- Graduate School of Medical Science and Engineering, Korea Advanced Institute of Science and Technology (KAIST), Daejeon, South Korea
An increasing number of studies have revealed that the progression of colorectal cancer (CRC) is related to gut microbiome composition. Under normal conditions, the gut microbiome acts as a barrier to other pathogens or infections in the intestine and modulates inflammation by affecting the host immune system. These gut microbiota are not only related to the intestinal inflammation associated with tumorigenesis but also modulation of the anti-cancer immune response. Thus, they are associated with tumor progression and anti-cancer treatment efficacy. Studies have shown that the gut microbiota can be used as biomarkers to predict the effect of immunotherapy and improve the efficacy of immunotherapy in treating CRC through modulation. In this review, we discuss the role of the gut microbiome as revealed by recent studies of the growth and progression of CRC along with its synergistic effect with anti-cancer treatment modalities.
Introduction
Colorectal cancer (CRC) is one of the most common types of cancer and is the third highest leading cause of death worldwide (1). Numerous epidemiological studies have demonstrated that the prevalence of CRC is related to a western diet and intake of dietary fiber, thus highlighting the important relationship between diet and CRC (2–5). In this context, the gut environment, including the microbiome, has been in the spotlight and has emerged as an important factor related to CRC (6).
A multitude of microorganisms live in the intestines of mammals. In the human intestine, there are more than 1000 species and 1014 microorganisms forming a colony (7). They play an important role in maintaining a normal physiologic environment, including energy metabolism, interacting with the normal gut barrier system, promoting the survival of epithelial cells, and, importantly, protecting our body against other external or opportunistic pathogens (8). Over the past few decades, studies have shown that the gut microbiome influences the host significantly (9–11). Dysbiosis in the intestines is known to be associated with the pathogenesis of a variety of diseases, including neurological, gastrointestinal, and metabolic diseases (12). Changes in the gut microbiome can be induced by eating habits or changes in environmental factors and studies have shown that changes in the gut microbiome induce CRC through inflammatory diseases, microbial metabolites, or virulence factors (13–15). The gut microbiome has been demonstrated to affect not only the generation of CRC, but also its progression. Furthermore, the gut microbiome has been associated with controlling the efficacy of cancer treatment and the toxicities of therapeutic agents. Thus, therapeutic agents, such as probiotics, that can control the gut microbiome are expected to among the most effective approaches for helping to fight CRC (16, 17).
Recent advances in our understanding of the role of the gut microbiome are due to the development of technologies, such as 16S rRNA sequencing, that enable the discovery of many microorganisms in the intestine that could not be identified previously (18). Many studies related to metabolomics and metagenomics describe the effects of these gut microbes on the human body, and some studies revealed their involvement in cancer prevention, tumorigenesis, and anti-cancer effects (19, 20). In particular, changes in gut microbial metabolites, such as short-chain fatty acids (SCFAs), polyphenols, vitamins, tryptophan catabolites, and polyamines produced or affected by the gut microbiota, may have a wide range of effects on the formation and progression of CRC and even metastasis (21). As our understanding of the role of the anti-cancer immune response in the tumor microenvironment during cancer progression and treatment increases, the effect of the gut microbiome on tumor immunity is also receiving greater attention (21). It is known that changes in the gut microbiota not only affect tumor immunotherapy, but also affect therapeutic toxicity (22). Thus, modulation of the gut microbiome can be used as a novel treatment modality.
The gut microbiome has emerged as an important factor in various diseases, and the relationship between the gut microbiome and CRC has become an important issue in several studies. In this review, the potential role of the gut microbiome will be reviewed with a focus on how the gut microbiome affects the tumorigenesis processes associated with CRC. Furthermore, we discuss methods of gut microbiome modulation that can be used to treat CRC.
Correlation Between CRC and Gut Microbiome
With changes in western dietary habits worldwide, the incidence of CRC is expected to increase steadily, resulting in 2.2 million new cases by 2030 (23). Studies have shown that approximately 90% of CRC occurs sporadically and the remainder is caused by genetic factors or exposure to specific environmental factors (24–27). In particular, lifestyle factors such as physical inactivity, smoking history, western diet, low fiber intake, alcohol intake, and obesity are major influences on CRC. It is important to note that most of these environmental factors can induce changes in the gut microbiota (26, 28, 29). Many studies have confirmed that susceptibility to CRC or tumor progression is affected by changes in the gut microbiome, which has been found to induce inflammation, DNA damage, or metabolites produced from microorganisms (30).
Evidence from several studies has suggested the existence of a close link between the gut microbiome and the host during the development of CRC (31–33). Studies using high-throughput microbiome sequencing have been conducted to investigate the microbiome community in tumor-formed and normal colon tissues (27), enabling a better understanding of the differences in gut microbiome between CRC and healthy patients. Reports have shown that the diversity and richness of the gut microbiome decreases in CRC patients (33, 34). In particular, analysis of the gut microbiome of CRC patients revealed that significant changes in specific microbial groups occurs, leading researchers to hypothesize that these changes might have a greater impact on the mucosal immune response of CRC patients compared to that of healthy individuals (34). A total of 11 operational taxonomic units (OTUs) belonging to the genera Enterococcus, Escherichia/Shigella, Klebsiella, Streptococcus, and Peptostreptococcus were significantly found to be more abundant in the gut microbiota of CRC patients, while 5 OTUs belonging to Roseburia and other butyrate-producing bacteria from the Lachnospiraceae family were less abundant (35). In addition, dysbiosis was observed in the gut microbiome of CRC patients as the balance between microorganisms was disrupted (36). Dysbiosis of gut microbiota and increased intestinal permeability induce colonic inflammation and may cause the promotion or progression of CRC (37). Fusobacterium nucleatum (F. nucleatum) is significantly increased in human CRC compared to healthy patients (38). Moreover, early-stage CRC patients (advanced adenoma) have a different microbiome composition compared to advanced-stage CRC patients (definitive CRC) (35, 39). These studies indicate a very close correlation between CRC and the gut microbiome; however, further investigation is still required to fully elucidate the effect of the gut microbiome on CRC.
Influence of the Gut Microbiome on CRC Formation
Although much is still unknown about the formation of CRC, chronic inflammation has been implicated in the initiation of malignancy. It is estimated that approximately 20% of malignant tumors occurring in the colon are preceded by chronic inflammation (40). During carcinogenesis, inflammatory cytokines and chemokines produced by cancerous cells attract immature myeloid cells or pro-inflammatory helper T cells. This pro-tumorigenic microenvironment is characterized by the synthesis of growth and angiogenic factors, as well as tissue remodeling enzymes, and the suppression of anti-tumor T-cell responses, favoring tumor progression (41).
Knowledge that the gut microbiome affects CRC formation was first obtained in the early 1970s. When the colon was exposed to a carcinogen called 1,2-dimethlylhydrazine in a germ-free mouse model, the degree of CRC formation was found to be significantly reduced (42). At the time, it was not possible to specify which bacteria caused this phenomenon. However, a similar experiment using various colon cancer models confirmed that the presence or absence of intestinal microbes had a significant effect on the formation of colon cancer (43, 44). Since then, many studies using high-throughput microbiome sequencing have identified the various intestinal microorganisms that affect CRC formation.
Streptococcus bovis (S. bovis) has been reported as one of the risk factors for CRC (45–47). S. bovis is normally colonized in the gastrointestinal tract. Thus, the occurrence of S. bovis-induced endocarditis or bacteremia was an early clue to its involvement in colon cancer (45). The association between inflammation and colon carcinogenesis was confirmed when the relationship between the pro-inflammatory potential of S. bovis proteins and their carcinogenic properties was observed (48, 49). S. bovis has been found to play an active role in CRC development, perhaps through an inflammation-based sequence of tumor development or propagation involving interleukin (IL)-1, cyclooxygenase-2 (COX-2), and IL-8 (48).
F. nucleatum is one of the most widely known strains related to CRC tumor formation (50). Metagenomic analysis showed that the commensal Fusobacterium spp. are associated with CRC in humans; however, it remains unclear whether this is indirect or causal (38). Castellarin and coworkers confirmed that the transcripts of the strain were increased approximately 400 times in CRC tumor tissue compared to normal tissue (50). In a study using the adenomatous polyposis coli (APC) +/- mouse CRC model, F. nucleatum developed a pro-inflammatory environment which induced neoplasia progression in intestinal epithelial cells and recruited tumor-infiltrating immune cells (38). In addition, studies demonstrated that IL-17a was highly expressed in CRC patients with abundant F. nucleatum (51). This strain induces early carcinogenesis through increased bacterial adherence in the mucosal surface (52). F. nucleatum produces a unique protein called Fusobacterium adhesin A (FadA), which induces activation of the β-catenin signaling pathway after binding to E-cadherin, which is a potent oncogenic stimulator.
