Viral Load in COVID-19 Patients: Implications for Prognosis and Vaccine Efficacy in the Context of Emerging SARS-CoV-2 Variants
- 1Laboratory of Virology and Experimental Therapy (LAVITE), Department of Virology, Aggeu Magalhães Institute (IAM), Oswaldo Cruz Foundation (Fiocruz), Recife, Brazil
- 2Leslie Dan Faculty of Pharmacy, University of Toronto, Toronto, ON, Canada
- 3MRC-University of Glasgow Centre for Virus Research, Glasgow, United Kingdom
The worldwide spread of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused an unprecedented public health crisis in the 21st century. As the pandemic evolves, the emergence of SARS-CoV-2 has been characterized by the emergence of new variants of concern (VOCs), which resulted in a catastrophic impact on SARS-CoV-2 infection. In light of this, research groups around the world are unraveling key aspects of the associated illness, coronavirus disease 2019 (COVID-19). A cumulative body of data has indicated that the SARS-CoV-2 viral load may be a determinant of the COVID-19 severity. Here we summarize the main characteristics of the emerging variants of SARS-CoV-2, discussing their impact on viral transmissibility, viral load, disease severity, vaccine breakthrough, and lethality among COVID-19 patients. We also provide a rundown of the rapidly expanding scientific evidence from clinical studies and animal models that indicate how viral load could be linked to COVID-19 prognosis and vaccine efficacy among vaccinated individuals, highlighting the differences compared to unvaccinated individuals.
Introduction
The emergence of coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has resulted in a worldwide emergency through rapid expansion of the virus (1). Clinically, most COVID-19 patients present mild or moderate symptoms, but ~15% of infected patients progress to pneumonia and 5% eventually develop more critical manifestations including acute respiratory distress syndrome (ARDS), and multiple organ dysfunction or failure (2, 3). Many studies have sought to elucidate predictors for COVID-19 severity in order to guide clinical management and prognosis of the disease and shed light into new therapeutic strategies (4). With this in mind, a growing body of evidence suggests that severe forms COVID-19 are associated with pronounced lymphopenia, lymphocyte dysfunction and activation, monocyte and granulocyte abnormalities, cytokine storm (increased levels of IL-1β, IL-6, IL-2, IL-8, IL-17, IP10, MCP1, MIP1α, G-CSF, GM-CSF, and TNF-α), high levels of C-reactive protein (CRP), D-dimer, immunoglobulin G (IgG), and total antibodies (4–7). Additionally, a number of reports have investigated the correlation between high viral loads and COVID-19 severity, where the results demonstrated high, little, or no statistical correlation with COVID-19 disease severity (6, 8–19).
As of December 28, 2021, SARS-CoV-2 accounted for more than 281.6 million infections and over 5.4 million deaths across the world-wide human population (1). Approximately 2 years have passed since the emergence of the virus, SARS-CoV-2 genomes are being routinely monitored through epidemiological investigations, virus genetic sequence-based surveillance, and shared at an unprecedented rate, with more than 6.5 million SARS-CoV-2 sequences available via the Global Initiative on Sharing All Influenza Data (GISAID), permitting near real-time surveillance and track the emergence of SARS-COV-2 mutations and new variants (20). Although most mutations in the SARS-CoV-2 genome are expected to be either neutral or deleterious, a proportion of these mutations will affect functional and viral properties in a way that confers a fitness advantage, which may alter infectivity, tropism, virulence, transmissibility, and/or interactions with host immunity (21). Beneficial mutations tend to occur in the minority of cases, when compared to negative effect or no effect “neutral” mutations (21). Notably, many of these beneficial mutations are due to non-synonymous nucleotide substitutions in key areas of the immunodominant spike protein of SARS-CoV-2, resulting in a change in amino acid. Five main variants of concern (VOCs) have emerged since the beginning of the pandemic and have attracted the most widespread attention: alpha (B.1.1.7) in the United Kingdom, beta (B.1.351) in South Africa, gamma (P.1) in Brazil, delta (B.1.617.2) in India, and omicron (B.1.1.529) in Botswana and South Africa (22–27). These variants have been associated with increased transmissibility, viral load, disease severity, evasion of immunity from infection and vaccinations, and reduced susceptibility to monoclonal antibody therapies (22, 23, 28–30), resulting in a catastrophic impact on SARS-COV-2 infection.
Based on the scientific knowledge published so far, we summarize the main characteristics of the emerging variants of SARS-CoV-2, including their impact on viral transmissibility, viral load, disease severity, vaccine breakthrough, and lethality among COVID-19 patients. We also discuss the rapidly expanding scientific evidence from clinical studies and animal models that indicate how viral load could be linked to COVID-19 prognosis and vaccine efficacy among vaccinated individuals, shedding light the differences compared to unvaccinated individuals (Figure 1).
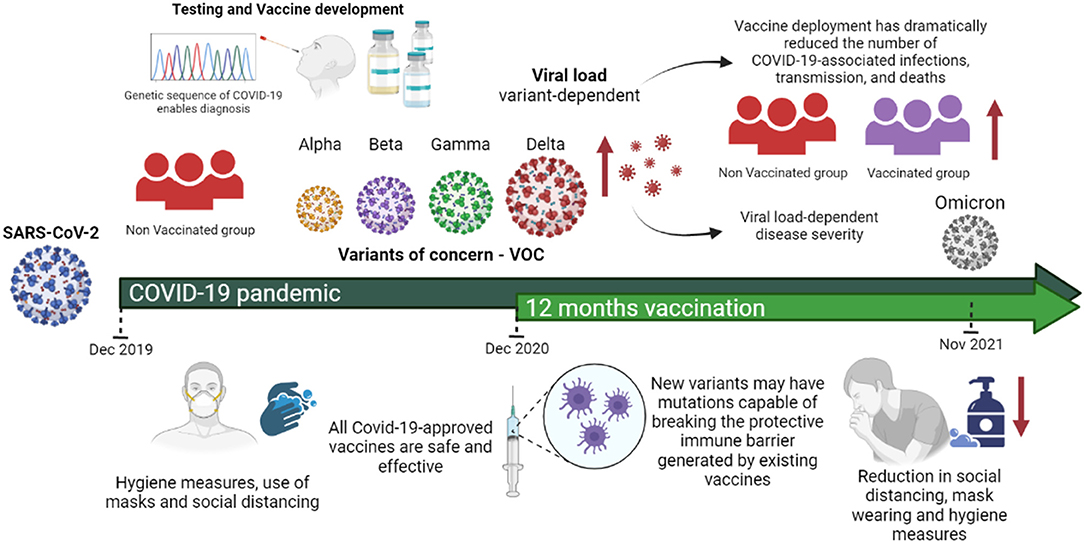
Figure 1. SARS-CoV-2 variants of concern and their impact on viral load, disease severity and vaccine efficacy. All approved COVID-19 vaccines are effective against the variants in circulation. However, in general the effectiveness is slightly lower against them and new variants may have mutations capable of breaking the protective immune barrier generated by existing vaccines. Vaccines together with mitigating measures such as social distance, mask wearing, ventilation and hygiene are important to counter the spread of these variants. Evidence from clinical studies and animal models has indicated that SARS-CoV-2 infectious dose is one likely determinant of ultimate COVID-19 severity and prognosis. However, reduction in hygiene care, mask wearing and social isolation further greater viral transmissibility. In summary, most studies have concluded that vaccination reduces the risk of infection by VOC infection and accelerated viral clearance. Figure 1 was created with Biorender.com under academic license.
Emerging SARS-CoV-2 Variants of Concern
During replication of SARS-CoV-2, the RNA-dependent RNA polymerase introduces mutations in the viral genome which may be subjected to selection pressures and then fixed in the population. It has been estimated that an average of 0.5 mutations are accumulated in every person during infection cycle (31). Given the high transmission rates of SARS-CoV-2 around the world, virtually every single base mutation is being generated de novo and transmitted daily to a new human host (31). To give rise to new variants, a SARS-CoV-2 mutant must overcome selection pressures and successfully establish a transmission chain among humans (31), which is the major bottleneck for SARS-CoV-2 inter-host dynamics, in which most mutated viruses do not transmit from their original host to another person (32). But despite this, several SARS-CoV-2 variants have been emerging and circulating widely since the beginning of the COVID-19 pandemic (Table 1).
Given the evolution of SARS-CoV-2 as the pandemic continues, a US government interagency group has developed a variant classification scheme to categorize mutant viruses that have arisen in the human population. The variants of concern (VOC) are the most relevant. According to their definition, a VOC is “a variant for which there is evidence of an increase in transmissibility, more severe disease (for example, increased hospitalizations or deaths), significant reduction in neutralization by antibodies generated during previous infection or vaccination, reduced effectiveness of treatments or vaccines, or diagnostic detection failures” (27).
The first SARS-CoV-2 VOC was the alpha variant, which emerged in September 2020 in the UK and rapidly become the dominant circulating variant spreading for 169 countries, presenting a cumulative world prevalence of 21% (https://outbreak.info/situation-reports). This variant belongs to Pango lineage B1.1.7, derived from clade 20I (V1) (91). The spike protein harbors several most mutations (Δ69–70 deletion, Δ144 deletion, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H) (33, 92). Phylogenetic analyses indicate that the alpha variant was associated with a growth rate estimated to be 40–70% higher than that of other SARS-CoV-2 variants (38). The N501Y and A570D mutations, located in the region binding domain (RBD), have been related to increase of viral binding affinity to ACE2 host receptor, contributing to transmissibility (33, 57, 93). Others such as P681H, exclusive of alpha VOC and localized adjacent to the furin cleavage site, may be involved in membrane fusion and immune escape (66). The Δ69–70 and Δ144 deletions may to affect the viral recognition, by neutralizing antibodies (Nabs), the testing kit failures (94, 95) and antibody escape (96). Previous studies in hamsters infected by SARS-CoV-2 and human for N501Y, Δ69 deletion and Δ70 deletion, observed a relation between these mutations with high viral load levels in nasal secretions and upper airway (in hamsters), and human airway epithelial cells (97). As well as the ancestor B.1 strain, the alpha variant could produce a high viral load, leading to a low Ct value in RT-qPCR-based diagnosis (47) while being ~50% more transmissible (33, 38, 39, 98). Previous analysis by Public Health England suggested an increase of death risk in patients infected by alpha VOC (99–101). The reinfection risk is very low (39). Mutations in alpha VOC have been related to reduce the neutralizing activity of monoclonal antibody-based therapies. Alpha VOC is refractory to neutralization by most monoclonal antibodies (mAbs) targeting the N-terminal domain (NTD) and is relatively resistant to mAbs against the RBD (102, 103). It is susceptible to neutralizing mAbs as well as by most plasma samples from previously infected and vaccinated individuals (29, 47, 63, 104–107). The sera from individuals who received the BNT162b2 (Pfizer) vaccine or convalescent sera of individuals who recovered from COVID-19 reduced neutralizing activities (47). Some studies have shown a 3-fold to 10-fold reduced susceptibility to 15% of plasma samples from recipients of an authorized mRNA vaccine (47, 104–106, 108, 109). Individuals vaccinated with mRNA vaccines [BNT162b2 (Pfizer) and mRNA-1273 (Moderna)] and adenovirus-vector (Ad26.COV2.S – Johnson & Johnson) may significantly neutralized the alpha VOC compared to the D614G variant (110). The BNT162b2 (Pfizer) vaccine demonstrated an effectiveness of 48.7% (CI, 45.5–51.7%) after one dose, while it was 93.7 (91.6–95.3%) after two doses among infected individuals with the alpha VOC (51). With the AZD1222 (AstraZeneca) vaccine, the effectiveness after one dose was similar to the results found with the BNT162b2 (Pfizer) vaccine, while it was 74.5% (95% CI, 68.4–79.4) after two doses among infected persons with the alpha VOC (51). For mRNA-1273 (Moderna) vaccine against RT-qPCR positive infections, the effectiveness was 88.1% (CI, 83.7–91.5%) and 100% (CI, 91.8–100%) after the first and second doses, respectively (64). Other clinical trials with the BNT162b2 (Pfizer) vaccine, conducted in Israel and Qatar, showed an efficacy > 90% against alpha variant (52, 111). When compared to NVX-CoV2373 clinical trial, the vaccine efficacy was 86.3% against alpha variant compared with 96.4% against non-alpha variants (53).
