ts) mutant cells (MQ456) growing at the permissive temperature of 30°C. (E) SecA-GFPuv4 in acpPD38V(Ts) mutant cells (MQ456) growing at the non-permissive temperature of 42°C. (F) SecY-GFPuv4 in wild-type cells (MQ319) growing at 30°C. (G) SecY-GFPuv4 in secA51(Ts) mutant cells (MQ748) growing at the permissive temperature of 30°C. (H) SecY-GFPuv4 in secA51(Ts) mutant cells (MQ748) growing at the non-permissive temperature of 42°C. (I) SecY-GFPuv4 in wild-type cells (MQ319) with 1 mM sodium azide growing at 30°C. The incubation time with azide was approximately 5 min. (J) SecY-GFPuv4 in secA204 azide-resistant mutant cells (MQ625) growing at 30°C (without sodium azide). (K) ParC-GFPuv4 in wild-type cells (MQ537) growing at 30°C. (L) ParE-GFPuv4 in wild-type cells (MQ539) growing at 30°C. (M) GyrA-GFPuv4 in wild-type cells (MQ323) growing at 30°C. (N) GyrB-GFPuv4 in wild-type cells (MQ324) growing at 30°C. (O) MukB-GFPuv4 in wild-type cells (MQ529) growing at 30°C. (P) GFPuv4 in wild-type cells (MQ668) growing at 30°C.
Surprisingly, GFPuv4-fused SecA protein molecules showed dynamic localization in living cells (Movie S25). SecA-GFPuv4 was observed as clusters that repeatedly accumulated and dissociated at one cell pole or the other, or at other cellular positions as density spots moving back and forth (Figure 2A; Movie S25). This is consistent with the result that SecA is present in both peripheral region of cell membrane and cytoplasm. SecY-GFPuv4 behaved in a similar manner as SecA-GFPuv4, with slightly more uniform distribution (Figure 2F; Movie S30).
Living cells with SecY-GFPuv4 grown in media C and L were observed with a conventional fluorescence microscope (Figure 3). In living cells grown in richer medium L, strong fluorescence foci of SecY-GFPuv4 were observed (Figure 3D). The strong foci appeared to localize in peripheral regions of cell membrane. The foci might be assembled clusters of many membrane protein translocation channels. The strong fluorescence signal from SecY-GFPuv4 (and also SecA-GFPuv4) foci are prominent in Figures 3C,D. However, in immunostaining images of Figure 1, the contrast was not so prominent, despite there are still vague fluorescence peaks supposed to be originated from the former foci in living cells. During permeablization of immunostaining procedure, some of the proteins might be dissociated from the corresponding foci closely located near cell membrane.
Figure 3. Living wild-type cells with SecA-GFPuv4/SecY-GFPuv4 were observed with a conventional fluorescence microscope. (A) Fluorescence signal of SecA-GFPuv4 in cells grown in medium C. (B) Fluorescence signal of SecY-GFPuv4 in cells grown in medium C. (C) Fluorescence signal of SecA-GFPuv4 in cells grown in medium L. (D) Fluorescence signal of SecY-GFPuv4 in cells grown in medium L.
ParC-GFPuv4 accumulated and dissociated in a similar manner to that of SecA-GFPuv4 (Figure 2K; Movie S35). ParE-GFPuv4 appeared to accumulate and dissociate within a near-polar or mid-cell region (Figure 2L; Movie S36). These results for ParC-GFPuv4 and ParE-GFPuv4 were consistent with previously reported results (Espeli et al., 2003).
GyrA and GyrB (subunits of DNA gyrase) fused to GFPuv4 showed dynamic localization in a similar manner as SecA-GFPuv4 (Figures 2M,N; Movies S37, S38). Previous transmission electron microscopy with immunogold particles indicates that Gyr protein molecules are predominantly localized at cytosol (~90%) mainly peripheral to nucleoids, suggesting their role in DNA segments unwinded from the nucleoid (Thronton et al., 1994). Since GFP would in principle localize outside the nucleoid with strong fluorescence from cell poles (Figure 1E), the localizations of Gyr proteins are different from GFP. This does not contradict with our analyses, since fluorescence signals all around the peripheral regions of nucleoid could result in broad signals that seem to surround the nucleoids, mimicking cytosol.
Live-cell imaging of MukB-GFPuv4 in strain MQ529 in which genomic mukB was replaced with mukB-gfpUV4 (Ohsumi et al., 2001) showed preferential localization of MukB-GFPuv4 as two clusters in the cell quarter positions. An individual cluster showed frequent oscillation in which it divided into two or dispersed and fused again to form one cluster (Figure 2O; Movie S39).
AcpP-GFPuv4 exhibited tortuous localization with condensed regions or net-like localization, but its dynamics were not clear (data not shown). The localization patterns of these proteins were similar when concentrations of IPTG lower than 0.1 mM were used. Dynamic localization of these fusion proteins was observed at high frequencies as summarized in Table 1. As a control, GFPuv4 (not fused to a protein) was observed as dispersed signal peaks with no clusters distributed throughout the entire cell and did not exhibit clear dynamic localization (Figure 2P; Movie S40). Assuming that the observed proteins described in this section are more or less associated with nucleoid, these results are consistent with previous reports that described density spots moving back and forth in bacterial nucleoids (Fisher et al., 2013).
