Temporal Changes in Microbial Communities Beneath Fish Farm Sediments Are Related to Organic Enrichment and Fish Biomass Over a Production Cycle
- 1Institute for Biological Resources and Marine Biotechnologies, National Research Council (IRBIM-CNR), Ancona, Italy
- 2Institute of Anthropic Impacts and Sustainability in Marine Environment, National Research Council (IAS-CNR), Palermo, Italy
The marine fish farming industry is growing at a significant rate, yet a number of concerns still remain with regards to environmental impacts on the surrounding coastal sea and its biota. Here, we assessed the impact of intensive farming on benthic prokaryotic communities at a Mediterranean sea bass and sea bream intensive aquaculture site over a period of 10 months, in relation to the increase in fish biomass within the cage together with the organic matter enrichment in the sediments. We report positive relationships between prokaryotic abundance and both organic matter and fish biomass, and a contextual decrease in prokaryotic diversity below the cages. A significant shift in microbial community composition occurred in fish farm sediments (FF) over time, indicating a likely impact of ongoing aquaculture activity on prokaryotic communities. Among the dominant taxa at the impacted site, we found Epsilonproteobacteria and Bacteroidetes, which showed a general increase with fish biomass. Analyses on specialist taxa underlined significant contributions of Clostridiales and Bacteroidales in the farmed sediments. Finally, sea bream and sea bass gut microbiome-related taxa were detected during the sampling period. Our results indicate that prokaryotic community composition underneath the cages is related to fish biomass and organic enrichment over the course of production, and confirms that the study of benthic microbial communities at aquaculture sites represents a useful tool to assess the impact of intensive mariculture on the surrounding environment.
Introduction
Over the last decade, fish farming has increased significantly, and it is predicted that more than half of fish consumed on a global scale will be produced by aquaculture by 2030 (FAO, 2016). In the Mediterranean Sea and North-Eastern Atlantic regions, gilthead sea bream (Sparus [S.] aurata) and European sea bass (Dicentrarchus [D.] labrax) aquaculture are among the most widely established fish farms. For these species, rearing is mainly performed in coastal net-pen facilities (Arechavala-Lopez et al., 2018).
The rapid expansion of aquaculture in the coastal sea has drawn increasing concern regarding its environmental impact (Pusceddu et al., 2007; Holmer et al., 2008). A number of concerns emerge from this increasing production, among which the dispersion of farming-related wastes into the surrounding waters and on the seafloor (Carroll et al., 2003) is particularly urgent. In fact, this organic enrichment often leads to eutrophication and bottom hypoxia, and can significantly impair biogeochemical processes, energy transfer through the food web and the overall functioning of the benthic ecosystem (Mirto et al., 2012; Keeley et al., 2013), often facilitating the establishment and spread of specifically adapted and more resilient non-indigenous species (Mangano et al., 2019). The problems caused by the high organic and nutrient load conflict with other uses of the coastal zone; additionally, the organic loading has been modeled using current and future forecast temperature increase scenarios for the whole Mediterranean basin in an attempt to inform more sustainable siting of aquaculture farms (Sarà et al., 2018). Other environmental concerns of this industry are related to the use of chemicals and their dispersal in the surrounding environment (e.g., antibiotics and antifoulants) as well to the introduction and spread of pathogens (Wu, 1995; Tamminen et al., 2011).
The impact of high loads of organic matter deriving from uneaten food, fecal material and other sources into the sediment below aquaculture farms (Holmer et al., 2008) on larger benthic organisms such as meio- and macrofauna has been widely studied (Apostolaki et al., 2007; Mirto et al., 2010), while much less is known about their effects on sediment microbial communities. Sediment prokaryotes play crucial roles in the degradation and remineralization of sedimentary organic matter, the carbon and energy transfer to higher trophic levels and the overall functioning of the benthic environment (Manini et al., 2003; Reimers et al., 2013), and respond quickly to environmental perturbations (Galand et al., 2016). However, to date, studies on the consequences of mariculture on sediment prokaryotes have focused mostly on evaluating changes in their abundance and biomass (Mirto et al., 2000; La Rosa et al., 2001a, 2004; Richardson et al., 2008), microbial metabolic activities (Vezzulli et al., 2002; Luna et al., 2013), and the presence or diffusion of antibiotic resistance (Chelossi et al., 2003; Dang et al., 2006; Chen et al., 2017). Studies addressing the effects on bacterial diversity and community composition have been more rare (Bissett et al., 2006) or have been performed using the low–resolution microbiological techniques available at that time (e.g., genetic fingerprinting or cloning and sequencing of ribosomal genes; Kawahara et al., 2009; Luna et al., 2013) that have provided the first evidence for mariculture effects on microbial diversity, without, however, allowing in-depth analyses of which microbial taxa are affected. More recent studies have applied the techniques of High-Throughput Sequencing (HTS) of 16S rRNA microbial genes (also known as metabarcoding) that allow detailed lists of microbial taxa to be produced (Sinclair et al., 2015), however, these studies have focused mostly on salmon aquaculture (Dowle et al., 2015; Hornick and Buschmann, 2018; Stoeck et al., 2018), while studies that addressed the effects of sea bream and sea bass farming have remained scarce. Different types of farmed fishes are likely to exert a specific impact on the sedimentary environment, as they differ in the diet and the feed used for their growth and have different gut microbiota, which could have different impacts on the seafloor (both in terms of microbiota that is dispersed via feces and of quantity and composition of the released organic matter). Thus, the effect of sea bream and sea bass farming on benthic microbial communities is worthy of further investigation.
Given the profound impact that mariculture has on the coastal sea, it is imperative to move toward a more sustainable aquaculture (Alexander et al., 2015) and to routinely perform environmental impact assessment and monitoring of the fish farm environment. Morphology-based identification of macrobenthic infauna species is nowadays used to perform the ecological impact of aquaculture (Klaoudatos et al., 2006; Apostolaki et al., 2007). However, morphology-based identification requires taxonomic expertise and is labor-intensive. A recent study on biomonitoring at fish farms (Pawlowski et al., 2014) has demonstrated the potential of using an HTS-based technique on the identification of foraminiferal populations, suggesting that HTS can be used as an alternative to morphology for benthic monitoring in marine systems, including aquaculture settings (Lejzerowicz et al., 2015). The application of a similar metabarcoding approach can prove useful also for prokaryotic microbes, which are potentially valuable bioindicators of fish farm impacts due to their rapid growth and fast response to environmental changes (La Rosa et al., 2001b; Vezzulli et al., 2002; Luna et al., 2013).
In the present study, we investigated the influence of sea bream and sea bass fish farming on the organic loading and the microbial communities beneath the fish cages. To do this, we monitored, over a period of 10 months of fish growth (October to July), that covered the four seasons, the concentration of sediment organic matter and the main sediment microbial variables (abundance, richness and community composition) under a fish farm and compared with a reference site. The diversity analysis was undertaken using the Illumina MiSeq platform on 16S rRNA gene amplicons.
Materials and Methods
Study Site and Sampling Strategy
The study was undertaken in the “Ittica San Giorgio” marine fish farm located at the opening of the harbor of Licata in Southern Sicily (Ionian Sea; geographic coordinates 37°06′36″N, 13°56′49″E; Figure 1), and lasted from October 2014 to July 2015. During the sampling period, seawater temperature ranged from 15°C to 24°C (in March and in July, respectively). The studied farm, as described by Ape et al. (2019), is composed of 23 floating cages arranged in two rows, which cover a total surface of ∼8,000 m2. The cages contain sea bass (D. labrax) or sea bream (S. aurata) for a total annual production that exceeds 300 tons. The farm is in operation from 1994, and is located in a semi-enclosed and sheltered area, characterized by limited hydrodynamism and by shallow depth (∼10 m). Consequently, uneaten food and feces of the reared fish cause the accumulation of a large amount of organic matter on the seafloor under the cages, a phenomenon that causes a progressive transformation of the benthic substrate into muddy black sediment (Holmer et al., 2008). Severe hypoxia phenomena affect the fish farm during the year, especially in the period from August to October, when the concentration of oxygen inside the cages can drop to as low as ca. 2 mg L–1.
