- 1Graduate School of Medical Science and Engineering, Korea Advanced Institute of Science and Technology (KAIST), Daejeon, South Korea
- 2Department of Dermatology, Ajou University School of Medicine, Suwon, South Korea
Microbiota is essential to the development and functional maturation of the immune system. The effects of the gut microbiota on myeloid cells remote from the gut, especially the skin remain unclear. Transcriptomic analysis revealed that type I interferon (IFN) signaling was down-regulated in the skin of germ-free mice compared to that in specific pathogen-free mice. The decrease in type I IFN signaling was closely related to the presence of microbiota and macrophage-specific marker CD169. The absence of CD169+ macrophages resulted in increased bacterial burden and impaired immune responses against Staphylococcus aureus skin infection. CD169+ macrophages mediated the recruitment of γδ T cells as well as the activation of γδ T cells via interleukin (IL)-23. Our findings demonstrate the role of the microbiota in establishment of a specific myeloid cell subset expressing CD169 in the skin and provide evidence of a specific mechanism by which this subset protects against bacterial skin infection.
Introduction
The microbiome actively affects the host’s immune system in several ways. Commensals residing in the gastrointestinal tract regulate immune responses in the gut and across distal sites other than the gut (1). There are three major mediators of microbiota affecting remote tissues: microbes, microbial products (metabolites), and circulating immune cells (2, 3). These mediators translocate from the intestines via circulations and affect immune responses at distal sites. Recently, the gut microbiota was found to influence type I IFN signaling at the remote tissues such as the lung, lymph nodes, and spleen (4, 5).
Type I IFNs and interferon stimulated genes (ISGs) protect the host against viral pathogens, bacteria and parasites (6, 7). Aberrantly expressed type I IFNs and/or ISGs are linked to autoimmune disorders and cancers (8, 9). Type I IFNs are constitutively secreted at low amounts in various organs (10), and the tonic IFN signaling maintains homeostasis of resident immune cells and primes cells to mount a rapid and robust immune response when challenged (4, 10).
Our analysis of a published data suggested that type I IFN signaling increases in the skin of specific pathogen free (SPF) mice compared to germ-free (GF) mice (11). Higher expression of genes related to multiple monocyte/macrophage lineage were also noted in SPF mice. Therefore, in this study, we investigated the possible relationship between increased type I IFN signaling and monocyte/macrophage lineage in the skin. We show that CD169 expression in skin macrophages is regulated by microbiota-dependent type I IFN signaling. Furthermore, we comprehensively explored the characteristics and roles of skin CD169 expressing cells in host defense.
Results
Reduced Type I IFN Signaling in GF Skin Alters Expression of the Macrophage-Specific Marker, CD169
We mined the data of a previous study on genes differentially expressed in the skin between GF and SPF mice using the GEO database (11). We pre-ranked 15,448 featured genes using log2FC values and analyzed them using GSEA. GSEA showed increase in type I IFN signature in the skin of SPF mice (Figure 1A). Gene Ontology (GO) analysis of the top 100 genes highly expressed in SPF mice revealed an increase in pathways related to type I IFN signaling (Figure 1B). There was also selective enrichment of tissue-resident macrophage markers, and monocytes in SPF condition (Figure S1). Thus, we hypothesized that there might be a relationship between type I IFN signaling and monocyte/macrophage lineage cells. Macrophage markers were investigated in Ifnar KO mice for evidence of this relationship. Mannose receptor CD206 is a well-known macrophage marker of the skin and is important for wound healing (12). CD169+ macrophages have been described in skin (13, 14), although their features are not identical to those of CD169+ cells in other tissues (15). Both CD206 and CD169, are ISGs, as queried using the Interferome database (16). We speculated that the decrease in type I IFN signaling would affect myeloid cells and result in decreased CD206 and/or CD169 expression. Flow cytometry data showed that CD169 expression was dramatically reduced in CD11b+ F4/80+ skin cells of Ifnar KO mice, but CD206 expression was not (Figure 1C).
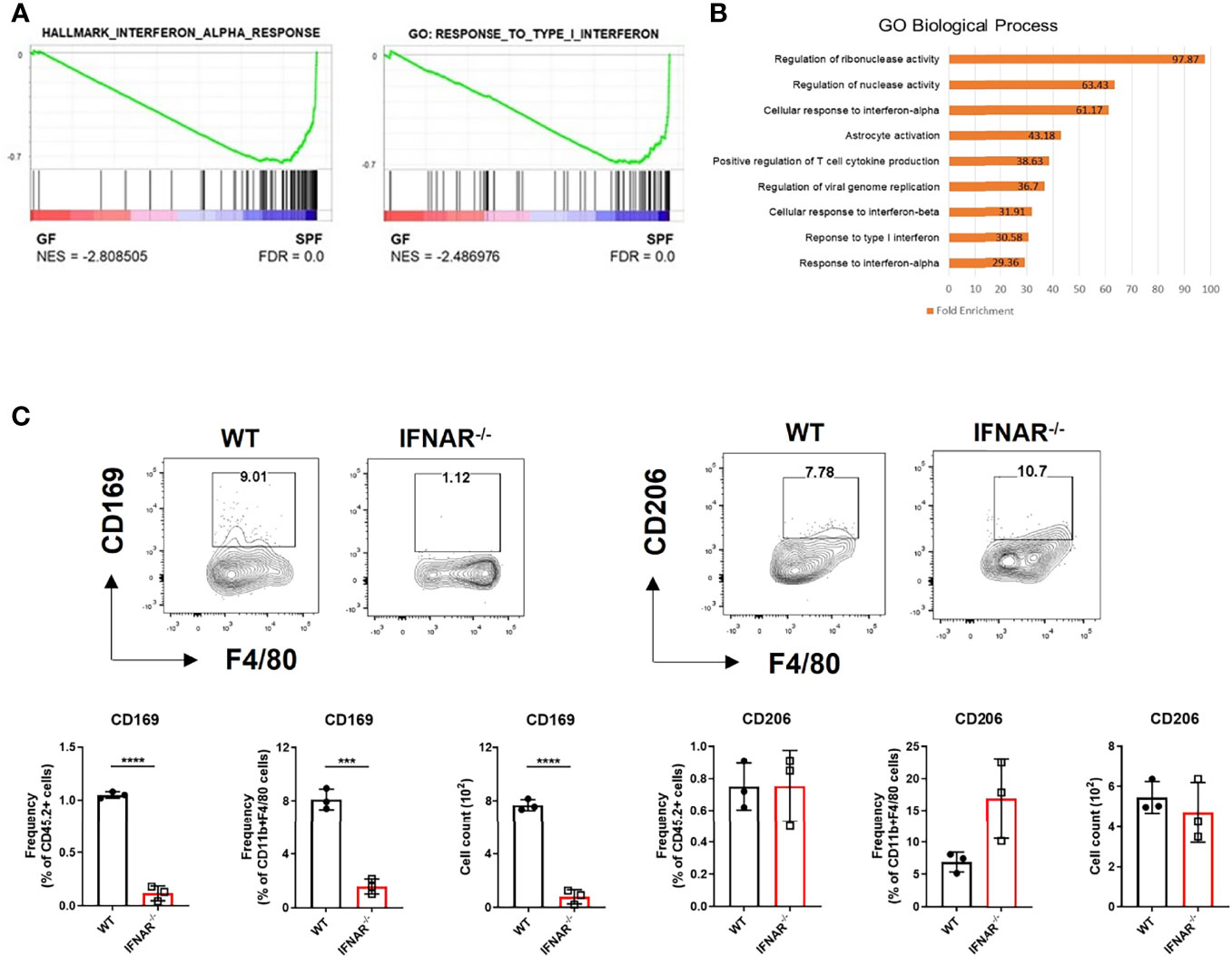
Figure 1 Diminished type I IFN signaling in germ-free skin alters expression of macrophage-specific marker, CD169. (A) Gene set enrichment analysis (GSEA) enrichment plots performed on two gene set collections (h.all.v7.2.symbols.gmt and c5.all.v7.2.symbols.gmt) in germ-free (GF) and specific pathogen-free (SPF) mice from data published in Meisel et al., 2018 (GSE98877 from GEO database). GSEA performed 1000 permutations. (B) Gene Ontology analysis of GSE98877. (C) Representative FACS plot and graphs showing frequency and cell counts of CD169+ and CD206+ myeloid cells in skin of WT and Ifnar KO mice (n = 3 per group). The cells were gated on FSC/SSC, PI-, singlet, CD45.2+, CD11b+, F4/80+ cells. Data represent two independent experiments. Data are expressed as means ± SEM. Horizontal lines above bars indicate statistical comparisons with statistical differences between categories (***p < 0.001, ****p < 0.0001).
