- 1STIMULUS (Research and Teaching Neuromodulation Uz Brussel) Research Group, Vrije Universiteit Brussel, Brussels, Belgium
- 2Department of Neurosurgery, Universitair Ziekenhuis Brussel, Brussels, Belgium
- 3Center for Neurosciences (C4N), Vrije Universiteit Brussel, Brussels, Belgium
- 4Pain in Motion (PAIN) Research Group, Department of Physiotherapy, Human Physiology and Anatomy, Faculty of Physical Education and Physiotherapy, Vrije Universiteit Brussel, Brussels, Belgium
- 5Research Foundation—Flanders (FWO), Brussels, Belgium
- 6Charles E. Schmidt College of Medicine, Florida Atlantic University, Boca Raton, FL, United States
- 7Department Microbiology and Infection Control, Universitair Ziekenhuis Brussel, Brussels, Belgium
- 8AIMS Lab, Center for Neurosciences, Faculty of Medicine and Pharmacy, Vrije Universiteit Brussel, Brussels, Belgium
- 9PRISMATICS Lab (Predictive Research in Spine/Neuromodulation Management and Thoracic Innovation/Cardiac Surgery), Poitiers University Hospital, Poitiers, France
- 10Department of Spine Surgery and Neuromodulation, Poitiers University Hospital, Poitiers, France
- 11Pprime Institute UPR 3346, CNRS, ISAE-ENSMA, University of Poitiers, Chasseneuil-du-Poitou, France
- 12Department of Radiology, Universitair Ziekenhuis Brussel, Brussels, Belgium
Introduction: Recent evidence supports the contribution of gut microbiota dysbiosis to the pathophysiology of rheumatic diseases, neuropathic pain, and neurodegenerative disorders. The bidirectional gut-brain communication network and the occurrence of chronic pain both involve contributions of the autonomic nervous system and the hypothalamic pituitary adrenal axis. Nevertheless, the current understanding of the association between gut microbiota and chronic pain is still not clear. Therefore, the aim of this study is to systematically evaluate the existing knowledge about gut microbiota alterations in chronic pain conditions.
Methods: Four databases were consulted for this systematic literature review: PubMed, Web of Science, Scopus, and Embase. The Newcastle-Ottawa Scale was used to assess the risk of bias. The study protocol was prospectively registered at the International prospective register of systematic reviews (PROSPERO, CRD42023430115). Alpha-diversity, β-diversity, and relative abundance at different taxonomic levels were summarized qualitatively, and quantitatively if possible.
Results: The initial database search identified a total of 3544 unique studies, of which 21 studies were eventually included in the systematic review and 11 in the meta-analysis. Decreases in alpha-diversity were revealed in chronic pain patients compared to controls for several metrics: observed species (SMD= -0.201, 95% CI from -0.04 to -0.36, p=0.01), Shannon index (SMD= -0.27, 95% CI from -0.11 to -0.43, p<0.001), and faith phylogenetic diversity (SMD -0.35, 95% CI from -0.08 to -0.61, p=0.01). Inconsistent results were revealed for beta-diversity. A decrease in the relative abundance of the Lachnospiraceae family, genus Faecalibacterium and Roseburia, and species of Faecalibacterium prausnitzii and Odoribacter splanchnicus, as well as an increase in Eggerthella spp., was revealed in chronic pain patients compared to controls.
Discussion: Indications for gut microbiota dysbiosis were revealed in chronic pain patients, with non-specific disease alterations of microbes.
Systematic review registration: https://www.crd.york.ac.uk/prospero/, identifier CRD42023430115.
1 Introduction
The gut microbiota refers to the dynamic community of microorganisms inhabiting the gastro-intestinal tract, whereby the genetic and functional profile of microbial species is denoted as the gut microbiome (1, 2). During the last decade, several studies pointed out associations between alterations in microbiota composition and diverse host disease conditions, among those gastrointestinal conditions [e.g., irritable bowel syndrome (3), gastroduodenal diseases (4)] as well as more physically remote conditions among which neurodegenerative diseases (e.g., Parkinson’s disease, Alzheimer’s disease, or multiple sclerosis) (5), or neuropsychiatric disorders (6). To accomplish these complex involvements, neuro-immune-endocrine mediators underlie the bidirectional communication network between the gut and the central nervous system, i.e. the gut-brain axis (7). As such, the gut-brain crosstalk ensures the proper maintenance of gastrointestinal homeostasis, while it also connects the emotional and cognitive centers of the brain with peripheral intestinal functions and mechanisms through immune activation, intestinal permeability, and entero-endocrine signaling (8).
The hypothalamic pituitary adrenal (HPA) axis, as part of the limbic system, is the core stress efferent axis that reacts with secretion of corticotropin-releasing factor from the hypothalamus in response to stressors of any kind (e.g., emotion or stress), consecutively leading to adrenocorticotropic hormone secretion from the pituitary gland, which in turn leads to cortisol release from the adrenal glands (9). While chronically elevated cortisol levels negatively affect brain function (10), HPA axis activation also alters the composition of the gut microbiota and increases gastrointestinal permeability (11), triggering an inflammatory response (12). Additionally, the autonomic nervous system drives both efferent signals from the central nervous system to the intestinal wall, mainly through vagal efferent fibers, and afferent signals from the lumen through enteric, spinal, and vagal pathways to the central nervous system (8). Unless the intestinal epithelium integrity is affected, whereby gut microbiota can directly interact with the vagal nerve, enteroendocrine cells recognize bacterial products or bacterial metabolites (e.g., short-chain fatty acids) to facilitate an indirect communication with vagal afferents through synaptic connections (13, 14). Additionally, production of bacterial metabolites (15), interference with the kynurenine pathway (16), and neuroendocrine signaling (17) contribute to the communication between the gut and the central nervous system.
Bidirectional interactions and connections between the pain regulatory system and the autonomic nervous system have been revealed (18), as well as altered sensitivity of the HPA axis in relation to chronic pain and stress (19), which are both suggestive of the involvement of the gut-brain axis in chronic pain due to shared pathways. Therefore, the aim of this study is to systematically evaluate the existing knowledge about gut microbiota alterations across a spectrum of chronic pain conditions.
2 Methods
2.1 Protocol registration
This systematic review was conducted according to the PRISMA statement (Preferred Reporting Items for Systematic Review and Meta-Analyses) (20). The protocol was a priori registered in PROSPERO under registration number CRD42023430115.
2.2 Search strategy
The search strategy was conducted in four databases: PubMed, Web of Science, Embase, and Scopus on June 3rd, 2023. All authors contributed to the development of the search strategy. The research question was created according to the PICOS (Population-Intervention-Control-Outcome-Study design) framework (21) to investigate perturbations in gut microbiota (Outcome) in chronic pain patients (Population). The final search strategy was built by combining both free and MeSH terms. Between each part of the PICO question, the Boolean operator AND was used. Within the components, search terms were combined using the Boolean operator OR. No limits were applied to this search strategy. The complete search strategy for PubMed can be found in Supplementary Datasheet 1. After building the search string in PubMed, it was individually adapted for the other three databases.
2.3 Eligibility criteria
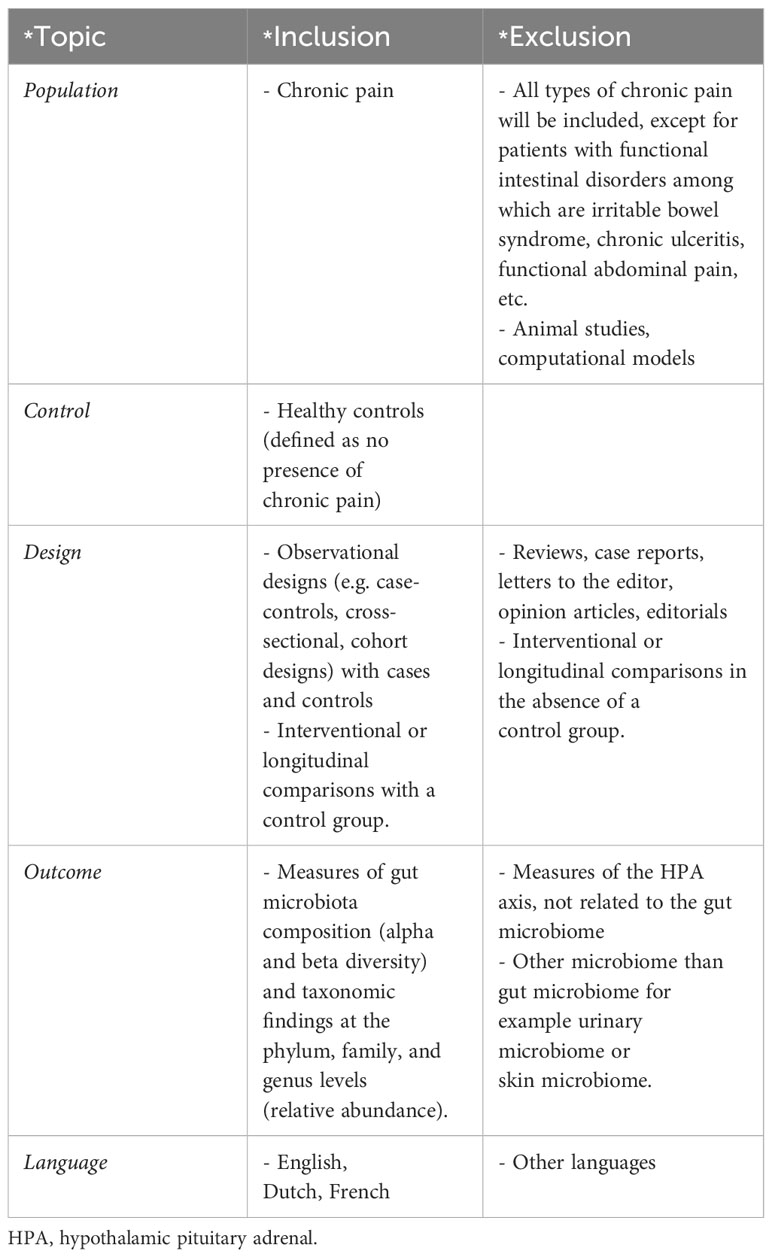
Studies evaluating gut microbiota in chronic pain patients, in comparison to controls, were eligible. All types of chronic pain [pain > 3 months according to ICD-11 criteria (22)] were included, with the exception of functional intestinal disorders. As study designs, both observational and experimental designs were allowed, as long as a control group of patients without chronic pain was included. Only studies exploring gut microbiota were incorporated. Studies reporting in languages other than English, Dutch, or French were excluded. Full eligibility criteria are presented in Table 1.