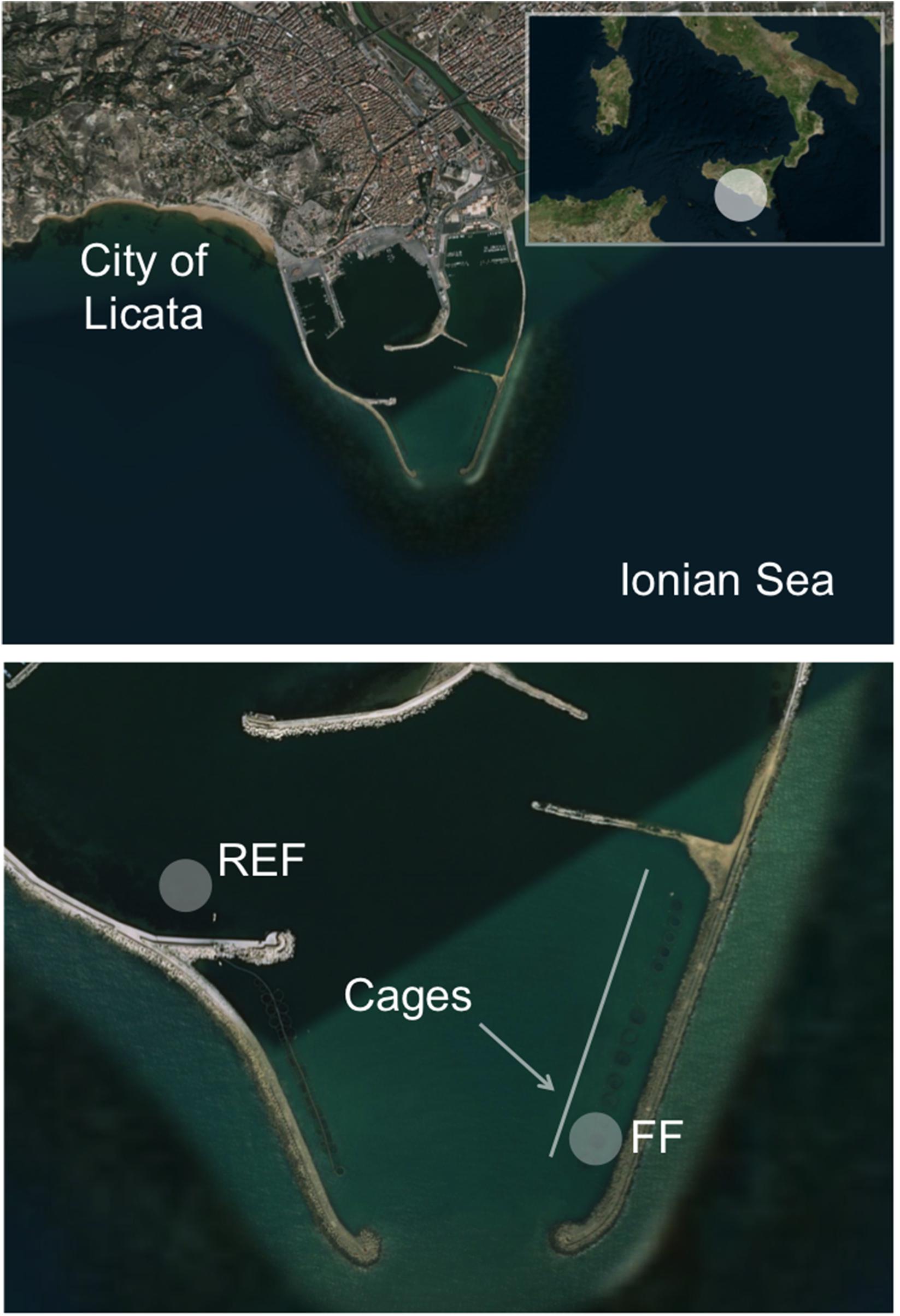
Figure 1. Location of the two sampling stations inside the harbor of Licata (Southern Sicily, Ionian Sea). The FF station was located below one cage inside the fish farm, while the REF station was located outside the farm. The rectangle in the upper panel shows the Southern Italy, where a white circle highlights the study area located in the Sicily region. The two arrows in the lower panel show the location of the fish farm cages. The figure has been drawn using Bing Maps.
Sampling was carried out in October (hereafter “Oct”), in December (“Dec”), February (“Feb”), May (“May”), and July (“Jul”). Along the sampled time points, fish biomass within the cage increased as follows: 11.2 kg/m3 in October, 13.8 kg/m3 in December, 14 kg/m3 in February, 14.8 kg/m3 in May, and 15.2 kg/m3 in July. Fish size increased accordingly from an average of 295 g to 440 g. Sediment samples were collected at ∼10 m below one cage containing sea bass (volume ca. 2,000 m3), hereafter named fish farm sediments (“FF”), and at an external control station (“REF”; ∼8 m deep) not influenced by the farm and chosen, after a preliminary survey of the bottom characteristics and circulation pattern, at ca. 700 m distance from the fish farm and at comparable depth, near the opening of the harbor (Figure 1).
Sediment samples were collected as described in Ape et al. (2019). Briefly, triplicate plexiglas corers (diameter 3,6 cm) were used for the analysis of the organic matter [specifically, biopolymeric organic carbon, or organic matter content (BPC); sensu Pusceddu et al., 2003], while sterile corers (sterile plastic syringes; diameter 3,5 cm) were used for microbial analyses (abundance, diversity). The corers for biochemical and microbiological sampling were collected within close proximity to each other in order to directly relate these datasets. After sampling, the corers were transported to the laboratory, and the top 1 cm of each corer was carefully extruded and stored at −20°C for the analyses of organic matter, prokaryotic abundance and diversity.
Concentration of Sedimentary Organic Matter
For the analysis of organic matter concentration, proteins, carbohydrates and lipids were analyzed in triplicate on all sediment samples by using spectrophotometric methods (Pusceddu et al., 2003) and their concentrations expressed as bovine serum albumin, glucose, and tripalmitine equivalents (mg g–1 of dry sediment), respectively. Carbohydrate, protein and lipid concentrations were converted into carbon (C) equivalents using 0.40, 0.49, and 0.75 mg C mg–1 as conversion factors (respectively), and their sum reported as BPC.
Prokaryotic Abundance
The total abundance of prokaryotic cells (TPA) was quantified using a staining technique with Acridine Orange, as described by Manini et al. (2003). Briefly, one gram of sediment (in triplicate) was placed in a sterile tube and fixed pre-filtered, 2% formalin solution, previously buffered with Na2B4O7 × 10H2O until a complete coverage of samples by the solution. Fixed samples were preserved overnight at 4°C. Subsequently, 100 μL of this suspension was diluted in 3 mL of filtered water, mixed with 500 μL of 0,025% acridine orange for 30 s and incubated for 5 min in the dark. Subsamples were diluted 500-fold, and filtered on black Nuclepore 0.2-μm-pore-size filters. The filters were analyzed using epifluorescence microscopy (1000× magnification). Total prokaryotic counts were normalized to sediment dry weight after desiccation (24 h at 60°C).
Prokaryotic Diversity
PowerSoil® DNA Isolation Kit (MoBio Laboratories Inc., California, United States) was used to extract DNA from one gram of each sediment sample, by following the manufacturer’s instructions with some slight modifications to increase the DNA yield and quality as described in Ape et al. (2019). Illumina Miseq V3 sequencing analyses were carried out on the hypervariable V3 and V4 regions of the 16S rRNA gene by amplifying using the 341F (5′-CCTACGGGNGGCWGCAG-3′) and 785R (5′-GACTACHVGGGTATCTAATCC-3′) universal bacterial primers (Eiler et al., 2012). Paired-end reads were quality checked (with default settings and minimum quality score of 20) and analyzed with QIIME v1.8.0 software package (Quantitative Insights Into Microbial Ecology; Caporaso et al., 2010) as described in Ape et al. (2019). Briefly, reads were clustered into OTUs by using UCLUST v1.2.22 (Edgar, 2010) with a >97% similarity threshold. USEARCH v6.1 was used to detect chimeras (Edgar, 2010) and Greengenes 13.8 was used as reference database for chimera checking and taxonomy assignment (DeSantis et al., 2006). In each sample, abundances were normalized on the number of sequences of sample with the lowest number of reads retained. The sequences are submitted to the SRA – Sequence Read Archive (BioProject PRJNA525837, accession numbers SRX5485548-57, and SRX5485570-71).
Statistical Analysis
To test for statistical differences in BPC, and prokaryotic abundances (TPA) between sites (FF and REF) and among times as well as the combined effect of both site and time, we performed a two-way ANOVA. Due to the lack of replication for OTU richness data, a t-test was used to compare OTU richness values between FF and REF, combining all sampling times. PERMANOVA (anosim function, vegan R package) was used to assess the difference between FF and REF prokaryotic community composition.
We used the CLAM test (Chazdon et al., 2011) as a statistical approach for classifying generalists and specialists in REF and FF. This test uses a multinomial model based on estimated species’ relative abundance in two “habitats” (i.e., fish farms and reference sediments), allowing a robust statistical classification of habitat specialists and generalists, without excluding rare species a priori. CLAM analysis was performed using the OTU table as input; based on CLAM results, which mainly identified taxa at the Order level, we collapsed all generalists belonging to the same taxonomical Order.
