Concise Review: Functional Roles and Therapeutic Potentials of Long Non-coding RNAs in Cholangiopathies
- 1Division of Gastroenterology and Hepatology, Department of Medicine, Indiana University School of Medicine, Indianapolis, IN, United States
- 2Department of Medical Physiology, Texas A&M University, College of Medicine, Bryan, TX, United States
- 3Richard L. Roudebush VA Medical Center, Indianapolis, IN, United States
Long non-coding RNAs (lncRNAs) are RNAs with lengths exceeding 200 nucleotides that are not translated into proteins. It is well-known that small non-coding RNAs, such as microRNAs (miRNAs), regulate gene expression and play an important role in cholangiopathies. Recent studies have demonstrated that lncRNAs may also play a key role in the pathophysiology of cholangiopathies. Patients with cholangiopathies often develop cholangiocarcinoma (CCA), which is cholangiocyte-derived cancer, in the later stage. Cholangiocytes are a primary target of therapies for cholangiopathies and CCA development. Previous studies have demonstrated that expression levels of lncRNAs are altered in the liver of cholangiopathies or CCA tissues. Some lncRNAs regulate gene expression by inhibiting functions of miRNAs leading to diseased liver conditions or CCA progression, suggesting that lncRNAs could be a novel therapeutic target for those disorders. This review summarizes current understandings of functional roles of lncRNAs in cholangiopathies and seek their potentials for novel therapies.
Introduction
It has been well-known since early studies that the human genome contains very small percentage (~1%) of exons of protein-coding genes (1). Although ~5-10% of the human genome is transcribed into RNAs, the large portions of RNA sequences do not code functional proteins (2). In recent years, these non-coding RNAs have been classified according to their lengths and characteristics, and especially small non-coding RNAs called microRNAs (miRNAs) have been studied to understand the pathophysiology of human diseases (3). Altered expression levels of miRNAs are a hallmark in diseased conditions, and the regulation of gene expression by miRNAs plays a critical role in pathogenesis of various human disorders including liver diseases (4). miRNAs could be useful as biomarkers to diagnose liver diseases including liver fibrosis and cancer, and could be a novel therapeutic target to regulate specific gene expression as well as cell events (5). Long non-coding RNAs (lncRNAs) are another class of non-coding RNAs that are >200 bp long. While the major function of miRNAs is to target mRNAs and regulate their expressions, various functions of lncRNAs have been suggested including regulation of gene expression, X-chromosome inactivation, telomere regulation, and chromatin structure regulation (3). Although functions of large numbers of lncRNAs are undefined, they could play a key role in the pathophysiology of liver diseases as well as miRNAs.
Cholangiopathies include bile duct disorders, such as primary sclerosing cholangitis (PSC), primary biliary cholangitis (PBC), and biliary atresia, which are characterized by a syndrome of biliary obstruction resulting from infection-related inflammation or autoimmune responses (6–8). Numbers of miRNAs have been identified in patients with cholangiopathies representing their potentials as novel diagnostic biomarkers or therapeutic targets (9–11). Recent, studies have also demonstrated that lncRNAs may be associated with pathogenesis and diseased conditions during cholestatic liver injury and could be another therapeutic target for cholangiopathies. This review summarizes current understandings of functional roles of lncRNAs and their potentials as therapeutic targets in cholangiopathies.
Long Non-Coding RNAs IN Cholangiopathies
Cholestatic Liver Injury and Primary Sclerosing Cholangitis
MEG3
Previous studies suggested the association of lncRNA maternally expressed gene 3 (MEG3) with liver fibrosis and hepatocellular carcinoma (12, 13). Another study has demonstrated that MEG3 interacts with RNA-binding protein polypyrimidine tract-binding protein 1 (PTBP1), which binds to small heterodimer partner (SHP) (14). SHP is a key regulator for bile acid synthesis by regulating cytochrome P450 family 7 subfamily A member 1 (Cyp7a1) and cytochrome P450 family 8 subfamily B member 1 (Cyp8b1), which are enzymes for bile acid synthesis from cholesterol (15). The PTBP1-MEG3 complex destabilizes SHP mRNA leading to its degradation and elevated Cyp7a1 and Cyp8b1 expression. Overexpression of MEG3 induced SHP degradation and elevated bile acid synthesis resulting in cholestatic liver injury in mice (14). These findings suggest that MEG3 is associated with pathogenesis of cholestatic liver injury and could be a therapeutic target to manage bile acid homeostasis and improve liver conditions.
H19
Zhang et al. have demonstrated that B-cell lymphoma protein 2 (Bcl2) is a key regulator of bile acid homeostasis, and overexpression of Bcl2 increases serum levels of bile acids leading to cholestatic liver injury in mice (16). Overexpression of Bcl2 induced SHP protein degradation as well as upregulation of lncRNA H19 (16). This study has demonstrated that SHP is a transcriptional repressor of H19, and overexpression of SHP and knockdown of H19 attenuated Bcl2-induced cholestatic liver injury in vivo, suggesting the association of H19 with SHP expression and cholestatic liver diseases (16). Bile duct ligation (BDL) is a surgical obstruction of common bile duct performed in rodents, which is utilized as an animal model of cholestatic liver injury (17). Song et al. have demonstrated that H19 expression is elevated in the liver after BDL, and overexpression of H19 exacerbates BDL-induced liver damage and fibrosis in mice (18). H19 deficient mice represented attenuated liver damage and fibrosis compared to wild-type mice after BDL, indicating the association of expression levels of H19 and liver conditions during cholestatic liver injury (18). Multidrug resistance 2 knockout (Mdr2−/−) mice are the most common transgenic mice that are utilized as the animal model of human PSC (19). Mdr2−/− mice represent liver damage and fibrosis as well as elevated H19 expression in the liver, especially in female mice (20). Downregulation of H19 attenuated liver damage and fibrosis in Mdr2−/− mice, suggesting that H19 could be a therapeutic target for the management of liver conditions in PSC (20).