Perturbation of the Dynamic Localization of SecY-GFPUV4 with the secA Conditional Mutation
We next examined effects of SecA on dynamic localization of SecY-GFPuv4. When temperature-sensitive secA51 mutant cells (MQ748) were incubated at the non-permissive temperature of 42°C for 2 h, the dynamic localization of SecY-GFPuv4 was remarkably affected. Almost all the molecules seemed to scatter throughout the cell, moving very rapidly without localization (Figure 2H; Movie S32), in contrast to the dynamic clusters that were seen at the permissive temperature of 30°C (Figure 2G; Movie S31). As a negative control, the dynamic localization of SecY-GFPuv4 was not affected in isogenic wild-type secA cells (MQ743) by incubation at 42°C (data not shown). Thus, the dynamic localization of SecY-GFPuv4 depends on SecA function.
Perturbation of the Dynamics of SecA-GFPUV4 with secY and acpP Conditional Mutations
We also found that the dynamic localization of SecA-GFPuv4 was affected in cold-sensitive secY39 mutant cells (MQ435) when incubated at the non-permissive temperature of 22°C for 5 min (Figure 2C; Movie S27), but not at the permissive temperature of 37°C (Figure 2B; Movie S26). In contrast, both dynamic localization of SecA-GFPuv4 was observed in the isogenic wild-type secY strain (MQ519) after incubation at 22°C (data not shown). These results show that SecY plays an important role in the dynamics of SecA-GFPuv4.
In addition, the dynamic localization of SecA-GFPuv4 was also affected in temperature-sensitive acpPD38V mutant cells (MQ456: amino acid substitution of V for D at amino acid position 38) at the non-permissive temperature of 42°C after incubation for 30 min (Figure 2E; Movie S29), but not at the permissive temperature of 30°C (Figure 2D; Movie S28). The fluorescence signal for SecA-GFPuv4 was observed even after incubation for 30 min at 42°C in the isogenic wild-type acpP strain (MQ516), suggesting that incubation at the higher temperature did not disrupt the dynamic localization of SecA-GFPuv4. Thus, the dynamic localization of SecA-GFPuv4 depends on the function of AcpP. Overall, these results demonstrate that the dynamic localization of SecA and SecY of the membrane protein translocation machinery and AcpP in cells was interdependent on each other.
Effect of Sodium Azide on Dynamic Localization of SecY-GFPUV4
We next utilized 1 mM sodium azide to inhibit SecA ATPase activity. SecA, but not other essential ATPases, is considered to be the functional target of this concentration of sodium azide because the growth kinetics and cellular and nucleoid morphologies are normal in the azide-resistant secA204 mutant (Adachi et al., 2014). The normal growth of the mutant indicates that respiratory metabolisms including cytochrome c oxidase activity are not significantly affected at this concentration. Perturbation of the dynamic localization of SecY-GFPuv4 was also observed in the presence of 1 mM sodium azide in wild-type strains (Figure 2I; Movie S33), indicating that the SecA ATPase activity is essential for the dynamics of these proteins. The dynamics of SecY-GFP was not clearly observed in the azide-resistant secA204 mutant (Figure 2J; Movie S34) in contrast to the wild-type secA strain (Figure 2F; Movie S30), suggesting that this mutant was defective (or leaky) in the unknown function of SecA acting on SecY-GFP dynamics. This result was consistent with the result that the positioning of oriC was abnormal in the mutant (Adachi et al., 2014). The effect of azide was not observed in the secA204 mutant (data not shown).
Perturbations of the Positionings of MukB and SeqA Foci with Sodium Azide
Because 1 mM sodium azide perturbs the proper positioning of the oriC region (Adachi et al., 2014), we examined whether the compound has similar effects on the positioning of MukB, a key protein in the positioning of oriC. MukB-GFPuv4 foci in the mukB-gfpUV4 strain (MQ525) are normally located at the mid-cell or cell quarter positions, implying a role for MukB in the chromosome reorganizing center by acting on condensation and proper patterning of the entire nucleoid (Ohsumi et al., 2001; Adachi et al., 2008). When the mukB-gfpUV4 cells (MQ525) were treated with 1 mM sodium azide in medium C for 4 h, abnormal localization of MukB-GFPuv4 foci at cell pole(s) was observed in approximately 30% (11/39) of cells (Figure 4A). Such abnormal polar localization was observed in less than 1% (2/201) of cells in the same strain without azide. When the isogenic azide-resistant secA204 mutant strain (MQ585) was grown in the presence of sodium azide, less than 1% (1/132) of the cells showed abnormal polar localization of MukB-GFPuv4 foci. This indicated that the SecA protein is the functional target protein of 1 mM sodium azide among many essential ATPases in E. coli.
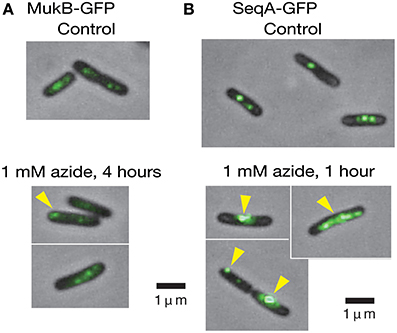
Figure 4. Living cells with MukB-GFPuv4/SeqA-GFPuv4 were observed with a conventional fluorescence microscope. (A) The effect of 1 mM sodium azide on the localization of MukB-GFPuv4 (MQ529). The yellow arrowhead shows abnormal localization of a fluorescent focus at the cell pole. (B) The effect of 1 mM sodium azide on localization of SeqA-GFPuv4 (MQ190). The yellow arrowheads show the abnormal localization of fluorescent foci at the cell pole and large fluorescent clusters.