CD169+ Cells Are Skin Macrophages With Unique Characteristics and Less Frequent in GF Skin
We found CD169+ cells primarily in the margins of the lower dermis and subcutaneous fat, otherwise termed dermal white adipose tissue (DWAT) (Figure 2A). The cells also overlapped with some F4/80+ cells, a marker of myeloid cells. In the horizontal sections, CD169+ cells were abundant in the lower dermis and were also mainly perivascular (Figure 2B). Most CD11b+/CD169+ cells were not Ly6chi monocytes or CD11chi/MHCIIhi dendritic cells, but were macrophages, defined by their expression of F4/80 and CD64 (Figure 2C). We also found CD169+ macrophages in the lower dermis and DWAT of human skin (Figure 2D). We presumed that the CD169+ macrophages would be less frequent in skin of GF mice due to its decreased type I IFN signaling. Our confocal microscope images showed lack of CD169+ cells in GF skin compared to substantial amount of CD169+ cells in SPF skin, as expected (Figure 2E). These cells were mostly bone marrow-derived (BMD), as confirmed by domination of CD45.1+ and CD169+ expressing cells in CD45.1/CD45.2 chimeric mice (Figure S2A). High C-X3-C motif chemokine receptor 1 (CX3CR1) expression accounts for tissue-residency and self-renewal with minimal contribution from blood monocytes (17–19). CX3CR1hi macrophages did not overlap with CD169+ cells in Cx3cr1+/gfp mice (Figure S2B). Nearly all CD169+ macrophages were CX3CR1lo (Figure S2C). These results indicate that CD169+ macrophages were replenished from BMD monocytes.
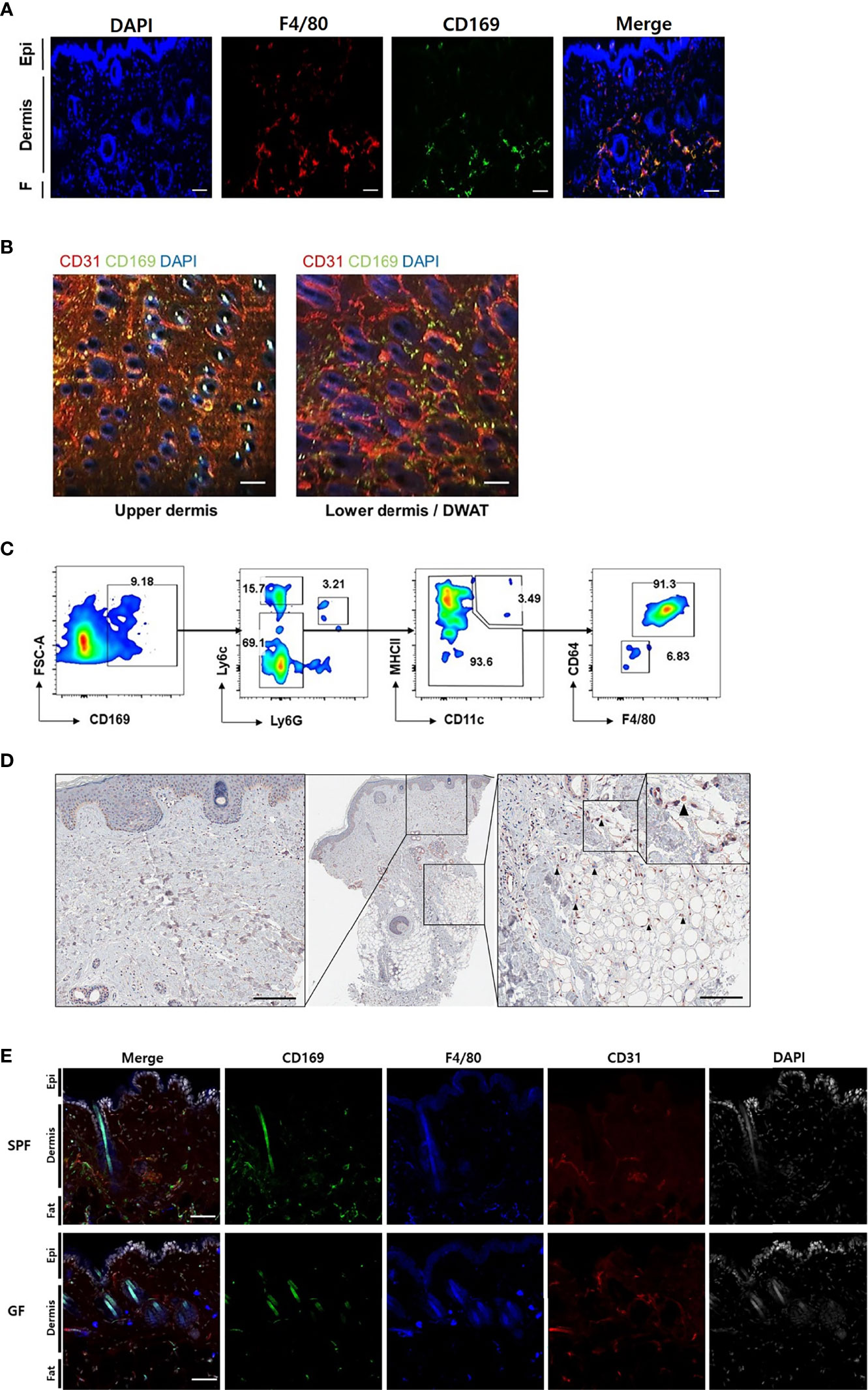
Figure 2 CD169+ cells are detectable in the skin of mouse and human, and less frequent in skin of germ-free mice compared to specific-pathogen-free mice. (A) Orthogonal projected image of a vertical skin section (100 µm) using confocal microscopy (F4/80-red, CD169-green, DAPI-blue color). Scale bar indicates 100 μm (Epi: epidermis, F: subcutaneous fat). (B) Merged images from a horizontal section (CD31-red, CD169-green, DAPI-blue color) of dermis and dermal white adipose tissue (DWAT) from WT mice. White spots indicate auto fluorescent hair shafts. Scale bar, 100 μm. Images represent three (A) and two (B) independent experiments. (C) Representative gating strategy and features of FACS-sorted CD169+ cells in skin. Gated on FSC/SSC, PI-, CD45.2 and CD11b. Plots represent two independent experiments. (D) Representative human skin sample immunostained with CD169 (black arrows indicate CD169+ cells). (E) Orthogonal projected images of vertical skin sections (100 µm) from SPF and GF mice (CD169-green, F4/80-blue, CD31-red, DAPI-white color). Scale bar indicates 100 μm. Images represent three independent experiments.
Type I IFN Signaling Dictates CD169+ Expression in BMDMs and Skin Macrophages
We investigated whether CD169 expression was induced by type I IFN in BMDMs. IFNα and IFNβ induced higher CD169 expression than media and lipopolysaccharide (LPS) (Figures 3A, B). Low levels of type I IFNs was capable of inducing CD169 expression, which was higher at 6 hours post stimulation and decreased after 24 hours. A challenge with a various dose of LPS (10 ng/mL and 100 ng/mL) or S. aureus also did not change CD169 expression (Figure S3A). The results demonstrate that type I IFN signaling in macrophages might specifically alter CD169 expression, but may not be dose-dependent, due to the change in type I IFN receptor (IFNAR) expression (Figure S3B). Our flow cytometry data was in accordance with image data (Figure 2E), as CD169 expression was diminished in F4/80+ cells of GF mice compared to SPF mice (Figure 3C). We speculated that antibiotic treatment might cause a similar decrease in CD169 expression, and we confirmed, albeit statistically insignificant (p=0.09), the decrease in skin macrophages (CD11b+/CD64+ cells) (Figure S3C). Finally, fecal transplantation, not topical S. epidermis application, was capable of partially restoring CD169 expression in skin macrophages (Figure 3D), possibly suggesting the importance of gut microbiota, rather than skin microbiota in CD169 expression.