H19 Carried in Extracellular Vesicles
Exosomes and microparticles are extracellular vesicles (EVs) that are secreted from cells. Exosomes are small EVs (~100 nm in diameter) formed and secreted through the endosomal network, and microparticles (0.1–1 μm) are larger EVs formed by outward budding of the plasma membrane (21). These membrane-bound vesicles contain cargo mediators including DNAs, RNAs, and proteins, and secreted EVs from donor cells can be transferred into recipient cells delivering those cargo mediators (22, 23). This EV-mediated cell-to-cell communication followed by the regulation of cellular events plays a key role in the pathophysiology of liver diseases. A previous study has demonstrated that expression levels of H19 are elevated in the liver of PSC patients as well as in the mouse livers after carbon tetrachloride (CCl4)-induced liver damage, and CCl4 administration also increases levels of H19 carried in EVs isolated from mouse serum (24). H19-enriched cholangiocyte-derived EVs decreased SHP expression in hepatocytes, and injection of serum EVs isolated from Mdr2−/− mice increased bile acid synthesis and exacerbated liver conditions in other Mdr2−/− mice, suggesting EV-mediated cell-to-cell communication via cargo H19 (24). Since patients with liver cirrhosis have serum EVs carrying elevated levels of H19 compared to those from healthy individuals, H19-carrying EVs may play a critical role in the pathogenesis of cholestatic liver diseases and liver cirrhosis (24). Another study has demonstrated the correlation between expression levels of H19 and fibrogenic markers including collagen I and alpha smooth muscle actin (αSMA) in patients with PSC and PBC as well as in BDL and Mdr2−/− mouse models (25). H19-enriched cholangiocyte-derived EVs induced proliferation and activation of hepatic stellate cells (HSCs) leading to fibrogenesis and cholestatic liver fibrosis in vivo (25). These studies suggest that EVs and cargo H19 delivery from cholangiocytes to other liver cells such as hepatocytes and HSCs are a critical step for pathogenesis of cholestatic liver injury, and H19 could be another therapeutic target for the treatment of liver fibrosis.
Primary Biliary Cholangitis
PBC is an autoimmune disorder which is characterized by bile duct obstruction and cholestasis caused by intrahepatic bile duct destruction and inflammation (26). The cause of autoimmunity against bile ducts and cholangiocytes is still unknown. Therefore, previous studies have performed genotyping and association studies to identify susceptible loci or genes. A previous study has performed fine-mapping and association studies using a cohort of 2,861 cases and have identified three candidate loci that are associated with PBC (27). Hrdlichova et al. have extracted RNAs from seven immune cell types (granulocytes, monocytes, NK cells, B cells, memory T cells, naïve CD4+ and naïve CD8+ T cells) to obtain RNA sequencing libraries for patients with autoimmune disorders including PBC (28). This study has demonstrated that various lncRNAs expressed in immune cells are shared between autoimmune disorders, and NK cells, memory T cells and CD8+ cells in PBC patients have enriched those shared lncRNAs (28). Although this study suggests that lncRNAs may contribute to autoimmunity and pathogenesis of PBC, current studies are limited and detailed mechanisms and functional roles of lncRNAs in PBC are largely unknown.
Biliary Atresia
Biliary atresia is a progressive bile duct disorder in infants representing cholestasis, jaundice, and liver fibrosis (29). Although previous studies has suggested the association between perinatal viral infection and biliary atresia development in infants, detailed mechanisms of pathogenesis in biliary atresia are still undefined (30). Chen et al. have performed genome-wide association study using a cohort of 343 non-related biliary atresia patients and 1,716 healthy controls to identify susceptible loci to biliary atresia (31). This study identified numbers of candidate loci, and one of significant SNPs was located in the gene ADD3-AS1, which encodes an lncRNA (31). Pseudogenes are DNA sequences that are related to genes but do not encode fully functional proteins. Therefore, transcripts of pseudogenes are recognized as lncRNAs. Pseudogenes and pseudogene-derived lncRNAs can be functional by regulating gene expression and could be a therapeutic target (32, 33). Annexin A2 (ANXA2) pseudogene 3 (ANXA2P3) is a pseudogene related to ANXA2. Previous studies have demonstrated that upregulation of ANXA2 is associated with liver fibrosis and can be useful as a biomarker for hepatitis B virus-related liver fibrosis (34, 35). Expression levels of ANXA2 as well as ANXA2P3 are also upregulated in liver tissues of biliary atresia patients, indicating that ANXA2P3 may be involved in the pathophysiology of biliary atresia development (36). As mentioned previously, lncRNA H19 is upregulated in the liver of patients with PSC and mouse models of PSC (24, 25). Another study analyzed H19 expression levels in biliary atresia patients and found that H19 was upregulated in the liver of biliary atresia patients compared to healthy individuals, and the expression of H19 was correlated with the expression of fibrogenic markers αSMA and transforming growth factor beta 1 (TGF-β1) (37). This study has demonstrated that H19 regulates functions of miRNAs let-7 families by binding them leading to elevated expression of the target of let-7, high-mobility group AT-hook 2 (HMGA2) (37). Decreased levels of let-7 are associated with ductular reaction and liver fibrosis during cholestatic liver injury (38). These studies suggest that lncRNAs are associated with PBC and biliary atresia although further studies are required to elucidate detailed mechanisms.
Cholangiocarcinoma
lncRNAs as Competing Endogenous RNAs in CCA
Cholangiocarcinoma (CCA) is a cancer that is derived from the biliary tree, and patients with PSC have a high risk for the development of CCA (39). Functions of lncRNAs have attracted interests in recent CCA studies because accumulating evidence suggests that lncRNAs may play a key role in cancer development, proliferation, and invasion of CCA. H19 binds to let-7 families and inhibit their functions like an let-7 sponge, as mentioned (37). lncRNAs function as competing endogenous RNAs (ceRNAs), which interrupt miRNA functions and alter protein expression, and this may be a characteristic hallmark in CCA. Genome-wide data analysis or RNA-Seq profiling identified various lncRNAs and ceRNA networks associated with CCA, and some candidate lncRNAs are significantly associated with survival rates (40–42). Recent studies have identified a number of lncRNAs that are associated with CCA progression and invasion. This review introduces selected studies of lncRNAs in CCA especially from recent studies. For other lncRNAs in CCA, see previous schematic reviews (43, 44).
