- 1State Key Laboratory of Biotherapy and Cancer Center, Collaborative Innovation Center for Biotherapy, West China Hospital, West China Medical School, Sichuan University, Chengdu, China
- 2College of Polymer Science and Engineering, Sichuan University, Chengdu, China
- 3Department of Medical Examination, Chengdu Seventh People’s Hospital, Chengdu, China
- 4Department of Biotherapy and Cancer Center, State Key Laboratory of Biotherapy, West China Hospital, Sichuan University, Chengdu, China
- 5Sichuan Aupone Pharmaceutical Co., Ltd, Chengdu, China
Cancer is a major disease endangering human health. More and more studies have shown that microorganisms play an extremely important role in the occurrence, development and treatment of tumors. As a very promising tumor treatment strategy, immunotherapy has also been proved to have a great relationship with microorganisms. Here, the authors review the contribution of the microbiota to cancer and the research on its impact on cancer immunotherapy. We also highlight the possible mechanism of their interaction and outlined the potential application of microbiota in tumor immunotherapy.
Introduction
The microbiota lives on all the epithelial surfaces of the human body, including the skin, respiratory tract, digestive tract, and urogenital tract, and their presence can be detected even within tumors (1, 2). Hundreds of millions of years of evolution have established a lasting relationship between the microbiome and the human body (3). Past studies have shown that the composition of the microbiota in the epithelial barrier affects systemic functions, including metabolism, nervous system, inflammation, and immunity (4–6). The intestine is the largest digestive organ in the human body. It is constantly exposed to foreign antigens and other environmental factors (7). Microorganisms are distributed along the intestine, and the number of microorganisms in the colon is the largest (8). Studies have found that the imbalance in the intestinal flora is related to the occurrence of many diseases, such as obesity, inflammatory bowel disease, autism and cancers (9). In addition to being closely related to cancer, the microbiome is closely related to the immune system, including innate and acquired immunity (10). Furthermore, many research results show that interventions targeting microbiota, especially intestinal microbes, have achieved gratifying results in cancer immunotherapy (11, 12).
It is worth noting that the relationship between microorganisms and cancer is complex, and there is also an intricate network of factors causing tumor immunotherapy effect by microorganisms (13). Here we review the relationship between carcinogenic microbial infections, microbial disorders, and carcinogenesis. The mechanism of the interaction between microorganisms and immune cells is discussed. We also review the latest reports on the impact of microorganisms on cancer immunotherapy, and finally outlined a new direction for improving the effect of tumor immunotherapy; that is an application of microorganisms.
Carcinogenic microorganism infections, microbial disorders, and carcinogenesis
Bacterial and viral infections
Although many microorganisms reside in the human epithelial barrier, only 12 (1 bacteria, 8 viruses and 3 parasites) are currently considered human carcinogens by the International Agency for Research on Cancer (IACR) (14). The most well-known microorganisms associated with cancer is Helicobacter pylori (H. pylori), which is considered the most common pathogen of infection-related cancers (15). H. pylori is a gram-negative bacterium that can selectively colonize the gastric epithelium (16). About half of the world’s population is infected by H. pylori (17). Colonization with it causes no symptoms in humans; however, long-term colonization with H. pylori significantly increases the risk of gastric cancer (18, 19). The carcinogenic virulence factors of gastric cancer are closely related to H. pylori virulence proteins such as CagA, VacA, and CagPAI (20, 21). Besides the virulence factors, H. pylori-induced oxidative stress, DNA damage, up-regulation of pro-inflammatory cytokines, and activation of multiple signaling pathways are all responsible for H. pylori-induced gastric cancer (22). These have been discussed in several articles (22–24). Notably, while H. pylori is the only bacterium currently considered to be a human carcinogen, many other bacteria have also been reported to be closely linked with cancer development. Studies have shown that pks+ strains of Escherichia coli (E. coli) can synthesize colibactin to induce DNA double-strand breaks (25). Besides, colibactin can not only induce the emergence of senescent cells, which promote tumor growth by the secretion of growth factors, but also change the immune microenvironment through impairment of antitumor T-cell response, leading to tumoral resistance to immunotherapy (26, 27). With the development of organoid technology, the mutation characteristics of colon cancer caused by colibactin are being gradually clarified (25, 28, 29). Enterotoxigenic Bacteroides fragilis toxin cleaves E-cadherin, resulting in Wnt/β-catenin signaling and alter the gene expression in colonic epithelial cells (14, 25, 30). These factors are all potential factors leading to colorectal cancer (CRC) (31). In addition, Propionibacterium acnes (P. acnes) stimulate prostate cells to secrete interleukin (IL)-6 and IL-8, which may be related to the occurrence and development of prostate cancer (32).
The mechanisms by which tumor viruses promote carcinogenesis are more diverse. For example, human papillomavirus (HPV), Epstein-Barr virus (EBV), and Merkel cell polyomavirus (MCPyV) cause tumors by encoding oncogenic proteins that can regulate cell proliferation, apoptosis, or blood vessels generated to promote the occurrence of cancer (33). Other human tumor viruses, such as hepatitis B virus (HBV) and hepatitis C virus (HCV), do not express definitive oncogenic proteins, but cause tumorigenesis primarily by inducing a chronic inflammatory state (34, 35). Simultaneously, sustained inflammatory and immune responses can lead to increased production of reactive oxygen species (ROS) and reactive nitrogen species (RNS); thus inducing gene mutations (36). The promotion of genomic instability by these factors is one of the mechanisms by which viral infection promotes cancer development.
Microbiota dysbiosis
Improvement in socioeconomic factors is often associated with detrimental lifestyle changes and environmental exposures that are major determinants of cancer (37). An interesting study shows that for all cancers and a large number of cancer types, there is a strong and positive correlation between cancer incidence and national socioeconomic level in both men and women (37). There is a growing recognition of another gene pool, the microbiome, that needs to be considered when assessing the impact of environmental factors on human health (38). Environmental and host-related factors can drive dysbiosis, which is defined as changes in the composition and function of the microbiota (39). There is increasing evidence that dysbiosis of the microbiota is associated with cancer development, and this relationship is particularly evident with the gut microbiota. Human studies have shown that compared with healthy individuals, patients with CRC have a less diverse gut microbiome (40). Furthermore, at different CRC stages, ranging from adenomatous polyps to early-stage cancer to metastatic disease, the microbiome undergoes specific changes, including a marked increase in the DNA and RNA levels of Fusobacterium nucleatum (F. nucleatum) in human CRC (40–42). In addition, the gut microbiota also affects normal intestinal stem cells (ISCs). Dysbiosis induces aberrant programming of ISCs through multiple mechanisms, leading to the transformation of ISCs into cancer stem cells, which are thought to initiate CRC (43). Dysregulation of gut microbes not only affects the occurrence and development of CRC locally, but also affects the occurrence of distant organ cancers. The majority of liver cancers occur in patients with cirrhosis, and these patients often exhibit leaky gut and dysbiosis, which are thought to be the main cause of liver cancer in patients with cirrhosis (44). Patients with hepatocellular carcinoma (HCC) have been reported to have higher levels of E. coli and other Gram-negative bacteria in the gut microbiota compared with healthy individuals (45). On the other hand, gut microbes in patients with HCC have reduced levels of Lactobacillus spp., Bifidobacterium spp., and Enterococcus spp (46).. A new study shows that gut microbes can even influence the production of male hormones to interfere with the development of prostate cancer in castrated mice (47). In addition to the gut microbiota, dysbiosis of other epithelium-distributed microbes is also closely linked to carcinogenesis at their colonization sites. Lactobacillus is the dominant genus of human vaginal microbiota at reproductive age (48). The cervicovaginal microbiota is dominated by Lactobacillus crispatus, Lactobacillus iners, Lactobacillus gasseri, or Lactobacillus jensenii, which help to maintain the pH of the vagina (49). Studies have reported that women with or at risk of developing ovarian cancer have an imbalance in the cervicovaginal microbiota, as manifested by a reduced ratio of Lactobacillus to total vaginal microbes (50).
The causes of microbe disorders in the human body are diverse, including diet, antibiotics, genetics, family transmission, and other factors (39). Here, we mainly discuss the impact of infection and inflammation on dysbiosis. As mentioned earlier, H. pylori is the strongest risk factor identified for gastric cancer, and studies have shown that H. pylori-negative individuals have a highly diverse gastric microbiome (51). When 1833 bacterial clones from 23 gastric biopsy samples were analyzed, sequencing identified 128 phylotypes within 8 bacterial phyla. In contrast, only 33 phylotypes were detected in three H. pylori-infected populations (52). These data suggest that H. pylori colonization greatly reduces the overall diversity of the gastric microbiota. However, in many studies, the composition of microorganisms varies greatly between individuals, and the mechanism of how H. pylori colonization affects other microorganisms in the stomach still needs to be further explored. F. nucleatum, an invasive and pro-inflammatory bacterium known to cause oral and gastrointestinal infections, has also been detected in tumors from CRC patients (53–56). F. nucleatum is a potent stimulator of the inflammatory cytokines, IL-6, IL-8, and TNF-α, and regarding dysbiosis, F. nucleatum induces increased inflammation, becoming a pathogen (57). In CRC, F. nucleatum not only activates the inflammatory response but also promotes colorectal carcinogenesis through its FadA adhesin regulation of E-cadherin/β-catenin signaling (58). Its regulation of autophagy also promotes chemoresistance in CRC (59).