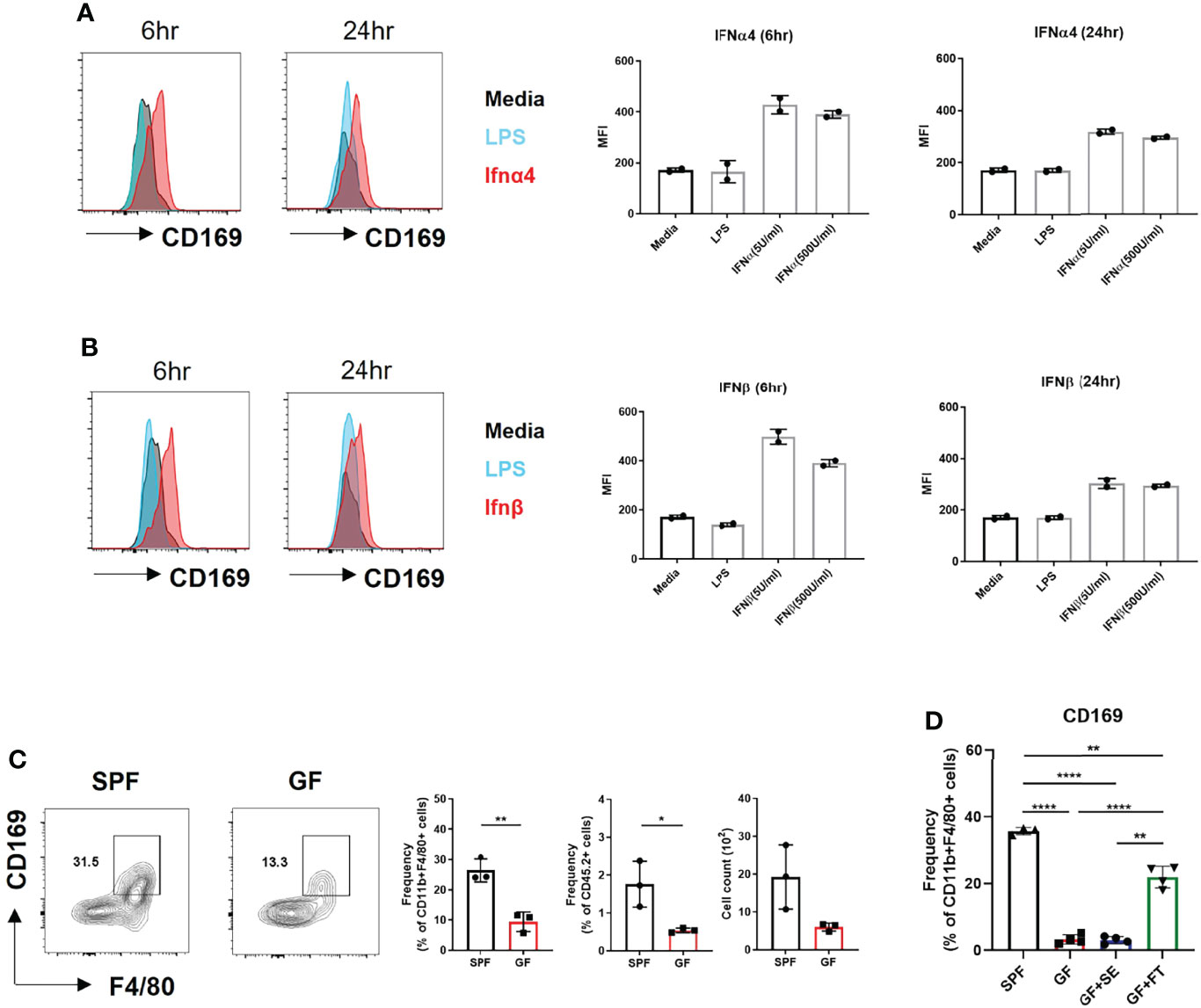
Figure 3 Microbiota-induced type I Interferon signaling dictates CD169+ expression in BMDMs and skin macrophages. (A) Representative FACS plot and bar graphs showing mean fluorescence intensity (MFI) of CD169 in BMDMs challenged with various doses of recombinant IFNα4 (5 U/mL and 500 U/mL) and (B) recombinant IFNβ (5 U/mL and 500 U/mL) at different time points (6 hours and 24 hours). Data represent two independent experiments. (C) Representative FACS plot showing frequency and cell counts of F4/80+ CD169+ cells in skin of SPF and GF mice (n = 3 per group). Gated on FSC/SSC, PI-, singlet, CD45.2+, CD11b+ cells. Data are expressed as means ± SEM (*p<0.05, **p<0.01). Data represent three independent experiments. (D) Frequency of CD169+ macrophages in skin of SPF mice (n = 3) with DPBS oral gavage (SPF), GF mice (n = 3) with DPBS oral gavage (GF), GF mice (n = 3) with S. epidermis topical application (GF+SE) and GF mice (n = 3) with fecal transplantation (GF+FT). The cells were gated on FSC/SSC, PI-, singlet, CD45.2+, CD11b+, F4/80+ cells. Combined data of three independent experiments are shown. Data are expressed as means ± SEM (**p < 0.01, ***p < 0.001, ****p < 0.0001).
CD169+ Cell-Deficient Mice Have an Impaired Immune Response Against S. aureus Skin Infection
To address whether such decrease of CD169+ macrophages might affect host immune response against pathogens, we measured the dermonecrotic area after intradermal challenge with methicillin-resistant S. aureus (MRSA) on SPF and GF mice. The size of the skin lesion was significantly larger in GF mice (Figures 4A, B). We presumed that the result might, at least in part, be due to decrease of CD169+ macrophages. To further elucidate the role of CD169+ macrophages, we used CD169-DTR mice, which have a diphtheria toxin (DT) receptor (DTR) on all cells expressing CD169, and CD169+ cells are readily depleted upon administration of DT (20, 21). Mice were treated with DT (25 ng/g bodyweight) on 3 days (D-3) and 1 day (D-1) before S. aureus infection. The dermonecrotic area caused by S. aureus was significantly larger in CD169-DTR mice compared to WT mice (Figures 4C, D). There were significantly more bacteria on day 7 (but not on day 4) in skin of deficient mice of CD169+ cells (Figure 4E). Taken together, the immune response against S. aureus was impaired in absence of CD169+ cells. To discern the cause of underlying the difference in immune response, we analyzed isolated immune cells (CD45+ cells) on day 0, day 2, and day 5 of S. aureus skin infection using scRNA-seq. Using UMAP and conventional cell markers, we determined the lineages of several immune cell clusters (Figure S4A and Figure 5A). The identified clusters contained fewer infiltrated immune cells in GF mice, confirming the impairment of the local immune reaction (Figure S4B).