Functional Roles of lncRNAs in CCA
Previous studies have demonstrated that expression levels of lncRNA H19 are elevated in PSC and biliary atresia patients as described previously (24, 37). A study using tissue samples from patients with perihilar, distal, or intrahepatic CCA (iCCA) has represented that H19 expression is upregulated in CCA tissues compared to corresponding non-tumor tissues, and expression levels of H19 are associated with poor survival rates of patients (45). This study also demonstrated that H19 induced cell proliferation and migration in CCA cell lines RBE and QBC939 cells (45). Wang et al. analyzed lncRNA profiles expressed in CCA cell lines, RBE, QBC939, and SK-cha-1 cells, and found that lncRNAs H19 and HULC were upregulated during hydrogen peroxidase-induced oxidative stress (46). This study has demonstrated that H19 disrupts functions of let-7a and let-7b, which inhibit interleukin-6 (IL-6) expression as a target, and HULK interferes miR-372 and miR-373 that target C-X-C motif chemokine receptor 4 (CXCR4) (46). Since IL-6 and CXCR4 are associated with proliferation, migration, and metastasis of CCA (47–49), upregulation of H19 and HULC may lead to aberrant expression of IL-6 and CXCR4 as well as poor survival rates of CCA patients although further studies are required (46). Microarray analysis for lncRNAs using samples of fifty two CCA patients has identified five candidate lncRNAs that are significantly upregulated in iCCA tissues compared to adjacent non-tumorous tissues (50). Expression levels of one of those candidate lncRNAs, SNHG3, represented correlation with TNM stages, and patients with high SNHG3 expression had lower survival rates compared with patients with low SNHG3 expression (50). Another study using sixty CCA patients (intrahepatic, extrahepatic, and perihilar) has identified lnc-PKD2-2-3 as a candidate lncRNA, and high lnc-PKD2-2-3 expression was correlated with poor survival rates and high TNM stages (51). Although functional roles and targets of SNHG3 and lnc-PKD2-2-3 are undefined, these studies indicate the correlation between lncRNAs and CCA prognosis, and these lncRNAs could be utilized as a diagnostic biomarker for CCA. Epithelial-mesenchymal transition (EMT) is a process that epithelial cells adopt structural and functional characteristics of mesenchymal cells and is an important phenomenon in carcinogenesis and metastases in cancers including CCA (52, 53). Previous report have demonstrated that lncRNA-NEF and runt-related transcription factor 1 (RUNX1) are associated with EMT in cancer (54, 55). Liang et al. analyzed expression levels of lncRNA-NEF and RUNX1 in 56 iCCA patients and 42 healthy individuals and found that lncRNA-NEF was downregulated and RUNX1 was upregulated in iCCA tissues (56). This study has demonstrated that low expression levels of lncRNA-NEF are associated with poor survival rates, and lncRNA-NEF expression is negatively correlated with RUNX1 expression in iCCA patients (56). FENDRR is a lncRNA, which is downregulated in various cancers such as breast cancer, prostate cancer, and hepatocellular carcinoma (57–59). A study using 60 CCA patients has found that expression of FENDRR is downregulated in CCA tissues compared to non-cancerous tissues, and FENDRR expression is negatively correlated with expression of survivin (60). Survivin is a protein that inhibits apoptosis and upregulated in cancers (61, 62). FENDRR repressed proliferation, migration, and invasion of CCA cell lines HuCCT1 and QBC939 cells via regulation of survivin (60). An in vitro study using CCA cell lines (HuCCT1, Huh-28, KKU-214, and RBE) has demonstrated that CCA cells express elevated levels of lncRNA LINC01061 (63). LINC01061 binds to miR-612 and inhibits functions of miR-612, which targets semaphoring-4D (SEMA4D) (63). Since SEMA4D promotes invasion and metastasis of cancers (64, 65), this study indicates that LINC01061 functions as ceRNA for SEMA4D by sponging miR-612 leading to cell proliferation and migration of CCA cell lines (63). These studies suggest that expression levels of lncRNAs are associated with cell proliferation, migration, and invasion of CCA, and lncRNAs play an important role in physiological events of CCA cells by regulating protein expression. Table S1 summarizes lncRNAs identified in cholangiopathies and CCA.
Candidate Therapeutic Approaches FOR lncRNAs
Current studies represent the association of lncRNAs with cholangiopathies and abnormal liver functions, such as excess bile acid synthesis and liver fibrosis as well as CCA characteristics, such as CCA cell migration and invasion, metastasis, or prognosis. These findings suggest that lncRNAs could be a novel therapeutic target to manage disease conditions in cholangiopathies.
RNA Interference Targeting lncRNA
The majority of lncRNAs associated with cholangiopathies is upregulated in the diseased liver. RNA interference technology using shRNA or siRNA can be utilized to manage liver conditions. For example, shRNA targeting LINC01061 decreased cell proliferation and increased apoptosis in CCA cell lines KKU-214 and RBE cells (63). Antisense oligonucleotides that inhibit lncRNA functions or induce lncRNA degradation by RNaseH can be utilized for lncRNA silencing. Treatments of antisense oligonucleotides for lncRNA MALAT1 decrease tumor volumes and metastases in the mouse model of lung cancer (66). Gene knockout targeting lncRNAs is another approach for cholangiopathies. H19 is upregulated during cholestatic liver injury, and H19−/− mice represent attenuated liver fibrosis during BDL compared to wild-type mice (25). Previous studies have introduced a technique for lncRNA silencing using zinc finger nucleases to induce lncRNA destabilization and degradation leading to 1,000-fold decreased expression of MALAT1 (66, 67). However, current studies are limited for cholangiopathies and CCA, and the majority of current studies using RNA interference is based on in vitro experiments. Further studies are required to establish the methodology for effective lncRNA silencing in vivo.