Intratumoral and local tumor microbes
With advances in sequencing technology, the presence of microbes within tumors has been identified. Studies show that each cancer subtype has a unique microbiome with specific metabolic functions, and the intratumor bacteria are mostly present in both cancer and immune cells (2, 60). Therefore, the intratumoral microbiome plays a crucial role in tumor development and treatment. Above, we mentioned various microorganisms that are in direct contact with gastrointestinal tumors, such as H. pylori, pks+ E. coli, and F. nucleatum, which are involved in the occurrence and development of gastric or CRC. Therefore, here we mainly focus on intratumoral and local tumor microbes in other tumors. In a spontaneous murine mammary tumor model, Fu et al. (61) found that the depletion of intratumoral bacteria via tail vein injection of mixed antibiotics that had no effect on the gut microbiota, significantly reduced lung metastasis without affecting primary tumor growth. Shi et al. (62) found that Bifidobacterium in the gut can accumulate in the tumor microenvironment, and intratumoral injection of very low doses of mixed antibiotics reduced the efficacy of anti-CD47 immunotherapy in tumor-bearing mice. Boesch et al. (63) found compared to healthy lung tissue that the lung tissue of patients with non-small cell lung cancer had a higher abundance of Gammaproteobacteria, which correlates with low programmed death-ligand 1 (PD-L1) expression and worse overall survival (OS) under immune checkpoint inhibitor (ICI) therapy. Ma et al. (64) analyzed microbial compositions of intratumor bacteria in prostate cancer to determine the influence of the microbiome on metastatic growth. They identified specific microbes that can significantly deter the development of prostate cancer (Listeria monocytogenes and Methylobacterium radiotolerans JCM 2831) or contribute to cancer aggressiveness (Stackebrandtia nassauensis DSM 44728 and Mycoplasma hyorhinis HUB-1). In terms of the mechanisms by which intratumoral microorganisms affect tumor progression, DNA damage, immunosuppression, drug metabolism, and activation of oncogenic pathways are still the main mechanisms (65). Recently, an interesting study showed that fungi within mouse pancreatic ductal adenocarcinoma (PDAC) tissue can drive IL-33 secretion, further recruit and activate TH2 cells and innate lymphoid cells 2 (ILC2) in tumor issue, ultimately leading to the inhibition of anti-tumor immune response and promotion of tumor progression (66). Accordingly, intratumoral and local tumor microbes remain a promising research direction for tumor progression.
In general, carcinogenic microbial infection and dysbiosis of microbiota are closely related to tumorigenesis (Figure 1), and intratumoral and local tumor microbes also have a significant contribution to tumor progression. However, the causal relationship among them still needs to be further studied.
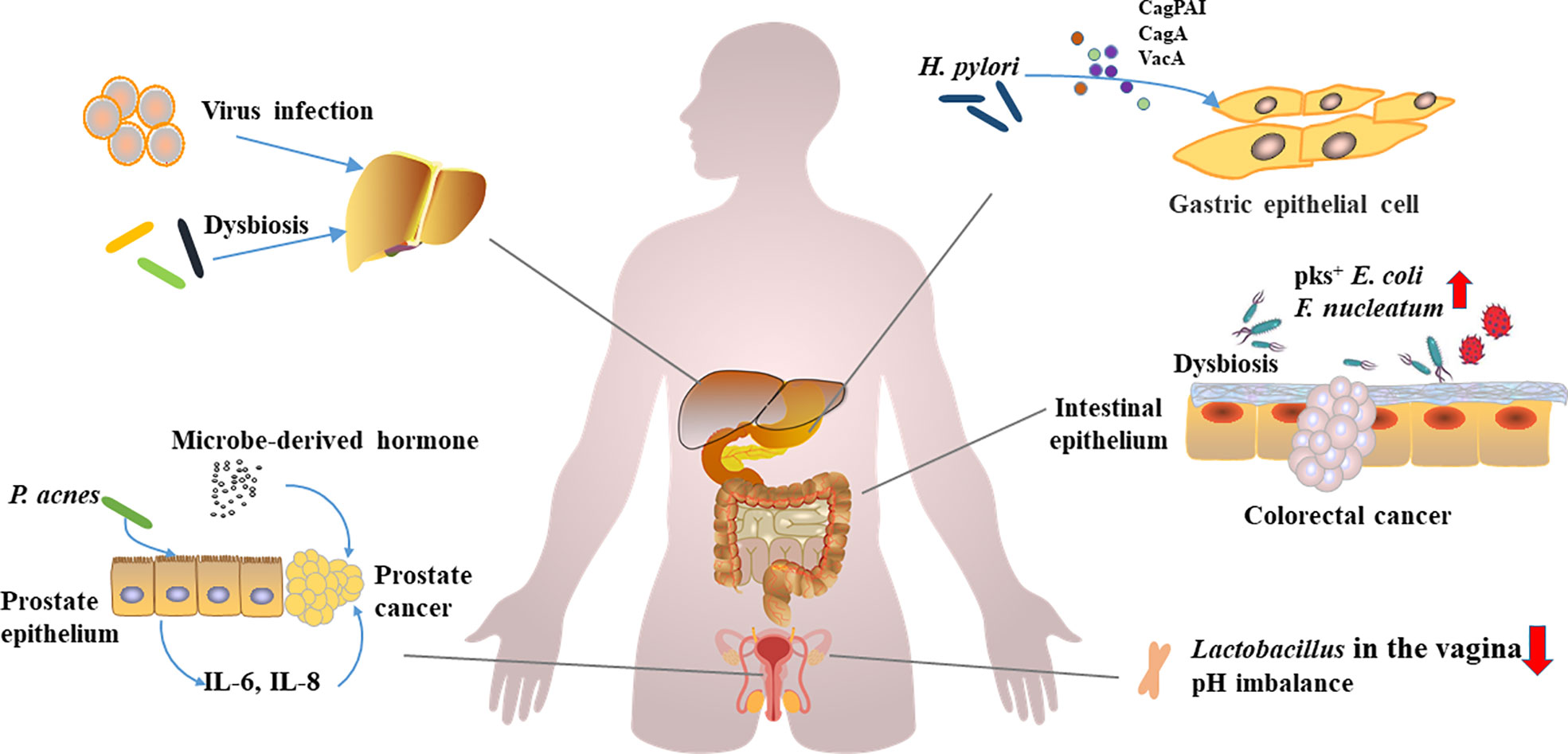
Figure 1 Carcinogenic microorganism infections, microbial disorders, and carcinogenesis.Viruses, oncogenic microorganisms infections, and dysbiosis of the microbiota have been implicated in the development of multiorgan cancers. H. pylori promotes gastric carcinogenesis through virulence factors (CagPAI, CagA, and VacA etc.). F. nucleatum and pks+ E. coli can promote the development of colorectal cancer. Lactobacillus, a vagina-dominant genus that helps regulate pH, is reduced in abundance in ovarian cancer patients. P. acnes induces prostate cancer by stimulating the production of IL-6 and IL-8 in prostate cells, and hormones derived from microorganisms promote the development of prostate cancer. Hepatitis virus induces hepatitis, and persistent inflammation and dysbiosis can affect the occurrence of liver cancer.
Microbiota and cancer immunotherapy
Cancer immunotherapy is an approach that harnesses the immune system to fight cancer (67). Current immunotherapy can be roughly divided into oncolytic virus therapies, cancer vaccines, cytokine therapies, adoptive cell transfer (ACT), and ICIs (68). Their common features are enhanced immune responses, including innate immunity and/or adaptive immunity to clear cancer cells. The microbiota and its metabolites provide key signals for the induction, development, and function of the host immune system (69, 70). Growing evidence suggests that the microbiome plays a key role in cancer immunotherapy, and here we link the microbiome and immunotherapy from three perspectives: adaptive immunity, innate immunity, and metabolism.
Linking microbiota and cancer immunotherapy from adaptive immunity perspective
Cytotoxic T lymphocyte-associated antigen 4 (CTLA-4), programmed cell death protein 1 (PD-1) and its ligand (PD-L1) are important immune checkpoints and are also important factors in regulating T cell immune function (71). Inhibition of these targets reactivates T cells more effectively, provides novel treatments for a variety of cancers including melanoma, non-small cell lung cancer (NSCLC), and significantly improves patients survival (72). However, these therapies targeting immune checkpoints are not effective in all patients. Pembrolizumab, an anti-PD-1 monoclonal antibody, has been shown in clinical studies to respond better in patients with lung metastases compared to patients with liver metastases (62% vs. 22%) (73). Anti-CTLA-4 blockade using ipilimumab is the first treatment to prolong OS in patients with advanced melanoma in a randomized setting (74). In a study of 30 patients with melanoma treated with ipilimumab, only 11 (37%) had their disease under control (75). Therefore, how to improve the patient’s response to monoclonal antibodies has become an important issue. In recent years, increasing number of studies have shown a significant impact of intestinal microbiota on treatment with ICIs.
Gopalakrishnan et al. (76) divided 112 melanoma patients receiving PD-1 immunotherapy into responder and non-responder groups to determine significant differences in the diversity and composition of the gut microbiome between them. Faecalibacterium was more abundant in fecal microbiome responders, while fecal microbiome non-responders had higher abundance of Bacteroides thetaiotaomicron, E. coli, and Anaerotruncus colihominis. More CD8+ T cells infiltration and stronger systemic antitumor immune responses were observed in the tumors of responders (76). Similar studies revealed that antibiotics inhibited the beneficial effects of ICIs in patients with advanced cancer and that patient response to ICIs was associated with a relative abundance of Akkermansia muciniphila (Akk) (12). Oral Akk supplementation restores the response to PD-1 blockade in an IL-12-dependent manner by increasing the recruitment of CCR9+ CXCR3+ CD4+ T lymphocytes in mouse tumor beds (12). Oral administration of Bifidobacterium modulates the activation of mouse DC cells, improves the effector function of CD8+ T cells, and enhances the efficacy of PD-L1 (77). In studies of CTLA-4, similar results were observed with PD-1. A study found that germ-free mice did not respond to CTLA-4 blockade, the efficacy of CTIA-4 blockade was affected by B. fragilis, B. thetaiotaomicron, and Burkholderiales, and these microorganisms also affected IL-12-dependent Th1 immune responses (78).
A large proportion of cancer patients do not benefit from ICIs therapy, and of those who do, some responders still relapse after a period of response (79). Absence of a relevant number of immunogenic tumor antigens, defects in the antigen processing and presenting machinery, and insufficient T cell infiltration are all mechanisms of the resistance (80). Interestingly, ICIs therapy seems to be more effective in tumors with a high tumor mutational burden (TMB), and the reason may be related to the neoantigens produced by the tumor (81).The cross-reactivity of T cells allows each T cell to recognize multiple antigens (82). The human microbiome is a huge gene pool, and neoantigens produced by tumors may be mimicked by peptides encoded by the microbiota. When tumor neoantigens appear, memory T cells can quickly provide protection (80, 83). Bessell et al. (84) found that T cells targeting an epitope called SVYRYYGL (SVY), expressed in the commensal bacterium, Bifidobacterium breve (B. breve), cross-react with a model neoantigen, SIYRYYGL (SIY). Moreover, B. Breve colonization can shape SVY- reactive T cell receptor library, influence T cell response, and then affect the growth of tumor that expresses neoantigens (84). Balachandran et al. (85) found that neoantigen-specific immunity gained during primary tumor outgrowth could be associated with decreased relapse and prolonged survival. Taken together, tumor antigen mimicry generated by the microbiota and cross-reactivity of T cells may be beneficial for tumor immunotherapy, and these are possible explanations for the large differences in response to checkpoint inhibitors in cancer patients (86).