Enterococcus faecalis (E. faecalis) is a gut commensal bacterium that produces a superoxide from the autoxidation of membrane-associated demethylmenaquinone (53). Infection with E. faecalis induces DNA damage to intestinal epithelial cells by forming the superoxide. Thus, the abundance of E. faecalis was shown to be significantly increased in CRC patients compared to healthy individuals (35, 54, 55). Moreover, in vitro and in vivo studies demonstrated that E. faecalis can produce hydroxyl radicals (56, 57), which are potent mutagens that cause DNA breaks, point mutations, and protein-DNA crosslinking, thereby contributing to chromosomal instability and CRC risk (58).
Enterotoxigenic Bacteroides fragilis (ETBF) is a bacterium that produces B. fragilis toxin (BFT) and causes diarrhea and inflammatory bowel disease (IBD) (59–62). This strain plays a role in promoting tumors by elevating signal transducer and activator of transcription 3 (STAT3) and the Th17 response during colon tumorigenesis (60). Although STAT3 activation is required for colon tumorigenesis, it alone is not sufficient to trigger colon tumorigenesis by ETBF. Notably, IL-17-dependent nuclear factor kappa B (NF-κB) activation results in the formation of a proximal to distal mucosal gradient of CXC chemokines, which mediates the recruitment of CXCR2-expressing polymorphonuclear immature myeloid cells to cause ETBF-mediated distal colon tumorigenesis in parallel (62).
Peptostreptococcus anaerobius (P. anaerobius) induces a pro-inflammatory immune microenvironment and promotes tumor formation in the intestine. This strain plays a role in tumor formation by increasing the expression of pro-inflammatory cytokines in a mouse model and recruiting tumor-infiltrating immune cells such as immunosuppressive myeloid-derived suppressor cells (63). P. anaerobius increases the levels of reactive oxidative species that interact with toll-like receptor (TLR) 2 and TLR4 in colon cells to promote cholesterol synthesis and cell proliferation, ultimately causing dysplasia of colon cells (64).
Salmonella infections and colonization can be chronic and increase the risk of chronic cholecystitis and other gastrointestinal diseases, including cancers (65). Salmonella promotes colon tumorigenesis by relying on AvrA protein, which can activate both the Wnt/b-catenin and STAT3 signaling pathways in colon tumor cells (66–68). Salmonella also produces a genotoxin called typhoid toxin, which damages DNA via the phosphoinositide 3-kinase (PI3K) pathway in colonic epithelial cells (69). The reduced DNA repair capacity and inability to activate appropriate checkpoint responses have been associated with increased genomic instability in APC-deficient cells exposed to genotoxin. Campylobacter jejuni produces a cytolethal distending toxin (CDT), a genetic toxin with DNAse activity that causes DNA double-strand breaks and promotes colorectal tumorigenesis (70). Rapamycin, which inhibits mammalian target of mTOR signaling in mammals, has been shown to attenuate C. jejuni-induced colitis and carcinogenesis (70, 71).
Sulfate-reducing bacteria (SRB) are a microbiome component that is of particular interest with respect to colitis (72). These microorganisms can produce hydrogen sulfide (H2S) by using methionine and cysteine as substrates. Studies have shown increased amounts of SRB in the stool of CRC patients compared to healthy individuals (73). H2S produced by SRB can stimulate CRC progression by inhibiting butyrate oxidation and destroying the gut barrier, as well as induce DNA damage through reactive oxygen species (ROS) (74, 75).
Research to understand the relationship between other intestinal microbes with CRC formation is still ongoing. Thus, the bacteria discussed above do not constitute all of the causative bacteria of CRC.
Influence of Gut the Microbiome on CRC Progression
The gut microbiome affects not only the formation of colon malignancy, but also its progression. Published literature related to the development of CRC has demonstrated that many bacteria affect tumor development and growth. In addition, it was observed that the progression of colon adenoma was promoted in a spontaneous CRC mouse model characterized by expression of mutated Apc, a tumor suppressor gene (76). This section will describe research findings associated with progression-related mechanisms rather than tumor formation. Figure 1 summarizes the bacteria and their mechanisms of involvement in CRC initiation and progression.
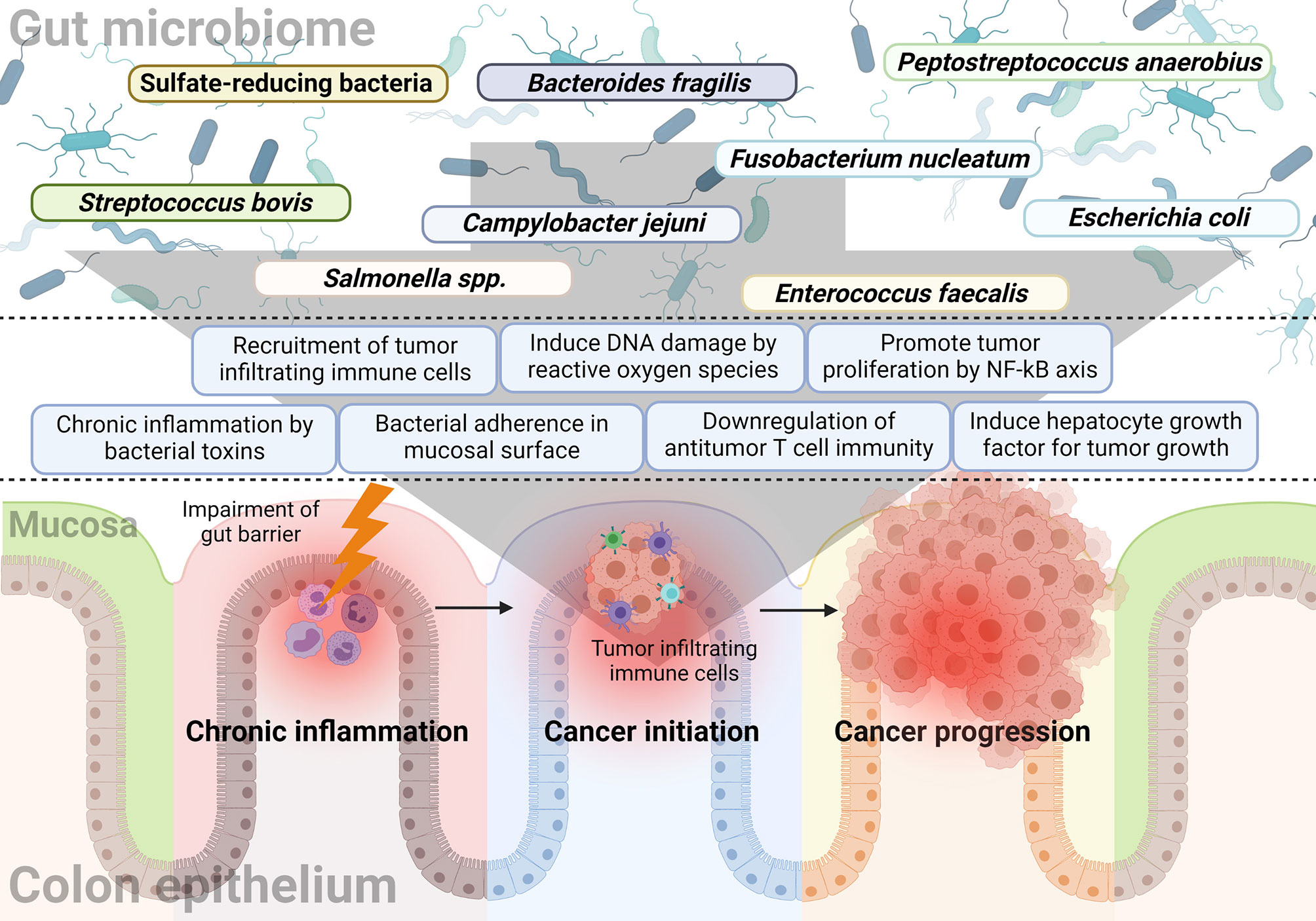
Figure 1 The relationship between the gut microbiome and sequential progression of colorectal carcinoma. Specific gut microorganisms induce chronic inflammation in the colorectal epithelium. For example, typhoid toxin or colibactin secreted by Salmonella or E. coli, respectively, leads to pro-inflammatory cytokine production and bacterial adherence. Chronic inflammation is one of the major causes of CRC and increased ROS with epithelial cell DNA damage also play a major role in cancer initiation by the gut microbiome. Some microorganisms like F. nucleatum and B. fragilis induce a tumor-favorable immune microenvironment by reducing CD3+ T cell density along with the recruitment and proliferation of CD4+CCR6+IL17A+ Th17 cells. Furthermore, bacterial components such as putative cell wall binding repeat 2 surface protein in P. anaerobius activate the NF-κB signaling pathway in CRC tumor cells and promote tumor cell proliferation. Colibactin-producing E. coli encodes enzymes responsible for HGF synthesis and induces senescence and tumor growth.
