Current Update on Intrinsic and Acquired Colistin Resistance Mechanisms in Bacteria
- Department of Biosciences, Jamia Millia Islamia, New Delhi, India
Colistin regained global interest as a consequence of the rising prevalence of multidrug-resistant Gram-negative Enterobacteriaceae. In parallel, colistin-resistant bacteria emerged in response to the unregulated use of this antibiotic. However, some Gram-negative species are intrinsically resistant to colistin activity, such as Neisseria meningitides, Burkholderia species, and Proteus mirabilis. Most identified colistin resistance usually involves modulation of lipid A that decreases or removes early charge-based interaction with colistin through up-regulation of multistep capsular polysaccharide expression. The membrane modifications occur by the addition of cationic phosphoethanolamine (pEtN) or 4-amino-L-arabinose on lipid A that results in decrease in the negative charge on the bacterial surface. Therefore, electrostatic interaction between polycationic colistin and lipopolysaccharide (LPS) is halted. It has been reported that these modifications on the bacterial surface occur due to overexpression of chromosomally mediated two-component system genes (PmrAB and PhoPQ) and mutation in lipid A biosynthesis genes that result in loss of the ability to produce lipid A and consequently LPS chain, thereafter recently identified variants of plasmid-borne genes (mcr-1 to mcr-10). It was hypothesized that mcr genes derived from intrinsically resistant environmental bacteria that carried chromosomal pmrC gene, a part of the pmrCAB operon, code three proteins viz. pEtN response regulator PmrA, sensor kinase protein PmrAB, and phosphotransferase PmrC. These plasmid-borne mcr genes become a serious concern as they assist in the dissemination of colistin resistance to other pathogenic bacteria. This review presents the progress of multiple strategies of colistin resistance mechanisms in bacteria, mainly focusing on surface changes of the outer membrane LPS structure and other resistance genetic determinants. New handier and versatile methods have been discussed for rapid detection of colistin resistance determinants and the latest approaches to revert colistin resistance that include the use of new drugs, drug combinations and inhibitors. Indeed, more investigations are required to identify the exact role of different colistin resistance determinants that will aid in developing new less toxic and potent drugs to treat bacterial infections. Therefore, colistin resistance should be considered a severe medical issue requiring multisectoral research with proper surveillance and suitable monitoring systems to report the dissemination rate of these resistant genes.
Overview of Colistin and Emergence of Resistance
The antibiotics have been widely used in human, animal husbandry, and aquaculture, aiming to fight bacterial infections. The unmonitored and continued use of antibiotics has led to contamination of diversified environments, results in selective pressure on bacteria, and subsequently increases in the prevalence of antibiotic resistance (1–3). A steady increase in antibiotic resistance coupled with the decline in the development of new drugs is leading the world toward the pre–antibiotic era (4). This global public health threat requires immediate multidisciplinary steps to achieve the sustainable development goals, which are a collection of 17 interlinked global goals designed to be a roadmap for achieving a better and more sustainable future for all. Among these, Goal 3, i.e., Good Health and Well-being, was set up by the United Nations General Assembly to ensure healthy lives and promote well-being for all ages (5). New antibiotics active against Gram-positive bacteria provided some extent of respite (6, 7), but infections caused by antibiotic-resistant Gram-negative bacteria are emerging as a greater threat. Antibiotic-resistant bacteria and associated antibiotic resistance genes are gradually considered as diverse environmental contaminants. These antibiotic resistance genes are no longer limited to point sources, e.g., hospitals, sewage, and farms, but can also get disseminated in other relatively pristine environments, including rivers, lakes, and soils (8, 9). The occurrence of extremely drug-resistant and multidrug-resistant (MDR) bacteria has led to the reuse of polymyxin, a last-resort drug against severe bacterial infections (10–12). Polymyxins, non-ribosomal, cyclic oligopeptides antimicrobials structurally comprised a cyclic heptapeptide with five major chemical compounds: polymyxins A, B, C, D, and E. These compounds are differentiated based on variation in their amino acid sequences and fatty acid side chains. The prime representatives of polymyxin that have been used in clinical practice are polymyxin B and polymyxin E (colistin) (Figure 1) (12–16). Colistin is a polypeptide antibiotic isolated in 1947 from the bacterium Paenibacillus polymyxa subspecies Colistinus (17). Thereafter, it was reported from Japan (1947) that colistin is a secondary metabolite of the Gram-positive bacteria P. polymyxa subspecies Colistinus (18). In the 1950s, colistin was used as an intravenous formulation. In 1959, the US Food and Drug Administration approved colistin to treat various types of diarrhea and urinary tract infections. Failure of carbapenems against Gram-negative bacteria has led to the unprecedented increase in the use of colistin (one of the last-resort drugs) and subsequent emergence and dissemination of colistin resistance (14). Resistance to polymyxins has mainly emerged against polymyxin E class (colistin), a cationic polypeptide drug with cyclic decapeptide ring attached by an amide linkage to a fatty acyl chain, which is differentiated by single-amino-acid phenylalanine (D-Phe) in polymyxin B peptide structure with a leucine (D-Leu) in colistin (Figure 1) (16, 19). Until 2015, colistin resistance was known to be caused by chromosomal genes (phoPQ, pmrAB, and mgrB) (19–21). After the first report of plasmid-mediated mcr-1 gene from China in 2015 (14), more than 27 bacterial species have been identified from six continents (Asia, Europe, Africa, North America, South America, and Oceania). It is interesting to note that after 2015 the new reports about plasmid-mediated mcr-1 were made in isolates dating back to as early as 1980. The rise in the number of reports may be attributed to the long-term use of colistin in veterinary medicine. Similarly, global trade and travel to countries such Canada, United States, Japan, and Tunisia and overprescription of colistin in human medicine to treat highly resistant bacterial infection are likely reasons for colistin resistance (22–27). There is a high prevalence of colistin resistance that requires further studies to evaluate the factors involved, mechanism of acquisition, and dissemination (28, 29). We present herein an overview of recognition of alternative mechanisms of colistin action, the spread of acquired colistin resistance determinants, and diverse strategies taken by bacteria to extend resistance against colistin antibiotic.
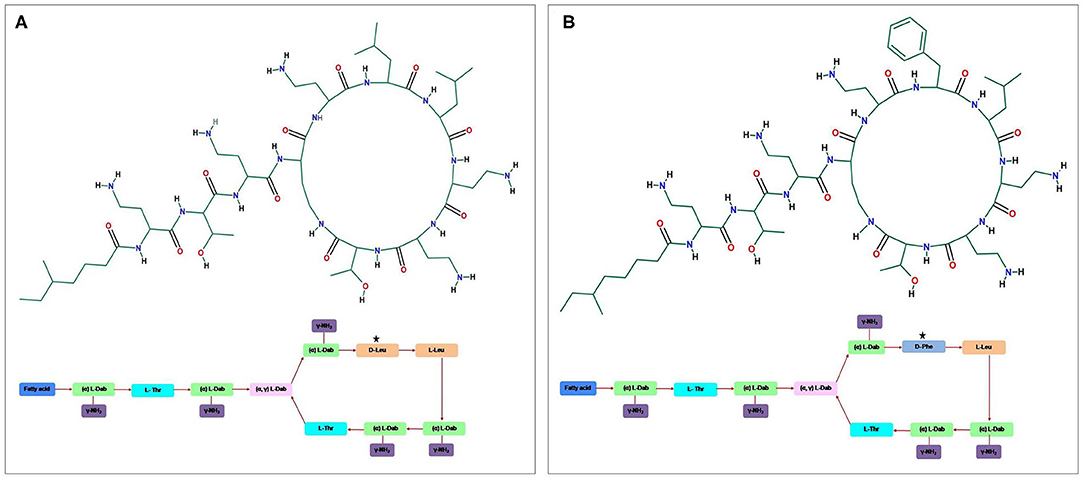
Figure 1. The general structure of the cyclic cationic peptide (A) polymyxin E (colistin) and (B) polymyxin B, phenylalanine (D-Phe) in polymyxin B peptide structure replaced with a leucine in colistin.
Mechanism of Colistin Activity
Antibacterial activity of colistin occurs on the outer membrane (OM) of Gram-negative bacteria. In addition, Gram-negative bacteria are well-characterized by the existence of an outer lipopolysaccharide (LPS) membrane that constitutes cell surface, which limits the entry of hydrophobic components and antibiotics (30, 31). Moreover, bacterial anionic LPS confers stability and integrity of the outer LPS membrane. But polymyxins are polycationic peptides crucial for their interaction with lipid A, a hydrophobic constituent of the LPS layer (32, 33). The antibacterial activity of colistin occurs through two-step mechanisms that are initial binding and employed permeabilization of the outer LPS membrane induces the displacement of Ca2+ and Mg2+ ions from the phosphate groups of LPS in a competitive way resulting in destabilizing cytoplasmic membrane, leading to disruption of the outer LPS and the loss of inner cellular contents, hence bacterial killing. The critical step of colistin action is based on the electrostatic interaction of cationic colistin peptide and anionic lipid A membrane also known as endotoxin component of LPS layer (Figure 2) (16, 19, 34, 35). Furthermore, It has been reported that bactericidal activity is independent of the passage of colistin into a bacterial cell (36) but inhibited in the presence of these divalent cations (33). However, LPS is the initial target for bacterial killing, but still, the exact mode of colistin action remains uncertain. Another antibacterial mechanism of colistin occurs by a potent antiendotoxin activity where the lipid A portion of LPS represents an endotoxin in Gram-negative bacteria. Therefore, colistin inhibits the endotoxin activity of lipid A by binding to and neutralizing the LPS molecules. This antibacterial activity mechanism occurs in vivo only (12, 37, 38). Moreover, another mechanism of action occurs by vital respiratory enzymes (type II NADH-quinone oxidoreductases NADH-2) inhibition by colistin drug in Gram-negative bacteria (39). The alternative strategy of colistin action occurs by induction of rapid cell death via hydroxyl radical production through colistin binding to the lipid membrane. The free radicals are generated when colistin crosses the OM and IM of LPS. The hydroxyl radical generation occurs via the production of the reactive oxygen species; hydroxyl radicals. (•OH), superoxide (), and hydrogen peroxide (H2O2), which cause oxidative stress. is generated when colistin enters into and crosses the OM and IM, followed by the conversion of into H2O2 by superoxide dismutase. After that, H2O2 oxidizes ferrous iron (Fe2+) into ferric iron (Fe3+), besides the formation of •OH; this process is known as Fenton reaction. This reaction can induce oxidative damage in bacterial DNA, proteins, and lipids, leading to cell death. This mechanism of killing has been shown to occur in the colistin-sensitive and MDR isolates of Acinetobacter baumannii and Escherichia coli but does not take place in polymyxin-resistant strains (40, 41).
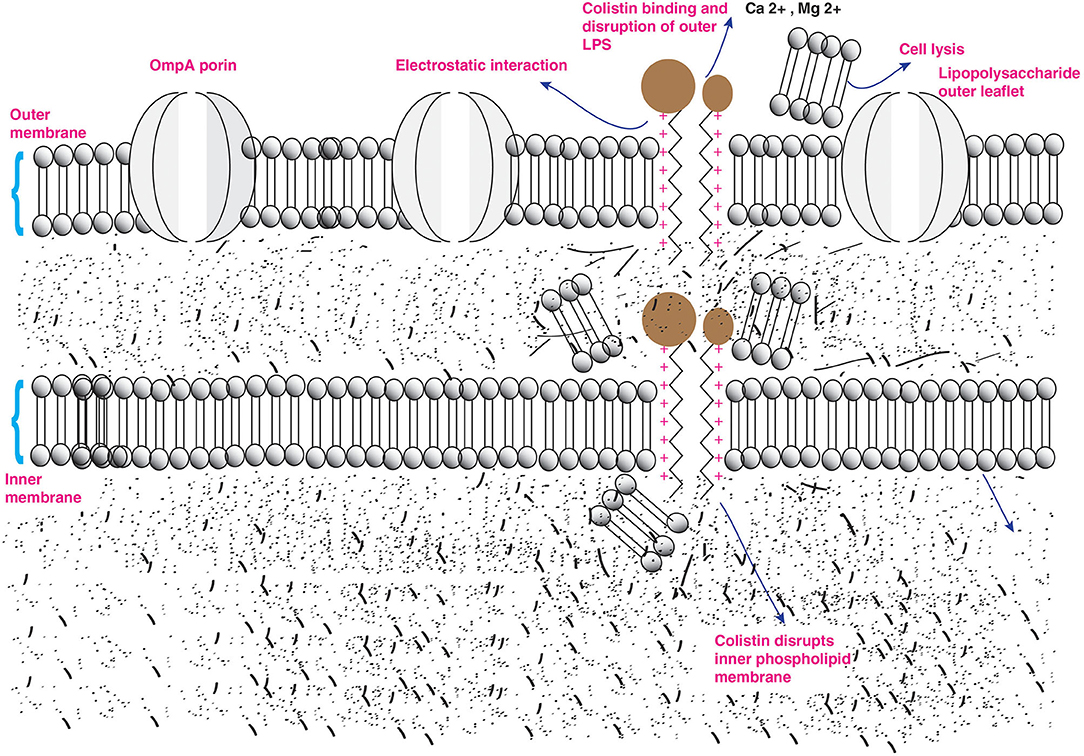
Figure 2. Action of colistin on bacterial membrane. The cationic cyclic decapeptide structure of colistin binds with the anionic LPS molecules by displacing calcium and magnesium from the outer cell membrane of Gram-negative bacteria, leading to permeability changes in the cell envelope and leakage of cell contents.
Spectrum of Activity
Colistin has a narrow bactericidal activity spectrum against the most common Gram-negative Enterobacteriaceae. Colistin has antibacterial activity against members of the Enterobacteriaceae family, including Klebsiella species, E. coli, Citrobacter species, Shigella species, Enterobacter species, and Salmonella species. Colistin also has significant bactericidal activity against prevalent non-fermentative Gram-negative bacteria, including Stenotrophomonas maltophilia, A. baumannii, and Pseudomonas aeruginosa (12, 42, 43). Moreover, some species are resistant naturally to colistin, such as Proteus species, Providencia species, Morganella morganii, Pseudomonas mallei, Chromobacterium species, Burkholderia cepacia, Serratia marcescens, Edwardsiella species, Campylobacter, Vibrio cholera, and Brucella, Legionella. Moreover, colistin antibiotic is not active against Gram-negative cocci (Neisseria species), anaerobic bacteria (12, 35), and Aeromonas species (except Aeromonas jandaei), whereas Aeromonas hydrophila was found to have inducible resistance (44, 45).
Pharmachemistry of Colistin
The pharmacokinetics (PK) and pharmacodynamics (PD) determine the related dosage and therapeutics of colistin. It is imperative to determine the effective dosage of a drug to treat dreadful infections. With increasing resistance, it has become necessary to study the PKPD relation of colistin to determine proper treatment regimen. Until now, PK and PD data of colistin are not well-reported, especially in patients suffering from renal replacement therapy (46, 47). Therefore, time-bound studies are needed further to understand the PD/PK relationship of colistin to determine fixed doses to critically ill patients. Randomized controlled trials are urgently required to clarify further the issues surrounding the efficacy and safety of colistin. Two decades ago, non-specific microbiological assays were applied to estimate the colistin concentrations in biological fluids (38). Moreover, it has been reported that bactericidal activity is dependent on colistin concentration (11, 48–50). The plasma colistin concentration of 2 μg/mL has been found to be a reasonable dose for bacteria having minimum inhibitory concentrations (MICs) of ≤1 μg/mL (51). Postantibiotic effect was noticed among P. aeruginosa, A. baumannii, and Klebsiella pneumoniae, although the main antibacterial activity is noticed only when susceptible strains are exposed to colistin. However, in A. baumannii and K. pneumoniae, regrowth has been reported for static time-kill studies (49, 50). It has also been reported that the emergence of colistin-resistant subpopulation is dependent on colistin heteroresistance that enables growth at ≥4 μg/mL of colistin within a sensitive population with a MIC of ≤2 μg/mL (52, 53), e.g., K. pneumonia (50, 54) P. aeruginosa, and A. baumannii (55).