Linking microbiota and cancer immunotherapy from innate immunity perspective
Cytotoxic T lymphocytes (CTL), especially CD8+ T lymphocytes, are the main anti-tumor effector cells and the main target cells for tumor immunotherapy (87). However, innate immune cells also play an important role in cancer immunotherapy. Current research shows that innate immunity not only indirectly affects anti-tumor immune responses by controlling T cells, but also directly and critically shapes the tumor microenvironment (85), which is an important part of tumor immunity (88). Crosstalk between the microbiota and innate immunity affects multiple aspects of body homeostasis, and this complex bilateral interaction is critical for human health (89). Therefore, some tumor immunotherapies targeting innate immune cells have been developed, and the influence of the microbiota on innate immune cells has also been confirmed to be relevant to a variety of cancer immunotherapies.
Pattern recognition receptors (PRRs) are an important part of innate immune defense, and they are expressed on a variety of immune cells such as leukocytes and macrophages (90). PRRs respond to a variety of bacterial and viral ligands, also known as pattern-associated molecular pattern (PAMP), including peptidoglycan (PGN), lipopolysaccharide (LPS), double-stranded RNA, and CpG DNA (90, 91). PRRs genes, including NOD1/2, NLRP3, and various toll-like receptor (TLR) genes, recognize PAMPs as non-self-entities and trigger intracellular signaling pathways that induce a variety of cytokines and chemokines that help maintain host response against infection (91, 92). Gram-negative bacterial cell wall component, LPS, is recognized by TLR4, and activation of TLR4 promotes prostate cancer development and induces nitric oxide and IL-6 production in CRC (93). TLR3 agonist, poly(I:C), was developed to mimic infection by pathogens and boost immune system activation to promote anti-cancer therapy (94). NOD2 receptor is an inflammatory pathway and microbiota modulator, and studies demonstrate that loss of NOD2 activity led to more severe colitis and a higher risk of adenoma and CRC in mouse models (95). In conclusion, the rich innate immune signaling pathways initiated by the microbiota through PRRs are important in infection, inflammation, and cancer development (96).
TME is a complex system that includes many different types of cells, abnormal vasculature, and immunosuppressive cytokines, and it is one of the important reasons why tumors evade immune surveillance (88, 97). Mononuclear phagocytes (MPs) (i.e., monocytes [Mo], macrophages [Macs], and dendritic cells [DCs]) are the major innate immune cells and important components of the TME (98). A recent study sheds light on the effect of the microbiota on MPs in TME and innovatively proposed that MPs in TME can be remodeled by microorganisms to improve ICIs efficacy. Lam et al. (98) demonstrated that microbiota-derived stimulator of interferon gene (STING) agonists such as c-di-AMP induce type I interferon (IFN-I) production by intratumoral Mo, which regulates their skewing and natural killer (NK)-DC crosstalk. The triggering of this mechanism can be achieved by a high-fiber diet, which enriches Akkermansia muciniphila, further produces c-di-AMP, and enhances the therapeutic effect of ICIs in melanoma patients. Monocytes are more inclined to differentiate into tumor-promoting Macs when the microbiota is adversely disrupted (98). Another study also found that Bifidobacterium colonized in the tumor microenvironment can effectively stimulate STING signaling and increase cross-priming of DCs after anti-CD47 treatment (62). Overall, these studies revealed possible mechanisms of the interaction between the microbiota and innate immune cells, and we believe that more specific mechanisms will be explored in the future.
ACT therapy is an immunotherapy method in which autoimmune cells, especially T and NK cells, are isolated, modified, amplified, and re-injected into a patient to eliminate cancer cells (68, 99). Chimeric antigen receptor-T (CAR-T) cell therapy is used to treat different malignant tumors including lymphoma and leukemia, and it is one of the promising ACT therapies (100). Like ICIs, CAR-T therapy is not effective in all patients. The complete response rate in patients with aggressive lymphoma is from 40% to 60%, and a large proportion of patients will relapse (101). Recent studies have shown that oral administration of vancomycin, an antibiotic mainly targeting Gram-positive bacteria, can improve the efficacy of CAR-T therapy in mice with cervical cancer. Mechanistically, vancomycin treatment induces an increase in systemic CD8α+ DCs, which sustains systemic adoptively transferred antitumor T cells in an IL-12–dependent manner (102). NK cells, as the name suggests, are non-specific tumor-killing cells in innate immunity. They do not need any antigenic priming before attacking the target, and can quickly kill tumor cells through a variety of mechanisms (103, 104). Therefore, CAR-NK has some significant advantages over CAR-T, such as multiple mechanisms of activating cytotoxic activity and better safety (103). Although there is no clear report on the association between CAR-NK and the microbiome, existing studies have shown that high-salt diet (HSD) increases the abundance of Bifidobacteria and leads to increased intestinal permeability, which further leads to Bifidobacteria colonization within tumors, enhancing NK cell function and promoting tumor regression (104). The use of mixed antibiotics has also been found to promote glioma growth in mice, which is associated with disruption of the gut microbiota and reduction of cytotoxic NK cell subsets (105). These evidence give us reason to believe that the microbiota may contribute to CAR-NK therapy.
Linking microbiota and cancer immunotherapy from metabolism perspective
The gut microbiota can ferment undigested food in the colon and can also utilize endogenous compounds produced by the host (70). Some of the diverse metabolites produced by microorganisms can enter and interact with host cells, thereby affecting immunity and disease risk (106). Multiple metabolites produced by the microbiota have also been shown to be relevant for tumor immunotherapy.
Short-chain fatty acids (SCFAs) are the main end-products of indigestible carbohydrates fermented by gut microbiota, mainly including formate, acetate, propionate, and butyrate (107). Among them, butyrate has been shown to have a potential role in immune regulation, inhibiting nuclear factor activation in macrophages and also inhibiting histone deacetylation in acute myeloid leukemia, while exerting an inhibitory effect on CRC (107, 108). Butyrate and propionate inhibit LPS-induced expression of cytokines such as IL-6 and IL-12p40, exhibiting strong anti-inflammatory effects (109). SCFAs have been recognized to maintain intestinal homeostasis by regulating different cells. SCFAs can enhance mucus production by goblet cells, while promoting the production of IL-22 by CD4+ T cells to maintain intestinal epithelial barrier function (110, 111). A growing number of studies have shown that the gut microbiota can influence tumor immunotherapy through SCFAs. A new study shows that valeric acid and butyric acid enhance the antitumor activity of CTL and CAR-T cells through metabolic and epigenetic reprogramming. The mechanism lies in the increased production of effector molecules such as CD25, IFN-γ, and TNF-α (112). Through oral administration of pectin, inulin, and other polysaccharide dietary fibers in mice, researchers have found that they can significantly improve the therapeutic effect of PD-1 mAb. All can increase the relative abundance of key symbiotic microorganisms (such as Akkermansia and Lactobacillus) and SCFAs, further promoting the invasion of CD8 + T cells into the tumor (113, 114). In addition, SCFAs were found to increase the memory potential of antigen-primed CD8+ T cells and trigger their differentiation into stem cell-like Tcf1+PD-1+CD8+ T cells, resulting in potent and long-term antitumor effects (113, 115). It is worth mentioning that in addition to SCFAs, other lipid metabolisms also have an impact on cancer immunotherapy, such as glycerophospholipid metabolism and sphingolipid metabolism (116). However, studies on the impact of lipid metabolism on tumor immunotherapy still focus on effector T cells (117).
Amino acid metabolism is also an important aspect of host and microbial metabolism, which also plays an important role in cancer immunity. Reacquiring durable immune memory is challenging in the setting of severe T cell exhaustion, and exhausted T cells exhibit distinct histone profiles and limit tumor immunotherapy (118, 119). Studies have found that tumor cells compete with CD8+T for methionine through the high expression of methionine transporter, which reduces the levels of methionine and methyl donor s-adenosylmethionine (SAM) in T cells, and inhibition of transporters enhances immune checkpoint-induced tumor immunity (118). Elevating L-arginine levels induces global metabolic changes including a shift from glycolysis to oxidative phosphorylation in activated T cells, promoting the production of central memory-like cells with antitumor activity in mice model (120). Moreover, blocking glutamine metabolism not only inhibits tumor growth, but also enhances the efficacy of ACT and PD-1 mAb. The mechanism involves the blocking of glutamine metabolism, which inhibits glucose metabolism through the tricarboxylic acid cycle and glycolysis-related pathways (121). L-tryptophan contributes much to maintaining the balance between the gut microbiota (122). Changes in the microbiota can also modulate tryptophan and its metabolites (including kynurenine) and thus affect the host immune system (123). Researchers found that ginseng polysaccharides (GPs, a polysaccharide extracted from ginseng) could significantly improve the therapeutic effect of PD-1 mAb in tumor-bearing mice. Mechanically, oral administration of GPs increases valeric acid produced by microbial metabolism and decreases L-kynurenine and Kyn/Trp ratios (124).
Notably, in addition to lipid metabolism and amino acid metabolism, other metabolic pathways of the microorganism and host also appear to have an impact on cancer immunotherapy. B. pseudolongum, an intestinal Lactobacillus bacterium, enhances immunotherapeutic responses by producing the metabolite, inosine. Specifically, inosine promotes Th1 cell activation in a context-dependent manner through T cell-specific A2A receptor signaling for immune enhancement (125). Purine metabolism, a downstream metabolic pathway of inosine, has also been shown to be involved in host immunity. A recent study demonstrated that priming of the purine nuclease FAMI in DC inhibits CD4+ and CD8+ T cell priming. DCs lacking FAMIN activity enhance antigen-specific cytotoxicity, IFN-γ secretion, and T cell expansion (126). Rhein can increase Lactobacillus levels, alter purine metabolism, and reduce uric acid levels in the gut and further alleviate dextran sulfate sodium (DSS)-induced enteritis in mice (127). These evidence suggest that microbe-mediated purine metabolism seems to have great research prospects in inflammatory to cancer transformation.