We next asked if inhibition of SecA with sodium azide affects the localization of SeqA protein. SeqA binds to hemimethylated GATC sequences of newly synthesized DNA immediately after the passage of replication forks that are not yet fully methylated (Lu et al., 1994), allowing the use of SeqA to detect replication forks. Interestingly, treatment of the SeqA-GFPuv4-expressing strain (MQ190) with 1 mM sodium azide for 1 h in medium C at 30°C led to a strong fluorescence signal from abnormally large clusters of SeqA-GFPuv4 in approximately 40% (7/18) of the cells (Figure 4B). This may reflect the defect in partitioning of SeqA-GFPuv4 foci. These abnormal types of cells were seen in less than 5% (2/43) of cells in a control subculture without azide, which is consistent with the results from Onogi et al. (1999). The isogenic azide-resistant secA204 strain (MQ337) exhibited fewer abnormalities (2/85), even in the presence of 1 mM sodium azide. These results show that inhibition of the SecA ATPase activity with sodium azide causes defects in the positioning of MukB-GFPuv4 and SeqA-GFPuv4 foci. This is in line with our assumption that SecA is involved in proper positioning of MukB and SeqA by uncharacterized mechanism.
FRAP Assay for SecA-GFPUV4, SecY-GFPUV4, AcpP-GFPUV4, and other Fluorescent Proteins
To gain further insight into the biochemical mechanisms underlying the interdependency of SecA, SecY, and AcpP, we performed a FRAP assay (Axelrod et al., 1976) and quantified the proportion of the immobile form of SecA-GFPuv4, SecY-GFPuv4, and AcpP-GFPuv4 in living cells (Figure 5; for the control experiments, see Supplementary Text).
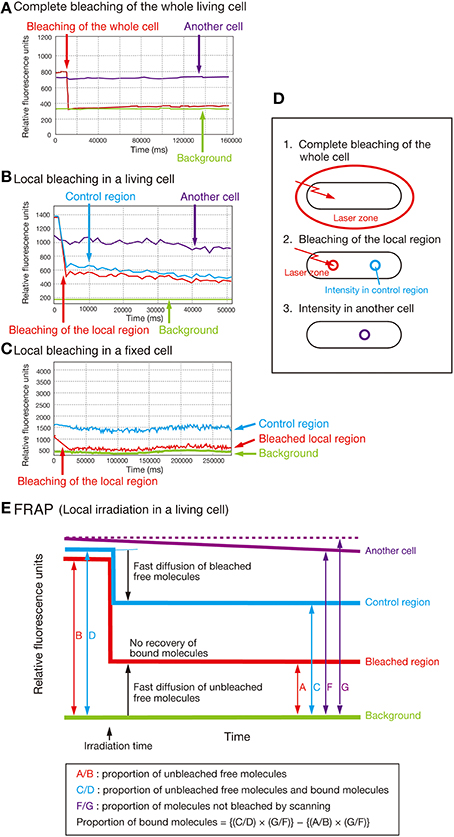
Figure 5. FRAP analysis. (A) Complete bleaching of the entire cell in the strain expressing SecA-GFPuv4 (MQ318). “Another cell” indicates another cell in exactly the same field that was not photobleached. (B) Typical FRAP data obtained with local bleaching in a cell of strain MQ318. (C) Effect of bleaching on the cells fixed with 1% formaldehyde in the strain MQ318 expressing SecA-GFPuv4. Note that the bleaching time was longer in fixed cells than in living cells. This is due to the reduced bleaching efficiency in fixed cells. (D,E) Concepts of the FRAP analysis.
The results of the FRAP assay are summarized in Table 2. The average proportion of the immobile form of SecA-GFPuv4 was 5–10% in wild-type cells (MQ318) but was increased to 15–20% in the presence of 1 mM sodium azide (Exp. 1). Similarly, the immobile form increased to 15–20% both in the cold-sensitive secY39(Cs) (a stimulator of the SecA ATPase activity) mutant and the temperature-sensitive acpPD38V mutant at the non-permissive temperatures of 22 and 42°C, respectively (Exps. 2 and 3). These results are consistent with the observation that the dynamic localization of SecA-GFPuv4 was affected in these conditions.
Note that the microscopy used in FRAP is entirely different from that used in time-lapse microscopy. For the FRAP assay, we imaged the cells with a sufficiently long exposure (~1 s) to ensure that all the signals could be detected in each image. On the other hand, for the time-lapse microscopy, we used a shorter exposure (~100 ms) for observing dynamic properties. Deletion of the protein encoded by secB, which is another stimulator of SecY ATPase, had no effect on the proportion of the immobile form of SecA-GFPuv4 (Exp. 4).
The average proportion of the immobile form of SecY-GFPuv4 was 7–8% in wild-type cells and 14% in the presence of 1 mM sodium azide (Exp. 5). On the other hand, addition of sodium azide had no effect on the immobile form of SecY-GFPuv4 in the azide-resistant secA204 mutant (Exp. 6). The discrepancy between the FRAP data and the time-lapse data might be due to the different microscopic systems to detect the fluorescence. Further explanation is described in the Discussion section. In contrast, addition of sodium azide had no remarkable effect on the proportion of the immobile form of AcpP-GFPuv4 (Exp. 7). For ParC-GFPuv4, ParE-GFPuv4 and GyrB-GFPuv4, the proportions of immobile molecules were 5–10%, similar to the other molecules studied (Exps. 8, 9, 11). For GyrA-GFPuv4, the proportion of immobile molecule was ~15%, slightly higher than the other molecules (Exp. 10). This might be the consequence of GyrA functions acting as a hub of protein–protein interaction network, involving MreB and PstB that interact with inner cell membrane proteins (Butland et al., 2005). For MukB, we were unable to bleach the small strong fluorescent foci without the effect on other regions with lower fluorescence because of the failure in making a small bleaching region in our system. Additionally, in wild-type strains, the proportion of the immobile form of SecA-GFPuv4 was the same at 37, 30, and 22°C, showing that the various temperatures did not affect the dynamics detected with FRAP.