Finally, within each site and sampling time, we recruited a suite of OTUs identified as belonging to the most common gut microbiome taxa in seabass and seabream as reported in the most recent literature (Kormas et al., 2014; Nikouli et al., 2018). The list of selected taxa is reported in Supplementary Table S1. As a further check, we performed BLASTN (Altschul et al., 1990) of our representative sequences against a custom reference database of 16S rRNA gene sequences of the taxa of interest, and filtered the “best hit” results with ≥99% of similarity and query coverage ≥85% of the sequence length.
Results
Sedimentary Organic Matter and Prokaryotic Abundance
Biopolymeric carbon differed between stations (Supplementary Table S2 and Supplementary Figure S1-A), and two-way ANOVA analysis indicated that the organic enrichment was significantly different between sites (p < 0.01), but not among sampling times; the combined effect of site and time was not significant.
The abundance of total prokaryotes in the sediments was, on average, two-fold higher in FF beneath the cage than in REF sediments (3.22 ± 0.18 and 1.51 ± 0.08 cell × 108 g–1, respectively, Supplementary Figure S1-B). Two-way ANOVA showed that prokaryotic abundance was significantly different between sites (p < 0.001), but not among sampling times. However, the combined effect of site and time was significant (p < 0.01).
As reported above, the organic enrichment below the cage increased over time, although not significantly along each of the sampled events. However, a significant, positive correlation was found between the BPC content and prokaryotic abundance (n = 30, r = 0.923; Supplementary Figure S2, left panel) considering both REF and FF, and when considering FF only (n = 15, r = 0.94). When considering the variations in fish biomass along the sampling period at the FF site, we also found a strongly positive relation between fish biomass and TPA (n = 12; r = 0.73) as well as with BPC (n = 12; r = 0.87; Supplementary Figure S2).
Microbial Diversity and Community Composition
More detailed results of the HTS of 16S rRNA gene are reported in Ape et al. (2019). Overall, benthic microbial richness showed significantly lower values (t-test, p < 0.01) in FF compared to REF sediments (Figure 2), with values at FF site in a range between 1,095 and 1,567 OTUs, as opposed to the range of 1,536–2,366 OTUs observed in the REF sediments. Prokaryotic richness and abundance were differently correlated to the enrichment in organic matter in the sediments. From one side, in the reference site, an overall weak, but positive, correlation was observed between richness and BPC (Figure 2B). On the other hand, in FF, we found that richness values significantly decreased with increasing organic load (Figure 2A). The same pattern was depicted when considering TPA, which, in REF site, was positively correlated to OTU richness (Figure 2D), conversely to what observed at the FF site, where richness strongly decreased with higher prokaryotic abundances (Figure 2C).
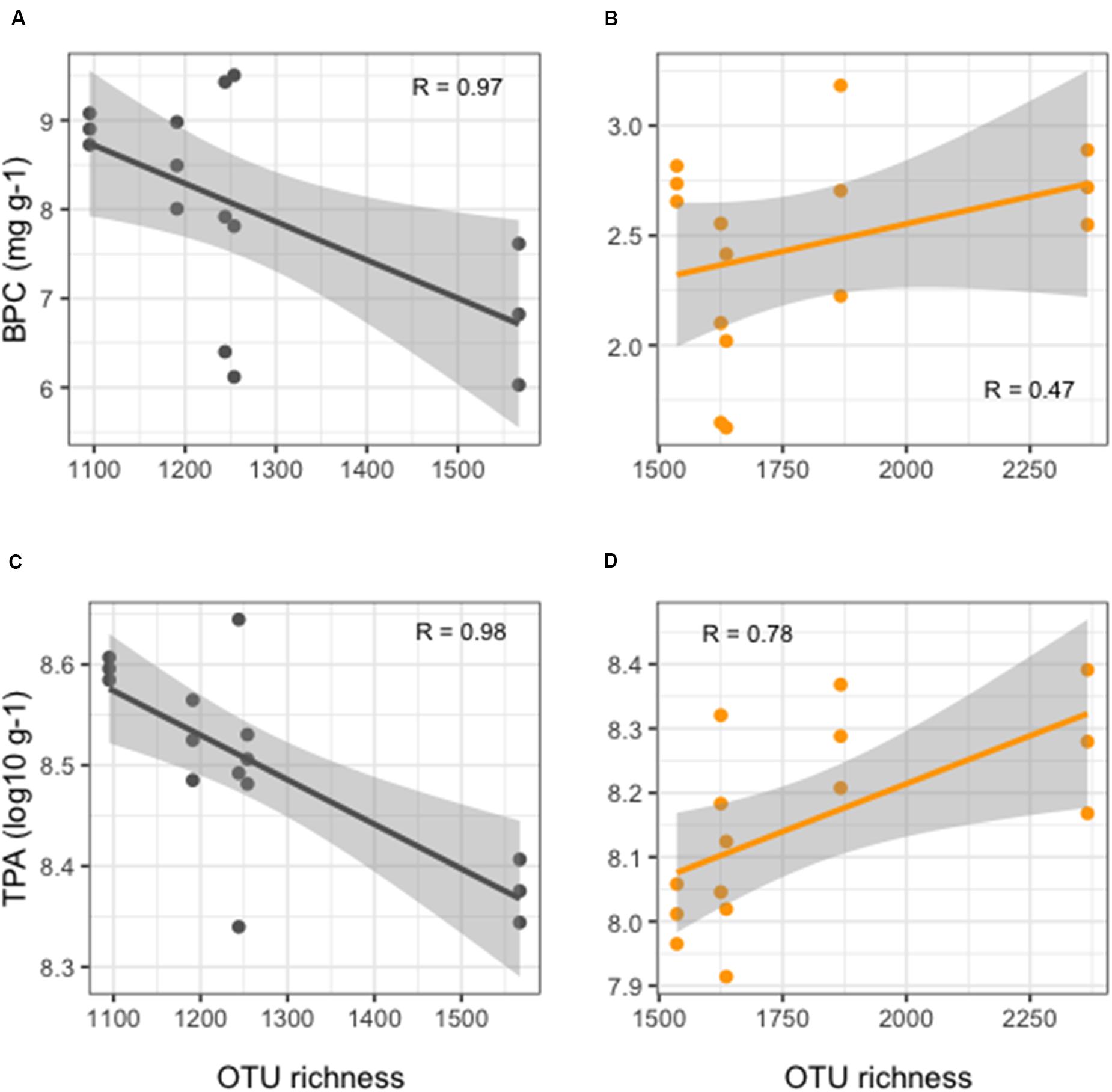
Figure 2. Linear regression curves showing relationships between OTU richness and organic enrichment (A,B) and TPA (C,D) in FF (black lines, left side of the figure) and in REF (orange lines, right side of the figure) sites. Each dot represents an individual sample; gray shaded area represents the 95% confidence interval.
To test whether fish biomass, which increases during the production cycle, is related to increasing levels of organic matter in the environment, and whether microbial diversity is influenced by organic matter content, we collected sediment samples along the time of the production chain from smaller (295 g) to bigger (440 g) fish size, and we tested our data for significant relationships between fish biomass and microbial OTU richness in the sediments. We found that, at the farmed site, no clear correlation was visible between farmed animal biomass and richness (Supplementary Figure S3).
Microbial community composition analysis (Supplementary Figure S4) indicated that a significant shift (ANOSIM, R = 0.717) in microbial diversity could be observed in FF compared to REF sediments. At the phylum level, Proteobacteria dominated both sediment types (range 38.7% in October to 66.7% in July at FF, and 52.7% in May to 56.5% in February at REF). Within this phylum, the main difference between fish farm and control sediments was evident for the Epsilonproteobacteria class, that dominated in FF (range 21.3–35.8% at FF) while representing only a minor constituent of the non-aquaculture sediments (0.8–4.4% at REF), which displayed, on the other side, dominance of Gammaproteobacteria (range 18.8–26.2% at REF), as opposed to FF (range 2.3–10%). Also, Epsilonproteobacteria and Bacteroidetes showed a slight increase at FF over time (Supplementary Figure S5).
The ratio of FF/REF (in terms of % of relative abundance) was calculated, for the most abundant phyla, to highlight the shift in community composition (Figure 3). The ratio was typically <1 for Proteobacteria, suggesting a slightly higher relative abundance of members of this phylum in REF sediments. Conversely, the ratio was always >1 for the phylum Firmicutes, with a peak of 87.2 observed in October. We found that for Bacteroidetes, Spirochaetes and Verrucomicrobia, FF/REF ratios also indicated a general higher proportion of these phyla in FF, with few exceptions. On the other hand, Acidobacteria, Chloroflexi, and Actinobacteria, although at different extents and with some exception, were almost constantly at higher relative abundance in REF. Planctomycetes and Gemmatimonadetes were found to be at similar abundances at both sites and over time (Figure 3).