Applications of microorganisms: A new strategy to improve cancer immunotherapy
The impact of microorganisms on immunity is multi-faceted, and microorganisms are increasingly being applied to various immunotherapy to improve immunotherapy. Even the microbes themselves are being used as new targets for immunotherapy.
Efficacy improvement: Diet, probiotic use, and fecal microbiota transplantation
Diet is a key factor in altering gut microbiota composition and function (128). Appropriate intake of dietary fiber and prebiotics has been recognized as a positive contribution to human health, including weight control, cardiovascular protection, blood sugar control, and brain health (129, 130). Based on the profound impact of the microbiota on tumor immunotherapy, an increasing number of preclinical studies have attempted to improve immunotherapy through dietary interventions. Previous studies have found that oral administration of inulin and pectin can enhance the efficacy of ICIs, which is related to the change in intestinal flora and metabolism (113, 114). We have described a specific mechanism in the last section. Messaoudene et al. (131) gavaged mice with polyphenol-rich berry camu-camu and found that the berry significantly enhanced the efficacy of ICIs. The main active component of this berry, castalagin, alters bile acid metabolism in mice and binds to the surface of Ruminococcus bromii, resulting in better antitumor activity of PD-1 antibody. Spencer et al. (132) conducted a high-fiber dietary intervention in patients receiving ICIs and found that higher dietary fiber was associated with significantly improved progression-free survival in patients on ICIs.
Probiotics are defined as live microorganisms that, when ingested in sufficient amounts, confer a health benefit to the host (133). Colonization with probiotics is beneficial to the host in the long run. However, it is a long and arduous process from oral probiotics to colonization of probiotics in the intestine, which is affected by various aspects such as colonization resistance, intestinal mucosa, and mucus layer (134). van Zyl et al. provide a detailed review of the in vivo kinetics of multiple probiotics following oral administration. By comparing the number of cells in feces before and after ingesting a particular strain, they found differences in survival and persistence between genera and even between strains (135). However, previous research has focused on the role of probiotics in intestinal diseases, especially intestinal inflammation and diarrhea (135, 136). With the rise in immunotherapy, in recent years, improving immunotherapy through probiotic supplementation has also become an emerging research direction. A new study found that microbial exopolysaccharide produced by Lactobacillus delbrueckii subsp. bulgaricus OLL1073R-1 (EPS-R1) induced CCR6+ CD8+ T cells in mice and humans. In mouse models of colon adenocarcinoma and breast cancer, ingestion of EPS-R1 augmented the antitumor effects of anti-CTLA-4 or anti-PD-1 mAb (137). Another study also found that Clostridioides butyricum MIYAIRI 588 strain significantly improved OS in patients with NSCLC treated with ICI therapy (138). Colonization of Bifidobacterium pseudolongum, Lactobacillus johnsonii, and Olsenella species in the gut enhances the efficacy of CTLA-4, which is associated with CD4+ and CD8+ T cell activation (125). High abundance of AKK appears to correlate with better efficacy of ICIs in both humans and mice (12). As a potential star probiotic, AKK has been proven to improve tumor immunotherapy, as well as improve obesity, anti-diabetes, and inhibit inflammation in mice (139). Interestingly, both live and inactivated AKK had positive health implications (140, 141). The adverse reactions of ICIs involve skin, gastrointestinal tract, thyroid, heart, and other organ systems (142). Some microbiota can also reduce the adverse reactions caused by ICIs. For example, Bifidobacterium can alleviate colitis induced by ipilimumab (CTLA-4 inhibitor) treatment by inhibiting the release of pro-inflammatory cytokines (143). However, it is worth mentioning that the focus on a certain immunotherapy-enhancing probiotic often occur after the use of ICIs, and precise probiotic supplementation (such as bulgaricus OLL1073R-1) is still an aspect that requires attention in basic research and preclinical trials, as well as areas that need to be expanded.
Fecal microbiota transplantation (FMT) is defined as the transplantation of gut microbiota from healthy donors to diseased patients via an upper or lower gastrointestinal route to restore gut microbial diversity (144). In human medicine, FMT was originally used to treat microbial-induced gastrointestinal diseases such as Clostridioides difficile infection and ulcerative colitis (145–147). FMT is now being used more widely, including for the treatment of metabolic syndrome, diabetes, Crohn’s disease, Parkinson’s disease, multiple sclerosis, psoriasis, anorexia nervosa, or Alzheimer’s disease (148). The impact of FMT on the microbiome has led researchers to see its potential in tumor immunotherapy. Baruch et al. found that combining FMT (from complete response donors) with reinduction anti-PD-1 therapy is safe, feasible, and potentially effective in patients with refractory metastatic melanoma (149).
In addition to ICIs, microbes have a facilitating role in other immunotherapies. Combination of oral Wilms’ tumor 1 (WT1) cancer vaccine and anti-PD-1 antibody treatment using a Bifidobacterium vector has been shown to eliminate tumor growth in a syngeneic mouse model of bladder cancer (150). Vaccine delivery based on the antigenic action of the microbiota may significantly inhibit tumor-associated microorganisms, such as H. pylori, which possesses a variety of bacterial toxins and proteins, and can serve as key candidates for H. pylori vaccine construction (151). Following radiation therapy, intratumoral injection of genetically attenuated Salmonella strains coated with antigen-adsorbing cationic polymer nanoparticles resulted in tumor antigen accumulation around the tumor (152). This enhances crosstalk between antigens and DCs, and the use of flagellated bacteria to transport tumor antigens around tumors to enhance DC activation may open up new strategies for in situ cancer vaccination (152). In cytokine therapy, beneficial commensal microorganisms, AKK, combined with IL-2, can enhance the antitumor efficacy of IL-2 and enhance immune surveillance. Mechanistically, the antitumor immune response elicited by AKK is partially mediated by Amuc, derived from the outer membrane protein of AKK, through activating TLR2 signaling pathway (153).
A new target for immunotherapy: The microbiome itself
Microbiota itself holds great promise as a new target for immunotherapy. Montalban-Arques et al. (154) found that four butyrate-producing Clostridioides species: Roseburia gutis, Eubacterium hallii, Faecalibacterium prausnitzii, and Anaerostipes caccae (CC4) can prevent tumor development, including CRC, melanoma, breast, and lung cancers. Specifically, CC4 supplementation increases the frequency and activity of tumor-infiltrating IFN-γ+ CD8+ T cells (154). In addition, some studies have found that Lactobacillus gallinarum can promote the apoptosis of CRC cells by secreting a protective metabolite indole-3-lactic acid, thereby preventing the occurrence of CRC (155). Lactobacillus reuteri metabolizes to produce reuterin, which inhibits CRC growth by inducing oxidative stress and inhibiting protein translation (156). Surgical castration is one of the main methods for the treatment of prostate cancer (157). However, castration resistance after castration is an important reason for the development of prostate cancer (158). Pernigoni et al. (47) found that treatment with a combination of broad-spectrum antibiotics slowed the development of prostate cancer in mice. Ruminococci enriched in the gut microbiota of castration-resistant mice have the ability to convert pregnenolone and hydroxypregnenolone to downstream androgenic steroids (dehydroepiandrosterone [DHEA] and testosterone).
In general, the modification and application of microorganisms have great contribution to tumor immunotherapy, including improving efficacy and reducing side effects (Figure 2). At the same time, the microbiota, as a therapeutic target, also plays an important role in tumor treatment and prevention. However, microbial-targeted measures have largely focused on gut microbes, while applications to colonization of other epithelial barrier microbes remain to be expanded.
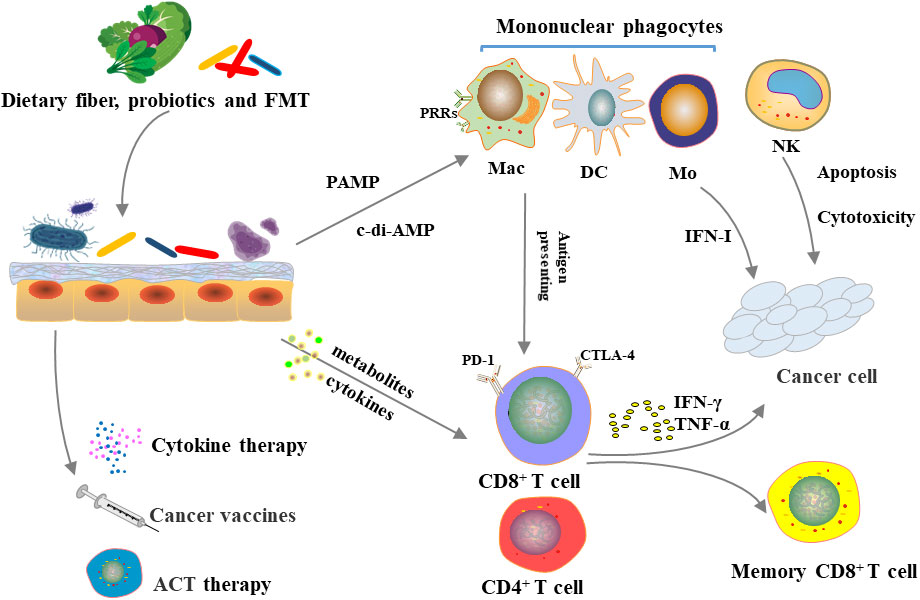
Figure 2 Microbiome, immune system and cancer immunotherapy.Diet, probiotic use and FMT can alter gut microbiota. The microbiota can directly influence innate and adaptive immunity or indirectly influence immune system through metabolism, which in turn affects the efficacy of immunosuppressive checkpoint inhibitors, cytokine therapy, tumor vaccines, and adoptive cell transfer therapy.
Conclusion
The microbiota directly or indirectly activates and regulates the host’s immune system. Cancer immunotherapy, as a strategy that relies on the autoimmune system to fight tumors, has been proved to be related to the microbiota by several studies. In this review, we summarize the relationship between oncogenic microbial infection, microbiota dysbiosis, and carcinogenesis, and describe the relevant mechanisms. We also link the microbiota and tumor immunity from three perspectives (innate immunity, adaptive immunity, and metabolism). The impact of crosstalk between the microbiota and its metabolites on innate immune cells (NK, macrophages, and DCs) and effector T cells (especially CD8+T) on immunotherapy is described. Finally, we summarize the role of microbial modification and application in various tumor immunotherapies (ICIs, ACT, cytokine therapy, and tumor vaccines), including the use of microorganisms themselves as targets to treat and prevent cancer.