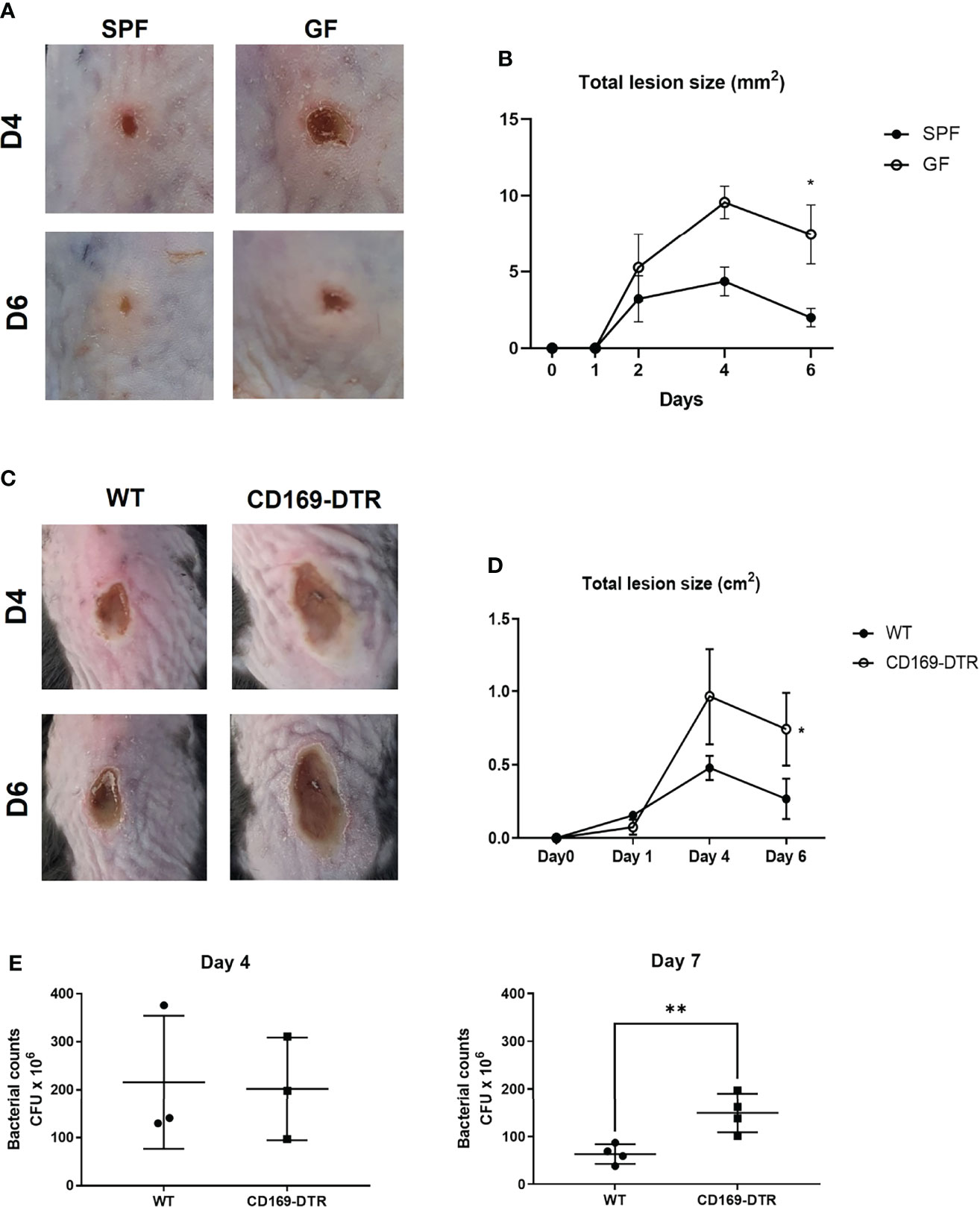
Figure 4 Impairment of immune response in germ-free and CD169+ cell-deficient mice against Staphylococcus aureus skin infection. (A) SPF and GF mice (n = 3 per group) were intradermally challenged with 2×107 CFUs of CA-MRSA (USA300). Representative pictures of skin lesions on day 4 (D4) and day 6 (D6) post infection. (B) Total lesion size of SPF and GF mice (n = 3 per group) over-time calculated by ImageJ program. The Images and data represent two independent experiments. (C) Representative pictures of skin lesions in WT and CD169-DTR mice (n = 3 per group) after intradermal S. aureus infection on D4 and D6 post infection. (D) Total lesion size of WT and CD160-DTR mice over-time. The Images (C) and data (D) represent three independent experiments. (E) CFU counts in skin lesion homogenates of WT and CD169-DTR mice after S. aureus intradermal infection on day 4 (n = 3 per group) and day 7 (n = 4 per group). Data represent three independent experiments. Data are expressed as means ± SEM (*p < 0.05, **p < 0.01).
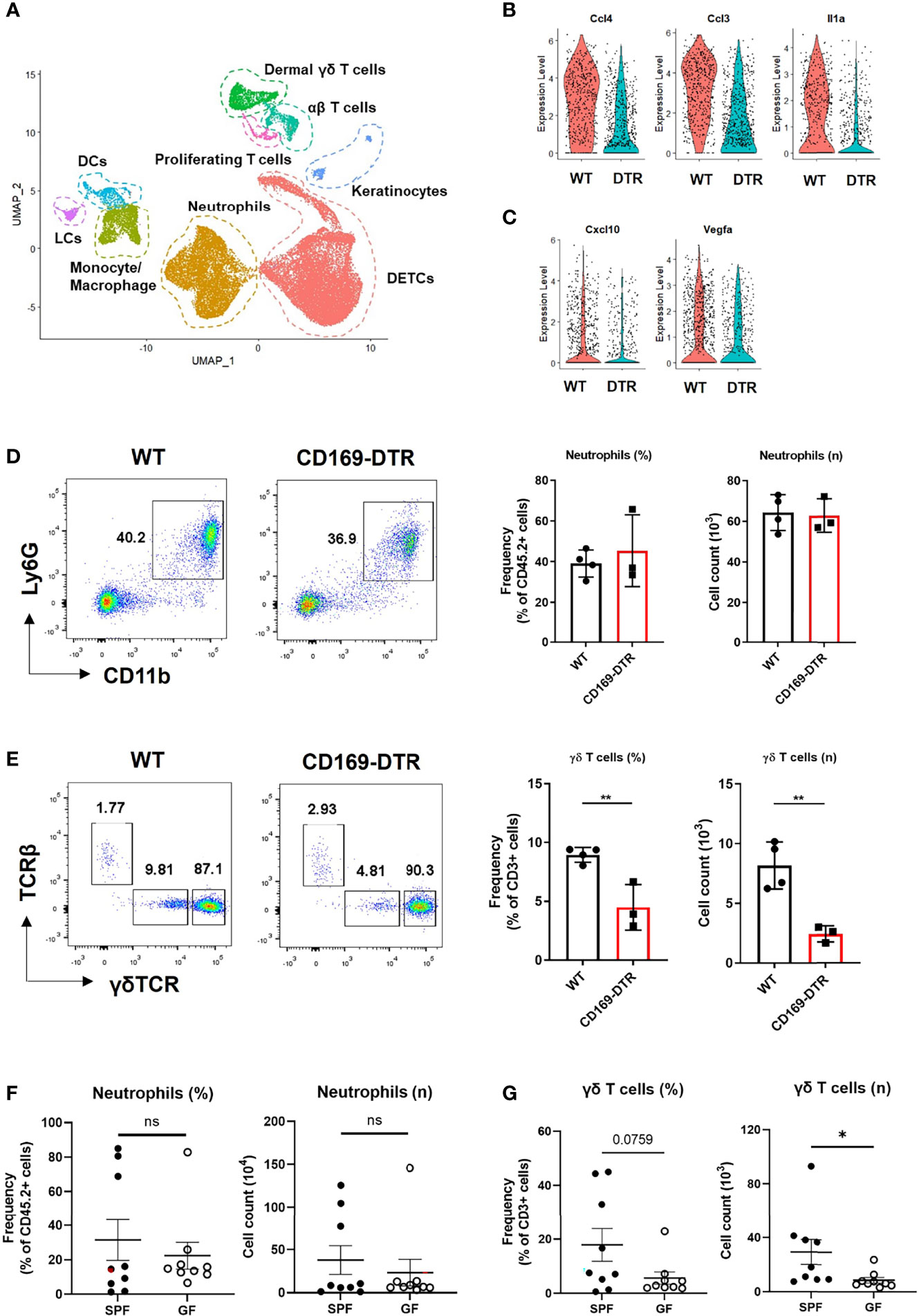
Figure 5 γδ T cells, not neutrophils, are affected by an absence of CD169+ cells at late time points. (A) UMAP projections of all skin-infiltrating immune cells. (B, C) Violin plot showing DEGs between WT and CD169-DTR of monocyte/macrophage cluster on day 2 (B) and day 5 (C). (D, E) Flow cytometry analysis of neutrophils (D) and dermal γδ T cells (E) in WT (n = 4) and CD169-DTR (n = 3) mice skin at day 6 post infection. The cells are gated on FSC/SSC, singlet, PI-, CD45.2+ (D) and FSC/SSC, singlet, PI-, CD45.2+, CD3+, CD11b-tolo (E) cells. Data are representative of three experiments. (F, G) Flow cytometry analysis of neutrophils (F) and dermal γδ T cells (G) in SPF (n = 9) and GF (n = 9) mice skin at day 6 post infection. The cells are gated on FSC/SSC, singlet, PI-, CD45.2+ (F) and FSC/SSC, singlet, PI-, CD45.2+, CD3+, CD11b-tolo (G) cells. Representative data of two independent experiments are shown. Data are expressed as means ± SEM (*p < 0.05, **p < 0.01, ns, not significant).