In October 2020, in South Africa, researchers found a new SARS-COV-2 B.1.351 variant, derived from the 20H (V2) clade and named beta variant by WHO (92, 93). Currently, this variant is present in more than 117 countries and has an accumulated worldwide prevalence of around 1% (https://outbreak.info/situation-reports). The beta variant has ten mutations (D80A, D215G, L241del, L242del, A243del, K417N, E484K, N501Y, D614G, and A701V) in the spike protein. Two new mutations (E484K and K417N) were identified in RBD region (25). The alpha and beta VOCs share two mutations (N501Y and D614G). Notably, these mutations in RBD region (N501Y, E484K, and K417N) may to enhance the binding affinity to human ACE2 receptor (33, 57), which can contribute as a critical role in the SARS-COV-2 transmission (58, 112). Mathematical modeling studies indicated that the beta variant is around 50% more transmissible than pre-existing SARS-CoV-2 variants (113). Reinfections have been reported, indicating immune evasion (114). Little is known whether the beta variant is associated with higher viral levels or disease severity, because once detected it was no longer co-circulating with other SARS-CoV-2 variants. In many Sub-Sahara Africa countries, the beta variant was responsible for more than 50% of infections (https://nextstrain.org/sars-cov-2/). In K18-hACE2 transgenic mice, alpha and beta VOCs induced pathogenic patterns and were 100-fold more lethal than early SARS-CoV-2 lineages (115). The European Centre for Disease Prevention and Control (ECDC), compared the COVID-19 severity cases between VOCs and non-VOCs, and observed that the beta variant was associated with a higher ratio for hospitalization (3.6) (60). The intensive care unit (ICU) and death risk data showed no difference in relation SARS-CoV-2 variants (60). The beta variant has been associated with reduced susceptibility to many mAbs and neutralizing Abs even in vaccinated or previously infected individuals, due to the E484K and K417N mutations (59, 60, 62). A differential susceptibility to neutralizing activities in beta VOC convalescent individuals has been observed. Indeed 46% of convalescent plasma samples displayed 3-fold to 10-fold reduced susceptibility, and 22% of these, >10-fold reduced susceptibility in comparison to early SARS-CoV-2 variants (29, 48, 59, 62, 106, 107, 109, 116–120). Vaccine neutralizing activities also have been evaluated. Individuals that received one of the mRNA vaccines [BNT162b2 (Pfizer) or mRNA-1273 (Moderna)] showed 3-fold to 10-fold (45% of plasma samples) and >10-fold (30% of plasma samples) reduced beta variant neutralizing activity (29, 46, 63, 69, 102, 107, 120–122). In relation to the adenovirus-based AZD1222 (AstraZeneca) vaccine, 42% of plasma samples had 3-fold to 10-fold, and 54% had >10-fold reduced beta variant neutralizing activity (81, 120, 123). With regard the efficacy of vaccines, a study carried out in Qatar-based individuals, revealed that the mRNA-1273 (Moderna) had an efficacy of 61.3 and 96.4% after the first and second doses against the beta variant, respectively (64). On the other hand, the NVX-CoV2373 (Novavax) vaccine showed an efficacy of 60% against the beta variant (54).
The gamma variant was first detected in four travelers returning to Japan from Amazonas state of Brazil in January 2021, but its emergence occurred in November 2020 (24, 28, 124). Currently, the gamma VOC is spread across 86 countries, with an accumulated worldwide prevalence of 2% (https://outbreak.info/situation-reports). The gamma variant has 12 missense mutations (L18F, T20N, P26S, D138Y, R190S, D614G, H655Y, T1027I, V1176F, K417T, E484K, and N501Y) in the spike protein, three of these (N501Y K417N and E484K) are located in the receptor-binding domain (RBD) (28). The triplet of K417T, E484K, and N501Y have been associated to increase virus binding affinity to human ACE2 receptor, which may contribute to increased transmissibility (28, 125). The gamma variant may be 1.7- to 2.4-fold more transmissible that previous SARS-CoV-2 variants (28). The emergence of this variant was associated with a resurgence of COVID-19 in Manaus, Brazil, resulting in an abrupt increase in the number of cases and deaths in this part of the world in January 2021 (22, 24, 28, 126). At the same time, the gamma variant was estimated to result in virus levels 3–4 times higher than earlier SARS-CoV-2 variants, being responsible for an estimated 1.1-fold to 1.8-fold higher mortality (28). Other studies reported a high proportion of COVID-19 infections in several South American and Caribbean countries and about 10% of USA infection rates in June 2021 (127). Additionally, a related study showed an increased risk for hospitalizations and ICU admission, comparing the disease severities among VOCs (B.1.1.7/SGTF, B.1.351 and P.1) and non-VOCs (60). Some mutations, such as L18F, may interfere in the binding of the NTD of spike-targeting neutralizing Abs (128). The resistance profile to the gamma to FDA EUA-approved mAbs is comparable to that of the beta variant (43, 129–131). Antibodies produced by natural infection or vaccines may be less likely to neutralize the gamma (69, 132). SARS-CoV-2 with the E484K might escape neutralization by Abs from convalescent plasma of recovered individual infected with earlier SARS-CoV-2 strains (74). A nationwide case-control study evaluating the protection at 7 days after the second dose of mRNA vaccine against the VOCs (alpha, beta, and gamma) and other non-SARS-CoV-2 variants in France estimated an effectiveness of 88% (alpha), 86% (beta) and 77% (gamma) against COVID-19 (70).
The fourth VOC, called delta by WHO, emerged in October 2020 in India (133) and it has spread to over 171 countries, becoming dominant with an accumulated worldwide prevalence of 54% (https://outbreak.info/situation-reports). Belonging to B.1.617.2 Pango lineage, derived from the 21A clade, the delta variant has 14 mutations (T19R, V70F, G142D, E156del, F157del, R158G, A222V, W258L, K417N, L452R, T478K, D614G, P681R, D950N) in the spike protein, among which only D614G is common to previous circulating SARS-CoV-2 variants (92). The spike RBD region carries 2 new non-synonymous mutations (L452R and T478K) and a deletion (del157). The L452R mutation may stabilize the interaction between the spike and the ACE2 receptor, increasing infectivity (74, 76, 134). P681, located near the furin cleavage site, may to optimize spike cleavage, which in turn may impact on transmissibility. Some studies have revealed that this variant has about 50% higher transmissibility compared to alpha variant (75, 92, 135). Despite differences between countries, Campbell et al. estimated a change in effective reproduction number of SARS-CoV-2 variants in 64 countries (data until 3 June 2021) (30). That study estimated the increase in transmissibility of VOC relative to non-VOC: alpha-29% (95% CI: 24–33), beta-25% (95% CI: 20–30), gamma-38% (95% CI: 29–48), and delta-97% (95% CI: 76–117) (30). In addition to enhanced transmissibility, individuals infected with the delta variant have higher viral load and sheds virus for longer periods (73), impacting the severity disease. The comparison between the virulence of the delta VOC and the non-VOC revealed a remarkable risk for disease severity associated to delta variant, with the increased hospitalizations, higher oxygen requirement, ICU admissions, and deaths (80). An in vitro study revealed that the delta variant (containing the mutations G124D) was 6-fold and 8-fold less sensitive to serum neutralizing Abs from recovered persons and vaccine-elicited Abs, respectively, compared to wild type D614G containing SARS-CoV-2 (77, 136). The delta variant may be resistant to neutralization by some anti-N-terminal domains and anti-RBD mAbs, including bamlanivimab (81). The presence of L452R in RBD decreased recognition by mAbs (76, 82, 137). The convalescent sera of individuals up to 12 months after the onset of symptoms were 4-fold less potent against the delta variant compared to alpha variant (81). Similarly, plasma samples obtained from recipients of BNT162b2 (Pfizer) and AZD1222 (AstraZeneca) vaccines displayed a reduction of neutralizing activity against the delta variant (65, 81, 121, 138). Even in the absence of the spike mutations, N501Y and E484K, the delta VOC was found to spread faster in the body and in vitro studies reveal lesser sensitivity to the BNT162b2 (Pfizer) vaccine (47, 121). Other studies have demonstrated that the BNT162b2 (Pfizer) and AZD1222 (AstraZeneca) vaccines were 85 and 60% effective against delta variant in the UK (83, 139). In the UK, previous studies have demonstrated that the effectiveness after the first dose of BNT162b2 (Pfizer) and AZD1222 (AstraZeneca) vaccines was 35.6 and 30.0% against symptomatic disease by delta variant. Following the second dose, the effectiveness was 88 and 67% for BNT162b2 (Pfizer) and AZD1222 (AstraZeneca) vaccines, respectively (51). In relation to COVID-19 hospitalization by alpha and delta VOCs vaccinated (mRNA-1273 or AZD1222), a Scottish study cohort observed among delta VOC individuals a strong vaccine effect in reducing the risk of hospital admission compared to unvaccinated individuals (83). The BNT162b2 (Pfizer) vaccine, at least 14 days after the second dose, offered 92 and 79% of protection against alpha and delta VOC infections, respectively (83), while the AZD1222 (AstraZeneca) vaccine offered 73 and 60% of protection against alpha and delta VOCs, respectively (83).
The fifth SARS-CoV-2 variant that has emerged so far is the omicron (B.1.1.529), which was first reported in Botswana and South Africa, November 2021 (26). This variant was detected on 6 continents within a month of its initial discovery and has raised concerns around the world. The omicron VOC is a heavily mutated SARS-CoV-2, with 30 amino acid substitutions, deletion of six residues, and insertion of three residues in the spike protein, mostly concentrated around the receptor binding motif (88). Among the mutations, an insertion (ins214EPE) in spike that was not been previously observed in other SARS-CoV-2 variants (140). It has been hypothesized that this insertion could have been acquired by template switching involving the genome of a low pathogenic coronavirus which can cause the common cold: HCoV-229R (140). A recent study has been estimated that the omicron variant is least ten times more infectious than the wild-SARS-CoV-2 and about twice as infectious as the delta variant (87). Analyzing the replication competence and cellular tropism of the wild-type virus, D614G, alpha, beta, delta and omicron variants in ex vivo explant cultures of human bronchus and lung, it was found that the omicron variant replicated faster than all other SARS-CoV-2 variants in the bronchus but less efficiently in the lung parenchyma (141). Based on in silico studies, it has been estimated that omicron may be twice more likely to escape immunity generated by current vaccines in comparison to the delta variant (87). It has been demonstrated that the omicron variant was associated with a substantial decrease in neutralization titer of vaccinated individuals with two doses [BNT162b2 (Pfizer) and AZD1222 (AstraZeneca)] (89). Similarly, by using the sera from 25 BNT162b2 (Pfizer) and 25 Coronavac (Sinovac) vaccine recipients, it was found that only 20% of BNT162b2 recipients had detectable neutralizing antibody against the omicron variant, while none of the Coronavac recipients had detectable neutralizing antibody titer against the Omicron VOC (90). These findings suggest that the omicron variant may be associated with lower COVID-19 vaccine effectiveness. Regarding the effect of neutralizing activity against monoclonal antibody-based therapies, recent findings showed that the omicron VOC substantially reduces neutralization by most of a large panel of potent monoclonal antibodies and antibodies under commercial development (88). The impact of omicron on disease severity, death and hospitalization is yet to be answered.