One Health Perspective for Colistin Resistance
Antimicrobial resistance is a public health problem of complex epidemiology suitable for a comprehensive study as part of the One Health perspective. One Health is defined as the collaborative effort of multiple disciplines working locally, nationally, and globally to attain optimal health for people, animals, and the environment through policy, research, education, and practice (56, 57). The drivers of antimicrobial resistance include antimicrobial use and abuse in humans, animals, and the environment followed by the dissemination of resistant bacteria and resistance determinants between and among these sectors around the globe. Major concerns in the animal health and agriculture sectors are mass medication of animals with antimicrobials that are critically important for humans, like colistin, one of the last-resort drugs to tackle Gram-negative bacterial infection. The unmonitored use of colistin in human and veterinary medicine results in the emergence of colistin resistance among Enterobacteriaceae. The emergence of mcr-1 has almost certainly been exacerbated by the use of colistin on Chinese and Southeast Asian farms (14) and subsequent spread to dozens of other countries (35, 58, 59). The colistin resistance has risen in Spain, Italy, and Greece with 31, 43, and 20.8%, respectively (60–62). Colistin resistance makes it difficult to use it as a therapeutic option for multidrug-resistant bacteria. Therefore, the use of colistin is dependent on the type of infection, sensitivity phenotype of bacteria, and the target of PK/PD antibiotic, including possible side effects (63). The most adverse effects of colistin are nephrotoxicity and neurotoxicity due to parenteral use (18, 64, 65). In addition to human medicine, colistin has been widely used to prevent and treat various veterinary infectious diseases. Colistin use in veterinary varies extensively among different countries, e.g., in Spain, it is used during lactation, gestation, and control of metaphylactic intestinal diseases (66). In France, Austria, and Sweden, colistin is used during the postweaning time in pig farms (67–69). A German and Netherlands study reported that colistin is the most widely used antibiotic in animal farming next to trimethoprim/sulfonamides, tetracyclines, lincosamides, and macrolides (70). In Asian countries, colistin and other antibiotics are used extensively for veterinary infections and agriculture purposes (71, 72). The increasing trend of colistin consumption is assumed to reach 16,000 tons in China by the end of 2021 (72). There are reports of colistin resistance in the Enterobacteriaceae from fruits and vegetable samples (73–77). Reports revealed the association between the incidence of food-borne diseases and the food production chain that occurs via eating contaminated raw vegetables (9, 73–75, 78).
The emergence and rapid geographic dissemination of colistin-resistant bacteria and colistin resistance genes have become a health concern. Therefore, an integrated and holistic multisectoral approach is the need of the hour to combat colistin resistance, in particular, better integration of human health, veterinary, and environment (79–81). Several countries and international agencies have included a One Health perspective within their action plans to address antimicrobial resistance. Necessary actions include improvements in antimicrobial use regulation and policy, surveillance, stewardship, infection control, sanitation, animal husbandry, and alternatives to antimicrobials. World Health Organization (WHO) has recently launched new guidelines on the use of medically important antimicrobials in food-producing animals, recommending that farmers and the food industry should stop using antimicrobials routinely to promote growth and to prevent disease in healthy animals. These guidelines aim to preserve the effectiveness of antimicrobials that are important for human medicine by reducing their use in animals (82–84). Different monitoring systems were established in European countries for surveillance to check colistin consumption and the emergence of resistance (85–87). Recent multinational strategies to address the urgency of AMR include the US National Action Plan for combating antibiotic-resistant bacteria and WHO Global Action Plan on Antimicrobial Resistance (88, 89). WHO's GLASS (Global Antimicrobial Resistance Surveillance System) is helping countries strengthen national surveillance systems and provides more comprehensive standardized AMR surveillance data (90). The countries and networks at the forefront of AMR efforts should engage additional stakeholders in developing an effective strategy that will have far-reaching benefits in minimizing the impact of this urgent problem on human and animal health, environment, global economy and national and global security.
Challenges of Testing and Detecting Colistin Resistance Determinants
It is critical to design phenotypic tests capable of detecting colistin resistance in Gram-negative bacteria. Until now, there was no agreement on the methodology for colistin susceptibility testing. Because of the weak diffusion of colistin in agar, the disc diffusion process and gradient tests were inaccurate (91–95). As a result, disc diffusion and gradient diffusion are ineffective in determining polymyxin susceptibility. Both the European Committee on Antimicrobial Susceptibility Testing (EUCAST) and the Clinical and Laboratory Standards Institute recommended the International Standard Organization 20776 standard broth dilution method for testing colistin MIC values (96, 97). The reference broth microdilution method, on the other hand, is difficult to apply in routine microbiological diagnostics. The EUCAST does not recommend using automated systems to determine the phenotype of bacterial sensitivity, such as Vitek 2 (bioMérieux, France), WalkAway (Beckman Coulter, USA) or BD Phoenix (Becton Dickinson, USA) for the analysis of Gram-negative bacteria sensitivity to colistin. This is because these systems' accuracy in determining colistin MIC is minimal compared to the reference method especially in the 2–4-mg/L range (98–101). There is currently insufficient understanding of acquired colistin resistance mechanisms to design a sensitive molecular test specific enough to be recommended as best practice. Genotypic methods, in particular, are unlikely to detect any of the chromosomal defects known to cause most phenotypic colistin resistance in clinical settings (20). A negative polymerase chain reaction (PCR) molecular test result cannot be used to predict colistin susceptibility because the test cannot rule out the existence of chromosomal mechanisms of resistance or even novel mcr genes not included in the test. As evidence of this limitation, high colistin resistance rates have been reported among K. pneumoniae strains that produce carbapenemase but lack mcr genes (102–105). In these circumstances, a negative PCR result for mcr genes would have poor predictive value for a colistin-susceptible phenotype. However, if the results are intended to guide clinical management, inference of phenotype based solely on a genotypic result may be valid only when the genotypic result is positive (i.e., mechanisms or genes detected) with the caveat that the resistance may not be detected.
Methods for Rapid Detection of Colistin Resistance
The rapid polymyxin NP is an innovative technique for identification of colistin resistance among Gram-negative bacteria (98). The researchers are currently working on tests to detect colistin resistance in non-fermenting bacilli. In the presence of a given concentration of polymyxin E and B, the rapid polymyxin NP test detects glucose fermentation associated with bacterial growth; the presence of acid metabolites is shown by a shift in pH and the color of the indicator (red phenol) turning from orange to yellow. The test's sensitivity and specificity are similar to the reference broth microdilution method (99.3 and 95.4%, respectively). This test is simple to perform and yields a result in <2 h (98). Chromogenic media are widely used for screening because they enable bacteria to develop as properly colored colonies. The super polymyxin screening medium was the first agar medium for detecting colistin-resistant Gram-negative rods from bacterial cultures and rectal swab samples (106). The commercial version of this medium is super polymyxin medium (ELITechGroup, Puteaux, France) for the identification of colistin-resistant Enterobacterales strains, including those with the low MIC values (mg/L) that contain the mcr-1 gene (107). It consists of methylene blue agar and includes colistin, daptomycin, and amphotericin B at concentrations (3.5, 10, and 5 g/mL, respectively). The other medium, CHROMagar COL-APSE medium was used to identify colistin-resistant bacteria (108); this medium distinguishes colistin-resistant Enterobacterales strains from non-fermenting rods. The LBJMR medium is a new polyvalent culture medium for the isolation and selection of colistin-resistant bacteria and vancomycin-resistant bacteria (109). This medium was developed by combining colistin sulfate salt (4 g/mL), vancomycin (50 g/mL), and a fermentation substrate (7.5 g/L of glucose) with purple agar base (31 g/L). Moreover, new chromogenic medium, CHROMID Colistin R agar (COLR; bioMérieux, France), was introduced in the market in early 2018, allowing the screening of colistin-resistant Enterobacteriaceae in clinical samples such as rectal swabs and stools. The COLR is a manual qualitative diagnostic test that distinguishes colistin-resistant isolates from susceptible isolates. Colistin-resistant strains form colored colonies on chromogenic media, with the color varying according to the species. However, colistin-susceptible isolates, on the other hand, do not grow on the COLR plate (110). The chromogenic method is based on agar dilution. Still, EUCAST does not recommend it for determining bacterial susceptibility to colistin because the detectability threshold increases with the growth of the bacterial inoculums (100). Moreover, Turlej-Rogacka et al. (111) reported that when compared to broth dilution methods, the agar dilution method yields more accurate results in evaluating colistin MIC values (111). Behera et al. (93) confirmed the strong correlation between the reference and agar dilution methods (93, 94). The biggest challenge is the adhesion of colistin to plastic during handling (112). According to the above authors, the agar dilution process reduces the colistin plastic–binding process significantly, and the MIC results obtained by the agar dilution method are exact (93, 112, 113). The COLR medium uses the borderline concentrations of colistin to qualify strains as susceptible or resistant. This chromogenic medium is a qualitative detection method for Enterobacteriaceae and does not permit colistin MIC values to be determined against the test bacterial strains; thus, it should be considered as a screening test only. The clinical interpretation is, by contrast, significant in the treatment of infections caused by colistin-resistant bacteria. This means that the colistin resistance is categorized rather than the MIC value determined as maximum dosages are prescribed irrespective of the precise sensitivity levels. However, MIC values are important to monitor the rise in colistin resistance in Gram-negative bacteria. Other new-generation methods for detecting colistin resistible strains were recently developed, e.g., loop-mediated isothermal amplification (LAMP) for nucleic acid detection (114) and CT103XL microarray (115). It has been demonstrated the sensitivity of the LAMP test is 10 times higher than conventional PCR and confirmed its usefulness in the detection of the mcr-1 gene from Enterobacterales (114). Similarly, the new microarray CT103xl has been demonstrated by simultaneously identifying mcr-1, mcr-2, and clinically important ESBL genes (115). Whole-genome sequencing would allow screening for mcr genes and known chromosomal mutations that confer colistin resistance. Bioinformatics analysis could be conducted by applying the Center for Genomic Epidemiology Web tools (116) and ResFinder 4.0 (117). Although the sensitivity and negative predictive value would be affected by the inclusion of strains with novel mechanisms of resistance, this is the most comprehensive method for detecting all currently known putative colistin resistance mechanisms. It will also enable a retrospective analysis of sequencing data as new resistance mechanisms are described. Whatever the molecular method used, it is critically important to ensure that either the PCR detects all currently known mcr genes or the databases used to impute resistance mechanisms from whole-genome sequencing data are up to date. As our understanding of colistin resistance mechanisms improves, so will the concordance between phenotypic and genotypic test results. As for many other antimicrobial agents, molecular testing may eventually offer an alternative to phenotypic testing for the surveillance of colistin resistance.
Resistance Mechanisms in Enterobacteriaceae
Mechanisms of Intrinsic Resistance
Resistance to colistin occurs naturally in S. marcescens and Proteus mirabilis by arnBCADTEF and eptB gene expression and consequently addition of phosphoethanolamine (pEtN) and 4-amino-4-deoxy-L-arabinose (L-Ara4N) cationic groups on LPS, respectively. This modulation increases the cationic charge on the LPS membrane, which is the initial target of the colistin. It therefore decreases colistin antibiotic binding results in intrinsic resistance of these bacterial strains (12, 28, 118, 119).
Acquired Resistance in Enterobacteriaceae
Chromosomal Modulation of PmrAB and PhoPQ Two-Component Systems
The acquired colistin resistance has been reported in some Gram-negative bacteria such as Enterobacter, E. coli, Salmonella, and K. pneumoniae but remains unclear for some other bacterial strains (35, 120). Colistin resistance mechanism occurs by chromosomal modulations similar to bacteria that are naturally resistant to colistin. The various molecular mechanisms have been determined, and the most common modifications occur via cationic groups (L-Ara4N and pEtN) to the lipid membrane of bacterial strains (14, 35). The several operons and genes are related to the LPS membrane modulations (Figure 3). The two-component systems PhoPQ and PmrAB are extensively responsible for LPS modifications by addition of cationic groups to the LPS membrane. The operons coding enzymes are responsible for modifications in pmrC gene, pmrE gene, and the pmrHFIJKLM operon regulatory genes. Moreover, two-component systems are regulated by the mgrB gene, which negatively controls the expression of the two-component PhoPQ system. The pmrCAB operon codes three proteins: (a) the pEtN response regulator PmrA, (b) sensor kinase protein PmrAB, and (c) phosphotransferase PmrC (121, 122). However, the synthesis of the L-aminoarabinose group on LPS occurs by activation of pmrHFIJKLM and pmrE gene expression (123). Likewise, PmrAB two-component regulatory system encoded by PmrA and pmrB is activated by various environmental stimuli such as low pH (5.5), ferric (Fe3+) iron, macrophage phagosomes aluminum(Al3+), etc., and results in PmrB activation via periplasmic domain (121). In turn, pmrB activates PmrA by phosphorylation through tyrosine kinase protein of the pmrB gene. The PmrA activates transcription of the pmrCAB operon and the attached pmrE gene result in LPS modifications with the addition of cationic pEtN and L-Ara4N moieties (Figure 3) (121, 124). Reportedly, Specific mutations within the PmrA and pmrB genes have been found responsible for acquired colistin antibiotic resistance in K. pneumoniae (13, 98, 125–127), Enterobacter aerogenes (128), and Salmonella enterica (129, 130) (Table 1). Another PhoPQ two-component system encoded by PhoP and PhoQ genes, which expresses two proteins: (a) regulator protein PhoP and (b) sensor protein kinase PhoQ. The PhoPQ system is activated in acidic conditions (low pH) by various environmental stimuli such as low magnesium (Mg2+) and macrophage phagosomes and intercede PhoQ activation through its periplasmic lipid domain (124). Moreover, transcription activation of the pmrHFIJKLM operon occurs by PhoP resulting in L-Ara4N addition to the LPS membrane (180, 181). The PmrA protein is also activated by the PhoP gene either directly or indirectly via the PmrD (connector protein), causing the addition of pEtN to the LPS. However, acquired colistin resistance was reported in K. pneumoniae by mutations in the PhoP and PhoQ genes (13, 98, 126, 140, 182, 183) (Table 1). It has been observed in E. coli that a potential mutation in PmrAB causes acquired colistin resistance (131). The sum of constitutive activation of PhoPQ through modulations causes overexpression of the pmrHFIJKLM operon, thus synthesizing lipid-modifying moieties L-Ara4N that binds to lipid A membrane.
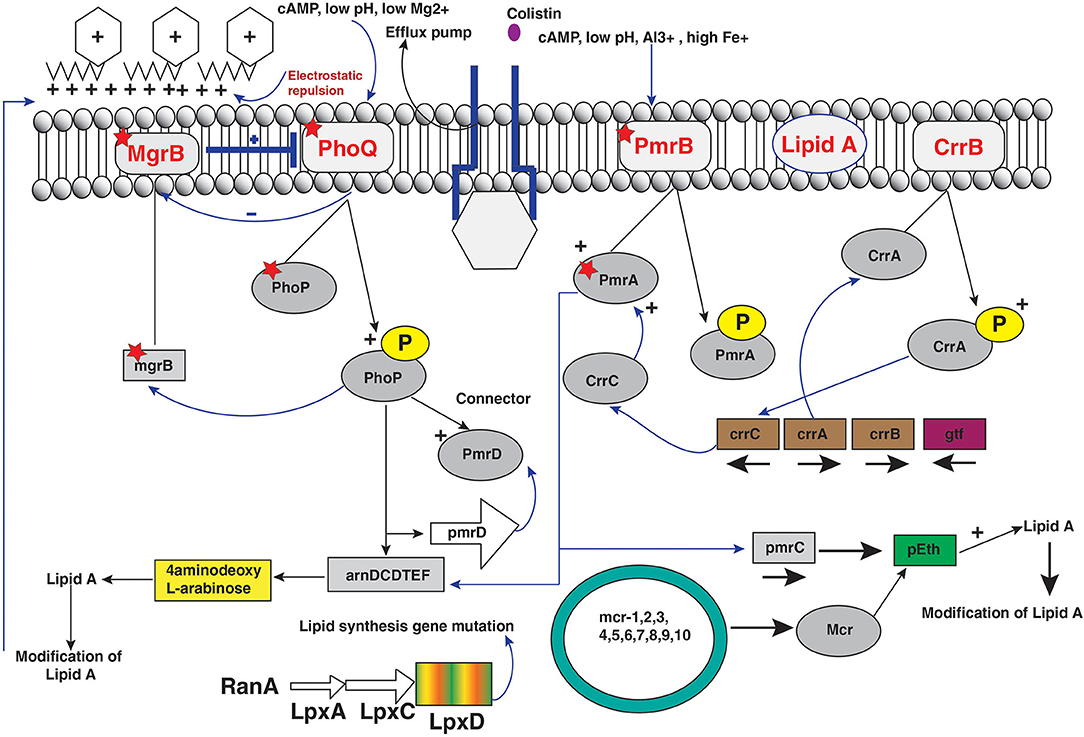
Figure 3. Regulators of colistin resistance mechanisms via chromosomal and plasmid-mediated pathways of lipopolysaccharide modifications in Enterobacteriaceae.