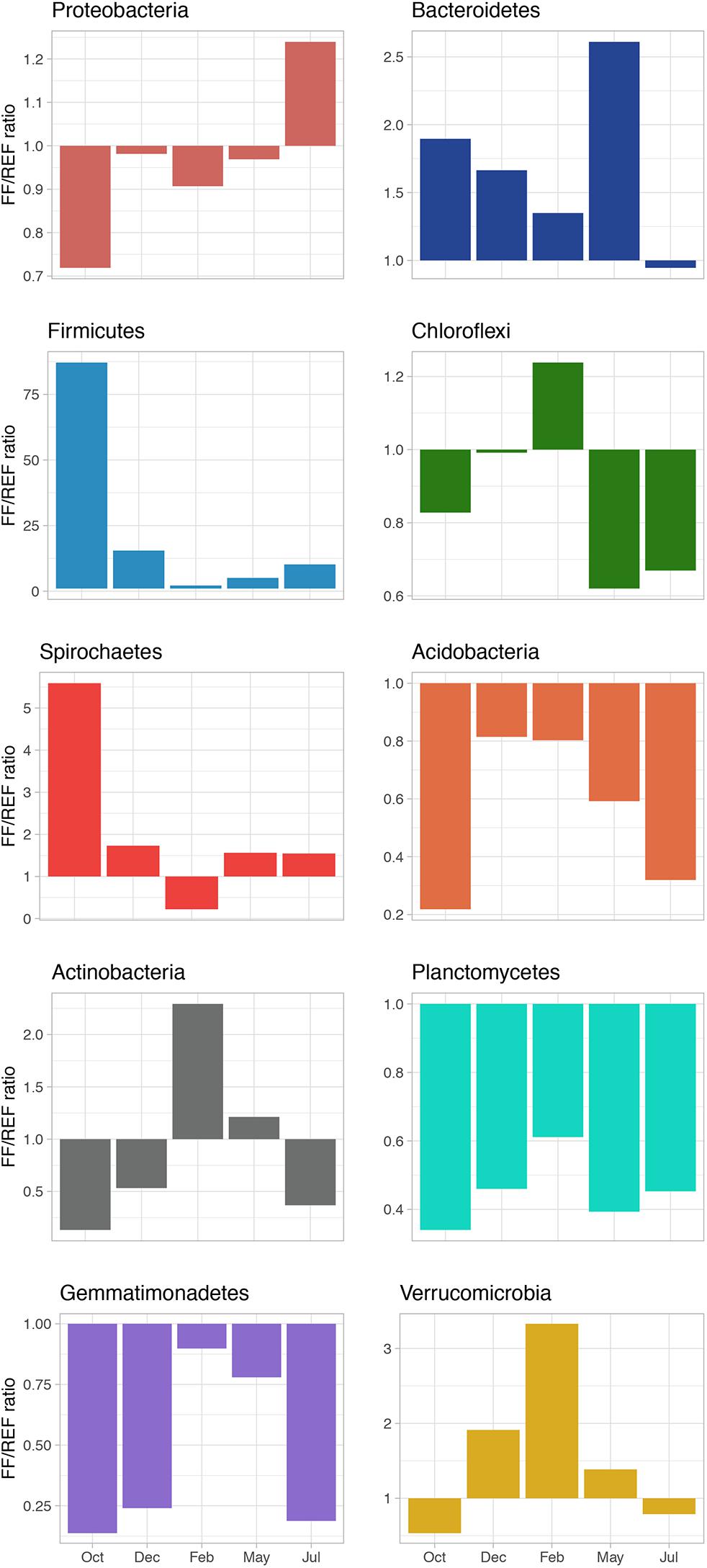
Figure 3. FF/REF ratios of the relative abundances of the main bacterial phyla identified in the prokaryotic communities within this study as obtained by HTS sequencing.
Specialist Taxa
Several taxa, at the Order taxonomic level, showed significant differences between FF and REF samples at each step of fish growth and were thus identified by the CLAM test as “specialist” taxa (Figure 4).
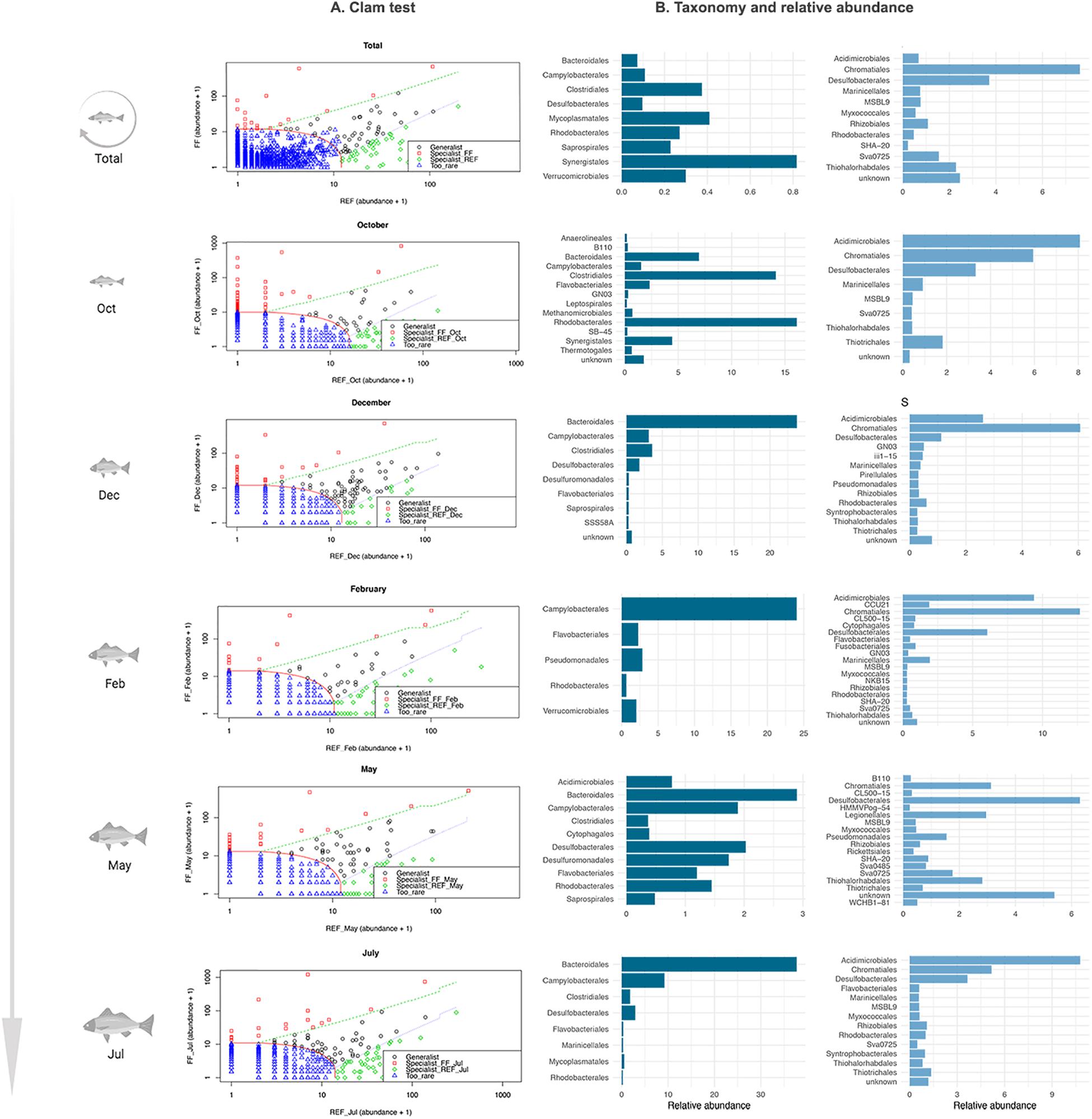
Figure 4. Results of the CLAM test analysis for the classification of specialist taxa in prokaryotic communities of FF and REF sites over the 10-months sampling points. (A) summary plot generated by the CLAM test in vegan, showing generalists, specialists and rare taxa in relation to two conditions tested (i.e., FF vs. REF) and calculated according to the estimated species relative abundance in FF and REF samples. The x and y axes represent, respectively, the abundance of different Orders in FF and REF samples. Counts were added by 1 to let the marginal taxa be evenly arranged in the plot space. Specialists for FF samples (red dots) map at the left side of the plot, whereas specialists for REF samples (green dots) at the right side. Generalists taxa (black dots) are located in the middle of the plot and rare taxa are at the bottom left corner (blue dots). Dotted lines indicate thresholds used for the classification into generalist, specialist and rare taxa. (B) taxonomic assignation and relative abundance of the specialist taxa in FF (dark blue) and REF (light blue) sites at the order level.
Over the ten-month monitoring, we found significant differences in the composition of specialist taxa (i.e., orders) in the FF site vs. the REF site (Figure 4). At the FF site, a number of taxa which are commonly considered to include pathogens or potential pathogens were identified as specialists. Among these, we found Bacteroidales, Clostridiales, Campylobacterales, and Synergistales (Figure 4). On the other hand, at the REF site, the specialist community was mainly composed of Chromatiales and Desulfobacterales.