γδ T Cell Recruitment Is Impaired in GF and CD169+ Cell-Deficient Mice
We analyzed the difference in monocyte/macrophage cluster at each time points with scRNA-seq. The monocyte/macrophage cluster was identified using markers Ly6c2, Fcgr1, and Adgre1. WT and CD169-deficient mice differed in a number of DEGs on day 2, which included multiple cytokines and chemokines (Ccl4, Ccl3, Il1a, Cxcl3, Cxcl2, and Cxcl1) critical to leukocyte recruitment (Figure 5B and Figure S5A). On day 5, monocyte/macrophage cluster of WT mice did not show any differences between these cytokines and chemokines (Figure S5B). Cxcl10 and Vegfa were the only DEGs noted on day 5 (Figure 5C). There were no differences in neutrophil number between the two groups on day 6 (Figure 5D), but significantly impaired recruitment of γδ T cells (Figure 5E), which are known for their involvement in S. aureus skin infection (22, 23). Similar significant compromise in recruitment of dermal γδ T cells was noted in GF mice challenged with S. aureus, which also did not show differences between SPF mice in neutrophil number on day 6 (Figures 5F, G). There were no other significant differences in skin T cells, except decreased frequency of dendritic epidermal γδ T cells (Figures S5C, D).
Activation of γδ T Cells by CD169+ Macrophages
Local activation of γδ T cells was suspected from GSEA of γδ T cells on day 2 (Figure 6A) and larger number of IL-17A+ γδ T cells in SPF mice compared to GF mice, despite statistically insignificant (Figure S6A). As γδ T cells can be activated without T cell receptor signaling, we investigated whether differences in IL-1β and IL-23 expression by CD169+ macrophages resulted in local activation of IL-17-producing γδ T cells. Interestingly, using scRNA-seq analysis, we found that monocyte/macrophage cluster was the most popular source of Il23a in myeloid clusters, whereas DCs expressed more Il12b (Figure 6B). Using BMDMs, we were able to detect IL-23p19 production from macrophages challenged with S. aureus and confirmed that CD169+ macrophages were the major IL-23p19 producers (Figure 6C). Intracellular staining (ICS) of skin immune cells on day 1 post infection revealed that CD169+ macrophages mostly produced IL-23p19 production in vivo (Figure 6D). The absence of CD169+ cells resulted in a decrease of IL-17-producing γδ T cells on day 6 (Figure 6E). The number of IFN-γ producing T cells was minimal and was not altered in absence of CD169+ macrophages (Figure S6B). Taken all together, we assumed that the absence of CD169+ macrophages result in the decreased number of IL-17-producing γδ T cells, which lead to the impairment of host defense response, as observed from the high CFU counts and a larger area of dermonecrosis.
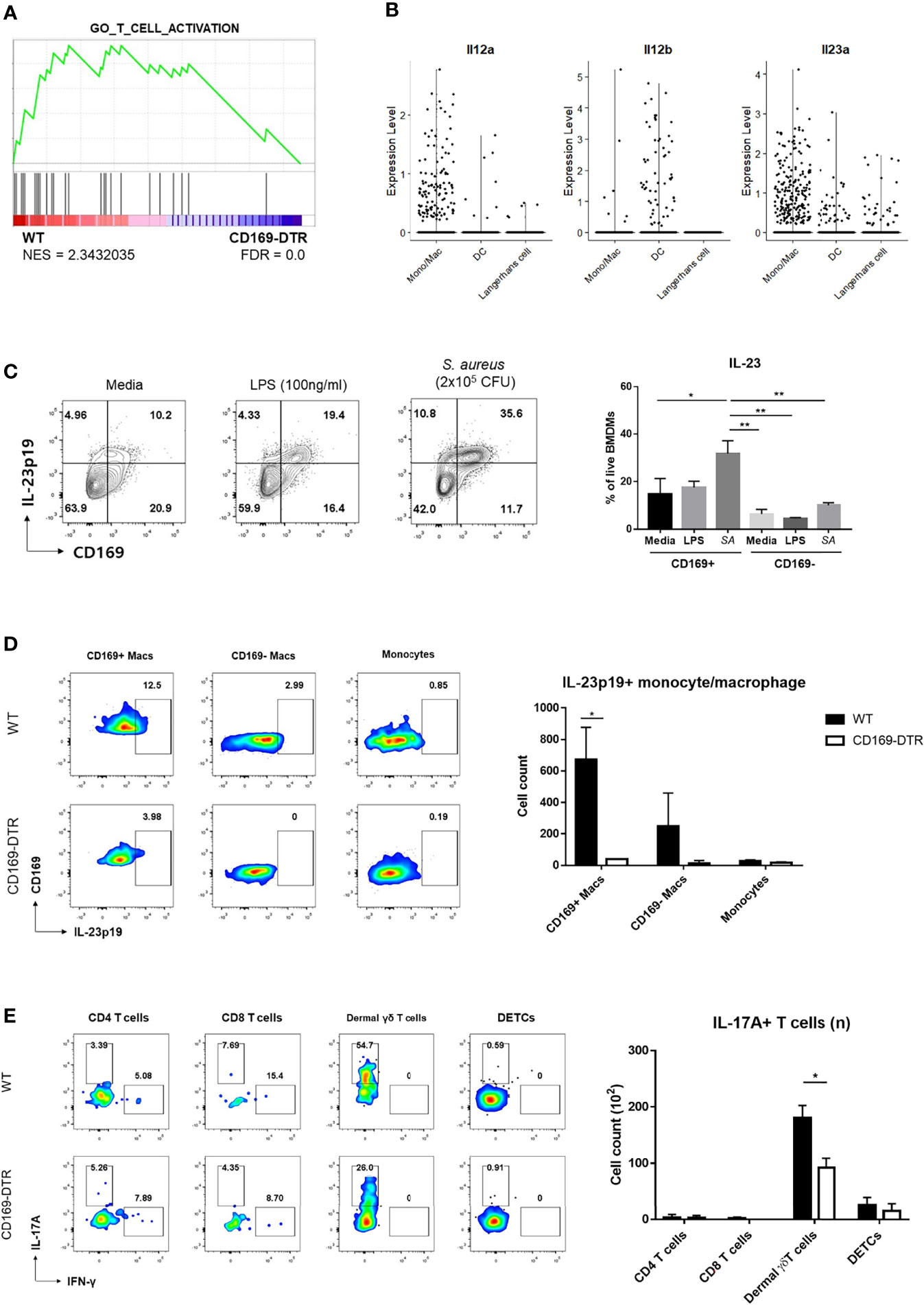
Figure 6 Activation of γδ T cells by CD169+ macrophages. (A) GSEA of DEGs from dermal γδ T cells of WT mice compared to CD169-DTR mice. (B) Violin plots of Il12a, Il12b, and Il23a expression in monocyte/macrophage, dendritic cell (DCs) and Langerhans cell clusters from scRNA-seq analysis data. (C) Representative flow cytometry plots and bar graph of BMDMs challenged with LPS (100 ng/mL) and S. aureus (SA, 2x105 CFU). (D) Representative plots and bar graph of IL-23p19 production by CD169+ macrophages (Macs), CD169- macrophages and monocytes on day 1 post skin S. aureus infection. Single-cell suspensions from two mice were pooled for both WT and CD169-DTR group. CD169+/- Macs were gated on FSC/SSC, PI-, singlet, CD45.2+, CD11b+, EpCAM-, Ly6c-tolo, Ly6G-, F4/80+ cells. Monocytes are gated on FSC/SSC, PI-, singlet, CD45.2+, CD11b+, EpCAM-, Ly6chi cells. (E) Representative plots and bar graph of IL-17A-producing T cells in skin 6 days post infection. T cells are gated on FSC/SSC, singlet, PI-, CD45.2+, CD3+, CD11b-tolo cells. Single-cell suspensions from two mice were pooled for both WT and CD169-DTR group. Data are representative of three independent experiments. Error bars show mean ± SEM (*p < 0.05; **p < 0.01).