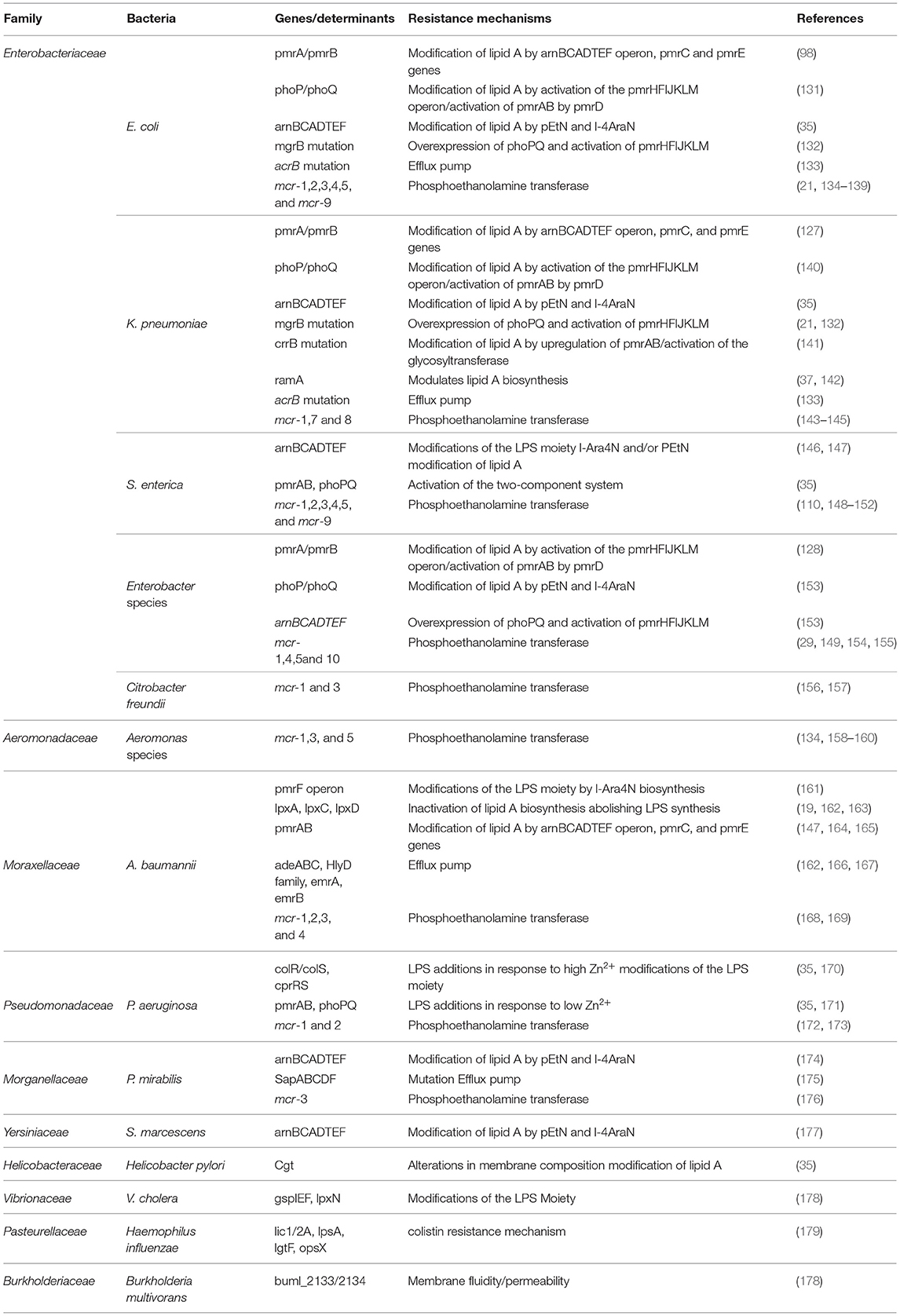
Table 1. Characteristics of colistin resistance mechanisms and modifications associated with most known bacteria.
CrrAB Two-Component System
CrrAB is a two-component regulatory system modulating the PmrAB system. It encodes two protein products as CrrA as a regulatory protein and CrrB as a sensor kinase protein. Colistin resistance in K. pneumoniae was reported by mutation of the crrB part of two-component systems (183). The glycosyltransferase-like protein was expressed through mutation of the CrrB protein that leads to modification on the outer LPS membrane (183). However, colistin resistance was observed by six amino acid substitutions in two-component crrB protein with MIC range 512–2,048 μg/mL. The expression analysis of pmrHFIJKLM operon with pmrC and pmrE leads to overexpression of PmrAB operon indirectly controlled by mutation of crrB gene and hence excess production of cationic pEtN and 4-amino-4-deoxy-L-arabinose to lipid A membrane that leads to colistin resistance (141, 183). Additionally, CrrAB and PmrAB two-component systems are indirectly connected via CrrC; mutations in the crrB gene system result in increased expression of CrrC. The amino acid substitution on the crrB protein leads to higher activation of protein by autophosphorylation that results in colistin resistance (141).
MgrB Regulator of the PmrAB and PhoPQ Two-Component Systems
The regulator of PmrAB and PhoPQ two-component system gene mgrB encodes a 47-amino-acid transmembrane protein that exerts negative feedback regulation of the PhoPQ two-component system and inhibits the kinase activity of PhoQ resulting in repression of the PhoQ gene (132, 184, 185). However, upregulation of PhoPQ operon occurs by inactivation and mutation of mgrB gene and consequent activation of pmrHFIJKLM operon, leading to excess production of cationic L-Ara4N that results in blocking colistin binding to LPS membrane. The various missense and non-sense mutations resulted in amino acid substitutions with truncated mgrB protein causing acquired resistance to colistin in Gram-negative bacteria, particularly K. pneumoniae. Moreover, it has been reported that other modes of modifications such as deletion or insertion in the mgrB gene sequence cause complete elimination of the mgrB locus (182, 184). In addition, insertional inactivation of the mgrB gene was found by several insertion sequences belonging to various families and inserted at different positions in the mgrB gene locus (13, 76, 98, 182, 184–188). Recently, colistin resistance was reported by transposition of genes encoding carbapenemase and extended-spectrum of β-lactamases, which leads to chromosomal mgrB gene disruption (189, 190). Moreover, reports suggest that coselection of colistin resistance with β-lactamase genes occurs with selection pressure, and deletion of the mgrB gene led to upregulated expression of the PhoP gene in E. coli resulting in colistin resistance (132).
Resistance by a Mutation in LPS Synthesis Genes
Resistance to colistin antibiotic was reported by loss of the initial colistin and membrane binding by electrostatic interactions with lipid A component of LPS in Gram-negative bacteria (14, 35). The binding is lost by complete loss of LPS membrane target site for colistin antibiotic. The LPS synthesis is governed by ramA gene locus containing three subgenes: ramA, romA, and ramR. The ramA and romA genes were downregulated by ramR; moreover, ramA regulator is known to be present in some Gram-negative bacteria such as Citrobacter species, Salmonella species, K. pneumoniae, and Enterobacter species. However, ramA modifies lipid A membrane biosynthesis that regulates permeability barriers (191). Recently, it has been reported that higher levels of RamA cause LPS modulations and hence increased colistin resistance (191). The loss of LPS results in colistin resistance in A. baumannii. The mutations in lipid A biosynthesis genes, lpxA, lpxC, and lpxD, cause total loss of LPS production halting colistin binding to membrane and hence colistin resistance. There are mutations in the first three genes of the LPS production and therefore complete loss of the LPS layer (19, 192).
Role of the Capsule in Colistin Resistance
The capsule in bacteria acts as a defensive and protective covering against an antimicrobial peptide including colistin (20, 193), and capsular polysaccharide is released by the bacteria from their surface (194). However, it has been reported that resistance pattern is dependent on the number of capsule layers that the bacteria can produce. Gram-negative bacteria such as K. pneumoniae with multiple capsule layers were found more resistant than bacteria with only a few CPS layers (10, 195). Moreover, the unregulated expression of CPS syntheses decreases colistin electrostatic interaction with the target site in K. pneumoniae, resulting in increased colistin antibiotic resistance (193). Subsequently, there are Cpx (conjugative pilus expression) and Rcs (regulator of capsule synthesis) regulators of capsule layer formation located on the LPS membrane (196). It has been reported that two-component PhoPQ and efflux pump KpnEF are activated by Rcs and Cpx, respectively (20, 197). Additionally, the ugd gene was found in CPS and L-Ara4N biosynthesis via phosphorylation causing assembly of capsules in bacterial strains and hence colistin resistance (198, 199).
Role of Efflux Pumps
There are reports of efflux pumps such as KpnEF, AcrAB, and Sap proteins systems involved in colistin resistance among bacterial isolates. The activation of these efflux pumps resulted in the increase of colistin resistance (20, 189, 200–203). The KpnEF pump is a member of the Cpx regulon and belongs to the SMR protein family (10). It has been revealed that efflux pumps are activated by colistin resistance to other related antibiotics such as rifampicin, ceftriaxone, and erythromycin (201). Moreover, the mutation in KpnEF results in more susceptibility and double reduction in the MICs with colistin antibiotic (201). From the same pipeline, efflux pump AcrAB is a small part of the AcrAB-TolC multifaceted structure involved in colistin resistance. Accordingly, AcrAB-mutant E. coli revealed an 8-fold increase in colistin antibiotic sensitivity. In addition, it has been observed that PhoPQ TCS is dependent on the expression of AcrAB pump proteins (204). Similarly, SapABCDF operon constitutes five proteins in P. mirabilis resulted in increasing sensitivity to colistin by a mutation in Sap ABCDEF operon (175, 204). It has been shown that the use of efflux-pump inhibitors in the test medium carbonyl cyanide 3-chlorophenylhydrazone leads to a reduction in MIC for colistin-resistant strains (203).
Emergence of Plasmid-mediated mcr-Like Genes
The acquired resistance from chromosomal to plasmid DNA coded on transposable genetic elements on plasmids with mcr-1 and several variants has been reported first in E. coli from China; after that, plasmid-mediated mcr-1 and variants have been detected in other Gram-negative bacterial isolates (134, 148). The resistance pattern was found to be the same as the chromosomal pmrC gene that codes mcr-1 protein pEtN transferase. It was hypothesized that mcr genes derived from intrinsically resistant environmental bacteria, e.g., Paenibacillus species, but mcr genes disseminated worldwide with an extremely transmissible plasmid (14). However, epidemiological and molecular studies have observed that mcr-1 in diversified Enterobacteriaceae family includes K. pneumoniae (14, 205), E. aerogenes (154), Shigella sonnei (206), Enterobacter cloacae (154), Salmonella (207, 208), Kluyvera species (209), Cronobacter sakazakii (210), Citrobacter species (159, 211), and Raoultella ornithinolytica (75) (Table 1). Additionally, mcr-1 harboring bacterial isolates exhibited complex resources including human linked environments and natural ecosystem (134, 212–214), food (26, 75, 148, 215), animals (216–218), and human (219–221). The LPS is modified by mcr-1 expression by adding cation pEtN transferase (pEtN transferase) (10). However, new variants of mcr-1 (mcr-1.0 to mcr-1.30) were reported with expression by modification of LPS membrane (Figure 4). Additional mcr variants were also reported such as mcr-2 (mcr-2.1 to mcr-2.7) (135). Phylogenetic studies observed that it is a new variant of mcr-1 with 80% identity. Subsequently, three more plasmid-mediated mcr-like gene variants were reported in E. coli and Salmonella as mcr-3 (mcr-3.1 to mcr-3.41) (136), mcr-4 (mcr-4.1 to mcr-4.6) (222), and mcr-5 (mcr-5.1 to mcr-5.4) (149). Phylogenetic studies reported mcr-3, mcr-4, and mcr-5 were descent genes of mcr-1/mcr−2. In 2018, new mcr gene variants, mcr-6 (mcr-6.1), mcr-7 (mcr-7.1), and mcr-8 (mcr-8.1-mcr-8.5), were identified that caused increasing spectrum of colistin resistance (143, 144, 223). Carrol et al. reported a novel mcr homolog, i.e., mcr-9 (mcr-9.1 to mcr-9.3), in multidrug-resistant colistin-susceptible S. enterica serovar Typhimurium isolates (150). Surprisingly, S. enterica serovar Typhimurium strain was phenotypically sensitive to colistin with an MIC value of 2 mg/L, according to (EUCAST) guidelines. Comparison analysis revealed that protein structures of all nine mcr homologs (mcr-1 to mcr-9) depicted that mcr-3, mcr-4, mcr-7, and mcr-9 genes have a high degree of resemblance at the structural level (150). Recently, mcr-10 (mcr-10.1) variant has been identified on an IncFIA plasmid of an Enterobacter roggenkampii clinical strain. This mcr variant has the highest nucleotide identity (79.69%) with mcr-9 and encodes mcr-10 with 82.93% amino acids identical to mcr-9 (29). The steep increase in plasmid-mediated mcr gene variants has raised a serious public health concern during the last few years.
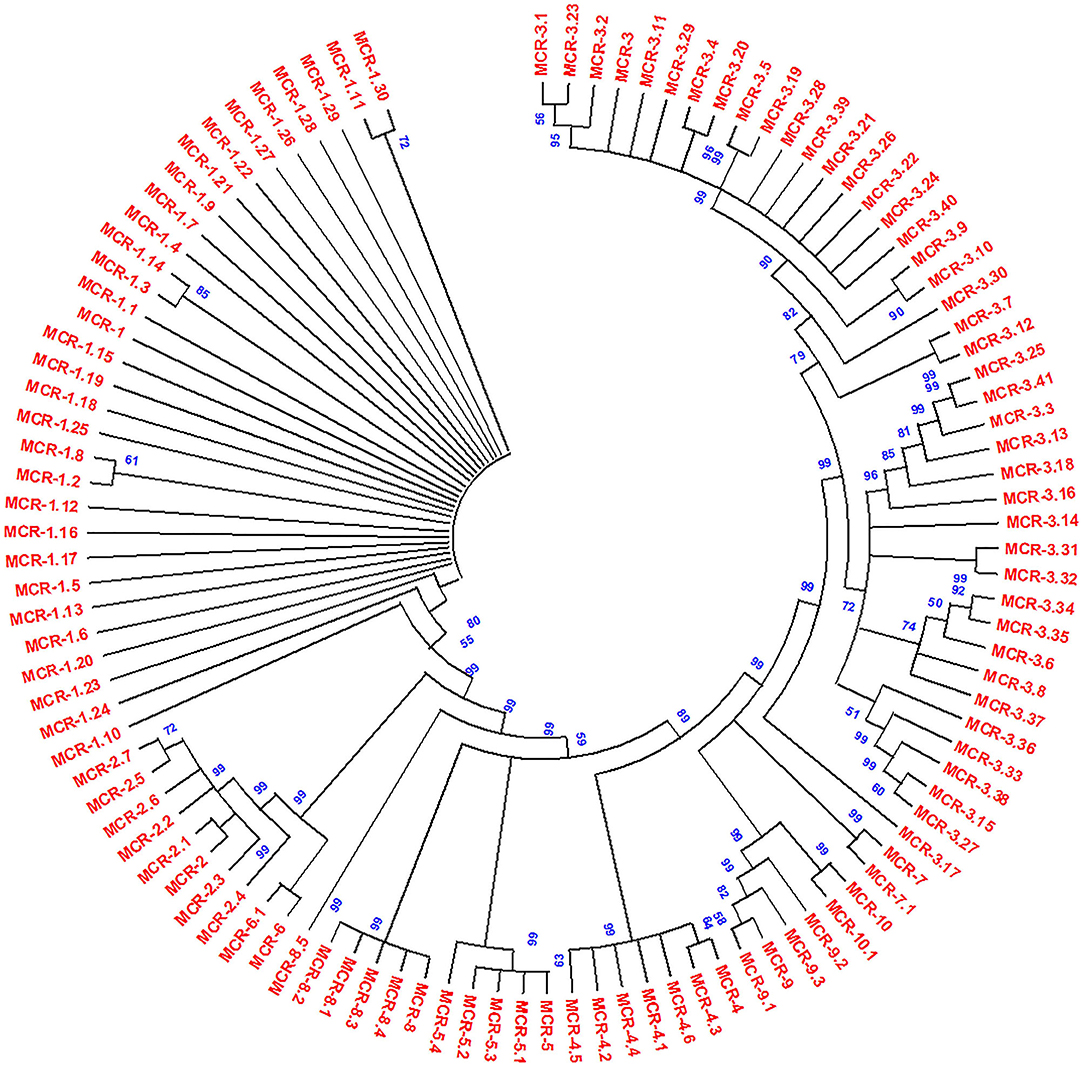
Figure 4. Phylogenetic tree of mcr gene variants, constructed by neighbor-joining method (bootstrap analysis with 500 replicates). All the proteins used for the phylogenetic tree were members of the PEA lipid A transferase family. Multiple sequence alignment was conducted using Clustal W, and resultant output was generated with Mega X.