It is worth noting that the interaction of microbiota, immune system, and immunotherapy is complex; therefore, some problems still persist regarding the participation of microbiota in immunotherapy (4, 146). As discussed in a previous section, precise probiotic supplementation requires expanded basic research and preclinical trials. Besides, existing studies have shown that the use of probiotics in cancer immunotherapy is not necessarily positive, and some probiotics may hinder the effect of immunotherapy and may even promote cancer progression (132, 159). Therefore, better preparation should be done before conducting human trials to study the effect of commercially available probiotics on cancer immunotherapy. Although FMT may have an effect on ICIs, the effect of FMT on the reinduction of anti–PD-1 immunotherapy in patients with refractory metastatic melanoma is suboptimal, with only 30% of patients benefiting from it in one clinical trial (149). Furthermore, there are many side effects of FMT, such as abdominal discomfort, cramping, bloating, diarrhea, or constipation, which emphasizes higher FMT donor requirements (160). In addition, the composition of the microbiota varies in different individuals, and this is affected by multiple factors such as age, diet, circadian rhythm, as well as medication exposure (161). The uncertainty brought about by these factors also brings challenges for microorganisms in tumor immunotherapy. Therefore, future research may be able to combine multi-omics analysis, such as carefully characterizing the biological characteristics of microorganisms through genome sequencing and biochemical/microbiological analyses, to develop combinations of specific bacterial strains to treat various diseases including tumors (162).
Collectively, our review elucidates some of the mechanisms by which the microbiome contributes to cancer and cancer immunotherapy. These mechanisms also provide novel strategies for microbe-based cancer immunotherapy.
Author contributions
Conceptualization: PZ and JL. Writing –original draft: PZ and XW. Writing –review & editing: PZ, YH, LS, and XL.Visualization: PZ, YH, JY, YJZ, FZ, YZ and HS. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (81472650, 81673061, 31271483, 81573050, 31872739, 30300313); National Science and Technology Major Project (2019ZX09201003-003, 2018ZX09733001-001-006, 2013ZX09301304001-003, 2012ZX10002006—003—001, 2009ZX09103-714); Sichuan Provincial Outstanding Youth Fund (2015JQO025); Key Research and Development Program of Sichuan Province (2020YFS0271); Applied Basic Research Program of Sichuan Province (2008SZ0093).
Conflict of interest
Author HS was employed by company Sichuan Aupone Pharmaceutical Co., Ltd,.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature (2012) 486(7402):207–14. doi: 10.1038/nature11234
2. Heymann CJF, Bard JM, Heymann MF, Heymann D, Bobin-Dubigeon C. The intratumoral microbiome: Characterization methods and functional impact. Cancer Lett (2021) 522:63–79. doi: 10.1016/j.canlet.2021.09.009
3. Dominguez-Bello MG, Godoy-Vitorino F, Knight R, Blaser MJ. Role of the microbiome in human development. Gut (2019) 68(6):1108–14. doi: 10.1136/gutjnl-2018-317503
4. Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell (2014) 157(1):121–41. doi: 10.1016/j.cell.2014.03.011
5. Cryan JF, Dinan TG. Mind-altering microorganisms: The impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci (2012) 13(10):701–12. doi: 10.1038/nrn3346
6. Blasco T, Perez-Burillo S, Balzerani F, Hinojosa-Nogueira D, Lerma-Aguilera A, Pastoriza S, et al. An extended reconstruction of human gut microbiota metabolism of dietary compounds. Nat Commun (2021) 12(1):4728. doi: 10.1038/s41467-021-25056-x
7. Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nat Rev Immunol (2014) 14(10):667–85. doi: 10.1038/nri3738
8. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature (2010) 464(7285):59–65. doi: 10.1038/nature08821
9. Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, Knight R. Current understanding of the human microbiome. Nat Med (2018) 24(4):392–400. doi: 10.1038/nm.4517
10. Zhang CX, Wang HY, Chen TX. Interactions between intestinal Microflora/Probiotics and the immune system. BioMed Res Int (2019) 2019:6764919. doi: 10.1155/2019/6764919
11. Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science (2013) 342(6161):967–70. doi: 10.1126/science.1240527
12. Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, et al. Gut microbiome influences efficacy of pd-1-Based immunotherapy against epithelial tumors. Science (2018) 359(6371):91–7. doi: 10.1126/science.aan3706
13. Garrett WS. Cancer and the microbiota. Science (2015) 348(6230):80–6. doi: 10.1126/science.aaa4972
14. DiMaio D, Emu B, Goodman AL, Mothes W, Justice A. Cancer microbiology. J Natl Cancer Inst (2021) 114(5):651–63. doi: 10.1093/jnci/djab212
15. de Martel C, Georges D, Bray F, Ferlay J, Clifford GM. Global burden of cancer attributable to infections in 2018: A worldwide incidence analysis. Lancet Glob Health (2020) 8(2):e180–e90. doi: 10.1016/S2214-109X(19)30488-7
16. Weeks DL, Eskandari S, Scott DR, Sachs G. A h+-gated urea channel: The link between helicobacter pylori urease and gastric colonization. Science (2000) 287(5452):482–5. doi: 10.1126/science.287.5452.482
17. Yang JC, Lu CW, Lin CJ. Treatment of helicobacter pylori infection: Current status and future concepts. World J Gastroenterol (2014) 20(18):5283–93. doi: 10.3748/wjg.v20.i18.5283
18. Peek RM Jr., Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer (2002) 2(1):28–37. doi: 10.1038/nrc703
19. Uemura N, Okamoto S, Yamamoto S, Matsumura N, Yamaguchi S, Yamakido M, et al. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med (2001) 345(11):784–9. doi: 10.1056/NEJMoa001999
20. Kidane D. Molecular mechanisms of h. pylori-induced dna double-strand breaks. Int J Mol Sci (2018) 19(10). doi: 10.3390/ijms19102891
21. Camilo V, Sugiyama T, Touati E. Pathogenesis of helicobacter pylori infection. Helicobacter (2017) 22 Suppl 1. doi: 10.1111/hel.12405
22. Wang F, Meng W, Wang B, Qiao L. Helicobacter pylori-induced gastric inflammation and gastric cancer. Cancer Lett (2014) 345(2):196–202. doi: 10.1016/j.canlet.2013.08.016
23. Bakhti SZ, Latifi-Navid S. Interplay and cooperation of helicobacter pylori and gut microbiota in gastric carcinogenesis. BMC Microbiol (2021) 21(1):258. doi: 10.1186/s12866-021-02315-x
24. Amieva M, Peek RM Jr. Pathobiology of helicobacter pylori-induced gastric cancer. Gastroenterology (2016) 150(1):64–78. doi: 10.1053/j.gastro.2015.09.004
25. Pleguezuelos-Manzano C, Puschhof J, Rosendahl Huber A, van Hoeck A, Wood HM, Nomburg J, et al. Mutational signature in colorectal cancer caused by genotoxic pks(+) e. Coli Nat (2020) 580(7802):269–73. doi: 10.1038/s41586-020-2080-8
26. Cougnoux A, Dalmasso G, Martinez R, Buc E, Delmas J, Gibold L, et al. Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype. Gut (2014) 63(12):1932–42. doi: 10.1136/gutjnl-2013-305257
27. Lopes A, Billard E, Casse AH, Villeger R, Veziant J, Roche G, et al. Colibactin-positive escherichia coli induce a procarcinogenic immune environment leading to immunotherapy resistance in colorectal cancer. Int J Cancer (2020) 146(11):3147–59. doi: 10.1002/ijc.32920
28. Veziant J, Villeger R, Barnich N, Bonnet M. Gut microbiota as potential biomarker and/or therapeutic target to improve the management of cancer: Focus on colibactin-producing escherichia coli in colorectal cancer. Cancers (Basel) (2021) 13(9). doi: 10.3390/cancers13092215
29. Iftekhar A, Berger H, Bouznad N, Heuberger J, Boccellato F, Dobrindt U, et al. Genomic aberrations after short-term exposure to colibactin-producing e. coli transform primary colon epithelial cells. Nat Commun (2021) 12(1):1003. doi: 10.1038/s41467-021-21162-y
30. Sears CL, Garrett WS. Microbes, microbiota, and colon cancer. Cell Host Microbe (2014) 15(3):317–28. doi: 10.1016/j.chom.2014.02.007