2.4 Study selection
Two reviewers independently screened all retrieved articles for title and abstract using online software Rayyan, after de-duplication in both EndNote X9 and Rayyan. During the next phase, two reviewers independently performed full text screening. In case of conflicts at each stage, they were resolved in a consensus meeting with a third reviewer.
2.5 Data extraction
The relevant data were selected by an a priori developed data extraction form with information on publication details, participant demographic and clinical characteristics, and methodological information. As outcomes of interest, community-level measures of gut microbiota composition (alpha- and beta-diversity) and taxonomic findings at the phylum, family, genus, and species levels (relative abundance) were extracted. The alpha-diversity refers to the variation within an individual sample (i.e. microbial community) with a differentiation between richness (i.e. number of species) and evenness (i.e. how well each species is represented), while beta-diversity refers to the variation between samples (2, 23). The data extraction table was composed by one reviewer and checked for correctness by another reviewer. Any sort of discrepancies were discussed in a consensus meeting between both reviewers.
2.6 Quality assessment
The methodological quality of the included studies was evaluated with the Newcastle-Ottawa Scale (NOS), a tool developed for the purposes of evaluating nonrandomized studies used in systematic reviews and meta-analyses (24, 25). This scale is designed to assess the selection of participants (four items), comparability (one item), and exposure (three items) domains. A total NOS score ≤ 5 was considered as low quality, a score of 6 or 7 as moderate quality, and a score of 8 or 9 as high quality (26).
2.7 Data synthesis
Differences in alpha-diversity, beta-diversity, and relative abundance were qualitatively presented for patients with chronic pain, compared to controls. Additionally, random-effect meta-analyses were performed for alpha-diversity metrics (e.g. observed species, Chao1, abundance coverage estimator, Pielou, Shannon index, Simpson index, inverse Simpson index, and faith phylogenetic diversity) between chronic pain patients and controls in case ≥2 effect sizes were available for a specific metric. Standardized mean difference (SMD) was selected as metric for the meta-analyses, with the following interpretation: SMD ≤ 0.2 as trivial, < 0.2 < SMD < 0.5 as small, 0.5 ≤ SMD < 0.8 as moderate, and SMD ≥ 0.8 as large (23, 27). In case the necessary information could not be extracted adequately, the study authors were contacted to request it. When the median with the first and third quartile or interquartile range was provided, the mean and standard deviation were calculated manually, according to formulas provided by Wan et al. (2014) (28). In addition, if data were expressed only as a graph (rather than numerical data within the text), the software Engauge Digitizer 12.1 was used to extract numerical values. Heterogeneity was evaluated with I² statistic and publication bias with Egger’s test. All analyses were performed in R Studio version 2022.07.2. P values <0.05 were considered statistically significant.
3 Results
3.1 Study selection
A total of 6285 articles were identified through the four selected databases (Figure 1). After removing all duplicates, 3544 articles were selected for screening. After screening on title and abstract, 37 articles remained eligible for full screening. The percentage of agreement on title and abstract screening between both reviewers was 99.8% (7 conflicts). The reasons for exclusion were wrong population (n=1693), followed by wrong study design (n=1331), wrong topic (n=224), wrong outcome (n=197), and to a lesser extent wrong publication type, foreign language, and no controls. Afterward, 2 articles were excluded because there was no full text available. Citation screening identified 12 additional articles of which 7 were deemed suitable for full text screening. After full-text screening (N=42), 21 articles were included in this systematic review. The percentage of agreement on full text screening between both reviewers was 83.78%.
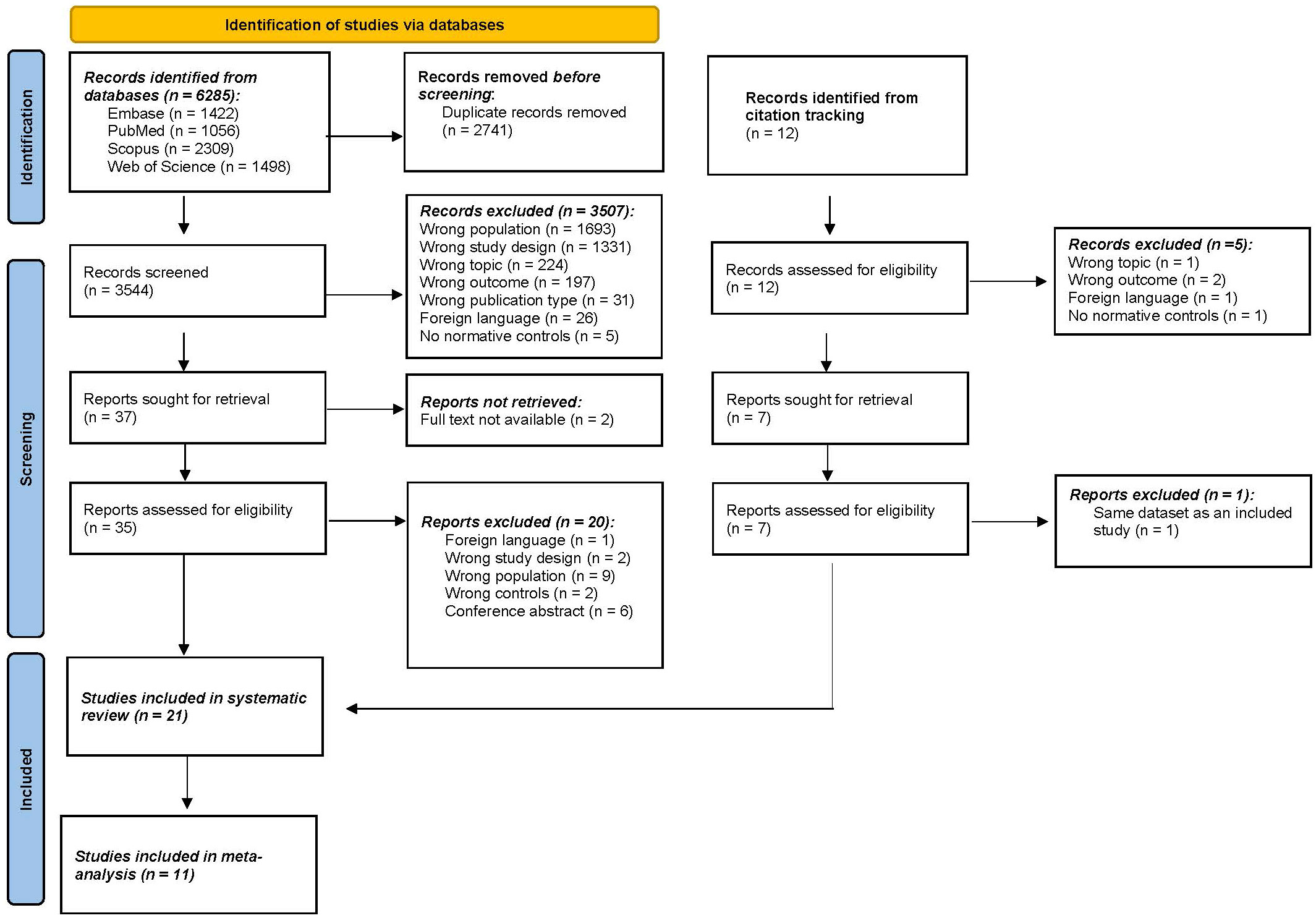
Figure 1 PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) flowchart. n, number.