Discussion
To briefly summarize, the present work demonstrates: (I) SecA, SecY, and AcpP localizations were non-uniform with condensed regions, embracing nucleoids; (II) SecA and SecY, together with MukB, Topo IV, and DNA gyrase, were dynamically localized in living cells; (III) Perturbation of SecY and AcpP resulted in abolishment of the dynamic localization of SecA; (IV) Conversely, perturbation of SecA results in abolishment of the dynamic localization of SecY; (V) The SecA ATPase activity was essential for the dynamic localization of SecY.
Chemical inhibition using 1 mM sodium azide (Figure 2I) or conditional genetic inhibition (Figure 2H) of SecA abolished the dynamic localization of SecY. These results indicate that SecA plays an essential role in the dynamics and that the rapid change in local patterns of these proteins is not due to random Brownian motion of protein molecules. Conversely, SecY or AcpP deficiency perturbed the normal dynamics of SecA (Figures 2C,E). These data suggest that SecA, SecY, and AcpP function in an interdependent manner and that the dynamics observed are not secondary effect of the mechanisms based on the other proteins, such as the secondary effects caused by aberrant membrane translocation of other proteins or proteins that localize dynamically besides SecA/SecY/AcpP. The results from the FRAP assay (Table 2) indicated that the addition of sodium azide caused an increase in the proportion of the immobile form (possibly the tortuous multimer along nucleoids, see Figure 1A) of SecA-GFPuv4 and that the immobile form of SecA-GFPuv4 was dependent on the ATP-bound immobile form of SecA (van der Wolk et al., 1997; Bauer et al., 2014) (Table 2, Exp. 1). Note that SecA exists both in peripheral of inner cell membrane and cytoplasm (Cabelli et al., 1991). SecA-ATP may be in an active polymeric state and form a stable bridge for chromosomes in this situation, as mentioned in Introduction section. SecY stimulates the ATPase activity of SecA, and therefore, SecY may stimulate release of the SecA bridge from the nucleoid by converting immobile SecA-ATP to mobile SecA-ADP (Bauer et al., 2014). In agreement with this speculation, the secY(Cs) mutation caused an increase in the proportion of the immobile form of SecA-GFPuv4 at the non-permissive temperature (Table 2, Exp. 2). SecB also stimulates the ATPase activity of SecA (Miller et al., 2002), but deletion of secB (MQ515) did not affect the proportion of the immobile form of SecA-GFPuv4 (Table 2, Exp. 4). Furthermore, the dynamic localizations of MukB, DNA gyrase and Topo IV are abolished in the presence of 1 mM sodium azide (our unpublished data). These results suggest that SecA/SecY, together with MukB and DNA topoisomerases, can act as a molecular “tether” for bacterial chromosomes by mediating the stressing (via the immobile form) and releasing (via the mobile form) cycles of chromosomes (Fisher et al., 2013). For further discussion of AcpP-GFPuv4, see the Supplementary Text.
As we briefly mentioned in the Results section, there is a discrepancy between the FRAP data and the time-lapse data: higher proportion (up to approximately 20%) of immobile forms in the mutants under non-permissive conditions or the azide treatment (defined as “disturbed conditions”) in FRAP data (Table 2), and reduced fluorescence under the disturbed conditions in the time-lapse imaging (Figures 2C,E,H). The discrepancy might be due to the different microscopic systems to detect the fluorescence. As one of the possible explanations, we speculate that the fluorescence molecules are classified into three forms: (1) immobile molecules centralized by SecA-ATP (Bauer et al., 2014); (2) molecules with medium values of diffusion coefficient; (3) molecules with the highest values of diffusion coefficient. The immobile form of SecA should be immobile SecA-ATP because in FRAP experiments, immobility is observed in the order of minutes, which is in agreement with “Push and Slide” mechanism proposed by Bauer et al. (2014). In the FRAP system (Table 2), the type 1 immobile molecules were 5–20% of the total fluorescence. The rest of fluorescence fraction might be classified into the two types, 2 and 3. On the other hand, in the time-lapse imaging, the type 3 molecules were unable to be detected because the diffusion speed of the type 3 molecules was too rapid. The major fraction of fluorescence detected in the time-lapse imaging of wild-type cells (Figures 2A,F) might be the type 2 molecules with medium values of diffusion coefficient, regarding that the fraction of type 1 molecule is less than 20%. In the mutants of time-lapse imaging under disturbed conditions (Figures 2C,E,H), the type 2 molecules might be reduced, while the type 3 molecules was remarkably increased, resulting in the intensive reduction of detectable fluorescence. As a whole, in the disturbed conditions, a large portion of the type 2 molecules might be converted to the type 3 molecules and a small portion might be converted to the type 1 molecules, resulting in the small increase of the type 1 molecules up to approximately 20% of fluorescence in FRAP. Fluorescence images of the time-lapse imaging thus resulted in the remarkable reduction of detectable fluorescence and disappearance of the dynamics of fluorescence localization. Therefore, the type 2 molecules act a prominent role on the observed dynamics in wild-type cells, which is performed by cooperative functions of SecA, SecY, and AcpP. The types 1, 2, and 3 of molecules presumably correspond to immobile core of the multimer interacting with other membrane proteins, dynamic multimer with medium values of diffusion coefficient, and monomer with the highest values of diffusion coefficient, respectively, similar to the case in eukaryotic actin filament.