When looking specifically at the single sampled time points, we found that, at the FF site, Bacteroidales, followed by Clostridiales, were almost constantly identified as specialist, except in February (Figure 4). At the REF site, an overall dominance of Acidimicrobiales, Chromatiales and Desulfobacterales as specialists was observed, except for May sampling, when Acidimicrobiales were not included as specialists, and Legionellales as well as a number of unknown taxa were among the most represented specialist taxa.
Sea Bass and Sea Bream Gut Microbes
The FF sediments showed an enrichment of certain taxa that are typically associated with sea bass and sea bream gut microbiomes. Among these, we found an increase of Propionibacterium acnes and Pelomonas aquatica along fish growth (Figure 5, panel FF), together with a peak of Acinetobacter spp. in February which was not retrieved anymore until the end of the experiment. Conversely, the selected taxa showed no or very low abundances at the REF site (Figure 5, panel REF), although signals of Acinetobacter, Corynebacterium, and Propionibacterium acnes were found from May to July at the control station.
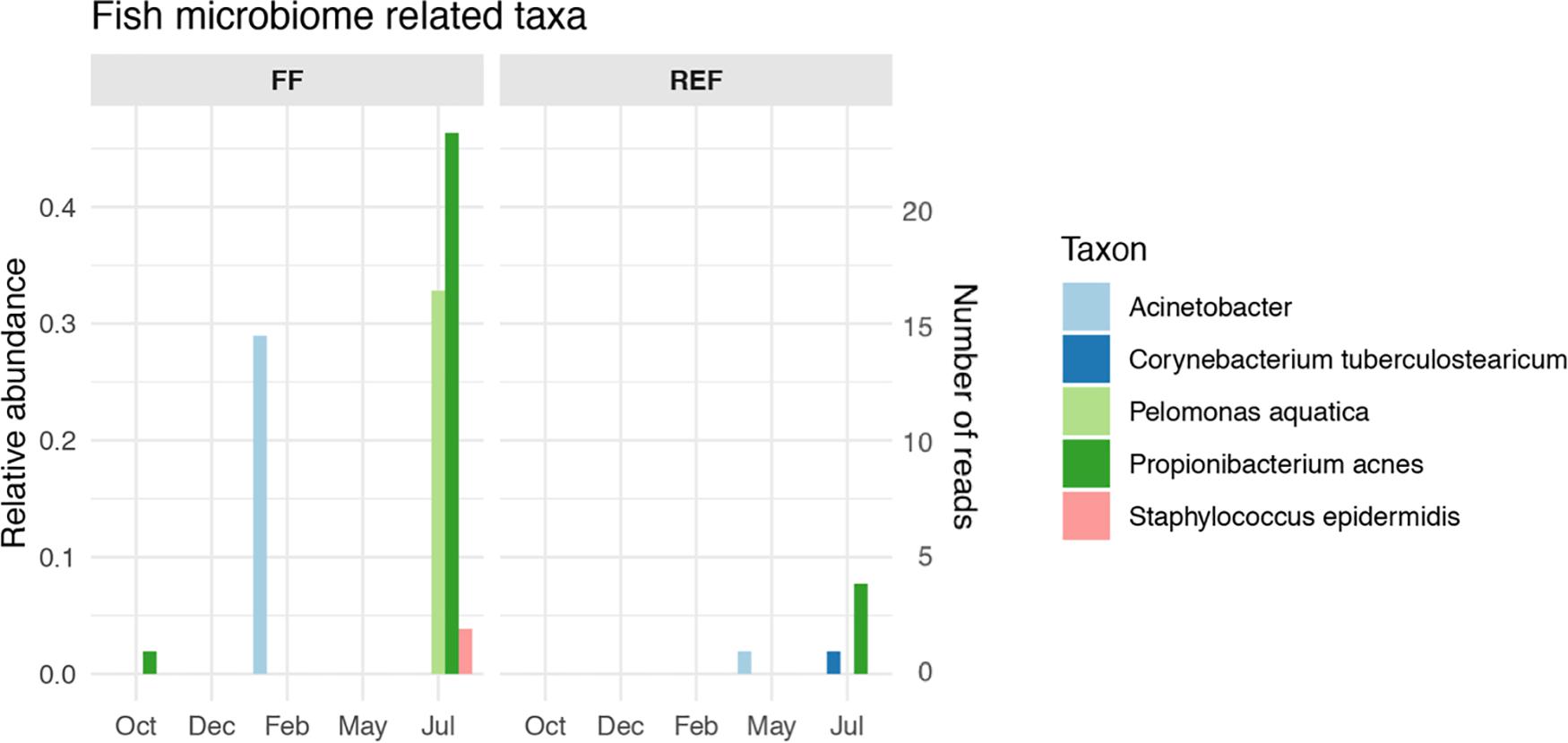
Figure 5. Relative abundance and number of reads of fish microbiome-related taxa in aquaculture (FF) and reference (REF) sites along the experiment. Taxa listed in Supplementary Table S1 and not in this Figure’s legend were not retrieved in our dataset.
Discussion
Because prokaryotes are sensitive to environmental change, and may shift in abundance, diversity and community composition according to environmental perturbations, they have been widely studied in aquaculture, and suggested as one of the most promising tools for biomonitoring of farmed areas (Verhoeven et al., 2018). However, most of the studies performed so far did not take into consideration the increase in fish biomass over the production cycle, and its influence on the accumulation of the organic matter on the fish farm sediment. In this study, we investigated the effects of intensive sea bream and sea bass farming along a 10-months period of fish growth and the consequent sediment organic enrichment on the microbial communities in the sediments that surround the fish cages. Previous studies have investigated such effects on the benthic microbes (Vezzulli et al., 2002; Dang et al., 2006; Luna et al., 2013; Chen et al., 2017), however, they have not addressed the differences occurring in abundance and community composition according with the increase in farmed fish biomass.
As expected, we observed a significant enrichment in organic matter under the farming cages, in contrast to what we observed at the control site. The accumulation of organic matter beneath fish cages, considered as one of the major impacts of aquaculture (Holmer et al., 2008), likely occurred as a consequence of waste products released from the fish farm, in particular fish feces and feed (Carroll et al., 2003; Mirto et al., 2012; Ape et al., 2019). As fish size increased (leading to an increase in biomass within the cage), differences – especially between the winter and summer months – in organic enrichment were measured, suggesting that the increase (about 37%) of biomass during the sampled 10-months was somehow reflected into an increased input of organic matter to the sediments. Correlation analyses between fish biomass and BPC content supported this hypothesis, although it must be also pointed out that other factors (e.g., hydrodynamics, sediment resuspension, accumulation of organic matter over time), may have contributed to explain this pattern.
In response to the increased accumulation of organic matter in the sediments, we observed an overall doubled total prokaryotic abundance beneath the fish cages. The TPA increase and its positive correlation with BPC content support the concept of increasing microbial abundances with the increased availability of organic matter. A higher availability of sediment organic matter, in fact, is well known to be able to stimulate prokaryotic growth, and the higher prokaryotic abundance is thus considered as an effect of biodeposition in fish farming plants (La Rosa et al., 2001a; Vezzulli et al., 2002; Caruso et al., 2003; Luna et al., 2013; Ape et al., 2019). The direct and positive correlations between prokaryotic abundance and both organic matter and fish biomass support the existence of relationships between benthic microbial communities and organic enrichment, with the latter increasing over the course of production as fish biomass increases.
Among the changes observed, a clear pattern that emerged in our study was the lower richness consistently observed below the cage with respect to the non-impacted sediments. This pattern has been reported previously in other fish farms (Bissett et al., 2006; Luna et al., 2013), suggesting that the organic enrichment lead to a decrease in microbial species. Also, Dowle et al. (2015) reported that bacterial OTUs richness, in both DNA- and RNA-based analyses, increased with distance from cage, with richness values under the cages of 143–189 as opposed to richness values 442–467 at 4,000 meters distance from the fish farm, thus confirming that fish farming activities significantly decrease diversity of benthic microbial communities. Shifts in prokaryotic diversity in response to aquaculture has been also reported in salmon farms, where a decline in bacterial diversity was observed in farm sediments beneath the cages. All these findings suggest that such decrease in microbial richness may have obvious deleterious consequences on the ecological functioning on the benthic food web (Hornick and Buschmann, 2018; Verhoeven et al., 2018), deserving more investigations.
As shown by the data of prokaryotic community structure and composition, over the 10-month period a significant shift in microbial community composition was observed in FF, as well as a significantly different overall community structure between FF and REF, indicating a possible impact of the fish farm and the fish biomass increase on prokaryotic communities. Previous studies have shown changes in bacterial community structure (Bissett et al., 2006, 2007, 2009; Garren et al., 2008), as well as in functionality (Christensen et al., 2000; Holmer et al., 2003; Bissett et al., 2009), in sediments below fish cages. Although community changes provide useful information regarding the ecosystem-wide response to aquaculture pollution and the potential biogeochemical process modifications (Hornick and Buschmann, 2018), to the best of our knowledge this is the first study focusing on such changes in response to fish biomass increases within the cages.