3.2 Study characteristics
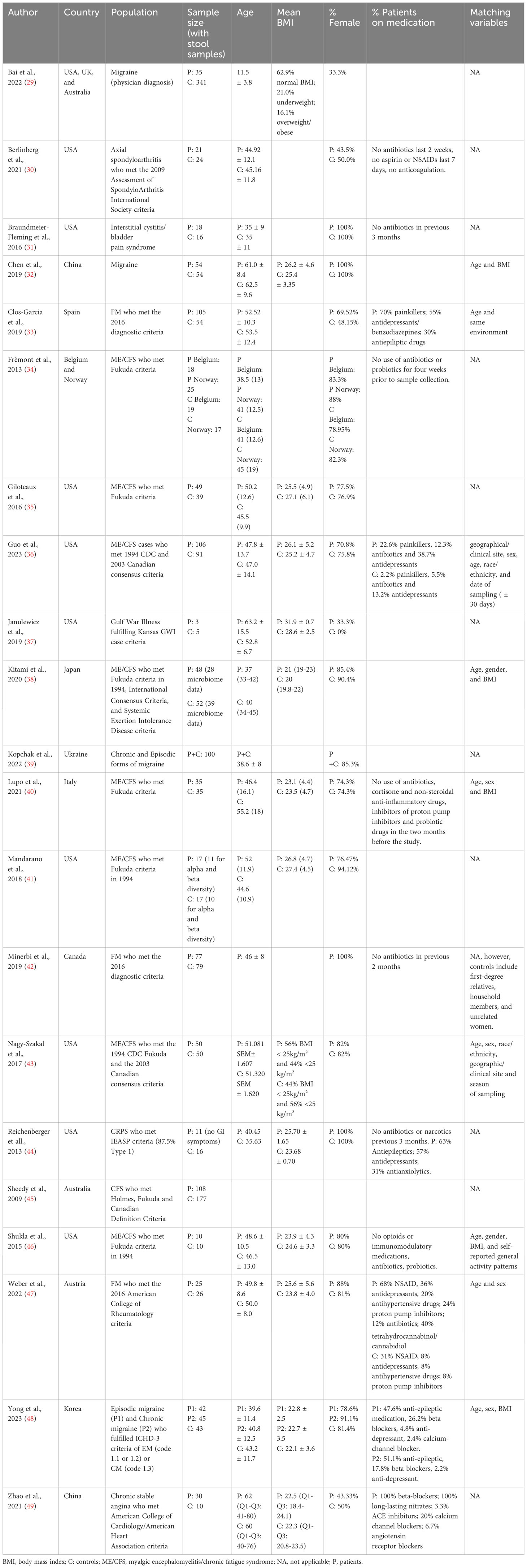
Characteristics of the included studies are presented in Table 2. Nine studies (42.8%) were conducted in the USA, four (19%) in Asian countries, four (19%) in European countries, one (4.8%) in Canada, one (4.8%) in Australia, one (4.8%) in Ukraine, and one in the USA, UK, and Australia (4.8%). In terms of chronic pain populations, 19 studies explored chronic primary pain syndromes (pain is conceived as a disease), while 2 evaluated chronic secondary pain syndromes (pain manifests as a symptom of another disease). Specifically, 9 (42.8%) studies evaluated myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS), 4 (19%) studies included patients with migraine, and 3 (14.3%) studies evaluated patients with fibromyalgia. The following conditions were explored in only one study: axial spondyloarthritis (4.8%), interstitial cystitis/bladder pain syndrome (4.8%), Gulf War Illness (4.8%), complex regional pain syndrome (CRPS) (4.8%), and chronic stable angina (4.8%). In total, data from 962 chronic pain patients and data from 1212 controls without chronic pain were included. Patients and controls were matched in 9 studies on the following variables: age (9 studies), sex (7 studies), BMI (5 studies), geographical site/environment (3 studies), race/ethnicity (2 studies), date of sampling (1 study), season of sampling (1 study), and general activity patterns (1 study). The NOS of the included studies ranged from 2-9, with 10 studies classified as low quality, 4 as moderate quality, and 7 as high quality (Supplementary Table 1).
3.3 Microbiome characteristics
After collection of samples, 14 studies (66.7%) froze the samples at -80°C until further use, 1 study (4.8%) at -70°C, 2 studies (9.5%) at -20°C, and it was not reported for 4 studies (19%). In terms of stool processing, a broad variety was observed (Supplementary Table 2). Only one study explored eukaryotes (41). In terms of sequencing, 14 studies conducted 16S sequencing, 3 studies shotgun metagenomics, 1 study paired-end metagenomic sequencing, 1 study 18S sequencing, and 2 studies did not report the sequencing. The 18S sequencing was performed at region V9, while the 16S sequencing was performed at regions V1-V2 (1 study), V2 (1 study), V3-V4 (4 studies), V3-V5 (1 study), V4 (4 studies), and V5-V6 (2 studies).
3.3.1 Alpha-diversity
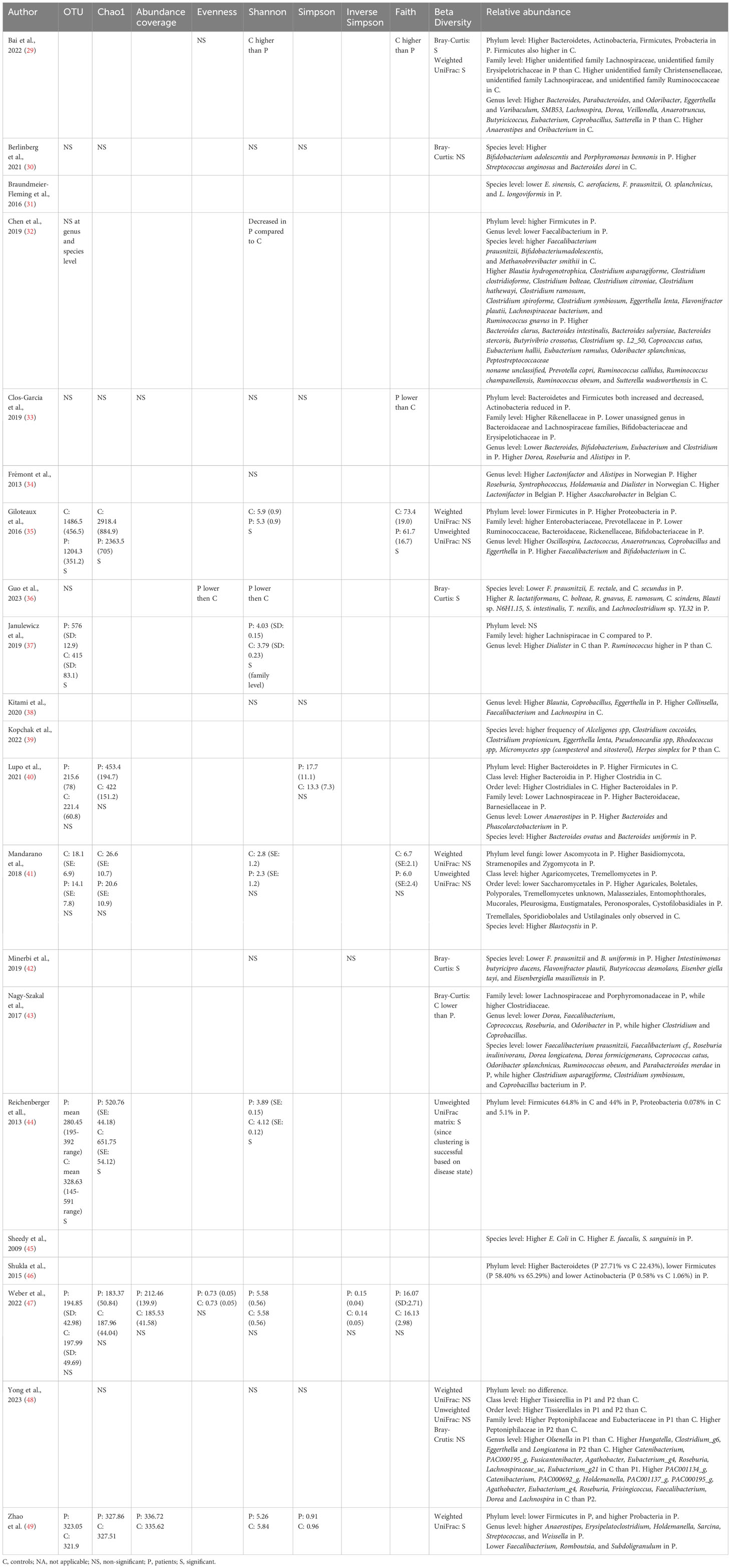
Sixteen studies provided data for alpha-diversity, evaluated through 8 different metrics. When evaluating richness through observed species, non-significant differences were revealed for patients with axial spondyloarthritis (30), ME/CFS (36, 40, 41), migraine (32), and fibromyalgia (33, 47) compared to controls. For patients with ME/CFS, only one study found significant differences with higher richness in controls compared to patients (35). Significantly increased values for observed species were found for patients with Gulf War Illness (37), while significantly decreased values for patients with CRPS (44) in relation to controls. Based on pooled estimates, a significant SMD of -0.201 (95% CI from -0.04 to -0.36, p=0.01, I²=41.9%, 11 effect sizes) was revealed, classified as a small effect size, pointing towards lower observed species numbers in chronic pain patients compared to controls (Figure 2). Egger’s test did not reveal indications for funnel plot asymmetry (t=0.9, df=9, p=0.39). For Chao1, significantly reduced values were obtained in patients with CRPS (44) and in one study with ME/CFS patients (35), while the other studies did not reveal significant differences between chronic pain patients and controls (30, 33, 40, 41, 47, 48). Non-significant results were revealed for the abundance coverage estimator (33, 47), as confirmed with a meta-analysis (SMD of -0.17 (95% CI from -0.44 to 0.10), p=0.22). For evenness, the Pielou metric resulted in significantly lower values in patients with ME/CFS compared to controls (36), while other reports did not reveal significant differences (29, 47). For richness/evenness, 15 studies explored the Shannon index with significant differences in favor of chronic pain patients (37), in favor of controls (29, 32, 35, 36, 44), and no significant difference between controls and chronic pain patients (30, 33, 34, 38, 41, 42, 47, 48). A random-effect meta-analysis resulted in a significantly decreased index in chronic pain patients compared to controls (p<0.001) with a small effect size (SMD -0.27, 95% CI from -0.11 to -0.43, 12 effect sizes, Egger’s Test t=0.25, df=10, p=0.81). Non-significant results were revealed for the Simpson index between chronic pain patients and controls (30, 33, 38, 40, 48), as was the case for the inverse Simpson index (42, 47). Faith phylogenetic diversity indicated increased values in controls in three studies (29, 33, 35), while two other studies revealed no significant differences (41, 47) between chronic pain patients and controls. A random-effect meta-analysis resulted in a significantly decreased index in chronic pain patients compared to controls (p=0.01) with a small effect size (SMD -0.35, 95% CI from -0.08 to -0.61, 4 effect sizes, Egger’s Test t=0.71, df=2, p=0.55). The meta-analysis for Chao1 and Pielou did not reveal significant differences between controls and chronic pain patients. The study of Zhao et al. (49) provided mean values for observed species, Chao1, abundance coverage, Shannon index, and Simpson index for patients with chronic stable angina compared to controls, however, it was not clear whether the results were significant. Therefore, these results were not qualitatively discussed, however, they are incorporated into the meta-analyses.