Discussion
CD169+ macrophages have been studied primarily in lymphoid organs due to their location, for example subcapsular sinus and medullary macrophages in lymph nodes (LNs) and marginal metallophilic macrophages in the marginal zone of the spleen (24, 25). In LNs, the presence of B cells is necessary to generate CD169+ macrophages, mediated by their production of lymphotoxin alpha beta (LTα1β2) (26, 27). CD169+ macrophages also reside in various peripheral tissues, such as the brain, gastrointestinal tract, liver, lung, kidney, and skin (13, 15, 21). CD169 expression in these tissues is possibly not due to B cells, as peripheral B cells are more infrequent and generally not adjacent to macrophages. Our results suggest that local tonic IFN signaling induced by microbiota might be a factor governing CD169 expression of macrophages in these tissues and yield additional interesting findings. First, CD169-expressing macrophages were present in the lower dermis and within DWAT. Second, these cells are replenished by monocytes. Third, the selective depletion of CD169+ macrophages resulted in increased bacterial burden and impaired immune response. This phenotype was possibly due to an impaired ability to recruit γδ T cells. The CD169+ macrophages also were capable of γδ T cell activation, which resulted in IL-17 production by γδ T cells. Based on these findings, we concluded that CD169+ cells are an indispensable for combating intradermal S. aureus infection.
Surprisingly low levels of type I IFNs were sufficient to induce CD169 in BMDMs, while a high amount and/or prolonged type I IFN did not upregulate CD169 expression. CD169 expression might be under the control of type I IFN receptor (IFNAR) expression, which is affected by the strength of type I IFN signaling. Our results, in part, support the notion that a specific tone of type I IFNs is required for homeostatic CD169 expression by macrophages (4, 28). Furthermore, we present the novel mechanism of how microbiota-induced tonic type I IFN signaling affects monocyte/macrophage lineage cells. However, we also found that CD169 expression of skin macrophages in vivo is not strictly consistent, which might suggest a possibility of other factors affecting the CD169 expression in more precise manner.
Without CD169+ cells, the skin was more susceptible to S. aureus infection, as shown by larger dermonecrotic areas and higher CFU counts in CD169+ cell-deficient mice than in WT mice. Also, the WT monocyte/macrophage cluster expressed more Cxcl10 and Vegfa, both chemokines important in proliferation phase and remodeling phase of wound healing (29), compared to the CD169-DTR monocyte/macrophage cluster at day 5. We speculated that the lesions of WT mice more successfully eliminated S. aureus and were heading towards resolution phase, whereas the CD169+ cell-deficient mice remained in the active inflammatory phase. Early excessive neutrophil accumulation, however, might also result in tissue damage and dermonecrosis (30). The area of dermonecrosis should be interpreted along with the data on bacterial burden.
Intradermal injection results in involvement of the whole skin layer including DWAT (31). In this regard, CD169+ can act as the initiator of immune responses in intradermal infection, although they are located distantly from the epidermis. A similar role was proposed in a dextran sulfate sodium-induced colitis model, as the colitis symptoms decreased in the absence of CD169+ cells (20). The number of RORγt+ cells (Th17, γδ T17, LTi, and ILC3) did not change in that study but showed diminished IL-17 and IL-22 mRNA expression. Our data suggested that the skin CD169+ macrophages not only affected the recruitment of IL-17A-producing γδ T cells, but also acted as activators of γδ T cells via local IL-23 production.
Our study has multiple limitations. First, we did not evaluate the phagocytic activity or previously reported antigen presentation activity of CD169+ macrophages (32). It is unlikely, however, that the possible increase in phagocytic activity of CD169+ macrophages resulted in such a difference in the phenotype, as the number of resident immune cells were much lower than those of infiltrating neutrophils, monocytes, and T cells. CD169+ can also cross-prime CD8 T cells or transfer antigens to CD8a+ DCs (32, 33), but acute S. aureus infection mostly involves IL-17-producing γδ T cells, in which the activation of the T cells do not require antigen presentation. Consequently, CD8 T cells are likely to have minimal effects on S. aureus infection. Second, we were not able to fully elucidate h microbiota enhance type I IFN signaling in skin remain to be elucidated. Previous research suggested that the lung stromal cells (28) and plasmacytoid DCs in lymphatic organs as the source of tonic type I IFN signals (4). These studies indicated that the gut microbiota was involved in the process of tonic signaling, while it is not clear where and how the cells received microbiota signals. We showed that gut microbiota is important in CD169 expression using the fecal transplantation experiment. However, we cannot be completely sure that gut microbiota is entirely responsible for CD169 expression, as not only did we not address the possibility of the act of oral gavage being an inducer of CD169, but also because we were not able to address specific microbiota signals. We also cannot fully exclude the possibility of skin commensals being involved in local type I IFN signaling, as S. epidermidis cannot represent the plethora of commensal microorganism skin harbors. Future work will need to explore the exact source and nature of signals leading to enhanced type I IFN and ISG signatures.
Materials and Methods
Ethics Statement
All animal experiments in this study were in accordance with the legal and ethical requirements and the guidelines and protocols for rodent research were approved by the institutional animal care and use committee of Korea Advance Institute Science and Technology (KAIST, KA2017-40).
Mice
SPF male C57BL/6 mice were purchased from the KAIST (Daejeon, Korea) or DBL Co. Ltd (Eumseong, Korea). Ifnar knock-out (KO) mice (stock number: 028288, B6(Cg)-Ifnar1tm1.2Ees/J) and CX3CR1-GFP (B6.129P2(Cg)-Cx3cr1tm1Litt/J, Stock number: 005582) were purchased from the Jackson Laboratory (Bar Harbor, ME, USA). CD169-DTR (Siglec1<tm1 (HBEGF) Mtka>) mice were purchased from RIKEN BioResource Research Center with the permission of Dr. Masato Tanaka of the Tokyo University of Pharmacy and Life Sciences (Tokyo, Japan). GF mice were obtained from animal care facility of Microbiome Core Research Support Center at Pohang University of Science and Technology (POSTECH, Pohang, Korea). All mice used in this study were maintained in a SPF facility at the KAIST Laboratory Animal Resource Center.
Bacteria Preparation
Community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) strain USA300 (ATCC® BAA-1717) and Staphylococcus epidermidis (ATCC®12228) were purchased from the ATCC (ATCC, Manassas, VA, USA) and Korean Collection for Type Cultures (KCTC), respectively. Overnight cultures of a single bacteria colony were pelleted and resuspended in a 15% glycerol solution, aliquoted and stored at -80°C. One hundred microliters of aliquoted bacteria were defrosted and grown overnight in tryptic soy broth (TSB) at 37°C in a shaking incubator (200 rpm). A 2-h subculture (1:20 dilution) was made subsequently. The bacteria were pelleted and resuspended in phosphate-buffered saline (PBS). Optical Density was measured to estimate the colony forming units (CFUs), which was verified after overnight culture on tryptic soy agar (TSA) plates.