31. Montalban-Arques A, Scharl M. Intestinal microbiota and colorectal carcinoma: Implications for pathogenesis, diagnosis, and therapy. EBioMedicine (2019) 48:648–55. doi: 10.1016/j.ebiom.2019.09.050
32. Fassi Fehri L, Mak TN, Laube B, Brinkmann V, Ogilvie LA, Mollenkopf H, et al. Prevalence of propionibacterium acnes in diseased prostates and its inflammatory and transforming activity on prostate epithelial cells. Int J Med Microbiol (2011) 301(1):69–78. doi: 10.1016/j.ijmm.2010.08.014
33. Ramalingam D, Kieffer-Kwon P, Ziegelbauer JM. Emerging themes from ebv and kshv microrna targets. Viruses (2012) 4(9):1687–710. doi: 10.3390/v4091687
34. Yuen MF, Chen DS, Dusheiko GM, Janssen HLA, Lau DTY, Locarnini SA, et al. Hepatitis b virus infection. Nat Rev Dis Primers (2018) 4:18035. doi: 10.1038/nrdp.2018.35
35. Morozov VA, Lagaye S. Hepatitis c virus: Morphogenesis, infection and therapy. World J Hepatol (2018) 10(2):186–212. doi: 10.4254/wjh.v10.i2.186
36. Gaglia MM, Munger K. More than just oncogenes: Mechanisms of tumorigenesis by human viruses. Curr Opin Virol (2018) 32:48–59. doi: 10.1016/j.coviro.2018.09.003
37. Lortet-Tieulent J, Georges D, Bray F, Vaccarella S. Profiling global cancer incidence and mortality by socioeconomic development. Int J Cancer (2020) 147(11):3029–36. doi: 10.1002/ijc.33114
38. Kelsen JR, Wu GD. The gut microbiota, environment and diseases of modern society. Gut Microbes (2012) 3(4):374–82. doi: 10.4161/gmic.21333
39. Levy M, Kolodziejczyk AA, Thaiss CA, Elinav E. Dysbiosis and the immune system. Nat Rev Immunol (2017) 17(4):219–32. doi: 10.1038/nri.2017.7
40. Ahn J, Sinha R, Pei Z, Dominianni C, Wu J, Shi J, et al. Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst (2013) 105(24):1907–11. doi: 10.1093/jnci/djt300
41. Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med (2019) 25(6):968–76. doi: 10.1038/s41591-019-0458-7
42. Song M, Chan AT, Sun J. Influence of the gut microbiome, diet, and environment on risk of colorectal cancer. Gastroenterology (2020) 158(2):322–40. doi: 10.1053/j.gastro.2019.06.048
43. Marzano M, Fosso B, Piancone E, Defazio G, Pesole G, De Robertis M. Stem cell impairment at the host-microbiota interface in colorectal cancer. Cancers (Basel) (2021) 13(5). doi: 10.3390/cancers13050996
44. Schwabe RF, Greten TF. Gut microbiome in hcc - mechanisms, diagnosis and therapy. J Hepatol (2020) 72(2):230–8. doi: 10.1016/j.jhep.2019.08.016
45. Milosevic I, Vujovic A, Barac A, Djelic M, Korac M, Radovanovic Spurnic A, et al. Gut-liver axis, gut microbiota, and its modulation in the management of liver diseases: A review of the literature. Int J Mol Sci (2019) 20(2). doi: 10.3390/ijms20020395
46. Zhang HL, Yu LX, Yang W, Tang L, Lin Y, Wu H, et al. Profound impact of gut homeostasis on chemically-induced pro-tumorigenic inflammation and hepatocarcinogenesis in rats. J Hepatol (2012) 57(4):803–12. doi: 10.1016/j.jhep.2012.06.011
47. Pernigoni N, Zagato E, Calcinotto A, Troiani M, Mestre RP, Cali B, et al. Commensal bacteria promote endocrine resistance in prostate cancer through androgen biosynthesis. Science (2021) 374(6564):216–24. doi: 10.1126/science.abf8403
48. Chee WJY, Chew SY, Than LTL. Vaginal microbiota and the potential of lactobacillus derivatives in maintaining vaginal health. Microb Cell Fact (2020) 19(1):203. doi: 10.1186/s12934-020-01464-4
49. Morikawa A, Kawabata A, Shirahige K, Akiyama T, Okamoto A, Sutani T. Altered cervicovaginal microbiota in premenopausal ovarian cancer patients. Gene (2022) 811:146083. doi: 10.1016/j.gene.2021.146083
50. Nene NR, Reisel D, Leimbach A, Franchi D, Jones A, Evans I, et al. Association between the cervicovaginal microbiome, Brca1 mutation status, and risk of ovarian cancer: A case-control study. Lancet Oncol (2019) 20(8):1171–82. doi: 10.1016/S1470-2045(19)30340-7
51. Andersson AF, Lindberg M, Jakobsson H, Backhed F, Nyren P, Engstrand L. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PloS One (2008) 3(7):e2836. doi: 10.1371/journal.pone.0002836
52. Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, et al. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci U.S.A. (2006) 103(3):732–7. doi: 10.1073/pnas.0506655103
53. Han YW, Shi W, Huang GT, Kinder Haake S, Park NH, Kuramitsu H, et al. Interactions between periodontal bacteria and human oral epithelial cells: Fusobacterium nucleatum adheres to and invades epithelial cells. Infect Immun (2000) 68(6):3140–6. doi: 10.1128/IAI.68.6.3140-3146.2000
54. Krisanaprakornkit S, Kimball JR, Weinberg A, Darveau RP, Bainbridge BW, Dale BA. Inducible expression of human beta-defensin 2 by fusobacterium nucleatum in oral epithelial cells: Multiple signaling pathways and role of commensal bacteria in innate immunity and the epithelial barrier. Infect Immun (2000) 68(5):2907–15. doi: 10.1128/IAI.68.5.2907-2915.2000
55. Swidsinski A, Dorffel Y, Loening-Baucke V, Theissig F, Ruckert JC, Ismail M, et al. Acute appendicitis is characterised by local invasion with fusobacterium Nucleatum/Necrophorum. Gut (2011) 60(1):34–40. doi: 10.1136/gut.2009.191320
56. Warren RL, Freeman DJ, Pleasance S, Watson P, Moore RA, Cochrane K, et al. Co-Occurrence of anaerobic bacteria in colorectal carcinomas. Microbiome (2013) 1(1):16. doi: 10.1186/2049-2618-1-16
57. Han YW. Fusobacterium nucleatum: A commensal-turned pathogen. Curr Opin Microbiol (2015) 23:141–7. doi: 10.1016/j.mib.2014.11.013
58. Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating e-Cadherin/Beta-Catenin signaling via its fada adhesin. Cell Host Microbe (2013) 14(2):195–206. doi: 10.1016/j.chom.2013.07.012
59. Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell (2017) 170(3):548–63 e16. doi: 10.1016/j.cell.2017.07.008
60. Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science (2020) 368(6494):973–80. doi: 10.1126/science.aay9189
61. Fu A, Yao B, Dong T, Chen Y, Yao J, Liu Y, et al. Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell (2022) 185(8):1356–72 e26. doi: 10.1016/j.cell.2022.02.027
62. Shi Y, Zheng W, Yang K, Harris KG, Ni K, Xue L, et al. Intratumoral accumulation of gut microbiota facilitates Cd47-based immunotherapy via sting signaling. J Exp Med (2020) 217(5). doi: 10.1084/jem.20192282
63. Boesch M, Baty F, Albrich WC, Flatz L, Rodriguez R, Rothschild SI, et al. Local tumor microbial signatures and response to checkpoint blockade in non-small cell lung cancer. Oncoimmunology (2021) 10(1):1988403. doi: 10.1080/2162402X.2021.1988403
64. Ma J, Gnanasekar A, Lee A, Li WT, Haas M, Wang-Rodriguez J, et al. Influence of intratumor microbiome on clinical outcome and immune processes in prostate cancer. Cancers (Basel) (2020) 12(9). doi: 10.3390/cancers12092524
65. Xie Y, Xie F, Zhou X, Zhang L, Yang B, Huang J, et al. Microbiota in tumors: From understanding to application. Adv Sci (Weinh) (2022) 9(21):e2200470. doi: 10.1002/advs.202200470
66. Alam A, Levanduski E, Denz P, Villavicencio HS, Bhatta M, Alhorebi L, et al. Fungal mycobiome drives il-33 secretion and type 2 immunity in pancreatic cancer. Cancer Cell (2022) 40(2):153–67 e11. doi: 10.1016/j.ccell.2022.01.003
67. Tan S, Li D, Zhu X. Cancer immunotherapy: Pros, cons and beyond. BioMed Pharmacother (2020) 124:109821. doi: 10.1016/j.biopha.2020.109821
68. Zhang Y, Zhang Z. The history and advances in cancer immunotherapy: Understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell Mol Immunol (2020) 17(8):807–21. doi: 10.1038/s41423-020-0488-6
69. Belkaid Y, Harrison OJ. Homeostatic immunity and the microbiota. Immunity (2017) 46(4):562–76. doi: 10.1016/j.immuni.2017.04.008
70. Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol (2016) 16(6):341–52. doi: 10.1038/nri.2016.42
71. Buchbinder EI, Desai A. Ctla-4 and pd-1 pathways: Similarities, differences, and implications of their inhibition. Am J Clin Oncol (2016) 39(1):98–106. doi: 10.1097/COC.0000000000000239