Current Development to Revert Colistin Resistance
Several approaches are being developed for the treatment of colistin-resistant superbugs (224). To date, three primary approaches to reducing mcr-1 associated colistin resistance have been investigated. The first solution is the development of new antibiotics against mcr positive organisms, such as eravacycline (225), plazomicin (226), and artilysin (226, 227). Another technique tends to be the standard strategy that involves effective colistin administration and the possible use of combination therapies with additional agents to produce synergistic associations. These agents can include antibiotics that are typically restricted for use against Gram-positive bacteria, such as amikacin (228, 229), aztreonam (229), rifampin (230), azithromycin (230, 231), clarithromycin (232), linezolid (230), azidothymidine (233), tigecycline (234), and derivatives of tryptamine (235). Natural products can also be used to act as adjuvants, some of which might interact with LPSs, such as pentamidine and meridianine D analogs, to disturb the outer bacterial membrane (236, 237). In contrast, other adjuvants do not have specific roles so far, such as resveratrol (238), pterostilbene (239), osthole (240), and niclosamide (241). The last but most important and focused direction is to identify specific drugs targeting mcr. There are several methods identified to reduce mcr expression at the gene level, such as the use of peptide conjugated phosphorodiamidate morpholino oligomers to target mcr-1 mRNA (224), peptide nucleic acid against the mcr-1 gene (242), and the CRISPR/Cas9 system to target mcr-1 harboring plasmids (243). However, few studies have examined specific drugs targeting mcr, with promising results having been observed only for 1-phenyl-2-(phenylamino) ethanone derivatives (244) and the lipid A analog ethanolamine (245). The inhibitory action of ethanolamine against bacteria that produce mcr-1 was also tested in vivo to further confirm that it can be used as an inhibitor of mcr-1 activity. In the presence of 4 mg/mL polymyxin B, the results clearly showed that ethanolamine could inhibit mcr-1 expression in a concentration-dependent manner. Furthermore, ethanolamine can be used as an inhibitor of mcr-1 activity in light of the structural model and functional unification within the mcr family. Reports depicted that ethanolamine acts as an inhibitor of other mcr members. However, this would require further experimentation to validate. EptA catalyzes the transfer of PEA from phosphatidylethanolamine to lipid A at 1 and/or 4′ head group positions. EptA is an integral membrane protein consisting of an N-terminal transmembrane domain and a C-terminal soluble periplasmic-facing domain (246, 247). Moreover, EptA enzymes are found in many Gram-negative pathogens, e.g., E. coli, S. enterica, K. pneumoniae, etc. (248). In-depth studies are being undertaken to identify and optimize potential EptA inhibitors that suppress expression (224). In addition, similar approaches to EptA inhibition in Neisseria species are helpful for developing new therapies. Promising therapies are currently under development to boost phagocytic cells bactericidal activities (249), which could be used as novel combination therapies combined with anti-EptA compounds to effectively decrease transmission of multidrug-resistant bacteria. One strategy to counter this problem is to develop novel antivirulence agents that inhibit lipid A modification by EptA. Inhibition of EptA will hopefully restore the efficacy of polymyxin, support the clearance of infection by the immune system, and minimize the proliferation of colistin resistance (250).
Future Prospects of Resistance
The emergence of colistin resistance occurs via various mechanisms against the last line defense among carbapenem-resistant Gram-negative bacteria. Several studies led detection of different colistin resistance mechanisms in Enterobacteriaceae. However, there is still a lack of knowledge regarding colistin binding and initiating bactericidal activity. As new resistant variants of plasmid-mediated genes emerge that express new mechanisms of LPS modifications, on another side, many bacterial strains have inbuilt intrinsic colistin antibiotic resistance. Such a type of drug resistance occurs by modification of LPS with LAra4N. Moreover, naturally occurring resistant bacteria against colistin were found to have an expression of some specific chromosomal mediated genes such as eptA of N. meningitides. It becomes a need of the hour to decipher other possible mechanisms of colistin resistance that are still unknown. Reports revealed that colistin resistance follows only when bacteria are exposed to colistin antibiotics, but other reports suggest that colistin resistance can occur without earlier colistin antibiotic exposure. This represents a serious threat that obstructs the application of colistin as the last line therapy against multidrug-resistant Gram-negative bacteria. A perspective of this phenomenon is crucial and fundamental to protect against the future possibility of the deadly development of bacteria conferring colistin resistance.
Conclusion
The current epidemiological situation with multiple drug resistance results in the reuse of last-resort antibiotic (colistin) to treat bacterial infection. Moreover, colistin in combination with other drugs becomes a therapy against pathogenic bacteria. The unprecedented use of colistin drug in human medicine, animal husbandry, aquaculture, and agriculture has a serious impact on the emergence and dissemination of colistin resistance among Gram-negative bacteria. The resistance to this lifesaving drug has become a serious public health problem. The primary mechanism of colistin resistance occurs by modulation of chromosomal two-component PmrAB and PhoPQ systems, resulting in modification of the bacterial OM. Additionally, rising risks occur in heteroresistance by colistin attributed in suboptimal colistin dosages and represent a mean potential source of colistin resistance. The emergence of the plasmid-mediated mcr-1 gene encoding colistin resistance in bacteria can transfer horizontally from one bacteria to another and further disseminate among animals, humans, and the environment. Moreover, identification of new mcr variants such as mcr-2, mcr-3, mcr-4, mcr-5, mcr-6, mcr-7, mcr-8, mcr-9, and recently detected mcr-10 becomes a more concern to health. There is a clear indication of the rapid spread of plasmid-mediated colistin resistance variants, which requires further studies to evaluate the factors involved, the mechanism of acquisition and dissemination among bacteria. The handier, versatile, and robotic methods are needed to identify different new deadly mcr determinants rapidly. Indeed, more investigations are required to identify the exact role of different colistin resistance determinants that will potentially aid in developing new less toxic and potent drugs to treat infections caused by resistant Gram-negative bacteria. Therefore, colistin resistance should be distinguished as a serious medical issue that requires multisectoral research with proper surveillance and suitable monitoring systems to report the rate of dissemination of these resistant genes.
Author Contributions
FG and QH conceived the idea. FG and MS searched and reviewed the literature and summarized. FG wrote the draft of the paper. FG, MS, and IS contributed to graphics and references. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
FG wishes to acknowledge Indian Council of Medical Research (ICMR), India, for financial support in the form of fellowship (File No. AMR/Fellowship/1/2019-ECD-II). Authors wish to acknowledge Mohd Younis Bhat, postdoctoral fellow, Karolinska Institutet, Sweden for assistance in graphics and designing.
References
1. Peterson E, Kaur P. Antibiotic resistance mechanisms in bacteria: relationships between resistance determinants of antibiotic producers, environmental bacteria, and clinical pathogens. Front Microbiol. (2018) 9:2928. doi: 10.3389/fmicb.2018.02928
2. Siddiqui MT, Mondal AH, Gogry FA, Husain FM, Alsalme A, Haq QMR. Plasmid-Mediated ampicillin, quinolone, and heavy metal co-resistance among ESBL-producing isolates from the Yamuna River, New Delhi, India. Antibiotics. (2020) 9:826. doi: 10.3390/antibiotics9110826
3. Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. (2010) 74:417–33. doi: 10.1128/MMBR.00016-10
4. Paterson DL, Lipman J. Returning to the pre-antibiotic era in the critically ill: the XDR problem. Crit Care Med. (2007) 35:1789–91. doi: 10.1097/01.CCM.0000269352.39174.A4
5. WHO. Antimicrobial Resistance. Geneva: World Health Organization (2020). Available online at: https://www.undp.org/content/undp/en/home/sustainable-development-goals.html (accessed June 20, 2021).
6. Steenbergen JN, Alder J, Thorne GM, Tally FP. Daptomycin: a lipopeptide antibiotic for the treatment of serious Gram-positive infections. J Antimicrob Chemother. (2005) 55:283–8. doi: 10.1093/jac/dkh546
7. Koulenti D, Xu E, Mok IYS, Song A, Karageorgopoulos DE, Armaganidis A, et al. Novel antibiotics for multidrug-resistant gram-positive microorganisms. Microorganisms. (2019) 7:270. doi: 10.3390/microorganisms7080270
8. Siddiqui MT, Mondal AH, Sultan I, Ali A, Haq QMR. Co-occurrence of ESBLs and silver resistance determinants among bacterial isolates inhabiting polluted stretch of river Yamuna, India. Int J Environ Sci Technol. (2019) 16:5611–22. doi: 10.1007/s13762-018-1939-9
9. Zurfuh K, Poirel L, Nordmann P, Nüesch-Inderbinen M, Hächler H, Stephan R. Occurrence of the plasmid-borne mcr-1 colistin resistance gene in extended-spectrum-β-lactamase-producing enterobacteriaceae in river water and imported vegetable samples in Switzerland. Antimicrob Agents Chemother. (2016) 60:2594–5. doi: 10.1128/AAC.00066-16
10. Baron S, Hadjadj L, Rolain JM, Olaitan AO. Molecular mechanisms of polymyxin resistance: knowns and unknowns. Int J Antimicrob Agents. (2016) 48:583–91. doi: 10.1016/j.ijantimicag.2016.06.023
11. Li J, Nation RL, Turnidge JD, Milne RW, Coulthard K, Rayner CR, et al. Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect Dis. (2006) 6:589–601. doi: 10.1016/S1473-3099(06)70580-1
12. Falagas ME, Kasiakou SK. Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin Infect Dis. (2005) 40:1333–41. doi: 10.1086/429323
13. Cheng YH, Lin TL, Pan YJ, Wang YP, Lin YT, Wang JT. Colistin resistance mechanisms in Klebsiella pneumoniae strains from Taiwan. Antimicrob Agents Chemother. (2015) 59:2909–13. doi: 10.1128/AAC.04763-14
14. Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. (2016) 16:161–8. doi: 10.1016/S1473-3099(15)00424-7
15. Vaara M. Polymyxins and their potential next generation as therapeutic antibiotics. (2019) Front Microbiol. 10:1689. doi: 10.3389/fmicb.2019.01689
16. Evans ME, Feola DJ, Rapp RP. Polymyxin B sulfate and colistin: old antibiotics for emerging multiresistant gram-negative bacteria. Ann Pharmacother. (1999) 33:960–7. doi: 10.1345/aph.18426
17. Storm DR, Rosenthal KS, Swanson PE. Polymyxin and related peptide antibiotics. Annu Rev Biochem. (1977) 46:723–63. doi: 10.1146/annurev.bi.46.070177.003451
18. Falagas ME, Kasiakou SK. Toxicity of polymyxins: a systematic review of the evidence from old and recent studies. Crit Care. (2006) 10:R27. doi: 10.1186/cc3995
19. Moffatt JH, Harper M, Harrison P, Hale JD, Vinogradov E, Seemann T, et al. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob Agents Chemother. (2010) 54:4971–7. doi: 10.1128/AAC.00834-10
20. Olaitan AO, Morand S, Rolain JM. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol. (2014) 5:643. doi: 10.3389/fmicb.2014.00643
21. Cannatelli A, D'Andrea MM, Giani T, Di Pilato V, Arena F, Ambretti S, et al. In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob Agents Chemother. (2013) 57:5521–6. doi: 10.1128/AAC.01480-13
22. Payne M, Croxen MA, Lee TD, Mayson B, Champagne S, Leung V, et al. mcr-1-Positive colistin-resistant Escherichia coli in traveler returning to Canada from China. Emerg Infect Dis. (2016) 22:1673–5. doi: 10.3201/eid2209.160177
23. Macesic N, Green D, Wang Z, Sullivan SB, Shim K, Park S, et al. Detection of mcr-1-carrying Escherichia coli causing bloodstream infection in a New York City hospital: avian origins, human concerns? Open Forum Infect Dis. (2017) 4:ofx115. doi: 10.1093/ofid/ofx115
24. Tada T, Uechi K, Nakasone I, Shimada K, Nakamatsu M, Kirikae T, et al. Emergence of a colistin-resistant Escherichia coli clinical isolate harboring mcr-1 in Japan. Int J Infect Dis. (2017) 63:21–2. doi: 10.1016/j.ijid.2017.07.023
25. Nishino Y, Shimojima Y, Suzuki Y, Ida M, Fukui R, Kuroda S, et al. Detection of the mcr-1 gene in colistin-resistant Escherichia coli from retail meat in Japan. Microbiol Immunol. (2017) 61:554–7. doi: 10.1111/1348-0421.12549
26. Grami R, Mansour W, Mehri W, Bouallègue O, Boujaâfar N, Madec JY, et al. Impact of food animal trade on the spread of mcr-1-mediated colistin resistance, Tunisia, July 2015. Euro Surveill. (2016) 21:30144. doi: 10.2807/1560-7917.ES.2016.21.8.30144
27. Theuretzbacher U. Global antimicrobial resistance in Gram-negative pathogens and clinical need. Curr Opin Microbiol. (2017) 39:106–12. doi: 10.1016/j.mib.2017.10.028
28. El-Sayed Ahmed MAE, Zhong LL, Shen C, Yang Y, Doi Y, Tian GB. Colistin and its role in the Era of antibiotic resistance: an extended review (2000-2019). Emerg Microbes Infect. (2020) 9:868–85. doi: 10.1080/22221751.2020.1754133
29. Wang C, Feng Y, Liu L, Wei L, Kang M, Zong Z. Identification of novel mobile colistin resistance gene mcr-10. Emerg Microbes Infect. (2020) 9:508–16. doi: 10.1080/22221751.2020.1732231
30. Vidaillac C, Benichou L, Duval RE. In vitro synergy of colistin combinations against colistin-resistant Acinetobacter baumannii, Pseudomonas aeruginosa, and Klebsiella pneumoniae isolates. Antimicrob Agents Chemother. (2012) 56:4856–61. doi: 10.1128/AAC.05996-11