Mouse S. aureus Skin Infection Model And Treatments
Mice were anesthetized with 2% isoflurane, and their backs were shaved and depilated a day before S. aureus injection. A suspension of 1–2 × 107 CFUs/100μL PBS was injected intradermally using a 29-gauge insulin syringe. Digital photographs of the backs of the injected mice and total lesion size (cm2) measurements were analyzed using the image analysis software program ImageJ (https://imagej.nih.gov/ij/). For additional mice treatments, please see the Supplementary Data.
Generation of Bone Marrow Chimeras
C57BL/6 CD45.2 recipient mice were irradiated with 600cGy twice and reconstituted with 5 × 106 BM cells from C57BL/6 CD45.1 donor mice via tail vein injection. Chimeric mice were supplied with antibiotic containing water. The transplanted mice were analyzed after 6 weeks.
Bone Marrow-Derived Macrophage Preparation
Cells were prepared by flushing the femurs and tibiae of mice and were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS) (HyClone, Logan, USA), 1% penicillin/streptomycin, and 30% macrophage colony stimulating factor (M-CSF) conditional medium (supernatants of M-CSF from L929 cell cultures). After 7 days, adherent bone marrow derived macrophages (BMDMs) were collected via pipetting.
Enumeration of S. aureus CFU
Skin specimens were obtained from the center of lesions using an 8mm-disposable punch (Kai Industries, Seki city, Japan), and homogenized in 1mL PBS using a TissueRuptor II (Qiagen, Germantown, MD, USA). The homogenates were passed through 100-µm cell strainers and plated at serial dilution onto TSA plates. The number of CFU was determined after overnight incubation at 37°C.
Single Cell Preparation From Skin
Whole skin cells were prepared from mouse back skin. 4 cm2 of back skin tissue were harvested. The skin tissue was minced by blade and treated with 2 mg/mL Dispase II (Roche, Basel, Switzerland) for 1 h at 37°C. The skin was further minced into small pieces using scissors and digested with DMEM containing 1% FBS, 1 mg/mL Collagenase IV (Worthington Biochemical Corp., NJ, USA), 0.1 mg/mL DNase I (Roche), and 1.2 mg/ml hyaluronidase (Sigma-Aldrich, St Louis, MO, USA) for 30 minutes at 37°C. The digested skin tissues were then plunged through 70 µm cell strainers. The cells were resuspended on a Percoll gradient (70%/30%) (GE Healthcare, Chicago, IL, USA) and centrifuged at 2000 rpm for 20 min at 25°C. Cells were treated with ACK lysis buffer for 5 min at RT to remove red blood cells.
Flow Cytometry
Single-cell suspensions were pretreated with anti-CD16/32 (clone 2.4G2; TONBO Biosciences, CA, USA) antibody to block Fc receptors. For additional flow cytometry methods, please see the Supplementary Data.
Single-Cell Transcriptomic Analysis
At 2 days (48 hours) and 5 days (128 hours) after intradermal injection of S. aureus, single-cell suspensions were obtained from skin as described above. Cells were treated with anti-CD16/32 to neutralize Fc receptors. Cells then were stained with anti-CD45.2. and 7-AAD, and live leukocytes were isolated using a FACS Aria II cell sorter (BD Biosciences). Single-cell RNA sequencing was performed using a Chromium Single Cell 3’ V3 Reagent kit (10X Genomics, Pleasanton, CA, USA) according to the manufacturer’s protocol. For additional information on single-cell RNA sequencing and its analysis methods, please see the Supplementary Data.
Histological Analysis and Image Acquisition
Skin samples were obtained from the center of the dorsal skin using an 8 mm-disposable biopsy punch (Kai Industries). Samples were fixed in 4% paraformaldehyde (Duksan General Science, Seoul, Korea) and dehydrated in a 30% sucrose solution for 24 h at 4°C. Tissues were embedded in OCT compound (4538, Sakura Finetek, Alphen aan den Rijin, South Holland, Netherlands). Frozen blocks were cut into 100-μm thick sections using a cryostat (Microm HM525, Thermo Fisher Scientific). For detailed information on immunofluorescence staining methods and image acquisition, please check the Supplementary Data.
Analysis of Publicly Curated Gene Expression Dataset
The gene datasets deposited by Grice and Meisel in 2018 were accessed from the Gene Expression Omnibus database (NCBI GEO database, accession GSE98877) (11). The functional classification for the previously published results was analyzed according to their functional similarity based on PANTHER (v15.0, www.pantherdb.org). For GSEA (34), 15,448 differentially expressed genes were annotated to the reference gene set based on MSigDB 7.2.
Statistical Analysis
All data are presented as means with standard deviations. Statistical analyses were performed using GraphPad Prism 9 (GraphPad Software, Inc., La Jolla, CA, USA) and R statistical program where relevant. An unpaired Student’s t-test was used for analysis between two groups with Welch’s correction when necessary, and unpaired one-way analysis of variance was used for multiple comparisons. A difference between groups was considered significant: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE98877.
Ethics Statement
The studies involving human participants were reviewed and approved by Ajou University Hospital (AJIRB-MED-KSP-21-376). The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by The institutional animal care and use committee of Korea Advance Institute Science and Technology.
Author Contributions
YJP, BHK and H-JK conducted the experiments. YJP, JEO and HKL analyzed the data. YJP and HKL wrote the manuscript. HKL supervised the project. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Research Foundation of Korea (NRF-2021M3A9D3026428 and NRF-2021M3A9H3015688) funded by the Ministry of Science and ICT of Korea.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank the members of the Laboratory of Host Defenses for their helpful advice and discussions. The authors thank Ji Ye Kim at the BioMedical Research Center for technical service.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.799598/full#supplementary-material
References
1. Molloy MJ, Bouladoux N, Belkaid Y. Intestinal Microbiota: Shaping Local and Systemic Immune Responses. Semin Immunol (2012) 24:58–66. doi: 10.1016/j.smim.2011.11.008
2. Constantinides MG, Link VM, Tamoutounour S, Wong AC, Perez-Chaparro PJ, Han SJ, et al. MAIT Cells are Imprinted by the Microbiota in Early Life and Promote Tissue Repair. Science (2019) 366. doi: 10.1126/science.aax6624
3. Zheng D, Liwinski T, Elinav E. Interaction Between Microbiota and Immunity in Health and Disease. Cell Res (2020) 30:492–506. doi: 10.1038/s41422-020-0332-7
4. Schaupp L, Muth S, Rogell L, Kofoed-Branzk M, Melchior F, Lienenklaus S, et al. Microbiota-Induced Type I Interferons Instruct a Poised Basal State of Dendritic Cells. Cell (2020) 181:1080–96.e19. doi: 10.1016/j.cell.2020.04.022
5. Steed AL, Christophi GP, Kaiko GE, Sun L, Goodwin VM, Jain U, et al. The Microbial Metabolite Desaminotyrosine Protects From Influenza Through Type I Interferon. Science (2017) 357:498–502. doi: 10.1126/science.aam5336