The presence of F. nucleatum is associated with worse prognosis in CRC patients (77, 78). Expression of tumor necrosis factor alpha, β-catenin, and NF-κB was increased in the F. nucleatum-abundant group and COX-2, matrix metallopeptidase 9, and NF-κB were highly expressed in the B. fragilis-abundance group. Immunohistochemical analysis showed that Kirsten rat sarcoma virus (KRAS) and proto-oncogene B-Raf (BRAF) expression were increased in the presence of F. nucleatum and B. fragilis (78). F. nucleatum-high cases were inversely associated with the density of CD3+ T cells (79). Experimental evidence suggests that F. nucleatum can promote colonic tumor development by downregulation of anti-tumor T cell-mediated adaptive immunity. Natural killer cells (NK cells) were also found to be affected by F. nucleatum in various carcinomas including CRC (80). Gur and colleagues found that the Fap2 protein of F. nucleatum directly interacts with T cell immunoreceptor with Ig and ITIM domains (TIGIT) to inhibit the cytotoxicity of NK cells.
ETBF was also revealed to support the progression of malignancy as well as tumorigenesis (81). This strain induces the secretion of exosome-like nanoparticles in intestinal epithelial cell lines and contains chemokine CC motif ligand 20 and prostaglandin E2 in the particle. Thus, ETBF induces the recruitment and proliferation of CD4+CCR6+IL17A+ Th17 cells via the IL-17 signaling pathway, thereby participating in tumorigenesis and cancer cell growth.
Long, et al. found that the surface protein of P. anaerobius, putative cell wall binding repeat 2 (PCWBR2), promotes CRC development in APC+/- mice (63). PCWBR2 initiates the oncogenic PI3K-Akt signaling pathway that directly binds to the intestinal epithelial cell receptor integrin α2/β1 and promotes tumor cell proliferation via the PCWBR2-integrin α2/β1-PI3K-Akt-NF-κB signaling axis.
Escherichia coli (E. coli), which is the most highly abundant strain residing in the intestine, is also closely related to the growth of CRC. Studies have shown that the level of mucosal-associated E. coli is increased in CRC tumor tissues compared with in normal colon tissues (82). The pathogenic E. coli strain showed a correlation with inflammation and ROS production, which may propagate tumor infiltration (83). E. coli has polyketide synthase which codes for production of colibactin, the polyketide-peptide genotoxin found to play a significant impact on tumor growth (84, 85). In a xenograft model, colibactin-producing E. coli indirectly promotes tumor growth by inducing hepatocyte growth factor (HGF) (86). HGF is the main mechanical link between pks+ (which encodes enzymes responsible for HGF synthesis) E. coli-induced senescence and tumor growth. Other factors, including microRNA-20a-5p, sentrin-specific protease 1 (SENP1), and activated HGF receptors, are also affected by the presence of pks+ E. coli in human CRC.
In contrast, the presence or enrichment of certain intestinal strains leads to anti-cancer effects on the growth of CRC. Numerous animal studies have shown several emerging chemical candidates as key mechanisms for probiotics to induce protective effects against CRC. Faecalibacterium prausnitzii is a potential probiotic that can downregulate the NF-kB pathway in gut epithelial cells by producing hydrophobic microbial anti-inflammatory molecules and prevent colitis in animal models (87). Lactobacillus rhamnosus GG and Bifidobacterium lactis Bb12 help to prevent abnormal epithelial proliferation in patients with a history of polyps and improve the intestinal epithelial tight junction barrier (88). Lactobacilli and Bifidobacteria were suggested to play a role in suppressing tumor progression and volume in a CRC mouse model (89, 90). The presence of these probiotics was confirmed to induce increasing SCFA production, thus inducing apoptosis and inhibiting tumor proliferation (91). Butyrate, one of the SCFA metabolites produced by probiotics, can induce the expansion of T reg lymphocytes for regulating the immune response in colorectal tissues and suppressing carcinogenesis and tumor growth (92).
Influence of the Gut Microbiome on CRC Treatment
Because the gut microbiome has been closely associated with CRC, numerous studies have been focused on investigating its effect on CRC treatment. Research related to the effect of gut microbiome on tumor treatment is the most important part of the cancer-microbiome research field and many studies are being conducted in combination with various treatment modalities to apply it to clinical cancer treatment. In addition to existing chemotherapeutic agents or radiotherapy, new discoveries are being made about the synergistic effects of the gut microbiome with immune checkpoint inhibitors (ICIs) (93). Figure 2 summarizes the research findings discussed below.
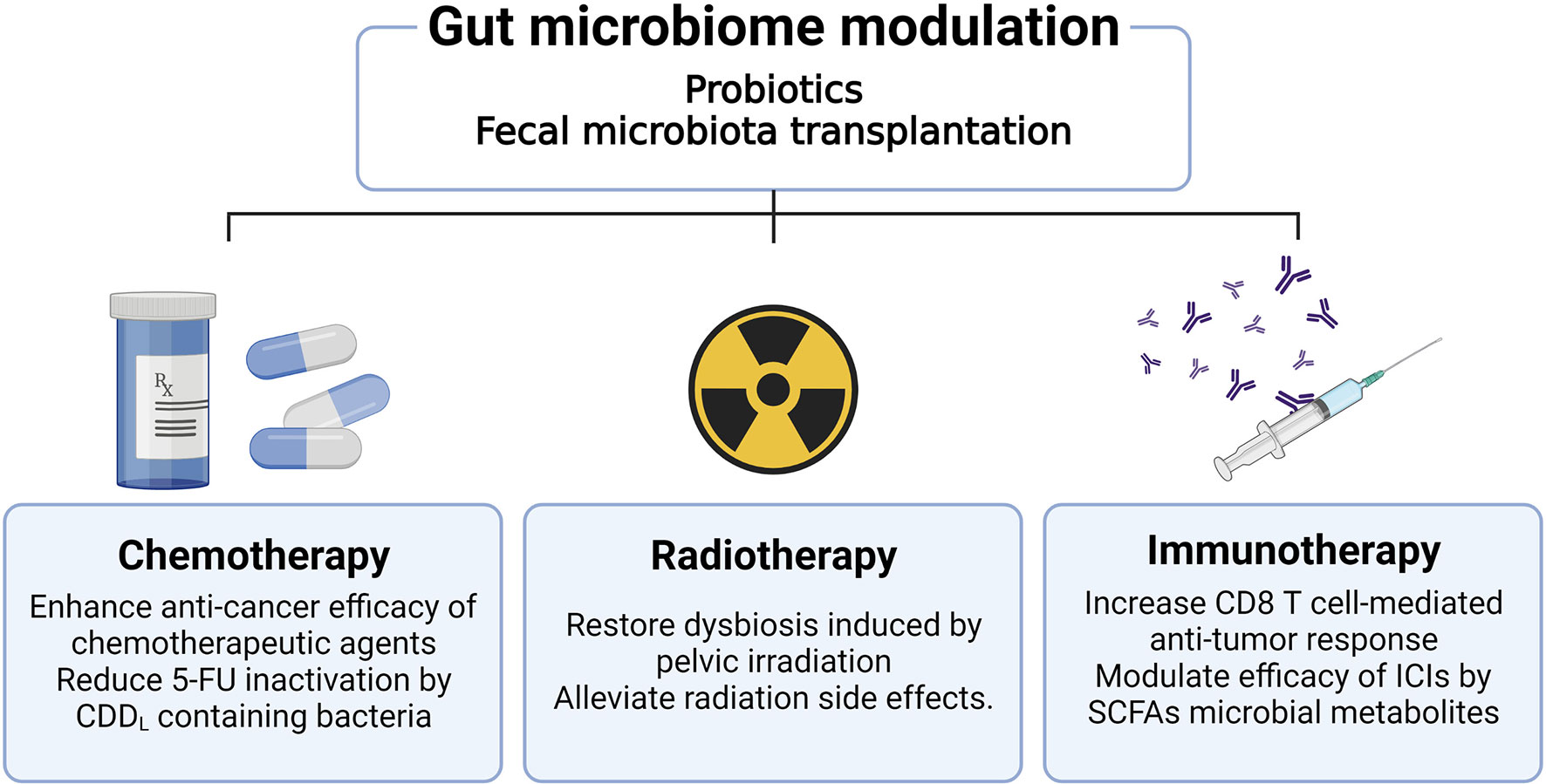
Figure 2 Effects of gut microbiome modulation on cancer treatment. Therapies which modulate the gut microbiome, including administration of probiotics or fecal microbiota transplantation, improve the efficacy of cancer treatment. Administration of antibiotics can reduce the efficacy of oxaliplatin and CpG oligodeoxynucleotides chemotherapeutic agents. The use of antibiotics increases pathogenic bacteria such as Escherichia shigella and Enterobacter, as well as reduces the anti-cancer effect of 5-FU. Radiation of the pelvic area causes dysbiosis and has the potential to affect the treatment modality of CRC. Furthermore, radiation-induced gut epithelial damage worsens the prognosis of CRC patients. These radiation side effects can be ameliorated through fecal microbiome transplantation as well as probiotics administration. The gut microbiota plays a role in modulating mucosal immunity in the colorectal region, acting to improve the efficacy of immunotherapy by enhancing the CD8+ T cell immune response or SCFA metabolite production.