Over the last decade, studies related to the impact of fish farming on prokaryotic communities have underlined a distinctive occurrence of bacteria linked to the sulfur cycling, which have been detected often, including SOBs within Epsilonproteobacteria. In accordance with these studies, in our study, at both sites, we also found that, overall, Epsilonproteobacteria dominated prokaryotic communities at the farming site, as opposed to the non-aquaculture sediments. The dominance of Epsilonproteobacteria is thus not novel in the study of aquaculture impact on benthic microbial communities, and their presence has been reported under fish cages even using different molecular approaches (Bissett et al., 2006; Kawahara et al., 2009; Fodelianakis et al., 2015). It has been hypothesized that the high relative importance of Epsilonproteobacteria indicate that they are stably associated with fish farm activities, and that they might even be used as biomarker for qualitative and quantitative assessment of aquaculture impact in coastal environments. Epsilonproteobacteria include a number of taxa having a significant role in the sulfur cycling in marine and terrestrial environments (Polz and Cavanaugh, 1995; Engel et al., 2004; Campbell et al., 2006), and members of this class are being increasingly associated, or detected at relatively high abundances, in human-impacted coastal sediments (Quero et al., 2015). Our result support previous studies showing accelerated sulfur cycle in sediments beneath intensive fish farming (Asami et al., 2005; Rubio-Portillo et al., 2019), due to the organic matter enrichment, the consequent oxygen depletion, and the enhanced activity of anaerobic bacteria leading to the increase of sulfide (Hargrave et al., 2008). Additionally, we observed significantly higher relative abundances of Bacteroidetes beneath the fish cages with respect to the reference site. Bacteroidetes are often associated with organic matter enriched environments (Fernández-Gomez et al., 2013; Aylagas et al., 2017) and are highly specialized in degrading high molecular weight organic compounds. They have been previously reported in relation to aquaculture impact studies (Dowle et al., 2015), and their increased relevance in fish farming impacted sediments led Bacteroidetes to be identified as biomarkers in the sediments lying under fish cages (Verhoeven et al., 2018). Overall, our results and previous studies reporting the dominance of Epsilonproteobacteria and Bacteroidetes exclusively in sediments under fish cages, strongly suggest that they originate from fish feces or feed products (Rubio-Portillo et al., 2019).
Higher relative abundances in Bacteroidetes, Firmicutes and Spirochaetes were observed in the farmed site, corroborating previous finding reporting the same taxa among the most represented in highly impacted sediments, also for other farmed species (Verhoeven et al., 2018). In our study, we observed that Proteobacteria (mainly Epsilonproteobacteria) and Bacteroidetes showed a general increasing pattern with time and fish biomass increase (Supplementary Figure S5), with an unexpected drop of the latter in July (Figure 3), which we hypothesize is due to a sudden change in environmental conditions at the sampling time. Interestingly, for other main phyla (Figure 3), FF/REF relative abundance ratios largely fluctuated over time and did not show consistent patterns over the course of production. We hypothesize that, in addition to biotic interactions, different factors that may include seasonal variations, accumulation of organic matter over time and sediment resuspension, occur differentially between FF and reference sites and may contribute to these shifts.
Analyses of specialist taxa underlined significant differences between the farmed and the reference sites (Figure 4). Corroborating our previous analyses, several taxa were identified as specialists at the farmed site. Members of these taxa (e.g., Clostridiales, Bacteroidales) are usually associated with the organic enrichment and anaerobic conditions, confirming that certain groups of microbes, previously reported to be major contributors of the differences in bacterial communities between aquaculture and non-aquaculture sediments (Li et al., 2013; Ape et al., 2019), play a fundamental role in the anaerobic degradation of organic material in aquatic systems (Schwarz et al., 2008), and particularly in intensive aquaculture. Moreover, Bacteroidales consistently increased at FF according to the increase in fish biomass (except in February). Both Bacteroidales and Clostridiales have been described as the most abundant orders in marine herbivore gut communities (Sullam et al., 2012); their presence in the sediments thus suggests an impact from fish feces release on benthic microbial communities at intensive aquaculture sites. In addition, it must be pointed out that, although rich in non-pathogenic members, both bacterial orders contain common potential pathogens, thus the possible role of farm sediments as environmental reservoirs for fish and human pathogens should also be taken into consideration.
Given the evidence for this high contribution of gut-related taxa to the benthic prokaryotic communities impacted by fish farms, we looked at taxa specifically associated with the gut microbiome of Mediterranean sea bass and sea bream. Even at this specific level, we could detect species-specific microbial biomarkers, with an overall increasing pattern with fish biomass increase (Figure 5), although, given their low abundances, it cannot be excluded that other factors, independent of fish growth, may have contributed to this result. Signals of some the gut-specific taxa were detected occasionally at the control site, and we speculate that fish gut microbes or traces of their DNA could have been transported to REF site by hydrodynamic processes.
Given the growth of the aquaculture industry, concerns remain about the environmental impact of intensive fish farming on the surrounding marine biota, and the assessment of factors determining such changes in response to the impact needs to be further explored. In this study, we aimed at assessing whether increasing organic enrichment over the course of production was related to shifts in prokaryotic abundance, diversity and community composition. Our results indicate that benthic communities were largely affected by the increased organic matter concentration below the cages as fish biomass increased. Shifts in community structure could be related to the size of farmed fishes, as fishes higher in size are expected to release larger amounts of fecal material and require more food to grow (which leads to an increased accumulation at the seafloor). Although other factors not investigated here, including seasonality, hydrodynamics, sediment resuspension, accumulation of organic matter over time as well as biotic interactions, may have contributed to affect prokaryotic assemblage’ variations, our study highlights changes in microbial communities below the cages over the course of fish production. Based on these results, we also underline the usefulness of studying benthic microbial communities as a tool to assess the impact of intensive mariculture activities on the surrounding environment.
Data Availability Statement
The datasets generated for this study can be found in the repository: SRA (Sequence Read Archive), accession numbers: SRX5485548, SRX5485549, SRX5485550, SRX5485551, SRX5485552, SRX5485553, SRX5485554, SRX5485555, SRX5485556, SRX5485557, SRX5485570, and SRX5485571.