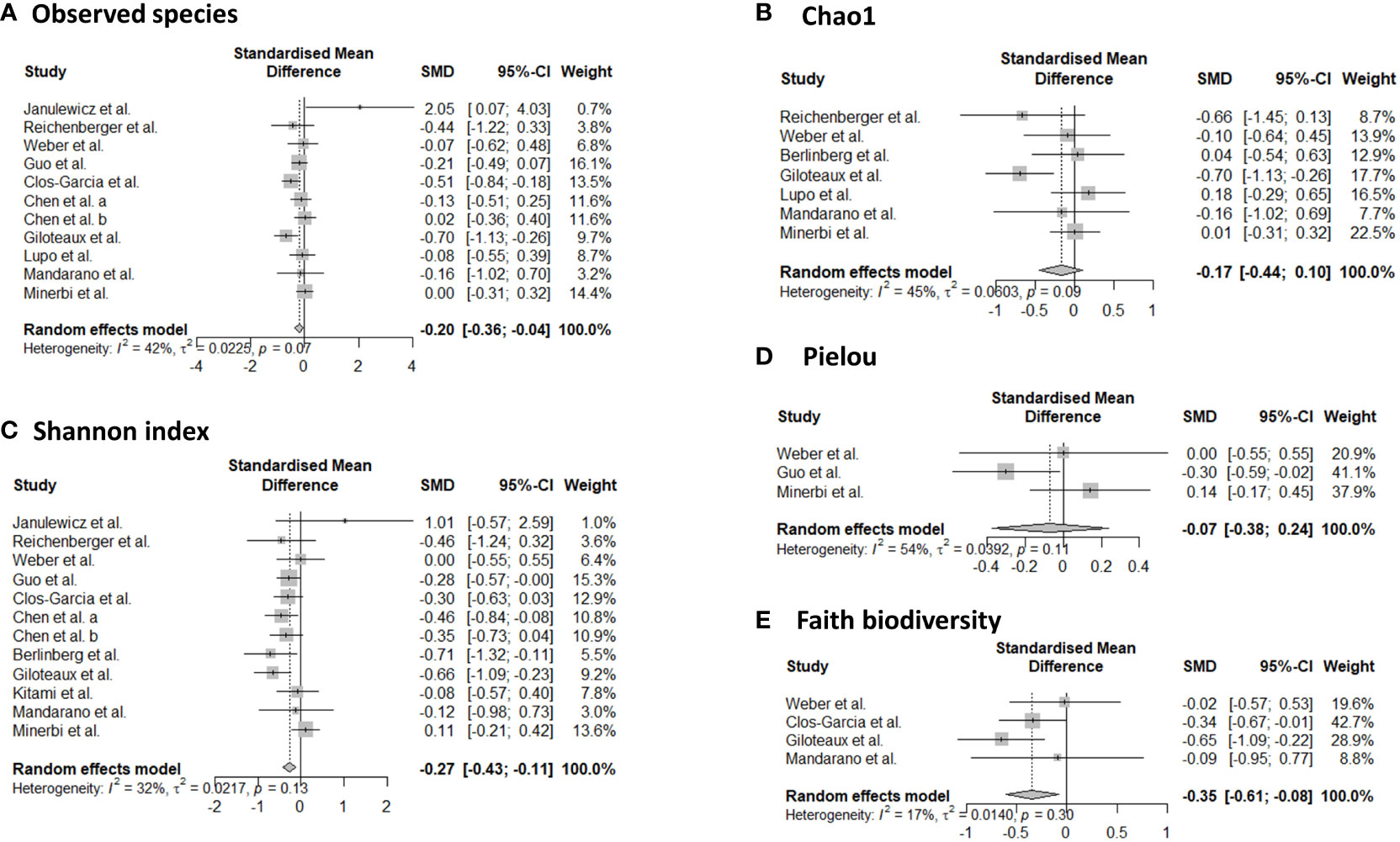
Figure 2 Forest plots of α-diversity metrics observed species (A), Chao1 (B), Shannon index (C), Pielou (D), and faith phylogenetic diversity (E). Standardized mean differences were used as effect sizes whereby a negative point estimate denotes a higher value in controls and a positive estimate a higher value in chronic pain patients.
3.3.2 Beta-diversity
Ten studies explored beta-diversity with the aid of three different metrics (Bray-Curtis, Weighted UniFrac, and Unweighted UniFrac) (29, 30, 35, 36, 41–44, 48, 49). In patients with migraine, inconsistent results were revealed with significant differences in beta-diversity according to Bai et al. (Bray-Curtis and Weighted UniFrac) (29) and non-significant results by Yong et al. (Bray-Curtis, Weighted UniFrac, and Unweighted UniFrac) (48). In patients with ME/CFS, two studies pointed towards significant differences in β-diversity, measured with Bray-Curtis, compared to healthy participants (36, 43), and two other studies did not reveal differences (35, 41). For patients with fibromyalgia (42), CRPS (44), and chronic stable angina (49), significant differences in beta-diversity were revealed, by one study for each condition. A non-significant result was revealed for patients with axial spondyloarthritis (30).
3.3.3 Differentially abundant microbes
Twenty out of twenty-one studies explored the relative abundance of gut microbes in chronic pain patients compared to controls (Table 3). Differences were found in 8 phyla, 14 families, 52 genera, and 73 species. An overview of the differences between the populations can be found in Table 4. At the phylum level, four main taxa were explored namely Actinobacteria (29, 33, 46), Bacteroidetes (29, 33, 40, 46), Firmicutes (29, 32, 33, 35, 40, 44, 46, 49), and Proteobacteria (29, 35, 44, 49). For Actinobacteria, Bacteroidetes, and Firmicutes both increases and decreases were revealed in chronic pain patients compared to controls, pointing towards inconsistent results. For Proteobacteria, a decrease was revealed in chronic pain patients compared to controls in all four studies (29, 35, 44, 49). Four fungal phyla were explored as well, with an increase in abundance in controls in Ascomycotae and decreased abundances in Basidiomycotae, Stramenopiles, and Zygomycota (41). At the family level, Lachnospiraceae were most often explored whereby 5 out of 6 studies indicated a decrease in relative abundance in chronic pain patients, compared to controls (29, 33, 37, 40, 43). At the genus level, Faecalibacterium spp. were most often explored, followed by Dorea spp., Eggerthella spp., and Roseburia spp. A decrease was found in Faecalibacterium spp. in patients with migraine (32, 48), ME/CFS (35, 38, 43) and chronic angina (49). For Dorea spp., inconsistent results were revealed for migraine patients (29, 48), an increase in patients with FM (33), and a decrease in patients with ME/CFS compared to controls (43). For Roseburia spp., 3 out of 4 studies revealed an increased relative abundance in controls (34, 43, 48), while one study revealed an increase in patients with fibromyalgia (33). In the genus Eggerthella, an increased relative abundance was found in patients with migraine (29, 48) and ME/CFS (35, 38). At the species level, a decrease in the relative abundance of Faecalibacterium prausnitzii was revealed for patients with migraine (32), ME/CFS (36, 43), fibromyalgia (42), and bladder pain syndrome (31). Odoribacter splanchnicus had a lower abundance in patients with migraine (32), ME/CFS (43), and bladder pain syndrome (31). Clostridium asparagiforme and Clostridium symbiosum increased in patients with migraine and ME/CFS, while Coprococcus catus and Ruminococcus obeum decreased in these patients (32, 43). Flavonifractor plautii had an increased abundance in patients with migraine and fibromyalgia (32, 42). Finally, Eggerthella lenta also increased in patients with migraine (32, 39).
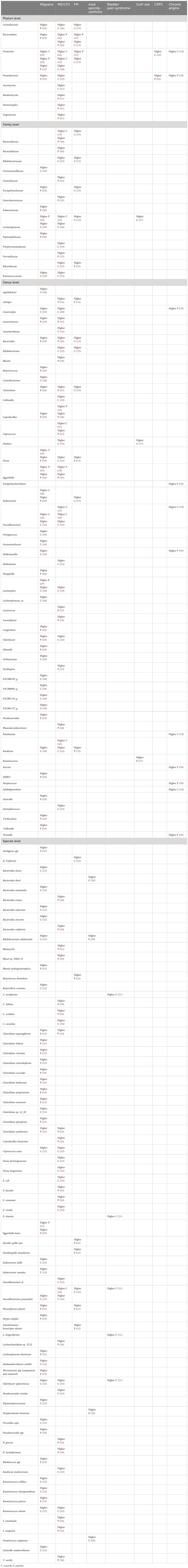
Table 4 Changes in relative abundance of microbes in chronic pain patients compared to controls at phylum, family, genus and species level.
4 Discussion
This study evaluated alterations in gut microbiota composition in chronic pain patients compared to controls. In terms of alpha-diversity, the richness metric observed species indicated a significantly decreased number of unique operational taxonomic units in chronic pain patients. Additionally, a lower Shannon index and faith phylogenetic diversity were revealed in patients compared to controls. For beta-diversity, inconclusive results were revealed. Finally, there was a decreased relative abundance of Lachnospiraceae in 83% of studies that evaluated this family in chronic pain patients compared to controls. A decreased abundance of Faecalibacterium prausnitzii and Odoribacter splanchnicus species was demonstrated in patients compared to controls. Based on this systematic review, with complementary meta-analyses, there are indications for dysbiosis of gut microbiota in chronic pain patients.