Chemotherapy
The gut microbiota can modulate the efficacy of conventional chemotherapy. For example, it is known that certain gut microbiota may play a role in regulating cytotoxicity by participating in the metabolic process of anti-cancer drugs. The anti-cancer effect of platinum-based chemotherapeutic agents such as oxaliplatin and CpG oligodeoxynucleotides was decreased in mice treated with antibiotics (94), which exhibited lower cytokine secretion and ROS production, resulting in reduced tumor necrosis following anti-tumor therapy in the MC38 mouse colon tumor transplant model.
Gemcitabine has been shown to convert into an inactivated form with reduced anti-cancer effect when a specific gammaproteobacteria is present in the tumor (95). Gammaproteobacteria contain a long isoform of the cytidine deaminase enzyme which converts gemcitabine into an inactivated form. The anti-cancer effect was shown to be suppressed when the bacteria were eliminated by antibiotic treatment in a mouse model of CRC (95). Even in mouse tumor experiments using 5-fluorouracil (5-FU), antibiotic administration reduced the anti-cancer effect of 5-FU administration in the CRC model (96). In 16S rRNA seq analysis, pathogenic bacteria such as Escherichia shigella and Enterobacter were significantly increased when antibiotics were administered, and these changes were restored by taking probiotics.
F. nucleatum, which was previously known to greatly influence tumor initiation and progression, affects CRC treatment outcomes as well as CRC risk and dysplasia. A qPCR analysis based on colorectal tissue samples from 122 CRC patients confirmed a better prognosis occurred in patients with low F. nucleatum levels (77, 97). The level of F. nucleatum enrichment was positively correlated with poor response to 5-FU and oxalipatin in CRC patients (98). F. nucleatum stimulates the TLR4 and Myd88 innate immune signals and interferes with apoptosis, contributing to activation of the autophagy pathway and CRC chemoresistance (98).
Radiotherapy
Dysbiosis caused by radiation therapy has the potential to adversely affect the other treatment modalities of CRC. Analysis of the gut microbiome after radiation treatment showed a decrease in commensal bacteria such as Bifidobacterium, Faecalibacterium, and Clostridium spp., as well as an increase in Bacteroides and Enterococcus spp (99). In addition, in the case of patients receiving radiation therapy to the pelvic region, there was a tendency for the Fusobacteria taxa to increase by about 3% (100). These changes show the potential for tumor-promoting capacities. These microbiota can pass through the impaired gut barrier as a result of epithelial inflammatory damage caused by radiation therapy, leading to an additional intestinal inflammatory response and tissue damage (101).
Immunotherapy
Certain intestinal microbes are involved in tumor growth by regulating the immune response. Studies have been conducted to elucidate the mechanism of intestinal microbes and how they affect the efficacy of immunotherapeutic agents. In 2015, it was reported that the commensal gut microbiome could enhance the anti-tumor efficacy of programmed death-ligand 1 (PD-L1) ICIs through two mouse studies. Cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) inhibitors are one of the most widely used ICIs in clinical practice. The efficacy of CTLA-4 inhibitors was demonstrated to be altered by the population of the gut microbiome (102). The literature has identified an important role for Bacteroides species in the immunostimulatory modulation of CTLA-4 blockade. The modulation of ICI efficacy mediated by bacterial species in the gut microbiome is not limited to CTLA-4 signaling. The efficacy of a PD-L1 inhibitor was also shown to be modulated by the gut microbiota composition in a mouse tumor model (103). Recent studies have indicated that the anti-tumor effect was found associated with various bacteria such as Akkermansia, Faecalibacterium, Clostridiales, and Bifidobacterium spp (104–106). Although some details remain to be understood, this anti-tumor effect has been partially attributed to SCFA microbial metabolites such as butyrate and propionate (107). Another mechanism for modulation of ICIs is that host immune cells can interact directly with specific bacteria. Akkermansia muciniphila improves the efficacy of immunotherapeutic agents in an IL-12-dependent manner through direct interaction with dendritic cells in the lymph node (106). Bacteroides spp. can also directly increase Th1 and CD8 T cell anti-tumor immune responses (102).
Microbiome Modulation for Colon Cancer Treatment
Growing evidence clearly illustrates the significant influence that the gut microbiome has on tumors. Thus, it is not surprising that attempts have been made to inhibit tumor growth or modulate the efficacy of tumor therapy by regulating the gut microbiota. Efforts are ongoing to increase the effectiveness of tumor treatment and reduce side effects through fecal microbiome transplantation as well as probiotic therapy.
We discussed results from studies showing that Lactobacilli and Bifidobacterium affect the occurrence and progression of CRC in animal models (89–91). Some probiotics can help to not only enhance the effects of anti-cancer therapeutic agents but also alleviate the side effects caused by conventional cancer treatments (108). However, these probiotics also have the potential to act as opportunistic pathogens that can easily penetrate the intestinal barrier and immune environment after weakening by intestinal tumors (109). Appropriate probiotics with appropriate administration methods that can enhance anti-cancer effects and alleviate side effects are needed.
Fecal microbiota transplantation (FMT) is an emerging biotherapeutic method for altering the microbiota by transplanting stool information from healthy donors to patients (110). FMT can be applied to various gastrointestinal diseases including C. difficile infection, IBD, and restored eubiosis (111, 112). Many efforts are being made to apply FMT in the clinic as a tumor treatment. Reports have shown that FMT could be used to overcome resistance to immunosuppressants in the CRC mouse model (113). In addition, FMT can be helpful in alleviating the side effects of ICIs such as immune checkpoint inhibitor-associated colitis (114). Complete resolution of colitis through FMT was sustained for 53 days after one dose and for 78 days after two doses. Although clinical application as a treatment for CRC is still unexplored, a recent FMT study of melanoma patients reported that FMT succeeded in overcoming resistance to immunotherapy in patients who did not respond to immunotherapy (115, 116). These results suggest that FMT can be effectively used in the treatment of CRC. However, since the gut microbiome environment consists of a very large network with many unknowns remaining, more research is needed before microbiome modulation can be administered as an anti-cancer treatment in CRC.
Conclusion
Various animal and clinical experiments have demonstrated that changes in the composition of the gut microbiota affect the initiation of precancerous cancer lesions and cancer progression. Because the colorectal region is a site where changes in the gut microbiota can influence the organs directly, CRC is considered to be affected by the gut microbiome more than other tumors. Studies of the gut microbiome revealed that dysbiosis occurred more frequently in CRC patients than in healthy people. The proportion of butyrate-producing bacteria was found to be reduced along with inflammation in the intestine while opportunistic pathogens were increased. Epidemiological studies have highlighted dietary factors such as western eating habits and reduced dietary fiber intake as risk factors for CRC, suggesting the gut microbiome as one of the mechanisms linking these factors to CRC. Dietary fiber can be fermented into SCFAs by intestinal bacteria and animal experiments demonstrated that various SCFAs such as butyrate could affect cancer initiation and progression. Finally, the use of antibiotics may also be a risk factor for CRC and studies of the gut microbiome demonstrate its involvement in this effect.