Induction of lncRNA Expression
Some lncRNAs could be therapeutic or protective against liver diseases or cancer. For example, expression levels of lncRNA-NEF and FENDRR are downregulated in CCA tissues compared to normal tissues (54, 60). Overexpression of these lncRNAs inhibited cell migration and invasion of CCA cell lines HuCCT1, QBC939, or TFK-1 cells, indicating the potentials of lncRNA induction as another therapeutic approach for CCA (54, 60). As well as lncRNA silencing, lncRNA induction has same limitations: (i) Current studies are limited in the use of in vitro cultured CCA cell lines; and (ii) Technical difficulties to induce specific lncRNAs expression in specific cell types such as CCA cells. Gene therapy using a plasmid encoding the target gene has been performed for breast cancer (68), and the methodology could be modified to target therapeutic/protective lncRNAs in cholangiopathies although further studies are needed to seek their potentials.
Small Molecule Inhibitors
Functions of lncRNAs could be impaired by small molecules. For example, some lncRNAs function as ceRNA by sponging miRNAs and regulating protein expression. Administration of small molecules that bind to the region for miRNA sponging may inhibit interaction between miRNAs and lncRNAs leading to effective inhibition of the target protein expression by miRNAs. Some lncRNAs interact with proteins to form a complex, and this lncRNA-protein complex function as an inhibitor that suppresses expression of the specific proteins. For example, lncRNA MEG3 interacts with PTBP1 to form a complex. This PTBP1-MEG3 complex binds to and destabilizes mRNA of SHP leading its degradation followed by elevated bile acid synthesis and cholestatic liver injury (14). Small molecules that interfere RNA-protein interaction between MEG3 and PTBP1 may have therapeutic effects for cholestatic liver injury induced by downregulated SHP and aberrant bile acid synthesis. Small molecules that bind to the specific region of lncRNAs and inhibit its correct folding could be utilized to induce lncRNA degradation and functional inhibition. Although these ideas may be theoretically possible, studies are still ongoing and no candidate molecules for cholangiopathies to be utilized for clinical trials are available to date.
Targeting or Utilization of EVs
Recent studies have demonstrated that EVs play a key role in cholangiopathies. H19 is upregulated in PSC patients, and cholangiocyte-derived EVs transfer cargo H19 to hepatocytes or HSCs in diseased conditions leading to bile acid synthesis or fibrogenesis, respectively (24, 25). Drugs that decrease EV production or secretion may inhibit fibrogenic cell-to-cell communication via H19-enriched EVs in PSC. High throughput screen assay has identified compounds that modulate EV biogenesis or release in prostate cancer cells (69). These compounds could also be effective on EV production or secretion in cholangiocytes or CCA cells leading to improved liver conditions although further studies are required.
EVs functions as a disease-inducing mediator carrier during cholestatic liver injury by delivering H19 from cholangiocytes to other liver cells (24, 25). This means that EVs could be utilized as a drug or therapeutic mediator carrier to manage liver conditions. A recent study has demonstrated that injection of EVs isolated from liver stem cells attenuates ductular reaction and liver fibrosis in Mdr2−/− mice via delivering cargo let-7, indicating the potentials of EVs as a therapeutic tool and an miRNA carrier (19). Injection of EVs carrying mediators, such as small molecules or nucleotides targeting lncRNAs could be performed to regulate lncRNA functions in vivo and manage liver conditions. EVs carrying candidate mediators such as miRNAs can be produced by cell transfection (70), and previous studies have also reported that modification of EV cargo mediators for miRNAs or miRNA inhibitors can be accomplished by electroporation (71, 72). Although further studies are required, these studies indicate that the methodology could be modified for lncRNAs or mediators targeting lncRNAs that are carried in EVs, and lncRNAs-targeting EVs could be useful to manage liver conditions and cancer progression.
Conclusion
Current studies have demonstrated that expression levels of lncRNAs are associated with diseased conditions of cholestatic liver diseases and CCA. lncRNAs function as ceRNAs by sponging miRNAs to regulate protein expression. Although there are various approaches available that are theoretically possible to regulate functions of lncRNAs leading to the management of cholangiopathies, further studies are required to understand detailed mechanisms of functions of lncRNAs and to develop the methodology for a novel therapy targeting lncRNAs. Figure 1 represents a diagram for the roles of lncRNAs in liver diseases.
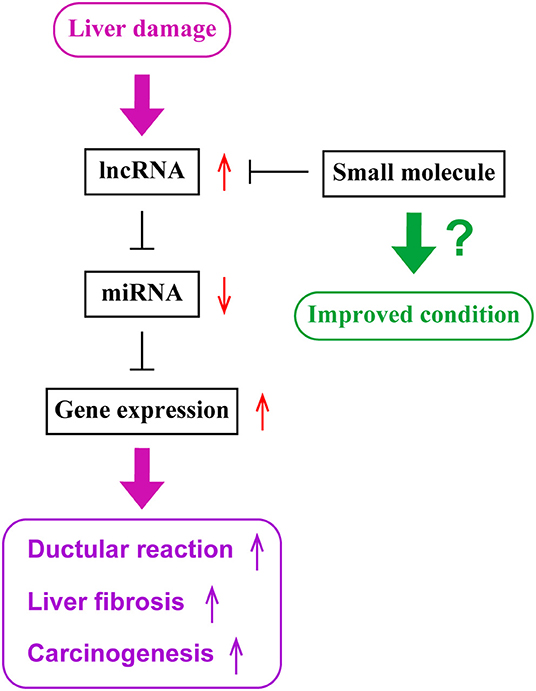
Figure 1. The role of lncRNAs in liver diseases. During liver damage, expression levels of long non-coding RNAs (lncRNAs), such as H19, are elevated in the liver. These lncRNAs sponge microRNAs (miRNAs), such as let-7 families, and inhibit their functions. Since miRNAs inhibit the expression of target genes, such as HMGA2, elevated levels of lncRNAs lead to enhanced gene expressions of target genes. Elevated gene expression is associated with ductular reaction, liver fibrogenesis and inflammation, or carcinogenesis or tumor progression. Small molecules targeting lncRNAs may be utilized as novel therapeutic tools to inhibit lncRNA functions and maintain liver homeostasis.