The interest in gut microbiota as a potential underlying factor of disease maintenance has drastically increased during the last decade. Gut dysbiosis is expected to contribute to the etiology of, e.g., inflammatory bowel disease (50, 51), type 2 diabetes (52), colorectal cancer (53, 54), hypertension (55), and rheumatic diseases (23), besides its modulating role in chronic pain (56). The mechanisms by which acute infectious pain becomes chronic are very diverse and can include, among others, molecular mimicry (structural similarity between microbial and host molecules which could induce autoimmune responses), bystander activation, or microbe invasion (57, 58). Specific microbes such as Borrelia species and Mycobacterium leprae or viruses (e.g., HIV, SARS-Cov-2) are associated with a high incidence of chronic pain (57). A cross-disease meta-analysis was previously performed, whereby consistent patterns characterizing disease-associated microbiome changes were revealed (59). Some diseases were characterized by the presence of potentially pathogenic microbes, whereas others revealed a depletion of health-associated bacteria (59). About half of the genera associated with individual studies were bacteria that respond to more than one disease, supporting the hypothesis of non-disease-specific alterations but shared alterations (i.e. non-specific response) to health and disease (59). Based on this hypothesis, the current systematic review and meta-analysis was conducted in patients with chronic pain, regardless of the underlying disease condition.
Gut microbiome alpha-diversity has been associated with human health, whereby reduced levels are indicative of acute and chronic diseases (60). Alpha-diversity metrics provide summary statistics that focus on summarizing the breadth of diversity present in an environment (61). The current study indicated a decrease in alpha-diversity in patients with chronic pain compared to controls, as reflected in several metrics namely, a decreased number of unique operational taxonomic units, a decreased Shannon index [which is a popular diversity index in the ecological field to reflect the richness of bacterial community (62)], and a decreased Faith’s phylogenetic diversity in chronic pain patients. Faith’s phylogenetic diversity accounts for the phylogenetic relatedness of community members and has been denoted as more sensitive to distinguishing disease factors relative to other alpha diversity metrics (63). Despite the small effect sizes, these alpha-diversity metrics all point towards a decreased richness in chronic pain patients, which may point out the need for nutritional interventions in patients with chronic pain. The gut microbiota produces polyamines, which in turn excites N-methyl-D-aspartate receptors, a crucial factor of central nervous system sensitization (64), which is common in patients with chronic pain (65).
A reduction in the relative abundance of the Lachnospiraceae family was found in patients with chronic pain. All Lachnospiraceae members are anaerobic, fermentative and chemoorganotrophic, and are already present in early infancy (66). Aging is associated with increases in Lachnospiraceae abundance (67). The genera Blautia and Roseburia, belonging to the Lachnospiraceae family, are often associated with a healthy state (68). These genera are the main short-chain fatty acid (SCFA) producers [whereby SCFA activity modulates the surrounding microbial environment and interacts with the host immune system (69)] and are involved in the control of gut inflammatory processes, and maturation of the immune system (66, 70). A higher relative abundance of Roseburia ssp. was revealed in controls compared to chronic pain patients, highlighting the value of this genus in health states. Additionally, a decrease in the relative abundance of Odoribacter splanchnicus, another common SCFA-producing member of the human intestinal microbiota (71), was found in chronic pain patients. This finding was previously also described in patients with inflammatory bowel disease (72, 73).
Another finding was a decreased relative abundance of the Faecalibacterium genus, belonging to the family Ruminococcaceae, which comprises only one validated species, namely Faecalibacterium prausnitzii (74). A decrease in Faecalibacterium prausnitzii was observed in chronic pain patients, a species known to play a crucial role in host wellbeing and gut physiology (75). It is one of the main butyrate producers in the intestine (76), whereby butyrate is involved in maintaining mucosal integrity, alleviating inflammation (via macrophage function as well as a reduction in proinflammatory cytokines), and increasing anti-inflammatory mediators (77). Thus, this species is known for its anti-inflammatory properties (75). In murine models, it was revealed that Faecalibacterium prausnitzii cells could reduce the severity of both acute, chronic, and chemical-induced inflammation (78–80). Faecalibacterium prausnitzii depletion has been reported in adults with Crohn’s disease, ulcerative colitis, and colorectal cancer (81–84), as well as in patients with rheumatic disorders (23, 85) and is proposed as a biomarker to discriminate between gut disorders and healthy subjects (75). This alteration may not be specific to inflammatory diseases and may be a more generic phenomenon of disease states since it is also revealed in chronic pain patients.
Combining these findings, it seems that SCFAs [mainly composed of acetic acid, propionic acid, and butyric acid (86)] play an important role in the context of chronic pain (Figure 3). There are two main mechanisms through which SCFAs can enter cells and consequently alter inflammation, namely cell signal transduction and passive diffusion combined with transport proteins. The latter functions through sodium-coupled monocarboxylate transport 1/2 (SMCT1/2), Na+ coupled transporters in the apical membrane of colonic epithelium, and monocarboxylate transporter 1/4 (MCT1/4), H+ coupled transporters mainly expressed in the apical and basolateral membrane of the colonic epithelium (89). Once SCFAs enter the cell through passive diffusion or transporters, they inhibit histone deacetylation (86). In dendritic cells and macrophages, inhibition of histone deacetylation is the main pathway to exert anti-inflammatory effects, while in neutrophils and monocytes, SCFAs inhibit tumor necrosis factor expression, the NF-κB signaling pathway, and histone deacetylase in addition to promoting interleukin-10 production as an anti-inflammatory cytokine. Cell signal transduction is realized by SCFAs through G protein-coupled cell membrane receptors GPR109A, GPR43, and GPR41 (90, 91). In macrophages, butyrate activates GPR41 to down-regulate pro-inflammatory factors among which are nitric oxide synthase, tumor necrosis factor, interleukin 6, and monocyte chemoattractant protein-1 (92). In macrophages and neutrophils, SCFAs down-regulate interleukin 8 expression through activation of GPR43 and GPR41 (93). Finally, SCFAs can also regulate inflammation by activating anti-inflammatory signaling pathways by inhibiting histone deacetylase (86). Besides the role of SCFAs in inflammation, they also regulate the differentiation of T cells and B cells and regulate the function of innate immune cells among which are macrophages, neutrophils, and dendritic cells (86).
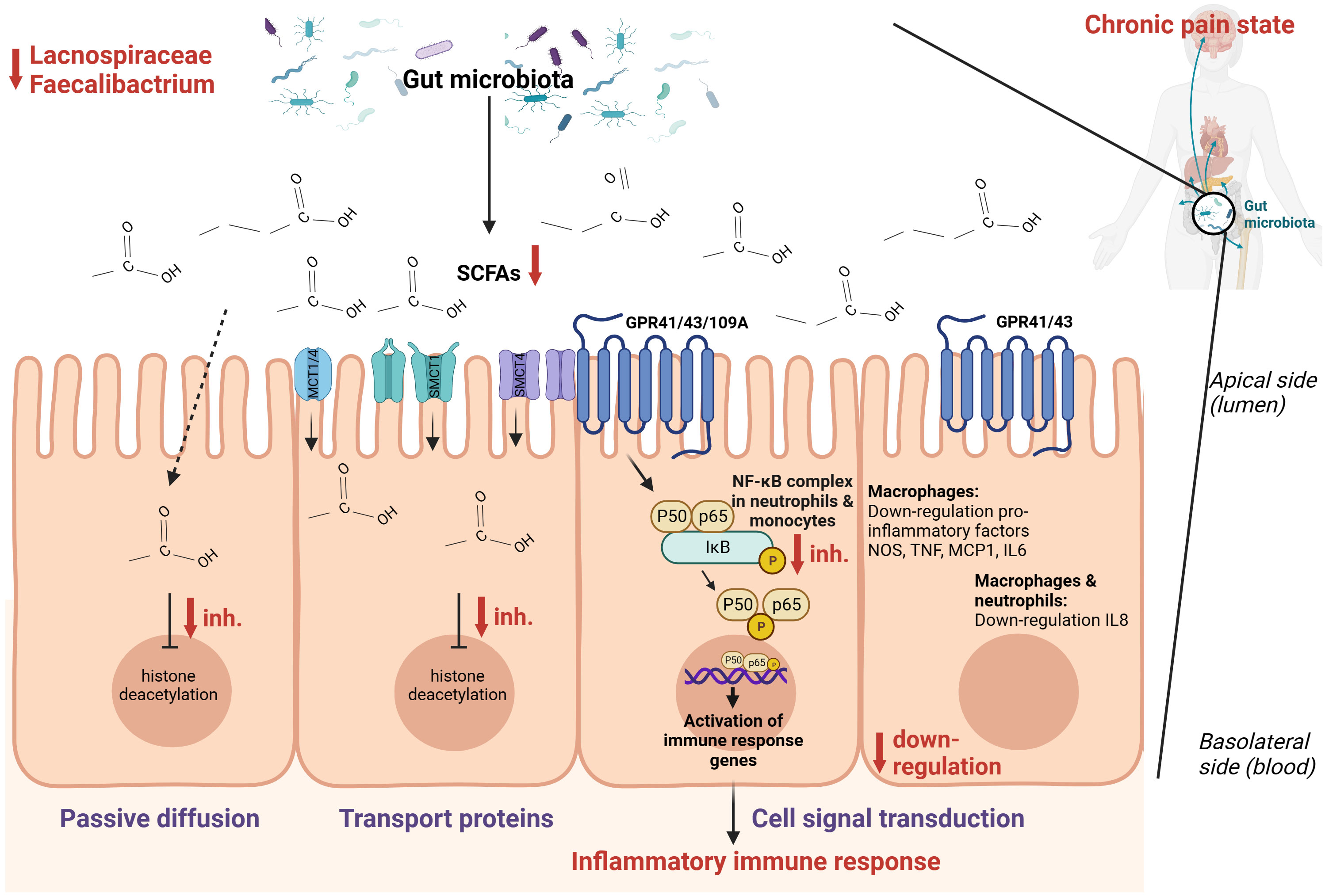
Figure 3 Hypothesized schematic representation of the role of short-chain fatty acids (SCFAs) in the regulation of gut and systemic immunity in relation to chronic pain (86–88). SCFAs can regulate inflammation through cell signal transduction by binding at G-protein coupled receptors GPR109A, GPR43, and GPR41 and down-regulate the NOS, TNF, MCP-1, IL-6, IL-8, and the NF-κB signaling pathway. Through passive diffusion and transport proteins (MCT1, MCT4, SMCT1, SMCT2), SCFAs can inhibit histone deacetylase. This is a simplified representation of the pathways involved in inflammation with the pathways expected to be relevant in the setting of chronic pain.