Many published results have demonstrated that the gut microbiome acts as an important key factor in the initiation and progression of carcinoma in the treatment of CRC. However, we still understand only a small part of the gut microbiome and further research is needed to elucidate the underlying mechanisms and to modulate the gut microbiome as an important strategy in the treatment and prevention of CRC. This review describes the gut microbiome strains that affect each stage of the tumorigenesis process, including the underlying mechanisms, supplying an overview of the microbiota species likely involved in future studies examining the associations between the gut microbiome and CRC.
Author Contributions
JK and HL wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Research Foundation of Korea (NRF-2021M3A9D3026428 and NRF-2021M3A9H3015688) funded by the Ministry of Science and ICT of Korea.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Figures were created with BioRender.com.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J Clin (2021) 71:(3):209–49. doi: 10.3322/caac.21660
2. Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, et al. Linking Long-Term Dietary Patterns With Gut Microbial Enterotypes. Science (2011) 334(6052):105–8. doi: 10.1126/science.1208344
3. De Almeida CV, de Camargo MR, Russo E, Amedei A. Role of Diet and Gut Microbiota on Colorectal Cancer Immunomodulation. World J Gastroenterol (2019) 25(2):151–62. doi: 10.3748/wjg.v25.i2.151
4. Murphy N, Moreno V, Hughes DJ, Vodicka L, Vodicka P, Aglago EK, et al. Lifestyle and Dietary Environmental Factors in Colorectal Cancer Susceptibility. Mol Aspects Med (2019) 69:2–9. doi: 10.1016/j.mam.2019.06.005
5. Zheng X, Hur J, Nguyen LH, Liu J, Song M, Wu K, et al. Comprehensive Assessment of Diet Quality and Risk of Precursors of Early-Onset Colorectal Cancer. JNCI: J Natl Cancer Inst (2020) 113(5):543–52. doi: 10.1093/jnci/djaa164
6. Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial Community Variation in Human Body Habitats Across Space and Time. Science (2009) 326(5960):1694–7. doi: 10.1126/science.1177486
7. Thursby E, Juge N. Introduction to the Human Gut Microbiota. Biochem J (2017) 474(11):1823–36. doi: 10.1042/BCJ20160510
8. Tremaroli V, Bäckhed F. Functional Interactions Between the Gut Microbiota and Host Metabolism. Nature (2012) 489(7415):242–9. doi: 10.1038/nature11552
9. Shreiner AB, Kao JY, Young VB. The Gut Microbiome in Health and in Disease. Curr Opin Gastroenterol (2015) 31(1):69. doi: 10.1097/MOG.0000000000000139
10. Busnelli M, Manzini S, Chiesa G. The Gut Microbiota Affects Host Pathophysiology as an Endocrine Organ: A Focus on Cardiovascular Disease. Nutrients (2019) 12(1):79. doi: 10.3390/nu12010079
11. Fan Y, Pedersen O. Gut Microbiota in Human Metabolic Health and Disease. Nat Rev Microbiol (2021) 19(1):55–71. doi: 10.1038/s41579-020-0433-9
12. Helmink BA, Khan MW, Hermann A, Gopalakrishnan V, Wargo JAJNm. The Microbiome, Cancer, and Cancer Therapy. Nat Med (2019) 25: (3):377–88. doi: 10.1038/s41591-019-0377-7
13. Song M, Chan AT, Sun JJ. Influence of the Gut Microbiome, Diet, and Environment on Risk of Colorectal Cancer. Gastroenterology (2020) 158(2):322–40. doi: 10.1053/j.gastro.2019.06.048
14. Wang J, Chen W-D, Wang Y-D. The Relationship Between Gut Microbiota and Inflammatory Diseases: The Role of Macrophages. Front Microbiol (2020) 11:1065. doi: 10.3389/fmicb.2020.01065
15. Lavelle A, Sokol H. Gut Microbiota-Derived Metabolites as Key Actors in Inflammatory Bowel Disease. Nat Rev Gastroenterol Hepatol (2020) 17(4):223–37. doi: 10.1038/s41575-019-0258-z
16. Allen J, Sears CL. Impact of the Gut Microbiome on the Genome and Epigenome of Colon Epithelial Cells: Contributions to Colorectal Cancer Development. Genome Med (2019) 11(1):1–18. doi: 10.1186/s13073-019-0621-2
17. Chattopadhyay I, Dhar R, Pethusamy K, Seethy A, Srivastava T, Sah R, et al. Exploring the Role of Gut Microbiome in Colon Cancer. Appl Biochem Biotechnol (2021) 193:1780–99. doi: 10.1007/s12010-021-03498-9
18. Schloss PD. Identifying and Overcoming Threats to Reproducibility, Replicability, Robustness, and Generalizability in Microbiome Research. mBio (2018) 9(3):e00525–18. doi: 10.1128/mBio.00525-18
19. Zhang X, Li L, Butcher J, Stintzi A, Figeys D. Advancing Functional and Translational Microbiome Research Using Meta-Omics Approaches. Microbiome (2019) 7(1):1–12. doi: 10.1186/s40168-019-0767-6
20. Cho I, Blaser MJ. The Human Microbiome: At the Interface of Health and Disease. Nat Rev Genet (2012) 13(4):260–70. doi: 10.1038/nrg3182
21. Johnson CH, Spilker ME, Goetz L, Peterson SN, Siuzdak G. Metabolite and Microbiome Interplay in Cancer Immunotherapy. Cancer Res (2016) 76(21):6146–52. doi: 10.1158/0008-5472.CAN-16-0309
22. Zitvogel L, Ma Y, Raoult D, Kroemer G, Gajewski TF. The Microbiome in Cancer Immunotherapy: Diagnostic Tools and Therapeutic Strategies. Science (2018) 359(6382):1366–70. doi: 10.1126/science.aar6918
23. Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Patterns and Trends in Colorectal Cancer Incidence and Mortality. Gut (2017) 66(4):683–91. doi: 10.1136/gutjnl-2015-310912
24. Sánchez-Alcoholado L, Ramos-Molina B, Otero A, Laborda-Illanes A, Ordóñez R, Medina JA, et al. The Role of the Gut Microbiome in Colorectal Cancer Development and Therapy Response. Cancers (Basel) (2020) 12(6):1406. doi: 10.3390/cancers12061406
25. Feng Q, Liang S, Jia H, Stadlmayr A, Tang L, Lan Z, et al. Gut Microbiome Development Along the Colorectal Adenoma-Carcinoma Sequence. Nat Commun (2015) 6:6528. doi: 10.1038/ncomms7528
26. Nakatsu G, Li X, Zhou H, Sheng J, Wong SH, Wu WKK, et al. Gut Mucosal Microbiome Across Stages of Colorectal Carcinogenesis. Nat Commun (2015) 6(1):8727. doi: 10.1038/ncomms9727
27. Yu J, Feng Q, Wong SH, Zhang D, Liang QY, Qin YW, et al. Metagenomic Analysis of Faecal Microbiome as a Tool Towards Targeted non-Invasive Biomarkers for Colorectal Cancer. Gut (2017) 66(1):70. doi: 10.1136/gutjnl-2015-309800
28. Feng Q, Liang S, Jia H, Stadlmayr A, Tang L, Lan Z, et al. Gut Microbiome Development Along the Colorectal Adenoma–Carcinoma Sequence. Nat Commun (2015) 6(1):6528. doi: 10.1038/ncomms7528
29. Garcia-Gonzalez AP, Ritter AD, Shrestha S, Andersen EC, Yilmaz LS, Walhout AJM. Bacterial Metabolism Affects the C. Elegans Response to Cancer Chemotherapeutics. Cell (2017) 169(3):431–41.e8. doi: 10.1016/j.cell.2017.03.046
30. Cheng Y, Ling Z, Li L. The Intestinal Microbiota and Colorectal Cancer. Front Immunol (2020) 11:615056. doi: 10.3389/fimmu.2020.615056
31. Cipe G, Idiz UO, Firat D, Bektasoglu H. Relationship Between Intestinal Microbiota and Colorectal Cancer. World J Gastroint Oncol (2015) 7(10):233–40. doi: 10.4251/wjgo.v7.i10.233