Viral Load as a Predictor of COVID-19 Severity
A cumulative body of data has indicated that the viral load is one likely factor of COVID-19 severity, as it is the case of other viral diseases. The relationship between SARS-CoV-2 viral load and risk of disease progression in COVID-19 patients remains undefined. In this section, we summarize the scientific evidence and the major findings from clinical studies, highlighting how viral load could be linked to a disease severity in COVID-19 patients.
In one of the first studies assessing the link between viral load and COVID-19 disease severity, Liu et al. analyzed the viral RNA shedding patterns by RT-qPCR in COVID-19 patients classified with mild and severe disease using samples from 76 patients (8). They found that viral load in nasopharyngeal specimens of severe cases was around 60 times higher than mild cases, and this positive correlation was maintained during the first 12 days of infection (8). In a further study, SARS-CoV-2 RNA viral shedding was evaluated in 3,497 samples (serum, respiratory, stool, and urine) from 96 consecutively admitted patients in a hospital in Zhejiang province, China (15). Viral load in respiratory samples, but not in stool and serum samples, of patients with severe disease was higher than in patients with mild disease. In severe ill patients, male gender and old age were associated with longer duration of virus shedding (15).
In a retrospective cohort study, the predictive power of several reported previously identified prognosis marker [circulating lymphocytes, IL-6, lactic acid, procalcitonin, CRP (C-reactive protein), and viral load] of 142 COVID-19 patients (6) were assessed. In this cohort, non-survivors had higher SARS-CoV-2 load in oropharyngeal swabs when compared to survivors. They authors of the study suggested that circulating lymphocytes, CRP, procalcitonin, IL-6, and viral load could serve as predictors for disease typing and guide classification of COVID-19 patients, and that circulating lymphocytes was the most reliable and sensitive predictor biomarker (6).
We analyzed the viral shedding patterns in nasopharyngeal specimens of 388 Brazilian patients with different forms of COVID-19 (10). Our results revealed that severe patients had higher viral load when compared to patients with mild disease, after 14 days of symptom onset. On the other hand, there was no statistically significant difference in viral load according to severity of COVID-19 in patients at early stage of infection (up to 14 days of symptoms onset) (10).
Pajudas et al. investigated the viral load of SARS-CoV-2 in nasopharyngeal swabs at time of diagnosis in a large cohort (n = 1,145) of hospitalized patients and found that survivors showed lower viral loads (n = 807; mean log10 viral load 5.2 copies per mL) than non-survivors (n = 338; 6.4 copies per mL) (142). Westblade et al. examined the SARS-CoV-2 viral load in 100 patients with cancer and 2,914 without cancer who were admitted to three New York City hospitals. In the overall cohort, the in-hospital mortality rate was 38.8% among patients with a high viral load, 24.1% among patients with a medium viral load, and 15.3% among patients with a low viral load (p < 0.001); and importantly, this association was also observed in patients with cancer (143). Fajnzylber et al. quantified SARS-CoV-2 viral load from the respiratory tract, plasma and urine of 231 patients with a diverse range of COVID-19 severity. They concluded that SARS-CoV-2 viral loads, especially in plasma, were associated with increased risk of mortality (144). In Spain, Calle et al. assessed the influence of viral load on the development of respiratory failure during admission in 455 sequential patients. They found that Ct value < 25 in nasopharyngeal samples was associated with increased risk of respiratory failure during admission (OR: 2.99, 95% IC: 1.57–5.69) and suggested that SARS-CoV-2 viral load at time of admission is a valuable predictor for COVID-19 severity (145).
However, the association between SARS-CoV-2 viral load and COVID-19 severity and outcome has not been consistently demonstrated in humans. A study in South Korea did not find any difference in viral load between asymptomatic vs. symptomatic patients (146). This was corroborated by a study in Turkish patients in which the viral load was not a critical factor for hospitalization and mortality among COVID-19 patients (147). The possibility of a type 2 error should be considered. However, it should be noted that not finding a statistical significance does not mean that no difference exists. Another factor that should be taken into account is that most studies that assess the viral load among COVID-19 patients just considered the Ct value for analysis, instead of the number of RNA copies/per milliliter (mL). In fact, Ct values are correlated with the amount of viral RNA in a patient sample (148). However, Ct values cannot be directly compared across RT-qPCR tests and, therefore, they must be interpreted with caution (148). Since many technical issues (differences in protocols, threshold values, viral target, enzymes and research kits, primers, calibration of RT-qPCR machine, period of sample collection, and type of biological specimens) that might impact and alter the Ct value during RT-qPCR reactions, this can represent a bias during the statistical analysis. We suggest that further studies consider using the RNA copies/per mL for viral load analysis among COVID-19 patients. A better comparative standard, combined with the evaluation of host-related factors (e.g., age, sex, comorbidities, etc.) (149) will be crucial to elucidate the real impact of SARS-CoV-2 viral load on COVID-19 disease severity.
Viral Load and Exposure Dose as a Predictor of COVID-19 Transmission and Severity in Animal Models
Because it would be unethical to assess how viral exposure dose would impact COVID-19 outcome in human controlled experiments, studies using animal models are being used to answer this question. Evidence from animal-based experiments has supported the notion that viral dose could impact disease outcome. The pathogenicity and replication of SARS-CoV-2 in hamsters infected with a low and high viral dose has given clues in that direction. No difference in viral replication titers in several organs were found. However, animals infected with high dose of SARS-CoV-2 had worse outcomes compared to those infected with the low dose (150). Similar results were obtained in mice, in which SARS-CoV-2 resulted in a dose-dependent lethal disease course of infection (151). A recent publication demonstrated that ferrets infected with a high (5 × 106 PFU) and medium (5 × 104 PFU) dose of SARS-CoV-2 had a more consistent upper respiratory tract viral RNA shedding and more severe lung pathology than animals infected with low viral dose (5 × 102 PFU) (152). Intranasal inoculation of ferrets with a high (5 × 106 PFU) or medium (5 × 104 PFU) doses of SARS-CoV-2 resulted in RNA viral shedding in nasal secretions of 100% of the animals; however, only 16.7% of the ferrets in the low challenge dose group (5 × 102 PFU) had detectable viral RNA in the nasal wash. In addition, live SARS-CoV-2 was not detected only in the nasal wash of the medium and low dose groups (152). Overall, a dose-dependent effect was observed in the clinical disease and histopathology (152). In non-human primates, aerosol exposure of cynomolgus macaques (Macaca fascicularis) with different doses of SARS-CoV-2 (5–906 TCID50) showed that the probability of infection and subsequent disease presentation were dose-dependent. The median infectious dose was 52 TCID50 (95% CI: 23–363 TCID50) for seroconversion and 256 TCID50 (95% CI: 102–603 TCID50) for fever development (153). The very low infectious dose of SARS-CoV-2 in this model supports the high transmissibility of the virus in humans.
The relative contribution of different transmission routes (intranasal, aerosol and fomite exposure) was evaluated in hamsters. Intranasal and aerosol exposure resulted in higher viral shedding and more severe disease compared to fomite exposure. Of the three routes studied, aerosol exposure resulted in more rapid virus replication in the lung and weight loss compared to intranasal inoculation and fomite exposure (154). The effects of mask wearing in reducing virus exposure dose and disease severity have been assessed in the hamster model. Hamsters placed in cages separated by a surgical mask partition were shown to be less likely to get infected by SARS-CoV-2 and if they did acquire the illness, it was milder than in animals not protected by a mask. Suggesting that the exposure dose is associated with disease severity (155).
Association Between Viral Load and Disease Severity in the Context of SARS-CoV-2 Variants
In the context of emerging variants of SARS-CoV-2, recent studies have investigated the impact of SARS-CoV-2 variants on disease severity. In a retrospective study, Ong et al. compared the outcomes of patients infected with alpha, beta, and delta with wild-type SARS-CoV-2 lineages from early 2020 (73). A total of 829 patients infected with these three VOCs in Singapore were enrolled in the study. After adjusting for age and sex, infection by the delta variant was linked with higher odds of oxygen requirement, ICU admission, or death [adjusted odds ratio (aOR), 4.90; 95% confidence interval (CI): 1.43–30.78], while these differences were not seen with alpha and beta variants. Vaccination status was associated with decreased severity. The delta variant was associated with significantly lower Ct values (≤30) and longer viral shedding (median duration 18 days for delta variant, while 13 days for wild type) (73). Taken together, these results suggest that infections with the delta variant feature higher peak viral loads than those in other SARS-CoV-2 variants. This finding also corroborates with outcomes obtained by other research groups, which showed that the delta SARS-CoV-2 variant has a higher viral load than beta and alpha variants in respiratory specimens obtained from COVID-19 patients (56). Within the same perspective, Teyssou et al. compared the relative viral load of the beta variant with alpha variant (156). Using a total of 643 RT-qPCR SARS-CoV-2 positive nasopharyngeal samples, they showed that the beta variant presented an intermediate relative viral load between the alpha other SARS-CoV-2 lineages in nasopharyngeal samples at diagnosis (156).
With regards to the relationship between disease severity and the emerging variants of SARS-CoV-2, a recent meta-analysis study investigated the relationship between SARS-CoV-2 variants and COVID-19 severity (157). Analyzing 26 studies from June 1, 2020, to October 15, 2021, they observed that alpha, beta, gamma, and delta SARS-CoV-2 variants were all more concerning than the wild-type virus in terms of hospitalization, ICU admission, and mortality (157). Interestingly, COVID-19 patients with beta and delta variants have a higher risk to develop severe clinical outcomes even death, when compared to patients with alpha and gamma variants (157).
In South Africa, a recent study investigated the breakthrough infections during periods of circulating beta, delta and omicron VOCs, among healthcare workers participating in the Sisonke phase 3B Ad26.COV2.S vaccine trial (158). Analyzing the data collected between 17 February and 15 December 2021, a total of 40,538 breakthrough infections were observed, resulting in 609 with beta, 22,279 with delta, and 17,650 with omicron. These findings revealed that the omicron variant was associated with a high number of breakthrough infections during the first 30-days of the omicron period in South Africa, while it was linked to less severe disease among COVID-19 patients (158). Interestingly, this finding also corroborate with recent insights achieved about the replication competence of the omicron variant in ex vivo explant cultures of human bronchus and lung, suggesting that the lower replication competence of omicron in human lung may be compatible with reduced severity in COVID-19 patients (141). However, further studies are required to confirm this hypothesis since the determinants of severe disease are multifactorial.