This study evaluated gut microbiome alterations in chronic pain patients compared to controls without chronic pain. Studies from different parts of the world were included among which were the USA, Europe, Asia, and Australia. There is no universal healthy gut microbiota (94, 95), since nationality and food preferences, among other factors, induce an influence on the gut microbiota. For example, the gut microbiome of a healthy European (including Slavic nationality) is characterized by the dominance of the phyla Firmicutes, Bacteroidota, Actinobacteria, Proteobacteria, Fusobacteria, and Verrucomicrobia, while the gut microbiome of Asians is very diverse and rich in members of the genera Prevotella, Bacteroides Lactobacillus, Faecalibacterium, Ruminococcus, Subdoligranulum, Coprococcus, Collinsella, Megasphaera, Bifidobacterium, and Phascolarctobacterium (96). Therefore, this study only included studies that compared gut microbiota to a control group to limit the influence of local differences in gut microbiota composition.
The field of chronic pain and gut microbiota composition is still in its infancy, wherefore condition-specific alterations remain to be elucidated when more research is available, in case the hypothesis of shared alterations is not valid in pain settings. The majority of studies explored chronic primary pain syndromes, wherefore gut dysbiosis in chronic secondary pain syndromes still needs to be explored in more detail. When interpreting the results of this study, it should be taken into account that medication was previously denoted as an important covariate, and more specifically antibiotics, osmotic laxatives, inflammatory bowel disease medication, female hormones, benzodiazepines, antidepressants, and antihistamines (60). Recently, a multi-omics analysis elaborated on the concept of opioid-induced dysbiosis in gut microbiota (97), which further supports the hypothesis of addressing the gut-brain axis in patients with chronic pain, especially in those patients who take opioids as pain medication. Medication use was reported for every individual study, however, it was not possible to take a numerical output for medication use into account in the conducted meta-analysis. As revealed by this review, there is no common pipeline to conduct laboratory analyses, statistical evaluations, or quality assurance for gut microbiome data. Future steps should be conducted towards harmonization of processing gut microbiome data to ensure better comparability of the results.
5 Conclusions
This review pointed towards the potential value of dysbiosis in chronic pain patients, with non-specific disease alterations of microbes.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
LG: Conceptualization, Formal analysis, Methodology, Writing – original draft, Writing – review & editing. TD: Formal analysis, Writing – review & editing. JP: Investigation, Writing – review & editing. MB: Investigation, Writing – review & editing. MR: Investigation, Writing – review & editing. PR: Investigation, Writing – review & editing. MM: Conceptualization, Formal analysis, Investigation, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
The authors would like to thank Jonas Callens for serving as the second reviewer during the screening of titles and abstracts.
Conflict of interest
LG is a postdoctoral research fellow funded by the Research Foundation Flanders FWO, Belgium project number 12ZF622N. JP received grant support from Medtronic, Boston Scientific, Abbott, NIH 2R01CA166379-06, NIH R01EB030324, NIH Blueprint 3U54EB015408 and NIH U44NS115111. She is part of the Board of Directors of Facial Pain Association, Board of Directors at Large of International Neuromodulation Society, President Elect of American Society of Stereotactic and Functional Neurosurgery and President of North American Neuromodulation Society. PR reports grants from Medtronic, Abbott and Boston Scientific and consultant fees and payments for lectures from Medtronic and Boston Scientific, outside the submitted work. MM has received speaker fees from Medtronic, Saluda and Nevro. STIMULUS received independent research grants from Medtronic.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1342833/full#supplementary-material
References
1. Ursell LK, Metcalf JL, Parfrey LW, Knight R. Defining the human microbiome. Nutr Rev (2012) 70 Suppl 1:S38–44. doi: 10.1111/j.1753-4887.2012.00493.x
2. Al Bander Z, Nitert MD, Mousa A, Naderpoor N. The gut microbiota and inflammation: an overview. Int J Environ Res Public Health (2020) 17(20):7618. doi: 10.3390/ijerph17207618
3. Zhao Y, Zou DW. Gut microbiota and irritable bowel syndrome. J Dig Dis (2023) 24(5):312–20. doi: 10.1111/1751-2980.13204
4. Sharma P, Phatak SM, Warikoo P, Mathur A, Mahant S, Das K, et al. Crosstalk between Helicobacter pylori and gastrointestinal microbiota in various gastroduodenal diseases-A systematic review. 3 Biotech (2023) 13:303. doi: 10.1007/s13205-023-03734-5
5. Yadav H, Jaldhi, Bhardwaj R, Anamika, Bakshi A, Gupta S, et al. Unveiling the role of gut-brain axis in regulating neurodegenerative diseases: A comprehensive review. Life Sci (2023) 330:122022. doi: 10.1016/j.lfs.2023.122022
6. Anand N, Gorantla VR, Chidambaram SB. The role of gut dysbiosis in the pathophysiology of neuropsychiatric disorders. Cells (2022) 12(1):54. doi: 10.3390/cells12010054
7. Montagnani M, Bottalico L, Potenza MA, Charitos IA, Topi S, Colella M, et al. The crosstalk between gut microbiota and nervous system: A bidirectional interaction between microorganisms and metabolome. Int J Mol Sci (2023) 24(12):10322. doi: 10.3390/ijms241210322
8. Carabotti M, Scirocco A, Maselli MA, Severi C. The gut-brain axis: interactions between enteric microbiota, central and enteric nervous systems. Ann Gastroenterol (2015) 28:203–9.
9. Farzi A, Fröhlich EE, Holzer P. Gut microbiota and the neuroendocrine system. Neurotherapeutics (2018) 15:5–22. doi: 10.1007/s13311-017-0600-5
10. Liu B, Liu J, Wang M, Zhang Y, Li L. From serotonin to neuroplasticity: evolvement of theories for major depressive disorder. Front Cell Neurosci (2017) 11:305. doi: 10.3389/fncel.2017.00305
11. de Punder K, Pruimboom L. Stress induces endotoxemia and low-grade inflammation by increasing barrier permeability. Front Immunol (2015) 6:223. doi: 10.3389/fimmu.2015.00223
12. Kelly JR, Kennedy PJ, Cryan JF, Dinan TG, Clarke G, Hyland NP. Breaking down the barriers: the gut microbiome, intestinal permeability and stress-related psychiatric disorders. Front Cell Neurosci (2015) 9:392. doi: 10.3389/fncel.2015.00392
13. Han Y, Wang B, Gao H, He C, Hua R, Liang C, et al. Vagus nerve and underlying impact on the gut microbiota-brain axis in behavior and neurodegenerative diseases. J Inflammation Res (2022) 15:6213–30. doi: 10.2147/JIR.S384949
14. Silva YP, Bernardi A, Frozza RL. The role of short-chain fatty acids from gut microbiota in gut-brain communication. Front Endocrinol (Lausanne) (2020) 11:25. doi: 10.3389/fendo.2020.00025
15. Masse KE, Lu VB. Short-chain fatty acids, secondary bile acids and indoles: gut microbial metabolites with effects on enteroendocrine cell function and their potential as therapies for metabolic disease. Front Endocrinol (Lausanne) (2023) 14:1169624. doi: 10.3389/fendo.2023.1169624
16. Kennedy PJ, Cryan JF, Dinan TG, Clarke G. Kynurenine pathway metabolism and the microbiota-gut-brain axis. Neuropharmacology (2017) 112:399–412. doi: 10.1016/j.neuropharm.2016.07.002
17. Yu Y, Yang W, Li Y, Cong Y. Enteroendocrine cells: sensing gut microbiota and regulating inflammatory bowel diseases. Inflamm Bowel Dis (2020) 26:11–20. doi: 10.1093/ibd/izz217
18. Forte G, Troisi G, Pazzaglia M, Pascalis V, Casagrande M. Heart rate variability and pain: A systematic review. Brain Sci (2022) 12(2):153. doi: 10.3390/brainsci12020153
19. Nees F, Löffler M, Usai K, Flor H. Hypothalamic-pituitary-adrenal axis feedback sensitivity in different states of back pain. Psychoneuroendocrinology (2019) 101:60–6. doi: 10.1016/j.psyneuen.2018.10.026
20. Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gøtzsche PC, Ioannidis JPA, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ (2009) 339:b2700. doi: 10.1136/bmj.b2700
21. O'Sullivan D, Wilk S, Michalowski W, Farion K. Using PICO to align medical evidence with MDs decision making models. Stud Health Technol Inform (2013) 192:1057.
22. Treede RD, Rief W, Barke A, Aziz Q, Bennett MI, Benoliel R, et al. A classification of chronic pain for ICD-11. Pain (2015) 156:1003–7. doi: 10.1097/j.pain.0000000000000160
23. Wang Y, Wei J, Zhang W, Doherty M, Zhang Y, Xie H, et al. Gut dysbiosis in rheumatic diseases: A systematic review and meta-analysis of 92 observational studies. EBioMedicine (2022) 80:104055. doi: 10.1016/j.ebiom.2022.104055
24. Sanderson S, Tatt ID, Higgins JP. Tools for assessing quality and susceptibility to bias in observational studies in epidemiology: a systematic review and annotated bibliography. Int J Epidemiol (2007) 36:666–76. doi: 10.1093/ije/dym018
25. Wells G, Shea B, O'connell D, Peterson J, Welch V, Losos M, et al. Quality assessment scales for observational studies. Ottawa Health Res Institute (2004).