Author Contributions
GQ and FA: conceptualization, sampling, analyses, writing, review, and editing. EM: analyses, review, and editing. SM: conceptualization, resources, review, and editing. GL: conceptualization, analyses, writing, review, editing, and resources. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the INNOVAQUA project financed by the National Operative Programme (PON) for Research and Competitiveness 2007e2013 (PON02_000451_ 3362185/1), by the Flagship Project RITMARE (Italian Research for the Sea), coordinated by the Italian National Research Council within the National Research Program 2011–2015 and by the CIRCLES (Controlling mIcRobiomes CircuLations for bEtter food Systems) project (Grant agreement no.: 818290) within HORIZON 2020 (H2020-SFS-2018-2020 – Sustainable Food Security).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Laura Perini for her precious help in the DNA extraction from the sediment samples.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2020.00524/full#supplementary-material
References
Alexander, K. A., Potts, T. P., Freeman, S., Israel, D., Johansen, J., Kletou, D., et al. (2015). The implications of aquaculture policy and regulation for the development of integrated multi-trophic aquaculture in Europe. Aquaculture 443, 16–23. doi: 10.1016/j.aquaculture.2015.03.005
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Ape, F., Manini, E., Quero, G. M., Luna, G. M., Sarà, G., Vecchio, P., et al. (2019). Biostimulation of in situ microbial degradation processes in organically-enriched sediments mitigates the impact of aquaculture. Chemosphere 226, 715–725. doi: 10.1016/j.chemosphere.2019.03.178
Apostolaki, E. T., Tsagaraki, T., Tsapakis, M., and Karakassis, I. (2007). Fish farming impact on sediments and macrofauna associated with seagrass meadows in the Mediterranean. Estuar. Coast. Shelf Sci. 75, 408–416. doi: 10.1016/j.ecss.2007.05.024
Arechavala-Lopez, P., Toledo-Guedes, K., Izquierdo-Gomez, D., Šegvi-Bubi, T., and Sanchez-Jerez, P. (2018). Implications of sea bream and sea bass escapes for sustainable aquaculture management: a review of interactions, risks and consequences. Rev. Fish. Sci. Aquac. 26, 214–234. doi: 10.1080/23308249.2017.1384789
Asami, H., Aida, M., and Watanabe, K. (2005). Accelerated sulfur cycle in coastal marine sediment beneath areas of intensive shellfish aquaculture. Appl. Environ. Microbiol. 71, 2925–2933. doi: 10.1128/AEM.71.6.2925-2933.2005
Aylagas, E., Borja, Á., Tangherlini, M., Dell’Anno, A., Corinaldesi, C., Michell, C. T., et al. (2017). A bacterial community-based index to assess the ecological status of estuarine and coastal environments. Mar. Pollut. Bull. 114, 679–688. doi: 10.1016/j.marpolbul.2016.10.050
Bissett, A., Bowman, J., and Burke, C. (2006). Bacterial diversity in organically-enriched fish farm sediments. FEMS Microbiol. Ecol. 55, 48–56. doi: 10.1111/j.1574-6941.2005.00012.x
Bissett, A., Burke, C., Cook, P. L., and Bowman, J. P. (2007). Bacterial community shifts in organically perturbed sediments. Environ. Microbiol. 9, 46–60. doi: 10.1111/j.1462-2920.2006.01110.x
Bissett, A., Cook, P. L. M., Macleod, C., Bowman, J. P., and Burke, C. (2009). Effects of organic perturbation on marine sediment betaproteobacterial ammonia oxidizers and on benthic nitrogen biogeochemistry. Mar. Ecol. Prog. Ser. 392, 17–32. doi: 10.3354/meps08244
Campbell, B. J., Engel, A. S., Porter, M. L., and Takai, K. (2006). The versatile ε- proteobacteria: key players in sulphidic habitats. Nat. Rev. Microbiol. 4, 458–468. doi: 10.1038/nrmicro1414
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Carroll, M. L., Cochrane, S., Fieler, R., Velvin, R., and White, P. (2003). Organic enrichment of sediments from salmon farming in Norway: environmental factors, management practices, and monitoring techniques. Aquaculture 226, 165–180. doi: 10.1016/S0044-8486(03)00475-7
Caruso, G., Genovese, L., Mancuso, M., and Modica, A. (2003). Effects of fish farming on microbial enzyme activities and densities: comparison between three Mediterranean sites. Lett. Appl. Microbiol. 37, 324–328. doi: 10.1046/j.1472-765X.2003.01401.x
Chazdon, R. L., Chao, A., Colwell, R. K., Lin, S. Y., Norden, N., Letcher, S. G., et al. (2011). A novel statistical method for classifying habitat generalists and specialists. Ecology 92, 1332–1343. doi: 10.1890/10-1345.1
Chelossi, E., Vezzulli, L., Milano, A., Branzoni, M., Fabiano, M., Riccardi, G., et al. (2003). Antibiotic resistance of benthic bacteria in fish-farm and control sediments of the Western Mediterranean. Aquaculture 219, 83–97. doi: 10.1016/S0044-8486(03)00016-4
Chen, C. Q., Zheng, L., Zhou, J. L., and Zhao, H. (2017). Persistence and risk of antibiotic residues and antibiotic resistance genes in major mariculture sites in Southeast China. Sci. Total Environ. 580, 1175–1184. doi: 10.1016/j.scitotenv.2016.12.075
Christensen, P. B., Rysgaard, S., Sloth, N. P., Dalsgaard, T., and Schwaerter, S. (2000). Sediment mineralization, nutrient fluxes, denitrification and dissimilatory nitrate reduction to ammonium in an estuarine fjord with sea cage trout farms. Aquat. Microb. Ecol. 21, 73–84. doi: 10.3354/ame021073
Dang, H., Zhang, X., Song, L., Chang, Y., and Yang, G. (2006). Molecular characterizations of oxytetracycline resistant bacteria and their resistance genes from mariculture waters of China. Mar. Pollut. Bull. 52, 1494–1503. doi: 10.1016/j.marpolbul.2006.05.011
DeSantis, T. Z., Hugenholtz, P., Larsen, N., Rojas, M., Brodie, E. L., Keller, K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. doi: 10.1128/AEM.03006-05
Dowle, E., Pochon, X., Keeley, N., and Wood, S. A. (2015). Assessing the effects of salmon farming seabed enrichment using bacterial community diversity and high-throughput sequencing. FEMS Microbiol. Ecol. 91:fiv089. doi: 10.1093/femsec/fiv089
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Eiler, A., Heinrich, F., and Bertilsson, S. (2012). Coherent dynamics and association networks among lake bacterioplankton taxa. ISME J. 6, 330–342. doi: 10.1038/ismej.2011.113
Engel, A. S., Porter, M. L., Stern, L. A., Quinlan, S., and Bennett, P. C. (2004). Bacterial diversity and ecosystem function of filamentous microbial mats from aphotic (cave) sulfidic springs dominated by chemolithoautotrophic “Epsilonproteobacteria”. FEMS Microbiol. Ecol. 51, 31–53. doi: 10.1016/j.femsec.2004.07.004
Fernández-Gomez, B., Richter, M., Schüler, M., Pinhassi, J., Acinas, S. G., González, J. M., et al. (2013). Ecology of marine Bacteroidetes: a comparative genomics approach. ISME J. 7, 1026–1037. doi: 10.1038/ismej.2012.169
Fodelianakis, S., Papageorgiou, N., Karakassis, I., and Ladoukakis, E. D. (2015). Community structure changes in sediment bacterial communities along an organic enrichment gradient associated with fish farming. Ann. Microbiol. 65, 331–338. doi: 10.1007/s13213-014-0865-4
Galand, P. E., Lucas, S., Fagervold, S. K., Peru, E., Pruski, A. M., Vétion, G., et al. (2016). Disturbance increases microbial community diversity and production in marine sediments. Front. Microbiol. 7:1950. doi: 10.3389/fmicb.2016.01950
Garren, M., Smriga, S., and Azam, F. (2008). Gradients of coastal fish farm effluents and their effect on coral reefs. Environ. Microbiol. 10, 2299–2312. doi: 10.1111/j.1462-2920.2008.01654.x
Hargrave, B. T., Holmer, M., and Newcombe, C. P. (2008). Towards a classification of organic enrichment in marine sediments based on biogeochemical indicators. Mar. Pollut. Bull. 56, 810–824. doi: 10.1016/j.marpolbul.2008.02.006
Holmer, M., Black, K., Duarte, C. M., Marbà, N., and Karakassis, I. (2008). Aquaculture in the Ecosystem. Berlin: Springer, 326.
Holmer, M., Duarte, C. M., Heilskov, A., Olesen, B., and Terrados, J. (2003). Biogeochemical conditions in sediments enriched by organic matter from net-pen fish farms in the Bolinao area, Philippines. Mar. Pollut. Bull. 46, 1470–1479. doi: 10.1016/S0025-326X(03)00281-9
Hornick, K. M., and Buschmann, A. H. (2018). Insights into the diversity and metabolic function of bacterial communities in sediments from Chilean salmon aquaculture sites. Ann. Microbiol. 68, 63–77. doi: 10.1007/s13213-017-1317-8
Kawahara, N., Shigematsu, K., Miyadai, T., and Kondo, R. (2009). Comparison of bacterial communities in fish farm sediments along an organic enrichment gradient. Aquaculture 287, 107–113. doi: 10.1016/j.aquaculture.2008.10.003
Keeley, N. B., Forrest, B. M., and Macleod, C. K. (2013). Novel observations of benthic enrichment in contrasting flow regimes with implications for marine farm monitoring and management. Mar. Pollut. Bull. 66, 105–116. doi: 10.1016/j.marpolbul.2012.10.024