Perturbation of the SecA ATPase by sodium azide disrupted the positioning of MukB-GFPuv4 and the proper organization of SeqA-GFPuv4. These results indicate that SecA/SecY/AcpP dynamics may be involved in the organization of MukB and SeqA. Furthermore, according to DNA decatenation and superhelicity data, SecA has epistasis over MukB and Topo IV/DNA gyrase (Adachi et al., 2014). These data are further supported by the protein–protein interactions. DNA-binding chromosome partitioning proteins such as MukB, Topo IV, and DNA gyrase appear to physically interact with SecA, SecY, and AcpP, among approximately 4300 proteins in E. coli (Butland et al., 2005). The results of the large-scale protein-protein interaction network suggest the presence of a sub-network of SecA-AcpP-MukB-ParC/ParE-GyrA/GyrB. In addition, AcpP co-purifies with MukB (Niki et al., 1992), and SeqA interacts with ParC (Kang et al., 2003). Stimulation of Topo IV activity by MukB and execution of MukB activity through the Topo IV-MukB interaction have been shown in previous studies (Hayama and Marians, 2010; Li et al., 2010; Hayama et al., 2013). Additionally, MukB is essential for the decatenation function of DNA gyrase and Topo IV in vivo (Adachi et al., 2014). According to the dynamic nature of the proteins, it may be possible that reaction-diffusion type positioning that is due to oscillating substances is achieved in the process, in which SecA is an activator and SecY is an inhibitor, similar to the bacterial Min system, which determines the plane of cell division (Rowlett and Margolin, 2013).
It may be possible that the observed deficiencies in dynamics of the proteins are secondary effects of inhibition of the membrane protein translocation. However, immediate inhibition of the dynamics (within 5 min) after the addition of 1 mM sodium azide or secY39(Cs) mutation suggests that this possibility is unlikely. secY39(Cs) mutant cells lose their translocation activity for membrane proteins within 1 min of being placed in a cold temperature (22°C) (Baba et al., 1990). In addition, wild-type cells with a normal secA allele grew exponentially at least for 2 h after the addition of the chemical compound, and then the growth was gradually inhibited, suggesting that growth defects due to abolishing protein translocation were not prominent during a time scale of minutes (Adachi et al., 2014). Furthermore, secA204 azide-resistant mutant, in which the membrane protein translocation activity of SecA is normal, exhibited abnormal localization of the oriC loci and a mild defect in SecY-GFPuv4 dynamics even in the absence of sodium azide (Adachi et al., 2014, this work). These results indicate that SecA is involved in proper positioning mechanism of oriC independently of the membrane protein translocation activity.
The apparent correlation between the dynamic properties of SecA/SecY/AcpP and the organization of the chromosome partitioning proteins may be important for these proteins to act as the molecular bridge between the inner cell membrane and chromosomes, possibly through molecular “tether” activity described in Fisher et al. (2013). Here, we can speculate that the tip of translocating multiple transmembrane protein can be immediately folded and interact with other membrane proteins that are often tightly linked with immobile protein multimers or cytoskeltons. This may make SecA-ATP in immobile form whereas SecA-ADP is in mobile form. Here, the mechanical force of SecA-ATP can result in pulling force (Bauer et al., 2014) of SecA-interacting AcpP-MukB or DNA topoisomerases (Adachi et al., 2014). From this speculation one may think that SecA might be involved in chromosome partitioning of E. coli. The present data on dynamics show that SecA and SecY may mediate the shorter scale (second order) dynamics of sister chromosomes described by Fisher et al. (2013). That is, these proteins may accumulate and release intra-nucleoid stress as the global molecular tether between the cell membrane and nucleoids acting on radial confinement and dynamic nucleoid density spots. On the other hand, an unknown programmed tether acts on a 20-min scale and promotes disposition of oriC, ter, and various loci in sister nucleoids at particular cell stages. Deficiency in SecA results in alteration of DNA topology such as superhelicity and decatenation (Adachi et al., 2014). This may be due to the absence of mechanical “greasing” activity of tethers as described in Fisher et al. (2013), resulting in a defect in separation of sister chromosomes. Chromosomal partitioning defects were observed in secA, secY, and acpP mutants (Adachi et al., 2014).
Our current results show that the dynamics of the membrane protein translocation machinery may be linked to chromosome partitioning and/or chromosome structure maintenance, possibly via bridging between SecA/SecY and AcpP/MukB/DNA gyrase/Topo IV/SeqA.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
For supplying bacterial strains and antibodies, we thank the E. coli Genetic Stock Center, National BioResource Project (NIG, Japan), and Drs. Koreaki Ito, Charles Rock, Masaaki Wachi, John Cronan, Junichi Kato, Ken Kreuzer, Akiko Nishimura, and Yuzuru Husimi. We thank Drs. Hiroshi Kimura and Naoki Watanabe for technical advice. We also thank Drs. Shunichi Takeda, Eiichiro Sonoda, Mitsuyoshi Yamazoe, Lawrence I. Rothfield, and Robert Lloyd for their kind advice and Dr. Toru Fukushima, Ayano Namazu, Takao Kumazawa, Yasuhiro Kuwata, Reo Hata, Ayano Tsukuda, and Akari Takagi for assistance. Shun Adachi thanks the Support Center for Advanced Medical Sciences, Institute of Health Biosciences, The University of Tokushima Graduate School for support and The Research Fellowship of The Japan Society for the Promotion of Science (JSPS) for Young Scientists for a fellowship and grant. Sota Hiraga thanks The Ministry of Education, Science, Sports, Culture and Technology of Japan for grants, and also The Center of Excellence (COE) of Kyoto University for financial support.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmicb.2015.00075/abstract
Movies S1–S24. Z-stacked deconvoluted images. For details, see the legend to Figure 1. Red, GFP/native immunofluorescnece. Green, DAPI staining.