26. Bechard LJ, Rothpletz-Puglia P, Touger-Decker R, Duggan C, Mehta NM. Influence of obesity on clinical outcomes in hospitalized children: a systematic review. JAMA Pediatr (2013) 167:476–82. doi: 10.1001/jamapediatrics.2013.13
27. Nikolova VL, Smith MRB, Hall LJ, Cleare AJ, Stone JM, Young AH. Perturbations in gut microbiota composition in psychiatric disorders: A review and meta-analysis. JAMA Psychiatry (2021) 78:1343–54. doi: 10.1001/jamapsychiatry.2021.2573
28. Wan X, Wang W, Liu J, Tong T. Estimating the sample mean and standard deviation from the sample size, median, range and/or interquartile range. BMC Med Res Methodol (2014) 14:135. doi: 10.1186/1471-2288-14-135
29. Bai J, Shen N, Liu Y. Associations between the gut microbiome and migraines in children aged 7-18 years: an analysis of the american gut project cohort. Pain Manage Nurs (2023) 24:35–43. doi: 10.1016/j.pmn.2022.06.002
30. Berlinberg AJ, Regner EH, Stahly A, Brar A, Reisz JA, Gerich ME, et al. Multi 'Omics analysis of intestinal tissue in ankylosing spondylitis identifies alterations in the tryptophan metabolism pathway. Front Immunol (2021) 12:587119. doi: 10.3389/fimmu.2021.587119
31. Braundmeier-Fleming A, Russell NT, Yang W, Nas MY, Yaggie RE, Berry M, et al. Stool-based biomarkers of interstitial cystitis/bladder pain syndrome. Sci Rep (2016) 6:26083. doi: 10.1038/srep26083
32. Chen J, Wang Q, Wang A, Lin Z. Structural and functional characterization of the gut microbiota in elderly women with migraine. Front Cell Infect Microbiol (2019) 9:470. doi: 10.3389/fcimb.2019.00470
33. Clos-Garcia M, Andrés-Marin N, Fernández-Eulate G, Abecia L, Lavín JL, van Liempd S, et al. Gut microbiome and serum metabolome analyses identify molecular biomarkers and altered glutamate metabolism in fibromyalgia. EBioMedicine (2019) 46:499–511. doi: 10.1016/j.ebiom.2019.07.031
34. Frémont M, Coomans D, Massart S, De Meirleir K. High-throughput 16S rRNA gene sequencing reveals alterations of intestinal microbiota in myalgic encephalomyelitis/chronic fatigue syndrome patients. Anaerobe (2013) 22:50–6. doi: 10.1016/j.anaerobe.2013.06.002
35. Giloteaux L, Goodrich JK, Walters WA, Levine SM, Ley RE, Hanson MR. Reduced diversity and altered composition of the gut microbiome in individuals with myalgic encephalomyelitis/chronic fatigue syndrome. Microbiome (2016) 4:30. doi: 10.1186/s40168-016-0171-4
36. Guo C, Che X, Briese T, Ranjan A, Allicock O, Yates RA, et al. Deficient butyrate-producing capacity in the gut microbiome is associated with bacterial network disturbances and fatigue symptoms in ME/CFS. Cell Host Microbe (2023) 31:288–304.e8. doi: 10.1016/j.chom.2023.01.004
37. Janulewicz PA, Seth RK, Carlson JM, Ajama J, Quinn E, Heeren T, et al. The gut-microbiome in gulf war veterans: A preliminary report. Int J Environ Res Public Health (2019) 16(19):3751. doi: 10.3390/ijerph16193751
38. Kitami T, Fukuda S, Kato T, Yamaguti K, Nakatomi Y, Yamano E, et al. Deep phenotyping of myalgic encephalomyelitis/chronic fatigue syndrome in Japanese population. Sci Rep (2020) 10:19933. doi: 10.1038/s41598-020-77105-y
39. Kopchak OO, Hrytsenko OY, Pulyk OR. Peculiarities of the gut microbiota in patients with migraine comparing to healthy individuals. Wiad Lek (2022) 75:2218–21. doi: 10.36740/WLek202209207
40. Lupo GFD, Rocchetti G, Lucini L, Lorusso L, Manara E, Bertelli M, et al. Potential role of microbiome in Chronic Fatigue Syndrome/Myalgic Encephalomyelits (CFS/ME). Sci Rep (2021) 11:7043. doi: 10.1038/s41598-021-86425-6
41. Mandarano AH, Giloteaux L, Keller BA, Levine SM, Hanson MR. Eukaryotes in the gut microbiota in myalgic encephalomyelitis/chronic fatigue syndrome. PeerJ (2018) 6:e4282. doi: 10.7717/peerj.4282
42. Minerbi A, Gonzalez E, Brereton NJB, Anjarkouchian A, Dewar K, Fitzcharles MA, et al. Altered microbiome composition in individuals with fibromyalgia. Pain (2019) 160:2589–602. doi: 10.1097/j.pain.0000000000001640
43. Nagy-Szakal D, Williams BL, Mishra N, Che X, Lee B, Bateman L, et al. Fecal metagenomic profiles in subgroups of patients with myalgic encephalomyelitis/chronic fatigue syndrome. Microbiome (2017) 5:44. doi: 10.1186/s40168-017-0261-y
44. Reichenberger ER, Alexander GM, Perreault MJ, Russell JA, Schwartzman RJ, Hershberg U, et al. Establishing a relationship between bacteria in the human gut and complex regional pain syndrome. Brain Behav Immun (2013) 29:62–9. doi: 10.1016/j.bbi.2012.12.005
45. Sheedy JR, Wettenhall RE, Scanlon D, Gooley PR, Lewis DP, McGregor N, et al. Increased d-lactic Acid intestinal bacteria in patients with chronic fatigue syndrome. In Vivo (2009) 23:621–8.
46. Shukla SK, Cook D, Meyer J, Vernon SD, Le T, Clevidence D, et al. Changes in gut and plasma microbiome following exercise challenge in myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS). PloS One (2015) 10:e0145453. doi: 10.1371/journal.pone.0145453
47. Weber T, Tatzl E, Kashofer K, Holter M, Trajanoski S, Berghold A, et al. Fibromyalgia-associated hyperalgesia is related to psychopathological alterations but not to gut microbiome changes. PloS One (2022) 17:e0274026. doi: 10.1371/journal.pone.0274026
48. Yong D, Lee H, Min HG, Kim K, Oh HS, Chu MK. Altered gut microbiota in individuals with episodic and chronic migraine. Sci Rep (2023) 13:626. doi: 10.1038/s41598-023-27586-4
49. Zhao X, Chen Y, Li L, Zhai J, Yu B, Wang H, et al. Effect of DLT-SML on chronic stable angina through ameliorating inflammation, correcting dyslipidemia, and regulating gut microbiota. J Cardiovasc Pharmacol (2021) 77:458–69. doi: 10.1097/FJC.0000000000000970
50. Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol (2012) 13:R79. doi: 10.1186/gb-2012-13-9-r79
51. Mottawea W, Chiang CK, Mühlbauer M, Starr AE, Butcher J, Abujamel T, et al. Altered intestinal microbiota-host mitochondria crosstalk in new onset Crohn's disease. Nat Commun (2016) 7:13419. doi: 10.1038/ncomms13419
52. Kootte RS, Levin E, Salojärvi J, Smits LP, Hartstra AV, Udayappan SD, et al. Improvement of insulin sensitivity after lean donor feces in metabolic syndrome is driven by baseline intestinal microbiota composition. Cell Metab (2017) 26:611–619.e6. doi: 10.1016/j.cmet.2017.09.008
53. Chen W, Liu F, Ling Z, Tong X, Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PloS One (2012) 7:e39743. doi: 10.1371/journal.pone.0039743
54. Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. Isme J (2012) 6:320–9. doi: 10.1038/ismej.2011.109
55. Yang T, Santisteban MM, Rodriguez V, Li E, Ahmari N, Carvajal JM, et al. Gut dysbiosis is linked to hypertension. Hypertension (2015) 65:1331–40. doi: 10.1161/HYPERTENSIONAHA.115.05315
56. Liu L, Wu Q, Chen Y, Ren H, Zhang Q, Yang H, et al. Gut microbiota in chronic pain: Novel insights into mechanisms and promising therapeutic strategies. Int Immunopharmacol (2023) 115:109685. doi: 10.1016/j.intimp.2023.109685
57. Cohen SP, Wang EJ, Doshi TL, Vase L, Cawcutt KA, Tontisirin N. Chronic pain and infection: mechanisms, causes, conditions, treatments, and controversies. BMJ Med (2022) 1:e000108. doi: 10.1136/bmjmed-2021-000108
58. Rojas M, Restrepo-Jiménez P, Monsalve DM, Pacheco Y, Acosta-Ampudia Y, Ramírez-Santana C, et al. Molecular mimicry and autoimmunity. J Autoimmun (2018) 95:100–23. doi: 10.1016/j.jaut.2018.10.012
59. Duvallet C, Gibbons SM, Gurry T, Irizarry RA, Alm EJ. Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nat Commun (2017) 8:1784. doi: 10.1038/s41467-017-01973-8
60. Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, et al. Population-level analysis of gut microbiome variation. Science (2016) 352:560–4. doi: 10.1126/science.aad3503
61. Armstrong G, Cantrell K, Huang S, McDonald D, Haiminen N, Carrieri AP, et al. Efficient computation of Faith's phylogenetic diversity with applications in characterizing microbiomes. Genome Res (2021) 31:2131–7. doi: 10.1101/gr.275777.121
62. Yin L, Wan YD, Pan XT, Zhou CY, Lin N, Ma CT, et al. Association between gut bacterial diversity and mortality in septic shock patients: A cohort study. Med Sci Monit (2019) 25:7376–82. doi: 10.12659/MSM.916808
63. Youngblut ND, de la Cuesta-Zuluaga J, Ley RE. Incorporating genome-based phylogeny and functional similarity into diversity assessments helps to resolve a global collection of human gut metagenomes. Environ Microbiola (2022) 24(9):3966–84. doi: 10.1111/1462-2920.15910