32. Zackular JP, Baxter NT, Iverson KD, Sadler WD, Petrosino JF, Chen GY, et al. The Gut Microbiome Modulates Colon Tumorigenesis. mBio (2013) 4(6):e00692–13. doi: 10.1128/mBio.00692-13
33. Chen W, Liu F, Ling Z, Tong X, Xiang C. Human Intestinal Lumen and Mucosa-Associated Microbiota in Patients With Colorectal Cancer. PloS One (2012) 7(6):e39743. doi: 10.1371/journal.pone.0039743
34. Saffarian A, Mulet C, Regnault B, Amiot A, Tran-Van-Nhieu J, Ravel J, et al. Crypt- and Mucosa-Associated Core Microbiotas in Humans and Their Alteration in Colon Cancer Patients. mBio (2019) 10(4):e01315–19. doi: 10.1128/mBio.01315-19
35. Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, et al. Structural Segregation of Gut Microbiota Between Colorectal Cancer Patients and Healthy Volunteers. ISME J (2012) 6(2):320–9. doi: 10.1038/ismej.2011.109
36. Coker OO, Nakatsu G, Dai RZ, Wu WKK, Wong SH, Ng SC, et al. Enteric Fungal Microbiota Dysbiosis and Ecological Alterations in Colorectal Cancer. Gut (2019) 68(4):654–62. doi: 10.1136/gutjnl-2018-317178
37. Dzutsev A, Goldszmid RS, Viaud S, Zitvogel L, Trinchieri G. The Role of the Microbiota in Inflammation, Carcinogenesis, and Cancer Therapy. Eur J Immunol (2015) 45(1):17–31. doi: 10.1002/eji.201444972
38. Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, et al. Fusobacterium Nucleatum Potentiates Intestinal Tumorigenesis and Modulates the Tumor-Immune Microenvironment. Cell Host Microbe (2013) 14(2):207–15. doi: 10.1016/j.chom.2013.07.007
39. Viljoen KS, Dakshinamurthy A, Goldberg P, Blackburn JM. Quantitative Profiling of Colorectal Cancer-Associated Bacteria Reveals Associations Between Fusobacterium Spp., Enterotoxigenic Bacteroides Fragilis (ETBF) and Clinicopathological Features of Colorectal Cancer. PloS One (2015) 10(3):e0119462. doi: 10.1371/journal.pone.0119462
40. Grivennikov SI. Inflammation and Colorectal Cancer: Colitis-Associated Neoplasia. Semin Immunopathol (2013) 35(2):229–44. doi: 10.1007/s00281-012-0352-6
41. Francescone R, Hou V, Grivennikov SI. Microbiome, Inflammation, and Cancer. Cancer J (2014) 20(3):181–9. doi: 10.1097/PPO.0000000000000048
42. Reddy BS, Narisawa T, Wright P, Vukusich D, Weisburger J, Wynder E. Colon Carcinogenesis With Azoxymethane and Dimethylhydrazine in Germ-Free Rats. Cancer Res (1975) 35(2):287–90.
43. Reddy BS, Narisawa T, Weisburger J. Colon Carcinogenesis in Germ-Free Rats With Intrarectal 1, 2-Dimethylhydrazine and Subcutaneous Azoxymethane. Cancer Res (1976) 36(8):2874–6.
44. Son JS, Khair S, Pettet DW III, Ouyang N, Tian X, Zhang Y, et al. Altered Interactions Between the Gut Microbiome and Colonic Mucosa Precede Polyposis in Apcmin/+ Mice. PloS One (2015) 10: (6):e0127985. doi: 10.1371/journal.pone.0127985
45. Gupta A, Madani R, Mukhtar HJ. Streptococcus Bovis Endocarditis, A Silent Sign for Colonic Tumour. Colorectal Dis (2010) 12(3):164–71. doi: 10.1111/j.1463-1318.2009.01814.x
46. Srivastava A, Walter N, Atkinson P. Streptococcus Bovis Infection of Total Hip Arthroplasty in Association With Carcinoma of Colon. J Surg Orthop Adv (2010) 19(2):125–8.
47. Boleij A, van Gelder MM, Swinkels DW, Tjalsma HJ. Clinical Importance of Streptococcus Gallolyticus Infection Among Colorectal Cancer Patients: Systematic Review and Meta-Analysis. Clin Infect Dis (2011) 53(9):870–8. doi: 10.1093/cid/cir609
48. Biarc J, Nguyen IS, Pini A, Gosse F, Richert S, Thierse D, et al. Carcinogenic Properties of Proteins With Pro-Inflammatory Activity From Streptococcus Infantarius (Formerly s. bovis). Carcinogenesis (2004) 25(8):1477–84. doi: 10.1093/carcin/bgh091
49. Abdulamir AS, Hafidh RR, Bakar FA. Molecular Detection, Quantification, and Isolation of Streptococcus Gallolyticus Bacteria Colonizing Colorectal Tumors: Inflammation-Driven Potential of Carcinogenesis via IL-1, COX-2, and IL-8. Mol Cancer (2010) 9(1):249. doi: 10.1186/1476-4598-9-249
50. Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, et al. Fusobacterium Nucleatum Infection is Prevalent in Human Colorectal Carcinoma. Genome Res (2012) 22(2):299–306. doi: 10.1101/gr.126516.111
51. Ye X, Wang R, Bhattacharya R, Boulbes DR, Fan F, Xia L, et al. Fusobacterium Nucleatum Subspecies Animalis Influences Proinflammatory Cytokine Expression and Monocyte Activation in Human Colorectal Tumors. Cancer Prev Res (Phila) (2017) 10:(7):398–409. doi: 10.1158/1940-6207.CAPR-16-0178
52. Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, YWJCh H, et al. Fusobacterium Nucleatum Promotes Colorectal Carcinogenesis by Modulating E-Cadherin/β-Catenin Signaling via its Fada Adhesin. Cell Host Microbe (2013) 14(2):195–206. doi: 10.1016/j.chom.2013.07.012
53. Winters MD, Schlinke TL, Joyce WA, Glore SR, Huycke MM. Prospective Case-Cohort Study of Intestinal Colonization With Enterococci That Produce Extracellular Superoxide and the Risk for Colorectal Adenomas or Cancer. Am J Gastroenterol (1998) 93(12):2491–500. doi: 10.1111/j.1572-0241.1998.00710.x
54. Wang X, Huycke MM. Extracellular Superoxide Production by Enterococcus Faecalis Promotes Chromosomal Instability in Mammalian Cells. Gastroenterology (2007) 132(2):551–61. doi: 10.1053/j.gastro.2006.11.040
55. Wang X, Allen TD, May RJ, Lightfoot S, Houchen CW, Huycke M. Enterococcus Faecalis Induces Aneuploidy and Tetraploidy in Colonic Epithelial Cells Through a Bystander Effect. Cancer Res (2008) 68(23):9909–17. doi: 10.1158/0008-5472.CAN-08-1551
56. Huycke MM, Moore D, Joyce W, Wise P, Shepard L, Kotake Y, et al. Extracellular Superoxide Production by Enterococcus Faecalis Requires Demethylmenaquinone and is Attenuated by Functional Terminal Quinol Oxidases. Mol Microbiol (2001) 42(3):729–40. doi: 10.1046/j.1365-2958.2001.02638.x
57. Huycke MM, Moore DR. In Vivo Production of Hydroxyl Radical by Enterococcus Faecalis Colonizing the Intestinal Tract Using Aromatic Hydroxylation. Free Radical Biol Med (2002) 33(6):818–26. doi: 10.1016/S0891-5849(02)00977-2
58. Evans MD, Dizdaroglu M, Cooke MS. Oxidative DNA Damage and Disease: Induction, Repair and Significance. Mutat Res (2004) 567(1):1–61. doi: 10.1016/j.mrrev.2003.11.001
59. Sears C. Enterotoxigenic Bacteroides Fragilis: A Rogue Among Symbiotes. Clin Microbiol Rev (2009) 22(2):349–69. doi: 10.1128/CMR.00053-08
60. Wu S, Rhee K-J, Albesiano E, Rabizadeh S, Wu X, Yen H-R, et al. A Human Colonic Commensal Promotes Colon Tumorigenesis via Activation of T Helper Type 17 T Cell Responses. Nat Med (2009) 15(9):1016–22. doi: 10.1038/nm.2015
61. Sears CL, Geis AL, Housseau F. Bacteroides Fragilis Subverts Mucosal Biology: From Symbiont to Colon Carcinogenesis. J Clin Invest (2014) 124(10):4166–72. doi: 10.1172/JCI72334