Movie S1. Nucleoid with tortuous structures (PA340).
Movie S2. SecA-GFPuv4 with tortuous localization (MQ318).
Movie S3. Nucleoid with tortuous localization (MQ318).
Movie S4. Merged movie with tortuous localization (MQ318).
Movie S5. SecY-GFPuv4 with tortuous localization (MQ319).
Movie S6. Nucleoid with tortuous localization (MQ319).
Movie S7. Merged movie with tortuous localization (MQ319).
Movie S8. AcpP-GFPuv4 with tortuous localization (MQ322).
Movie S9. Nucleoid with tortuous localization (MQ322).
Movie S10. Merged movie with tortuous localization (MQ322).
Movie S11. GFPuv4 (MQ668).
Movie S12. Nucleoid with tortuous localization (MQ668).
Movie S13. Merged movie with tortuous localization (MQ668).
Movie S14. A control for GFPuv4 (non-fusion) staining (PA340).
Movie S15. Nucleoid with tortuous localization (PA340).
Movie S16. SecA with tortuous localization (PA340).
Movie S17. Nucleoid with tortuous localization (PA340).
Movie S18. Merged movie of S16 and S17 with tortuous localization (PA340).
Movie S19. SecY with tortuous localization (PA340).
Movie S20. Nucleoid with tortuous localization (PA340).
Movie S21. Merged movie of S19 and S20 with tortuous localization (PA340).
Movie S22. AcpP with tortuous localization (PA340).
Movie S23. Nucleoid with tortuous localization (PA340).
Movie S24. Merged movie of S22 and S23 with tortuous localization (PA340).
Movies S25–S40. Time-lapse movies of GFPuv4-fused proteins. Time-lapse images were taken by exposure for 100 ms, and the interval of each frame is 3 s. For details, see legend to Figure 2.
Movie S25. SecA-GFPuv4 in wild-type cells (MQ318) growing at 30°C.
Movie S26. SecA-GFPuv4 in secY39(Cs) mutant cells (MQ435) growing at the permissive temperature of 37°C.
Movie S27. SecA-GFPuv4 in secY39(Cs) mutant cells (MQ435) growing at the non-permissive temperature of 22°C for 5 min.
Movie S28. SecA-GFPuv4 in acpPD38V(Ts) mutant cells (MQ456) growing at the permissive temperature of 30°C.
Movie S29. SecA-GFPuv4 in acpPD38V(Ts) mutant cells (MQ456) growing at the non-permissive temperature of 42°C for 3 h.
Movie S30. SecY-GFPuv4 in wild-type cells (MQ319) growing at 30°C.
Movie S31. SecY-GFPuv4 in secA51(Ts) mutant cells (MQ748) growing at the permissive temperature of 30°C.
Movie S32. SecY-GFPuv4 in secA51(Ts) mutant cells (MQ748) growing at the non-permissive temperature of 42°C for 2 h.
Movie S33. SecY-GFPuv4 in wild-type cells (MQ319) growing at 30°C in the presence of 1 mM sodium azide. The incubation time with azide was approximately 5 min.
Movie S34. SecY-GFPuv4 in secA204 mutant cells (MQ625) growing at 30°C (without sodium azide).
Movie S35. ParC-GFPuv4 in wild-type cells (MQ537) growing at 30°C.
Movie S36. ParE-GFPuv4 in wild-type cells (MQ539) growing at 30°C.
Movie S37. GyrA-GFPuv4 in wild-type cells (MQ323) growing at 30°C.
Movie S38. GyrB-GFPuv4 in wild-type cells (MQ324) growing at 30°C.
Movie S39. MukB-GFPuv4 in wild-type cells (MQ529) growing at 30°C.
Movie S40. GFPuv4 in wild-type cells (MQ668) growing at 30°C.
References
Adachi, S., Fukushima, T., and Hiraga, S. (2008). Dynamic events of sister chromosomes in the cell cycle of Escherichia coli. Genes Cells 13, 181–197. doi: 10.1111/j.1365-2443.2007.01157.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Adachi, S., Murakawa, Y., and Hiraga, S. (2014). SecA defects are accompanied by dysregulation of MukB, DNA gyrase, chromosome partitioning and DNA superhelicity in Escherichia coli. Microbiology 160, 1648–1658. doi: 10.1099/mic.0.077685-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Axelrod, A., Koppel, D. E., Schlessinger, J., Elson, E., and Webb, W. W. (1976). Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys. J. 16, 1055–1069. doi: 10.1016/S0006-3495(76)85755-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Baba, T., Jacq, A., Brickman, E., Beckwith, J., Taura, T., Uegichi, C., et al. (1990). Characterization of cold-sensitive secY mutants of Escherichia coli. J. Bacteriol. 172, 7005–7010.
Badrinarayanan, A., Reyes-Lamothe, R., Uphoff, S., Leake, M. C., and Sherratt, D. J. (2012). In vivo architecture and action of bacterial structural maintenance of chromosome proteins. Science 338, 528–531. doi: 10.1126/science.1227126
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bates, D., and Kleckner, N. (2005). Chromosome and replisome dynamics in E. coli: loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell 121, 899–911. doi: 10.1016/j.cell.2005.04.013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bauer, B. W., Shemesh, T., Chen, Y., and Rapoport, T. A. (2014). A “push and slide” mechanism allows sequence-insensitive translocation of secretory proteins by the SecA ATPase. Cell 157, 1416–1429. doi: 10.1016/j.cell.2014.03.063
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Butland, G., Peregrin-Alvarez, J. M., Li, J., Yang, W., Yang, X., Canadien, V., et al. (2005). Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 433, 531–537. doi: 10.1038/nature03239
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cabelli, R. J., Dolan, K. M., Qian, L., and Oliver, D. B. (1991). Characterization of membrane-associated and soluble states of SecA protein from wild-type and secA51(TS) mutant strains of Escherichia coli. J. Biol. Chem. 266, 24420–24427.