Author Contributions
KS designed the study and wrote the manuscript. SG and HF critically reviewed the manuscript. GA conducted and designed the project, and critically reviewed the manuscript.
Funding
This work was supported by the Senior Research Career Scientist Award to GA and the VA Merit awards to SG (5I01BX002192), HF (1I01BX003031), and GA (5I01BX000574) from the United States Department of Veteran's Affairs Biomedical Laboratory Research and Development Service, U.S. National Institutes of Health (NIH) National Institute of Diabetes and Digestive and Kidney Diseases Grants DK108959, DK119421, DK115184, DK054811, DK076898, DK107310, DK110035, and DK062975 to SG, HF, and GA, and NIH National Institute on Alcohol Abuse and Alcoholism Grants AA025997 and AA025157 to SG and GA, The Hickam Endowed Chair, Division of Gastroenterology and Hepatology, Department of Medicine, Indiana University School of Medicine. The project described was supported by the Indiana University Health - Indiana University School of Medicine Strategic Research Initiative. GA acknowledged the support from PSC Partners Seeking a Cure. This material is the result of work supported by resources at the Central Texas Veterans Health Care System, Temple, TX, Richard L. Roudebush VA Medical Center, Indianapolis, IN, and Medical Physiology, Medical Research Building, Temple, TX.
Disclaimer
The views expressed in this article are those of the authors and do not necessarily represent the views of the Department of Veterans Affairs.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2020.00048/full#supplementary-material
Abbreviations
ANXA2, Annexin A2; ANXA2P3, Annexin A2 pseudogene 3; αSMA, alpha smooth muscle actin; Bcl2, B-cell lymphoma protein 2; BDL, bile duct ligation; CCl4, carbon tetrachloride; ceRNAs, competing endogenous RNAs; Cyp7a1, cytochrome P450 family 7 subfamily A member 1; Cyp8b1, cytochrome P450 family 8 subfamily B member 1; CXCR4, C-X-C motif chemokine receptor 4; EMT, epithelial-mesenchymal transition; HMGA2, high-mobility group AT-hook 2; HSCs, hepatic stellate cells; iCCA, intrahepatic CCA; IL-6, interleukin-6; long non-coding RNAs, lncRNAs; Mdr2, multidrug resistance 2; MEG3, maternally expressed gene 3; miRNAs, microRNAs; PBC, primary biliary cholangitis; PSC, primary sclerosing cholangitis; PTB1, polypyrimidine tract-binding protein 1; RUNX1, runt-related transcription factor 1; SHP, small heterodimer partner; SEMA4D, semaphoring-4D; TGF-β1, transforming growth factor beta 1.
References
1. Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. (2007) 316:1484–8. doi: 10.1126/science.1138341
2. DiStefano JK. The emerging role of long noncoding RNAs in human disease. Methods Mol Biol. (2018) 1706:91–110. doi: 10.1007/978-1-4939-7471-9_6
3. Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. (2011) 12:861–74. doi: 10.1038/nrg3074
4. Szabo G, Bala S. MicroRNAs in liver disease. Nat Rev Gastroenterol Hepatol. (2013) 10:542–52. doi: 10.1038/nrgastro.2013.87
5. Roderburg C, Luedde T. Circulating microRNAs as markers of liver inflammation, fibrosis and cancer. J Hepatol. (2014) 61:1434–7. doi: 10.1016/j.jhep.2014.07.017
6. Dyson JK, Beuers U, Jones DEJ, Lohse AW, Hudson M. Primary sclerosing cholangitis. Lancet. (2018) 391:2547–59. doi: 10.1016/S0140-6736(18)30300-3
7. Lleo A, Marzorati S, Anaya JM, Gershwin ME. Primary biliary cholangitis: a comprehensive overview. Hepatol Int. (2017) 11:485–99. doi: 10.1007/s12072-017-9830-1
8. Lakshminarayanan B, Davenport M. Biliary atresia:a comprehensive review. J Autoimmun. (2016) 73:1–9. doi: 10.1016/j.jaut.2016.06.005
9. Munoz-Garrido P, Garcia-Fernandez de Barrena M, Hijona E, Carracedo M, Marin JJ, Bujanda L, et al. MicroRNAs in biliary diseases. World J Gastroenterol. (2012) 18:6189–96. doi: 10.3748/wjg.v18.i43.6189
10. O'Hara SP, Gradilone SA, Masyuk TV, Tabibian JH, LaRusso NF. MicroRNAs in cholangiopathies. Curr Pathobiol Rep. (2014) 2:133–42. doi: 10.1007/s40139-014-0048-9
11. Pisarello MJ, Loarca L, Ivanics T, Morton L, LaRusso N. MicroRNAs in the cholangiopathies: pathogenesis, diagnosis, and treatment. J Clin Med. (2015) 4:1688–712. doi: 10.3390/jcm4091688