Klaoudatos, S. D., Klaoudatos, D. S., Smith, J., Bogdanos, K., and Papageorgiou, E. (2006). Assessment of site specific benthic impact of floating cage farming in the eastern Hios island, Eastern Aegean Sea, Greece. J. Exp. Mar. Biol. Ecol. 338, 96–111. doi: 10.1016/j.jembe.2006.07.002
Kormas, K. A., Meziti, A., Mente, E., and Frentzos, A. (2014). Dietary differences are reflected on the gut prokaryotic community structure of wild and commercially reared sea bream (Sparus aurata). Microbiologyopen 3, 718–728. doi: 10.1002/mbo3.202
La Rosa, T., Mirto, S., Marino, A., Alonzo, V., and Maugeri, T. L. (2001a). Heterotrophic bacteria community and pollution indicators of mussel-farm impact in the Gulf of Gaeta (Tyrrhenian Sea). Mar. Environ. Res. 52, 301–321. doi: 10.1016/S0141-1136(00)00272-5
La Rosa, T., Mirto, S., Mazzola, A., and Danovaro, R. (2001b). Differential responses of benthic microbes and meiofauna to fish-farm disturbance in coastal sediments. Environ. Pollut. 112, 427–434. doi: 10.1016/S0269-7491(00)00141-X
La Rosa, T., Mirto, S., Mazzola, A., and Maugeri, T. L. (2004). Benthic microbial indicators of fish farm impact in a coastal area of the Tyrrhenian Sea. Aquaculture 230, 153–167. doi: 10.1016/S0044-8486(03)00433-2
Lejzerowicz, F., Esling, P., Pillet, L., Wilding, T. A., Black, K. D., and Pawlowski, J. (2015). High-throughput sequencing and morphology perform equally well for benthic monitoring of marine ecosystems. Sci. Rep. 5:13932. doi: 10.1038/srep13932
Li, J., Li, F., Yu, S., Qin, S., and Wang, G. (2013). Impacts of mariculture on the diversity of bacterial communities within intertidal sediments in the northeast of China. Microb. Ecol. 66, 861–870. doi: 10.1007/s00248-013-0272-6
Luna, G. M., Corinaldesi, C., Dell’Anno, A., Pusceddu, A., and Danovaro, R. (2013). Impact of aquaculture on benthic virus-prokaryote interactions in the Mediterranean Sea. Water Res. 47, 1156–1168. doi: 10.1016/j.watres.2012.11.036
Mangano, M. C., Ape, F., and Mirto, S. (2019). The role of two non-indigenous serpulid tube worms in shaping artificial hard substrata communities: case study of a fish farm in the central Mediterranean Sea. Aquac. Environ. Interact. 11, 41–51. doi: 10.3354/aei00291
Manini, E., Fiordelmondo, C., Gambi, C., Pusceddu, A., and Danovaro, R. (2003). Benthic microbial loop functioning in coastal lagoons: a comparative approach. Oceanol. Acta 26, 27–38. doi: 10.1016/S0399-1784(02)01227-6
Mirto, S., Bianchelli, S., Gambi, C., Krzelj, M., Pusceddu, A., Scopa, M., et al. (2010). Fish-farm impact on metazoan meiofauna in the Mediterranean Sea: analysis of regional vs. habitat effects. Mar. Environ. Res. 69, 38–47. doi: 10.1016/j.marenvres.2009.07.005
Mirto, S., Gristina, M., Sinopoli, M., Maricchiolo, G., Genovese, L., Vizzini, S., et al. (2012). Meiofauna as an indicator for assessing the impact of fish farming at an exposed marine site. Ecol. Indic. 18, 468–476. doi: 10.1016/j.ecolind.2011.12.015
Mirto, S., La Rosa, T., Danovaro, R., and Mazzola, A. (2000). Microbial and meiofaunal response to intensive mussel-farm biodeposition in coastal sediments of the Western Mediterranean. Mar. Pollut. Bull. 40, 244–252. doi: 10.1016/S0025-326X(99)00209-X
Nikouli, E., Meziti, A., Antonopoulou, E., Mente, E., and Kormas, K. (2018). Gut bacterial communities in geographically distant populations of farmed sea bream (Sparus aurata) and sea bass (Dicentrarchus labrax). Microorganisms 6:92. doi: 10.3390/microorganisms6030092
Pawlowski, J., Esling, P., Lejzerowicz, F., Cedhagen, T., and Wilding, T. A. (2014). Environmental monitoring through protist next-generation sequencing metabarcoding: assessing the impact of fish farming on benthic foraminifera communities. Mol. Ecol. Resour. 14, 1129–1140. doi: 10.1111/1755-0998.12261
Polz, M. F., and Cavanaugh, C. M. (1995). Dominance of one bacterial phylotype at a Mid-Atlantic Ridge hydrothermal vent site. Proc. Natl. Acad. Sci. U.S.A. 92, 7232–7236. doi: 10.1073/pnas.92.16.7232
Pusceddu, A., Dell’Anno, A., Danovaro, R., Manini, E., Sara, G., and Fabiano, M. (2003). Enzymatically hydrolyzable protein and carbohydrate sedimentary pools as indicators of the trophic state of detritus sink systems: a case study in a Mediterranean coastal lagoon. Estuaries 26, 641–650. doi: 10.1007/BF02711976
Pusceddu, A., Fraschetti, S., Mirto, S., Holmer, M., and Danovaro, R. (2007). Effects of intensive mariculture on sediment biochemistry. Ecol. Appl. 17, 1366–1378. doi: 10.1890/06-2028.1
Quero, G. M., Cassin, D., Botter, M., Perini, L., and Luna, G. M. (2015). Patterns of benthic bacterial diversity in coastal areas contaminated by heavy metals, polycyclic aromatic hydrocarbons (PAHs) and polychlorinated biphenyls (PCBs). Front. Microbiol. 6:1053. doi: 10.3389/fmicb.2015.01053
Reimers, C. E., Alleau, Y., Bauer, J. E., Delaney, J., Girguis, P. R., Schrader, P. S., et al. (2013). Redox effects on the microbial degradation of refractory organic matter in marine sediments. Geochim. Cosmochim. Acta 121, 582–598. doi: 10.1016/j.gca.2013.08.004
Richardson, N. F., Ruesink, J. L., Naeem, S., Hacker, S. D., Tallis, H. M., Dumbauld, B. R., et al. (2008). Bacterial abundance and aerobic microbial activity across natural and oyster aquaculture habitats during summer conditions in a northeastern Pacific estuary. Hydrobiologia 596, 269–278. doi: 10.1007/s10750-007-9102-5
Rubio-Portillo, E., Villamor, A., Fernandez-Gonzalez, V., Antón, J., and Sanchez-Jerez, P. (2019). Exploring changes in bacterial communities to assess the influence of fish farming on marine sediments. Aquaculture 506, 459–464. doi: 10.1016/j.aquaculture.2019.03.051
Sarà, G., Gouhier, T. C., Brigolin, D., Porporato, E. M. D., Mangano, M. C., Mirto, S., et al. (2018). Predicting shifting sustainability trade-offs in marine finfish aquaculture under climate change. Glob. Change Biol. 24, 3654–3665. doi: 10.1111/gcb.14296
Schwarz, J. I., Eckert, W., and Conrad, R. (2008). Response of the methanogenic microbial community of a profundal lake sediment (Lake Kinneret, Israel) to algal deposition. Limnol. Oceanogr. 53, 113–121. doi: 10.4319/lo.2008.53.1.0113
Sinclair, L., Osman, O. A., Bertilsson, S., and Eiler, A. (2015). Microbial community composition and diversity via 16S rRNA gene amplicons: evaluating the Illumina platform. PLoS One 10:e0116955. doi: 10.1371/journal.pone.0116955
Stoeck, T., Frühe, L., Forster, D., Cordier, T., Martins, C. I., and Pawlowski, J. (2018). Environmental DNA metabarcoding of benthic bacterial communities indicates the benthic footprint of salmon aquaculture. Mar. Pollut. Bull. 127, 139–149. doi: 10.1016/j.marpolbul.2017.11.065
Sullam, K. E., Essinger, S. D., Lozupone, C. A., O’Connor, M. P., Rosen, G. L., Knight, R., et al. (2012). Environmental and ecological factors that shape the gut bacterial communities of fish: a meta-analysis. Mol. Ecol. 21, 3363–3378. doi: 10.1111/j.1365-294x.2012.05552.x
Tamminen, M., Karkman, A., Lohmus, A., Muziasari, W. I., Takasu, H., Wada, S., et al. (2011). Tetracycline resistance genes persist at aquaculture farms in the absence of selection pressure. Environ. Sci. Technol. 45, 386–391. doi: 10.1021/es102725n
Verhoeven, J. T. P., Salvo, F., Knight, R., Hamoutene, D., and Dufour, S. C. (2018). Temporal bacterial surveillance of salmon aquaculture sites indicates a long lasting benthic impact with minimal recovery. Front. Microbiol. 9:3054. doi: 10.3389/fmicb.2018.03054
Vezzulli, L., Chelossi, E., Riccardi, G., and Fabiano, M. (2002). Bacterial community structure and activity in fish farm sediments of the Ligurian Sea (Western Mediterranean). Aquac. Int. 10, 123–141. doi: 10.1023/A:1021365829687
Keywords: fish farm, seabream, seabass, microbial community, high-throughput sequencing, fish biomass, organic enrichment
Citation: Quero GM, Ape F, Manini E, Mirto S and Luna GM (2020) Temporal Changes in Microbial Communities Beneath Fish Farm Sediments Are Related to Organic Enrichment and Fish Biomass Over a Production Cycle. Front. Mar. Sci. 7:524. doi: 10.3389/fmars.2020.00524
Received: 31 October 2019; Accepted: 09 June 2020;
Published: 30 June 2020.
Edited by:
Jonathan Richir, Fonds National de la Recherche Scientifique (FNRS), BelgiumReviewed by:
M. Cristina Mangano, University of Naples Federico II, ItalySuzanne Dufour, Memorial University of Newfoundland, Canada
Copyright © 2020 Quero, Ape, Manini, Mirto and Luna. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Simone Mirto, simone.mirto@cnr.it
†These authors have contributed equally to this work
 Grazia Marina Quero
Grazia Marina Quero Francesca Ape
Francesca Ape Elena Manini1
Elena Manini1  Simone Mirto
Simone Mirto Gian Marco Luna
Gian Marco Luna