31. Zgurskaya HI, Löpez CA, Gnanakaran S. Permeability barrier of gram-negative cell envelopes and approaches to bypass it. ACS Infect Dis. (2015) 1:512–22. doi: 10.1021/acsinfecdis.5b00097
32. Beceiro A, Moreno A, Fernández N, Vallejo JA, Aranda J, Adler B, et al. Biological cost of different mechanisms of colistin resistance and their impact on virulence in Acinetobacter baumannii. Antimicrob Agents Chemother. (2014) 58:518–26. doi: 10.1128/AAC.01597-13
33. Falagas ME, Rafailidis PI, Matthaiou DK. Resistance to polymyxins: mechanisms, frequency and treatment options. Drug Resist Updat. (2010) 13:132–8. doi: 10.1016/j.drup.2010.05.002
34. Mendes CA, Burdmann EA. [Polymyxins - review with emphasis on nephrotoxicity]. Rev Assoc Med Bras. (1992) 55:752–9. doi: 10.1590/S0104-42302009000600023
35. Poirel L, Jayol A, Nordmann P. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev. (2017) 30:557–96. doi: 10.1128/CMR.00064-16
36. Gales AC, Jones RN, Sader HS. Global assessment of the antimicrobial activity of polymyxin B against 54 731 clinical isolates of Gram-negative bacilli: report from the SENTRY antimicrobial surveillance programme (2001-2004). Clin Microbiol Infect. (2006) 12:315–21. doi: 10.1111/j.1469-0691.2005.01351.x
37. Sherry N, Howden B. Emerging Gram negative resistance to last-line antimicrobial agents fosfomycin, colistin and ceftazidime-avibactam - epidemiology, laboratory detection and treatment implications. Expert Rev Anti Infect Ther. (2018) 16:289–306. doi: 10.1080/14787210.2018.1453807
38. Li J, Nation RL, Milne RW, Turnidge JD, Coulthard K. Evaluation of colistin as an agent against multi-resistant Gram-negative bacteria. Int J Antimicrob Agents. (2005) 25:11–25. doi: 10.1016/j.ijantimicag.2004.10.001
39. Deris ZZ, Akter J, Sivanesan S, Roberts KD, Thompson PE, Nation RL, et al. A secondary mode of action of polymyxins against Gram-negative bacteria involves the inhibition of NADH-quinone oxidoreductase activity. J Antibiot. (2014) 67:147–51. doi: 10.1038/ja.2013.111
40. Velkov T, Roberts KD, Nation RL, Thompson PE, Li J. Pharmacology of polymyxins: new insights into an 'old' class of antibiotics. Future Microbiol. (2013) 8:711–24. doi: 10.2217/fmb.13.39
41. Talbot GH, Bradley J, Edwards JE Jr, Gilbert D, Scheld M, et al. Bad bugs need drugs: an update on the development pipeline from the antimicrobial availability task force of the infectious diseases society of America. Clin Infect Dis. (2006) 42:657–68. doi: 10.1086/499819
42. Lim LM, Ly N, Anderson D, Yang JC, Macander L, Jarkowski A, et al. Resurgence of colistin: a review of resistance, toxicity, pharmacodynamics, and dosing. Pharmacotherapy. (2010) 30:1279–91. doi: 10.1592/phco.30.12.1279
43. Sultan I, Ali A, Gogry FA, Rather IA, Sabir JSM, Haq QMR. Bacterial isolates harboring antibiotics and heavy-metal resistance genes co-existing with mobile genetic elements in natural aquatic water bodies. Saudi J Biol Sci. (2020) 27:2660–8. doi: 10.1016/j.sjbs.2020.06.002
44. Fosse T, Giraud-Morin C, Madinier I. Induced colistin resistance as an identifying marker for Aeromonas phenospecies groups. Lett Appl Microbiol. (2003) 36:25–9. doi: 10.1046/j.1472-765X.2003.01257.x
45. Yahav D, Farbman L, Leibovici L, Paul M. Colistin: new lessons on an old antibiotic. Clin Microbiol Infect. (2012) 18:18–29. doi: 10.1111/j.1469-0691.2011.03734.x
46. Garonzik SM, Li J, Thamlikitkul V, Paterson DL, Shoham S, Jacob J, et al. Population pharmacokinetics of colistin methanesulfonate and formed colistin in critically ill patients from a multicenter study provide dosing suggestions for various categories of patients. Antimicrob Agents Chemother. (2011) 55:3284–94. doi: 10.1128/AAC.01733-10
47. Tran TB, Velkov T, Nation RL, Forrest A, Tsuji BT, Bergen PJ, et al. Pharmacokinetics/pharmacodynamics of colistin and polymyxin B: are we there yet? Int J Antimicrob Agents. (2016) 48:592–7. doi: 10.1016/j.ijantimicag.2016.09.010
48. Bulitta JB, Yang JC, Yohonn L, Ly NS, Brown SV, D'Hondt RE, et al. Attenuation of colistin bactericidal activity by high inoculum of Pseudomonas aeruginosa characterized by a new mechanism-based population pharmacodynamic model. Antimicrob Agents Chemother. (2010) 54:2051–62. doi: 10.1128/AAC.00881-09
49. Owen RJ, Li J, Nation RL, Spelman D. In vitro pharmacodynamics of colistin against Acinetobacter baumannii clinical isolates. J Antimicrob Chemother. (2007) 59:473–7. doi: 10.1093/jac/dkl512
50. Poudyal A, Howden BP, Bell JM, Gao W, Owen RJ, Turnidge JD, et al. In vitro pharmacodynamics of colistin against multidrug-resistant Klebsiella pneumoniae. J Antimicrob Chemother. (2008) 62:1311–8. doi: 10.1093/jac/dkn425
51. Landersdorfer CB, Nation RL. Colistin: how should it be dosed for the critically ill? Semin Respir Crit Care Med. (2015) 36:126–35. doi: 10.1055/s-0034-1398390
52. Li J, Rayner CR, Nation RL, Owen RJ, Spelman D, Tan KE, et al. Heteroresistance to colistin in multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother. (2006) 50:2946–50. doi: 10.1128/AAC.00103-06
53. Yau W, Owen RJ, Poudyal A, Bell JM, Turnidge JD, Yu HH, et al. Colistin hetero-resistance in multidrug-resistant Acinetobacter baumannii clinical isolates from the Western Pacific region in the SENTRY antimicrobial surveillance programme. J Infect. (2009) 58:138–44. doi: 10.1016/j.jinf.2008.11.002
54. Meletis G, Tzampaz E, Sianou E, Tzavaras I, Sofianou D. Colistin heteroresistance in carbapenemase-producing Klebsiella pneumoniae. J Antimicrob Chemother. (2011) 66:946–7. doi: 10.1093/jac/dkr007
55. Bergen PJ, Li J, Nation RL. Dosing of colistin-back to basic PK/PD. Curr Opin Pharmacol. (2011) 11:464–9. doi: 10.1016/j.coph.2011.07.004
56. Rubin C, Myers T, Stokes W, Dunham B, Harris S, Lautner B, et al. Review of institute of medicine and national research council recommendations for One Health initiative. Emerg Infect Dis. (2013) 19:1913–7. doi: 10.3201/eid1912.121659
57. Gronvall G, Boddie C, Knutsson R, Colby M. One Health security: an important component of the global health security agenda. Biosecur Bioterror. (2014) 12:221–4. doi: 10.1089/bsp.2014.0044
58. Wang R, van Dorp L, Shaw LP, Bradley P, Wang Q, Wang X, et al. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat Commun. (2018) 9:1179. doi: 10.1038/s41467-018-03205-z
59. Marston HD, Dixon DM, Knisely JM, Palmore TN, Fauci AS. Antimicrobial resistance. JAMA. (2016) 316:1193–204. doi: 10.1001/jama.2016.11764
60. Monaco M, Giani T, Raffone M, Arena F, Garcia-Fernandez A, Pollini S, et al. Colistin resistance superimposed to endemic carbapenem-resistant Klebsiella pneumoniae: a rapidly evolving problem in Italy, November 2013 to April 2014. Euro Surveill. (2014) 19:pii=20939. doi: 10.2807/1560-7917.ES2014.19.42.20939
61. Pena I, Picazo JJ, Rodríguez-Avial C, Rodríguez-Avial I. Carbapenemase-producing Enterobacteriaceae in a tertiary hospital in Madrid, Spain: high percentage of colistin resistance among VIM-1-producing Klebsiella pneumoniae ST11 isolates. Int J Antimicrob Agents. (2014) 43:460–4. doi: 10.1016/j.ijantimicag.2014.01.021
62. Meletis G, Oustas E, Botziori C, Kakasi E, Koteli A. Containment of carbapenem resistance rates of Klebsiella pneumoniae and Acinetobacter baumannii in a Greek hospital with a concomitant increase in colistin, gentamicin and tigecycline resistance. New Microbiol. (2015) 38:417–21.
63. Petrosillo N, Taglietti F, Granata G. Treatment options for colistin resistant Klebsiella pneumoniae: present and future. J Clin Med. (2019) 8:934. doi: 10.3390/jcm8070934
64. Ordooei Javan A, Shokouhi S, Sahraei Z. A review on colistin nephrotoxicity. Eur J Clin Pharmacol. (2015) 71:801–10. doi: 10.1007/s00228-015-1865-4
65. Sorlí L, Luque S, Grau S, Berenguer N, Segura C, Montero MM, et al. Trough colistin plasma level is an independent risk factor for nephrotoxicity: a prospective observational cohort study. BMC Infect Dis. (2013) 13:380. doi: 10.1186/1471-2334-13-380
66. Casal J, Mateu E, Mejía W, Martín M. Factors associated with routine mass antimicrobial usage in fattening pig units in a high pig-density area. Vet Res. (2007) 38:481–92. doi: 10.1051/vetres:2007010
67. Kempf I, Fleury MA, Drider D, Bruneau M, Sanders P, Chauvin C, et al. What do we know about resistance to colistin in Enterobacteriaceae in avian and pig production in Europe? Int J Antimicrob Agents. (2013) 42:379–83. doi: 10.1016/j.ijantimicag.2013.06.012
68. Trauffler M, Griesbacher A, Fuchs K, Köfer J. Antimicrobial drug use in Austrian pig farms: plausibility check of electronic on-farm records and estimation of consumption. Vet Rec. (2014) 175:402. doi: 10.1136/vr.102520
69. Sjölund M, Backhans A, Greko C, Emanuelson U, Lindberg A. Antimicrobial usage in 60 Swedish farrow-to-finish pig herds. Prev Vet Med. (2015) 121:257–64. doi: 10.1016/j.prevetmed.2015.07.005
70. Bos ME, Taverne FJ, van Geijlswijk IM, Mouton JW, Mevius DJ, Heederik DJ. Consumption of antimicrobials in pigs, veal calves, and broilers in the Netherlands: quantitative results of nationwide collection of data in 2011. PLoS ONE. (2013) 8:e77525. doi: 10.1371/journal.pone.0077525
71. Catry B, Cavaleri M, Baptiste K, Grave K, Grein K, Holm A, et al. Use of colistin-containing products within the European Union and European Economic Area (EU/EEA): development of resistance in animals and possible impact on human and animal health. Int J Antimicrob Agents. (2015) 46:297–306. doi: 10.1016/j.ijantimicag.2015.06.005
72. Centre QMR. The Global Polymyxin Industry Report Ottawa (Canada) (2015). Available online at: http://www.qyresearch.com (accessed February 19, 2021).
73. Liu BT, Li X, Zhang Q, Shan H, Zou M, Song FJ. Colistin-Resistant mcr-Positive Enterobacteriaceae in fresh vegetables, an increasing infectious threat in China. Int J Antimicrob Agents. (2019) 54:89–94. doi: 10.1016/j.ijantimicag.2019.04.013
74. Jones-Dias D, Manageiro V, Ferreira E, Barreiro P, Vieira L, Moura IB, et al. Architecture of class 1, 2, and 3 integrons from gram negative bacteria recovered among fruits and vegetables. Front Microbiol. (2016) 7:1400. doi: 10.3389/fmicb.2016.01400
75. Luo J, Yao X, Lv L, Doi Y, Huang X, Huang S, et al. Emergence of mcr-1 in Raoultella ornithinolytica and Escherichia coli Isolates from retail vegetables in China. Antimicrob Agents Chemother. (2017) 61(10). doi: 10.1128/AAC.01139-17
76. Ghafur A, Shankar C, GnanaSoundari P, Venkatesan M, Mani D, Thirunarayanan MA, et al. Detection of chromosomal and plasmid-mediated mechanisms of colistin resistance in Escherichia coli and Klebsiella pneumoniae from Indian food samples. J Glob Antimicrob Resist. (2019) 16:48–52. doi: 10.1016/j.jgar.2018.09.005
77. Zhou HW, Zhang T, Ma JH, Fang Y, Wang HY, Huang ZX, et al. Occurrence of plasmid- and chromosome-carried mcr-1 in waterborne enterobacteriaceae in China. Antimicrob Agents Chemother. (2017) 61:e00017-17. doi: 10.1128/AAC.00017-17
78. Jung Y, Jang H, Matthews KR. Effect of the food production chain from farm practices to vegetable processing on outbreak incidence. Microb Biotechnol. (2014) 7:517–27. doi: 10.1111/1751-7915.12178
79. Andersson DI, Hughes D. Microbiological effects of sublethal levels of antibiotics. Nat Rev Microbiol. (2014) 12:465–78. doi: 10.1038/nrmicro3270
80. Robinson TP, Bu DP, Carrique-Mas J, Fèvre EM, Gilbert M, Grace D, et al. Antibiotic resistance is the quintessential One Health issue. Trans R Soc Trop Med Hyg. (2016) 110:377–80. doi: 10.1093/trstmh/trw048
81. Larsson DGJ. Pollution from drug manufacturing: review and perspectives. Philos Trans R Soc Lond B Biol Sci. (2014) 369:20130571. doi: 10.1098/rstb.2013.0571
82. Hoelzer K, Wong N, Thomas J, Talkington K, Jungman E, Coukell A. Antimicrobial drug use in food-producing animals and associated human health risks: what, and how strong, is the evidence? BMC Vet Res. (2017) 13:211. doi: 10.1186/s12917-017-1131-3
83. Tang KL, Caffrey NP, Nóbrega DB, Cork SC, Ronksley PE, Barkema HW, et al. Restricting the use of antibiotics in food-producing animals and its associations with antibiotic resistance in food-producing animals and human beings: a systematic review and meta-analysis. Lancet Planetary health. (2017) 1:e316–27. doi: 10.1016/S2542-5196(17)30141-9
84. Scott AM, Beller E, Glasziou P, Clark J, Ranakusuma RW, Byambasuren O, et al. Is antimicrobial administration to food animals a direct threat to human health? A rapid systematic review. Int J Antimicrob Agents. (2018) 52:316–23. doi: 10.1016/j.ijantimicag.2018.04.005
85. Borck Høg K, Helle Bisgaard; Wolff Sönksen, Uti; Bager, Flemming; Bortolaia, Valeria; Ellis-Iversen, Johanne; Hendriksen, et al. DANMAP 2016 - Use of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, food and humans in Denmark. In: Statens Serum Institut, National Veterinary Institute, Technical University of Denmark National Food Institute, Technical University of Denmark. (Lyngby) (2017). 130 p.
86. Veldman K, Mevius D, Wit B, Pelt W, Franz E, Heederik D. MARAN 2019: Monitoring of antimicrobial resistance and antibiotic usage in animals in the Netherlands in 2018. In: Combined with NETHMAP-2019: Consumption of Antimicrobial Agents and Antimicrobial Resistance Among Medically Important Bacteria in the Netherlands (Leiden) (2019).
87. SWEDRES/SVARM. Consumption of Antibiotics and Occurrence of Antibiotic Resistance in Sweden. Solna/Uppsala (Sweden): PublicHealth Agency of Sweden and National Veterinary Institute (2018). Available online at: https://www.folkhalsomyn-digheten.se/contentassets/d8f6b3d187a94682a1d50a48f0a4fb3d/swedres-svarm-2018.pdf (accessed June 20, 2021).
88. White House. National Action Plan for Combating Antibiotic-Resistant Bacteria. Washington, DC: White House. (2015). Available online at: https://obamawhitehouse.archives.gov/sites/default/files/docs/national_action_plan_for_combating_antibotic-resistant_bacteria.pdf (accessed June 20, 2021).