Viral Load in Vaccinated and Unvaccinated Individuals
The primary aim of COVID-19 vaccination is to protect individuals against clinical disease and death, ideally also reducing SARS-CoV-2 transmission in the human population. Although the approved COVID-19 vaccines are safe and effective, they do not provide sterilizing immunity, i.e., viral shedding can occur in vaccinated persons upon exposure to SARS-CoV-2. In addition, breakthrough infections have been reported in vaccinated individuals, although these cases tend to be much milder than in naïve individuals (34, 159, 160). Overall, real world data from several countries have unequivocally shown that vaccine deployment has dramatically reduced the number of COVID-19 associated infections, hospitalizations, and deaths (161, 162).
Some studies have analyzed breakthrough infections mRNA-based (BNT162b2—Pfizer) vaccine recipients, which is being used in many countries around the world. In Israel, its efficiency was 90% in preventing asymptomatic infection, suggesting a potential for halting virus spread (162). Previous studies done in Tel Aviv and Pittsburgh have ratified the role of BNT162b2 (Pfizer) vaccine in the control of the COVID-19 pandemic. In both studies, it was observed a lower viral load in nasal secretions vaccinated compared to non-vaccinated individuals was observed (163, 164). Evaluation of viral shedding after the first dose of the BNT162b2 (Pfizer) vaccine demonstrated that the viral load in nasal secretions was substantially reduced in vaccinated individuals (n = 1,888) compared to demographically matched unvaccinated controls (n = 1,888) (165).
On the other hand, SARS-CoV-2 shedding did not differ between vaccinated (BNT162b2 vaccine) and non-vaccinated healthcare workers infected with the alpha variant, suggesting potentially reduced efficacy of BNT162b2 in preventing transmission of this variant (166). Similar findings were reported in studies conducted in California (167), Wisconsin (168), Massachusetts (80), and Singapore (169) against the delta variant.
A study done with 3720 Italian healthcare workers fully vaccinated with the BNT162b2 vaccine showed that the 100-day cumulative incidence of vaccine breakthrough infections by the alpha variant was 0.93% in vaccinated vs. 5.78% in non-vaccinated individuals (170). Furthermore, antibody and T-cell responses are not reduced in subjects with breakthrough infection (170). Other studies have been characterized SARS-CoV-2 breakthrough infections in individuals fully vaccinated with mRNA vaccines (BNT162b2 and mRNA-1273). More recently, 14 breakthrough infections have been reported among vaccinated individuals by Deng et al. (159). In that study, half of the cases were immunosuppressed subjects who developed severe disease and required hospitalization (159). Sequencing analysis of infecting virus revealed four distinct SARS-CoV-2 variants, including the alpha and gamma (159). High viral load was detected in symptomatic and asymptomatic patients regardless of disease severity, highlighting the vulnerability of immunosuppressed individuals to post-vaccination infections by diverse variants of SARS-CoV-2 (159).
SARS-CoV-2 viral dynamics were investigated in a prospective, longitudinal study with 173 participants (37 vaccinated and 136 unvaccinated) in the USA using 19,941 patient samples (171). Among them, 36 participants were infected with the alpha variant, 36 participants with the delta variant, and 41 participants with a non-VOC SARS-CoV-2 (171). They found no meaningful difference in the mean peak viral load, shedding duration, clearance duration, or duration of acute infection of either the variants as compared with non VOC (171). Breakthrough infections in vaccinated individuals showed a faster clearance time (5.5 days) compared to non-vaccinated individuals (7.5 days), demonstrating a shorter overall shedding duration among vaccine recipients (171).
A similar prospective and longitudinal study was conducted in the UK in order to investigate the delta variant viral load kinetics in vaccinated [BNT162b2 (Pfizer), AZD1222 (AstraZeneca) and CoronaVac (Sinovac)] and unvaccinated individuals (172). The results revealed that the fully vaccinated individuals with delta variant had a faster mean rate of viral load decline when compared to non-vaccinated individuals with pre-alpha, alpha, or delta variant infections (172). Moreover, it was found that the faster viral load growth was correlated with higher peak viral load and slower decline among individuals (172). In summary, the authors suggested that vaccination reduced the risk of delta variant infection and accelerated viral clearance (172).
A retrospective multicenter cohort study of 17 hospitals in Israel detailed 152 breakthrough infections in fully vaccinated with the BNT162b2 vaccine who developed COVID-19 disease more than 7 days after the second vaccine dose and required hospitalization (173). The cohort was characterized by a large proportion of patients with comorbidities (96%) and immunodepression (40%) (173). They found that higher SARS-CoV-2 load was associated with a significant risk for poor outcome (173). By sequencing analysis, SARS-CoV-2 sequences indicated that most breakthrough infections were caused by the alpha variant (89%) followed by the wild-type virus (7%), and the beta variant (4%) (173).
More recently, another study conducted in Israel analyzed the viral load of 16,000 infections during delta-VOC after vaccination and booster with BNT162b2 vaccine (174). The breakthrough infections in recently fully vaccinated individuals by delta variant showed a lower viral load in comparison to non-vaccinated individuals, but this effect started to decline 2 months after vaccination and vanished 6 months or longer after vaccination (174). In addition, it was found that the effect of the BNT162b2 vaccine on reducing breakthrough infections viral loads is restored after a booster dose (174). Taken together, these findings suggest that BNT162b2 vaccine might decrease the infectiousness of breakthrough infections even with the delta variant (174). However, this protective effect declines over time, but it can be restored with a third vaccine dose (174).
Final Considerations and Public Health Perspectives
In conclusion, a cumulative body of evidence from animal models and clinical studies has indicated that the SARS-CoV-2 viral load is one likely determinant of ultimate COVID-19 severity and prognosis. The viral exposure dose is also a key determinant to a vaccine protective efficacy (175) and should be kept in mind as vaccine coverage of COVID-19 expands around the world. Despite the current approved COVID-19 vaccines are effective against severe forms of COVID-19, but they are not 100% effective in preventing infection. With is mind, breakthrough infections have been reported in vaccinated individuals, although these cases tend to be much milder than in naïve individuals, especially by the delta variant, which is more transmissible when compared to other SARS-CoV-2 variants. Overall, real world findings from several countries have unequivocally shown that vaccine deployment has dramatically reduced the number of COVID-19 cases, ICU admissions, and deaths. Thus, strategies to reduce viral exposure dose such as masking, frequent hand washing, avoid close contact, mouth and nose covering when coughing, frequently cleaning and disinfecting touched surfaces are crucial to prevent new cases of COVID-19 in the human population. These measures, combined with high coverage vaccination and booster shots, will be crucial to control the COVID-19 and prevent the emergence and spread of new SARS-CoV-2 variants.
In the context of emerging SARS-CoV-2 variants, the recent findings suggest that alpha, beta, gamma, and delta SARS-CoV-2 variants are more serious than the wild-type virus in terms of hospitalization, ICU admission, mortality, and are associated with higher viral loads. Within the SARS-CoV-2 VOCs, patients with beta and delta variants have a higher risk to develop severe clinical outcomes even death, when compared to infected patients with alpha and gamma variants.
Author Contributions
LP, AK, and SS conceived the work. SS, SL, RS, and LP wrote the original draft. SS, SL, RS, AK, and LP reviewed the final draft. LP supervised the work. All authors contributed to the article and approved the submitted version.
Funding
The work in LP's lab was funded by the Fiocruz Inova Program and the Foundation for Science and Technology of Pernambuco – FACEPE, Brazil (Grant No. APQ-0560-2.12/19). SS was recipient of a Ph.D. fellowship sponsored by the Foundation for Science and Technology of Pernambuco (FACEPE) Brazil, (Grant No. IBPG-1321-2.12/18). AK was supported by UK Medical Research Council (Grant No. MC_UU_12014/8). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. (2020) 20:533–4. doi: 10.1016/S1473-3099(20)30120-1
2. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. (2020) 395:497–506. doi: 10.1016/S0140-6736(20)30183-5
3. Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. (2020) 8:420–2. doi: 10.1016/S2213-2600(20)30076-X
4. Yang L, Liu S, Liu J, Zhang Z, Wan X, Huang B, et al. COVID-19: immunopathogenesis and immunotherapeutics. Signal Transduct Target Ther. (2020) 5:128. doi: 10.1038/s41392-020-00243-2
5. Cao X. COVID-19: immunopathology and its implications for therapy. Nat Rev Immunol. (2020) 20:269–70. doi: 10.1038/s41577-020-0308-3
6. Tan L, Kang X, Ji X, Li G, Wang Q, Li Y, et al. Validation of predictors of disease severity and outcomes in COVID-19 patients: a descriptive and retrospective study. Med. (2020) 1:128–38.e3. doi: 10.1016/j.medj.2020.05.002
7. Zheng Z, Peng F, Xu B, Zhao J, Liu H, Peng J, et al. Risk factors of critical & mortal COVID-19 cases: a systematic literature review and meta-analysis. J Infect. (2020) 81:e16–e25. doi: 10.1016/j.jinf.2020.04.021
8. Liu Y, Yan LM, Wan L, Xiang TX, Le A, Liu JM, et al. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect Dis. (2020) 20:656–7. doi: 10.1016/S1473-3099(20)30232-2
9. Lescure FX, Bouadma L, Nguyen D, Parisey M, Wicky PH, Behillil S, et al. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect Dis. (2020) 20:697–706. doi: 10.1016/S1473-3099(20)30200-0
10. Ferraz de Magalhães JJ, Germano Mendes RP, Alves da Silva CT, Ribeiro da Silva SJ, Guarines KM, Pena L, et al. Epidemiological and clinical characteristics of the first 557 successive patients with COVID-19 in pernambuco state, Northeast Brazil. Travel Med Infect Dis. (2020) 38:101884. doi: 10.1016/j.tmaid.2020.101884
11. Pan Y, Zhang D, Yang P, Poon LLM, Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis. (2020) 20:411–2. doi: 10.1016/S1473-3099(20)30113-4
12. Shi F, Wu T, Zhu X, Ge Y, Zeng X, Chi Y, et al. Association of viral load with serum biomakers among COVID-19 cases. Virology. (2020) 546:122–6. doi: 10.1016/j.virol.2020.04.011
13. To KK, Tsang OT, Leung WS, Tam AR, Wu TC, Lung DC, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. (2020) 20:565–74. doi: 10.1016/S1473-3099(20)30196-1