12. He Y, Wu YT, Huang C, Meng XM, Ma TT, Wu BM, et al. Inhibitory effects of long noncoding RNA MEG3 on hepatic stellate cells activation and liver fibrogenesis. Biochim Biophys Acta. (2014) 1842:2204–15. doi: 10.1016/j.bbadis.2014.08.015
13. Anwar SL, Krech T, Hasemeier B, Schipper E, Schweitzer N, Vogel A, et al. Loss of imprinting and allelic switching at the DLK1-MEG3 locus in human hepatocellular carcinoma. PLoS ONE. (2012) 7:e49462. doi: 10.1371/journal.pone.0049462
14. Zhang L, Yang Z, Trottier J, Barbier O, Wang L. Long noncoding RNA MEG3 induces cholestatic liver injury by interaction with PTBP1 to facilitate shp mRNA decay. Hepatology. (2017) 65:604–15. doi: 10.1002/hep.28882
15. Chiang JY. Bile acids: regulation of synthesis. J Lipid Res. (2009) 50:1955–66. doi: 10.1194/jlr.R900010-JLR200
16. Zhang Y, Liu C, Barbier O, Smalling R, Tsuchiya H, Lee S, et al. Bcl2 is a critical regulator of bile acid homeostasis by dictating Shp and lncRNA H19 function. Sci Rep. (2016) 6:20559. doi: 10.1038/srep20559
17. Wan Y, Ceci L, Wu N, Zhou T, Chen L, Venter J, et al. Knockout of α-calcitonin gene-related peptide attenuates cholestatic liver injury by differentially regulating cellular senescence of hepatic stellate cells and cholangiocytes. Lab Invest. (2019) 99:764–76. doi: 10.1038/s41374-018-0178-5
18. Song Y, Liu C, Liu X, Trottier J, Beaudoin M, Zhang L, et al. H19 promotes cholestatic liver fibrosis by preventing ZEB1-mediated inhibition of epithelial cell adhesion molecule. Hepatology. (2017) 66:1183–96. doi: 10.1002/hep.29209
19. McDaniel K, Wu N, Zhou T, Huang L, Sato K, Venter J, et al. Amelioration of ductular reaction by stem cell derived extracellular vesicles in MDR2 knockout mice via lethal-7 microRNA. Hepatology. (2019) 69:2562–78. doi: 10.1002/hep.30542
20. Li X, Liu R, Yang J, Sun L, Zhang L, Jiang Z, et al. The role of long noncoding RNA H19 in gender disparity of cholestatic liver injury in multidrug resistance 2 gene knockout mice. Hepatology. (2017) 66:869–84. doi: 10.1002/hep.29145
21. Yanez-Mo M, Siljander PR, Andreu Z, Zavec AB, Borras FE, Buzas EI, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. (2015) 4:27066. doi: 10.3402/jev.v4.27066
22. Sato K, Meng F, Glaser S, Alpini G. Exosomes in liver pathology. J Hepatol. (2016) 65:213–21. doi: 10.1016/j.jhep.2016.03.004
23. Hirsova P, Ibrahim SH, Verma VK, Morton LA, Shah VH, LaRusso NF, et al. Extracellular vesicles in liver pathobiology: small particles with big impact. Hepatology. (2016) 64:2219–33. doi: 10.1002/hep.28814
24. Li X, Liu R, Huang Z, Gurley EC, Wang X, Wang J, et al. Cholangiocyte-derived exosomal long noncoding RNA H19 promotes cholestatic liver injury in mouse and humans. Hepatology. (2018) 68:599–615. doi: 10.1002/hep.29838
25. Liu R, Li X, Zhu W, Wang Y, Zhao D, Wang X, et al. Cholangiocyte-derived exosomal long noncoding RNA H19 promotes hepatic stellate cell activation and cholestatic liver fibrosis. Hepatology. (2019) 70:1317–35. doi: 10.1002/hep.30662
26. Reshetnyak VI. Primary biliary cirrhosis: clinical and laboratory criteria for its diagnosis. World J Gastroenterol. (2015) 21:7683–708. doi: 10.3748/wjg.v21.i25.7683
27. Liu JZ, Almarri MA, Gaffney DJ, Mells GF, Jostins L, Cordell HJ, et al. Dense fine-mapping study identifies new susceptibility loci for primary biliary cirrhosis. Nat Genet. (2012) 44:1137–41. doi: 10.1038/ng.2395
28. Hrdlickova B, Kumar V, Kanduri K, Zhernakova DV, Tripathi S, Karjalainen J, et al. Expression profiles of long non-coding RNAs located in autoimmune disease-associated regions reveal immune cell-type specificity. Genome Med. (2014) 6:88. doi: 10.1186/s13073-014-0088-0
29. Nizery L, Chardot C, Sissaoui S, Capito C, Henrion-Caude A, Debray D, et al. Biliary atresia: clinical advances and perspectives. Clin Res Hepatol Gastroenterol. (2016) 40:281–7. doi: 10.1016/j.clinre.2015.11.010
30. Averbukh LD, Wu GY. Evidence for viral induction of biliary atresia: a review. J Clin Transl Hepatol. (2018) 6:410–9. doi: 10.14218/JCTH.2018.00046
31. Chen Y, Gilbert MA, Grochowski CM, McEldrew D, Llewellyn J, Waisbourd-Zinman O, et al. A genome-wide association study identifies a susceptibility locus for biliary atresia on 2p16.1 within the gene EFEMP1. PLoS Genet. (2018) 14:e1007532. doi: 10.1371/journal.pgen.1007532