89. World Health Organization. Global Action Plan on Antimicrobial Resistance. World Health Organization (2015). Available online at: http://www.who.int/iris/handle/10665/193736 (accessed June 20, 2021).
90. Tornimbene B, Eremin S, Escher M, Griskeviciene J, Manglani S, Pessoa-Silva CL. WHO global antimicrobial resistance surveillance system early implementation 2016-17. Lancet Infect Dis. (2018) 18:241–2. doi: 10.1016/S1473-3099(18)30060-4
91. Giske CG, Kahlmeter G. Colistin antimicrobial susceptibility testing-can the slow and challenging be replaced by the rapid and convenient? Clin Microbiol Infect. (2018) 24:93–4. doi: 10.1016/j.cmi.2017.10.007
92. Green DA, Macesic N, Uhlemann A-C, Lopez M, Stump S, Whittier S, et al. Evaluation of calcium-enhanced media for colistin susceptibility testing by gradient agar diffusion and broth microdilution. J Clin Microbiol. (2020) 58:e01522-19. doi: 10.1128/JCM.01522-19
93. Behera B, Mathur P, Das A, Kapil A, Gupta B, Bhoi S, et al. Evaluation of susceptibility testing methods for polymyxin. Int J Infect Dis. (2010) 14:e596–601. doi: 10.1016/j.ijid.2009.09.001
94. Dafopoulou K, Zarkotou O, Dimitroulia E, Hadjichristodoulou C, Gennimata V, Pournaras S, et al. Comparative evaluation of colistin susceptibility testing methods among carbapenem-nonsusceptible Klebsiella pneumoniae and Acinetobacter baumannii clinical isolates. Antimicrob Agents Chemother. (2015) 59:4625–30. doi: 10.1128/AAC.00868-15
95. Chew KL, La MV, Lin RTP, Teo JWP. Colistin and Polymyxin B susceptibility testing for carbapenem-resistant and mcr-Positive enterobacteriaceae: comparison of sensititre, MicroScan, Vitek 2, and Etest with broth microdilution. J Clin Microbiol. (2017) 55:2609–16. doi: 10.1128/JCM.00268-17
96. CLSI. Performance standards for antimicrobial susceptibility testing. 26th ed CLSI supplement M100S. Wayane, PA: Clinical and Laboratory Standards Institute (2016).
97. EUCAST. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 6.0 2016. Basel: The European Committee on Antimicrobial Susceptibility Testing (2016). Available online at: https://www.eucast.org (accessed June 11, 2019).
98. Nordmann P, Jayol A, Poirel L. Rapid detection of polymyxin resistance in enterobacteriaceae. Emerg Infect Dis. (2016) 22:1038–43. doi: 10.3201/eid2206.151840
99. Lellouche J, Schwartz D, Elmalech N, Ben Dalak MA, Temkin E, Paul M, et al. Combining VITEK® 2 with colistin agar dilution screening assist timely reporting of colistin susceptibility. Clin Microbiol Infect. (2019) 25:711–6. doi: 10.1016/j.cmi.2018.09.014
100. Matuschek E DL, Ahman J, Kahlmeter G, Wootton M. Can agar dilution be used for colistin MIC determination? In: Session: Colistin antimicrobial susceptibility testing – which methods are available? Madrid: ECCMID (2018).
101. Bosacka K, Kozińska A, Stefaniuk E, Rybicka J, Mikołajczyk A, Młodzińska E, et al. Colistin antimicrobial susceptibility testing of Gram-negative bacteria – evaluation of tests available in Poland. In: ECCMID 2018 Abstract Publication E0116, 21–24 (Madrid) (2018).
102. Cheong HS, Kim SY, Seo J, Wi YM, Peck KR, Ko KS. Colistin resistance and extensive genetic variations in PmrAB and PhoPQ in Klebsiella pneumoniae isolates from South Korea. Curr Microbiol. (2020) 77:2307–11. doi: 10.1007/s00284-020-02074-4
103. Di Tella D, Tamburro M, Guerrizio G, Fanelli I, Sammarco ML, Ripabelli G. Molecular epidemiological insights into colistin-resistant and carbapenemases-producing clinical Klebsiella pneumoniae isolates. Infect Drug Resist. (2019) 12:3783–95. doi: 10.2147/IDR.S226416
104. Janssen AB, Doorduijn DJ, Mills G, Rogers MRC, Bonten MJM, Rooijakkers SHM, et al. Evolution of colistin resistance in the Klebsiella pneumoniae complex follows multiple evolutionary trajectories with variable effects on fitness and virulence characteristics. Antimicrob Agents Chemother. (2020) 65:e01958-20. doi: 10.1128/AAC.00754-21
105. Rojas LJ, Salim M, Cober E, Richter SS, Perez F, Salata RA, et al. Colistin resistance in carbapenem-resistant Klebsiella pneumoniae: laboratory detection and impact on mortality. Clin Infect Dis. (2016) 64:711–8. doi: 10.1093/cid/ciw805
106. Nordmann P, Jayol A, Poirel L. A universal culture medium for screening polymyxin-resistant gram-negative isolates. J Clin Microbiol. (2016) 54:1395–9. doi: 10.1128/JCM.00446-16
107. Jayol A, Poirel L, André C, Dubois V, Nordmann P. Detection of colistin-resistant Gram-negative rods by using the SuperPolymyxin medium. Diagn Microbiol Infect Dis. (2018) 92:95–101. doi: 10.1016/j.diagmicrobio.2018.05.008
108. Abdul Momin MHF, Bean DC, Hendriksen RS, Haenni M, Phee LM, Wareham DW. CHROMagar COL-APSE: a selective bacterial culture medium for the isolation and differentiation of colistin-resistant Gram-negative pathogens. J Med Microbiol. (2017) 66:1554–61. doi: 10.1099/jmm.0.000602
109. Bardet L, Le Page S, Leangapichart T, Rolain JM. LBJMR medium: a new polyvalent culture medium for isolating and selecting vancomycin and colistin-resistant bacteria. BMC Microbiol. (2017) 17:220. doi: 10.1186/s12866-017-1128-x
110. Garcia-Graells C, De Keersmaecker SCJ, Vanneste K, Pochet B, Vermeersch K, Roosens N, et al. Detection of plasmid-mediated colistin resistance, mcr-1 and mcr-2 Genes, in Salmonella spp. isolated from Food at Retail in Belgium from 2012 to 2015. Foodborne Pathog Dis. (2018) 15:114–7. doi: 10.1089/fpd.2017.2329
111. Turlej-Rogacka A, Xavier BB, Janssens L, Lammens C, Zarkotou O, Pournaras S, et al. Evaluation of colistin stability in agar and comparison of four methods for MIC testing of colistin. Eur J Clin Microbiol Infect Dis. (2018) 37:345–53. doi: 10.1007/s10096-017-3140-3
112. Humphries RM. Susceptibility testing of the polymyxins: where are we now? Pharmacotherapy. (2015) 35:22–7. doi: 10.1002/phar.1505
113. Matuschek E, Åhman J, Webster C, Kahlmeter G. Antimicrobial susceptibility testing of colistin - evaluation of seven commercial MIC products against standard broth microdilution for Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter spp. Clin Microbiol Infect. (2018) 24:865–70. doi: 10.1016/j.cmi.2017.11.020
114. Zou D, Huang S, Lei H, Yang Z, Su Y, He X, et al. Sensitive and rapid detection of the plasmid-encoded colistin-resistance gene mcr-1 in enterobacteriaceae isolates by loop-mediated isothermal amplification. Front Microbiol. (2017) 8:2356. doi: 10.3389/fmicb.2017.02356
115. Bernasconi OJ, Principe L, Tinguely R, Karczmarek A, Perreten V, Luzzaro F, et al. Evaluation of a new commercial microarray platform for the simultaneous detection of β-Lactamase and mcr-1 and mcr-2 genes in enterobacteriaceae. J Clin Microbiol. (2017) 55:3138–41. doi: 10.1128/JCM.01056-17
116. Zankari E, Hasman H, Kaas RS, Seyfarth AM, Agersø Y, Lund O, et al. Genotyping using whole-genome sequencing is a realistic alternative to surveillance based on phenotypic antimicrobial susceptibility testing. J Antimicrob Chemother. (2013) 68:771–7. doi: 10.1093/jac/dks496
117. Bortolaia V, Kaas RS, Ruppe E, Roberts MC, Schwarz S, Cattoir V, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. (2020) 75:3491–500. doi: 10.1093/jac/dkaa345
118. Li J, Milne RW, Nation RL, Turnidge JD, Smeaton TC, Coulthard K. Pharmacokinetics of colistin methanesulphonate and colistin in rats following an intravenous dose of colistin methanesulphonate. J Antimicrob Chemother. (2004) 53:837–40. doi: 10.1093/jac/dkh167
119. Dixon RA, Chopra I. Leakage of periplasmic proteins from Escherichia coli mediated by polymyxin B nonapeptide. Antimicrob Agents Chemother. (1986) 29:781–8. doi: 10.1128/AAC.29.5.781
120. Stefaniuk EM, Tyski S. Colistin resistance in enterobacterales strains - a current view. Pol J Microbiol. (2019) 68:417–27. doi: 10.33073/pjm-2019-055
121. Gunn JS. The Salmonella PmrAB regulon: lipopolysaccharide modifications, antimicrobial peptide resistance and more. Trends Microbiol. (2008) 16:284–90. doi: 10.1016/j.tim.2008.03.007
122. Hua J, Jia X, Zhang L, Li Y. The characterization of two-component system PmrA/PmrB in Cronobacter sakazakii. Front Microbiol. (2020) 11:903. doi: 10.3389/fmicb.2020.00903
123. Yan A, Guan Z, Raetz CR. An undecaprenyl phosphate-aminoarabinose flippase required for polymyxin resistance in Escherichia coli. J Biol Chem. (2007) 282:36077–89. doi: 10.1074/jbc.M706172200
124. Chin C-Y, Gregg K, Napier B, Ernst R, Weiss D. A PmrB-regulated deacetylase required for lipid a modification and polymyxin resistance in Acinetobacter baumannii. Antimicrob Agents Chemother. (2015) 59:7911–4. doi: 10.1128/AAC.00515-15
125. Cannatelli A, Di Pilato V, Giani T, Arena F, Ambretti S, Gaibani P, et al. In vivo evolution to colistin resistance by PmrB sensor kinase mutation in KPC-producing Klebsiella pneumoniae is associated with low-dosage colistin treatment. Antimicrob Agents Chemother. (2014) 58:4399–403. doi: 10.1128/AAC.02555-14
126. Choi MJ, Ko KS. Mutant prevention concentrations of colistin for Acinetobacter baumannii, Pseudomonas aeruginosa and Klebsiella pneumoniae clinical isolates. J Antimicrob Chemother. (2014) 69:275–7. doi: 10.1093/jac/dkt315
127. Jayol A, Poirel L, Brink A, Villegas MV, Yilmaz M, Nordmann P. Resistance to colistin associated with a single amino acid change in protein PmrB among Klebsiella pneumoniae isolates of worldwide origin. Antimicrob Agents Chemother. (2014) 58:4762–6. doi: 10.1128/AAC.00084-14
128. Diene SM, Merhej V, Henry M, El Filali A, Roux V, Robert C, et al. The rhizome of the multidrug-resistant Enterobacter aerogenes genome reveals how new “killer bugs” are created because of a sympatric lifestyle. Mol Biol Evol. (2013) 30:369–83. doi: 10.1093/molbev/mss236
129. Olaitan AO, Dia NM, Gautret P, Benkouiten S, Belhouchat K, Drali T, et al. Acquisition of extended-spectrum cephalosporin- and colistin-resistant Salmonella enterica subsp. enterica serotype Newport by pilgrims during Hajj. Int J Antimicrob Agents. (2015) 45:600–4. doi: 10.1016/j.ijantimicag.2015.01.010
130. Sun S, Negrea A, Rhen M, Andersson DI. Genetic analysis of colistin resistance in Salmonella enterica serovar Typhimurium. Antimicrob Agents Chemother. (2009) 53:2298–305. doi: 10.1128/AAC.01016-08
131. Olaitan AO, Thongmalayvong B, Akkhavong K, Somphavong S, Paboriboune P, Khounsy S, et al. Clonal transmission of a colistin-resistant Escherichia coli from a domesticated pig to a human in Laos. J Antimicrob Chemother. (2015) 70:3402–4. doi: 10.1093/jac/dkv252
132. Lippa AM, Goulian M. Feedback inhibition in the PhoQ/PhoP signaling system by a membrane peptide. PLoS Genet. (2009) 5:e1000788. doi: 10.1371/journal.pgen.1000788
133. Padilla E, Llobet E, Doménech-Sánchez A, Martínez-Martínez L, Bengoechea JA, Albertí S. Klebsiella pneumoniae AcrAB efflux pump contributes to antimicrobial resistance and virulence. Antimicrob Agents Chemother. (2010) 54:177–83. doi: 10.1128/AAC.00715-09
134. Gogry FA, Siddiqui MT, Haq QMR. Emergence of mcr-1 conferred colistin resistance among bacterial isolates from urban sewage water in India. Environ Sci Pollut Res Int. (2019) 26:33715–7. doi: 10.1007/s11356-019-06561-5
135. Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, Goossens H, et al. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill. (2016) 21:30280. doi: 10.2807/1560-7917.ES.2016.21.27.30280
136. Yin W, Li H, Shen Y, Liu Z, Wang S, Shen Z, et al. Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. MBio. (2017) 8:e00543-17. doi: 10.1128/mBio.00543-17
137. Hasman H, Hammerum AM, Hansen F, Hendriksen RS, Olesen B, Agersø Y, et al. Detection of mcr-1 encoding plasmid-mediated colistin-resistant Escherichia coli isolates from human bloodstream infection and imported chicken meat, Denmark 2015. Euro Surveill. (2015) 20:30085. doi: 10.2807/1560-7917.ES.2015.20.49.30085
138. Fukuda A, Sato T, Shinagawa M, Takahashi S, Asai T, Yokota SI, et al. High prevalence of mcr-1, mcr-3 and mcr-5 in Escherichia coli derived from diseased pigs in Japan. Int J Antimicrob Agents. (2018) 51:163–4. doi: 10.1016/j.ijantimicag.2017.11.010
139. Tyson GH, Li C, Hsu CH, Ayers S, Borenstein S, Mukherjee S, et al. The mcr-9 gene of salmonella and Escherichia coli is not associated with colistin resistance in the United States. Antimicrob Agents Chemother. (2020) 64:e00573-20. doi: 10.1128/AAC.00573-20
140. Jayol A, Nordmann P, Brink A, Poirel L. Heteroresistance to colistin in Klebsiella pneumoniae associated with alterations in the PhoPQ regulatory system. Antimicrob Agents Chemother. (2015) 59:2780–4. doi: 10.1128/AAC.05055-14
141. Cheng YH, Lin TL, Lin YT, Wang JT. Amino acid substitutions of CrrB responsible for resistance to colistin through CrrC in Klebsiella pneumoniae. Antimicrob Agents Chemother. (2016) 60:3709–16. doi: 10.1128/AAC.00009-16
142. De Majumdar S, Yu J, Fookes M, McAteer SP, Llobet E, Finn S, et al. Elucidation of the RamA regulon in Klebsiella pneumoniae reveals a role in LPS regulation. PLoS Pathog. (2015) 11:e1004627. doi: 10.1371/journal.ppat.1004627
143. Wang X, Wang Y, Zhou Y, Li J, Yin W, Wang S, et al. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg Microbes Infect. (2018) 7:122. doi: 10.1038/s41426-018-0124-z