14. Yu F, Yan L, Wang N, Yang S, Wang L, Tang Y, et al. Quantitative detection and viral load analysis of SARS-CoV-2 in infected patients. Clin Infect Dis. (2020) 71:793–8. doi: 10.1093/cid/ciaa345
15. Zheng S, Fan J, Yu F, Feng B, Lou B, Zou Q, et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. (2020) 369:m1443. doi: 10.1136/bmj.m1443
16. Abdulrahman A, Mallah SI, Alqahtani M. COVID-19 viral load not associated with disease severity: findings from a retrospective cohort study. BMC Infect Dis. (2021) 21:688. doi: 10.1186/s12879-021-06376-1
17. Knudtzen FC, Jensen TG, Lindvig SO, Rasmussen LD, Madsen LW, Hoegh SV, et al. SARS-CoV-2 viral load as a predictor for disease severity in outpatients and hospitalised patients with COVID-19: a prospective cohort study. PLoS ONE. (2021) 16:e0258421. doi: 10.1371/journal.pone.0258421
18. El Zein S, Chehab O, Kanj A, Akrawe S, Alkassis S, Mishra T, et al. SARS-CoV-2 infection: initial viral load (iVL) predicts severity of illness/outcome, and declining trend of iVL in hospitalized patients corresponds with slowing of the pandemic. PLoS ONE. (2021) 16:e0255981. doi: 10.1371/journal.pone.0255981
19. Pérez-García F, Martin-Vicente M, Rojas-García RL, Castilla-García L, Muñoz-Gomez MJ, Hervás Fernández I, et al. High SARS-CoV-2 viral load and low CCL5 expression levels in the upper respiratory tract are associated with COVID-19 severity. J Infect Dis. (2021) 1–19. doi: 10.1093/infdis/jiab604
20. Meredith LW, Hamilton WL, Warne B, Houldcroft CJ, Hosmillo M, Jahun AS, et al. Rapid implementation of SARS-CoV-2 sequencing to investigate cases of health-care associated COVID-19: a prospective genomic surveillance study. Lancet Infect Dis. (2020) 20:1263–71. doi: 10.1016/S1473-3099(20)30562-4
21. Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. (2021) 19:409–24. doi: 10.1038/s41579-021-00573-0
22. Silva SJRD, Pena L. Collapse of the public health system and the emergence of new variants during the second wave of the COVID-19 pandemic in Brazil. One Health. (2021) 13:100287. doi: 10.1016/j.onehlt.2021.100287
23. Abdool Karim SS, de Oliveira T. New SARS-CoV-2 variants - clinical, public health, and vaccine implications. N Engl J Med. (2021) 384:1866–8. doi: 10.1056/NEJMc2100362
24. Naveca FG, Nascimento V, de Souza VC, Corado AL, Nascimento F, Silva G, et al. COVID-19 in Amazonas, Brazil, was driven by the persistence of endemic lineages and P.1 emergence. Nat Med. (2021) 27:1230–8. doi: 10.1038/s41591-021-01378-7
25. Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. (2021) 592:438–43. doi: 10.1038/s41586-021-03402-9
26. Callaway E. Heavily mutated Omicron variant puts scientists on alert. Nature. (2021) 600:21. doi: 10.1038/d41586-021-03552-w
28. Faria NR, Mellan TA, Whittaker C, Claro IM, Candido DDS, Mishra S, et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science. (2021) 372:815–21. doi: 10.1126/science.abh2644
29. Wang P, Nair MS, Liu L, Iketani S, Luo Y, Guo Y, et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. (2021) 593:130–5. doi: 10.1038/s41586-021-03398-2
30. Campbell F, Archer B, Laurenson-Schafer H, Jinnai Y, Konings F, Batra N, et al. Increased transmissibility and global spread of SARS-CoV-2 variants of concern as at June 2021. Euro Surveill. (2021) 26:2100509. doi: 10.2807/1560-7917.ES.2021.26.24.2100509
31. Sender R, Bar-On Bar-On YM, Gleizer S, Bernshtein B, Flamholz A, Phillips R, et al. The total number and mass of SARS-CoV-2 virions. Proc Natl Acad Sci USA. (2021) 118:1–9. doi: 10.1073/pnas.2024815118
32. Braun KM, Moreno GK, Wagner C, Accola MA, Rehrauer WM, Baker DA, et al. Acute SARS-CoV-2 infections harbor limited within-host diversity and transmit via tight transmission bottlenecks. PLoS Pathog. (2021) 17:e1009849. doi: 10.1371/journal.ppat.1009849
33. Davies NG, Abbott S, Barnard RC, Jarvis CI, Kucharski AJ, Munday JD, et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. (2021) 372:eabg3055. doi: 10.1126/science.abg3055
34. Calistri P, Amato L, Puglia I, Cito F, Di Giuseppe A, Danzetta ML, et al. Infection sustained by lineage B.1.1.7 of SARS-CoV-2 is characterised by longer persistence and higher viral RNA loads in nasopharyngeal swabs. Int J Infect Dis. (2021) 105:753–55. doi: 10.1016/j.ijid.2021.03.005
35. Costa R, Bueno F, Giménez E, Bracho A, Albert E, Carretero D, et al. Initial viral load and decay kinetics of SARS-CoV-2 lineage B.1.1.7 in the upper respiratory tract of adults and children. J Infect. (2021) 83:496–522. doi: 10.1016/j.jinf.2021.08.015
36. Frampton D, Rampling T, Cross A, Bailey H, Heaney J, Byott M, et al. Genomic characteristics and clinical effect of the emergent SARS-CoV-2 B.1.1.7 lineage in London, UK: a whole-genome sequencing and hospital-based cohort study. Lancet Infect Dis. (2021) 21:1246–56. doi: 10.1016/S1473-3099(21)00170-5
37. Kidd M, Richter A, Best A, Cumley N, Mirza J, Percival B, et al. S-variant SARS-CoV-2 lineage B1.1.7 is associated with significantly higher viral load in samples tested by TaqPath polymerase chain reaction. J Infect Dis. (2021) 223:1666–70. doi: 10.1093/infdis/jiab082
38. Volz E, Mishra S, Chand M, Barrett JC, Johnson R, Geidelberg L, et al. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature. (2021) 593:266–9. doi: 10.1038/s41586-021-03470-x
39. Graham MS, Sudre CH, May A, Antonelli M, Murray B, Varsavsky T, et al. Changes in symptomatology, reinfection, and transmissibility associated with the SARS-CoV-2 variant B.1.1.7: an ecological study. Lancet Public Health. (2021) 6:e335–45. doi: 10.1101/2021.01.28.21250680
40. Mukherjee R, Satardekar R. Why are some coronavirus variants more infectious? J Biosci. (2021) 46:1–10. doi: 10.1007/s12038-021-00221-y
41. Gómez CE, Perdiguero B, Esteban M. Emerging SARS-CoV-2 variants and impact in global vaccination programs against SARS-CoV-2/COVID-19. Vaccines. (2021) 9:243. doi: 10.3390/vaccines9030243
42. Soh SM, Kim Y, Kim C, Jang US, Lee HR. The rapid adaptation of SARS-CoV-2-rise of the variants: transmission and resistance. J Microbiol. (2021) 59:807–18. doi: 10.1007/s12275-021-1348-5
43. Hoffmann M, Arora P, Groß R, Seidel A, Hörnich BF, Hahn AS, et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. (2021) 184:2384–93.e2312. doi: 10.1016/j.cell.2021.03.036
44. Gallais F, Gantner P, Bruel T, Velay A, Planas D, Wendling M, et al. Evolution of antibody responses up to 13 months after SARS-CoV-2 infection and risk of reinfection. EBioMedicine. (2021) 71:1–11. doi: 10.1016/j.ebiom.2021.103561
45. Stirrup O, Boshier F, Venturini C, Guerra-Assunção JA, Alcolea-Medina A, Beckett A, et al. SARS-CoV-2 lineage B.1.1.7 is associated with greater disease severity among hospitalised women but not men: multicentre cohort study. BMJ Open Respir Res. (2021) 8:e001029. doi: 10.1136/bmjresp-2021-001029
46. Wu K, Werner AP, Koch M, Choi A, Narayanan E, Stewart-Jones GBE, et al. Serum neutralizing activity elicited by mRNA-1273 vaccine. N Engl J Med. (2021) 384:1468–70. doi: 10.1056/NEJMc2102179
47. Collier DA, De Marco A, Ferreira IATM, Meng B, Datir RP, Walls AC, et al. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature. (2021) 593:136–41. doi: 10.1038/s41586-021-03412-7
48. Edara VV, Norwood C, Floyd K, Lai L, Davis-Gardner ME, Hudson WH, et al. Infection- and vaccine-induced antibody binding and neutralization of the B.1.351 SARS-CoV-2 variant. Cell Host Microbe. (2021) 29:516–21.e513. doi: 10.1016/j.chom.2021.03.009
49. Emary KRW, Golubchik T, Aley PK, Ariani CV, Angus B, Bibi S, et al. Efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine against SARS-CoV-2 variant of concern 202012/01 (B.1.1.7): an exploratory analysis of a randomised controlled trial. Lancet. (2021) 397:1351–62. doi: 10.1016/S0140-6736(21)00628-0
50. Shen X, Tang H, McDanal C, Wagh K, Fischer W, Theiler J, et al. SARS-CoV-2 variant B.1.1.7 is susceptible to neutralizing antibodies elicited by ancestral spike vaccines. Cell Host Microbe. (2021) 29:529–39.e523. doi: 10.1016/j.chom.2021.03.002
51. Lopez Bernal J, Andrews N, Gower C, Gallagher E, Simmons R, Thelwall S, et al. Effectiveness of Covid-19 vaccines against the B.1.617.2 (Delta) variant. N Engl J Med. (2021) 385:585–94. doi: 10.1056/NEJMoa2108891
52. Abu-Raddad LJ, Chemaitelly H, Butt AA, Vaccination NSGC. Effectiveness of the BNT162b2 Covid-19 vaccine against the B.1.1.7 and B.1.351 variants. N Engl J Med. (2021) 385:187–9. doi: 10.1056/NEJMc2104974
53. Heath PT, Galiza EP, Baxter DN, Boffito M, Browne D, Burns F, et al. Safety and efficacy of NVX-CoV2373 covid-19 vaccine. N Engl J Med. (2021) 385:1172–83. doi: 10.1056/NEJMoa2107659
54. Mahase E. Covid-19: novavax vaccine efficacy is 86% against UK variant and 60% against South African variant. BMJ. (2021) 372:n296. doi: 10.1136/bmj.n296
55. Sadoff J, Gray G, Vandebosch A, Cárdenas V, Shukarev G, Grinsztejn B, et al. Safety and efficacy of single-dose Ad26.COV2.S vaccine against Covid-19. N Engl J Med. (2021) 384:2187–201. doi: 10.1056/NEJMoa2101544
56. Teyssou E, Delagrèverie H, Visseaux B, Lambert-Niclot S, Brichler S, Ferre V, et al. The delta SARS-CoV-2 variant has a higher viral load than the Beta and the historical variants in nasopharyngeal samples from newly diagnosed COVID-19 patients. J Infect. (2021) 83:e1–3. doi: 10.1016/j.jinf.2021.08.027
57. Starr TN, Greaney AJ, Hilton SK, Ellis D, Crawford KHD, Dingens AS, et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. (2020) 182:1295–310.e1220. doi: 10.1016/j.cell.2020.08.012
58. Ramanathan M, Ferguson ID, Miao W, Khavari PA. SARS-CoV-2 B.1.1.7 and B.1.351 spike variants bind human ACE2 with increased affinity. Lancet Infect Dis. (2021) 21:1070. doi: 10.1016/S1473-3099(21)00262-0
59. Wibmer CK, Ayres F, Hermanus T, Madzivhandila M, Kgagudi P, Oosthuysen B, et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat Med. (2021) 27:622–5. doi: 10.1038/s41591-021-01285-x
60. Funk T, Pharris A, Spiteri G, Bundle N, Melidou A, Carr M, et al. Characteristics of SARS-CoV-2 variants of concern B.1.1.7, B.1.351 or P.1: data from seven EU/EEA countries, weeks 38/2020 to 10/2021. Euro Surveill. (2021) 26:2100348. doi: 10.2807/1560-7917.ES.2021.26.16.2100348
61. Jassat W, Mudara C, Ozougwu L, Tempia S, Blumberg L, Davies M, et al. Increased mortality among individuals hospitalised with COVID-19 during the second wave in South Africa. MedRxiv. (2021) 1–26. doi: 10.1101/2021.03.09.21253184
62. Cele S, Gazy I, Jackson L, Hwa SH, Tegally H, Lustig G, et al. Escape of SARS-CoV-2 501Y.V2 from neutralization by convalescent plasma. Nature. (2021) 593:142–6. doi: 10.1038/s41586-021-03471-w
63. Shen X, Tang H, Pajon R, Smith G, Glenn GM, Shi W, et al. Neutralization of SARS-CoV-2 variants B.1.429 and B.1.351. N Engl J Med. (2021) 384:2352–4. doi: 10.1056/NEJMc2103740
64. Chemaitelly H, Yassine HM, Benslimane FM, Al Khatib HA, Tang P, Hasan MR, et al. mRNA-1273 COVID-19 vaccine effectiveness against the B.1.1.7 and B.1.351 variants and severe COVID-19 disease in Qatar. Nat Med. (2021) 27:1614–21. doi: 10.1038/s41591-021-01446-y