32. Milligan MJ, Lipovich L. Pseudogene-derived lncRNAs: emerging regulators of gene expression. Front Genet. (2014) 5:476. doi: 10.3389/fgene.2014.00476
33. Roberts TC, Morris KV. Not so pseudo anymore: pseudogenes as therapeutic targets. Pharmacogenomics. (2013) 14:2023–34. doi: 10.2217/pgs.13.172
34. Kolgelier S, Demir NA, Inkaya AC, Sumer S, Ozcimen S, Demir LS, et al. Serum levels of annexin A2 as a candidate biomarker for hepatic fibrosis in patients with chronic hepatitis B. Hepat Mon. (2015) 15:e30655. doi: 10.5812/hepatmon.30655
35. Yang M, Wang C, Li S, Xv X, She S, Ran X, et al. Annexin A2 promotes liver fibrosis by mediating von willebrand factor secretion. Dig Liver Dis. (2017) 49:780–8. doi: 10.1016/j.dld.2017.02.013
36. Nuerzhati Y, Dong R, Song Z, Zheng S. Role of the long noncoding RNA: annexin A2 pseudogene 3/annexin A2 signaling pathway in biliary atresia-associated hepatic injury. Int J Mol Med. (2019) 43:739–48. doi: 10.3892/ijmm.2018.4023
37. Xiao Y, Liu R, Li X, Gurley EC, Hylemon PB, Lu Y, et al. Long noncoding RNA H19 contributes to cholangiocyte proliferation and cholestatic liver fibrosis in biliary atresia. Hepatology. (2019) 70:1658–73. doi: 10.1002/hep.30698
38. Glaser S, Meng F, Han Y, Onori P, Chow BK, Francis H, et al. Secretin stimulates biliary cell proliferation by regulating expression of microRNA 125b and microRNA let7a in mice. Gastroenterology. (2014) 146:1795–808. doi: 10.1053/j.gastro.2014.02.030
39. Taghavi SA, Eshraghian A, Niknam R, Sivandzadeh GR, Bagheri Lankarani K. Diagnosis of cholangiocarcinoma in primary sclerosing cholangitis. Expert Rev Gastroenterol Hepatol. (2018) 12:575–84. doi: 10.1080/17474124.2018.1473761
40. Li G, Liu T, Zhang B, Chen W, Ding Z. Genome-wide identification of a competing endogenous RNA network in cholangiocarcinoma. J Cell Biochem. (2019) 120:18995–9003. doi: 10.1002/jcb.29222
41. Song W, Miao DL, Chen L. Comprehensive analysis of long noncoding RNA-associated competing endogenous RNA network in cholangiocarcinoma. Biochem Biophys Res Commun. (2018) 506:1004–12. doi: 10.1016/j.bbrc.2018.10.186
42. Zhang C, Ge C. A simple competing endogenous rna network identifies novel mRNA, miRNA, and lncRNA markers in human cholangiocarcinoma. Biomed Res Int. (2019) 2019:3526407. doi: 10.1155/2019/3526407
43. Dai K, Quan J, Yan F, Jin X, Pan X, Song X, et al. lncRNAs as potential molecular biomarkers in the clinicopathology and prognosis of cholangiocarcinoma: a systematic review and meta-analysis. Onco Targets Ther. (2019) 12:1905–15. doi: 10.2147/OTT.S188134
44. Jiang F, Ling X. The advancement of long non-coding RNAs in cholangiocarcinoma development. J Cancer. (2019) 10:2407–14. doi: 10.7150/jca.32411
45. Xu Y, Wang Z, Jiang X, Cui Y. Overexpression of long noncoding RNA H19 indicates a poor prognosis for cholangiocarcinoma and promotes cell migration and invasion by affecting epithelial-mesenchymal transition. Biomed Pharmacother. (2017) 92:17–23. doi: 10.1016/j.biopha.2017.05.061
46. Wang WT, Ye H, Wei PP, Han BW, He B, Chen ZH, et al. LncRNAs H19 and HULC, activated by oxidative stress, promote cell migration and invasion in cholangiocarcinoma through a ceRNA manner. J Hematol Oncol. (2016) 9:117. doi: 10.1186/s13045-016-0348-0
47. Wehbe H, Henson R, Meng F, Mize-Berge J, Patel T. Interleukin-6 contributes to growth in cholangiocarcinoma cells by aberrant promoter methylation and gene expression. Cancer Res. (2006) 66:10517–24. doi: 10.1158/0008-5472.CAN-06-2130
48. Zhao S, Wang J, Qin C. Blockade of CXCL12/CXCR4 signaling inhibits intrahepatic cholangiocarcinoma progression and metastasis via inactivation of canonical wnt pathway. J Exp Clin Cancer Res. (2014) 33:103. doi: 10.1186/s13046-014-0103-8
49. Kaemmerer D, Schindler R, Mussbach F, Dahmen U, Altendorf-Hofmann A, Dirsch O, et al. Somatostatin and CXCR4 chemokine receptor expression in hepatocellular and cholangiocellular carcinomas: tumor capillaries as promising targets. BMC Cancer. (2017) 17:896. doi: 10.1186/s12885-017-3911-3
50. Tian DG, Wei XP, Zhu H, Zhu L, Li TH, Li W. LncRNA-SNHG3 is an independent prognostic biomarker of intrahepatic cholangiocarcinoma. Int J Clin Exp Pathol. (2019) 12:2706–12.