72. Han Y, Liu D, Li L. Pd-1/Pd-L1 pathway: Current researches in cancer. Am J Cancer Res (2020) 10(3):727–42.
73. Joseph RW, Elassaiss-Schaap J, Kefford R, Hwu WJ, Wolchok JD, Joshua AM, et al. Correction: Baseline tumor size is an independent prognostic factor for overall survival in patients with melanoma treated with pembrolizumab. Clin Cancer Res (2018) 24(23):6098. doi: 10.1158/1078-0432.CCR-18-3340
74. Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med (2010) 363(8):711–23. doi: 10.1056/NEJMoa1003466
75. Farolfi A, Ridolfi L, Guidoboni M, Nicoletti SV, Piciucchi S, Valmorri L, et al. Ipilimumab in advanced melanoma: Reports of long-lasting responses. Melanoma Res (2012) 22(3):263–70. doi: 10.1097/CMR.0b013e328353e65c
76. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti-Pd-1 immunotherapy in melanoma patients. Science (2018) 359(6371):97–103. doi: 10.1126/science.aan4236
77. Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal bifidobacterium promotes antitumor immunity and facilitates anti-Pd-L1 efficacy. Science (2015) 350(6264):1084–9. doi: 10.1126/science.aac4255
78. Vetizou M, Pitt JM, Daillere R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer immunotherapy by ctla-4 blockade relies on the gut microbiota. Science (2015) 350(6264):1079–84. doi: 10.1126/science.aad1329
79. Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell (2017) 168(4):707–23. doi: 10.1016/j.cell.2017.01.017
80. Boesch M, Baty F, Rothschild SI, Tamm M, Joerger M, Fruh M, et al. Tumour neoantigen mimicry by microbial species in cancer immunotherapy. Br J Cancer (2021) 125(3):313–23. doi: 10.1038/s41416-021-01365-2
81. Chan TA, Yarchoan M, Jaffee E, Swanton C, Quezada SA, Stenzinger A, et al. Development of tumor mutation burden as an immunotherapy biomarker: Utility for the oncology clinic. Ann Oncol (2019) 30(1):44–56. doi: 10.1093/annonc/mdy495
82. Xu J, Jo J. Broad cross-reactivity of the T-cell repertoire achieves specific and sufficiently rapid target searching. J Theor Biol (2019) 466:119–27. doi: 10.1016/j.jtbi.2019.01.025
83. Selin LK, Brehm MA, Naumov YN, Cornberg M, Kim SK, Clute SC, et al. Memory of mice and men: Cd8+ T-cell cross-reactivity and heterologous immunity. Immunol Rev (2006) 211:164–81. doi: 10.1111/j.0105-2896.2006.00394.x
84. Bessell CA, Isser A, Havel JJ, Lee S, Bell DR, Hickey JW, et al. Commensal bacteria stimulate antitumor responses via T cell cross-reactivity. JCI Insight (2020) 5(8). doi: 10.1172/jci.insight.135597
85. Balachandran VP, Luksza M, Zhao JN, Makarov V, Moral JA, Remark R, et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature (2017) 551(7681):512–6. doi: 10.1038/nature24462
86. Sioud M. T-Cell cross-reactivity may explain the Large variation in how cancer patients respond to checkpoint inhibitors. Scand J Immunol (2018) 87(3). doi: 10.1111/sji.12643
87. Mami-Chouaib F, Blanc C, Corgnac S, Hans S, Malenica I, Granier C, et al. Resident memory T cells, critical components in tumor immunology. J Immunother Cancer (2018) 6(1):87. doi: 10.1186/s40425-018-0399-6
88. Hinshaw DC, Shevde LA. The tumor microenvironment innately modulates cancer progression. Cancer Res (2019) 79(18):4557–66. doi: 10.1158/0008-5472.CAN-18-3962
89. Thaiss CA, Zmora N, Levy M, Elinav E. The microbiome and innate immunity. Nature (2016) 535(7610):65–74. doi: 10.1038/nature18847
90. Keogh CE, Rude KM, Gareau MG. Role of pattern recognition receptors and the microbiota in neurological disorders. J Physiol (2021) 599(5):1379–89. doi: 10.1113/JP279771
91. Fawkner-Corbett D, Simmons A, Parikh K. Microbiome, pattern recognition receptor function in health and inflammation. Best Pract Res Clin Gastroenterol (2017) 31(6):683–91. doi: 10.1016/j.bpg.2017.11.001
92. Maloy KJ, Powrie F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature (2011) 474(7351):298–306. doi: 10.1038/nature10208
93. Chen CY, Kao CL, Liu CM. The cancer prevention, anti-inflammatory and anti-oxidation of bioactive phytochemicals targeting the Tlr4 signaling pathway. Int J Mol Sci (2018) 19(9). doi: 10.3390/ijms19092729
94. Bianchi F, Pretto S, Tagliabue E, Balsari A, Sfondrini L. Exploiting Poly(I:C) to induce cancer cell apoptosis. Cancer Biol Ther (2017) 18(10):747–56. doi: 10.1080/15384047.2017.1373220
95. Branquinho D, Freire P, Sofia C. Nod2 mutations and colorectal cancer - where do we stand? World J Gastrointest Surg (2016) 8(4):284–93. doi: 10.4240/wjgs.v8.i4.284
96. Cui J, Chen Y, Wang HY, Wang RF. Mechanisms and pathways of innate immune activation and regulation in health and cancer. Hum Vaccin Immunother (2014) 10(11):3270–85. doi: 10.4161/21645515.2014.979640
97. Qiu Q, Lin Y, Ma Y, Li X, Liang J, Chen Z, et al. Exploring the emerging role of the gut microbiota and tumor microenvironment in cancer immunotherapy. Front Immunol (2020) 11:612202. doi: 10.3389/fimmu.2020.612202
98. Lam KC, Araya RE, Huang A, Chen Q, Di Modica M, Rodrigues RR, et al. Microbiota triggers sting-type I ifn-dependent monocyte reprogramming of the tumor microenvironment. Cell (2021) 184(21):5338–56 e21. doi: 10.1016/j.cell.2021.09.019
99. Morgan MA, Buning H, Sauer M, Schambach A. Use of cell and genome modification technologies to generate improved “Off-the-Shelf” car T and car nk cells. Front Immunol (2020) 11:1965. doi: 10.3389/fimmu.2020.01965
100. Nair R, Westin J. Car T-cells. Adv Exp Med Biol (2020) 1244:215–33. doi: 10.1007/978-3-030-41008-7_10
101. Sermer D, Brentjens R. Car T-cell therapy: Full speed ahead. Hematol Oncol (2019) 37 Suppl 1:95–100. doi: 10.1002/hon.2591
102. Uribe-Herranz M, Bittinger K, Rafail S, Guedan S, Pierini S, Tanes C, et al. Gut microbiota modulates adoptive cell therapy via Cd8alpha dendritic cells and il-12. JCI Insight (2018) 3(4). doi: 10.1172/jci.insight.94952
103. Xie G, Dong H, Liang Y, Ham JD, Rizwan R, Chen J. Car-nk cells: A promising cellular immunotherapy for cancer. EBioMedicine (2020) 59:102975. doi: 10.1016/j.ebiom.2020.102975
104. Daher M, Rezvani K. Outlook for new car-based therapies with a focus on car nk cells: What lies beyond car-engineered T cells in the race against cancer. Cancer Discovery (2021) 11(1):45–58. doi: 10.1158/2159-8290.CD-20-0556
105. D’Alessandro G, Antonangeli F, Marrocco F, Porzia A, Lauro C, Santoni A, et al. Gut microbiota alterations affect glioma growth and innate immune cells involved in tumor immunosurveillance in mice. Eur J Immunol (2020) 50(5):705–11. doi: 10.1002/eji.201948354
106. Kim M, Qie Y, Park J, Kim CH. Gut microbial metabolites fuel host antibody responses. Cell Host Microbe (2016) 20(2):202–14. doi: 10.1016/j.chom.2016.07.001
107. Morrison DJ, Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes (2016) 7(3):189–200. doi: 10.1080/19490976.2015.1134082
108. McNabney SM, Henagan TM. Short chain fatty acids in the colon and peripheral tissues: A focus on butyrate, colon cancer, obesity and insulin resistance. Nutrients (2017) 9(12). doi: 10.3390/nu9121348
109. Nastasi C, Candela M, Bonefeld CM, Geisler C, Hansen M, Krejsgaard T, et al. The effect of short-chain fatty acids on human monocyte-derived dendritic cells. Sci Rep (2015) 5:16148. doi: 10.1038/srep16148
110. Yang W, Yu T, Huang X, Bilotta AJ, Xu L, Lu Y, et al. Intestinal microbiota-derived short-chain fatty acids regulation of immune cell il-22 production and gut immunity. Nat Commun (2020) 11(1):4457. doi: 10.1038/s41467-020-18262-6
111. Ranjbar R, Vahdati SN, Tavakoli S, Khodaie R, Behboudi H. Immunomodulatory roles of microbiota-derived short-chain fatty acids in bacterial infections. BioMed Pharmacother (2021) 141:111817. doi: 10.1016/j.biopha.2021.111817
112. Luu M, Riester Z, Baldrich A, Reichardt N, Yuille S, Busetti A, et al. Microbial short-chain fatty acids modulate Cd8(+) T cell responses and improve adoptive immunotherapy for cancer. Nat Commun (2021) 12(1):4077. doi: 10.1038/s41467-021-24331-1
113. Han K, Nam J, Xu J, Sun X, Huang X, Animasahun O, et al. Generation of systemic antitumour immunity via the in situ modulation of the gut microbiome by an orally administered inulin gel. Nat BioMed Eng (2021) 5(11):1377–88. doi: 10.1038/s41551-021-00749-2
114. Zhang SL, Mao YQ, Zhang ZY, Li ZM, Kong CY, Chen HL, et al. Pectin supplement significantly enhanced the anti-Pd-1 efficacy in tumor-bearing mice humanized with gut microbiota from patients with colorectal cancer. Theranostics (2021) 11(9):4155–70. doi: 10.7150/thno.54476
115. Bachem A, Makhlouf C, Binger KJ, de Souza DP, Tull D, Hochheiser K, et al. Microbiota-derived short-chain fatty acids promote the memory potential of antigen-activated Cd8(+) T cells. Immunity (2019) 51(2):285–97 e5. doi: 10.1016/j.immuni.2019.06.002
116. Lv J, Jia Y, Li J, Kuai W, Li Y, Guo F, et al. Gegen qinlian decoction enhances the effect of pd-1 blockade in colorectal cancer with microsatellite stability by remodelling the gut microbiota and the tumour microenvironment. Cell Death Dis (2019) 10(6):415. doi: 10.1038/s41419-019-1638-6
117. Liu X, Hartman CL, Li L, Albert CJ, Si F, Gao A, et al. Reprogramming lipid metabolism prevents effector T cell senescence and enhances tumor immunotherapy. Sci Transl Med (2021) 13(587). doi: 10.1126/scitranslmed.aaz6314
118. Bian Y, Li W, Kremer DM, Sajjakulnukit P, Li S, Crespo J, et al. Cancer Slc43a2 alters T cell methionine metabolism and histone methylation. Nature (2020) 585(7824):277–82. doi: 10.1038/s41586-020-2682-1
119. Pauken KE, Sammons MA, Odorizzi PM, Manne S, Godec J, Khan O, et al. Epigenetic stability of exhausted T cells limits durability of reinvigoration by pd-1 blockade. Science (2016) 354(6316):1160–5. doi: 10.1126/science.aaf2807
120. Geiger R, Rieckmann JC, Wolf T, Basso C, Feng Y, Fuhrer T, et al. L-arginine modulates T cell metabolism and enhances survival and anti-tumor activity. Cell (2016) 167(3):829–42 e13. doi: 10.1016/j.cell.2016.09.031