65. Liu J, Liu Y, Xia H, Zou J, Weaver SC, Swanson KA, et al. BNT162b2-elicited neutralization of B.1.617 and other SARS-CoV-2 variants. Nature. (2021) 596:273–5. doi: 10.1038/s41586-021-03693-y
66. Peacock TP, Goldhill DH, Zhou J, Baillon L, Frise R, Swann OC, et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat Microbiol. (2021) 6:899–909. doi: 10.1038/s41564-021-00908-w
67. Nascimento VAD, Corado ALG, Nascimento FOD, Costa Á, Duarte DCG, Luz SLB, et al. Genomic and phylogenetic characterisation of an imported case of SARS-CoV-2 in Amazonas State, Brazil. Mem Inst Oswaldo Cruz. (2020) 115:e200310. doi: 10.1590/0074-02760200310
68. Souza WM, Amorim MR, Sesti-Costa R, Coimbra LD, Brunetti NS, Toledo-Teixeira DA, et al. Neutralisation of SARS-CoV-2 lineage P.1 by antibodies elicited through natural SARS-CoV-2 infection or vaccination with an inactivated SARS-CoV-2 vaccine: an immunological study. Lancet Microbe. (2021) 2:e527–35. doi: 10.1016/S2666-5247(21)00129-4
69. Garcia-Beltran WF, Lam EC, St Denis K, Nitido AD, Garcia ZH, Hauser BM, et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. (2021) 184:2372–83.e2379. doi: 10.1016/j.cell.2021.03.013
70. Charmet T, Schaeffer L, Grant R, Galmiche S, Chény O, Von Platen C, et al. Impact of original, B.1.1.7, and B.1.351/P.1 SARS-CoV-2 lineages on vaccine effectiveness of two doses of COVID-19 mRNA vaccines: results from a nationwide case-control study in France. Lancet Reg Health Eur. (2021) 8:100171. doi: 10.1016/j.lanepe.2021.100171
71. Hitchings MDT, Ranzani OT, Dorion M, D'Agostini TL, de Paula RC, de Paula OFP, et al. Effectiveness of ChAdOx1 vaccine in older adults during SARS-CoV-2 Gamma variant circulation in São Paulo. Nat Commun. (2021) 12:6220. doi: 10.1038/s41467-021-26459-6
72. Hitchings MDT, Ranzani OT, Torres MSS, de Oliveira SB, Almiron M, Said R, et al. Effectiveness of CoronaVac among healthcare workers in the setting of high SARS-CoV-2 Gamma variant transmission in Manaus, Brazil: A test-negative case-control study. Lancet Reg Health Am. (2021) 1:100025. doi: 10.1016/j.lana.2021.100025
73. Ong SWX, Chiew CJ, Ang LW, Mak TM, Cui L, Toh MPHS, et al. Clinical and virological features of SARS-CoV-2 variants of concern: a retrospective cohort study comparing B.1.1.7 (Alpha), B.1.315 (Beta), and B.1.617.2 (Delta). Clin Infect Dis. (2021) 1–24. doi: 10.1093/cid/ciab721
74. Deng X, Garcia-Knight MA, Khalid MM, Servellita V, Wang C, Morris MK, et al. Transmission, infectivity, and neutralization of a spike L452R SARS-CoV-2 variant. Cell. (2021) 184:3426–37.e3428. doi: 10.1016/j.cell.2021.04.025
75. Dougherty K, Mannell M, Naqvi O, Matson D, Stone J. SARS-CoV-2 B.1.617.2 (Delta) variant COVID-19 outbreak associated with a gymnastics facility - Oklahoma, April-May 2021. MMWR Morb Mortal Wkly Rep. (2021) 70:1004–7. doi: 10.15585/mmwr.mm7028e2
76. Chen J, Wang R, Wang M, Wei GW. Mutations strengthened SARS-CoV-2 infectivity. J Mol Biol. (2020) 432:5212–26. doi: 10.1016/j.jmb.2020.07.009
77. Suryadevara N, Shrihari S, Gilchuk P, VanBlargan LA, Binshtein E, Zost SJ, et al. Neutralizing and protective human monoclonal antibodies recognizing the N-terminal domain of the SARS-CoV-2 spike protein. Cell. (2021) 184:2316–31.e2315. doi: 10.1016/j.cell.2021.03.029
78. Cherian S, Potdar V, Jadhav S, Yadav P, Gupta N, Das M, et al. SARS-CoV-2 Spike mutations, L452R, T478K, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. Microorganisms. (2021) 9:1542. doi: 10.3390/microorganisms9071542
79. Bast E, Tang F, Dahn J, Palacio A. Increased risk of hospitalisation and death with the delta variant in the USA. Lancet Infect Dis. (2021) 21:1629–30. doi: 10.1016/S1473-3099(21)00685-X
80. Chia PY, Xiang Ong SW, Chiew CJ, Ang LW, Chavatte JM, Mak TM, et al. Virological and serological kinetics of SARS-CoV-2 Delta variant vaccine-breakthrough infections: a multi-center cohort study. Clin Microbiol Infect. (2021) 1–7. doi: 10.1016/j.cmi.2021.11.010
81. Planas D, Veyer D, Baidaliuk A, Staropoli I, Guivel-Benhassine F, Rajah MM, et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. (2021) 596:276–80. doi: 10.1038/s41586-021-03777-9
82. Wang R, Chen J, Gao K, Wei GW. Vaccine-escape and fast-growing mutations in the United Kingdom, the United States, Singapore, Spain, India, and other COVID-19-devastated countries. Genomics. (2021) 113:2158–70. doi: 10.1016/j.ygeno.2021.05.006
83. Sheikh A, McMenamin J, Taylor B, Robertson C, Public Health Scotland and the EAVE II Collaborators. SARS-CoV-2 Delta VOC in Scotland: demographics, risk of hospital admission, and vaccine effectiveness. Lancet. (2021) 397:2461–2. doi: 10.1016/S0140-6736(21)01358-1
84. Gao SJ, Guo H, Luo G. Omicron variant (B.1.1.529) of SARS-CoV-2, a global urgent public health alert! J Med Virol. (2021) 1–2. doi: 10.1002/jmv.27491
85. Gu H, Krishnan P, Ng DYM, Chang LDJ, Liu GYZ, Cheng SSM, et al. Probable transmission of SARS-CoV-2 omicron variant in quarantine hotel, Hong Kong, China, November 2021. Emerg Infect Dis. (2021) 28. doi: 10.3201/eid2802.212422
86. Karim SSA, Karim QA. Omicron SARS-CoV-2 variant: a new chapter in the COVID-19 pandemic. Lancet. (2021) 398:2126–8. doi: 10.1016/S0140-6736(21)02758-6
87. Chen J, Wang R, Gilby NB, Wei GW. Omicron (B.1.1.529): infectivity, vaccine breakthrough, and antibody resistance. J Chem Inf Model. (2021) 1–11. doi: 10.1021/acs.jcim.1c01451
88. Dejnirattisai W, Huo J, Zhou D, Zahradník J, Supasa P, Liu C, et al. Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell. (2021) 1–64. doi: 10.1016/j.cell.2021.12.046
89. Dejnirattisai W, Shaw RH, Supasa P, Liu C, Stuart AS, Pollard AJ, et al. Reduced neutralisation of SARS-CoV-2 omicron B.1.1.529 variant by post-immunisation serum. Lancet. (2021) 1–3. doi: 10.1016/S0140-6736(21)02844-0
90. Lu L, Mok BW, Chen LL, Chan JM, Tsang OT, Lam BH, et al. Neutralization of SARS-CoV-2 Omicron variant by sera from BNT162b2 or Coronavac vaccine recipients. Clin Infect Dis. (2021) 1–21. doi: 10.1093/cid/ciab1041
91. Chaillon A, Smith DM. Phylogenetic analyses of SARS-CoV-2 B.1.1.7 lineage suggest a single origin followed by multiple exportation events versus convergent evolution. Clin Infect Dis. (2021) 73:2314–7. doi: 10.1093/cid/ciab265
92. Choi JY, Smith DM. SARS-CoV-2 variants of concern. Yonsei Med J. (2021) 62:961–8. doi: 10.3349/ymj.2021.62.11.961
93. Tatsi EB, Filippatos F, Michos A. SARS-CoV-2 variants and effectiveness of vaccines: a review of current evidence. Epidemiol Infect. (2021) 149:e237. doi: 10.1017/S0950268821002430
94. Meng B, Kemp SA, Papa G, Datir R, Ferreira IATM, Marelli S, et al. Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the Alpha variant B.1.1.7. Cell Rep. (2021) 35:109292. doi: 10.1016/j.celrep.2021.109292
95. Kemp SA, Collier DA, Datir RP, Ferreira IATM, Gayed S, Jahun A, et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature. (2021) 592:277–82. doi: 10.1038/s41586-021-03291-y
96. Yang TJ, Yu PY, Chang YC, Liang KH, Tso HC, Ho MR, et al. Effect of SARS-CoV-2 B.1.1.7 mutations on spike protein structure and function. Nat Struct Mol Biol. (2021) 28:731–9. doi: 10.1038/s41594-021-00652-z
97. Liu Y, Liu J, Plante KS, Plante JA, Xie X, Zhang X, et al. The N501Y spike substitution enhances SARS-CoV-2 infection and transmission. Nature. (2021) 1–24. doi: 10.1101/2021.03.08.434499
98. Leung K, Shum MH, Leung GM, Lam TT, Wu JT. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Euro Surveill. (2021) 26:2002106. doi: 10.2807/1560-7917.ES.2020.26.1.2002106
99. Challen R, Brooks-Pollock E, Read JM, Dyson L, Tsaneva-Atanasova K, Danon L. Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: matched cohort study. BMJ. (2021) 372:n579. doi: 10.1136/bmj.n579
100. Davies NG, Jarvis CI, Edmunds WJ, Jewell NP, Diaz-Ordaz K, Keogh RH, et al. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature. (2021) 593:270–4. doi: 10.1038/s41586-021-03426-1
101. Grint DJ, Wing K, Williamson E, McDonald HI, Bhaskaran K, Evans D, et al. Case fatality risk of the SARS-CoV-2 variant of concern B.1.1.7 in England, 16 November to 5 February. Euro Surveill. (2021) 26:2100256. doi: 10.2807/1560-7917.ES.2021.26.11.2100256
102. Chen RE, Zhang X, Case JB, Winkler ES, Liu Y, VanBlargan LA, et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat Med. (2021) 27:717–26. doi: 10.1038/s41591-021-01294-w
103. Ho D, Wang P, Liu L, Iketani S, Luo Y, Guo Y, et al. Increased resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7 to antibody neutralization. Res Sq. (2021) 1–23. doi: 10.21203/rs.3.rs-155394/v1
104. Supasa P, Zhou D, Dejnirattisai W, Liu C, Mentzer AJ, Ginn HM, et al. Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera. Cell. (2021) 184:2201–11.e2207. doi: 10.1016/j.cell.2021.02.033
105. Rees-Spear C, Muir L, Griffith SA, Heaney J, Aldon Y, Snitselaar JL, et al. The effect of spike mutations on SARS-CoV-2 neutralization. Cell Rep. (2021) 34:108890. doi: 10.1016/j.celrep.2021.108890
106. Betton M, Livrozet M, Planas D, Fayol A, Monel B, Védie B, et al. Sera neutralizing activities against severe acute respiratory syndrome coronavirus 2 and multiple variants 6 months after hospitalization for coronavirus disease 2019. Clin Infect Dis. (2021) 73:e1337–44. doi: 10.1093/cid/ciab308
107. Planas D, Bruel T, Grzelak L, Guivel-Benhassine F, Staropoli I, Porrot F, et al. Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat Med. (2021) 27:917–24. doi: 10.1038/s41591-021-01318-5
108. Wang P, Nair M, Liu L, Iketani S, Luo Y, Guo Y, et al. Antibody resistance of SARS-CoV-2 variants B.I.I.351 and B.I.I.7. bioRxiv. (2021) 1–24. doi: 10.1101/2021.01.25.428137
109. Chen X, Chen Z, Azman AS, Sun R, Lu W, Zheng N, et al. Neutralizing antibodies against SARS-CoV-2 variants induced by natural infection or vaccination: a systematic review and pooled meta-analysis. Clin Infect Dis. (2021) 1–9. doi: 10.1093/cid/ciab646
110. Tada T, Zhou H, Samanovic MI, Dcosta BM, Cornelius A, Mulligan MJ, et al. Comparison of neutralizing antibody titers elicited by mRNA and adenoviral vector vaccine against SARS-CoV-2 variants. bioRxiv. (2021) 1–29. doi: 10.1101/2021.07.19.452771
111. Haas EJ, Angulo FJ, McLaughlin JM, Anis E, Singer SR, Khan F, et al. Impact and effectiveness of mRNA BNT162b2 vaccine against SARS-CoV-2 infections and COVID-19 cases, hospitalisations, and deaths following a nationwide vaccination campaign in Israel: an observational study using national surveillance data. Lancet. (2021) 397:1819–29. doi: 10.1016/S0140-6736(21)00947-8
112. Khan A, Zia T, Suleman M, Khan T, Ali SS, Abbasi AA, et al. Higher infectivity of the SARS-CoV-2 new variants is associated with K417N/T, E484K, and N501Y mutants: an insight from structural data. J Cell Physiol. (2021) 236:7045–57. doi: 10.1002/jcp.30367
113. Pearson C, Russell T, Davies N, Kucharski A, CMMID COVID-19 working group, John Edmunds W, et al. Estimates of severity and transmissibility of novel SARS-CoV-2 variant 501Y.V2 in South Africa. CMMID Repository. (2021) 1–4.