Cronan, J. E. Jr., and Rock, C. O. (1996). “Biosynthesis of membrane lipids,” in Escherichia coli and Salmonella: Cellular and Molecular Biology, 2nd Edn., ed F. C. Neidhardt (Washington, DC: ASM Press), 612–636.
Cunha, S., Woldringh, C. L., and Odijk, T. (2001). Polymer-mediated compaction and internal dynamics of isolated Escherichia coli nucleoids. J. Struct. Biol. 136, 53–66. doi: 10.1006/jsbi.2001.4420
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Espeli, O., Levine, C., Hassing, H., and Marians, K. J. (2003). Temporal regulation of topoisomerase IV activity in E. coli. Mol. Cell 11, 189–201. doi: 10.1016/S1097-2765(03)00013-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fisher, J., Bourniquel, A., Witz, G., Weiner, B., Prentiss, M., and Kleckner, N. (2013). Four-dimensional imaging of E. coli nucleoid organization and dynamics in living cells. Cell 153, 882–895. doi: 10.1016/j.cell.2013.04.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hadizadeh Yazdi, N., Guet, C. C., Johnson, R. C., and Marko, J. F. (2012). Variation of the folding and dynamics of the Escherichia coli chromosome with growth conditions. Mol. Microbiol. 86, 1318–1333. doi: 10.1111/mmi.12071
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hayama, R., Bahng, S., Karasu, M., and Marians, K. J. (2013). The MukB-ParC interaction affects the intramolecular, not intermolecular, activities of Topoisomerase IV. J. Biol. Chem. 288, 7653–7661. doi: 10.1074/jbc.M112.418087
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hayama, R., and Marians, K. J. (2010). Physical and functional interaction between the condensin MukB and the decatenase topoisomerase IV in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 107, 18826–18831. doi: 10.1073/pnas.1008140107
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hiraga, S. (2000). Dynamic localization of bacterial and plasmid chromosomes. Annu. Rev. Genet. 34, 21–59. doi: 10.1146/annurev.genet.34.1.21
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hiraga, S., Ichinose, C., Niki, H., and Yamazoe, M. (1998). Cell cycle-dependent duplication and bidirectional migration of SeqA-associated DNA-protein complexes in E. coli. Mol. Cell 1, 381–387. doi: 10.1016/S1097-2765(00)80038-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hiraga, S., Ichinose, C., Onogi, T., Niki, H., and Yamazoe, M. (2000). Bidirectional migration of SeqA-bound hemimethylated DNA clusters and pairing of oriC copies in Escherichia coli. Genes Cells 5, 327–341. doi: 10.1046/j.1365-2443.2000.00334.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hiraga, S., Niki, H., Ogura, T., Ichinose, C., Mori, H., Ezaki, B., et al. (1989). Chromosome partitioning in Escherichia coli: novel mutants producing anucleate cells. J. Bacteriol. 171, 1496–1505.
Hirano, M., Mori, H., Onogi, T., Yamazoe, M., Niki, H., Ogura, T., et al. (1998). Autoregulation of the partition genes of the mini-F plasmid and the intracellular localization of their products in Escherichia coli. Mol. Gen. Genet. 257, 392–403. doi: 10.1007/s004380050663
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Issartel, J.-P., Koronakis, V., and Hughes, C. (1991). Activation of Escherichia coli prohaemolysin to the mature toxin by acyl carrier protein-dependent fatty acylation. Nature 351, 7559–7761. doi: 10.1038/351759a0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ito, Y., Suzuki, M., and Husimi, Y. (1999). A novel mutant of green fluorescent protein with enhanced sensitivity for microanalysis at 488 nm excitation. Biochem. Biophys. Res. Commun. 264, 556–560. doi: 10.1006/bbrc.1999.1541
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jun, S., and Mulder, B. (2006). Entropy-driven spatial organization of highly confined polymers: lessons for the bacterial chromosome. Proc. Natl. Acad. Sci. U.S.A. 103, 12388–12393. doi: 10.1073/pnas.0605305103
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kang, S., Han, J. S., Park, J. H., Skarstad, K., and Hwang, D. S. (2003). SeqA protein stimulates the relaxing and decatenating activities of topoisomerase IV. J. Biol. Chem. 278, 48779–48785. doi: 10.1074/jbc.M308843200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, Y., Stewart, N. K., Berger, A. J., Vos, S., Schoeffler, A. J., Berger, J. M., et al. (2010). Escherichia coli condensin MukB stimulates topoisomerase IV activity by a direct physical interaction. Proc. Natl. Acad. Sci. U.S.A. 107, 18832–18837. doi: 10.1073/pnas.1008678107
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lu, M., Campbell, J. L., Boye, E., and Kleckner, N. (1994). SeqA: A negative modulator of replication initiation in E. coli. Cell 77, 413–426. doi: 10.1016/0092-8674(94)90156-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Miller, A., Wang, L., and Kendall, D. A. (2002). SecB modulates the nucleotide-bound state of SecA and stimulates ATPase activity. Biochemistry 41, 5325–5332. doi: 10.1021/bi025639p
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Miller, J. H. (ed.). (1992). “Minimal salts,” in A Short Course in Bacterial Genetics, (New York, NY: Cold Spring Harbor Laboratory Press), 25.3.