62. Chung L, Orberg ET, Geis AL, Chan JL, Fu K, Shields CED, et al. Bacteroides Fragilis Toxin Coordinates a Pro-Carcinogenic Inflammatory Cascade via Targeting of Colonic Epithelial Cells. Cell Host Microbe (2018) 23(2):203–14.e5. doi: 10.1016/j.chom.2018.01.007
63. Long X, Wong CC, Tong L, Chu ES, Szeto CH, Go MY, et al. Peptostreptococcus Anaerobius Promotes Colorectal Carcinogenesis and Modulates Tumour Immunity. Nat Microbiol (2019) 4(12):2319–30. doi: 10.1038/s41564-019-0541-3
64. Tsoi H, Chu ESH, Zhang X, Sheng J, Nakatsu G, Ng SC, et al. Peptostreptococcus Anaerobius Induces Intracellular Cholesterol Biosynthesis in Colon Cells to Induce Proliferation and Causes Dysplasia in Mice. Gastroenterology (2017) 152(6):1419–33.e5. doi: 10.1053/j.gastro.2017.01.009
65. Gradel KO, Nielsen HL, Schønheyder HC, Ejlertsen T, Kristensen B, Nielsen H. Increased Short- and Long-Term Risk of Inflammatory Bowel Disease After Salmonella or Campylobacter Gastroenteritis. Gastroenterology (2009) 137(2):495–501. doi: 10.1053/j.gastro.2009.04.001
66. Lu R, Wu S, Zhang YG, Xia Y, Liu X, Zheng Y, et al. Enteric Bacterial Protein Avra Promotes Colonic Tumorigenesis and Activates Colonic Beta-Catenin Signaling Pathway. Oncogenesis (2014) 3(6):e105. doi: 10.1038/oncsis.2014.20
67. Lu R, Wu S, Y-g Z, Xia Y, Zhou Z, Kato I, et al. Salmonella Protein Avra Activates the STAT3 Signaling Pathway in Colon Cancer. Neoplasia (2016) 18(5):307–16. doi: 10.1016/j.neo.2016.04.001
68. Wang J, Lu R, Fu X, Dan Z, Zhang Y-G, Chang X, et al. Novel Regulatory Roles of Wnt1 in Infection-Associated Colorectal Cancer. Neoplasia (2018) 20(5):499–509. doi: 10.1016/j.neo.2018.03.001
69. Martin OC, Bergonzini A, d’Amico F, Chen P, Shay JW, Dupuy J, et al. Infection With Genotoxin-Producing Salmonella Enterica Synergises With Loss of the Tumour Suppressor APC in Promoting Genomic Instability via the PI3K Pathway in Colonic Epithelial Cells. Cell Microbiol (2019) 21(12):e13099. doi: 10.1111/cmi.13099
70. He Z, Gharaibeh RZ, Newsome RC, Pope JL, Dougherty MW, Tomkovich S, et al. Campylobacter Jejuni Promotes Colorectal Tumorigenesis Through the Action of Cytolethal Distending Toxin. Gut (2019) 68(2):289–300. doi: 10.1136/gutjnl-2018-317200
71. Sun X, Threadgill D, Jobin C. Campylobacter Jejuni Induces Colitis Through Activation of Mammalian Target of Rapamycin Signaling. Gastroenterology (2012) 142(1):86–95.e5. doi: 10.1053/j.gastro.2011.09.042
72. Figliuolo VR, dos Santos LM, Abalo A, Nanini H, Santos A, Brittes NM, et al. Sulfate-Reducing Bacteria Stimulate Gut Immune Responses and Contribute to Inflammation in Experimental Colitis. Life Sci (2017) 189:29–38. doi: 10.1016/j.lfs.2017.09.014
73. Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, et al. Metagenomic and Metabolomic Analyses Reveal Distinct Stage-Specific Phenotypes of the Gut Microbiota in Colorectal Cancer. Nat Med (2019) 25(6):968–76. doi: 10.1038/s41591-019-0458-7
74. Louis P, Hold GL, Flint HJ. The Gut Microbiota, Bacterial Metabolites and Colorectal Cancer. Nat Rev Microbiol (2014) 12(10):661–72. doi: 10.1038/nrmicro3344
75. Marquet P, Duncan SH, Chassard C, Bernalier-Donadille A, Flin T. Lactate Has the Potential to Promote Hydrogen Sulphide Formation in the Human Colon. FEMS Microbiol Lett (2009) 299(2):128–34. doi: 10.1111/j.1574-6968.2009.01750.x
76. Li L, Li X, Zhong W, Yang M, Xu M, Sun Y, et al. Gut Microbiota From Colorectal Cancer Patients Enhances the Progression of Intestinal Adenoma in Apcmin/+ Mice. EBioMedicine (2019) 48:301–15. doi: 10.1016/j.ebiom.2019.09.021
77. Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, et al. Fusobacterium Nucleatum in Colorectal Carcinoma Tissue and Patient Prognosis. Gut (2016) 65(12):1973–80. doi: 10.1136/gutjnl-2015-310101
78. Wei Z, Cao S, Liu S, Yao Z, Sun T, Li Y, et al. Could Gut Microbiota Serve as Prognostic Biomarker Associated With Colorectal Cancer Patients’ Survival? A Pilot Study on Relevant Mechanism. Oncotarget (2016) 7(29):46158. doi: 10.18632/oncotarget.10064
79. Mima K, Sukawa Y, Nishihara R, Qian ZR, Yamauchi M, Inamura K, et al. Fusobacterium Nucleatum and T Cells in Colorectal Carcinoma. JAMA Oncol (2015) 1(5):653–61. doi: 10.1001/jamaoncol.2015.1377
80. Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, et al. Binding of the Fap2 Protein of Fusobacterium Nucleatum to Human Inhibitory Receptor TIGIT Protects Tumors From Immune Cell Attack. Immunity (2015) 42(2):344–55. doi: 10.1016/j.immuni.2015.01.010
81. Deng Z, Mu J, Tseng M, Wattenberg B, Zhuang X, Egilmez NK, et al. Enterobacteria-Secreted Particles Induce Production of Exosome-Like S1P-Containing Particles by Intestinal Epithelium to Drive Th17-Mediated Tumorigenesis. Nat Commun (2015) 6(1):6956. doi: 10.1038/ncomms7956
82. Bonnet M, Buc E, Sauvanet P, Darcha C, Dubois D, Pereira B, et al. Colonization of the Human Gut by E. Coli and Colorectal Cancer Risk. Clin Cancer Res (2014) 20(4):859–67. doi: 10.1158/1078-0432.CCR-13-1343
83. Veziant J, Gagnière J, Jouberton E, Bonnin V, Sauvanet P, Pezet D, et al. Association of Colorectal Cancer With Pathogenic Escherichia Coli: Focus on Mechanisms Using Optical Imaging. World J Clin Oncol (2016) 7(3):293–301. doi: 10.5306/wjco.v7.i3.293
84. Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, Nougayrède J-P. Escherichia Coli Induces DNA Damage In Vivo and Triggers Genomic Instability in Mammalian Cells. Proc Natl Acad Sci USA (2010) 107(25):11537–42. doi: 10.1073/pnas.1001261107
85. Arthur JC, Gharaibeh RZ, Mühlbauer M, Perez-Chanona E, Uronis JM, McCafferty J, et al. Microbial Genomic Analysis Reveals the Essential Role of Inflammation in Bacteria-Induced Colorectal Cancer. Nat Commun (2014) 5:4724. doi: 10.1038/ncomms5724
86. Cougnoux A, Dalmasso G, Martinez R, Buc E, Delmas J, Gibold L, et al. Bacterial Genotoxin Colibactin Promotes Colon Tumour Growth by Inducing a Senescence-Associated Secretory Phenotype. Gut (2014) 63(12):1932–42. doi: 10.1136/gutjnl-2013-305257
87. Quévrain E, Maubert MA, Michon C, Chain F, Marquant R, Tailhades J, et al. Identification of an Anti-Inflammatory Protein From Faecalibacterium Prausnitzii, a Commensal Bacterium Deficient in Crohn’s Disease. Gut (2016) 65(3):415–25. doi: 10.1136/gutjnl-2014-307649
88. Rafter J, Bennett M, Caderni G, Clune Y, Hughes R, Karlsson PC, et al. Dietary Synbiotics Reduce Cancer Risk Factors in Polypectomized and Colon Cancer Patients. Am J Clin Nutr (2007) 85(2):488–96. doi: 10.1093/ajcn/85.2.488
89. Lee HA, Kim H, Lee K-W, Park K-Y. Dead Nano-Sized Lactobacillus Plantarum Inhibits Azoxymethane/Dextran Sulfate Sodium-Induced Colon Cancer in Balb/C Mice. J Med Food (2015) 18(12):1400–5. doi: 10.1089/jmf.2015.3577