114. Nonaka CKV, Franco MM, Gräf T, de Lorenzo Barcia CA, de Ávila Mendonça RN, de Sousa KAF, et al. Genomic evidence of SARS-CoV-2 reinfection involving E484K Spike mutation, Brazil. Emerg Infect Dis. (2021) 27:1522–4. doi: 10.3201/eid2705.210191
115. Radvak P, Kwon HJ, Kosikova M, Ortega-Rodriguez U, Xiang R, Phue JN, et al. SARS-CoV-2 B.1.1.7 (alpha) and B.1.351 (beta) variants induce pathogenic patterns in K18-hACE2 transgenic mice distinct from early strains. Nat Commun. (2021) 12:6559. doi: 10.1038/s41467-021-26803-w
116. Li Q, Nie J, Wu J, Zhang L, Ding R, Wang H, et al. SARS-CoV-2 501Y.V2 variants lack higher infectivity but do have immune escape. Cell. (2021) 184:2362–71.e2369. doi: 10.1016/j.cell.2021.02.042
117. McCallum M, Bassi J, De Marco A, Chen A, Walls AC, Di Iulio J, et al. SARS-CoV-2 immune evasion by the B.1.427/B.1.429 variant of concern. Science. (2021) 373:648–54. doi: 10.1126/science.abi7994
118. Faulkner N, Ng KW, Wu MY, Harvey R, Margaritis M, Paraskevopoulou S, et al. Reduced antibody cross-reactivity following infection with B.1.1.7 than with parental SARS-CoV-2 strains. Elife. (2021) 10:e69317. doi: 10.7554/eLife.69317
119. Moyo-Gwete T, Madzivhandila M, Makhado Z, Ayres F, Mhlanga D, Oosthuysen B, et al. Cross-reactive neutralizing antibody responses elicited by SARS-CoV-2 501Y.V2 (B.1.351). N Engl J Med. (2021) 384:2161–3. doi: 10.1056/NEJMc2104192
120. Zhou D, Dejnirattisai W, Supasa P, Liu C, Mentzer AJ, Ginn HM, et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell. (2021) 184:2348–61.e2346. doi: 10.1016/j.cell.2021.02.037
121. Wall EC, Wu M, Harvey R, Kelly G, Warchal S, Sawyer C, et al. Neutralising antibody activity against SARS-CoV-2 VOCs B.1.617.2 and B.1.351 by BNT162b2 vaccination. Lancet. (2021) 397:2331–3. doi: 10.1016/S0140-6736(21)01290-3
122. Goel RR, Apostolidis SA, Painter MM, Mathew D, Pattekar A, Kuthuru O, et al. Distinct antibody and memory B cell responses in SARS-CoV-2 naïve and recovered individuals following mRNA vaccination. Sci Immunol. (2021) 6:eabi6950. doi: 10.1126/sciimmunol.abi6950
123. Madhi SA, Baillie V, Cutland CL, Voysey M, Koen AL, Fairlie L, et al. Efficacy of the ChAdOx1 nCoV-19 Covid-19 vaccine against the B.1.351 variant. N Engl J Med. (2021) 384:1885–98. doi: 10.1056/NEJMc2110093
124. Fujino T, Nomoto H, Kutsuna S, Ujiie M, Suzuki T, Sato R, et al. Novel SARS-CoV-2 variant in travelers from Brazil to Japan. Emerg Infect Dis. (2021) 27:1243–5. doi: 10.3201/eid2704.210138
125. Sabino EC, Buss LF, Carvalho MPS, Prete CA, Crispim MAE, Fraiji NA, et al. Resurgence of COVID-19 in Manaus Brazil, despite high seroprevalence. Lancet. (2021) 397:452–5. doi: 10.1016/S0140-6736(21)00183-5
126. Buss LF, Prete CA, Abrahim CMM, Mendrone A, Salomon T, de Almeida-Neto C, et al. Three-quarters attack rate of SARS-CoV-2 in the Brazilian Amazon during a largely unmitigated epidemic. Science. (2021) 371:288–92. doi: 10.1126/science.abe9728
127. Bolze A, Cirulli E, Luo S, White S, Cassens T, Jacobs S, et al. Rapid displacement of SARS-CoV-2 variant B.1.1.7 by B.1.617.2 and P.1 in the United States. MedRxiv. (2021) 1–9. doi: 10.1101/2021.06.20.21259195
128. Chaqroun A, Hartard C, Schvoerer E. Anti-SARS-CoV-2 vaccines and monoclonal antibodies facing viral variants. Viruses. (2021) 13:1171. doi: 10.3390/v13061171
129. Dejnirattisai W, Zhou D, Supasa P, Liu C, Mentzer AJ, Ginn HM, et al. Antibody evasion by the P.1 strain of SARS-CoV-2. Cell. (2021) 184:2939–54.e2939. doi: 10.1016/j.cell.2021.03.055
130. Copin R, Baum A, Wloga E, Pascal KE, Giordano S, Fulton BO, et al. The monoclonal antibody combination REGEN-COV protects against SARS-CoV-2 mutational escape in preclinical and human studies. Cell. (2021) 184:3949–61.e3911. doi: 10.1016/j.cell.2021.06.002
131. Wang P, Casner RG, Nair MS, Wang M, Yu J, Cerutti G, et al. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell Host Microbe. (2021) 29:747–51.e744. doi: 10.1016/j.chom.2021.04.007
132. Chen LL, Lu L, Choi CY, Cai JP, Tsoi HW, Chu AW, et al. Impact of SARS-CoV-2 variant-associated RBD mutations on the susceptibility to serum antibodies elicited by COVID-19 infection or vaccination. Clin Infect Dis. (2021) 1–8. doi: 10.1093/cid/ciab656
133. Ferreira IATM, Kemp SA, Datir R, Saito A, Meng B, Rakshit P, et al. SARS-CoV-2 B.1.617 mutations L452R and E484Q are not synergistic for antibody evasion. J Infect Dis. (2021) 224:989–94. doi: 10.1093/infdis/jiab368
134. Teng S, Sobitan A, Rhoades R, Liu D, Tang Q. Systemic effects of missense mutations on SARS-CoV-2 spike glycoprotein stability and receptor-binding affinity. Brief Bioinform. (2021) 22:1239–53. doi: 10.1093/bib/bbaa233
135. Madewell ZJ, Yang Y, Longini IM, Halloran ME, Dean NE. Household transmission of SARS-CoV-2: a systematic review and meta-analysis. JAMA Netw Open. (2020) 3:e2031756. doi: 10.1001/jamanetworkopen.2020.31756
136. Mlcochova P, Kemp SA, Dhar MS, Papa G, Meng B, Ferreira IATM, et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature. (2021) 599:114–9. doi: 10.21203/rs.3.rs-637724/v1
137. Starr TN, Greaney AJ, Dingens AS, Bloom JD. Complete map of SARS-CoV-2 RBD mutations that escape the monoclonal antibody LY-CoV555 and its cocktail with LY-CoV016. Cell Rep Med. (2021) 2:100255. doi: 10.1016/j.xcrm.2021.100255
138. Liu C, Ginn HM, Dejnirattisai W, Supasa P, Wang B, Tuekprakhon A, et al. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell. (2021) 184:4220–36.e4213. doi: 10.1016/j.cell.2021.06.020
139. Hoffmann M, Hofmann-Winkler H, Krüger N, Kempf A, Nehlmeier I, Graichen L, et al. SARS-CoV-2 variant B.1.617 is resistant to bamlanivimab and evades antibodies induced by infection and vaccination. Cell Rep. (2021) 36:109415. doi: 10.1016/j.celrep.2021.109415
140. Venkatakrishnan A, Anand P, Lenehan P, Suratekar R, Raghunathan B, Niesen M, et al. Omicron variant of SARS-CoV-2 harbors a unique insertion mutation of putative viral or human genomic origin. OSF Preprints. (2021) 1–16. doi: 10.31219/osf.io/f7txy
141. Chan M, Hui K, Ho J, Cheung M, Ng K, Ching R, et al. SARS-CoV-2 Omicron variant replication in human respiratory tract ex vivo. Research Square. (2021) 1–20. doi: 10.21203/rs.3.rs-1189219/v1
142. Pujadas E, Chaudhry F, McBride R, Richter F, Zhao S, Wajnberg A, et al. SARS-CoV-2 viral load predicts COVID-19 mortality. Lancet. Respiratory Med. (2020) 8:e70. doi: 10.1016/S2213-2600(20)30354-4
143. Westblade LF, Brar G, Pinheiro LC, Paidoussis D, Rajan M, Martin P, et al. SARS-CoV-2 viral load predicts mortality in patients with and without cancer who are hospitalized with COVID-19. Cancer cell. (2020) 38:661–71.e662. doi: 10.1016/j.ccell.2020.09.007
144. Fajnzylber J, Regan J, Coxen K, Corry H, Wong C, Rosenthal A, et al. SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat Commun. (2020) 11:5493. doi: 10.1038/s41467-020-19057-5
145. de la Calle C, Lalueza A, Mancheno-Losa M, Maestro-de la Calle G, Lora-Tamayo J, Arrieta E, et al. Impact of viral load at admission on the development of respiratory failure in hospitalized patients with SARS-CoV-2 infection. Eur J Clin Microbiol Infect Dis. (2021) 40:1209–16. doi: 10.1007/s10096-020-04150-w
146. Lee S, Kim T, Lee E, Lee C, Kim H, Rhee H, et al. Clinical course and molecular viral shedding among asymptomatic and symptomatic patients with SARS-CoV-2 infection in a community treatment center in the Republic of Korea. JAMA Intern Med. (2020) 180:1447–52. doi: 10.1001/jamainternmed.2020.3862
147. Karahasan Yagci A, Can Sarinoglu R, Bilgin H, Yanilmaz Ö, Sayin E, Guneser D, et al. Relationship of the cycle threshold values of SARS-CoV-2 polymerase chain reaction and total severity score of computerized tomography in patients with COVID 19. Int J Infect Dis. (2020) 101:160–6. doi: 10.1016/j.ijid.2020.09.1449