64. Nijs J, Tumkaya Yilmaz S, Elma Ö, Tatta J, Mullie P, Vanderweeën L, et al. Nutritional intervention in chronic pain: an innovative way of targeting central nervous system sensitization? Expert Opin Ther Targets (2020) 24:793–803. doi: 10.1080/14728222.2020.1784142
65. Nijs J, Leysen L, Vanlauwe J, Logghe T, Ickmans K, Polli A, et al. Treatment of central sensitization in patients with chronic pain: time for change? Expert Opin Pharmacother (2019) 20:1961–70. doi: 10.1080/14656566.2019.1647166
66. Vacca M, Celano G, Calabrese FM, Portincasa P, Gobbetti M, De Angelis M. The controversial role of human gut lachnospiraceae. Microorganisms (2020) 8(4):573. doi: 10.3390/microorganisms8040573
67. Odamaki T, Kato K, Sugahara H, Hashikura N, Takahashi S, Xiao JZ, et al. Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC Microbiol (2016) 16:90. doi: 10.1186/s12866-016-0708-5
68. Koh A, De Vadder F, Kovatcheva-Datchary P, Bäckhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell (2016) 165:1332–45. doi: 10.1016/j.cell.2016.05.041
69. Steinmeyer S, Lee K, Jayaraman A, Alaniz RC. Microbiota metabolite regulation of host immune homeostasis: a mechanistic missing link. Curr Allergy Asthma Rep (2015) 15:24. doi: 10.1007/s11882-015-0524-2
70. Kasahara K, Krautkramer KA, Org E, Romano KA, Kerby RL, Vivas EI, et al. Interactions between Roseburia intestinalis and diet modulate atherogenesis in a murine model. Nat Microbiol (2018) 3:1461–71. doi: 10.1038/s41564-018-0272-x
71. Hiippala K, Barreto G, Burrello C, Diaz-Basabe A, Suutarinen M, Kainulainen V, et al. Novel odoribacter splanchnicus strain and its outer membrane vesicles exert immunoregulatory effects in vitro. Front Microbiol (2020) 11:575455. doi: 10.3389/fmicb.2020.575455
72. Wang Y, Gao X, Ghozlane A, Hu H, Li X, Xiao Y, et al. Characteristics of faecal microbiota in paediatric crohn's disease and their dynamic changes during infliximab therapy. J Crohns Colitis (2018) 12:337–46. doi: 10.1093/ecco-jcc/jjx153
73. Li F, Sun G, Wang Z, Wu W, Guo H, Peng L, et al. Characteristics of fecal microbiota in non-alcoholic fatty liver disease patients. Sci China Life Sci (2018) 61:770–8. doi: 10.1007/s11427-017-9303-9
74. Zou Y, Lin X, Xue W, Tuo L, Chen M-S, Chen X-H, et al. Characterization and description of Faecalibacterium butyricigenerans sp. nov. and F. longum sp. nov., isolated from human faeces. Sci Rep (2021) 11:11340. doi: 10.1038/s41598-021-90786-3
75. Lopez-Siles M, Duncan SH, Garcia-Gil LJ, Martinez-Medina M. Faecalibacterium prausnitzii: from microbiology to diagnostics and prognostics. ISME J (2017) 11:841–52. doi: 10.1038/ismej.2016.176
76. Duncan SH, Hold GL, Harmsen HJM, Stewart CS, Flint HJ. Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov., comb. nov. Int J Syst Evol Microbiol (2002) 52(Pt 6):2141–6. doi: 10.1099/00207713-52-6-2141
77. Miquel S, Martín R, Bridonneau C, Robert V, Sokol H, Bermúdez-Humarán LG, et al. Ecology and metabolism of the beneficial intestinal commensal bacterium Faecalibacterium prausnitzii. Gut Microbes (2014) 5:146–51. doi: 10.4161/gmic.27651
78. Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux JJ, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA (2008) 105:16731–6. doi: 10.1073/pnas.0804812105
79. Martín R, Miquel S, Chain F, Natividad JM, Jury J, Lu J, et al. Faecalibacterium prausnitzii prevents physiological damages in a chronic low-grade inflammation murine model. BMC Microbiol (2015) 15:67. doi: 10.1186/s12866-015-0400-1
80. Martín R, Chain F, Miquel S, Lu J, Gratadoux JJ, Sokol H, et al. The commensal bacterium Faecalibacterium prausnitzii is protective in DNBS-induced chronic moderate and severe colitis models. Inflamm Bowel Dis (2014) 20:417–30. doi: 10.1097/01.MIB.0000440815.76627.64
81. Balamurugan R, Rajendiran E, George S, Samuel GV, Ramakrishna BS. Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, Desulfovibrio and Enterococcus faecalis in the feces of patients with colorectal cancer. J Gastroenterol Hepatol (2008) 23:1298–303. doi: 10.1111/j.1440-1746.2008.05490.x
82. Miquel S, Leclerc M, Martin R, Chain F, Lenoir M, Raguideau S, et al. Identification of metabolic signatures linked to anti-inflammatory effects of Faecalibacterium prausnitzii. mBio (2015) 6(2):e00300-15. doi: 10.1128/mBio.00300-15
83. Miquel S, Martín R, Rossi O, Bermúdez-Humarán LG, Chatel JM, Sokol H, et al. Faecalibacterium prausnitzii and human intestinal health. Curr Opin Microbiol (2013) 16:255–61. doi: 10.1016/j.mib.2013.06.003
84. Lopez-Siles M, Martinez-Medina M, Surís-Valls R, Aldeguer X, Sabat-Mir M, Duncan SH, et al. Changes in the abundance of faecalibacterium prausnitzii phylogroups I and II in the intestinal mucosa of inflammatory bowel disease and patients with colorectal cancer. Inflamm Bowel Dis (2016) 22:28–41. doi: 10.1097/MIB.0000000000000590
85. Chu XJ, Cao NW, Zhou HY, Meng X, Guo B, Zhang HY, et al. The oral and gut microbiome in rheumatoid arthritis patients: a systematic review. Rheumatol (Oxford) (2021) 60:1054–66. doi: 10.1093/rheumatology/keaa835
86. Yao Y, Cai X, Fei W, Ye Y, Zhao M, Zheng C. The role of short-chain fatty acids in immunity, inflammation and metabolism. Crit Rev Food Sci Nutr (2022) 62:1–12. doi: 10.1080/10408398.2020.1854675
87. Śliżewska K, Markowiak-Kopeć P, Śliżewska W. The role of probiotics in cancer prevention. Cancers (Basel) (2020) 13(1):20. doi: 10.3390/cancers13010020
88. Peng C, Ouyang Y, Lu N, Li N. The NF-κB signaling pathway, the microbiota, and gastrointestinal tumorigenesis: recent advances. Front Immunol (2020) 11:1387. doi: 10.3389/fimmu.2020.01387
89. Sivaprakasam S, Bhutia YD, Yang S, Ganapathy V. Short-chain fatty acid transporters: role in colonic homeostasis. Compr Physiol (2017) 8:299–314. doi: 10.1002/cphy.c170014
90. Tolhurst G, Heffron H, Lam YS, Parker HE, Habib AM, Diakogiannaki E, et al. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes (2012) 61:364–71. doi: 10.2337/db11-1019
91. Le Poul E, Loison C, Struyf S, Springael JY, Lannoy V, Decobecq ME, et al. Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. J Biol Chem (2003) 278:25481–9. doi: 10.1074/jbc.M301403200
92. Ohira H, Fujioka Y, Katagiri C, Mamoto R, Aoyama-Ishikawa M, Amako K, et al. Butyrate attenuates inflammation and lipolysis generated by the interaction of adipocytes and macrophages. J Atheroscler Thromb (2013) 20:425–42. doi: 10.5551/jat.15065
93. Halnes I, Baines KJ, Berthon BS, MacDonald-Wicks LK, Gibson PG, Wood LG. Soluble fibre meal challenge reduces airway inflammation and expression of GPR43 and GPR41 in asthma. Nutrients (2017) 9(1):57. doi: 10.3390/nu9010057
94. McBurney MI, Davis C, Fraser CM, Schneeman BO, Huttenhower C, Verbeke K, et al. Establishing what constitutes a healthy human gut microbiome: state of the science, regulatory considerations, and future directions. J Nutr (2019) 149:1882–95. doi: 10.1093/jn/nxz154
95. Piquer-Esteban S, Ruiz-Ruiz S, Arnau V, Diaz W, Moya A. Exploring the universal healthy human gut microbiota around the World. Comput Struct Biotechnol J (2022) 20:421–33. doi: 10.1016/j.csbj.2021.12.035
96. Syromyatnikov M, Nesterova E, Gladkikh M, Smirnova Y, Gryaznova M, Popov V. Characteristics of the gut bacterial composition in people of different nationalities and religions. Microorganisms (2022) 10(9):1866. doi: 10.3390/microorganisms10091866
Keywords: microbiota, gut-brain axis, persistent pain, biomarker, gut composition, stool samples
Citation: Goudman L, Demuyser T, Pilitsis JG, Billot M, Roulaud M, Rigoard P and Moens M (2024) Gut dysbiosis in patients with chronic pain: a systematic review and meta-analysis. Front. Immunol. 15:1342833. doi: 10.3389/fimmu.2024.1342833
Received: 22 November 2023; Accepted: 08 January 2024;
Published: 30 January 2024.
Edited by:
Youcef Shahali, Centre Hospitalier Universitaire de Besançon, FranceReviewed by:
Shaoyi Zhang, University of California, San Francisco, United StatesWerner Solbach, University of Lübeck, Germany
Copyright © 2024 Goudman, Demuyser, Pilitsis, Billot, Roulaud, Rigoard and Moens. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lisa Goudman, lisa.goudman@gmail.com
 Lisa Goudman
Lisa Goudman Thomas Demuyser
Thomas Demuyser Julie G. Pilitsis
Julie G. Pilitsis Maxime Billot
Maxime Billot Manuel Roulaud9
Manuel Roulaud9