121. Leone RD, Zhao L, Englert JM, Sun IM, Oh MH, Sun IH, et al. Glutamine blockade induces divergent metabolic programs to overcome tumor immune evasion. Science (2019) 366(6468):1013–21. doi: 10.1126/science.aav2588
122. Gao J, Xu K, Liu H, Liu G, Bai M, Peng C, et al. Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism. Front Cell Infect Microbiol (2018) 8:13. doi: 10.3389/fcimb.2018.00013
123. Kennedy PJ, Cryan JF, Dinan TG, Clarke G. Kynurenine pathway metabolism and the microbiota-Gut-Brain axis. Neuropharmacology (2017) 112(Pt B):399–4121. doi: 10.1016/j.neuropharm.2016.07.002
124. Huang J, Liu D, Wang Y, Liu L, Li J, Yuan J, et al. Ginseng polysaccharides alter the gut microbiota and Kynurenine/Tryptophan ratio, potentiating the antitumour effect of antiprogrammed cell death 1/Programmed cell death ligand 1 (Anti-Pd-1/Pd-L1) immunotherapy. Gut (2021) 71(4):734–45. doi: 10.1136/gutjnl-2020-321031
125. Mager LF, Burkhard R, Pett N, Cooke NCA, Brown K, Ramay H, et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science (2020) 369(6510):1481–9. doi: 10.1126/science.abc3421
126. Saveljeva S, Sewell GW, Ramshorn K, Cader MZ, West JA, Clare S, et al. A purine metabolic checkpoint that prevents autoimmunity and autoinflammation. Cell Metab (2022) 34(1):106–24 e10. doi: 10.1016/j.cmet.2021.12.009
127. Wu J, Wei Z, Cheng P, Qian C, Xu F, Yang Y, et al. Rhein modulates host purine metabolism in intestine through gut microbiota and ameliorates experimental colitis. Theranostics (2020) 10(23):10665–79. doi: 10.7150/thno.43528
128. Holscher HD. Diet affects the gastrointestinal microbiota and health. J Acad Nutr Diet (2020) 120(4):495–9. doi: 10.1016/j.jand.2019.12.016
129. Slavin J. Fiber and prebiotics: Mechanisms and health benefits. Nutrients (2013) 5(4):1417–35. doi: 10.3390/nu5041417
130. Gonzalez Olmo BM, Butler MJ, Barrientos RM. Evolution of the human diet and its impact on gut microbiota, immune responses, and brain health. Nutrients (2021) 13(1). doi: 10.3390/nu13010196
131. Messaoudene M, Pidgeon R, Richard C, Ponce M, Diop K, Benlaifaoui M, et al. A natural polyphenol exerts antitumor activity and circumvents anti-Pd-1 resistance through effects on the gut microbiota. Cancer Discovery (2022) 12(4):1070–87. doi: 10.1158/2159-8290.CD-21-0808
132. Spencer CN, McQuade JL, Gopalakrishnan V, McCulloch JA, Vetizou M, Cogdill AP, et al. Dietary fiber and probiotics influence the gut microbiome and melanoma immunotherapy response. Science (2021) 374(6575):1632–40. doi: 10.1126/science.aaz7015
133. Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, et al. Expert consensus document. the international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol (2014) 11(8):506–14. doi: 10.1038/nrgastro.2014.66
134. Han S, Lu Y, Xie J, Fei Y, Zheng G, Wang Z, et al. Probiotic gastrointestinal transit and colonization after oral administration: A long journey. Front Cell Infect Microbiol (2021) 11:609722. doi: 10.3389/fcimb.2021.609722
135. van Zyl WF, Deane SM, Dicks LMT. Molecular insights into probiotic mechanisms of action employed against intestinal pathogenic bacteria. Gut Microbes (2020) 12(1):1831339. doi: 10.1080/19490976.2020.1831339
136. Kim SK, Guevarra RB, Kim YT, Kwon J, Kim H, Cho JH, et al. Role of probiotics in human gut microbiome-associated diseases. J Microbiol Biotechnol (2019) 29(9):1335–40. doi: 10.4014/jmb.1906.06064
137. Kawanabe-Matsuda H, Takeda K, Nakamura M, Makino S, Karasaki T, Kakimi K, et al. Dietary lactobacillus-derived exopolysaccharide enhances immune-checkpoint blockade therapy. Cancer Discovery (2022) 12(5):1336–55. doi: 10.1158/2159-8290.CD-21-0929
138. Tomita Y, Ikeda T, Sakata S, Saruwatari K, Sato R, Iyama S, et al. Association of probiotic clostridium butyricum therapy with survival and response to immune checkpoint blockade in patients with lung cancer. Cancer Immunol Res (2020) 8(10):1236–42. doi: 10.1158/2326-6066.CIR-20-0051
139. Grajeda-Iglesias C, Durand S, Daillere R, Iribarren K, Lemaitre F, Derosa L, et al. Oral administration of akkermansia muciniphila elevates systemic antiaging and anticancer metabolites. Aging (Albany NY) (2021) 13(5):6375–405. doi: 10.18632/aging.202739
140. Cani PD, Depommier C, Derrien M, Everard A, de Vos WM. Akkermansia muciniphila: Paradigm for next-generation beneficial microorganisms. Nat Rev Gastroenterol Hepatol (2022). doi: 10.1038/s41575-022-00631-9
141. Zhao Q, Yu J, Hao Y, Zhou H, Hu Y, Zhang C, et al. Akkermansia muciniphila plays critical roles in host health. Crit Rev Microbiol (2022), 1–19. doi: 10.1080/1040841X.2022.2037506
142. Wu M, Bai J, Ma C, Wei J, Du X. The role of gut microbiota in tumor immunotherapy. J Immunol Res (2021) 2021:5061570. doi: 10.1155/2021/5061570
143. Badgeley A, Anwar H, Modi K, Murphy P, Lakshmikuttyamma A. Effect of probiotics and gut microbiota on anti-cancer drugs: Mechanistic perspectives. Biochim Biophys Acta Rev Cancer (2021) 1875(1):188494. doi: 10.1016/j.bbcan.2020.188494
144. Chen D, Wu J, Jin D, Wang B, Cao H. Fecal microbiota transplantation in cancer management: Current status and perspectives. Int J Cancer (2019) 145(8):2021–31. doi: 10.1002/ijc.32003
145. Vindigni SM, Surawicz CM. Fecal microbiota transplantation. Gastroenterol Clin North Am (2017) 46(1):171–85. doi: 10.1016/j.gtc.2016.09.012
146. Gupta S, Mullish BH, Allegretti JR. Fecal microbiota transplantation: The evolving risk landscape. Am J Gastroenterol (2021) 116(4):647–56. doi: 10.14309/ajg.0000000000001075
147. Costello SP, Hughes PA, Waters O, Bryant RV, Vincent AD, Blatchford P, et al. Effect of fecal microbiota transplantation on 8-week remission in patients with ulcerative colitis: A randomized clinical trial. JAMA (2019) 321(2):156–64. doi: 10.1001/jama.2018.20046
148. Antushevich H. Fecal microbiota transplantation in disease therapy. Clin Chim Acta (2020) 503:90–8. doi: 10.1016/j.cca.2019.12.010
149. Baruch EN, Youngster I, Ben-Betzalel G, Ortenberg R, Lahat A, Katz L, et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science (2021) 371(6529):602–9. doi: 10.1126/science.abb5920
150. Kitagawa K, Tatsumi M, Kato M, Komai S, Doi H, Hashii Y, et al. An oral cancer vaccine using a bifidobacterium vector suppresses tumor growth in a syngeneic mouse bladder cancer model. Mol Ther Oncolytics (2021) 22:592–603. doi: 10.1016/j.omto.2021.08.009
151. Jiang J, Mei J, Yi S, Feng C, Ma Y, Liu Y, et al. Tumor associated macrophage and microbe: The potential targets of tumor vaccine delivery. Adv Drug Delivery Rev (2022) 180:114046. doi: 10.1016/j.addr.2021.114046
152. Wang W, Xu H, Ye Q, Tao F, Wheeldon I, Yuan A, et al. Systemic immune responses to irradiated tumours via the transport of antigens to the tumour periphery by injected flagellate bacteria. Nat BioMed Eng (2022) 6(1):44–53. doi: 10.1038/s41551-021-00834-6
153. Shi L, Sheng J, Chen G, Zhu P, Shi C, Li B, et al. Combining il-2-Based immunotherapy with commensal probiotics produces enhanced antitumor immune response and tumor clearance. J Immunother Cancer (2020) 8(2). doi: 10.1136/jitc-2020-000973
154. Montalban-Arques A, Katkeviciute E, Busenhart P, Bircher A, Wirbel J, Zeller G, et al. Commensal clostridiales strains mediate effective anti-cancer immune response against solid tumors. Cell Host Microbe (2021) 29(10):1573–88 e7. doi: 10.1016/j.chom.2021.08.001
155. Sugimura N, Li Q, Chu ESH, Lau HCH, Fong W, Liu W, et al. Lactobacillus gallinarum modulates the gut microbiota and produces anti-cancer metabolites to protect against colorectal tumourigenesis. Gut (2021). doi: 10.1136/gutjnl-2020-323951
156. Bell HN, Rebernick RJ, Goyert J, Singhal R, Kuljanin M, Kerk SA, et al. Reuterin in the healthy gut microbiome suppresses colorectal cancer growth through altering redox balance. Cancer Cell (2021) 40(2):185–200 e6. doi: 10.1016/j.ccell.2021.12.001
157. Gamat M, McNeel DG. Androgen deprivation and immunotherapy for the treatment of prostate cancer. Endocr Relat Cancer (2017) 24(12):T297–T310. doi: 10.1530/ERC-17-0145
158. Chang AJ, Autio KA, Roach M 3rd, Scher HI. High-risk prostate cancer-classification and therapy. Nat Rev Clin Oncol (2014) 11(6):308–23. doi: 10.1038/nrclinonc.2014.68
159. Arthur JC, Gharaibeh RZ, Uronis JM, Perez-Chanona E, Sha W, Tomkovich S, et al. Vsl3 probiotic modifies mucosal microbial composition but does not reduce colitis-associated colorectal cancer. Sci Rep (2013) 3:2868. doi: 10.1038/srep02868
160. Gupta A, Khanna S. Fecal microbiota transplantation. JAMA (2017) 318(1):102. doi: 10.1001/jama.2017.6466
161. Cresci GA, Bawden E. Gut microbiome: What we do and don’t know. Nutr Clin Pract (2015) 30(6):734–46. doi: 10.1177/0884533615609899
Keywords: oncomicrobes, microbiota disorders, gut microbiota, immune cells, cancer immunotherapy, metabolism
Citation: Zhou P, Hu Y, Wang X, Shen L, Liao X, Zhu Y, Yu J, Zhao F, Zhou Y, Shen H and Li J (2022) Microbiome in cancer: An exploration of carcinogenesis, immune responses and immunotherapy. Front. Immunol. 13:877939. doi: 10.3389/fimmu.2022.877939
Received: 17 February 2022; Accepted: 20 July 2022;
Published: 08 August 2022.
Edited by:
JoAnn M. Sekiguchi, Michigan Medicine, University of Michigan, United StatesReviewed by:
Maximilian Boesch, Cantonal Hospital St. Gallen, SwitzerlandMichael Sigal, Charité Universitätsmedizin Berlin, Germany
Karolina Skonieczna-Żydecka, Pomeranian Medical University, Poland
Copyright © 2022 Zhou, Hu, Wang, Shen, Liao, Zhu, Yu, Zhao, Zhou, Shen and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiong Li, lijionghh@scu.edu.cn
 Pei Zhou
Pei Zhou Yawen Hu1
Yawen Hu1 Yajuan Zhu
Yajuan Zhu Jiong Li
Jiong Li