Natale, P., Swaving, J., van der Does, C., de Keyzer, J., and Driessen, A. J. M. (2004). Binding of SecA to the SecYEG complex accelerates the rate of nucleotide exchange on SecA. J. Biol. Chem. 279, 13769–13777. doi: 10.1074/jbc.M312892200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Niki, H., Imamura, R., Kitaoka, M., Yamanaka, K., Ogura, T., and Hiraga, S. (1992). E. coli MukB protein involved in chromosome partition forms a homodimer with a rod-and-hinge structure having DNA binding and ATP/GTP binding activities. EMBO J. 11, 5101–5109.
Nolivos, S., and Sherratt, D. (2014). The bacterial chromosome: architecture and action of bacterial SMC and SMC-like complexes. FEMS Microbiol. Rev. 38, 380–392. doi: 10.1111/1574-6976.12045
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Odijk, T. (1998). Osmotic compaction of supercoiled DNA into a bacterial nucleoid. Biophys. Chem. 73, 23–29. doi: 10.1016/S0301-4622(98)00115-X
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ohsumi, K., Yamazoe, M., and Hiraga, S. (2001). Different localization of SeqA-bound nascent DNA clusters and MukF-MukE-MukB complex in Escherichia coli cells. Mol. Microbiol. 40, 835–845. doi: 10.1046/j.1365-2958.2001.02447.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Onogi, T., Niki, H., Yamazoe, M., and Hiraga, S. (1999). The assembly and migration of SeqA-Gfp fusion in living cells of Escherichia coli. Mol. Microbiol. 31, 1775–1782. doi: 10.1046/j.1365-2958.1999.01313.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Onogi, T., Ohsumi, K., Katayama, T., and Hiraga, S. (2002). Replication-dependent recruitment of ß-subunit of DNA polymerase III from cytosolic spaces to replication forks in Escherichia coli. J. Bacteriol. 184, 867–870. doi: 10.1128/JB.184.3.867-870.2002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Robson, A., and Collinson, I. (2006). The structure of the Sec complex and the problem of protein translocation. EMBO Rep. 7, 1099–1103. doi: 10.1038/sj.embor.7400832
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rowlett, V. W., and Margolin, W. (2013). The bacterial Min system. Curr. Biol. 23, R553–R556. doi: 10.1016/j.cub.2013.05.024
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Swulius, M. Y., and Jensen, G. J. (2012). The helical MreB cytoskeleton in Escherichia coli MC1000/pLE7 is artifact of the N-terminal Yellow Fluorescent Protein Tag. J. Bacteriol. 194, 6382–6386. doi: 10.1128/JB.00505-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Thronton, M., Armitage, M., Maxwell, A., Dosanjh, B., Howells, A. J., Norris, V., et al. (1994). Immunogold localization of GyrA and GyrB proteins in Escherichia coli. Microbiology 140, 2371–2382. doi: 10.1099/13500872-140-9-2371
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Towler, D. A., and Gordon, J. I. (1988). The biology and enzymology of eukaryotic protein acylation. Ann. Rev. Biochem. 57, 69–99. doi: 10.1146/annurev.bi.57.070188.000441
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
van der Wolk, J. P. W., de Wit, J. G., and Driessen, A. J. M. (1997). The catalytic cycle of the Escherichia coli SecA ATPase comprises two distinct preprotein translocation events. EMBO J. 16, 7297–7304. doi: 10.1093/emboj/16.24.7297
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wiggins, P. A., Cheveralls, K. C., Martin, J. S., Lintner, R., and Kondev, J. (2010). Strong intranucleoid interactions organize the Escherichia coli chromosome into a nucleoid filament. Proc. Natl. Acad. Sci. U.S.A. 107, 4991–4995. doi: 10.1073/pnas.0912062107
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yamazoe, M., Adachi, S., Kanaya, S., Ohsumi, K., and Hiraga, S. (2005). Sequential binding of SeqA protein to nascent DNA segments at replication forks in synchronized cultures of Escherichia coli. Mol. Microbiol. 55, 289–298. doi: 10.1111/j.1365-2958.2004.04389.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: chromosome partition, SecA, SecY, AcpP, MukB, DNA topoisomerase
Citation: Adachi S, Murakawa Y and Hiraga S (2015) Dynamic nature of SecA and its associated proteins in Escherichia coli. Front. Microbiol. 6:75. doi: 10.3389/fmicb.2015.00075
Received: 29 October 2014; Accepted: 21 January 2015;
Published online: 10 February 2015.
Edited by:
Mickael Desvaux, Institut National de la Recherche Agronomique, FranceReviewed by:
Anne Galinier, Centre National de la Recherche Scientifique, FranceOlivier Espeli, Centre National de la Recherche Scientifique, France
Tassos Economou, Katholieke Universiteit Leuven, Belgium
Copyright © 2015 Adachi, Murakawa and Hiraga. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sota Hiraga, Department of Radiation Genetics, Graduate School of Medicine, Kyoto University, Konoe-cho, Yoshida, Sakyo-ku, Kyoto 606-8501, Japan e-mail: hiraga64@grape.plala.or.jp
†Present address: Shun Adachi, Medical Research Project, Institute of Health Biosciences, The University of Tokushima Graduate School, Tokushima, Japan;
Yasuhiro Murakawa, Berlin Institute for Medical Systems Biology, Max Delbrück Center for Molecular Medicine Berlin-Buch, Berlin, Germany
 Shun Adachi
Shun Adachi Yasuhiro Murakawa
Yasuhiro Murakawa Sota Hiraga
Sota Hiraga