90. Talero E, Bolivar S, Ávila-Román J, Alcaide A, Fiorucci S, Motilva V. Inhibition of Chronic Ulcerative Colitis-Associated Adenocarcinoma Development in Mice by VSL3. Inflamm Bowel Dis (2015) 21(5):1027–37. doi: 10.1097/MIB.0000000000000346
91. Gamallat Y, Meyiah A, Kuugbee ED, Hago AM, Chiwala G, Awadasseid A, et al. Lactobacillus Rhamnosus Induced Epithelial Cell Apoptosis, Ameliorates Inflammation and Prevents Colon Cancer Development in an Animal Model. Biomed Pharmacother (2016) 83:536–41. doi: 10.1016/j.biopha.2016.07.001
92. Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, et al. Activation of Gpr109a, Receptor for Niacin and the Commensal Metabolite Butyrate, Suppresses Colonic Inflammation and Carcinogenesis. Immunity (2014) 40(1):128–39. doi: 10.1016/j.immuni.2013.12.007
93. Roy S, Trinchieri G. Microbiota: A Key Orchestrator of Cancer Therapy. Nat Rev Cancer (2017) 17(5):271–85. doi: 10.1038/nrc.2017.13
94. Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, et al. Commensal Bacteria Control Cancer Response to Therapy by Modulating the Tumor Microenvironment. Science (2013) 342(6161):967–70. doi: 10.1126/science.1240527
95. Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, et al. Potential Role of Intratumor Bacteria in Mediating Tumor Resistance to the Chemotherapeutic Drug Gemcitabine. Science (2017) 357(6356):1156–60. doi: 10.1126/science.aah5043
96. Yuan L, Zhang S, Li H, Yang F, Mushtaq N, Ullah S, et al. The Influence of Gut Microbiota Dysbiosis to the Efficacy of 5-Fluorouracil Treatment on Colorectal Cancer. Biomed Pharmacother = Biomed Pharmacother (2018) 108:184–93. doi: 10.1016/j.biopha.2018.08.165
97. Flanagan L, Schmid J, Ebert M, Soucek P, Kunicka T, Liska V, et al. Fusobacterium Nucleatum Associates With Stages of Colorectal Neoplasia Development, Colorectal Cancer and Disease Outcome. Eur J Clin Microbiol Infect Dis (2014) 33(8):1381–90. doi: 10.1007/s10096-014-2081-3
98. Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, et al. Fusobacterium Nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell (2017) 170(3):548–63.e16. doi: 10.1016/j.cell.2017.07.008
99. Touchefeu Y, Montassier E, Nieman K, Gastinne T, Potel G, Bruley Des Varannes S, et al. Systematic Review: The Role of the Gut Microbiota in Chemotherapy- or Radiation-Induced Gastrointestinal Mucositis - Current Evidence and Potential Clinical Applications. Alimentary Pharmacol Ther (2014) 40(5):409–21. doi: 10.1111/apt.12878
100. Nam YD, Kim HJ, Seo JG, Kang SW, Bae JW. Impact of Pelvic Radiotherapy on Gut Microbiota of Gynecological Cancer Patients Revealed by Massive Pyrosequencing. PloS One (2013) 8(12):e82659. doi: 10.1371/journal.pone.0082659
101. Al-Qadami G, Van Sebille Y, Le H, Bowen J. Gut Microbiota: Implications for Radiotherapy Response and Radiotherapy-Induced Mucositis. Expert Rev Gastroenterol Hepatol (2019) 13(5):485–96. doi: 10.1080/17474124.2019.1595586
102. Vetizou M, Pitt JM, Daillere R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer Immunotherapy by CTLA-4 Blockade Relies on the Gut Microbiota. Science (2015) 350(6264):1079. doi: 10.1126/science.aad1329
103. Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal Bifidobacterium Promotes Antitumor Immunity and Facilitates Anti-PD-L1 Efficacy. Science (2015) 350(6264):1084–9. doi: 10.1126/science.aac4255
104. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut Microbiome Modulates Response to Anti-PD-1 Immunotherapy in Melanoma Patients. Science (2018) 359(6371):97–103. doi: 10.1126/science.aan4236
105. Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre M-L, et al. The Commensal Microbiome is Associated With Anti–PD-1 Efficacy in Metastatic Melanoma Patients. Science (2018) 359(6371):104–8. doi: 10.1126/science.aao3290
106. Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, et al. Gut Microbiome Influences Efficacy of PD-1-Based Immunotherapy Against Epithelial Tumors. Science (2018) 359(6371):91–7. doi: 10.1126/science.aan3706
107. Hayase E, Jenq RR. Role of the Intestinal Microbiome and Microbial-Derived Metabolites in Immune Checkpoint Blockade Immunotherapy of Cancer. Genome Med (2021) 13(1), 107. doi: 10.1186/s13073-021-00923-w
108. Rodriguez-Arrastia M, Martinez-Ortigosa A, Rueda-Ruzafa L, Folch Ayora A, Ropero-Padilla C. Probiotic Supplements on Oncology Patients’ Treatment-Related Side Effects: A Systematic Review of Randomized Controlled Trials. Int J Environ Res Public Health (2021) 18(8):4265. doi: 10.3390/ijerph18084265
109. Veiga P, Suez J, Derrien M, Elinav E. Moving From Probiotics to Precision Probiotics. Nat Microbiol (2020) 5(7):878–80. doi: 10.1038/s41564-020-0721-1
110. Aroniadis OC, Brandt L. Fecal Microbiota Transplantation: Past, Present and Future. Curr Opin Gastroenterol (2013) 29(1):79–84. doi: 10.1097/MOG.0b013e32835a4b3e
111. Van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, De Vos WM, et al. Duodenal Infusion of Donor Feces for Recurrent Clostridium Difficile. N Engl J Med (2013) 368(5):407–15. doi: 10.1056/NEJMoa1205037
112. Paramsothy S, Kamm MA, Kaakoush NO, Walsh AJ, van den Bogaerde J, Samuel D, et al. Multidonor Intensive Faecal Microbiota Transplantation for Active Ulcerative Colitis: A Randomised Placebo-Controlled Trial. Lancet (2017) 389(10075):1218–28. doi: 10.1016/S0140-6736(17)30182-4
113. Rosshart SP, Vassallo BG, Angeletti D, Hutchinson DS, Morgan AP, Takeda K, et al. Wild Mouse Gut Microbiota Promotes Host Fitness and Improves Disease Resistance. Cell (2017) 171(5):1015–28.e13. doi: 10.1016/j.cell.2017.09.016
114. Wang Y, Wiesnoski DH, Helmink BA, Gopalakrishnan V, Choi K, DuPont HL, et al. Fecal Microbiota Transplantation for Refractory Immune Checkpoint Inhibitor-Associated Colitis. Nat Med (2018) 24(12):1804–8. doi: 10.1038/s41591-018-0238-9
115. Davar D, Dzutsev AK, McCulloch JA, Rodrigues RR, Chauvin J-M, Morrison RM, et al. Fecal Microbiota Transplant Overcomes Resistance to Anti–PD-1 Therapy in Melanoma Patients. Science (2021) 371(6529):595–602. doi: 10.1126/science.abf3363
Keywords: gut microbiota, colorectal cancer, immunotherapy, chemotherapy, immune checkpoint inhibitors
Citation: Kim J and Lee HK (2022) Potential Role of the Gut Microbiome In Colorectal Cancer Progression. Front. Immunol. 12:807648. doi: 10.3389/fimmu.2021.807648
Received: 02 November 2021; Accepted: 17 December 2021;
Published: 07 January 2022.
Edited by:
Ti Wen, The First Affiliated Hospital of China Medical University, ChinaReviewed by:
Philip Brandon Busbee, University of South Carolina, United StatesAna Maria Caetano Faria, Federal University of Minas Gerais, Brazil
Copyright © 2022 Kim and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Heung Kyu Lee, heungkyu.lee@kaist.ac.kr
 Jaeho Kim
Jaeho Kim Heung Kyu Lee
Heung Kyu Lee