51. Qiu G, Ma D, Li F, Sun D, Zeng Z. lnc-PKD2–2-3, identified by long non-coding RNA expression profiling, is associated with pejorative tumor features and poor prognosis, enhances cancer stemness and may serve as cancer stem-cell marker in cholangiocarcinoma. Int J Oncol. (2019) 55:45–58. doi: 10.3892/ijo.2019.4798
52. Brabletz T, Kalluri R, Nieto MA, Weinberg RA. EMT in cancer. Nat Rev Cancer. (2018) 18:128–34. doi: 10.1038/nrc.2017.118
53. Vaquero J, Guedj N, Claperon A, Nguyen Ho-Bouldoires TH, Paradis V, Fouassier L. Epithelial-mesenchymal transition in cholangiocarcinoma: from clinical evidence to regulatory networks. J Hepatol. (2017) 66:424–41. doi: 10.1016/j.jhep.2016.09.010
54. Liang WC, Ren JL, Wong CW, Chan SO, Waye MM, Fu WM, et al. LncRNA-NEF antagonized epithelial to mesenchymal transition and cancer metastasis via cis-regulating FOXA2 and inactivating Wnt/beta-catenin signaling. Oncogene. (2018) 37:1445–56. doi: 10.1038/s41388-017-0041-y
55. Zhou T, Luo M, Cai W, Zhou S, Feng D, Xu C, et al. Runt-related transcription factor 1 (RUNX1) promotes tgf-beta-induced renal tubular epithelial-to-mesenchymal transition (EMT) and renal fibrosis through the PI3K subunit p110delta. EBioMedicine. (2018) 31:217–25. doi: 10.1016/j.ebiom.2018.04.023
56. Liang Z, Zhu B, Meng D, Shen X, Li X, Wang Z, et al. Down-regulation of lncRNA-NEF indicates poor prognosis in intrahepatic cholangiocarcinoma. Biosci Rep. (2019) 39:BSR20181573. doi: 10.1042/BSR20181573
57. Li Y, Zhang W, Liu P, Xu Y, Tang L, Chen W, et al. Long non-coding RNA FENDRR inhibits cell proliferation and is associated with good prognosis in breast cancer. Onco Targets Ther. (2018) 11:1403–12. doi: 10.2147/OTT.S149511
58. Wang B, Xian J, Zang J, Xiao L, Li Y, Sha M, et al. Long non-coding RNA FENDRR inhibits proliferation and invasion of hepatocellular carcinoma by down-regulating glypican-3 expression. Biochem Biophys Res Commun. (2019) 509:143–7. doi: 10.1016/j.bbrc.2018.12.091
59. Zhang G, Han G, Zhang X, Yu Q, Li Z, Li Z, et al. Long non-coding RNA FENDRR reduces prostate cancer malignancy by competitively binding miR-18a-5p with RUNX1. Biomarkers. (2018) 23:435–45. doi: 10.1080/1354750X.2018.1443509
60. Qin X, Lu M, Zhou Y, Li G, Liu Z. LncRNA FENDRR represses proliferation, migration and invasion through suppression of survivin in cholangiocarcinoma cells. Cell Cycle. (2019) 18:889–97. doi: 10.1080/15384101.2019.1598726
61. Yang R, Liu M, Liang H, Guo S, Guo X, Yuan M, et al. miR-138–5p contributes to cell proliferation and invasion by targeting survivin in bladder cancer cells. Mol Cancer. (2016) 15:82. doi: 10.1186/s12943-016-0569-4
62. Sankpal UT, Ingersoll SB, Ahmad S, Holloway RW, Bhat VB, Simecka JW, et al. Association of Sp1 and survivin in epithelial ovarian cancer: Sp1 inhibitor and cisplatin, a novel combination for inhibiting epithelial ovarian cancer cell proliferation. Tumour Biol. (2016) 37:14259–69. doi: 10.1007/s13277-016-5290-9
63. Yu A, Zhao L, Kang Q, Chen K, Li J, Fu H, et al. LncRNA LINC01061 sponges miR-612 to regulate the oncogenic role of SEMA4D in cholangiocarcinoma. Biochem Biophys Res Commun. (2019) 513:465–71. doi: 10.1016/j.bbrc.2019.03.125
64. Zhou H, Kann MG, Mallory EK, Yang YH, Bugshan A, Binmadi NO, et al. Recruitment of Tiam1 to semaphorin 4D activates rac and enhances proliferation, invasion, and metastasis in oral squamous cell carcinoma. Neoplasia. (2017) 19:65–74. doi: 10.1016/j.neo.2016.12.004
65. Takada H, Ibaragi S, Eguchi T, Okui T, Obata K, Masui M, et al. Semaphorin 4D promotes bone invasion in head and neck squamous cell carcinoma. Int J Oncol. (2017) 51:625–32. doi: 10.3892/ijo.2017.4050
66. Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. (2013) 73:1180–9. doi: 10.1158/0008-5472.CAN-12-2850
67. Gutschner T, Baas M, Diederichs S. Noncoding RNA gene silencing through genomic integration of RNA destabilizing elements using zinc finger nucleases. Genome Res. (2011) 21:1944–54. doi: 10.1101/gr.122358.111
68. Kim H, Danishmalik SN, Hwang H, Sin JI, Oh J, Cho Y, et al. Gene therapy using plasmid DNA-encoded anti-HER2 antibody for cancers that overexpress HER2. Cancer Gene Ther. (2016) 23:341–7. doi: 10.1038/cgt.2016.37
69. Datta A, Kim H, McGee L, Johnson AE, Talwar S, Marugan J, et al. High-throughput screening identified selective inhibitors of exosome biogenesis and secretion: a drug repurposing strategy for advanced cancer. Sci Rep. (2018) 8:8161. doi: 10.1038/s41598-018-26411-7
70. Mathiyalagan P, Sahoo S. Exosomes-based gene therapy for MicroRNA delivery. Methods Mol Biol. (2017) 1521:139–52. doi: 10.1007/978-1-4939-6588-5_9
71. Bala S, Csak T, Momen-Heravi F, Lippai D, Kodys K, Catalano D, et al. Biodistribution and function of extracellular miRNA-155 in mice. Sci Rep. (2015) 5:10721. doi: 10.1038/srep10721
Keywords: cholangiocytes, bile duct, microRNAs, long non-coding RNAs, cholangiocarcinoma
Citation: Sato K, Glaser S, Francis H and Alpini G (2020) Concise Review: Functional Roles and Therapeutic Potentials of Long Non-coding RNAs in Cholangiopathies. Front. Med. 7:48. doi: 10.3389/fmed.2020.00048
Received: 18 September 2019; Accepted: 31 January 2020;
Published: 20 February 2020.
Edited by:
Pedro M. Baptista, University of Zaragoza, SpainReviewed by:
Roberto Gramignoli, Karolinska Institutet (KI), SwedenHuiping Zhou, Virginia Commonwealth University, United States
Jose J. G. Marin, University of Salamanca, Spain
Copyright © 2020 Sato, Glaser, Francis and Alpini. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gianfranco Alpini, galpini@iu.edu; gianfranco.alpini@va.gov
 Keisaku Sato
Keisaku Sato Shannon Glaser
Shannon Glaser Heather Francis
Heather Francis Gianfranco Alpini
Gianfranco Alpini