6. Kovarik P, Castiglia V, Ivin M, Ebner F. Type I Interferons in Bacterial Infections: A Balancing Act. Front Immunol (2016) 7:652. doi: 10.3389/fimmu.2016.00652
7. McNab F, Mayer-Barber K, Sher A, Wack A, O'Garra A. Type I Interferons in Infectious Disease. Nat Rev Immunol (2015) 15:87–103. doi: 10.1038/nri3787
8. Jiang J, Zhao M, Chang C, Wu H, Lu Q. Type I Interferons in the Pathogenesis and Treatment of Autoimmune Diseases. Clin Rev Allergy Immunol (2020) 59:248–72. doi: 10.1007/s12016-020-08798-2
9. Trinchieri G. Type I Interferon: Friend or Foe? J Exp Med (2010) 207:2053–63. doi: 10.1084/jem.20101664
10. Gough DJ, Messina NL, Clarke CJ, Johnstone RW, Levy DE. Constitutive Type I Interferon Modulates Homeostatic Balance Through Tonic Signaling. Immunity (2012) 36:166–74. doi: 10.1016/j.immuni.2012.01.011
11. Meisel JS, Sfyroera G, Bartow-McKenney C, Gimblet C, Bugayev J, Horwinski J, et al. Commensal Microbiota Modulate Gene Expression in the Skin. Microbiome (2018) 6:20. doi: 10.1186/s40168-018-0404-9
12. Hesketh M, Sahin KB, West ZE, Murray RZ. Macrophage Phenotypes Regulate Scar Formation and Chronic Wound Healing. Int J Mol Sci (2017) 18. doi: 10.3390/ijms18071545
13. Thornley TB, Fang Z, Balasubramanian S, Larocca RA, Gong W, Gupta S, et al. Fragile TIM-4-Expressing Tissue Resident Macrophages are Migratory and Immunoregulatory. J Clin Invest (2014) 124:3443–54. doi: 10.1172/JCI73527
14. Yanagihashi Y, Segawa K, Maeda R, Nabeshima YI, Nagata S. Mouse Macrophages Show Different Requirements for Phosphatidylserine Receptor Tim4 in Efferocytosis. Proc Natl Acad Sci U S A (2017) 114:8800–5. doi: 10.1073/pnas.1705365114
15. Sheng J, Chen Q, Soncin I, Ng SL, Karjalainen K, Ruedl C. A Discrete Subset of Monocyte-Derived Cells Among Typical Conventional Type 2 Dendritic Cells Can Efficiently Cross-Present. Cell Rep (2017) 21:1203–14. doi: 10.1016/j.celrep.2017.10.024
16. Rusinova I, Forster S, Yu S, Kannan A, Masse M, Cumming H, et al. Interferome V2.0: An Updated Database of Annotated Interferon-Regulated Genes. Nucleic Acids Res (2013) 41:D1040–6. doi: 10.1093/nar/gks1215
17. Chorro L, Geissmann F. Development and Homeostasis of 'Resident' Myeloid Cells: The Case of the Langerhans Cell. Trends Immunol (2010) 31:438–45. doi: 10.1016/j.it.2010.09.003
18. Burgess M, Wicks K, Gardasevic M, Mace KA. Cx3CR1 Expression Identifies Distinct Macrophage Populations That Contribute Differentially to Inflammation and Repair. Immunohorizons (2019) 3:262–73. doi: 10.4049/immunohorizons.1900038
19. Kolter J, Feuerstein R, Zeis P, Hagemeyer N, Paterson N, d'Errico P, et al. A Subset of Skin Macrophages Contributes to the Surveillance and Regeneration of Local Nerves. Immunity (2019) 50:1482–97.e7. doi: 10.1016/j.immuni.2019.05.009
20. Asano K, Takahashi N, Ushiki M, Monya M, Aihara F, Kuboki E, et al. Intestinal CD169(+) Macrophages Initiate Mucosal Inflammation by Secreting CCL8 That Recruits Inflammatory Monocytes. Nat Commun (2015) 6:7802. doi: 10.1038/ncomms8802
21. Gupta P, Lai SM, Sheng J, Tetlak P, Balachander A, Claser C, et al. Tissue-Resident CD169(+) Macrophages Form a Crucial Front Line Against Plasmodium Infection. Cell Rep (2016) 16:1749–61. doi: 10.1016/j.celrep.2016.07.010
22. Cho JS, Pietras EM, Garcia NC, Ramos RI, Farzam DM, Monroe HR, et al. IL-17 Is Essential for Host Defense Against Cutaneous Staphylococcus Aureus Infection in Mice. J Clin Invest (2010) 120:1762–73. doi: 10.1172/JCI40891
23. Marchitto MC, Dillen CA, Liu H, Miller RJ, Archer NK, Ortines RV, et al. Clonal Vgamma6(+)Vdelta4(+) T Cells Promote IL-17-Mediated Immunity Against Staphylococcus Aureus Skin Infection. Proc Natl Acad Sci USA (2019) 116:10917–26. doi: 10.1073/pnas.1818256116
24. Saunderson SC, Dunn AC, Crocker PR, McLellan AD. CD169 Mediates the Capture of Exosomes in Spleen and Lymph Node. Blood (2014) 123:208–16. doi: 10.1182/blood-2013-03-489732
25. Uchil PD, Pi R, Haugh KA, Ladinsky MS, Ventura JD, Barrett BS, et al. A Protective Role for the Lectin CD169/Siglec-1 Against a Pathogenic Murine Retrovirus. Cell Host Microbe (2019) 25:87–100.e10. doi: 10.1016/j.chom.2018.11.011
26. Moseman EA, Iannacone M, Bosurgi L, Tonti E, Chevrier N, Tumanov A, et al. B Cell Maintenance of Subcapsular Sinus Macrophages Protects Against a Fatal Viral Infection Independent of Adaptive Immunity. Immunity (2012) 36:415–26. doi: 10.1016/j.immuni.2012.01.013
27. Xu HC, Huang J, Khairnar V, Duhan V, Pandyra AA, Grusdat M, et al. Deficiency of the B Cell-Activating Factor Receptor Results in Limited CD169+ Macrophage Function During Viral Infection. J Virol (2015) 89:4748–59. doi: 10.1128/JVI.02976-14
28. Bradley KC, Finsterbusch K, Schnepf D, Crotta S, Llorian M, Davidson S, et al. Microbiota-Driven Tonic Interferon Signals in Lung Stromal Cells Protect From Influenza Virus Infection. Cell Rep (2019) 28:245–56.e4. doi: 10.1016/j.celrep.2019.05.105
29. Ridiandries A, Tan JTM, Bursill CA. The Role of Chemokines in Wound Healing. Int J Mol Sci (2018) 19. doi: 10.3390/ijms19103217
30. Castleman MJ, Febbraio M, Hall PR. CD36 Is Essential for Regulation of the Host Innate Response to Staphylococcus Aureus Alpha-Toxin-Mediated Dermonecrosis. J Immunol (2015) 195:2294–302. doi: 10.4049/jimmunol.1500500
31. Yasuda T, Ura T, Taniguchi M, Yoshida H. Intradermal Delivery of Antigens Enhances Specific IgG and Diminishes IgE Production: Potential Use for Vaccination and Allergy Immunotherapy. PLoS One (2016) 11:e0167952. doi: 10.1371/journal.pone.0167952
32. Bernhard CA, Ried C, Kochanek S, Brocker T. CD169+ Macrophages are Sufficient for Priming of CTLs With Specificities Left Out by Cross-Priming Dendritic Cells. Proc Natl Acad Sci USA (2015) 112:5461–6. doi: 10.1073/pnas.1423356112
33. van Dinther D, Veninga H, Iborra S, Borg EGF, Hoogterp L, Olesek K, et al. Functional CD169 on Macrophages Mediates Interaction With Dendritic Cells for CD8(+) T Cell Cross-Priming. Cell Rep (2018) 22:1484–95. doi: 10.1016/j.celrep.2018.01.021
Keywords: microbiota, Siglec-1, CD169, macrophages, interferon, γδ T cells, IL-17, S. aureus
Citation: Park YJ, Kang BH, Kim H-J, Oh JE and Lee HK (2022) A Microbiota-Dependent Subset of Skin Macrophages Protects Against Cutaneous Bacterial Infection. Front. Immunol. 13:799598. doi: 10.3389/fimmu.2022.799598
Received: 21 October 2021; Accepted: 06 May 2022;
Published: 09 June 2022.
Edited by:
Laurel L. Lenz, University of Colorado, United StatesReviewed by:
Tao Jin, Sahlgrenska University Hospital, SwedenMathieu Paul Rodero, UMR8601 Laboratoire de Chimie et Biochimie Pharmacologiques et Toxicologiques, France
Copyright © 2022 Park, Kang, Kim, Oh and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Heung Kyu Lee, heungkyu.lee@kaist.ac.kr; orcid.org 0000-0002-3977-1510
 Young Joon Park
Young Joon Park Byeong Hoon Kang
Byeong Hoon Kang Hyun-Jin Kim
Hyun-Jin Kim Ji Eun Oh
Ji Eun Oh Heung Kyu Lee
Heung Kyu Lee