144. Yang YQ, Li YX, Lei CW, Zhang AY, Wang HN. Novel plasmid-mediated colistin resistance gene mcr-7.1 in Klebsiella pneumoniae. J Antimicrob Chemother. (2018) 73:1791–5. doi: 10.1093/jac/dky111
145. Du H, Chen L, Tang YW, Kreiswirth BN. Emergence of the mcr-1 colistin resistance gene in carbapenem-resistant Enterobacteriaceae. Lancet Infect Dis. (2016) 16:287–8. doi: 10.1016/S1473-3099(16)00056-6
146. Rhouma M, Beaudry F, Thériault W, Letellier A. Colistin in pig production: chemistry, mechanism of antibacterial action, microbial resistance emergence, and one health perspectives. Front Microbiol. (2016) 7:1789. doi: 10.3389/fmicb.2016.01789
147. Lima WG, Alves MC, Cruz WS, Paiva MC. Chromosomally encoded and plasmid-mediated polymyxins resistance in Acinetobacter baumannii: a huge public health threat. Eur J Clin Microbiol Infect Dis. (2018) 37:1009–19. doi: 10.1007/s10096-018-3223-9
148. Doumith M, Godbole G, Ashton P, Larkin L, Dallman T, Day M, et al. Detection of the plasmid-mediated mcr-1 gene conferring colistin resistance in human and food isolates of Salmonella enterica and Escherichia coli in England and Wales. J Antimicrob Chemother. (2016) 71:2300–5. doi: 10.1093/jac/dkw093
149. Borowiak M, Fischer J, Hammerl JA, Hendriksen RS, Szabo I, Malorny B. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J Antimicrob Chemother. (2017) 72:3317–24. doi: 10.1093/jac/dkx327
150. Carroll LM, Gaballa A, Guldimann C, Sullivan G, Henderson LO, Wiedmann M. Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype typhimurium isolate. MBio. (2019) 10:e00853-19. doi: 10.1128/mBio.00853-19
151. Litrup E, Kiil K, Hammerum AM, Roer L, Nielsen EM, Torpdahl M. Plasmid-borne colistin resistance gene mcr-3 in Salmonella isolates from human infections, Denmark, 2009-17. Euro Surveill. (2017) 22:30587. doi: 10.2807/1560-7917.ES.2017.22.31.30587
152. Carretto E, Brovarone F, Nardini P, Russello G, Barbarini D, Pongolini S, et al. Detection of mcr-4 positive Salmonella enterica serovar Typhimurium in clinical isolates of human origin, Italy, October to November 2016. Euro Surveill. (2018) 23:1700821. doi: 10.2807/1560-7917.ES.2018.23.2.17-00821
153. Kang KN, Klein DR, Kazi MI, Guérin F, Cattoir V, Brodbelt JS, et al. Colistin heteroresistance in Enterobacter cloacae is regulated by PhoPQ-dependent 4-amino-4-deoxy-l-arabinose addition to lipid A. Mol Microbiol. (2019) 111:1604–16. doi: 10.1111/mmi.14240
154. Zeng KJ, Doi Y, Patil S, Huang X, Tian GB. Emergence of the plasmid-mediated mcr-1 gene in colistin-resistant Enterobacter aerogenes and Enterobacter cloacae. Antimicrob Agents Chemother. (2016) 60:3862–3. doi: 10.1128/AAC.00345-16
155. Chavda B, Lv J, Hou M, Chavda KD, Kreiswirth BN, Feng Y, et al. Coidentification of mcr-4.3 and bla(NDM-1) in a Clinical Enterobacter cloacae Isolate from China. Antimicrob Agents Chemother. (2018) 62:e00649-18. doi: 10.1128/AAC.00649-18
156. Li XP, Fang LX, Jiang P, Pan D, Xia J, Liao XP, et al. Emergence of the colistin resistance gene mcr-1 in Citrobacter freundii. Int J Antimicrob Agents. (2017) 49:786–7. doi: 10.1016/j.ijantimicag.2017.04.004
157. Börjesson S, Greko C, Myrenås M, Landén A, Nilsson O, Pedersen K. A link between the newly described colistin resistance gene mcr-9 and clinical Enterobacteriaceae isolates carrying blaSHV-12 from horses in Sweden. J Global Antimicrob Resist. (2020) 20:285–9. doi: 10.1016/j.jgar.2019.08.007
158. Ragupathi NKD, Sethuvel DPM, Anandan S, Murugan D, Asokan K, Neethi Mohan RG, et al. First hybrid complete genome of Aeromonas veronii reveals chromosome-mediated novel structural variant mcr-3.30 from a human clinical sample. Access Microbiol. (2020) 2:acmi000103. doi: 10.1099/acmi.0.000103
159. Ma S, Sun C, Hulth A, Li J, Nilsson LE, Zhou Y, et al. Mobile colistin resistance gene mcr-5 in porcine Aeromonas hydrophila. J Antimicrob Chemother. (2018) 73:1777–80. doi: 10.1093/jac/dky110
160. Shen Y, Xu C, Sun Q, Schwarz S, Ou Y, Yang L, et al. Prevalence and genetic analysis of mcr-3-Positive Aeromonas species from humans, retail meat, and environmental water samples. Antimicrob Agents Chemother. (2018) 62:e00404-18. doi: 10.1128/AAC.00404-18
161. Jeannot K, Bolard A, Plésiat P. Resistance to polymyxins in Gram-negative organisms. Int J Antimicrob Agents. (2017) 49:526–35. doi: 10.1016/j.ijantimicag.2016.11.029
162. Hood MI, Becker KW, Roux CM, Dunman PM, Skaar EP. Genetic determinants of intrinsic colistin tolerance in Acinetobacter baumannii. Infect Immun. (2013) 81:542–51. doi: 10.1128/IAI.00704-12
163. Bojkovic J, Richie DL, Six DA, Rath CM, Sawyer WS, Hu Q, et al. Characterization of an Acinetobacter baumannii lptD deletion strain: permeability defects and response to inhibition of lipopolysaccharide and fatty acid biosynthesis. J Bacteriol. (2015) 198:731–41. doi: 10.1128/JB.00639-15
164. Adams MD, Nickel GC, Bajaksouzian S, Lavender H, Murthy AR, Jacobs MR, et al. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob Agents Chemother. (2009) 53:3628–34. doi: 10.1128/AAC.00284-09
165. Cheah SE, Johnson MD, Zhu Y, Tsuji BT, Forrest A, Bulitta JB, et al. Polymyxin resistance in Acinetobacter baumannii: genetic mutations and transcriptomic changes in response to clinically relevant dosage regimens. Sci Rep. (2016) 6:26233. doi: 10.1038/srep26233
166. Thi Khanh Nhu N, Riordan DW, Do Hoang Nhu T, Thanh DP, Thwaites G, Huong Lan NP, et al. The induction and identification of novel Colistin resistance mutations in Acinetobacter baumannii and their implications. Sci Rep. (2016) 6:28291. doi: 10.1038/srep28291
167. Lin MF, Lin YY, Lan CY. Contribution of EmrAB efflux pumps to colistin resistance in Acinetobacter baumannii. J Microbiol. (2017) 55:130–6. doi: 10.1007/s12275-017-6408-5
168. Al-Kadmy IMS, Ibrahim SA, Al-Saryi N, Aziz SN, Besinis A, Hetta HF. Prevalence of Genes Involved in colistin resistance in Acinetobacter baumannii: First Report from Iraq. Microbial Drug Resistance. (2019) 26:616–22. doi: 10.1089/mdr.2019.0243
169. Ma F, Shen C, Zheng X, Liu Y, Chen H, Zhong L, et al. Identification of a Novel Plasmid Carrying mcr-4.3 in an Acinetobacter baumannii Strain in China. Antimicrob Agents Chemother. (2019) 63:e00133-19. doi: 10.1128/AAC.00133-19
170. Kaye KS, Pogue JM, Tran TB, Nation RL, Li J. Agents of last resort: polymyxin resistance. Infect Dis Clin North Am. (2016) 30:391–414. doi: 10.1016/j.idc.2016.02.005
171. Miller AK, Brannon MK, Stevens L, Johansen HK, Selgrade SE, Miller SI, et al. PhoQ mutations promote lipid A modification and polymyxin resistance of Pseudomonas aeruginosa found in colistin-treated cystic fibrosis patients. Antimicrob Agents Chemother. (2011) 55:5761–9. doi: 10.1128/AAC.05391-11
172. Caselli E, D'Accolti M, Soffritti I, Piffanelli M, Mazzacane S. Spread of mcr-1-driven colistin resistance on hospital surfaces, Italy. Emerg Infect Dis. (2018) 24:1752–3. doi: 10.3201/eid2409.171386
173. Azimi L, Lari AR. Colistin-resistant Pseudomonas aeruginosa clinical strains with defective biofilm formation. GMS Hyg Infect Control. (2019) 14:Doc12. doi: 10.3205/dgkh000328
174. Olaitan AO, Morand S, Rolain JM. Emergence of colistin-resistant bacteria in humans without colistin usage: a new worry and cause for vigilance. Int J Antimicrob Agents. (2016) 47:1–3. doi: 10.1016/j.ijantimicag.2015.11.009
175. McCoy AJ, Liu H, Falla TJ, Gunn JS. Identification of Proteus mirabilis mutants with increased sensitivity to antimicrobial peptides. Antimicrob Agents Chemother. (2001) 45:2030–7. doi: 10.1128/AAC.45.7.2030-2037.2001
176. Wang X, Zhai W, Li J, Liu D, Zhang Q, Shen Z, et al. Presence of an mcr-3 Variant in Aeromonas caviae, Proteus mirabilis, and Escherichia coli from one domestic duck. Antimicrob Agents Chemother. (2018) 62:e02106–17. doi: 10.1128/AAC.02106-17
177. Lin QY, Tsai YL, Liu MC, Lin WC, Hsueh PR, Liaw SJ. Serratia marcescens arn, a PhoP-regulated locus necessary for polymyxin B resistance. Antimicrob Agents Chemother. (2014) 58:5181–90. doi: 10.1128/AAC.00013-14
178. Mlynarcik P, Kolar M. Molecular mechanisms of polymyxin resistance and detection of mcr genes. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. (2019) 163:28–38. doi: 10.5507/bp.2018.070
179. Morey P, Viadas C, Euba B, Hood DW, Barberán M, Gil C, et al. Relative contributions of lipooligosaccharide inner and outer core modifications to nontypeable Haemophilus influenzae pathogenesis. Infect Immun. (2013) 81:4100–11. doi: 10.1128/IAI.00492-13
180. Groisman EA. The pleiotropic two-component regulatory system PhoP-PhoQ. J Bacteriol. (2001) 183:1835–42. doi: 10.1128/JB.183.6.1835-1842.2001
181. Park SY, Groisman EA. Signal-specific temporal response by the Salmonella PhoP/PhoQ regulatory system. Mol Microbiol. (2014) 91:135–44. doi: 10.1111/mmi.12449
182. Olaitan AO, Diene SM, Kempf M, Berrazeg M, Bakour S, Gupta SK, et al. Worldwide emergence of colistin resistance in Klebsiella pneumoniae from healthy humans and patients in Lao PDR, Thailand, Israel, Nigeria and France owing to inactivation of the PhoP/PhoQ regulator mgrB: an epidemiological and molecular study. Int J Antimicrob Agents. (2014) 44:500–7. doi: 10.1016/j.ijantimicag.2014.07.020
183. Wright MS, Suzuki Y, Jones MB, Marshall SH, Rudin SD, van Duin D, et al. Genomic and transcriptomic analyses of colistin-resistant clinical isolates of Klebsiella pneumoniae reveal multiple pathways of resistance. Antimicrob Agents Chemother. (2015) 59:536–43. doi: 10.1128/AAC.04037-14
184. Cannatelli A, Giani T, D'Andrea MM, Di Pilato V, Arena F, Conte V, et al. MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob Agents Chemother. (2014) 58:5696–703. doi: 10.1128/AAC.03110-14
185. Poirel L, Jayol A, Bontron S, Villegas MV, Ozdamar M, Türkoglu S, et al. The mgrB gene as a key target for acquired resistance to colistin in Klebsiella pneumoniae. J Antimicrob Chemother. (2015) 70:75–80. doi: 10.1093/jac/dku323
186. López-Camacho E, Gómez-Gil R, Tobes R, Manrique M, Lorenzo M, Galván B, et al. Genomic analysis of the emergence and evolution of multidrug resistance during a Klebsiella pneumoniae outbreak including carbapenem and colistin resistance. J Antimicrob Chemother. (2014) 69:632–6. doi: 10.1093/jac/dkt419
187. Jayol A, Poirel L, Villegas MV, Nordmann P. Modulation of mgrB gene expression as a source of colistin resistance in Klebsiella oxytoca. Int J Antimicrob Agents. (2015) 46:108–10. doi: 10.1016/j.ijantimicag.2015.02.015
188. Olaitan AO, Rolain JM. Interruption of mgrB in the mediation of colistin resistance in Klebsiella oxytoca. Int J Antimicrob Agents. (2015) 46:354–5. doi: 10.1016/j.ijantimicag.2015.06.003
189. Jayol A, Nordmann P, Desroches M, Decousser JW, Poirel L. Acquisition of broad-spectrum cephalosporin resistance leading to colistin resistance in Klebsiella pneumoniae. Antimicrob Agents Chemother. (2016) 60:3199–201. doi: 10.1128/AAC.00237-16
190. Zowawi HM, Forde BM, Alfaresi M, Alzarouni A, Farahat Y, Chong TM, et al. Stepwise evolution of pandrug-resistance in Klebsiella pneumoniae. Sci Rep. (2015) 5:15082. doi: 10.1038/srep15082
191. De Majumdar S, Yu J, Fookes M, McAteer SP, Llobet E, Spence SF, et al. Correction: elucidation of the RamA regulon in Klebsiella pneumoniae reveals a role in LPS regulation. PLoS Pathog. (2016) 12:e1005649. doi: 10.1371/journal.ppat.1005649
192. Saleh NM, Hesham MS, Amin MA, Samir Mohamed R. Acquisition of colistin resistance links cell membrane thickness alteration with a point mutation in the lpxD gene in Acinetobacter baumannii. Antibiotics. (2020) 9:164. doi: 10.3390/antibiotics9040164
193. Campos MA, Vargas MA, Regueiro V, Llompart CM, Albertí S, Bengoechea JA. Capsule polysaccharide mediates bacterial resistance to antimicrobial peptides. Infect Immun. (2004) 72:7107–14. doi: 10.1128/IAI.72.12.7107-7114.2004
194. Llobet E, Tomás JM, Bengoechea JA. Capsule polysaccharide is a bacterial decoy for antimicrobial peptides. Microbiology. (2008) 154:3877–86. doi: 10.1099/mic.0.2008/022301-0
195. Formosa C, Herold M, Vidaillac C, Duval RE, Dague E. Unravelling of a mechanism of resistance to colistin in Klebsiella pneumoniae using atomic force microscopy. J Antimicrob Chemother. (2015) 70:2261–70. doi: 10.1093/jac/dkv118
196. Hirakawa H, Nishino K, Hirata T, Yamaguchi A. Comprehensive studies of drug resistance mediated by overexpression of response regulators of two-component signal transduction systems in Escherichia coli. J Bacteriol. (2003) 185:1851–6. doi: 10.1128/JB.185.6.1851-1856.2003
197. Zhai YJ, Huang H, Liu J, Sun HR, He D, Pan YS, et al. CpxR overexpression increases the susceptibility of acrB and cpxR double-deleted Salmonella enterica serovar Typhimurium to colistin. J Antimicrob Chemother. (2018) 73:3016–24. doi: 10.1093/jac/dky320
198. Lacour S, Doublet P, Obadia B, Cozzone AJ, Grangeasse C. A novel role for protein-tyrosine kinase Etk from Escherichia coli K-12 related to polymyxin resistance. Res Microbiol. (2006) 157:637–41. doi: 10.1016/j.resmic.2006.01.003
199. Lacour S, Bechet E, Cozzone AJ, Mijakovic I, Grangeasse C. Tyrosine phosphorylation of the UDP-glucose dehydrogenase of Escherichia coli is at the crossroads of colanic acid synthesis and polymyxin resistance. PLoS ONE. (2008) 3:e3053. doi: 10.1371/journal.pone.0003053