148. Shah VP, Farah WH, Hill JC, Hassett LC, Binnicker MJ, Yao JD, et al. Association between SARS-CoV-2 cycle threshold values and clinical outcomes in patients with COVID-19: a systematic review and meta-analysis. Open Forum Infect Dis. (2021) 8:ofab453. doi: 10.1093/ofid/ofab453
149. Barek MA, Aziz MA, Islam MS. Impact of age, sex, comorbidities and clinical symptoms on the severity of COVID-19 cases: a meta-analysis with 55 studies and 10014 cases. Heliyon. (2020) 6:e05684. doi: 10.1016/j.heliyon.2020.e05684
150. Imai M, Iwatsuki-Horimoto K, Hatta M, Loeber S, Halfmann PJ, Nakajima N, et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc Natl Acad Sci USA. (2020) 117:16587–95. doi: 10.1073/pnas.2009799117
151. Yinda CK, Port JR, Bushmaker T, Owusu IO, Avanzato VA, Fischer RJ, et al. K18-hACE2 mice develop respiratory disease resembling severe COVID-19. bioRxiv. (2020) 1–30. doi: 10.1101/2020.08.11.246314
152. Ryan KA, Bewley KR, Fotheringham SA, Slack GS, Brown P, Hall Y, et al. Dose-dependent response to infection with SARS-CoV-2 in the ferret model and evidence of protective immunity. Nat Commun. (2021) 12:81. doi: 10.1038/s41467-020-20439-y
153. Dabisch PA, Biryukov J, Beck K, Boydston JA, Sanjak JS, Herzog A, et al. Seroconversion and fever are dose-dependent in a nonhuman primate model of inhalational COVID-19. PLoS Pathog. (2021) 17:e1009865. doi: 10.1371/journal.ppat.1009865
154. Port JR, Yinda CK, Owusu IO, Holbrook M, Fischer R, Bushmaker T, et al. SARS-CoV-2 disease severity and transmission efficiency is increased for airborne compared to fomite exposure in Syrian hamsters. Nat Commun. (2021) 12:4985. doi: 10.1038/s41467-021-25156-8
155. Chan JF, Yuan S, Zhang AJ, Poon VK, Chan CC, Lee AC, et al. Surgical mask partition reduces the risk of non-contact transmission in a golden Syrian hamster model for Coronavirus Disease 2019 (COVID-19). Clin Infect Dis. (2020) 71:2139–49. doi: 10.1093/cid/ciaa644
156. Teyssou E, Soulie C, Visseaux B, Lambert-Niclot S, Ferre V, Marot S, et al. The 501Y.V2 SARS-CoV-2 variant has an intermediate viral load between the 501Y.V1 and the historical variants in nasopharyngeal samples from newly diagnosed COVID-19 patients. J Infect. (2021) 83:119–45. doi: 10.1016/j.jinf.2021.04.023
157. Lin L, Liu Y, Tang X, He D. The disease severity and clinical outcomes of the SARS-CoV-2 variants of concern. Front Med. (2021) 9:775224. doi: 10.3389/fpubh.2021.775224
158. Goga A, Bekker L, Garrett N, Reddy T, Yende-Zuma N, Fairall L, et al. Breakthrough Covid-19 infections during periods of circulating Beta, Delta and Omicron variants of concern, among health care workers in the Sisonke Ad26.COV2.S vaccine trial, South Africa. MedRxiv. (2021). doi: 10.1101/2021.12.21.21268171
159. Deng X, Evdokimova M, O'Brien A, Rowe CL, Clark NM, Harrington A, et al. Breakthrough infections with multiple lineages of SARS-CoV-2 variants reveals continued risk of severe disease in immunosuppressed patients. Viruses. (2021) 13:1743. doi: 10.3390/v13091743
160. Bian L, Gao F, Zhang J, He Q, Mao Q, Xu M, et al. Effects of SARS-CoV-2 variants on vaccine efficacy and response strategies. Expert Rev Vaccines. (2021) 20:365–73. doi: 10.1080/14760584.2021.1903879
161. Tartof SY, Slezak JM, Fischer H, Hong V, Ackerson BK, Ranasinghe ON, et al. Effectiveness of mRNA BNT162b2 COVID-19 vaccine up to 6 months in a large integrated health system in the USA: a retrospective cohort study. Lancet. (2021) 398:1407–16. doi: 10.1016/S0140-6736(21)02183-8
162. Dagan N, Barda N, Kepten E, Miron O, Perchik S, Katz MA, et al. BNT162b2 mRNA Covid-19 vaccine in a nationwide mass vaccination setting. N Engl J Med. (2021) 384:1412–23. doi: 10.1056/NEJMoa2101765
163. Levine-Tiefenbrun M, Yelin I, Katz R, Herzel E, Golan Z, Schreiber L, et al. Initial report of decreased SARS-CoV-2 viral load after inoculation with the BNT162b2 vaccine. Nat Med. (2021) 27:790–2. doi: 10.1038/s41591-021-01316-7
164. McEllistrem MC, Clancy CJ, Buehrle DJ, Lucas A, Decker BK. Single dose of an mRNA severe acute respiratory syndrome coronavirus 2 (SARS-Cov-2) vaccine is associated with lower nasopharyngeal viral load among nursing home residents with asymptomatic coronavirus disease 2019 (COVID-19). Clin Infect Dis. (2021) 73:e1365–7. doi: 10.1093/cid/ciab263
165. Elliott P, Haw D, Wang H, Eales O, Walters CE, Ainslie KEC, et al. Exponential growth, high prevalence of SARS-CoV-2, and vaccine effectiveness associated with the Delta variant. Science. (2021) 374:eabl9551. doi: 10.1126/science.abl9551
166. Ioannou P, Karakonstantis S, Astrinaki E, Saplamidou S, Vitsaxaki E, Hamilos G, et al. Transmission of SARS-CoV-2 variant B.1.1.7 among vaccinated health care workers. Infect Dis. (2021) 53:876–9. doi: 10.1080/23744235.2021.1945139
167. Acharya C, Schrom J, Mitchell A, Coil D, Marquez C, Rojas S, et al. No significant difference in viral load between vaccinated and unvaccinated, asymptomatic and symptomatic groups when infected with SARS-CoV-2 delta variant. MedRxiv. (2021) 1–13. doi: 10.1101/2021.09.28.21264262
168. Riemersma K, Grogan B, Kita-Yarbro A, Halfmann P, Segaloff H, Kocharian A, et al. Shedding of infectious SARS-CoV-2 despite vaccination. MedRxiv. (2021) 1–12. doi: 10.1101/2021.07.31.21261387
169. Brown CM, Vostok J, Johnson H, Burns M, Gharpure R, Sami S, et al. Outbreak of SARS-CoV-2 infections, including COVID-19 vaccine breakthrough infections, associated with large public gatherings - barnstable county, Massachusetts, July 2021. MMWR Morb Mortal Wkly Rep. (2021) 70:1059–62. doi: 10.15585/mmwr.mm7031e2
170. Rovida F, Cassaniti I, Paolucci S, Percivalle E, Sarasini A, Piralla A, et al. SARS-CoV-2 vaccine breakthrough infections with the alpha variant are asymptomatic or mildly symptomatic among health care workers. Nat Commun. (2021) 12:6032. doi: 10.1038/s41467-021-26154-6
171. Kissler SM, Fauver JR, Mack C, Tai CG, Breban MI, Watkins AE, et al. Viral dynamics of SARS-CoV-2 variants in vaccinated and unvaccinated persons. New Engl J Med. (2021) 26:2489–91. doi: 10.1101/2021.02.16.21251535
172. Singanayagam A, Hakki S, Dunning J, Madon KJ, Crone MA, Koycheva A, et al. Community transmission and viral load kinetics of the SARS-CoV-2 delta (B.1.617.2) variant in vaccinated and unvaccinated individuals in the UK: a prospective, longitudinal, cohort study. Lancet Infect Dis. (2021) 1–13. doi: 10.1016/S1473-3099(21)00648-4
173. Brosh-Nissimov T, Orenbuch-Harroch E, Chowers M, Elbaz M, Nesher L, Stein M, et al. BNT162b2 vaccine breakthrough: clinical characteristics of 152 fully vaccinated hospitalized COVID-19 patients in Israel. Clin Microbiol Infect. (2021) 27:1652–7. doi: 10.1016/j.cmi.2021.06.036
174. Levine-Tiefenbrun M, Yelin I, Alapi H, Katz R, Herzel E, Kuint J, et al. Viral loads of Delta-variant SARS-CoV-2 breakthrough infections after vaccination and booster with BNT162b2. Nat Med. (2021) 27:2108–10. doi: 10.1038/s41591-021-01575-4
Keywords: SARS-CoV-2, prognosis, disease severity, emerging variants, vaccines, transmissibility, lethality, viral load
Citation: Silva SJRd, Lima SCd, Silva RCd, Kohl A and Pena L (2022) Viral Load in COVID-19 Patients: Implications for Prognosis and Vaccine Efficacy in the Context of Emerging SARS-CoV-2 Variants. Front. Med. 8:836826. doi: 10.3389/fmed.2021.836826
Received: 15 December 2021; Accepted: 29 December 2021;
Published: 31 January 2022.
Edited by:
Pragya Dhruv Yadav, National Institute of Virology (ICMR), IndiaReviewed by:
Amita Jain, King George's Medical University, IndiaScott Burkholz, Flow Pharma, United States
Copyright © 2022 Silva, Lima, Silva, Kohl and Pena. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Severino Jefferson Ribeiro da Silva, jeffersonbiotecviro@gmail.com; Lindomar Pena, lindomar.pena@fiocruz.br
 Severino Jefferson Ribeiro da Silva
Severino Jefferson Ribeiro da Silva Suelen Cristina de Lima
Suelen Cristina de Lima Ronaldo Celerino da Silva
Ronaldo Celerino da Silva Alain Kohl
Alain Kohl Lindomar Pena1*
Lindomar Pena1*