200. Chambers JR, Sauer K. The MerR-like regulator BrlR impairs Pseudomonas aeruginosa biofilm tolerance to colistin by repressing PhoPQ. J Bacteriol. (2013) 195:4678–88. doi: 10.1128/JB.00834-13
201. Srinivasan VB, Rajamohan G. KpnEF, a new member of the Klebsiella pneumoniae cell envelope stress response regulon, is an SMR-type efflux pump involved in broad-spectrum antimicrobial resistance. Antimicrob Agents Chemother. (2013) 57:4449–62. doi: 10.1128/AAC.02284-12
202. Machado D, Antunes J, Simões A, Perdigão J, Couto I, McCusker M, et al. Contribution of efflux to colistin heteroresistance in a multidrug resistant Acinetobacter baumannii clinical isolate. J Med Microbiol. (2018) 67:740–9. doi: 10.1099/jmm.0.000741
203. Ni W, Li Y, Guan J, Zhao J, Cui J, Wang R, et al. effects of efflux pump inhibitors on colistin resistance in multidrug-resistant gram-negative bacteria. Antimicrob Agents Chemother. (2016) 60:AAC.00248-16. doi: 10.1128/AAC.00248-16
204. Parra-Lopez C, Baer MT, Groisman EA. Molecular genetic analysis of a locus required for resistance to antimicrobial peptides in Salmonella typhimurium. EMBO J. (1993) 12:4053–62. doi: 10.1002/j.1460-2075.1993.tb06089.x
205. Stoesser N, Mathers AJ, Moore CE, Day NP, Crook DW. Colistin resistance gene mcr-1 and pHNSHP45 plasmid in human isolates of Escherichia coli and Klebsiella pneumoniae. Lancet Infect Dis. (2016) 16:285–6. doi: 10.1016/S1473-3099(16)00010-4
206. Pham Thanh D, Thanh Tuyen H, Nguyen Thi Nguyen T, Chung The H, Wick RR, Thwaites GE. Inducible colistin resistance via a disrupted plasmid-borne mcr-1 gene in a 2008 Vietnamese Shigella sonnei isolate. J Antimicrob Chemother. (2016) 71:2314–7. doi: 10.1093/jac/dkw173
207. Campos J, Cristino L, Peixe L, Antunes P. MCR-1 in multidrug-resistant and copper-tolerant clinically relevant Salmonella 1,4,[5],12:i:- and S. Rissen clones in Portugal, 2011 to 2015. Euro Surveill. (2016) 21:30270. doi: 10.2807/1560-7917.ES.2016.21.26.30270
208. Webb HE, Granier SA, Marault M, Millemann Y, den Bakker HC, Nightingale KK, et al. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis. (2016) 16:144–5. doi: 10.1016/S1473-3099(15)00538-1
209. Zhao F, Zong Z. Kluyvera ascorbata strain from hospital sewage carrying the mcr-1 colistin resistance gene. Antimicrob Agents Chemother. (2016) 60:7498–501. doi: 10.1128/AAC.01165-16
210. Liu BT, Song FJ, Zou M, Hao ZH, Shan H. Emergence of colistin resistance gene mcr-1 in Cronobacter sakazakii producing NDM-9 and in Escherichia coli from the same animal. Antimicrob Agents Chemother. (2017) 61:e01444-16. doi: 10.1128/AAC.01444-16
211. Sennati S, Di Pilato V, Riccobono E, Di Maggio T, Villagran AL, Pallecchi L, et al. Citrobacter braakii carrying plasmid-borne mcr-1 colistin resistance gene from ready-to-eat food from a market in the Chaco region of Bolivia. J Antimicrob Chemother. (2017) 72:2127–9. doi: 10.1093/jac/dkx078
212. Trung NV, Matamoros S, Carrique-Mas JJ, Nghia NH, Nhung NT, Chieu TT, et al. Zoonotic transmission of mcr-1 colistin resistance gene from small-scale poultry farms, vietnam. Emerg Infect Dis. (2017) 23:529–32. doi: 10.3201/eid2303.161553
213. Fernandes MR, Sellera FP, Esposito F, Sabino CP, Cerdeira L, Lincopan N. Colistin-Resistant mcr-1-Positive Escherichia coli on public beaches, an infectious threat emerging in recreational waters. Antimicrob Agents Chemother. (2017) 61:e00234-17. doi: 10.1128/AAC.00234-17
214. Guenther S, Falgenhauer L, Semmler T, Imirzalioglu C, Chakraborty T, Roesler U, et al. Environmental emission of multiresistant Escherichia coli carrying the colistin resistance gene mcr-1 from German swine farms. J Antimicrob Chemother. (2017) 72:1289–92. doi: 10.1093/jac/dkw585
215. Monte DF, Mem A, Fernandes MR, Cerdeira L, Esposito F, Galvão JA, et al. Chicken meat as a reservoir of colistin-resistant Escherichia coli strains carrying mcr-1 genes in South America. Antimicrob Agents Chemother. (2017) 61:e02718-16. doi: 10.1128/AAC.02718-16
216. Wang Q, Sun J, Li J, Ding Y, Li XP, Lin J, et al. Expanding landscapes of the diversified mcr-1-bearing plasmid reservoirs. Microbiome. (2017) 5:70. doi: 10.1186/s40168-017-0288-0
217. Chiou CS, Chen YT, Wang YW, Liu YY, Kuo HC, Tu YH, et al. Dissemination of mcr-1-carrying plasmids among colistin-resistant salmonella strains from humans and food-producing animals in Taiwan. Antimicrob Agents Chemother. (2017) 61:e00338-17. doi: 10.1128/AAC.00338-17
218. Duggett NA, Sayers E, AbuOun M, Ellis RJ, Nunez-Garcia J, Randall L, et al. Occurrence and characterization of mcr-1-harbouring Escherichia coli isolated from pigs in Great Britain from 2013 to 2015. J Antimicrob Chemother. (2017) 72:691–5.
219. Zhang XF, Doi Y, Huang X, Li HY, Zhong LL, Zeng KJ, et al. Possible transmission of mcr-1-Harboring Escherichia coli between companion animals and human. Emerg Infect Dis. (2016) 22:1679–81. doi: 10.3201/eid2209.160464
220. Ye H, Li Y, Li Z, Gao R, Zhang H, Wen R, et al. Diversified mcr-1-harbouring plasmid reservoirs confer resistance to colistin in human gut microbiota. MBio. (2016) 7:e00177. doi: 10.1128/mBio.00177-16
221. Wang Y, Tian GB, Zhang R, Shen Y, Tyrrell JM, Huang X, et al. Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1-positive Enterobacteriaceae in patients and healthy adults from China: an epidemiological and clinical study. Lancet Infect Dis. (2017) 17:390–9. doi: 10.1016/S1473-3099(16)30527-8
222. Carattoli A, Villa L, Feudi C, Curcio L, Orsini S, Luppi A, et al. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill. (2017) 22:30589. doi: 10.2807/1560-7917.ES.2017.22.31.30589
223. AbuOun M, Stubberfield EJ, Duggett NA, Kirchner M, Dormer L, Nunez-Garcia J, et al. mcr-1 and mcr-2 variant genes identified in Moraxella species isolated from pigs in Great Britain from 2014 to 2015. J Antimicrob Chemother. (2017) 72:2745–9. doi: 10.1093/jac/dkx286
224. Daly SM, Sturge CR, Felder-Scott CF, Geller BL, Greenberg DE. MCR-1 inhibition with peptide-conjugated phosphorodiamidate morpholino oligomers restores sensitivity to polymyxin in Escherichia coli. MBio. (2017) 8:e01315-17. doi: 10.1128/mBio.01315-17
225. Fyfe C, LeBlanc G, Close B, Nordmann P, Dumas J, Grossman TH. Eravacycline is active against bacterial isolates expressing the polymyxin resistance gene mcr-1. Antimicrob Agents Chemother. (2016) 60:6989–90. doi: 10.1128/AAC.01646-16
226. Denervaud-Tendon V, Poirel L, Connolly LE, Krause KM, Nordmann P. Plazomicin activity against polymyxin-resistant Enterobacteriaceae, including MCR-1-producing isolates. J Antimicrob Chemother. (2017) 72:2787–91. doi: 10.1093/jac/dkx239
227. Schirmeier E, Zimmermann P, Hofmann V, Biebl M, Gerstmans H, Maervoet VET, et al. Inhibitory and bactericidal effect of Artilysin(®) Art-175 against colistin-resistant mcr-1-positive Escherichia coli isolates. Int J Antimicrob Agents. (2018) 51:528–9. doi: 10.1016/j.ijantimicag.2017.08.027
228. Zhou YF, Tao MT, Feng Y, Yang RS, Liao XP, Liu YH, et al. Increased activity of colistin in combination with amikacin against Escherichia coli co-producing NDM-5 and MCR-1. J Antimicrob Chemother. (2017) 72:1723–30. doi: 10.1093/jac/dkx038
229. Bulman ZP, Chen L, Walsh TJ, Satlin MJ, Qian Y, Bulitta JB, et al. Polymyxin combinations combat Escherichia coli harboring mcr-1 and bla(NDM-5): preparation for a Postantibiotic Era. MBio. (2017) 8:e00540-17. doi: 10.1128/mBio.00540-17
230. Brennan-Krohn T, Pironti A, Kirby JE. Synergistic activity of colistin-containing combinations against colistin-resistant Enterobacteriaceae. Antimicrob Agents Chemother. (2018) 62:e00873-18. doi: 10.1128/AAC.00873-18
231. Li Y, Lin X, Yao X, Huang Y, Liu W, Ma T, et al. Synergistic antimicrobial activity of colistin in combination with rifampin and azithromycin against Escherichia coli producing MCR-1. Antimicrob Agents Chemother. (2018) 62:e01631-18. doi: 10.1128/AAC.01631-18
232. MacNair CR, Stokes JM, Carfrae LA, Fiebig-Comyn AA, Coombes BK, Mulvey MR, et al. Overcoming mcr-1 mediated colistin resistance with colistin in combination with other antibiotics. Nat Commun. (2018) 9:458. doi: 10.1038/s41467-018-02875-z
233. Hu Y, Liu Y, Coates A. Azidothymidine produces synergistic activity in combination with colistin against antibiotic-resistant Enterobacteriaceae. Antimicrob Agents Chemother. (2019) 63:e01630-18. doi: 10.1128/AAC.01630-18
234. Zhou Y-F, Liu P, Zhang C-J, Liao X-P, Sun J, Liu Y-H. Colistin combined with tigecycline: a promising alternative strategy to combat Escherichia coli Harboring blaNDM−5 and mcr-1. Front Microbiol. (2020) 10:2957. doi: 10.3389/fmicb.2019.02957
235. Barker WT, Chandler CE, Melander RJ, Ernst RK, Melander C. Tryptamine derivatives disarm colistin resistance in polymyxin-resistant gram-negative bacteria. Bioorg Med Chem. (2019) 27:1776–88. doi: 10.1016/j.bmc.2019.03.019
236. Stokes JM, MacNair CR, Ilyas B, French S, Côté JP, Bouwman C, et al. Pentamidine sensitizes Gram-negative pathogens to antibiotics and overcomes acquired colistin resistance. Nat Microbiol. (2017) 2:17028. doi: 10.1038/nmicrobiol.2017.28
237. Huggins WM, Barker WT, Baker JT, Hahn NA, Melander RJ, Melander C. Meridianin D analogues display antibiofilm activity against MRSA and increase colistin efficacy in gram-negative bacteria. ACS Med Chem Lett. (2018) 9:702–7. doi: 10.1021/acsmedchemlett.8b00161
238. Cannatelli A, Principato S, Colavecchio OL, Pallecchi L, Rossolini GM. Synergistic activity of colistin in combination with resveratrol against colistin-resistant gram-negative pathogens. Front Microbiol. (2018) 9:1808. doi: 10.3389/fmicb.2018.01808
239. Zhou Y, Liu S, Wang T, Li H, Tang S, Wang J, et al. Pterostilbene, a potential MCR-1 inhibitor that enhances the efficacy of polymyxin B. Antimicrob Agents Chemother. (2018) 62:e02146-17. doi: 10.1128/AAC.02146-17
240. Zhou Y, Wang J, Guo Y, Liu X, Liu S, Niu X, et al. Discovery of a potential MCR-1 inhibitor that reverses polymyxin activity against clinical mcr-1-positive Enterobacteriaceae. J Infect. (2019) 78:364–72. doi: 10.1016/j.jinf.2019.03.004
241. Domalaon R, Silva PMD, Kumar A, Zhanel GG, Schweizer F. The Anthelmintic Drug Niclosamide Synergizes with Colistin and Reverses Colistin Resistance in Gram-Negative Bacilli. Antimicrob Agents Chemother. (2019) 63:e02574-18. doi: 10.1128/AAC.02574-18
242. Nezhadi J, Narenji H, Soroush Barhaghi MH, Rezaee MA, Ghotaslou R, Pirzadeh T, et al. Peptide nucleic acid-mediated re-sensitization of colistin resistance Escherichia coli KP81 harboring mcr-1 plasmid. Microb Pathog. (2019) 135:103646. doi: 10.1016/j.micpath.2019.103646
243. Dong H, Xiang H, Mu D, Wang D, Wang T. Exploiting a conjugative CRISPR/Cas9 system to eliminate plasmid harbouring the mcr-1 gene from Escherichia coli. Int J Antimicrob Agents. (2019) 53:1–8. doi: 10.1016/j.ijantimicag.2018.09.017
244. Lan XJ, Yan HT, Lin F, Hou S, Li CC, Wang GS, et al. Design, synthesis and biological evaluation of 1-Phenyl-2-(phenylamino) ethanone derivatives as Novel MCR-1 inhibitors. Molecules. (2019) 24:2719. doi: 10.3390/molecules24152719
245. Wei P, Song G, Shi M, Zhou Y, Liu Y, Lei J, et al. Substrate analog interaction with MCR-1 offers insight into the rising threat of the plasmid-mediated transferable colistin resistance. FASEB J. (2018) 32:1085–98. doi: 10.1096/fj.201700705R
246. Anandan A, Piek S, Kahler CM, Vrielink A. Cloning, expression, purification and crystallization of an endotoxin-biosynthesis enzyme from Neisseria meningitidis. Acta Crystallogr Sect F Struct Biol Cryst Commun. (2012) 68:1494–7. doi: 10.1107/S1744309112042236
247. Wanty C, Anandan A, Piek S, Walshe J, Ganguly J, Carlson RW, et al. The structure of the neisserial lipooligosaccharide phosphoethanolamine transferase A (LptA) required for resistance to polymyxin. J Mol Biol. (2013) 425:3389–402. doi: 10.1016/j.jmb.2013.06.029
248. Gao R, Hu Y, Li Z, Sun J, Wang Q, Lin J, et al. Dissemination and mechanism for the MCR-1 colistin resistance. PLoS Pathog. (2016) 12:e1005957. doi: 10.1371/journal.ppat.1005957
249. Munguia J, Nizet V. Pharmacological targeting of the host-pathogen interaction: alternatives to classical antibiotics to combat drug-resistant superbugs. Trends Pharmacol Sci. (2017) 38:473–88. doi: 10.1016/j.tips.2017.02.003
Keywords: chromosomal genes, mcr, electrostatic interaction, lipopolysaccharide, colistin resistance, Enterobacteriaceae
Citation: Gogry FA, Siddiqui MT, Sultan I and Haq QMR (2021) Current Update on Intrinsic and Acquired Colistin Resistance Mechanisms in Bacteria. Front. Med. 8:677720. doi: 10.3389/fmed.2021.677720
Received: 08 March 2021; Accepted: 09 July 2021;
Published: 12 August 2021.
Edited by:
Filipa Grosso, University of Porto, PortugalReviewed by:
Swaminath Srinivas, University of Illinois at Urbana-Champaign, United StatesGerald Larrouy-Maumus, Imperial College London, United Kingdom
Copyright © 2021 Gogry, Siddiqui, Sultan and Haq. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qazi Mohd. Rizwanul Haq, haqqmr@gmail.com; qhaque@jmi.ac.in
 Firdoos Ahmad Gogry
Firdoos Ahmad Gogry Mohammad Tahir Siddiqui
Mohammad Tahir Siddiqui Insha Sultan
Insha Sultan Qazi Mohd. Rizwanul Haq
Qazi Mohd. Rizwanul Haq